- Institute of Quality Standard and Testing Technology for Agro-Products, Sichuan Academy of Agricultural Sciences, Chengdu, China
Cadmium is a hazardous heavy metal that is commonly found in the soil and poses significant risks to soil organisms. The toxic effects of Cd on soil microorganisms and earthworms (Eisenia foetida) have been extensively studied, but most studies focused on high Cd pollution levels. Therefore, this study aims to investigate the different responses of soil organisms to moderate and low levels of Cd contamination. According to the study, the presence of 2.5 mg/kg of Cd had a significant impact on the microbial community’s composition and diversity. The relative abundance of most microbes decreased, while the abundance of Firmicutes and Actinobacteriota showed a considerable increase. The LEFSE analysis revealed that the Bacillus genus of the Firmicutes phylum can serve as a biomarker in soil contaminated with 2.5 mg/kg of Cd. At the same time, the functional analysis of PICRUSt 2 shows that microorganisms found in polluted soil have a noticeable decrease in their ability to metabolize lipids. On the other hand, our findings indicate that Cd has a detrimental effect on the biomass of earthworms and induces oxidative stress in these animals. The activation of superoxide dismutase (SOD) and catalase (CAT) enzymes in earthworms was carried out to mitigate oxidative stress. The study found a strong positive relationship between SOD and both time and Cd pollution. However, CAT exhibited inhibition throughout the later stages of the experiment, particularly when exposed to relatively higher levels of pollution. The analysis of RNA in earthworms revealed that soil Cd pollution at a concentration of 2.5 mg/kg primarily impacts the cellular structure and function of earthworms. This pollution disrupts the integrity of the cytoskeleton structure, hampers DNA replication, and compromises the precision of cell signaling. Simultaneously, when compared to the control group, several metabolic pathways exhibited abnormalities.
1 Introduction
Soil is an intricate system that exists at the intersection of the atmosphere, lithosphere, hydrosphere, and biosphere. Soil plays a critical role as a primary source of essential nutrients for plants and supports biodiversity preservation. The global issue of soil pollution by heavy metals has arisen due to their persistence, concealment, and mobility (Peng et al., 2022; Zhang and Wang, 2020). Cd is one of the most toxic metals. The presence of Cd in soil originates from a multitude of sources. Parent materials are the primary source, followed by industrial and life sources (Khan et al., 2022). The average concentration of soil Cd is 0.36 mg/kg globally (Kubier et al., 2019). However, because of geological features and various anthropogenic activities, the concentration can increase local concentrations by several orders of magnitude (Wang et al., 2015). According to the official report (Ministry of Environmental Protection of the People's Republic of China, 2014), Cd has emerged as a significant inorganic pollutant in China’s contaminated soil. The percentage of Cd pollution exceeding the legal limit is currently at 7.0%, making it the highest among heavy metal pollutants that exceed the statutory limit. This ubiquitous element inevitably impacts a diverse array of soil organisms (Huang et al., 2020; Li et al., 2021). Primarily impacted are the microorganisms, a crucial component of terrestrial ecosystems. They actively circulate soil organic matter, nitrogen, phosphorus, and other elements, regulate numerous ecological processes, and have a close relationship with soil health and crop production (Haj-Amor et al., 2022; Hemkemeyer et al., 2021; Lazcano et al., 2021). Plenty of research has demonstrated that introducing Cd into soil has a significant impact on various aspects of soil microbiology. This includes altering enzyme activity, microbial population, microbe community structure, and diversity (Du et al., 2021; Luo et al., 2019; Zhao et al., 2021). Currently, there is a lack of comprehensive understanding regarding soil microorganisms that can serve as a reliable indicator for monitoring soil Cd pollution, particularly in cases of mild pollution (Huang et al., 2023; Sun et al., 2022).
Earthworms, making up approximately 80% of the total soil biomass, are a highly prevalent and significant soil organism. They play a vital function in decomposing organic matter, circulating mineral elements, and upholding the general wellbeing of the soil ecosystem (Zeb et al., 2020). Multiple studies have demonstrated that the presence of Cd in soil has a detrimental impact on earthworm growth and development. For instance, exposure to Cd stress can trigger an overproduction of reactive oxygen species and aldehydes in earthworms. This disrupts the redox balance, resulting in oxidative stress and abnormal gene expression. It also leads to growth retardation, reduced reproductive capacity, behavioral changes, and ultimately a decline in animal population and biodiversity (Sinkakarimi et al., 2020; Urionabarrenetxea et al., 2020; Žaltauskaitė and Sodienė, 2014). Nevertheless, the majority of these endeavors have concentrated on investigating the impacts of elevated levels (ranging from 10 to 1,000 mg/kg) of Cd pollution (Du et al., 2014; Urionabarrenetxea et al., 2020) on various aspects of organisms, such as their avoidance behavior, growth, survival, and reproduction. These endpoints may be insensitive to mid-level Cd pollution toxicity. Nowadays, the advancement of molecular biology has facilitated a more profound understanding of the intricate reactions exhibited by organisms when subjected to environmental stress. RNA-Seq is a very efficient and rapid high-throughput sequencing approach that generates comprehensive gene expression data (Costa-Silva et al., 2023; Shi et al., 2021). It is a valuable tool for identifying numerous differently expressed genes in organisms of interest. Regrettably, there is a lack of valuable data about the transcriptome and expressed sequence of earthworms exposed to mild Cd contamination (Chai et al., 2020; Zhang et al., 2017).
Therefore, the primary objective of this work was to investigate the impact of mild Cd pollution on the composition, function, and biomarkers of soil microbiota in the presence of earthworms. Additionally, the study utilized the transcriptome analysis method for profiling gene expression in earthworms (Eisenia foetida) to examine the impact on gene expression in earthworms. This project aims to comprehensively elucidate the ecotoxicity response of key species (microorganisms and earthworms) in soil under moderate Cd pollution. It will investigate the impact on genes and physiological processes, providing insights into future strategies to manage and mitigate soil Cd pollution.
2 Materials and methods
2.1 Soil and earthworms
We obtained soil samples from the top 20 cm of the soil in an agricultural farmland located outside Chengdu City (Sichuan, China), ground it, and sieved it to 2 mm. The total Cd concentration was determined according to HJ803-2016 (Ministry of Environmental Protection of the People's Republic of China, 2016), which is 0.25 mg/kg. Other properties were determined according to the method of Lu (2000). pH is 6.14, organic matter is 21.2 g/kg, total N is 1.12 g/kg, total P is 0.81 g/kg, and total K is 24.4 g/kg. To simulate typical mid Cd contamination status in agricultural soil, gradient amounts of CdCl2 were mixed into experiment soil (the detail was described below). In this experiment, Cd-certified reference materials were used as additional Cd sources purchased from Inorganic Ventures (Virginia, United States).
The earthworms (Eisenia foetida) were acquired from a commercial earthworm breeding enterprise located in Jiangsu, China. Each healthy adult earthworm with a fully developed clitellum was delicately cleansed using ultrapure water and then placed in a small dish covered with moistened filter papers for starving for 48 h to empty its digestive system before exposure experiments.
2.2 Experimental set up
The experiment contained 4 treatments. Upon the addition of the requisite quantity of CdCl2 solution, the soil’s Cd concentration reached 0.25 mg/kg (CK), 1 mg/kg (T1), 1.5 mg/kg (T2), and 2.5 mg/kg (T3), respectively. The experiment was replicated three times for each treatment. Following a stabilization period of 4 weeks, 2 kg of treated soil were placed into a polypropylene pot with a height of 15 cm and a diameter of 20 cm. The soil was then spiked with deionized water until it reached 60% of its maximum water holding capacity. Later, 30 starving earthworms were transferred to each pot. The pot is covered by a lid full of holes and placed in an artificial climate chamber set to 21.0°C (temperature) and 80% (air humidity), and deionized water was supplied irregularly to make sure soil kept 60% of the maximum water holding capacity. However, no additional food was given to the earthworms, relying solely on the soil as a source of nutrition. The experiment lasted for 28 days. Six earthworms were sampled from each box on the 0 d, 7 d, 14 d, and 28 d, respectively, to measure biomass, Cd concentration, and enzyme activity. At the end of the experiment, apart from previous parameters, 3 earthworms were additionally selected from CK and T3 to measure transcriptomes. Simultaneously, soil samples from CK and T3 were also collected and stored at −80°C to measure microbial communities.
