
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 25 July 2024
Sec. Toxicology, Pollution and the Environment
Volume 12 - 2024 | https://doi.org/10.3389/fenvs.2024.1423339
This article is part of the Research Topic Biological Contaminants of Concern in Water and Wastewater: An Environmental Health Perspective View all 15 articles
Toxic cyanobacterial species occur in aquatic ecosystems when favourable environmental conditions prevail. These bacteria can produce natural hepatotoxic metabolites called microcystins that can affect the quality of water. Human exposure to microcystins results from ingesting contaminated drinking water and therefore cyanobacterial species producing these toxins should be monitored in these waters. The present study aimed to trace and identify cyanobacterial strains that potentially produce microcystins in drinking water. To achieve this objective, advanced digital flow cytometry and polymerized chain reaction were used for the detection and identification of cyanobacterial strains in water samples collected from water storage containers in Mawoni village. Full-length 16S rRNA genes from cultured cyanobacteria were amplified and sequenced using the 16S primers. Three novel strains of Chroococcus sp. (m64187e-7881, m64187e-2143, and m64187e-0930) and two strains of Microcystis aeruginosa (m64187e-6729 and m64187e-1069) were detected and identified in drinking water samples. The presence of these strains could indicate the potential of microcystins occurrence in drinking water, which therefore, could present potential human health risk due to exposure to such cyanotoxins.
The cyanobacteria phylum of the photosynthetic prokaryotes has been in existence for over 3.5 billion years and can be found in various habitats including freshwater bodies (Chia et al., 2022; Wang et al., 2024). They are considered the oldest organisms in the evolutionary process which are capable of performing oxygenic photosynthesis and are known to oxygenate the atmosphere of the earth (Balasooriya, 2019). Favourable physicochemical conditions support the growth of these phyla (Chia et al., 2022). According to Chia et al. (2022); Melaram et al. (2022), factors influenced by global warming, such as eutrophication, high temperatures and increased solar irradiation can influence the excessive growth of harmful cyanobacteria in aquatic systems. Amongst the cyanobacteria species are those that produce and release a copious number of metabolites called cyanotoxin-microcystin. Currently, cyanobacteria have more than 60 species that are known as microcystin producers worldwide (Usman et al., 2022), and some of these species include Microcystis and Chroococcus sp. (Magonono et al., 2018; Chen et al., 2021; Mirasbekoc et al., 2021; Valadez-Cano et al., 2022; Thawabteh et al., 2023). Apart from being the water quality problem such as affecting the aquatic ecosystem and aquatic animals, the occurrence of cyanobacterial species and their toxins can also pose severe health threats to humans and animals (Chen et al., 2021; Jia et al., 2024). These toxins have been reported to comprise potent neurotoxic, hepatotoxic, dermatoxic and cytotoxic agents that they produce (Nowruzi et al., 2023) as well as causing gastro-intestinal toxicity (Li et al., 2024). Many studies have been focused on microcystins globally, due to their extensive occurrence, steadiness and resilience to chemical and biological breakdown, as well as their ability to reach high concentrations in scums and blooms (Dai et al., 2016; Welten et al., 2020).
According to Carmichael et al. (2013), microcystins are clyclic heptapeptides tha consist five common amino acids and pairs of L-amino acids as variants; with the most known acids being methyl aspartic acid, N-methyldehydroalanine, alanine, glutamic acid, and Adda (3-amino-9-methoxy-2,6,8-trimethyl-10- phenyldeca-4,6 dienoic acid) (Pearson et al., 2010). It was further reported that microcystins fall within a group of small molecular weight causing human and animal poisoning that involves acute hepatotoxicosis (Sarma, 2013). Previous studies have reported that most human health problems are linked with continuous low microcystin concentrations exposure (Drobac et al., 2013), of which one of the common exposure routes is through drinking microcystins-contaminated water (Mutoti et al., 2022; Melaram et al., 2022). The health threats caused by microcystins have led the World Health Organization (WHO) to set the standards for drinking water quality for these toxins; for example, a guideline value for total MCs is 1 μg/L in drinking water (Farrer et al., 2015; WHO, 2017). A liver is one of the variety of organs that can be targeted by microcystins. Acute exposure to these toxins can result in headaches, eye pain, blurred vision, nausea, and vomiting (Lad et al., 2022; Lim et al., 2023). With many studies focused on the cyanotoxins impact on animal health, there is limited information on the human health impact of cyanotoxins. Therefore, the human health effects of microcystins are still not well understood in some cases, mostly chronic health impacts (O’Keeffe, 2019). Potential associations have been identified by various studies between liver disease and microcystin exposure. The tissue of dialysis patients in Brazil dialysis centre were found to be accumulated by microcystins, causing death to many patients due to liver failure (O’Keeffe, 2019). Similarly, correlation was observed between freshwater microcystins-producing cyanobacteria with incident of liver cancer and mortalities in central Serbia (Melaran et al., 2022).
Most drinking water authorities have been facing the challenges of treating water contaminated with cyanobacteria. Although a guideline document to assist portable water producers in dealing with toxic cyanobacteria in water sources has been developed (WHO, 2017), Mutoti et al. (2022) reported that there is still a lack of personnel who are skilled enough to develop protocols and programmes to monitor cyanobacteria toxins, consequently becoming the key challenge faced by water treatment works (DWTW) in Southern Africa.
Currently, numerous analytical methods have been used for the assessment of microbiological characteristics of the water samples globally. A polyphasic approach has been utilized as a system of cyanobacterial species description that combines 16S rRNA, morphology, phycology, and ecology in trying to acknowledge the need to incorporate phylogenetics into cyanobacterial taxonomy (Willis and Woodhouse, 2020). This approach has been supported by taxonomists such as Komárek et al. (2014) and Komárek (2020) who pointed out that the polyphasic approach provides the best means of conducting taxonomic practice. Reviews have been conducted on DNA sequencing that is used for the phylogenetic and taxonomic analysis of cyanobacterial isolates (Mutoti et al., 2022). Many studies have utilized these approaches for the classification and identification of cyanobacteria species in drinking water (Mutoti et al., 2022; Valade-Cano et al., 2022). Moreover, FlowCAM has been used that capture images of cyanobacteria that are then identified based on their morphological parameters (area, length, width, equivalent spherical diameter, and area-based diameter) and shape; whereas, the PCR uses 16S rRNA gene sequencing to identify cyanobacterial to their species level (Wilson et al., 2000). Both FlowCAM and PCR techniques are taxonomic methods that enable the identification of cyanobacterial species in natural habitats (Lee et al., 2017) based on morphological characteristics and genomic information (Willis and Woodhouse, 2020). Komárek et al. (2014) reported that there is very limited data for most genera such as Chroococcus within the Chroococcaceae family; whereas, the well-known genus Microcystis is constantly monophyletic, but the species level classification within the genus can be controversial. This therefore triggers studies of this nature to try and fill up this vital data gap.
