- Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, USA
Baja California disjuncts, which are marine species that are present in the northern Sea of Cortez and along the Pacific Coast of California and Baja California, but absent from the south, have been shown to be interesting models of allopatric speciation. While at least 19 species of fish are Baja California disjuncts, no study has ever evaluated the genetic patterns of closely related species. Here, we used three sympatric California blennies (genus Hypsoblennius), where two species have disjunct populations and one does not. Based on one mitochondrial and one nuclear molecular marker, we found that the two disjunct species exhibit different genetic patterns, one species showing disjunct populations as reciprocally monophyletic assemblages, while the other species showed evidence of gene flow between disjunct populations. In addition, the non-disjunct species, H. gilberti, exhibits high gene flow along the California and Baja California coasts. I hypothesized that adult habitat and historical vicariant events, more so than pelagic larval duration, played major roles in shaping the currently observed distribution and genetic patterns for these species.
Introduction
Phylogeographic approaches provide insight in understanding the processes leading to speciation (Avise, 2000; Bernardi, 2013). Such approaches are particularly powerful when used in comparative ways either by employing different molecular markers, or several species that share common geographic distributions (Bernardi et al., 2003; Eble et al., 2009; Toonen et al., 2011; Von der Heyden et al., 2011; Dawson, 2012). Most comparative phylogeographic studies based on a panel of unrelated species interpret shared patterns as a likely signature of shared histories. In contrast, dissimilar patterns are assumed to be due to scenarios that depend on historical, ecological, or life history characteristics of the organisms (Avise, 2000). An alternative way to address complex scenarios is to use comparative methods with closely related species. This is rarely done because there are few cases where related species that are amenable to a comparative phylogeographic approach live in sympatry (Crow et al., 2010; Bird et al., 2011). Here, we use three congeneric blennies (genus Hypsoblennius) as a model system to assess patterns of population genetic structuring and speciation. The three focal species live sympatrically along the Pacific coast of California and Baja California. In addition, two of those species have Baja California disjunct populations, which are isolated in the northern Sea of Cortez (Gulf of California).
Baja California Disjunction
The Baja California peninsula is a geologically active region originally formed by a budding peninsula and several islands that slowly merged and gradually drifted away from the mainland over time, a process that started approximately 5.3 Mya (Grismer, 2000; Riddle et al., 2000). While the process was underway, at different times, marine seaways cut across the peninsula, separating terrestrial populations on the continent, and at the same time offering a conduit for marine species to migrate across the Peninsula between the Sea of Cortez and the open Pacific Ocean (Upton and Murphy, 1997; Grismer, 2000; Riddle et al., 2000; Bernardi et al., 2003; Recuero et al., 2006).
Some Baja California disjunct populations are the results of such seaways, where marine organisms exhibit distributions restricted to the upper Sea of Cortez and the northern part of the Pacific coast of the Baja California Peninsula, but are absent from the southern region of Baja California. Similarly to other population disjunctions, such as the Trans-Isthmian geminate species (Lessios, 2008), Baja California disjunctions resulted in different levels of genetic separation between Pacific Ocean and Sea of Cortez populations (Bernardi et al., 2003). Indeed, a combination of historical and ecological factors played an important role in shaping the population genetics that are observed today (Terry et al., 2000; Huang and Bernardi, 2001; Stepien et al., 2001; Bernardi et al., 2003; Balart et al., 2004; Bernardi and Lape, 2005; Muñiz-Salazar et al., 2005; Gaffney et al., 2007; Hurtado et al., 2010; Schinske et al., 2010; Peterson et al., 2013; Poortvliet et al., 2013; Castillo-Páez et al., 2014). In the case of fishes, approximately 19 species are considered Baja California disjuncts (Bernardi et al., 2003). Information on the genetics of fish disjunct populations is available for 14 of these species (Stepien et al., 2001; Bernardi et al., 2003; Gaffney et al., 2007; Schinske et al., 2010; Castillo-Páez et al., 2014), yet it is still difficult to draw a comprehensive image of the processes that have shaped the genetics of those populations. For example, in the case of the longjaw mudsucker, Gillichthys mirabilis, where disjunct populations are genetically distinct (reciprocally monophyletic assemblages), the absence of tides in the southern Sea of Cortez (an ecological requirement for this intertidal species) is likely to have reinforced the separation that may have been initiated by historical events, thus both ecological and historical factors have played a role in initiating and maintaining the pattern of genetic divergence in this species (Huang and Bernardi, 2001).
Most phylogeographic studies of marine organisms have traditionally focused on abiotic factors, such as oceanographic characteristics that affect larval dispersal, and historical factors that produce vicariant events (Avise, 2000; White et al., 2010; Bowen et al., 2014). However, recently, in part due to a better understanding of larval behavior (Jones et al., 1999; Swearer et al., 1999), the role of larval dispersal has been de-emphasized, and the role of adult ecology has taken a more prominent role (Luiz et al., 2011, 2013). The taxonomic diversity of disjunct species is an asset when using a comparative phylogeographic approach, however the 14 species studied so far belong to 14 different genera and 13 families, potentially precluding an understanding of the role that ecological factors play in the system and in particular on adult individuals. In contrast, important clues could be obtained by using species that are closely related. Among the 19 disjunct species, only two are congeneric, the bay blenny Hypsoblennius gentilis and the mussel blenny Hypsoblennius jenkinsi. Therefore, our goal in this study was to capitalize on the presence of two congeneric disjunct blennies to shed some light on the mechanisms that lead to genetic disjunction.
The genus Hypsoblennius
Hypsoblennius comprises 16 species that are found in the Tropical Western Atlantic (TWA, 4 species) and the Tropical Eastern Pacific (TEP, 12 species) (Froese and Pauly, 2000), including two presumed trans-Isthmian geminate species, Hypsoblennius invemar, from the TWA, and H. brevipinnis, from the TEP, which are suggested to have been separated by the rise of the Isthmus of Panama, 2.8–3.5 Mya (Almada et al., 2009; Hastings, 2009). In our study area, 3 sympatric species are found: the bay blenny, H. gentilis, the rockpool blenny, H. gilberti, and the mussel blenny, H. jenkinsi. The distribution of the three species mostly overlap along the California and Baja California coasts. Hypsoblennius gentilis is found from Monterey to Bahia Magdalena, H. gilberti from Morro Bay to Bahia Magdalena, and H. jenkinsi from Morro Bay to Puerto Marquis (Punta Marquez), a few kilometers south of Bahia Magdalena. Importantly to our study, disjunct populations of H. gentilis and H. jenkinsi are also found in the northern Sea of Cortez (Miller and Lea, 1972; Allen and Robertson, 1994; Thomson et al., 2000; Love, 2011). For all species, the male produces pheromones to attract females and after benthic spawning, guards a nest (Losey, 1969). Hatching occurs within 4–18 days, larvae are pelagic and remain in the water column between 44 (H. jenkinsi) and 66 days (H. gentilis and H. gilberti), suggesting that all three species have large dispersal potentials (Riginos and Victor, 2001; Shanks and Eckert, 2005; Selkoe and Toonen, 2011).
