- 1Soil Microbial Ecology Group, NEIKER-Tecnalia, Department of Conservation of Natural Resources, Derio, Spain
- 2Department of Biochemistry and Molecular Biology, Instituto BIOFISIKA (CSIC, UPV/EHU), University of the Basque Country, Bilbao, Spain
The use of nanoscale zero-valent iron (nZVI) particles for soil remediation is gaining increased attention. However, there are concerns about the potential adverse effects of nZVI on soil microbial communities and, hence, soil quality. The objective of this study was to assess the impact of the application of nZVI on soil microbial parameters (as bioindicators of soil quality) during the nanoremediation of soil artificially contaminated with lindane (10 mg γ-HCH kg−1 DW soil) and zinc (1,500 mg Zn kg−1 DW soil). nZVI particles were also applied to non-contaminated soil. The following nZVI doses were applied twice: 0, 0.25, 0.5, 1, and 2 mg nZVI g−1 DW soil. Nine weeks after nZVI application, the following parameters were determined in soil samples: lindane concentration, extractable Zn concentration, microbial biomass carbon (CMB), bacterial and fungal abundance (gene copy numbers by qPCR), enzyme activities (β-glucosidase, β-glucosaminidase, xylosidase, acid phosphatase, arylsulphatase, and Leu-aminopeptidase) and bacterial richness by ARISA profiles. The application of nZVI reduced lindane and extractable Zn concentrations following a dose-dependent pattern. The presence of contaminants reduced soil microbial biomass and activity. The application of nZVI negatively affected the microbial quality of the contaminated soil but not of the non-contaminated soil. This observation might reflect a “stress-on-stress” effect, i.e., the already stressed microbial populations present in the contaminated soil were more sensitive to the application of nZVI (a second stress) than those present in the non-contaminated soil.
Introduction
Microorganisms are known to play key roles in soil and other ecosystems (Epelde et al., 2015). Pesticides, such as lindane, can negatively impact soil microbial communities (Hussain et al., 2009; Imfeld and Vuilleumier, 2012). Although microorganisms can degrade lindane, this option presents serious limitations (lindane is a recalcitrant contaminant), especially in mixed-contaminated soils (Kumar and Pannu, 2018). Among the newer methods of remediation, nanoremediation is becoming a promising technology. Owing to their high reactivity, nanoparticles, such as nanoscale zero-valent iron particles (nZVI), are being extensively used for environmental remediation (Tosco et al., 2014). The majority of microbiological studies on nZVI toxicity have been performed with single bacterial pure cultures and short-term exposures (Xie et al., 2017). These studies have reported contrasting results regarding the effect of nZVI on bacteria and fungi (Lefevre et al., 2016; Semerád and Cajthaml, 2016). Much research is needed on the ecotoxicity of nanoparticles in complex environmental matrices such as the soil.
The objective of this study was to assess the impact of nZVI application on soil microbial parameters during the nanoremediation of soil artificially contaminated with lindane (10 mg kg−1 DW soil) and zinc (1,500 mg kg−1 DW soil). Microbial parameters are increasingly used as bioindicators of soil quality due to their high sensitivity and rapid response to disturbances, which make them ideal tools to assess short-term effects (Mijangos et al., 2010). We hypothesized that the application of nZVI will reduce lindane concentration through nZVI-induced reductive dechlorination followed by microbial degradation and, hence, will lead to a recovery of soil quality. We also hypothesized that the effect of nZVI on soil microbial quality will be different in non-contaminated vs. contaminated soil, as microorganisms in the latter are already subjected to a disturbance (i.e., contamination).
