- 1Department of Clinical Laboratory, The First Affiliated Hospital of Henan University of Science and Technology, Luoyang, Henan, China
- 2Department of Clinical Laboratory, Second Affiliated Hospital of Lanzhou University, Lanzhou, Gansu, China
- 3Department of Clinical Laboratory, Zibo First Hospital, Zibo, Shandong, China
- 4Department of Clinical Laboratory, Second Affiliated Hospital of Zhejiang University, Hangzhou, Zhejiang, China
Carbapenem-resistant Enterobacterales (CRE) is a global concern. This study investigated the prevalence of fecal colonization carriage and clonal dissemination of CRE among population in four provinces of China. A total of 685 stool samples were collected from four provinces in China. Among these samples, 141 and 544 were obtained from healthy and hospitalized individuals, respectively. The overall fecal carriage rate was 9.6% (65/685) with 4.26% (95% CI: 0.9–7.6) in healthy individuals and 10.84% (95% CI: 8.2–13.5) in hospitalized patients. The highest prevalence was in Henan province (18.35%, 95% CI: 9%–18.7%). Sixty-six CRE isolates were identified in Escherichia coli (56.06%, 37/66), Klebsiella (15.15%, 10/66), Citrobacter (13.63%, 9/66), Enterobacter (12.12%, 8/66), and Atlantibacter (1.51%, 1/66). All CRE strains carried carbapenemase genes and multiple antibiotics resistance genes, blaNDM−5 (77.27%, 51/66) was the most common carbapenemase gene, followed by blaNDM−1 (19.69%, 13/66). Antibiotic resistance genes, including blaIMP−4, and the colistin colistin resistance (mcr-1) gene were also identified. All CRE isolates belonged to different sequence types (STs). ST206 (36.84%, 14/38) in E. coli and ST2270 (60%, 6/10) in Klebsiella were significantly dominant clones. The results indicated the prevalence of CRE fecal carriage among adults of China, mostly blaNDM-producing E coli, which pose significant challenges for clinical management. Screening for CRE colonization is necessary to control infection.
1. Introduction
Carbapenem-resistant Enterobacterales (CRE) is experiencing rapid global dissemination. According to data from the China Antimicrobial Surveillance Network (CHINET), the rates of carbapenem resistance in Escherichia coli and Klebsiella pneumoniae in China increased from 0% and 2.9% in 2005 to 2% and 21.4% in 2022, respectively. In Japan, the yearly number of reported cases of CRE infections has remained rather stable since 2015, with a stool CRE carriage rate of 12.2% (1). However, this rate drastically increased in 2018. In Taiwan, the prevalence rates of CRE among all clinical Enterobacteriaceae isolates have risen to approximately 7% in 2013; 2017 data from in intensive care units (ICUs) revealed a prevalence rate of 15.3% (2). Because of the increasing prevalence and limitation of treatment options, CRE infection leads to higher resistance rate and a considerable mortality risk (3).
Carbapenemases consist of five major enzymes, including K. pneumoniae carbapenemases (KPC), imipenemase metallo-β-lactamase (IMP), veronaintegron-encoded metallo-β-lactamase (VIM), New Delhi metallo-β-lactamase (NDM), and oxacillinase-48-type carbapenemases (OXA-48) (4). The production of these enzymes constitutes the primary resistance mechanism and is found in approximately 85% of CRE cases worldwide (5). Carbapenemase-encoding genes are usually present on mobile genetic elements, which are driving the increasing rapid spread and extensive distribution of CRE through multiple routes (6, 7). Therefore, the infection control risk of CRE is likely higher (8). CRE colonization is significantly associated with CRE infection.
Recently, rectal or perirectal swabs have been utilized for screening based on Centers for Disease Control and Prevention and Healthcare Infection Control Practices Advisory Committee recommendations. These swabs might produce higher yields compared to testing of other body sites (9). Early identification of CRE and detection of asymptomatic carriers through rectal surveillance is exceedingly important and can reduce dissemination (10, 11). This fecal carriage is an important concern for the infection control practitioner because the gut can be considered a reservoir for exchanging resistance genes among bacteria. As a strategy to control cross-transmission, screening of CRE intestinal carriage is used by many hospitals, especially in ICUs. A previous study reported CRE carriage rates of 0.5%–12.2% among hospital patients in different areas of the world (12–16), with rates of 75.5% in patients with hematological malignancies (17) and 6.79% in those with liver disease (18).
However, minimal data regarding the prevalence and mobile resistance elements of CRE fecal carriage colonization among population with various regions are available. The dearth of reliable information on the CRE-mediated infection impacts patient health. Increased knowledge would aid the understanding of CRE transmission characteristics. Therefore, addressing the prevalence and molecular epidemiological of CRE requires further attention.
In this study, we isolated and analyzed stool samples in various regions in China, including Gansu, Zhejiang, Henan, and Shandong provinces. The data provided up-to-data knowledge on epidemiological and genomic levels of CRE fecal carriage.
2. Materials and methods
2.1. Retrospective screening of CRE isolates
A total of 685 fecal individual samples were collected from five hospitals in four distinct provinces. Convenient sampling was used. Healthy individual stool samples were also collected. No duplicate samples were isolated from the same individual. The number of specimens for each region was as follows: Zhejiang (n = 274), Henan (n = 195), Shandong (n = 116), and Gansu (n = 100). Among the 685 fecal swabs, 141 samples were obtained from healthy individuals. All strains were identified by matrix-assisted laser desorption/ionization- time-of-flight mass spectrometry (MALDI-TOF MS) using a Microflex LT instrument (Bruker Daltonik GmbH, Bremen, Germany).