2.3 Sample analysis
2.3.1 Soil microbial DNA extraction, PCR amplification and illumina miseq sequencing analysis
The soil’s total DNA was extracted using the OMEGA Soil DNA Kit (M5635-02) from Omega Bio-Tek, located in Norcross, GA, United States. The extraction process followed the directions provided by the manufacturer. PCR amplification was performed using specific primers targeting the V3-V4 region of the bacterial 16S rRNA. The forward primer used was 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and the reverse primer used was 806R (5′-GGACTACHVGGG TWTCTAAT-3′). The amplified products underwent sequencing analysis utilizing the Illumina NovaSeq platform at Shanghai Personal Biotechnology Co., Ltd. in Shanghai, China. The specific information can be found in the Supplementary Material.
2.3.2 Mortality and biomass of earthworms
Before exposure, every earthworm was weighted and recorded after starving for 48 h. On the 7 d, 14 d, and 28 d, we randomly selected 6 earthworms from each box. After 48 h of starvation, the fresh weight was recorded. The mortality rate was calculated at the end of the experiment.
2.3.3 The Cd concentration of earthworm
On the 0 d, 7 d, 14 d, and 28 d, 6 earthworms were thoroughly washed with deionized water and starved for 48 h to clean the intestines. After purification, the earthworm was snap-frozen in liquid nitrogen and ground into powder. A portion of the sample (1 g) was weighted and placed into a porcelain crucible. It was then subjected to dry-ashing in a muffle furnace as the method described by Lv et al. (2021). Inductively coupled plasma mass spectrometry (NexION 300D, Perkin Elmer, United States) was employed to quantify the concentration of Cd in earthworms.
2.3.4 The activity of enzyme in earthworms
The biomarkers associated with oxidative stress in earthworms, specifically SOD and CAT, were determined to follow the protocols of commercial ELISA kits provided by Meimian Industry Co., Ltd. (Jiangsu, China). The data were expressed as the relative activity with respect to CK, which was set as 100%.
2.3.5 Transcriptome analysis of earthworm
The Trizol Reagent (Invitrogen Life Technologies) was used to isolate the total RNA of earthworms. Subsequently, the concentration, purity, and integrity of the RNA were assessed using a NanoDrop spectrophotometer (Thermo Scientific). The sequencing library was further processed using the NovaSeq 6000 platform, manufactured by Illumina and operated by Shanghai Personal Biotechnology Cp. Ltd. The detail can be found in the Supplementary Material.
2.4 Statistical analyses
SPSS 26.0 and the R software package were conducted for general data analysis. We assessed statistical distinctions using one-way ANOVA tests.
High-through sequence data analyses were primarily conducted using QIIME2 and R programs (v3.2.0). Alpha diversity metrics at the ASV level were computed in QIIME2. Linear discriminant analysis (LDA) effect size (LEfSe) (LDA >2, p < 0.05) was also conducted (http://huttenhower.sph.harvard.edu/LEfSe) to identify biomarkers with statistical differences between groups. Function and metabolic pathway prediction of soil microorganisms were analyzed using PICRUSt2 software, based on the website (https://github.com/picrust/picrust2).
The earthworm’s reference genome and gene annotation files were obtained from the genome website. The processed reads were aligned to the reference genome using HISAT2 (v 2.1.0). We utilized HTSeq (v 0.9.1) statistics to perform a comparative analysis of the read count values for each gene. FPKM was subsequently employed to normalize the gene expression. The gene expression was assessed using DESeq (v 1.38.3) under the specified conditions: A significant difference in expression is determined when the absolute value of the log2foldChange exceeds 1 and the P-value is below 0.05. Simultaneously, we assigned all the genes to corresponding terms in the Gene Ontology database and computed the quantities of genes that were differentially enriched in each term. The ClusterProfiler program (v 4.6.0) was also utilized to perform an enrichment analysis of the KEGG pathway for the differentially expressed genes.
3 Result
3.1 Soil microbial community composition and diversity
The 16SrRNA MiSeq sequencing analyses generated 88,939 ± 4,445 raw sequence and 92,495 ± 1,170 raw sequence in CK and T3 soil. After demultiplexing the raw sequence data, the primers were removed using the cutadapt plugin. The data then underwent quality filtering, denoising, splicing, and chimera removal using DADA2. This process resulted in the generation of 42,579 ± 3,166 and 51,988 ± 3,218 high-quality sequences in CK and T3, respectively (Supplementary Table S1). The sequences obtained were merged according to 100% sequence similarity to generate characteristic sequence ASVs.
The taxonomic classification of whole qualified ASVs revealed 30 phyla, 72 classes, 154 orders, 221 families, 308 genera, and 109 species in the CK sample, and 34 phyla, 83 classes, 174 orders, 244 families, 355 genera, and 110 species in the T3 sample. Figure 1 exhibits the dominant bacteria at the phylum level. The predominant phyla (relative abundance >4%) in CK soil were Proteobacteria, Bacteroidota, Gemmatimonadota, Actinobacteria, and Verrucomicrobiota, with relative abundances of 54.7%, 12.4%, 8.3%, 7.0%, and 6.4%, respectively. In contrast, the highest bacterial phylum was Proteobacteria (51.4%), followed by Bacteroidota (12.3%), Actinobacteria (9.1%), Gemmatimonadota (7.0%), and Verrucomicrobiota (5.8%) in T3 treatment.
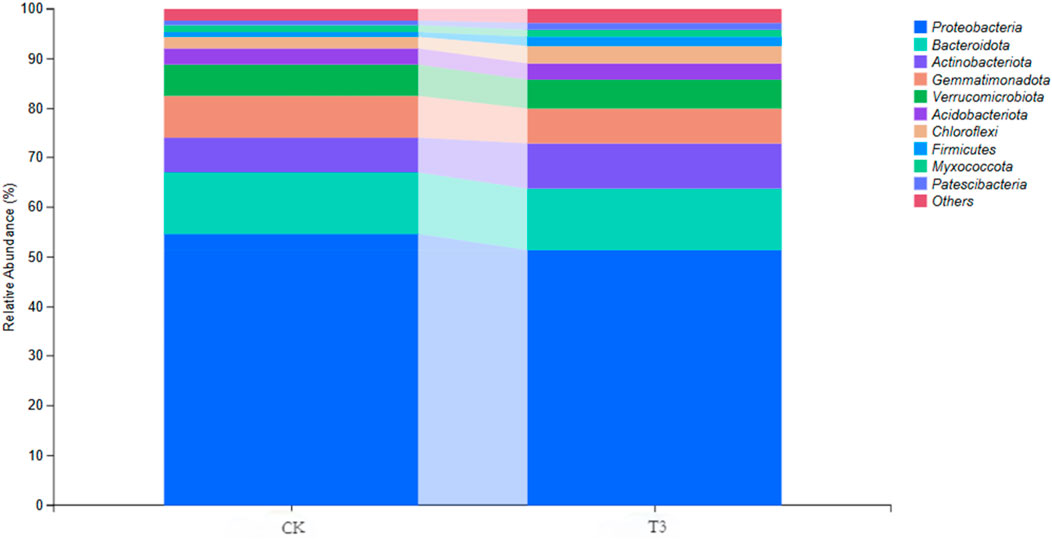
Figure 1. Relative abundance of microbial taxa at phylum level of bacterial communities in CK and T3 soil sample.
As shown in Figure 2, microbial diversities were further analyzed by Alpha-diversity (indexes of Chao1, Observed_species, Simpon, Shannon, and Faith_pd). Firstly, the examination of the rarefaction curve suggested that the abundance of high-quality sequences was sufficient to capture the majority of microbial diversity information and, to some extent, reflects that the microbial diversity in T3 is higher than that in CK (Supplementary Figure S2). Chao1, Observed_species, and Shannon index did not exhibit significant differences between the CK and T3 treatments. However, the Simpson and Faith_pd indices of T3 were substantially greater than those of CK.
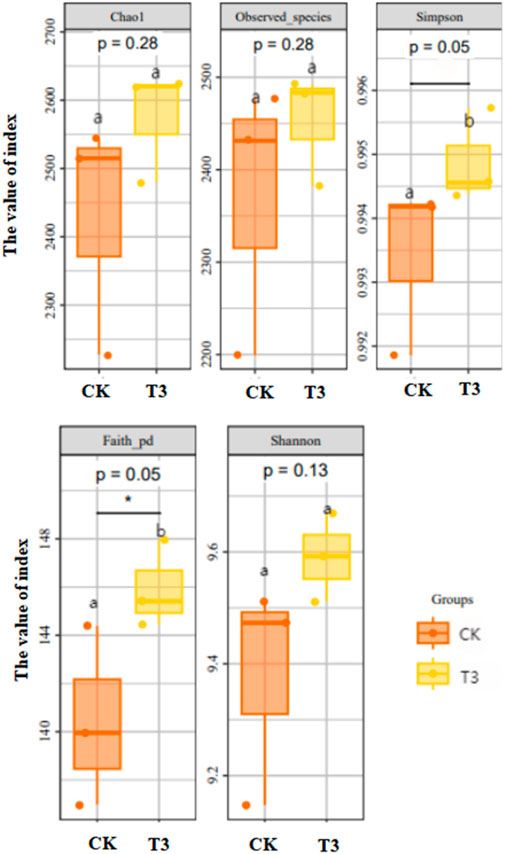
Figure 2. Estimation of bacterial Alpha-diversity in CK and T3 soil sample (different letter reflects significant difference, p < 0.05).
