
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Epidemiol. , 15 February 2024
Sec. Infectious Disease Epidemiology
Volume 4 - 2024 | https://doi.org/10.3389/fepid.2024.1279835
This article is part of the Research Topic Malaria molecular epidemiology current situation in Africa View all 5 articles
 Sena Adzoa Matrevi1,2
Sena Adzoa Matrevi1,2 Tryphena Adams2
Tryphena Adams2 Kwesi Zandoh Tandoh1,2
Kwesi Zandoh Tandoh1,2 Philip Opoku-Agyeman2
Philip Opoku-Agyeman2 Selassie Bruku2
Selassie Bruku2 Nana Aba Ennuson2
Nana Aba Ennuson2 Paa Kwesi Apau-Danso3
Paa Kwesi Apau-Danso3 Emmanuel Fiagbedzi3
Emmanuel Fiagbedzi3 Mary Avornyo3
Mary Avornyo3 Charles James Myers3
Charles James Myers3 Joy Futagbi3
Joy Futagbi3 Oheneba Charles Hagan1
Oheneba Charles Hagan1 Benjamin Abuaku2
Benjamin Abuaku2 Kwadwo Ansah Koram2
Kwadwo Ansah Koram2 Gordon Awandare1
Gordon Awandare1 Neils Ben Quashie1,2,4
Neils Ben Quashie1,2,4 Nancy Odurowah Duah-Quashie1,2*
Nancy Odurowah Duah-Quashie1,2*
Introduction: Antimalarial drugs including artemisinin-based combination therapy (ACT) regimens and sulphadoxine-pyrimethamine (SP) are used in Ghana for malaria therapeutics and prophylaxis respectively. The genetic basis of Plasmodium falciparum development of drug resistance involves single nucleotide polymorphisms in genes encoding proteins for multiple cellular and metabolic processes. The prevalence of single nucleotide polymorphisms in nine P. falciparum genes linked to ACT and SP resistance in the malaria parasite population was determined.
Methods: Archived filter paper blood blot samples from patients aged 9 years and below with uncomplicated malaria reporting at 10 sentinel sites located in three ecological zones for the Malaria Therapeutic Efficacy Studies were used. The samples used were collected from 2007-2018 malaria transmission seasons and mutations in the genes were detected using PCR and Sanger sequencing.
Results: In all 1,142 samples were used for the study. For falcipain-2 gene (pffp2), Sanger sequencing was successful for 872 samples and were further analysed. The prevalence of the mutants was 45% (392/872) with pffp2 markers V51I and S59F occurring in 15.0% (128/872) and 3.0% (26/872) of the samples respectively. Prevalence of other P. falciparum gene mutations: coronin (pfcoronin) was 44.8% (37/90); cysteine desulfurase (pfnfs) was 73.9% (68/92); apicoplast ribosomal protein S10 (pfarps10) was 36.8% (35/95); ferredoxin (pffd) was 8.8% (8/91); multidrug resistance protein-1 (pfmrp1) was 95.2.0% (80/84); multidrug resistance protein-2 (pfmrp2) was 91.4% (32/35); dihydrofolate reductase (pfdhfr) was 99.0% (84/85); dihydropteroate synthase (pfdhps) was 72% (68/95).
Discussion: The observation of numerous mutations in these genes of interest in the Ghanaian isolates, some of which have been implicated in delayed parasite clearance is of great interest. The presence of these genotypes may account for the decline in the efficacies of ACT regimens being used to treat uncomplicated malaria in the country. The need for continuous monitoring of these genetic markers to give first-hand information on parasite susceptibility to antimalarial drugs to inform policy makers and stakeholders in malaria elimination in the country is further discussed.
Malaria continues to be a major public health problem globally with high rates of morbidity and mortality in disease-endemic areas of the world. In 2022, 249 million cases and 608,000 estimated deaths occurred worldwide (1). The World Health Organization (WHO) African Malaria Region accounted for 94% and 95% respectively of the morbidity and mortality estimates stated above (1). Chemotherapy remains the mainstay of malaria control and elimination with artemisinin-based combination therapy (ACT) in disease-endemic areas (2). The reported occurrence of Plasmodium falciparum resistance to artemisinin (ART) in Southeast Asia (SEA) and a few countries in Africa has led to active surveillance through Therapeutic Efficacy Studies (TES) investigating clinical, in vitro, molecular, and pharmacokinetic analyses of antimalarial drug resistance (3).
Molecular analysis of the P. falciparum genome revealed mutations in the kelch propeller gene (pfk13) to be associated with the development of parasite tolerance to ART and are being monitored in parasite populations as markers of drug resistance (2, 4). ART resistance pfk13 candidate mutations and novel ones have been reported in Ghanaian isolates (5–7). A study conducted in Cambodia has also shown parasites with increased ring stage survival but lacking the pfk13 mutations which implies that there could be other genes involved and/or different resistance mechanisms (8). Some studies have suggested the contribution of multiple cellular and metabolic processes in ART resistance including hemoglobin degradation, proteotoxic/unfolded protein stress response, vesicular biogenesis as well as oxidative stress response, and mitochondrial functions (9). These studies have implicated single nucleotide polymorphisms (SNPs) in multiple genes in antimalarial drug resistance development. The antimalarial drug resistance markers are effective tools for assessing the drug resistance status of parasites for malaria control measures (3, 10, 11).
Studies have linked parasite genetic loci with in vivo and in vitro studies to ART resistance and these include missense SNPs that lead to loss of function in genetic alleles of actin-binding protein coronin (pfcoronin G50E, V62M, R100K, E107V, N112N/Y/D) and falcipain 2 (pffp2 N4H, S35*, V51I, S59F, S69*, K255R) (12–15). In addition, the co-inheritance of several mutations including P. falciparum apicoplast ribosomal protein S10 (pfarps10 V127M), ferredoxin (pffd D193Y), chloroquine resistance transporter (pfcrt I356T) and the multidrug resistance gene 2 (pfmdr2 T484I) were shown to constitute a parasite genetic background (PGB) that allows for the emergence of mutations in the pfk13 (16). Mutations in other genes, cysteine desulfurase (pfnfs S62N, K65Q, E67G) and multidrug resistance protein 1 and 2 (pfmrp1 I876V and K1466R; pfmrp2 N622D, I1775V, and c2397t) have also been implicated in ACT resistance (17–19). Sulphadoxine-pyrimethamine (SP) is still used for intermittent preventive treatment in pregnant women (IPTp) and seasonal malaria chemoprevention (SMC) in children in Ghana. Mutations in P. falciparum dihydrofolate reductase (pfdhfr N51I, C59R, S108N, and I164l) and dihydropteroate synthase (pfdhps S436A, A437G, and K540E) are implicated in SP resistance (20). More recently a mutation, pfdhps I431V was observed exclusively in parasites from pregnant women from Nigeria (21) and Cameroon (22, 23), however, its role in resistance is not known.
In Ghana, ACT regimens used in the treatment of uncomplicated malaria are artemether-lumefantrine (AL), artesunate-amodiaquine (AS-AQ), artesunate-pyronaridine (AS-PD), and dihydroartemisinin-piperaquine (DHAP) whilst SP is used IPTp and SMC. AS-AQ was introduced in 2005, and AL and DHAP were added in 2008 for the treatment of uncomplicated malaria. Currently, the regimens for first-line treatment are AL, AS-AQ, and AS-PD whilst DHAP is the second-line drug (24). This study assessed the prevalence of mutations in nine resistance markers (pffp2, pfcoronin, pfnfs, pfarps10, pffd, pfmrp1, pfmrp2, pfdhfr, and pfdhps) in the P. falciparum population in Ghana as part of ongoing antimalaria drug efficacy/resistance surveillance studies.
There are 10 sentinel sites established by the National Malaria Elimination Program (NMEP) in collaboration with the Noguchi Memorial Institute for Medical Research (NMIMR) for Treatment Efficacy Studies (TES) on antimalarial drugs in Ghana (25). The sentinel sites are located in three ecological zones of the country, Accra and Cape Coast in the coastal savannah zone (CS); Begoro, Bekwai, Hohoe, Sunyani, and Tarkwa in the forest zone (FZ); Navrongo, Wa, and Yendi in the Guinea savannah zone (GS) (Figure 1). Archived samples taken from children aged 9 years and below with uncomplicated malaria reporting to health facilities at the sentinel sites were used for this study (25). The samples were taken in the transmission seasons of time points, 2007–2008, 2010–2012, 2013–2014, 2015–2016, and 2018.
