
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci., 30 June 2023
Sec. Toxicology, Pollution and the Environment
Volume 11 - 2023 | https://doi.org/10.3389/fenvs.2023.1195643
Extensive arsenic contamination of groundwater, rivers, and agricultural farms has led to widespread arsenic poisoning and consequent problems associated with health and economy. In recent years, many studies have been performed to understand bacterial arsenic detoxification and metabolism which has paved the way for bioremediation applications. This study attempted to isolate and characterize arsenic-resistant bacteria from the drying Urmia lake. As a result of the 16S rDNA analysis, six arsenic-resistant strains were identified as the members of Shouchella, Salipaludibacillus, and Evansella genera. For some of the strains, the maximum tolerance concentration for either arsenate or arsenite was considerably high, 320 and 16 mM, respectively. All the strains harbored the arsenate reductase gene (arsC). The arsenate permease (arsB) gene was identified in all strains except in strains S1, S12, and E15. The metabolic genes of respiratory arsenate reductase (arrB) and arsenite oxidase (arxA) were identified in none of these strains. This feature of the strains with the arsC gene region can be exploited to bioremediate arsenic from contaminated areas by using a two-step process. The second step can be carried out by the adsorption of arsenite to iron oxide or by precipitation with sulfide. Today, more and more ecosystems are being destroyed due to anthropogenic pollution, and it is important to reveal the negative effects and solutions of this situation on the ecosystems. So, the current study provides a potential source of bacteria for such studies that implement bioremediation practices to prevent arsenic catastrophe in vicinal territories, and reveals arsenic pollution in Urmia lake.
Arsenic is known to be a toxic metalloid and is naturally present in the Earth’s crust, water, soil, and air. Natural geological activities and some human-based activities such as the use of pesticides and wood preservatives, mining activities, and coal burning can lead to arsenic contamination (Wang and Mulligan, 2006). There are two forms of inorganic arsenic available in the environment. More toxic forms are arsenite [As(III)] and arsenate [As(V)], a phosphate analog (Firrincieli et al., 2019). Especially due to the contamination of these two forms of arsenic, several environmental pollution problems may arise. Arsenic contamination of water and agricultural areas from different sources has led to serious problems in many countries worldwide. Bangladesh, India, China, and the United States are experiencing very serious arsenic problems (Kao et al., 2013). This situation is becoming a disaster especially in West Bengal and Bangladesh, where underground waters are the most important drinking water resources and consequently millions of people are being exposed to arsenic (Smedley and Kinniburgh, 2002). In addition to water sources used as drinking and utility water, food and various aquatic sources also cause arsenic exposure. One of the factors is the salty lakes, which are about to dry up with the effect of global warming. Urmia Salt Lake is one of these lakes, which is one of the 59 biospheres under the protection by UNESCO. Lake Urmia, the second largest salt lake in the world, is located between the East and West Azerbaijan provinces of Iran and the northwest of the country. The drying Urmia lake was declared as a wetland of international importance with the Ramsar Convention in 1971 and a UNESCO biosphere reserve as designated in 1976 (Soudi et al., 2017). In recent years, Lake Urmia has experienced severe droughts due to climate change and the construction of dams around the lake (77 dams). On the other hand, it has been massively polluted as a consequence of the industrial sites and factories around the lake (Taghipour et al., 2013; Ouria and Sevinc, 2016). The drying up of the lake and increase in pollution rates due to anthropogenic effects negatively affect not only the lake ecosystem but also the entire environment, especially through aerosols. It has been reported that fine aerosol particles accumulate many elements, especially arsenic (As), cadmium (Cd), copper (Cu), iron (Fe), sodium (Na), nickel (Ni), and lead (Pb), on the northern and southern shores of Lake Urmia, and it has been emphasized that this situation endangers public health (Gholampour et al., 2017). So, unfortunately, high levels of salt and heavy metals are spreading in the region due to wind and consequently cause soil and water pollution. In addition, this ecological catastrophe in the region also threatens neighboring countries like Turkey, and the situation is becoming a major issue (Mohammadi et al., 2018). A large number of organisms living in areas that are likely to be affected by this require very long evolutionary selections in order to adapt to these conditions and not be harmed. Microorganisms, on the other hand, represent an advantageous group in the adaptation process due to their small surface/volume ratio. So, during the evolutionary process, arsenic defense mechanisms have been developed in a wide variety of microorganisms, which are exposed to high levels of the arsenic derivatives (Ben Fekih et al., 2018). As in every biochemical reaction, some genetic arrangements occur for the necessary reactions to take place, and there are genes responsible for the production of specific proteins. These genes mediate the exclusion of arsenic from the cell in order to neutralize its toxic effects in bacteria (Sá-Pereira et al., 2007). Some genes may exist in gene clusters to enable organisms to more rapidly regulate the expression of various genes depending on the environmental conditions. The genes required for arsenic metabolism are also arranged in this framework. So, four types of arsenic-related operons have been discovered. One is the ars operon that is involved in detoxification mechanism, dissimilatory arsenate reduction by arr operon, and aio/arx for respiratory As(III) oxidation (Andres and Bertin, 2016). The ars operon, which confers arsenic resistance to microorganisms, is widespread among microorganisms. Although ars operons vary between strains, three central genes are always present. As in Escherichia coli, the three-gene operon (arsRBC) provides the basic arsenic resistance (Carlin et al., 1995). The five-gene operon (arsRDABC) found in E. coli plasmid R773 ensures elevated arsenic resistance (Chen et al., 1986).
As(V) reductase (arsC), As(III) permease (arsB), and regulatory protein (arsR) are clustered in the ars operon. As(V) enters the cell through the phosphate transport PiT system and is reduced to As(III) by As(V) reductase and effluxed out of the cell via the transmembrane protein arsB (Rosen, 2002). Detecting these gene regions in arsenic resistance and the bacteria that have them is necessary for the protection of ecosystems that are about to disappear. Urmia Salt Lake, which is about to dry up, is one of these ecosystems. Although Urmia Salt Lake is located in the north-west of Iran and is the largest lake in the Middle East, it is, however, one of the largest permanent hypersaline lakes on Earth. When evaluated in terms of the area it covers in the world, it is seen that the lake, which initially covered an area of 5,000–6,000 km2, now covers an area of 1,000 km2 (Mardi et al., 2018). As the most important factors in the 90% reduction of the lake area, global warming and climate change, spring water movements, increased agricultural activities, and many anthropogenic activities are considered (Tourian et al., 2015; Ghale et al., 2017). As a result of the decrease in the amount of lake water, the surface of the resulting lake bed becomes active salt and dust sources that are sensitive to wind erosion. The Aral Sea is one of the most obvious examples of this. Studies carried out on people in residential areas around such dried-up lakes show that there is an increase in cases of risk of thyroid disease and nodules (Mazhitova et al., 1998; Houshyar et al., 2022). Apart from this, many problems that might appear in the long term are already awaited, and when we evaluate this within the scope of a single health approach, it can be seen that many components are in danger. Considering that every lake will dry up one day, it is necessary to determine the factors that endanger all living forms in such habitats and to identify potential living groups for further studies.
