- 1Laboratory of Natural-Focal Viral Infections, M.Aikymbayev's National Scientific Center for Especially Dangerous Infections, Almaty, Kazakhstan
- 2Department of Epizootology of Especially Dangerous Infections with the Museum and Insectarium, M.Aikymbayev's National Scientific Center for Especially Dangerous Infections, Almaty, Kazakhstan
- 3Department of Biostatistics and GIS, M.Aikymbayev's National Scientific Center for Especially Dangerous Infections, Almaty, Kazakhstan
- 4Laboratory of Applied Genetics, National Center for Biotechnology, Astana, Kazakhstan
- 5M.Aikymbayev's National Scientific Center for Especially Dangerous Infections, Almaty, Kazakhstan
The natural foci of Crimean-Congo haemorrhagic fever (CCHF) in Kazakhstan are geographically located in the southern regions of the country (Kyzylorda, Turkestan and Zhambyl regions), where the infection of ticks with the CCHF virus predominantly reside, tick species composition and the number of vectors are monitored annually. The objective of our research was to investigate the genetic variants of the CCHF virus in the southern endemic regions, as well as to monitor the spread of the CCHF virus in the western regions of the country (Aktobe, Atyrau and Mangystau regions). In total, 974 (216 pools) ticks from the western regions and 3527 (583 pools) ticks from the southern regions collected during 2021–2022 were investigated. The presence of CCHF virus was detected by real-time reverse transcription PCR (qRT- PCR) in 1 pool out of 799 pools (0.12%) with Hyalomma scupense ticks captured in the CCHF-endemic Kyzylorda region. In the western regions, CCHF virus was not detected in ticks. The sequencing of incomplete fragments of the S, M and L segments of the CCHF virus in the detected virus was identified as genotype Asia - I. Phylogenetic analysis showed that the isolate obtained in this study is grouped with the isolate from a patient with CCHF, which we reported in 2015 (KX129738 Genbank). Our findings highlight the importance of including sequencing in the annual monitoring system for better understanding the evolution of the CCHF virus in the study areas of our country.
Introduction
Emerging and re-emerging human viral infections are one of the global health challenges (1–3). The gravity of such infections is due to the sudden appearance, rapid spread and often the lack of specific protection and prevention. The main source of new human viruses are zoonotic viruses, which, under the influence of a combination of factors, acquire human-pathogenic properties and epidemic potential (4, 5). A significant proportion of the causative agents of these infections are arboviruses, mainly containing RNA, which are transmitted to humans through blood-sucking arthropod vectors. The Crimean-Congo haemorrhagic fever virus (CCHF) belongs to the genus Orthonairovirus, family Nairoviridae, order Bunyavirales (6). Due to the fact that the majority of modern vaccines are at the stage of development and clinical trials (7), and the strategy for creating effective antiviral drugs is experiencing certain difficulties along the way (8), according to the WHO, this disease is considered to be a potential public health emergency. There are several classes of drugs with antiviral activities against different viruses, including CCHF virus, such as nucleoside analogues (ribavirin, favipiravir, 2′-deoxy-2′-fluorocytidine), ovarian tumor (OTU) domain proteases (phenanthrenequinone compounds), interferons, immunotherapy (convalescent serum, monoclonal antibodies) and a few anti-malarial and anti-psychotic drugs (chloroquine and chlorpromazine) (8). Ribavirin is widely used in Kazakhstan as anti-CCHF viral medicine due to its low cost and accessibility, and is considered the drug of choice in our country.
Crimean-Congo haemorrhagic fever is one of the most significant natural focal viral infections in Kazakhstan. Outbreaks of the disease are annually reported in three endemic regions (Kyzylorda, Turkestan and Zhambyl regions) geographically located in the southern part of the country. The main vectors of CCHF virus in Kazakhstan are ticks of the Hyalomma genus (Hyalomma asiaticum, H.scupense, H.anatolicum). These tick species are pasture-stall arthropods with a two- or three-host life cycle. The natural habitat of the Hyalomma spp. in Kazakhstan are deserts and steppes. These ticks are also successfully adapted to inhabit in populated areas and their environs. These arthropods actively parasitize both domestic cattle and small ruminants, as well as wild animals (kulans, saigas, goitered gazelles, etc.) in Kazakhstan (9).
The committee for veterinary control and supervision of the Ministry of Agriculture and national centers of expertise together with anti-plague stations of the Ministry of Health are responsible for monitoring of situations around CCHF in Kazakhstan. The annual treatment of farm animals with pesticides in CCHF foci does not give the proper level of infection reduction, and the increased economic activity of the local population in CCHF natural foci in recent years has led to an increase in human contacts with arthropod vectors that are not only the vectors but also the reservoirs of the virus. As a result, human cases of CCHF are being reported in areas where the disease has not been seen for many decades. We set the following tasks for the implementation of the national scientific program: to collect ticks in the southern regions endemic for CCHF (Kyzylorda, Turkestan and Zhambyl regions) and non-endemic western regions (Aktobe, Atyrau and Mangystau regions), to determine the species composition of captured ticks, to monitor the infection of ticks with the CCHF virus by PCR, followed by sequencing of isolated virus isolates. The aim of our study was to investigate the ticks, collected from the southern endemic regions and western non-endemic regions for the presence of the genetic variants of the CCHF virus.
