- 1State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3State Key Laboratory of Pollution Control and Resource Reuse, School of the Environment, Nanjing University, Nanjing, China
- 4Jiangxi Institute of Red Soil, National Engineering and Technology Research Center for Red Soil Improvement, Nanchang, China
The effects of land use and fertilization regimes on soil denitrification potential (SDP) are often investigated, but less is known about the responses of SDP to different types of land use with different fertilization regimes in soils derived from the same parent material. We conducted a study using soil samples from 2 long-term (over 30 years) fertilization experiments to determine the difference in SDP between paddy soil and upland soil derived from the same quaternary red clay parent material. The results showed that the SDP in paddy soil was 6.82 times higher than the upland soil, which was due to the higher abundances of narG, nirS, and nirK genes and nirS-denitrifying bacteria (Bradyrhizobium, Cupriavidus, and Herbaspirillum) in paddy soil. Inorganic fertilization regimes did not significantly affect the SDP of upland soil over the control group, whereas SDP in the NPK and 2NPK treatments in paddy soil were reduced relative the control group by 26.48% and 75.65%, respectively. Compared with the control, the NPKOM treatment consistently yielded the highest SDP in both soils, with 2.47 times and 2.86 times higher for paddy soil and upland soil, respectively. The SDP of paddy soil was significantly correlated with narG and nirS genes mainly regulated by amorphous aluminum oxide, whereas the treatment effect on SDP for upland soil largely depended on differences in nirS-denitrifying bacteria at the genus level (Herbaspirillum, Sulfuritalea, and Cupriavidus) and species level, which were mainly controlled by soil pH. Partial least squares path models further demonstrated that the direct effects of functional genes on SDP were greatest for paddy soil, whereas nirS-denitrifying bacterial communities played a larger role in upland soil. The results presented herein represent a key step toward understanding the mechanisms that govern SDP under land use and long-term fertilization.
Introduction
Soil denitrification is an alternative respiratory microbial process under oxygen-limited conditions (Philippot and Hallin, 2005), which is an important process of the nitrogen cycle in upland and paddy fields (Galloway et al., 2004; Butterbach-Bahl et al., 2013; Yin et al., 2015). Denitrifying bacterial communities, the main organisms driving soil denitrification (Hallin et al., 2009; Stein, 2011), are complex systems widely distributed in unrelated phylogenetic species, and genera, rather than a specific taxonomic group. Thus, 16SrDNA gene-based analysis is not suitable for investigating denitrifying bacterial communities (Balows et al., 2013). Instead, key functional genes, such as narG, nirS, nirK, and nosZ, involved in the denitrification process, have been commonly used to detect the abundance or diversity of denitrifying bacterial flora (Wolsing and Prieme, 2004; Sun et al., 2015). During this process, the nitrogen oxides (NO3– and NO2–) were reduced stepwise to gaseous oxides (NO, N2O, and/or N2) by particular groups of ubiquitous microorganisms under limiting oxygen (Henry et al., 2004; Philippot et al., 2007). The reaction of NO2– to NO has been considered to be the process distinguishing denitrifiers from other nitrate-respiring bacteria (Zumft, 1997) and the speed-limiting step in the denitrification process (Cutruzzola et al., 2001) and is catalyzed by nirS/nirK gene (Throback et al., 2004). Although copper-containing reductase (nirK) genes and cd1-containing reductase (nirS) have often been employed as molecular biomarkers to detect denitrifier communities, nirS is more commonly distributed than nirK in the environment (Braker et al., 1998). Since 1970, the concentration of N2O in the atmosphere has been increasing at approximately 750 ppt year–1 (IPCC, 2014), and more than half of the global greenhouse gas N2O comes from agricultural land (Hu et al., 2015). Therefore, a better understanding of the mechanisms by which denitrifying bacterial communities drive soil denitrification will be essential for developing management practices to regulate the soil N cycle.
Fertilization is one of the most common practices for enhancing soil quality and crop productivity (Zhang and Xu, 2005; Bassouny and Chen, 2016), which is also able to influence soil biological and chemical processes (Williams et al., 2013; Leff et al., 2015). Previous study showed that long-term fertilization with compost alone or with inorganic fertilizers markedly enhanced soil denitrification potential (SDP) by inducing changes in soil properties and altering soil denitrifying bacterial community in a fluvo-aquic soil (Ding et al., 2013). Sun et al. (2015) reported that long-term organic–inorganic combination fertilization significantly increased the abundance of nirS and nosZ genes. Similarly, Cui et al. (2016) showed that N2O emission was significantly strengthened by long-term applications of inorganic fertilizer, organic manure, or inorganic plus organic manure fertilizers via changes in the abundance and structure of nirS-denitrifying bacteria in black soil. In addition, organic fertilization was found to result in greater diversity of 13C-labeled nirS-denitrifying taxa including Rhodobacteraceae and unclassified Burkholderiales and a greater number of unknown 13C-labeled nirK-denitrifying taxa than the inorganic fertilization in a 32-year experiment of paddy soil (Hou et al., 2018). However, Yin et al. (2015) found that long-term organic fertilization had no effect on the abundance of nirS and nirK genes, and soil potential denitrification had significantly correlated with nirK denitrifiers community structure but not with nirS denitrifiers community structure. Although impacts of fertilization regimes on the abundance, diversity, and structure of the denitrifier communities have been extensively emphasized, limited information was available on the responses of the denitrifier communities to inorganic and inorganic–organic combination fertilization regimes in both paddy and upland soil that derived from the same parent material.
Paddy and upland soils are two different types of land uses in the red soil region of China (Zhang et al., 2009), which were the major sources of oxides of nitrogen (NO and N2O) (Mosier et al., 2004; Ruser et al., 2006). In this study, two long-term fertilization experiments (paddy and upland soils derived from quaternary red clay) were selected to investigate the relationships among SDP, abundance of denitrifying functional genes, and diversities of nirS-denitrifying bacterial communities under different land uses with long-term fertilization regimes [unamended control (control); chemical N, P, and K fertilizers (NPK); twofold chemical NPK fertilizers (2NPK); and chemical NPK fertilizers plus swine manure as the organic manure (NPKOM)] through real-time quantitative polymerase chain reaction (qPCR) and high-throughput sequencing technology. The aims of the study were (1) to investigate differences in SDP among different land uses under long-term fertilization regimes, (2) to identify the nirS-denitrifying bacterial communities that participate in SDP, and (3) to distinguish the key factors driving SDP under different land uses and fertilization regimes.
Materials and Methods
Soil Description and Soil Sampling
The soil samples were collected in December from two long-term fertilization experiments (paddy soil and upland soil) located at Jinxian Red Experimental Station in Jiangxi Province of China (28°37′ N, 116°26′ E). The two long-term experiments are 150 m apart, and both paddy soil and upland soil derived from Quaternary red clay. Detailed information on the long-term experiment is provided in the Supplementary Material. In the experiment, only four fertilization treatments were chosen from the two long-term fertilization experiments: unamended control (control); chemical NPK; 2NPK; and chemical NPKOM. All the fertilizers applied in two seasons. The doses of N, P, K, and the amount of organic manure were based on the farmers’ fertilization practices. Detailed information on the different fertilization treatments is provided in Supplementary Table S1.
Five subsamples from each replicate field plot for each treatment were collected at the depth of 0 to 20 cm in December 2015 and then mixed as a composite sample. The initial soil properties (0–20 cm) for the two experiments are listed in Supplementary Table S2. The soil samples were divided into three parts. Approximately 100 g of each fresh soil sample was stored at −80°C until measurements were made of the levels of narG, nirS, nirK, and nosZ- and nirS-type denitrifiers. The second part of each sample was stored at 4°C for the measurement of available N (NO3–-N and NH4+-N). The remaining parts were air dried at room temperature (25°C) and passed through a 2-mm sieve, after which all the visible leaves and roots were removed to determine additional chemical properties and conduct the incubation experiment.
Chemical Analysis
Soil pH was measured at a soil-to-water ratio of 1:5 (wt/wt) using a pH meter equipped with a composite electrode (FE20; Mettler Toledo, Switzerland Zurich). Soil organic matter (SOM) was determined by dichromate oxidation (Page et al., 1982). Total N (TN) was measured using Kjeldahl method (Page et al., 1982). Nitrate (NO3–-N) and NH4+-N were extracted with 2 M KCl (Page et al., 1982) and determined using a flow injection autoanalyzer (Skalar Analytic B.V., De Breda, Netherlands). Available P (AP) was extracted by 0.5 M NaHCO3 and measured using the molybdenum blue method (Olsen, 1954). Free iron oxide (Fed) and aluminum oxide (Ald) were extracted using dithionite–citrate–bicarbonate method and the amorphous iron oxide (Feo) and amorphous aluminum oxide (Alo) by acidic ammonium oxalate method (Pansu and Gautheyrou, 2006). The iron and aluminum in extracted solutions were measured by inductively coupled plasma mass spectrometry (NexION 300; PerkinElmer, Waltham, MA, United States).
Design of Incubation Experiment
Soil denitrification potential is commonly used to indicate soil denitrification, which can usually be determined by the simple and affordable method of the acetylene (C2H2) inhibition technique (AIT), as the acetylene inhibits N2O conversion into N2 (Miller et al., 1986; Qin et al., 2012). The SDP that occurred under anaerobic conditions to determine the difference among different land uses and fertilization regimes was expressed as the total N2O-N gas production per hour during the whole incubation periods (Zhang et al., 2009). Prior to the experiment, the soils were adjusted to 40% water-filled pore space (WFPS) and preincubated at 25°C under dark conditions for 1 week.
The experimental procedure followed the method of Pell et al. (1996) and the modified method of Adouani et al. (2010). The incubation experiment procedure was as follows: 6 g of each preincubated soil sample (oven-dry equivalent soil) was placed into a 120-mL serum culture flask, and then 8.658 mg KNO3 (N 200 μg g–1 dried soil) and 20.508 mg CH3COONa (C 1 mg g–1 dried soil) were added into the flask. The moisture content was adjusted to 70% WFPS. The flask was sealed with a dark butyl stopper and an aluminum cap and vacuumed for 2 min with a vacuum pump, and then 120 mL pure argon (99.999%) was injected to create anaerobic conditions; each flask was evacuated and flushed three times. Finally, 12 mL (10% vol/vol) acetylene and 108 mL pure argon (99.999%) were injected into the headspace of each flask, and all the flasks were shaken at 180 revolutions/min and 25°C for 15 min and then incubated at 0.1 KPa and 25°C in an incubator under dark conditions for 6 h. Gas samples were collected from the headspace of each flask with a gas-tight syringe. Incubated flasks were vacuumed and injected with pure argon (99.999%) three times to ensure anaerobic conditions and filled with acetylene and 108 mL pure argon for the next 6 h of incubation after each gas collection. The entire incubation experiment was conducted under a constant temperature of 25°C for 24 h. The gas samples were transferred into pre-evacuated 20-mL glass vials sealed with butyl rubber stoppers.