In this context, within the scope of this study, it was aimed to determine the arsenic content of Urmia Salt Lake, identify bacteria that have adapted to these conditions in the evolutionary process, and perform arsenic resistance profiling. In this way, new data would have been added to the studies reporting arsenic contents of different parts of lakes in the last few years (Kazemi et al., 2019; Alvyar et al., 2022) and at the same time, data on the isolation and identification of arsenic-resistant bacteria from Lake Urmia will be presented for the first time. Thereby, potential bacterial source candidates will be presented for other studies to determine their potential for pollution removal.
Water and sediment samples were obtained from a region close to the Tabriz–Urmia causeway bridge in East Azerbaijan province (7.76° 45' 41.3' N–45.3° 18' 28.6'' E) during the summer of 2018 (Figure 1). The summer season was chosen as the sampling time because the lake water level decreases more in summer and the anthropogenic pollution effects are more evident in this season. The region of the lake close to agricultural areas and industrial sites was chosen as the sampling point. The samples were taken to the laboratory in sampling bottles embedded in ice packs for bacterial isolation and heavy metal analysis using the inductively coupled plasma mass spectrometer (ICP-MS). The water and sediment samples were examined in terms of 24 different heavy metals. All heavy metal analyses were performed using the EPA method 3051 analysis method. The amount of heavy metals was determined by microwave-assisted nitric acid digestion using 10 mL HNO3 (Yang et al., 2007). In situ water pH and temperature measurements (HANNA HI991001) were done.
Isolation of arsenic-resistant bacteria was carried out by using the spread plate method by diluting the samples of water and sediment. The isolation medium used was Luria–Bertani (LB) medium modified with 10 mM sodium arsenate, 7.5% sodium chloride, and pH adjusted at 9.5 using carbonate buffer. The samples were incubated at 30°C for 3–7 days (Sunitha et al., 2015). At the end of the incubation period, colonies with different morphologies and different colony colors were taken for the purification processes. After purification, glycerol stocks were prepared with a 25% final glycerol concentration and stored at −80°C for all arsenic-resistant strains.
For phenotypic characterization, briefly, Gram reaction, the KOH test, endospore and capsule staining, the catalase test, starch hydrolysis (1% soluble starch), caseinase, gelatinase (12% gelatine), esterase (1% tributyrin), the motility test, and nitrate reduction were carried out. For all biochemical assays, the bacterial density was adjusted to correspond to 1 McFarland. To perform the carbohydrate fermentation test, 1% of glucose, fructose, mannose, sorbitol, arabinose, and maltose carbohydrates was prepared. A solution of 0.008% (w/v) thymol blue was added as the indicator (Yumoto et al., 2008; Alfred and Heidi, 2016). The incubation conditions were determined to be 30°C and 48 h. At the same time, in all media, the pH and NaCl concentration of the medium were adjusted to 9.5 and 7.5%, respectively.
For genomic DNA extraction, bacterial pellets were collected in the exponential phase and the PureLink™ Genomic DNA extraction kit (ThermoFisher Scientific, K182001) was used for DNA extraction. For the 16S rDNA amplification of isolates, fD1 5′-AGAGTTTGATCCTGGCTCAG-3′ and rD1 5′-AAGGAGGT GATCCAGCC-3′ primers were used (Weisburg et al., 1991) and the PCR conditions were first denaturation at 94°C for 15 min followed by 35 cycles at 94°C for 30 s, 55°C for 1 min, 2 min at 72°C, and final elongation at 72°C for 10 min (Weisburg et al., 1991). The bidirectional sequence analysis of purified amplicons (with the Sephadex® G-50 kit, Sigma-Aldrich G5080, Merck KGaA, Darmstadt, Germany) was conducted with the ABI 3130xl Genetic Analyzer (Applied Biosystems, Waltham, MA, United States) using the same primers. Chromatograms were analyzed with the FinchTV, ApE, and BioEdit programs. The phylogenetic relationship of organisms was then determined by the comparison of the individual 16S rDNA sequence with other existing sequences in the public database (GenBank http:www.ncbi.nlm.nih.gov/). The sequences of strains were deposited in the NCBI GenBank database to acquire the accession numbers (Ersoy Omeroglu et al., 2022). The phylogenetic tree was constructed by the neighbor-joining method using the Geneious 6.0 software.
In order to determine the cytoplasmic arsenate reductase gene arsC (370 bp), the primers of 5′-GTA ATA CGC TGG AGA TG ATC CG-3′ and reverse 5′-TTT TCC TGC TTC ATC AAC GAC-3′ were used (Saltikov and Olson, 2002). The reaction was carried out in a total volume of 25 µL using the Geneaid PCR kit, and the PCR conditions were first denaturation at 94°C for 3 min followed by 30 cycles of denaturation at 94°C for 30 s, elongation at 56°C for 30 s, annealing at 72°C for 30 s, and final extension at 72°C for 7 min (Saltikov and Olson, 2002). For arsenate permease gene (arsB), the forward 5′-CGGTGGTGTGGAATATTGTC-3′ and 5′- GTCAGAATAA-GAGCCGCACC-3′ reverse primers were used (Mazhitova et al., 1998). The PCR reaction was done in a total volume of 25 µL. The PCR conditions were first denaturation at 94°C for 3 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, elongation at 72°C for 30 s, and the final extension at 72°C for 7 min (Saltikov and Olson, 2002).
The respiratory arsenate reductase (arrB) gene was amplified using the forward primer 5′-AAC ACG AAC GAC GGT ATT CAC TGG-3′ and reverse primer 5′-ATA CCT TGC TCT GTG GAT CAT CTA-3′ (Chang et al., 2007). The PCR conditions were first denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 40 s, annealing at 62°C, elongatıon at 72°C for 1 min, and final extension at 72°C for 7 min (Chang et al., 2007).
The arsenite oxidase (arxA) gene was amplified using the forward 5′-GGACTAGTCCAACTCGCG GTAGTAGTCC-3′ and reverse 5′-GGACTAGTGATCGGACCCACCGTATAGA-3′ primers (Zargar et al., 2012). The PCR reaction was carried out using the PfuTurbo Cx Hotstart Polymerase (Agilent, #600410) for a total volume of 50 µL. The PCR conditions were denaturation at 95°C for 1 min, 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, elongation at 72°C for 1 min, and final extension at 72°C for 7 min (Zargar et al., 2012). For all PCR assays, Shewanella ANA-3 (Saltikov et al., 2003), Halomonas sp. BSL (Conrad, 2014), Staphylococcus aureus ATCC 25923 (Rosdahl and Rosendal, 1980), and Enterobacter sp. strain L MF000792 (strain from our other studies) were used as the positive controls.
Since the preferred temperature for bacterial isolations from habitats with similar characteristics is 30°C, it was selected as the incubation temperature in order to determine the maximum tolerable concentration (MTC) values and physicochemical properties of the strains. To find out As(V) and As(III) MTCs, 0–320 mM sodium arsenate and 0–32 mM sodium arsenite were used. The LB medium (7.5% NaCl and pH 9.5) containing different concentrations of A(V) and As(III) was prepared and 250 µL was added in each well of flat bottom 96-well plates (Bruvoll, 2017) (see Supplementary Material, Supplementary Figure S1).
