- 1Department of Biological Sciences, The University of Alabama, Tuscaloosa, AL, United States
- 2Alabama Water Institute, The University of Alabama, Tuscaloosa, AL, United States
- 3Center for Complex Hydrosystems Research, The University of Alabama, Tuscaloosa, AL, United States
- 4Division of Natural Sciences and Mathematics, Miles College, Fairfield, AL, United States
- 5Department of Environmental Sciences, Louisiana State University, Baton Rouge, LA, United States
Coastal salt marshes provide valuable ecosystem services but are subjected to multiple concomitant stressors that may impact their ability to provide those services. Global climate change has led to the poleward expansion of mangroves into salt marshes on each continent where mangroves and marshes co-occur. In the northern Gulf of Mexico, warming winter temperatures have resulted in the expansion of Avicennia germinans (black mangrove) into forb-dominated salt marshes, resulting in a shift in ecosystem structure that can impact the ecosystem services marshes provide, including biogeochemical processes such as nitrogen removal. There have been limited studies addressing how mangrove expansion impacts nitrogen removal rates in salt marshes, but it is possible that mangroves enhance microbial nitrogen removal capacity through more efficient oxygen translocation to sediments. However, mangroves are more sensitive to oiling (such as occurred during the 2010 Deepwater Horizon spill) than marsh plants, such as Spartina alterniflora, which have a higher turnover. Thus, even if they enhance nitrogen removal, if they cannot withstand disturbances such as oiling, there still may be a loss of function associated with woody encroachment. We conducted a field study to assess the impact of woody encroachment in mediating biogeochemical recovery 7 to 8 years after the Deepwater Horizon oil spill. We collected sediments from S. alterniflora- and A. germinans-dominated plots in the Chandeleur Islands (LA, United States), a chain of barrier islands in the northern Gulf of Mexico subjected to a range of oiling following the spill. We compared nitrate reduction rates (denitrification and dissimilatory nitrate reduction to ammonium), microbial community composition, and denitrifier marker gene abundance at sites subjected to light and moderate oiling using a combination of isotope pairing on sediment slurries, 16S sequencing, and qPCR. We predicted that overall, denitrification rates and microbial functional capacity would be enhanced in mangrove-dominated sediments. We also predicted that these enhancements would be diminished at the more intensely oiled site due to the higher susceptibility of A. germinans to oiling. Denitrification potential rates were higher in mangrove sediments at the lightly oiled site, whereas dissimilatory nitrate reduction to ammonium potential rates were higher in marsh sediments. Indicator analysis of 16S rRNA data selected putative sulfur cycling taxa as indicators of marsh sediments, suggesting that changes in oxygen availability associated with encroachment may be driving the differences in process rates. There was no difference in process rates between plant types at the moderately oiled site, where heavily weathered oil residue was still present. Sediment nutrient stocks were lower in moderately oiled mangrove plots than in lightly oiled mangrove plots, suggesting that sediment fertility recovery following the spill may have been slower in the mangroves, contributing to a change in ecosystem function. This study shows that woody encroachment has the potential to impact both the biogeochemical services that marshes provide and their response to and recovery from disturbances.
Introduction
Humans have dramatically increased the amount of reactive nitrogen (N) in the environment, with negative ecological consequences (Paerl and Whitall 2010; Fowler et al., 2013). Wetlands, including salt marshes, intercept and remove as much as a third of the excess N they receive through uptake, burial, and microbially mediated processes (Jordan et al., 2011), protecting vulnerable coastal ecosystems (Valiela and Cole 2002). However, disturbances such as sea-level rise, eutrophication, and warming temperatures have altered the composition and distribution of wetland vegetation (e.g., Donnelly and Bertness, 2001; Saintilan et al., 2014), with subsequent impacts on the valuable biogeochemical services wetlands provide (Kelleway et al., 2017; Charles et al., 2019). One widespread shift in marsh vegetation composition is the woody encroachment of mangroves into coastal salt marshes. Warming winter temperatures associated with climate change have resulted in the poleward migration of mangroves on all continents where marshes and mangroves co-occur (Osland et al., 2013; Armitage et al., 2015; Kelleway et al., 2017), with subsequent changes in faunal diversity (Smee et al., 2017; Scheffel et al., 2018), habitat structure (Guo et al., 2017), ecosystem nutrient stocks (Doughty et al., 2016; Simpson et al., 2019; Macy et al., 2020), and microbial community composition (Barreto et al., 2018). Despite the myriad impacts of woody encroachment on marsh structure and function, we still have a limited understanding of how mangrove expansion impacts the N cycle (but see Henry 2012; Steinmuller et al., 2020; Macy et al., 2020), particularly the removal of excess anthropogenic N.
Nitrogen cycling is tightly coupled to vegetation in coastal wetlands (Koop-Jakobsen and Giblin, 2009; Giblin et al., 2013). Plant root exudates provide a source of organic carbon (OC) for heterotrophic microbes (Spivak and Reeve, 2015). Additionally, oxygen (O2) translocation to the rhizosphere alters redoximorphic conditions, promoting processes such as nitrification (Hamersley and Howes, 2005; Koop-Jakobsen and Giblin, 2009) and limiting sulfate reduction (Holmer et al., 2002). Mangrove expansion into marshes results in multiple physicochemical changes to sediments that could impact microbially driven biogeochemical processes (Perry and Mendelssohn, 2009). Pneumatophores are efficient at translocating O2 to the subsurface, potentially altering subsurface redox conditions (Perry and Mendelssohn, 2009; Comeaux et al., 2012). Enhanced O2 translocation also impacts microbial community structure, altering the functional capacity of sediment microbial communities (Barreto et al., 2018). Mangrove expansion can also impact the availability of N in sediments. Mangroves competitively utilize N compared to marsh plants (Simpson et al., 2013; Dangremond et al., 2020), potentially reducing porewater N concentrations (Macy et al., 2019). However, mangroves can have a higher leaf N content than marsh plants (Macy et al., 2020) that can accelerate litter decomposition and N cycling (Simpson et al., 2021). Therefore, it is likely that woody encroachment affects N removal rates in marsh sediments.
The dominant pathway for permanent N removal in coastal wetlands is denitrification, the microbially mediated stepwise reduction of nitrate (NO3−) to nitrous oxide (N2O) and dinitrogen (N2) gases (Knowles, 1982). Dissimilatory NO3− reduction to ammonium (DNRA) is a competing process for NO3− reduction in which N is retained as ammonium (NH4+) rather than removed (Burgin and Hamilton, 2007). The composition and productivity of marsh vegetation can influence whether N is removed by denitrification or retained by DNRA (Ledford et al., 2020). Increased oxygen translocation to the rhizosphere promotes coupled nitrification–denitrification (Reddy et al., 1989; Koop-Jakobsen and Giblin, 2009) and reduces sulfide accumulation, which inhibits both nitrification and denitrification (Sorensen et al., 1980; Joye and Hollibaugh, 1995). It is possible that the greater root O2 translocation by mangroves compared to marshes (Perry and Mendelssohn, 2009) would promote denitrification over DNRA following encroachment. OC availability is another important factor that controls whether NO3− is removed or retained. Due to the higher energetic demand of DNRA, retention is favored under high sediment OC: NO3− ratios (Hardison et al., 2015; Kessler et al., 2018). Thus, if mangroves rapidly increase sediment OC pools, as has sometimes been observed (Doughty et al., 2016; Simpson et al., 2019), encroachment may promote DNRA over denitrification.
In addition to altering the physicochemical characteristics of marshes, mangrove encroachment may also affect ecosystem response to disturbance (e.g., Lin and Mendelssohn, 2012; McKee and Vervaeke, 2018; Armitage et al., 2020). Increased ecosystem susceptibility to disturbance has important implications for biogeochemical processes such as N removal, as salt marsh damage and loss can lead to a loss of biogeochemical function (Hinshaw et al., 2017). One type of disturbance that mangroves can be particularly vulnerable to is oil spills. The pneumatophore root structure easily traps oil residues in the sediment, which, coupled with the relatively slow turnover of aboveground biomass, contributes to long-term negative impacts on plant health (Lewis et al., 2011; Duke, 2016). Oiling also suffocates mangroves by coating leaves and pneumatophores, which can weaken and kill plants for years following initial oiling (Duke, 2016). Following the 2010 Deepwater Horizon (DWH) oil spill, ecosystem recovery in mixed mangrove–marsh systems was dominated by marsh vegetation (Lin and Mendelssohn, 2012; Shapiro et al., 2016), suggesting enhanced oil sensitivity in mangroves. Although oiling has a neutral (Kleinhuizen et al., 2017) or even positive (Levine et al., 2017a) effect on N removal in marsh sediments, it can lead to long-term losses in marsh N removal capacity (Hinshaw et al., 2017; Tatariw et al., 2021), highlighting a need to understand potential differences in oiling response in marshes and mangroves.
We measured NO3− reduction (i.e., denitrification and DNRA) rates and the microbial community structure (16S rRNA) in mangrove (Avicennia germinans)- and marsh (Spartina alterniflora)-dominated sediments at sites subjected to light and moderate oiling during the DWH spill. We hypothesized that denitrification would be favored over DNRA in A. germinans plots because 1) A. germinans is more efficient at translocating oxygen to the sediment, reducing sulfide inhibition and promoting coupled nitrification/denitrification, and 2) A. germinans litter has a lower C:N ratio, which favors denitrification over DNRA. We predicted that denitrification and DNRA rates would be lower at the moderately oiled site due to the detrimental effects of oiling on plant function and loss of plant C, with a greater impact in A. germinans, which is more sensitive to oiling than S. alterniflora.
Materials and methods
Site description
The Chandeleur Islands are a chain of low-lying (<2 m) barrier islands situated approximately 60 km south of Biloxi, Mississippi, in the Gulf of Mexico (Figure 1). The islands run north to south for 80 km and range 2–10 km in width. The dominant vegetation is Spartina alterniflora (smooth cordgrass) and Avicennia germinans (black mangrove). During the DWH oil spill, the islands were subjected to a range of very light to heavy oiling, as determined by the Shoreline Cleanup and Assessment Technique (SCAT) data from the summer of 2010 (EMRA, 2015). SCAT oiling intensities are determined by the distribution, width, and thickness of oil bands (Michel et al., 2013; Nixon et al., 2016). There were no clean-up efforts on the islands.
