- 1Department of Animal Ecology and Systematics, Justus Liebig University Giessen, Giessen, Germany
- 2Department of Zoology and Wildlife Conservation, University of Dar es Salaam, Dar es Salaam, Tanzania
- 3Zoology Department, Invertebrate Zoology Section, National Museums of Kenya, Nairobi, Kenya
- 4Department of Biology, Mbarara University of Science and Technology, Mbarara, Uganda
Mountains are considered island-like systems often characterized by exceptional biodiversity and endemism. There are many highly isolated mountain ranges in Africa that collectively have been called the Afromontane archipelago. Freshwater snails of the genus Bulinus are composed of many veterinary and biomedical important species. These intermediate hosts for schistosomiasis parasites are wide spread and some of the species are considered to be highly adapted to very harsh environmental conditions such as droughts or low temperatures. However, the extent to which the Bulinus have adapted to live in high-altitudes and factors influencing these range shifts are not well investigated. In this study, we analyzed pan-African Bulinus species from various habitats across different altitudinal ranges, focusing on the high mountains or “sky islands” to examine how the contemporary climate and historical geological factors affect species distributions and evolutionary processes. Using a fossil-calibrated multigene phylogeny composed of two mitochondrial genes (cox1, 16S) and two nuclear genes (ITS2 and H3), we tested: 1) how often and when extreme altitudes were colonized, 2) what are the biogeographical affinities and degree of isolation of high-altitude species, 3) which lineages diversified and evolved endemism in the Afromontane environments, and 4) whether the Afromontane regions represent “sky islands”. Bayesian phylogenetic inference employing a fossil-calibrated molecular clock resulted in a strongly supported phylogeny resolving the relationships between the four Bulinus groups. High-altitude colonization exists exclusively within the Bulinus truncatus/tropicus complex. Several independent colonization events occurred in the Pliocene and Pleistocene throughout Africa, mostly from nearby regions of the respective mountain ranges. Most species evolved in low to mid-altitudinal ranges. Endemism is pronounced in the Ethiopian Highlands and those of Kenya and Lesotho. A previously unknown species was found at an extreme altitude (∼4,000 m a.s.l.) on Mt. Elgon/Uganda extending the formerly known altitudinal maximum of the genus by roughly 900 m. The endemic species has already diverged in the Pliocene (∼4 myr) and is currently characterized by low genetic diversity. There is further cryptic diversity in mountain ranges of Lesotho. Our findings are discussed in a biogeographical, conservation and biomedical context.
Introduction
Mountains worldwide are well-known cradles of biodiversity additionally characterized by elevated levels of endemism (Perrigo et al., 2020). Mountains are also island-like systems, surrounded by low-lying land masses (Itescu, 2019). A number of mountains are thus termed “sky islands” (e.g., McCormack et al., 2009; Mairal et al., 2017). The complex nature of these sky islands and the isolation between them restrict dispersal, thus affecting species distribution, richness and abundance (Gillespie et al., 2009). Colonisation for sky islands, unlike real islands is through immigration from low lands (altitudinal range niche shifts) or short and long dispersal from the nearby sky islands (latitudinal range shift) (McCormack et al., 2009). These immigration events, followed by in-situ speciation may give rise to cryptic species and in some cases species endemism. Consequently, these resulting narrow-range species are often under threat (Mairal et al., 2017; Martín-Queller et al., 2017). Besides historical events such as mountain-building, other factors have been “implicated” in shaping species richness (Georgopoulou et al., 2016). These are primarily temperature, species richness, isolation and area (Steinbauer et al., 2016). Varying climates along steep altitudinal gradients are often seen as prime drivers of isolation of populations, eventually leading to speciation on mountain tops (Rahbek et al., 2019). Mountain species are regularly characterized by pronounced niche conservatism (Antonelli et al., 2018). As isolated geographical features with characteristic species composition and distribution along pronounced altitudinal and thus climatic gradients, they are often used as models in climate change research (e.g., Fischer et al., 2011).
There are many highly isolated mountain ranges in Africa, a topographically diverse continent significantly shaped by the rifting processes involving volcanism. These processes, in turn, produced several prominent volcanic mountains such as iconic Kilimanjaro, Mt. Kenya, Mt. Elgon, or the Cameroon Volcanic line, a long chain of volcanoes. The Ethiopian Highlands, Maloti-Drakensberge, Kenyan-Tanzanian Highlands, Eastern Arc Mts., and the Angolan Highlands represent other important mountain ranges that collectively have been called the Afromontane archipelago, i.e., a collection of widely scattered sky islands (Figure 1). They have been studied repeatedly regarding their diversity, endemicity and interconnectivity (e.g., Measey and Tolley, 2011; Mairal et al., 2017). Remarkably, they represent both cradles and refugia for biodiversity (Perrigo et al., 2020). Contrasting hypotheses have been proposed as to corridors connecting these sky islands in close proximity but also across the whole continent, e.g., from the Kenya-Tanzania Highlands to the Cameroon Volcanic line (Allen et al., 2021). Pleistocene corridors of Afromontane forest belts have been proposed (Mairal et al., 2021) but remain to be tested for most taxa, as the results of hitherto conducted studies are equivocal (e.g., Brühl, 1997; Cox et al., 2014). For mountain biota in Africa, montane refugia are often invoked as speciation mechanism, followed by montane gradient speciation and, to a lesser extent, peripatric speciation and rapid adaptive radiation, whereas the role of polyploidization as a genomic mechanism is not well understood (Couvreur et al., 2021).
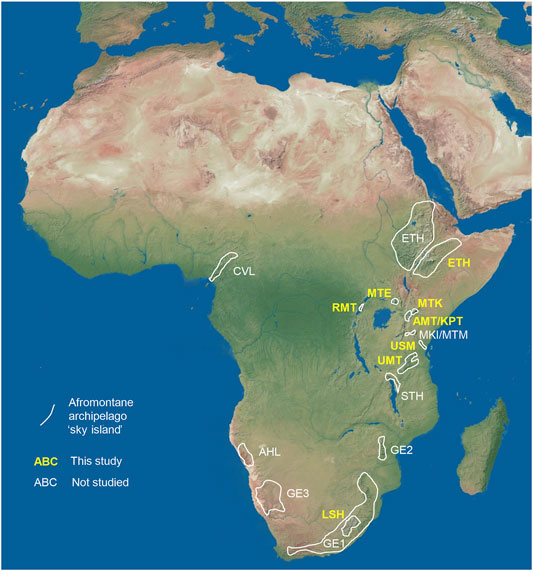
FIGURE 1. Afromontane “sky islands” studied. The mountain ranges in sub-Saharan Africa covered in the current study are italicized and colored in yellow. Abbreviations: AHL, Angolan Highlands; AMT, Aberdares Mts.; CVL, Cameroon Volcanic Line; ETH, Ethiopian Highlands; GE1–3, Great Escarpment; Kinangop Plateau (KPT)-; LSH, Lesotho Highlands; MKI, Mt. Kilimanjaro; MTK, Mt. Kenya; MTM, Mt. Meru; MTE, Mt. Elgon; STH, Southern Tanzania Highlands; USM, Usambara Mts.; UMT, Udzungwa Mts.
Many patterns seen in biodiversity of the Afromontane archipelago have been attributed to changing topography and ecology triggered by geological rifting (e.g., Kingdon, 1989; Menegon et al., 2014). In addition, Pleistocene climatic changes produced multiple refugia, including cold-climate areas such as in sky-islands. These often acted as cradles for diversity on the population (Mairal et al., 2017) and species levels (Cox et al., 2014). The African Rift System and associated mountain ranges and beyond are thus an ideal model to study the interacting forces of climatic changes and geographic barriers on genetic biodiversity patterns over evolutionary times (Platts et al., 2013). Such studies are also critical in times of enormous pressure on the remaining montane biodiversity hotspots throughout the continent (White, 1981; Burgess et al., 2007). Even though these outstanding biodiversity hotspots have long been recognized (Mittermeier et al., 2011), most knowledge stems from either plants such as grasses (Mairal et al., 2021) or trees (DeBusk, 1998) and vertebrates such as small mammals, birds and amphibians (e.g., Cox et al., 2014; Loader et al., 2014). Invertebrates are less frequently studied and if so, mostly mobile organisms such as butterflies (but see flightless insects; Brühl, 1997). The overwhelming majority though represent terrestrial taxa, freshwater organisms are exceptionally scarcely studied (e.g., Daniels et al., 2020; Musonge et al., 2020).
Freshwater molluscs are very suitable for biogeographical studies of high-altitude ecosystems since they are less mobile, often highly habitat specific and have a moderate level of diversity but are comparatively well-known taxonomically (Brown, 1994), unlike most other invertebrates groups in tropical regions. African sky islands are only inhabited by a limited number of freshwater mollusc genera, namely pea-clams (Sphaeriidae; e.g., Kuiper, 1966; Clewing et al., 2022) and pulmonate snails (some Bulinidae, Planorbidae, Lymnaeidae; Brown, 1994). Mollusc species living in high-altitudes (roughly above 2,500 m above sea level) may experience extreme environmental conditions. These include, for example, higher radiation, desiccation of habitat, short periods for reproduction, shortage of food or nutrient availability, cold temperatures including freezing and low abundances (Bößneck, 2012).