For physicochemical parameters, in order to determine the salt concentrations at which the isolates are metabolically active, LB media (pH 9.5) containing 0–25% NaCl were prepared. Then, for each of the different media, 250 µL of the medium was added to each well of the flat bottom 96-well plates. To determine the reproductive abilities at different pH levels, LB media (7.5% NaCl) were prepared at pH values varying between 4.5 and 10.5. Different buffer solutions (acetate buffer, phosphate buffer, and carbonate buffer for pH 4.5–5.5, 6.5–7.5, and 8.5–10.5, respectively) were used to adjust for different pH levels. For all different conditions, 250 µL of media was added to each well of the 96-well microplates to determine the pH growth intervals. For the growth temperature, the LB liquid medium (7.5% NaCl and pH 9.5) was dispersed into the wells (250 µL) and the temperature values of 25°C, 30°C, 35°C, 45°C, and 55°C were selected to examine the growth. To equalize the amount of bacteria in all wells, 1 McFarland density (3.0 × 108 cfu/mL) of bacterial solutions was prepared, and for inoculation to wells, 10 µL of bacterial suspension was added to each wells. All microplates were incubated at 30°C for 24 h. Then, 0.1% triphenyltetrazolium chloride (TTC) was prepared as a vitality indicator and 20 µL was added to each well. After that, bacterial incubation was continued during the extra 24 h. The formation of red-pink triphenylformazan (TPF) was evaluated as positive for bacterial growth. While the value of the well in which TPF did not occur was considered as the minimum inhibitory concentration, the highest value at which TPF occurred was determined as the MTC (Bruvoll, 2017). All bacterial growths were determined via optical density measurement at OD620nm and bacterial vitality was corrected by using TTC.
An arsenic transformation assay was performed using a silver nitrate test (Sarkar et al., 2013). For optimization, three different agar media [chemically defined medium (CDM), Basal Mineral Salt Medium (BSM), and LB] were used. For arsenic-resistant bacteria that live optimally under aerobic and neutral conditions, the arsenic transformation test is usually performed using the LB medium. However, since the experiment was conducted with halophilic and alkaliphilic bacteria in this study, the experiment was carried out in three media with different contents in order to obtain the real result. All media were supplemented with sodium arsenate (Na3AsO4, 10 mM) and sodium arsenite (NaAsO₂, 1 mM). After streaking onto agar plates, all cultures were incubated at 30°C for 5 days. As soon as the incubation period had elapsed, all plates were flooded with a 0.1 M silver nitrate (AgNO3) solution and conserved in dark conditions. Color precipitations were examined to evaluate the results. A brownish bright yellow precipitate indicated the presence of silver arsenate (Ag3AsO4) (arsenite-oxidizing bacteria) and silver arsenite (Ag3AsO3) (arsenate-reducing bacteria), respectively. As positive controls, Shewanella ANA-3 (Saltikov et al., 2003) and Halomonas sp. BSL-1 (Conrad, 2014) were used.
In order to determine the growth potential of the isolates under anaerobic conditions, 18–24 h fresh microorganisms were adjusted to 0.5 McFarland concentration and 10 µL of each was placed on LB and Brewer Anaerobic Agar media (HiMedia) inside the anaerobic jar and incubated for 48 h at 30°C (Alfred and Heidi, 2016).
Under aerobic conditions, the BSM [NaCl (20.0 g/L), KH2PO4 (0.48 g/L), K2HPO4 (0.6 g/L), (NH4)2SO4 (0.45 g/L), MgSO4 (0.20 g/L), 2 mg/mL vitamin B12 (200 μL/L), SL-10 trace element solution (2 mL/L), and 1,000× vitamins solution (20 mL/L)] was prepared containing an electron donor to determine the growth of isolates. BSM was supplemented with 10 mM As(V) and sodium lactate (10 mM), sodium acetate (10 mM), and sodium citrate (10 mM). Then, the isolates were prepared at 0.5 McFarland concentration (1.5 × 108 cfu/mL) and spotted on petri plates. After this, they were incubated at 30°C for 48 h (Hoeft et al., 2004).
The pH and temperature of the lake water was measured to be 8.6 and 33°C, respectively. The concentrations of arsenic were found to be 8.739 and 0.230 mg/kg for sediment and water samples, respectively. The results of 24 different heavy metal analyses of water and sediment samples are given in Table 1. As expected, it was determined that there was more arsenic in the sediment samples due to excessive decrease in lake water.
In total, six isolates designated as S1, S2, S4, S12, and E15 for the sediment sample and UW1 for the water sample were selected due to the differences in colony morphology. The phenotypic characteristics of the isolates are given in Table 3. It was determined that all strains were gram-positive and rod shaped, had endospores and capsules, showed a positive reaction in the catalase enzyme test, assimilated fructose, mannose, maltose, arabinose, and sorbitol, and were not metabolically active under anaerobic conditions. It is seen that there are different characteristics between species in terms of mobility, and amylase and esterase activities (Supplementary Material; Supplementary Figure S2).
The phylogenetic analysis based on the 16S rDNA gene sequence indicated that the isolates S1 and S2 were closely related to Alkalihalobacillus sp. and Alkalihalobacillus patagoniensis, S4 and S12 isolates were related to Salipaludibacillus sp. and Salipaludibacillus neizhouensis, UW1 was related to Evansella sp. and Evansella cellulosilytica, and finally E15 was related to Alkalibacillus filiformis and Alkalibacillus haloalkaliphilus. Based on the partial 16S rDNA sequences, the Geneious 6.0 program was used and a phylogenetic tree was created using the neighbor-joining method (Figure 2; Table 2).
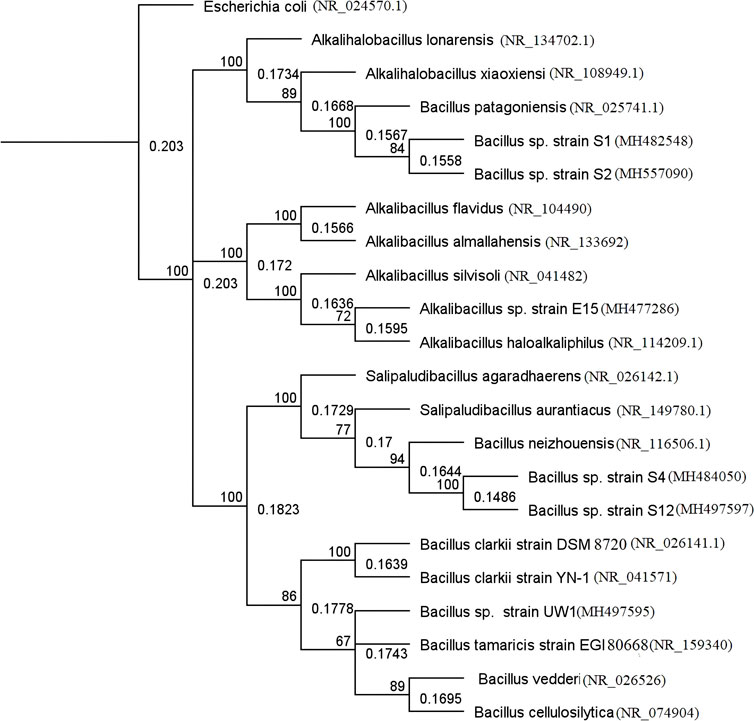
FIGURE 2. Phylogenetic tree was constructed based on the neighbor-joining method using Geneious 6.0 software. 16 s rDNA sequences were deposited in NCBI GenBank by the following accession numbers: S1 (MH482548.1), S2 (MH557090.1), S4 (MH484050.1), S12 (MH497597.1), UW1 (MH497595.1), and E15 (MH477286.1). Bootstrap analysis was carried out as 1,000 replications. Numerals above branches indicate statistical bootstrap values. The correct names of the strain Alkalihalobacillus lonarensis, Alkalihalobacillus xiaoxiensis, Bacillus patagoniensis, Bacillus neizhouensis, Bacillus clarkii, Bacillus tamaricis, Bacillus vedderi, and Bacillus cellulosilytica are Shouchella lonarensis, Shouchella xiaoxiensis, Shouchella patagoniensis, Salipaludibacillus neizhouensis, Evansella clarkia, Evansella tamaricis, Evansella vedderi, and Evansella cellulosilytica, respectively.