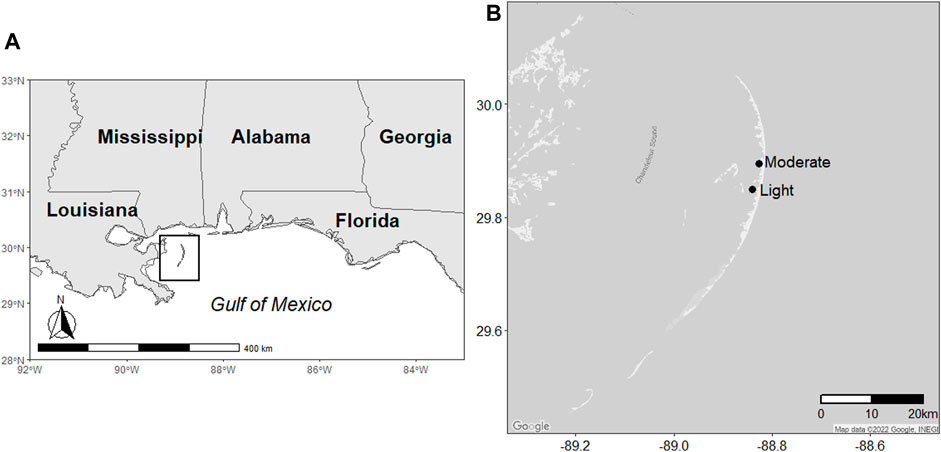
FIGURE 1. Map depicting the location of (A) the Chandeleur Islands and (B) the locations of the study sites.
We used SCAT data to establish two sampling sites subjected to different oiling intensities during the DWH spill (Figure 1). Both sites were located along the western shore of the islands and had both S. alterniflora and A. germinans present. The southern site was subjected to light oiling during the spill (“lightly oiled”; 29.863750° −88.841466°), and the northern site was subjected to moderate oiling during the DWH spill (“moderately oiled”; 29.895448° -88.827780°). Three 1 × 1-m plots were established within each vegetation type at each site. Samples were collected from each plot on five dates (30 May 2017, 03 July 2017, 02 August 2017, 07 May 2018, and 13 June 2018). Salinities and nutrient concentrations from the water column adjacent to the shore were comparable between sites.
On each sampling date, triplicate sediment samples (0–5 cm) were collected for nitrate reduction potentials, microbial analysis, and oil residue analysis using a 60-ml syringe corer (i.d. 2.6 cm). A 10-ml syringe corer (i.d. 1.3 cm) was used to collect triplicate sediment samples (0–5 cm) for porewater nutrient extraction. Single cores were collected from each plot on each date to test for sediment C and N content and sediment chlorophyll a (chl-a) (0–1 cm). All samples were transported to the lab on ice.
Plant metrics
A. germinans height was measured on all dates but 30 May 2017 and 03 July 2017. The average plant height (m) for each plot was determined by measuring six points along the length of the plant perpendicular to the shoreline. This transect included the highest point of the plant (maximum plant height, m). Sediment cores (7.9 cm i.d.) were collected to a depth of 5 cm to determine plant belowground biomass for all dates but 30 May 2017 and 03 July 2017. Roots were separated from the sediment by rinsing with tap water through a 2-mm sieve. The remaining plant material was oven-dried at 60 °C to a constant weight, and the belowground biomass was determined on an areal basis (kg m−2).
Sediment physicochemical analysis
A core (0–5 cm, i.d. 1.3 cm) was collected from each plot to determine porewater extractable NH4+. Porewater NH4+ was extracted with 2N potassium chloride (KCl) by incubation on a shaker table. After 24 h, the slurries were centrifuged, and the supernatant was filtered (0.45 µm nylon membrane filter) and frozen until analysis. Porewater NH4+ was determined fluorometrically (Holmes et al., 1999: Protocol B). Sediment chlorophyll a (chl-a) inventories were determined from the top 2 cm of the sediment from each plot (i.d. 1.3 cm). Sediments were freeze-dried, sediment dry weight was recorded, and chl-a was extracted with 90% acetone for 24 h. Chl-a concentrations were determined fluorometrically (Welschmeyer 1994). To determine the molar carbon to nitrogen ratio (C:N) of sediments, cores (5 cm × 1.3 cm) were oven-dried at 60°C, ground with a mortar and pestle, and then fumigated with 12N HCl overnight to remove carbonates. Following fumigation, samples were oven-dried again and ground prior to analysis on a Costech 4010 CHN analyzer.
Oil residue analysis
Oil-source fingerprinting was conducted on the leftover frozen microbial sediments. Profiles were determined from petroleum biomarker compounds, as measured via GC/MS analytical methods at Louisiana State University (Iqbal et al., 2008; Meyer et al., 2018). Source matching was done by visually comparing hopane and sterane biomarker compound concentrations in their respective m/z191 (hopanes), m/z 217 (sterane), and m/z 218 (sterane) ion chromatograms. The ion chromatograms of extracted field samples were compared to the equivalent data for biomarker compounds in the MC252 source oil.
Nitrate reduction potentials
Triplicate samples were collected from each plot for nitrate reduction (i.e., denitrification, anammox, and DNRA) potential assays as described in Tatariw et al. (2021). Two cores (0–5 cm, i.d. 2.6 cm) were homogenized for each replicate. Following overnight incubation at ambient water temperature, roughly one half of each sample was slurried with artificial sea water (ASW) adjusted to the average salinity of the sitewater at the time of sampling. The remaining sediments were stored at −80°C. Slurries were bubbled with dinitrogen (N2) gas to produce anoxic conditions and siphoned into Exetainer vials (Labco). Following overnight incubation at the sitewater temperature to remove ambient NO3− and oxygen (O2), slurries were spiked to ∼50 µM Na15NO3 (98 atom %; Cambridge Isotope Laboratories, Inc.). One half of the slurry tubes were immediately spiked with zinc chloride (ZnCl2, 50% W/V) to stop biological activity. The other half were incubated for ∼6 h at ambient water temperature and then amended with ZnCl2.
Denitrification and anaerobic ammonium oxidation (annamox) were measured based on the concentrations of 29N2 and 30N2 in slurry water using a membrane inlet mass spectrometer (MIMS) outfitted with a copper reduction column to remove excess oxygen (Nielsen, 1992; Kana et al., 1994; Eyre et al., 2002). DNRA was measured based on 15NH4 production by means of hypobromite reduction (Thamdrup and Dalsgaard, 2002; Yin et al., 2014). Briefly, DNRA tubes were bubbled with N2 to purge the 29N2 and 30N2 produced by denitrification and anammox. Samples were then amended with 200 µL sodium hypobromite, which converts NH4+ to N2. The resulting 29N2 and 30N2 concentrations were measured on the MIMS. Following analysis, sediments in the tubes were dried to a constant weight to calculate NO3− reduction rates as μmol N kg dry weight−1 h−1.
To determine the isotope enrichment of the slurries, slurry tubes with and without 15NO3− addition were filtered and frozen for NO2+3− analysis as described earlier. Ambient NO2+3− was extremely low, and samples were 15N enriched at >95% across all dates. The contribution of anammox to potential nitrate reduction was very small (<1%) and was excluded from further analysis.
DNA isolation and 16S rRNA amplicon sequencing
Total genomic DNA was extracted in triplicate from each core with an MP FastDNA™ Spin Kit for soil (MP Biomedicals, LLC) according to the manufacturer’s instructions with the addition of 2-min ice incubations following the homogenization and 4°C centrifugation steps. The triplicate extractions were then pooled and purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research) and eluted in sterile ddH2O (total volume 50 µL). Total DNA concentration was quantified spectrophotometrically using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) and stored at -20°C.
One of the three cores from each plot and date was selected for 16S rRNA sequencing for microbial community analysis (total 59 samples). Briefly, the hypervariable V3–V4 region of the 16S rRNA gene from total DNA was amplified with region-specific primers that included Illumina flowcell adapter sequences. Following library preparation, a 2 × 250-bp amplification run was conducted using Illumina MiSeq 2000 technology by the University of Tennessee Genomics Core (Knoxville, TN, United States).
Sequence processing
Raw paired-end FASTQ files were trimmed (cutadapt), dereplicated, denoised, and merged using DADA2 in QIIME2 (version 2018.8). Chimeric sequences were identified and removed using the consensus method in DADA2 (Callahan et al., 2016). Taxonomic assignment of amplicon sequence variants (ASVs) was performed using the QIIME2 q2-feature-classifier plugin using a SILVA 13_5 99% trained Naïve Bayes classifier for the V3–V4 region of the 16S rRNA gene (Quast et al., 2013). Multiple sequence alignment and phylogenetic reconstruction were carried out using MAFFT and FastTree. To account for the diverse sequence distribution across samples, ASV abundance data were transformed into relative abundance by dividing each ASV by the sum of a given sample. Amplicon sequences were randomly subsampled to an even depth of 53,963 reads per sample. Pielou’s evenness and Shannon’s diversity were calculated to assess alpha (i.e., within sample) diversity using the vegan package in R (Oksanen et al., 2015).
Indicator analysis
Multipatt and random forest modeling (RFM) were used to identify the ASVs driving community differences between oiling intensity (i.e., sites) and vegetation types. The taxonomic assignment for each identifier is listed in Supplementary Table S3. To reduce the noise caused by rare features (Hackstadt and Hess, 2009), ASVs consisting of more than 80% zeros were removed with METAGENassist (Arndt et al., 2012). ASVs that were selected by both indicator species and random forest modeling were designated as important species similar to Kolton et al. (2020). The multipatt function was used to identify indicator species using correlation to identify ASVs based on fidelity (present in all samples of a habitat) and exclusivity (only present in a habitat) in the indicspecies package in R with 999 permutations (De Cáceres and Legendre, 2009). A classification random forest analysis (Breiman, 2001) was used to identify significant ASVs by vegetation type and oiling intensity with the randomForest package in R (Liaw and Wiener, 2002). The RFM was run with 501 trees and cross-validated with leave-one-out validation. The 15 ASVs with the greatest mean decrease in accuracy were considered the most important.
qPCR on N cycling genes
Quantitative PCR (qPCR) was performed to assess the abundance of two genes in the denitrification pathway: nirS (nitrite reductase) and nosZ (nitrous oxide reductase). Total community abundance was determined by qPCR analysis of 16S rRNA. Each reaction included Platinum SYBR Green qPCR SuperMix-UDG with ROX (12.5 µl) (Invitrogen), 1 µl of forward and reverse primer (5 μM, IDT DNA Technology), 0.5 µl MgCl2 (50 mM), and 5 ng of DNA template. The total reaction volume was adjusted to 25 µl with PCR grade water (ThermoFisher). Genomic DNA isolated from boil preparations of Vibrio fischeri and Pseudomonas stutzeri were used for PCR amplification to generate single-copy gene plasmids for nirS, nosZ, and 16S rRNA standard curves. Samples were run in duplicate on a 7000 Sequence Detection System (ABI Prism) with the primer combinations and qPCR conditions listed in Supplementary Table S1. Following qPCR, random samples were run on a 1.2% sodium boric acid (SB) gel to assess correctly sized amplicons. PCR inhibition due to co-extracted humic material was tested by standard addition during reaction setup. DNA copy numbers were expressed as gene copies per gram of wet sediment as previously described in Lindemann et al. (2016) and Warneke et al. (2011).
