
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 17 August 2023
Sec. Conservation and Restoration Ecology
Volume 11 - 2023 | https://doi.org/10.3389/fenvs.2023.1130210
This article is part of the Research Topic The Role of Community and Industry Surveillance in Managing Invasive Species: a Review of Current Knowledge View all 7 articles
 Kara K. Lanning1*
Kara K. Lanning1* Norma Kline2
Norma Kline2 Marianne Elliott3
Marianne Elliott3 Elizabeth Stamm4
Elizabeth Stamm4 Taylor Warnick1
Taylor Warnick1 Jared M. LeBoldus2,4
Jared M. LeBoldus2,4 Matteo Garbelotto5
Matteo Garbelotto5 Gary Chastagner3
Gary Chastagner3 Joseph M. Hulbert3
Joseph M. Hulbert3Phytophthora species are plant pathogens responsible for many notable biological invasions in agricultural, forests, and natural ecosystems. Detection and monitoring for invasive introductions of Phytophthora spp. is time and resource intensive. Development of citizen science detection and monitoring programs can aid in these efforts focused on reducing Phythophthora impacts. There are multiple methods for monitoring and detecting Phytophthora invasions suitable for citizen science approaches such as, leaf sampling, stream baiting or soil collections. Here we summarize five active projects in western North America where citizen scientists are aiding the monitoring and research efforts surrounding Phytophthora species and their impacts. Projects varied in scope, scale, methods, and capacity, but each project increased citizen scientists’ abilities for surveillance and advanced detection or knowledge of Phytophthora species. Some projects were integrated with school programs, others involved hands-on training with small groups, and another approach invited mass participation from interested citizens. Overall, all projects had positive outcomes multiplied across education, monitoring, and research. Together these case studies demonstrate how citizen scientists can amplify surveillance efforts, advance baseline knowledge, and reduce the impacts of biological invasions.
The genus Phytophthora contains several important plant pathogens causing blights, damping off, and downy mildews. As part of the phylum Oomycota, Phytophthora species are also known as water molds and have fungal-like morphology. Like fungi, they produce spore stages that aid in reproduction, dispersal, and survival. A unique characteristic of oomycetes is the production of motile zoospores that can swim in steams or water films between hosts using flagella (Hansen et al., 2019). Throughout history, introductions of Phytophthora species have had devastating impacts on both natural and agricultural ecosystems (Cooke et al., 2007), the most notable example being Phytophthora infestans, the causal agent of the Late Blight of Potato which resulted in the Irish Potato Famine of the 1840s (Saville and Ristaino, 2021).
More recently, the introduction of Phytophthora lateralis to the northwestern United States in the 1920s caused extensive mortality of Port-Orford-cedar (Chamaecyprus lawsoniana), an agriculturally, economically, and ecologically important tree, across its native range. Phytophthora lateralis spreads readily through streams and along roads infecting the host roots, disrupting water absorption and nutrient uptake (Hansen et al., 2000). Several decades later, the introduction of P. ramorum in the western United States during the mid 1990s killed millions of tanoak trees in coastal mixed hardwood forests (Rizzo et al., 2005) by diseases known as Sudden Oak Death (SOD) and Ramorum Blight, both caused by the same pathogen (Grünwald et al., 2008). Extensive aerial monitoring of Phytophthora species by aircraft of state and federal agencies began following the P. ramorum outbreak (Meentemeyer et al., 2015; Goheen et al., 2017), but the large scope of the problem surpasses the capacity and resources for monitoring in most states.
Early detection of Phytophthora species is critical for minimizing the impact of their introduction (Meentemeyer et al., 2015). Land managers and regulatory agencies use many strategies to detect and identify potentially harmful Phytophthora species.
Ground and aerial surveys from aircraft can be used to detect symptomatic hosts (Rizzo et al., 2005) and symptomatic tissue can be directly plated on oomycete-selective media for isolation and morphological identification of the pathogen (Figure 1) (Shew and Bensen, 1982). Symptomatic tissue can also be analyzed using DNA-based methods to detect the presence of Phytophthora species (Calmin et al., 2007). Rapid field-based diagnostic assays, such as Enzyme-Linked Immunosorbent Assay (ELISA) test strips, lateral flow devices (LFD), and Loop Mediated Isothermal Amplification (LAMP) assays have also been developed specifically for detection of Phytophthora species (Kox et al., 2007).
A method known as baiting can be used to detect infestations in watersheds. When baiting in a water source, leaves from susceptible hosts are secured in a mesh bag then placed in the water source of interest, which may include a high-risk stream or an irrigation pond (Figure 2). After 1–3 weeks, the bag is removed and leaves are evaluated for symptomatic lesions (Sutton et al., 2009). Oomycetes can also be isolated from soil using baiting methods. First the soil is collected and placed in a container that is then flooded with water (Figure 2). Leaf baits float on the surface of the water and develop lesions if they are infected by oomycetes (Shew and Bensen, 1982). An additional leaf baiting method includes leaf baits in buckets placed under the canopy of tanoaks to capture rain drip. This bucket-bait method allows monitoring for P. ramorum in the canopy of individual trees (Hansen et al., 2008). In all cases, infected tissue from baits can be cultured on oomycete-selective media and the species identified using morphological or DNA-based methods.
Early detection is key for land managers to effectively react and manage new outbreaks or infested areas. If detected early, land managers can conduct treatments to eradicate or contain the infestation. For example, the management of P. ramorum in Oregon has demonstrated the effectiveness of eradication on local scales at several sites (Hansen et al., 2019). Despite advances in Phytophthora detection, there is a need to increase monitoring capacity to detect new invasions.
Common obstacles faced by Phytophthora monitoring programs include lack of funding for aerial reconnaissance programs, access and time requirements to monitor large geographic areas, limited field personnel available for on-the-ground evaluations, and complex ownership patterns on the landscape (Cunniffe et al., 2016). One of the critical limitations of large-scale surveys is caused by the modest symptoms of Phytophthora disease in its early stages of establishment, mostly limited to foliar spots or branch die-backs which cannot be detected through aerial surveys or by cursory ground surveys. Citizen scientists can play a role in early detection (Crowl et al., 2008) and offset many of these limitations associated with the detection of invasive agents in ecological settings (Larson et al., 2020; Encarnação et al., 2021).
Citizen scientists have been increasingly recruited for invasive species detection (Pocock et al., 2017) and their engagement in Phytophthora surveys has been demonstrated in a few projects internationally (Hulbert et al., 2017). One example of a successful citizen science disease detection program is the California-based Sudden Oak Death (SOD) Blitz project (Garbelotto Lab, UC Berkeley, CA). Initiated in 2008, SOD Blitz monitors the spread of P. ramorum, the invasive agent that causes SOD and threatens the survival of tanoak and many oak species in California and Oregon (Grünwald et al., 2008; Goheen et al., 2017; LeBoldus et al., 2022). The program engages citizen scientists from 24 coastal communities, and trains them on data collection using a workshop format before monitoring. Data collected by SOD Blitz citizen scientists led to remarkable increases in data collection representing multiple communities, yielding greater accuracy of disease prediction models (Meentemeyer et al., 2015).
Citizen science projects can also provide benefits beyond data collection. Citizen scientists have reported increases in knowledge with respect to invasive plant species (Jordan et al., 2011), and exotic tree-pest identification (Norman-Burgdolf and Rieske, 2021). For example, in Minnesota, citizen scientists collected data on invasive oak pests despite the risks of regulatory impacts of these species on their own land (Andow et al., 2016). Engaging citizen scientists in invasive species monitoring can provide educational benefits in addition to enhanced capacity for monitoring.
This review highlights five projects that use different approaches to engage citizen scientists to survey Phytophthora species in California, Washington, and Oregon (Table 1). The synthesis of the different approaches, scale, and engagement strategies resulted in a diversity of lessons learned and recommendations. Below we highlight and discuss these different approaches for engaging communities in surveillance and research of Phytophthora species, with citizen science programs listed in order of establishment.
The idea of enlisting citizen scientists to help with the statewide SOD survey was born from a meeting between Carmel Valley, CA citizens and UC Berkeley Forest Pathologists. Recruiting citizens would relieve an existing gap in surveillance capability, while creating a meaningful large-scale collaboration between the public, researchers, and governmental agencies. The SOD Blitz program emerged from a true research need, distinguishing it from other projects in which a citizen science component is added primarily to educate the public, rather than project success and completion (Bonney et al., 2009).
The SOD Blitzes were driven by scientific and community outcomes. The name “SOD Blitz” was coined for the project: initially, its purpose was to organize fine scale surveys to map the distribution of SOD symptoms on California bay laurel and tanoak leaves using 100 m intervals. This project would benefit from citizen science engagement specifically by leveraging their access and familiarity of their own neighborhoods supporting surveying goals. Quickly, additional goals were added, including the discovery of new outbreaks, monitoring the apparent disappearance of extant outbreaks, calculation of local disease incidence, identification of new hosts, and the estimation of oak and tanoak mortality. Over a decade later, the project purpose expanded again with the emergence of multiple lineages of the SOD pathogen, Phytophthora ramorum in California forests (Garbelotto et al., 2021a).
The first ever emergence of SOD occurred on the California coast in the mid 1990s (Garbelotto et al., 2001). The discovery of the causal agent, P. ramorum, was a pivotal moment for scientists, forest managers, individual landowners, and the public who were finally able to put a name on the agent that killed tens of thousands of tanoaks and oaks in a period of 5 years (Garbelotto et al., 2001; Werres et al., 2001; Rizzo et al., 2002). Although the discovery of P. ramorum was an irrefutable advancement, little was known about how the invasive pathogen was introduced to California, how it spread, and how to control it.
