
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Environ. Sci. , 25 February 2022
Sec. Freshwater Science
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.725360
This article is part of the Research Topic Digital Science and Technology for Sustainable Water Futures View all 8 articles
 Yingchun Xing1*
Yingchun Xing1* Wanru Gao1,2,3
Wanru Gao1,2,3 Zhixin Shen4
Zhixin Shen4 Yuanyuan Zhang5
Yuanyuan Zhang5 Jie Bai1,2,3
Jie Bai1,2,3 Xingwei Cai4
Xingwei Cai4 Jilong Ouyang6
Jilong Ouyang6 Yahui Zhao2*
Yahui Zhao2*Environmental DNA (eDNA) has been used in research relevant to fish ecology such as species diversity and conservation studies, threatened and invasive species monitoring, and analyses of population structure and distribution. How to choose the optimal laboratory protocols on the basis of the research targets is the first question to be considered when conducting an eDNA study. In this review, we searched 554 published articles using the topic subject ((eDNA or environmental DNA) and (fish)) within the time span 2011–2021 via Thompson Reuters Web of Science (WoS) and China National Knowledge Infrastructure (CNKI) literature databases, and screened 371 articles related to eDNA research on fish ecology. These articles were categorized into “article (334)”, “review (36)”, and “letter (1)” based on the type, and “article” was divided into “article (method research)” and “article (eDNA application)” in line with the study objectives. The experimental methods adopted in each study were reviewed, and advantages and disadvantages of the main protocols were analyzed for each step. We recommend a set of optimal protocols for regular eDNA-based fish diversity detection and present the following suggestions for water sample collection and subsequent sample processing and experiments. Sample size is suggested to be 2 L regardless of the type of water bodies; three water replicates are recommended per sampling site, and water collection sites should be designed to cover various water layers and micro-habitats within research areas. Filtration is the best method for collecting eDNA from the larger water samples; 0.45 μm glass fiber/glass microfiber (GF) filters and mixed cellulose acetate and nitrate (MCE) filters are recommended for use, and MCE filters are suitable for use in turbid waters; pre-filtration (>10 μm filtering membranes) can be used to prevent clogging. Freezing temperature storage can slow eDNA degradation, and this is the optimal way to store DNA no matter what filtering method is applied. The Qiagen DNeasy Blood and Tissue DNA extraction kit was the most economical and efficient DNA extraction method compared to other commercial kits. The 12S rRNA gene is the first choice for detecting interspecies variation in fishes, and five 12s primer sets, Ac12S, MDB07, Mi-Fish, Vert-12SV5, and Teleo, are recommended. The TruSeq DNA PCR-free LT Sample Prep kit and NEBNext DNA Library Prep Master Mix Set for the 454 kit can be chosen. The Illumina HiSeq platform can obtain sufficient data depth for fish species detection. QIIME and OBITools are independent software packages used for eDNA sequences analysis of fishes, and bioinformatic analyses include several indispensable steps such as filtering raw reads, clustering filtered reads into molecular operational taxonomic units (MOTUs) or amplicon sequence variants (ASVs), and completing taxon annotation. Contamination, inhibition, lack of reference DNA data, and bioinformatic analysis are key challenges in future eDNA research, and we should develop effective experimental techniques and analysis software regarding these aspects. This review intends to help eDNA beginners to quickly understand laboratory protocols applied in fish ecological research; the information will be useful for the improvement and development of eDNA techniques in the future.
Environmental DNA (eDNA) refers to DNA obtained from environmental samples such as water, soil, and sediments as well as DNA released through skin flakes, urine, and feces (Pedersen et al., 2015). With the increasing development of primers for multicellular organisms, eDNA has been widely applied to detect the presence of plants and animals, and fishes have become major research objects in current studies (Thomsen et al., 2012; Bista et al., 2017; Ficetola et al., 2018). Owning to fish releasing DNA into water (such as through feces, tissues, or secretions), eDNA can be used to monitor essential aspects of fish ecology, including taxa composition and distribution, species abundance, and population dynamics (Takahara et al., 2012; Lacoursière-Roussel et al., 2016; Zhang et al., 2020a; Fukaya et al., 2020; Griffiths et al., 2020; Hänfling, 2021; Milhau et al., 2021). eDNA is a noninvasive, efficient, and easily standardized research approach compared to traditional fish sampling methods. Therefore, application of eDNA allows us to explore aquatic ecosystems without capturing biological specimens and thereby reduces the possibility of diminishing wild fish populations and biomass through sampling, especially for threatened or endangered species.
Experiments using eDNA consist of three key steps: 1) collection and preservation; 2) extraction and purification; and 3) detection of fish taxa (Rees et al., 2014; Tsuji et al., 2019). Almost every mature eDNA research group has developed independent laboratory protocols used for eDNA-based detection of fish taxa. Consequently, there are many protocols for eDNA experiments, and thus it may be difficult to identify the most appropriate laboratory protocols for a fish ecological research project. Recently, several articles have reviewed eDNA experimental techniques and methodology for aquatic animal research (Bohmann et al., 2014; Rees et al., 2014; Goldberg et al., 2016; Zhang et al., 2020b; Wang et al., 2021). However, there are only few reviews for eDNA research in fish ecology. This study presents a comprehensive search of the published academic literature on fish ecology from 2011 to 2021, and the major aims are to organize information on eDNA field and laboratory protocols, recommend optimal protocols, and propose methodological challenges. We also provide information on future trends in eDNA methodology used in the field of fish ecology. We hope to help eDNA beginners fundamentally understand regular methods and present researchers with the advantages and disadvantages of major laboratory protocols and developmental trends in this field.
The eDNA articles on fish ecology during the past 10 years (2011–2021) were searched and synthesized using the software CiteSpace 5.7.2 R. We obtained data on references in Chinese and other languages from the universal Chinese literature database China National Knowledge Infrastructure (CNKI) and the Thompson Reuters Web of Science (WoS), respectively, using the search type “Topic Subject (TS) = [(eDNA or environmental DNA) and (fish)]” within the time span 2011–2021 and retrieved a total of 554 articles published or available as early access by December 2021. We screened the articles by hand to obtain 371 articles most relevant to fish ecology (including species diversity, alien species, biomass, and population dynamics) published in formal academic journals. The results comprised three document types: “article” (334), “review” (36), and “letter” (1). We divided “article” into two types according to the following criteria: “article (method research)” represented the articles that described the development of eDNA techniques and compared eDNA with other investigation methods, and “article (eDNA application)” represented the articles that involved specific studies using eDNA methods.
The eDNA research on fish ecology began in 2011, when a relevant article demonstrated that the DNA of fish released into water could be detected for research on freshwater ecosystems (Dejean et al., 2011). Before 2014, the number of publications in this field remained low (Figure 1), indicating that the eDNA method had not been completely established and was only accepted by a fraction of researchers, and its use by a minority of fish ecologists. The studies focused on detecting invasive and threatened species in this stage (Thomsen et al., 2012). Since 2014, the number of published eDNA articles has largely increased, and fish eDNA has become a frequent research topic (Keskin et al., 2014; Klymus et al., 2015; Sigsgaard et al., 2015). As an emerging DNA technology, the development and improvement of eDNA laboratory protocols are still ongoing, and most of the relevant research appeared after 2014 (Figure 1). The application of eDNA to the research field of fish ecology exhibited a rapid growth in quantity in 2016 when numerous articles on fish diversity based on eDNA appeared. Subsequently, the amount of research on fish ecology using eDNA has steadily increased through 2021 (Figure 1). From 10 to nearly 30 articles on eDNA methods were published every year between 2016 and 2021, and the number of articles on eDNA application increased yearly during this period (Figure 1). Review articles about eDNA used in fish ecological research were concentrated from 2018 to 2020; these articles included reviews of fish monitoring by eDNA, and a few involved the experimental protocols.
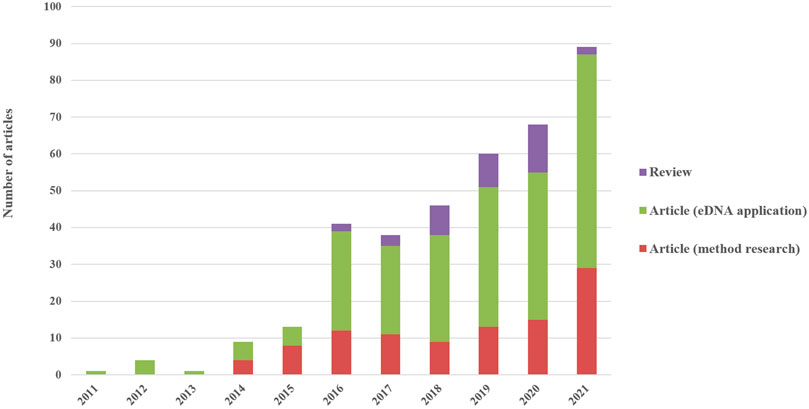
FIGURE 1. Change trend of number of articles published in formal academic journals from 2011 to 2021.
To detect fish species using eDNA for ecological analysis, water samples are obtained from lakes, rivers, streams, seas, artificial ponds/reservoirs/canals/ditches, lagoons, and lab tanks that contain fish. Sample volume depends on the water type and individual research target (Schumer et al., 2019; Dressler et al., 2020; Fraija-Fernández et al., 2020; Sigsgaard et al., 2020; Brys et al., 2021). Considering that eDNA may degrade after sample collection due to microbial activity (nucleases) and oxygenation reactions (Lindahl, 1993), sampling effectiveness is a factor that must be considered when planning the volume of each sample. On the one hand, enough eDNA should be obtained; on the other hand, not too much time spent in sampling, especially at large spatial scales.
Water sample volumes from 15 ml to 45 L used for eDNA collection for fish ecological research have been recorded (Takahara et al., 2012; Thomsen et al., 2012; Sigsgaard et al., 2015; Civade et al., 2016; McDevitt et al., 2019; Brys et al., 2021; Milhau et al., 2021; Sugiura et al., 2021). In experimental conditions such as lab tanks or experimental enclosures, the sample volume will depend on the volume of the containers, often from 20 ml to 2 L (Takahara et al., 2012; Capo et al., 2019). Regular sample volumes in the field have varied from 1 to 2 L in natural water bodies (Takahara et al., 2012; Li et al., 2018; Bagley et al., 2019; Zou et al., 2019; Griffiths et al., 2020; Riaz et al., 2020; Brys et al., 2021). With the development of sampling equipment, researchers have tended to collect large amounts of water (>10 L) to study species diversity, distribution patterns, seasonal dynamics, and conservation of fish communities in rivers and lakes (Civade et al., 2016; Pont et al., 2018; Goutte et al., 2020; Milhau et al., 2021). The commonly accepted assumption is that we can obtain higher amounts of DNA and detect more eDNA-based species from larger volumes of water. Muha et al. (2019) detected the highest amount of eDNA from 2 L water samples compared to 15 ml, 100 ml, 250 ml, and 1 L samples collected from two lentic and one lotic freshwater bodies located in Wales, United Kingdom. Cantera et al. (2019) compared change trends of species richness detected through filtered eDNA from 34 L to 340 L water samples collected in the tropical streams and rivers of French Guiana and found that 34–68 L sample sizes were suitable to detect most fauna, and there was no further advantage above 68 L. At present, 45 L is the largest water sample volume used for fish ecological research (Civade et al., 2016).