N2O Emission Measurement
N2O emission rate was determined using a gas chromatograph (Shimadzu GC-14B, Kyoto, Japan). The gas chromatograph was equipped with a 63Ni electron capture detector, and the temperature of the detector was 350°C. The velocity of the carrier gas (95% argon and 5% methane) was 55 mL min–1. The gas standard for N2O (303 ppbv) was supplied by the National Institute for Agro-Environmental Sciences, Japan. The response value of the gas chromatograph was linear within 200 to 5,000 ppbv. The N2O emission rate during the incubation experiments was calculated using the following equation:
where F is the emission rate of N2O (μg N kg–1 h–1, N2O-N), dc/dt is increase in gas concentration within a unit time (ppm h–1), M is molar mass of N2O (28 g mol–1, N2O-N), Vm is molar volume (22.4 L mol–1), V is the volume of the culture flask (0.12 L), T is the incubation temperature (25°C), and M is the weight of the oven-dried soil in the culture flask (kg).
DNA Extraction and Quantitative Real-Time PCR
Soil DNA was extracted from 0.5 g fresh soil with a Fast® DNA spin kit for soil (MP Biomedicals, Santa Ana, CA, United States), according to the manufacturer’s instructions. To reduce the bias in DNA extraction, we mixed three successive extractions of each sample together (Feinstein et al., 2009). Quantity and quality of soil DNA were checked by a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). The extracted DNA was stored at -80°C for further assays.
Amplification of denitrification genes was performed with a CFX96 Optical Real-Time Detection System (Bio-Rad Laboratories, Hercules, CA, United States). The narG, nirK, and nosZ gene amplification systems were 20 μL qPCR reaction mixtures containing of 10 μL SYBR® Premix Ex Taq (TliRNaseH Plus, 2×; Takara Bio, Otsu, Shiga, Japan), 0.4 μL PCR forward and reverse primers (both 10 μM), 2 μL DNA template (concentration of DNA template < 20 ng μL–1), and 7.2 μL double-distilled water (ddH2O). The amplification system for the nirS gene was a total volume of 20 μL containing 10 μL SYBR® Premix Ex Taq (TliRNaseH Plus, 2×; Takara Bio, Otsu, Shiga, Japan), 0.5 μL PCR forward and reverse primers (both 10 μM), 1 μL DNA template (concentration of DNA template < 20 ng μL–1), and 8 μL ddH2O. Primers and amplification conditions are detailed in Supplementary Table S3. Standard curves of narG, nirS, nirK, and nosZ were generated using three replicates of 10-fold serial dilutions of linearized plasmid containing the sequences as the templates, under the cycling conditions mentioned above. Performing the amplification procedure with ddH2O instead of DNA was the negative control. The amplification efficiencies of narG, nirS, nirK, and nosZ genes were 90.4 to 93.1%, 84.1 to 92.8%, 86.0 to 87.8%, and 89.8 to 89.0%, respectively.
Illumina MiSeq Sequencing and Analyses
Polymerase chain reaction amplicons of the nirS gene were sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, United States). The pair of primers for the nirS gene was cd3aF/R3cd. All the PCRs were conducted in a 25-μL reaction mixture containing 5 μL 5 × reaction buffer, 5 μL 5 × GC buffer, 2 μL dNTP (2.5 mM), 1 μL forward primer/reverse primer10 μM, 2 μL DNA (20 ng/μL) template, 8.75 μL ddH2O, and 0.25 μL Q5 DNA polymerase (M0491L, NEB). The PCR amplification was performed under the following conditions: 2 min at 98°C for initial denaturing; 30 cycles of 98°C for 15 s, 55°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. The PCR products were gel-purified using Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN, United States) after being detected by 2% agarose gel electrophoresis and then quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, United States). The amplicons were pooled in equal amounts after the individual quantification step, and paired-end 2 × 300 bp sequencing was performed using the Illlumina MiSeq platform with the MiSeq Reagent Kit v3 at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). The sequence data were processed using Quantitative Insights Into Microbial Ecology (Caporaso et al., 2010).
Briefly, the raw sequence reads with exact matches to the barcodes were assigned to respective samples and identified as valid sequences, and then low-quality sequences were filtered through the following criteria: sequences had low average quality score of <Q20, sequences had a length of <150 bp, and sequences contained ambiguous base pairs (Gill et al., 2006; Chen and Jiang, 2014). Paired-end reads were assembled using FLASH (Magoc and Salzberg, 2011). The obtained high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity by UCLUST (Edgar, 2010). The sequence was chosen from each OTU using default parameters. OTUs containing less than 0.001% of total sequences across all samples were discarded. All the samples were rarefied to the same sequencing depth for the data analysis. The relative abundances of each group at different taxonomic levels (phylum, class, order, family, and genus) were employed for the subsequent analysis. The sequences in the main OTUs were analyzed using BLAST with the NCBI database to obtain the most similar published sequences (Gu et al., 2019). All the sequence data have been submitted to the NCBI Sequence Read Archive with accession number PRJNA601844.
Data Analysis
Statistical analysis and data processing were accomplished using SPSS 20.0 (Chicago, IL, United States) and R 3.6.1. Principal component analysis (PCA) and correspondence analysis (CA) were performed to investigate the influences of different fertilization regimes on the structure of denitrifying bacterial communities. Canonical correlation analysis (CCA) and redundancy analysis (RDA) were used to establish the relationships between nirS-type denitrifying bacterial taxa, SDP, and soil properties. The correlation coefficients between soil properties and nirS-type denitrifying communities were calculated using 1,000 Monte Carlo permutations. STAMP was employed to identify the difference in genera among different treatments. The volcano plot (R package “DESeq2”) was used to identify significantly different OTUs among different treatments (Edwards et al., 2015). Spearman rank correlation coefficients (R package “corrplot”) were used to test the relationships between soil properties and SDP and denitrifying functional genes. In addition, random forest (rf) modeling was used to predict the most important soil properties inducing changes in SDP and the functional genes (Liu et al., 2017; Wang et al., 2019). Multivariate regression tree (MRT; De’ath, 2006) analysis was performed to detect the predominant factors influencing the nirS-denitrifying bacterial community composition. R 3.6.1 was employed to construct partial least squares path models (Sanchez, 2013). Treatment means and standard errors presented in the figures were calculated based on untransformed data. Significance was accepted at P < 0.05 in all cases.
Results
Soil Properties
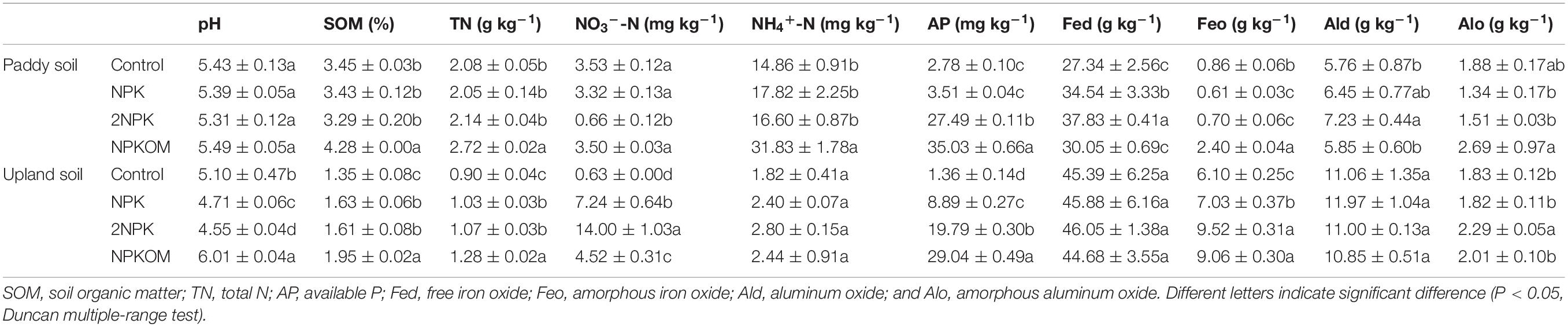
The chemical characteristics of soil subjected to long-term addition of organic and inorganic fertilizers exhibited great differences between paddy and upland soils (Table 1). Although both paddy and upland soils developed from the Quaternary red clay, different land uses have led to significant differences in SOM, TN, NO3–, NH4+, AP, Fed, Feo, and Ald for the control sites (Supplementary Table S4). In paddy soil, NPKOM application significantly increased contents of SOM, TN, and NH4+-N compared with the control, but no obvious differences among control, NPK, and 2NPK were found. The 2NPK treatment notably reduced the content of NO3–-N from 3.53 ± 0.12 mg kg–1 to 0.66 ± 0.12 mg kg–1. Compared with the control site, soil AP was higher in the NPKOM and 2NPK treatments, changing from 2.78 ± 0.10 mg kg–1 to 27.49 ± 0.11 mg kg–1, and 35.03 ± 0.66 mg kg–1, respectively. The soil contained the highest Fed and Ald contents (37.83 ± 0.41 and 7.23 ± 0.44 g kg–1) under the application of 2NPK, followed by the NPK and NPKOM treatments. Unlike the Fed, the content of Feo was much less under NPK and 2NPK treatments, while significantly elevated in NPKOM. The Alo concentration in the NPKOM treatment was higher than that in control, averaging 2.69 ± 0.97 and 1.88 ± 0.17 g kg–1, respectively.
In upland soil, soil pH was significantly depressed in NPK and 2NPK treatments but elevated in the NPKOM treatment (Table 1). The highest Feo and Alo contents were observed under 2NPK at 9.52 ± 0.31 and 2.29 ± 0.05 g kg–1, respectively, whereas no obvious difference was found across the treatments for Fed or Ald contents. Relative to the control, the SOM, TN, NO3–-N, and AP from the fertilized treatments were higher by 19.3 to 44.4%, 14.4 to 42.2%, 617 to 2122%, and 553 to 2035%, respectively.
