- 1Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, China
- 2College of Agronomy, Qingdao Agricultural University, Qingdao, China
- 3Longyan City Company of Fujian Tobacco Company, Longyan, China
- 4China Tobacco Shandong Industrial Co., Ltd., Longyan, China
- 5Fujian Branch of China Tobacco Company, Fuzhou, China
Ammonia-oxidizing archaea (AOA) and bacteria (AOB) are the most important ammonia oxidation functional community, while the coastal environment just provides a different oxygen environment for the ammonia oxidation process. However, few surveys concentrated on the influence of oxygen concentration on the niche specialization of AOA and AOB in the ocean intertidal zones. Here, high-throughput sequencing by Illumina MiSeq and qPCR were applied to detect the change of abundance, diversity as well as community structure of both AOA and AOB with 0–60 cm sediments depth in the intertidal zone in Qingdao, China. Results showed that the AOA/AOB amoA gene copy numbers and AOA/AOB OTU numbers rate increased as sediment depth went more profound, which indicated that AOA was more adaptive to oxygen-limited niches compared to AOB. Oxygen indeed led to the niche specialization of AOA and AOB in intertidal sediments. The dominant AOA and AOB were the clusters of Nitrosopumilus and Nitrosospira, respectively, which indicated ecological success in the intertidal zone. A significant and positive correlation (p < 0.01) between AOB abundance/AOB OTU numbers and Oxidation-reduction potential (ORP) was observed. In addition, both total nitrogen (TN) (p < 0.01) and pH (p < 0.05) were significantly negatively correlated to AOB abundance. TN was also significantly negatively correlated to AOB OTU numbers (p < 0.05). Hence, oxygen led to niche specialization of AOA and AOB, especially under anoxic conditions, AOA played a dominant role in the process of ammonia oxidation. The Nitrosopumilus and Nitrosospira clusters were the dominant AOA and AOB, respectively, representing an ecological success in the intertidal zone.
Introduction
Nitrification, a vital role in the ecosystem nitrogen cycle, converts ammonia to nitrate through nitrite (Beeckman et al., 2018). Three kinds of microorganisms of ammonia oxidizers, nitrite oxidizers, and complete ammonia oxidizers were involved in the nitrification process (Stein and Klotz, 2016). Ammonia-oxidizing bacteria (AOB) were long thought to be the only microorganism that performed ammonia oxidation, the first rate-limiting step in nitrification. However, the ammonia oxidation theory was changed after discovering ammonia-oxidizing archaea (AOA) (Konneke et al., 2005). To date, several strains of AOA, such as Candidatus Nitrososphaera gargensis (Hatzenpichler et al., 2008), Candidatus Nitrosocaldus yellowstonii (de la Torre et al., 2008), Candidatus Nitrosoarchaeum limnia SFB1 (Blainey et al., 2011), and Candidatus Nitrosotalea devanaterra (Lehtovirta-Morley et al., 2011), were enriched or isolated from various ecosystems. Until now, both AOA and AOB have been shown to perform ammonia-oxidizing (de la Torre et al., 2008; Hatzenpichler et al., 2008; Jia and Conrad, 2009). Various AOA and AOB functional gene abundance, community structures, and activity patterns were reported in different biotopes (Wang et al., 2020). Environmental factors play a crucial role in the niche differentiation between AOA and AOB. Moreover, the AOA and AOB relative contribution to ammonia oxidation in various ecosystems gradually become the hot spots in the field of ammonia oxidation.
As reported previously, a series of environmental factors may cause the niche specialization between AOA and AOB, such as pH, temperature, salinity, ammonia concentrations, and oxygen concentrations. AOA seems to be more adaptive in niches with lower pH and more likely to dominate the process of ammonia oxidation in acidic niches (Gubry-Rangin et al., 2011; Prosser and Nicol, 2012). AOB was more adaptable than AOA with increasing abundance but no composition alteration at elevated temperature (Zhang et al., 2019). Microcosm test also verified that pH and temperature were vital factors that led to the niche specialization of AOA and AOB (Aigle et al., 2020). Ammonia, as one of the substrates for ammonia oxidation, could cause niche specialization of AOA and AOB. The concentration of ammonia half-saturation constant of some AOB strains (Martens-Habbena et al., 2009; Jung et al., 2011; Kim et al., 2012) was much higher than AOA strains, indicated that AOA had a higher affinity for ammonia than AOB. The half-saturation constants of Candidatus Nitrosoarchaeum koreensis MY1, Candidatus Nitrososphaera sp. JG1 and Nitrosopumilus maritimus SCM1 were 0.69 , 2.15 μM, and 133 nM, respectively, which were much lower than that of Nitrosomonas europaea (Km = 553 μM) (Martens-Habbena et al., 2009). Therefore, AOA is more competitive than AOB in the oligotrophic environment (Beman et al., 2008; Verhamme et al., 2011). In other substrates for ammonia oxidation, oxygen could also lead to niche specialization of AOA and AOB. The half-saturation constants for oxygen of AOA, such as Candidatus Nitrosoarchaeu mkoreensis MY1, Candidatus Nitrososphaera sp. JG1 and N. maritimus SCM1 were 10.38, 4.67, and 3.90 μM, separately (Martens-Habbena et al., 2009; Jung et al., 2011; Kim et al., 2012), which were much lower than that of Nitrosomonas oligotropha NL7 (76.3 μM) and N. europaea C-31 (183.3 μM) (Park et al., 2010; Park and Noguera, 2007). The affinities of AOA for oxygen were much higher than that of AOB, which means that AOA had competitive advantages over AOB in oxygen-limited environments. The AOA lived better in many oxygen-limited environments. AOA dominated the transcriptome and probably dominated the ammonia oxidation process in the oxygen minimum zone (Stewart et al., 2012). The increasing AOA OTU number and abundance ratio (AOA: AOB) in flooded soil proved that AOA could better adapt to low oxygen conditions (Liu et al., 2015).
The intertidal zone is an ecological crisscross zone with significant environmental and ecological functions affected by marine and terrestrial ecosystems (Community structure and organization of tidepools). The daily tidal rhythm made the sediment is submerged or exposed to air. As one of the biggest wharves in Qingdao, the Shazikou wharf is significantly affected by human activities and the decayed fish, which provided enough ammonia nitrogen for the ammonia-oxidizing microbes in nearby intertidal zones. The ecological distribution, including the community structure and AOA/AOB abundance in intertidal zones, have been discussed previously (Bernhard et al., 2005; Zhang et al., 2015; Hu et al., 2019). However, the relationships between oxygen concentration and niche specialization of AOA and AOB have not been investigated, especially, the ecological distribution of AOA and AOB under oxy-gen-limited habitats under different layers/depth of the sediments. So, the main objectives of this research are: 1) to study the difference of abundances and community structure of AOA and AOB in different layers/depth of the sediments in the intertidal zones, 2) to explore which, AOA or AOB, was the main driver performing ammonia oxidation in different layers/depth of the sediments in intertidal zones, 3) to evaluate how the oxygen concentration influences the niche specialization of AOA and AOB in intertidal zones.