Containers (20–25 L) are used in rural areas such as Mawoni for collection and storing water for future use. The presence of pathogenic bacteria, including cyanobacteria in this water has been assumed. There has been some observation of cyanobacteria development inside these containers forming biofilm, which potentially pose threats to the households that use such water. Studies on the occurrence of cyanobacteria blooms have been seldom conducted in African aquatic resources (Chia et al., 2022). In Mawoni village and the surrounding areas located in the North part of South Africa in Limpopo, nothing or limited information has been reported regarding the underlying genetic characterization, morphology, and geographical origin among the strains of cyanobacteria. This gap in knowledge limits our understanding of cyanobacteria diversity, distribution, toxin production, and the nature of their congeners, highlighting the risks of exposure through potable water resource contamination. Therefore, to fill-up this gap, the present study is the first of its kind to assess the presence of cyanobacteria strains that potentially produce and release microcystins in drinking water collected from water storage containers in Mawoni village. This information is fundamental to the water supply authorities that require critical control and management approaches to prevent health problems that are associated with these bioactive metabolites.
In the present study, sixteen water samples (four batch of four samples: n = 16) were collected using Latex Free, Polypropylene Medline Basic Specimen containers with screw lids (manufactured by Medline, US). Samples were collected from water containers that contain green seeded biomass on their side walls and are used to store drinking water in Mawoni village. Sampling was conducted ones during summer season. During samples collection, physicochemical parameters were measured insitu using the Accsen PC 70 multimeter (manufactured by Accsen Instrumental in Italy) and the Turbicheck WL portable turbidity meter (TB 250 WL) (manufactured by Lovibond Water Testing, Germany) was used for turbidity measurements. Instruments were calibrated before use and all the physicochemical parameters were measured in triplicates from all the samples collected. Cyanobacterial strains from the collected water samples were cultured and grown in sterile shake flasks containing liquid medium (BG11). Before the examination, these cultures were incubated for 4 weeks at room temperature (25°C) and elevated temperature (40°C) under 24 h of white fluorescent lamp light (at 27 μmol m−2 s−1) as described by Magonono et al. (2018); Mutoti et al. (2022). Since the taxonomic system of cyanobacteria according to Komárek et al. (2014) has radically changed due to the introduction of electron microscopy; and molecular and genetic methods for the characterization of cyanobacterial taxa; this study further utilizes a Leica DM3000 semi-automated laboratory microscope (Leica, Germany) to observe morphological characteristics of filaments and cells. Furthermore, the bench-top FlowCAM (8100-C, Fluid Imaging Technologies Inc., Scarborough., ME, United States) was used to generate images of cyanobacterial strains. Microscope and bench-top FlowCAM were adopted for preliminary analysis of water samples. The FlowCAM captures cyanobacterial cells as the water sample flows past the filter tube inside the FlowCAM device. The instrument was equipped with software (VisualSpreadsheet V3.2.2) used to view and select images of interest. During analysis, the instrument was on auto-image mode (capture rate of 20 frame/sec) and auto-focused for ×10 objective with the flow rate of 0.4 mL/min. Although the primary morphological characteristics that are diagnostic of the families were first given in Anagnostidis and Komárek (1990) and updated in Komárek (2013), the morphology of captured cyanobacterial strains in the present study was further identified according to van Vuuren et al. (2006); Balsooriya (2019) descriptions.
Furthermore, cyanobacterial cells harvested from cultures of four samples (each sample from each batch) by filtering were further used for molecular characterization and identification using PCR. The included analytical procedures were DNA extraction, amplification using PCR, sequencing, and phylogenetic analysis, and these procedures are described below.
Standard procedures for extraction of total gDNA were performed according to the instructions supplied by the manufacturer. Quick-DNA/RNA™ Miniprep Plus Kit (supplied by Inqaba Biotech Laboratories, South Africa) was used for the isolation of DNA. The first step of extraction began with cyanobacteria cell lysis, where an equal mixture of 400 µL of 2X concentrate with 400 µL of nuclease-free water was homogenized in a Lysis Tubes to get 1X DNA/RNA Shield™, followed by 10 min of centrifugation at 1600 × g and transfer of supernatant into an RNase-free tube. An equal volume of DNA/RNA Lysis Buffer was added, mixed well and then transferred into a Spin-Away™ Filter in collection tubes and centrifuged for 1 min. The Spin-Away™ Filter was again transferred into a new collection tube, 400 µL DNA/RNA Prep Buffer was added to the column and centrifuged for 30 s and the flow-through was discarded. 700 μL of DNA/RNA Wash Buffer was then added and centrifuged for 30 s and again the flow-through was discarded. 400 μL of DNA/RNA Wash Buffer was added and centrifuged for 2 min to ensure complete removal of wash buffer then the column was carefully transferred into clean microcentrifuge tubes. A volume of 100 µL of DNase/RNase-Free Water was directly added to the column matrix, let stand for 5 min, and then centrifuged to elute DNA from the respective column. The eluted DNA was stored at −70°C and then sent to Inqaba Biotech Laboratories for further analysis.
The basic molecular characterization of strains were performed after 16S rRNA gene fragments amplification with PCR using the primers: 27F (/5AmMC6/gcagtcgaacatgtagctgactcaggtcac-AGAGTTTGATCCTGGCTCAG) and 1492R (/5AmMC6/tggatcacttgtgcaagcatcacatcgtag-TACGGYTACCTTGTTACGACTT), which were adopted from Meirkhanova et al. (2023). The methods for the multi-gene analysis and sequencing were carried out as per Mutoti et al. (2022). The Barcoded Universal Primer approach was used for multiplexing 16S amplicons using two rounds of PCR workflow (Supplementary Table S1). The first round used the universal primer-tailed 16S primers (targeting the bacterial 16S rRNA genes) and the PacBio Barcoded Universal Primers were used in the second round (Gueidan et al., 2019). DNA bands of amplified PCR products were clearly shown on agarose gel sizing at base pairs of ≥1100 (Figure 1). A PacBio (www.pacb.com) sequel system was used for sample sequencing. Version 9.0 Single Molecular Real Time (SMRT) link was used for processing raw subreads to produce highly accurate reads. The V-search (https://github.com/torognes/vsearch) was then used to process these highly accurate reads and based on QIMME2 the taxonomic information was determined.
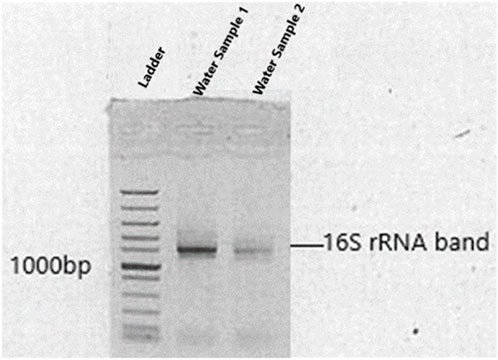
Figure 1. Amplified 16S rRNA of bacterial isolates shown on agarose gel electrophoresis. Levels of gene migration are expressed by lanes (water samples 1 and 2).
