
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 06 October 2015
Sec. Chemical Ecology
Volume 3 - 2015 | https://doi.org/10.3389/fevo.2015.00115
This article is part of the Research Topic Functional Characterization of Insect Chemoreceptors: Receptivity Range and Expression View all 14 articles
Insect odorant receptors (ORs) are key sensors of environmental odors and members of the lepidopteran pheromone receptor subfamily are thought to play important roles in mate finding by recognizing sex pheromones. Much research has been done to identify putative pheromone receptors in lepidopteran males, but little attention has been given to female counterparts. In this study, degenerate oligonucleotide primers designed against a conserved amino acid region in the C-terminus of lepidopteran pheromone receptors were used in 3′ RACE reactions to identify candidate pheromone receptors expressed in the antennae of female navel orangeworm. Two near full-length transcripts of 1469 and 1302 nt encoding the complete open reading frames for proteins of 446 and 425 amino acids, respectively, were identified. Based on BLAST homology and phylogenetic analyses, the putative proteins encoded by these transcripts are members of the lepidopteran pheromone receptor subfamily. Characterization of these transcripts indicates that they are alternatively spliced products of a single gene. Tissue expression studies indicate that the transcripts are female-biased with detection mainly in female antennae. To the best of our knowledge, these transcripts represent the first detection of alternatively spliced female-biased members of the lepidopteran pheromone receptor subfamily.
The insect chemosensory system is critical for the detection of chemical cues in the environment, processing these signals in the central nervous system and eliciting behavioral responses to these stimuli (Smith, 2007). Within the chemosensory system, olfaction plays an important role in mate and host plant seeking behaviors (Depetris-Chauvin et al., 2015). Two major molecular components of the olfactory system are odorant binding proteins, which serve as a link between the external environment to shuttle hydrophobic volatile compounds through the sensillar lymph, and odorant receptors (ORs), which serve as key detectors of olfactants present in the environment and when bound by ligand, play a role in transducing signals along the olfactory neuron (Leal, 2013). ORs are part of a diverse family of seven transmembrane domain proteins, which are located on the dendritic membranes of olfactory neurons. The first insect ORs were identified from the Drosophila genome (Clyne et al., 1999; Gao and Chess, 1999; Vosshall et al., 1999) and their structure is quite different from their mammalian counterparts (Bargmann, 2006; Benton, 2006). Because ORs show poor sequence homologies both within and between species, they have been identified mainly through bioinformatic algorithms to scan completed genomes or transcriptome sequences.
For the Lepidoptera, the first ORs were identified from Bombyx mori and Heliothis virescens through analysis of sequenced genomes (Krieger et al., 2004, 2005; Sakurai et al., 2004; Nakagawa et al., 2005). Further mining of the B. mori genome led to the identification of 48 ORs, and a subfamily, grouped by relatedness, contained putative pheromone receptors from both B.mori and H.virescens (Wanner et al., 2007). Subsequently, members of this subfamily have been referred to as male-biased sex pheromone receptor clade (Jordan et al., 2009), sex pheromone receptor subfamily (Miura et al., 2009, 2010; Wanner et al., 2010), pheromone receptors (Patch et al., 2009; Groe-Wilde et al., 2010), and male-specific OR subfamily (Mitsuno et al., 2008). To gain a better understanding of how lepidopteran sex pheromones work at the molecular level, research to identify and characterize pheromone receptors in males has been the topic of much research (Krieger et al., 2004, 2005; Sakurai et al., 2004; Nakagawa et al., 2005; Groe-Wilde et al., 2007, 2010; Wanner et al., 2007, 2010; Mitsuno et al., 2008; Jordan et al., 2009; Miura et al., 2009, 2010; Patch et al., 2009; Xu et al., 2012). However, some members of the pheromone receptor subfamily are also activated by plant kairomones (Jordan et al., 2009; Bengtsson et al., 2014), indicating that not all of these ORs respond to pheromones. Because of the importance of male sex pheromone receptors in enhancing mating disruption in lepidopteran pest control programs, much research has gone into their discovery and characterization, however, female attractants and their ORs remain an untapped resource for future lepidopteran control efforts.
The navel orangeworm, Amyelois transitella Walker (Lepidoptera:Pyralidae), is the major insect pest of almonds and pistachios in California, as well as a pest of other crops including walnuts and figures. Control of navel orangeworm is mainly achieved with sanitation techniques and by using pyrethroids and insect growth regulators, but other control methods using sex pheromones for mating disruption, and female attractants or oviposition attractants are sorely needed (Higbee et al., 2014). Recently, two pheromone subfamily receptors from navel orangeworm males have been identified and characterized (Xu et al., 2012). In this current study, we describe the identification of two transcripts expressed in antennae of navel orangeworm females that encode ORs with high similarity to those belonging to the lepidopteran pheromone receptor subfamily. A degenerate primer approach was used to identify the expressed transcripts (Garczynski et al., 2012) and the cDNAs encoding the full open reading frames were obtained using 5′RACE. Comparison of these sequences indicated that they may be produced by alternative splicing. Alternative splicing is a mechanism by which multiple mRNAs can be produced from a single gene, thereby increasing an organism's proteome (Nilsen and Graveley, 2010; Kornblihtt et al., 2013). To characterize the navel orangeworm transcripts, we included comparison of the cDNA transcript sequences to a partial cloned gene sequence and determined their tissue expression profiles. Implications of these results are discussed.