Materials and methods
Description of sampling sites
The sampling sites were distributed in Shazikou wharf coast (36°6′N, 120°33′E). The Shazikou wharf was built in 1976 and was one of the biggest wharves in Qingdao, China. The region is located in the north temperate monsoon region, with a temperate monsoon climate. Regulated by the marine environment, it also has significant marine climate characteristics. The mean annual temperature is 12.6°C, and the mean annual rainfall is 734.3 mm. The sunshine duration is 2281.4 h, with a frost-free period of 179 days. Coastal lowlands or coastal beaches are typical saline soils.
Collection and physicochemical properties analysis of sediment
The sediment samples were collected using a cylindrical soil sample from the ocean intertidal zone near the Shazikou wharf in Qingdao, Shandong Province. Three sampling sites (biological triplicates) along the sandy beach were selected. For each site, different depths (0–10, 10–20, 20–30, 30–40, 40–50, and 50–60 cm, namely S0–10, S10–20, S20–30, S30–40, S40–50, S50–60, respectively) were sampled, and the sediments in the same depth were mixed. Totally six sediment samples were obtained. Each sample was divided into two subsamples: one, stored at 4°C, was used to analyze physicochemical properties; another, frozen at −80°C, was applied to molecular analysis. The soil pH was determined at a soil: water ratio of 1:5 with a conductivity meter (FE38-FiveEasyPlus™, Mettler-Toledo, Zurich, Switzerland). The oxidation-reduction potential (ORP) was measured by an ORP meter (ORP-2096, Boqu, China). The content of moisture content (MC) was measured at the same time with soil samples collecting, and the method of oven-drying at 105°C for 10 h. Total nitrogen (TN) was measured by Kjeldahl method. The ammonium (NH4+-N) and nitrate (NO3−-N) were leached with KCl solution (1 mol L−1), and the filtrate was taken and measured by a continuous flow analyzer (Seal-AA3, Germany). The nitrite (NO2−-N) was determined by ultraviolet spectrophotometry (Liu et al., 2013).
DNA extraction and quantitative PCR of amoA genes
Total genomic DNA was extracted from the fresh soil samples by employing the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, United States) as the protocols of manufacturer. The final DNA concentration of extracts was determined using a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, United States). The extracted DNA quality was assessed on 1% agarose gel using a nanometer droplet spectrophotometer (ND-1000; Isogen Life Science, Netherlands) to measure the concentration of DNA.
For the quantification of genes (AOB/AOA amoA) copy numbers, the Abi7300 fluorescent quantitative PCR instrument (Applied Biosystems, United States) was applied to perform real-time PCR. Specific primers were used to amplify target genes. The forward primer sequence of AOA-amoA is STAATGGTCTGGCTTAGACG (5′–3′), and the reverse primer is GCGGCCATCCATCTGTATGT (5′–3′). The forward primer sequence of AOB-amoA is GGGGTTTCTACTGGTGGT (5′–3′), and the reverse primer is CCCCTCKGS AAAGCCTTCTTC (5′–3′) (Park et al., 2008). The reaction system (Supplementary Table S1) and reaction conditions (Supplementary Table S2) are shown in the supporting materials. The constructed plasmid was identified by sequencing. Then the plasmid value was measured at 260 nm by UV spectrophotometer (NanoDrop2000, Thermo Fisher Scientific, United States), and converted into gene copies (copies g−1 dry soil) by formula. The standard curve was prepared by 10-fold gradient dilution of the constructed plasmids (10−1–10−7 for AOB amoA, 10−2–10−8 for AOA amoA) (Hu et al., 2019).
High-throughput sequencing and bioinformatic analysis for the amoA genes
The AOB and AOA amoA genes were amplified using the two primer sets mentioned above, and the protocols used to amplify have been previously reported (Shen et al., 2008; Pester et al., 2012). Illumina MiSeq sequencing, carried out by Personal (Shanghai, China), was applied to ensure the excellent coverage of each clone library. Bar code oligonucleotides were connected to the ends of two pairs of primers to distinguish amoA amplicons of different samples. For each sample, triplicate PCR products were obtained and then mixed. PCR products length was subsequently determined by electrophoresis in a 1.5% agarose gel. Bioinformatic analysis was conducted with the Mothur software package (Schloss et al., 2009). After the screening, trimming, and chimeras checking procedures, the high-quality reads were left for further analysis. Both AOA and AOB amoA genes were grouped into OTUs using an 85% similarity as a previously recommended cut-off value (Pester et al., 2012). The obtained sequences were submitted to the RDP pipeline using the FrameBot tool to remove non-amoA sequences (Wang et al., 2013). The OTUs were taxonomically categorized using neighbor-joining phylogenetic trees constructed from representative sequences with the longest length of the amoA genes. AOA amoA representatives were matched with an existing high-quality amoA database (Pester et al., 2012), while AOB amoA representatives were matched with GeneBank reference sequences. The taxonomy-derived reference sequences were combined with the Kimura 2-parameter distance (MEGA 6.0), and the bootstrap values were calculated using 1,000 repetitions (Kumar et al., 2016). In addition, the diversity indices of ACE, Chao1, Shannon, and Simpson for each OTU were also calculated.
Statistical analysis
Pearson correlation analysis with a 0.05 significance level was used to determine the correlation between the AOA/AOB diversities or abundances and environmental factors. In addition, the representative sequences with the longest length were selected for the following phylogenetic analyses, which were conducted with MEGA 6 through a neighbor-joining tree using Kimura 2-parameter distance with 1,000 bootstrap replicates (Tamura et al., 2013). Redundancy analysis (RDA) was performed to investigate the relationship between AOA/AOB microbial community structure and the environmental factors with CANOCO 5.0 (CANOCO, Microcomputer Power Inc., Ithaca, NY, United States).
