
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 03 August 2016
Sec. Behavioral and Evolutionary Ecology
Volume 4 - 2016 | https://doi.org/10.3389/fevo.2016.00090
This article is part of the Research Topic The Importance of Olfaction in Intra- and Interspecific Communication View all 7 articles
 Danielle J. Whittaker1,2
Danielle J. Whittaker1,2 Nicole M. Gerlach3
Nicole M. Gerlach3 Samuel P. Slowinski4
Samuel P. Slowinski4 Kyle P. Corcoran5
Kyle P. Corcoran5 Andrew D. Winters6
Andrew D. Winters6 Helena A. Soini5
Helena A. Soini5 Milos V. Novotny5
Milos V. Novotny5 Ellen D. Ketterson4
Ellen D. Ketterson4 Kevin R. Theis1,6*
Kevin R. Theis1,6*Chemical signaling is an underappreciated means of communication among birds, as may be the potential contributions of symbiotic microbes to animal chemical communication in general. The dark-eyed junco (Junco hyemalis) produces and detects volatile compounds that may be important in reproductive behavior. These compounds are found in preen oil secreted by the uropygial gland, and this gland supports diverse bacterial communities including genera known to produce some of these volatile compounds. We investigated the relative contributions of shared environments and genetic relatedness in shaping juncos' symbiotic bacterial communities, and investigated whether these bacterial communities underlie juncos' chemical signaling behavior. We sampled parents and nestlings at 9 junco nests during one breeding season at Mountain Lake Biological Station in Virginia, USA. From each individual, we collected swabs of the uropygial gland and the cloaca, preen oil, and a small blood sample for paternity testing. We characterized junco bacterial communities through 16S rRNA gene surveys and preen oil volatile compounds via gas chromatography-mass spectrometry. Nest membership and age class had the strongest influence on the structure of bacterial and volatile profiles. We compared father-offspring similarity based on paternity, and nestling similarity in nests containing full siblings and half siblings, and found that relatedness did not noticeably affect bacterial or volatile profiles. While we cannot rule out an influence of genetic relatedness on these profiles, it is clear that shared environments are more influential in shaping bacterial and volatile profiles among juncos. We did not find significant covariation between individual bacterial and volatile profiles. Possible explanations for this result include: (1) bacteria do not underlie volatile production; (2) ample redundancy in volatile production among bacterial types obscures covariation; or (3) the relationship is confounded by the fact that, unlike glands exclusively dedicated to chemical communication, uropygial glands have multiple functions, and symbiotic bacteria are hypothesized to contribute to each of these. Therefore, different bacteria may contribute to different phenotypes of the avian holobiont. Future work will include cultivation, metabolomic, genomic, and behavioral assay approaches to tease these scenarios apart.
Animals ubiquitously interact with symbiotic (i.e., resident) microbes, many of which provide substantial benefits to their hosts (Gilbert et al., 2012; McFall-Ngai et al., 2013). For example, symbiotic microbes provide their hosts with access to otherwise inaccessible energy and nutrients (Tremaroli and Backhed, 2012; Hacquard et al., 2015), they prime and bolster host immune systems (Hooper et al., 2012; Lee and Hase, 2014), they catalyze the effective development and functioning of host organs (McFall-Ngai, 2002; Fraune and Bosch, 2010), and they can directly contribute to their hosts' behavioral phenotypes (Archie and Theis, 2011; Ezenwa et al., 2012), including chemical signals used in intraspecific communication (Archie and Theis, 2011; Ezenwa and Williams, 2014). A key line of inquiry in contemporary host-microbial ecology is determining the relative influence of the environment and host genotype on the development of an individual host's microbiota (Spor et al., 2011; Goodrich et al., 2014; Org et al., 2015). Teasing apart these effects is particularly intriguing when considering the potential contribution of symbiotic microbes to intraspecific chemical signaling because animals' chemical signals are often thought to be indicative of individual genetic quality (Johansson and Jones, 2007), particularly when they are related to immune function or parasite load (Penn and Potts, 1998). That symbiotic microbial communities, and consequently animals' chemical signals, could in fact be strongly influenced by animals' environments raises interesting questions regarding signal fidelity, honesty, and selection.
Although it is logically apparent that variation in both environments and host genotypes could shape host microbiota, the extent to which these factors actually do so, and the scales at which they operate, remain largely unknown. Among humans, despite high inter-individual variation in the composition and structure of gut microbiota, microbial community signatures are still generally evident at the level of family households and even nations (Turnbaugh et al., 2009; Lee et al., 2011; Yatsunenko et al., 2012; Goodrich et al., 2014). These patterns could be due to either variation in shared environments or genetic relatedness (Spor et al., 2011; Goodrich et al., 2014; Org et al., 2015). Most available data suggest a strong environmental component in shaping the human gut microbiota: cohabiting parents have more similar gut microbiota than do men and women not living together (Yatsunenko et al., 2012), and monozygotic and dizygotic twin pairs generally have gut microbiota that differ to similar degrees (Turnbaugh et al., 2009; Lee et al., 2011; Yatsunenko et al., 2012). However, a recent study demonstrated a role for genetic relatedness, calling these prior twin studies into question: they found that monozygotic twins do indeed have more similar gut microbiota than do dizygotic twins, and further demonstrated high heritability of some gut bacterial types—specifically, members of the family Christensenellaceae (a recently described taxon associated with health and leanness in humans) appear to constitute the hub of a module of co-occurring heritable microbial families (Goodrich et al., 2014).
Equivocal evidence exists for the factors shaping most other animal microbiota as well. At a broad level, host-microbiota specificity across different environments suggests that host genetics outweigh environmental effects with respect to which microbes may colonize animal species. For example, epithelial microbiota differ significantly between two congeners of the cnidarian Hydra, and the microbiota of individuals in hydra lines maintained in the lab for over three decades remained similar to those of their wild conspecifics (Fraune and Bosch, 2007). A few studies have even identified specific, single host genes that affect the composition of an individual's microbiota; they are primarily genes involved in immune function, but a few have metabolic roles (reviewed in Spor et al., 2011). However, environmental effects still appear to account for a great deal of variation in the composition and structure of animals' microbiota. Notably, an individual's environment includes conspecifics, especially in social species. Across the animal kingdom, vertical and/or horizontal transmission of microbes from mother to offspring appears universal (Funkhouser and Bordenstein, 2013). In mammals, vaginal birth and lactation are major mechanisms for inoculating neonates with microbes, while in birds egg-laying, incubation, and brooding can effectively transmit microbes to chicks (Ruiz-de-Castañeda et al., 2011a). Non-relatives can also be key contributors to an individual's microbiota. For example, in yellow baboons (Papio cynocephalus), social group membership and grooming networks are the strongest predictors of gut microbiota structure (Tung et al., 2015), and adult zebra finches (Taeniopygia guttata) share cloacal microbes through copulation and allopreening (Kulkarni and Heeb, 2007). Other environmental factors that could markedly affect microbiota include variation in diet (Wu et al., 2011; David et al., 2014) and stochastic variability in the colonization process of different individuals (Sloan et al., 2006; Spor et al., 2011).
The most broadly studied relationships between symbiotic microbes and animal behavior so far are in the area of intraspecific chemical communication (Archie and Theis, 2011; Davis et al., 2013; Ezenwa and Williams, 2014). The fermentation hypothesis for animal chemical communication (Albone, 1984) suggests that some components of animals' chemical signals are products of symbiotic microbial metabolism, and that variation in these signals, both among and within species, is due in part to underlying variation in the structure of animals' odor-producing microbial communities. Symbiotic microbes have been posited to contribute odors signaling a great deal of information about their host animals, including their species and individual identity, group membership, sex, reproductive state, and general quality (Archie and Theis, 2011; Davis et al., 2013; Ezenwa and Williams, 2014).
Here we evaluate the effects of variation in environment and host genotype in shaping the microbiota and chemical signals of a songbird, the Carolina subspecies of the dark-eyed junco (Junco hyemalis carolinensis). Previous studies have suggested that there are strong environmental influences on avian microbiota, in both the cloaca and the uropygial gland (Lucas and Heeb, 2005; Klomp et al., 2008; Ruiz-Rodriguez et al., 2014). Cross infection is a likely explanation for these patterns, as symbiotic bacteria can be transmitted to mates via copulation and to other social partners via cohabitation and allopreening (Westneat and Rambo, 2000; Kulkarni and Heeb, 2007; White et al., 2010), and symbiotic bacteria can also be readily transmitted from mothers to offspring during egg-laying, brooding, and grooming (Ruiz-de-Castañeda et al., 2011a,b). The uropygial gland secretes preen oil which birds spread on their feathers with their bills to clean and maintain the feathers and to manage the feather-degrading bacterial communities that populate them (Jacob and Ziswiler, 1982; Shawkey et al., 2003; Martín-Vivaldi et al., 2009, 2010). This oil also contains volatile compounds that can communicate information about the individual's species identity (Soini et al., 2013), sex (Soini et al., 2007; Whittaker et al., 2010), age (Shaw et al., 2011), and breeding condition (Whittaker et al., 2011), and that can even be predictive of an individual's reproductive success (Whittaker et al., 2013). Recently, we demonstrated that the dark-eyed junco's uropygial gland harbors a diverse and rich bacterial community, and that several of the bacterial genera identified are known to produce volatile compounds present in junco preen oil (Whittaker and Theis, 2016). To our knowledge, no study has yet demonstrated that symbiotic bacteria produce avian chemical signals.
