
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 09 April 2021
Sec. Water and Wastewater Management
Volume 9 - 2021 | https://doi.org/10.3389/fenvs.2021.632089
This article is part of the Research Topic The Interplay between Freshwater Microorganisms and Water Quality View all 7 articles
Water eutrophication is a global ecological issue, and thermal stratification of water bodies can enable eutrophication. We examined bacterial communities in the stratified water columns and sediments in two different trophic reservoirs along the Wujiang River using quantitative real-time PCR and high-throughput sequencing. Bacterial 16S rRNA gene copies varied from 3.70 × 107 to 5.27 × 108 copies/L in the water column of Hongjiadu (HJD) Reservoir (60 m water depth) with slightly stratified variation; while in Wujiangdu (WJD) Reservoir (70 m water depth), bacterial abundance decreased markedly from the surface to the bottom(1.74 × 109 to 2.38 × 107 copies/L). The vertical distribution patterns of bacteria in both reservoirs resembled those of water Chlorophyll a (Chla) concentrations. The abundance was negatively correlated with water depth (D), total nitrogen (TN), nitrate (NO3–-N), and positively correlated with water temperature (T) and dissolved oxygen (DO) level. In contrast, the alpha diversity of bacteria showed the opposite trend in the vertical water column. Proteobacteria, Actinobacteria, and Bacteroidetes were the predominant phyla in the water column of both reservoirs. Compared to WJD Reservoir, HJD Reservoir displayed marked vertical spatial difference in bacterial community structure during thermal stratification. In particular, Pseudomonas was frequently detected at the bottom of the HJD Reservoir. These results were consistent with predictive metagenomic profiling that revealed different vertical functional variation patterns of the bacterial communities in the two reservoirs. The bacterial community structure of HJD Reservoir was associated with water D, ammonium (NH4+-N), nitrite (NO2–-N), and total phosphorus (TP). The community structure of WJD Reservoir was related to water T, Chla, NO3–-N, and TN. The findings highlighted the important roles played by thermal stratification and nutrients in shaping the water bacterial community structure. Additionally, the absolute abundance of water nitrifiers (AOB gene copies) and denitrifiers (narG, nirS, norB, and nosZ gene copies) displayed significant vertical differences in the water columns of both reservoirs. Gene copies involved in denitrification were significantly higher than those involved in nitrification. Water phosphorus and nitrogen contents were important variables influencing the absolute abundance of ammonia oxidizers and denitrifying bacteria, respectively. Our study revealed that the emergence of thermal stratification was responsible for the vertical stratification of bacteria in water and affected the bacterial community structure together with nutrients.
Water eutrophication has become one of the major environmental issues affecting lakes and reservoirs on a global scale (Paerl et al., 2016; Wang et al., 2019). Compared to natural rivers, artificial reservoirs generally exhibit hydrodynamic variations and longer exchange periods (Jardim et al., 2020). Additionally, numerous nutrients, such as nitrogen (N) and phosphorus (P) are intercepted, which alter the nutrient cycle, drive the rapid propagation of algae and other plankton, and decrease the dissolved oxygen (DO) content in the water body (Van Cappellen and Maavara, 2016; Shi et al., 2017; Petty et al., 2020), thereby worsening the eutrophication of the water body. According to 2018 data from the United Nations Water Resources Organization, over 75% of closed water bodies (such as lakes and reservoirs) are eutrophic to a certain extent, with deterioration in water quality.
Thermal stratification is a common phenomenon after reservoir completion. Because of the exchange of material and energy by the atmosphere and solar radiation, the temperature difference between the upper and lower layers of the water body leads to a difference in water density, which results in the formation of an epilimnion – thermocline – hypolimnion vertical profile from the top to the bottom of the water column (Tuo et al., 2014; Kirillin and Shatwell, 2016). The thermocline hinders the mixing of upper and lower waters and slows the vertical transfer of DO and particulates in the epilimnion layer (Zhang et al., 2015), which causes anoxia in the bottom water and the release of nutrients from sediments, and aggravates the eutrophication (Conley et al., 2009; Rigosi and Rueda, 2012; Müller et al., 2016). With the appearance of thermal stratification, the vertical changes in pH, DO, turbidity, and Chla concentration are highly consistent and display marked stratification. Water stratification may be one of the major factors determining the vertical distribution of phytoplankton, which leads to the death of aquatic organisms, such as fish, in affected lakes. However, little is known about the effect of water thermal stratification on the abundance of bacteria in reservoirs (Yu et al., 2014; Wang et al., 2015; Yang et al., 2015).
In aquatic ecosystems, microorganisms are highly sensitive to changes in the water environment and are affected by a variety of environmental factors, including organic matter, nitrogen, phosphorus, pH, DO, and temperature (Zhao et al., 2012; Daggett et al., 2015; Ruiz-González et al., 2015; Sinistro et al., 2015; Li et al., 2017). Moreover, microbial activity drives nutrient cycling (Salcher et al., 2010) and influences water quality (Houshmand et al., 2020) to maintain the structure and function of aquatic ecosystems. Nitrogen serves as a limiting factor for water eutrophication (Canfield et al., 2010). Denitrification plays a critical role in nitrogen removal (Peng et al., 2016), and denitrifying bacteria contribute to over 87% of the nitrogen loss in freshwater (Schubert et al., 2006; Annika and Christopher, 2010; Chen et al., 2012). Denitrifiers are facultative anaerobes that are affected by DO (Oshiki et al., 2016), organic matter (Cornwell et al., 2014), and N availability (Smith et al., 2015). Examining the expression of the genes encoding for denitrifiers (such as NAR/NAP, NIR, NOR, and NOS) in the bacterial communities can help clarify the denitrification potential in freshwater ecosystems (Braker et al., 2015; Smith et al., 2015; Tatti et al., 2015; Gao et al., 2016; Mao et al., 2017).
In this study, we focused on the bacterial communities in two reservoirs of the Wujiang River in Southwestern China. Water columns and surface sediments from the lacustrine zone were chosen for comparison. The two major objectives were to (1) evaluate the effects of water thermal stratification on bacterial communities with various levels of eutrophication, and to (2) investigate the vertical dynamic distribution of nitrification and denitrification in the water columns.
The sampling areas of HJD and WJD reservoirs were located in the Wujiang River. The Wujiang River is the largest tributary south of the upper reaches of the Yangtze River. HJD Reservoir was built in 2004, with a total reservoir capacity of 49.3 × 108 m3, the highest water level of 1140 m, the normal water level area of 80.5 km2, and the retention time of the water body is 360 days, which suggests it is a regulating reservoir in the year of oligotrophic nutrition (Zhao et al., 2017); WJD Reservoir was built in 1979, with a drainage area of 27,790 km2, a total reservoir capacity of 21.4 × 108m3, and the normal water level of 47.8 km2. The reservoir is a seasonal regulating reservoir with eutrophic nutrition (Zhao et al., 2017) and appreciably affected by human development and use. The samples were collected from the lacustrine zones of the two reservoirs of the Wujiang River in April 2017.
Water samples from the vertical profiles in both reservoirs (HJD and WJD) were collected at a range of depths (within 0, 5, 15, 30, and 50 m for HJD; within 0, 5, 15, 30, 45, and 60 m for WJD) using a Niskin water sampler (Model 1010, General Oceanics, United States) (Figure 1). About 500 mL sample volumes were filtered through a 0.22 μm sterilized filter membrane (MF-Millipore, United States) and stored in 50 mL sterile centrifuge tubes at −80 °C. The remaining water samples were stored at 4 °C for physicochemical analyses. Sediment samples (top 0–20 cm) were collected using a Van Veen grab sampler (35 cm × 25 cm × 30 cm), and three adjacent sub-samples were collected at each site to form a composite sample. Each sample campaign included two replicates and was stored at −20 °C for DNA extraction and molecular analyses.
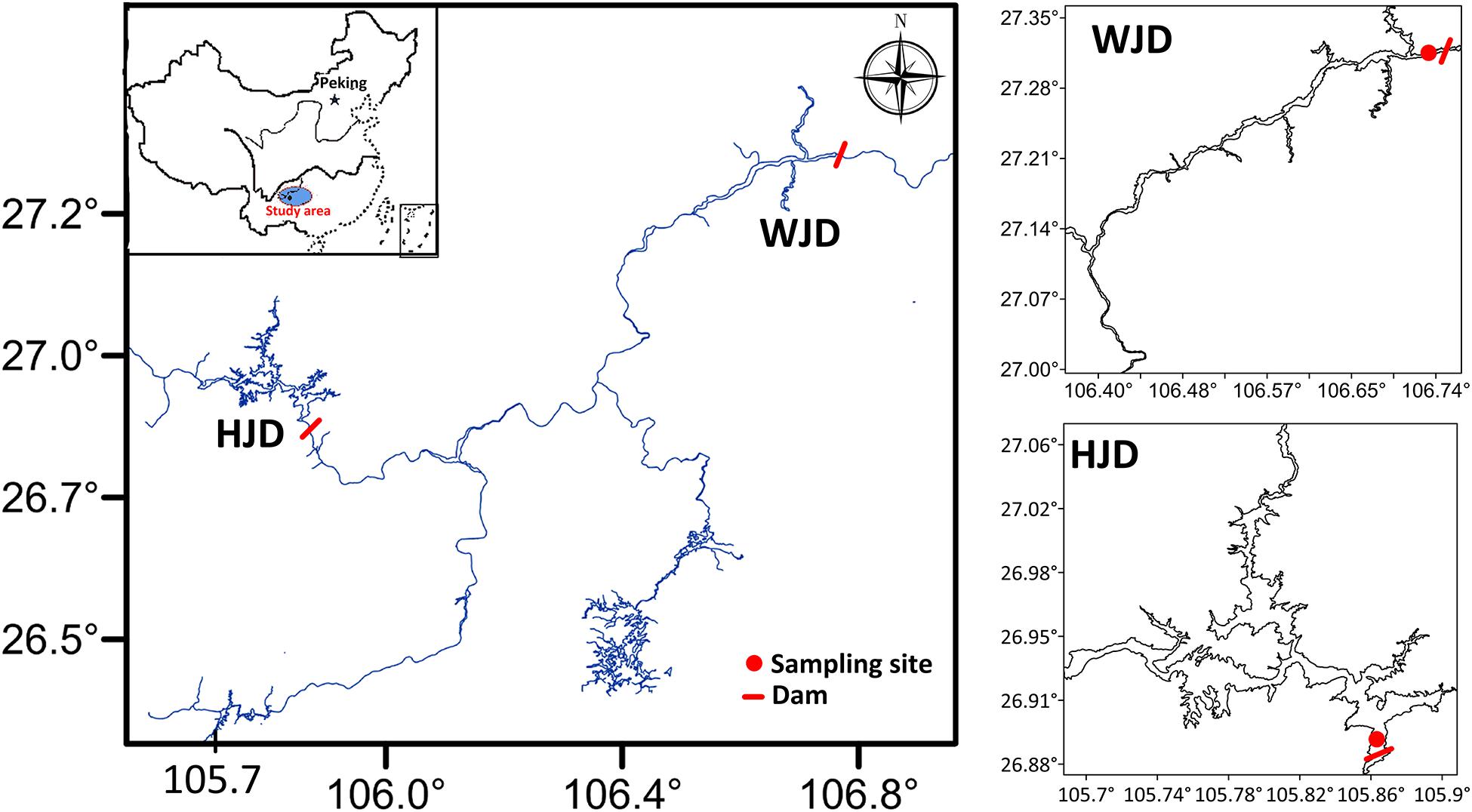
Figure 1. Sampling sites and locations of the Hongjiadu (HJD) and Wujiangdu (WJD) reservoirs along the Wujiang River, China.
