- 1Hubei Provincial Key Laboratory for Protection and Application of Special Plants in Wuling Area, College of Life Sciences, South-Central University for Nationalities, Wuhan, China
- 2Research Center for Ecology and Environment of Qinghai-Tibetan Plateau, Tibet University, Lhasa, China
- 3State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 4College of Life Science, University of Chinese Academy of Sciences, Beijing, China
- 5CAS Key Laboratory of Aquatic Botany and Watershed Ecology, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 6Hubei Key Laboratory of Wetland Evolution and Ecological Restoration, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
Microorganisms played important roles in nutrient removal in Pond-ditch circulation system (PDCS). However, dynamics of microbial community in the PDCS, and responses of rhizosphere and non-rhizosphere microbial community to rural wastewater remains unclear. In this paper, average operational taxonomic units numbers of sediment microbial varied from 10,254 to 17,112, and values in rhizosphere were higher than those of the non-rhizosphere (p < 0.05). Bacillus, Clostridium sensu stricto 1, and Geobacter were the predominant genera in PDCS sediment with relative abundances of 0.52–17.61%, 0.26–8.08%, and 0.20–4.58%, respectively. However, Bacillus, Clostridium sensu stricto 1, and Geobacter genera in rhizosphere were more abundant than those in non-rhizosphere at day 30. Chao 1 index ranged from 10,225 to 17,033 and showed significant positive correlations with all sediment properties (p < 0.05). Chao 1 and Shannon indices in rhizosphere were significant positively related to tartaric acid and total organic carbon, respectively; while significant correlation between Shannon and Simpson indices in non-rhizosphere and oxidation-reduction potential were detected (p < 0.05). Redundancy analysis suggested that lactic acids, proteins, and amino acids had strong positive effects on Geobacter and Clostridiu sensu stricto 12 in the rhizosphere; while Bacillus and Clostridium in non-rhizosphere were significantly affected by sediment ammonia nitrogen and nitrate nitrogen. Environmental variables accounted for 66.9 and 60.3% of the total variation for the microbial community of non-rhizosphere and rhizosphere sediments, respectively. Our results highlight that root exudates and sediment available N alter predominant genera in the rhizosphere and non-rhizosphere, respectively, which is benefit for optimizing removal efficiency of PDCSs in large-scale applications.
Introduction
With the rapid development of economy and industrialization, the problem of water pollution in rural areas of China is getting worse. Large amounts of untreated domestic wastewater were discharged into the ponds, ditches, lakes, and rivers, which resulted in the deterioration of water environment and threatened the health of people (Bowes et al., 2015). In order to solve this problem, many types of wastewater treatment systems have been developed and widely used, such as constructed wetland system (Childers, 2020; Torrens et al., 2021), high-rate algal ponds (Evans et al., 2005), mineral-based small-scale active filter system (Gustafsson et al., 2008), sequencing batch reactor-biofilm system (Yin et al., 2015). Recently, our group have developed a new rural wastewater treatment system, pond-ditch circulation system (PDCS), which exhibits the advantages of high efficiency, cheap, and simple management and is an appropriate alternative for rural wastewater remediation (Ma et al., 2015a; Ma et al., 2015b).
In the PDCS, microorganisms played important roles in nitrogen (N) and phosphorus (P) removal. For example, approximately 79.5% of the total nitrogen removal was attributed to microbial process, especially nitrification and denitrification, which were mediated by a range of microbes, such as amoA, arch-amoA, nirS, and nirK genes (Ma et al., 2016). Meanwhile, anaerobic anammox bacteria have been widely used in nitrogen removal reactors due to their ability of converting ammonium into nitrogen gas under the anoxic conditions (Hu et al., 2010; Liu et al., 2020). The anaerobic anammox processes also performed a crucial role in nitrogen removal of the PDCS due to 41.3–50.3% contribution to nitrogen gas production (Ma et al., 2019). Similarly, some bacteria such as denitrifying phosphate-accumulating organisms can utilize nitrate (NO3−-N) or nitrite (NO2−-N) as the terminal electron receptor under the anoxic condition for the simultaneous removal of N and P (Li et al., 2019; Chen et al., 2021). Many studies reported that structures and diversities of microorganisms were affected by a number of abiotic and biotic factors, such as pH, temperature, oxygen concentration, carbon availability, hydraulic retention time, and plants (Khammar et al., 2005; Tomaszewski et al., 2017; Li et al., 2018; Liu et al., 2020).
Our previous studies have demonstrated that in the PDCS, aquatic plants participated in nutrient removal, were responsible for 10.1% N and 50% P reduction, respectively (Ma et al., 2016; Ma et al., 2019). Besides direct absorption, plants can also regulate the nutrient removal via root exudates such as sugars, polysaccharide, amino acids, and organic acids. These exudates can be used as the carbon and nitrogen sources, or stimulating signals by the rhizosphere microorganisms (Bais et al., 2006). This will in turn lead to the gathering of more microorganisms around the plant rhizosphere and result in “rhizosphere effect” (Egamberdieva et al., 2008). This “rhizosphere effect” is beneficial for the removal of nutrient in the rhizosphere microenvironment (Nie et al., 2015; Chen et al., 2016). and can exert significant impacts on the rhizosphere microorganisms (Li et al., 2016). Therefore, the compositions, diversities of both rhizosphere and non-rhizosphere sediment bacteria vary greatly. Although several studies have correlated environmental factors to the microbial communities in decentralized treatment approaches, how rhizosphere and non-rhizosphere sediment bacteria in the PDCS responds to abiotic environmental factors still remain unclear. Based on our previous studies, we assumed that biotic factors such as plants and microbial communities rather than abiotic environmental factors were the dominant factors for nutrient removal in the PDCSs.
In this study, we have built two small-scale PDCS systems to treat domestic wastewater in rural areas of Southern China. These systems continuously operated for 2 months. This study aimed to elucidate dynamic changes of microbial community structures and diversities between rhizosphere and non-rhizosphere sediments and their responses to the environmental factors. The main contents of this study are as follows: (1) the total efficiency of N and P removal in the PDCS; (2) bacterial clustering information in rhizosphere and non-rhizosphere microenvironments; (3) relative abundances and diversities of bacteria in rhizosphere and non-rhizosphere environments at phylum and genus levels; (4) spearman’s correlation coefficient analysis between rhizosphere and non-rhizosphere microbial diversities, water quality, sediment properties, and root exudates; (5) redundancy analysis (RDA) of dominant bacterial species and environmental factors in the rhizosphere and non-rhizosphere of the PDCSs. Based on this research, we have elucidated the dynamic changes of microbial communities in the PDCSs and the relations between environmental variables and microbial communities. These findings will be helpful in the design and the optimization of nutrient removal in PDCSs and other nutrient removal systems in large-scale applications.
Materials and Methods
Experimental Water and Sediments
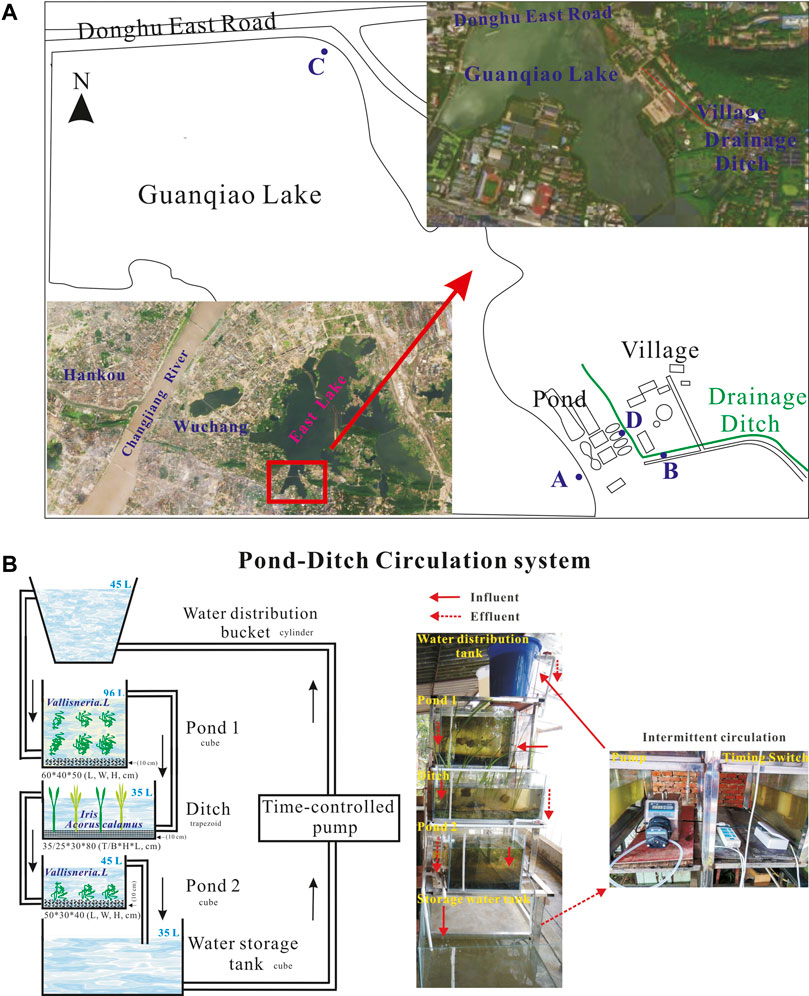
The experimental sediments and water were collected and used as we previously described (Ma et al., 2015a). Briefly, four different types of water were collected from sites A, B, C, and D, respectively; two different types of sediments were sampled from sites A and C (Figure 1A). Initial total nitrogen concentrations in these four types of water were 3.28 mg L−1, 24.22 mg L−1, 4.16 mg L−1, and 0.81 mg L−1, respectively; While initial total phosphorus contents in the overlying water of sites A, B, C, and D were 0.26 mg L−1, 2.0 mg L−1, 0.4 mg L−1, and 0.07 mg L−1, respectively. The initial physic-chemical indices of water and sediments in pond 1, ditch, pond 2, and water distribution storage apparatuses were analyzed and presented in Supplementary Tables S1, S2.
