- 1UMR8576, Centre National de la Recherche Scientifique, INRA USC 1409, Lille University of Science and Technology, Unité de Glycobiologie Structurale et Fonctionnelle, Lille, France
- 2Miniaturisation Pour la Synthèse, l'Analyse et la Protéomique, Centre National de la Recherche Scientifique USR 3290, Lille University of Science and Technology, Lille, France
- 3ALLICE, Paris, France
- 4INRA, UEPAO, Unité Expérimentale de Physiologie Animale de l'Orfrasière, Nouzilly, France
Odorant-Binding Proteins (OBP) are major players of perireceptor events in olfaction. Despite their importance, a molecular mechanism explaining their specificity for odors and pheromones has yet to be proposed. A new approach is provided by the analysis of the pig olfactory secretome that is mainly composed of OBP isoforms, generated from 3 gene products by two types of post-translational modifications (PTM): (i) phosphorylation and (ii) O-β-N-acetylglucosaminylation (O-GlcNAcylation), which are unusual for secreted proteins. Although both types of PTM can be demonstrated on OBP isoforms by specific antibodies, they have to be identified by mass spectrometry (MS), as localizing PTM sites and identifying PTM patterns can help predict binding affinities. In this paper, we report the identification of phosphorylation and O-GlcNAcylation sites on peptides coming from trypsin digestion of only OBP (sensu stricto) by nanoLC-nanoESI-HCD-MS/MS-Orbitrap. These PTM were not present on VEG and SAL. PEAKS software analysis of raw MS data allowed selecting spectra that were analyzed manually to identify PTMs. Four peptides corresponding to two different portions of OBP sequence were modified either by a phosphate group or by a hexNAc moiety. Due to the high energy used in HCD, the data did not allow precise localization of the modified sites. The new findings contribute to a better understanding of the mechanisms by which OBP isoforms could extend the binding repertoire of the secreted OBPs. Data are available via ProteomeXchange with identifier PXD007955.
Introduction
Odorant-binding proteins (OBPs) are one of the molecular players involved in perireceptor events of olfaction (Pelosi, 1996). These globular proteins bind odorant molecules into a hydrophobic pocket (Spinelli et al., 1998) and transport them to olfactory receptors, ultimately responsible for olfactory transduction (Buck and Axel, 1991). Whether the odorant molecule or the complex OBP/odorant acts as a receptor ligand is still an open question, as none of the two hypotheses have received direct experimental evidence. The recent discovery of OBP isoform diversity in pig (Sus scrofa) nasal mucus (Nagnan-Le Meillour et al., 2014) favors the hypothesis that OBPs could be more than passive carriers of odorants, being more likely involved in the selection of chemical structures to be presented to a given olfactory receptor. Proteomic analysis of pig nasal mucus revealed that the olfactory secretome (Nagnan-Le Meillour et al., 2014) is mainly composed of around 30 OBP-, VEG- (Von Ebner's Gland protein), and SAL- (Salivary Lipocalin) isoforms, resulting from post-translational modifications (PTMs) of these three gene products and variants. The pig genome contains three genes coding for odorant-binding proteins: (i) the OBP gene sensu stricto, (ii) a second OBP transcript encoded by the porcine VEG (Von Ebner's gland protein) gene, LCN1, (iii) a third OBP, the salivary lipocalin (SAL) encoded by SAL1 gene. OBP and LCN1 genes have two transcripts (OBPX1 and OBPX2, VEG-VNO and VEG-RM, respectively), whereas SAL1 leads to a single transcript. In addition to their splicing variants, protein isoforms for these three OBP genes vary between tissues, and in the nasal mucus, around 9 protein isoforms have been previously identified for VEG, 7 for SAL and 12 for OBP (Nagnan-Le Meillour et al., 2014), due to post-translational modifications targeting the secreted proteins. Immunodetection with specific antibodies indicated that only OBP sensu stricto isoforms are modified by phosphorylation and O-linked-β-N-acetylglucosaminylation (O-GlcNAcylation).
If phosphorylation of proteins following the secretion pathway has recently been documented (Tagliabracci et al., 2015), few data report on O-GlcNAcylation of secreted proteins (Alfaro et al., 2012). Indeed, typical glycosylation takes place in cellular compartments processing secreted proteins, such as the Endoplasmic Reticulum (ER) and Golgi apparatus (Schwarz and Aebi, 2011). It consists in the addition of complex glycan chains on asparagine (N-linked) or serine/threonine (O-linked mucin type) residues, whereas O-GlcNAc moieties attached on nuclear and cytoplasmic proteins are not elongated to form complex glycans. Since its discovery (Torres and Hart, 1984), O-GlcNAcylation has been shown to regulate almost all intracellular functions by a dynamic interplay together with phosphorylation on the same S/T residues or on adjacent sites (for a full review see Hart and Akimoto, 2009). In the same way that nucleocytoplasmic glycosylation was absent from the Glycobiology dogma until its discovery, many publications also claimed that O-GlcNAcylation cannot occur within the secretion pathway. However, it was recently reported in Drosophila that EGF repeats of extracellular domains of Notch1 are O-GlcNAcylated by a newly identified glycosyltransferase, EOGT (EGF domain specific O-Linked N-acetylglucosamine transferase; Sakaidani et al., 2011), ER-resident and totally unrelated to its intracellular counterpart, OGT (O-linked N-acetylglucosamine transferase). The new enzyme has—for obvious reasons—been named “EGF-domain specific O-GlcNAc transferase,” even if its substrate specificity has not been fully investigated. Although the EOGT gene is well conserved throughout evolution (Nagnan-Le Meillour et al., 2014), the role of extracellular O-GlcNAcylation in Mammals remains to be elucidated, despite identification of an increasing number of modified proteins (Alfaro et al., 2012; Hoffman et al., 2012). A first opening on possible roles of this process is the report of a mutation in the EOGT gene associated with a phenotype of Adams-Oliver syndrome, a human congenital disorder (Shaheen et al., 2013). In pigs, the odorant binding properties of OBP (sensu stricto) isoforms seem to be driven at least by phosphorylation (Brimau et al., 2010) and we hypothesize that O-GlcNAcylation patterns could have a similar function. Before initiating functional assays, we need to identify OBP O-GlcNAcylation by high-resolution mass spectrometry. In this paper we report on the identification of phosphorylation and O-GlcNAcylation of OBP (sensu stricto) peptides, by nanoLC-nanoESI-HCD-MS/MS-Orbitrap from 2-DE spots of total pig olfactory secretome.