Soil Denitrification Potential
Significant differences (P < 0.05) in SDP were observed among soil samples (Figure 1). SDP in the paddy soil control was 6.82 times higher than that in upland soil. In paddy soil, NPK, and 2NPK reduced the SDP by 26.48% and 75.65%, respectively, whereas addition of NPKOM increased the SDP by 147.06%, with the highest value of 201.74 ± 1.64 μg N2O-N kg–1 dry soil–1. However, there were no significant differences in the SDP of upland soil among control, NPK, and 2NPK treatments, averaging 13.41 ± 0.81, 14.56 ± 0.66, and 19.78 ± 3.03 μg N2O-N kg–1 dry soil–1, respectively. Similar to the result with the paddy soil, SDP in the NPKOM treatment of the upland soil was also clearly highest across all the fertilization regimes.
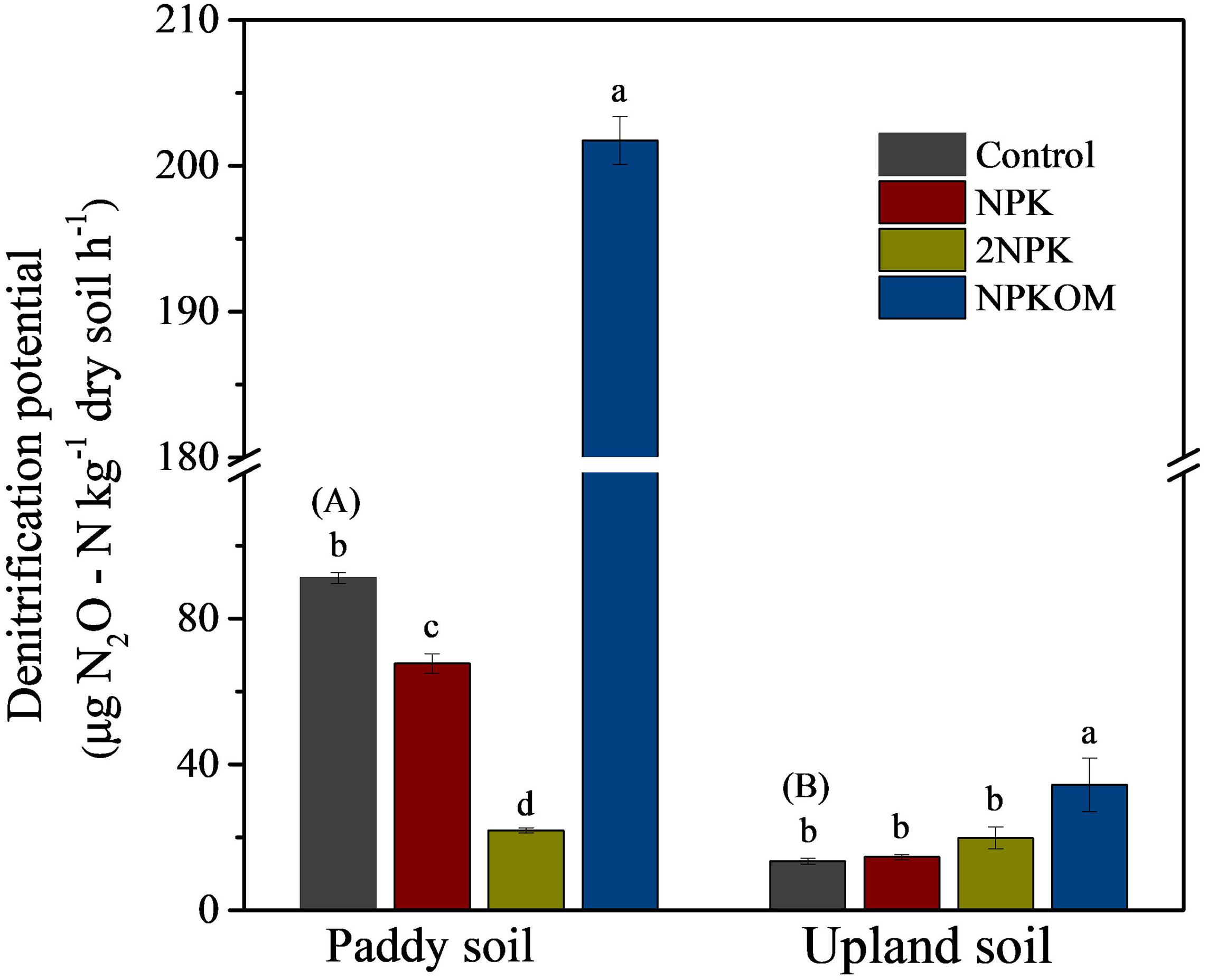
Figure 1. Soil denitrification potential among different fertilization regimes in the paddy soil and upland soil. Different uppercase letters in brackets indicate significant differences for the treatment of control between paddy soil and upland soil by independent-sample t test, and different lowercase letters indicate significant differences among fertilizer treatments for each soil type (P < 0.05, Duncan multiple-range test).
Abundances of Functional Genes Involved in the Denitrification
The variations of levels of related functional genes involved in SDP (narG, nirS, nirK, and nosZ) were analyzed by qPCR. To guarantee the comparison of levels of each gene among different fertilization regimes, the amplification efficiency of the same gene was similar for all treatments. The levels of the narG, nirS, nirK, and nosZ genes in the paddy soil control were significantly higher than values in upland soil (Figure 2), which is consistent with results for SDP (Figure 1). Compared with the control, NPKOM significantly increased levels of narG, nirS, and nirK in paddy and upland soils (Figures 2A–C). In upland soil, NPK markedly increased the abundance of nirK and reduced the abundance of nosZ compared with the control (Figures 2C,D). The application of 2NPK clearly reduced the level of narG in paddy soil, while enhancing levels of nirS in upland soil and nirK in paddy soil.
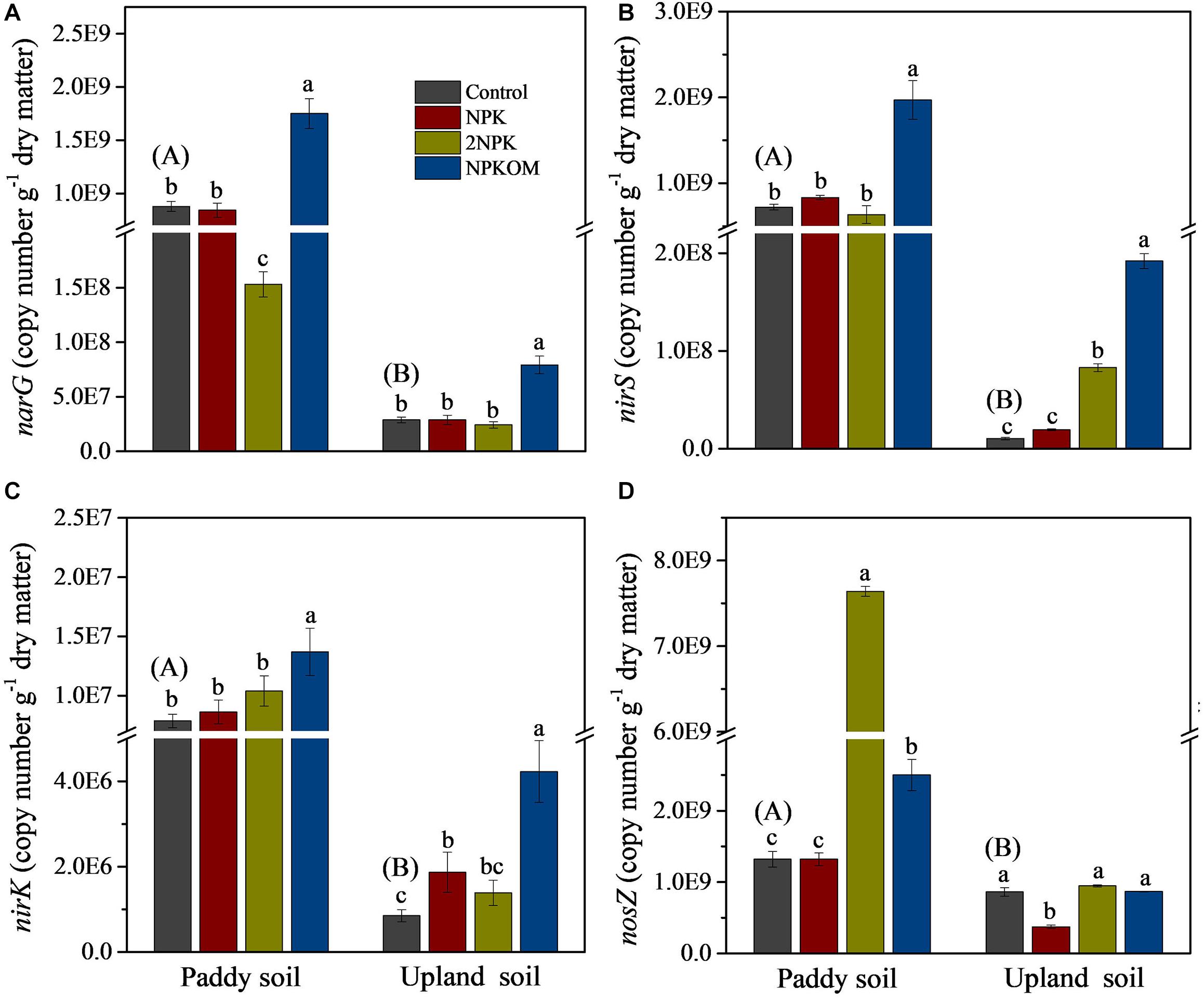
Figure 2. Abundances of narG (A), nirS (B), nirK (C), and nosZ (D) genes among different fertilization regimes. Different uppercase letters in brackets indicate significant differences for the treatment of control between paddy soil and upland soil, and different lowercase letters indicate significant differences among fertilizer treatments for each soil type (P < 0.05, Duncan multiple-range test).
nirS-Denitrifying Bacterial Communities
A total of 597,359 high-quality sequences were obtained from the 24 samples, and all the sequences were clustered into 5866 OTUs at 97% identity. Nearly 95.53% (paddy soil) and 96.83% (upland soil) of nirS-denitrifying sequences came from the phylum Proteobacteria.
