- 1Institue for Chemistry and Biology of the Marine Environment, University of Oldenburg, Oldenburg, Germany
- 2German Centre for Marine Biodiversity, Senckenberg am Meer, Wilhelmshaven, Germany
Subterranean estuaries (STEs) play an important role in linking nutrient cycling between marine and terrestrial systems. As being the primary drivers of nutrient cycling, the composition of microbial communities and their adaptation toward both, terrestrial and marine conditions are of special interest. While bacterial communities of STEs have received increasing scientific attention, archaeal and meiofaunal diversity was mostly neglected. Previous studies at the investigated sampling site, the STE of a mesotidal beach at the German North Sea island of Spiekeroog, focused on spatial and seasonal patterns of geochemical and bacterial diversity. By additionally investigating the archaeal and meiofaunal diversity and distribution, we now aimed to fill this gap of knowledge to understand the microbial response to submarine groundwater discharge (SGD). The topography of Spiekeroog beach and associated geochemical gradients in porewater displayed a distinct cross-shore zonation, with seawater infiltration on the upper beach at the high water line (HWL), and saline and brackish porewater exfiltration (SGD) at the ridge-runnel structure and the low water line (LWL) on the lower beach. This led to a higher evenness of prokaryotic communities in lower beach areas impacted by SGD compared to unimpacted areas. Archaea contributed 1–4% to the 16S rRNA gene sequence dataset. Those were dominated by Nitrosopumilaceae, corresponding well to higher concentrations of NH4+ in the discharge area of the STE. The unimpacted sites had elevated abundances of Wosearchaeia, which were also detected previously in impacted areas of an STE at Mobile Bay (Gulf of Mexico). While a large proportion of prokaryotes were present in the entire intertidal area, meiofaunal community compositions were site specific and dominated by nematodes. Nematode communities of the high-water line differed distinctively from the other sites. Overall, our data indicates that the three domains of life display distinctly different adaptations when facing the same conditions within the STE. Therefore, distribution patterns of any domain can only be understood if all of them, together with basic environmental information are investigated in an integrated context.
Introduction
Sandy beaches are highly permeable, advection driven systems (Anschutz et al., 2009), receiving labile organic matter (OM), oxygen, sulfate (Huettel et al., 2014) and seasonally nitrate (Ahrens et al., 2020) by seawater infiltration. Therefore, beaches have been compared to bioreactors exhibiting high microbial activity, despite their relatively low OM content (Anschutz et al., 2009). An extreme example are high energy beaches, which are prone to strong physical forcing by waves and tides, enhancing the recharge of nutrients and substrates to deeper sediment depths. This increases the magnitude of exchange between sediments and the coastal ocean compared to less permeable sediments, or those with a lower energy input (Webster and Taylor, 1992; Reckhardt et al., 2015). Additionally, constant sediment reallocation prevents stable biogeochemical conditions, resulting in a weaker redox zonation than usually observed along the depth profile of marine sediments (Froelich et al., 1979).
Another process impacting the redox zonation of coastal sediments around the globe is submarine groundwater discharge (SGD) (Moore, 1996; Paerl, 1997; Santos et al., 2009; Rodellas et al., 2014; Anschutz et al., 2016; Cho et al., 2018). SGD introduces fresh meteoric groundwater as well as recirculated sea water into the marine sediments, lowering the salinity and introducing reduced chemical compounds like ammonium and dissolved iron (Moore et al., 2008; Kwon et al., 2014). These compounds are either re-oxidized along the flow path or discharged into the coastal ocean (Riedel et al., 2011; Reckhardt et al., 2015; Beck et al., 2017; Kim et al., 2017). The outflow marks the discharge area of the so called subterranean estuary (STE) which exhibits a horizontal redox zonation toward the low water line (Moore, 1999). The constant resupply of oxygen and labile OM from the ocean in combination with a steady supply of reduced chemicals by SGD creates niches for many microorganisms. Thus, mixing of terrestrial and marine constituents within the STE leads to elevated microbial activities (Anschutz et al., 2009; Gao et al., 2012; Charbonnier et al., 2013) and result in physiological diversity maxima in the oxic-anoxic transition zone (Hong et al., 2018).
Several microbiological studies have been conducted on sandy beaches from regional (Boehm et al., 2014) to global scales (Staley and Sadowsky, 2016), detecting a cosmopolitan core within the bacterial communities of sandy beaches. Site specific studies on bacterial diversity in sandy STE’s reported contrasting findings mostly caused by differences in the physical energy input. On the one hand, STE’s discharging at low energy beaches exhibit horizontal redox zonation with bacterial communities adapted to the different zones (McAllister et al., 2015). On the other hand, at high energy beaches, this zonation is constantly disrupted and thus pronounced weaker. Most likely, this physical reworking also overprints visible effects of SGD on the bacterial communities. In our previous study on the bacterial diversity at the high energy beach of Spiekeroog Island (Northern Germany), we have shown that these dynamics appear to result in bacterial communities of adapted generalists (Degenhardt et al., 2020).
Except for a recent study by Adyasari et al. (2020), almost all previous investigations focused on bacterial communities of sandy beaches and STEs, neglecting archaeal diversity. In the last decade, knowledge about archaeal diversity was greatly enhanced by cultivation independent techniques like single-amplified genomes (SAG) or metagenomics (Brochier-Armanet et al., 2011; Rinke et al., 2013). Ecological studies started to shed light on distribution patterns (Auguet et al., 2010; Hugoni et al., 2015; MacLeod et al., 2019), yet knowledge about the ecological roles of most Archaea remains scarce (Meng et al., 2014; Castelle et al., 2015). Aquatic Archaea are known from a wide variety of habitats like costal (Rios-Del Toro et al., 2018) and deep sea sediments (Lloyd et al., 2013), freshwater (Ortiz-Alvarez and Casamayor, 2016) and pelagic environments (Karner et al., 2001). In most benthic environments, they are less abundant than bacteria, yet their numbers seem to increase with sediment depth (Lipp et al., 2008). Physiologically, they are known for their role in processes like methanogenesis (Thauer et al., 2008) but also in ammonia oxidation (Könneke et al., 2005), anaerobic ammonia oxidation (Rios-Del Toro et al., 2018), or anaerobic oxidation of methane (Boetius et al., 2000). Their involvement in the nitrogen cycle and elevated abundances in oxygen minimum zones (Rios-Del Toro et al., 2018; Muck et al., 2019) might make them important players at the oxic-anoxic interface of STE’s. Information on archaeal diversity of Spiekeroog beach will broaden the knowledge about their role in shaping biogeochemical processes within the STE and the intertidal area of a high energy beach.
Another often overlooked fraction of interstitial life are eukaryotes belonging to the so called meiofauna, which is one of the most diverse group of small organisms, unicellular protists and multicellular metazoans that live in aquatic sediments (Giere, 2009). Meiofauna is a collective name for organisms of 20 different phyla that pass through a 1 mm mesh sieve and are retained in a 32 μm sieve. Inhabiting various marine, brackish and freshwater environments (Higgins and Thiel, 1988; Giere, 2009), they are the most suitable candidates in terms of monitoring ecological changes (Zeppilli et al., 2014) and serve as indicators for environmental variabilities (Jessup et al., 2004; Nascimento et al., 2012; Bonaglia et al., 2014). Limited body size and mobility make those organisms very susceptible to changes in sediment and porewater biochemistry, which may influence their abundance and diversity (Kennedy and Jacoby, 1999). The majority of meiobenthic communities in most marine habitats consists of nematodes and harpacticoid copepods (Gallucci et al., 2012; Losi et al., 2013; Semprucci et al., 2015). Their communities are uniquely sensitive to temperature, pH, dissolved oxygen, redox potential and heavy metal content of the sediment matrix (Moreno et al., 2011; Vanaverbeke et al., 2011; Meadows et al., 2015). Thus, nematodes have been identified as indicators of environmental pollution and dynamic changes in marine systems (Losi et al., 2013; Semprucci et al., 2015). They were frequently reported by both, morphological and molecular studies, as the most abundant taxa among marine meiobenthos communities followed by harpacticoids (Huys et al., 1992; Karanovic and Cooper, 2012; Zeppilli et al., 2015; de Faria et al., 2018). Despite their importance, meiofauna were included only recently into ecosystem functioning investigations, due to difficulties arising from the small size of the examined animals. This limits differentiation on the species level to a few highly skilled taxonomists (Huys, 1996). Furthermore, due to their high abundance and diversity, working through each sample is very time-consuming, making a high sample throughput difficult. Additionally, high-throughput sequencing techniques for meiofauna have only emerged in the past few years and lack behind on respective studies on prokaryotic diversity (Taberlet et al., 2012; Haenel et al., 2017).
As SGD shapes the physical environment by altering (i) grain size distribution of bottom sediments and (ii) altering the vertical position of redox and pH gradients, it affects the distribution of meiofauna within the sediments (Miller and Ullman, 2004; Zipperle and Reise, 2005). However, investigations of SGD-affected areas have shown variable community responses with regards to of diversity and abundance. Studies from the coast of France and Portugal e.g., found no impact of SGD on meiofaunal abundance (Ouisse et al., 2011), but enhanced metabolism (Migné et al., 2011) and increased diversity (Encarnação et al., 2013). Contrastingly, in a study from the Polish coast lower abundances were found in areas influenced by SGD as well as distinctively different communities between affected and unaffected sites (Kotwicki et al., 2014). In an investigation at the coast of Long Island, a lower diversity among nematode genera was detected at impacted sites compared to a generally higher diversity in areas unimpacted by SGD (Grzelak et al., 2018). Overall, these studies show that SGD does influence meiofaunal community structure, yet in different ways depending on the characteristics of the respective sites. However, all abovementioned studies from SGD affected areas were based on morphological investigations, while no molecular studies are available, yet.
In the present study, we attempt to cover the entire microbial diversity in the discharge area of a shallow STE and to unravel putative interactions between all three domains of life. The study site is located at Spiekeroog Island, which harbors a freshwater lens that fuels the STE. The discharge area at Spiekeroog beach has previously been subject of intensive multidisciplinary research (Beck et al., 2017; Waska et al., 2019b; Degenhardt et al., 2020; Grünenbaum et al., 2020a,b). Investigations on its hydrogeology, porewater geochemistry and bacterial diversity gave a solid insight into the coupling of benthic processes. The strongest gradients of porewater chemistry within the discharge area of the STE were found in cross-shore direction (Waska et al., 2019b). The whole discharge area of the beach was divided into multiple zones of pore water in- and exfiltration which were not fixed in position throughout the year. The high water line (HWL) and the low water line (LWL) were physico-chemically most distinct, likely due to relatively stable in- and exfiltrating conditions. The zones in between showed strong spatial and temporal heterogeneity, possibly due to extensive morphological changes of the beach, which in turn influenced their hydrochemistry (Waska et al., 2019b). Seasonal variations of seawater constituents were found to influence the pathways of bacterial organic matter degradation (Ahrens et al., 2020). During colder seasons, porewater conditions within the STE were predominantly oxic or nitrate reducing and shifted to manganese and iron reduction during warmer seasons. As a result, the discharge area changed from a dissolved inorganic nitrogen (DIN) sink in spring to a source in late summer. Bacterial communities within those sediments were shown to be highly uniform down to a depth of 1 m and over the whole intertidal area (Degenhardt et al., 2020). Since this was observable throughout the year, this assemblage has been dubbed the bacterial core community of the investigated sediments.
