
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 14 January 2019
Sec. Microbiological Chemistry and Geomicrobiology
Volume 6 - 2018 | https://doi.org/10.3389/fenvs.2018.00162
This article is part of the Research Topic Microbial Sulfur Metabolism - From Symbiosis to Global Nutrient Cycling View all 37 articles
Anaerobic oxidation of methane (AOM) coupled to sulfate reduction is mediated by, respectively, anaerobic methanotrophic archaea (ANME) and sulfate reducing bacteria (SRB). When a microbial community from coastal marine Lake Grevelingen sediment, containing ANME-3 as the most abundant type of ANME, was incubated under a pressure gradient (0.1–40 MPa) for 77 days, ANME-3 was more pressure sensitive than the SRB. ANME-3 activity was higher at lower (0.1, 0.45 MPa) over higher (10, 20, and 40 MPa) CH4 total pressures. Moreover, the sulfur metabolism was shifted upon changing the incubation pressure: only at 0.1 MPa elemental sulfur was detected in a considerable amount and SRB of the Desulfosarcina/Desulfococcus genera were more enriched at elevated pressures than the Desulfobulbus. This study provides evidence that ANME-3 can be constrained at shallow environments (45 m depth), despite the scarce bioavailable energy, because of its pressure sensitivity. Besides, the association between ANME-3 and SRB can be steered by changing solely the incubation pressure. The ANME-3 cells present in the marine Lake Grevelingen possess high specific AOM-SR rates and thus, can be of great potential to be applied in the industry after enrichment.
Microbially-mediated anaerobic oxidation of methane (AOM) coupled to sulfate reduction (SR) is a major sink in the oceanic methane (CH4) budget. AOM-SR is mediated by two different groups of microorganisms, namely anaerobic methanotrophs (ANME) and sulfate reducing bacteria (SRB). ANME are grouped into three distinct clades, i.e., ANME-1, ANME-2 and ANME-3 based on the phylogenetic analysis of their 16S rRNA genes (Hinrichs et al., 1999; Boetius et al., 2000; Knittel et al., 2005).
The net stoichiometry of the AOM-SR reaction is shown in Equation 1 (Reeburgh, 2007):
The thermodynamics of this reaction depend on the concentration of dissolved CH4. CH4 is poorly soluble: 1.3 mM is its concentration in sea water at ambient pressure and at 15°C (Yamamoto et al., 1976). Theoretically, an elevated CH4 pressure favors the AOM coupled to the SR (AOM-SR) bioconversion since (i) the Gibbs free energy becomes more negative at higher CH4 partial pressures and (ii) the dissolved CH4 concentration increases and is thus more bioavailable (Table 1). Thus, the activity and the growth of the microorganisms mediating the process, are expected to be higher at elevated pressures. In vitro incubations of ANME-1 and ANME-2 dominated microbial communities from deep sea sediments in high-pressure reactors showed indeed a strong positive relation of the activity of the microorganisms capable of the AOM-SR process with the CH4 partial pressure, up to 12 MPa (Nauhaus et al., 2002; Krüger et al., 2005; Niemann et al., 2006; Deusner et al., 2009; Zhang et al., 2010).
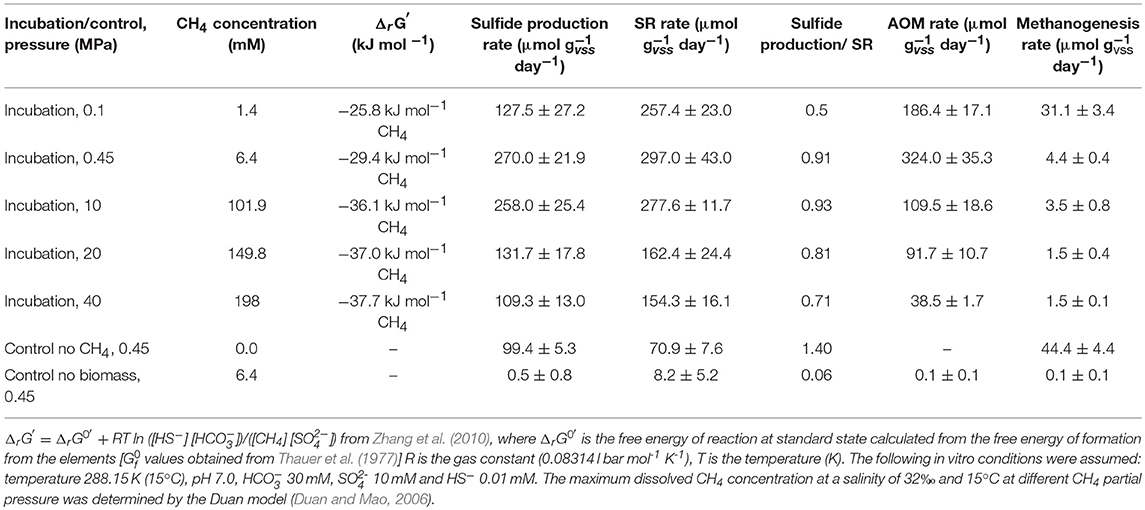
Table 1. Gibbs free energy of reaction (ΔrG′) for AOM coupled to SR at different CH4 total pressures, AOM, SR, sulfide production and methanogenesis rates, and the ratios between the sulfate consumed and sulfide produced determined in this study by marine Lake Grevelingen sediment.
The AOM dependent SR rate of Black Sea sediment was enhanced from 1.3 mmol l−1 day−1 at 0.2 MPa to 10 to 15-fold after increasing the CH4 partial pressure to 10 MPa (Deusner et al., 2009). Similarly, the AOM-SR rate of Hydrate Ridge sediment was also higher at elevated CH4 partial pressures: the AOM rate increased from 7 μmol (g dry mass sediment)−1 day−1 at 0.15 MPa to 17 μmol (g dry mass sediment)−1 day−1 at 1.5 MPa (Krüger et al., 2008) and after 24 months incubation at 1.4 MPa, the AOM rate increased from 20 to 230 μmol (g dry mass sediment)−1 day−1 (Nauhaus et al., 2002).