Materials and Methods
Animals and Tissues
Animals (Large White Sus scrofa) were bred at the experimental farm of INRA (UEPAO, Nouzilly, France). Individuals coming from the same offspring (brothers) were slaughtered by a licensed butcher at UEPAO slaughterhouse (authorization # A37801 E37-175-2) in agreement with EU directives about animal welfare. Two adult males were used in this study (300014 - M14 and 300039 - M39). Respiratory mucosa was collected immediately after death from each animal and stored in tubes at −80°C before protein extraction.
Protein Extraction
Proteins were extracted from pig tissues by phase partition using chloroform/methanol (v/v, 2/1) on ice. After soft grinding in this solution, samples were centrifuged (15,000 g for 15 min at 4°C) and the methanol phase was collected and evaporated in a Speed-Vac concentrator (Eppendorf). Dried samples were stored at −20°C. Aliquots representing 1/250 and 1/500 of each tissue were tested by native-PAGE as already described (Guiraudie et al., 2003) and compared with an abacus previously obtained with recombinant porcine OBP (Brimau et al., 2010) in order to estimate the quantity of proteins in each tissue for the standardization of sample loading in 2-DE.
Two-Dimensional Gel Electrophoresis (2-DE)
All chemicals and reagents were from Sigma-Aldrich. Each sample (M14 and M39) was divided into six identical aliquots for Coomassie-blue staining, immunodetection with anti-OBP, anti-VEG, anti-SAL, anti-phosphoS/Y/T, and anti-O-GlcNAc CTD110.6. For the first separation in IEF (isoelectric focalization), each aliquot of dried proteins (15 μg) representing 1/6 of each sample (M14 and M39) was solubilized in 150 μL of rehydration buffer [8 M urea, 2 M thiourea, 2% (w/v) CHAPS, 10 mM dithiothreitol (DTT), 1.2% (v/v) IPG (immobilized pH gradient) buffer (pH 3-5.6, GE Healthcare) and bromophenol blue]. After vigorous shaking, proteins were loaded onto 7-cm IPG strip (non-linear pH 3–5.6, GE Healthcare) by overnight passive rehydration at room temperature (RT). The first dimensional IEF was carried out on a PROTEAN IEF Cell (Bio-Rad) using the following program: 300 V for 30 min, 1,000 V for 1 h, 5,000 V for 2 h and 500 V for 3 h, with a current limited to 50 μA/gel. All steps were performed at rapid ramped-voltage. When IEF was complete (10,000 V.h final), strips were incubated twice for 15 min in the equilibration buffer [375 mM tris-HCl pH 8.8, 6 M urea, 2% (w/v) SDS (sodium dodecyl sulfate) and 30% (v/v) glycerol] containing first 1.5% (w/v) DTT then 2% (w/v) iodoacetamide. The second dimension separation was performed using 16.8% SDS-PAGE. The 12 gels were run at the same time in Criterion® Dodeca™ Cell (Bio-Rad) to minimize differences in migration in order to superimpose the scanned images with ImageJ® software.
Staining and Western-Blot
After 2-DE, gels were either stained with colloidal Coomassie blue R solution (12% trichloroacetic acid, 5% ethanolic solution of 0.035% Serva blue R-250), or transferred onto nitrocellulose membrane (Hybond C-Extra, GE Healthcare) for PTM characterization, or PVDF membranes (Immobilon P, Millipore) for primary sequence characterization. For immunodetection of O-GlcNAc modified proteins, we used the Thermo Scientific Pierce “O-GlcNAc western-blot detection kit,” following manufacturer's instructions. For other immunodetections, membranes were blocked overnight at 4°C either in 5% (w/v) non-fat dry milk in tris-buffered saline-0.05% tween-20 (TBS-T) for probing with polyclonal antibodies (anti-OBP, anti-SAL and anti-VEG) or in 3% bovine serum albumin (BSA) for probing with anti-phosphoserine/threonine/tyrosine monoclonal antibodies (SPM101 Mab, Fisher Scientific). Membranes were then incubated with antibodies in PBS-T overnight at 4°C (CTD110.6 of the kit mentioned above, Thermo Scientific Pierce, 1:5,000) or TBS-T 1 h at room temperature (RT) (SPM101, 1:500; Anti-OBP, 1:50,000; Anti-VEG, 1:5,000; Anti-SAL, 1:10,000). After six washes in PBS-T or TBS-T, membranes were incubated 1 h at RT with the appropriate horseradish peroxidase-conjugated secondary antibody [anti-mouse IgM-HRP for CTD110.6, 1:10,000; anti-rabbit IgG-HRP for polyclonal antibodies, 1:30,000; anti-mouse IgG-HRP for SPM101 (all secondary antibodies were from Fisher Scientific)]. After 6 final washes in PBS-T or TBS-T, blots were developed using enhanced chemiluminescence (ECL Plus and ECL Prime reagents, GE Healthcare; Super-Signal™ West Dura, Fisher Scientific). Scanned images of stained gels and blots were merged using Image J software.
Digestion of Protein Spots
Spots of interest were excised from the Coomassie-stained gels and rinsed with a mixture of ACN 50%/50 mM ammonium bicarbonate (v/v), then dehydrated with ACN (acetonitrile). The gel slices were subjected to reduction of disulfide bonds by 10 mM DTT at 45°C for 1 h. Alkylation step was then performed with 50 mM iodoacetamide for 1 h at room temperature in dark. Before trypsin digestion, the gel slices were washed with 50 mM ammonium bicarbonate and dehydrated with ACN. Gel slices were then incubated overnight with 100 ng of trypsin in 50 mM ammonium bicarbonate (Trypsin Gold; Promega) at 37°C. Peptides were extracted by two incubations in formic acid 10%/ACN at 30°C for 15 min, then in formic acid 5%/ACN at RT for 15 min. Supernatants were pooled and dried in Speed-Vac concentrator (Eppendorf).