The relative abundances of nirS-denitrifying bacteria at the genus level are shown in Figure 3 and Supplementary Table S5. The significant differences in relative abundance among major genera (>1.00%) in paddy and upland soils under different fertilization regimes were tested (Supplementary Figures S1, S2). By comparing the control groups for the two soils, it was determined that relative abundances of Bradyrhizobium, Cupriavidus, and Herbaspirillum were higher in the paddy soil than in the upland soil, whereas relative abundances of Rhodanobacter, Ideonella, and Pseudomonas were lower in the paddy soil than in the upland soil (Supplementary Figure S1 and Supplementary Table S5). In paddy soil, there were no significant differences at the genus level between the control and NPK, 2NPK, and NPKOM treatments. In upland soil, compared with the control, NPK markedly increased the relative abundances of Herbaspirillum and Cupriavidus, but clearly decreased the relative abundance of Rubrivivax (Supplementary Figure S2a); 2NPK significantly enhanced the relative abundance of Sulfuritalea, while markedly reducing Rubrivivax and Bradyrhizobium (Supplementary Figure S2b); and NPKOM significantly increased the relative abundance of Herbaspirillum, whereas it decreased the relative abundances of Rubrivivax and Pseudomonas (Supplementary Figure S2c).
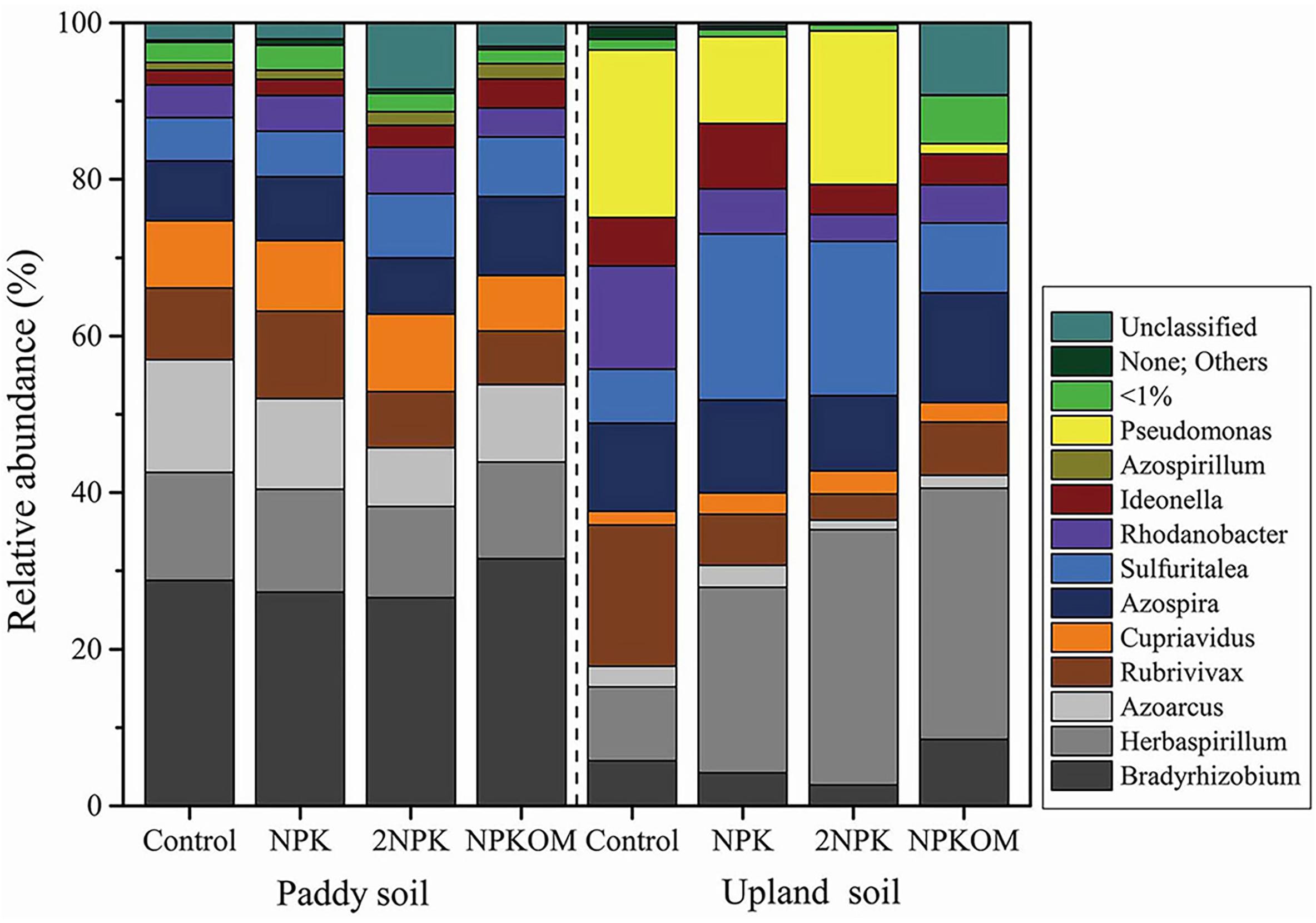
Figure 3. Taxonomic composition at the genus level of the nirS-denitrifying bacterial communities in paddy soil and upland soil. None; others indicate that a large degree of nirS-type diversity is not represented in the current genome database.
Principal component analusis (CA) and PERMANOVA were used to analyze the structure of nirS-denitrifying bacterial communities, revealing groups stratified by land use and fertilization regimes (Supplementary Figure S3 and Supplementary Table S6). In paddy soil, PCA analysis demonstrated the statistical differentiation of nirS-denitrifying bacterial communities among control, 2NPK, and NPKOM, and there were no significant differences between control and NPK (Supplementary Figure S3a). PERMANOVA further confirmed the differences between different fertilization treatments (Supplementary Table S6). In upland soil, CA analysis, and PERMANOVA indicated that control was significantly separated from NPK, 2NPK, and NPKOM (Supplementary Figure S3b and Supplementary Table S6).
Compared with the upland soil control, the enriched, depleted, and stable OTUs for the paddy soil control accounted for approximately 13.03, 2.83, and 84.14% of the total sequences (Supplementary Figure S4a), respectively, suggesting that a few nirS-denitrifying bacteria were sensitive to land use. In paddy soil, no enriched OTU (with relative abundance > 0.1%) was found to be positively associated with SDP. In upland soil, some OTUs (with relative abundance > 0.1%) were directly associated with SDP compared to the control (Supplementary Figures S4e–g and Supplementary Table S7). A significant positive association between SDP and Herbaspirillum (TSO47-2) was observed in NPK; Azospira (NC3H-14), Rubrivivax (Gelatinosus), and Herbaspirillum (TSO47-2) in 2NPK; Sulfuritalea (Hydrogenivorans), Bradyrhizobium (UNPF333 and TSA26), Rhodanobacter (D208a), Azospira (NC3H-14), Rubrivivax (Gelatinosus), and Herbaspirillum (TSO47-2) in NPKOM. The above results suggest that, under different fertilization regimes, the difference in SDP in upland soil was influenced more by nirS-denitrifying bacteria.
Relationships Between nirS-Denitrifying Bacterial Communities and Soil Properties
The relationships between nirS-denitrifying bacterial communities and soil properties were assessed with RDA and MRT (Figure 4 and Supplementary Figure S4). Monte Carlo permutation test analysis revealed that the soil properties of SOM, TN, NO3–, NH4+, AP, Fed, Feo, and Alo notably correlated with nirS-denitrifying bacterial communities in paddy soil (Figure 4A and Supplementary Table S8). The properties selected in paddy soil explained greater than 88.04% of the variation in the nirS-denitrifying bacterial community, and RDA1 and RDA2 explained 35.02 and 16.40%, respectively. Combined with the MRT analysis (Supplementary Figure S5a), the content of NO3– was the most important factor determining the nirS-denitrifying bacterial community, explaining 35.20% of the variance in the first split, and the content of SOM was a second important factor, explaining 16.40% in the second split.
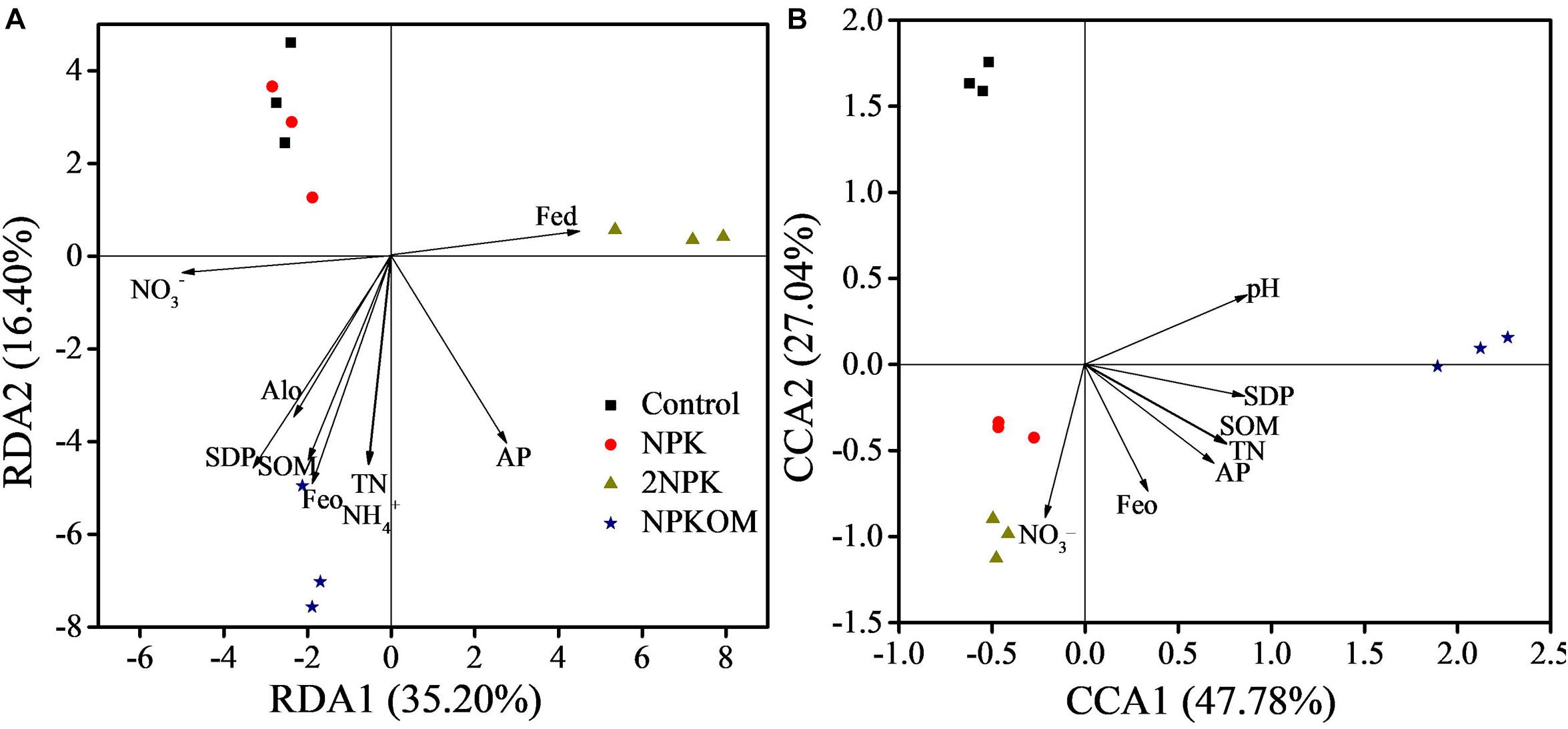
Figure 4. Redundancy analysis (RDA) of the correlation between nirS-denitrifying bacterial communities and soil properties and SDP in the paddy soil (A). Canonical correlation analysis (CCA) of correlations between nirS-denitrifying bacterial communities and soil properties and SDP in the upland soil (B). Soil properties presented in (A,B) significantly affected the nirS-denitrifying bacterial communities assessed by the Monte Carlo test.
