
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 01 March 2024
Sec. Water and Wastewater Management
Volume 12 - 2024 | https://doi.org/10.3389/fenvs.2024.1336318
This article is part of the Research Topic Biological Contaminants of Concern in Water and Wastewater: An Environmental Health Perspective View all 15 articles
Several studies have investigated the effects of swimming in sewage-polluted recreational beach water, highlighting the associated health hazards. To mitigate potential pathogen transmission, it is imperative that the polluted water is released away from recreational waters and foreshores, where children tend to play. At present, domestic sewage in the Faroe Islands solely undergoes primary wastewater treatment within primary settling tanks before being discharged into the ocean. Effluents are a major anthropogenic source of antibiotic resistance genes and antibiotic resistant bacteria, which are released into the environment. The aim of this study was to investigate antibiotic resistant Gram-negative bacteria and antibiotic resistance genes in influents and effluents of wastewater subjected solely to primary treatment, along with their release into the environment during both summer and winter. Water samples were collected from influents and effluents as well as with increasing distance away from the wastewater outlet and from nearby tidepools. Samples were cultured on MacConkey agar with four different antibiotics for detection of antibiotic-resistant bacteria and antibiotic resistance genes were quantified by droplet digital PCR. All multi-drug resistant bacteria were identified using the API 20E kit. We observed an overall decrease of the abundance of Gram-negative bacteria from the effluents compared to influents, however, we observed the opposite trend in the antibiotic resistance genes. Antibiotic resistant bacteria and antibiotic resistance genes in addition to multi-drug resistant bacteria were found in the surrounding oceanic and several terrestrial tidepool samples. Of the multi-drug resistant bacteria, we found, e.g., Escherichia coli, P. aeruginosa, and A. hydrophila species, which can be pathogenic, potentially causing an infection if encountering a host. These results indicate a relatively wide pollution range of the effluents from the septic tank and treated sewage released into the environment, posing a potential hazard for both humans and wildlife.
Recent research has unveiled new insight into the composition of wastewater and its environmental implications globally (J. Li and Zhang, 2019; Pendergraft et al., 2023). Of particular concern is the presence of antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) in wastewater and its release into the environment (Da Silva et al., 2006; Munk et al., 2022). This concern stems in part from the fact that antibiotic resistance represents a significant and pressing threat to global health (WHO, 2020) and was estimated to directly cause 1.27 million deaths and associated with ˜ 5 million deaths globally in 2019 (Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, 2022).
Municipal wastewater systems have been identified as significant reservoirs and hotspots for ARB and ARGs (Da Silva et al., 2006; Berendonk et al., 2015; Cesare et al., 2016; Fouz et al., 2020). These systems receive wastewater from various sources, including households, hospitals, and industries, which can contain a wide range of antibiotics and human gut bacteria. While bacterial concentrations typically diminish throughout wastewater treatment plants, a significant load can persist, ultimately being discharged into the environment and posing potential exposure to nearby animals and humans (Akiyama and Savin, 2010). Studies have also illustrated the mobilization of ARGs in wastewater, even spreading to divergent bacterial phyla and potentially undergoing further evolution (Berendonk et al., 2015; Shi et al., 2023; Ochman et al., 2024). A recent study demonstrated that reduction in tetracycline resistance genes after primary treatment seemed to be insignificant but reduced after secondary treatment (Ochman et al., 2024), highlighting the importance of properly treating wastewater before its release into the environment.
One consequence of the spread of ARGs and ARB into the environment is that pathogenic bacteria that previously were thought to be controlled, are resurfacing in new, more resistant forms, rendering traditional antibiotic therapies ineffective (Levy and Marshall, 2004). Unlike many chemical contaminants that tend to decrease in concentration through processes like degradation, dilution or sorption, ARB and mobile genetic elements containing ARGs can persist and even spread in the environment (Berendonk et al., 2015).
This investigation focused on specific ARGs deliberately chosen to typify genes commonly present in wastewater, demonstrating resistance across multiple antibiotic classes, β-lactams (blaOXA-like genes), sulfonamides (sul2) and tetracyclines (tetA) (Auerbach et al., 2007; Du et al., 2007; Zieliński et al., 2019; Munk et al., 2022).
Several studies have investigated the effects of swimming in sewage polluted recreational beach water, demonstrating a health hazard and increased cases of GI (gastrointestinal) illnesses such as vomiting and diarrhoea (Dwight et al., 2004; J; Li and Zhang, 2019; Wade et al., 2010). Thus, it is imperative that the polluted water is released away from recreational waters and foreshores, where children tend to play. However, pollution from wastewater is not only limited to the coastal waters where it is discharged, bacteria has also been found to transfer from the polluted ocean to the atmosphere in sea spray aerosol, affecting more individuals than previously anticipated (Pendergraft et al., 2023).
For densely populated areas, large wastewater treatment plants (WWTPs) use treatments such as biological treatment, chemical treatment, and radiation to eliminate most contaminants (Lira et al., 2020). Currently, household sewage in the Faroe Islands exclusively undergoes primary wastewater treatment through individual or communal septic tanks (Umhvørvisstovan, 2023). Substances denser and lighter than water remain in the primary settling tanks and the wastewater is subsequently discharged through outlets into the ocean; in certain locations discharge is only 3 m from the shoreline (The municipality of Tórshavn, 2021; Kunngerð um spillivatn, 2009). As the Faroe Islands are surrounded by turbid waters, the general idea is that the ocean dilutes wastewater to such a degree that it poses little to no risk to the environment or humans. However, more recent reports show that in some areas the concentrations of faecal indicators are higher than the recommended threshold for recreational purposes (The municipality of Tórshavn, 2023).
Antibiotics can enter the sewage system by inappropriate disposal or in urine and excrements as parent compounds or metabolites (Kümmerer, 2009; Leung et al., 2012). Sub-inhibitory concentrations of antibiotics present in sewage serve as an optimal environment for development of antibiotic resistance, partly due to selection pressure (Da Silva et al., 2006; Auerbach et al., 2007). The effect of antibiotics in wastewater is not fully understood but it can cause alterations in bacteria, such as changes in the expression of virulence genes, in the transfer of antibiotic resistance and affect cell functions (Ohlsen et al., 1998; Goh et al., 2002; Scornec et al., 2017) potentially causing great environmental damage of unknown proportions.
No studies have investigated whether bacteria from wastewater can be detected on land and in the ocean surrounding the wastewater outlets in the Faroe Islands and its possible effect on wildlife as well as humans on land. The aim of this study is to evaluate presence and abundance of ARB and ARGs at three locations within Tórshavn municipality. In particular, we want to investigate if there is an increase in the concentration of antibiotic resistant Gram-negative bacteria and of specific ARGs in the effluents compared to the influents to provide insight into the effect of the primary treatment processes on ARB and ARG concentrations. Additionally, we seek to assess the extent of dissemination into the marine ecosystem and identify multi-drug resistant bacteria present.
Combined, there are 20 outlets releasing wastewater in Tórshavn, Argir, and Hoyvík. Three of these outlets Skansin (Station S), Argir (Station A), and Havnará (Station H) are subjected to primary treatment and subsequently discharged into the ocean. Station S and Station A have smaller and larger primary settling tanks, where sewage from multiple households and businesses are combined and undergo primary treatment. The Skansin (Station S) outlet is situated in the North-eastern part of Tórshavn and releases wastewater from >550 citizens ˜ 3 m from land (Figure 1A). In Argir, the South-eastern part of Tórshavn, wastewater is collected from ˜ 3,100 individuals and is discharged ˜ 50 m away from the shore, within the pier (Figure 1B).
