
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Environ. Sci., 05 July 2022
Sec. Water and Wastewater Management
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.887093
This article is part of the Research TopicSustainable Technologies for the Treatment of Sewage SludgeView all 5 articles
With the increasing use of animal and plant proteins, pollution due to nitrogen sources is attracting increasing attention. In particular, the amount of nitrogen-containing sewage discharged into the environment has increased significantly, causing eutrophication of water bodies and environmental degradation of water quality. Traditionally, nitrifying bacteria perform ammonia nitrification under aerobic conditions, while denitrifying bacteria perform nitrate/nitrite denitrification under anaerobic conditions. However, heterotrophic nitrifying and aerobic denitrifying microorganisms (HNADs) perform ammonia nitrification and nitrate/nitrite denitrification under the same aerobic conditions using an organic carbon source, which is a much simpler and more efficient process. In this review, the distribution and evolutionary relationships of novel HNADs strains are presented, and the influencing factors, metabolic pathways, key enzymes, and practical applications of HNADs are reviewed.
With the increasing use of animal and plant proteins in the farming industry, pollution due to nitrogen sources is attracting increasing attention. In particular, the amount of nitrogen-containing sewage discharged into the environment has increased significantly, and this has led to various ecological health and environmental safety issues, such as the eutrophication of water bodies (Zhu et al., 2012). Eutrophication causes algal blooms that threaten the ecosystem function of freshwater, aquaculture safety, and public health, and it is becoming a major environmental problem (Tang et al., 2019). Therefore, treating nitrogen-containing wastewater has become an important element of environmental management related to sustainable development of the environment.
Conventionally, high-concentration nitrogen-containing wastewater is treated using physical, chemical, or biological methods (Zhang et al., 2011). The physical method mainly uses physical adsorption, which only adsorbs ammonia nitrogen and nitrite in the water body on the solid surface, and does not fundamentally reduce the content of ammonia nitrogen and nitrite in the water body. Chemical method in the use of the same high cost, small range, low security, and other disadvantages. Biological method mainly uses nitrifying bacteria and denitrifying bacteria to convert ammonia nitrogen and nitrite into nitrogen gas through nitrification and denitrification, because the reaction conditions are mild and can reduce the total nitrogen content in the water body, and is the safest and most effective method (Khardenavis et al., 2007). In particular, heterotrophic nitrifying and aerobic denitrifying microorganisms (hereinafter referred to as HNADs) have been studied increasingly since Thiosphaera pantotropha was isolated in the 1980s (Robertson and Kuenen, 1983), and these include Acinetobacter (An et al., 2020), Alcaligenes (Chen et al., 2021), Arthrobacter (He et al., 2017), Klebsiella (Padhi et al., 2013), Bacillus (Rout et al., 2017), Enterobacter (Guo et al., 2016), Paracoccus (Zhang et al., 2018a), and Pseudomonas (He et al., 2016). Heterotrophic nitrifying and aerobic denitrifying microorganisms (HNADs) can use organic matter as a carbon source to oxidize ammonia to nitrite and nitrate, while converting nitrate and nitrite to gaseous nitrogen under aerobic conditions, resulting in both nitrification and denitrification (He X. et al., 2019). Compared with the conventional autotrophic nitrification and anaerobic denitrification processes, HNADs are more streamlined and offer smaller reactor size, lower cost, and higher nitrogen-removal efficiency (Duan et al., 2015).
Heterotrophic nitrification–aerobic denitrification is a biological nitrogen removal technology with unique advantages compared with traditional anaerobic denitrification and autotrophic aerobic denitrification proposed in recent years. On the one hand, denitrification is carried out under aerobic conditions, nitrification and denitrification can be carried out simultaneously in one reactor, and equipment and operation costs are significantly reduced; on the other hand, compared with the traditional autotrophic nitrifying bacteria, heterotrophic nitrifying bacteria have a short generation cycle and rapid growth, which can be applied to the environment of organic wastewater. The products of nitrification can be directly used as substrate for denitrification, which avoids the inhibition of nitrification and strengthens the nitrification and denitrification processes.
This review summarizes the recent research on HNAD strains and their physiological characteristics and metabolic pathways. The distribution and evolutionary relationship of novel HNAD strains are presented, and the influencing factors, metabolic pathways, and critical enzymes of HNADs are reviewed.
Recently, new advances have been made in studying HNADs from various environmental sources, and many new microbial strains have been screened and identified. The domain, strain, nitrogen source, and isolation environment of each of these reported HNADs are given in Table 1. HNADs are found mostly in soil, sludge, wastewater, and lakes because these environments contain abundant nitrogen sources. However, despite different HNADs being isolated from different environments, their HNAD function is the same. The reported HNADs include not only bacteria but also fungi and yeasts, and their scope is thus relatively wide. Figure 1 shows partial 16S rRNA sequences of HNAD bacteria downloaded from GenBank, with their evolutionary relationship tree plotted.
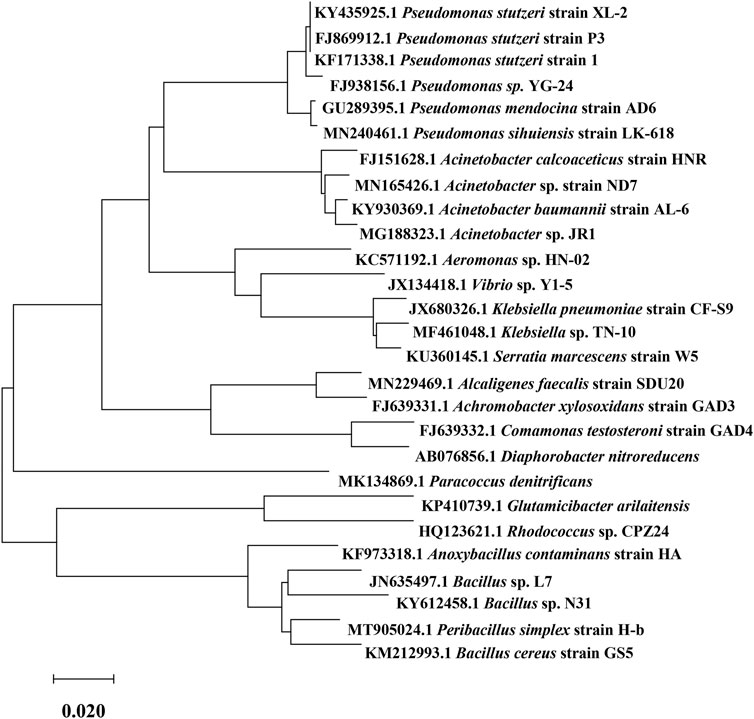
FIGURE 1. Evolutionary relationship tree of heterotrophic nitrifying and aerobic denitrifying (HNAD) bacteria.
Compared to traditional denitrifying bacteria that can only use nitrate, many HNADs can also use ammonia and/or nitrite and convert them to gaseous nitrogen and biomass through the aerobic denitrification pathway and assimilation process (Chen and Ni, 2012; Xia et al., 2020). Under anaerobic conditions, denitrifying bacteria usually use nitrate as the final electron acceptor, but HNADs use oxygen as the final electron acceptor, thereby replacing nitrate for electron transfer in the aerobic respiratory chain and generating more energy. Therefore, HNADs are better in terms of growth rate and nitrogen-removal efficiency. When Pseudomonas tolaasii Y-11was cultivated with an initial nitrate concentration of over 200 mg/L, 93.5% of the nitrate was removed in 4 days without nitrite accumulation via the denitrification pathway (He et al., 2016). In Pseudomonas stutzeri ZF31, calculated by nitrogen balance, the results showed that about 75% of the initial nitrate was converted to gaseous nitrogen (Huang et al., 2015). Ammonia nitrogen can also be converted to gaseous nitrogen via the denitrification pathway by HNADs. For example, Acinetobacter sp. JR1 can use ammonium nitrogen as the sole nitrogen source for conversion into gaseous nitrogen under aerobic conditions (Yang et al., 2019). Similar results were reported for P. stutzeri YZN-001 (Zhang et al., 2011), Acinetobacter sp. YS2 (Lang et al., 2020), and mixed Bacillus strains (Kim et al., 2005). In addition, some HNADs such as Rhodococcus erythropolis Y10 (Ma et al., 2021) remove ammonia nitrogen by assimilation, converting the nitrogen source into biological nitrogen for their own growth.
The conventional ammonium nitrification pathway is coordinated by two types of bacteria: (i) ammonia-oxidizing bacteria, which are responsible for oxidizing ammonium to nitrite; and (ii) nitrite-oxidizing bacteria, which are responsible for oxidizing nitrite to nitrate. Nitrate is finally converted to gaseous nitrogen by heterotrophic denitrifying bacteria via the denitrification pathway under anaerobic conditions (Jaroszynski and Oleszkiewicz, 2011). The conversion efficiency of HNADs is better than that of the conventional process of ammonium removal because there are differences in intermediate products and key catalytic enzymes.
In the 1990s, a classical coupled model of ammonium nitrification in HNAD bacteria was reported (Wehrfritz et al., 1993). Ammonium is oxidized to hydroxylamine in the periplasm by ammonia monooxygenase (AMO), followed by oxidation to nitrite by hydroxylamine oxidoreductase (HAO), and finally to nitrate in the cytoplasm (Song et al., 2021). In HNADs, nitrate is reduced via denitrification by a series of oxidoreductases under aerobic conditions, and stable isotopes and enzyme inhibitors are used for more-accurate detection of intermediate metabolites in the study of nitrogen conversion pathways in HNADs.
Alcaligenes faecalis NR can convert ammonium to N2O and N2 with ammonium as the sole nitrogen source, and intermediate nitrate and nitrite are not detected under aerobic conditions (Zhao et al., 2012). When hydroxylamine is the sole nitrogen source, gaseous nitrogen is still detected, but only hydroxylamine oxidase activity is detected, and nitrate and nitrite reductase activity is undetected (Zhao et al., 2012). Hydroxylamine is converted directly to nitrous oxide replacing nitrite, and a similar finding has been reported for Acinetobacter calcoaceticus HNR (Zhao et al., 2010). It has been speculated that the ammonium-removal pathway of A. calcoaceticus HNR and A. faecalis NR is NH4+-N→NH2OH→N2O→N2 (a shortcut ammonium metabolic pathway), and experiments have confirmed that Photobacterium sp. NNA4 can transform hydroxylamine directly to N2O when hydroxylamine is the sole nitrogen source (Liu et al., 2019).
Enzyme activation inhibitors can block metabolic pathways, resulting in the accumulation of intermediate products or the disappearance of end products, and they are often used to study the metabolic pathways of nitrogen conversion. In Alcaligenes sp. TB, when nitrite reductase inhibitor (pb2+) is present, the accumulation of nitrate and nitrite is detected, but gaseous nitrogen is not detected, whereas when nitrate reductase inhibitor (Na2WO4) is present, the accumulation of nitrate and gaseous nitrogen is detected simultaneously; these results show that the strain TB carries out ammonium removal via the pathway NH4+-N→NH2OH→NO2−-N→N2O→N2 (an incomplete ammonium metabolic pathway) (Chen et al., 2016). The production of gaseous nitrogen is found to be accompanied by the interconversion of nitrite and nitrate, which has similarly been reported for Agrobacterium sp. LAD9 (Chen and Ni, 2012). In addition, Alcaligenes sp. TB can convert approximately 35.7% of the ammonium to gaseous nitrogen without enzyme inhibitors via another pathway, that is, NH4+-N→NH2OH→NO2−-N→NO3−-N→NO2−-N→N2O→N2 (a complete ammonium metabolic pathway) (Chen et al., 2016). The same nitrogen-removal pathway has been reported for B. subtilis A1 (Yang et al., 2011), K. pneumoniae CF-S9 (Padhi et al., 2013), B. cereus GS-5 (Rout et al., 2017), and Rhodococcus sp. CPZ24 (Chen et al., 2012).
