- 1Department of Laboratory Medicine, Beijing Hospital, National Center of Gerontology; Institute of Geriatric Medicine, Chinese Academy of Medical Sciences, Beijing, China
- 2Clinical Laboratory Medicine, Peking University Ninth School of Clinical Medicine, Beijing, China
- 3National Center for Clinical Laboratories, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences, Beijing Hospital/National Center of Gerontology, Beijing, China
Combined with human-to-human contact, the potential for SARS-CoV-2 virus transmission via aerosols and feces raises the urgency for effective treatment of hospital wastewater (HWW), which has been assumed as an important source of pathogenic microorganisms spreading into aquatic environments. However, there are few reviews discussing the presence and removal of pathogens in hospital wastewaters matrices. This review summarizes the pathogenic microorganisms including bacteria, fungi, viruses and parasites present in hospital wastewater, the related diseases and treatment processes. In addition, the removal of pathogens during hospital wastewater treatment process is reviewed, including the preliminary process, secondary process and tertiary process. Due to the growing concerns over the effects of the current global pandemic on hospital wastewater treatment process, further research is necessary to investigate the actual fate of pathogens in hospital wastewater and optimize disinfection processes.
1 Introduction
Hospital wastewater (HWW), being hazardous and infectious, is different from the domestic wastewater. The effluent discharged from operating rooms, diagnostic laboratories, radiology and infectious wards (Saguti et al., 2021) contains pathogenic bacteria and viruses, toxic organic pollutants, radioactive elements and pharmaceutical compounds (PhCs) such as antibiotics and psychiatric drugs, etc. The presence of numerous pathogenic microorganisms in HWW highlights the potential threat to public health posed by HWW discharge to receiving water (Cai and Zhang, 2013; Azuma and Hayashi, 2021). In China, hospital wastewater is an important reservoir for the spread of antimicrobial resistance genes and variants. In particular, the COVID-19 pandemic has further increased hospital waste, especially high concentrations of pharmaceuticals and antibiotic compounds, posing a greater threat to human health and the ecological environment.
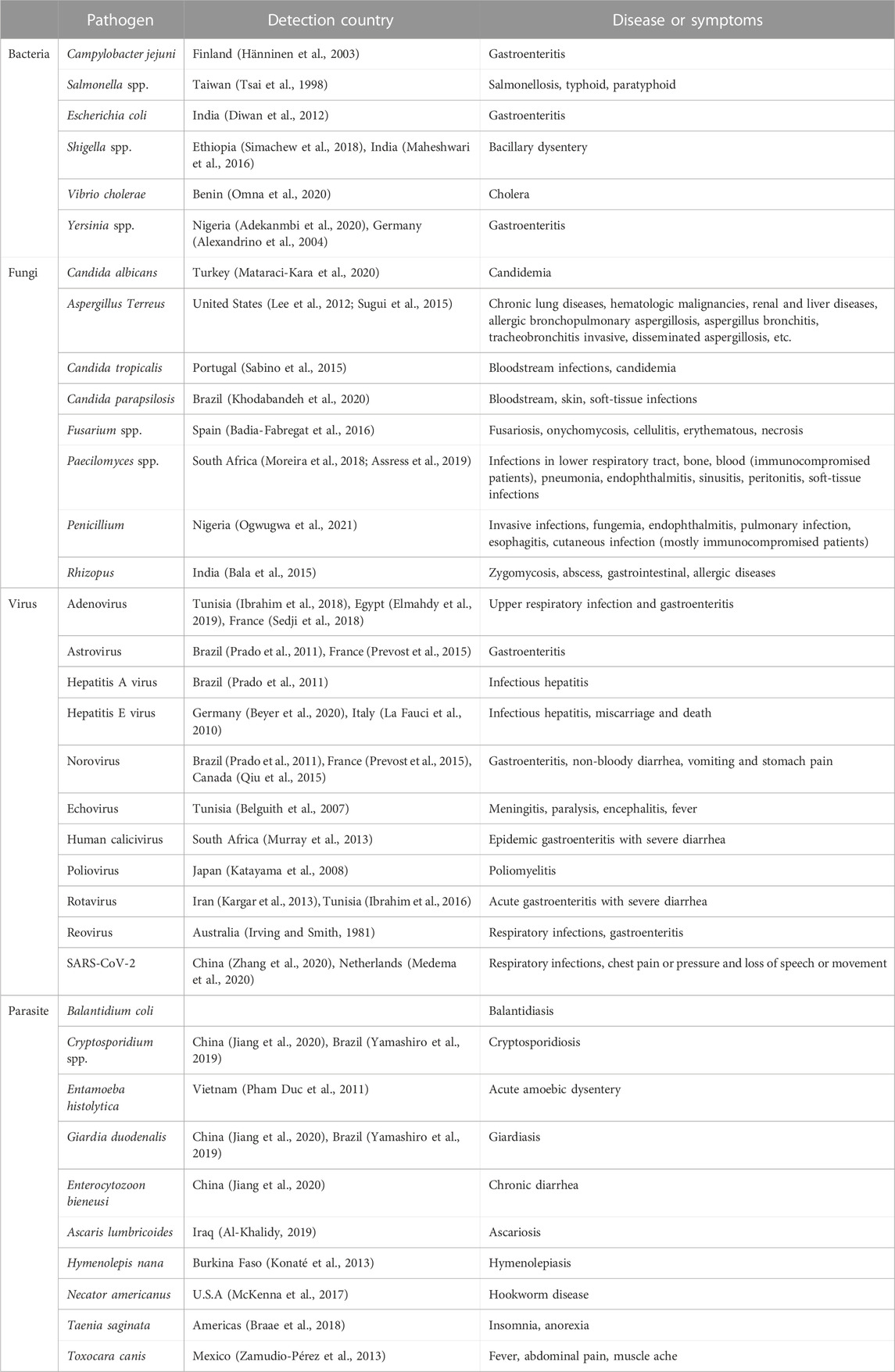
HWW provides an ideal environment for pathogenic microbes including bacteria, viruses, fungi and parasites. Thus, wastewater and sludge from HWW treatment facilities are considered as hotspots of infectious microorganisms. It was reported that the average microorganisms numbers in HWW sludge were detected to be: total count of dry weight of sludge 8.1 × 107 CFU·g−1 and 1.4 × 106, 3.6×105, 1.6 × 105, 2.2 × 105 and 5.5 × 104 CFU·g−1 for total coliforms, faecal coliforms, faecal streptococci, Pseudomonas aeruginosa, and Salmonella spp., respectively (Tsai et al., 1998). Moreover, resistant bacteria and antibiotic residues were widely observed in HWW, resulting in growth suppression of susceptible bacteria and population increase of resistant bacteria in the receiving water (Hänninen et al., 2003). Overall, the biological risk of HWW is a variety of pathogenic microorganisms, which are from the feces of infected humans. The identified pathogens of concern in HWW and related diseases is shown in Table 1.