At each sample site, an automated multiparameter profiler (model YSI EXO) was used to measure water temperature (T), conductivity (Cond), total dissolved solids (TDS), pH, dissolved oxygen (DO), and chlorophyll a (Chla) in situ. Total nitrogen (TN) was tested using ion chromatography (EPA 300.0, revision 2.1, 1993) with prior persulfate oxidation for the water samples (Hosomi and Sudo, 1986). Samples for nitrate (NO3–) and ammonium (NH4+) analyses were acidified by addition of H2SO4 (pH < 2) and stored at <4°C in the dark prior to analysis. Concentrations of NO3– and NH4+ were measured with an automatic flow analyzer (SKALAR Sans Plus Systems, Breda, Netherlands). NO3– was tested using ion chromatography (EPA 300.0, revision 2.1, 1993). The indophenol colorimetric method was used to analyze NH4+ (EPA 350.1, revision 2.0, 1993). The ascorbic acid colorimetric method with prior oxidation used to determine total phosphorus (TP) (EPA 365.3, 1978). The main physical properties of each water sample are shown in Supplementary Table 1.
Genomic DNA was extracted from the filter and sediment (0.5 g) samples using the FastDNA SPIN Kit for Soil (MP, United States), following the manufacturer’s instructions. The V3 − V4 region of bacterial 16S rRNA genes was amplified using the forward primer 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and reverse primer 806R (5′-GGACTACHVGGGTWTCTA AT-3′) (Xu et al., 2016). The PCR was performed using the following protocol: initial denaturation at 94°C for 5 min, followed by 25 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s; and a final extension step at 72°C for 5 min. Each sample was amplified in triplicate, pooled, and purified using the Agencourt AMPure XP Beads (Backman, United States). DNA amplicons were quantified using the QubitTM dsDNA Assay Kit (Invitrogen, Carlsbad, CA, United States). Subsequently, an equi molar concentration of each DNA amplicon was mixed and then sequenced using the MiSeq PE300 platform (Illumina, San Diego, CA, United States) at Majorbio Bio-Pharm Technology Co. Ltd., Shanghai, China1.
The absolute abundance of bacterial 16S rRNA, ammonia monooxygenase of ammonia oxidizing bacteria (AOB), narG from nitrate reductase (narG), nirS from nitrite reductase (nirS), norB from reductase (norB), and nosZ from nitrous oxide reductase (nosZ) were measured by qPCR using bacteria-specific primer pairs. Primers used for the amplification of these genes are listed in Supplementary Table 2. A 10-fold serial dilution of standard DNA plasmids was prepared by cloning the target gene fragment amplified from samples using the same primer used for qPCR. The PCR reaction mixtures contained 10 μL of 2 × SYBR® Premix Ex TaqTM II (Takara, Japan), 1 μL (10 pmol/μL) of both forward and reverse primers, and 1 μL template DNA in a final volume of 20 μL. All qPCR procedures were performed on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, United States) with an initial denaturation at 95°C for 2 min, followed by 40 cycles lasting 20 s each at 95°C, 60 s at 63°C, and 60 s at 68°C. The specificity of each amplicon was checked by melt curve analysis when PCR amplification was completed. A standard curve was constructed by plotting the relative fluorescent units at a threshold fluorescence value (CT) versus the logarithm of copy number of the standard plasmid DNA. The efficiencies of qPCR ranged from 85 to 98% (R2 > 0.997). Each sample DNA and standard DNA were analyzed in triplicate.
For sequencing data, the forward and reverse reads were joined, assigned to samples, and truncated by splicing the barcode and primer sequence based on the overlap relationship between paired-end reads. The quality of reads and the effect of merging were filtered by QIIME (version 1.9.12) to obtain the optimized data. The effective sequences were grouped into operational taxonomic units (OTUs) at a 97% sequence identity level using Usearch (version 7.03). Subsequently, Mothur (version 1.30.24) was used to determine the richness index (Chao1 and ACE), coverage index (Good’s coverage), phylogenetic diversity index (Faith’s PD), and diversity index (Shannon and Simpson) for alpha diversity analysis. Taxonomic classification was assigned using the SILVA132 database (Release 1325) to calculate the community composition of each sample. Based on the unweighted-UniFrac distance matrices, Principal coordinate analysis (PCoA) was created by Program R (version 3.0.1) to study the similarity or difference among samples. Analysis of similarity (ANOSIM) was performed to verify the significance of community compositions from different environmental categories of samples. The longest gradient lengths were less than 3.5 in preliminary detrended correspondence analysis. Thus redundancy analysis (RDA) was used to explore the relationship between bacterial community and environmental variables. The environmental factors subset was obtained by the maximum Pearson correlation coefficient of the distribution difference between the environmental factors and the sample communities. The samples and environmental factors were reflected on the same two-dimensional ranking chart by CANOCO software (version 4.5, Wageningen, Netherlands), and a Monte Carlo permutation test with 999 permutations was used to analyze the significance of the parameters (Zhang et al., 2021b). Based on bacterial 16S rRNA gene sequences, the metabolic functions were predicted using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) version 1.1.06. A co-occurrence network of bacterial community at the genus level was generated based on Spearman’s correlation significant analysis (Zhang et al.,2021a,b). Calculation of the correlation between species established a connection between species at a certain threshold correlation coefficient. The species correlation network diagram was generated. One-way ANOVA was performed using SPSS Statistics 20 (IBM, United States). Graphs were generated using GraphPad Prism 5 software.
The 16S rRNA gene sequences from this study have been deposited in the public NCBI database7 under the accession numbers PRJNA680428.
The water Chla concentration of HJD Reservoir ranged from 0.21 μg/L to 2.36 μg/L and corresponded with the characteristics of an oligotrophic reservoir. The water temperature decreased from 18.3°C at the surface to 12.1°C at the bottom (Supplementary Table 1). TN and TP concentrations fluctuated from 3.74 mg/L to 4.05 mg/L and from 0.041 mg/L to 0.085 mg/L, respectively. WJD Reservoir was in a eutrophic state, with a maximum Chl-a concentration of 15.5 μg/L on the surface. The water temperature ranged from 12.8 to 20.2°C. TN and TP concentrations followed similar trends to those of HJD Reservoir, ranging from 3.63 mg/L to 4.41 mg/L and from 0.065 mg/L to 0.296 mg/L, respectively. The DO concentration remained high throughout the water column in two reservoirs, varying from 6.35 mg/L to 9.42 mg/L and from 6.34 mg/L to 13.1 mg/L, respectively. The water bodies in reservoirs were stratified based on the measurement of water temperature (T) at different water depths. In HJD Reservoir, 0 m was the epilimnion, 5–15 m was the thermocline, and below 15 m was the hypolimnion. In WJD Reservoir, 0 m was the epilimnion, 5–15 m was the thermocline, and below 15 m was the hypolimnion.
With increasing water depth, copy numbers of bacterial 16S rRNA genes slightly increased at 5 m and then slightly decreased at 30 m in HJD Reservoir, while the decrease was greater in WJD Reservoir (Figure 2). In reservoir profile water, the average 16S rRNA gene copy numbers varied from 2.38 × 107 to 1.74 × 109 copies/L. The copy numbers of bacterial 16S rRNA genes were 7.84 × 1011 to 9.67 × 1012 copies/L on average in sediments, which were based on the density of the solid at approximately 2.5 g/cm3 (Xu et al., 2006) (Supplementary Figure 1). The lowest number of 16S rRNA gene copies was found in the hypolimnion, followed by the epilimnion and thermocline. The highest number was present in the sediment. The vertical distribution patterns of bacterial abundance in the two reservoirs were consistent with the Chla concentrations. The abundance of bacterial 16S rRNA genes in the sediment was significantly different from water (p < 0.05). The bacterial 16S rRNA copies were negatively correlated with D (p < 0.01), TN (p < 0.01), and NO3–-N (p < 0.01), and positively correlated with T (p < 0.01), DO (p < 0.01), pH-w (p < 0.01), and Chla (p < 0.01) in water samples (Table 1).
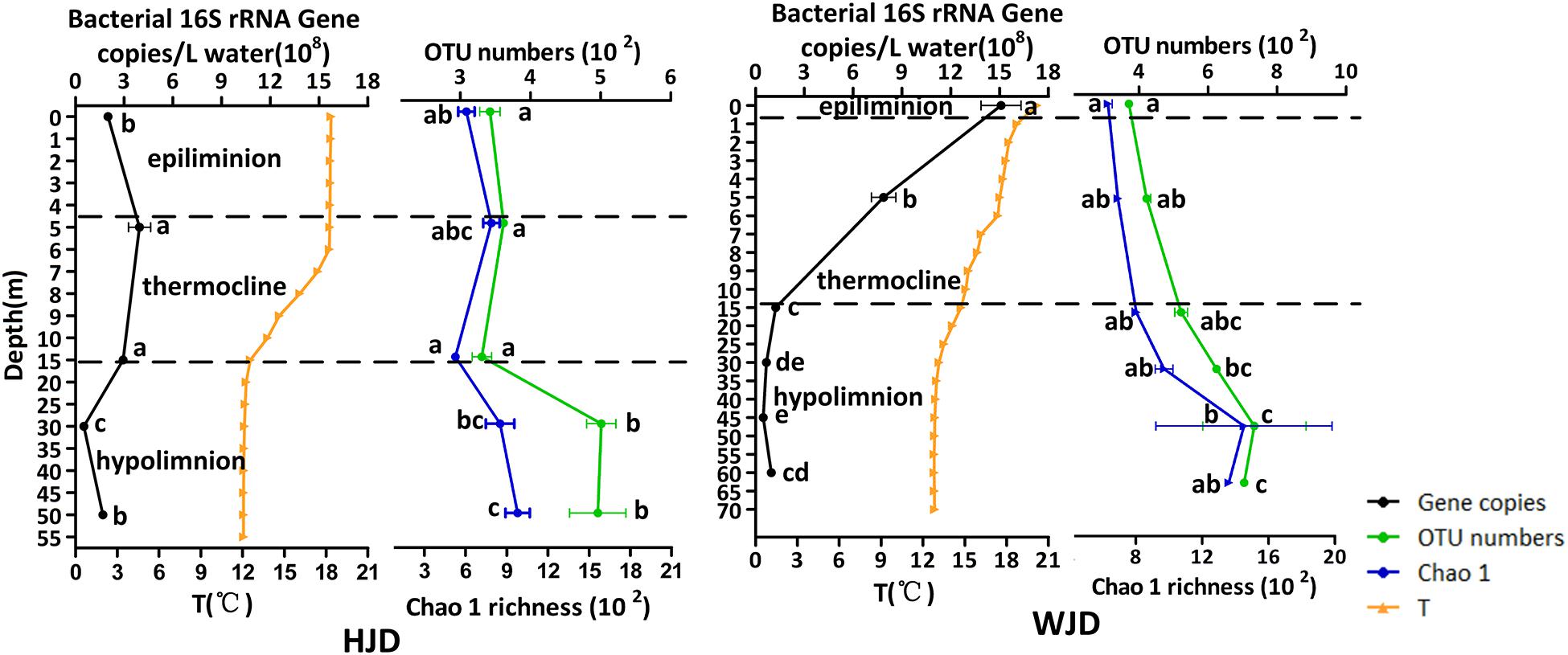
Figure 2. Variations in bacterial abundance, operational taxonomic units (OTUs), Chao1, and water temperature (T) at different water depths in Hongjiadu (HJD) and Wujiangdu (WJD) reservoirs. Error bars represent standard deviation of the mean (n ≥ 2). Different letters above the columns indicate significant differences at p < 0.05.