In this study, we determine the relative influence of social environment and genetic relatedness among siblings and their parents in shaping the composition and structure of avian bacterial and odor profiles in a wild population of dark-eyed juncos. The dark-eyed junco is a widespread North American emberizid sparrow that has been the subject of behavioral, physiological, and evolutionary field and captive studies for over a century (Nolan et al., 2002). With respect to behavioral ecology, the junco has been an important study system for investigating a wide range of topics, including the biological timing of migration and reproduction (Rowan, 1925; Ball and Ketterson, 2007), the role of hormones and responsive tissues in the modulation and evolution of behavior (Ketterson and Nolan, 1992; Ketterson et al., 2001; Bergeon Burns et al., 2013), and adaptation to novel environments (Atwell et al., 2014). In fact, the dark-eyed junco has been referred to as an ecological model organism (Peterson et al., 2012).
One of the traits that makes dark-eyed juncos such intriguing study subjects is that although they are socially monogamous (Nolan et al., 2002), they exhibit appreciable levels of extra-pair paternity (EPP)—about 30% of offspring in all populations studied to date were sired by an extra-pair male (Ketterson et al., 1998; Gerlach et al., 2012a,b; Atwell et al., 2014). This variation in paternity creates a kind of natural experiment, allowing for comparisons among parent-offspring dyads and sibling groups with different levels of genetic relatedness. Females incubate eggs for 12 days, and brood nestlings for 11–12 days, at which time nestlings typically fledge (Nolan et al., 2002). Both males and females provision nestlings, but since only females brood eggs and nestlings, we can also compare the effects of physical contact between offspring and the two parents on bacterial and odor similarity.
Our specific objectives in this study were to (1) assess the effects of sex, age (i.e., adult vs. nestling), and nest identity on junco cloacal and uropygial microbiota; (2) assess the effects of the same variables on junco preen oil volatile odor profiles; (3) test for covariation between junco microbial and volatile profiles; and (4) elucidate the relative influence that social environment (e.g., degree of physical contact) and genetic relatedness (i.e., related vs. unrelated parent-offspring pairs; full vs. half siblings) have in shaping juncos' developing microbiota and volatile odor profiles.
We conducted this study on the Carolina subspecies of the dark-eyed junco at Mountain Lake Biological Station (MLBS) near Pembroke, Virginia, USA (37°22′N, 80°32′W). MLBS is the site of a long-term study on dark-eyed juncos led by Ellen Ketterson of Indiana University since 1983. As part of the long-term study, adult juncos are captured every year using baited mist nets and traps from mid-April to mid-May, in order to census returning individuals and to band new birds with United States Fish and Wildlife Service bands. Study investigators also take morphological measurements of all birds, and determine sex by the presence of a brood patch (female) or cloacal protuberance (male), as well as by plumage and wing length (Nolan et al., 2002). For paternity testing, a small blood sample (50–100 μL) is collected from the alar vein of each bird and stored in Longmire's lysis buffer solution (Longmire et al., 1992) at 4°C. All birds are released at the site where they were captured, typically less than 1 h after capture.
Samples for this particular study were collected in May-June 2012. We searched intensively for nests daily throughout May, and once we located a nest, we monitored it every other day until hatching (day 0). On day 3 and 6 we weighed the nestlings, and on day 6 we banded each nestling and took a small blood sample for paternity testing and sex determination. On day 11–12 (the time at which junco nestlings typically leave the nest, or “fledge”), we captured the nestlings as well as both adults at all nests to verify the identity of the social father and to collect blood samples if they had not previously been obtained. We then sampled each individual's cloacal and uropygial bacterial communities, and took samples of preen oil. We sampled cloacal bacterial communities by gently swabbing the opening of the cloaca. We sampled uropygial gland bacterial communities by rubbing a sterile cotton swab over the tip of the gland, which stimulates preen oil production and ensured that our sampled solution was similar to the mixture that juncos collect on their bills for preening. All swabs were stored in cryogenic vials at −80°C until analysis. Preen oil was collected by gently rubbing the uropygial gland with a 100 μL glass capillary tube (Drummond Scientific, Broomall, PA). This action stimulates the gland to secrete 1–3 mg of preen oil (Whittaker et al., 2010). We stored preen oil samples on ice immediately after collection, at −20°C within 1 h, and long-term at −80°C until they were analyzed by gas chromatography-mass spectrometry (GC-MS).
After sampling, the adults were released at the site of capture and nestlings were returned to the nest. In total, we sampled 9 adult females, 8 adult males (one nest was not attended by an adult male), and 27 nestlings from 9 nests (range: 2–4 nestlings/nest) at MLBS (Figure 1). This study builds upon a preliminary study in which we characterized the uropygial gland microbiota of adult juncos from 13 nests (Whittaker and Theis, 2016). Here we additionally include data from junco nestlings and data on the cloacal microbiota, uropygial gland volatile profiles, and paternity. The current study focuses on 9 nests because we did not have microbiota profiles, volatile compound profiles, and/or paternity data for all nestlings of 4 nests sampled in 2012. This study was conducted in compliance with the Bloomington Institutional Animal Care and Use Committee guidelines (BIACUC protocol 12-050), US Fish and Wildlife Service permit MB093279-0, and Virginia Department of Game and Inland Fisheries permit 041506.
Figure 1. Relative locations of the nine dark-eyed junco nests simultaneously sampled at Mountain Lake Biological Station (MLBS), Virginia. The nest-specific color-coding persists in figures throughout the paper, as applicable.
We extracted junco DNA from blood samples using standard phenol-chloroform techniques (Sambrook et al., 1989) and IBI Scientific MINI Genomic DNA kits (Peosta, IA). We genotyped adult birds and nestlings at eight microsatellite loci (Gerlach et al., 2012a), and determined paternity using the program CERVUS 3.0 (Kalinowski et al., 2007). We determined the sex of each nestling by amplifying the CHD gene on the W and Z chromosomes; males and females are homogametic and heterogametic, respectively (Griffiths et al., 1998).
Of the 9 sampled nests, 3 contained only within-pair young (WPY), 2 contained only extra-pair young (EPY), and 4 were of mixed paternity, containing both WPY and EPY young. Eleven of the 27 nestlings (41%) were EPY. One of the EPY-only nests contained young sired by multiple different males. One mixed paternity nest contained young sired by 3 different males. Therefore, 4 nests contained only full siblings, and 5 contained half-siblings sired by multiple males. Henceforth, we will use “parents” and “father” to refer to the attendant male at the nest (which may or may not be the genetic sire).
We extracted DNA from the bacteria on swabs using MO BIO PowerSoil® DNA Isolation kits (MO BIO Laboratories, Inc., Carlsbad, CA), following the manufacturer's recommended protocol, with two additional steps: we incubated the swab in Bead Solution within the Bead Tube for 10 min, and subsequently vigorously vortexed the Bead Tube for 1 min, before removing the swab aseptically and proceeding with the extraction. The order of extractions was randomized with respect to individual and nest identity, sex, organ, and age group. Sample extractions yielded amplifiable 16S rDNA evident via gel electrophoresis, while blank control extractions did not. However, this does not preclude the amplification and ultimate sequencing of trace background DNA in extraction kits and/or reagents (Salter et al., 2014). Therefore, the breadth of bacterial taxonomic diversities observed in this study should be viewed as a preliminary observation requiring further validation via metagenomic sequencing and targeted cultivation. The V4 region of the bacterial 16S rRNA gene was targeted for sequencing on the Illumina MiSeq platform at the Michigan State University Research Technology Support Facility's Genomics Core. Sample preparation, nucleotide sequencing, and preliminary quality filtering were completed using previously published and widely adopted Illumina protocols (Caporaso et al., 2012).
Sample-specific MiSeq run files, which have been deposited on the NCBI Sequence Read Archive (BioProject ID PRJNA328228; Accession SRP078320), were processed using mothur software, v. 1.31.2 (Schloss et al., 2009; Kozich et al., 2013). We removed all sequences that (1) contained ambiguous base calls, (2) had homopolymer runs exceeding 8 bases, (3) did not start and end at the appropriate primer positions, (4) were labeled chimeric by the uchime tool, or (5) were taxonomically classified as mitochondria, chloroplasts, archaea, eukaryotes, or unknown, using the Ribosomal Database Project's trainset9_032012 reference database (Wang et al., 2007; Claesson et al., 2009). This process removed 25% of the total sequences. One cloacal sample, from a male nestling, was not successfully sequenced due to low DNA recovery, so data from that sample were discarded. Each of the 87 remaining samples was subsampled to a depth of 6,000 sequences, which was the number of sequences in the least represented sample. These sequences were binned into operational taxonomic units (OTUs) using mothur's average neighbor, split-clustering algorithm (level 3) and a 97% sequence similarity cutoff. Sample coverage was high, as indicated by Good's coverage scores averaging 97.8 ± 0.5 and 97.6 ± 2.5% for cloaca and uropygial glands, respectively. All alpha diversity analyses were completed using the full OTU data set. Specifically, for each sample, we used the number of OTUs observed (OTUobs = OTUs observed out of each sample's 6000 sequences) as an indicator of bacterial richness, and calculated the non-parametric Shannon Index (Ĥ) to indicate broader alpha diversity (i.e., both richness and evenness) (Chao and Shen, 2003). Due to non-normal distributions of many of the alpha diversity data sets, we used Kruskal-Wallis, Mann-Whitney, and Wilcoxon signed rank tests to evaluate the effects of sex, age, nest identity, and organ on microbiota richness and evenness (PAST, v.3.11) (Hammer et al., 2001). Given that many background factors could vary among individual nests (e.g., degree of extrapair mating behavior, variation in habitat quality and/or paternal investment), we treated adult males and females within nests, as well as nestlings and adults within nests, as matched pair observations when evaluating effects of sex and age. Correlations were assessed using Spearman's correlation coefficients (rs). Raw p-values are presented for univariate analyses (Gotelli and Ellison, 2004).