Materials and methods
Tick collection and species identification
Trapping of ticks was carried out in open areas of the countryside where there is vegetation, using the “flag” method in the southern (Kyzylorda, Turkestan and Zhambyl) and western (Aktobe, Atyrau and Mangystau) regions of Kazakhstan in April, May and June of 2021–2022. As a flag, a cotton fabric measuring 0.6 × 1.0 m was used, the narrow side of which was attached to a stick 1.25–1.50 m long. Every 5 m, the fabric was inspected for the presence of ticks. The collection of ticks from farm animals was carried out mainly in the yards of the local population and in private livestock farms. To do this, the animal was fixed and examined in typical places of tick bites: in the udder, on the inside of the thighs and in the groin. Ticks were removed with tweezers and collected into plastic tubes with a tightly screw lid. Animal housing facilities were also inspected for the presence of ticks, mainly fenced pens where animals are kept after they return from pasture. In the western regions of Kazakhstan (Aktobe, Atyrau and Mangystau regions), ticks were mainly removed from farm animals due to unstable weather in spring, accompanied by a sharp decrease and increase in temperature, and precipitation. The collected ticks were stored in the tubes at −20°C until further analysis. Species identification of ticks was carried out according to manuals (10–12) using a binocular stereoscopic microscope and by morphological features such as the pattern and shape of the scutum, festoons, capitulum, hypostome, palps, eyes, peritreme, legs etc. After that the ticks were grouped into pools of 5–10 ticks of the same species each. Geographic information system technologies were used for mapping the locations where ticks were collected. Electronic maps of Kazakhstan were applied as a topographic basis and included the following base layers: mathematical elements, hydrography and administrative structure. The obtained information on ticks was digitized by creating electronic databases using Excel program, and then adapted to work in the ArcMap program.
Viral RNA extraction from ticks
Tick pools were homogenized using a “TissueLyser II” (Qiagen, Hilden, Germany) homogenizer in 6 min at 300 Hz with the addition of stainless steel beads and 1 ml of cell growth medium (DMEM, Gibco™, Thermo Fisher Scientific, Waltham, MA, USA) to each sample. RNA was extracted from 140 μl tick homogenates using the commercial kit (QIAamp Viral RNAMini Kit; Qiagen, Hilden, Germany), according to the manufacturer's instruction and stored at −80°C.
Real-time reverse transcription PCR (qRT-PCR)
All PCR procedures were conducted in a three- room regime. The presence of CCHF viral RNA was determined according to the protocol from B. Atkinson et al. (13) for qRT-PCR using the LightCycler 2.0 instrument (Roche Applied Science, Switzerland). We used SuperScript III (SSIII) Platinum One-step qRT-PCR Kit (Invitrogen) to prepare the reaction mixture. The volume of the master mix (15 μl) included 10 μl of 2× reaction mixture, 1.7 μl of PCR-grade water, 1 μl of each primer (for 18 µM working concentration), 0.5 µl probe (for 25 µM working concentration) and 0.8 µl SuperScrip Taq Mix. After that, 5 μl of RNAwas added to the master mix to obtain a final reaction volume of 20 μl. The applied amplification program: at 50°C for 10 min., at 95°C for 2 min., then 45 cycles of 95°C for 10 s and at 60°C for 40 s (with fluorescence quantitation at the end of each step at 60°C) and the final stage of cooling at 40°C for 30 s.
Reverse transcription and sequencing
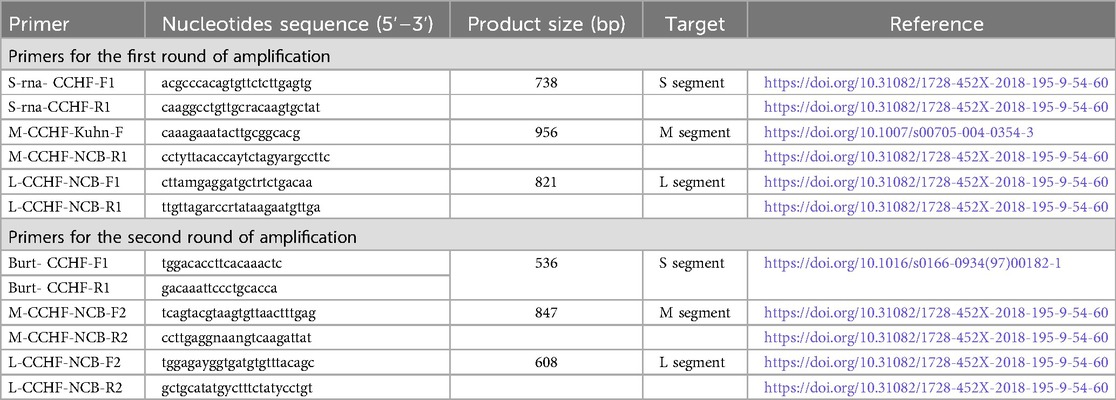
The extracted RNAsamples were converted into cDNAby reverse transcription using the commercial kit (Reverta L; AmpliSens, Russia). The S, L and M segments fragments were amplified using two-round PCR with CCHF virus-specific primers (Table 1) (14, 15). PCR was conducted in a volume of 30 µl. The reaction mixture contained 0.65-µm concentration of segment-specific primers for each round, 200-µm concentration of each deoxynucleoside triphosphate, 2.5 mm MgCl2 1× PCR buffer, 1× Q-Solution 1.5 U HotstarTaq plus DNA polymerase (Qiagen, Valencia, CA, USA), and 5 µl of cDNA or 5 µl of the product obtained from the first round of PCR. PCR was carried out with a “T100™ Thermal Cycler” (Bio-Rad, USA) by using the following temperature program—at 95°C for 5 min; 35 cycles: at 95°C for 25 s, at 58°C (at 49°C for the second round of PCR) for 1 min; and final polymerization at 72°C for 10 min. PCR products of the first and second stages were subjected to electrophoretic separation in 1.5% agarose. All amplified target fragments were sequenced. PCR products were purified with exonuclease I (Exo I, Thermo Fisher Scientific Baltics UAB) and alkaline phosphatase (Shrimp Alkaline Phosphatase, Thermo Fisher Scientific Baltics UAB) as described earlier (16). Purified PCR products were labeled with fluorescent dyes using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems™, USA). Samples were sequenced using an ABI 3730xl DNA Analyzer (Applied Biosystems™, USA). SeqMan II software (Lasergene, version 6.1; DNAstar) was used to analyze electrophorerograms and assemble contigs.
