
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci., 11 July 2022
Sec. Soil Processes
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.932402
This article is part of the Research TopicSoil-Plant-Microbe Interactions: An Innovative Approach Towards Improving Soil Health and Plant GrowthView all 26 articles
The present investigation was carried out to isolate, identify, and characterize sulfur-oxidizing bacteria (SOB) from coal mines and to evaluate the efficient strains for their ability to influence plant growth and S uptake in pigeonpea. Thirteen bacterial isolates belonging to Stenotrophomonas maltophilia (2), Stenotrophomonas pavanii (2), Rhizobium pusense (5), Bacillus velezensis (2), and Paenibacillus massiliensis (2) were obtained. Among these, seven strains that could reduce the pH of thiosulfate broth were further characterized for sulfur oxidation, plant growth-promoting (PGP) attributes, and in planta studies. Among the seven strains characterized, maximum sulfate ion was recorded for S. maltophilia DRC-18-7A (311.43 mg L−1) closely followed by S. pavanii DRC-18-7B (273.44 mg L−1) and S. maltophilia DRC-18-10 (265.75 mg L−1) after 21 days of inoculation. Among the PGP attributes quantified, maximum P solubilization was recorded in case of S. maltophilia DRC-18-7A (24.39 μg ml−1), while highest siderophore production and IAA production were recorded in S. maltophilia DRC-18-10 (14.25%) and R. pusense DRC-18-25 (15.21 μg ml−1), respectively. S. maltophilia DRC-18-7A closely followed by S. pavanii DRC-18-7B outperformed others in enhancing seed germination (%) and vigour indices. Results clearly indicated that microbial inoculants colonized the plant roots and developed biofilm on the root surface. It was further observed that plants treated with microbial inoculants induce an early formation of secondary and tertiary roots in the pigeonpea compared to the untreated control which was further confirmed by assessing the root architecture using the root scanner. Inoculation of these two strains to pigeonpea significantly enhanced plant growth parameters, the activity of reactive oxygen scavenging (ROS) enzymes, and accumulation of flavonoids, carotenoids, and proline both under sterilized and non-sterilized growth medium (sand and soil in 1:3 ratio). The application of microbial inoculants significantly increased the uptake of nitrogen, phosphorus, potassium, and sulfur in plant shoots. Further, transcript level of phosphate, potassium, and sulfur transporter genes significantly increases upon microbial inoculation leading to increased uptake and translocation of P, K, and S in the pigeonpea. The results indicate that S. maltophilia DRC-18-7A and S. pavanii DRC-18-7B could be recommended as inoculants for pigeonpea to improve its growth and sulfur nutrition.
Sulfur (S) is an important plant nutrient that contributes to the growth and yield of different crops (Vidhyalaxmi and Sridar, 2007). Sulfur is involved in different metabolic and enzymatic activities in plants. The role of S in protein synthesis is important as they are the constituent of amino acids like methionine, cystine, and cysteine (Vidyalakshmi et al., 2009). The deficiency of sulfur in soils leads to stunting of plant growth and the typical symptom is the yellowing of young leaves (Natesan et al., 1985; Tandon, 1991a,b). In India, on an average, about 11.4, 29.4, and 17.8% of soils were acute deficient, deficient, and latent deficient with the mean concentrations of 27.0 ± 29.9 mg kg−1 for available S (Shukla et al., 2021). Large-scale deficiency of available sulfur in soils necessitates the management of this nutrient through chemical or biological means to support optimum growth and yield of plants. In comparison to cereal crops, pulse crops have a high requirement for sulfur. Application of 20–30 kg S ha−1 as gypsum or sometimes as ammonium sulfate gives a mean yield increase of 168–428 kg ha−1 for pulse crops (Rakesh et al., 2020). Inoculation of SOB along with elemental sulfur and gypsum has also been reported to enhance the growth, nodulation, and uptake of S in cowpea in sodic soils (Stamford et al., 2013). Inoculation of Proteus mirabilis, an SOB isolated from buffalo dung enhanced the growth and yield of Foeniculum vulgare (Dhiman et al., 2019).
Sulfur-containing minerals are the major source of sulfur in soils. Among them, gypsum, epsomite, anhydrite, iron pyrite, arsenopyrite, sphalerite, and galena are some of the important S-containing minerals in rocks and soils (Kumar et al., 2018; 2022). In the natural ecosystem, sulfur occurs in organic fractions, particularly in combination with carbon and nitrogen (Tabatabai, 1984). In general, plants take up sulfur in the form of sulfate ions. Microorganisms play a key role in the transformation of sulfate ions from organic and inorganic sulfur-containing compounds. They oxidize the elemental and other forms of sulfur to produce sulfate ions in soils (Sridar et al., 2013). In general, the majority of SOB utilize CO2 as their primary carbon source and sulfur as an electron donor (Brune, 1989; Friedrich et al., 2005). Among the diverse groups of soil bacteria, the sulfur-oxidizing bacteria (SOB) are metabolically and nutritionally diverse which include autotrophs, photoautotrophs, chemolithotrophs (obligate and facultative), heterotrophs, and mixotrophs. Photoautotrophs include green sulfur bacteria (such as Chlorobium) and purple sulfur bacteria (such as Allochromatium, Chromatium, and Thiocystis). Among obligate chemolithotrophs, species of Thiobacillus (T. thiooxidans, T. ferrooxidans, T. thioparus, T. denitirificans) and Thiomicrospira are well characterized and reported by several researchers (Vidyalakshmi et al., 2009; Kawai et al., 2019). The sulfur oxidation in soil is not restricted to obligate chemolithotrophs; it is observed that several species of heterotrophs and mixotrophs have the capability to oxidize sulfur in agricultural soils. They exhibit chemolithotrophic growth in presence of inorganic sulfur. The heterotrophs include species of Achromobacter, Arthrobacter, Brevibacterium, Flavobacterium, Klebsiella, Micrococcus, Mycobacterium, Paracoccus, Streptomyces, Thiosphaera, and Xanthobacter (Wainwight, 1984; Kuenen et al., 1992, Ito et al., 2004; Ryan et al., 2009; Sajjad et al., 2016; Chaudhary et al., 2017, 2021). However, mixotrophs including species of Stenotrophomonas, Bacillus, Pseudomonas, Paenibacillus, Micrococcus, Pseudoclavibacter, Diaphorobacter, Aeromonas, Alcaligenes, Citrobacter, Rhizobium, and Bordetella are the key bacterial species playing important roles in nutrient mineralization and promoting plant growth (Sultan and Faisal, 2016; Sanwani et al., 2022). Sulfur-oxidizing bacteria are isolated from different habitats including not only rhizosphere soils but also soil and water as sampled from coal mines; copper ore leaching; and sewage water, biogas slurry, tannery effluent, hydrothermal vents, and crude oil mine soils (Kodama and Watanabe, 2003; Anandham et al., 2005; Vidyalakshmi and Sridar, 2007; Marvi et al., 2016; Sultan and Faisal, 2016; Chaudhary et al., 2017; Meier et al., 2017). Apart from the S oxidation, some of the SOB isolated from different niches have been characterized for other plant growth-promoting attributes like production of IAA, siderophores, ACC deaminase, and antibiotics; solubilization of phosphorus, potassium, and zinc; and inhibition of fungal pathogens (Grayston and Germida, 1991; Veerender et al., 2014; Sajjad et al., 2016; Reddy et al., 2018; Dhiman et al., 2019; Yousef et al., 2019; Kumar et al., 2022). The strains with multiple plant-promoting attributes have been reported to enhance the growth and yield in many crop plants (Grayston and Germida, 1991; Dhiman et al., 2019). Application of rhizospheric and endophytic bacteria significantly enhances plant growth directly and/or indirectly in many crops. The direct mechanism includes production of phytohormones, elicitors, mineralization/solubilization of mineral nutrients, N fixation, etc., while indirect mechanisms include modulation of physio-biochemical pathways, gene expression, scavenging the reactive oxygen species, etc., (Singh et al., 2016a, Singh et al., 2016b; Singh et al., 2019, Singh et al., 2021a,b; Yadav et al., 2022).
These plant growth-promoting rhizobacteria employ their key mechanisms by colonizing the plant rhizosphere to combat environmental stresses, improve nutrient uptake, and translocation preventing subsequent yield penalties. Strategies adopted by these microbial inoculants include de novo synthesis of osmolytes for cellular osmotic adjustment, activation of reactive oxygen species scavenging defence systems of plants to cope with deleterious effects of oxidative stress, regulation of ionic transporters, and maintenance of homeostasis to reduce ill effects of mineral deficiency. On the other hand, they modulate the transport activity within and among plant organs. These transporters regulate nutrient uptake into root cells and subsequent translocation within the plant system. Scientific evidence specify that plants have evolved diverse transporters with distinct substrate specificities, transport affinities, cell type expression, and subcellular localization to ensure appropriate flux and compartmentalization of nutrients in the plant. These transporters are playing a key role in the initial uptake, vascular translocation of mineral nutrients, and further release of vacuolar sulfate to sustain proficient utilization of S-pools in the plant system (Yoshimoto et al., 2002; 2007). Pigeonpea is the most important rainy season pulse crop in India and faces the problem of S deficiency in different parts of the country. The package and practices for pigeonpea cultivation in different parts of India include inoculation with specific rhizobia to meet the nitrogen requirement. It will also be worthwhile to select sulfur-oxidizing bacteria that can perform well in the pigeonpea rhizosphere and fulfil the S requirement of the crop. The selection of efficient strains of SOB could further lead to the development of a consortium of rhizobia and SOB so that both N and S requirements of the crop can be met with no or minimal chemical inputs. Looking at the importance of sulfur in pulse production, the present study aimed to isolate, identify, and characterize SOB from coal mines and to evaluate the efficient strains for their ability to influence plant growth and S uptake in pigeonpea. The fossil fuel, coal mine drainage, and coal are the rich source of sulfur and sulfur-containing inorganic and organic compounds (Harrison, 1978; Williams and Cloete 2008). It was hypothesized that the microbial community harbouring in the niches for long has more potential to oxidize the sulfur as compared to the community present in the normal soil and plant rhizosphere (Harrison, 1978; Kumar et al., 2018). This was the main reason behind selecting the coal mine for isolation of SOB.
The sediment, water, coal, and coal mixed soil samples (10 samples) were collected from three different open-cast coal mines (West Mudidih Colliery, Singhpur Opencast mine, and Sirka Opencast mine) from the state of Jharkhand, India (Figure 1). All these mines are the oldest coal mines in India surrounded by natural vegetation. A detailed description of the sites along with the key biogeochemical characteristics of the samples including pH, EC, and temperature are given in Table 1. Sampling was done in October 2018. Coal drainage water and coal mud from water bodies were collected in sterile polypropylene screw-capped bottles. However, coal and coal mixed soil samples were collected in zipper bags (polythene) and brought to the laboratory.
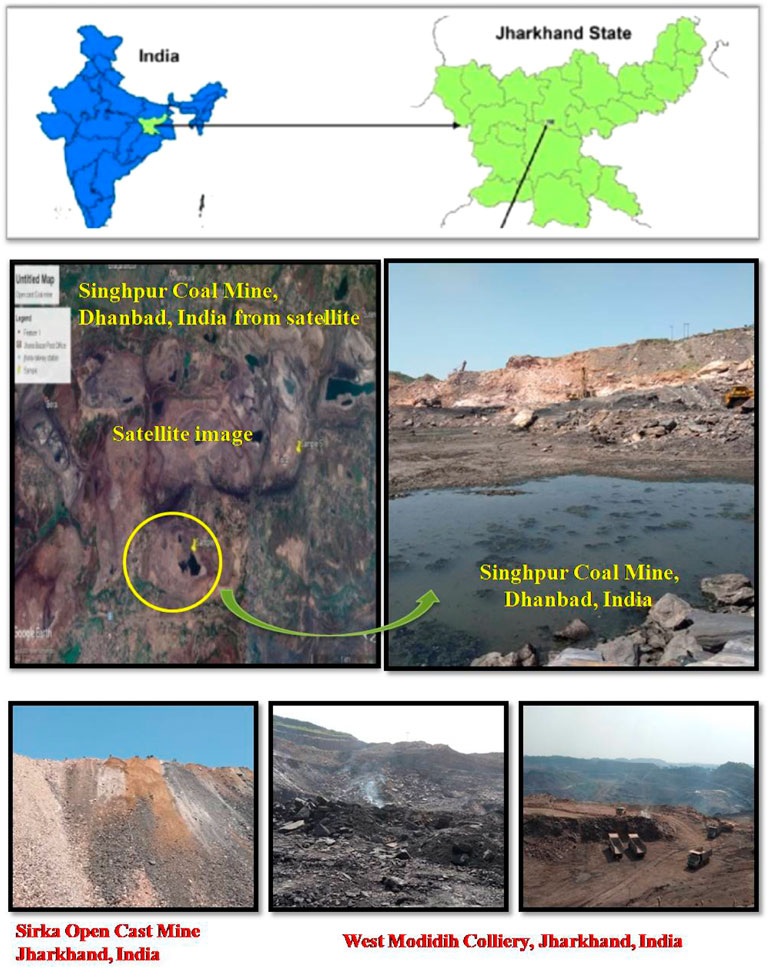
FIGURE 1. Sites for collection of soil, coal, and sediment samples for isolation of sulfur-oxidizing bacteria.
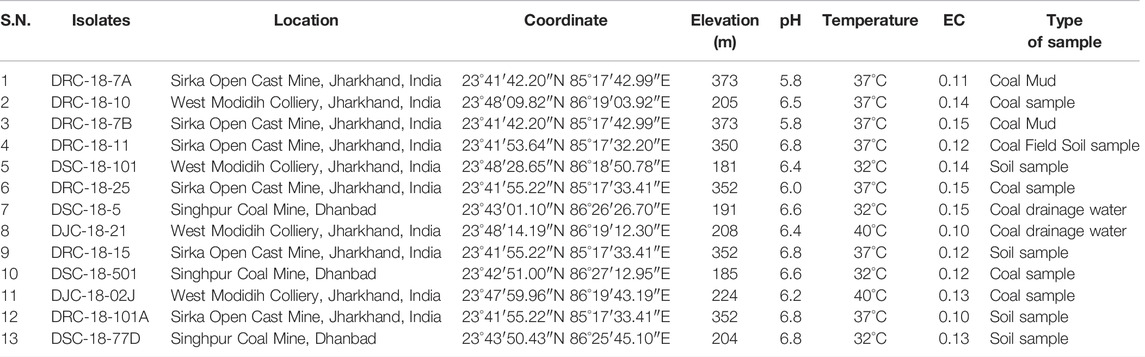
TABLE 1. Geographical locations of different coal mines in Jharkhand province of India for collection of samples for isolation of sulfur-oxidizing bacteria.
Coal and coal mixed soil samples were crushed and homogenized using pestle and mortar. Coal and coal mixed soil samples (1 g) were taken and inoculated into screw-capped glass tubes (Borosil, India) containing sterile thiosulfate, Starkey, and NCL broth separately and incubated at 28°C for 21 days. The composition of each medium is given in Supplementary Table S1. The pH of the media was recorded at 3 days intervals. After 12 days of inoculation, the colour of the thiosulfate broth started changing from purple to pale yellow which indicated a reduction in the pH. After 21 days of inoculation, the pH of thiosulfate broth was reduced from 8 to 2. However, no change in the pH was recorded in the Starkey and NCL broth amended with soil samples even after 21 days of inoculation (Starkey, 1934). Similar procedures were followed for water and mud samples collected from different sites. After 21 days of inoculation, thiosulfate broth that showed a reduction in pH from 8 to 2 was mixed, and 100 µl was spread plated on thiosulfate medium. The plates were incubated at 28°C for 10–15 days. After 7 days of inoculation, tiny pinhead growth was observed and each colony was purified by re-streaking on the thiosulfate medium and the pure cultures were maintained at 4°C until further use.