For beta diversity analyses, singleton (i.e., OTUs represented by a single sequence) and doubleton (i.e., OTUs represented by two sequences) OTUs were removed from the data set, and a consensus taxonomy was determined for each of the remaining OTUs (N = 2825) using a conservative 80% confidence threshold (Claesson et al., 2009). Taxonomic designations and raw count data for each of these OTUs by sample are provided as Supplementary Material (Supplemental Data 1). Microbiota composition and structure were characterized using Dice (i.e., Sørensen) and Bray-Curtis ecological similarity indices, respectively (PAST, v.3.11). Principal coordinates (PCoA) plots were generated to visualize microbiota variation among samples, and the effects of sex, age, nest identity, and organ on microbiota composition and structure were evaluated using both analyses of similarity (ANOSIM) and non-parametric MANOVA (PAST, v.3.11; 9999 permutations). Differences in dispersion among groups were evaluated using deviations from spatial medians and permutations of least absolute deviations (PERMDISP2; 9999 permutations) (Anderson, 2006). When appropriate, similarity percentage (SIMPER) analyses were conducted to ascertain the contribution of specific OTUs to observed community-level variation. Mantel tests were used to evaluate the strength of covariation between the microbiota of cloacae and uropygial glands, as well as between the bacterial and volatile profiles of uropygial glands (PAST, v.3.11; 9999 permutations). Raw p-values are presented for all multivariate permutation tests (Gotelli and Ellison, 2004; Hammer, 2015). Heat maps were created using Matrix2png (Pavlidis and Noble, 2003).
Our GC-MS methods have been described previously (Whittaker et al., 2010, 2011; Soini et al., 2013). We extracted volatile compounds from the preen oil samples using a Twister® stir bar and performed quantitative analysis with an Agilent 6890N gas chromatograph connected to a 5973i MSD mass spectrometer (Agilent Technologies, Inc., Wilmington, DE) with a Thermal Desorption Autosampler and Cooled Injection System (TDSA-CIS 4 from Gerstel GmbH, Mülheim an der Ruhr, Germany). All major compounds were identified by comparison to standards from Sigma-Aldrich Chemical Company (St. Louis, MO), using mass spectra and retention times. Peak areas of the compounds of interest were normalized by dividing each peak area by that of the internal standard (7-tridecanone) in corresponding runs, yielding relative concentrations (i.e., relative amounts per 1.0 mg of preen oil). Relative standard deviation (RSD, a measure of reproducibility) of the internal standard peak area was 13% (N = 12).
Preen oil volatile compound concentration varies in the short term in response to factors such as breeding condition and hormone level changes (Whittaker et al., 2011). In contrast, the proportions of those compounds that make up the total blend are highly repeatable and reflect individual identity (Whittaker et al., 2010). We converted the measurements to proportions by dividing the observed peak area of each compound by the sum of the peaks of all of the compounds. PCoA plots were used to visualize volatile profile variation among samples, and the effects of sex, age, and nest identity on profile structure were evaluated using analyses of similarity (ANOSIM) and non-parametric MANOVA. SIMPER analyses were conducted to ascertain the contribution of specific volatile compounds to observed profile differences.
The alpha diversity of adults' cloacal microbiota was greater than their uropygial gland microbiota (Wilcoxon test; N = 17; OTUobs: W = 148, p = 0.0002; Ĥ: W = 145, p = 0.0004). No such differences were observed among nestlings (N = 26; OTUobs: W = 201, p = 0.517; Ĥ: W = 199, p = 0.551). The alpha diversity of cloacal and uropygial gland microbiota did not differ between males and females among either adults (Wilcoxon test; Nm = 8, Nf = 8; OTUobs: W = 23, p = 0.549; Ĥ: W = 22, p = 0.642) or nestlings (Mann-Whitney test; Nm = 11, Nf = 15; OTUobs: U = 78.5, p = 0.849; Ĥ: U = 56, p = 0.180). There was no effect of age on the alpha diversity of cloacal microbiota (Wilcoxon test on average values by age class per nest, N = 9; OTUobs: W = 24, p = 0.910; Ĥ: W = 30, p = 0.427; Figure 2). However, the richness (OTUobs: W = 44, p = 0.010) of the uropygial gland microbiota differed between adults and nestlings, with those of nestlings being higher than those of adults (Figure 2).
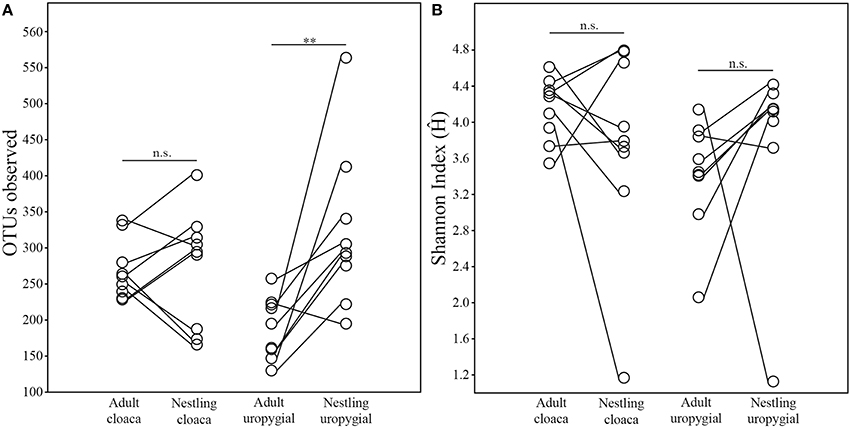
Figure 2. Comparisons of similarities between the (A) richness (OTUs observed) and (B) broader alpha diversity (nonparametric Shannon Index) of the cloacal and uropygial gland microbiota of adult and nestling juncos. Comparisons between age classes reflect average values paired by nest (**p ≤ 0.01).
Among adults, the alpha diversity of cloacal and uropygial gland microbiota did not consistently vary among nests (Kruskal-Wallis test; cloaca: OTUobs: H = 9.201, p = 0.239; Ĥ: H = 7.5, p = 0.379; uropygial gland: OTUobs: H = 12.05, p = 0.099; Ĥ: H = 11.74, p = 0.110). Among nestlings, the alpha diversity of uropygial gland microbiota also did not vary among nests (OTUobs: H = 8.003, p = 0.433; Ĥ: H = 8.239, p = 0.410). However, richness and evenness of cloacal microbiota were influenced by nest identity (OTUobs: H = 18.24, p = 0.019; Ĥ: H = 20.41, p = 0.009). Nestling cloacal microbiota richness (OTUobs: rs = 0.589, p = 0.002) was positively correlated with that of their mother, but not their father (OTUobs: rs = 0.330, p = 0.133).
The composition (ANOSIM: R = 0.107, p = 0.002; NPMANOVA: F = 1.595, p = 0.002; Figure 3), but not the structure (ANOSIM: R = 0.039, p = 0.098; NPMANOVA: F = 1.332, p = 0.123), of cloacal and uropygial gland microbiota differed for adults. Twenty-nine OTUs were present in at least half of the cloacae and half of the uropygial glands of adults, 28 OTUs were present in at least half of the cloacae but not in half of the uropygial glands, and only one OTU was present in at least half the uropygial glands but not in half of the cloacae (Proteobacteria: Salinisphaera; Supplemental Data 1). Coupled with the above results on greater cloacal than uropygial microbiota alpha diversity, this suggests that the uropygial gland microbiota is a subset of the broader cloacal microbiota for adult juncos. Neither the composition (ANOSIM: R = −0.034, p = 0.970; NPMANOVA: F = 0.705, p = 0.981) nor structure (ANOSIM: R = −0.032, p = 0.976; NPMANOVA: F = 0.483, p = 0.994) of cloacal and uropygial gland microbiota differed for nestlings.
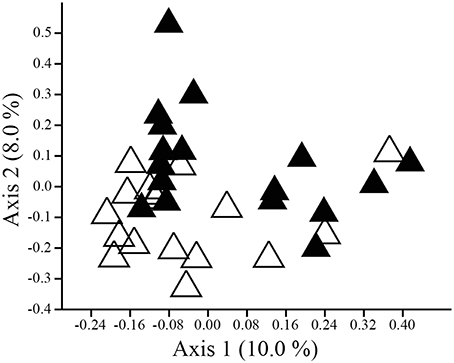
Figure 3. PCoA plot illustrating separation in composition (Dice similarity) between the cloacal (open triangles) and uropygial gland (closed triangles) microbiota of adult juncos.
The composition and structure of cloacal and uropygial gland microbiota did not differ between males and females among either adults or nestlings (Table S1). There were, however, effects of age and nest identity on the beta diversities of cloacal and uropygial gland microbiota (Table 1; Figure 4). SIMPER analyses indicated that there were no particular OTUs responsible for the majority of observed variation between adults and nestlings; rather, many OTUs had modest influence (Supplemental Data 2; Figures S1, S2). The composition of glandular microbiota was not more variable among nestlings than adults (PERMDISP; cloaca: p = 0.168; uropygial: p = 0.166; Figures 4A,B).