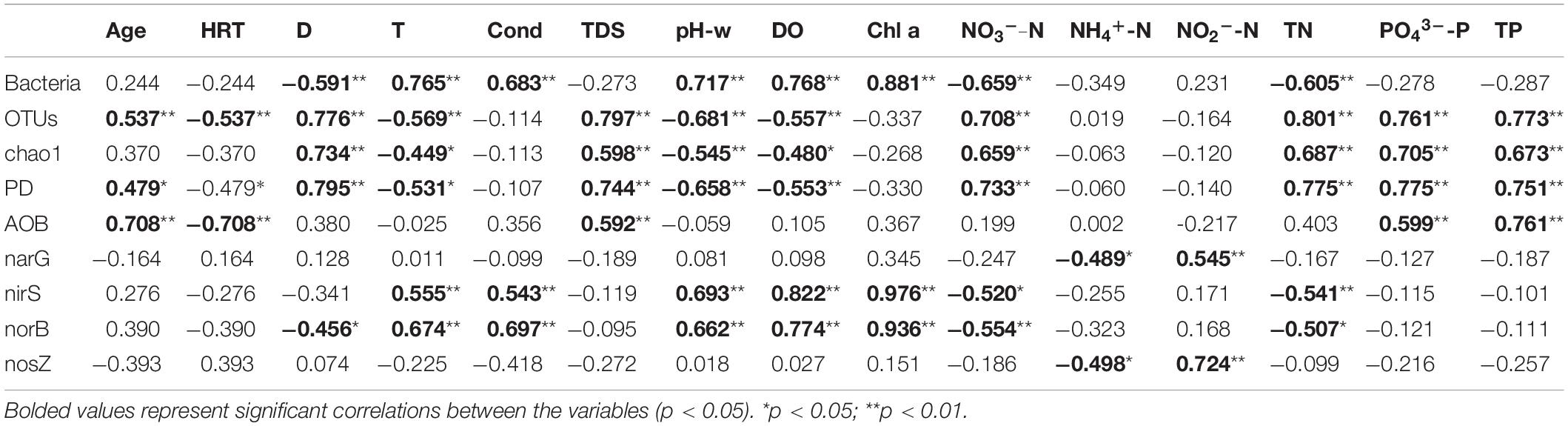
Table 1. Correlation analysis of water bacterial abundance, diversity indexes with the environmental factors.
Generally, 1,422,843 raw sequences were obtained after quality filtering for 26 libraries. The high quality reads of each sample ranged from 30,121 to 44,687 (Supplementary Table 3). Based on the 97% identity threshold (Figure 2), the OTU numbers in HJD and WJD reservoirs ranged from 317 to 536, and 369 to 884, respectively. The Chao 1 index and Faith’s PD of water samples were 473–1,920, and 48.0–134, respectively. Both OTUs and the Chao 1 index were significantly higher in the HJD Reservoir than those in the WJD Reservoir. In water samples, the results in the Chao 1 index corresponded well with those of OTUs, and continued to increase along with the water depth of the reservoirs. In the epilimnion and thermocline of the HJD Reservoir, OTUs and Chao1 showed no significant change, but the hypolimnion exhibited significantly higher changes than the epilimnion and thermocline. However, in the WJD Reservoir, OTUs and Chao1 were not affected by the thermocline, but were inverse to the trend observed for bacterial abundance. The OTU numbers and Chao 1 index of sediments in the two reservoirs varied from 2938 to 3027, and 4,833 to 4,982, respectively, which were significantly higher (3–10 times) than those in the water samples. These measurements suggested that sediments harbored more diverse bacteria than the water within each site. Both water and sediment of WJD Reservoir had greater bacterial diversity and richness. Correlation analysis demonstrated that OTUs and Chao 1 index were positively correlated with D (p < 0.01), TN (p < 0.01), NO3–-N (p < 0.01), TP (p < 0.01), and PO43–-P (p < 0.01) and negatively correlated with T (p < 0.01), DO (p < 0.01), and pH-w (p < 0.01) in water samples from the two reservoirs (Table 1).
The bacterial community among the 26 samples was analyzed by PCoA based on the unweighted-unifrac matrix at the OTU level (Figure 3A). The variance of 37.5% was attributed to PC1, whereas PC2 captured 13.4% of the variance. The whole water communities were found to be separated into six distinct groups and the sediment samples from the two reservoirs clustered together. Corresponding to the water stratification, the epilimnion, thermocline, and hypolimnion of the two reservoirs clustered together. Consistent with the results of bacterial abundance and diversity, significant differences in bacterial communities were observed between sediments and water as well as in different water layers.
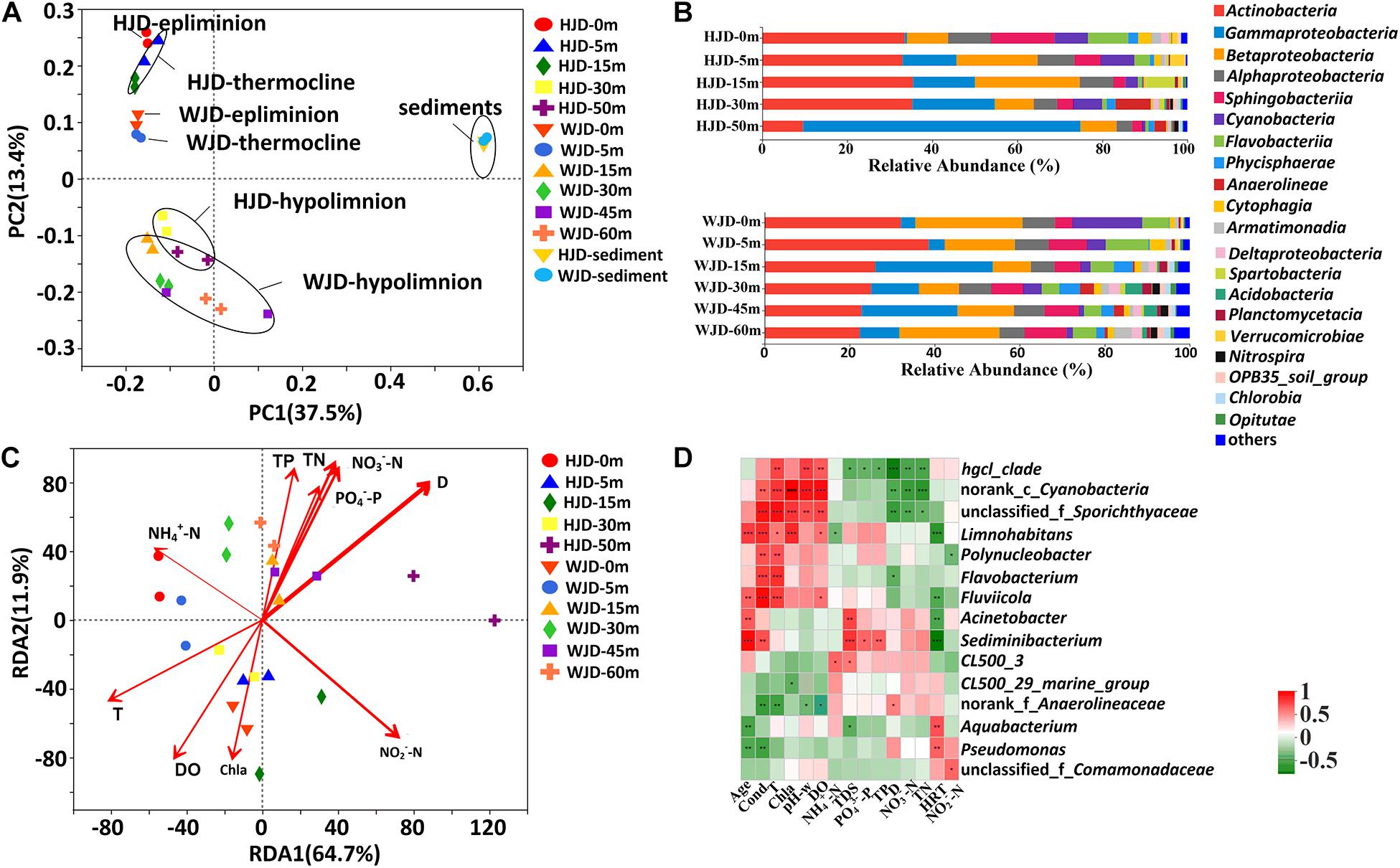
Figure 3. Beta diversity analysis, taxonomical distribution, and redundancy analysis (RDA) of the bacterial community composition in Hongjiadu (HJD) and Wujiangdu (WJD) reservoirs. (A) Principal coordinates analysis (PCoA) of each sample at the OTU level based on the unweighted-unifrac matrix between samples. Percentages of the variation explained by each axis are indicated on the figure. Different circles represent the samples that were collected from the different groups. (B) The relative abundance of bacteria at class level in water samples. Only the classes that have relative abundance > 1% are shown. (C) RDA biplot of the community composition and environmental variables in water. The red arrows denote the environmental variables, including D (water depth), DO (water dissolved oxygen), T (water temperature), Chla (water Chlorophyll a), NO3–-N (water nitrate), NH4+-N (water ammonium), NO2–-N (water nitrite), TN (water total nitrogen), TP (water total phosphorus) and PO43–-P (water orthophosphate). (D) Heatmap of environmental factors affecting bacterial community structure at genus level among reservoir water samples. D (water depth), DO (water dissolved oxygen), T (water temperature), Chla (water Chlorophyll a), NO3–-N (water nitrate), NH4+-N (water ammonium), NO2–-N (water nitrite), TN (water total nitrogen), TP (water total phosphorus) and PO43–-P (water orthophosphate) were measured in water.