Ecological studies done in California revealed that the three Hypsoblennius species have very similar feeding habits, yet they also show important ecological differences (Stephens et al., 1970). The rockpool blenny, H. gilberti, lives mostly in the upper intertidal in rocky pools, and occasionally in the very shallow subtidal, while the mussel blenny, H. jenkinsi, lives exclusively subtidally in mussel, barnacle, and boring clam burrows. The mussel blenny is a sedentary species, while the rockpool blenny has much larger territories. This is accompanied by differences in homing behavior, where rockpool blennies that are displaced by as much as 45 m can return to their territories, while mussel blennies displaced by as little as 5 m usually do not (Stephens et al., 1970). In contrast to the two species discussed above, the bay blenny, H. gentilis, lives mostly in shallow subtidal sand, and occasionally in large intertidal pools. Overall, the rockpool blenny, H. gilberti and the mussel blenny, H. jenkinsi, seem more specialized than the bay blenny, H. gentilis, which was therefore assumed to be an ancestral form Stephens et al. (1970).
Allozyme work showed that disjunct H. jenkinsi populations are genetically divergent (Present, 1987). Additional work based on one mitochondrial marker showed that Pacific and Sea of Cortez populations of H. jenkinsi formed reciprocally monophyletic groups (Bernardi et al., 2003). No genetic information is available for H. gentilis or H. gilberti.
The goal of this study was to use a phylogeographic approach to evaluate genetic patterns in three sympatric species of Hypsoblennius based on a mitochondrial and a nuclear molecular marker. Our study capitalizes on the distribution differences of the species, with one species restricted to the Pacific coast of California and Baja California and two Baja California disjunct species. Other studies have shown that Baja California disjunct populations provide an excellent system to study the early stages of speciation (Terry et al., 2000; Huang and Bernardi, 2001; Stepien et al., 2001; Bernardi et al., 2003; Balart et al., 2004; Bernardi and Lape, 2005; Hurtado et al., 2010; Schinske et al., 2010). In addition, we used two Trans-Isthmian geminate congeneric species, H. invemar (Atlantic) and H. brevipinnis (Pacific) to internally calibrate a molecular clock to estimate the timing of historical events.
Materials and Methods
Sample Collections
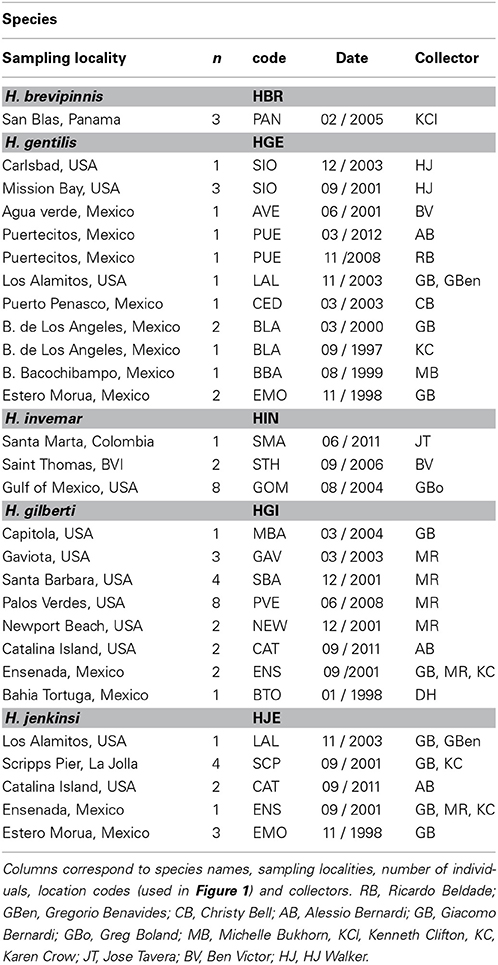
Samples were collected with hand nets in tidepools or while scuba diving. For Hypsoblennius gentilis, which lives over sandy habitats, some samples were collected with beach seines. We collected samples along a large portion of the range of each species, from Monterey, California to Bahia Tortuga, Baja California, and in the northern and central Sea of Cortez, as described in Table 1 and Figure 1. Interestingly, we collected one H. gilberti in Capitola, northern Monterey Bay, which is approximately 200 km north of its published range (Morro Bay) (Miller and Lea, 1972; Love, 2011). For outgroups, we used two Trans-Isthmian geminate species, Hypsoblennius brevipinnis and H. invemar (Table 1). One individual of H. brevipinnis, the species from the Tropical Eastern Pacific, has been recorded in the Atlantic, near the entrance of the Panama Canal, possibly following a recent migration through the Canal, thus casting doubt about the possibility of using those species to calibrate a molecular clock due to potential hybridizations between the two geminate species. That fish, however, was a misidentified Hypleurochilus pseudoaequipinnis (J. Williams, pers. comm.), and there is, to date, no evidence of any Hypsoblennius migrating across the Panama Canal (McCosker and Dawson, 1975). Hypsoblennius brevipinnis was collected at the San Blas islands in Panama, H. invemar was collected at the type locality (INVEMAR, Santa Marta, Colombia), in the British Virgin Islands, and on oil rig pilings off Texas in the Gulf of Mexico (Table 1).