Moreover, a higher CH4 partial pressure (10 MPa) showed higher AOM-SR activity in in vitro incubations of ANME-2 dominated sediments. For instance, in vitro high pressure incubations using an enriched ANME-2 inoculum from the deep marine Gulf of Cadiz sediment (900 m water depth at ~10 MPa pressure and 15°C temperature) showed that the highest AOM rate was obtained at 10 MPa (0.7 mmol day−1) (Bhattarai et al., 2018). Zhang et al. (2010) determined that the affinity constant (Km) for CH4 of the ANME-2 enriched Gulf of Cadiz sediment is about 37 mM, which is equivalent to 3 MPa CH4 partial pressure.
Similarly, batch incubations using the shallow marine sediment of Eckernförde Bay (25 m water depth, ~0.2 MPa pressure, 15°C temperature), dominated by ANME-2, showed a linear increase of the AOM-SR rate with the CH4 partial pressure from 0.00 to 0.15 MPa (Meulepas et al., 2009b) and from 0.1 to 10.1 MPa (Timmers et al., 2015a). The measured Km for CH4 was higher than 0.075 MPa (1.1 mM) (Meulepas et al., 2009b). Moreover, the Eckernförde Bay sediment was incubated for 240 days in a high-pressure membrane capsule bioreactor, showing that the CH4 partial pressure influenced the growth of different subtypes of ANME-2 and SRB, i.e., at 10.1 MPa only the ANME-2c and SEEP-SRB2 subtypes were enriched (Timmers et al., 2015a).
Studying the effect of pressure on ANME and SRB will thus help understand the growth of the different ANME clades and their SRB partner. Finding ANME-SRB consortia that can grow fast at ambient pressure is of great importance for the application of AOM-SR in the desulfurization of industrial wastewater. Sulfate and other sulfur oxyanions, such as thiosulfate, sulfite or dithionite, are contaminants discharged in fresh water by industrial activities such as food processing, fermentation, coal mining, tannery and paper processing. Biological desulfurization under anaerobic conditions is a well-known biological treatment, in which these sulfur oxyanions are anaerobically reduced to sulfide (Weijma et al., 2006; Liamleam and Annachhatre, 2007; Sievert et al., 2007). The produced sulfide precipitates with the metals, thus enabling their recovery (Meulepas et al., 2010b). In the process of desulfurization of groundwater, mining or inorganic wastewater, electron donor for sulfate reduction needs to be supplied externally. Electron donors such as ethanol, hydrogen, methanol, acetate, lactate and propionate (Liamleam and Annachhatre, 2007) are usually supplied, but these increase the operational and investment costs (Meulepas et al., 2010b). The use of easily accessible and low-priced electron donors such as CH4 is therefore appealing for field-scale applications (Gonzalez-Gil et al., 2011). Moreover, from a logistic, economical and safety view point, bioreactors operating at ambient conditions are preferred over those operated at high pressures.
Coastal marine sediment from Lake Grevelingen (the Netherlands) hosts both ANME and SRB (Bhattarai et al., 2017). Among the ANME types, ANME-3 is predominant, which makes this sediment a beneficial inoculum to investigate the effects of pressure on ANME-3. ANME-3 is often found in cold seep areas and mud volcanoes with high CH4 partial pressures and relatively low temperatures (Niemann et al., 2006; Lösekann et al., 2007; Vigneron et al., 2013).
In this study, the shallow marine sediment was incubated at different pressures (0.1, 0.45, 10, 20, and 40 MPa) to study the influence of pressure on the AOM-SR activity. In addition, methanogenic activity and the potential formation of carbon, e.g., acetate or methanethiol (Valentine et al., 2000) and sulfur, e.g., elemental sulfur and polysulfides (Milucka et al., 2012) intermediates were also evaluated at different pressures. Moreover, phylogenetic analysis and visualization of microorganisms by fluorescence in situ hybridization (FISH) were used to study the shifts in community composition upon incubation of marine Lake Grevelingen sediment at different pressures in batches for 77 days.
The sediment was obtained from the Scharendijke Basin in the marine Lake Grevelingen (water depth of 45 m: position 51° 44.541′ N; 3° 50.969′ E), which is a former estuary in the southwestern part of the Netherlands. A detailed description of the biogeochemical chracteristics and microbial community can be found in Hagens et al. (2015), Egger et al. (2016), Sulu-Gambari et al. (2016), and Bhattarai et al. (2017). Coring was done in November 2013 on the vessel R/V Luctor by the Royal Netherlands Institute for Sea Research (Yerseke, the Netherlands). The sediment layers at 5–15 cm depth in each core were sampled under a nitrogen atmosphere and mixed together, and a homogenized slurry mix was prepared as described in Bhattarai et al. (2017). The sediment was kept at 4°C in the dark in 500-ml serum bottles with a headspace of CH4 at 0.15 MPa until use.
The effect of the pressure on the CH4 oxidation, SR and CH4 production rate of the marine Lake Grevelingen sediment was assessed in 200 ml pressure vessels incubated in triplicates at 0.1 MPa, 0.45 MPa (mimicking the in situ conditions), 10, 20, and 40 MPa. Volatile suspended solids (VSS) were measured in the collected sediment. The sediment was homogenized and 0.07 (± 0.01) gVSS was distributed in each vessel in order to quantify the biomass added to each vessel and being able to compare it to previous studies. The vessels were flushed and pressurized with 100 % CH4, from which about 20% was 13C-labeled CH4 (13CH4). The marine Lake Grevelingen sediment used as inoculum was incubated with anaerobic artificial saline mineral medium with sulfate (10 mM) at 15°C for 77 days. Two different controls were prepared in triplicates at 0.45 MPa: (1) without biomass with CH4 in the headspace and (2) with biomass and nitrogen in the headspace.
Slurry samples were taken every week for chemical analysis. Approximately 1 mL sample was taken by attaching a connector and a vacuum tube to the exit port while gently opening the tap. Weight and pressure were measured in the vacuum tube before and after sampling. Pressure in each vessel was restored by adding fresh basal medium using a high performance liquid chromatography (HPLC) pump (SSI, USA). For molecular analysis, slurry samples (approx. 5 mL) were withdrawn at the start (immediately after the distribution of sediment in each vessel) and at the end (77 days) of the incubation period. 200 μL of slurry from each vessel was used for FISH analysis and the rest for DNA extraction.