Male and female pupae were obtained from Bradley Higbee, Research Entomologist (Paramount Farming Co., Bakersfield, CA) and antennae were dissected from 50 adults within 24 h of emergence and placed directly into 1.5 ml microfuge tubes containing 100 μl RNAlater® (Ambion, Austin, TX). Antennae in RNAlater® were stored for up to 4 weeks at 4°C until RNA was extracted (see below). Insects for expression studies were obtained from a lab colony maintained at University of California, Davis. For expression studies, antennae, heads, thoraces, and abdomens were dissected from newly eclosed adults (0–24 h old), placed into tubes containing 100 μl RNAlater® and stored at 4°C until the RNA was extracted.
Total RNA was extracted from dissected antennae, heads, thoraces and abdomens using the RNeasy® Plus Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol for purification of total RNA from animal tissues. SuperScript® III First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA) was used to generate cDNA from total RNA extracted from male and female antennae using the CDSIII/3′PCR primer and SMART II™ A oligonucleotide (Clontech, Mountain View, CA). Double-strand (ds) cDNAs and cDNA amplification were performed using components and procedures of the SMART™ PCR cDNA Synthesis Kit (Clontech, Mountain View, CA). Amplified cDNAs were purified using the High Pure PCR Product Purification Kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. Genomic DNA was prepared from adult male and female abdomens using the DNeasy® Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol for purification of total DNA from animal tissue. Total DNA was quantitated using the Quant-iT™ PicoGreen® dsDNA assay kit (Invitrogen, Carlsbad, CA) according to manufacturer's instructions.
Identification of putative members of the pheromone receptor family was achieved by amplifying cDNA using degenerate primers designed against a conserved amino acid region in the C-terminus of lepidopteran pheromone receptors and a 3′Rapid amplification of cDNA ends (RACE) protocol as previously described (Garczynski et al., 2012). PCR reactions were set up using amplified antennal cDNA as template and the forward primer PR0 (5′-GTNCCNTGGGARTAYATGGAYAC-3′; Garczynski et al., 2012) and CDSIII/3′PCR as the reverse primer using the following conditions: initial denaturation for 5 min at 94°C, then amplification for 30 s at 94°C, 30 s at 62°C, 3 min at 72°C for 40 cycles, followed by a final 5 min 72°C incubation. PCR products were separated on 1.2% agarose gels containing ethidium bromide and visualized on a UV light box. Excised bands were extracted and purified using GenElute minus EtBr spin columns (Sigma, St. Louis, MO), and the purified PCR products were cloned using the TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA) with TOP 10 Escherichia coli chemically competent cells. Plasmid DNA was extracted from picked colonies using the QIAprep spin mini prep kit (Qiagen, Valencia, CA). The cDNA clones were sequenced at MC Laboratories (MCLab, San Francisco, CA).
To amplify the 5′ end of the female ORs identified above, a 5′RACE protocol was performed using the SMART™ RACE cDNA amplification kit (Clontech, Mountain View, CA) according to manufacturer supplied protocols. For 5′RACE, first strand cDNA was prepared as above except that total RNA was converted to cDNA using the 5′-RACE CDS Primer A and SMART II™ A oligonucleotide (Clontech, Mountain View, CA). From the cDNA sequences obtained above, reverse primers were designed for use in 5′RACE reactions using the Primer3 (v 0.4.0; http://frodo.wi.mit.edu/primer3/). Amplification was performed using the 10X Universal Primer A Mix as forward primer and sequence specific primers (AtraOR4 5′RACE Rev1: 5′- GAAAAGTGAAATACGACGGCGATG-3′ and AtraOR4 5′RACE Rev2: 5′- CCATATCCGTAACTCCGAGAGCC-3′) and the following conditions; initial denaturation for 3 min at 94°C, then amplification for 30 s at 94°C, 2 min at 72°C for 40 cycles, followed by a final 5 min 72°C incubation. PCR products were separated on 1% agarose gels and excised, TA cloned and sequenced as above. To obtain the full length sequences of the open reading frames for each receptor, PCR reactions were done with sequence specific primers (AtraOR4 ORF Fwd: 5′-GTATTTAAAATGGATATCAGTGCACAAAATAGAGC -3′ and AtraOR4 ORF Rev: 5′- CTACTTGCATAAATTAATCAATTTTTCGTAGAAAAG TG-3′) and the reaction conditions above. PCR products were cloned and sequenced as above, and the consensus sequence of three clones was generated in Geneious (version 7.1.9 created by Biomatters, available from http://www.geneious.com/). To determine identity of encoded proteins, tblastn on the National Center for Biotechnology Information website was used (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Phylogenetic and evolutionary analyses were conducted using MEGA5 software (Tamura et al., 2011) to classify the AtraOR4 pheromone subfamily receptors. Amino acid sequences for ORs identified from B. mori (Wanner et al., 2007) and H. virescens (Krieger et al., 2002, 2004) along with odorant receptor members of the pheromone receptor subfamily from Ostrinia nubialis (Wanner et al., 2010), Manduca sexta (Patch et al., 2009), Cydia pomonella (Bengtsson et al., 2012; Garczynski, unpublished), Plutella xylostella (Mitsuno et al., 2008), Mythimna separata (Mitsuno et al., 2008), Diaphania indica (Mitsuno et al., 2008), and A. transitella (this study) were aligned using the Clustal W program (Thompson et al., 1994) built into the MEGA5 software. The bootstrap consensus tree was inferred from 500 replicates (Felsenstein, 1985) and branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The analysis involved 78 amino acid sequences. All positions with less than 50% site coverage were eliminated. That is, fewer than 50% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 406 positions in the final dataset.