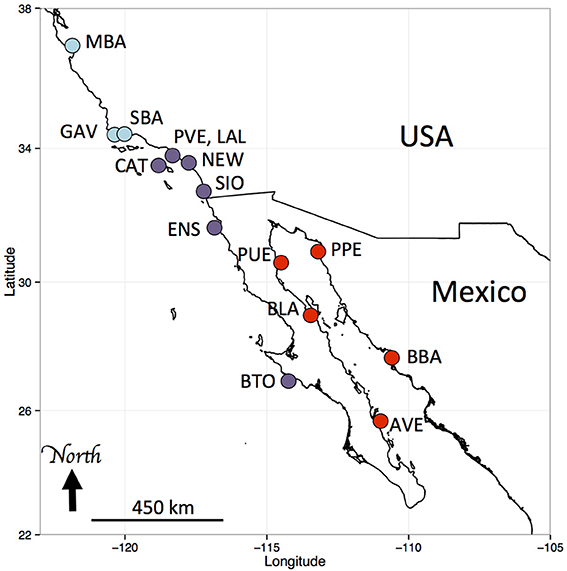
Figure 1. Sampling locations of Hypsoblennius spp. Blue and purple dots indicate Pacific coast locations (central and southern California, respectively), red dots indicate Sea of Cortez locations. MBA, Monterey Bay, GAV, Gaviota; SBA, Santa Barbara; CAT, Santa Catalina Island; PVE, Palos Verdes; LAL, Los Alamitos; NEW, Newport Beach; SIO, Scripps Pier; ENS, Ensenada; BTO, Bahia Tortuga; AVE, Agua Verde; BBA, Bahia Bacochibampo; BLA, Bahia de Los Angeles; PUE, Puertecitos; PPE, Puerto Penasco (sample numbers are provided in Table 1).
PCR and Sequence Analysis
Amplifications of the highly variable 5′ end of the mitochondrial control region and the first intron of the nuclear S7 ribosomal protein gene were accomplished with the universal CR-A and CR-E and the S7RPEX1F and S7RPEX2R primers, respectively, following published protocols (Lee et al., 1995; Chow and Hazama, 1998). After purification following the manufacturer's protocol (Applied Biosystems, Foster City, CA), sequencing was performed using the forward primers after having checked a subset of sequences obtained by sequencing in both directions.
Sequence Diversity, and Variability
Sequences were trimmed and aligned using the MAFFT routine (Katoh et al., 2002) implemented in Geneious 5.0 (Biomatters). Mitochondrial DNA variability was estimated by computing haplotype diversity, Hd, and nucleotide diversity π (Nei, 1987) using DNAsp (Librado and Rozas, 2009). Analyses were performed independently on each marker and jModeltest 2.1.4 (Posada and Crandall, 1998; Darriba et al., 2012) was used to determine the substitution model that best fit the data based on the corrected Akaike Information Criterion. For nuclear markers, ambiguous sites corresponding to heterozygous positions were labeled using IUPAC codes.
Phylogenetic Relationships
Phylogenetic relationships were assessed using Neighbor Joining (NJ), Maximum Likelihood (ML) and Bayesian inference (BI). Neighbor Joining analyses were generated in R using the APE package (Paradis et al., 2004). Maximum Likelihood analyses were performed in GARLI 2.0 (Zwickl, 2006). The GARLI search was performed in 5 independent runs each automatically terminated after 1117103 generations without improving the topology and, specifying the substitution model previously obtained by jModeltest allowing the parameters to be re-estimated during the run. The support was evaluated with 100 bootstrap replicates. Bayesian Inference was assessed in MrBayes 3.2.2 (Ronquist et al., 2012) setting priors to fit the evolutionary model suggested by jModeltest but allowing the parameters to be recalculated during the run. Four Markov chains were used to sample the probability space in two simultaneous but completely independent runs starting from different random trees (default option); the number of generations fluctuated depending on the convergence of chains, a sample frequency every 100 generations was performed. The two runs were combined and 25% of the initial trees and parameters sampled were discarded as the burn-in phase. Likelihood ratio test (LRT) were used to compare phylogenetic topologies and to test the null hypothesis that data evolved under a strict molecular clock as implemented in PAUP.
Population Structure
Population structure was evaluated using fixation indexes (Φst values) implemented in ARLEQUIN (Excoffier and Lischer, 2010). Deviations from values expected under the null hypothesis of genetic homogeneity were tested using a non-parametric permutation approach using 1000 replicates (Excoffier et al., 1992).
Historical Demography
Historical demography (population fluctuations based on coalescent models) was evaluated using the program LAMARC (Kuhner, 2006). Population parameters Θ theta = 2Nμ, where μ is the mutation rate for mtDNA and g (the exponential growth parameter in units of μ) were estimated, the parameter Θ being estimated with population growth (parameters are estimated jointly) or with growth kept constant (g = 0). Both estimates were obtained by running 5 replicates, which generated a mean value and its associated standard deviation. Analysis of each data set was done with 10 short Monte Carlo chains of 4000 steps each and 5 long chains of length 20,000, with a sampling increment of 20.
Coalescence times were estimated by assuming that coalescence was reached when the population size was reduced to 1% of its present-day value (Wares and Cunningham, 2001). In order to estimate coalescence time, we used a mutation rate (μ) as μ = substitutions per site per generation obtained for the pairs of Trans-Isthmian geminates Hypsoblennius brevipinnis and H. invemar (assuming their generation time being one year), thus providing a window of mutation rates (3.2 × 10−8 to 4.0 × 10−8). This rate is similar to the rate provided for other fish species (Bernardi and Lape, 2005; Domingues et al., 2005).
Results
Sequence Characteristics
Mitochondrial control regions (Dloop, DL) were obtained for a total of 83 individuals. Of those, 7 sequences from Hypsoblennius jenkinsi were from a previous study (Bernardi et al., 2003). Aligned sequences of DL included 447 base pairs (bp), with 182 variable sites, and 173 parsimony informative sites. Nuclear ribosomal protein S7 (S7) sequences were obtained for 61 individuals and as expected were less variable than the mitochondrial ones. Aligned sequences for S7 included 616 bp, with 237 variable sites, and 226 parsimony informative sites. Sequences were deposited in GenBank (accession numbers KM266661-KM266804).
While Hypsoblennius jenkinsi included the smallest number of individuals, its haplotype diversity (0.96) and nucleotide diversity (0.035) were the highest (Table 2). In contrast, H. gilberti, which is restricted to the Pacific Coast, showed the lowest values (0.26 and 0.001, respectively).