The gas composition was measured on a gas chromatograph-mass spectrometer (GC-MS Agilent 7890A-5975C). The GC-MS system was composed of a Trace GC equipped with a GC-GasPro column (30 m by 0.32 mm; J & W Scientific, Folsom, CA) and an Ion-Trap MS. Helium was the carrier gas at a flow rate of 1.7 ml min−1. The column temperature was 30°C. The fractions of CH4 and CO2 in the headspace were derived from the peak areas in the gas chromatograph, while the fractions of 13CH4, 12CH4, 12CO2, and 13CO2 were derived from the mass spectrum as described by Shigematsu et al. (2004).
Total dissolved sulfide was measured by using the methylene blue method (Hach Lange method 8131) and a DR5000 Spectrophotometer (Hach Lange GMBH, Düsseldorf, Germany). Samples for sulfate and thiosulfate analysis were first diluted in a solution of zinc acetate (5 g/L) and centrifuged at 13,200 g for 3 min to remove insoluble zinc sulfide, and filtrated through 0.45 μm membrane filters. Sulfate and thiosulfate concentrations were then determined by ion chromatography (Metrohm 732 IC Detector) with a METROSEP A SUPP 5–250 column. The pH was checked by means of pH paper.
Polysulfides were methylated using the protocol by Kamyshny et al. (2006) and analyzed by reversed-phase HPLC. Elemental sulfur from the slurry sample was extracted using methanol following the method described by Kamyshny et al. (2009), but modified for small volumes. Dimethylpolysulfanes and extracted elemental sulfur were analyzed by an HPLC (HPLC 1200 Series, Agilent Technologies, USA) with diode array and multiple wavelength detector. A mixture of 90% MeOH and 10% water was used as eluent. A reversed phase C-18 column (Hypersil ODS, 125 × 4.0 mm, 5 μm, Agilent Technologies, USA) was used for separation. Concentrations of dimethylpolysulfanes from Me2S3 to Me2S7 were calculated from calibration curves of polysulfides standards prepared following the protocol of Milucka et al. (2012). UV detector response to Me2S8 was calculated by the algorithm discussed in Kamyshny et al. (2004).
The VSS was estimated at the beginning of the experiment on the basis of the difference between the dry weight total suspended solids and the ash weight of the sediment according to the procedure outlined in Standard Method (APHA, 2012).
Both AOM and SR rates were expressed as μmol of sulfide or dissolved inorganic carbon (DIC) production per gram of VSS per day (μmol gvss−1 d−1). For the AOM rate calculation, the total production of 13C-carbonate species (13C-DIC), i.e., 13CO2 in both liquid and gas phases, H13CO and 13CO in liquid phase, were first calculated. Considering that only 20% of CH4 was 13CH4, the total 13C-DIC was divided by the fractional abundance of 13C in the CH4 measured and used for each batch to determine the total amount of DIC produced from CH4 oxidation (Zhang et al., 2014). For methanogenesis and for the formation of carbonate species from other carbon sources than CH4, 12CH4, and H12CO were taken respectively, and divided by the 12C fractional abundance. The rate was calculated over the linear decrease or increase of the different compounds (12CH4, 13CH4, H12CO, H13CO, total dissolved sulfide and sulfate), over at least four consecutive points (Meulepas et al., 2010a). The rates were divided by the biomass content in the vessels (0.07 ± 0.01 gVSS).
DNA was extracted by using a FastDNA® SPIN Kit for soil (MP Biomedicals, Solon, OH, USA) by following the manufacturer's protocol. Approximately 0.5 g of the sediment was used for DNA extraction from the initial inoculum and from the enriched slurry (77 days). The extracted DNA was quantified and quality was checked as described by Bhattarai et al. (2017).
The DNA was amplified using bar coded archaea specific primer pair Arc516F and reverse Arc855R. The PCR reaction mixture was prepared as described by Bhattarai et al. (2017), however, the PCR amplification was modified using a touch-down temperature program. PCR conditions consisted of a pre-denaturation step of 5 min at 95°C, followed by 10 touch-down cycles of 95°C for 30 s, annealing at 68°C for 30 s with a decrement per cycle to reach the optimized annealing temperature of 63°C and extension at 72°C. This was followed by 25 cycles of denaturation at 95°C for 30 s and 30 s of annealing and extension at 72°C. The final elongation step was extended for 10 min.
The primer pairs used for bacteria were forward bac520F 5′-3′ AYT GGG YDT AAA GNG and reverse Bac802R 5′-3′ TAC NNG GGT ATC TAA TCC (Song et al., 2013). The following program was used: initial denaturation step at 94°C for 5 min, followed by denaturation at 94°C for 40 s, annealing at 42°C for 55 s and elongation at 72°C for 40 s (30 cycles). The final elongation step was extended to 10 min. Five micro liter of the amplicons were visualized by standard agarose gel electrophoresis (1% agarose gel, a running voltage of 120 V for 30 min, stained by gel red) and documented using a UV transilluminator with Gel Doc XR System (Bio-Rad, USA).
After checking the correct band size, 150 μl of PCR amplicons were loaded in 1% agarose gel and electrophoresis was performed for 120 min at 120 V. The gel bands were excited under UV light and the PCR amplicons were cleaned using E.Z.N.A.® Gel Extraction Kit by following the manufacturer's protocol (Omega Biotek, USA). The purified DNA amplicons were sequenced by an Illumina HiSeq 2000 (Illumina, San Diego, USA) and analyzed according to the procedure described in Bhattarai et al. (2017).
A total of 34,000 (± 5,000) and 36,000 (± 5,000) sequence reads were assigned to archaea and bacteria, respectively, examining the tags assigned to the amplicons. More details on the analytical procedure have been described by Bhattarai et al. (2017). After eliminating the chimeras, 30,000 and 32,000 sequences for archaea and bacteria, respectively, were analyzed and classified in MOTHUR (Schloss and Westcott, 2011). In short, the faulty sequences with mismatch tags or primers and with a size <200 bp were removed by using the shhh.flows command. Then, the putative chimeric sequences were identified and removed by the chimera.uchime command using the most abundant reads in the respective sequence data sets as references. Assembled reads that passed the chimera checking were clustered into de novo operational taxonomic units (OTUs) at a cut off of 97% sequence similarity. Finally, the sequence reads were classified according to the SILVA taxonomy database version 121 (Pruesse et al., 2007) using the classify.seqs command, and the relative sequence abundance of each phylotype was estimated.
