- 1Department of Environmental and Chemical Engineering, Shanghai University, Shanghai, China
- 2Department of Ocean Science, The Hong Kong University of Science and Technology, Kowloon, China
- 3Department of Environmental Science, Xi’an Jiaotong-Liverpool University, Suzhou, China
- 4Key Laboratory of Gas Hydrate, Guangzhou Institute of Energy Conversion, Chinese Academy of Sciences, Guangzhou, China
Reservoir sediments harbor abundant bacterial communities that are sensitive to substances transferred from the water column and might record historic water quality in environmental DNA (eDNA). The unique bacterial community along the sediment profile were retrieved based on eDNA in a reservoir to investigate potential links between water quality and the microbial population on a long-time scale. Bacterial communities in sediment samples gathered into three clusters along the depth (depths of 18–38, 8–18, and 1–7 cm). These three sections accumulated during three periods in which water quality was recorded in history (the pristine stage, degraded stage and remediated stage). Sediment samples from the degraded stage had lower microbial community evenness and diversity and higher microgAMBI indices than the other two sections, suggesting that poor water quality during that period was recorded in sedimental eDNA. After decades of biogeochemical cycles, statistical analysis revealed that the main factors affecting the microbial community were bromine, chlorine, and high molecular-weight PAHs in sediments from the degraded stage. The relevant functional groups Dehalococcoidia, Gemmatimonadales, Sva0485, Burkholderiales and Xanthomonadales might be indicators of the historical loading of these pollutants. Amending the microgAMBI index with our functional group of pollution can better illustrate the significant long-term environmental changes caused by historic anthropogenic activities. In sediments from the pristine stage with less pollution input, DIC (dissolved inorganic carbon) from the karst landform was the dominant factor controlling microbial communities. Whereas, the surface sediments, which accumulated during the remediated stage, had more correlation with chemistry, such as sulfate and heavy metals, in the overlying water. Our research revealed that historical changes in the water condition, that can be affected by anthropogenic activities, can be depicted by changes in the bacterial communities stored in the sediment using sedimental eDNA. Assessments of the bacterial communities in the sediments, either by describing their biodiversity or using particular species as indicators, would be potential proxies to describe historical environmental development of microbial communities.
Introduction
Human activities have profoundly impacted the nutrient supply and contaminants of inland waters (Williamson et al., 2008;Gao et al., 2020). To improve water quality, the Chinese government has taken a series of actions to remediate pollution and protect water resources (Shi et al., 2015; Yu et al., 2019). Evaluation of critical chemical criteria in water showed that these multiple efforts had remedied the water quality to a certain extent (Wang et al., 2017; Ma et al., 2020). However, can this strategy of action administration after contamination erase early contamination in history? The long-term effects of anthropogenic pollution and protection activities on the whole water system are still unclear.
As a crucial part of the water ecosystem, sediments tend to accumulate inputs derived from sources associated with human activities in river basins (Taylor and Owens, 2009; Ariztegui et al., 2015). Sediments in reservoirs are especially characterized by their ability to accumulate pollutants and nutrients sequestered due to dam retention effects and relatively longer water residence times (Kunz et al., 2011;Michalec and Cupak, 2021). During accumulation, sediments incessantly reserve nutrients, pollutants and particles in water bodies over a long time period. Highly accumulated substances cause two- to tenfold higher microbial abundance in sediments than in water bodies (Vignesh et al., 2014). These microorganisms were recognized to play a prominent role in mobilizing and regenerating nutrients and pollutants in freshwater (Newton et al., 2011). The close contact and frequent exchanges between sediments and the overlying water raised a question about the sediment deposition as keeping environmental information that the corresponding chemical and bacterial profile can record in sediment. Jorgensen et al. compared the profile of the microbial community and geochemical data in the sediments from the Arctic Mid-Ocean Ridge. Their results suggested that organic carbon and minerals can change microbial communities (Jorgensen et al., 2012). Because natural and anthropogenic activities are the main sources of these chemical substances, we speculated that the present profile of geochemical and microbiological activities could reflect the past environment. However, most studies on sediment profiles have focused on diagenetic processes, such as oxygen availability, nitrate reduction zones and sulfate reduction zones (Gu et al., 2007; Li et al., 2012; Bertran et al., 2020). The possibility of sedimental chemical substances and microbial communities keeping messages of historical water quality remains undiscovered.
Advances in high-throughput sequencing technologies allow for studying microbial communities in situ with high taxonomic resolution. Environmental DNA (eDNA) is DNA isolated from environmental samples, such as water or sediment samples from the aquatic ecosystem (Pawlowski et al., 2020). Buried sedimental eDNA is a powerful source of continuous temporal data for research on long-term ecological variations (Balint et al., 2018). The occurrence and abundance of specific taxonomic groups were proposed to biologically assess aquatic ecosystems (Pawlowski et al., 2018; Sun et al., 2021). During the past decade, an increasing number of studies have applied aquatic sediment DNA to trace the impacts of pollution (Rodriguez et al., 2021), eutrophication (Liu et al., 2020), anthropogenic activities (Li et al., 2018a). However, studies on the long-term impact of human activities on ecosystems using sedimental eDNA are rare (Balint et al., 2018). Even though microbial eDNA can persist for only 1–14 days before degradation, the microbial community does not change until environmental factors change (Jorgensen et al., 2012; Dornelas et al., 2013). Aquatic sediments are heterogeneous because of the geochemical substances accumulated at different times. Patterns of how historical environmental changes impact biodiversity may be traced-validated with sedimental eDNA. Furthermore, the microbial indices in sediments under different water qualities in history can help us to understand the ecological attributes of microbial communities and predict the future variation in aquatic ecosystems (Pawlowski et al., 2018).
Hongfeng Reservoir is a typical reservoir in China that has been polluted and remediated throughout history. With the development and management of the Hongfeng Reservoir, the water quality was recorded as three distinct periods according to the environmental quality standard for surface water in China (GB3838-2006, divided water quality into five classes: Class I-V, good to poor). The Hongfeng Reservoir served as a source of drinking water when dams were initially constructed in the 1960s. During that period, the water quality was good and met the standard of class II (An et al., 2009). Since the 1990s, with the development of the industry, agriculture, fishery and tourism around Hongfeng Reservoir, the water body has been seriously polluted with pollutants from factories, mining, farming and more. It was recorded that the chemical oxygen demand (COD) concentration was higher than the Class V assessment criteria of water quality in GB 3838-2002 (Chen et al., 2009; Yang et al., 2014; Dong and Wen, 2015). In the 2010s, to protect the quality of aquatic environments in China, the government and scientists endeavored to control pollution. The water quality monitoring results showed that the water quality had been improved to class III (Chen et al., 2009; Yang et al., 2014). Here, we name these three periods the pristine stage (1960–1990s), degraded stage (1990s–2007) and remediated stage (2008–2018). Furthermore, many studies have been performed in the Hongfeng Reservoir, leaving us abundant information about the geological characteristics, especially the sedimentation rates, of this site (Wei et al., 2005; Zheng et al., 2008; Guo et al., 2011). These historical changes in water quality caused by anthropogenic pressures in Hongfeng Reservoir provide us with an ideal setting to study the possibility of tracing historical water quality through sedimental chemical substances and microbial communities.
We hypothesized that sedimental geochemical properties and microbial communities at different depths have recorded information about historical water quality, and we can trace the changes in historical water quality through sedimental eDNA. In this study, the microbial and physicochemical properties along the sediment depth were analyzed synchronously to explore the microbial and physicochemical indices under different aquatic environmental qualities in history. This study can improve our understanding of the long-term effects of anthropogenic activities on water systems.
Materials and Methods
Sampling Site and Sample Collection
The Hongfeng Reservoir (106°19′–106°28′E, 26°26′–26°35′N) is located on the Guizhou Plateau of southwestern China. It was built in the 1960s and served as one of the sizable artificial reservoirs in southwestern China. It has a surface area of 57.2 km2, a mean water depth of 10.5 m (max. 45 m) and a water capacity of 6.01 × 108 m3. The area has a typical karst landform. In this area, carbonate rocks are widely distributed, and water is near neutral to slightly alkaline (pH 7.47–8.67).
The sediment cores were collected in the central area of the Hongfeng Reservoir (106°25′E, 26°29′N), which had a water depth of ∼15 m. Three sediment cores were collected by a gravity sampler (Wang and Huang, 1998) in January 2016. These three sediment cores have the same diameter of 80 mm, and the bottom of the sediments was observed as the initial soil before flooding with yellow compacting characteristics. The upper water layer in the sampling column was collected immediately into sampling bottles to test chemical properties in the water body. All the columns were split and then put into sterile zip lock bags in the field immediately. For each column, the surface 0–20 cm was sliced at 1 cm intervals. The other part was sliced at 2 cm intervals. Pore water was collected immediately in vacuum blood collection tubes by a Rhizon soil moisture sampler (Rhizosphere Research Products B.V., Netherlands) and stored at 4°C for testing. Some of the sediment samples were stored at −80°C for DNA extraction and sequencing, and others were stored at 4°C for soil property testing. We only collected samples at one site in this study, more comparation with other researches in Hongfeng Reservoir should be conducted when we analyze the results.