2.2. Detection of carbapenem genes
All CRE were isolated according to a previously described method (19, 20). Briefly, approximately five grams of each fresh fecal specimen was directly cultured in 5 ml of LB broth (Luqiao, Beijing, China) and enriched at 35°C for 22–24 h. Ten microliter aliquots of the culture were inoculated onto two selective China Blue agar plates to screen for non-susceptible Enterobacterales isolates. One contained 0.3 μg/ml meropenem and the other 8 μg/ml Ceftazidime-avibactam. The plates were incubated at 35°C for 16–18 h. Colonies on the plate were selected for further purification and confirmation as Enterobacterales. Isolates from the same patient exhibiting different colonial morphotypes were also studied. Only one suspected colony was analyzed per sample. Colonies were screened for the presence of five common carbapenemase genes (blaKPC, blaNDM, blaOXA−48−like, blaIMP, and blaVIM) using the NG-Test Carba5 assay (NG Biotech, Guipry, France) as previously described (21). According to the manufacturer's instructions, a colony of a pure cultivated strain was mixed with five drops of lysis buffer. After vortexing, the mixture was left at 20–25°C for 10 min. Once hundred microliters of the mixture was transferred to the NG-Test Carba5 cassette. The results were read after 15 min of incubation.
2.3. Antimicrobial susceptibility testing
Susceptibility to antimicrobials was tested by the broth microdilution method. The tested antimicrobials included amikacin, imipenem, meropenem, ertapenem, cefmetazole, cefotaxime, aztreonam, ceftazidime, tazobactam and piperacillin, cefepime, ciprofloxacin, colistin, tigecycline, ceftazidime-avibactam, and cefoperazone-sulbactam. Interpretation of minimum inhibitory concentration (MIC) breakpoints was based on guidelines given by the CLSI criteria (22). European Committee on Antimicrobial Susceptibility Testing breakpoints were used for tigecycline (23).
2.4. Whole genome sequencing and bioinformatics analysis
Total genomic DNA was extracted from overnight cultures of 66 carbapenem-resistant Enterobacterales isolates using the PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA) according to the provided instructions. All isolates harboring carbapenem genes underwent whole genome sequencing using the HiSeq platform (Illumina, San Diego, CA, USA). De novo assembly was performed using SPAdes version 3.15.1 (24). The locations of carbapenem genes were determined by NCBI databases. Multilocus sequence typing (MLST) of strains was performed using the MLST tool (https://github.com/tseemann/mlst) and contigs were assembled. Sequence types (STs) and corresponding MLST gene allele profiles were recorded in BioNumerics v7.6.3 (Applied Maths, Sint-Martens-Latem, Belgium). The assembled sequences were analyzed using the ResFinder bioinformatic database (25) and ISFinder (https://www-is.biotoul.fr/) bioinformatic tools. Advanced Heatmap Plots for antimicrobial resistance (AMR) genes were performed using the OmicStudio tools (https://www.omicstudio.cn). The molecular features of each strain were visualized using the iTOL online tool (26) and Phyloviz (http://www.phyloviz.net/) for eBURST analysis.
Accession numbers
All sequences have been deposited at GenBank under BioProject number PRJNA1023209.
2.5. Statistical analysis
Descriptive statistics were used to summarize the epidemiologic characteristics of CRE strains. For categorical variables, the percentage of CRE strains in each category was calculated. All analyses were performed using the SPSS Statistics (version 21; IBM, Armonk, NY, USA).
3. Results
3.1. Prevalence characterization of CRE strains in four provinces in China
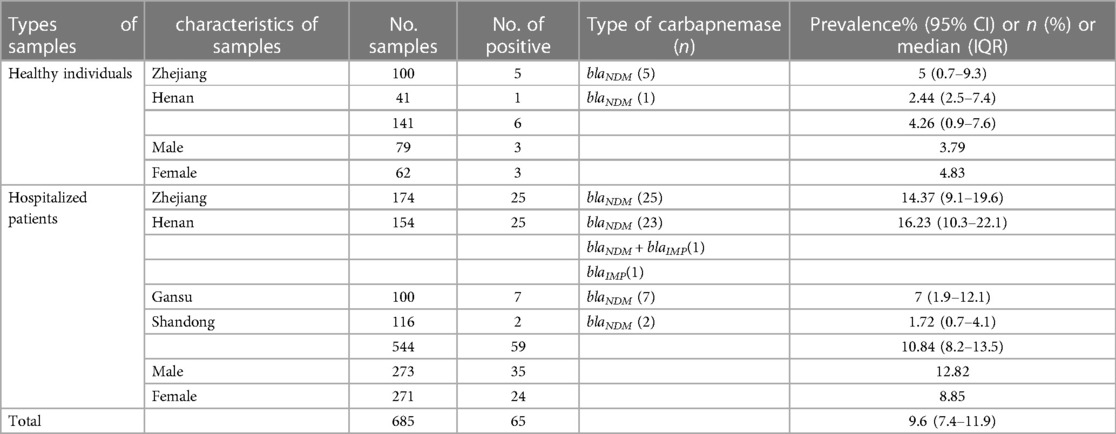
Fecal colonization rates of CRE in samples collected in four different provinces were determined. The largest number of samples were from the provinces of Zhejiang (n = 274) and Henan (n = 195). Of the 685 human fecal samples, 65 tested positive for CRE. Sixty-six CRE strains were isolated from 65 patient, representing a carriage rate of 9.6% (95% CI: 7.4%–11.9%).
A total of 141 fecal samples were obtained from healthy persons. From these samples, six (4.26%, 95% CI: 0.9–7.6) CRE strains were identified, which were isolated from three males and three females with a median age (IQR) of 58.5 (54.25–65) years. Five strains were isolated from 100 samples (5%, 95% CI: 0.7%–9.3%) from Zhejiang and 1 strain was isolated from 41 samples (2.44%, 95% CI: 2.5%–7.4%) from Henan.
