- 1Department of Ecology and Evolutionary Biology, University of California, Irvine, Irvine, CA, United States
- 2Department of Earth System Science, University of California, Irvine, Irvine, CA, United States
Microorganisms exhibit shifts in elemental stoichiometry in response to short-term temperature increases due to varying growth rate, biochemical reactions, and protein degradation. Yet, it is unknown how an organism's elemental stoichiometry will respond to temperature change on evolutionary timescales. Here we ask how cellular elemental stoichiometry and physiology change in Escherichia coli that have adapted to high temperature over 2,000 generations compared to their low temperature adapted ancestor. Cell lines evolved to a temperature of 42.2°C via two, negatively epistatic adaptive pathways leading to mutations in either the RNA polymerase complex or the termination factor rho. Compared to the ancestor, high temperature adapted cell lines overall had 14% higher N:P ratios, but did not differ significantly in C:N or C:P. However, cell lines with mutations in the rho gene had 13% lower C:N and 34% higher N:P. Furthermore, the two adaptive strategies of the rho and RNA polymerase mutations varied significantly from one another, cell lines with the rho mutation had lower C:N, higher N:P and higher protein content compared to cell lines with the RNA polymerase mutation. Thus, specific adaptive pathways modulate the effect of temperature on the cellular elemental stoichiometry and may explain why the elemental composition of specific lineages is differentially affected by temperature changes.
Introduction
The possibility of stoichiometric changes, or changes in cellular elemental composition, has important implications for understanding how evolving organisms will respond to anthropogenic change (Arrigo, 2005; Dijkstra et al., 2012). Microorganisms, especially, present an interesting group capable of responding through natural selection to environmental change on ecological timescales due to their short generation times and generally large populations (Lenski et al., 1991). Prokaryotic life has adapted to a vast range of different temperatures over the course of evolutionary history (Price and Sowers, 2004). While various microorganisms are adapted to different ranges of temperatures, many biological and biochemical processes are directly impacted by temperature and are disrupted when temperatures change extensively (Burra et al., 2010). To deal with short-term changes, Escherichia coli is capable of quickly modifying gene expression with a heat shock response when thermally stressed (Arsene et al., 2000). Moreover, studies have shown that microorganisms are capable of evolving to environmental factors such as temperature, pH, etc. in a relatively rapid period of time (Bennett et al., 1990; Hughes et al., 2007). The evolutionary microbial response to temperatures above their growth optimum includes shifts in gene expression, mutations in transcription, and translation-related genes, maintaining increased heat shock proteins in the cell, etc. (Mongold et al., 1996; Riehle et al., 2003; Rudolph et al., 2010). Adaptive changes in macromolecular content and overall resource allocation can drive shifts in cellular stoichiometry (Sterner and Elser, 2002). Thus, changes in temperature can affect the elemental composition of bacteria through mechanisms of adaptation.
A previous hypothesis for the impact of temperature on stoichiometry predicts a lower phosphorus cell quota and higher carbon-to-phosphorus and nitrogen-to-phosphorus ratios for growth at elevated temperature (Toseland et al., 2013). Ribosomes and protein biosynthesis in bacteria are more efficient at elevated temperature (Broeze et al., 1978; VanBogelen and Neidhardt, 1990). This improved efficiency can result in in a lower requirement for P-rich ribosomes to maintain a specific biosynthesis rate (Toseland et al., 2013; Yvon-Durocher et al., 2015). Assuming the same growth rate, this should lead to higher C:P and N:P in warming environments. This theory applies to both acclimation (i.e., cells growing across a temperature gradient) and adaptation (cells adapted to different temperature levels) (Moreno and Martiny, 2018). Accordingly, E. coli grown at higher temperatures show higher levels of cellular C and N and decreased P, yielding these predicted results (Cotner et al., 2006). However, there are a multitude of ways that microbes adapt to increased temperature (Mongold et al., 1996; Riehle et al., 2003; Rudolph et al., 2010). Several bacterial strains, including psychrophiles, show variation in biomass stoichiometry independent of phylogenetic relatedness, suggesting small genetic differences can yield variable stoichiometry (Zimmerman et al., 2014). Moreover, Prochlorococcus ecotypes with different temperature optima (Johnson et al., 2006) are variable in their elemental ratios at the strain level (Martiny et al., 2016). Thus, it is still unclear if there is a consistent relationship between adaptation to temperature and how microbes alter their allocation of chemical resources.