Different nitrogen-source combinations also affect the denitrification pathway of ammonium. When ammonium and nitrite are present simultaneously, the ammonium is transformed directly to gaseous nitrogen via a shortcut ammonium metabolic pathway, but when ammonium and nitrate are present simultaneously, the ammonium is first transformed to nitrate and then reduced to gaseous nitrogen via a complete ammonium metabolic pathway (Wei et al., 2021). Furthermore, compared with the traditional autotrophic nitrification pathway, the hydroxylamine pathway is simpler, which makes it more advantageous to use HNADs in treating high-concentration nitrogenous wastewater (Peng and Zhu, 2006).
The reduction of nitrate to gaseous nitrogen by HNADs is accomplished via four reductases under aerobic conditions in four processes, that is, NO3−-N→NO2−-N→NO→N2O→N2 (Song et al., 2021). Nitrite reductase is also a bottleneck in the denitrification pathway of HNADs because it is as sensitive to oxygen as are traditional anaerobic denitrifying bacteria. The ability of P. stutzeri T13 to convert nitrate efficiently with nitrate as the sole nitrogen source has been investigated, finding that the conversion efficiency of intermediate nitrite metabolites is significantly higher at low levels of dissolved oxygen (DO) than at high levels (Sun et al., 2015). This sensitivity to oxygen inhibits the reduction of nitrite, and it accumulates during denitrification with high DO levels. Some HNAD bacteria have different properties and are effective in converting nitrite to gaseous nitrogen at high DO levels, such as P. stutzeri ZF31, P. mendocina X49, and Vibrio sp. AD2 (Huang et al., 2015; Ren et al., 2021; Xie et al., 2021).
Nitrite is an intermediate product of the denitrification metabolic pathway, and because of its cytotoxicity, few HNADs can use nitrite directly for denitrification conversion. It has been reported that Pseudomonas sp. yy7 can use nitrite via assimilation and denitrification (Wan et al., 2011), and isotope-tracking experiments have shown that P. stutzeri converts nitrous oxide (N2O)—a precursor of denitrification—to nitrogen gas (N2) under aerobic conditions (Desloover et al., 2014). The denitrification pathway with nitrite or nitrous oxide as the initial nitrogen source is an incomplete metabolic pathway.
Assimilation synthesizes proteins and nucleic acids required for microbial growth, which are important for both cell growth and nitrogen conversion. It has been reported that R. erythropolis Y10 can use ammonium, nitrate, and nitrite via the assimilation pathway; 98.23% of the nitrogen is assimilated to biomass nitrogen with ammonium as the sole nitrogen source, much higher than the 52.73 and 56.22% for nitrate and nitrite, respectively (Ma et al., 2021). Vibrio sp. Y1-5 can assimilate nitrate to biomass nitrogen under aerobic conditions, and the assimilation efficiency increases with increasing DO concentration (Li et al., 2017).
Transcriptomics and the expression of critical enzyme genes have been used to study the assimilation of metabolic pathways in Klebsiella sp. KSND when using different nitrogen sources under aerobic conditions, and the results showed that the transcription levels of NADP-glutamate dehydrogenase increased by a factor of approximately 8.6 and glutamine synthetase was not detected. This suggests that NH4+-N assimilation is through the NADP-glutamate dehydrogenase pathway in Klebsiella sp. KSND (Jin et al., 2019).
A new nitrate-dissimilation conversion pathway (NO3−-N→NO2−-N→NH4+-N) for Pseudomonas putida Y-9 was reported via 15N isotope experiments in which nitrate was reduced to ammonium under aerobic conditions (Huang X. et al., 2020). This pathway occurs under not only aerobic conditions but also anaerobic ones (Yoon et al., 2015).
The nitrogen conversion pathways of HNADs are shown in Figure 2 and include mainly ammonia nitrification, nitrate denitrification, nitrite incomplete denitrification, and nitrate assimilation/dissimilation under aerobic conditions.
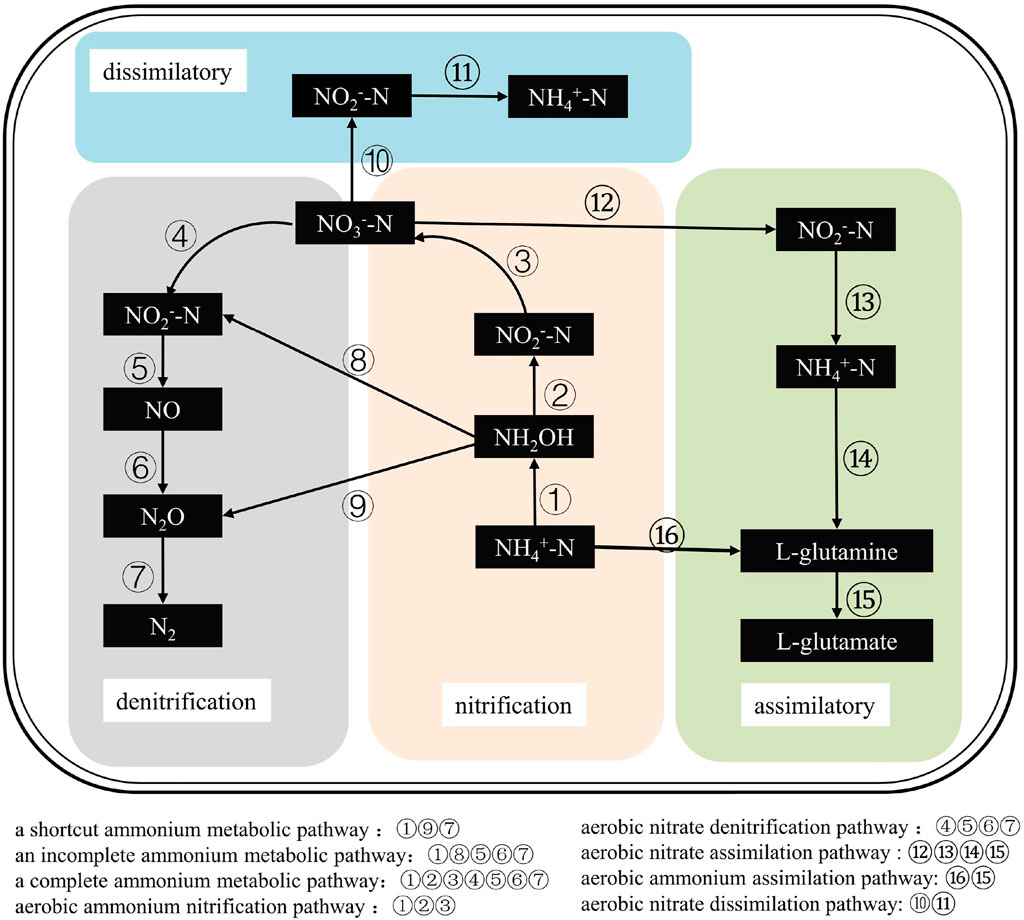
FIGURE 2. The nitrogen conversion pathways of HNADs, which include mainly ammonia nitrification, nitrate denitrification, nitrite incomplete denitrification, and nitrate assimilation/dissimilation under aerobic conditions.
The reduction of nitrate to nitrite is the start of the denitrification reaction and is carried out by NR. Outside the cell is periplasmic nitrate reductase (Nap), and bound to the cell membrane is nitrate reductase (Nar) (Figure 3) (Vivián et al., 1999).
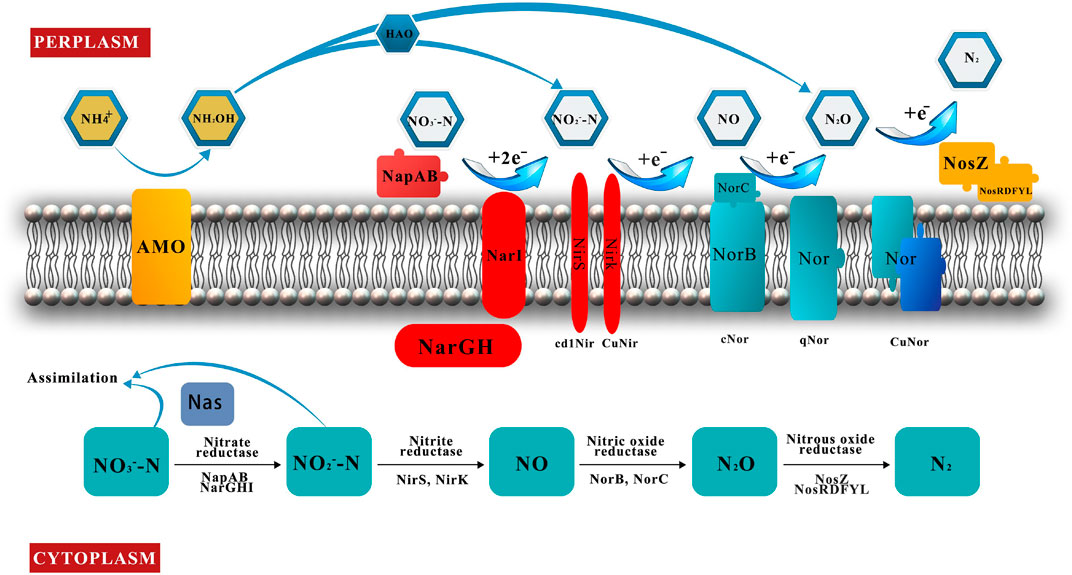
FIGURE 3. Schematic of nitrification, denitrification, and assimilation reactions under aerobic conditions.
Nar is a Mo-containing enzyme that has three catalytic subunits: NarG, NarH, and NarI. NarG is the largest catalytic subunit, comprising a [4Fe-4S] center (FS0) and a Moco active site. NarH binds to NarG and is located in the cytoplasm, containing three [4Fe-4S] centers (FS1, FS2, and FS3) and one structurally distinct [3Fe-4S] center (FS4); it is mainly responsible for electron transfer. NarI is the smallest subunit; it is anchored to the cell membrane and is responsible for the transmembrane movement of protons (Coelho and Romao, 2015). The expression of Nar has been detected in B. cereus GS-5 under aerobic conditions and was encoded by 561 bp nar (Rout et al., 2017).
Nap is an extracellular enzyme that reduces nitrate in the periplasm under aerobic conditions and plays a critical role in HNADs; it was first purified from Paracoccus pantotrophus (Sears et al., 1995; Yang J. et al., 2020). Nap may be a key factor for nitrogen conversion by HNAD bacteria under aerobic conditions. The first crystal structures of periplasmic nitrate reductases were reported in Desulfovibrio desulfuricans at 1.9-Å resolution (Dias et al., 1999). Nap contains two subunits, that is, NapA and NapB, and the genes encoding these are napA and napB, respectively; napA has been successfully amplified in P. stutzeri XL-2 (Zhao et al., 2018).