Based on these findings, we revisited Spiekeroog beach in March 2019 and sampled the same cross-shore transect as investigated previously (Waska et al., 2019b; Ahrens et al., 2020; Degenhardt et al., 2020), in order to fill the knowledge gaps on the entire microbial diversity. We hypothesize that (i) there is a large archaeal and meiofaunal core community and (ii) the conditions in the discharge area of the STE favor the presence of those Archaea involved in nitrogen cycling. We further hypothesize that (iii) the harsh high-energy conditions at Spiekeroog beach result in a nematode dominated community over a copepod dominated community. In general, our investigation will enhance the understanding of correlations between all three microbial domains within the intertidal area.
Methods
Site Description
The Island of Spiekeroog is part of the East Frisian Barrier Island chain located in the German North Sea, with the Wadden Sea in its back barrier region. The island harbors a freshwater lens, which is formed by rain and separated from the surrounding sea water by a density gradient. The freshwater lens partially discharges through a subterranean estuary within the intertidal area along the northern shore of the island. This intertidal area is located on a sandy, mesotidal, high energy beach with a runnel-ridge system. A schematic of the investigated area is shown in the supplements (Supplementary Figure 1). The runnel-ridge system is not stationary but frequently relocated within the intertidal area due to wind and wave action. Areas above the mean sea-level are characterized as sea-water infiltration zones, while areas below the sea level are net discharge areas of the STE (Grünenbaum et al., 2020a). Due to the aforementioned strong morphodynamics, these in- and exfiltration areas are not static and the discharge is more diffusive than observed for other STEs. Detailed descriptions of hydrogeochemistry (Beck et al., 2017; Waska et al., 2019b; Grünenbaum et al., 2020a,b), geochemistry (Ahrens et al., 2020) and bacterial diversity (Degenhardt et al., 2020) can be found in previous studies investigating the Spiekeroog STE.
Sediment Sampling
Samples were collected in March 2019 at the North beach of Spiekeroog Island. Sediment cores were recovered with aluminum core liners (80 mm diameter) along a transect between the high- and low water line. Four cores were taken along the transect at the specific topographical features: the HWL, runnel, ridge and LWL. The cores were subsampled for prokaryotic diversity analyses at 0–2, 10–12, 30–32, 50–52, and 100–102 cm depth using sterile cut-off syringes. The different depths will be referred hereafter as 0, 10, 30, 50, and 100 cm. Samples were stored frozen at −20°C until further processing. Meiofauna was sampled individually by transferring sediments of the upper 10 cm taken in close vicinity of the long sediment cores into plastic containers. Five replicates were fixed with ethanol, another five were fixed with a solution containing dimethyl sulfoxide, disodium EDTA, and saturated NaCl (DESS) (Yoder et al., 2006). The ethanol fixed samples were re-fixed after 12 h and all samples were stored at 4°C until further processing.
Porewater samples were obtained by using stainless steel push-point samplers and pre-rinsed polyethylene syringes, and were processed according to Ahrens et al. (2020) for NO3–, NO2–, NH4+, Fe, and Si. PO43– was determined photometrically, following the methods of Itaya and Ui (1966) for samples with concentrations below 2.1 μM and Laskov et al. (2007) for samples exceeding 2.1 μM PO43–. DOC samples were treated and processed according to Waska et al. (2019a).
In order to extract the meiofauna, sediment samples were sieved through a 32 μm sieve and centrifuged using Kaolin and Levasil (Kurt Obermeier GmbH & Co., KG, Bad Berleburg, Germany) following McIntyre and Warwick (1984). A total number of 150 nematode specimens (when available) were sorted out from each meiofauna sample in one separate vial for each sample. In case of samples with less than 150 specimens, all nematodes were taken into account. The remaining meiofauna was filtered using glass filters (1.6 μm particle size). Ethanol was used to wash the extracted meiofauna from DESS residues. The filters (containing meiofauna) were dried using a speed-vacuum system for 1 h at 45°C and stored in a sterile 1.5 ml Eppendorf mini-tube for DNA extraction.
DNA-Extraction, Amplification, and Sequencing
Genomic DNA for prokaryotic samples was extracted by using a modified phenol-chloroform protocol (Gabor et al., 2003; Lueders et al., 2004). A one step PCR attaching Illumina adaptors and barcodes was performed in 50 μl setups. The master mix contained 34.4 μl PCR grade water, 10 μl Phusion-Polymerase Buffer, 0.75 μl (10 pmol/μl) of each primer published by Parada et al. (2016) 0.2 μL MgCl2 (50 mM), 1 μl BSA (25 mg/ml), 0.4 μl dNTPs (10 mM each) and 0.5 μl Phusion Polymerase (1 U/μl) per reaction. Reactions were performed in 96 well plates (Eppendorf) using 2 μl template. Cycler settings included an activation step at 98°C for 1 min, followed by 32 cycles of denaturation at 98°C or 45 s, annealing at 50°C for 45 s, elongation at 72°C for 30 s and a final elongation step at 72°C for 5 min.
DNA extraction from meiofauna and nematode samples was carried out using the E.Z.N.A.® Mollusc DNA Kit (Omega BIO-TEK) following their protocol. The genomic DNA was eluted in 100 μl of nuclease-free water for each library (sample). Two PCRs were performed to amplify the V1 and V2 hypervariable regions of the 18S rRNA from each meiofauna and nematode library and bind the Illumina Unique Dual Nextera XT Indexes and compatible adapters. The first PCR was performed using the SSU universal primers F04 and R22 (Blaxter et al., 1998) tagged by the first part of the Illumina adapters in total volume of 20 μl containing 10 μl of Phusion Green Hot Start II High-Fidelity PCR Master Mix (ThermoFisher), 0.5 μl of each primer (10 pmol/μl), 2 μl of genomic DNA and 7 μl of molecular grade water. Cycler settings for the first PCR encompassed an initial denaturation step at 98°C for 2 min, 20 cycles of denaturation at 98°C for 15 s, annealing at 60°C for 30 s, elongation at 72°C for 30 s and a final elongation at 72°C for 2 min. Five microliter of the first PCR products were purified using 2 μl of ExoSAP-IT PCR Product Cleanup Reagent (ThermoFisher). A two-step PCR was carried out to bind the Nextera XT Indexes and Illumina adapter overhang according to Illumina 16S Metagenomic Sequencing Library Preparation guide (15044223Rev.B), using 7 μl of DNA template (purified first PCR product), 10 μl of Phusion Green Hot Start II High-Fidelity PCR Master Mix, 1 μl of molecular grade water and 1 μl of each index primers from Nextera XT Indexes. The cycler setting contained an initial denaturation step at 98°C for 2 min, 15 cycles of denaturation at 98°C for 15 s, annealing and elongation at 72°C for 35 s, and a final elongation at 72°C for 2 min. The PCR products were checked on a 1.5% agarose gel containing 1% GelRed to check the amplification and target fragment length. Two to ten microliter of the 2nd PCR products have been pooled into one Eppendorf tube and purified using AMPure XP Beads for PCR Purification (Beckman Coulter) in the volume of 60%. The pooled library was denatured and 20% PhiX control was added before performing a test run using the MiSeq Reagent Nanokit v2 (150-cycles paired end), and a final sequencing run with the MiSeq Reagent kit v3 (300-cycles paired end). Sequencing was performed at the DZMB Metabarcoding lab in Wilhelmshaven (Germany) on an Illumina MiSeq platform. The sequences were deposited at the European Nucleotide Archive under the accession number PRJEB42988.
Bioinformatics and Statistical Analysis of Prokaryotic Sequence Data
Prokaryotic 16S rRNA gene sequences were processed according to the pipeline of Yeh et al. (2019). First, primers were trimmed from reads using cutadapt and all reads missing forward or reverse primer sequences were discarded. The error threshold within the primer sequence was set to 20% to account for mismatches to the primers, primer sequences including higher error rates were discarded. Trimmed reads were matched against a curated 16S/18S database originating from SILVA132 (Quast et al., 2013) and PR2 (Guillou et al., 2013). Eukaryotic sequences were discarded.
All 16S rRNA gene sequences were further processed using QIIME2 (Bolyen et al., 2019) and its q2-dada2 plugin. Low quality ends of the reads were removed and denoised using DADA2 (Callahan et al., 2016). After removal of chimeric reads, prokaryotic sequences were classified using the classify-sklearn plugin of QIIME2 against a curated SILVA132 database, which was fitted to the V4-V5 primer region. Plastidal sequences were reclassified using the PhytoRef database (Decelle et al., 2015). Finally, count tables were exported from QIIME for further statistical analysis. To account for erroneous sequences, amplicon sequence variants (ASVs) were filtered according to Milici et al. (2016): Only those ASVs that had an abundance of >0.001% within the complete dataset and (i) were present in relative abundance of >1% in at least one sample or (ii) had an abundance of >0.1% within a sample in at least 2% of all samples or (iii) were present in >5% of all samples in any abundance were kept. This strict abundance filter was mainly used because DADA2 retained lots of very rare sequences that inflated the amount of ASVs found in each sample. All further analyses were based on this dataset. Samples were rarefied to 14,885 counts for alpha-diversity analyses, which was the lowest count number found among all samples. Barplots were drawn with an abundance threshold of 0.5% by which groups were sorted into “others.” Principal coordinate analysis (PCoA) was performed using Bray-Curtis transformed community distances. Venn-Diagrams were used to show the separation of ASV richness between locations. All analyses were done using R statistical software (R Core Team, 2019) and its package vegan (Oksanen et al., 2019).
Bioinformatics and Statistical Analysis of Eukaryotic Sequence Data
The NGS reads were demultiplexed by indexes and trimmed from the sequencing primers prior to analyses using BBmap. The DADA2 pipeline (Callahan et al., 2016), a divisive amplicon de-noising algorithm for correcting Illumina errors, was used to analyze the sequence reads from meiofauna and nematode libraries in which the Illumina reads were de-noised based on the error rate of the data. They were filtered by length and quality scores, chimera detected and de-replicated to high resolution ASVs. The ASVs were further processed using a custom script (SGN Metabarcoding pipeline) where all sequences shorter than 320 bp were eliminated and each ASV was blasted against the database NCBI in which the 10 best blast hits were retrieved and pooled with our own 18S reference library (available from DNA barcode archive of the DZMB institute). This merged dataset was used as final blast database to assign the best and closest taxonomical assignment to each ASV. The ASV table containing taxonomical assignments, percentage identities, query coverage and number of reads per library were analyzed using a custom post-processing script Dada2pp implemented in the R statistical software and by using the vegan package for eliminating contaminations, short or non-target sequences, determining target meiofaunal groups and detailed diversity analyses and comparisons. Above mentioned analyses were performed on nematode sequence libraries at the genus level.
Results
A total of 20 sediment samples from four designated stations of the intertidal area of the North beach of Spiekeroog Island were analyzed to identify the bacterial and archaeal diversity by next generation sequencing (NGS). Those were taken in cross-shore direction at the HWL, the runnel, ridge and the LWL at five different depths (0, 10, 30, 50, and 100 cm). Additionally, shorter cores (0–10 cm) were taken in five replicates at each location to investigate meiofaunal diversity. As microscopic investigation revealed the dominance of nematodes within the meiofaunal community, those were analyzed separately.
Porewater Geochemistry of the Beach
The discharge areas of the Spiekeroog beach STE were marked by brackish salinities at the ridge and the LWL (Figure 1). This was complemented by elevated concentrations in total dissolved nitrogen (TDN), NH4+, PO43–, Si and Mn at the LWL in comparison to the other sampling sites. Additionally, porewaters from deeper sediment depths of the ridge (30 and 100 cm) had elevated concentrations of Fe(II), indicating reducing conditions. Porewaters from the LWL and the HWL were generally low in Fe(II), as well as in NO3– and NO2–. The shallow porewaters from the runnel (0 and 10 cm) were characterized by higher amounts of NO3– and slightly elevated concentrations of DOC in comparison to the other sites. This can be interpreted as a sign for infiltrating conditions. Since some sediments were too dry to obtain porewaters for the respective analyses, they were marked by gray bars in the heatmap (Figure 1).