GenBank was used to deposit sequences of novel strains found in this study under accession numbers OP323090, OP323094, and OP323097 for Chroococcus sp. and accession numbers OP323093 and OP323094 for Microcystis aeruginosa. The alignment of sequences obtained was performed using a program (BioEdit software) for DNA sequencing and a ClustalW (MEGA-X Version 4.0). To validate the origin of cyanobacteria of sequenced samples, NCBI/BLAST (obtainable from http://www.ncbi.nlm.nih.gov/BLAST/) was used for comparison of 16S rRNA gene sequences compiled from available databases with sequences obtained in this study; therefore, these sequences were used to construct phylogenetic trees. The p-distance method (Nei and Kumar, 2000) and the neighbour-joining method (Saitou and Nei, 1987) were used to calculate and construct evolutionary distances and phylogenetic trees, respectively, using 1000 replicates for bootstrap analysis and only bootstrap above 50%. Bacillus subtilis strain was used as an out-group taxon when contracting the phylogenetic tree.
Pearson’s correlation coefficient analysis was utilized in the present study to determine the aspects of a linear relationship between various physical and chemical parameters observed during water sample collection.
The physicochemical parameters that were determined in this study indicated values that met World Health Organization recommended standards for domestic requirements (WHO, 2022). The mean and standard deviation (STDEV) for seven important physicochemical parameters were determined and shown in Table 1. Furthermore, Pearson’s correlation analysis was conducted in the present study (Table 2); and this analysis revealed a strong correlation between parameters with values highlighted in blue and further indicated moderate positive correlations between parameters with values highlighted in orange at a 0.01 level of significance.
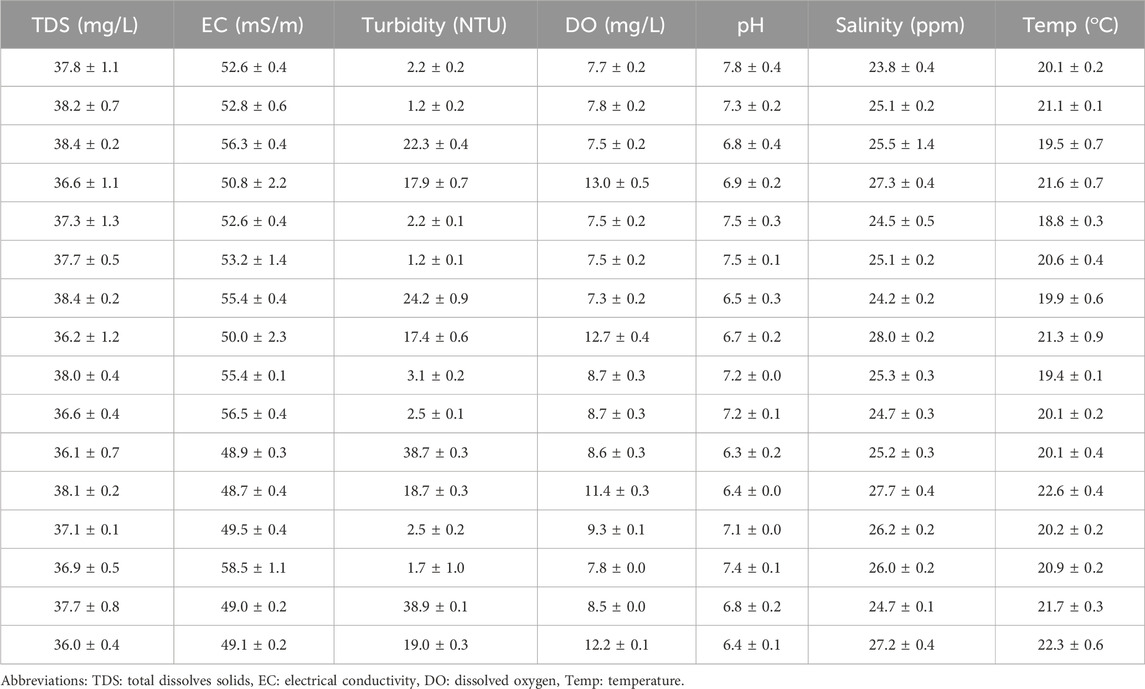
Table 1. Physicochemical parameters (mean and STDEV) status of water determined during the collection of water sample (n = 16).
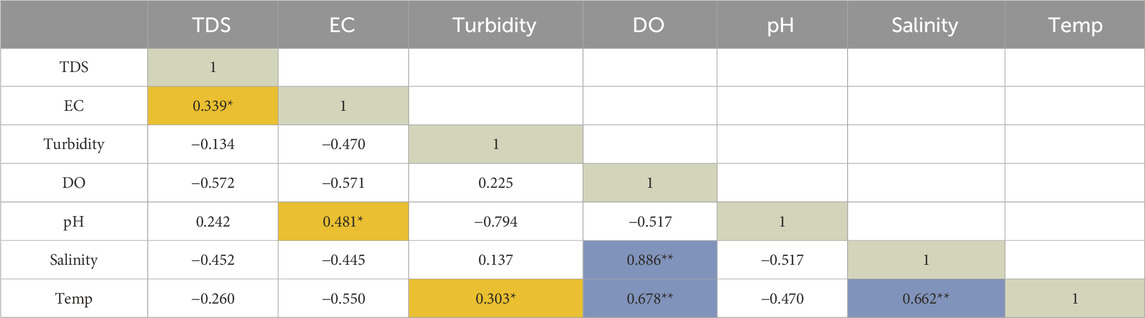
Table 2. Relationship between physicochemical parameters determined by the Pearson’s correlation analysis (n = 16).
Morphological characteristics were used for the identification of two cyanobacteria species Microcystis and Chroococcus. The isolated Chroococcus sp. (Figure 2) was observed as a group of two and/or four cells enclosed with clear and an amorphous mucilage sheath, with each cell having an individual envelope. Chroococcus sp. are prokaryotic organisms that have chlorophyll pigment, are unicellular, have a coccus cell shape, and form a mucous membrane (Goshtasbi et al., 2022; Fendiyanto et al., 2023). Chroococcus are egg to rod-shaped unicellular algae with a diameter of 0.4–50 µm. It is an autotrophic organism capable of surviving without oxygen. The mucus layers of different colonies stick together so that they can form a biofilm visible to the naked eye. Chroococcus sp. is classified as division cyanophyta, class cyanophyceae, order chroococcales, and genus Chroococcus. In the new classification system, this species, based on microalgae phylogeny studies by Shayler and Siver (2006), is classified closer to bacteria than other eukaryotic algae organisms (Fendiyanto et al., 2023).