There are two broad conceptual models for how adaptation might affect cellular biochemistry and stoichiometry: adaptations could restore the organism to it normal pre-stressed physiological state or acclimation phenotypes could be reinforced by adaptations (Hug and Gaut, 2015). Physiologically in E. coli, protein degradation as well as peptide elongation rates increase at high temperature, resulting in a higher turnover of peptides (Farewell and Neidhardt, 1998). However, their genome contains a number of heat inducible genes, encoding for chaperone proteins and proteases, which prevent aggregation of proteins at high temperature and refold denatured proteins (Arsene et al., 2000). While the heat-shock response of E. coli to short-term changes in temperature has been well-studied, recent research has also emerged concerning the long-term evolutionary response of microbes to high temperature. E. coli grown at 42.2°C had significant fitness gains after only 200 generations, compared to its ancestor (Bennett et al., 1990). An expanded experiment analyzed E. coli after being exposed to 42.2°C over 2,000 generations and subsequently sequenced a clonal genome from 115 adapted cell lines. Here, a total of 1,331 mutations were present (Tenaillon et al., 2012) and two of the most frequent mutations were within the rho and RNA polymerase β subunit (rpoB) genes. Out of the 115 cell lines sequenced, 17 lines had mutations in rho codon 15 and 18 had mutations in rpoB codon 966. Every cell line in the study was found to have either a mutation in the rho gene or the rpoBC operon. Additionally, mutations in these genes were in negative epistasis with one another, meaning mutations occurring in both genes at the same time happened less often than by random chance (Tenaillon et al., 2012). Nonetheless, there were some lines that did have mutations in both genes, although they did not include either of the most prevalent rho and rpoB mutations. Phenotypic differences between cells with rho and rpoB mutations were identified using Biolog plates (Hug and Gaut, 2015). The strongest differences between rho and rpoB lines were among chemical sensitivity, but also included differential amino acid usage, suggesting there may be differences in their resource requirements (Hug and Gaut, 2015). The gene rpoB encodes the β subunit of RNA polymerase and may increase the capacity of cell lines to adapt (Barrick et al., 2010). Rho is a ubiquitous prokaryotic termination factor that ends transcription, but also functions in maintaining chromosomal integrity in some circumstances (Washburn and Gottesman, 2011; Grylak-Mielnicka et al., 2016). Mutations in both of these transcriptional regulators have reduced susceptibility to antibiotics in other bacterial genera (Taniguchi et al., 1996; Lee and Helmann, 2014). While their broad cellular function may be a part of the reason for their adaptive role in new environments, the elemental resource allocation outcomes of these adaptations to growth at high temperature are currently unknown.
We propose two competing hypotheses for how cellular macromolecule content and C:N:P may shift in cell lines with adaptations to high temperature (Figure 1). The first is a compensation mechanism, which suggests cells produce a large amount of macromolecules (protein and ribosomes) to offset the heat related degradation (Farewell and Neidhardt, 1998). If cells compensate for degradation through mutations allowing the cell to elevate macromolecular levels, this would predict lower carbon-to-nutrient ratios (C:N and C:P) and an elevated N:P ratio in adapted lines due to high amounts of N-rich protein (Figure 1). Here a fitness tradeoff occurs because the increase in growth rate through production of additional ribosomes or other proteins will incur a heavy energetic cost. Alternatively, adaptation of enzymes and ribosomes may increase their efficiency at higher temperature leading to a lower need for proteins and ribosomes in the adapted lines compared to the ancestor at 42.2°C. This would lead to increased C:N and C:P ratios in high temperature adapted cells (Figure 1). A fitness cost occurs because the increased efficiency that comes from tuning an enzyme to a specific environment however, may reduce an organism's phenotypic plasticity.
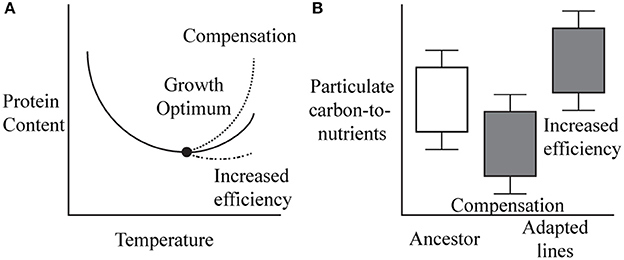
Figure 1. (A) The compensation model predicts that cells will adapt to high temperature by producing a larger amount of protein and ribosome compared to the ancestor. (B) This would likely result in a depressed carbon to nutrients (nitrogen and phosphorus) ratio due to higher levels of nitrogen rich protein or phosphorus rich ribosomes. The increased efficiency model predicts macromolecule content to be lower in adapted cell lines than in the ancestor, due to adaptations that allow cells to better maintain proteins at a higher temperature or increase the efficiency of ribosomes. This would correspond to an increased carbon to nutrients (nitrogen and phosphorus) ratio.
To identify how adaptation to elevated temperature affects the elemental stoichiometry of heterotrophic bacteria, we analyzed E. coli lines experimentally adapted to 42.2°C (Tenaillon et al., 2012). We first ask if lines adapted to higher temperature have different C:N, C:P, and N:P ratios compared to their ancestral lineage. Second, we ask whether changes in the C:N:P ratio correspond to changes in total cellular protein content as changes in protein levels could alter cellular nitrogen. Third, we ask if different genetic pathways (rpoB vs. rho) for adaption to high temperature lead to differences in C:N:P ratio and protein content.