Arsenate reductase gene (arsC) was detected in all of the strains, while arsenate permease gene (arsB) was not detected in S1, S12, and E15 strains. The arrB and arxA genes were detected in none of the strains. Although strains S1 and S2 were of the same species, it was determined that strain S2, unlike S1, had the arsB gene region. A similar situation was encountered in strains S4 and S12 (Table 3).
All strains could grow between 9.5 and 10.5 pH values. The bacterial growth temperature ranged from 25 to 35°C. The maximum tolerance concentration was determined to be 320 mM As (V) for the S1, S4, and UW1 strains and 16 mM As(III) for the E15 strain. Also, the E15 strain was evaluated as the only strain that could grow in 25% NaCl medium (Table 3) (see Supplementary Material, Supplementary Figure S3).
The AgNO3 screening technique was used to detect the oxidation of As(III) to As(V) or the reduction of As(V) to As(III). Although we tried different environments and conditions that have been stated in the literature in order to obtain results, no results could be obtained for alkaliphilic organisms (Ersoy Omeroglu et al., 2022).
All strains failed to grow under anaerobic conditions and were considered as strict aerobes. Except for strain S4, all strains could grow in the medium that included As(V) and sodium lactate, sodium citrate, and sodium acetate (Table 4). The strains probably transformed arsenic oxyanions in a detoxification fashion, while sodium lactate, sodium citrate, and sodium acetate could be used as carbon sources.
Urmia Salt Lake (northwest of Iran, South Azerbaijan) can be considered as one of the world’s largest salt lakes, and there is a similarity in morphological and chemical properties between Lake Urmia and the Great Salt Lake in the western United States (Kelts and Shahrabi, 1986). It is the second largest hypersaline lake with an active food chain. It is saltier than the Aral Sea (>100 g/L) and Garabogazköl, Kara-Bogaz-Gol and also Black Strait Lake (Leroy et al., 2006). However, a progressive drought caused many dramatical changes to its physicochemical properties and volume of water (Hoseinpour, 2010): the salinity exceeded ˃300 g/L and its surface area decreased to less than 2,366 km2. The level of the lake decreased to 1,271 m and the water volume decreased from 42 billion to 223 km3 between 1995 and 2010, respectively. As a result of all these negative effects, the water depth decreased to 6 m (Hoseinpour, 2010), and because of negative changes in the physicochemical properties of the lake, not only was the lake ecosystem but also its surrounding habitats adversely affected by the situation. Several studies have reported high heavy metal contaminations such as arsenic in the nearby rivers, agricultural soil, and plant leaves (Saatloo et al., 2014; Mohammadi et al., 2018). This situation was also seen to be very effective on the lake microbiota.
Several microorganisms such as green sulfur bacteria, purple sulfur bacteria, and iron bacteria are found in the sediments of Lake Urmia (Ghaheri et al., 1999). Of course, the diversity and abundance of the existing species cannot remain stable as a result of changes in the ecosystem. There are naturally high amounts of heavy metals in a few hypersaline systems. For instance, the Searles Lake and Mono Lake (Western US) contain high As(V) levels. However, the majority of heavy metal–rich saline systems are due to industrial and human activities (Voica et al., 2016).
Based on this information, Lake Urmia is also an important habitat facing arsenic pollution and destruction of biodiversity. To the best of our knowledge, there is some information about arsenic-resistant microorganisms in Lake Urmia. In a study, Haloarcula sp. was isolated from Lake Urmia and analyzed for its bioremediation potentials (Taran et al., 2013). When we evaluate in general, based on their physiological characteristics, the Bacillus species appeared to be commonly found in such habitats. For this reason, by determining the genotypic and phenotypic characteristics of the Bacillus species isolated from such habitats, it has been revealed that there are different characteristics between the members of the same species. For this reason, updates were accelerated in the Bacilllaceae systematics in the 2000s. The order Bacillales within the class Bacilli (or Firmibacteria) represents an extremely heterogeneous bacterial taxon. The family Bacillaceae is one of the largest taxa in the order Bacillales, in which several novel genera have been reported (Sultanpuram and Mothe, 2016). Bacillus patagoniensis, Bacillus neizhouensis, and Bacillus cellulosilyticus species were proposed under new genera names [Shouchella patagoniensis (Joshi et al., 2012), Salipaludibacillus neizhouensis (Sultanpuram and Mothe, 2016), and Evansella cellulosilytica] and caused changes in the systematics. Especially for the Bacillus patagoniensis species, a change in taxonomy was first suggested for Alkalihalobacillus patagoniensis, but it was updated as Shouchella patagoniensis as a result of recent studies (Joshi et al., 2012). When the species obtained within the scope of this study are examined based on their partial 16S rDNA sequences, it can be said that they are the members of Bacillus. However, despite 98% sequence homology, different species may also emerge. In this context, considering the current systematics, it can be said that the strains obtained belong to the genera Shouchella, Salipaludibacillus, and Evansella. The best approach to classify an isolate is by sequencing its genome. Also, it should be evaluated in terms of the many features within the scope of the study aiming for this purpose. For example, strain UW1 shows the same degree of sequence similarity to both Evansella cellulosilytica and Evansella clarkia. However, in terms of phenotypic characteristics, while it is stated in the literature that Evansella cellulosilytica strains are not motile (Liu et al., 2022), the UW1 strain was found to be active, unlike this feature.
Bacillus members are usually found in arsenic-contaminated environments (Magar et al., 2022). They have recently attracted much interest due to their role in arsenic bioremediation and have been shown to carry chromosomal or plasmid-borne arsC genes, and these genes may remove arsenic from contaminated environments. The abundance of Bacillus isolates harboring arsC and also aioA genes plays an important role in the degradation and oxidation of arsenic. This also shows their potential in future bioremediation processes (Anderson and Cook, 2004). In this context, it is seen that the arsC gene is commonly found in strains that are specified as the Bacillus sp. in the old systematics but are now classified under the new genus. The fact that six strains obtained within the scope of the study have this gene region appears as the supporting data. Arsenic-resistant Bacillus species such as B. infantis and B. solimangrovi impact the alkaline crater lakes in India (Bagade et al., 2016). Furthermore, a hyper-tolerant B. firmus L-148 strain (updated as Cytobacillus firmus) was reported to have tolerated 3.3 M As(III) and 4 M As(V). This strain, which is the most known tolerant strain, contains both ars and aio operons with the ability of As(III) oxidation under alkaline conditions (Bagade et al., 2020). Another study reported the isolation of arsenic-resistant B. cellulosilyticus (updated as Evansella cellulosilytica) and Salipaludibacillus aurantiacus from Maharloo Lake which could tolerate up to 600 mM sodium As(V) (Fatemi et al., 2021). As a result of our study, it was determined that a single strain (Alkalibacillus sp. strain E-15) was tolerant to As(III) concentration as high as 16 mM. The highest As(V) resistance (320 mM) was found in Alkalihalobacillus sp. strain S1, Salipaludibacillus sp. strain S4, and Evansella sp. strain UW1. Interestingly, although S1 and S2 strains were both defined as the same genus (Alkalihalobacillus sp.), the arsB gene region was absent in S1 but present in S2. It was observed that this situation caused a difference in As(V) resistance. While S1 strain could tolerate 320 mM As(V), S2 strain could not survive in the environment containing more than 40 mM As(V) (Table 2; Table 3). When all these data are evaluated, it becomes clear how important it is to measure arsenic from different areas contaminated with arsenic and to isolate resistant bacteria. There are differences in resistance profiles and gene regions between the members of the same genus, depending on the strain difference. Due to increasing global warming and anthropogenic pollution inputs, more resistant bacteria with different gene region combinations are needed for the bioremediation of arsenic from these ecosystems.
