
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci., 26 August 2022
Sec. Toxicology, Pollution and the Environment
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.989267
This article is part of the Research TopicHazardous Contaminants Associated with Plastics: Occurrence and Environmental EffectsView all 7 articles
 Susanne Kublik1*
Susanne Kublik1* Silvia Gschwendtner1
Silvia Gschwendtner1 Tobias Magritsch1
Tobias Magritsch1 Viviane Radl1
Viviane Radl1 Matthias C. Rillig2
Matthias C. Rillig2 Michael Schloter1,3
Michael Schloter1,3Microplastic (MP) pollution poses a threat to agricultural soils and may induce a significant loss of the soil quality and services provided by these ecosystems. Studies in marine environments suggest that this impact is mediated by shifts in the microbiome. However, studies on the mode of action of MP materials on the soil microbiome are rare, particularly when comparing the effects of different MP materials. In this study, we characterized the microbiota colonizing two different MP materials, granules made of polypropylene (PP) and expanded polystyrene (ePS), introduced into arable soil and incubated for 8 weeks using a molecular barcoding approach. We further assessed the consequences on the microbiome of bulk soil. The complexity of the bacterial communities colonizing MP materials was significantly higher on ePS compared to PP. Many of the detected genera colonizing the MP materials belonged to taxa, that are known to degrade polymeric substances, including TM7a, Phenylobacterium, Nocardia, Arthrobacter and Streptomyces. Interestingly, in bulk soil samples amended with MP materials, microbial diversity was higher after 8 weeks compared to the control soil, which was incubated without MP materials. The composition of bacterial communities colonizing the MP materials and bulk soil differed. Mainly Acidobacteria were mostly found in bulk soil, whereas they were rare colonizers of the MP materials. Differences in diversity and community composition between the MP affected bulk soil samples were not found. Overall, our data indicate that MP materials form a new niche for microbes in soil, with a specific community composition depending on the materials used, strongly influencing the bulk soil microbiota in the short term. Long-term consequences for the soil microbiome and associated functions including different soils need to be further elaborated in the future for a proper risk assessment of the mode of action of MP materials in terrestrial ecosystems.
Plastic particles < 5 mm in diameter are defined as microplastic (MP) (Frias and Nash, 2019). MP pollution is present all over the globe, potentially affecting all ecosystems, including marine and freshwater environments as well as soils (Mahon et al., 2017; Corradini et al., 2019; Rillig et al., 2019; Oliveira et al., 2020; Rillig and Lehmann, 2020). Therefore analyzing the impact of MP on various organisms has become an important topic of research in the last decades. MP materials have an impact on biota of all trophic levels and may accumulate in the gut of animals, including mammals, as a result of the complex food web structures (Thiel et al., 2018; Cverenkárová et al., 2021; Ugwu et al., 2021). Recently MP has been discovered and quantified even in human blood (Leslie et al., 2022). Many MP materials enter the human body by consumption of food, derived from terrestrial ecosystems. Estimates indicated that the input of MP materials into terrestrial ecosystems might be 4- to 23-fold higher than the release into aquatic environments (Horton et al., 2017). Especially arable soils are an important sink for MP materials, due to the use of plastic mulch films and sewage sludge as fertilizer (Mahon et al., 2017). Another main source of MP contamination in terrestrial environments is tire abrasion (Sommer et al., 2018; Luo et al., 2021). It has been shown, that MP derived from tires supported pathogenic bacteria in urban water environments (Wang et al., 2020). If MP materials accumulate in the soil, they directly or indirectly affect the function of soil ecosystems. They could alter the physical properties of soil (e.g., increase water holding capacity or decrease bulk density) (de Souza Machado et al., 2018) influence the soil structure, soil fertility and affect soil biodiversity and functioning (Rillig, 2012). Effects of MP on the soil microbiome have been clearly demonstrated (Huang et al., 2019; Zhou et al., 2020; Hou et al., 2021; Luo et al., 2022; Shi et al., 2022; Zhu et al., 2022), affecting microbial activities and associated important functions including nutrient turnover, bacterial transport and the decomposition of organic matter. MP induced microbiome composition disruption, immune response, enzyme activity and gene expression changes (Santos et al., 2022). Ng et al. (2021) nicely demonstrated that MP materials act as selective niche for bacteria and fungi. The bacterial biofilms colonizing the MP materials change the surface properties of the MP. They affect its bioavailability, degradability and mobility. However, the knowledge on the effects of MP materials on the soil microbiome is still limited as they may strongly vary within different polymer types, shapes, sizes and concentrations, as well as in different soil types (Awet et al., 2018; De Souza Machado et al., 2018; De Souza Machado et al., 2019; Zhang et al., 2019; Rüthi et al., 2020; Sun et al., 2022). Defining MP as a single compound, as media articles or policy reports often did, led to confusions. Studying the fate and effects of a single MP material with a specific chemical composition, size and shape does not tell the fate and effects of MP in general (Rochman et al., 2019). It has been shown in the marine environment that different MP materials are colonized by different microorganisms (Rosato et al., 2020). In contrast to that many studies in the soil environment were based on field experiments of MP contaminated areas, e.g., Zhang et al., 2019; Chai et al., 2020; Shan et al., 2022. Those studies addressed different questions and did not focus on the microbial colonization of defined MP materials. It is important to fully understand the mechanism behind the colonization by performing controlled lab experiments using defined MP materials. To address this, a pot experiment using an agricultural soil was established to study the colonization of two different polymer types of MP materials by soil microbiota and the subsequent feedback loops on the bacterial community composition of the bulk soil. The MP materials polypropylene (PP) and expanded polystyrene (ePS) in granule shape were selected. PP and ePS are thermoplasts, but differ in their backbones. PP belongs to the group of polyolefins, is crystalline and non- polar. ePS is a non- polar, synthetic polymer made from monomers of the aromatic hydrocarbon styrene and it is foamed. Those polymer types are wide spread all over the world and represent 22% of the plastic material in Germany (Heinrich-Bö;ll-Stiftung, 2019).2 Both of them are frequently used as packing material and moved into almost every household. Given the fact, they are relevant contaminants of agricultural soils the knowledge on their effects on soil microbial communities is limited. As MPs concentrations in soil are increasing (Kim et al., 2020b) the results gained in this study would enhance the understanding of the ecological risk of PP and ePS MP contamination in agroecosystems and will be of importance for developing targeted mitigation and management strategies for affected sites to maintain soil quality.
