
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 10 October 2022
Sec. Toxicology, Pollution and the Environment
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.1008900
This article is part of the Research Topic Soil and Sediment Pollution, Processes and Remediation, Volume II View all 17 articles
Dioxins (PCDD/Fs) are one of the most toxic environmental pollutants known to date. Due to their structural stability and extreme hydrophobicity dioxins persist in the ecosystems and can be bioaccumulated to critical levels in both human and animal food chains. Soils are the most important reservoirs of dioxins, thus soil microbes are highly exposed to dioxins, impacting their diversity, genetics and functional characteristics. To experimentally evaluate these effects, the diversity and functionality of soil microbes were assessed in seven local sites potentially exposed to PCDD/Fs.
Concentration of dioxins in soils samples was firstly determined and the soils cultivable microbes were identified and molecularly characterized as a function of their in vitro ability to degrade the TCDD. Our results revealed that the diversity of microbial communities largely varied among the sites and was likely inversely proportional to their level of contamination with PCDD/Fs. Furthermore, the genetics profiling of dioxin-degrading bacteria revealed that the Cytochrome P450 CYPBM3-positive species largely belong to the genus Bacillus and were randomly distributed among the soils samples, while the angular dioxygenase (AD)-positive species were mainly found in highly polluted soils with a major presence of the genus Pseudomonas. Finally, the functionality of dioxin-biodegrading genes (AD or CYPBM3), was confirmed by the ability of bacteria to consume 2,3,7,8-TCDD, and this was synchronized with an induced level of both pathways. Our results suggest that different dioxin-metabolizing pathways exist under the same environmental conditions and work differentially for an effective removal of PCDD/Fs.
Polychlorinated dibenzo-p-dioxins (PCDDs) and polychlorinated dibenzofurans (PCDFs), collectively termed “dioxins” are highly potent class of persistent organic pollutants (POPs) that are covered by the Stockholm Convention (Stockholm, 2001). According to chlorination degree (P = 1–8), dioxins comprise 75 PCDD congeners and 135 PCDF congeners with different toxicities. Specifically, PCDD/F congeners possessing chlorine atoms in 2′, 3′, 7′ and 8′ positions of the aromatic rings are the most toxic. Typically, the toxicity of PCDD/Fs is defined as Toxicological Equivalence or TEQ units. The sum of TEQ for a given environmental sample is depending on the Toxic Equivalency Factors (TEFs). The TEF values indicate the level of toxicity compared to 2′,3′,7′,8′-tetrachlorinated dibenzo-p-dioxin (2′,3′,7′,8′-TCDD), the most toxic congener of dioxins, that has been given a reference TEF value of 1 (World Health Organization, 2016).
Naturally, dioxins can be released into the environment through volcanic eruptions and forest fires (Hay, 1981). Such scenarios are becoming more frequent with the actual increases of forest fires (Salamanca et al., 2016; Oliveira et al., 2020). Dioxins can also enter the environment through domestic and municipal incinerations (Tuppurainen et al., 2003), by various manufacturing processes including the synthesis of chlorinated aromatic products, such as herbicides, pesticide and paper processing. Electronic waste (e-waste) is also an important and active source of PCDD/Fs (Jin et al., 2020). Once emitted into the environment, dioxins contaminate specific environmental compartments, bioaccumulate, and therefore, transmit through the food chain due to their structural stability and high lipid-solubility (Geyer et al., 1993). Their adverse effects on human health are now well established, e.g., immunotoxicity (Marshall and Kerkvliet, 2010), wasting syndrome (Huuskonen et al., 1994), dysfunction of immune and reproductive systems (Carney et al., 2006), carcinogenicity (Toth et al., 1979), and teratogenicity (Baker et al., 1995).
Due to their high affinity to soil organic matter, PCDD/Fs have a high value of organic carbon–water partition coefficient (KOC). This confers a low mobility together with a low water solubility to such compounds, thus enabling them to accumulate in soil, affecting plants as well as soil microorganisms (Chrostowski and Foster, 1996; Hanano et al., 2014b; Hanano et al., 2015a; Hanano et al., 2018a; Hanano et al., 2018b). Once accumulated in the soil, PCDD/Fs affect “soil health” (Gul et al., 2021). Soil microbiota plays determinant roles in the maintenance of soil health and in the detoxification of detrimental chemicals, including PCDD/Fs (Cerniglia et al., 1979). Soil microbes respond to dioxin exposure by a set of biological modulations that impact their diversity and functionality. In this context, multiple parameters, e.g., density, diversity and enzymatic activities, are now used as indicators for monitoring and assessing the exposure levels of soil microbial communities to stressors (Yao et al., 2018; Mahfouz et al., 2020). Consequently, diverse bacterial and fungal species were identified and characterized as potential biodegraders of dioxins (Magan et al., 2010; Stella et al., 2017; Hanano et al., 2019a).
So far, diverse bacterial enzymatic pathways have been characterized with respect to the biodegradation of dioxins such as the angular dioxygenases (Sato et al., 1997; Armengaud et al., 1998; Habe et al., 2001), certain microbial peroxidases and anaerobic dehalogenases (Bumpus et al., 1985; Bunge et al., 2003). Furthermore, it was shown that specific enzymes of bacterial P450s, initially identified as homologs of xenobiotic-mammalian metabolizing P450s, exhibited similar activities towards dioxins (Narhi and Fulco, 1987; Boddupalli et al., 1992). The first bacterial P450 was characterized in Bacillus megaterium ATCC 14581 by the group of Fulco et al. (Matson et al., 1977), conducting a detailed characterization of three distinguished isoforms of P450 from B. megaterium, referred as to P450BM-1, P450BM-2 and P450BM-3 (Kim and Fulco, 1983; Schwalb et al., 1985). Beyond their original activities as fatty acids oxygenases, both native or engineered P450BM-1 and P450BM-3 have shown remarkable activities to oxidize an emergent range of exogenous substrates including certain drugs such as phenacetin and methoxyresorufin (Kim et al., 2010), certain chlorinated insecticides (Seralathan et al., 2014; Meena et al., 2016) and even more certain dioxins notably 2,3,7-trichloro-dibenzo-p-dioxin (Sulistyaningdyah et al., 2004).
The main objective of this work is firstly to demonstrate whether the diversity, genetics and functional signatures of soil microbial community are specifically affected by the contamination of soil with dioxins. Secondly, if these signatures vary according to the concentration of dioxins in soil. Our work presents a new set of data on the composition, genetics and functional properties of microbial communities in soil samples contaminated with PCDD/Fs. Soil microbial communities were subjected to a detailed characterization in terms of density and diversity linked with contamination level of soils with PCDD/Fs. Moreover, large-scale analyses were performed for genes transcripts of specific dioxin-degradation pathways in Gram-positive and Gram-negative bacterial species. Our report suggests that the most characterized bacterial pathways responsible for degradation of dioxins, angular dioxygenase (AD) and cytochrome P450 BM3 (CYPBM3), are found in both Gram-negative and Gram-positive bacteria. Effectively, this new statement raises questions on a possible functional connection between these two distinct dioxin-degradation pathways when both are found in such bacteria.
Supplementary Table S1 presents a set of informative data about the sampling sites (A1, A2, A3, A4, B, C, and D) that have been targeted in this study. The sites A1 to A3 are located at 10 km (6.2 mi) Southwest of Damascus, the Syrian capital, corresponding to waste incineration stations. The site A4 corresponds to an olive grove located next to waste incineration sites (A1, A2, and A3). The site B is an open site of industrial and domestic wastewater collection located in Deir al-Asafir, 12 km Southeast of Damascus. The site C is nearby Homs’s refinery, located in Western Syria, 162 km North of Damascus. The site D corresponds to an open waste incineration site in Al Suwayda city located in southwestern of Damascus. Two soil samples were collected form the site A4 and three soil samples were respectively collected form the sites A1, A2, A3, B, C, and D. The samples (S1-S20) (about 500 g each) were taken from the surface of soil (0–5 cm depth), put in sterile bags and promptly stored at + 4°C until further use.
The soil samples were air-dried, sieved using a 1 mm-sieve to eliminate rough materials. Total organic nitrogen, NO3 and NH4 were determined as described before (Kjeldahl, 1883). Briefly, the Kjeldahl method consists of three successive steps: 1) the soil sample is digested by sulfuric acid in the presence of a catalyst that helps in converting the amine nitrogen to ammonium ions (NH4+); 2) the NH4+ ions are then converted into ammonia gas that is heated, distilled and trapped into a solution where it is dissolved again; 2) finally the amount of the ammonia that has been trapped is determined by titration with a standard solution.