Estimation of Sedimentation Rate
The sedimentation rate of the Hongfeng Reservoir was reported to be 0.81–1.09 cm yr−1 (Wei et al., 2005; Zheng et al., 2008; Guo et al., 2011). Zheng et al. used the Pu activity to analyze the profile of sedimentation rate and relative depositional time from the buildup year (1960) to their sampling time (2004) at Hongfeng Reservoir (Supplementary Table S1A). Our sediment cores contained initial soil from the 1960s at the bottom, and it is reasonable to estimate that sediments in deeper sections have a similar depositional time to Zheng et al.’s result (Supplementary Table S1B, depositional time of sediment from 18–38 cm was estimated as 1960–1990s). However, we observed a seasonal change in the length of the sediment core, which was 38 cm in January 2016 and 60 cm in June of the same year. The thicker sediment in summer may be due to the high gas production, for example, methane and carbon dioxide, at that time, causing higher porosity and less compaction. (Xiao et al., 2013; Gruca-Rokosz and Tomaszek, 2015). Wilkinson et al. reported that summer methane ebullition in the sediments could produce 4–10 times more methane than winter methane ebullition, and sediment methane formation was dominated by the layer above 20 cm (Wilkinson et al., 2015). Zheng et al.’s study also supported this idea. Their sediment core was collected in July 2004, which was 37 cm and had a higher porosity and sedimentation rate in the upper layer (∼1.3–3 cm yr−1). In this study, we conducted our sampling in January 2016, which only had a similar length (38 cm) as Zheng’s research 12 years ago (Zheng et al., 2008). Thus, for our present study, the average sedimentation rates [17 cm/ (2017–1990) yr = 0.63 cm yr−1] in the upper section (0–17 cm) should be smaller than those in Zheng’s research (1.58 cm yr−1). Following the pattern that the sedimentation rate should decrease with depth, we estimated sedimentation rates of 0–7 and 8–17 cm as 0.78–0.88 cm yr−1 and 0.60–0.63 cm yr−1, respectively (Supplementary Table S1).
Properties of Pore Water and Sediment
All the upper water and pore water samples were filtered using filter paper with a 0.22 μm pore size before testing. After diluting the filtered samples 100 times with deionized water, the concentrations of NO3−, SO42−, Cl−, and Br− were determined by ion chromatography (Dionex ICS-1100 RFIC system, Thermo Scientific Dionex, United States) with a Dionex IonPac AG19-250/4.0 column (Dionex, United States), and the flow rate was 1.0 ml min−1. The total concentrations of Fe, Mn, Ni, As, Pb and Cr in pore water were analyzed by inductively coupled plasma−mass spectrometry (ICP−MS, iCAP Q ICP-MS, Thermo, United States). Dissolved inorganic carbon (DIC) was analyzed by a Gasbench II-isotope ratio mass spectrometer (Gasbench II-IRMS, Delta V Advantage, Thermo Scientific, United States). The concentration of total organic carbon (TOC) was analyzed with a TOC analyser (Multi N/C 3100 with solid module HT1300, Analytik Jena AG, Germany). Pre-weighed sediment samples were dried at 40°C for 48 h to a steady weight. The water content was calculated based on the difference between the wet weight and dry weight. We measured all the chemical characteristics of water samples, including the pore water of sediments and the upper water. The chemical characteristics of sediment were overlooked in this study, which needs more support from published researches when we analyze the results.
DNA Extraction and Sequencing
Total genomic DNA from each sample was extracted by the UltraClean Soil DNA Isolation Kit (Mobio Laboratories Inc., United States) following the manufacturer’s protocol. The quantity of DNA was checked by a UV–vis spectrophotometer (Q5000, Quawell, United States) and verified by electrophoresis in 1% (w/v) agarose gels. The conserved V4−V5 region of the bacterial 16S rRNA gene was amplified by using primers tag-515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). A volume of 25 μL reaction system contained 2.5 μL reaction buffer 10x, 1 μL dNTPs (10 mM), 0.5 μL bovine serum albumin (BSA; 1 mg ml-1), 1 μL MgCl2 (50 mM), 0.3 μL of each primer (10 μM), 10 ng of template DNA, and 0.5 μL of Taq DNA Polymerase (5 U μL1) (rTaq, TaKaRa, Dalian, China). The thermocycling program was followed by initial denaturation for 3 min at 95°C, 30 cycles of 30 s at 95°C, 30 s at 55°C for bacteria, and 30 s at 72°C, and a final extension for 10 min at 72°C. High-throughput sequencing was processed with standard Illumina MiSeq protocols (Majorbio Technology, Shanghai, China) to generate paired-end reads.
The sequence data were processed by the QIIME (version 1.9.1) pipeline (Lai et al., 2014). The raw reads were subsequently preprocessed by removing bad sequences, including artificial, ambiguous and length less than 200 bp sequences. After chimaera removal (Edgar, 2010), the preprocessed reads were aligned using PyNAST (Caporaso et al., 2010) with the SILVA database (https://www.arb-silva.de), and operational units (OTUs) were clustered with a 97% similarity cut-off. Then, the phylogenetic affiliation of typical sequences was analyzed by RDP Classifier against the Silva (SSU115) 16S rRNA database with a confidence threshold of 70%.
Alpha Diversity Analysis
The alpha diversity indices were calculated with MOUTHER to analyze the complexity of species diversity for each sample (versionv.1.30.1 http://www.mothur.org/wiki/Schloss_SOP#Alpha_diversity). The Sobs indices can reflect the observed community richness of each sample, and higher Sobs values demonstrate higher community richness. The heip indices can reflect the community evenness with Heip’s metric of each sample, and a higher heip value demonstrates higher community evenness. The Shannon indices can reflect community diversity, and a higher Shannon value demonstrates higher community diversity (Mendes et al., 2015; Xie et al., 2016).
MicrogAMBI Index
MicrogAMBI is a bacterial community-based index that uses bacterial composition to assess the ecological status of the environment (Aylagas et al., 2017; Borja, 2018). According to the taxa list provided by the method, microbes are classified into two ecological groups: EGI, as taxa not associated with pollution inputs, and EGIII, as taxa associated with pollution inputs. By comparing the microbial community of our samples to the taxa list, we calculated the microgAMBI index as:
A higher value of the microgAMBI index means a larger proportion of pollution-related microbes and more inferior environmental status. In this method, they classified the environmental ecological status into five groups according to the microgAMBI index: high ecological status (0< microgAMBI ≤1.2), good ecological status (1.3< microgAMBI ≤2.4), moderate ecological status (2.5< microgAMBI ≤3.6), poor ecological status (3.7< microgAMBI ≤4.8) and bad ecological status (4.9< microgAMBI ≤6). We calculated indices of microgAMBI according to the taxa list summarized by Borja (Borja, 2018).
Statistical Analysis
Principal component analysis (PCA) was conducted with R language using the corresponding analysis and plot packaging to reveal the difference and similarity between microbial communities. Redundancy analysis (RDA) was performed by the vegan package of R Language to evaluate the correlation between the microbe community and environmental factors. Partial least squares discriminant analysis (PLS-DA) was conducted with the mix omics package of R Language to show the reasonableness of grouping. The heatmap showing the correlation between microbes in the top 50 most abundant taxa and environmental factors was constructed with the pheatmap package of R language using the Spearman correlation coefficient. The significance of the ecological matrix of the microbial community was calculated with SPSS (Statistical Product and Service Solutions).
Accession Numbers
The OTU sequences are publicly available at the BioSample database of NCBI (BioProject: PRJNA408013, Biosamples: SAMN07673091- SAMN07673145). These data can be found here: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA408013.
Results
Depositional Time of Sediment Profile
We estimated that sediments at depths of 1–7 cm were deposited from 2007 until the sampling time, and sediments at depths of 8–17 and 18–38 cm were deposited from 1990–2007 and 1960–1990, respectively (Supplementary Table S1). Therefore, correlating the sediment depth and water qualities of the three periods, sediments at depths of 1–7 cm were deposited from the remediated stage, sediments at depths of 8–17 cm accumulated during the degraded stage, and sediments at depths of 18–38 cm settled from the pristine stage.
Furthermore, the percentages of TOC in the sediment profile are listed in Table 1. The results revealed higher TOC concentrations in sediment at depths of 8–17 cm (2.36–4.03% at depths of 8–17 cm, compared to 1.92–2.35% at depths of 1–7 cm and 1.20–1.52% at depths of 18–38 cm), which accumulated due to high pollutant inputs during the degraded stage. This consistency further verified our estimation of the depositional time of the sediment profile.