2 Microorganisms and pathogens originating from HWW
2.1 Bacteria
HWW is thought to be one of the major environmental reservoirs of pathogenic bacteria, which are the most diverse class of human pathogens found in HWW. Many different types of bacteria colonize the human intestine and are excreted in feces. Although many of these bacteria flora are host-commensal and host-beneficial, the majority of bacterial pathogens in wastewater are enteric bacterial pathogens. Salmonella spp., Escherichia spp., Shigella spp., Yersinia spp., Klebsiella spp., Aeromonas hydrophila, Vibrio cholerae, Legionella pneumophila, Mycobacterium spp., Leptospira spp., and Pseudomonas were identified in HWW. By 16S rRNA gene fingerprinting, the pathogenic bacteria abundance in the total bacteria population was estimated to be .06%–3.20% in general (Cai and Zhang, 2013). In the Nordic countries of Sweden, Norway, and Finland, waterborne outbreaks caused by Campylobacter jejuni contaminated drinking water are observed (Hänninen et al., 2003). Azuma and Hayashi (2021) found that sewage effluent from the hospital comprised 40 bacterial phyla, 123 classes, 217 orders, 366 families, and 684 genera, with Proteobacteria being the most common phylum in the hospital effluent (51%), followed by Bacteroidetes (34%), Firmicutes (11%), Actinobacteria (1%), and TM7 (1%) before treatment.
2.2 Fungi
Fungi have simpler nutritional requirements compared with bacteria, and can grow at lower water activity levels. The microbiological investigation of several HWW revealed the prevalence of fungal, bacterial and coliform species. The fungal populations can easily spread their spores into the surrounding environment, thereby directly affecting humans. The severity of fungal infectious disease depends on the site of infection and host immunity. Invasive candidiasis caused by Candida spp. nowadays serves the 4th common nosocomial infection in numerous regions, demonstrating mortality of up to 40% (Thompson et al., 2019). Candida albicans is the major species responsible for infections of medical origin. As shown in Table 1, C. albicans, Aspergillus Terreus, Candida tropicalis, Candida parapsilosis, Fusarium spp., Paecilomyces spp., Penicillium and Rhizopus were observed in HWW, which could induce severe diseases and illness to human beings.
2.3 Viruses
Viruses are one of common waterborne pathogens responsible for the sporadic outbreak of waterborne illnesses. It was reported that raw domestic wastewater can contain as many as 105–107 virus L−1 wastewater. Enteric viruses including enterovirus (EVs), adenoviruses (AdVs), hepatitis A viruses (HAV), noroviruses (NoVs) and coronaviruses (including SARS-CoV-2) among others could be detected in wastewater. Clinical epidemiology has verified enteric viruses to be frequently the source of acute gastroenteritis in the human beings. Both the United States Environmental Protection Agency (USEPA) and World Health Organization (WHO) have proposed enteric viruses as powerful contaminant candidates and health risks. Discharges of faeces, vomit and urine of infected humans could introduce viruses into HWW, with risk of virus being transmitted to healthy people via drinking contaminated water. Adenovirus, astrovirus, Hepatitis A virus, Hepatitis E virus, norovirus, echovirus, human calicivirus, poliovirus, rotavirus, reovirus, and SARS-CoV-2 have been identified in HWW, indicating their potential transmission via ineffective treatment and disinfection of HWW.
2.4 Parasites
The direct release of HWW into aquatic bodies, heavily loaded with pathogens could be directly demonstrated by intestinal parasites and cutaneous infections. Protozoa are another important microbial taxon in HWW, with 10 times the size of bacteria. Protozoa was considered to be more resistant to disinfection than the enteric bacteria (Okojokwu et al., 2014). By methods of PCR and DNA sequencing, Jiang examined 164 raw wastewater samples and found that the positive rates of Cryptosporidium, Giardia duodenalis, and Enterocytozoon bieneusi in HWW were 18.9%, 27.4%, and 74.4%, respectively (Jiang et al., 2020). In 1993, more than 4,00,000 people became ill and 100 died in Wisconsin, by the waterborne protozoan parasite. Cryptosporidium is transmitted into raw water bodies through oocysts and cysts which are excreted in abundance in the feces of warm-blooded animal or infected hosts (Ashbolt, 2004). Moreover, the small spores are resistant to environmental stresses, making them highly resistant to disposal and inactivation during HWW treatment. Several studies have detected average Cryptosporidium oocysts concentration of 10–200 oocysts L−1 in treated and untreated wastewater (Plutzer et al., 2010; Nasser, 2016). Since the oocysts of Cryptosporidium is highly resistant to chemical disinfectants, the water quality may compile with the fecal coliform guideline while Cryptosporidium outbreak was observed (Craun et al., 1998). As shown in Table 1, up to now, Cryptosporidium spp., Entamoeba histolytica, G. duodenalis, E. bieneusi, Ascaris lumbricoides, Necator americanus, Hymenolepis nana, Taenia saginata, and Toxocara canis were successfully identified in HWW, which should be paid enough attention.
3 HWW treatment process
As the contact with non-treated HWW and fecal wastes can increase the potential of pathogen transmission, it is recommended that routine environmental investigation and efficient treatment and disinfection of pathogens should be incorporated into the treatment of HWW to reduce the risk of pathogen transmission. HWW treatment facilities are engineered to take away nutrients and pollutants in sewage via the performance of activated sludge. Some pathogenic microorganisms may survive and reproduce under advantaged environmental conditions, especially in HWW.