This apparent jump from tanoaks to oaks was pivotal in the history of the disease: tanoaks, regarded as sacred by many California Indigenous Peoples, were largely unknown to the larger public, while oaks were and are a major and familiar component of landscapes throughout California (Garbelotto and Rizzo, 2005). Oak mortality was visible, leading to public pressure on policymakers to fund multiple research efforts. By 2004, scientists had reached major milestones on SOD research yielding partial answers regarding disease epidemiology, diagnosis, and control. Results fostered continued public interest (Garbelotto and Rizzo, 2005). However, because the issue was on a regional scale and affecting multiple counties and jurisdictions, agencies responsible for the management of California woodlands generally found it too costly or complex to apply the findings to curb the SOD epidemic.
Between 2000 and 2008 strong evidence was obtained that P. ramorum moves short distances (Mascheretti et al., 2008), and that the main transmissive hosts were California bay laurels and tanoaks, and that oaks were non-transmissive dead-end hosts (Garbelotto and Rizzo, 2005). This fundamentally shifted disease diagnostic, quantification and control strategies. Testing California bay laurels and tanoaks leaves was an easier task than testing bark cankers on oaks and tanoaks, but the abundance of California bay laurels was orders of magnitude higher than that of oaks. Finally, the short-distance movement of the pathogen (Mascheretti et al., 2008; Mascheretti et al., 2009), combined with the vast size of the zone of infestation, required a shift in the surveying approach. The risk of infection for oaks existed only when the pathogen was within a few hundred meters requiring SOD outbreaks be confirmed at a much finer scale. The magnitude of the problem, insufficient labor and funding led to the recruitment of citizen scientists to aid disease monitoring efforts.
SOD Blitz was designed with local communities as the main participants; with each community recruiting volunteers via extant community-based environmental programs (see Pandya, 2012). In this case, the term “community” is narrow and refers to people inhabiting the same area or people identifying as part of an historical community, such as a First Nation, and does not include online communities (Aristeidou et al., 2020). To increase interest, local environmentalists, land managers, and educators shared SOD Blitz information and benefits in ways tailored to their groups. Programs were invited to integrate SOD-blitzes into the scope of their existing work.
SOD Blitz field surveys occurred, and still occur, annually in the Spring when SOD foliar symptoms are most visible (Eyre et al., 2013). In-person training sessions are organized in each participating community and communities are assigned a specific long weekend for the SOD Blitz. Training content cautioned participants about the movement of infected material and laid out careful instructions for submitting samples directly and securely to UC Berkley. Volunteers are tasked with identifying symptomatic foliage, collecting 6–10 leaves from hosts, recording all pertinent information using pencil and paper, and delivering the samples to the local organizer. At U.C. Berkeley, samples are logged and freeze dried. DNA is extracted using a ROSE extraction method (Osmundson et al., 2013), and SOD is confirmed by the Taqman assay of Hayden et al. (2006). Geolocation of each sample and results of the P. ramorum PCR assay are uploaded on a Google Earth Platform and shared with the public online at SODblitz.org and SODmap.org and on tablets or cell phones using the SODmap mobile App (Garbelotto et al., 2014). In-person or online town hall meetings are organized for all participating communities. Results are communicated with the media thanks to News Releases coordinated by U.C. Berkeley.
Some results of the SOD blitzes are summarized in Table 2. Given the very large number of samples collected and tested, it is no surprise that the SOD blitzes identified innumerable new outbreaks, although only one was in a county where P. ramorum had yet to be identified, resulting in a change in State and Federal policies (Garbelotto et al., 2021a). Likewise, several Arctostaphylos species were added to the list of federally regulated plant species thanks to the SOD blitzes (Garbelotto et al., 2020). The 2020 SOD blitzes identified the EU1 lineage, previously unreported in California forests, in a forest in Del Norte County (Garbelotto et al., 2021a). Phytopthhora ramorum lineages have distinctively different phenotypes, including virulence, and different mating types (Garbelotto et al., 2021b), hence the discovery of the EU1 lineage led to an eradication effort. Unfortunately, subsequent SOD blitzes have detected trees infected with the EU1 lineage, despite continuing eradication efforts. Although identifying SOD outbreaks is the main goal of the program, an equally important result has been the consistent negative results of tests performed on a large number of samples coming from San Luis Obispo County, which is still not federally regulated, in part thanks to such negative results.
The above results are all notable but the main consequence of the SOD blitzes has been the implementation of recommended disease management practices in areas with confirmed SOD. In the 3 years between 2020 and 2022, 143 of 793 SOD Blitz volunteers participated in a post SOD blitz survey (SODquest.org) and reported to have treated 19,054 trees, in part motivated by the SOD blitz results. The Arctostaphylos species, newly identified as hosts for P. ramorum, were largely rare and threatened, and the discovery of SOD resulted in the timely removal of infected plants and plant parts, in order to curb spread of the disease. A less known outcome of the SOD blitzes, one which highlights the added value of its community-based organization, is the eradication of an isolated outbreak in Atherton (San Mateo County), possible thanks to the joint effort to remove multiple bay laurels located in different properties. To our knowledge, this is the only known successful eradication of P. ramorum facilitated by citizen scientists in California woodlands, and it was possible only thanks to the timely detection of the pathogen in an area several miles away from the closest outbreak.
The SOD blitzes have generated an extensive database on SOD distribution in wildlands and urban areas. Thanks to its size, we have been able to leverage the inclusion of all SOD distribution data, including those obtained by academia and government, in an individual database called SODmap, which probably represents one of the largest single databases in the world of lab-confirmed distribution of an invasive forest disease (Garbelotto et al., 2014). This SOD map database has also led to the development of a prediction model for the spread of SOD in mixed oak woodlands (Meentemeyer et al., 2015). Another important outcome has been the identification of additional variables that are correlated with reversal of California bay trees from infected to a non-infected status, such as proximity to other bay laurels and conducive weather conditions that promote sporulation (Lione et al., 2017). This information is critical for long-term disease spread predictions.
First and foremost, we believe that structuring the SOD blitzes as a community-based program has ensured the long-term survival of the program. This approach is more likely to build a long-term relationship between volunteers and the program than an approach based mostly on online recruitment of volunteers (Pandya, 2012). Obviously, the online approach makes up for its shortcomings because it can reach a wider audience, but it cannot guarantee return participation, and is less suitable to the adoption of different, but equally acceptable, sampling approaches. To support the above statements about the importance of fostering diversity and about the value of return participation, we present two sets of results: First, our study identified significant differences between rural and urban communities when comparing actions taken to curb SOD (e.g., chemical treatments were more common in urban areas) and when comparing how volunteers heard about the program (e.g., email was more effective in urban areas, while word of mouth and community programs were more effective in rural communities). The second study indicated volunteers who participated in the program 3 or more times were as effective as an elite group of 12 top-performing volunteers. As of 2022, 34.8% of participants were “return” volunteers, further bolstering the continued success of the program.
The quality of the data collected by volunteers has been one of the top priorities of the SOD blitzes. Meentemeyer et al. (2015) have shown convincingly that trained SOD blitz participants contributed a higher percentage of positive detections (33%) compared to professionals (19%). This result is of paramount importance and we believe it is due both to the nature of the task requested, involving the relatively simple, yet project-specific identification of SOD symptoms on leaves of two tree species, and to the quality of the training program. We believe that simpler tasks, such as pinpointing the location of oaks, may be too simple for sustained interest, while other tasks such as determining the severity of SOD infection in trees, may be too complex for a large-scale citizen science program. Additionally, the use of pencil and paper was much more attractive to various audiences. This may appear as counterintuitive in our highly digital world, but pencil-and-paper was embraced by older, very young, tech-naive or tech-adverse communities equally, while the opposite may not be true for digital platforms. One shortcoming of long hand data collection is that it does require the added step of electronic data entry, and there is a risk that paper sheets can be lost or damaged before the data is digitized.
High quality data can also be ensured when, as it happens in the SOD blitz, all samples collected by volunteers are further processed in a laboratory using a standard method. We strongly support this design, which is truly cooperative between citizens who are trained by experts, but perform all of the collections, and researchers, who perform all of the lab analyses.
From the beginning, the SOD blitzes included the sharing of the data with citizen scientists participating in the program. Sharing the data in a fast, simple, intuitive and online accessible way, rather than in technical reports, has built trust between researchers and volunteers and has solidified the base of participants. We believe that the lengthy scientific peer review publishing process, although acceptable and appreciated in the scientific community, is not compatible with a Citizen Science program in which Citizens are co-owners of the collected data and desire a quicker communication timeline.
The huge attention given by the media to the SOD blitz results was unexpected, but welcomed. Media interfacing requires a completely different set of skills and a person dedicated to the task, but we quickly learned that it greatly fostered program participation. More importantly, through analysis of website visits, App usage, and readership of newspapers, we were able to estimate that the data collected in 15 years by approximately 7,000 participants well surpassed a user audience of 3 million people. More research to understand the role of media in motivating participation and volunteer retention would be valuable as the media coverage likely benefitted the SOD blitz program over time.
The SOD blitzes have had their share of issues to resolve, and as a result, the approaches somewhat changed in time. Three issues emerged early on in this process: 1- depending on the communities’ goals, each SOD Blitz had to be conducted following different sampling schemes, ranging from all sampling being performed by volunteers in their own property, to all sampling being performed on key sentinel trees identified in various locations, independent of ownership. The need for adaptation of Citizen Science programs to the needs of different communities has been discussed by Pandya (2012); 2 Citizens concerned for trees’ survival wanted results to be shared quickly so that disease management options could be quickly implemented, as needed; 3 the only unified and equitable sampling method acceptable across all communities was one that did not involve digital technology U.C. organizers responded to expressed needs by offering training tailored for the specific community group, provided all necessary collection materials (Figure 3), and distributed location-specific information when the data was verified and analyzed. In order to respond to the above needs and requirements, the SOD blitzes were organized as explained in the methods.