To amplify the 3′ end of the AtraOR4 gene, gene specific primers derived from cDNA sequences were used (AtraOR4 3′UTR Fwd1: 5′- GAACAGGGAGAATCGATTCATCG-3′ and AtraOR4 ORF Rev: see above; 200 nM final concentration) in PCR reactions with genomic DNA as template and using Advantage Taq polymerase (Clontech). PCR reactions were carried out using the following conditions; initial denaturation for 3 min at 94°C, then amplification for 20 s at 94°C, 20 s at 62°C, 3 min at 72°C for 40 cycles, followed by a final 5 min 72°C incubation. PCR products were separated on 1% agarose gels and excised and TA cloned as described above. Resultant gene clones were sequenced and the consensus sequence of three clones was generated in Geneious (version 7.1.9 created by Biomatters, available from http://www.geneious.com/).
To determine which tissues express AtraOR4, reverse transcription PCR (RT-PCR) was used. Total RNA was extracted from antennae, heads, thoraces, and abdomens as above, and quantitated using the Quant-iT™ RiboGreen® RNA assay kit (Invitrogen, Carlsbad, CA). First strand cDNA synthesis was performed as above, using 100 ng of total RNA and primed with 5′-RACE CDS Primer A. PCR amplification was done using gene specific primers (200 nM final concentration) to detect AtraOR4 (AtraOR4 3′UTR Fwd1 and AtraOR4 ORF Rev; sequences are above), AtraOrco (AtraOrco Fwd: 5′- AGATGTTGGCTCGTTCTGCT-3′ and AtraOrco Rev: 5′- AAGCCGCTTCCATTACTGAC-3′) and Actin (Actin 1F: 5′- GGTCGCGATCTCACAGACTA-3′ and Actin 1R: 5′- TCGAGTTGTAGGTGGTTTCG-3′), and cDNA template equivalent to 5 ng of input RNA using the following conditions; initial denaturation for 3 min at 94°C, then amplification for 20 s at 94°C, 20 s at 62°C, 30 s at 72°C for 35 cycles, followed by a final 5 min 72°C incubation. PCR products were separated by loading 1/4th of the total reaction onto 1.5% agarose gels and visualized on a UV light box. To confirm identity of PCR products, bands were excised, TA cloned and sequenced as above.
To detect potential pheromone receptor subfamily members expressed in the antennae of female navel orangeworm, degenerate primers and a 3′ RACE technique was used (Garczynski et al., 2012). The 3′ end of cDNAs of transcripts present in female antennae were amplified and PCR products visualized on agarose gels were excised and TA cloned (data not shown). The cDNA sequence of one clone encoded for a putative pheromone receptor subfamily member (named AtraOR4) based on homology of TBLASTN searches using the deduced amino acid sequence against those present in the NCBI non-redundant nucleotide database (data not shown).
The cDNA sequence obtained by 5′ RACE encompassed the putative start methionine of AtraOR4 (data not shown). An oligonucleotide primer encompassing the nucleotides surrounding the start methionine was designed and used in a 3′ RACE reaction to amplify the cDNA encoding the full open reading frame. Two PCR products (~1550 and 1400 bp) were visualized on agarose gels (Figure 1A), excised and TA cloned. The nucleotide sequences of the PCR products were determined, yielding actual transcript lengths of 1469 (GenBank ID: JN701807) and 1302 nt (GenBank ID: JN701806) encoding for 446 and 425 amino acids (Figure S1B and Figure S1A, respectively). The 1302 nt transcript was designated AtraOR4 and the 1469 nt transcript, AtraOR4A. Alignment of the nucleotide sequences of these transcripts showed that they were nearly identical except for an additional 167 nt internal sequence in the 3′ end of the longer AtraOR4A transcript (Figure S2, Supplementary Material). Alignment of the deduced amino acid sequences revealed that the two putative proteins are nearly identical for the first 349 amino acids, and then diverge at the C-terminus (Figure S3, Supplementary Material).