Top soil (0–20 cm) was obtained from an agricultural field in Karlskron, Southern Germany (latitude 48.658142, longitude 11.381436, 407 m above sea level) in April 2019. The field was cultivated using principles of ecological farming. The crop rotation included alfalfa in 2018, triticale in 2017 and potatoes in 2016. The soil was characterized as loamy sand with a soil texture consisting of 14% clay, 19% silt, 67% sand and pH of 7.4. The soil contained 1.1% total carbon and 0.1% total nitrogen, resulting in a C: N ratio of 11. Ammonia and nitrate concentrations were 1.2 mg/g (NH4+) and 16.6 mg/g (NO3−) and dissolved organic carbon (DOC) content was 64.1 ± 2.5 µg g−1 dry weight (dw) at the time point of sampling. The soil was air-dried, sieved (2 mm mesh size) and homogenized before the experiment.
Two different polymer types were used in the experiment. The applied MP materials treatment consisted either of polypropylene granules (PP) of 4 mm nominal granule size (Merck, Darmstadt, Germany; item number GF63935860) or of expanded polystyrene granules (ePS) of 1–3 mm (SitJoy® Sitzsackfüllung, frago GmbH, Schenefeld, Germany; item number SW10361). The MP materials were sterilized for 1 week under ultra violet light. Sterilized MP granules were placed on LB-agar (Merck KGaA, Darmstadt, Germany) plates to verify the success of the sterilization. MP materials where no colony growth was observed after 1 week of incubation at room temperature were considered as sterile and used for the subsequent experiment.
Five replicates, each with 100 g of sieved soil were incubated at room temperature (RT) in seedling pots (size: 8 cm × 7 cm) for 8 weeks without MP materials, with 0.5% (w/w) pre-sterilized PP or 0.5% (w/w) pre-sterilized ePS. A homogeneous distribution of the MP materials granules was obtained by mixing them into the sieved soil using a sterile spoon. Control samples received the same amount of disturbance. Afterwards the water holding capacity of the soil was adjusted to 50% of the maximum water holding capacity separately for each treatment. After 8 weeks of incubation soil samples were taken. This time point was chosen based on a previous study published by Rüthi et al. (2020), who reported changes in the microbial community structure in soils after 8 weeks of incubation with MP materials. The DOC content in soil increased during the incubation period and was 81.0 ± 1.3 µg g−1 dw for control soil (without MP materials), 87.8 ± 10.4 µg g−1 dw for ePS soil and 198.5 ± 3.6 µg g−1 dw for PP soil after 8 weeks of MP materials incubation.
At the time point of sampling, the MP materials were separated from the soil using sterile tweezers (Gkoutselis et al., 2021) and carefully washed in 50 ml sterile 1 × PBS. The MP materials were therefore put into a sterile kitchen strainer “Teela-Spoon 1” fine mesh (mesh size 0.5 mm, diameter 3.5 cm) (Tee Gschwendner, Meckenheim, Germany) and dipped into the washing buffer twice for 1 minute and thoroughly washed by shaking. The adhering soil particles were removed, but the biofilm on the MP materials was not affected, that has been demonstrated in pre-experiments (data not shown). To verify the minimal disturbance of the washing procedure, the wash buffer was collected after use and subjected to molecular metabarcoding, as described below. Our data indicated that the observed number of amplicon sequence variants (ASVs) was in the range of 1%–4% of the ASVs, that were found on the MP and only a small overlap of taxa (Supplementary Figure S2) was present, indicating that the washing procedure did not affect the biofilms developed on the MP. MP free soil samples, control soil and washed MP materials were frozen at −80°C.
Genomic DNA was extracted from 0.5 g of soil, 0.25 g of PP, 0.15 g of ePS or 500 μl of PBS wash buffer to ensure best cell lysing conditions for each sample type by using the NucleoSpin® Soil Kit (Macherey-Nagel, Düren, Germany). For cell lysis of soil and plastic- associated microbes, buffer SL1 was used. As an extraction control (blank), an extraction without soil or MP materials was processed. DNA was quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific, Darmstadt, Germany). For the assessment of bacterial communities the “16S Metagenomics Sequencing Library Preparation” protocol (Illumina, San Diego, CA, United States) and quality guidelines by Schöler et al. (2017) were used. The v4 region of the 16S rRNA gene was amplified using primer pair 515F (Parada et al., 2016) and 806R (Apprill et al., 2015). PCR reaction mixtures contained 10 ng of DNA for soil and 3 μl of DNA for MP materials, PBS wash and blanks, 0.5 μl of 10 pmol of each primer, 2.5 μl of 3% BSA, 12.5 μl of NEBNext High-Fidelity 2x PCR Master Mix (New England Biolabs, Frankfurt am Main, Germany) and DEPC- treated water up to 25 μl. The amplification program for the 16S rRNA gene was initiated at 98°C for 1 min, followed by 25 cycles of 98°C for 10 s, 55°C for 30 s and 72°C for 30 s, and terminated at 72°C for 5 min. PCR products were purified using MagSi NGSprep Plus beads (Steinbrenner, Wiesenbach, Germany) and quantified and quality checked using the Fragment Analyzer (Agilent Technologies, Santa Clara, CA, United States) using the NGS Fragment Kit (1–6,000 bp) (Agilent Technologies, Santa Clara, CA, United States). Indexing PCR was performed in a reaction mix (25 μl) consisting of 10 ng of the purified amplicon, 2.5 μl of each indexing primer (Nextera® XT Index Kit v2 Set C or D; Illumina, San Diego, CA, United States), 12.5 μl NEBNext High-Fidelity 2 × PCR Master Mix, and 6.5 μl DEPC-treated water. Afterwards amplicons were purified, quantity and quality checked as previously described. The library was diluted to 4 nM and equimolar pooled. For sequencing the MiSeq® Reagent kit v3 (600 cycles) (Illumina, San Diego, CA, United States) was used for paired- end sequencing on the MiSeq® instrument (Illumina, San Diego, CA, United States).
Sequences were analyzed on the Galaxy web platform (www.usegalaxy.org; Afgan et al., 2016). FASTQ files were trimmed with a minimum read length of 50 using Cutadapt (Martin, 2011). Quality control was performed via FastQC (Andrews, 2010).1 For subsequent data analysis DADA2 pipeline (Galaxy Version 1.20) (Callahan et al., 2016) was used with the following trimming and filtering parameters: 20 bp were removed n- terminally and reads were truncated at position 240 (forward) and 200 (reverse), respectively, with expected error of 3 (forward) and 4 (reverse). Resulting unique amplicon sequence variants (ASVs) are biological sequences discriminated from errors, allowing the detection of single-nucleotide differences over the sequenced gene. Taxonomic analysis was performed using SILVA v138.1 release 99%. To exclude potential contamination, ASVs occurring in b, unassigned, mitochondrial and chloroplast reads and singletons (ASVs represented by only one read) were removed from the dataset.
The raw sequence data obtained in this study are deposited in the short read archive of NCBI under accession number PRJNA819581.