An unexpected and occasional problem was later encountered with the diagnostic assay employed in the program. The Hayden et al. (2006) assay is based on the nuclear ITS and had been tested in a ring trial and vetted as one of the better performing PCR diagnostics assays (Martin et al., 2009); however, in 2016, multiple positives were detected in San Luis Obispo County, but no samples from the same County could be confirmed as positive by PCR or culturing in the following year. A careful comparison of results from both years identified all San Luis Obispo positives from 2016 and other 2016 and 2017 positives from elsewhere, as having a positive but weaker signal than all other samples. The use of a second PCR assays, based on the mitochondrial locus COXI as described by Kroon et al. (2004) and modified using SYBR green to determine the melt curve of the PCR amplicon, confirmed that the 2016 San Luis Obispo positives in fact were negative for P. ramorum but positive for P. pseudosyringae, which was present in unusually large amounts, while the vast majority of all other questionable samples was positive. Since then, the COXI assay resolves the infection status of samples with questionable diagnostic results using the assay of Hayden et al. (2006). Additionally, all samples coming from putatively noninfested counties were also cultured after 2016, in order to make sure notifications to California Department of Food and Agriculture are fully supported by results. This approach proved very successful when P. ramorum was first detected in Del Norte County (Garbelotto et al., 2021a). Finally, the lab component of the project can be modified without significantly affecting the citizen science component of it. This means that novel analyses can be added if necessary: this was the case for the tests necessary to differentiate among the three lineages of the pathogen, which were started with success since the discovery of the EU1 lineage in Del Norte County in 2020.
Another issue emerged in 2020 when, due to the COVID pandemic, all field research programs were suspended in California. In a matter of 3 weeks, we were able to capitalize on the popularity of the program and succeeded in having the project declared as essential to the State. However, in order for the program to be allowed to run, we had to develop a totally contactless protocol. Instead of in-person meetings, during which training and necessary materials were given, we shifted to online training thanks to a very effective website (sodblitz.org), a training video, and use of outdoor SOD blitz stations (Figure 4) where people could obtain and later return sample and data collection materials. Results were shared by online postings and through a webinar during which data were presented and interpreted as previously done during town hall meetings. In spite of the recent extensive online component, the SOD blitzes have remained a community-based program and, as such, the program continues to be advertised by local organizers and survey and sampling occur on long weekends, each preassigned to different communities.

FIGURE 4. An outdoor SOD Blitz station allowed for a contactless collection of materials and return of samples.
Overall, the SOD blitz program engaged many communities to advance knowledge and understanding about the distribution and presence of Phytophthora ramorum across California’s landscapes. Citizen participation increased the detection and monitoring of P. ramorum populations, but more research to understand how the training and instructions reduced human mediated spread would be valuable. The SOD blitz program was careful to caution about the biohazards and risk of collecting and moving SOD infected materials but it is unclear how this translated into individual sanitation behaviors such as boot cleaning and movement of firewood. Additional research into these social impacts would be valuable. Furthermore, much of the results were dependent on positive recoveries and more work to understand and analyze the negative detections is needed. Further analyses in quality and control measures, particularly assessing rates of false-positives or false-negatives, is needed to better understand the pros and cons of the different approaches implemented throughout the duration of the project. Future projects aimed at monitoring invasions of potentially regulated species should be designed to include controls and measure social impacts would be valuable next steps for advancing approaches to engage communities in similar bio monitoring projects.
Starting in 2010, Washington State University (WSU) initiated a project to engage citizen scientists and students in the collection and study of Phytophthora species in many streams throughout western Washington. The purpose of the project was to monitor for P. ramorum, but also document other species present in the waterways.
Stream baiting is an effective way to monitor for the early detection and the spread of P. ramorum (Sutton et al., 2009). Identifying where P. ramorum is present can inform decisions for eradication efforts to prevent the further spread and establishment. The Phytophthora stream monitoring project was started in 2010 to expand state and federal monitoring efforts to ultimately reduce the risk of P. ramorum spreading into forests in Washington. The project, designed to provide baseline information for other Oomycete species present in western Washington streams, also provides educational opportunities to the public, land management professionals, and students about the importance of these organisms and their effects on ecosystems.
Stream monitoring sites were selected based on input from state and federal agencies, or on previous findings. For example, two monitoring sites were included from a Puget Sound estuary containing brackish water because P. ramorum was detected there previously (Washington State Dept. of Agriculture, pers. comm.). Other streams were selected based on the likelihood of an infestation and access, such as upstream from ornamental plant nurseries. In total 28 streams were sampled by citizen scientists and 20 streams were sampled by state agencies during the duration of the project.
Citizen scientists were recruited from stewardship groups, Master Gardeners, schools, and interested individuals in the counties containing chosen streams. Recruitment efforts of citizen scientists varied and included solicitation for volunteers at community P. ramorum educational workshops, or by direct invitation of entire-class participation from high school and community college groups. Citizen scientists were provided with kits containing the materials needed, such as bait bags, a thermometer for measuring water temperature, data sheets, mailing materials for baits, leaf baits, and an instruction manual.
During some years, students from local high-schools and colleges assisted with the lab work either as part of the class project or as an independent study project. Stream monitoring for oomycetes was developed into a module for a community college biology class encompassing the field and lab components (Figure 5). In addition to stream monitoring for Phytophthora, class projects included baiting in local stormwater retention ponds and testing several different bait materials for recovering oomycetes.

FIGURE 5. Students from Pierce College participating in Washington Phytophthora Stream Monitoring project.
Stream monitoring began in late February and continued at 2-week intervals for 6 weeks until early May in most cases. At the first sampling time, WSU researchers met with citizen scientists at their stream site and worked with them on deploying the bait bags and sampling techniques. Two mesh bait bags (Figure 2A), kindly assembled by Master Gardener volunteers, were deployed in each stream containing five Rhododendron leaves. Leaf baits were collected from plants grown at WSU Puyallup Research and Extension Center and shipped to citizen scientists, along with mailers for return shipments. In some cases, citizen scientists used one leaf from a different plant species of their choice in addition to four Rhododendron leaves. At the end of the 6 weeks, WSU researchers met with citizen scientists at their stream sites to collect the final set of samples and the rest of the materials.
Laboratory methods for isolating and identifying Phytophthora species were similar to the laboratory methods in the Oregon P. ramorum monitoring project below. Leaf baits were rinsed, and excised sections of symptomatic tissues were cultured on PARPH-V8 media (Ferguson and Jeffers, 1999). Colonies were then selected based on morphological characteristics to capture the variability in species isolated. A subset of samples from each morphological group was identified after extracting and amplifying DNA with PCR and sequencing the ITS regions of the rDNA using primers ITS6 and ITS4 (Cooke et al., 2000). Preliminary identification of species was completed by querying the ITS sequences against the GenBank database using BLAST. Identification was confirmed by examination of morphological characteristics such as oospores, chlamydospores, and other structures and sequence comparisons with voucher sequences in a phylogenetic tree (Abad et al., 2022). Selected isolates were maintained on 1/3CV8A slants at 15C in the WSU Phytophthora culture collection.
The Washington Phytophthora stream monitoring project provided presence/absence data about the distribution of P. ramorum in a total of 48 Washington streams sampled over 10 years, adding to state and federal monitoring efforts. P. ramorum was only detected once by a citizen scientist sample near a known positive plant nursery. However, many other Phytophthora species were recovered through the monitoring efforts.
Phytophthora chlamydospora, P. gonapodyides, and P. lacustris were commonly recovered from streams (Supplementary Table S1) and sampling from a single stream in Puyallup, WA indicated they can persist in these streams for 10 years. Important agricultural pathogens such as species P. cactorum, P. citricola, and P. cryptogea were also occasionally recovered from streams in areas baited near agricultural inputs and runoff. Monitoring in the Puget Sound estuaries also yielded many isolates of undescribed Halophytophthora spp. (Man in ’t Veld et al., 2019). In addition, P. cambivora, P. pseudosyringae, and P. siskiyouensis were recovered from streams and a recent soil survey with students from Bellarmine Preparatory High school isolated them from soils in a declining red alder stand. This finding has led to further research to complete pathogenicity testing on red alder.
The Washington Phytophthora stream monitoring project has expanded surveillance efforts by monitoring additional streams and engaging with citizen scientists and communities. The results of the project have advanced knowledge of the diversity and commonality of Phytophthora species living in western Washington streams. Many results, such as the discovery of the Halophytophthora spp., would not have been made without engagement with citizen scientists.
The Washington Phytophthora stream monitoring project demonstrated there is interest and value for engaging citizen scientists to monitor for invasive species and collect baseline diversity information about Phytophthora species from streams. Although research and diagnostics about Phytophthora species requires technology such as microscopes and thermocyclers, the collection and monitoring approaches—primarily methods involving baiting—were great activities for individuals and groups, including large classes (Figure 4). The results are valuable because they survey vulnerable sites, improve methodologies, inform research priorities, reveal new research areas, and enhance science communication to diverse audiences.
The educational outcomes of the project, particularly with early college students, has inspired continued partnership development with higher education faculty. Participating in the project offered many students a first introduction to plant pathology and environmental microbiology and informed decisions about career paths. Many of these students subsequently joined the lab as hourly workers, volunteers, and interns and developed laboratory skills, including sterile technique, DNA extraction, and PCR. Students use the experience on their resume when applying to college and for jobs in the field. Some students involved even pursued graduate education and careers in plant pathology.