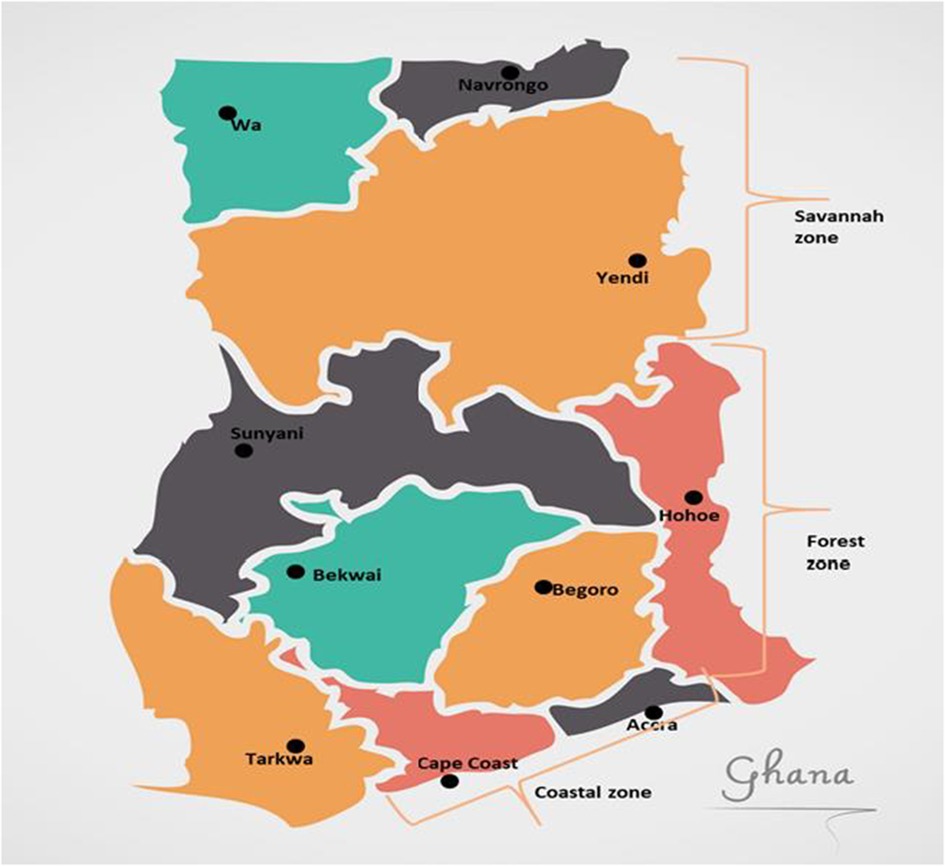
Figure 1. Map of Ghana showing the location of the sentinel sites in the three ecological zones. The sentinel sites for monitoring antimalarial drug efficacy/resistance in Ghana are in the 10 regions of the country. These sites have hospitals and health centres for treatment efficacy studies in malaria and were set up by NMIMR in collaboration with the National Malaria Elimination Program (NMEP).
Parasite DNA was extracted from filter paper blood blots using QIAamp DNA minikit (QIAGEN, Hilden Germany) following the manufacturer's protocol. Polymerase chain reaction (PCR) was carried out to amplify all the genes of interest for this study (pffp2, pfcoronin, pfnfs, pffd, pfarps10, pfmrp1, pfmrp2, pfdhfr, and pfdhps). The references of the protocols used, primer sequences, and cycling conditions for all the genes are shown in Supplementary Table S1.
For pffp2, the primary reaction was in a total volume of 15 µl and the secondary reaction was done in 50 µl with final reagent concentrations of 160 mM deoxynucleoside triphosphates (dNTPs), 160 nM primers, 1U Taq DNA Polymerase and 1X standard Taq Buffer [20 mM Tris-HCl (pH 8.9,), 1.8 mM MgCl2, 22 mM NH4Cl, 22 mM KCl, 0.06% IGEPAL CA-630, and 0.05% Tween 20] and 0.5 µl of genomic DNA. Amplification of pffd and pfarps10 was conducted in a 30 µl reaction volume consisting of 1X PCR reaction buffer, 4 mM of MgCl2, 0.4 μM of each of the primers, 0.2 mM of dNTPs and 1 U of Taq polymerase. For pfcoronin and pfnfs amplification, a 25 μl reaction volume contained 12.5 μl 2X Kapa Hifi Hotstart ready master mix (Roche, SA) and 0.2 µM, primers (forward and reverse for pfcoronin and pfnfs) were used (Eurofins Genomics). The amplification of pfmrp1 and pfmrp2 was done in a total volume of 25 µl containing 0.2 µM of each primer, 0.2 mM deoxynucleotides triphosphate (dNTPs), and 2U One-Taq polymerase (New England Biolabs, Massachusetts, USA). Nested PCR was used to amplify the pfdhps gene with a primary amplification in 10 μl reaction mixture consisting of 0.4 μM of each primer and 5 μl 2X Amplitaq Gold DNA polymerase (Applied Biosystems; Thermofisher Scientific) master mix. 1 μl of the template was used in a 30 μl secondary reaction of 0.4 μM primers and 15 μl 2X Amplitaq Gold DNA polymerase. The pfdhfr was amplified in a single PCR run containing 0.4 μM of each primer and 15 μl 2X Amplitaq Gold DNA polymerase in a total volume of 30 μl. PCR products were detected using agarose gels stained with Gel red dye (Biotium, California, USA) or SYBR safe (Invitrogen, USA).
All Amplicons from the PCRs of the various genes were sequenced at Macrogen Europe (Amsterdam, Netherlands).
There were variable sample sizes analyzed for each gene due to the availability of archived samples and funds, therefore computational analysis was done to determine haplotype and nucleotide diversities for pffp2 only with samples for four time points. The chi-squared test for trend analysis was used to determine the trend of the prevalence of the pffp2 mutations over the time points with StatCalc in Epi InfoTM 7.2.5.0 (Centres for Disease Control and Prevention, Atlanta, USA). Sequences were verified using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/) upon receipt from Macrogen Europe. This was used to determine the authenticity of the sequences of the genes. Multiple sequences were aligned with MAFFT (EMBL.EBI, Hinxton, Cambridge, UK) using the 3D7 wild-type sequences as references: pffp2 - PF3D7_1115700; pfarps10 - PF3D7_1460900; pffd - PF3D7_1318100; pfdhps - PF3D7_0810800; pfdhfr - PF3D7_0417200; pfnfs - PF3D7_0727200; pfmrp1 - PF3D7_0112200; pfmrp2 - PF3D7_1229100; pfcoronin - PF3D7_1251200. Consensus sequence editing and single nucleotide polymorphisms (SNP) determination were carried out using the CLC main workbench 7.9.1 (Qiagen) and Benchling.com (California, CA, USA). Computational methods were employed for the pffp2 gene to determine haplotype and nucleotide diversities. Base-calling, alignment, and deconvolution of Sanger chromatogram trace files were done using the command-line version of the application Tracy. The output binary variant call format (bcf) files for each sample were converted to human-readable variant call format (vcf) files using custom bash scripts. Low-quality variants (<40) and indels were filtered out from the vcf file after which the files were merged and variants extracted and annotated into a text file using custom bash scripts, SnpEff (v4.1), and vcf tools. Fasta files were generated using custom bash scripts and fed into DnaSP6.0 to determine the DNA polymorphism metrics and Tajima's D.
A total of 1,170 filter paper blood blot samples obtained from children aged ≤ 9 years with uncomplicated P. falciparum malaria were used for the study. The study population median age was 50 months (4 years and 2 months). The minimum parasite density observed among the study participants was 1,000 parasites/µl of blood and was recorded from the FZ and the maximum of 249,960 parasites µl/ of blood from the CS. There was a significant difference between the parasite counts from the CS and FZ (P ≤ 0.05) only (Figure 2).
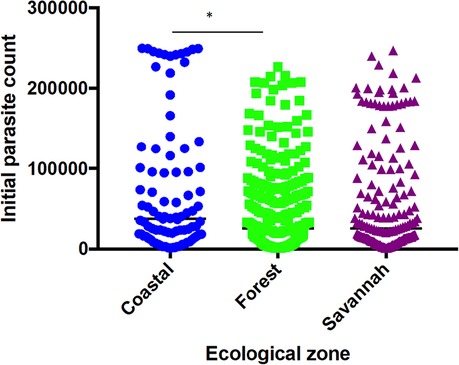
Figure 2. Parasite densities of study participants at the sentinel sites located in the three ecological zones. Each dot represents the parasite density determined per patient. There was a significant difference in parasite densities of participants from the coastal and forest zones, p ≤ 0.05 (*) using Dunn's test.
A total of 872 good sequences were analyzed for polymorphisms in the pffp2 and the proportion of mutant genotypes was 44.95% (392/872) of the isolates. The number of samples analyzed for each ecological zone and the proportion of mutants are shown in Table 1. Of the 392 mutants, there were 575 mutations with 81.6% (469/575) being non-synonymous (NS) mutations. Samples from time point, 2007–2008, showed 100 mutations at 110 codons, 2010–2012 had 28 mutations at 39 codons, 2013–2014 had 62 mutations at 68 codons, and 2015–2016 had 450 mutations at 245 codons. Mutations varied from a single mutation per codon to ten mutations per codon. Some of the mutations at specific codons were observed at various time points. The amino acid variants observed at codon 114 ranged from 1 to 10. The S35* mutation implicated in slow parasite clearance was not observed, however, five mutant variants, S35l/N/P/S/T, were observed. The synonymous S35S was observed in 19 samples. The frequency of other relevant mutations at codon 59 was 4.5% (26/575), codon 68 was 1.6% (9/575), and codon 69 was 0.9% (5/575). Other reported mutations implicated in ART resistance, V51I, S59F, S69P, and V70F were observed in 32.6% (128/392), 6.6% (26/392), 0.3% (1/392), and 0.5% (2/392) respectively of the mutants. Other mutations A8A, R27K, N32K, V21V, S43S, V47V, and V51I were observed in samples from all the ecological zones. Some zone-specific mutations observed were: GS: Y3D, M5l, Q16K/R, V21F/G, S43T, R60I, V70I, and K225Q; FZ: Y3N, D6N, H10Q/N, S14F, S35l, V51F, T56A/S/F, T65S, S68F, M167I, Y207F and H220l/P; CS: A8I, P9P, T56P, S68P, S69P, K255G, F260S, and R301K. For the time points, the polymorphisms, V51I and S59F were highest among the 2015–2016 samples and also observed at all the four time points. Others such as D151E were only observed in samples from 2010 to 2012, 2013–2014, and 2015–2016 whereas K255R was observed in parasite isolates from 2015 to 2016 only. The highest prevalence of mutations was observed in 2010–2012 at 81.8% for CS, and for the FZ in 2015–2016 at 63.4% whilst GS had the highest prevalence in 2015–2016 and was 65.3% (Figure 3). The prevalence of pffp2 mutations showed an increasing trend over the time points (χ2 = 16.7647, p = 0.00004) as shown in Figure 4. All the observed pffp2 mutations are listed in Supplementary Table S2.