Construction of the PDCS Systems
Two sets of rural wastewater treatment system, was designed and operated as we previously described elsewhere (Ma et al., 2021b). Briefly, the small-scale system consisted of six parts: a water distribution bucket, pond 1, ditch, pond 2, a water-storage tank, and a water pump (Figure 1B). The two ponds were laid with 10 cm sediments sampled from the sites A and C and planted with Vallisneria natans L., respectively. Moreover, in the ditch, a 10 cm thick gravel layer was evenly spread and planted with Iris tectorum L. and Acorus calamus L. Then, these five microcosms were filled with 45 L, 96 L, 35 L, 45 L, and 35 L water sampled from sites D, A, B, C, and D, respectively. In the PDCS system, the running parameters was circulating every other 4 h with a water flow of 3.6 Lh-1, thus resulting in 0.30 m3m−2d−1, 0.43 m3m−2d−1, and 0.37 m3m−2d−1 hydraulic loading rates for pond 1, ditch, and pond 2, respectively. Meanwhile, in the static group, the velocity of water flow was zero.
Analysis of Water and Sediments
The system continuously ran for 60 days since the beginning of the operation. At days 1, 4, 7, 11, 15, 18, 22, 26, 30, 40, 50, and 60, 500 ml water samples were collected from pond 1, ditch, and pond 2 from the control (static) and PDCS systems, respectively. For these water samples, the 15 indices were analyzed as following: water salinity, dissolved oxygen (DO), pH, temperature (W-temp), oxidation-reduction potential (ORP), turbidity, total phosphorus (TP), total nitrogen (TN), inorganic nitrogen (IP), ammonia nitrogen (NH4+-N), nitrate nitrogen (NO3−-N), nitrite nitrogen (NO2−-N), total suspended solids (TSS) (State EPA of China, 2002), chemical oxygen demand (CODcr) and Chl-a (Holm-Hansen and Riemann, 1978).
Approximate 50 g rhizosphere and non-rhizosphere sediments in pond 1, ditch, and pond 2 of these two systems were also collected at days 15, 30, and 60, representing the early, middle, and late stages of the experiment, respectively (Zou et al., 2021). About 10 g of fresh sediment samples was used to measure water contents (WC) of each sediment sample gravimetrically. The contents of organic matter were determined gravimetrically using approximate 5 g fresh sediment sample dried at 450°C for 3 h. The contents of total organic carbon (TOC) and sediment total nitrogen (STN) in air-dried and sieved sediment samples were determined using an elemental analyzer (Vario TOC cube, Hanau, Germany). The contents of NH4+-N, NO3−-N, NO2−-N in sediments were determined using an automatic nutrient analyzer (EasyChem plus, Systea, Italy). Sediment P fractions were measured using the SMT harmonized P extraction protocol, and the four main forms of P in sediment total phosphorus (STP), sediment inorganic phosphorus (SIP), NaOH-P, and HCl-P were analyzed as previously described (Ruban et al., 2001). The left sediments were mixed thoroughly and frozen in −20°C for future analysis. The dynamic changes of physic-chemical characteristics for the non-rhizosphere sediments for the PDCS systems were shown in Supplementary Tables S3, S4, respectively.
Analysis of Root Exudates
The contents of five types of root exudates proteins, polysaccharides, amino acids, lactic acids and tartaric acids were measured (Ma et al., 2021a). Briefly, 10 strains of Vallisneria natas L., 1 strain of Acorus calamus L. and Iris tectorum L. were sampled from the ponds and ditches at days 15, 30, and 60, respectively. However, in the pond 1 of the static group, all Vallisneria natans L. died due to the deterioration of water quality in the early stage of the experiment. Therefore, root exudates for this microcosm were missed. After careful rinsing in the water, plant roots were immersed in 500 ml ultra-pure water for 24 h. The values of pH for these solutions were measured. Then, 400 ml culture solution was concentrated to 20 ml using a vacuum circumgyration evaporator. The contents of proteins and polysaccharides were analyzed using the coomassie brilliant blue method (Spector, 1978) and the anthrone-sulfuric acid chromatometry method (Grandy et al., 2000), respectively. The contents of amino acids were determined by an ICS 5000 + amino acid analyzer (model 120A, Thermo Fisher, United States). Meanwhile, the contents of tartaric acids in root exudates were measured using a DIONEX ICS-5000 + DP chromatograph (Thermo Fisher, United States). The initial and changes of proteins, polysaccharides, amino acids, lactic acids, tartaric acids, and pH of root exudates of aquatic plants were presented in Supplementary Tables S5, S4, respectively.
Construction of 16S rDNA Library
Bacterial DNA were extracted and used for 16S rDNA gene sequencing as previously described with some modifications (Poret-Peterson et al., 2019). Briefly, the total DNA of sediment samples was extracted using the PowerSoil DNA Isolation Kit according the manufacturer’s instructions (MoBio, Carlsbad, CA, United States). Then, the concentrations and qualities of DNA samples were spectrophotometrically measured (Nanodrop 2000, NanoDrop Technology, Wilmington, DE, United States). The hypervariable V3-V4 regions of bacterial 16S rDNA were amplified using the specific bar-coded forward primer 5’-ACTCCTACGGGAGGCAGCAG-3’ and reverse primer 518R, 5’-ATTACCGCGGCTGCTGG-3’. Polymerase chain reaction (PCR) amplification procedures were as follows: denaturation at 95°C for 3 min, denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s, 35 cycles. Following the pre-amplification, the qualified DNA samples were used in the next PCR amplification. Then, the amplified PCR products were purified and used to construct the 16S rDNA library.
Sequencing of 16S rDNA Gene and Data Analysis
Sequencing of PCR products were performed using the Illumina paired-end sequencing technology (Mega Genomics Corp. Beijing, China). Briefly, the raw data of bacterial 16S rDNA gene sequencing were demultiplexed, qualify-filtered by Trimmomatic using the specific filtering criteria of QIIME (Caporaso et al., 2010) and merged using the FLASH software. Chimeric sequences were determined and deleted using the UCHIME algorithm (Edgar et al., 2011). The operational taxonomic units (OTUs) were clustered with a threshold of 97% sequence similarity using UPARSE (version 7.1, http://drive5.com/uparse/), annotated and classified using the SILVA database. Sediment bacterial α-diversity was estimated via the observed chao1, Shannon, and Simpson indices.
Statistical Analysis
All data were presented as mean ± SEM. One-way ANOVA with Turkey’s post hoc tests were performed to analyze the differences in water quality, sediment physic-chemical parameters, α-diversity and richness indices of sediment bacteria, root exudates in these two systems. Sediment bacterial community richness (presented as observed species and goods coverage) and α-diversity (presented as Chao 1, Simpson, and Shannon) in rhizosphere and non-rhizosphere microenvironments in these two systems were calculated in R version 3.5.2 using the vegan package (Zhou et al., 2020). Spearman’s correlation coefficients were calculated to determine the associations among sediment microbial diversity, water quality, sediment properties, and root exudates. RDA was performed to determine the correlations between the top 10 sediments bacterial and abiotic environmental variables using the Canoco software (Version 5.0, Microcomputer, Ithaca, NY, United States). The RDA will be picked up and used as the appropriate ordination form when the longest gradient length of detrended correspondence analysis was <2. All statistical analyses were done in SPSS software (version 20.0, SPSS Inc., Chicago, United States). p < 0.05 was considered statistically significant.
Results
Nutrient Removal Rates in PDCS Systems
The efficiencies of N and P removal in the two systems were demonstrated in Figure 2. In the PDCS, the concentrations of TN, NH4+-N, and NO2−-N in the three microcosms pond 1, ditch, and pond 2 declined from day 4 and reached a plateau level at day 30. Contents of NO3−-N in the PDCS remained at a low level before day 20, and then significantly increased, peaking at day 40, and gradually declined to a low level. The final removal efficiencies for TN and NH4+-N in the PDCS system ranged from 72.7 ± 2.3 to 94.5 ± 1.5%, and 72.3 ± 2.6 to 99.1 ± 0.3%, respectively. For the P, in the PDCS, the concentrations of TP and IP in the three microcosm’s pond 1, ditch, and pond 2 significantly declined from day 4 and reached a stable low level at day 30. The final removal efficiencies of TP in pond 1, ditch, and pond 2 in the PDCS system were 81.7 ± 2.3, 97.4 ± 1.1, and 77.8 ± 2.1%, respectively. Significant differences were detected in TN, NH4+-N, NO3−-N, NO2−-N, TP, and IP between these two systems (p < 0.05).