These sequence data have been submitted to the NCBI GenBank database under BioProject accession number PRJNA415004 (direct link: http://www.ncbi.nlm.nih.gov/bioproject/415004).
Archaeal and bacterial plasmids were used to prepare Q-PCR standard. Plasmids were isolated using the Plasmid Kit (Omega Biotek, USA). The plasmid was digested with the EcoR I enzyme. After digestion purification was done by gel extraction (Gel extraction Kit, Omega Biotek, USA). The copy number was calculated from the total mass and the nucleic acid concentration. Extracted DNA from the sediment at the start and at the end of the incubation period (77 days) was used for qPCR analysis to quantify archaea and bacteria. Amplifications were done in triplicates in a 7500 Real-Time PCR System (Applied Biosystem). Each reaction (20 μl) contained 1 × Power SYBR-Green PCR MasterMix (Applied Biosystems), 0.4 μM of each primer, and 5 ng template DNA. The 16S rRNA genes of bacterial origin were amplified using the primers Bac331f (5′-TCCTACGGGAGGCAGCAGT3′) and Bac797r (5′-GGACTACCAGGGTCTAATCCTGTT-3′) (Nadkarni et al., 2002). Cycling conditions were 95°C for 10 min; and 40 cycles at 95°C for 30 s and 60°C for 30 s and 72°C for 30 s. Archaea were quantified using the primer set Arch349f (5′-GYGCASCAGKCGMGAAW-3′) and Arch806r (5′-GGACTACVSGGGTATCTAAT-3′) (Takai and Horikoshi, 2000). Cycling conditions were 95°C for 10 min; and 40 cycles at 95°C for 30 s and 50°C for 30 s and 72°C for 30 s. Triplicate standard curves were obtained with 10-fold serial dilutions ranged between 107 and 10−2 copies per μl of plasmids. The efficiency of the reactions was up to 100% and the R2 of the standard curves were up to 0.999.
At the start and at the end of the incubation period (77 days), samples were withdrawn from each vessel for cell counting and FISH analysis. Hundred micro liter of sample was filtered on 0.2 μm membrane filters, 200–300 μL of 20 × SYBR green solution (fluorescent dye, Takara, Japan) was added on top of the filter and incubated in the dark at room temperature for 30 min. Cells on membrane filters were then counted by observation under a fluorescence microscope.
For FISH analysis, 200 μL of sample from each vessel was fixed in a final 2% paraformaldehyde solution for 4 h on ice. The samples were washed twice with 1 × phosphate buffer saline solution (PBS). Then, it was stored in a mixture of PBS and ethanol (EtOH), with a PBS/EtOH ratio of 1:1 at −20°C as previously described by Boetius et al. (2000).
Nuclease free water was used to dilute 100 μL of stored sample, which was then sonicated for 40 s and then filtered on 0.2 μm membrane filters. The filters were dried and mounted on a glass slide with 100 μL glycerol 10%. For FISH analysis, the filtrated sample was hybridized with the archaeal probe ARCH915 (Stahl, 1991) and the bacterial probe EUB I-III (Daims et al., 1999). Different CY3-labeled ANME probes were used: ANME-1 350 (Boetius et al., 2000), ANME-2 538 (Treude et al., 2005), ANME-3 1249 (Niemann et al., 2006). The following Cy5-labeled SRB specific probes were used: Desulfosarcina/Desulfococcus (DSS) DSS658 (Boetius et al., 2000) and Desulfobulbus (DBB) DBB660 (Daly et al., 2000). Negative control counts were performed with a NonEUB (nonsense EUB338, negative control) probe (Wallner et al., 1993). The hybridization of the samples and microscopic visualization of the hybridized cells were performed as described previously (Snaidr et al., 1997).
All the cells were counterstained with 4', 6-diamidino-2-phenylindole (DAPI) (Wagner et al., 1993) and visualized using an epifluorescence microscope (Carl Zeiss, Germany). For cell counting, single hybridization was performed. 700–1,000 DAPI-stained cells (30–50 microscopy grids) and their corresponding probe fluorescent signals for each probe per filter section were counted as described previously in the literature (Musat et al., 2008; Siegert et al., 2011; Kleindienst et al., 2012). Double hybridization was performed to visualize the preferential archeal-bacterial aggregates. The detection limit was 0.003 of EUBmix-positive cells over DAPI stained cells as determined with the Cy5- and Cy3-labeled NonEUB probe.
The results are given as the average ± standard deviation of triplicates. The statistical data analysis was performed using SPSS 25.0 (IBM, USA). Significant differences were determined by one-way ANOVA and post-hoc analysis for multiple group comparison (Tukey HSD). Differences were considered to be significant at p ≤ 0.05.The homogeneity of the variance of the parameters was evaluated using a Levene test.
The highest sulfide production rates of the coastal marine Lake Grevelingen sediment was in the incubations at the in situ pressure (0.45 MPa) and 10 MPa: 270 (± 27) and 258 (± 25) μmol gvss−1 d−1, respectively (Table 1). The sulfide production rate at 20 MPa was 132 (± 18), while at 40 MPa was 109 (± 13) μmol gvss−1 d−1, comparable to the rate in the control without CH4 but N2 in the headspace, 99 (± 5) μmol gvss−1 d−1 (Table 1). Similarly, high SR rates were recorded for the incubations at 0.45 MPa and 10 MPa (Table 1): 297 (± 43) and 278 (± 12) μmol gvss−1 d−1, respectively. In contrast, the SR rate at 0.1 MPa was 257 (± 23) μmol gvss−1 d−1, while the sulfide production was only 127 (± 27) μmol gvss−1 d−1 (Figures 1A,B). The lowest SR for the incubations were obtained at high pressures (20 and 40 MPa) 162 (± 24) and 154 (± 16) μmol gVSS−1 d−, respectively.