Table 1. Analyses evaluating variation in the composition (Dice similarity) and structure (Bray-Curtis similarity) of cloacal and uropygial gland microbiota based on age class and nest identity.
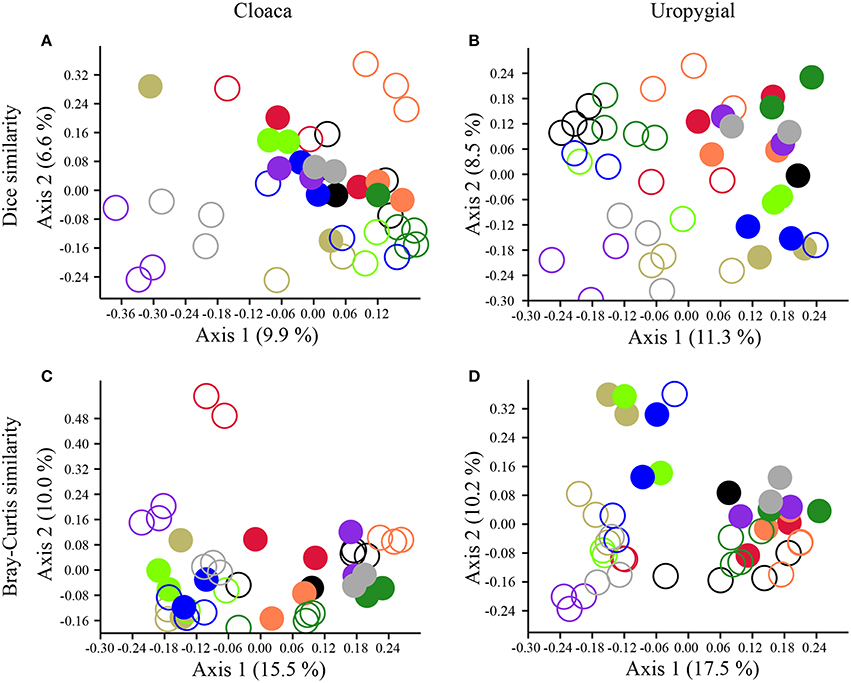
Figure 4. PCoA plots illustrating separation in the (A,B) composition (Dice similarity) and (C,D) structure (Bray-Curtis similarity) of cloacal and uropygial gland microbiota of juncos based on age class and nest identity. Closed circles denote adults, while open circles denote nestlings. Color-coding reflects nest identity.
Nestlings' cloacal microbiota were more similar to those of their parents in both composition and structure than were nestlings' uropygial gland microbiota to the respective microbiota of their parents (Wilcoxon test; mother: composition: W = 275, p = 0.011, structure: W = 262, p = 0.028; father: composition: W = 209, p = 0.006, structure: W = 207, p = 0.007). The composition of nestlings' cloacal microbiota was equally similar to those of their mother and social father (Wilcoxon test; N = 22; W = 140, p = 0.679; Figure 5A), but there was a tendency for the structure of nestlings' cloacal microbiota to be more similar to that of their mother than father (W = 185, p = 0.059; Figure 5B). Overall, nestlings' cloacal microbiota were more similar, in composition (N = 26; W = 345, p < 0.0001) and structure (W = 286, p = 0.005), to those of adult female than adult male juncos in the broader population (i.e., excluding nestlings' parents; Figure 5). Nevertheless, nestlings' cloacal microbiota were more similar to that of their mother than other adult females (composition: W = 282, p = 0.007; structure: W = 328, p = 0.0001), and more similar to that of their father than other adult males (composition: W = 197, p = 0.021; structure: W = 212, p = 0.004; Figure 5). Whether an attendant adult male was the sire or not of its nestlings did not affect the compositional (Mann-Whitney test; Nsire = 11, Nnotsire = 11; U = 57, p = 0.847) or structural (U = 56, p = 0.793) similarity of their cloacal microbiota. Furthermore, the average similarity of nestlings' cloacal microbiota did not differ between full-sibling nests and those containing half-siblings (Mann-Whitney test; Nfull = 4, Nhalf = 5; composition: U = 9, p = 0.905; structure: U = 7, p = 0.556).
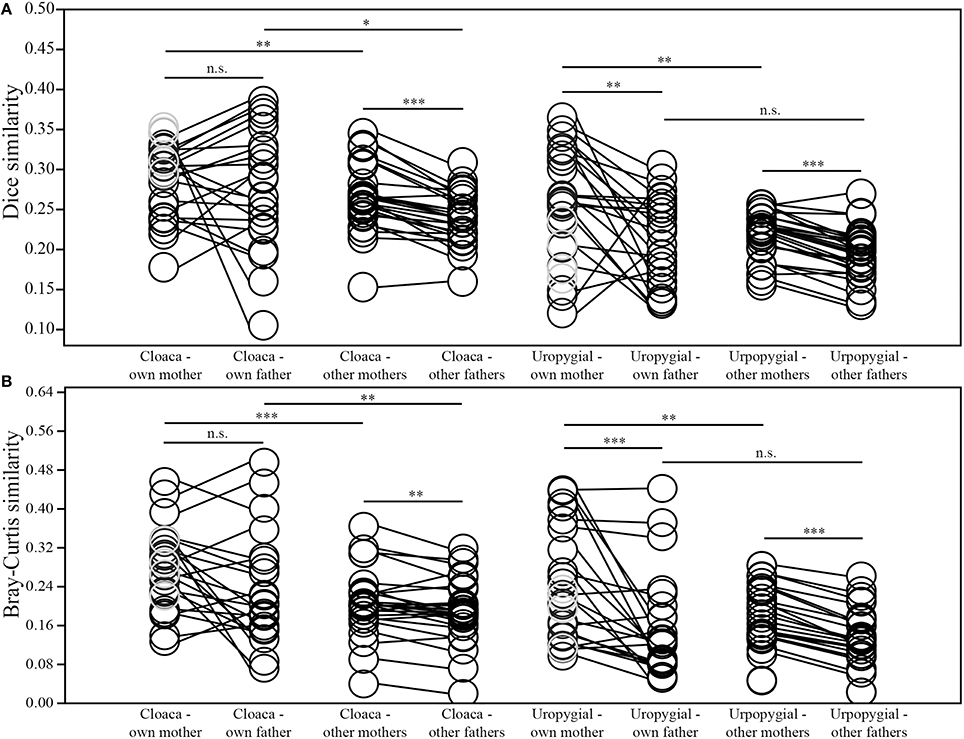
Figure 5. Comparisons of similarities between the (A) composition and (B) structure of the cloacal and uropygial gland microbiota of nestling juncos with those of their own mother and father, and those of other adult females and males in the population (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001). The samples colored gray denote the 4 nestlings that did not have an attendant father.
Both the composition (Wilcoxon test; N = 23; W = 234, p = 0.002) and structure (W = 256, p < 0.0001) of nestlings' uropygial gland microbiota were more similar to the respective microbiota of their mother than father (Figure 5). As with cloacal microbiota, nestlings' uropygial microbiota were more similar, in composition (N = 27; W = 370, p < 0.0001) and structure (W = 378, p < 0.0001), to those of adult female than male juncos in the broader population (Figure 5). Nestlings' uropygial microbiota were more similar to the respective microbiota of their mother than to those of other adult females in the population (N = 27; composition: W = 320, p = 0.002; structure: W = 321, p = 0.002; Figure 5). This was not the case with their father and other adult males (N = 23; composition: W = 185, p = 0.160; structure: W = 172, p = 0.315). As with cloacal microbiota, whether an attendant male was the sire or not of its young did not affect the compositional or structural similarity of their uropygial microbiota (Mann-Whitney test; Nsire = 12, Nnotsire = 11; composition: U = 47, p = 0.260; structure: U = 53, p = 0.449), and there was not a difference in the similarity of the composition or structure of uropygial microbiota between full-sibling and half-sibling nests (Mann-Whitney test; Nfull = 4, Nhalf = 5; composition: U = 10, p = 1.000; structure: U = 9, p = 0.905).
The structure of preen oil volatile profiles did not consistently differ between males and females among either adults (Nm = 8, Nf = 9; ANOSIM: R = −0.076, p = 0.979; NPMANOVA: F = 0.666, p = 0.880) or nestlings (Nm = 12, Nf = 15; ANOSIM: R = −0.048, p = 0.874; NPMANOVA: F = 1.422, p = 0.217). However, similar to the microbiota, there were effects of age and nest identity on the structure of juncos' volatile profiles (ANOSIM: Age: R = 0.55, p = 0.0002, Nest: R = 0.11, p = 0.059; NPMANOVA: Age: F = 5.863, p = 0.0001, Nest: F = 1.162, p = 0.042, Interaction: F = 0.006, p = 0.408; Figure 6). The nest effect was modest; the age effect robust. SIMPER analyses indicated that nearly half (44%) of the observed variation in the structure of volatile profiles between adults and nestlings was due to higher proportions of 1-undecanol in adult preen oil (Mann-Whitney test; U = 42, p < 0.0001; 19.1%), and higher levels of 1-hexadecanol (U = 96, p = 0.001; 15.8%) and 1-pentadecanol (U = 53, p < 0.0001; 9.1%) in nestling oil (Figure 7; Supplemental Data 2).