Materials and Methods
Lines and Experimental Design
A lab adapted E. coli strain REL1206 and eight lines adapted from this common ancestor at 42.2°C were obtained from Dr. Brandon Gaut's Lab (Tenaillon et al., 2012) (Supplementary Tables 1, 2). Lines 5, 7, and 70 had mutations in both rpoB and rho genes, lines 75 and 135 had the prevalent mutation in rpoB and lines 20, 60, and 73 had the prevalent mutation in rho (Tenaillon et al., 2012). All adapted cell lines had additional background mutations, some of which were shared across lines, for example the cation/proton antiporter (ybaL) or a cardiolipin synthase (cls) were found in 5 of the 9 assayed lines (Supplementary Table 3). All cell lines, including the ancestor, were frozen from single colony cultures at −80°C storage then revived in lysogeny broth for 24 h at 37°C to acclimate cells to culture conditions. Next, they were transferred into a modified Davis Mingioli (MDM) media with a C:N:P ratio of 220:16:1 to yield a higher density of cells, for 24 h at 37°C. Finally, they were transferred into MDM media for 48 h at 42.2°C, the same temperature at which they were adapted for 2,000 generations, The length of time for growth after the second MDM transfer was increased to 48 instead of 24 h to reach higher density. It is possible that mutations may have arisen during the course of this experiment. Several biological replicates were used to reduce this noise. With separate populations new mutations will have a smaller effect on the overall signal.
Cellular Composition Analyses
After reaching sufficient population density of around 107 cells/mL, 5 mL of bacterial culture were filtered onto pre-combusted 25 mm glass fiber filters and rinsed with 5 mL of 0.17 M anhydrous Na2SO4 for particulate organic phosphorus (POP) and particulate organic carbon (POC) plus particulate organic nitrogen (PON) assays. Culture subsamples were preserved in glutaraldehyde and frozen for flow cytometry analysis at the time of filtration. Cells were stained with SYBR green and read at a wavelength of 488 nm with a BD Accuri C6 flow cytometer for cell counts.
Particulate organic phosphorus was determined in each sample using a modified ash-hydrolysis method (Lomas et al., 2010). Filters were placed in glass vials with 2 mL 0.017 M MgSO4 and dried at 80–90°C overnight. Vials were transferred to a muffle furnace and combusted for 2 h at 500°C. After cooling, 5 mL of 0.2 M HCl were added and samples were heated at 80°C for 30 min. A mixed reagent of 2:5:1:2 ratio of 0.024 M (NH4)6Mo7O24, 5N H2SO4, 0.002 M K2Sb2(C4H2O6)2·3 H2O, and 0.3 M C6H8O6 was added to samples. After 30 min the samples were read in a spectrophotometer at a wavelength of 885 nm. A standard curve of KH2PO4 was used to calculate the concentration of phosphate in samples based on their absorbance.
Particulate organic carbon and particulate organic nitrogen assays were dried overnight and packed into tin capsules. Standards were created by packing pre-weighed quantities of atropine and peach leaf into capsules. POC and PON concentrations were determined using a FlashEA1112 nitrogen and carbon analyzer (Sharp, 1974).
Protein content was quantified using a Coomassie (Bradford) assay kit with bovine serum albumin standards (Bradford, 1976). One milliliter culture samples were aliquoted into microtubes with 0.2 mm glass beads and beat in a bead-beater for 5 min to disrupt cell membranes. These tubes were spun in a centrifuge and the supernatant was collected. Coomassie dye was added to bovine serum albumin standards and samples in a microplate. The microplate was read at 595 nm in a plate reader and protein concentration was calculated based on standards and absorbance.
Statistical Analyses
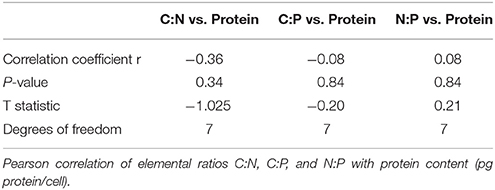
Carbon to nitrogen, nitrogen to phosphate, and carbon to phosphate ratios were calculated from POC, PON, and POP results. Outliers were eliminated using Tukey's method and the average of two technical replicates for each sample was calculated. Data were square-root transformed before all statistical analyses. A Welch's t-test was performed to determine the differences in mean C:P, C:N, and N:P ratios and protein content between the ancestral line and all adapted lines. Levene's test was performed to determine homogeneity of variances. A single factor ANOVA test was used to analyze the differences in mean C:P, C:N, and N:P ratios and protein content between groups of cells sharing the same mutation and between cell lines. Post-hoc analysis was performed using a Tukey's HSD test to analyze the relative contributions of each pair of means to the overall variation. The Pearson product-moment correlation coefficient was calculated for the protein content vs. C:N, N:P, and C:P ratios of individual lines and to compare epimutations with the rho and rpoB mutations. All analyses were done in R (R Development Core Team, 2016).