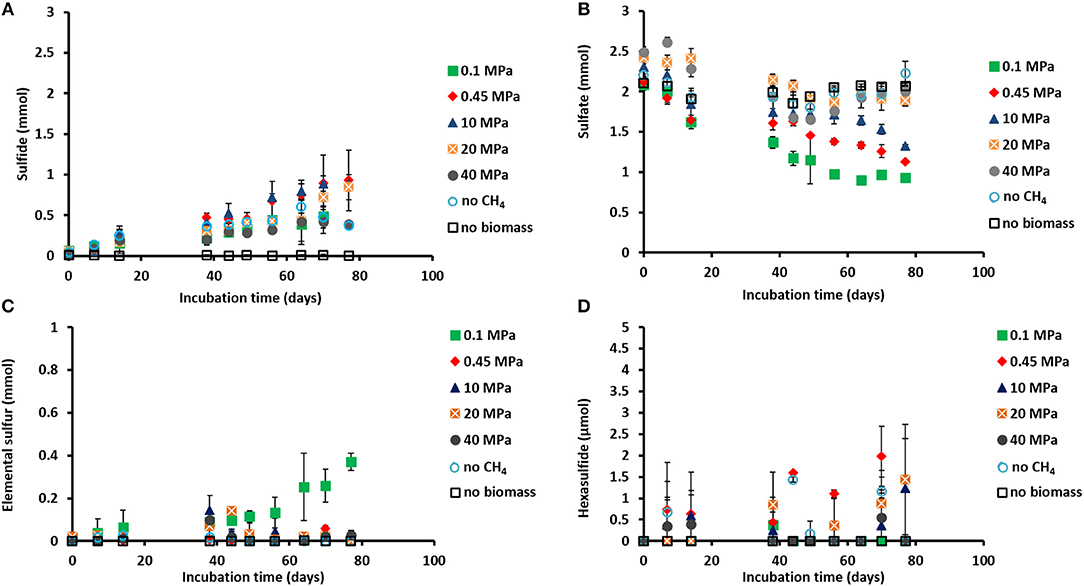
Figure 1. Concentration profiles of (A) total dissolved sulfide, (B) sulfate, (C) elemental sulfur, and (D) hexasulfide (S) during the incubations of Grevelingen sediment at different pressures, controls without CH4 but with N2 in the headspace and controls without biomass. Error bars indicate the standard deviation (n = 3).
Sulfide was produced in almost all the incubations (Figure 1A), with the exception of the control without biomass (Figure 1A). The sulfate concentration profiles varied with initial incubation pressure: at 0.45 MPa sulfate was reduced to sulfide in a 1:1 ratio, closing the sulfur balance (Figure 1B; Table 1), whereas at 40 MPa sulfate was reduced in a 1:1 ratio only during the first 40 days of incubation (Figures 1A,B). In the incubation at 0.1 MPa, 0.37 mmol of elemental sulfur was produced along with 0.54 mmol of sulfide (Figures 1A,C). In the other incubations at different pressures, hardly any elemental sulfur was formed (Figure 1C). Instead, long chain polysulfides (S) were formed along the incubation depending on the sulfide concentration and pH (Figure 1D and Supplementary Figure S1), but in small amounts (≤2 μmol per vessel): 2 μmol of hexasulfide (S) per vessel was determined at 0.45 MPa CH4 pressure (Figure 1D) and 1.2 and 1.4 μmol S per vessel at 10 MPa and 20 MPa, respectively (Figure 1D).
The AOM rates were calculated from the DIC produced from 13CH4. The Km for CH4 of the marine Lake Grevelingen sediment was determined to be around 1.7 mM. The DIC production rates followed a similar trend as the sulfide production rates: the highest rate was found at 0.45 MPa and the lowest rate at 40 MPa: 324 ± 35 and 38 ± 2 μmol gvss−1 d−1, respectively (Table 1). In the incubation at 0.45 MPa (in situ pressure), the total DIC produced from CH4 was similar to the sulfide produced: ~0.9 mmol per vessel (Supplementary Figure S2A and Figure 1A). However, sulfide was produced from the start for all the incubations, while the total DIC from CH4 was mainly produced only after 40 days of incubation (Figure 1A and Supplementary Figure S2A).
CH4 was produced in all the incubations, with the exception of the batches without biomass (Table 1; Supplementary Figure S2B). The highest amount of CH4 formed was recorded in the vessel at 0.1 MPa (Supplementary Figure S2B). The highest methanogenic rate was determined in the control vessel without CH4 (N2 in the headspace) and in the incubation at 0.1 MPa: 44 and 31 μmol gvss−1 d−1, respectively, while it was below 5 μmol gvss−1 d−1 in all the other batch incubations (Table 1). Assuming that all the total 12C-DIC was produced from the oxidation of other carbon sources than CH4, its production rate was low in almost all the incubations: lower than 3 μmol gvss−1 d−1, except for the incubation without CH4 (64 μmol gvss−1 d−1, data not shown).
The total cell counts (Figure 2A) performed using SYBR green showed that the only two statistically significant increases in number of cells were found in the incubation at the in situ pressure of 0.45 MPa, from 6 to 8 × 107 cells ml−1, and at 10 MPa, from 6.7 to 7.9 × 107 cells ml−1 (determined by one-way ANOVA, p = 0.02). There was no statistically significant difference in the other incubations and controls.
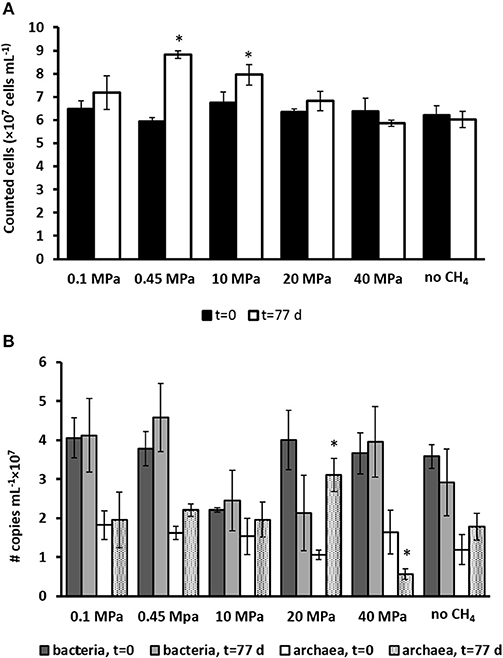
Figure 2. (A) Total number of active cells and (B) number of copies of archaea and bacteria from Q-PCR analysis per ml of wet sediment in each pressurized vessel at the start (t = 0 days) and at the end of the incubation (t = 77 days) for all the incubations and control without CH4 but with N2 in the headspace. Error bars indicate the standard deviation (n = 3). Significant results (determined by one-way ANOVA, p < 0.05) were highlighted with a star (*).