One Gram of each soil sample was added into 5 ml of one X PBS (Phosphate buffer Saline, pH 7.2) and vigorously shaken for 5 min. The suspensions were diluted by the same buffer, then 100 µL of dilutions from 10−3 to 10−6 were cultured on Luria–Bertani (LB) agar plates. The plates were incubated at 28 ± 2°C until the appearance of distinct colonies (Hanano et al., 2014c). Single colonies were transferred onto fresh LB plates and kept at 4°C for further analysis. The density of cultured bacteria was estimated by measuring colony forming unit (CFU) per Gram dry weight soil and expressed as CFU g−1 DW. Finally, all bacterial isolates were stored at—80°C in LB with 20% glycerol.
Before proceeding, soil samples were dried at room temperature, ground and sieved. PCDD/Fs extraction was performed as described previously (Hanano et al., 2014c). Briefly, 5 g of soil were mixed with 20 ml of hexane containing 20% acetone and the mix was horizontally shaken at 200 rpm for 1 h at room temperature. The organic phase was separated by a brief centrifugation at 4,000 rpm for 5 min, then carefully recovered and promptly mixed with 8 ml of sulfuric acid (∼0.9 mM) and briefly shaken for 2 min in the same conditions. Subsequently, the upper organic phase, corresponding to hexane extract, was carefully transferred into a clean 40-ml vial. The cleaned up of fractions was performed with a column composed of 0.5 g anhydrous Na2SO4 on top and 1.0 g of florisil at the bottom. This column was first activated with 3 ml of dichloromethane/hexane/methanol (50:45:5). PCDD/Fs were then eluted with 5 ml of the same solvents mix. The extract volume was reduced to 1 ml under nitrogen flow. One hundred microlitres of Dimethyl sulfoxide (DMSO) was added to the extract and the mix was dried to remove all trace of solvents. 500 μL of DMSO was added and the final volume of the extracts was adjusted to 1 ml by deionized H2O. Dilution of 1:10 with 50% DMSO in deionized water was used for analysis of dioxin by enzyme-linked immunosorbent assay (ELISA). To evaluate the quality of extraction protocol, a TCDD-free soil sample was spiked with 100 ng L−1 of 2,3,7,8-TCDD and the same extraction protocol was applied.
An Abraxis TCDD-ELISA kit was used to determine the PCDD/Fs concentration in the extracts according to the manufacturer’s instructions (Abraxis LLC, United States ). The absorbance was measured at 450 nm by a microplate reader (Multiskan EX, Thermo/Labsystems, United States ).
The extraction of bacterial genomic DNA was done using a Genomic DNA extraction kit (Qiagen-Germany) according to the manufacturer’s manual. The isolated gDNA was eluted in 50 μL of distilled water and the concentration was adjusted to 200 ng μL−1 and kept at −20°C. A 1450 bp-fragment of 16S rRNA gene was amplified by PCR using the primers 27F and 1492R (Supplementary Table S2, Supplementary Material S1) (Marcial Gomes et al., 2008). The PCR was performed as described before by (Hanano et al., 2014c). The 25 μL final volume reaction was composed of 3 mM MgSO4, 200 μM each of the four dNTPs, 10 μM of each primers, 2.5 U Taq DNA polymerase and 200 ng of gDNA. PCR conditions were 94°C for 4 min, 35 cycles at 94°C for 30 s, 56°C for 30 s, 72°C for 1 min, followed by a final extension step at 72°C for 10 min.
The PCR products of 16S rRNA were sequenced using an ABI 310 Genetic Analyzer (Applied Biosystems). 16S rRNA PCR products were sequenced in both direction forward and reverse. The quality of sequencing was confirmed by sequencing a 16s rRNA amplicon of reference strain of B. megaterium (Hanano et al., 2019b). The reads of 16S rRNA sequence were analysed using the BLAST search program from GenBank-NCBI database (https://www.ncbi.nlm.nih.gov/). Bacterial genus and species were identified with a score of similarity of (≥99%). The 16S rRNA sequences reported in this study were submitted to the GenBank-NCBI (GenBank ID: from MW475085 to MW475154).
Transcripts of dioxin-degrading genes were quantified by reverse-transcription quantitative PCR (RT-qPCR) according to (Hanano et al., 2015b). RNAs were extracted from cultured bacteria as previously described (Hanano et al., 2019b). The extracted RNA was diluted to 50 ng μL−1 using RNase-free water and conserved at –80°C. One μg total RNA were used for first-strand cDNA synthesis (Hanano et al., 2014c). Real-time PCR was carried out in 96-well plates using an AriaMx Real-time PCR System (Agilent technologies, United States ). Where, 25 μL-reaction mixtures were composed of 0.5 μM of each primers (Supplementary Table S2), 12.5 μL of SYBR Green PCR mix (Bio-Rad, United States) and 100 ng cDNA. Fragments of 168, 144, and 433 bp from the coding sequence of bacterial angular dioxygenase (ADα-subunit), CYPBM1 and 16S rRNA, were respectively amplified using gene-specific primers as described before (Hanano et al., 2019b). The relative quantification (RQ = 2(−∆∆CT)) of target genes was determined by the AriaMx qPCR system.
Bacterial isolates that harbor dioxin-degradation genes were assessed for their ability to metabolize 2,3,7,8-TCDD in a laboratory-scale experiment. A pure colony of the bacterial isolate was pre-cultured in LB medium and incubated overnight at 28 ± 2°C. Five hundred microlitres (500 µL) of bacterial culture having an OD600 = 1 was taken, centrifuged and washed with 1 ml of mineral salt medium (MSM) to eliminate all traces of LB medium. Mineral salt medium was composed as reported before (Hanano et al., 2014c). Isolates were cultured into 10 ml of MSM supplemented with 100 ng L−1 (0.1 ppb) of 2,3,7,8-TCDD (final concentration), as a sole carbon source. The cultures were incubated for 6 weeks at 28 ± 2°C. A negative control, the same culture without 2,3,7,8-TCDD was performed. Bacterial growth was measured and expressed as CFU mL−1.
Data were expressed as means ± standard deviation (SD). The comparisons between control and treatments were statistically confirmed by one-way analysis of variance (ANOVA) SPSS Statistics software. Differences between control and treatments were significant as p < 0.05 or very significant as p < 0.01. Microbial diversity for each soil samples was assessed using two diversity indexes; Shannon’s diversity index (H) and Simpson’s diversity index (D). Shannon’s diversity index (H), an informative statistic index, which means that all species present in a sample are randomly sampled. Shannon’s Index (H) is calculated by the following equal: H' = ─ ∑ S i=1 pi ln pi, where p is the proportion (n/N) of individuals of one given species found (n) divided by the total number of individuals found (N), ln is the natural log, ∑ is the sum of the calculations, and s is the number of species. Simpson’s diversity index (D) is calculated by the following equal: D = 1/∑ S i=1 pi2, where p is the proportion (n/N) of individuals of one specific species found (n) divided by the total number of individuals found (N), ∑ is still the sum of the calculations, and s is the number of species. Bacterial species richness was evaluated by Margalef’s index DMg and calculated by the following equal: DMg = (S-1)/(ln N), where S is the number of species recorded, N is the total number of individuals in the sample and ln is the natural log. DMg, H′ and D indices were calculated using an online calculator for species richness and biodiversity at http://www.alyoung.com/labs/biodiversity_calculator.html.
The analysis of PCDD/Fs content in the soils samples (S1-S20) showed that they vary significantly in respect to the concentrations of PCDD/Fs, compared to a reference soil sample (SDF; dioxin-free soil) (Hanano et al., 2014c). As shown in Figure 1A, the concentration of dioxins in soil samples ranged from 5 to ∼50 ng L−1. Accordingly, the concentration of PCDD/Fs was categorized into three levels. The highest one, Level I (LI) corresponds to a concentration of 50 ng L−1 of PCDD/Fs, and comprises soils samples S15, S16 and S17, collected from the site C. The Level 2 (L2) categorizes the soils samples that contained 12 and 20 ng L−1 of PCDD/Fs, including S1 to S9, S18, S19 and S20 collected from the sites A1, A2, A3 and D. Whilst, the lowest level, Level 3 (L3), represents concentrations ranged from 5 to ∼10 ng L−1 of PCDD/Fs, comprising soil samples S12, S13, S14, S10 and S11 collected from the sites B and A4, respectively. These data indicate that the soils samples of the site C, situated nearby Homs refinery showed the highest level of dioxin contamination. Soils samples of the sites A1, A2, A3, and D, located around the waste incineration stations, were moderately contaminated with dioxins. While, the lowest contaminated soil samples belonged to the site A4, corresponding to the olive grove located close to a waste incineration sites as well as to the site B, corresponding to an open site of industrial and domestic wastewater collection.