Nitrite is converted to nitric oxide catalyzed via NIR, which contains two types of catalase: cd1Nir and CuNir (Figure 3). These two enzymes are nonhomologous isozymes and usually cannot coexist in the same microorganism (Zumft et al., 1994). However, the coexistence of two enzymes has been reported in the same microorganism, such as in Bradyrhizobium oligotrophicum S58 (Sanchez and Minamisawa, 2018) and Rhodothermus marinus (Graf et al., 2014).
Cd1Nir contains two identical subunits with cofactors heme d1 and heme c; it is a homologous dimer protein and is encoded by nirS (Nojiri et al., 2009). In Agrobacterium sp. LAD9, 799 bp of the nirS gene fragment have been amplified under aerobic conditions (Chen and Ni, 2012).
CuNir is a homologous trimeric protein containing three identical subunits with two Cu-binding sites on each (Godden et al., 1991). CuNir is usually encoded by the nirK gene and sometimes by the nirV gene (Yang J. et al., 2020). The nirK gene of nitrite reductases in S. pararoseus Y1 and in Alcaligenes faecalis NR has been amplified (Huang et al., 2018; Zeng et al., 2020).
NOR is embedded in the cell membrane with the catalytic site in the periplasm and is responsible for the reduction of nitric oxide to nitrous oxide (Figure 3). NOR is divided into three types according to the differences in proton and electron transfer centers: cNor, qNor, and CuANor (Hino et al., 2010; Matsumoto et al., 2012).
Two structurally distinct subunits (NorB and NorC) constitute cNor. NorC is a smaller subunit that is on the periplasmic side and has one c-type heme as its electron acceptor. NorB is a larger subunit containing three metal catalytic centers, that is, heme b, heme b3, and nonheme FeB. A segment of NOR-encoding gene (2,904 bp norB) that can be translated into a 759 amino acid sequence has been amplified successfully in Alcaligenes faecalis NR (Huang et al., 2018). qNor contains only one subunit without heme, and CuANor has two subunits and has been reported in Bacillus (Suharti and De Vries, 2005).
NOS reduces nitrous oxide (N2O) to nitrogen (N2), with its site of action located in the periplasm (Figure 3). Two copper-containing enzymatic catalytic sites, that is, CuA and CuZ, are located on the same monomer (CuA and CuZ) (Brown et al., 2000a; Brown et al., 2000b). The intermediate product (N2O) is a more harmful greenhouse gas than carbon dioxide (CO2), and emissions of nitrous oxide must be reduced (Lee et al., 2019), but some microorganisms also emit nitrous oxide into the atmosphere as a terminal gaseous nitrogen (Gaimster et al., 2018). Therefore, reducing nitrogen oxide emissions is a very important issue in protecting the atmosphere. NOS is encoded by the nosZ gene and the accessory gene nosRDFYL (Honisch and Zumft, 2003), and the expression of the nosZ gene has been detected in B. californica K1 (Fang et al., 2021).
The conversion of ammonium into hydroxylamine via AMO is the first step in the HNAD nitrification process, and hydroxylamine is an important intermediate metabolite (Hooper et al., 1997). AMO-containing quinone oxidase has two subunits, which are inhibited by light and chelating agents during oxidation, but copper ions can increase the enzyme activity (Moir et al., 1996). The encoding gene of AMO is amoA, which was amplified and 397 bp in S. pararoseus Y1. Strain Y1 can convert ammonia into hydroxylamine under aerobic conditions (Table 2) (Zeng et al., 2020).
Oxidation of hydroxylamine to nitrite by HNADs occurs in the periplasm via HAO. The HAO encoding gene has been expressed successfully in B. cereus GS-5 (Rout et al., 2017). Recent reports suggest that HAO catalyzes hydroxylamine to produce nitric oxide rather than nitrite as found in previous studies, and nitric oxide is re-oxidized to nitrite (Caranto and Lancaster, 2017). In addition, hydroxylamine has been found to be oxidized to N2O by P460 (Caranto et al., 2016; White and Lehnert, 2016) under anaerobic conditions and HAO enzymes (Zhao et al., 2019) under aerobic conditions in both ammonia oxidizing bacteria and heterotrophic denitrifying bacteria, respectively.
Under aerobic conditions, ammonium/nitrate/nitrite assimilation by heterotrophic denitrifying bacteria via NAS is an important driving force for nitrogen removal. Nitrate first enters the cell interior through ion channels and then enters the assimilation pathway. Based on the protein sequence and biochemical properties of the catalytic subunits, NAS is classified into three types, that is, NasA, NasB, and NarC (Sparacino-Watkins et al., 2014). nasABGHC has been expressed in Paracoccus denitrificans PD1222 when nitrate was used as the nitrogen source (Luque-Almagro et al., 2017).
The organic carbon source provides nutrients and energy for growth while also being responsible for acting as an electron donor during nitrification and denitrification by HNADs. During the catabolic and anabolic processes of microorganisms, carbon sources provide the necessary nutrients and energy for bacteria (Nancharaiah and Kiran Kumar Reddy, 2018). However, a given HNAD strain exhibits a significantly different growth state, growth rate, and nitrogen-removal rate when using different organic carbon sources (Ren et al., 2014).
Different carbon sources—such as glucose, acetic acid, succinic acid, citric acid, methanol, and sucrose—have different effects on the growth and nitrogen removal of HNADs, as given in Table 3. The molecular weight and chemical structure of the carbon sources vary greatly and have a significant impact on nitrogen removal, and HNAD bacteria usually prefer carboxylates such as acetate to carbohydrates such as glucose (Jia et al., 2019). The reason for this may be that carboxylate has a simple chemical structure and a low molecular weight, making it more conducive to absorption and utilization through the tricarboxylic acid cycle. When performing nitrogen removal under aerobic conditions, sodium acetate is the optimal carbon source for B. subtilis A1 and V. diabolicus SF16 (Yang et al., 2011; Duan et al., 2015), and sodium pyruvate and succinate are the best recommended carbon sources for Klebsiella sp. TN-10 and B. methylotrophicus L7 (Zhang et al., 2012; Li et al., 2019). However, other strains such as P. aeruginosa P-1, A. contaminans HA, and E. cloacae HNR prefer glucose to carboxylate (Wei et al., 2021; Chen et al., 2015; Guo et al., 2016).
As well as conventional carbon sources, some solid ones have been tried in denitrification applications. As a by-product of tea extraction, tea residue has been used as a solid carbon source to cultivate the fungus F. solani RADF-77 and can be used as a biofilm attachment carrier in denitrification systems (Cheng et al., 2020). Wood-chip leachate has also been used as a carbon source in the nitrogen removal of P. tropicum IS0293 under aerobic conditions (Yao et al., 2020).
The C/N ratio affects the growth metabolism, energy conversion, and denitrification efficiency of HNADs, and it is a parameter that reflects the electron donor and acceptor requirements (Huang and Tseng, 2001; Rajta et al., 2020). A low carbon-source concentration affects not only the growth requirements of the strain but also the uptake and conversion efficiency of the microorganisms regarding the nitrogen source, and the optimal C/N ratio for most HNADs is between 8 and 15 (Table 4).
In Acinetobacter sp. FYF8, it has been reported that the nitrogen-gas conversion ratio is 39.88, 68.85, and 78.79% at a C/N ratio of 2.0, 2.5, and 3.0, respectively (Fan et al., 2021); these results show that the C/N ratio has a significant effect on nitrogen conversion by a given strain under particular incubation conditions. In addition, different nitrogen conversion pathways also require different C/N ratios; for B. methylotrophicus L7, the optimal C/N ratio is six for heterotrophic nitrification but 20 for aerobic denitrification (Zhang et al., 2012).
Within certain ranges of carbon-source concentration and C/N ratio, the higher the C/N ratio, the faster the bacterial growth and the higher the nitrogen-removal efficiency (Patureau et al., 2000). It has been found that the denitrification rate of A. junii YB increases with increasing C/N ratio in the range of 2–15, and nitrogen-balance analysis has shown that more ammonium is converted to biological nitrogen for cell growth at high C/N ratio (Ren et al., 2014). However, although it has been reported that a higher C/N ratio is more favorable for microbial growth and ammonium removal (Chen and Ni 2012), the latter can be inhibited if the C/N ratio is too high (Wang et al., 2016; Zhao et al., 2020). Most HNADs prefer a high C/N ratio, but some, such as Acinetobacter sp. Y16, can have high nitrogen-removal capacity at low C/N ratio (i.e., 2), which is a great advantage when treating wastewater with a low C/N ratio (Huang et al., 2013).
The inorganic ions Cu2+, Mg2+, Ni2+, Pb2+, Mn2+, Zn2+, Fe3+, and Cr6+, along with other heavy metal ions, are ubiquitous in wastewater and are either toxic or potent and affect the nitrogen-removal efficiency of HNADs (He T. et al., 2019). The nitrogen-removal efficiency of Arthrobacter arilaitensis Y-10 is significantly higher—by 10.88%—when compared with the control in the presence of copper at 0.1 mg/L but is strongly inhibited in the presence of copper at 0.25 mg/L (He T. et al., 2019). These results show that a low concentration of copper ions helps in ammonia–nitrogen conversion, but a high concentration reduces the removal efficiency.
Metal ions affect not only the growth of the strain but also its ability to remove nitrogen, and the type and concentration of ions have significantly different effects. When metal-ion addition experiments were conducted with A. aeruginosa HN-02, a low concentration of Cu2+ (0.5 mg/L) barely affected its ammonium-removal efficiency, but it decreased by ∼83% at 1.0 mg/L and became almost zero at 1.5 mg/L; compared to Cu2+, the ammonium-removal efficiency with the addition of Zn2+ (two to eight mg/L) was essentially the same as that of the control, and the toxicity of Cu2+ and Zn2+ in co-existence was much higher than that of single-addition Cu2+ (Chen et al., 2014). However, certain metal ions—such as Fe3+—can improve the functionality of the microorganism by stimulating its metabolism. For P. stutzeri T13, it has been reported that the nitrogen-assimilation capacity and maximum nitrate reduction rate increase significantly upon adding Fe3+ (Feng et al., 2021).
Compared with other nitrifying bacteria, Cupriavidus sp. S1 shows high nitrogen-removal capacity and excellent resistance to metal ions when various highly concentrated metal ions are added (Sun et al., 2016). In the presence of a high concentration of copper ions, Pseudomonas aeruginosa ZN1 has a copper-resistance gene and protein that allow the strain to grow and remove nitrogen at a copper–ion concentration of 800 mg/L (Zhang N. et al., 2018).
Salinity affects the denitrification capacity and cell growth of HNADs. High osmotic pressure may lead to strain disintegration, resulting in the loss of either cellular or enzymatic activity; therefore, high concentrations of salt inhibit cell growth and nitrogen removal. Previous studies have shown that most HNADs tolerate salinity below 40 g/L NaCl (Table 5), and halotolerant HNADs (Ventosa et al., 1998) may be more advantageous for treating wastewater with high salinity.