Mass Spectrometry (MS) Analysis
A nanoflow liquid chromatography (LC) instrument (nanoLC U3000 RSLC, Thermo Fisher Scientific) was coupled on-line to a Q Exactive plus (Orbitrap, Thermo Scientific) with a nanoelectrospray ion source (nanoLC-nanoESI). Peptides were resuspended in 10 μL of nano-HPLC buffer A (5% ACN/0.1% formic acid) and 1 μL (corresponding to 200 ng of proteins) was loaded onto the pre-concentration trap (Thermo Scientific, Acclaim PepMap100 C18, 5 μm, 300 μm i.d × 5 mm) using partial loop injection, for 5 min at a flow rate of 10 μL.min-1 with buffer A, then separated on column (Acclaim PepMap100 C18, 3 μm, 75 mm i.d. × 500 mm) with a linear gradient of 5–50% buffer B (75% ACN/0.1% formic acid) at a flow rate of 250 nL.min−1 and 45°C. The total time for a LC MS/MS (tandem mass spectrometry) run was about 240 min long and each sample was injected three times.
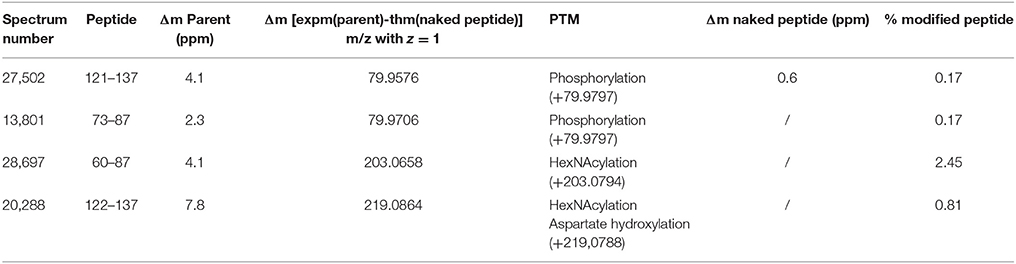
MS data were acquired on Q Exactive plus using a data-dependent top 20 method dynamically choosing the most abundant precursor ions from the survey scan (400–1,600 m/z) for HCD fragmentation (High-energy Collision Dissociation). Dynamic exclusion duration was 60 s. Isolation of precursors was performed with a 1.6 m/z window and MS/MS scans were acquired with a starting mass of 80 m/z. Survey scans were acquired at a resolution of 70,000 at m/z 400 (AGC set to 1,106 ions with a maximum fill time of 120 ms). Resolution for HCD spectra was set to 35,500 at m/z 200 (AGC set to 5.105 ions with a maximum fill time of 180 ms). Normalized collision energy was 28 eV. The underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.3%. The instrument was run with peptide recognition mode (i.e., from 2 to 8 charges), exclusion of singly charged and of unassigned precursor ions enabled. The acquired raw files were analyzed with PEAKS 7 studio software (Bioinformatics Solutions Inc., Waterloo, Canada) using a custom-made OBP database (Table 1). The full sequences and details are provided in Supplementary Data 1. The peptide mass tolerance was set to 10 ppm and 0.01 Da for MS/MS. Variable modifications included were as follows: Oxidation of M, Y, H, deamidation of N, Q, carbamidomethylation of C, phosphorylation of Y, S, T, pyro-G, and O-N-acetylhexosaminylation of S, T. For high-confidence peptide identification a FDR (False Discover Rate) of 1% and a minimum of 2 ranked peptides were used for peptide filtering.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD007955.
Results
Identification of OBP-, VEG-, and SAL- Isoforms Contained in 2-DE Spots by Western-Blot with Specific Antibodies
With the extraction method used in this study, only secreted proteins (OBP sensu stricto, VEG, and SAL) of the nasal mucus were extracted and, as revealed by 2-DE, compose the pig olfactory secretome. Coomassie-blue staining revealed similar profiles for the two male secretomes (Figures 1, 2A,E). The distribution pattern shows that OBP (Figures 2B,F), VEG (Figures 2C,G), and SAL (Figures 2D,H) isoforms segregate into 3 well-separated groups with no apparent overlapping. SAL isoforms are not visible with Coomassie-blue staining (Figures 2A,E), but are well labeled by specific antibodies (Figures 2D,H). Western-blot with anti-O-GlcNAc and anti-phosphoS/Y/T labeling co-localized only with OBP sensu stricto-containing spots (Figures 3A–D, respectively) in both samples. From the Coomassie-stained gel of M14 sample, 42 spots were cut (Figure 4) and analyzed by nanoLC-nanoESI-HCD-MS/MS-Orbitrap.
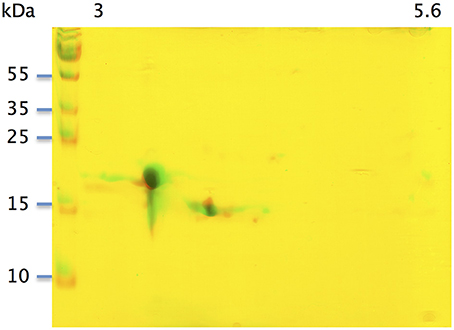
Figure 1. Two-dimensional electrophoresis of soluble proteins extracted from two pubertal male pigs M14 and M39. Merge of Coomassie-blue staining of M14 and M39 secretomes obtained by 2-DE. X-axis: pI, Y-axis: molecular weight.