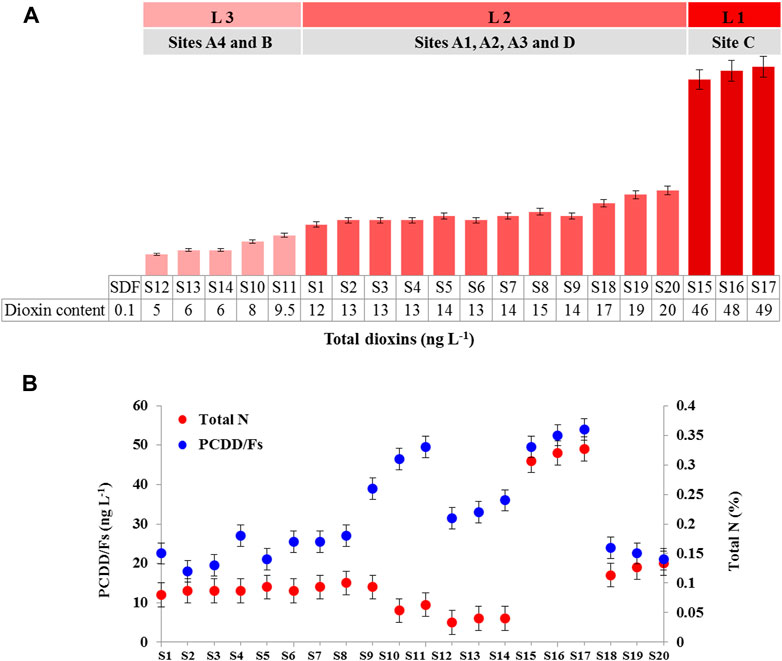
FIGURE 1. Level of PCDD/Fs in soil samples. (A) PCDD/Fs were extracted from soil samples and the total PCCD/Fs in each sample was determined by a PCDD/Fs-specific ELISA kit. Dioxins levels in soil samples were categorized into three levels; level L 1 (∼50 ng L−1), the level L2 (from 12 to 20 ng L−1), and level L3 (from 5 to ∼10 ng L−1). (B) The relation between the total nitrogen content in soil samples and their levels of contamination with PCDD/Fs. All measurement were done in triplicate. Values are means ± S.D (n = 9).
Total N2 in soils samples of the sites A1, A2, and A3, presented in Supplementary Table S3, ranged between 0.12% and 0.18%, which was relatively similar to the total N at site D (from 0.14% to 0.16%). Whereas, the highest values of the total N were detected in the soil samples of site C with a range of 0.33%–0.36%. Also, high levels of total N were found in the samples of site A4 that ranged between 0.26% and 0.33%. Furthermore, the NH4/NO3 ratio considerably varied among soil samples, the highest NH4/NO3 ratios (8.4–9.6) were found in soil samples from the site A4, while the lowest ones (1.9–2.7) were in soil samples from the sites A1, A2, and A3. However, the NH4/NO3 ratio was similar (5.5–7.9) for the sites B and D and lower in the site C (3.5–5.1). Next, the total N content of soil samples was evaluated as a function of their level of contamination with dioxins, and as Figure 1B shows, there is no correlation between the total N content in soil samples and their level of contamination with dioxins. In exception of that, the highest polluted soil samples with PCDD/Fs (S15, S16 and S17 from site C) showed a significant correlation (p < 0.01) with the content of total nitrogen. Altogether, these data indicate that soils from the site C, situated nearby Homs refinery, showed the highest level of contamination with dioxins and this was significantly correlated with a high level of total nitrogen.
The bacterial population densities, evaluated for each soil samples and expressed as CFU g−1 of dry weight, differed considerably among soil samples. As Table 1 shows, the highest bacterial densities were found in soil samples S7, S17, S16, S9, S12, S8 and S13 (from sites C, A3, A4 and B) corresponding to 159 × 107, 212 × 107, 135 × 107, 64 × 107, 24.5 × 107, 23 × 107 and 10 × 107 CFU g−1dw, respectively. Whilst, the bacterial densities in soil samples S10, S11, S1, S15, S6 and S14 were ranged between 15 × 106 and 84 × 106 CFU g−1 dw. However, the samples of sites A1, A2 and D (S2, S3, S4, S5, S18, S19, and S20), with densities ranging between 92 × 105 and 75 × 104 CFU g−1dw, were less populated compared to other sites. In respect to their level of contamination with PCDD/Fs, two of the highest bacterial densities were found in the highest contaminated soil samples (S16 and S17) (Figure 2). This was supported a relatively strong positive Pearson correlation (r = 0.7759), which means that high bacterial densities scores go with high PCDD/Fs level scores (and vice versa). These data indicate that the highest densities of bacterial populations were found in soil samples from sites A3, A4, B and C and the lowest were found in soil samples from the site D.
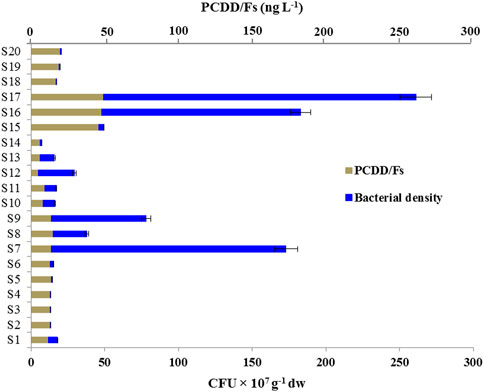
FIGURE 2. Density of microbial communities in soil samples with respect to PCDD/Fs concentrations. The densities of soil microbial communities are expressed as CFU × 107 per g of dry soil (dw). Level of PCCD/Fs in soil samples was determined as described before. CFUs were determined in triplicate. Values are means ± S.D (n = 3).
We noted that Bacillus genus was the most abundant in soil samples with 37.3% of total bacterial genera identified, followed by the genera Pseudomonas and Acinetobacter that represent 21.4% and 14%, respectively (Table 2). The genera Enterobacter and Klebsiella constitutes about 4.6% to 3.7% of the whole bacterial community. Beyond the top five, the bacterial genera e.g., Massilia, Staphylococcus, Arthrobacter and Cronobacter were also identified and constitutes 8.83% of the total bacterial community. Bacteria genera with less than 1% contribution to the bacterial community include Solibacillus, Lysinibacillus, Brevibacillus, Brevundimonas, Oxalicibacterium, Microbacterium, Macrococcus, Chryseobacterium, Pantoea, Citrobacter and Cellulosimicrobium.
Supplementary Table S4 shows that the bacterial community of site A1 (S1, S2 and S3) was predominantly composed of Bacillus genus with B. subtilis and B. cereus being the most abundant. Also, the genus Pseudomonas was interestingly represented by P. stutzeri and P. putida. While Bacillus and Pseudomonas genera also dominated at site A2 (S4, S5, and S6), different species of the genera were recovered at the site. These include B. mycoides, B. circulans, B. mobilis, P. bauzanensis and P. saudiphocaensis. The composition of bacterial community in the site A3 (S7, S8 and S9) was remarkably typified by the presence of Acinetobacter genus, where A. calcoaceticus, A. tjernbergiae, A. radioresistence and A. pittii were the major species of the bacterial community in this site. Although the structure of bacterial community at the site A4 (S10 and S11) was comparatively similar to those of sites A1 and A2, the bacterial species, Arthrobacter crystallopoietes, was exclusively detected at the site. Bacterial community in the site B (S12, S13 and S14) was proportionally composed of Bacillus, Pseudomonas, Acinetobacter and Klebsiella genera with a particular appearance of K. aerogenes and K. pneumonia. The structure of bacterial community of the site C (S15, S16, and S17) was limited in term of species number, these species were mainly belonging to Enterobacter and Cronobacter. Finally, the composition of bacterial community of the site D (S18, S19, and S20) was predominantly consisting of Bacillus and Pseudomonas genera, and the species Bacillus licheniformis and Pseudomonas stutzeri were the most abundant in this site. The bacterial strains identified by sequencing of 16S rRNA with a high score of certainty (72 strains) were submitted in the NCBI GeneBank databases under the accession numbers from MW475085 to MW475154 (please refer to Supplementary Table S4).
Microbial diversity for each site was determined using two diversity indices, the Shannon’s diversity Index (H) and the Simpson’s diversity index (D), however, the microbial species richness was determined by Margalef’s index DMg (please refer to Supplementary Tables S5–S11). Figure 3A shows the variation in H index according to sites, while this index ranged between 2.5 and 2.2 for the sites A1, A2, A3, A4, and B, it showed its lowest value (1.49) in the site C, and was ∼2.0 in the site D. In a similar tendency, the index D held a record (∼13.8) in the site A1, then lowered in the sites A2, A3, A4 and B, and was about 4.0 in the site C (Figure 3B). In parallel, the Margalef’s index DMg, expressing the species richness of microbial communities varied according the sites and showed high values (4.59 and 4.78) in the sites A1 and A2, intermediated values in the sites A3, A4 and B, and a low value (1.92) in the site C (Figure 3C). In connection with the level of PCDD/Fs detected in the sites, it is worth noting that the values of H, D and DMg indices were significantly lower in soil samples that are contaminated with the highest concentration of PCDD/Fs (p < 0.01). While, the highest values of H, D and DMg (2.5, 13.8, 4.78) were found in the sites with low levels of PCDD/Fs (A1 and A2), the lowest values of H, D and DMg (1.49, 4.0, 1.92) were found in the heavily contaminated sites with PCDD/Fs, C and D (Figures 3D–F). Together, our data show that the indices of biodiversity and species richness of microbial communities largely varied according to the sites and are inversely proportioned to their level of contamination with PCDD/Fs.