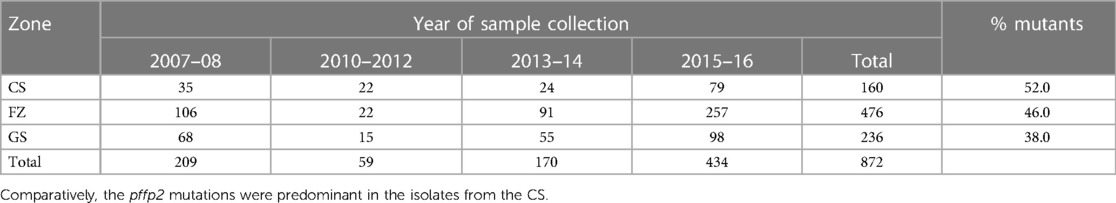
Table 1. Number of samples per ecological zone at the time points and the proportion with pffp2 mutations.
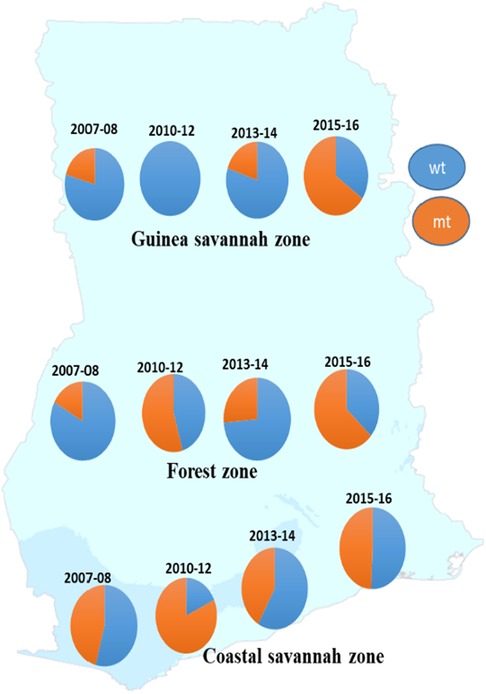
Figure 3. Distribution of the prevalence of pffp2 wild-type and mutant alleles in parasite populations in the ecological zones at four time points. The prevalence of the wildtype and mutant alleles for the pffp2 varied with time and the three ecological zones, however, the timepoint 2015–16 showed the highest prevalence of the mutation for the FZ and GS.
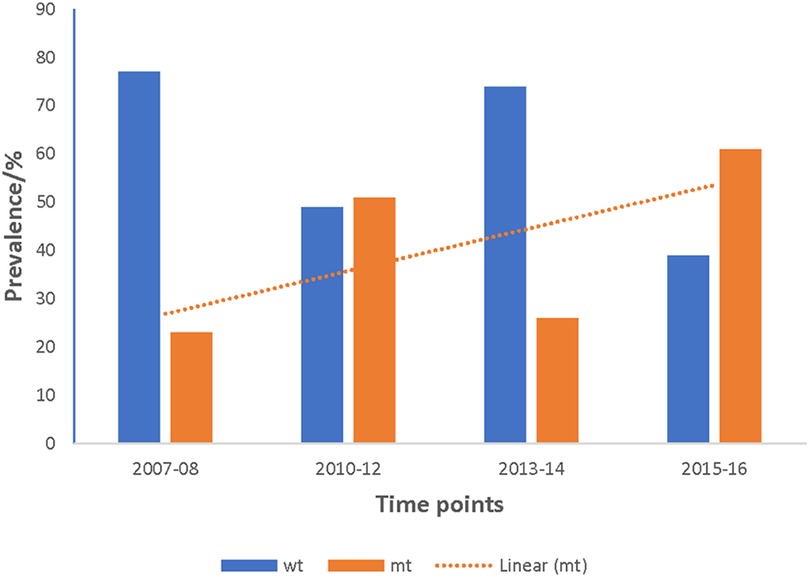
Figure 4. Increasing trend of the prevalence of pffp2 mutant alleles in Ghanaian isolates over the four time points. The pooled data from all the sites for the four time points showed an increasing trend for the pffp2 mutant alleles using the chi-square test for trends (χ2 = 16.7647, p = 0.00004).
Haplotypes of the pffp2 linked to delayed clearance in the SEA (26). were observed in Ghanaian isolates at various time points and are shown in Table 2. These include the mutant haplotype V47-I51 (VI) which was seen in isolates from all time points and the highest prevalence was 31.7% (126/398) and mostly in samples from the 2015–2016 time point. Other mutant haplotypes S105-K107-N108 (SKN) were not seen in samples from the 2013–2014 time point but were observed at the other time points. For K255-F260-Y265-D266 (KFYD) it was observed only in the 2007–2008 and 2015–2016 time points (Table 2).
Analysis for the distribution of SNPs across the genome showed that 2.4% were found in the intergenic region, 13.2% were missense variants, 29.4% were non-coding transcript variants, 9.7% were synonymous variants and 6.4% were frameshift variants (Supplementary Figure S1). The biological effect of the SNPs analyzed resulted mostly in missense variants. The time point 2015–2016 gave the highest number of haplotypes of 150 with 826 segregating sites. The FZ also gave the highest haplotype of 172 with 1116 segregating sites. Nucleotide diversity was low within the pffp2. The negative values for Tajima's D indicate purifying or negative selection. The mutation rates and the number of haplotypes observed within the pffp2 for the ecological zones and time points are shown in Table 3.
Ninety-six samples including post-treatment recurrent parasites collected in 2018 were amplified and sequenced for pfcoronin. Of the 90 good sequences analyzed, 53 (58.9%) were wildtype sequences and the mutants were 37 (41.1%). The number of mutations seen in the 37 mutants was 90, of which the majority were NS mutations (77.8%, 70/90). Of the 90 samples, the number of samples from the FZ, CS, and GS were 22, 19, and 49 respectively and the prevalence of mutants was 18% (4/22), 52.3% (10/19), and 46.9% (23/49) in the same order. The proportions of the 90 mutations observed in the samples were distributed in the zones as 6.7% (6/90), 24.4 (22/90), and 68.9% (62/90) for FZ, CS, and GS respectively. The most prevalent mutation was P76S observed in 24% (22/90: 6 - CS; 2 - FZ; 14 - GS) of the isolates. It is a published mutation but with no link to ACT delayed clearance and characterized by a nucleotide base change from CCC-TCC. The known mutations, G50E, R100K, and E107V, linked to delayed parasite clearance with ACT use were not observed in the Ghanaian isolates. However, a variant of R100K which is R100R (AGA-AGG) was observed in only one isolate in the CS. The variants of some other published mutations, V62M, N112D/N/Y, and V128V were observed as; V62l, N112K, and V128E respectively. For pre-treatment and post-treatment isolates, 38.9% (35/90) and 61.1% (55/90) were mutants respectively. Statistical analysis showed a significant difference in the occurrence of SNPs between pre-treatment and post-treatment isolates at the three ecological zones (p = 0.00003). The mutations observed in the pfcoronin gene of all the isolates are shown in Supplementary Table S3.
The total number of samples analyzed for the pfnfs was 96 samples collected in 2018 which includes recurrent samples. Ninety-two good-quality sequences were analyzed for the identification of mutations, of which 26.1% (24/92) were wild-type genotypes. The 92 samples are distributed as 22, 20, and 50 for the FZ, CS, and GS respectively. The 68 mutants had a total of 356 mutations and the majority were NS (88.2%, 314/356). Of the 356 mutations, 62.1% (221/356) SNPs were detected in the pre-treatment samples and 37.9% (135/356) in the post-treatment samples. The highest occurring mutation, K65Q, was 14.9% (53/356) of all mutations observed followed by S62N in 14.6% (52/356) and E67G in 14.3% (51/356). These are the three known SNPs linked to delayed parasite clearance for lumefantrine. The shared and most dominant mutations are shown in Figure 5. Analysis of the proportion of pre-treatment and post-treatment SNPs from the three ecological zones revealed no significant difference (p = 0.76). There were some mutations shared between both the pre-treatment and post-treatment samples. The K65Q occurred the most in the pre-treatment samples with a prevalence of 15.4%. It is interesting to note that this mutation together with S62N and E67G had the same prevalence (14.1%) in the post-treatment samples.