Statistical analyses
Differences in NO3− reduction rates, sediment characteristics, and 16S alpha diversity between vegetation types and sites were tested using a linear mixed-effects model (LME; package nlme, Pinheiro et al., 2015) with site x vegetation type x sampling year as main factors and plot as a random factor. A. germinans height was tested with LME with site x year as a main factor and plot as a random factor. Significant interactions were tested using tests of simple effects. All variables except for nirS abundance were natural log-transformed to meet the assumption of normality. NirS abundance was square-root transformed. To determine the effect of site and vegetation type on beta diversity, we conducted a permutational multivariate analysis of variance (PERMANOVA) using the adonis() function in vegan (Oksanen et al., 2015).
Multivariate homogeneity of group dispersions was used to test for differences in variances between land-use types (function: Betadisper). PERMANOVA was performed on the distance matrix to test for differences in β-diversity between land-use types (function: Pairwise.adonis2) (Martinez Arbizu, 2017). Bioenv was used with a mantel test to identify the best environmental characteristics that correlated with community dissimilarities using Spearman’s correlation (Clarke and Ainsworth, 1993). Non-metric multidimensional (NMDS) scaling was used to visualize differences in microbial communities between sites and vegetation types. NMDS was performed on a Bray–Curtis distance matrix using metaMDS. Vectors of environmental variables were fitted to the NMDS coordinates with envfit.
Results
Plant and sediment characteristics
The average A. germinans height was 1.2 ± 0.3 m and did not differ between sites (LME, p = 0.391) or years (LME, p = 0.156). Belowground biomass did not differ between sites for either plant type (LME, p = 0.941). The belowground biomass was 2X greater in S. alterniflora plots (1.1 ± 0.4 kg m−2) than in A. germinans plots (0.5 ± 0.3 kg m−2) (LME, p < 0.001).
On average, extractable NH4+ was nearly twice as high at the lightly oiled site than the moderately oiled site (0.24 ± 0.20 vs. 0.15 ± 0.11 μmol g−1) (LME, p = 0.011), and the effect of vegetation type depended on site (LME, p = 0.006). Extractable NH4+ concentrations were significantly lower (2.3X) in A. germinans plots at the moderately oiled site than in the lightly oiled site (TSE, p < 0.001), whereas in S. alterniflora plots there were no differences in concentrations between sites (TSE, p = 0.719). Temporal effects were also dependent on site (LME, p = 0.039). In 2017, NH4+ concentrations at the lightly oiled site were more than double those at the moderately oiled site (0.30 ± 0.23 vs. 0.13 ± 0.09 μmol g−1) (TSE, p < 0.001), whereas there were no differences between sites in 2018 (TSE, p = 0.985).
The effect of vegetation type was dependent on site for both sediment C and N (LME, p = 0.005; Supplementary Table S3). In A. germinans plots, sediments C and N were, respectively, 2.3X and 3X lower at the moderately oiled site (TSE, p < 0.001 and p < 0.001). In contrast, neither sediment C nor sediment N differed between sites in S. alterniflora plots (TSE, p = 0.480 and p = 0.151). Both C and N were higher in 2017 than 2018 (131 ± 95 vs. 36 ± 22 μmol C g sediment−1 and 8 ± 7 vs. 2 ± 1 μmol N g sediment−1) (LME, p < 0.001 for both).
Sediment chl-a inventories differed between vegetation types; on average, inventories were 2.1X higher in S. alterniflora plots than in A. germinans plots (LME, p < 0.001). The effect of site on sediment chl-a depended on vegetation type (LME, p < 0.001). Chl-a inventories in A. germinans plots were lower at the moderately oiled site than at the lightly oiled site (38 ± 13 vs 83 ± 66 mg m−2) (TSE, p < 0.001), whereas there were no differences in chl-a inventories between moderately and lightly oiled sites in S. alterniflora plots (144 ± 46 vs 105 ± 53 mg m−2) (TSE, p = 0.090). Sediment chl-a inventories were consistent across years (LME, p = 0.794).
Oil residue analysis
Heavily weathered oil residues were detected in 10 of the 60 samples analyzed (Supplementary Table S2). All but one of the samples with detectable residues were from the moderately oiled site. The majority (7) of the samples with detectable residues were collected from A. germinans plots. Each collection date had at least one sample with detectable residues.
Potential nitrate reduction rates
Average denitrification rates were 1.6X higher at the lightly oiled site than at the moderately oiled site (Figure 2A, LME, p = 0.013), although the effect was largely driven by differences in A. germinans plots (site*vegetation, LME, p = 0.006). At the lightly oiled site, denitrification rates were 1.6X higher in A. germinans plots that in S. alterniflora plots (TSE, p = 0.034). There was no difference in denitrification rates between vegetation types at the moderately oiled site (TSE, p = 0.319).
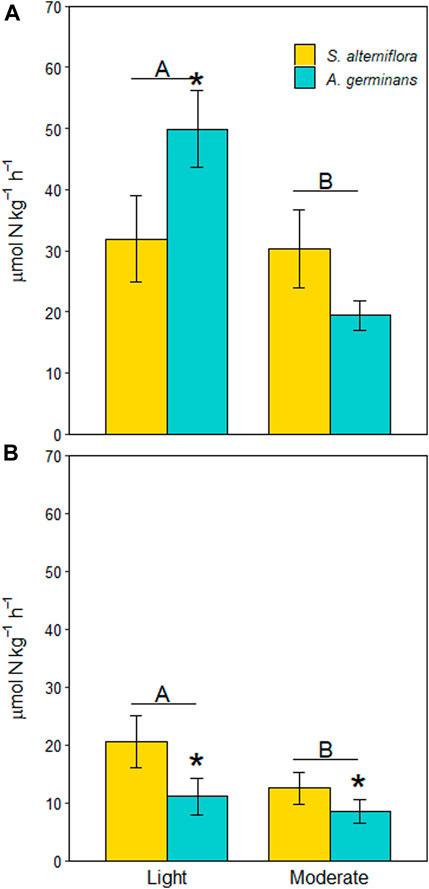
FIGURE 2. Denitrification (A) and DNRA (B) potential rates for each site and vegetation type (indicated by color). Significant differences between sites are indicated by capital letters. Significant differences between vegetation types within sites are indicated with an asterisk (*). Bars are standard error.
DNRA rates were 1.5X higher at the lightly oiled site that at the moderately oiled site (Figure 1B, LME, p = 0.037). Unlike denitrification rates, the effect of vegetation type was independent of site (site*vegetation, LME, p = 0.559). Rather, DNRA rates were on average 1.7X higher in S. alterniflora plots than A. germinans plots across both sites (Figure 2B, LME, p = 0.004).
N removal dominated over DNRA, accounting for 71 ± 2% of NO3− reduction on average (Figure 3). The effect of site on percent denitrification depended on vegetation type (site*vegetation, LME, p = 0.004). At the lightly oiled site, percent denitrification was 1.4X higher in A. germinans plots than S. alterniflora plots (TSE, p < 0.001), whereas there was no difference between vegetation types at the moderately oiled site (TSE, p = 0.370).
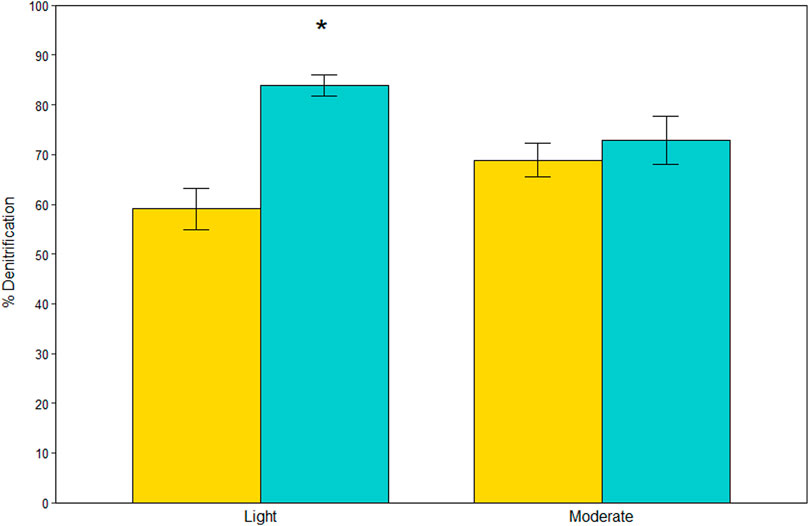
FIGURE 3. Percent (%) contribution of denitrification to total nitrate reduction for each site and vegetation type (indicated by color). Significant differences between vegetation types within sites are indicated with an asterisk (*). Bars are standard error.
On average, denitrification potential rates were 1.3X higher in 2018 than 2017 across both sites and vegetation types (LME, p = 0.026). Similarly, DNRA potential rates were significantly higher (2.2X) in 2018 than 2017 (LME, p < 0.001). Percent denitrification was 1.2X higher in 2017 than 2018.
16S rRNA community diversity
16S rRNA gene sequencing resulted in a total of 18,733,736 sequences in 59 samples. Among the 8,345,939 quality-filtered sequences, 7,942,475 (95.3%) were assigned to bacterial 16S rRNA ASV IDs. Quality-filtered reads ranging from 53,963 to 273,796 reads per sample accounted for a total of 796 16S rRNA ASVs. Fourteen of the 64 assigned phyla contributed to greater than 1% of relative abundance. Proteobacteria, Chloroflexi, and Bacteroidetes were the three most abundant phyla, accounting for 62% of ASVs.