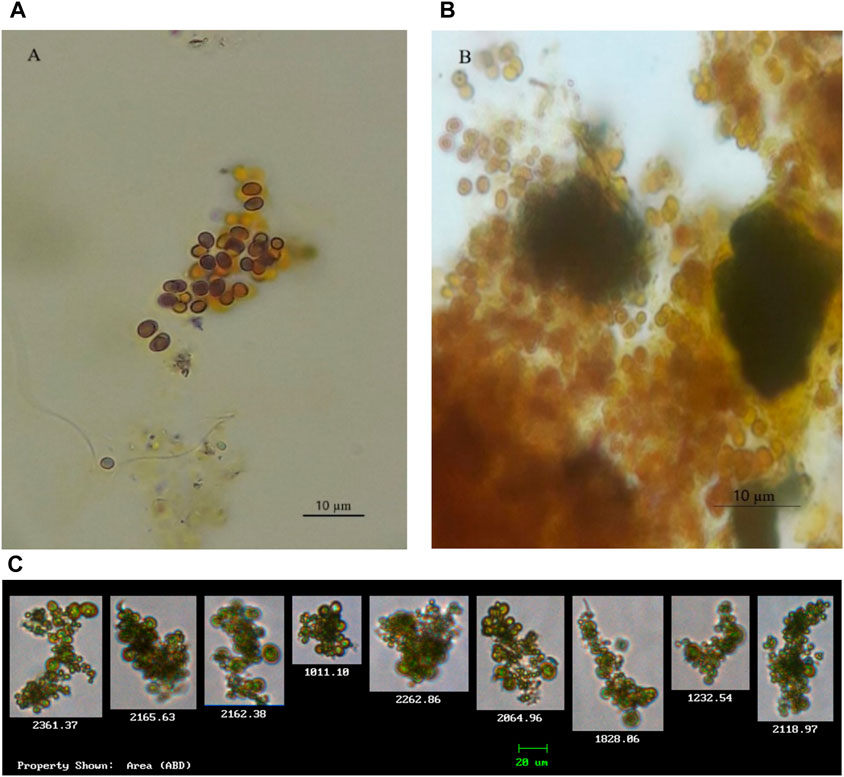
Figure 2. Isolated cyanobacterial species and their morphological features. (A,B) Chroococcus strains (observed with a microscope); (C) Chroococcus sp. (observed and captured with FlowCAM).
Species with cells that are green in colour and spherical were observed and captured using the FlowCAM. Characteristics of such species resemble that of Microcystis (Figure 3). According to Balsooriya (2019), Mutoti et al. (2022), and Thawabteh et al. (2023), cells of this species are spherical and green in colour and strains are classified as large since their diameter varies from 0.5 to 9 μm. They further reported that the cells of Microcystis are densely or sparsely, and irregularly arranged with fine and colourless mucilage that sometimes forms a wide margin around the cells. Cells of Microcystis are pale-blue–green and are spherical or hemispherical after division, but they appear brownish due to aerotopes that mask the blue–green colour of the protoplast. Due to vacuoles that are found within the cells, when they are viewed through a light microscope, they often have a black appearance (Mutoti et al., 2022).
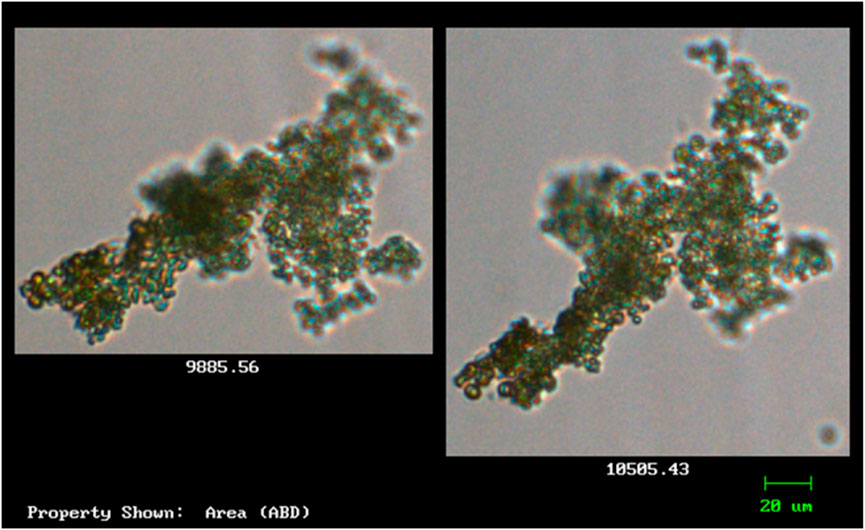
Figure 3. The morphological features of the isolated cyanobacteria species Microcystis (observed and captured with FlowCAM).
Sequences of cyanobacteria were targeted by primers set that were adopted in this study to target the 16S rDNA genes. Amongst these, were the sequences that resemble that of M. aeruginosa and Chroococcus sp. from the order chroococcales with sequences of Chroococcus sp. dominating the water samples. Table 3 shows strains of cyanobacteria that could not be identified using morphological characters, instead, were identified using molecular characters performed after culturing and isolation of samples. Sequence homology search performed by use of the BLAST program shows strains of cyanobacteria (Table 3) aligning most confidently to their closest related sequences from the NCBI database. Table 3 shows that strains m64187e-7881, m64187e-2143, and m64187e-0930 were closely related to Chroococcus sp. JJCM with percentage similarities of 95%, 96%, and 94%, respectively. On the other hand, cyanobacterial species M. aeruginosa NIES-1062 and strain m64187e-6729 had a similarity of 93%. Strain m64187e-1069 and species M. aeruginosa NIES-933 were found with a similarity of 93%.
In Figures 4, 5, the neighbour-joining method (Saitou and Nei, 1986) was used to construct the topology of sequences isolated from the present study and their closely related sequences from the NCBI database. The percentage of replicates for the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). Five base substitutions for nucleotide positions are represented by the scale bar. An outgroup taxon is shown by red dot.