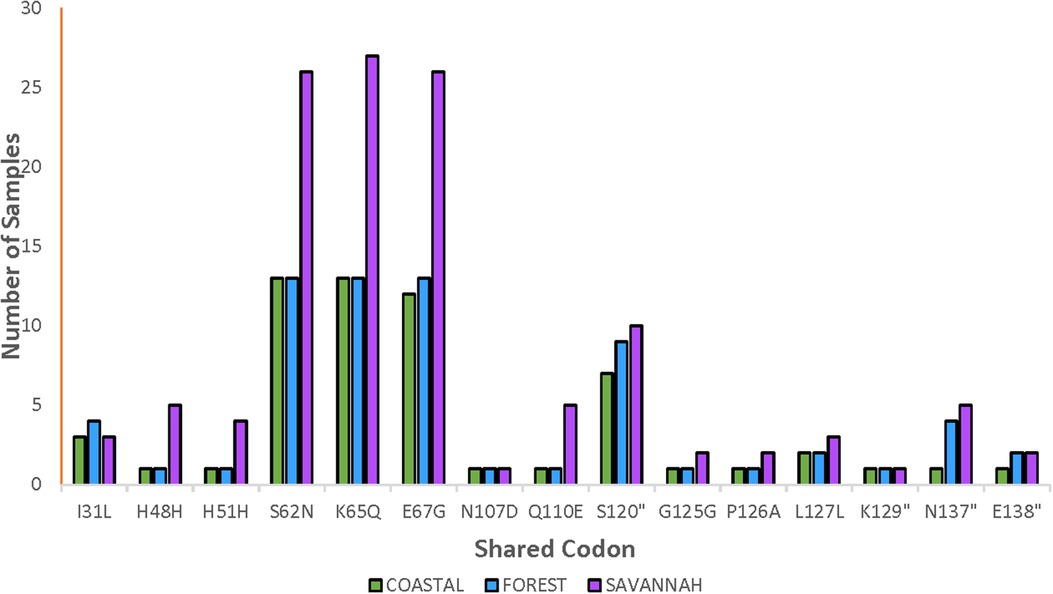
Figure 5. Distribution of shared pfnfs mutations in the three ecological zones. Some observed mutations of the pfnfs were seen in isolates from the ecological zones and were termed as shared mutations and are being displayed with the number of samples they were observed in at the three zones.
Ninety-five samples collected in 2018 were analyzed for the pfarps10 for the detection of mutations. The wildtype parasites were 63.2% (60/95) of the isolates with ecological representations as 62.5% (20/32) from CS, 65.6% (21/32) from FZ, and 61.3% (19/31) from GS. Codon 127 had the highest number of variants, 13.7% (13/95) among the isolates. V127W was the highest occurring mutation observed in 6.3% (6/95) of the isolates followed by H126M in 5.3% (5/95) and then D128I in 4.2% (4/95). V127M is one of the PGB mutations of ART resistance and was seen in one sample from the GS. Other variants V127D, V127G, and V127C, were observed in one, two, and two samples respectively. The observed mutations in all isolates are listed in Supplementary Table S4.
A total of 96 samples collected in 2018 were sequenced of which 91 were of good quality and used for analysis. The proportion of wild-types in these samples was 91.2% (83/91). No mutation of this gene was observed in samples from the FZ, however, the CS and the GS zones had two and six mutants respectively. In all, 15 mutations were observed in the eight mutants and the majority were NS (73.3%, 11/15). V148G was the most prevalent mutation; occurring in 3 isolates. Apart from the shared mutations by all the zones, of the 15 mutations, 13 (86.7%) were from GS. SYN mutations recorded were V126V, V182V, T174T, and P177P. There was a change from lysine to a stop codon at codon 146 (K146*) in one isolate. Other observed mutations, A145S, E149K, K187N, D189H, M194V, D161Y, E162D, E163Y, Q164H, and K146* were from isolates from GS. The published marker, D193Y was not observed in the samples analysed. All observed pffd mutations in the isolates are shown in Supplementary Table S5.
Of the 84 good-quality sequences, the proportion of wild-type was 4.8% (4/84). The 80 mutants had a total of 58 mutations, with 89.7% (52/58) being NS. It was observed that 75% (63/84) of the mutants had two or more mutations. The following mutations; I876V, S885K, S885N, S885S, S885T, and K886K were detected in all mutants. Figure 6 shows the distribution of the prominent mutations detected across the zones. At least one SNP was detected in 96.3% (26/27), 95.1% (39/41), and 93.8% (15/16) of the analyzed sequences from CS, FZ, and GS respectively. There was no statistical difference in the number of SNPs between the three zones (p = 0.620815). S885T was the most frequent mutation and was found in 63.1% (53/84) of the isolates and was highest in FZ with a prevalence of 78% (32/41). The I876V mutation which is reportedly linked to AL resistance was observed in in 9.5% (8/84) of the isolates. GS isolates had the highest number of this mutation 50% (4/8). All observed pfmrp1 mutations in the isolates analyzed are shown in Supplementary Table S6.
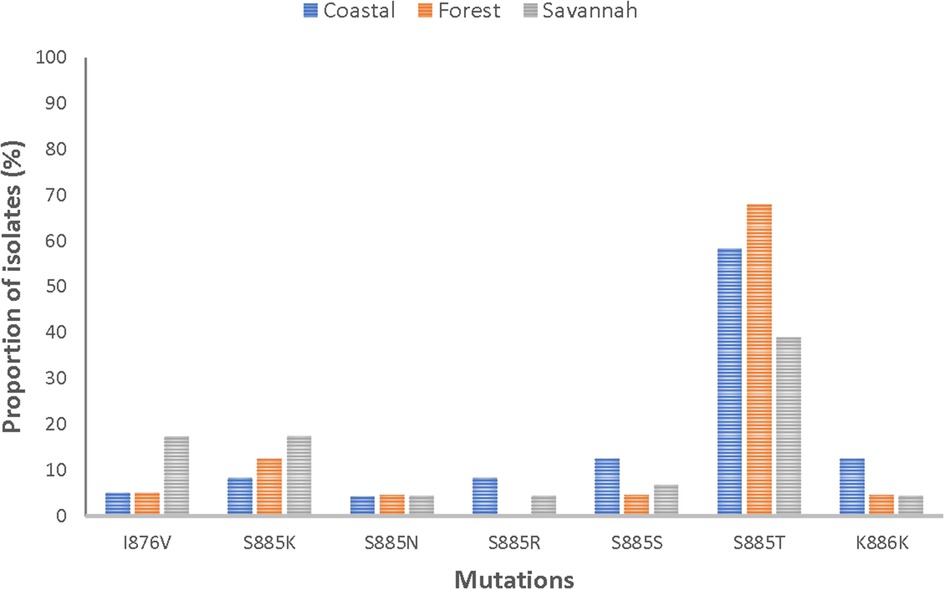
Figure 6. Proportion of isolates with shared pfmrp1 mutations in the three ecological zones. Some of the observed mutations were seen in isolates from the three ecological zones and have been depicted as proportions of isolates they were observed in.
The total number of samples analyzed for pfmrp2 was 105. Thirty-five good sequences were analyzed. The prevalence of wild-type was 8.57% (3/35) and the 32 mutants had a total of 126 mutations, of which 94.4% (119/126) were SNPs and 5.6% (7/126) were insertions. Out of the 119 SNPs, 87.4% (104/119) were NS mutations with 2.9% (3/104) being nonsense mutations i.e., a change from AA to a stop codon. Of all the mutants observed, 87.5% (28/32) of the isolates had multiple mutations while 12.5% (4/32) had a single mutation. Figure 7 shows the prevalence of the single or multiple SNPs detected in samples from each ecological zone. The most abundant mutation detected was N645D (8.0%, 10/126). The SNPs D631G and K714I followed with a frequency of 5.5% (7/126) each. The most prevalent SYN mutation was P566P which was detected in 8.6% (3/35) of the isolates. None of the SNPs was shared across all the zones. SNPs detected multiple times in isolates from FZ are I533l, R534K, T550P, P566P, E574K, K600K, N622S, D629N, D633N, Y636C, D637N, D649N, M652V, and Q694H. The SNP N622D which was reported by Veiga and others was not detected, however a variant N622S was seen. All observed pfmrp2 mutations in the isolates analyzed are shown in Supplementary Table S7.
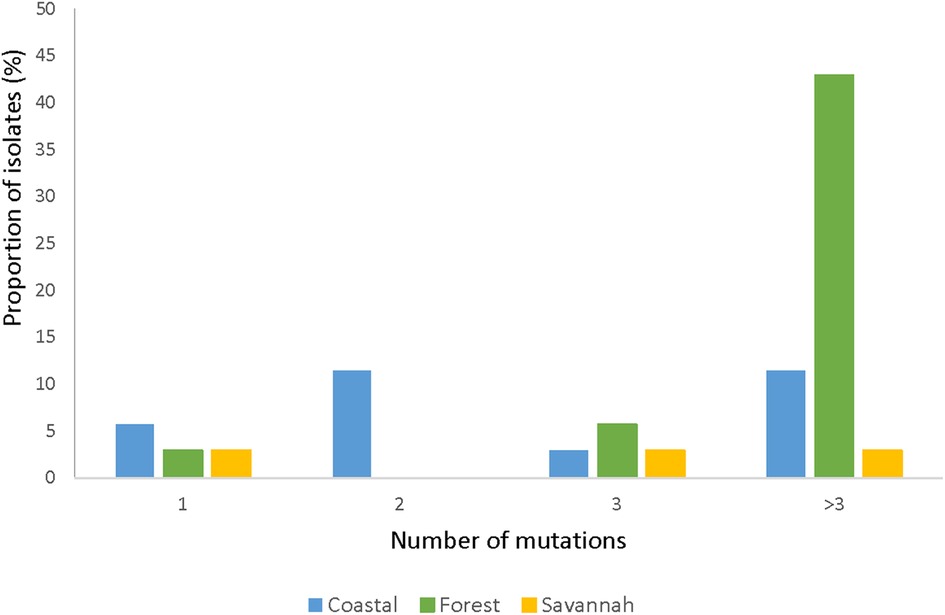
Figure 7. Proportion of isolates with single and multiple pfmrp2 mutations in isolates from the ecological zones. For the pfmrp2 mutations, it was observed that one isolate could have up to three mutations, and the proportion of such isolates at the three ecological zones is portrayed by the histogram.