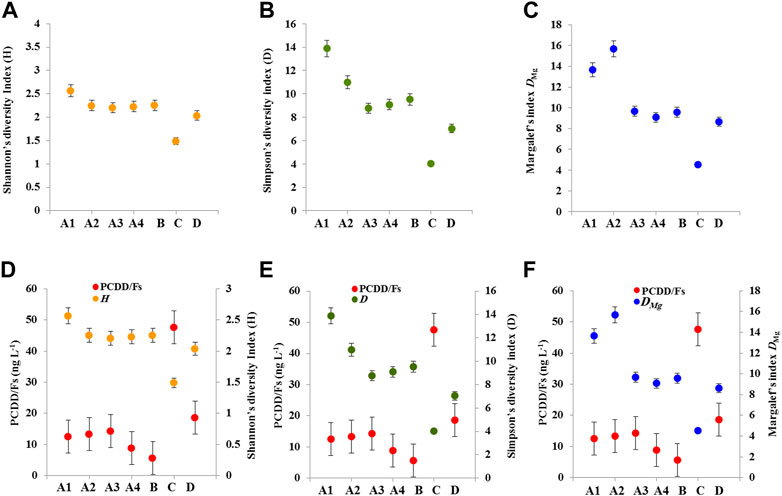
FIGURE 3. Diversity indices of microbial communities as a function of contamination level with PCDD/Fs in different sites. (A–C) Diversity and bacterial species richness were evaluated by The Shannon’s diversity Index (H), The Simpson’s diversity index (D) and Margalef’s index DMg. (D–F) The relations between diversity and richness species, H, D and DMg, and the concentration of PCDD/Fs in different sites. Values are means ± S.D (n = 3). Diversity indices were significantly lower in soil samples that are contaminated with the highest concentration of PCDD/Fs (p < 0.01).
The bacterial communities of soil samples were genetically screened for the presence of angular dioxygenases (AD) and the cytochrome P450 BM3 (CYPBM3) genes, the most characterized bacterial pathways responsible for degradation of dioxins. We identified 22 Gram-negative isolates possessing an AD-encoding gene and 14 Gram-positive isolates with a CYPBM3-encoding gene. Figure 4A shows that although there is random distribution of AD-positive species among soils samples, about 31% of them were found in the highest PCDD/Fs-polluted soils (S15-S20). A moderate positive Pearson correlation (r = 0.6427) was found, which means there is a tendency for high AD-positive species number scores go with high PCDD/Fs level scores (and vice versa). The identities of AD-positive species were determined for each soil samples, and as Figure 4C shows, the AD-positive bacterial species that found in the highest PCDD/Fs-contaminated soils (S15-S20) belonged mainly to the genus Pseudomonas (57.14%), and to the genera Citrobacter, Cronobacte and Pantoea with 14.25% for each. A different scenario was observed for the CYPBM3-positive species, while the highest abundance of these species was detected in the soil samples collected from the sites A1, A2, A3 and A4, showing an intermediate level of contamination with PCDD/Fs. These species were absent in the highest contaminated soils samples (S15-S20) (Figure 4B). The most abundant CYPBM3-positive species belonged to the genus Bacillus (85.71%), while one species belong to Arthrobacter and another to Solibacillus (Figure 4D). Our data indicate that most of bacterial species that harbor the AD encoding gene were found in the highest polluted soils with a major presence of the genus Pseudomonas.
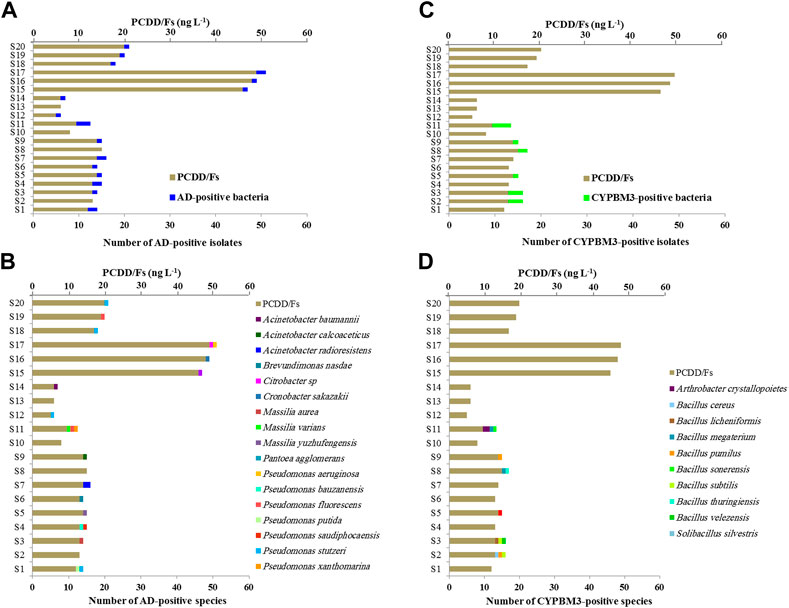
FIGURE 4. Genetic screening for dioxin-degrading genes in bacterial communities. Screening for dioxin-metabolizing genes in bacterial isolates was done using gene-specific primers against the bacterial angular dioxygenase (AD) and cytochrome P450 CYPBM3. (A,C) The number in AD-positive or CYPBM3-positive bacterial isolates in connection with the concentration of PCDD/Fs in soil samples are presented respectively. (B,D) Distribution of bacterial species among the soil samples. The number of isolates from each species found in the soil samples are presented.
Laboratory-scale experiments were conducted to evaluate the ability of 36 bacterial isolates, harbouring the dioxin-degrading genes, to degrade 2,3.7.8-TCDD using the pollutant as the sole carbon source. Results showed that only six bacterial strains grew on the 2,3,7,8-TCDD, with different abilities. Figure 5A shows the growth curves of these stains expressed as CFU mL−1. Of them, the strain A4-2 d of Bacillus megaterium grew more actively in comparison with others strains and reached about of 15.7 × 106 CFU ml−1 after 5 weeks. Bacillus pumilus (A1-2c and A4-2d) showed a weak growth curve with a maximum CFU mL−1 of 7.2 × 106 and 5.4 × 106, respectively. On the other hand, the strain Pseudomonas stutzeri D2e effectively grew in the presence of 2,3,7,8-TCDD plateauing to 11.44 × 106 CFU ml−1 at week 5, which is relatively higher than the growth of Pseudomonas putida A1-1d, while Pseudomonas fluorescens A4-3d, reached 9.52 × 106 and 7.28 × 106 CFU ml−1 at the same time point.
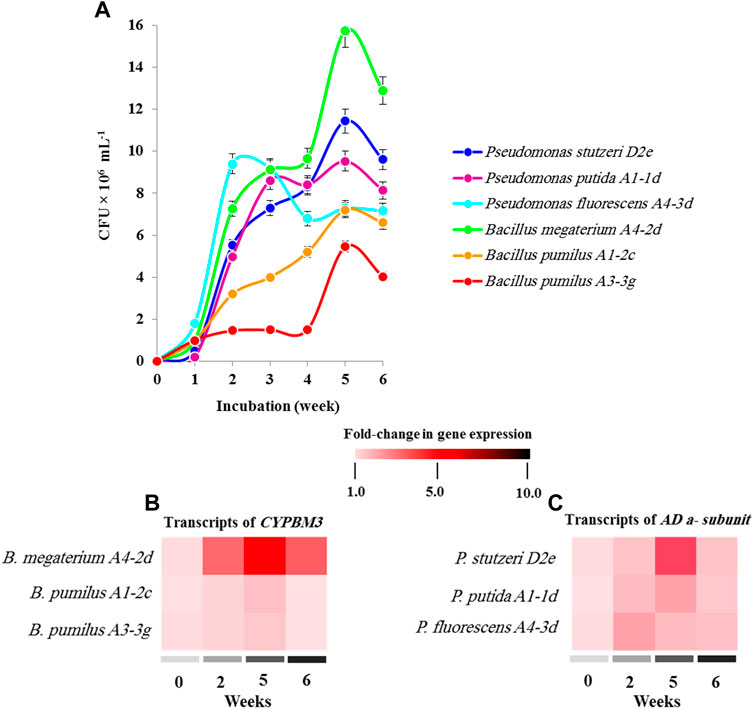
FIGURE 5. Functional identification of 2,3,7,8-TCDD-degrading bacteria. (A) The functionality of dioxin-biodegrading genes (AD and CYPBM3) was assayed by evaluating bacterial growth on 2,3,7,8-TCDD, as sole carbon source in a laboratory-scale experiment. The growth of each strain was expressed as CFU × 106 ml−1 per week for a period of 6 weeks. (B,C) The transcripts levels of CYPBM3 and AD α-subunit genes in different strains growing in the presence of 2,3,7,8-TCDD at 2, 5 and 6 weeks. The colour scale (white-red-black) indicates relative changes of transcript abundance of 1, 5 and 10 fold, respectively. All measurement were performed in triplicates. Values are means ± S.D (n = 9).