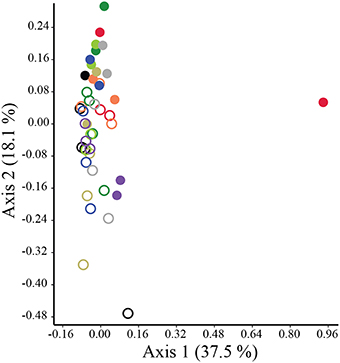
Figure 6. PCoA plot illustrating separation in the structure of the uropygial gland volatile profiles of adult and nestling juncos. Closed circles denote adults, while open circles denote nestlings. Color-coding reflects nest identity.
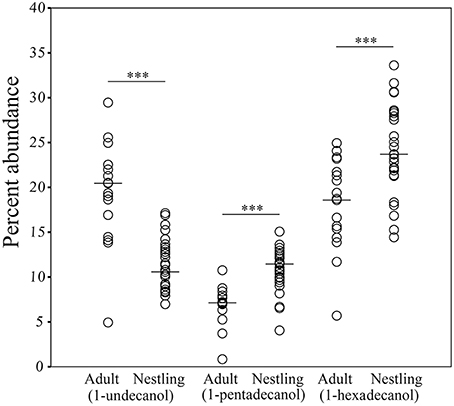
Figure 7. Comparisons of differences in the percent abundance of 1-undecanol, 1-pentadecanol, and 1-hexadecanol in the volatile profiles of adult and nestling uropygial gland oils (***p ≤ 0.001). Lines indicate median values.
The volatile profile of the preen oil of nestlings was generally more similar to that of their father than their mother (N = 23; Wilcoxon test; W = 223, p = 0.008; Figure 8). Furthermore, nestlings' volatile profiles were markedly more similar to those of adult males than females in the broader population (N = 27; W = 378, p < 0.0001; Figure 8). Nevertheless, the volatile profiles of the preen oil of nestlings were more similar to those of their mother than to those of other adult females in the population (N = 27; W = 299, p = 0.008). This was not the case with respect to their fathers—nestlings' volatile profiles were not more similar to that of their father than to those of other adult males in the population (N = 23; W = 175, p = 0.273). Whether an attendant father sired a nestling or not had no influence on their volatile profile similarity (Mann-Whitney test; Nsire = 12, Nnotsire = 11; U = 59, p = 0.695). Lastly, average volatile profile similarity among nestlings did not differ between nests containing only full siblings and those containing mixed paternity broods (Nfull = 4, Nhalf = 5; U = 9, p = 0.905).
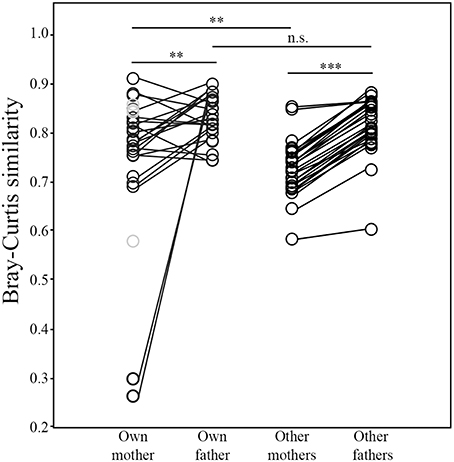
Figure 8. Comparisons of similarities in the structure of the uropygial gland volatile profiles between nestling juncos with those of their own mother and father, and those of other adult females and males in the population (**p ≤ 0.01; ***p ≤ 0.001). The samples colored gray denote the 4 nestlings that did not have an attendant father.
There was no indication that preen oil volatile profiles covaried with the composition (Mantel test; N = 44; R = 0.101, p = 0.155) or structure (R = −0.085, p = 0.793) of uropygial gland microbiota. This result held when considering adults (N = 17; composition: R = −0.145, p = 0.711; structure: R = −0.129, p = 0.602) and nestlings (N = 27; composition: R = 0.077, p = 0.207; structure: R = −0.002, p = 0.447) separately as well.
The objectives of this study were to assess the effects of sex, age class, and nest identity on cloacal and uropygial gland microbiota as well as uropygial volatile profiles, to elucidate the relative influence that variation in environment and genetic relatedness have in shaping juncos' microbiota and volatile profiles, and to test for covariation between these profiles among juncos. The phylum-level results of our characterization of junco microbiota were consistent with studies of avian microbiota in general: they were typified by Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes (Hird et al., 2014; Waite and Taylor, 2015). More specifically, our genus-level data agreed with a recent cultivation-based survey of junco ventral feather bacteria, which included Proteobacteria (Brevundimonas, Methylobacterium, Pseudomonas, Rhizobium, and Sphingomonas) and Firmicutes (Staphylococcus) that were widespread and abundant in our dataset (Supplemental Data 1) (Dille et al., 2016).
We did not find an effect of sex on cloacal or uropygial microbiota, or on uropygial volatile profiles, among either adults or nestlings. This result was surprising given that significant sex differences in junco volatile profiles have been previously reported (Whittaker et al., 2010) and replicated (Whittaker et al., 2013). However, a potential explanation is that the juncos in the previous studies had not yet reproduced that season, while the adult juncos in the current study were sampled on the day their offspring fledged from the nest. It is also possible that our smaller adult sample size (n = 17 here, compared to 34 and 26 in the previous studies) may have lacked the statistical power to detect sex differences in the volatile profiles. Juncos do not possess sex-specific volatile compounds but instead differ in the relative proportions of shared compounds (Whittaker et al., 2010). The lack of sex differences in microbiota could reflect microbe sharing between pair mates, as observed in captive zebra finches (Kulkarni and Heeb, 2007) and free-living barn swallows, Hirundo rustica (Kreisinger et al., 2015). Underlying influences of sex, and potentially genotype (discussed below), may be masked by the much stronger influence of environmental variation, including social behavior. For example, genetic and sex effects on the intestinal microbiota of broiler chickens were evident under common diet and husbandry conditions (Zhao et al., 2013), but those same effects may not have been evident in less controlled scenarios.
A central question in animal-microbial ecology is how the composition and structure of symbiotic microbial communities varies with age and how these communities are acquired and shaped (van Dongen et al., 2013). In this study, there were effects of age and nest identity on juncos' microbial and odor profiles. Although the cloacal and uropygial microbiota of nestlings and adults differed significantly, the differences were modest and not consistent across nests. Variation in the composition of nestling microbiota was similar to that of adults, and many OTUs were shared between parents and offspring—there were no OTUs that stood out as being nestling-specific. Nevertheless, nestling uropygial gland microbiota were more OTU-rich than those of adults, an observation consistent with the idea that nestlings, and young animals in general, host more transient and rare symbionts than adult animals (Palmer et al., 2007; van Dongen et al., 2013). Notably, while the richness and composition of cloacal and uropygial gland microbiota differed in adults, with uropygial gland bacterial communities appearing to be a subset of those found in the cloaca, the microbiota of the two glands did not differ in nestlings, suggesting that the cloaca and uropygial gland of nestlings do not yet afford dissimilar niches for symbiotic bacteria. One likely factor here is that the uropygial gland develops throughout the nestling period and does not begin secreting preen oil until about day 6 or later (personal observation; DJW).
Acute age-specific differences were evident in the volatile profiles of junco preen oil. Nestlings had significantly lower proportions of 1-undecanol than adults. This compound is typically higher in adult females than adult males (Whittaker et al., 2010), and increased in response to exogenous testosterone (Whittaker et al., 2011). Nestlings also had significantly higher proportions of 1-pentadecanol and 1-hexadecanol, neither of which have been found to relate to any aspect of adult phenotype, and could instead be important for kin recognition (Célérier et al., 2011; Bonadonna and Sanz-Aguilar, 2012), or stimulating parental care (Mas and Kolliker, 2008; Morales and Velando, 2013).
Nest-specific microbial and volatile odor profiles could reflect influences of shared environments and/or genetic relatedness, and teasing these influences apart is one of the most pressing lines of inquiry in host microbial ecology (Spor et al., 2011; Goodrich et al., 2014; Org et al., 2015). Dark-eyed juncos afford an opportunity to do so because they are socially monogamous yet exhibit appreciable levels of extra-pair paternity (Ketterson et al., 1998; Gerlach et al., 2012a,b; Atwell et al., 2014), and although they biparentally provision their offspring, only mothers brood the eggs and nestlings (Nolan et al., 2002). Therefore, there is variation in genetic relatedness among siblings as well as between nestlings and attendant fathers among nests, and there are differences in degree of physical contact with nestlings between junco mothers and fathers.
There were pronounced effects of nest identity on the cloacal and uropygial microbiota of adults and nestlings. With respect to both cloacal and uropygial microbiota, nestlings bore greater similarity to their mother than other adult females in the population. Among nestlings, cloacal microbiota richness values were positively correlated with those of their mothers but not their social fathers, and there was a tendency for the structure of nestlings' cloacal microbiota to be more similar to those of their mother than father. Both the composition and structure of nestlings' uropygial gland microbiota were markedly more similar to those of their mother than their father. Whether or not a nestling's attendant father was their genetic father did not affect the similarity of their microbiota, nor did variation in genetic relatedness among nestlings influence their microbiota similarities. Collectively, these results suggest that characteristics of nestlings' microbiota strongly reflect their immediate environments, especially the social partners with whom they interact with frequently, most notably their mother (Funkhouser and Bordenstein, 2013). A caveat that must be included here is that we were unable to account for possible direct maternal genetic effects on offspring microbiota as well. Notably, however, such effects, even if prominent, cannot explain the observed phenotypic similarities between mated pairs or between nestlings and their social fathers, both of which show nest-specific patterns.