A total of 85 good sequences were analyzed for the pfdhfr gene with 99% (84/85) having mutations. The known mutations N51I, C59R, and S108N were observed in a majority of the samples. C59R was observed in 89.4% (76/85), S108N in 87.1% (74/85), and N51I in 78.8% (67/85) of the isolates. Two rare variants, N51F and S108D were observed in one parasite isolate from GS. Rare variants of the I164l occurred in isolates, I164R from GS and I164K from CS. The most prevalent haplotype for the pfdhfr gene was the triple mutant IRN observed in 72.9% (62/85) of the isolates. The double mutant RN haplotype was also observed in 5.9% (5/85) of the isolates. The proportions of pfdhfr mutations and haplotypes in the isolates for the ecological zones are shown in Figure 8.
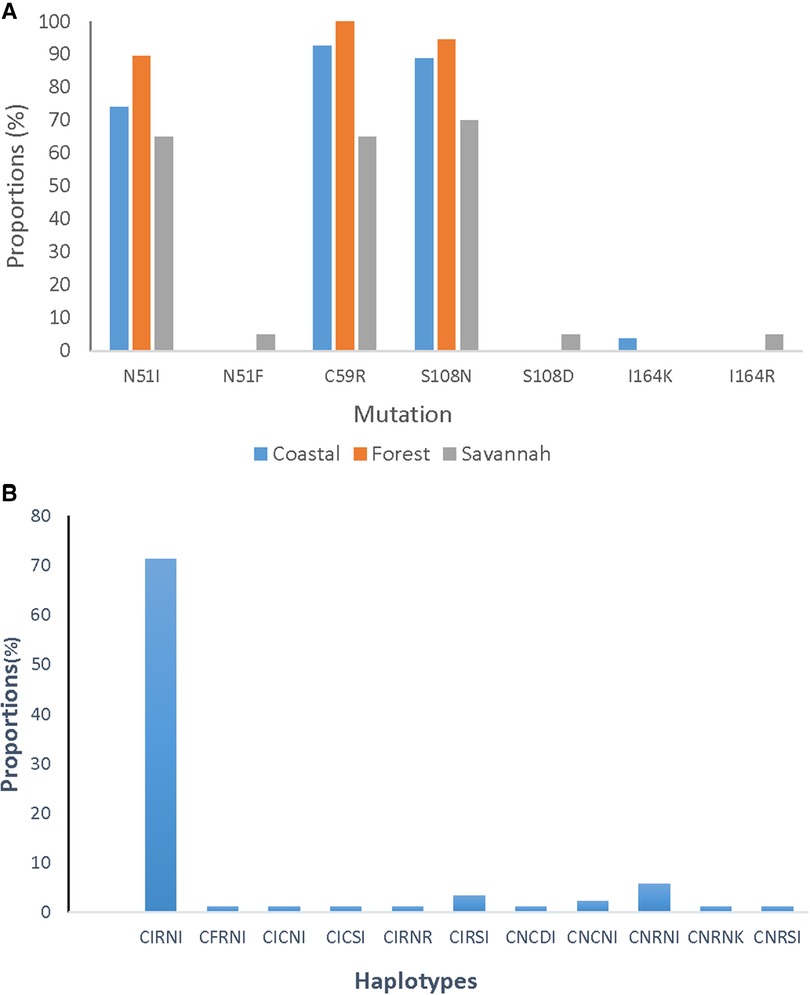
Figure 8. Proportion of pfdhfr mutations and haplotypes from Ghana. (A) The proportion of isolates with the mutations I51, R59, and N108 are still dominating in the parasites but were seen as highest in the isolates from the FZ followed by CS and GS. (B) The CIRNI haplotype which is a mutant genotype still dominates in the parasite population in Ghana with a prevalence above 70%.
In all, 95 good sequences were analyzed for the pfdhps and 72% (68/95) had mutations. The most observed mutation was A581A (GCG to GCA) in 78.0% (53/68), followed by K540N at 57.3% (39/68) of the mutants. The aforementioned mutations are variants of A581G and K540E associated with sulphadoxine resistance, however, the K540E was not observed in the Ghanaian samples. There was also no detection of the novel I431V mutation, seen in pregnant women from Cameroon and Nigeria in the Ghanaian isolates. The proportion of isolates with the different mutations from the three zones as well as the haplotypes are shown in Figure 9. There were zone-specific mutations observed such that S436A was observed in 4.2% (4/95) of the samples from FZ (3) and GS (1). In addition, A437G was only observed in 1.1% (1/95) of the isolates from GS.
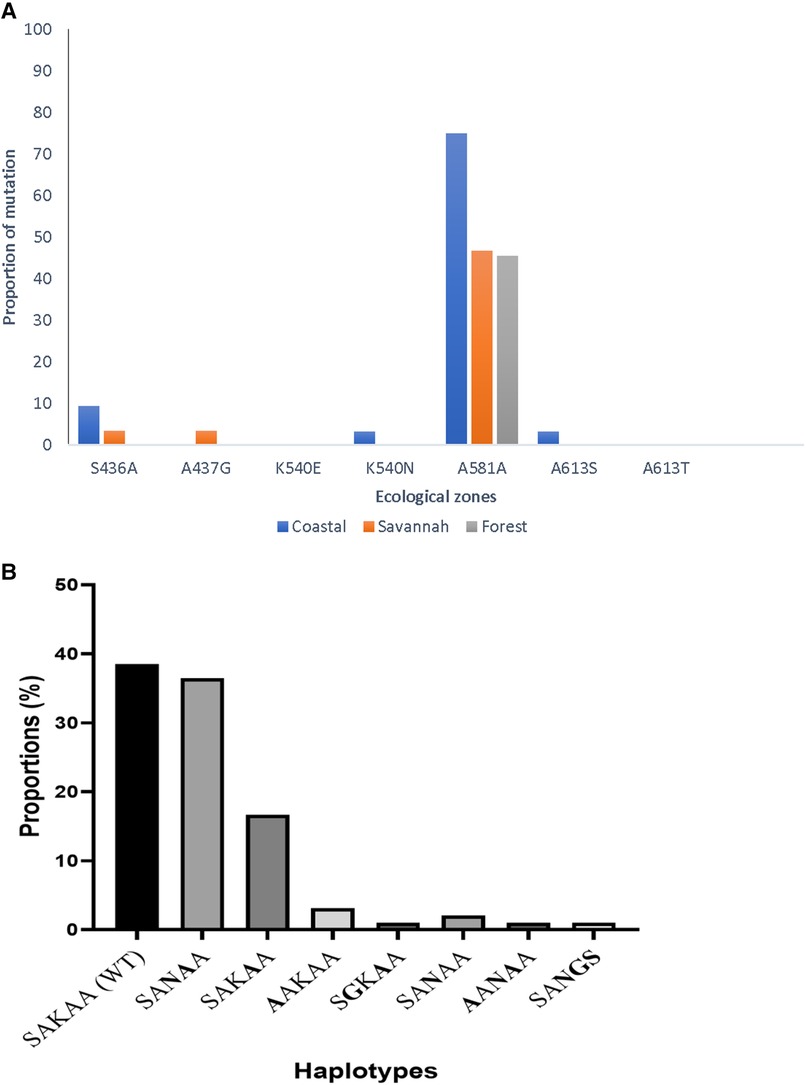
Figure 9. Proportion of pfdhps mutations and haplotypes in isolates from Ghana. (A) The proportion of isolates with both known and novel pfdhps mutations are shown. The K540E mutant for sulphadoxine resistance was absent, however, a new synonymous mutation A581A was seen to predominate in isolates from all the ecological zones. (B) The wild-type haplotype, SAKAA, predominated the mutant haplotypes observed in the isolates. The amino acids in bold are the SNPs.
In summary, all the genetic mutations of known relevance in ACT and SP resistance as well as novel mutations in the nine genes investigated in the Ghanaian P. falciparum isolates from symptomatic patients are shown in Table 4.
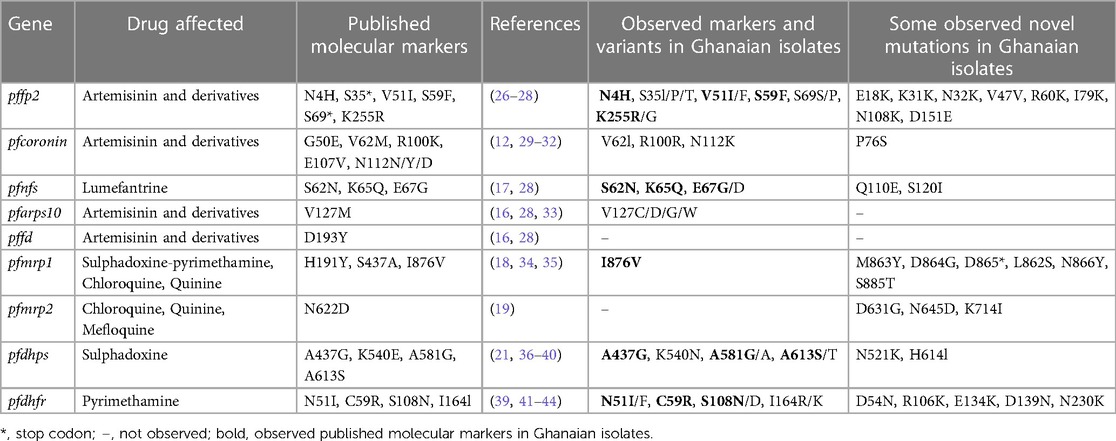
Table 4. Summary of observed markers of antimalarial drug resistance and variants in malaria parasites from Ghana.