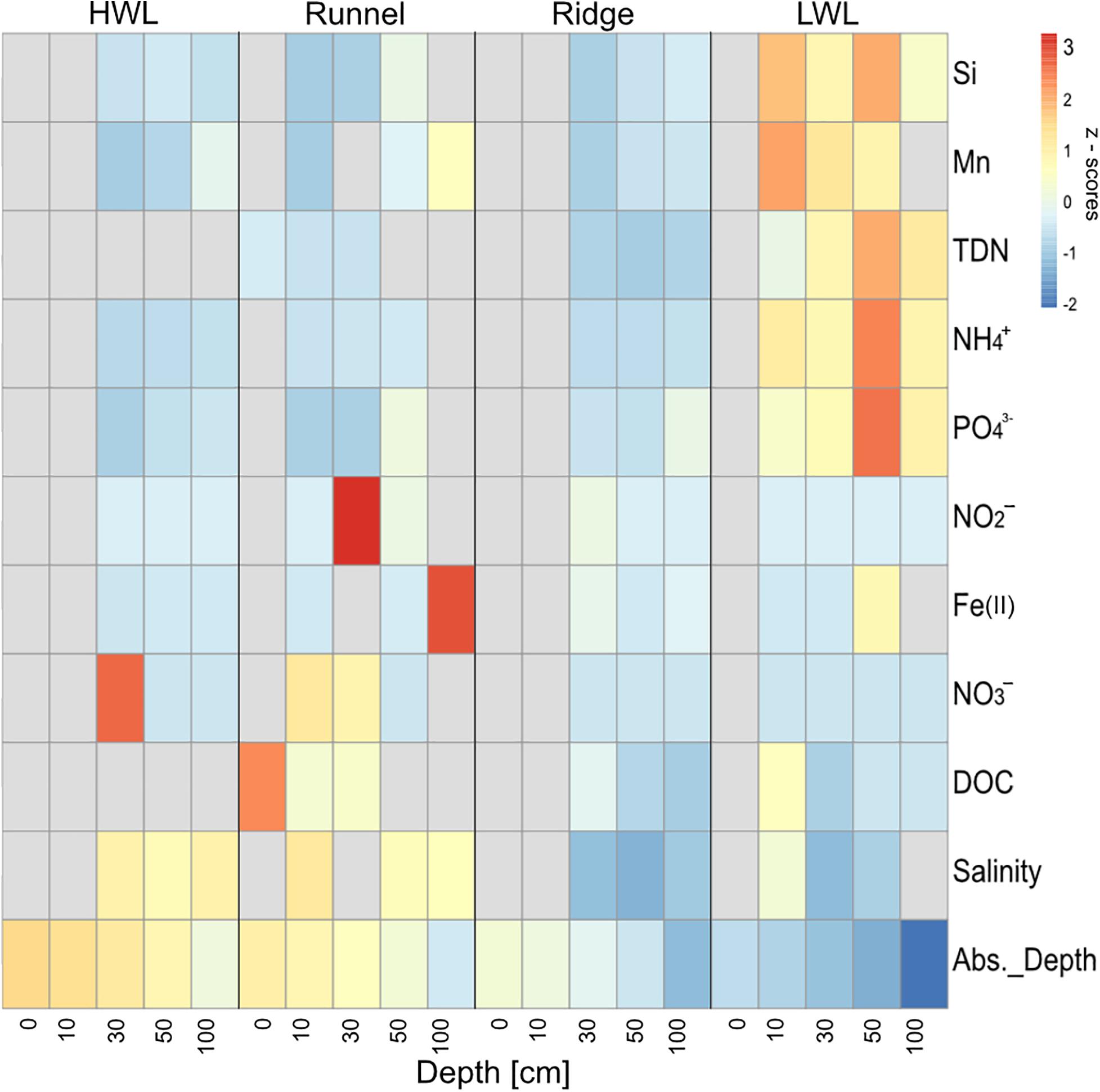
Figure 1. Normalized porewater constituents (z scores) across the transect. Colder colors indicate lower, warmer colors higher z scores, respectively. Gray bars indicate missing values caused by porewater scarcity. depth, absolute depth above or below sea level; DOC, dissolved organic carbon; TDN, total dissolved nitrogen.
Composition of the Sequence Data Set
Sequencing of prokaryote samples resulted in a total of 3,079,547 reads, recovering a total diversity of 13,568 different ASVs. After applying the abundance filter, the majority of 2,967,880 reads was left representing a final diversity of 6944 ASVs within the dataset. Samples were deeply sequenced with a mean of 148,394 reads per sample. Amplicon sequencing of the V1 and V2 hypervariable region of 18S rRNA gene yielded a total of 3,537,703 reads and 1,263 ASVs belonging to 50 eukaryote groups based on NCBI taxonomic assignment. After filtering a total of 3,408,572 reads and 846 ASVs were classified as 11 target meiofaunal groups. From the nematode samples, 18 NGS libraries yielded a total of 2,214,455 reads and 1,455 ASVs where 1,680,256 reads and 255 ASVs were detected as nematode encompassing 43 different species and 25 genera. Two libraries were excluded from the nematode dataset as they did not contain enough sequencing reads.
Microbial Diversity Patterns
The presence and absence of ASVs at the different stations were compared to identify horizontal patterns among prokaryotic and eukaryotic community compositions along the investigated transect (Figure 2). For prokaryotes, a large proportion of ASVs was shared among all stations (14%), which is in accordance with the previously detected bacterial core community (Degenhardt et al., 2020). Yet, every station still harbored a number of site specific ASVs (10–16%), which was largest among the HWL samples (16%). In contrast, meiofaunal communities only shared a small amount of ASVs among all stations (3%) and large proportions of site specific ASVs at every station. The highest number of unique ASVs was detected in the runnel (26%) followed by the ridge (24%), the LWL (16%) and the HWL (15%), respectively. In comparison to all other sites, the percentage of ASVs shared between runnel and ridge was comparably high (6%).
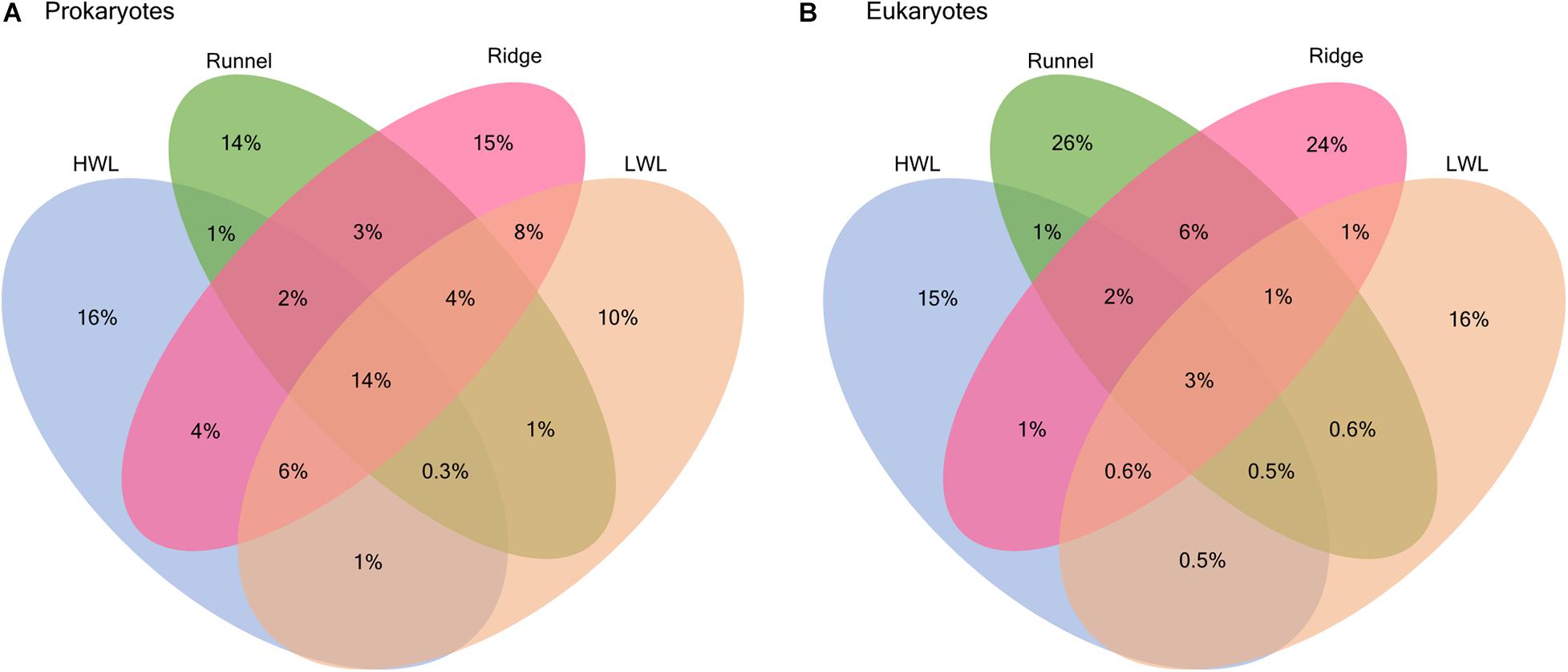
Figure 2. Venn diagrams showing the percentage of shared ASVs between different stations for (A) prokaryotic and (B) eukaryotic communities.
Principle coordinate analysis (PCoA) distance matrices were used to compare community compositions across the intertidal area (Figure 3). The prokaryotic community structures differed between the individual sampling sites. Especially, all samples taken at the runnel clustered apart from the other sites yet were quite dissimilar among each other. In contrast, the LWL and the ridge communities were highly similar. The HWL community exhibited large differences with depth, while all other sites showed no coherent vertical patterns. The eukaryotic communities also showed a strong site-specific ordination within the PCoA. Here, all communities of the LWL replicates were rather similar, which was also the case for the communities of the runnel replicates, yet they were forming groups clearly separated from the rest. Both, the ridge and HWL communities showed a higher scatter among the replicates.