A sample with a fixed volume of water collected from multiple subsites at one sampling location or taking sample replicates from one sampling site can help to obtain enough DNA for detecting fish taxa, because a random distribution of eDNA is usually assumed (Goldberg et al., 2016; Griffiths et al., 2020). Hunter et al. (2019) compared eDNA amounts between 200 ml of water filtered through one filter and 800 ml through four filters, and the results indicated that the average DNA concentration was 1.4 copies/μl in the former and 6.39 copies/μl in the latter. There is no explicit standard for the number of subsites that should be collected for each sample; one 1 L sample from 10 different subsites was the case with the highest number of subsites for sampling that we could find up to the present (Lacoursière-Roussel et al., 2016). Previous research used three to six water replicates at a single site; for example, Sigsgaard et al. (2015) collected 3 × 15 ml water samples per site in Jutland, Denmark, to monitor the European endangered fish species weather loach (Misgurnus fossilis). West et al. (2020) sampled 6 × 1 L water replicates in the lagoons around the Cocos Islands in the eastern Indian Ocean aiming to detect coral reef fishes, and Griffiths et al. (2020) collected 2 L water samples composed of 5 × 400 ml subsamples taken from pumped waterways and drainage ditches in the Fens, United Kingdom, to research community structure of the European eel (Anguilla anguilla). A recent study on coral fish biodiversity based on environmental DNA suggested that about 25 replicate water samples could often reflect enough species richness in the tropical oceans (Stauffer et al., 2021). Indeed, spatial sampling designs have expressed correlations with fish community composition. Previous research has confirmed that more fish species were detected in shallow water than in deep water, and eDNA may accumulate in nearshore areas so that shoreline samples generally captured the majority of the fish diversity compared to interior samples in lakes (Hänfling et al., 2016; Zhang et al., 2020a). Therefore, it is suggested that more water collection sites should be designed at the surface and nearshore areas of lentic water bodies. For lotic water bodies such as streams and oceans, water movement leads to diffusion and advection of eDNA, so multiple locations and known habitat components should be targeted (Goldberg et al., 2016). For the tropical ocean ecosystems such as coral reef environments, it has been suggested that sampling should be performed not only at the surfaces of water bodies but also along a depth gradient (Stauffer et al., 2021).
The eDNA collection methods include filtration, precipitation, and centrifugation, each of which has individual operation steps (Figure 2). Filtration is the most common method used to collect eDNA for fish ecological research, and this method was used in more than 90% of our analyzed studies. Compared with other methods, filtration has been determined to obtain the highest eDNA yields (Eichmiller et al., 2015). In the filtration method, water samples are pumped through a filter with a certain pore size to collect eDNA. In summary, the pore size used for filtering water samples varies from 0.22 to 5 μm, and filter types generally include glass fiber/glass microfiber (GF) filters, nylon filters (NFs), cellulose nitrate (CN) filters, polycarbonate (PC) filters, polyethersulfone (PES) filters, polyvinylidene difluoride (PVD) filters, mixed cellulose acetate and nitrate (MCE) filters, and filter capsules (Sterivex enclosure filter, GP). Different pore sizes and filter types can affect DNA yields. Generally, 0.45 and 0.7 μm filters are the most used for fish detection (Shu et al., 2020; Wang et al., 2021). Compared to filters with larger pore sizes (>1 μm), the smaller pore-size filters performed better for fish species detection. The 0.2 μm filters were recommended to capture DNA of common carp (Cyprinus carpio), and the 0.45 μm filters could collect the highest amount of DNA and fish species information compared to 0.8 and 1.2 μm filters (Li et al., 2018). The GF filter performed better than other types of filters for eDNA capture from water samples, and the MCE filter was suitable for turbid and high species-density water bodies (Eichmiller et al., 2015; Lacoursière-Roussel et al., 2016; Muha et al., 2019).
Although the smaller pore size filters performed better at capturing more DNA yields from water samples, clogging is always a problem, especially in turbid and high-biodiversity water bodies (Li et al., 2018; Wang et al., 2021). Pre-filtration and increasing filtration volume are two optional solutions in addition to the selection of the larger pore size filters. Takahara et al. (2012) used 0.8 μm filters to collect fish eDNA and conducted pre-filtration using 12 μm filters before main filtration to preventing clogging. Li et al. (2018) confirmed that 20 μm pre-filtration was essential to save filtration time and improve eDNA capture by removing suspended particulate matter.
In the precipitation method, proportions of ethanol and sodium acetate are added to obtain eDNA, and thus this method is therefore usually called the ethanol precipitation method. This method has been used regularly for eDNA capture from small water samples (<30 ml) (Sigsgaard et al., 2015; Wang et al., 2021) and volumes up to 100 ml as an exception (Eichmiller et al., 2015; Minamoto et al., 2016). Minamoto et al. (2016) demonstrated that the highest amount of eDNA from Cyprinus carpio could be obtained through ethanol precipitation compared with phenol extraction and ultrafiltration methods. Recently, some researchers have used a combination of precipitation and filtration methods to collect eDNA from large-volume water samples (e.g., 30 L). Pont et al. (2018) and Goutte et al. (2020) both used a 0.45 μm filtration device connected to a peristaltic pump to filter 30 L water samples for approximately 30 min and added CL1 conservation buffer to the filter capsules after emptying them, and then stored the samples at room temperature; ethanol (33.5 ml) and sodium acetate (1.5 ml, 3M) were then added after centrifugation of the CL1 buffer, and the supernatant was removed and stored at −20°C until DNA extraction. Muha et al. (2019) proposed that using a combination of ethanol-sodium acetate precipitation and filtration methods could reduce the volume filtered and the contamination risk when the smallest water samples were tested.
In addition to the ethanol precipitation method, the isopropanol precipitation method is also an alternative for fish eDNA collection from larger than 50 ml volume water samples. Doi et al. (2017) compared the effects of eDNA capture using the ethanol and isopropanol precipitation methods in both experimental and field conditions and found that isopropanol precipitation could collect more common carp (Cyprinus carpio) eDNA copies than ethanol precipitation when using 50 ml of water. However, ethanol precipitation could obtain more eDNA from 15 ml water samples compared with the isopropanol method in the mesocosm sampling. In contrast, there was no difference between the two precipitation methods in the field experiments. The isopropanol precipitation method has two advantages compared with the ethanol method: 1) it can yield a higher DNA concentration when processing higher volume water samples (>50 ml); 2) it is cheaper and easier to operate.
Centrifugation is the least commonly used method for collecting eDNA in studies of fish ecology because it is not suitable for large volumes of water. Eichmiller et al. (2015) estimated common carp (Cyprinus carpio) DNA concentrations by comparing centrifugation with filtration and precipitation methods; the results indicated that centrifugation obtained a higher eDNA yield than precipitation and a lower yield than filtration. Recent research indicates that the centrifugation method has been rarely used for eDNA collection from water samples.
Appropriate methods of eDNA storage can slow the degradation rate and improve the extraction efficiency of eDNA when conducting fish ecological research. Keeping eDNA at a freezing temperature (e.g., using ice, liquid nitrogen, or freezing the samples) and adding a fixed buffer (e.g., ethanol, or lysis buffer) to eDNA samples are common preservation methods used in recent research (Bagley et al., 2019; Zou et al., 2019; Alam et al., 2020; Juhel et al., 2020; Brys et al., 2021; Milhau et al., 2021; Sugiura et al., 2021). Freezing is the most commonly method used and is the best for eDNA precipitation by centrifugation (Thomsen et al., 2012; Sigsgaard et al., 2015), and freezing and lysis buffers can be used for preserving filtered eDNA samples (Civade et al., 2016; Sigsgaard et al., 2017; Bagley et al., 2019). Although freezing at −20°C has excellent performance for eDNA storage, it is difficult to achieve these conditions in most field environments. Therefore, preservation buffers can be a practical alternative. Renshaw et al. (2015) studied the preservation time of filtered eDNA samples using two lysis buffers (CTAB and Longmire’s) at room, freezing, and higher temperatures and concluded that eDNA could be preserved in these two lysis buffers for more than 2 weeks at room temperature. Hinlo et al. (2017) used the oriental weather loach (Misgurnus anguillicaudatus) as a research animal to study how to optimize eDNA preservation, and they suggested that refrigeration may be a better choice than freezing for storage lasting three to 5 days. Majaneva et al. (2018) stored filtered samples using ethanol, lysis buffer, ice, or drying by silica gel and demonstrated that the drying method may be a good option if DNA was extracted and used within a short time, and lysis buffer could preserve eDNA for several weeks. In general, CL1 conservation buffer is regularly used to store water samples at room temperature, and then the precipitation method is applied to collect the eDNA (Pont et al., 2018; Goutte et al., 2020; Milhau et al., 2021). In addition to common preservation buffers such as ethanol, lysis buffer, and CL1 conservation buffer, several new buffers have been used in recent fish ecological research. Yamanaka et al. (2017) tested the storage ability of benzalkonium chloride (BAC) for both short-term and long-term periods, and 0.01% BAC was found to effectively maintain 92% of the eDNA of bluegill sunfish (Lepomis macrochirus) for 1 day, 50–70% for 1–10 days, and 20% after 10 days at ambient temperature.
eDNA extraction is a process used to release DNA from intact cells and organelles present in environmental samples and to enable researchers to effectively remove polymerase chain reaction (PCR) inhibitors such as humic acids or humic substances through purification (Eichmiller et al., 2015). The traditional DNA extraction method is the phenol-chloroform-isoamylol (PCI) method. Deiner et al. (2015) modified the PCI protocol to perform eDNA extraction. Although PCI provided better eDNA yields and lower amounts of inhibitors combined with specific filters and preservation methods when fish ecological research is conducted (Eichmiller et al., 2015; Renshaw et al., 2015; Hunter et al., 2019), commercial extraction kits are more often recommended because harmful substances such as phenol and chloroform are used during PCI extraction. The types of commercial kits used for eDNA extraction are diverse, including the Qiagen DNeasy Blood and Tissue Kit (DNeasy kit), the MoBio PowerWater DNA Extraction Kit (PowerWater kit), the MoBio PowerSoil DNA Extraction Kit (PowerSoil kit), the MP Biomedicals FastDNA SPIN Kit (Biomedicals kit), the MP Biomedicals FastDNA SPIN Kit for soil kit (Biomedicals soil kit), the PowerMax Soil kit (PowerMax kit), the Qiagen QIAamp DNA Stool Mini Kit (Stool Mini kit), and the DNeasy Mericon Food Kit (Mericon Food Kit).
An ideal eDNA extraction kit is assessed by its ability to reduce inhibitors while also obtaining a high DNA concentration and saving as much cost as possible. Based on a review of the literature and our research experience, the eDNA yields, experimental expenses, and levels of inhibitors of several of the most-used eDNA extraction kits were compared (Table 1). The DNeasy kit was the most well-established, economical, and efficient choice, although it does not include “inhibitor removal” steps. The PowerWater kit had the better performance for DNA yield and inhibitor removal; however, experimental expenses are high, nearly four times the cost of the DNeasy kit. Eichmilleret al. (2015) also compared six commercial kits (the PowerSoil, PowerWater, Biomedicals, Biomedicals soil, DNeasy, and Stool Mini) and suggested that the Biomedicals kit was most sensitive to detect the DNA of common carp (Cyprinus carpio), and the PowerSoil kit had the lowest variation between lake and well water and did not produce inhibitors.