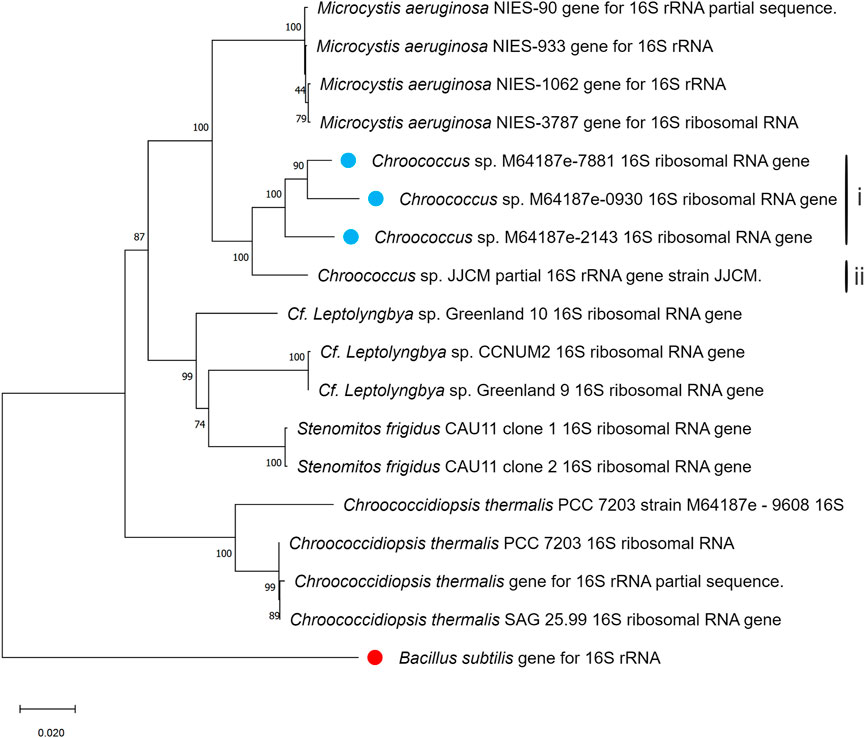
Figure 4. Phylogenetic tree of isolated Chroococcus sp. based on sequences isolated from the present study and sequences of its neighbouring species from the NCBI database. The tree was calculated using a maximum-likelihood algorithm with the bootstrap values given at the nodes. The cyanobacterial strains from the current study are indicated by a blue dot.
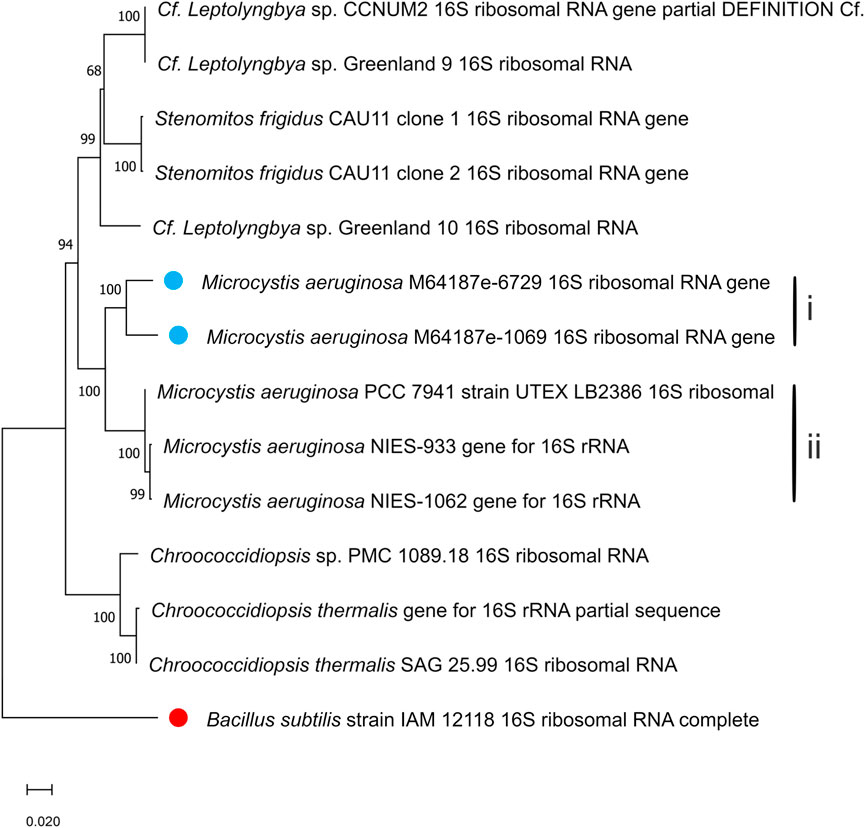
Figure 5. Phylogenetic tree of isolated Microcystis aeruginosa based on sequences isolated from the present study and sequences of its neighbouring species from the NCBI database. The tree was calculated using a maximum-likelihood algorithm with the bootstrap values given at the nodes. The cyanobacterial strains from the current study are indicated by a blue dot.
This study aimed at detecting and identifying the presence of cyanobacteria strains producing microcystin in drinking water. Leica DM3000 Semi-Automated Laboratory Microscope and FlowCAM were used for morphological characterization of cyanobacteria species detected in the present study. Captured images of cyanobacterial species were identified through comparison based on available literature. Cyanobacteria species (Microcystis and Chroococcus) from order Chroococcales were detected and identified, showing diverse morphological characteristics and phylogenetic relationships. Their morphological characteristics resemble that of Balsooriya (2019); Wang et al. (2021); Fendiyanto et al. (2023). Moreover, further morphological description of Chroococcus sp. in the present study was also consistent with previous descriptions by Balasooriya (2019); Goshtasbi et al. (2022). In a study conducted by Fendiyanto et al. (2023) in Dramaga Bogor, water samples were collected from freshwater found in high and low light-intensity environments. In their study, they aimed to identify the morphological diversity level of microalgae-based on their environment (high light and low light environment). Their results indicated that cyanobacterial species Chroococcus and Microcystis from order chroococcales were found among the most dominating species in freshwater samples collected from high-light intensity environment. They concluded that light was the main environmental factor influencing the growth and production of these microalgae in freshwater, posing a threat to humans and the aquatic environment.
In addition, the BLAST program (NCBI) was further utilized in the present study to search for strains of cyanobacteria, which are closely related to the strains of cyanobacteria found in this study. The BLAST search confirmed that the stains of cyanobacteria found in this study were closely related to species of cyanobacteria that have the potential to produce microcystins, such as Microcystis and Chroococcus (Mirazbekov et al., 2021). Borowitzka (2018) reported that the genus Microcystis contains several toxins-producing species, with M. aeruginosa being the best-known and globally distributed. They further highlighted that microcystins produced by Microcystis are a family of monocyclic heptapeptides with a characteristic feature, the unusual β-amino acid, Adda (3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4E,6E-dienoic acid). Numerous research studies that have been conducted previously have reported on microcystins released by Microcystis and Chroococcus species in freshwater bodies. A review study by Chia et al. (2022) reports on the presence of potentially toxic Microcystis strains in natural lakes found in the Middle Atlas Mountains in Morocco. In their study, they detected microcystin synthetase mcy genes using PCR. They further reported a study that characterized 27 potentially toxic cyanobacterial strains including Microcystis from seven reservoirs in Tunisia, which also detected microcystin synthetase mcy cluster (mcyA, -B, -C, -D, -E, and -G). Apart from these findings, there are numerous studies that have been conducted previously across the world and have reported on microcystins-producing cyanobacteria in drinking water (Table 4).