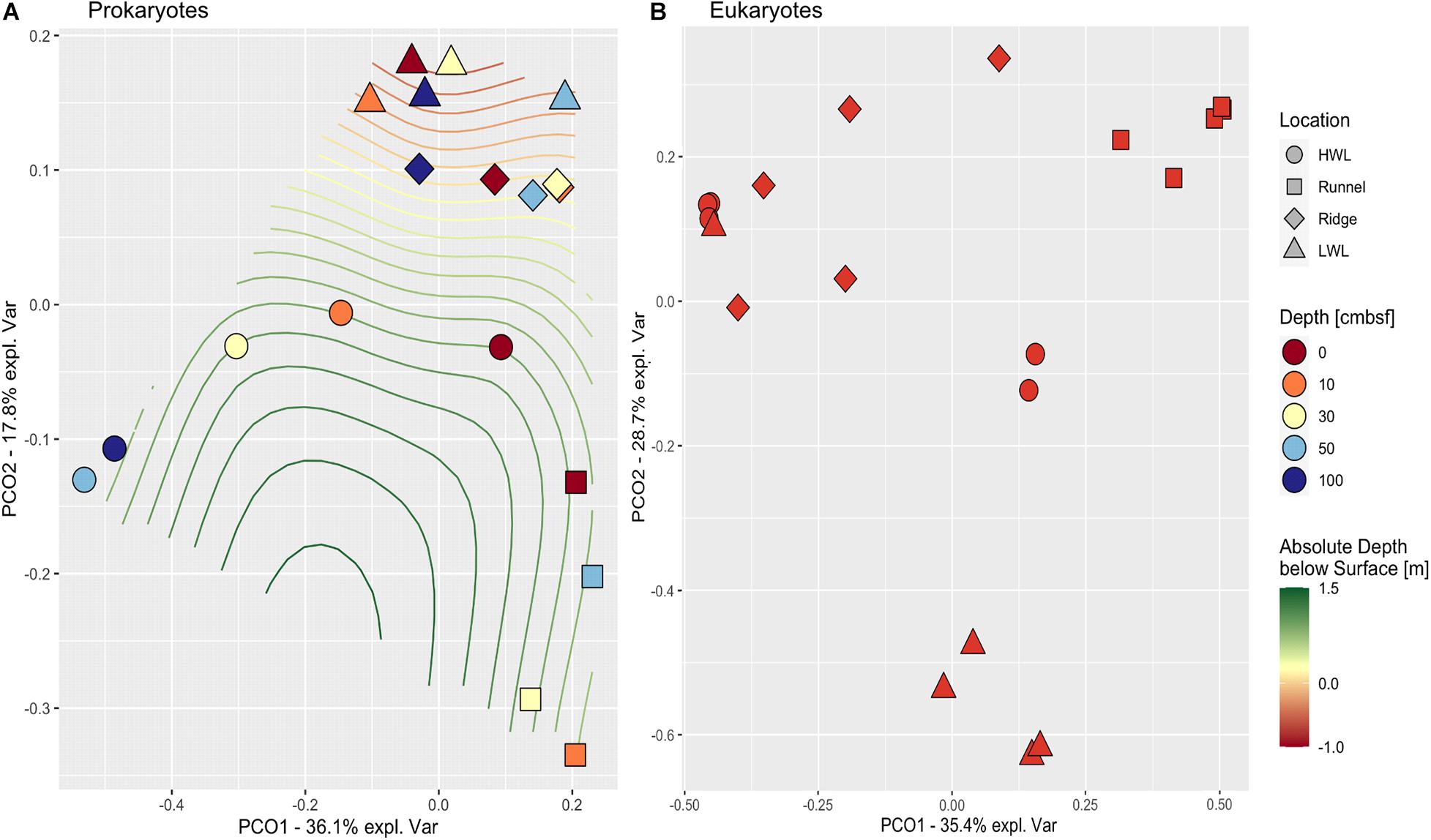
Figure 3. PCoA of prokaryotic (A) and eukaryotic (B) communities. The PCoA plots depict the similarities between samples based on community composition comparing the four locations (symbols) and depths (colors). Absolute depth above or below sea level is indicated by isoclines. Depth is showed in centimeters below sea floor (cmbsf).
Prokaryotic Community Composition
The dominant part of the prokaryotic community was composed of 11 different bacterial phyla. Yet, 25–40% of the community were attributed to rare taxa, here summarized as “others” (Figure 4A). Those include most Archaea due to their low abundances within the dataset. Summarized, archaeal relative abundances reached 1–4%. The dominant phyla exhibiting by far the largest amount of internal diversity, were the Bacteroidetes and the Proteobacteria, represented by 12 genera, each. Within the Proteobacteria, most ASVs were related to Deltaproteobacteria, indicating sub- to anoxic conditions. The only other phyla represented by more than one genus were the Acidobacteria and Planctomycetes, represented by two genera, each. The other bacterial phyla detected were Actinobacteria, Calditrichaeota, Gemmatimonadetes, Kiritimatiellaeota, Latescibacteria, and Nitrospirae. The only archaeal phylum detected with an abundance above our filter criteria were Thaumarchaeota of the family Nitrosopumilaceae. Differences between the sampling sites could be explained by varying abundances of individual taxa. The HWL differed mostly due to higher relative abundances of Bacteroidetes, which additionally increased in abundance with depth. This increase was attributed to the genera Muriicola, Portibacter and unknown members of the Rhodotermaceae and Saprospiraceae. These genera were also detected at the other sites, but in lower relative abundances. The distinct community composition of the runnel samples can be attributed to increased relative abundances of members of the Proteobacteria and Sediminicola (Bacteriodetes). Surprisingly, most members of this class were represented by ASVs of unknown genera with the exceptions of Woseia and Cupriavidus. The latter was present in high relative abundances, which was unexpected since that genus is not typically found in marine sediments. The community composition of the LWL and the ridge samples appeared to be an intermediate version of the two other sites.
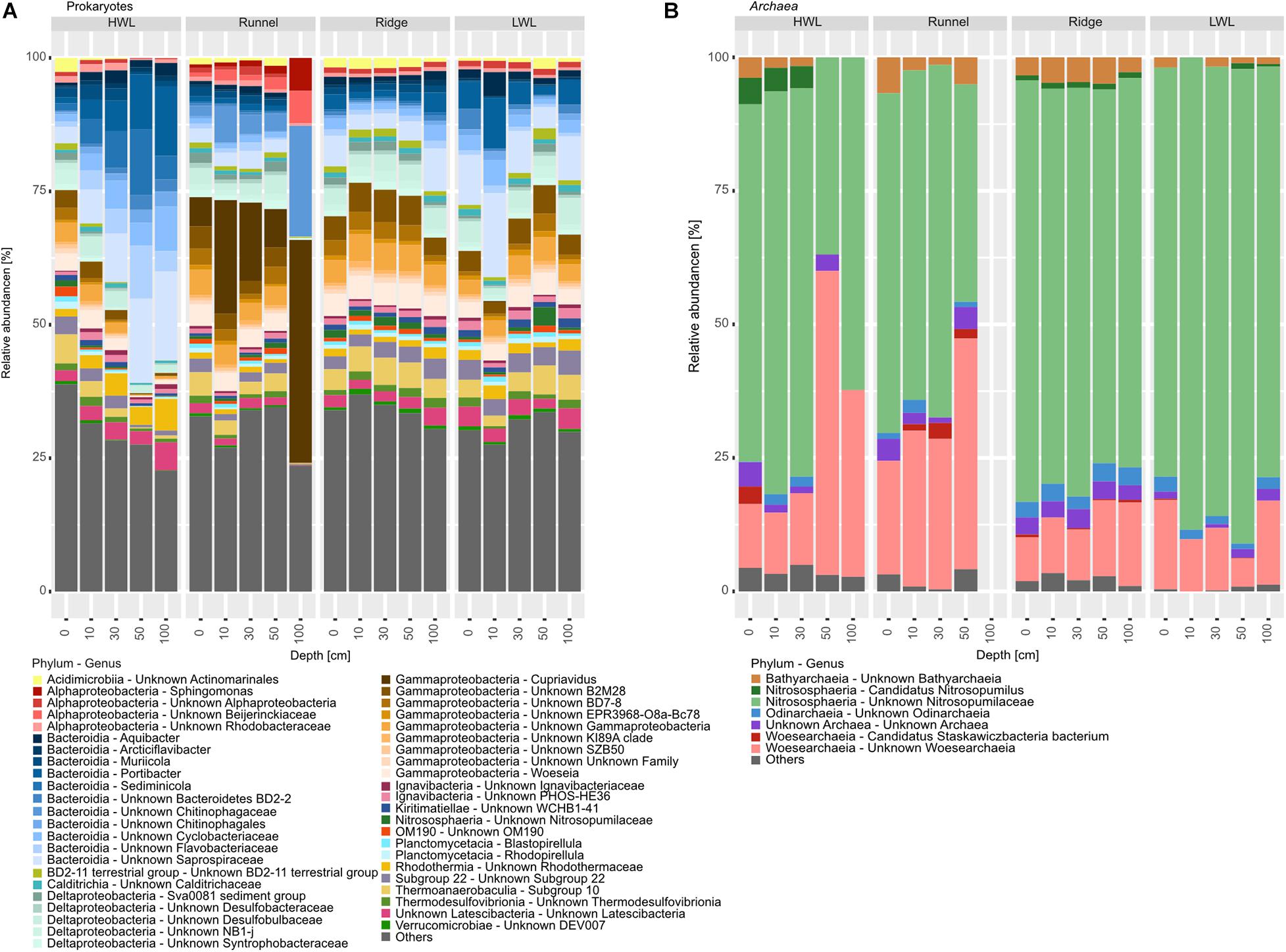
Figure 4. Barplots of prokaryotic communities along the transect depicted on phylum and genus level. (A) Shows total prokaryotic diversity, while (B) zooms into the Archaeal diversity, due to their low relative abundances. Taxa below the abundance threshold were summarized as “others.”
Despite their low relative abundances, the intra-archaeal diversity could be unraveled, due to deep sequencing (Figure 4B). The community was dominated by ASVs related to unknown Nitrosopumilaceae, with relative abundances between 60 and 80% of all detected Archaea. The Woesearchaeia, represented by Candidatus Stazkawiczbacteria and unknown Wosearchaeia, were far less abundant. The phyla Bathyarchaeota and Odinarchaeota were also detected in most samples in relatively low abundances. The amount of completely unknown archaeal ASVs or those grouped into “others” was surprisingly low with about 5%, only. Relative abundances of unknown Wosearchaeia were higher at the HWL and runnel (infiltration areas) than at the LWL and Ridge (exfiltration area). Additionally, they were increasing with depth at the HWL. The shallower depths of the HWL as well as the runnel and Ridge sites showed elevated abundances of unknown Bathyarchaeia. The distribution of Odinarchaeia was patchy with elevated relative abundances at the Ridge.
Eukaryotic Community Composition
A total number of 10 meiofaunal groups was detected in this study comprising Nematoda, Crustacea, Platyhelminthes, Annelida, Gastrotricha, Chaetognatha, Mollusca, Bryozoa, Tardigrada and Nemertea (Figure 5A; rare taxa are categorized as others). Considering the diversity and relative number of ASVs per taxon (Figure 5A), meiofaunal composition differed greatly among the studied sites. In general, the HWL appeared to be the least diverse site. Interestingly, two of the HWL replicates were dominated by Annelida, while with nematodes being the most abundant taxa in the other three replicates. While all replicates of the runnel were dominated by the genus Archiola de Beauchamp, 1910, four replicates of the LWL showed a high relative abundance of Nematoplana, 1938, both Platyhelminthes. In contrast, the ridge was again dominated by nematodes, with two replicates also showing increased relative abundances of Platyhelminthes.