3.2 The biomarkers and potential function of the microbial community
LEFSE was employed to determine significant biomarkers in soil microbes (Figure 3). At genus level, the CK soil bacterial group was enriched in Sphingomonas, Lysobacter, Comamonas, Roseisolibacter, and Arenimonas. In contrast, in T3 soil, under Cd stress, bacterial groups spread into more genera. For instance, Adhaeribacter, Luteimonas, Bacillus, Saccharimonadales, Limnobacter, Microvirga, JG30_KF_CM45, and Pedobacter. The distinct reaction of different soil bacterial goups indicates that Cd can induce shifts in the composition of bacterial communities. These distinct groups could serve as indicators for Cd contamination.
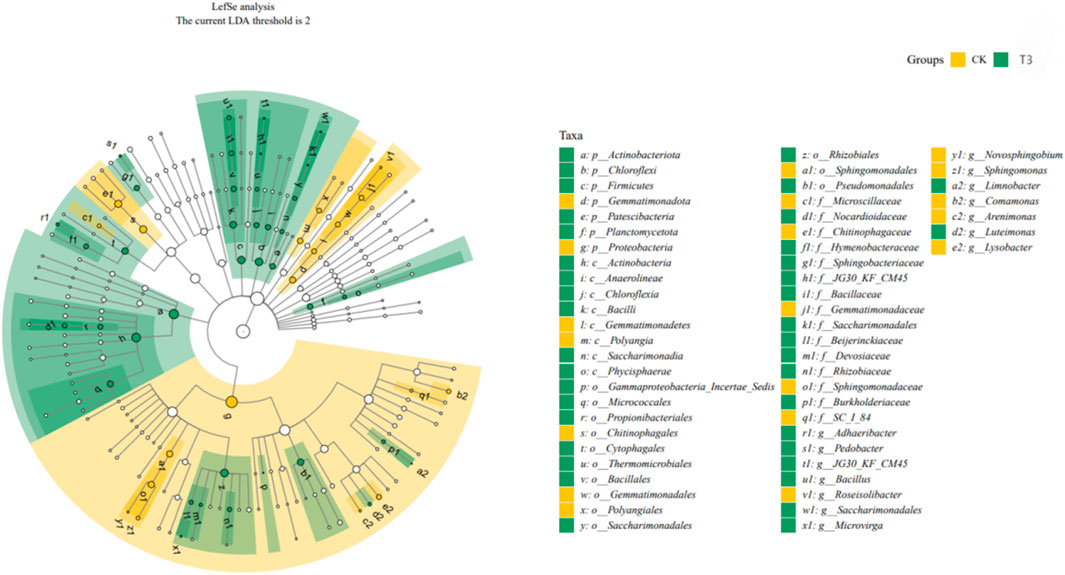
Figure 3. Linear discriminant analysis (LDA) effect size (LEfSe) (LDA >2, p < 0.05) to identify biomarkers with statistical differences between CK and T3. The five circles from the center represent phylum, class, order, family and genus levels. The significant biomarkers in CK an T3 are shown by dots in yellow and green.
Microbial functions were predicted by PICRUSt2. As shown in Figure 4, six primary functional layer biological pathways were obtained, mainly composed of cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and the organismal system. The functions related to metabolism were most abundant in soil bacteria. Further analysis of the biological metabolic pathways in the secondary functional layer revealed that metabolism consists of amino acid metabolism, biosynthesis of other secondary metabolites, carbohydrate metabolism, et al. And among these functions, amino acid metabolism, carbohydrate metabolism, and metabolism of cofactors and vitamins are the three most active functions in metabolism. Through analysis of variance, it was found that only lipid metabolism showed significant differences under different treatments. The lipid metabolism function of bacteria in Cd-contaminated soil was significantly reduced.
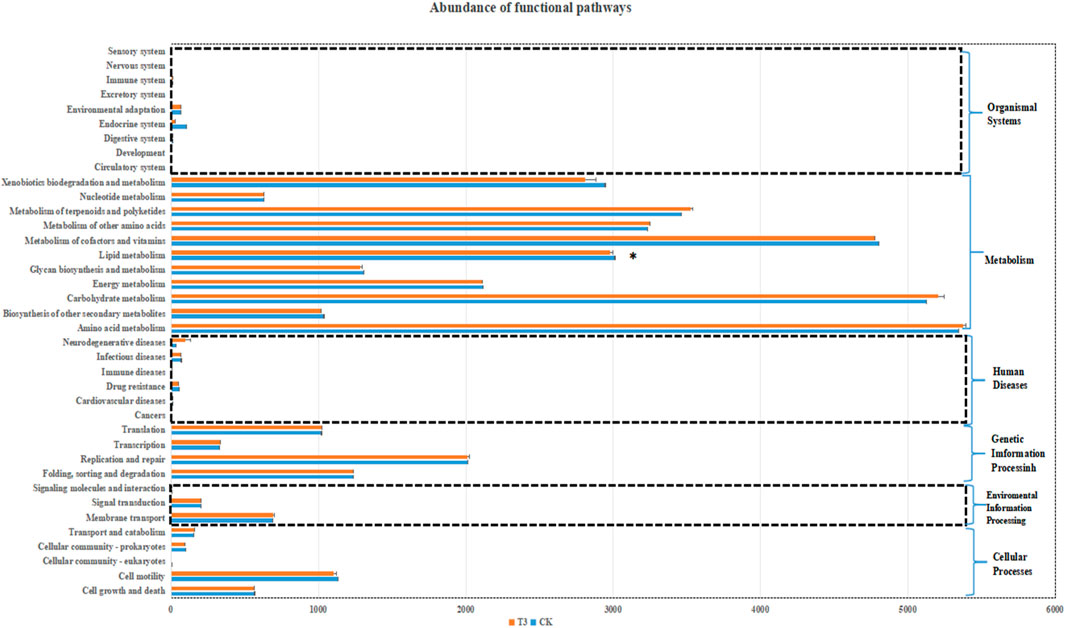
Figure 4. PICRUSt predicted KEGG pathways in soil microbial community (group with * indicates a significant difference, p < 0.05).
3.3 Mortality and biomass of earthworms
There were no deaths recorded for the whole duration of the experiment across all treatment groups. Figure 5 shows that the average weight of CK grew from 371 mg/earthworm to 504 mg/earthworm, with a growth rate of 35.8%. In both the T1 and T2 treatments, the weight of earthworms also consistently increased over time. The average weight values rose from 370 mg to 467 mg and 369 mg–430 mg, respectively, representing a 26.2% and 16.5% increase. Whileless, the maximum average weight of T3 was found on day 21, reaching 400 mg/kg, followed by a little decrease to 381 mg on day 28.
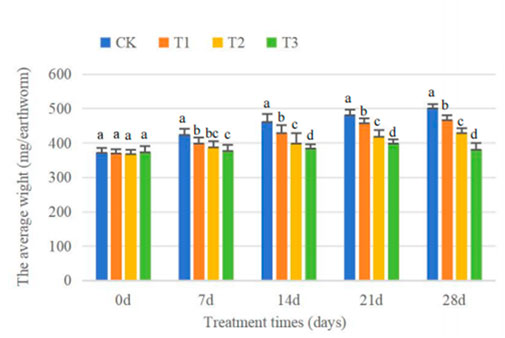
Figure 5. Effect of Cd concentration on earthworm average body weight with the respect to the time. (Different letters represent significant differences among different treatments at the same sampling time p < 0.05).