Downstream analyses were performed in R 4.0.5. Alpha diversity was calculated using species richness based on ASV number, Pielou evenness and Shannon diversity index. Beta diversity was analyzed via unweighted and weighted UniFrac distance matrix. For statistical purpose, KruskalWallis test, Wilcoxon-rank sum test and PERMANOVA with Benjamini-Hochberg p value correction for multiple comparison was used. For identification of biomarker taxa, two generalized linear models (R packages MASS and pscl) were used. Additionally, differences between log2fold changes were calculated and LEfSe (Linear discriminant analysis Effect Size) was used to validate the results. Multiple test correction was performed by p value adjustment via Benjamini-Hochberg method. Plots were created in R using ggplot2, ggpubr and metacoder. Venn diagrams for core microbiome analysis were created using Venny (Oliveros, 2020), with a minimum ASV threshold of 80% (ASV must be present in 80% of the samples to be considered for the group).
Sequencing of the generated amplicons resulted in 33,108 to 95,235 reads per sample after quality filtering. To compare samples without statistical bias, a subsampling strategy of 33,108 reads per sample was performed, reflecting the lowest observed read number. Rarefaction analysis indicated that the resulting sampling depth was sufficient for further analysis of all samples (Supplementary Figure S1). Overall, alpha diversity of bacterial communities, which developed on MP materials, was significantly lower compared to bulk soil. However, bacterial communities colonizing ePS showed a significantly higher alpha diversity compared to PP, while the alpha diversity of bacterial communities in the respective bulk soil samples did not differ (Figure 1A). Interestingly, alpha diversity of the control soil was lower compared to soils treated with MP materials. Beta diversity confirmed different bacterial community structures colonizing the two MP materials and indicated differences in bacterial diversity between bulk soil and MP materials. Differences between bulk soil samples treated with MP materials and control soils became obvious (Figure 1B).
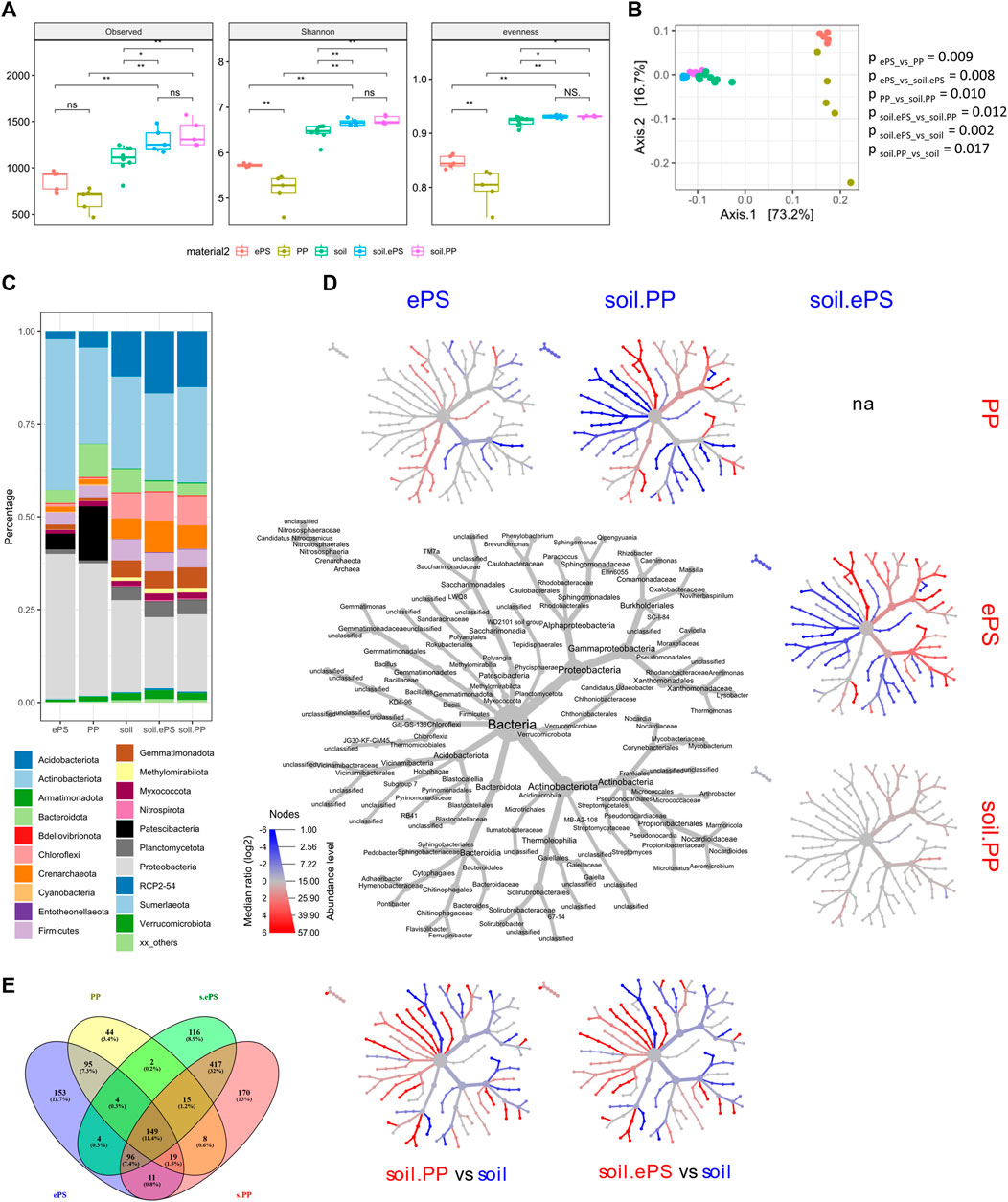
FIGURE 1. Community structure of bacteria colonizing different MP materials (ePS, PP) after application to soil and bulk soil samples treated with MP materials as well as untreated control soils. (A) Box plots of different alpha diversity indices. Statistical analysis was performed using Kruskal-Wallis and Wilcoxon Rank-Sum test with Benjamini-Hochberg correction for multiple comparisons, respectively. (B) PCoA plot of weighted Unifrac distances. Statistical analysis was performed using PERMANOVA with Benjamini-Hochberg correction for multiple comparisons. Differences were considered as significant with (p < 0.05). (C) Barplot showing the relative abundance of top 20 phyla on MP materials and in MP- treated and untreated bulk soil. (D) Heat tree including genera ≥ 1% of all reads in at least one sample. The labelled tree in the middle shows the taxonomic information (domain to genus) and is the key for the unlabelled smaller trees. Smaller trees represent a comparison between MP and soil in the columns and rows. Coloured taxa are more abundant (based on log2-transformed ratio of median proportions) in the samples indicated in the respective column or row. Only significant changes (p < 0.05) are coloured according to the legend. Statistical analysis was performed using Wilcoxon Rank-Sum test with Benjamini-Hochberg correction for multiple comparisons. (E) Venn diagram of shared ASVs between MP materials and MP treated bulk soil.