Evolving antimalarial drug resistance in the malaria parasite, especially for the ACT regimens which are currently characterized by slow parasite clearance, is of great concern because it threatens malaria elimination at the country level as well as the global disease eradication plan. One important recommendation for guaranteeing country-level disease elimination is increasing molecular surveillance of the parasite population in disease-endemic areas which involves monitoring the prevalence of molecular markers of antimalaria drug resistance. These markers are polymorphisms occurring in genes encoding important proteins for multiple cellular and metabolic processes for parasite survival. The findings from this investigation revealed numerous polymorphisms in the nine genes studied (pffp2, pfcoronin, pfnfs, pfarps10, pffd, pfmrp1, pfmrp2, pfdhfr, and pfdhps), and most of the known markers of drug resistance were identified in the Ghanaian isolates. Variants of some of the known markers of antimalarial drug resistance were also observed. One of the genes (pffp2) investigated is under purifying selection and showed increasing trends in the prevalence of mutations over the years.
Polymorphisms observed in the pffp2 gene were diverse and the S69* mutation linked to slow parasite clearance by Ariey and others (4) was not observed in our study. This observation is similar to studies carried out in Uganda and Kenya (27, 28). The mutations, N4H, V51I, and S59F, also linked to slow parasite clearance (26, 27) were observed in our isolates and occurred in isolates from the three ecological zones. The study in Kenya also reported the observation of these mutations, S59F, N4H, A8I, P9P, H10N, and E11E as seen in the Ghanaian isolates (28). Most of the pffp2 mutations were shared among the isolates from the different ecological zones, signifying similar selective forces such as host immunity and drug pressure (Zhu et al. 2016). The HIF (15-51-59) haplotype observed in this study has been seen in isolates from the SEA region (26). The parasites with the HIF haplotype were reported as having a high Michaelis–Menten constant (Km) which implies that a higher substrate (Z-LR-AMC, a preferred substrate for falcipain 2) concentration was needed to enhance enzyme activity (26). The observation of the increasing prevalence of pffp2 mutations in the circulating parasite population could be indicative of emerging ART-resistant parasites in Ghana and may contribute to the 5% decrease in the efficacy of both AS-AQ and 3% for DHAP (25, 45). The nucleotide diversity (π) in the pffp2 gene was very low which suggests a decreased probability for selection in the gene. The estimated negative Tajima's D values may also be suggestive of a recent population expansion with multiple low-frequency variants.
The pfcoronin mutations, G50E, R100K, and E107V, which have been associated with reduced P. falciparum susceptibility to ART (12), were not observed in both pre-treatment and post-treatment isolates. However, a SYN variant R100R of the R100K was observed in an isolate from CS. The mutation, P76S, was seen in isolates from all three ecological zones at a prevalence of 24%, which is higher than those observed in studies conducted in Senegal, Gabon, Ghana, Kenya, and Congo, where the prevalence ranged from 4% to 17% (29, 30). The observation of mutations in this study is not in accordance with reports from Senegal and Pakistan where no pfcoronin mutants were observed (46, 47). The pfcoronin mutations in the pre and post-treatment isolates had equal proportions of the known SNP P76S. The same samples were used for the detection of mutations in pfnfs and the K65Q mutation was the most dominant mutation (14.9%) which occurred in isolates from all three zones. In addition, the S62N (14.6%) and E67G (14.3%) were also observed which is similar to reports from a study by Wamae and others (2019), where the prevalence of mutations was reported to be high. In that study, the K65Q was in high linkage disequilibrium with S62N and E67G (28). The pfnfs mutation K65Q, was more prevalent in the pre-treatment than post-treatment isolates.
The pffd gene forms part of the PGB associated with pfk13 mutations in enhancing ART resistance. In this study, the D193Y associated with slow parasite clearance (16) was not observed in the isolates which is contrary to a report from Sudan where 46.3% of isolates carried this mutation (48). The majority of the isolates studied were wild-type genotypes (91.2%) which is supportive of the low rate of spontaneous mutations in pffd as reported in a study in Kenya (28). The pfarps10 mutation, V127M implicated in ART resistance (Miotto et al. 2015), and other variants (V127C/D/G/W/*) were detected but at low frequencies. The V127M was observed in one sample just as reported by a Ugandan study (33), however, another study in Asia reported observing the mutation in 41% of the isolates investigated (49). The Ugandan study also reported the D128H mutation which was detected in one isolate, however, its relevance to resistance remains unknown (33). Other variants of this SNP (D128G/I/K/N) were also observed but their relevance to drug resistance is yet to be investigated. The low prevalence of the pfarps10 gene mutations that were identified in this study was in agreement with previously investigated PGB mutations, however, studies carried out in Kenya, Mali, and Tanzania observed no pfarps10 mutations (28, 50, 51).
Pfmrp1 and pfmrp2 are 41% identical at the amino acid level and therefore have similar antimalarial drug targets (34, 52). The pfmrp1 I876V mutation was present in 7.7% of the isolates and this is interesting because the mutant genotype has been reported to be highly selected under AL pressure in Tanzania and Zanzibar (53). Mutations at codon 885 were five unique variants (S885G/K/N/R/T) and these were observed in all three ecological zones at varied frequencies. This codon would be monitored in subsequent TES to detect the selection pressure and its relevance in drug resistance in the parasite population. The pfmrp2 K714I observed by Veiga and colleagues in Thai isolates and reported to be linked to slow parasite clearance (19) was detected in Ghanaian isolates. Another mutation reported from the same study, N622D, was not observed in our samples, however a variant N622S was detected. There were several other pfmrp2 novel mutations observed in our isolates which have not been linked to slow parasite clearance currently and need functional characterization.
The study showed a high prevalence of P. falciparum isolates with pfdhfr mutations (IRN) in Ghana as reported in previous studies over the years and is approaching saturation (7, 54, 55). Comparatively, the pfdhps mutations are in low prevalence with the key mutation, K540E for resistance to sulfadoxine still not detected in Ghanaian isolates. Interestingly, 41.1% of the isolates had a variant mutation K540N. The relevance of this mutation is yet to be ascertained. Nevertheless, this does not prevent the likelihood of sulfadoxine resistance in the population since some of the isolates had both S436A and A437G mutant alleles. This is similar to a study conducted in Burkina Faso, where none of the isolates carried the K540E mutation (56). The A581G and A613S mutants observed for failure of SP prophylaxis in pregnancy (7, 57) were also detected in this study. The pfdhps I431V mutation observed in pregnant women in Cameroon and Nigeria (36, 37) was not observed in any of the isolates which could be due to the small sample size compared to those used in the two previous studies.
The distribution of the mutations in the various genes investigated at the three ecological zones showed interesting patterns. It was detected that mutations observed in the pffd, pfnfs, pfcoronin, and pff2 were highest in the GS, whilst those of pfmrp2, pfdhps, pfdhfr, and pfmrp1 dominated in the FZ and pfarps10 mutations were predominant in the CS. The observation of shared, zone-specific, and novel mutations in the ecological zones for these genes may be suggestive that malaria transmission intensity as well as human migratory patterns, may have played a role in the distribution of mutations (5, 58–60). Typically, areas with high transmission intensity show increased exchange of genetic material amongst the parasite population which subsequently leads to the development of spontaneous mutations that can confer resistance to antimalarial drugs (5, 61). In addition, the observation of both zone-specific and shared mutations across the zones may indicate gene flow amongst individuals in the population through intra-country migration (5, 6, 62).
Broadly translated, our findings indicate that P. falciparum genes may be under drug pressure in the country following over a decade of use of ACT regimens and SP. Sustained drug pressure, aided by reduced sensitivity to ACT partner drugs may lead to strong selection for the de novo evolution of resistance in Africa. The observation of novel mutations requires further surveillance efforts to determine their roles in the tolerance or susceptibility of the Ghanaian P. falciparum populations to ACTs. Our data strongly supports the relevance of continuous molecular surveillance of markers of antimalarial drug resistance in Ghana and other malaria-endemic countries of the world to aid in the collation of evidence-based scientific data for global malaria eradication efforts.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: [https://www.ncbi.nlm.nih.gov/ from OR498143 to OR498204].
The studies involving humans were approved by Noguchi Memorial Institute for Medical Research -IRB (IRB CPN 032/05-06a amed.2021). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
SAM: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. TA: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. KT: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. PO-A: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. SB: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. NE: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. PA-D: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. EF: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. MA: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. CM: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. JF: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. OH: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. BA: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Supervision, Validation, Visualization, Writing – review & editing. KK: Conceptualization, Investigation, Supervision, Validation, Visualization, Writing – review & editing. GA: Conceptualization, Funding acquisition, Investigation, Supervision, Validation, Visualization, Writing – review & editing. NQ: Conceptualization, Investigation, Methodology, Supervision, Validation, Visualization, Writing – review & editing. ND-Q: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
SAM and the work reported were supported by funds for her PhD fellowship award from the World Bank African Centres of Excellence Grant (WACCBIP, Awandare, PI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fepid.2024.1279835/full#supplementary-material
1. WHO. Malaria Fact Sheet 2023 (2023). Available online at: Available at: https://www.who.int/news-room/fact-sheets/detail/malaria (accessed 19th January, 2024).
