- 1Food Security and Safety Focus Area, Faculty of Natural and Agricultural Sciences, North-West University, Mmabatho, South Africa
- 2Department of Microbiology, Faculty of Science, Kings University, Odeomu, Osun State, Nigeria
- 3Environmental Pollution Science and Technology (ENPOST), Ido-Ijesha, Ilesha, Nigeria
- 4Department of Biological Sciences, Microbiology Unit, School of Science, Olusegun Agagu University of Science and Technology, Okitipupa, Nigeria
- 5Department of Criminology and Security Studies, Faculty of Humanities, Management and Social Sciences, Kings University, Odeomu, Osun State, Nigeria
- 6Department of Medical Microbiology and Parasitology, School of Medicine and Pharmacy, College of Medicine and Health Sciences, University of Rwanda, Butare, Rwanda
- 7Department of Agricultural Technology, Ekiti State Polytechnic, Isan-Ekiti, Nigeria
- 8Department of Agricultural Economics and Farm Management, Faculty of Agriculture, University of Ilorin, Ilorin, Nigeria
- 9Institute of Mountain Hazards and Environment, University of Chinese Academy of Sciences, Beijing, China
- 10Department of Microbiology, Obafemi Awolowo University, Ile-Ife, Nigeria
- 11Microbiology Unit, Department of Applied Sciences, Osun State College of Technology, Esa-Oke, Nigeria
- 12Pan African University Life and Earth Sciences Institute, University of Ibadan, Ibadan, Nigeria
- 13Department of Biotechnology, Baze University, Abuja, Nigeria
The uncontrolled use of chemicals, urban wastes, nuclear resources, mining, petrochemicals and disposal of sewage sludge only a few anthropogenic activities that have contributed to the rapid industrialization and severe heavy metal contamination of soils and waterways. Both inorganic and organic pollutants, such as heavy metals, pesticides, petroleum hydrocarbons, and polycyclic aromatic hydrocarbons, can impact the composition and functionality of soils. Soils and plants are affected by pollution, thus, pose a dire threat to food security. This directly renders the soil unuseful for agricultural purposes, destroys the beneficial microbes in the soil, reduces the soil organic matter content, causes the imbalance of soil nutrients, affects plant growth and the interaction between the plants and microbes, subsequently affecting the soil and crop productivity. In addition, environmental pollutants affect human health, leading to different illnesses such as headaches, allergies, coughs, depression, chest pain, nausea, diabetes, liver problems, cancers, eye problems, and so on. Remediation (physical, chemical or biological) is therefore necessary to reduce the impacts of these pollutants in the environment. Bioremediations involve using natural products from plants, microbes, and so on, to detoxify the environment and make it useful or productive again. A key type of remediation is the Remediation by Enhanced Natural Attenuation (RENA) which involves the turning of soil to promote microbial proliferation, aeration, nutrient availability, moisture and consequently, the degradation of pollutants. This review discusses the technology of RENA, the associated microbes, the mechanism of its action, challenges associated with its usage and recommendations to advance the use of RENA for a sustainable environment.
1 Introduction
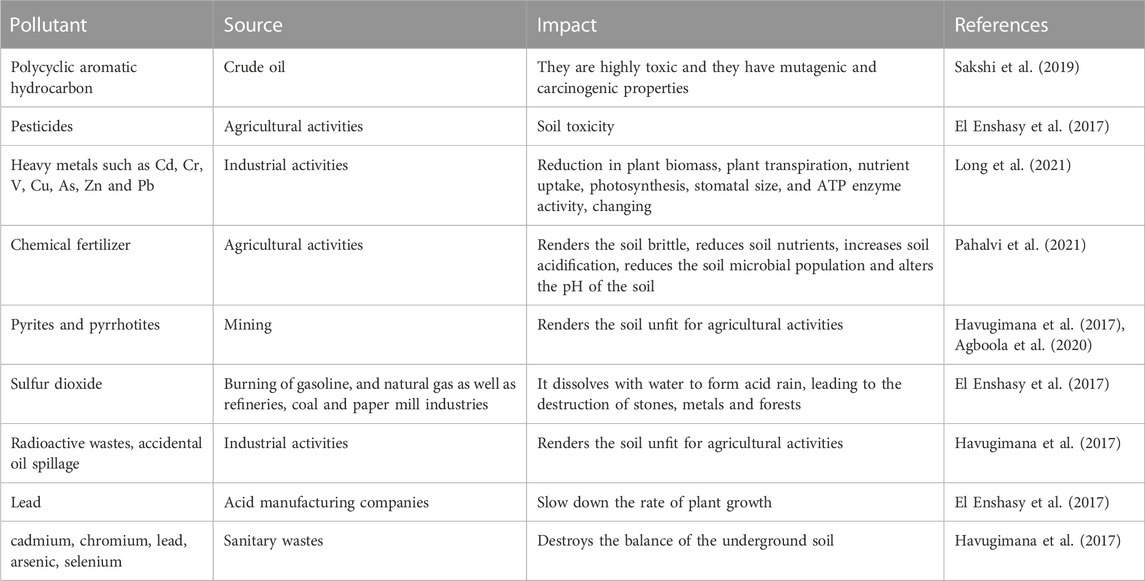
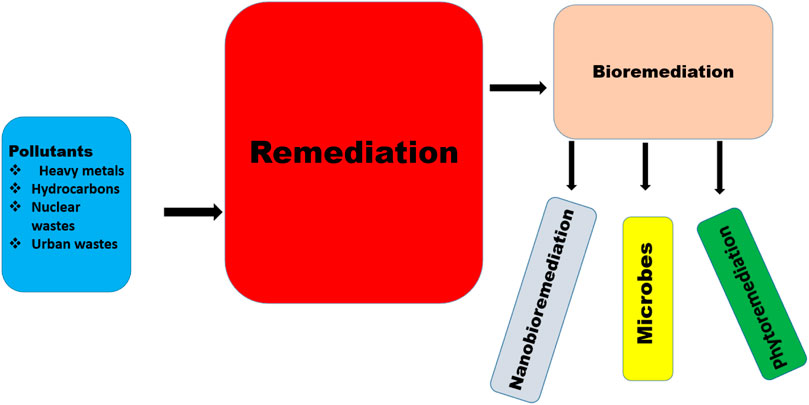
The soil is often polluted by different agents, such as heavy metals from mining, agricultural, petrochemical and other industries, including radiological, nuclear and other anthropogenic pollutants (Li et al., 2019b; Chaudhary et al., 2022) (Table 1). These agents negatively affect the soil, plants, organisms and humans, and have necessitated the need for remediation (Tyagi and Kumar, 2021). Many compounds arising from different pollutants have been reported to be very dangerous to humans. Acetaldehyde causes lesions to the nervous system, benzaldehyde causes irritation to the respiratory system, eyes, and skin and reduces the functioning ability of the brain, while polychlorinated dibenzo-dioxin causes cancers, respiratory system, eye, and skin irritation (Alabi et al., 2019). In animals, plastic wastes have been reported to disrupt the digestive systems of cattle (Evode et al., 2021). Equally, plant growth and root development have been reduced as a result of hydrocarbon pollutants (Hussain et al., 2019). These have called for a need for remediation. Remediation is the removal or reduction of the effects of pollutants in the environment; remediation can be carried out physically, chemically or biologically (Ayangbenro et al., 2018) (Figure 1). The physical and chemical methods involve soil washing, chemical extraction, soil treatment, supercritical fluid oxidation, encapsulation, stream extraction, chemical treatment, thermal treatment and volatilization (Riser-Roberts, 2020). The biological method, referred to as bioremediation, is a process by which wastes and toxic materials are organically removed or rendered less harmful into the environment (Ojuederie and Babalola, 2017; Ayangbenro and Babalola, 2018). This utilizes agents such as plants, microbes and nanoparticles of biological sources (Figure 1), which are more cost-effective compared to the other methods (Tyagi and Kumar, 2021; Chaudhary et al., 2023b; Bhandari et al., 2023). Remediation by enhanced natural attenuation (RENA) is a type of remediation process used to control soil pollutants by turning the soil to promote microbial proliferation, aeration, nutrient, moisture and degradation (Okoye et al., 2019). RENA is mainly used to control pollution caused by crude oil through different microbes such as those belonging to the genera Achromobacter, Azospirillus, Ochrobactrum, Bacillus, Alcaligenes, Lysinibacillus, Pusillimonas, and Proteus (Chikere et al., 2017). These microbes reduce the environmental pollutants by using them as carbon sources or by immobilizing them, thus making them unavailable for plant uptake (Chikere et al., 2017). Different organic and inorganic substrates are applied to improve the ability of microbes used in RENA (Kumar et al., 2021b; Mafiana et al., 2021; Parveen et al., 2022). Microbes produce different enzymes such as lipase, hydrogenase, laccase, etc., which help to degrade a wide range of pollutants (Bhandari et al., 2021). This is a very important mechanism of pollutants which should be well explored, especially in cases where an environment is polluted with more than one contaminant. The efficiency of RENA bioremediation can be altered by different factors which include the environmental pH, oxygen, temperature, and nutrient (Al-Hawash et al., 2018b). These factors affect the microorganisms directly and in cases where they are unfavorable the microbes that are expected to carry the bioremediation process die. RENA can be applied on both dry and swampy land areas, in cases where bioremediation is carried out in a swampy area, different steps are taken. Firstly, the stumps on the land has to be removed before the baseline studies are carried out (this is to ensure that the proper method of bioremediation is utilized (Orji et al., 2012). These procedures are followed by soil tillage, nutrient application and the monitoring of remediation. In cases where baseline studies are not carried out on the soil, the extent of pollution might not be known. For instance, in cases of underground water pollution, which leads to an assumption that RENA is not an efficient bioremediation procedure (Adesipo et al., 2020). The aim of this review is to discuss the technology of RENA, the different microbes associated with its usage, its mechanism of action, challenges associated with its application and recommendations to advance its usage to promote a sustainable environment.
2 Environmental pollutants and their negative impacts
There are different types of soil pollutants which have different detrimental effects on the environment.
3 Types of bioremediation
To remove pollutants arising from heavy metals such ascadmium, copper, nickel, mercury, organic pollutants and hydrocarbons from the soil, different plants, microbes, and nanobiomaterials have been utilized (Masowa et al., 2022). Therefore, according to their agents, bioremediation can be classified as a plant, microbial and nanobiomaterial remediation.
3.1 Phytobioremediation
Phytobioremediation (phytoremediation), is a process by which plants and the microbes associated with them (the plants) helps to reduce toxic pollutants in the environment. This mechanism of pollutant reduction is a cost-effective and environment friendly, it stabilizes, immobilizes, degrades and uptake pollutants from the environment (Kafle et al., 2022). Plants are capable of removing antibiotics, heavy metals, pesticides, radionuclides and organic pollutants from the environment (Kafle et al., 2022). Different plants have been reported to be involved in phytoremediation, these include Parthenium hysterophorus, Melilotus indicus, Cnicus benedictus, Anagallis arvensis, Verbesina encelioides, Dalbergia sissoo, Conyza canadensis, Lathyrus aphaca, Stellaria media, Xanthium strumarium, Chenopodium album, Medicago polymorpha, Amaranthus viridis, Mirabilis jalapa, Chenopodiastrum murale, Prosopis juliflora, Chrozophora tinctoria, C. tinctoria, Cerastium dichotomum and Arenaria serpyllifolia (Naz et al., 2022).
Jiang et al. (2019) and Parsamanesh and Sadeghi (2019) reported that plant species, Medicago scutellata and Mulberry sp. were able to bioremediate cadmium successfully from the soil. Similarly, other plants have been reported to bioremediate heavy metals; for instance, Cassia tora was reported to remediate chromium (Patra et al., 2021), while Polypogon monspeliensis and Rumex dentatus remediated nickel from the soil (Samreen et al., 2021). The different types of phytoremediation that exist include phytoextraction, phytostabilization, phytofiltration and phytovolatilization (Shen et al., 2022). Phytoextraction is the process by which metalloids are extracted from contaminated soil, with the aid of accumulator plants (Yu et al., 2022). The phytostabilization process, uses plants that are resistant to metals to reduce he availability of pollutants in the environment (Kumar et al., 2023). Phytofiltration is the application of plants and the microbes associated with their roots in the removal of heavy metals from the environment (Akhtar et al., 2019). Phytovolatization is the process by which wastes are taken off the environment through plants and they are transformed into gaseous state, which is released into the atmosphere (Pidlisnyuk et al., 2021). These processes are affected by the bioavailability of heavy metals and biomass of plants (Shen et al., 2022). The efficiency of phytoremediation is affected by stomatal conductance, the species of the microorganism present, intensity of light, plant species involved (its metabolism, photosynthesis and absorption rate) and temperature (Wei et al., 2021).