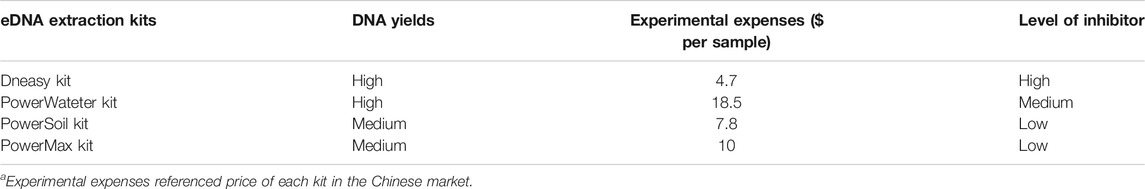
TABLE 1. Comparison of several of the most-used eDNA extraction kits in the fish ecological research.
eDNA detection in fish ecological studies can be divided into two categories: species-specific detection and multi-species detection (Thomsen and Willerslev, 2015). With regard to the number of published articles concerning fish ecology studies on species-specific detection and multi-species detection on the basis of eDNA between 2011 and 2021 (Figure 3), the number of articles on both research directions were basically the same. There were more articles published on species-specific detection in 2013 and 2014. Year 2015 was a turning point when the number of articles on multi-species detection dramatically increased, and the number of articles on detection of specific species was clearly less than that of the former. Different protocols were chosen to conduct detection of fish using eDNA for specific purposes.
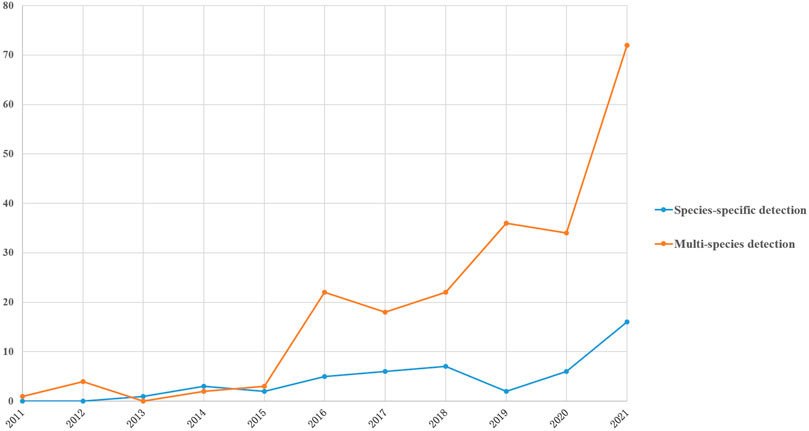
FIGURE 3. Trends of the number of studies on species-specific detection and multispecies detection in 2011–2021.
Species-specific detection using eDNA was mostly applied in monitoring threatened or invasive fish species in analyses of the distribution, population size, and population dynamics of these target species. The information on relevant research is shown in Table 2. Targeted species detection using eDNA has shown that it can fulfil the requirements of fishery management and ecosystem monitoring and can be used as an alternative approach to answer relevant questions for managers (Gilbey et al., 2021).
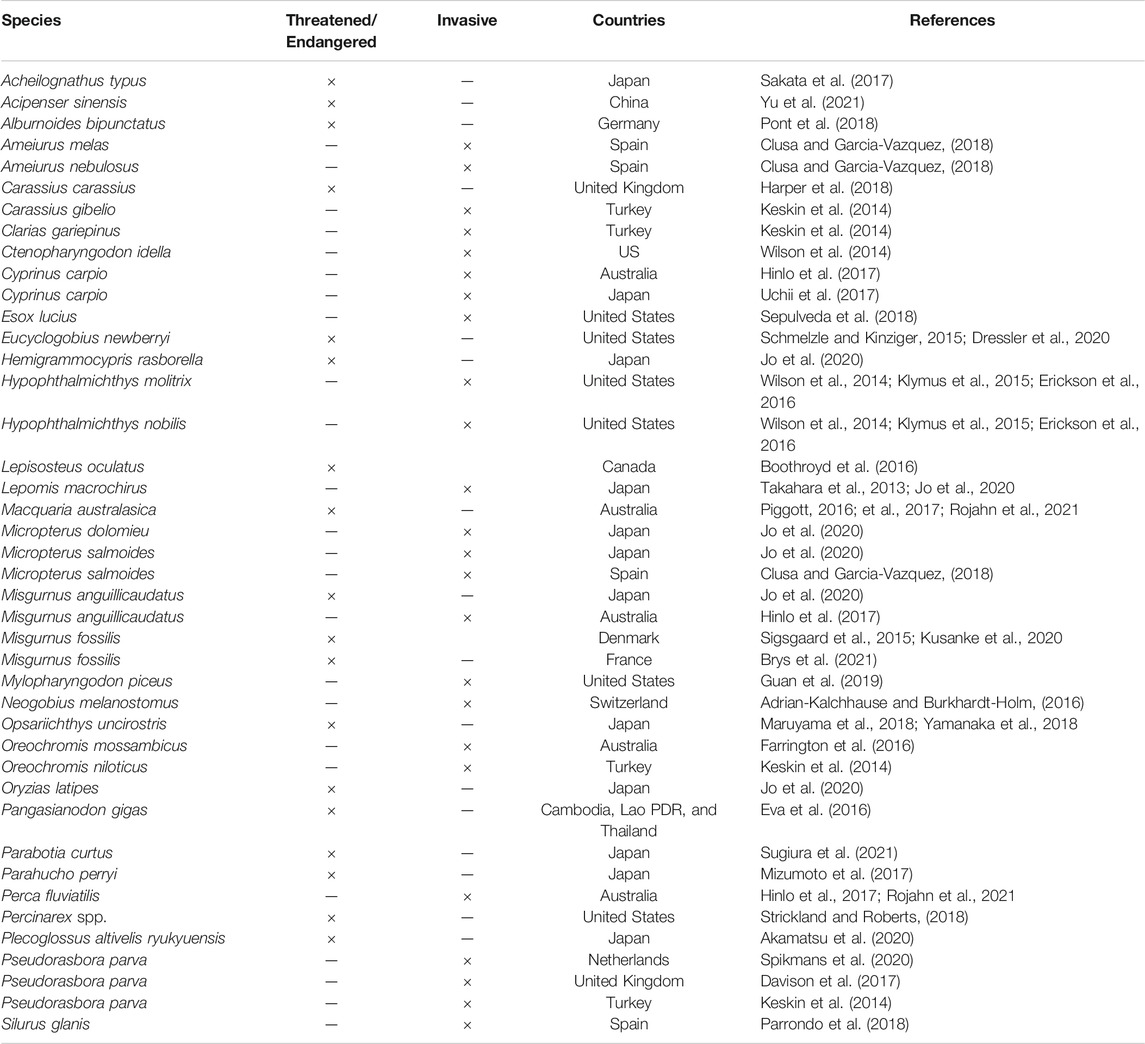
TABLE 2. Summary of articles describing the use of eDNA for detection of threatened and endangered fish species or for detecting invasive fish species.
Species-specific detection was carried out by designing specific primers and identifying target species using PCR technology (Thomsen and Willerslev, 2015). Mitochondrial DNA (mt-DNA) fragments such as 12S ribosomal RNA (12S), 16S ribosomal RNA (16S), cytochrome c oxidase subunit 1 (COI), cytochrome b (Cytb), and D-loop are suitable for single-species detection compared with nuclear DNA because these types of mt-DNA contain more copy numbers per cell enables easier detection, and conserved interspecific sequence variations of mtDNA allow reliable species identification (Erickson et al., 2016; Minamoto et al., 2016; Brys et al., 2021). The target region of molecular markers for eDNA detection is usually 80–120 bp (Thomsen et al., 2012; Jerde et al., 2013; Deiner et al., 2015; Edwards et al., 2018). According to the statistics of relevant articles (there were a total of 36 articles on specific fish detection; Figure 4), Cytb was used most in species-specific detection (n = 12, 33.33%) followed by COI (n = 9, 25.00%); this was attributed to the fact that the sequences of these two genes of fish species were more easily found on NCBI, so that species-specific primers for the fragments could be designed. Specific primers were generally designed using professional software such as Primer version 3 or PrimerHunter (Dougherty et al., 2016). Recent research compared the particle size distribution of nuclear eDNA (nu-eDNA) of Japanese Jack Mackerel (Trachurus japonicus) in experimental tanks with that of mt-eDNA and found that the concentration of nu-eDNA was higher than that of mt-eDNA in the 3–10 μm size fraction, and 0.8 to 3 μm and 0.4–0.8 μm size eDNA fragment concentrations of both types increased with higher temperature (Jo et al., 2019a). This trend may suggest that nu-eDNA could be used for fish specific species detection in the next step.
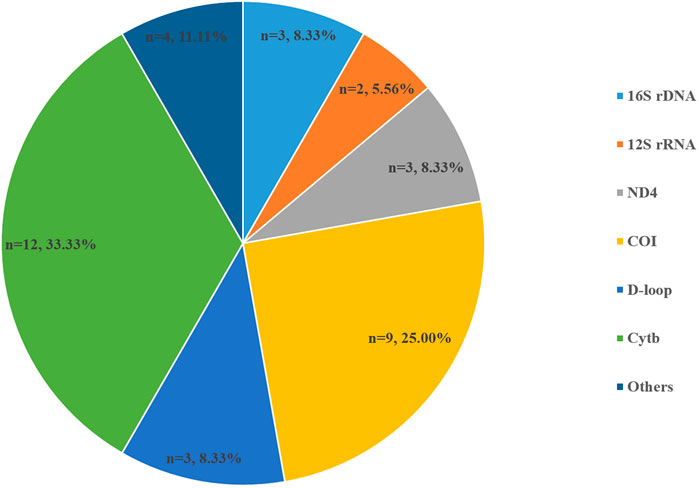
FIGURE 4. Composition and percentage of target primer fragments for PCR (figures on the map refer to the frequency of occurrence of each molecular marker in a total of 36 articles).
PCR, real-time quantitative PCR (qPCR), and droplet digital PCR (ddPCR) are generally used for fish species-specific detection. These methods have individual advantages and disadvantages (Nathan et al., 2014; Dressler et al., 2020; Griffiths et al., 2020). PCR amplification is the traditional method used to detect target fragments, and measured results should be confirmed using electrophoresis (Atkinson et al., 2018). The PCR method is low-cost and simple to operate, and gel electrophoresis can be used to visualize results of PCR products. However, electrophoresis also easily increases the possibility of contamination if gel-cutting recovery is needed. Unlike regular PCR, a single qPCR can simultaneously detect several species (typically two to five species) labeled with unequal fluorescent dyes (Tsuji et al., 2018). This method has its own characteristics of specificity, sensitivity, and quantification ability as well as reduced necessary reaction time. Therefore, qPCR has become the main method for eDNA-based fish species-specific detection in recent years. TaqMan probe-based qPCR is the most effective way for eDNA detection of single species (Thomsen and Willerslev, 2015; Wang et al., 2021). ddPCR, as “third-generation” PCR, is also a quantitative PCR method like qPCR and can provide absolute quantification of target DNA. Compared with qPCR, ddPCR does not require a standard curve of the reference relevant to target DNA (Hindson et al., 2011). When using ddPCR to detect fish species, lesser numbers of species can be estimated compared to qPCR (Hindson et al., 2011; Nathan et al., 2014). Doi et al. (2015a) documented that ddPCR was better for measuring eDNA concentration in water bodies, because it provided more accurate results related to the abundance and biomass of the target species compared with qPCR. However, the disadvantage of this method is that ddPCR is more expensive. In addition to these three common PCR methods, new PCR strategies have been developed. For example, Stoeckle et al. (2018) developed nested PCR (GoFish) to detect the eDNA of a single fish species. GoFish has the advantages of both Illumina MiSeq metabarcoding and qPCR and also has individual merits; for example, target tissue materials are not required, and a basic thermal cycler is sufficient. Doi et al. (2021) developed an innovative novel method that uses an ultra-rapid mobile PCR platform to detect on-site eDNA, and it has been successfully applied to eDNA measurements of the fish species Hypophthalmichthys molitrix. The method reduced the measurement time to 30 min and provided high detectability.