The pH reduction test was carried out by inoculating pure cultures into thiosulfate broth, incubating for 21 days, and recording the pH at 3 days intervals. The bacterial isolates that showed a significant reduction in pH were further evaluated for the production of sulfate ions in the thiosulfate broth. The amount of sulfate ions (SO42-) produced was determined spectrophotometrically. Each bacterial isolate was inoculated into 10 ml of thiosulfate broth and the inoculated tubes were incubated at 28°C for 7 days. Following incubation, the broths were centrifuged at 10,000 rpm for 10 min, the supernatant was collected and mixed vigorously with an equal volume of barium chloride solution (10%, w/v) (Cha et al., 1999) with slight modifications (Roy and Roy, 2019). The white turbidity developed due to the formation of barium sulfate was measured at 450 nm. The amount of sulfate ions produced was derived by following comparison with the standard curve. The growth curve for each culture was developed using Growth Kinetic analyser (Bioscreen Pvt. Ltd., India). The intrinsic antibiotic resistance profile was developed using an antibiotic sensitivity disc (HiMedia, Mumbai, India) according to Khan et al. (2019). These strains were further characterized for plant growth-promoting traits such as IAA production (Brick et al., 1991), phosphate solubilization (Nautiyal, 1999), siderophore production (Schwyn and Neilands, 1987), and ammonia production (Dey et al., 2004). The H2O2 production, catalase, amylase, and starch hydrolysis tests were performed according to Whitman et al. (2012). Estimation of the activity of chitinase and protease was carried out following the protocols given by Boller and Mauch (1988).
Quantitative estimation of phosphate solubilization was done in NBRIP liquid medium according to Fernández et al. (2007). The available P fraction was estimated spectrophotometrically by the molybdenum blue method using a spectrophotometer at 880 nm and P concentrations were expressed as μg ml−1. The quantitative estimation of IAA was done spectrophotometrically at 535 nm (Barra et al., 2016). IAA concentration was expressed as μg ml−1. For quantitative estimation of siderophore, bacterial isolates were sub-cultured in thiosulfate broth for 5 days at 200 r min−1 and 28°C. An aliquot of 1.5 ml of each culture was centrifuged at 10,000 r min−1 for 10 min. The relative level of siderophore was estimated in a fixed volume of supernatant (10 µl) using the CAS assay method (Schywan and Neilands, 1987) with slight modifications (Abo-Zaid et al., 2020). Briefly, 0.5 ml CAS assay solution was added to 10 µl of culture supernatant and mixed properly. Thereafter, 10 µl of shuttle solution was added, mixed, and kept at ambient room temperature for a few minutes. The disappearance of the blue colour relates to the presence of siderophores. The absorbance was measured at 630 nm using the media as blank. The relative level of siderophores (%) was calculated by using the formula given below:
Ar refers to the absorption of CAS solution plus media plus shuttle solution, whereas As refers to the absorption of CAS solution plus culture supernatant plus shuttle solution.
DNA was extracted from pure cultures using Nucleo-pore® gDNA Fungal Bacterial Mini kit (Cat. No. NP-7006D, Genetics Biotech Asia Pvt. Ltd., India) following the manufacturer’s protocol. The 16S rRNA gene was amplified using universal primers pair 27F: AGAGTTTGATCCTGGCTCAG and 1492R: AAGGAGGTGATCCAGCCGCA (Edward et al., 1989), and sequenced by using amplicon sequencing technology (Eurofins Pvt. Ltd., India) and the sequence similarity was matched using EzBiocloud database for correct identification. 16S rRNA gene sequences were submitted to NCBI GenBank and accession numbers were obtained. Phylogenetic analysis was carried out using the Molecular Evolutionary Genetics Analysis (MEGA X) tools.
In planta assay was carried out in earthen pots using sterile or non-sterile sand:soil mixture (1:3 ratio). The experiment was laid out in a Completely Randomized Design (CRD) under glasshouse conditions. The experiment was conducted with eight different treatments: T1—Stenotrophomonas maltophilia DRC-18-7A, T2—S. Maltophilia DRC-18-10, T3—S. Pavanii DRC-18-7B, T4—S. pavanii DRC-18-11, T5—Rhizobium pusense DRC-18-25, T6—R. pusense DSC-18-5, T7—R. pusense DJC-18-21, and T8—untreated control (no inoculation). Each treatment was replicated 10 times. The pots containing sterile growth medium were irrigated with sterile water, while pots with non-sterile soil were irrigated with non-sterile water on alternate days to maintain the moisture content at field capacity level.
Experimental soil was collected from the Research farm, ICAR-Indian Institute of Seed Sciences, Kushmaur, Uttar Pradesh (India), sieved (2 mm), and dried. The soil was analysed for its physico-biochemical properties (Supplementary Table S2). The soil was mixed with sand in a ratio of 3:1, amended with nitrogen, phosphorous, potash, and sulfur at 120, 60, 40, and 20 kg ha−1, respectively, and autoclaved twice at 121°C (15 psi) for 30 min at 12 h interval.
Pigeonpea seeds (cv. Malviya Chamatkar or MAL-13) were obtained from the Department of Genetics and Plant Breeding, Banaras Hindu University, Varanasi (India). Five seeds were sown in each pot containing sterile or non-sterile sand:soil mixture (1:3 ratio). One ml of broth culture (2 × 108 colony-forming unit ml−1) of selected strains was inoculated over seeds in each pot. The uninoculated pots were maintained as control. The average mean temperature and relative humidity during the experimentation were 27°C and 85%, respectively.
An experiment was conducted to see the effects of seed inoculation on seed germination in the pigeonpea grown in pots at 15 days of sowing following the International Seed Testing Association (ISTA, 2003) protocols (Singh et al., 2016b). Seeds were surface sterilized with mercuric chloride (0.1%) for 3 min followed by ethanol (70%) for 30 s. The surface-sterilized seeds were washed thoroughly with sterile distilled water (consequently three times). Surface-sterilized seeds were treated with respective bacterial cultures and sown in the pots containing sterilized sand–soil mixture (1:3 ratio) under glasshouse conditions. For the seed germination test, 400 seeds were taken and sown in pots (20 seeds pot−1). The germinated seeds were recorded after 15 days of sowing and seed germination (%) was calculated using the formula given below:
Five replicates of each treatment were harvested after 30 days to record the effects of seed inoculation on vigour indices (vigour index I and II). Observations were recorded on root and shoot length and root and shoot dry weight. Vigour indices were calculated according to the International Seed Testing Association protocols (ISTA, 2003) using the formulae given below:
Vigour Index I = (mean root length + mean shoot length) × germination (%).
Vigour Index II = (mean dry wt. of root + mean dry wt. of shoot) × germination (%).
The root colonization potential was studied using Scanning Electron Microscope (Hitachi S-3400N, United States) following the methods described by Singh S. et al. (2021). Briefly, the treated seeds were sown in small pots containing a sterile sand-soil mixture (1:3 ratio, 1 kg) under glasshouse conditions. Plants were uprooted gently after 15 days of sowing and washed in running tap water. Thereafter, root samples were fixed in a mixture of formaldehyde (37%) and glutaraldehyde (2.5%) (1:1 ratio) for 24 h at 4°C. The second fixation was done using osmium tetraoxide solution (HiMedia, Mumbai, India) for 12 h at ambient room temperature. The pre-fixed root samples were dehydrated using a gradient of ethyl alcohol (30, 50, 70, 90, and 100%) for 30 min each and dried under vacuum. After proper drying, the samples were coated with gold (20 nm) and visualized under Scanning Electron Microscope.
To see the effect of microbial inoculation on root architecture and root development, plants were uprooted carefully and brought to the laboratory after 30 days of sowing. Root samples were washed in gentle running tap water. Thereafter, root scanning was done and clean roots were scanned using a root scanner (Regent Instrument, Canada). The scanned images were analysed using image analysis software “WinRhizo Pro 2017” (Client# IN1803202) to study the different parameters of root architecture.
After 30 and 60 days of sowing, the plants from five replications of each treatment were uprooted. Plant growth parameters like root and shoot length and fresh and dry root and shoot biomass were recorded.
The total chlorophyll, carotene, total soluble sugar, total protein, proline, and phenolic content in the plant leaves were estimated spectrophotometrically by following the methods/protocols described by Sadasivam and Manickem (1996) at 30 DAS. Total flavonoid content and activity of antioxidative enzymes such as phenylalanine ammonia lyase (PAL), peroxidase (Pox), ascorbate peroxidase (Apx), catalase (CAT), and superoxide dismutase (SOD) were estimated quantitatively as per methods described by Thimmaiah (2012) at 30 DAS.
To see the effects of microbial inoculation on up- and down-regulation of P, K, and S transporters in pigeonpea grown under sterilized sand–soil mixture, gene expression analyses were performed by using quantitative real-time PCR (qRT-PCR) at 30 and 60 DAS. Plant roots were sampled from each treatment, washed in running tap water, and brought to the laboratory. Thereafter, samples were quick frozen in liquid nitrogen. Total RNA was isolated using the Total RNA Isolation kit (Bangalore GeNi, Bengaluru, India) according to the manufacturer’s protocols/instructions (Singh S. et al., 2021). The qualitative and quantitative assessments of these RNA samples were conducted using Bio-analyzer (Agilent 2100, Agilent Technologies, Mumbai, India). RNA samples with RNA integrity (RIN) value ≥ 8 were taken for the synthesis of cDNA using iScript cDNA Synthesis kit (BioRAD, India) following the manufacturer’s protocols/instructions. The quality and concentration of cDNA were determined using Nanodrop 2000c (Thermo Scientific, United States). There were three biological replicates for each treatment. The qRT-PCR was performed using the SYBR Green master mix (Thermo Scientific, United States) on the BioRAD Real-Time PCR System (MJ MiniOpticon, BioRAD, India). The housekeeping gene actin was used as an endogenous standard to normalize the quantitative expression data. The expression of key transporters (P, K, and S Transporters) genes was analysed using gene-specific primers. The primers used in the expression study are listed in Supplementary Table S3. Their specificity was confirmed by analysis of the melting curves. The 2–△△CT method (Livak and Schmittgen, 2001) was used to quantify the value of every sample using Actin as an internal reference.
To see the effects of microbial inoculation on N, P, K, and S content, plant samples were harvested at 60 DAS and brought to the laboratory. The nitrogen content in the plant leaves was estimated by using Kjeldahl method following the procedure described by Bilbao et al. (1999). For estimation of phosphorous content in the plant leaves, samples were digested in the tri-acid mixture (HClO4 + H2SO4 + HNO3; 3:1:10). The 5 ml extract was taken in a 50-ml volumetric flask, added 5 ml 5N HNO3 and 5 ml ammonium metavanadate solution (0.25%), and mixed thoroughly. Thereafter, 5 ml ammonium molybdate (5%) was added and absorbance was recorded at 470 nm wavelength using UV–Vis spectrophotometer (Jackson, 1973). However, potassium content was estimated quantitatively by using a flame photometer (Jackson, 1973). The total sulfur in plant samples was estimated by using the barium sulfate turbidimetry method (Lachicaa Garrido, 1964). Sulfur present in the plant tissue is converted into sulfate ions and precipitated as barium sulfate after treatment with barium chloride. The observance was recorded at 420 nm using UV–Vis spectrophotometer and S content (%) in the plant tissue was calculated.
Glasshouse experiments were laid out in Completely Randomized Design (CRD) with 10 replications. Data were analysed using Statistical Package for Social Science (SPSS), Version 18.0. Graphs were made using statistical software, OriginPro 9.0.
During the course of isolation, 13 bacterial isolates were obtained from different samples collected from Open Cast Projects of Jharkhand (India). These isolates were nominated as DRC-18-7A, DRC-18-10, DRC-18-7B, DRC-18-11, DSC-18-101, DRC-18-25, DSC-18-5, DJC-18-21, DRC-18-15, DSC-18-501, DJC-18-02J, DRC-18-101A, and DSC-18-77D (Table 1). Molecular identification of 13 isolates was carried out based on 16S rRNA gene sequences and identification was done based on per cent similarity (EzBiocloud, a public database of type strains) by BLAST homology. These sequences were submitted to NCBI GenBank and accession numbers were obtained. Based on BLAST homology results, DRC-18-7A and DRC-18-10 were identified as Stenotrophomonas maltophilia, while DRC-18-7B and DRC-18-11 showed maximum BLAST homology with Stenotrophomonas pavanii. Further, isolates DSC-18-101, DRC-18-25, DSC-18-5, DJC-18-21, and DRC-18-15 were identified as Rhizobium pusense. Two isolates, DSC-18-501 and DJC-18-02J were identified as Bacillus velezensis, whereas DRC-18-101A and DSC-18-77D were identified as Paenibacillus massiliensis (Figure 2, Supplementary Table S4).
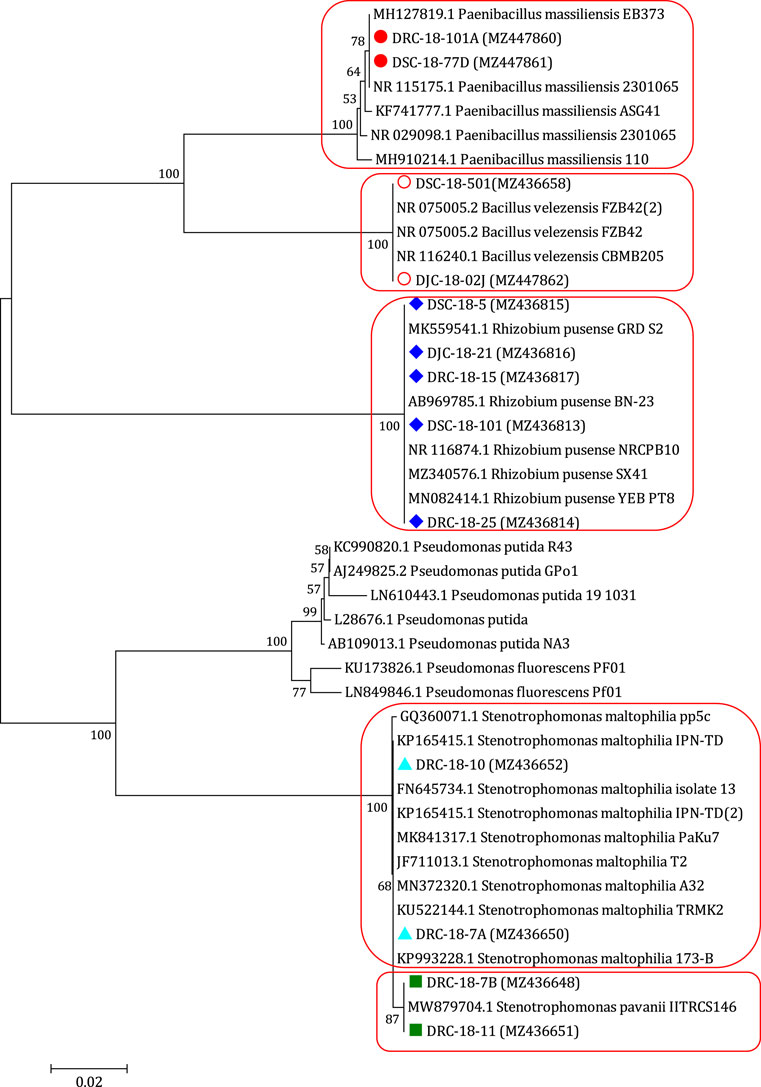
FIGURE 2. Neighbour joining tree derived by CLUSTAL W and MEGA X using analysis of 16S rRNA gene sequences of bacterial strains isolated from coal mines. The numbers at nodes indicate bootstrap support values, as calculated by MEGA X.
All these strains were further characterized for pH reduction using thiosulfate broth. Among the 13 bacterial strains tested, 6 strains (Rhizobium pusense DRC-18-15 and DSC-18-101; Bacillus velezensis DSC-18-501 and DJC-18-02J; and Paenibacillus massiliensis DRC-18-101A and DSC-18-77D) could not reduce the pH of thiosulfate broth after 21 days of incubation, and hence were not included for further studies (Figure 3A). Among the seven strains characterized, maximum sulfate ion was recorded for S. maltophilia DRC-18-7A (311.43 mg L−1) closely followed by S. pavanii DRC-18-7B (273.44 mg L−1) and S. maltophilia DRC-18-10 (265.75 mg L−1) after 21 days of inoculation (Figure 3B). Growth kinetic studies revealed that maximum growth was achieved by S. pavanii DRC-18-7B (3.13 × 106) followed by S. maltophilia DRC-18-7A (2.92 × 106) and S. pavanii DRC-18-11 (2.22 × 106) after 21 days of inoculation (Figure 3C).