The results of this study add to a growing body of literature indicating that environments, including social environments, can have pronounced effects on avian cloacal and uropygial gland microbiota (Waite and Taylor, 2015). For example, in free-living barn swallows mated pairs share similarities in their cloacal microbiota (Kreisinger et al., 2015), and experiments with wild kittiwakes, Rissa tridactyla, and captive zebra finches have demonstrated that symbiotic bacteria are readily transmitted between the cloacae of mated pairs (Kulkarni and Heeb, 2007). Furthermore, cross-fostering experiments with tits, Parus major and P. caeruleus, and hoopoes, Upupa epops, have shown that rearing environment can trump species identity and genetic relatedness in shaping the composition of cloacal and uropygial gland microbiota, respectively (Lucas and Heeb, 2005; Ruiz-Rodriguez et al., 2014). Natural observations of an avian brood parasite, the brown-headed cowbird, Molothrus ater, also indicated the importance of nest environment, as well as diet, in shaping developing avian microbiota (Hird et al., 2014). The influence of diet on mammalian intestinal microbiota is well-documented (Ley et al., 2008; Xu and Knight, 2015). Although junco parents provision their offspring, the diets of nestlings and adults are often different: adult diets consist of seeds supplemented with arthropods, while nestling diets are comprised primarily of arthropods, particularly larvae (Nolan et al., 2002). However, adult diets can vary seasonally based on food availability, with adults consuming more arthropods in the breeding season (Gashwiler and Ward, 1968). Therefore, although it is unlikely that variation in diet alone could explain the nest-specific variation in cloacal and uropygial gland microbiota found in this study, it is certainly reasonable to assume that it explains some.
There was also a nest-specific effect on the volatile odor profiles of juncos. However, in contrast to observed patterns in the microbiota, nestlings' volatile profiles were more similar to those of their father than mother, and were also more similar to those adult males than adult females. As noted above, this is likely due to adult females having higher proportions of 1-undecanol in their volatile profiles than either adult males or nestlings (Whittaker et al., 2010). Nevertheless, nestlings' volatile profiles were more similar to those of their mother than other adult females. As with uropygial gland microbiota, whether or not a nestling's attendant father was their genetic father did not affect their preen oil volatile profile similarity, and genetic relatedness among nestlings also had no evident influence of volatile profile similarity. Clearly, as with the microbiota, nest-specific environments were influential in shaping nestlings' volatile profiles. However, it is important to note that simply because an underlying influence of variation in genetic relatedness is not evident against the pronounced influence of environmental variation does not mean that variation in genotype is not also influencing the microbial and odor phenotypes of junco nestlings—it simply means its influence is of much less magnitude.
It has been suggested that contributions to animal chemical communication may be an underappreciated benefit of symbiotic microbes (Albone, 1984; Archie and Theis, 2011; Ezenwa and Williams, 2014). Although this hypothesis has been increasingly investigated in scent-marking mammals (Zomer et al., 2009; Sin et al., 2012; Theis et al., 2013; Leclaire et al., 2014), it has received less attention with respect to chemical signaling in birds (Whittaker and Theis, 2016). If symbiotic microbes contribute to their hosts' odor profiles, then host organs emitting the odors should be populated by communities of odor-producing microbes, and the microbial and volatile odor profiles of the organs should covary. We previously illustrated that the uropygial glands of adult dark-eyed juncos support bacterial communities that include genera (e.g., Burkholderia, Pseudomonas) known to produce many of the volatile compounds in preen oil (Whittaker and Theis, 2016), including two compounds, 2-tridecanone and 2-pentadecanone, with sex-specific relationships to reproductive success in this species (Whittaker et al., 2013). However, here we have additionally considered the cloacal microbiota, and found that the bacterial communities populating the uropygial gland are a subset of the communities inhabiting the cloaca. For example, Burkholderia and Pseudomonas are common to both the cloaca and the uropygial gland—they are not specific to preen oil. This finding does not preclude that these and other symbiotic microbes contribute to the volatile profiles of both glands, although this potentiality has not been addressed.
Our results also indicated that the microbial and volatile odor profiles of junco uropygial glands did not covary—patterns in the composition and structure of uropygial microbial profiles were not consistently similar to patterns in the structure of uropygial volatile profiles. It is therefore logical to deduce that changes in the bacterial communities of the uropygial gland do not result in changes to community-level metabolism that consequently shift glandular odor profile (Archie and Theis, 2011; Ezenwa and Williams, 2014). However, there are other potential explanations. First, our sample sizes may have been too low to detect covariation. Second, there may be ample redundancy in the production of volatile odorants among discrete bacterial types in these communities, thereby obscuring covariation. 16S rRNA gene surveys afford a phylogenetic snapshot of environmental bacterial communities, but information about their metabolic potential must be inferred (Carlos et al., 2012; Langille et al., 2013). Finally, the relationships between uropygial gland bacterial types and volatile odorants may be confounded by the fact that, unlike animal glands devoted exclusively to chemical signaling, avian uropygial glands have multiple functions, including parasite and pathogen defense, feather protection, and thermoregulation—symbiotic bacteria have been broadly hypothesized to contribute to each (Jacob and Ziswiler, 1982; Moyer et al., 2003; Shawkey et al., 2003; Giraudeau et al., 2010; Martín-Vivaldi et al., 2010; Soler et al., 2010). Using cultivation, metabolomic, and metagenomic approaches as a complement to the 16S rRNA gene surveys employed here would afford greater detail of the metabolic potential of these symbiotic communities (Waite and Taylor, 2015), permitting a more nuanced evaluation of the hypothesis that uropygial gland microbiota contribute to junco chemical signaling.
This study used concurrent molecular microbial surveys and chemical analyses of the glands of a natural population of dark-eyed juncos to tease apart the relative influence of environmental and genetic variation in shaping juncos' microbial and volatile odor profiles. Our data suggest that social environment is a primary driver of individual microbiota and odor profile structure among juncos: birds who are in close proximity and in frequent contact develop similar profiles. This includes mated pairs and their nestlings. Although significantly influential on each, environmental variation (e.g., exposure to microbiota of dietary items, nest materials, siblings, and parents) most strongly affected the cloacal and uropygial gland microbiota; preen oil volatile profiles seemed less affected. Although underlying variation in host genotype may be influencing the composition and structure of juncos' microbial and odor profiles, the influence is not evident against the background influence of environmental variation. We did not find evidence that variation in microbial profiles translates to variation in odor profiles, and therefore a direct contribution of symbiotic microbes to avian chemical communication remains to be demonstrated. However, definitive evaluation of this hypothesis will require a more comprehensive investigation that affords a detailed, non-inferential analysis of the metabolic capabilities of the bacterial communities associated with the uropygial gland of juncos.
DW co-conceived the study, conducted the fieldwork, conducted statistical analyses of volatile compounds, and co-wrote the manuscript. NG conducted the paternity analyses and assisted with the fieldwork. SS assisted with the fieldwork and GC-MS analyses. KC assisted with the GC-MS analyses. AW assisted with the sequence analyses and figure creation. HS and MN led the GC-MS analyses. EK oversaw the fieldwork and the long-term junco project. KT co-conceived the study, led the sequence analyses, and co-wrote the manuscript. All authors read, edited, and approved the manuscript.