Materials and Methods
Soil (Hypocalcic Calcisol) was collected (0–20 cm depth) from local grasslands, left to dry at ambient temperature for 5 days and sieved to < 6 mm. Half of the soil was artificially contaminated with 10 mg kg−1 dry weight (DW) soil of lindane (gamma-hexachlorocyclohexane, γ-HCH) and 1,500 mg Zn kg−1 DW soil. A stock solution of 100 mg lindane in 250 mL of acetone was prepared and then diluted 8-fold in acetone. Two hundred mL of this solution was sprayed per kilogram of soil and, subsequently, the soil was left in the hood for 2 h to allow acetone evaporation. For the artificial contamination with Zn, 68.22 g of zinc nitrate hexahydrate was diluted in 1 L of distilled water; then, 100 mL of this solution was sprayed per kilogram of soil. The soil was left to equilibrate for one month. The remaining half of the soil was left uncontaminated as control.
Afterwards, 200 g of soil (contaminated or non-contaminated) was placed in 0.5 L beaker flasks and maintained at 60% water holding capacity. The following concentrations of nZVI [Nanofer 25S, aqueous dispersion of Fe(0) nanoparticles, NANO IRON s.r.o., Czech Republic] were added as slurry to both soils: 0, 0.25, 0.5, 1, and 2 mg nZVI g−1 DW soil (5 replicates per treatment). nZVI-spiked soils were thoroughly mixed by hand. Three days later, a second application of nZVI was performed as described above. Final nZVI doses (i.e., treatments) were as follows: 0, 0.5, 1, 2, and 4 mg nZVI g−1 DW soil. Soils were incubated at room temperature for 9 weeks and, finally, soil samples were collected from the flasks and stored at 4°C for analysis.
Lindane concentration was measured in a Gas Chromatograph (Agilent 6890N) with a μECD detector and an HP-ULTRA 2 column. The extraction was carried out by sonication (40 kHz, 100 W; 5 min, 3 cycles) of 1.5 g DW soil in 5 mL of hexane/acetone (50:50 v/v). Extractable Zn concentrations (as an estimation of bioavailable Zn) were determined in 0.01 M CaCl2 extracts by Atomic Absorption following Houba et al. (2000).
The soil was characterized according to standard methods (Ministerio de Agricultura, Pesca y Alimentación, 1994), obtaining the following values: total organic carbon = 2.3%; pH (in water, 1:2.5 w/v) = 8.1; and texture = clay-loam. The content (%) of dry matter was measured at 105°C for 24 h.
Microbial biomass carbon (CMB) was determined by the fumigation-extraction method (Vance et al., 1987). β-glucosidase (GLU), acid phosphatase (PHO), arylsulphatase (SUL), β-glucosaminidase (GLM), xylosidase (XYL) and Leu-aminopeptidase (AMP) activities were determined following ISO/TS-22939 (2010), Parham and Deng (2000), and Taylor et al. (2002).
DNA was extracted from soil (0.25 g) samples using the DNA PowerSoil™ Isolation Kit (MO Bio Laboratories). Prior to DNA extraction, soil samples were washed twice in 120 mM K2HPO4 (pH 8.0) to wash away extracellular DNA (Kowalchuk et al., 1997). The concentration of extracted DNA was determined with Nanodrop, ND-1000. For the determination of bacterial and fungal abundances, real time q-PCR analyses were performed following Dhanasekaran et al. (2010). Primers used to determine gene copy numbers for total bacteria (16S rRNA) were Ba519f and Ba907r (Lueders et al., 2004) and for total fungi (18S rRNA) FF390r and Fung5f (Vainio and Hantula, 2000). Community-level genetic profiles for bacteria were obtained using ARISA (Cardinale et al., 2004), with primers ITSF (GTCGTAACAAGGTAGCCGTA) and ITSReub (GCCAAGGCATCCACC). Data were analyzed with GeneMarker Software (Softgenetics LCC) (Welkie et al., 2010). With all the microbial parameters, a Soil Quality Index (SQI) was determined following Bloem et al. (2006).
Differences between treatments at a 5% significance level were calculated using parametric one-way ANOVA followed by Duncan's test (p < 0.05) using the R Core Team program (v.3.5.0). Multivariate analyses were performed to explore relationships between microbial properties and nZVI treatments: Principal Component Analysis (PCA) in non-contaminated soil, Redundancy Analysis (RDA) in contaminated soil and, finally, RDA for both soils, using Canono for Windows 5.0 (Ter Braak and Šmilauer, 2002).