A total of 544 fecal samples were gathered from hospitalized patients, from which 59 (10.84%, 95% CI: 8.2%–13.5%) CRE strains were isolated. The prevalence of CRE in the hospitalized patients was significantly higher than the healthy individuals (P = 0.001). These patients included 35 males and 24 females with a median age (IQR) of 57 (48–67) years. The prevalence rate was highest in Henan province (16.23%, 95% CI: 10.3–22.1%), followed by Zhejiang province (14.37%, 95% CI: 9.1%–19.6%). Seven samples tested positive in Gansu province, yielding a prevalence rate of 7% (95% CI: 1.9%–12.1%). Only two samples from Shandong province were positive, indicating a prevalence rate of 1.72% (95% CI: 0.7%–4.1%) (Table 1).
3.2. Antibiotic resistance patterns of CRE strains
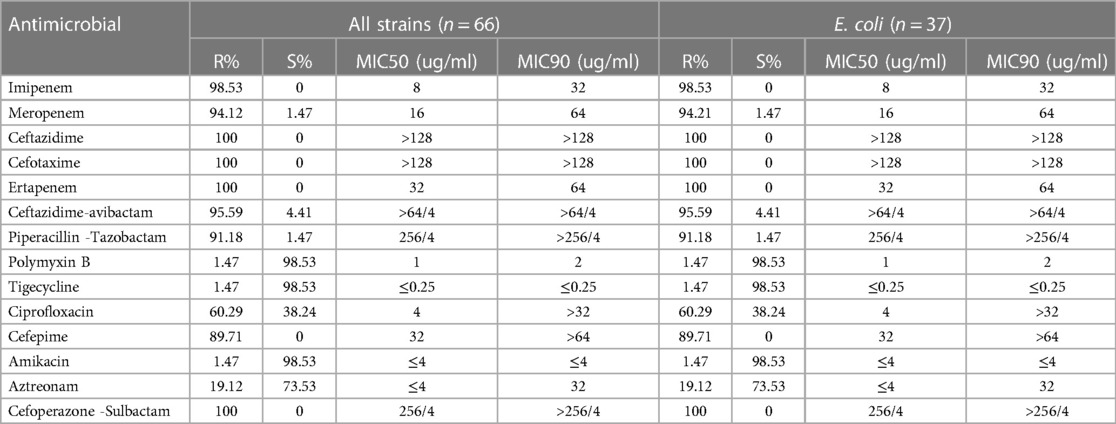
Antimicrobial susceptibility testing was performed on all carbapenem-resistant strains using 14 clinical antibiotics. The CRE strains exhibited 100% resistance to multiple antibiotics that included ceftazidime, cefotaxime, ertapenem, and cefoperazone-sulbactam. Additionally, notable high resistance rates were evident for imipenem (98.53%), meropenem (94.12%), ceftazidime-avibactam (95.59%), piperacillin-tazobactam (91.18%), and cefepime (89.71%). Low resistance rates were observed for polymyxin B (1.47%), tigecycline (1.47%), and amikacin (1.47%). A comprehensive overview of resistance profiles of the 66 CRE positive strains against 14 antimicrobial agents is presented in Table 2. E. coli strains displayed the same resistance to antibiotics agents.
3.3. Diversity of carbapenem resistance genes among CRE
Whole genome sequencing analysis revealed that the CRE comprised different genera. Escherichia was the most prevalent genus (57.57%, 38/66), followed by Klebsiella (15.15%, 10/66), Citrobacter (13.63%, 9/66), Enterobacter (12.12%, 8/66), and Atlantibacter (1.51%, 1/66). All Escherichia and Enterobacter were identified as E. coli and E. hormaechei, respectively. In contrast, Klebsiella consisted of various species, including seven K. pneumoniae isolates, one K. quasipneumoniae, one K. pasteurii, and one K. planticola. Citrobacter isolates included three C. braakii, two C. portucalensis, one C. freundii and three other Citrobacter isolates (Figure 1).
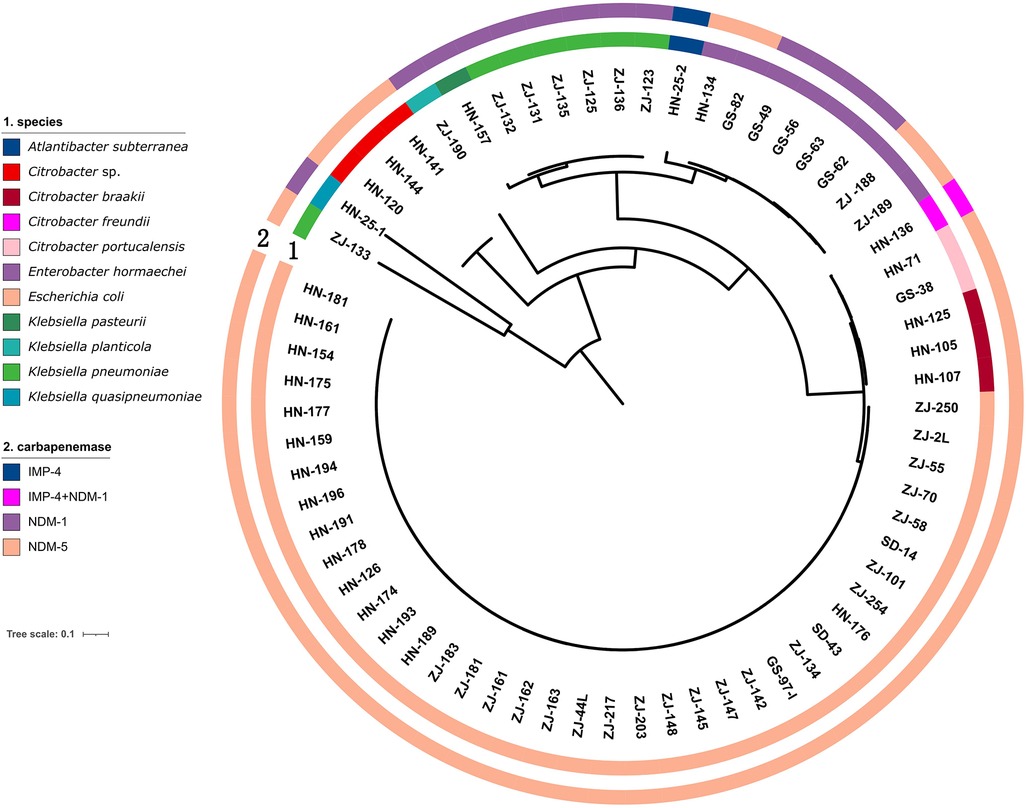
Figure 1. Genetic analysis of 66 CRE isolates. Circles 1 and 2 indicate the isolates and the carbapenemase genes, respectively.