The pulmonate gastropod genus Bulinus is, arguably, the most widespread pulmonate snail species in Afromontane regions. The genus is composed of many veterinary and biomedical important species. Many Bulinus species act as intermediate hosts for Schistosoma trematodes that cause, especially, human urogenital schistosomiasis, a disease affecting at least 130 million people in Africa alone (Brindley and Hotez, 2013). Factors that affect species distributions in Africa have not been investigated for many of the approximately 35 extant Bulinus species (Brown, 1994), especially in high-altitude freshwater habitats. Generally, climate change has been predicted to cause changes in species distribution and consequently changes in endemism, evolutionary processes such as speciation and extinction dynamics (Hua and Wiens, 2013), including molluscs (Böhm et al., 2021). Some species of the genus Bulinus are considered to be highly adapted to very harsh environmental conditions such as desiccation (e.g., B. globosus) or low temperatures (e.g., B. africanus) (Brown, 1994). The role of polyploidization has been discussed in this context (Brown, 1994). However, the extent to which Bulinus snails have adapted to high-altitudes in general and extreme altitude above 3,000 m a.s.l., in particular, is not yet accounted for. Here, we explore how and when species of Bulinus have evolved altitudinal ranges. This is important in order to understand the climate-related dynamics of host-parasite interactions. Furthermore, it is a prerequisite for potential predictions of how the ongoing climate change throughout sub-Saharan Africa will affect occurrence of intermediate host snails (Stensgaard et al., 2019; De Leo et al., 2020). In turn, this has direct implications for parasite prevalence and infections of both humans and livestock in areas where diseases have previously been absent (Stanton et al., 2017). Predictions for schistosomiasis in Africa foresee disappearance regionally in lowlands due to extended droughts (shortage of water and thus habitat). Contemporarily, however, increase in schistosomiasis in mountainous regions is predicted due to the expansion of suitable conditions, given the warming experienced (Stensgaard et al., 2019). As a consequence, the altitudinal threshold for schistosomiasis is expected to rise, potentially putting more people and an unrecognized part of the populations at risk (Stanton et al., 2017). Currently, there are no empirical studies available on this subject.
In this study, we analyzed pan-African Bulinus species from various habitats across different altitudinal ranges, focusing on the high mountains or “sky islands” to examine how the contemporary climate and historical geological factors affect species distributions and evolutionary processes. High-altitude samples from most Afromontane regions were collected and altitudinal ranges compiled from our own continent-wide sampling, the literature and databases.
Using a multigene phylogeny of three mitochondrial and two nuclear markers, molecular clock and character evolution analyses, we tested several evolutionary and biogeographical hypotheses for the evolution of the altitudinal niches for all species groups of Bulinus.
Specifically, we tested:
1) how often and when extreme altitudes were colonized,
2) what are the biogeographical affinities and degree of isolation of high-altitude species,
3) which lineages diversified and evolved endemism in the Afromontane environments, and
4) whether the Afromontane regions represent “sky islands”.
Our findings are discussed in the context of biogeographical and diversification history and the dominant mode of speciation. Potential future developments are evaluated given changing climate predictions as well as biomedical and conservation implications.
Materials and Methods
Sampling
Specimens were collected from mountains and highland regions throughout Africa between 2010 and 2018. These areas included the Lesotho Highlands (Maloti-Drakensberge), Ethiopian Highlands (Abyssinian Massif), Kenyan Highlands (Aberdares range including Kinangop Plateau, Mt. Kenya/Laikipia), Tanzanian Eastern Arc Mts. (Udzungwa, East Usambara), and Rwenzori Foothills (Supplementary Table S1; Figure 1). We also obtained samples from lower altitudes, primarily from Uganda, Malawi, Tanzania, the Democratic Republic of Congo, and Cameroon, for comparative purposes and maximized taxonomic representation. Sampling was done using mainly a scoop net, and in some occasions, handpicking from pools, wetlands marshes, lotic and lentic waterbodies. The material was fixed and preserved in 70%–80% ethanol. Coordinates and the altitude records were obtained using a hand-held Garmin etrex V global position system (GPS) device, and were later verified with Google Earth Pro version 7.3.4.
DNA Extraction, Amplification and Sequencing
At least two snails of each of the Bulinus population were selected for DNA isolation, resulting in a total of 104 Bulinus specimens. Prior to DNA isolation, each of the selected snails was photographed using a Keyence digital microscope system (KEYENCE VHX-2000; Keyence Deutschland GmbH, Neu-Isenburg, Germany). Genomic DNA was then extracted from a small piece of the foot muscle (∼2 mg) using a CTAB method (Wilke et al., 2006) or a DNeasy Blood and Tissue Kit (Qiagen, Mississauga, ON, Canada) following the manufacturer’s protocol. The specimen vouchers (shells and DNA) are stored in the University of Giessen Systematics and Biodiversity collection (UGSB; Diehl et al., 2018). DNA amount and quality was checked using a NanoDrop 2000 (Thermo Fisher Scientific Inc., Waltham, MA, United States). Mitochondrial cytochrome c oxidase subunit I (cox1; “Folmer” and “Asmit” regions) and large subunit ribosomal RNA (16S) gene fragments were amplified using primers LCO1490 and HCO2198 (Folmer et al., 1994) or COR722b (Wilke and Davis, 2000) for the “Folmer” region, Asmit1, Asmit2 for the “Asmit” region (Bowles et al., 1992), and 16Sar and 16Sbr for 16S (Palumbi et al., 1991). The internal transcribed spacer 2 (ITS2) and histone 3 (H3) were amplified using primers LT1 (Bargues et al., 2001), ITS2-RIXO (Almeyda-Artigas et al., 2000) and H3F and H3R (Colgan et al., 2000), respectively.
PCR conditions and primer details are described in the Supplementary Table S4. Sanger sequencing was performed on an ABI 3730xl DNA analyzer using the BigDye Terminator Kit (Life Technologies, LGC Genomics GmbH, Berlin, Germany).
Dataset Compilation, Alignment, and Substitution Models
DNA sequences were edited and aligned using BioEdit version 7.0.5 (Hall, 1999). For phylogenetic analyses, additional sequences for all five genetic markers were downloaded from the NCBI database, resulting in a dataset composed of 145 ingroup taxa. After the sequences were reduced to unique haplotypes (Supplementary Figure S1 and Supplementary Table S1), the final dataset consisted of 96 taxa and was used for subsequent analyses. Indoplanorbis exustus, the sister species to Bulinus spp. (Albrecht et al., 2007; Albrecht et al., 2019), was used as an outgroup. Multiple sequence alignment for cox1 (both the Folmer and Asmit regions) and H3 was conducted using ClustalW tool implemented in BioEdit software program while the gap forming partitions 16S and ITS2 were aligned using MAFFT an online alignment tool (Katoh et al., 2019). Gblocks program version 0.91b (http://molevol.cmima.csic.es/castresana/Gblocks_server.html), was used to remove poorly aligned regions, applying less stringent settings. The final alignments contained 655 bp, 390 bp, 432 bp, 452 bp and 328 bp for cox1 (Folmer region), cox1 (“Asmit” region), 16S, ITS2, and H3, respectively. All new sequences have been deposited in NCBI GenBank (Supplementary Table S1).
The concatenated sequence dataset was created using SequenceMatrix version 1.7.8 (Vaidya et al., 2011). The software jModelTest version 2.1.10 (Darriba et al., 2012) was used to select the best-fit substitution models for each partition, with the number of substitution schemes set to 3.
Fossil-Calibrated Molecular Phylogeny
The estimated substitution models were used for the subsequent Bayesian inference analysis employing a molecular-clock approach for estimating species divergence using BEAST version 1.8.4 (Drummond et al., 2012). The analyses were run on the CIPRES Science Gateway V 3.3 (Miller et al., 2010). A fossil calibration was used to estimate divergence times in Bulinus using BEAST version 1.8.4 (Drummond et al., 2012). We used the oldest Bulinus fossil (c. 20 myr; Pickford, 2008) to date the most recent common ancestor (MRCA) of the ingroup.
Based on the Akaike information criterion (AIC), the best-fit substitution models for the concatenated dataset were: HKY + I + Γ, HKY + I + Γ, GTR + I+Γ, GTR + I + Γ and SYM + I for cox1 (Folmer region), cox1 (“Asmit” region), 16S, ITS2, and H3, respectively.
The Markov chain Monte Carlo (MCMC) in BEAST analysis was run for 40 million generations, sampling every 1000th tree, using both a strict-clock and a relaxed-clock model with a birth–death tree prior (Gernhard, 2008) and a gamma prior for the fossil calibration point with the settings: offset = 19, shape = 1.0, and scale = 2.0. The log file was checked in Tracer version 1.5 (http://tree.bio.ed.ac.uk/software/tracer) to examine the parameter convergence. The maximum clade credibility (MCC) tree was generated using TreeAnnotator version 1.8.4 (BEAST package) with a burnin of 50%.
Whereas most of the parameters revealed effective sampling size (ESS) values for the strict-clock model slightly better (ESS of most parameters >300) than with the relaxed-clock model (ESS of most parameters <150), convergence for some of the strict clock model parameters was not reached (i.e., cases in which ESS values were <200). Therefore, the maximum clade credibility (MCC) tree obtained from the strict-clock analyses was used for subsequent analyses, with adjusted substitution models as follows: HKY + I + Γ, HKY + I + Γ, HKY + I + Γ, HKY + I + Γ and HKY (all base frequencies equal), for cox1 (Folmer region), cox1 (“Asmit” region), 16S, ITS2, and H3 respectively, resulting in sufficient ESS values (>200). We also tested a general invertebrate molecular substitution rate (mitochondrial) clock (Wilke et al., 2009) for the HKY + I + Γ, model of sequence evolution (1.57% My−1) in the preliminary analyses (hence data not shown), to check the suitability of the fossil in estimating the divergence time of the Bulinus phylogeny., and this did not result in much difference.
Species Delimitation
Specimens were preliminarily determined based on their shell morphology and ecological setting information such as habitat and geographical position. However, we also used approaches for molecular species delimitation recently conducted (Chibwana et al., 2020; Clewing et al., 2020; Mahulu et al., 2021). Molecular operational taxonomic units (MOTUs) were identified in the BEAST MCC tree using this approach and computational methods for species delimitation; specifically the Generalized Mixed Yule Coalescent (GMYC) for multiple thresholds (Fujisawa and Barraclough, 2013; Pons et al., 2006) and Poisson Tree Process (PTP; both maximum likelihood and highest Bayesian supported solutions; Zhang et al., 2013). The MOTUs were finally assessed in an integrated approach considering the preliminary identifications and named by integrating as many nominal taxa as possible. Doubtful cases or apparently new lineages were simply labelled as Bulinus sp., sometimes with a qualifier (e.g., Bulinus sp. 8 MTE for Mt. Elgon).
Reconstruction of Ancestral Altitudinal Ranges
Two sets of altitudinal information were compiled. Each specimen had a specific sampling altitude, either measured by ourselves in the field or taken from the literature for the GenBank sequences. Google Earth Pro was used to determine the altitudes in cases when only locality names were available in the original literature. In the cases of BtroAY18 and BtruAY19 (Supplementary Table S1), the altitude ranges of their countries of origin were used since no more specific locality information was available.