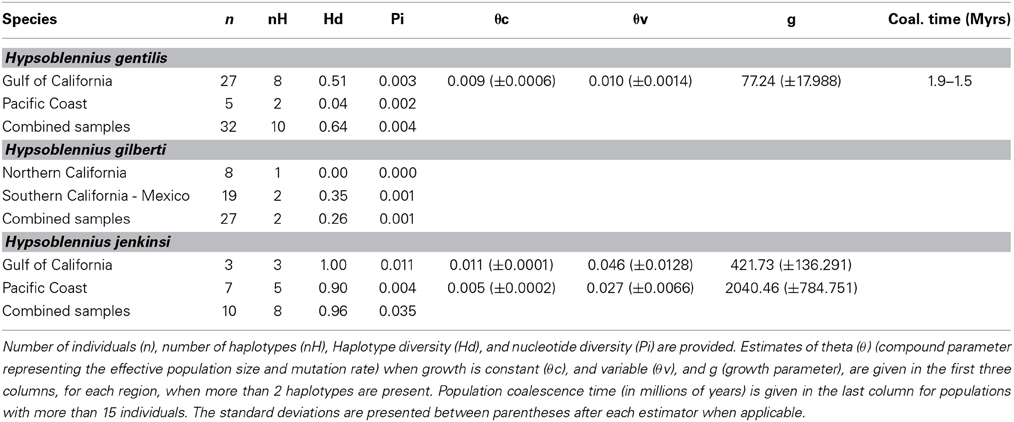
Table 2. Demographic parameters for Hypsoblennius gentilis, H. gilberti, and H. jenkinsi based on the mitochondrial control region.
Phylogenetic Reconstructions
Substitution models estimated by jModeltest based on the mitochondrial control region, nuclear S7, and combined markers were the HKY+G, TrN+G, and Trn+G models, respectively. Phylogenetic reconstructions based on Neighbor-Joining, Maximum Likelihood, and Bayesian Inference methods were statistically indistinguishable (Figure 2). When considering the relative position of each species (global topology of the trees), topologies based on mitochondrial and nuclear markers were statistically indistinguishable, however when including all taxa and their relative positions, mitochondrial and nuclear markers produced topologies that were statistically different (LRT test, p < 0.05). The phylogenetic topologies at the species level were identical, however the relative arrangements of each individual within a species occasionally differed. We therefore decided not to combine molecular marker types but rather to show each tree separately, where these differences can better be judged (Figures 2A,B). In general, phylogenetic trees were very well supported with most nodes showing bootstrap or posterior probability values above 90% for all methods and all markers (Figure 2). Both markers showed H. gentilis and H. jenkinsi as sister species, and H. gilberti as the sister species to this assemblage. Pacific Ocean and Sea of Cortez populations of Hypsoblennius gentilis did not partition in separate groups for either types of markers. In contrast, while nuclear markers did not separate Pacific Ocean and Sea of Cortez populations of H. jenkinsi, these populations formed distinct groups based on the mitochondrial marker. Differences between nuclear and mitochondrial data have been assigned to differences in dispersal between male and female lineages (Prugnolle and de Meeus, 2002). There is no evidence, however, that males and females disperse differently in H. jenkinsi. Instead, a more likely explanation is that this result is due to incomplete lineage sorting in the nuclear data (which have a longer sorting time compared to mitochondrial markers) (e.g., Egger et al., 2007).
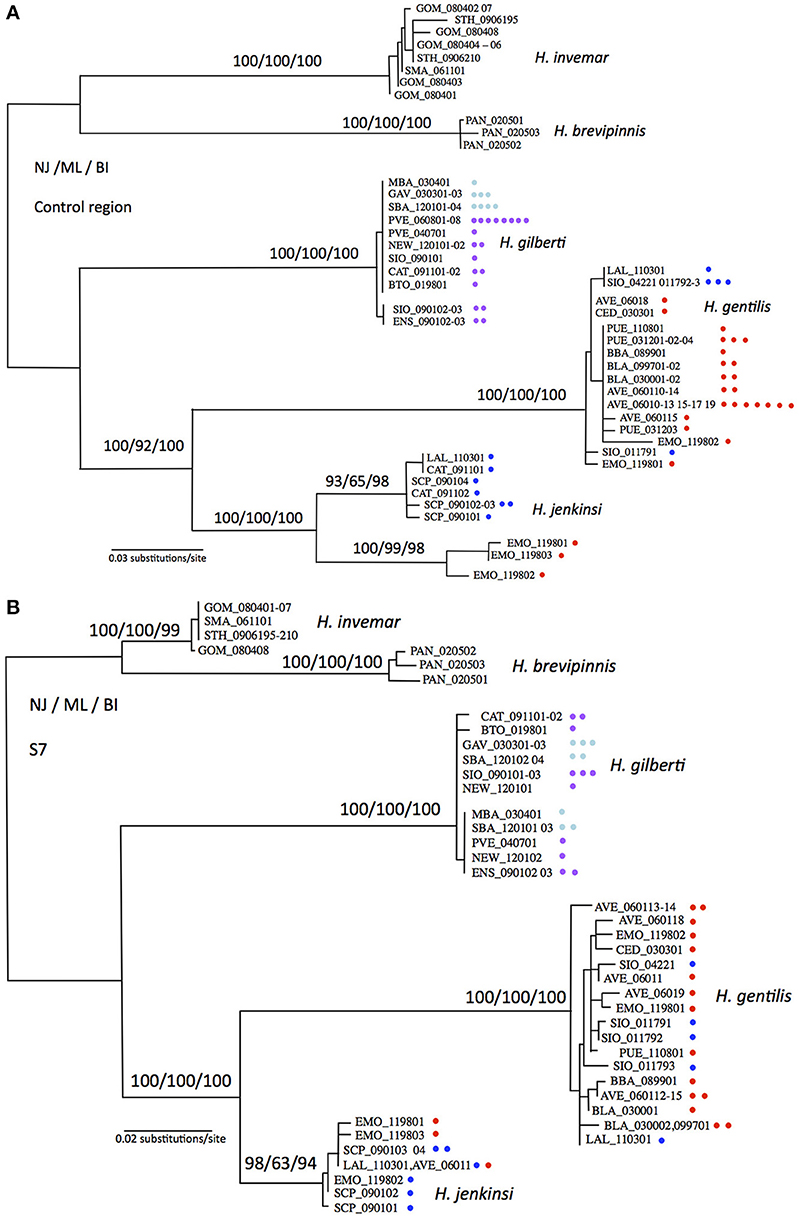
Figure 2. Maximum likelihood phylogenetic reconstructions for Hypsoblennius spp. Support for major nodes is given in percent bootstrap or posterior probability for Neighbor Joining, Maximum Likelihood and Bayesian Inference above the nodes (NJ/ML/BI). One branch is shown per haplotype, each individual is indicated by a dot, as to show the total number of individuals sequenced. Phylogenetic trees are based on the mitochondrial control region (A), and the nuclear ribosomal protein S7 (B). Hypsoblennius gentilis and Hypsoblennius jenkinsi individuals collected in the Sea of Cortez and the Pacific coast are labeled with red and blue circles, respectively. Hypsoblennius gilberti individuals collected in northern California and southern California + Mexico are labeled with blue and purple circles, respectively (see Figure 1 for label colors).