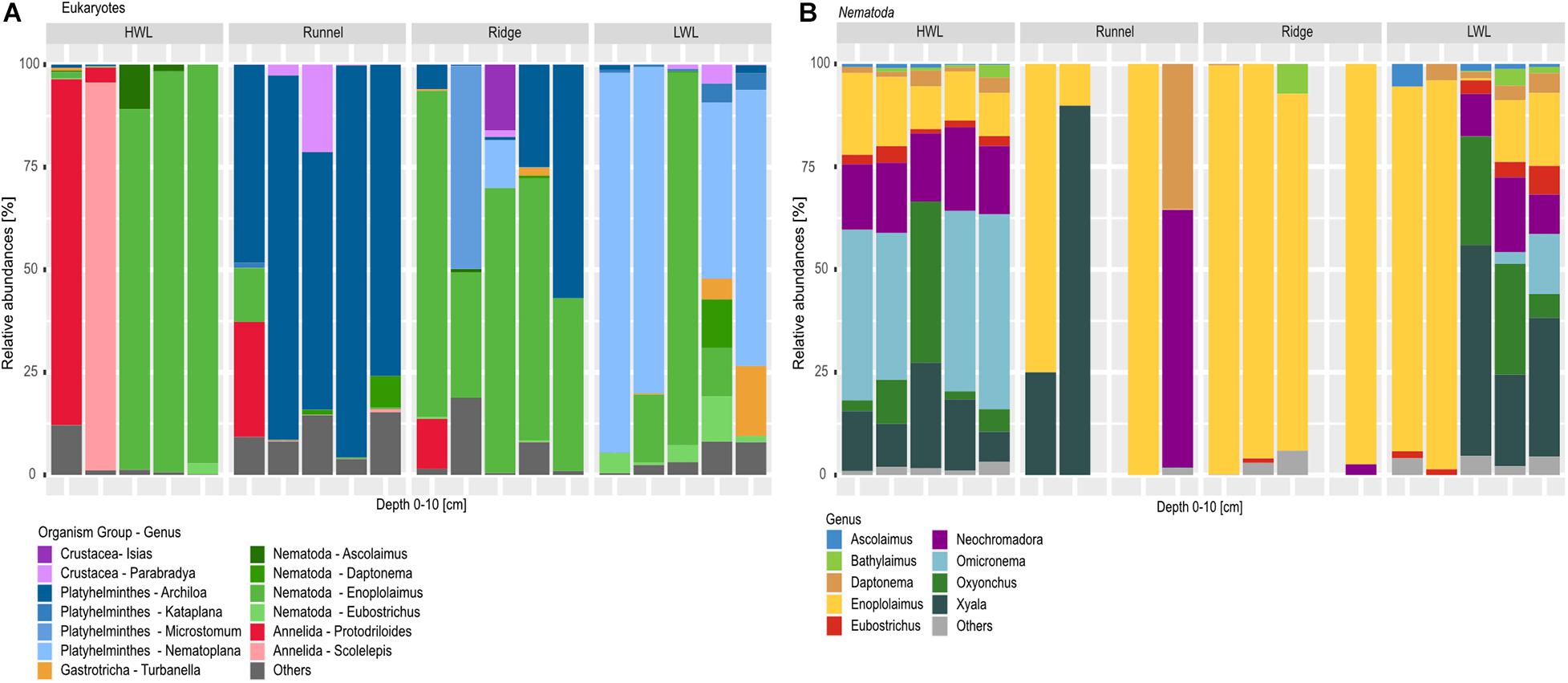
Figure 5. Barplots of eukaryotic communities along the transect depicted on phylum and genus level. (A) Shows total meiofauna diversity, while (B) zooms into nematode diversity, due to their high relative abundances within the dataset and during morphological investigation. Taxa below the abundance threshold were summarized as “others.” One replicate from the runneland Ridge were excluded from the nematode libraries analyses due to low numbers of reads.
Detailed community analysis of the relative number of picked nematode ASVs (Figure 5B) showed certain differences among the HWL and LWL compared to the runnel and ridge at the genus level. All HWL replicates and three LWL samples revealed a high nematode diversity with the most dominant genera being Xyala Cobb, 1920 or Omicronema Cobb, 1920 followed by Oxyonchus Filipjev, 1918 and Neochromadora Micoletzky, 1924. All ridge samples showed a low nematode diversity at genus level with 4–6 genera present in each library, greatly dominated by Enoplolaimus de Man, 1893. The runnel stations, however, were represented by a maximum of two nematode genera in each replicate, being Enoplolaimus and Xyala within three replicates Neochromadora and Daptonema Cobb, 1920 in one of them. In general, a low nematode diversity and a prevalence of specimens of the genus Enoplolaimus may indicate their successful adaptation to this habitat.
Discussion
Adaptation of Prokaryotic Communities to the Effect of SGD
The prokaryotic community composition within the exfiltration areas of the STE was more evenly distributed compared to the seawater infiltration sites of the beach. Furthermore, the archaeal community at the exfiltration sites was dominated by members the ammonia oxidizing Nitrosopumilaceae, which can be interpreted as an effect of increased ammonium supply through the STE. In our previous investigation on the same cross-shore transect, we described a dominant bacterial core community across all sites, depths and seasons, which was disrupted by a subsurface bloom during spring (Degenhardt et al., 2020). In the current study, we could confirm the presence of a prokaryotic core community, but in a weaker pronunciation. Surprisingly, distinctive communities were observed at every site with almost equal proportions. This might be due to different environmental conditions such as changing beach topographies and a stronger impact of SGD, reflected by distinctively lower salinities in comparison to those measured in our previous sampling campaigns (Waska et al., 2019b; Ahrens et al., 2020; Degenhardt et al., 2020). Furthermore, the chosen sequencing approach allowed to simultaneously investigate bacterial and archaeal diversity (Parada et al., 2016), yet direct comparisons with our previous findings might not be valid.
Due to the effects of SGD, environmental conditions across the intertidal area showed a drop in salinity at the ridge and LWL compared to the HWL and runnel. Additionally, enhanced concentrations of reduced compounds clearly mark these two stations as discharge areas of the STE. This is reflected in the prokaryotic community structures, exhibiting similarities between the ridge and LWL communities were more similar to each other, despite differences over all sites. In our preceding study we could not detect an adaptation of the bacterial community to the effects of SGD, concluding that those were overprinted by the high energy conditions of Spiekeroog beach. Since the discharge area of the STE is chemically much clearer defined in the present study, its effect may previously have been underestimated (Waska et al., 2019b; Degenhardt et al., 2020). Instead of being negligible due to being overprinted, SGD might be an additional factor in forming stable and diverse communities. The significance of this effect could vary depending on differences in time scales for physical disturbances, porewater flowpaths, and community adaptations.
All detected bacterial genera have originally been reported to thrive in marine sediments, except for Cupriavidus, which reached high relative abundances within the runnel samples. Isolates of this genus are highly versatile and were cultured from different habitats, such as soils (Cuadrado et al., 2010) plants (Platero et al., 2016), anthropogenic environments (Mergeay, 2015), but also from freshwater sediments (Qian et al., 2010; Nguyen and Lee, 2014). Since the salinity at the runnel was clearly marine, a possible explanation for their high relative abundance could be the detection of an uncultured marine representative of this genus. Environmental disturbances like the burial of plant material by winter storms might be an alternative explanation for their occurrence. The second genus that distinguished the prokaryotic community of the runnel from the other sites of the transect was Sediminicola. This bacterium is a facultative anaerobe, capable of nitrate reduction and known from coastal and deep sea sediments (Khan et al., 2006). The HWL community differed from the other sites by high relative abundances of Muriicola (Kahng et al., 2010) and Portibacter (Yoon et al., 2012) which are both aerobes, with Muriicola also being capable of nitrate reduction. The common features of both sites, the HWL and runnel, are a higher elevation above sea-level and higher salinity values, indicating marine conditions, which are typically more oxygenated. These characteristics have been described for those sites at Spiekeroog beach (Waska et al., 2019b; Ahrens et al., 2020; Degenhardt et al., 2020). The dominant genera found at the HWL and runnel could also be detected at the ridge and LWL, yet in lower relative abundances. Overall, the communities of the latter sites, located in the discharge area of the STE, showed a higher evenness in the relative abundances of the different taxa.
During our previous investigation, we detected increased relative abundances of members of the Bacteroidetes (Gillisia) and Firmicutes (Planococcus and Planomicrobium) at the same month and sampling sites, which we described as a “subsurface bloom.” In the current investigation, members of the Firmicutes could only be detected in relative abundances ranging from 0.1 to 0.2%, only. While we did not detect the genus Gilisia in high numbers, other Bacteroidetes were present in every sample and elevated relative abundances at the HWL. Known relatives of the genera with increased abundances are aerobes, capable of nitrate reduction. In our preceding study we attributed the “subsurface bloom” to elevated concentrations and penetration depths of oxygenated compounds like nitrate. As these conditions were not found during this sampling campaign, the right settings for a bloom might not have been present. We conclude that we either sampled the same month but under different geochemical conditions, or that we previously sampled an irregular, rather than a regularly occurring event (Degenhardt et al., 2020).
Adaptation of Archaeal Communities
Within the archaeal community, cross-shore differences appeared as a decrease in relative abundances of Wosearchaeia and an increase of Nitrosopumilaceae. The dominance of Nitrosopumilaceae within the discharge area of the STE is very likely related to increased ammonium concentrations, as members of this phylum are known for ammonia oxidation (Nakagawa and Stahl, 2013; Shafiee et al., 2019). Thus, Nitrosopumilus might outcompete other archaea when high ammonium concentrations are present. On the other hand, Wosearchaeia are known for having very small genomes and a symbiotic or fermentation based lifestyle (Castelle et al., 2015). This might be beneficial at high energy beaches a steady resupply of seawater constituents and metabolites, explaining why they are the second most abundant archaea at Spiekeroog beach. Especially in the infiltration zones, this constant resupply might foster competitive advantages they may have due to their streamlined genomes. Another archaeal group observed within most of our samples, but in low abundances are the Bathyarchaeia. They are known to be globally distributed generalists, which are often found in anoxic sediments (Zhou et al., 2018). Additionally, a rather enigmatic archaeal taxon, the Odinarchaeia, were detected in very low, but increased abundances at the ridge. Since not much is known about their lifestyle, their role within the discharge area of the STE remains to be investigated.
In general, our results on the diversity and relative abundances of Archaea at the Spiekeroog STE are in accordance with findings by Adyasari et al. (2020), the only study that also investigated archaeal diversity within an STE at several geochemically different discharge areas. Within the porewaters of two SGD hotspots in Mobile Bay (Gulf of Mexico), the authors found different archaeal communities with varying abundances, depending on the investigated site. At an anoxic site, the archaeal community was dominated by Wosearchaeia and constituted 4–8% of the microbial community. Another discharge area within that study was more similar to our sampling site. It was characterized by coarse sand and less reducing conditions, caused by higher hydraulic conductivity. Under these conditions, the Archaea made up 1% of the microbial community and were dominated by ammonia oxidizing Thaumarchaea, which fits to our observations.
Meiofaunal Communities Are Structured Site-Specifically
In contrast to the prokaryotes, meiofauna exhibited almost no core community across all sampling sites, but large site-specific assemblages and a high spatial distribution between the replicates. A possible explanation for this could be their narrower tolerance range for different environmental conditions in comparison to prokaryotes, such as oxygen concentrations and salinity. Additionally, being much more mobile than prokaryotes, they possibly migrate toward preferred conditions, forming site specific communities.
We detected a large variance in community composition between replicates taken at the respective sites. However, cross shore differences could be observed between the HWL and the other sites due to the distinct differences in the community compositions. The relative number of ASVs per group are indicative for a high abundance of nematodes in three replicates from the HWL, whereas the runnel was mainly dominated by platyhelminthes. The LWL and ridge, however, showed a difference within the meiofaunal community composition of the two other sites, where either platyhelminthes or nematodes were the most abundant taxa in the different replicates. These results are in agreement with the study of Grzelak et al. (2018), in which the diversity of nematodes was significantly higher at the saline HWL at the coast of Long Island compared to the areas with lower salinities. Although the LWL and ridge (as the discharge area of the STE) showed similar prokaryotic communities, this effect is not pronounced for the investigated eukaryotes. Different composition of meiofaunal communities within SGD affected and non-affected areas may be an indication of a non-adaptation toward the effect of SGD. The presence of crustaceans (mainly copepods) was only perceptible in the runnel replicates. Also, morphological observations did not recognize many copepod specimens within the samples. This may also be an indication of their seasonal variabilities as the sampling took place in early spring. Seasonal variations of seawater alter the organic matter input and the associated degradation by bacteria. Therefore, the STE areas are lower in the amount of inorganic nitrogen in early spring compared to late summer (Ahrens et al., 2020) which could explain the overall low abundance and diversity of harpacticoid copepods in our study.