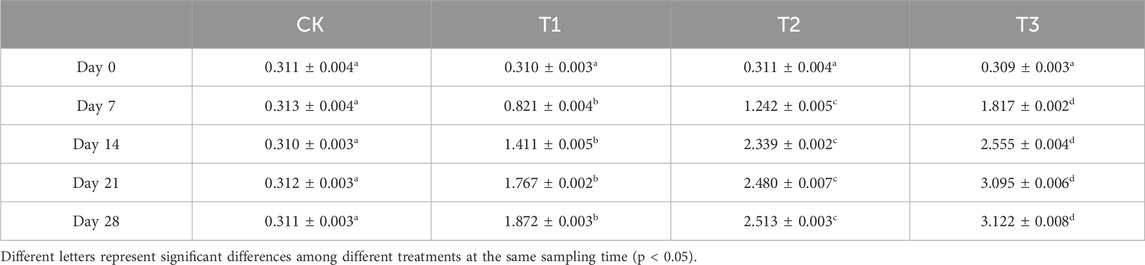
3.4 Cd level in earthworm
Table 1 shows the concentration of Cd in earthworms under different treatments. There was no significant difference in the Cd concentration in earthworm samples obtained under various treatments on day 0, all of which were approximately 0.31 mg/kg. However, on the seventh day, the Cd levels in earthworms have shown obvious differences, ranging from 0.313 mg/kg to 1.817 mg/kg. At the end of the experiment, the Cd concentrations of CK, T1, T2, and T3 reached 0.311 mg/kg, 1.872 mg/kg, 2.513 mg/kg, and 3.122 mg/kg. The Cd content in earthworms increased by 5, 7, and 10-fold following T1, T2, and T3 treatments, respectively, except for the control group, where it remained unaltered.
3.5 Temporal activities of SOD, CAT of earthworms
SOD activity responses in earthworms among CK and Cd treatments are depicted in Figure 6A. On the 7 d, the SOD activity in earthworms subjected to the T1 treatment was not substantially different from that in the CK. The T2 and T3 groups demonstrated a significant rise in SOD activity. Over time, specifically starting from the 14 d, all of the groups treated with Cd exhibited significantly increased SOD activity compared to CK. Furthermore, there was a noticeable positive relationship between SOD activity and both time and dosage. At the end of this experiment, compared to CK, the SOD activity increased 13%, 17%, and 19% at T1, T2, T3, respectively.
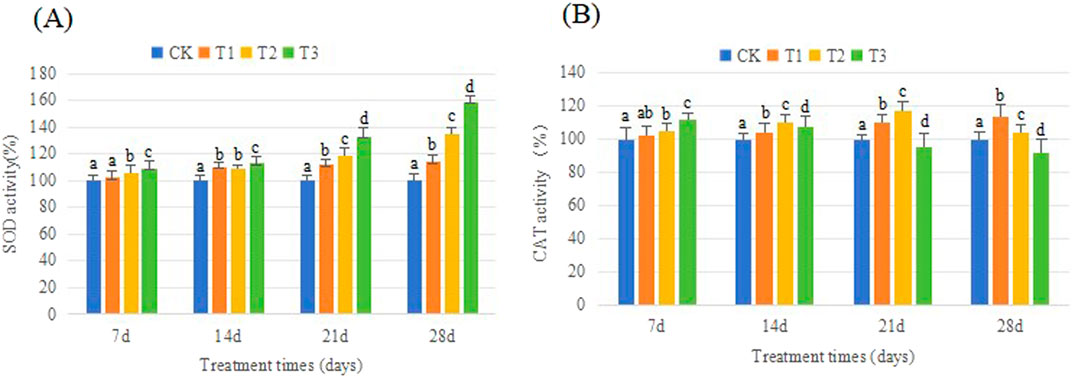
Figure 6. Effect of Cd on the SOD, CAT activity of earthworm. Different letter represents significant difference (p < 0.05).
Figure 6B shows the effects of Cd exposure on earthworm CAT activity. The CAT content in T1 and T2 treatments was always not lower than that in CK. At the end of the experiment, the CAT activity in the T1 group was 13% higher than in the control group, while the T2 group showed a 4% increase. In contrast, the CAT activity in earthworms in the T3 group initially increased and then decreased. Ultimately, at the end of the experiment, the enzyme activity in the T3 group was reduced by 8% compared to the control group.
3.6 Tanscriptomic analysis
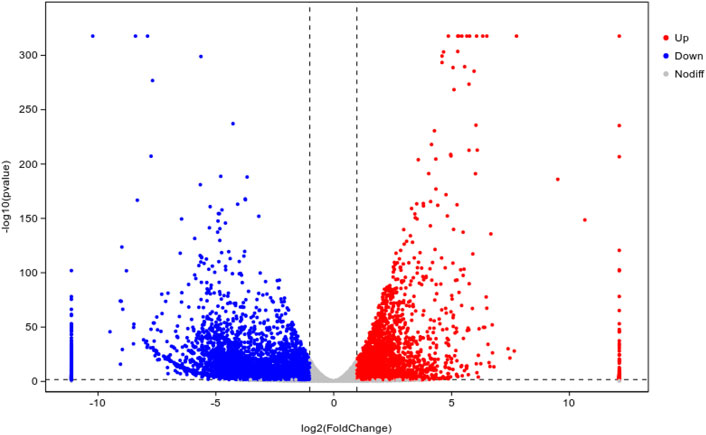
To reveal the potential toxicity mechanisms of Cd to earthworms, tanscriptomic analysis was performed. The number of differentially expressed genes (DEGs) in earthworms under CK and T3 treatment after 28-day exposure is shown in Figure 7. The total number of DEGs between CK and T3 was 8,294 (detail in Supplementary Table S3). Among them, 3,207 genes were upregulated and 5,087 genes were downregulated (CK vs. T3).
The GO enrichment analysis classified the DEGs into three primary functional groups, based on a P-value of 0.05. Supplementary Table S3 displays the difference between CK and T3, revealing 43 GO terms for biological process (BP), 1 term for molecular function (MF), and 8 terms for cellular component (CC). The top 20 GO terms with the most downregulated genes are shown in Figure 8A. These terms are mostly related to the structure and function of cells, such as the cytoskeleton, cytoskeletal components, DNA replication, and the nucleoside diphosphate metabolic process. In contrast, the upregulated genes were primarily involved in the stimulation of many enzymes, such as oxidoreductase, catalytic, and dioxygenase. They also played a role in various metabolic processes in living organisms, including the metabolism of the aromatic amino acid family, the catabolism of organic acids, the catabolism of carboxylic acids, and other processes (Figure 8B).
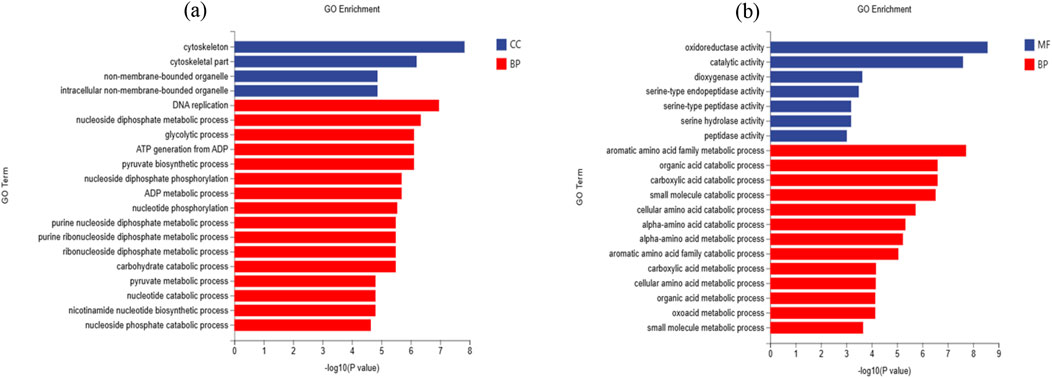
Figure 8. Gene Ontology (GO) classification of top 20 (A) downregulated genes (B) upregulated genes in CK vs. T3 groups of earthworms.
To further evaluate the biological significance of DEGs, the enrichment analysis of the KEGG pathway was performed. After mapping to the KEGG database, the results in CK vs. T3 were successfully annotated and assigned to 247 pathways (Supplementary Table S4). The top 20 pathways with the most downregulated genes were shown in Figure 9A. These pathways were mainly involved in how the extracellular matrix (ECM) and receptors interacted, as well as focal adhesion, phagosomes, protein digestion and absorption, DNA replication, and purine metabolism. Figure 9B displayed the top 20 pathways that were enriched with the highest number of upregulated genes. Most of these pathways are related to metabolism, particularly retinol metabolism, and some metabolic processes are associated with cytochrome P450.

Figure 9. KEGG enrichment of top 20 (A) downregulated genes (B) upregulated genes in CK vs. T3 groups of earthworms.