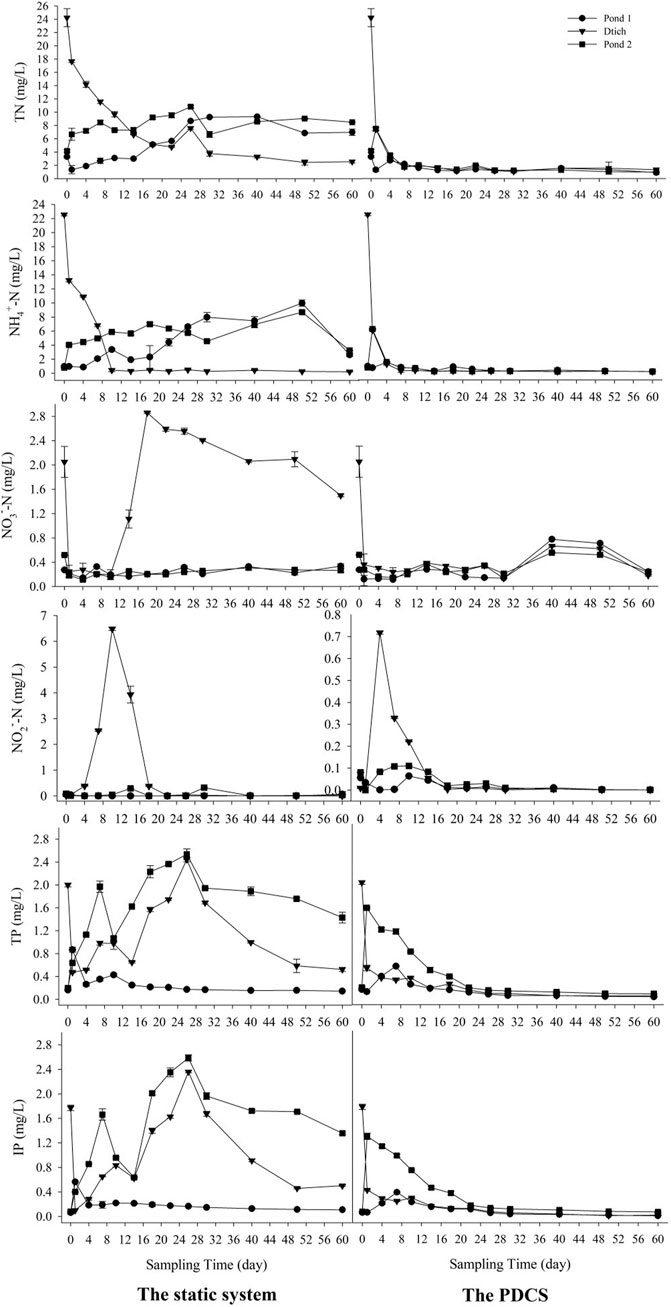
FIGURE 2. Changes of TN, NH4+-N, NO3−-N, NO2−-N, TP, and IP in overlying water for two systems within 60 sampling days. In the circulation PDCS system, the water was circulated 3.6 L/h every other 4 h, while the control group was static.
The values of the other six indices ORP, W-temp, pH, DO, turbidity, and salinity for water were presented in Supplementary Figure S1. The ORPs for the two systems increased from day 1 to day 15, and then decreased. The W-temp for these two systems fluctuated from 25.5 ± 0.3°C to 32.6 ± 0.2°C.The values of pH generally increased from 7.16 ± 0.13 from day 0 to 8.19 ± 0.15 at day 60. The values of DO also increased from 0.05 ± 0.01 to 5.4 ± 0.1 mg L−1. For the turbidity, their values fluctuated two times and generally decreased. For the salinity, the change trend for the static system were increasing, the values in the PDCS remained at a stable level. Significant differences in pH, DO, salinity, and turbidity were observed between these two systems (p < 0.05).
OTU Analysis in Single Microcosms of the PDCS System
Both rhizosphere and non-rhizosphere sediments were sampled for analyzing the compositions of bacteria in two systems at days 15, 30, and 60, respectively. After quality control and filtering, twenty-four OTU numbers for eight microcosms were obtained (Figure 3A). Due to the death of plants in the pond 1 of the static system and the existence of two different types of plants in the ditches of the two systems, there were 452,989 ± 3,426 bacterial OTUs observed in the 36 rhizosphere/non-rhizosphere sediment samples. The OTU numbers in the S-ditch, S-pond 2, ditch, and pond 2 microcosms were all significantly lower than those of their corresponding rhizosphere counterparts, respectively (p < 0.05). Meanwhile, the OTU numbers in the microcosm of R-pond 1 was statistically insignificantly higher compared with that in the microcosm of pond 1 (p < 0.05). In order to show the unique and shared OTUs between the static and PDCS systems, a petal diagram was drawn based on the OTU analysis results (Figure 3B). The ten petals represented ten different microcosms in these two systems. There were 1,360 OTUs shared by the 10 microcosms. The numbers of unique OTUs in the S-ditch, ditch, and pond 2 microcosms were significantly lower than those of their corresponding counterparts (p < 0.05).
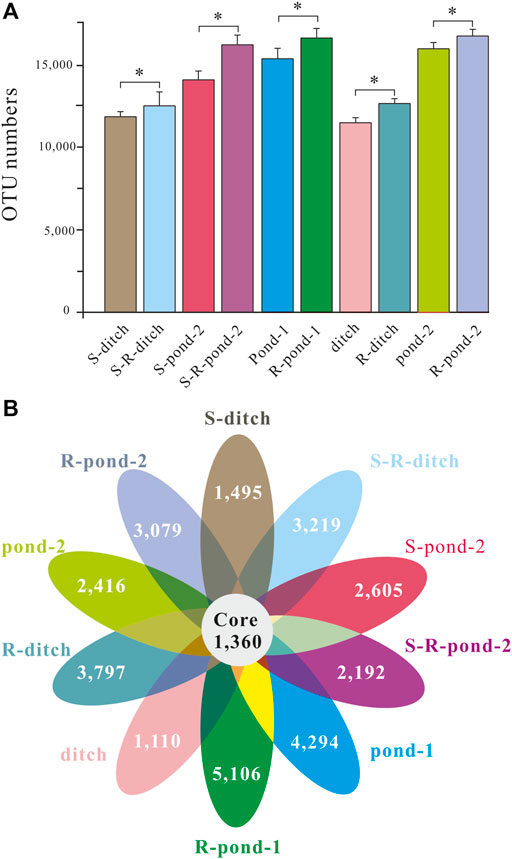
FIGURE 3. OTU numbers and Venn diagram for two systems. (A) OTU numbers in the pond-1, R-pond-1, ditch, R-ditch, pond-2, R-pond-2 of the PDCS and S-ditch, S-R-ditch, S-pond-2, S-R-pond-2 of the control static group, respectively. Due to the death of plants in the pond-1 of the control static group died at the early stage, the data for the S-pond-1 and S-Rpond-1 were omitted. (B) Each circle represents a single microcosm of the PDCS systems. The numbers in the overlapping area of the circles represent the numbers of OTUs shared between the microcosms, while the numbers without overlaps represent the numbers of OTUs unique to these single microcosms.
Heatmap of the Relative Abundances of Bacterial Communities
Relative abundances of top 35 bacteria at phylum and genus levels were statistically analyzed (Figure 4). The utmost dominant bacterial phyla in pond 2 of the two systems were Firmicutes, Deinococcus, and Armatimonadetes. The frequencies of these three bacterial phyla in the rhizosphere of pond 2 were more abundant in those of the non-rhizosphere. For the ditch of two systems, the dominant bacterial phyla were Bacteroidetes and Proteobacteria, their values in the rhizosphere were significantly higher than those of their corresponding non-rhizosphere groups. In addition, the frequencies of Nitrospirae, and Acidobacteria in the rhizosphere of pond 1 for two systems were significantly higher than those of their corresponding non-rhizosphere groups. At the genus level, Geobacter and Pseudomanas genera were in relatively higher abundances in the rhizosphere of pond 1 for the PDCS than those in the non-rhizosphere. The frequencies of Clostridium_sensu stricto 1, Romboutsia, and Clostridium_sensu stricto 13 genera in the rhizosphere of pond 1 for two systems were more abundant in those in non-rhizosphere. For the pond 2 of the PDCS, the dominant bacterial phyla were Methenoregula and Methanosaeta, their values in the rhizosphere were significantly higher than those of their corresponding non-rhizosphere groups.
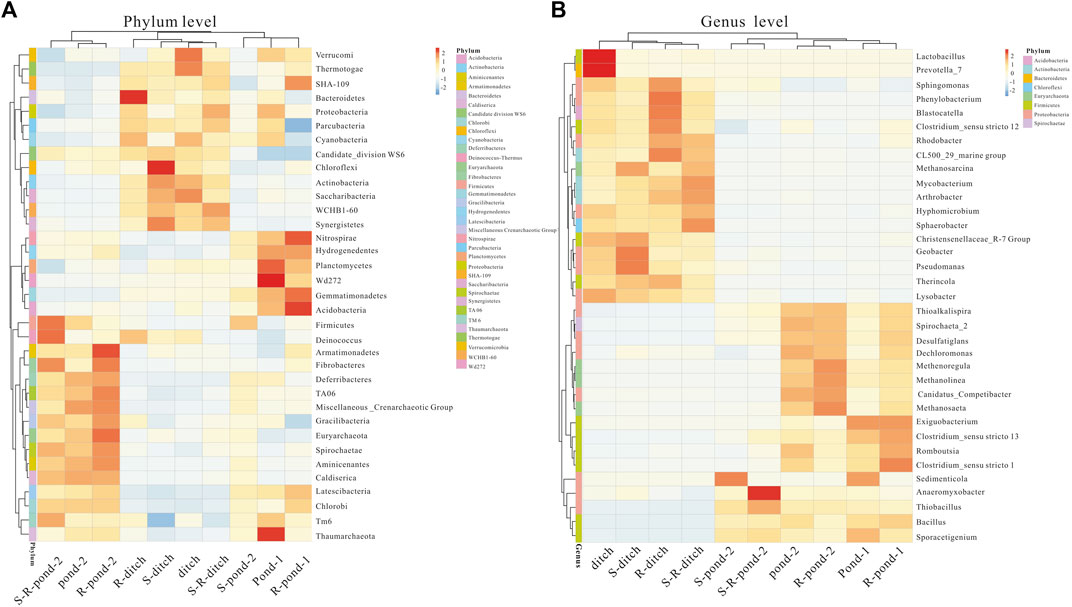
FIGURE 4. Relative abundance heatmap of top 35 bacteria in rhizosphere and non-rhizosphere sediments in two systems at the phylum (A) and genus (B) levels.
