- 1Department of Environmental Microbiology, Helmholtz Centre for Environmental Research—UFZ, Leipzig, Germany
- 2Avram and Stella Goldstein-Goren Department of Biotechnology Engineering, Ben-Gurion University of the Negev, Be'er Sheva, Israel
- 3Department of Plant Pathology and Microbiology, Hebrew University of Jerusalem, Jerusalem, Israel
- 4German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Leipzig, Germany
Complex ecosystems harbor multiple predators and prey species whose direct and indirect interactions are under study. In particular, the combined effects of predator diversity and resource preference on prey removal are not known. To understand the effect of interspecies interactions, combinations of micro-predators—i.e., protists (generalists), predatory bacteria (semi-specialists), and phages (specialists)—and bacterial prey were tracked over a 72-h period in miniature membrane bioreactors. While specialist predators alone drove their preferred prey to extinction, the inclusion of a generalist resulted in uniform losses among prey species. Most importantly, presence of a generalist predator enabled coexistence of all predators and prey. As the generalist predator also negatively affected the other predators, we suggest that resource partitioning between predators and the constant availability of resources for bacterial growth due to protist predation stabilizes the system and keeps its diversity high. The appearance of resistant prey strains and subsequent evolution of specialist predators unable to infect the ancestral prey implies that multitrophic communities are able to persist and stabilize themselves. Interestingly, the appearance of BALOs and phages unable to infect their prey was only observed for the BALO or phage in the absence of additional predators or prey species indicating that competition between predators might influence coevolutionary dynamics.
Introduction
Predatory interactions among organisms of different trophic levels are key processes that determine the coexistence in ecological communities. Although top-down and bottom-up effects that depend upon the specialization level of the consumer (Jiang and Morin, 2005; Filip et al., 2014) strongly affect system productivity and stability (Duffy et al., 2007; Jiang et al., 2009), a mechanistic understanding of these phenomena is still lacking. Early investigations focused on pairwise predator–prey interactions that were then generalized to complete communities under the assumption that multiple pairwise interactions had additive effects (Oksanen et al., 1981; Wootton, 1994). Later, laboratory and field studies focused on predation by multiple species and mostly observed the effects of a pair of predators on one shared prey (Sih et al., 1998; Schmitz, 2007; Carey and Wahl, 2010). However, trophic interactions in nature are highly complex (Polis, 1991; Martinez, 1992; Lafferty et al., 2006), and even in environments consisting of only a limited number of species, a prey may interact with about 10 different predators (Polis, 1991). Additive pairwise experiments may be limited in discerning reciprocal effects. In contrast, integrating multiple predators of different trophic levels in a single experiment may better reflect the effects of multitrophic food webs on ecosystem functioning (Griffin et al., 2008), pest control (Philpott et al., 2012), or bacterial adaptation to stress gradients (Friman et al., 2015). Modeling and experimental approaches in which different trophic levels were combined (Duffy et al., 2007; Ellis et al., 2011) revealed a positive effect on net ecosystem productivity. It further showed that food web complexity could dampen trophic cascades, resulting to a decreased impact of predators on herbivores (Finke and Denno, 2004). In addition, species-specific traits, such as resource specialization, strongly affected prey biomass and diversity (Jiang and Morin, 2005; Filip et al., 2014) and induced shifts in trophic cascades (Steiner, 2001). Studies focusing on the combined effect of multiple predators with different resource utilization are unfortunately rare (Jiang and Morin, 2005; Diehl et al., 2013), but most likely better reflect natural communities and enable the direct observation of a major mediator of coexistence: resource partitioning (Chesson, 1991). Narwani and Mazumder (2010) showed that the overall consumption of specialist predators decreases at higher resource diversities, whereas generalist predators show an increase in consumption (Narwani and Mazumder, 2010). Species not directly involved in consumptive interactions are the key to survival of susceptible prey and increased persistence of prey species in more complex food webs (Hammill et al., 2015).
In order to gain more insights into the effects of multiple predators with partially overlapping resource preferences, we investigated three potentially interacting micro-predator groups, namely protists, predatory bacteria, and phages. These micro-predators exert substantial top-down control on bacteria in environmental systems and are characterized by a different prey range (Johnke et al., 2014). Predation by protists is the leading contributor to bacterial turnover in aquatic systems (Sherr and Sherr, 2002) and is a key process in engineered systems such as wastewater treatment plants (Curds, 1973). Protists are known to exploit a relatively large spectrum of bacterial species and are, therefore, classified as generalist predators. However, grazing-resistant bacteria can develop in response to protist predation. Common resistance mechanisms can act either before ingestion or afterwards and include the formation of inedible cells or microcolonies, increased motility, surface masking, or toxin release (reviewed in Matz and Kjelleberg, 2005).
A further group of microbial consumers is the predatory bacteria, such as Bdellovibrio and like organisms (BALOs), which are obligate predators of Gram-negative bacteria. BALO predation pressure can result in the evolution of prey resistance (Varon, 1979) followed by antagonistic coevolution of the predatory bacterium (Gallet et al., 2009). The appearance of coevolved BALOs was shown to be dependent on the experimentally applied ecological conditions: bacterial prey exposed to higher disturbances evolved super-resistance to which the predator could not counter adapt (Gallet et al., 2009). The mechanisms of resistance evolution and coevolution of the BALO are so far unknown. Furthermore, phenotypic plastic resistance of prey bacteria appears to be widespread as the co-cultivation of Bdellovibrio bacteriovorus and a single prey species does not lead to the extinction of the prey, whereas prey cultured in the absence of BALOs return to the ancestral susceptible state (Kadouri and O'Toole, 2005). So far, information on the ecological impact of BALO predation in natural communities is limited. Microcosm experiments indicate that BALO predation significantly contributes to bacterial mortality in ocean water where they even may respond faster than viruses to the presence of, or a sudden increase in, potential prey bacteria (Chen et al., 2011, 2012; Williams et al., 2016).
Another important driver of bacterial mortality in the environment is the phages. Although current knowledge about the extent of phage specificity in natural systems is quite incomplete (Koskella and Meaden, 2013), it is assumed that their host range is restricted and in some cases even strain-specific (Hyman and Abedon, 2010). Interactions of bacteria with phages usually quickly result in the formation of resistant bacteria (Middelboe et al., 2001). Mechanisms leading to resistance include the prevention of phage attachment, blockage of DNA entry, restriction-modification systems, infection abortion, assembly interference, and CRISPR-Cas (reviewed by Labrie et al., 2010; Seed, 2015). Due to their rapid multiplication rates, phages have different strategies to overcome these bacterial defense mechanisms (reviewed by Samson et al., 2013).
Recently, it was shown that the simultaneous presence of protists and phages strongly affects the community response to antibiotics (Friman et al., 2015), bacterial virulence (Friman and Buckling, 2014), and diversification (Friman and Buckling, 2013). Depending upon their feeding strategies, protists may indirectly remove phages by lowering the susceptibility of the shared prey to phages (Deng et al., 2014; Ormala-Odegrip et al., 2015). However, detailed information on the impact of micro-predator interactions on microbial communities is missing (Johnke et al., 2014). Particularly, the combined effects of protists, BALOs, and phages (i.e., predators exhibiting different levels of specificity) acting on a prey community are unknown. The diversity among micro-predators and their competition may influence prey community assembly and coexistence (Chase et al., 2002; Ryberg et al., 2012). In addition, interactions among different predators are likely to occur and might have a positive (Simek et al., 2001; Sime-Ngando and Ram, 2005; Weinbauer et al., 2007) or a negative impact on the predator growth (Diehl and Feissel, 2001). For instance, a decreased growth in the presence of additional predators might be due to intraguild predation (predation between resource competitor, Holt and Polis, 1997), which, under certain conditions, enables coexistence in prey–predator systems (Kang and Wedekin, 2013).