Xinjiang province, one of the most polluted areas of China, is affected by pollutants, especially with arsenic. Bacillus sp. XS2 strain at a concentration of 6,400 mg/L As(III) can grow and convert As(III) to As(V). In the study by Chang et al. (2007), the Bacillus sp. strain, which has a high tolerance to sodium As(III) of up to 50 mM and successfully transformed/oxidized up to a concentration of 1 mM, was isolated; however, the Bacillus sp. XS2 can oxidize As(III) up to 4,000 mg/L and is the most potent As(III) oxidation seen in a Bacillus. Arsenic-resistant B. indicus (updated as Metabacillus indicus), which was isolated from an aquifer in West Bengal, can tolerate As(V) and As(III) by 20 and 3 mM, respectively (Suresh et al., 2004). Other studies have also reported the characterization of hyper-tolerant strains such as Microbacterium sp. strain A33 and Rhodococcus erythropolis OS1 (Drewniak et al., 2008; Achour-Rokbani et al., 2010). However, since these studies were carried out before the changes in the systematics, it is seen that the highest resistance is in the members of the Bacillus genus. Therefore, it is important to isolate and identify from new sources in order to increase the available data in updated systematics.
B. subtilis skin element gives resistance to As(V) and As(III). The ars set in the skin element contains four genes in the order arsR, Orf2, arsB, and arsC. The arsR and arsC products are homologous to the products of S. aureus and E. coli, respectively. B. subtilis arsB product is also homologous with S. aureus arsB product. However, the arsB protein exhibits only limited sequence identity (24%) to S. aureus arsB. Furthermore, there is no homolog of the Orf1 product in S. aureus. The Bacillus sp. UWC, which was isolated from fly ash in acid mine drainage, was shown to contain five genes arsRBCDA which are arranged unusually when compared to the E. coli plasmid R773 arsRDABC operon (Musingarimi et al., 2010). The arsenite oxidase gene (aox) has been described in many Bacillus strains, e.g., B. flexus, isolated from rice fields in India (Majumder et al., 2013).
Although S1 and S2, as well as S4 and S12, strains obtained within the scope of the study showed similar sequence homology among themselves, it was observed that the arsB gene region was detected in one of the strains, while being absent in the other (Table 3). In a study, it had been seen that enteric bacteria obtained from samples taken from raw sewage were resistant to As(III) even though they did not have the arsB gene region and the studied ars genes (Saltikov and Olson, 2002). As a matter of fact, in our study, it has been seen that the E15 strain gave negative results in the PCR study of the arsB gene region but had the highest As(III) resistance (16 mM) among the strains (Table 3). At the same time, the fact that the E15 strain can grow in the medium containing 25% NaCl (Supplementary Material; Supplementary Figure S3), unlike other strains, reveals that the physiological and genotypic characteristics of this strain should be studied in more detail. Much less is known about the second family of As(III) transporters, Acr3p. The members of this family are found in bacteria, archaea, and fungi but have been functionally characterized in only a few species (Achour et al., 2007). In strains that are resistant to As(III) but do not have arsB gene region, this gene region may be effective in providing resistance. Therefore, screening of all gene regions and even the discovery of new gene regions should be studied. However, it is necessary to first carry out studies on the extent of arsenic pollution and existing bacteria. In this context, Shewanella sp. O23S strain attracts much attention. As a result of the study conducted by Staicu et al. (2022a), this strain isolated from an old gold mine in Poland was found to have both ars and arr genes. It has been found to be quite effective in bioremediation studies from real industrial wastes rich in arsenic. Studies on this strain have also shown that AsS biominerals are formed using a separate biochemical pathway to selenium biomineralization (Staicu et al., 2022b). It is very important to determine the gene regions that they have in order to reveal their potential for use in bioremediation studies. However, it is also important to determine the products released as a result of biochemical transformations. Therefore, a silver nitrate assay is used for arsenic biotransformation test. However, since many biochemical tests such as this test are pH dependent, they cause problems in alkaliphilic studies. Moreover, since the arsenic biotransformation test is both salt and pH dependent, it may cause false negative results in experiments performed with halophilic and alkaliphilic bacteria in the study. As a matter of fact, although trials were carried out with different media in the study, no results were obtained for any of the strains. In this direction, we come across a study conducted by Bahar et al. (2020). In this study, a new spectrophotometric method with high potential for screening arsenite-oxidizing bacteria was developed. In the experiment, which is carried out depending on the color intensity formed by mixing arsenic and potassium permanganate, a distinction can be made since arsenate and potassium permanganate do not react, and the pH can be studied in the range 4–9. The inability of the strains to grow under pH 9.5 and also the fact that these strains are halophiles caused negative results in many trials. However, with this new test technique (Bahar et al., 2020) and other tests to be developed, biotransformation trials of arsenic-resistant bacteria isolated from especially extreme habitats should be performed.
In this context, within the scope of this study, it was aimed to determine the bacteria in the area with arsenic pollution and intense anthropogenic pollution. Heavy metal analysis results from tree leaves found around Lake Urmia, one of the largest salt lakes in the world, showed that it accumulated many metals, such as arsenic. Since there is a large amount of agricultural land around the sampling point and the amount of arsenic accumulated in these areas is high, the consumption of food produced from these areas by all living forms, especially humans, causes a risk to the health of these. However, as a result of anthropogenic pollution from these areas to the lake, the amount of arsenic in the lake is also increasing. With the effect of global warming, the arsenic contamination of agricultural lands and living spaces increases with the effect of winds from the lake, and this has become concentrated in terms of arsenic content with the gradual decrease of lake water. Both the deterioration of the ecological balance and the health of all living forms are in danger. As a result of this, our essential resources for many lives, which are in the status of renewable resources, are becoming depleted resources. In this context, it is necessary to ensure the sustainability of the environment, humans, animals, and microorganisms with the “One Health” approach. For this reason, it is important to determine arsenic contamination levels, which is one of the most important anthropogenic pollution sources, for the sustainability of Lake Urmia—which was defined as a protected area in 1967 and declared as a national park in 1976—and its local people. In addition, phenotypic and molecular characterization of the obtained arsenic-resistant bacteria should be made and their presence in the ecosystem should be revealed. Pathways to metabolize this toxic metalloid and resistance profiles should also be determined and announced to the scientific world in terms of their bioremediation potential.