Bacterial Community Structure Analysis of Rhizosphere/Non-Rhizosphere Sediment
Relative abundances of top 10 bacteria in rhizosphere and non-rhizosphere sediment samples at days 15, 30, and 60 at phylum and genus levels were analyzed and demonstrated in Supplementary Figure S2 and Figure 5, respectively. At the phylum level, the most abundant bacteria in both rhizosphere and non-rhizosphere sediments of the PDCS were Proteobacteria, Firmicutes, Chloroflexi, Bacteroidetes, and Actinobacteria. Relative abundances for these five bacterial phyla in the rhizosphere of the PDCS ranged from 25.52 ± 2.68 to 53.44 ± 2.56%, 3.62 ± 0.37 to 45.2 ± 2.15%, 5.97 ± 0.67 to 15.06 ± 1.03%, 2.69 ± 0.28 to 9.01 ± 0.98%, and 1.92 ± 0.35 to 11.74 ± 0.51%, respectively. Meanwhile, relative abundances of these five bacterial phyla in the non-rhizosphere ranged from 20.76 ± 1.36 to 46.88 ± 1.57%, 2.12 ± 0.26 to 25.75 ± 1.18%, 6.63 ± 0.34 to 14.59 ± 0.43%, 2.32 ± 0.10 to 7.12 ± 0.23%, and 2.07 ± 0.16 to 14.26 ± 0.24%, respectively. Relative abundances of Firmicutes in two ponds of two systems increased from day 15 to day 30. At the middle of experiment (day 30), Firmicutes was found to be more enriched in the rhizosphere of ponds for two systems than those in the non-rhizosphere of two ponds. Meanwhile, the phylum Proteobacteria was more abundant in the rhizosphere of the ditch for two systems than those in the non-rhizosphere of the ditch at day 30.
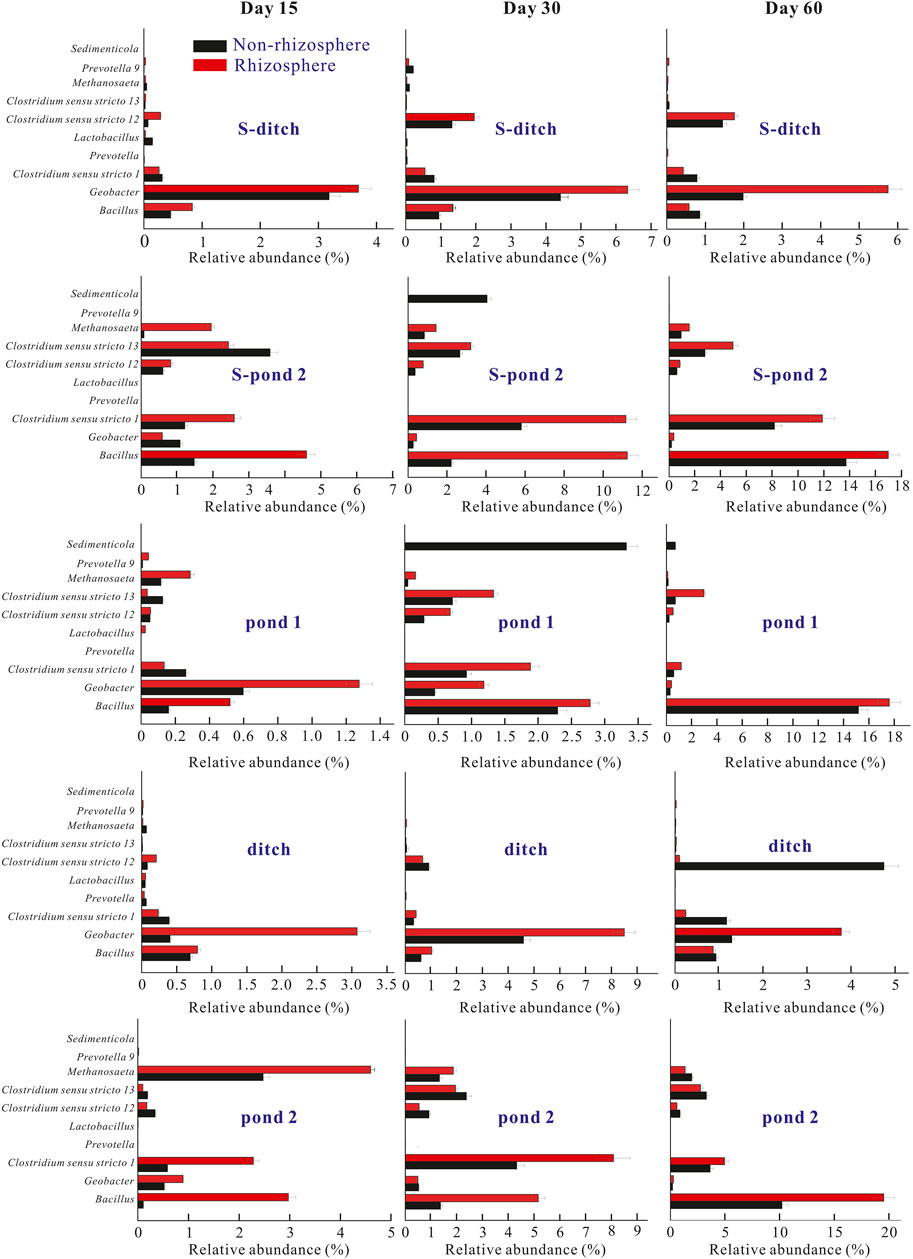
FIGURE 5. Relative abundances of top 10 bacteria at the genus level in the two systems. The relative abundances of top 10 bacteria at the genus level at days 15, 30, and 60 were calculated. Abbreviations: In the static systems, S-ditch, S-R-ditch, S-pond-2, and S-R-pond-2 mean the sediments collected from the ditch, plant rhizosphere in the ditch, pond 2, plant rhizosphere in pond 2, respectively. The plants in the pond 1 of the static system died at the early stage of the experiment. In the PDCS, Pond-1, R-pond-1, ditch, R-ditch, pond-2, and R-pond-2 represented the sediments collected from the pond 1, plant rhizosphere of the pond 1, the ditch, plant rhizosphere in the ditch, pond 2, plant rhizosphere in the pond 2, respectively.
At the genus level, the most abundant bacteria in both rhizosphere and non-rhizosphere sediments of the PDCS were Bacillus, Geobacter, and Clostridium sensu stricto 1 genera. The relative abundances of these three bacterial genera in the rhizosphere of the PDCS ranged from 0.56 ± 0.02 to 19.52 ± 1.95%, 0.28 ± 0.03 to 8.50 ± 2.13%, and 0.13 ± 0.02 to 11.89 ± 0.36%, respectively. Meanwhile, the relative abundances of these three bacterial genera in the non-rhizosphere ranged from 0.12 ± 0.01 to 15.61 ± 0.68%, 0.20 ± 0.02 to 4.58 ± 0.32%, and 0.26 ± 0.01 to 8.08 ± 1.35%, respectively. Dominant genera in ditches of two systems were Geobacter. Relative abundances of ditches for two systems increased from day 15 to day 30, and then decreased (Figure 5). Meanwhile, the frequencies of Geobacter in genus Proteobacteria in the rhizosphere sediments of these two ditches at day 30 were significantly higher than those of their non-rhizosphere sediments (p < 0.05). Moreover, in the microcosms pond 1 and pond 2, the most dominant bacterial genera were Bacillus and Clostridium sensus stricto 1, and relative abundances of these two genera in two ponds of the PDCS remarkably increased from day 15 to 30. The frequencies of Bacillus and Clostridium sensus stricto 1 in the rhizosphere of ponds for the PDCS were higher than those in the non-rhizosphere (Figure 5).
Richness and α-Diversity Analysis of Rhizosphere and Non-Rhizosphere Sediment Bacteria
To further explore the effects of root exudates on bacterial community in the PDCS, the richness (presented as observed species, goods coverage) and α-diversity (presented as Chao1, Shannon and Simpson) of rhizosphere and non-rhizosphere sediment bacteria were performed. As shown in Figure 6, the values for the observed species in the rhizosphere and non-rhizosphere sediments in the PDCS ranged from 9,560 ± 1,235 to 12,500 ± 1,230 and 9,080 ± 1,254 to 12,100 ± 1,035, respectively. The numbers of observed species in the rhizosphere groups S-R-ditch, S-R-pond-2, R-ditch, and R-pond-2 were significantly higher than those of their corresponding non-rhizosphere groups S-ditch, S-pond-2, ditch, and pond-2, respectively. Meanwhile, the values for goods coverage in the rhizosphere and non-rhizosphere sediments in the PDCS ranged from 0.90 to 0.94 and 0.9 to 0.94, respectively. Moreover, Chao 1, Shannon, and Simpson indices in the PDCS ranged from 11,600 ± 1,396 to 16,400 ± 1,543, 11.3 ± 0.3 to 12.0 ± 0.3, and 0.9971 ± 0.0021 to 0.9979 ± 0.0003 in the rhizosphere sediments and ranged from 11,300 ± 286 to 15,900 ± 879, 11.1 ± 0.2 to 12.0 ± 0.2, and 0.9970 ± 0.0023 to 0.9977 ± 0.0002 in the non-rhizosphere sediment, respectively. Chao1 values in the five rhizosphere groups S-R-ditch, S-R-pond-2, R-pond-1, R-ditch, and R-pond-2 were all significantly higher than their corresponding non-rhizosphere groups S-ditch, S-pond-2, pond-1, ditch, and pond-2, respectively (p < 0.05). Similarly, the values of Shannon in the S-R-ditch, R-ditch and R-pond-2 groups were significantly higher than those of their corresponding non-rhizosphere groups S-ditch, ditch, and pond-2, respectively (p < 0.05). Moreover, Simpson values in the S-R-ditch, R-pond-1, and R-pond-2 were significantly higher than those of their corresponding non-rhizosphere groups S-ditch, pond-1, and pond-2, respectively (p < 0.05).