In humans, exposure to low doses of microcystin may lead to liver and kidney failure (Mutoti et al., 2022; Chia et al., 2022); on the other hand, exposure to high doses may lead to immediate liver necrosis and intrahepatic hemorrhage (Chia et al., 2022). Therefore, understanding the potential health effects of microcystins exposure underscore the importance of implementing comprehensive measures to prevent and minimize the presence of these toxins in drinking water. Hence, standards that serve as benchmarks for water quality management have been established by regulatory agencies and health organizations such as WHO and DWS (Mutoti et al., 2022; Mutoti et al., 2022). Maximum permissible levels of microcystins in portable water have been established to protect consumers from the potential health risks associated with these toxins (WHO, 2017). They safe guide public health and ensure the quality of water supplies. Moreover, these standards guide the implementation of monitoring and treatment measures to mitigate microcystin contamination.
Furthermore, sequences of different nucleotides together with those observed in the present study were utilized for the construction of the phylogenetic trees, where B. subtilis strain was included as an external group. The phylogenetic trees generated based on 16S rDNA displayed the phylogenetic position of the strains among their closely related cyanobacteria (Figures 4, 5). According to the phylogenetic tree in Figure 4, the maximum likelihood tree of the 16S rDNA sequences showed a tight cluster of Chroococcus sp. strains separated from all the other strains of the order chroococcales included in the present study. The whole cluster was supported by a bootstrap value of 100%. Two separate sub-clusters within the Chroococcus sp. cluster could be distinguished in the phylogenetic tree (Figure 4). The first cluster (I) comprised the strains m64187e-7881, m64187e-2143, and m64187e-0930, and the second cluster (II), which was separated from cluster I comprised Chroococcus sp. JJCM. Cluster I was supported by a bootstrap value of 100%. All the strains within cluster I, that is, m64187e-7881 (now referred to as Chroococcus sp. M64187e-7881 16S ribosomal RNA gene), m64187e-2143 (now referred to as Chroococcus sp. M64187e-2143 16S ribosomal RNA gene), and m64187e-0930 (now referred to as Chroococcus sp. M64187e-0930 16S ribosomal RNA gene) showed identical 16S rDNA sequences similarities of 94%–95% with strain Chroococcus sp. JJCM (with accession number AM710384) in cluster II isolated from the freshwater reservoir in South Bohemia (Jezberova, 2006).
Another tight cluster of M. aeruginosa is shown by the maximum likelihood three of the 16S rDNA sequences in Figure 5, supported by a bootstrap of 100%. Figure 5 also distinguished two separated clusters within the Microcystis cluster, with cluster I comprising strains m64187e-6729 and m64187e-1069, and cluster II comprising three different strains of M. aeruginosa; that is, M. aeruginosa PCC 7941, M. aeruginosa NIES-933, and M. aeruginosa NIES-1062. Both cluster I and II were supported by 100% bootstrap. Strain m64187e-6729 (thereafter called M. aeruginosa M64187e-6729 16S ribosomal RNA gene) had identical 16S rDNA sequences similarities of 93% with strain M. aeruginosa NIES-1062 gene (KX014841) isolated from farm ponds in USA by Yuan et al. (2020). Moreover, another identical 16S rDNA sequence similarity of 93% was observed between strain m64187e-1069 (thereafter called M. aeruginosa M64187e-1069 16S ribosomal RNA gene) and strain M. aeruginosa NIES-933 gene (LC557455) isolated from Japan by Suzuki et al. (2022).
In conclusion, this study has successfully identified and characterized two microcystin-producing cyanobacteria, M. aeruginosa and Chroococcus sp., isolated from drinking water samples in Mawoni village, South Africa. Utilizing a polyphasic approach combining morphological and molecular techniques, we employed FlowCAM imaging and PCR analysis to confirm the presence of these cyanobacteria species, which gave positive results for the identification and classification of members of the genus from the order Chroococcales, supported by BLAST analysis and phylogenetic tree construction. Notably, this study reports for the first time the presence of these biofilm-producing strains in drinking water, highlighting potential health risks associated with stored water. The findings of this study suggest that the potential for treated water stored in storage containers to support a range of toxic cyanobacteria may currently be underestimated, and therefore, isolation and screening of more cultured strains from treated water is required to further investigate the diversity of toxin-producing cyanobacteria. As such, future research should prioritize the analysis of microcystin synthetase genes and quantification of microcystins in potable water to assess health risks accurately. This approach could lead to improved monitoring and management strategies for cyanobacterial toxins in drinking water supplies. By focusing on these advancements, researchers and public health authorities can better safeguard water quality and protect community health. Furthermore, future research should also be focused on conducting cytotoxicity studies in these microcystin-producing species and more experimental studies to understand the detailed change in microbial diversity considering the variation in temperature and pH range in freshwater.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, OP323090, OP323093, OP323097, OP323100, and OP323094.
MM: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Validation, Writing–original draft, Writing–review and editing. JG: Conceptualization, Methodology, Project administration, Resources, Supervision, Validation, Writing–review and editing. AK: Data curation, Formal Analysis, Methodology, Validation, Writing–review and editing. AJ: Conceptualization, Formal Analysis, Supervision, Validation, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Research Foundation (South Africa) through a Doctoral scholarship, grant number UID 102072.
The authors thank the National Research Foundation for funding this project through Doctoral Scholarship (UID 102072) for MM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2024.1423339/full#supplementary-material
Anagnostidis, K., and Komárek, J. (1990). Modern approach to the classification system of the cyanophytes: stigonematales. Algol. Stud. 86, 1–74.
Balasooriya, B. L. W. K. (2019). Culture collection of cyanobacteria and microalgae at department of biotechnology wayamba university of Sri Lanka. Sri Lanka: Nethwin Printers Kandy. 978-624-5327-00-3.
Borges, H. L., Branco, L. H., Martins, M. D., Lima, C. S., Barbosa, P. T., Lira, G. A., et al. (2015). Cyanotoxin production and phylogeny of benthic cyanobacterial strains isolated from the northeast of Brazil. Harmful Algae 43, 46–57. doi:10.1016/j.hal.2015.01.003
Borowitzka, M. A., Schembri, M. A., Baker, P. D., and Saint, C. P. (2018). Molecular characterization of the toxic cyanobacterium Cylindrospermopsis raciborskii and design of a species-specific PCR. Appl. Environ. Microbiol. 66, 332–338. doi:10.1128/aem.66.1.332-338.2000
Carmichael, W. W., Billing, L. M., Blais, S., Hyde, J. B., Masonbrink, L. M., Palmer, M., et al. (2013). Human health effects from harmful algal blooms: a synthesis submitted by the HPAB to the International Joint Commission. Ottawa, Canada: International Joint Commission.