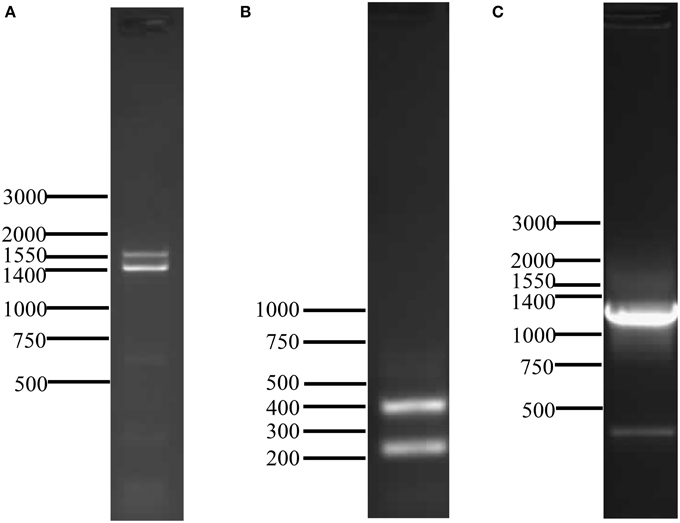
Figure 1. PCR detection of AtraOR4 and AtraOR4A transcripts and 3′ end of AtraOR4 gene. (A) Full-length transcripts amplified by 3′ RACE. Amplification products were generated by 3′ RACE and visualized on 1% agarose gels by ethidium bromide staining and UV illumination. (B) RT-PCR detection of the 3′ end of AtraOR4 and AtraOR4A transcripts. Amplification using RT-PCR and transcript specific primers. PCR products were visualized on 1.5% agarose gels by ethidium bromide staining and UV illumination. (C) PCR detection of the 3′ end of AtraOR4 gene. Amplification was done using transcript specific primers and PCR product was visualized on 1% agarose gels by ethidium bromide staining and UV illumination.
To determine if AtraOR4 and AtraOR4A are putative members of the lepidopteran pheromone receptor subfamily, TBLASTN searches (Altschul et al., 1990) were done using the deduced amino acid sequences against the NCBI non-redundant nucleotide database limited to arthropods. AtraOR4 and AtraOR4A displayed 34–41% identity with the top 15 published blast hits (E-values ranging from 1.00−77 to 2.00−105) and all of the similar sequences were members of the pheromone receptor subfamily (data not shown). These results indicate that on the basis of amino acid sequence homology, AtraOR4, and AtraOR4A are members of the lepidopteran pheromone receptor subfamily. To confirm these results, a phylogenetic analysis was performed and a tree was constructed. The amino acid sequences of 78 lepidopteran ORs, including 26 putative pheromone subfamily receptors, were aligned using ClustalW and the resulting Neighbor Joining tree shows that AtraOR4 and AtraOR4A cluster within the pheromone receptor subfamily (Figure 2).
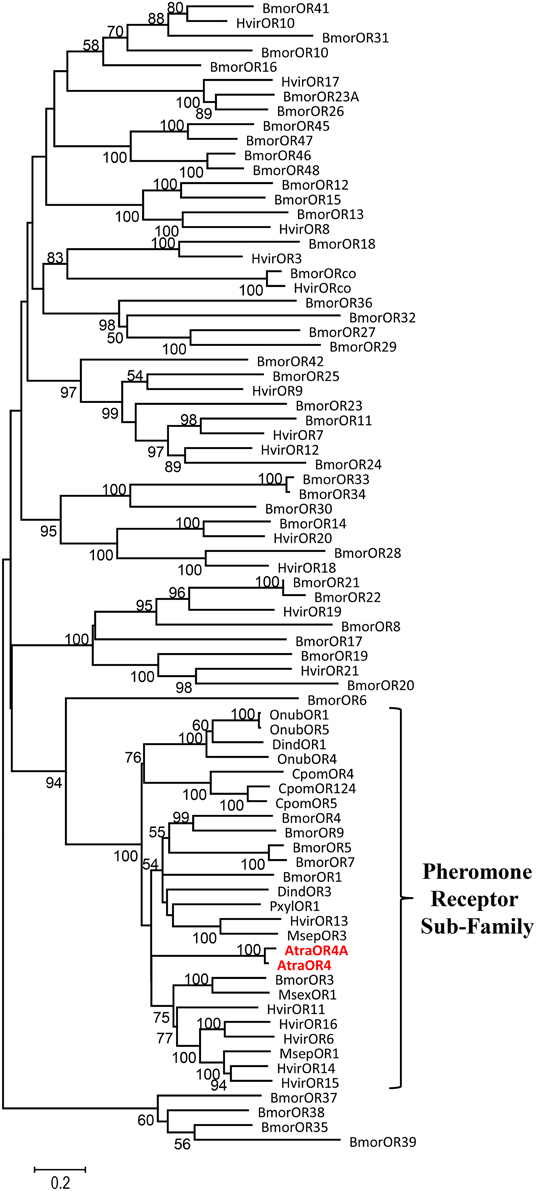
Figure 2. Neighbor joining tree of AtraOR4 and AtraOR4A with 78 lepidopteran ORs, including 26 putative pheromone subfamily receptors from the following insects: Bombyx mori (Bmor), Heliothis virescens (Hvir), Ostrinia nubialis (Onub), Manduca sexta (Msex), Cydia pomonella (Cpom), Plutella xylostella (Pxyl), Mythimna separata (Msep), Diaphania indica (Dind), and Amyelois transitella (Atra).