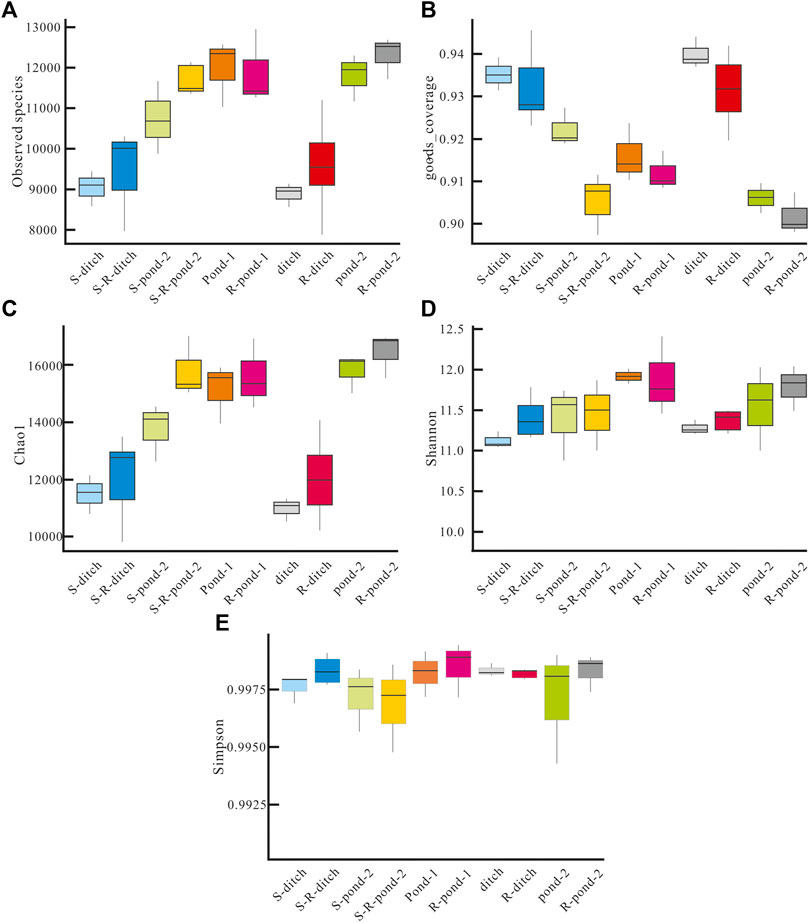
FIGURE 6. Richness and diversity of rhizosphere and non-rhizosphere sediment bacteria in two systems. (A) Numbers of observed species, (B) the values of goods coverage, (C) the Chao1 index, (D) the Shannon index, and (E) the Simpson index of non-rhizosphere and rhizosphere sediment bacteria in different microcosms of two systems. S-ditch and S-R-ditch meant non-rhizosphere and rhizosphere ditch groups in the static system, respectively; Similarly, S-pond-2, and S-R-pond-2 meant non-rhizosphere and rhizosphere pond-2 groups in the static system, respectively. The same naming rules were applicable in pond-1, R-pond-1, ditch, R-ditch, pond-2, and R-pond-2 in the PDCS.
Spearman’s correlation coefficients between microbial diversity indices and water quality were presented in Table 1. In the rhizosphere, Chao 1 index showed significant and positive correlations with NH4+-N and turbidity (p < 0.05). Meanwhile, turbidity was significantly positively correlated with the Shannon index (p < 0.01). In the non-rhizosphere, significant positive correlations between the ORP and the Shannon and Simpson indices were observed (p < 0.05); while Simpson index was significantly negatively correlated with W-temp, and was significantly positively associated with salinity (p < 0.05).
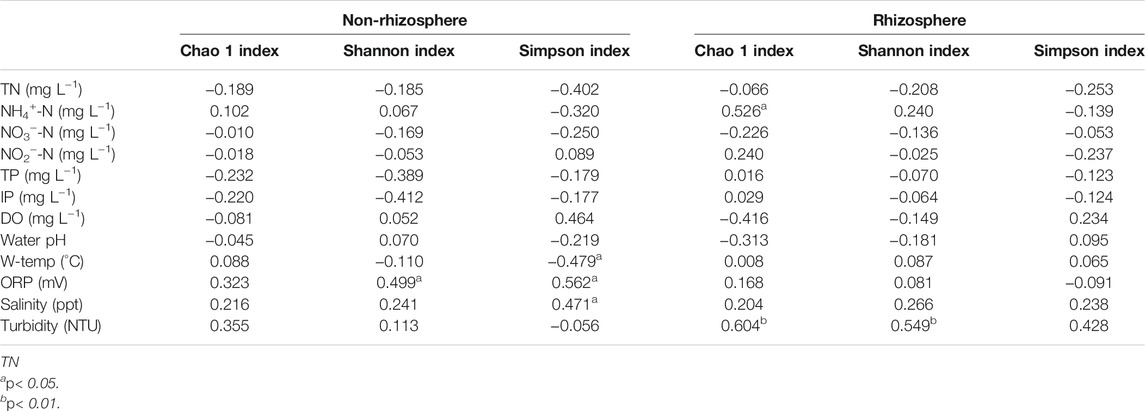
TABLE 1. Spearman’s correlation coefficients between sediment microbial diversity and water quality.
Spearman’s correlation coefficients between microbial diversity indices, sediment properties, and root exudates were demonstrated in Table 2. In the rhizosphere, Chao 1 index showed significant correlation with sediment total nitrogen (STN), sediment ammonia nitrogen (SNH4+-N), sediment nitrate nitrogen (SNO3−-N), STP, SIP, OM, TOC, WC, and tartaric acids respectively (p < 0.01). Meanwhile, TOC was significantly positively related to Shannon index (p < 0.05). Similarly, in the non-rhizosphere sediments, Chao 1 index showed significant correlations with all measured sediment properties (p < 0.05).
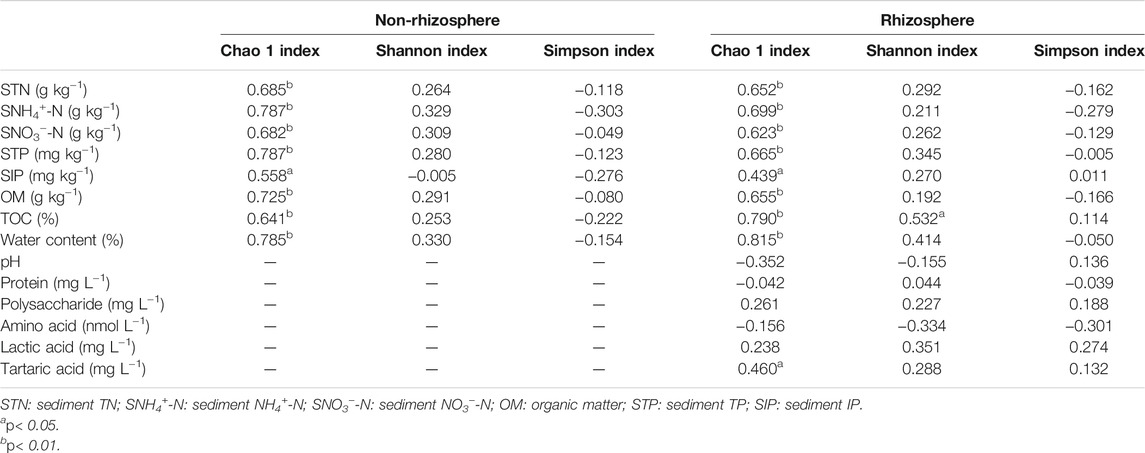
TABLE 2. Spearman’s correlation coefficients between microbial diversity indices, sediment properties and root exudates.
Relationship Between Sediment Bacteria and Environmental Factors
As demonstrated in Figure 7A, the first two eigenvalues could explain 48.97 and 13.32% of the total variation at the genus level in the non-rhizosphere sediments, respectively. The environmental variables accounted for 66.9% of the total variation for the microbial community of non-rhizosphere sediments. In the non-rhizosphere sediment groups, SNH4+-N and SNO3−-N exhibited significant positive effects on the dominant bacterial genera Clostridium sensu stricto1, Clostridium sensu stricto 13, and significant negative effects on Arthrobacter and Geobacter. Moreover, pH, DO, and ORP had strong positive effects on Clostridium sensu stricto 12 and Bacillus.