The observed number of ASVs ranged from 1849 to 5,937 ASVs per sample. There was no difference in Shannon diversity between sites (LME, p = 0.434; Supplementary Figure S1), but microbial communities in A. germinans plots were more diverse than those in S. alterniflora plots across both sites (LME, p = 0.018). There was a marginal interaction between site and vegetation type (LME, p = 0.052) that showed a trend of higher diversity in A. germinans plots at the lightly oiled site. Like Shannon diversity, Pielou’s evenness was significantly higher in A. germinans plots than S. alterniflora plots (LME, p = 0.0247; Supplementary Figure S1). The effect of site on Pielou’s evenness depended on vegetation type (LME, site x vegetation, p = 0.037). In A. germinans plots, evenness was higher at the lightly oiled site (TSE, p = 0.037), whereas there was no difference in evenness between sites in S. alterniflora plots (TSE, p = 0.970). Shannon diversity was higher in 2018 than 2017 (LME, p = 0.008), but there was no difference in Pielou’s evenness between years (LME, p = 0.088).
PERMANOVA revealed a significant site and vegetation interaction for beta diversity (PERMANOVA, p < 0.004), and pairwise comparisons (pairwise.adonis) between each site and vegetation combination confirmed that beta diversity differed between all site–vegetation combinations (PERMANOVA, p < 0.006; Figure 4). Beta dispersion did not differ between sites (p = 0.111) and vegetation types (p = 0.752), indicating that community differences between sites and vegetation types were driven by changes in microbial community structure and not changes in variance between the groups.
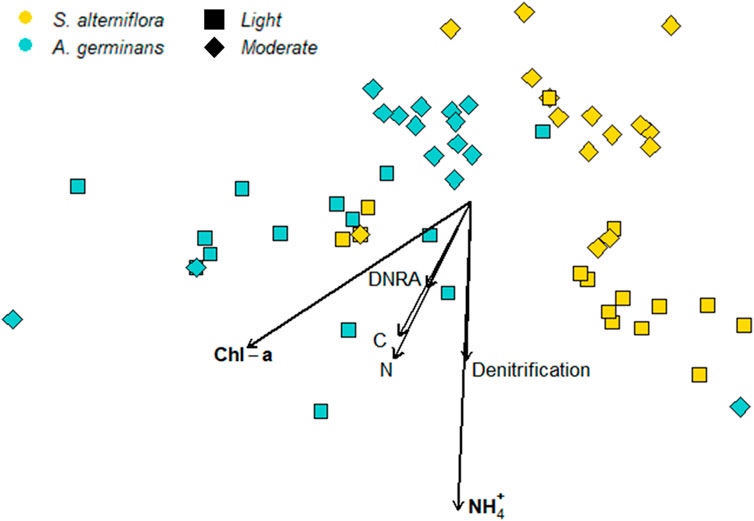
FIGURE 4. NMDS of the 16S microbial community with bioenv variables as vectors. Oiling intensity is indicated by symbol shape, with lightly oiled = squares, moderately oiled = diamonds. Turquoise symbols indicate A. germinans communities and goldenrod symbols indicate S. alterniflora communities.
Bioenv selected chl-a, extractable NH4+, bulk N, and denitrification rates as the best sediment variables to correlate with community distances (Mantel test, ρ = 0.19, p = 0.004). Two of the fitted vectors for sediment variables were significantly correlated with community structure (Table 1). Sediment chl-a correlated with both NMDS axes (R2 = 0.170, p = 0.014), and extractable NH4+ correlated with NMDS axis 2 (R2 = 0.227, p = 0.002).
Indicator analysis
Seven of the 15 taxa were selected as indicators of the lightly oiled site (Figure 5). Four of these were indicators for S. alterniflora sediments. The S. alterniflora indicators included two taxa from the phylum Proteobacteria: genus Erythrobacter and family Desulfobulbaceae. S. alterniflora indicator taxa at the lightly oiled site also included Eudoraea, a genus in Bacteroidetes, and Gemm-2, a class in Gemmatimonadetes. Of the three indicator taxa selected for A. germinans at the lightly oiled site: two were from Proteobacteria; one was from the genus Rhodovibrio in Alphaproteobacteria; and the other was from Thiohalorhabdales in Gammaproteobacteria. The third A. germinans indicator at the lightly oiled site was the class AT-s2-57 in Acidobacteria.
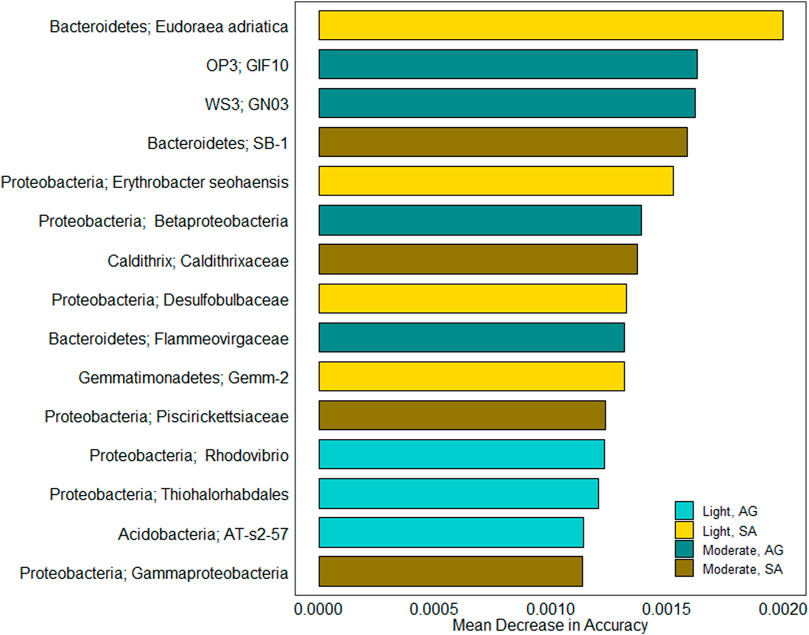
FIGURE 5. Indicator taxa driving community differences between oiling intensity and vegetation types. Bars depict mean decrease in accuracy for the 15 most important features as determined by random forest analysis. The color of each bar indicates for which combination of oiling intensity and vegetation type each taxon was an indicator.
The eight indicator taxa selected for the moderately oiled site were evenly divided between S. alterniflora and A. germinans. Two S. alterniflora taxa were Gammaproteobacteria, including one from the family Piscirickettsiaceae. The remaining two S. alterniflora taxa were from Bacteroidetes (family SB-1) and Caldithrix (family Caldithrixaceae). The indicator taxa for A. germinans at the moderately oiled site were from four different phyla: Proteobacteria (class Betaproteobacteria), Bacteroidetes (family Flammeovirgaceae), OP3 (order GIF10), and WS3 (order GNO3).
qPCR
The effect of vegetation type on 16S abundance depended on site (LME, p = 0.044; Figure 6A). There was no difference in 16S abundance between A. germinans and S. alterniflora sediments at the lightly oiled site (TSE, p = 0.981), but at the moderately oiled site, 16S abundance was 1.3X lower in A. germinans plots than in S. alterniflora sediments (TSE, p = 0.013; Figure 6A). There was no difference in 16S rRNA abundance between sites in S. alterniflora plots, but 16S abundance was significantly lower (2.4X) at the moderately oiled site in A. germinans plots (TSE, p = 0.018). As with denitrification and DNRA potential rates, 16S amplicon abundance differed between years; however, in contrast to process rates, 16S abundances were 2X higher in 2017 than 2018 (LME, p = 0.001).
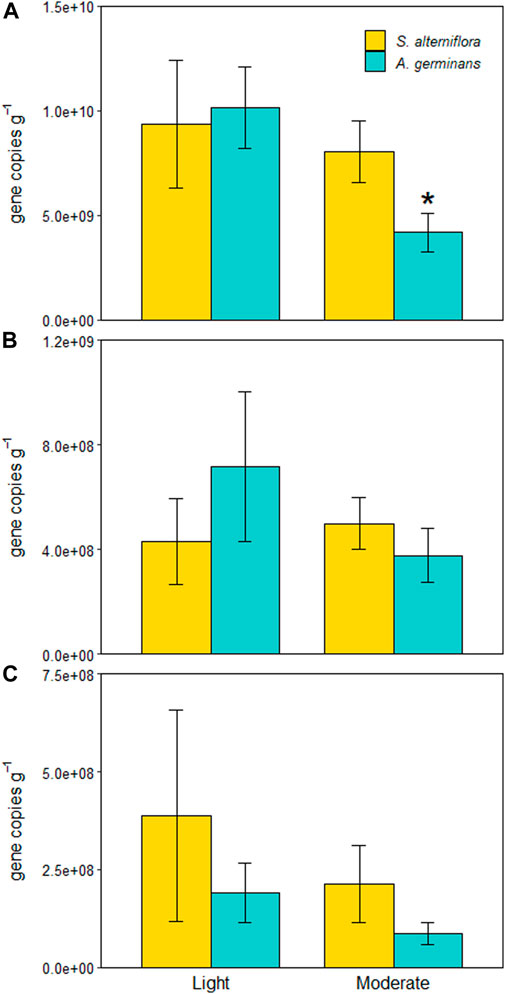
FIGURE 6. 16S (A), nirS (B), and nosZ (C) marker abundances for each site and vegetation type (indicated by color). Significant differences between vegetation types within sites are indicated with an asterisk (*). Bars are standard error.
Neither nitrite reductase (nirS) nor nitrous oxide reductase (nosZ) functional markers differed significantly between sites (LME, p = 0.723 and p = 0.378) or vegetation types (LME, p = 0.955 and p = 0.616) (Figure 5B). Although not statistically significant (LME, p = 0.290 and p = 0.817), trends in nirS and nosZ abundances between sites and vegetation types mirrored those of denitrification potentials showing a decrease from the lightly oiled to the moderately oiled site in A. germinans plots compared to similar abundances at both sites in S. alterniflora plots. Neither nitrite reductase (nirS) nor nitrous oxide reductase (nosZ) functional markers differed significantly between years (LME, p = 0.529 and p = 0.439).