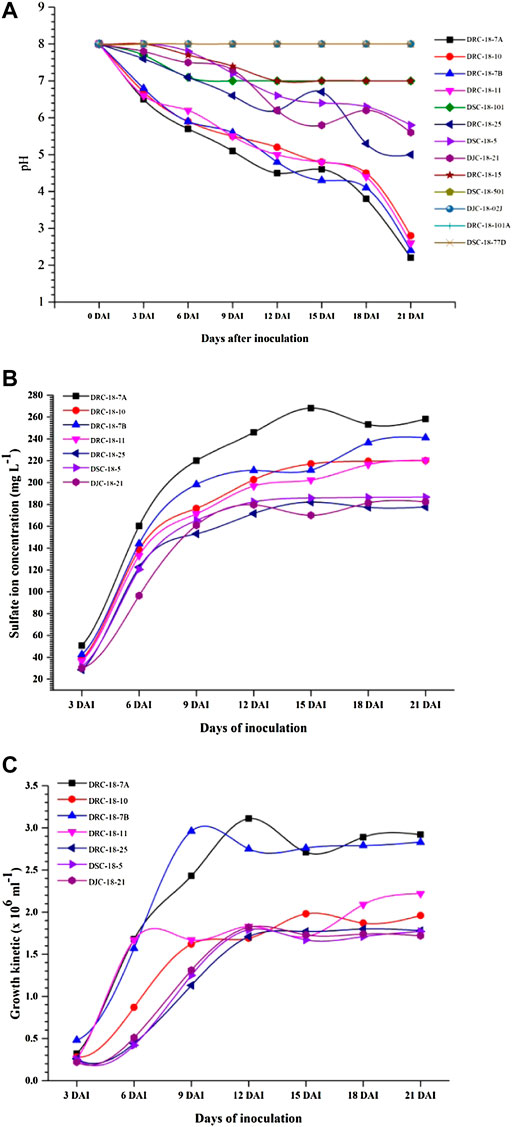
FIGURE 3. Characterization of sulfur-oxidizing bacterial isolates for (A) pH reduction, (B) sulfate ion production, and (C) growth kinetics under controlled laboratory conditions.
All the seven strains selected were further screened for antibiotic sensitivity and PGP traits. Results indicated that these strains showed different reactions to antibiotics tested under controlled laboratory conditions (Supplementary Table S5). Similarly, most of the strains tested were found positive for different PGP traits analysed. Specifically, strain S. maltophilia DRC-18-7A showed a positive reaction for all the PGP attributes tested except for H2O2 and chitinase activity. Strains S. maltophilia DRC-18-10 and S. pavanii DRC-18-7B showed negative responses for H2O2 production, catalase, and chitinase (Supplementary Table S6). The quantitative estimation indicated that maximum P solubilization was recorded for S. maltophilia DRC-18-7A (24.39 μg ml−1) after 10 days of inoculation (Supplementary Table S7). However, maximum siderophore production was achieved in broth inoculated with S. maltophilia DRC-18-10, whereas maximum IAA production was achieved for R. pusense DRC-18-25 (15.25 μg ml−1) (Supplementary Table S7).
Glasshouse experiments were conducted to see the effect of inoculation on seed germination (%) and vigour indices in sterilized and non-sterilized soils. Results revealed a significant increase in the seed germination (90.20%) (being maximum) upon inoculation of S. maltophilia DRC-18-7A, followed by S. pavanii DRC-18-7B (87.10%) and S. maltophilia DRC-18-10 (85.25%), respectively, in the sterilized soil. The least germination was recorded in the untreated control (75.90%). A more or less similar pattern was recorded in the case with non-sterilized soil (Table 2).
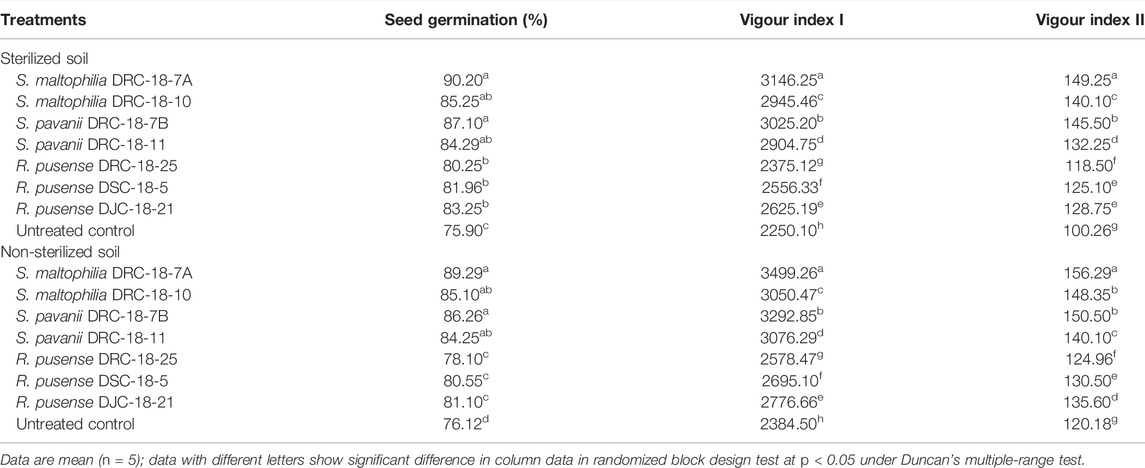
TABLE 2. Effect of seed inoculation of SOB on germination (%) and vigour indices of pigeonpea grown in pots after 15 and 30 days of sowing, respectively.
In the case of vigour indices, significantly higher vigour index I and vigour index II were recorded in the plants inoculated with S. maltophilia DRC-18-7A (3146.25 and 149.25, respectively), followed by S. pavanii DRC-18-7B (3025.20 and 145.50, respectively). However, the least value of vigour indices was recorded in untreated control plants (vigour index I: 3146.25 and vigour index II: 149.25) in sterilized soil after 30 days of sowing (Table 2). A similar trend was recorded in the non-sterilized soil. However, the values were slightly higher than those in sterilized soil (Table 2).
Scanning electron microscopic photographs clearly indicated that selected strains exhibited the differential potential to colonize the pigeonpea root at 15 days of sowing (Figure 4). S. Maltophilia DRC-18-7A colonized pigeonpea roots at a very high population density which is clearly visible in scanning electron microphotographs. The cells were anchored to the root surfaces and themselves by a network of fibrillar material, the exo-polysaccharide produced by them on the root surface (Figure 4). S. maltophilia DRC-18-7A produced primarily microaggregates and later on converted into macroaggregates on the root surface after 15 days of inoculation (Figure 4). In contrast, the other strains yielded mainly single cells embedded in the root epidermis and rarely formed microaggregates (Figure 4).
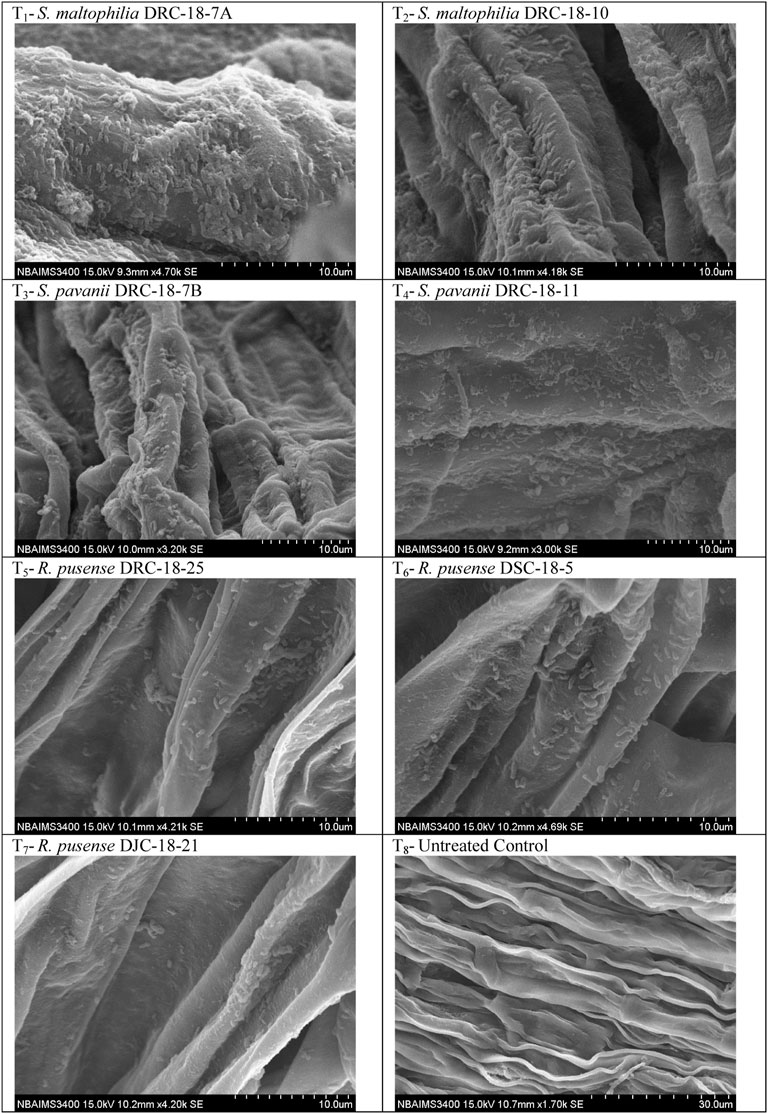
FIGURE 4. Scanning electron microphotographs showing root colonization of selected strains of SOB at 15 days of sowing.
Seed inoculation with sulfur-oxidizing bacterial strains significantly impacts root architecture and the development of secondary and tertiary roots. A significant increase was seen in the root surface area (67.50 cm2), average diameter (0.66 mm), total root volume (3.45), root length (196.25 cm), and the number of tops (346.25), forks (745.25), crossings (805.15), and links (1215.66) in the plant inoculated with S. Maltophilia DRC-18-7A followed by those inoculated with S. pavanii DRC-18-7B [root surface area (52.47 cm2), average diameter (0.54 mm), total root volume (2.96), root length (179.70 cm), and the number of tops (310.66) and forks (675.24)] as compared to other treatments and untreated control plants after 30 days of sowing (Table 3; Figure 5).
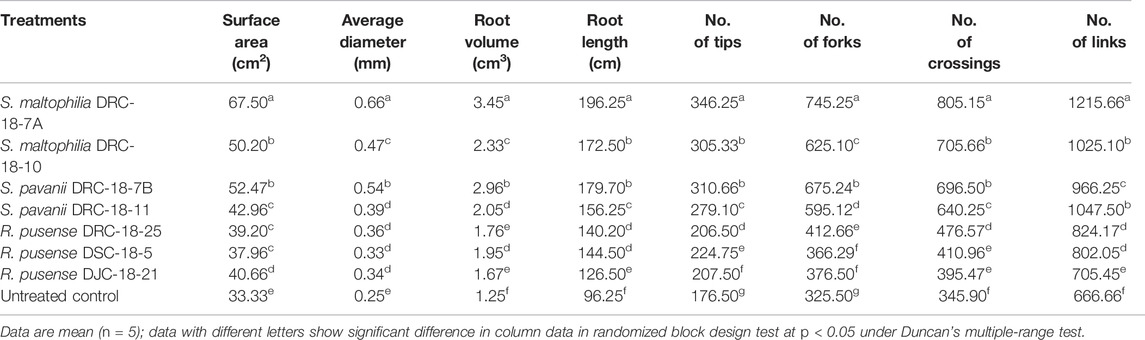
TABLE 3. Effect of microbial inoculation on root development and attributes in pigeonpea grown in pots at 30 days of sowing.
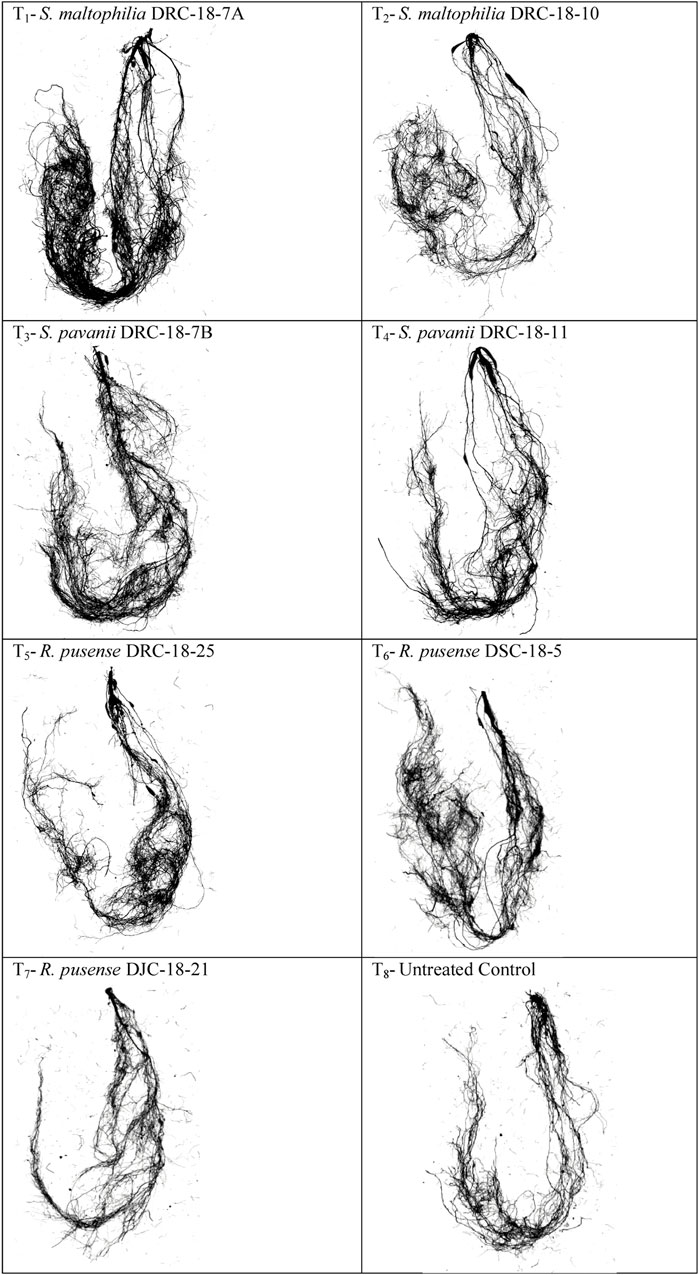
FIGURE 5. Effects of seed inoculation of SOB on root architecture in the pigeonpea leaves at 30 days of sowing.
Seed inoculation with sulfur-oxidizing bacterial strains tends to overproduce total chlorophyll, antioxidative enzymes, and biomolecules in the pigeonpea grown in sterilized soil. Results of glasshouse experiments clearly showed that maximum synthesis and accumulation of total chlorophyll (10.25 mg g−1 fresh wt.), total carotenoids (0.36 mg g−1 fresh wt.), total soluble sugar (19.67 mg g−1 dry wt.), and total protein content (16.26 mg g−1 dry wt.) were recorded in the leaves of plant inoculated with S. maltophilia DRC-18-7A followed by S. pavanii DRC-18-7B. These values were significantly higher than untreated control plants at 30 days of sowing (Figure 6). Similarly, the maximum accumulation of total phenolics (18.26 μmol g−1 fresh wt.), flavonoids (1.10 μmol g−1 fresh wt.), and proline (3.79 mg g−1 dry wt.) was recorded in the plants treated with S. maltophilia DRC-18-7A followed by S. pavanii DRC-18-7B (Figure 6).