The number of copies for bacteria and archaea were assessed by Q-PCR data performed on samples at the start (t = 0 days) and after 77 days of incubation (Figure 2B). Based on Q-PCR results, the only two statistically significant differences in archaeal copy numbers were obtained for the incubation at 20 and 40 MPa (determined by one-way ANOVA, at p = 0.01). There was no statistically significant difference in all other incubations and controls. The bacterial copy numbers were statistically similar in all the incubations and controls (Figure 2B).
Based on the 16S rRNA gene analysis, the relative sequence abundance of both archaeal and bacterial communities differs along the 77 days incubation at different pressures (Figure 3 and Supplementary Figures S3, S4). The most abundant OTUs (operational taxonomic unit) with archaeal signature (relative abundance ≥0.001 on average) are shown in the Supplementary Figure S3. Specifically, the relative sequence abundance of ANME-3 among all the archaea (Figure 3A) is significantly higher after 77 days in incubations at 0.45 MPa than at the start of the incubation (one-way ANOVA: ANME-3, p = 0.00). On the other hand, ANME-2 sequence reads are significantly the highest at 20 MPa (Figure 3A, one-way ANOVA: ANME-2, p = 0.00). Sequences of methanogens, specifically belonging to the Methanomicrobiales, were significantly more “abundant” after the incubation at 0.45 MPa, rather than at higher partial pressures (Figure 3A, one-way ANOVA: Methanomicrobiales, p = 0.02).
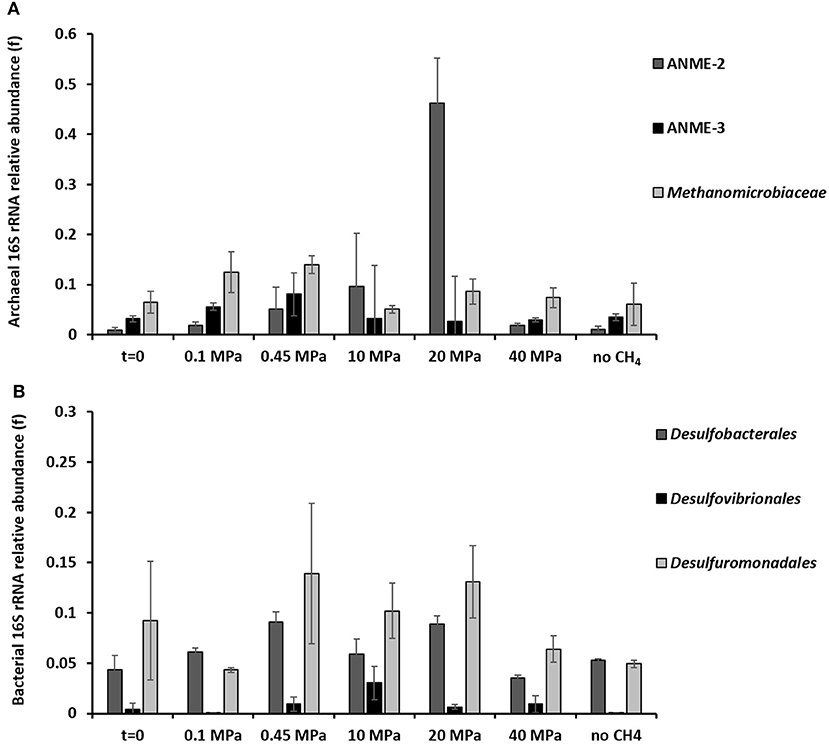
Figure 3. 16S rRNA (A) archaeal and (B) bacterial sequences showing the phylogenetic affiliation up to order level of the microorganisms putatively mediating the AOM-SR process as derived by high throughput sequencing. The graphs are showing the archaeal and bacterial sequences at the beginning (t = 0) and at the end (t = 77 days) of the incubations of the marine Lake Grevelingen sediment at 0.1, 0.45, 10, 20, 40 MPa, and control without CH4 but N2 in the headspace. Error bars indicate the standard deviation (n ≥ 3).
The bacterial communities were very diverse in all the incubations, the ones with the highest percentage of sequence reads (relative abundance ≥0.001 on average) are shown in the Supplementary Figure S4. The relative sequence abundance for most of the bacterial species did not differ significantly between incubations. However, the relative sequence abundance of the Desulfobacterales was significantly higher at 0.45 and 20 MPa than at the start of the incubations (Figure 3B, one-way ANOVA: Desulfobacterales, p = 0.00), while the Desulfovibrionales sequence reads were only significantly higher at 10 MPa (Figure 3B, one-way ANOVA: Desulfovibrionales at 10 MPa, p = 0.00).
The relative sequence abundance of sulfur reducing bacteria, e.g., Desulfovibrionales or Desulforomonodales, is lower in the incubation at 0.1 MPa than at CH4 partial pressures higher than 0.45 MPa (Figure 3B), whereas the relative sequence abundance of sulfate reducing Desulfobacterales is high also at lower CH4 partial pressure (0.1 and 0.45 MPa, Figure 3B), where the highest AOM-SR rates were found (Table 1).
ANME-3, ANME-2, DSS, and DBB were visualized in all the batch incubations (Figures 4, 5), while ANME-1 were hardly visualized in any of the incubations and controls, always below the detection limit (< 0.003 of ANME-1 cells over DAPI stained cells). At the start of the incubation (t = 0 days), the percentage of visualized ANME-3, ANME-2 and DSS cells was not significantly different among them (Figure 4, one-way ANOVA: t = 0, p = 0.97). Moreover, the controls without CH4 showed that only the number of DBB cells significantly increased from the start of the incubation (Figure 4, one-way ANOVA: DBB, p = 0.00). However, FISH analysis after 77 days of incubation showed variations in the ANME and SRB abundance depending on the incubation pressure (Figures 4, 5).