Discussion
We found that at the less oil-impacted site, mangrove sediments had a significantly higher N removal potential than marsh sediments, both in terms of denitrification potential rates and the proportion of NO3− being removed. Woody encroachment has been shown to alter the chemical, microbial, and redox characteristics of marsh sediments (Perry and Mendelssohn, 2009; Guo et al., 2017; Barreto et al., 2018; Simpson et al., 2019), all of which are important drivers of biogeochemical processes. Our data suggest that a combination of redox and microbial community changes associated with woody encroachment, rather than changes in nutrient availability, resulted in increased denitrification potential in mangrove sediments. Vegetation is an important driver of N removal in coastal wetlands, as root O2 translocation to the sediment alleviates sulfide inhibition of coupled nitrification–denitrification (Koop-Jakobsen and Giblin, 2009). The mangrove pneumatophore root structure is efficient at O2 translocation to the subsurface and can increase sediment redox potential (Andersen and Kristensen, 1988; Perry and Mendelssohn, 2009; Comeaux et al., 2012). Additionally, A. germinans prefers higher elevations, and better drainage can also increase sediment redox potential (Perry and Mendelssohn, 2009). Although we did not directly measure changes in redox, the results of our microbial indicator analysis suggest that O2 availability may differ between Avicennia and Spartina sediments. A. germinans indicator taxa included Acidobacteria, which are commonly found in soil and sediments and consist mainly of aerobic heterotrophs (Ward et al., 2009), whereas S. alterniflora indicators included Caldithrixales (phylum Caldithrix), a putative anaerobe that has been found in Spartina marshes (Bulseco et al., 2019). This is consistent with the findings of Barreto et al. (2018), who found that mangrove indicator taxa were more likely to be putative aerobes, whereas marsh indicator taxa were more likely to be putative anaerobes (Barreto et al., 2018). Furthermore, they also found that Acidobacteria and Caldithrix were indicators for mangrove and marsh sediments, respectively (Barreto et al., 2018).
The higher DNRA rates in S. alterniflora sediments also support the idea that higher redox potential in A. germinans sediments favors denitrification. DNRA is favored under highly reducing conditions or when there is a high organic C: NO3− ratio (Gardner and McCarthy, 2009; Hardison et al., 2015; Kessler et al., 2018). We did not observe differences in sediment C and N availability in sediments dominated by different vegetation types, possibly because high sediment deposition rates in the northern Gulf of Mexico can outweigh the influence of woody encroachment on nutrient stocks (Henry and Twilley, 2013). Rather, the selection of taxa associated with sulfur cycling suggests that sulfide accumulation associated with more reducing conditions in marsh sediments may be the driving factor of N retention via DRNA. Desulfobulbaceae (Deltaproteobacteria) was an indicator taxon for S. alterniflora that includes sulfate-reducing bacteria (Dyksma et al., 2018) and cable bacteria (Kjeldsen et al., 2019). Cable bacteria are filamentous bacteria capable of sulfide oxidation using oxygen or nitrate as electron acceptors (Murphy et al., 2020). They are found where sulfide accumulates, and using long-distance electron transport, they create sulfide-free suboxic zones (Larsen et al., 2015; Kjeldsen et al., 2019). Significantly, they can promote DNRA by increasing the Fe2+ pool by iron sulfide dissolution (Kessler et al., 2019). Another putative sulfide oxidizer selected as an indicator for S. alterniflora was Piscirickettsiaceae (Proteobacteria) (Borin et al., 2009). Piscirickettsiaceae has been shown to thrive in sulfidic environments (Pavlovska et al., 2021).
The two vegetation types also had different associated sediment microbial communities that were correlated with sediment-extractable NH4+ and chl-a inventories. While there were no vegetation effects on NH4+, chl-a was significantly lower in A. germinans, likely due to increased canopy shading (Guo et al., 2017). This suggests that even though there were no differences in substrate quantity (e.g., bulk C and N), there may have been differences in substrate quality that impacted microbial diversity and activity. A. germinans leaves have a higher N content than S. alterniflora leaves (Macy et al., 2019), and changes in litter quality associated with woody encroachment can increase decomposition rates in A germinans-dominated sediments compared to S. alterniflora-dominated sediments (Simpson et al., 2021). While we did not directly test for changes in litter quality, the microbial indicator analysis selected taxa associated with litter degradation in S. alterniflora. Gemmatimonadetes have metabolic pathways for carbohydrate and protein degradation (Baker et al., 2015) and have been found in S. alterniflora salt marshes (Bowen et al., 2012). Erythrobacter is a saprophyte that has been associated with S. alterniflora litter degradation (Buchan et al., 2003). Additionally, Bacteroidales SB-1 (phylum Bacteroidetes) and Caldithrixales (phylum Caldithrix) have been previously associated with S. alterniflora sediments (Bulseco et al., 2019; Emery et al., 2019). Thus, we suggest that the changes in litter quality associated with woody encroachment resulted in microbial community shifts that can influence other ecosystem functions, such as N removal.
Although woody encroachment enhanced N removal at the lightly oiled site, the effect did not persist at the moderately oiled site. We measured a similar effect on microbial diversity and sediment nutrient inventories in mangrove sediments that did not co-occur in marsh sediments, suggesting that ecosystem recovery, and by extension functional recovery, is slower in marshes impacted by woody encroachment. Avicennia species of mangroves are generally more sensitive to oil than other mangrove species (Lewis et al., 2011), whereas S. alterniflora is less sensitive to oiling than other marsh grasses (Michel and Rutherford, 2014). Although there were no differences in mangrove height or belowground biomass between the two sites, the majority (70%) of oil residues were detected in mangrove sediments at the moderately oiled site, possibly because pneumatophores trap oil in sediments (Duke, 2016). Re-suspension of oil residues, such as occurs during the slurrying process, can reduce denitrification rates (Levine et al., 2017a). Following the DWH spill, S. alternifora was faster to recover than A. germinans (Lin and Mendelssohn, 2012; Shapiro et al., 2016). The higher turnover and litter production of S. alterniflora (Mckee and Rooth, 2008) may have contributed to fewer differences in process rates and microbial community composition between sites in marsh sediments.
Another mechanism through which oiling may have impacted denitrification rates in A. germinans sediments is by altering sediment redox conditions. Buried oil residues can decrease the redox potential of rhizosphere sediments (Levine et al., 2017b). We found that the indicator taxa for A. germinans at the moderately oiled site selected more anaerobic taxa than the lightly oiled site. GIF10 (phylum OP3) is found in anoxic environments (Glöckner et al., 2010) and is associated with sulfate cycling (Liu et al., 2013). GNO3 is a member of the phylum Latescibacteria (WS3) found in marine environments (Baker et al., 2015; Farag et al., 2017). Latescibacteria are largely saprophytic (Farag et al., 2017) and have metabolic potential for anaerobic fermentation (Youssef et al., 2015). Thus, it is possible that altered redox conditions associated with oiling affected the N removal capacity of A. germinans-dominated sediments. While future research is necessary to elucidate the direct mechanism, our research indicates that oiling has a more prolonged negative effect on N removal rates in mangrove than marsh systems.
Enrichment of hydrocarbon-degrading microbial communities was documented following the DWH spill (Beazley et al., 2012; King et al., 2015; Engel et al., 2017). This was also reflected in our indicator analysis, as putative hydrocarbon degraders were selected as indicator taxa for both vegetation types, regardless of oiling intensity. These included Erythrobacter seohaensis (phylum Alphaproteobacteria) and Rhodovibrio (phylum Proteobacteria) (Yoon et al., 2005; Liu and Liu, 2013; Godoy-Lozano et al., 2018; Machado et al., 2019) at the lightly oiled site and the class Gammaproteobacteria (Proteobacteria), the family Piscirickettsiaceae (Proteobacteria), the family SB-1 (Bacteroidetes), and the class Betaproteobacteria (Hazen et al., 2010; Rivers et al., 2013; Hamdan et al., 2018; Emery et al., 2019; Tan and Parales, 2019) at the moderately oiled site. The indicator analysis also selected microbial taxa that have been identified as indicators of oiling recovery. At the lightly oiled site, a selected indicator was Group Gemm-2 in Gemmatimonadetes. Gemm-2 was found to be in decline after oiling in beach sediments and then returned to pre-oiling numbers following cleanup (Huettel et al. 2018). Flammeovirgaceae (phylum Bacteroidetes) was selected at the moderately oiled site and is a widely distributed, salt-tolerant, aerobic taxa that has been known to decline in abundance after oiling (Koo et al., 2015; Mishamandani et al., 2016; Rath et al., 2018). Regardless of whether these taxa reflect long-term microbial community shifts as a result of the DWH spill or the representation of indigenous hydrocarbon-degrading communities, these results indicate that both marsh and mangrove sediment communities have the functional capacity to degrade oil. This is important, as while the DWH spill was an extreme event, natural oil seeps are highly prevalent in the Gulf of Mexico (MacDonald et al., 2015), and the persistence of hydrocarbon-degrading communities in A. germinans sediments may enhance resilience to future oiling events.
Conclusion
This is one of the first studies to compare nitrogen removal rates between A. germinans-dominated sediments and the S. alterniflora marsh sediments they are replacing. We found that A. germinans expansion may enhance marsh N removal capacity, possibly through changes in sediment redox potential or litter quality, although additional research is needed to directly test these mechanisms. We also found that this effect did not occur in sediments subjected to moderate oiling from the DWH spill. Results from our indicator taxa analysis suggest that this may be due to changes in sediment redox potential or shifts in the functional microbial community following oiling. Both S. alterniflora and A. germinans sediment microbial communities included putative hydrocarbon-degrading taxa, regardless of oiling intensity, suggesting that these systems have the functional capacity to break down oil. However, the higher sensitivity of A. germinans to oiling may result in an overall slower recovery following a spill and therefore a lag in biogeochemical functional recovery that may not co-occur in salt marshes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://data.gulfresearchinitiative.org, 10.7266/Z5MG1AFS, https://data.gulfresearchinitiative.org, 10.7266/4BSSB0AZ.
Author contributions
CT, AK, and BM were responsible for study conception and design. CT was responsible for manuscript preparation. CT and AK were responsible for fieldwork, sediment analyses, and nitrate reduction rates. PC was responsible for nucleic acid isolation. NF was responsible for 16S RNA sequence processing, and NF and CT were responsible for 16S data analysis. NF and PC were responsible for qPCR. EO was responsible for oil residue analysis. BM and PS received funding for the project.