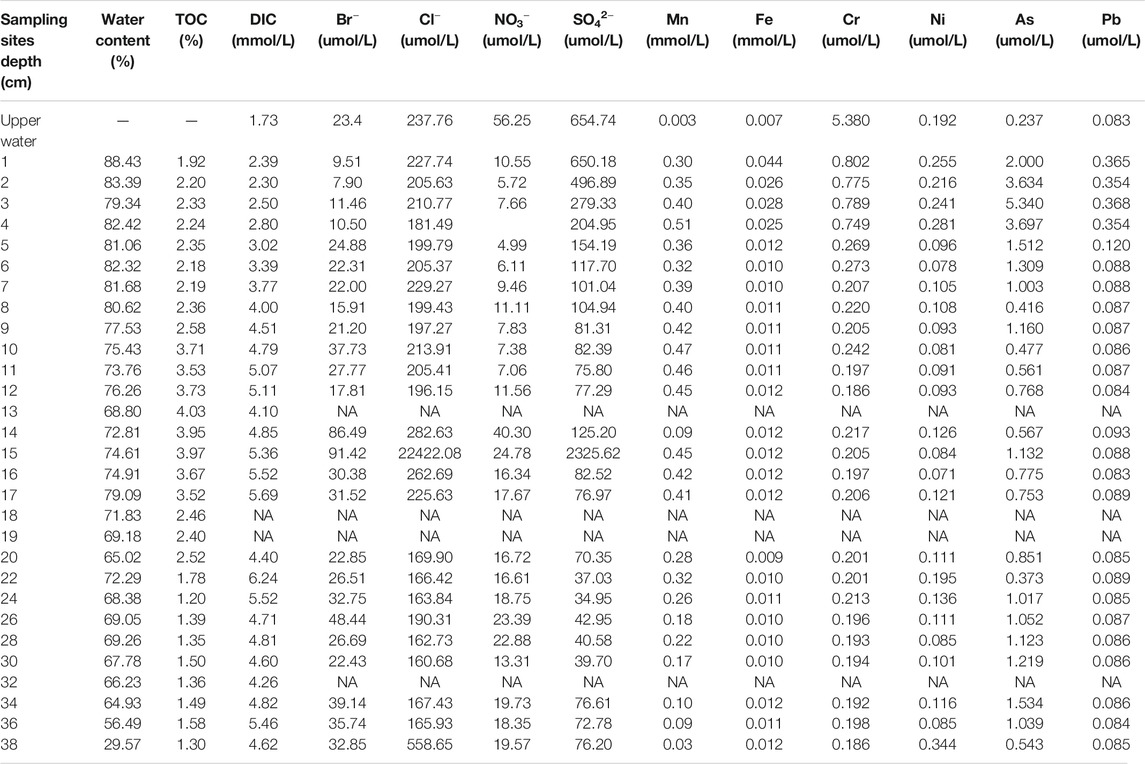
TABLE 1. Characteristics of upper water, pore water and sediment. Water content was measured with sediment, other chemicals were measured with pore water or upper water.
Taxonomic Structure and Diversity of the Bacterial Community
The constant changes in the microbial community in the reservoir sediment profile are demonstrated in Figure 1A. The microbial communities of the sediment profile apparently gathered into three clusters (remediated stage, degraded stage and pristine stage) by PCA analysis (Figure 1B, PLS-DA and RDA analyses showed the same results in Supplementary Figure S1 and Figure 2). The microbial community changed dynamically with the depth of the sediment and was quite distinct among the three groups (Figure 1C). The dominant classes of Deltaprotebacteria, Betaprotebacteria and Gammaprotebacteria together accounted for 20.3–57.7% in the degraded stage, which was higher than the other depths of the column. Specifically, a higher content of Gammaprotebacteria was observed in the degraded stage (16.4–21.4%), while the relative abundances of Betaprotebacteria and Alphaprotebacteria were much lower in the pristine stage (∼4% and 1.1–1.7%, respectively).
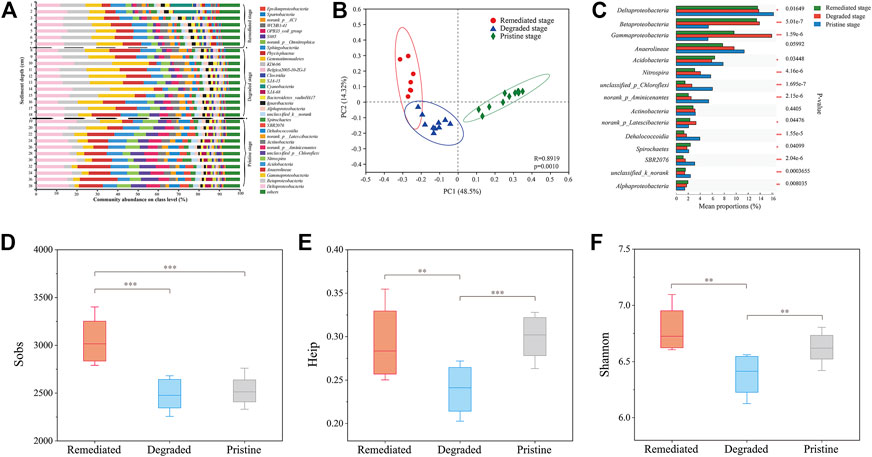
FIGURE 1. Analysis of microbial community in the Hongfeng reservoir sediment profile. Samples from different depths are clustered into three stages which are related to the up-layer water quality in corresponding historical period (remediated stage: depth 0–7 cm, degraded stage: depth 8–17 cm, pristine stage: 18–38 cm). The microbial community structure at class level along the sediment profile (A); PCA analysis showing the differences and distance of microbial communities between sediment samples from different depth (B); microbial distribution in sediment of different stages (C); community richness (D), community evenness (E) and community diversity (F) of sediment from these three stages.
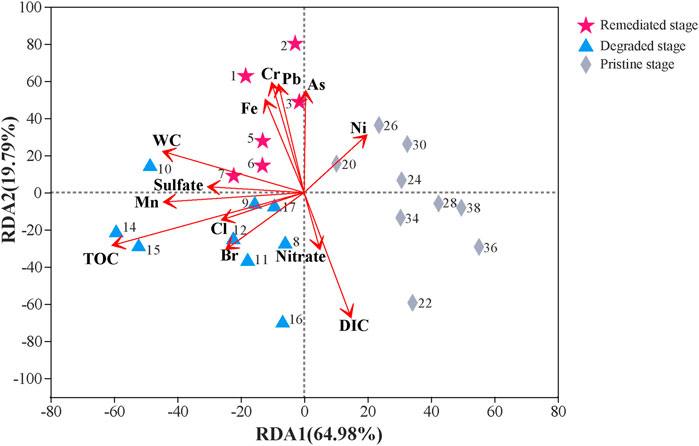
FIGURE 2. RDA analysis (Phylum level) for microbial distribution and environmental factors of sediment from different depth. The red arrows represent environmental factors which can affect the microbial communities in sediment samples. Pink pentagrams are sediment from remediated stage (1–7 cm). Blue triangles are sediment from degraded stage (8–17 cm). And gray rhombs are sediment from pristine stage (18–38 cm). Numbers next to the symbols are the depth of each sample (cm).
Accordingly, microbial biodiversity varies at different stages of sediment depth. The Sobs index showed that the community richness of the remediated stage in the surface sediment was significantly higher than that at deeper depths (Figure 1D), indicating noticeable improvement in water quality under the protection efforts. The Heip index indicated lower community evenness in the sediment of the degraded stage (Figure 1E), and the Shannon index revealed a relatively lower community diversity in the sediment of the degraded stage (Figure 1F). These results showed poorer ecological conditions in the degraded stage.
Environmental Factors Associated With Indicator Species
The characteristics of pore water and sediment at different sedimental depths are listed in Table 1. The results of RDA indicated that the microbial communities from the three periods (pristine stage, degraded stage and remediated stage) were different from each other, and the main factors causing these differences were TOC (r2 = 0.8325, p = 0.001), DIC (r2 = 0.6833, p = 0.001), Cr (r2 = 0.5132, p = 0.001), and Pb (r2 = 0.4899, p = 0.001) (Figure 2; Supplementary Table S2). Within these four factors, the Cr and Pb concentrations had apparent effects on the sedimental microbial communities from the remediated stage (0–7 cm). TOC had a stronger correlation with microbial communities in the degraded stage (8–17 cm), and DIC affected microbial communities in the pristine stage (18–38 cm). Other factors also influenced the microbial communities within these three stages to some extent, including water content (r2 = 0.437, p = 0.002), As (r2 = 0.4242, p = 0.004), Fe (r2 = 0.3763, p = 0.005), Mn (r2 = 0.3743, p = 0.012), and Br (r2 = 0.2489, p = 0.043) (Figure 2; Supplementary Table S2).
The surface 0–7 cm sediment, which met the historical period of the remediated stage (2008-sampling time), had an obvious correlation with the water content and the concentrations of several heavy metals, including Cr, Pb, As, and Fe (Figure 2). The water content decreased from 88.43 to 56.49% with increasing sediment depth, and the initial sediment at a depth of 38 cm had the lowest water content of 29.57% (Table 1). In the 0–7 cm sediment, the order of Hydrogenophilales and Sva0485 was significantly negatively correlated with the water content (Figure 3A, r = −0.94, p = 0.005). However, we could not see this significant correlation in the other two sediment sections (Figures 3B,C). The concentrations of Cr, Pb, As, and Fe were 2–3 times higher in the 0–7 cm sediment than in the lower sections (Table 1). Compared to the upper water, the concentrations of Pb, As, and Fe in the sediment pore water were higher, while the concentration of Cr was lower in the sediment pore water (Table 1). TM6-Dependentiae showed obvious negative correlations with the concentrations of Pb (r = −0.94, p = 0.005), As (r = −0.94, p = 0.005), and Fe (r = −0.82, p = 0.041) (Figure 3A). The abundance rank of TM6-Dependentiae was below 50 in the other two sections (Figures 3B,C). Additionally, sulfate also had an impact on the microbial communities of the remediated stage (Figure 2). The sulfate concentration decreased dramatically with depth, indicating that sulfate was the main electron acceptor in this layer. Several microbes had a close correlation with the concentration of sulfate. For example, an order of Cyanobacteria (r = 0.94, p = 0.005) showed a strong positive correlation with sulfate concentration. Sphingobateriales (r = 0.89, p = 0.019) was significantly positively correlated with sulfate, while Rhodocyclales (r = −0.94, p = 0.0059), Chloroflexi (r = −0.94, p = 0.004) and Anaerolineale (r = −0.89, p = 0.019) were significantly negatively correlated with sulfate (Figure 3A).