The second set of altitudinal information consisted of published altitudinal occurrences for all nominal species of Bulinus, which we screened for minima and maxima (Supplementary Table S2). This information was compiled through literature research in comprehensive publications (Mandahl-Barth, 1957; Mandahl-Barth, 1965; Brown, 1994) or species-specific accounts (e.g., De Kock and Wolmarans, 2005a; De Kock and Wolmarans, 2005b). In addition, online databases such as Global Biodiversity Information Facility (GBIF), International Union for Conservation of Nature (IUCN) Redlist, NHM London Zoological Specimen database were checked. A character matrix with six categories was created: A) 0–500 m, B) 501–1,000 m, C) 1,001–1,500, D) 1,501–2,000 m, E) 2,001–2,500, and F) > 2,500 m. The altitude ranges of all the MOTUs were coded as present (1) or absent (0) in these categories. In very few cases, our own sampling served for character coding. Character evolution of the altitudinal range distribution was traced along the phylogenetic tree using stochastic character mapping (Huelsenbeck et al., 2003) as implemented in phytools 1.0-1 (Revell, 2012) for the R statistical environment 4.1.2 (R Core Team, 2021). Three models of character evolution, ARD (all rates different), SYM (symmetric rates), and ER (equal rates), were tested by calculating 100 stochastic character maps and finally compared with AIC.
Results
Species Diversity, Phylogenetic Relationships, and Divergence Times
The automated species delimitation methods yielded 24 and 38 MOTUs for bPTP and 33 for GMYC (Supplementary Figure S1). Our integrative approach resulted in a final set of 30 MOTUs. Bulinus wrighti is the sole representative of the B. reticulatus group. The B. forskalii group comprises B. forskalii, B. bavayi, B. cernicus, B. barthi, and Bulinus sp. 6. The B. africanus group primarily consists of B. globosus and B. nasutus, but also B. africanus and B. obtusispira. In addition, it contains Bulinus sp. 2 (of Chibwana et al., 2020) and a MOTU which could not be assigned to nominal names (Bulinus sp. 3). Most species belong to the B. truncatus/tropicus complex, of which a total of 18 MOTUs correspond to species-level taxa. Names cannot be applied to all of them, but instead nine nominal taxa could be assigned. Bulinus sp. 1 was previously reported in Chibwana et al. (2020). A further two have been labelled Bulinus sp. 4 and Bulinus sp. 5 for convenience. Bulinus sp. 7 LSH (= Lesotho) and Bulinus sp. 8 MTE (= Mt. Elgon) were previously unknown species. The MOTU called Bulinus sp. 9 MTK (= Mt. Kenya) might represent a small species complex of closely related taxa (B. rumrutiensis/laikipiensis). Both B. truncatus and B. tropicus were not found monophyletic, instead there where other MOTUs resembling them morphologically (B. cf. truncatus, B. cf. tropicus).
The phylogenetic reconstruction (Figure 2) revealed four major clades, which are strongly supported. These clades correspond to the four traditional species groups of Bulinus, i.e., the B. forskalii, B. africanus, B. reticulatus groups, and the B. truncatus/tropicus complex. Bulinus reticulatus and the B. truncatus/tropicus complex are well supported sister-groups (PP 1.0) and B. africanus is the sister group to them (PP 1.0). The B. forskalii group is ancestral to the other three groups (PP 1.0).
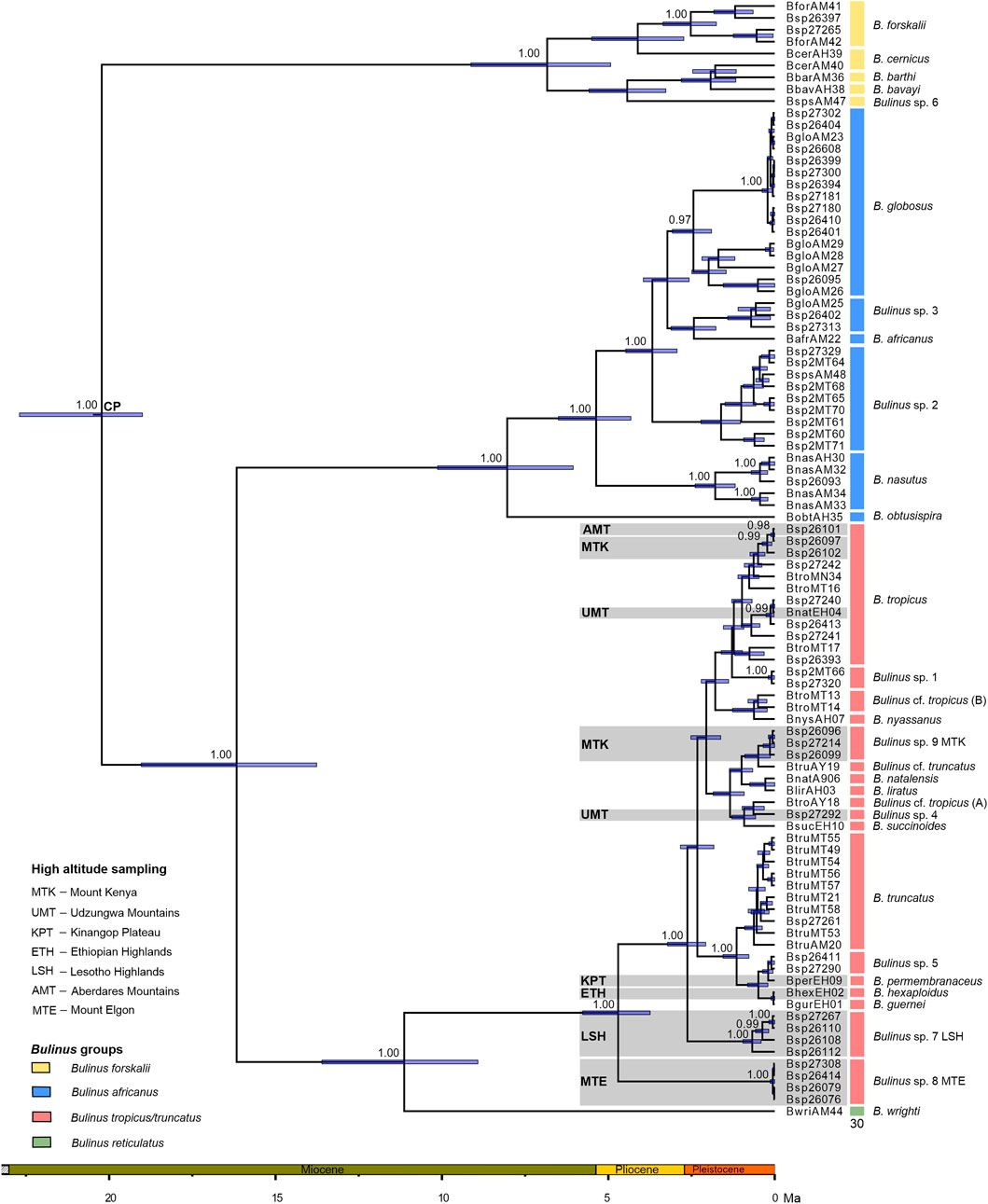
FIGURE 2. BEAST MCC tree with the posterior probabilities above 0.95 given. 95% highest probability densities (blue bars) and the calibration point (CP) are provided. The outgroup was removed a posteriori. Specimen details can be seen in Supplementary Table S1.
High-mountain lineages from the Aberdares Mts. and the Mt. Kenya region are sister groups in B. tropicus, whereas the Udzungwa Mts. lineage is not closely related to it. It clusters with regional lineages (Figure 2; Supplementary Figure S1). Bulinus sp. 9 MTK is sister to a specimen from Malawi (BtruAY19), but this relationship is not supported. This is also the case for Bulinus sp. 4 the sister of which is BtroAY18, a specimen from Zimbabwe. Bulinus permembranaceus from the Kinangop Plateau is closest to Bulinus sp. 5, represented by specimens from Cameroon and the D.R. Congo. Bulinus hexaploidus from the Ethiopian Highlands is sister to B. guernei.
According to our molecular-clock analysis, Bulinus started to diversify in the Early Miocene, which is also when the B. forskalii group diverged from the remaining groups. In the mid-Miocene (∼16 Ma), the B. africanus group diverged from the B. reticulatus group and the B. truncatus/tropicus complex. The latter two diverged from each other around 11 Ma. Within the B. truncatus/tropicus complex, Bulinus sp. 8 MTE from Mt. Elgon originated in the Pliocene (4.71 Ma) (Figure 2). A series of divergence events leading to all other lineages in the complex happened near the Plio-Pleistocene border at 2.62 Ma (Figure 2). This marks the split of Bulinus sp. 7 LSH from the remaining B. truncatus/tropicus complex. All other divergence events for the B. truncatus/tropicus complex are dated to the Pleistocene, concerning all high mountain linages and also B. liratus from Madagascar. Much more lineages were accumulated in the Pliocene in both the B. africanus and B. forskalii groups. Here, the B. obtusispira from Madagascar is sister to the remaining members of the B. africanus group from which it split 8.05 Ma (Figure 2; Supplementary Figure S1).
Biogeographical Patterns
Extreme altitudes (>2,500 m) have been colonized by Bulinus at least three times independently and exclusively in the B. truncatus/tropicus complex: 1) The highlands of Lesotho are inhabited by an apparently endemic lineage (Bulinus sp. 7 LSH, up to 2,528 m a.s.l.); 2) Bulinus sp. 8 MTE (up to ∼4,000 m a.s.l.) represents an absolute extreme for Bulinus (Figure 3); 3) The Ethiopian Highlands are represented by a specimen of B. hexaploidus from 2,592 m (note that B. octoploidus is missing in the phylogeny).