As shown in Figure 1, three conventional stages of primary, secondary and tertiary wastewater treatment are also applicable to HWW. Among these processes, suspended solids, biodegradable organics and pathogenic microbes were removed through physical-chemical and biological responses. In accordance with WHO’s safe management of healthcare waste, the direct discharge of harmful liquids and chemical waste into sewers is strictly prohibited. For economic reasons and inadequate analytical resources of pathogens, some bacteria such as coliforms have been used as indicators to assess the degree of HWW contamination. It is important to note that the removal of coliforms does not directly correlated with the removal of fungi, viruses and protozoa from the water.
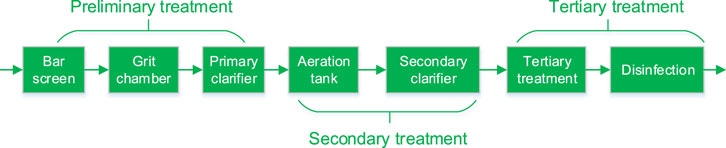
FIGURE 1. Schematic diagram of HWW treatment process including preliminary, secondary and tertiary treatment processes.
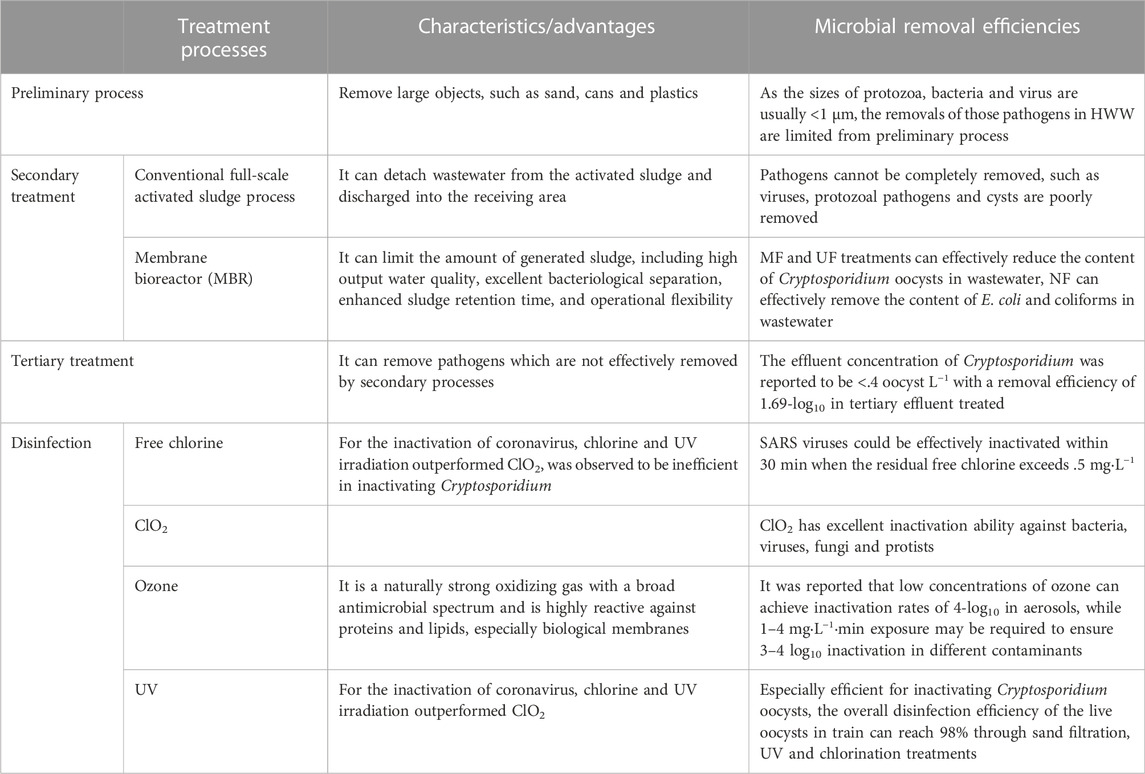
Different wastewater treatment processes may produce different log pathogen reductions, resulting in different concentrations of pathogens in the treated wastewater (Rachmadi et al., 2018). It was reported that membrane bioreactor (MBR)-based treatment facility could remove enteric viruses up to 7-log10, while the activated sludge unit and drip filter can only remove 4-log10 enteroviruses (Kitajima et al., 2014; O’Brien and Xagoraraki, 2020). Therefore, it is necessary to evaluate the removal of pathogens in the preliminary process, secondary process and tertiary process to ensure the effective disinfection of pathogens from HWW.
3.1 Preliminary process
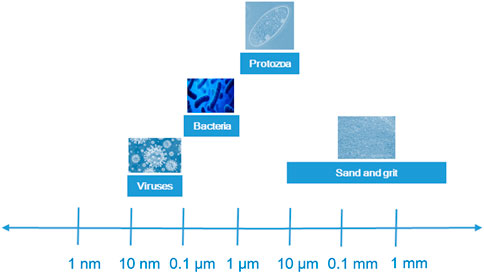
The purpose of the preliminary treatment stage is to remove large objects (such as sand, cans, and plastics), including sieving and grit removal, either using strip, tumbler, cut or belt screens that slant the inflow of water and trap objects as the effluent flows through. As shown in Figure 2, the particulate matter in HWW is either inert or biogenic, and they vary in size, shape and density. Influent particles can be classified according to size as follows: dissolved (<.001 μm), colloidal (.001–1 μm), supracolloidal (1–100 μm), or settleable (>100 μm). The removal of pathogens in HWW treatment process could be affected by physical, chemical and biological processes. The conventional gravity settling method used for primary treatment can remove particles smaller than 50 μm, whereas the chemically enhanced primary treatment process and flocculation method can remove smaller particles more effectively. Dense, easily settled inorganic particles, like sand and grit larger than 10 microns, can be removed in the preliminary treatment stage, and lower density organic and inorganic particles between .1 and 35 µm in size are removed in the primary settling tank. As the sizes of protozoa, bacteria and virus are usually <1 μm, the removals of those pathogens in HWW are limited from preliminary process.