Phylogenetic analysis
The nucleotide sequences were aligned using the MUSCLE algorithm (17). Phylogenetic analysis was performed using the MEGA-X software (version 10.2.6) (18), and the evolutionary history was determined by the maximum likelihood (ML) method (19) with 1,000 bootstrap replicates. The identification of clades was carried out as previously described (20, 21). The Africa 4 clade was added to the S gene phylogenetic tree in accordance with the data of M. P. Sánchez-Seco et al. (22).
Results, discussion
All investigated ticks were collected in the field or removed from agricultural and wild animals over two seasons in the spring and fall of 2021–2022. The spring and fall tick collections were planned depending on weather conditions. We tried to choose days with the most optimal ambient temperature, when the greatest emergence of famished ticks was observed. It is considered the most favorable weather conditions for the emergence of ixodid ticks when the average ambient temperature is 16°C–26°C, especially for ticks of the genus Dermacentor (23). As for ticks of the genus Hyalomma, they are active throughout warm period of the year, from April to October, reaching maximum numbers in May–June (9). Consequently, the increase in the incidence of CCHF in Kazakhstan is observed precisely in May and June (24).
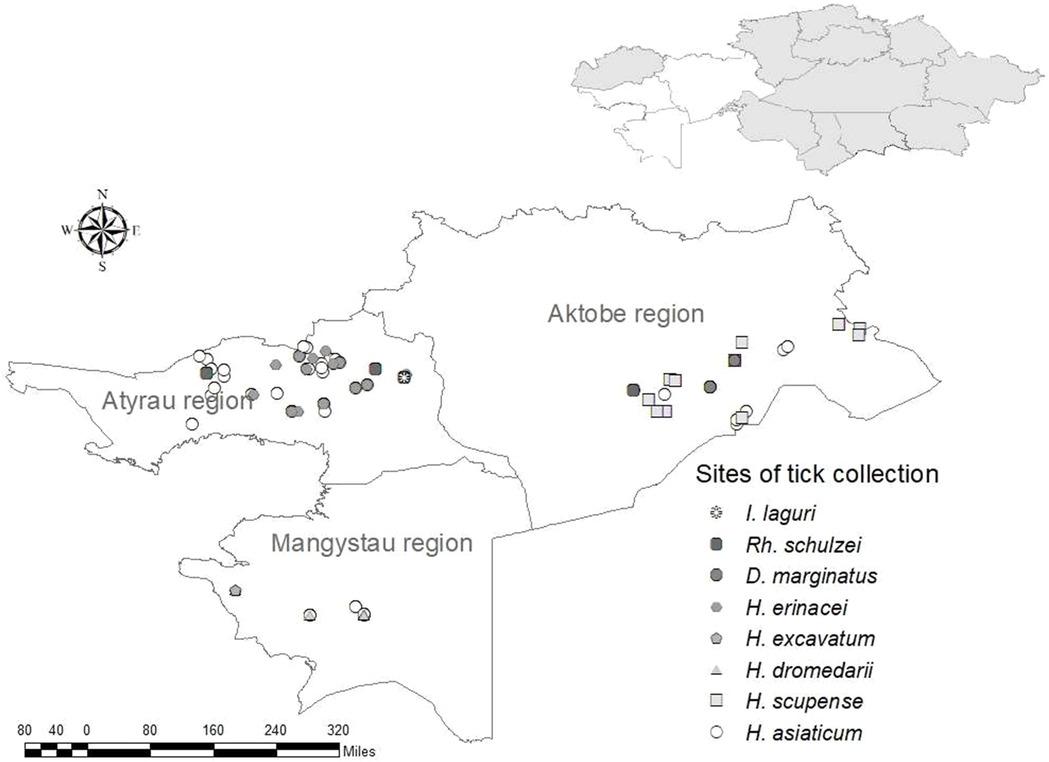
Out of the 974 ticks that were collected in the western area of Kazakhstan (Aktobe, Atyrau, Mangystau regions): 628 ticks were removed from camels, 90 ticks—from cows, 248 ticks—from large gerbils and their burrows and only 8 ticks in the field. Ticks of the genus Hyalomma were dominant in these three investigated regions. The number of collected ticks, species and pools are presented in Table 2.