Results
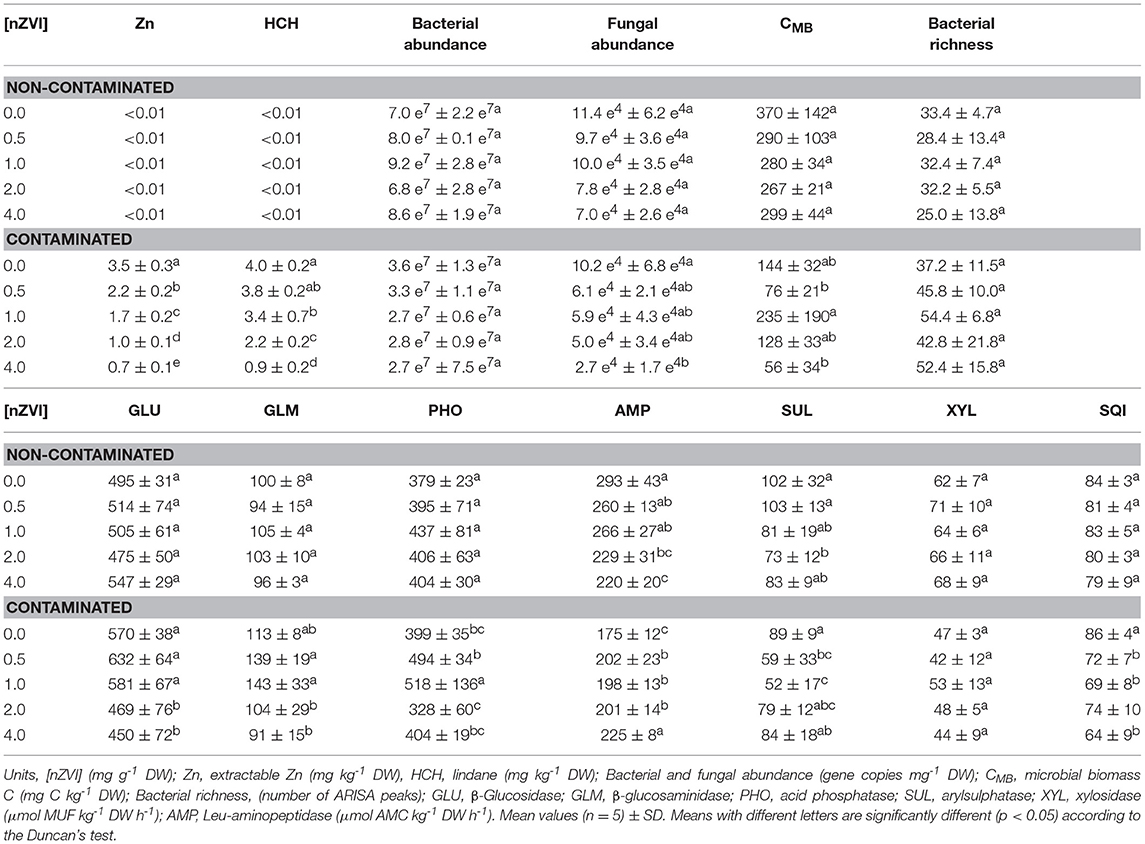
After the 9-week incubation, lindane and extractable Zn concentrations were progressively lower at increasing nZVI concentrations (Table 1). Bacterial abundance in both soils (contaminated, non-contaminated) was not significantly affected by nZVI. Fungal abundance in contaminated soil was significantly lower at 4 mg nZVI g−1 DW soil, compared to soil without nZVI (no significant differences among nZVI treatments were observed in non-contaminated soil). In contaminated soil, CMB values did not show a clear pattern in relation to nZVI treatments (again, no significant differences among nZVI treatments were observed in non-contaminated soil). In both soils, bacterial richness of dominant taxa, obtained from ARISA, was not significantly altered by nZVI.