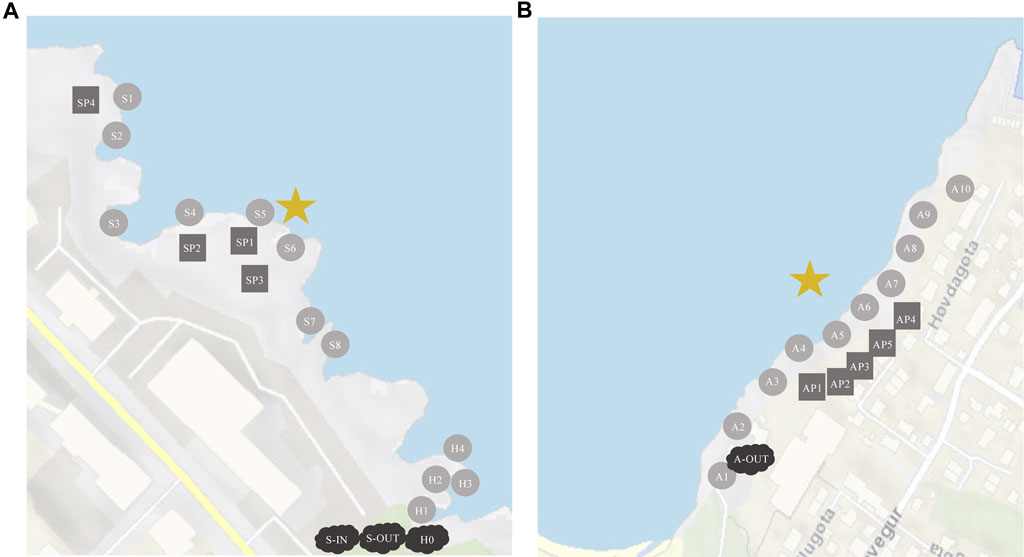
FIGURE 1. Sampling locations at Station S and H (A), and at Station A (B). Circles are oceanic samples, rectangles are tidepools on land, clouds are samplings from the well. The star is where the effluents are released into the ocean. (A) S-1N = influents, S-OUT = effluents, H0 = effluents. (B) A-OUT = effluents, no influent samples were accessible for this location.
On the shore right next to Skansin there is an outlet that originates from the main river that flows, mostly under the streets, through the centre of Tórshavn (Supplementary Figure S1). This river (Havnará, Station H) carries waste from some relatively large corporations, institutions, and approximately 6,000–7,000 citizens (Supplementary Table S1). The wastewater from these three outlets (Station S, A and H) and its distribution in the surrounding areas will be the focus of this study.
Thirty-three samples were collected on 17 August 2022 and 31 samples on 7 February 2023 from all stations (S, A, and H, Figure 1). In addition, samples were collected from Gomlurætt Station G at the same dates as a negative control site, situated relatively far away from any settlements approx. 15 min drive from Tórshavn. We filtered 250–300 mL of water from all samples collected with increasing distances away from the wastewater outlets (source).
At Station S (Figure 1A), we collected influents and effluents from the primary settling tank (S-IN and S-OUT) as well as eight marine water samples (S1-S8) from the ocean close to the shore. In addition, samples were collected from four different tidepools (SP1-SP4). In the same area, effluents from Havnará (H0) and four marine water samples (H1-H4), with increasing distance from H0, were collected (Figure 1A).
At Argir (Figure 1B), effluents (A-OUT) and ten marine water samples (A1-A10) were collected from the shore with increasing distance from the wastewater outlet, as well as samples from five different tidepools (AP1-AP5) on the foreshore. All samples from effluents and influents were tested in triplicates.
All samples were transported back to the laboratory and kept cold until further processing done on the same day of collection. All samples were cultivated and tested for gene copy numbers of blaOXA-like genes, tetA, and sul2. Multi-drug resistant (MDR) colony forming units (CFUs) were species identified. Samples for cultivation were preserved in glycerol by adding 7–8 mL of each water sample in 15 mL tubes with autoclaved glycerol, with the ratio of water sample to glycerol being approx. 1:4. The tubes were frozen at 80°C prior to further processing (Islam, n.d, 2023). Samples to be analyzed for ARGs were immediately filtered using 0.22 μm Sterivex™ (Millipore). Approx. 300 mL of each water sample was filtered by forcing the liquid through each filter, using a sterile syringe. With some highly concentrated samples, less than 100 mL could be filtered. The sterivex filters were frozen at −80°C until further analysis.
Frozen samples were thawed prior to cultivation. An amount corresponding to 100 µL of water sample was pipetted onto Petri dishes containing just MacConkey agar (MacConkey agar No. 2, Oxoid) and Petri dishes containing MacConkey agar and ampicillin. Concentrated samples of influents and effluents were diluted through a serial dilution to 10−2 and 10−3, before being added onto agar plates.
To spread the sample on the agar plates, a spread plate technique called “Copacabana” was performed, using sterile glass beads (Dahal, 2022). Beads were added to the Petri dish before putting the lid back on the plate. The agar plate was then slid back and forth to distribute the sample on the plate. The beads were removed, and agar plates incubated for 48–72 h at 37 °C. Colony forming units (CFUs) were counted after 24 h, 48 h and 72 h because colonies from the thawed samples required a longer time to develop.
Antibiotics in powder form were dissolved in autoclaved, distilled water and diluted to achieve minimum inhibitory concentration (MIC) for the respective antibiotics in agar (Supplementary Table S2). The Minimum Inhibitory Concentrations (MICs) were derived from antibiotic susceptibility testing performed on enteric bacteria by the National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS). The concentrations considered were specifically those relevant to Shigella, Salmonella, and E. coli (Escherichia coli). These bacteria, being opportunistic pathogens, are commonly employed for the assessment of microbial quality in water.
A few drops of HCl were added to the ciprofloxacin solution to make it dissolve completely. The antibiotic solutions were filtered through 0.22 μm Sterivex™ filters (Millipore). Antibiotic solutions were then added to liquid MacConkey agar that had cooled to 50°C and poured into Petri dishes.
Colonies resistant to ampicillin were subsequently tested for multi-drug resistance. They were subjected to tetracycline, ciprofloxacin, kanamycin, and ampicillin in agar at concentrations corresponding to their respective MIC (Supplementary Table S2). Ampicillin agar plates were a positive control.
Triplicate samples were inoculated, however due to the high number of CFUs on each agar plate, the one with the highest amount of CFUs was selected for MDR testing using the API 20E system. In the assessment of MDR, the CFUs that were resistant were further tested for identification (Supplementary Figure S2). To identify the MDR bacteria, API 20 E kit (bioMérieux) was used as described by the manufacturer. API 20E kit is used to identify bacteria belonging to the Enterobacteriaceae family and other non-fastidious Gram-negative bacteria. Before the identification using the API 20 E kit, the individual MDR colonies were re-streaked 4 times on MacConkey agar and incubated at 37°C to get pure colonies that were no older than 24 h.
Freshly isolated colonies were also subjected to an oxidase test. A drop of oxidase reagent (bioMérieux) was added onto clean filter paper placed in a Petri dish and a sterile inoculation loop was used to transfer bacteria onto the filter paper. A positive reaction turned the colony purple within approximately 10 seconds and a negative reaction left the colour unchanged (UK Standards, 2009).
DNA was extracted from the sterivex filters using DNeasy® PowerWater® Sterivex™ Kit (Qiagen) as described by the manufacturer. The concentration of the DNA was measured using Qubit (Invitrogen) as described by the manufacturer.
Absolute quantification of the genes blaOXA, tetA, and sul2 (Table 1) was performed using ddPCR and EvaGreen Supermix (BioRad) according to the manufacturer’s protocol. Threshold was set based on the number of positive droplets in the negatives for each of the genes (Supplementary Figure S5).
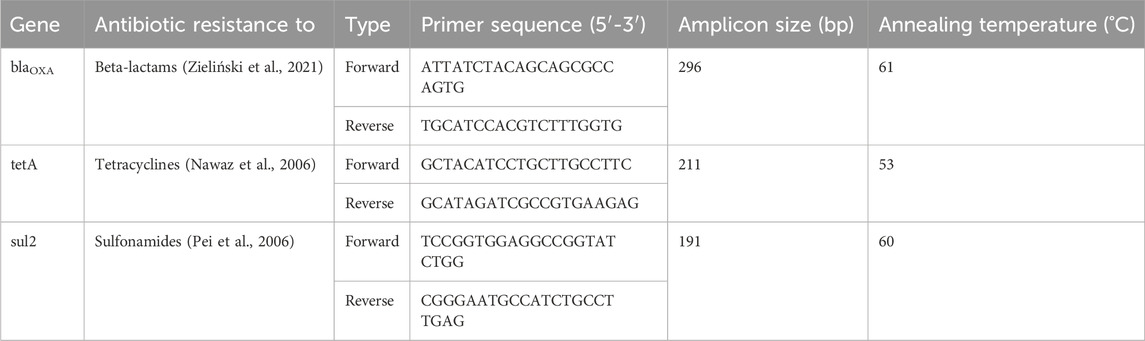
TABLE 1. Primers used for quantification of blaOXA-like genes, tetA gene and sul2 gene in the samples.