The relative abundance of the bacterial community at the phylum level is shown in Supplementary Figure 3A. Overall, the two reservoirs contained 59 bacterial phyla, including 11 and 17 dominant phyla (relative abundance > 1% of the total in each sample) in the water column and sediment (Supplementary Figure 2), respectively. Proteobacteria, Actinobacteria, and Bacteroidetes were the predominant phyla across the water columns of the two reservoirs. In HJD Reservoir, Proteobacteria ranged from 22.31 to 78.04% and exhibited significant vertical differences. The relative abundance of Proteobacteria in bottom water had a high degree of outbreak during the stratification period, accounting for more than 78% of the bacterial community at 50 m. This value was much higher than that the values in the middle and upper waters, with reduced proportions of other phyla, including Actinobacteria, Bacteroidea, Cyanobacteria, and Verrucomicrobia. Actinobacteria represented 32.99–35.47% of the phyla, with no vertical difference in 0 to 30 m, but minimum proportion (9.60%) in 50 m. The relative abundance of Bacteroidetes decreased with increasing water depth. Chloroflexi, Acidobacteria, Nitrospirae, and Chlorobi were higher from 30 to 50 m than from 0 to 15 m. In WJD Reservoir, Actinobacteria, Bacteroidetes, and Verrucomicrobia represented 22.39 to 38.60%, 11.62 to 22.72%, and 1.67 to 4.21%, of the phyla, respectively, with no vertical difference. Cyanobacteria ranged from 1.11 to 16.46% with the maximum abundance (16.46%) at 0 m. The proportion of Planctomycetes, Chloroflexi, Armatimonadetes, Acidobacteria, Nitrospirae, and Chlorobi showed significant vertical differences and was higher from 15 to 60 m than from 0 to 5 m.
The bacterial classes were also investigated, and twenty dominant classes (relative abundance > 1%) were screened among samples (Figure 3B). In HJD Reservoir, both γ- and β-Proteobacteria exhibited significant vertical differences. γ-Proteobacteria increased with increasing water depth, with the maximum abundance (65.24%) at 50 m and minimum proportion (0.53%) at 0 m. β-Proteobacteria were more abundant at 5 and 15 m than at 0, 30, and 50 m. Sphingobacteriia, Flavobacteriia, Cytophagia, Armatimonadia, and Verrucomicrobia were higher from 0 to 5 m than from 15 to 50 m, while that of Planctomycetacia, Nitrospira, and Chlorobia was higher from 30 to 50 m than from 0 to 15 m. In WJD Reservoir, γ- and β-Proteobacteria showed inverse trends, accounting for 3.41–27.64% and 8.95–25.19%, respectively. δ-Proteobacteria were higher from 15 to 60 m than from 0 to 5 m and α-Proteobacteria showed no vertical difference. Flavobacteriia were higher from 0 to 5 m than from 15 to 60 m, whereas Anaerolineae, Nitrospira, and Acidobacteria were higher from 30 to 60 m than from 0 to 15 m. Phycisphacrae and Chlorobia abundance was higher from 15 to 60 m than from 0 to 5 m.
At the genus level, the dominant genera were defined as a relative abundance of more than 5% (Supplementary Figure 3B). In HJD Reservoir, the hgcl_clade was the dominant genus at 0, 5, and 15 m, hgcl_clade, Pseudomonas and CL500-29-marine-group were dominant at 30 m. The proportion of Pseudomonas increased with increasing water depth, especially at 50 m, and Pseudomonas was the absolute dominant genus, resulting in the proportional compression of the other genera. Limnohabitans showed no vertical differences. The abundances of Unclassified_f_Sporichthyaceae, Flavobacterium. and Polynucleobacter were higher from 0 to 5 m than from 15 to 60 m, whereas those of Norank_f_Anaerolineaccae were higher from 30 to 50 m than from 0 to 15 m. Unclassified_f_Comamonadaceae and Terrimicrobium were more abundant at 15 m than at other water depths, accounting for 14.05 and 6.45%, respectively. In WJD Reservoir, hgcl_clade, Limnohabitans, and Cyanobacteria were dominant at 0 m, hgcl_clade was dominant at 5 and 30 m, Acinetobacter was dominant at 15 and 45 m, and hgcl_clade and Limnohabitans were dominant at 60 m. Acinetobacter and Limnohabitans represented 2.86–25.08% and 2.22–17.08%, respectively, with significant vertical differences. Unclassified_f_Sporichthyaceae, Flavobacterium and unclassified_f_Comamonadaceae were more abundance from 0 to 5 m than from 15 to 60 m. The distributions of the bacterial community at the phylum, class, and genus levels clearly exhibited significant vertical differences in the stratification of reservoir water.
Redundancy analysis (RDA) evaluated the influences of environmental factors on bacterial communities in water (Figure 3C). The eigenvalues of the first two principal components closely approached 73.7%. The cumulative contribution ratios of RDA 1 and RDA 2 were 47.5 and 26.2%, respectively. The results indicated that TN (p < 0.01), NO2–-N (p < 0.01), NO3–-N (p < 0.05), TP (p < 0.05), and PO43–-P (p < 0.05) were significantly positively correlated with the variations in bacterial communities in water. Overall, nutrient nitrogen was a crucial driver in the water bacterial communities. Based on the Pearson correlation analysis, the relationship between environmental parameters and the bacterial community was investigated (Figure 3D and Supplementary Figure 4). At the phylum level, Bacteroidetes, Cyanobacteria, Chloroflexi, Armatimonadetes and Nitrospirae were significantly correlated with T. Actinobacteria, Armatimonadetes, TM6_Dependentiae_, Nitrospirae, Chlorobi, and Acidobacteria were significantly correlated with DO (Supplementary Figure 4A). At the class level, Flavobacteriia, Phycisphaerae, Anaerolineae, and Cytophagia were significantly positively correlated for NO3–-N, NH4+-N, TN, PO43–-P, and TP. γ-Proteobacteria and β-Proteobacteria exhibited a negative relationship with TN and NH4+-N (Supplementary Figure 4B). At the genus level, Limnohabitans, Fluviicola, Acinetobacter, and Sediminibacterium showed a positive relationship with age, but a negative relationship with HRT (Figure 3D). Pseudomonas and Aquabacterium were significantly positively related with HRT, but negatively related with age. Hgcl_clade, norank_c_Cyanobacteria, unclassified_f_Sporichthyaceae, Limnohabitans, Polynucleobacter, Flavobacterium and Fluviicola showed a positive relationship with T.
To understand the biological features and functional contributions of the microbial population at different water depths, metagenomes were predicted by PICRUSt using the 16S rRNA gene amplicon data. Metabolism pathways of microbial community predicted by PICRUSt were further analyzed as shown in Figure 4. The relative abundance of the metabolism pathway was the highest among the level 1 Kyoto Encyclopedia of Genes and Genomes (KEGG) categories in two reservoirs, accounting for 73.0–78.9% in HJD Reservoir and from 77.4 to 78.4% in WJD Reservoir (Figure 4A). Carbohydrate metabolism and amino acid metabolism were the top two pathways among the metabolism categories at level 2 (Figure 4B). Carbohydrate metabolism including pyruvate metabolism, glyoxylate and dicarboxylate metabolism, propanoate metabolism, butanoate metabolism, and others, was relatively highly abundant at 50 m in HJD Reservoir. At the same depth, amino acid metabolism pathways were also highly abundant. These included glycine, serine and threonine metabolism, cysteine and methionine metabolism, valine, leucine and isoleucine degradation, alanine, aspartate and glutamate metabolism, arginine and proline metabolism, and others. The top 30 metabolic pathways among the level 3 KEGG categories are listed in Figure 4C. Generally, the biosynthesis of amino acids and carbon metabolism were the most abundant pathways in the two reservoirs. Interestingly, environmental information processing related pathways, such as ABC transporters and two-component system, displayed high relative abundance in HJD Reservoir compared to that in WJD Reservoir, especially at 50 m, indicating different enriched KEGG pathway patterns between these reservoirs.
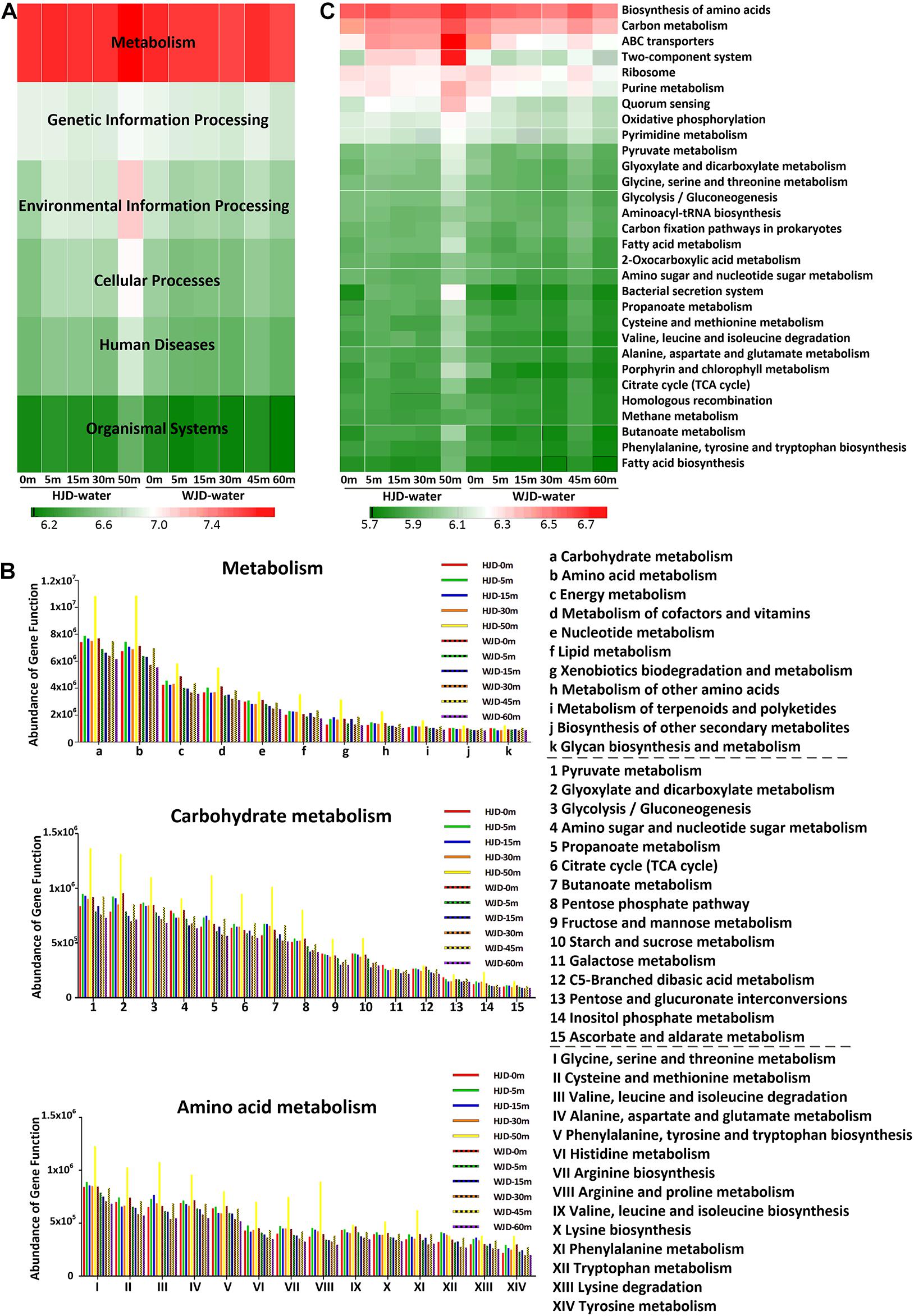
Figure 4. Functional prediction of the bacterial communities at different depths in two reservoirs. (A) Relative abundances of metabolic pathways on KEGG categories (level 1). (B) Relative abundances of metabolism (level 2), carbohydrate metabolism and amino acid metabolism (level 3) based on KEGG categories. (C) Relative abundances of top 30 metabolic pathways based on KEGG categories (level 3).