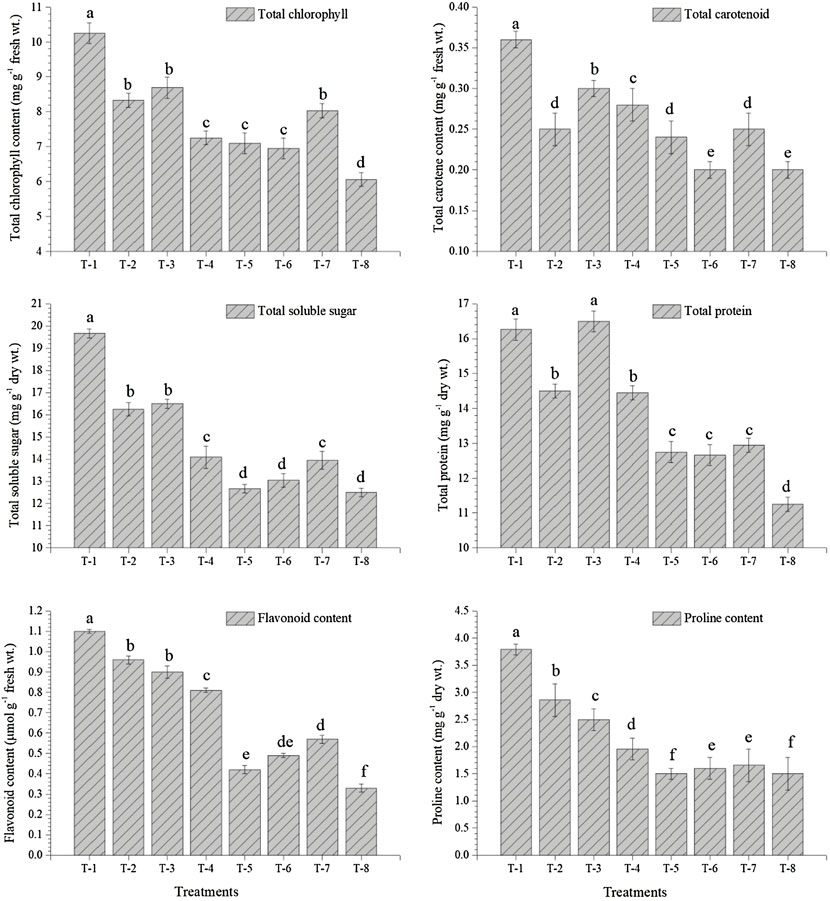
FIGURE 6. Effects of seed inoculation of SOB on accumulation of total chlorophyll, total carotenoids, total soluble sugar, total protein, flavonoids, and proline in the pigeonpea leaves at 30 days of sowing. Treatments were as follows: T1—Stenotrophomonas maltophilia DRC-18-7A, T2—S. maltophilia DRC-18-10, T3—S. pavanii DRC-18-7B, T4—S. pavanii DRC-18-11, T5—R. pusense DRC-18-25, T6—R. pusense DSC-18-5, T7—R. pusense DJC-18-21, and T8—Untreated control (No inoculation). Data are mean (n = 5) and vertical bar represents the standard deviation. Data with different letters show significant difference in column data in randomized block design test at p < 0.05 under Duncan’s multiple-range test.
The data presented in the current study reveal significant differences in the activity of PAL, POx, APx, CAT, and SOD in the leaves of pigeonpea plants inoculated with either of the strains as compared to control plants. Significantly higher activities of PAL (16.25 mM trans-cinnamic acid h−1 g−1 fresh wt.), POx (8.50 unit g−1 fresh wt.), APx (12.75 unit g−1 fresh wt.), CAT (19.25 unit g−1 fresh wt.), and SOD (15.39 unit g−1 fresh wt.) were recorded in the leaves of pigeonpea inoculated with S. maltophilia DRC-18-7A followed by the plants inoculated with S. pavanii DRC-18-7B at 30 DAS under glasshouse conditions (Figure 7). However, the least value for the activities of PAL (4.50 mM trans-cinnamic acid h−1 g−1 fresh wt.), POx (2.50 unit g−1 fresh wt.), APx (5.29 unit g−1 fresh wt.), CAT (8.50 unit g−1 fresh wt.), and SOD (6.50 unit g−1 fresh wt.) was recorded in the leaves of untreated control plants at 30 DAS (Figure 7).
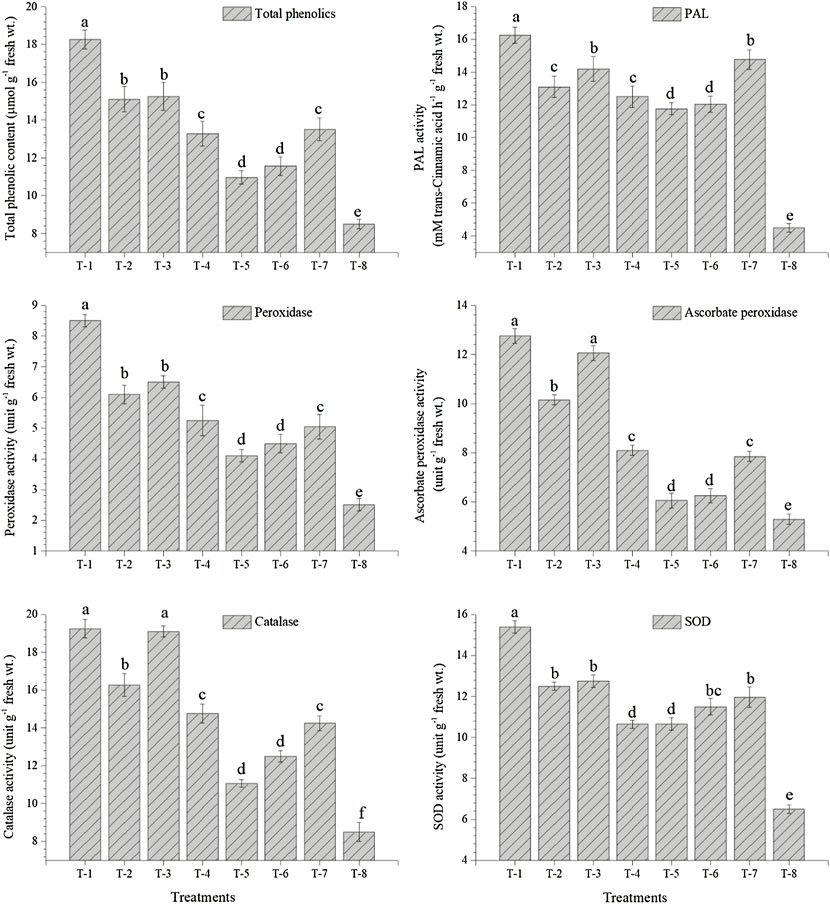
FIGURE 7. Effects of seed inoculation of SOB on accumulation of total phenolics and activities of PAL, peroxidase, ascorbate peroxidase, catalase, and SOD in the pigeonpea leaves at 30 days of sowing. Treatments were as follows: T1—Stenotrophomonas maltophilia DRC-18-7A, T2—S. maltophilia DRC-18-10, T3—S. pavanii DRC-18-7B, T4—S. pavanii DRC-18-11, T5—R. pusense DRC-18-25, T6—R. pusense DSC-18-5, T7—R. pusense DJC-18-21, and T8—Untreated control (No inoculation). Data are mean (n = 5) and vertical bar represents the standard deviation. Data with different letters show significant difference in column data in randomized block design test at p < 0.05 under Duncan’s multiple-range test.
Seed inoculation with sulfur-oxidizing bacterial strains significantly enhances the plant growth attributes in pigeonpea grown in sterilized and non-sterilized soil under glasshouse conditions at 30 and 60 days of sowing (Tables 4, 5). Results of the present investigation revealed maximum shoot length, root length, and fresh and dry weight of shoot and root in the plants inoculated with S. maltophilia DRC-18-7A followed by those inoculated with S. pavanii DRC-18-7B at 30 and 60 DAS in the sterilized soil as compared to other treatments under glasshouse conditions (Table 4 and 5, respectively). A similar trend was recorded in the plants grown in non-sterilized soil at 30 and 60 DAS. However, the values were significantly higher than the plant grown in sterilized soil (Tables 4, 5, respectively).
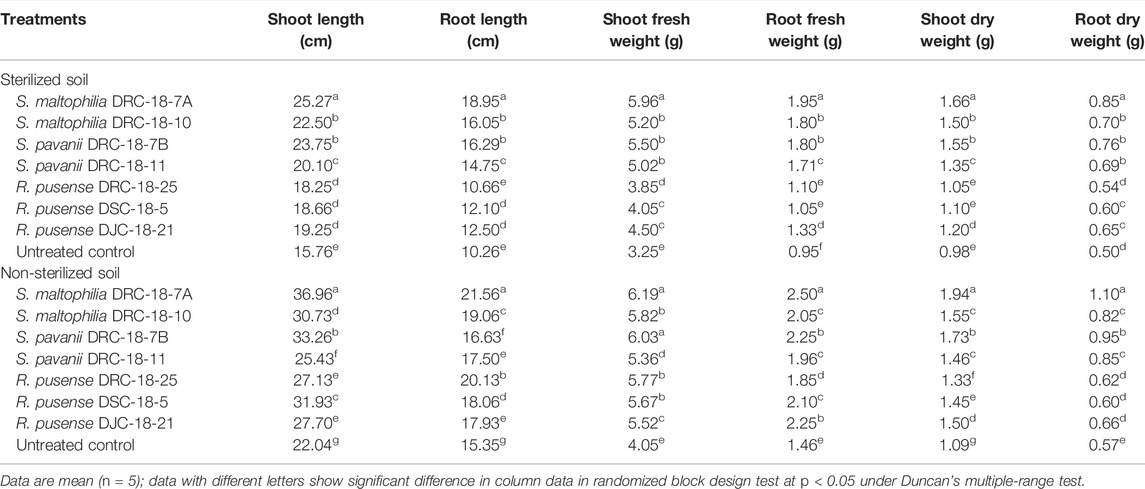
TABLE 4. Effect of seed inoculation on plant growth parameters of pigeonpea grown in pots after 30 days of sowing.
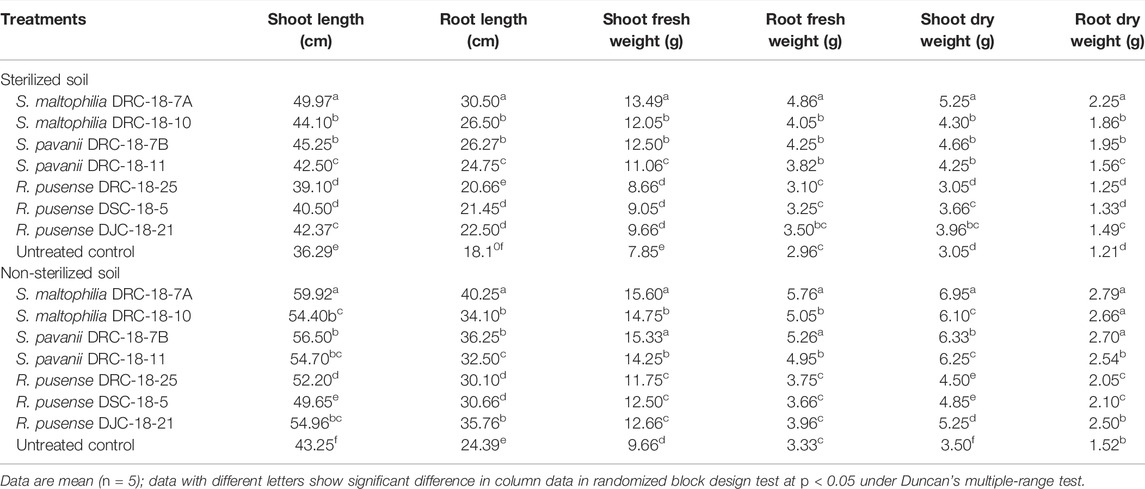
TABLE 5. Effect of seed inoculation on plant growth parameters of pigeonpea grown in pots after 60 days of sowing.
To see the effects of inoculation on the expression profile of phosphorous, potassium, and sulfur transporters, qPCR analyses were conducted. We selected a few key transporters that have been reported from each subfamily. As shown in Figure 8, Cajanus cajan-probable inorganic phosphate transporter 1-3 (CcPh(i)T 1-3) and Cajanus cajan phosphate transporter PHO1 homolog 9 (CcPhT PHO1-9) were highly expressed in the root as compared to other transporters, viz., Cajanus cajan phosphate transporter PHO1 homolog 3 (CcPhT PHO1-3), Cajanus cajan phosphate transporter (CcPhT PHO1), and Cajanus cajan inorganic phosphate transporters 1–4 (CcPh(i)T 1-4), at 30 DAS. In general, significantly higher expression was reported in the plant roots inoculated with S. maltophilia DRC-18-7A followed by S. pavanii DRC-18-7B. However, the least expression was recorded in untreated control plants (Figure 8).
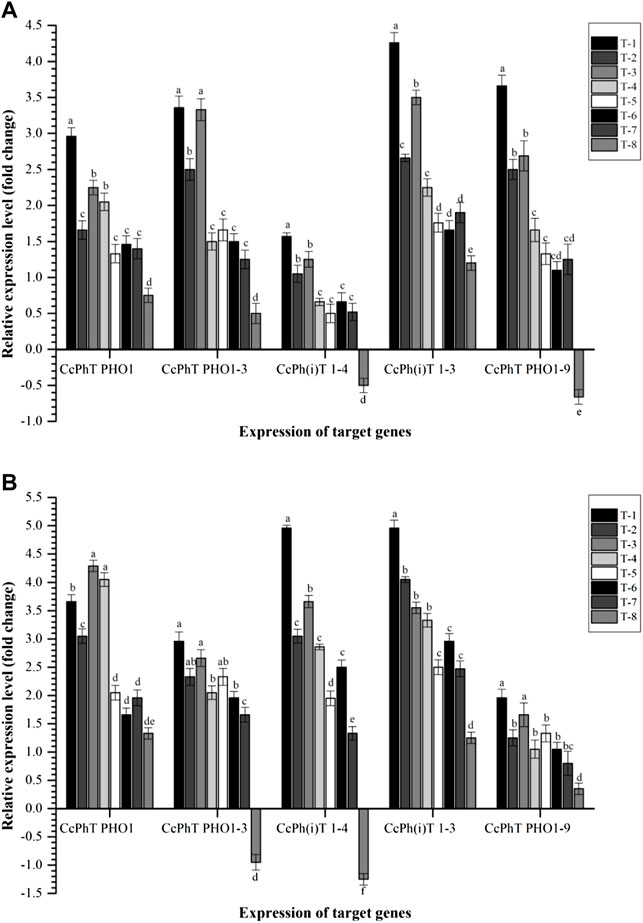
FIGURE 8. Effects of seed inoculation of SOB on expression of phosphate transporters in the pigeonpea leaves at (A) 30 and (B) 60 days of sowing. Treatments were as follows: T1—Stenotrophomonas maltophilia DRC-18-7A, T2—S. maltophilia DRC-18-10, T3—S. pavanii DRC-18-7B, T4—S. pavanii DRC-18-11, T5—R. pusense DRC-18-25, T6—R. pusense DSC-18-5, T7—R. pusense DJC-18-21, and T8—Untreated control (No inoculation). Data are mean (n = 5) and vertical bar represents the standard deviation. Data with different letters show significant difference in column data in randomized block design test at p < 0.05 under Duncan’s multiple-range test.
In contrast, CcPh(i)T 1-3 and CcPh(i)T 1-4 transporters were found to be highly expressed in the roots of pigeonpea at 60 DAS across the treatments. Significantly higher expression was recorded in the roots of pigeonpea inoculated with S. maltophilia DRC-18-7A followed by S. pavanii DRC-18-7B in general. However, results indicated that P transporters genes had differential expressions in root tissues. As indicated in the Figure 8, maximum expression of CcPhT PHO1-3 was recorded in the plants inoculated with S. pavanii DRC-18-7B (4.29-fold) followed by S. pavanii DRC-18-11 (4.05-fold) and S. maltophilia DRC-18-7A (3.66-fold) at 60 DAS (Figure 8). This suggested that CcPh(i)T 1-3 and CcPhT PHO1-9 genes may play a key role in transporting P at the early stage of plant growth, while CcPh(i)T 1-4, CcPh(i)T 1-3, and CcPhT PHO1-3 play role at the advance stage of plant growth.