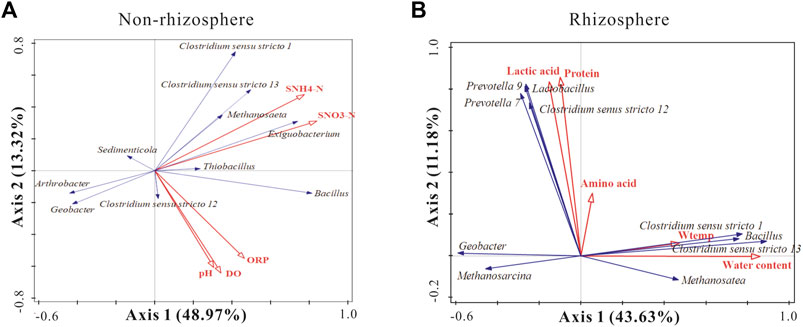
FIGURE 7. RDA analysis of species composition of sediment bacterial taxa associated with significant environmental factors. The blue and red arrows indicate top 10 functional bacteria genus and significant environmental factors in the non-rhizosphere sediments (A) and the rhizosphere sediments (B).
As demonstrated in Figure 7B, the first two eigenvalues could explain 43.63 and 11.18% of the total variation at the genus level in the rhizosphere sediments, respectively. Environmental variables accounted for 60.3% of the total variation for the microbial community of rhizosphere sediments. In the rhizosphere, lactic acids, proteins, and amino acids showed significant positive effects on Lactobacillus, Geobacter and Clostridiu sensu stricto 12. Moreover, Bacillus, Clostridium sensu stricto 1, and Clostridium sensu stricto 13 were significantly positively affected by W-temp and WC.
Discussion
The Role of Bacteria in Nutrient Removal of the PDCS
In the present study, the averaged final TN and TP removal efficiencies of the PDCS at day 60 were mostly higher than that in IVCWs (51.6%) (Chang et al., 2015), in line with the mean value in algal pond combined with constructed wetlands (69.74%) (Zhao et al., 2016), yet lower than that in SFCWs (96.14%) (Li et al., 2020), suggesting the PDCS system was a valuable choice for rural wastewater remediation. Our previous study have demonstrated that microorganisms play important role in N and P removal in the PDCS (Ma et al., 2019). The removal of N was mainly contributed to simultaneous nitrification, anaerobic ammonium oxidation, and denitrification processes via the nitrogen-related bacteria (Zhao et al., 2019). In this paper, Bacillus was one of the three predominant genera in ponds and the ditch for the two systems, respectively. This finding was in line with the previous study reported that Bacillus was one of the six predominant bacterial genera for heterotrophic nitrification and aerobic denitrification in aquatic ecosystems (Qiao et al., 2020). Previous studies reported that Bacillus. sp. participated in aerobic denitrification in the PDCS, and Clostridium sp. was used for anaerobic ammonium-oxidizing in the sludge fermentation reactor (Yang et al., 2011; Ma et al., 2021b). A earlier study reported that rhizosphere bacteria played a pivotal role in regulating the P transformation and can utilize various P forms in wetland plants (Teng et al., 2018). Geobacter and Bacillus. sp. were known to act as polyphosphate-accumulating organisms, showing a strong capacity of poly-P accumulation and P storage (Schelfhout et al., 2015; Wang et al., 2019).
Comparison of Microbial Community Structure and Diversity Between Rhizosphere and Non-Rhizosphere Bacteria
The “rhizosphere effect” can lead to greatly variations in compositions and diversities of rhizosphere and non-rhizosphere sediment bacteria, where the impacts were influenced by seasons (Dewedar et al., 2009), soil types (Zhao et al., 2017), and plant species (Yin et al., 2018). The OTU numbers in the rhizosphere sediments of five microcosms were all higher than those of their counterparts in the non-rhizosphere, suggesting the presence of more bacteria and more complex microbial communities in the rhizosphere sediments. Moreover, α-diversities of rhizosphere groups S-R-ditch, S-R-pond-2, R-pond-1, R-ditch, and R-pond-2 were all significantly higher than their corresponding counterparts (p < 0.05). These results indicated that compared with the non-rhizosphere, both the richnesses and diversities of microorganisms in the rhizosphere were enhanced due to the exudates from the plant roots (Stringlis et al., 2018). These effects will indirectly promote the roles of plants in nutrient removal in the PDCS. This was in line with the previous study in wetland plants which reported that “rhizosphere effects” affected the removal efficiency of nutrient via regulating the density and diversity of the rhizosphere microbes (Chen et al., 2016). Similarly, a recent study have reported that “rhizosphere effects” can result in the increase of relative abundances of some beneficial bacteria such as Streptomycetaceae and Bacillaceae, which exhibited significantly positive correlations with the uptake of N (Wu et al., 2021).
Both non-rhizosphere and rhizosphere groups shared five predominant bacterial genera, Bacillus, Geobacter, Clostridium sensu stricto 1, Clostridium sensu stricto 12, and Clostridium sensu stricto 13. These findings were consistent with the previous research in tidal marsh soil reported that relative abundances of Geobacter, Bacillus, Clostridium and Shewanella in the rhizosphere were higher than those in the bulk soil (Luo et al., 2018). However, at day 60, the relative abundance of Geobacter in the ditch of the PDCS was richer than those of two ponds; while frequencies of Bacillus and Clostridium sensu stricto 1 in two ponds of the PDCS at day 60 were significantly higher than those in the ditch. This was mainly due to that differences in environmental factors such as soil types (Bakker et al., 2015) and plant species (Yin et al., 2018), resulting in varying root exudates composition and contents (Ma et al., 2021b), thereby affected the community structure of bacteria. Moreover, the frequencies of Bacillus and Clostridium sensus stricto 1, and Geobacter in the rhizosphere of ponds and the ditch for two systems at day 30 were higher than those in the non-rhizosphere (Figure 5). This was consistent with our results that protein and amino acids contents of Vallisneria natans L. in PDCS’s two ponds increased from day 15 to 30 (Supplementary Table S5). This suggested that plants could adjust the secretion to cope with abiotic environmental stress (Edayilam et al., 2018). Relative higher proteins and amino acids values were conductive to increment of rhizosphere bacteria, which would be helpful in promoting the removal of nutrients (Bakker et al., 2015).
Responses of Rhizosphere and Non-Rhizosphere Bacteria to Abiotic Environmental Stress
Root exudates such as amino acids, organic acids, and sugars could provide carbon source for microbial growth and can drive sediment bacterial population richness and activities (Raaijmakers et al., 2009; Berendsen et al., 2012). Meanwhile, root exudates excreted by plants distributed in strong gradient manners from root surface into the soil. These root exudate gradients exert selective pressures, which affected the local microbial community structure such as their abundance and composition (Nunes da Rocha et al., 2009). Meanwhile, WC and W-temp played important roles in modulating the community structure of bacteria (Liddell et al., 2007; Gaumont-Guay et al., 2008). This was confirmed by our observations that Chao 1 indices of the rhizosphere and non-rhizosphere bacteria were both significantly correlated with water content (Table 2); while significant negative relationship between Simpson index and W-temp was only detected in the non-rhizosphere (Table 1). Bacillus and Clostridium sp. in the rhizosphere were significantly affected by water content and W-temp. Higher water content could enhance oxygen availability and diffusion rates in the sediment, which was benefit for Bacillus growth (Ma et al., 2021b). This was confirmed by our finding that the relative abundance of Bacillus was positively associated with DO in the non-rhizosphere (Figure 7A). In addition, Chao 1 index showed significant positive correlations with all measured sediment properties in the rhizosphere and non-rhizosphere (Table 2). Compared to the rhizosphere, Bacillus and Clostridium sp. in the non-rhizosphere were mainly affected by SNH4+-N and SNO3−-N (Figure 7A). Nitrification and denitrification always occurred in adjacent area resulting in the NO3− formed by nitrification diffusing towards an anaerobic zone where it was the terminal electron receptor for the denitrification process (Lu et al., 2012).
Conclusion
The PDCS system exhibited great capabilities in reducing nutrients N and P from rural wastewater, with the removal efficiencies ranging from 72.7 to 97.4%. Microorganisms were largely responsible for N and P removal, however, the community structures and diversities of rhizosphere and non-rhizosphere sediment bacteria varied greatly. The average numbers of bacterial OTUs in the rhizosphere sediments were significantly higher than their corresponding counterparts (p < 0.05). In the PDCS, the different microcosms in both rhizosphere and non-rhizosphere sediments shared dominant bacterial genera, such as Bacillus and Clostridium sensu stricto 1 for the ponds, and Geobacter for the ditch, respectively. Meanwhile, the contents of these three bacterial genera in the rhizosphere were higher than those of the non-rhizosphere at day 30. Moreover, Chao 1 index in both rhizosphere and non-rhizosphere was significantly positively correlated with all measured sediment properties, such as STN, STP, and OM. In the rhizosphere, Chao 1 index showed significant positive associations with NH4+-N, turbidity, tartaric acid, and Shannon index was significantly positively correlated with turbidity and TOC (p < 0.05). Similarly, in the non-rhizosphere, the Shannon and Simpson indices were associated with ORP (p < 0.05), respectively. RDA analysis demonstrated that exudates such as lactic acids, proteins, and amino acids exhibited strong positive effects on Geobacter and Clostridiu sensu stricto 12 in the rhizosphere; while Bacillus and Clostridium were significantly associated with SNH4+-N and SNO3−-N in non-rhizosphere. Together, regulating root exudates and sediment available N will increase the richness and diversity of dominant microbial species in the rhizosphere and non-rhizosphere. These findings are beneficial for the optimization and design of PDCSs and other nutrient removal systems in large-scale wastewater treatment applications.