The purpose of the Oregon-based Phytophthora ramorum monitoring project is to engage landowners in surveillance approaches and provide educational opportunities for early detection and treatment strategies. Volunteer landowners deployed bucket and stream baits in this project for the early detection of P. ramorum near known infested sites in southern Oregon. Sampling strategies for the citizen science project were designed to add resolution to surveys conducted by Oregon’s interagency SOD program. Project coordination is by Oregon State University (OSU) county-based Extension faculty in partnership with the OSU forest pathology lab and Oregon’s interagency SOD program.
Since SOD was first detected in Oregon in 2001 a quarantine area was established and eradication efforts have been initiated (Goheen et al., 2017). Disease spread has resulted in several expansions of the quarantine area, and multiple clonal lineages have been detected since the disease was first discovered (Grünwald et al., 2016; Peterson et al., 2022). Sites on state and private ownerships continue to be treated under state regulatory authority (LeBoldus et al., 2022), but more monitoring was needed to help contain the spread of the pathogen.
Citizen scientists were initially contacted during SOD educational events focused on general community awareness of the disease or other types of extension and outreach. Interested participants attended citizen science training covering a detailed sampling process. Follow-up site visits were scheduled with each citizen scientist to help select monitoring sites and review sampling procedures in the field.
One or two sample sites were located on the volunteer’s property or with signed permission from another property owner in easily accessible locations, typically in proximity to homes or roads. Most sites were located near the leading edge of SOD infestations. Additionally, when available, control sites were sampled in known infested areas.
Two monitoring techniques were utilized. Bucket bait monitoring stations were deployed under tanoak trees in the rainy season to detect infestations at the individual tree scale. In-stream bait bags were placed in the late spring/summer for monitoring at the drainage level (Hansen et al., 2008; Sutton et al., 2009). For bucket sampling, two buckets were placed beneath each tree, and bait leaves (2 rhododendron, 2 tanoak) were placed in each bucket. Bucket kits included 2-gallon utility buckets, bucket liners, screens, elastic, and detailed procedure booklets. Stream kits included mesh bags and sampling booklets. For both projects, sampled leaves were retrieved at a 2-week frequency, double-bagged and mailed to the OSU forest pathology lab in prepaid envelopes along with data sheets. Bait leaves were provided from sites located outside of a 6-mile distance from the quarantine boundary. From 2019 to 2022 a total of 52 buckets and 7 streams were sampled and submitted for lab analysis. Buckets were deployed during three sampling seasons, and streams were sampled for two seasons.
Commercially purchased rapid tests designed to screen for Phytophthora spp. provided practice in hands-on bait inspection and testing procedures. Designated bait leaves were inspected for symptomatic lesions and prepared for rapid testing. A positive result was recorded when two lateral lines became apparent on the test strip within the allocated time period. Rapid testing for stream sites was discontinued after the first season because of landowner concern regarding non-target species detected in streams.
Upon being received at the OSU forest pathology, bait leaves were immediately rinsed in tap water and towel dried. Leaves were then wrapped in a dry paper towel, placed in a clean zip lock bag, and stored at 4°C until plating (no longer than 48 h). Leaves were surface sterilized with 50% ethanol, and petioles, midribs, and any obviously symptomatic tissue were excised from each leaf using sterile instruments. Excised leaf sections were plated on corn meal agar amended with 200 ppm Na-ampicillin, 10 ppm rifamycin, 25 ppm hymexazol, 30 ppm benzimidazole, and 10 ppm natamycin. Plates were then monitored for 4 weeks, and Phytophthora species were identified by morphology.
There have been no confirmed positive results for P. ramorum outside control areas at either bucket or stream bait sites. Lab testing has resulted in positive lab results from samples collected at a control site within a known infested area. One positive lab result originated from a bucket outside a known infestation, but the field site could not be confirmed with field inspection. There have been no positive rapid test results from designated leaves in paired buckets.
Incidental (non-target Phytophthora spp.) species have been detected from lab analysis of stream samples. Citizen scientists also recorded positive rapid test results (for the Phytophthora genus) from stream sites during the first season; separate samples from the same site were subsequently tested in the lab to confirm the presence of other species such as P. lateralis. These results indicate that citizen scientists could help collect baseline Phytophthora stream assemblage data if it was a goal of the project.
Citizen scientists have generally had good follow-through with sampling, recording, and mailing procedures. In many instances, family and friends have participated in on-site training sessions and assisted with sampling. However, data sheets were not always included with samples and delays in sampling and mailing occasionally occurred due to inclement weather or landowner absence. While the engagement of citizens increases capacity for monitoring and provides educational benefits it is unlikely a substitute for statutory monitoring, at the current scale.
Sampling for a regulated invasive species requires careful communication regarding intended uses for collected data. In this study, data may be used to prioritize SOD treatment including tanoak removal within a prescribed radius around infested trees. Therefore, a positive sample result is followed up by the SOD program with field review, additional sampling and testing for confirmation. Likewise, a negative citizen science sample result does not conclusively determine an area is free of infestation.
Additional concerns include the detection of non-target Phytophthora species in the lab. The impacts of these species are often unknown and their presence can be worrisome to citizen scientists. However, positive results provided excellent opportunities to discuss other Phytophthora species in the landscape. Similarly, positive rapid tests can cause anxiety as well. The delay between immediate rapid test results and analysis of lab samples leads to a period of uncertainty regarding the presence of P. ramorum.
Expanding the citizen science efforts to include other methods of detection may also increase the value of the project. In collaboration with USDA-FS and Oregon State University, the Oregon Department of Forestry extensively monitors infested and at-risk areas by utilizing aerial surveys, ground surveys, and stream baiting. Samples collected with these methods are evaluated at OSU with culture-based diagnostics and pathogen identification by qPCR. Currently, the Oregon P. ramorum monitoring project primarily utilizes culture-based diagnostics, but it may be advantageous to include molecular identification methods in the future.
Undergraduate plant pathology students at Pacific University, in Forest Grove, Oregon collaborated with Washington State University (WSU) to analyze microbial communities in soils collected from Western redcedar (Thuja plicata) trees exhibiting dieback symptoms. This course provided undergraduate students, who had limited exposure to plant biology topics, an opportunity to explore the concepts of disease and ecological health and function. The goals of this program were to introduce plant pathology concepts, introduce common soil-pathogen detection methods, and to determine if Oomycete species are associated with symptomatic trees. The project was supported by the Department of Biology at Pacific University and the Ornamental Plant Pathology Program at WSU Puyallup Research and Extension Center.
Collaboration between undergraduate science programs and research labs provides an opportunity for students to gain access to high-impact learning experiences. These experiences are defined as pedagogical methods that enhance student learning outcomes, including problem-based learning, experiential learning, and undergraduate research experiences (Kuh and Schneider, 2008). Such experiences increase student performance, collaboration, critical thinking, and academic retention in various student populations. This retention includes students representing minoritized populations in ecology (Ganesh and Smith, 2017; Awad and Brown, 2021). In contrast to the traditional lab curriculum, where the methods and expected outcomes are known, integrating citizen science projects as part of, or all of the laboratory curriculum, exposes students to the realities of scientific exploration while increasing student access to research and mentoring opportunities.
A research collaboration between Pacific University and WSU resulted when a conservation district newsletter issued a call for citizen scientists (Hulbert, 2021). A course-based undergraduate research experience was designed for the plant pathology class to study the oomycete communities under western redcedar. Dieback of western redcedar, an important species in Pacific Northwest forests, has increased in recent years (Buhl et al., 2019). The cause of the dieback is unknown, though climate change was suspected as the primary factor, because the symptoms were commonly associated with water stress. Investigation into the possible role of plant pathogens such as Phytophthora species is needed.
Fifteen undergraduate students enrolled in a plant pathology course during the 2021 Fall semester were introduced to the emerging problem of western redcedar dieback. They read scientific literature on invasive Phytophthora species in forest ecosystems to help put the current situation into context (e.g., Hansen et al., 2000; Cooke et al., 2007; Hansen, 2008). Before collection, students were trained to identify western redcedars using morphological characteristics, how to collect soil samples, and how to assess severity of dieback symptoms.
Assessment of Western redcedar dieback and subsequent soil analysis took place primarily at the John Blodgett Pacific University Arboretum. Two other sites in Washington and Yamhill counties, Oregon were assessed as part of an independent undergraduate research project.
The John Blodgett Arboretum includes 33 acres of coniferous forests and Gales Creek is a permanent waterway that runs through it. Fifteen Western redcedars were identified and evaluated for symptoms of dieback. Symptoms included: percentage of canopy transparency and the number of additional unhealthy trees within sight. Students assessed other factors, including insect damage, physical injury, and other pathogen signs. An undergraduate research student also recorded geospatial information for each tree.
Soil was collected approximately 1 m from the base of the trunk of symptomatic trees after forest debris was removed. Approximately three 100 g soil samples were collected at a depth of 10 cm around the perimeter of each tree and were combined in a zip top bag resulting in one bulk sample per tree. Shovels were cleaned using bleach wipes in between each tree. Samples were transported to the university lab via vehicle and stored at room temperature for 1 week.