T-tests, means, and variance was calculated to assess statistical differences between: Summer and winter samples, influents (S-IN) and effluents (S-OUT), blaOXA-like genes, tetA, and sul2. When comparing temporal data, only samples with data from all relevant dates were used. When cultivating bacteria on agar, marine water samples and tidepools were not made into triplicates and were for that reason not used to make statistical tests. All other samples were done in triplicates and could be used for statistical analyses. A few of these samples had CFU counts fewer than ten, but the triplicates had little variation in CFUs, indicating consistency in pipetting technique.
Culturable bacterial concentrations (grown on MacConkey agar) in source samples (S-IN, S-OUT, A-OUT and H0) ranged from ∼5 × 104–1.6×105 bacteria/mL bacteria/mL in summer and ∼1.2 × 104–9.9×104 bacteria/mL in winter (Figure 2; Supplementary Table S3), with a significant difference between the seasons for all source locations at stations S, A and H (p = 0.019).
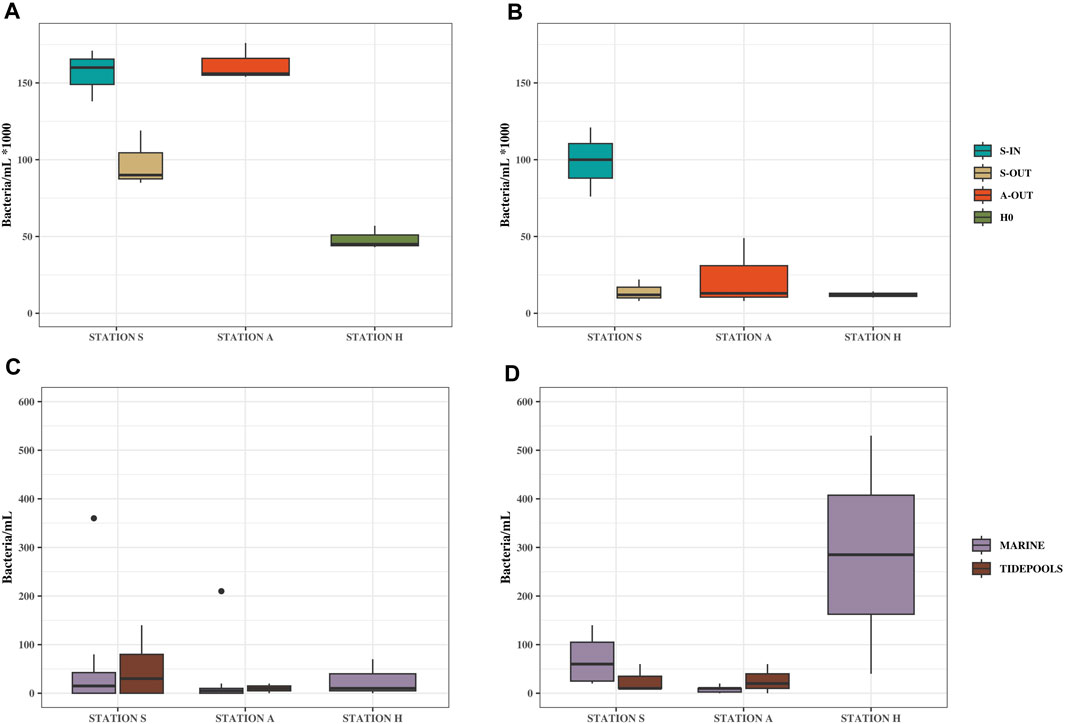
FIGURE 2. Culturable bacteria/mL from source, tidepools and marine water samples. Samples from Station S, A and H, summer (August 17, 2022) and winter (February 7, 2023). (A) Bacterial concentrations from source samples in summer (B) Bacterial concentrations from source samples in winter. (C) Bacterial concentrations from tidepools and marine water samples in summer. (D) Bacterial concentrations from tidepools and marine water samples in winter.
The ARG concentrations in source samples from stations S, A and H showed similar trends to the culturable bacteria with higher concentrations in summer than winter. In the summer tetA, blaOXA-like genes, and sul2 had concentrations of ∼2.2 × 104–1×105, ∼1.8 × 104–6.4×104, and ∼1.2 × 104–6×104 gene copies/mL, respectively, and winter concentrations of ∼5 × 103–1.1×104, ∼3 × 103–6×103, and ∼5 × 103–1.5×104 gene copies/mL, respectively (tetA p = 0.032, blaOXA–like genes p = 0.033, and sul2 p = 0.037).
In addition, both culturable bacteria and ARGs, showed opposite trends between influent and effluent concentrations (S-IN and S-OUT), for summer especially. There were significantly fewer culturable bacteria released into the environment (t-test, p = 0.028) but relatively higher concentrations of ARGs. This was especially pronounced during the summer while ARG concentrations were approximately the same for S-OUT and S-IN in the winter samples for tetA and sul2.
Compared to the source samples, relatively low concentrations of both culturable bacteria and ARGs were found in the environment (Figure 2; Figure 3). Concentrations of bacteria ranged from ∼0 to 400 bacteria/mL in the summer and ∼0–500 bacteria/mL in the winter (Figure 1; Supplementary Table S3). All ARGs also showed this same trend with ∼0–1.5×103 gene copies/mL in the summer and ∼0–3.8×103 in the winter. Station A generally had lower bacterial and ARG concentrations in the environment (A1-A10) and tidepools (AP1-AP5), compared to the environmental samples (S1-S8) and tidepools (SP1-SP4) from Station S (Supplementary Table S3). Environmental samples from Station H (H2-H4), had higher bacterial and gene concentrations, where winter had the highest concentrations measured in any environmental sample at 530 bacteria/mL, ∼2×103 blaOXA-like gene copies/mL, ∼3.8×103 tetA gene copies/mL, and ∼3.2×103 sul2 gene copies/mL (Figure 2D; Figure 3B).
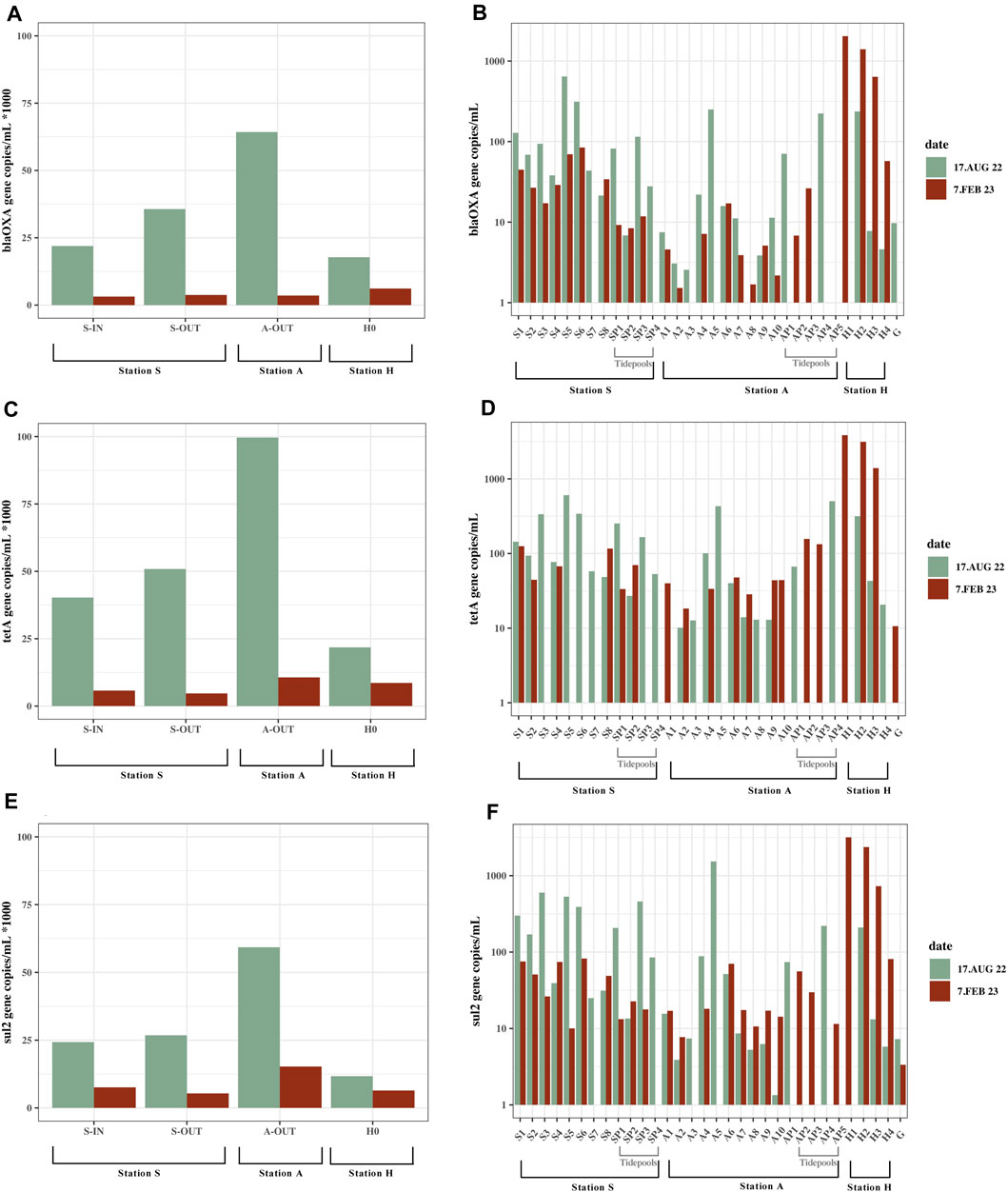
FIGURE 3. Gene copies/mL from all locations; seawater, tidepools and source. Samples from Station S, A and H, summer (August 17, 2022) and winter (February 7, 2023). Data does not exist for all locations on both dates (Supplementary Table S3). (A) blaOxA—like gene concentrations in source samples. Shown as 1/1,000 of the actual concentrations. (B) blaOxA—like gene concentrations in marine water samples and tidepools. Values are on a logarithmic scale. (C) tetA concentrations in source samples. Shown as 1/1000 of the actual concentrations. (D) tetA concentrations in marine water samples and tidepools. Values are on a logarithmic scale. (E) sul2 concentrations in source samples. Shown as 1/1000 of the actual concentrations. (F) sul2 concentrations in marine water samples and tidepools. Values are on a logarithmic scale.