3.2 Nanoremediation
Nanoremediation is the application of nanomaterials in the treatment of environmental pollutants, especially on the soil and in water (El-Ramady et al., 2020). The different types of nanoparticles used in bioremediation, they include zinc nanoparticles, iron nanoparticles, aluminum nanoparticles, gold nanoparticles, titanium nanoparticles, carbon nanoparticles, and silver nanoparticles (Alazaiza et al., 2021). Nanoparticles, especially iron nanoparticles, are very active in soil bioremediation. The mechanism of nanoremediation include catalysis, adsorption, photodegradation and filtration (Mukhopadhyay et al., 2022). In a study carried out by Ji et al. (2023), diatomaceous earth nanoparticles were combined with polyethyleneimine nanoparticles to remove copper pollutants arising from acid mine drainages. Equally, nano biosurfactants have been reported to be capable of cleaning up toxic wastes from the soil arising from fertilizers, pesticides, herbicides, insecticides and heavy metals (Boregowda et al., 2022). Research carried out by Cao et al. (2022) showed that iron oxide nanoparticles could bioremediate cadmium and lead. These nanobiomaterials have been proven to be very efficient in the removal of toxic chemicals and heavy metals from the environment, particularly, the soil (Torimiro et al., 2021 (Chaudhary et al., 2023a)). However, a few drawbacks like adverse effects on soil microorganisms and the reduction in the activity of these nanoparticles as they age have been reported (Cecchin et al., 2017). Since the reduction in the efficiency of these nanoparticles with age can impact negatively their shelf life when made into commercial products, it is necessary that more research should be channeled toward strategies to increase their stability and lessen their harmful impacts on beneficial soil microbes to enhance their applicability as agents of bioremediation. The efficiency of nanoparticle as agents of bioremediation can be improved by fortifying them with polymers, zeolites, biochar, activated carbon and clay minerals (Mukhopadhyay et al., 2022).
3.2.1 Microbial bioremediation
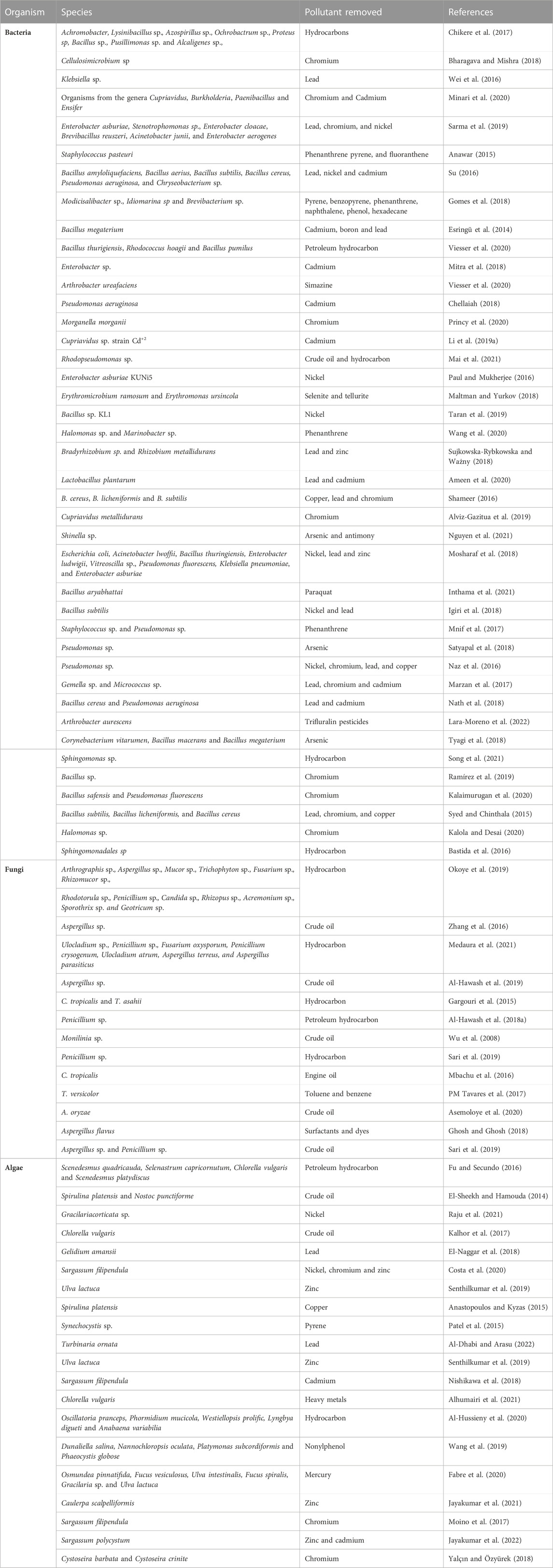
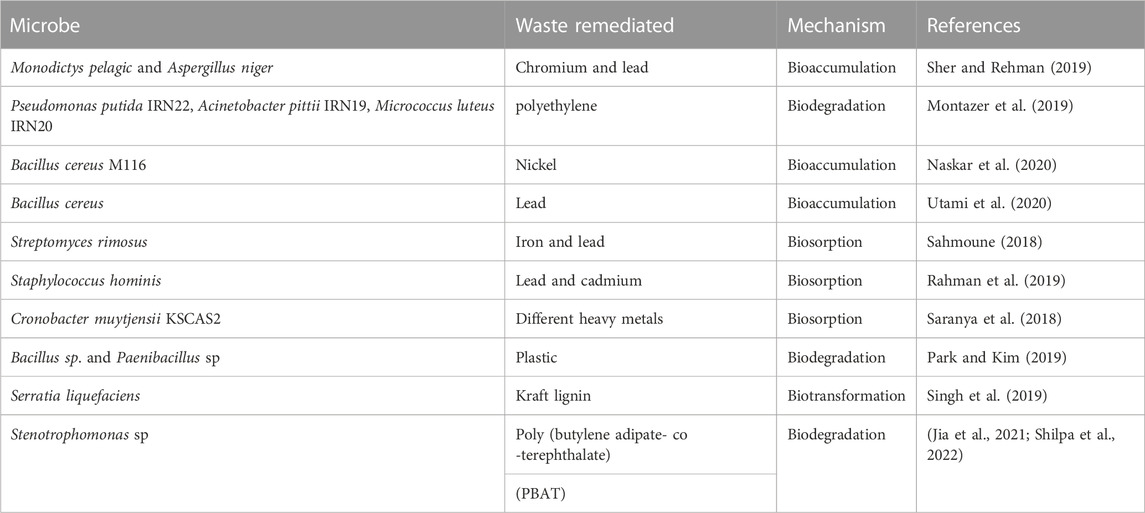
Microbial remediation is the use of microbes to reduce the concentration of heavy metal pollutants in the environment (Jin et al., 2018). Microorganisms such as fungi, algae and bacteria have been used to bioremediate polluted soils (Ndeddy Aka and Babalola, 2016; Karthika et al., 2017; Ndeddy Aka and Babalola, 2017; Thesai et al., 2021; Kumar et al., 2022) (Table 2). The efficiency of these organisms is affected by the temperature of the environment, substrate where the microbes are getting their nutrient from and the pH of the environment (Jin et al., 2018). A report by Ghosh et al. (2021) revealed that generally, bacteria species like Brevibacterium iodinum, Pseudomonas aeruginosa, Pseudomonas florescens and Alcaligenes faecalis, as well as the fungi species like Saccharomyces cerevisiae are very active in the bioremediation of the soil; the researchers equally reported the effectiveness of Anaeromyxobacter sp., Comamonas sp., Saccharibacteria sp., Desulfomicrobium sp., Acinetobacter sp., Zoogloea sp., Sphingobiun sp., Terrimonas sp. and Thiobacillus sp. in the bioremediation of organic compounds such as pyridine, indole and quinolone. El-Ansary and Ahmed-Farid (2021) as well reported the ability of algae species such as Scenedesmus obliquus, Nostoc muscorum, Chlorella vulgaris and Anabaena oryza to degrade oxyl nematicide. These organisms usually use these pollutants as carbon and energy sources and convert them into water, microbial biomass, metabolites and carbon dioxide, which are generally not as toxic as the initial pollutants but could be useful to the soil health and plants alike (Tyagi and Kumar, 2021).
Some microbes utilize pollutants as their energy source during remediation, while some may immobilize or transform them and make them unavailable for plant uptake. RENA has been reported to be effective in the remediation of coal tar; Telesiński and Kiepas-Kokot (2021) reported that there was a decrease in the contents of phenol and naphthalene by 98%–100%, when the RENA technology was used.
3.2.1.1 Bacterial bioremediation
Bacteria are important agents of bioremediation, they are capable of removing polyaromatic, aromatic and aliphatic hydrocarbons (Table 2). The optimum conditions required for bacterial remediation include a temperature ranging between 30°C and 40°C, a carbon/nitrogen and phosphorus ratio of 100; 20; 1, a pH range of between five to eight and an oxygen level of between 10 and 40 percent (Kebede et al., 2021). Bacteria involved in bioremediation can live in a cooperative or competitive relationship, when in a cooperative relationship, the biodegradation process is enhanced; however, when they are in a competitive relationship, the biodegradation process is reduced (Kebede et al., 2021). Resident bacteria are more competitive in hydrocarbon degradation compared to the introduced species, especially in a long term (Kaminsky et al., 2019). Bacteria undergo genetic modifications to maximally remediate hydrocarbons, otherwise, they produce enzymes. Therefore, more research should be carried out to focus on the discovery of more enzymes that can successfully bioremediate complex pollutants.
3.2.2 Fungal bioremediation
Different fungal species have been successfully used as agents of bioremediation (Table 2). Fungi degrade pollutants by intracellular compartmentalization, organic acid precipitation, metal-binding proteins, active transport, metabolite, and inorganic acid precipitation (Li et al., 2020). When different metabolites and compounds are released by fungi, they help to immobilize and mobilize metal pollutants in the soil. In addition, fungi produce melanin and polymers which have oxygen groups which include carbonyl, carboxyl, phenolic hydroxyl, methoxyl, and alcoholic hydroxyl which are used to clean up pollutants in the environment (Li et al., 2020). They are capable of degrading a wide range of substrates such as pesticides and hydrocarbons, due to their ability to tolerate and survive in adverse environments (Mostafa et al., 2022).
3.2.2.1 Algal bioremediation
Algaebioremediation happens majorly through two different mechanisms, the first is adsorption, while the second is adsorption (Dwivedi, 2012). Adsorption involves the adherence of pollutants to the surface of algae, the process takes place very fast and is independent of the cell metabolism. Adsorption is a two-way process, initially, the pollutants get adsorbed to the surface of the cell, and subsequently, they are moved into the cytoplasm, in a process called chemisorption (Liu et al., 2021). Interestingly, algae can bioremediate pollutants both in their living and dead states, but the living cells have the ability to bioremediate pollutants better than the dead ones (Salama et al., 2019). The ability of algae in bioremediation is affected by different factors such as pH, the effect of counter ion, temperature, ionic strength and contact time (Salama et al., 2019). If these factors are optimized, the maximum potentials in the bioremediation process of algae would be tapped into.
4 Mechanism of RENA
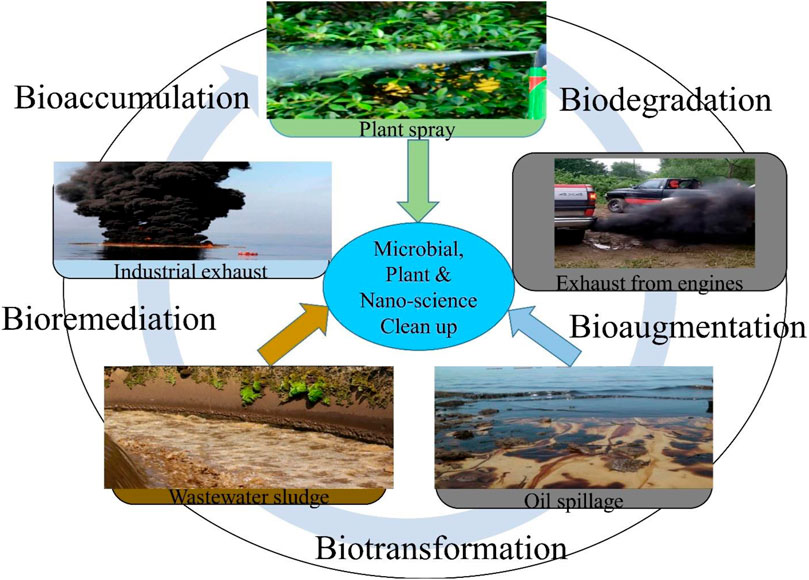
RENA is a modern concept of microbial-assisted remediation, and it is gaining more insights because it enhances the function and the ability of the microbes in bioremediation. It is a more affordable method when compared to other remediation methods which include washing of soils and burning and helps to ensure maximum remediation as the microbes involved are able to completely break down pollutants such as hydrocarbons (Kanwal et al., 2022). Pollutants in the soil can be degraded by the RENA technique through different mechanisms, which include biodegradation, biotransformation (bioaugmentation) and bioaccumulation (bioassimilation) (Oghoje et al., 2021) (Figure 2).
4.1 Bioassimilation
Bioassimilation, which is also referred to as bioaccumulation is the process by which microbes accumulate heavy metals in their body, making them unavailable for uptake by the plant; for instance, the sorption of chromium, cadmium and lead by Panteoa species and Pseudomonas koreensis have been reported (Ayangbenro and Babalola, 2017; Ayangbenro et al., 2019; Oghoje and Ukpebor, 2020) (Table 3). Bioaccumulation is a bioremediation process that consumes energy and also serves as a basis for methylation and redox in microbial remediation (Yin et al., 2022). When bacterial cells are used, bioaccumulation includes transport mediated by carriers, ion pumps, lipid infiltration and endocytosis (Yin et al., 2022). The pollutants removed from the environment through this method is attached to the cellulose derivatives, chitin and polysaccharides active sites of microorganisms through the physical and chemical binding with biofunctional groups (Tarfeen et al., 2022). This method involves forces such as the ion exchange processes, electrostatic attraction, covalent bonding, Van der Waal’s forces, and microprecipitation, while the functional groups involved include the sulfhydryl, hydroxyl, phosphonate, carboxyl and amine on the active cell component (Tarfeen et al., 2022).