Co-occurrence network analysis clarified the co-existence relationship among different bacterial genera (Figure 5A). Pseudomonas and Aquabacterium exhibited a significant symbiotic relationship. These two genera were significantly repellent to other dominant bacterial genera. In addition, a significant symbiotic relationship was evident between unclassified_Cyanobacteria, hgcI_clade, unclassified_Comamonadaceae, Sediminibacterium, Limnoh abita, Fluviicola, Polynucleobacter, unclassified_Sporichthyaceae and Flavobacterium. Correlations between the bacterial relative abundance and the enriched KEGG orthologous (KO) gene families are presented in Figure 5B. In brief, 41 KOs were significantly positively correlated with Pseudomonas abundance (p < 0.01 or 0.05), including K02003 (putative ABC transport system ATP-binding protein) and K02004 (putative ABC transport system permease protein) involved in the environmental information processing related pathways. Unclassified_Cyanobacteria and hgcI_clade showed positive correlations with K01952 (phosphoribosylformylglycinamidine synthase), K03657 (DNA helicase II/ATP-dependent DNA helicase PcrA) and K01358 (ATP-dependent Clp protease, protease subunit) (p < 0.01 or 0.05). The abundance of Sediminibacterium and Fluviicola was negatively associated with 23 KOs and 11 KOs, respectively (p < 0.01 or 0.05). Fluviicola, unclassified_Sporichthyaceae and Flavobacterium exhibited positive correlations with both K01952 (phosphoribosylformylglycinamidine synthase) and K00826 (branched-chain amino acid aminotransferase). CL500-3 and CL500-29 marine group exhibited strong negative correlations with K01996 (branched-chain amino acid transport system ATP-binding protein) and K01998 (branched-chain amino acid transport system permease protein), respectively (p < 0.01 or 0.05).
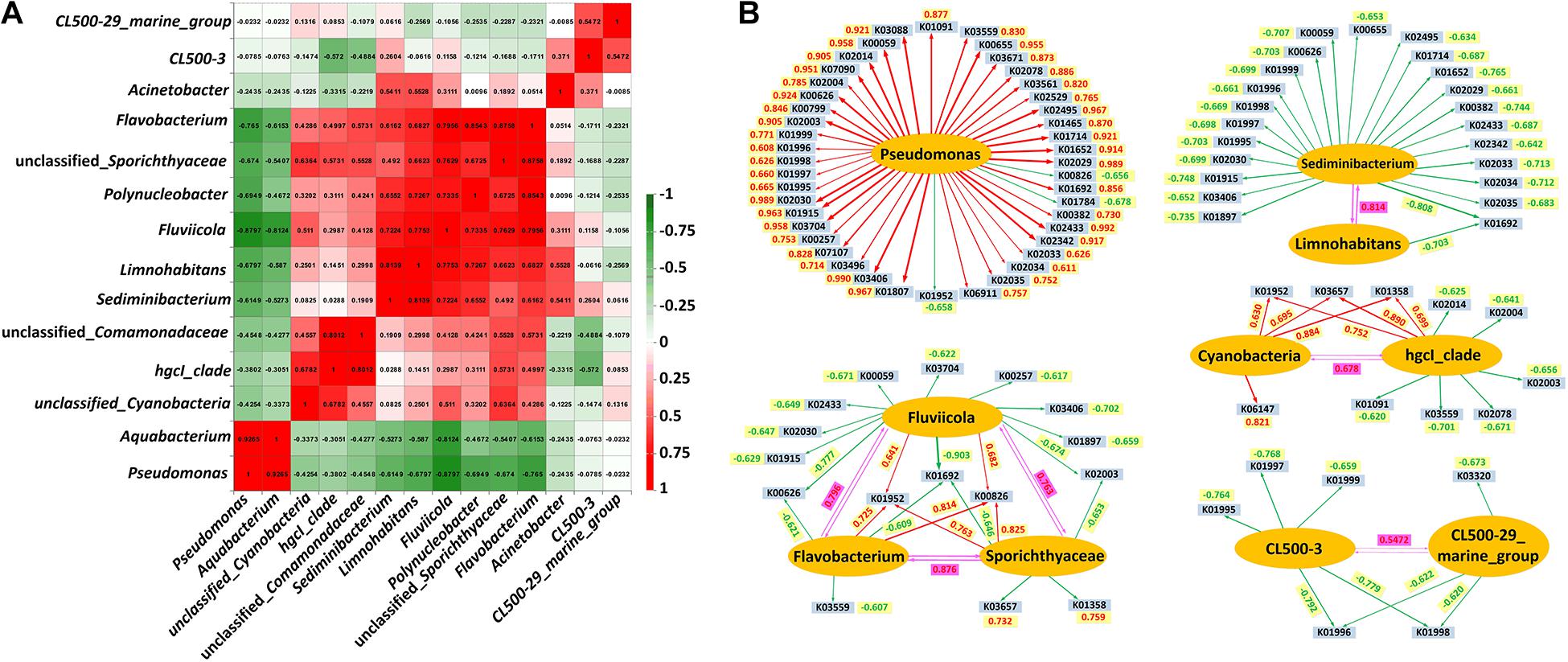
Figure 5. Co-occurrence network analysis of the bacterial communities at different depths in the two reservoirs. (A) Analysis of co-occurrence network among different dominant genera. Only the genera that have relative abundances > 0.5% are shown. (B) Pearson correlations between the KEGG orthologous (KO) gene families (top 50 KOs) and bacterial relative abundances. Only the genera that have relative abundances > 0.5% are shown. Bacterial genera are presented in ellipses with orange background. KOs are depicted in rectangles with light blue background. The correlation between different genera are indicated by a purple background. Positive and negative associations are indicated by red and green arrow lines respectively. Red and green values represented the correlation coefficients of positive and negative associations, respectively.
In HJD Reservoir, the gene abundance of the nitrification gene (AOB) ranged from 9,731 to 1.02 × 105 copies/L (Figure 6A). The AOB abundance decreased from 0 to 15 m, and then increased from 15 to 50 m. However, these changes were not statistically significant. Gene copies of narG and nosZ genes ranged from 9.50 × 104 to 3.21 × 107 copies/L and from 1.55 × 104 to 5.99 × 106 copies/L, respectively. The abundance of narG and nosZ genes increased from 0 to 15 m, decreased sharply at 30 m, and then significantly increased at 50 m. The abundance of nirS and norB genes ranged from 2.14 × 105 to 6.88 × 106 copies/L and from 3.31 × 104 to 8.24 × 105 copies/L, respectively. No significant difference in the nirS and norB genes was observed among the water layers. In WJD Reservoir, the copy number of the AOB gene ranged from 1.16 × 104 to 3.83 × 106 copies/L. The temporal changes in the abundance of this gene were similar to those observed in HJD Reservoir. The copy numbers of narG and nosZ genes ranged from 5.44 × 105 to 1.76 × 107 copies/L and from 1.03 × 104 to 3.07 × 106 copies/L. The abundance was greatest at 0 m and then decreased from 5 to 60 m (although not significantly). The copy numbers of nirS and norB genes ranged from 1.40 × 105 to 2.97 × 107 copies/L and from 8.80 × 104 to 1.18 × 107 copies/L, respectively. The abundance of nirS and norB genes was the highest at 0 m, and no significant difference was observed between 5 and 60 m. Correlation analysis (Figure 6B) revealed the key physicochemical factors explaining changes in the expression of all selected genes. AOB abundance was significantly positively correlated with PO43–-P and TP; narG and nosZ copy numbers were significantly negatively correlated with NH4+-N and positively correlated with NO2–-N; nirS and norB gene copies were significantly negatively correlated with NO3–-N and TN.