The highest concentrations of culturable bacteria found in the tidepools were in Station S in SP1 at 140 bacteria/mL. Highest number of gene copies were in AP4 and SP3. AP4 had 500 tetA gene copies/mL, ∼220 blaOXA-like gene copies/mL. SP3 had ∼460 sul2 gene copies/mL (Figure 3; Supplementary Table S3). Several of the tidepools had higher ARG concentrations than the marine water samples close to the effluent outlets (Supplementary Table S3).
Gomlurætt (Station G) was used as background sample and had zero culturable bacteria, and gene copies ranged from 0–11 copies/mL (Supplementary Table S3). Of all the samples collected for gene copy numbers, 18 samples of blaOXA-like genes, 9 samples of tetA and 12 samples of sul2 contained a greater amount of gene copies/mL in summer compared to winter, however the difference was only statistically significant for sul2 (blaOXAlike genes p = 0.059, tetA p = 0.067, sul2 p = 0.048).
At both Station S and Station A, the highest environmental bacterial and gene concentrations were found in the locations nearest the effluent outlets in both summer and winter, around S5 and S6 (Station S) and at A5 and A6 (Station A), compared to the other marine samples (Supplementary Table S3). The concentrations of bacteria/mL and blaOXA-like genes, tetA and sul2 gene copies/mL decreased drastically within the first 40 m from the effluent outlet (Figure 4).
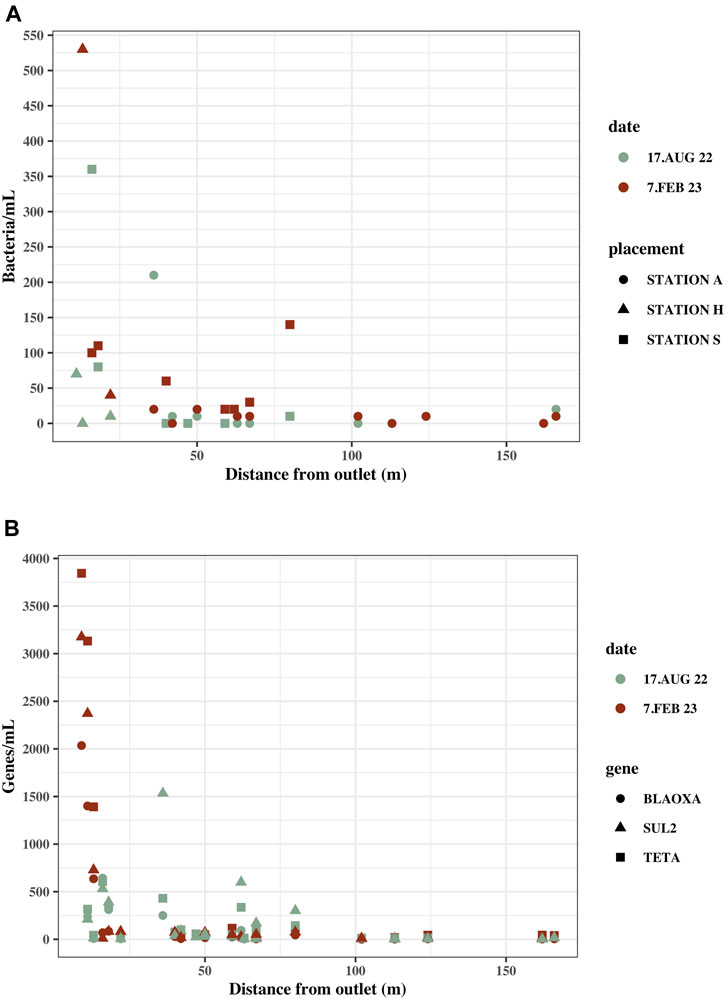
FIGURE 4. Concentrations in marine water samples with distance from the effluent outlet in the ocean. Distance from sample location to effluent outlet was measured on kortal.fo. (A) Culturable bacteria/mL in marine water samples at their distance from the effluent outlet. Samples are from Station S, A and H. (B) blaOxA—like genes, tetA and sul2 gene copies/mL in marine water samples at their distance from effluent outlet. Samples are from Station S, A and H.
The highest ARG concentrations were measured at Station H and Station S (Supplementary Table S3). The samples closest to the effluent outlet at Station S and H were within 20 m from the effluent outlet in contrast to the closest sample at Station A, which was located 36 m from the effluent outlet.
Sampling locations furthest from the effluent outlet still contain bacteria and ARGs exceeding the background concentration. S8, which is the sampling location furthest from the effluent outlet at Station S (59 m), had 20 culturable bacteria/mL in winter and zero in summer and ARG concentrations all exceeding the background concentration ranging from 21 to 116 gene copies. A10 at Station A is 162 m from the effluent outlet and had zero culturable bacteria but all ARG concentrations, except sul2 in summer, exceeded the background concentrations.
In all source samples (S-IN, S-OUT, A-OUT and H0), there were fewer CFUs growing on MacConkey agar with ampicillin compared to CFUs growing on MacConkey agar without ampicillin (Figure 5). Concentrations of culturable bacteria on McConkey agar with ampicillin from the source ranged from ∼4.3 × 104–9×104 ampicillin-resistant bacteria/mL in the summer, and ∼5 × 103–1.3×103 ampicillin-resistant bacteria/mL in the winter (p = 0.013). The collective samples exhibit an average resistance of 61% to ampicillin. The highest (90%) and lowest (45%) percentage of ampicillin resistant bacteria were found in H0 in summer and winter, respectively (Figure 5). A higher number of ampicillin resistant bacteria was found in most tidepools from Stations S compared to Station A.