One week post-collection, soil samples were assessed for the presence of oomycetes using a leaf bait method (Shew and Bensen, 1982). Bait material included English ivy (Hedera helix), Rhododendron and common red rose (Rosa rubiginosa) petals. After 7 days, symptomatic bait tissue was cut and cultured on PARPH-V8 agar in the dark at 20 C for 7 days. Two agar plugs per plate were subsequently plated using PARPH-V8 agar plates, incubated for an additional 7 days, and replated on V8-agar media (Ferguson and Jeffers, 1999). Colony morphology characteristics were recorded. DNA was extracted from colonies and the ITS region was amplified using the ITS6 and ITS4 PCR primers (Calmin et al., 2007). The amplicons were run on a 1.5% TAE agarose gel and sequenced (Eurofin Genomics, LLC). Sequence homology was compared using the NCBI data base and BLAST searches.
The impacts of the undergraduate research experience on student education, attitudes and behaviors were assessed using a course-based undergraduate research survey (Shortlidge and Brownell, 2016).
Students identified 7 Western redcedar trees as asymptomatic and 8 trees as symptomatic within a small area of the arboretum. Trees considered symptomatic had between 40 and 100 percent canopy transparency (Figure 6).

FIGURE 6. Western redcedar dieback at John Blodgett Pacific University Arboretum, Gales Creek, Oregon.
The site was reassessed in Spring 2022 as part of an independent undergraduate research project that further evaluated the presence of Western redcedar dieback at the arboretum expanding the number of Western redcedars assessed to 60. Twenty-eight trees were asymptomatic, and the remaining 32 were symptomatic of dieback, having between 20 and 100 percent canopy transparency (Figure 7).
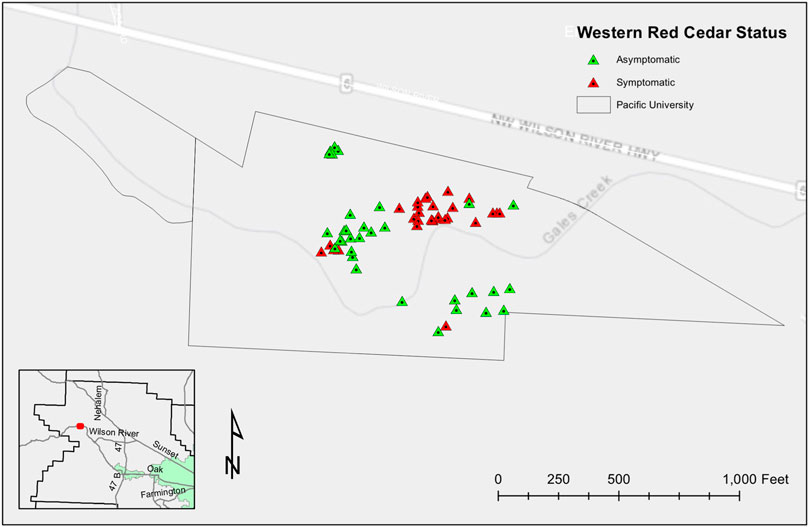
FIGURE 7. Distribution of WRC dieback at Pacific University’s John W. Blodgett Arboretum. Sixty trees were mapped using ArcGIS, 28/60 symptomatic WRC and 32/60 asymptomatic WRC. Black lines represent the property boundaries.
Students were successful in isolating Oomycetes using a soil bait method from both symptomatic and asymptomatic Western redcedars at all sites, though Phytophthora species were only recovered from soil collected during the independent student project at a separate site. Phytophthora cambivora was recovered from soils collected from asymptomatic and symptomatic trees at a site outside of the arboretum (Supplementary Table S1).
The post-course survey captured a 100 percent response rate and indicated that most students positively benefited from the whole-class project. Ten out of fifteen students reported having learned “a lot” or an “extensive” amount about integrating scientific theory into practice. Thirteen out of fifteen students indicated that they had learned “a lot” or an “extensive” amount about data collection, and their general understanding of the scientific process. Lastly, ten out of fifteen students reported that their post-graduation plans had changed to pursue either a Masters or Doctorate degrees in a science-related field, including fields in forestry and plant pathology.
Collaboration with undergraduate science programs can increase research capacity, especially at the early stages of exploration. The whole class project expanded the survey area of the primary lab, allowing researchers to gain access to field data that was not in an established survey area. While the class project did not recover Phytophthora species, they did isolate Pythium species, another pathogen group belonging to phylum Oomycota. This finding may warrant further investigation should Pythium spp. continue to be isolated from soil surrounding Western redcedar exhibiting dieback symptoms.
Engagement with undergraduate researchers increases student confidence in the research process, leads to improved student outcomes, and can lead to increased interest in research-based careers. It is crucial to prepare the students for the ambiguities associated with the research process and to develop mechanisms to confirm data quality. Students had to be flexible exploring a new project with little known information. Working in collaboration with research labs allowed the faculty-educator to expand their research program while providing an impactful educational experience for students who may not have access to research mentoring opportunities otherwise.
Citizen scientists in the Forest Health Watch program were invited to participate in a preliminary study about the diversity of oomycetes associated with western redcedar trees in 2021. Citizen scientists collected soil samples under healthy and unhealthy trees and the soils were baited to assess and compare oomycete communities. The purpose of this study was to investigate the possible association between the dieback of western redcedar and the presence of oomycetes.
The Forest Health Watch program was designed as an umbrella for multiple citizen science projects starting in 2020. The first project was initiated to study the factors associated with the dieback of western redcedar. Citizen scientists shared observations of healthy and unhealthy trees using iNaturalist (Fraisl et al., 2022). Notable variation was observed within sites, often with healthy trees next to unhealthy trees, stimulating questions about the possible roles of plant pathogens. Citizen scientists were then invited to collect soils to investigate the possible associations with soil oomycete communities.
Project leaders, students, and citizen scientists collected soil samples independently and in groups as part of a preliminary oomycete survey in the Forest Health Watch program (Figure 8). Soil was collected from four points within 3 m of a tree and then mixed in a single sample bag. The soil was collected after removing the litter and duff layers and digging down to a depth of 10 cm. Fine roots were also included in soil samples.
Soil samples were baited for 10 days at room temperature. Baits included leaves of rhododendron, lilac, and rose petals (Figure 2B). Bait leaves were removed from soil samples when symptomatic, with visible margins of leaf necrosis. Primary isolations were made by cutting bait tissues and adding them to petri-plates containing PARPH V8 media (Ferguson and Jeffers, 1999). Secondary isolations were conducted after 2–4 days by hyphal tipping. Isolate collections were grouped based on morphology and 2–4 representative isolates were prepared for sequencing.
DNA was extracted and the ITS region was amplified as described in the Washington Stream Survey project above using ITS4 and ITS6 primers (Cooke et al., 2000). Forward and reverse directions of the ITS region were sequenced by GeneWiz (Azenta Life Sciences, New Jersey) and aligned using Geneious Prime® version 2022.2.2. Amplicons were compared to NCBI Genbank and internal databases using BLAST and identities were confirmed by comparing sequences to published sequences with phylogenetic trees.
Ninety-two samples were collected under redcedar trees from 26 sites at 9 locations by researchers and 7 citizen scientists in the preliminary study. Forty-five of the redcedar trees were healthy, 38 trees were unhealthy (i.e., trees had crown dieback or thinning foliage), and 9 trees were dead. Oomycetes were recovered from 59 soil samples (collected under 27 healthy, 28 unhealthy and 4 dead trees). Of those soil samples, Phytophthora species were collected from soils surrounding 4 healthy, 6 unhealthy and 2 dead trees).
Nine Phytophthora species were recovered, including four Clade 6 species (P. bilorbang, P. chlamydospora, P. crassamura, and P. megasperma), two Clade 2 species (P. occultans and P. plurivora), one Clade 3 species (P. pseudosyringae), one Clade 7 species (P. cambivora) and one Clade 8 species (P. syringae). Phytophthora plurivora was recovered from soils surrounding 5 trees, P. megasperma was recovered from soils surrounding 4 trees, and P. chlamydospora and P. syringae were both recovered from soils surrounding 2 trees. The remaining species were only recovered from soils surrounding one tree each.
This preliminary research conducted while engaging citizen scientists has generated new research needs. The general low recovery of Phytophthora species may indicate they are not primary factors in the dieback of Western redcedar, but their presence in these systems warrants further research and exploration to understand their function and impacts. Special attention for the species recovered under multiple trees and locations (e.g., P. plurivora and P. megasperma) would be especially valuable. More robust surveys and collections around individual trees may reveal higher levels of presence and would add confidence in accepting or rejecting their possible role in the dieback of western redcedar.
The results of the Forest Health Watch soil sampling activities demonstrate the potential to engage community scientists to collect Phytophthora species from soils. Although Phytophthora species recovery was only detected in 13% of total samples, the project demonstrates citizen scientists can help collect enough samples for surveying difficult to isolate Phytophthora communities. Furthermore, the results of this project indicate more research is needed to understand the possible roles of these species, especially in association with the dieback of western redcedar.
Preparing citizen scientists for the generally low recovery and providing real-time sample process feedback may be valuable for setting expectations and fostering continued motivation to participate. Citizen scientists generally want to know the outcomes and results of their work. Because soil sampling requires thorough post-collection laboratory procedures, developing a real-time sample process update may help citizen scientists stay engaged. For example, a system to indicate ‘samples flooded, ‘baits added’, primary isolations completed, ‘secondary isolations completed’, ‘DNA extracted’ may satisfy citizen scientists curiosities about the laboratory process.
Recognizing variation in preferences for participation may also ensure a broader community of citizen scientists are included. While some citizen scientists have continued to collect samples independently after initial training, other citizen scientists preferred to collect samples in a group as part of an organized hike. For example, one citizen scientist indicated they participated “to get out for a purpose” when referring to a 1-day group sampling effort organized by partners of the Forest Health Watch. However, this citizen scientist has not collected samples independently since the activity. Additionally, in some cases, citizen scientists may be most engaged if they are networking or building community with other citizen scientists. Therefore, organizing multiple types of activities or methods to engage with other citizen scientists may improve project inclusion.