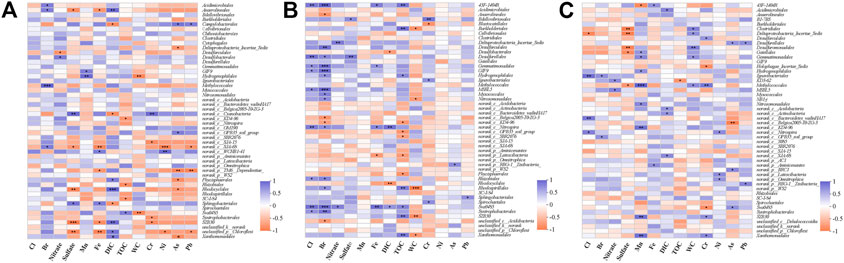
FIGURE 3. Heatmaps showing Spearman correlation of microbe distribution (Order) and environment factors whthin three stages [remediated stage: (A); degrade stage: (B); pristine stage: (C)]. The color illustrated the r-value according to the color bar. The P-value was marked with asterisks (*), *: 0.01 < p ≤ 0.05; **: 0.001 < p ≤ 0.01; ***p ≤ 0.001.
In the degraded stage, TOC had a great influence on the microbial communities (Figure 2). The TOC percentages were 2–3 times higher in this section than in the other two sections (Table 1). Several microbes showed a strong positive correlation with the concentration of TOC, including 43F-1404R (r = 0.83, p = 0.005), Burkholderiales (r = 0.87, p = 0.002), Rhodospirillales (r = 0.80, p = 0.009) and Xanthomonadales (r = 0.81, p = 0.008) (Figure 3B). Similarly, these microbes did not show obvious correlations with TOC in the other two sections (Figures 3A,C). The concentration of Mn also showed a correlation with the microbial communities in this stage, but we did not find microbes presenting an obvious correlation with this factor (Figures 2, 3B). Meanwhile, the concentrations of bromide (Br) and chlorine (Cl) also impacted the microbial community of the degraded stage (Figure 2). Spearman correlation analysis uncovered more microbes that had quite a close positive correlation with the concentration of Br and Cl, including two orders of class Dehalococcoidia MSBL5 (Br: r = 0.95, p = 0.0001, Cl: r = 0.74, p = 0.021) and GIF9 (Br: r = 0.92, p = 0.0005, Cl: r = 0.67, p = 0.050), 43F-1404R (Br: r = 0.95, p = 0.00009, Cl: r = 0.82, p = 0.007), Gemmatimonadales (Br: r = 0.9, p = 0.0009, Cl: r = 0.7, p = 0.036), Sva0485 (Br: r = 0.90, p = 0.0009, Cl: r = 0.87, p = 0.002), and an order of class Nitrospira (Br: r = 0.70, p = 0.036, Cl: r = 0.80, p = 0.010) (Figure 3B).
The concentration of DIC in the pore water was relatively higher (4.26–6.24 mM) in pristine sediment (Table 1), and DIC became the key factor impacting the community of microbes (Figure 2). In addition, Actinobacteria (r = 0.68, p = 0.042), Acidobacteria (r = 0.68, p = 0.042), Acidimicrobiales (r = 0.77, p = 0.016) and SJA-68 (r = 0.73, p = 0.025) showed a relatively high correlation with DIC concentration in the pristine stage, but these correlations were not evident in the degraded stage and remediated stage (Figure 3).
Microbial Indicators
According to the taxa list summarized by Borja (Borja, 2018), microgAMBI indices showed a moderate ecological status of the whole sediment core at the sampling time (Figure 4A, microgAMBI: 2.02–3.22). However, the microgAMBI indices of sediment from the degraded stage were slightly higher than those of the others, suggesting poor ecological status in this section (microgAMBI in the degraded stage: 2.49–3.12, microgAMBI in the remediated stage: 2.03–2.80, and microgAMBI in the pristine stage: 2.43–32.96). Two upper sediment samples from the remediated stage had good ecological status (microgAMBI was 2.03 and 2.27 in sediment from 1 to 2 cm depths).
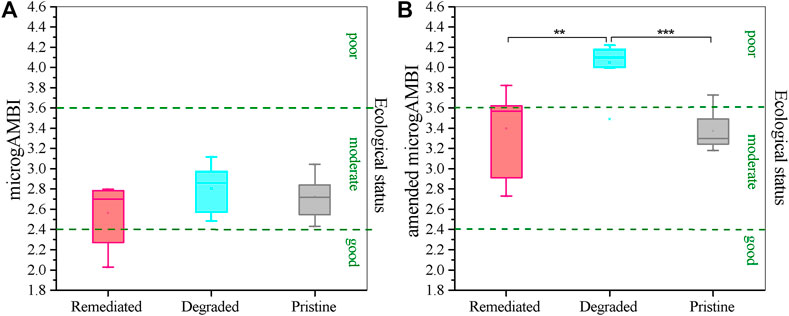
FIGURE 4. Microbial indicators for the ecological status of sediment from different historical environment. MicrogAMBI index showed a slightly poor ecological status for sediment from degraded stage, but the differences were not significant (A). Amended MicrogAMBI index illustrate significant difference among these three sections (B).
We made some amendments to Borja’s list and obtained indices of amended microgAMBI (Figure 4B). After analysing the microbial taxa in the heatmaps (Figure 3), we found three kinds of taxa showed correlations with pollution but were not listed in Borja’s list. Thus, we added Dehalococcoidia to the list and set it to EGIII (taxa associated with pollution inputs). We also changed Gemmatimonadales and Xanthomonadales’s ecological group from EGI to EGIII. The amended microgAMBI showed that sediment in the section of the degraded stage (microgAMBI: 3.49–4.22) had poor ecological status and sediment from remediated (microgAMBI: 2.73–3.82) and pristine stage (microgAMBI: 3.18–3.73) had moderated ecological status (Figure 4B).
Discussion
Historical Water Quality Can Be Recorded in the Sediment and Reflected by Microbial Community
This is the first study to illustrate that the constant changes in the microbial community in reservoir sediment profiles are linked to the water quality and some specialized pollutants recorded in history. Layered sediments were supposed to sequester and accumulate substrates and pollutants from the overlying water during settling down. The sedimentary indigenous microbial community may shift its composition and structure and its function to increase its adaptability and/or resistance to nutrients or pollutants from the overlying water (Chen et al., 2018; Zhang et al., 2021). In this study, microbial communities changed dynamically along depth of the sediment and apparently gathered into three groups corresponding to the three periods of water quality (Figures 1A,B): the pristine stage (sediment from 18–38 cm), degraded stage (sediment from 8–17 cm) and remediated stage (sediment from 0–7 cm). Sediments from these three periods deposited different substrates and pollutants in the relevant layers, leading to the variation of microbial communities between layers. Pollution in the environment can result in a decrease in microbial diversity and a change in microbial community structure (Xie et al., 2016). Our results suggested that some pollutants from water bodies can profoundly influence the richness and diversity of benthic prokaryotic assemblages even after decades (Figures 1D–F).
In environments with less anthropogenic disturbance, sedimentary microbial communities should correlate better with geological processes, which are the main source of their nutrients. In our study, for sediments from the pristine stage, the microbial communities were mainly controlled by the concentration of DIC in the pore water. This high concentration of DIC, which was the karst geological feature, served as the main carbon source in this section. For example, Actinobacteria and Acidobacteria are active in CO2 fixation by ribulose bisphosphate carboxylase/oxygenase (RuBisCO) in the sediment (Norris et al., 2011; Li et al., 2018), and they showed a positive correlation with DIC concentration in this study. SJA-68, which belongs to the phylum Chloroflexi, also showed a positive correlation with DIC concentration in the pristine stage. This might be explained by the ability of Chloroflexi to utilize cell decay materials from autotrophs in this layer (Yang et al., 2018).