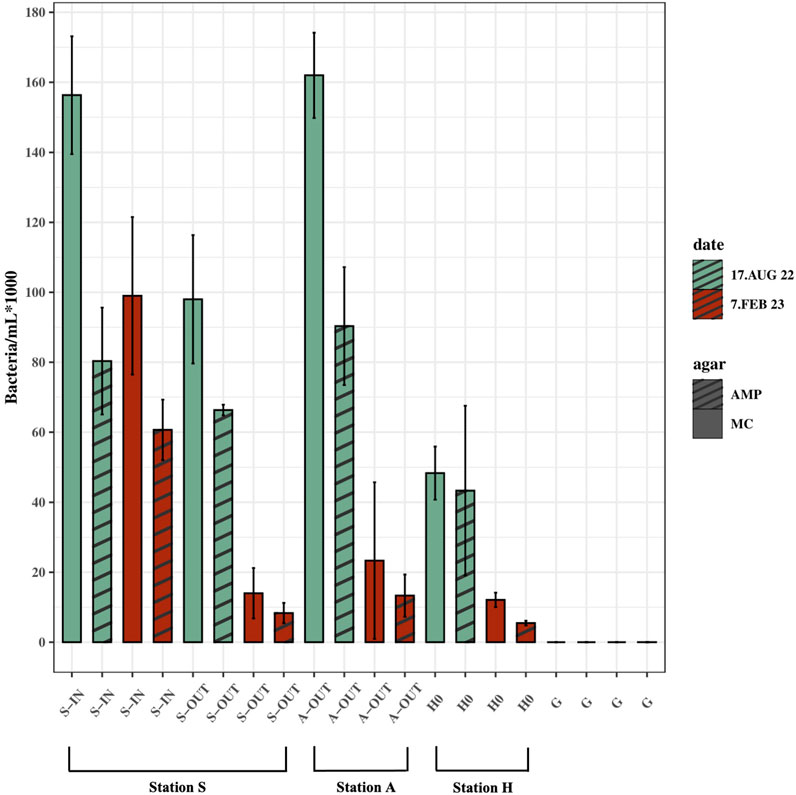
FIGURE 5. Ampicillin-resistant culturable bacteria/mL (AMP) vs. culturable bacteria/mL grown on agar without ampicillin (MC). Samples from source (S-IN, S-OUT, A-OUT and H0) and background sample from Gomlurætt (G) grown on agar with ampicillin (AMP) and without ampicillin (MC), situated side by side in the figure. Samples are from summer (August 17, 2022) and winter (February 7, 2023). Concentrations are based on the mean of triplicates.
Of all 565 ampicillin-resistant bacteria tested for multi-drug resistance, summer, and winter, 39% of them were resistant to tetracycline, 33% to kanamycin and the least resistance was to ciprofloxacin at 19% (Figure 6B). Similar pattern of resistance was observed at Station A and S, but not for Station H, where resistance to kanamycin rather than tetracycline was highest (Figure 6B). Only bacteria from the replicate with the highest number of colonies were identified, resulting in 87 bacteria in total from winter and summer. Of 87 only 67 bacteria could be identified, 54 of which belonged to the Enterobacteriaceae family, while 13 belonged to other bacterial families (Figure 6A). Specific locations of the identified bacteria are listed in the Supplementary Table S4. 41 out of 87 were identified as Klebsiella (47%). 20 of these were Klebsiella pneumoniae spp. 1 and 2, while 20 were K. oxytoca, and one was only identified as Klebsiella. 15 out of 87 (17%) tested MDR bacteria tested positive for oxidase. Ten of these were not identified, while the other five were: Pseudomonas species and Aeromonas and Elizabethkingia belonging to the classes Gammaproteobacteria and Flavobacteria, respectively.
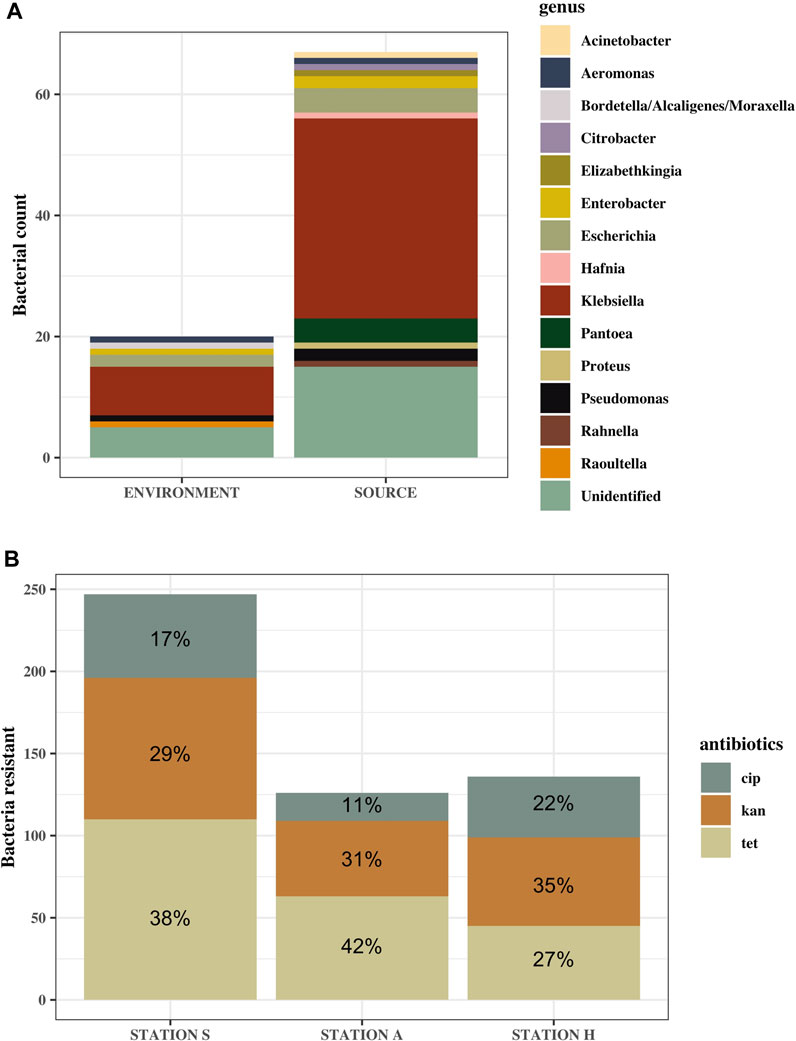
FIGURE 6. Identified genus of MDR bacteria (A) and the ratio of antibiotic resistance to the different antibiotics transferred from agar containing ampicillin (B). (A) Genus of identified bacteria from source samples (S-IN, S-OUT, A-OUT and H0) and the environmental samples, including tidepools and seawater. “Unidentified” are MDR bacteria that could not be identified using the API 20 E kit. More bacteria from source were subjected to MDR testing, hence the larger difference in number of bacteria in this category. (B) n = 564. Ratio is based on the mean of how many of the ampicillin-resistant bacteria were resistant to ciprofloxacin (cip), kanamycin (kan) and tetracycline (tet).
When comparing influents and effluents from Station S (S-IN and S-OUT respectively), S-IN contained a significantly higher concentration of bacteria, when cultivated on agar, compared to S-OUT, demonstrating that the primary treatment is reducing the number of culturable bacteria being released into the environment. Bacteria transferred from their niche to an environment with different temperatures and conditions, such as gut bacteria transferred to sewage, can also enter the viable but non-culturable (VBNC) state due to the stressful conditions, which could also affect the bacterial count on agar (Du et al., 2007; Li et al., 2014). This could presumably account for a small decrease seen from S-IN to S-OUT, while much of the decrease in bacterial concentration could be due to bacteria dying in the well or being withheld in the well along with the solids.
In contrast to the tendency of culturable bacterial concentration, blaOXA-like genes, tetA and sul2 concentrations were generally higher in S-OUT compared to S-IN, especially in summer. This could indicate that horizontal gene transfer occurs in the well at Station S. This is in accordance with other studies, demonstrating that wastewater treatment plants could function as horizontal gene transfer hotspots (Berendonk et al., 2015). Instead of reducing the number of antibiotic resistance genes released into the environment, in this instance, the concentrations are increased.
Horizontal gene transfer most likely happens through conjugation, as the resistant bacteria are in close contact with other non-resistant bacteria in the wells (Norman et al., 2009; Wintersdorff et al., 2016). Wastewater treatment in the Faroe Islands does not consist of the removal of chemicals, which means that antibiotics also end up in the effluents. Broadly speaking, antibiotic concentrations in untreated sewage are typically around 1 ug/L, which can cause selection pressure for antibiotic resistance (Larsson and Flach, 2022).
The presence of antibiotics has also been shown to induce competence in several species of bacteria, competence being a physiological state that is necessary for most bacteria to take up free DNA (Thomas and Nielsen, 2005; Huddleston, 2014). The HGT could therefore also happen through transformation. Antibiotics are then not only selecting for resistance but also act as a catalyst for transformation of antibiotic resistant genes.