Similar to P transporters, K transporters were also expressed differently at 30 and 60 days after sowing. Results indicated that maximum expression of Cajanus cajan-probable potassium transporter 17 (CcPoT 17) was recorded at 30 DAS across the treatments (Figure 9). However, Cajanus cajan potassium transporter 3 (CcPoT 3) and Cajanus cajan-putative potassium transporter 12 (CcPoT 12) expressed highly at 60 DAS (Figure 9). Among different treatments, maximum expression of CcPoT 3, CcPoT 6, CcPoT 12, and CcPoT 17 was reported in the plants inoculated with S. maltophilia DRC-18-7A (2.50-, 2.25-, 1.25-, and 3.50-fold, respectively) at 30 DAS (Figure 9). In contrast, CcPoT 3 and CcPoT 17 were expressed highly in the plants inoculated with S. pavanii DRC-18-7B while CcPoT 6 and CcPoT 12 were found to be expressed highly in the plants inoculated with S. maltophilia DRC-18-7A at 60 DAS (Figure 9).
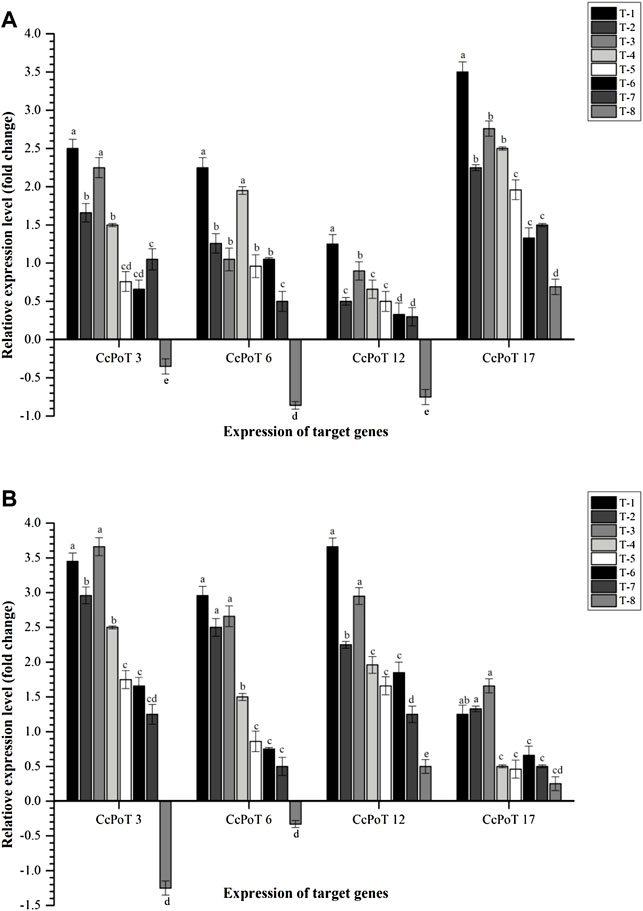
FIGURE 9. Effects of seed inoculation of SOB on expression of potassium transporters in the pigeonpea leaves at (A) 30 and (B) 60 days of sowing. Treatments were as follows: T1—Stenotrophomonas maltophilia DRC-18-7A, T2—S. maltophilia DRC-18-10, T3—S. pavanii DRC-18-7B, T4—S. pavanii DRC-18-11, T5—R. pusense DRC-18-25, T6—R. pusense DSC-18-5, T7—R. pusense DJC-18-21, and T8—Untreated control (No inoculation). Data are mean (n = 5) and vertical bar represents the standard deviation. Data with different letters show significant difference in column data in randomized block design test at p < 0.05 under Duncan’s multiple-range test.
In the present investigation, six key sulfate transporters, viz., Cajanus cajan sulfate transporter 1.1 (CcSULTR1;1), Cajanus cajan sulfate transporter 1.2 (CcSULTR1;2), Cajanus cajan high affinity sulfate transporter 2.1 (CcSULTR2;1), Cajanus cajan-probable sulfate transporter 3.4 (CcSULTR3;4), Cajanus cajan sulfate transporter 4.1 (CcSULTR4;1), and Cajanus cajan sulfate transporter 4.2 (CcSULTR4;2) were taken. Results indicated that all these genes were highly expressed in the plant root inoculated with the S. maltophilia DRC-18-7A followed by S. pavanii DRC-18-7B at 30 DAS. Among different S transporters, CcSULTR4; 2, CcSULTR3;4, and CcSULTR4;1, were found to be expressed highly across the treatments at 30 DAS (Figure 10). In contrast, CcSULTR3;4 was the highly expressed S transporter at 60 DAS followed by CcSULTR2;1 gene. Among different treatments, a more or less similar trend was observed as recorded in the case of 30 DAS (Figure 10).
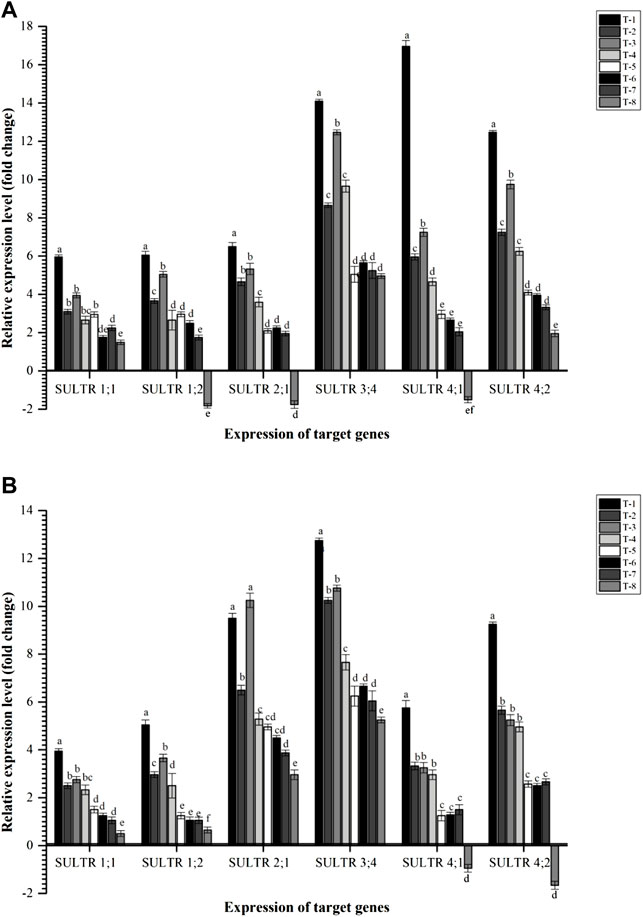
FIGURE 10. Effects of seed inoculation of SOB on expression of sulfur transporters in the pigeonpea leaves at (A) 30 and (B) 60 days of sowing. Treatments were as follows: T1—Stenotrophomonas maltophilia DRC-18-7A, T2—S. maltophilia DRC-18-10, T3—S. pavanii DRC-18-7B, T4—S. pavanii DRC-18-11, T5—R. pusense DRC-18-25, T6—R. pusense DSC-18-5, T7—R. pusense DJC-18-21, and T8—Untreated control (No inoculation). Data are mean (n = 5) and vertical bar represents the standard deviation. Data with different letters show significant difference in column data in randomized block design test at p < 0.05 under Duncan’s multiple-range test.
To see the effects of seed inoculation on the N, P, K, and S content in pigeonpea, plant samples from different treatments were digested and analysed to estimate the N, P, K, and S content in pigeonpea grown in the sterilized and non-sterilized soil at 60 DAS. Under glasshouse conditions, N content in the leaves of pigeonpea was significantly higher in the plants treated with S. maltophilia DRC-18-7A (2.54%) followed by the plants inoculated with S. pavanii DRC-18-7B (2.37%) grown in sterilized soil as compared to other treatments (Table 6). However, in non-sterilized soil, N content in the leaves of pigeonpea was significantly higher in the plants treated with S. pavanii DRC-18-7B (2.91%) followed by the plants inoculated with S. maltophilia DRC-18-7A (2.80%). In contrast, significantly higher P and K contents were recorded in the plants grown in sterile soil treated with S. pavanii DRC-18-7B (0.46 and 0.55%, respectively) as compared to plants treated with S. maltophilia DRC-18-7A (0.33 and 0.48%, respectively) and other inoculants (Table 6). Similar to N content, maximum S content was recorded in the plants grown in non-sterilized soil and treated with S. maltophilia DRC-18-7A (0.62%) followed by S. pavanii DRC-18-7B (0.60%). The least N, P, K, and S contents were recorded in untreated control plants grown in sterilized and non-sterilized soil (Table 6).
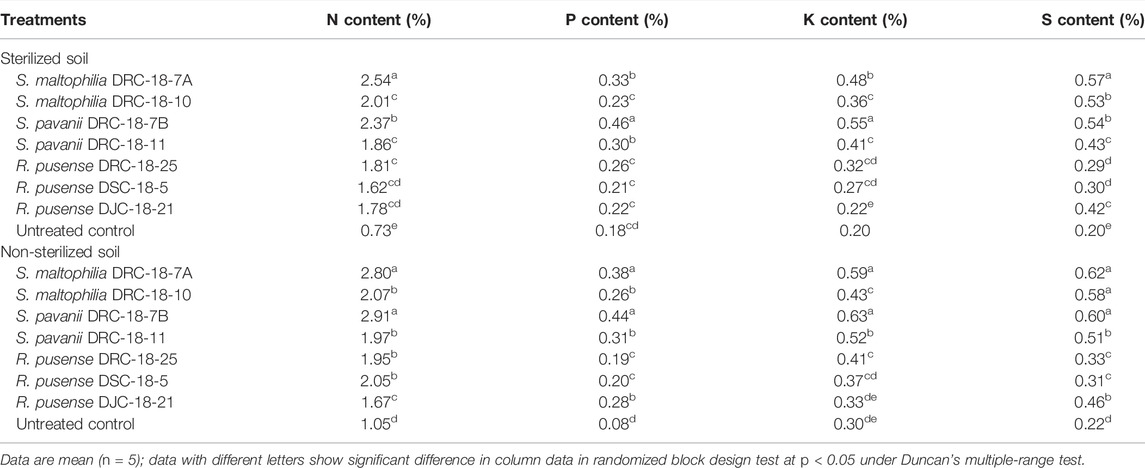
TABLE 6. Effect of seed inoculation of SOB on nitrogen, phosphorous, potassium, and sulfur content in the leaves of pigeonpea after 60 days of sowing.
In general, sulfur is one of the essential and indispensable nutrients for the plant growth and metabolism. Naturally, sulfur is oxidized into the soil by sulfur-oxidizing bacteria promoting S uptake and plant health (Grayston and Germida, 1991; Banerjee and Yesmin, 2004; Anandham et al., 2014). The aim of the present investigation was to isolate, identify, and characterize SOB from coal mines and to evaluate the efficient strains for their ability to influence plant growth and S uptake in pigeonpea. In the earlier studies, several attempts had been made to isolate SOB from the rhizosphere of mustard, paddy, black gram, and sesame (Vidyalakshmi and Sridar, 2007; Anandham et al., 2010; Chaudhary et al., 2017), as well as black shale and acid mine drainage (Sajjad et al., 2016). Anandam et al. (2005) isolated chemolithotrophic SOB using enrichment technique from different niches. Vidyalakshmi and Sridar (2007) also published a report on the isolation of SOB from biogas slurry. To the best of our knowledge and literature available so far, no significant investigations have been made on the isolation of SOB from coal mines and their effects on sulfur uptake and translocation in pigeonpea. In the present study, 13 bacterial isolates were obtained from different samples collected from Open Cast Coal Mines Projects (West Mudidih Colliery, Singhpur Opencast mine, and Sirka Opencast mine, Jharkhand, India). Coal, petroleum, and natural gas contain a high level of organic and inorganic sulfur and are supposed to be a rich source of SOB by and large (Sultan and Faisal, 2016). Several heterotrophic, mixotrophic, chemolithotrophic, and chemoorganotrophic microorganisms are associated with coal and accelerate the bioleaching of different metals. Harrison (1978) suggested that organic sulfur present in coal may first be attacked by heterotrophs and the sulfur released may undergo further oxidation by Thiobacillus, particularly T. ferrooxidans. Further, acidophilic pyrite-oxidizing bacteria metabolize both sulfur and pyrite in coal and utilize the energy released to support their growth (Sultan and Faisal, 2016). Based on 16S rRNA sequencing, these isolates were identified as Stenotrophomonas maltophilia (2), Stenotrophomonas pavanii (2), Rhizobium pusense (5), Bacillus velezensis (2), and Paenibacillus massiliensis (2). SOB isolated from acid mine drainage and black shale were identified as Stenotrophomonas, Alcaligenes, Bordetella spp., and Pseudomonas (Sajjad et al., 2016). Sulfur-oxidizing Pseudomonas spp. have also been isolated from soil of Bhitarakanika, Odisha, India (Thatoi et al., 2012). El-Tarabily et al. (2006) published the first report on sulfur-oxidizing Rhizobium sp. from calcareous soils of the United Arab Emirates. Vidyalakshmi and Sridar (2007) reported isolation of only Thiobacillus sp. from soil, sewage, biogas slurry, mine, soil, and tannery effluent. Although chemoautotrophs are considered to be chiefly responsible for microbial oxidation of reduced sulfur compounds, in the present study all the SOB isolates identified were mixotrophs. These findings are in line of other researchers (Lopez-Aguirre et al., 1999; El-Tarabily et al., 2006; Veerender et al., 2014; Sajjad et al., 2016; Reddy et al., 2018; Yousef et al., 2019). It has been documented that the population densities of mixotrophic sulfur oxidizers are high in agricultural soils and they play an important role in the oxidation of reduced sulfur to sulfate and make it available to the plants (Lawerence and Germida, 1991a; Lawerence and Germida, 1991b; Lawerence and Germida, 1991c; Grayston and Germida, 1991; Germida and Janzen, 1993; Sajjad et al., 2016).
Among the 13 strains obtained in the present study, 6 (Rhizobium pusense DRC-18-15 and DSC-18-101; Bacillus velezensis DSC-18-501 and DJC-18-02J; and Paenibacillus massiliensis DRC-18-101A and DSC-18-77D) could not reduce the pH of the growth medium, and hence were not included for further characterization. The selected seven strains efficiently oxidized sulfur and sulfide into sulfate ions and resulted in a reduction in pH of thiosulfate broth medium up to 2.0 from the initial pH 8 after 21 days of incubation. Various researchers in their reports have already discussed pH reduction due to the oxidation of sulfur and subsequent production of acid in the medium by sulfur-oxidizing bacteria (Khalid et al., 1993; Donati et al., 1996; Vidyalakshmi and Sridar, 2007). The strains could produce sulfate ions in the range 160–311.43 mg L−1, the maximum being recorded for S. maltophilia DRC-18-7A. Vimala and Sridar (2009) recorded 32.7 mg of sulfate per 100 ml of the broth, by MSA2C4 isolate. Anandham et al. (2006) reported the production of sulfate ions in growth medium ranging from 31.7 mg 100 ml−1 to 118.0 mg 100 ml−1 by SOB. The concentration of sulfate ions produced by different SOB was estimated in the range 179–272 mg L−1 (Sajjad et al., 2016). Sulfate–sulfur over the range 100–200 μg ml−1 was obtained on the oxidation of elemental sulfur by Thiobacillus thiooxidans (Vogler and Umbreit, 1941). Proteus mirabilis BUFF14 could release sulfate ions in the range 80 mg ml−1–130.50 mg ml−1 till 40 days of incubation (Dhiman et al., 2019). The SOB were further characterized for different plant growth-promoting attributes so as to select potent inoculants for pigeonpea that could improve plant growth besides providing sulfur nutrition to the plants. Plant growth-promoting microorganisms support nutrient uptake through solubilization of phosphorus and production of auxins and siderophore. Four strains belonging to S. maltophilia and S. pavanii were positive for solubilization of phosphorus and production of siderophore. Auxin production was not exhibited by S. pavanii DRC-18-11. None of the three strains belonging to R. pusense could solubilize phosphorus. R. pusense DSC-18-5 and DRC-18-25 were positive for the production of siderophore and auxin. R. pusense DJC-18-21 failed to solubilize P and produce siderophore and auxins. S. maltophilia and S. pavanii have been reported to possess different PGP attributes and have also been implicated to alleviate biotic and abiotic stresses (Stamford et al., 2002; Singh and Jha, 2017; Alexander et al., 2019; Singh S. et al., 2020; Roy Chowdhury, 2020). The potential of R. pusense as PGPR has also been reported by different workers (Sukweenadhi et al., 2019; Chaudhary et al., 2021; Mir et al., 2021). Since the bioinoculants performed well upon seed inoculation, it represents high degree of rhizosphere competence which is the first and foremost requirement for developing a successful microbial inoculant (Singh DP. et al., 2020, Singh et al., 2021a, b, Yadav et al., 2022). Results of the present investigation indicated that SOB colonized the pigeonpea roots and develop biofilm on the root surface (Figure 4).