Developing projects as part of a larger program can help build a long-lasting network of citizen scientists. The soil sampling efforts were only a small and preliminary part of a larger project to better understand the factors associated with the dieback of western redcedar, and this project is only one of many in the Forest Health Watch program. The relationships developed and the growing network of citizen scientists engaged in each project can be leveraged for future research and called to action if other Phytophthora species or forest pests emerge as issues.
The results of these case studies demonstrate there is merit to engaging citizen scientists in oomycete surveys. Here we summarized five projects that involved community groups in monitoring and research of Phytophthora, the important plant pathogen group that often has devastating ecological impacts. Although these organisms are complex and generally require technical laboratory methods and expertise to study, citizen scientists can recognize their impacts and can aid in their collection.
Project approaches varied in terms of research, engagement, and education outcomes. Notable differences included the geographic scope, audience, participant motivations, sampling methods, and whether the projects were designed for monitoring, exploration, or both. Additional sources of variation were noted in recruitment strategies, training approaches, lab capacity, target organisms, needs for education, and partnerships with regulatory agencies.
Including citizens increased the sampling distributions of surveys in each case study on multiple scales and scopes geographically. For example, in the SOD Blitz program the severity of the problem caused by SOD in California, coupled with a string of discoveries related to the causal agent within a short period of time, generated a strong collaboration between the public and researchers. Citizen Scientists collected a large quantity of high-quality data over many SOD blitzes becoming the backbone of the Statewide SOD survey efforts in California. This dataset informed numerous research pursuits and resulted in multiple peer reviewed publications. Another strength of the program has been the repeated participation by a large percentage of volunteers. SOD blitzes are a true example of how to generate large amounts of solid, crowd-sourced scientific data while engaging and educating a large proportion of the public (Dickinson et al., 2012).
Citizen scientists involved with the P. ramorum monitoring project in Oregon increased the resolution of sampling efforts by baiting local streams and trees that would not have been sampled otherwise. In contrast to this local effort, the SOD Blitz program has demonstrated the merit of engaging the public to identify hotspots of disease emergence on a regional scale (Meentemeyer et al., 2015). Engagement of citizen scientists may also increase access to private lands. For example, all of the projects discussed engaged citizen scientists or students to monitor or collect samples on private lands, especially for early detection. Projects with larger geographic scopes unsurprisingly had notably large citizen scientist pools in contrast to smaller site-specific projects. Likewise, projects with longer sampling periods requiring more time commitment had fewer volunteers.
The audiences and participants also varied between projects and each project differed in objectives. Three of the projects were initiated to monitor for P. ramorum where the other two were more exploratory and attempted to find associations with the dieback of western redcedar. These differences demonstrate the breadth of citizen science applications, including both investigation and discovery. Local projects tended to engage specific audiences such as landowners, retired professionals, or student groups. In contrast, the SOD Blitz project benefited from mass participation where anyone could participate. Although the audience did vary, some groups such as Master Gardeners, participated in multiple case studies. These findings were consistent with Master Gardener volunteer engagement in citizen science monitoring efforts for alien arthropod species (O'Callaghan and Skelly, 2013) and in applied research projects (Davenport-Hagen et al., 2019). The Pacific University Phytophthora monitoring is a great example of a project that reaches a different audience and demonstrates the value of engaging students in tertiary education systems.
Each case study had different techniques for training citizen scientists. While a critical component to project success, training and educational programs are time consuming. Some projects included guest lectures or guidance from researchers and laboratory technicians in the region. Most projects also included in-person sampling activities tailored for small to large groups. The Forest Health Watch program also produced field guides and included virtual office hours for citizen scientists. The P. ramorum monitoring project in Oregon utilized webinar recordings, originally necessitated by Covid-19 shutdowns, but later found to be efficient for initial training and review. These methods may also be suitable for newer teaching pedagogies. For example, Larkin et al. (2018) assessed flipping the classroom, a pedagogical method where students review lecture content before coming to class and then engaged in problem-based learning during class, to provide education more efficiently in citizen science programs. When possible, the programs also trained individuals to train other groups. In all cases, training and outreach opportunities tended to improve data quality.
In each case study, citizen scientists were involved in the sample collection, but generally not the lab diagnostics. However, these projects do provide opportunities to engage and train interested citizen scientists in laboratory protocols leading to heightened project commitment and possible pathways into microbiology careers. Being a part of a science-based monitoring approach provides learning opportunities as well as contributing to monitoring efforts, both seen as important motivations for citizen scientists (Domroese and Johnson, 2017).
The Pacific University Phytophthora monitoring also demonstrates a ‘sweet spot’ where educational outcomes and research can formally overlap. While some students participated solely because of the educational outcomes rather than the scientific findings, the project introduced the students to a group of organisms that would not have been covered typically. The partnerships between educators, students, and researchers ensured the student engagement was valuable for both research and educational outcomes simultaneously.
Although most of the case studies here attempted to isolate Phytophthora from bait leaves, there may be more capacity for direct isolation from symptomatic plant tissues. For example, the OSU Forest Pathology Lab, which supports the Oregon P. ramorum monitoring program, receives petri plates with symptomatic plant tissues for regulatory monitoring. This approach may be feasible with arborists or other professionals, especially in circumstances with individual species and consistent host symptoms associated with P. ramorum such as tanoak bleeds.
Involving citizen scientists and students expanded research priorities of many of the case studies. Engaging students in the Washington stream monitoring project revealed the presence of a novel species of Halophytophthora, stimulating interest about the role of oomycetes in brackish water. These findings also led to the discovery of a new Phagomyxid parasite on eelgrass (Zostera marina) (Elliott et al., 2019). In the P. ramorum monitoring project in Oregon, where incidental Phytophthora species were not necessarily identified, the finding of other Phytophthora species during stream baiting near disease resistant Port-Orford-cedar plantings prompted discussions on broadening the identification to include P. lateralis. Understanding pathogen pressures on disease resistant plantings is helpful to understanding overall Port-Orford-cedar disease resistant durability in the environment (Sniezko et al., 2020). Discussions to include P. lateralis in future surveys was an example of expanded research priorities.
Although engaging citizen scientists increased capacity for sample collection, sample processing and laboratory capacity was still a general limitation. Each case study had different sample processing capacity and some projects explored options for streamlining processing times. The P. ramorum monitoring project in Oregon explored the use of newer technologies like ELISA kits, which are accessible enough for citizen scientists to use independently with the potential for enhanced sample screening before laboratory diagnostics. The continued development of field based rapid tests will be advantageous for any type of monitoring program (Cooke et al., 2007) and are directly applicable to citizen science programs.
Positive findings can be a possible drawback when monitoring regulated pathogens with citizen scientists because they can be ecologically and economically costly. Because citizen scientists in the Oregon and California projects surveyed for a regulated pathogen, the possible finding made some participants anxious about the results. Regulatory aspects may deter some landowners from revealing information if it results in changes to the property or costs in responding (e.g., destroying infested material or tree felling). These dilemmas may limit the retention of citizen scientists or decrease engagement, and require careful program design to maintain trust (Pocock et al., 2020). In contrast, the potential to strongly affect policy and land management decisions may be a motivating factor for some citizen scientists to participate. For example, in Minnesota citizen scientists collected complex data on invasive oak pests despite knowing the possible outcomes and risks of regulatory control on their own land (Andow et al., 2016).
Positive samples collected by citizen scientists must be confirmed by trained personnel. The use of field-based diagnostics presents the risk of inaccurate test interpretation or unsubstantiated detections. All positive detections must be verified on-site and with laboratory confirmation. Oftentimes, detections are only the start of a project and once a detection is made, the citizen science project may need to shift or adapt its approach. For example, while stream sampling provides information on the species present in a watershed, identifying the source of potential infestations requires ground survey follow-up (Sutton et al., 2009). In the P. ramorum monitoring project in Oregon, project leaders generally tried to engage citizen scientists on the leading margin of the quarantine zone to prioritize early detections. To retain citizen scientists, post detection, options can be explored to adapt projects for further engagement. These include the possibility of revisiting sites where P. ramorum was previously recovered to investigate pathogen persistence or potential movement into neighboring ecosystems.
When designing citizen science opportunities, it is important to consider the lessons learned from related citizen science projects and adapt the methods for future opportunities. For example, targeted sampling can efficiently address localized issues and reduce data fragmentation. Ensure that there is lab capacity to produce timely results. Otherwise, explore partnerships with area collaborators. These may be private or public entities that have different objectives but have lab capacity to provide diagnostics for associated monitoring projects or baseline diversity studies. Additionally, involving collaborators across institutions can expand monitoring networks and may lead to longer-term research projects.
Effective approaches in preparing citizen scientists for data collection include providing training on specific applied methods utilized in the project. This should incorporate demonstrations of techniques that the citizen scientists will later deploy on their own. During this training, it is crucial to communicate the impacts of regulated pathogens. It is also essential to clearly communicate points of contact for questions regarding the project as a whole or sampling methods. In-person and virtual training are possible methods for training citizen scientists; this relies heavily on choosing the appropriate approach for the audience and the project. During training, data collection kits could be provided. Providing kits to citizen scientists may enable more participation and increase data quality because sampling is more standardized.
During the entire duration of a citizen scientist project, ongoing engagement with participants is a key element for project success. Motivating citizens to continue participation throughout the project can be done through media campaigns, news articles or blog posts. These are all ways to publicly highlight citizen scientist accomplishments and in addition will provide updates to participants on project outcomes. Post-project communication is also important as it allows the citizen scientists to see the collective effort of all involved, access results, and understand the significance of their data. There is also an opportunity after the project, to continue to engage volunteers by providing educational updates and new opportunities for involvement.