The presumptuous discharge of pollutants into the water body can change both the geological and microbiological properties of the sediments. Our results suggested that these changes can be recorded in the sediments for decades. One reason for this phenomenon should be the stability of some chemical pollutants. In the sediment section from the degraded stage, the concentration of TOC was extremely high, which was caused by audacious pollution discharging decades ago. The high concentrations of Br and Cl in the pore water of this section also indicated the dumping of halogenated organic compounds during that period. For example, the Hongfeng Reservoir was seriously polluted by polycyclic aromatic hydrocarbons (PAHs) from domestic coal combustion, biomass burning and fuel combustion at that time, and some PAHs might contain Br, such as brominated flame retardants, including polybrominated diphenyl ethers (PBDEs) and tetrabromobisphenol A (TBBA) (Guo et al., 2011; Zhang et al., 2019). Some pesticides and herbicides, such as deltamethrin, profenofos and linuron, contain bromide or chloride and have been used in agriculture. The presumptuous discharge of these chemical pollutants led to the enrichment of TOC and Br or Cl in the sediment. Guo et al. found that PAHs in the sediments of the Hongfeng Reservoir were dominated by low molecular-weight components, but high molecular-weight PAHs increased significantly in the upper segments in 2006, which corresponded to the degraded stage in our study (Guo et al., 2011). The high molecular-weight PAHs have low bioavailability and high persistence, which can last approximately 10–20 years in the sediment, contributing to the high concentration of TOC in this section (Wei et al., 2005; Li et al., 2009). Some low molecular-weight PAHs could degrade to 50% within three months (Johnsen and Karlson, 2005), which can release Br in sediment pore water. These anthropogenic sources Br and Cl can serve as geochemical proxies for superior water intrusion (especially for pollutants) into the sediments (Reddy et al., 2000; Moreno et al., 2015). Our results showed that after decades of biogeochemical cycles, the concentrations of TOC, Br and Cl turned out to be main factors affecting the microbial communities in sediments from the degraded stage (Figure 2). Another possible reason for the “records” might be the weak resilience of microbial communities under anthropogenic disturbance (Beattie et al., 2020). Chen et al. reported geological evidence for the fragility of Hongfeng Reservoir and another karst reservoir that can hardly recover from pollution even after a long period (Chen et al., 2020). We found some specific microbes related to anthropogenic discharge in this study. Spearman correlation analysis showed that two orders of Dehalococcoidia classes (MSBL5 and GIF9) had close correlations with Br and Cl concentrations. Dehalococcoides, which is a genus of bacteria within the class Dehalococcoidia, can dehalogenate halogenated organic compounds in a mode of anaerobic respiration (Loffler et al., 2013) and are tolerant to many pollutants (such as benzenes, PCBs and recalcitrant pollutants) (MaymoGatell et al., 1997; Maphosa et al., 2012; Mayer-Blackwell et al., 2017). The taxa Gemmatimonadales and Sva0485, which also showed a significant positive correlation with the concentration of Br and Cl, were reported to be possibly active in the degradation of bromated or chlorinated PAHs such as PBDEs, TBBA and linuron (Dealtry et al., 2016; Feng et al., 2019; Jiang et al., 2020). Burkholderiales and Xanthomonadales, which showed a significant positive correlation with the concentration of TOC, were reported to have a strong degradation effect on organic matters, especially for PAHs or PBDEs (Nunez et al., 2012; Crampon et al., 2018; Feng et al., 2019). Our results further indicated that Dehalococcoidia, Gemmatimonadales and Sva0485 would potentially be candidate microbial signals for persistent bromide pollutants in the sediment, and Burkholderiales, Xanthomonadales, Gemmatimonadales and Sva0485 would potentially be candidate microbial signals for persistent PAH pollutants in the sediment.
The surface sediments, a hotspot for frequently exchanging nutrients and pollutants at the water-sediment interface, were impacted mostly by the overlying water. In the 2010s, to protect the quality of aquatic environments in China, the government and scientists endeavored to control pollution from agriculture, domesticity and industry. Heavy metals in water body of Hongfeng Reservoir decreased under effective protection measures (Wu et al., 2014). However, there were still relatively high concentrations of heavy metals (Cr, Pb, As, and Fe) in the pore water of the surface sediments, which had significant impacts on the microbial communities (Figure 2; Table 1). For example, our results showed a negative correlation between TM6-Dependentiae and the concentrations of Pb and As. This is in agreement with Luo et al.’s research, and they claimed that TM6-Dependentiae could be an indicator of Pb pollution (Luo et al., 2018). Tian et al. studied the heavy metals in overlying water, pore water and sediment of Hongfeng Reservoir, and they found that the concentration of heavy metals was highest in sediment and that in pore water was higher than that in overlying water (Tian et al., 2011). Our results regarding the concentrations of Pb and As were consistent with Tian et al.’s reports, which had higher concentrations in pore water than in upper water. These results further indicated the accumulation of heavy metals in sediment and the risk of sediment releasing heavy metals into the overlying water (Reddy et al., 2004; Birch et al., 2020). Wu et al. claimed that the As in the pore water were from lithogenic sources, and Pb and Cr might originate from anthropogenic activities, including traffic pollution and residual effects of former industrial practices (Wu et al., 2014). Our results also demonstrated that sulfate was a main factor driving the changes in the microbial community in this section. For example, Cyanobacteria, which may promote sulfate reduction (Chen et al., 2014), showed a significantly positive correlation with the concentration of sulfate. Another order of Campylobacterales was significantly correlated with sulfate, which was identified to have thioredoxin reductase activities (Kaakoush et al., 2007). The chemical profile indicated that sulfate was an important electron acceptor in this layer, consistent with Wei et al.’s observation in the same reservoir (Wei et al., 2005). The sedimental sulfate in this period mainly (60–70%) came from acid deposition and coal mining drainage (Song et al., 2008; Wang et al., 2011). This sulfate reduction process can produce sulfide, which can react with As and Pb in pore water to form sulfide precipitates or coprecipitate it with iron (Kirk et al., 2004; Hoa et al., 2007; Xu et al., 2019). Consequently, sulfate reduction can control the release of heavy metals from sediment to the upper water.
Our results showed that the microbial communities in the sediment profile had a strong correlation with the water quality in the corresponding historical time. This further confirmed the feasibility of using sediment profiles as a data source to study the long-term impacts of human activities on ecosystems. A similar method was also used to trace changes in microbial communities caused by eutrophication and climate change throughout history (Capo et al., 2016; Monchamp et al., 2017). We combined the geochemistry characteristics with microbial data, which can give us a better explanation of the heterogeneous composition of sediments. And this result can improve our understanding of the long-term feedback of protection/pollution actions to the water ecosystem.
Microbial Index of Ecological Status in Sediment Profile
Sedimental eDNA can provide information about microbial communities on a long-time scale (Balint et al., 2018). We speculated that the present ecological statuses of sediments accumulated from different environments should be different and are historically associated with the environment. We used microgAMBI indices to assess the ecological status of each section based on microbial communities (Aylagas et al., 2017; Borja, 2018). Even though the microgAMBI indices indicated the slightly poor ecological status of the degrade stage, we failed to see any significant difference among these three stages, all of which were predicted as moderate ecological status (Figure 4A). One possible reason for this unexpected result might be the different ecological niches of these three sections. The sediment profile is heterogeneous even in the steady state, which does not have a human disturbance. The oxygen concentration drops sharply in the top profile, and other electron acceptors, for example, sulfate in the upper layer in this research, become dominant oxidants in anaerobic sediment according to the free energy needed for the reaction (Gu et al., 2007; Li et al., 2012; Bertran et al., 2020). While the mircogAMBI did not count this situation in, the long-term biogeochemical cycle can degrade a large portion of pollutants and keep only refractory substances, for example, bromine, chlorine and high molecular-weight PAHs, in the deep sediment in this study, causing relatively indistinctive changes in microgAMBI indices. Our results suggested that the microbial indicators for long-term environmental quality should be partially different from the original microgAMBI. A new system of microbial indicators for long-term environmental quality remains to be established (Pawlowski et al., 2018).
We found some unexpected differences between the taxa list of Borja’s method and our results (Borja, 2018). After revising three possible candidate microbial indicators for pollution based on our results, the amended microgAMBI indices were able to reflect the significant differences in ecological status, as we expected (Figure 4B). In Borja’s method, Gemmatimonadales and Xanthomonadales were recognized as taxa not associated with pollution inputs according to Sun et al.’s research (Sun et al., 2013; Borja, 2018). Sun et al. found that in the surface layer of sediment, the outer zone of an uncontaminated estuary correlated with a high abundance of Xanthomonadales. This was consistent with studies arguing that Xanthomonadales can exist extensively in sediments with wood substrates and plant residues, which may indicate less anthropogenic disturbance (Schmalenberger et al., 2013; Herve et al., 2014). However, many studies have claimed that Xanthomonadales can be simulated in PAH- or PBDE-polluted environments and can improve the degradation of these pollutants (Nunez et al., 2012; Crampon et al., 2018; Feng et al., 2019). There was no clear statement about Gemmatimonadales’s potential to reveal a polluted environment in Sun et al.’s paper (Sun et al., 2013). They found a negative correlation between chloroplasts and a species of Gemmatimonadetes, which is also the phylum Gemmatimonadales, and the author suggested further research about this correlation (Sun et al., 2013). However, Gemmatimonadales was reported to be possibly active in degrading bromated or chlorinated PAHs such as PBDEs, TBBA and linuron in subsequent studies (Dealtry et al., 2016; Feng et al., 2019; Jiang et al., 2020). Dehalococcoidia, which was missing in Borja’s list, was tolerant to many pollutants, including PCBs and benzenes (MaymoGatell et al., 1997; Maphosa et al., 2012; Mayer-Blackwell et al., 2017). Based on our results, we therefore propose to amend and further develop the original microgAMBI index.
Our research further implied the remaining challenges about using eDNA to study microbial communities. The development of high-throughput sequencing technologies provides rapid and abundant taxonomic data, including many uncultured species (Pawlowski et al., 2020). However, the knowledge of these taxa’s metabolic functions and ecological characteristics is far from sufficient when we analyze this high diversity of taxa (Jones, 2008; Pawlowski et al., 2018). In this study, we recommend Dehalococcoidia, Gemmatimonadales and Sva0485 as potential candidate microbial signals for persistent bromide pollutants and Burkholderiales, Xanthomonadales, Gemmatimonadales and Sva0485 as potential candidate microbial signals for persistent PAH pollutants in sediment. However, the ecology of these species and their response to pollution still need to be further studied. For example, can these microbial indicators respond to pollution directly, or do the interactions between microbial species contribute more to the tolerance? Although some researchers also declared that some specific microbes were always inhabited in the polluted environment to improve the tolerance of the ecosystem (Tolkkinen et al., 2015; Garcia et al., 2018), we are still dubious about connecting these specialized microbes to the pollution directly. Sagova-Mareckova et al. argued that because of the close biotic interactions including trophic relationships, symbiosis, parasitism or chemical communication in microbial communities, calculation considering the multi-species and community approaches should be more reliable for the microbial index of ecological status (Sagova-Mareckova et al., 2021). To figure out this question, we need more knowledge about the metabolic and physiological properties of these microbes. Complement of the taxa reference database are urgent for assessing the biological status with microbial indices.