Soil pH, SOM, TN, NH4+, AP, and the Feo significantly affected the nirS-denitrifying bacterial communities in upland soil (Figure 4B and Supplementary Table S8). Selected properties were able to explain greater than 88.97% of the variation in the denitrifying community, and RDA1 and RDA2 explained 47.78 and 27.04%, respectively. Soil pH was the main factor determining the nirS-denitrifying bacterial community composition in upland soil (Supplementary Figure S5b), explaining 50.67, 22.54, and 15.71% of the variance in the first, second, and third split, respectively.
Discussion
Associations of Land Use and Long-Term Fertilization Regimes With SDP
To avoid any interaction effects from the land use type and fertilization regime, results from the control were used to compare SDP between the paddy soil and upland soil. It was found that SDP of control from paddy soil was higher than that from upland soil (Figure 1). The increasing soil water content reduced O2 diffusion into the soil (Abbasi and Adams, 2000; Ullah and Faulkner, 2006; Huang et al., 2007), and it was expected that the hypoxic conditions were more beneficial to the growth of denitrifiers in the paddy soil than the upland soil. Several studies have demonstrated that land use types with higher average soil moisture values show greater SDP than those with relatively low soil water content (Luo et al., 2000; Hunt et al., 2007; Xu and Cai, 2007; Liu et al., 2012). In addition, SOM and NO3– contents are the key factors that determine differences in denitrification rate (Liang and MacKenzie, 1997; Xu and Cai, 2007). In the present study, the paddy soil had higher SOM and NO3– contents than the upland soil (Table 1 and Figure 1).
The SDP was significantly affected by fertilization regimes (Figure 1). In the paddy soil, when compared with the control, the SDPs from the NPK and 2NPK treatments were significantly reduced, agreeing with results of some previous studies (Yin et al., 2014; Cui et al., 2016). Spearman rank correlation analysis showed that SDP had significantly positive correlation with Alo and NO3–, but negative correlation with AP and Fed (P < 0.05). The NPK and 2NPK treatments had lower Alo and NO3– while higher AP and Fed than the control (Table 1). The SDP in the NPKOM treatment was clearly increased (Figure 1), agreeing with results of previous studies (Enwall et al., 2005; Chen et al., 2012; Yin et al., 2014; Cui et al., 2016; Rahman et al., 2018), because organic manure can lead to soil anaerobiosis and create a relatively suitable environment for denitrification (Dobbie and Smith, 2001; Butterbach-Bahl et al., 2013). Consistent with the SDP result, the higher Alo was observed in the NPKOM than NPK treatment (Table 1). Based on the correlation and the random forest analyses, Alo content was the important variable for adjusting denitrification, followed by Fed and AP (Supplementary Figure S6). Our results were consistent with those reported by Wang et al. (2019), showing that Alo was the important variable for adjusting the nitrogen cycle in the paddy soil of southern China. Generally, the bacterial cell surface carries a net negative charge (Poortinga et al., 2002). Amorphous aluminum oxide can simultaneously adsorb the NO3–/NO2– and denitrifiers because of high positive charge on the surface (Liu et al., 2015; Wu et al., 2019). We inferred that higher Alo content may enhance the contact of the denitrifiers with NO3–/NO2– and then affect SDP.
Unlike the paddy soil, the NPK and 2NPK treatments had no significant effect on SDP in the upland soil, whereas the NPKOM significantly enhanced the SDP, consistent with the results observed in the paddy soil (Figure 1). Spearman rank correlation analysis showed that SDP in the control, NPK, and 2NPK significantly and positively correlated with NO3–, NH4+, AP, Feo, TN, and Alo, but negatively correlated with pH (P < 0.05). And that the higher SOM, AP, and pH in NPKOM than NPK may be responsible for the increase in SDP (Table 1). Although most denitrifiers have optimal pH values for growth and activity between 6 and 8 (Aulakh et al., 1992), adaptation of denitrifying microorganisms to low pH occurred over the long-term fertilization (Parkin et al., 1985; Šimek and Hopkins, 1999). Furthermore, soil pH has been called the “master variable,” which can influence the characteristics of soil physics, chemistry, and biology (Brady et al., 2008). The TN, NO3–, NH4+, AP, and Feo in upland soil were strongly affected by pH (P < 0.05). Based on the correlation and the random forest analyses, we also found that pH was the important factor for regulating SDP in upland soil, followed by AP and SOM (Supplementary Figure S6).
Associations of Abundance of Functional Genes With SDP
Denitrification functional genes, encoding enzymes responsible for the stepwise reductive pathway, are preferable and allow for both characterization and quantitative determination of function that is polyphyletically dispersed within a community (Braker et al., 1998; Throback et al., 2004). Previous investigation has also demonstrated that narG, nirS, and nirK genes can influence N2O emission (Throback et al., 2007; Chen et al., 2012; Liu et al., 2012; Wang et al., 2013). The higher levels of narG, nirS, and nirK in paddy soil may be responsible for the finding that SDP was higher in paddy soil than in upland soil (Figures 1, 2).
In the paddy soil, positive correlations between SDP and abundance of narG and nirS genes were found. Spearman rank correlation analysis demonstrated that the abundance of narG gene positively correlated with contents of Feo and Alo, while it negatively correlated with Fed. Furthermore, the abundance of nirS gene showed significantly positive correlation with Alo. NPK and 2NPK enhanced the contents of Fed, whereas they reduced those of Feo and Alo. Therefore, the changes of Alo and Feo may be the factors explaining the effects of NPK and 2NPK on SDP in the present study. NPKOM treatment exhibited the highest contents of Feo and Alo (Table 1). Meanwhile, the application of organic manure was able to increase the abundances of narG and nirS (Kandeler et al., 2006; Tatti et al., 2013; Cui et al., 2016), consistent with our result (Figure 2). We believe that the highest SDP observed in the NPKOM treatment (Figure 1) was likely attributable to the highest abundances of narG and nirS genes, resulting from the highest contents of Alo, Feo, and SOM (Figure 2). In addition, random forest analysis demonstrated that narG (13.88% IncMSE) and nirS (12.74% IncMSE) were more important than nirK (8.69% IncMSE) and nosZ (10.80% IncMSE) in influencing the SDP of paddy soil. Therefore, the variation in SDP in paddy soil under different fertilization regimes likely resulted from the differences in narG and nirS genes caused by the varied soil properties (Fed, Feo, and Alo), and Alo was also the dominant factor influencing levels of narG and nirS genes.
In upland soil, NPK and 2NPK clearly decreased soil pH compared to the control (Table 1), agreeing with previous studies (Aref and Wander, 1997; Barak et al., 1997; Qu et al., 2014; Cai et al., 2015). In strongly acidic soil (pH < 5.0), the level of nirS was the main factor that affected SDP, and pH was significantly negatively related with abundance of nirS. In addition, positive relationships between nirS and other soil properties, including TN, NO3–, NH4+, AP, and Feo, were found. Similar to the pattern observed with paddy soil, NPKOM significantly enhanced the abundance of related functional genes (narG, nirS, and nirK). And that may be one of the reasons that NPKOM in upland soil showed the highest SDP among different treatments. Furthermore, random forest analysis revealed that nirS (13.60% IncMSE) and narG (11.47% IncMSE) were also more important than nirK (9.63% IncMSE) or nosZ (6.13% IncMSE) in controlling SDP.