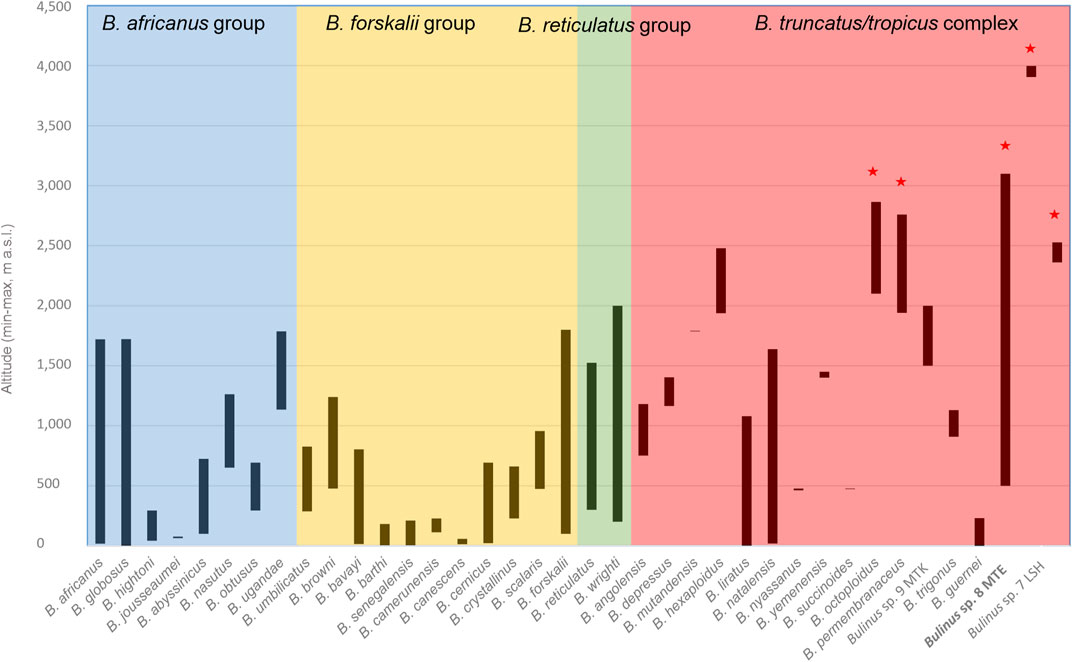
FIGURE 3. Altitudinal distribution of members of the Bulinus groups (highlighted with colors) throughout their entire range compiled from various literature and collection sources. Red stars mark species occurring in extreme altitudes above 2,500 m a.s.l. Note the extraordinary high occurrence of Bulinus sp. 8 MTE on Mt. Elgon, Uganda. This species is new to science and is only distantly related to the B. truncatus/tropicus complex. Further details on altitudinal ranges can be found in the Supplementary Tables S2, S3.
Distinct colonizations occurred on Mt. Elgon and the Lesotho Highlands, spatially and temporally independent from one another and the other Afromontane regions. Diversification on Mt. Elgon is very recent though (0.06 Ma), whereas populations in Lesotho are roughly 0.67 myr old. The colonization patterns of the Aberdares Mts. and other Kenyan Highlands as well as the Udzungwa Mts. (Eastern Arc Mts.) are more complex with the respective species and lineages being widely scattered among the B. truncatus/tropicus complex. Bulinus hexaploidus is closely related to B. permembranaceus from the Kinangop Plateau in Kenya, though they are not sister species. There are two distinct MOTUs from the Mt. Kenya region, representing independent colonizations of the region (B. tropicus and Bulinus sp. 9 MTK). The Udzungwa Mts. as part of the old Eastern Arc Mts. complex are inhabited by at least two MOTUs (B. tropicus and Bulinus sp. 4) (Figure 2). All Ethiopian, Kenyan, and Tanzania MOTUs occurring in higher altitudes represent comparatively young lineages, having diversified in the late Pleistocene (Figure 2). No species of either the B. forskalii, nor in the B. africanus group occur in the second highest altitudinal band from 2,001 to 2,500 m. Bulinus wrighti has been described from low altitudes up to 2,000 m.
Altitudinal Range Evolution
The MOTUs in this study range in altitudinal distribution from sea level to almost 4,000 m (Figure 3; Supplementary Table S2). However, the majority occurs from sea level to 1,500 m with 22 MOTUs in 0–500 m, 15 in 501–1,000 m band and 17 MOTUs found in band 1,001–1,500 m. Much fewer occur in band 1,501–2,000 m (10 MOTUs) and 2,001–2,500 m (six MOTUs), whereas only four MOTUs are found at altitudes above 2,500 m (Supplementary Table S3). Considering altitudinal ranges of all Bulinus spp. including the new MOTUs of this study, the average range is 639 m (5–2,600 m). The average minimum altitude is 678 m (0–3,905 m) and the average maximum is 1,317 m (57–3,997 m) (Figure 3 and Supplementary Table S2).
Most species evolved in low to mid-altitudinal ranges and only approximately nine times evolved adaptations to Afromontane regions. The Equal rates model (ER) was the best-fit model, assuming equal rates for the transition between altitudinal range bands (AIC: ARD: 148.99, ER: 109.51; SYM: 136.27). As many species of Bulinus occur in more than a single altitudinal range band, estimation of ancestral ranges is equivocal for each node in the phylogeny (Figure 4). The MRCA of the B. forskalii group most likely lived in low to intermediate altitudes (0–2,000 m) whereas the MRCA of the B. africanus group most likely lived in altitudes from 0 to 1,500 m, except for B. globosus which extends its distribution up to 2,000 m. Bulinus reticulatus group was only represented by B. wright, and the MRCA of this group has already lived in altitudes covering four bands from low- to mid-altitudes (0–2,000 m). The MRCA of B. truncatus/tropicus complex is shown to have lived in all the six altitudinal bands, and is from which the Bulinus sp. 8 MTE in the extreme altitude at Mt. Elgon has evolved (Figure 4).
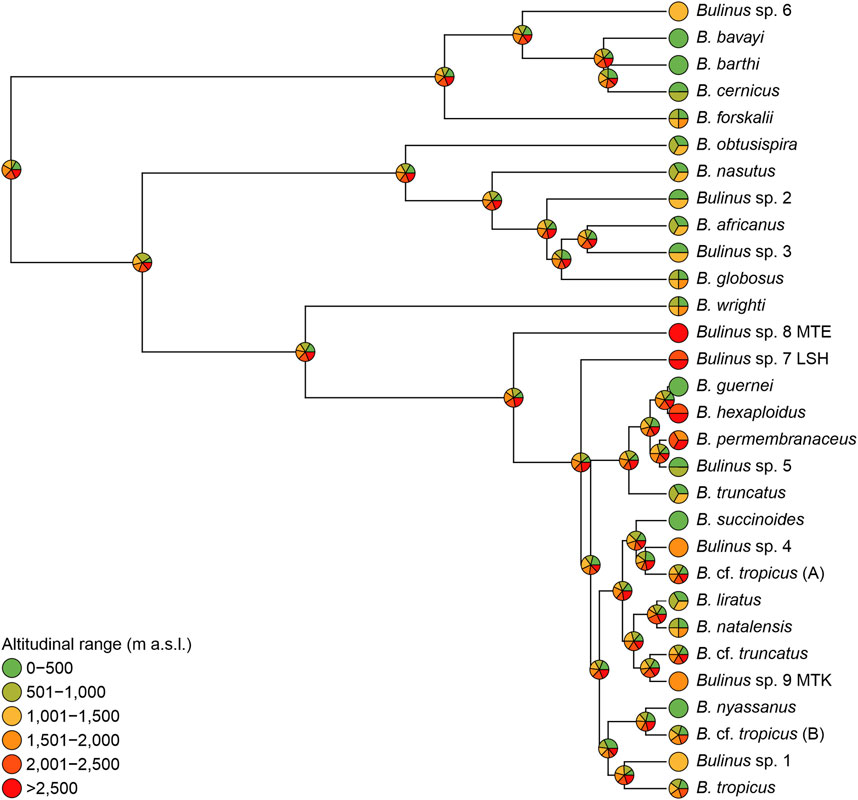
FIGURE 4. Character evolution of altitudinal bands along the BEAST MCC MOTU-like tree. Six classes of altitudinal occurrences are used and pie charts represent probabilities of ancestral states at the respective nodes (see also Supplementary Tables S2, S3).
Discussion
Phylogenetic Relationships and Species Delimitation
Based on the increased taxon and character sampling of the new multi-gene phylogeny, all previously defined subgroups of Bulinus spp. were recovered. Unlike in previous phylogenies based on single markers and less extensive sampling (Kane et al., 2008; Jørgensen et al., 2007; Jørgensen et al., 2011; Jørgensen et al., 2013), intergroup relationships were also well supported in our phylogeny. This is important since the relationship among the Bulinus groups has been extensively debated in the past (e.g., Jørgensen et al., 2011; Jørgensen et al., 2013). Our findings strongly support that the B. reticulatus and the B. truncatus/tropicus complex are sister-groups and B. africanus forms the sister group to the first two whereas the B. forskalii group is ancestral to the other three groups. A sister relationship of the B. reticulatus group and the B. truncatus/tropicus complex has been well supported before (Kane et al., 2008; Jørgensen et al., 2011; Jørgensen et al., 2013). Various constellations of this group relative to the B. africanus and B. forskalii groups had been proposed and none conforms with the one found here (Stothard et al., 1996; Stothard et al., 2001; Jørgensen et al., 2007; Kane et al., 2008; Nalugwa et al., 2010; Jørgensen et al., 2011; Jørgensen et al., 2013; Zein-Eddine et al., 2014; Tumwebaze et al., 2019; Clewing et al., 2020).
The species delimitation overall found a match of morphologically identified species and MOTUs in many cases, however, there were also several cases of non-monophyly and mismatches. Most often this occurred in the B. tropicus/truncatus complex, which has been known for notorious difficulties in species assignments (e.g., Brown, 1994; Kane et al., 2008). Bulinus globosus was another case, known to be an “umbrella” name for various lineages of similar shell morphology (Pennance, 2020). In our conservative approach, we preferred to label unclear specimens as “sp.” rather than assigning names to these lineages in order to not further complicate the situation of potentially wrongly labelled sequence information in GenBank. A major obstacle in Bulinus systematics is to disentangle wrongly identified specimens from cases where indeed cryptic species are involved in potentially rapidly diverging lineages (Tumwebaze et al., 2019). The complexity of the species level systematics stems from different taxonomic values of character used and even species concepts applied by the various authors. Here, either shell characters or anatomy, chromosome numbers, parasite susceptibility and the pre-condition of hermaphroditic reproduction mode have been implemented to various extents over time (Mandahl-Barth, 1957; Mandahl-Barth, 1965; Brown, 1994). Given these circumstances, a comparatively consistent level of MOTUs that correspond to nominal taxa has been found in this study. It should be further substantiated in future studies, which should strictly focus on type or topotypic material of all recognized species. In order to disentangle the real diversity of Bulinus and also the roles that certain processes play in diversification (such as polyploidization), genomic approaches should be used, especially since mitochondrial and nuclear genomes are already available for B. truncatus providing the much needed baseline (Young et al., 2021; Young et al., 2022; Zhang et al., 2022).