In this study, we experimentally tested whether predators with different prey ranges maintain the diversity of predator–prey communities and consequently affect species coexistence in a controlled laboratory system. We exposed different bacterial prey species to a protist (generalist predator), a BALO (semi-specific predator), and a phage (highly specific predator). We hypothesized that the potential trophic complementarity of predators would result in a reduction of all prey species. We further expected direct interactions between the predator species since protists are able to graze directly upon phages (Gonzalez and Suttle, 1993; Hennemuth et al., 2008) and BALOs (Johnke et al., 2017). Finally, the high grazing pressure exerted by specialist predators should induce the emergence of predation-resistant bacteria. Resistance to phages and BALOs is known to induce fitness costs (Bohannan and Lenski, 1999; Bohannan et al., 2002; Meyer and Kassen, 2007; Gallet et al., 2009), and the subsequent appearance of counter-adapted predators might have direct consequences for the coexistence of the community members.
Materials and Methods
Organisms
Three different micro-predators and three bacterial species were used in the study: a ciliate protist (Tetrahymena pyriformis), a BALO (B. bacteriovorus strain 109J), and a Klebsiella phage originally isolated from a wastewater treatment plant (WWTP) in Langenreichenbach, Germany. Two of the prey strains were isolated from the inflow of the same WWTP and identified as Klebsiella sp. and as Staphylococcus sp. by 16S rRNA gene sequencing. Pseudomonas putida KT2440 from our laboratory collection was also used as a prey. The micro-predators are characterized by different prey ranges: while the protist feeds on all three prey species, the BALO only preys upon the two Gram-negative bacteria (Klebsiella sp. and P. putida), and the phage infects only the Klebsiella strain (Figure 1).
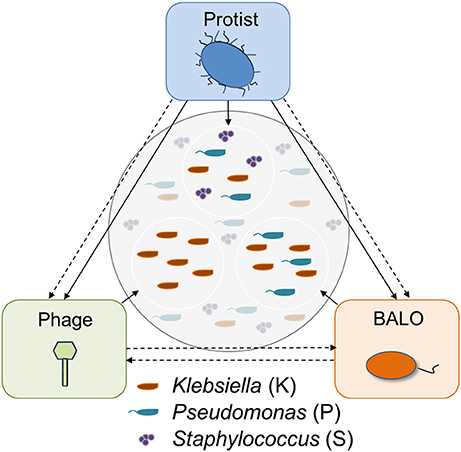
Figure 1. Possible trophic interactions between the predators in the system. Direct (straight) and indirect (dashed) interactions exist between all predators. Direct predatory interactions are known for protists and BALOs (Johnke et al., 2017); and protists and phages (Deng et al., 2014). Additionally, superinfection of a prey bacterium by a BALO and a phage was shown by Chen and Williams (2012).
The Miniature Membrane Bioreactor System (mMBR)
Experiments were conducted in custom-built mMBRs (Supplementary Figure 1; Segev et al., 2013). We used a 0.2-μm membrane filter that enables the washout of the phages, whereas all other microorganisms remain in the system. Each reactor was filled with 100 ml of artificial wastewater (Patil et al., 2010). An inflow and outflow rate of 10 ml d−1 was kept constant over the course of the experiment. Ambient air was introduced continuously into the system through a 0.2-μm filter and diffused into the medium, thus allowing a steady mixing (~0.5 l h−1).
Culturing and Experimental Procedure
T. pyriformis was cultured axenically (i.e., without any bacterial prey) in proteose peptone yeast extract medium (PPY, 20 g l−1 proteose peptone, 2.5 g l−1 yeast extract, Smith and Doerder, 1992) prior to the experiments. For each experiment, cells from the stock culture were washed by centrifugation for 10 min at 1,000 × g and resuspension in artificial wastewater. Approximately 1 × 103 cells ml−1 were added to the reactor at the start of an experiment (minimum amount: 517 cells ml−1 and maximum amount: 1,133 cells ml−1).
Phages were obtained after the infection of the host Klebsiella strain and subsequent cultivation in LB broth over night at 37°C. On the day of the experiment, the culture was filtered through a 0.2-μm filter to remove all the remaining host bacteria. At least 1.7 × 105 PFU ml−1 (maximum amount: 2.51 × 106 PFU ml−1) were added to a reactor at the beginning of an experiment.
B. bacteriovorus strain 109J was maintained as previously described (Jurkevitch, 2005) and with Klebsiella as host. On the day of the experiment, BALOs were passed through a 0.45-μm filter to remove the remaining prey cells and subsequently washed with artificial wastewater. At least 3.1 × 103 PFU ml−1 (maximum amount: 1.25 × 106 PFU ml−1) were added to a reactor at the beginning of an experiment.
Single colonies of bacterial prey cultures were re-suspended in 5 ml of LB and incubated over night at 37°C with gentle shaking. Cells were washed in equivalent amount of artificial wastewater. At the start of each experiment, 1.5 ml of prey culture adjusted to an optical density (OD600) of 1.5 was added to each reactor. In experiments including all three prey organisms, 0.5 ml of each prey species were added to maintain an equal total volume.
Different predator–prey diversity levels (Figure 2) were established in triplicates, and experiments were run for 72 h. To control bacterial growth without predation, single prey (C I) as well as multiple prey controls (C II) were used in duplicates. This constrain in control replication was mainly due to the limited amount of reactors that we were able to run at the same time but was considered appropriate as the control reactors showed almost no variation.
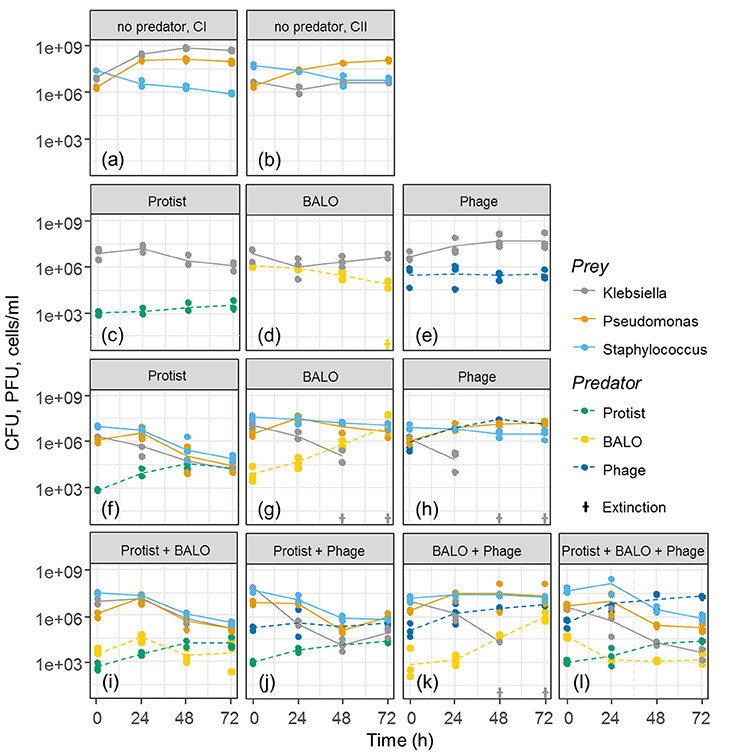
Figure 2. Growth curves of predators and prey in experiments with different diversity levels. CI: single prey control without predators (a), CII: multiple prey control without predators (b). (c–l) represent the results of the experiments that contained one or more predator/s. Extinctions are indicated by crosses. Protist abundance is shown as cells/ml, BALO and Phage abundances as PFU/ml, and Klebsiella, Pseudomonas, and Staphylococcus abundances as CFU/ml.