Population Structure
Phylogenetic approach: As mentioned above, divergence between Sea of Cortez and Pacific populations has been shown in Hypsoblennius jenkinsi based on allozymes and mitochondrial markers (Present, 1987; Bernardi et al., 2003). Here we report the same results for the mitochondrial marker, where Sea of Cortez and Pacific populations show no evidence of gene flow by being reciprocally monophyletic. The nuclear marker, in contrast, does not show genetic divergence between the two populations and reduction of gene flow between these populations was not observed. This is not unusual, since the effective population size, and consequently lineage sorting, are very different between mitochondrial and nuclear markers (Avise, 2004; Bernardi and Lape, 2005). For the other disjunct species, H. gentilis, Pacific and Sea of Cortez populations did not sort in reciprocally monophyletic clades for either types of molecular markers.
Gene flow approach. Calculated gene flow based on mitochondrial markers between disjunct populations of H. gentilis was very low (Φst = 0.1379, p = 0.045). Similarly to H. jenkinsi, there was no evidence of reduced gene flow based on the nuclear marker. For the third species that is found along the Pacific coast, H. gilberti, we did not find evidence of reduced gene flow along the California and Baja California coast. Indeed, we evaluated the amount of gene flow across a known genetic break, the Los Angeles Area (Bernardi, 2000, 2005; Dawson, 2001; Dawson et al., 2002), between the northern populations of Monterey, Gaviota, and Santa Barbara vs. all the remaining populations to the South of Los Angeles (Φst = 0.0939, p = 0.153).
Historical Demography
While molecular clocks need to be used with caution, they provide insight into the history of populations (Crandall et al., 2012). Here we used the geminate species H. brevipinnis and H. invemar for internal calibration of the Hypsoblennius molecular clock. Indeed Transisthmian species separated between approximately 3.5 and 2.8 Mya, and their genetic divergence has been used as a yardstick to assess the remainder of a given phylogenetic tree (Lessios, 2008). The genetic divergence (model HKY+G) between the two geminate Hypsoblennius species was found to be 22.46%. This corresponds to an average divergence of 6.4–8.0% sequence divergence per million year (My). Sequence divergence between the Pacific and Sea of Cortez populations of H. jenkinsi was 5.61%. Using a molecular clock calibrated as described above, the divergence between Pacific and Sea of Cortez populations of H. jenkinsi was estimated to have occurred between 8,76,000 and 7,01,000 years ago.
Since disjunct populations of H. gentilis are not reciprocally monophyletic, we could not use the same method as described above for H. jenkinsi. Instead, we estimated coalescence time for the Sea of Cortez population of H. gentilis, as was done for other fish species (Bernardi and Lape, 2005). Mutation rates based on the Trans-Isthmian geminates Hypsoblennius brevipinnis and H. invemar, and assuming their generation time being one year, were estimated at 3.2 × 10−8 to 4.0 × 10−8 substitutions per site per generation. This rate is similar to the one provided for other fish species (Bernardi and Lape, 2005; Domingues et al., 2005). Using this substitution rate, we found that the coalescence time of the Sea of Cortez population of H. gentilis ranged between 1.9 and 1.5 Mya (Table 2).
Discussion
While the three focal Hypsoblennius species seem superficially very similar in most respects, some unique characteristics must have played a role in defining differences in distribution and population genetics. Three main features characterize the species. First, the presence of disjunct populations for Hypsoblennius gentilis and H. jenkinsi, second the population structure of the disjunct populations is different between those two species, and lastly for H. gilberti, the lack of population structure along a very large stretch of coast, from Monterey to the southern Pacific coast of Baja California, which corresponds to approximately 2000 km of coastline.
Baja California Disjunction
Habitat preference differs in the two disjunct species. Hypsoblennius jenkinsi is mostly subtidal, while H. gentilis is found both in the intertidal, as well as the subtidal over sandy bottoms. While the northern Sea of Cortez experiences extreme tidal fluxes and harbors long stretches of suitable habitat, the southern part of the Sea of Cortez has nearly no tides, thus becoming unsuitable for species that are relying on intertidal habitats. The divergence between disjunct populations of H. jenkinsi is estimated to have occurred approximately 7,00,000–9,00,000 years ago, while the Sea of Cortez population of H. gentilis is approximately 1.5–1.9 My old. Therefore, the divergence between disjunct populations is ancient and is likely to have occurred during on the Baja California seaway opening events, such as the midpeninsular seaway which was present around 1Mya (Upton and Murphy, 1997; Bernardi et al., 2003). Yet the reciprocal monophyly in H. jenkinsi and the lack of reciprocal monophyly in H. gentilis disjunct populations may be a reflection of a complete cessation of gene flow in the former species and a continued (but limited) gene flow in the latter species. These results would be consistent with the idea that the absence of suitable habitat in the south prevents H. jenkinsi to ever round the southern tip of the Baja California Peninsula, while it may not be the case for H. gentilis, which prefers sandy habitats. This result is also consistent with ecological characteristics of H. jenkinsi mentioned above, where individuals that were moved by just a few meters could not find their way back to their original pools (Stephens et al., 1970). Alternative hypotheses are also possible. For example, differences in effective population size, or the number of colonists that founded disjunct populations may have had a strong influence on the genetic background observed today. With additional demographic data, these possibilities could indeed be teased out, but dealing with cryptic species (in the ecological sense, small, well camouflaged individuals) it is easy to make major errors in estimating population sizes in the field.