Regarding enzyme activities (Table 1), in non-contaminated soil, no significant differences were observed among treatments for GLU, GLM, PHO, and XYL. AMP values were significantly lower at 2 and 4 mg nZVI g−1 DW soil, compared to soil without nZVI. SUL values were significantly lower at 2 mg nZVI g−1 DW soil, compared to soil without nZVI.
In contaminated soil, GLU values were significantly lower at 2 and 4 mg nZVI g−1 DW soil, compared to soil without nZVI. XYL activity was not affected by nZVI. AMP values were significantly higher in ZVI-treated soils, compared to soil without nZVI.
In contaminated soil, values of the SQI were significantly lower in ZVI-treated soils, compared to soil without nZVI (no significant differences among treatments were observed in non-contaminated soil).
In non-contaminated soil, the Monte Carlo permutation test was not significant (pseudo-F = 1.3, p = 0.138) and, therefore, PCA was performed (Figure 1A). PC1 and PC2 explained 29 and 16% of the variability, respectively. However, in non-contaminated soil, no clear trend was observed between microbial parameter values and nZVI concentrations. In contaminated soil (pseudo-F = 4.8, p = 0.002), axes 1 and 2 of the RDA explained 32 and 12% of the variability, respectively (Figure 1B). Extractable Zn and lindane concentrations were lower at increasing nZVI concentrations. According to this RDA, some microbial biomass parameters (fungal abundance and, to a lesser extent, bacterial abundance) were lower at increasing nZVI concentrations.
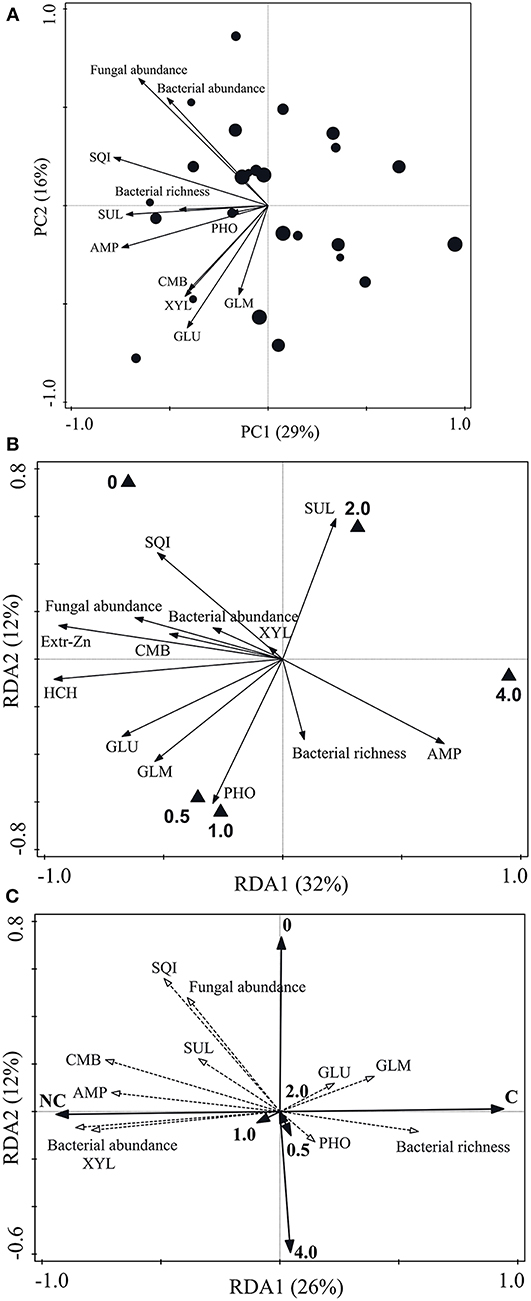
Figure 1. Multivariate analyses of relationships between nZVI concentrations and microbial parameters. (A) PCA of non-contaminated soil: the larger the circle size, the higher the nZVI concentration (from 0 to 4 mg g−1 DW soil); (B) RDA of contaminated soil; (C) RDA of both non-contaminated (NC) and contaminated (C) soil. Abbreviations as in Table 1.