Sampling
Samples were taken every 24 h for a total of 72 h by injecting a sterile needle into a rubber septum located at the side of each reactor. For counting bacterial colony forming units (CFU), 200 μl samples were serially diluted and 20 μl drops (four times for each dilution) were incubated on LB plates over night at 37°C. Klebsiella and Pseudomonas colonies were distinguished on chromogenic E. coli/Coliform agar (Fluka). Since Staphylococcus colonies cannot grow on this medium, cell numbers were obtained by subtracting the number of Klebsiella and Pseudomonas colonies from the total number of colonies observed on LB plates. In addition, Staphylococcus appeared as distinctive yellow colonies on the LB plates; counting these colonies served as a control for the subtraction method. All plates were incubated overnight at 37°C and examined for changes of the colony morphology.
Protists were quantified in a Sedgwick-Rafter chamber with 500 μl samples which were fixed with 5% Lugol solution. At least 2 × 20 squares of the chamber were counted.
Phage and BALO numbers were quantified in plaque assays. Five hundred micro liter samples were filtered through a 0.2-μm filter (phages) and a 0.45-μm filter (BALOs), respectively. Phage plaque forming units (PFU) were obtained using the double agar technique with LB plates (1.5% agar) and LB soft agar (0.7% agar; Kropinski et al., 2009). Triplicates of multiple dilutions were plated for proper counting. Phage samples were diluted with saline (0.9%) and plates were incubated at 37°C overnight. BALO plaque assays were performed as described elsewhere (Jurkevitch, 2005). Plates were incubated at 30°C until plaque formation was observed.
Generation Time
To investigate fitness costs of acquired resistances, we recorded and compared the growth kinetics of prey bacteria for the ancestral bacterial strains (t = 0 h) and the evolved bacteria (t = 72 h). Single colonies of prey bacteria were incubated in 5 ml LB overnight at 37°C with gentle shaking to provide a starter culture. Growth measurements were then performed by preparing cultures with an initial OD600 of 0.05 and then measuring OD every 30 min for 24 h. To connect the growth data with the colony doubling time, multiple dilutions of the respective prey culture were incubated on LB plates for 24 h and colonies were counted to calculate the prey CFU per ml of the culture used in the OD measurement.
Cross-Infectivity Tests
We re-isolated Klebsiella cells, phages, and BALO cells at the end of the experiments from each reactor. Resistance of potentially evolved Klebsiella cells to ancestral phages or BALOs and of ancestral Klebsiella cells to potentially evolved phages or BALOs was tested in cross-infectivity tests (adapted from Friman and Buckling, 2014). A pure phage or BALO filtrate was obtained by filtering the sample through 0.2 or 0.45 μm filter, respectively. The phage filtrate was streaked out in lines on LB agar plates to dry. Ten micro liter of the respective bacterial overnight culture was streaked across the phage lines, cultivated at 37°C overnight, and subsequently checked for bacterial growth.
The BALO filtrates were tested against the respective overnight prey cultures in a plaque assay as described earlier. Assays with evolved Klebsiella strains and ancestral phages or BALOs were performed with at least 10 cultures from single Klebsiella colonies to calculate the percentage of resistant strains.
Statistical Analysis
All statistical analyses were performed within the R software environment (R. Core Team, 2016) using the package “vegan” (Oksanen et al., 2007) or the included “stats” package. Possible factors accounting for the changes in the prey removal rates were analyzed with ANOVA as the variances in prey removal between groups of data were similar and normally distributed. The prey removal rate per hour was calculated as the average log slope between all sampling points. Using the aov() function, we tested the correlation between the degree of predator specialization (“generalist,” “semi-specialist,” “specialist,” “generalist + semi-specialist,” “generalist + specialist,” “semi-specialist + specialist,” or “generalist + semi-specialist + specialist”) and the prey removal rate per hour. The same function was used to correlate the number of additional predators (“none,” “one,” or “two”) and the predator removal rate due to normal distribution of the data and variance differences that still allow a robust analysis.
The differences between the BALO and phage growth rates with one additional predator were tested with the two-sample t-test as well as the differences in prey removal of all three bacteria individually in the presence or absence of a generalist predator, since the data were normally distributed and showed little variance differences. Significant differences in the generation times of the respective prey bacteria compared to the ancestral Klebsiella generation time were tested with the one sample t-test due to the normal distribution and low variance differences of the data.
Results
None of the bacterial colonies appeared morphologically different during the experiment. The control treatments containing single (Figure 2a, CI) and multiple prey species (Figure 2b, CII) indicated that all prey species were able to survive in the reactor and coexisted when together. The Staphylococcus strain exhibited a slight reduction in cell numbers over time.
Prey Dynamics
All prey from the reactors containing the generalist protist predator showed a reduction in cell numbers to a more or less similar extent and independent of the presence of additional predators (Figures 2c,f,i,j,l). Both specialized predators reduced only the concentration of Klebsiella cells when all three prey species were present. Klebsiella went extinct in these assemblages after 48 and 72 h, respectively (Figures 2g,h,k), even though the BALO has the potential to grow on Pseudomonas (Figure 1). Reactors containing one or two specialists as well as the generalist predator showed reductions in the cell numbers of all three prey species. However, no extinction of any prey species was observed (Figures 2i,j,l), suggesting that the presence of a generalist predator allows for the coexistence of all prey bacteria and their predators. Contrary to reactors containing all prey strains (Figures 2f–h), the presence of only one predator and one prey (Figures 2c–e) did not lead to a similarly strong reduction of Klebsiella, indicating the appearance of resistant prey strains.
For all three prey species, independently we found a highly significant correlation between the prey removal rate and the degree of predator specialization [ANOVA: Klebsiella F(6, 14) = 216.86, P < 0.001; Pseudomonas F(6, 14) = 28.82, P < 0.001; Staphylococcus F(6, 14) = 46.569, P < 0.001]. However, the relationship between specialization and prey removal rate differed for the three prey bacteria. Klebsiella removal by specialist and semi-specialist predators occurred at a higher rate than in the presence of a generalist (P < 0.001, two-sample t-test), whereas Pseudomonas and Staphylococcus removal rates were higher in the presence of the generalist than in the absence of a generalist (P < 0.001, two-sample t-test, Supplementary Figure 2).
Predator Growth
Presence of additional predators had no effect on the growth of protists [Figure 3a, ANOVA: F(2, 9) = 0.081, P = 0.923]. Phage proliferation was also not significantly influenced by additional predators [Figure 3c, ANOVA: F(2, 9) = 1.037, P = 0.393]. Note, however, that our measurements of phage growth rate are likely to underestimate the true rate since phage particles are small enough to be washed out of the reactor.