Although a low salt concentration is more conducive to cell growth and reproduction, some bacteria isolated from specific environments such as seawater and high-saline effluents have higher nitrogen-removal efficiency with appropriate salinity. In B. litoralis N31 isolated from the marine environment, it has been shown that an appropriate salinity (30–40 g/L) is beneficial for removing ammonium, probably because this is more suitable for enzyme activity (Huang F. et al., 2017). Similar phenomena have also been reported for Marinobacter NNA5 (Liu et al., 2016).
It has been found that pH and salinity also interact in nitrogen removal. In salinity tests, S. marcescens W5 showed a significant difference in ammonium removal between pH seven and pH 10 (Wang et al., 2016). However, studies on the mechanisms that produce this phenomenon are very limited.
Most HNADs are more adapted to neutral-to-alkaline environments (pH 6.0–9.0), and the optimal pH range for nitrogen removal is 7.0–8.0 (Table 6) (Chen et al., 2014; Liu et al., 2016; Hou et al., 2021). Some bacteria such as Acinetobacter sp. JR1 and S. marcescens W5 have a broad spectrum of pH adaptability (pH 4.5–10.0) (Wang et al., 2016; Yang et al., 2019). For S. marcescens W5 and P. stutzeri PCN-1, high ammonium-removal efficiency has been reported under both neutral and alkaline conditions (Zheng et al., 2014; Wang et al., 2016). Few strains can grow at extreme pH values, but strain HN-02 shows excellent acid adaptability and can grow cells and remove nitrogen at a pH value as low as 2.3 (Chen et al., 2014). Fungi are more advantageous than bacteria in converting organic nitrogen sources because of their better acid tolerance and denitrification ability (Cheng et al., 2020). HNADs have a broad spectrum of pH adaptability and can be used in various wastewater treatments.
The best growth state and highest nitrogen-removal efficiency of HNADs are usually found in the optimum pH range, such as for Acinetobacter sp. T1, Klebsiella sp. TN-10, and Marinobacter NNA5 (Liu et al., 2016; Chen et al., 2019; Li et al., 2019). However, the optimal cell-proliferation pH and the highest nitrogen-removal pH of Photobacterium sp. NNA4 are inconsistent, at 5.0–6.0 and 7.0, respectively (Liu et al., 2019).
Temperature affects microbial growth and the catalytic efficiency of enzymatic activity, and the optimal temperature range for HNADs is 30–37°C (Table 7) (Li et al., 2019; Liu et al., 2019; Xia et al., 2020). The higher the temperature in the enzyme-activity temperature range, the higher the enzyme reaction rate; the nitrification efficiency doubles for each 10 °C increase, and the denitrification efficiency doubles for each 4 °C increase (Zaitsev et al., 2008; Wu et al., 2019). The growth of most HNADs is inhibited at low temperature, but at 2 °C, Acinetobacter sp. Y16 can still achieve half of its nitrogen-removal efficiency at the optimal temperature (20 °C), with good low-temperature adaptability (Huang et al., 2013). The optimal temperature for B. simplex H-b is 10 °C, which is lower than that for other HNADs (Yang Q. et al., 2021). The growth trend of microorganisms is usually consistent with nitrogen removal, but at lower temperatures, microbial growth requires a longer lag period. Low-temperature nitrogen-removal HNADs can be used in wastewater treatment in cold areas.
In contrast to denitrifying bacteria, DO affects HNAD microbial colonization and nitrogen-removal efficiency and is a control parameter of the denitrification pathway (Hocaoglu et al., 2011). Conventional anaerobic denitrifying bacteria use nitrate as an electron acceptor to obtain energy, but the aerobic HNADs require the participation of oxygen to act as an electron acceptor for the denitrification process (Yang J. et al., 2020). In nitrogen removal by the denitrification pathway, nitrite reductase is sensitive to DO and can be inhibited by high DO concentration (Ka et al., 1997). It has been found that nitrite removal by P. stutzeri T13 is increased significantly by reducing the DO concentration and adjusting the rotational speed: Low rotational speed with low DO concentration is more useful for removing nitrite (Sun et al., 2015). HNADs have adapted to high-DO conditions for nitrogen removal and have higher efficiency than anaerobic denitrifying bacteria, with the optimal speeds given in Table 8.
The DO level is usually achieved by adjusting the rotation rate, but the DO concentration differs under different experimental conditions. The optimal shaking speed for both Acinetobacter sp. T1 and Photobacterium sp. NNA4 is 160 rpm, but their respective DO values are 5.1 mg/L and 5.89 mg/L (Chen et al., 2019; Liu et al., 2019).
Acinetobacter sp. ND7 requires different levels of DO for ammonium removal via the nitrification pathway and for nitrate removal via the denitrification pathway, with high DO favoring ammonium removal and low DO favoring nitrate removal (Xia et al., 2020).
With the widespread use of antibiotics in aquaculture, agriculture, and medicine worldwide, antibiotic residues affect the removal of nitrogen. Sulfamethoxazole (>2 μg/L) damages denitrifying cells, reduce bacterial electron-transport activity, and inhibit N2O reduction, leading to a reduction in aerobic denitrification nitrogen-removal efficiency in P. stutzeri PCN-1 (Gui et al., 2017). However, the nitrogen-removal efficiency of P. balearica RAD-17 through the aerobic denitrification pathway is insensitive to low and high concentrations of antibiotics such as ciprofloxacin and hygromycin (Ruan et al., 2021).
In the past decade, ZnO and CuO nanoparticles (NPs) have been used widely in manufacturing industries and have diffused into the environment, posing a potential risk to the nitrogen-removal process (Huang et al., 2010). In P. aeruginosa PCN-1, ZnO NPs inhibit the gene expression of nitrate and nitrite reductases, thereby affecting the catalytic activity of the enzymes and leading to a significant accumulation of nitrite (Chen et al., 2017). Uncoincidentally, the presence of ZnO NPs inhibits NR and NIR activities, reduces the denitrification efficiency of Clostridium perfringens HNR, and leads to increased production of extracellular polymeric substances (Ma et al., 2020). In addition, in P. tolaasii Y-11, CuO NPs have been found to not only affect nitrate reduction through their own action but also cause Cu2+-inhibited ammonium conversion (Yang Y. et al., 2020).
By contrast, the growth and nitrogen-removal capacities of P. tolaasii Y-11 are increased significantly in the presence of Fe2+ because of the polymerization of ZnO and CuO NPs promoted by Fe2+ (Yang Y. et al., 2020; Yang Y. et al., 2021). Also, the presence of phosphate reduces significantly the concentration of toxic Zn(II) released from ZnO NPs in the medium, and it weakens the effect of ZnO NPs on nitrogen removal (Cheng et al., 2019).
The use of HNADs in practical wastewater treatment has been investigated. P. denitrificans Z195 was inoculated into raw wastewater obtained from wastewater treatment plants, and the bacteria had excellent total nitrogen removal and denitrification performance (Zhang et al., 2020). Single HNAD strains have been studied widely, but mixed cultures of aerobic denitrifying bacteria have attracted less attention. A30, D10, and Z40 are three new bacteria isolated from sediment and inoculated into actual wastewater; 86% of total nitrogen and 93% of chemical oxygen demand (COD) were removed (Zhang H. et al., 2019). Huang F. et al. (2020) reported that a consortium of four novel bacteria (Marinomonas, Marinobacterium, Halomonas, and Cobetia) could convert ammonium into biological (∼60%) and gaseous (36–38%) nitrogen, with significantly lower denitrification by a single strain alone than by the mixture.
The use of HNADs in agricultural and industrial wastewater treatment has also been investigated. In swine wastewater treatment, the highest removal of ammonium and COD by P. stutzeri SDU10 can reach 97.6 and 94.2%, respectively, with the presence of sodium acetate to adjust the C/N ratio to 10 (Chen et al., 2020). Similar results have been reported for Acinetobacter sp. T1, which can significantly improve the nitrogen removal of pig-farm wastewater when compared with traditional activated sludge (Chen et al., 2019). In addition, S. marcescens CL1502 isolated from North Atlantic deep-sea sediments has shown efficient conversion of total nitrogen, ammonium, and COD from actual tannery-industry wastewater (Huang G. et al., 2017).
The use of feed has promoted the rapid development of aquaculture, but it also brings significant environmental problems, especially nitrogen pollution. The presence of P. stutzeri SC221-M in an experimental aquaculture system reduced pollutant production, improved water quality, and influenced the microbial community structure (Deng et al., 2014).
HNADs are isolated from soil, sludge, and sewage; they are mostly bacteria, but a few are fungi. Nitrogen is not only assimilated into organic nitrogen required for cell growth but also converted into gaseous nitrogen by HNADs under aerobic conditions. Ammonium conversion is achieved through three different metabolic pathways, and hydroxylamine is an important intermediary therein. The critical enzymes and encoding genes of the nitrification and denitrification metabolic pathways are also studied, as is optimizing the carbon source, C/N, metal ions, salinity, pH, temperature, and DO to improve the nitrogen-removal efficiency. Fe2+ and phosphoric acid reduce the effect of NPs on nitrogen removal. HNAD bacteria have excellent application potential in treating breeding and leather wastewater, removing nitrogen sources, and improving water quality.
Currently, most research into HNADs is focused on isolation and strain identification. Research into metabolic transformation mechanisms and the transformation of key enzyme encoding genes is still very limited, and the use of HNADs in sewage treatment needs to be improved.
WF: investigation, visualization, writing—review and editing; GS: project administration and supervision; YW: investigation and visualization; QW: writing—review and editing; PD: investigation; CL: investigation; XZ: conceptualization and project administration; ZR: conceptualization, supervision, and funding acquisition.
This work was supported by the National Key Research and Development Program of China (Grant No. 2021YFC2100900), the National Natural Science Foundation of China (Grant No. 32171471), the Key Research and Development Program of Ningxia Hui Autonomous Region (Grant No. 2020BFH01001), the Foundation of Fujian Key Laboratory of Functional Aquafeed and Culture Environment Control (Grant No. FACE20200003), the Program of the Key Laboratory of Industrial Biotechnology, Ministry of Education, China (Grant No. KLIB-KF202103), the 111 Project (No. 111-2-06), and the “Collaborative Innovation Center for Advanced Industrial Fermentation” industry development program of Jiangsu province.