Chen, L., Giesy, J. P., Adamovsky, O., Svirčev, Z., Meriluoto, J., Codd, G. A., et al. (2021). Challenges of using blooms of Microcystis spp. in animal feeds: a comprehensive review of nutritional, toxicological and microbial health evaluation. Sci. Total Environ. 764, 142319. doi:10.1016/j.scitotenv.2020.142319
Chia, M. A., Ameh, I., George, K. C., Balogun, E. O., Akinyemi, S. A., and Lorenzi, A. S. (2022). Genetic diversity of microcystin producers (cyanobacteria) and microcystin congeners in aquatic resources across africa: a review paper. Toxics 10, 772. doi:10.3390/toxics10120772
Christoffersen, K., and Kaas, H. (2010). Toxic cyanobacteria in water. A guide to their public health consequences, monitoring and management. Limnol. Oceanogr. 45, 1212. doi:10.4319/lo.2000.45.5.1212
Dai, R., Wang, P., Jia, P., Zhang, Y., Chu, X., and Wang, Y. (2016). A review on factors affecting microcystins production by algae in aquatic environments. World J. Microbiol. Biotechnol. 32, 51–57. doi:10.1007/s11274-015-2003-2
Drobac, D., Tokodi, N., Simeunović, J., Baltić, V., Stanić, D., and Svirčev, Z. (2013). Human exposure to cyanotoxins and their effects on health. Arh. za Hig. rada i Toksikol. 64, 305–316. doi:10.2478/10004-1254-64-2013-2320
Farrer, D., Counter, M., Hillwig, R., and Cude, C. (2015). Health-based cyanotoxin guideline values allow for cyanotoxin-based monitoring and efficient public health response to cyanobacterial blooms. Toxins 7 (2), 457–477. doi:10.3390/toxins7020457
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. evolution 39, 783–791. doi:10.2307/2408678
Fendiyanto, M. H., Pratami, M. P., Satrio, R. D., Nikmah, I. A., Sari, P., Intan, N., et al. (2023). Species diversity of freshwater microalgae in Dramaga, bogor based on morpho-ecological identification between low and high light intensity environment. Jordan J. Biol. Sci. 16.
Frazão, B., Martins, R., and Vasconcelos, V. (2010). Are known cyanotoxins involved in the toxicity of picoplanktonic and filamentous North Atlantic marine cyanobacteria? Mar. Drugs 8 (6), 1908–1919. doi:10.3390/md8061908
Goshtasbi, H., Atazadeh, E., and Movafeghi, A. (2022). Polyphasic study of three cyanobacteria species from Kani Barazan international wetland in the northwest of Iran using morphological, molecular, biochemical, and bioinformatics approaches. Biologia. 77, 503–516. doi:10.1007/s11756-021-00940-5
Graham, J. L., Dubrovsky, N. M., Foster, G. M., King, L. R., Loftin, K. A., Rosen, B. H., et al. (2020). Cyanotoxin occurrence in large rivers of the United States. Inland Waters 10 (1), 109–117. doi:10.1080/20442041.2019.1700749
Gueidan, C., Elix, J. A., McCarthy, P. M., Roux, C., Mallen-Cooper, M., and Kantvilas, G. (2019). PacBio amplicon sequencing for metabarcoding of mixed DNA samples from lichen herbarium specimens. MycoKeys 53, 73–91. doi:10.3897/mycokeys.53.34761
Ivanov, D., Yaneva, G., Potoroko, I., and Ivanova, D. G. (2021). Contribution of cyanotoxins to the ecotoxicological role of lichens. Toxins 13 (5), 321. doi:10.3390/toxins13050321
Jezberová, J. (2006). Phenotypic diversity and phylogeny of picocyanobacteria in mesotrophic and eutrophic freshwater reservoirs investigated by a cultivation-dependent polyphasic approach. Czechia: University of Bohemia.
Jia, Y., Huang, Y., Ma, J., Zhang, S., Liu, J., Li, T., et al. (2024). Toxicity of the disinfectant benzalkonium chloride (C14) towards cyanobacterium Microcystis results from its impact on the photosynthetic apparatus and cell metabolism. J. Environ. Sci. 135, 198–209. doi:10.1016/j.jes.2022.11.007
Komárek, J. (2013). “Cyanoprokaryota and heterocytous genera,” in Freshwater flora of central europe. Editors B. Büdel, G. Gärtner, L. Krienitz, and M. Schagerl (Berlin, Heidelberg: Springer Spektrum).
Komárek, J. (2020). Quo vadis, taxonomy of cyanobacteria. Fottea 20, 104–110. doi:10.5507/fot.2019.020
Komárek, J., Kaštovský, J., Mareš, J., and Johansen, J. R. (2014). Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 86, 295–335.
Lad, A., Breidenbach, J. D., Su, R. C., Murray, J., Kuang, R., Mascarenhas, A., et al. (2022). As we drink and breathe: adverse health effects of microcystins and other harmful algal bloom toxins in the liver, gut, lungs and beyond. Life 12 (3), 418. doi:10.3390/life12030418
Lee, E., Khurana, M. S., Whiteley, A. S., Monis, P. T., Bath, A., Gordon, C., et al. (2017). Novel primer sets for next generation sequencing-based analyses of water quality. PLoS One 12, e0170008. doi:10.1371/journal.pone.0170008
Li, S. C., Gu, L. H., Wang, Y. F., Wang, L. M., Chen, L., Giesy, J. P., et al. (2024). A proteomic study on gastric impairment in rats caused by microcystin-LR. Sci. Total Environ. 917, 169306. doi:10.1016/j.scitotenv.2023.169306
Lim, C. C., Yoon, J., Reynolds, K., Gerald, L. B., Ault, A. P., Heo, S., et al. (2023). Harmful algal bloom aerosols and human health. EBioMedicine 93, 104604. doi:10.1016/j.ebiom.2023.104604
Magonono, M., Oberholster, P. J., Shonhai, A., Makumire, S., and Gumbo, J. R. (2018). The presence of toxic and non-toxic cyanobacteria in the sediments of the Limpopo River Basin: implications for human health. Toxins 10, 269. doi:10.3390/toxins10070269
Meirkhanova, A., Zhumakhanova, A., Len, P., Schoenbach, C., Levi, E. E., Jeppesen, E., et al. (2023). Dynamics of associated microbiomes during algal bloom development: to see and to be seeing. bioRxiv, 2023–2109.
Melaram, R., Newton, A. R., and Chafin, J. (2022). Microcystin contamination and toxicity: implications for agriculture and public health. Toxins 14, 350. doi:10.3390/toxins14050350
Mioni, C., Kudela, R., Baxa, D., Sullivan, M., Hayashi, K., Smythe, U. T., et al. (2011). Harmful cyanobacteria blooms and their toxins in clear lake and the sacramento-san joaquin delta (California). Delta (California) 10, 058–150.