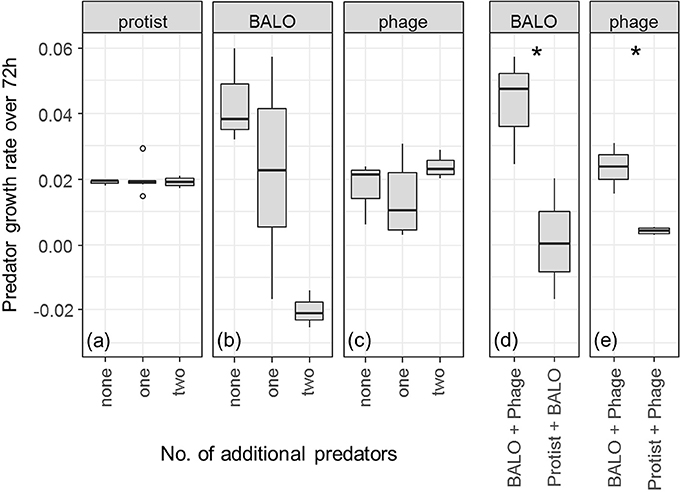
Figure 3. (a–c) Changes in the predator growth rate over the full course of the experiment with increasing predator diversity. Only reactors that contained all three prey species were considered. P-values are given in Supplementary Table 2. Three replicates (six in case of “one” additional predator) are shown. (d,e) Breakdown of the two possible combinations of a reactor that contains a BALO or phage with one additional predator. Significances were calculated with a two-sample t-test. Three replicates are shown. Circles represent outlier, asterisks represent significant difference with 0.011 > p < 0.05.
We found a negative relationship between the number of predators and BALO growth [Figure 3b, ANOVA, F(2, 9) = 6.597, P = 0.0172]. In addition, the range of growth rates of BALOs and phages in the presence of one additional predator was rather broad and depended on the type of predator present in the system (Figures 3b,c). Both BALO and phage growth rates were significantly lowered by the presence of the protist (Figure 3d, P = 0.044, two-sample t-test, and Figure 3e, P = 0.013, two-sample t-test, respectively).
Potential Antagonistic Coevolution of Klebsiella and Specialist Predators
As the prey dynamics of some reactors indicated the presence of Klebsiella strains resistant to predation (Figures 2d,e), we performed cross-infectivity tests to identify if bacteria developed resistance to the two specialized predators and if the predators subsequently adapted; the latter is implied by the loss of the ability to infect ancestral prey strains (Friman and Buckling, 2013).
Only the phage and Klebsiella from the reactors containing no additional predator or prey species showed evidence of potential short-term coevolutionary dynamics, i.e., bacteria became resistant against sympatric phages, whereas the phages showed reduced infectivity against ancestral Klebsiella (Figure 4a). We found resistant Klebsiella strains in reactors that contained protists, phages, and all three bacteria (Figure 4c), and in reactors containing all predators and prey species (Figure 4e). In contrast, we did not obtain any resistant Klebsiella strain in reactors containing either phages and all three prey bacteria (Figure 4b) or phages, BALOs, and all three prey bacteria (Figure 4d). Furthermore, none of the Klebsiella strains developed resistance against the ancestral BALO (Figures 4f–h). However, infectivity of BALOs from the reactors that only contained BALOs and Klebsiella was reduced (Figure 4f).
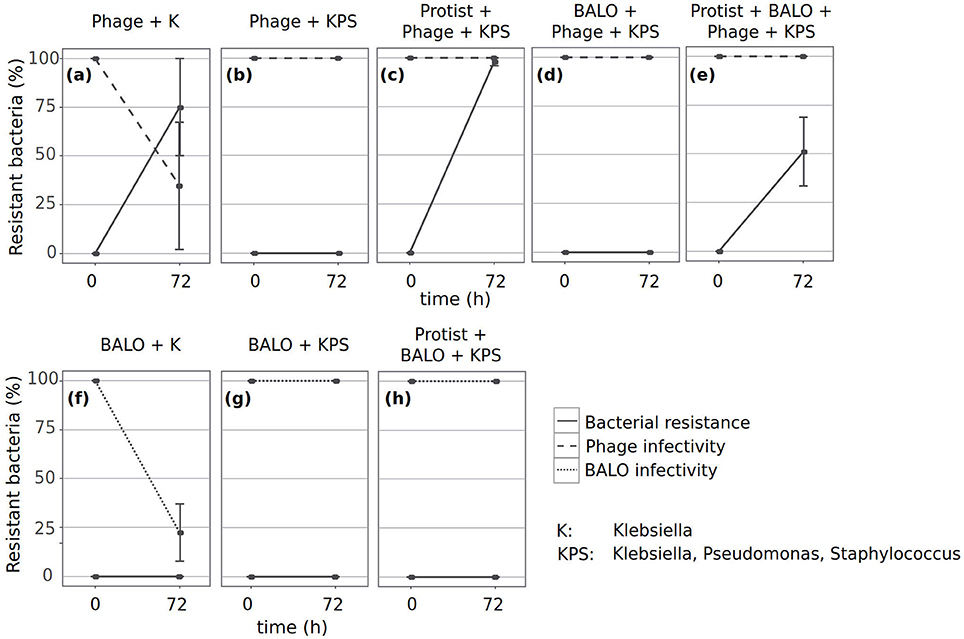
Figure 4. Resistance dynamics of the phage and Klebsiella (a–e) and of the BALO and Klebsiella (f–h). Cross-infectivity tests were performed with ancestral strains (0 on x-axis) and strains isolated from the different reactors after 72 h (72 on x-axis). Dashed lines indicate the predator infectivity of ancestral Klebsiella strains. Straight lines indicate the resistance of Klebsiella strains to ancestral predators.
Unfortunately, we were not able to evaluate the resistance of ancestral Klebsiella to BALOs that were in the reactor with phages, as the presence of large numbers of phage plaques prevented the clear identification of BALO plaques.
Inability to infect ancestral prey by evolved predators was only observed when no additional predators and prey species were present (Figures 4a,f). The dynamics in these reactors indicate coevolution of the phage in response to resistant Klebsiella (Figures 2e, 4a) and potential coevolution of BALOs as response to a plastic resistance phenotype of Klebsiella (Figures 2d, 4f).
Cost of Resistance to Predation
We compared the generation time of evolved and ancestral Klebsiella in order to identify the costs of acquired resistances in the fitness of the prey bacteria (Figure 5). In general, bacteria did not show reduced fitness in terms of a prolonged generation time. In contrast, we found a significant reduction in the generation time of Klebsiella strains from reactors that contained (i) additional prey species (P = 0.021, one sample t-test), (ii) the protist and Klebsiella (P = 0.004, one sample t-test), (iii) the phage and Klebsiella (P = 0.003, one sample t-test), and (iv) the protist, BALO, and all prey species (P = 0.033, one sample t-test).
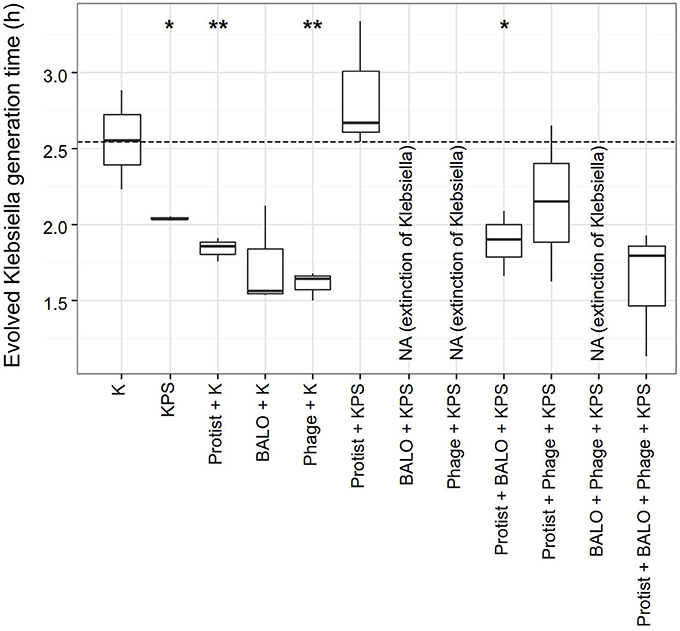
Figure 5. Generation times in hours of Klebsiella strains after the different 72 h experiments. The generation time of the ancestral Klebsiella strain is indicated by the dashed line (2.55 h). P-values are given in the Supplementary Table 3. P-values: *0.011 > p < 0.05, **0.0011 > p < 0.01.