Furthermore, the transcripts of bacterial angular dioxygenase (AD a-subunit) and the cytochrome CYPBM3 was quantified using RT-qPCR following the growth of the studied strains in the presence of 2,3,7,8-TCDD. As shown in Figure 5B, the level of CYPBM3 gene transcripts in the strain B. megaterium A4-2d was significantly (p < 0.01) increased and reached about 5-fold at week 5 compared with first week of inoculation. While, the transcripts level of CYPBM3 gene did not changed significantly in both strain of B. pumilus (A1-2c and A4-2d). On the other hand, a significant abundance (p < 0.05) (∼3-fold) for the AD α-subunit gene transcripts was measured in P. stutzeri D2e growing on 2,3,7,8-TCDD at week 5. Whereas a tight change in AD α-subunit gene expression (1.65-fold) was detected in P. putida A1-1d and P. fluorescens A4-3d at week 5 and week 2, respectively (Figure 5C). Our data suggest that both AD- and CYPBM3-mediating pathways for degradation of dioxin were identified in the microbial communities isolated from soil samples polluted with dioxins.
Dioxins are extremely potent environmental toxins with proven toxicological effects in humans, animals, plants and microorganisms (Wu et al., 2002; Kong et al., 2018; Hanano et al., 2019a; Hanano et al., 2019b; González and Domingo, 2021; Hanano et al., 2021; Li et al., 2021; Sun et al., 2021). Nowadays, it is well known that the main pathway by which mammalians are exposed to dioxins is occurring via the consuming of dioxin-contaminated food, thus it is worth to avoid, as downstream as possible, these contaminants to go into the food chain. Following this logic, the investigation of interaction between dioxins and soil microbes should be prioritized because soils, together with sediments, are considered the major reservoirs of such contaminants in most ecosystems. In this context, the contaminated soils with dioxin have been considered as rich sources for the isolation of microorganisms with potential abilities in metabolizing such compounds (Hiraishi, 2003; Jacques et al., 2009; Hanano et al., 2014c; Hanano et al., 2019b; Nguyenag et al., 2021). Therefore, the current study highlights the interrelationship between the contamination level of soils with dioxin and the composition, genetic and functional characteristics of soil microbial communities, and follows up our previous works for identifying bacterial strains as potential dioxins degraders (Hanano et al., 2014c; Hanano et al., 2019b; Almnehlawi et al., 2021; Nguyenag et al., 2021).
Compared to our pervious study, selectively conducted on environmental sites of coastal and middle regions in Syria with potential contamination sources of PCDD/Fs, the current study particularly targets sites that historically were dedicated as waste incineration stations around the capital, Damascus. Our results indicated that the soils samples of the sites located around the waste incineration stations, were moderately contaminated with dioxins while the soil samples collected from a site nearby Homs refinery showed the highest level of contamination with dioxins (Hanano et al., 2014c). Although the total N content of soil samples did not correlate with their level of contamination with dioxins, it is well worth noting that the highest polluted soil samples with PCDD/Fs had also the highest content of total nitrogen. This statement could be possibly supported by the high affinity of dioxin towards soil organic materials (Huyen et al., 2015; Li et al., 2021; Yuan et al., 2021).
Next, the soil microbial communities of the selected sites were characterized in terms of population densities, diversity and species richness. Our results showed that although the density of microbial communities varied largely between soil samples, the highest densities of cultivable bacteria were found in the highest contaminated soil samples which comes in line with previous reports (Hanano et al., 2014c). Regarding the composition of microbial communities, it was observed that the five most abundant genera in soil samples were Bacillus, Pseudomonas, Acinetobacter, Enterobacter and Klebsiella. The presence of such genera was reported in soils contaminated with dioxins, thus suggesting the implication of certain Bacillus species (B. megaterium and B. pumilus), Pseudomonas sp. and Klebsiella sp. in the transformation of PCCD/Fs in the environment (Choi et al., 2003; Hong et al., 2004; Hanano et al., 2019b). In this context, it was suggested that the contaminants can induce an immediate increase in the tolerance of soil microbial community due to their initial toxicity followed by a more gradual enhancement (Van Der Meer, 1994). Expectedly, our data show also that the indices of biodiversity and species richness of microbial communities largely varied according to the sites and inversely proportioned to their level of contamination with PCDD/Fs. These results can be supported by the event that any stressor will lead to a preliminary decrease in biodiversity of soil microbes (Van Bruggen and Semenov, 2000).
Microbial biodegradation of dioxins is now well established for various microorganisms. In bacteria, several mechanisms involved in the biodegradation of dioxins have been described in detail, including but not limited to oxidative degradation by the angular dioxygenases (AD) of aerobic bacteria (Habe et al., 2001) and the cytochrome P450 (CYPBM3) (Sulistyaningdyah et al., 2004) or reductive dechlorination by anaerobic bacteria (Bunge et al., 2003). As we showed, plenty of bacterial strains that harbor the AD encoding gene were found in the highest polluted soils. While, the B. megaterium CYPBM3 was randomly distribution among soils. The genetic screening of microbial communities for AD α-subunit and CYPBM3 encoding gene revealed that the AD-positive bacteria found in the highest PCDD/Fs-contaminated soils belonged mainly to the genera Pseudomonas, Citrobacter, Cronobacte and Pantoea. While, the CYPBM3-positive species were abundant in the soil samples having an intermediate level of contamination with PCDD/Fs. A similar scenario was also found on a previous study, where the abundance of bacteria possessing the AD α-subunit encoding gene was more abundant in the highly polluted soils (Hanano et al., 2014c). In this regard, it was reported that the functional profiling of dioxin-degrading pathways in soils microbes could possibly be affected by the chlorination degree of PCDD/Fs substituents. Apparently, the angular dioxygenases (AD) attack the position 1,10a in dibenzo-p-dioxin and the position 4,4a in dibenzofuran without preference for the level of chloride substituent (Habe et al., 2001; Desta et al., 2021). Therefore, three types of angular dioxygensases have been characterized; a carbazole 1,9a dioxygenase (CARDO) from Pseudomonas sp. strain CA10 (Habe et al., 2001); a dibenzofuran 4,4a dioxygenase (DFDO) from Terrabacter sp. strain DBF63 (Kasuga et al., 2001); and a dibenzo-p-dioxygenase 1,10a from Sphingomonas sp. strain RW1 (Wittich et al., 1992; Armengaud et al., 1998; Armengaud and Timmis, 1998). In a different fashion, a particular link between the CYPBM3 activities and the number of chloride substituent of PCDDs has been documented. While, the highest activity of CYPBM3 was found with substrates with one or two chloride substituents (Sulistyaningdyah et al., 2004), other isoforms of CYPBM3 actively metabolized tetrachlorinated substituents (Hanano et al., 2019b).
The functional abilities of AD and CYPBM3-positive strains were assayed by evaluating the bacterial growth in the presence of 2,3,7,8-TCDD. Of particular interest, the strain B. megaterium A4-2 d grew more actively in comparison with others strains and reached the highest values of CFU mL−1 at week 5 after inoculation and this was likely correlated to a remarkable induction of CYPBM3 gene expression. While two other stains of B. pumilus (A1-2c and A4-2d) showed a weak growth. In line with this, other strains of B. megaterium were identified as potential metabolizers of environmental pollutants such as dioxins, petroleum hydrocarbons, agrochemicals and polystyrene (Quensen and Matsumura, 1983; Thavasi et al., 2011; Bardot et al., 2015; Achiles Do Prado et al., 2021; Meng et al., 2021). Interestingly, these catalytic activities are likely synchronized to specific biological adaptations assisting microorganisms to uptake such hydrophobic contaminants in an efficient manner. e.g., enhancing biosurfactant production, modulating of surface hydrophobicity and composition of the cell membrane in B. megaterium (Thavasi et al., 2008; Bouassida et al., 2018; Hanano et al., 2019b; Singh et al., 2021) or in other microorganisms (Hanano et al., 2014a; Hanano et al., 2015b; Plaza et al., 2016; Hanano et al., 2017; Paraszkiewicz et al., 2018). Intriguingly, the PCDDs-biotransforming activity of CYPBM3 has been experimentally demonstrated for the native or recombinant P450BM-3 protein, where the purified enzymes can hydroxylate 2,3-dichloro-, 2,3,7-trichloro-dibenzo-p-dioxin and 2,3,7,8-tetrachloro-dibenzo-p-dioxin into less toxic intermediates (Sulistyaningdyah et al., 2004; Hanano et al., 2019b). On the other hand, our result also showed that certain strains of P. stutzeri, P.putida and P. fluorescens effectively grew on 2,3,7,8-TCDD with a significant increase of the AD α-subunit gene transcripts in the P. stutzeri D2e. Indeed, several earlier studies reported that such species, and others from the genus Pseudomonas were remarkable degraders of carbazole and dioxins (Shintani et al., 2003; Larentis et al., 2011).