Results
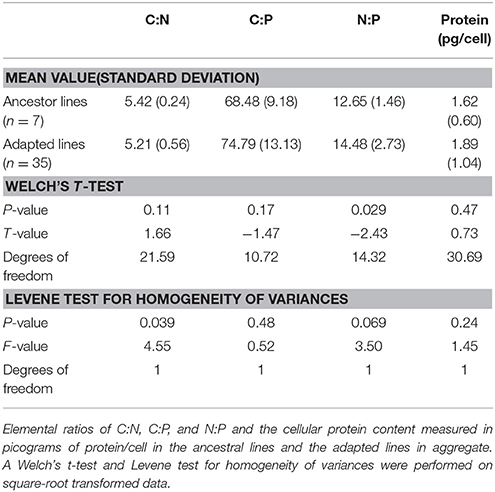
To identify the elemental outcome of adaptation to high temperature, we compared the C:N:P of the ancestral and adapted E. coli lines. We found that adaptation to high temperature did not have a single consistent effect on C:N, C:P, and N:P ratios (Figure 2). Although the C:N ratio of the adapted lines was slightly lower than the ancestral line (5.21 vs. 5.41), it was not significantly different (Welch's t-test, Table 1). The average N:P ratio of the adapted lines was significantly higher than the ancestor (14.48 vs. 12.65, Welch's t-test, Table 1). C:N variance was significantly different between ancestor and adapted lines and N:P variance was marginally but not significantly different (Levene's test, Table 1). To determine whether or not the differences in stoichiometric ratios could be explained by differences in protein level, the average protein content per cell was compared. Protein content was slightly higher in adapted cell lines (1.89 compared to 1.62 pg/cell), although not significantly (Welch's t-test, Table 1). Protein content vs. C:N had the strongest relationship of the elemental ratios with a negative correlation (r = −0.36, Table 2), but was not significant.
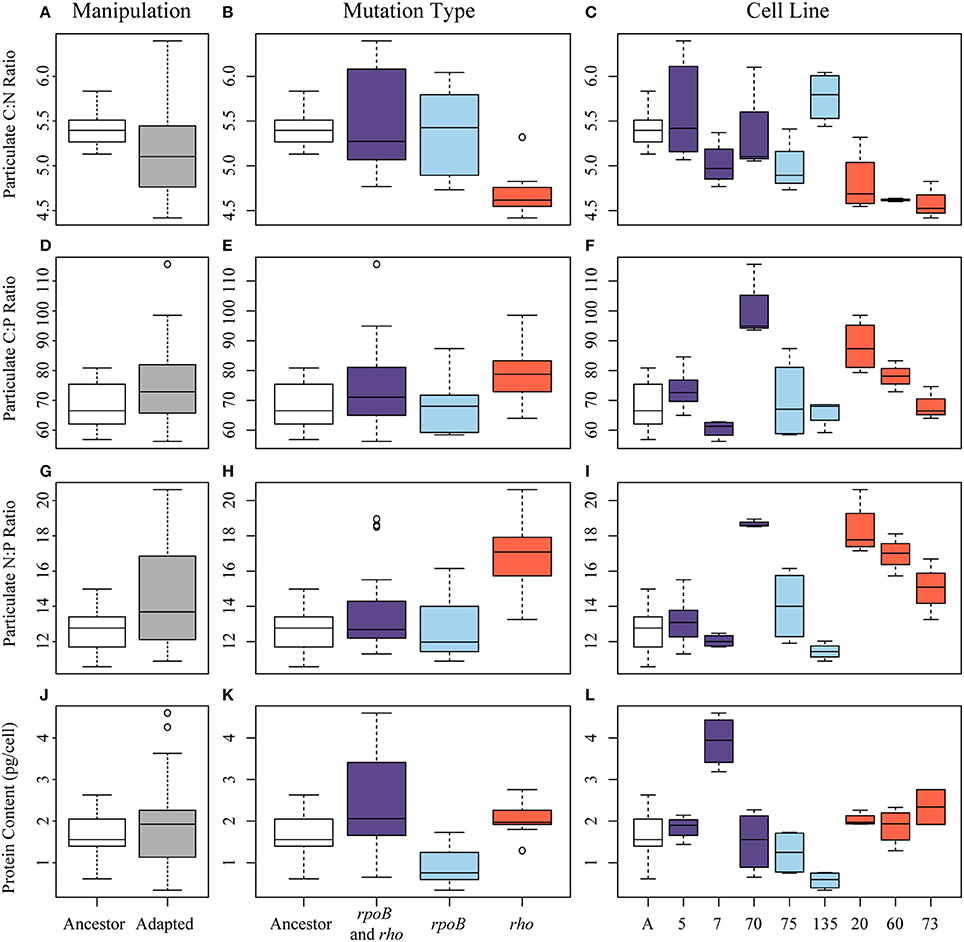
Figure 2. Elemental ratios and protein content of high temperature adapted E. coli. Boxplots representing the median value and upper and lower quartiles for particulate C:N of (A) the ancestor (white) and all adapted cell lines (gray); (B) by the mutation type either the ancestor, having both rho and rpoB, only rpoB, or only rho mutations; (C) or by line and colored by mutation type (white-ancestor, purple-both rpoB and rho mutations, light blue-rpoB mutation, red-rho mutation. Similarly for particulate C:P ratios in (D–F), particulate N:P ratios in (G–I), and protein content measured as picograms/cell in (J–L).