Ticks of the Hyalomma genus are widespread in Kazakhstan and play an important role in the transfer of pathogens of many types of human and animal infections, including the CCHF virus (24, 25). Hyalomma asiaticum is the most widespread of the ticks of this genus known for the western area of the countryand was discovered there and collected by us in all three regions. This species is widely spread not only in Kazakhstan, but also in the adjacent territories of Kyrgyzstan, Uzbekistan, as well as in other countries of Central Asia (26–28), where it is a vector of the pathogens of many infectious diseases (9, 29, 30). H. excavatum and H. dromedari were collected only in the Mangystau region, and H. scupense—only in Aktobe region. Dermacentor marginatus was also collected by us in all three western regions, but only one specimen was found in the Mangystau region. This find is likely accidental, as this species is not usually live in this region. Haemaphysalis erinacei, Ixodes laguri and Rhipicephaluss schulzei are burrowing ticks that were found and collected from wild animals, mainly from rodents, their number was low and they were collected only in the Atyrau region (Figure 1).
The southern area of Kazakhstan is endemic for CCHF, where human cases of this disease are reported annually, and includes three regions: Kyzylorda, Turkestan and Zhambyl. The continued circulation of the CCHF virus in these regions is facilitated by favorable climatic conditions for vectors and their hosts and intensive breeding of farm animals.
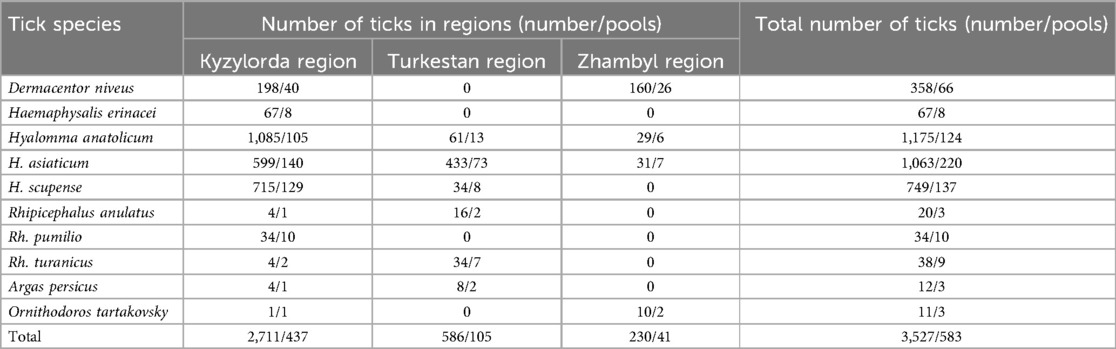
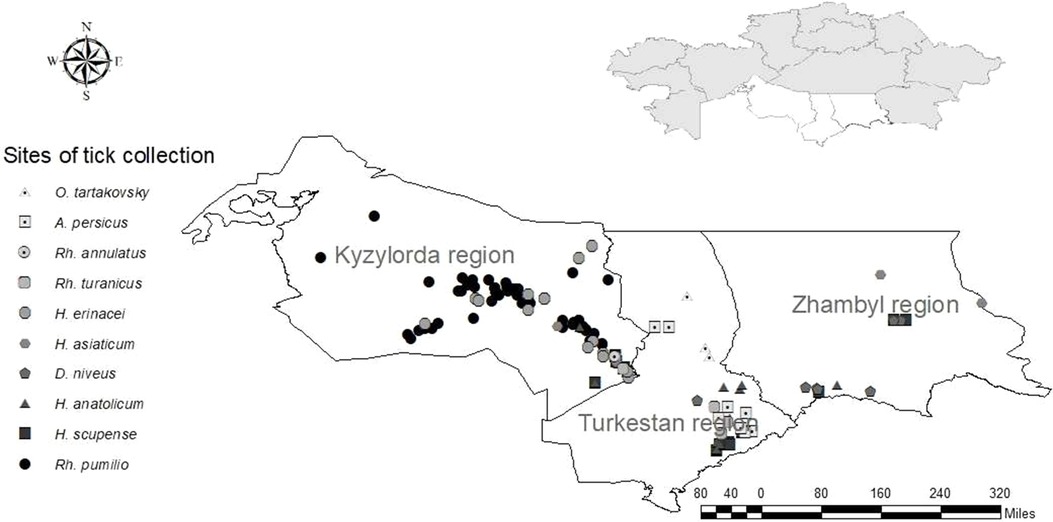
Out of the 3,527 ticks that were investigated from the southern area of the country (Kyzylorda, Turkestan and Zhambyl regions): 566 ticks were removed from agricultural animals, 1,190 ticks—from large gerbils and their burrows, 153 ticks were collected from animal housing facilities and 1,618 ticks—in the field. The diversity of the local natural and climatic landscape provides habitat for a large number of tick species. Ticks of the genus Hyalomma were also the most numerous in the southern area of Kazakhstan (Table 3).
H. asiaticum and H. anatolicum occupy a dominant position in distribution and were also discovered by us in all three southern regions of the country. H. scupense, which is also widespread in the south of Kazakhstan, was collected in the Kyzylorda and Turkestan regions and was inferior in number to the two previous species. Dermacentor niveus was collected in the Kyzylorda and Zhambyl regions, and ticks of the genus Rhipicephalus were collected in the Turkestan and Kyzylorda regions (Figure 2). In addition to ixodid ticks, we collected and investigated for the presence of the CCHF virus two species of argasid ticks—Argas persicus, collected from poultry keeping facilities, and Ornithodoros tartakovskiy—from rodent's burrows.