A more detailed analysis of the bacterial taxa, which were able to colonize the two different MP materials, revealed in total, 41 phyla, with most of the annotated reads in bulk soil samples belonging to the phyla Actinobacteriota (25%), Proteobacteria (22%), and Acidobacteriota (14%), whereas on MP materials Proteobacteria (37%), Actinobacteriota (33%) and Patescibacteria (9%) were dominant (Figure 1C).
Clear differences in community structure were observed when comparing bacteria colonizing the two MP materials, namely for Acidobacteriota, Bacteroidota, Myxococcota, Planctomycota and Verrucomicrobiota, which were significantly enriched on PP compared to ePS (Figures 1C,D). On the level of genera Adhaeribacter, Bacteroides, Ferruginibacter, Flavisolibacter and Pedobacter (Bacteroidota), Rhizobacter (Proteobacteria) and TM7a as well as LWQ8_unclassified (Patescibacteria) were increased in relative abundance on PP, whereas many members of Proteobacteria (Cavicella, Massilia, Noviherbaspirillum and Phenylobacterium), Arthrobacter and Streptomyces (both Actinobacteriota) were enriched on ePS (Figure 1D).
Clear consequences of the applied MP materials were detected for the community structure of the bulk soil. Crenarchaeota, Acidobacteriota, Chloroflexi, Myxococcota, Gemmatimonadota, Planctomycota and Verrucomicrobiota, were enriched in MP treated soil samples compared to the control soil (Figures 1C,D). Most genera enriched on MP materials, e.g., Bacteroides, Nocardia, Rhizobacter and TM7a showed a lower abundance in the MP treated bulk soil samples compared to the control soil. Some genera, which were enriched on MP materials (e.g., Streptomyces, Aeromicrobium), were also enriched in the MP treated bulk soil samples compared to the control soil. This result was independent from the applied MP material, even on the level of genera, with the exception of Arthrobacter, which showed a response in bulk soil samples depending on the applied MP material.
A comparison between MP colonizers and bacteria in bulk soil samples on the level of ASVs revealed 1303 ASVs in total, with 149 ASVs shared by all groups, representing the core microbiome (Figure 1E). While 703 ASVs were found in soil (417 in both soils) but not on the MP materials, 292 ASVs were observed only on MP materials and not in bulk soil samples. Bacteria colonizing both MP materials were represented by 95 ASVs, which comprise 44 ASVs assigned to Actinobacteria (e.g., Aeromicrobium, Nocardia, Nocardioides and Streptomyces), 14 ASVs assigned to Bacteroidota (e.g., Bacteroides and Ferruginibacter), 8 ASVs assigned to Patescibacteria (e.g., TM7a) and 19 ASVs assigned to Proteobacteria (e.g., Cavicella and Rhizobacter) (Supplementary Table S1).
Soils are a particle-rich environment and the addition of a surface and the increase of the available surface is proportionally less important than in aquatic environments. Therefore it is still under debate if data from marine systems can be used to describe the colonization of MP materials in soil (Rillig and Bonkowski, 2018). However MP particles have specific surfaces and shapes, creating a new ecological niche in the soil environment for microbiota and can easily be colonized due to their hydrophobicity, resulting in the formation of biofilms, which are well adapted to the surface-related lifestyle of a number of bacterial groups (Chai et al., 2020; Zhu et al., 2022). This was nicely demonstrated in this study. PP and ePS are common MP materials and are wide spread over the globe. We assume that they correspond to a huge part of the MP contamination in agricultural soils. Nevertheless, the results of this study cannot be extrapolated to the effects of other, than the tested MP materials on the bacterial community composition in agricultural soils, as they are extremely variable in their chemical composition, size and shape. Further experiments using different MP materials will be necessary to investigate those effects more in detail. Sterilized MP materials were used in this study to follow the colonization abilities of the materials by the soil microbiome. Further, a random inoculation of bacteria with the plastic materials to soil, which derive from the handling of the materials and the storage conditions, was excluded and thus it can be considered as reproducible. A constant water content (reflecting 50% of the max water holding capacity) in the soil was used to ensure an equal distribution of pores in soil with differing water content and thus redox conditions, which allow the survival of microbiota with differing abilities for the use of various electron acceptors.
In this study, almost half of the shared microbiome between PP and ePS ASVs belong to Actinobacteria, indicating a possible role of this phylum in the biodegradation of different MP materials in soil, or the utilization of leachates of the MP particles. Actinobacteria are present in almost all ecological habitats, like soils, marine environments and freshwater (Valan et al., 2012) and play an important role in the C cycle of the soils by degrading recalcitrant C sources (Ventura et al., 2007; Mohammadipanah and Wink, 2016). From pure culture studies, Actinobacteria are known together with fungi as the most active taxa amongst plastic biodegraders (Tosin et al., 2012) and have been described as degraders for a wide range of plastic materials including polylactic acid or polyethylene (Butbunchu and Pathom-Aree, 2019). Especially Streptomyces and Rhodococcus have been identified as having a high degradation potential of low density polyethylene (LDPE) biofilms (Soleimani et al., 2021). Nocardia and Arthrobacter are also well known to be involved into PE degradation (Mohan Pathak, 2017). However most studies so far regarding the degradation of MP materials were performed in aquatic environments (Rillig, 2012; Horton et al., 2017), where MP materials can be considered as an additional surface in the water body. Recently genera of Actinobacteria involved in bioremediation and biodegradation of plastic materials in marine environments were reported (Rathore et al., 2021). Especially Rhodococcus, Streptomyces, Nocardioides, Arthrobacter, and Aeromicrobium are major drivers for the biodegradation. Interestingly we detected all of these genera as colonizers of the MP materials in our study, with Arthrobacter and Streptomyces being significantly higher abundant on ePS than on PP. However it must be taken into account that marine Actinobacteria are effective in using polymers as sole carbon source (Oliveira et al., 2020), which might be different in soils, as MP materials are not the only available carbon source present.
Proteobacteria were the most abundant bacterial phylum detected on the MP materials in our study. 19 ASVs were found on both materials. Same trends were already reported for the “plastisphere” of polybutylene adipate terephthalate, polylactic acid and polyethylene (Rüthi et al., 2020) as well as polyvinyl chloride, polyamide, and polystyrene (Zhu et al., 2022). We observed Rhizobacter in higher abundances on PP than on ePS, while on ePS Cavicella, Massilia, Noviherbaspirillum and Phenylobacterium were higher abundant. Of most interest Rhizobacter has been described recently as an important bacterial genus involved in the mobilization of macro- and micronutrients in the rhizosphere soil of Zea mays (Fu et al., 2022), which may indicate an indirect influence of MP materials in soil on plant growth mediated by the microbiome associated to MP materials. For other proteobacterial groups the link to the degradation of MP materials has been described. For example, Massilia was positively associated with unknown polymer particles in the Rhine river (Mughini-Gras et al., 2021). Members of the genus Phenylobacterium were found to be enriched in soil after plastic mulching treatments (Luo et al., 2022) and these bacteria are capable of degrading polycyclic aromatic hydrocarbons (Yang et al., 2014). Only for Cavicella a link to MP materials in soils has not be described, but the genus is known as hydrocarbonoclastic (Assil et al., 2021) and has at least the potential to utilize the carbon of the MP materials.
