- 1SAMRC Microbial Water Quality Monitoring Center, University of Fort Hare, Alice, South Africa
- 2Applied and Environmental Microbiology Research Group (AEMREG), Department of Biochemistry and Microbiology, University of Fort Hare, Alice, SouthAfrica
- 3Department of Environmental Health Sciences, College of Health Sciences, University of Sharjah, Sharjah, United Arab Emirates
- 4Department of Clinical Nutrition and Dietetics, College of Health Sciences, University of Sharjah, Sharjah, United Arab Emirates
- 5Department of Nutrition and Food Technology, Faculty of Agriculture, Jordan University of Science and Technology, Irbid, Jordan
- 6Department of Biology, Albion College, Albion, MI, United States
The occurrence and antibiogram profiles of Listeria monocytogenes in environmental waters in the Eastern Cape Province, South Africa, were investigated. Seventy-eight samples from rivers, wastewater, and irrigation water were collected at different geographical locations within the province from February to September 2019. The procedure of the International Organization for Standardization EN ISO 11290:2017 part 1 and 2 was adopted for the isolation of Lm. The counts of presumptive Lm ranged from 2.0 × 103 CFU/100 ml to 3.6 × 105 CFU/100 ml. About 39.74% of the samples were positive for Lm, and the isolates were confirmed as serotype 1/2a (78.95%) and serotype 4b (21.05%). About 68.42% of the isolates demonstrated biofilm-forming potentials. Ten virulence determinants including the plcA, inlA, and inlB were detected in all the isolates; however, inlC (85.97%), inlJ (80.70%), actA (59.65%), prfA (92.98%), plcB (89.47%), hly (80.70%), and mpl (73.68%) were less prevalent. The antibiogram profiles of confirmed Lm isolates revealed high susceptibilities (>50%) to all antibiotics ranging from 52.63% (cefotetan) to 100% (ampicillin) except for sulfamethoxazole (35.09%), erythromycin (26.32%), streptomycin (38.60%), oxytetracycline (45.61%), and amoxicillin (49.12%). Conversely, high resistance rates against sulfamethoxazole (63.16%), oxytetracycline (54.39%), and amoxicillin (50.88%) were observed. Isolates exhibited 52 resistance patterns against 22 antibiotics tested ranging from 1 to 15 antibiotics, and 82.46% showed multidrug-resistance phenotypes against the antibiotics. The antibiotic resistance index (ARI) of river water (0.18) was less than the permissible (0.2) Krumperman threshold, whereas that of irrigation and wastewater were higher than the Krumperman threshold. The multiple/antibiotic resistance index ((M)ARI) of each of the isolates ranged from 0.05–0.68. Resistance determinants (21) encoding resistance against sulphonamides, β-lactamase, phenicols, aminoglycosides, and tetracyclines were detected among the phenotypically resistant Lm isolates investigated which justifies the phenotypic resistance observed in this study. The abundance of resistance determinants in Lm recovered from environmental waters in this study suggests that the aquatic environments may serve as a channel for the dissemination of antimicrobial-resistant Lm to other niches including the food chain.
Introduction
The cost of managing infectious diseases is gradually rising as more life-threatening diseases than ever encountered are emerging (Ho and Hendi, 2018). This could be attributed to the exponential increase and ascendancy of antibiotic-resistant bacteria/antibiotic resistance genes (ARB/ARGs) due to the residual impact of antimicrobial agents in the environment (Zhang et al., 2009; Titilawo et al., 2015; Wang et al., 2019). This menace might obliterate humans’ successes in antibiotics discovery and advancements. Although ARB/ARGs predate antibiotics, the unrestricted spread is primarily dependent on the indiscriminate use of antibiotics in humans and animal husbandry (Titilawo et al., 2015). Consequently, ARB/ARGs could impair the efficacy of antibiotics currently used in alleviating microbial infections (Titilawo et al., 2015). Thus, this scenario if not addressed could lead to serious health concerns and the future emergence of superbugs in the environment including the aquatic ecosystem.
Natural water sources are essential resources endowed by nature for biological and human activities including domestic, industrial, recreational, and agricultural purposes (Chigor et al., 2013; Hassan Omer, 2020). However, anthropogenic impacts have immensely impaired their quality and immeasurably contributed to the abundance of pathogenic microbial species in the aquatic milieu (Chigor et al., 2013). As such, contaminated water was identified as one of the prominent reservoirs of pathogenic microbes commonly responsible for about 4 billion diarrhea cases, about 2.2 million deaths in most developing nations, and contributing about 90% of the global disease burden (Ahmed et al., 2020). Despite that ensuring proper sanitation and equal universal access to safe water remains one of the cardinal agendas of the United Nations Sustainable Development Goals (SDGs) (Ait-Kadi, 2016), yet water bodies are still under threats of constant pollution. The World Health Organization identified that a wide range of health-related challenges including gastroenteritis, dermal infections, microbial waterborne infections, and toxic metal–related infections could be attributed to direct or indirect human exposure to contaminated water (Sweileh et al., 2016). Waterborne diseases (infections and intoxications) are often acquired through the consumption of contaminated water; direct person-to-person and fecal-oral contamination could also play significant roles in transmission (Ahmed et al., 2020). More worrisome is the increasing spread of ARB/ARGs in environmental waters (including sewage, wastewater from animal husbandry, hospital effluents, and not limited to the wastewater treatment plant (WWTTP)). These sources may serve as a channel for dissemination of ARB/ARGs to other aquatic ecosystems including streams, lakes, rivers, ponds, or leached into the soil (Zhang et al., 2009; Titilawo et al., 2015). ARB/ARGs could be transmitted to humans via the food chain or in drinking water sources considering that clinical resistance has been linked to various environmental resistant bacteria (Zhang et al., 2009).
Several antibiotic resistance determinants have been identified among microbial communities in water. The detection of tetracycline resistance genes among microorganisms isolated from water (Jacobs and Chenia, 2007; Rahman et al., 2008; Titilawo et al., 2015) and sulphonamide ARGs was reported (Pei et al., 2006; Titilawo et al., 2015). Also, various types of resistance genes including aac (Lee et al., 1998), aad, aph, and str (Mohapatra et al., 2008; Titilawo et al., 2015) were detected in surface water isolates. Furthermore, the β-lactamase (bla and ampC) genes from surface water have been reported (Henriques et al., 2006). Globally, ARB/ARGs are a major environmental contaminant of interest and have been accorded increased attention in recent times. As such, sufficient information on the prevalence of ARB/ARGs of medically important pathogens including Listeria monocytogenes (Lm) would serve as a good rationale for the choice of antimicrobial therapy when required (Okeke et al., 2005; Titilawo et al., 2015).
Lm, a Gram-positive pathogenic bacterium of the family Listeriaceae, is commonly found adapted and isolated from various natural environmental niches (Kayode et al., 2020). Members of the genus Listeria are currently 21, but only two (Lm and Listeria ivanovii) are often regarded as pathogenic species known to infect both humans and animals (Kayode et al., 2020; Quereda et al., 2020). Lm is known for sporadic food-borne outbreaks characterized by high fatalities ranging from 20–30% (Drolia and Bhunia, 2019). Usually, symptoms manifest after food contaminated by the pathogen is ingested. Studies have also shown that the severity of these symptoms could be strain dependent based on the virulence factors including actA, hly, iap, plcB, plcC, prfA, and mpl, inlA, inlcB, and inlC harboured by infecting strains (Montero et al., 2015).
Recent decades have recorded episodes of listeriosis outbreaks globally which might be attributable in part to increasing recreational and tourist activities and high demand for and consumption of wide arrays of fresh produce, especially those that require little or no listericidal treatments before consumption (Aureli et al., 2000; Gaul et al., 2013; Zhu et al., 2017; World Health Organization, 2018; Olanya et al., 2019). For instance, South Africa reported one of the largest listeriosis outbreaks worldwide in 2018 involving 1,060 laboratory-confirmed cases and 216 deaths (Drolia and Bhunia, 2019; Kayode et al., 2020). In this particular reference, this pathogen could have found its way into the food chain as farmers use contaminated water sources for irrigation due to the shortage of natural safe water supply threatened by climatic change and inconsistent rainfall patterns in the region. In this study, the prevalence and antibiogram signatures of Lm in selected environmental waters within the Eastern Cape Province, South Africa (ECPSA), were examined.
Methodology and Materials
Study Area, Sampling, and Counts of Presumptive Listeria monocytogenes
The study area included the Amathole, Chris Hani, and Sarah Baartman District Municipalities (DMs) in the ECPSA located within the 32.5842°S, 27.3616°E, 31.8743°S, 26.7968°E, and 33°57′S 25°36′E on the map, respectively. The Amathole DM occupies the central coastal part of the ECPSA, Chris Hani DM is a landlocked district situated at the center of the ECPSA, and the Sarah Baartman DM is situated at the western part of the ECPSA. Environmental water samples including wastewater (n = 21), river water (n = 37 s), and untreated surface water used for irrigation (n = 20) were fetched in 1 L sterile bottles. Wastewater was collected before treatment and downstream of the river serving as a receiving watershed for the final effluent at different geographical locations in the study area. Environmental water samples were collected on a once-off regime between February and September 2019. Anthropogenic activities including animal grazing and waste/refuse disposal were observed on some of the river courses. Samples collected were labelled and conveyed to the laboratory for analyses in an insulated ice-packed container. For the enumeration of presumptive plate counts of Lm, three series of ten-fold (10–1, 10–2, and 10–3) dilution of the environmental water samples were carried out (Adefisoye and Okoh, 2016). A vacuum pump filtration of 100 ml of each dilution through a membrane filter (0.45 μm pores Merck, South Africa) was performed. The filters were aseptically placed on the Chromogenic Listeria Agar (ISO) Base (CM1084 Oxoid Ltd., United Kingdom) supplemented with (ISO OCLA) the differential supplement (SR0244E Oxoid Ltd., United Kingdom) and selective supplements (SR0226E Oxoid Ltd., United Kingdom) and the Brilliance Listeria Agar Base (CM1080 Oxoid Ltd., United Kingdom) supplemented with the Brilliance differential supplement (SR0228E Oxoid Ltd., United Kingdom) and selective supplements (SR0227E Oxoid Ltd., United Kingdom). Incubation was performed at 37°C for 48 (±2) hours in aerobic conditions. Presumptive colonies of Lm (blue/green colonies) obtained were counted and expressed as CFU/100 ml (colony-forming units per 100 ml).