3.2 Secondary treatment
3.2.1 Conventional full-scale activated sludge process
Activated sludge process (ASP) is the most popular secondary treatment process for HWW, which removes carbon and nutrients by treating the raw wastewater with numerous microorganisms. Through secondary sedimentation tanks, the treated wastewater is eventually detached from the activated sludge and discharged into the receiving area. Previous studies showed that the conventional activated sludge process can not completely remove pathogens. For example, norovirus and enterovirus have been found in river basins formed by municipal wastewater treatment discharges in Netherlands (van Beek et al., 2016). The removals of many viruses are also poorly by secondary treatment processes (Cheng et al., 2012). Protozoal pathogens are widespread in HWW of United States, where traditional activated sludge process has little success at removing cysts (Kitajima et al., 2014).
3.2.2 Membrane bioreactor (MBR)
The MBR has a great potential to improve effluent water quality as the treatment processes include microfiltration (MF), ultrafiltration (UF), nanofiltration (NF), and reverse osmosis (RO). In term of the separation ranges, 100–1,000 nm is for MF; 5–100 nm is for UF; 1–5 nm is for NF; .1–1 nm is for RO. In particular, by using MF and UF range of membranes for physical screening, MBR can achieve efficient secondary treatment. Compared with conventional activated sludge treatment process, MBR could limit the amount of generated sludge, which is compact and has many advantages over traditional biological activated sludge process, including high output water quality, excellent bacteriological separation, enhanced sludge retention time, and operational flexibility. MBR thus have shown promising results for the pathogen removal from HWW and do so with greater efficiency than conventional activated sludge treatment process.
Among a variety of factors that can influence the effectiveness of MBR in removing pathogens, the presence of biofilm is the most important. The balance between the presence of biofilm and regular membrane backwashing and cleaning to maintain a stable flow rate remains a key consideration in MBR operations. After ultrafiltration treatment, the reduction of Cryptosporidium oocysts in the effluent reached 1.8-log10, such that the concentration of Cryptosporidium oocysts was below the detection limit. The removal of Cryptosporidium oocysts by MF and UF could reach >7-log10 (Hirata and Hashimoto, 1998; Fu et al., 2010). A medical wastewater treatment system in Vietnam that combined an MBR system with NF was evaluated and found to have log removal values higher than four of Escherichia coli and coliforms throughout the study period (Tran et al., 2019).
3.3 Tertiary treatment and disinfection
3.3.1 Tertiary treatment
As conventional HWW treatment processes cannot control pathogens completely, high influent pathogens may induce an insufficient reduction of pathogens before its discharge into sewer or receiving water (Nasser, 2016). Tertiary HWW treatment is tentatively adopted to remove pathogens which are not effectively removed by secondary processes and subsequently pose a public health risk. To enhance the effluent water quality in HWW treatment facility, a treatment package including first chemical coagulation of secondary effluent, then sedimentation, quick sand filtration, advanced oxidation and final disinfection may be adopted to polish the secondary effluent. The effluent concentration of Cryptosporidium was reported to be <.4 oocyst L−1 with a removal efficiency of 1.69-log10 in tertiary effluent treated by flocculation and sand filtration (Fu et al., 2010). Thus, tertiary treatment process is an effective guarantee for the effective clearance of pathogens from HWW. With further removals of turbidity and water matrices, the tertiary treatment of HWW could enhance the removal of pathogenic microorganism simultaneously.
3.3.2 Disinfection
Disinfection is the final barrier to prevent the release of pathogenic microorganisms from HWW into sewer or receiving water (Hänninen et al., 2003). For chemical disinfection, disinfectants including free chlorine, chlorine dioxide (ClO2), ozone and peroxyacetic acid are widely used for HWW disinfection. Chlorine-based wastewater disinfection is widely used worldwide because of its broad disinfection efficiency, high inactivation rate, low price and ease of decomposition with free chlorine and monochloramine being the most widely used disinfectants (Rachmadi et al., 2020). A proposal was made to operate direct chlorination of HWW flowed into municipal wastewater treatment plant immediately after introduction of sewage (Azuma and Hayashi, 2021). When the residual free chlorine exceeds .5 mg·L−1, SARS viruses could be effectively inactivated within 30 min (Chen et al., 2006). For the inactivation of coronavirus, chlorine and ultraviolet (UV) irradiation outperformed ClO2. Chlorine disinfection was observed to be inefficient in inactivating Cryptosporidium, while UV irradiation was revealed to be very effective in inactivating Cryptosporidium oocysts (Nasser, 2016). As HWW contains high-concentration suspended solids, higher doses of free chlorine might be needed to avoid shielding pathogens from suspended solids (Mandal et al., 2020). Thus, the maintenance of free chlorine during disinfection of HWW is vital to achieve the effective inactivation of pathogens.
ClO2, an alternative to free chlorine, has been widely used in water and wastewater disinfection. ClO2 has excellent inactivation ability against bacteria (e.g., E. coli and Staphylococcus aureus (Huang et al., 1997b)), viruses (e.g., poliovirus and adenovirus (Huang et al., 1997a)), fungi (e.g., Penicillium chrysogenum and Stachybotrys chartarum) and protists (e.g., Cryptosporidium parvum (Chauret et al., 2001; Wilson et al., 2005)). It was reported that the inactivation kinetics of ClO2 against pathogens consists of two phases: an initial rapid inactivation phase and a subsequent dragging phase. Moreover, the disinfection rate increased with increasing pH or temperature, but exhibited divergent trends with increasing concentration of dissolved organic substances.
With oxidation potential being 2.07 V, ozone is a naturally strong oxidizing gas with a broad antimicrobial spectrum and is highly reactive against proteins and lipids, especially biological membranes. Ozone is considered to be one of the best biocides against pathogenic microorganisms and has been widely used for water disinfection since the 20th century (Blanco et al., 2021). The ozone inactivation of pathogens involves a reaction between the ozone and the biological molecules of the target pathogen. It was reported that low concentrations of ozone can achieve inactivation rates of 4-log10 in aerosols, while 1–4 mg·L-1 min exposure may be required to ensure 3–4 log10 inactivation in different contaminants (Bayarri et al., 2021). A two-stage response curve of ozone-inactivated type 1 poliovirus was reported. In Phase I, 99% of the viruses could be inactivated within seconds with an ozone dosage of 1.24 mg·L−1, while in Phase II the effective inactivation of remainder viruses could last several minutes (Katzenelson et al., 1979).