The sparse fossil record of Bulinidae has been discussed elsewhere (Jørgensen et al., 2013; Albrecht et al., 2019). The fossil-calibrated tree might potentially underestimate the real age of the group. Given that external substitution rates are often taxon- and gene-specific and may be saturated over time (see, e.g., Wilke et al., 2009), it was still worthwhile to explore it in the case of Bulinus. It should be noted though that the fossil-independent dating strategy using a universal invertebrate molecular clock rate did not yield substantially different age estimates for the major splits in the phylogeny (data not shown). As such, the ages of the endemics on Madagascar (B. liratus and B. obtusispira, B. bavayi) are interesting. Although B. bavayi and B. obtusipira represent comparatively old lineages in their respective Bulinus groups, their splitting from other lineages by far postdates the separation of Madagascar from East Africa and the Indian subcontinent (e.g., Masters et al., 2021). The Bulinus spp. on Madagascar thus do likely not represent vicariant forms (Wright, 1971; Stothard et al., 2001; Jørgensen et al., 2011). Interestingly, a similar pattern has been found for another gastropod species of the genus Lanistes endemic to Madagascar (Mahulu et al., 2021). The now available temporal framework of Bulinus evolution also allows specific colonization patterns and evolutionary patterns of endemism to be evaluated.
Colonization History and Evolution of Endemism
Members of the genus Bulinus have colonized altitudes from sea level to real alpine altitudes in some Afromontane regions. This study has extended the known altitudinal range of the genus by 900 m to around 4,000 m on the top of Mt. Elgon, where a new species of Bulinus has been characterized for the first time from such an extreme environment. Interestingly only four species (14% of the currently recognized biodiversity), all belonging to the B. truncatus/tropicus complex, evolved very high altitude occurrences (>2,500 m). All except B. tropicus are “true” high-altitude species whereas B. tropicus has an exceptional wide altitudinal range (500–3,100 m), the widest of all Bulinus species. To which extent the high-altitude populations of this species indeed belong to B. tropicus or also represent independent lineages needs to be shown in future studies. Bulinus sp. 7 LSH, previously considered to belong to B. tropicus, is a good example of how cryptic morphology can mask true phylogenetic and biogeographical patterns. The Bokong bogs (Lesotho) at 3,100 m, was recorded as the highest place for B. tropicus so far (Brown, 1994). It remains to be seen whether this indeed represents B. tropicus or rather Bulinus sp. 7 LSH as identified in this study. Although the sampling was certainly limited, it is noteworthy that no B. tropicus lineage was found in the highlands of Lesotho. Colonization might have occurred via a continuous upwards dispersal since Bulinus populations are usually found in all intermediate altitudes if suitable habitats are present and the slopes are not too steep. A notable exception it’s the conically shaped Mt. Elgon, where intermediate altitudes are free of Bulinus (Howell et al., 2012).
Altitudinal endemism is pronounced in the case of the Ethiopian species as well as the Kenyan Highlands. Here, phylogeographical sampling within these mountain ranges has to further disentangle the species status of several nominal taxa such as B. octoploidus in Ethiopia and B. permembranaceus in Kenya. This specifically applies to the Bulinus sp. 9 MTK which potentially includes the taxa B. laikipiensis, alluaudi, rumrutiensis (currently fallen in synonymy). It would also be interesting to test the hypothesis that polyploidisation might be linked to the ability to adapt to extreme altitudes (Brown, 1994). A striking finding of this study is the fact that high altitude Bulinus sp. 8 MTE from Mt. Elgon and Bulinus sp. 7 LSH from Lesotho are representing old lineages (= long branches), very isolated in the B. truncatus/tropicus complex with no immediate sister species currently known. The genetic distinctness of Bulinus sp. 8 MTE is very pronounced and might even warrant separation from the B. truncatus/tropicus complex. Even though this “stand alone” situation could potentially be a sampling artefact, it is the East African and Southern African regions that are comparatively well represented in previous phylogenetic studies of Bulinus spp. (Kane et al., 2008; Jørgensen et al., 2011; Jørgensen et al., 2013; Pennance, 2020), which render missing major lineages less likely. Given the phylogenetic pattern and acknowledging the sampling available to date, it appears as if living in high-altitudes is a condition already present in the MRCA of the B. truncatus/tropicus complex. It was an equally likely ancestral condition in the MRCA of the B. reticulatus group and the B. truncatus/tropicus complex. The character tracing revealed an equally likely colonization from altitudes from 0 to 2,000 m for Bulinus sp. 8 MTE. The Lesotho Highlands in contrast were colonized from either low (0–500 m) or high (>2,500 m) altitudes. Given the wide altitudinal ranges many Bulinus species occupy and their easy colonization capabilities, it is challenging to reconstruct ancestral conditions using phylogenetic approaches. Here, phylogeographical studies of species or species-groups might help to further elucidate the evolution of rare adaptations to extreme altitudes.
Role of Paleogeography and Paleoclimate
The Afromontane mountains are recognized as the most isolated mountain ranges in the world (see Mairal et al., 2021). This isolation has led to speciation and endemism in floras and faunas (Levin et al., 2020; Cuypers et al., 2022) and is also seen in Bulinus spp. It is noteworthy that isolation is apparently more pronounced in the Lesotho Mts. and on Mt. Elgon, where the oldest lineages of the B. truncatus/tropicus complex are found. There is no indication of hopping dispersal from “sky island” to “sky island” in these areas. Similarity in floras and faunas of Afromontane regions has been attributed to the existence of habitat corridors in the Holocene potentially facilitating dispersal between the various montane regions (Cooper, 2021). In Bulinus, this has likely not happened between the southern mountains and East Africa. To what extent such biotic exchange occurred between the Ethiopian Highlands and the Kenyan and Eastern Arc Mts. cannot be answered conclusively with the data at hand. Judging from the branching patterns and the molecular clock estimates it is less likely that such exchange happened as recently as after the last glacial maximum. Inter-mountain long distance dispersal has been shown for various taxa, the majority of which are terrestrial (Mairal et al., 2017; Mairal et al., 2021). This is also less likely in the case of Bulinus since no direct sister-group relationships were found. However, it is interesting since in a similar case of a lymnaeid gastropod (Galba mweruensis) such a pattern connecting widely isolated Afromontane populations was demonstrated, though the actual dispersal mechanism remained unknown (Mahulu et al., 2019). In the case of Bulinus it is likely that adaptation to cold climates (= high-altitudes) represents a form of niche conservatism which makes these lineages or species trapped in their specific environment with subsequent in situ speciation. This has been shown for pulmonate snails in other high-altitude regions (Albrecht et al., 2022). It is again noteworthy that such adaptations happened considerably earlier in the southern African Mts. and Mt. Elgon, i.e., in the Pliocene or Plio-Pleistocene transition times. Interestingly, this older or longer separation of populations from Lesotho and Mt. Elgon has also been found in Galba (Mahulu et al., 2019), with those populations from Mt. Elgon to be the most distinct ones as well. A striking feature remains regarding the roles of volcanic activities and glaciation of Mt. Elgon. Extensive icefields and glaciers existed on Mt. Elgon in the Late Pleistocene (Osmaston, 2004). Earlier (around 3 Ma), volcanic eruptions occurred on Mt. Elgon (Scott, 1998). Both conditions argue against a Pliocene colonization and uninterrupted existence of Bulinus populations on Mt. Elgon. Alternatively, a post-glacial long dispersal from a yet unknown source must be assumed, possibly located in East Africa. The current study clearly demonstrated that Afromontane regions represent “sky islands” for freshwater organism, here Bulinus gastropods. Rifting processes and associated climatic fluctuations have impacted the distribution patterns of Bulinus lineages at least since mid-Pliocene times. It is likely that recurrent climatic changes (Kohler et al., 2014) might lead to range shifts, reduction or even extinctions in high mountain populations. These populations have been postulated to be particularly sensitive to changing climates (Trew and Maclean, 2021). Such range shifts might also have significant implications for species of Bulinus that act as intermediate host for trematode diseases.
Implications for Schistosomiasis
Whereas Bulinus snails adapt to cold conditions, this is not necessarily the case for their parasites. Though our knowledge is still scanty for most trematode parasites, species of the genus Schistosoma received considerable attention due to their role in causing, e.g., cancerogenic urogenital schistosomiasis in humans (e.g., Rollinson, 2009). With warming climates, regions in altitudes where schistosomiasis was previously unknown, might become suitable for thermally restricted schistosomes. Warming means that known intermediate hosts might occur in high places in the future and that species not yet susceptible to schistosomes might become susceptible or get into contact with the parasites. A multiplex-PCR based screening of the specimens of Bulinus sp. 8 MTE did not detect infections (data not shown). These populations at altitudes of around 4,000 m are less likely to be visited by final hosts. Lower altitudes between 2,000 and 3,000 m are much more frequented in Afromontane regions. Bulinus permembranaceus and B. hexaploidus are currently not known to be intermediate hosts for human schistosomes (Brown, 1994). Bulinus octoploidus on the other hand could be experimentally infected with S. haematobium (Lo et al., 1970) and is naturally infected with S. bovis a major parasite causing livestock schistosomiasis (Brown, 1994). Schistosoma bovis is also known from B. tropicus and thus potentially also from Bulinus sp. 7 LSH. Future assessments of high-altitude species should use the newly developed xenomonitoring approaches (Pennance et al., 2020; Hammoud et al., 2022) to enhance our knowledge of snail-trematode communities in these particular species. This is relevant since the above mentioned shift of prevalence towards higher altitudes have already been documented for the Biomphalaria-S. mansoni system (John et al., 2008; Stanton et al., 2017). It is also desirable to carry out transect assessments along altitudinal gradients in all Afromontane regions (Howell et al., 2012; Stanton et al., 2017) as a baseline for tracing future shift of both snail and parasite communities. It would also help to more precisely define the actual and potential altitudinal limits of the disease that is so pertinent for both humans and livestock throughout Africa.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GenBank accession numbers of newly sequenced samples are provided in the Supplementary Table S1.