Isolation of Presumptive Listeria monocytogenes
For enrichment purpose, the EN ISO 11290–1:2017 standard method for Lm was adopted in this study (ISO, 2017; Public Health England, 2019). Twenty-five (25 ml) each, of the samples, were aseptically preenriched in 225 ml of the Half Fraser Broth base (CM0895) supplemented with the Half Fraser selective supplement (SR0166E Oxoid Ltd., United Kingdom) and incubated at 30°C for 24 h to resuscitate the cells. After that, 0.1 ml of preenriched broth was added to a 10 ml Fraser broth for secondary enrichment in the selective medium for 48 (±2) hours at 37°C. After incubation, the broths were surface plated on the Chromogenic Listeria Agar (ISO) Base (CM1084 Oxoid Ltd., United Kingdom) supplemented with the OCLA (ISO) differential supplement (SR0244E Oxoid Ltd., United Kingdom) and selective supplement (SR0226E Oxoid Ltd., United Kingdom) and the Brilliance Listeria Agar Base (CM1080 Oxoid Ltd., United Kingdom) supplemented with the Brilliance differential supplement (SR0228E Oxoid Ltd., United Kingdom) and Brilliance selective supplement (SR0227E Oxoid Ltd., United Kingdom). The plates were incubated aerobically at 37°C for 48 (±2) hours. Each sample was analyzed in triplicates, and representative distinct colonies (blue/green colonies with halos) obtained were subcultured on a nutrient agar for purity. Pure cultures of presumptive isolates were stored at −80°C in the tryptic soy broth (TSB) made up of 25% glycerol.
Molecular Identification and Characterization
Extraction of Genomic DNA
DNA extraction was carried out using the direct boiling method (Garrido-Maestu et al., 2020) with modifications. Presumptive Listeria isolates were resuscitated from the glycerol stock in 5 ml of tryptic soy broth (CM0129 Oxoid Ltd., United Kingdom) at 37°C for 18 h. The broths were centrifuged at 16,000 rpm for 5 min in 2 ml Eppendorf tubes using a minispin microcentrifuge. The supernatants were removed, and the pellets were washed in normal saline and resuspended in 300 μL nuclease-free water to boil in a heating block (TECHNE Digital Dri-Block DB-3D, United Kingdom) at 100°C for 10 min and were left to cool on ice for 10 min. The cell debris was removed by centrifugation at 16,000 rpm for 5 min, and the DNA template was transferred into a clean sterile Eppendorf tube and preserved at −20°C for further analysis. DNA was quantified to determine its concentration in a fluorometer (Invitrogen Qubit Fluorometer, Turner BioSystems).
DNA Amplification and Multiplex PCR
A 370 bp segment of the 16S rRNA prs gene was amplified by the polymerase chain reaction (PCR) in a thermal cycler (BIO-RAD T100) using the primer sets F-GCTGAAGAGATTGCGAAAGAAG and R-CAAAGAAACCTTGGATTTGCGG (Doumith et al., 2004). Further classification of the isolates using specific primer sets F-ACAAGCTGCACCTGTTGCAG and R-TGACAGCGTGTGTAGTAGCA that targets the iap gene for Lm was performed (Furrer et al., 1991). The multiplex PCR technique previously documented by Doumith et al. (2004) was adopted for the molecular classification of Lm to various serotypes. Referenced strains of Lm (ATCC 19118 and ATCC 7644) were used as a positive control, while sterile nuclease-free water served as a no-template control.
Detection of Virulence Genes
Ten virulence-associated (inlA, inlB, inlC, inlJ, actA, prfA, hlyA, plcA, and mpl) genes of the confirmed Lm were amplified using the primer sets (as shown in Supplementary Table S1) reported in previous studies by Jung et al. (2009) and Du et al. (2017). The PCR products generated were separated by loading 5 µL amplified DNA products in 1.5% agarose gel stained with ethidium bromide resolved at 100 V for 45 min suspended in a 5 × TBE buffer using an agarose gel electrophoresis system (ADVANCE Mupid™ One Takara, Japan) and detected with an Alliance 4.7 UV transilluminator (Alliance XD-79.WL/26MX, France).
Assessment of Biofilm-Forming Potential
Lm strains were assessed for the ability to form biofilm using the crystal violet (CV) techniques. Freshly grown overnight cultures were centrifuged for 2 min at 7,000 rpm after which the cells recovered were washed and suspended in sterile phosphate-buffered saline (PBS, pH 7.2) and at 0.5 McFarland standards (Basson et al., 2008). The bacterial cell adherence was assessed in a final 200 μL volume comprising sterile 180 μL TSB and 20 μL standardized cell suspensions inoculated into sterile 96-well polystyrene microtiter plates with lids in triplicates (Igbinosa et al., 2015). The TSB or PBS served as negative controls, while Lm strains (ATCC 19118 MediMark ® Europe and ATCC 7644 Mast diagnostics group Ltd., Merseyside, U.K.) served as the positive control. The microtiter plates were incubated covered at 37°C for 72 h. Following incubation, the contents of the microtiter plates were aspirated and washed in PBS (200 μL) thrice to remove cells unattached to the microtiter plates. Cells attached to the wells of the microtiter plates were fixed for 15 min with 98% ethanol and allowed to air dry. The fixed cells (biofilm) were stained for 30 min with 200 μL 2% crystal violet, and the wells were washed in distilled water to remove excess stains and air-dried. After this, 200 μL of 35% acetic acid was added to each well to resolubilize the crystal violet for 30 min on a shaker (Orbit™ 1900 High-Capacity Lab Shaker, Labnet International, Inc., United States) before the absorbance was read at 595 nm (Abs@595) by using a microtiter photometer (SynergyTMMx Monochromator-Based MultiMode Reader w/Time-resolved fluorescence, BioTek Instruments, United States). Abs@595 nm recorded for all positive and negative controls (AbsNC) was computed to obtain the mean and standard deviation. The results obtained were used to categorize Lm isolates as either weak = (AbsNC < Abs@595 nm ≤ 2 × AbsNC), moderate = (2 × AbsNC < Abs@595 nm ≤ 4 × AbsNC), or strong = (4 × AbsNC < Abs@595 nm) biofilm formers (Stepanović et al., 2000).
Antimicrobial Susceptibility Profiling
The Kirby Bauer disc diffusion method was employed according to the standard procedure described by the Clinical and Laboratory Standards Institute (CLSI, 2017) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2020). The isolates were tested against a panel of 22 antibiotic (listed in Supplementary Table S2) discs (Mast Diagnostics, Oxoid, United Kingdom) commonly used for the treatment of microbial infections. A 100 μL fresh culture of each bacterial cell suspension was transferred into a sterile 0.89% saline solution adjusted to 0.5 Mc Farland standard (Mcfarland, 1907) and spread plated on prepared Mueller-Hinton agar plates. Antibiotic discs were dispensed on the surfaces of the inoculated plates, and the plates were incubated at 37°C for 24 h. The zones of inhibition were measured to the nearest millimeters (mm) after incubation. The classification of isolates as resistant (R), intermediate (I), or susceptible (S) to a particular antibiotic was based on the result obtained using standard reference values according to CLSI, 2017; EUCAST, 2020 documents.
Multiple/Antibiotic Resistance Phenotypes and Multiple/Antibiotic Resistance Index
The multiple antibiotic resistance patterns (MARPs) of Lm in respect of the antibiotics tested were applied to each isolate that showed phenotypic resistance against three or more antibiotics and indexed for MARI (Multiple Antibiotic Resistance Index) scores (Krumperman, 1983). The MARI was, thus, computed as follows:
Furthermore, the antibiotic resistance index (ARI) was calculated for each of the environmental water samples as described by Krumperman (1983). Thus, ARI for the samples was computed as follows:
The multidrug resistance resistance (MDR) pattern, the frequency of resistance, and the total sum of antibiotics to which the isolates exhibited resistance were described.
Detection of Antimicrobial Resistance Genes Among Listeria monocytogenes Isolates
Various resistance determinants (44) that encode the expression of tetracycline (tetA, B, C, D, E, G, K, L, and M), chloramphenicol (catI, catII, and cmlA1), sulphonamides (sul1 and sul2), and aminoglycosides (strA, aadA, aac (3)-IIa (aacC2)a, aph (3)-Ia(aphA1)a, and aph (3)-IIa (aphA2)a) resistance were screened by the simplex/multiplex PCR techniques. The sequences of primers, PCR protocols, and the amplicon sizes (Supplementary Table S5) were as described in our previous report (Titilawo et al., 2015). Also, genes (22) that encode ampC β-lactamases and extended spectrum of β-lactamases variants (ESBLs) and carbapenemases and the blaTEM and blaZ genes (Supplementary Table S6) were also screened for using the simplex and multiplex PCR techniques as described elsewhere (Dallenne et al., 2010). The PCR protocol for the amplification of the β-lactamase ampC gene includes the following: 94°C–4 min; 30 (94°C–45 s; 60°C–45 s; and 72°C–45 s); and 72°C–7 min. ESBL genes were detected using the following: 94°C–10 min; 30 (94°C–40 s; 60°C–40 s; and 72°C–1 min); and 72°C–7 min. The carbapenem-optimized annealing temperature for the amplification of blaGES and blaOxa-48 genes was 57°C, while blaVIM, blaIMP, and blaKPC genes were amplified at 55°C. The blaTEM gene was amplified using the following outlined PCR protocol: 94°C–4 min; 30 (94°C–45 s; 60°C–45 s; and 72°C 45 s); and 72°C–7 min, while the blaZ PCR protocol reads as follows: 94°C–5 min; 30 (94°C–30 s; 60°C–30 s; and 72°C 1½ min); and 72°C–5 min. All PCR reactions were prepared in a 25 µL final volume including 12.5 µL of master mix (Quick-Load BioLabs, United Kingdom) and 1 µL each of the reverse and forward primers (Inqaba Biotech, SA), 3 µL aliquots of DNA template, 0.5 µL of MgCl2 buffer, and 6.5 µL of sterile nuclease-free water and amplified using a thermal cycler (BIO-RAD T100™ Thermal Cycler, Singapore). The PCR products generated were separated by loading 5 µL amplified DNA products in 1.5% agarose gel (Merck, SA) stained with ethidium bromide (Sigma-Aldrich, United States) suspended in a 5 × TBE buffer and resolved at 100 V for 45 min using an agarose gel electrophoresis system (ADVANCE Mupid™ One Takara, Japan) and detected with an Alliance 4.7 UV transilluminator (Alliance XD-79.WL/26MX, France).
Typing of Listeria monocytogenes by Enterobacterial Repetitive Intergenic Consensus PCR
The molecular diversity of the Lm isolates was determined by ERIC-PCR using the primer sets ERIC-1 5′-ATGTAAGCTCCTGGGGATTCAC-3′ and ERIC-2 5′-AAGTAAGTGACTGGGGTGAGCG-3′ (Dorneles et al., 2014). The reaction products (5–10 µL) were separated in a 1.5% agarose gel stained with ethidium bromide (3 µL) and suspended in a 5 × buffer (buffer) subjected to 90 V for 240 min using the Bio-Rad electrophoresis machine (Bio-Rad® PowerPac™ Basic Power Supply, Singapore). Five microliters volume of DNA ladder ranging from 100 bp and 10 kb was put in the gel. The gel image was snapped and recorded using an Alliance 4.7 UV transilluminator (Alliance XD-79.WL/26MX, France).