UV irradiation was green disinfection process without the addition of chemical disinfectant. It was reported that UV process is especially efficient for inactivating Cryptosporidium oocysts. Previous studies have verified the effectiveness of low-dose UV irradiation in inactivating Cryptosporidium oocysts. Morita et al. (2002) found that with a UV dosage of 1.0 mW·cm−2, a 2-log10 reduction efficiency could be achieved. The overall disinfection efficiency of the live oocysts in train can reach 98% through sand filtration, UV and chlorination treatments (Montemayor et al., 2005). Although UV radiation is the most effective disinfection technique for inactivating Cryptosporidium, its inactivation efficiency is easily impacted by the water matrices, temperature and fouling. Thus, a real time monitoring of UV dose is necessary to achieve a stable inactivation of pathogens. Since the final step in HWW treatment is normally disinfection, it is recommended that combined disinfection process should be adopted to inactivate the pathogen effectively. To sum up, the removal efficiencies of different microorganisms in different treatment processes are summarized in Table 2.
4 Conclusion
It has been verified HWW is a hotspot for various pathogenic microorganisms. To manage the health hazards posed by pathogens transmission, HWW needs to be treated sufficiently to reduce the release of pathogens. As conventional wastewater treatment process (e.g., chlorination, UV, ozone, etc.) cannot effectively remove the pathogen and especially the organic pollutants (e.g., antivirals, antibacterial, and other toxic substances, etc.) from HWW, hospital sewage treatment via combining tertiary and disinfection processes should be prioritized to ensure public health. Some treatment technologies for hospital wastewater like integrated MBR-membrane systems and electron beam technology possesse distinct advantages and should be further promoted. In addition, as an increasing global concern about the impact of the COVID-19 pandemic (Shi and Dong, 2021), more systematic research is necessary to focusing on the occurrence and fate of pathogens in HWW. Through combing preliminary, secondary, tertiary and disinfection processes, it is expected to control the human health risk to the pathogenic micro-organisms exposure in HWW (Omna et al., 2020, Simachew et al., 2018, Zamudio-Pérez et al., 2013).
Author contributions
YTT, writing-original draft. PYY, revised, corrected, edited, funding and validate the manuscript.
Funding
This work was supported by Beijing Natural Science Foundation (5222034).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adekanmbi, A., Adejoba, A. T., Banjo, O. A., and Saki, M. (2020). Detection of sul1 and sul2 genes in sulfonamide-resistant bacteria (SRB) from sewage, aquaculture sources, animal wastes and hospital wastewater in South-West Nigeria. Gene Rep. 20, 100742–100747. doi:10.1016/j.genrep.2020.100742
Al-Khalidy, K. A. H. Molecular detection of enterobius vermicularis and ASCARIS lumbricoides in sewage water for al-diwaniyah city hospitals. Qadisiyah, Iraq, Middle East, 7791–7794. (2020).
Alexandrino, M., Grohmann, E., and Szewzyk, U. (2004). Optimization of PCR-based methods for rapid detection of Campylobacter jejuni, Campylobacter coli and Yersinia enterocolitica serovar 0:3 in wastewater samples. Water Res. 38 (5), 1340–1346. doi:10.1016/j.watres.2003.10.036
Ashbolt, N. J. (2004). Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 198 (1-3), 229–238. doi:10.1016/j.tox.2004.01.030
Assress, H. A., Selvarajan, R., Nyoni, H., Ntushelo, K., Mamba, B. B., and Msagati, T. A. M. (2019). Diversity, Co-occurrence and implications of fungal communities in wastewater treatment plants. Sci. Rep. 9 (1), doi:14056doi:10.1038/s41598-019-50624-z
Azuma, T., and Hayashi, T. (2021). On-site chlorination responsible for effective disinfection of wastewater from hospital. Sci. Total Environ. 776, doi:145951doi:10.1016/j.scitotenv.2021.145951
Badia-Fabregat, M., Lucas, D., Pereira, M. A., Alves, M., Pennanen, T., Fritze, H., et al. (2016). Continuous fungal treatment of non-sterile veterinary hospital effluent: Pharmaceuticals removal and microbial community assessment. Appl. Microbiol. Biotechnol. 100 (5), 2401–2415. doi:10.1007/s00253-015-7105-0
Bala, K., Chander, J., Handa, U., Punia, R. S., and Attri, A. K. (2015). A prospective study of mucormycosis in north India: Experience from a tertiary care hospital. Med. Mycol. 53 (3), 248–257. doi:10.1093/mmy/myu086
Bayarri, B., Cruz-Alcalde, A., López-Vinent, N., Micó, M. M., and Sans, C. (2021). Can ozone inactivate SARS-CoV-2? A review of mechanisms and performance on viruses. J. Hazard. Mater. 415, doi:125658doi:10.1016/j.jhazmat.2021.125658
Belguith, K., Hassen, A., Bouslama, L., Khira, S., and Aouni, M. (2007). Enterovirus circulation in wastewater and behavior of some serotypes during sewage treatment in Monastir, Tunisia. J. Environ. Health 69 (10), 52–56.