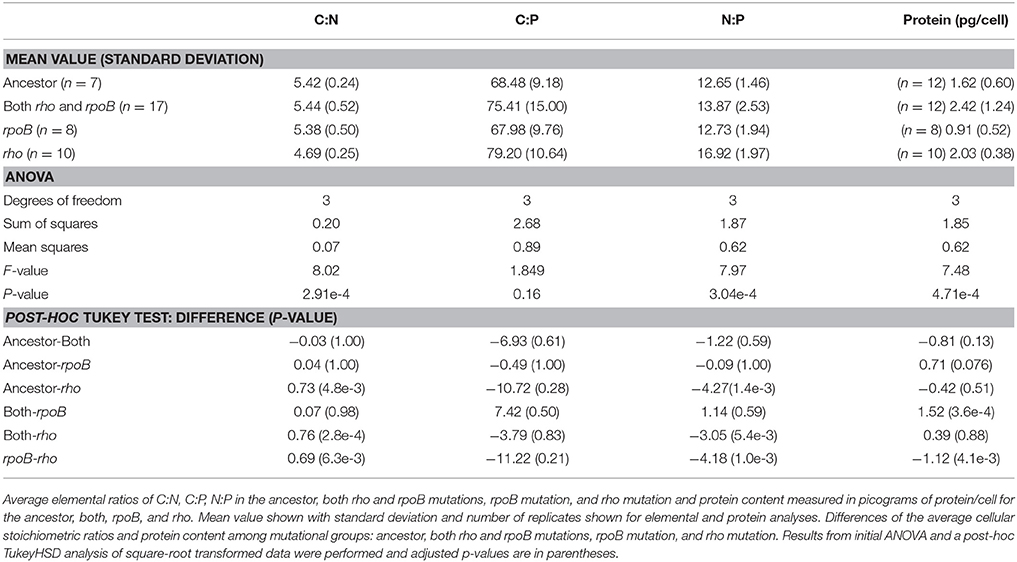
The specific genetic pathways for adaptation to high temperature had a significant effect on the elemental stoichiometry of E. coli (ANOVA, Table 3). Cell lines with the rpoB mutation had C:N, N:P, and C:P ratios nearly identical to the ancestor (Table 3). In contrast, cell lines with the rho mutation had significantly decreased C:N (4.69 vs. 5.42, TukeyHSD, Table 3) and increased N:P (16.92 vs. 12.65, TukeyHSD, Table 3) compared to the ancestor. Cell lines with both the rho and rpoB mutations had average stoichiometric ratios with values in between the rho and rpoB lines, but did not differ significantly from the ancestor. Protein content varied significantly between the mutation groups (ANOVA, Table 3; Figure 2). Cell lines with the rpoB mutation had the lowest protein content (0.91 pg/cell), while cell lines with both mutations had the highest (2.42 pg/cell) (Table 3). The rho group had significantly higher average protein content compared to the rpoB group (2.03 vs. 0.91 pg/cell, TukeyHSD, Table 3) and rho protein content was also higher than the ancestor, but not significantly higher (Table 3).
There was substantial variation in stoichiometric ratios among cell lines with the same mutation type. While cell lines with the rho mutation were consistent in the direction in which they varied from the ancestor, cell lines with the rpoB mutation or both mutations were sometimes higher than the ancestor and sometimes lower (Figures 2C,F,I,L). Thus, when pooled by mutation, those cell lines had average ratios similar to the ancestor. All lines have several additional mutations, 9 of which were shared between at least two lines (Supplementary Table 3). Some mutations were significantly associated with rho or rpoB mutations, e.g., line 75 and 135 both have mutations in yif B, a gene encoding a magnesium chelatase family protein, while other epimutations were less specific (Supplementary Table 4). Thus, the specific underlying genetic makeup of adaptation to high temperature can influence the cellular elemental ratios.
Discussion
Overall, there was strong variation in the complex phenotype of cellular stoichiometry between the high temperature adapted cell lines derived from a common ancestor. Thus, this study quantifies how the specific adaptive pathways underlying adaptation to high temperature are important for the stoichiometric outcome of the cell. Cells may adapt in many ways to increased temperature (Mongold et al., 1996; Riehle et al., 2003; Rudolph et al., 2010). Thus, adaptive mechanisms (i.e., compensation for degradation or increased macromolecular efficiency) may introduce unique changes in cellular stoichiometry. This work supports the idea that rho cell lines may compensate for the lower efficiency of proteins by accumulating a higher protein concentration than the ancestor and the rpoB type. There is a strong relationship between cellular nitrogen and protein content (Jones, 1941; Mariotti et al., 2008). Thus, a shift in protein concentration is likely the underlying cause for how the C:N and N:P ratio changes as a response to adaptation to high temperature in rho lines. The increase of N:P in rho lines relative to the ancestor suggests both mechanisms could be operating simultaneously. In contrast, lines with the rpoB mutation had significantly lower cellular amounts of protein compared to rho and slightly lower than the ancestor, although not significantly different. Considering how the stoichiometric ratios of these cell lines were nearly identical to the ancestor, this result does not follow either of the two initial hypotheses. Additionally, cell lines with both the rho and rpoB mutation did not vary much from the ancestor in stoichiometric ratios nor protein content. It is therefore difficult to align them with either model. Moreover, it suggests that stoichiometry need not change as a consequence of temperature adaptation.