Data Availability Statement
The data presented in the study are deposited in the National Genomics Data Center repository, accession number CRA004345.
Author Contributions
MY, YP, LY, and WL performed the experiments. FH and LM designed the experiments and wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant numbers. 82070031, 51709255) and Hubei Provincial Key Laboratory of River Basin Water Resources and Eco-environmental Sciences, Changjiang River Scientific Research Institute open research program (Grant number. CKWV2019769/KY).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Weijun Tong (Institute of hydrobiology, Chinese Academy of Sciences) for his help in sampling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.717458/full#supplementary-material
References
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The Role of Root Exudates in Rhizosphere Interactions with Plants and Other Organisms. Annu. Rev. Plant Biol. 57 (1), 233–266. doi:10.1146/annurev.arplant.57.032905.105159
Bakker, M., Chaparro, J., Manter, D., and Vivanco, J. (2015). Impacts of Bulk Soil Microbial Community Structure on Rhizosphere Microbiomes of Zea mays. Plant Soil 392, 115–126. doi:10.1007/s11104-015-2446-0
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 17 (8), 478–486. doi:10.1016/j.tplants.2012.04.001
Bowes, M. J., Jarvie, H. P., Halliday, S. J., Skeffington, R. A., Wade, A. J., Loewenthal, M., et al. (2015). Characterising Phosphorus and Nitrate Inputs to a Rural River Using High-Frequency Concentration-Flow Relationships. Sci. Total Environ. 511, 608–620. doi:10.1016/j.scitotenv.2014.12.086
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 7 (5), 335–336. doi:10.1038/nmeth.f.303
Chang, J. J., Wu, S. Q., Zhang, S. Y., Zhang, S. H., and Liang, W. (2015). Comparative Evaluation of Total Phosphorus Removal Performances for Treatment of Domestic and Secondary Wastewater Using Integrated Vertical-Flow Constructed Wetlands: Two Years' Experience. Desalin Water Treat. 56 (5), 1379–1388. doi:10.1080/19443994.2014.950343
Chen, H., Zhou, W., Zhu, S., Liu, F., Qin, L., Xu, C., et al. (2021). Biological Nitrogen and Phosphorus Removal by a Phosphorus-Accumulating Bacteria Acinetobacter Sp. Strain C-13 with the Ability of Heterotrophic Nitrification–Aerobic Denitrification. Bioresour. Technol. 322, 124507. doi:10.1016/j.biortech.2020.124507
Chen, Z.-J., Tian, Y.-H., Zhang, Y., Song, B.-R., Li, H.-C., and Chen, Z.-H. (2016). Effects of Root Organic Exudates on Rhizosphere Microbes and Nutrient Removal in the Constructed Wetlands. Ecol. Eng. 92, 243–250. doi:10.1016/j.ecoleng.2016.04.001
Childers, D. L. (2020). A Decade of Ecosystem-Scale Research at an Aridland Constructed Treatment Wetland. Front. Env Sci-switz. 8, 576936. doi:10.3389/fenvs.2020.576936
Dewedar, A., Bahgat, M., and Shabana, E. E. (2009). Seasonal Distribution and Activity of Nitrogen-Cycling Bacteria in Bardawil Lagoon, Egypt. Int. J. Clin. Exp. Med. 34 (2), 183–188. doi:10.2989/AJAS.2009.34.2.9.896
Edayilam, N., Montgomery, D., Ferguson, B., Maroli, A. S., Martinez, N., Powell, B. A., et al. (2018). Phosphorus Stress-Induced Changes in Plant Root Exudation Could Potentially Facilitate Uranium Mobilization from Stable Mineral Forms. Environ. Sci. Technol. 52 (14), 7652–7662. doi:10.1021/acs.est.7b05836
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 27 (16), 2194–2200. doi:10.1093/bioinformatics/btr381
Egamberdieva, D., Kamilova, F., Validov, S., Gafurova, L., Kucharova, Z., and Lugtenberg, B. (2008). High Incidence of Plant Growth-Stimulating Bacteria Associated with the Rhizosphere of Wheat Grown on Salinated Soil in Uzbekistan. Environ. Microbiol. 10 (1), 1–9. doi:10.1111/j.1462-2920.2007.01424.x
Evans, R. A., Cromar, N. J., and Fallowfield, H. J. (2005). Performance of a Pilot-Scale High Rate Algal Pond System Treating Abattoir Wastewater in Rural South Australia: Nitrification and Denitrification. Water Sci. Technol. 51 (12), 117–124. doi:10.1016/j.watres.2004.10.012
Gaumont-Guay, D., Black, T. A., Barr, A. G., Jassal, R. S., and Nesic, Z. (2008). Biophysical Controls on Rhizospheric and Heterotrophic Components of Soil Respiration in a Boreal Black spruce Stand. Tree Physiol. 28 (2), 161–171. doi:10.1093/treephys/28.2.161
Grandy, A. S., Erich, M. S., and Porter, G. A. (2000). Suitability of the Anthrone–Sulfuric Acid Reagent for Determining Water Soluble Carbohydrates in Soil Water Extracts. Soil Biol. Biochem. 32 (5), 725–727. doi:10.1016/S0038-0717(99)00203-5
Gustafsson, J. P., Renman, A., Renman, G., and Poll, K. (2008). Phosphate Removal by mineral-based Sorbents Used in Filters for Small-Scale Wastewater Treatment. Water Res. 42 (1-2), 189–197. doi:10.1016/j.watres.2007.06.058
Holm-Hansen, O., and Riemann, B. (1978). Chlorophyll a Determination: Improvements in Methodology. Oikos 30 (3), 438–447. doi:10.2307/3543338
Hu, B.-l., Zheng, P., Tang, C.-j., Chen, J.-w., van der Biezen, E., Zhang, L., et al. (2010). Identification and Quantification of Anammox Bacteria in Eight Nitrogen Removal Reactors. Water Res. 44 (17), 5014–5020. doi:10.1016/j.watres.2010.07.021
Khammar, N., Malhautier, L., Degrange, V., Lensi, R., Godon, J. J., and Fanlo, J. L. (2005). Link between Spatial Structure of Microbial Communities and Degradation of a Complex Mixture of Volatile Organic Compounds in Peat Biofilters. J. Appl. Microbiol. 98 (2), 476–490. doi:10.1111/j.1365-2672.2004.02474.x
Li, H., Yang, X., Weng, B., Su, J., Nie, S. a., Gilbert, J. A., et al. (2016). The Phenological Stage of rice Growth Determines Anaerobic Ammonium Oxidation Activity in Rhizosphere Soil. Soil Biol. Biochem. 100, 59–65. doi:10.1016/j.soilbio.2016.05.015
Li, J., Li, J., Gao, R., Wang, M., Yang, L., Wang, X., et al. (2018). A Critical Review of One-Stage Anammox Processes for Treating Industrial Wastewater: Optimization Strategies Based on Key Functional Microorganisms. Bioresour. Technol. 265, 498–505. doi:10.1016/j.biortech.2018.07.013
Li, X., Li, Y., Lv, D., Li, Y., and Wu, J. (2020). Nitrogen and Phosphorus Removal Performance and Bacterial Communities in a Multi-Stage Surface Flow Constructed Wetland Treating Rural Domestic Sewage. Sci. Total Environ. 709, 136235. doi:10.1016/j.scitotenv.2019.136235
Li, Y., Zhao, S., Zhang, J., He, Y., Zhang, J., and Ge, R. (2019). Screening and Diversity Analysis of Aerobic Denitrifying Phosphate Accumulating Bacteria Cultivated from A2O Activated Sludge. Processes 7 (11), 827. doi:10.3390/pr7110827
Liddell, K., Krivtsov, V., Staines, H., Brendler, A., Garside, A., and Griffiths, B. (2007). A Study of Population Numbers and Ecological Interactions of Soil and forest Floor Microfauna. Anim. Biol. 57 (4), 467–484. doi:10.1163/157075607782232189
Liu, L., Ji, M., Wang, F., Wang, S., and Qin, G. (2020). Insight into the Influence of Microbial Aggregate Types on Nitrogen Removal Performance and Microbial Community in the Anammox Process - A Review and Meta-Analysis. Sci. Total Environ. 714, 136571. doi:10.1016/j.scitotenv.2020.136571
Lu, X., Yan, Y., Fan, J., and Wang, X. (2012). Gross Nitrification and Denitrification in Alpine Grassland Ecosystems on the Tibetan Plateau. Arct Antarct Alp Res. 44 (2), 188–196. doi:10.1657/1938-4246-44.2.188
Luo, M., Liu, Y., Huang, J., Xiao, L., Zhu, W., Duan, X., et al. (2018). Rhizosphere Processes Induce Changes in Dissimilatory Iron Reduction in a Tidal Marsh Soil: a Rhizobox Study. Plant Soil 433 (1), 83–100. doi:10.1007/s11104-018-3827-y
Ma, L., He, F., Huang, T., Zhou, Q., Zhang, Y., and Wu, Z. (2016). Nitrogen and Phosphorus Transformations and Balance in a Pond-Ditch Circulation System for Rural Polluted Water Treatment. Ecol. Eng. 94, 117–126. doi:10.1016/j.ecoleng.2016.05.051