Beyer, S., Szewzyk, R., Gnirss, R., Johne, R., and Selinka, H. C. (2020). Detection and characterization of hepatitis E virus genotype 3 in wastewater and urban surface waters in Germany. Food Environ. Virol. 12 (2), 137–147. doi:10.1007/s12560-020-09424-2
Blanco, A., Ojembarrena, F. B., Clavo, B., and Negro, C. (2021). Ozone potential to fight against SAR-COV-2 pandemic: Facts and research needs. Environ. Sci. Pollut. Res. 28 (13), 16517–16531. doi:10.1007/s11356-020-12036-9
Braae, U. C., Thomas, L. F., Robertson, L. J., Dermauw, V., Dorny, P., Willingham, A. L., et al. (2018). Epidemiology of Taenia saginata taeniosis/cysticercosis: A systematic review of the distribution in the americas. Parasites Vectors 11 (1), 518. doi:10.1186/s13071-018-3079-y
Cai, L., and Zhang, T. (2013). Detecting human bacterial pathogens in wastewater treatment plants by a high-throughput shotgun sequencing technique. Environ. Sci. Technol. 47 (10), 5433–5441. doi:10.1021/es400275r
Chauret, C. P., Radziminski, C. Z., Lepuil, M., Creason, R., and Andrews, R. C. (2001). Chlorine dioxide inactivation of Cryptosporidium parvum oocysts and bacterial spore indicators. Appl. Environ. Microbiol. 67 (7), 2993–3001. doi:10.1128/aem.67.7.2993-3001.2001
Chen, C., Zhang, X. J., Wang, Y., Zhu, L. X., and Liu, J. (2006). Waste water disinfection during SARS epidemic for microbiological and toxicological control. Biomed. Environ. Sci. 19 (3), 173–178.
Cheng, H. W., Lucy, F. E., Broaders, M. A., Mastitsky, S. E., Chen, C. H., and Murray, A. (2012). Municipal wastewater treatment plants as pathogen removal systems and as a contamination source of noroviruses and Enterococcus faecalis. J. Water Health 10 (3), 380–389. doi:10.2166/wh.2012.138
Craun, G. F., Hubbs, S. A., Frost, F., Calderon, R. L., and Via, S. H. (1998). Waterborne outbreaks of cryptosporidiosis. J. Am. Water Works Assoc. 90 (9), 81–91. doi:10.1002/j.1551-8833.1998.tb08500.x
Diwan, V., Chandran, S. P., Tamhankar, A. J., Stålsby Lundborg, C., and Macaden, R. (2012). Identification of extended-spectrum β-lactamase and quinolone resistance genes in Escherichia coli isolated from hospital wastewater from central India. J. Antimicrob. Chemother. 67 (4), 857–859. doi:10.1093/jac/dkr564
Elmahdy, E. M., Ahmed, N. I., Shaheen, M. N. F., Mohamed, E. B., and Loutfy, S. A. (2019). Molecular detection of human adenovirus in urban wastewater in Egypt and among children suffering from acute gastroenteritis. J. Water Health 17 (2), 287–294. doi:10.2166/wh.2019.303
Fu, C. Y., Xie, X., Huang, J. J., Zhang, T., Wu, Q. Y., Chen, J. N., et al. (2010). Monitoring and evaluation of removal of pathogens at municipal wastewater treatment plants. Water Sci. Technol. 61 (6), 1589–1599. doi:10.2166/wst.2010.757
Hänninen, M. L., Haajanen, H., Pummi, T., Wermundsen, K., Katila, M. L., Sarkkinen, H., et al. (2003). Detection and typing of Campylobacter jejuni and Campylobacter coli and analysis of indicator organisms in three waterborne outbreaks in Finland. Appl. Environ. Microbiol. 69 (3), 1391–1396. doi:10.1128/aem.69.3.1391-1396.2003
Hirata, T., and Hashimoto, A. (1998). Experimental assessment of the efficacy of microfiltration and ultrafiltration for Cryptosporidium removal. Water Sci. Technol. 38 (12), 103–107. doi:10.2166/wst.1998.0515
Huang, J., Li, W., Ren, N., Li, L. X., Yang, G., and Guanle, Y. (1997). Disinfection effect of chlorine dioxide on viruses, algae and animal planktons in water. Water Res. 31 (3), 455–460. doi:10.1016/S0043-1354(96)00276-X
Huang, J., Wang, L., Ren, N., Ma, F., and Juli, (1996). Disinfection effect of chlorine dioxide on bacteria in water. Water Res. 31 (3), 607–613. doi:10.1016/S0043-1354(96)00275-8
Ibrahim, C., Cherif, N., Hammami, S., Pothier, P., and Hassen, A. (2016). Quantification and genotyping of rotavirus A within two wastewater treatment processes. Clean. Soil Air Water 44 (4), 393–401. doi:10.1002/clen.201400588
Ibrahim, C., Hassen, A., Pothier, P., Mejri, S., and Hammami, S. (2018). Molecular detection and genotypic characterization of enteric adenoviruses in a hospital wastewater. Environ. Sci. Pollut. Res. 25 (11), 10977–10987. doi:10.1007/s11356-018-1399-2
Irving, L. G., and Smith, F. A. (1981). One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl. Environ. Microbiol. 41 (1), 51–59. doi:10.1128/aem.41.1.51-59.1981
Jiang, W., Roellig, D. M., Li, N., Wang, L., Guo, Y., Feng, Y., et al. (2020). Contribution of hospitals to the occurrence of enteric protists in urban wastewater. Parasitol. Res. 119 (9), 3033–3040. doi:10.1007/s00436-020-06834-w
Kargar, M., Javdani, N., Najafi, A., and Tahamtan, Y. (2013). First molecular detection of group A rotavirus in urban and hospital sewage systems by nested-RT PCR in Shiraz, Iran. J. Environ. Health Sci. Eng. 11 (1), 4. doi:10.1186/2052-336x-11-4
Katayama, H., Haramoto, E., Oguma, K., Yamashita, H., Tajima, A., Nakajima, H., et al. (2008). One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 42 (6-7), 1441–1448. doi:10.1016/j.watres.2007.10.029
Katzenelson, E., Koerner, G., Biedermann, N., Peleg, M., and Shuval, H. I. (1979). Measurement of the inactivation kinetics of poliovirus by ozone in a fast-flow mixer. Appl. Environ. Microbiol. 37 (4), 715–718. doi:10.1128/aem.37.4.715-718.1979
Khodabandeh, M., Shirani-Bidabadi, L., Madani, M., and Zahraei-Ramazani, A. (2020). Study on Periplaneta americana (blattodea: Blattidae) fungal infections in hospital sewer system, esfahan city, Iran, 2017. J. Pathogens 2020, doi:42967201–8. doi:10.1155/2020/4296720
Kitajima, M., Haramoto, E., Iker, B. C., and Gerba, C. P. (2014). Occurrence of Cryptosporidium, Giardia, and Cyclospora in influent and effluent water at wastewater treatment plants in Arizona. Sci. Total Environ. 484, 129–136. doi:10.1016/j.scitotenv.2014.03.036
Konaté, Y., Maiga, A. H., Basset, D., Picot, B., and Casellas, C. (2013). Occurrence, removal and accumulation in sludge of protozoan cysts and helminth eggs in a full-scale anaerobic pond in Burkina Faso. Water Sci. Technol. 67 (1), 193–200. doi:10.2166/wst.2012.557
La Fauci, V., Sindoni, D., Grillo, O. C., Calimeri, S., Lo Giudice, D., and Squeri, R. (2010). Hepatitis E virus (HEV) in sewage from treatment plants of messina university hospital and of messina city council. J. Prev. Med. Hyg. 51 (1), 28–30.