5 Biotransformation
Biotransformation, also called bioaugmentation, is the conversion of a pollutant from one oxidation state to another. In bioaugmentation, microbes that are capable of degrading hydrocarbons but are not residents in the soil are introduced from external sources to supplement the resident organism, mostly bacteria and fungi (Essabri et al., 2019; Sayed et al., 2021) (Table 3). Sulfur-reducing bacteria such as Sulfolobus sp., Acidiphilium cryptum, T. thioparus, A. cryptum, S. acidophilus, Citrobacter sp., Thiobacillus denitrificans, Acidithiobacillus ferrooxidans, Sulfobacillus thermosulfidooxidans, At. caldus, Gallionella ferruginea, At. thiooxidans, Ferroplasma acidiphilum, Clostridium sp., Acidianus brierleyi, and Leptospirillum ferrooxidans have been reported to be potent in the mobilization and elimination of heavy metals from mines by dissolution, precipitation and retrieving safer metals from them (Schippers et al., 2010; Martins et al., 2011; Anawar, 2015; Ayangbenro et al., 2018). The bioaugmentation procedure is a fast, easy and effective method of RENA using different agents such as bacteria, algae, archaea and fungi (Ghosal et al., 2016; Kong et al., 2018).
6 Biodegradation
Biodegradation is the process by which microbes use the carbon in petroleum pollutants as a source of energy to disintegrate the hydrocarbon compounds into an environment-friendly form, such as carbon dioxide and water, with the aid of genes and enzymes produced by the microbes (Arumugam et al., 2017; Ahmed et al., 2018; Arumugam et al., 2018; Masowa et al., 2018; Karthika et al., 2020; Masowa et al., 2021; Priya et al., 2022) (Table 3). Adlan et al. (2020) reported the biodegradation of crude oil paraffin wax using Anoxybacillus geothermalis, Geobacillus kaustophilus, Parageobacillus caldoxylostilyticus, Geobacillus jurassicus, Geobacillus stearothermophilus and Geobacillus thermocatenulatus where around 70% degradation efficiency was observed in crude oil; the organisms involved in the degradation produced enzymes such as esterase, alkane monooxygenase, lipase and alcohol dehydrogenase.
When microbes use pollutants as carbon sources, they do so through three different techniques, namely, biostimulation, bioaugmentation and bio-facilitation (Azubuike et al., 2016; Oghoje et al., 2021). Bio-facilitation is a process by which the physiochemical content, and oxygen level, among others, in the soil are altered to make the inherent soil microbes more active to access pollutants. An example of this technique is land farming which involves the spread of evacuated soil and the tilling of soils where pollutants are found to increase oxidation (Oghoje et al., 2021). For instance, nutrients from organic wastes, synthetic fertilizers, humic acids, nanomaterials and so on, can be added to enhance microbial growth (Bianco et al., 2020). Bianco et al. (2020) carried out research to assess the ability of fresh organic municipal solid wastes, combined macro and micronutrients and digestate to increase the metabolism of microbes in the remediation of pyrene, anthracene, fluoranthene and phenanthrene. Other agents of biostimulation include blood meal which is a form of fertilizer that releases nutrients slowly. It is highly rich in nutrients such as tryptophan, lysine, histidine, leucine and valine, and can be used in the degradations of dichlorodiphenyltrichloroethane and polycyclic hydrocarbons (Wang et al., 2017). Biostimulation can be carried out using composting; it is cost friendly as it employs agents like manures, activated sludge and maple leaves, which will increase the population of microbes in the soil; compost from green forest debris and sewage sludge was able to reduce polycyclic hydrocarbons pollution (Guo et al., 2020).
7 Enzymes used in RENA
Scanty information is available on the microbial enzymes used in RENA; however, enzymes like laccase, hydrolase, cytochrome P450, protease, dehalogenase, lipase, dehydrogenase and so on, have been reported to be useful (Adlan et al., 2020). Proteases enhance the disintegration of peptide bonds in a protein. Their ability to degrade a wide range of substrates and their unique catalytic mechanism makes them a very effective method of bioremediation (Bhunia and Basak, 2014). Examples of microbes that produce proteinases are Aspergillus sp., Cladosporium sp., Trichoderma sp., Bacillus sp. and Penicillium sp. (Kumar and Jain, 2020). Cytochrome P450 enzymes are monooxygenases (haem) that can bioremediate pollutant compounds by reducing an atom to water and inserting an atom from molecular oxygen into the substrate (Behrendorff, 2021). White rot fungi have recently been used extensively in degrading pollutants, owing to the cytochrome enzyme produced by them (Lin et al., 2022); cytochrome P450 degrades xenobiotics using chemical transformation techniques such as dehalogenation, epoxidation, dealkylation and aliphatic hydroxylation (Bhandari et al., 2021). Laccases are enzymes that oxidize compounds in pollutants, leading to degradation and bioremediation. They are majorly observed in plants and fungi but lately, researches are emerging about their production by bacteria (because of their ability to survive at high temperatures and in different organic compounds), although their efficiency is determined by their substrate tolerance, selectivity and the center of their catalysis (Zhang et al., 2020). Dai et al. (2020) reported the ability of laccase from bacteria to degrade pollution resulting from heavy oil by 66.5% in 100 days. Remediation using dehydrogenase happens through reductive, oxygenlytic and hydrolytic mechanisms; oxygenlytic mechanism occurs when one or two atoms of oxygen is incorporated into a substrate, hydrolytic mechanism occurs with the water molecule acts as a cofactor when the hydroxyl group replaces the halogen substituent in SN reaction. In contrast, in the reductive mechanism, under aerobic conditions, hydrogen substitutes the halogen using the organohalides as the terminal electron acceptor (Bhandari et al., 2021). Li et al. (2019c) unraveled a novel hydrolase enzyme from Bacillus amyloliquefaciens to degrade soil and food polluted by carbendazim. Equally, Ugochukwu et al. (2008) carried out a research and reported that Bacterium aliphaticum, Candida tropicalis, Bacillus megaterium, Pseudomonas maltiphilia, Bacillus cereus, Botryodiphodia thiobroma, Edwardsiella tarda, Fusarium vertiaculloide, Cryptococcus neofomas, Aspergillus niger and Fusiarum oxysporum can bioremediate crude oil pollution due to the production of lipase.
Six Esterases which are produced by Bacillus sp. are useful in degrading polyesters, plastics and polyurethane (Bhatt et al., 2019). Nitrilases were used to remediate cyanide and can be produced by different yeast, bacteria and fungi (Gong et al., 2012; Park et al., 2017). Similarly, peroxidases produced by Sphingobium sp. are used to degrade phenols, lignin, methoxybenzenes and manganese oxide (Wang et al., 2009; Sharma et al., 2019). Dehydrogenase which is an active enzyme utilized for respiration by microbes was observed to be very helpful in the measurement of the total oxidation activity during the bioremediation of diesel (Lee et al., 2011). Manganese peroxidase which was sourced from fungi was reported to be positively correlated with the complete biodegradation of petroleum hydrocarbon (Košnář et al., 2019). Out of all the enzymes produced by fungi, oxidoreductases and hydrolases have a broad spectrum, with the ability to degrade a wide variation of pollutants (Kumar et al., 2021a). In their natural environment, microbes produce enzymes which are very active in the degradation of environment pollutants, however, the further utilization of these microbes has proved to be very challenging (Sharma et al., 2018). Sometimes, compaenzyme production in the lab can be complicated, as microbes tend to act differently in the lab red to the when they are in their natural environment. In situations where they are able to produce the enzymes in the laboratory, technologies such as enzyme immobilization, nanozymes and recombinant technology are used to multiply and/or stabilize the enzymes so that they can be further used on the field (Meng et al., 2019; Kumar et al., 2021a). Practical application of RENA at the field level.
On the field scale, RENA is used to remove pollutants from the soil and water, using methods such as stabilization, chemical transformation, dilution, volatilization dispersion and biodegradation (Naeem and Qazi, 2020). RENA has been used to clean up different oil spillage in groundwater and soil. In Nigeria, RENA was reported to be used in the remediation of oil spillage in Emohua community, Rivers State by adding top soils to the polluted site and frequent aeration (Chikere et al., 2019). A significant reduction was observed in the petroleum hydrocarbon from 8,635.68 mg/kg to 677.2 mg/kg after 56 days on this site. The bacteria involved were Pseudomonas sp., Xenorhabdus sp., Bacillus sp., Myroides sp., Proteus sp., Staphylococcus sp., Pectobacterium sp., and Providencia sp., while the fungi species which include Fusarium sp., Penicillium sp., Meyerozyma sp., and Candida sp. were used. Generally, topsoil contains many bacteria and fungi species and nutrients irrespective of organic and inorganic nutrient amendments. Hence, mixed plowing of nutrient-rich topsoil with contaminated soil can increase soil nutrient parameters necessary for microbial growth (Celestina et al., 2019) Similarly, in the Ikarama community, Bayelsa state, Nigeria RENA was used to clear oil spillage in a period of 60 days, by burning the polluted site’s vegetation and plowing with a tractor (Ezekoye et al., 2018). This process utilized different organisms such as Acremonium sp., Phoma sp., Candida sp., Scopulariosis sp., Aspergillus sp., Sepedonium sp., Cladophialophora carrionii, Gliocladium sp., Paecilomyces variotii, Trichophyton tonsurans and Geotrichum cardidum (Ezekoye et al., 2018). The effective usage of RENA on the field has been ascertained; however, the technology might be ineffective when practices such as fertilizer application, tilling and windrow are used, this is as a result of the presence of non-biodegradable residues in the soil beyond where there is aeration. Other challenges with this technology include a delayed response to oil spillage emergencies, which enhances the penetration of oil to a region beyond the soil which can be reached by turning (Mafiana et al., 2021).
Chicken manure digestates have been reported to serve as a source of nutrients when RENA technology is used in the removal of diesel on farmland (Oghoje et al., 2021). When 10% and 20% of the chicken manure digestate were used, about 50% and 58% of the diesel were removed from the environment. Spent mushroom substrate also have been reported to be used to serve as a source nutrients for four different fungal species, namely, Agaricus bisporus, Pleurotus eryngii, Pleurotus ostreatus, and Lentinula edodes, which were used to remediate petroleum hydrocarbon. The remediation process was evaluated for 40 days and the aliphatic and aromatic hydrocarbon were observed to reduce from C10 to C35 (Antón-Herrero et al., 2022).
7.1 Molecular mechanism of RENA
Identification of the microbes involved in RENA bioremediation is very necessary as it will help to understand better, the mechanism and enzymes used in the remediation of different heavy metals, organic pollutants, and hydrocarbons and also enhance the remediation process. Different molecular methods are used to study the microbes involved in the bioremediation processes such as RENA, as the methods (proteomics, transcriptomics, metagenomics and metabolomics) help to elucidate non-culturable organism, and also reveals the genes involved in bioremediation process (Rawat and Rangarajan, 2019). Each of these methods have an advantage of others, for instance, in a case where the quantity of the total mRNA is required, transcriptomics is used; however, this method does not reveal other expressed protein as well as their biological activity and expressed protein. Metagenomics is a method used to study the taxonomic and functional structure of different microbes (Pande et al., 2020; Hualpa-Cutipa et al., 2023). Proteomics is the study of the entire protein that are produced or modified by different microorganism (Nascimento et al., 2022). Transcriptomics deals with use of the total set of mRNA and the noncoding RNA transcripts which are produced by microbial cells, it controls the physical expression and acts as a connector between protein and DNA (Bogati and Walczak, 2022). Metabolomics is the study of all the primary and secondary metabolites produced by microbes, and in RENA, it involves microbes that are involved in RENA remediation (Wu et al., 2022). Hence, to have a detailed knowledge of the microbes present during RENA remediation, it would be helpful to combine different omics processes instead of just one approach.
8 Factors affecting the efficiency of RENA
The application of RENA to biodegrade pollutants is hindered by several factors ranging from the availability of microbes capable of degrading the pollutants to pH, temperature, nutrients and oxygen. When the diversity of microbes that can degrade microbes in the soil is limited, the biodegradation process is limited (Al-Hawash et al., 2018b). This is why sometimes nutrients are added to the environment, which will promote the growth and activities of the desired organism.
Bioremediation using RENA can be carried out in the presence and absence of oxygen, while in the presence of oxygen, it is referred to as aerobic condition the lack of oxygen, it is termed anaerobic condition; in the latter, microbes utilizes iron, carbon dioxide, sulfate and nitrate to exchange electron, thereby, forming methane and carbon (Patel et al., 2020). An oxygen percentage of 10%–40% was reported to be optimum for the biodegradation of hydrocarbon because, at this temperature, microbial activity and degradative enzymes are promoted (Kebede et al., 2021).
The mobility of pollutants (e.g., oil) can be affected by the moisture content in the soil, which consequently affects the allocation, existence, movement and activities of microbes in the soil (Al-Hawash et al., 2018b). Some pollutants are more soluble in aqueous solutions, while some are not, and this affects their remediation (Fu and Secundo, 2016). The chemical constituent of hydrocarbons or waste to be degraded is an important factor to be considered when hydrocarbons are degraded, and specific microorganisms are just capable of degrading a single chemical compound, while some are capable of degrading multiple hydrocarbons or pollutants (Fu and Secundo, 2016).
The pH of the soil is paramount in the survival of microbes in the soil, unfavorable pH can be dangerous to the survival of microbes in the soil. Those that survive in acidic pH are termed acidophiles. Those that live in alkaline pH are called alkaliphiles, while those that survive neutral pH are called neutrophils (Schröder et al., 2020). The optimum pH observed when A. niger was used to bioremediate Hg, Cu, Co., Zn, and Ag was between 4–5.5 (Acosta-Rodríguez et al., 2018). On the other hand, Pawar (2015) reported that a pH of 7.5 was optimum for the degradation of hydrocarbons when Penicillium freii and A. niger were used.