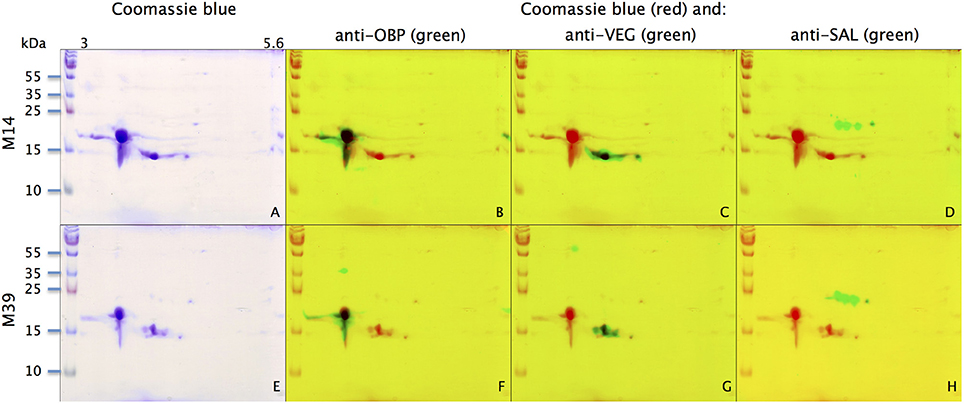
Figure 2. Identification of OBP, VEG, and SAL with specific antibodies in M14 (A–D) and M39 (E–H) secretomes. (A,E) Coomassie-blue staining. (B,F) Merge of Coomassie-blue staining (red) and western-blot with anti-OBP (green). (C,G) Merge of Coomassie-blue staining (red) and western-blot with anti-VEG (green). (D,H) Merge of Coomassie-blue staining (red) and western-blot with anti-SAL (green). X-axis: pI, Y-axis: molecular weight.
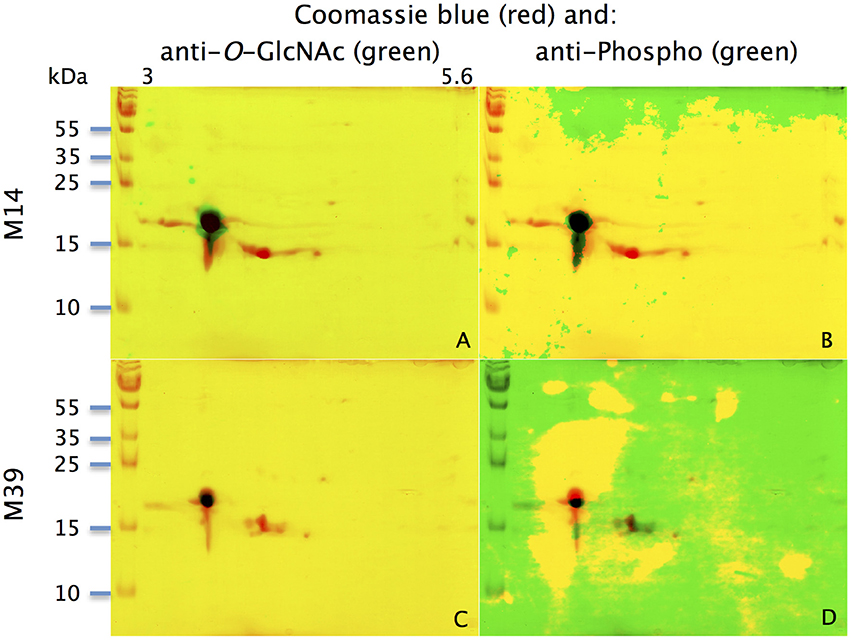
Figure 3. Identification of potential phosphorylation and O-GlcNAcylation of M14 (A,B) and M39 (C,D) protein spots. M14: (A,C) represent the merging of Coomassie-blue staining (red) and western-blot with anti-O-GlcNAc (CTD110.6, green) for individuals M14 and M39, respectively. (B,D) represent the merging of Coomassie-blue staining (red) and western-blot with anti-phosphoS/Y/T (green) for M14 and M39, respectively. X-axis: pI, Y-axis: molecular weight
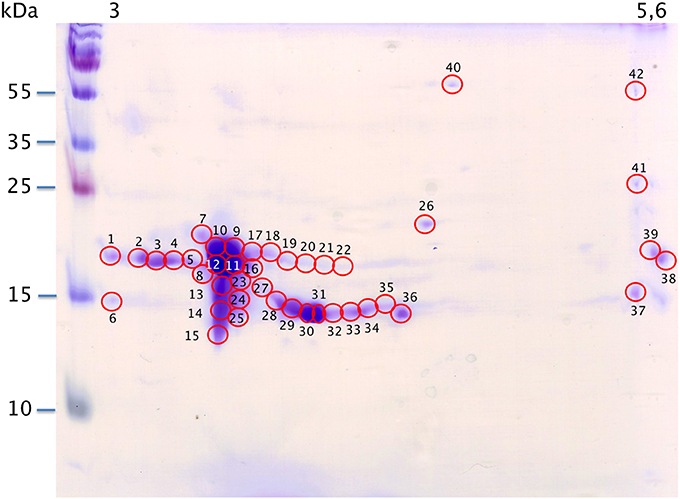
Figure 4. Map of M14 sample spotting (Coomassie blue staining) for nanoLC-nanoESI-HCD-MS/MS-Orbitrap analysis. X-axis: pI, Y-axis: molecular weight. Numbering corresponds with data in Supplementary Table S1.
Characterization of OBP-, VEG-, and SAL-Isoforms from 2-DE Spots by NanoLC-NanoESI-HCD-MS/MS-Orbitrap
Raw files were analyzed by PEAKS software using the custom-made OBP database (Table 1) and resulting data are given in Supplementary Table S1. PEAKS software considers as relevant those identifications with score higher than 67. A few identifications were below this score (in peaks 32, 34, 37, and 41) and correspond to spots of faint intensity. Nonetheless, accurate identification does not depend on spot intensity. For example in spot 1, the OBP sequence is covered at 52% (PEAKS score of 307.18, 13 peptides) although spot 38 displaying the same intensity gave only 8% of sequence coverage (PEAKS score 67.14, 1 peptide). OBP was characterized in all spots, except spot 6 that contains only a VEG peptide. OBP is alone in spots 1–5, 7–12, 15–19, 21–23, and 38–42. OBP is present with VEG in spots 13, 14, 20, 24–25, 27–37, and with SAL in spot 26. Other SAL isoforms, labeled with anti-SAL antibodies were not visible with Coomassie blue staining and were not analyzed. Data from spot 11 displayed the best PEAKS score (603.29) with 285 peptides and 93% of sequence coverage for the four OBP isoforms (X1A, X1J, X2A, X2J). Peptides from OBPX2 were only retrieved in spots 9 and 11. This analysis did not allow reconstruction of the isoform sequences due to the complexity remaining in each spot, despite the resolution of 2-DE separation.