4 Discussion
4.1 Soil bacterial community response
When differentiating between soil degradation or improvement, it is more dependable to detect structural changes in microbial populations and activities rather than directly detecting soil physicochemical properties (Garau et al., 2007), due to their significant role in the soil nutrition cycle and their sensitivity to environmental change. Recent advancements in methodology have opened up possibilities for mapping intricate structures of microbial communities and understanding the underlying mechanisms behind their interactions with environmental factors (Adomako et al., 2022; Liu et al., 2020). Method like 16S rRNA-based high-throughput sequencing have significantly enhanced our understanding of the soil microbiome by providing detailed analysis of its diversity, composition, and function (Fierer, 2017). After checking the quality of the extracted DNA, we did PCR amplification, library construction, and other steps for high-throughput sequencing to get the raw sequencing data. Instead of the traditional OTU sequence clustering method, the ASV noise reduction method was used in our study. ASV employs an error correction technique to mitigate sequencing errors, resulting in more precise detection of sequence variants. It has high sensitivity towards single nucleotide changes and enables a more detailed examination of microbial diversity. In other words, the ASV provides a more reliable and replicable identifier for each variety, which proves highly advantageous when comparing the composition of microbial communities across several studies (García-López et al., 2021; Jeske and Gallert, 2022). The dominant soil phylum exhibited variations between CK and T3 treatment. It is important to mention that the addition of Cd resulted in a decrease in the abundance of several dominant bacterial phyla, including Proteobacteria (decreased by 6.0%) and Gemmatimonadota (decreased by 15.7%). Nevertheless, certain phyla demonstrated an increased tendency. In particular, the relative abundance of Firmicutes increased from 1.05% to 1.85% (an increase of 76%), and the relative abundance of Actinobacteria increased by 30%, rising from 7.0% to 9.1%. This alteration demonstrates the impact of Cd on the microbial ecology. Heavy metals inhibited the growth of metal-sensitive bacteria while promoting the growth of metal-tolerant bacteria. Our study’s results agree with those of Luo et al. (2019), who found that as the concentration of Cd rose, the amounts of Proteobacteria, Verrucomicrobia, and Nitrospirae decreased. Conversely, Actinobacteria and Firmicutes exhibited an increase in abundance with higher Cd concentrations. Some papers (Li et al., 2019; Xu et al., 2023) have confirmed Actinobacteria’s Cd tolerance, and they have even isolated them to remediate Cd-contaminated soils (Pathom-aree et al., 2021). The resistance mechanism of Actinobacteria is linked to their capacity to produce a diverse array of surface-active molecules that are designed to bind and eliminate mono- and divalent cations, such as heavy metals, through chemical interactions between the amphiphiles and the metal ions (Remenár et al., 2014).
The diversity of microorganisms in the soil is a crucial component that influences the composition and operation of subterranean ecosystems. Alpha-diversity indexes were used to judge the diversity of the soil microbial community. The results of Chao1, Observed_species suggest that there is no significant difference in species richness between CK and T3. Nevertheless, the Simpson and Faith_pd indices revealed that T3 exhibited much more bacterial diversity and variety in bacterial evolution compared to CK. This finding contrasts with the research conducted by Luo et al. (2019), which showed that within the soil-microorganism system, the alpha diversity of bacteria, as measured by the Chao1, Shannon, and Simpson indexes, had notable inverse associations with the concentration of Cd in the soil. This could be attributed to the existence of earthworms in our experiment. The eating habit of earthworms impacts the diversity of microorganisms in the soil. Our trials did not include any external fertilizers, which meant that earthworms could only rely on soil organic matter as their source of nutrients. High soil Cd concentrations have a major impact on earthworm digestion and absorption abilities (see details below). After 28 days, the organic matter level in T3 soil would be higher than that in CK soil. Total N (TN) and available N (AN) are crucial to the cellular metabolism process of bacteria, including synthetic amino acids and cell division. For example, Wu et al. (2017) reported that total P (TP) and total organic C (TOC) had a greater impact on microbial communities in the e-waste recycling region than heavy metals. This could be why microbial diversity is higher in contaminated soil, at least at the Cd pollution level we established. In addition, Oladipo et al. (2018) revealed that some heavy elements (Fe, Hg, Mn et al.) could accelerate microbial metabolism and simulate the growth of specific bacterial strains, especially the stain Bacillus kochii, which showed a significantly enhanced growth rate in 25 mg/L Cd Mueller-Hinton broth (MHB).
LEfSe is a data analysis technique that integrates the non-parametric Kruskal–Wallis and Wilcoxon rank sum tests with the linear discriminant analysis (LDA) effect size. The LEfSe approach enables simultaneous differential analysis across all categorization levels. Additionally, LEfSe focuses on identifying reliable differential species between groups, namely, marker species or biomarkers. One of its key characteristics is its ability to not only analyze variations in community composition among different sample groups, but also to delve into specific subgroups and identify microbial groupings that consistently exhibit distinctive characteristics throughout these subgroups. It has been extensively utilized in the areas of microbial amplicon analysis and metagenomic analysis. The taxonomic cladogram illustrates the hierarchical relationship of the primary taxa within the sample community, ranging from phylum to genus. The taxa are arranged in concentric circles, with the inner circle representing the phylum and the outer circle representing the genus. The size of the node corresponds to the average relative abundance of the taxon. Nodes that are open represent taxa with negligible differences between groups, whereas nodes in other hues (such as green and yellow) represent taxa that exhibit significant variations between groups Letters serve to designate the names of taxa that exhibit notable differences between groupings. In our study, the Bacillus of Bacillaceae of Bacillales of Bacilli of Firmicutes has most high LDA score (3.39). It showed the most significant difference between the CK and T3 groups and can become a marker in Cd-contaminated soil.
Besides the analysis of diversity and species composition of the microbiome, we are also cared the potential functions of these of microbiomes. With the development of data analysis technology, we can approach it by comparing 16S rRNA with known microbial genome data. In this research, the metabolic functions of the microbial community were predicted by PICRUSt2. We found that regardless of the treatment, the main functions of soil bacteria were concentrated in metabolism, especially the metabolism of amino acids and carbohydrate. The findings of this study align with the research conducted by Ren et al. (2021), which revealed that amino acid metabolism accounted for the highest proportion (about 11%) of microbial biological metabolic pathways in CK and Cd-contaminated soil. Amino acid metabolism and carbohydrate metabolism are essential prerequisites for bacteria to sustain vital functions. It is expected that a significant number of genes associated with these functions were identified. At a Cd pollution level of 2.5 mg/kg, the lipid metabolism of bacteria in the soil was significantly impaired Cd has a high affinity for -SH and -COOH groups, leading to the disruption of lipid metabolism (Rajakumar et al., 2020). Additionally, Zn, a trace element, is crucial for the process of maturation, advancement, and cellular operation (Singh et al., 2016). It acts as a cofactor for important proteins involved in lipid metabolism (Regalla and Lyons, 2006). Cd shares comparable physical and chemical characteristics with Zn. In some life activities, Cd replaces Zn in cells, leading to a decrease in intracellular Zn levels and disrupting Zn homeostasis (Rajakumar et al., 2016).
What we should notice is that 16S rRNA data reveal potential biomarkers in Cd-contaminated soil and indirectly reflect the function of microorganisms in this research. However, these findings require more verification. Due to its focus on a restricted set of universally present genes, 16S rRNA provides a narrow scope of analysis. Hence, additional investigations employing alternative methodologies, such as transcriptomics and non-target metabolomics, are necessary to comprehensively and impartially evaluate.
4.2 The toxicity on earthworms
Mortality and biomass serve as the most easily understandable measures of biological growth and development. During the experimental period, the earthworms in the control treatment showed excellent survival rates and consistently gained in weight. This indicates that the experimental settings were satisfactory in terms of providing a conducive atmosphere and suitable medium for the earthworms. The average weight of earthworms of T1 and T2 also showed an increasing trend, but their increasing level decreases with the increase of Cd concentration. Additionally, the mean weight of T3 exhibited a pattern of initially increasing and subsequently decreasing. This result is consistent with Elyamine et al. (2018), who observed that the weight of earthworms (Eisenia fetida) decreased by 11% within 30 days in 2.5 mg/kg Cd-contaminated soil. The reduction in body weight caused by Cd treatments is likely associated with the earthworm’s response to stress, wherein it decreases food consumption to escape the poisons. Earthworms frequently employ this method to prevent poisoning, not only from heavy metals but also from organic compounds (Wu et al., 2013). When earthworms are exposed to a polluted environment, they can reduce the harmful effects of the toxin by adjusting their internal metabolic activity, which includes losing body weight (Parihar et al., 2019).
The concentration of Cd in earthworm tissue increases proportionally with the concentration of Cd in the soil. And the Cd enrichment continued with time. At the end of the experiment, the Cd concentration of T3 is 3.12 mg/kg which is 10 times that of CK (0.311 mg/kg). This finding is consistent with the findings by Liang et al. (2022), who similarly detected a more than tenfold increase in Cd concentration in earthworms living in soil contaminated with 2.5 mg/kg of Cd. It is noteworthy that the rate of Cd accumulation in earthworms decreased dramatically in the later stage of the experiment compared to the early stage. The growth rate of Cd is 162%, 297%, and 481% for T1, T2, and T3, respectively, from day 0 to day 7. However, the rate decreases to 5.9%, 1.3%, and 0.87% from day 21 to day 28. This phenomenon could be attributed to the inherent capacity of organisms to adjust and thrive in response to pressure in their surroundings. During the later phase of the experiment, earthworms effectively decreased the intake of contaminants in the environment by employing diverse detoxifying mechanisms, either actively or passively.