The case of H. gilberti
It is puzzling that while three sympatric species of Hypsoblennius are found on the Pacific coast, two have disjunct populations and one does not. Hypsoblennius gilberti, which is restricted to the Pacific coast of California and Baja California, exhibits large territories, and also displays high levels of gene flow (this study), suggesting large dispersal potential. Yet, strong homing behavior even when displaced by large distances, may be an indication of philopatric behavior that counters easy dispersal. In addition, its preference for the higher intertidal restricts this species to very particular habitats that are neither continuous, nor conducive to dispersal. Altogether, these ecological characteristics may have played a role in preventing dispersal of this species across the Baja California peninsula when an opportunity arose.
Speciation
Baja California disjunctions have been used as a model to understand the early steps of speciation. Dispersal in marine fishes has traditionally been equated to the duration of the pelagic larval stage (Waples, 1987; Doherty et al., 1995; Shulman and Bermingham, 1995; Riginos and Victor, 2001), yet recent work has shown that adult dispersal and ecology are often better predictors of phylogeographic patterns (Schinske et al., 2010; Luiz et al., 2013). In this study, the use of phylogenetically closely related species allows for comparisons that remove a number of ecological variables. The pelagic larval duration (PLD) of the species is shorter in H. jenkinsi that in the other two species, which may provide some explanation for the difference in patterns between the two disjunct species, H. jenkinsi and H. gentilis, yet H. gentilis and H, gilberti are supposed to have identical PLDs and show very different patterns (the former has disjunct populations while the latter does not). Therefore, we do not consider that PLDs are good predictors for phylogeographic patterns in this case also. The lack of intertidal habitat in the south of our area of investigation and its associated lack of suitable habitat is likely to have played a role in maintaining the disjunction that was probably started during a vicariant event that happened approximately 1–1.5 Mya (a time that matches the presence of a midpeninsular seaway).
In this study, the use of phylogenetically closely related species allowed to critically assess the role of adult habitat in shaping the evolutionary history of Hypsoblennius. Ecological requirements of the different species seem to have played an essential role in shaping the currently observed distribution and genetic patterns of those species.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is dedicated to the memory of our dear friend and colleague Vitor Almada. We would like to thank all the colleagues that provided samples for this study over the course of almost two decades, Christy Bell, Ricardo Beldade, Gregorio Benavides, Greg Boland, Michelle Buckhorn, Ken Clifton, Karen Crow, Marina Ramon, José Tavera, Ben Victor, and Alessio Bernardi. We would like to thank the Scripps Institution of Oceanography and HJ Walker for giving us access to samples from their collection. This work was funded by a UCSC COR grant.
References
Allen, G. R., and Robertson, D. R. (1994). Fishes of the Tropical Eastern Pacific. Honolulu: University of Hawaii Press.
Almada, V. C., Robalo, J. I., Levy, A., Freyhof, J., Bernardi, G., and Doadrio, I. (2009). Phylogenetic analysis of peri-Mediterranean blennies of the genus Salaria: molecular insights on the colonization of freshwaters. Mol. Phylogenet. Evol. 52, 424–431. doi: 10.1016/j.ympev.2009.03.029
Avise, J. C. (2000). Phylogeography: The History and Formation of Species. Harvard University Press, Cambridge, MA.
Avise, J. C. (2004). Molecular Markers, Natural History, and Evolution. 2nd Edn. Sunderland, MA: Sinauer.
Balart, C. V. Æ. E., Sandoval-Castillo, J., Rocha-Olivares, A., Villavicencio-Garayzar, C., and Balart, E. (2004). Cryptic isolation of Sea of Cortez shovelnose guitarfish evidenced by mitochondrial DNA. Mar. Biol. 145, 983–988. doi: 10.1007/s00227-004-1378-7
Bernardi, G. (2000). Barriers to gene flow in Embiotoca jacksoni, a marine fish lacking a pelagic larval stage. Evolution 54, 226–237. doi: 10.1554/0014-3820(2000)054%5B0226:BTGFIE%5D2.0.CO;2
Bernardi, G. (2005). Phylogeography and demography of sympatric sister surfperch species, Embiotoca jacksoni and E. lateralis along the California coast: historical versus ecological factors. Evolution 59, 386–394. doi: 10.1111/j.0014-3820.2005.tb00997.x
Bernardi, G., Findley, L., and Rocha-Olivares, A. (2003). Vicariance and dispersal across Baja California in disjunct marine fish populations. Evolution 57, 1599–1609. doi: 10.1111/j.0014-3820.2003.tb00367.x
Bernardi, G., and Lape, J. (2005). Tempo and mode of speciation in the Baja California disjunct fish species Anisotremus davidsonii. Mol. Ecol. 14, 4085–4096. doi: 10.1111/j.1365-294X.2005.02729.x
Bird, C. E., Holland, B. S., Bowen, B. W., and Toonen, R. J. (2011). Diversification of sympatric broadcast-spawning limpets (Cellana spp.) within the Hawaiian archipelago. Mol. Ecol. 20, 2128–2141. doi: 10.1111/j.1365-294X.2011.05081.x
Bowen, B. W., Shanker, K., Yasuda, N., Celia, M., Malay, M. C. (Machel) D., von der Heyden, S., et al. (2014). Phylogeography unplugged: comparative surveys in the genomic era. Bull. Mar. Sci. 90, 13–46. doi: 10.5343/bms.2013.1007
Castillo-Páez, A., Sosa-Nishizaki, O., Sandoval-Castillo, J., Galván-Magaña, F., Blanco-Parra, M. P., and Rocha-Olivares, A. (2014). Strong population structure and shallow mitochondrial phylogeny in the banded guitarfish, Zapteryx exasperata (Jordan y Gilbert, 1880), from the Northern Mexican Pacific. J. Hered. 105, 91–100. doi: 10.1093/jhered/est067
Chow, S., and Hazama, K. (1998). Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol. 7, 1255–1256.