All CRE isolates encoded carbapenemase genes (Figure 2). Among 66 CRE isolates, 77.27% (51/66) harbored blaNDM−5 and 19.69% (13/66) harbored blaNDM−1. Both blaIMP−4 and blaNDM−1 were detected in one C. freundii isolated from a male, 59-year-old patient with a tumor. The patient had no history of travel in the preceding 6 months. The blaIMP−4 gene was detected in one Atlantibacter subterranean isolated from Henan. One E. coli strain harbored both blaNDM−5 and mcr-1 gene; it exhibited resistance to colistin. The prevalence of the blaNDM genes varied among different strains of CRE. Specifically, the blaNDM−5 gene was detectable in all tested E. coli isolates, while it was present in only 10% of Klebsiella isolates. blaNDM−1 was detected in all Enterobacter isolates and 90% of Klebsiella isolates.
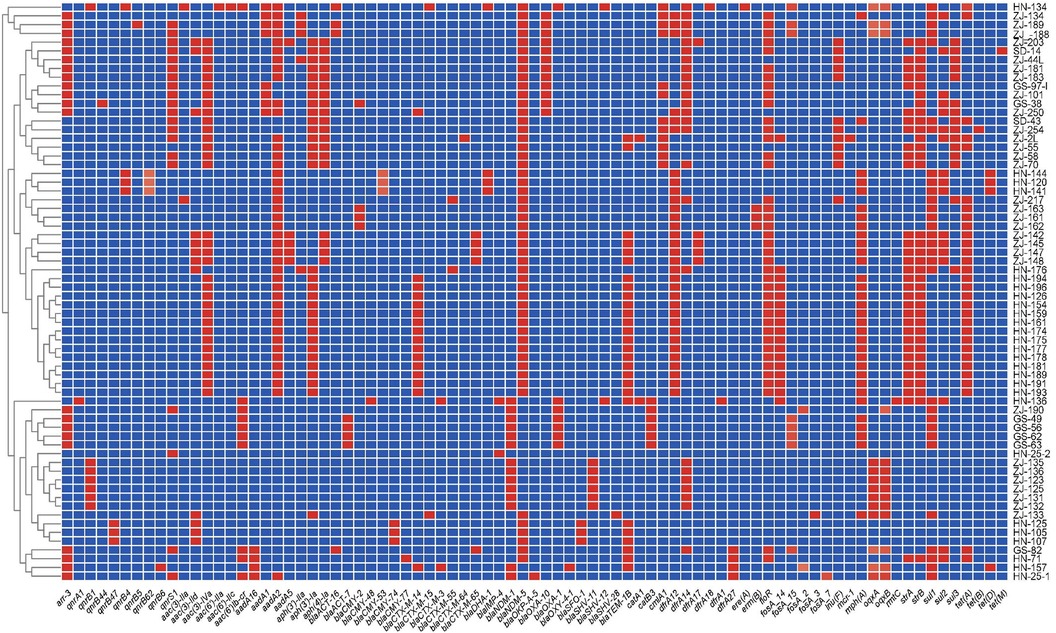
Figure 2. Heatmap of antimicrobial resistance genes of 66 CRE strains collected in the four provinces in China. The X-axis denotes the antimicrobial resistance genes harbored by each strain. The Y-axis denotes the CRE strains. Labels in the Y-axis represent the source of the strain. ZJ, Zhejiang; GS, Gansu; HN, Henan; and SD, Shandong. Red and blue colors indicate the presence and absence of the corresponding antimicrobial resistance genes, respectively.
Sequence analysis identified various acquired drug resistance genes among the 66 CRE isolates (Figure 2). A number of diverse acquired resistance genes were identified, including aminoglycosides resistance genes (aac(3)-IIa, aac(3)-IId, aac(3)-IV, aac(6')-Ib-cr, aac(6')-IIa, aac(6’)-IIc, aadA16, aadA2, aadA5, aph(3')-Ia, aph(3’)-IIa, and aph(4)-Ia), quinolone resistance genes (qnrS1, qnrA1, qnrB4, qnrB44, qnrB47, qnrB5, qnrB62, qnrB6, and qnrS1), extended spectrum beta-lactamase (ESBL) resistance genes [CTX-M-3, SHV-12, CTX-M-14, OXY-4-1, SFO-1, CTX-M-65, CTX-M-15, tetracycline resistance genes (tetA, tetD, tetB, and tetM), AmpC β-lactamase gene(blaCMY−48, blaACT−17, blaCMY−157, blaCMY−82, blaACT−25, blaCMY−2, blaDHA−1], and other narrow-spectrum β-lactam resistance genes (blaOXA−1, blaTEM−1D, blaOXA−10, and blaOKP−A−5, blaSHV−11, blaSHV−28). In the 66 CRE isolates, aminoglycoside genes were identified in 58 strains, the most prevalent aac(3)-Iv (n = 33). Thirty-seven CRE isolates also carried AmpC β-lactamase genes, including blaCMY (n = 14), blaACT (n = 8), blaDHA−1 (n = 4). Thirty-seven isolates harbored ESBL genes, including blaCTX−M (n = 27), blaSFO−1 (n = 3), and blaSHV−12 (n = 2). Finally, 38 strains harbored tetracycline resistance genes, mostly tetA (n = 32).