Funding
This research was made possible by a grant from the Gulf of Mexico Research Initiative (RFP-IV) awarded to the Alabama Center for Ecological Resilience (ACER).
Acknowledgments
We thank T. Ledford and D. Tollette for their assistance in the field and laboratory. We thank L. Hu for assistance with data management. We thank D. Goettert for assistance with oil residue analysis and P. Whitehurst and J. Gilkeson for assistance with DNA isolation. We thank Dr. C. Staudhammer for assistance with statistical analyses. The authors gratefully acknowledge use of the resources of the Alabama Water Institute at The University of Alabama. This publication was made possible with the help from The University of Alabama — Alabama Water Institute.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.951365/full#supplementary-material
References
Andersen, F., and Kristensen, E. (1988). Oxygen microgradients in the rhizosphere of the mangrove Avicennia marina. Mar. Ecol. Prog. Ser. 44, 201–204. doi:10.3354/meps044201
Armitage, A. R., Highfield, W. E., Brody, S. D., and Louchouarn, P. (2015). The contribution of mangrove expansion to salt marsh loss on the Texas Gulf Coast. PLoS ONE 10, e0125404–17. doi:10.1371/journal.pone.0125404
Armitage, A. R., Weaver, C. A., Kominoski, J. S., and Pennings, S. C. (2020). Resistance to hurricane effects varies among wetland vegetation types in the marsh–mangrove ecotone. Estuaries Coasts 43, 960–970. doi:10.1007/s12237-019-00577-3
Arndt, D., Xia, J., Liu, Y., Zhou, Y., Guo, A. C., Cruz, J. A., et al. (2012). METAGENassist: A comprehensive web server for comparative metagenomics. Nucleic Acids Res. 40 (W1), 88–95. doi:10.1093/nar/gks497
Baker, B. J., Lazar, C. S., Teske, A. P., and Dick, G. J. (2015). Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3, 14–12. doi:10.1186/s40168-015-0077-6
Barreto, C. R., Morrissey, E. M., Wykoff, D. D., and Chapman, S. K. (2018). Co-occurring mangroves and salt marshes differ in microbial community composition. Wetlands 38, 497–508. doi:10.1007/s13157-018-0994-9
Beazley, M. J., Martinez, R. J., Rajan, S., Powell, J., Piceno, Y. M., Tom, L. M., et al. (2012). Microbial community analysis of a coastal salt marsh affected by the Deepwater Horizon oil spill. PLoS ONE 7, e41305. doi:10.1371/journal.pone.0041305
Borin, S., Brusetti, L., Mapelli, F., D’Auria, G., Brusa, T., Marzorati, M., et al. (2009). Sulfur cycling and methanogenesis primarily drive microbial colonization of the highly sulfidic Urania deep hypersaline basin. Proc. Natl. Acad. Sci. U. S. A. 106, 9151–9156. doi:10.1073/pnas.0811984106
Bowen, J. L., Morrison, H. G., Hobbie, J. E., and Sogin, M. L. (2012). Salt marsh sediment diversity: A test of the variability of the rare biosphere among environmental replicates. ISME J. 6, 2014–2023. doi:10.1038/ismej.2012.47
Buchan, A., Newell, S. Y., Butler, M., Biers, E. J., Hollibaugh, J. T., and Moran, M. A. (2003). Dynamics of bacterial and fungal communities on decaying salt marsh grass? Appl. Environ. Microbiol. 69, 6676–6687. doi:10.1128/aem.69.11.6676-6687.2003
Bulseco, A. N., Giblin, A. E., Tucker, J., Murphy, A. E., Sanderman, J., Hiller‐Bittrolff, K., et al. (2019). Nitrate addition stimulates microbial decomposition of organic matter in salt marsh sediments. Glob. Change Biol. 25, 3224–3241. doi:10.1111/gcb.14726
Burgin, A. J., and Hamilton, S. K. (2007). Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front. Ecol. Environ. 5, 89–96. doi:10.1890/1540-9295(2007)5[89:hwotro]2.0.co;2
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi:10.1038/nmeth.3869
Charles, S. P., Kominoski, J. S., Troxler, T. G., Gaiser, E. E., Servais, S., Wilson, B. J., et al. (2019). Experimental saltwater intrusion drives rapid soil elevation and carbon loss in freshwater and brackish everglades marshes. Estuaries Coasts 42, 1868–1881. doi:10.1007/s12237-019-00620-3
Clarke, K. R., and Ainsworth, M. (1993). A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser. 92 (3), 205–219. doi:10.3354/meps092205
Comeaux, R. S., Allison, M. A., and Bianchi, T. S. (2012). Mangrove expansion in the Gulf of Mexico with climate change: Implications for wetland health and resistance to rising sea levels. Estuar. Coast. Shelf Sci. 96, 81–95. doi:10.1016/j.ecss.2011.10.003
Dangremond, E. M., Simpson, L. T., Osborne, T. Z., and Feller, I. C. (2020). Nitrogen enrichment accelerates mangrove range expansion in the temperate–tropical ecotone. Ecosystems 23, 703–714. doi:10.1007/s10021-019-00441-2
de Cáceres, M., and Legendre, P. (2009). Associations between species and groups of sites: Indices and statistical inference. Ecology 90 (12), 3566–3574. doi:10.1890/08-1823.1
Donnelly, J. P., and Bertness, M. D. (2001). Rapid shoreward encroachment of salt marsh cordgrass in response to accelerated sea-level rise. Proc. Natl. Acad. Sci. U. S. A. 98, 14218–14223. doi:10.1073/pnas.251209298
Doughty, C. L., Langley, J. A., Walker, W. S., Feller, I. C., Schaub, R., and Chapman, S. K. (2016). Mangrove range expansion rapidly increases coastal wetland carbon storage. Estuaries Coasts 39, 385–396. doi:10.1007/s12237-015-9993-8
Duke, N. C. (2016). Oil spill impacts on mangroves: Recommendations for operational planning and action based on a global review. Mar. Pollut. Bull. 109, 700–715. doi:10.1016/j.marpolbul.2016.06.082
Dyksma, S., Lenk, S., Sawicka, J. E., and Mußmann, M. (2018). Uncultured Gammaproteobacteria and Desulfobacteraceae account for major acetate assimilation in a coastal marine sediment. Front. Microbiol. 9, 1–10. doi:10.3389/fmicb.2018.03124
Emery, H. E., Angell, J. H., and Fulweiler, R. W. (2019). Salt marsh greenhouse gas fluxes and microbial communities are not sensitive to the first year of precipitation change. J. Geophys. Res. Biogeosci. 124, 1071–1087. doi:10.1029/2018jg004788
EMRA. (2015). Web application: Gulf of Mexico environmental response management application. National Oceanic and Atmospheric Administration. Available at: http://erma.noaa.gov/gulfofmexico. (Accessed December 1, 2016).
Engel, A. S., Liu, C., Paterson, A. T., Anderson, L. C., Turner, R. E., and Overton, E. B. (2017). Salt marsh bacterial communities before and after the Deepwater Horizon oil spill. Appl. Environ. Microbiol. 83, e00784-17–17. doi:10.1128/aem.00784-17
Eyre, B. D., Rysgaard, S., Dalsgaard, T., and Christensen, P. B. (2002). Comparison of isotope pairing and N2:Ar methods for measuring sediment denitrification—assumptions, modifications, and implications. Estuaries 25 (6A), 1077–1087. doi:10.1007/BF02692205
Farag, I. F., Youssef, N. H., and Elshahed, M. S. (2017). Global distribution patterns and pangenomic diversity of the candidate phylum “Latescibacteria” (WS3). Appl. Environ. Microbiol. 83, e00521-17. doi:10.1128/aem.00521-17
Fowler, D., Coyle, M., Skiba, U., Sutton, M. A., Cape, J. N., Reis, S., et al. (2013). The global nitrogen cycle in the twenty-first century. Phil. Trans. R. Soc. B 368, 20130164. doi:10.1098/rstb.2013.0164
Gardner, W. S., and McCarthy, M. J. (2009). Nitrogen dynamics at the sediment-water interface in shallow, sub-tropical Florida Bay: Why denitrification efficiency may decrease with increased eutrophication. Biogeochemistry 95, 185–198. doi:10.1007/s10533-009-9329-5
Giblin, A., Tobias, C., Song, B., Weston, N., Banta, G., and Rivera-Monroy, V. (2013). The importance of dissimilatory nitrate reduction to ammonium (DNRA) in the nitrogen cycle of coastal ecosystems. Oceanogr. Wash. D. C). 26, 124–131. doi:10.5670/oceanog.2013.54
Glöckner, J., Kube, M., Shrestha, P. M., Weber, M., Glöckner, F. O., Reinhardt, R., et al. (2010). Phylogenetic diversity and metagenomics of candidate division OP3. Environ. Microbiol. 12, 1218–1229. doi:10.1111/j.1462-2920.2010.02164.x
Godoy-Lozano, E. E., Escobar-Zepeda, A., Raggi, L., Merino, E., Gutierrez-Rios, R. M., Juarez, K., et al. (2018). Bacterial diversity and the geochemical landscape in the southwestern Gulf of Mexico. Front. Microbiol. 9, 1–15. doi:10.3389/fmicb.2018.02528
Guo, H., Weaver, C., Charles, S. P., Whitt, A., Dastidar, S., D’Odorico, P., et al. (2017). Coastal regime shifts: rapid responses of coastal wetlands to changes in mangrove cover. Ecology 98, 762–772. doi:10.1002/ecy.1698
Hackstadt, A. J., and Hess, A. M. (2009). Filtering for increased power for microarray data analysis. BMC Bioinform. 10, 1–12. doi:10.1186/1471-2105-10-11
Hamdan, L. J., Salerno, J. L., Reed, A., Joye, S. B., and Damour, M. (2018). The impact of the Deepwater Horizon blowout on historic shipwreck-associated sediment microbiomes in the northern Gulf of Mexico. Sci. Rep. 8, 1–14. doi:10.1038/s41598-018-27350-z
Hamersley, M. R., and Howes, B. L. (2005). Coupled nitrification-denitrification measured in situ in a Spartina alterniflora marsh with a 15NH4+ tracer. Mar. Ecol. Prog. Ser. 299, 123–135. doi:10.3354/meps299123
Hardison, A. K., Algar, C. K., Giblin, A. E., and Rich, J. J. (2015). Influence of organic carbon and nitrate loading on partitioning between dissimilatory nitrate reduction to ammonium (DNRA) and N2 production. Geochimica Cosmochimica Acta 164, 146–160. doi:10.1016/j.gca.2015.04.049
Hazen, T. C., Dubinsky, E. A., DeSantis, T. Z., Andersen, G. L., Piceno, Y. M., Singh, N., et al. (2010). Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330, 204–208. doi:10.1126/science.1195979
Henry, K. M. (2012). Linking nitrogen biogeochemistry to different stages of wetland soil development in the Mississippi river delta. Louisiana: Lousiana State University and Agricultural and Mechanical College.