In the RDA carried out simultaneously with microbial parameter data from both soils (non-contaminated and contaminated) (Figure 1C), it can be observed that contamination had a greater effect on microbial properties (data distribution along axis 1) than nZVI treatments (data distribution along axis 2). Microbial biomass values (CMB, bacterial and fungal abundance) were generally higher in non-contaminated vs. contaminated soil. In most cases, values of some microbial activity parameters (AMP, XYL, and SUL) were higher in non-contaminated vs. contaminated soil.
Discussion
A considerable portion of the lindane spiked to soil during the process of artificial contamination was lost either through volatilization or biodegradation (Table 1). The high rate of lindane removal was probably due to the fact that, after artificial contamination, the soil was left to equilibrate for just 1 month, implying that the amount of non-recalcitrant lindane might still be high. We left the soil to equilibrate for only 1 month because our main interest was to detect short-term effects of nZVI on soil quality (in other words, our study was not aimed at studying contaminant removal). After 1 month of soil equilibration (“aging”), only 0.22% of the total Zn was present in a bioavailable form (CaCl2-extractable Zn), probably due to the relatively high pH of our soil (pH = 8.1). There was a clear relationship between lindane and extractable Zn concentration and nZVI dose (Table 1). At the highest nZVI dose, an 80 and 78% reduction in extractable Zn and lindane, respectively, was observed, compared to soil without nZVI.
Many studies on lindane-contaminated soils have focused on the search for degrading strains (Garg et al., 2016; Saez et al., 2018). On the other hand, the impact of lindane and other HCH isomers on soil microorganisms has also been studied (Anza et al., 2016). Soil microbial parameters have regularly been used to assess the impact of contaminants on soil quality (Epelde et al., 2010; Barrutia et al., 2011; Pardo et al., 2014; Burges et al., 2015). Instead, the impact of nZVI on the quality of lindane-contaminated soil in the presence of other contaminants (mixed contamination) has, to our knowledge, not been studied. Although our soil was artificially contaminated with a high Zn concentration, at the end of the experiment, the concentration of bioavailable Zn was quite low. This suggests that, as far as contaminant effects are concerned, lindane may have had a greater influence on soil microbial parameters than Zn. In any case, the process of artificial contamination resulted in lower values of microbial biomass and, to a lesser extent, microbial activity. On the contrary, an increase in bacterial richness, as reflected by ARISA, was observed in contaminated soil compared to non-contaminated soil, suggesting that a higher number of dominant bacterial taxa were selected under contaminant-induced stressing conditions. ARISA data provide information only on dominant taxa, i.e., they cannot be used to estimate total richness.
In non-contaminated soil, no negative effects of nZVI on microbial parameters and, concomitantly, soil microbial quality were detected. In contaminated soil, despite the observed reduction in contaminant concentrations, soil microbial quality was reduced by nZVI treatments. This observation might reflect a “stress-on-stress” effect, i.e., the already stressed microbial populations present in contaminated soil were more sensitive to the application of nZVI (a second stress) than those present in non-contaminated soil.
Author Contributions
MA and OS carried out the laboratory analyses. MA, OS, and LE contributed to different aspects of data analysis and interpretation. MA, OS, IA, and CG wrote the paper. LE and CG contributed to the conception and design of the study. All remaining authors revised it critically.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Spanish Ministry of Economy, Industry and Competitiveness through NANORRIZORREM-2 Project (AGL2016-76592-R).
References
Anza, M., Epelde, L., Martín-Sánchez, I., Blanco, F., and Garbisu, C. (2016). Microbial indicators for assessing the adverse impact of technical-grade hexachlorocyclohexane on soil quality. Span. J. Soil Sci. 6, 1–14. doi: 10.3232/SJSS.2016.V6.N1.01
Barrutia, O., Garbisu, C., Epelde, L., Sampedro, M. C., Goicolea, M. A., and Becerril, J. M. (2011). Plant tolerance to diesel minimizes its impact on soil microbial characteristics during rhizoremediation of diesel-contaminated soils. Sci. Total Environ. 409, 4087–4093. doi: 10.1016/j.scitotenv.2011.06.025
Bloem, J., Schouten, A., Sørensen, S., Rutgers, M., van der Werf, A., and Breure, A. (2006). “Monitoring and evaluating soil quality,” in Microbiological Methods for Assessing Soil Quality, eds J. Bloem, D. Hopkins and A. Benedetti (Wallingford, CT: CAB International), 23–49.