PTM Characterization by NanoLC-NanoESI-HCD-MS/MS-Orbitrap
Analysis by PEAKS software of the 30,617 peptide spectra of spot n°11 revealed PTMs along almost all the OBP sequence. After a rapid spectrum screening, only spectra with a PEAKS score above 67 were kept in order to focus on relevant identifications. This allowed identification of 4 OBP peptides (Table 2) modified by phosphorylation (+79.9797) or hexNAcylation (+203.0794). Spectrum n°20288 also revealed an aspartate hydroxylation, a rarely described PTM known as β-hydroxyaspartate, which remains intact and observable under precise conditions of hydrolysis (Castellino et al., 2008). Its role appears to inhibit fucosylation of adjacent threonine (Harris et al., 1992). In all cases, we were unable to precisely assign these PTMs to a particular amino acid, due to the fragmentation method and the number of potential sites on each peptide. However, phosphorylation and hexNAcylation revealed by spectra n°13801 and 28697, respectively, modify peptides of OBPX1 isoform only (see isoform sequences in Supplementary Data S1). Considering that no PTMs were found on peptides in this region without the specific OBPX1 peptide sequence NYAGNN, phosphorylation and hexNAcylation are potentially localized on the Y82 residue even if tyrosine glycosylation is rarely described (Jank et al., 2015).
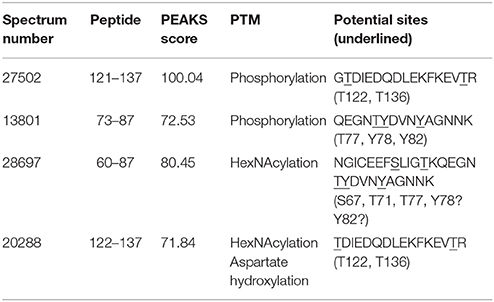
Table 2. Identification of the peptides modified by phosphorylation and hexNAcylation: PEAKS analysis of PTMs in spot 11 from M14 sample.
The raw data of these four MS/MS spectra (Figure 5) and their corresponding MS spectra (Figure 6) were analyzed (Table 3) in order to confirm the conclusions of the PEAKS approach. The manual assignment of each MS/MS spectrum peak led us to the same peptide identification. The most intense peak in spectrum n°27,502 (m/z = 1,012.002, Δm = 0.6 ppm) corresponds to the presence of the naked peptide, resulting from the first fragmentation of the parent ion, given that PTM are readily removed by HCD. When the HCD energy is too high, the naked peptide is fragmented just after losing PTM, leaving no time to detect it, which explains its absence in the other spectra. However, neither PEAKS nor manual analysis were able to attribute all peaks. This could be explained by ions of the same mass in mixtures as supported by the distributions of parent ions isotopes (Figure 6). Indeed, experimental isotopic distributions of parent ions showed some differences with respect to the theoretical ones. For each supposed PTM, the difference between the parent ions experimental mass and the theoretical mass of the corresponding naked peptide was calculated. Results showed mass differences close to the theoretical mass of PTM (Phosphorylation: +79.9797; HexNAcylation: +203.0794). Furthermore, in each case the mass accuracy of the modified parent ion signal is below 10 ppm, providing strong evidence of its supposed PTM. Finally, the intensity of each modified parent ion was compared to the intensity of the corresponding unmodified peptide to determine a PTM-modified peptides ratio. This ratio is very low in each case and far from the predicted average of 10% for phosphorylation (Ma and Hart, 2014). Indeed, these PTMs are extremely labile, making their investigation very hard in real-world samples, due to losses during sample treatment and the limited quantities of material available. In the case of OBP, there is also probably strong ion suppression for detecting modified peptides in the presence of naked peptides, which are much more abundant (Wang and Hart, 2008).
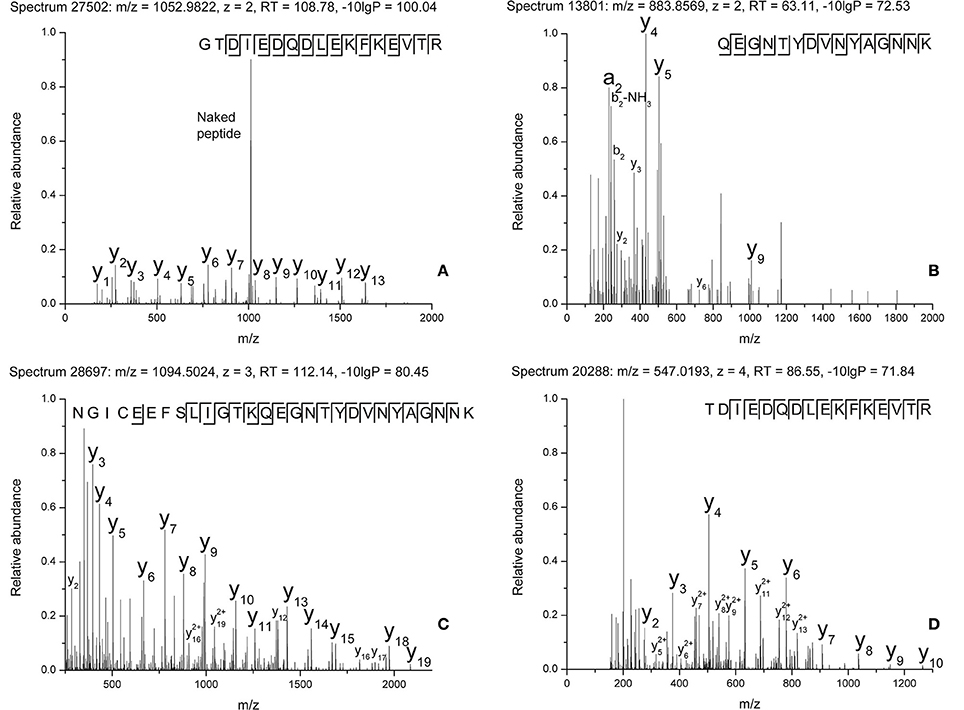
Figure 5. MS/MS spectra of modified peptides and the relevant fragments for their attribution. (A) Spectrum n°27502. (B) Spectrum n°13801. (C) Spectrum n°28697. (D) Spectrum n°20288.