To further analyze the 3′ regions of the two nearly identical transcripts encoding AtraOR4 and AtraOR4A, specific oligonucleotide primers upstream of the divergent 3′ nucleotide sequences were designed for use in 3′ RACE. Consistent with the results above (Figure 1A), two PCR products of approximately 250 and 400 nt were detected in the 3′ RACE reaction performed with the transcript specific primers (Figure 1B). The DNA sequence of these products indicated, as above, that they are nearly identical except for an additional 167 nt of internal sequence in the AtraOR4A product (Figure S2, Supplementary Material). Because the dinucleotide AG was at the end of the additional 167 nt internal sequence and directly preceded the end of the shorter transcript we considered the possibility that these two transcripts were alternatively spliced products of the same gene. To test this, oligonucleotide primers encompassing the 3′ end of the transcripts were used in PCR reactions to amplify genomic DNA. A single product of approximately 1300 bp was detected on agarose gels (Figure 1C) from the PCR amplification using the transcript specific primers. These results indicated that the AtraOR4 and AtraOR4A transcripts were produced from the same gene.
To determine if the AtraOR4 and AtraOR4A transcripts were generated from the same gene, the PCR product of the genomic DNA amplification was cloned and sequenced. The cloned gene product was 1271 bp (Figure 3; GenBank ID: JN701808). Alignment of the 3′ regions of the AtraOR4 and AtraOR4A transcripts with the genomic DNA showed that their sequences are identical and contained within the gene sequence (Figure 3). The AtraOR4 gene sequence contains an intron with a canonical dinucleotide GT splice site shared by both the AtraOR4 and AtraOR4A transcripts. For AtraOR4A, a canonical dinucleotide AG splice site is located 931 bp downstream yielding the 3′ end of its transcript and a canonical dinucleotide AG splice site is located 1098 bp downstream for the AtraOR4 transcript (Figure 3).

Figure 3. Partial genomic DNA sequence encoding 3′ end of AtraOR4 and alignment of AtraOR4 and AtraOR4A transcripts. AtraOR4 genomic DNA sequence is in black, AtraOR4A is in red, and AtraOR4 is in blue. Putative splice sites are  for excision and
for excision and  insertion.
insertion.
To determine where the AtraOR4 and AtraOR4A transcripts are expressed in adult males and females, a tissue expression profile using RT-PCR was performed with cDNA prepared from total RNA extracted from dissected antennae, heads (without antennae), thoraces, and abdomens (Figure 4). Expression of AtraOR4 and AtraOR4A was female-biased, with PCR products detected mainly in female antennae (Figure 4, top). AtraOrco, a conserved co-receptor that forms heteromers with ORs in insects (Larsson et al., 2004; Orco has previously been called OR83b in Drosophila, OR2 in lepidopterans and OR7 in mosquitoes), expression was detected in male antennae and to a lesser extent in heads, and in female antennae and to a lesser extent in heads and abdomens (Figure 4, middle). As a control, primers to detect AtraActin were used and it was detected in all tissues (Figure 4, bottom).
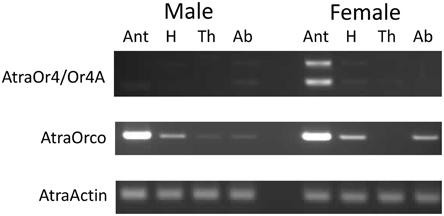
Figure 4. AtraOR4 and AtraOR4A expression profile in cDNA prepared from total RNA extracted from male and female antennae (Ant), heads (H), thoraces (Th) and abdomens (Ab). Top: Detection of AtraOR4 and AtraOR4A using transcript specific primers in reverse transcription PCR (RT-PCR) reactions. Middle: Detection of AtraOrco using specific primers in RT-PCR reactions. Bottom: Detection of AtraActin using specific primers in RT-PCR reactions. For each panel, PCR products were visualized on 1.5% agarose gels by ethidium bromide staining and UV illumination. Bands produced were of the size predicted for the transcript specific primer pairs.