Results
Physicochemical properties of sediment samples
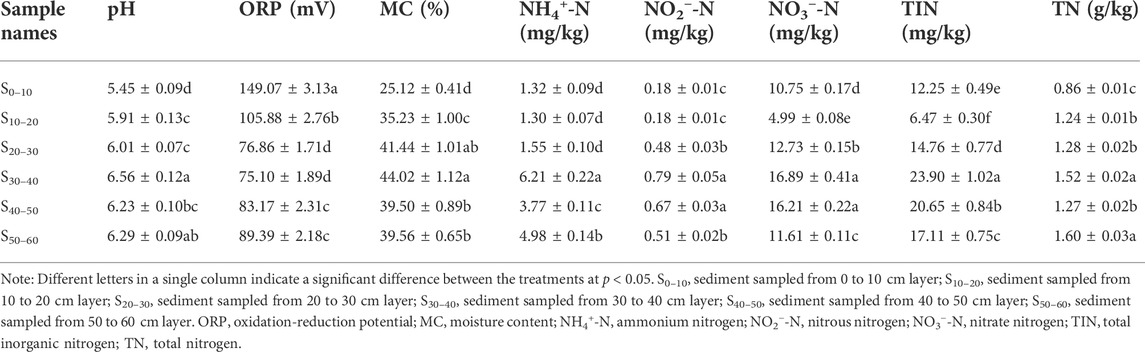
Determination of physicochemical properties of sediment samples are showed in Table 1. All the sediments had acidic pH (i.e., 5.45–6.56). The pH value of the upper layer sediment samples (S0–10) was relatively lower than the other five layers. Middle and deep layers (S30–40, S40–50, and S50–60) had higher pH values, and S30–40 showed the highest pH of 6.56, which probably resulted from the higher nitrification rates in the upper layer sediments. However, the ORP of the upper layer sediments was much higher than the rest of the samples by 40.79%–98.50%. The moisture varied from 25.12% to 44.02%, and the middle layer (S20–30, S30–40) was the highest of other layer sediments. The NH4+-N content of the six sediment samples ranged from 1.32 to 6.21 mg kg−1, and the peak was found in the sample of S30–40, which was significantly higher than the other five sediment samples by 24.70%–377.70%. All the sediment samples had low NO2−-N content, and the upper layer sediment samples (S0–10, S20–30) were the lowest. The NO3−-N content ranged from 4.99 to 16.89 mg/kg, and the NO3−-N content in S30–40 and S40–50 layers was higher than that of the other layers. The TIN of middle and deep layers was higher than the upper layers, and S30–40 was higher than the rest samples by 15.74%–269.40%. As for the TN, the content ranged from 857.77 to 1604.05 mg kg−1, and the TN content increased as the depth went deeper.
Abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria
The AOA amoA gene copy numbers in the upper layers (S0–10, S10–20) were much lower than that in the middle (S20–30, S30–40) and deep (S40–50, S50–60) layer (Figure 1). On the contrary to AOA amoA genes, the number of AOB amoA genes in the upper layers (S0–10, S10–20) was much higher than that in the middle (S20–30, S30–40) and deep (S40–50, S50–60) layer (Figure 1). The ratio of AOA/AOB amoA gene copy numbers ranged from 0.04 to 150.75 in the six sediment samples, and the ratio increased dramatically from upper layers to deep layers. The ratios of AOA/AOB amoA genes in the six sediment samples were 0.04 (S0–10), 0.11 (S10–20), 8.20 (S20–30), 7.07(S30–40), 86.77 (S40–50) and 150.75 (S50–60), respectively. In the upper layers (S0–10, S10–20), where the oxygen was sufficient, AOB won the competition with AOA and maybe the main driver of ammonia oxidation. However, in the middle (S20–30, S30–40) and deep layer (S40–50, S50–60), where the environment became anoxic, the higher affinities of AOA for oxygen could help them to win the competition with AOB. In the middle tidal zone, where the oxygen was relatively sufficient, the number of AOB amoA genes was higher than AOA. In the subtidal zones where oxygen was limited, AOA outnumbered AOB (Hu et al., 2019). From the point of abundance, oxygen concentration indeed led to the niche specialization of AOA and AOB.
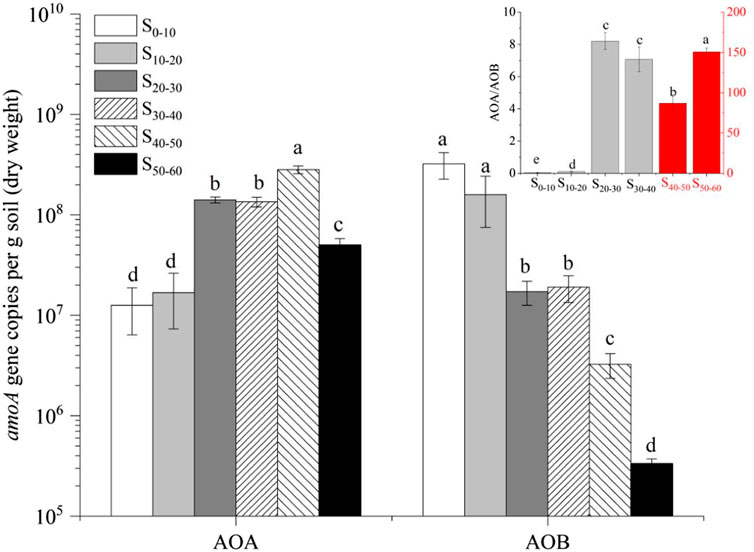
FIGURE 1. Quantitative analysis of AOA and AOB in the six sediment samples (means, SEs displayed with errors bars, n = 3). S0–10, sediment sampled from 0 to 10 cm layer; S10–20, sediment sampled from 10 to 20 cm layer; S20–30, sediment sampled from 20 to 30 cm layer; S30–40, sediment sampled from 30 to 40 cm layer; S40–50, sediment sampled from 40 to 50 cm layer; S50–60, sediment sampled from 50 to 60 cm layer. AOA, ammonia-oxidizing archaea; AOB, ammonia-oxidizing bacteria. In the figure in the upper right corner, four gray columns corresponded to the principal coordinate axis, and two red columns corresponding to the sub coordinate axis.