Author Contributions
IT and CA designed the study, CA and IT collected in the field and compiled data and wrote the draft manuscript. IT conducted lab work. IT and CC performed the analyses. FC organized field work in Tanzania and helped in the lab. JK facilitated work in Kenya. CA secured funding and did project administration. All authors read, contributed to the initial drafts, and approved the final version of the manuscript.
Funding
Funding was provided by the German research Foundation (DFG) to CA (Lesotho AL 1076/12-1, Kenya AL 1076/14-1, RiftLink AL1076/5-2). IT received a PhD scholarship provided by the German Academic Exchange Service (DAAD) whereas FC benefited from a post-doctoral fellowship of the Alexander von Humboldt Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the help of all collaborators in the field and for specimen donations. Particularly gratefully acknowledged are the following with countries they provided samples from in brackets: Ulrich Bößneck † (Lesotho, Tanzania), Anna Mahulu (Tanzania), Julius Tumusiime, Naboth Ainomugisha, Daniel Engelhard and Roland Schultheiß (Uganda), Thies Geertz (Uganda, Kenya), Russell Stothard (Uganda, Cameroon, Tanzania), Oscar Wembo Ndeo (DR Congo). Charles Lange facilitated the 2010 field campaign in Kenya. Help in the lab came from Silvia Nachtigall. We appreciate the fruitful discussion on phylogenies and biogeographical patterns with Björn Stelbrink and the helpful comments of the reviewers.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.902900/full#supplementary-material
References
Albrecht, C., Kuhn, K., and Streit, B. (2007). A Molecular Phylogeny of Planorboidea (Gastropoda, Pulmonata): Insights from Enhanced Taxon Sampling. Zool. Scr. 36 (1), 27–39. doi:10.1111/j.1463-6409.2006.00258.x
Albrecht, C., Stelbrink, B., and Clewing, C. (2019). “Bulinidae P. Fischer & Crosse, 1880,” in Freshwater Mollusks of the World (Baltimore, USA: Johns Hopkins University Press), 167–171.
Albrecht, C., Stelbrink, B., and Clewing, C. (2022). “Patterns and Processes of Speciation in Lymnaeidae,” in Vasquez: The Lymnaeidae: A Handbook on Their Natural History and Parasitological Significance. Editor M. A. A. Vinarski (Springer Books Series).
Allen, K. E., Tapondjou, W. P., Freeman, B., Cooper, J. C., Brown, R. M., and Peterson, A. T. (2021). Modelling Potential Pleistocene Habitat Corridors between Afromontane Forest Regions. Biodivers. Conserv. 30 (8), 2361–2375. doi:10.1007/s10531-021-02198-4
Almeyda-Artigas, R. J., Bargues, M. D., and Mas-Coma, S. (2000). Its-2 Rdna Sequencing of Gnathostoma Species (Nematoda) and Elucidation of the Species Causing Human Gnathostomiasis in the Americas. J. Parasitol. 86 (3), 537–544. doi:10.1645/0022-3395(2000)086[0537:irsogs]2.0.co;2
Antonelli, A., Kissling, W. D., Flantua, S. G. A., Bermúdez, M. A., Mulch, A., Muellner-Riehl, A. N., et al. (2018). Geological and Climatic Influences on Mountain Biodiversity. Nat. Geosci. 11 (10), 718–725. doi:10.1038/s41561-018-0236-z
Bargues, M. D., Vigo, M., Horak, P., Dvorak, J., Patzner, R. A., Pointier, J. P., et al. (2001). European Lymnaeidae (Mollusca: Gastropoda), Intermediate Hosts of Trematodiases, Based on Nuclear Ribosomal DNA ITS-2 Sequences. Infect. Genet. Evol. 1 (2), 85–107. doi:10.1016/S1567-1348(01)00019-3
Böhm, M., Dewhurst-Richman, N. I., Seddon, M., Ledger, S. E., Albrecht, C., Allen, D., et al. (2021). The Conservation Status of the World’s Freshwater Molluscs. Hydrobiologia 848, 3231–3254. doi:10.1007/s10750-020-04385-w
Bowles, J., Blair, D., and McManus, D. (1992). Genetic Variants within the Genus Echinococcus Identified by Mitochondrial DNA Sequencing. Mol. Biochem. Parasitol. 54 (2), 165–173. doi:10.1016/0166-6851(92)90109-W
Bößneck, U. (2012). “Leben am Limit: Besiedlung von Süßwasser-Habitaten extremer Hochlagen Asiens, Amerikas und Afrikas durch Mollusken (Mollusca: Bivalvia & Gastropoda),” in Biodiversität und Naturausstattung im Himalaya IV’. Editors M. Hartmann, and J. Weipert, 103–106.
Brindley, P. J., and Hotez, P. J. (2013). Break Out: Urogenital Schistosomiasis and Schistosoma haematobium Infection in the Post-genomic Era. Plos Negl. Trop. Dis. 7 (3), e1961. doi:10.1371/journal.pntd.0001961
Brown, D. S. (1994). Freshwater Snails of Africa and Their Medical Importance. London: Taylor & Francis.
Brühl, C. (1997). Flightless Insects: a Test Case for Historical Relationships of African Mountains. J. Biogeogr. 24 (2), 233–250. doi:10.1046/j.1365-2699.1997.00073.x
Burgess, N. D., Butynski, T. M., Cordeiro, N. J., Doggart, N. H., Fjeldså, J., Howell, K. M., et al. (2007). The Biological Importance of the Eastern Arc Mountains of Tanzania and Kenya. Biol. Conserv. 134 (2), 209–231. doi:10.1016/j.biocon.2006.08.015
Chibwana, F. D., Tumwebaze, I., Mahulu, A., Sands, A. F., and Albrecht, C. (2020). Assessing the Diversity and Distribution of Potential Intermediate Hosts Snails for Urogenital Schistosomiasis: Bulinus spp. (Gastropoda: Planorbidae) of Lake Victoria. Parasites Vectors 13 (1), 1–18. doi:10.1186/s13071-020-04281-1
Clewing, C., Geertz, T., Rassam, H., Woldekiros, T. H., and Albrecht, C. (2022). Freshwater Diversity at a Biogeographic Edge Zone: the High-Mountain Pea-Clams of Ethiopia. Syst. Biodivers. 20 (1), 1–15. doi:10.1080/14772000.2021.2005706
Clewing, C., Van Bocxlaer, B., and Albrecht, C. (2020). First Report of Extraordinary Corkscrew Gastropods of the Genus Bulinus in Lake Malawi. J. Gt. Lakes. Res. 46 (5), 1168–1175. doi:10.1016/j.jglr.2020.05.014
Colgan, D. J., Ponder, W. F., and Eggler, P. E. (2000). Gastropod Evolutionary Rates and Phylogenetic Relationships Assessed Using Partial 28S rDNA and Histone H3 Sequences. Zool. Scr. 29 (1), 29–63. doi:10.1046/j.1463-6409.2000.00021.x
Cooper, J. C. (2021). Hierarchical Analyses of Community Biogeography in the Afromontane Highlands. Front. Biogeogr. 13 (4), e51310. doi:10.21425/F5FBG51310
Couvreur, T. L. P., Dauby, G., Blach‐Overgaard, A., Deblauwe, V., Dessein, S., Droissart, V., et al. (2021). Tectonics, Climate and the Diversification of the Tropical African Terrestrial Flora and Fauna. Biol. Rev. 96 (1), 16–51. doi:10.1111/brv.12644
Cox, S. C., Prys-Jones, R. P., Habel, J. C., Amakobe, B. A., and Day, J. J. (2014). Niche Divergence Promotes Rapid Diversification of East African Sky Island White-Eyes (Aves: Zosteropidae). Mol. Ecol. 23 (16), 4103–4118. doi:10.1111/mec.12840
Cuypers, L. N., Sabuni, C., Šumbera, R., Aghová, T., Lišková, E., Leirs, H., et al. (2022). Biogeographical Importance of the Livingstone Mountains in Southern Tanzania: Comparative Genetic Structure of Small Non-Volant Mammals. Front. Ecol. Evol. 9, 1–11. doi:10.3389/fevo.2021.742851
Daniels, S. R., Bittencourt-Silva, G. B., Muianga, V., and Bayliss, J. (2020). Phylogenetics of the Freshwater Crab (Potamonautes MacLeay, 1838) Fauna from ‘sky Islands’ in Mozambique with the Description of a New Species (Brachyura: Potamoidea: Potamonautidae). Eur. J. Taxon. 716, 1. doi:10.5852/ejt.2020.716
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods. 9 (8), 772. doi:10.1038/nmeth.2109
De Kock, K., and Wolmarans, C. (2005a). Distribution and Habitats of Bulinus depressus and Possible Role as Intermediate Host of Economically Important Helminth Parasites in South Africa. Water sa. 31 (4), 491–496. doi:10.4314/wsa.v31i4.5140
De Kock, K., and Wolmarans, C. (2005b). Distribution and Habitats of the Bulinus africanus Species group, Snail Intermediate Hosts of Schistosoma haematobium and S. mattheei. Water sa. 31 (1), 117–125. doi:10.4314/wsa.v31i1.5128
De Leo, G. A., Stensgaard, A.-S., Sokolow, S. H., N’Goran, E. K., Chamberlin, A. J., Yang, G.-J., et al. (2020). Schistosomiasis and Climate Change. BMJ 371, m4324. doi:10.1136/bmj.m4324
DeBusk, G. H. (1998). A 37,500-year Pollen Record from Lake Malawi and Implications for the Biogeography of Afromontane Forests. J. Biogeogr. 25 (3), 479–500. doi:10.1046/j.1365-2699.1998.2530479.x
Diehl, E., Jauker, B., Albrecht, C., Wilke, T., and Wolters, V. (2018). “GIEßEN: University Collections: Justus Liebig University Gießen,” in Zoological Collections of Germany. Natural History Collections. Editor L. Beck (Cham: Springer), 373–381. doi:10.1007/978-3-319-44321-8_29
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29 (8), 1969–1973. doi:10.1093/molbev/mss075
Fischer, A., Blaschke, M., and Bässler, C. (2011). Altitudinal Gradients in Biodiversity Research: the State of the Art and Future Perspectives under Climate Change Aspects. Waldökologie, Landschaftsforsch. Naturschutz. 11, 35–47.