Using degenerate oligonucleotide primers and 3′ RACE (Garczynski et al., 2012), we have identified two transcripts expressed in antennae of female navel orangeworm that encode putative members of the lepidopteran pheromone receptor subfamily. Analysis of cDNAs containing full-length ORFs indicate that these transcripts appear to be the products of alternative splicing from a single gene. Comparison of the transcript nucleotide sequences with genomic DNA sequence support that AtraOR4 and AtraOR4A are produced from the same gene by alternative splicing. Phylogenetic analysis supports that the proteins encoded by the AtraOR4 and AtraOR4A transcripts are members of the lepidopteran pheromone receptor subfamily. Tissue expression profiles indicate that these transcripts are mainly found in female antennae. While female-biased ORs have been previously identified in B. mori (Wanner et al., 2007; Anderson et al., 2009), these ORs were not homologous to members of the lepidopteran pheromone receptor subfamily. We believe AtraOR4 and AtraOR4A to be the first example of alternatively spliced, female-biased members of the lepidopteran pheromone receptor subfamily.
Alternative splicing is a common mechanism used to produce multiple proteins from a single gene and this process can significantly increase the size of an organism's proteome. For mammalian ORs, alternative splicing is extensive, but rarely occurs within the coding region (Volz et al., 2003; Young et al., 2003). Alternative splicing at the N-termini of ORs and mainly, gustatory receptors has been detected for Drosophila, Anopheles, Aedes, and Tribolium (Clyne et al., 2000; Hill et al., 2002; Robertson et al., 2003; Abdel-Latief, 2007; Kent et al., 2008), however, this is the first evidence of alternative splicing changing the C-terminal amino acid sequence in an OR from insects. The extent of alternative splicing of insect OR genes is unknown. Because most lepidopteran (and other insects) ORs are being identified from de novo assembled transcriptomes, detection of alternate spliced products may be difficult because of the limitations of bioinformatic resources (Florea, 2006). Until bioinformatics resources become sufficient enough to assemble and predict alternative spliced products, traditional methods of sequencing RACE amplified transcripts can still be used to identify alternately spliced OR gene products.
The predicted protein products for AtraOR4 and AtraOR4A align most closely with members of the pheromone receptor subfamily. In this current study, we did not attempt to determine odorant ligands for the protein products encoded by the AtraOR4 and AtraOR4A transcripts, but we expect that they will be either pheromone(s) or host-plant volatile(s) in nature. For some lepidopteran pheromone receptor subfamily members, the ligands for these ORs are sex pheromones (for examples see Sakurai et al., 2004; Wanner et al., 2010; Xu et al., 2012 among others). However, not all pheromone receptor subfamily members respond to sex pheromones. For B. mori, odorants that activate only OR1 (bombykol) and OR3 (bombykal) have been identified while OR4, OR5, and OR6 remain orphans (Nakagawa et al., 2005). Furthermore, two ORs belonging to the pheromone receptor subfamily clade are activated by plant volatiles. In the light brown apple moth, Epiphyas postvittana, a pheromone receptor subfamily member, EpOR1, is stimulated by several plant volatiles with the strongest binding to methyl salicylate (1.8 × 10−12 M) and geraniol (5.8 × 10−11 M) (Jordan et al., 2009). More recently, a codling moth, C. pomonella, pheromone receptor subfamily member, CpomOR3, was shown to be activated by pear ester, an important host plant kairomone for this insect (Bengtsson et al., 2014). Perhaps the categorization of the pheromone receptor subfamily (aka male-biased sex pheromone receptor clade, sex pheromone receptor subfamily, pheromone receptors, and male-specific OR subfamily) should be reconsidered until a significant number of receptors has been de-orphanized.
There is not much information on the structure/function relationships of ligand binding for insect ORs. Much work is being done to determine which ligands activate different ORs, but this line of research does not tell us where on the receptors the odorants are binding. Recently, it has been determined that the C-terminal amino acids are important for odor specificity, namely conferring enantiomeric selectivity (Hill et al., 2015). Because alternate spliced products of AtraOR4 and AtraOR4A results in changes of the C-terminal amino acids of these proteins, it is possible that these changes enable these receptors to recognize different ligands or discriminate different enantiomers of their ligands. However, until further characterizations of ORs are completed the role of the C-terminal amino acids in ligand binding remains speculative.
This article reports the results of research only. Mention of a commercial or proprietary product does not constitute an endorsement of the product by the United States Department of Agriculture. USDA is an equal opportunity provider and employer.