Microbes required for the degradation of toxic heavy metals can die in the absence of optimum nutrients. Hence the presence of desired nutrients is needed for them to metabolize and remediate pollutants effectively. For example, Aspergillus sydowii bioremediated heavy metals and pesticides when mineral salt was used as the medium, as the nutrient was optimum for the growth and metabolism of the fungi (Zhang et al., 2019).
Some organisms are capable of surviving in extreme conditions; some organisms which live in extreme temperatures are termed extremophiles; those capable of living in cold regions are termed psychrophiles, and those who live in high temperatures are termed thermophiles (Malakootian et al., 2018). Research carried out by Acosta-Rodríguez et al. (2018) where A. niger was used to remove heavy metal pollutants in the environment revealed that the optimum remediation process occurred at a temperature of 28°C (Acosta-Rodríguez et al., 2018). Hence, it is important to carry out more research to understand the different pH levels, temperature, nutrient and oxygen requirements which will enable the survival of other microbes that can degrade various pollutants in the environment.
9 Challenges associated with the adoption of RENA
RENA, as technology has proven useful in the degradation of pollutants in the soil, and in ensuring the effective bioremediation of soil, but, the RENA process comes with some challenges which will be highlighted in this section. First, the RENA technology cannot degrade all waste. For example, RENA is highly specific in nature, hence, if there is the presence of more than one waste on the site, different microbes must be recruited, which probably might not be compatible to survive together (Sharma, 2020). Second, the biodegradation process of RENA sometimes brings about new products that may be more toxic compared to the initial pollutant, and the process is often time-consuming (Sharma, 2020). Third, the introduction of external nutrients or the turning of the normal soil structure of the soil during RENA to enhance the activity of microbes from another perspective can be seen as a negative move, as the normal eco-balance of the soil could shift and the soil structure can be destroyed by the movement of the soil (Chikere et al., 2017). Still, the addition of nutrients, especially from synthetic sources can lead to air and water pollution during rainy seasons (Chikere et al., 2017). Turning of the soil during RENA can as well promote the leaching of soil pollutants, as the pollutants which were at the surface are moved downwards, leading to underground water pollution (Chikere et al., 2017). Fourth, when the environment is altered, such as in the case of RENA, the expression of the gene by microbes might be altered (Smith et al., 2018). This may eventually affect the activity of the microbes either positively or negatively. It would not be appropriate to leave this to a game of chance, more research should be done to optimize the alteration in gene expression to be favorable to the microbes.
Lastly, the inhabitants of many communities do not trust the RENA technology, owing to the fact that in cases of severe pollution, a rapid method is always preferred. Since RENA is most times slow, it is assumed that it is not effective (Council, 2000). As a buttress, in some cases where a soil sample is taken from the environment to the laboratory or greenhouse to demonstrate the RENA technology, a false positive result might emerge because other factors might favor the technique in the greenhouse and might be otherwise on the field, resulting in lack of trust from the populace (O’Brien et al., 2021).
10 Conclusion and future prospects
RENA is an adequate, sustainable, non-toxic, cost-effective process which can be carried out at the site of pollution without posing any major health threat to the land, microbes and humans. This method removes the pollutants permanently, and not just transfer them to another environment (Sharma, 2020; Nuhu et al., 2022). Methods of RENA, such as the introduction of aeration and moisture to the soil which does not involve the movement of the soil, should be encouraged. Also, in cases where external nutrients would be added to the soil, organic nutrients in optimum quantity should be added as they are non-toxic to the environment, and if applied in optimum quantity, they will help to reduce the risk of eutrophication in water bodies.
RENA technology has proven to be effective and has successfully bioremediated some wastes, it comes with some drawbacks. This includes the inability to degrade some pollutants and the fact that RENA occurs mainly at the topsoil (0–5 m). Hence pollutants that go beyond this depth are leached into underground water (Ebuehi et al., 2005; Orji et al., 2012; Bolade et al., 2021)); therefore, it is recommended that research should be intensified to unravel organisms with strong abilities to bioremediate complex wastes. Also anaerobic microorganisms can be utilized through a deep injection technique to give microbes’ access to wastes that are beyond 5 m where microbes used in RENA cannot access. Alternatively, RENA should be combined with other safe bioremediation techniques such as phytoremediation and nanoremediation to ensure a safe and more sustainable environment.
Since remediation using RENA has come with some challenges, it is therefore advised to carry out more research to beat the challenges associated with it to improve the utilization of the technology. For instance, when using RENA, the microbes used are highly specific, that is, they remove one compound at a time from the environment which makes the process complicated in a situation where different compounds pollute a particular environment. Hence, it is recommended that more studies should be carried out to discover different microbes that have no antagonistic effects on each other and are capable of remediating different organic compounds and pollutants in a favourable manner. In addition, more studies should be conducted to recruit more microbes, especially from untapped resources such as the endophytes, rhizosphere and rhizoplane of underutilized legumes, since the underutilized legumes can survive in extreme conditions and the microbes capable of surviving in such environments could possess the same ability as well. Furthermore, not much work has been done on the different microbial enzymes and their ability to produce enzymes in different environments. An insight on this could help develop these enzymes into stable products for commercial purposes, which could be applied directly on the soil for bioremediation. To promote the use and acceptability of RENA, it has become important that more attention should be paid to local communities. Informal and formal education in secondary schools on the benefits of a sustainable environment should be conducted. Hesitancy and other cultural quirks that could slow the adoption of RENA technology should be broken through proper enlightenment. This will in turn, help to foster homegrown technological expertise, researchers, stake holders, investors, and grant donors, thereby, further reducing the cost of bioremediation in the long run.
Experiments which have been tested in the laboratory or greenhouse should be well tested on the field in different environments to ascertain their functionality and effectiveness across diverse soil conditions, to earn the trust of the populace. Governmental policies could also be amended to promote the use of bioremediation techniques like RENA, compared to the physical and chemical methods, which are more disadvantageous and environmentally unsustainable. Finally, it is necessary to understand the mechanism of RENA when combined with other bioremediation methods, as this would help to improve these two technologies and make them more sustainable.
Author contributions
MA and OB conceived the idea, wrote and revised the manuscript. BA, MA, SA, CF, FO, UA, LG, RO, RJ, and ME contributed to the writing of the manuscript and the final version of the work was approved by all authors for publication and production. All authors contributed to the article and approved the submitted version.
Funding
This study was funded through research grants from the National Research Foundation, South Africa (UID: 123634).
Acknowledgments
This OB appreciates the research grants from the National Research Foundation, South Africa.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acosta-Rodríguez, I., Cárdenas-González, J. F., Rodríguez Pérez, A. S., Oviedo, J. T., and Martínez-Juárez, V. M. (2018). Bioremoval of different heavy metals by the resistant fungal strain Aspergillus Niger. Bioinorganic Chemistry Applications.
Adesipo, A. A., Freese, D., and Nwadinigwe, A. O. (2020). Prospects of in-situ remediation of crude oil contaminated lands in Nigeria. Sci. Afr. 8, e00403. doi:10.1016/j.sciaf.2020.e00403
Adlan, N. A., Sabri, S., Masomian, M., Ali, M. S. M., and Rahman, R. N. Z. R. A. (2020). Microbial biodegradation of paraffin wax in Malaysian crude oil mediated by degradative enzymes. Front. Microbiol. 11, 565608. doi:10.3389/fmicb.2020.565608
Agboola, O., Babatunde, D. E., Fayomi, O. S. I., Sadiku, E. R., Popoola, P., Moropeng, L., et al. (2020). A review on the impact of mining operation: Monitoring, assessment and management. Results Eng. 8, 100181. doi:10.1016/j.rineng.2020.100181
Ahmed, A. A. Q., Babalola, O. O., and Mckay, T. (2018). Cellulase-and xylanase-producing bacterial isolates with the ability to saccharify wheat straw and their potential use in the production of pharmaceuticals and chemicals from lignocellulosic materials. Waste biomass valorization 9, 765–775. doi:10.1007/s12649-017-9849-5
Akhtar, M., Oki, Y., Bich, B., and Nakashima, Y. (2019). Estimation of phytofiltration potential for Cu and Zn and relative growth response of Azolla japonica and Azolla pinnata. J. Agric. Sci. Technol. 21, 895–909.
Al-Dhabi, N. A., and Arasu, M. V. (2022). Biosorption of hazardous waste from the municipal wastewater by marine algal biomass. Environ. Res. 204, 112115. doi:10.1016/j.envres.2021.112115
Al-Hawash, A. B., Alkooranee, J. T., Abbood, H. A., Zhang, J., Sun, J., Zhang, X., et al. (2018a). Isolation and characterization of two crude oil-degrading fungi strains from Rumaila oil field, Iraq. Biotechnol. Rep. 17, 104–109. doi:10.1016/j.btre.2017.12.006
Al-Hawash, A. B., Dragh, M. A., Li, S., Alhujaily, A., Abbood, H. A., Zhang, X., et al. (2018b). Principles of microbial degradation of petroleum hydrocarbons in the environment. Egypt. J. Aquatic Res. 44, 71–76. doi:10.1016/j.ejar.2018.06.001
Al-Hussieny, A. A., Imran, S. G., and Jabur, Z. A. (2020). The use of local blue-green algae in the bioremediation of hydrocarbon pollutants in wastewater from oil refineries. Plant Arch. 20, 797–802.
Alabi, O. A., Ologbonjaye, K. I., Awosolu, O., and Alalade, O. E. (2019). Public and environmental health effects of plastic wastes disposal: A review. J. Toxicol. Risk Assess. 5, 1–13.
Alazaiza, M. Y., Albahnasawi, A., Ali, G. A., Bashir, M. J., Copty, N. K., Amr, S. S. A., et al. (2021). Recent advances of nanoremediation technologies for soil and groundwater remediation: A review. Water 13, 2186. doi:10.3390/w13162186
Alhumairi, A. M., Hamouda, R. A., and Saddiq, A. A. (2021). Bio-remediation of most contaminated sites by heavy metals and hydrocarbons in dhiba port kingdom of Saudi arabia using Chlorella vulgaris. Res. Square 1, 1–17.
Alviz-Gazitua, P., Fuentes-Alburquenque, S., Rojas, L. A., Turner, R. J., Guiliani, N., and Seeger, M. (2019). The response of Cupriavidus metallidurans CH34 to cadmium involves inhibition of the initiation of biofilm formation, decrease in intracellular c-di-GMP levels, and a novel metal regulated phosphodiesterase. Front. Microbiol. 10, 1499. doi:10.3389/fmicb.2019.01499
Al-Hawash, A. B., Zhang, X., and Ma, F. (2019). Removal and biodegradation of different petroleum hydrocarbons using the filamentous fungus Aspergillus sp. RFC-1. Microbiologyopen 8, e00619. doi:10.1002/mbo3.619
Ameen, F. A., Hamdan, A. M., and El-Naggar, M. Y. (2020). Assessment of the heavy metal bioremediation efficiency of the novel marine lactic acid bacterium, Lactobacillus plantarum MF042018. Sci. Rep. 10, 314–411. doi:10.1038/s41598-019-57210-3
Anastopoulos, I., and Kyzas, G. Z. J. J. O. M. L. (2015). Progress in batch biosorption of heavy metals onto algae. J. Mol. Liq. 209, 77–86. doi:10.1016/j.molliq.2015.05.023
Anawar, H. M. (2015). Sustainable rehabilitation of mining waste and acid mine drainage using geochemistry, mine type, mineralogy, texture, ore extraction and climate knowledge. J. Environ. Manag. 158, 111–121. doi:10.1016/j.jenvman.2015.04.045
Antón-Herrero, R., García-Delgado, C., Baena, N., Mayans, B., Delgado-Moreno, L., and Eymar, E. (2022). Assessment of different spent mushroom substrates to bioremediate soils contaminated with petroleum hydrocarbons. Appl. Sci. 12, 7720. doi:10.3390/app12157720
Arumugam, K., Renganathan, S., Babalola, O. O., and Muthunarayanan, V. (2018). Investigation on paper cup waste degradation by bacterial consortium and Eudrillus eugeinea through vermicomposting. Waste Manag. 74, 185–193. doi:10.1016/j.wasman.2017.11.009
Arumugam, K., Seenivasagan, R., Kasimani, R., Sharma, N., and Babalola, O. (2017). Enhancing the post consumer waste management through vermicomposting along with bioinoculumn. Int. J. Eng. Trends Technol. 44, 179–182. doi:10.14445/22315381/ijett-v44p235
Asemoloye, M. D., Tosi, S., Daccò, C., Wang, X., Xu, S., Marchisio, M. A., et al. (2020). Hydrocarbon degradation and enzyme activities of Aspergillus oryzae and Mucor irregularis isolated from nigerian crude oil-polluted sites. Microorganisms 8, 1912. doi:10.3390/microorganisms8121912
Ayangbenro, A. S., and Babalola, O. O. (2017). A new strategy for heavy metal polluted environments: A review of microbial biosorbents. Int. J. Environ. Res. public health 14, 94–16. doi:10.3390/ijerph14010094
Ayangbenro, A. S., Babalola, O. O., and Aremu, O. S. (2019). Bioflocculant production and heavy metal sorption by metal resistant bacterial isolates from gold mining soil. Chemosphere 231, 113–120. doi:10.1016/j.chemosphere.2019.05.092
Ayangbenro, A. S., and Babalola, O. O. (2018). Metal(loid) bioremediation: Strategies employed by microbial polymers. Sustainability 10, 3028. doi:10.3390/su10093028
Ayangbenro, A. S., Olanrewaju, O. S., and Babalola, O. O. (2018). Sulfate-reducing bacteria as an effective tool for sustainable acid mine bioremediation. Front. Microbiol. 1, 1986. doi:10.3389/fmicb.2018.01986
Azubuike, C. C., Chikere, C. B., and Okpokwasili, G. C. (2016). Bioremediation techniques–classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 32, 180–218. doi:10.1007/s11274-016-2137-x
Bastida, F., Jehmlich, N., Lima, K., Morris, B., Richnow, H., Hernández, T., et al. (2016). The ecological and physiological responses of the microbial community from a semiarid soil to hydrocarbon contamination and its bioremediation using compost amendment. J. Proteomics 135, 162–169. doi:10.1016/j.jprot.2015.07.023
Behrendorff, J. B. (2021). Reductive cytochrome P450 reactions and their potential role in bioremediation. Front. Microbiol. 12, 649273. doi:10.3389/fmicb.2021.649273
Bhandari, G., Dhasmana, A., Chaudhary, P., Gupta, S., Gangola, S., Gupta, A., et al. (2023). A perspective review on green nanotechnology in agro-ecosystems: Opportunities for sustainable agricultural practices and environmental remediation. Agriculture 13, 668. doi:10.3390/agriculture13030668
Bhandari, S., Poudel, D. K., Marahatha, R., Dawadi, S., Khadayat, K., Phuyal, S., et al. (2021). Microbial enzymes used in bioremediation. J. Chem. 2021, 1–17. doi:10.1155/2021/8849512
Bharagava, R. N., and Mishra, S. (2018). Hexavalent chromium reduction potential of Cellulosimicrobium sp. isolated from common effluent treatment plant of tannery industries. Ecotoxicol. Environ. Saf. 147, 102–109. doi:10.1016/j.ecoenv.2017.08.040
Bhatt, P., Gangola, S., Chaudhary, P., Khati, P., Kumar, G., Sharma, A., et al. (2019). Pesticide induced up-regulation of esterase and aldehyde dehydrogenase in indigenous Bacillus spp. Bioremediation J. 23, 42–52. doi:10.1080/10889868.2019.1569586
Bhunia, B., and Basak, B. (2014). A review on application of microbial protease in bioremediation. Industrial Environ. Biotechnol. 1, 217–228.