Patescibacteria were the third most abundant phylum on MP materials in our study. TM7a and LWQ8_unclassified were significantly higher abundant on PP. Patescibacteria seem to be an important phylum interacting with MP materials in soils and were recently reported in two studies (Rüthi et al., 2020; Sun et al., 2022). It has been proposed that these bacteria live in close interaction with other microorganisms depending on their co-metabolism on the MP materials (Lemos et al., 2019). In the TM7 cluster strain for RAAC3 and other Saccharibacteria genes involved in the catabolism of complex carbon sources were identified and metagenomics data indicate that representatives of the phylum are able to degrade a variety of polymers (Kantor et al., 2013; Starr et al., 2018). TM7 ASVs has been described as associated to different MP materials and were are also reported on MP materials from different studies of marine environments (Scales et al., 2021). For TM7a a positive correlation between relative abundance and MP concentration in soil was recently reported, which indicates that this group of bacteria is very effective in growing on MP surfaces (Li et al., 2022) and may induce biofilm formation.
Bacterial communities in bulk soil samples of MP treated and control soil were dominated by Actinobacteria, Proteobacteria and Acidobacteria representing common soil microbiota (Fierer, 2017). Sun et al., 2022 reviewed 14 studies on the effects of MP materials on the microbial alpha diversity of terrestrial environments and only three of them reported that MP materials could significantly increase or decrease the alpha diversity of soil bacteria. We could not confirm this trend, as in our study the amendment of MP materials significantly increased the alpha diversity of bacterial communities in soil. One reason might be the amendment of MP materials as an additional carbon source to the soil samples without plants, generating a new microbial hotspot (Zhou et al., 2021) due to missing rhizodeposits and root exudates. Rillig et al. (2021) additionally discussed that changes in microbial activity, which subsequently may induce also changes in microbial community structure can be linked to altered physiochemical soil properties (De Souza Machado et al., 2019), direct toxic effects of MP materials, its additives, or sorbed contaminants. Another reason might be the leaching of additives of MP materials, which occurs in relative short time scales (Kim et al., 2020a). Additives are potentially toxic to microbes and can affect soil aggregation processes, as microbial exopolysaccharides function as “glue” and promote soil stability (Dorioz et al., 1993; Caesar-Tonthat, 2002; Tisdall et al., 2012). For marine environments it was shown that the additives associated to MP materials can be beneficial, deleterious or both to bacteria. The responses are highly species-specific and are depending on the type and concentration of the additives (Fernández-Juárez et al., 2021). Interestingly, we could not observe significant differences in the alpha diversity between the ePS- and the PP- soil, indicating that the effect was not depending on the polymer type.
In our experiment, Gemmatimonadates were more abundant in MP- soils compared to the control soil. This phylum is known for its catalytic ability and the decomposition of complex organic matters. Furthermore, bacteria of this taxa are involved in the C and N turnover and in the transformation of phosphorus (Cesarano et al., 2017; Li et al., 2017; Chee-Sanford et al., 2019; Hou et al., 2021), which might influence plant growth. Gemmatimonadates and Chloroflexi were recently considered as keystone species in soils contaminated with MP materials (Zhang et al., 2019). Our data confirmed these findings, as we found them in higher abundance in the MP treated soils than in the control soil. Chloroflexi play a role in the chlorine cycle and can metabolize organohalides (Krzmarzick et al., 2012).
Of interest might be Acidobacteria, which we found in significantly higher abundance in MP treated soil than in the control soil. Acidobacteria are known to have a beneficial role during the recovery of soils from pollution (Huang et al., 2015) and thus might support the recovery of soils after MP contamination.
Other bacterial taxa like Bacteroidota, Patescibacteria and Proteobacteria were reduced in their relative abundance as a result of the MP addition in bulk soil. Mainly for Bacteroidota this observation is quite surprising as they are usually more abundant in soils with high carbon content. They are considered to be specialized in degrading complex organic matter in the biosphere (Fierer, 2017; Wolińska et al., 2017). Apparently, the quality of the DOC, which includes also potentially toxic compounds and leachates, in soils contaminated with MP materials interferes negatively with Bacteroidota. However, this needs further mechanistic studies using also pure cultures to clarify the role of DOC released from MP materials for selected bacterial taxa.
The microbiome, which colonized PP and ePS MP materials introduced in an arable soil, was characterized in this study. Clear differences between the bacterial community structure on the MP materials as well as in bulk soil were detected. The results suggest that MP materials form a new habitat for microbial assemblages distinct from bulk soil. Most likely, the microbes colonizing the MP materials have their own metabolic pathways and lifestyles and might interact with other microorganisms like fungi. Some phyla were enriched on both MP materials (Actinobacteria, Proteobacteria and Patescibacteria) and might be considered as core microbiome of the “plastisphere”. We found several candidates, which might be able to degrade plastics like TM7a, Phenylobacterium, Nocardia, Arthrobacter and Streptomyces. Nevertheless, future studies must take into account how agricultural management influences the observed pattern of colonization of MP materials. This needs to be considered for the development of further management strategies, as some of the identified bacterial taxa enriched on MP materials might influence plant growth and ecosystem services provided by soils.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SK, TM, VR, and MS contributed to the conception and design of the study. SK and TM performed the experiments. SG analyzed the data. SK, VR, SG, MR, and MS wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
We thank the Federal Ministry of Education and Research (BMBF), Germany for funding the project µPlastic (grant number 031B0907 B) in frame of the Rhizo4Bio initiative.
We thank the technicians Cornelia Galonska and Gudrun Hufnagel of the Research Unit for Comparative Microbiome Analysis at Helmholtz Zentrum München for their help in laboratory work and sampling.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.989267/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Rarefaction curves (number of reads vs. number of detected ASVs) for soil, microplastic and PBS wash samples show saturation at subsamplin depth of 33108 reads.
SUPPLEMENTARY FIGURE S2 | Venn diagram of shared ASVs between microplastic (ePS, PP) and PBS wash (w.ePS, w.PP) samples indicate that diversity loss by washing is neglectable.
SUPPLEMENTARY TABLE S1 | List of ASVs shared by all groups (core), MP-treated and control soil and both microplastic materials (ePS and PP).