SG and WL conceived and designed the work, and supplied materials. SG performed the work and wrote the manuscript. WL and SG revised the manuscript to its final submitted form.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Drs. Tom Unruh and Tonja Fisher for their review of this manuscript and helpful suggestions. Laura Willett, Alexandria Portelli and Lindsay Portelli are thanked for their technical assistance. This project is supported by a grant from the Washington Tree Fruit Research Commission (awarded to SG) and National Research Initiative (NRI) Arthropod and Nematode Biology and Management Program of the USDA National Institute of Food and Agriculture, Grant no. 2008-35302-18839 (awarded to SG).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fevo.2015.00115
Abdel-Latief, M. (2007). A family of chemoreceptors in Tribolium castaneum (Tenebrionidae: Coleoptera). PLoS ONE 2:e1319. doi: 10.1371/journal.pone.0001319
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anderson, A. R., Wanner, K. W., Trowell, S. C., Warr, C. G., Jaquin-Joly, E., Zagatti, P., et al. (2009). Molecular basis of female-specific odorant responses in Bombyx mori. Ins. Biochem. Mol. Biol. 39, 189–197. doi: 10.1016/j.ibmb.2008.11.002
Bargmann, C. I. (2006). Comparative chemosensation from receptors to ecology. Nature 444, 295–301. doi: 10.1038/nature05402
Bengtsson, J. M., Gonzalez, F., Cattaneo, A. M., Montagne, N., Walker, W. B., Bengtsson, M., et al. (2014). A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2:33. doi: 10.3389/fevo.2014.00033
Bengtsson, J. M., Trona, F., Montagne, N., Anfora, G., Ignell, R., Witzgall, P., et al. (2012). Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE. 7:e31620. doi: 10.1371/journal.pone.0031620
Benton, R. (2006). On the ORigin of smell: odorant receptors in insects. Cell. Mol. Life Sci. 63, 1579–1585. doi: 10.1007/s00018-006-6130-7
Clyne, P. J., Warr, C. G., Freeman, M. R., Lessing, D., Kim, J., and Carlson, J. R. (1999). A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22, 327–338.
Clyne, P. J., Warr, C. G., and Carlson, J. R. (2000). Candidate taste receptors in Drosophila. Science 287, 1830–1834. doi: 10.1126/science.287.5459.1830
Depetris-Chauvin, A., Galagovsky, D., and Grosjean, Y. (2015). Chemicals and chemoreceptors: ecologically relevant signals driving behavior in Drosophila. Front. Ecol. Evol. 3:41. doi: 10.3389/fevo.2015.00041
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Florea, L. (2006). Bioinformatics of alternative splicing and its regulation. Brief. Bioinformatics. 7, 55–69. doi: 10.1093/bib/bbk005
Gao, Q., and Chess, A. (1999). Identification of candidate Drosophila olfactory receptors from genomic DNA. Genomics 60, 31–39.
Garczynski, S. F., Wanner, K. W., and Unruh, T. R. (2012). Identification and initial characterization of the 3′ end of gene transcripts encoding putative members of the pheromone receptor subfamily in Lepidoptera. Insect Sci. 19, 64–74. doi: 10.1111/j.1744-7917.2011.01423.x
Groe-Wilde, E., Gohl, T., Bouché, E., Breer, H., and Krieger, J. (2007). Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromone compounds. Eur. J. Neurosci. 25, 2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x
Groe-Wilde, E., Stieber, R., Forstner, M., Krieger, J., Wicher, D., and Hansson, B. S. (2010). Sex-specific odorant receptors of the tobacco hornworm Manduca sexta. Front. Cell. Neurosci. 4:22. doi: 10.3389/fncel.2010.00022
Higbee, B. S., Burks, C. S., and Larsen, T. E. (2014). Demonstration and characterization of a persistent pheromone lure for the navel orangeworm, Amyelois transitella (Lepidoptera: Pyralidae). Insects 5, 596–608. doi: 10.3390/insects5030596
Hill, C. A., Fox, A. N., Pitts, R. J., Kent, L. B., Tan, P. L., Chrustal, M. A., et al. (2002). G protein-couple receptors in Anopheles gambiae. Science 298, 176–178. doi: 10.1126/science.1076196
Hill, S. R., Majeed, S., and Ignell, R. (2015). Molecular basis for odorant receptor tuning: a short C-terminal sequence is necessary and sufficient for selectivity of mosquito OR8. Insect Mol. Biol. 24, 491–501. doi: 10.1111/imb.12176
Jordan, M. D., Anderson, A., Begum, D., Carraher, C., Authier, A., Marshall, S. D. G., et al. (2009). Odorant receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem. Senses 34, 383–394. doi: 10.1093/chemse/bjp010
Kent, L. B., Walden, K. K. O., and Robertson, H. M. (2008). The Gr family of candidate gustatory and olfactory receptors in the yellow-fever mosquito Aedes aegypti. Chem. Senses 33, 79–93. doi: 10.1093/chemse/bjm067
Kornblihtt, A. R., Schor, I. E., Allo, M., Dujardin, G., Petrillo, E., and Munoz, M. J. (2013). Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 13, 153–165. doi: 10.1038/nrm3525