In conclusion, we proposed that bacterial community composition is vertically heterogeneous with sediment depth and is impacted significantly by historical changes in the water condition which were affected by anthropogenic activities. Our research revealed the long-term feedback of protection/pollution actions to the water ecosystem using sedimental eDNA. New microbial indices that can cover historical impacts should be developed. These results contribute to water resource management aimed at protecting aquatic ecosystems and restoring degraded systems.
Data Availability Statement
The data has been uploaded to NCBI and can be accessed using the accession number PRJNA408013.
Author Contributions
JS and XC designed research; JS, XC, JY, ZC, YY and ZF performed research; JS, XC, MY and LL analyzed data; JS, XC, MY, ZC and FW wrote the paper.
Funding
This research was funded by the National Key Research and Development Project by Most of China (2016YFA0601000), the National Natural Science Foundation of China (41776071).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate Williamson Gustave for helping us polishing the Language.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.669582/full#supplementary-material
References
Ariztegui, D., Thomas, C., and Vuillemin, A. (2015). Present and Future of Subsurface Biosphere Studies in Lacustrine Sediments through Scientific Drilling. Int. J. Earth Sci. (Geol Rundsch) 104 (6), 1655–1665. doi:10.1007/s00531-015-1148-4
Aylagas, E., Borja, Á., Tangherlini, M., Dell'Anno, A., Corinaldesi, C., Michell, C. T., et al. (2017). A Bacterial Community-Based index to Assess the Ecological Status of Estuarine and Coastal Environments. Mar. Pollut. Bull. 114 (2), 679–688. doi:10.1016/j.marpolbul.2016.10.050
Bálint, M., Pfenninger, M., Grossart, H.-P., Taberlet, P., Vellend, M., Leibold, M. A., et al. (2018). Environmental DNA Time Series in Ecology. Trends Ecol. Evol. 33 (12), 945–957. doi:10.1016/j.tree.2018.09.003
Beattie, R. E., Bandla, A., Swarup, S., and Hristova, K. R. (2020). Freshwater Sediment Microbial Communities Are Not Resilient to Disturbance from Agricultural Land Runoff. Front. Microbiol. 11, 539921. doi:10.3389/fmicb.2020.539921
Bertran, E., Waldeck, A., Wing, B. A., Halevy, I., Leavitt, W. D., Bradley, A. S., et al. (2020). Oxygen Isotope Effects during Microbial Sulfate Reduction: Applications to Sediment Cell Abundances. Isme J. 14 (6), 1508–1519. doi:10.1038/s41396-020-0618-2
Birch, G. F., Lee, J. H., Tanner, E., Fortune, J., Munksgaard, N., Whitehead, J., et al. (2020). Sediment Metal Enrichment and Ecological Risk Assessment of Ten Ports and Estuaries in the World Harbours Project. Mar. Pollut. Bull. 155. doi:10.1016/j.marpolbul.2020.111129
Borja, A. (2018). Testing the Efficiency of a Bacterial Community-Based index (microgAMBI) to Assess Distinct Impact Sources in Six Locations Around the World. Ecol. Indicators 85, 594–602. doi:10.1016/j.ecolind.2017.11.018
Capo, E., Debroas, D., Arnaud, F., Guillemot, T., Bichet, V., Millet, L., et al. (2016). Long-term Dynamics in Microbial Eukaryotes Communities: a Palaeolimnological View Based on Sedimentary DNA. Mol. Ecol. 25 (23), 5925–5943. doi:10.1111/mec.13893
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010). PyNAST: a Flexible Tool for Aligning Sequences to a Template Alignment. Bioinformatics 26 (2), 266–267. doi:10.1093/bioinformatics/btp636
An, H. P., and Zou, B. (2009). Current Situation and Pollution protection Method of Drinking Water Sources Quality of Hongfeng Lake and Baihua Lake in Guiyang Municipality. Res. Develop. Market. 25 (10). doi:10.3969/j.issn.1005-8141.2009.10.019
Chen, J. G., Yu, J., Bai, X. Y., Zeng, Y., and Wang, J. F. (2020). Fragility of Karst Ecosystem and Environment: Long-Term Evidence from lake Sediments. Agric. Ecosyst. Environ. 294, 106862. doi:10.1016/j.agee.2020.106862
Chen, M., Ye, T. R., Krumholz, L. R., and Jiang, H. L. (2014). Temperature and Cyanobacterial Bloom Biomass Influence Phosphorous Cycling in Eutrophic lake Sediments. Plos One 9 (3), e93130. doi:10.1371/journal.pone.0093130
Chen, X.-P., Chen, H.-Y., Sun, J., Zhang, X., He, C.-Q., Liu, X.-Y., et al. (2018). Shifts in the Structure and Function of the Microbial Community in Response to Metal Pollution of Fresh Water Sediments in Finland. J. Soils. Sediments. 18 (11), 3324–3333. doi:10.1007/s11368-017-1782-5
Crampon, M., Bodilis, J., and Portet-Koltalo, F. (2018). Linking Initial Soil Bacterial Diversity and Polycyclic Aromatic Hydrocarbons (PAHs) Degradation Potential. J. Hazard. Mater. 359, 500–509. doi:10.1016/j.jhazmat.2018.07.088
Dealtry, S., Nour, E. H., Holmsgaard, P. N., Ding, G. C., Weichelt, V., Dunon, V., et al. (2016). Exploring the Complex Response to Linuron of Bacterial Communities from Biopurification Systems by Means of Cultivation-independent Methods. Fems Microbiol. Ecol. 92 (2), fiv157. doi:10.1093/femsec/fiv157
Dong, L. L., and Wen, T. C. (2015). Analysis of Water Quality in Hongfeng Lake, Guizhou. Res. Economi. Environ. Prot. 9, 190–191. doi:10.3969/j.issn.1673-2251.2015.09.156
Dornelas, M., Magurran, A. E., Buckland, S. T., Chao, A., Chazdon, R. L., Colwell, R. K., et al. (2013). Quantifying Temporal Change in Biodiversity: Challenges and Opportunities. P. Roy. Soc. B-biol. Sci. 280, 1750. doi:10.1098/rspb.2012.1931
Edgar, R. C. (2010). Search and Clustering Orders of Magnitude Faster Than BLAST. Bioinformatics 26 (19), 2460–2461. doi:10.1093/bioinformatics/btq461
Feng, J., Shen, X., Chen, J., Shi, J., Xu, J., Tang, C., et al. (2019). Improved Rhizoremediation for Decabromodiphenyl Ether (BDE-209) in E-Waste Contaminated Soils. Soil Ecol. Lett. 1 (3), 157–173. doi:10.1007/s42832-019-0007-9
Gao, X. B., Li, X., Wang, W. Z., and Li, C. C. (2020). Human Activity and Hydrogeochemical Processes Relating to Groundwater Quality Degradation in the Yuncheng basin, Northern china. Int. J. Environ. Res. Public Health 17 (3), 867. doi:10.3390/ijerph17030867
Garcia, S. L., Stevens, S. L. R., Crary, B., Martinez-Garcia, M., Stepanauskas, R., Woyke, T., et al. (2018). Contrasting Patterns of Genome-Level Diversity across Distinct Co-occurring Bacterial Populations. Isme J. 12 (3), 742–755. doi:10.1038/s41396-017-0001-0
Gruca-Rokosz, R., and Tomaszek, J. A. (2015). Methane and Carbon Dioxide in the Sediment of a Eutrophic Reservoir: Production Pathways and Diffusion Fluxes at the Sediment-Water Interface. Water Air Soil Poll 226 (2), 16. doi:10.1007/s11270-014-2268-3
Gu, C. H., Hornberger, G. M., Mills, A. L., Herman, J. S., and Flewelling, S. A. (2007). Nitrate Reduction in Streambed Sediments: Effects of Flow and Biogeochemical Kinetics. Water Resour. Res. 43 (12). doi:10.1029/2007wr006027
Guo, J.-Y., Wu, F.-C., Zhang, L., Liao, H.-Q., Zhang, R.-Y., Li, W., et al. (2011). Screening Level of PAHs in Sediment Core from Lake Hongfeng, Southwest China. Arch. Environ. Contam. Toxicol. 60 (4), 590–596. doi:10.1007/s00244-010-9568-4
Hervé, V., Le Roux, X., Uroz, S., Gelhaye, E., and Frey-Klett, P. (2014). Diversity and Structure of Bacterial Communities Associated withPhanerochaete Chrysosporiumduring wood Decay. Environ. Microbiol. 16 (7), 2238–2252. doi:10.1111/1462-2920.12347
Hoa, T. T. H., Liamleam, W., and Annachhatre, A. P. (2007). Lead Removal through Biological Sulfate Reduction Process. Bioresour. Technol. 98 (13), 2538–2548. doi:10.1016/j.biortech.2006.09.060