In conclusion, the current study presents a set of biological indicators on the density, diversity, species richness and functional genetics of microbial communities in contaminated soils with dioxins. This was with the objective of identifying new bacteria species with remarkable metabolizing activities against dioxins. This led to identify two bacterial strains with remarkable abilities to metabolize 2,3,7,8-TCDD in laboratory-scale experiment, B. megaterium A4-2d and P. stutzeri D2e with distinct catalytic pathways for the biotransformation of TCDD, the angular dioxygenase (AD α-subunit) and cytochrome P450 (CYPBM3). Based on the different composition of PCDD/Fs and subsequently of their toxicities, the microbial biodegradation of dioxins seems to be a complicated process, requiring different metabolizing pathways that work cooperatively for an effective removal of PCDD/Fs from contaminated soil. As future work, it will be important to focus on certain Gram-positive isolates, mainly those from Bacillus megaterium, with more detailed molecular and biochemical characterization of their angular dioxygenases (AD), which are typically found in Gram-negative bacteria. Moreover, it would be interesting to determine the functional aspect of these distinct dioxin-degradation pathways (AD and CYPBM3) when both are found in such bacteria.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession numbers can be found below: www.ncbi.nlm.nih.gov/nuccore/, from MW475085 to MW475154.
SM and GM collected related materials. SM performed all experimental work. AH conceptualized and wrote the manuscript. All authors read and approved the final manuscript.
We would like to thank Prof. I. OTHMAN, Director General of the AECS and to the Head of Molecular Biology and Biotechnology Department for their crucial support. Also, we thank the International Network on Microbial Ecotoxicology (EcotoxicoMic) (https://ecotoxicomic.org/) for its crucial support throughout our research activities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.1008900/full#supplementary-material
AD, angular dioxygenase; CYP450BM3, cytochrome P450 from B. megaterium; Koa, octanol: air partition coefficients; KOC, organic carbon–water partition; Kow, Octanol/Water partition coefficients; PCBs, polychlorinated biphenyls; PCDDs, polychlorinated dibenzodioxins; PCDFs, polychlorinated dibenzofurans; POPs, Persistent Organic Pollutants; TCDD, Tetrachlorodibenzo-p-dioxin; TEF, toxic equivalency factor; TEQ, toxic equivalent.
Achiles Do Prado, C. C., Pereira, R. M., Durrant, L. R., Scorza Júnior, R. P., Raquel, M., and Bonfá, L. (2021). Fipronil biodegradation and metabolization by Bacillus megaterium strain E1. J. Chem. Technol. Biotechnol. 17, 474–481.
Almnehlawi, H., Dean, R. K., Capozzi, S., Rodenburg, L. A., Zylstra, G. J., and Fennell, D. E. (2021). Agromyces and Arthrobacter isolates from surficial sediments of the Passaic River degrade dibenzofuran, dibenzo-p-dioxin and 2-monochlorodibenzo-p-dioxin. Bioremediation J. 25, 204–224. doi:10.1080/10889868.2021.1892027
Armengaud, J., Happe, B., and Timmis, K. N. (1998). Genetic analysis of dioxin dioxygenase of Sphingomonas sp. Strain RW1: Catabolic genes dispersed on the genome. J. Bacteriol. 180, 3954–3966. doi:10.1128/jb.180.15.3954-3966.1998
Armengaud, J., and Timmis, K. N. (1998). The reductase RedA2 of the multi-component dioxin dioxygenase system of Sphingomonas sp. RW1 is related to class-I cytochrome P450-type reductases. Eur. J. Biochem. 253, 437–444. doi:10.1046/j.1432-1327.1998.2530437.x
Baker, T. K., Kwiatkowski, A. P., Madhukar, B. V., and Klaunig, J. E. (1995). Inhibition of gap junctional intercellular communication by 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) in rat hepatocytes. Carcinogenesis 16, 2321–2326. doi:10.1093/carcin/16.10.2321
Bardot, C., Besse-Hoggan, P., Carles, L., Le Gall, M., Clary, G., Chafey, P., et al. (2015). How the edaphic Bacillus megaterium strain Mes11 adapts its metabolism to the herbicide mesotrione pressure. Environ. Pollut. 199, 198–208. doi:10.1016/j.envpol.2015.01.029
Boddupalli, S. S., Hasemann, C. A., Ravichandran, K. G., Lu, J. Y., Goldsmith, E. J., Deisenhofer, J., et al. (1992). Crystallization and preliminary x-ray diffraction analysis of P450terp and the hemoprotein domain of P450BM-3, enzymes belonging to two distinct classes of the cytochrome P450 superfamily. Proc. Natl. Acad. Sci. U. S. A. 89, 5567–5571. doi:10.1073/pnas.89.12.5567
Bouassida, M., Ghazala, I., Ellouze-Chaabouni, S., and Ghribi, D. (2018). Improved biosurfactant production by Bacillus subtilis SPB1 mutant obtained by random Mutagenesis and its Application in enhanced oil Recovery in a Sand System. J. Microbiol. Biotechnol. 28, 95–104. doi:10.4014/jmb.1701.01033
Bumpus, J. A., Tien, M., Wright, D., and Aust, S. D. (1985). Oxidation of persistent environmental pollutants by a white rot fungus. Science 228, 1434–1436. doi:10.1126/science.3925550
Bunge, M., Adrian, L., Kraus, A., Opel, M., Lorenz, W. G., Andreesen, J. R., et al. (2003). Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421, 357–360. doi:10.1038/nature01237
Carney, S. A., Prasch, A. L., Heideman, W., and Peterson, R. E. (2006). Understanding dioxin developmental toxicity using the zebrafish model. Birth Defect. Res. A 76, 7–18. doi:10.1002/bdra.20216
Cerniglia, C. E., Morgan, J. C., and Gibson, D. T. (1979). Bacterial and fungal oxidation of dibenzofuran. Biochem. J. 180, 175–185. doi:10.1042/bj1800175
Choi, S. D., Hong, H. B., and Chang, Y. S. (2003). Adsorption of halogenated aromatic pollutants by a protein released from Bacillus pumilus. Water Res. 37, 4004–4010. doi:10.1016/s0043-1354(03)00308-7
Chrostowski, C. P., and Foster, A. S. (1996). A methodology for assessing congener-specific partitioning and plant uptake of dioxins and dioxin-like compounds. Chemosphere 32, 2285–2304. doi:10.1016/0045-6535(96)00118-x
Desta, M., Zhang, L., Wang, W., Xu, P., and Tang, H. (2021). Molecular mechanisms and biochemical analysis of fluorene degradation by the Pseudomonas sp. SMT-1 strain. 3 Biotech. 11, 416. doi:10.1007/s13205-021-02946-x
Geyer, H. J., Scheunert, I., Rapp, K., Gebefügi, I., Steinberg, C., and Kettrup, A. (1993). The relevance of fat content in toxicity of lipophilic chemicals to terrestrial animals with special reference to dieldrin and 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD). Ecotoxicol. Environ. Saf. 26, 45–60. doi:10.1006/eesa.1993.1040
González, N., and Domingo, J. L. (2021). Polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/Fs) in food and human dietary intake: An update of the scientific literature. Food Chem. Toxicol. 157, 112585. doi:10.1016/j.fct.2021.112585
Gul, N., Khan, B., Khan, H., Muhammad, S., Ahmad, I., and Gul, N. (2021). Levels of polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/Fs) in municipal waste dumping site, incinerator and brick kiln residues: Evaluation for potential risk assessment. Arab. J. Geosci. 14, 741. doi:10.1007/s12517-021-07108-0
Habe, H., Chung, J. S., Lee, J. H., Kasuga, K., Yoshida, T., Nojiri, H., et al. (2001). Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by two types of bacteria having angular dioxygenases with different features. Appl. Environ. Microbiol. 67, 3610–3617. doi:10.1128/aem.67.8.3610-3617.2001
Hanano, A., Al-Arfi, M., Shaban, M., Daher, A., and Shamma, M. (2014a). Removal of petroleum-crude oil from aqueous solution by Saccharomyces cerevisiae SHSY strain necessitates at least an inducible CYP450ALK homolog gene. J. Basic Microbiol. 54, 358–368. doi:10.1002/jobm.201200525
