- 1Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2College of Life Science, Northeast Agricultural University, Harbin, China
- 3Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 4Key Laboratory of Plant Resources Conservation and Utilization, College of Biology and Environmental Sciences, Jishou University, Jishou, China
- 5Research and Collections Center, Illinois State Museum, Springfield, IL, United States
Adenocaulon (Asteraceae) is a small genus with only five species but has a broad amphi-Pacific distribution pattern with three species distributed disjunctly in South America, Central America, and North America and two endemic species spanning from eastern Asia to the Himalayas. To trace the biogeographic pattern of the genus, we reconstructed its phylogenetic relationships and diversification history based on one nuclear and eight plastid gene regions. Our results showed that Adenocaulon is monophyletic and may have originated in Central America during the Miocene, dispersed into North America and finally reached the Himalayas via the Bering Land Bridge. The hypothesized trajectory implies that long-distance dispersal may have played an important role in the formation of the distribution of this group of species. This hypothesis seems to have gained support from the special morphological structure of fruits of the genus.
Introduction
The genus Adenocaulon Hook., consisting of five species, is a small genus in the angiosperm family Asteraceae, and has a broad amphi-Pacific distribution (Bittmann, 1990). Two species are endemic to East Asia while the other three occur in North and South America. Among them, Adenocaulon nepalense Bittmann is native to Bhutan, Nepal and India, and Adenocaulon himalaicum Edgew is distributed from the Himalayas to northeastern Asia and Japan (Bittmann, 1990; Funk and Hind, 2016). In the New World, Adenocaulon bicolor Hook. is distributed in North America, Adenocaulon lyratum S. F. Blake in Guatemala and Chiapas of Mexico, and Adenocaulon chilense Less. in Chile and Argentina (Figure 1; Funk and Hind, 2016). Thus the five species generally present a disjunct distribution pattern, with no species of Adenocaulon present in Africa, the Mediterranean, tropical Asia or Oceania.
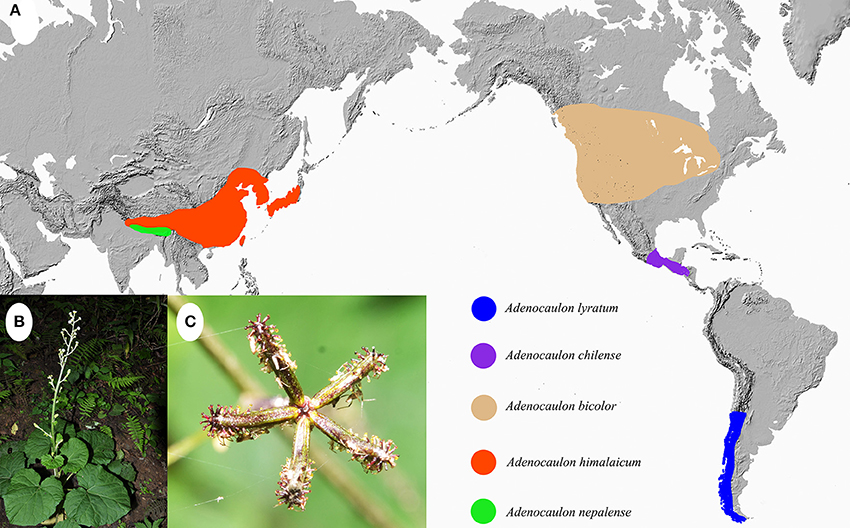
Figure 1. (A) Distribution of Adenocaulon species; (B,C), Habit (B) and achenes (C) of A. himalaicum.
Historically the taxonomic position of Adenocaulon has undergone extensive circumscription. Jensen and Kim (1996) placed it in the tribe Mutisieae, Katinas (2000) placed it in Cichorioideae, and Kim et al. (2002) transferred the genus to the Nassauvieae. A recent molecular phylogenetic study strongly supports the two American species, A. bicolor and A. chilense, forming a single lineage within Mutisieae (Panero and Funk, 2008), with the relationship of Adenocaulon to the other genera in Mutisieae being unresolved. The phylogenetic relationship and geographic pattern of the genus are still unclear. The tribe Mutisieae, as well as other members of the early diverging branches of Asteraceae (e.g., Barnadesioideae and Mutisioideae s.l.), has a predominant center of diversity in South America, which is believed to be the most possible ancestral area of these lineages (Katinas et al., 2009; Ortiz et al., 2009).
A distinct feature of members of Asteraceae is one-seeded cypselae fruit. Further, the majority of extant Asteraceae species have a modified calyx above the inferior ovary structure called a pappus, allowing for seed dispersal by wind and biological agents over great distances (Katinas et al., 2013). However, this specialized morphological trait is not evident in Adenocaulon. Members of this genus usually grow in under-stories of temperate moist forests (Katinas, 2000), which may well be a good case study of the biogeographic link between the Old World (specifically East Asia) and the New World by inferring the dispersal mechanism and biogeographic patterns across the Pan-Pacific areas.
Although the East Asian and North, Central and South American disjunct pattern is not unusual in angiosperms, and in fact has been investigated in several studies (e.g., Wen et al., 2010), the species-rich family Asteraceae provides a good system to probe the biogeographic pattern of taxa with distinct distribution patterns around the globe (Panero and Funk, 2008). Specifically members of Adenocaulon may provide an excellent opportunity to examine species adaptation in a recent diversification. Fossil and phylogenetic evidence demonstrates that Asteraceae may be of South American origin and underwent a major diversification event, followed by an African explosion (DeVore and Stuessy, 1995; Funk et al., 2005; Barreda et al., 2010; Stuessy, 2010) and subsequently moved to various continents (Nie et al., 2016). However, this model does not apply to the small genus Adenocaulon.