GS, YW, and PD were employed by Fujian Dabeinong Huayou Aquatic Science and Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
An, Q., Zhou, Y., Zhao, B., and Huang, X. L. (2020). Efficient Ammonium Removal through Heterotrophic Nitrification-Aerobic Denitrification by Acinetobacter Baumannii Strain AL-6 in the Presence of Cr(VI). J. Biosci. Bioeng. 130, 622–629. doi:10.1016/j.jbiosc.2020.07.010
Brown, K., Djinovic-Carugo, K., Haltia, T., Cabrito, I., Saraste, M., Moura, J. G., et al. (2000a). Revisiting the Catalytic CuZ Cluster of Nitrous Oxide (N2O) Reductase. J. Biol. Chem. 275, 41133–41136. doi:10.1074/jbc.m008617200
Brown, K., Tegoni, M., Prudêncio, M., Pereira, A. S., Besson, S., Moura, J. J., et al. (2000b). A Novel Type of Catalytic Copper Cluster in Nitrous Oxide Reductase. Nat. Struct. Biol. 7 (3), 191–195. doi:10.1038/73288
Caranto, J. D., and Lancaster, K. M. (2017). Nitric Oxide is an Obligate Bacterial Nitrification Intermediate Produced by Hydroxylamine Oxidoreductase. Proc. Natl. Acad. Sci. U. S. A. 114, 8217–8222. doi:10.1073/pnas.1704504114
Caranto, J. D., Vilbert, A. C., and Lancaster, K. M. (2016). Nitrosomonas Europaea Cytochrome P460 is a Direct Link between Nitrification and Nitrous Oxide Emission. Proc. Natl. Acad. Sci. U. S. A. 113, 14704–14709. doi:10.1073/pnas.1611051113
Chen, Q., and Ni, J. (2010). Heterotrophic Nitrification-Aerobic Denitrification by Novel Isolated Bacteria. J. Ind. Microbiol. Biotechnol. 38, 1305–1310. doi:10.1007/s10295-010-0911-6
Chen, Q., and Ni, J. (2012). Ammonium Removal by Agrobacterium sp. LAD9 Capable of Heterotrophic Nitrification-Aerobic Denitrification. J. Biosci. Bioeng. 113, 619–623. doi:10.1016/j.jbiosc.2011.12.012
Chen, P., Li, J., Li, Q. X., Wang, Y., Li, S., Ren, T., et al. (2012). Simultaneous Heterotrophic Nitrification and Aerobic Denitrification by Bacterium Rhodococcus sp. CPZ24. Bioresour. Technol. 116, 266–270. doi:10.1016/j.biortech.2012.02.050
Chen, M., Wang, W., Feng, Y., Zhu, X., Zhou, H., Tan, Z., et al. (2014). Impact Resistance of Different Factors on Ammonia Removal by Heterotrophic Nitrification-Aerobic Denitrification Bacterium Aeromonas sp. HN-02. Bioresour. Technol. 167, 456–461. doi:10.1016/j.biortech.2014.06.001
Chen, J., Zheng, J., Li, Y., Hao, H.-H., and Chen, J.-M. (2015). Characteristics of a Novel Thermophilic Heterotrophic Bacterium, Anoxybacillus Contaminans HA, for Nitrification-Aerobic Denitrification. Appl. Microbiol. Biotechnol. 99, 10695–10702. doi:10.1007/s00253-015-6870-0
Chen, J., Gu, S., Hao, H., and Chen, J. (2016). Characteristics and Metabolic Pathway of Alcaligenes sp. TB for Simultaneous Heterotrophic Nitrification-Aerobic Denitrification. Appl. Microbiol. Biotechnol. 100, 9787–9794. doi:10.1007/s00253-016-7840-x
Chen, Q., Li, T., Gui, M., Liu, S., Zheng, M., and Ni, J. (2017). Effects of ZnO Nanoparticles on Aerobic Denitrification by Strain Pseudomonas Stutzeri PCN-1. Bioresour. Technol. 239, 21–27. doi:10.1016/j.biortech.2017.04.123
Chen, S., He, S., Wu, C., and Du, D. (2019). Characteristics of Heterotrophic Nitrification and Aerobic Denitrification Bacterium Acinetobacter sp. T1 and its Application for Pig Farm Wastewater Treatment. J. Biosci. Bioeng. 127, 201–205. doi:10.1016/j.jbiosc.2018.07.025
Chen, L., Lin, J., Pan, D., Ren, Y., Zhang, J., Zhou, B., et al. (2020). Ammonium Removal by a Newly Isolated Heterotrophic Nitrification-Aerobic Denitrification Bacteria Pseudomonas Stutzeri SDU10 and its Potential in Treatment of Piggery Wastewater. Curr. Microbiol. 77, 2792–2801. doi:10.1007/s00284-020-02085-1
Chen, L., Chen, L., Pan, D., Lin, H., Ren, Y., Zhang, J., et al. (2021). Heterotrophic Nitrification and Related Functional Gene Expression Characteristics of Alcaligenes Faecalis SDU20 with the Potential Use in Swine Wastewater Treatment. Bioprocess. Biosyst. Eng. 44, 2035–2050. doi:10.1007/s00449-021-02581-z
Cheng, Y.-F., Zhang, Z.-Z., Li, G.-F., Zhu, B.-Q., Zhang, Q., Liu, Y.-Y., et al. (2019). Effects of ZnO Nanoparticles on High-Rate Denitrifying Granular Sludge and the Role of Phosphate in Toxicity Attenuation. Environ. Pollut. 251, 166–174. doi:10.1016/j.envpol.2019.04.138
Cheng, H.-Y., Xu, A.-A., Kumar Awasthi, M., Kong, D.-D., Chen, J.-S., Wang, Y.-F., et al. (2020). Aerobic Denitrification Performance and Nitrate Removal Pathway Analysis of a Novel Fungus Fusarium Solani RADF-77. Bioresour. Technol. 295, 122250. doi:10.1016/j.biortech.2019.122250
Coelho, C., and Romão, M. J. (2015). Structural and Mechanistic Insights on Nitrate Reductases. Protein Sci. 24, 1901–1911. doi:10.1002/pro.2801
Deng, B., Fu, L., Zhang, X., Zheng, J., Peng, L., Sun, J., et al. (2014). The Denitrification Characteristics of Pseudomonas Stutzeri SC221-M and its Application to Water Quality Control in Grass Carp Aquaculture. PLoS One 9, e114886. doi:10.1371/journal.pone.0114886
Desloover, J., Roobroeck, D., Heylen, K., Puig, S., Boeckx, P., Verstraete, W., et al. (2014). Pathway of Nitrous Oxide Consumption in Isolated Pseudomonas Stutzeri Strains under Anoxic and Oxic Conditions. Environ. Microbiol. 16, 3143–3152. doi:10.1111/1462-2920.12404
Dias, J. M., Than, M. E., Humm, A., Huber, R., Bourenkov, G. P., Bartunik, H. D., et al. (1999). Crystal Structure of the First Dissimilatory Nitrate Reductase at 1.9 Å Solved by MAD Methods. Structure 7, 65–79. doi:10.1016/s0969-2126(99)80010-0
Duan, J., Fang, H., Su, B., Chen, J., and Lin, J. (2015). Characterization of a Halophilic Heterotrophic Nitrification-Aerobic Denitrification Bacterium and its Application on Treatment of Saline Wastewater. Bioresour. Technol. 179, 421–428. doi:10.1016/j.biortech.2014.12.057
Fan, Y., Su, J., Zheng, Z., Gao, J., and Ali, A. (2021). Denitrification Performance and Mechanism of a Novel Isolated Acinetobacter sp. FYF8 in Oligotrophic Ecosystem. Bioresour. Technol. 320, 124280. doi:10.1016/j.biortech.2020.124280
Fang, J., Liao, S., Zhang, S., Li, L., Tan, S., Li, W., et al. (2021). Characteristics of a Novel Heterotrophic Nitrification-Aerobic Denitrification Yeast, Barnettozyma Californica K1. Bioresour. Technol. 339, 125665. doi:10.1016/j.biortech.2021.125665
Feng, Y., Feng, J., and Shu, Q. L. (2018). Isolation and Characterization of Heterotrophic Nitrifying and Aerobic Denitrifying Klebsiella pneumoniae and Klebsiella Variicola Strains from Various Environments. J. Appl. Microbiol. 124, 1195–1211. doi:10.1111/jam.13703
Feng, L., Yang, J., Ma, F., Xing, L., Pi, S., Cui, D., et al. (2021). Biological Stimulation with Fe(III) Promotes the Growth and Aerobic Denitrification of Pseudomonas Stutzeri T13. Sci. Total Environ. 776, 145939. doi:10.1016/j.scitotenv.2021.145939
Fu, G., Zhao, L., Huangshen, L., and Wu, J. (2019). Isolation and Identification of a Salt-Tolerant Aerobic Denitrifying Bacterial Strain and its Application to Saline Wastewater Treatment in Constructed Wetlands. Bioresour. Technol. 290, 121725. doi:10.1016/j.biortech.2019.121725
Gaimster, H., Alston, M., Richardson, D. J., Gates, A. J., and Rowley, G. (2018). Transcriptional and Environmental Control of Bacterial Denitrification and N2O Emissions. FEMS Microbiol. Lett. 365, fnx277. doi:10.1093/femsle/fnx277
Gao, J., Zhu, T., Liu, C., Zhang, J., Gao, J., Zhang, J., et al. (2020). Ammonium Removal Characteristics of Heterotrophic Nitrifying Bacterium Pseudomonas Stutzeri GEP-01 with Potential for Treatment of Ammonium-Rich Wastewater. Bioprocess. Biosyst. Eng. 43, 959–969. doi:10.1007/s00449-020-02292-x
Godden, J. W., Turley, S., Teller, D. C., Adman, E. T., Liu, M. Y., Payne, W. J., et al. (1991). The 2.3 Angstrom X-Ray Structure of Nitrite Reductase from Achromobacter Cycloclastes. Science 253, 438–442. doi:10.1126/science.1862344
Graf, D. R. H., Jones, C. M., and Hallin, S. (2014). Intergenomic Comparisons Highlight Modularity of the Denitrification Pathway and Underpin the Importance of Community Structure for N2O Emissions. PLoS One 9, e114118. doi:10.1371/journal.pone.0114118
Gui, M., Chen, Q., and Ni, J. (2017). Effect of Sulfamethoxazole on Aerobic Denitrification by Strain Pseudomonas Stutzeri PCN-1. Bioresour. Technol. 235, 325–331. doi:10.1016/j.biortech.2017.03.131
Guo, L.-J., Zhao, B., An, Q., and Tian, M. (2016). Characteristics of a Novel Aerobic Denitrifying Bacterium, Enterobacter Cloacae Strain HNR. Appl. Biochem. Biotechnol. 178, 947–959. doi:10.1007/s12010-015-1920-8