Hanano, A., Almousally, I., and Shaban, M. (2019a). Exposure of Aspergillus flavus NRRL 3357 to the environmental toxin, 2, 3, 7, 8-tetrachlorinated dibenzo-p-dioxin, results in a Hyper Aflatoxicogenic phenotype: A possible role for caleosin/peroxygenase (AfPXG). Front. Microbiol. 10, 2338. doi:10.3389/fmicb.2019.02338
Hanano, A., Almousally, I., Shaban, M., Moursel, N., Shahadeh, A., and Alhajji, E. (2015a). Differential tissue accumulation of 2, 3, 7, 8-Tetrachlorinated dibenzo-p-dioxin in Arabidopsis thaliana affects plant chronology, lipid metabolism and seed yield. BMC Plant Biol. 15, 193. doi:10.1186/s12870-015-0583-5
Hanano, A., Almousally, I., Shaban, M., and Murphy, D. J. (2018a). Arabidopsis plants exposed to dioxin result in a WRINKLED seed phenotype due to 20S proteasomal degradation of WRI1. J. Exp. Bot. 69, 1781–1794. doi:10.1093/jxb/ery027
Hanano, A., Almousally, I., and Shaban, M. (2014b). Phytotoxicity effects and biological responses of Arabidopsis thaliana to 2, 3, 7, 8-tetrachlorinated dibenzo-p-dioxin exposure. Chemosphere 104, 76–84. doi:10.1016/j.chemosphere.2013.10.060
Hanano, A., Ammouneh, H., Almousally, I., Alorr, A., Shaban, M., Alnaser, A. A., et al. (2014c). Traceability of polychlorinated dibenzo-dioxins/furans pollutants in soil and their ecotoxicological effects on genetics, functions and composition of bacterial community. Chemosphere 108, 326–333. doi:10.1016/j.chemosphere.2014.01.061
Hanano, A., Shaban, M., Almousally, I., and Al-Ktaifani, M. (2015b). Saccharomyces cerevisiae SHSY detoxifies petroleum n-alkanes by an induced CYP52A58 and an enhanced order in cell surface hydrophobicity. Chemosphere 135, 418–426. doi:10.1016/j.chemosphere.2014.11.011
Hanano, A., Shaban, M., and Almousally, I. (2017). Biochemical, molecular, and transcriptional highlights of the Biosynthesis of an effective biosurfactant produced by Bacillus safensis PHA3, a petroleum-Dwelling bacteria. Front. Microbiol. 8, 77. doi:10.3389/fmicb.2017.00077
Hanano, A., Shaban, M., Almousally, I., and Murphy, D. J. (2018b). Identification of a dioxin-responsive oxylipin signature in roots of date palm: Involvement of a 9-hydroperoxide fatty acid reductase, caleosin/peroxygenase PdPXG2. Sci. Rep. 8, 13181. doi:10.1038/s41598-018-31342-4
Hanano, A., Shaban, M., Almutlk, D., and Almousally, I. (2019b). The cytochrome P450BM-1 of Bacillus megaterium A14K is induced by 2, 3, 7, 8-Tetrachlorinated dibenzo-p-dioxin: Biophysical, molecular and biochemical determinants. Chemosphere 216, 258–270. doi:10.1016/j.chemosphere.2018.10.103
Hanano, A., Shaban, M., and Murphy, D. J. (2021). Functional involvement of caleosin/peroxygenase PdPXG4 in the accumulation of date palm leaf lipid droplets after exposure to dioxins. Environ. Pollut. 281, 116966. doi:10.1016/j.envpol.2021.116966
Hiraishi, A. (2003). Biodiversity of dioxin-degrading microorganisms and potential utilization in bioremediation. Microbes Environ. 18, 105–125. doi:10.1264/jsme2.18.105
Hong, H. B., Nam, I. H., Murugesan, K., Kim, Y. M., and Chang, Y. S. (2004). Biodegradation of dibenzo-p-dioxin, dibenzofuran, and chlorodibenzo-p-dioxins by Pseudomonas veronii PH-03. Biodegradation 15, 303–313. doi:10.1023/b:biod.0000042185.04905.0d
Huuskonen, H., Unkila, M., Pohjanvirta, R., and Tuomisto, J. (1994). Developmental toxicity of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) in the most TCDD-resistant and -susceptible rat strains. Toxicol. Appl. Pharmacol. 124, 174–180. doi:10.1006/taap.1994.1021
Huyen, D. T., Igarashi, T., and Shiraiwa, T. (2015). Vertical distribution of dioxins in soil of Bien Hoa airbase, Vietnam. SpringerPlus 4, 300. doi:10.1186/s40064-015-1064-x
Jacques, R. J., Okeke, B. C., Bento, F. M., Peralba, M. C., and Camargo, F. A. (2009). Improved enrichment and isolation of polycyclic aromatic hydrocarbons (PAH)-degrading microorganisms in soil using anthracene as a model PAH. Curr. Microbiol. 58, 628–634. doi:10.1007/s00284-009-9381-3
Jin, R., Bu, D., Liu, G., Zheng, M., Lammel, G., Fu, J., et al. (2020). New classes of organic pollutants in the remote continental environment—Chlorinated and brominated polycyclic aromatic hydrocarbons on the Tibetan plateau. Environ. Int. 137, 105574. doi:10.1016/j.envint.2020.105574
Kasuga, K., Habe, H., Chung, J. S., Yoshida, T., Nojiri, H., Yamane, H., et al. (2001). Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem. Biophysical Res. Commun. 283, 195–204. doi:10.1006/bbrc.2001.4763
Kim, B. H., and Fulco, A. J. (1983). Induction by barbiturates of a cytochrome P-450-dependent fatty acid monooxygenase in Bacillus megaterium: Relationship between barbiturate structure and inducer activity. Biochem. Biophysical Res. Commun. 116, 843–850. doi:10.1016/s0006-291x(83)80219-8
Kim, D. H., Kim, K. H., Kim, D., Jung, H. C., Pan, J. G., Chi, Y. T., et al. (2010). Oxidation of human cytochrome P450 1A2 substrates by Bacillus megaterium cytochrome P450 BM3. J. Mol. Catal. B Enzym. 63, 179–187. doi:10.1016/j.molcatb.2010.01.017
Kjeldahl, J. A. (1883). A new method for the estimation of nitrogen in organic compounds. Anal. Biochem. 22, 366–382.
Kong, F. X., Sun, G. D., and Liu, Z. P. (2018). Degradation of polycyclic aromatic hydrocarbons in soil mesocosms by microbial/plant bioaugmentation: Performance and mechanism. Chemosphere 198, 83–91. doi:10.1016/j.chemosphere.2018.01.097
Larentis, A. L., Sampaio, H. C. C., Carneiro, C. C., Martins, O. B., and Alves, T. L. M. (2011). Evaluation of growth, carbazole biodegradation and anthranilic acid production by Pseudomonas stutzeri. Braz. J. Chem. Eng. 28, 37–44. doi:10.1590/s0104-66322011000100005
Li, M., He, W., and Li, Y. (2021). Enhanced plant-microbe remediation of PCBs in soil using enzyme modification technique combined with molecular docking and molecular dynamics. Biochem. J. 478, 1921–1941. doi:10.1042/bcj20210104
Magan, N., Fragoeiro, S., and Bastos, C. (2010). Environmental factors and bioremediation of xenobiotics using white rot fungi. Mycobiology 38, 238–248. doi:10.4489/myco.2010.38.4.238
Mahfouz, S., Mansour, G., Murphy, J. D., and Hanano, A. (2020). Dioxin impacts on lipid metabolism of soil microbes: Towards effective detection and bioassessment strategies. Bioresour. Bioprocess. 7, 59. doi:10.1186/s40643-020-00347-1
Marcial Gomes, N. C., Borges, L. R., Paranhos, R., Pinto, F. N., Mendonca-Hagler, L. C., and Smalla, K. (2008). Exploring the diversity of bacterial communities in sediments of urban mangrove forests. FEMS Microbiol. Ecol. 66, 96–109. doi:10.1111/j.1574-6941.2008.00519.x
Marshall, N. B., and Kerkvliet, N. I. (2010). Dioxin and immune regulation: Emerging role of aryl hydrocarbon receptor in the generation of regulatory T cells. Ann. N. Y. Acad. Sci. 1183, 25–37. doi:10.1111/j.1749-6632.2009.05125.x
Matson, R. S., Hare, R. S., and Fulco, A. J. (1977). Characteristics of a cytochrome P-450-dependent fatty acid omega-2 hydroxylase from bacillus megaterium. Biochimica Biophysica Acta - Lipids Lipid Metabolism 487, 487–494. doi:10.1016/0005-2760(77)90218-1
Meena, S. S., Sharma, R. S., Gupta, P., Karmakar, S., and Aggarwal, K. K. (2016). Isolation and identification of Bacillus megaterium YB3 from an effluent contaminated site efficiently degrades pyrene. J. Basic Microbiol. 56, 369–378. doi:10.1002/jobm.201500533