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 3 (5), 294–299.
Fujisawa, T., and Barraclough, T. G. (2013). Delimiting Species Using Single-Locus Data and the Generalized Mixed Yule Coalescent Approach: A Revised Method and Evaluation on Simulated Data Sets. Syst. Biol. 62 (5), 707–724. doi:10.1093/sysbio/syt033
Georgopoulou, E., Neubauer, T. A., Harzhauser, M., Kroh, A., and Mandic, O. (2016). Distribution Patterns of European Lacustrine Gastropods: a Result of Environmental Factors and Deglaciation History. Hydrobiologia 775 (1), 69–82. doi:10.1007/s10750-016-2713-y
Gernhard, T. (2008). The Conditioned Reconstructed Process. J. Theor. Biol. 253 (4), 769–778. doi:10.1016/j.jtbi.2008.04.005
Gillespie, R., Gillespie, R. G., and Clague, D. A. (2009). Encyclopedia of Islands (No. 2). Berkeley: University of California Press.
Hall, T. (1999). BioEdit: a User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hammoud, C., Mulero, S., Van Bocxlaer, B., Boissier, J., Verschuren, D., Albrecht, C., et al. (2022). Simultaneous Genotyping of Snails and Infecting Trematode Parasites Using High‐throughput Amplicon Sequencing. Mol. Ecol. Resour. 22 (2), 567–586. doi:10.1111/1755-0998.13492
Howell, A., Mugisha, L., Davies, J., LaCourse, E. J., Claridge, J., Williams, D. J., et al. (2012). Bovine Fasciolosis at Increasing Altitudes: Parasitological and Malacological Sampling on the Slopes of Mount Elgon, Uganda. Parasites Vectors 5 (1), 1–10. doi:10.1186/1756-3305-5-196
Hua, X., and Wiens, J. J. (2013). How Does Climate Influence Speciation? Am. Nat. 182 (1), 1–12. doi:10.1086/670690
Huelsenbeck, J. P., Nielsen, R., and Bollback, J. P. (2003). Stochastic Mapping of Morphological Characters Syst. Biol. 52 (2), 131–158. doi:10.1080/10635150390192780
Itescu, Y. (2019). Are island-Like Systems Biologically Similar to Islands? A Review of the Evidence Ecography. 42 (7), 1298–1314. doi:10.1111/ECOG.03951
John, R., Ezekiel, M., Philbert, C., and Andrew, A. (2008). Schistosomiasis Transmission at High Altitude Crater Lakes in Western Uganda. BMC Infect. Dis. 8 (1), 1–6. doi:10.1186/1471-2334-8-110
Jørgensen, A., Stothard, J. R., Madsen, H., Nalugwa, A., Nyakaana, S., and Rollinson, D. (2013). The ITS2 of the Genus Bulinus: Novel Secondary Structure Among Freshwater Snails and Potential New Taxonomic Markers. Acta Trop. 128 (2), 218–225. doi:10.1016/j.actatropica.2012.05.009
Jørgensen, A., Jørgensen, L. V. G., Kristensen, T. K., Madsen, H., and Stothard, J. R. (2007). Molecular Phylogenetic Investigations of Bulinus (Gastropoda: Planorbidae) in Lake Malawi with Comments on the Topological Incongruence between DNA Loci. Zool. Scr. 36 (6), 577–585. doi:10.1111/j.1463-6409.2007.00298.x
Jørgensen, A., Madsen, H., Nalugwa, A., Nyakaana, S., Rollinson, D., Stothard, J. R., et al. (2011). A Molecular Phylogenetic Analysis of Bulinus (Gastropoda: Planorbidae) with Conserved Nuclear Genes. Zool. Scr. 40 (2), 126–136. doi:10.1111/j.1463-6409.2010.00458.x
Kane, R. A., Stothard, J. R., Emery, A. M., and Rollinson, D. (2008). Molecular Characterization of Freshwater Snails in the Genus Bulinus: a Role for Barcodes? Parasites Vectors 1 (1), 15. doi:10.1186/1756-3305-1-15
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Briefings Bioinforma. 20 (4), 1160–1166. doi:10.1093/bib/bbx108
Kohler, T., Wehrli, A., and Jurek, M. (2014). Mountains and Climate Change: A Global Concern. Sustainable Mountain Development Series, 136. Bern, Switzerland: Centre for Development and Environment (CDE), Swiss Agency for Development and Cooperation (SDC) and Geographica Bernensia.
Kuiper, J. G. J. (1966). Les especes africaines du genre Pisidium, leur synonymie et leur distribution (Mollusca, Lamellibranchiata, Sphaeriidae). Tervuren 151, 1–77.
Levin, B. A., Simonov, E., Dgebuadze, Y. Y., Levina, M., and Golubtsov, A. S. (2020). In the Rivers: Multiple Adaptive Radiations of Cyprinid Fishes (Labeobarbus) in Ethiopian Highlands. Sci. Rep. 10 (1), 1–13. doi:10.1038/s41598-020-64350-4
Lo, C. T., Burch, J. B., and Schutte, C. H. J. (1970). Infection of Diploid Bulinus s.s with Schistosoma haematobium. Malacol. Rev. 3 (2), 121–126.
Loader, S. P., Sara Ceccarelli, F., Menegon, M., Howell, K. M., Kassahun, R., Mengistu, A. A., et al. (2014). Persistence and Stability of Eastern Afromontane Forests: Evidence from Brevicipitid Frogs. J. Biogeogr. 41 (9), 1781–1792. doi:10.1111/jbi.12331
Mahulu, A., Clewing, C., Stelbrink, B., Chibwana, F. D., Tumwebaze, I., Russell Stothard, J., et al. (2019). Cryptic Intermediate Snail Host of the Liver Fluke Fasciola hepatica in Africa. Parasites Vectors 12 (1), 1–11. doi:10.1186/s13071-019-3825-9
Mahulu, A., Stelbrink, B., Van Bocxlaer, B., Riedel, F., and Albrecht, C. (2021). Going with the Flow? Diversification of Gastropods Reflects Drainage Evolution in Africa. J. Biogeogr. 48 (7), 1579–1593. doi:10.1111/jbi.14096
Mairal, M., Namaganda, M., Gizaw, A., Chala, D., Brochmann, C., and Catalán, P. (2021). Multiple Mountain‐hopping Colonization of Sky‐islands on the Two Sides of Tropical Africa during the Pleistocene: The Afroalpine Festuca Grasses. J. Biogeogr. 48 (8), 1858–1874. doi:10.1111/jbi.14117
Mairal, M., Sanmartín, I., Herrero, A., Pokorny, L., Vargas, P., Aldasoro, J. J., et al. (2017). Geographic Barriers and Pleistocene Climate Change Shaped Patterns of Genetic Variation in the Eastern Afromontane Biodiversity Hotspot. Sci. Rep. 7 (1), 1–13. doi:10.1038/srep45749
Mandahl-Barth, G. (1957). Intermediate Hosts of Schistosoma; African Biomphalaria and Bulinus. I. Monogr. Ser. World Health Organ 57 (1), 1–131.
Mandahl-Barth, G. (1965). The Species of the Genus Bulinus, Intermediate Hosts of Schistosoma. Bull. World Health Organ 33 (1), 33–44.
Martín-Queller, E., Albert, C. H., Dumas, P.-J., and Saatkamp, A. (2017). Islands, Mainland, and Terrestrial Fragments: How Isolation Shapes Plant Diversity. Ecol. Evol. 7 (17), 6904–6917. doi:10.1002/ece3.3150
Masters, J. C., Génin, F., Zhang, Y., Pellen, R., Huck, T., Mazza, P. P. A., et al. (2021). Biogeographic Mechanisms Involved in the Colonization of Madagascar by African Vertebrates: Rifting, Rafting and Runways. J. Biogeogr. 48 (3), 492–510. doi:10.1111/jbi.14032
McCormack, J. E., Huang, H., Knowles, L. L., Gillespie, R., and Clague, D. (2009). Sky Islands. Encycl. Isl. 4, 841–843.
Measey, G. J., and Tolley, K. A. (2011). Sequential Fragmentation of Pleistocene Forests in an East Africa Biodiversity Hotspot: Chameleons as a Model to Track Forest History. Plos one 6 (10), e26606. doi:10.1371/journal.pone.0026606
Menegon, M., Loader, S. P., Marsden, S. J., Branch, W. R., Davenport, T. R. B., and Ursenbacher, S. (2014). The Genus Atheris (Serpentes: Viperidae) in East Africa: Phylogeny and the Role of Rifting and Climate in Shaping the Current Pattern of Species Diversity. Mol. Phylogenetics Evol. 79, 12–22. doi:10.1016/j.ympev.2014.06.007
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Gateway Computing Environments Workshop. New Orleans: GCE, 1–8. doi:10.1109/GCE.2010.5676129
Mittermeier, R. A., Turner, W. R., Larsen, F. W., Brooks, T. M., and Gascon, C. (2011). “Global Biodiversity Conservation: The Critical Role of Hotspots,” in Biodiversity Hotspots. Editors F. Zachos, and J. Habel (Berlin, Heidelberg: Springer), 3–22. doi:10.1007/978-3-642-20992-5_1
Musonge, P. S. L., Boets, P., Lock, K., and Goethals, P. L. M. (2020). Drivers of Benthic Macroinvertebrate Assemblages in Equatorial Alpine Rivers of the Rwenzoris (Uganda). Water 12 (6), 1668. doi:10.3390/w12061668
Nalugwa, A., Joslash, A., Nyakaana, S., and Kristensen, T. K. (2010). Molecular Phylogeny of Bulinus (Gastropoda: Planorbidae) Reveals the Presence of Three Species Complexes in the Albertine Rift Freshwater Bodies. IJGMB 2 (7), 130–139.