This material is based in part upon work supported by the National Science Foundation under Cooperative Agreement No. DBI-0939454. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. Fieldwork was supported by a Mountain Lake Biological Station Early Career Fellowship to DW.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the University of Virginia, Mountain Lake Biological Station, Mountain Lake Hotel, and the Dolinger family for allowing research on their grounds. We thank Abby Kimmitt, Dustin Reichard, Sarah Wanamaker, and Joseph Welklin for assistance in the field, and John Dover and Arvind Venkataraman for assistance with DNA extraction and sequence processing, respectively.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fevo.2016.00090
Anderson, M. J. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253. doi: 10.1111/j.1541-0420.2005.00440.x
Archie, E. A., and Theis, K. R. (2011). Animal behaviour meets microbial ecology. Anim. Behav. 82, 425–436. doi: 10.1016/j.anbehav.2011.05.029
Atwell, J. W., Cardoso, G. C., Whittaker, D. J., Price, T. D., and Ketterson, E. D. (2014). Hormonal, behavioral and life-history traits exhibit correlated shifts in relation to population establishment in a novel environment. Am. Nat. 184, E147–E160. doi: 10.1086/678398
Ball, G. F., and Ketterson, E. D. (2007). Sex differences in the response to environmental cues regulating seasonal reproduction in birds. Philos. Trans. R. Soc. Lond. B 363, 231–246. doi: 10.1098/rstb.2007.2137
Bergeon Burns, C., Rosvall, K. A., and Ketterson, E. D. (2013). Neural steroid sensitivity and aggression: comparing individuals of two songbird subspecies. J. Evol. Biol. 26, 820–831. doi: 10.1111/jeb.12094
Bonadonna, F., and Sanz-Aguilar, A. (2012). Kin recognition and inbreeding avoidance in wild birds: the first evidence for individual kin-related odour recognition. Anim. Behav. 84, 509–513. doi: 10.1016/j.anbehav.2012.06.014
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Carlos, N., Tang, Y. W., and Pei, Z. H. (2012). Pearls and pitfalls of genomics-based microbiome analysis. Emerg. Microbes Infect. 1:e45. doi: 10.1038/emi.2012.41
Célérier, A., Bon, C., Malapert, A., Palmas, P., and Bonadonna, F. (2011). Chemical kin label in seabirds. Biol. Lett. 7, 807–810. doi: 10.1098/rsbl.2011.0340
Chao, A., and Shen, T. J. (2003). Nonparametric estimation of Shannon's index of diversity when there are unseen species in sample. Environ. Ecol. Stat. 10, 429–443. doi: 10.1023/A:1026096204727
Claesson, M. J., O'Sullivan, O., Wang, Q., Nikkilä, J., Marchesi, J. R., Smidt, H., et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 4:e6669. doi: 10.1371/journal.pone.0006669
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Davis, T. S., Crippen, T. L., Hofstetter, R. W., and Tomberlin, J. K. (2013). Microbial volatile emissions as insect semiochemicals. J. Chem. Ecol. 39, 840–859. doi: 10.1007/s10886-013-0306-z
Dille, J. W., Rogers, C. M., and Schneegurt, M. A. (2016). Isolation and characterization of bacteria from the feathers of wild Dark-eyed Juncos (Junco hyemalis). Auk 133, 155–167. doi: 10.1642/AUK-15-126.1
Ezenwa, V. O., Gerardo, N. M., Inouye, D. W., Medina, M., and Xavier, J. B. (2012). Animal behavior and the microbiome. Science 338, 198–199. doi: 10.1126/science.1227412
Ezenwa, V. O., and Williams, A. E. (2014). Microbes and animal olfactory communication: where do we go from here? Bioessays 36, 847–854. doi: 10.1002/bies.201400016
Fraune, S., and Bosch, T. C. G. (2007). Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. U.S.A. 104, 13146–13151. doi: 10.1073/pnas.0703375104
Fraune, S., and Bosch, T. C. G. (2010). Why bacteria matter in animal development and evolution. Bioessays 32, 571–580. doi: 10.1002/bies.200900192
Funkhouser, L. J., and Bordenstein, S. R. (2013). Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 11:e1001631. doi: 10.1371/journal.pbio.1001631
Gashwiler, J. S., and Ward, A. L. (1968). Oregon junco foods in coniferous forests. Murrelet 49, 29–36. doi: 10.2307/3534086
Gerlach, N. M., McGlothlin, J. W., Parker, P. G., and Ketterson, E. D. (2012a). Promiscuous mating produces offspring with higher lifetime fitness. Proc. R. Soc. B 279, 860–866. doi: 10.1098/rspb.2011.1547
Gerlach, N. M., McGlothlin, J. W., Parker, P. G., and Ketterson, E. D. (2012b). Reinterpreting Bateman gradients: multiple mating and selection in both sexes of a songbird species. Behav. Ecol. 23, 1078–1088. doi: 10.1093/beheco/ars077
Gilbert, S. F., Sapp, J., and Tauber, A. I. (2012). A symbiotic view of life: we have never been individuals. Q. Rev. Biol. 87, 325–341. doi: 10.1086/668166
Giraudeau, M., Czirjak, G. A., Duval, C., Bretagnolle, V., Eraud, C., McGraw, K. J., et al. (2010). Effect of restricted preen-gland access on maternal self maintenance and reproductive investment in mallards. PLoS ONE 5:e13555. doi: 10.1371/journal.pone.0013555
Goodrich, J. K., Waters, J. L., Poole, A. C., Sutter, J. L., Koren, O., Blekhman, R., et al. (2014). Human genetics shape the gut microbiome. Cell 159, 789–799. doi: 10.1016/j.cell.2014.09.053
Gotelli, N. J., and Ellison, A. M. (2004). A Primer of Ecological Statistics. Sunderland: Sinauer Associates, Inc.
Griffiths, R., Double, M. C., Orr, K., and Dawson, R. J. G. (1998). A DNA test to sex most birds. Mol. Ecol. 7, 1071–1075. doi: 10.1046/j.1365-294x.1998.00389.x
Hacquard, S., Garrido-Oter, R., González, A., Spaepen, S., Ackermann, G., Lebeis, S., et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616. doi: 10.1016/j.chom.2015.04.009
Hammer, O. (2015). PAST: PAleontological STatistics reference manual, Version 3.06. Oslo: Natural History Museum.
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST: PAleontological STatistics software package for education and data analysis. Palaeontol. Electron 4, 1–9.
Hird, S. M., Carstens, B. C., Cardiff, S. W., Dittmann, D. L., and Brumfield, R. T. (2014). Sampling locality is more detectable than taxonomy or ecology in the gut microbiota of the brood-parasitic Brown-headed Cowbird (Molothrus ater). Peer J 2:e321. doi: 10.7717/peerj.321
Hooper, L. V., Littman, D. R., and Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. doi: 10.1126/science.1223490
Jacob, J. P., and Ziswiler, V. (1982). “The uropygial gland,” in Avian Biology, eds D. S. Farner, J. R. King, and K. C. Parkes (New York, NY: Academic Press), 199–324.
Johansson, B. G., and Jones, T. M. (2007). The role of chemical communication in mate choice. Biol Rev 82, 265–289. doi: 10.1111/j.1469-185X.2007.00009.x
Kalinowski, S., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Ketterson, E. D., and Nolan, V. Jr. (1992). Hormones and life histories: an integrative approach. Am. Nat. 140, S33–S62. doi: 10.1086/285396
Ketterson, E. D., Nolan, V. Jr., Casto, J. M., Buerkle, C. A., Clotfelter, E. D., Grindstaff, J. L., et al. (2001). “Testosterone, phenotype, and fitness: a research program in evolutionary behavioral endocrinology,” in Avian Endocrinology, eds A. Dawson and M. Chaturvedi (New Delhi: Narosa Publishing House), 19–40.
Ketterson, E. D., Parker, P. G., Raouf, S. A., Nolan, V. Jr., Ziegenfus, C., and Chandler, C. R. (1998). “Relative importance of extra-pair fertilizations to male and female reproductive success in dark-eyed juncos (Junco hyemalis),” in Avian Reproductive Tactics: Female and Male Perspectives, eds P.G. Parker and N. T. Burley (Lawrence, KS: American Ornithologists' Union), 81–101.
Klomp, J. E., Murphy, M. T., Bartos Smith, S., McKay, J. E., Ferrera, I., and Reysenbach, A., L. (2008). Cloacal microbial communities of female spotted towhees Pipilo maculatus: microgeographic variation and individual sources of variability. J. Avian Biol. 39, 530–538. doi: 10.1111/j.0908-8857.2008.04333.x
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Kreisinger, J., Cizkova, D., Kropackova, L., and Albrecht, T. (2015). Cloacal microbiome structure in a long-distance migratory bird assessed using deep 16sRNA pyrosequencing. PLoS ONE 10:e0137401. doi: 10.1371/journal.pone.0137401
Kulkarni, S., and Heeb, P. (2007). Social and sexual behaviours aid transmission of bacteria in birds. Behav. Proc. 74, 88–92. doi: 10.1016/j.beproc.2006.10.005
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814. doi: 10.1038/nbt.2676
Leclaire, S., Nielsen, J. F., and Drea, C. M. (2014). Bacterial communities in meerkat anal scent secretions vary with host sex, age, and group membership. Behav. Ecol. 25, 996–1004. doi: 10.1093/beheco/aru074
Lee, S., Sung, J., Lee, J., and Ko, G. (2011). Comparison of the gut microbiotas of healthy adult twins living in South Korea and the United States. Appl. Environ. Microb. 77, 7433–7437. doi: 10.1128/AEM.05490-11
Lee, W. J., and Hase, K. (2014). Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 10, 416–424. doi: 10.1038/nchembio.1535
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R., and Gordon, J. I. (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. doi: 10.1038/nrmicro1978
Longmire, J. L., Gee, G. F., Handenkipf, O. L., and Mark, G. A. (1992). Establishing paternity in whooping cranes, Grus americana, by DNA analysis. Auk 109, 522–529.
Lucas, F. S., and Heeb, P. (2005). Environmental factors shape cloacal bacterial assemblages in great tit Parus major and blue tit P. caeruleus nestlings. J. Avian Biol. 36, 510–516. doi: 10.1111/j.0908-8857.2005.03479.x
Martín-Vivaldi, M., Peña, A., Peralta-Sánchez, J. M., Sánchez, L., Ananou, S., Ruiz-Rodríguez, M., et al. (2010). Antimicrobial chemicals in hoopoe preen secretions are produced by symbiotic bacteria. Proc. R. Soc. B 277, 123–130.
Martín-Vivaldi, M., Ruiz-Rodríguez, M., Soler, J. J., Peralta-Sánchez, J. M., Méndez, M., Valdivia, E., et al. (2009). Seasonal, sexual and developmental differences in hoopoe Upupa epops preen gland morphology and secretions: evidence for a role of bacteria. J. Avian Biol. 40, 191–205. doi: 10.1111/j.1600-048X.2009.04393.x
Mas, F., and Kolliker, M. (2008). Maternal care and offspring begging in social insects: chemical signalling, hormonal regulation and evolution. Anim. Behav. 76, 1121–1131. doi: 10.1016/j.anbehav.2008.06.011
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
McFall-Ngai, M. J. (2002). Unseen forces: the influence of bacteria on animal development. Dev. Biol. 242, 1–14. doi: 10.1006/dbio.2001.0522
Morales, J., and Velando, A. (2013). Signals in family conflicts. Anim. Behav. 86, 11–16. doi: 10.1016/j.anbehav.2013.04.001
Moyer, B. R., Rock, A. N., and Clayton, D. H. (2003). Experimental test of the importance of preen oil in rock doves (Columba livia). Auk 120, 490–496. doi: 10.1642/0004-8038(2003)120[0490:ETOTIO]2.0.CO;2
Nolan, V. Jr., Ketterson, E. D., Cristol, D. A., Rogers, C. M., Clotfelter, E. D., Titus, R., et al. (2002). Dark-Eyed Junco (Junco hyemalis), Vol. 716. Philadelphia, PA: The Birds of North America, Inc.