Multiple ARGs are often located on the same resistance plasmids, giving resistance to multiple classes of antibiotics at once during HGT. The copy number of plasmids in each bacterium can vary, depending on the type of plasmid (Rozwandowicz et al., 2018). The generally higher tetA concentrations compared to blaOXA-like genes and sul2 does not necessarily indicate more tetracycline-resistant bacteria, but rather serve as an indication of ARG pollution, potentially leading to increased resistance in the surrounding environment. High levels of ARGs in the environment can also be an indicator of large amounts of wastewater reaching the specific location, which is the case in this study.
Both the ARG concentration and the ratio of ampicillin-resistant bacteria increased in S-OUT compared to S-IN in summer, indicate selection pressure on the culturable bacteria (Figure 3; Figure 5). However, more samples are required to give more insight into the seasonal and daily fluctuation of ampicillin resistance, due to the large variation between sampling events. Looking at bacterial counts from cultivation on agar, a significant difference was seen in the mean bacterial concentration of source samples between summer (17 August 2022) and winter (7 February 2023), with over threefold as many bacteria in summer. Weather conditions might have influenced these results. As storm sewers are connected to the sewage pipes, precipitation might dilute the wastewater, leading to lower bacterial concentrations/mL. In February, it had rained the previous day, compared to August, when it had been sunny and dry for days prior to the sampling event (Supplementary Figure S3; Figure 4). Temperature could also be a factor influencing the viability of the bacteria as well as the ARGs (Gutiérrez-Cacciabue et al., 2016; Yang et al., 2020). A study has demonstrated a strong correlation between temperature and the abundance of ARGs, where abundance was lower in winter, when temperatures were low (Yang et al., 2020). This is in accordance with our findings from the gene quantification of tetA, blaOXA-like genes and sul2, where there also was a statistically significant decrease in the concentration of gene copies/mL in winter compared to summer in the source samples.
Through cultivation of samples on MacConkey agar and quantification of blaOXA-like and tetA genes, there was a general tendency of lower concentrations in the marine water samples from Station A (Supplementary Table S3), where the wastewater outlet is situated farther away from land (50 m), than the wastewater outlets at Station S (3 m) and Station H (0 m). The concentration of bacteria and blaOXA-like genes, tetA and sul2 in the effluents from Station A were approximately the same as in the effluents from Station H and Station S, or higher, and does therefore not account for the low bacterial and ARG concentrations in the environment at Station A. In some instances, the concentrations in effluents from Station A were, in fact, the highest of the effluents measured on certain dates. This gives an indication, that the distance from wastewater release to land affects the dilution of wastewater and contamination reaching the foreshore, which is important to consider for city planning and potentially hazardous recreational areas. The relatively high concentrations in marine water samples closest to the wastewater outlets, both at Station A and Station S, further support the tendency of decreasing concentrations with distance from the effluent discharge.
At Station S, S1 had a higher bacterial concentration in winter than S2 and S3, which are situated closer to the wastewater outlet (Figure 1A). This tendency was also seen for blaOXA-like genes, tetA and sul2 concentrations in these locations (Figure 3). On the collection date, the ocean current was flowing towards the sampling locations and was predominately directed towards S1 (Supplementary Figure S4). This could indicate that not only distance is of importance, but that currents could also affect the spread of wastewater contamination.
Most tidepools had ARG concentrations exceeding the background concentration. In addition, at Station S, culturable bacteria were detected in three tidepools, indicating that treated sewage reaches the tidepools through sea spray aerosols or by surf, which would be in accordance with research regarding the transfer of wastewater in aerosols (Pendergraft et al., 2023). Two tidepools at Station A and one at Station S only contained ARGs and no culturable bacteria. ARGs can persist longer in the environment (especially those carried on plasmids) than bacteria and could have been transferred either as free ARGs or with bacteria by surf during turbulent weather or aerosols, persisting after the bacteria died (Zhang et al., 2018). However, the bacteria and ARGs could also have been transferred from wildlife, such as rats, in the nearby environment. The normally nocturnal rat species (Rattus Norwegicus) present in the Faroe Islands, was spotted multiple times while sampling in the daytime, indicating an abundant rat population on the foreshore (Puckett et al., 2020). Rats were observed at Station A, although more frequently at Station S. The rats are in close contact with wastewater from the well, being situated only 3 m from land and with wastewater from Station H, which is an open cave, both serving as an abundant and easily accessible food source. Rats can contaminate the surrounding area with their own urine and faeces and are also plausible vectors and reservoirs of contaminants from sewage such as pathogenic and multi-drug resistant bacteria, spreading them to tidepools and terrestrial ground (Byers et al., 2021).
Station H is the location, where the effluent outlet is closest to land. The reason for the high concentrations of bacteria and ARGs at this location compared to the other locations is presumably also because the area is relatively isolated, decelerating the dispersion of wastewater (Supplementary Figure S1). Without specific metrics in the guidelines, stating that there should be no accumulation of wastewater in the environment (Kunngerð um spillivatn, 2009), the meaning of accumulation becomes a subjective assessment. However, with constant flow of wastewater and bacteria, there is no need for accumulation to maintain high bacterial concentrations.
To ensure statistical accuracy of bacterial numbers, it is common practice to plate the samples in triplicates and count the CFUs from plates that contain a visible colony amount ranging from ∼20 to 200. However, in several of the environmental samples in our study, this criterion was not fulfilled, resulting in the low bacterial abundances, and the exclusion of the environmental samples from statistical tests. Nevertheless, despite their unquantifiable nature, they still provide valuable information by indicating the presence of bacteria and have therefore been included in Supplementary Table S3, as well as in Figure 2D. Similar tendencies were generally seen in the concentrations of blaOXA-like genes, tetA and sul2 in the marine water samples as in the cultivated samples. ddPCR allows for absolute quantification of samples containing low concentrations of ARGs (Taylor et al., 2017), adding to the reliability of the concentrations in the cultivated samples. To increase the reliability of the results from statistical tests, biological triplicates of all samples and more frequent sampling could further strengthen the tendencies observed in our study. However, the background sampling from Station G was made into triplicates and indirectly served as a control for contamination, as the background samples did not have any CFUs (apart from 1 single CFU on one plate out of several, Supplementary Table S3), we still deem our data reliable. The low concentration in the background sample from Station G gives an indication of the bacteria found at Stations S, A and H being a result of wastewater contamination. If this were not the case, the background samples would likely have had a higher bacterial concentration or a concentration in the same range as those seen at Station S, A and H. This gives further confidence to the CFUs from environmental samples not being due to contamination or due to ubiquitous bacteria.
Wastewater contamination in Tórshavn has been investigated previously, but little is known about how winds and currents affect the advection, diffusion, and dispersion of effluents in the ocean (The Municipality of Tórshavn, 2021). Attempts have been made abroad to predict and simulate effluent release and spread and could determine, that wastewater released many meters under the ocean surface tends to spread upwards in all directions, without contaminating the deeper layers due to the lower density of the treated sewage water compared to the surrounding ocean (Thupaki et al., 2010; Dumasdelage and Delestre, 2020). So even though the wastewater outlet at Station A is situated around 5 m below the surface, the wastewater is not necessarily dispersed at that depth predominately.
Seabirds were observed to cluster around the plume where wastewater is released in the ocean, presumably feeding off the effluents. Small birds were also seen in the cave, where effluents from Station H are led into the ocean. Birds that have been in contact with sewage or sewage contaminated waters have been shown to contain a higher amount of resistance genes than other birds, serving as vectors and reservoirs of ARGs (Marcelino et al., 2019). Northern fulmars (Fulmaris glacialis) are frequently hunted for food by Faroese people and these birds were in particular seen gathering around the wastewater. The close contact during hunting could possibly cause pathogenic and multi-drug resistant bacteria to be transmitted to humans. The birds can travel long distances, possibly spreading the contaminants even further. Our study gives an indication of the bacterial and ARG concentrations that humans and wildlife can be exposed to at the foreshore near wastewater outlets in Tórshavn.
Out of all the multi-drug resistant (MDR) bacteria tested using API 20 E, ∼62% belonged to the Enterobacteriaceae family, which are common inhabitants of the human GI tract. In fact, E. coli, which belongs to this family, is an indicator of faecal contamination (Holcomb and Stewart, 2020).