DNA meta-barcoding is an approach used to simultaneously identify multiple taxa (Taberlet et al., 2012). Although DNA meta-barcoding is theoretically similar to classical DNA barcoding, there are differences in practical operation and target DNA regions (Hebert et al., 2003). Both barcoding methods rely on short DNA regions amplified by PCR, typically utilizing mitochondrial or ribosomal RNA (rRNA) genes. DNA meta-barcoding depends on universal primers for multiple taxa amplified by PCR. At present, a total of 25 primer sets have been recorded for the target genes 12S, 16S, Cytb, and COI that can be used for meta-barcoding analysis of fish (Bylemans et al., 2018; Zhang et al., 2020b; Wang et al., 2021). Previous in silico and in vitro tests revealed that almost all fish primers amplified relatively large proportions of the sequences of non-fish organisms in analyses of the primer performance regarding taxonomic specificity (Zhang et al., 2020b). Compared to other primer sets, 12S appears to have highly effective fish coverage and species resolution, and Cytb primers have generally provided fewer fish taxa than the 12S and 16S primers, while COI appears to be a less common target for fish than other mitochondrial genes (Zhang et al., 2020b). This may be due to universal primers being impeded by interspecific genetic variation (Deagle et al., 2014; Thomsen and Willerslev, 2015). Among 12S primer sets, Ac12S and AcMDB07 displayed the best performance for detecting fish species in lakes, and MiFish-U and AcMDB07 primers provided a higher taxonomic resolution and more accurate species-level assignments in research on freshwater fish biodiversity using eDNA meta-barcoding (Bylemans et al., 2018; Zhang et al., 2020b). Since the longer eDNA fragments can be amplified to provide more sequence information (Deiner et al., 2017; Bylemans et al., 2018), Ac12S and AcMDB07 (target DNA fragment approximately 300–400 bp) have provided the highest numbers of fish taxa and species-level assignments (Zhang et al., 2020b). Miya et al. (2015) developed a set of universal PCR primers (MiFish) for eDNA metabarcoding of fishes that have been used for monitoring multispecies fish eDNA in the Huddersfield Narrow Canal in the United Kingdom (McDevitt et al., 2019), a coastal wetland of the Pearl River Estuary (Zou et al., 2019), four Korean rivers (Alam et al., 2020), and coastal marine ecosystems of Japan (Ushio et al., 2017).
Environmental variation also can affect eDNA detectability. (Barnes and Turner, 2016). have shown that fish eDNA degradation increased under lower aerobic activity and pH. Seymour et al. (2018) used four sets of upland stream mesocosms with an acid–base gradient to assess the temporal and environmental degradation of multispecies eDNA (e.g., zooplankton, plants, and fish) and suggested that acidic environments could accelerate the degradation of eDNA.
High-throughput sequencing (HTS) can be used for species identification. With the development of HTS technology, meta-barcoding used for eDNA research has become more efficient and less expensive. The Illumina sequencing platforms are commonly used, including HiSeq and MiSeq sequencers, and choice of the sequencer is determined by the sequencing depth needed. Through HTS, molecular operational taxonomic units (MOTUs), short barcode sequences for species identification, can be obtained to yield taxonomic information at the species level (Bienert et al., 2012; Thomsen and Willerslev, 2015). Amplicon sequence variant (ASV) is an alternative approach for species level detection (Callahan et al., 2017). Forster et al. (2019) compared the performance of eDNA-based ASVs and MOTUs sequenced on the Illumina sequencing platform; ASVs were much closer to the known number of species than MOTUs, and thus the authors recommended ASVs as replacement for MOTUs. ASVs have been used for fish monitoring based on eDNA. Antognazza et al. (2020) used ASVs to detect fish species composition in a fragmented lowland river and compared eDNA-based and long-term fishing monitoring results; the results showed that eDNA could detect species such as European shads (Alosa spp.) with high conservation value that were never sampled by capture techniques.
Species identification on the basis of meta-barcoding can be conducted through various bioinformatics software programs such as PhyloPythia, Mothur, Usearch, QIIME, OBITools, and Cutadapt (Kalyuzhnaya et al., 2008; Schloss et al., 2009; Caporaso et al., 2010; Edgar, 2010; Sigsgaard et al., 2017). QIIME and OBITools are usually the independent software packages used for eDNA HTS sequences analysis of fishes, and both were developed in Python. QIIME can be run in many environments (Caporaso et al., 2010; Kuczynski et al., 2011), and OBITOOLS can only be operated under the UNIX environment (Biggs et al., 2015; Boyer et al., 2016). Pipelines of sequences analysis include the following: 1) Filtering raw reads to discard low frequency and error reads from further analysis, a process that comprises the three main procedures of demultiplexing raw reads, alignment of paired-end reads, and quality control of aligned reads. Errors can be reduced by quality filtering, and the parameters can be set according to needs of individual research. Choice of parameters should be in accordance with target fragments. 2) Clustering filtered reads into MOTUs or ASVs at a number of different similarity cutoffs, where the cutoff value is a parameter used in the clustering algorithm (Balasingham et al., 2017). Recent studies on fish eDNA have adopted a minimum 97% similarity cutoff (Williams et al., 2017; Majaneva et al., 2018). 3) Comparison of MOTUs or ASVs and taxon annotation using specific DNA sequence databases. Comparing MOTUs or ASVs to classical taxonomy is an important step in eDNA research, and NCBI-Blast is the most commonly used tool. Using relevant sequence analysis programs requires researchers to have certain knowledge and operating experience with computer programs, a situation that has made these approaches difficult for many ecologists. To solve this problem, HTS data analysis platforms were developed that are simpler and more user-friendly for researchers unfamiliar with computer programs. MitoFish and MiFish Pipeline were developed for analysis of mitochondrial genome databases of fish for eDNA. The programs are key sequence analysis platforms for studies of fish evolution, ecology, and conservation (Sato et al., 2018).
Undoubtedly, a variety of eDNA protocols have already been applied aiming at different research objectives and study areas. However, an eDNA beginner or an ecological monitor may feel puzzled facing such a variety of protocols. The above review on sampling and experimental methods used in the articles we screened combined with our own eDNA research experience allows us to propose a set of basic eDNA field and laboratory protocols for the majority of research situations (Table 3). The suitability, portability, efficiency, and cost are considerations for setting up a general protocol to decrease investment in equipment, reduce manpower, shorten sampling and filtering time, and save experimental expenses.

TABLE 3. Major steps and optimal protocols on field and laboratory of regular eDNA-base fish ecological research.
Choice of an appropriate volume of water sample needs to consider whether it can supply enough taxonomic information and whether it can complete filtration within a limited sampling time. A 2 L water volume appears to be the proper size for sample replicates from lakes, rivers, and offshore seawater, as this volume can reveal more fish species compare to ≤1 L volumes while saving filtering time (Muha et al., 2019). In contrast, sampling a larger volume of water may extend the time needed for filtering and increase the probability of DNA degradation. Water sample replication per sampling site is aimed to eliminate randomness, and three replicates is the lowest statistical standard. Spatial design of sampling sites needs to cover all water layers and various micro-habitats to collect eDNA of all species as much as possible (Table 3; Hänfling et al., 2016; Zhang et al., 2020a).
Filtration is the most recommended eDNA collection method, because it can collect eDNA from larger water samples of 2 L, while precipitation and centrifugation methods have been shown to be suitable to collect eDNA from samples of 30 ml or less (Sigsgaard et al., 2015; Wang et al., 2021). The 0.45 μm GF and MCE filters generally have best performance on DNA yield and species detection in clear and general turbid water bodies. The MCE filters with the larger pore size (i.e., >1 μm) could be adopted for collecting water samples from extremely turbid waters. Although 0.22–0.26 μm filters may obtain higher DNA yields compared to 0.45 μm filters, those with the smaller pore size are very easy to be clogged by suspended matter and algae, even in relatively clear water bodies, and thus we do not recommend the use of the smaller pore-size filters. Pre-filtration using >10 μm filtering membranes is recommended to prevent clogging. eDNA storage at freezing temperature is still the best choice no matter what filtration methods applied, and ordinarily the portable car or ship refrigerator can provide storage conditions in the field. Because we found that ethanol and lysis buffers led to unstable storage effects under different temperature conditions in our research, these two methods are not considered as the best options for routine eDNA research. For eDNA extraction, the DNeasy kit and the PowerWater kit are most often used for total DNA extracted from water samples, and they have similar effects on DNA concentration and inhibitor removal, and the PowerWater kit has the better performance. However, the DNeasy kit is more economical, because its price is only one fourth of the price of the PowerWater kit. Therefore, the former kit is more recommended for regular research, especially in the case of limited funds. The remaining several kinds of commercial kits all have features not ideal for DNA yields from water samples, even though “inhibitor removal” steps are included. When using the DNeasy kit, the spin column protocols should be modified to be suitable for water samples rather than tissue samples (Table 3; Sigsgaard et al., 2017).
PCR is an indispensable step for fish species diversity detection. The 12S rRNA gene is preferred to detect interspecies variation of fish, and one of five most-used 12S primer sets can be chosen (Table 4; Zhang et al., 2020b; Wang et al., 2021). PCR system ratio and reaction conditions are not completely fixed due to different DNA template concentrations and primer sets used, and even using different PCR machines. Therefore, one must explore the most suitable reaction system and conditions in individual research.
The Illumina HiSeq sequencer platform is the better choice for eDNA meta-barcoding HTS, because it can obtain sufficient data depth for sequence analysis. Methods and specific software for bioinformatic analysis of eDNA data are being continuously developed and updated, and algorithm choice is closely related to the targets of the research. QIIME, OBITools, Vsearch, and Trimmomatic are software programs we have used, and they can effectively analyze and cluster sequence reads into MOTUs and complete taxon annotation using NCBI-blast. Filtered reads are usually clustered into MOTUs with a >97% similarity cutoff. The GenBank database of NCBI is also the first choice of reference database for eDNA-based fish taxa identification because it contains a wide variety of global fish species nucleotide sequences.
Contamination is a serious challenge for eDNA research, because it can occur in all stages of the laboratory protocols, including eDNA collection, extraction, amplification, and library building. Sampling from the field is the experimental step most at risk of cross-contamination. According to previous studies, 10% bleach can be used to clean sampling appliances, including the ship or boat used, and mouth masks and disposable latex gloves should be worn by samplers to prevent DNA contamination (Sigsgaard et al., 2017; Muha et al., 2019; Kusanke et al., 2020; Riaz et al., 2020). Unmanned aerial vehicles (UAVs) have been used as a new technique to collect environmental samples for eDNA studies and avoid contamination (Doi et al., 2017). UAVs were used to collect water samples from the dam reservoir of the Joge River, Japan, and the results were compared with traditional boat sampling; the quantitative DNA of two species (bluegill sunfish and largemouth bass) were analyzed by qPCR. The results showed that the concentration of DNA with the UAV sampling method was equal to or higher than with the boat sampling method, and the contamination risk was lower.