Henry, K. M., and Twilley, R. R. (2013). Soil development in a coastal Louisiana wetland during a climate-induced vegetation shift from salt marsh to mangrove. J. Coast. Res. 292, 1273–1283. doi:10.2112/JCOASTRES-D-12-00184.1
Hinshaw, S. E., Tatariw, C., Flournoy, N., Kleinhuizen, A., Taylor, C. B., Sobecky, P., et al. (2017). Vegetation loss decreases salt marsh denitrification capacity: Implications for marsh erosion. Environ. Sci. Technol. 51, 8245–8253. doi:10.1021/acs.est.7b00618
Holmer, M., Gribsholt, B., and Kristensen, E. (2002). Effects of sea level rise on growth of Spartina anglica and oxygen dynamics in rhizosphere and salt marsh sediments. Mar. Ecol. Prog. Ser. 225, 197–204. doi:10.3354/meps225197
Holmes, R. M., Aminot, A., Kérouel, R., Hooker, B. A., and Peterson, B. J. (1999). A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can. J. Fish. Aquat. Sci. 56, 1801–1808. doi:10.1139/f99-128
Huettel, M., Overholt, W. A., Kostka, J. E., Hagan, C., Kaba, J., Wells, W. B., et al. (2018). Degradation of Deepwater Horizon oil buried in a Florida beach influenced by tidal pumping. Mar. Pollut. Bull. 126 (October 2017), 488–500. doi:10.1016/j.marpolbul.2017.10.061
Iqbal, J., Overton, E. B., and Gisclair, D. (2008). Polycyclic aromatic hydrocarbons in Louisiana rivers and coastal environments: Source fingerprinting and forensic analysis. Environ. Forensics 9, 63–74. doi:10.1080/15275920801888301
Jordan, S. J., Stoffer, J., and Nestlerode, J. A. (2011). Wetlands as sinks for reactive nitrogen at continental and global scales: A meta-analysis. Ecosystems 14, 144–155. doi:10.1007/s10021-010-9400-z
Joye, S. B., and Hollibaugh, J. T. (1995). Influence of sulfide inhibition of nitrification on nitrogen regeneration in sediments. Science 270, 623–625. doi:10.1126/science.270.5236.623
Kana, T. M., Darkangelo, C., Hunt, M. D., Oldham, J. B., Bennett, G. E., and Cornwell, J. C. (1994). Membrane inlet mass spectrometer for rapid high-precision determination of N2, O2, and Ar in environmental water samples. Anal. Chem. 66 (23), 4166–4170. doi:10.1021/ac00095a009
Kelleway, J. J., Cavanaugh, K., Rogers, K., Feller, I. C., Ens, E., Doughty, C., et al. (2017). Review of the ecosystem service implications of mangrove encroachment into salt marshes. Glob. Chang. Biol. 23, 3967–3983. doi:10.1111/gcb.13727
Kessler, A. J., Roberts, K. L., Bissett, A., and Cook, P. L. M. (2018). Biogeochemical controls on the relative importance of denitrification and dissimilatory nitrate reduction to ammonium in estuaries. Glob. Biogeochem. Cycles 32, 1045–1057. doi:10.1029/2018gb005908
Kessler, A. J., Wawryk, M., Marzocchi, U., Roberts, K. L., Wong, W. W., Risgaard-Petersen, N., et al. (2019). Cable bacteria promote DNRA through iron sulfide dissolution. Limnol. Oceanogr. 64, 1228–1238. doi:10.1002/lno.11110
King, G. M., Kostka, J. E., Hazen, T. C., and Sobecky, P. A. (2015). Microbial responses to the Deepwater Horizon oil spill: From coastal wetlands to the deep sea. Ann. Rev. Mar. Sci. 7, 377–401. doi:10.1146/annurev-marine-010814-015543
Kjeldsen, K. U., Schreiber, L., Thorup, C. A., Boesen, T., Bjerg, J. T., Yang, T., et al. (2019). On the evolution and physiology of cable bacteria. Proc. Natl. Acad. Sci. U. S. A. 116, 19116–19125. doi:10.1073/pnas.1903514116
Kleinhuizen, A. A., Bernard, R. J., and Mortazavi, B. (2017). Influence of crude oil on denitrification in subtidal sediments in the northern Gulf of Mexico. Clean. - Soil, Air, Water 45, 1600546. doi:10.1002/clen.201600546
Kolton, M., Rolando, J. L., and Kostka, J. E. (2020). Elucidation of the rhizosphere microbiome linked to Spartina alterniflora phenotype in a salt marsh on Skidaway Island, Georgia, USA. FEMS Microbiol. Ecol. 96 (4), 1–19. doi:10.1093/femsec/fiaa026
Koo, H., Mojib, N., Huang, J. P., Donahoe, R. J., and Bej, A. K. (2015). Bacterial community shift in the coastal Gulf of Mexico salt-marsh sediment microcosm in vitro following exposure to the Mississippi Canyon Block 252 oil (MC252). 3 Biotech. 5, 379–392. doi:10.1007/s13205-014-0233-x
Koop-Jakobsen, K., and Giblin, A. E. (2009). New approach for measuring denitrification in the rhizosphere of vegetated marsh sediments. Limnol. Oceanogr. Methods 7, 626–637. doi:10.4319/lom.2009.7.626
Larsen, S., Nielsen, L. P., and Schramm, A. (2015). Cable bacteria associated with long-distance electron transport in New England salt marsh sediment. Environ. Microbiol. Rep. 7, 175–179. doi:10.1111/1758-2229.12216
Ledford, T. C., Mortazavi, B., Tatariw, C., and Mason, O. U. (2020). Elevated nutrient inputs to marshes differentially impact carbon and nitrogen cycling in two northern Gulf of Mexico saltmarsh plants. Biogeochemistry 149, 1–16. doi:10.1007/s10533-020-00656-9
Levine, B. M., White, J. R., and DeLaune, R. D. (2017a). Impacts of the long-term presence of buried crude oil on salt marsh soil denitrification in Barataria Bay, Louisiana. Ecol. Eng. 99, 454–461. doi:10.1016/j.ecoleng.2016.11.017
Levine, B. M., White, J. R., DeLaune, R. D., and Maiti, K. (2017b). Crude oil effects on redox status of salt marsh soil in Louisiana. Soil Sci. Soc. Am. J. 81, 647–653. doi:10.2136/sssaj2016.12.0398
Lewis, M., Pryor, R., and Wilking, L. (2011). Fate and effects of anthropogenic chemicals in mangrove ecosystems: A review. Environ. Pollut. 159, 2328–2346. doi:10.1016/j.envpol.2011.04.027
Liaw, A., and Wiener, M. (2002). Classification and Regression by randomForest. R News 2 (3), 18022.
Lin, Q., and Mendelssohn, I. A. (2012). Impacts and recovery of the Deepwater Horizon oil spill on vegetation structure and function of coastal salt marshes in the northern Gulf of Mexico. Environ. Sci. Technol. 46, 3737–3743. doi:10.1021/es203552p
Lindemann, S., Zarnoch, C. B., Castignetti, D., and Hoellein, T. J. (2016). Effect of eastern oysters (Crassostrea virginica) and seasonality on nitrite reductase gene abundance (nirS, nirK, nrfA) in an urban estuary. Estuaries Coasts 39, 218–232. doi:10.1007/s12237-015-9989-4
Liu, G., Ling, F. Q., Magic-Knezev, A., Liu, W. T., Verberk, J. Q. J. C., and Van Dijk, J. C. (2013). Quantification and identification of particle-associated bacteria in unchlorinated drinking water from three treatment plants by cultivation-independent methods. Water Res. 47, 3523–3533. doi:10.1016/j.watres.2013.03.058
Liu, Z., and Liu, J. (2013). Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf of Mexico after the Deepwater Horizon oil spill. MicrobiologyOpen 2, 715–504. doi:10.1002/mbo3.117
MacDonald, I. R., Garcia-Pineda, O., Beet, A., Daneshgar Asl, S., Feng, L., Graettinger, G., et al. (2015). Natural and unnatural oil slicks in the Gulf of Mexico. J. Geophys. Res. Oceans 120, 8364–8380. doi:10.1002/2015jc011062
Machado, L. F., de Assis Leite, D. C., da Costa Rachid, C. T., Paes, J. E., Martins, E. F., Peixoto, R. S., et al. (2019). Tracking mangrove oil bioremediation approaches and bacterial diversity at different depths in an in situ mesocosms system. Front. Microbiol. 10, 2107–2114. doi:10.3389/fmicb.2019.02107
Macy, A., Osland, M. J., Cherry, J. A., and Cebrian, J. (2020). Changes in ecosystem nitrogen and carbon allocation with black mangrove (Avicennia germinans) encroachment into Spartina alterniflora salt marsh. Ecosystems 24, 1007–1023. doi:10.1007/s10021-020-00565-w
Macy, A., Sharma, S., Sparks, E., Goff, J., Heck, K. L., Johnson, M. W., et al. (2019). Tropicalization of the barrier islands of the northern Gulf of Mexico: A comparison of herbivory and decomposition rates between smooth cordgrass (Spartina alterniflora) and black mangrove (Avicennia germinans). PLOS ONE 14, e0210144. doi:10.1371/journal.pone.0210144
Martinez Arbizu, P. (2020). pairwiseAdonis: Pairwise multilevel comparison using adonis. R package version 0.4.