Diversity of ammonia-oxidizing archaea and ammonia-oxidizing bacteria
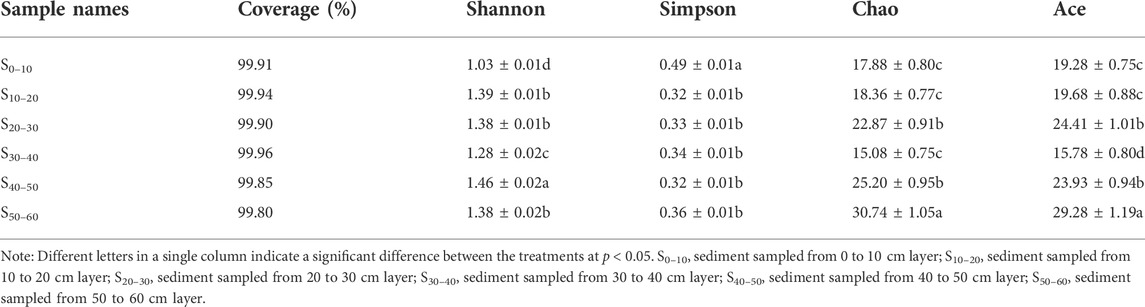
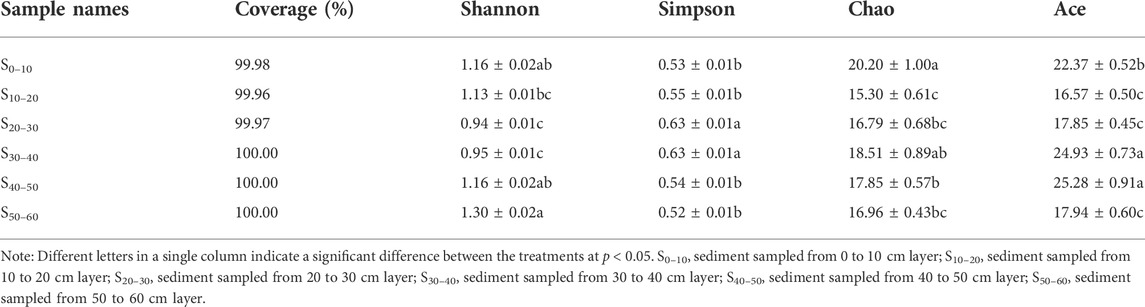
High-quality sequences of AOA and AOB, after quality control procedures, were applied to the analysis of diversity. In AOA, 17144 high-quality sequences were obtained for the six samples of sediment. The coverages of the six AOA amoA gene clone libraries were all higher than 99.0%, ranging from 99.80% to 99.96% among the 18 OTUs obtained from the AOA amoA gene (Table 2). The AOA OTU numbers of the six sediment samples ranged from 10 to 15, with S30–40 (10 OTUs) and S50–60 (15 OTUs) displayed the lowest and highest diversity, respectively (Table 2). The OTU numbers in deep layers were higher than those in the upper and middle layers (Figure 2). The coverage of the six AOB amoA gene clone libraries was ranged from 99.96 to 100.00% (Table 3). The sample S0–10 showed the highest diversity, holding 11 OTUs in total, which was higher than the rest five sediment samples. The OTU numbers in the rest of the five samples were all the same, holding 8 OTUs. Similar to the change rules of abundance, the AOB OTU numbers decreased as sediment depth went deeper, which indicated that AOB was not tolerant or adaptable to hypoxic environment. The AOB OTU numbers were higher in the non-flooded zones where the oxygen concentration was sufficient than in the flooded zones where the environment was anoxic. The diversity of AOA was always higher than AOB in all the sediment samples (Table 3), and the ratio of AOA/AOB OTU numbers increased from 1.09 to 1.88 as the sediment depth went deeper, which was similar to the changing pattern of the abundance.
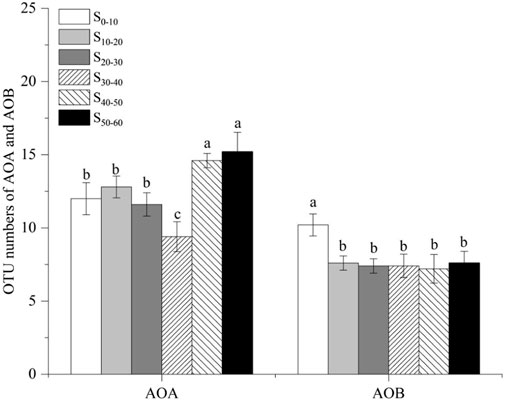
FIGURE 2. The OTU numbers of AOA and AOB in the six sediment samples (means, SE displayed with errors bars, n = 3). S0–10, sediment sampled from 0 to 10 cm layer; S10–20, sediment sampled from 10 to 20 cm layer; S20–30, sediment sampled from 20 to 30 cm layer; S30–40, sediment sampled from 30 to 40 cm layer; S40–50, sediment sampled from 40 to 50 cm layer; S50–60, sediment sampled from 50 to 60 cm layer. AOA, ammonia-oxidizing archaea; AOB, ammonia-oxidizing bacteria.
Phylogenetic analysis and community structure of ammonia-oxidizing archaea/ammonia-oxidizing bacteria
For AOA, 17144 high-quality sequences after quality control procedures were obtained for the six sediment samples. As showed in Figure 3, the representative sequences of the 18 OTUs, according to the amoA genes classification of AOA (Pester et al., 2012), have been grouped into four different clusters. Neither Nitrosotalea nor Thermal-related AOA was found in all the sediment samples. Nitrosophaera cluster contained 3 OTUs (OTU 12, OUT 15, OTU 18), a total of 9 sequences. OTU 12 (containing 3 seqs) belonging to Nitrosophaera cluster was retrieved from samples S20–30 and S40–50, OTU 15 (containing 2 seqs) was detected in sample S0–10 and S50–60, and OTU 18 (containing 4 seqs) was only retrieved from deep layer samples (S40–50, S50–60). The rest of the 15 OTUs all belonged to the Nitrosopumilus cluster. Among the 17144 sequences, 17135 accounted for 99.94% of all obtained sequences belonged to the Nitrosopumilus cluster. AOB obtained 18865 sequences from 6 sediment samples by the same quality control procedure as AOA, and then these sequences were assigned to 11 OTUs using the 15% cut-off recommended previously (Purkhold et al., 2000). The representative sequences of the 11 OTUs were subsequently grouped into two clusters, as showed in Figure 4. OTU 9 and OTU 10 were clustered into the Nitrosomonas, and the remaining 9 OTUs belonged to the Nitrosospira cluster. Only 35 sequences belonged to the cluster of Nitrosomonas. All six AOB communities were primarily composed of Nitrosospira-related sequences.