The overwhelming majority of meiofauna detected during morphological investigation were nematodes. Thus, the relatively high proportions of reads belonging to other groups such as annelids and platyhelminthes may be caused by the higher biomass of those taxa, the presence of their appendages or environmental DNA. Although the abundance of certain meiofaunal groups such as nematodes was higher at the HWL, the relative diversity was greater in the runnel, ridge and LWL samples. This was caused by the presence of rare taxa such as marine Tardigrada, Chaetognatha, Nemertea and Bryozoa (included in “others” in Figure 5) which were not detected at the HWL. This was probably due to the lower salinity that turns the environmental condition intolerable for the addressed taxa. In general, we detected different meiofaunal community compositions between all four study sites. This is in agreement with Kotwicki et al. (2014), indicating the strong effects of environmental conditions on meiofaunal communities even within SGD-impacted areas of high-energy beaches.
In contrast to the overall meiofaunal diversity, nematode diversity did not follow the described cross-shore trend. At the HWL and LWL, the nematode communities were much more diverse in comparison to the runnel and ridge, where the dominating genera were Enoplolaimus and Xyala. We detected Xyala in high relative abundance at every site, implying an adaptation to the conditions of the intertidal area. When investigating three beaches impacted by SGD at the Long Island peninsula (California, United States), at every station Grzelak et al. (2018) also detected Xyalidae Chitwood, 1951, which includes the genera Xyala and Theristus Bastian, 1865. Furthermore, the authors described the presence of Enoplolaimus sp., an omnitrophic predatory genus, which was distinctive for areas unaffected by SGD at one of the beaches studied. Yet, it was also found at impacted sites of another beach they investigated.
In our study, we detected Enoplolaimus at the LWL, the discharge area of the STE, with clearly brackish salinities. Thus, SGD alone may not affect the presence of this genus. This is in accordance to another study on nematode diversity within the intertidal area of two sandy beaches in Italy and Poland (Gheskiere et al., 2005). Furthermore, in our study, Enoplolaimus ASVs showed high relative abundances at every zone of the beach, agreeing with Gheskiere et al. (2005) where 7–10% of the detected nematodes were Enoplolaimus, with a more consistent presence at the beach exhibiting higher energy input. Since Spiekeroog beach is characterized by even stronger physical forcing, the high relative abundances of Enoplolaimus ASVs in our dataset could indicate an adaptation to high energy environments. Furthermore, we detected a lower nematode diversity at the runnel and ridge sites of Spiekeroog beach. Our findigs again fit to those of Gheskiere et al. (2005), also indicating a lower nematode diversity and evenness at the investigated swash and breaker zones, which are comparable to our runnel and ridge sites. A possible explanation for this might be the greater physical stress due to the high wave action in these zones. In both, our and their study, a maximum of nematode diversity was detected at the HWL. This is probably caused by the presence of taxa which are related to more terrestrial environments within the transition zone between the terrestrial and the marine environment.
Genera typical for brackish conditions like Bathylaimus Cobb, 1894 and Tryploides de Man, 1886 (Gheskiere et al., 2005) were also detected at the HWL and LWL of Spiekeroog beach with low relative abundances (both genera are marked as “others” in Figure 5). At the same stations, we also found Microlaimus de Man, 1880 (categorized as “others” in Figure 5) which have been found exclusively at sites unaffected or affected by SGD, respectively (Grzelak et al., 2018). While the flowpath and exfiltration zones of the Spiekeroog STE are usually patchy and heterogeneous, beaches investigated by Grzelak et al. (2018) had distinct areas affected or unaffected by SGD. Yet, especially during our campaign, the LWL showed a reliable chemical signal of SGD and brackish salinities, probably caused by a beach topography which was unusually stable for the stormy winter/spring season.
Potential Influence of Nematode Diversity on Prokaryotic Diversity
We found that nematode communities significantly correlated with changes in bacterial communities, while the overall meiofauna did not. However, due to the different sampling strategies for prokaryote and eukaryote diversity analyses, a statistically valid testing could not be applied. Since only a few studies have targeted community level interactions between these domains of life and none of them investigated the specific role of STE’s, our study could be a starting point for this kind of investigations. Co-occurrences between bacteria and nematodes e.g., were detected in submarine deep-sea canyons and correlated to physiological needs of the respective bacterial taxa (Rzeznik-Orignac et al., 2018). For example, taxa like Eubostrichus, which has also been detected in our study, might serve as “shuttles” fur sulfur- and ammonia oxidizers between oxic and anoxic sediment layers, due to their tolerance for anoxic conditions (Schiemer et al., 1990). This form of interaction is probably less prevalent at Spiekeroog beach since it exhibits much stronger recharge of seawater constituents and a less stable redox zonation compared to deep-sea sediments. Yet, the authors also found several predatory taxa, which are expected to have an influence on bacterial diversity. Furthermore, while there is no direct study on interactions between Archaea and nematodes, members of Nitrosopumilus were detected within single nematode individuals in an extensive study on the microbiomes of free living marine nematodes (Schuelke et al., 2018). Even though we could not detect significant correlations between the variances of nematode and archaeal communities in Spiekeroog beach sediments, Nitrosopumilus was still the dominant archaeal taxon.
Concluding Remarks
In this study we expected to (i) detect a large archaeal and meiofaunal core community within the discharge area of the Spiekeroog beach STE. We detected a core-community of prokaryotes, yet less dominant than previously observed. This is possibly related to a previously unobserved stability of the beach morphology, which has been shown to govern the hydrogeochemical conditions within the STE. Regarding the meiofauna, we did not find a core community, but dominant site-specific communities and a cross shore difference between the HWL and all other sites. We attribute this to the higher motility and narrower tolerance range of meiofauna to environmental parameters. We further hypothesized, that (ii) the conditions in the discharge area of the STE favor the presence of those Archaea involved in nitrogen cycling. We could confirm this by detecting an overwhelming dominance of Nitrosopumliaceae ASVs in the entire intertidal area, which increased at the ridge and LWL, marked as exfiltration zones by geochemical (NH4, salinity) and topography data. Lastly, we hypothesized, that (iii) the harsh high-energy conditions a Spiekeroog beach result in a nematode dominated community over a copepod dominated community. This also holds true, as the vast majority of meiofauna detected by morphological analysis were nematodes, which also dominated the sequence dataset. Contrary to a previous investigation of the Spiekeroog STE we could detect an influence of SGD on prokaryotic communities, which was visible as a higher evenness at SGD-impacted sites (ridge, LWL) compared to unimpacted sites (HWL, runnel). This influence is probably related to the weaker pronunciation of the runnel-ridge system and a rather dissipative morphology of the beach. Yet, correlations between prokaryotic and meiofaunal communities remain to be explored in greater detail.
Data Availability Statement
The data presented in the study are deposited in the European Nucleotide Archive (https://www.ebi.ac.uk/ena), accession number PRJEB42988.
Author Contributions
JD performed sampling, molecular biological lab work, and wrote the manuscript. SK performed Illumina sequencing, bioinformatics, and writing. FM did bioinformatics. HW performed geochemical analysis. BE secured funding and wrote the manuscript. PM did eukaryotic data analysis and writing. All authors read and approved the manuscript.
Funding
This study was financed by the Niedersächsisches Ministerium für Wissenschaft und Kultur (MWK) in the scope of project “BIME” (ZN3184).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We want to thank the BIME research consortium and especially Frank Meyerjürgens, for their help during sampling. We would also like to thank Helmo Nikolai and Gerrit Behrends for providing logistics and equipment, and Heike Simon and Ina Ulber for support with sampling and DOC analyses. Further, we thank Jannik Schnier for sorting and picking meiofauna samples and preparing them for sequencing. Also, we thank Carola Lehners, Eleonore Gründken, Heike Simon, Ina Ulber and Matthias Friebe for their support with porewater nutrient and DOC analysis. Likewise, we would like to acknowledge the Niedersächsischer Landesbetrieb für Wasserwirtschaft, Küsten- und Naturschutz (NLWKN), Svante Pitters, the Umweltzentrum Wittbülten, and the Quellerdünen hostel, for on-site logistics and infrastructure. This is publication No 75 of the Senckenberg am Meer Metabarcoding Laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.642098/full#supplementary-material
Supplementary Figure 1 | Location of the sampling site at Spiekeroog beach, modified after Ahrens et al. (2020) and Degenhardt et al. (2020). Displayed is a cross-section through the beach, including hydrological flow paths and beach morphology. The squares mark the sampling sites across the intertidal area. MHWL, mean high water line; MSL, mean sea-level; MLWL, mean low water line.
References
Adyasari, D., Hassenruck, C., Montiel, D., and Dimova, N. (2020). Microbial community composition across a coastal hydrological system affected by submarine groundwater discharge (SGD). PLoS One 15:e0235235. doi: 10.1371/journal.pone.0235235
Ahrens, J., Beck, M., Marchant Hannah, K., Ahmerkamp, S., Schnetger, B., and Brumsack, H. J. (2020). Seasonality of organic matter degradation regulates nutrient and metal net fluxes in a high energy sandy beach. J. Geophys. Res. Biogeosci. 125:e2019JG005399.
Anschutz, P., Charbonnier, C., Deborde, J., Deirmendjian, L., Poirier, D., Mouret, A., et al. (2016). Terrestrial groundwater and nutrient discharge along the 240-km-long Aquitanian coast. Mar. Chem. 185, 38–47. doi: 10.1016/j.marchem.2016.04.002
Anschutz, P., Smith, T., Mouret, A., Deborde, J., Bujan, S., Poirier, D., et al. (2009). Tidal sands as biogeochemical reactors. Estuar. Coast. Shelf Sci. 84, 84–90. doi: 10.1016/j.ecss.2009.06.015
Auguet, J. C., Barberan, A., and Casamayor, E. O. (2010). Global ecological patterns in uncultured Archaea. ISME J. 4, 182–190. doi: 10.1038/ismej.2009.109
Beck, M., Reckhardt, A., Amelsberg, J., Bartholoma, A., Brumsack, H. J., Cypionka, H., et al. (2017). The drivers of biogeochemistry in beach ecosystems: a cross-shore transect from the dunes to the low-water line. Mar. Chem. 190, 35–50. doi: 10.1016/j.marchem.2017.01.001
Blaxter, M. L., De Ley, P., Garey, J. R., Liu, L. X., Scheldeman, P., Vierstraete, A., et al. (1998). A molecular evolutionary framework for the phylum Nematoda. Nature 392, 71–75. doi: 10.1038/32160
Boehm, A. B., Yamahara, K. M., and Sassoubre, L. M. (2014). Diversity and transport of microorganisms in intertidal sands of the California coast. Appl. Environ. Microbiol. 80, 3943–3951. doi: 10.1128/aem.00513-14
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857.
Bonaglia, S., Nascimento, F. J., Bartoli, M., Klawonn, I., and Bruchert, V. (2014). Meiofauna increases bacterial denitrification in marine sediments. Nat. Commun. 5:5133.
Brochier-Armanet, C., Forterre, P., and Gribaldo, S. (2011). Phylogeny and evolution of the archaea: one hundred genomes later. Curr. Opin. Microbiol. 14, 274–281. doi: 10.1016/j.mib.2011.04.015
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Castelle, C. J., Wrighton, K. C., Thomas, B. C., Hug, L. A., Brown, C. T., Wilkins, M. J., et al. (2015). Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr. Biol. 25, 690–701. doi: 10.1016/j.cub.2015.01.014
Charbonnier, C., Anschutz, P., Poirier, D., Bujan, S., and Lecroart, P. (2013). Aerobic respiration in a high-energy sandy beach. Mar. Chem. 155, 10–21. doi: 10.1016/j.marchem.2013.05.003
Cho, H. M., Kim, G., Kwon, E. Y., Moosdorf, N., Garcia-Orellana, J., and Santos, I. R. (2018). Radium tracing nutrient inputs through submarine groundwater discharge in the global ocean. Sci. Rep. 8:2439.