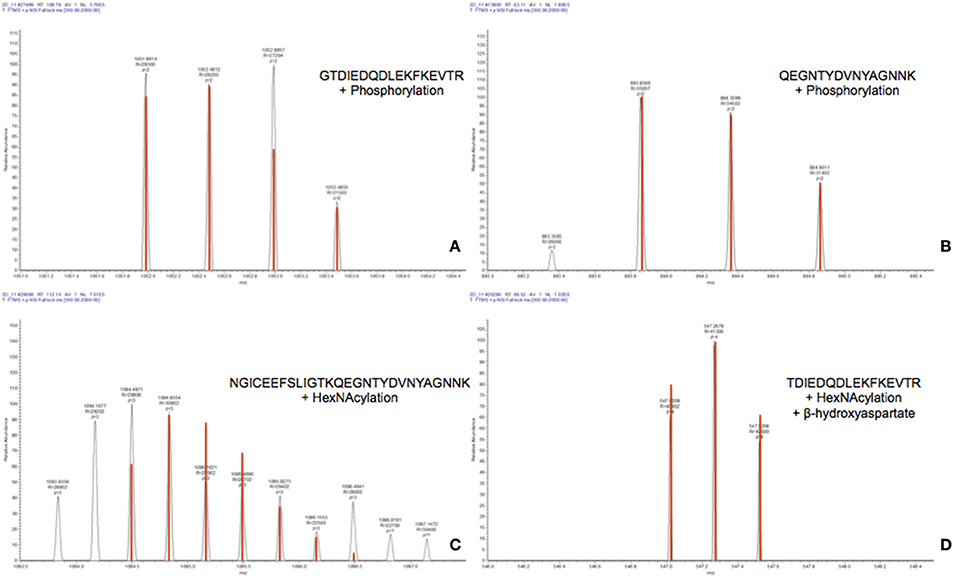
Figure 6. MS spectra of modified peptides and their theoretical isotopic distribution (in red). (A) Spectrum n°27502. (B) Spectrum n°13801. (C) Spectrum n°28697. (D) Spectrum n°20288.
Discussion
We have shown in a previous work (Nagnan-Le Meillour et al., 2014) that around 30 OBP-, VEG-, and SAL- isoforms compose the pig olfactory secretome. Among them, only OBP sensu stricto isoforms were labeled with specific antibodies raised against phosphorylation and O-GlcNAcylation. These two types of PTM, although unusual for secreted proteins, could be involved in odorant and pheromone binding specificity of OBP isoforms. To better understand how PTM could drive OBP binding properties, it is necessary to identify PTM sites and patterns. Indeed, identification by using antibodies raised some concerns because CTD110.6, although specific to O-GlcNAc moieties, recognize a large range of modified proteins (Tashima and Stanley, 2014). In particular, they can bind to terminal β-GlcNAc on complex N-glycans, but with careful controls, they can be used to detect proteins processed in the secretory pathway that are modified by O-GlcNAc. Several arguments strongly support that OBP labeling obtained here is specific: (i) The same OBP isoforms were recognized by both RL2 and CTD110.6 (Nagnan-Le Meillour et al., 2014) and there is no specificity concern for RL2. (ii) In the pig olfactory secretome, only SAL bears N-glycan chains on N53 (Loëbel et al., 2000; Scaloni et al., 2001), whereas OBP and VEG do not (Paolini et al., 1998; Scaloni et al., 2001). In this study, only one SAL isoform was detectable with Coomassie blue staining (spot 26), but was not labeled by CTD 110.6. (iii) False positive would not be possible in the case of OBP, which is neither N-glycosylated nor O-glycosylated (mucin type).
The MS analysis described here confirmed that only OBP sensu stricto is modified by both phosphorylation and O-GlcNAcylation, on the same peptides. If BEMAD method already assessed OBP phosphorylation (Brimau et al., 2010), this is the first time that OBP O-GlcNAcylation is confirmed by mass spectrometry.
The identification of PTM was derived from PEAKS data, but assigned and controlled manually on MS/MS spectra. Only spot 11 provided enough material to identify PTM without ambiguity. For each peptide, several sites are potentially modified. For sequence 121-137, two threonines (T122 and T136) could be phosphorylated and/or hexNAcylated (Table 2). These two sites were previously identified for phosphorylation by using the BEMAD method (Brimau et al., 2010), T122 in recombinant OBP produced by CHO cells, and T136 in recombinant OBP produced by the yeast Pichia pastoris. The peptide of sequence 73–87 could be modified at three positions, T77, Y78, Y82 (Table 2), all 3 potentially modified when using BEMAD (Brimau et al., 2010). In this previous paper, the results of BEMAD were misinterpreted concerning the phosphorylation of Y78 and Y82, because the cycle of tyrosine cannot be opened by beta-elimination. The MS analysis conducted here does not bring any additional information about site assignment. Concerning hexNAcylation, this is the first time that such a modification is assessed by mass spectrometry on OBP peptides, confirming the data obtained by 2-DE and labeling with specific antibodies. In the sequence 60–87, three amino acids, S67, T71, and T77 could bear a GlcNAc moiety (Table 2). We cannot totally exclude that Y78 and Y82 could also be modified even if intracellular O-GlcNAcylation is assumed to only occur on serine and threonine. Indeed, Jank et al. (2015) reported the transfer of UDP-GlcNAc onto tyrosine by a new bacterial glycosyltransferase involved in the pathogenicity of Yersinia species. The O-GlcNAcylation of secreted proteins is new and not well understood, and the function of EOGT has not been investigated in mammals, but could differ from that of OGT, the intracellular enzyme. Indeed, there is no consensus sequence described until now, which could predict the nature of amino acid residues targeted by this PTM. The hypothesis of tyrosine O-GlcNAcylation should be kept in mind for further studies.