Osmaston, H. (2004). Quaternary Glaciations in the East African Mountains. Dev. Quat. Sci. 2, 139–150. doi:10.1016/s1571-0866(04)80119-2
Palumbi, S. R., Martin, A., Romano, S., McMillan, W. O., Stice, L., and Grabowski, G. (1991). The Simple Fool’s Guide to PCR. Honolulu: University of Hawaii.
Pennance, T., Archer, J., Lugli, E. B., Rostron, P., Llanwarne, F., Ali, S. M., et al. (2020). Development of a Molecular Snail Xenomonitoring Assay to Detect Schistosoma haematobium and Schistosoma bovis Infections in Their Bulinus Snail Hosts. Molecules 25 (17), 4011. doi:10.3390/molecules25174011
Pennance, T. (2020). Genetic Diversity and Evolution within the Genus Bulinus and Species-Level Interactions with the Transmission of Schistosoma Haematobium Group Parasites. Cardiff: Doctoral dissertation, Cardiff University.
Perrigo, A., Hoorn, C., and Antonelli, A. (2020). Why Mountains Matter for Biodiversity. J. Biogeogr. 47 (2), 315–325. doi:10.1111/jbi.13731
Pickford, M. (2008). Freshwater and Terrestrial Mollusca from the Early Miocene Deposits of the Northern Sperrgebiet, Namibia. Memoirs Geol. Surv. Namibi 20, 65–74.
Platts, P. J., Gereau, R. E., Burgess, N. D., and Marchant, R. (2013). Spatial Heterogeneity of Climate Change in an Afromontane Centre of Endemism. Ecography 36 (4), 518–530. doi:10.1111/j.1600-0587.2012.07805.x
Pons, J., Barraclough, T. G., Gomez-Zurita, J., Cardoso, A., Duran, D. P., Hazell, S., et al. (2006). Sequence-based Species Delimitation for the DNA Taxonomy of Undescribed Insects. Syst. Biol. 55 (4), 595–609. doi:10.1080/10635150600852011
R Core Team (2021). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: http://www.R-project.org.
Rahbek, C., Borregaard, M. K., Antonelli, A., Colwell, R. K., Holt, B. G., Nogues-Bravo, D., et al. (2019). Building Mountain Biodiversity: Geological and Evolutionary Processes. Science 365 (6458), 1114–1119. doi:10.1126/science.aax0151
Revell, L. J. (2012). Phytools: an R Package for Phylogenetic Comparative Biology (And Other Things). Methods Ecol. Evol. 3, 217–223. doi:10.1111/j.2041-210X.2011.00169.x
Rollinson, D. (2009). A Wake up Call for Urinary Schistosomiasis: Reconciling Research Effort with Public Health Importance. Parasitology 136 (12), 1593–1610. doi:10.1017/S0031182009990552
Scott, P. (1998). From Conflict to Collaboration: People and Forests at Mount Elgon, Uganda. Gland, Switzerland and Cambridge, UK: IUCN, 158.
Stanton, M. C., Adriko, M., Arinaitwe, M., Howell, A., Davies, J., Allison, G., et al. (2017). Intestinal Schistosomiasis in Uganda at High Altitude (>1400 M): Malacological and Epidemiological Surveys on Mount Elgon and in Fort Portal Crater Lakes Reveal Extra Preventive Chemotherapy Needs. Infect. Dis. Poverty 6 (1), 1–10. doi:10.1186/s40249-017-0248-8
Steinbauer, M. J., Field, R., Grytnes, J.-A., Trigas, P., Ah-Peng, C., Attorre, F., et al. (2016). Topography-driven Isolation, Speciation and a Global Increase of Endemism with Elevation. Glob. Ecol. Biogeogr. 25 (9), 1097–1107. doi:10.1111/geb.12469
Stensgaard, A.-S., Vounatsou, P., Sengupta, M. E., and Utzinger, J. (2019). Schistosomes, Snails and Climate Change: Current Trends and Future Expectations. Acta Trop. 190, 257–268. doi:10.1016/j.actatropica.2018.09.013
Stothard, J. R., Brémond, P., Andriamaro, L., Sellin, B., Sellin, E., and Rollinson, D. (2001). Bulinus Species on Madagascar: Molecular Evolution, Genetic Markers and Compatibility with Schistosoma Haematobium. Parasitology 123 (7), 261–275. doi:10.1017/S003118200100806X
Stothard, J. R., Hughes, S., and Rollinson, D. (1996). Variation within the Internal Transcribed Spacer (ITS) of Ribosomal DNA Genes of Intermediate Snail Hosts within the Genus Bulinus (Gastropoda: Planorbidae). Acta Trop. 61 (1), 19–29. doi:10.1016/0001-706X(95)00137-4
Trew, B. T., and Maclean, I. M. D. (2021). Vulnerability of Global Biodiversity Hotspots to Climate Change. Glob. Ecol. Biogeogr. 30 (4), 768–783. doi:10.1111/geb.13272
Tumwebaze, I., Clewing, C., Dusabe, M. C., Tumusiime, J., Kagoro-Rugunda, G., Hammoud, C., et al. (2019). Molecular Identification of Bulinus spp. Intermediate Host Snails of Schistosoma spp. In Crater Lakes of Western Uganda with Implications for the Transmission of the Schistosoma haematobium Group Parasites. Parasites Vectors 12 (1), 1–23. doi:10.1186/s13071-019-3811-2
Vaidya, G., Lohman, D. J., and Meier, R. (2011). Sequence Matrix: Concatenation Software for the Fast Assembly of Multi-Gene Datasets with Character Set and Codon Information. Cladistics 27 (2), 171–180. doi:10.1111/j.1096-0031.2010.00329.x
White, F. (1981). The History of the Afromontane Archipelago and the Scientific Need for its Conservation. Afr. J. Ecol. 19 (1‐2), 33–54. doi:10.1111/j.1365-2028.1981.tb00651.x
Wilke, T., and Davis, G. M. (2000). Infraspecific Mitochondrial Sequence Diversity in Hydrobia ulvae and Hydrobia Ventrosa (Hydrobiidae: Rissooidea: Gastropoda): Do Their Different Life Histories Affect Biogeographic Patterns and Gene Flow? Biol. J. Linn. Soc. 70 (1), 89–105. doi:10.1111/j.1095-8312.2000.tb00202.x
Wilke, T., Davis, G. M., Qiu, D., and Spear, R. C. (2006). Extreme Mitochondrial Sequence Diversity in the Intermediate Schistosomiasis Host Oncomelania hupensis robertsoni: Another Case of Ancestral Polymorphism? Malacologia 48 (1/2), 143–157.
Wilke, T., Schultheiß, R., and Albrecht, C. (2009). As Time Goes by: A Simple Fool's Guide to Molecular Clock Approaches in Invertebrates*. Am. Malacol. Bull. 27 (1/2), 25–45. doi:10.4003/006.027.0203
Wright, C. A. (1971). Bulinus on Aldabra and the Subfamily Bulininae in the Indian Ocean Area. Phil. Trans. R. Soc. Lond. B 260, 299–313. doi:10.1098/rstb.1971.0016
Young, N. D., Kinkar, L., Stroehlein, A. J., Korhonen, P. K., Stothard, J. R., Rollinson, D., et al. (2021). Mitochondrial Genome of Bulinus truncatus (Gastropoda: Lymnaeoidea): Implications for Snail Systematics and Schistosome Epidemiology. Curr. Res. Parasitol. Vector-Borne Dis. 1, 100017. doi:10.1016/j.crpvbd.2021.100017
Young, N. D., Stroehlein, A. J., Wang, T., Korhonen, P. K., Mentink-Kane, M., Stothard, J. R., et al. (2022). Nuclear Genome of Bulinust truncatus, an Intermediate Host of the Carcinogenic Human Blood Fluke Schistosoma haematobium. Nat. Commun. 13 (1), 1–15. doi:10.1038/s41467-022-28634-9
Zein-Eddine, R., Djuikwo-Teukeng, F. F., Al-Jawhari, M., Senghor, B., Huyse, T., and Dreyfuss, G. (2014). Phylogeny of Seven Bulinus Species Originating from Endemic Areas in Three African Countries, in Relation to the Human Blood Fluke Schistosoma Haematobium. BMC Evol. Biol. 14 (1), 1–12. doi:10.1186/s12862-014-0271-3
Zhang, J., Kapli, P., Pavlidis, P., and Stamatakis, A. (2013). A General Species Delimitation Method with Applications to Phylogenetic Placements. Bioinformatics 29 (22), 2869–2876. doi:10.1093/bioinformatics/btt499
Zhang, S. M., Bu, L., Lu, L., Babbitt, C., Adema, C. M., and Loker, E. S. (2022). Comparative Mitogenomics of Freshwater Snails of the Genus Bulinus, Obligatory Vectors of Schistosoma haematobium, Causative Agent of Human Urogenital Schistosomiasis. Sci. Rep. 12 (1), 1–10. doi:10.1038/s41598-022-09305-7
Keywords: Afrotropics, endemism, sky islands, biodiversity hotspots, schistosomiasis, climate change, high-altitude, intermediate host snails
Citation: Tumwebaze I, Clewing C, Chibwana FD, Kipyegon JK and Albrecht C (2022) Evolution and Biogeography of Freshwater Snails of the Genus Bulinus (Gastropoda) in Afromontane Extreme Environments. Front. Environ. Sci. 10:902900. doi: 10.3389/fenvs.2022.902900
Received: 23 March 2022; Accepted: 19 April 2022;
Published: 17 May 2022.
Edited by:
Tatenda Dalu, University of Mpumalanga, South AfricaReviewed by:
Dilian Georgiev, Plovdiv University “Paisii Hilendarski”, BulgariaZolile Maseko, Rhodes University, South Africa
Copyright © 2022 Tumwebaze, Clewing, Chibwana, Kipyegon and Albrecht. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: I. Tumwebaze, dHVtd2ViYXplaW1tYWN1bGF0ZUBnbWFpbC5jb20=, SW1tYWN1bGF0ZS5UdW13ZWJhemVAYmlvLnVuaS1naWVzc2VuLmRl
 I. Tumwebaze
I. Tumwebaze C. Clewing
C. Clewing F. D. Chibwana2
F. D. Chibwana2 J. K. Kipyegon
J. K. Kipyegon C. Albrecht
C. Albrecht