Evaluation of Intraspecies/Strain Multiplicity of Listeria monocytogenes Isolates
Fingerprint images of Lm strains were digitized by a computer-assisted software analysis (GelJ version 2.0). The determination of the molecular weight and occurrence matrices of the ERIC-PCR bands was performed by the UPGMA algorithm (unweighted pair group arithmetic mean) at 1.0% tolerance level for control of quality. The presence/absence of a fingerprint across the isolates formed the basis for the strain homogeneity (associations) assessment. The dendrograms of two matrices were generated by neighbor joining (NJ) through a Jaccard similarity index, and the relative abundance of strains forming a clade was determined.
Statistical Analysis
Data obtained were analyzed statistically to compare Lm counts in water using one-way analysis of variance (ANOVA). The statistical significance of mean ± SD was considered at (p ≤ 0.05). Spearman’s chi-square test was used to measure the prevalence and association between categorical variables (serotypes and genetic virulence determinants). The relationship between the distribution of phenotypic and genotypic resistance of Lm in environmental water was determined using Spearman’s correlation. Significant differences were identified at (p ≤ 0.01) and (p ≤ 0.05) as appropriate. The statistical analysis was performed using IBM SPSS statistical software version 25 and Microsoft Excel spreadsheet.
Results
Presumptive Counts and Confirmation of Listeria monocytogenes
A sum of 78 samples from rivers, wastewater, and irrigation water were collected at different geographical locations within the study area. The presumptive Lm counts of water samples ranged from 2.0 × 103 CFU/100 ml to 3.6 × 105 CFU/100 ml. A comparison of the counts (mean ± SD) using the one-way analysis of variance (ANOVA) revealed that water types significantly influenced the counts (p < 0.05).
Two hundred and seventeen presumptive Lm isolates were recovered from the water samples, and 57 were confirmed as Lm from the presumptive isolates. The largest proportion of Lm was confirmed in wastewater (n = 30, 52.63%), while the distribution of Lm in river water (n = 15, 26.32%) and irrigation water (n = 12, 21.05%) was lower. Lm was detected in (n = 31) samples representing 39.74% of the total environmental water samples collected. The prevalence and distribution of Lm isolates were summarized in Table 1.
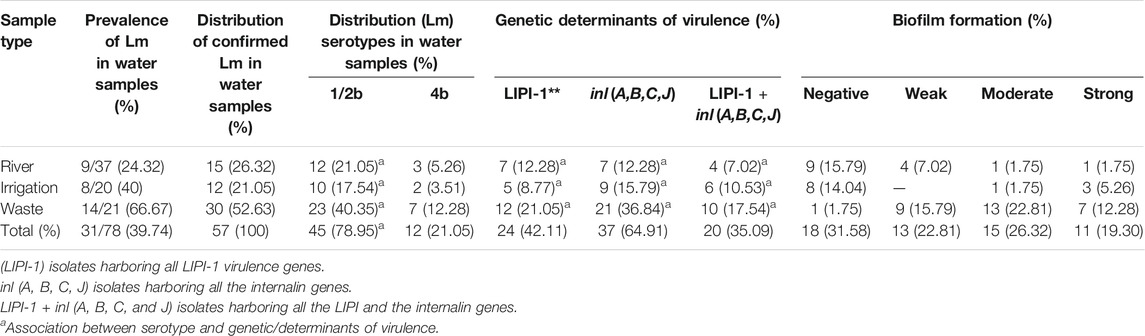
TABLE 1. Occurrence, serotypes, phenotypic, and genotypic determinants of Listeria monocytogenes virulence recovered from environmental waters.
Serotypes and Virulence Determinants of Listeria monocytogenes Isolates
Serotypes (4b and 1/2b) were detected among the Lm isolates (Table 2). Serotype 1/2b was the most prevalent (n = 45, 78.95%), while serotype 4b (n = 12, 21.05%) was less prevalent. Also, the prevalence of 10 virulence determinants among the isolates was screened in this study. The plcA, inlA, and inlB were detected in all isolates; the prevalence of inlC (85.97%), inlJ (80.70%), actA (59.65%), prfA (92.98%), plcB (89.47%), hly (80.70%), and mpl (73.68%) varied among the isolates. The genotypic virulence profile of the listeriosis agent recovered from the samples is presented in Table 2. All the LIPI-1 (Listeria pathogenic island 1) virulence genes (including plcA, actA, prfA, plcB, hly, and mpl) were detected among 24 (42.11%) Lm isolates, all internalin virulence genes (including inlA, InlB, inlC, and inlJ) were detected among 37 (64.91%) Lm, and all the LIPI-1 + inl (A, B, C, and J) genes were detected among 20 (35.09%) Lm isolates recovered from environmental water samples. The data obtained were further subjected to a chi-square test to ascertain the frequencies and association of genetic determinants (Table 1). The association between the genetic determinants of serotype 1/2b observed was statistically significant (p > 0.01).
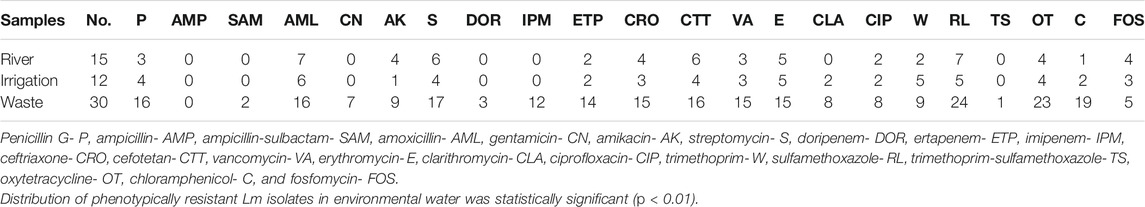
TABLE 2. Distribution of antibiotic-resistant Listeria monocytogenes isolates recovered from environmental waters.
Biofilm-Forming Potential of Listeria monocytogenes Isolates
We observed that 39 (68.42%) of the Lm isolates demonstrated potential for biofilm formation. The classification as weak (n = 13, 22.81%), medium (n = 15, 26.32%), and strong (n = 11, 19.30%) was carried out based on the strength of biofilm forming. Isolates recovered from wastewater were predominantly potential biofilm formers (96.67%). However, biofilm formation among Lm isolates from river water and irrigation water was less prevalent. A detailed description of the distribution of the biofilm-forming isolates in environmental waters was provided in Table 1.
Antibiotic Susceptibility and Distribution of Antimicrobial-Resistant Listeria monocytogenes Recovered From Environmental Waters
Antibiogram profiles and susceptibility patterns of each of the Lm isolates tested against 22 antibiotics were summarized in Supplementary Table S2. High susceptibilities (>50%) were observed to all antibiotics ranging from 52.63% (cefotetan) to 100% (ampicillin) except for sulfamethoxazole (35.09%), erythromycin (26.32%), streptomycin (38.60%), oxytetracycline (45.61%), and amoxicillin (49.12%). Conversely, a high resistance rate against sulfamethoxazole (63.16%), oxytetracycline (54.39%), and amoxicillin (50.88%) was observed. Intermediate resistance (>15%) was recorded for erythromycin (29.82%), clarithromycin (17.54%), and amikacin (15.79%). No intermediate resistance was observed among the β-lactams, carbapenems, glycopeptides, and the tetracyclines antibiotics. Figure 1 shows a heatmap representing the phenotypic antibiotic susceptibility pattern and the variations among each of the isolates examined in the study. This pattern revealed the effectiveness of each antibiotic toward the individual isolate, and it could reflect the genetic characteristics of the isolates. The frequency of Lm isolates that showed phenotypic resistance against various antibiotics tested ranged between 1 and 24 in environmental waters. High frequencies of resistant Lm against penicillin, amoxicillin, streptomycin, cefotetan, sulfamethoxazole, oxytetracycline, and chloramphenicol were observed, particularly in isolates recovered from wastewater (Table 2). The results indicated a statistically significant (p < 0.01) relationship in the distribution of phenotypically antimicrobial-resistant Lm isolates in environmental water samples examined.
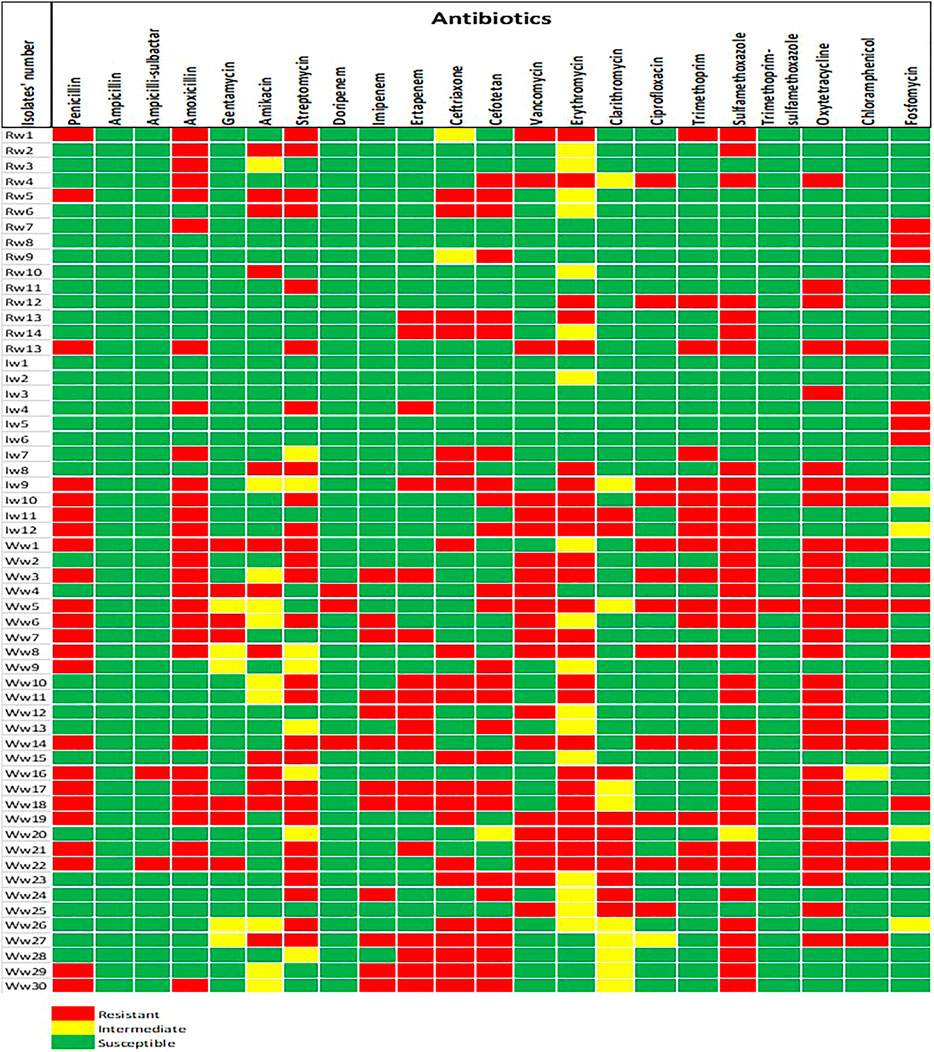
FIGURE 1. The phenotypic antibiotic resistance profiles of Listeria monocytogenes isolates recovered from environmental waters. The color codes indicated resistant, intermediate, and susceptible phenotypes to 22 antibiotics. The code (Rw) denotes isolates recovered from river water, (Iw) for irrigation water, and (Ww) for wastewater.