Bianco, F., Race, M., Papirio, S., and Esposito, G. (2020). Removal of polycyclic aromatic hydrocarbons during anaerobic biostimulation of marine sediments. Sci. Total Environ. 709, 136141. doi:10.1016/j.scitotenv.2019.136141
Bogati, K., and Walczak, M. (2022). The impact of drought stress on soil microbial community, enzyme activities and plants. Agronomy 12, 189. doi:10.3390/agronomy12010189
Bolade, O. P., Akinsiku, A. A., Oluwafemi, O. S., Williams, A. B., and Benson, N. U. (2021). Biogenic iron oxide nanoparticles and activated sodium persulphate for hydrocarbon remediation in contaminated soil. Environ. Technol. Innov. 23, 101719. doi:10.1016/j.eti.2021.101719
Boregowda, N., Jogigowda, S. C., Bhavya, G., Sunilkumar, C. R., Geetha, N., Udikeri, S. S., et al. (2022). Recent advances in nanoremediation: Carving sustainable solution to clean-up polluted agriculture soils. Environ. Pollut. 297, 118728. doi:10.1016/j.envpol.2021.118728
Cao, X., Xu, L., Chen, Y. P., Decho, A. W., Cui, Z., and Lead, J. R. A. (2022). Contribution, composition, and structure of eps by in vivo exposure to elucidate the mechanisms of nanoparticle-enhanced bioremediation to metals. Environ. Sci. Technol. 56, 896–906. doi:10.1021/acs.est.1c05326
Cecchin, I., Reddy, K. R., Thomé, A., Tessaro, E. F., and Schnaid, F. (2017). Nanobioremediation: Integration of nanoparticles and bioremediation for sustainable remediation of chlorinated organic contaminants in soils. Int. Biodeterior. Biodegrad. 119, 419–428. doi:10.1016/j.ibiod.2016.09.027
Celestina, C., Wood, J. L., Manson, J. B., Wang, X., Sale, P. W., Tang, C., et al. (2019). Microbial communities in top-and subsoil of repacked soil columns respond differently to amendments but their diversity is negatively correlated with plant productivity. Scientific Reports 9 (1), 8890.
Chaudhary, P., Ahamad, L., Chaudhary, A., Kumar, G., Chen, W.-J., and Chen, S. (2023a). Nanoparticle-mediated bioremediation as a powerful weapon in the removal of environmental pollutants. J. Environ. Chem. Eng. 11, 109591. doi:10.1016/j.jece.2023.109591
Chaudhary, P., Singh, S., Chaudhary, A., Agri, U., and Bhandari, G. (2022). Agrochemicals and their effects on soil microbial population. Plant Prot. Chem. Biol. 1, 45.
Chaudhary, P., Xu, M., Ahamad, L., Chaudhary, A., Kumar, G., Adeleke, B. S., et al. (2023b). Application of synthetic consortia for improvement of soil fertility, pollution remediation, and agricultural productivity: A review. Agronomy 13, 643. doi:10.3390/agronomy13030643
Chellaiah, E. R. (2018). Cadmium (heavy metals) bioremediation by Pseudomonas aeruginosa: A minireview. Appl. water Sci. 8, 154–210. doi:10.1007/s13201-018-0796-5
Chikere, C. B., Azubuike, C. C., and Fubara, E. M. (2017). Shift in microbial group during remediation by enhanced natural attenuation (RENA) of a crude oil-impacted soil: A case study of Ikarama community, Bayelsa, Nigeria. Biotech 7, 152–211. doi:10.1007/s13205-017-0782-x
Chikere, C. B., Tekere, M., and Adeleke, R. (2019). Enhanced microbial hydrocarbon biodegradation as stimulated during field-scale landfarming of crude oil-impacted soil. Sustain. Chem. Pharm. 14, 100177. doi:10.1016/j.scp.2019.100177
Costa, C. S. D., Bertagnolli, C., Boos, A., Da Silva, M. G. C., and Vieira, M. G. A. (2020). Application of a dealginated seaweed derivative for the simultaneous metal ions removal from real and synthetic effluents. J. Water Process Eng. 37, 101546. doi:10.1016/j.jwpe.2020.101546
Dai, X., Lv, J., Yan, G., Chen, C., Guo, S., and Fu, P. (2020). Bioremediation of intertidal zones polluted by heavy oil spilling using immobilized laccase-bacteria consortium. Bioresour. Technol. 309, 123305. doi:10.1016/j.biortech.2020.123305
Dwivedi, S. (2012). Bioremediation of heavy metal by algae: Current and future perspective. J. Adv. Lab. Res. Biol. 3, 195–199.
Ebuehi, O., Abibo, I., Shekwolo, P., Sigismund, K., Adoki, A., and Okoro, I. (2005). Remediation of crude oil contaminated soil by enhanced natural attenuation technique. Jasem 9, 103–106.
El Enshasy, H. A., Hanapi, S. Z., Abdelgalil, S. A., Malek, R. A., and Pareek, A. (2017). Mycoremediation: Decolourization potential of fungal ligninolytic enzymes. Mycoremediation Environ. Sustain. 1 (1), 69–104.
El-Ansary, M. S. M., and Ahmed-Farid, O. A. (2021). Bioremediation of oxyl compounds by algae: Description and traits of root-knot nematode control. Waste Biomass Valorization 12, 251–261. doi:10.1007/s12649-020-00950-5
El-Naggar, N. E.-A., Hamouda, R. A., Mousa, I. E., Abdel-Hamid, M. S., and Rabei, N. H. (2018). Biosorption optimization, characterization, immobilization and application of Gelidium amansii biomass for complete Pb2+ removal from aqueous solutions. Sci. Rep. 8, 13456–13519. doi:10.1038/s41598-018-31660-7
El-Ramady, H., El-Henawy, A., Amer, M., Omara, A. E.-D., Elsakhawy, T., Salama, A.-M., et al. (2020). Agro-pollutants and their nano-remediation from soil and water: A mini-review. Environ. Biodivers. Soil Secur. 4, 0–375. doi:10.21608/jenvbs.2020.47751.1111
El-Sheekh, M., and Hamouda, R. (2014). Biodegradation of crude oil by some cyanobacteria under heterotrophic conditions. Desalination Water Treat. 52, 1448–1454. doi:10.1080/19443994.2013.794008
Esringü, A., Turan, M., Güneş, A., and Karaman, M. R. (2014). Roles of Bacillus megaterium in remediation of boron, lead, and cadmium from contaminated soil. Commun. soil Sci. plant analysis 45, 1741–1759. doi:10.1080/00103624.2013.875194
Essabri, A., Aydinlik, N. P., and Williams, N. E. (2019). Bioaugmentation and biostimulation of total petroleum hydrocarbon degradation in a petroleum-contaminated soil with fungi isolated from olive oil effluent. Water, Air, Soil Pollut. 230, 76–16. doi:10.1007/s11270-019-4127-8
Evode, N., Qamar, S. A., Bilal, M., Barceló, D., and Iqbal, H. M. (2021). Plastic waste and its management strategies for environmental sustainability. Case Stud. Chem. Environ. Eng. 4, 100142. doi:10.1016/j.cscee.2021.100142
Ezekoye, C., Chikere, C., and Okpokwasili, G. (2018). Fungal diversity associated with crude oil-impacted soil undergoing in-situ bioremediation. Sustain. Chem. Pharm. 10, 148–152. doi:10.1016/j.scp.2018.11.003
Fabre, E., Dias, M., Costa, M., Henriques, B., Vale, C., Lopes, C. B., et al. (2020). Negligible effect of potentially toxic elements and rare Earth elements on mercury removal from contaminated waters by green, Brown and red living marine macroalgae. Sci. Total Environ. 724, 138133. doi:10.1016/j.scitotenv.2020.138133
Fu, P., and Secundo, F. (2016). Algae and their bacterial consortia for soil bioremediation. Chem. Eng. Trans. 49, 427–432.
Gargouri, B., Mhiri, N., Karray, F., Aloui, F., and Sayadi, S. (2015). Isolation and characterization of hydrocarbon-degrading yeast strains from petroleum contaminated industrial wastewater. BioMed Res. Int. 2015, 1–11. doi:10.1155/2015/929424
Ghosal, D., Ghosh, S., Dutta, T. K., and Ahn, Y. (2016). Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A review. Front. Microbiol. 7, 1369. doi:10.3389/fmicb.2016.01369
Ghosh, P., and Ghosh, U. (2018). Bioconversion of agro-waste to value-added product through solid-state fermentation by a potent fungal strain Aspergillus flavus PUF5. Utilization and Management of Bioresources. Springer.
Ghosh, S., Sharma, I., Nath, S., and Webster, T. J. (2021). Bioremediation—the natural solution. Microbial ecology of wastewater treatment plants. Elsevier.
Gomes, M., Gonzales-Limache, E., Sousa, S., Dellagnezze, B., Sartoratto, A., Silva, L., et al. (2018). Exploring the potential of halophilic bacteria from oil terminal environments for biosurfactant production and hydrocarbon degradation under high-salinity conditions. Int. Biodeterior. Biodegrad. 126, 231–242. doi:10.1016/j.ibiod.2016.08.014
Gong, J.-S., Lu, Z.-M., Li, H., Shi, J.-S., Zhou, Z.-M., and Xu, Z.-H. (2012). Nitrilases in nitrile biocatalysis: Recent progress and forthcoming research. Microb. Cell factories 11, 142–218. doi:10.1186/1475-2859-11-142
Guo, Y., Rene, E. R., Wang, J., and Ma, W. (2020). Biodegradation of polyaromatic hydrocarbons and the influence of environmental factors during the co-composting of sewage sludge and green forest waste. Bioresour. Technol. 297, 122434. doi:10.1016/j.biortech.2019.122434
Havugimana, E., Bhople, B. S., Kumar, A., Byiringiro, E., Mugabo, J. P., and Kumar, A. (2017). Soil pollution–major sources and types of soil pollutants. Environ. Sci. Eng. 11, 53–86.
Hualpa-Cutipa, E., Acosta, R. A. S., Cariga, O. J. M., Espinoza-Medina, M. A., Chavez-Rojas, D. C., Medina-Cerna, D., et al. (2023). Metagenomic approach role of psychrotrophic and psychrophilic microbes in bioremediation. Metagenomics to Bioremediation. Elsevier.