In other efforts to characterize these pathways, the fitness tradeoffs of growing at different temperatures were assessed. A niche shift was observed associated with mutations in rpoB, while a niche expansion was observed in lines with a mutation in rho (Rodríguez-Verdugo et al., 2014). The pleiotropic effects of mutations in rpoB vs. rho may explain some of the results in this study. RNA polymerase affects every single gene during its function while rho only affects the transcription termination of a subset of genes (20–50%) (Zhu and Von Hippel, 1998; Peters et al., 2009). If a rpoB mutation influences multiple processes, the net effect on stoichiometry may result in the similarity we see between these lines and the ancestor. Single amino acid changes within rpoB have caused significant gene expression shifts. Specifically, changes in codon 572 of rpoB restored gene expression back toward the ancestral pre-stress conditions (Rodríguez-Verdugo et al., 2016). This supports a “compensation” response and may underlie the limited stoichiometric differences between the ancestor and the rpoB mutation lines.
There is a strong fitness advantage of the evolved lines compared to the ancestor when grown at 42.2°C. The ancestor also displays a strong difference in gene expression when grown at its optimal temperature of 37°C compared to 42.2°C (Rodríguez-Verdugo et al., 2016). Moreover, gene expression of engineered lines carrying either the rho or rpoB mutations in isolation also display differential gene expression from one another and from the ancestor (González-González et al., 2017) also some of the rpoB mutations appeared to be heading toward a pre-stressed ancestral state (Rodríguez-Verdugo et al., 2016). Yet the fact that different mutation types display differential CNP suggests that there is more happening than simply growth restoration. Some of the difference may have to do with co-occurring mutations, e.g., cardiolipin synthetase (cls) association with rho or rod-shape genes, which affect cell shape, and their association with rpoB (Tenaillon et al., 2012). Size of the cell may directly impact the cell stoichiometry, as seen in marine bacteria (Garcia et al., 2016). These affiliated mutations may be affecting the composition of cell membranes, for example, resulting in differential C:N:P.
Temperature influences both microbial community composition and physiology (Bennett et al., 1990; Zinser et al., 2007; Bradford et al., 2008), but we currently have a limited understanding of the stoichiometric outcomes of adaptation vs. acclimation to this environmental factor (Yvon-Durocher et al., 2015; Martiny et al., 2016). The work presented here uses a laboratory strain but demonstrates that unique evolutionary “biochemical outcomes” of short-term adaptation to a specific temperature regime can have a significant impact on the elemental composition. Microbes do experience sustained environmental changes, for example migration from one environment to another or movement from the external environment to mammalian intestines. Different shifts in environment will likely differ in the resource and stoichiometric mechanisms that are favored. Studies with other organisms are needed to evaluate if our observations apply more broadly to other microorganisms. Due to the broad effect of temperature on most, if not all, cellular processes, perhaps this variation in outcomes should be expected. It may further partly explain the extensive variation in how temperature affects closely related bacterial lineages (Martiny et al., 2016). Thus, there may be some overall trends in how different temperature conditions influence stoichiometry (Yvon-Durocher et al., 2015), but our data suggest that specific microbial lineages adapted to a particular temperature condition will likely have varying elemental stoichiometry. As such, there is not a uniform link between adaptation to elevated temperature and stoichiometry. Instead, the specific genetic and biochemical modifications underlying adaptation to a temperature regime need to be considered.
Author Contributions
AK and AM were involved in the conception and design of the project. KL conducted, analyzed, and interpreted the results. All authors helped write and revise the manuscript and provided important intellectual content.
Funding
KL was supported by the Undergraduate Research Opportunities Program at the University of California, Irvine. AK was supported by the National Science Foundation-Graduate Research Fellowship Program (DGE-1321846) and the National Institute of Biomedical Imaging and Bioengineering, National Research Service Award EB009418 from the University of California, Irvine, Center for Complex Biological Systems. AM was supported by the National Science Foundation (OCE-0928544 and OCE-1046297).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Rich Puxty for assisting with flow cytometry, Brandon Gaut for many helpful comments on the manuscript, and other members of the Martiny lab for their support and encouragement.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2017.00173/full#supplementary-material
References
Arrigo, K. R. (2005). Marine microorganisms and global nutrient cycles. Nature 437, 343–348. doi: 10.1038/nature04159
Arsene, F., Tomoyasu, T., and Bukau, B. (2000). The heat shock response of Escherichia coli. Int. J. Food Microbiol. 55, 3–9. doi: 10.1016/S0168-1605(00)00206-3
Barrick, J. E., Kauth, M. R., Strelioff, C. C., and Lenski, R. E. (2010). Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evol. 27, 1338–1347. doi: 10.1093/molbev/msq024
Bennett, A. F., Dao, K. M., and Lenski, R. E. (1990). Rapid evolution in response to high-temperature selection. Nature 346, 79–81. doi: 10.1038/346079a0
Bradford, M. A., Davies, C. A., Frey, S. D., Maddox, T. R., Melillo, J. M., Mohan, J. E., et al. (2008). Thermal adaptation of soil microbial respiration to elevated temperature. Ecol. Lett. 11, 1316–1327. doi: 10.1111/j.1461-0248.2008.01251.x
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Broeze, R. J., Solomon, C. J., and Pope, D. H. (1978). Effects of low temperature on in vivo and in vitro protein synthesis in Escherichia coli and Pseudomonas fluorescens. J. Bacteriol. 134, 861–874.