Lab contamination is unavoidable, especially when the frequency of PCR is very high during eDNA lab work. Therefore, clean labs should be built only for eDNA experiments, and strict decontamination procedures and the separation of pre- and post-PCR labs should be followed to limit cross-contamination during experimental stages (Thomsen and Willerslev, 2015; Goldberg et al., 2016). Negative controls should be made to monitor contamination at the links of water sampling, eDNA collection, extraction, amplification, and library building (Goldberg et al., 2016). During HTS, many cross-contamination target fragments are produced, and these fragments should be removed when sequence analyses are conducted using specific computer software.
Factors such as humic acids, humus, and organic compounds contained in environmental samples are extracted together with the target DNA, and these have an inhibitory effect on subsequent PCR. Inhibition reduction depends on DNA extraction dilution and purification (Biggs et al., 2015; Balasingham et al., 2017; Williams et al., 2017; Harper et al., 2019). Since eDNA samples may contain many impurities, we chose to dilute eDNA templates 10 times before PCR. PCR reaction enhancers such as BSA, RSA, DMSO, and Tween 200 as well as reducing the amount of DNA template added are also able to decrease the impact of inhibitors on PCR (Takahara et al., 2015; Sidstedt et al., 2017; King et al., 2019). qPCR and ddPCR can be used to make quantitative analysis of eDNA, although inhibition exists (Hindson et al., 2011; Nathan et al., 2014; Doi et al., 2015b). In addition, PCR inhibition can be tested through qPCR to define “Ct shift” (Jane et al., 2015).
Species identification derived from eDNA fragments depends of necessity on a reference DNA database, and related DNA sequence databases still need to be established, although global gene databases have been built, including NCBI (www.ncbi.nlm.nih.gov/genbank), databases of DNA barcodes targeting specific biological taxa such as World’s Fish (www.fishbol.org), the mammal DNA database (http://www.mammaliabol.org), and the bird species DNA database (http://www.barcodingbirds.org) as well as databases for specific regions such as the European Molecular Biology Laboratory (https://www.embl.org). China harbors high biological diversity and 9% of global freshwater fish species (Xing et al., 2016). China itself, however, has no DNA database. When studying the species diversity of cave fish using eDNA, we found that less than 50% of the DNA barcodes of Chinese cave fishes were collected in the NCBI database, a situation that hindered our research and prevented it from proceeding smoothly (Bai et al., 2020). Therefore, the continuous improvement of DNA reference databases will be crucial for the development of eDNA research, including DNA barcodes and even complete mitochondrial DNA for wider application.
Sequencing errors and low-quality sequences are inevitable, although sequencing technology is always evolving, and such errors are important factors leading to inaccurate detection results based on eDNA meta-barcoding (Kunin et al., 2010; Thomsen and Willerslev, 2015). Therefore, improving bioinformatic pipelines and developing useful computer programs for raw sequences filtering and quality control will be beneficial for removing interfering sequences and increasing identification rates of MOTUs, in addition to improving reference databases. Trimmomatic is a popular tool at present that performs a variety of useful trimming tasks for Illumina sequencing platform results to control raw sequences quality (Bolger et al., 2014). Undoubtedly, bioinformatic analysis and pipeline optimization will be important aspects of future eDNA meta-barcoding analyses.
eDNA has been widely accepted and adopted as a useful tool in research on fish ecology and water environmental health. In this review, we have attempted to recommend optimal field and laboratory protocols, presenting the merits and drawbacks of each protocol at each step in the process to help general users of eDNA to understand the outline of this method and how to perform it in their research. However, it is difficult to evaluate efficiencies of every protocol or provide a set of definite eDNA protocols available for different fish taxa, habitats, and research targets only through comparative study of the literature. Therefore, we recommend the protocols that are consistent with most common eDNA sampling and experimental steps used in recent fish ecological research. eDNA techniques and analysis methods are still developing and improving. This method is not only used in countries such as the United States, Europe, Japan, and Australia but also is widely applied in countries that are facing water environmental degradation and decreasing biodiversity, and thus the future of eDNA indicates the methods becoming routine measurement for biodiversity changes and water ecosystem evaluation in fish ecology. We hope that eDNA methods will become more standardized and portable so that users besides professional researchers can master this method.
YX. and YZ. reviewed and compared protocols from the literature and setup the optimal protocols. WG. searched and screened the literature. All co-authors drafted the manuscript and provided comments and intellectual contributions to this manuscript and have approved it for publication.
This research was funded by the National Natural Science Foundation of China (No. 32172975), the Central Public-Interest Scientific Institution Basal Research Fund, CAFS (No. 2017B001 and 2020A001), the Hainan Provincial Joint Project of Sanya Yazhou Bay Science and Technology City Grant No. 320LH069, the Bureau of Agriculture and Rural Affairs of Wenchang City Grant No. WCCYY202205 and Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO.2020TD11) to YX, the National Natural Science Foundation of China (No. 31972868), the Sino BON-Inland Water Fish Diversity Observation Network, and the National Science and Technology Fundamental Resources Investigation Program of China (Grant No. 2019FY101800) to YZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adrian-Kalchhauser, I., and Burkhardt-Holm, P. (2016). An eDNA Assay to Monitor a Globally Invasive Fish Species from Flowing Freshwater. PLoS One 11 (1), e0147558. doi:10.1371/journal.pone.0147558
Akamatsu, Y., Kume, G., Gotou, M., Kono, T., Fujii, T., Inui, R., et al. (20202020). Using Environmental DNA Analyses to Assess the Occurrence and Abundance of the Endangered Amphidromous Fish Plecoglossus Altivelis Ryukyuensis. Biodivers Data J. 8, e39679. doi:10.3897/BDJ.8.e39679
Alam, M. J., Kim, N.-K., Andriyono, S., Choi, H.-k., Lee, J.-H., and Kim, H.-W. (2020). Assessment of Fish Biodiversity in Four Korean Rivers Using Environmental DNA Metabarcoding. PeerJ 8, e9508. doi:10.7717/peerj.9508
Antognazza, C. M., Britton, R. J., Read, D. S., Goodall, T., Mantzouratou, A., De Santis, V., et al. (2020). Application of eDNA Metabarcoding in a Fragmented lowland River: Spatial and Methodological Comparison of Fish Species Composition. Environ. DNA 3 (2), 458–471. doi:10.1002/edn3.136
Atkinson, S., Carlsson, J. E. L., Ball, B., Egan, D., Kelly-Quinn, M., Whelan, K., et al. (2018). A Quantitative PCR-Based Environmental DNA Assay for Detecting Atlantic salmon (Salmo salar L.). Aquat. Conserv: Mar Freshw Ecosyst 28 (5), 1238–1243. doi:10.1002/aqc.2931
Bagley, M., Pilgrim, E., Knapp, M., Yoder, C., Santo Domingo, J., and Banerji, A. (2019). High-throughput Environmental DNA Analysis Informs a Biological Assessment of an Urban Stream. Ecol. Indicators 104, 378–389. doi:10.1016/j.ecolind.2019.04.088
Bai, J., Xing, Y. C., Gao, W. R., and Zhao, Y. H. (2020). Environmental DNA: an Emerging Tool in Studying Cave Organisms. Carsologica Sinica. Available at: https://kns.cnki.net/kcms/detail/45.1157.P.20201214.1134.004.html.
Balasingham, K. D., Walter, R. P., and Heath, D. D. (2017). Residual eDNA Detection Sensitivity Assessed by Quantitative Real-Time PCR in a River Ecosystem. Mol. Ecol. Resour. 17 (3), 523–532. doi:10.1111/1755-0998.12598
Barnes, M. A., and Turner, C. R. (2016). The Ecology of Environmental DNA and Implications for Conservation Genetics. Conserv Genet. 17, 1–17. doi:10.1007/s10592-015-0775-4
Bienert, F., De Danieli, S., Miquel, C., Coissac, E., Poillot, C., Brun, J.-J., et al. (2012). Tracking Earthworm Communities from Soil DNA. Mol. Ecol. 21 (80), 2017–2030. doi:10.1111/j.1365-294X.2011.05407.x
Biggs, J., Ewald, N., Valentini, A., Gaboriaud, C., Dejean, T., Griffiths, R. A., et al. (2015). Using eDNA to Develop a National Citizen Science-Based Monitoring Programme for the Great Crested Newt (Triturus Cristatus). Biol. Conservation 183, 19–28. doi:10.1016/j.biocon.2014.11.029
Bista, I., Carvalho, G. R., Walsh, K., Seymour, M., Hajibabaei, M., Lallias, D., et al. (2017). Annual Time-Series Analysis of Aqueous eDNA Reveals Ecologically Relevant Dynamics of lake Ecosystem Biodiversity. Nat. Commun. 8, 14087. doi:10.1038/ncomms14087
Bohmann, K., Evans, A., Gilbert, M. T. P., Carvalho, G. R., Creer, S., Knapp, M., et al. (2014). Environmental DNA for Wildlife Biology and Biodiversity Monitoring. Trends Ecol. Evol. 29, 358–367. doi:10.1016/j.tree.2014.04.003
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30 (15), 2114–2120. doi:10.1093/bioinformatics/btu170
Boothroyd, M., Mandrak, N. E., Fox, M., and Wilson, C. C. (2016). Environmental DNA (eDNA) Detection and Habitat Occupancy of Threatened Spotted Gar (Lepisosteus oculatus). Aquat. Conserv: Mar. Freshw. Ecosyst. 26 (6), 1107–1119. doi:10.1002/aqc.2617