Assessment of the Multiple/Antibiotic Resistance Phenotypes and Multiple/Antibiotic Resistance Index of Listeria monocytogenes Isolates
Patterns of antibiotic resistance phenotypes was presented in Table 3. Fifty-two different phenotypic resistance patterns ranging from 1 to 15 antibiotics were observed among resistant Lm isolates. Two isolates (3.51%) from the environmental water samples were not resistant against any of the antibiotics, six (10.53%) showed resistance against one antibiotic, 3 of the Lm isolates (5.26%) were resistant against two antibiotics, and 47 (82.46%) showed multidrug-resistant phenotypes (resistance against more than 2 antibiotics) against various antibiotics tested. The ARI of river water (0.18) was less than the permissible (0.2) Krumperman threshold, whereas that of irrigation and wastewater was higher than the Krumperman threshold. This could be an indication of higher antibiotic selection pressures in irrigation and wastewater compared to river water. Isolates with MARI >0.2 were more abundant in wastewater. The (M)ARI of each of the isolates ranged from 0.05 to 0.68 (Supplementary Table S3).
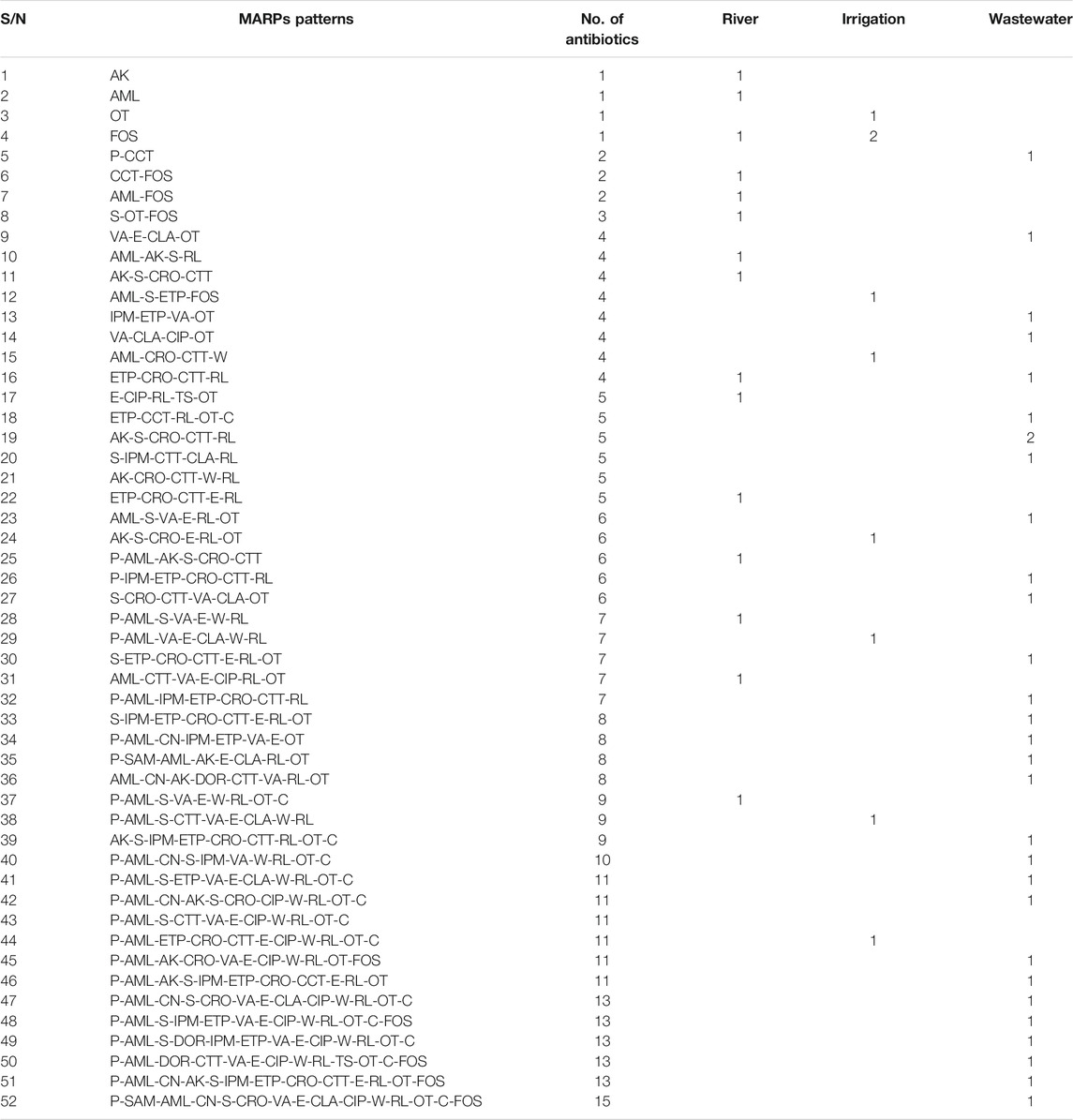
TABLE 3. Distribution of Multiple/Antibiotic Resistance Phenotypes ((M)ARPs) of Listeria monocytogenes isolates from environmental waters.
Distribution of Antimicrobial Resistance Genes Among Listeria monocytogenes Isolates
The genotypic resistance determinants of Lm strains were investigated, and twenty-one (21) resistance genes were detected among environmental water isolates out of 44 genes screened (Table 4). Sulphonamide’s resistance genes were detected in 19 (55.88%) of 34 isolates. The sul2 gene was more prevalent among the isolates (n = 14, 41.18%) compared with the sul1 gene (n = 13, 38.24%). The β-lactamase genes were screened for in about 20 isolates that expressed phenotypic resistance against β-lactam antibiotics, and 15 (75%) were positive for the genes. The isolates were further screened for the ampC, various variants of ESBLs, and carbapenem genes. The blaTEM variants (n = 7, 35%) and blaSHV (15%) were more prevalent among the β-lactam genes encoding resistance against β-lactam antibiotics. Genes encoding tetracycline, phenicols, and aminoglycosides antibiotic resistance were detected among isolates from environmental water. Among the tetracycline resistance genes (tetA, B, C, D, E, G, K, L, and M) screened, the tetA (n = 23, 85.19%) and tetM (19, 70.37%) were more prevalent among the environmental water isolates while tetE and tetL were not detected among tetracycline-resistant isolates. The catI resistance gene was detected among isolates from river (n = 1, 6.67%) and wastewater (n = 7, 46.67%). The prevalence of strA (n = 13, 43.33%), aadA (n = 6, 20%), and aph (3)-IIa (aphA2)a (n = 4, 13.33%) genes was detected among the isolates. The ARGs were more prevalent among isolates recovered from wastewater. The statistical analysis revealed a significant association between the distribution of resistant genes among Lm isolates in environmental water including sul1, sul2, blaTEM, blaZ, tetA, tetM, catI, and strA, at (p < 0.01) and tetG, catI, and blaTEM variants (TEM-1 and TEM-2) at (p < 0.05).
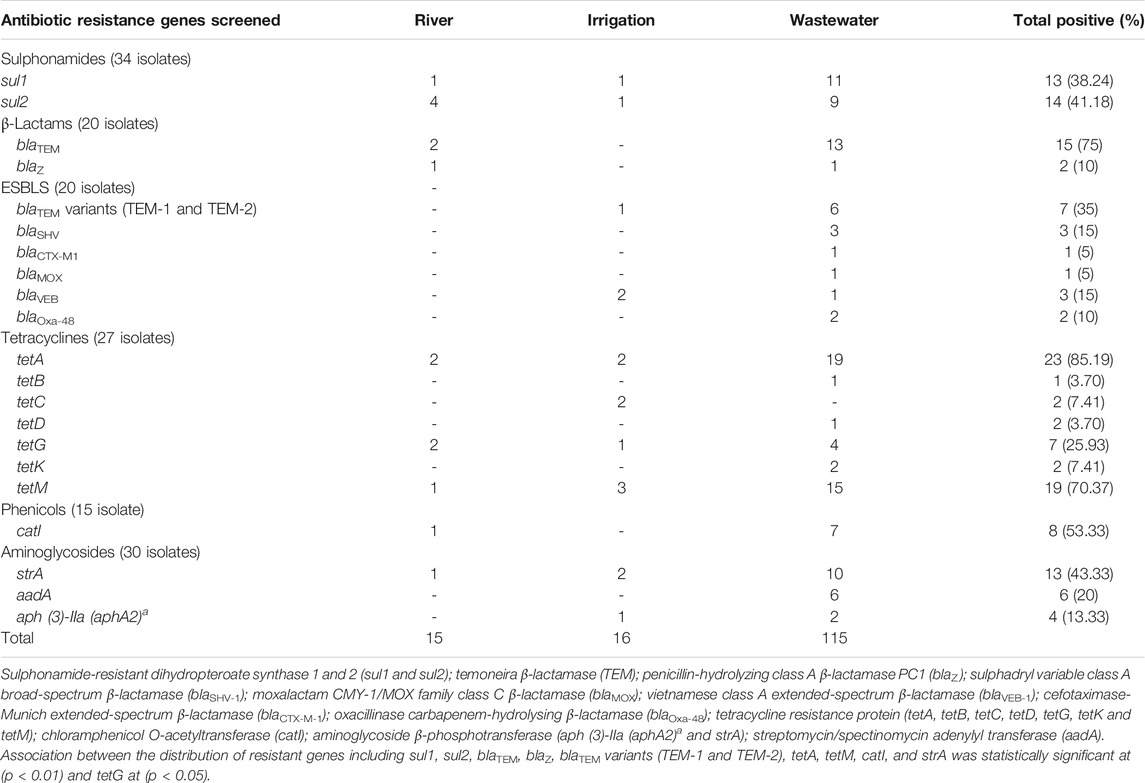
TABLE 4. Description of the occurrence of resistance genes of Listeria monocytogenes from environmental waters.