As a result of Real-time qPCR, 1 positive pool for the CCHF virus was obtained out of 799 investigated pools of ticks. The positive pool was from the ticks of Hyalomma scupense, which were collected in the endemic for CCHF Kyzylorda region. It was early reported that ticks of this species can transmit the CCHF virus in this region (31–33). The partial sequences of the S, M and L segments were obtained for this sample (GenBank accession numbers OR633376, OR633377 and OR633378). Phylogenetic analysisclustered the analyzed samples into the Asia 1 clade (Figures 3–5). The S-segment sequences (GenBank accession number OR633376) showed 99.43% identity at the nucleotide and aminoacid level, respectively, to the strain (GenBank accession number KX129738) previously isolated froma patient with CCHF in the Turkestan region (34) (Figure 3).
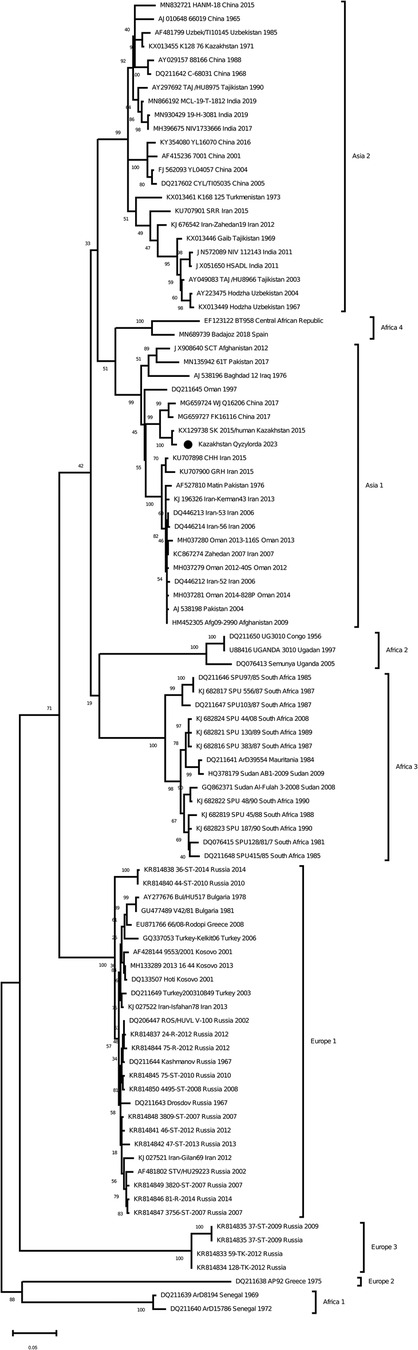
Figure 3. Phylogenetic analysis of the CCHF virus (32, 33). The phylogenetic tree was constructed using oligonucleotide sequences of the S gene of the CCHF virus from the ticks of Hyalomma scupense, collected in the southern regions of Kazakhstan, and shown as a black dot. Oligonucleotide sequences were aligned using the software MEGA version 6.0.
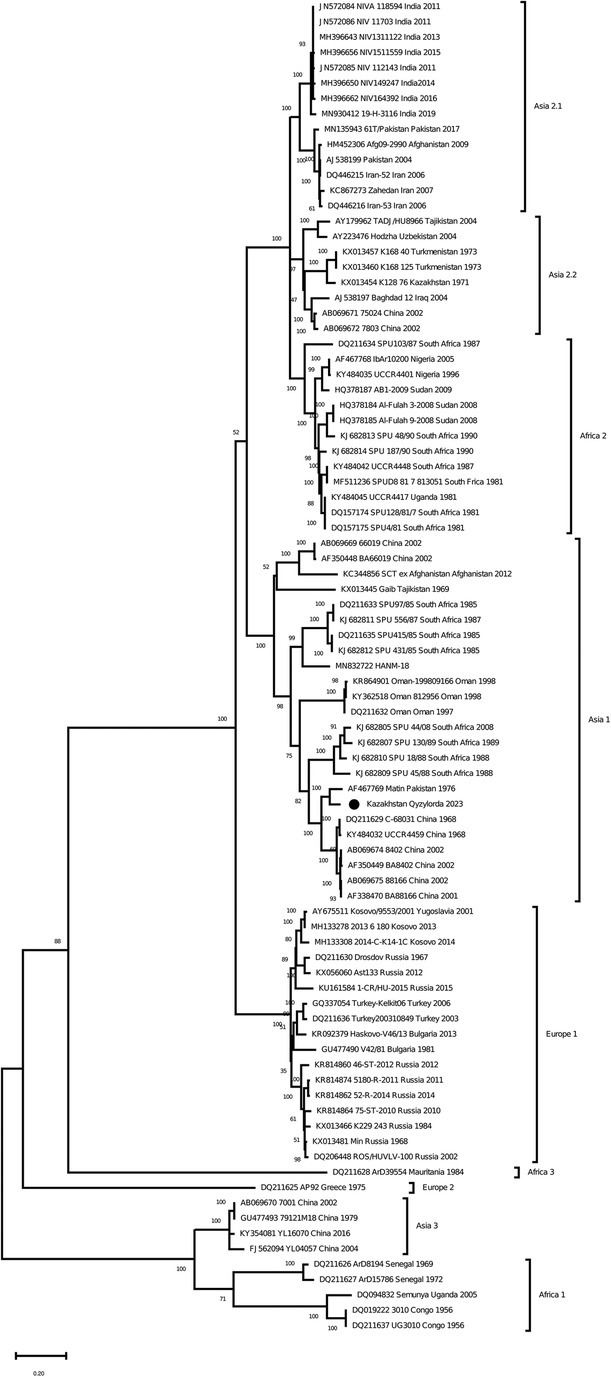
Figure 4. Phylogenetic analysis of the CCHF virus. The phylogenetic tree was constructed using oligonucleotide sequences of the M gene of the CCHF virus from the ticks of Hyalomma scupense, collected in the southern regions of Kazakhstan, and shown as a black dot. Oligonucleotide sequences were aligned using the software MEGA version 6.0.