Associations of nirS-Denitrifying Bacterial Communities With SDP
Based on STAMP analysis, the genera Bradyrhizobium, Cupriavidus, and Herbaspirillum were significantly higher in the paddy soil control than in the upland soil (Supplementary Figure S1) and were responsible for the higher SDP in paddy soil (Figure 1). The main genera remained stable in paddy soil with different fertilization regimes, whereas some specific genera in upland soil under the applications NPK, 2NPK, and NPKOM were significantly different from the control (Supplementary Figure S2). It was noted that Rubrivivax in the control was notably higher than in the NPK, 2NPK, and NPKOM treatments in upland soil (Supplementary Figure S2). Spearman rank correlation (Supplementary Table S9) and random forest suggest that NO3– and Feo were the most important factors for determining Rubrivivax, and soil pH, NH4+, and Alo were the second most important factors. Qian et al. (2019) demonstrated that high pH could improve nitrite accumulation and stimulated the growth of Rubrivivax. Herbaspirillum in NPK and NPKOM was notably higher than its abundance in the control (Supplementary Figure S2), which showed a positive correlation with content of SOM, TN, NO3–, NH4+, AP, Feo, and Alo (Supplementary Table S9). Previous researchers have demonstrated that nitrogen and ammonium can influence Herbaspirillum species (Fu and Burris, 1989; Souza et al., 2000). Cupriavidus in NPK was higher than that in the control (Supplementary Figure S2a), which was mainly influenced by NO3– content (Supplementary Table S9). Compared with the control, 2NPK treatment enhanced the abundance of Sulfuritalea and reduced that of Bradyrhizobium (Supplementary Figure S2b). Bradyrhizobium was positively correlated with soil pH but negatively correlated with NO3– and Fed (Supplementary Table S9). Graham et al. (1994) and Appunu and Dhar (2006) reported that Bradyrhizobium species were positively correlated with pH under acidic conditions (4.5 < pH < 7.0). Therefore, the lower pH in 2NPK does not fit for the growth of Bradyrhizobium. In the NPKOM treatment, Herbaspirillum significantly increased, whereas Rubrivivax and Pseudomonas decreased compared with the control (Supplementary Figure S2c). OTU 47279 (Pseudomonas grimontii) was the dominant species (48.82%) of Pseudomonas in upland soil and had a negative correlation with pH (P < 0.05) and SOM (P < 0.05). Although P. grimontii is an extremely aerobic Gram-negative bacterium, it can also grow under anaerobic conditions (Baida, 2002; Gomes et al., 2017). The characteristics of P. grimontii resulted in more Pseudomonas in upland soil. Application of pig manure may increase O2 consumption and decrease redox potentials (Gardiner and James, 2012), thus inhibiting the growth of P. grimontii.
The composition of nirS-denitrifying bacterial communities greatly varied among different long-term fertilization regimes (Yin et al., 2014; Cui et al., 2016). However, Chen et al. (2010) found that nirS denitrifiers were stable to different fertilizer regimes. Therefore, response of nirS-denitrifying bacterial community to fertilization regimes is inconsistent. In paddy soil, the nirS-denitrifying bacterial community was stable against long-term NPK application (Supplementary Figure S3a and Supplementary Table S6), whereas 2NPK and NPKOM substantially changed the structure of nirS-denitrifying bacterial communities (Supplementary Figure S3a and Supplementary Table S6). In most cases, content of NO3– was a decisive factor influencing denitrifying bacterial communities. Francis et al. (2013) reported that NO3– was the reaction substrate for denitrification, markedly influencing the structure of denitrifying bacteria communities. In upland soil, composition of nirS-denitrifying bacterial communities greatly changed with different long-term fertilization regimes (Supplementary Figure S3b and Supplementary Table S6). This result agreed with findings of previous research (Yu et al., 2018). Soil pH was the main factor determining nirS-denitrifying bacterial community composition. Enwall et al. (2005) also discovered that pH can affect the composition of the denitrifying communities in soil. In general, variations in soil properties resulted in differences in denitrifying bacterial community composition (Enwall et al., 2005; Chen et al., 2010; Sun et al., 2015; He et al., 2019; Qian et al., 2019).
Potential Pathways Influencing SDP
In paddy soil, soil properties (SOM, NO3–, Fed, Feo, Ald, and Alo), functional genes (narG and nirS), and the value of PC1 were retained. Partial least squares path modeling explained 99.42% of SDP variation (Figure 5A). The direct effect of functional genes (0.890) on SDP was larger than the value for soil parameters (0.486) and the structure of nirS-denitrifying bacterial communities (0.119). In upland soil, soil properties (pH, SOM, NO3–, AP, Feo, and Alo), functional genes (nirS and nirK), and the value of CA1 were retained. Partial least squares path modeling explained 86.8% of SDP (Figure 5B). The direct effect of the structure of nirS-denitrifying bacterial communities (1.001) on SDP was larger than those of soil parameters (0.591) and functional genes (-0.622). Despite the fact that higher levels of nirS and nirK usually caused greater SDP, nirS, and nirK appear to have direct negative effects on SDP in upland soil. However, with larger indirect effects (1.415), the negative effects of nirS and nirK on SDP can be offset.
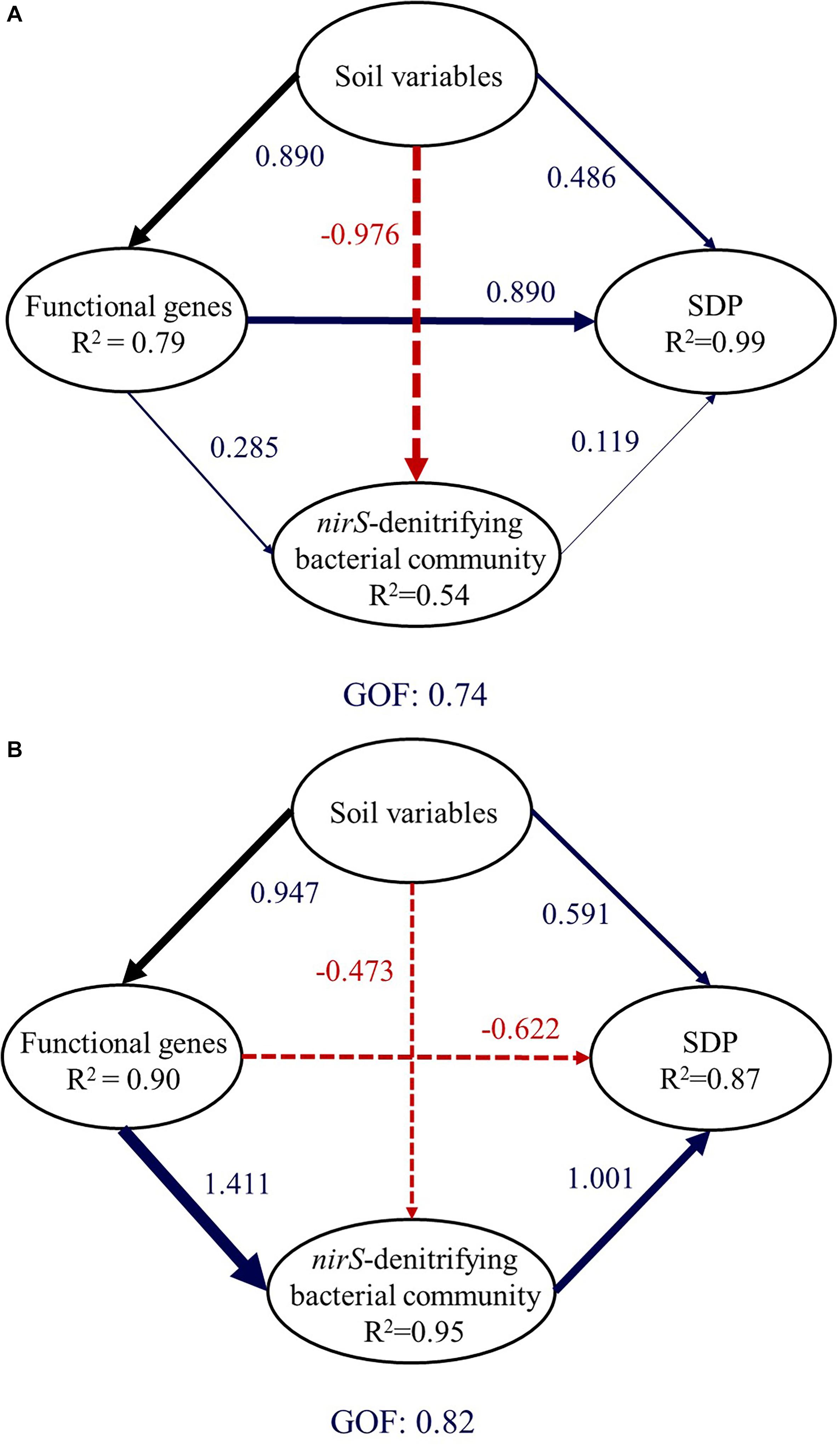
Figure 5. Directed graph of the partial least squares path model for paddy soil (A) and upland soil (B). Larger path coefficients are reflected in the width of the arrow, with blue indicating a positive effect, and red, a negative effect. Path coefficients are calculated after 1,000 bootstraps. The model is assessed using the goodness-of-fit statistic, a measure of the overall prediction performance. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Conclusion
Our study revealed that the SDP was higher in paddy soil than the upland soil, mainly due to the higher abundances of narG, nirS, and nirK genes and nirS-denitrifying bacteria (Bradyrhizobium, Cupriavidus, and Herbaspirillum), resulting from higher contents of moisture, SOM, and NO3– in paddy soil than the upland soil. Land use differently mediated the impacts of fertilization regimes on SDP. In the paddy soil, inorganic fertilization significantly decreased SDP, whereas organic–inorganic combination fertilization obviously increased SDP. The SDP in the paddy soil showed positive association with the narG and nirS genes, which were mainly regulated by Alo. In the upland soil, inorganic fertilization had no significant effect on SDP, whereas organic–inorganic combination fertilization clearly enhanced SDP. The trend in SDP of upland soil largely depended on differences in structure of nirS-denitrifying bacteria, which was mainly controlled by soil pH.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Sequence Read Archive with accession number PRJNA601844 (http://www.ncbi.nlm.nih.gov/).
Author Contributions
HW, BZ, and JZ designed the experiments. HW, LM, DL, KL, and QH performed the experiments. HW drafted the manuscript. BZ and ZL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Funding for the work was provided by the National Natural Science Foundation of China (41977102), National Key Research and Development Program of China (2016YFD0300802), and the China Agriculture Research System (CARS-03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2020.00105/full#supplementary-material
References
Abbasi, M. K., and Adams, W. A. (2000). Estimation of simultaneous nitrification and denitrification in grassland soil associated with urea-N using15N and nitrification inhibitor. Biol. Fertil. Soils 31, 38–44. doi: 10.1007/s003740050621
Adouani, N., Lendormi, T., Limousy, L., and Sire, O. (2010). Effect of the carbon source on N2O emissions during biological denitrification. Resourc. Conserv. Recycl. 54, 299–302. doi: 10.1016/j.resconrec.2009.07.011
Appunu, C., and Dhar, B. (2006). Symbiotic effectiveness of acid-tolerant Bradyrhizobium strains with soybean in low pH soil. Afr. J. Biotechnol. 5, 842–845.