3.4. Distribution of STs among carbapenemase-producing CRE isolates
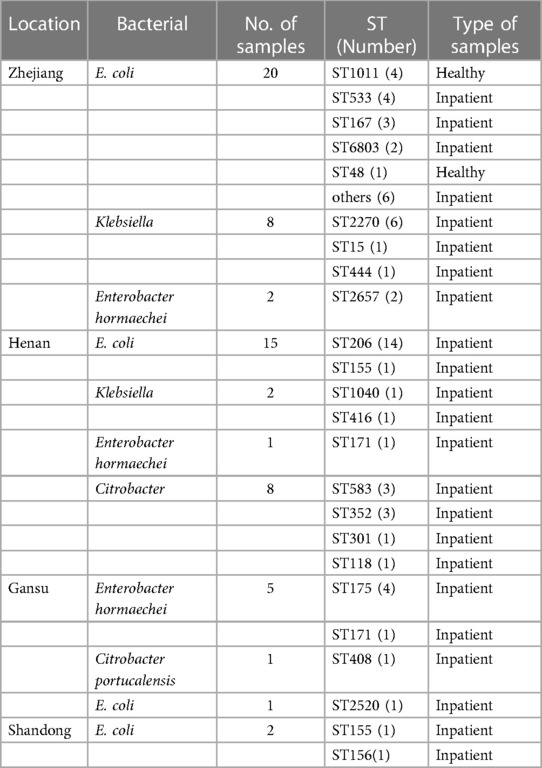
Among the 38 E. coli harboring carbapenemase genes, 15 distinct STs were identified. Notably, ST206 (36.84%, 14/38) was the predominant type, followed by ST1011 (10.52%, 4/38), ST533 (10.52%, 4/38), and ST167 (7.89%, 3/38) (Figure 3). ST206 remained the primary type in Henan province, suggesting a notable role of clonal dissemination in the propagation of carbapenemase-producing E. coli strains. Figure 3 provides further insight into the top 3 STs of E. coli in Zhejiang province. The results suggest the important role of non-clonal dissemination. Interestingly, ST155 was detected in both Henan and Shandong provinces. Regarding the 10 carbapenemase-producing Klebsiella isolates, five distinct STs were identified. ST2270 (60%, 6/10) was the most dominant, with a single isolate each of ST15, ST444, ST1040, and ST416. Another noteworthy observation was the close association between specific ST types and the presence of particular carbapenemase genes. Importantly, all ST types of E. coli harbored blaNDM−5. A similar phenomenon was observed in carbapenem-resistant Klebsiella isolates; ST2270, ST444, ST1040 and ST416 predominantly harbored blaNDM−1, while ST15 harbored blaNDM−5 (Table 3).
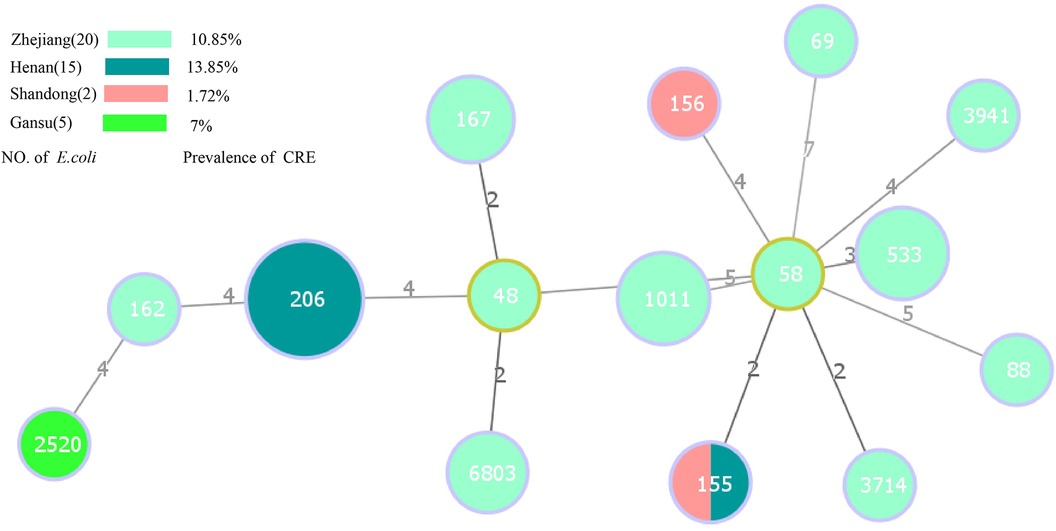
Figure 3. Minimal spanning tree based on multilocus sequence typing of carbapenem-resistant E. coli. Colored circles represent different location. The circle sizes are proportional to the number of STs. The length of the connecting lines represents the number of target genes with different alleles. Red dots indicate ST48 and ST1011 E. coli strains isolated from healthy individuals, the remaining strains are isolated from inpatient.
A surprising observation concerned carbapenem-resistant Citrobacter freundii, ST118 collected from Henan province harbored two carbapenemase genes, blaNDM−1 and blaIMP−4. Furthermore, ST175 and ST171 were observed among the eight carbapenemase-producing E. hormaechei in Gansu province, which harbored blaNDM−1 gene. Conversely, an uncertain classification of A. subteranean isolate harbored blaIMP−4.
4. Discussion
CRE is a global crisis that is associated with high mortality rates and poses a severe threat to public health (27, 28). Comprehensive understanding of the epidemiology and symptoms of CRE are vital, especially in location active surveillance. This knowledge would help facilities determine control measures.