Assessment of Antibiotic Resistance Genotype Patterns of Listeria monocytogenes
The genotypic resistance profile of listeriosis agents recovered from environmental waters was presented in Supplementary Table S4. Antibiotic resistance genotypic patterns of Lm isolates exhibited 32 patterns ranging from 1–13 resistance genes (Table 5). The sul1, tetA-tetM, and the tetA-tetG-tetM genotypes occurring thrice were the frequently occurring genotype pattern observed. Generally, resistance determinants were detected in 40 (70.12%) of the environmental water isolates. Isolates recovered from wastewater harbor more dual or multiple ARGs compared with irrigation and river water.
Genetic Diversity of Listeria monocytogenes
The genetic diversity of the isolates recovered from environmental waters was studied using the ERIC-PCR assay. The classification of the genetic diversity of the isolates by the dendrogram (Figure 2) was generated from the ERIC-PCR fingerprints using the jelJ software (version 2.0). The dendrogram was clustered into 5 clades (A, B, C, D, and E) of Lm strains. Three of the Lm strains were not clustered with other strains. The clusters formed by isolates in each clade could suggest the origin of the isolates. The clonality cutoff of 50% was used for the diversity study of Lm strains.
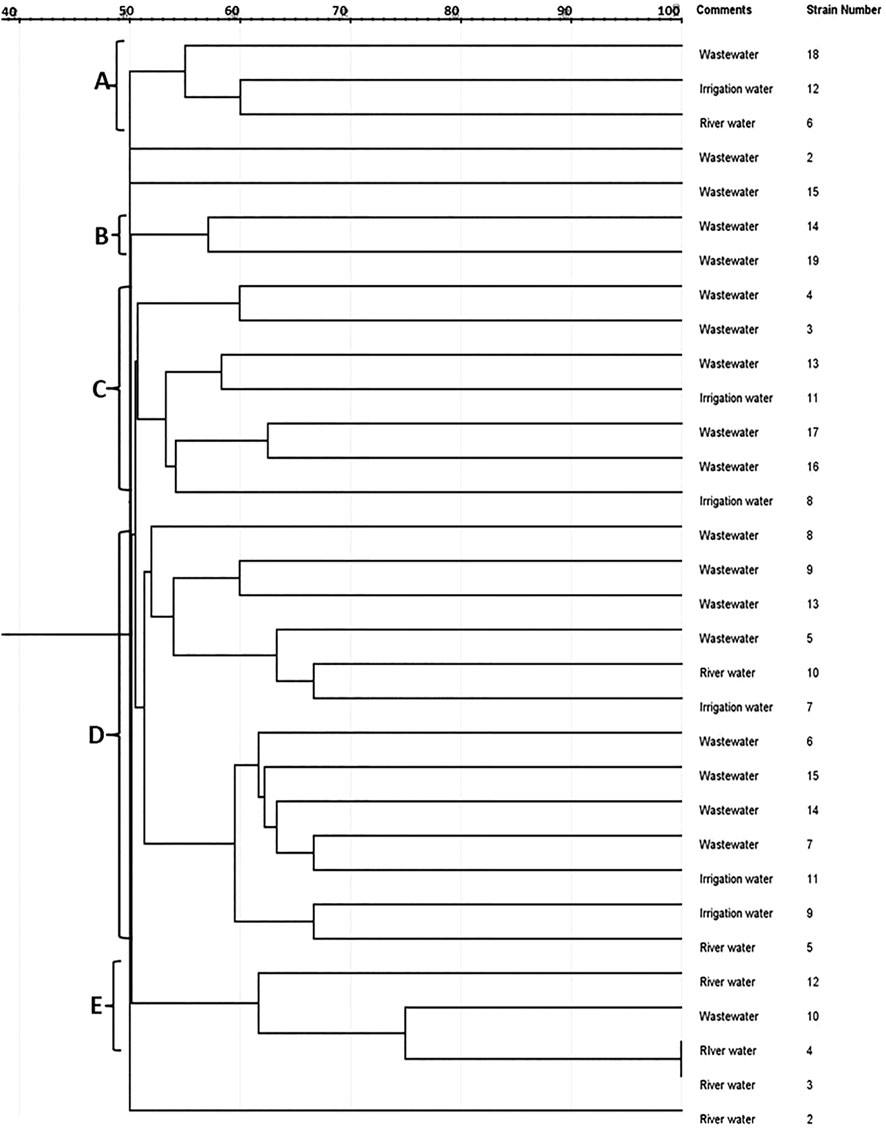
FIGURE 2. A neighbor-joining dendrogram of ERIC-PCR fingerprints of Listeria monocytogenes strains indicating similarities between clustered isolates and their sources.
Discussion
Lm is an important pathogen of medical importance known for sporadic food-borne outbreaks. In this study, we observed a wide distribution of Lm in environmental waters, and this highlights the potential health risk to humans. Although, there is currently no standard guideline regarding the benchmark for Lm counts in environmental waters in South Africa. As such, the estimated infectious dose of 102–1010 CFUs of oral exposures derived from a dose-response model that employed rhesus monkeys as a surrogate since exposure and etiology of listeriosis infection are the same in humans and primates was adopted (Smith et al., 2008). Most of the environmental waters, especially wastewaters, sampled fall short of this limit. The use of such water for irrigation purposes is not encouraged as it could increase the chance of produce contamination.
The prevalence of the pathogen was evaluated, and about 39.74% of the environmental water samples were positive for Lm. The prevalence of Lm observed in our study is lower compared to 59.4% prevalence in environmental waters reported by Mpondo et al. (2021) in a previous study in South Africa. Also, a higher prevalence of 84% free-living and 59–75% planktonic Lm recovered from the effluent of wastewater treatment plant (WWTTP) in South Africa was previously reported by Odjadjare et al. (2010). The prevalence of Lm in environmental waters documented in other regions varies. Lyautey et al. earlier reported a lower prevalence (10%) of Lm recovered from surface water in Ontario, Canada (Lyautey et al., 2007), and 13% was reported in New York (Weller et al., 2015). In Switzerland, Raschle, (2021) documented that 13% of surface water including streams, rivers, and inland canal samples contained Lm. However, a higher prevalence (31%) was reported in Mid Atlantic United States (Sharma et al., 2020). In present study, the prevalence of Lm in wastewater (66.67%) was the highest observed, while the prevalence of Lm in irrigation water (40%) and river (24.32%) was lower. Also, the distribution of confirmed Lm isolates was higher in wastewater (52.14%) compared to the river (26.32%) and irrigation water (21.05%). This observation is consistent with expectations for environmental waters not impacted with wastewater. A similar observation was reported by Dijkstra (1982) noting a higher prevalence of Lm in the effluent of WWTTP and surface waters near cities and towns compared to 21% observed in surface waters in the Netherlands. Furthermore, a previous study reported an 18.5% prevalence of Lm in river water, 83% influent, and 50% in the effluent of WWTTP in Iran (Jalali et al., 2013). The high prevalence of Lm observed in our study could be reflective of the anthropogenic impact in environmental waters.
The serotypes and virulence determinants of Lm isolates were investigated. Serotype 1/2a was the most prevalent (78.95%) compared to serotype 4b (21.05%). Our findings corroborated a similar report of Lm isolates recovered from surface water samples in Canada and Switzerland where strains were either serotype 1/2a or 4b (Lyautey et al., 2007; Raschle et al., 2021). By contrast, another study carried out on a municipal water supply watershed in Nova Scotia, Canada, found serotypes 4b/4d/4e as the most prevalent strains recovered from water (Stea et al., 2015). Our study is in support of previous studies stating that Lm serotypes usually found in various cases of human listeriosis were also found in environmental waters (Raschle et al., 2021). Notably, about 95% of Lm isolated in clinical samples belongs to serotypes 1/2a, 1/2b, 1/2c, and 4b (Doumith et al., 2004; Pontello et al., 2012). This indicates that environmental waters in the studied municipalities could constitute a potential health risk in humans.
Multiple virulence factors enhance Lm cellular invasion and survival considering its survival in the gastrointestinal tract (Raschle et al., 2021). Virulence determinants including the plcA, inlA, and inlB were detected in all environmental water isolates; the prevalence of inlC (85.97%), inlJ (80.70%), actA (59.65%), prfA (92.98%), plcB (89.47%), hly (80.70%), and mpl (73.68%) varied among the isolates. Similarly, the presence of inlA and B among all Lm isolates recovered from water in Switzerland was reported (Raschle et al., 2021). A higher rate (100%) of the internalin C and J genes were recovered from irrigation water (Iwu and Okoh, 2020). Also, a previous study reported the presence of prfA gene in all Lm isolates from water sources (Raschle et al., 2021). The prevalence of actA and plcA reported by Olaniran et al. (2015) was much lower compared to our report. However, a previous study observed a higher rate of the plcB and hly genes (Iwu and Okoh, 2020). The LIPI-1 virulence genes coordinate the major steps of intracellular infections, and the presence of these virulence genes in Lm is known to influence the virulence potential of the pathogen. For instance, cellular adhesion and internalization within the epithelial cell of the host are coordinated by the internalin genes. Metalloprotease (mpl) regulates the activation of plcB and epithelial cell infection and vacuole escape. Furthermore, the actA gene enhances the polymerization of actin and cell-to-cell spread. The hly gene ensures the release of bacterial cells into the host cytoplasm (Alvarez and Agaisse, 2016; Kayode et al., 2020). The presence of the virulence genes strongly suggests that Lm isolates from environmental waters investigated in our study possessed the potentials to cause listeriosis in humans.
Biofilm formation is known to be responsible for the resilience of Lm in food processing plants (Piercey et al., 2016). The biofilm profiles revealed that 68.42% of the isolates were potential biofilm-forming Lm. The classification as weak, medium, and strong was carried out to determine the strength of the biofilm formed. The biofilm formation by Lm in tap water was demonstrated as reported in a previous study by Gião and Keevil (2014). Also, a previous study posited that the capability of Lm isolates for biofilm formation could be attributed to the level of expression of the prfA and agr genes (Gandra et al., 2019; Grudlewska-Buda et al., 2020). In addition, Lm ability for biofilm formation is known to enhance its persistence in the environment and also contribute to higher antimicrobial resistance (Pan et al., 2006; Grudlewska-Buda et al., 2020; Bjergbæk et al., 2021). Although, Herbert-Guillou et al. (2001) put forward that biofilm is usually formed wherever there is water in the environment. However, the prevalence of Lm observed in river water was low, and it could be attributed to the turbulence flow of rivers which do not favor biofilm formation.