The goal of this study is to elucidate the phylogenetic relationships within the genus and to infer its historical biogeography utilizing chloroplast and nuclear DNA in conjunction with fossil evidence. Knowing details about how and when the genus originated and dispersed can provide insight into understanding the plausible biogeographic affinity between East Asia and America. Our study would shed new lights on the formation of intercontinental disjunction of plants.
Materials and Methods
Taxa Sampling and Data Collection
Ten samples (accessions) were used for each of the 5 species of Adenocaulon (Table 1). Based on the results from previous phylogenetic analyses of Asteraceae (Panero and Funk, 2008; Ortiz et al., 2009), additional samples were used from eight genera that are closely related to Adenocaulon (Gerbera, Leibnitzia, Chaptalia, Trichocline, Brachyclados, Pachylaena, Mutisia, and Chaetanthera of the tribe Mutisieae of the subfamily Mutisioideae) and 15 distantly related genera of the subfamily (Trixis, Dolichlasium, Jungia, Perezia, Nassauvia, Acourtia, Leucheria, Proustia, and Lophopappus in the tribe Nassauvieae of the subfamily Mutisioideae; Onoseris, Urmenetea, Lycoseris, Aphyllocladus, Gypothamnium, and Plazia from the tribe Onoserideae of the subfamily Mutisioideae). We also included nine species from Wunderlichioideae and five species from Barnadesioideae. We used Acicarpha spathulata (Calyceraceae) to serve as the outgroup, following Panero and Funk (2008) (Table 1).
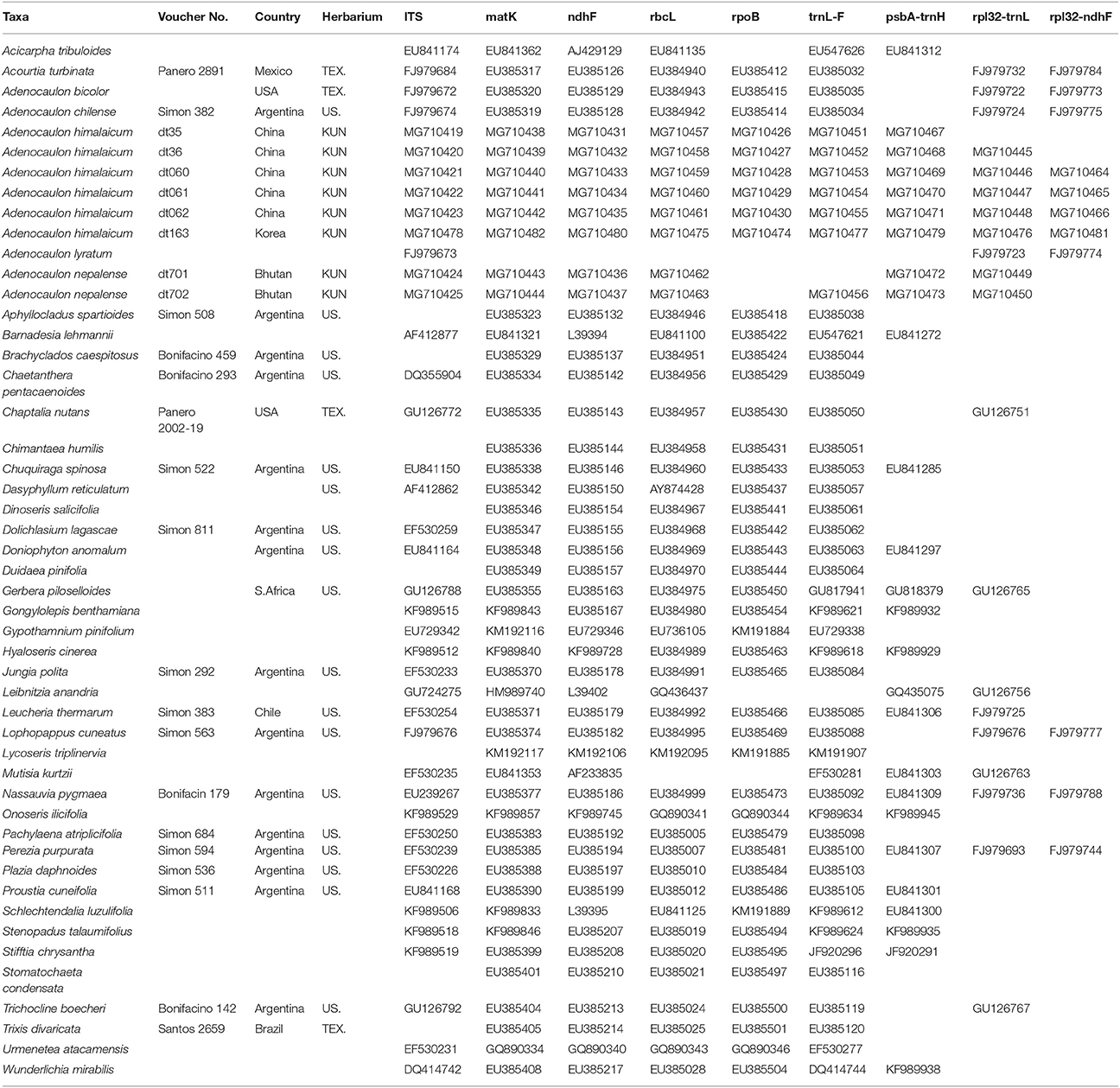
Table 1. Species names, localities, voucher deposition, and GenBank accession numbers used in the present study.