In this study, 9.6% of samples were characterized as CRE, with carriage rates of 4.26% from healthy individuals and 10.84% from hospitalized inpatients. These rates are higher than the rates of 6.6% and 8.5% in 2014 and 2019 studies from China, respectively (29, 30), but lower than the rates of 12.2% in Japan (31) and 37.9% in Iran (32). Previous studies among non-hospitalized human rectal swabs reported carriage rates of CRE of 0.6% in Vietnam (33), 0.4% in Spain (34), and 6.1% in India (35). However, the prevalence of CRE (10.84%) among hospitalized patients was a much lower than the 13% reported from Vietnam (36), and higher than the 2.9% rate in Spain (34). The increased prevalence may also be associated with failure to control CRE and broad-spectrum antibiotic use in hospitals. The previous studies were performed in hospitalized patients, acute rehabilitation units, long-term care hospitals, and acute-care hospitals. In the present study, we detected stool samples in healthy people and inpatients from four different provinces. It is pertinent to point out that a direct comparison of these studies should be done with caution, given the different settings, selective methods, and patients. Our results suggested that Henan province had a higher positive rate, which is consistent with previous study (37). We also observed that the rate of CRE was higher than the previous reported rate (38), because this study was conducted in ICUs.
The most commonly identified CRE was E. coli followed by Klebsiella and Citrobacter. A previous study reported different results (39). In another study, Citrobacter was the third most common isolates following Klebsiella and E. coli, similar to this study (40). blaNDM is the predominant resistance gene in E. coli strains. Other carbapenemase genes, such as blaIMP−4, were notably rare and detected only in a single isolate of A. subterranean. Simultaneously, one C. freundii strain, isolated from Henan province, harbored both blaNDM−1 and blaIMP−4. These findings may indicate that blaNDM was the key carbapenemase gene responsible for mediating development of the carbapenem resistance phenotypes in China. Previous studies suggested blaNDM−5 was mostly located on a 46-kb IncX3 plasmids, which are increasingly associated with the dissemination of CRE (41, 42). Because of rapid and widespread of blaNDM, there is a pressing need to develop novel therapies to treat CRE producing NDM carbapenemase. However, current new antibiotics like ceftazidime-avibactam may not be suitable to remedy CRE in China (8).
MLST analysis showed that STs of CRE isolated in this study were genetically highly diverse. E. coli included 15 ST types, ST206 was the major type, followed by ST533 and ST1011. All strains harbored blaNDM−5. Other strain types were relatively rare and more sporadic. As for Klebsiella, ST2270, ST444, ST1040 and ST416 predominantly harbored blaNDM−1, while ST15 harbored blaNDM−5. A previous study suggested that ST11 of K. pneumoniae (mainly harboring blakpc−2) was a major strain type in China (19). ST11, ST15, ST258, and ST512 are reportedly the main types in Europe and America (18). For E. coli, ST174 and ST648 that harbor blaNDM−5 are prevalent in India (43). However, in Korea, ST101 harboring blaNDM−1 is most prevalent (44). This variance may also be associated with the sample numbers and different regions in the different studies. Our findings showed the clonal and non-clonal transmission of E. coli harboring blaNDM in Henan and Zhejiang provinces, which reflects the high risk of specific clones disseminating.
There were several limitations in our study. These included the small sample size. The lack of discussion of the regional differences was the biggest flaw of this study. In addition, there were inconsistencies in collection of fecal samples from healthy and hospitalized individuals in provinces where fecal samples were collected. These inconsistencies may have biased the findings. Therefore, the true proportion of CRE in China was not able to be determined. Further studies should be performed.
In conclusion, a surveillance of healthy persons and hospitalized patients rectal carriage by CRE in four provinces in China was performed. We describe the prevalence of fecal colonization with CRE in these locations. The CRE positive rate in Henan province was highest, followed by Zhejiang province. ST206 of E. coli harboring blaNDM−5was most prevalent. The findings emphasized that early screening of CRE fecal colonization carriage in populations could clarify the prevalence of local CRE, and highlighted the importance for adopting measures to mitigate spread of CRE.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: [https://www.ncbi.nlm.nih.gov/bioproject / PRJNA1023209].
Ethics statement
The studies involving humans were approved by Second Affiliated Hospital of Zhejiang University. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because The specimen used in this study was obtained in previous clinical diagnosis and treatment.