Aref, S., and Wander, M. M. (1997). Long-term trends of corn yield and soil organic matter in different crop sequences and soil fertility treatments on the morrow plots. Adv. Agron. 62, 153–197. doi: 10.1016/s0065-2113(08)60568-4
Aulakh, M., Doran, J., and Mosier, A. (1992). “Soil denitrification—significance, measurement, and effects of management,” in Advances in Soil Science, ed. B.A. Stewart, (New York, NY: Springer-Verlag), 1–57. doi: 10.1007/978-1-4612-2844-8_1
Baida, N. (2002). Pseudomonas grimontii sp. nov. Intern. J. Syst. Evol. Microbiol. 52, 1497–1503. doi: 10.1099/ijs.0.01971-0
Balows, A., Trüper, H. G., Dworkin, M., Harder, W., and Schleifer, K.-H. (2013). The Prokaryotes: A Handbook On The Biology Of Bacteria: Ecophysiology, Isolation, Identification, Applications. Cham: Springer.
Barak, P., Jobe, B. O., Krueger, A. R., Peterson, L. A., and Laird, D. A. (1997). Effects of long-term soil acidification due to nitrogen fertilizer inputs in Wisconsin. Plant Soil 197, 61–69.
Bassouny, M., and Chen, J. (2016). Effect of long-term organic and mineral fertilizer on physical properties in root zone of a clayey Ultisol. Archiv. Agron. Soil Sci. 62, 819–828. doi: 10.1080/03650340.2015.1085649
Brady, N. C., Weil, R. R., and Weil, R. R. (2008). The Nature And Properties Of Soils. Upper Saddle River, NJ: Prentice Hall.
Braker, G., Fesefeldt, A., and Witzel, K.-P. (1998). Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64, 3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998
Butterbach-Bahl, K., Baggs, E. M., Dannenmann, M., Kiese, R., and Zechmeister-Boltenstern, S. (2013). Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20130122. doi: 10.1098/rstb.2013.0122
Cai, Z. J., Wang, B. R., Xu, M. G., Zhang, H. M., He, X. H., Zhao, L., et al. (2015). Intensified soil acidification from chemical N fertilization and prevention by manure in an 18-year field experiment in the red soil of southern China. J. Soils Sediments 15, 260–270. doi: 10.1007/s11368-014-0989-y
Caporaso, J., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F., Costello, E., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Chen, H., and Jiang, W. (2014). Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front. Microbiol. 5:508. doi: 10.3389/fmicb.2014.00508
Chen, Z., Liu, J., Wu, M., Xie, X., Wu, J., and Wei, W. (2012). Differentiated response of denitrifying communities to fertilization regime in paddy soil. Microb. Ecol. 63, 446–459. doi: 10.1007/s00248-011-9909-5
Chen, Z., Luo, X., Hu, R., Wu, M., Wu, J., and Wei, W. (2010). Impact of long-term fertilization on the composition of denitrifier communities based on nitrite reductase analyses in a paddy soil. Microb. Ecol. 60, 850–861. doi: 10.1007/s00248-010-9700-z
Cui, P. Y., Fan, F. L., Yin, C., Song, A. L., Huang, P. R., Tang, Y. J., et al. (2016). Long-term organic and inorganic fertilization alters temperature sensitivity of potential N2O emissions and associated microbes. Soil Biol. Biochem. 93, 131–141. doi: 10.1016/j.soilbio.2015.11.005
Cutruzzola, F., Brown, K., Wilson, E. K., Bellelli, A., Arese, M., Tegoni, M., et al. (2001). The nitrite reductase from Pseudomonas aeruginosa: essential role of two active-site histidines in the catalytic and structural properties. Proc. Natl. Acad. Sci. U.S.A. 98, 2232–2237. doi: 10.1073/pnas.041365298
Ding, W., Luo, J., Li, J., Yu, H., Fan, J., and Liu, D. (2013). Effect of long-term compost and inorganic fertilizer application on background N2O and fertilizer-induced N2O emissions from an intensively cultivated soil. Sci. Total Environ. 465, 115–124. doi: 10.1016/j.scitotenv.2012.11.020
Dobbie, K. E., and Smith, K. A. (2001). The effects of temperature, water-filled pore space and land use on N2O emissions from an imperfectly drained gleysol. Eur. J. Soil Sci. 52, 667–673. doi: 10.1046/j.1365-2389.2001.00395.x
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edwards, J., Johnson, C., Santos-Medellin, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920.
Enwall, K., Philippot, L., and Hallin, S. (2005). Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 71, 8335–8343. doi: 10.1128/aem.71.12.8335-8343.2005
Feinstein, L. M., Sul, W. J., and Blackwood, C. B. (2009). Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl. Environ. Microbiol. 75, 5428–5433. doi: 10.1128/aem.00120-09
Francis, C. A., O’mullan, G. D., Cornwell, J. C., and Ward, B. B. (2013). Transitions in nirS-type denitrifier diversity, community composition, and biogeochemical activity along the Chesapeake Bay estuary. Front. Microbiol. 4:237. doi: 10.3389/fmicb.2014.00237
Fu, H., and Burris, R. H. (1989). Ammonium inhibition of nitrogenase activity in Herbaspirillum seropedicae. J. Bacteriol. 171, 3168–3175. doi: 10.1128/jb.171.6.3168-3175.1989
Galloway, J. N., Dentener, F. J., Capone, D. G., Boyer, E. W., Howarth, R. W., Seitzinger, S. P., et al. (2004). Nitrogen cycles: past, present, and future. Biogeochemistry 70, 153–226.
Gardiner, D. T., and James, S. (2012). Wet soil redox chemistry as affected by organic matter and nitrate. Am. J. Clim. Chang. 01, 205–209. doi: 10.4236/ajcc.2012.14017
Gill, S. R., Pop, M., Deboy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359.
Gomes, L. C., Piard, J. C., Briandet, R., and Mergulhão, F. J. (2017). Pseudomonas grimontii biofilm protects food contact surfaces from Escherichia coli colonization. LWT Food Sci. Technol. 85, 309–315. doi: 10.1016/j.lwt.2017.03.005
Graham, P. H., Draeger, K. J., Ferrey, M. L., Conroy, M. L., Hammer, B. E., Martinez, E., et al. (1994). Acid pH tolerance in strains of Rhizobium and Bradyrhizobium, and initial studies on the basis for acid tolerance of Rhizobium tropici UMR1899. J. Microbiol. 40, 198–207. doi: 10.1139/m94-033
Gu, Y., Mi, W. H., Xie, Y. N., Ma, Q. X., Wu, L. H., Hu, Z. P., et al. (2019). Nitrapyrin affects the abundance of ammonia oxidizers rather than community structure in a yellow clay paddy soil. J. Soils Sediments 19, 872–882. doi: 10.1007/s11368-018-2075-3
Hallin, S., Jones, C. M., Schloter, M., and Philippot, L. (2009). Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3, 597–605. doi: 10.1038/ismej.2008.128
He, L. L., Shan, J., Zhao, X., Wang, S. Q., and Yan, X. Y. (2019). Variable responses of nitrification and denitrification in a paddy soil to long-term biochar amendment and short-term biochar addition. Chemosphere 234, 558–567. doi: 10.1016/j.chemosphere.2019.06.038
Henry, S., Baudoin, E., Lopez-Gutierrez, J. C., Martin-Laurent, F., Brauman, A., and Philippot, L. (2004). Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59, 327–335. doi: 10.1016/j.mimet.2004.07.002
Hou, S. P., Ai, C., Zhou, W., Liang, G. Q., and He, P. (2018). Structure and assembly cues for rhizospheric nirK- and nirS-type denitrifier communities in long-term fertilized soils. Soil Biol. Biochem. 119, 32–40. doi: 10.1016/j.soilbio.2018.01.007
Hu, H. W., Chen, D., and He, J. Z. (2015). Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol. Rev. 39, 729–749. doi: 10.1093/femsre/fuv021
Huang, S. H., Pant, H. K., and Lu, J. (2007). Effects of water regimes on nitrous oxide emission from soils. Ecol. Eng. 31, 9–15. doi: 10.1016/j.ecoleng.2007.04.001
Hunt, P. G., Matheny, T. A., and Ro, K. S. (2007). Nitrous oxide accumulation in soils from riparian buffers of a coastal plain watershed-carbon/nitrogen ratio control. J. Environ. Q. 36, 1368–1376. doi: 10.2134/jeq2006.0255
IPCC (2014). Climate Change 2014: Mitigation Of Climate Change, Summary For Policymakers. Contribution of Working Group III to the Fifth Assessment Report Of The Intergovernmental Panel On Climate Change. New York, NY: Cambridge University Press.
Kandeler, E., Deiglmayr, K., Tscherko, D., Bru, D., and Philippot, L. (2006). Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 72, 5957–5962. doi: 10.1128/aem.00439-06
Leff, J. W., Jones, S. E., Prober, S. M., Barberán, A., Borer, E. T., Firn, J. L., et al. (2015). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. U.S.A. 112, 10967–10972. doi: 10.1073/pnas.1508382112
Liang, B. C., and MacKenzie, A. F. (1997). Seasonal denitrification rates under corn (Zea mays L.) in two Quebec soils. Can. J. Soil Sci. 77, 21–25. doi: 10.4141/s96-018
Liu, J., Hou, H., Sheng, R., Chen, Z., Zhu, Y., Qin, H., et al. (2012). Denitrifying communities differentially respond to flooding drying cycles in paddy soils. Appl. Soil Ecol. 62, 155–162. doi: 10.1016/j.apsoil.2012.06.010
Liu, Y.-R., Delgado-Baquerizo, M., Trivedi, P., He, J.-Z., Wang, J.-T., and Singh, B. K. (2017). Identity of biocrust species and microbial communities drive the response of soil multifunctionality to simulated global change. Soil Biol. Biochem. 107, 208–217. doi: 10.1016/j.soilbio.2016.12.003
Liu, Z., Wang, H., Li, J., Hong, Z., and Xu, R. (2015). Adhesion of Escherichia coli and Bacillus subtilis to amorphous Fe and Al hydroxides and their effects on the surface charges of the hydroxides. J. Soils Sediments 15, 2293–2303. doi: 10.1007/s11368-015-1147-x
Luo, J., Tillman, R. W., and Ball, P. R. (2000). Nitrogen loss through denitrification in a soil under pasture in New Zealand. Soil Biol. Biochem. 32, 497–509. doi: 10.1016/s0038-0717(99)00179-0
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Miller, L. G., Oremland, R. S., and Paulsen, S. (1986). Measurement of nitrous oxide reductase activity in aquatic sediments. Appl. Environ. Microbiol. 51, 18–24. doi: 10.1128/aem.51.1.18-24.1986
Mosier, A., Wassmann, R., Verchot, L., King, J., and Palm, C. (2004). Methane and nitrogen oxide fluxes in tropical agricultural soils. Environ. Dev. Sustain. 6, 11–49. doi: 10.1007/978-94-017-3604-6_2
Olsen, S. R. (1954). Estimation of Available Phosphorus In Soils By Extraction With Sodium Bicarbonate. Washington, DC: US Department of Agriculture.