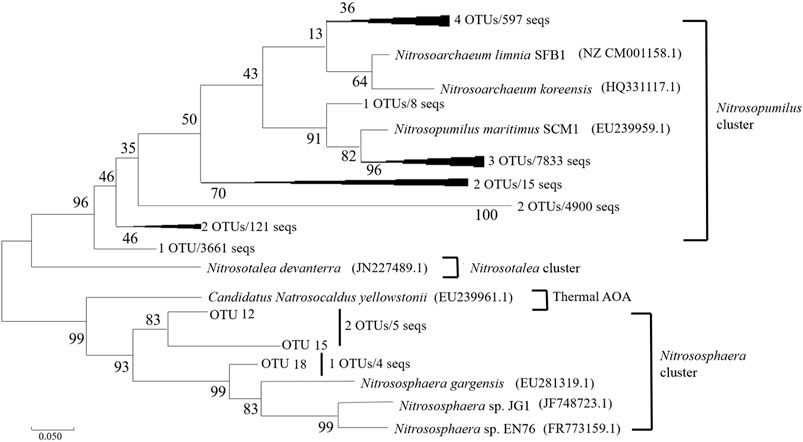
FIGURE 3. Maximum-Likelihood phylogenetic tree showing the phylogenetic affiliations of the AOA sequences recovered from the sediment samples. The numbers at the nodes are percentages that indicate the levels of bootstrap support from 2,000 replicates. The scale bar represents 0.05 nucleic acid substitutions per nucleotide position.
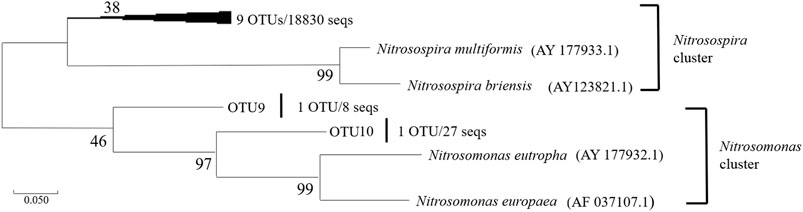
FIGURE 4. Maximum-Likelihood phylogenetic tree showing the phylogenetic affiliations of the AOB sequences recovered from the sediment samples. The numbers at the nodes are percentages that indicate the levels of bootstrap support from 2,000 replicates. The scale bar represents 0.05 nucleic acid substitutions per nucleotide position.
Environmental factors that influence the diversity and communities of ammonia-oxidizing archaea and ammonia-oxidizing bacteria
As shown in Table 4, the linear relationship between various environmental factors and OTU number, amoA gene abundance as well as AOA/AOB diversity index was determined by the Pearson correlation coefficient. ORP was positively correlated with AOB’s abundance and OTU numbers (p < 0.01), indicating that AOB preferred the niches with higher oxygen concentrations. On the contrary, a negative correlation between ORP and AOA abundance was observed in Table 4, implying that AOA preferred the niches with lower oxygen concentrations. Moreover, these results could be due to the higher oxygen affinity of AOA than that of AOB. Significantly negative correlation between MC and AOB abundance (p < 0.01)/AOB OTU numbers (p < 0.05) was also observed. The AOB abundance was significantly negatively correlated to the TN (p < 0.01) and pH (p < 0.05). In addition, TN was also significantly negatively with AOB OTU numbers (p < 0.05).
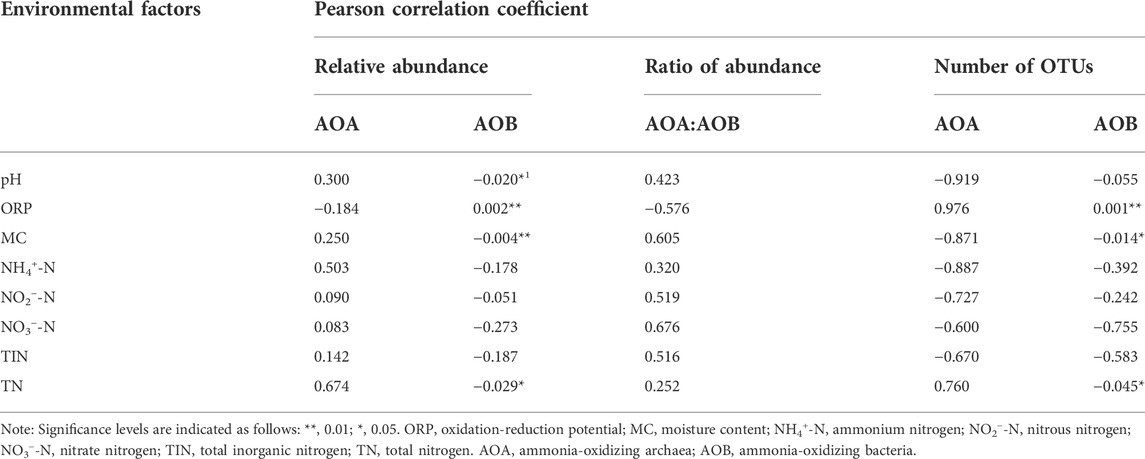
TABLE 4. Correlation analysis of environmental factors and AOA, AOB abundance, OTU numbers, and diversity index.
In order to further study the dominant role of the environmental factors of different sediment site in AOA and AOB community structure, the redundancy analysis (RDA) was used to rank the influencing factors (Figure 5). Figure 5A performed the relationship between AOA community structure and environmental factors. The explanation degree of each factor for AOA community structure distribution on the horizontal axis was 54.06%, and on the vertical axis was 22.25%. Therefore, it mainly referred to the vector projection length of each factor vector on the horizontal axis. NH4+-N, pH, and NO2−-N were the main factors affected the AOA community structure distribution because of their long projection (>0.05) on the horizontal axis. ORP had the opposite performance with other environmental factors, which was also consistent with the results in Table 4. Figure 5B performed the relationship between AOB community structure and environmental factors. The explanation degree of each factor for AOB community structure distribution on the horizontal axis was 42.52%, and on the vertical axis was 38.43%. ORP and MC were the important environmental factors that affected the AOB community structure distribution because of their long projection on the horizontal and vertical axis. The results of RDA were consistent with those of correlation analysis (Table 4) to a certain extent, which confirmed the significant effects of OPR and MC on the distribution of AOB community.