The increase in antibiotic-resistant Lm in environmental waters is a threat to humans’ health. Antibiotics act differently in their action against microorganisms; for instance, sulphonamides function by inhibiting folic acid synthesis and tetracyclines inhibit protein synthesis, while β-lactams inhibit the final step of bacterial cell wall synthesis (Stuart and Marshall, 2004). These antibiotics are extremely less toxic, hence their overuse for alleviating microbial infections. The binding of the aminoglycosides to bacterial ribosomes inhibits protein synthesis. Chloramphenicol inhibits the processes of protein translation during protein synthesis. It was banned because of the side effect causing aplastic anemia. The relatively high resistance level against antimicrobial agents is a true reflection of the excessive and indiscriminate usage (Lateef, 2004). Although, Lm was previously susceptible to a wide variety of antibiotics active against Gram-positive bacteria (Charpentier and Courvalin, 1999). Lm strains in this study exhibited a variable level of susceptibility to antibiotics tested. Susceptibility (100%) recorded for ampicillin is encouraging as it ascertains the therapeutic use of ampicillin, the drug of choice recommended for alleviating listeriosis infection is still effective. Susceptibilities >50% were observed to other antibiotics tested ranging from 52.63% (cefotetan) to trimethoprim-sulfamethoxazole (98.25%) except for erythromycin (26.32%), sulfamethoxazole (35.09%), streptomycin (38.60%), oxytetracycline (45.61%), and amoxicillin (49.12%). Susceptibilities to imipenem and vancomycin in our study were lower compared to the report of Manjur et al. (2018). Lm isolates showed varying percentages of resistance against all antibiotics tested except ampicillin for which no resistance was recorded similar to the report by Keet and Rip, (2021). High resistance rates against sulfamethoxazole (63.16%), oxytetracycline (54.39%), and amoxicillin (50.88%) were observed. Contrary to our report, Manjur et al. (2018) reported a high resistance of Lm from surface water in Bangladesh against ampicillin, trimethoprim-sulfamethoxazole, and penicillin and 100% resistance against erythromycin. A higher resistance against tetracycline (a class of oxytetracycline) was documented by Rodas-Suarez et al. (2006). The intermediate resistance (>15%) observed for erythromycin, clarithromycin, and amikacin could be an indication of a gradual increase of Lm resistance against the antibiotics (Supplementary Table S1). Previous studies had earlier reported varying susceptibility patterns to antibiotics tested against Lm recovered from environmental waters (Odjadjare et al., 2010; Olaniran et al., 2015; Iwu and Okoh, 2020; Mpondo et al., 2021). The varying susceptibility patterns of Lm strains in environmental waters may be dependent on antibiotic use in humans and animals and geographical variations. In this study, the distribution of phenotypically resistant Lm isolates in environmental waters was statistically significant (p < 0.01). Fifty-two patterns of antibiotic resistance were observed in our study, and 82.46% showed multidrug resistance (MDR) phenotypes against the antibiotics. Similar trends were observed among Lm isolates recovered from environmental waters in previous studies (Odjadjare et al., 2010; Iwu and Okoh, 2020; Mpondo et al., 2021). MDR in Lm reflected misuse of antibiotics and could lead to antibiotic selection pressures and encourage the emergence of resistance in Lm. Also, industrial and human activities contribute to MDR patterns because environmental waters are primary reservoirs of industrial effluents (Titilawo et al., 2015). As such, community infection due to Lm could result from the aerosolization of environmental waters, particularly wastewater. An earlier report by Fuhrimann et al. (2017) revealed that farmers using wastewater from the To Lich River in Vietnam had a higher burden of gastrointestinal disease. The ARI of river water (0.18) was less than the permissible (0.2) Krumperman threshold, whereas that of irrigation and wastewater was higher than the Krumperman threshold. A high frequency of resistance against penicillin, amoxicillin, streptomycin, cefotetan, sulfamethoxazole, oxytetracycline, and chloramphenicol was observed among Lm, particularly isolates recovered from wastewater. Coupled with the highest ARI score (0.37) observed in wastewater is an indication that wastewater could serve as a reservoir and channel for the dissemination of antibiotic-resistant Lm in the aquatic ecosystem.
The emergence of ARGs in the aquatic milieu is becoming a worldwide challenge. The antibiotic use and the acquisition of resistance genes could speed up the development of antibiotic resistance genes (ARGs) in Lm. Several ARGs encoding resistance against different varieties of antibiotics have been found in microorganisms distributed in drinking water, surface water, groundwater, sewage, wastewater, wastewater from animal husbandry, hospital effluents, and not limited to WWTTP (Zhang et al., 2009). Resistance determinants encoding resistance against such antibiotics as sulphonamides, β-lactams, phenicols, aminoglycosides, and tetracyclines were detected among the phenotypically resistant Lm isolates that were investigated. Sulphonamide resistance genes were detected in 55.88% of the isolates. The sul2 gene was more prevalent among the isolates (41.18%) compared with the sul1 gene (38.24%). The high frequency of these genes among Lm isolates in environmental water suggested a penitential misuse of the drug in the region. The β-lactamase resistance genes including blaZ, blaTEM, blaSHV, blaCTX-M1, blaMOX, blaVEB, and blaOxa-48 were detected among the resistant isolates. The blaTEM variants were the most prevalent (35%) among the β-lactamase resistance genes screened. The prevalence of β-lactamase resistance genes could be traced to their wide therapeutic use as characterized by their low toxicity. The potential health effect of the abundance of these ARGs in Lm cannot be overemphasized. Genes encoding tetracycline (tetA, B, C, D, G, K, and M) were detected, and the tetA gene (85.19%) was the most prevalent among resistant isolates. The abundance of the tetracycline ARGs among Lm isolates in environmental water could be attributed to the wide use of tetracycline antibiotics in human and animal husbandry. The catI, catII, and cmlA1 genes were screened, and only one isolate harbors catI gene. This could reflect the restriction on chloramphenicol antibiotics and the limitation on their excessive use. Furthermore, the aminoglycosides resistance genes strA (43.33%) encoding resistance against streptomycin, aadA (20%), and aph (3)-IIa (aphA2)a (13.33%) genes encoding resistance against neomycin and kanamycin were detected, respectively, among the isolates. Previous studies earlier documented the varying prevalence of ARGs among Lm isolates in environmental waters similar to the prevalence of ARGs observed in our study (Zhang et al., 2009; Iwu and Okoh, 2020; Mpondo et al., 2021). Generally, ARGs were detected in 70.12% Lm isolates recovered from environmental waters, and more worrisome is the fact that some of the isolates harbored more than 1 ARGs including both the ESBLs and other ARGs. The sul1, tetA-tetG-tetM, and the tetA-tetM genotypes occurring thrice were the frequently observed genotype pattern. This scenario observed among MDR Lm calls for serious attention as it could lead to the emergence of superbugs with severe consequences in the health sector. The statistical analysis revealed significant association between the distribution of resistant genes among Lm isolates in environmental water including sul1, sul2, blaTEM, blaZ, tetA, tetM, catI, and strA at (p < 0.01) and tetG, catI, and blaTEM variants (TEM-1 and TEM-2) at (p < 0.05). The abundance of these resistance determinants in Lm suggests that the aquatic environment, especially wastewater, may serve as a reservoir and channel for the dissemination of resistant Lm to other environmental niches including the food chain.
ERIC-PCR typing revealed high genetic diversity among Lm isolates studied. The dendrogram produced by the computer-assisted analysis generating five clades is an indication of the high genetic diversity of the environmental water isolates. Clusters formed by Lm strains from the environmental water samples showed an evolutionary relationship among the isolates. Evidence of genetic diversity among Lm strains isolated from environmental waters was also documented by Maćkiw et al. (2021). The genetic diversity observed among isolates obtained from the environmental water samples collected from different sources revealed the relevance and potential application of this typing technique in source tracking Lm, particularly during outbreak investigations.
Conclusion
This study revealed a high prevalence of resistant Lm and ARGs in environmental waters, and this could enhance the dissemination of the pathogen to other environments, including the food chain as farmers employ unconventional water sources in farming operations, especially in developing countries threatened by water scarcity. The presumptive plate counts and the high prevalence of virulence genes among Lm isolates are of epidemiological significance. This highlighted the potential health threat of Lm isolates recovered from environmental water in the catchment area. The prevalence of multidrug-resistant Lm could suggest the emergence of superbugs of clinical relevance and poses a concern to both human and animal health in the future. We, therefore, advise here that sensitization on the health implication of the indiscriminate use of antibiotics should be properly implemented. Also, wastewater should be properly treated before they are discharged into the environment to limit the rate of emergence of ARB/ARGs in environmental waters. Although, our study did not collect treated wastewater samples before discharge or at the point of discharge into receiving watershed as well as river sediments known to act as reservoir of antimicrobial-resistant bacteria. Furthermore, intense research effort should be deployed to investigate the best possible ways to alleviate the residual effects of antibiotic occurrence in the natural environment. Also, adequate regulation of antibiotic usage is necessary to ameliorate and/or reduce the burden of environmental pollution by these agents.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
Conceptualization: AO and AK; Writing — original draft preparation: AK Review and editing: AO and AK, LS, TO, OO; Statistical analysis: AK. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
TWAS-NRF and SAMRC supported the research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to the South African Medical Research Council, The World Academy of Science, and the National Research Foundation (110811 and 130765) of South Africa for financial support. Opinions and conclusions of findings in this article are those of the authors and should not be automatically credited to SAMRC or NRF-TWAS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.737435/full#supplementary-material
References
Adefisoye, M. A., and Okoh, A. I. (2016). Identification and Antimicrobial Resistance Prevalence of Pathogenic Escherichia coli Strains from Treated Wastewater Effluents in Eastern Cape, South Africa. Microbiologyopen 5, 143–151. doi:10.1002/mbo3.319
Ahmed, J., Wong, L. P., Chua, Y. P., Channa, N., Mahar, R. B., Yasmin, A., et al. (2020). Quantitative Microbial Risk Assessment of Drinking Water Quality to Predict the Risk of Waterborne Diseases in Primary-School Children. Ijerph 17, 2774. doi:10.3390/ijerph17082774
Ait-Kadi, M. (2016). Water for Development and Development for Water: Realizing the Sustainable Development Goals (SDGs) Vision. Aquat. Proced. 6, 106–110. doi:10.1016/j.aqpro.2016.06.013
Alvarez, D. E., and Agaisse, H. (2016). The Metalloprotease Mpl Supports Listeria Monocytogenes Dissemination through Resolution of Membrane Protrusions into Vacuoles. Infect. Immun. 84, 1806–1814. doi:10.1128/IAI.00130-16
Aureli, P., Fiorucci, G. C., Caroli, D., Marchiaro, G., Novara, O., Leone, L., et al. (2000). An Outbreak of Febrile Gastroenteritis Associated with Corn Contaminated byListeria Monocytogenes. N. Engl. J. Med. 342, 1236–1241. doi:10.1056/NEJM200004273421702
Basson, A., Flemming, L. A., and Chenia, H. Y. (2008). Evaluation of Adherence, Hydrophobicity, Aggregation, and Biofilm Development of Flavobacterium Johnsoniae-like Isolates. Microb. Ecol. 55, 1–14. doi:10.1007/s00248-007-9245-y
Bjergbæk, L. A., Hesselsoe, M., Pagh, S., and Roslev, P. (2021). Survival of Animal and Human-Associated Listeria Monocytogenes in Drinking Water and Biofilms. Water Supply., 1–11. doi:10.2166/ws.2021.116
Charpentier, E., and Courvalin, P. (1999). Antibiotic Resistance in Listeria Spp. Antimicrob. Agents Chemother. 43, 2103–2108. doi:10.1128/aac.43.9.2103
Chigor, V. N., Sibanda, T., and Okoh, A. I. (2013). Studies on the Bacteriological Qualities of the Buffalo River and Three Source Water Dams along its Course in the Eastern Cape Province of South Africa. Environ. Sci. Pollut. Res. 20, 4125–4136. doi:10.1007/s11356-012-1348-4
CLSI (2017). “Performance Standards for Antimicrobial Susceptibility Testing,” in CLSI Supplement M100. 27th ed. (Wayne, PA: Clinical and Laboratory Standards Institute).