Author contributions
YL: Writing – original draft, Writing – review & editing. RZ: Funding acquisition, Writing – original draft, Writing – review & editing. LM: Writing – original draft. XD: Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Akeda Y. Current situation of carbapenem-resistant enterobacteriaceae and acinetobacter in Japan and Southeast Asia. Microbiol Immunol. (2021) 65(6):229–37. doi: 10.1111/1348-0421.12887
2. Jean SS, Lee NY, Tang HJ, Lu MC, Ko WC, Hsueh PR. Carbapenem-resistant enterobacteriaceae infections: Taiwan aspects. Front Microbiol. (2018) 9:2888. doi: 10.3389/fmicb.2018.02888
3. Li Y, Sun QL, Shen Y, Zhang Y, Yang JW, Shu LB, et al. Rapid increase in prevalence of carbapenem-resistant enterobacteriaceae (CRE) and emergence of Colistin resistance gene mcr-1 in CRE in a hospital in Henan, China. J Clin Microbiol. (2018) 56(4):e01932–17. doi: 10.1128/JCM.01932-17
4. Singh-Moodley A, Perovic O. Phenotypic and genotypic correlation of carbapenememase-producing enterobacteriaceae and problems experienced in routine screening. S Afr Med J. (2018) 108(6):495–501. doi: 10.7196/SAMJ.2018.v108i6.12878
5. Tompkins K, van Duin D. Treatment for carbapenem-resistant enterobacterales infections: recent advances and future directions. Eur J Clin Microbiol Infect Dis. (2021) 40(10):2053–68. doi: 10.1007/s10096-021-04296-1
6. van Duin D, Doi Y. The global epidemiology of carbapenemase-producing enterobacteriaceae. Virulence. (2017) 8(4):460–9. doi: 10.1080/21505594.2016.1222343
7. Zhang Y, Wang Q, Yin Y, Chen H, Jin L, Gu B, et al. Epidemiology of carbapenem-resistant enterobacteriaceae infections: report from the China CRE network. Antimicrob Agents Chemother. (2018) 62(2):e01882–17. doi: 10.1128/AAC.01882-17
8. Lutgring JD. Carbapenem-resistant enterobacteriaceae: an emerging bacterial threat. Semin Diagn Pathol. (2019) 36(3):182–6. doi: 10.1053/j.semdp.2019.04.011
9. Centers for Disease Control and Prevention (CDC). Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb Mortal Wkly Rep. (2009) 58(10):256–60.19300408
10. Ben-David D, Maor Y, Keller N, Regev-Yochay G, Tal I, Shachar D, et al. Potential role of active surveillance in the control of a hospital-wide outbreak of carbapenem-resistant klebsiella pneumoniae infection. Infect Control Hosp Epidemiol. (2010) 31(6):620–6. doi: 10.1086/652528
11. Karampatakis T, Tsergouli K, Iosifidis E, Antachopoulos C, Karapanagiotou A, Karyoti A, et al. Impact of active surveillance and infection control measures on carbapenem-resistant gram-negative bacterial colonization and infections in intensive care. J Hosp Infect. (2018) 99(4):396–404. doi: 10.1016/j.jhin.2018.05.010
12. Ben-David D, Masarwa S, Navon-Venezia S, Mishali H, Fridental I, Rubinovitch B, et al. Carbapenem-resistant Klebsiella pneumoniae in post-acute-care facilities in Israel. Infect Control Hosp Epidemiol. (2011) 32(9):845–53. doi: 10.1086/661279
13. Swaminathan M, Sharma S, Poliansky Blash S, Patel G, Banach DB, Phillips M, et al. Prevalence and risk factors for acquisition of carbapenem-resistant enterobacteriaceae in the setting of endemicity. Infect Control Hosp Epidemiol. (2013) 34(8):809–17. doi: 10.1086/671270
14. Viale P, Tumietto F, Giannella M, Bartoletti M, Tedeschi S, Ambretti S, et al. Impact of a hospital-wide multifaceted programme for reducing carbapenem-resistant enterobacteriaceae infections in a large teaching hospital in northern Italy. Clin Microbiol Infect. (2015) 21(3):242–7. doi: 10.1016/j.cmi.2014.10.020
15. Goodman KE, Simner PJ, Klein EY, Kazmi AQ, Gadala A, Toerper MF, et al. Predicting probability of perirectal colonization with carbapenem-resistant enterobacteriaceae (CRE) and other carbapenem-resistant organisms (CROs) at hospital unit admission. Infect Control Hosp Epidemiol. (2019) 40(5):541–50. doi: 10.1017/ice.2019.42
16. Al Fadhli AH, Jamal WY, Rotimi VO. Prevalence of carbapenem-resistant enterobacteriaceae and emergence of high rectal colonization rates of blaOXA-181-positive isolates in patients admitted to two major hospital intensive care units in Kuwait. PLoS One. (2020) 15(11):e0241971. doi: 10.1371/journal.pone.0241971
17. Kumar A, Mohapatra S, Bir R, Tyagi S, Bakhshi S, Mahapatra M, et al. Intestinal colonization due to carbapenem-resistant enterobacteriaceae among hematological malignancy patients in India: prevalence and molecular charecterisation. Indian J Hematol Blood Transfus. (2022) 38(1):1–7. doi: 10.1007/s12288-021-01415-y
18. Tian F, Li Y, Wang Y, Yu B, Song J, Ning Q, et al. Risk factors and molecular epidemiology of fecal carriage of carbapenem resistant enterobacteriaceae in patients with liver disease. Ann Clin Microbiol Antimicrob. (2023) 22(1):10. doi: 10.1186/s12941-023-00560-8
19. Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, et al. Nationwide surveillance of clinical carbapenem-resistant enterobacteriaceae (CRE) strains in China. EBioMedicine. (2017) 19:98–106. doi: 10.1016/j.ebiom.2017.04.032
20. Davari N, Khashei R, Pourabbas B, Nikbin VS, Zand F. High frequency of carbapenem-resistant enterobacteriaceae fecal carriage among ICU hospitalized patients from southern Iran. Iran J Basic Med Sci. (2022) 25(12):1416–23. doi: 10.22038/IJBMS.2022.63099.13938
21. Gu D, Yan Z, Cai C, Li J, Zhang Y, Wu Y, et al. Comparison of the NG-test carba 5, colloidal gold immunoassay (CGI) test, and Xpert carba-R for the rapid detection of carbapenemases in carbapenemase-producing organisms. Antibiotics (Basel). (2023) 12(2):300. doi: 10.3390/antibiotics12020300