He, T., Li, Z., Sun, Q., Xu, Y., and Ye, Q. (2016). Heterotrophic Nitrification and Aerobic Denitrification by Pseudomonas Tolaasii Y-11 without Nitrite Accumulation during Nitrogen Conversion. Bioresour. Technol. 200, 493–499. doi:10.1016/j.biortech.2015.10.064
He, T., Xie, D., Li, Z., Ni, J., and Sun, Q. (2017). Ammonium Stimulates Nitrate Reduction during Simultaneous Nitrification and Denitrification Process by Arthrobacter Arilaitensis Y-10. Bioresour. Technol. 239, 66–73. doi:10.1016/j.biortech.2017.04.125
He, T., Xie, D., Ni, J., Cai, X., and Li, Z. (2019a). Investigating the Effect of Copper and Magnesium Ions on Nitrogen Removal Capacity of Pure Cultures by Modified Non-Competitive Inhibition Model. Ecotoxicol. Environ. Saf. 170, 479–487. doi:10.1016/j.ecoenv.2018.12.019
He, X., Sun, Q., Xu, T., Dai, M., and Wei, D. (2019b). Removal of Nitrogen by Heterotrophic Nitrification-Aerobic Denitrification of a Novel Halotolerant Bacterium Pseudomonas Mendocina TJPU04. Bioprocess. Biosyst. Eng. 42, 853–866. doi:10.1007/s00449-019-02088-8
Hino, T., Matsumoto, Y., Nagano, S., Sugimoto, H., Fukumori, Y., Murata, T., et al. (2010). Structural Basis of Biological N2O Generation by Bacterial Nitric Oxide Reductase. Science 330, 1666–1670. doi:10.1126/science.1195591
Hocaoglu, S. M., Insel, G., Cokgor, E. U., and Orhon, D. (2011). Effect of Low Dissolved Oxygen on Simultaneous Nitrification and Denitrification in a Membrane Bioreactor Treating Black Water. Bioresour. Technol. 102, 4333–4340. doi:10.1016/j.biortech.2010.11.096
Hong, P., Huang, Y., Chen, M., and Xiao, B. (2021). Efficacy of Inorganic Nitrogen Removal by a Salt-Tolerant Aerobic Denitrifying Bacterium, Pseudomonas Sihuiensis LK-618. Bioprocess. Biosyst. Eng. 44, 1227–1235. doi:10.1007/s00449-021-02525-7
Honisch, U., and Zumft, W. G. (2003). Operon Structure and Regulation of the Nos Gene Region of Pseudomonas Stutzeri, Encoding an ABC-Type ATPase for Maturation of Nitrous Oxide Reductase. J. Bacteriol. 185, 1895–1902. doi:10.1128/jb.185.6.1895-1902.2003
Hooper, A. B., Vannelli, T., Bergmann, D. J., and Arciero, D. M. (1997). Enzymology of the Oxidation of Ammonia to Nitrite by Bacteria. Ant. Van Leeuwenhoek 71, 59–67. doi:10.1023/a:1000133919203
Hou, P., Sun, X., Fang, Z., Feng, Y., Guo, Y., Wang, Q., et al. (2021). Simultaneous Removal of Phosphorous and Nitrogen by Ammonium Assimilation and Aerobic Denitrification of Novel Phosphate-Accumulating Organism Pseudomonas Chloritidismutans K14. Bioresour. Technol. 340, 125621. doi:10.1016/j.biortech.2021.125621
Huang, H. K., and Tseng, S. K. (2001). Nitrate Reduction by Citrobacter Diversus under Aerobic Environment. Appl. Microbiol. Biotechnol. 55, 90–94. doi:10.1007/s002530000363
Huang, Y.-W., Wu, C.-H., and Aronstam, R. S. (2010). Toxicity of Transition Metal Oxide Nanoparticles: Recent Insights from In Vitro Studies. Materials 3, 4842–4859. doi:10.3390/ma3104842
Huang, X., Li, W., Zhang, D., and Qin, W. (2013). Ammonium Removal by a Novel Oligotrophic Acinetobacter sp. Y16 Capable of Heterotrophic Nitrification-Aerobic Denitrification at Low Temperature. Bioresour. Technol. 146, 44–50. doi:10.1016/j.biortech.2013.07.046
Huang, T., Guo, L., Zhang, H., Su, J., Wen, G., and Zhang, K. (2015). Nitrogen-Removal Efficiency of a Novel Aerobic Denitrifying Bacterium, Pseudomonas Stutzeri Strain ZF31, Isolated from a Drinking-Water Reservoir. Bioresour. Technol. 196, 209–216. doi:10.1016/j.biortech.2015.07.059
Huang, F., Pan, L., Lv, N., and Tang, X. (2017a). Characterization of Novel Bacillus Strain N31 from Mariculture Water Capable of Halophilic Heterotrophic Nitrification-Aerobic Denitrification. J. Biosci. Bioeng. 124, 564–571. doi:10.1016/j.jbiosc.2017.06.008
Huang, G., Ou, L., Pan, F., Wang, Y., Fan, G., Liu, G., et al. (2017b). Isolation of a Novel Heterotrophic Nitrification–Aerobic Denitrification Bacterium Serratia Marcescens CL1502 from Deep-Sea Sediment. Environ. Eng. Sci. 34, 453–459. doi:10.1089/ees.2016.0363
Huang, Y. S., An, Q., Zhao, B., Lv, Q. H., and Guo, J. S. (2018). Potential for Aerobic NO2- Reduction and Corresponding Key Enzyme Genes Involved in Alcaligenes Faecalis Strain NR. Arch. Microbiol. 200, 147–158. doi:10.1007/s00203-017-1428-4
Huang, F., Pan, L., He, Z., Zhang, M., and Zhang, M. (2020a). Identification, Interactions, Nitrogen Removal Pathways and Performances of Culturable Heterotrophic Nitrification-Aerobic Denitrification Bacteria from Mariculture Water by Using Cell Culture and Metagenomics. Sci. Total Environ. 732, 139268. doi:10.1016/j.scitotenv.2020.139268
Huang, X., Weisener, C. G., Ni, J., He, B., Xie, D., and Li, Z. (2020b). Nitrate Assimilation, Dissimilatory Nitrate Reduction to Ammonium, and Denitrification Coexist in Pseudomonas Putida Y-9 under Aerobic Conditions. Bioresour. Technol. 312, 123597. doi:10.1016/j.biortech.2020.123597
Jaroszynski, L. W., and Oleszkiewicz, J. A. (2011). Autotrophic Ammonium Removal from Reject Water: Partial Nitrification and Anammox in One‐Reactor Versus Two‐Reactor Systems. Environ. Technol. 32, 289–294. doi:10.1080/09593330.2010.497500
Jia, Y., Zhou, M., Chen, Y., Luo, J., and Hu, Y. (2019). Carbon Selection for Nitrogen Degradation Pathway by Stenotrophomonas Maltophilia: Based on the Balances of Nitrogen, Carbon and Electron. Bioresour. Technol. 294, 122114. doi:10.1016/j.biortech.2019.122114
Jin, P., Chen, Y., Yao, R., Zheng, Z., and Du, Q. (2019). New Insight into the Nitrogen Metabolism of Simultaneous Heterotrophic Nitrification-Aerobic Denitrification Bacterium in mRNA Expression. J. Hazard. Mat. 371, 295–303. doi:10.1016/j.jhazmat.2019.03.023
Joo, H.-S., Hirai, M., and Shoda, M. (2005). Nitrification and Denitrification in High-Strength Ammonium by Alcaligenes Faecalis. Biotechnol. Lett. 27, 773–778. doi:10.1007/s10529-005-5634-9
Ka, J., Urbance, J., Ye, R., Ahn, T., and Tiedje, J. (1997). Diversity of Oxygen and N-Oxide Regulation of Nitrite Reductases in Denitrifying Bacteria. FEMS Microbiol. Lett. 156 (1), 55–60. doi:10.1016/s0378-1097(97)00404-7
Khardenavis, A. A., Kapley, A., and Purohit, H. J. (2007). Simultaneous Nitrification and Denitrification by Diverse Diaphorobacter sp. Appl. Microbiol. Biotechnol. 77, 403–409. doi:10.1007/s00253-007-1176-5
Kim, J. K., Park, K. J., Cho, K. S., Nam, S.-W., Park, T.-J., and Bajpai, R. (2005). Aerobic Nitrification-Denitrification by Heterotrophic Bacillus Strains. Bioresour. Technol. 96, 1897–1906. doi:10.1016/j.biortech.2005.01.040
Lang, X., Li, Q., Ji, M., Yan, G., and Guo, S. (2020). Isolation and Niche Characteristics in Simultaneous Nitrification and Denitrification Application of an Aerobic Denitrifier, Acinetobacter Sp. YS2. Bioresour. Technol. 302, 122799. doi:10.1016/j.biortech.2020.122799
Lee, Y.-Y., Choi, H., and Cho, K.-S. (2019). Effects of Carbon Source, C/N Ratio, Nitrate, Temperature, and pH on N2O Emission and Functional Denitrifying Genes during Heterotrophic Denitrification. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 54, 16–29. doi:10.1080/10934529.2018.1503903
Li, A., Gai, Z., Cui, D., Ma, F., Yang, J., Zhang, X., et al. (2012). Genome Sequence of a Highly Efficient Aerobic Denitrifying Bacterium, Pseudomonas Stutzeri T13. J. Bacteriol. 194 (20), 5720. doi:10.1128/jb.01376-12
Li, C., Yang, J., Wang, X., Wang, E., Li, B., He, R., et al. (2015). Removal of Nitrogen by Heterotrophic Nitrification-Aerobic Denitrification of a Phosphate Accumulating Bacterium Pseudomonas Stutzeri YG-24. Bioresour. Technol. 182, 18–25. doi:10.1016/j.biortech.2015.01.100
Li, Y., Wang, Y., Fu, L., Gao, Y., Zhao, H., and Zhou, W. (2017). Aerobic-Heterotrophic Nitrogen Removal through Nitrate Reduction and Ammonium Assimilation by Marine Bacterium Vibrio sp. Y1-5. Bioresour. Technol. 230, 103–111. doi:10.1016/j.biortech.2017.01.049
Li, D., Liang, X., Jin, Y., Wu, C., and Zhou, R. (2019). Isolation and Nitrogen Removal Characteristics of an Aerobic Heterotrophic Nitrifying-Denitrifying Bacterium, Klebsiella sp. TN-10. Appl. Biochem. Biotechnol. 188, 540–554. doi:10.1007/s12010-018-02932-9
Liu, Y., Ai, G.-M., Miao, L.-L., and Liu, Z.-P. (2016). Marinobacter Strain NNA5, a Newly Isolated and Highly Efficient Aerobic Denitrifier with Zero N2O Emission. Bioresour. Technol. 206, 9–15. doi:10.1016/j.biortech.2016.01.066