When exposed to environmental stressors like drought, salt, or metal toxicity, the levels of reactive oxygen species (ROS) in an organism can significantly escalate. To lessen oxidative stress damage induced by exogenous pollutants and scavenge excess ROS, various antioxidant enzymes were triggered in earthworms, such as SOD, CAT, and MDA. SOD is an enzyme that converts the superoxide anion radical (O2−) into regular molecular oxygen (O2) and hydrogen peroxide (H2O2), serving as the first line of defense against oxidative stress. Catalase (CAT) is an essential enzyme that breaks down the byproducts of the superoxide dismutase (SOD) enzyme, specifically hydrogen peroxide (H2O2), into water and molecular oxygen. When comparing with CK, it was found that on the 7d, SOD activity showed considerable increases in T2 and T3, but no significant change was noticed in the slightly polluted treatment (T1). This could be attributed to the delayed onset of oxidative stress in earthworms caused by low levels of Cd contamination, and the resulting physiological harm to organisms is not substantial within a short timeframe. Unlike the initial reaction, a substantial increase in SOD activity was observed in all Cd-treated samples after 14 days of exposure. The rate of increase is positively correlated with the soil Cd level. This finding aligns with prior research, which similarly documented elevated SOD activity in the presence of heavy metal contamination (Hu et al., 2016). CAT is another key enzyme involved in the removal of toxic peroxides which can decompose products of SOD enzyme (H2O2) to water and molecular oxygen. In our study the CAT activity was not in accord with the activity of SOD. During the initial phase, when exposed to oxidative stress, the catalase enzyme was stimulated. The activity of the CAT enzyme in each group treated with Cd was greater than that in the control group, and the level of activity was directly related to the concentration of Cd. Nevertheless, it exhibited a decline of varying magnitudes in the subsequent phase, particularly in T3. On the 28th day, the concentration of CAT was lower than that of CK. CAT is present in peroxisomes and mitochondria. The destructive effect of Cd on CAT enzyme in earthworms is mainly reflected in the following aspects: (1) Cd-induced impairment of mitochondrial function. Cd could bind to specific protein thiols on the mitochondrial membrane, changing mitochondrial permeability and causing membrane lipid peroxidation (Dorta et al., 2003). Additionally, Cd could impair the function of mitochondrial enzymes such as citrate synthase and nicotinamide adenine dinucleotide phosphate-dependent isocitrate dehydrogenase, both of which are involved in the tricarboxylic acid cycle. (2) Cd triggers peroxisomal senescence by increasing the activity of the native proteases and glyoxylate cycle enzymes (del Río and López-Huertas 2016).
In order to further reveal the effects of Cd on various physiological functions of earthworms at the genetic level, we conducted transcriptome analysis on earthworms treated with CK and T3, and classified the genes with significant differences in the GO and KEGG databases. Mapping all downregulated genes in the GO database revealed that Cd exposure has a significantly negative influence on the structure and function of cells. In eukaryotes, microtubules are one of the major components of the cytoskeleton and play a role in many processes, including structural support, intracellular transport, and DNA segregation. Pizzaia et al. (2019) found that tubulin-based structures are Cd targets. Exposure to Cd not only affects the movement of substances within cells due to disruptions in the cytoskeleton, but also reduces cells’ ability to communicate with each other by decreasing plasmodesmata permeability (O’Lexy et al., 2018). Furthermore, the expression of genes related to DNA replication was significantly downregulated in Cd pollution treatment. DNA replication is crucial to ensuring the accurate and complete transmission of genetic information. The significantly downregulated related genes suggest that Cd pollution poses a threat to gene expression stability. On the other hand, there was a notable increase in the expression of genes associated with enzyme activity in earthworms, particularly those involved in antioxidant functions. This aligns with our prior finding that the concentration of SOD enzymes in earthworms inhabiting Cd-contaminated soil exhibited a substantial increase. The upregulated antioxidant-related genes at transcriptional levels in eathworms also suggested oxidative stress generated by Cd pollution.
KEGG is a comprehensive database that combines genomic, chemical, and systematic functional information. The gene catalog is derived from the fully sequenced genome. One of the characteristics of the KEGG database is its correlation with higher-level system functions at the cellular, species, and ecosystem levels. We utilized clusterprofiler to conduct KEGG enrichment analysis, employing the differentially expressed genes annotated by KEGG pathways to assess the gene list associated with each pathway. Identify the KEGG pathway in which differentially expressed genes are significantly enriched compared to the entire genome background. The most obvious effect of downregulated genes is ECM-receptor interaction. The ECM is a sophisticated network structure of proteins and polysaccharides assembled by cell secretion. Its primary functions are to provide attachment sites and structural support for cells, as well as to regulate cell activity and organ and tissue function by conveying important biological information. Deregulation is a common occurrence in disease situations and has a direct impact on cancer progression by affecting cellular transformation (Kolluru et al., 2019). Additionally, focal adhesions anchor the cytoskeleton to the ECM and play a critical role in regulating cell proliferation, apoptosis, and migration. Choong et al. (2013) reported that Cd disrupts focal adhesions at the ends of organized actin structures, particularly the loss of vinculin and focal adhesion kinase (FAK) from the contacts. In our study, genes related to ECM-receptor interaction and focal adhesion were both downregulated under Cd stress. This suggests issues with signal transmission among cells. Phagocytosis refers to the act of engulfing large solid entities, such as bacteria and cellular pieces. This process consists of two rather autonomous processes: adsorption and swallowing. The engulfed particles first attach to the cell surface. Afterwards, the cell membrane at the adsorption area curves inward to create a concave capsule, and the membrane at the opening of the capsule merges and seals to form a vesicle. It detaches from the cell membrane and enters the cytoplasm to move. A phagosome is the term used to describe the vesicle that is created through the process of engulfment. The phagosome undergoes fusion with the lysosome within the cell, resulting in the decomposition of the material by the enzymes present in the lysosome. Protozoa commonly exhibit phagocytosis. The primary purpose of the reticuloendothelial system is to remove foreign substances and contribute to the anti-counterfeiting mechanism in animal organisms. This system consists of macrophages, monocytes, and neutrophils, which work together to protect against microbial invasion and eliminate aging and deceased cells. The notable decrease in the expression of Phagosome-related genes suggests that earthworms have a diminished capacity to defend against microbial invasion and eliminate old and deceased cells.
Several gene families associated with metabolism, such as retinol metabolism, cytochrome P450-related metabolism, exhibited upregulation under Cd stress. The study conducted by Cui et al. (2009) found that the signaling of retinoic acid increased in mice cells after being exposed to Cd concentrations of 5 and 10 μm for 24 h. Analysis of gene expression revealed that the activation of retinoic acid signaling by Cd might be facilitated by the excessive expression of Bcmo1. In addition, Cd suppressed the expression of Cyp26a1 and Cyp26b1, which play a role in the breakdown of retinoic acid. The results demonstrated that Cd induces retinoic acid signaling by regulating the expression of genes involved in retinoic acid metabolism.