DNA Extraction, Amplification, and Sequencing
Total genomic DNA was isolated from silica-dried leaf materials and herbarium specimens using a Universal Genomic DNA Extraction Kit (Takara, Dalian, China). Nine gene regions, including eight plastid DNA regions (ndhF, matK, rpoB, rbcL, trnL-F, rpl32-trnL, rpl32-ndhF, and psbA-trnH) and the nuclear ITS, were employed. According to Shaw et al. (2007) and Timme et al. (2007), both rpl32-trnL and rpl32-ndhF have faster molecular evolution rate than other chloroplast regions and can provide adequate resolution in phylogenetic reconstructions of Asteraceae. The ITS data were generated from single amplifications using primers ITS4 and ITS5 (White et al., 1990). Primers used for amplification and sequencing were tabe and tabf for trnL-F region (Taberlet et al., 1991), Z1 and 1204R for rbcL (Zurawski et al., 1981), and psbA_F and trnH_R for psbA-trnH intergenic spacer (Sang et al., 1997). The primer sets for ndhF, matK, and rpoB regions were from Panero and Funk (2008), while the primers for rpl32-trnL and rpl32-ndhF were from Shaw et al. (2007). Amplified products were purified using a Qiaquick gel extraction kit (Qiagen, Inc., Valencia, California, USA) and sequenced in both directions by an ABI 3730 automated sequencer (Applied Biosystems, Foster City, California, USA). The resulting sequences were edited using Sequencher (version 4.1.4) and aligned with MUSCLE (version 3.6; Edgar, 2004), followed by manual adjustment in Se-Al (v2.0a11; Rambaut, 2002). All sequences generated for this study were deposited in GenBank under accession numbers shown in Table 1.
Phylogenetic Analysis
The nuclear and the plastid datasets were analyzed both separately and simultaneously using maximum parsimony (MP), Bayesian inference (BI), and maximum likelihood (ML). The topologies of the data sets were compared with each other to detect any incongruence. Because no significant incongruence was observed, we chose to combine the nrDNA and cpDNA data sets. For the maximum parsimony analysis we used PAUP/(version 4.0b10; Swofford, 2003). All characters were weighted equally and unordered. Heuristic searches were conducted using 100 random-taxon-addition replicates with tree bisection-reconnection (TBR) branch swapping, using MulTrees option in effect, and a maximum of 10,000 trees. Bootstrap analyses (1,000 pseudoreplicates) were conducted to examine the relative level of support for individual clades on the cladograms of each search (Felsenstein, 1985).
Models of nucleotide substitution were selected based on the Akaike Information Criterion (AIC) as determined by MrModelTest (version 2.3; Nylander, 2004). Maximum-likelihood searches and bootstrap analyses were performed on the XSEDE online computing cluster accessed via the CIPRES Science Gateway (Miller et al., 2010) using RAxML-HPC2 (version 7.4.2; Stamatakis et al., 2008). Data were partitioned into gene regions, allowing for independent parameter estimates on each partition, with branch length estimates optimized across all gene regions. The GTR + Γ model was employed for all analyses.
Bayesian inference was conducted using MrBayes (version 3.2.2, Ronquist and Huelsenbeck, 2003). The Markov chain Monte Carlo (MCMC) algorithm was run for 10,000,000 generations with one cold and three heated chains, starting from random trees and sampling one out of every 1,000 generations. The burn-in and convergence diagnostics were graphically assessed using AWTY (Nylander et al., 2008). The burn-in trees were excluded, and the remaining trees were assumed to be representative of the posterior probability (PP) distribution.
Divergence Time Estimation
The combined nuclear and plastid sequences were used to estimate the divergence time of Adenocaulon using BEAST (version 1.8.0; Drummond and Rambaut, 2007). BEAST employs a Bayesian MCMC approach to co-estimate topology, substitution rates and node ages (Drummond et al., 2002). BEAUti was used to set criteria for the analysis, in which we applied a general time reversible (GTR) nucleotide-substitution model with Gamma + Invariant sites, gamma shape distribution (with four categories) and proportion of invariant sites. A Yule tree prior model was implemented in the analysis, with rate variation across branches assumed to be uncorrelated and lognormally distributed (Drummond et al., 2006). Posterior distributions of parameters were approximated using two independent MCMC analyses of 50,000,000 generations (sampling once for every 5,000 generations) after a 10% burn-in for each. Convergence of the chains was checked using Tracer (version 1.5; Rambaut and Drummond, 2007).
For the fossil calibrations, we followed the strategy adopted by Nie et al. (2016). The first calibration point was the Raiguenrayun cura Barreda, Katinas, Passalia & Palazzesi capitulescence from the Patagonia described as belonging to the crown Asteraceae and dated by radiometric methods at 47.5 Ma (von Nickisch-Rosenegk et al., 1999). This fossil was used as a minimum age constraint for the split between Barnadesioideae and the rest of family and with an applied lognormal prior distribution with an offset of 47.5 Ma, a mean of 1.0 and a standard deviation of 0.7. As a minimum age constraint for the crown group of the subfamily Barnadesioideae, we used the fossil pollen Quilembaypollis sp., from the latest Oligocene/earliest Miocene, dated at 23 Ma (Chiang et al., 1998). Following Bergh and Linder (2009), the 95% confidence interval for this prior lies between 16.9 and 44.1 Ma with the mean at 22.3 Ma.