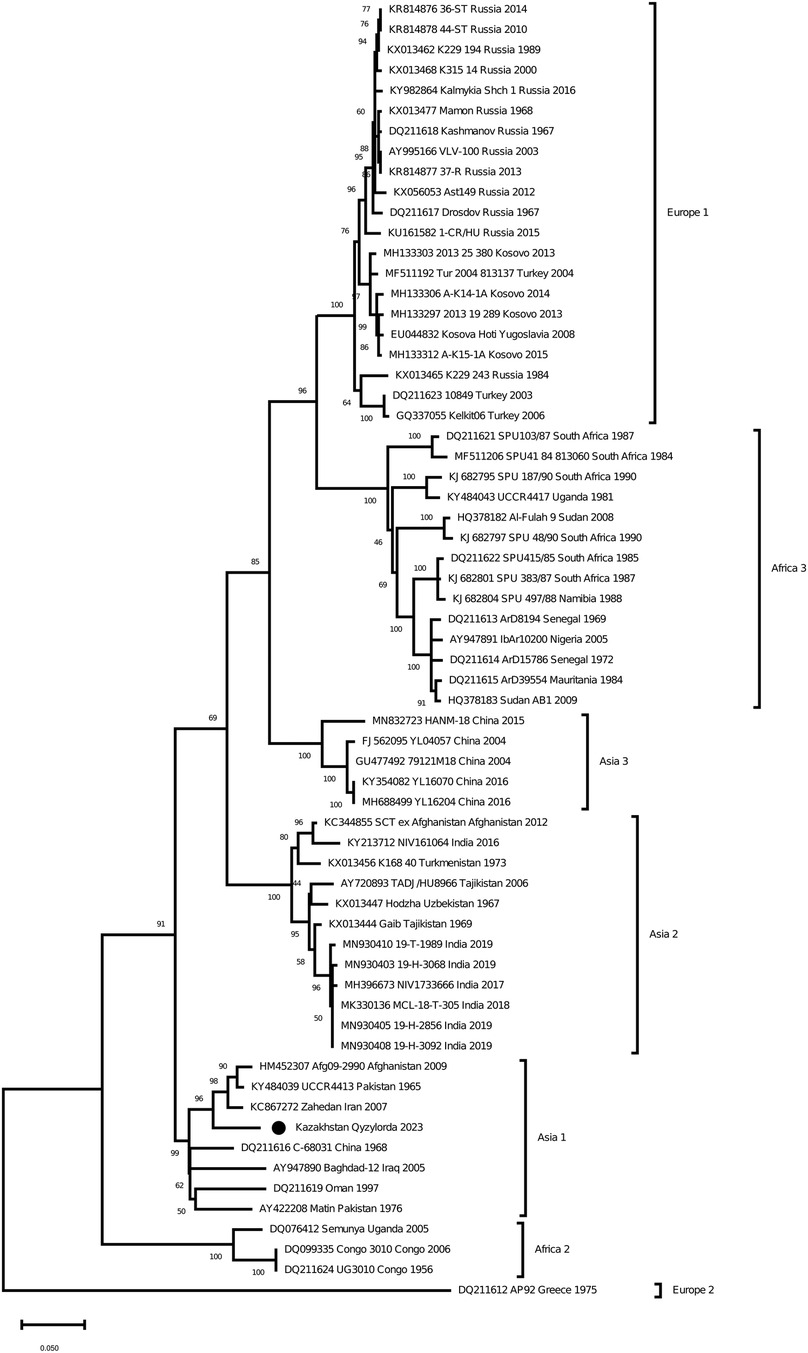
Figure 5. Phylogenetic analysis of the CCHF virus. The phylogenetic tree was constructed using oligonucleotide sequences of the L gene of the CCHF virus from the ticks of Hyalomma scupense, collected in the southern regions of Kazakhstan, and shown as a black dot. Oligonucleotide sequences were aligned using the software MEGA version 6.0.
The results confirm the circulation of the CCHF virus in the investigated endemic areas (34). The detection of the virus in the ticks of Hyalomma scupense has important epidemiological significance, since this species is a pasture-stall ectoparasite with a one- or two-host development cycle. Ticks of this species spend their life cycle in animal housing facilities and in the vicinity of populated areas. Their larvae and nymphs can feed on blood and molt on the same animal, as a result of which the emerging imagos, having had enough, are moved to the ground for reproduction in populated areas or its environs, where an exchange of ectoparasites between animals occurs. The indicated features of the life cycle of this species of ticks must be taken into account when carrying out anti-tick treatments.
The presence of geographically distant but genetically similar strains suggests that the CCHF viruses are spread either through the trade in livestock or through migratory birds. This hypothesis requires further in-depth investigation to confirm or disprove it.