Burra, P. V., Kalmar, L., and Tompa, P. (2010). Reduction in structural disorder and functional complexity in the thermal adaptation of prokaryotes. PLoS ONE 5:e12069. doi: 10.1371/journal.pone.0012069
Cotner, J. B., Makino, W., and Biddanda, B. A. (2006). Temperature affects stoichiometry and biochemical composition of Escherichia coli. Microb. Ecol. 52, 26–33. doi: 10.1007/s00248-006-9040-1
Dijkstra, F. A., Pendall, E., Morgan, J. A., Blumenthal, D. M., Carrillo, Y., Lecain, D. R., et al. (2012). Climate change alters stoichiometry of phosphorus and nitrogen in a semiarid grassland. New Phytol. 196, 807–815. doi: 10.1111/j.1469-8137.2012.04349.x
Farewell, A., and Neidhardt, F. C. (1998). Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J. Bacteriol. 180, 4704–4710.
Garcia, N. S., Bonachela, J. A., and Martiny, A. C. (2016). Interactions between growth-dependent changes in cell size, nutrient supply and cellular elemental stoichiometry of marine Synechococcus. ISME J. 10, 2715–2724. doi: 10.1038/ismej.2016.50
González-González, A., Hug, S. M., Rodríguez-Verdugo, A., Patel, J. S., and Gaut, B. S. (2017). Adaptive mutations in RNA polymerase and the transcriptional terminator Rho have similar effects on Escherichia coli gene expression. Mol. Biol. Evol. 34, 2839–2855. doi: 10.1093/molbev/msx216
Grylak-Mielnicka, A., Bidnenko, V., Bardowski, J., and Bidnenko, E. (2016). Transcription termination factor Rho: a hub linking diverse physiological processes in bacteria. Microbiology 162, 433–447. doi: 10.1099/mic.0.000244
Hug, S. M., and Gaut, B. S. (2015). The phenotypic signature of adaptation to thermal stress in Escherichia coli. BMC Evol. Biol. 15:177. doi: 10.1186/s12862-015-0457-3
Hughes, B. S., Cullum, A. J., and Bennett, A. F. (2007). Evolutionary adaptation to environmental pH in experimental lineages of Escherichia coli. Evolution 61, 1725–1734. doi: 10.1111/j.1558-5646.2007.00139.x
Johnson, Z. I., Zinser, E. R., Coe, A., McNulty, N. P., Woodward, E. M. S., and Chisholm, S. W. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311, 1737–1740. doi: 10.1126/science.1118052
Jones, D. B. (1941). Factors for converting percentages of nitrogen in foods and feeds into percentages of proteins. Circ. US Dep. Agric. 183, 1–22.
Lee, Y. H., and Helmann, D. (2014). Mutations in the primary sigma factor σ A and termination factor Rho that reduce susceptibility to cell wall. Antibiotics 196, 3700–3711. doi: 10.1128/JB.02022-14
Lenski, R. E., Rose, M. R., Simpson, S. C., and Tadler, S. C. (1991). Long-term experimental evolution in Escherichia coli. I. adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341. doi: 10.1086/285289
Lomas, M. W., Burke, A. L., Lomas, D. A., Bell, D. W., Shen, C., Dyhrman, S. T., et al. (2010). Sargasso 321Sea phosphorus biogeochemistry: an important role for dissolved organic phosphorus (DOP). Biogeosciences 7, 695–710. doi: 10.5194/bg-7-695-2010
Mariotti, F., Tomé, D., and Mirand, P. P. (2008). Converting nitrogen into protein–beyond 6.25 and Jones' factors. Crit. Rev. Food Sci. Nutr. 48, 177–184. doi: 10.1080/10408390701279749
Martiny, A. C., Ma, L., Mouginot, C., Chandler, J. W., and Zinser, E. R. (2016). Interactions between thermal acclimation, growth rate, and phylogeny influence Prochlorococcus elemental stoichiometry. PLoS ONE 11:e0168291. doi: 10.1371/journal.pone.0168291
Mongold, J. A., Bennett, A. F., and Lenski, R. E. (1996). Evolutionary adaptation to temperature. IV. Adaptation of Escherichia coli at a niche boundary. Evolution 50, 35–43.