Jiang, Y. C., Lu, H. L., Xia, K., Wang, Q., Yang, J. J., Hong, H. L., et al. (2020). Effect of Mangrove Species on Removal of Tetrabromobisphenol A from Contaminated Sediments. Chemosphere 244, 125385. doi:10.1016/j.chemosphere.2019.125385
Johnsen, A. R., and Karlson, U. (2005). PAH Degradation Capacity of Soil Microbial Communities-Does it Depend on PAH Exposure? Microb. Ecol. 50 (4), 488–495. doi:10.1007/s00248-005-0022-5
Jones, F. C. (2008). Taxonomic Sufficiency: The Influence of Taxonomic Resolution on Freshwater Bioassessments Using Benthic Macroinvertebrates. Environ. Rev. 16, 45–69. doi:10.1139/A07-010
Jorgensen, S. L., Hannisdal, B., Lanzen, A., Baumberger, T., Flesland, K., Fonseca, R., et al. (2012). Correlating Microbial Community Profiles with Geochemical Data in Highly Stratified Sediments from the Arctic Mid-ocean Ridge. Proc. Natl. Acad. Sci. 109 (42), E2846–E2855. doi:10.1073/pnas.1207574109
Kaakoush, N. O., Sterzenbach, T., Miller, W. G., Suerbaum, S., and Mendz, G. L. (2007). Identification of Disulfide Reductases in Campylobacterales: a Bioinformatics Investigation. Antonie Van Leeuwenhoek 92 (4), 429–441. doi:10.1007/s10482-007-9171-5
Kirk, M. F., Holm, T. R., Park, J., Jin, Q., Sanford, R. A., Fouke, B. W., et al. (2004). Bacterial Sulfate Reduction Limits Natural Arsenic Contamination in Groundwater. Geol. 32 (11), 953–956. doi:10.1130/G20842.1
Kunz, M. J., Anselmetti, F. S., Wuest, A., Wehrli, B., Vollenweider, A., Thuring, S., et al. (2011). Sediment Accumulation and Carbon, Nitrogen, and Phosphorus Deposition in the Large Tropical Reservoir Lake Kariba (Zambia/Zimbabwe). J. Geophys. Res-biogeo. 116. doi:10.1029/2010jg001538
Lai, C.-Y., Yang, X., Tang, Y., Rittmann, B. E., and Zhao, H.-P. (2014). Nitrate Shaped the Selenate-Reducing Microbial Community in a Hydrogen-Based Biofilm Reactor. Environ. Sci. Technol. 48 (6), 3395–3402. doi:10.1021/es4053939
Li, A., Rockne, K. J., Sturchio, N., Song, W., Ford, J. C., and Wei, H. (2009). PCBs in Sediments of the Great Lakes - Distribution and Trends, Homolog and Chlorine Patterns, and In Situ Degradation. Environ. Pollut. 157 (1), 141–147. doi:10.1016/j.envpol.2008.07.014
Li, F., Peng, Y., Fang, W., Altermatt, F., Xie, Y., Yang, J., et al. (2018a). Application of Environmental DNA Metabarcoding for Predicting Anthropogenic Pollution in Rivers. Environ. Sci. Technol. 52 (20), 11708–11719. doi:10.1021/acs.est.8b03869
Li, J., Crowe, S. A., Miklesh, D., Kistner, M., Canfield, D. E., and Katsev, S. (2012). Carbon Mineralization and Oxygen Dynamics in Sediments with Deep Oxygen Penetration, Lake Superior. Limnol. Oceanogr. 57 (6), 1634–1650. doi:10.4319/lo.2012.57.6.1634
Li, Y., Liang, X., Tang, C., Li, Y., Chen, Z., Chang, S. X., et al. (2018b). Moso Bamboo Invasion into Broadleaf Forests Is Associated with Greater Abundance and Activity of Soil Autotrophic Bacteria. Plant Soil 428 (1-2), 163–177. doi:10.1007/s11104-018-3648-z
Liu, Q., Zhang, Y., Wu, H., Liu, F. W., Peng, W., Zhang, X. N., et al. (2020). A Review and Perspective of eDNA Application to Eutrophication and HAB Control in Freshwater and marine Ecosystems. Microorganisms 8 (3), 417. doi:10.3390/microorganisms8030417
Löffler, F. E., Yan, J., Ritalahti, K. M., Adrian, L., Edwards, E. A., Konstantinidis, K. T., et al. (2013). Dehalococcoides Mccartyi Gen. nov., Sp. nov., Obligately Organohalide-Respiring Anaerobic Bacteria Relevant to Halogen Cycling and Bioremediation, Belong to a Novel Bacterial Class, Dehalococcoidia Classis nov., Order Dehalococcoidales ord. Nov. And Family Dehalococcoidaceae Fam. nov., within the Phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63, 625–635. doi:10.1099/ijs.0.034926-0
Luo, Z. B., Ma, J., Chen, F., Li, X. X., and Zhang, S. L. (2018). Effects of Pb Smelting on the Soil Bacterial Community Near a Secondary lead Plant. Int. J. Env. Res. Pub. He. 15 (5), 1030. doi:10.3390/ijerph15051030
Ma, T., Zhao, N., Ni, Y., Yi, J. W., Wilson, J. P., He, L. H., et al. (2020). China's Improving Inland Surface Water Quality since 2003. Sci. Adv. 6 (1), eaau3798. doi:10.1126/sciadv.aau3798
Maphosa, F., Lieten, S. H., Dinkla, I., Stams, A. J., Smidt, H., and Fennell, D. E. (2012). Ecogenomics of Microbial Communities in Bioremediation of Chlorinated Contaminated Sites. Front. Microbio. 3, 351. doi:10.3389/fmicb.2012.00351
Mayer-Blackwell, K., Azizian, M. F., Green, J. K., Spormann, A. M., and Semprini, L. (2017). Survival of Vinyl Chloride RespiringDehalococcoides Mccartyiunder Long-Term Electron Donor Limitation. Environ. Sci. Technol. 51 (3), 1635–1642. doi:10.1021/acs.est.6b05050
Maymo-Gatell, X., Chien, Y. T., Gossett, J. M., and Zinder, S. H. (1997). Isolation of a Bacterium that Reductively Dechlorinates Tetrachloroethene to Ethene. Science 276 (5318), 1568–1571. doi:10.1126/science.276.5318.1568
Mendes, L. W., Tsai, S. M., Navarrete, A. A., de Hollander, M., van Veen, J. A., and Kuramae, E. E. (2015). Soil-borne Microbiome: Linking Diversity to Function. Microb. Ecol. 70 (1), 255–265. doi:10.1007/s00248-014-0559-2
Michalec, B., and Cupak, A. (2021). Assessment of Quality of Water and Sediments in Small Reservoirs in Southern Poland - A Case Study. Environ. Eng. Res. 27, 200660. doi:10.4491/eer.2020.660
Monchamp, M.-E., Enache, I., Turko, P., Pomati, F., Rîşnoveanu, G., and Spaak, P. (2017). Sedimentary and Egg-Bank DNA from 3 European Lakes Reveal Concurrent Changes in the Composition and Diversity of Cyanobacterial and Daphnia Communities. Hydrobiologia 800 (1), 155–172. doi:10.1007/s10750-017-3247-7
Moreno, J., Fatela, F., Leorri, E., Araújo, M. F., Moreno, F., De la Rosa, J., et al. (2015). Bromine Enrichment in Marsh Sediments as a Marker of Environmental Changes Driven by Grand Solar Minima and Anthropogenic Activity (Caminha, NW of Portugal). Sci. Total Environ. 506-507, 554–566. doi:10.1016/j.scitotenv.2014.11.062
Newton, R. J., Jones, S. E., Eiler, A., McMahon, K. D., and Bertilsson, S. (2011). A Guide to the Natural History of Freshwater lake Bacteria. Microbiol. Mol. Biol. Rev. 75 (1), 14–49. doi:10.1128/Mmbr.00028-10
Norris, P. R., Davis-Belmar, C. S., Brown, C. F., and Calvo-Bado, L. A. (2011). Autotrophic, Sulfur-Oxidizing Actinobacteria in Acidic Environments. Extremophiles 15 (2), 155–163. doi:10.1007/s00792-011-0358-3
Núñez, E. V., Valenzuela-Encinas, C., Alcántara-Hernández, R. J., Navarro-Noya, Y. E., Luna-Guido, M., Marsch, R., et al. (2012). Modifications of Bacterial Populations in Anthracene Contaminated Soil. Appl. Soil Ecol. 61, 113–126. doi:10.1016/j.apsoil.2012.04.005
Pawlowski, J., Apothéloz‐Perret‐Gentil, L., and Altermatt, F. (2020). Environmental DNA: What's behind the Term? Clarifying the Terminology and Recommendations for its Future Use in Biomonitoring. Mol. Ecol. 29 (22), 4258–4264. doi:10.1111/mec.15643
Pawlowski, J., Kelly-Quinn, M., Altermatt, F., Apothéloz-Perret-Gentil, L., Beja, P., Boggero, A., et al. (2018). The future of biotic indices in the ecogenomic era: Integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 637-638, 1295–1310. doi:10.1016/j.scitotenv.2018.05.002
Reddy, C. M., Heraty, L. J., Holt, B. D., Sturchio, N. C., Eglinton, T. I., Drenzek, N. J., et al. (2000). Stable Chlorine Isotopic Compositions of Aroclors and Aroclor-Contaminated Sediments. Environ. Sci. Technol. 34 (13), 2866–2870. doi:10.1021/es9908220