Escherichia coli and Klebsiella, which belong to Enterobacteriaceae, are opportunistic pathogens known to cause disease in humans (Brinas et al., 2002; Paterson, 2006). These were found in several sample locations, also far away from the wastewater outlets such as S1 and A9, meaning, that they can survive 90 m and 120 m from the wastewater outlets. P. aeruginosa, and A. hydrophila were also detected in oceanic samples and are known to cause disease in humans, such as necrotizing fasciitis (Tsai et al., 2012). It is not clear, how the bacteria natural to the GI tract, can survive the extreme changes from gut to wastewater to oceanic waters (Cohen et al., 2020). However, it has been speculated that the spatial and temporal heterogeneity in osmolarity within the human gut has led to an adaption, enabling the gut bacteria to cope with varying salinity levels, which might improve their chances of survival in the marine environment (Cohen et al., 2020).
Several of the cultivated environmental samples from our study contained few CFUs, it is however noteworthy, that from these few, MDR bacteria were found that are associated with disease in humans (Brinas et al., 2002; Paterson, 2006; Aiello et al., 2008; Kim et al., 2009; Rizzo et al., 2013; Akbari et al., 2016; Prokesch et al., 2016; DavinRegli et al., 2019; CDC, 2023).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
AM: Writing–original draft, Investigation, Conceptualization, Data curation, Formal Analysis, Methodology, Visualization. SP: Writing–original draft, Investigation, Conceptualization, Data curation, Formal Analysis, Methodology, Visualization. MB: Writing–review and editing, Data curation, Methodology, Resources, Validation. AD: Supervision, Writing–review and editing, Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The municipality of Tórshavn.
Special thanks to the Municipality of Tórshavn for financially supporting this project and in particular the Technical Department, for general interest and assistance. Thanks also to The University of Faroe Islands for use of facilities as well as financial support. We are also grateful to Marjun í Túni Mortensen for great assistance, advice in the laboratory and moral support. Lastly, we would like to thank Jóhanna Jørgensen for good advice.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2024.1336318/full#supplementary-material
Aiello, A. E., Larson, E., and Sedlak, R. (2008). Hidden heroes of the health revolution Sanitation and personal hygiene. Am. J. Infect. Control 36 (10), S128–S151. doi:10.1016/j.ajic.2008.09.008
Akbari, M., Bakhshi, B., and Peerayeh, S. N. (2016). Particular distribution of Enterobacter cloacae strains isolated from urinary tract infection within clonal complexes. PubMed 20 (1), 49–55. doi:10.7508/ibj.2016.01.007
Akiyama, T., and Savin, M. C. (2010). Populations of antibiotic-resistant coliform bacteria change rapidly in a wastewater effluent dominated stream. Sci. Total Environ. 408 (24), 6192–6201. doi:10.1016/j.scitotenv.2010.08.055
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399 (10325), 629–655. doi:10.1016/S0140-6736(21)02724-0
Auerbach, E., Seyfried, E. E., and McMahon, K. D. (2007). Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 41 (5), 1143–1151. doi:10.1016/j.watres.2006.11.045
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13 (5), 310–317. doi:10.1038/nrmicro3439
Briñas, L., Zarazaga, M., Saenz, Y., Ruiz-Larrea, F., and Torres, C. (2002). β-Lactamases in ampicillin-resistant Escherichia coliisolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 46 (10), 3156–3163. doi:10.1128/aac.46.10.31563163.2002
Byers, K. A., Booker, T. R., Combs, M., Himsworth, C. G., Munshi-South, J., Patrick, D. M., et al. (2020). Using genetic relatedness to understand heterogeneous distributions of urban rat-associated pathogens. Evol. Appl. 14 (1), 198–209. doi:10.1111/eva.13049
CDC (2023). CDC (no date) Antibiotics Tested by NARMS. Available at: https://www.cdc.gov/narms/antibioticstested.html (Accessed May 20, 2023).
Cohen, R., Paikin, S., Rokney, A., Rubin-Blum, M., and Astrahan, P. (2020). Multidrug-resistant enterobacteriaceae in coastal water: an emerging threat. Antimicrob. Resist. Infect. Control 9 (1), 169. doi:10.1186/s13756-020-00826-2
Dahal, P. (2023). Spread Plate method- definition, principle, procedure, uses. Available at: https://microbenotes.com/spread-plate-technique/.
Da Silva, M. F., Tiago, I., Veríssimo, A., Boaventura, R. A. R., Nunes, O. C., and Manaia, C. M. (2006). Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 55 (2), 322–329. doi:10.1111/j.1574-6941.2005.00032.x
Davin-Régli, A., Lavigne, J. P., and Pagès, J. (2019). Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 32 (4), e00002-19. doi:10.1128/cmr.00002-19
Di Cesare, A., Eckert, E. M., D'Urso, S., Bertoni, R., Gillan, D. C., Wattiez, R., et al. (2016). Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 94, 208–214. doi:10.1016/j.watres.2016.02.049
Du, M., Chen, J., Zhang, X., Li, Y., and Wang, Y. (2007). Retention of virulence in a viable but nonculturable Edwardsiella tarda isolate. Appl. Environ. Microbiol. 73 (4), 1349–1354. doi:10.1128/aem.02243-06
Dumasdelage, R., and Delestre, O. (2020). Simulating coliform transport and decay from 3D hydrodynamics model and in situ observation in Nice area. SN Appl. Sci. 2 (8), 1348. doi:10.1007/s42452-020-3122-4
Dwight, R. H., Baker, D. B., Semenza, J. C., and Olson, B. H. (2004). Health effects associated with recreational coastal water use: urban versus Rural California. Am. J. Public Health 94 (4), 565–567. doi:10.2105/ajph.94.4.565
Fouz, N., Pangesti, K. N. A., Yasir, M., Al-Malki, A. L., Azhar, E. I., Hill-Cawthorne, G. A., et al. (2020). The contribution of wastewater to the transmission of antimicrobial resistance in the environment: implications of mass gathering settings. Trop. Med. Infect. Dis. 5 (1), 33. doi:10.3390/tropicalmed5010033
Goh, E.-B., Yim, G., Tsui, W., McClure, J., Surette, M. G., and Davies, J. (2002). Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. U. S. A. 99 (26), 17025–17030. doi:10.1073/pnas.252607699
Gutiérrez-Cacciabue, D., Cid, A. G., and Rajal, V. B. (2016). How long can culturable bacteria and total DNA persist in environmental waters? The role of sunlight and solid particles. Sci. Total Environ. 539, 494–502. doi:10.1016/j.scitotenv.2015.07.138
Holcomb, D., and Stewart, J. R. (2020). Microbial Indicators of fecal pollution: recent progress and challenges in assessing water quality. Curr. Environ. Health Rep. 7 (3), 311–324. doi:10.1007/s40572-020-00278-1
Huddleston, J. R. (2014). Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect. Drug Resist. 167, 167–176. doi:10.2147/idr.s48820
Islam, P. (2023). Creating bacterial glycerol stocks for long-term storage. Available at: https://www.protocols.io/view/creating-bacterial-glycerol-stocks-for-long-terms-bf5qjq5w.pdf (Accessed May 21, 2023).
Kim, J., Jeon, S., Rhie, H., Lee, B., Park, M., Lee, H., et al. (2009). Rapid detection of extended spectrum β-lactamase (ESBL) for enterobacteriaceae by use of a multiplex PCR-based method. Infect. Chemother. 41 (3), 181. doi:10.3947/ic.2009.41.3.181
Kümmerer, K. (2009). Antibiotics in the aquatic environment – a review – Part I. Chemosphere 75 (4), 417–434. doi:10.1016/j.chemosphere.2008.11.086
Kunngerð um spillivatn (2009). Kunngerð nr. 111 frá 7. september 2009 um spillivatn (no date). Available at: https://logir.fo/Kunngerd/111-fra-07-09-2009-um-spillivatn.