Biogeographic Analyses
We compiled distribution data for the Adenocaulon species and assigned the included taxa to respective ranges. Considering the areas of endemism in Adenocaulon and tectonic history of continents, we used four areas for the Adenocaulon taxa: (A) South America, (B) Central America, (C) North America, and (D) East Asia. Each sample was assigned to its respective area according to its contemporary distribution range.
Biogeographic inferences were obtained by applying both statistical dispersal–vicariance analysis (S-DIVA) and dispersal-extinction-cladogenesis (DEC) model calculated by Lagrange (Ree and Smith, 2008) implemented in RASP (version 3.0) using the default settings (Yu et al., 2010). A subset of 1,000 randomly selected trees from the posterior distribution output of BEAST was used and the maximum number of individual unit areas was set to two. The probability of dispersal between areas was maintained as equal.
Results
Phylogenetic Analyses
The incongruence length difference and Shimodaira and Hasegawa (SH) tests failed to reveal significant incongruence, allowing the individual datasets to be combined. There were 12,416 characters of combined data set, of which 1,571 were variable, and 1,266 parsimony informative sites. In general, the combined data matrix provided greater resolution and stronger support for phylogenetic relationships than did the individual datasets (Figure 2).
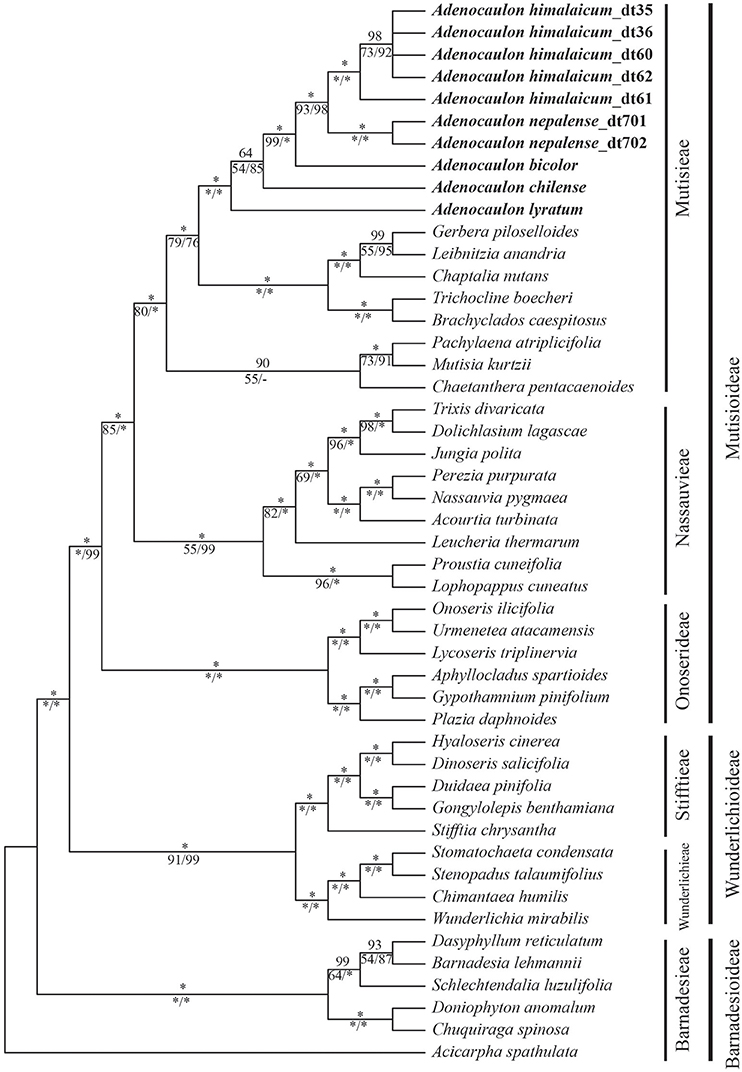
Figure 2. Phylogeny of the basal grade of the Asteraceae, including Mutisioideae, Wunderlichioideae and Barnadesioideae, based on eight chloroplast DNA regions (ndhF, matK, rpoB, rbcL, trnL-F, rpl32-trnL, rpl32-ndhF, and psbA-trnH) and nuclear ITS. Numbers above the nodes are Bayesian posterior probabilities and below the nodes are bootstrap values obtained from MP analysis and posterior probabilities support values from ML analysis. “*” indicates full support for the respective analysis.
The results from MP and BI analysis, as well as ML analysis, strongly support Adenocaulon being a monophyletic clade (PP = 100, BP = 100, PL = 100). Within Adenocaulon, the Central American species, A. lyratum, diverged first, followed by A. chilense being sister to a clade including the remaining species from North America and East Asia albeit with low support (PP = 64, BP = 54, PL = 85). The two species found in East Asia, A. himalaicum and A. nepalense, form a clade with high support values (PP = 100, BP = 93, PL = 98), which is sister to A. bicolor (North America) (Figure 2).
Biogeographic Analysis
The chronogram and results of divergence-time estimation based on the nrDNA and cpDNA data showed that the crown age of Adenocaulon was estimated at 9.05 Ma with a 95% highest posterior density (HPD) of 4.73–14.99 Ma. The stem age, that is the divergence time between Adenocaulon and its close relatives, was estimated at 14.74 Ma (with 95% HPD: 9.6–23.7 Ma). The split between the South American species A. chilense from the remaining species of Adenocaulon was at 6.92 Ma (95%HPD: 3.55–12.05 Ma), whilst the disjunction between North American A. bicolor and East Asian species at 4.79 Ma (95% HPD: 2.11–8.68) (Figure 3). S-DIVA analysis demonstrated that Adenocaulon may have originated in the tropical America and then dispersed to North America and East Asia (Figure 3).