5 Conclusion
This paper investigated the ecotoxicological impacts of gentle Cd pollution on soil microorganisms and earthworms Eisenia foetida. An analysis of 16S rRNA amplicon sequencing revealed that the introduction of 0.25 mg/kg of Cd in the earthworm-soil-microorganism system resulted in an enhanced diversity of microorganisms. This abundance of some Cd-tolerant microorganisms obviously increased, such as Firmicutes and Actinobacteriota. Specifically, the Bacillus genus of the Firmicutes phylum exhibited a significant difference between the CK and the Cd-contaminated soil, suggesting that it could serve as a biomarker for soil Cd contamination. Additionally, the introduction of 2.5 mg/kg Cd had a significant inhibitory effect on the lipid metabolism of microorganisms. On the other hand, varying levels of Cd contamination will negatively impact the biomass of earthworms. Cd exhibits a pronounced enrichment impact in the bodies of earthworms and triggers the activation of their antioxidant mechanism. The activity of SOD enzyme in earthworms was positively correlated with time and Cd dose, and CAT enzyme showed a trend of first increasing and then decreasing in the 0.25 mg/kg treatment. The stability of earthworm cell structure, the accuracy of intracellular DNA replication, and the information transmission between cells were affected under Cd treatment. Multiple metabolic pathways were upregulated under Cd stimulation, which may be the result of Cd acting on different target organs in the earthworm. Later pathological sections are needed in further research to identify whether there are lesions in different organs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
RU: Writing–original draft. HL: Writing–original draft, Software, Methodology. XL: Writing–review and editing, Data curation, Conceptualization. LL: Writing–review and editing, Validation, Formal Analysis. PW: Writing–review and editing, Methodology, Conceptualization. HX: Writing–review and editing, Validation, Software. YZ: Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by Sichuan Provincial Finance Independent Innovation Special Project: Soil Pollutants and Food Security Risk Assessment (2022ZZCX039).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2024.1479500/full#supplementary-material
References
Adomako, M. O., Roiloa, S., and Yu, F.-H. (2022). Potential roles of soil microorganisms in regulating the effect of soil nutrient heterogeneity on plant performance. Microorganisms 10, 2399. doi:10.3390/microorganisms10122399
Chai, L., Yang, Y., Yang, H., Zhao, Y., and Wang, H. (2020). Transcriptome analysis of genes expressed in the earthworm eisenia fetida in response to cadmium exposure. Chemosphere 240, 124902. doi:10.1016/j.chemosphere.2019.124902
Choong, G., Liu, Y., and Templeton, D. M. (2013). Cadmium affects focal adhesion kinase (fak) in mesangial cells: involvement of camk-ii and the actin cytoskeleton. J. Cell. Biochem. 114, 1832–1842. doi:10.1002/jcb.24529
Costa-Silva, J., Domingues, D. S., Menotti, D., Hungria, M., and Lopes, F. M. (2023). Temporal progress of gene expression analysis with rna-seq data: a review on the relationship between computational methods. Comput. Struct. Biotechnol. J. 21, 86–98. doi:10.1016/j.csbj.2022.11.051
Cui, Y., and Freedman, J. H. (2009). Cadmium induces retinoic acid signaling by regulating retinoic acid metabolic gene expression. J. Biol. Chem. 284, 24925–24932. doi:10.1074/jbc.m109.026609
del Río, L. A., and López-Huertas, E. (2016). Ros generation in peroxisomes and its role in cell signaling. Plant Cell. Physiology 57, 1364–1376. doi:10.1093/pcp/pcw076
Dorta, D. J., Leite, S., DeMarco, K. C., Prado, I. M., Rodrigues, T., Mingatto, F. E., et al. (2003). A proposed sequence of events for cadmium-induced mitochondrial impairment. J. Inorg. Biochem. 97, 251–257. doi:10.1016/s0162-0134(03)00314-3
Du, Y., Zhang, D., Zhou, D., Liu, L., Wu, J., Chen, H., et al. (2021). The growth of plants and indigenous bacterial community were significantly affected by cadmium contamination in soil–plant system. Amb. Express 11, 103. doi:10.1186/s13568-021-01264-y
Du, Y.-L., He, M.-M., Xu, M., Yan, Z.-G., Zhou, Y.-Y., Guo, G.-L., et al. (2014). Interactive effects between earthworms and maize plants on the accumulation and toxicity of soil cadmium. Soil Biol. Biochem. 72, 193–202. doi:10.1016/j.soilbio.2014.02.004
Elyamine, A. M., Afzal, J., Rana, M. S., Imran, M., Cai, M., and Hu, C. (2018). Phenanthrene mitigates cadmium toxicity in earthworms eisenia fetida (epigeic specie) and aporrectodea caliginosa (endogeic specie) in soil. Int. J. Environ. Res. Public Health 15, 2384. doi:10.3390/ijerph15112384
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi:10.1038/nrmicro.2017.87
Garau, G., Castaldi, P., Santona, L., Deiana, P., and Melis, P. (2007). Influence of red mud, zeolite and lime on heavy metal immobilization, culturable heterotrophic microbial populations and enzyme activities in a contaminated soil. Geoderma 142, 47–57. doi:10.1016/j.geoderma.2007.07.011
García-López, R., Cornejo-Granados, F., Lopez-Zavala, A. A., Cota-Huízar, A., Sotelo-Mundo, R. R., Gómez-Gil, B., et al. (2021). Otus and asvs produce comparable taxonomic and diversity from shrimp microbiota 16s profiles using tailored abundance filters. Genes. 12, 564. doi:10.3390/genes12040564
Haj-Amor, Z., Araya, T., Kim, D.-G., Bouri, S., Lee, J., Ghiloufi, W., et al. (2022). Soil salinity and its associated effects on soil microorganisms, greenhouse gas emissions, crop yield, biodiversity and desertification: a review. Sci. Total Environ. 843, 156946. doi:10.1016/j.scitotenv.2022.156946
Hemkemeyer, M., Schwalb, S. A., Heinze, S., Joergensen, R. G., and Wichern, F. (2021). Functions of elements in soil microorganisms. Microbiol. Res. 252, 126832. doi:10.1016/j.micres.2021.126832
Hu, S., Zhang, W., Li, J., Lin, K., and Ji, R. (2016). Antioxidant and gene expression responses of eisenia fetida following repeated exposure to bde209 and pb in a soil-earthworm system. Sci. Total Environ. 556, 163–168. doi:10.1016/j.scitotenv.2016.02.194
Huang, L., Wang, Q., Zhou, Q., Ma, L., Wu, Y., Liu, Q., et al. (2020). Cadmium uptake from soil and transport by leafy vegetables: a meta-analysis. Environ. Pollut. 264, 114677. doi:10.1016/j.envpol.2020.114677
Huang, Y., Huang, Y., Hou, J., Wu, L., Christie, P., and Liu, W. (2023). Microbial community assembly of the hyperaccumulator plant sedum plumbizincicola in two contrasting soil types with three levels of cadmium contamination. Sci. Total Environ. 863, 160917. doi:10.1016/j.scitotenv.2022.160917
Jeske, J. T., and Gallert, C. (2022). Microbiome analysis via otu and asv-based pipelines—a comparative interpretation of ecological data in wwtp systems. Bioengineering 9, 146. doi:10.3390/bioengineering9040146
Khan, Z., Elahi, A., Bukhari, D. A., and Rehman, A. (2022). Cadmium sources, toxicity, resistance and removal by microorganisms-a potential strategy for cadmium eradication. J. Saudi Chem. Soc. 26, 101569. doi:10.1016/j.jscs.2022.101569
Kolluru, V., Tyagi, A., Chandrasekaran, B., and Damodaran, C. (2019). Profiling of differentially expressed genes in cadmium-induced prostate carcinogenesis. Toxicol. Appl. Pharmacol. 375, 57–63. doi:10.1016/j.taap.2019.05.008
Kubier, A., Wilkin, R. T., and Pichler, T. (2019). Cadmium in soils and groundwater: a review. Appl. Geochem. 108, 104388. doi:10.1016/j.apgeochem.2019.104388
Lazcano, C., Zhu-Barker, X., and Decock, C. (2021). Effects of organic fertilizers on the soil microorganisms responsible for n2o emissions: a review. Microorganisms 9, 983. doi:10.3390/microorganisms9050983
Li, K., Tang, X., Zhao, J., Guo, Y., Tang, Y., and Gao, J. (2019). Streptomyces cadmiisoli sp. Nov., a novel actinomycete isolated from cadmium-contaminated soil. Int. J. Syst. Evol. Microbiol. 69, 1024–1029. doi:10.1099/ijsem.0.003262
Li, Z., Liang, Y., Hu, H., Shaheen, S. M., Zhong, H., Tack, F. M. G., et al. (2021). Speciation, transportation, and pathways of cadmium in soil-rice systems: a review on the environmental implications and remediation approaches for food safety. Environ. Int. 156, 106749. doi:10.1016/j.envint.2021.106749
Liang, X., Zhou, D., Wang, J., Li, Y., Liu, Y., and Ning, Y. (2022). Evaluation of the toxicity effects of microplastics and cadmium on earthworms. Sci. Total Environ. 836, 155747. doi:10.1016/j.scitotenv.2022.155747
Liu, H., Wang, C., Xie, Y., Luo, Y., Sheng, M., Xu, F., et al. (2020). Ecological responses of soil microbial abundance and diversity to cadmium and soil properties in farmland around an enterprise-intensive region. J. Hazard. Mater. 392, 122478. doi:10.1016/j.jhazmat.2020.122478
Lu, R. (2000). The chemical analysis of agricultural soil; agriculture science and technique. Beijing, China: Press.