The seven characterized strains were initially evaluated for their ability to enhance seed germination and vigour. Seed biopriming with all the seven SOB characterized in the present study led to enhanced seed germination and vigour as compared to the untreated control. S. maltophilia DRC-18-7A closely followed by S. pavanii DRC-18-7B outperformed, however, others in enhancing seed germination and vigour indices. There are many reports showing an increase in seed viability and vigour due to inoculation of PGPR in different crops like rice, maize, cocoa, etc., (Kade and Jusoff, 2013; Hanapi et al., 2014; Elekhtyar, 2015; Sutariati et al., 2021). Sulfur-oxidizing bacteria with additional PGP traits have been reported to enhance the growth of different crops like canola (Grayston and Germida, 1991) and maize (Anandham et al., 2014) as it is a constituent of amino acids like cysteine, cystine, and methionine. It is also involved in the regulation of activities of various reactive oxygen scavenging (ROS) enzymes like phenylalanine ammonia lyase (PAL), peroxidase (POx), ascorbate peroxidase (APx), catalase (CAT), and superoxide dismutase (SOD). In the present study, inoculation of SOB, particularly S. maltophilia DRC-18-7A and S. pavanii DRC-18-7B with multiple PGP traits enhanced the plant growth parameters of pigeonpea after 30 days of sowing both under sterilized and non-sterilized conditions. These increases are concomitant with the uptake of nitrogen, phosphorus, potassium, and sulfur in higher concentrations. In addition, inoculation of SOB enhanced the activity of reactive oxygen species (ROS)-scavenging enzymes like phenylalanine ammonia lyase (PAL), peroxidase (POx), ascorbate peroxidase (APx), catalase (CAT), and superoxide dismutase (SOD). In addition to the activity of ROS-scavenging enzymes, there was a significant increase in the accumulation of flavonoids, carotenoids, and proline due to inoculation of SOB. Further, the accumulation of proteins, soluble sugars, and chlorophyll was significantly influenced due to seed priming with SOB resulting in higher biomass accumulation. It has been reported that deficiency of sulfur can lead to chlorosis with lower plant growth and yield (Saha et al., 2018). In field studies, inoculation of Thiobacillus and amendment of different levels of sulfur to inter-cropped sesame and mung bean resulted in enhancement of chlorophyll a, chlorophyll b, total chlorophyll, plant growth parameters, and yield (Gilani et al., 2021).
The plants have developed an efficient antioxidant system for their survival and optimum growth even under any kind of biotic and/or abiotic stresses. These antioxidant systems have both enzymatic and non-enzymatic components. The enzymatic component includes POx, APx, CAT, GR, and SOD. The non-enzymatic antioxidants include ascorbic acid (AA), reduced glutathione (GSH), α-tocopherol, carotenoids, flavonoids, and the osmolyte proline (Das and Roychaudhary, 2014). Increased plant height, nitrogen uptake, and grain yield were recorded for maize due to inoculation of Thiobacillus sp. (Pourbabaee et al., 2020). Inoculation of chemoautotrophic (Thiobacillus ferrooxidans) and heterotrophic sulfur-oxidizing bacteria (M2 and A12) along with pyrite amendment to canola and wheat enhanced the nitrogen uptake as well as and straw and grain yields (Joseph et al., 2014). Besides grain yield, inoculation of Thiobacillus thiooxidans along with different levels of sulfur to sunflower enhanced the oil yield (Pujar et al., 2014; Singh R. K. et al., 2020). The significant increase in plant growth parameters of pigeonpea in non-sterilized growth medium indicates that inoculated strains were able to compete with the native population and could colonize the rhizosphere efficiently. The root colonization studies with SOB confirm the same.
Nitrogen, phosphorus, potassium, and sulfur (N P K S) are the limiting nutrient factors for plant growth and play a crucial role in the physiological processes of the plant (Mmbaga et al., 2014). They are the main components for building a plant cell including proteins, genes, and chromosomes (Mmbaga et al., 2014). Although significantly higher concentrations of these elements are present in the atmosphere (nitrogen 78%) and soil (P 0.05%, K 0.03%; and S 0.0029%), plants are unable to utilize them directly as nutrients and they remain in bind/fixed form or complexes (Singh et al., 2021a,b). The soil-dwelling beneficial microbes play a significant role in the circulation of plant nutrients, which ultimately minimizes the use of chemical fertilizers. Further, plants have evolved sophisticated and complex signalling mechanisms/response pathways resulting in adaptive responses through genetical and physio-biochemical changes (Malviya et al., 2020; Singh S. et al., 2021). In the past few years, several transporters for phosphorous, potassium, sulfur, zinc, irons, etc. were identified in a large number of plant species (Yoshimoto et al., 2002, 2003; Howarth et al., 2003; Zeng et al., 2017). The presence and site of action may vary in different plant species. Some of them are responsible for the acquisition of minerals and nutrients from soil solutes, while others play important role in the transport of minerals from root hairs to sieve tubes and xylem bundles and subsequent transport to shoots. In plants, some of the key phosphate transporters are phosphate transporter PHO1 homolog 3, phosphate transporter PHO, inorganic phosphate transporters 1–4, probable inorganic phosphate transporters 1–3, and phosphate transporter PHO1 homolog 9 (Kulcheski et al., 2015; Suleman et al., 2018; Pattnaik et al., 2021). Several reports indicated the role of potassium transporters in the acquisition and transport of potassium from soil to plant roots and then shoot. However, potassium can be easily acquired by roots via Na+/K+ flux and other ion channels (Kulcheski et al., 2015; Pattnaik et al., 2021). Similarly, in the recent past, a number of sulfate transporters were identified in a large number of plant species. In Arabidopsis alone, 12 sulfate transporters belonging to 4 different groups (SULTR1, SULTR2, SULTR3, and SULTR4) were identified and characterized based on the similarity in their protein sequences (Smith et al., 1997; Bolchi et al., 1999; Takahashi et al., 2000; Vidmar et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002, 2003; Howarth et al., 2003). However, the role of these transporters and exact mechanisms of P, K, and S uptake and transport in pigeonpea via these transporters are not well established. Further, the role of microbial inoculants in the regulation of these transporters is not documented yet. To the best of my knowledge, this may be the first report on the microbe-mediated, especially via SOB regulation, P, K, and S transport in pigeonpea.
In the present investigation, results indicated that inoculation of SOB up-regulates these transporters in the root of pigeonpea, which is clearly evidenced from transcript accumulation of these transporters in the plants inoculated with SOB as compared to uninoculated control plants. The sulfate transporters SULTR1;1 and SULTR1;2 are highly expressed in the epidermis and cortical tissues of the root and transcripts accumulated under sulfate deprivation, indicating that these transporters have a specialized function to import and facilitate the initial uptake of sulfate from the soil to the roots (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). The results of the present investigation are in line with that. In legumes, inoculation of SOB has shown significant influence on nodulation, nitrogen fixation, sulfur uptake, enhanced plant growth, and yield (Vidyalakshmi et al., 2009). In pigeonpea, increased uptake of N, P, and K have been reported due to sulfur nutrition (Umarani et al., 1994). Interaction in the rhizosphere between microbes and plant roots not only influences the growth of roots but also influences the soil nutrient transformation, mobilization, and their efficient use by plants Shen et al. (2013). Microbial inoculants have the capability to improve nutrient uptake, increase nutrient availability, or stimulate plant growth (Harman et al., 2004). A large number of studies have reported that PGPRs have the potential to mineralize organic compounds, solubilize mineral nutrients, and fix the atmospheric nitrogen (Mia et al., 2013; Mmbaga et al., 2014). It is clearly evidenced that selected SOB, Stenotrophomonas spp., and R. pusense do not produce nodules in pigeonpea. However, the application of SOB enhances the N, P, K, and S contents in the pigeonpea as compared to untreated control plants. They might increase the N mineralization in the rhizosphere and thereby increase the nitrogen use efficiency of plants along with soil fertility which is reflected in the form of increased N content in the plants. On the other hand, co-inoculation of SOB and specific root nodule bacteria in cowpea along with amendments significantly enhanced the dry biomass and uptake of different minerals (S, K, N, P, Fe, Mn, Zn, and Cu) (Mohamed and Gomaa, 2005). Bhagwan (2017) reported that uptake of N, P, and K for pigeonpea was significantly influenced by the application of graded levels of sulfur and SOB. The combined application of SOB and Rhizobium enhanced the pod yield of groundnut (Hanif and Krishnamoorthi, 2016).
Some studies showed that the application of sulfur significantly increases the yield and nutrient uptake in pigeonpea as compared to absolute control (Palsaniya and Ahlawat, 2009; Jat and Ahlawat, 2010; Kumar et al., 2012). Similarly, Deshbhratar et al. (2010) reported significant increment in grain yield (14.81 q ha−1) and straw yield (41.26 q ha−1) of pigeonpea incorporated with 20 kg S ha−1 and 50 kg P2O5 ha−1 along with a recommended dose of N (30 kg ha−1) as compare to control. In the present investigation also seed inoculation by different strains of Stenotrophomonas spp. and R. pusense significantly increased the shoot and root length, as well as fresh and dry biomass in the pigeonpea at 30 and 60 days of sowing. Moreover, sulfur oxidation is an important biogeochemical reaction that improves the sulfur availability in the rhizosphere and cannot be overlooked. However, sulfur oxidation by PGPR is not a new approach, but none of the studies have yet reported the potential of SOB for their use as biofertilizers to overcome sulfur deficiency in pigeonpea crops. Further in-depth investigations are needed to verify the exact role of these sulfur-oxidizing microbial inoculants along with other putative mechanisms participating in microbe-mediated expression of S transporters and regulation of physiological and metabolic pathways in pigeonpea. It would be interesting to explore how the overexpression of S transporters, and other antioxidant pathways could be used to enhance tolerance level under sulfur-deficient conditions and improve crop growth in pigeonpea, similar to achievements made in A. thaliana and other model plants.
In the present investigation, 13 strains were isolated from coal mines and identified as S. maltophilia, S. pavanii, R. pusense, B. velezensis, and P. massiliensis on the basis of 16S rRNA gene sequences. Out of 13 strains, 7 strains were found to reduce the pH and produce sulfate ions and were taken in the present study. Based on the results obtained, S. maltophilia DRC-18-7A and S. pavanii DRC-18-7B colonized the plant roots profusely and were found to be promising SOB which could increase the seed germination, vigour indices, and root architecture in pigeonpea at the early stage of plant growth under sterilized and non-sterilized conditions. Further, these strains up-regulated the P, K, and S transporters in pigeonpea and this is going to be a first report where S. maltophilia, S. pavanii, and R. pusense could regulate the P, K, and S transporters. These results are substantiated with the data obtained on P, K, and S content. Based on the results, it was confirmed that selected strains of SOB could performed well under sterilized and non-sterilized sand–soil mixture. The study has led to the identification of two potential SOB (S. maltophilia DRC-18-7A and S. pavanii DRC-18-7B) that could be used as inoculants for pigeonpea to enhance its growth and yield. Furthermore, SOB-mediated sulfur uptake and translocation, and molecular mechanisms/insights behind the plant growth and development with special reference to pigeonpea need to be explored in future courses of research. In order to achieve maximum benefit, the work is in progress to arrive at compatible combinations with pigeonpea-specific rhizobia and to develop consortium formulations that can be recommended in a package of practices.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
DM, AS, US, and AV conceived and designed the experiments. DM, SS, and US performed the experiments. DM, US, and HS analysed the data. DM and US wrote the manuscript. AS and AV have edited and given final touch to the manuscript. All authors have reviewed the manuscript and have given approval to the final version.
This research was supported by the Indian Council of Agricultural Research, New Delhi (India).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Authors gratefully acknowledge the Mr. Krishna Kumar Choudhary, Electrical Foreman, West Mudidih Colliery, Sijua (Dhanbad), and Mr. Subhas Yadav, Assistant Manager (Mining), Sirka Open Cast Project, Ramgarh (Jharkhand), and other higher officials for providing all necessary logistic and technical supports during the entire sampling process. The authors gratefully acknowledge Mr. Manish Roy, ICAR-NBAIM, for providing technical support to carry out scanning electron microscopy. Authors duly acknowledge Dr. Jai P. Rai, Banaras Hindu University, Varanasi, for editing the final version of revised manuscript. The authors gratefully acknowledge the Indian Council of Agricultural Research, Ministry of Agriculture and Farmers Welfare, Government of India, for providing financial support to the study and providing fellowship to the first author.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.932402/full#supplementary-material
Abo-Zaid, G. A., Soliman, N. A.-M., Abdullah, A. S., El-Sharouny, E. E., Matar, S. M., and Sabry, S. A.-F. (2020). Maximization of Siderophores Production from Biocontrol Agents, Pseudomonas aeruginosa F2 and Pseudomonas Fluorescens JY3 Using Batch and Exponential Fed-Batch Fermentation. Processes 8, 455. doi:10.3390/pr8040455
Alexander, A., Singh, V. K., Mishra, A., and Jha, B. (2019). Plant Growth Promoting Rhizobacterium Stenotrophomonas Maltophilia BJ01 Augments Endurance against N2 Starvation by Modulating Physiology and Biochemical Activities of Arachis Hypogea. Plos one 14, e0222405. doi:10.1371/journal.pone.0222405
Anandham, R., Sridar, R., Nalayini, P., Poonguzhali, S., Madhaiyan, M., and Sa, T. (2006). Potential for Plant Growth Promotion in Groundnut (Arachis hypogaea L.) Cv. ALR-2 by Co-inoculation of Sulfur-Oxidizing Bacteria and Rhizobium. Microbiol. Res. 162, 139–153. doi:10.1016/j.micres.2006.02.005
Anandham, R., Indiragandhi, P., Kwon, S. W., Sa, T. M., Jeon, C. O., Kim, Y. K., et al. (2010). Pandoraea thiooxydans sp. nov., a Facultatively Chemolithotrophic, Thiosulfate-Oxidizing Bacterium Isolated from Rhizosphere Soils of Sesame (Sesamum indicum L.). Int. J. Syst. Evol. Microbiol. 60, 21–26. doi:10.1099/ijs.0.012823-0
Anandham, R., Janahiraman, V., Gandhi, P. I., and Kwon, S. W. (2014). Early Plant Growth Promotion of Maize by Various Sulphur Oxidizing Bacteria that Uses Different Thiosulfate Oxidation Pathway. Afr. J. Microbiol. Res. 8, 19–27.
Anandham, R., Sridar, R., Nalayini, P., Madhaiyan, M., Gandhi, P. I., Choi, K., et al. (2005). Isolation of Sulfur Oxidizing Bacteria from Different Ecological Niches. Korean J. Soil Sci. ferti. 38, 1–6.
Banerjee, M. R., and Yesmin, L. (2004). BioBoost: a New Sulphur-Oxidizing Bacterial Inoculant for Canola. Proceedings for the 4th international crop science congress. Brisbane, Australia.
Barra, P. J., Inostroza, N. G., Acuña, J. J., Mora, M. L., Crowley, D. E., and Jorquera, M. A. (2016). Formulation of Bacterial Consortia from Avocado (Persea Americana Mill.) and Their Effect on Growth, Biomass and Superoxide Dismutase Activity of Wheat Seedlings under Salt Stress. Appl. Soil Ecol. 102, 80–91. doi:10.1016/j.apsoil.2016.02.014
Bhagwan, G. H. (2017). Effect of Sulphur Oxidizing Bacteria and Sulphur Levels on Growth, Yield and Nutrient Uptake by Pigeonpea. Parbhani (M.S), India: Vasantrao Naik Marathwada Krishi Vidyapeeth. Master’s thesis.