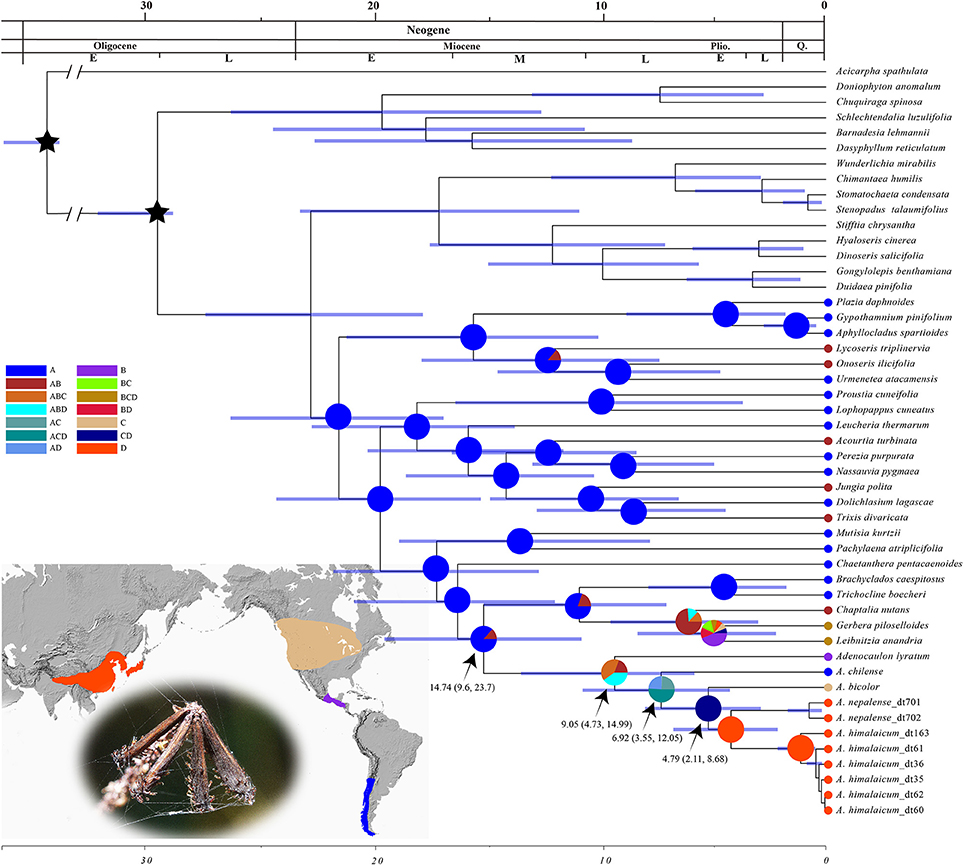
Figure 3. The results of BEAST analysis and RASP ancestral area reconstruction analysis based on combined chloroplast DNA and nrITS. Blue bars represent the 95% highest posterior density intervals for node ages. Fossil calibrations are marked with black stars. Four areas of endemism are defined: A (blue), South America; B (purple), Central America; C (yellow), North America; D (red), eastern Asia. Pie charts show probabilities of ancestral area reconstructions.
Discussion
Phylogenetic Relationship of Adenocaulon with Other Genera within Mutisieae
Our study showed that Adenocaulon is monophyletic within the Mutisieae (PP = 100, BP = 100, PL = 100) and is sister to a clade including Gerbera, Leibnitzia, Chaptalia, Trichocline, and Brachyclados (Figure 2). The genus is characterized by a distinct set of synapomorphy including basal constriction of anther appendages, small anthers, disciform capitula, and typical glandular achenes without pappus (Bittmann, 1990). The placement of Adenocaulon has long been disputed among numerous taxonomists, going as far as calling it an anomalous genus, and being unplaced in the tribe (Bremer, 1994). Based on our broad analysis of the early diverging clades of Asteraceae, Adenocaulon is confirmed to belong to the tribe Mutisieae (Mutisioideae) and to be sister to the Gerbera-complex (PP = 100, BP = 79, PL = 76) (Figure 2).
Our results indicate that the Central American A. lyratum diverged early. This species differs from the other four species in a stem winged by decurrently leaf bases, lyrate-pinnatifid sessile leaves and distinct club-shaped achene. Representatives of A. chilense from southern Chile and Argentina show petiolate entire leaves in a rosulate order. The pistillate flowers have a distinct bilabiate corolla and the scabrate pollen exhibit two layers of columellae with a larger inner one. The other three species from the Northern Hemisphere form a clade with high support. They share common features, e.g., leaves concentrated at the base, leaves triangular with cordate leaf base, heart- or kidney-shaped, with dentate or sinuate margin. Within this clade, the North American A. bicolor is sister to a clade with the two Asian species A. bicolor and A. himalaicum; these two species can be easily distinguished from each other by the glandular hairs: the former being pin-shaped, occurring on both vegetative and synflorescence branches, whereas the latter is nail-shaped but restricted to the synflorescence branches (Bittmann, 1990). A. nepalense is a recently described new species (Bittmann, 1990) endemic to Bhutan, Nepal, and India, and having distinct features such as winged by decurrently leaf-based stems and the bearing whip hairs achenes.