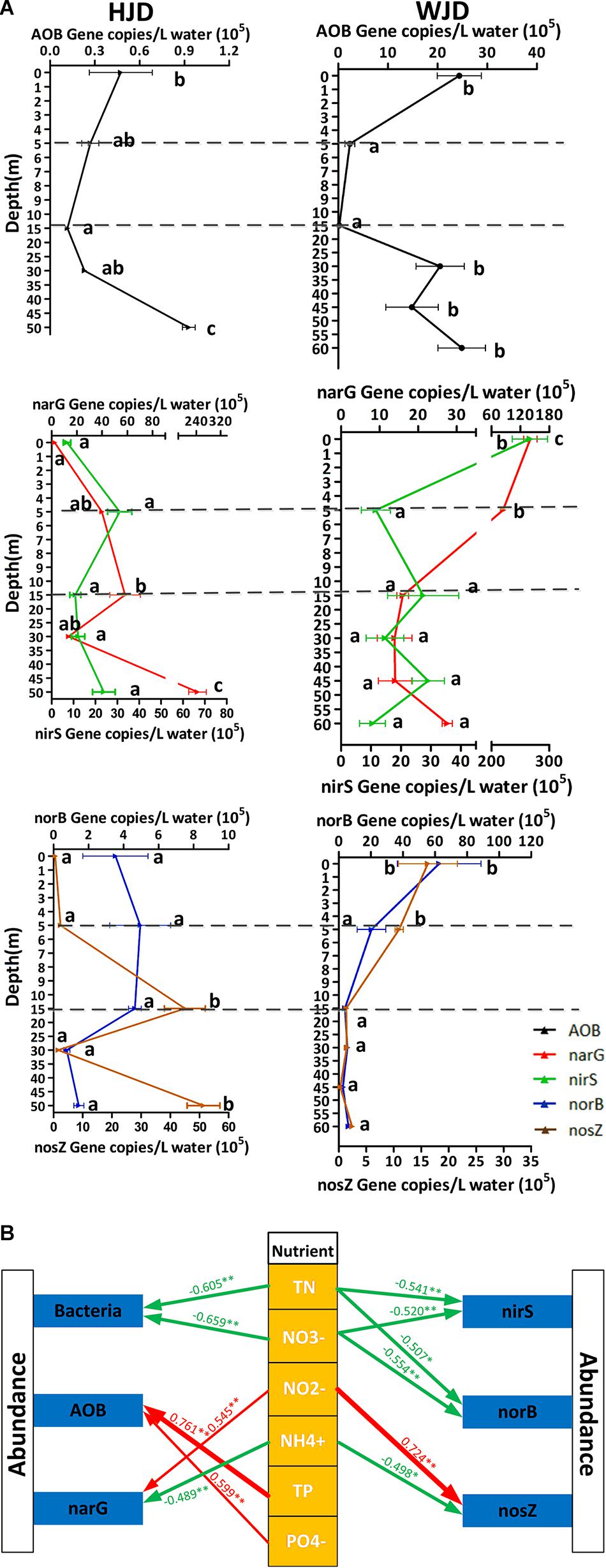
Figure 6. Abundance of nitrogen functional genes and the correlation of environmental factors. (A) The absolute abundance of nitrification and denitrification genes in water stratification of the two reservoirs. Error bars represent the standard deviation (n ≥ 4). Different letters indicate significant differences at p < 0.05 level (one-way ANOVA). (B) SPSS correlation analyses of the abundance, diversity, and environmental nutrient factors. Arrows represent positive (red) or negative (green) correlation between the variables, respectively. The arrow width is proportional to the correlation strength. Numbers on the arrows are the correlation coefficients. ∗p < 0.05; ∗∗p < 0.01.
The trends in variation of bacterial abundance did not correspond with different trophic reservoirs. The spatial change in bacterial abundance in HJD Reservoir was relatively slight. HJD Reservoir was reported as an oligotrophic reservoir (Zhao et al., 2017) with long hydraulic residence time and high light intensity. Bacteria selectively grew in the water layer with appropriate light, and displayed increased abundance with increasing water depth in the epilimnion layer. The appearance of a thermocline reduces the levels of particulate matter (Wilhelm and Adrian, 2008), which provides an epiphytic habitat for microorganisms, resulting in bacterial accumulation. The lack of nutrients and decrease in DO vertical migration (Yu et al., 2014; Zhang et al., 2015) limited the bacterial growth in the hypolimnion layer.
A remarkable spatial dynamic distribution of bacterial abundance has been observed in WJD Reservoir (Figure 2). WJD Reservoir has been facing a serious threat to eutrophication (Zhao et al., 2017). The trophic status might be a key driving force for increased bacterio-plankton abundance in freshwater (Lindstrm, 2000; Dai et al., 2016). Excessive nutrients and weak light transmittance can promote the growth and reproduction of phytoplankton in surface water (Rother, 1977; Feng et al., 2019). In this study, Chla and cyanobacteria contents were 15.5 μg/L and 16.5%, respectively, WJD surface water. Furthermore, the growth and decomposition of phytoplankton could induce the growth of bacteria (Paver et al., 2013). This may be the reason for the higher bacterial abundance in surface water of the more eutrophic WJD Reservoir than that in the surface water of the oligotrophic HJD Reservoir. However, excessive oxygen consumption by algae and the release of harmful algal toxins (Berg et al., 2009) might lead to extensive bacterial death (Nitin et al., 2017). In addition, the thermocline hindered the mixing of the water column, limited the vertical migration of DO and particulate matter, increased the heterogeneity of the water column (Selmeczy et al., 2016), and resulted in the generation of an environment that was less conducive to bacterial growth. Meanwhile, low DO stimulated the release of nutrients from the sediment to the overlying water (Rigosi and Rueda, 2012), which would worsen the water quality (Elçi, 2008) and might aggravate the degree of eutrophication of WJD Reservoir.
In this study, OTUs, richness, and diversity of bacteria in the sediments of the two reservoirs were found to be significantly higher than those in the water, consistent with the results of previous studies (Dai et al., 2016; Feng et al., 2019; Yue et al., 2021). In the vertical distribution of the water column, the OTUs, richness, and diversity of bacteria in eutrophic reservoirs were also significantly higher than those in oligotrophic reservoirs and were significantly correlated with variables related to the nutritional status. Recent studies have shown that the nutritional status might be an important driving force for increased bacterial community diversity (Li et al., 2017; Sun et al., 2020). In our study, bacterial OTUs, richness, and diversity of the hypolimnion were found to be significantly greater than those of the epilimnion and thermocline in the same reservoir. It was possible that the thermal stratification caused hypoxia in the hypolimnion and stimulated nutrient decomposition in the sediment, which increased the number of microorganisms entering the hypolimnion.
Actinobacteria (8.77–37.3%) was the dominant phylum at most water depths in the two reservoirs (Allgaier and Grossart, 2006). The finding indicates that Actinobacteria was unaffected by the nutritional status and mixed water environment (Nitin et al., 2017). However, compared to WJD Reservoir (the eutrophic reservoir), HJD Reservoir (the oligotrophic reservoir) displayed a more pronounced vertical spatial difference in terms of bacterial community structure during thermal stratification. Correspondingly, predictive metagenomic profiling revealed different vertical functional variation patterns of the bacterial community in the two reservoirs. Bacteroidetes (26.1–28.6%), β-Proteobacteria (16.7–28.1%), and γ-Proteobacteria (16.8–66.2%) were dominant in the epilimnion, thermocline, and hypolimnion of HJD Reservoir, respectively. Pseudomonas was the most dominant genus at 50 m, and was significantly involved in 44 KOs (Figure 5B and Supplementary Table 4), such as the functional genes related to metabolism and environmental information processing pathways. Considering that the competition among the bacterial communities for the nutrient resource might be intense, given the oligotrophic nature of this reservoir, the high abundance of Pseudomonas at 50 m might be associated with the multiple roles of these bacteria in the water column.
Nutritional status is considered an important determinant of various aspects of bacterial community structure (Wu et al., 2010; Jankowski et al., 2014; Davis et al., 2015; Zwirglmaier et al., 2015). It has been reported that TP and TN significantly contribute to the bacterial community in the Qingliu River that had been affected by domestic sewage (Zhang et al., 2020). Li et al. (2017) also showed that the nutrients (especially total carbon) play a more important role rather than the sample locations in shifting bacterial structure in the shallow small lakes. Our RDA results showed that the bacterial community structure in the two reservoirs was significantly correlated with nutrient parameters, in agreement with the results from previous studies. Furthermore, the co-occurrence network analysis based on our dataset revealed the significant symbolic relationships among Cyanobacteria, Actinobacteria (hgcl_clade, Sporichthyaceae) and β-Proteobacteria (Comamonadaceae, Limnohabita, Polynucleobacter), consistent with the reported observations from other studies. For instance, α-Proteobacteria (Berg et al., 2009), β-Proteobacteria (Eiler et al., 2012) and Actinobacteria (Zhao et al., 2016) are respectively in physical association with some members of Cyanobacteria in freshwater systems. Similarly, Chloroflexi, Acidobacteria, Cyanobacteria, and Planctomycetes have significant co-occurrence relationships with many phyla in the surface water in Xukou Bay of Lake Taihu (Cao et al., 2018).
Proteobacteria (Hou et al., 2018), Bacteroidetes (Wu et al., 2018), Actinobacteria (Cetin et al., 2018), Cyanobacteria (Yu et al., 2014), Planctomycetes (Han and Li, 2016), and Flavobacterium (Hou et al., 2017) are involved in aspects of the nitrogen cycle that include nitrification, denitrification, nitrogen fixation, and nitrate reduction. β-Proteobacteria are responsible for degrading pollutants, which are positively correlated with total nitrogen (Bai et al., 2012) and contribute to nitrogen removal (Jie et al., 2018). γ-Proteobacteria and δ-Proteobacteria are negatively correlated with NH4+-N and NO3–-N, respectively (Zhang et al., 2015). Chloroflexi (Wang et al., 2017) and Flavobacteria (Vu et al., 2019) are important for nutrient removal and organic carbon decomposition, respectively. HgcI_clade (Garcia et al., 2012), Limnohabitans (Chai et al., 2017), and Comamonadaceae (Ge et al., 2015) are responsible for phosphorous pathways that include accumulation and removal. Therefore, thermal stratification (Zhou et al., 2020) and nutrients might play important roles in shaping the bacterial community structure of reservoirs with different nutritional status.
In this study, qPCR was used to estimate the vertical distribution of water column nitrification and denitrification abundance in HJD and WJD reservoirs. In HJD Reservoir, nitrification and denitrification in the hypolimnion were much higher than those in the thermocline. DO was the main limiting factor in denitrification owing to the lack of nutrients in the water column. The emergence of a thermocline reduced the DO content and increased denitrification. This occurred because the DO content in the upper water could not supply the hypolimnion, which stimulated an increase in denitrification. Despite the general assumption that aerobic conditions are conducive to nitrification, nitrification was greatest in the hypolimnion. Correlation analysis of the process of nitrification revealed a positive correlation between amoA and nirS and norB. These findings suggested that nitrification and denitrification had coupled and exhibited synergistic effects in the water column. In the hypolimnion, denitrification resulted in the production of inorganic substances and provided NH4+ to facilitate nitrification. Meanwhile, with the increased nitrification in the hypolimnion, DO was continuously consumed and promoted denitrification.
However, in WJD Reservoir, no significant difference was observed in the nitrification and denitrification processes in the different thermal strata of the water column, indicating that the sensitivity of nitrification and denitrification was less affected by heterogeneity in the vertical direction of the reservoir’s water column. The correlation analysis also revealed that NO3–-N, NH4+-N, NO2–-N, TN, TP, and PO43–-P were significantly correlated with nitrification gene (AOB) and denitrification genes (nirS, norB). These findings suggest that nutrient sufficiency is the main limiting factor for nitrification and denitrification processes in eutrophication reservoirs (Tang et al., 2016). The sudden reduction in DO (Yu et al., 2014; Zhang et al., 2015) and the slow migration of particulate matter (Wilhelm and Adrian, 2008) caused by thermal stratification had little effect on nitrification and denitrification.