The discovery of Asia 1 (clade VI) serogroup of CCHF virus in ticks in the Kyzylorda region, which has genetic similarity to strains isolated from a patient in 2015 in the Turkestan region, is important in understanding the evolution of this virus in the study areas.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent from the [patients/ participants OR patients/participants legal guardian/next of kin] was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
TN: Data curation, Investigation, Methodology, Supervision, Writing – original draft. NT: Data curation, Investigation, Writing – review & editing. ZS: Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. VS: Formal Analysis, Visualization, Writing – review & editing. AS: Investigation, Writing – review & editing. GT: Project administration, Supervision, Writing – review & editing. NT: Data curation, Investigation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
The research was carried out within the framework of program-targeted funding on the topic “Development and scientific substantiation of public health technologies, biological safety for the impact on the prevention of dangerous infectious diseases”. Research project IRN - BR11065207. Source of funding - Ministry of Health of the Republic of Kazakhstan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ma B, Hang C, Papa A. Sequencing and comparative analysis of the complete glycoprotein gene of three Crimean—Congo hemorrhagic fever virus Chinese isolates. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. (2001) 15:105–11. PMID: 11436635.11436635
2. L’vov DK. Emerging and re-emerging infections—a dozing volcano. Prob Espec Danger Infect. (2008) 96:5–8. (in Russian). doi: 10.21055/0370-1069-2008-2(96)-5-8
3. Shestakova IV. The infectious diseases morbidity in the Russian federation, 2000–2015: success or failure? Infect Dis. (2017) 3:11–20. (in Russian). doi: 10.24411/2305-3496-2017-00045
4. Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Philos Trans R Soc B. (2001) 356(1411):983–9. doi: 10.1098/rstb.2001.0888
5. Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. (2008) 451(7181):990–3. doi: 10.1038/nature06536
6. Garrison AR, Alkhovsky SV, Avšič-Županc T, Bente DA, Bergeron É, Burt F, et al. ICTV virus taxonomy profile: nairoviridae. J Gen Virol. (2020) 101:798–9. doi: 10.1099/jgv.0.001485
7. Dowall SD, Carroll MW, Hewson R. Development of vaccines against Crimean-Congo haemorrhagic fever virus. Vaccine. (2017) 35(44):6015–23. doi: 10.1016/j.vaccine.2017.05.031
8. Dai S, Deng F, Wang H, Ning Y. Crimean-Congo hemorrhagic fever virus: current advances and future prospects of antiviral strategies. Viruses. (2021) 13:1195. doi: 10.3390/v13071195
9. Kulemin MV, Rapoport LP, Vasilenko AV, Kobeshov ZV, Shukputov TM, Sailaubekuly R, et al. Ixodid ticks of agricultural animals in Southern Kazakhstan: the fauna structure, abundance, epizootological significance. Parasitologia. (2020) 54(1):25–31. (in Russian).
10. Walker AR, Bouattour A, Camicas J-L, Estrada-Peña A, Horak IG, Latif AA, et al. Ticks of Domestic Animals in Africa: A Guide to Identification of Species. Edinburgh: Bioscience Reports (2003). p. 221.
11. Apanaskevich DA. The role of preimaginal phases in the taxonomy of ixodid ticks of the genus hyalomma koch—vectors of pathogens. [thesis of a candidate of biological sciences]. St. Petersburg (2004). p. 274. (in Russian).
12. Sayakova ZZ. Identification of Ixodid Ticks in Kazakhstan. Methodical Manual. Almaty: Kazakh University (2020). p. 144. (in Kazakh).
13. Atkinson B, Chamberlain J, Logue CH, Cook N, Bruce C, Dowall SD, Hewson R. Development of a real-time RT-PCR assay for the detection of Crimean-Congo hemorrhagic fever virus. Vector Borne Zoonotic Dis. Larchmt. (2012) 12:786–93. doi: 10.1089/vbz.2011.0770
14. Kuhn JH, Seregin SV, Morzunov SP, Petrova ID, Vyshemirskii OI, Lvov DK, et al. Genetic analysis of the M RNA segment of Crimean Congo hemorrhagic fever virus strains involved in the recent outbreaks in Russia. Arch Virol. (2004) 149(11):2199–213. doi: 10.1007/s00705-004-0354-3
15. Burt FJ, Leman PA, Smith JF, Swanepoel R. The use of a reverse transcription-polymerase chain reaction for the detection of viral nucleic acid in the diagnosis of Crimean-Congo haemorrhagic fever. J Virol Methods. (1998) 70(2):129–37. doi: 10.1016/s0166-0934(97)00182-1
16. Lins TC, Nogueira LR, Lima RM, Gentil P, Oliveira RJ, Pereira RW. A multiplex single-base extension protocol for genotyping Cdx2, FokI, BsmI, ApaI, and TaqI polymorphisms of the vitamin D receptor gene. Genet Mol Res. (2007) 6(2):316–24. PMID: 17573662.17573662
17. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. (2013) 30(12):2725–9. doi: 10.1093/molbev/mst197
18. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
19. Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. (1980) 16:111–20. doi: 10.1007/BF01731581
20. Kajihara M, Simuunza M, Saasa N, Dautu G, Mori-Kajihara A, Qiu Y, et al. Serologic and molecular evidence for circulation of Crimean-Congo hemorrhagic fever virus in ticks and cattle in Zambia. PLoS Negl Trop Dis. (2021) 15(6):e0009452. doi: 10.1371/journal.pntd.0009452
21. Kong Y, Yan C, Liu D, Jiang L, Zhang G, He B, et al. Phylogenetic analysis of Crimean-Congo hemorrhagic fever virus in inner Mongolia, China. Ticks Tick Borne Dis. (2022) 13(1):101856. doi: 10.1016/j.ttbdis.2021.101856
22. Sánchez-Seco MP, Sierra MJ, Estrada-Peña A, Valcárcel F, Molina R, de Arellano ER, et al. Widespread detection of multiple strains of Crimean-Congo hemorrhagic fever virus in ticks, Spain. Emerg Infect Dis. (2021) 28(2):394–402. doi: 10.3201/eid2802.211308
23. Glazunov Yu V. Abiotic factors influencing the number of ixodid ticks of the genus dermacentor in the northern trans-urals. Young Sci. (2016) 26(130):167–70. (in Russian).