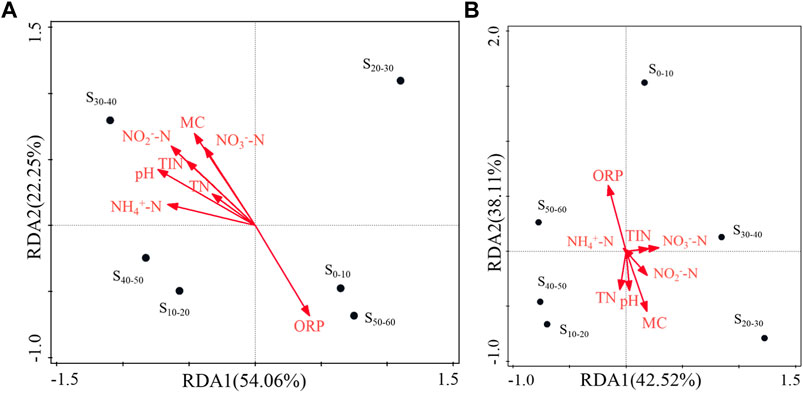
FIGURE 5. Redundancy analysis between AOA (A) and AOB (B) flora structure and environmental factors. S0–10, sediment sampled from 0 to 10 cm layer; S10–20, sediment sampled from 10 to 20 cm layer; S20–30, sediment sampled from 20 to 30 cm layer; S30–40, sediment sampled from 30 to 40 cm layer; S40–50, sediment sampled from 40 to 50 cm layer; S50–60, sediment sampled from 50 to 60 cm layer. ORP, oxidation-reduction potential; MC, moisture content; NH4+-N, ammonium nitrogen; NO2−-N, nitrous nitrogen; NO3−-N, nitrate nitrogen; TIN, total inorganic nitrogen; TN, total nitrogen.
Discussion
The difference of abundances and community structure of ammonia-oxidizing archaea and ammonia-oxidizing bacteria
The number of AOA and AOB amoA genes observed in this work was similar toa previous study in the intertidal zones in Zhoushan, Zhejiang province (Hu et al., 2019). Hu found that the AOA and AOB amoA gene copy numbers in the intertidal zone ranged from 107 to 108. The AOB amoA gene copy numbers in this study were relatively lower since samples S40–50 and S50–60 were collected in the deep layers of the sediments. The ratios of AOA/AOB amoA genes increased as the depth of the sediments went deeper, indicating that AOA may dominate the ammonia oxidation in the deep layer. This founding was in accordance with the previous work focusing on the ammonia-oxidizing microbes in soils (Leininger et al., 2006). Leininger reported that the ratios of AOA/AOB amoA genes increased from 55 to 842 with soil depth went deeper. This phenomenon could be explained by the higher affinities of AOA for oxygen compared to AOB (Martens-Habbena et al., 2009; Jung et al., 2011; Kim et al., 2012). In the subtidal zones where oxygen was limited, AOA outnumbered AOB (Hu et al., 2019). From the point of abundance, oxygen concentration indeed led to the niche specialization of AOA and AOB.
Overall, the change rule of the AOA OTU numbers was similar to the AOA abundance. Both of them increased with sediment depth. AOA showed higher diversity as oxygen concentration became lower, according to previous work in water-level-fluctuating zones in Three Gorges Reservoir (Liu et al., 2015). At higher oxygen concentrations of non-flooded areas, AOA OTU values were lower. However, AOA OTU values were higher in the relatively low oxygen concentration in flooded areas. This phenomenon represented that AOA was more adaptive to the oxygen-limited environments. A total of 18865 high-quality AOA sequences were obtained from 6 sediment samples, and these sequences were assigned to 11 OTUs using 15% as cut-off value (Purkhold et al., 2000). All the six sediment samples were predominantly composed of sequences that belonged to the Nitrosopumilus cluster. The observation of the Nitrosopumilus cluster dominance over the Nitrosophaera cluster in this research was consistent with the previous reportsin the intertidal zones in Zhoushan Island (Hu et al., 2019) and Chongming eastern intertidal sediments (Zheng et al., 2013). The most AOA belongs to the cluster of Nitrosopumilus (73%) confirmed by the phylogeny and meta-data analyses of AOA amoA sequences, which was nearly twice as much as the Nitrososphaera cluster (37%) (Alves et al., 2018). These results of the dominance of the Nitrosospira cluster over the Nitrosomonas cluster were consistent with the previous report in the coastal Pearl River estuary (Cao et al., 2011). The lower concentration of ammonia nitrogen (0.03–0.11 mm NH4+) in the intertidal zone of Qingdao may help to explain the absolute dominance of Nitrosoma. The cluster of Nitrosomonas has a lower substrate affinity which leads to higher substrate concentration adaption. Conversely, the Nitrosospira cluster has relatively higher substrate affinity and is more likely to survive in environments with a low concentration of substrate (Zheng et al., 2014; Yu et al., 2016).
Environmental factors
Ammonia oxidizing microorganisms (AOA and AOB) were the most critical microorganisms involved in nitrification and were the first rate-limiting step in the nitrification process. Studies showed that the abundance of AOA and AOB were significantly related to nitrogen forms and soil pH (Segal et al., 2017). Our results (Figure 5) also showed that NH4+-N, pH, and NO2−-N were the main factors affected the AOA community structure distribution. AOB was the main driver of nitrification in alkaline soils, while AOA was the main driver of nitrification in acidic soils (Hu et al., 2013; Zhang et al., 2012; Lu et al., 2012). Thus, a significantly negative correlation was shown between AOB and pH in our study (Table 4). These results consistently confirmed the characteristics of niche differentiation of AOA and AOB. AOB tended to play a leading role in neutral or alkaline environments with high nitrogen and pH value, while AOA was more suitable to dominate in acidic and low nitrogen soil (Ke et al., 2013; Qin et al., 2013). However, our consequence showed that AOB was significantly negatively correlated with TN and ORP, which was the opposite of some other studies. For example, studies of Di et al. (2010) and Pratscher et al. (2011) showed that a high nutrient environment was more conducive to the growth of AOB, while AOA was more suitable to grow in a low nitrogen environment. Most of these studies were aimed at the upper soil. However, our research range varied from 0 to 60 cm soil layer. The differential changes of ammonia oxidation bacteria in different soil layers might be an important reason for the correlation with environmental factors. Some studies showed that the reduction of ORP was conducive to improve the abundance of AOB (Ke and Lu, 2012), and AOA was easier to fix in a low CO2 environment (Martens-Habbena et al., 2009). This was contrary to the result that AOB likes niches with high oxygen concentration in our study. On the one hand, it was related to the different pH and salinity of different tested soils (Min et al., 2016). On the other hand, it was also related to the different flooding conditions between different soil layers (Ke and Lu, 2012). Therefore, it was necessary to study further the effects of multiple environmental factors on ammonia-oxidizing bacteria in the future.
Conclusion
In summary, our results showed the changing pattern of AOA and AOB abundance and diversity with sediment depth. In upper layers, the AOB abundance was higher compared to that of AOA. In contrast, AOA abundance outnumbered AOB in deep layers. Both the AOA/AOB amoA gene copy numbers rate and AOA/AOB OTU numbers rate increased as sediment depth went deeper, which indicated that oxygen indeed led to the niche specialization of AOA and AOB. AOA won the competition with AOB in the oxygen-limited niches. AOA may be dominant in the ammonia oxidation process in the deep layers. The Nitrosopumilus cluster and Nitrosospira cluster were the dominant AOA and AOB, respectively, representing ecological success in the intertidal zone.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
Conceptualization, JD; methodology, LM; formal analysis, PC; data curation, MQ; writing—original draft preparation, SC and BZ; writing—review and editing, WS; supervision, XZ. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the the Agricultural Science and Technology Innovation Program (ASTIP No. CAAS-ZDRW202201) and the Provincial Natural Science Foundation of Shandong (ZR2021QD036).