Crandall, E. D., Sbrocco, E. J., Deboer, T. S., Barber, P. H., and Carpenter, K. E. (2012). Expansion dating: calibrating molecular clocks in marine species from expansions onto the Sunda Shelf Following the Last Glacial Maximum. Mol. Biol. Evol. 29, 707–719. doi: 10.1093/molbev/msr227
Crow, K. D., Munehara, H., and Bernardi, G. (2010). Sympatric speciation in a genus of marine reef fishes. Mol. Ecol. 19, 2089–2105. doi: 10.1111/j.1365-294X.2010.04611.x
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
Dawson, M. N. (2001). Phylogeography in coastal marine animals: a solution from California? J. Biogeogr. 28, 723–736. doi: 10.1046/j.1365-2699.2001.00572.x
Dawson, M. N. (2012). Parallel phylogeographic structure in ecologically similar sympatric sister taxa. Mol. Ecol. 21, 987–1004. doi: 10.1111/j.1365-294X.2011.05417.x
Dawson, M. N., Louie, K. D., Barlow, M., Jacobs, D. K., and Swift, C. C. (2002). Comparative phylogeography of sympatric sister species, Clevelandia ios and Eucyclogobius newberryi (Teleostei, Gobiidae), across the California Transition Zone. Mol. Ecol. 11, 1065–1075. doi: 10.1046/j.1365-294X.2002.01503.x
Doherty, P. J., Planes, S., and Mather, P. (1995). Gene flow and larval duration in seven species of fish from the Great Barrier Reef. Ecology 76, 2373–2391. doi: 10.2307/2265814
Domingues, V. S., Bucciarelli, G., Almada, V. C., and Bernardi, G. (2005). Historical colonization and demography of the Mediterranean damselfish, Chromis chromis. Mol. Ecol. 14, 4051–4063. doi: 10.1111/j.1365-294X.2005.02723.x
Eble, J. A., Toonen, R. J., and Bowen, B. W. (2009). Endemism and dispersal: comparative phylogeography of three surgeonfishes across the Hawaiian Archipelago. Mar. Biol. 156, 689–698. doi: 10.1007/s00227-008-1119-4
Egger, B., Koblmüller, S., Sturmbauer, C., and Sefc, K. M. (2007). Nuclear and mitochondrial data reveal different evolutionary processes in the Lake Tanganyika cichlid genus Tropheus. BMC Evol. Biol. 7:137. doi: 10.1186/1471-2148-7-137
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
Froese, R., and Pauly, D. (2000). FishBase 2000: Concepts, Design and Data Sources. Los Baños: ICLARM.
Gaffney, P. M., Rupnow, J., and Domeier, M. L. (2007). Genetic similarity of disjunct populations of the giant sea bass Stereolepis gigas. J. Fish Biol. 70, 111–124. doi: 10.1111/j.1095-8649.2007.01393.x
Grismer, L. L. (2000). Evolutionary biogeography on Mexico's Baja California peninsula: a synthesis of molecules and historical geology. Syst. Zool. 97, 14017–14018. doi: 10.1073/pnas.260509697
Hastings, P. (2009). “Biogeography of New World Blennies,” in The Biology of Blennies, eds R. A. Patzner, E. J. Gonçlaves, P. A. Hastings, and B. G. Kapoor (Enfield, NH: Science Publishers), 95–118.
Huang, D., and Bernardi, G. (2001). Disjunct Sea of Cortez-Pacific Ocean Gillichthys mirabilis populations and the evolutionary origin of their Sea of Cortez endemic relative, Gillichthys seta. Mar. Biol. 138, 421–428. doi: 10.1007/s002270000454
Hurtado, L. A., Mateos, M., and Santamaria, C. A. (2010). Phylogeography of supralittoral rocky intertidal Ligia isopods in the pacific region from central California to central Mexico. PLoS ONE 5:e11633. doi: 10.1371/journal.pone.0011633
Jones, G. P., Milicich, M. J., Emslie, M. J., and Lunow, C. (1999). Self-recruitment in a coral reef fish population. Nature 402, 802–804. doi: 10.1038/45538
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment basedon fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kuhner, M. K. (2006). LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22, 768–770. doi: 10.1093/bioinformatics/btk051
Lee, W. J., Conroy, J., Howell, W. H., and Kocher, T. D. (1995). Structure and evolution of teleost mitochondrial control regions. J. Mol. Evol. 41, 54–66. doi: 10.1007/BF00174041
Lessios, H. A. (2008). The great American schism: divergence of marine organisms after the rise of the Central American Isthmus. Annu. Rev. Ecol. Evol. Syst. 39, 63–91. doi: 10.1146/annurev.ecolsys.38.091206.095815
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Losey, G. S. (1969). Sexual pheromone in some fishes of the genus Hypsoblennius. Science 163, 181–183. doi: 10.1126/science.163.3863.181
Love, M. S. (2011). Certainly more than you Want to Know About the Fishes of the Pacific Coast. Santa Barbara, CA: Really Big Press.
Luiz, O. J., Allen, A. P., Robertson, D. R., Floeter, S. R., Kulbicki, M., Vigliola, L., et al. (2013). Adult and larval traits as determinants of geographic range size among tropical reef fishes. Proc. Natl. Acad. Sci. U.S.A. 110, 16498–16502. doi: 10.1073/pnas.1304074110
Luiz, O. J., Madin, J. S., Robertson, D. R., Rocha, L. A., Wirtz, P., and Floeter, S. R. (2011). Ecological traits influencing range expansion across large oceanic dispersal barriers: insights from tropical Atlantic reef fishes. Proc. R. Soc. B Biol. Sci. 279, 1033–1040. doi: 10.1098/rspb.2011.1525
McCosker, J. E., and Dawson, C. E. (1975). Biotic passage through the Panama Canal, with particular reference to fishes. Mar. Biol. 30, 343–351. doi: 10.1007/BF00390639
Miller, D. J., and Lea, R. N. (1972). “Guide to coastal marine fishes of California,” in Fish Bulletin 157 (Oakland, CA: Department of Fish and Game), 249.