Dallenne, C., da Costa, A., Decré, D., Favier, C., and Arlet, G. (2010). Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi:10.1093/jac/dkp498
Dijkstra, R. G. (1982). The Occurrence of Listeria Monocytogenes in Surface Water of Canals and Lakes, in Ditches of One Big Polder and in the Effluents and Canals of a Sewage Treatment Plant. Zentralbl. Bakteriol. Mikrobiol. Hyg. - Abt. 1 Orig. B. Hyg. 176, 202.
Dorneles, E. M. S., Santana, J. A., Ribeiro, D., Dorella, F. A., Guimarães, A. S., Moawad, M. S., et al. (2014). Evaluation of ERIC-PCR as Genotyping Method for Corynebacterium Pseudotuberculosis Isolates. PLoS One 9, e98758. doi:10.1371/journal.pone.0098758
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the Major listeria Monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi:10.1128/JCM.42.8.3819-3822.2004
Drolia, R., and Bhunia, A. K. (2019). Crossing the Intestinal Barrier via Listeria Adhesion Protein and Internalin A. Trends Microbiol. 27, 408–425. doi:10.1016/j.tim.2018.12.007
Du, X.-j., Zhang, X., Wang, X.-y., Su, Y.-l., Li, P., and Wang, S. (2017). Isolation and Characterization of Listeria Monocytogenes in Chinese Food Obtained from the central Area of China. Food Control. 74, 9–16. doi:10.1016/j.foodcont.2016.11.024
Eucast (2020). Breakpoint Tables for Interpretation of MICs for Antifungal Agents, Version 10.0, 2020. Basel, Switzerland: European Committee on Antimicrobial Susceptibility Testing. Available at: http://www.eucast.org/clinical_breakpoints/.
Fuhrimann, S., Nauta, M., Pham-Duc, P., Tram, N. T., Nguyen-Viet, H., Utzinger, J., et al. (2017). Disease burden Due to Gastrointestinal Infections Among People Living along the Major Wastewater System in Hanoi, Vietnam. Adv. Water Resour. 108, 439–449. doi:10.1016/j.advwatres.2016.12.010
Furrer, B., Candrian, U., Hoefelein, C., and Luethy, J. (1991). Detection and Identification ofListeria Monocytogenesin Cooked Sausage Products and in Milk Byin Vitroamplification of Haemolysin Gene Fragments. J. Appl. Bacteriol. 70, 372–379. doi:10.1111/j.1365-2672.1991.tb02951.x
Gandra, T. K. V., Volcan, D., Kroning, I. S., Marini, N., de Oliveira, A. C., Bastos, C. P., et al. (2019). Expression Levels of the Agr Locus and prfA Gene during Biofilm Formation by Listeria Monocytogenes on Stainless Steel and Polystyrene during 8 to 48 H of Incubation 10 to 37 °C. Int. J. Food Microbiol. 300, 1–7. doi:10.1016/j.ijfoodmicro.2019.03.021
Garrido-Maestu, A., Azinheiro, S., Fuciños, P., Carvalho, J., and Prado, M. (2020). Comparative Study of Multiplex Real-Time Recombinase Polymerase Amplification and ISO 11290-1 Methods for the Detection of Listeria Monocytogenes in Dairy Products. Food Microbiol. 92, 103570. doi:10.1016/j.fm.2020.103570
Gaul, L. K., Farag, N. H., Shim, T., Kingsley, M. A., Silk, B. J., and Hyytia-Trees, E. (2013). Hospital-acquired Listeriosis Outbreak Caused by Contaminated Diced celery-texas, 2010. Clin. Infect. Dis. 56, 20–26. doi:10.1093/cid/cis817
Gião, M. S., and Keevil, C. W. (2014). Listeria Monocytogenes Can Form Biofilms in Tap Water and Enter into the Viable but Non-cultivable State. Microb. Ecol. 67, 603–611. doi:10.1007/s00248-013-0364-3
Grudlewska-Buda, K., Skowron, K., and Gospodarek-Komkowska, E. (2020). Comparison of the Intensity of Biofilm Formation by Listeria Monocytogenes Using Classical Culture-Based Method and Digital Droplet PCR. AMB Expr. 10. doi:10.1186/s13568-020-01007-5
Hassan Omer, N. (2020). “Water Quality Parameters,” in Water Quality - Science, Assessments and Policy. London, United Kingdom: Intechopen Limited. doi:10.5772/intechopen.89657
Henriques, I. S., Fonseca, F., Alves, A., Saavedra, M. J., and Correia, A. (2006). Occurrence and Diversity of Integrons and β-lactamase Genes Among Ampicillin-Resistant Isolates from Estuarine Waters. Res. Microbiol. 157, 938–947. doi:10.1016/j.resmic.2006.09.003
Herbert-Guillou, D., Tribollet, B., and Festy, D. (2001). Influence of the Hydrodynamics on the Biofilm Formation by Mass Transport Analysis. Bioelectrochemistry 53, 119. doi:10.1016/S0302-4598(00)00121-5
Ho, J. Y., and Hendi, A. S. (2018). Recent Trends in Life Expectancy across High Income Countries: Retrospective Observational Study. BMJ 362, k2562. doi:10.1136/bmj.k2562
Igbinosa, I. H., Igbinosa, E. O., and Okoh, A. I. (2015). Detection of Antibiotic Resistance, Virulence Gene Determinants and Biofilm Formation in Aeromonas Species Isolated from Cattle. Environ. Sci. Pollut. Res. 22, 17596–17605. doi:10.1007/s11356-015-4934-4
ISO (2017). ISO 11290-1:2017 Microbiology of the Food Chain — Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria Spp. — Part 1: Detection Method. Vernier, Geneva, Switzerland: ISO.
Iwu, C. D., and Okoh, A. I. (2020). Characterization of Antibiogram Fingerprints in Listeria Monocytogenes Recovered from Irrigation Water and Agricultural Soil Samples. PLoS One 15, e0228956. doi:10.1371/journal.pone.0228956
Jacobs, L., and Chenia, H. Y. (2007). Characterization of Integrons and Tetracycline Resistance Determinants in Aeromonas Spp. Isolated from South African Aquaculture Systems. Int. J. Food Microbiol. 114, 295–306. doi:10.1016/j.ijfoodmicro.2006.09.030
Jalali, M., Taherkhani, A., Attar, H., Moazzam, M. M., and Mirzaee, S. (2013). Prevalence of Listeria Monocytogenes in the River Receiving the Effluent of Municipal Wastewater Treatment Plant. Int. J. Env Health Eng. 2, 49. doi:10.4103/2277-9183.122447
Jung, H.-J., Park, S.-H., Ha, S.-D., Lee, K.-H., Chung, D. H., Kim, C.-H., et al. (2009). Species-Specific Detection ofListeria monocytogenesUsing Polymerase Chain Reaction Assays Targeting theprfAVirulence Gene Cluster. Biosci. Biotechnol. Biochem. 73, 1412–1415. doi:10.1271/bbb.70252
Kayode, A. J., Igbinosa, E. O., and Okoh, A. I. (2020). Overview of Listeriosis in the Southern African Hemisphere-Review. J. Food Saf. 40, e12732. doi:10.1111/jfs.12732
Keet, R., Rip, D., and Rip, D. (2021). Listeria Monocytogenes Isolates from Western Cape, south africa Exhibit Resistance to Multiple Antibiotics and Contradicts Certain Global Resistance Patterns. AIMS Microbiol. 7, 40–58. doi:10.3934/microbiol.2021004
Krumperman, P. H. (1983). Multiple Antibiotic Resistance Indexing of Escherichia coli to Identify High-Risk Sources of Fecal Contamination of Foods. Appl. Environ. Microbiol. 46, 165–170. doi:10.1128/aem.46.1.165-170.1983
Lateef, A. (2004). The Microbiology of a Pharmaceutical Effluent and its Public Health Implications. World J. Microbiol. Biotechnol. 20, 167–171. doi:10.1023/B:WIBI.0000021752.29468.4e
Lee, Y. J., Han, H. S., Seong, C. N., Lee, H. Y., and Jung, J. S. (1998). Distribution of Genes Coding for Aminoglycoside Acetyltransferases in Gentamicin Resistant Bacteria Isolated from Aquatic Environment. J. Microbiol. 36 (4), 249–255.