Conflicts of interest
Author MQ was employed by Longyan City Company of Fujian Tobacco Company. Author SC was employed by China Tobacco Shandong Industrial Co., Ltd. Author BZ was employed by Fujian Branch of China Tobacco Company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.976618/full#supplementary-material
References
Aigle, A., Gubry-Rangin, C., Thion, C., Estera-Molina, K. Y., Richmond, H., Pett-Ridge, J., et al. (2020). Experimental testing of hypotheses for temperature- andpH-based niche specialization of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 22, 4032–4045. doi:10.1111/1462-2920.15192
Alves, R. J. E., Minh, B. Q., Urich, T., von Haeseler, A., and Schleper, C. (2018). Unifying the global phylogeny and environmental distribution of ammonia-oxidising archaea based on amoA genes. Nat. Commun. 9, 1517. doi:10.1038/s41467-018-03861-1
Beeckman, F., Motte, H., and Beeckman, T. (2018). Nitrification in agricultural soils: Impact, actors and mitigation. Curr. Opin. Biotechnol. 50, 166–173. doi:10.1016/j.copbio.2018.01.014
Beman, J. M., Popp, B. N., and Francis, C. A. (2008). Molecular and biogeochemical evidence for ammonia oxidation by marine crenarchaeota in the gulf of California. ISME J. 2, 429–441. doi:10.1038/ismej.2007.118
Bernhard, A. E., Donn, T., Giblin, A. E., and Stahl, D. A. (2005). Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7, 1289–1297. doi:10.1111/j.1462-2920.2005.00808.x
Blainey, P. C., Mosier, A. C., Potanina, A., Francis, C. A., and Quake, S. R. (2011). Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLOS ONE 6, e16626. doi:10.1371/journal.pone.0016626
Cao, H., Hong, Y., Li, M., and Gu, J. (2011). Diversity and abundance of ammonia-oxidizing prokaryotes in sediments from the coastal pearl river estuary to the south China sea. Antonie Leeuwenhoek 100, 545–556. doi:10.1007/s10482-011-9610-1
de la Torre, J. R., Walker, C. B., Ingalls, A. E., Koenneke, M., and Stahl, D. A. (2008). Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10, 810–818. doi:10.1111/j.1462-2920.2007.01506.x
Di, H., Cameron, K. C., Shen, J., Winefield, C. S., O'Callaghan, M., Bowatte, S., et al. (2010). Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol. Ecol. 72, 386–394. doi:10.1111/j.1574-6941.2010.00861.x
Gubry-Rangin, C., Hai, B., Quince, C., Engel, M., Thomson, B. C., James, P., et al. (2011). Niche specialization of terrestrial archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108, 21206–21211. doi:10.1073/pnas.1109000108
Hatzenpichler, R., Lebedeva, E. V., Spieck, E., Stoecker, K., Richter, A., Daims, H., et al. (2008). A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105, 2134–2139. doi:10.1073/pnas.0708857105
Hu, H., Zhang, L., Dai, Y., Di, H., and He, J. (2013). pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J. Soils Sediments 13, 1439–1449. doi:10.1007/s11368-013-0726-y
Hu, J., Liu, S., Yang, W., He, Z., Wang, J., Liu, H., et al. (2019). Ecological success of the nitrosopumilus and nitrosospira clusters in the intertidal zone. Microb. Ecol. 78, 555–564. doi:10.1007/s00248-019-01359-x
Jia, Z., and Conrad, R. (2009). Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11, 1658–1671. doi:10.1111/j.1462-2920.2009.01891.x
Jung, M., Park, S., Min, D., Kim, J., Rijpstra, W. C., Damste, J. S. S., et al. (2011). Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl. Environ. Microbiol. 77, 8635–8647. doi:10.1128/aem.05787-11
Ke, X., Angel, R., Lu, Y., and Conrad, R. (2013). Niche differentiation of ammonia oxidizers and nitrite oxidizers in rice paddy soil. Environ. Microbiol. 15, 2275–2292. doi:10.1111/1462-2920.12098
Ke, X., and Lu, Y. (2012). Adaptation of ammonia-oxidizing microorganisms to environment shift of paddy field soil. FEMS Microbiol. Ecol. 80, 87–97. doi:10.1111/j.1574-6941.2011.01271.x
Kim, J. G., Jung, M. Y., Park, S. J., Rijpstra, W. I. C., Damste, J. S. S., Madsen, E. L., et al. (2012). Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I.1b from an agricultural soil. Environ. Microbiol. 14, 1528–1543. doi:10.1111/j.1462-2920.2012.02740.x
Konneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi:10.1038/nature03911
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi:10.1093/molbev/msw054
Lehtovirta-Morley, L. E., Stoecker, K., Vilcinskas, A., Prosser, J. I., and Nicol, G. W. (2011). Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. U. S. A. 108, 15892–15897. doi:10.1073/pnas.1107196108
Leininger, S., Urich, T., Schloter, M., Schwark, L., Qi, J., Nicol, G. W., et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809. doi:10.1038/nature04983
Liu, S., Shen, L., Lou, L., Tian, G., Zheng, P., and Hu, B. (2013). Spatial distribution and factors shaping the niche segregation of ammonia-oxidizing microorganisms in the qiantang river, China. Appl. Environ. Microbiol. 79, 4065–4071. doi:10.1128/aem.00543-13
Liu, S., Hu, B., He, Z., Zhang, B., Tian, G., Zheng, P., et al. (2015). Ammonia-oxidizing archaea have better adaptability in oxygenated/hypoxic alternant conditions compared to ammonia-oxidizing bacteria. Appl. Microbiol. Biotechnol. 99, 8587–8596. doi:10.1007/s00253-015-6750-7
Lu, L., Han, W., Zhang, J., Wu, Y., Wang, B., Lin, X., et al. (2012). Nitrification of archaeal ammonia oxidizers in acid soils is supported by hydrolysis of urea. ISME J. 6, 1978–1984. doi:10.1038/ismej.2012.45
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J. R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979. doi:10.1038/nature08465