Mckee, K. L., and Rooth, J. E. (2008). Where temperate meets tropical: Multi-factorial effects of elevated CO2, nitrogen enrichment, and competition on a mangrove-salt marsh community. Glob. Change Biol. 14, 971–984. doi:10.1111/j.1365-2486.2008.01547.x
McKee, K. L., and Vervaeke, W. C. (2018). Will fluctuations in salt marsh–mangrove dominance alter vulnerability of a subtropical wetland to sea‐level rise? Glob. Chang. Biol. 24, 1224–1238. doi:10.1111/gcb.13945
Meyer, B. M., Adhikari, P. L., Olson, G. M., Overton, E. ., and Miles, M. S. (2018). “Louisiana coastal marsh environments and MC252 oil biomarker chemistry,” in Oil spill environmental forensics case studies. Editors Z. Wang, and S. A. Stout (Oxford, UK: Butterworth-Heinemann).
Michel, J., Owens, E. H., Zengel, S., Graham, A., Nixon, Z., Allard, T., et al. (2013). Extent and degree of shoreline oiling: Deepwater Horizon oil spill, Gulf of Mexico, USA. PLoS ONE 8, e65087. doi:10.1371/journal.pone.0065087
Michel, J., and Rutherford, N. (2014). Impacts, recovery rates, and treatment options for spilled oil in marshes. Mar. Pollut. Bull. 82, 19–25. doi:10.1016/j.marpolbul.2014.03.030
Mishamandani, S., Gutierrez, T., Berry, D., and Aitken, M. D. (2016). Response of the bacterial community associated with a cosmopolitan marine diatom to crude oil shows a preference for the biodegradation of aromatic hydrocarbons. Environ. Microbiol. 18, 1817–1833. doi:10.1111/1462-2920.12988
Murphy, A. E., Bulseco, A. N., Ackerman, R., Vineis, J. H., and Bowen, J. L. (2020). Sulphide addition favours respiratory ammonification (DNRA) over complete denitrification and alters the active microbial community in salt marsh sediments. Environ. Microbiol. 22, 2124–2139. doi:10.1111/1462-2920.14969
Nielsen, L. P. (1992). Denitrification in sediment determined from nitrogen isotope pairing technique. FEMS Microbiol. Lett. 86, 357–362.
Nixon, Z., Zengel, S., Baker, M., Steinhoff, M., Fricano, G., Rouhani, S., et al. (2016). Shoreline oiling from the Deepwater Horizon oil spill. Mar. Pollut. Bull. 107, 170–178. doi:10.1016/j.marpolbul.2016.04.003
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2015). vegan: Community Ecology Package. R package version 2, 3–1.
Osland, M. J., Enwright, N., Day, R. H., and Doyle, T. W. (2013). Winter climate change and coastal wetland foundation species: Salt marshes vs. mangrove forests in the southeastern United States. Glob. Chang. Biol. 19, 1482–1494. doi:10.1111/gcb.12126
Paerl, H. W., and Whitall, D. R. (2010). Anthropogenically-derived atmospheric nitrogen deposition, marine eutrophication and harmful algal bloom expansion : Is there a link? Ambio 28, 307–311.
Pavlovska, M., Prekrasna, I., Dykyi, E., Zotov, A., Dzhulai, A., Frolova, A., et al. (2021). Niche partitioning of bacterial communities along the stratified water column in the Black Sea. MicrobiologyOpen 10, e1195–24. doi:10.1002/mbo3.1195
Perry, C. L., and Mendelssohn, I. A. (2009). Ecosystem effects of expanding populations of Avicennia germinans in a Louisiana salt marsh. Wetlands 29, 396–406. doi:10.1672/08-100.1
Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D., and Team Core, R. (2015). nlme: Linear and nonlinear mixed effects models. R package version 3.1-122. Available at: http://CRAN.R-project.org/package=nlme
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi:10.1093/nar/gks1219
Rath, K. M., Fierer, N., Murphy, D. V., and Rousk, J. (2018). Linking bacterial community composition to soil salinity along environmental gradients. ISME J. 13, 836–846. doi:10.1038/s41396-018-0313-8
Reddy, K. R., Patrick, W. H., and Lindau, C. W. (1989). Nitrification-denitrification at the plant root-sediment interface in wetlands. Limnol. Oceanogr. 34, 1004–1013. doi:10.4319/lo.1989.34.6.1004
Rivers, A. R., Sharma, S., Tringe, S. G., Martin, J., Joye, S. B., and Moran, M. A. (2013). Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 7, 2315–2329. doi:10.1038/ismej.2013.129
Saintilan, N., Wilson, N. C., Rogers, K., Rajkaran, A., and Krauss, K. W. (2014). Mangrove expansion and salt marsh decline at mangrove poleward limits. Glob. Chang. Biol. 20, 147–157. doi:10.1111/gcb.12341
Scheffel, W. A., Heck, K. L., and Johnson, M. W. (2018). Tropicalization of the northern Gulf of Mexico: Impacts of salt marsh transition to black mangrove dominance on faunal communities. Estuaries Coasts 41, 1193–1205. doi:10.1007/s12237-017-0334-y
Shapiro, K., Khanna, S., and Ustin, S. L. (2016). Vegetation impact and recovery from oil-induced stress on three ecologically distinct wetland sites in the Gulf of Mexico. J. Mar. Sci. Eng. 4, 33–19. doi:10.3390/jmse4020033
Simpson, L. T., Cherry, J. A., Smith, R. S., and Feller, I. C. (2021). Mangrove encroachment alters decomposition rate in saltmarsh through changes in litter quality. Ecosystems 24, 840–854. doi:10.1007/s10021-020-00554-z
Simpson, L. T., Feller, I. C., and Chapman, S. K. (2013). Effects of competition and nutrient enrichemnt on Avicennia germinans in the salt marsh-mangrove ecotone. Aquat. Bot. 104, 55–59. doi:10.1016/j.aquabot.2012.09.006
Simpson, L. T., Stein, C. M., Osborne, T. Z., and Feller, I. C. (2019). Mangroves dramatically increase carbon storage after 3 years of encroachment. Hydrobiologia 834, 13–26. doi:10.1007/s10750-019-3905-z
Smee, D. L., Sanchez, J. A., Diskin, M., and Trettin, C. (2017). Mangrove expansion into salt marshes alters associated faunal communities. Estuar. Coast. Shelf Sci. 187, 306–313. doi:10.1016/j.ecss.2017.02.005
Sorensen, J., Tiedje, J. M., and Firestone, R. B. (1980). Inhibition by sulfide of nitric and nitrous oxide reduction by denitrifying Pseudomonas fluorescens. Appl. Environ. Microbiol. 39, 105–108. doi:10.1128/aem.39.1.105-108.1980
Spivak, A. C., and Reeve, J. (2015). Rapid cycling of recently fixed carbon in a Spartina alterniflora system: a stable isotope tracer experiment. Biogeochemistry 125, 97–114. doi:10.1007/s10533-015-0115-2
Steinmuller, H. E., Foster, T. E., Boudreau, P., Ross Hinkle, C., and Chambers, L. G. (2020). Tipping points in the mangrove march: Characterization of biogeochemical cycling along the mangrove–salt marsh ecotone. Ecosystems 23, 417–434. doi:10.1007/s10021-019-00411-8
Tan, W. A., and Parales, R. E. (2019). Taxonomy, genomics and ecophysiology of hydrocarbon-degrading microbes, 125–141.
Tatariw, C., Mortazavi, B., Ledford, T. C., Starr, S. F., Smyth, E., Griffin Wood, A., et al. (2021). Nitrate reduction capacity is limited by belowground plant recovery in a 32-year-old created salt marsh. Restor. Ecol. 29, 1–9. doi:10.1111/rec.13300
Thamdrup, B., and Dalsgaard, T. (2002). Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68, 1312–1318. doi:10.1128/aem.68.3.1312-1318.2002
Valiela, I., and Cole, M. L. (2002). Comparative evidence that salt marshes and mangroves may protect seagrass meadows from land-derived nitrogen loads. Ecosystems 5, 92–102. doi:10.1007/s10021-001-0058-4
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75, 2046–2056. doi:10.1128/aem.02294-08
Warneke, S., Schipper, L. A., Matiasek, M. G., Scow, K. M., Cameron, S., Bruesewitz, D. A., et al. (2011). Nitrate removal, communities of denitrifiers and adverse effects in different carbon substrates for use in denitrification beds. Water Res. 45, 5463–5475. doi:10.1016/j.watres.2011.08.007
Welschmeyer, N. A. (1994). Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and pheopigments. Limnol. Oceanogr. 39, 1985–1992. doi:10.4319/lo.1994.39.8.1985
Yin, G., Hou, L., Liu, M., Liu, Z., and Gardner, W. S. (2014). A novel membrane inlet mass spectrometer method to measure 15NH4+ for isotope-enrichment experiments in aquatic ecosystems. Environ. Sci. Technol. 48, 9555–9562. doi:10.1021/es501261s
Yoon, J. H., Oh, T. K., and Park, Y. H. (2005). Erythrobacter seohaensis sp. nov. and Erythrobacter gaetbuli sp. nov., isolated from a tidal flat of the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 55, 71–75. doi:10.1099/ijs.0.63233-0
Keywords: woody encroachment, denitrification, oil spill, S. alterniflora, A. germinans, DNRA, 16S rRNA, microbial diversity
Citation: Tatariw C, Mortazavi B, Flournoy N, Kleinhuizen AA, Crawford P, Overton EB and Sobecky PA (2022) Enhanced susceptibility to oiling may limit denitrification recovery in marshes subjected to woody encroachment. Front. Environ. Sci. 10:951365. doi: 10.3389/fenvs.2022.951365
Received: 23 May 2022; Accepted: 25 July 2022;
Published: 16 September 2022.
Edited by:
Charles William Martin, University of Florida, United StatesReviewed by:
Anne Ellen Giblin, Marine Biological Laboratory (MBL), United StatesKevin Yeager, University of Kentucky, United States
Copyright © 2022 Tatariw, Mortazavi, Flournoy, Kleinhuizen, Crawford, Overton and Sobecky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Corianne Tatariw, Y3RhdGFyaXdAdWEuZWR1
 Corianne Tatariw
Corianne Tatariw Behzad Mortazavi
Behzad Mortazavi Nikaela Flournoy
Nikaela Flournoy Alice A. Kleinhuizen1
Alice A. Kleinhuizen1 Patrice Crawford
Patrice Crawford Edward B. Overton
Edward B. Overton Patricia A. Sobecky
Patricia A. Sobecky