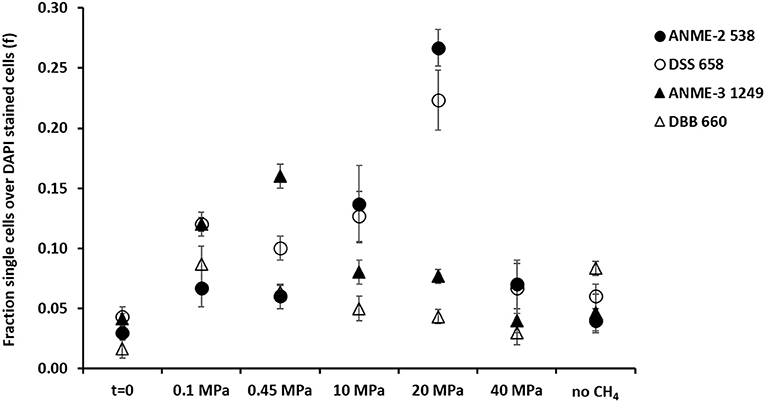
Figure 4. Fraction of FISH cell counts over DAPI stained cells at the beginning (t = 0) and at the end (t = 77 days) of the incubations of the marine Lake Grevelingen sediment at 0.1, 0.1, 10, 20, 40 MPa, and control without CH4 but N2 in the headspace. Error bars indicate the standard deviation (n = 3). FISH probes used: ANME-2 538 (Treude et al., 2005), ANME-3 1249 (Niemann et al., 2006), DSS658 (Boetius et al., 2000) and DBB660 (Daly et al., 2000).
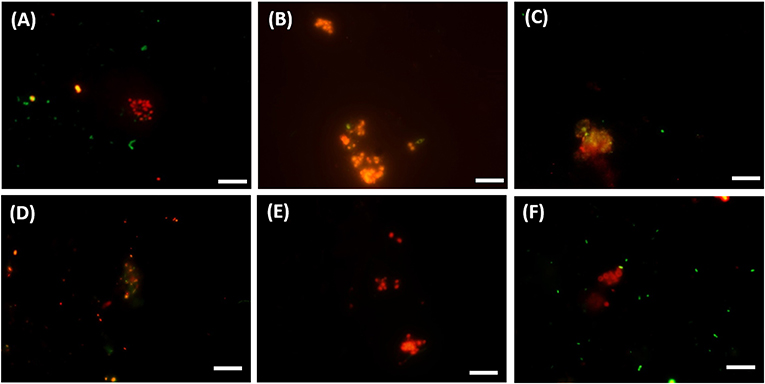
Figure 5. FISH images (A–C) from CY3-labeled ANME-3 in red color, CY5-labeled Desulfobulbus (DBB) in green after 77 days of incubation at (A) 0.1 MPa, (B) 0.45 MPa, and (C) 20 MPa total CH4 pressure. FISH images (D–F) from CY3-labeled ANME-2 in red color, CY5-labeled Desulforsarcina/Desulfococcus group (DSS) in green after 77 days of incubation at (D) 0.45 MPa, (E) 20 MPa, and (F) 40 MPa total CH4 pressure. White scale bar representing 10 μm.
At 0.1 and 0.45 MPa, ANME-3 cells were significantly more abundant than in the other incubations (one-way ANOVA: at 0.1 and 0.45 MPa, p = 0.00): ~10 % more cells were stained with the ANME-3 probe (Figure 4). The DBB cells were not as high in number as the ANME-3 cells in any of the incubations (Figure 4), but they also significantly increased in the incubations at 0.1 and 0.45 MPa compared to the start of the incubation (Figure 4, one-way ANOVA: at 0.1 and 0.45 MPa, p = 0.00). In the incubations at 10 and 20 MPa, ANME-3 cells were significantly less abundant than in the incubation at 0.45 MPa (One-way ANOVA: at 10 and 20 MPa, p = 0.00) and DBB cells were even more rarely pictured (Figure 5). At 40 MPa, ANME-3 and DBB were the least abundant and not significantly different than at the start of the incubation (Figure 4, one-way ANOVA: ANME-3, p = 1 and DBB, p = 0.41).
The DBB cells were not always found concomitant to the ANME-3 cells at 0.1 and 0.45 MPa, (Figures 5A,B) and, even if present, the ANME-3 cells outnumbered the DBB cells (Figures 5A,B). Differently, at 20 MPa, ANME-3 and DBB cells were rare (Figure 4), but the stained cells formed tight ANME-3/DBB aggregates (Figure 5C).
Differently than ANME-3 and DBB, ANME-2, and DSS cells were significantly higher after 77 days incubations at 10 and 20 MPa than at the start of the incubations (Figure 4, One-way ANOVA: ANME-2 at 10 and 20 MPa, p = 0.00 and DSS at 20 MPa, p = 0.00). At lower pressures (0.1 and 0.45 MPa), ANME-2 cells were significantly lower in number than ANME-3 cells (Figure 4, one-way ANOVA: at 0.1 and 0.45 MPa, p = 0.00).
At 0.45 MPa incubation pressure, the DSS cells were mainly visualized together with ANME-2 (Figure 5D), while at 20 MPa clusters of ANME-2 cells were visualized without DSS (Figure 5E). At 40 MPa there was no significant difference between the number of cells of the different archaeal and bacterial groups:number of ANME-2 cells at 40 MPa not significantly different from DSS (p = 0.99), ANME-3 (p = 0.18), and DBB (p = 0.062) cells (Figure 4).