Moreno, A. R., and Martiny, A. C. (2018). Ecological stoichiometry of ocean plankton. Annu. Rev. Mar. Sci. 10, 1–27. doi: 10.1146/annurev-marine-121916-063126
Peters, J. M., Mooney, R. A., Kuan, P. F., Rowland, J. L., Keles, S., and Landick, R. (2009). Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 106, 15406–15411. doi: 10.1073/pnas.0903846106
Price, P. B., and Sowers, T. (2004). Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc. Natl. Acad. Sci. U.S.A. 101, 4631–4636. doi: 10.1073/pnas.0400522101
R Development Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org/
Riehle, M. M., Bennett, A. F., Lenski, R. E., and Long, A. D. (2003). Evolutionary changes in heat-inducible gene expression in lines of Escherichia coli adapted to high temperature. Physiol. Genomics 14, 47–58. doi: 10.1152/physiolgenomics.00034.2002
Rodríguez-Verdugo, A., Carrillo-Cisneros, D., Gonzalez-Gonzalez, A., Gaut, B. S., and Bennett, A. F. (2014). Different tradeoffs result from alternate genetic adaptations to a common environment. Proc. Natl. Acad. Sci. U.S.A. 111, 12121–12126. doi: 10.1073/pnas.1406886111
Rodríguez-Verdugo, A., Tenaillon, O., and Gaut, B. S. (2016). First-Step mutations during adaptation restore the expression of hundreds of genes. Mol. Biol. Evol. 33, 25–39. doi: 10.1093/molbev/msv228
Rudolph, B., Gebendorfer, K. M., Buchner, J., and Winter, J. (2010). Evolution of Escherichia coli for growth at high temperatures. J. Biol. Chem. 285, 19029–19034. doi: 10.1074/jbc.M110.103374
Sharp, J. H. (1974). Improved analysis for “particulate” organic carbon and nitrogen from seawater. Limnol. Oceangr. 19, 984–989.
Sterner, R. W., and Elser, J. J. (2002). Ecological Stoichiometry: the Biology of Elements from Molecules to the Biosphere. Princeton, NJ: Princeton University Press.
Taniguchi, H., Aramaki, H., Nikaido, Y., Mizuguchi, Y., Nakamura, M., Koga, T., et al. (1996). Rifampicin resistance and mutation of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 144, 103–108. doi: 10.1111/j.1574-6968.1996.tb08515.x
Tenaillon, O., Rodríguez-Verdugo, A., Gaut, R. L., McDonald, P., Bennett, A. F., Long, A. D., et al. (2012). The molecular diversity of adaptive convergence. Science 335, 457–461. doi: 10.1126/science.1212986
Toseland, A., Daines, S. J., Clark, J. R., Kirkham, A., Strauss, J., Uhlig, C., et al. (2013). The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Chang. 3, 979–984. doi: 10.1038/nclimate1989
VanBogelen, R., a, and Neidhardt, F. C. (1990). Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87, 5589–5593. doi: 10.1073/pnas.87.15.5589
Washburn, R. S., and Gottesman, M. E. (2011). Transcription termination maintains chromosome integrity. Proc. Natl. Acad. Sci. U.S.A. 108, 792–797. doi: 10.1073/pnas.1009564108
Yvon-Durocher, G., Dossena, M., Trimmer, M., Woodward, G., and Allen, A. P. (2015). Temperature and the biogeography of algal stoichiometry. Glob. Ecol. Biogeogr. 24, 562–570. doi: 10.1111/geb.12280
Zhu, A. Q., and Von Hippel, P. H. (1998). Rho-dependent termination within the trp t' terminator. I. Effects of Rho loading and template sequence. Biochemistry 37, 11202–11214.
Zimmerman, A. E., Allison, S. D., and Martiny, A. C. (2014). Phylogenetic constraints on elemental stoichiometry and resource allocation in heterotrophic marine bacteria. Environ. Microbiol. 16, 1398–1410. doi: 10.1111/1462-2920.12329
Keywords: stoichiometry, experimental evolution, adaptation, temperature, Escherichia coli
Citation: Linzner KA, Kent AG and Martiny AC (2018) Evolutionary Pathway Determines the Stoichiometric Response of Escherichia coli Adapted to High Temperature. Front. Ecol. Evol. 5:173. doi: 10.3389/fevo.2017.00173
Received: 01 March 2017; Accepted: 22 December 2017;
Published: 11 January 2018.
Edited by:
Patrick S. Fitze, Museo Nacional de Ciencias Naturales (CSIC), SpainReviewed by:
Bopi Biddanda, Grand Valley State University, United StatesOlivier A. Tenaillon, Institut National de la Santé et de la Recherche Médicale, France
Copyright © 2018 Linzner, Kent and Martiny. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alyssa G. Kent, YWdrZW50QHVjaS5lZHU=
 Krista A. Linzner
Krista A. Linzner Alyssa G. Kent
Alyssa G. Kent Adam C. Martiny
Adam C. Martiny