Discussion
Predator Specialization and Species Coexistence
We studied how generalist and specialist micro-predators influence the dynamics of a bacterial community consisting of three different species. Due to the predators' partially distinct prey range and a potential trophic complementarity, we expected the greatest prey removal in the presence of all three predators (Poisot et al., 2013). However, we did not find the number of different predators to be a trigger for increased prey removal, but rather the prey range of a predator. The generalist predator decreased all three prey species, whereas a specialist predator removed only one prey species, i.e., its preferred prey in case of the BALO and the only susceptible prey in case of the phage (Figure 2). Presence of the protist prevented the extinction of the most preferred prey species (Figures 2d,e,g). In particular, Klebsiella, which can be preyed upon by all predators, coexisted with the other prey bacteria in the presence of the generalist and specialist predators. In contrast, we found that the occurrence of predators of greater specialization and the concurrent absence of a generalist predator led to the extinction of the shared prey Klebsiella. This is contrary to the expected resource complementarity (Loreau et al., 2001; Hooper et al., 2002) that would posit conservation of the diversity. However, it seems that the BALO preferred Klebsiella over Pseudomonas despite its experimentally confirmed ability to infect Pseudomonas (Supplementary Table 1). Preferential predation by B. bacteriovorus 109J was also described for a mixture of two potential prey species resulting in significant shifts of each prey's abundance (Rogosky et al., 2006).
As expected, only the protist grazed upon all prey species and decreased their cell concentrations to a similar extent. This effect appears to be strong enough to dampen the effect of Klebsiella predation by the BALO and the phage and might, therefore, be the reason for the observed coexistence of all predator and prey species. Our results indicate that loss of a generalist predator can have a large impact on the diversity of a prey community and might lead to the extinction of prey species that are shared by more specialized predators. The strong stabilizing effects of generalist predators on predator–prey cycles were already described in a theoretical study using a reaction-diffusion-advection modeling framework (Vitense et al., 2016). In addition, a meta-analysis of 60 predator-exclusion experiments with different specialized aphid predators showed similar results (Diehl et al., 2013). Assemblages that contained either only specialists or specialists and generalists had the greatest effect on the reduction of the aphid population compared to an assemblage of only generalists.
Overall, the results indicate that predator–prey dynamics depend on the resource specialization of the predators. Predator–prey dynamics can either result in the extinction of a prey species in case of the specialist predator and Klebsiella or enable coexistence of all prey and predator species in the presence of a generalist predator.
We compared the growth rates of the different predators alone and in the presence of one or two more predators (Figure 3). As expected from its broad prey range, the protist growth rate was not affected by the presence of additional predators (Figure 3a). BALO and phage growth, however, significantly decreased in the presence of the protist (Figures 3d,e) and also in the case of BALO when both protist and phage were present (Figure 3b). This indicates that the protist either ingested free BALOs (Johnke et al., 2017), phages (Deng et al., 2014), or BALO- or phage-infected prey, or was the strongest resource competitor. Another possibility is an indirect effect of the overall prey density reduction by protist predation, which might have substantially lowered the resource availability for the BALO and phage and consequently reduced the growth of the latter. However, the shared resource bacterium Klebsiella was able to survive in reactors containing all three predators and we, therefore, can assume a continuous resource availability for the two specialists. Even though these results may indicate direct negative interactions between the different predators, such as, intraguild predation (Holt and Polis, 1997), further experiments are required to draw definitive conclusions. Independent of the ultimate cause, the presence of a protist had a negative impact on the growth of the more specialized predators possibly resulting in decreased predation pressure on Klebsiella and coexistence of all species.
Predator-Prey Coevolutionary Dynamics
We observed Klebsiella strains resistant to ancestral phage predation after 72 h in multiple reactors. Bacterial resistance to phage predation can be acquired by multiple mechanisms that either prevent phage attachment (e.g., blockage of phage receptors, production of extracellular matrix, or production of competitive inhibitors) or phage DNA entry (e.g., the Sis system); degrade phage genetic material after entry (e.g., restriction-modification systems); or induce death of phage-infected cells in order to protect the remaining population (e.g., abortive infection systems; Labrie et al., 2010). The mechanism responsible for the resistance of Klebsiella in our experiments and the mechanisms leading to the resistance of bacteria against BALOs are currently unknown. Resistant Klebsiella strains appeared only in reactors that contained only the phage and Klebsiella (Figure 4a), the protist and phage together with all prey bacteria (Figure 4c), or all predators and all prey bacteria (Figure 4e). One explanation for the occurrence of resistant Klebsiella strains in only those treatments might be the availability of resources that allow a bacterium to develop resistance, a theory adapted from Thingstad et al. (2014). This resource availability is reduced in reactors that include all prey bacteria due to resource competition between them. However, the presence of a generalist predator might counterbalance this effect by reducing the amount of competing bacteria. Contrary to another study that investigated the effect of protist and phage predation on resistance evolution, we did not observe a negative association between the appearance of resistant prey bacteria and the number of predators (Friman and Buckling, 2013). However, this study was performed with a single prey bacterium and this may explain the differences in our results. The authors argued that the reduced appearance of resistant prey in experimental set-ups that contained both predators vs. set-ups that only contained the phage was potentially governed by reduced prey densities due to protist predation. As an outcome, prey encounter rates with predators were lower and might have resulted in decreased mutation rates (de Visser et al., 1999). Less encounters between the phage and its prey decrease the selection strength for prey resistance and phage infectivity (Hochberg and van Baalen, 1998; Brockhurst et al., 2003; Lopez-Pascua and Buckling, 2008). Since the protist was able to graze on all three prey bacteria in our set-up, this density-mediated effect was most likely much weaker and might have had no influence on the resistance evolution of Klebsiella.
We did not find any BALO-resistant Klebsiella strains. However, BALO predation can result in a more resistant, but plastic prey phenotype (Shemesh and Jurkevitch, 2004), that can be rapidly lost when the bacterium is re-grown in the absence of BALOs. Unfortunately, our experimental setup required a BALO-free cultivation step for Klebsiella in order to obtain sufficient material for the sensitivity assay, and this step may have resulted in the loss of any resistance phenotype. This hypothesis is supported by the fact that we found BALOs that have lost the ability to infect ancestral Klebsiella strains in reactors that only contained BALO and Klebsiella. Plastic phenotypic resistance may, thus, explain the observation that Klebsiella cell numbers remained stable in the reactors containing BALO as the sole predator (Figure 2d). From the three replicate reactors containing BALOs and Klebsiella, BALO extinction during the last day of the experiment was observed in only one. This also suggests the development of resistance mechanisms by Klebsiella as there was no concurrent decrease in Klebsiella cell numbers. Similarly, we observed phages that lost infectivity of ancestral Klebsiella in reactors that only contained Klebsiella. However, the concentration of phages did not decrease even though a proportion of phages was most likely washed out of the reactors due to their small size. Selection, therefore, was not only by the appearance of resistant prey strains but also by the continuous loss of phage particles due to the washout.