Based on the data obtained from such studies, feasibility reports should be prepared and urgent preventive and cleaning approaches should be put into action. Otherwise, the existing high salt and heavy metal concentration starts to negatively affect the all living things in the surrounding countries. As a result of the pollution of water resources and agricultural lands, health problems will gradually increase and the agriculture and live-stock sector will be adversely affected by this situation. As a result of all this, all life forms such as humans will be forced to migrate and the ecological diversity of regions will be completely destroyed. Therefore, isolation and identification should be made, especially from extreme habitats. All microbiota members should be determined by the metagenomic techniques, especially due to the limitations of cultural methods. Arsenic metabolism pathways should be determined in terms of genetic and biochemical characteristics and their potential in bioremediation studies should be revealed.
This study was carried out to isolate and characterize arsenic-resistant bacteria from the drying Lake Urmia. This study could be remarkable of its kind in Lake Urmia because it is relatively the first study. The strains which are resistant to arsenic belong to the Shouchella, Salipaludibacillus, and Evansella genera. These new strains, previously classified as Bacillus genus, are commonly found in arsenic-containing environments based on publications on the initial identification data. The presence of arsenic-related genes (arsC, arsB, arrB, and arxA) was investigated. All of the strains were negative for arrB and arxA genes. All strains were also tolerant to high concentrations of arsenic. The current study provides data for studies conducting bioremediation applications in order to prevent arsenic disaster in vicinal territories. A two-stage bioremediation process was applied to bioremediate As(V)-contaminated habitats. In the first stage, As(III) was formed due to the activity of the arsC gene region, and in the second stage, the reaction was completed by the adsorption of As(III) to iron oxide or precipitation with sulfide. Another process performed was in converting the released As(III) to trimethyl arsine gas by methylation or evaporation. However, several factors should be carefully evaluated in choosing the methylation technique for the bioremediation of arsenic because the physiological significance and biochemical basis of arsenic methylation has uncertain points. Although at first it was thought that methylation was associated with arsenic detoxification as the pentavalent species were relatively harmless, it was later found that the process could increase arsenic toxicity since the trivalent species were more toxic than inorganic As(III). The toxicity of inorganic and organic arsenic is dimethylarsinous acid [DMA(III)], monomethylarsonous acid [MMA(III)] ˃ As(III) ˃ As(V) ˃ dimethylarsinic acid [DMA(V)], monomethylarsinic acid [MMA(V)], and trimethylarsine oxide (TMAO). The main pentavalent products DMA(V) and TMAO are approximately 100 and 1,000 times less toxic, respectively, and less toxic than As(III). MMA(III), DMA(III), and trimethylarsine [TMA(III)] are more toxic than As(III). In this way, they prevent its accumulation in the cell. However, here, the cell protects itself, but a toxic gas is released into the environment. For this reason, in case of bioremediation studies from arsenic-contaminated areas using the methylation technique, the system should be operated in a controlled room and arsine gas should be captured and disposed of instead of being released into the environment.
Since bacteria with the arsB gene region will form As(V), which is 100 times less toxic as a result of the reaction, these can be used in bioremediation studies with the same two-step process. These strains also have the arsC gene region. Undoubtedly, further transcriptomic studies will be useful in order to understand the exact mechanism and to identify the products released. In this way, important steps will be taken to propose new arsenic bioremediation processes by clarifying other arsenic-related processes for oxidation and respiratory reduction reactions.
Species that have adapted to live in arsenic concentrations that are toxic to other living forms in the evolutionary process have a high potential for arsenic bioremediation. In this process, they have been subjected to various evolutionary adjustments to metabolize arsenic. Thanks to these features, arsenic has to be removed for the sustainability of habitats that are in need of protection at risk, and therefore the species that can metabolize arsenic emerge as potential sources. By carrying out such studies, the pathway by which arsenic is metabolized is determined at the gene level. As a result, the data on which resistant strain can be used in areas contaminated with different forms of arsenic are obtained.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
MH: literature research, isolation method development, isolation of bacteria, data analysis, molecular genetic work, and writing of the manuscript; SD: isolation of bacteria and molecular genetic work; IK: writing of the manuscript; and EE: isolation method development, molecular genetic work, revised the manuscript, critically reviewed the manuscript, and is the senior author. All authors contributed to the article and approved the submitted version.
This study was supported by the EGE University Scientific Research Fund (BAP; 15-FBE-002).
The authors would like to thank EGE University Scientific Research Fund (BAP) for financially supporting this research study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, editors, and reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1195643/full#supplementary-material
Achour, A. R., Bauda, P., and Billard, P. (2007). Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res. Microbiol. 158 (2), 128–137. doi:10.1016/j.resmic.2006.11.006
Achour-Rokbani, A., Cordi, A., Poupin, P., Bauda, P., and Billard, P. (2010). Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. strain A33. Appl. Environ. Microbiol. 76 (3), 948–955. doi:10.1128/aem.01738-09
Alfred, E. B., and Heidi, R. S. (2016). Benson’s microbiological applications: Laboratory manual in general microbiology. 13th Editi. McGraw-Hill Education, 616. Complete Version 13th Edition.
Alvyar, Z., Shahbazi, F., Oustan, S., Dengiz, O., and Minasny, B. (2022). Digital mapping of potentially toxic elements enrichment in soils of Urmia Lake due to water level decline. Sci.Total Environ. 808, 152086. doi:10.1016/j.scitotenv.2021.152086
Anderson, C. R., and Cook, G. M. (2004). Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr. Microbiol. 48 (5), 341–347. doi:10.1007/s00284-003-4205-3
Andres, J., and Bertin, P. N. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 40 (2), 299–322. doi:10.1093/femsre/fuv050
Bagade, A., Nandre, V., Paul, D., Patil, Y., Sharma, N., Giri, A., et al. (2020). Characterisation of hyper tolerant Bacillus firmus L-148 for arsenic oxidation. Environ. Pollut. 261, 114124. doi:10.1016/j.envpol.2020.114124
Bagade, A. V., Paul, D., Rikame, T., Giri, A. P., Dhotre, D., Pawar, S., et al. (2016). “Diversity of arsenic resistant bacteria from lonar lake: A meteorite impact alkaline Crater Lake in India,” in Arsenic Research and Global Sustainability: Proceedings of the Sixth International Congress on Arsenic in the Environment (As2016), Stockholm, Sweden, June 19-23, 2016 (CRC Press), 113.
Bahar, M. M., Mahbub, K. R., Naidu, R., and Megharaj, M. (2020). A simple spectrophotometric method for rapid quantitative screening of arsenic bio-transforming bacteria. Environ. Technol. Inno. 19, 100840. doi:10.1016/j.eti.2020.100840
Ben Fekih, I., Zhang, C., Li, Y. P., Zhao, Y., Alwathnani, H. A., Saquib, Q., et al. (2018). Distribution of arsenic resistance genes in prokaryotes. Front. Microbiol. 9, 2473–2511. doi:10.3389/fmicb.2018.02473
Bruvoll, A. (2017). Characterization of the biofilm forming ability of Listeria monocytogenes and evaluation of the TTC assay as a method to assess the effect of disinfectants on biofilm bacteria. Master's thesis. Ås: Norwegian University of Life Sciences.
Carlin, A., Shi, W., Dey, S., and Rosen, B. P. (1995). The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 177 (4), 981–986. doi:10.1128/jb.177.4.981-986.1995
Chang, J. S., Yoon, I. H., and Kim, K. W. (2007). Isolation and ars detoxification of arsenite-oxidizing bacteria from abandoned arsenic-contaminated mines. J. Microbiol. Biotechnol. 17, 812–821.