Larsson, D. G. J., and Flach, C.-F. (2021). Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20 (5), 257–269. doi:10.1038/s41579-021-00649-x
Leung, H. W., Minh, T., Murphy, M., Lam, J. C., So, M., Martin, M., et al. (2012). Distribution, fate and risk assessment of antibiotics in sewage treatment plants in Hong Kong, South China. South China Environ. Int. 42, 1–9. doi:10.1016/j.envint.2011.03.004
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10 (S12), S122–S129. doi:10.1038/nm1145
Li, J., and Zhang, X. (2019). Beach pollution effects on health and productivity in California. Int. J. Environ. Res. Public Health 16 (11), 1987. doi:10.3390/ijerph16111987
Li, L., Mendis, N., Trigui, H., Oliver, J. D., and Faucher, S. P. (2014). The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 5, 258. doi:10.3389/fmicb.2014.00258
Lira, F., Vaz-Moreira, I., Tamames, J., Manaia, C. M., and Martínez, J. L. (2020). Metagenomic analysis of an urban resistome before and after wastewater treatment. Sci. Rep. 10 (1), 8174. doi:10.1038/s41598020-65031-y
Marcelino, V. R., Wille, M., Hurt, A. C., González-Acuña, D., Klaassen, M., Schlub, T. E., et al. (2019). Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. 17 (1), 31. doi:10.1186/s12915-019-0649-1
Munk, P., Brinch, C., Møller, F. D., Petersen, T. N., Hendriksen, R. S., Seyfarth, A. M., et al. (2022). Genomic analysis of sewage from 101 countries reveals global landscape of antimicrobial resistance. Nat. Commun. 13 (1), 7251. doi:10.1038/s41467-022-34312-7
Nawaz, M. S., Sung, K., Khan, S. A., Khan, A. A., and Steele, R. (2006). Biochemical and molecular characterization of tetracycline-resistant Aeromonas veronii isolates from catfish. Appl. Environ. Microbiol. 72 (10), 6461–6466. doi:10.1128/aem.00271-06
Norman, A., Hansen, L. H., and Sørensen, S. J. (2009). Conjugative plasmids: vessels of the communal gene pool. Philosophical Trans. R. Soc. B 364 (1527), 2275–2289. doi:10.1098/rstb.2009.0037
Ochman, H., Quandt, E. M., Gottell, N., and Gilbert, J. A. (2024). Examining the taxonomic distribution of tetracycline resistance in a wastewater plant. Sustain. Microbiol. 1, qvad003. doi:10.1093/sumbio/qvad003
Ohlsen, K., Ziebuhr, W., Koller, K. P., Hell, W., Wichelhaus, T. A., and Hacker, J. (1998). Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 42 (11), 2817–2823. doi:10.1128/aac.42.11.2817
Paterson, D. L. (2006). Resistance in gram-negative bacteria: enterobacteriaceae. Am. J. Infect. Control 34 (5), S20–S28. doi:10.1016/j.ajic.2006.05.238
Pei, R., Kim, S. C., Carlson, K. H., and Pruden, A. (2006). Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 40 (12), 2427–2435. doi:10.1016/j.watres.2006.04.017
Pendergraft, M. A., Belda-Ferre, P., Petras, D., Morris, C. K., Mitts, B. A., Aron, A. T., et al. (2023). Bacterial and chemical evidence of coastal water pollution from the tijuana river in sea spray aerosol. Environ. Sci. Technol. 57 (10), 4071–4081. doi:10.1021/acs.est.2c02312
Prokesch, B. C., TeKippe, M., Kim, J., Raj, P., TeKippe, E. M., and Greenberg, D. E. (2016). Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 16 (9), e190–e195. doi:10.1016/s1473-3099(16)30021-4
Puckett, E. E., Magnussen, E., Khlyap, L. A., Strand, T. M., Lundkvist, Å., and Munshi-South, J. (2019). Genomic analyses reveal three independent introductions of the invasive brown rat (Rattus norvegicus) to the Faroe Islands. Heredity 124 (1), 15–27. doi:10.1038/s41437-019-0255-6
Rizzo, L., Manaia, C., Merlin, C., Schwartz, T., Dagot, C., Ploy, M., et al. (2013). 'Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447, 345–360. doi:10.1016/j.scitotenv.2013.01.032
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73 (5), 1121–1137. doi:10.1093/jac/dkx488
Scornec, H., Bellanger, X., Guilloteau, H., Groshenry, G., and Merlin, C. (2017). Inducibility of Tn916 conjugative transfer in Enterococcus faecalis by subinhibitory concentrations of ribosome-targeting antibiotics. J. Antimicrob. Chemother. 72 (10), 2722–2728. doi:10.1093/jac/dkx202
Shi, B., Zhao, R., Su, G., Liu, B., Liu, W., Xu, J., et al. (2023). Metagenomic surveillance of antibiotic resistome in influent and effluent of wastewater treatment plants located on the Qinghai-Tibetan Plateau. Sci. Total Environ. 870, 162031. doi:10.1016/j.scitotenv.2023.162031
Taylor, S. C., Laperriere, G., and Germain, H. (2017). Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 7 (1), 2409. doi:10.1038/s41598-017-02217-x
The municipality of Tórshavn (2023). Soleiðis er vatngóðskan til útisvimjing í løtuni. Available at: https://www.torshavn.fo/nattura-fritid-og-mentan/upplivingar-inatturuni/soleidis-er-vatngodskan-til-utisvimjing-i-loetuni (Accessed September 15, 2023).
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3 (9), 711–721. doi:10.1038/nrmicro1234
Thupaki, P., Phanikumar, M. S., Beletsky, D., Schwab, D. J., Nevers, M. B., and Whitman, R. L. (2009). Budget analysis of Escherichia coli at a southern lake Michigan beach. Environ. Sci. Technol. 44 (3), 1010–1016. doi:10.1021/es902232a
The Municipality of Tórshavn (2021). Umhvørviskanning av Havnarvág. Available at: https://www.torshavn.fo/media/hmekw0qm/vatng%C3%B3%C3%B0skukanning-havnarv%C3%A1g-2021.pdf (Accessed March 4, 2023).
Tsai, Y.-H., Huang, K. C., Shen, S. H., Hsu, W. H., Peng, K. T., and Huang, T. J. (2012). Microbiology and surgical indicators of necrotizing fasciitis in a tertiary hospital of southwest Taiwan. Int. J. Infect. Dis. 16 (3), e159–e165. doi:10.1016/j.ijid.2011.11.001
UK Standards (2009). UK Standards for microbiology investigations. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/771781/TP_26i4.pdf (Accessed February 5, 2023).
Umhvørvisstovan (2023). Rottangar og reinsiverk. Available at: https://www.us.fo/Umhvørvisstovan/Umhvørvisstovan%20og%20dalking/A%20sjonum/Spillivatn/Rottangar%20og%20reinsiverk (Accessed May 28, 2023).
Von Wintersdorff, C. J. H., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 173. doi:10.3389/fmicb.2016.00173
Wade, T. J., Sams, E., Brenner, K. P., Haugland, R., Chern, E., Beach, M., et al. (2010). Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ. Health 9 (1), 66. doi:10.1186/1476-069x-9-66
World Health Organization (2020). Antibiotic resistance. Available at: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance.
Yang, J., Wang, H., Roberts, D. J., Du, H. N., Yu, X. F., Zhu, N. Z., et al. (2020). Persistence of antibiotic resistance genes from river water to tap water in the Yangtze River Delta. Sci. Total Environ. 742, 140592. doi:10.1016/j.scitotenv.2020.140592
Zhang, Y., Li, A., Dai, T., Li, F., Xie, H., Chen, L., et al. (2017). Cell-free DNA: a neglected source for antibiotic resistance genes spreading from WWTPs. Environ. Sci. Technol. 52 (1), 248–257. doi:10.1021/acs.est.7b04283
Zieliński, W., Buta, M., Hubeny, J., Korzeniewska, E., Harnisz, M., Nowrotek, M., et al. (2019). Prevalence of beta lactamases genes in sewage and sludge treated in Mechanical-Biological Wastewater treatment plants. J. Ecol. Eng. 20 (9), 80–86. doi:10.12911/22998993/112506
Keywords: sewage, gram-negative bacteria, multi-drug resistance, enterobacteriaceae, antibiotic resistance genes, wastewater
Citation: Mortensen AMS, Poulsen SJ, Berbisá MáF and Djurhuus A (2024) Distribution of antibiotic resistant bacteria and genes in sewage and surrounding environment of Tórshavn, Faroe Islands. Front. Environ. Sci. 12:1336318. doi: 10.3389/fenvs.2024.1336318
Received: 10 November 2023; Accepted: 15 February 2024;
Published: 01 March 2024.
Edited by:
Maria Elisa Magri, Federal University of Santa Catarina, BrazilReviewed by:
Niti B Jadeja, University of Virginia, United StatesCopyright © 2024 Mortensen, Poulsen, Berbisá and Djurhuus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anni Djurhuus, YW5uaS5kanVyaHV1c0BnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.