Boyer, F., Mercier, C., Bonin, A., Le Bras, Y., Taberlet, P., and Coissac, E. (2016). Obitools: Aunix-Inspired Software Package for DNA Metabarcoding. Mol. Ecol. Resour. 16, 176–182. doi:10.1111/1755-0998.12428
Brys, R., Halfmaerten, D., Neyrinck, S., Mauvisseau, Q., Auwerx, J., Sweet, M., et al. (2021). Reliable eDNA Detection and Quantification of the European Weather Loach ( Misgurnus fossilis ). J. Fish. Biol. 98, 399–414. doi:10.1111/jfb.14315
Bylemans, J., Gleeson, D. M., Hardy, C. M., and Furlan, E. (2018). Toward an Ecoregion Scale Evaluation of eDNA Metabarcoding Primers: A Case Study for the Freshwater Fish Biodiversity of the Murray-Darling Basin (Australia). Ecol. Evol. 8 (17), 8697–8712. doi:10.1002/ece3.4387
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 11, 2639–2643. doi:10.1038/ismej.2017.119
Cantera, I., Cilleros, K., Valentini, A., Cerdan, A., Dejean, T., Iribar, A., et al. (2019). Optimizing Environmental DNA Sampling Effort for Fish Inventories in Tropical Streams and Rivers. Sci. Rep. 9 (1), 3085. doi:10.1038/s41598-019-39399-5
Capo, E., Spong, G., Norman, S., Königsson, H., Bartels, P., and Byström, P. (2019). Droplet Digital PCR Assays for the Quantification of Brown trout (Salmo trutta) and Arctic Char (Salvelinus alpinus) from Environmental DNA Collected in the Water of mountain lakes. PLoS One 14 (12), e0226638. doi:10.1371/journal.pone.0226638
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 7 (5), 335–336. doi:10.1038/nmeth.f.303
Civade, R., Dejean, T., Valentini, A., Roset, N., Raymond, J.-C., Bonin, A., et al. (2016). Spatial Representativeness of Environmental DNA Metabarcoding Signal for Fish Biodiversity Assessment in a Natural Freshwater System. PLoS One 11, e0157366. doi:10.1371/journal.pone.0157366
Clusa, L., and García-Vázquez, E. (2018). A Simple, Rapid Method for Detecting Seven Common Invasive Fish Species in Europe from Environmental DNA. Aquat. Conserv: Mar Freshw Ecosyst 28 (3), 619–629. doi:10.1002/aqc.2890
Davison, P. I., Copp, G. H., Créach, V., Vilizzi, L., and Britton, J. R. (2017). Application of Environmental DNA Analysis to Inform Invasive Fish Eradication Operations. Sci. Nat. 104 (3-4), 35. doi:10.1007/s00114-017-1453-9
Deagle, B. E., Jarman, S. N., Coissac, E., Pompanon, F., and Taberlet, P. (2014). DNA Metabarcoding and the Cytochrome C Oxidase Subunit I Marker: Not a Perfect Match. Biol. Lett. 10, 20140562. doi:10.1098/rsbl.2014.0562
Deiner, K., Renshaw, M. A., Li, Y., Olds, B. P., Lodge, D. M., and Pfrender, M. E. (2017). Long‐range PCR Allows Sequencing of Mitochondrial Genomes from Environmental DNA. Methods Ecol. Evol. 8, 1888–1898. doi:10.1111/2041-210X.12836
Deiner, K., Walser, J.-C., Mächler, E., and Altermatt, F. (2015). Choice of Capture and Extraction Methods Affect Detection of Freshwater Biodiversity from Environmental DNA. Biol. Conservation 183, 53–63. doi:10.1016/j.biocon.2014.11.018
Dejean, T., Valentini, A., Duparc, A., Pellier-Cuit, S., Pompanon, F., Taberlet, P., et al. (2011). Persistence of Environmental DNA in Freshwater Ecosystems. PLoS One 6, e23398. doi:10.1371/journal.pone.0023398
Doi, H., Takahara, T., Minamoto, T., Matsuhashi, S., Uchii, K., and Yamanaka, H. (2015a). Droplet Digital Polymerase Chain Reaction (PCR) Outperforms Real-Time PCR in the Detection of Environmental DNA from an Invasive Fish Species. Environ. Sci. Technol. 49, 5601–5608. doi:10.1021/acs.est.5b00253
Doi, H., Uchii, K., Matsuhashi, S., Takahara, T., Yamanaka, H., and Minamoto, T. (2017). Isopropanol Precipitation Method for Collecting Fish Environmental DNA. Limnol. Oceanogr. Methods 15, 212–218. doi:10.1002/lom3.10161
Doi, H., Uchii, K., Takahara, T., Matsuhashi, S., Yamanaka, H., and Minamoto, T. (2015b). Use of Droplet Digital PCR for Estimation of Fish Abundance and Biomass in Environmental DNA Surveys. PLoS One 10, e0122763. doi:10.1371/journal.pone.0122763
Doi, H., Watanabe, T., Nishizawa, N., Saito, T., Nagata, H., Kameda, Y., et al. (2021). On‐site Environmental DNA Detection of Species Using Ultrarapid mobile PCR. Mol. Ecol. Resour. 21 (7), 2364–2368. doi:10.1111/1755-0998.13448
Dougherty, M. M., Larson, E. R., Renshaw, M. A., Gantz, C. A., Egan, S. P., Erickson, D. M., et al. (2016). Environmental DNA (eDNA) Detects the Invasive Rusty crayfishOrconectes Rusticusat Low Abundances. J. Appl. Ecol. 53, 722–732. doi:10.1111/1365-2664.12621
Dressler, T. L., Lafferty, K. D., Jerde, C. L., and Dudley, T. (2020). Looking where It's Hard to See: a Case Study Documenting Rare Eucyclogobius Newberryi Presence in a California Lagoon. J. Fish. Biol. 97 (2), 572–576. doi:10.1111/jfb.14401
Edgar, R. C. (2010). Search and Clustering Orders of Magnitude Faster Than BLAST. Bioinformatics 26 (19), 2460–2461. doi:10.1093/bioinformatics/btq461
Edwards, M. E., Alsos, I. G., Yoccoz, N., Coissac, E., Goslar, T., Gielly, L., et al. (2018). Metabarcoding of Modern Soil DNA Gives a Highly Local Vegetation Signal in Svalbard Tundra. The Holocene 28, 2006–2016. doi:10.1177/0959683618798095
Eichmiller, J. J., Miller, L. M., and Sorensen, P. W. (2015). Optimizing Techniques to Capture and Extract Environmental DNA for Detection and Quantification of Fish. Mol. Ecol. Resour. 16 (1), 56–68. doi:10.1111/1755-0998.12421
Erickson, R. A., Rees, C. B., Coulter, A. A., Merkes, C. M., Mccalla, S. G., Touzinsky, K. F., et al. (2016). Detecting the Movement and Spawning Activity of Bigheaded Carps with Environmental DNA. Mol. Ecol. Resour. 16, 957–965. doi:10.1111/1755-0998.12533
Eva, B., Harmony, P., Thomas, G., Francois, G., Alice, V., Claude, M., et al. (2016). Trails of River Monsters: Detecting Critically Endangered Mekong Giant Catfish Pangasianodon Gigas Using Environmental DNA. Glob. Ecol. Conservation 7, 148–156. doi:10.1016/j.gecco.2016.06.007
Evans, N. T., Olds, B. P., Renshaw, M. A., Turner, C. R., Li, Y., Jerde, C. L., et al. (2016). Quantification of Mesocosm Fish and Amphibian Species Diversity via Environmental DNA Metabarcoding. Mol. Ecol. Resour. 16, 29–41. doi:10.1111/1755-0998.12433
Farrington, H. L., Edwards, C. E., Guan, X., Carr, M. R., Baerwaldt, K., and Lance, R. F. (2016). Mitochondrial Genome Sequencing and Development of Genetic Markers for the Detection of DNA of Invasive Bighead and Silver Carp (Hypophthalmichthys Nobilis and H. molitrix) in Environmental Water Samples from the United States. PLoS One 10, e0117803. doi:10.1371/journal.pone.0117803
Ficetola, G. F., Miaud, C., Pompanon, F., and Taberlet, P. (2008). Species Detection Using Environmental DNA from Water Samples. Biol. Lett. 4, 423–425. doi:10.1098/rsbl.2008.0118
Forster, D., Lentendu, G., Filker, S., Dubois, E., Wilding, T. A., and Stoeck, T. (2019). Improving eDNA‐based Protist Diversity Assessments Using Networks of Amplicon Sequence Variants. Environ. Microbiol. 21 (11), 4109–4124. doi:10.1111/1462-2920.14764
Fraija-Fernández, N., Bouquieaux, M.-C., Rey, A., Mendibil, I., Cotano, U., Irigoien, X., et al. (2020). Marine Water Environmental DNA Metabarcoding Provides a Comprehensive Fish Diversity Assessment and Reveals Spatial Patterns in a Large Oceanic Area. Ecol. Evol. 10 (14), 7560–7584. doi:10.1101/864710
Fukaya, K., Murakami, H., Yoon, S., Minami, K., Osada, Y., Yamamoto, S., et al. (20202020). Estimating Fish Population Abundance by Integrating Quantitative Data on Environmental DNA and Hydrodynamic Modelling. Mol. Ecol. 30, 3057–3067. doi:10.1111/mec.15530
Gilbey, J., Carvalho, G., Castilho, R., Coscia, I., Coulson, M. W., Dahle, G., et al. (2021). Life in a Drop: Sampling Environmental DNA for marine Fishery Management and Ecosystem Monitoring. Mar. Pol. 124, 104331. doi:10.1016/j.marpol.2020.104331
Goldberg, C. S., Turner, C. R., Deiner, K., Klymus, K. E., Thomsen, P. F., Murphy, M. A., et al. (2016). Critical Considerations for the Application of Environmental DNA Methods to Detect Aquatic Species. Methods Ecol. Evol. 7, 1299–1307. doi:10.1111/2041-210X.12595
Goutte, A., Molbert, N., Guérin, S., Richoux, R., and Rocher, V. (2020). Monitoring Freshwater Fish Communities in Large Rivers Using Environmental DNA Metabarcoding and a Long‐term Electrofishing Survey. J. Fish. Biol. 97 (2), 444–452. doi:10.1111/jfb.14383
Griffiths, N. P., Bolland, J. D., Wright, R. M., Murphy, L. A., Donnelly, R. K., Watson, H. V., et al. (2020). Environmental DNA Metabarcoding Provides Enhanced Detection of the European Eel Anguilla anguilla and Fish Community Structure in Pumped River Catchments. J. Fish. Biol. 97, 1375–1384. doi:10.1111/jfb.14497
Guan, X., Monroe, E. M., Bockrath, K. D., Mize, E. L., Rees, C. B., Lindsay, D. L., et al. (2019). EnvironmentalDNA(eDNA) Assays for Invasive Populations of Black Carp in North America. Trans. Am. Fish. Soc. 148 (6), 1043–1055. doi:10.1002/tafs.10195
Hänfling, B., Lawson Handley, L., Read, D. S., Hahn, C., Li, J., Nichols, P., et al. (2016). Environmental DNA Metabarcoding of lake Fish Communities Reflects Long-Term Data from Established Survey Methods. Mol. Ecol. 25, 3101–3119. doi:10.1111/mec.13660
Hänfling, B., and Lawson‐Handley, L. (2021). EnvironmentalDNA‐based Approaches for the Monitoring of Fish Populations Have Come of Age. J. Fish. Biol. 98, 339–340. doi:10.1111/jfb.14660
Harper, L. R., Buxton, A. S., Rees, H. C., Bruce, K., Brys, R., Halfmaerten, D., et al. (2019). Prospects and Challenges of Environmental DNA (eDNA) Monitoring in Freshwater Ponds. Hydrobiologia 826, 25–41. doi:10.1007/s10750-018-3750-5
Harper, L. R., Griffiths, N. P., Lawson Handley, L., Sayer, C. D., Read, D. S., Harper, K. J., et al. (2018). Development and Application of Environmental DNA Surveillance for the Threatened Crucian Carp (Carassius carassius ). Freshw. Biol. 64, 93–107. doi:10.1111/fwb.13197