Liu, Y., Ai, G.-M., Wu, M.-R., Li, S.-S., Miao, L.-L., and Liu, Z.-P. (2019). Photobacterium Sp. NNA4, an Efficient Hydroxylamine-Transforming Heterotrophic Nitrifier/Aerobic Denitrifier. J. Biosci. Bioeng. 128, 64–71. doi:10.1016/j.jbiosc.2018.12.014
Luque-Almagro, V. M., Manso, I., Sullivan, M. J., Rowley, G., Ferguson, S. J., Moreno-Vivián, C., et al. (2017). Transcriptional and Translational Adaptation to Aerobic Nitrate Anabolism in the Denitrifier Paracoccus Denitrificans. Biochem. J. 474, 1769–1787. doi:10.1042/bcj20170115
Ma, T.-F., Chen, Y.-P., Fang, F., Yan, P., Shen, Y., Kang, J., et al. (2020). Effects of ZnO Nanoparticles on Aerobic Denitrifying Bacteria Enterobacter Cloacae Strain HNR. Sci. Total Environ. 725, 138284. doi:10.1016/j.scitotenv.2020.138284
Ma, S., Huang, S., Tian, Y., and Lu, X. (2021). Heterotrophic Ammonium Assimilation: An Important Driving Force for Aerobic Denitrification of Rhodococcus Erythropolis Strain Y10. Chemosphere 291 (3), 132910. doi:10.1016/j.chemosphere.2021.132910
Matsumoto, Y., Tosha, T., Pisliakov, A. V., Hino, T., Sugimoto, H., Nagano, S., et al. (2012). Crystal Structure of Quinol-Dependent Nitric Oxide Reductase from Geobacillus Stearothermophilus. Nat. Struct. Mol. Biol. 19, 238–245. doi:10.1038/nsmb.2213
Moir, J. W. B., Crossman, L. C., Spiro, S., and Richardson, D. J. (1996). The Purification of Ammonia Monooxygenase from Paracoccus Denitrificans. FEBS Lett. 387, 71–74. doi:10.1016/0014-5793(96)00463-2
Nancharaiah, Y. V., and Kiran Kumar Reddy, G. (2018). Aerobic Granular Sludge Technology: Mechanisms of Granulation and Biotechnological Applications. Bioresour. Technol. 247, 1128–1143. doi:10.1016/j.biortech.2017.09.131
Nojiri, M., Koteishi, H., Nakagami, T., Kobayashi, K., Inoue, T., Yamaguchi, K., et al. (2009). Structural Basis of Inter-protein Electron Transfer for Nitrite Reduction in Denitrification. Nature 462, 117–120. doi:10.1038/nature08507
Padhi, S. K., Tripathy, S., Sen, R., Mahapatra, A. S., Mohanty, S., and Maiti, N. K. (2013). Characterisation of Heterotrophic Nitrifying and Aerobic Denitrifying Klebsiella Pneumoniae CF-S9 Strain for Bioremediation of Wastewater. Int. Biodeterior. Biodegr. 78, 67–73. doi:10.1016/j.ibiod.2013.01.001
Patureau, D., Bernet, N., Delgenès, J. P., and Moletta, R. (2000). Effect of Dissolved Oxygen and Carbon-Nitrogen Loads on Denitrification by an Aerobic Consortium. Appl. Microbiol. Biotechnol. 54 (4), 535–542. doi:10.1007/s002530000386
Peng, Y., and Zhu, G. (2006). Biological Nitrogen Removal with Nitrification and Denitrification via Nitrite Pathway. Appl. Microbiol. Biotechnol. 73, 15–26. doi:10.1007/s00253-006-0534-z
Rajta, A., Bhatia, R., Setia, H., and Pathania, P. (2020). Role of Heterotrophic Aerobic Denitrifying Bacteria in Nitrate Removal from Wastewater. J. Appl. Microbiol. 128, 1261–1278. doi:10.1111/jam.14476
Ren, Y.-X., Yang, L., and Liang, X. (2014). The Characteristics of a Novel Heterotrophic Nitrifying and Aerobic Denitrifying Bacterium, Acinetobacter Junii YB. Bioresour. Technol. 171, 1–9. doi:10.1016/j.biortech.2014.08.058
Ren, J., Wei, C., Ma, H., Dai, M., Fan, J., Liu, Y., et al. (2019). The Nitrogen-Removal Efficiency of a Novel High-Efficiency Salt-Tolerant Aerobic Denitrifier, Halomonas Alkaliphile HRL-9, Isolated from a Seawater Biofilter. Int. J. Environ. Res. Public Health 16, 4451. doi:10.3390/ijerph16224451
Ren, J., Ma, H., Liu, Y., Ruan, Y., Wei, C., Song, J., et al. (2021). Characterization of a Novel Marine Aerobic Denitrifier Vibrio spp. AD2 for Efficient Nitrate Reduction without Nitrite Accumulation. Environ. Sci. Pollut. Res. Int. 28, 30807–30820. doi:10.1007/s11356-021-12673-8
Robertson, L. A., and Kuenen, J. G. (1983). Thiosphaera Pantotropha Gen. Nov. sp. nov., a Facultatively Anaerobic, Facultatively Autotrophic Sulphur Bacterium. Microbiology 129, 2847–2855. doi:10.1099/00221287-129-9-2847
Rout, P. R., Bhunia, P., and Dash, R. R. (2017). Simultaneous Removal of Nitrogen and Phosphorous from Domestic Wastewater Using Bacillus Cereus GS-5 Strain Exhibiting Heterotrophic Nitrification, Aerobic Denitrification and Denitrifying Phosphorous Removal. Bioresour. Technol. 244, 484–495. doi:10.1016/j.biortech.2017.07.186
Ruan, Y., Taherzadeh, M. J., Kong, D., Lu, H., Zhao, H., Xu, X., et al. (2020). Nitrogen Removal Performance and Metabolic Pathways Analysis of a Novel Aerobic Denitrifying Halotolerant Pseudomonas Balearica Strain RAD-17. Microorganisms 8, 72. doi:10.3390/microorganisms8010072
Ruan, Y., Cai, L., Lu, H., Zhang, M., Xu, X., and Li, W. (2021). Performance of Aerobic Denitrification by the Strain Pseudomonas Balearica RAD-17 in the Presence of Antibiotics. Microorganisms 9, 1584. doi:10.3390/microorganisms9081584
Sánchez, C., and Minamisawa, K. (2018). Redundant Roles of Bradyrhizobium Oligotrophicum Cu-Type (NirK) and cd1-Type (NirS) Nitrite Reductase Genes under Denitrifying Conditions. FEMS Microbiol. Lett. 365 (5), fny015. doi:10.1093/femsle/fny015
Sears, H. J., Bennett, B., Spiro, S., Thomson, A. J., and Richardson, D. J. (1995). Identification of Periplasmic Nitrate Reductase Mo(V) EPR Signals in Intact Cells of Paracoccus Denitrificans. Biochem. J. 310, 311–314. doi:10.1042/bj3100311
Silva, L. C. F., Lima, H. S., Mendes, T. A. d. O., Sartoratto, A., Sousa, M. P., De Souza, R. S., et al. (2020). Physicochemical Characterization of Pseudomonas Stutzeri UFV5 and Analysis of its Transcriptome under Heterotrophic Nitrification/Aerobic Denitrification Pathway Induction Condition. Sci. Rep. 10, 2215. doi:10.1038/s41598-020-59279-7
Song, T., Zhang, X., Li, J., Wu, X., Feng, H., and Dong, W. (2021). A Review of Research Progress of Heterotrophic Nitrification and Aerobic Denitrification Microorganisms (HNADMs). Sci. Total Environ. 801, 149319. doi:10.1016/j.scitotenv.2021.149319
Sparacino-Watkins, C., Stolz, J. F., and Basu, P. (2014). Nitrate and Periplasmic Nitrate Reductases. Chem. Soc. Rev. 43, 676–706. doi:10.1039/c3cs60249d
Su, J. f., Shi, J. x., and Ma, F. (2017). Aerobic Denitrification and Biomineralization by a Novel Heterotrophic Bacterium, Acinetobacter sp. H36. Mar. Pollut. Bull. 116, 209–215. doi:10.1016/j.marpolbul.2017.01.014
Su, Z., Li, Y., Pan, L., He, Z., Le Dou, D., Liu, L., et al. (2021). Nitrogen Removal Performance, Quantitative Detection and Potential Application of a Novel Aerobic Denitrifying Strain, Pseudomonas sp. GZWN4 Isolated from Aquaculture Water. Bioprocess. Biosyst. Eng. 44, 1237–1251. doi:10.1007/s00449-021-02523-9
Suharti, , and De Vries, S. (2005). Membrane-Bound Denitrification in the Gram-Positive Bacterium Bacillus Azotoformans. Biochem. Soc. Trans. 33, 130–133. doi:10.1042/bst0330130
Sun, Y., Li, A., Zhang, X., and Ma, F. (2015). Regulation of Dissolved Oxygen from Accumulated Nitrite during the Heterotrophic Nitrification and Aerobic Denitrification of Pseudomonas Stutzeri T13. Appl. Microbiol. Biotechnol. 99, 3243–3248. doi:10.1007/s00253-014-6221-6
Sun, Z., Lv, Y., Liu, Y., and Ren, R. (2016). Removal of Nitrogen by Heterotrophic Nitrification-Aerobic Denitrification of a Novel Metal Resistant Bacterium Cupriavidus sp. S1. Bioresour. Technol. 220, 142–150. doi:10.1016/j.biortech.2016.07.110
Sun, Y., Feng, L., Li, A., Zhang, X., Yang, J., and Ma, F. (2017). Ammonium Assimilation: An Important Accessory during Aerobic Denitrification of Pseudomonas Stutzeri T13. Bioresour. Technol. 234, 264–272. doi:10.1016/j.biortech.2017.03.053
Takaya, N., Catalan-Sakairi, M. A. B., Sakaguchi, Y., Kato, I., Zhou, Z., and Shoun, H. (2003). Aerobic Denitrifying Bacteria that Produce Low Levels of Nitrous Oxide. Appl. Environ. Microbiol. 69, 3152–3157. doi:10.1128/aem.69.6.3152-3157.2003
Tang, Y., Zhang, M., Sun, G., and Pan, G. (2019). Impact of Eutrophication on Arsenic Cycling in Freshwaters. Water Res. 150, 191–199. doi:10.1016/j.watres.2018.11.046
Ventosa, A., Nieto, J. J., and Oren, A. (1998). Biology of Moderately Halophilic Aerobic Bacteria. Microbiol. Mol. Biol. Rev. 62 (2), 504–544. doi:10.1128/mmbr.62.2.504-544.1998
Vivián, C. M., Cabello, P., Luque, M. M., Blasco, R., and Castillo, F. (1999). Prokaryotic Nitrate Reduction: Molecular Properties and Functional Distinction Among Bacterial Nitrate Reductases. J. Bacteriol. 181 (21), 6573–6584.