Hussain, I., Puschenreiter, M., Gerhard, S., Sani, S. G. A. S., Khan, W.-U.-D., and Reichenauer, T. G. (2019). Differentiation between physical and chemical effects of oil presence in freshly spiked soil during rhizoremediation trial. Environ. Sci. Pollut. Res. 26, 18451–18464. doi:10.1007/s11356-019-04819-6
Igiri, B. E., Okoduwa, S. I., Idoko, G. O., Akabuogu, E. P., Adeyi, A. O., and Ejiogu, I. K. (2018). Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: A review. J. Toxicol., 2018, 1–16. doi:10.1155/2018/2568038
Inthama, P., Pumas, P., Pekkoh, J., Pathom-Aree, W., and Pumas, C. (2021). Plant growth and drought tolerance-promoting bacterium for bioremediation of paraquat pesticide residues in agriculture soils. Front. Microbiol. 12, 604662. doi:10.3389/fmicb.2021.604662
Jayakumar, V., Govindaradjane, S., Rajamohan, N., and Rajasimman, M. (2022). Biosorption potential of Brown algae, Sargassum polycystum, for the removal of toxic metals, cadmium and zinc. Environ. Sci. Pollut. Res. 29, 41909–41922. doi:10.1007/s11356-021-15185-7
Jayakumar, V., Govindaradjane, S., and Rajasimman, (2021). Efficient adsorptive removal of Zinc by green marine macro alga Caulerpa scalpelliformis–characterization, optimization, modeling, isotherm, kinetic, thermodynamic, desorption and regeneration studies. Surfaces Interfaces 22, 100798. doi:10.1016/j.surfin.2020.100798
Ji, M., Li, B., Majdi, A., Alkhalifah, T., Alturise, F., and Ali, H. E. (2023). Application of nano remediation of mine polluted in acid mine drainage water using machine learning model. Chemosphere 311, 136926. doi:10.1016/j.chemosphere.2022.136926
Jia, H., Zhang, M., Weng, Y., Zhao, Y., Li, C., and Kanwal, A. (2021). Degradation of poly (butylene adipate-co-terephthalate) by Stenotrophomonas sp. YCJ1 isolated from farmland soil. J. Environ. Sci. 103, 50–58. doi:10.1016/j.jes.2020.10.001
Jiang, Y., Jiang, S., Li, Z., Yan, X., Qin, Z., and Huang, R. (2019). Field scale remediation of Cd and Pb contaminated paddy soil using three mulberry (Morus alba L) cultivars. Ecol. Eng. 129, 38–44. doi:10.1016/j.ecoleng.2019.01.009
Jin, Y., Luan, Y., Ning, Y., and Wang, L. (2018). Effects and mechanisms of microbial remediation of heavy metals in soil: A critical review. Appl. Sci. 8, 1336. doi:10.3390/app8081336
Kafle, A., Timilsina, A., Gautam, A., Adhikari, K., Bhattarai, A., and Aryal, N. (2022). Phytoremediation: Mechanisms, plant selection and enhancement by natural and synthetic agents. Environ. Adv. 8, 100203. doi:10.1016/j.envadv.2022.100203
Kalaimurugan, D., Balamuralikrishnan, B., Durairaj, K., Vasudhevan, P., Shivakumar, M., Kaul, T., et al. (2020). Isolation and characterization of heavy-metal-resistant bacteria and their applications in environmental bioremediation. Int. J. Environ. Sci. Technol. 17, 1455–1462. doi:10.1007/s13762-019-02563-5
Kalhor, A. X., Movafeghi, A., Mohammadi-Nassab, A. D., Abedi, E., and Bahrami, A. (2017). Potential of the green alga Chlorella vulgaris for biodegradation of crude oil hydrocarbons. Mar. Pollut. Bull. 123, 286–290. doi:10.1016/j.marpolbul.2017.08.045
Kalola, V., and Desai, C. (2020). Biosorption of Cr (VI) by Halomonas sp. DK4, a halotolerant bacterium isolated from chrome electroplating sludge. Environ. Sci. Pollut. Res. 27, 27330–27344. doi:10.1007/s11356-019-05942-0
Kaminsky, L. M., Trexler, R. V., Malik, R. J., Hockett, K. L., and Bell, T. H. (2019). The inherent conflicts in developing soil microbial inoculants. Trends Biotechnol. 37, 140–151. doi:10.1016/j.tibtech.2018.11.011
Kanwal, M., Ullah, H., Gulzar, A., Sadiq, T., Gul, Z., Ullah, M., et al. (2022). Biodegradation of petroleum hydrocarbons and the factors effecting rate of biodegradation. Am. J. Biomed. Sci. Res. 16, 6–15. doi:10.34297/ajbsr.2022.16.002182
Karthika, A., Seenivasagan, R., Kasimani, R., Babalola, O. O., and Vasanthy, M. (2017). The role of Eudrillus eugenia in the degradation of paper cup waste and the morphological, physiological and histological changes in the organism. Bioremediation Sustain. Technol. Clean. Environ. 1, 65–76.
Karthika, A., Seenivasagan, R., Kasimani, R., Babalola, O., and Vasanthy, M. (2020). Cellulolytic bacteria isolation, screening and optimization of enzyme production from vermicompost of paper cup waste. Waste Manag. 116, 58–65. doi:10.1016/j.wasman.2020.06.036
Kebede, G., Tafese, T., Abda, E. M., Kamaraj, M., and Assefa, F. (2021). Factors influencing the bacterial bioremediation of hydrocarbon contaminants in the soil: Mechanisms and impacts. J. Chem. 2021, 1–17. doi:10.1155/2021/9823362
Kong, F.-X., Sun, G.-D., and Liu, Z.-P. (2018). Degradation of polycyclic aromatic hydrocarbons in soil mesocosms by microbial/plant bioaugmentation: Performance and mechanism. Chemosphere 198, 83–91. doi:10.1016/j.chemosphere.2018.01.097
Košnář, Z., Částková, T., Wiesnerová, L., Praus, L., Jablonský, I., Koudela, M., et al. (2019). Comparing the removal of polycyclic aromatic hydrocarbons in soil after different bioremediation approaches in relationto the extracellular enzyme activities. J. Environ. Sci. 76, 249–258. doi:10.1016/j.jes.2018.05.007
Kumar, A., Das, S. K., Nainegali, L., and Reddy, K. R. (2023). Phytostabilization of coalmine overburden waste rock dump slopes: Current status, challenges, and perspectives. Bull. Eng. Geol. Environ. 82, 130. doi:10.1007/s10064-023-03159-7
Kumar, A., Yadav, A. N., Mondal, R., Kour, D., Subrahmanyam, G., Shabnam, A. A., et al. (2021a). Myco-remediation: A mechanistic understanding of contaminants alleviation from natural environment and future prospect. Chemosphere 284, 131325. doi:10.1016/j.chemosphere.2021.131325
Kumar, G., Lal, S., Maurya, S. K., Bhattacherjee, A., Chaudhary, P., Gangola, S., et al. (2021b). Exploration of Klebsiella pneumoniae M6 for paclobutrazol degradation, plant growth attributes, and biocontrol action under subtropical ecosystem. Plos one 16, e0261338. doi:10.1371/journal.pone.0261338
Kumar, G., Lal, S., Soni, S. K., Maurya, S. K., Shukla, P. K., Chaudhary, P., et al. (2022). Mechanism and kinetics of chlorpyrifos co-metabolism by using environment restoring microbes isolated from rhizosphere of horticultural crops under subtropics. Front. Microbiol. 13, 891870. doi:10.3389/fmicb.2022.891870
Kumar, L., and Jain, S. K. (2020). Role of proteases in bioremediation of temple protein-containing waste with special reference to mangalnath, ujjain (MP)–India. Indian J. Pure Appl. Biosci. 8, 602–607. doi:10.18782/2582-2845.8178
Lara-Moreno, A., Morillo, E., Merchán, F., Madrid, F., and Villaverde, J. (2022). Bioremediation of a trifluralin contaminated soil using bioaugmentation with novel isolated bacterial strains and cyclodextrin. Sci. Total Environ. 840, 156695. doi:10.1016/j.scitotenv.2022.156695
Lee, E.-H., Kang, Y.-S., and Cho, K.-S. (2011). Bioremediation of diesel-contaminated soils by natural attenuation, biostimulation and bioaugmentation employing Rhodococcus sp. EH831. Microbiol. Biotechnol. Lett. 39, 86–92.
Li, F., Zheng, Y., Tian, J., Ge, F., Liu, X., Tang, Y., et al. (2019a). Cupriavidus sp. strain Cd02-mediated pH increase favoring bioprecipitation of Cd2+ in medium and reduction of cadmium bioavailability in paddy soil. Ecotoxicol. Environ. Saf. 184, 109655. doi:10.1016/j.ecoenv.2019.109655
Li, Q., Liu, J., and Gadd, G. M. (2020). Fungal bioremediation of soil co-contaminated with petroleum hydrocarbons and toxic metals. Appl. Microbiol. Biotechnol. 104, 8999–9008. doi:10.1007/s00253-020-10854-y
Li, T., Liu, Y., Lin, S., Liu, Y., and Xie, Y. (2019b). Soil pollution management in China: A brief introduction. Sustainability 11, 556–615. doi:10.3390/su11030556
Li, Y., Chi, M., and Ge, X. (2019c). Identification of a novel hydrolase encoded by hy-1 from Bacillus amyloliquefaciens for bioremediation of carbendazim contaminated soil and food. Int. J. Agric. Biol. Eng. 12, 218–224. doi:10.25165/j.ijabe.20191202.4190
Lin, S., Wei, J., Yang, B., Zhang, M., and Zhuo, R. (2022). Bioremediation of organic pollutants by white rot fungal cytochrome P450: The role and mechanism of CYP450 in biodegradation. Chemosphere 301, 134776. doi:10.1016/j.chemosphere.2022.134776
Liu, P., Rao, D., Zou, L., Teng, Y., and Yu, H. (2021). Capacity and potential mechanisms of Cd (II) adsorption from aqueous solution by blue algae-derived biochars. Sci. Total Environ. 767, 145447. doi:10.1016/j.scitotenv.2021.145447
Long, Z., Huang, Y., Zhang, W., Shi, Z., Yu, D., Chen, Y., et al. (2021). Effect of different industrial activities on soil heavy metal pollution, ecological risk, and health risk. Environ. Monit. Assess. 193, 20–12. doi:10.1007/s10661-020-08807-z
Mafiana, M. O., Bashiru, M. D., Erhunmwunsee, F., Dirisu, C. G., and Li, S.-W. (2021). An insight into the current oil spills and on-site bioremediation approaches to contaminated sites in Nigeria. Environ. Sci. Pollut. Res. 28, 4073–4094. doi:10.1007/s11356-020-11533-1
Mai, C. T. N., Linh, N. V., Lich, N. Q., Ha, H. P., Van Quyen, D., Tang, D. Y. Y., et al. (2021). Advanced materials for immobilization of purple phototrophic bacteria in bioremediation of oil-polluted wastewater. Chemosphere 278, 130464. doi:10.1016/j.chemosphere.2021.130464
Malakootian, M., Mahdizadeh, H., Nasiri, A., Mirzaienia, F., Hajhoseini, M., and Amirmahani, N. (2018). Investigation of the efficiency of microbial desalination cell in removal of arsenic from aqueous solutions. Desalination 438, 19–23. doi:10.1016/j.desal.2018.03.025
Maltman, C., and Yurkov, V. (2018). Bioremediation potential of bacteria able to reduce high levels of selenium and tellurium oxyanions. Archives Microbiol. 200, 1411–1417. doi:10.1007/s00203-018-1555-6
Martins, M., Santos, E. S., Faleiro, M. L., Chaves, S., Tenreiro, R., Barros, R. J., et al. (2011). Performance and bacterial community shifts during bioremediation of acid mine drainage from two Portuguese mines. Int. Biodeterior. Biodegrad. 65, 972–981. doi:10.1016/j.ibiod.2011.07.006
Marzan, L. W., Hossain, M., Mina, S. A., Akter, Y., and Chowdhury, A. M. A. (2017). Isolation and biochemical characterization of heavy-metal resistant bacteria from tannery effluent in Chittagong city, Bangladesh: Bioremediation viewpoint. Egypt. J. Aquatic Res. 43, 65–74. doi:10.1016/j.ejar.2016.11.002
Masowa, M., Kutu, F., Babalola, O., and Mulidzi, A. (2022). Optimizing application rate of winery solid waste compost for improving the performance of maize (zea mays L) grown on luvisol and cambisol. Appl. Ecol. Environ. Res. 20, 815–828. doi:10.15666/aeer/2001_815828
Masowa, M., Kutu, F., Babalola, O., and Mulidzi, A. (2018). Physico-chemical properties and phyto-toxicity assessment of co-composted winery solid wastes with and without effective microorganism inoculation. Res. Crops 19, 549–559.