Hebert, P. D. N., Cywinska, A., Ball, S. L., and DeWaard, J. R. (2003). Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B 270 (1512), 313–321. doi:10.1098/rspb.2002.2218
Hindson, B. J., Ness, K. D., Masquelier, D. A., Belgrader, P., Heredia, N. J., Makarewicz, A. J., et al. (2011). High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 83 (22), 8604–8610. doi:10.1021/ac202028g
Hinlo, R., Gleeson, D., Lintermans, M., and Furlan, E. (2017). Methods to Maximise Recovery of Environmental DNA from Water Samples. PLoS One 12, e0179251. doi:10.1371/journal.pone.0179251
Hunter, M. E., Ferrante, J. A., Meigs-Friend, G., and Ulmer, A. (2019). Improving eDNA Yield and Inhibitor Reduction through Increased Water Volumes and Multi-Filter Isolation Techniques. Sci. Rep. 9, 5259. doi:10.1038/s41598-019-40977-w
Jane, S. F., Wilcox, T. M., McKelvey, K. S., Young, M. K., Schwartz, M. K, Lowe, W. H., et al. (2015). Distance, Flow and PCR Inhibition: eDNA Dynamics in Two Headwater Streams. Mol. Ecol. Resour. 15 (4), 216–27. doi:10.1111/1755-0998.12285
Jerde, C. L., Chadderton, W. L., Mahon, A. R., Renshaw, M. A., Corush, J., Budny, M. L., et al. (2013). Detection of Asian Carp DNA as Part of a Great Lakes basin-wide Surveillance Program. Can. J. Fish. Aquat. Sci. 70, 522–526. doi:10.1139/cjfas-2012-0478
Jo, T., Arimoto, M., Murakami, H., Masuda, R., and Minamoto, T. (2019a). Particle Size Distribution of Environmental DNA from the Nuclei of marine Fish. Environ. Sci. Technol. 53 (16), 9947–9956. doi:10.1021/acs.est.9b02833
Jo, T., Fukuoka, A., Uchida, K., Ushimaru, A., and Minamoto, T. (2020). Multiplex Real-Time PCR Enables the Simultaneous Detection of Environmental DNA from Freshwater Fishes: a Case Study of Three Exotic and Three Threatened Native Fishes in Japan. Biol. Invasions 22, 455–471. doi:10.1007/s10530-019-02102-w
Juhel, J.-B., Utama, R. S., Marques, V., Vimono, I. B., Sugeha, H. Y., Kadarusman, , et al. (2020). Accumulation Curves of Environmental DNA Sequences Predict Coastal Fish Diversity in the Coral triangle. Proc. R. Soc. B. 287, 20200248. doi:10.1098/rspb.2020.0248
Kalyuzhnaya, M. G., Lapidus, A., Ivanova, N., Copeland, A. C., McHardy, A. C., Szeto, E., et al. (2008). High-resolution Metagenomics Targets Specific Functional Types in Complex Microbial Communities. Nat. Biotechnol. 26, 1029–1034. doi:10.1038/nbt.1488
Keskin, E., Unal, E. M., and Atar, H. H. (2016). Detection of Rare and Invasive Freshwater Fish Species Using eDNA Pyrosequencing: Lake Iznik Ichthyofauna Revised. Biochem. Syst. Ecol. 67, 29–36. doi:10.1016/j.bse.2016.05.020
King, C. E., Debruyne, R., Kuch, M., Schwarz, C., and Poinar, H. N. (2009). A Quantitative Approach to Detect and Overcome PCR Inhibition in Ancient DNA Extracts. Biotechniques 47, 941–949. doi:10.2144/000113244
Klymus, K. E., Richter, C. A., Chapman, D. C., and Paukert, C. (2015). Quantification of eDNA Shedding Rates from Invasive Bighead Carp Hypophthalmichthys Nobilis and Silver Carp Hypophthalmichthys Molitrix. Biol. Conservation 183, 77–84. doi:10.1016/j.biocon.2014.11.020
Kuczynski, J., Stombaugh, J., Walters, W. A., González, A., Caporaso, J. G., and Knight, R. (2011). Using QIIME to Analyze 16S rRNA Gene Sequences from Microbial Communities. Curr. Protoc. Bioinformatics 36, e1007. doi:10.1002/0471250953.bi1007s36
Kunin, V., Engelbrektson, A., Ochman, H., and Hugenholtz, P. (2010). Wrinkles in the Rare Biosphere: Pyrosequencing Errors Can lead to Artificial Inflation of Diversity Estimates. Environ. Microbiol. 12, 118–123. doi:10.1111/j.1462-2920.2009.02051.x
Kusanke, L. M., Panteleit, J., Stoll, S., Korte, E., Sünger, E., Schulz, R., et al. (2020). Detection of the Endangered European Weather Loach ( Misgurnus fossilis ) via Water and Sediment Samples: Testing Multiple eDNA Workflows. Ecol. Evol. 10 (15), 8331–8344. doi:10.1002/ece3.6540
Lacoursière-Roussel, A., Rosabal, M., and Bernatchez, L. (2016). Estimating Fish Abundance and Biomass from eDNA Concentrations: Variability Among Capture Methods and Environmental Conditions. Mol. Ecol. Resour. 16, 1401–1414. doi:10.1111/1755-0998.12522
Li, J., Lawson Handley, L.-J., Read, D. S., and Hänfling, B. (2018). The Effect of Filtration Method on the Efficiency of Environmental DNA Capture and Quantification via Metabarcoding. Mol. Ecol. Resour. 18 (5), 1102–1114. doi:10.1111/1755-0998.12899
Lindahl, T. (1993). Instability and Decay of the Primary Structure of DNA. Nature 362, 709–715. doi:10.1038/362709a0
Majaneva, M., Diserud, O. H., Eagle, S. H. C., Boström, E., Hajibabaei, M., and Ekrem, T. (2018). Environmental DNA Filtration Techniques Affect Recovered Biodiversity. Sci. Rep. 8, 4682. doi:10.1038/s41598-018-23052-8
Maruyama, A., Sugatani, K., Watanabe, K., Yamanaka, H., and Imamura, A. (2018). Environmental DNA Analysis as a Non-invasive Quantitative Tool for Reproductive Migration of a Threatened Endemic Fish in Rivers. Ecol. Evol. 8, 11964–11974. doi:10.1002/ece3.4653
McDevitt, A. D., Sales, N. G., Browett, S. S., Sparnenn, A. O., Mariani, S., Wangensteen, O. S., et al. (2019). Environmental DNA Metabarcoding as an Effective and Rapid Tool for Fish Monitoring in Canals. J. Fish. Biol. 95 (2), 679–682. doi:10.1111/jfb.14053
Milhau, T., Valentini, A., Poulet, N., Roset, N., Jean, P., Gaboriaud, C., et al. (2021). Seasonal Dynamics of Riverine Fish Communities Using eDNA. J. Fish. Biol. 98, 387–398. doi:10.1111/jfb.14190
Minamoto, T., Naka, T., Moji, K., and Maruyama, A. (2016). Techniques for the Practical Collection of Environmental DNA: Filter Selection, Preservation, and Extraction. Limnology 17, 23–32. doi:10.1007/s10201-015-0457-4
Miya, M., Sato, Y., Fukunaga, T., Sado, T., Poulsen, J. Y., Sato, K., et al. (2015). MiFish, a Set of Universal PCR Primers for Metabarcoding Environmental DNA from Fishes: Detection of More Than 230 Subtropical marine Species. R. Soc. Open Sci. 2 (7), 150088. doi:10.1098/rsos.150088
Mizumoto, H., Urabe, H., Kanbe, T., Fukushima, M., and Araki, H. (2017). Establishing an Environmental DNA Method to Detect and Estimate the Biomass of Sakhalin Taimen, a Critically Endangered Asian Salmonid. Limnology 19, 219–227. doi:10.1007/s10201-017-0535-x
Muha, T. P., Robinson, C. V., Garcia de Leaniz, C., and Consuegra, S. (2019). An Optimised eDNA Protocol for Detecting Fish in Lentic and Lotic Freshwaters Using a Small Water Volume. PLoS One 14 (7), e0219218. doi:10.1371/journal.pone.0219218
Nathan, L. M., Simmons, M., Wegleitner, B. J., Jerde, C. L., and Mahon, A. R. (2014). Quantifying Environmental DNA Signals for Aquatic Invasive Species across Multiple Detection Platforms. Environ. Sci. Technol. 48 (21), 12800–12806. doi:10.1021/es5034052
Parrondo, M., Clusa, L., Mauvisseau, Q., and Borrell, Y. J. (2018). Citizen Warnings and post Checkout Molecular Confirmations Using eDNA as a Combined Strategy for Updating Invasive Species Distributions. J. Nat. Conservation 43, 95–103. doi:10.1016/j.jnc.2018.02.006
Pedersen, M. W., Overballe-Petersen, S., Ermini, L., Sarkissian, C. D., Haile, J., Hellstrom, M., et al. (2015). Ancient and Modern Environmental DNA. Phil. Trans. R. Soc. B 370, 1–11. doi:10.1098/rstb.2013.0383
Piggott, M. P. (2016). Evaluating the Effects of Laboratory Protocols on eDNA Detection Probability for an Endangered Freshwater Fish. Ecol. Evol. 6 (9), 2739–2750. doi:10.1002/ece3.2083
Pont, D., Rocle, M., Valentini, A., Civade, R., Jean, P., Maire, A., et al. (2018). Environmental DNA Reveals Quantitative Patterns of Fish Biodiversity in Large Rivers Despite its Downstream Transportation. Sci. Rep. 8 (1), 10361. doi:10.1038/s41598-018-28424-8
Rees, H. C., Maddison, B. C., Middleditch, D. J., Patmore, J. R. M., and Gough, K. C. (2014). REVIEW: The Detection of Aquatic Animal Species Using Environmental DNA - a Review of eDNA as a Survey Tool in Ecology. J. Appl. Ecol. 51, 1450–1459. doi:10.1111/1365-2664.12306
Renshaw, M. A., Olds, B. P., Jerde, C. L., McVeigh, M. M., and Lodge, D. M. (2015). The Room Temperature Preservation of Filtered Environmental DNA Samples and Assimilation into a Phenol-Chloroform-Isoamyl Alcohol DNA Extraction. Mol. Ecol. Resour. 15, 168–176. doi:10.1111/1755-0998.12281
Riaz, M., Kuemmerlen, M., Wittwer, C., Cocchiararo, B., Khaliq, I., Pfenninger, M., et al. (2020). Combining Environmental DNA and Species Distribution Modeling to Evaluate Reintroduction success of a Freshwater Fish. Ecol. Appl. 30 (2), e02034. doi:10.1002/eap.2034
Riaz, T., Shehzad, W., Viari, A., Pompanon, F., Taberlet, P., and Coissac, E. (2011). ecoPrimers: Inference of New DNA Barcode Markers from Whole Genome Sequence Analysis. Nucleic Acids Res. 39 (21), e145. doi:10.1093/nar/gkr732
Rojahn, J., Pearce, L., Gleeson, D. M., Duncan, R. P., Gilligan, D. M., and Bylemans, J. (2021). The Value of Quantitative Environmental DNA Analyses for the Management of Invasive and Endangered Native Fish. Freshw. Biol. 66 (8), 1619–1629. doi:10.1111/fwb.13779
Sakata, M. K., Maki, N., Sugiyama, H., and Minamoto, T. (2017). Identifying a Breeding Habitat of a Critically Endangered Fish, Acheilognathus typus, in a Natural River in Japan. Sci. Nat. 104, 100. doi:10.1007/s00114-017-1521-1
Sato, Y., Miya, M., Fukunaga, T., Sado, T., and Iwasaki, W. (2018). MitoFish and MiFish Pipeline: A Mitochondrial Genome Database of Fish with an Analysis Pipeline for Environmental DNA Metabarcoding. Mol. Biol. Evol. 35 (6), 1553–1555. doi:10.1093/molbev/msy074