The evolution of resistance in prey is often accompanied by trade-offs. A common trade-off for the development of bacterial defense mechanisms is a reduction in the competitive ability of bacteria, e.g., a decline in growth rate (Bohannan and Lenski, 1999; Bohannan et al., 2002; Meyer and Kassen, 2007). Surprisingly, we observed the exact opposite, namely a significant decrease in the generation time of some of the bacteria (Figure 5). All experiments were conducted in a resource-rich environment that may lead to selection for increased growth rates in small populations (Hairston et al., 1970). Exploitation of new resources had been already theoretically concluded to be a necessity for the evolution of bacterial strains under phage predation (Thingstad et al., 2015). It would be, therefore, interesting to see if the evolved Klebsiella strains would show reduced growth in less complex media. In one case, we saw both a significant reduction in Klebsiella generation time and resistance evolution of the Klebsiella in contrast to the usually observed trade-off. This unexpected result questions the benefit of resistance evolution for a fast growing strain. However, we found this to be the case only in Klebsiella from reactors that also contained phages. Here, predation pressure might have been particularly strong, since no other organisms were present. We, therefore, expect that phage–host encounters almost always led to an infection and the faster growth rate might have not induced a strong enough effect in order to prevent extinction. Another explanation might be related to what was observed by Hewlett (2015): bacterial resistance to a phage with an additional increase in growth rate was induced by a mutation within a protein that led to both resistance and an increased rate of sugar uptake (Hewlett, 2015). Interestingly, resistance without costs has already been shown in a number of studies on plants and in Daphnia (Bergelson and Purrington, 1996; Labbé et al., 2010).
Altogether, our results led to a theory of coexistence in a micro-predator system as described in the Supplementary Figure 3. The final cause that determines if a prey species indeed faces extinction seems to depend on the availability of enough resources. An environment containing multiple resource competitors can be made accessible by a generalist predator that reduces the overall density of competing bacteria. Our study indicates that coevolution of a specialist predator might be affected by the presence of additional competing predators in resource-rich environments. That is, the presence of specialist predators increases the predation pressure on shared prey, but only in the absence of a generalist predator. As a consequence, coevolution in multi-predator communities including a generalist might be less common or at least require more time due to competition effects between predators. Even though this hypothesis needs further testing, it could have a great impact on the design of augmentation trials used to combat pathogens in the context of agriculture or wastewater treatment. Further experiments over longer time scales and in more realistic environments should, therefore, be conducted.
Author Contributions
All authors gave the final approval to the current version for publication and agreed to be accountable for all aspects of the work. Questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors were part of the revision process of the draft manuscript and the interpretation of the data. All data were acquired and analyzed by JJ and MB. The mMBR system was developed and tested by AK; MdL supported the set-up of the mMBR systems during the experiment. The experimental design was developed by AC, EJ, and AK. The manuscript was written by JJ.
Funding
This work was funded by the German Research Foundation (DFG) grant CH 731/2-1.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jack Aidley for his support on the revision of the manuscript. This work was funded by the German Research Foundation (DFG) grant CH 731/2-1.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2017.00124/full#supplementary-material
References
Bergelson, J., and Purrington, C. B. (1996). Surveying patterns in the cost of resistance in plants. Am. Nat. 148, 536–558. doi: 10.1086/285938
Bohannan, B. J. M., Kerr, B., Jessup, C. M., Hughes, J. B., and Sandvik, G. (2002). Trade-offs and coexistence in microbial microcosms. Antonie Van Leeuwenhoek 81, 107–115. doi: 10.1023/A:1020585711378
Bohannan, B. J. M., and Lenski, R. E. (1999). Effect of prey heterogeneity on the response of a model food chain to resource enrichment. Am. Nat. 153, 73–82. doi: 10.1086/303151
Brockhurst, M. A., Morgan, A. D., Rainey, P. B., and Buckling, A. (2003). Population mixing accelerates coevolution. Ecol. Lett. 6, 975–979. doi: 10.1046/j.1461-0248.2003.00531.x
Carey, M. P., and Wahl, D. H. (2010). Interactions of multiple predators with different foraging modes in an aquatic food web. Oecologia 162, 443–452. doi: 10.1007/s00442-009-1461-3
Chase, J. M., Abrams, P. A., Grover, J. P., Diehl, S., Chesson, P., Holt, R. D., et al. (2002). The interaction between predation and competition: a review and synthesis. Ecol. Lett. 5, 302–315. doi: 10.1046/j.1461-0248.2002.00315.x
Chen, H., Athar, R., Zheng, G., and Williams, H. N. (2011). Prey bacteria shape the community structure of their predators. ISME J. 5, 1314–1322. doi: 10.1038/ismej.2011.4
Chen, H., and Williams, H. N. (2012). Sharing of prey: coinfection of a bacterium by a virus and a prokaryotic predator. Mbio 3:e00051-12. doi: 10.1128/mBio.00051-12
Chen, H., Young, S., Berhane, T. K., and Williams, H. N. (2012). Predatory bacteriovorax communities ordered by various prey species. PLoS ONE 7:e34174. doi: 10.1371/journal.pone.0034174
Chesson, P. (1991). A need for niches. Trends Ecol. Evol. 6, 26–28. doi: 10.1016/0169-5347(91)90144-M
Curds, C. R. (1973). The role of protozoa in the activated-sludge process. Am. Zool. 13, 161–169. doi: 10.1093/icb/13.1.161
Deng, L., Krauss, S., Feichtmayer, J., Hofmann, R., Arndt, H., and Griebler, C. (2014). Grazing of heterotrophic flagellates on viruses is driven by feeding behaviour. Environ. Microbiol. Rep. 6, 325–330. doi: 10.1111/1758-2229.12119
de Visser, J. A. G. M., Zeyl, C. W., Gerrish, P. J., Blanchard, J. L., and Lenski, R. E. (1999). Diminishing returns from mutation supply rate in asexual populations. Science 283, 404–406. doi: 10.1126/science.283.5400.404
Diehl, E., Sereda, E., Wolters, V., and Birkhofer, K. (2013). Effects of predator specialization, host plant and climate on biological control of aphids by natural enemies: a meta-analysis. J. Appl. Ecol. 50, 262–270. doi: 10.1111/1365-2664.12032
Diehl, S., and Feissel, M. (2001). Intraguild prey suffer from enrichment of their resources: a microcosm experiment with ciliates. Ecology 82, 2977–2983. doi: 10.1890/0012-9658(2001)082[2977:IPSFEO]2.0.CO;2
Duffy, J. E., Cardinale, B. J., France, K. E., McIntyre, P. B., Thebault, E., and Loreau, M. (2007). The functional role of biodiversity in ecosystems: incorporating trophic complexity. Ecol. Lett. 10, 522–538. doi: 10.1111/j.1461-0248.2007.01037.x