The Origin and Early Dispersal of Adenocaulon
Our biogeographic reconstruction suggests that Adenocaulon may have originated in Central America and then dispersed to North America and South America during the Miocene (14.74 Ma, 95% HPD: 9.6–23.7) (Figure 3). The dispersal of biota between Central America, North America and South America (commonly called the Great American Biotic Interchange) is one of the most significant events in neotropical biogeography. The Great American Biotic Interchange was caused by the closure of the Isthmus of Panama ca. 3 Ma (Burnham and Graham, 1999; Cody et al., 2010), resulting in the exchange of plants and animals between the two once separated continents. However, some authors suggest that an earlier land connection made possible the exchange of biota within the Americas. A land bridge may have existed between North and South Americas 3–7 Ma (Bermingham and Martin, 1998). Moreover, some plants could have successfully crossed the Isthmus of Panama before it closed (Cody et al., 2010; Bacon et al., 2015). Here, we propose that Adenocaulon may have dispersed into South America from Central America during the Miocene via the land bridge link. Hypotheses of long distance dispersal events have gained popularity in the last decade as a way of explaining many disjunct distributions of plant lineages (Givnish et al., 2004; Popp et al., 2011; Nie et al., 2013).
Dispersal of Adenocaulon may have been enhanced by birds due to its special fruit structures. The prominent glandular hairs on the cypselae are very sticky and cling readily to fabrics, fur, and feathers. The intercontinental disjunction within the range of Adenocaulon is possibly a result of dispersal of cypselae on the feathers of birds. The bird dispersal pattern may also account for the intracontinental disjunction of A. bicolor (in the Pacific Northwest, the Black Hills, and the northern Great Lakes region). Thus, we do not postulate vicariance as a factor that shaped the disjunct pattern of Adenocaulon.
The Intercontinental Dispersal within Northern Hemisphere
The intercontinental dispersal of plants in the Northern Hemisphere has been a focus of biogeography for a long time (Donoghue et al., 2001; Donoghue and Smith, 2004). During the Cenozoic, two routes for plant dispersal between the New World and the Old World were the Bering Land Bridge and the North Atlantic Land Bridge (Milne and Abbott, 2002; Wen et al., 2010). These two links have played significant roles in the formation of the modern flora of the Northern Hemisphere but happened in different geologic times (Tiffney, 1985a,b). The Bering Land Bridge is assumed to have existed from the Paleocene to Miocene (Tiffney and Manchester, 2001) but later this dispersal route was blocked due to global cooling (Milne, 2006). We suggest that Adenocaulon most likely migrated from North America to East Asia via the Bering Land Bridge. The estimated divergence time between the North American and East Asian groups is ca. 11.91 Ma, which coincided the cooling of Northern Hemisphere in which the continuous distribution of the Boreotropical flora was disrupted (Tiffney and Manchester, 2001). Our results suggest Adenocaulon may have originated in Central America, dispersed into North America and further into East Asia via the Bering Land Bridge, and finally reached the Himalayas.
Role of Fruits in Adenocaulon Dispersibility
Our results suggest that the biogeographic history of Adenocaulon species have been influenced by dispersal events, which could be facilitated by their unusual floral features. The pappus and the fruit trichome can prevent desiccation, and other characteristics of the family lack in Adenocaulon. Instead, the fruits of Adenocaulon are covered by glands with a sticky secretion. An umbrella-like “infructescence” emerges from the otherwise short plants usually embedded in other under-story vascular plants or mosses, and exposes the sticky fruits. Pervious researches indicated that viscid fruits can adhere by bird feathers (Carlquist, 1967, 1983). Also, the possibility that birds could have been the dispersal vectors for Adenocaulon fruits was proposed by Bittmann (1990) to explain some of the extant distributions of the genus. Alternatively, successful over-water dispersal events could be postulated for Adenocaulon; the oily fruit cover is suggested to aid in flotation, as was the case for other families. Thus, the absence of pappus in Adenocaulon does not mean that this genus lacks mechanisms for dispersal. The sticky fruits, with the “infructescence,” can act as an alternative dispersal mechanism involving several potential dispersal vectors.
Author Contributions
HS: conceived this research project; TD, YC, HW, XZ, SV, ZY, and HQ: collected the data; TD and YC: analyzed the data; TD, YC, SV, HW, and HS: led the writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Major Program of National Natural Science Foundation of China (31590823) and the National Key R&D Program of China (2017YFC0505200) to HS, and the National Natural Science Foundation of China (31700165) and CAS “Light of West China” Program to TD. We thank Dr. Jacob Landis of University of California, Riverside in English editing and Dr. Rinchen Yangzom of National Herbarium, National Biodiversity Center of Bhutan for providing leaf materials.