Lee, L. D., Hachem, R. Y., Berkheiser, M., Hackett, B., Jiang, Y., and Raad, Y. II (2012). Hospital environment and invasive aspergillosis in patients with hematologic malignancy. Am. J. Infect. Control 40 (3), 247–249. doi:10.1016/j.ajic.2011.03.031
Maheshwari, M., Yaser, N. H., Naz, S., Fatima, M., and Ahmad, I. (2016). Emergence of ciprofloxacin-resistant extended-spectrum β-lactamase-producing enteric bacteria in hospital wastewater and clinical sources. J. Glob. Antimicrob. Resist. 5, 22–25. doi:10.1016/j.jgar.2016.01.008
Mandal, P., Gupta, A. K., and Dubey, B. K. (2020). A review on presence, survival, disinfection/removal methods of coronavirus in wastewater and progress of wastewater-based epidemiology. J. Environ. Chem. Eng. 8 (5), doi:104317doi:10.1016/j.jece.2020.104317
Mataraci-Kara, E., Ataman, M., Yilmaz, G., and Ozbek-Celik, B. (2020). Evaluation of antifungal and disinfectant-resistant Candida species isolated from hospital wastewater. Arch. Microbiol. 202 (9), 2543–2550. doi:10.1007/s00203-020-01975-z
McKenna, M. L., McAtee, S., Bryan, P. E., Jeun, R., Ward, T., Kraus, J., et al. (2017). Human intestinal parasite burden and poor sanitation in rural Alabama. Am. J. Trop. Med. Hyg. 97 (5), 1623–1628. doi:10.4269/ajtmh.17-0396
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., and Brouwer, A. (2020). Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 7 (7), 511–516. doi:10.1021/acs.estlett.0c00357
Montemayor, M., Valero, F., Jofre, J., and Lucena, F. (2005). Occurrence of Cryptosporidium spp. oocysts in raw and treated sewage and river water in north-eastern Spain. J. Appl. Microbiol. 99 (6), 1455–1462. doi:10.1111/j.1365-2672.2005.02737.x
Moreira, D. C., Oliveira, M. M. E., and Borba, C. M. (2018). Human pathogenic Paecilomyces from food. Microorganisms 6 (3), 64. doi:10.3390/microorganisms6030064
Morita, S., Namikoshi, A., Hirata, T., Oguma, K., Katayama, H., Ohgaki, S., et al. (2002). Efficacy of UV irradiation in inactivating Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 68 (11), 5387–5393. doi:10.1128/aem.68.11.5387-5393.2002
Murray, T. Y., Mans, J., and Taylor, M. B. (2013). Human calicivirus diversity in wastewater in South Africa. J. Appl. Microbiol. 114 (6), 1843–1853. doi:10.1111/jam.12167
Nasser, A. M. (2016). Removal of Cryptosporidium by wastewater treatment processes: A review. J. Water Health 14 (1), 1–13. doi:10.2166/wh.2015.131
O’Brien, E., and Xagoraraki, I. (2020). Removal of viruses in membrane bioreactors. J. Environ. Eng. 146 (7). doi:10.1061/(asce)ee.1943-7870.0001743
Ogwugwa, V. H., Oyetibo, G. O., and Amund, O. O. (2021). Taxonomic profiling of bacteria and fungi in freshwater sewer receiving hospital wastewater. Environ. Res. 192, doi:110319doi:10.1016/j.envres.2020.110319
Okojokwu, O. J., Inabo, H. I., and Yakubu, S. E. (2014). Parasitological profile of raw wastewater and the efficacy of biosand filter in reduction of parasite ova and cysts. J. Appl. Sci. Environ. Manag. 18 (1), 5. doi:10.4314/jasem.v18i1.1
Omna, B., Jkfa, C., Taa, B., Sao, A., and Taa, C. (2020). Performance of Moringa oleifera seeds protein and Moringa oleifera seeds protein-polyaluminum chloride composite coagulant in removing organic matter and antibiotic resistant bacteria from hospital wastewater. J. Water Process Eng. 33 (101103), doi:101103doi:10.1016/j.jwpe.2019.101103
Pham Duc, P., Nguyen-Viet, H., Hattendorf, J., Zinsstag, J., Dac Cam, P., and Odermatt, P. (2011). Risk factors for Entamoeba histolytica infection in an agricultural community in Hanam province, Vietnam. Parasites Vectors 4, 102. doi:10.1186/1756-3305-4-102
Plutzer, J., Ongerth, J., and Karanis, P. (2010). Giardia taxonomy, phylogeny and epidemiology: Facts and open questions. Int. J. Hyg. Environ. Health 213 (5), 321–333. doi:10.1016/j.ijheh.2010.06.005
Prado, T., Silva, D. M., Guilayn, W. C., Rose, T. L., Gaspar, A. M., and Miagostovich, M. P. (2011). Quantification and molecular characterization of enteric viruses detected in effluents from two hospital wastewater treatment plants. Water Res. 45 (3), 1287–1297. doi:10.1016/j.watres.2010.10.012
Prevost, B., Lucas, F. S., Ambert-Balay, K., Pothier, P., Moulin, L., and Wurtzer, S. (2015). Deciphering the diversities of astroviruses and noroviruses in wastewater treatment plant effluents by a high-throughput sequencing method. Appl. Environ. Microbiol. 81 (20), 7215–7222. doi:10.1128/aem.02076-15
Qiu, Y., Lee, B. E., Neumann, N., Ashbolt, N., Craik, S., Maal-Bared, R., et al. (2015). Assessment of human virus removal during municipal wastewater treatment in Edmonton, Canada. J. Appl. Microbiol. 119 (6), 1729–1739. doi:10.1111/jam.12971