Meng, T. K., Mohd Kassim, A. S. B., Bin a Razak, A. H., and Binti Mohd Fauzi, N. A. (2021). Bacillus megaterium: A potential and an efficient BioDegrader of polystyrene. Braz. Arch. Biol. Technol. 64, e21190321. doi:10.1590/1678-4324-2021190321
Narhi, L. O., and Fulco, A. J. (1987). Identification and characterization of two functional domains in cytochrome P-450BM-3, a catalytically self-sufficient monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 262, 6683–6690. doi:10.1016/s0021-9258(18)48296-8
Nguyenag, B. T., Hsieh, J. L., Lo, S. C., Wang, S. Y., Hung, C. H., Huang, E., et al. (2021). Biodegradation of dioxins by Burkholderia cenocepacia strain 869T2: Role of 2-haloacid dehalogenase. J. Hazard. Mater. 401, 123347. doi:10.1016/j.jhazmat.2020.123347
Oliveira, M., Costa, S., Vaz, J., Fernandes, A., Slezakova, K., Delerue-Matos, C., et al. (2020). Firefighters exposure to fire emissions: Impact on levels of biomarkers of exposure to polycyclic aromatic hydrocarbons and genotoxic/oxidative-effects. J. Hazard. Mater. 383, 121179. doi:10.1016/j.jhazmat.2019.121179
Paraszkiewicz, K., Bernat, P., Kusmierska, A., Chojniak, J., and Plaza, G. (2018). Structural identification of lipopeptide biosurfactants produced by Bacillus subtilis strains grown on the media obtained from renewable natural resources. J. Environ. Manag. 209, 65–70. doi:10.1016/j.jenvman.2017.12.033
Plaza, G. A., Chojniak, J., Mendrek, B., Trzebicka, B., Kvitek, L., Panacek, A., et al. (2016). Synthesis of silver nanoparticles by Bacillus subtilis T-1 growing on agro-industrial wastes and producing biosurfactant. IET Nanobiotechnol. 10, 62–68. doi:10.1049/iet-nbt.2015.0016
Quensen, J. F., and Matsumura, F. (1983). Oxidative degradation of 2, 3, 7, 8-tetrachlorodibenzo-P-dioxin by microorganisms. Environ. Toxicol. Chem. 2, 261–226. doi:10.1897/1552-8618(1983)2[261:odotbm]2.0.co;2
Salamanca, M., Chandia, C., and Hernandez, A. (2016). Impact of forest fires on the concentrations of polychlorinated dibenzo-p-dioxin and dibenzofurans in coastal waters of central Chile. Sci. Total Environ. 573, 1397–1405. doi:10.1016/j.scitotenv.2016.07.113
Sato, S. I., Nam, J. W., Kasuga, K., Nojiri, H., Yamane, H., and Omori, T. (1997). Identification and characterization of genes encoding carbazole 1, 9a-dioxygenase in Pseudomonas sp. strain CA10. J. Bacteriol. 179, 4850–4858. doi:10.1128/jb.179.15.4850-4858.1997
Schwalb, H., Narhi, L. O., and Fulco, A. J. (1985). Purification and characterization of pentobarbital-induced cytochrome P-450BM-1 from Bacillus megaterium ATCC 14581. Biochimica Biophysica Acta - General Subj. 838, 302–311. doi:10.1016/0304-4165(85)90227-2
Seralathan, M. V., Sivanesan, S., Bafana, A., Kashyap, S. M., Patrizio, A., Krishnamurthi, K., et al. (2014). Cytochrome P450 BM3 of Bacillus megaterium - a possible endosulfan biotransforming gene. J. Environ. Sci. 26, 2307–2314. doi:10.1016/j.jes.2014.09.016
Shintani, M., Nojiri, H., Yoshida, T., Habe, H., and Omori, T. (2003). Carbazole/dioxin-degrading car gene cluster is located on the chromosome of Pseudomonas stutzeri strain OM1 in a form different from the simple transposition of Tn4676. Biotechnol. Lett. 25, 1255–1261. doi:10.1023/a:1025079027730
Singh, V., Waris, Z., Banat, I. M., Saha, S., and Padmanabhan, P. (2021). Assessment of Rheological Behaviour of water-in-oil Emulsions mediated by glycolipid biosurfactant produced by Bacillus megaterium SPSW1001. Appl. Biochem. Biotechnol. 194, 1310–1326. doi:10.1007/s12010-021-03717-3
Stella, T., Covino, S., Cvancarova, M., Filipova, A., Petruccioli, M., D'annibale, A., et al. (2017). Bioremediation of long-term PCB-contaminated soil by white-rot fungi. J. Hazard. Mater. 324, 701–710. doi:10.1016/j.jhazmat.2016.11.044
Stockholm, C. (2001). Stockholm convention on persistent organic pollutants (POPs). Geneva: Secretariat of the Stockholm Convention. http://chm.pops.int.
Sulistyaningdyah, W. T., Ogawa, J., Li, Q. S., Shinkyo, R., Sakaki, T., Inouye, K., et al. (2004). Metabolism of polychlorinated dibenzo-p-dioxins by cytochrome P450 BM-3 and its mutant. Biotechnol. Lett. 26, 1857–1860. doi:10.1007/s10529-004-5317-y
Sun, S., Cao, R., Jin, J., Zhang, Y., Gao, Y., Lu, X., et al. (2021). Accumulation characteristics and estimated dietary intakes of polychlorinated dibenzo-p-dioxins, polychlorinated dibenzofurans and polychlorinated biphenyls in plant-origin foodstuffs from Chinese markets. Sci. Total Environ. 775, 145830. doi:10.1016/j.scitotenv.2021.145830
Thavasi, R., Jayalakshmi, S., Balasubramanian, T., and Banat, I. M. (2008). Production and characterization of a glycolipid biosurfactant from Bacillus megaterium using economically cheaper sources. World J. Microbiol. Biotechnol. 24, 917–925. doi:10.1007/s11274-007-9609-y
Thavasi, R., Jayalakshmi, S., and Banat, I. M. (2011). Effect of biosurfactant and fertilizer on biodegradation of crude oil by marine isolates of Bacillus megaterium, Corynebacterium kutscheri and Pseudomonas aeruginosa. Bioresour. Technol. 102, 772–778. doi:10.1016/j.biortech.2010.08.099
Toth, K., Somfai-Relle, S., Sugar, J., and Bence, J. (1979). Carcinogenicity testing of herbicide 2, 4, 5-trichlorophenoxyethanol containing dioxin and of pure dioxin in Swiss mice. Nature 278, 548–549. doi:10.1038/278548a0
Tuppurainen, K., Asikainen, A., Ruokojarvi, P., and Ruuskanen, J. (2003). Perspectives on the formation of polychlorinated dibenzo-p-dioxins and dibenzofurans during municipal solid waste (MSW) incineration and other combustion processes. Acc. Chem. Res. 36, 652–658. doi:10.1021/ar020104+
Van Bruggen, A. H. C., and Semenov, A. M. (2000). In search of biological indicators for soil health and disease suppression. Appl. Soil Ecol. 15, 13–24. doi:10.1016/s0929-1393(00)00068-8
Van Der Meer, J. R. (1994). Genetic adaptation of bacteria to chlorinated aromatic compounds. FEMS Microbiol. Rev. 15, 239–249. doi:10.1016/0168-6445(94)90115-5
Wittich, R. M., Wilkes, H., Sinnwell, V., Francke, W., and Fortnagel, P. (1992). Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 58, 1005–1010. doi:10.1128/aem.58.3.1005-1010.1992
World Health Organization, (2016). Executive Summary on the Assessment of the health risk of dioxins. WHO Consultation. May 25-29 1998 www.who.int/pcs/docs/dioxin-exec-sum-final.
Wu, Q., Ohsako, S., Baba, T., Miyamoto, K., and Tohyama, C. (2002). Effects of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) on preimplantation mouse embryos. Toxicology 174, 119–129. doi:10.1016/s0300-483x(02)00047-1
Yao, H., Lu, S., Zhang, X., Pei, J., and Lu, Y. T. (2018). Pollution status and risks of dioxin-like polychlorinated biphenyls in the soil of the yellow river. Huan jing ke xue 39, 123–129. doi:10.13227/j.hjkx.201704100
Keywords: dioxins impact soil microbial communities soil microbial community, polychlorinated dibenzo-p-dioxins/furans (PCDD/Fs), angular dioxygenase (AD), biodiversity, cytochrome P450 (CYPBM3), biodegradation
Citation: Mahfouz S, Mansour G and Hanano A (2022) Compositional, genetic and functional characterization of soil culturable microbial communities in polychlorinated dibenzo-p-dioxins/furans contaminated soil. Front. Environ. Sci. 10:1008900. doi: 10.3389/fenvs.2022.1008900
Received: 01 August 2022; Accepted: 26 September 2022;
Published: 10 October 2022.
Edited by:
Chunhao Gu, University of Delaware, United StatesReviewed by:
Sadia Bibi, University of Agriculture, Faisalabad, PakistanCopyright © 2022 Mahfouz, Mansour and Hanano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdulsamie Hanano, YXNoYW5hbm9AYWVjLm9yZy5zeQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.