Cuadrado, V., Gomila, M., Merini, L., Giulietti, A. M., and Moore, E. R. B. (2010). Cupriavidus pampae sp. nov., a novel herbicide-degrading bacterium isolated from agricultural soil. Int. J. Syst. Evol. Microbiol. 60, 2606–2612. doi: 10.1099/ijs.0.018341-0
de Faria, L. C., Di Domenico, M., Andrade, S. C. S., dos Santos, M. C., Fonseca, G., Zanol, J., et al. (2018). The use of metabarcoding for meiofauna ecological patterns assessment. Mar. Environ. Res. 140, 160–168. doi: 10.1016/j.marenvres.2018.06.013
Decelle, J., Romac, S., Stern, R. F., Bendif, E. M., Zingone, A., Audic, S., et al. (2015). PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol. Ecol. Resour. 15, 1435–1445. doi: 10.1111/1755-0998.12401
Degenhardt, J., Dlugosch, L., Ahrens, J., Beck, M., Waska, H., and Engelen, B. (2020). Seasonal dynamics of microbial diversity at a sandy high energy beach reveal a resilient core community. Front. Mar. Sci. 7:573570.
Encarnação, J., Leitão, F., Range, P., Piló, D., Chícharo, M. A., and Chícharo, L. (2013). The influence of submarine groundwater discharges on subtidal meiofauna assemblages in south Portugal (Algarve). Estuar. Coast. Shelf Sci. 130, 202–208. doi: 10.1016/j.ecss.2013.04.013
Froelich, P. N., Klinkhammer, G. P., Bender, M. L., Luedtke, N. A., Heath, G. R., Cullen, D., et al. (1979). Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: suboxic diagenesis. Geochimica et Cosmochimica Acta 43, 1075–1090. doi: 10.1016/0016-7037(79)90095-4
Gabor, E. M., Vries, E. J., and Janssen, D. B. (2003). Efficient recovery of environmental DNA for expression cloning by indirect extraction methods. FEMS Microbiol. Ecol. 44, 153–163. doi: 10.1016/s0168-6496(02)00462-2
Gallucci, F., Hutchings, P., Gribben, P., and Fonseca, G. (2012). Habitat alteration and community-level effects of an invasive ecosystem engineer: a case study along the coast of NSW. Australia. Mar. Ecol. Prog. Ser. 449, 95–108. doi: 10.3354/meps09547
Gao, H., Matyka, M., Liu, B., Khalili, A., Kostka, J. E., Collins, G., et al. (2012). Intensive and extensive nitrogen loss from intertidal permeable sediments of the Wadden Sea. Limnol. Oceanogr. 57, 185–198. doi: 10.4319/lo.2012.57.1.0185
Gheskiere, T., Vincx, M., Urban-Malinga, B., Rossano, C., Scapini, F., and Degraer, S. (2005). Nematodes from wave-dominated sandy beaches: diversity, zonation patterns and testing of the isocommunities concept. Estuar. Coast. Shelf Sci. 62, 365–375. doi: 10.1016/j.ecss.2004.09.024
Giere, O. (2009). Synecological perspectives in meiobenthology. Meiobenthology 373–415. doi: 10.1007/978-3-540-68661-3_9
Grünenbaum, N., Ahrens, J., Beck, M., Gilfedder, B. S., Greskowiak, J., Kossack, M., et al. (2020a). A multi-method approach for quantification of in- and exfiltration rates from the subterranean estuary of a high energy beach. Front. Earth Sci. 8:571310.
Grünenbaum, N., Greskowiak, J., Sültenfuß, J., and Massmann, G. (2020b). Groundwater flow and residence times below a meso-tidal high-energy beach: a model-based analyses of salinity patterns and 3H-3He groundwater ages. J. Hydrol. 587:124948. doi: 10.1016/j.jhydrol.2020.124948
Grzelak, K., Tamborski, J., Kotwicki, L., and Bokuniewicz, H. (2018). Ecostructuring of marine nematode communities by submarine groundwater discharge. Mar. Environ. Res. 136, 106–119. doi: 10.1016/j.marenvres.2018.01.013
Guillou, L., Bachar, D., Audic, S., Bass, D., Berney, C., Bittner, L., et al. (2013). The protist ribosomal reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–604. doi: 10.1093/nar/gks1160
Haenel, Q., Holovachov, O., Jondelius, U., Sundberg, P., and Bourlat, S. J. (2017). NGS-based biodiversity and community structure analysis of meiofaunal eukaryotes in shell sand from Hallo island, Smogen, and soft mud from Gullmarn Fjord, Sweden. Biodivers. Data J. 5:e12731. doi: 10.3897/bdj.5.e12731
Higgins, R. P., and Thiel, H. (1988). Introduction to the Study of Meiofauna. Washington, DC: Smithsonian Institution Press.
Hong, Y., Wu, J., Wilson, S., and Song, B. (2018). Vertical stratification of sediment microbial communities along geochemical gradients of a subterranean estuary located at the gloucester beach of virginia, united states. Front. Microbiol. 9:3343.
Huettel, M., Berg, P., and Kostka, J. E. (2014). Benthic exchange and biogeochemical cycling in permeable sediments. Ann. Rev. Mar. Sci. 6, 23–51. doi: 10.1146/annurev-marine-051413-012706
Hugoni, M., Domaizon, I., Taib, N., Biderre-Petit, C., Agogue, H., Galand, P. E., et al. (2015). Temporal dynamics of active archaea in oxygen-depleted zones of two deep lakes. Environ. Microbiol. Rep. 7, 321–329. doi: 10.1111/1758-2229.12251
Huys, R. (1996). Marine and Brackish Water Harpacticoid Copepods. Shrewsbury: Field Studies Council.
Huys, R., Herman, P. M. J., Heip, C. H. R., and Soetaert, K. (1992). The meiobenthos of the North Sea: density, biomass trends and distribution of copepod communities. ICES J. Mar. Sci. 49, 23–44. doi: 10.1093/icesjms/49.1.23
Itaya, K., and Ui, M. (1966). A new micromethod for the colorimetric determination of inorganic phosphate. Clin. Chim. Acta 14, 361–366. doi: 10.1016/0009-8981(66)90114-8
Jessup, C. M., Kassen, R., Forde, S. E., Kerr, B., Buckling, A., Rainey, P. B., et al. (2004). Big questions, small worlds: microbial model systems in ecology. Trends Ecol. Evol. 19, 189–197. doi: 10.1016/j.tree.2004.01.008
Kahng, H. Y., Lee, S. S., Kim, J. M., Jung, J. Y., Lee, M. Y., Park, W., et al. (2010). Muriicola jejuensis gen. nov., sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 60, 1644–1648. doi: 10.1099/ijs.0.015313-0
Karanovic, T., and Cooper, S. J. B. (2012). Explosive radiation of the genus Schizopera on a small subterranean island in Western Australia (Copepoda:Harpacticoida): unravelling the cases of cryptic speciation, size differentiation and multiple invasions. Invertebrate Syst. 26, 115–192. doi: 10.1071/is11027
Karner, M. B., DeLong, E. F., and Karl, D. M. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510. doi: 10.1038/35054051
Kennedy, A. D., and Jacoby, C. A. (1999). Biological indicators of marine environmental health: meiofauna - a neglected benthic component? Environ. Monit. Assess. 54, 47–68.
Khan, S. T., Nakagawa, Y., and Harayama, S. (2006). Sediminicola luteus gen. nov., sp. nov., a novel member of the family flavobacteriaceae. Int. J. Syst. Evol. Microbiol. 56, 841–845. doi: 10.1099/ijs.0.64047-0
Kim, K. H., Heiss, J. W., Michael, H. A., Cai, W.-J., Laattoe, T., Post, V. E. A., et al. (2017). Spatial patterns of groundwater biogeochemical reactivity in an intertidal beach aquifer. J. Geophys. Res. Biogeosci. 122, 2548–2562. doi: 10.1002/2017jg003943
Könneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Kotwicki, L., Grzelak, K., Czub, M., Dellwig, O., Gentz, T., Szymczycha, B., et al. (2014). Submarine groundwater discharge to the Baltic coastal zone: impacts on the meiofaunal community. J. Mar. Syst. 129, 118–126. doi: 10.1016/j.jmarsys.2013.06.009
Kwon, E. Y., Kim, G., Primeau, F., Moore, W. S., Cho, H. M., DeVries, T., et al. (2014). Global estimate of submarine groundwater discharge based on an observationally constrained radium isotope model. Geophys. Res. Lett. 41, 8438–8444. doi: 10.1002/2014gl061574
Laskov, C., Herzog, C., Lewandowski, J., and Hupfe, M. (2007). Miniaturized photometrical methods for the rapid analysis of phosphate, ammonium, ferrous iron, and sulfate in pore water of freshwater sediments. Limnol. Oceanogr. Methods 4, 63–71. doi: 10.4319/lom.2007.5.63
Lipp, J. S., Morono, Y., Inagaki, F., and Hinrichs, K. U. (2008). Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454, 991–994. doi: 10.1038/nature07174
Lloyd, K. G., Schreiber, L., Petersen, D. G., Kjeldsen, K. U., Lever, M. A., Steen, A. D., et al. (2013). Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218. doi: 10.1038/nature12033
Losi, V., Ferrero, T. J., Moreno, M., Gaozza, L., Rovere, A., Firpo, M., et al. (2013). The use of nematodes in assessing ecological conditions in shallow waters surrounding a Mediterranean harbour facility. Estuar. Coast. Shelf Sci. 130, 209–221. doi: 10.1016/j.ecss.2013.02.017
Lueders, T., Manefield, M., and Friedrich, M. W. (2004). Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6, 73–78. doi: 10.1046/j.1462-2920.2003.00536.x
MacLeod, F., Kindler, G. S., Wong, H. L., Chen, R., and Burns, B. P. (2019). Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol. 5, 48–61. doi: 10.3934/microbiol.2019.1.48
McAllister, S. M., Barnett, J. M., Heiss, J. W., Findlay, A. J., MacDonald, D. J., Dow, C. L., et al. (2015). Dynamic hydrologic and biogeochemical processes drive microbially enhanced iron and sulfur cycling within the intertidal mixing zone of a beach aquifer. Limnol. Oceanogr. 60, 329–345. doi: 10.1002/lno.10029
Meadows, A. S., Ingels, J., Widdicombe, S., Hale, R., and Rundle, S. D. (2015). Effects of elevated CO2 and temperature on an intertidal meiobenthic community. J. Exp. Mar. Biol. Ecol. 469, 44–56. doi: 10.1016/j.jembe.2015.04.001
Meng, J., Xu, J., Qin, D., He, Y., Xiao, X., and Wang, F. (2014). Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J. 8, 650–659. doi: 10.1038/ismej.2013.174
Mergeay, M. (2015). The History of Cupriavidus Metallidurans Strains Isolated from Anthropogenic Environments. Berlin: Springer.