Krieger, J., Große-Wilde, E., Gohl, T., and Breer, H. (2005). Candidate pheromone receptors of the silkmoth Bombyx mori. Eur. J. Neurosci. 21, 2167–2176. doi: 10.1111/j.1460-9568.2005.04058.x
Krieger, J., Große-Wilde, E., Gohl, T., Dewer, Y. M. E., Raming, K., and Breer, H. (2004). Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc. Nat. Acad. Sci. U.S.A. 101, 11845–11850. doi: 10.1073/pnas.0403052101
Krieger, J., Raming, K., Dewer, Y. M. E., Bette, S., Conzelmann, S., and Breer, H. (2002). A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur. J. Neurosci. 16, 619–628. doi: 10.1046/j.1460-9568.2002.02109.x
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Leal, W. S. (2013). Odorant Reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Mitsuno, H., Sakurai, T., Murai, M., Yasuda, T., Kugimiya, S., Ozawa, R., et al. (2008). Identification of receptors of main sex-pheromone components of three lepidopteran species. Eur. J. Neurosci. 28, 893–902. doi: 10.1111/j.1460-9568.2008.06429.x
Miura, N., Nakagawa, T., Tatsuki, S., Touhara, K., and Ishikawa, Y. (2009). A male-specific odorant receptor conserved through the evolution of sex pheromones in Ostrinia moth species. Int. J. Biol. Sci. 5, 319–330. doi: 10.7150/ijbs.5.319
Miura, N., Nakagawa, T., Touhara, K., and Ishikawa, Y. (2010). Broadly and narrowly tuned odorant receptors are involved in female sex pheromone reception in Ostrinia moths. In. Biochem. Mol. Biol. 40, 64–73. doi: 10.1016/j.ibmb.2009.12.011
Nakagawa, T., Sakurai, T., Nishioka, T., and Touhara, K. (2005). Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–1642. doi: 10.1126/science.1106267
Nilsen, T. W., and Graveley, B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463. doi: 10.1038/nature08909
Patch, H. M., Velarde, R. A., Walden, K. K. O., and Robertson, H. M. (2009). A candidate pheromone receptor and two odorant receptors of the hawkmoth Manduca sexta. Chem. Senses 34, 305–316. doi: 10.1093/chemse/bjp002
Robertson, H. M., Warr, C. G., and Carlson, J. R. (2003). Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 100, 14537–14542. doi: 10.1073/pnas.2335847100
Sakurai, T., Nakagawa, T., Mitsuno, H., Mori, H., Endo, Y., Tanoue, S., et al. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 101, 16653–16658. doi: 10.1073/pnas.0407596101
Smith, D. P. (2007). Odor and pheromone detection in Drosophila melanogaster. Pflügers Arch. Eur. J. Physiol. 454, 749–758. doi: 10.1007/s00424-006-0190-2
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Volz, A., Ehlers, A., Younger, R., Forbes, S., Trowsdale, J., Schnorr, D., et al. (2003). Complex transcription and splicing of odorant receptor genes. J. Biol. Chem. 278, 19691–19701. doi: 10.1074/jbc.M212424200
Vosshall, L. B., Amrein, H., Morozov, P. S., Rzhetsky, A., and Axel, R. (1999). A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96, 725–736.
Wanner, K. W., Anderson, A. R., Trowell, S. C., Theilmann, D. A., Robertson, H. M., and Newcomb, R. D. (2007). Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, Bombyx mori. In. Mol. Biol. 16, 107–119. doi: 10.1111/j.1365-2583.2007.00708.x
Wanner, K. W., Nichols, A. S., Allen, J. E., Bunger, P. L., Garczynski, S. F., Linn, C. E., et al. (2010). Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS ONE 5:e8685. doi: 10.1371/journal.pone.0008685
Xu, P., Garczynski, S. F., Atungulu, E., Syed, Z., Choo, Y.-M., Vidal, D. M., et al. (2012). Moth sex pheromone receptors and deceitful parapheromones. PLoS ONE 7:e41653. doi: 10.1371/journal.pone.0041653
Young, J. M., Shykind, B. M., Lane, R. P., Tonnes-Priddy, L., Ross, J. A., Walker, M., et al. (2003). Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 4:R71. doi: 10.1186/gb-2003-4-11-r71
Keywords: odorant receptor, pheromone receptor subfamily, alternative splicing, navel orangeworm, 3′ RACE
Citation: Garczynski SF and Leal WS (2015) Alternative splicing produces two transcripts encoding female-biased pheromone subfamily receptors in the navel orangeworm, Amyelois transitella. Front. Ecol. Evol. 3:115. doi: 10.3389/fevo.2015.00115
Received: 09 July 2015; Accepted: 21 September 2015;
Published: 06 October 2015.
Edited by:
Sharon Rose Hill, Swedish University of Agricultural Sciences, SwedenReviewed by:
Jeremy J. Heath, North Carolina State University, USACopyright © 2015 Garczynski and Leal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen F. Garczynski, Yakima Agricultural Research Laboratory, United States Department of Agriculture—Agricultural Research Service, 5230 Konnowac Pass Rd., Wapato, WA 98951, USA,c3RldmUuZ2FyY3p5bnNraUBhcnMudXNkYS5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.