Implementing citizen scientist projects is the next step towards successfully engaging communities and classrooms in scientific research. There is a significant opportunity to develop undergraduate researchers as valuable participants for the early detection or discovery of plant pathogens. Bringing citizen science projects into classrooms can help retain non-traditional and underrepresented science students in plant pathology fields.
Citizen science initiatives increase outreach on complex topics. The case studies presented involved the inclusion of multiple groups, from students, landowners, master gardeners, to a diversity of other community members leading to high-impact educational opportunities. Our projects demonstrate that involvement with citizen scientists can lead to increased surveillance of current and future invasive Phytophthora species. Samples collected by citizen scientists have increased sampling distribution, increased knowledge of species distribution, contributed to the monitoring of quarantine zones, and the detection of new species. The advantages gained by the collaboration with citizens have ultimately led to many new research ventures and priorities. Lastly, our work demonstrates that citizen science projects provide the potential to expand research networks between separate labs and institutions.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
MG was supported by USFS Region 5 State and Private Forestry, Gordon and Betty Moore Foundation, NSF Ecology of Infectious Disease Program, PG & E Foundation, Mid Peninsula Open Space. NK was supported by USDA Animal and Plant Health Inspection Service and the USFWS OR Coastal Program. JH was supported by USDA National Institute of Food and Agriculture through a Postdoctoral Fellowship agreement #2023-00353 and funding provided by the Albert Victor Ravenholt Fund. KL and TW were supported by the Pacific University College of Arts and Sciences Undergraduate Research Funds.
The authors would like to acknowledge the many citizen scientists and students who have participated in the monitoring and detection of invasive Phytophthora species in the western United States.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor SH is currently organizing a Research Topic with the author JH.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1130210/full#supplementary-material
Abad, Z. G., Burgess, T., Redford, A. J., Bienapfl, J. C., Mathew, R., Srivastava, S. K., et al. (2022). IDphy: an international online resource for molecular and morphological identification of Phytophthora. Plant Dis. 107, 987–998. doi:10.1094/PDIS-02-22-0448-FE
Andow, D. A., Borgida, E., Hurley, T. M., and Williams, A. L. (2016). Recruitment and retention of volunteers in a citizen science network to detect invasive species on private lands. Environ. Manag. 58, 606–618. doi:10.1007/s00267-016-0746-7
Aristeidou, M., Scanlon, E., and Sharples, M. (2020). Learning outcomes in online citizen science communities designed for inquiry. Int. J. Sci. Educ. Part B 10, 277–294. doi:10.1080/21548455.2020.1836689
Awad, M. M., and Brown, M. A. (2021). Undergraduate research as a high-impact practice for engaging students of color in ecology. Bull. Ecol. Soc. Am. 102, 1816. doi:10.1002/bes2.1816
Bonney, R., Cooper, C. B., Dickinson, J., Kelling, S., Phillips, T., Rosenberg, K. V., et al. (2009). Citizen science: a developing tool for expanding science knowledge and scientific literacy. BioScience 59, 977–984. doi:10.1525/bio.2009.59.11.9
Buhl, C., Navarro, S., Norlander, D., Williams, W., Heath, Z., Ripley, K., et al. (2019). Forest health highlights in Oregon. Salem, OR: United States Department of Agriculture.
Calmin, G., Belbahri, L., and Lefort, F. (2007). Direct PCR for DNA barcoding in the genera phytophopthora and Pythium. Biotechnol. Biotechnol. Equip. 21, 40–42. doi:10.1080/13102818.2007.10817410
Cooke, D. E. L., Drenth, A., Duncan, J. M., Wagels, G., and Brasier, C. M. (2000). A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 30, 17–32. doi:10.1006/fgbi.2000.1202
Cooke, D. E. L., Schena, L., and Cacciola, S. O. (2007). Tools to detect, identify and monitor Phytophthora species in natural ecosystems. J. Plant Pathology 89, 13–28.
Crowl, T. A., Crist, T. O., Parmenter, R. R., Belovsky, G., and Lugo, A. E. (2008). The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 6, 238–246. doi:10.1890/070151
Cunniffe, N. J., Cobb, R. C., Meentemeyer, R. K., Rizzo, D. M., and Gilligan, C. A. (2016). Modeling when, where, and how to manage a forest epidemic, motivated by sudden oak death in California. Proc. Natl. Acad. Sci. U.S.A. 113, 5640–5645. doi:10.1073/pnas.1602153113
Davenport-Hagen, L., Weisenhorn, J., and Meyer, M. H. (2019). Applied research engages extension master gardener volunteers. J. Ext. 54, 6.
Dickinson, J., Shirk, J., Bonter, D., Bonney, R., Crain, R. L., Martin, J., et al. (2012). The current state of citizen science as a tool for ecological research and public engagement. Front. Ecol. Environ. 10, 291–297. doi:10.1890/110236
Domroese, M. C., and Johnson, E. A. (2017). Why watch bees? Motivations of citizen science volunteers in the great pollinator project. Biol. Conserv. 208, 40–47. doi:10.1016/j.biocon.2016.08.020
Elliott, J. K., Simpson, H., Teesdale, A., Replogle, A., Elliott, M., Coats, K., et al. (2019). A novel Phagomyxid parasite produces sporangia in root hair galls of Eelgrass (Zostera marina). Protist 170, 64–81. doi:10.1016/j.protis.2018.12.001
Encarnação, J., Teodósio, M. A., and Morais, P. (2021). Citizen science and biological invasions: a review. Front. Environ. Sci. 8, 602980. doi:10.3389/fenvs.2020.602980
Eyre, C. A., Kozanitas, M., and Garbelotto, M. (2013). Population dynamics of aerial and terrestrial populations of Phytophthora ramorum in a California forest under different climatic conditions. Phytopathology® 103, 1141–1152. doi:10.1094/PHYTO-11-12-0290-R
Ferguson, A. J., and Jeffers, S. N. (1999). Detecting multiple species of Phytophthora in container mixes from ornamental crop nurseries. Plant Dis. 83, 1129–1136. doi:10.1094/PDIS.1999.83.12.1129
Fraisl, D., Hager, G., Bedessem, B., Gold, M., Hsing, P. Y., Danielsen, F., et al. (2022). Citizen science in environmental and ecological sciences. Nat. Rev. Methods Prim. 2, 64. doi:10.1038/s43586-022-00144-4
Ganesh, C., and Smith, J. (2017). Using multiple high-impact practices to reduce bottlenecks and improve student learning in an undergraduate health science program. JoSoTL 17, 74–84. doi:10.14434/josotl.v17i2.20852
Garbelotto, M., Dovana, F., Schmidt, D., Chee, C., Lee, C., Fieland, V. J., et al. (2021a). First reports of Phytophthora ramorum clonal lineages NA1 and EU1 causing sudden oak death on tanoaks in Del Norte county, California. Plant Dis. 105, 2737. doi:10.1094/PDIS-12-20-2633-PDN
Garbelotto, M., Maddison, E. R., and Schmidt, D. (2014). SODmap and SODmap mobile: two tools to monitor the spread of sudden oak death. For. Phytophthoras 4, 3560. doi:10.5399/osu/fp.4.1.3560
Garbelotto, M., Popenuck, T., Hall, B., Schweigkofler, W., Dovana, F., Goldstein de Salazar, R., et al. (2020). Citizen science uncovers Phytophthora ramorum as a threat to several rare or endangered California manzanita species. Plant Dis. 104, 3173–3182. doi:10.1094/PDIS-03-20-0619-RE
Garbelotto, M., and Rizzo, D. M. (2005). A California-based chronological review (1995–2004) of research on Phytophthora ramorum, the causal agent of sudden oak death. Phytopathol. Mediterr. 44, 17.
Garbelotto, M., Schmidt, D., and Popenuck, T. (2021b). Pathogenicity and infectivity of Phytophthora ramorum vary depending on host species, infected plant part, inoculum potential, pathogen genotype, and temperature. Plant Pathol. 70, 287–304. doi:10.1111/ppa.13297
Garbelotto, M., Svihra, P., and Rizzo, D. M. (2001). New pests and diseases: sudden oak death syndrome fells 3 oak species. Calif. Agric. 55, 9–19. doi:10.3733/ca.v055n01p9
Goheen, E., Kanaskie, A., Navarro, S., and Hansen, E. (2017). Sudden oak death management in Oregon tanoak forests. Phytophthoras 7. doi:10.5399/osu/fp.7.1.4030
Grünwald, N. J., Goss, E. M., and Press, C. M. (2008). Phytophthora ramorum: a pathogen with a remarkably wide host range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol. Plant Pathol. 9, 729–740. doi:10.1111/j.1364-3703.2008.00500.x
Grünwald, N. J., Larsen, M. M., Kamvar, Z. N., Reeser, P. W., Kanaskie, A., Laine, J., et al. (2016). First report of the EU1 clonal lineage of Phytophthora ramorum on tanoak in an Oregon forest. Plant Dis. 100, 1024. doi:10.1094/PDIS-10-15-1169-PDN
Hansen, E. M., ReeserSuttonKanaskie, P. W. A., Navarro, S., Goheen, E. M., and Goheen, E. M. (2019). Efficacy of local eradication treatments against the sudden oak death epidemic in Oregon tanoak forests. For. Pathol. 49 (4), 12530. doi:10.1111/efp.12530
Hansen, E. M. (2008). Alien forest pathogens: phytophthora species are changing world forests. Boreal Environ. Res. 13, 33–41.