References
Bacon, C. D., Silvestro, D., Jaramillo, C., Smith, B. T., Chakrabarty, P., and Antonelli, A. (2015). Biological evidence supports an early and complex emergence of the Isthmus of Panama. Proc. Natl. Acad. Sci. U.S.A. 112, 6110–6115. doi: 10.1073/pnas.1423853112
Barreda, V., Palazzesi, L., Telleria, M. C., Katinas, L., and Crisci, J. V. (2010). Fossil pollen indicates an explosive radiation of basal Asteracean lineages and allied families during Oligocene and Miocene times in the Southern Hemisphere. Rev. Palaeobot. Palynol. 160, 102–110. doi: 10.1016/j.revpalbo.2010.02.004
Bergh, N. G., and Linder, H. P. (2009). Cape diversification and repeated out-of-southern-Africa dispersal in paper daisies (Asteraceae–Gnaphalieae). Mol. Phylogenet. Evol. 51, 5–18. doi: 10.1016/j.ympev.2008.09.001
Bermingham, E., and Martin, A. P. (1998). Comparative mtDNA phylogeography of neotropical freshwater fishes: testing shared history to infer the evolutionary landscape of lower Central America. Mol. Ecol. 7, 499–517. doi: 10.1046/j.1365-294x.1998.00358.x
Burnham, R. J., and Graham, A. (1999). The history of neotropical vegetation: new developments and status. Ann. M. Bot. Gard. 86, 546–589. doi: 10.2307/2666185
Carlquist, S. (1967). The biota of long-distance dispersal. V. Plant dispersal to Pacific Islands. Bull. Torrey Bot. Club 94, 129–162. doi: 10.2307/2484044
Chiang, T. Y., Schaal, B. A., and Peng, C. I. (1998). Universal primers for amplification and sequencing a noncoding spacer between the atpB and rbcL genes of chloroplast DNA. Bot. Bull.Acad. Sin. 39, 245–250.
Cody, S., Richardson, J. E., Rull, V., Ellis, C., and Pennington, R. T. (2010). The great American biotic interchange revisited. Ecography 33, 326–332. doi: 10.1111/j.1600-0587.2010.06327.x
DeVore, M. L., and Stuessy, T. F. (1995). “The place and time of origin of the Asteraceae, with additional comments on the Calyceraceae and Goodeniaceae,” in Advances in Compositae Systematics, eds D. J. N. Hind, C. Jeffrey, and G. V. Pope (Kew: Royal Botanic Gardens), 23–40.
Donoghue, M. J., Bell, C. D., and Li, J. (2001). Phylogenetic patterns in Northern Hemisphere plant geography. Int. J. Plant Sci. 162, S41–S52. doi: 10.1086/323278
Donoghue, M. J., and Smith, S. A. (2004). Patterns in the assembly of temperate forests around the Northern Hemisphere. Philos. T. R. Soc. B 359, 1633–1644. doi: 10.1098/rstb.2004.1538
Drummond, A. J., Ho, S. Y., Phillips, M. J., and Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. doi: 10.1371/journal.pbio.0040088
Drummond, A. J., Nicholls, G. K., Rodrigo, A. G., and Solomon, W. (2002). Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161, 1307–1320.
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids. Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Funk, V. A., Bayer, R. J., Keeley, S., Chan, R., Watson, L., Gemeinholzer, B., et al. (2005). Everywhere but Antarctica: using a supertree to understand the diversity and distribution of the Compositae. Biol. Skr. 55, 343–374.
Funk, V. A., and Hind, D. J. N. (2016). Typification of species names in Adenocaulon and Eriachaenium (Compositae/Asteraceae, Subfamily Mutisioideae, Tribe Mutisieae, Subtribe Adenocaulinae). PhytoKeys 69, 121–128. doi: 10.3897/phytokeys.69.9779
Givnish, T. J., Millam, K. C., Evans, T. M., et al. (2004). Ancient vicariance or recent long-distance dispersal? Inferences about phylogeny and South American–African disjunctions in Rapateaceae and Bromeliaceae based on ndhF sequence data. Ini. J. Plant Sci. 165, s35–s54. doi: 10.1086/421067
Jensen, L. H., and Kim, K. J. (1996). “Implications of chloroplast DNA for the classification and phylogeny of the Asteraceae,” in Proceedings of the International Compositae Conference, Vol. 1, eds D. J. N. Hind and H. Beentje (Kew: Royal Botanic Gardens), 317–339.
Katinas, L. (2000). Implications of morphological phylogenetics for the placement of the genera Adenocaulon and Eriachaenium (Asteraceae). Plant Syst. Evol. 223, 229–250. doi: 10.1007/BF00985282
Katinas, L., Crisci, J. V., Hoch, P., Tellería, M. C., and Apodaca, M. J. (2013). Trans-oceanic dispersal and evolution of early composites (Asteraceae). Perspect. Plant Ecol. 15, 269–280. doi: 10.1016/j.ppees.2013.07.003
Katinas, L., Sancho, G., Tellería, M. T., and Crisci, J. V. (2009). “Mutisieae sensu stricto (Mutisioideae sensu stricto),” Systematics, Evolution, and Biogeography of Compositae, in eds V. A. Funk, A. Susanna, T. F. Stuessy, and R. J. Bayer (Vienna: International Association for Plant Taxonomy), 229–248.
Kim, H.-G., Loockerman, D. J., and Jansen, R. K. (2002). Systematic implications of ndhF sequence variation in the Mutisieae (Asteraceae). Syst. Bot. 27, 598–609. doi: 10.1043/0363-6445-27.3.598
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. New Orleans, LA.