Wan, C., Yang, X., Lee, D.-J., Du, M., Wan, F., and Chen, C. (2011). Aerobic Denitrification by Novel Isolated Strain Using NO2--N as Nitrogen Source. Bioresour. Technol. 102, 7244–7248. doi:10.1016/j.biortech.2011.04.101
Wang, T., Dang, Q., Liu, C., Yan, J., Fan, B., Cha, D., et al. (2016). Heterotrophic Nitrogen Removal by a Newly-Isolated Alkalitolerant Microorganism, Serratia Marcescens W5. Bioresour. Technol. 211, 618–627. doi:10.1016/j.biortech.2016.03.142
Wehrfritz, J.-M., Reilly, A., Spiro, S., and Richardson, D. J. (1993). Purification of Hydroxylamine Oxidase from Thiosphaera Pantotropha: Identification of Electron Acceptors that Couple Heterotrophic Nitrification to Aerobic Denitrification. FEBS Lett. 335, 246–250. doi:10.1016/0014-5793(93)80739-h
Wei, R., Hui, C., Zhang, Y., Jiang, H., Zhao, Y., and Du, L. (2021). Nitrogen Removal Characteristics and Predicted Conversion Pathways of a Heterotrophic Nitrification-Aerobic Denitrification Bacterium, Pseudomonas Aeruginosa P-1. Environ. Sci. Pollut. Res. Int. 28, 7503–7514. doi:10.1007/s11356-020-11066-7
White, C. J., and Lehnert, N. (2016). Is There a Pathway for N2O Production from Hydroxylamine Oxidoreductase in Ammonia-Oxidizing Bacteria? Proc. Natl. Acad. Sci. U.S.A. 113, 14474–14476. doi:10.1073/pnas.1617953114
Wu, Z., Xu, F., Yang, C., Su, X., Guo, F., Xu, Q., et al. (2019). Highly Efficient Nitrate Removal in a Heterotrophic Denitrification System Amended with Redox-Active Biochar: A Molecular and Electrochemical Mechanism. Bioresour. Technol. 275, 297–306. doi:10.1016/j.biortech.2018.12.058
Xia, L., Li, X., Fan, W., and Wang, J. (2020). Heterotrophic Nitrification and Aerobic Denitrification by a Novel Acinetobacter Sp. ND7 Isolated from Municipal Activated Sludge. Bioresour. Technol. 301, 122749. doi:10.1016/j.biortech.2020.122749
Xie, F., Thiri, M., and Wang, H. (2021). Simultaneous Heterotrophic Nitrification and Aerobic Denitrification by a Novel Isolated Pseudomonas Mendocina X49. Bioresour. Technol. 319, 124198. doi:10.1016/j.biortech.2020.124198
Yang, X.-P., Wang, S.-M., Zhang, D.-W., and Zhou, L.-X. (2011). Isolation and Nitrogen Removal Characteristics of an Aerobic Heterotrophic Nitrifying-Denitrifying Bacterium, Bacillus Subtilis A1. Bioresour. Technol. 102, 854–862. doi:10.1016/j.biortech.2010.09.007
Yang, X., Wang, S., and Zhou, L. (2012). Effect of Carbon Source, C/N Ratio, Nitrate and Dissolved Oxygen Concentration on Nitrite and Ammonium Production from Denitrification Process by Pseudomonas Stutzeri D6. Bioresour. Technol. 104, 65–72. doi:10.1016/j.biortech.2011.10.026
Yang, Y., Lin, E., and Huang, S. (2017). Heterotrophic Nitrogen Removal in Bacillus Sp. K5: Involvement of a Novel Hydroxylamine Oxidase. Water Sci. Technol. 76, 3461–3467. doi:10.2166/wst.2017.510
Yang, J.-R., Wang, Y., Chen, H., and Lyu, Y.-K. (2019). Ammonium Removal Characteristics of an Acid-Resistant Bacterium Acinetobacter sp. JR1 from Pharmaceutical Wastewater Capable of Heterotrophic Nitrification-Aerobic Denitrification. Bioresour. Technol. 274, 56–64. doi:10.1016/j.biortech.2018.10.052
Yang, J., Feng, L., Pi, S., Cui, D., Ma, F., Zhao, H.-P., et al. (2020a). A Critical Review of Aerobic Denitrification: Insights into the Intracellular Electron Transfer. Sci. Total Environ. 731, 139080. doi:10.1016/j.scitotenv.2020.139080
Yang, Y., Zhang, C., Huang, X., Gui, X., Luo, Y., and Li, Z. (2020b). Exogenous Fe2+ Alleviated the Toxicity of CuO Nanoparticles on Pseudomonas Tolaasii Y-11 under Different Nitrogen Sources. PeerJ 8, e10351. doi:10.7717/peerj.10351
Yang, Q., Yang, T., Shi, Y., Xin, Y., Zhang, L., Gu, Z., et al. (2021a). The Nitrogen Removal Characterization of a Cold-Adapted Bacterium: Bacillus Simplex H-b. Bioresour. Technol. 323, 124554. doi:10.1016/j.biortech.2020.124554
Yang, Y., Zhang, C., Li, K., and Li, Z. (2021b). Fe2+ Alleviated the Toxicity of ZnO Nanoparticles to pseudomonas Tolaasii Y-11 by Changing Nanoparticles Behavior in Solution. Microorganisms 9, 2189. doi:10.3390/microorganisms9112189
Yao, Z., Yang, L., Wang, F., Tian, L., Song, N., and Jiang, H. (2020). Enhanced Nitrate Removal from Surface Water in a Denitrifying Woodchip Bioreactor with a Heterotrophic Nitrifying and Aerobic Denitrifying Fungus. Bioresour. Technol. 303, 122948. doi:10.1016/j.biortech.2020.122948
Yoon, S., Cruz-García, C., Sanford, R., Ritalahti, K. M., and Löffler, F. E. (2015). Denitrification versus Respiratory Ammonification: Environmental Controls of Two Competing Dissimilatory NO3-/NO2- Reduction Pathways in Shewanella Loihica Strain PV-4. ISME J. 9, 1093–1104. doi:10.1038/ismej.2014.201
Zaitsev, G., Mettänen, T., and Langwaldt, J. (2008). Removal of Ammonium and Nitrate from Cold Inorganic Mine Water by Fixed-Bed Biofilm Reactors. Min. Eng. 21, 10–15. doi:10.1016/j.mineng.2007.08.014
Zeng, J., Liao, S., Qiu, M., Chen, M., Ye, J., Zeng, J., et al. (2020). Effects of Carbon Sources on the Removal of Ammonium, Nitrite and Nitrate Nitrogen by the Red Yeast Sporidiobolus Pararoseus Y1. Bioresour. Technol. 312, 123593. doi:10.1016/j.biortech.2020.123593
Zhang, J., Wu, P., Hao, B., and Yu, Z. (2011). Heterotrophic Nitrification and Aerobic Denitrification by the Bacterium Pseudomonas Stutzeri YZN-001. Bioresour. Technol. 102, 9866–9869. doi:10.1016/j.biortech.2011.07.118
Zhang, Q.-L., Liu, Y., Ai, G.-M., Miao, L.-L., Zheng, H.-Y., and Liu, Z.-P. (2012). The Characteristics of a Novel Heterotrophic Nitrification-Aerobic Denitrification Bacterium, Bacillus Methylotrophicus Strain L7. Bioresour. Technol. 108, 35–44. doi:10.1016/j.biortech.2011.12.139
Zhang, H., Zhao, Z., Chen, S., Kang, P., Wang, Y., Feng, J., et al. (2018a). Paracoccus Versutus KS293 Adaptation to Aerobic and Anaerobic Denitrification: Insights from Nitrogen Removal, Functional Gene Abundance, and Proteomic Profiling Analysis. Bioresour. Technol. 260, 321–328. doi:10.1016/j.biortech.2018.03.123
Zhang, H., Zhao, Z., Kang, P., Wang, Y., Feng, J., Jia, J., et al. (2018b). Biological Nitrogen Removal and Metabolic Characteristics of a Novel Aerobic Denitrifying Fungus Hanseniaspora Uvarum Strain KPL108. Bioresour. Technol. 267, 569–577. doi:10.1016/j.biortech.2018.07.073
Zhang, N., Chen, H., Lyu, Y., and Wang, Y. (2018c). Nitrogen Removal by a Metal-Resistant Bacterium, Pseudomonas Putida ZN1, Capable of Heterotrophic Nitrification–Aerobic Denitrification. J. Chem. Technol. Biotechnol. 94, 1165–1175. doi:10.1002/jctb.5863
Zhang, H., Zhao, Z., Li, S., Chen, S., Huang, T., Li, N., et al. (2019a). Nitrogen Removal by Mix-Cultured Aerobic Denitrifying Bacteria Isolated by Ultrasound: Performance, Co-Occurrence Pattern and Wastewater Treatment. Chem. Eng. J. 372, 26–36. doi:10.1016/j.cej.2019.04.114
Zhang, Y., Xu, Z., Li, J., Liu, D., Yuan, Y., Chen, Z., et al. (2019b). Cooperation between Two Strains of Enterobacter and Klebsiella in the Simultaneous Nitrogen Removal and Phosphate Accumulation Processes. Bioresour. Technol. 291, 121854. doi:10.1016/j.biortech.2019.121854
Zhang, H., Li, S., Ma, B., Huang, T., Qiu, H., Zhao, Z., et al. (2020). Nitrate Removal Characteristics and 13C Metabolic Pathways of Aerobic Denitrifying Bacterium Paracoccus Denitrificans Z195. Bioresour. Technol. 307, 123230. doi:10.1016/j.biortech.2020.123230
Zhao, B., He, Y. L., Hughes, J., and Zhang, X. F. (2010). Heterotrophic Nitrogen Removal by a Newly Isolated Acinetobacter Calcoaceticus HNR. Bioresour. Technol. 101, 5194–5200. doi:10.1016/j.biortech.2010.02.043
Zhao, B., An, Q., He, Y. L., and Guo, J. S. (2012). N2O and N2 Production during Heterotrophic Nitrification by Alcaligenes Faecalis Strain NR. Bioresour. Technol. 116, 379–385. doi:10.1016/j.biortech.2012.03.113
Zhao, B., Cheng, D. Y., Tan, P., An, Q., and Guo, J. S. (2018). Characterization of an Aerobic Denitrifier Pseudomonas Stutzeri Strain XL-2 to Achieve Efficient Nitrate Removal. Bioresour. Technol. 250, 564–573. doi:10.1016/j.biortech.2017.11.038
Zhao, B., Ran, X. C., An, Q., Huang, Y. S., Lv, Q. H., and Dan, Q. (2019). N2O Production from Hydroxylamine Oxidation and Corresponding Hydroxylamine Oxidoreductase Involved in a Heterotrophic Nitrifier A. Faecalis Strain NR. Bioprocess. Biosyst. Eng. 42, 1983–1992. doi:10.1007/s00449-019-02191-w
Zhao, Y., Lu, W., Liu, Y., Wang, J., Zhou, S., Mao, Y., et al. (2020). Efficient Total Nitrogen Removal from Wastewater by Paracoccus Denitrificans DYTN-1. Lett. Appl. Microbiol. 70, 263–273. doi:10.1111/lam.13268
Zheng, M., He, D., Ma, T., Chen, Q., Liu, S., Ahmad, M., et al. (2014). Reducing NO and N2O Emission during Aerobic Denitrification by Newly Isolated Pseudomonas Stutzeri PCN-1. Bioresour. Technol. 162, 80–88. doi:10.1016/j.biortech.2014.03.125
Zhu, L., Ding, W., Feng, L.-j., Kong, Y., Xu, J., and Xu, X.-y. (2012). Isolation of Aerobic Denitrifiers and Characterization for Their Potential Application in the Bioremediation of Oligotrophic Ecosystem. Bioresour. Technol. 108, 1–7. doi:10.1016/j.biortech.2011.12.033
Keywords: heterotrophic nitrifying, aerobic denitrifying, nitrogen biotransformation pathway, influencing factors, HNADs
Citation: Fu W, Song G, Wang Y, Wang Q, Duan P, Liu C, Zhang X and Rao Z (2022) Advances in Research Into and Applications of Heterotrophic Nitrifying and Aerobic Denitrifying Microorganisms. Front. Environ. Sci. 10:887093. doi: 10.3389/fenvs.2022.887093
Received: 01 March 2022; Accepted: 30 May 2022;
Published: 05 July 2022.
Edited by:
Mi Yan, Zhejiang University of Technology, ChinaReviewed by:
Jose Navarro Pedreno, Miguel Hernández University of Elche, SpainCopyright © 2022 Fu, Song, Wang, Wang, Duan, Liu, Zhang and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian Zhang, enhAamlhbmduYW4uZWR1LmNu; Zhiming Rao, cmFvemhtQGppYW5nbmFuLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.