Page, A. L., Miller, R. H., and Keeney, D. R. (1982). Methods of Soil Analysis, part 2, Chemical and Microbiological Properties. Madison, WI.
Pansu, M., and Gautheyrou, J. (2006). Handbook of Soil Analysis: Mineralogical, Organic And Inorganic Methods. Heidelberg: Springer.
Parkin, T. B., Sexstone, A. J., and Tiedje, J. M. (1985). Adaptation of denitrifying populations to low soil pH. Appl. Environ. Microbiol. 49, 1053–1056. doi: 10.1128/aem.49.5.1053-1056.1985
Pell, M., Stenberg, B., Stenström, J., and Torstensson, L. (1996). Potential denitrification activity assay in soil—with or without chloramphenicol? Soil Biol. Biochem. 28, 393–398. doi: 10.1016/0038-0717(95)00149-2
Philippot, L., and Hallin, S. (2005). Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr. Opin. Microbiol. 8, 234–239. doi: 10.1016/j.mib.2005.04.003
Philippot, L., Hallin, S., and Schloter, M. (2007). Ecology of denitrifying prokaryotes in agricultural soil. Adv. Agron. 96, 249–305. doi: 10.1016/s0065-2113(07)96003-4
Poortinga, A. T., Bos, R., Norde, W., and Busscher, H. J. (2002). Electric double layer interactions in bacterial adhesion to surfaces. Surf. Sci. Rep. 47, 3–32.
Qian, W., Ma, B., Li, X., Zhang, Q., and Peng, Y. (2019). Long-term effect of pH on denitrification: high pH benefits achieving partial-denitrification. Bioresour. Technol. 278, 444–449. doi: 10.1016/j.biortech.2019.01.105
Qin, S., Hu, C., and Oenema, O. (2012). Quantifying the underestimation of soil denitrification potential as determined by the acetylene inhibition method. Soil Biol. Biochem. 47, 14–17. doi: 10.1016/j.soilbio.2011.12.019
Qu, Z., Wang, J., Almoy, T., and Bakken, L. R. (2014). Excessive use of nitrogen in Chinese agriculture results in high N2O/(N2O+N2) product ratio of denitrification, primarily due to acidification of the soils. Glob. Chang. Biol. 20, 1685–1698. doi: 10.1111/gcb.12461
Rahman, M. M., Shan, J., Yang, P., Shang, X., Xia, Y., and Yan, X. (2018). Effects of long-term pig manure application on antibiotics, abundance of antibiotic resistance genes (ARGs), anammox and denitrification rates in paddy soils. Environ. Pollut. 240, 368–377. doi: 10.1016/j.envpol.2018.04.135
Ruser, R., Flessa, H., Russow, R., Schmidt, G., Buegger, F., and Munch, J. C. (2006). Emission of N2O, N2 and CO2 from soil fertilized with nitrate: effect of compaction, soil moisture and rewetting. Soil Biol. Biochem. 38, 263–274. doi: 10.1016/j.soilbio.2005.05.005
Sanchez, G. (2013). PLS Path Modeling With R. Trowchez Editions. Berkeley: University of California.
Šimek, M., and Hopkins, D. (1999). Regulation of potential denitrification by soil pH in long-term fertilized arable soils. Biol. Fertil. Soils 30, 41–47. doi: 10.1007/s003740050585
Souza, E. M., Peodrosa, F. O., Rigo, L. U., and Machado, H. B. (2000). Expression of the nifA gene of Herbaspirillum seropedicae: role of the NtrC and NifA binding sites and of the-24/-12 promoter element. Microbiology 2000, 1407–1418. doi: 10.1099/00221287-146-6-1407
Stein, L. Y. (2011). Surveying N2O-producing pathways in bacteria. Methods Enzymol. 486, 131–152. doi: 10.1016/b978-0-12-381294-0.00006-7
Sun, R. B., Guo, X. S., Wang, D. Z., and Chu, H. Y. (2015). Effects of long-term application of chemical and organic fertilizers on the abundance of microbial communities involved in the nitrogen cycle. Appl. Soil Ecol. 95, 171–178. doi: 10.1016/j.apsoil.2015.06.010
Tatti, E., Goyer, C., Zebarth, B. J., Burton, D. L., Giovannetti, L., and Viti, C. (2013). Short-term effects of mineral and organic fertilizer on denitrifiers, nitrous oxide emissions and denitrification in long-term amended vineyard soils. Soil Sci. Soc. Am. J. 77, 113–122. doi: 10.2136/sssaj2012.0096
Throback, I. N., Enwall, K., Jarvis, A., and Hallin, S. (2004). Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49, 401–417. doi: 10.1016/j.femsec.2004.04.011
Throback, I. N., Johansson, M., Rosenquist, M., Pell, M., Hansson, M., and Hallin, S. (2007). Silver (Ag+) reduces denitrification and induces enrichment of novel nirK genotypes in soil. FEMS Microbiol. Lett. 270, 189–194. doi: 10.1111/j.1574-6968.2007.00632.x
Ullah, S., and Faulkner, S. P. (2006). Denitrification potential of different land-use types in an agricultural watershed, lower Mississippi valley. Cell 28, 131–140. doi: 10.1016/j.ecoleng.2006.05.007
Wang, C., Lu, H., Dong, D., Deng, H., Strong, P. J., Wang, H., et al. (2013). Insight into the effects of biochar on manure composting: evidence supporting the relationship between N2O emission and denitrifying community. Environ. Sci. Technol. 47, 7341–7349. doi: 10.1021/es305293h
Wang, X., Liu, B., Ma, J., Zhang, Y., Hu, T., Zhang, H., et al. (2019). Soil aluminum oxides determine biological nitrogen fixation and diazotrophic communities across major types of paddy soils in China. Soil Biol. Biochem. 131, 81–89. doi: 10.1016/j.soilbio.2018.12.028
Williams, A., Börjesson, G., and Hedlund, K. (2013). The effects of 55 years of different inorganic fertiliser regimes on soil properties and microbial community composition. Soil Biol. Biochem. 67, 41–46. doi: 10.1016/j.soilbio.2013.08.008
Wolsing, M., and Prieme, A. (2004). Observation of high seasonal variation in community structure of denitrifying bacteria in arable soil receiving artificial fertilizer and cattle manure by determining T-RFLP of nir gene fragments. FEMS Microbiol. Ecol. 48, 261–271. doi: 10.1016/j.femsec.2004.02.002
Wu, K., Li, Y., Liu, T., Huang, Q., Yang, S., Wang, W., et al. (2019). The simultaneous adsorption of nitrate and phosphate by an organic-modified aluminum-manganese bimetal oxide: adsorption properties and mechanisms. Appl. Surf. Sci. 478, 539–551. doi: 10.1016/j.apsusc.2019.01.194
Xu, Y. B., and Cai, Z. C. (2007). Denitrification characteristics of subtropical soils in China affected by soil parent material and land use. Eur. J. Soil Sci. 58, 1293–1303. doi: 10.1111/j.1365-2389.2007.00923.x
Yin, C., Fan, F., Song, A., Cui, P., Li, T., and Liang, Y. (2015). Denitrification potential under different fertilization regimes is closely coupled with changes in the denitrifying community in a black soil. Appl. Microbiol. Biotechnol. 99, 5719–5729. doi: 10.1007/s00253-015-6461-0
Yin, C., Fan, F. L., Song, A. L., Li, Z. J., Yu, W. T., and Liang, Y. C. (2014). Different denitrification potential of aquic brown soil in Northeast China under inorganic and organic fertilization accompanied by distinct changes of nirS- and nirK-denitrifying bacterial community. Eur. J. Soil Biol. 65, 47–56. doi: 10.1016/j.ejsobi.2014.09.003
Yu, Z. H., Liu, J. J., Li, Y. S., Jin, J., Liu, X. B., and Wang, G. H. (2018). Impact of land use, fertilization and seasonal variation on the abundance and diversity of nirS-type denitrifying bacterial communities in a Mollisol in Northeast China. Eur. J. Soil Biol. 85, 4–11. doi: 10.1016/j.ejsobi.2017.12.001
Zhang, H., Wang, B., Xu, M., and Fan, T. (2009). Crop yield and soil responses to long- term fertilization on a red soil in southern China. Pedosphere 19, 199–207. doi: 10.1016/s1002-0160(09)60109-0
Zhang, M. K., and Xu, J. M. (2005). Restoration of surface soil fertility of an eroded red soil in southern China. Soil Tillage Res. 80, 13–21. doi: 10.1016/j.still.2004.02.019
Keywords: soil denitrification, land use, long-term fertilization, functional genes, nirS-denitrifying bacteria
Citation: Wang H, Liu Z, Ma L, Li D, Liu K, Huang Q, Zhao B and Zhang J (2020) Denitrification Potential of Paddy and Upland Soils Derived From the Same Parent Material Respond Differently to Long-Term Fertilization. Front. Environ. Sci. 8:105. doi: 10.3389/fenvs.2020.00105
Received: 11 March 2020; Accepted: 18 June 2020;
Published: 23 July 2020.
Edited by:
Hannes Schmidt, University of Vienna, AustriaReviewed by:
Andrew T. Giguere, Aalborg University, DenmarkMeihua Deng, ZheJiang Academy of Agricultural Sciences, China
Copyright © 2020 Wang, Liu, Ma, Li, Liu, Huang, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingzi Zhao, YnpoYW9AaXNzYXMuYWMuY24=; Jiabao Zhang, amJ6aGFuZ0Bpc3Nhcy5hYy5jbg==
 Haicui Wang
Haicui Wang Zhaodong Liu
Zhaodong Liu Lei Ma
Lei Ma Dandan Li1,2
Dandan Li1,2 Bingzi Zhao
Bingzi Zhao