Milne, R. I. (2006). Northern Hemisphere plant disjunctions: a window on tertiary land bridges and climate change? Ann. Bot. 98, 465–472. doi: 10.1093/aob/mcl148
Milne, R. I., and Abbott, R. J. (2002). The origin and evolution of Tertiary relict floras. Adv. Bot. Res. 38, 281–314. doi: 10.1016/S0065-2296(02)38033-9
Nie, Z. L., Deng, T., Meng, Y., Sun, H., and Wen, J. (2013). Post-Boreotropical dispersals explain the pantropical disjunction in Paederia (Rubiaceae). Ann. Bot. 111, 873–886. doi: 10.1093/aob/mct053
Nie, Z. L., Funk, V. A., Meng, Y., Deng, T., Sun, H., and Wen, J. (2016). Recent assembly of the global herbaceous flora: evidence from the paper daisies (Asteraceae: Gnaphalieae). New Phytol. 209, 1795–1806. doi: 10.1111/nph.13740
Nylander, J. A. A. (2004). MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
Nylander, J. A. A., Wilgenbusch, J. C., Warren, D. L., and Swofford, D. L. (2008). AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583. doi: 10.1093/bioinformatics/btm388
Ortiz, S., Bonifacino, J. M., Crisci, J. V., Funk, V. A., Hansen, H. V., Nicholas Hind, D. J., et al. (2009). “The basal grade of Compositae: Mutisieae (sensu Cabrera) and Carduoideae,” in Systematics, Evolution, and Biogeography of Compositae, eds V. A. Funk, A. Susanna, T. F. Stuessy, and R. J. Bayer (Vienna, International Association for Plant Taxonomy), 193–213.
Panero, J. L., and Funk, V. A. (2008). The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol. Phylogenet. Evol. 47, 757–782. doi: 10.1016/j.ympev.2008.02.011
Popp, M., Mirre, V., and Brochmann, C. (2011). A single Mid-Pleistocene long-distance dispersal by a bird can explain the extreme bipolar disjunction in crowberries (Empetrum). Proc. Natl. Acad. Sci. U.S.A. 108, 6520–6525. doi: 10.1073/pnas.1012249108
Rambaut, A. (2002). Se-Al: Sequence Alignment Editor, version 2.0a11. Department of Zoology, University of Oxford. Available online at: http://tree.bio.ed.ac.uk/software/seal/
Rambaut, A., and Drummond, A. J. (2007). Tracer v1.4. Available online at: http://beast.bio.ed.ac.uk/Tracer
Ree, R. H., and Smith, S. A. (2008). Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14. doi: 10.1080/10635150701883881
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sang, T., Crawford, D. J., and Stuessy, T. F. (1997). Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 84, 1120–1136. doi: 10.2307/2446155
Shaw, J., Lickey, E. B., Schilling, E. E., and Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose non-coding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288. doi: 10.3732/ajb.94.3.275
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Swofford, D. L. (2003). PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10. Sunderland, MA: Sinauer Associates.
Taberlet, P., Gielly, L., Pautou, G., and Bouvet, J. (1991). Universal primers for amplification of 3 noncoding regions of chloroplast DNA. Plant Mol. Biol. 17:1105–1109. doi: 10.1007/BF00037152
Tiffney, B. H. (1985a). The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. J. Arno. Arbo. 66, 243–273. doi: 10.5962/bhl.part.13183
Tiffney, B. H. (1985b). Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. J. Arno. Arbo. 66, 73–94. doi: 10.5962/bhl.part.13179
Tiffney, B. H., and Manchester, S. R. (2001). The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere Tertiary. In. J. Plant Sci. 162, S3–S17. doi: 10.1086/323880
Timme, R. E., Kuehl, J. V., Boore, J. L., and Jansen, R. K. (2007). A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am. J. Bot. 94, 302–312. doi: 10.3732/ajb.94.3.302
von Nickisch-Rosenegk, M., Silva-Gonzalez, R., and Lucius, R. (1999). Modification of universal 12S rDNA primers for specific amplification of contaminated Taenia spp. (Cestoda) gDNA enabling phylogenetic studies. Parasitol. Res. 85, 819–825. doi: 10.1007/s004360050638
Wen, J., Ickert-Bond, S., Nie, Z. L., and Li, R. (2010). “Timing and modes of evolution of eastern Asian-North American biogeographic disjunctions in seed plants,” in Darwin's Heritage Today: Proceedings of the Darwin 200 Beijing International Conference eds M. Long, H. Gu, and Z. Zhou (Beijing: Higher Education Press).
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols: A Guide to Methods and Applications, M. A. Innis, D. H. Gelfand, J. J. Shinsky, and T. J. White (San Diego, CA: Academic Press), 315–322.
Yu, Y., Harris, A. J., and He, X. J. (2010). S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 56, 848–850. doi: 10.1016/j.ympev.2010.04.011
Keywords: Adenocaulon, Asteraceae, East Asia, America, herbaceous flora
Citation: Deng T, Chen Y, Wang H, Zhang X, Volis S, Yusupov Z, Qian H and Sun H (2018) Molecular Phylogeny and Biogeography of Adenocaulon Highlight the Biogeographic Links between New World and Old World. Front. Ecol. Evol. 5:162. doi: 10.3389/fevo.2017.00162
Received: 26 August 2017; Accepted: 29 November 2017;
Published: 04 January 2018.
Edited by:
Daniel M. Griffith, Oregon State University, United StatesReviewed by:
Tao Su, Xishuangbanna Tropical Botanical Garden (CAS), ChinaXiaolei Huang, Fujian Agriculture and Forestry University, China
Copyright © 2018 Deng, Chen, Wang, Zhang, Volis, Yusupov, Qian and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hang Sun, c3VuaGFuZ0BtYWlsLmtpYi5hYy5jbg==
†These authors have contributed equally to this work.
 Tao Deng1†
Tao Deng1† Hengchang Wang
Hengchang Wang Sergei Volis
Sergei Volis Ziyoviddin Yusupov
Ziyoviddin Yusupov Hang Sun
Hang Sun