Muñiz-Salazar, R., Talbot, S. L., Sage, G. K., Ward, D. H., and Cabello-Pasini, A. (2005). Population genetic structure of annual and perennial populations of Zostera marina L. along the Pacific coast of Baja California and the Sea of Cortez. Mol. Ecol. 14, 711–722. doi: 10.1111/j.1365-294X.2005.02454.x
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Peterson, D. L., Kubow, K. B., Connolly, M. J., Kaplan, L. R., Wetkowski, M. M., Leong, W., et al. (2013). Reproductive and phylogenetic divergence of tidepool copepod populations across a narrow geographical boundary in Baja California. J. Biogeogr. 40, 1664–1675. doi: 10.1111/jbi.12107
Poortvliet, M., Longo, G. C., Selkoe, K., Barber, P. H., Crow, W., Caselle, J. E., et al. (2013). Phylogeography of the California sheephead, Semicossyphus pulcher: the role of deep reefs as stepping stones and pathways to antitropicality. Ecol. Evol. 3, 4558–4571. doi: 10.1002/ece3.840
Posada, D., and Crandall, K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Present, T. M. C. (1987). Genetic differentiation of disjunct Sea of Cortez and Pacific outer coast populations of Hypsoblennius jenkinsi. Copeia 1987, 1010–1024. doi: 10.2307/1445565
Prugnolle, F., and de Meeus, T. (2002). Inferring sex-biased dispersal from population genetic tools: a review. Heredity 88, 161–165. doi: 10.1038/sj.hdy.6800060
Recuero, E., Martínez-Solano, I., Parra-Olea, G., and García-París, M. (2006). Phylogeography of Pseudacris regilla (Anura: Hylidae) in western North America, with a proposal for a new taxonomic rearrangement. Mol. Phylogenet. Evol. 39, 293–304. doi: 10.1016/j.ympev.2005.10.011
Riddle, B. R., Hafner, D. J., Alexander, L. F., and Jaeger, J. R. (2000). Cryptic vicariance in the historical assembly of a Baja California peninsular desert biota. Proc. Natl. Acad. Sci. U.S.A. 97, 14438–14443. doi: 10.1073/pnas.250413397
Riginos, C., and Victor, B. C. (2001). Larval spatial distributions and other early life-history characteristics predict genetic differentiation in eastern Pacific blennioid fishes. Proc. R. Soc. B Biol. Sci. Lond. 268, 1931–1936. doi: 10.1098/rspb.2001.1748
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schinske, J. N., Bernardi, G., Jacobs, D. K., and Routman, E. J. (2010). Phylogeography of the diamond turbot (Hypsopsetta guttulata) across the Baja California Peninsula. Mar. Biol. 157, 123–134. doi: 10.1007/s00227-009-1302-2
Selkoe, K., and Toonen, R. (2011). Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Mar. Ecol. Prog. Ser. 436, 291–305. doi: 10.3354/meps09238
Shanks, A. L., and Eckert, G. L. (2005). Population persistence of California current fishes and benthic crustaceans: a marine drift paradox. Ecol. Monogr. 75, 505–524. doi: 10.1890/05-0309
Shulman, M. J., and Bermingham, E. (1995). Early life histories, ocean currents, and the population genetics of Caribbean reef fishes. Evolution 49, 897–910. doi: 10.2307/2410412
Stephens, J. S. Jr., Johnson, R. K., Key, G. S., and McCosker, J. E. (1970). The comparative ecology of three sympatric species of California blennies of the genus Hypsoblennius Gill (Teleostomi, Blenniidae). Ecol. Monogr. 40, 213–233. doi: 10.2307/1942296
Stepien, C. A., Rosenblatt, R. H., and Bargmeyer, B. A. (2001). Phylogeography of the spotted sand bass, Paralabrax maculatiofasciatus: divergence of Sea of Cortez and Pacific coast populations. Evolution 55, 1852–1862. doi: 10.1111/j.0014-3820.2001.tb00834.x
Swearer, S. E., Caselle, J. E., Lea, D. W., and Warner, R. R. (1999). Larval retention and recruitment in an island population of a coral-reef fish. Nature 402, 799–802. doi: 10.1038/45533
Terry, A., Bucciarelli, G., and Bernardi, G. (2000). Restricted gene flow and incipient speciation in disjunct Pacific Ocean and Sea of Cortez populations of a reef fish species, Girella nigricans. Evolution (N.Y.) 54, 652–659. doi: 10.1111/j.0014-3820.2000.tb00066.x
Thomson, D. A., Findley, L. T., and Kerstitch, A. N. (2000). Reef fishes of the Sea of Cortez. The Rocky-shore Fishes of the Sea of Cortez. Tucson, AZ: The University of Arizona Press.
Toonen, R. J., Andrews, K., R., Baums, I., B., Christopher, E., Concepcion, G. T., et al. (2011). Defining boundaries for ecosystem-based management: a multispecies case study of marine connectivity across the Hawaiian archipelago. J. Mar. Biol. 2011, 13. doi: 10.1155/2011/460173
Upton, D. E., and Murphy, R. W. (1997). Phylogeny of the side-blotched lizards (Phrynosomatidae:Uta) based on mtDNA sequences: support for midpeninsular seaway in Baja California. Mol. Phylogenet. Evol. 8, 104–113. doi: 10.1006/mpev.1996.0392
Von der Heyden, S., Bowie, R. C. K., Prochazka, K., Bloomer, P., Crane, N. L., and Bernardi, G. (2011). Phylogeographic patterns and cryptic speciation across oceanographic barriers in South African intertidal fishes. J. Evol. Biol. 24, 2505–2519. doi: 10.1111/j.1420-9101.2011.02382.x
Waples, R. S. (1987). A multispecies approach to the analysis of gene flow in marine shore fishes. Evolution (N.Y.) 41, 385–400. doi: 10.2307/2409146
Wares, J. P., and Cunningham, C. W. (2001). Phylogeography and historical ecology of the North Atlantic intertidal. Evolution 55, 2455–2469. doi: 10.1111/j.0014-3820.2001.tb00760.x
Keywords: Baja California, population disjunctions, phylogeography, speciation, control region, ribosomal S7 protein
Citation: Bernardi G (2014) Baja California disjunctions and phylogeographic patterns in sympatric California blennies. Front. Ecol. Evol. 2:53. doi: 10.3389/fevo.2014.00053
Received: 10 June 2014; Accepted: 13 August 2014;
Published online: 29 August 2014.
Edited by:
Laura M. Boykin, The University of Western Australia, AustraliaReviewed by:
Octavio Salgueiro Paulo, Universidade de Lisboa, PortugalRyan C. Garrick, University of Mississippi, USA
Copyright © 2014 Bernardi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giacomo Bernardi, Department of Ecology and Evolutionary Biology, University of California Santa Cruz, 100 Shaffer Road, Santa Cruz, CA 95060, USA e-mail:YmVybmFyZGlAYmlvbG9neS51Y3NjLmVkdQ==
 Giacomo Bernardi
Giacomo Bernardi