Mirasbekov, Y., Abdimanova, A., Sarkytbayev, K., Samarkhanov, K., Abilkas, A., Potashnikova, D., et al. (2021). Combining imaging flow cytometry and molecular biological methods to reveal presence of potentially toxic algae at the Ural River in Kazakhstan. Front. Mar. Sci. 8, 680482. doi:10.3389/fmars.2021.680482
Mohamed, Z. A. (2016). Breakthrough of Oscillatoria limnetica and microcystin toxins into drinking water treatment plants-examples from the Nile River, Egypt. Water sa. 42 (1), 161–165. doi:10.4314/wsa.v42i1.16
Mutoti, M., Gumbo, J., and Jideani, A. I. (2022a). Occurrence of cyanobacteria in water used for food production: a review. Phys. Chem. Earth 125, 103101. doi:10.1016/j.pce.2021.103101
Mutoti, M. I., Jideani, A. I., and Gumbo, J. R. (2022b). Using FlowCam and molecular techniques to assess the diversity of Cyanobacteria species in water used for food production. Sci. Rep. 12, 18995. doi:10.1038/s41598-022-23818-1
Nowruzi, B., Becerra-Absalón, I., and Metcalf, J. S. (2023). A novel microcystin-producing cyanobacterial species from the genus desmonostoc, desmonostoc alborizicum sp. nov., isolated from a water supply system of Iran. Curr. Microbiol. 80, 49. doi:10.1007/s00284-022-03144-5
O’Keeffe, J. (2019). Cyanobacteria and drinking water: occurrence, risks, management and knowledge gaps for public health. Vancouver, BC, Canada: National Collaborating Centre for Environmental Health.
Pearson, L., Mihali, T., Moffitt, M., Kellmann, R., and Neilan, B. (2010). On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystin, nodularin, saxitoxin and cylindrospermopsin. Mar. Drugs. 8, 1650–1680.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi:10.1093/oxfordjournals.molbev.a040454
Sarma, T. A. (2013). Cyanobacterial toxins. In: Handbook of Cyanobacteria. Boca Raton, FL: CRC Press, Taylor and Francis Group, 487–606.
Shayler, H. A., and Siver, P. A. (2006). Key to Freshwater Algae: a web-based tool to enhance understanding of microscopic biodiversity. J. Sci. Educ. Technol. 15, 298–303. doi:10.1007/s10956-006-9016-4
Somdee, T., Kaewsan, T., and Somdee, A. (2013). Monitoring toxic cyanobacteria and cyanotoxins (microcystins and cylindrospermopsins) in four recreational reservoirs (Khon Kaen, Thailand). Environ. Monit. Assess. 185, 9521–9529. doi:10.1007/s10661-013-3270-8
Suzuki, S., Yamaguchi, H., and Kawachi, M. (2022) Potential genetic diversity of synechococcus-related strains maintained in the microbial culture collection of the national institute for environmental studies, Japan. Microbial resources and systematics, 38, 63–74.
Thawabteh, A. M., Naseef, H. A., Karaman, D., Bufo, S. A., Scrano, L., and Karaman, R. (2023). Understanding the risks of diffusion of cyanobacteria toxins in rivers, lakes, and potable water. Toxins 15, 582. doi:10.3390/toxins15090582
Usman, A. S., Merican, F., Zaki, S., Broady, P., Convey, P., and Muangmai, N. (2022). Microcystin production by oscillatorialean cyanobacteria isolated from cryopreserved Antarctic mats. Harmful algae. 120, 102336. doi:10.1016/j.hal.2022.102336
Valadez-Cano, C., Hawkes, K., Calvaruso, R., Reyes-Prieto, A., and Lawrence, J. (2022). Amplicon-based and metagenomic approaches provide insights into toxigenic potential in understudied Atlantic Canadian lakes. Facets 7, 194–214. doi:10.1139/facets-2021-0109
van Vuuren, S. J., Taylor, J., Gerber, A., and van Ginkel, C. (2006) “Easy identification of the most common freshwater algae. A guide for the identification of microscopic algae in South African freshwater,” in Brendan hohls: resource quality services.
Wang, X., Luo, Y., Zhang, S., and Zhou, L. (2024). Acetylacetone effectively controlled the secondary metabolites of Microcystis aeruginosa under simulated sunlight irradiation. J. Environ. Sci. 135, 285–295. doi:10.1016/j.jes.2022.12.004
Wang, Y., Jia, N., Geng, R., Yu, G., and Li, R. (2021). Phylogenetic insights into chroococcus-like taxa (Chroococcales, Cyanobacteria), describing Cryptochroococcus tibeticus gen. nov. sp. nov. and Limnococcus fonticola sp. nov. from Qinghai-Tibet plateau. J. Phycol. 57, 1739–1748. doi:10.1111/jpy.13205
Welten, R. D., Meneely, J. P., and Elliott, C. T. (2020). A comparative review of the effect of microcystin-LR on the proteome. Expo. Health 12, 111–129. doi:10.1007/s12403-019-00303-1
Willis, A., and Woodhouse, J. N. (2020). Defining cyanobacterial species: diversity and description through genomics. Crit. Rev. Plant Sci. 39, 101–124. doi:10.1080/07352689.2020.1763541
Wilson, K. M., Schembri, M. A., Baker, P. D., and Saint, C. P. (2000). Molecular characterization of the toxic cyanobacterium Cylindrospermopsis raciborskii and design of a species-specific PCR. Appl. Environ. Microbiol. 66, 332–338. doi:10.1128/aem.66.1.332-338.2000
Wiltsie, D., Schnetzer, A., Green, J., Borgh, M. V., and Fensin, E. (2018). Algal blooms and cyanotoxins in Jordan Lake, North Carolina. Toxins 10, 92. doi:10.3390/toxins10020092
Wood, S. A., Puddick, J., Fleming, R., and Heussner, A. H. (2017). Detection of anatoxin-producing Phormidium in a New Zealand farm pond and an associated dog death. N. Z. J. Bot. 55 (1), 36–46. doi:10.1080/0028825x.2016.1231122
World Health Organization (WHO) (2017). Guidelines for drinking-water quality: fourth edition incorporating the first addendum. Geneva: World Health Organization.
World Health Organization (WHO) (2022). Guidelines for drinking-water quality: fourth edition incorporating the first and second addenda. Geneva: CC BY-NC-SA 3.0 IGO. License.
Keywords: cyanotoxins, drinking water, human health, molecular identification, microcystins
Citation: Mutoti MI, Gumbo JR, Khwathisi A and Jideani AIO (2024) Newly isolated strains of potentially microcystin-producing cyanobacteria in potable water: case study of Mawoni village, South Africa. Front. Environ. Sci. 12:1423339. doi: 10.3389/fenvs.2024.1423339
Received: 25 April 2024; Accepted: 08 July 2024;
Published: 25 July 2024.
Edited by:
Visva Bharati Barua, University of North Carolina at Charlotte, United StatesReviewed by:
Sayanti Ghosh, University of Hawaii at Manoa, United StatesCopyright © 2024 Mutoti, Gumbo, Khwathisi and Jideani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mulalo I. Mutoti, bXV0b3RpbWlAdHV0LmFjLnph
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.