Ma, L., He, F., Sun, J., Huang, T., Xu, D., Zhang, Y., et al. (2015a). Effects of Flow Speed and Circulation Interval on Water Quality and Zooplankton in a Pond–Ditch Circulation System. Environ. Sci. Pollut. R. 22 (13), 10166–10178. doi:10.1007/s11356-015-4195-2
Ma, L., He, F., Sun, J., Wang, L., Xu, D., and Wu, Z. (2015b). Remediation Effect of Pond–Ditch Circulation on Rural Wastewater in Southern China. Ecol. Eng. 77, 363–372. doi:10.1016/j.ecoleng.2014.11.036
Ma, L., Liu, W., Tan, Q., Zhou, Q., Wu, Z., and He, F. (2019). Quantitative Response of Nitrogen Dynamic Processes to Functional Gene Abundances in a Pond-Ditch Circulation System for Rural Wastewater Treatment. Ecol. Eng. 134, 101–111. doi:10.1016/j.ecoleng.2019.05.008
Ma, L., Yang, L., Liu, W., Zhang, Y., Zhou, Q., Wu, Z., et al. (2021a). Effects of Root Exudates on Rhizosphere Bacteria and Nutrient Removal in Pond-Ditch Circulation Systems (PDCSs) for Rural Wastewater Treatment. Sci. Total Environ. 785, 147282. doi:10.1016/j.scitotenv.2021.147282
Ma, L., Yang, L., Liu, W., Zhang, Y., Zhou, Q., Wu, Z., et al. (2021b). Environmental Factors and Microbial Communities Jointly Regulate Biological Dephosphorization Process in Pond-Ditch Circulation Systems (PDCSs) for Rural Wastewater Treatment. Sci. Total Environ. 758, 143629. doi:10.1016/j.scitotenv.2020.143629
Nie, S. a., Li, H., Yang, X., Zhang, Z., Weng, B., Huang, F., et al. (2015). Nitrogen Loss by Anaerobic Oxidation of Ammonium in rice Rhizosphere. Isme J. 9 (9), 2059–2067. doi:10.1038/ismej.2015.25
Nunes da Rocha, U., Van Overbeek, L., and Van Elsas, J. D. (2009). Exploration of Hitherto-Uncultured Bacteria from the Rhizosphere. Fems Microbiol. Ecol. 69 (3), 313–328. doi:10.1111/j.1574-6941.2009.00702.x
Poret-Peterson, A. T., Albu, S., McClean, A. E., and Kluepfel, D. A. (2019). Shifts in Soil Bacterial Communities as a Function of Carbon Source Used during Anaerobic Soil Disinfestation. Front. Env Sci-switz. 6, 160. doi:10.3389/fenvs.2018.00160
Qiao, Z., Sun, R., Wu, Y., Hu, S., Liu, X., Chan, J., et al. (2020). Characteristics and Metabolic Pathway of the Bacteria for Heterotrophic Nitrification and Aerobic Denitrification in Aquatic Ecosystems. Environ. Res. 191, 110069. doi:10.1016/j.envres.2020.110069
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2009). The Rhizosphere: a Playground and Battlefield for Soilborne Pathogens and Beneficial Microorganisms. Plant Soil 321 (1), 341–361. doi:10.1007/s11104-008-9568-6
Ruban, V., López-Sánchez, J. F., Pardo, P., Rauret, G., Muntau, H., and Quevauviller, P. (2001). Harmonized Protocol and Certified Reference Material for the Determination of Extractable Contents of Phosphorus in Freshwater Sediments – A Synthesis of Recent Works. Fresen J. Anal. Chem. 370 (2), 224–228. doi:10.1007/s002160100753
Schelfhout, S., De Schrijver, A., De Bolle, S., De Gelder, L., Demey, A., Du Pré, T., et al. (2015). Phosphorus Mining for Ecological Restoration on Former Agricultural Land. Restor Ecol. 23 (6), 842–851. doi:10.1111/rec.12264
Spector, T. (1978). Refinement of the Coomassie Blue Method of Protein Quantitation: A Simple and Linear Spectrophotometric Assay for ≤0.5 to 50 μg of Protein. Anal. Biochem. 86 (1), 142–146. doi:10.1016/0003-2697(78)90327-5
State EPA of China (2002). Monitoring and Determination Methods for Water and Wastewater. 4th ed. Beijing: China Environmental Science Press. (in Chinese).
Stringlis, I. A., Yu, K., Feussner, K., de Jonge, R., Van Bentum, S., Van Verk, M. C., et al. (2018). MYB72-dependent Coumarin Exudation Shapes Root Microbiome Assembly to Promote Plant Health. P Natl. Acad. Sci. USA 115 (22), E5213. doi:10.1073/pnas.1722335115
Teng, Z., Zhu, Y., Li, M., and Whelan, M. J. (2018). Microbial Community Composition and Activity Controls Phosphorus Transformation in Rhizosphere Soils of the Yeyahu Wetland in Beijing, China. Sci. Total Environ. 628-629, 1266–1277. doi:10.1016/j.scitotenv.2018.02.115
Tomaszewski, M., Cema, G., and Ziembińska-Buczyńska, A. (2017). Influence of Temperature and pH on the Anammox Process: A Review and Meta-Analysis. Chemosphere 182, 203–214. doi:10.1016/j.chemosphere.2017.05.003
Torrens, A., Folch, M., and Salgot, M. (2021). Design and Performance of an Innovative Hybrid Constructed Wetland for Sustainable Pig Slurry Treatment in Small Farms. Front. Env Sci-switz. 8 (304), 577186. doi:10.3389/fenvs.2020.577186
Wang, B., Guo, Y., Zhao, M., Li, B., and Peng, Y. (2019). Achieving Energy-Efficient Nitrogen Removal and Excess Sludge Reutilization by Partial Nitritation and Simultaneous Anammox Denitrification and Sludge Fermentation Process. Chemosphere 218, 705–714. doi:10.1016/j.chemosphere.2018.11.168
Wu, A.-L., Jiao, X.-Y., Wang, J.-S., Dong, E.-W., Guo, J., Wang, L.-G., et al. (2021). Sorghum Rhizosphere Effects Reduced Soil Bacterial Diversity by Recruiting Specific Bacterial Species under Low Nitrogen Stress. Sci. Total Environ. 770, 144742. doi:10.1016/j.scitotenv.2020.144742
Yang, X.-P., Wang, S.-M., Zhang, D.-W., and Zhou, L.-X. (2011). Isolation and Nitrogen Removal Characteristics of an Aerobic Heterotrophic Nitrifying–Denitrifying Bacterium, Bacillus Subtilis A1. Bioresour. Technol. 102 (2), 854–862. doi:10.1016/j.biortech.2010.09.007
Yin, J., Zhang, P., Li, F., Li, G., and Hai, B. (2015). Simultaneous Biological Nitrogen and Phosphorus Removal with a Sequencing Batch Reactor–Biofilm System. Int. Biodeter Biodegr 103, 221–226. doi:10.1016/j.ibiod.2015.02.019
Yin, X., Liu, G., Peng, L., Hua, Y., Wan, X., Zhou, W., et al. (2018). Microbial Community of Nitrogen Cycle-Related Genes in Aquatic Plant Rhizospheres of Lake Liangzi in winter. J. Basic Microbiol. 58 (11), 998–1006. doi:10.1002/jobm.201800220
Zhao, D., Wang, S., Huang, R., Zeng, J., Huang, F., and Yu, Z. (2017). Diversity and Composition of Bacterial Community in the Rhizosphere Sediments of Submerged Macrophytes Revealed by 454 Pyrosequencing. Ann. Microbiol. 67 (4), 313–319. doi:10.1007/s13213-017-1262-6
Zhao, Z., Song, X., Xiao, Y., Zhao, Y., Gong, Z., Lin, F., et al. (2016). Influences of Seasons, N/P Ratios and Chemical Compounds on Phosphorus Removal Performance in Algal Pond Combined with Constructed Wetlands. Sci. Total Environ. 573, 906–914. doi:10.1016/j.scitotenv.2016.08.148
Zhao, Z., Zhang, X., Wang, Z., Song, X., Cheng, M., Cheng, M., et al. (2019). Enhancing the Pollutant Removal Performance and Biological Mechanisms by Adding Ferrous Ions into Aquaculture Wastewater in Constructed Wetland. Bioresour. Technol. 293, 122003. doi:10.1016/j.biortech.2019.122003
Zhou, W., Jiang, X., Ouyang, J., Lu, B., Liu, W., and Liu, G. (2020). Environmental Factors, More Than Spatial Distance, Explain Community Structure of Soil Ammonia-Oxidizers in Wetlands on the Qinghai–Tibetan Plateau. Microorganisms 8 (6), 933. doi:10.3390/microorganisms8060933
Keywords: pond-ditch circulation system, rhizosphere bacteria, microbial community structure, diversity, redundancy analysis
Citation: Yu M, Pan Y, Yang L, Liu W, He F and Ma L (2021) Response of Sediment Microbial Communities to the Rural Wastewater in the Pond-Ditch Circulation System. Front. Environ. Sci. 9:717458. doi: 10.3389/fenvs.2021.717458
Received: 31 May 2021; Accepted: 20 July 2021;
Published: 24 September 2021.
Edited by:
Yuanchun Zou, Northeast Institute of Geography and Agroecology (CAS), ChinaReviewed by:
JI An Sun, Central and Southern China Minicipal Engineering Design and Research Institute Cor., Ltd., ChinaWeiqi Wang, Fujian Normal University, China
Copyright © 2021 Yu, Pan, Yang, Liu, He and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Ma, bWFsaW5Ad2JnY2FzLmNu; Feng He, aGVmZW5nQGloYi5hYy5jbg==
†These authors have contributed equally to this work and share first authorship
 Mengfei Yu
Mengfei Yu Yongtai Pan2†
Yongtai Pan2† Lin Ma
Lin Ma