24. Nurmakhanov T, Sansyzbaev Y, Atshabar B, Kazakov S, Deryabin P, Zholshorinov A, et al. Crimean-Congo hemorrhagic fever virus in Kazakhstan (1948–2013). Int J Infect Dis. (2015) 38:19–23. doi: 10.1016/j.ijid.2015.07.007
25. Nurmakhanov TI, Sansyzbaev EB, Yeskhojayev OU, Vilkova AN, Sajlaubekuly R, Kulemin MV, et al. Genetic variants of the Crimean-Congo virus of hemorrhagic fever circulating in the territory of the south-Kazakhstan region. Medicine (Almaty). (2018) 9(195):54–60. (In Russ.). doi: 10.31082/1728-452X-2018-195-9-54-60
26. Mirzayeva AU, Azimov DA, Akramova FD, Mirkasimova K, Shakarbaev UA, Saidova S. Species diversity of ticks of the family ixodidae (acari: parasitiformes)—vectors of vector- borne diseases of farm animals and humans in the syrdarya region. The Path of Science. (2021) 6(88):35–8. (in Russian).
27. Iskandarov EKh. Ixodofauna of some species of wild animals of central Tajikistan. News Natl Acad Sci Tajikistan. (2022) 3(218):55–8. (in Russian).
28. Jalali MHR, Alborzi A, Hamidinejat H, Asadollahi Z, Boroujeni MP, Sazmand A. Study of cattle ixodid ticks in Khoozestan province, South-West of Iran. Acarina. (2014) 22(2):157–60.
29. Nurmakhanov TI, Erubayev TK, Sansyzbayev EB, Turebekov NA, Abdiyeva KS, Usenbekova DS, et al. Results oftick testing for detection of viruses Karshi, Tamdy, Issyk-kul fever, Syrdarya valley fever. Bull Karaganda Univ. (2021) 2(102):43–50. (in Russian). doi: 10.31489/2021BMG2/43-50
30. Kulemin MV, Abuova GN, Sarypbekova LL, Polukchi TV, Aliyev DS, Sadykhova DK, et al. Prevalence ofticks, vectors of the Crimean Congo hemorrhagic fever virus in the territory of Kazakhstan. Med Bull Bashkortostan. (2023) 2(104):84–7. (in Russian).
31. Nurmakhanov TI, Sansyzbayev EB, Daniyarova AB, Sayakova ZZ, Vilkova AN, Yeskhodzhayev OU, et al. Results of research of prevalence of Crimean Congo hemorrhagic fever virus in the south Kazakhstan region. Bull Kazakh Natl Med Univ. (2017) 2:56–60. (in Russian).
32. Deyde VM, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol. (2006) 80(17):8834–42. doi: 10.1128/JVI.00752-06
33. Papa A, Marklewitz M, Paraskevopoulou S, Garrison AR, Alkhovsky SV, Avšič-Županc T, et al. History and classification of aigai virus (formerly Crimean- Congo haemorrhagic fever virus genotype VI). J Gen Virol. (2022) 103(4):001734. doi: 10.1099/jgv.0.001734
34. Nurmakhanov TI, Sansyzbayev EB, Yeskhodzhayev OU, Vilkova AN, Sajlaubekuly R, Kulemin MV, et al. Genetic variants of the Crimean-Congo virus of hemorrhagic fever circulating in the territory of the South-Kazakhstan region. Medicine (Almaty). (2018) 9(195):54–60. (In Russ.). doi: 10.31082/1728-452X-2018-195-3079-54-60
Keywords: ticks, Crimean - Congo haemorrhagic fever virus, S, M and L segments, phylogenetic analysis, natural foci, Kazakhstan
Citation: Nurmakhanov T, Tukhanova N, Sayakova Z, Sadovskaya V, Shevtsov A, Tokmurziyeva G and Turebekov N (2024) Outcome of the entomological monitoring for Crimean-Congo haemorrhagic fever virus in the western and southern regions of Kazakhstan in 2021–2022. Front. Epidemiol. 4:1310071. doi: 10.3389/fepid.2024.1310071
Received: 9 October 2023; Accepted: 22 July 2024;
Published: 22 August 2024.
Edited by:
Shailendra Saxena, King George's Medical University, IndiaReviewed by:
Dominique Goedhals, Federal University of São Carlos, BrazilRoger Hewson, University of London, United Kingdom
Copyright: © 2024 Nurmakhanov, Tukhanova, Sayakova, Sadovskaya, Shevtsov, Tokmurziyeva and Turebekov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: N. Turebekov, bnR1cmViZWtvdjIzQGdtYWlsLmNvbQ==
 T. Nurmakhanov1
T. Nurmakhanov1 N. Tukhanova
N. Tukhanova Z. Sayakova
Z. Sayakova A. Shevtsov
A. Shevtsov N. Turebekov
N. Turebekov