Burges, A., Epelde, L., and Garbisu, C. (2015). Impact of repeated single-metal and multi-metal pollution events on soil quality. Chemosphere 120, 8–15. doi: 10.1016/j.chemosphere.2014.05.037
Cardinale, M., Brusetti, L., Quatrini, P., Borin, S., Puglia, A. M., Rizzi, A., et al. (2004). Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70, 6147–6156. doi: 10.1128/AEM.70.10.6147-6156.2004
Dhanasekaran, S., Doherty, T. M., and Kenneth, J. (2010). Comparison of different standards for real-time PCR based absolute quantification. J. Immunol. Methods 354, 34–39. doi: 10.1016/j.jim.2010.01.004
Epelde, L., Becerril, J. M., Kowalchuk, G. A., Deng, Y., Zhou, J. Z., and Garbisu, C. (2010). Impact of metal pollution and Thlaspi caerulescens growth on soil microbial communities. Appl. Environ. Microbiol. 76, 7843–7853. doi: 10.1128/AEM.01045-10
Epelde, L., Lanzén, A., Blanco, F., Urich, T., and Garbisu, C. (2015). Adaptation of soil microbial community structure and function to chronic metal contamination at an abandoned Pb-Zn mine. FEMS Microbiol. Ecol. 91, 1–11. doi: 10.1093/femsec/fiu007
Garg, N., Lata, P., Jit, S., Sangwan, N., Singh, A. K., Dwivedi, V., et al. (2016). Laboratory and field scale bioremediation of hexachlorocyclohexane (HCH) contaminated soils by means of bioaugmentation and biostimulation. Biodegradation 27, 179–193. doi: 10.1007/s10532-016-9765-6
Houba, V. J. G., Temminghoff, E. J. M., Gaikhorst, G. A., and van Vark, W. (2000). Soil analysis procedures using 0.01 M calcium chloride as extraction reagent. Commun. Soil Sci. Plant Anal. 31, 1299–1396. doi: 10.1080/00103620009370514
Hussain, S., Siddique, T., Saleem, M., Arshad, M., and Khalid, A. (2009). “Impact of pesticides on soil microbial diversity, enzymes and biochemical reactions,” in Advances in Agronomy, ed D. L. Sparks (San Diego, CA: Academic Press), 159–200.
Imfeld, G., and Vuilleumier, S. (2012). Measuring the effects of pesticides on bacterial communities in soil: a critical review. Eur. J. Soil Biol. 49, 22–30. doi: 10.1016/j.ejsobi.2011.11.010
ISO/TS-22939 (2010). Soil Quality — Measurement of Enzyme Activity Patterns in Soil Samples Using Fluorogenic Substrates in Micro-Well Plates.
Kowalchuk, G. A., Stephen, J., DeBoer, W., Prosser, J. I., Embley, T. M., and Woldendorp, J. W. (1997). Analysis of ammonia oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63, 489–1497.
Kumar, D., and Pannu, R. (2018). Perspectives of lindane (γ-hexachlorocyclohexane) biodegradation from the environment: a review. Bioresour. Bioprocess. 5:29. doi: 10.1186/s40643-018-0213-9
Lefevre, E., Bossa, N., Wiesner, M. R., and Gunsch, C. K. (2016). A review of the environmental implications of in situ remediation by nanoscale zero valent iron (nZVI): behavior, transport and impacts on microbial communities. Sci. Total Environ. 565, 889–901. doi: 10.1016/j.scitotenv.2016.02.003
Lueders, T., Manefield, M., and Friedrich, M. W. (2004). Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6, 73–78. doi: 10.1046/j.1462-2920.2003.00536.x
Mijangos, I., Albizu, I., Epelde, L., Amezaga, I., Mendarte, S., and Garbisu, C. (2010). Effects of liming on soil properties and plant performance of temperate mountainous grasslands. J. Environ. Manage. 91, 2066–2074. doi: 10.1016/j.jenvman.2010.05.011
Ministerio de Agricultura Pesca y Alimentación. (1994). Métodos Oficiales de Análisis de Suelos y Aguas Para Riego. Madrid: Secretaría General de Alimentación.