Rachmadi, A. T., Kitajima, M., Kato, T., Kato, H., Okabe, S., and Sano, D. (2020). Required chlorination doses to fulfill the credit value for disinfection of enteric viruses in water: A critical review. Environ. Sci. Technol. 54 (4), 2068–2077. doi:10.1021/acs.est.9b01685
Rachmadi, A. T., Kitajima, M., Watanabe, K., Yaegashi, S., Serrana, J., Nakamura, A., et al. (2018). Free-chlorine disinfection as a selection pressure on norovirus. Appl. Environ. Microbiol. 84 (13). doi:10.1128/aem.00244-18
Sabino, R., Sampaio, P., Rosado, L., Videira, Z., Grenouillet, F., and Pais, C. (2015). Analysis of clinical and environmental Candida parapsilosis isolates by microsatellite genotyping--a tool for hospital infection surveillance. Clin. Microbiol. Infect. 21 (10), 954.e1–954.e8. doi:10.1016/j.cmi.2015.06.001
Saguti, F., Magnil, E., Enache, L., Churqui, M. P., Johansson, A., Lumley, D., et al. (2021). Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res. 189, doi:116620doi:10.1016/j.watres.2020.116620
Sedji, M. I., Varbanov, M., Meo, M., Colin, M., Mathieu, L., and Bertrand, I. (2018). Quantification of human adenovirus and norovirus in river water in the north-east of France. Environ. Sci. Pollut. Res. 25 (30), 30497–30507. doi:10.1007/s11356-018-3045-4
Shi, Q., and Dong, X. P. (2021). Rapid global spread of the SARS-CoV-2 Delta (B.1.617.2) variant: Spatiotemporal variation and public health impact. Zoonoses1, 3. doi:10.15212/ZOONOSES-2021-0005
Simachew, D., Tarekegn, B., Argaw, A., and Geremew, S. (2018). Antibiotic resistant bacteria removal of subsurface flow constructed wetlands from hospital wastewater. J. Environ. Chem. Eng. 6 (4), 4265–4272. doi:10.1016/j.jece.2018.06.034
Sugui, J. A., Kwon-Chung, K. J., Juvvadi, P. R., Latgé, J. P., and Steinbach, W. J. (2014). Aspergillus fumigatus and related species. Cold Spring Harb. Perspect. Med. 5 (2), doi:a019786doi:10.1101/cshperspect.a019786
Thompson, A., Davies, L. C., Liao, C. T., da Fonseca, D. M., Griffiths, J. S., Andrews, R., et al. (2019). The protective effect of inflammatory monocytes during systemic C. albicans infection is dependent on collaboration between C-type lectin-like receptors. PLoS Pathog. 15 (6), doi:e1007850doi:10.1371/journal.ppat.1007850
Tran, T., Nguyen, T., Ho, H., Le, D., Lam, T., Nguyen, D., et al. (2019). Integration of membrane bioreactor and nanofiltration for the treatment process of real hospital wastewater in Ho chi minh city, Vietnam. Processes 7 (3), 123. doi:10.3390/pr7030123
Tsai, C. T., Lai, J. S., and Lin, S. T. (1998). Quantification of pathogenic micro-organisms in the sludge from treated hospital wastewater. J. Appl. Microbiol. 85 (1), 171–176. doi:10.1046/j.1365-2672.1998.00491.x
van Beek, J., de Graaf, M., Xia, M., Jiang, X., Vinjé, J., Beersma, M., et al. (2016). Comparison of norovirus genogroup I, II and IV seroprevalence among children in The Netherlands, 1963, 1983 and 2006. J. Gen. Virol. 97 (9), 2255–2264. doi:10.1099/jgv.0.000533
Wilson, S. C., Wu, C., Andriychuk, L. A., Martin, J. M., Brasel, T. L., Jumper, C. A., et al. (2005). Effect of chlorine dioxide gas on fungi and mycotoxins associated with sick building syndrome. Appl. Environ. Microbiol. 71 (9), 5399–5403. doi:10.1128/aem.71.9.5399-5403.2005
Yamashiro, S., Foco, M. L. R., Pineda, C. O., José, J., Nour, E. A. A., Siqueira-Castro, I. C. V., et al. (2019). Giardia spp. and Cryptosporidium spp. removal efficiency of a combined fixed-film system treating domestic wastewater receiving hospital effluent. Environ. Sci. Pollut. Res. 26 (22), 22756–22771. doi:10.1007/s11356-019-05500-8
Zamudio-Pérez, E., Rojas-Valencia, N., Chairez, I., and Torres, L. G. (2013). Coliforms and helminth eggs removals by coagulation-flocculation treatment based on natural polymers. J. Water Resour. Prot. 05 (11), 1027–1036. doi:10.4236/jwarp.2013.511108
Zhang, D., Ling, H., Huang, X., Li, J., Li, W., Yi, C., et al. (2020). Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 741, doi:140445. doi:10.1016/j.scitotenv.2020.140445
Keywords: hospital wastewater, pathogen, occurrence, removal, disinfection
Citation: Yuan T and Pian Y (2023) Hospital wastewater as hotspots for pathogenic microorganisms spread into aquatic environment: A review. Front. Environ. Sci. 10:1091734. doi: 10.3389/fenvs.2022.1091734
Received: 07 November 2022; Accepted: 05 December 2022;
Published: 04 January 2023.
Edited by:
Huiyu Dong, Research Center for Eco-Environmental Sciences (CAS), ChinaReviewed by:
Jun Hu, Zhejiang University of Technology, ChinaXiangjuan Yuan, Wuhan Textile University, China
Copyright © 2023 Yuan and Pian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaya Pian, cGlhbnlheWEwOTE5QDE2My5jb20=
 Tingting Yuan
Tingting Yuan Yaya Pian
Yaya Pian