2. WHO. Malaria Fact Sheet 2022 (2022). Available online at: Available at: https://www.who.int/news-room/fact-sheets/detail/malaria (Accessed 26th April, 2022).
3. WHO. World Malaria Report 2021 (2021). Available online at: Available at: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021 (Accessed 26th April, 2022).
4. Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant plasmodium falciparum malaria. Nature. (2014) 505(7481):50–5. doi: 10.1038/nature12876
5. Matrevi SA, Opoku-Agyeman P, Quashie NB, Bruku S, Abuaku B, Koram KA, et al. Plasmodium falciparum kelch propeller polymorphisms in clinical isolates from Ghana from 2007 to 2016. Antimicrob Agents Chemother. (2019) 63(11):e00802–19. doi: 10.1128/AAC.00802-19
6. Matrevi SA, Tandoh KZ, Bruku S, Opoku-Agyeman P, Adams T, Ennuson NA, et al. Novel pfk13 polymorphisms in plasmodium falciparum population in Ghana. Sci Rep. (2022) 12(1):7797. doi: 10.1038/s41598-022-11790-9
7. Mensah BA, Aydemir O, Myers-Hansen JL, Opoku M, Hathaway NJ, Marsh PW, et al. Antimalarial drug resistance profiling of plasmodium falciparum infections in Ghana using molecular inversion probes and next-generation sequencing. Antimicrob Agents Chemother. (2020) 64(4):e01423–19. doi: 10.1128/AAC.01423-19
8. Mukherjee A, Bopp S, Magistrado P, Wong W, Daniels R, Demas A, et al. Artemisinin resistance without pfkelch13 mutations in plasmodium falciparum isolates from Cambodia. Malar J. (2017) 16(1):195. doi: 10.1186/s12936-017-1845-5
9. Zhu L, van der Pluijm RW, Kucharski M, Nayak S, Tripathi J, White NJ, et al. Artemisinin resistance in the malaria parasite, plasmodium falciparum, originates from its initial transcriptional response. Commun Biol. (2022) 5(1):274. doi: 10.1038/s42003-022-03215-0
10. Menard D, Dondorp A. Antimalarial drug resistance: a threat to malaria elimination. Cold Spring Harbor Perspect Med. (2017) 7(7):a025619. doi: 10.1101/cshperspect.a025619
11. Rocamora F, Winzeler EA. Genomic approaches to drug resistance in malaria. Annu Rev Microbiol. (2020) 74:761–86. doi: 10.1146/annurev-micro-012220-064343
12. Demas AR, Sharma AI, Wong W, Early AM, Redmond S, Bopp S, et al. Mutations in plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc Natl Acad Sci U S A. (2018) 115(50):12799–804. doi: 10.1073/pnas.1812317115
13. Henrici RC, Edwards RL, Zoltner M, van Schalkwyk DA, Hart MN, Mohring F, et al. The plasmodium falciparum artemisinin susceptibility-associated AP-2 adaptin mu subunit is clathrin independent and essential for schizont maturation. mBio. (2020) 11(1):e02918–19. doi: 10.1128/mBio.02918-19
14. Klonis N, Crespo-Ortiz MP, Bottova I, Abu-Bakar N, Kenny S, Rosenthal PJ, et al. Artemisinin activity against plasmodium falciparum requires hemoglobin uptake and digestion. Proc Natl Acad Sci U S A. (2011) 108(28):11405–10. doi: 10.1073/pnas.1104063108
15. Wang Z, Cabrera M, Yang J, Yuan L, Gupta B, Liang X, et al. Genome-wide association analysis identifies genetic loci associated with resistance to multiple antimalarials in plasmodium falciparum from China-Myanmar border. Sci Rep. (2016) 6:33891. doi: 10.1038/srep33891
16. Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, et al. Genetic architecture of artemisinin-resistant plasmodium falciparum. Nat Genet. (2015) 47(3):226–34. doi: 10.1038/ng.3189
17. Amambua-Ngwa A, Jeffries D, Amato R, Worwui A, Karim M, Ceesay S, et al. Consistent signatures of selection from genomic analysis of pairs of temporal and spatial plasmodium falciparum populations from the Gambia. Sci Rep. (2018) 8(1):9687. doi: 10.1038/s41598-018-28017-5
18. Dahlstrom S, Veiga MI, Martensson A, Bjorkman A, Gil JP. Polymorphism in PfMRP1 (plasmodium falciparum multidrug resistance protein 1) amino acid 1466 associated with resistance to sulfadoxine-pyrimethamine treatment. Antimicrob Agents Chemother. (2009) 53(6):2553–6. doi: 10.1128/AAC.00091-09
19. Veiga MI, Osorio NS, Ferreira PE, Franzen O, Dahlstrom S, Lum JK, et al. Complex polymorphisms in the plasmodium falciparum multidrug resistance protein 2 gene and its contribution to antimalarial response. Antimicrob Agents Chemother. (2014) 58(12):7390–7. doi: 10.1128/AAC.03337-14
20. Plowe CV, Cortese JF, Djimde A, Wellems TE. Mutations in plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. (1997) 176:1590–6. doi: 10.1086/514159
21. Sutherland CJ, Fifer H, Pearce RJ, bin Reza F, Nicholas M, Haustein T, et al. Novel pfdhps haplotypes among imported cases of plasmodium falciparum malaria in the United Kingdom. Antimicrob Agents Chemother. (2009) 53(8):3405–10. doi: 10.1128/AAC.00024-09
22. Mikomangwa WP, Minzi O, Mutagonda R, Baraka V, Mlugu EM, Aklillu E, et al. Effect of sulfadoxine-pyrimethamine doses for prevention of malaria during pregnancy in hypoendemic area in Tanzania. Malar J. (2020) 19(1):160. doi: 10.1186/s12936-020-03234-4
23. Plowe CV. The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg. (2009) 103(Suppl 1):S11–4. doi: 10.1016/j.trstmh.2008.11.002
25. Abuaku B, Duah-Quashie NO, Quashie N, Gyasi A, Afriyie PO, Owusu-Antwi F, et al. Trends and predictive factors for treatment failure following artemisinin-based combination therapy among children with uncomplicated malaria in Ghana: 2005–2018. BMC Infect Dis. (2021) 21(1):1255. doi: 10.1186/s12879-021-06961-4
26. Siddiqui FA, Cabrera M, Wang M, Brashear A, Kemirembe K, Wang Z, et al. Plasmodium falciparum falcipain-2a polymorphisms in Southeast Asia and their association with artemisinin resistance. J Infect Dis. (2018) 218(3):434–42. doi: 10.1093/infdis/jiy188
27. Conrad MD, Bigira V, Kapisi J, Muhindo M, Kamya MR, Havlir DV, et al. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in plasmodium falciparum isolated from Ugandan children. PloS One. (2014) 9(8):e105690. doi: 10.1371/journal.pone.0105690
28. Wamae K, Okanda D, Ndwiga L, Osoti V, Kimenyi KM, Abdi AI, et al. No evidence of P. Falciparum K13 artemisinin conferring mutations over a 24-year analysis in coastal Kenya, but a near complete reversion to chloroquine wild type parasites. Antimicrob Agents Chemother. (2019) 63(12):e01067–19. doi: 10.1128/AAC.01067-19
29. Delandre O, Daffe SM, Gendrot M, Diallo MN, Madamet M, Kounta MB, et al. Absence of association between polymorphisms in the pfcoronin and pfk13 genes and the presence of plasmodium falciparum parasites after treatment with artemisinin derivatives in Senegal. Int J Antimicrob Agents. (2020) 56(6):106190. doi: 10.1016/j.ijantimicag.2020.106190
30. Velavan TP, Nderu D, Agbenyega T, Ntoumi F, Kremsner PG. An alternative dogma on reduced artemisinin susceptibility: a new shadow from east to west. Proc Natl Acad Sci U S A. (2019) 116(26):12611–2. doi: 10.1073/pnas.1907142116
31. Owoloye A, Olufemi M, Idowu ET, Oyebola KM. Prevalence of potential mediators of artemisinin resistance in African isolates of plasmodium falciparum. Malar J. (2021) 20(1):451. doi: 10.1186/s12936-021-03987-6
32. Delandre O, Gendrot M, Fonta I, Mosnier J, Benoit N, Amalvict R, et al. Prevalence of mutations in the pfcoronin gene and association with ex vivo susceptibility to common quinoline drugs against Plasmodium falciparum. Pharmaceutics. (2021) 13(8):1273. doi: 10.3390/pharmaceutics13081273
33. Ocan M, Ashaba FK, Mwesigwa S, Edgar K, Kamya MR, Nsobya SL. Prevalence of arps10, fd, pfmdr-2, pfcrt and pfkelch13 gene mutations in plasmodium falciparum parasite population in Uganda. PloS One. (2022) 17(5):e0268095. doi: 10.1371/journal.pone.0268095
34. Gupta B, Xu S, Wang Z, Sun L, Miao J, Cui L, et al. Plasmodium falciparum multidrug resistance protein 1 (pfmrp1) gene and its association with in vitro drug susceptibility of parasite isolates from north-east Myanmar. J Antimicrob Chemother. (2014) 69(8):2110–7. doi: 10.1093/jac/dku125