Reddy, M. S., Basha, S., Sravan Kumar, V. G., Joshi, H. V., and Ramachandraiah, G. (2004). Distribution, Enrichment and Accumulation of Heavy Metals in Coastal Sediments of Alang-Sosiya Ship Scrapping Yard, India. Mar. Pollut. Bull. 48 (11-12), 1055–1059. doi:10.1016/j.marpolbul.2003.12.011
Rodriguez, J., Gallampois, C. M. J., Haglund, P., Timonen, S., and Rowe, O. (2021). Bacterial Communities as Indicators of Environmental Pollution by POPs in marine Sediments. Environ. Pollut. 268, 115690. doi:10.1016/j.envpol.2020.115690
Sagova-Mareckova, M., Boenigk, J., Bouchez, A., Cermakova, K., Chonova, T., Cordier, T., et al. (2021). Expanding Ecological Assessment by Integrating Microorganisms into Routine Freshwater Biomonitoring. Water Res. 191, 116767. doi:10.1016/j.watres.2020.116767
Schmalenberger, A., O’Sullivan, O., Gahan, J., Cotter, P. D., and Courtney, R. (2013). Bacterial Communities Established in bauxite Residues with Different Restoration Histories. Environ. Sci. Technol. 47 (13), 7110–7119. doi:10.1021/es401124w
Shi, X. J., Li, L., and Zhang, T. (2015). Water Pollution Control Action Plan, a Realistic and Pragmatic Plan—An Interpretation of Water Pollution Control Action Plan. Envir. Prot. Sci. 41, 1–3. doi:10.16803/j.cnki.issn.1004-6216.2015.03.002
Song, L. T., Liu, C. Q., Wang, Z. L., and Liang, L. L. (2008). Stable Sulfur Isotopic Geochemistry to Investigate Potential Sources and Cycling Behavior of Sulfate in Lake Hongfeng,Guizhou Province. Geochim 37 (6). doi:10.3321/j.issn:0379-1726.2008.06.005
Sun, M. Y., Dafforn, K. A., Johnston, E. L., and Brown, M. V. (2013). Core Sediment Bacteria Drive Community Response to Anthropogenic Contamination over Multiple Environmental Gradients. Environ. Microbiol. 15 (9), 2517–2531. doi:10.1111/1462-2920.12133
Sun, X., Cao, X., Zhao, D., Zeng, J., Huang, R., Duan, M., et al. (2021). The Pattern of Sedimentary Bacterial Communities Varies with Latitude within a Large Eutrophic lake. Limnologica 87, 125860. doi:10.1016/j.limno.2021.125860
Taylor, K. G., and Owens, P. N. (2009). Sediments in Urban River Basins: a Review of Sediment-Contaminant Dynamics in an Environmental System Conditioned by Human Activities. J. Soils Sediments 9 (4), 281–303. doi:10.1007/s11368-009-0103-z
Tian, L., Hu, J., Qin, F., Huang, X., Liu, F., Luo, G., et al. (2011). Distribution of Heavy Metal Elements in the Water Body from Lake Hongfeng. China Environ. Sci. 31 (3), 481–489.
Tolkkinen, M., Mykrä, H., Annala, M., Markkola, A. M., Vuori, K. M., and Muotka, T. (2015). Multi-stressor Impacts on Fungal Diversity and Ecosystem Functions in Streams: Natural vs. Anthropogenic Stress. Ecology 96 (3), 672–683. doi:10.1890/14-0743.1
Vignesh, S., Dahms, H.-U., Emmanuel, K. V., Gokul, M. S., Muthukumar, K., Kim, B.-R., et al. (2014). Physicochemical Parameters Aid Microbial Community? A Case Study from marine Recreational Beaches, Southern India. Environ. Monit. Assess. 186 (3), 1875–1887. doi:10.1007/s10661-013-3501-z
Wang, C., and Huang, G. (1998). Development of SWB-1 Portable Lake Sediment-Interface Water Sampler. Geology-Geochemistry 1, 94–96.
Wang, T., Zhou, Y., Bi, C., Lu, Y., He, G., and Giesy, J. P. (2017). Determination of Water Environment Standards Based on Water Quality Criteria in China: Limitations and Feasibilities. J. Environ. Sci. 57, 127–136. doi:10.1016/j.jes.2016.11.010
Wang, Z. L., Yang, X., and Bai, L. (2011). Isotopic Composition and Potential Sources of Sulfate in Two Water Supply Reservoirs of the Guiyang City, Southwestern China. Fresen. Environ. Bull. 20 (11a), 2984–2991.
Wei, Z., Liu, C., Liang, X., Wang, F., and Wang, S. (2005). Degradation of Organic Matter in the Sediments of Hongfeng Reservoir. Chin.Sci.Bull. 50 (20), 2377–2380. doi:10.1007/BF03183750
Wilkinson, J., Maeck, A., Alshboul, Z., and Lorke, A. (2015). Continuous Seasonal River Ebullition Measurements Linked to Sediment Methane Formation. Environ. Sci. Technol. 49 (22), 13121–13129. doi:10.1021/acs.est.5b01525
Williamson, C. E., Dodds, W., Kratz, T. K., and Palmer, M. A. (2008). Lakes and Streams as Sentinels of Environmental Change in Terrestrial and Atmospheric Processes. Front. Ecol. Environ. 6 (5), 247–254. doi:10.1890/070140
Wu, B. B., Wang, G. Q., Wu, J., Fu, Q., and Liu, C. M. (2014). Sources of Heavy Metals in Surface Sediments and an Ecological Risk Assessment from Two Adjacent Plateau Reservoirs. Plos One 9 (7), e102101. doi:10.1371/journal.pone.0102101
Xiao, S., Wang, Y., Liu, D., Yang, Z., Lei, D., and Zhang, C. (2013). Diel and Seasonal Variation of Methane and Carbon Dioxide Fluxes at Site Guojiaba, the Three Gorges Reservoir. J. Environ. Sci. 25 (10), 2065–2071. doi:10.1016/S1001-0742(12)60269-1
Xie, Y., Fan, J. B., Zhu, W. X., Amombo, E., Lou, Y. H., Chen, L., et al. (2016). Effect of Heavy Metals Pollution on Soil Microbial Diversity and Bermudagrass Genetic Variation. Front. Plant Sci. 7, 755. doi:10.3389/fpls.2016.00755
Xu, X., Wang, P., Zhang, J., Chen, C., Wang, Z., Kopittke, P. M., et al. (2019). Microbial Sulfate Reduction Decreases Arsenic Mobilization in Flooded Paddy Soils with High Potential for Microbial Fe Reduction. Environ. Pollut. 251, 952–960. doi:10.1016/j.envpol.2019.05.086
Yang, T. Q., Liu, H. Y., and Yu, Y. H. (2014). Variation Trend of Water Quality and its Causing Effect of the Hongfneg Lake. Resour. Environ. Yangtze Basin. 23 (s). doi:10.11870/cjlyzyyhj2014z1014
Yang, Y., Zhang, L., Cheng, J., Zhang, S., Li, X., and Peng, Y. (2018). Microbial Community Evolution in Partial Nitritation/anammox Process: From Sidestream to Mainstream. Bioresour. Tech. 251, 327–333. doi:10.1016/j.biortech.2017.12.079
Yu, C., Huang, X., Chen, H., Godfray, H. C. J., Wright, J. S., Hall, J. W., et al. (2019). Managing Nitrogen to Restore Water Quality in China. Nature 567 (7749), 516–520. doi:10.1038/s41586-019-1001-1
Zhang, L., Wang, Z., Cai, H., Lu, W., and Li, J. (2021). Long-term Agricultural Contamination Shaped Diversity Response of Sediment Microbiome. J. Environ. Sci. 99, 90–99. doi:10.1016/j.jes.2020.06.013
Zhang, Y., Zhang, L., Huang, Z., Li, Y., Li, J., Wu, N., et al. (2019). Pollution of Polycyclic Aromatic Hydrocarbons (PAHs) in Drinking Water of China: Composition, Distribution and Influencing Factors. Ecotoxicology Environ. Saf. 177, 108–116. doi:10.1016/j.ecoenv.2019.03.119
Keywords: bacterial community, sediment profile, bacterial index, bacterial indicator, historical water quality
Citation: Sun J, Chen X, Yu J, Chen Z, Liu L, Yue Y, Fu Z, Yang M and Wang F (2021) Deciphering Historical Water-Quality Changes Recorded in Sediments Using eDNA. Front. Environ. Sci. 9:669582. doi: 10.3389/fenvs.2021.669582
Received: 19 February 2021; Accepted: 17 May 2021;
Published: 15 June 2021.
Edited by:
José Francisco Gonçalves Júnior, University of Brasilia, BrazilReviewed by:
Rafael Dettogni Guariento, Federal University of Mato Grosso do Sul, BrazilEllard R. Hunting, University of Bristol, United Kingdom
Simone Jaqueline Cardoso, Juiz de Fora Federal University, Brazil
Copyright © 2021 Sun, Chen, Yu, Chen, Liu, Yue, Fu, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueping Chen, eHBjaGVuQHNodS5lZHUuY24=; Ming Yang, bWluZ3lhbmdAc2h1LmVkdS5jbg==
 Jing Sun1,2
Jing Sun1,2 Xueping Chen
Xueping Chen Zheng Chen
Zheng Chen Yihong Yue
Yihong Yue Ming Yang
Ming Yang