Bilbao, B., Giraldo, D., and Hevia, P. (1999). Quantitative Determination of Nitrogen Content in Plant Tissue by a Colorimetric Method. Commun. Soil Sci. Plant Analysis 30, 1997–2005. doi:10.1080/00103629909370348
Bolchi, A., Petrucco, S., Luigi Tenca, P., Foroni, C., and Ottonello, S. (1999). Coordinate Modulation of Maize Sulfate Permease and ATP Sulfurylase mRNAs in Response to Variations in Sulfur Nutritional Status: Stereospecific Down-Regulation by L-Cysteine. Plant Mol. Biol. 39, 527–537. doi:10.1023/a:1006148815106
Boller, T., and Mauch, F. (1988). Colorimetric Assay for Chitinase. Methods Enzymol. 161, 430–435. doi:10.1016/0076-6879(88)61052-4
Bric, J. M., Bostock, R. M., and Silverstone, S. E. (1991). Rapid In Situ Assay for Indoleacetic Acid Production by Bacteria Immobilized on a Nitrocellulose Membrane. Appl. Environ. Microbiol. 57, 535–538. doi:10.1128/aem.57.2.535-538.1991
Brune, D. C. (1989). Sulfur Oxidation by Phototrophic Bacteria. Biochim. Biophys. Acta. 975, 189–221. doi:10.1016/S0005-2728(89)80251-8
Chaudhary, S., Dhanker, R., Tanvi, , and Goyal, S. (2017). Characterization and Optimization of Culture Condition for Sulphur Oxidizing Bacteria after Isolation from Thizospheric Mustard Soil, Decomposing Sites and Pit House. Int. J. Biol. Biomol. Agril. Food Biotechnol. Engin. 11, 379–383.
Chaudhary, T., Gera, R., and Shukla, P. (2021). Deciphering the Potential of Rhizobium Pusense MB-17a, a Plant Growth-Promoting Root Endophyte, and Functional Annotation of the Genes Involved in the Metabolic Pathway. Front. Bioeng. Biotechnol. 1463. doi:10.3389/fbioe.2020.617034
Das, K., and Roychoudhury, A. (2014). Reactive Oxygen Species (ROS) and Response of Antioxidants as ROS-Scavengers during Environmental Stress in Plants. Front. Environ. Sci. 2. doi:10.3389/fenvs.2014.00053
Deshbhratar, P. B., Singh, P. K., Jambhulkar, A. P., and Ramteke, D. S. (2010). Effect of Sulphur and Phosphorus on Yield, Quality and Nutrient Status of Pigeonpea (Cajanus Cajan). J. Environ. Biol. 31, 933–937.
Dey, R., Pal, K. K., Bhatt, D. M., and Chauhan, S. M. (2004). Growth Promotion and Yield Enhancement of Peanut (Arachis hypogaea L.) by Application of Plant Growth-Promoting Rhizobacteria. Microbiol. Res. 159, 371–394. doi:10.1016/j.micres.2004.08.004
Dhiman, S., Dubey, R. C., Maheshwari, D. K., and Kumar, S. (2019). Sulfur-oxidizing Buffalo Dung Bacteria Enhance Growth and Yield of Foeniculum Vulgare Mill. Can. J. Microbiol. 65, 377–386. doi:10.1139/cjm-2018-0476
Donati, E., Curutchet, G., Pogliani, C., and Tedesco, P. (1996). Bioleaching of Covellite Using Pure and Mixed Cultures of Thiobacillus ferrooxidans and Thiobacillus thiooxidans. Process Biochem. 31, 129–134. doi:10.1016/0032-9592(95)00037-2
Edwards, U., Rogall, T., Blöcker, H., Emde, M., and Böttger, E. C. (1989). Isolation and Direct Complete Nucleotide Determination of Entire Genes. Characterization of a Gene Coding for 16S Ribosomal RNA. Nucl. Acids Res. 17, 7843–7853. doi:10.1093/nar/17.19.7843
El-Tarabily, K. A., Soaud, A. A., Saleh, M. E., and Matsumoto, S. (2006). Isolation and Characterisation of Sulfur-Oxidising Bacteria, Including Strains of Rhizobium, from Calcareous Sandy Soils and Their Effects on Nutrient Uptake and Growth of Maize (Zea mays L.). Aust. J. Agric. Res. 57, 101–111. doi:10.1071/ar04237
Elekhtyar, N. M. (2015). Efficiency of Pseudomonas Fluorescens as Plant Growth-Promoting Rhizobacteria (PGPR) for the Enhancement of Seedling Vigor, Nitrogen Uptake, Yield and its Attributes of Rice (Oryza Sativa L.). Int. J. Sci. Res. Agric. Sci. 2, 57–67.
Fernández, L. A., Zalba, P., Gómez, M. A., and Sagardoy, M. A. (2007). Phosphate-solubilization Activity of Bacterial Strains in Soil and Their Effect on Soybean Growth under Greenhouse Conditions. Biol. Fertil. Soils 43, 805–809.
Friedrich, C. G., Bardischewsky, F., Rother, D., Quentmeier, A., and Fischer, J. (2005). Prokaryotic Sulfur Oxidation. Curr. Opin. Microbiol. 8, 253–259. doi:10.1016/j.mib.2005.04.005
Garrido, M. L. (1964). Determination of Sulphur in Plant Material. Analyst 89, 61–66. doi:10.1039/an9648900061
Germida, J. J., and Janzen, H. H. (1993). Factors Affecting the Oxidation of Elemental Sulfur in Soils. Fertilizer Res. 35, 101–114. doi:10.1007/bf00750224
Gilani, A., Abbasdokht, H., and Gholami, A. (2021). Effects of Thiobacillus and Different Levels of Sulfur Fertilizer on Growth and Physiological Indices in Intercropping of Sesame (Sesamum Indicum L.) and Mung Bean (Vigna Radiata L.). Gesunde Pflanz. 73, 317–333. doi:10.1007/s10343-021-00554-6
Grayston, S. J., and Germida, J. J. (1991). Sulfur-oxidizing Bacteria as Plant Growth Promoting Rhizobacteria for Canola. Can. J. Microbiol. 37, 521–529. doi:10.1139/m91-088
Hanapi, S. Z., Supari, N., Alam., S. A. Z., Javed, M. A., Din, A. R. J. M., Tin, L. C., et al. (2014). Microbial Effects on Seed Germination in Malaysian Rice (Oryza Sativa L.). Proc. Asia-Pasific Adv. Netw. 37, 42–51.
Hanif, N. A. K. A., and Krishnamoorthi, V. (2016). Assessment of Sulphur Oxidising Bacterial Inoculums on Groundnut Yield in Pudukkottai District of Tamil Nadu. Jour. Krish. Vigy. 4, 1–4. doi:10.5958/2349-4433.2016.00001.5
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma Species - Opportunistic, Avirulent Plant Symbionts. Nat. Rev. Microbiol. 2, 43–56. doi:10.1038/nrmicro797
Harrison, A. P. (1978). Microbial Succession and Mineral Leaching in an Artificial Coal Spoil. Appl. Environ. Microbiol. 36, 861–869. doi:10.1128/aem.36.6.861-869.1978
Howarth, J. R., Fourcroy, P., Davidian, J.-C., Smith, F. W., and Hawkesford, M. J. (2003). Cloning of Two Contrasting High-Affinity Sulfate Transporters from Tomato Induced by Low Sulfate and Infection by the Vascular Pathogen Verticillium dahliae. Planta 218, 58–64. doi:10.1007/s00425-003-1085-5
Ito, T., Sugita, K., and Okabe, S. (2004). Isolation, Characterization, and In Situ Detection of a Novel Chemolithoautotrophic Sulfur-Oxidizing Bacterium in Wastewater Biofilms Growing under Microaerophilic Conditions. Appl. Environ. Microbiol. 70, 3122–3129. doi:10.1128/aem.70.5.3122-3129.2004
Jat, R. A., and Ahlawat, I. P. S. (2010). Effect of Organic Manure and Sulphur Fertilization in Pigeonpea (Cajanus Cajan) + Groundnut (Arachis hypogaea) Intercropping System. Indian J. Agron. 55, 276–281.
Joseph, A. R., Kavimandan, S. K., Tilak, K. V. B. R., and Nain, L. (2014). Response of Canola and Wheat to Amendment of Pyrite and Sulphur-Oxidizing Bacteria in Soil. Archives Agron. Soil Sci. 60, 367–375. doi:10.1080/03650340.2013.799275
Kade, S. G. A., and Jusoff, K. (2013). Effectiveness of Bioinvigoration Technologies on Seed Viability and Vigor of Cocoa (Theobroma Cacao L.). World Appl. Sci. J. 26, 31–36.
Kawai, S., Kamiya, N., Matsuura, K., and Haruta, S. (2019). Symbiotic Growth of a Thermophilic Sulfide-Oxidizing Photoautotroph and an Elemental Sulfur-Disproportionating Chemolithoautotroph and Cooperative Dissimilatory Oxidation of Sulfide to Sulfate. Front. Microbiol. 10, 1150. doi:10.3389/fmicb.2019.01150
Khalid, A. M., Bhatti, T. M., and Umar, M. (1993). An Improved Solid Medium for Isolation, Enumeration and Genetic Investigations of Autotrophic Iron-And Sulphur-Oxidizing Bacteria. Appl. Microbiol. Biotechnol. 39, 259–263. doi:10.1007/bf00228616
Khan, Z. A., Siddiqui, M. F., and Park, S. (2019). Current and Emerging Methods of Antibiotic Susceptibility Testing. Diagnostics 9, 49. doi:10.3390/diagnostics9020049
Kodama, Y., and Watanabe, K. (2003). Isolation and Characterization of a Sulfur-Oxidizing Chemolithotroph Growing on Crude Oil under Anaerobic Conditions. Appl. Environ. Microbiol. 69, 107–112. doi:10.1128/aem.69.1.107-112.2003
Kuenen, L. G., Robertson, L. A., and Tuovinen, O. H. (1992). “The Genera Thiobacillus, Thiomicrospira and Thiosphera,”. The Prokaryotes. Editors A. Ballows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schlefier (New york, NY: Springer).
Kulcheski, F. R., Côrrea, R., Gomes, I. A., de Lima, J. C., and Margis, R. (2015). NPK Macronutrients and microRNA Homeostasis. Front. Plant Sci. 6, 451. doi:10.3389/fpls.2015.00451
Kumar, S., Tomar, J., Kishore, G. R., Kumar, A., and Singh, S. (2012). Effect of Phosphorus and Sulphur on Growth and Yield of Pigeon Pea (Cajanus Cajan). Adv. Res. J. Crop Improv. 3, 50–52.
Kumar, U., Kaviraj, M., Panneerselvam, P., and Nayak, A. K. (2022). Conversion of Mangroves into Rice Cultivation Alters Functional Soil Microbial Community in Sub-humid Tropical Paddy Soil. Front. Environ. Sci. 10, 858028. doi:10.3389/fenvs.2022.858028
Kumar, U., Panneerselvam, P., Gupta, V. V. S. R., Manjunath, M., Priyadarshinee, P., Sahoo, A., et al. (2018). “Diversity of Sulfur-Oxidizing and Sulfur-Reducing Microbes in Diverse Ecosystems,” in Advances in Soil Microbiology: Recent Trends and Future Prospects. Editor T. K. Adhya (Singapore: Springer), 65–89. doi:10.1007/978-981-10-6178-3_4
Lawerence, J. R., and Germida, J. J. (1991a). Microbial and Chemical Characteristics of Elemental Sulphur Beads in Agricultural Soils. Soil Biol. biochem. 23, 617–622.
Lawerence, J. R., and Germida, J. J. (1991b). Relationship between Microbial Biomass and Elemental Sulfur Oxidation in Agricultural Soils. Soil Sci. Soc. Am. J. 52, 672–677.
Lawrence, J. R., and Germida, J. J. (1991c). Enumeration of Sulfur-Oxidizing Populations in Saskatchewan Agricultural Soils. Can. J. Soil. Sci. 71, 127–136. doi:10.4141/cjss91-011
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lopez-Aguirre, J. G., Farias-Larios, J., Guzman-Gonzalez, S., Michel-Rosales, A., and De Freitas, J. R. (1999). Effect of Sulphur Application on Chemical Properties and Microbial Populations in a Tropical Alkaline Soil. Pedobiologia 43, 183–191.
Malviya, D., Singh, U. B., Singh, S., Sahu, P. K., Pandiyan, K., Kashyap, A. S., et al. (2020). “Microbial Interactions in the Rhizosphere Contributing Crop Resilience to Biotic and Abiotic Stresses,” in Rhizosphere Microbes (Singapore: Springer), 1–33. doi:10.1007/978-981-15-9154-9_1
Meier, D. V., Pjevac, P., Bach, W., Hourdez, S., Girguis, P. R., Vidoudez, C., et al. (2017). Niche Partitioning of Diverse Sulfur-Oxidizing Bacteria at Hydrothermal Vents. ISME J. 11, 1545–1558. doi:10.1038/ismej.2017.37
Mia, M. A. B., Hossain, M. M., Shamsuddin, Z. H., Islam, M. T., and Islam, M. T. (2013). Plant-associated Bacteria in Nitrogen Nutrition in Crops, with Special Reference to Rice and Banana. Bact. Agrobiology Crop Prod., 97–126. doi:10.1007/978-3-642-37241-4_5
Mir, M. I., Kumar, B. K., Gopalakrishnan, S., Vadlamudi, S., and Hameeda, B. (2021). Characterization of Rhizobia Isolated from Leguminous Plants and Their Impact on the Growth of ICCV 2 Variety of Chickpea (Cicer Arietinum L.). Heliyon 7, e08321. doi:10.1016/j.heliyon.2021.e08321
Mmbaga, G. W., Mtei, K. M., and Ndakidemi, P. A. (2014). Extrapolations on the Use of Rhizobium Inoculants Supplemented with Phosphorus (P) and Potassium (K) on Growth and Nutrition of Legumes. As 05, 1207–1226. doi:10.4236/as.2014.512130
Mohamed, H. A., and Gomaa, A. M. (2005). Faba Bean Growth and Green Yield and its Quality as Influenced by the Application of Bio-Organic Farming System. J. Appl. Sci. Res. 1, 350–385.
Myung Cha, J., Suk Cha, W., and Lee, J.-h. (1999). Removal of Organo-Sulphur Odour Compounds by Thiobacillus Novellus SRM, Sulphur-Oxidizing Microorganisms. Process Biochem. 34, 659–665. doi:10.1016/s0032-9592(98)00139-3
Natesan, S., Palani, N., and Ranganathan, V. (1985). Sulfur for Tea. Proc. of TNAU-FACT National seminar on Sulfur in agriculture. Oct. 18-19, 1985. Coimbatore, Tamil Nadu, India: TNAU, 129.
Nautiyal, C. S. (1999). An Efficient Microbiological Growth Medium for Screening Phosphate Solubilizing Microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi:10.1111/j.1574-6968.1999.tb13383.x
Palsaniya, D. R., and Ahlawat, I. P. S. (2009). Sulphur Management in Pigeonpea (Cajanus Cajan)–Wheat (Triticum aestivum) Cropping System. Indian J. Agron. 54, 272–277.
Pattnaik, S., Mohapatra, B., and Gupta, A. (2021). Plant Growth-Promoting Microbe Mediated Uptake of Essential Nutrients (Fe, P, K) for Crop Stress Management: Microbe-Soil-Plant Continuum. Front. Agron. 3, 689972. doi:10.3389/fagro.2021.689972
Pourbabaee, A. A., Koohbori Dinekaboodi, S., Seyed Hosseini, H. M., Alikhani, H. A., and Emami, S. (2020). Potential Application of Selected Sulfur-Oxidizing Bacteria and Different Sources of Sulfur in Plant Growth Promotion under Different Moisture Conditions. Commun. Soil Sci. Plant Analysis 51, 735–745. doi:10.1080/00103624.2020.1729377
Pujar, A. M., Kumar, B. N. A., and Geeta, G. S. (2014). Response of Sunflower (Helianthus Annuus L.) to Gradedlevels of Sulphur and Sulphur Oxidising Biofertilizer (Thiobacillus Thiooxidans). Biochem. Cell. Arch. 14, 339–342.