22. CLSI. Performance standards for antimicrobial susceptibility testing, 31st ed. Institute CaLS (2021).
23. TECoAS T. Breakpoint Tables for Interpretation of MICs and Zone Diameters. (2020). Version 10.0.
24. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19(5):455–77. doi: 10.1089/cmb.2012.0021
25. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. (2012) 67(11):2640–4. doi: 10.1093/jac/dks261
26. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. (2016) 44(W1):W242–5. doi: 10.1093/nar/gkw290
27. Landry J, Hurst H. The emerging threat from carbapenem-resistant enterobacteriaceae. Nurs Womens Health. (2013) 17(6):519–24. doi: 10.1111/1751-486X.12080
28. Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. (2022) 399(10325):629–55. doi: 10.1016/S0140-6736(21)02724-0
29. Zhao ZC, Xu XH, Liu MB, Wu J, Lin J, Li B. Fecal carriage of carbapenem-resistant enterobacteriaceae in a Chinese university hospital. Am J Infect Control. (2014) 42(5):e61–4. doi: 10.1016/j.ajic.2014.01.024
30. Liu Q, Liu L, Li Y, Chen X, Yan Q, Liu WE. Fecal carriage and epidemiology of carbapenem-resistant enterobacteriaceae among hospitalized patients in a university hospital. Infect Drug Resist. (2019) 12:3935–42. doi: 10.2147/IDR.S233795
31. Yamamoto N, Asada R, Kawahara R, Hagiya H, Akeda Y, Shanmugakani RK, et al. Prevalence of, and risk factors for, carriage of carbapenem-resistant enterobacteriaceae among hospitalized patients in Japan. J Hosp Infect. (2017) 97(3):212–7. doi: 10.1016/j.jhin.2017.07.015
32. Solgi H, Badmasti F, Aminzadeh Z, Giske CG, Pourahmad M, Vaziri F, et al. Molecular characterization of intestinal carriage of carbapenem-resistant enterobacteriaceae among inpatients at two Iranian university hospitals: first report of co-production of bla (NDM-7) and bla (OXA-48). Eur J Clin Microbiol Infect Dis. (2017) 36(11):2127–35. doi: 10.1007/s10096-017-3035-3
33. Yen NTP, Nhung NT, Phu DH, Dung NTT, Van NTB, Kiet BT, et al. Prevalence of carbapenem resistance and its potential association with antimicrobial use in humans and animals in rural communities in Vietnam. JAC Antimicrob Resist. (2022) 4(2):dlac038. doi: 10.1093/jacamr/dlac038
34. Gijón D, Curiao T, Baquero F, Coque TM, Cantón R. Fecal carriage of carbapenemase-producing enterobacteriaceae: a hidden reservoir in hospitalized and nonhospitalized patients. J Clin Microbiol. (2012) 50(5):1558–63. doi: 10.1128/JCM.00020-12
35. Arum N, Ghafur A, Kazi M, Rao R, Rodrigues C, Ratnamani MS, et al. Prevalence of faecal carriage of carbapenemase producing enterobacteriaceae in healthy Indian subjects from the community. Indian J Med Microbiol. (2022) 40(3):374–7. doi: 10.1016/j.ijmmb.2022.05.010
36. Tran DM, Larsson M, Olson L, Hoang NTB, Le NK, Khu DTK, et al. High prevalence of colonisation with carbapenem-resistant Enterobacteriaceae among patients admitted to Vietnamese hospitals: risk factors and burden of disease. J Infect. (2019) 79(2):115–22. doi: 10.1016/j.jinf.2019.05.013
37. CHINET[EB/OL]. http://www.chinets.com/Data/GermYear
38. Guo B, Guo Z, Zhang H, Shi C, Qin B, Wang S, et al. Prevalence and risk factors of carbapenem-resistant enterobacterales positivity by active screening in intensive care units in the Henan province of China: a multi-center cross-sectional study. Front Microbiol. (2022) 13:894341. doi: 10.3389/fmicb.2022.894341
39. Jaiswal SR, Gupta S, Kumar RS, Sherawat A, Rajoreya A, Dash SK, et al. Gut colonization with carbapenem-resistant enterobacteriaceae adversely impacts the outcome in patients with hematological malignancies: results of a prospective surveillance study. Mediterr J Hematol Infect Dis. (2018) 10(1):e2018025. doi: 10.4084/mjhid.2018.025
40. Ma J, Song X, Li M, Yu Z, Cheng W, Yu Z, et al. Global spread of carbapenem-resistant enterobacteriaceae: epidemiological features, resistance mechanisms, detection and therapy. Microbiol Res. (2023) 266:127249. doi: 10.1016/j.micres.2022.127249
41. Hu X, Yang L, Dong N, Lin Y, Zhang L, Wang X, et al. Dissemination of bla(NDM-5) in Escherichia coli through the IncX3 plasmid from different regions in China. Microb. Drug Resist. (2022) 28(4):453–60. doi: 10.1089/mdr.2021.0202
42. Tian D, Wang B, Zhang H, Pan F, Wang C, Shi Y, et al. Dissemination of the bla(NDM-5) gene via IncX3-type plasmid among enterobacteriaceae in children. mSphere. (2020) 5(1):e00699–19. doi: 10.1128/mSphere.00699-19
43. Paul D, Babenko D, Toleman MA. Human carriage of cefotaxime-resistant Escherichia coli in north-east India: an analysis of STs and associated resistance mechanisms. J Antimicrob Chemother. (2020) 75(1):72–6. doi: 10.1093/jac/dkz416
Keywords: carbapenem-resistant enterobacterales, blaNDM, carbapenemase genes, fecal carriage, dissemination
Citation: Li Y, Ma L, Ding X and Zhang R (2024) Fecal carriage and genetic characteristics of carbapenem-resistant enterobacterales among adults from four provinces of China. Front. Epidemiol. 3:1304324. doi: 10.3389/fepid.2023.1304324
Received: 29 September 2023; Accepted: 12 December 2023;
Published: 3 January 2024.
Edited by:
Shailendra Saxena, King George’s Medical University, IndiaReviewed by:
Shangxin (Shaun) Yang, University of California, Los Angeles, United StatesLinus Olson, Karolinska Institutet (KI), Sweden
© 2024 Li, Ma, Ding and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Zhang zhang-rong@zju.edu.cn
 Yuanyuan Li
Yuanyuan Li