We examined the bacterial community structures based on quantitative real-time PCR and high-throughput sequencing in two different trophic reservoirs. During water stratification in the water column, bacterial abundance in HJD Reservoir was slightly affected by the thermocline. In WJD Reservoir, bacterial gene copies sharply decreased with increasing water depth and exhibited obvious spatial differences. Proteobacteria, Actinobacteria, and Bacteroidetes were the dominant phyla. The dominant classes and genera were influenced by changes in water stratification and trophic levels. RDA and heatmap analysis indicated that thermal stratification and nutrients were derived from variations in the bacterial community structure. Moreover, significant differences were evident in nitrification and denitrification. The latter was markedly more extensive than nitrification, regardless of nutritional status. These results provide valuable insights into mechanisms by which water stratification and trophic levels control bacterial abundance and structure in cascade reservoirs. A limitation of the study was that it was conducted in April and stratification is more pronounced during summer. A study conducted in summer may reveal more interesting variations in bacterial structure.
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/, PRJNA680428.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was sponsored by National Key Research and Development Project by MOST of China (grant no. 2016YFA0601001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Jie Pan (Institute for Advanced Study, Shenzhen University, China) for the assistance during high-throughput sequencing analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.632089/full#supplementary-material
Allgaier, M., and Grossart, H. P. (2006). Diversity and seasonal dynamics of actinobacteria populations in four lakes in northeastern germany. Appl. Environ. Microbiol. 72, 3489–3497. doi: 10.1128/AEM.72.5.3489-3497.2006
Annika, C. M., and Christopher, A. F. (2010). Denitrifier abundance and activity across the San Francisco Bay estuary. Environ. Microbiol. Rep. 2, 667–676.
Bai, Y. H., Shi, Q., Wen, D. H., Li, Z. X., Jefferson, W. A., Feng, C., et al. (2012). Bacterial communities in the sediments of Dianchi Lake, a partitioned eutrophic waterbody in China. PLoS One 7:e37796. doi: 10.1371/journal.pone.0037796
Berg, K. A., Lyra, C., Sivonen, K., Paulin, L., Suomalainen, S., Tuomi, P., et al. (2009). High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J. 3, 314–325. doi: 10.1038/ismej.2008.110
Braker, G., Matthies, D., Hannig, M., Brandt, F. B., Brenzinger, K., and Grongroft, A. (2015). Impact of land use management and soil properties on denitrifier communities of Namibian Savannas. Microb. Ecol. 70, 981–992.
Canfield, D. E., Glazer, A. N., and Falkowski, P. G. (2010). The evolution and future of Earth’s nitrogen cycle. Science 330, 192–196. doi: 10.1126/science.1186120
Cao, X. Y., Zhao, D. Y., Xu, H. M., Huang, R., Zeng, J., and Yu, Z. B. (2018). Heterogeneity of interactions of microbial communities in regions of Taihu Lake with different nutrient loadings: a network analysis. Sci. Rep. 8:8890. doi: 10.1038/s41598-018-27172-z
Cetin, E., Karakas, E., Dulekgurgen, E., Ovez, S., Kolukirik, M., and Yilmaz, G. (2018). Effects of high-concentration influent suspended solids on aerobic granulation in pilot-scale sequencing batch reactors treating real domestic wastewater. Water Res. 131, 74–89. doi: 10.1016/j.watres.2017.12.014
Chai, X., Wu, B., Xu, Z., Yang, N., Song, L., Mai, J., et al. (2017). Ecosystem activation system (EAS) technology for remediation of eutrophic freshwater. Sci. Rep. U.K. 7:4818. doi: 10.1038/s41598-017-04306-3
Chen, X., Yang, L., Xiao, L., Miao, A., and Xi, B. (2012). Nitrogen removal by denitrification during cyanobacterial bloom in Lake Taihu. J. Freshw. Ecol. 27, 243–258. doi: 10.1080/02705060.2011.644405
Conley, D. J., Paerl, H. W., Howarth, R. W., Boesch, D. F., Seitzinger, S. P., Havens, K. E., et al. (2009). Controlling eutrophication:Nitrogen and phosphorus. Science 323, 1014–1015. doi: 10.1126/science.1167755
Cornwell, J. C., Glibert, P. M., and Owens, M. S. (2014). Nutrient fluxes from sediments in the San Francisco Bay Delta. Estuar. Coast. 37, 1120–1133. doi: 10.1007/s12237-013-9755-4
Daggett, C. T., Saros, J. E., Lafrancois, B. M., Simon, K. S., and Amirbahman, A. (2015). Effects of increased concentrations of inorganic nitrogen and dissolved organic matter on phytoplankton in boreal lakes with differing nutrient limitation patterns. Aquat Sci. 77, 511–521. doi: 10.1007/s00027-015-0396-5
Dai, Y., Yang, Y., Wu, Z., Feng, Q., Xie, S., and Liu, Y. (2016). Spatiotemporal variation of planktonic and sediment bacterial assemblages in two plateau freshwater lakes at different trophic status. Appl. Microbiol. Biot. 100, 4161–4175. doi: 10.1007/s00253-015-7253-2
Davis, T. W., Bullerjahn, G. S., Tuttle, T., Mckay, R. M., and Watson, S. B. (2015). Effects of increasing nitrogen and phosphorus concentrations on phytoplankton community growth and toxicity during planktothrix blooms in sandusky bay, lake erie. Environ. Sci. Technol. 49, 7197–7207. doi: 10.1021/acs.est.5b00799
Eiler, A., Heinrich, F., and Bertilsson, S. (2012). Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 6, 330–342. doi: 10.1038/ismej.2011.113
Elçi, Ş (2008). Effects of thermal stratification and mixing on reservoir water quality. Limnology 9, 135–142. doi: 10.1007/s10201-008-0240-x
Feng, C., Jia, J., Wang, C., Han, M., Dong, C., Huo, B., et al. (2019). Phytoplankton and bacterial community structure in Two Chinese Lakes of different trophic status. Microorganisms 7:621. doi: 10.3390/microorganisms7120621
Gao, J., Hou, L. J., Zheng, Y. L., Liu, M., Yin, G. Y., Li, X. F., et al. (2016). nirS-encoding denitrifier community composition, distribution, and abundance along the coastal wetlands of China. Appl. Microbiol. Biot. 100, 8573–8582.
Garcia, S. L., McMahon, K. D., Martinez-Garcia, M., Srivastava, A., Sczyrba, A., Stepanauskas, R., et al. (2012). Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J. 7, 137–417. doi: 10.1038/ismej.2012.86
Ge, H., Batstone, D. J., and Keller, J. (2015). Biological phosphorus removal from abattoir wastewater at very short sludge ages mediated by novel PAO clade Comamonadaceae. Water Res. 69, 173–182. doi: 10.1016/j.watres.2014.11.026
Han, H., and Li, Z. (2016). Effects of macrophyte-associated nitrogen cycling bacteria on ANAMMOX and denitrification in river sediments in the Taihu Lake region of China. Ecol. Eng. 93, 82–90. doi: 10.1016/j.ecoleng.2016.05.015
Hosomi, M., and Sudo, R. (1986). Simultaneous determination of total nitrogen and total phosphorus in freshwater samples using persulfate digestion. Int. J. Environ. Stud. 27, 267–275. doi: 10.1080/00207238608710296
Hou, D. W., Huang, Z. J., Zeng, S. Z., Liu, J., Wei, D. D., Deng, X. S., et al. (2017). Environmental factors shape water microbial community structure and function in shrimp cultural enclosure ecosystems. Front. Microbiol. 8:2359. doi: 10.3389/fmicb.2017.02359
Hou, L., Zhou, Q., Wu, Q., Gu, Q., Sun, M., and Zhang, J. (2018). Spatiotemporal changes in bacterial community and microbial activity in a full-scale drinking water treatment plant. Sci. Total Environ. 625, 449–459. doi: 10.1016/j.scitotenv.2017.12.301
Houshmand, H., Ahangarzadeh, M., Dehghan, M. S., Mortezaei, S., and Reza, S. (2020). Evaluation of SEYMAREH dam reservoir water quality by bacterial indices and relationship-with some water physical and chemical parameters. ISFJ. 28, 89–97.
Jankowski, K., Schindler, D. E., and Horner-Devine, M. C. (2014). Resource availability and spatial heterogeneity control bacterial community response to nutrient enrichment in lakes. PLoS One 9:e86991. doi: 10.1371/journal.pone.0086991
Jardim, P. F., Melo, M. M. M., Ribeiro, L. C., Collischonn, W., and Paz, A. R. (2020). A modeling assessment of large-scale hydrologic alteration in South American pantanal due to upstream dam operation. Front. Environ. Sci. 8:567450. doi: 10.3389/fenvs.2020.567450
Jie, G., Yu, D., Ying, L., Zhuang, X., Liu, Y., Bai, Z., et al. (2018). Long- and short-chain AHLs affect AOA and AOB microbial community composition and ammonia oxidation rate in activated sludge. J. Environ. Sci. 78, 53–62. doi: 10.1016/j.jes.2018.06.022
Kirillin, G., and Shatwell, T. (2016). Generalized scaling of seasonal thermal stratification in lakes. Earth Sci. Rev. 161, 179–190.
Li, J. Y., Zhang, Y. F., Yang, Z., and Wang, M. (2017). Bacterial diversity in shahu lake, northwest China is significantly affected by nutrient composition rather than location. Ann. Microbiol. 67, 469–478.
Lindstrm, E. S. (2000). Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb. Ecol. 40, 104–113. doi: 10.1007/s002480000036
Mao, G. Z., Chen, L., Yang, Y. Y., Wu, Z., Tong, T. L., Liu, Y., et al. (2017). Vertical profiles of water and sediment denitrifiers in two plateau freshwater lakes. Appl. Microbiol. Biot. 101, 3361–3370.
Müller, S., Mitrovic, S. M., and Baldwin, D. S. (2016). Oxygen and dissolvedorganic carbon control release of N, P and Fe from the sediments of a shallow, polymictic lake. J. Soil. Sediment 16, 1109–1120.
Nitin, P. N., Pandurang, K., Andrew, J., Synne, K., Hans, U., Anette, J., et al. (2017). Characterization of bacterial community associated with phytoplankton bloom in a eutrophic lake in south norway using 16s rrna gene amplicon sequence analysis. PLoS One 12:e0173408. doi: 10.1371/journal.pone.0173408
Oshiki, M., Satoh, H., and Okabe, S. (2016). Ecology and physiology of anaerobic ammonium oxidizing bacteria. Environ. Microbiol. 18, 2784– 2796.
Paerl, H. W., Scott, J. T., McCarthy, M. J., Newell, S. E., Gardner, W. S., Havens, K. E., et al. (2016). It takes two to tango: when and where dual nutrient (N & P) reductions are needed to protect lakes and downstream ecosystems. Environ. Sci. Technol. 50, 10805–10813. doi: 10.1021/acs.est.6b02575
Paver, S. F., Hayek, K. R., Gano, K. A., Fagen, J. R., Brown, C. T., Davis-Richardson, A. G., et al. (2013). Interactions between specific phytoplankton and bacteria affect lake bacterial community succession. Environ. Microbiol. 15, 2489–2504.