Lyautey, E., Lapen, D. R., Wilkes, G., McCleary, K., Pagotto, F., Tyler, K., et al. (2007). Distribution and Characteristics of Listeria Monocytogenes Isolates from Surface Waters of the South Nation River Watershed, Ontario, Canada. Appl. Environ. Microbiol. 73, 5401–5410. doi:10.1128/AEM.00354-07
Maćkiw, E., Korsak, D., Kowalska, J., Felix, B., Stasiak, M., Kucharek, K., et al. (2021). Incidence and Genetic Variability of Listeria Monocytogenes Isolated from Vegetables in Poland. Int. J. Food Microbiol. 339, 109023. doi:10.1016/j.ijfoodmicro.2020.109023
Manjur, M. S. E., Siddique, S., and Ahmed, S. (2018). Multi-drug Resistant Pathogenic Listeria Monocytogenes in Surface Water and Soil Samples of Dhaka City. Bangla. J. Microbiol. 33, 39–42. doi:10.3329/bjm.v33i1.39602
Mcfarland, J. (1907). The Nephelometer:an Instrument for Estimating the Number of Bacteria in Suspensions Used for Calculating the Opsonic Index and for Vaccines. Jama XLIX, 1176. doi:10.1001/jama.1907.25320140022001f
Mohapatra, H., Mohapatra, S. S., Mantri, C. K., Colwell, R. R., and Singh, D. V. (2008). Vibrio cholerae Non-O1, Non-o139 Strains Isolated before 1992 from Varanasi, India Are Multiple Drug Resistant, Contain intSXT, Dfr18 and aadA5 Genes. Environ. Microbiol. 10, 866–873. doi:10.1111/j.1462-2920.2007.01502.x
Montero, D., Bodero, M., Riveros, G., Lapierre, L., Gaggero, A., Vidal, R. M., et al. (2015). Molecular Epidemiology and Genetic Diversity of Listeria Monocytogenes Isolates from a Wide Variety of Ready-To-Eat Foods and Their Relationship to Clinical Strains from Listeriosis Outbreaks in Chile. Front. Microbiol. 6, 1–8. doi:10.3389/fmicb.2015.00384
Mpondo, L., Ebomah, K. E., and Okoh, A. I. (2021). Multidrug-resistant listeria Species Shows Abundance in Environmental Waters of a Key District Municipality in South Africa. Ijerph 18, 481. doi:10.3390/ijerph18020481
Odjadjare, E. E. O., Obi, L. C., and Okoh, A. I. (2010). Municipal Wastewater Effluents as a Source of Listerial Pathogens in the Aquatic Milieu of the Eastern Cape Province of South Africa: A Concern of Public Health Importance. Ijerph 7, 2376–2394. doi:10.3390/ijerph7052376
Okeke, I. N., Klugman, K. P., Bhutta, Z. A., Duse, A. G., Jenkins, P., O’Brien, T. F., et al. (2005). Antimicrobial Resistance in Developing Countries. Part II: Strategies for Containment. Lancet Infect. Dis. 5 (9), 568–580. doi:10.1016/S1473-3099(05)70217-6
Olaniran, A. O., Nzimande, S. B. T., and Mkize, N. G. (2015). Antimicrobial Resistance and Virulence Signatures of Listeria and Aeromonas Species Recovered from Treated Wastewater Effluent and Receiving Surface Water in Durban, South Africa. BMC Microbiol. 15, 1–10. doi:10.1186/s12866-015-0570-x
Olanya, O. M., Hoshide, A. K., Ijabadeniyi, O. A., Ukuku, D. O., Mukhopadhyay, S., Niemira, B. A., et al. (2019). Cost Estimation of Listeriosis (Listeria Monocytogenes) Occurrence in South Africa in 2017 and its Food Safety Implications. Food Control. 102, 231–239. doi:10.1016/j.foodcont.2019.02.007
Pan, Y., Breidt, F., and Kathariou, S. (2006). Resistance of Listeria Monocytogenes Biofilms to Sanitizing Agents in a Simulated Food Processing Environment. Appl. Environ. Microbiol. 72, 7711–7717. doi:10.1128/AEM.01065-06
Pei, R., Kim, S.-C., Carlson, K. H., and Pruden, A. (2006). Effect of River Landscape on the Sediment Concentrations of Antibiotics and Corresponding Antibiotic Resistance Genes (ARG). Water Res. 40, 2427–2435. doi:10.1016/j.watres.2006.04.017
Piercey, M. J., Hingston, P. A., and Truelstrup Hansen, L. (2016). Genes Involved in Listeria Monocytogenes Biofilm Formation at a Simulated Food Processing Plant Temperature of 15°C. Int. J. Food Microbiol. 223, 63–74. doi:10.1016/j.ijfoodmicro.2016.02.009
Pontello, M., Guaita, A., Sala, G., Cipolla, M., Gattuso, A., Sonnessa, M., et al. (2012). Listeria Monocytogenes Serotypes in Human Infections (Italy, 2000-2010). Ann. Ist. Super. Sanita. 48, 146–150. doi:10.4415/ANN_12_02_07
Public Health England (2019). Detection and Enumeration of Listeria Monocytogenes and Other Listeria Species. National Infection Service Food, Water and Environmental Microbiology Methods. London: National infection service food, water and environmental microbiology methods working group. Doc. Number FNES22 Version number 4 Eff. Date 28.06.18 UNCONTROLLED. Public Health England Wellington House 133-155 Waterloo Road L 22, 1–28. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/768777/detection_and_enumeration_of_listeria_monocytogenes_and_other_listeria_species.pdf.
Quereda, J. J., Leclercq, A., Moura, A., Vales, G., Gómez-Martín, Á., García-Muñoz, Á., et al. (2020). Listeria Valentina Sp. Nov., Isolated from a Water Trough and the Faeces of Healthy Sheep. Int. J. Syst. Evol. Microbiol. 70, 5868–5879. doi:10.1099/ijsem.0.004494
Rahman, M. H., Nonaka, L., Tago, R., and Suzuki, S. (2008). Occurrence of Two Genotypes of Tetracycline (TC) Resistance Genetet(M) in the TC-Resistant Bacteria in Marine Sediments of Japan. Environ. Sci. Technol. 42, 5055–5061. doi:10.1021/es702986y
Raschle, S., Stephan, R., Stevens, M. J. A., Cernela, N., Zurfluh, K., Muchaamba, F., et al. (2021). Environmental Dissemination of Pathogenic Listeria Monocytogenes in Flowing Surface Waters in Switzerland. Sci. Rep. 11, 1–11. doi:10.1038/s41598-021-88514-y
Rodas-Suárez, O. R., Flores-Pedroche, J. F., Betancourt-Rule, J. M., Quiñones-Ramírez, E. I., and Vázquez-Salinas, C. (2006). Occurrence and Antibiotic Sensitivity of Listeria Monocytogenes Strains Isolated from Oysters, Fish, and Estuarine Water. Appl. Environ. Microbiol. 72, 7410–7412. doi:10.1128/AEM.00956-06
Sharma, M., Handy, E. T., East, C. L., Kim, S., Jiang, C., Callahan, M. T., et al. (2020). Prevalence of Salmonella and Listeria Monocytogenes in Non-traditional Irrigation Waters in the Mid-Atlantic United States Is Affected by Water Type, Season, and Recovery Method. PLoS One 15, e0229365. doi:10.1371/journal.pone.0229365
Smith, M. A., Takeuchi, K., Anderson, G., Ware, G. O., McClure, H. M., Raybourne, R. B., et al. (2008). Dose-Response Model for Listeria Monocytogenes -Induced Stillbirths in Nonhuman Primates. Infect. Immun. 76, 726–731. doi:10.1128/IAI.01366-06
Stea, E. C., Purdue, L. M., Jamieson, R. C., Yost, C. K., and Truelstrup Hansen, L. (2015). Comparison of the Prevalences and Diversities of Listeria Species and Listeria Monocytogenes in an Urban and a Rural Agricultural Watershed. Appl. Environ. Microbiol. 81, 3812–3822. doi:10.1128/AEM.00416-15
Stepanović, S., Vuković, D., Dakić, I., Savić, B., and Švabić-Vlahović, M. (2000). A Modified Microtiter-Plate Test for Quantification of Staphylococcal Biofilm Formation. J. Microbiol. Methods 40 (2), 175–179. doi:10.1016/S0167-7012(00)00122-6
Stuart, L., and Marshall, B. (2004). Antibacterial Resistance Worldwide: Causes, Challenges and Responses. Nat. Med. 10, S122.
Sweileh, W. M., Zyoud, S. e. H., Al-Jabi, S. W., Sawalha, A. F., and Shraim, N. Y. (2016). Drinking and Recreational Water-Related Diseases: A Bibliometric Analysis (1980-2015). Ann. Occup. Environ. Med. 28, 1–11. doi:10.1186/s40557-016-0128-x
Titilawo, Y., Obi, L., and Okoh, A. (2015). Antimicrobial Resistance Determinants of Escherichia coli Isolates Recovered from Some Rivers in Osun State, South-Western Nigeria: Implications for Public Health. Sci. Total Environ. 523, 82–94. doi:10.1016/j.scitotenv.2015.03.095
Wang, R.-N., Zhang, Y., Cao, Z.-H., Wang, X.-Y., Ma, B., Wu, W.-B., et al. (2019). Occurrence of Super Antibiotic Resistance Genes in the Downstream of the Yangtze River in China: Prevalence and Antibiotic Resistance Profiles. Sci. Total Environ. 651, 1946–1957. doi:10.1016/j.scitotenv.2018.10.111
Weller, D., Wiedmann, M., and Strawn, L. K. (2015). Irrigation Is Significantly Associated with an Increased Prevalence of listeria Monocytogenes in Produce Production Environments in New York State. J. Food Prot. 78, 1132–1141. doi:10.4315/0362-028X.JFP-14-584
World Health Organization (2018). INFOSAN in Action to Prevent Cases of Listeriosis Caused by Contaminated Frozen Vegetables. Available at: https://www.who.int/news-room/detail/20-07-2018-infosan-in-action-to-prevent-cases-of-listeriosis-caused-by-contaminated-frozen-vegetables (Accessed June 14, 2020).
Zhang, X.-X., Zhang, T., and Fang, H. H. P. (2009). Antibiotic Resistance Genes in Water Environment. Appl. Microbiol. Biotechnol. 82, 397–414. doi:10.1007/s00253-008-1829-z
Keywords: listeriosis, antibiotic resistance, superbugs, environmental niches, drug misuse, genetic diversity, serotypes
Citation: Kayode AJ, Semerjian L, Osaili T, Olapade O and Okoh AI (2021) Occurrence of Multidrug-Resistant Listeria monocytogenes in Environmental Waters: A Menace of Environmental and Public Health Concern. Front. Environ. Sci. 9:737435. doi: 10.3389/fenvs.2021.737435
Received: 07 July 2021; Accepted: 18 August 2021;
Published: 12 October 2021.
Edited by:
Luther King Abia Akebe, University of KwaZulu-Natal, South AfricaReviewed by:
Cornelius Ssemakalu, Vaal University of Technology, South AfricaIsaac Dennis Amoah, Durban University of Technology, South Africa
James Owusu-Kwarteng, University of Energy and Natural Resources, Ghana
Copyright © 2021 Kayode, Semerjian, Osaili, Olapade and Okoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adeoye John Kayode, MjAxODIyMjE4QHVmaC5hYy56YQ==, am9obmtheXVzMzc0OUBnbWFpbC5jb20=
 Adeoye John Kayode
Adeoye John Kayode Lucy Semerjian
Lucy Semerjian Tareq Osaili4,5
Tareq Osaili4,5 Anthony Ifeanyi Okoh
Anthony Ifeanyi Okoh