Org, E., Parks, B. W., Joo, J. W. J., Emert, B., Schwartzman, W., Kang, E. Y., et al. (2015). Genetic and environmental control of host-gut microbiota interactions. Genome Res. 25, 1558–1569. doi: 10.1101/gr.194118.115
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A., and Brown, P. O. (2007). Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. doi: 10.1371/journal.pbio.0050177
Pavlidis, P., and Noble, W. S. (2003). Matrix2png: a utility for visualizing matrix data. Bioinformatics 19, 295–296. doi: 10.1093/bioinformatics/19.2.295
Penn, D., and Potts, W. K. (1998). Chemical signals and parasite-mediated sexual selection. Trends Ecol. Evol. (Amst). 13, 391–396. doi: 10.1016/S0169-5347(98)01473-6
Peterson, M. P., Whittaker, D. J., Ambreth, S., Sureschandra, S., Buechlein, A., Podicheti, R., et al. (2012). De novo transcriptome sequencing in a songbird, the dark-eyed junco (Junco hyemalis): genomic tools for an ecological model system. BMC Genomics 13:305. doi: 10.1186/1471-2164-13-305
Rowan, W. (1925). Relation of light to bird migration and developmental changes. Nature 115, 494–495. doi: 10.1038/115494b0
Ruiz-de-Castañeda, R., Vela, A. I., Lobato, E., Briones, V., and Moreno, J. (2011a). Bacterial loads on eggshells of the pied flycatcher: environmental and maternal factors. Condor 113, 200–208.
Ruiz-de-Castañeda, R., Vela, A. I., Lobato, E., Briones, V., and Moreno, J. (2011b). Prevalence of potentially pathogenic culturable bacteria on eggshells and in cloacae of female Pied Flycatchers in a temperate habitat in central Spain. J. Field Ornithol. 82, 215–224.
Ruiz-Rodriguez, M., Soler, J. J., Martín-Vivaldi, M., Martín-Platero, A. M., Méndez, M., Peralta-Sánchez, J. M., et al. (2014). Environmental factors shape the community of symbionts in the hoopoe uropygial gland more than genetic factors. Appl. Environ. Microb. 80, 6714–6723. doi: 10.1128/AEM.02242-14
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., Cookson, W. O., Moffatt, M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:87. doi: 10.1186/s12915-014-0087-z
Sambrook, E., Fritsch, E. P., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shaw, C. L., Rutter, J. E., Austin, A. L., Garvin, M. C., and Whelan, R. J. (2011). Volatile and semivolatile compounds in gray catbird uropygial secretions vary with age and between breeding and wintering grounds. J. Chem. Ecol. 37, 329–339. doi: 10.1007/s10886-011-9931-6
Shawkey, M. D., Pillai, S. R., and Hill, G. E. (2003). Chemical warfare? Effects of uropygial oil on feather-degrading bacteria. J. Avian Biol. 34, 345–349. doi: 10.1111/j.0908-8857.2003.03193.x
Sin, Y. W., Buesching, C. D., Burke, T., and Macdonald, D. W. (2012). Molecular characterization of the microbial communities in the subcaudal gland secretion of the European badger (Meles meles). FEMS Microbiol. Ecol. 81, 648–659. doi: 10.1111/j.1574-6941.2012.01396.x
Sloan, W. T., Lunn, M., Woodcock, S., Head, I. M., Nee, S., and Curtis, T. P. (2006). Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 8, 732–740. doi: 10.1111/j.1462-2920.2005.00956.x
Soini, H. A., Schrock, S. E., Bruce, K. E., Wiesler, D., Ketterson, E. D., and Novotny, M. V. (2007). Seasonal variation in volatile compound profiles of preen gland secretions of the dark-eyed junco (Junco hyemalis). J. Chem. Ecol. 33, 183–198. doi: 10.1007/s10886-006-9210-0
Soini, H. A., Whittaker, D. J., Wiesler, D., Ketterson, E. D., and Novotny, M. (2013). Chemosignaling diversity in songbirds: chromatographic profiling of preen oil volatiles in different species. J. Chromatogr. A 1317, 186–192. doi: 10.1016/j.chroma.2013.08.006
Soler, J. J., Martín-Vivaldi, M., Peralta-Sánchez, J. M., and Ruiz-Rodríguez, M. (2010). Antibiotic-producing bacteria as a possible defence of birds against pathogenic microorganisms. Open Ornithol. J. 3, 93–100. doi: 10.2174/1874453201003010093
Spor, A., Koren, O., and Ley, R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279–290. doi: 10.1038/nrmicro2540
Theis, K. R., Venkataraman, A., Dycus, J. A., Koonter, K. D., Schmitt-Matzen, E. N., Wagner, A. P., et al. (2013). Symbiotic bacteria appear to mediate hyena social odors. Proc. Natl. Acad. Sci. U.S.A. 110, 19832–19837. doi: 10.1073/pnas.1306477110
Tremaroli, V., and Backhed, F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. doi: 10.1038/nature11552
Tung, J., Barreiro, L. B., Burns, M. B., Grenier, J.-C., Lynch, J., Grieneisen, L. E., et al. (2015). Social networks predict gut microbiome composition in wild baboons. eLife 4:e05224. doi: 10.7554/elife.05224
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. doi: 10.1038/nature07540
van Dongen, W. F. D., White, J., Brandl, H. B., Moodley, Y., Merkling, T., Leclaire, S., et al. (2013). Age-related differences in the cloacal microbiota of a wild bird species. BMC Ecol. 13:11. doi: 10.1186/1472-6785-13-11
Waite, D. W., and Taylor, M. (2015). Exploring the avian gut microbiota: current trends and future directions. Front. Microbiol. 6:673. doi: 10.3389/fmicb.2015.00673
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Westneat, D. F., and Rambo, T. B. (2000). Copulation exposes female Red-winged Blackbirds to bacteria in male semen. J. Avian Biol. 31, 1–7. doi: 10.1034/j.1600-048x.2000.310101.x
White, J., Mirleau, P., Danchin, E., Mulard, H., Hatch, S. A., Heeb, P., et al. (2010). Sexually transmitted bacteria affect female cloacal assemblages in a wild bird. Ecol. Lett. 13, 1515–1524. doi: 10.1111/j.1461-0248.2010.01542.x
Whittaker, D. J., Gerlach, N. M., Soini, H. A., and Novotny, M. V. (2013). Bird odour predicts reproductive success. Anim. Behav. 86, 697–703. doi: 10.1016/j.anbehav.2013.07.025
Whittaker, D. J., Soini, H. A., Atwell, J. W., Hollars, C., Novotny, M. V., and Ketterson, E. D. (2010). Songbird chemosignals: volatile compounds in preen gland secretions vary among individuals, sexes, and populations. Behav. Ecol. 21, 608–614. doi: 10.1093/beheco/arq033
Whittaker, D. J., Soini, H. A., Gerlach, N. M., Posto, A. L., Novotny, M. V., and Ketterson, E. D. (2011). Role of testosterone in stimulating seasonal changes in a potential avian chemosignal. J. Chem. Ecol. 37, 1349–1357. doi: 10.1007/s10886-011-0050-1
Whittaker, D. J., and Theis, K. R. (2016). “Bacterial communities associated with junco preen glands: preliminary ramifications for chemical signaling,” in Chemical Signals in Vertebrates 13, eds B. A. Schulte, T. E. Goodwin, and M. H. Ferkin (Cham: Springer International Publishing), 105–117. doi: 10.1007/978-3-319-22026-0_8
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y.-Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Xu, Z. J., and Knight, R. (2015). Dietary effects on human gut microbiome diversity. Brit. J. Nutr. 113, S1–S5. doi: 10.1017/S0007114514004127
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Zhao, L., Wang, G., Siegel, P., He, C., Wang, H., Zhao, W., et al. (2013). Quantitative genetic background of the host influences gut microbiomes in chickens. Sci. Rep. 3:1163. doi: 10.1038/srep01163
Zomer, S., Dixon, S. J., Xu, Y., Jensen, S. P., Wang, H. T., Lanyon, C. V., et al. (2009). Consensus multivariate methods in gas chromatography mass spectrometry and denaturing gradient gel electrophoresis: MHC-congenic and other strains of mice can be classified according to the profiles of volatiles and microflora in their scent-marks. Analyst 134, 114–123. doi: 10.1039/B807061J
Keywords: preen oil, 16S rRNA gene sequencing, microbiome, uropygial gland, chemical communication, symbiotic bacteria
Citation: Whittaker DJ, Gerlach NM, Slowinski SP, Corcoran KP, Winters AD, Soini HA, Novotny MV, Ketterson ED and Theis KR (2016) Social Environment Has a Primary Influence on the Microbial and Odor Profiles of a Chemically Signaling Songbird. Front. Ecol. Evol. 4:90. doi: 10.3389/fevo.2016.00090
Received: 30 April 2016; Accepted: 15 July 2016;
Published: 03 August 2016.
Edited by:
Luisa Amo, Spanish National Research Council, SpainReviewed by:
Matthew R. E. Symonds, Deakin University, AustraliaCopyright © 2016 Whittaker, Gerlach, Slowinski, Corcoran, Winters, Soini, Novotny, Ketterson and Theis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kevin R. Theis, a3RoZWlzQG1lZC53YXluZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.