Chen, C. M., Misra, T., Silver, S., and Rosen, B. P. (1986). Nucleotide sequence of the structural genes for an anion pump. The plasmid-encoded arsenical resistance operon. J. Biol. Chem. 261 (32), 15030–15038. doi:10.1016/s0021-9258(18)66824-3
Conrad, A. (2014). The arx anaerobic arsenite-oxidization pathway is conserved in Halomonas and Ectothiorhodospira strains isolated from Big Soda Lake, Nevada. University of California, Santa Cruz.
Drewniak, L., Styczek, A., Majder-Lopatka, M., and Sklodowska, A. (2008). Bacteria, hypertolerant to arsenic in the rocks of an ancient gold mine, and their potential role in dissemination of arsenic pollution. Environ. Pollut. 156 (3), 1069–1074. doi:10.1016/j.envpol.2008.04.019
Ersoy Omeroglu, E., Sudagidan, M., and Ogun, E. (2022). Arsenic pollution and anaerobic arsenic metabolizing bacteria in lake van, the world’s largest soda lake. Life 12 (11), 1900. doi:10.3390/life12111900
Fatemi, A., Tabatabaei, M., Derakhshandeh, A., and Borghei, S. M. (2021). New isolated extremophiles arsenic oxidizing bacteria for the removal of arsenic from high and low COD media. WWJ 32 (5), 61–69. doi:10.22093/WWJ.2021.314968.3196
Firrincieli, A., Presentato, A., Favoino, G., Marabottini, R., Allevato, E., Stazi, S. R., et al. (2019). Identification of resistance genes and response to arsenic in Rhodococcus aetherivorans BCP1. Front. Microbiol. 10, 888–913. doi:10.3389/fmicb.2019.00888
Ghaheri, M., Baghal-Vayjooee, M. H., and Naziri, J. (1999). Lake Urmia, Iran: A summary review. Int. J. Salt Lake Res. 8 (1), 19–22. doi:10.1007/bf02442134
Ghale, A. G. Y., Baykara, M., and Unal, A. (2017). Analysis of decadal land cover changes and salinization in Urmia Lake Basin using remote sensing techniques. Nat. Hazards Earth Syst. Sci. Discuss. doi:10.5194/nhess-2017-212
Gholampour, A., Nabizadeh, R., Hassanvand, M. S., Nazmara, S., and Mahvi, A. H. (2017). Elemental composition of particulate matters around Urmia Lake, Iran. Toxicol. Environ. Chem. 99 (1), 17–31. doi:10.1080/02772248.2016.1166226
Hoeft, S. E., Kulp, T. R., Stolz, J. F., Hollibaugh, J. T., and Oremland, R. S. (2004). Dissimilatory arsenate reduction with sulfide as electron donor: Experiments with Mono Lake water and isolation of strain MLMS-1, a chemoautotrophic arsenate respirer. Appl. Environ. Microbiol. 70 (5), 2741–2747. doi:10.1128/aem.70.5.2741-2747.2004
Hoseinpour, M. (2010). “Death of Urmia Lake, a silent disaster investigating causes, results and solutions of Urmia Lake drying,” in 1st International Applied Geological Congress, Department of Geology, Islamic Azad University, Islamic Azad University-Mashad Branch, Iran, 26-28 April 2010, 700–704.
Houshyar, J., Ostadrahimi, A., Pourmoradian, S., Faramarzi, E., Tutunchi, H., and Mobasseri, M. (2022). Associations between Lake Urmia disaster and the prevalence of thyroid nodules and metabolic syndrome: The AZAR cohort survey. Health promot. Perspect. 12 (3), 310–314. doi:10.34172/hpp.2022.40
Joshi, A., Thite, S., Karodi, P., Joseph, N., and Lodha, T. (2012). Alkalihalobacterium elongatum gen. nov. sp. nov. An antibiotic-producing bacterium isolated from Lonar Lake and reclassification of the genus Alkalihalobacillus into seven novel genera. Front. Microbiol. 12, 722369. doi:10.3389/fmicb.2021.722369
Kao, A. C., Chu, Y. J., Hsu, F. L., and Liao, V. H. C. (2013). Removal of arsenic from groundwater by using a native isolated arsenite-oxidizing bacterium. J. Contam. Hydrol. 155, 1–8. doi:10.1016/j.jconhyd.2013.09.001
Kazemi, M., Chamani, A., and Agh, N. (2019). The assessment of arsenic contamination in Urmia Lake sediments and its effect on human health. J. Environ. Stud. 45 (3), 485–497. doi:10.22059/JES.2019.284975.1007892
Kelts, K., and Shahrabi, M. (1986). Holocene sedimentology of hypersaline Lake Urmia, northwestern Iran. Palaeogeogr. Palaeoclimatol. Palaeoecol. 54, 105–130. doi:10.1016/0031-0182(86)90120-3
Leroy, S. A., Marret, F., Giralt, S., and Bulatov, S. A. (2006). Natural and anthropogenic rapid changes in the Kara-Bogaz Gol over the last two centuries reconstructed from palynological analyses and a comparison to instrumental records. Quat. Int. 150 (1), 52–70. doi:10.1016/j.quaint.2006.01.007
Liu, G. H., Narsing Rao, M. P., Chen, Q. Q., Che, J. M., Shi, H., Liu, B., et al. (2022). Evansella halocellulosilytica sp. nov., an alkali halotolerant and cellulose-dissolving bacterium isolated from bauxite residue. Extremophiles 26 (2), 19–27. doi:10.1007/s00792-022-01267-y
Magar, L. B., Rayamajhee, B., Khadka, S., Karki, G., Thapa, A., Yasir, M., et al. (2022). Detection of Bacillus Species with arsenic resistance and plant growth promoting efficacy from agricultural soils of Nepal. Scientifica 2022, 9675041–9675110. doi:10.1155/2022/9675041
Majumder, A., Bhattacharyya, K., Bhattacharyya, S., and Kole, S. C. (2013). Arsenic-tolerant, arsenite-oxidising bacterial strains in the contaminated soils of West Bengal, India. Sci. Total Environ. 463, 1006–1014. doi:10.1016/j.scitotenv.2013.06.068
Mardi, A. H., Khaghani, A., MacDonald, A. B., Nguyen, P., Karimi, N., Heidary, P., et al. (2018). The Lake Urmia environmental disaster in Iran: A look at aerosol pollution. Sci. Total Environ. 633, 42–49. doi:10.1016/j.scitotenv.2018.03.148
Mazhitova, Z., Jensen, S., Ritzen, M., and Zetterström, R. (1998). Chlorinated contaminants, growth and thyroid function in schoolchildren from the Aral Sea region in Kazakhstan. Acta Paediatr. 87 (9), 991–995. doi:10.1111/j.1651-2227.1998.tb01771.x
Mohammadi, A., Hajizadeh, Y., Taghipour, H., Mosleh Arani, A., Mokhtari, M., and Fallahzadeh, H. (2018). Assessment of metals in agricultural soil of surrounding areas of Urmia Lake, northwest Iran: A preliminary ecological risk assessment and source identification. Hum. Ecol. Risk. Assess. 24 (8), 2070–2087. doi:10.1080/10807039.2018.1438173
Musingarimi, W., Tuffin, M., and Cowan, D. (2010). Characterization of the arsenic resistance genes in Bacillus sp. UWC isolated from maturing fly ash acid mine drainage Neutralised solids. S. Afr. J. Sci. 106 (1–2), 59–63.