Masowa, M. M., Kutu, F. R., Babalola, O. O., Mulidzi, A. R., and Dlamini, P. (2021). Effects of complementary and sole applications of inorganic fertilizers and winery solid waste compost on maize yield and soil health indices. Emir. J. Food Agric. 1, 565–574. doi:10.9755/ejfa.2021.v33.i7.2721
Mbachu, A., Chukwura, E., and Mbachu, N. (2016). Isolation and characterization of hydrocarbon degrading fungi from used (spent) engine oil polluted soil and their use for polycyclic aromatic hydrocarbons (PAHs) degradation. Univers. J. Microbiol. Res. 4, 31–37. doi:10.13189/ujmr.2016.040105
Medaura, M. C., Guivernau, M., Moreno-Ventas, X., Prenafeta-Boldú, F. X., and Viñas, M. (2021). Bioaugmentation of native fungi, an efficient strategy for the bioremediation of an aged industrially polluted soil with heavy hydrocarbons. Front. Microbiol. 12, 626436. doi:10.3389/fmicb.2021.626436
Meng, X., Fan, K., and Yan, X. (2019). Nanozymes: An emerging field bridging nanotechnology and enzymology. Sci. China Life Sci. 62, 1543–1546. doi:10.1007/s11427-019-1557-8
Minari, G. D., Saran, L. M., Constancio, M. T. L., Da Silva, R. C., Rosalen, D. L., De Melo, W. J., et al. (2020). Bioremediation potential of new cadmium, chromium, and nickel-resistant bacteria isolated from tropical agricultural soil. Ecotoxicol. Environ. Saf. 204, 111038. doi:10.1016/j.ecoenv.2020.111038
Mitra, S., Pramanik, K., Sarkar, A., Ghosh, P. K., Soren, T., and Maiti, T. K. (2018). Bioaccumulation of cadmium by Enterobacter sp. and enhancement of rice seedling growth under cadmium stress. Ecotoxicol. Environ. Saf. 156, 183–196. doi:10.1016/j.ecoenv.2018.03.001
Mnif, S., Chebbi, A., Mhiri, N., Sayadi, S., and Chamkha, M. (2017). Biodegradation of phenanthrene by a bacterial consortium enriched from Sercina oilfield. Process Saf. Environ. Prot. 107, 44–53. doi:10.1016/j.psep.2017.01.023
Moino, B. P., Costa, C. S., Da Silva, M. G., and Vieira, M. G. (2017). Removal of nickel ions on residue of alginate extraction from Sargassum <scp>f</scp>ilipendula seaweed in packed bed. Can. J. Chem. Eng. 95, 2120–2128. doi:10.1002/cjce.22859
Montazer, Z., Habibi Najafi, M. B., and Levin, D. B. (2019). Microbial degradation of low-density polyethylene and synthesis of polyhydroxyalkanoate polymers. Can. J. Microbiol. 65, 224–234. doi:10.1139/cjm-2018-0335
Mosharaf, M., Tanvir, M., Haque, M., Haque, M., Khan, M., Molla, A. H., et al. (2018). Metal-adapted bacteria isolated from wastewaters produce biofilms by expressing proteinaceous curli fimbriae and cellulose nanofibers. Front. Microbiol. 9, 1334. doi:10.3389/fmicb.2018.01334
Mostafa, A. A.-F., Yassin, M. T., Dawoud, T. M., Al-Otibi, F. O., and Sayed, S. R. (2022). Mycodegradation of diazinon pesticide utilizing fungal strains isolated from polluted soil. Environ. Res. 212, 113421. doi:10.1016/j.envres.2022.113421
Mukhopadhyay, R., Sarkar, B., Khan, E., Alessi, D. S., Biswas, J. K., Manjaiah, K., et al. (2022). Nanomaterials for sustainable remediation of chemical contaminants in water and soil. Crit. Rev. Environ. Sci. Technol. 52, 2611–2660. doi:10.1080/10643389.2021.1886891
Naeem, U., and Qazi, M. A. (2020). Leading edges in bioremediation technologies for removal of petroleum hydrocarbons. Environ. Sci. Pollut. Res. 27, 27370–27382. doi:10.1007/s11356-019-06124-8
Nascimento, S. V. D., Costa, P. H. D. O., Herrera, H., Caldeira, C. F., Gastauer, M., Ramos, S. J., et al. (2022). Proteomic profiling and rhizosphere-associated microbial communities reveal adaptive mechanisms of dioclea apurensis kunth in eastern amazon’s rehabilitating minelands. Plants 11, 712. doi:10.3390/plants11050712
Naskar, A., Majumder, R., and Goswami, M. (2020). Bioaccumulation of Ni (II) on growing cells of Bacillus sp.: Response surface modeling and mechanistic insight. Environ. Technol. Innovation 20, 101057. doi:10.1016/j.eti.2020.101057
Nath, S., Deb, B., and Sharma, I. (2018). Isolation of toxic metal-tolerant bacteria from soil and examination of their bioaugmentation potentiality by pot studies in cadmium-and lead-contaminated soil. Int. Microbiol. 21, 35–45. doi:10.1007/s10123-018-0003-4
Naz, R., Khan, M., Hafeez, A., Fazil, M., Khan, M., Ali, B., et al. (2022). Assessment of phytoremediation potential of native plant species naturally growing in a heavy metal-polluted industrial soils. Braz. J. Biol. 84, e264473. doi:10.1590/1519-6984.264473
Naz, T., Khan, M. D., Ahmed, I., Rehman, S. U., Rha, E. S., Malook, I., et al. (2016). Biosorption of heavy metals by Pseudomonas species isolated from sugar industry. Toxicol. Ind. health 32, 1619–1627. doi:10.1177/0748233715569900
Ndeddy Aka, R. J., and Babalola, O. O. (2016). Effect of bacterial inoculation of strains of Pseudomonas aeruginosa, Alcaligenes feacalis and Bacillus subtilis on germination, growth and heavy metal (Cd, Cr, and Ni) uptake of Brassica juncea. Int. J. Phytoremediation 18, 200–209. doi:10.1080/15226514.2015.1073671
Ndeddy Aka, R. J., and Babalola, O. O. (2017). Identification and characterization of Cr-Cd-and Ni-tolerant bacteria isolated from mine tailings. Bioremediation J. 21, 1–19. doi:10.1080/10889868.2017.1282933
Nguyen, D. D., Ha, M.-G., and Kang, H. Y. (2021). Potential of versatile bacteria isolated from activated sludge for the bioremediation of arsenic and antimony. J. Water Process Eng. 39, 101890. doi:10.1016/j.jwpe.2020.101890
Nishikawa, E., Da Silva, M. G. C., and Vieira, M. G. A. (2018). Cadmium biosorption by alginate extraction waste and process overview in life cycle assessment context. J. Clean. Prod. 178, 166–175. doi:10.1016/j.jclepro.2018.01.025
Nuhu, M. M., Rene, E. R., and Ishaq, A. (2022). Remediation of crude oil spill sites in Nigeria: Problems, technologies, and future prospects. Environ. Qual. Manag. 31, 165–175.
O’Brien, R. M., Phelan, T. J., Smith, N. M., and Smits, K. M. (2021). Remediation in developing countries: A review of previously implemented projects and analysis of stakeholder participation efforts. Crit. Rev. Environ. Sci. Technol. 51, 1259–1280. doi:10.1080/10643389.2020.1755203
Oghoje, S., and Ukpebor, J. (2020). The effects and efficacy of chicken manure digestates on bioremediation of petroleum hydrocarbons polluted soils. Niger. Res. J. Chem. Sci. 8, 311–328.
Oghoje, S., Ukpebor, J., and Ukpebor, E. (2021). The effects of chicken manure digestates on the removal of diesel range organics from petroleum products polluted soils. Bulg. J. Soil Sci. 6, 78–95. doi:10.5281/zenodo.4887779
Ojuederie, O. B., and Babalola, O. O. (2017). Microbial and plant-assisted bioremediation of heavy metal polluted environments: A review. Int. J. Environ. Res. public health 14, 1504. doi:10.3390/ijerph14121504
Okoye, A., Chikere, C., and Okpokwasili, G. (2019). Fungal population dynamics associated with active-phase of hydrocarbon degradation in oil-polluted soil. J. Adv. Microbiol. 19, 1–12. doi:10.9734/jamb/2019/v19i230190
Orji, F., Ibiene, A., and Ugbogu, O. (2012). Petroleum hydrocarbon pollution of mangrove swamps: The promises of remediation by enhanced natural attenuation. Am. J. Agric. Biol. Sci. 7, 207–216. doi:10.3844/ajabssp.2012.207.216
Pahalvi, H. N., Rafiya, L., Rashid, S., Nisar, B., and Kamili, A. N. (2021). Chemical fertilizers and their impact on soil health. Microbiota Biofertilizers Ecofriendly Tools Reclam. Degraded Soil Environs 2, 1–20.
Pande, V., Pandey, S. C., Sati, D., Pande, V., and Samant, M. (2020). Bioremediation: An emerging effective approach towards environment restoration. Environ. Sustain. 3, 91–103. doi:10.1007/s42398-020-00099-w
Park, J. M., Trevor Sewell, B., and Benedik, M. J. (2017). Cyanide bioremediation: The potential of engineered nitrilases. Appl. Microbiol. 101, 3029–3042. doi:10.1007/s00253-017-8204-x
Park, S. Y., and Kim, C. G. (2019). Biodegradation of micro-polyethylene particles by bacterial colonization of a mixed microbial consortium isolated from a landfill site. Chemosphere 222, 527–533. doi:10.1016/j.chemosphere.2019.01.159
Parsamanesh, S., and Sadeghi, H. (2019). The phytoremediation effect of Medicago scutellata (L) mill. On soils under Cd–water stress: A good choice for contaminated dry lands. Environ. Sci. Pollut. Res. 26, 29065–29073. doi:10.1007/s11356-019-05989-z
Parveen, H., Chaudhary, A., Chaudhary, P., Sultana, R., Kumar, G., Khati, P., et al. (2022). Omics approaches for the production of the microbial enzymes and applications. Industrial Applications of microbial enzymes. CRC Press.
Patel, A. B., Shaikh, S., Jain, K. R., Desai, C., and Madamwar, D. (2020). Polycyclic aromatic hydrocarbons: Sources, toxicity, and remediation approaches. Front. Microbiol. 11, 562813. doi:10.3389/fmicb.2020.562813
Patel, J. G., Nirmal Kumar, J., Kumar, R. N., and Khan, S. R. (2015). Enhancement of pyrene degradation efficacy of Synechocystis sp., by construction of an artificial microalgal-bacterial consortium. Cogent Chem. 1, 1064193. doi:10.1080/23312009.2015.1064193
Patra, D. K., Grahacharya, A., Pradhan, C., and Patra, H. K. (2021). Phytoremediation potential of coffee pod (Cassia tora): An in situ approach for attenuation of chromium from overburden soil of sukinda chromite mine, India. Environ. Prog. Sustain. Energy 40, e13510.
Paul, A., and Mukherjee, S. K. (2016). Enterobacter asburiae KUNi5, a nickel resistant bacterium for possible bioremediation of nickel contaminated sites. Pol. J. Microbiol. 65, 115–118. doi:10.5604/17331331.1197284
Pawar, R. (2015). The effect of soil pH on bioremediation of polycyclic aromatic hydrocarbons (PAHS). Bioremediat Biodegr. 2015, 291–304. doi:10.4172/2155-6199.1000291
Pidlisnyuk, V., Hettiarachchi, G. M., Zgorelec, Z., Prelac, M., Bilandžija, N., Davis, L. C., et al. (2021). Phytotechnologies for site remediation. Phytotechnology with biomass production. CRC Press.
Pm Tavares, A., R Pereira, S., and Mrb Xavier, A. (2017). Biotechnological applications of Trametes versicolor and their enzymes. Curr. Biotechnol. 6, 78–88. doi:10.2174/2211550105666160510113212
Princy, S., Sathish, S. S., Cibichakravarthy, B., and Prabagaran, S. R. (2020). Hexavalent chromium reduction by Morganella morganii (1Ab1) isolated from tannery effluent contaminated sites of Tamil Nadu, India. Biocatal. Agric. Biotechnol. 23, 101469. doi:10.1016/j.bcab.2019.101469
Priya, A., Dutta, K., and Daverey, A. (2022). A comprehensive biotechnological and molecular insight into plastic degradation by microbial community. J. Chem. Technol. Biotechnol. 97, 381–390. doi:10.1002/jctb.6675
Rahman, Z., Thomas, L., and Singh, V. P. (2019). Biosorption of heavy metals by a lead (Pb) resistant bacterium, Staphylococcus hominis strain AMB-2. J. basic Microbiol. 59, 477–486. doi:10.1002/jobm.201900024
Raju, C. A., Anitha, J., Kalyani, R. M., Satyanandam, K., and Jagadeesh, P. (2021). Sorption of cobalt using marine macro seaweed graciliariacorticatared algae powder. Mater. Today Proc. 44, 1816–1827. doi:10.1016/j.matpr.2020.12.009
Ramírez, V., Baez, A., López, P., Bustillos, R., Villalobos, M. Á., Carreño, R., et al. (2019). Chromium hyper-tolerant Bacillus sp. MH778713 assists phytoremediation of heavy metals by mesquite trees (Prosopis laevigata). Front. Microbiol. 10, 1833. doi:10.3389/fmicb.2019.01833
Rawat, M., and Rangarajan, S. (2019). Omics approaches for elucidating molecular mechanisms of microbial bioremediation. Smart bioremediation technologies. Elsevier.
Riser-Roberts, E. (2020). Remediation of petroleum contaminated soils: Biological, physical, and chemical processes. Boca Raton, Florida, United State: CRC Press.