Luo, L. Y., Xie, L. L., Jin, D. C., Mi, B. B., Wang, D. H., Li, X. F., et al. (2019). Bacterial community response to cadmium contamination of agricultural paddy soil. Appl. Soil Ecol. 139, 100–106. doi:10.1016/j.apsoil.2019.03.022
Lv, W.-X., Yin, H.-M., Liu, M.-S., Huang, F., and Yu, H.-M. (2021). Effect of the dry ashing method on cadmium isotope measurements in soil and plant samples. Geostand. Geoanalytical Res. 45, 245–256. doi:10.1111/ggr.12357
Ministry of Environmental Protection of the People's Republic of China (MEP-PRC) (2014). National survey report of soil contamination status of China.
Ministry of Environmental Protection of the People's Republic of China (MEP-PRC) (2016). Solid and sediment-Determination of aqua regia extracts of 12 metal elements-inductively coupled plasma mass spectrometry (HJ 803-2016).
Oladipo, O. G., Ezeokoli, O. T., Maboeta, M. S., Bezuidenhout, J. J., Tiedt, L. R., Jordaan, A., et al. (2018). Tolerance and growth kinetics of bacteria isolated from gold and gemstone mining sites in response to heavy metal concentrations. J. Environ. Manag. 212, 357–366. doi:10.1016/j.jenvman.2018.01.038
O’Lexy, R., Kasai, K., Clark, N., Fujiwara, T., Sozzani, R., and Gallagher, K. L. (2018). Exposure to heavy metal stress triggers changes in plasmodesmatal permeability via deposition and breakdown of callose. J. Exp. Bot. 69, 3715–3728. doi:10.1093/jxb/ery171
Parihar, K., Kumar, R., and Sankhla, M. S. (2019). Impact of heavy metals on survivability of earthworms. Int. medico-legal Report. J. 2.
Pathom-aree, W., Matako, A., Rangseekaew, P., Seesuriyachan, P., and Srinuanpan, S. (2021). Performance of actinobacteria isolated from rhizosphere soils on plant growth promotion under cadmium toxicity. Int. J. Phytoremediation 23, 1497–1505. doi:10.1080/15226514.2021.1913992
Peng, J.-y., Zhang, S., Han, Y., Bate, B., Ke, H., and Chen, Y. (2022). Soil heavy metal pollution of industrial legacies in China and health risk assessment. Sci. Total Environ. 816, 151632. doi:10.1016/j.scitotenv.2021.151632
Pizzaia, D., Nogueira, M. L., Mondin, M., Carvalho, M. E. A., Piotto, F. A., Rosario, M. F., et al. (2019). Cadmium toxicity and its relationship with disturbances in the cytoskeleton, cell cycle and chromosome stability. Ecotoxicology 28, 1046–1055. doi:10.1007/s10646-019-02096-0
Rajakumar, S., Abhishek, A., Selvam, G. S., and Nachiappan, V. (2020). Effect of cadmium on essential metals and their impact on lipid metabolism in saccharomyces cerevisiae. Cell. Stress Chaperones 25, 19–33. doi:10.1007/s12192-019-01058-z
Rajakumar, S., Ravi, C., and Nachiappan, V. (2016). Defect of zinc transporter zrt1 ameliorates cadmium induced lipid accumulation in saccharomyces cerevisiae. Metallomics 8, 453–460. doi:10.1039/c6mt00005c
Regalla, L. M., and Lyons, T. J. (2006). “Zinc in yeast: mechanisms involved in homeostasis,” in Molecular biology of metal homeostasis and detoxification: from microbes to man, 37–58.
Remenár, M., Karelová, E., Harichová, J., Zámocký, M., Krčová, K., and Ferianc, P. (2014). Actinobacteria occurrence and their metabolic characteristics in the nickel-contaminated soil sample. Biologia 69, 1453–1463. doi:10.2478/s11756-014-0451-z
Ren, C., Teng, Y., Chen, X., Shen, Y., Xiao, H., and Wang, H. (2021). Impacts of earthworm introduction and cadmium on microbial communities composition and function in soil. Environ. Toxicol. Pharmacol. 83, 103606. doi:10.1016/j.etap.2021.103606
Shi, H., Zhou, Y., Jia, E., Pan, M., Bai, Y., and Ge, Q. (2021). Bias in rna-seq library preparation: current challenges and solutions. BioMed Res. Int. 2021, 1–11. doi:10.1155/2021/6647597
Singh, N., Yadav, K. K., and Rajasekharan, R. (2016). Zap1-mediated modulation of triacylglycerol levels in yeast by transcriptional control of mitochondrial fatty acid biosynthesis. Mol. Microbiol. 100, 55–75. doi:10.1111/mmi.13298
Sinkakarimi, M. H., Solgi, E., and Hosseinzadeh, C. A. (2020). Interspecific differences in toxicological response and subcellular partitioning of cadmium and lead in three earthworm species. Chemosphere 238, 124595. doi:10.1016/j.chemosphere.2019.124595
Sun, H., Shao, C., Jin, Q., Li, M., Zhang, Z., Liang, H., et al. (2022). Effects of cadmium contamination on bacterial and fungal communities in panax ginseng-growing soil. BMC Microbiol. 22, 77. doi:10.1186/s12866-022-02488-z
Urionabarrenetxea, E., Garcia-Velasco, N., Marigómez, I., and Soto, M. (2020). Effects of elevated temperatures and cadmium exposure on stress biomarkers at different biological complexity levels in eisenia fetida earthworms. Comp. Biochem. Physiology Part C Toxicol. and Pharmacol. 231, 108735. doi:10.1016/j.cbpc.2020.108735
Wang, L., Cui, X., Cheng, H., Chen, F., Wang, J., Zhao, X., et al. (2015). A review of soil cadmium contamination in China including a health risk assessment. Environ. Sci. Pollut. Res. 22, 16441–16452. doi:10.1007/s11356-015-5273-1
Wu, S., Xu, X., Zhao, S., Shen, F., and Chen, J. (2013). Evaluation of phenanthrene toxicity on earthworm (eisenia fetida): an ecotoxicoproteomics approach. Chemosphere 93, 963–971. doi:10.1016/j.chemosphere.2013.05.062
Wu, W., Dong, C., Wu, J., Liu, X., Wu, Y., Chen, X., et al. (2017). Ecological effects of soil properties and metal concentrations on the composition and diversity of microbial communities associated with land use patterns in an electronic waste recycling region. Sci. Total Environ. 601-602, 57–65. doi:10.1016/j.scitotenv.2017.05.165
Xu, T., Xi, J., Ke, J., Wang, Y., Chen, X., Zhang, Z., et al. (2023). Deciphering soil amendments and actinomycetes for remediation of cadmium (cd) contaminated farmland. Ecotoxicol. Environ. Saf. 249, 114388. doi:10.1016/j.ecoenv.2022.114388
Žaltauskaitė, J., and Sodienė, I. (2014). Effects of cadmium and lead on the life-cycle parameters of juvenile earthworm eisenia fetida. Ecotoxicol. Environ. Saf. 103, 9–16. doi:10.1016/j.ecoenv.2014.01.036
Zeb, A., Li, S., Wu, J., Lian, J., Liu, W., and Sun, Y. (2020). Insights into the mechanisms underlying the remediation potential of earthworms in contaminated soil: a critical review of research progress and prospects. Sci. Total Environ. 740, 140145. doi:10.1016/j.scitotenv.2020.140145
Zhang, L., Duan, X., He, N., Chen, X., Shi, J., Li, W., et al. (2017). Exposure to lethal levels of benzo[a]pyrene or cadmium trigger distinct protein expression patterns in earthworms (eisenia fetida). Sci. Total Environ. 595, 733–742. doi:10.1016/j.scitotenv.2017.04.003
Zhang, Q., and Wang, C. (2020). Natural and human factors affect the distribution of soil heavy metal pollution: a review. Water, Air, and Soil Pollut. 231, 350. doi:10.1007/s11270-020-04728-2
Keywords: Cd, soil microorganisms, earthworm, metabolism pathway, gene expression, oxidative stress
Citation: You R, Li H, Li X, Luo L, Wang P, Xia H and Zhou Y (2024) Ecotoxicological impacts of cadmium on soil microorganisms and earthworms Eisenia foetida: from gene regulation to physiological processes. Front. Environ. Sci. 12:1479500. doi: 10.3389/fenvs.2024.1479500
Received: 12 August 2024; Accepted: 27 September 2024;
Published: 09 October 2024.
Edited by:
Jifu Ma, Yan’an University, ChinaReviewed by:
Deliang Yin, Guizhou University, ChinaZhenya Tang, Kunming University of Science and Technology, China
Xiaodong Zhang, Tianjin University, China
Copyright © 2024 You, Li, Li, Luo, Wang, Xia and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ya Zhou, emhvdXlhMjRAaG90bWFpbC5jb20=
 Rui You
Rui You Hui Li
Hui Li