1http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
2https://www.boell.de/de/2019/05/14/plastikatlas
Afgan, E., Baker, D., van den Beek, M., Blankenberg, D., Bouvier, D., Čech, M., et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10. doi:10.1093/nar/gkw343
Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Apprill, A., Mcnally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi:10.3354/ame01753
Assil, Z., Esegbue, O., Mašek, O., Gutierrez, T., and Free, A. (2021). Specific enrichment of hydrocarbonclastic bacteria from diesel-amended soil on biochar particles. Sci. Total Environ. 762, 143084. doi:10.1016/J.SCITOTENV.2020.143084
Awet, T. T., Kohl, Y., Meier, F., Straskraba, S., Grün, A. L., Ruf, T., et al. (2018). Effects of polystyrene nanoparticles on the microbiota and functional diversity of enzymes in soil. Environ. Sci. Eur. 30, 11. doi:10.1186/s12302-018-0140-6
Butbunchu, N., and Pathom-Aree, W. (2019). Actinobacteria as promising candidate for polylactic acid type bioplastic degradation. Front. Microbiol. 10, 2834. doi:10.3389/fmicb.2019.02834
Caesar-Tonthat, T. C. (2002). Soil binding properties of mucilage produced by a basidiomycete fungus in a model system. Mycol. Res. 106, 930–937. doi:10.1017/S0953756202006330
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi:10.1038/nmeth.3869
Cesarano, G., De Filippis, F., La Storia, A., Scala, F., and Bonanomi, G. (2017). Organic amendment type and application frequency affect crop yields, soil fertility and microbiome composition. Appl. Soil Ecol. 120, 254–264. doi:10.1016/j.apsoil.2017.08.017
Chai, B., Li, X., Liu, H., Lu, G., Dang, Z., and Yin, H. (2020). Bacterial communities on soil microplastic at Guiyu, an E-Waste dismantling zone of China. Ecotoxicol. Environ. Saf. 195, 110521. doi:10.1016/j.ecoenv.2020.110521
Chee-Sanford, J., Tian, D., and Sanford, R. (2019). Consumption of N2O and other n-cycle intermediates by gemmatimonas aurantiaca strain T-27. Microbiol. (United Kingdom) 165, 1345–1354. doi:10.1099/mic.0.000847
Corradini, F., Meza, P., Eguiluz, R., Casado, F., Huerta-Lwanga, E., and Geissen, V. (2019). Evidence of microplastic accumulation in agricultural soils from sewage sludge disposal. Sci. Total Environ. 671, 411–420. doi:10.1016/j.scitotenv.2019.03.368
Cverenkárová, K., Valachovičová, M., Mackul’ak, T., Žemlička, L., and Bírošová, L. (2021). Microplastics in the food chain. Life 11, 1349. doi:10.3390/life11121349
de Souza Machado, A. A., Kloas, W., Zarfl, C., Hempel, S., and Rillig, M. C. (2018a). Microplastics as an emerging threat to terrestrial ecosystems. Glob. Chang. Biol. 24, 1405–1416. doi:10.1111/gcb.14020
De Souza Machado, A. A., Lau, C. W., Kloas, W., Bergmann, J., Bachelier, J. B., Faltin, E., et al. (2019). Microplastics can change soil properties and affect plant performance. Environ. Sci. Technol. 53, 6044–6052. doi:10.1021/acs.est.9b01339
De Souza MacHado, A. A., Lau, C. W., Till, J., Kloas, W., Lehmann, A., Becker, R., et al. (2018b). Impacts of microplastics on the soil biophysical environment. Environ. Sci. Technol. 52, 9656–9665. doi:10.1021/acs.est.8b02212
Dorioz, J. M., Robert, M., and Chenu, C. (1993). The role of roots, fungi and bacteria on clay particle organization. An experimental approach. Geoderma 56, 179–194. doi:10.1016/0016-7061(93)90109-X
Fernández-Juárez, V., López-Alforja, X., Frank-Comas, A., Echeveste, P., Bennasar-Figueras, A., Ramis-Munar, G., et al. (2021). The good, the bad and the double-sword” effects of microplastics and their organic additives in marine bacteria. Front. Microbiol. 11, 581118. doi:10.3389/fmicb.2020.581118
Fierer, N. (2017). Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi:10.1038/nrmicro.2017.87
Frias, J. P. G. L., and Nash, R. (2019). Microplastics: Finding a consensus on the definition. Mar. Pollut. Bull. 138, 145–147. doi:10.1016/j.marpolbul.2018.11.022
Fu, Q., Lai, J., long, J. X., hui, L. Z., Wu, G., and Luo, X. G. (2022). Alterations of the rhizosphere soil microbial community composition and metabolite profiles of Zea mays by polyethylene-particles of different molecular weights. J. Hazard. Mat. 423, 127062. doi:10.1016/J.JHAZMAT.2021.127062
Gkoutselis, G., Rohrbach, S., Harjes, J., Obst, M., Brachmann, A., Horn, M. A., et al. (2021). Microplastics accumulate fungal pathogens in terrestrial ecosystems. Sci. Rep. 11, 13214. doi:10.1038/s41598-021-92405-7
Heinrich, B. S. (2019). Plastikatlas: Daten und Fakten über eine Welt voller Kunststoff. Available at: https://www.boell.de/de/2019/05/14/plastikatlas.