Min, W., Guo, H., Zhang, W., Zhou, G., Ma, L., Ye, J., et al. (2016). Irrigation water salinity and N fertilization: Effects on ammonia oxidizer abundance, enzyme activity and cotton growth in a drip irrigated cotton field. J. Integr. Agric. 15, 1121–1131. doi:10.1016/s2095-3119(15)61158-3
Park, H. D., and Noguera, D. R. (2007). Characterization of two ammonia-oxidizing bacteria isolated from reactors operated with low dissolved oxygen concentrations. J. Appl. Microbiol. 102, 1401–1417. doi:10.1111/j.1365-2672.2006.03176.x
Park, S. J., Park, B. J., and Rhee, S. K. (2008). Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 12, 605–615. doi:10.1007/s00792-008-0165-7
Park, B. J., Park, S. J., Yoon, D. N., Schouten, S., Damste, J. S. S., and Rhee, S. K. (2010). Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing Bacteria. Appl. Environ. Microbiol. 76, 7575–7587. doi:10.1128/aem.01478-10
Pester, M., Rattei, T., Flechl, S., Groengroeft, A., Richter, A., Overmann, J., et al. (2012). amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ. Microbiol. 14, 525–539. doi:10.1111/j.1462-2920.2011.02666.x
Pratscher, J., Dumont, M. G., and Conrad, R. (2011). Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc. Natl. Acad. Sci. U. S. A. 108, 4170–4175. doi:10.1073/pnas.1010981108
Prosser, J. I., and Nicol, G. W. (2012). Archaeal and bacterial ammonia-oxidisers in soil: The quest for niche specialisation and differentiation. Trends Microbiol. 20, 523–531. doi:10.1016/j.tim.2012.08.001
Purkhold, U., Pommerening-Roser, A., Juretschko, S., Schmid, M. C., Koops, H. P., and Wagner, M. (2000). Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: Implications for molecular diversity surveys. Appl. Environ. Microbiol. 66, 5368–5382. doi:10.1128/aem.66.12.5368-5382.2000
Qin, H., Yuan, H., Zhang, H., Zhu, Y., Yin, C., Tan, Z., et al. (2013). Ammonia-oxidizing archaea are more important than ammonia-oxidizing bacteria in nitrification and NO3 (-)-N loss in acidic soil of sloped land. Biol. Fertil. Soils 49, 767–776. doi:10.1007/s00374-012-0767-1
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi:10.1128/aem.01541-09
Segal, L. M., Miller, D. N., McGhee, R. P., Loecke, T. D., Cook, K. L., Shapiro, C. A., et al. (2017). Bacterial and archaeal ammonia oxidizers respond differently to long-term tillage and fertilizer management at a continuous maize site. Soil Tillage Res. 168, 110–117. doi:10.1016/j.still.2016.12.014
Shen, J., Zhang, L., Zhu, Y., Zhang, J., and He, J. (2008). Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ. Microbiol. 10, 1601–1611. doi:10.1111/j.1462-2920.2008.01578.x
Stein, L. Y., and Klotz, M. G. (2016). The nitrogen cycle. Curr. Biol. 26, 94–98. doi:10.1016/j.cub.2015.12.021
Stewart, F. J., Ulloa, O., and DeLong, E. F. (2012). Microbial meta transcriptomics in a permanent marine oxygen minimum zone. Environ. Microbiol. 14, 23–40. doi:10.1111/j.1462-2920.2010.02400.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi:10.1093/molbev/mst197
Verhamme, D. T., Prosser, J. I., and Nicol, G. W. (2011). Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 5, 1067–1071. doi:10.1038/ismej.2010.191
Wang, Q., Quensen, J. F., Fish, J. A., Lee, T. K., Sun, Y., Tiedje, J. M., et al. (2013). Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. MBio 4, e00513. doi:10.1128/mbio.00592-13
Wang, C., Tang, S., He, X., and Ji, G. (2020). The abundance and community structure of active ammonia-oxidizing archaea and ammonia-oxidizing bacteria shape their activities and contributions in coastal wetlands. Water Res. 171, 115464. doi:10.1016/j.watres.2019.115464
Yu, S., Yao, P., Liu, J., Zhao, B., Zhang, G., Zhao, M., et al. (2016). Diversity, abundance, and niche differentiation of ammonia-oxidizing prokaryotes in mud deposits of the eastern China marginal seas. Front. Microbiol. 7, 137. doi:10.3389/fmicb.2016.00137
Zhang, L., Hu, H., Shen, J., and He, J. (2012). Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6, 1032–1045. doi:10.1038/ismej.2011.168
Zhang, Y., Chen, L. J., Dai, T. J., Sun, R. H., and Wen, D. H. (2015). Ammonia manipulates the ammonia-oxidizing archaea and bacteria in the coastal sediment-water microcosms. Appl. Microbiol. Biotechnol. 99, 6481–6491. doi:10.1007/s00253-015-6524-2
Zhang, Q., Li, Y., He, Y., Brookes, P. C., and Xu, J. (2019). Elevated temperature increased nitrification activity by stimulating AOB growth and activity in an acidic paddy soil. Plant Soil 445, 71–83. doi:10.1007/s11104-019-04052-7
Zheng, Y., Hou, L., Liu, M., Lu, M., Zhao, H., Yin, G., et al. (2013). Diversity, abundance, and activity of ammonia-oxidizing bacteria and archaea in Chongming eastern intertidal sediments. Appl. Microbiol. Biotechnol. 97, 8351–8363. doi:10.1007/s00253-012-4512-3
Keywords: AOA, AOB, niche specialization, intertidal zone, high-throughput sequencing
Citation: Du J, Meng L, Qiu M, Chen S, Zhang B, Song W, Cong P and Zheng X (2022) Ammonia-oxidizing archaea and ammonia-oxidizing bacteria communities respond differently in oxy-gen-limited habitats. Front. Environ. Sci. 10:976618. doi: 10.3389/fenvs.2022.976618
Received: 23 June 2022; Accepted: 27 July 2022;
Published: 24 August 2022.
Edited by:
Hongbiao Cui, Anhui University of Science and Technology, ChinaReviewed by:
Kailou Liu, Jiangxi Institute of Red Soil, ChinaYuji Jiang, Institute of Soil Science (CAS), China
Yanfang Feng, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2022 Du, Meng, Qiu, Chen, Zhang, Song, Cong and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Cong, Y29uZ3BpbmdAY2Fhcy5jbg==; Xuebo Zheng, emhlbmd4dWVib0BjYWFzLmNu
†These authors have contributed equally to this work
 Jialin Du1,2†
Jialin Du1,2† Lin Meng
Lin Meng Wenjing Song
Wenjing Song Xuebo Zheng
Xuebo Zheng