It is remarkable that the same peptides were identified as modified by either phosphorylation or hexNAcylation, and particularly on the sole peptide that differs between the products of the two splicing variants (OBPX1 81NYAGNNK87 and OBPX2 81NCNNK85). Only the peptide specific to the OBPX1 isoform was identified as modified by both PTM. The fact that the same peptides could be modified by phosphorylation and O-GlcNAcylation raises questions about the possibility of a phospho/GlcNAc balance on extracellular proteins, as well as on nuclear and cytoplasmic ones. But to date, no ER-resident or extracellular counterpart of O-GlcNAcase, the enzyme that removes GlcNAc residues from intracellular modified proteins (OGA), has been identified. There is another hypothesis that does not involve an OGA-like enzyme. In this scenario, OBP could be modified in the ER, secreted in the extracellular compartment, and internalized after binding with an odorant and/or olfactory receptor, this latter event being evidenced by Strotmann and Breer (2011) in the mouse sustentacular cells of olfactory epithelium. This scheme fits well with the rapid turnover of OBPs described in the nasal mucus. If secreted together, phosphorylated and O-GlcNAcylated OBP isoforms could have different binding properties toward ligands, extending their binding repertoire, thereby supporting an active role of OBPs in odorant discrimination.
Functional studies (binding assays) are needed to address this question. However, before realizing site-directed mutagenesis for this purpose, it is necessary to identify O-GlcNAc sites and this remains a challenging task, especially when several amino acids of the same peptide could be modified. Indeed, the study of native proteins in complex mixture could explain why we did not find the oxonium ions (204.08) for HexNAcylation, typical of the HCD method (Zhao et al., 2011). These ions are usually detected in glycoprotein samples enriched in O-GlcNAcylated peptides (Toghi Eshghi et al., 2016) or exclusively composed of synthetic O-GlcNAcylated peptides (Chalkley and Burlingame, 2001; Malaker et al., 2017). In this study, identification of modified peptides was made by a manual assignment approach instead of using software as it is done in almost all PTM identification/localization papers. PEAKS software is useful to decrease the spectra number to a few spectra of interest, but should mostly be used as a first approach of investigation. When ones looks carefully at most of the spectra attributed to phosphorylated/hexNAcylated peptides by PEAKS software, there is no obvious proof of such modifications according to the MS/MS spectrum as the modification is the first to be lost in the collision activated dissociation. We therefore recommend that the software should be best used to limit the essential manual assignment work to a reduced number of relevant spectra instead of using it blindly. ESI-ETD-MS/MS has been reported to be the only method able to confidently localize the modification sites (Alfaro et al., 2012; Ma and Hart, 2014) because the energy of fragmentation is low and preserves the link between the protein and labile PTMs. But ETD (Electron Transfer Dissociation) is less sensitive than HCD and less useful for complex mixtures than for single proteins. In the absence of an ETD-MS/MS, we were able to characterize four peptides modified by PTMs without any enrichment. The first step to have all proofs of hexNAcylation is to enrich our sample in modified peptides by the use of specific columns to observe oxonium ions with HCD fragmentation. However, HCD fragmentation removes PTMs in priority making their localization possible only if one S/T/Y residue is present on the characterized modified peptide. The second step could be the use of ETD fragmentation on modified peptides characterized with HCD fragmentation. However, ETD requires more modified peptides overall quantity than HCD, making enrichment a crucial step for our future studies.
In conclusion, this work is the first step in PTM localization on OBP isoforms from a complex sample of whole secretome. It opens the way to molecular and cellular studies of this new metabolic pathway involved in olfaction.
Author Contributions
PN-L, CT, and CR conceptualized the project. 2-DE analyses were performed by JB and PN-L. CL and JB identified isoforms. High-resolution mass spectrometry was performed by FB. FB, JB, and CR analyzed Peaks data. All authors have participated in the preparation of the manuscript.
Funding
This work was funded by French INRA (Institut National de Recherche Agronomique) and CNRS (Centre National de Recherche Scientifique).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2017.00142/full#supplementary-material
References
Alfaro, J. F., Gong, C. X., Monroe, M. E., Aldrich, J. T., Clauss, T. R., Purvine, S. O., et al. (2012). Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U.S.A. 109, 7280–7285. doi: 10.1073/pnas.1200425109
Brimau, F., Cornard, J. P., Le Danvic, C., Lagant, P., Vergoten, G., Grebert, D., et al. (2010). Binding specificity of recombinant Odorant Binding Protein isoforms is driven by phosphorylation. J. Chem. Ecol. 36, 801–813. doi: 10.1007/s10886-010-9820-4
Buck, L., and Axel, R. (1991). A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187. doi: 10.1016/0092-8674(91)90418-X
Castellino, F. J., Ploplis, V. A., and Zhang, L. (2008). Gamma-glutamate and beta-hydroxyaspartate in proteins. Methods Mol. Biol. 446, 85–94. doi: 10.1007/978-1-60327-084-7_6
Chalkley, R. J., and Burlingame, A. L. (2001). Identification of GlcNAcylation sites of peptides and α-crystallin using Q-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 12, 1106–1113. doi: 10.1016/S1044-0305(01)00295-1
Guiraudie, G., Pageat, P., Cain, A. H., Madec, I., and Nagnan-Le Meillour, P. (2003). Functional characterization of olfactory binding proteins for appeasing compounds and molecular cloning in the vomeronasal organ of pre-pubertal pigs. Chem. Senses 28, 609–619. doi: 10.1093/chemse/bjg052
Harris, R. J., Ling, V. T., and Spellman, M. W. (1992). O-linked fucose is present in the first epidermal growth factor domain of factor XII but not in protein C. J. Biol. Chem. 267, 5102–5107.