Ellis, B. K., Stanford, J. A., Goodman, D., Stafford, C. P., Gustafson, D. L., Beauchamp, D. A., et al. (2011). Long-term effects of a trophic cascade in a large lake ecosystem. Proc. Natl. Acad. Sci. U.S.A. 108, 1070–1075. doi: 10.1073/pnas.1013006108
Filip, J., Bauer, B., Hillebrand, H., Beniermann, A., Gaedke, U., and Moorthi, S. D. (2014). Multitrophic diversity effects depend on consumer specialization and species-specific growth and grazing rates. Oikos 123, 912–922. doi: 10.1111/oik.01219
Finke, D. L., and Denno, R. F. (2004). Predator diversity dampens trophic cascades. Nature 429, 407–410. doi: 10.1038/nature02554
Friman, V. P., and Buckling, A. (2014). Phages can constrain protist predation-driven attenuation of Pseudomonas aeruginosa virulence in multienemy communities. ISME J. 8, 1820–1830. doi: 10.1038/ismej.2014.40
Friman, V.-P., and Buckling, A. (2013). Effects of predation on real-time host–parasite coevolutionary dynamics. Ecol. Lett. 16, 39–46. doi: 10.1111/ele.12010
Friman, V. P., Guzman, L. M., Reuman, D. C., and Bell, T. (2015). Bacterial adaptation to sublethal antibiotic gradients can change the ecological properties of multitrophic microbial communities. Proc. Biol. Sci. 282:20142920. doi: 10.1098/rspb.2014.2920
Gallet, R., Tully, T., and Evans, M. E. K. (2009). Ecological conditions affect evolutionary trajectory in a predator-prey system. Evolution 63, 641–651. doi: 10.1111/j.1558-5646.2008.00559.x
Gonzalez, J., and Suttle, C. (1993). Grazing by marine nanoflagellates on viruses and virus-sized particles: ingestion and digestion. Mar. Ecol. Prog. Ser. 94, 1–10. doi: 10.3354/meps094001
Griffin, J. N., De la Haye, K. L., Hawkins, S. J., Thompson, R. C., and Jenkins, S. R. (2008). Predator diversity and ecosystem functioning: density modifies the effect of resource partitioning. Ecology 89, 298–305. doi: 10.1890/07-1220.1
Hairston, N. G., Tinkle, D. W., and Wilbur, H. M. (1970). Natural selection and the parameters of population growth. J. Wildl. Manage 34, 681–690. doi: 10.2307/3799132
Hammill, E., Kratina, P., Vos, M., Petchey, O. L., and Anholt, B. R. (2015). Food web persistence is enhanced by non-trophic interactions. Oecologia 178, 549–556. doi: 10.1007/s00442-015-3244-3
Hennemuth, W., Rhoads, L. S., Eichelberger, H., Watanabe, M., Van Bell, K. M., Ke, L. E. I., et al. (2008). Ingestion and inactivation of bacteriophages by Tetrahymena. J. Eukaryot. Microbiol. 55, 44–50. doi: 10.1111/j.1550-7408.2007.00303.x
Hewlett, M. (2015). The Evolution of Resistance to Multidrug Antibiotic Therapies. Available online at: https://ore.exeter.ac.uk/repository/handle/10871/21596 (Accessed June 23, 2017).
Hochberg, M. E., and van Baalen, M. (1998). Antagonistic coevolution over productivity gradients. Am. Nat. 152, 620–634. doi: 10.1086/286194
Holt, R. D., and Polis, G. A. (1997). A theoretical framework for intraguild predation. Am. Nat. 149, 745–764. doi: 10.1086/286018
Hooper, D. U., Solan, M., Symstad, A., Gessner, N., Buchmann, V., Degrange, P., et al. (2002). “Species diversity, functional diversity, and ecosystem functioning,” in Biodiversity and Ecosystem Functioning. Synthesis and Perspectives, eds M. Loreau, S. Naeem, and P. Inchausti (Oxford, UK: Oxford University Press), 195–208.
Hyman, P., and Abedon, S. T. (2010). Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70, 217–248. doi: 10.1016/S0065-2164(10)70007-1
Jiang, L., Joshi, H., and Patel, S. N. (2009). Predation alters relationships between biodiversity and temporal stability. Am. Nat. 173, 389–399. doi: 10.1086/596540
Jiang, L., and Morin, P. J. (2005). Predator diet breadth influences the relative importance of bottom-up and top-down control of prey biomass and diversity. Am. Nat. 165, 350–363. doi: 10.1086/428300
Johnke, J., Boenigk, J., Harms, H., and Chatzinotas, A. (2017). Killing the killer: predation between protists and predatory bacteria. FEMS Microbiol. Lett. 364:fnx089. doi: 10.1093/femsle/fnx089
Johnke, A. (2014). Multiple micro-predators controlling bacterial communities in the environment. Curr. Opin. Biotechnol. 27, 185–190. doi: 10.1016/j.copbio.2014.02.003
Jurkevitch, E. (2005). “Isolation and classification of Bdellovibrio and like organisms,” in Current Protocols in Microbiology (John Wiley & Sons, Inc.). Available online at: http://onlinelibrary.wiley.com/doi/10.1002/9780471729259.mc07b01s26/abstract (Accessed September 14, 2016).
Kadouri, D., and O'Toole, G. A. (2005). Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Env. Microb. 71, 4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005
Kang, Y., and Wedekin, L. (2013). Dynamics of a intraguild predation model with generalist or specialist predator. J. Math. Biol. 67, 1227–1259. doi: 10.1007/s00285-012-0584-z
Koskella, B., and Meaden, S. (2013). Understanding bacteriophage specificity in natural microbial communities. Viruses 5, 806–823. doi: 10.3390/v5030806
Kropinski, A., Mazzocco, A., Waddell, T., Lingohr, E., and Johnson, R. (2009). “Enumeration of bacteriophages by double agar overlay plaque assay,” in Bacteriophages Methods in Molecular BiologyTM, eds M. J. Clokie and A. Kropinski (Humana Press), 69–76.
Labbé, P., Vale, P. F., and Little, T. J. (2010). Successfully resisting a pathogen is rarely costly in Daphnia magna. BMC Evol. Biol. 10:355. doi: 10.1186/1471-2148-10-355
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Micro. 8, 317–327. doi: 10.1038/nrmicro2315
Lafferty, K. D., Dobson, A. P., and Kuris, A. M. (2006). Parasites dominate food web links. Proc. Natl. Acad. Sci. U.S.A. 103, 11211–11216. doi: 10.1073/pnas.0604755103
Lopez-Pascua, L. D. C., and Buckling, A. (2008). Increasing productivity accelerates host-parasite coevolution. J. Evol. Biol. 21, 853–860. doi: 10.1111/j.1420-9101.2008.01501.x
Loreau, M., Naeem, S., Inchausti, P., Bengtsson, J., Grime, J., Hector, A., et al. (2001). Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294, 804–808. doi: 10.1126/science.1064088
Martinez, N. D. (1992). Constant connectance in community food webs. Am. Nat. 139, 1208–1218. doi: 10.1086/285382
Matz, C., and Kjelleberg, S. (2005). Off the hook - how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307. doi: 10.1016/j.tim.2005.05.009
Meyer, J. R., and Kassen, R. (2007). The effects of competition and predation on diversification in a model adaptive radiation. Nature 446, 432–435. doi: 10.1038/nature05599
Middelboe, M., Hagström, A., Blackburn, N., Sinn, B., Fischer, U., Borch, N. H., et al. (2001). Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb. Ecol. 42, 395–406. doi: 10.1007/s00248-001-0012-1
Narwani, A., and Mazumder, A. (2010). Community composition and consumer identity determine the effect of resource species diversity on rates of consumption. Ecology 91, 3441–3447. doi: 10.1890/10-0850.1
Oksanen, J., Kindt, R., Legendre, P., O'Hara, B., Stevens, M. H. H., Oksanen, M. J., et al. (2007). The vegan package. Community Ecol. Package 10, 631–637.