Horton, A. A., Walton, A., Spurgeon, D. J., Lahive, E., and Svendsen, C. (2017). Microplastics in freshwater and terrestrial environments: Evaluating the current understanding to identify the knowledge gaps and future research priorities. Sci. Total Environ. 586, 127–141. doi:10.1016/j.scitotenv.2017.01.190
Hou, J., Xu, X., Yu, H., Xi, B., and Tan, W. (2021). Comparing the long-term responses of soil microbial structures and diversities to polyethylene microplastics in different aggregate fractions. Environ. Int. 149, 106398. doi:10.1016/j.envint.2021.106398
Huang, X., Liu, L., Wen, T., Zhu, R., Zhang, J., and Cai, Z. (2015). Illumina MiSeq investigations on the changes of microbial community in the Fusarium oxysporum f.sp. cubense infected soil during and after reductive soil disinfestation. Microbiol. Res. 181, 33–42. doi:10.1016/j.micres.2015.08.004
Huang, Y., Zhao, Y., Wang, J., Zhang, M., Jia, W., and Qin, X. (2019). LDPE microplastic films alter microbial community composition and enzymatic activities in soil, Environ. Pollut., 254:112983. doi:10.1016/j.envpol.2019.112983
Kantor, R. S., Wrighton, K. C., Handley, K. M., Sharon, I., Hug, L. A., Castelle, C. J., et al. (2013). Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio 4, e00708–e00713. doi:10.1128/mBio.00708-13
Kim, S. W., Waldman, W. R., Kim, T.-Y., and Rillig, M. C. (2020a). Effects of different microplastics on nematodes in the soil environment: Tracking the extractable Additives using an ecotoxicological approach. Environ. Sci. Technol. 54, 13868–13878. doi:10.1021/acs.est.0c04641
Kim, Y. N., Yoon, J. H., and Kim, K. H. (2020b). Microplastic contamination in soil environment - a review. Soil Sci. Ann. 71, 300–308. doi:10.37501/soilsa/131646
Krzmarzick, M. J., Crary, B. B., Harding, J. J., Oyerinde, O. O., Leri, A. C., Myneni, S. C. B., et al. (2012). Natural niche for organohalide-respiring chloroflexi. Appl. Environ. Microbiol. 78, 393–401. doi:10.1128/AEM.06510-11
Lemos, L. N., Medeiros, J. D., Dini-Andreote, F., Fernandes, G. R., Varani, A. M., Oliveira, G., et al. (2019). Genomic signatures and co-occurrence patterns of the ultra-small Saccharimonadia (phylum CPR/Patescibacteria) suggest a symbiotic lifestyle. Mol. Ecol. 28, 4259–4271. doi:10.1111/mec.15208
Leslie, H. A., van Velzen, M. J. M., Brandsma, S. H., Vethaak, A. D., Garcia-Vallejo, J. J., and Lamoree, M. H. (2022). Discovery and quantification of plastic particle pollution in human blood. Environ. Int. 163, 107199. doi:10.1016/j.envint.2022.107199
Li, C., Cui, Q., Li, Y., Zhang, K., Lu, X., and Zhang, Y. (2022). Effect of LDPE and biodegradable PBAT primary microplastics on bacterial community after four months of soil incubation. J. Hazard. Mat. 429, 128353. doi:10.1016/j.jhazmat.2022.128353
Li, F., Chen, L., Zhang, J., Yin, J., Huang, S., Yuan, Z., et al. (2017). Rumen bacteria communities and performances of fattening lambs with a lower or greater subacute ruminal acidosis risk. Front. Microbiol. 8, 2506. doi:10.3389/fmicb.2017.02506
Luo, S., Wang, S., Zhang, H., Zhang, J., and Tian, C. (2022). Plastic film mulching reduces microbial interactions in black soil of northeastern China. Appl. Soil Ecol. 169, 104187. doi:10.1016/J.APSOIL.2021.104187
Luo, Z., Zhou, X., Su, Y., Wang, H., Yu, R., Zhou, S., et al. (2021). Environmental occurrence, fate, impact, and potential solution of tire microplastics: Similarities and differences with tire wear particles. Sci. Total Environ. 795, 148902. doi:10.1016/J.SCITOTENV.2021.148902
Mahon, A. M., O’Connell, B., Healy, M. G., O’Connor, I., Officer, R., Nash, R., et al. (2017). Microplastics in sewage sludge: Effects of treatment. Environ. Sci. Technol. 51, 810–818. doi:10.1021/acs.est.6b04048
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi:10.14806/ej.17.1.200
Mohammadipanah, F., and Wink, J. (2016). Actinobacteria from arid and desert habitats:Diversity and biological activity. Front. Microbiol. 6, 1541. doi:10.3389/fmicb.2015.01541
Mohan Pathak, V., and Navneet, (2017). Review on the current status of polymer degradation: A microbial approach. Bioresour. Bioprocess. 4, 15. doi:10.1186/s40643-017-0145-9
Mughini-Gras, L., van der Plaats, R. Q. J., van der Wielen, P. W. J. J., Bauerlein, P. S., and de Roda Husman, A. M. (2021). Riverine microplastic and microbial community compositions: A field study in The Netherlands. Water Res. 192, 116852. doi:10.1016/J.WATRES.2021.116852
Ng, E. L., Lin, S. Y., Dungan, A. M., Colwell, J. M., Ede, S., Huerta Lwanga, E., et al. (2021). Microplastic pollution alters forest soil microbiome. J. Hazard. Mat. 409, 124606. doi:10.1016/J.JHAZMAT.2020.124606
Oliveira, J., Belchior, A., da Silva, V. D., Rotter, A., Petrovski, Ž., Almeida, P. L., et al. (2020). Marine environmental plastic pollution: Mitigation by microorganism degradation and recycling valorization. Front. Mar. Sci. 7, 567126. doi:10.3389/fmars.2020.567126
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi:10.1111/1462-2920.13023
Rathore, D. S., Sheikh, M., and Singh, S. P. (2021). “Marine Actinobacteria: New horizons in bioremediation,” in Recent Developments in Microbial Technologies. Environmental and Microbial Biotechnology. Editors R. Prasad, V. Kumar, J. Singh, and C. P. Upadhyaya (Singapore: Springer), 425–449. doi:10.1007/978-981-15-4439-2_20
Rillig, M. C., and Bonkowski, M. (2018). Microplastic and soil protists:A call for research. Environ. Pollut. 241, 1128–1131. doi:10.1016/j.envpol.2018.04.147
Rillig, M. C., De Souza Machado, A. A., Lehmann, A., and Klümper, U. (2019). Evolutionary implications of microplastics for soil biota. Environ. Chem. 16, 3–7. doi:10.1071/EN18118
Rillig, M. C., and Lehmann, A. (2020). Microplastic in terrestrial ecosystems. Science 368, 1430–1431. doi:10.1126/science.abb5979
Rillig, M. C., Leifheit, E., and Lehmann, J. (2021). Microplastic effects on carbon cycling processes in soils. PLoS Biol. 19, e3001130. doi:10.1371/JOURNAL.PBIO.3001130
Rillig, M. C. (2012). Microplastic in terrestrial ecosystems and the soil? Environ. Sci. Technol. 46, 6453–6454. doi:10.1021/es302011r
Rochman, C. M., Brookson, C., Bikker, J., Djuric, N., Earn, A., Bucci, K., et al. (2019). Rethinking microplastics as a diverse contaminant suite. Environ. Toxicol. Chem. 38, 703–711. doi:10.1002/etc.4371