Hart, G. W., and Akimoto, Y. (2009). “Chapter 18, The O-GlcNAc modification,” in Essentials of Glycobiology. 2nd Edn. Copyright © 2009, The Consortium of Glycobiology Edtors, eds A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, C. R. Bertozzi, G. W. Hart, and M. E. Etzler (La Jolla, CA; Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
Hoffman, B. R., Liu, Y., and Mosher, D. F. (2012). Modification of EGF-like module 1 of thrombospondin-1, an animal extracellular protein, by O-linked N-acetylglucosamine. PLoS ONE 7:e32762. doi: 10.1371/journal.pone.0032762
Jank, T., Eckerle, S., Steinemann, M., Trillhaase, C., Schimpl, M., Wiese, S., et al. (2015). Tyrosine glycosylation of Rho by Yersinia toxin impairs blastomere cell behaviour in Zebrafish embryos. Nat. Commun. 6:7807. doi: 10.1038/ncomms8807
Loëbel, D., Scaloni, A., Paolini, S., Fini, C., Ferrara, L., Breer, H., et al. (2000). Cloning, post-translational modifications, heterologous expression and ligand-binding of boar salivary lipocalin. Biochem. J. 350, 369–379. doi: 10.1042/bj3500369
Ma, J., and Hart, G. W. (2014). O-GlcNAc profiling: from proteins to proteomes. Clin. Proteomics 11:8. doi: 10.1186/1559-0275-11-8
Malaker, S. A., Penny, S. A., Steadman, L. G., Myers, P. T., Loke, J. C., Raghavan, M., et al. (2017). Identification of glycopeptides as posttranslationally modified neoantigens in leukemia. Cancer Immunol. Res. 5, 376–384. doi: 10.1158/2326-6066.CIR-16-0280
Nagnan-Le Meillour, P., Vercoutter-Edouart, A. S., Hilliou, F., Le Danvic, C., and Lévy, F. (2014). Proteomic analysis of pig (Sus scrofa) olfactory soluble proteome reveals O-linked-N-acetylglucosaminylation of secreted odorant-binding proteins. Front. Endocrinol. 5:202. doi: 10.3389/fendo.2014.00202
Paolini, S., Scaloni, A., Amoresano, A., Marchese, S., Napolitano, E., and Pelosi, P. (1998). Amino acid sequence, post-translational modifications, binding and labelling of porcine odorant-binding protein. Chem. Senses 23, 689–698. doi: 10.1093/chemse/23.6.689
Pelosi, P. (1996). Perireceptor events in olfaction. J. Neurobiol. 30, 3–19. doi: 10.1002/(SICI)1097-4695(199605)30:1<3::AID-NEU2>3.0.CO;2-A
Sakaidani, Y., Nomura, T., Matsuura, A., Ito, M., Suzuki, E., Murakami, K., et al. (2011). O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat. Commun. 2:583. doi: 10.1038/ncomms1591
Scaloni, A., Paolini, S., Brandazza, A., Fantacci, M., Bottiglieri, C., Marchese, S., et al. (2001). Purification, cloning and characterisation of odorant-binding proteins from pig nasal epithelium. Cell. Mol. Life Sci. 58, 823–834. doi: 10.1007/PL00000903
Schwarz, F., and Aebi, M. (2011). Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582. doi: 10.1016/j.sbi.2011.08.005
Shaheen, R., Aglan, M., Keppler-Noreuil, K., Faqeih, E., Ansari, S., Horton, K., et al. (2013). Mutations in EOGT confirm the genetic heterogeneity of autosomal-recessive Adams-Oliver syndrome. Am. J. Hum. Genet. 92, 598–604. doi: 10.1016/j.ajhg.2013.02.012
Spinelli, S., Ramoni, R., Grolli, S., Bonicel, J., Cambillau, C., and Tegoni, M. (1998). The structure of the monomeric porcine odorant binding protein sheds light on the domain swapping mechanism. Biochemistry 37, 7913–7918. doi: 10.1021/bi980179e
Strotmann, R., and Breer, H. (2011). Internalization of odorant-binding proteins into the mouse olfactory epithelium. Histochem. Cell Biol. 136, 357–369. doi: 10.1007/s00418-011-0850-y
Tagliabracci, V. S., Wiley, S. E., Guo, X., Kinch, L. N., Durrant, E., Wen, J., et al. (2015). A single kinase generates the majority of the secreted phosphoproteome. Cell 161, 1619–1632. doi: 10.1016/j.cell.2015.05.028
Tashima, Y., and Stanley, P. (2014). Antibodies that detect O-GlcNAc on the extracellular domain of cell surface glycoproteins. J. Biol. Chem. 289, 11132–11142. doi: 10.1074/jbc.M113.492512
Toghi Eshghi, S., Yang, W., Hu, Y., Shah, P., Sun, S., Li, X., et al. (2016). Classification of tandem mass spectra for identification of N- and O-linked glycopeptides. Sci. Rep. 6:37189. doi: 10.1038/srep37189
Torres, C. R., and Hart, G. W. (1984). Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surface of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317.
Vizcaino, J. A., Csordas, A., del-Toro, N., Dianes, J. A., Griss, J., Lavidas, I., et al. (2016). 2016 update of the PRIDE database and related tools. Nucleic Acid Res. 44, D447–D456. doi: 10.1093/nar/gkv1145
Wang, Z., and Hart, G. W. (2008). Glycomic approaches to study O-GlcNAcylation: protein identification, site-mapping, and site-specific O-GlcNAc quantitation. Clin. Proteomics 4, 728–729. doi: 10.1007/s12014-008-9008-x
Keywords: odorant-binding protein, nanoLC-nanoESI-HCD-MS/MS-Orbitrap, O-GlcNAc, phosphorylation, 2-dimensional gel electrophoresis
Citation: Bouclon J, Le Danvic C, Guettier E, Bray F, Tokarski C, Rolando C and Nagnan-Le Meillour P (2017) Identification of Post-translational Modifications on Odorant-Binding Protein Isoforms from Pig Olfactory Secretome by High-Resolution Mass Spectrometry: O-β-N-acetylglucosaminylation and Phosphorylation. Front. Ecol. Evol. 5:142. doi: 10.3389/fevo.2017.00142
Received: 18 August 2017; Accepted: 07 November 2017;
Published: 22 November 2017.
Edited by:
Carla Mucignat, Università degli Studi di Padova, ItalyReviewed by:
Archunan Govindaraju, Bharathidasan University, IndiaMarjorie A. Lienard, Harvard University, United States
Copyright © 2017 Bouclon, Le Danvic, Guettier, Bray, Tokarski, Rolando and Nagnan-Le Meillour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patricia Nagnan-Le Meillour, cGF0cmljaWEubGUtbWVpbGxvdXJAdW5pdi1saWxsZTEuZnI=
†Present Address: Julien Bouclon, LBM, UMR 7203 Centre National de la Recherche Scientifique, Ecole Normale Supérieure, Paris, France
 Julien Bouclon
Julien Bouclon Chrystelle Le Danvic
Chrystelle Le Danvic Elodie Guettier4
Elodie Guettier4 Fabrice Bray
Fabrice Bray Christian Rolando
Christian Rolando Patricia Nagnan-Le Meillour
Patricia Nagnan-Le Meillour