Sahmoune, M. N. (2018). Performance of Streptomyces rimosus biomass in biosorption of heavy metals from aqueous solutions. Microchem. J. 141, 87–95. doi:10.1016/j.microc.2018.05.009
Salama, E.-S., Roh, H.-S., Dev, S., Khan, M. A., Abou-Shanab, R. A., Chang, S. W., et al. (2019). Algae as a green technology for heavy metals removal from various wastewater. World J. Microbiol. Biotechnol. 35, 75–19. doi:10.1007/s11274-019-2648-3
Samreen, S., Khan, A. A., Khan, M. R., Ansari, S. A., and Khan, A. (2021). Assessment of phytoremediation potential of seven weed plants growing in chromium-and nickel-contaminated soil. Water, Air, Soil Pollut. 232, 209–218. doi:10.1007/s11270-021-05124-0
Saranya, K., Sundaramanickam, A., Shekhar, S., Meena, M., Sathishkumar, R. S., and Balasubramanian, T. (2018). Biosorption of multi-heavy metals by coral associated phosphate solubilising bacteria Cronobacter muytjensii KSCAS2. J. Environ. Manag. 222, 396–401. doi:10.1016/j.jenvman.2018.05.083
Sari, E. M., Novianty, R., Awaluddin, A., and Pratiwi, N. W. (2019). Effectiveness of crude oil degrading fungi isolated from petroleum hydrocarbon contaminated soil in Siak, Riau. Acta Biochim. Indones. 2, 15–22. doi:10.32889/actabioina.v2i1.35
Sarma, H., Sonowal, S., and Prasad, M. (2019). Plant-microbiome assisted and biochar-amended remediation of heavy metals and polyaromatic compounds─ a microcosmic study. Ecotoxicol. Environ. Saf. 176, 288–299. doi:10.1016/j.ecoenv.2019.03.081
Satyapal, G. K., Mishra, S. K., Srivastava, A., Ranjan, R. K., Prakash, K., Haque, R., et al. (2018). Possible bioremediation of arsenic toxicity by isolating indigenous bacteria from the middle Gangetic plain of Bihar, India. Biotechnol. Rep. 17, 117–125. doi:10.1016/j.btre.2018.02.002
Sayed, K., Baloo, L., and Sharma, N. K. (2021). Bioremediation of total petroleum hydrocarbons (TPH) by bioaugmentation and biostimulation in water with floating oil spill containment booms as bioreactor basin. Int. J. Environ. Res. Public Health 18, 2226. doi:10.3390/ijerph18052226
Schippers, A., Breuker, A., Blazejak, A., Bosecker, K., Kock, D., and Wright, T. (2010). The biogeochemistry and microbiology of sulfidic mine waste and bioleaching dumps and heaps, and novel Fe (II)-oxidizing bacteria. Hydrometallurgy 104, 342–350. doi:10.1016/j.hydromet.2010.01.012
Schröder, C., Burkhardt, C., and Antranikian, G. (2020). What we learn from extremophiles. ChemTexts 6, 8–6. doi:10.1007/s40828-020-0103-6
Senthilkumar, R., Prasad, D. R., Govindarajan, L., Saravanakumar, K., and Prasad, B. N. J. E. T. (2019). Green alga-mediated treatment process for removal of zinc from synthetic solution and industrial effluent. Environ. Technol. 40, 1262–1270. doi:10.1080/09593330.2017.1420696
Shameer, S. (2016). Biosorption of lead, copper and cadmium using the extracellular polysaccharides (EPS) of Bacillus sp., from solar salterns. Biotech 6, 194–210. doi:10.1007/s13205-016-0498-3
Sharma, A., Sharma, T., Sharma, T., Sharma, S., and Kanwar, S. S. (2019). Role of microbial hydrolases in bioremediation. Microbes Enzym. Soil Health Bioremediation J. 16, 149–164.
Sharma, B., Dangi, A. K., and Shukla, P. (2018). Contemporary enzyme based technologies for bioremediation: A review. J. Environ. Manag. 210, 10–22. doi:10.1016/j.jenvman.2017.12.075
Sharma, I. (2020). “Bioremediation techniques for polluted environment: Concept, advantages, limitations, and prospects,” in Trace metals in the environment-new approaches and recent advances. Editors M. A. MURILLO-TOVAR, H. SALDARRIAGA-NORENA, and A. SAEID (London: IntechOpen).
Shen, X., Dai, M., Yang, J., Sun, L., Tan, X., Peng, C., et al. (2022). A critical review on the phytoremediation of heavy metals from environment: Performance and challenges. Chemosphere 291, 132979. doi:10.1016/j.chemosphere.2021.132979
Sher, S., and Rehman, A. (2019). Use of heavy metals resistant bacteria—A strategy for arsenic bioremediation. Appl. Microbiol. Biotechnol. 103, 6007–6021. doi:10.1007/s00253-019-09933-6
Shilpa, , Basak, N., and Meena, S. S. (2022). Microbial biodegradation of plastics: Challenges, opportunities, and a critical perspective. Front. Environ. Sci. Eng. 16, 161. doi:10.1007/s11783-022-1596-6
Singh, A. K., Yadav, P., Bharagava, R. N., Saratale, G. D., and Raj, A. (2019). Biotransformation and cytotoxicity evaluation of kraft lignin degraded by ligninolytic Serratia liquefaciens. Front. Microbiol. 10, 2364. doi:10.3389/fmicb.2019.02364
SAKSHI Singh, S., and Haritash, A. (2019). Polycyclic aromatic hydrocarbons: Soil pollution and remediation. Int. J. Environ. Sci. Technol. 16, 6489–6512. doi:10.1007/s13762-019-02414-3
Smith, A., Kaczmar, A., Bamford, R. A., Smith, C., Frustaci, S., Kovacs-Simon, A., et al. (2018). The culture environment influences both gene regulation and phenotypic heterogeneity in Escherichia coli. Front. Microbiol. 9, 1739. doi:10.3389/fmicb.2018.01739
Song, L., Niu, X., Zhang, N., and Li, T. J. C. (2021). Effect of biochar-immobilized Sphingomonas sp. PJ2 on bioremediation of PAHs and bacterial community composition in saline soil. Chemosphere 279, 130427. doi:10.1016/j.chemosphere.2021.130427
Su, L. S. (2016). Isolation and identification of heavy metal-tolerant bacteria from an industrial site as a possible source for bioremediation of cadmium, lead, and nickel. Adv. Environ. Biol. 10, 10–15.
Sujkowska-Rybkowska, M., and Ważny, R. (2018). Metal resistant rhizobia and ultrastructure of Anthyllis vulneraria nodules from zinc and lead contaminated tailing in Poland. Int. J. phytoremediation 20, 709–720. doi:10.1080/15226514.2017.1413336
Syed, S., and Chinthala, P. (2015). Heavy metal detoxification by different Bacillus species isolated from solar salterns. Scientifica 2015, 1–8. doi:10.1155/2015/319760
Taran, M., Fateh, R., Rezaei, S., and Gholi, M. K. (2019). Isolation of arsenic accumulating bacteria from garbage leachates for possible application in bioremediation. Iran. J. Microbiol. 11, 60–66. doi:10.18502/ijm.v11i1.707
Tarfeen, N., Nisa, K. U., Hamid, B., Bashir, Z., Yatoo, A. M., Dar, M. A., et al. (2022). Microbial remediation: A promising tool for reclamation of contaminated sites with special emphasis on heavy metal and pesticide pollution: A review. Processes 10, 1358. doi:10.3390/pr10071358
Telesiński, A., and Kiepas-Kokot, A. (2021). Five-year enhanced natural attenuation of historically coal-tar-contaminated soil: Analysis of polycyclic aromatic hydrocarbon and phenol contents. Int. J. Environ. Res. Public Health 18, 2265. doi:10.3390/ijerph18052265
Thesai, A. S., Nagarajan, G., Rajakumar, S., Pugazhendhi, A., and Ayyasamy, P. M. (2021). Bioaccumulation of fluoride from aqueous system and genotoxicity study on Allium cepa using Bacillus licheniformis. J. Hazard. Mater. 407, 124367. doi:10.1016/j.jhazmat.2020.124367
Tyagi, B., and Kumar, N. (2021). Bioremediation: Principles and applications in environmental management. Bioremediation for environmental sustainability. Elsevier.
Tyagi, D., Tyagi, S., and Narayan, R. (2018). Bacterial bioremediation of arsenic from contaminated groundwater and soil. Eur. J. Biotechnol. Biosci. 6, 45–51.
Ugochukwu, K. C., Agha, N., and Ogbulie, J. (2008). Lipase activities of microbial isolates from soil contaminated with crude oil after bioremediation. Afr. J. Biotechnol. 7.
Utami, U., Harianie, L., and Dunyana, N.ROMAIDI (2020). Lead-resistant bacteria isolated from oil wastewater sample for bioremediation of lead. Water Sci. Technol. 81, 2244–2249. doi:10.2166/wst.2020.281
Viesser, J. A., Sugai-Guerios, M. H., Malucelli, L. C., Pincerati, M. R., Karp, S. G., and Maranho, L. T. (2020). Petroleum-tolerant rhizospheric bacteria: Isolation, characterization and bioremediation potential. Sci. Rep. 10, 2060–2111. doi:10.1038/s41598-020-59029-9
Wang, B.-Z., Guo, P., Hang, B.-J., Li, L., He, J., and Li, S.-P. (2009). Cloning of a novel pyrethroid-hydrolyzing carboxylesterase gene from Sphingobium sp. strain JZ-1 and characterization of the gene product. Appl. Environ. Microbiol. 75, 5496–5500. doi:10.1128/aem.01298-09
Wang, C., Huang, Y., Zhang, Z., Hao, H., and Wang, H. (2020). Absence of the nahG-like gene caused the syntrophic interaction between Marinobacter and other microbes in PAH-degrading process. J. Hazard. Mater. 384, 121387. doi:10.1016/j.jhazmat.2019.121387
Wang, H., Wang, X., Liu, C., Wang, Y., Rong, L., Sun, L., et al. (2017). In-situ bioremediation of DDTs and PAH contaminated aging farmland soil using blood meal. Soil Sediment Contam. Int. J. 26, 623–635. doi:10.1080/15320383.2017.1385593
Wang, L., Xiao, H., He, N., Sun, D., and Duan, S. (2019). Biosorption and biodegradation of the environmental hormone nonylphenol by four marine microalgae. Sci. Rep. 9, 5277–5311. doi:10.1038/s41598-019-41808-8
Wei, W., Wang, Q., Li, A., Yang, J., Ma, F., Pi, S., et al. (2016). Biosorption of Pb (II) from aqueous solution by extracellular polymeric substances extracted from Klebsiella sp. J1: Adsorption behavior and mechanism assessment. J. Sci. Rep. 6, 31575–31610. doi:10.1038/srep31575
Wei, Z., Van Le, Q., Peng, W., Yang, Y., Yang, H., Gu, H., et al. (2021). A review on phytoremediation of contaminants in air, water and soil. J. Hazard. Mater. 403, 123658. doi:10.1016/j.jhazmat.2020.123658
Wu, C., Ma, Y., Wang, D., Shan, Y., Song, X., Hu, H., et al. (2022). Integrated microbiology and metabolomics analysis reveal plastic mulch film residue affects soil microorganisms and their metabolic functions. J. Hazard. Mater. 423, 127258. doi:10.1016/j.jhazmat.2021.127258
Wu, Y., Luo, Y., Zou, D., Ni, J., Liu, W., Teng, Y., et al. (2008). Bioremediation of polycyclic aromatic hydrocarbons contaminated soil with monilinia sp.: Degradation and microbial community analysis. Biodegradation 19, 247–257. doi:10.1007/s10532-007-9131-9
YalçıN, S., and Özyürek, M. (2018). Biosorption potential of two Brown seaweeds in the removal of chromium. Water Sci. Technol. 78, 2564–2576. doi:10.2166/wst.2019.007
Yin, S., Zhang, X., Yin, H., and Zhang, X. (2022). Current knowledge on molecular mechanisms of microorganism-mediated bioremediation for arsenic contamination: A review. Microbiol. Res. 258, 126990. doi:10.1016/j.micres.2022.126990
Yu, F., Tang, S., Shi, X., Liang, X., Liu, K., Huang, Y., et al. (2022). Phytoextraction of metal(loid)s from contaminated soils by six plant species: A field study. Sci. Total Environ. 804, 150282. doi:10.1016/j.scitotenv.2021.150282
Zhang, C., Tao, Y., Li, S., Tian, J., Ke, T., Wei, S., et al. (2019). Simultaneous degradation of trichlorfon and removal of Cd (II) by Aspergillus sydowii strain PA F-2. Environ. Sci. Pollut. Res. 26, 26844–26854. doi:10.1007/s11356-019-05811-w
Zhang, J. H., Xue, Q. H., Gao, H., Ma, X., and Wang, P. (2016). Degradation of crude oil by fungal enzyme preparations from Aspergillus spp. for potential use in enhanced oil recovery. J. Chem. Technol. Biotechnol. 91, 865–875. doi:10.1002/jctb.4650
Keywords: bioremediation, nanoremediation, phytoremediation, microbial enzymes, bioaugmentation
Citation: Ayilara MS, Adeleke BS, Adebajo MT, Akinola SA, Fayose CA, Adeyemi UT, Gbadegesin LA, Omole RK, Johnson RM, Edhemuino M, Ogundolie FA and Babalola OO (2023) Remediation by enhanced natural attenuation; an environment-friendly remediation approach. Front. Environ. Sci. 11:1182586. doi: 10.3389/fenvs.2023.1182586
Received: 09 March 2023; Accepted: 07 July 2023;
Published: 31 July 2023.
Edited by:
Parul Chaudhary, Graphic Era Hill University, IndiaReviewed by:
Geeta Bhandari, Swami Rama Himalayan University, IndiaRaman Kumar, Maharishi Markandeshwar University, Mullana, India
Viabhav Kumar Upadhayay, Dr. Rajendra Prasad Central Agricultural University, India
Copyright © 2023 Ayilara, Adeleke, Adebajo, Akinola, Fayose, Adeyemi, Gbadegesin, Omole, Johnson, Edhemuino, Ogundolie and Babalola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olubukola O. Babalola, b2x1YnVrb2xhLmJhYmFsb2xhQG53dS5hYy56YQ==
 Modupe S. Ayilara
Modupe S. Ayilara Bartholomew S. Adeleke
Bartholomew S. Adeleke Mosimininuoluwa T. Adebajo5
Mosimininuoluwa T. Adebajo5 Saheed A. Akinola
Saheed A. Akinola Chris A. Fayose
Chris A. Fayose Richard K. Omole
Richard K. Omole Mary Edhemuino
Mary Edhemuino Olubukola O. Babalola
Olubukola O. Babalola