Rosato, A., Barone, M., Negroni, A., Brigidi, P., Fava, F., Xu, P., et al. (2020). Microbial colonization of different microplastic types and biotransformation of sorbed PCBs by a marine anaerobic bacterial community. Sci. Total Environ. 705, 135790. doi:10.1016/J.SCITOTENV.2019.135790
Rüthi, J., Bölsterli, D., Pardi-Comensoli, L., Brunner, I., and Frey, B. (2020). The “plastisphere” of biodegradable plastics is characterized by specific microbial taxa of alpine and arctic soils. Front. Environ. Sci, 562263. 8. doi:10.3389/fenvs.2020.562263
Santos, A. L., Rodrigues, C. C., Oliveira, M., and Rocha, T. L. (2022). Microbiome: A forgotten target of environmental micro(nano)plastics? Sci. Total Environ. 822, 153628. doi:10.1016/j.scitotenv.2022.153628
Scales, B. S., Cable, R. N., Duhaime, M. B., Gerdts, G., Fischer, F., Fischer, D., et al. (2021). Cross-hemisphere study reveals geographically ubiquitous, plastic-specific bacteria emerging from the rare and unexplored biosphere. mSphere 6, e0085120. doi:10.1128/msphere.00851-20
Schöler, A., Jacquiod, S., Vestergaard, G., Schulz, S., and Schloter, M. (2017). Analysis of soil microbial communities based on amplicon sequencing of marker genes. Biol. Fertil. Soils 53, 485–489. doi:10.1007/s00374-017-1205-1
Shan, X., Zhang, W., Dai, Z., Li, J., Mao, W., Yu, F., et al. (2022). Comparative analysis of the effects of plastic mulch films on soil nutrient, yields and soil microbiome in three vegetable fields. Agronomy 12, 506. doi:10.3390/agronomy12020506
Shi, J., Sun, Y., Wang, X., and Wang, J. (2022). Microplastics reduce soil microbial network complexity and ecological deterministic selection. Environ. Microbiol. 24, 2157–2169. doi:10.1111/14622920.15955
Soleimani, Z., Gharavi, S., Soudi, M., and Moosavi-Nejad, Z. (2021). A survey of intact lowdensity polyethylene film biodegradation by terrestrial Actinobacterial species. Int. Microbiol. 24, 65–73. doi:10.1007/s10123-020-00142-0
Sommer, F., Dietze, V., Baum, A., Sauer, J., Gilge, S., Maschowski, C., et al. (2018). Tire abrasion as a major source of microplastics in the environment. Aerosol Air Qual. Res. 18, 2014–2028. doi:10.4209/aaqr.2018.03.0099
Starr, E. P., Shi, S., Blazewicz, S. J., Probst, A. J., Herman, D. J., Firestone, M. K., et al. (2018). Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome 6, 122. doi:10.1186/s40168018-0499-z
Sun, Y., Duan, C., Cao, N., Li, X., Li, X., Chen, Y., et al. (2022). Effects of microplastics on soil microbiome: The impacts of polymer type, shape, and concentration. Sci. Total Environ. 806, 150516. doi:10.1016/J.SCITOTENV.2021.150516
Thiel, M., Luna-Jorquera, G., álvarez-Varas, R., Gallardo, C., Hinojosa, I. A., Luna, N., et al. (2018). Impacts of marine plastic pollution from continental coasts to subtropical gyres-fish, seabirds, and other vertebrates in the SE Pacific. Front. Mar. Sci. 5, 238. doi:10.3389/fmars.2018.00238
Tisdall, J. M., Nelson, S. E., Wilkinson, K. G., Smith, S. E., and McKenzie, B. M. (2012). Stabilisation of soil against wind erosion by six saprotrophic fungi. Soil Biol. Biochem. 50, 134–141. doi:10.1016/J.SOILBIO.2012.02.035
Tosin, M., Weber, M., Siotto, M., Lott, C., and Innocenti, F. D. (2012). Laboratory test methods to determine the degradation of plastics in marine environmental conditions. Front. Microbiol. 3, 225. doi:10.3389/fmicb.2012.00225
Ugwu, K., Herrera, A., and Gómez, M. (2021). Microplastics in marine biota: A review. Mar. Pollut. Bull. 169, 112540. doi:10.1016/j.marpolbul.2021.112540
Valan, A. M., Asha, K. R. T., Duraipandiyan, V., Ignacimuthu, S., and Agastian, P. (2012). Characterization and phylogenetic analysis of novel polyene type antimicrobial metabolite producing actinomycetes from marine sediments: Bay of Bengal India. Asian pac. J. Trop. Biomed. 2, 803–810. doi:10.1016/S2221-1691(12)60233-0
Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G. F., Chater, K. F., et al. (2007). Genomics of Actinobacteria : Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548. doi:10.1128/mmbr.00005-07
Wang, L., Luo, Z., Zhen, Z., Yan, Y., Yan, C., Ma, X., et al. (2020). Bacterial community colonization on tire microplastics in typical urban water environments and associated impacting factors. Environ. Pollut. 265, 114922. doi:10.1016/J.ENVPOL.2020.114922
Wolińska, A., Kuźniar, A., Zielenkiewicz, U., Izak, D., Szafranek-Nakonieczna, A., Banach, A., et al. (2017). Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by a culture-independent approach. Appl. Soil Ecol. 119, 128–137. doi:10.1016/j.apsoil.2017.06.009
Yang, S., Wen, X., Zhao, L., Shi, Y., and Jin, H. (2014). Crude oil treatment leads to shift of bacterial communities in soils from the deep active layer and upper permafrost along the ChinaRussia crude oil pipeline route. PLoS One 9, e96552. doi:10.1371/journal.pone.0096552
Zhang, M., Zhao, Y., Qin, X., Jia, W., Chai, L., Huang, M., et al. (2019). Microplastics from mulching film is a distinct habitat for bacteria in farmland soil. Sci. Total Environ. 688, 470–478. doi:10.1016/j.scitotenv.2019.06.108
Zhou, B., Wang, J., Zhang, H., Shi, H., Fei, Y., Huang, S., et al. (2020). Microplastics in agricultural soils on the coastal plain of Hangzhou Bay, east China: Multiple sources other than plastic mulching film. J. Hazard. Mat. 388, 121814. doi:10.1016/j.jhazmat.2019.121814
Zhou, J., Wen, Y., Marshall, M. R., Zhao, J., Gui, H., Yang, Y., et al. (2021). Microplastics as an emerging threat to plant and soil health in agroecosystems. Sci. Total Environ. 787, 147444. doi:10.1016/j.scitotenv.2021.147444
Keywords: microplastics, polypropylene, expanded polystyrene, soil, microbiome, 16S rRNA gene sequencing
Citation: Kublik S, Gschwendtner S, Magritsch T, Radl V, Rillig MC and Schloter M (2022) Microplastics in soil induce a new microbial habitat, with consequences for bulk soil microbiomes. Front. Environ. Sci. 10:989267. doi: 10.3389/fenvs.2022.989267
Received: 08 July 2022; Accepted: 02 August 2022;
Published: 26 August 2022.
Edited by:
Rajeev Pratap Singh, Banaras Hindu University, IndiaReviewed by:
Ajar Nath Yadav, Eternal University, IndiaCopyright © 2022 Kublik, Gschwendtner, Magritsch, Radl, Rillig and Schloter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susanne Kublik, c3VzYW5uZS5rdWJsaWtAaGVsbWhvbHR6LW11ZW5jaGVuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.