Oksanen, L., Fretwell, S. D., Arruda, J., and Niemela, P. (1981). Exploitation ecosystems in gradients of primary productivity. Am. Nat. 118, 240–261. doi: 10.1086/283817
Ormala-Odegrip, A. M., Ojala, V., Hiltunen, T., Zhang, J., Bamford, J. K. H., and Laakso, J. (2015). Protist predation can select for bacteria with lowered susceptibility to infection by lytic phages. BMC Evol. Biol. 15:81. doi: 10.1186/s12862-015-0341-1
Patil, S., Harnisch, F., and Schröder, U. (2010). Toxicity response of electroactive microbial biofilms—a decisive feature for potential biosensor and power source applications. Chemphyschem 11, 2834–2837. doi: 10.1002/cphc.201000218
Philpott, S. M., Pardee, G. L., and Gonthier, D. J. (2012). Cryptic biodiversity effects: importance of functional redundancy revealed through addition of food web complexity. Ecology 93, 992–1001. doi: 10.1890/11-1431.1
Poisot, T., Mouquet, N., and Gravel, D. (2013). Trophic complementarity drives the biodiversity-ecosystem functioning relationship in food webs. Ecol. Lett. 16, 853–861. doi: 10.1111/ele.12118
Polis, G. A. (1991). Complex trophic interactions in deserts: an empirical critique of food-web theory. Am. Nat. 138, 123–155. doi: 10.1086/285208
R. Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Rogosky, A. M., Moak, P. L., and Emmert, E. A. (2006). Differential predation by Bdellovibrio bacteriovorus 109J. Curr. Microbiol. 52, 81–85. doi: 10.1007/s00284-005-0038-6
Ryberg, W. A., Smith, K. G., and Chase, J. M. (2012). Predators alter the scaling of diversity in prey metacommunities. Oikos 121, 1995–2000. doi: 10.1111/j.1600-0706.2012.19620.x
Samson, J. E., Magadan, A. H., Sabri, M., and Moineau, S. (2013). Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687. doi: 10.1038/nrmicro3096
Schmitz, O. J. (2007). Predator diversity and trophic interactions. Ecology 88, 2415–2426. doi: 10.1890/06-0937.1
Seed, K. D. (2015). Battling phages: how bacteria defend against viral attack. PLoS Pathog. 11:e1004847. doi: 10.1371/journal.ppat.1004847
Segev, O., Shapiro, O. H., Brenner, A., and Kushmaro, A. (2013). Application of a unique miniature MBR for screening the biodegradation of brominated flame retardants. Desalin. Water Treat. 51, 5909–5917. doi: 10.1080/19443994.2013.792139
Shemesh, Y., and Jurkevitch, E. (2004). Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ. Microbiol. 6, 8–12. doi: 10.1046/j.1462-2920.2003.00530.x
Sherr, E. B., and Sherr, B. F. (2002). Significance of predation by protists in aquatic food webs. Antonie Van Leeuwenhoek. 81, 293–308. doi: 10.1023/A:1020591307260
Sih, A., Englund, G., and Wooster, D. (1998). Emergent impacts of multiple predators on prey. Trends Ecol. Evol. 13, 350–355. doi: 10.1016/S0169-5347(98)01437-2
Simek, K., Pernthaler, J., Weinbauer, M. G., Hornak, K., Dolan, J. R., Nedoma, J., et al. (2001). Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67, 2723–2733. doi: 10.1128/AEM.67.6.2723-2733.2001
Sime-Ngando, T., and Ram, A. S. P. (2005). Grazer effects on prokaryotes and viruses in a freshwater microcosm experiment. Aquat. Microb. Ecol. 41, 115–124. doi: 10.3354/ame041115
Smith, D. L., and Doerder, F. P. (1992). Multiple effects of mutation on expression of alternative cell surface protein genes in Tetrahymena thermophila. Genetics 130, 97–104.
Steiner, C. F. (2001). The effects of prey heterogeneity and consumer identity on the limination of trophic-level biomass. Ecology 82, 2495–2506. doi: 10.1890/0012-9658(2001)082[2495:TEOPHA]2.0.CO;2
Thingstad, T. F., Pree, B., Giske, J., and Vage, S. (2015). What difference does it make if viruses are strain-, rather than species-specific? Front. Microbiol. 6:320. doi: 10.3389/fmicb.2015.00320
Thingstad, T. F., Våge, S., Storesund, J. E., Sandaa, R.-A., and Giske, J. (2014). A theoretical analysis of how strain-specific viruses can control microbial species diversity. Proc. Natl. Acad. Sci. U.S.A. 111, 7813–7818. doi: 10.1073/pnas.1400909111
Varon, M. (1979). Selection of predation-resistant bacteria in continuous culture. Nature 277, 386–388. doi: 10.1038/277386a0
Vitense, K., Wirsing, A. J., Tyson, R. C., and Anderson, J. J. (2016). Theoretical impacts of habitat loss and generalist predation on predator–prey cycles. Ecol. Model. 327, 85–94. doi: 10.1016/j.ecolmodel.2016.02.002
Weinbauer, M. G., Hornák, K., Jezbera, J., Nedoma, J., Dolan, J. R., and Šimek, K. (2007). Synergistic and antagonistic effects of viral lysis and protistan grazing on bacterial biomass, production and diversity. Environ. Microbiol. 9, 777–788. doi: 10.1111/j.1462-2920.2006.01200.x
Williams, H. N., Lymperopoulou, D. S., Athar, R., Chauhan, A., Dickerson, T. L., Chen, H., et al. (2016). Halobacteriovorax, an underestimated predator on bacteria: potential impact relative to viruses on bacterial mortality. ISME J. 10, 491–499. doi: 10.1038/ismej.2015.129
Keywords: Bdellovibrio bacteriovorus, predation, coexistence, predator-prey interactions, arms race, virus-host interactions
Citation: Johnke J, Baron M, de Leeuw M, Kushmaro A, Jurkevitch E, Harms H and Chatzinotas A (2017) A Generalist Protist Predator Enables Coexistence in Multitrophic Predator-Prey Systems Containing a Phage and the Bacterial Predator Bdellovibrio. Front. Ecol. Evol. 5:124. doi: 10.3389/fevo.2017.00124
Received: 27 June 2017; Accepted: 22 September 2017;
Published: 10 October 2017.
Edited by:
Mauricio Lima, Pontificia Universidad Católica de Chile, ChileReviewed by:
Sergio Andrés Estay, Universidad Austral de Chile, ChilePavel Kratina, Queen Mary University of London, United Kingdom
Copyright © 2017 Johnke, Baron, de Leeuw, Kushmaro, Jurkevitch, Harms and Chatzinotas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonis Chatzinotas, YW50b25pcy5jaGF0emlub3Rhc0B1ZnouZGU=
†Present Address: Julia Johnke, Department of Evolutionary Ecology and Genetics, Christian-Albrechts-Universität zu Kiel, Kiel, Germany
 Julia Johnke
Julia Johnke Maayan Baron
Maayan Baron Marina de Leeuw2
Marina de Leeuw2 Ariel Kushmaro
Ariel Kushmaro Edouard Jurkevitch
Edouard Jurkevitch Antonis Chatzinotas
Antonis Chatzinotas