Ouria, M., and Sevinc, H. (2016). The role of dams in drying up Lake Urmia and its environmental impacts on Azerbaijani districts of Iran. Saussurea 6 (1), 373–2525.
Rosdahl, V. T., and Rosendal, K. (1980). Resistance to cadmium, arsenate and mercury among Danish strains of Staphylococcus aureus isolates from cases of bacteriaemia, 1957–1974. J. Med. Microbiol. 13, 383–391. doi:10.1099/00222615-13-3-383
Rosen, B. P. (2002). Biochemistry of arsenic detoxification. FEBS Lett. 529, 86–92. doi:10.1016/s0014-5793(02)03186-1
Sá-Pereira, P., Rodrigues, M., Castro, E., and Simões, F. (2007). Identification of an arsenic resistance mechanism in rhizobial strains. World J. Microbiol. Biotechnol. 23 (10), 1351–1356. doi:10.1007/s11274-007-9370-2
Saatloo, S. J. E., Saatlo, M. E., Siosemarde, M., and Merufinia, E. (2014). Investigation and measurement of heavy metals amount (As, Pb, Cd, Hg) within rivers estuaries located in the west side of Urmia Lake. J. Civ. Eng. Urban. 4 (3), 233–238.
Saltikov, C. W., Cifuentes, A., Venkateswaran, K., and Newman, D. K. (2003). The ars detoxification system is advantageous but not required for as (V) respiration by the genetically tractable Shewanella species strain ANA-3. Appl. Environ. Microbiol. 69 (5), 2800–2809. doi:10.1128/aem.69.5.2800-2809.2003
Saltikov, C. W., and Olson, B. H. (2002). Homology of Escherichia coli R773 arsA, arsB, and arsC genes in arsenic-resistant bacteria isolated from raw sewage and arsenic-enriched Creek Waters. Appl. Environ. Microbiol. 68 (1), 280–288. doi:10.1128/aem.68.1.280-288.2002
Sarkar, A., Kazy, S. F., and Sar, P. (2013). Characterization of arsenic resistant bacteria from arsenic rich groundwater of West Bengal, India. Ecotoxicol 22, 363–376. doi:10.1007/s10646-012-1031-z
Smedley, P. L., and Kinniburgh, D. G. (2002). A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 17, 517–568. doi:10.1016/s0883-2927(02)00018-5
Soudi, M., Ahmadi, H., Yasi, M., and Hamidi, S. A. (2017). Sustainable restoration of the Urmia Lake: History, threats, opportunities and challenges. Eur. Water. 60, 341–347.
Staicu, L. C., Wójtowicz, P. J., Molnár, Z., Ruiz-Agudo, E., Gallego, J. L. R., Baragaño, D., et al. (2022b). Interplay between arsenic and selenium biomineralization in Shewanella sp. O23S. Environ. Poll. 306, 119451. doi:10.1016/j.envpol.2022.119451
Staicu, L. C., Wόjtowicz, P. J., Baragaño, D., Pόsfai, M., Molnár, Z., Ruiz-Agudo, E., et al. (2022a). Bioremediation of a polymetallic, arsenic-dominated reverse osmosis reject stream. Lett. Appl. Microb. 75 (5), 1084–1092. doi:10.1111/lam.13578
Sultanpuram, V. R., and Mothe, T. (2016). Salipaludibacillus aurantiacus gen. nov., sp. nov. a novel alkali tolerant bacterium, reclassification of Bacillus agaradhaerens as Salipaludibacillus agaradhaerens comb. nov. and Bacillus neizhouensis as Salipaludibacillus neizhouensis comb. nov. Int. J. Syst. Evol. Microbiol. 66 (7), 2747–2753. doi:10.1099/ijsem.0.001117
Sunitha, M., Prashanth, S., and Kavi Kishor, P. B. (2015). Characterization of arsenic-resistant bacteria and their ars genotype for metal bioremediation. Int. J. Sci. Eng. Res. 6 (2), 304–309. doi:10.1016/j.envpol.2022.119451
Suresh, K., Prabagaran, S. R., Sengupta, S., and Shivaji, S. (2004). Bacillus indicus sp. nov., an arsenic-resistant bacterium isolated from an aquifer in West Bengal, India. Int. J. Syst. Evol. Microbiol. 54 (4), 1369–1375. doi:10.1099/ijs.0.03047-0
Taghipour, H., Mosaferi, M., Armanfar, F., and Gaemmagami, S. J. (2013). Heavy metals pollution in the soils of suburban areas in big cities: A case study. Int. J. Environ. Sci. Technol. 10 (2), 243–250. doi:10.1007/s13762-012-0143-6
Taran, M., Safari, M., Monaza, A., Reza, J. Z., and Bakhtiyari, S. (2013). Optimal conditions for the biological removal of arsenic by a novel halophilic archaea in different conditions and its process optimization. Pol. J. Chem. Technol. 15 (2), 7–9. doi:10.2478/pjct-2013-0017
Tourian, M., Elmi, O., Chen, Q., Devaraju, B., Roohi, S., and Sneeuw, N. (2015). A spaceborne multisensor approach to monitor the desiccation of Lake Urmia in Iran. Remote Sens. Environ. 156, 349–360. doi:10.1016/j.rse.2014.10.006
Voica, D. M., Bartha, L., Banciu, H. L., and Oren, A. (2016). Heavy metal resistance in halophilic bacteria and archaea. FEMS Microbiol. Lett. 363 (14), fnw146–9. doi:10.1093/femsle/fnw146
Wang, S., and Mulligan, C. N. (2006). Occurrence of arsenic contamination in Canada: Sources, behavior and distribution. Sci. Total Environ. 366 (2–3), 701–721. doi:10.1016/j.scitotenv.2005.09.005
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173 (2), 697–703. doi:10.1128/jb.173.2.697-703.1991
Yang, L., Donahoe, R. J., and Redwine, J. C. (2007). In situ chemical fixation of arsenic-contaminated soils: An experimental study. Sci. Total Environ. 387, 28–41. doi:10.1016/j.scitotenv.2007.06.024
Yumoto, I., Hirota, K., Nodasaka, Y., Tokiwa, Y., and Nakajima, K. (2008). Alkalibacterium indicireducens sp. nov., an obligate alkaliphile that reduces indigo dye. Int. J. Syst. Evol. Microbiol. 58 (4), 901–905. doi:10.1099/ijs.0.64995-0
Keywords: arsenic, Urmia lake, bacteria, pollution, arsenate, arsenite
Citation: Haghi M, Diznabi SH, Karaboz I and Ersoy Omeroglu E (2023) Arsenic pollution and arsenic-resistant bacteria of drying Urmia Salt Lake. Front. Environ. Sci. 11:1195643. doi: 10.3389/fenvs.2023.1195643
Received: 30 March 2023; Accepted: 19 June 2023;
Published: 30 June 2023.
Edited by:
Mallavarapu Megharaj, The University of Newcastle, AustraliaReviewed by:
Mezbaul Bahar, The University of Newcastle, AustraliaCopyright © 2023 Haghi, Diznabi, Karaboz and Ersoy Omeroglu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Esra Ersoy Omeroglu, ZXNyYWVyc29AZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.