Rakesh, A. P., Kumari, V., Shekhar, D., and Prasad, R. P. (2020). Soil Sulphur Status and Response of Crops to Sulphur Application in Indian Soils: A Review. J. Pharmacogn. Phytochem. 9, 1406–1410. doi:10.20546/ijcmas.2020.904.322
Reddy, A. N., Gopal, A. V., Lakshmipathy, R., and Padma, V. (2018). Isolation, Characterization and Screening of Sulphur Oxidizing Bacteria from Rhizosphere Soils of Groundnut. Int. J. Curr. Microbiol. App. Sci. 7, 2639–2644. doi:10.20546/ijcmas.2018.708.272
Roy Chowdhury, A. (2020). A Study on the Role of Stenotrophomonas Maltophilia and Stenotrophomonas Pavanii in Sustainable Agriculture. Life Sci. Leafl. 129.
Roy, S., and Roy, M. (2019). Characterization of Plant Growth Promoting Feature of a Neutromesophilic, Facultatively Chemolithoautotrophic, Sulphur Oxidizing Bacterium Delftia sp. Strain SR4 Isolated from Coal Mine Spoil. Int. J. Phytoremediation 21, 531–540. doi:10.1080/15226514.2018.1537238
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The Versatility and Adaptation of Bacteria from the Genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi:10.1038/nrmicro2163
Sadeghi Pour Marvi, M., Pourbabaee, A. A., Alikhani, H. A., Haidari, A., and Manafi, Z. (2016). The Diversity of Sulfur-Oxidizing Bacterial Populations at an Iranian Copper Mine and the Surrounding Agricultural Soils. Appl. Ecol. Env. Res. 14, 509–533. doi:10.15666/aeer/1403_509533
Saha, B., Saha, S., Roy, P. D., Padhan, D., Pati, S., and Hazra, G. C. (2018). “Microbial Transformation of Sulphur: An Approach to Combat the Sulphur Deficiencies in Agricultural Soils,” in Role of Rhizospheric Microbes in Soil (Singapore: Springer), 77–97. doi:10.1007/978-981-13-0044-8_3
Sajjad, W., Bhatti, T. M., Hasan, F., Khan, S., Badshah, M., Naseem, A. A., et al. (2016). Characterization of Sulfur-Oxidizing Bacteria Isolated from Acid Mine Drainage and Black Shale Samples. Pak. J. Bot. 48, 1253–1262.
Sanwani, E., Jeremy, E., Chaerun, S. K., Mufakhir, F. R., and Astuti, W. (2022). Use of Mixotrophic Bacteria as Flocculating Agents to Separate Iron from Red Mud (Alumina Refinery Residue). J. Sustain. Metall. 8, 443–457. doi:10.1007/s40831-021-00479-4
Schwyn, B., and Neilands, J. B. (1987). Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 160, 47–56. doi:10.1016/0003-2697(87)90612-9
Shen, J., Li, C., Mi, G., Li, L., Yuan, L., Jiang, R., et al. (2013). Maximizing Root/rhizosphere Efficiency to Improve Crop Productivity and Nutrient Use Efficiency in Intensive Agriculture of China. J. Exp. Bot. 64, 1181–1192. doi:10.1093/jxb/ers342
Shibagaki, N., Rose, A., McDermott, J. P., Fujiwara, T., Hayashi, H., Yoneyama, T., et al. (2002). Selenate-resistant Mutants of Arabidopsis thaliana Identify Sultr1;2, a Sulfate Transporter Required for Efficient Transport of Sulfate into Roots. Plant J. 29, 475–486. doi:10.1046/j.0960-7412.2001.01232.x
Shukla, A. K., Behera, S. K., Prakash, C., Tripathi, A., Patra, A. K., and Dwivedi, B. S. (2021). Deficiency of Phyto-Available Sulfur, Zinc, Boron, Iron, Copper and Manganese in Soils of India. Sci. Rep. 11, 1–13. doi:10.1038/s41598-021-99040-2
Singh, D. P., Singh, V., Gupta, V. K., Shukla, R., Prabha, R., Sarma, B. K., et al. (2020a). Microbial Inoculation in Rice Regulates Antioxidative Reactions and Defense Related Genes to Mitigate Drought Stress. Sci. Rep. 10, 4818. doi:10.1038/s41598-020-61140-w
Singh, R. K., Singh, P., Li, H.-B., Guo, D.-J., Song, Q.-Q., Yang, L.-T., et al. (2020b). Plant-PGPR Interaction Study of Plant Growth-Promoting Diazotrophs Kosakonia Radicincitans BA1 and Stenotrophomonas Maltophilia COA2 to Enhance Growth and Stress-Related Gene Expression in Saccharum Spp. J. Plant Interact. 15, 427–445. doi:10.1080/17429145.2020.1857857
Singh, R. P., and Jha, P. N. (2017). The PGPR Stenotrophomonas Maltophilia SBP-9 Augments Resistance against Biotic and Abiotic Stress in Wheat Plants. Front. Microbiol. 8, 1945. doi:10.3389/fmicb.2017.01945
Singh, S., Singh, U. B., Malviya, D., Paul, S., Sahu, P. K., Trivedi, M., et al. (2020c). Seed Biopriming with Microbial Inoculant Triggers Local and Systemic Defense Responses against Rhizoctonia solani Causing Banded Leaf and Sheath Blight in Maize (Zea mays L.). Ijerph 17, 1396. doi:10.3390/ijerph17041396
Singh, S., Singh, U. B., Trivdi, M., Malviya, D., Sahu, P. K., Roy, M., et al. (2021a). Restructuring the Cellular Responses: Connecting Microbial Intervention with Ecological Fitness and Adaptiveness to the Maize (Zea mays L.) Grown in Saline-Sodic Soil. Front. Microbiol. 11, 568325. doi:10.3389/fmicb.2020.568325
Singh, S., Singh, U. B., Trivedi, M., Sahu, P. K., Paul, S., Paul, D., et al. (2019). Seed Biopriming with Salt-Tolerant Endophytic Pseudomonas Geniculata-Modulated Biochemical Responses Provide Ecological Fitness in Maize (Zea mays L.) Grown in Saline Sodic Soil. Ijerph 17, 253. doi:10.3390/ijerph17010253
Singh, U. B., Malviya, D., Singh, S., Singh, P., Ghatak, A., Imran, M., et al. (2021b). Salt-Tolerant Compatible Microbial Inoculants Modulate Physio-Biochemical Responses Enhance Plant Growth, Zn Biofortification and Yield of Wheat Grown in Saline-Sodic Soil. Ijerph 18, 9936. doi:10.3390/ijerph18189936
Singh, U. B., Malviya, D., Wasiullah, S., Singh, S., Imran, M., Pathak, N., et al. (2016a). Compatible Salt-Tolerant Rhizosphere Microbe-Mediated Induction of Phenylpropanoid Cascade and Induced Systemic Responses against Bipolaris sorokiniana (Sacc.) Shoemaker Causing Spot Blotch Disease in Wheat ( Triticum aestivum L.). Appl. Soil Ecol. 108, 300–306. doi:10.1016/j.apsoil.2016.09.014
Singh, U. B., Malviya, D., Wasiullah, S., Singh, S., Pradhan, J. K., Singh, B. P., et al. (2016b). Bio-protective Microbial Agents from Rhizosphere Eco-Systems Trigger Plant Defense Responses Provide Protection against Sheath Blight Disease in Rice (Oryza sativa L.). Microbiol. Res. 192, 300–312. doi:10.1016/j.micres.2016.08.007
Smith, F. W., Hawkesford, M. J., Ealing, P. M., Clarkson, D. T., Vanden Berg, P. J., Belcher, A. R., et al. (1997). Regulation of Expression of a cDNA from Barley Roots Encoding a High Affinity Sulphate Transporter. Plant J. 12, 875–884. doi:10.1046/j.1365-313x.1997.12040875.x
Sridar, R., Raveendran, M., Sivaji, M., and Gayathri, R. (2013). Isolation of Autotrophic Sulphur Oxidizer from Different Ecological Niches. J. Soil Biol. Ecol. 33, 66–76.
Stamford, N. P., Silva, A. J. N., Freitas, A. D. S., and Araújo Filho, J. T. (2002). Effect of Sulphur Inoculated with Thiobacillus on Soil Salinity and Growth of Tropical Tree Legumes. Bioresour. Technol. 81, 53–59. doi:10.1016/s0960-8524(01)00099-2
Stamford, N. P., Silva Junior, S. D., Rosália, C. E. D., Santos, S., Freitas, A. D. S. D., and Lira Júnior, M. D. A. (2013). Cowpea Nodulation, Biomass Yield and Nutrient Uptake, as Affected by Biofertilizers and Rhizobia, in a Sodic Soil Amended with Acidithiobacillus. Acta Sci. Agron. 35, 453–459. doi:10.4025/actasciagron.v35i4.16994
Starkey, R. L. (1934). Cultivation of Organisms Concerned in the Oxidation of Thiosulfate. J. Bacteriol. 28, 365–386. doi:10.1128/jb.28.4.365-386.1934
Sukweenadhi, J., Purwanto, M. G. M., Hardjo, P. H., Kurniawan, G., and Artadana, I. B. M. (2019). Isolation and In Vitro Screening of Plant Growth Promoting Rhizobacteria from Barak Cenana Red Rice. AIP Publ. LLC 2155 (No. 1), 020037. doi:10.1063/1.5125541
Suleman, M., Yasmin, S., Rasul, M., Yahya, M., Atta, B. M., and Mirza, M. S. (2018). Phosphate Solubilizing Bacteria with Glucose Dehydrogenase Gene for Phosphorus Uptake and Beneficial Effects on Wheat. PLoS one 13, e0204408. doi:10.1371/journal.pone.0204408
Sultan, S., and Faisal, M. (2016). Isolation and Characterization of Iron and Sulphur Oxidizing Bacteria from Coal Mines. J. Environ. Earth Sci. 6, 153–157.
Sutariati, G. A. K., Rakian, T. C., Muhidin, A., Aji, C. K., La Mudi, A., Khaeruni, G. N. A. W., et al. (2021). “Seed Biopriming Using Rhizobacterial Isolated Mixture on Increasing Growth and Yield of Shallots (Allium ascalonicum L.),” in International Seminar on Promoting Local Resources for Sustainable Agriculture and Development (ISPLRSAD 2020) (Dordrecht, Netherlands: Atlantis Press), 66–70.
Tabatabai, M. A. (1984). Importance of Sulphur in Crop Production. Biogeochemistry 1, 45–62. doi:10.1007/bf02181120
Takahashi, H., Watanabe-Takahashi, A., Smith, F. W., Blake-Kalff, M., Hawkesford, M. J., and Saito, K. (2000). The Roles of Three Functional Sulphate Transporters Involved in Uptake and Translocation of Sulphate inArabidopsis Thaliana. Plant J. 23, 171–182. doi:10.1046/j.1365-313x.2000.00768.x
Tandon, H. L. S. (1991b). Secondary and Micronutrients in Agriculture-Guide Book-Cum Directory. 2ndEd. New Delhi, India: FDCO, 122.
Tandon, H. L. S. (1991a). Sulfur Research and Agriculture Production in India. 3rd edition. New Delhi, India: FDCO.
Thatoi, H. N., Behera, B. C., Dangar, T. K., and Mishra, R. R. (2012). Microbial Biodiversity in Mangrove Soil of Bhitarakanika, Odisha, India. Int. J. Environ. Biol. 2, 50–58.
Thimmaiah, S. R. (2012). Standard Methods of Biochemical Analysis. New Delhi: Kalyani Publishers, 421–426.
Umarani, N., Kapoor, K. K., and Mishra, M. M. (1994). Solubilization of Mussoorie Rock and ratooningMSc (Ag.) Thesis. Tamil Nadu Phosphate by Use of Pyrite and Thiobacilli. Environ. Response of red gram genotypes 94. Coimbatore: Agricultural University.R.S. Malik and to varying plant densities, sulfur application K.C. Banger.
Veerender, K., Sridar, R., and Sivaji, M. (2014). Isolation and Characterization of Sulphur Oxidizing Bacteria from Different Ecosystems. Prog. Res. 9, 104–107.
Vidmar, J. J., Tagmount, A., Cathala, N., Touraine, B., and Davidian, J.-C. E. (2000). Cloning and Characterization of a Root Specific High-Affinity Sulfate Transporter from Arabidopsis thaliana. FEBS Lett. 475, 65–69. doi:10.1016/s0014-5793(00)01615-x
Vidyalakshmi, R., Paranthaman, R., and Bhakyaraj, R. (2009). Sulfur Oxidizing Bacteria and Pulse Nutrition- A Review. World J. Agric. Sci. 5, 270–278.
Vidyalakshmi, R., and Sridar, R. (2007). Isolation and Characterization of Sulfur Oxidizing Bacteria. J. Cult. Collect. 5, 73–77.
Vimala, P., and Sridar, R. (2009). Studies on the Use of Sulfur Oxidizing Bacteria (SOB) to Enhace Sulfur Nutrition, Productivity and Oil Content in Groundnut (Arachis hypogaea L.). J. Soil Biol. Ecol. 29, 30–41.
Vogler, K. G., and Umbreit, W. W. (1941). The Necessity for Direct Contact in Sulfur Oxidation by Thiobacillus Thiooxidans. Soil Sci. 51, 331–338. doi:10.1097/00010694-194105000-00001
Whitman, W. B., Goodfellow, M., Kämpfer, P., Busse, H. J., Trujillo, M. E., Ludwig, W., et al. (2012). Bergey’s Manual of Systematic Bacteriology Parts A and B. 2 Edn. New York, NY: Springer-Verlag.
Williams, P. J., and Cloete, T. E. (2008). Microbial Community Study of the Iron Ore Concentrate of the Sishen Iron Ore Mine, South Africa. World J. Microbiol. Biotechnol. 24, 2531–2538. doi:10.1007/s11274-008-9777-4
Yadav, R. C., Sharma, S. K., Varma, A., Rajawat, M. V. S., Khan, M. S., Sharma, P. K., et al. (2022). Modulation in Biofertilization and Biofortification of Wheat Crop by Inoculation of Zinc-Solubilizing Rhizobacteria. Front. Plant Sci. 13, 777771. doi:10.3389/fpls.2022.777771
Yoshimoto, N., Inoue, E., Saito, K., Yamaya, T., and Takahashi, H. (2003). Phloem-Localizing Sulfate Transporter, Sultr1;3, Mediates Re-distribution of Sulfur from Source to Sink Organs in Arabidopsis. Plant Physiol. 131, 1511–1517. doi:10.1104/pp.014712
Yoshimoto, N., Inoue, E., Watanabe-Takahashi, A., Saito, K., and Takahashi, H. (2007). Posttranscriptional Regulation of High-Affinity Sulfate Transporters in Arabidopsis by Sulfur Nutrition. Plant Physiol. 145, 378–388. doi:10.1104/pp.107.105742
Yoshimoto, N., Takahashi, H., Smith, F. W., Yamaya, T., and Saito, K. (2002). Two Distinct High-Affinity Sulfate Transporters with Different Inducibilities Mediate Uptake of Sulfate in Arabidopsis Roots. Plant J. 29, 465–473. doi:10.1046/j.0960-7412.2001.01231.x
Yousef, N., Mawad, A., and Hassanein, M. (2019). Isolation of Sulfur Oxidizing Bacteria from Polluted Water and Screening for Their Efficiency of Sulfide Oxidase Production. Glob. Nest J. 21, 259–264.
Keywords: sulfur-oxidizing bacteria, pigeonpea (Cajanus cajan), P solubilization, vigour indices, root colonization, nutrient transporters, nutrient uptake and translocation
Citation: Malviya D, Varma A, Singh UB, Singh S, Singh HV and Saxena AK (2022) Sulfur-Oxidizing Bacteria From Coal Mine Enhance Sulfur Nutrition in Pigeonpea (Cajanus cajan L.). Front. Environ. Sci. 10:932402. doi: 10.3389/fenvs.2022.932402
Received: 29 April 2022; Accepted: 06 June 2022;
Published: 11 July 2022.
Edited by:
Upendra Kumar, National Rice Research Institute (ICAR), IndiaReviewed by:
Himani Priya, Indian Council of Agricultural Research (ICAR), IndiaCopyright © 2022 Malviya, Varma, Singh, Singh, Singh and Saxena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anil K. Saxena, c2F4ZW5hNDYxQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.