Hansen, E. M., Goheen, D. J., Jules, E. S., and Ullian, B. (2000). Managing port-orford-cedar and the introduced pathogen Phytophthora lateralis. Plant Dis. 84, 4–14. doi:10.1094/PDIS.2000.84.1.4
Hansen, E. M., Kanaskie, A., Prospero, S., McWilliams, M., Goheen, E. M., Osterbauer, N., et al. (2008). Epidemiology of Phytophthora ramorum in Oregon tanoak forests. Can. J. For. Res. 38, 1133–1143. doi:10.1139/X07-217
Hayden, K., Ivors, K., Wilkinson, C., and Garbelotto, M. (2006). TaqMan chemistry for Phytophthora ramorum detection and quantification, with a comparison of diagnostic methods. Phytopathology 96, 846–854. doi:10.1094/PHYTO-96-0846
Hulbert, J. M., Agne, M. C., Burgess, T. I., Roets, F., and Wingfield, M. J. (2017). Urban environments provide opportunities for early detections of Phytophthora invasions. Biol. Invasions 19, 3629–3644. doi:10.1007/s10530-017-1585-z
Hulbert, J. (2021). Western redcedar trees need your help tualatin soil and water conservation district. Available at: https://tualatinswcd.org/western-redcedar-trees-need-your-help/.
Jordan, R. C., Gray, S. A., Howe, D. V., Brooks, W. R., and Ehrenfeld, J. G. (2011). Knowledge gain and behavioral change in citizen-science programs: citizen-scientist knowledge gain. Conserv. Biol. 25, 1148–1154. doi:10.1111/j.1523-1739.2011.01745.x
Kox, L. F. F., Brouwershaven, I. R. V., Vossenberg, B. T. L. H. V., Beld, H. E. V., Bonants, P. J. M., and Gruyter, J. D. (2007). Diagnostic values and utility of immunological, morphological, and molecular methods for in planta detection of Phytophthora ramorum. Phytopathology 97, 1119–1129. doi:10.1094/PHYTO-97-9-1119
Kroon, L. P. N. M., Bakker, F. T., van den Bosch, G. B. M., Bonants, P. J. M., and Flier, W. G. (2004). Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet. Biol. 41, 766–782. doi:10.1016/j.fgb.2004.03.007
Kuh, G. D., and Schneider, C. G. (2008). High-impact educational practices: what they are, who has access to them, and why they matter. Washington, DC: Association of American Colleges and Universities.
Larkin, D., Weber, M., Galatowitsch, S., and Gupta, A. (2018). Flipping the classroom to train citizen scientists in invasive species detection and response. J. Ext. 56, 4. doi:10.34068/joe.56.05.04
Larson, E. R., Graham, B. M., Achury, R., Coon, J. J., Daniels, M. K., Gambrell, D. K., et al. (2020). From eDNA to citizen science: emerging tools for the early detection of invasive species. Front. Ecol. Environ. 18, 194–202. doi:10.1002/fee.2162
LeBoldus, J. M., Navarro, S. M., Kline, N., Ritokova, G., and Grünwald, N. J. (2022). Repeated emergence of sudden oak death in Oregon: chronology, impact, and management. Plant Dis. 106, 3013–3021. doi:10.1094/PDIS-02-22-0294-FE
Lione, G., Gonthier, P., and Garbelotto, M. (2017). Environmental factors driving the recovery of bay laurels from Phytophthora ramorum infections: an application of numerical ecology to citizen science. Forests 8, 293–324. doi:10.3390/f8080293
Man in ’t Veld, W. A., Rosendahl, K. C. H. M., van Rijswick, P. C. J., Meffert, J. P., Boer, E., Westenberg, M., et al. (2019). Multiple Halophytophthora spp. and Phytophthora spp. including P. gemini, P. inundata and P. chesapeakensis sp. nov. isolated from the seagrass Zostera marina in the Northern hemisphere. Eur. J. Plant Pathol. 153, 341–357. doi:10.1007/s10658-018-1561-1
Martin, F. N., Coffey, M. D., Zeller, K., Hamelin, R. C., Tooley, P., Garbelotto, M., et al. (2009). Evaluation of molecular markers for Phytophthora ramorum detection and identification: testing for specificity using a standardized library of isolates. Phytopathology® 99, 390–403. doi:10.1094/PHYTO-99-4-0390
Mascheretti, S., Croucher, P. J. P., Kozanitas, M., Baker, L., and Garbelotto, M. (2009). Genetic epidemiology of the sudden oak death pathogen Phytophthora ramorum in California. Mol. Ecol. 18, 4577–4590. doi:10.1111/j.1365-294X.2009.04379.x
Mascheretti, S., Croucher, P. J. P., Vettraino, A., Prospero, S., and Garbelotto, M. (2008). Reconstruction of the Sudden Oak Death epidemic in California through microsatellite analysis of the pathogen Phytophthora ramorum. Mol. Ecol. 17, 2755–2768. doi:10.1111/j.1365-294X.2008.03773.x
Meentemeyer, R. K., Dorning, M. A., Vogler, J. B., Schmidt, D., and Garbelotto, M. (2015). Citizen science helps predict risk of emerging infectious disease. Front. Ecol. Environ. 13, 189–194. doi:10.1890/140299
Norman-Burgdolf, H., and Rieske, L. K. (2021). Healthy trees – healthy people: a model for engaging citizen scientists in exotic pest detection in urban parks. Urban For. Urban Green. 60, 127067. doi:10.1016/j.ufug.2021.127067
O'Callaghan, A. M., and Skelly, J. (2013). Community involvement to reduce insect threats to urban forests. J. Ext. 51 (6).
Osmundson, T. W., Robert, V. A., Schoch, C. L., Baker, L. J., Smith, A., Robich, G., et al. (2013). Filling gaps in biodiversity knowledge for macrofungi: Contributions and assessment of an herbarium collection DNA barcode sequencing project. PLoS One 8 (4), e62419. doi:10.1371/journal.pone.0062419
Pandya, R. E. (2012). A framework for engaging diverse communities in citizen science in the US. Front. Ecol. Environ. 10, 314–317. doi:10.1890/120007
Peterson, E. K., Søndreli, K. L., Reeser, P., Navarro, S., Nichols, C., Wiese, R., et al. (2022). First report of the NA2 clonal lineage of the sudden oak death pathogen, Phytophthora ramorum, infecting tanoak in Oregon forests. Plant Dis. 106, 2537. doi:10.1094/PDIS-10-21-2152-PDN
Pocock, M. J. O., Marzano, M., Bullas-Appleton, E., Dyke, A., de Groot, M., Shuttleworth, C. M., et al. (2020). Ethical dilemmas when using citizen science for early detection of invasive tree pests and diseases. Manag. Biol. Invasions 11, 720–732. doi:10.3391/mbi.2020.11.4.07
Pocock, M. J. O., Tweddle, J. C., Savage, J., Robinson, L. D., and Roy, H. E. (2017). The diversity and evolution of ecological and environmental citizen science. PLoS ONE 12, e0172579. doi:10.1371/journal.pone.0172579
Rizzo, D. M., Garbelotto, M., Davidson, J. M., Slaughter, G. W., and Koike, S. T. (2002). Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Dis. 86, 205–214. doi:10.1094/PDIS.2002.86.3.205
Rizzo, D. M., Garbelotto, M., and Hansen, E. M. (2005). Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests. Annu. Rev. Phytopathol. 43, 309–335. doi:10.1146/annurev.phyto.42.040803.140418
Saville, A. C., and Ristaino, J. B. (2021). Global historic pandemics caused by the FAM-1 genotype of Phytophthora infestans on six continents. Sci. Rep. 11, 12335. doi:10.1038/s41598-021-90937-6
Shew, H. D., and Bensen, D. M. (1982). Qualitative and quantitative soil assays for Phytophthora cinnamomi. Phytopathology 72, 1029. doi:10.1094/Phyto-72-1029
Shortlidge, E. E., and Brownell, S. E. (2016). How to assess your cure: a practical guide for instructors of course-based undergraduate research experiences. J. Microbiol. Biol. Educ. 17, 399–408. doi:10.1128/jmbe.v17i3.1103
Sniezko, R. A., Johnson, J. S., Reeser, P., Kegley, A., Hansen, E. M., Sutton, W., et al. (2020). Genetic resistance to Phytophthora lateralis in Port-Orford-cedar (Chamaecyparis lawsoniana) – basic building blocks for a resistance program. Plants People Planet 2, 69–83. doi:10.1002/ppp3.10081
Sutton, W., Hansen, E. M., Reeser, P. W., and Kanaskie, A. (2009). Stream monitoring for detection of Phytophthora ramorum in Oregon tanoak forests. Plant Dis. 93, 1182–1186. doi:10.1094/PDIS-93-11-1182
Keywords: citizen science, phytophthora, plant disease, monitoring, public engagement, invasive species
Citation: Lanning KK, Kline N, Elliott M, Stamm E, Warnick T, LeBoldus JM, Garbelotto M, Chastagner G and Hulbert JM (2023) Citizen science can add value to Phytophthora monitoring: five case studies from western North America. Front. Environ. Sci. 11:1130210. doi: 10.3389/fenvs.2023.1130210
Received: 22 December 2022; Accepted: 04 August 2023;
Published: 17 August 2023.
Edited by:
Susan M. Hester, The University of Melbourne, AustraliaReviewed by:
Edith Arndt, University of Melbourne, AustraliaCopyright © 2023 Lanning, Kline, Elliott, Stamm, Warnick, LeBoldus, Garbelotto, Chastagner and Hulbert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kara K. Lanning, a2FyYS5sYW5uaW5nQHBhY2lmaWN1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.