35. Lo E, Nguyen J, Oo W, Hemming-Schroeder E, Zhou G, Yang Z, et al. Examining plasmodium falciparum and P. vivax clearance subsequent to antimalarial drug treatment in the Myanmar-China border area based on quantitative real-time polymerase chain reaction. BMC Infect Dis. (2016) 16:154. doi: 10.1186/s12879-016-1482-6
36. Chauvin P, Menard S, Iriart X, Nsango SE, Tchioffo MT, Abate L, et al. Prevalence of plasmodium falciparum parasites resistant to sulfadoxine/pyrimethamine in pregnant women in yaounde, Cameroon: emergence of highly resistant pfdhfr/pfdhps alleles. J Antimicrob Chemother. (2015) 70(9):2566–71. doi: 10.1093/jac/dkv160
37. Oguike MC, Falade CO, Shu E, Enato IG, Watila I, Baba ES, et al. Molecular determinants of sulfadoxine-pyrimethamine resistance in plasmodium falciparum in Nigeria and the regional emergence of dhps 431V. Int J Parasitol Drugs Drug Resist. (2016) 6(3):220–9. doi: 10.1016/j.ijpddr.2016.08.004
38. Brooks DR, Wang P, Read M, Watkins WM, Sims PF, Hyde JE. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. (1994) 224(2):397–405. doi: 10.1111/j.1432-1033.1994.00397.x
39. Gregson A, Plowe CV. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev. (2005) 57(1):117–45. doi: 10.1124/pr.57.1.4
40. Triglia T, Cowman AF. Primary structure and expression of the dihydropteroate synthetase gene of plasmodium falciparum. Proc Natl Acad Sci U S A. (1994) 91(15):7149–53. doi: 10.1073/pnas.91.15.7149
41. Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of plasmodium falciparum. Proc Natl Acad Sci U S A. (1988) 85(23):9109–13. doi: 10.1073/pnas.85.23.9109
42. A-Elbasit IE, Alifrangis M, Khalil IF, Bygbjerg IC, Masuadi EM, Elbashir MI, et al. The implication of dihydrofolate reductase and dihydropteroate synthetase gene mutations in modification of plasmodium falciparum characteristics. Malar J. (2007) 6:108. doi: 10.1186/1475-2875-6-108
43. Menard D, Andriantsoanirina V, Jahevitra M, Barnadas C, Tichit M, Bouchier C, et al. Dihydrofolate reductase I164l mutation in plasmodium falciparum, Madagascar. Emerg Infect Dis. (2008) 14(7):1166–7. doi: 10.3201/eid1407.071498
44. Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A. (1988) 85(23):9114–8. doi: 10.1073/pnas.85.23.9114
45. Abuaku B, Boateng P, Peprah NY, Asamoah A, Duah-Quashie NO, Matrevi SA, et al. Therapeutic efficacy of dihydroartemisinin-piperaquine combination for the treatment of uncomplicated malaria in Ghana. Front Cell Infect Microbiol. (2022) 12:1058660. doi: 10.3389/fcimb.2022.1058660
46. Diallo MA, Yade MS, Ndiaye YD, Diallo I, Diongue K, Sy SA, et al. Efficacy and safety of artemisinin-based combination therapy and the implications of Pfkelch13 and pfcoronin molecular markers in treatment failure in Senegal. Sci Rep. (2020) 10(1):8907. doi: 10.1038/s41598-020-65553-5
47. Khan AQ, Pernaute-Lau L, Khattak AA, Luijcx S, Aydin-Schmidt B, Hussain M, et al. Surveillance of genetic markers associated with plasmodium falciparum resistance to artemisinin-based combination therapy in Pakistan, 2018-2019. Malar J. (2020) 19(1):206. doi: 10.1186/s12936-020-03276-8
48. Hussien M, Abdel Hamid MM, Elamin EA, Hassan AO, Elaagip AH, Salama AHA, et al. Antimalarial drug resistance molecular makers of plasmodium falciparum isolates from Sudan during 2015–2017. PloS One. (2020) 15(8):e0235401. doi: 10.1371/journal.pone.0235401
49. Nyunt MH, Wang B, Aye KM, Aye KH, Han JH, Lee SK, et al. Molecular surveillance of artemisinin resistance falciparum malaria among migrant goldmine workers in Myanmar. Malar J. (2017) 16(1):97. doi: 10.1186/s12936-017-1753-8
50. Bwire GM, Ngasala B, Mikomangwa WP, Kilonzi M, Kamuhabwa AAR. Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in southeast of Tanzania. Sci Rep. (2020) 10(1):3500. doi: 10.1038/s41598-020-60549-7
51. Diakite SAS, Traore K, Sanogo I, Clark TG, Campino S, Sangare M, et al. A comprehensive analysis of drug resistance molecular markers and plasmodium falciparum genetic diversity in two malaria endemic sites in Mali. Malar J. (2019) 18(1):361. doi: 10.1186/s12936-019-2986-5
52. Rijpma SR, van der Velden M, Bilos A, Jansen RS, Mahakena S, Russel FG, et al. MRP1 Mediates folate transport and antifolate sensitivity in plasmodium falciparum. FEBS Lett. (2016) 590(4):482–92. doi: 10.1002/1873-3468.12079
53. Dahlstrom S, Ferreira PE, Veiga MI, Sedighi N, Wiklund L, Martensson A, et al. Plasmodium falciparum multidrug resistance protein 1 and artemisinin-based combination therapy in Africa. J Infect Dis. (2009) 200(9):1456–64. doi: 10.1086/606009
54. Duah NO, Quashie NB, Abuaku BK, Sebeny PJ, Kronmann KC, Koram KA. Surveillance of molecular markers of plasmodium falciparum resistance to sulphadoxine-pyrimethamine 5 years after the change of malaria treatment policy in Ghana. Am J Trop Med Hyg. (2012) 87(6):996–1003. doi: 10.4269/ajtmh.2012.12-0202
55. Osei M, Ansah F, Matrevi SA, Asante KP, Awandare GA, Quashie NB, et al. Amplification of GTP-cyclohydrolase 1 gene in plasmodium falciparum isolates with the quadruple mutant of dihydrofolate reductase and dihydropteroate synthase genes in Ghana. PloS One. (2018) 13(9):e0204871. doi: 10.1371/journal.pone.0204871
56. Tahita MC, Tinto H, Erhart A, Kazienga A, Fitzhenry R, VanOvermeir C, et al. Prevalence of the dhfr and dhps mutations among pregnant women in rural Burkina Faso five years after the Introduction of intermittent preventive treatment with sulfadoxine-pyrimethamine. PloS One. (2015) 10(9):e0137440. doi: 10.1371/journal.pone.0137440
57. Yan H, Feng J, Yin JH, Huang F, Kong XL, Lin KM, et al. High frequency mutations in pfdhfr and pfdhps of plasmodium falciparum in response to sulfadoxine-pyrimethamine: a cross-sectional survey in returning Chinese migrants from Africa. Front Cell Infect Microbiol. (2021) 11:673194. doi: 10.3389/fcimb.2021.673194
58. Dery DB, Brown C, Asante KP, Adams M, Dosoo D, Amenga-Etego S, et al. Patterns and seasonality of malaria transmission in the forest-savannah transitional zones of Ghana. Malar J. (2010) 9:314. doi: 10.1186/1475-2875-9-314
59. Kasasa S, Asoala V, Gosoniu L, Anto F, Adjuik M, Tindana C, et al. Spatio-temporal malaria transmission patterns in navrongo demographic surveillance site, northern Ghana. Malar J. (2013) 12:63. doi: 10.1186/1475-2875-12-63
60. Tchouassi DP, Quakyi IA, Addison EA, Bosompem KM, Wilson MD, Appawu MA, et al. Characterization of malaria transmission by vector populations for improved interventions during the dry season in the kpone-on-sea area of coastal Ghana. Parasit Vectors. (2012) 5:212. doi: 10.1186/1756-3305-5-212
61. MalariaGen. Malariagen plasmodium falciparum community project. Genomic epidemiology of artemisinin resistant malaria. eLIFE. (2016) 5:e08714. doi: 10.7554/eLife.08714
Keywords: malaria, artemisinin-based combination therapy, sulphadoxine-pyrimethamine, antimalarial drug resistance, molecular markers
Citation: Matrevi SA, Adams T, Tandoh KZ, Opoku-Agyeman P, Bruku S, Ennuson NA, Apau-Danso PK, Fiagbedzi E, Avornyo M, Myers CJ, Futagbi J, Hagan OC, Abuaku B, Koram KA, Awandare G, Quashie NB and Duah-Quashie NO (2024) Putative molecular markers of Plasmodium falciparum resistance to antimalarial drugs in malaria parasites from Ghana. Front. Epidemiol. 4:1279835. doi: 10.3389/fepid.2024.1279835
Received: 18 August 2023; Accepted: 29 January 2024;
Published: 15 February 2024.
Edited by:
Ruchi Singh, National Institute of Pathology (ICMR), IndiaReviewed by:
Praveen K. Bharti, National Institute of Malaria Research (ICMR), India© 2024 Matrevi, Adams, Tandoh, Opoku-Agyeman, Bruku, Ennuson, Apau-Danso, Fiagbedzi, Avornyo, Myers, Futagbi, Hagan, Abuaku, Koram, Awandare, Quashie and Duah-Quashie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nancy Odurowah Duah-Quashie bmR1YWhAbm9ndWNoaS51Zy5lZHUuZ2g=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.