Peng, X., Ji, Q., Angell, J. H., Kearns, P. J., Yang, H. J., Bowen, J. L., et al. (2016). Long-term fertilization alters the relative importance of nitrate reduction pathways in salt marsh sediments. J. Geophys. Res-Biogeo. 121, 2082–2095. doi: 10.1002/2016JG003484
Petty, E. L., Obrecht, D. V., and North, R. L. (2020). Filling in the flyover zone: high phosphorus in midwestern (usa) reservoirs results in high phytoplankton biomass but not high primary productivity. Front. Environ. Sci. 8:111. doi: 10.3389/fenvs.2020.00111
Rigosi, A., and Rueda, F. J. (2012). Hydraulic control of short-term successional changes in the phytoplankton assemblage in stratified reservoirs. Ecol. Eng. 44, 216–226. doi: 10.1016/j.ecoleng.2012.04.012
Rother, A. J. (1977). The Annual Cycle of Growth and Reproduction of Planktonic Blue-Green Algae in the Salopian meres. Bethnal Green: Queen Mary University Lond.
Ruiz-González, C., Niño-García, J. P., Lapierre, J. F., and del Giorgio, P. A. (2015). The quality of organic matter shapes the functional biogeography of bacterioplankton across boreal freshwater ecosystems. Glob. Ecol. Biogeogr. 24, 1487–1498. doi: 10.1111/geb.12356
Salcher, M. M., Pernthaler, J., and Posch, T. (2010). Spatiotemporal distribution and activity patterns of bacteria from three phylogenetic groups in an oligomesotrophic lake. Limnol. Oceanogr. 55, 846–856.
Schubert, C. J., Durisch-Kaiser, E., Wehrli, B., Thamdrup, B., Lam, P., and Kuypers, M. M. (2006). Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environ. Microbiol. 8, 1857–1863.
Selmeczy, G. B., Tapolczai, K., Casper, P., Krienitz, L., and Padisák, J. (2016). Spatial- and niche segregation of DCM-forming cyanobacteria in Lake Stechlin (Germany). Hydrobiologia 764, 229–240. doi: 10.1007/s10750-015-2282-5
Shi, W., Chen, Q., Yi, Q., Yu, J., Ji, Y., Hu, L., et al. (2017). Carbon emission from cascade reservoirs: spatial heterogeneity and mechanisms. Environ. Sci. Technol. 51, 12175–12181. doi: 10.1021/acs.est.7b03590
Sinistro, R., Sánchez, M. L., Unrein, F., Schiaffino, M. R., Izaguirre, I., and Allende, L. (2015). Responses of phytoplankton and related microbial communities to changes in the limnological conditions of shallow lakes: a short-term cross-transplant experiment. Hydrobiologia 752, 139–153.
Smith, J. M., Mosier, A. C., and Francis, C. A. (2015). Spatiotemporal relationships between the abundance, distribution, and potential activities of ammonia-oxidizing and denitrifying microorganisms in intertidal sediments. Microb. Ecol. 69, 13–24. doi: 10.1007/s00248-014-0450-1
Sun, R., Tu, Z., and Fan, L. (2020). The correlation analyses of bacterial community composition and spatial factors between freshwater and sediment in Poyang Lake wetland by using artificial neural network (ANN) modeling. Braz. J. Microbiol. 51, 1191–1207. doi: 10.1007/s42770-020-00285-2
Tang, Y. Q., Zhang, X. Y., Li, D. D., Wang, H. M., Chen, F. S., Fu, X. L., et al. (2016). Impacts of nitrogen and phosphorus additions on the abundance and community structure of ammonia oxidizers and denitrifying bacteria in Chinese fir plantations. Soil. Biol. Biochem. 103, 284–293.
Tatti, E., Goyer, C., Burton, D. L., Wertz, S., Zebarth, B. J., Chantigny, M., et al. (2015). Tillage management and seasonal effects on denitrifier community abundance, gene expression and structure over winter. Microb. Ecol. 70, 795–808. doi: 10.1007/s00248-015-0591-x
Tuo, Y. C., Liu, Z. G., and Deng, Y. (2014). Water temperature of the Fengman reservoir with seasonal ice cover. Adv. Water Sci. 25, 731–738. doi: 10.14042/j.cnki.32.1309.2014.05.017
Van Cappellen, P., and Maavara, T. (2016). Rivers in the Anthropocene: global scale modifications of riverine nutrient fluxes by damming. Ecohydrol. Hydrobiol. 16, 106–111. doi: 10.1016/j.ecohyd.2016.04.001
Vu, C. H. T., Chun, S. J., Seo, S. H., Cui, Y., Ahn, C. Y., and Oh, H. M. (2019). Bacterial community enhances flocculation efficiency of Ettlia sp. by altering extracellular polymeric substances profile. Bioresour. Technol. 281, 56–65. doi: 10.1016/j.biortech.2019.02.062
Wang, J., Fu, Z., Qiao, H., and Liu, F. (2019). Assessment of eutrophication and water quality in the estuarine area of Lake Wuli, Lake Taihu, China. Sci. Total. Environ. 650, 1392–1402. doi: 10.1016/j.scitotenv.2018.09.137
Wang, J., Gong, B., Huang, W., Wang, Y., and Zhou, J. (2017). Bacterial community structure in simultaneous nitrification, denitrification and organic matter removal process treating saline mustard tuber wastewater as revealed by 16S rRNA sequencing. Bioresour. Technol. 228, 31–38.
Wang, L. N., Yu, Z., Yang, J., and Zhou, J. (2015). Diazotrophic bacterial community variability in a subtropical deep reservoir is correlated with seasonal changes in nitrogen. Environ. Sci. Pollut. R. 22, 19695–19705.
Wilhelm, S., and Adrian, R. (2008). Impact of summer warming on the thermal characteristics of a polymictic lake and consequences for oxygen, nutrients and phytoplankton. Freshw. Biol. 53, 226–237.
Wu, D., Zhang, Z., Yu, Z., and Zhu, L. (2018). Optimization of F/M ratio for stability of aerobic granular process via quantitative sludge discharge. Bioresour. Technol. 252, 150–156. doi: 10.1016/j.biortech.2017.12.094
Wu, L., Yu, Y. Y., Zhang, T. L., Feng, W. S., Zhang, X., and Li, W. (2010). Pcr-dgge fingerprinting analysis of plankton communities and its relationship to lake trophic status. Int. Rev. Hydrobiol. 94, 528–541. doi: 10.1002/iroh.200911129
Xu, H., Ai, L., Tan, L. C., and An, Z. S. (2006). Geochronology of a surface core in the northern basin of lake qinghai: evidence from 210pb and 137cs radionuclides. Chin. J. Geochem. 25, 301–306.
Xu, N., Tan, G., Wang, H., and Gai, X. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. doi: 10.1016/j.ejsobi.2016.02.004
Yang, X., Huang, T. L., and Zhang, H. H. (2015). Effects of seasonal thermal stratification on the functional diversity and composition of themicrobial community in a drinking water reservoir. Water 7, 5525–5546. doi: 10.3390/w7105525
Yu, Z., Yang, J., Amalfitano, S., Yu, X., and Liu, L. (2014). Effects of water stratification and mixing on microbial community structure in a subtropical deep reservoir. Sci. Rep. 4:5821. doi: 10.1038/srep05821
Yue, Y. H., Wang, F. S., Fu, Z. W., Tang, Y., Ma, J., Qin, Y., et al. (2021). Differential response of microbial diversity and abundance to hydrological residual time and age in cascade reservoirs. J. Soil Sediment. 21, 1290–1301. doi: 10.1007/s11368-020-02800-2
Zhang, H. H., Xu, L., Huang, T. L., Yan, M. M., Liu, K. W., Miao, Y. T., et al. (2021a). Combined effects of seasonality and stagnation on tap water quality: changes in chemical parameters, metabolic activity and co-existence in bacterial community. J. Hazard. Mater. 403:124018. doi: 10.1016/j.jhazmat.2020.124018
Zhang, H. H., Zong, R. R., He, H. Y., Liu, K. W., Yan, M. M., Miao, Y. T., et al. (2021b). Biogeographic distribution pattern of algal community structure in different urban lakes of China: insights into dynamics and co-existence. J. Environ. Sci. 100, 216–227. doi: 10.1016/j.jes.2020.07.024
Zhang, L., Zhong, M. M., Li, X. C., Lu, W. X., and Li, J. (2020). River bacterial community structure and co-occurrence patterns under the influence of different domestic sewage types. J. Environ. Manag. 266:110590. doi: 10.1016/j.jenvman.2020.110590
Zhang, Y. L., Wu, Z. X., Liu, M. L., He, J. B., Shi, K., Zhou, Y. Q., et al. (2015). Dissolved oxygen stratification and response to thermal structure and long-term climate change in a large and deep subtropical reservoir (Lake Qiandaohu, China). Water Res. 75, 249–258. doi: 10.1016/j.watres.2015.02.052
Zhao, D., Shen, F., Zeng, J., Huang, R., Yu, Z., and Wu, Q. L. (2016). Network analysis reveals seasonal variation of co-occurrence correlations between cyanobacteria and other bacterioplankton. Sci. Total Environ. 573, 817–825. doi: 10.1016/j.scitotenv.2016.08.150
Zhao, G., Du, J., Jia, Y., Lv, Y., Han, G., and Tian, X. (2012). The importance of bacteria in promoting algal growth in eutrophic lakes with limited available phosphorus. Ecol. Eng. 42, 107–111. doi: 10.1016/j.ecoleng.2012.02.007
Zhao, L., Guo, Y., Meng, B., Yao, H., and Feng, X. (2017). Effects of damming on the distribution and methylation of mercury in Wujiang River, Southwest China. Chemosphere 185, 780–788. doi: 10.1016/j.chemosphere.2017.07.077
Zhou, S. L., Sun, Y., Huang, T. L., Cheng, Y., Yang, X., Zhou, Z. Z., et al. (2020). Reservoir water stratification and mixing affects microbial community structure and functional community composition in a stratified drinking reservoir. J. Environ. Manag. 267:110456.
Keywords: bacterial community, water stratification, nutrient, reservoir, nitrification, denitrification
Citation: Yue Y, Cai L, Tang Y, Zhang Y, Yang M and Wang F (2021) Vertical Distribution of Bacterial Community in Water Columns of Reservoirs With Different Trophic Conditions During Thermal Stratification. Front. Environ. Sci. 9:632089. doi: 10.3389/fenvs.2021.632089
Received: 22 November 2020; Accepted: 18 March 2021;
Published: 09 April 2021.
Edited by:
Peng Bao, Institute of Urban Environment (CAS), ChinaReviewed by:
Quan Yuan, Institute of Geochemistry (CAS), ChinaCopyright © 2021 Yue, Cai, Tang, Zhang, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Yang, bWluZ3lhbmdAc2h1LmVkdS5jbg==; Fushun Wang, ZnN3YW5nQHNodS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.