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing Mothur: Open-Source, Platform-independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 75, 7537–7541. doi:10.1128/AEM.01541-09
Schmelzle, M. C., and Kinziger, A. P. (2016). Using Occupancy Modelling to Compare Environmental DNA to Traditional Field Methods for Regional-Scale Monitoring of an Endangered Aquatic Species. Mol. Ecol. Resour. 16, 895–908. doi:10.1111/1755-0998.12501
Schumer, G., Crowley, K., Maltz, E., Johnston, M., Anders, P., and Blankenship, S. (2019). Utilizing Environmental DNA for Fish Eradication Effectiveness Monitoring in Streams. Biol. Invasions 21, 3415–3426. doi:10.1007/s10530-019-02056-z
Sepulveda, A., Hutchins, P., Massengill, R., and Dunker, K. (2018). Tradeoffs of a Portable, Field-Based Environmental DNA Platform for Detecting Invasive Northern pike (Esox lucius) in Alaska. Mbi 9, 253–258. doi:10.3391/mbi.2018.9.3.07
Seymour, M., Durance, I., Cosby, B. J., Ransom-Jones, E., Deiner, K., Ormerod, S. J., et al. (2018). Acidity Promotes Degradation of Multi-Species Environmental DNA in Lotic Mesocosms. Commun. Biol. 1, 4. doi:10.1038/s42003-017-0005-3
Shu, L., Ludwig, A., and Peng, Z. (2020). Standards for Methods Utilizing Environmental DNA for Detection of Fish Species. Genes 11 (3), 296. doi:10.3390/genes11030296
Sidstedt, M., Romsos, E. L., Hedell, R., Ansell, R., Steffen, C. R., Vallone, P. M., et al. (2017). Accurate Digital Polymerase Chain Reaction Quantification of Challenging Samples Applying Inhibitor-Tolerant DNA Polymerases. Anal. Chem. 89, 1642–1649. doi:10.1021/acs.analchem.6b03746
Sigsgaard, E. E., Carl, H., Møller, P. R., and Thomsen, P. F. (2015). Monitoring the Near-Extinct European Weather Loach in Denmark Based on Environmental DNA from Water Samples. Biol. Conservation 183, 46–52. doi:10.1016/j.biocon.2014.11.023
Sigsgaard, E. E., Nielsen, I. B., Carl, H., Krag, M. A., Knudsen, S. W., Xing, Y., et al. (2017). Seawater Environmental DNA Reflects Seasonality of a Coastal Fish Community. Mar. Biol. 164, 128. doi:10.1007/s00227-017-3147-4
Sigsgaard, E. E., Torquato, F., Frøslev, T. G., Moore, A. B. M., Sørensen, J. M., Range, P., et al. (2020). Using Vertebrate Environmental DNA from Seawater in Biomonitoring of marine Habitats. Conservation Biol. 34 (3), 697–710. doi:10.1111/cobi.13437
Spikmans, F., Lemmers, P., op den Camp, H. J. M., van Haren, E., Kappen, F., Blaakmeer, A., et al. (2020). Impact of the Invasive Alien Topmouth Gudgeon (Pseudorasbora parva) and its Associated Parasite Sphaerothecum Destruens on Native Fish Species. Biol. Invasions 22, 587–601. doi:10.1007/s10530-019-02114-6
Stauffer, S., Jucker, M., Keggin, T., Marques, V., Andrello, M., Bessudo, S., et al. (2021). How many Replicates to Accurately Estimate Fish Biodiversity Using Environmental DNA on Coral Reefs. Ecol. Evol. 11 (21), 14630–14643. doi:10.1002/ece3.8150
Stoeckle, M. Y., Das Mishu, M., and Charlop-Powers, Z. (2018). GoFish: A Versatile Nested PCR Strategy for Environmental DNA Assays for marine Vertebrates. PLoS One 13 (12), e0198717. doi:10.1371/journal.pone.0198717
Strickland, G. J., and Roberts, J. H. (2018). Utility of eDNA and Occupancy Models for Monitoring an Endangered Fish across Diverse Riverine Habitats. Hydrobiologia 826, 129–144. doi:10.1007/s10750-018-3723-8
Sugiura, K., Tomita, S., Minamoto, T., Mishina, T., Iwata, A., Abe, T., et al. (2021). Characterizing the Spatial and Temporal Occurrence Patterns of the Endangered Botiid Loach Parabotia Curtus by Environmental DNA Analysis Using a Newly Developed Species-specific Primer Set. Ichthyol. Res. 68, 152–157. doi:10.1007/s10228-020-00756-4
Taberlet, P., Coissac, E., Hajibabaei, M., and Rieseberg, L. H. (2012). Environmental DNA. Mol. Ecol. 21, 1789–1793. doi:10.1111/j.1365-294x.2012.05542.x
Takahara, T., Minamoto, T., and Doi, H. (2015). Effects of Sample Processing on the Detection Rate of Environmental DNA from the Common Carp (Cyprinus carpio). Biol. Conservation 183, 64–69. doi:10.1016/j.biocon.2014.11.014
Takahara, T., Minamoto, T., and Doi, H. (2013). Using Environmental DNA to Estimate the Distribution of an Invasive Fish Species in Ponds. PLoS One 8 (2), e56584. doi:10.1371/journal.pone.0056584
Takahara, T., Minamoto, T., Yamanaka, H., Doi, H., and Kawabata, Z. i. (2012). Estimation of Fish Biomass Using Environmental DNA. PLoS One 7, e35868. doi:10.1371/journal.pone.0035868
Thomsen, P. F., Kielgast, J., Iversen, L. L., Wiuf, C., Rasmussen, M., Gilbert, M. T. P., et al. (2012). Monitoring Endangered Freshwater Biodiversity Using Environmental DNA. Mol. Ecol. 21, 2565–2573. doi:10.1111/j.1365-294X.2011.05418.x
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA - an Emerging Tool in Conservation for Monitoring Past and Present Biodiversity. Biol. Conservation 183, 4–18. doi:10.1016/j.biocon.2014.11.019
Tsuji, S., Iguchi, Y., Shibata, N., Teramura, I., Kitagawa, T., and Yamanaka, H. (2018). Real-time Multiplex PCR for Simultaneous Detection of Multiple Species from Environmental DNA: an Application on Two Japanese Medaka Species. Sci. Rep. 8, 9138. doi:10.1038/s41598-018-27434-w
Tsuji, S., Takahara, T., Doi, H., Shibata, N., and Yamanaka, H. (2019). The Detection of Aquatic Macroorganisms Using Environmental DNA Analysis-A Review of Methods for Collection, Extraction, and Detection. Environ. DNA 1, 99–108. doi:10.1002/edn3.21
Uchii, K., Doi, H., Yamanaka, H., and Minamoto, T. (2017). Distinct Seasonal Migration Patterns of Japanese Native and Non-native Genotypes of Common Carp Estimated by Environmental DNA. Ecol. Evol. 7, 8515–8522. doi:10.1002/ece3.3346
Ushio, M., Fukuda, H., Inoue, T., Makoto, K., Kishida, O., Sato, K., et al. (2017). Environmental DNA Enables Detection of Terrestrial Mammals from forest Pond Water. Mol. Ecol. Resour. 17 (6), e63–e75. doi:10.1111/1755-0998.12690
Valentini, A., Taberlet, P., Miaud, C., Civade, R., Herder, J., Thomsen, P. F., et al. (2016). Next-generation Monitoring of Aquatic Biodiversity Using Environmental DNA Metabarcoding. Mol. Ecol. 25 (4), 929–942. doi:10.1111/mec.13428
Wang, S., Yan, Z., Hänfling, B., Zheng, X., Wang, P., Fan, J., et al. (2021). Methodology of Fish eDNA and its Applications in Ecology and Environment. Sci. Total Environ. 755, 142622. doi:10.1016/j.scitotenv.2020.142622
West, K. M., Stat, M., Harvey, E. S., Skepper, C. L., DiBattista, J. D., Richards, Z. T., et al. (2020). eDNA Metabarcoding Survey Reveals fine‐scale Coral Reef Community Variation across a Remote, Tropical Island Ecosystem. Mol. Ecol. 29 (6), 1069–1086. doi:10.1111/mec.15382
Williams, M. R., Stedtfeld, R. D., Engle, C., Salach, P., Fakher, U., Stedtfeld, T., et al. (2017). Isothermal Amplification of Environmental DNA (eDNA) for Direct Field-Based Monitoring and Laboratory Confirmation of Dreissena Sp. PLoS One 12 (10), e0186462. doi:10.1371/journal.pone.0186462
Wilson, C., Wright, E., Bronnenhuber, J., MacDonald, F., Belore, M., and Locke, B. (2014). Tracking Ghosts: Combined Electrofishing and Environmental DNA Surveillance Efforts for Asian Carps in Ontario Waters of Lake Erie. Mbi 5 (3), 225–231. doi:10.3391/mbi.2014.5.3.05
Xing, Y., Zhang, C., Fan, E., and Zhao, Y. (2016). Freshwater Fishes of China: Species Richness, Endemism, Threatened Species and Conservation. Divers. Distrib. 22, 358–370. doi:10.1111/ddi.12399
Yamanaka, H., Minamoto, T., Matsuura, J., Sakurai, S., Tsuji, S., Motozawa, H., et al. (2017). A Simple Method for Preserving Environmental DNA in Water Samples at Ambient Temperature by Addition of Cationic Surfactant. Limnology 18, 233–241. doi:10.1007/s10201-016-0508-5
Yamanaka, H., Takao, D., Maruyama, A., and Imamura, A. (2018). Species-specific Detection of the Endangered Piscivorous Cyprinid Fish Opsariichthys uncirostris Uncirostris, Three-Lips, Using Environmental DNA Analysis. Ecol. Res. 33, 1075–1078. doi:10.1007/s11284-018-1612-2
Yu, D., Shen, Z., Chang, T., Li, S., and Liu, H. (2021). Using Environmental DNA Methods to Improve Detectability in an Endangered sturgeon (Acipenser Sinensis) Monitoring Program. BMC Ecol. Evo 21 (1), 216. doi:10.1186/s12862-021-01948-w
Zhang, S., Lu, Q., Wang, Y., Wang, X., Zhao, J., and Yao, M. (2020a). Assessment of Fish Communities Using Environmental DNA: Effect of Spatial Sampling Design in Lentic Systems of Different Sizes. Mol. Ecol. Resour. 20, 242–255. doi:10.1111/1755-0998.13105
Zhang, S., Zhao, J., and Yao, M. (2020b). A Comprehensive and Comparative Evaluation of Primers for Metabarcoding eDNA from Fish. Methods Ecol. Evol. 11, 1609–1625. doi:10.1111/2041-210X.13485
Zou, K., Chen, J., Ruan, H., Li, Z., Guo, W., Li, M., et al. (2019). eDNA Metabarcoding as a Promising Conservation Tool for Monitoring Fish Diversity in a Coastal Wetland of the Pearl River Estuary Compared to Bottom Trawling. Sci. Total Environ. 702, 134704. doi:10.1016/j.scitotenv10.1016/j.scitotenv.2019.134704
Keywords: eDNA, fish biodiversity and conservation, fish monitoring, analysis methods, experimental techniques, challenges and prospects
Citation: Xing Y, Gao W, Shen Z, Zhang Y, Bai J, Cai X, Ouyang J and Zhao Y (2022) A Review of Environmental DNA Field and Laboratory Protocols Applied in Fish Ecology and Environmental Health. Front. Environ. Sci. 10:725360. doi: 10.3389/fenvs.2022.725360
Received: 13 July 2021; Accepted: 24 January 2022;
Published: 25 February 2022.
Edited by:
Anik Bhaduri, Griffith University, AustraliaReviewed by:
Jeyaraj Antony Johnson, Wildlife Institute of India, IndiaCopyright © 2022 Xing, Gao, Shen, Zhang, Bai, Cai, Ouyang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingchun Xing, eGluZ3ljaEBjYWZzLmFjLmNu; Yahui Zhao, emhhb3loQGlvei5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.