Pardo, T., Clemente, R., Epelde, L., Garbisu, C., and Bernal, M. P. (2014). Evaluation of the phytostabilisation efficiency in a trace elements contaminated soil using soil health indicators. J. Hazard. Mater. 268, 68–76. doi: 10.1016/j.jhazmat.2014.01.003
Parham, J. A., and Deng, S. P. (2000). Detection, quantification and characterization of beta-glucosaminidase activity in soil. Soil Biol. Biochem. 32, 1183–1190. doi: 10.1016/S0038-0717(00)00034-1
Saez, J. M., Bigliardo, A. L., Raimondo, E. E., Briceño, G. E., Polti, M. A., and Benimeli, C. S. (2018). Lindane dissipation in a biomixture: effect of soil properties and bioaugmentation. Ecotox. Environ. Safety 156, 97–105. doi: 10.1016/j.ecoenv.2018.03.011
Semerád, J., and Cajthaml, T. (2016). Ecotoxicity and environmental safety related to nano-scale zerovalent iron remediation applications. Appl. Microbiol. Biotechnol. 100, 9809–9819. doi: 10.1007/s00253-016-7901-1
Taylor, J., Wilson, B., Mills, M., and Burns, R. (2002). Comparison of microbial numbers and enzymatic activities in surface soils and subsoils using various techniques. Soil Biol. Biochem. 34, 387–401. doi: 10.1016/S0038-0717(01)00199-7
Ter Braak, C. J. F., and Šmilauer, P. (2002). CANOCO Reference Manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination. Ithaca, NY: Microcomputer Power.
Tosco, T., Petrangeli, M., Cruz, C., and Sethi, R. (2014). Nanoscale zerovalent iron particles for groundwater remediation: a review. J. Clean. Prod. 77, 10–21. doi: 10.1016/j.jclepro.2013.12.026
Vainio, E. J., and Hantula, J. (2000). Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104, 927–936. doi: 10.1017/S0953756200002471
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19, 703–707. doi: 10.1016/0038-0717(87)90052-6
Welkie, D. G., Stevenson, D. M., and Weimer, P. J. (2010). ARISA analysis of ruminal bacterial community dynamics in lactating dairy cows during the feeding cycle. Anaerobe 16, 94–100. doi: 10.1016/j.anaerobe.2009.07.002
Keywords: bioindicators, nanoremediation, pollution, soil health, soil microorganisms
Citation: Anza M, Salazar O, Epelde L, Alkorta I and Garbisu C (2019) The Application of Nanoscale Zero-Valent Iron Promotes Soil Remediation While Negatively Affecting Soil Microbial Biomass and Activity. Front. Environ. Sci. 7:19. doi: 10.3389/fenvs.2019.00019
Received: 08 November 2018; Accepted: 30 January 2019;
Published: 19 February 2019.
Edited by:
Mohiuddin Md. Taimur Khan, Washington State University, United StatesReviewed by:
Juan M. Gonzalez, Spanish National Research Council (CSIC), SpainMd. Asaduzzaman Shishir, University of Dhaka, Bangladesh
Copyright © 2019 Anza, Salazar, Epelde, Alkorta and Garbisu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mikel Anza, bWFuemFAbmVpa2VyLmV1cw==
 Mikel Anza
Mikel Anza Oihane Salazar1
Oihane Salazar1 Lur Epelde
Lur Epelde Itziar Alkorta
Itziar Alkorta Carlos Garbisu
Carlos Garbisu