Migné, A., Ouisse, V., Hubas, C., and Davoult, D. (2011). Freshwater seepages and ephemeral macroalgae proliferation in an intertidal bay: II. effect on benthic biomass and metabolism. Estuar. Coast. Shelf Sci. 92, 161–168. doi: 10.1016/j.ecss.2010.12.023
Milici, M., Deng, Z. L., Tomasch, J., Decelle, J., Wos-Oxley, M. L., Wang, H., et al. (2016). Co-occurrence analysis of microbial taxa in the atlantic ocean reveals high connectivity in the free-living bacterioplankton. Front. Microbiol. 7:649.
Miller, D. C., and Ullman, W. J. (2004). Ecological consequences of ground water discharge to delaware bay, united states. Ground Water 42, 959–970. doi: 10.1111/j.1745-6584.2004.tb02635.x
Moore, W. S. (1996). Large groundwater inputs to coastal waters revealed by 226Ra enrichments. Nature 380, 612–614. doi: 10.1038/380612a0
Moore, W. S. (1999). The subterranean estuary: a reaction zone of ground water and sea water. Mar. Chem. 65, 111–125. doi: 10.1016/s0304-4203(99)00014-6
Moore, W. S., Sarmiento, J. L., and Key, R. M. (2008). Submarine groundwater discharge revealed by 228Ra distribution in the upper Atlantic Ocean. Nat. Geosci. 1, 309–311. doi: 10.1038/ngeo183
Moreno, M., Semprucci, F., Vezzulli, L., Balsamo, M., Fabiano, M., and Albertelli, G. (2011). The use of nematodes in assessing ecological quality status in the Mediterranean coastal ecosystems. Ecol. Indic. 11, 328–336. doi: 10.1016/j.ecolind.2010.05.011
Muck, S., De Corte, D., Clifford, E. L., Bayer, B., Herndl, G. J., and Sintes, E. (2019). Niche differentiation of aerobic and anaerobic ammonia oxidizers in a high latitude deep oxygen minimum zone. Front. Microbiol. 10:2141.
Nakagawa, T., and Stahl, D. A. (2013). Transcriptional response of the archaeal ammonia oxidizer nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl. Environ. Microbiol. 79, 6911–6916. doi: 10.1128/AEM.02028-13
Nascimento, F. J. A., Näslund, J., and Elmgren, R. (2012). Meiofauna enhances organic matter mineralization in soft sediment ecosystems. Limnol. Oceanogr. 57, 338–346. doi: 10.4319/lo.2012.57.1.0338
Nguyen, V. K., and Lee, J.-U. (2014). Antimony-oxidizing bacteria isolated from antimony-contaminated sediment – a phylogenetic study. Geomicrobiol. J. 32, 50–58. doi: 10.1080/01490451.2014.925009
Ortiz-Alvarez, R., and Casamayor, E. O. (2016). High occurrence of Pacearchaeota and Woesearchaeota (Archaea superphylum DPANN) in the surface waters of oligotrophic high-altitude lakes. Environ. Microbiol. Rep. 8, 210–217. doi: 10.1111/1758-2229.12370
Ouisse, V., Riera, P., Migné, A., Leroux, C., and Davoult, D. (2011). Freshwater seepages and ephemeral macroalgae proliferation in an intertidal bay: i effect on benthic community structure and food web. Estuar. Coast. Shelf Sci. 91, 272–281. doi: 10.1016/j.ecss.2010.10.034
Paerl, H. W. (1997). Coastal eutrophication and harmful algal blooms: importance of atmospheric deposition and groundwater as “new” nitrogen and other nutrient sources. Limnol. Oceanogr. 42, 1154–1165. doi: 10.4319/lo.1997.42.5_part_2.1154
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Platero, R., James, E. K., Rios, C., Iriarte, A., Sandes, L., Zabaleta, M., et al. (2016). Novel cupriavidus strains isolated from root nodules of native uruguayan mimosa species. Appl. Environ. Microbiol. 82, 3150–3164. doi: 10.1128/aem.04142-15
Qian, Y., Shi, J., Chen, Y., Lou, L., Cui, X., Cao, R., et al. (2010). Characterization of phosphate solubilizing bacteria in sediments from a shallow eutrophic lake and a wetland: isolation, molecular identification and phosphorus release ability determination. Molecules 15, 8518–8533. doi: 10.3390/molecules15118518
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596.
R Core Team (2019). R: a Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reckhardt, A., Beck, M., Seidel, M., Riedel, T., Wehrmann, A., Bartholomä, A., et al. (2015). Carbon, nutrient and trace metal cycling in sandy sediments: a comparison of high-energy beaches and backbarrier tidal flats. Estuar. Coast. Shelf Sci. 159, 1–14. doi: 10.1016/j.ecss.2015.03.025
Riedel, T., Lettmann, K., Schnetger, B., Beck, M., and Brumsack, H.-J. (2011). Rates of trace metal and nutrient diagenesis in an intertidal creek bank. Geochimica et Cosmochimica Acta 75, 134–147. doi: 10.1016/j.gca.2010.09.040
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J. F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Rios-Del Toro, E. E., Valenzuela, E. I., Ramírez, J. E., López-Lozano, N. E., and Cervantes, F. J. (2018). Anaerobic ammonium oxidation linked to microbial reduction of natural organic matter in marine sediments. Environ. Sci. Technol. Lett. 5, 571–577. doi: 10.1021/acs.estlett.8b00330
Rodellas, V., Garcia-Orellana, J., Tovar-Sánchez, A., Basterretxea, G., López-Garcia, J. M., Sánchez-Quiles, D., et al. (2014). Submarine groundwater discharge as a source of nutrients and trace metals in a Mediterranean bay (Palma Beach, Balearic Islands). Mar. Chem. 160, 56–66. doi: 10.1016/j.marchem.2014.01.007
Rzeznik-Orignac, J., Puisay, A., Derelle, E., Peru, E., Le Bris, N., and Galand, P. E. (2018). Co-occurring nematodes and bacteria in submarine canyon sediments. PeerJ 6:e5396. doi: 10.7717/peerj.5396
Santos, I. R., Burnett, W. C., Dittmar, T., Suryaputra, I. G. N. A., and Chanton, J. (2009). Tidal pumping drives nutrient and dissolved organic matter dynamics in a Gulf of Mexico subterranean estuary. Geochimica et Cosmochimica Acta 73, 1325–1339. doi: 10.1016/j.gca.2008.11.029
Schiemer, E., Novak, R., and Ott, J. (1990). Metabolic studies on thiobiotic free-living nematodes and their symbiotic microorganisms. Mar. Biol. 106, 129–137. doi: 10.1007/bf02114683
Schuelke, T., Pereira, T. J., Hardy, S. M., and Bik, H. M. (2018). Nematode-associated microbial taxa do not correlate with host phylogeny, geographic region or feeding morphology in marine sediment habitats. Mol. Ecol. 27, 1930–1951. doi: 10.1111/mec.14539
Semprucci, F., Losi, V., and Moreno, M. (2015). A review of Italian research on free-living marine nematodes and the future perspectives on their use as ecological indicators (EcoInds). Mediterr. Mar. Sci. 16, 352–365. doi: 10.12681/mms.1072
Shafiee, R. T., Snow, J. T., Zhang, Q., and Rickaby, R. E. M. (2019). Iron requirements and uptake strategies of the globally abundant marine ammonia-oxidising archaeon, Nitrosopumilus maritimus SCM1. ISME J. 13, 2295–2305. doi: 10.1038/s41396-019-0434-8
Staley, C., and Sadowsky, M. J. (2016). Regional similarities and consistent patterns of local variation in beach sand bacterial communities throughout the northern hemisphere. Appl. Environ. Microbiol. 82, 2751–2762. doi: 10.1128/aem.00247-16
Taberlet, P., Coissac, E., Pompanon, F., Brochmann, C., and Willerslev, E. (2012). Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. doi: 10.1111/j.1365-294x.2012.05470.x
Thauer, R. K., Kaster, A. K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591. doi: 10.1038/nrmicro1931
Vanaverbeke, J., Merckx, B., Degraer, S., and Vincx, M. (2011). Sediment-related distribution patterns of nematodes and macrofauna: two sides of the benthic coin? Mar. Environ. Res. 71, 31–40. doi: 10.1016/j.marenvres.2010.09.006
Waska, H., Brumsack, H.-J., Massmann, G., Koschinsky, A., Schnetger, B., Simon, H., et al. (2019a). Inorganic and organic iron and copper species of the subterranean estuary: origins and fate. Geochimica et Cosmochimica Acta 259, 211–232. doi: 10.1016/j.gca.2019.06.004
Waska, H., Greskowiak, J., Ahrens, J., Beck, M., Ahmerkamp, S., and Boning, P. (2019b). Spatial and temporal patterns of pore water chemistry in the inter-tidal zone of a high energy beach. Front. Mar. Sci. 6:154.
Webster, I. T., and Taylor, J. H. (1992). Rotational dispersion in porous media due to fluctuating flows. Water Resour. Res. 28, 109–119. doi: 10.1029/91wr02323
Yeh, Y.-C., McNichol, J. C., Needham, D. M., Fichot, E. B., and Fuhrman, J. A. (2019). Comprehensive single-PCR 16S and 18S rRNA community analysis validated with mock communities and denoising algorithms. bioRxiv [preprint] doi: 10.1101/866731
Yoder, M., De Ley, I. T., King, I. W., Mundo-Ocampo, M., Mann, J., Blaxter, M., et al. (2006). DESS: a versatile solution for preserving morphology and extractable DNA of nematodes. Nematology 8, 367–376. doi: 10.1163/156854106778493448
Yoon, J., Matsuo, Y., Kasai, H., and Yokota, A. (2012). Portibacter lacus gen. nov., sp. nov., a new member of the family Saprospiraceae isolated from a saline lake. J. Gen. Appl. Microbiol. 58, 191–197.
Zeppilli, D., Bongiorni, L., Serrao Santos, R., and Vanreusel, A. (2014). Changes in nematode communities in different physiographic sites of the condor seamount (north-East atlantic ocean) and adjacent sediments. PLoS One 9:e115601. doi: 10.1371/journal.pone.0115601
Zeppilli, D., Sarrazin, J., Leduc, D., Arbizu, P. M., Fontaneto, D., Fontaneto, C., et al. (2015). Is the meiofauna a good indicator for climate change and anthropogenic impacts? Mar. Biodivers. 45, 505–535.
Zhou, Z., Pan, J., Wang, F., Gu, J. D., and Li, M. (2018). Bathyarchaeota: globally distributed metabolic generalists in anoxic environments. FEMS Microbiol. Rev. 42, 639–655. doi: 10.1093/femsre/fuy023
Keywords: Archaea, bacteria, intertidal area, meiofauna, microbial diversity, North Sea, sandy beach, SGD
Citation: Degenhardt J, Khodami S, Milke F, Waska H, Engelen B and Martinez Arbizu P (2021) The Three Domains of Life Within the Discharge Area of a Shallow Subterranean Estuary at a High Energy Beach. Front. Environ. Sci. 9:642098. doi: 10.3389/fenvs.2021.642098
Received: 16 December 2020; Accepted: 07 April 2021;
Published: 22 April 2021.
Edited by:
Ross Thompson, University of Canberra, AustraliaReviewed by:
Guillaume Grosbois, Université du Québec en Abitibi Témiscamingue, CanadaDaniele Kasper, Federal University of Rio de Janeiro, Brazil
Copyright © 2021 Degenhardt, Khodami, Milke, Waska, Engelen and Martinez Arbizu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bert Engelen, ZW5nZWxlbkBpY2JtLmRl
 Julius Degenhardt
Julius Degenhardt Sahar Khodami
Sahar Khodami Felix Milke
Felix Milke Hannelore Waska
Hannelore Waska Bert Engelen
Bert Engelen Pedro Martinez Arbizu2
Pedro Martinez Arbizu2