This study showed that AOM and SR processes in Lake Grevelingen sediment depend on the CH4 partial pressure. According to Equation 1, the reaction rate is expected to be stimulated by the elevated CH4 partial pressure when the other parameters remain the same (Table 1). This expectation has been commonly accepted and has been shown in communities dominated by ANME-1, i.e., hydrocarbon seep in the Monterey canyon sediment (Girguis et al., 2005) and ANME-2, i.e., Eckernförde Bay (Meulepas et al., 2010a; Timmers et al., 2015b) and Gulf of Cadiz sediment (Bhattarai et al., 2018). In contrast, the AOM-SR process by the ANME-3 dominated marine Lake Grevelingen sediment has an optimal pressure at 0.45 MPa among all tested conditions (Table 1). This is in accordance with their natural habitat (Bhattarai et al., 2017), i.e., the in situ hydrostatic pressure of marine Lake Grevelingen is ~0.45 MPa (in-situ CH4 concentration of ~6 mM). However, this is in contrast with the theoretical thermodynamic calculation (Table 1), that predicts a higher CH4 solubility and thus a higher activity based on reported Km values, i.e., 37 mM as calculated from an ANME-2 predominant enrichment originated from the Gulf of Cadiz (Zhang et al., 2010). The calculated Km value for CH4, based on our ANME-3 dominated inoculum, is much lower than previously reported: around 1.7 mM. Thus, the ANME cells from Grevelingen marine sediment have a higher affinity for CH4 than the ANME-2 from the Gulf of Cadiz, explaining why a higher pressure, i.e., higher dissolved CH4 concentration, did not result in a higher AOM rate (Table 1). As a matter of fact, higher pressure resulted in a lower number of ANME-3 cells and ANME-3 proliferated only at lower pressures incubation (0.1 and 0.45 MPa). This might indicate ANME-3 cells of marine Lake Grevelingen are non-piezophilic. Marine Lake Grevelingen is a shallow sediment (45 m depth), but with high abundance of ANME-3, closely affiliated to the ANME-3 from the Haakon Mosby Mud Volcano (Bhattarai et al., 2017). This sediment is characterized by high CH4 partial pressures (~10 MPa, water depth > 1,000 m) and relatively low temperatures (Niemann et al., 2006; Lösekann et al., 2007; Vigneron et al., 2013). A recent study shows that gas eruptions and mud mixing are influencing the abundance of different ANME phylotypes in Haakon Mosby Mud Volcano: ANME-3 proliferate also in disturbed sediment differently than ANME-2a (Ruff et al., 2018). Marine Lake Grevelingen sediment is a relative disturbed sediment due to its high sedimentation rates and high methane fluxes (Egger et al., 2016), which could explain the higher abundance of ANME-3 than ANME-2 cells in situ (Bhattarai et al., 2017) and in incubations at CH4 partial pressure similar to in situ pressures (Figure 4). The number of ANME-2 cells increased only in ex-situ incubation (not disturbed) at high CH4 partial pressures (Figure 4).
This study showed that CH4 and sulfate were an effective energy source supporting AOM-SR in the microbial ecosystem from the marine Lake Grevelingen sediment (Figures 2, 4, and Supplementary Table S1). In vitro biomass growth was observed, especially at 0.45 MPa which is the in situ pressure (Figure 2A), with CH4 and sulfate supplied as the sole energy sources. At 0.45 MPa the AOM and SR rates reached 0.3 mmol gvss−1 d−1 (Table 1). These rates are comparable or even higher than the in vitro AOM rates of ANME-1 or ANME-2 dominated biomass (Supplementary Table S2). The rate obtained after the enrichment of Eckernförde Bay sediment dominated by ANME-2 type cells for more than 800 days in a continuous membrane bioreactor was 0.2 mmol gvss−1 d−1 (Meulepas et al., 2009a). Moreover, the AOM-SR rate measured in this study at 0.45 MPa is even higher than the AOM rate coupled to denitrification (1.8 to 3.6 nmol day-1 (ml sediment)−1), which is thermodynamically much more favorable (ΔG0′ = −924 kJ mol−1 CH4) (Deutzmann and Schink, 2011) than AOM-SR (ΔG0′ = −16.6 kJ mol−1 CH4, Equation 1).
The ANME-3 type is usually visualized in association with DBB as sulfate reducing partner (Niemann et al., 2006; Lösekann et al., 2007). In a recent study describing the microbial ecology of Lake Grevelingen sediment (incubation pressure = 0.1 MPa), the two species (ANME-3 and DBB) could not be visualized together and the DBB cells were much less abundant than ANME-3 (Bhattarai et al., 2017), similarly to this study (Figures 4, 5A). At 0.1 and 0.45 MPa, ANME-3 cells were visualized in aggregates mainly detached from DBB cells (Figures 5A,B). ANME-3 cells, similar to ANME-1 and ANME-2 (Orphan et al., 2002; Ruff et al., 2016), have been visualized without bacterial partner before (Omoregie et al., 2008; Vigneron et al., 2013), suggesting that this ANME type is supporting a metabolism independent of an obligatory bacterial association.
In contrast, as ANME-3 and DBB decreased in number at higher pressures, most of the ANME-3 and DBB visualized at 20 MPa were forming small ANME-3/DBB clusters (Figure 5C), suggesting that they possibly have mutual benefits at this pressure. High pressure can impact the structure of several cellular components and functions, such as membrane fluidity, protein activity, and structure (Jebbar et al., 2015). Only 25% of the isolated bacterial and archaeal strains from deep-sea environments with elevated hydrostatic pressure were found to be true piezophiles (Jebbar et al., 2015). However, many microorganisms can survive with the minimum energy requirement in high pressure conditions (Zhang et al., 2015). In view of that, ANME-3 and DBB, less proliferated at higher pressure (Figure 4), form these visualized tight aggregates. These aggregates were formed to have less impact on their cellular structure or because at higher pressure a direct electron transfer or shared intermediate between the microorganisms is necessary to facilitate the respiration in such harsh conditions. However, both ANME-3 and DBB cannot proliferate at very high pressure, as shown in the incubation at 40 MPa (Figure 4). It should be noted that even after 2 months incubation at 0.45 MPa (the most favorable pressure), the percentage of counted ANME was relatively low, i.e., 0.16 of ANME-3 cells over DAPI stained cells (Figure 4), showing that a relative small percentage of cells is responsible for a relative high AOM rate (Table 1).
A cooperative interaction between ANME-2 and DSS was suggested by their visualization in aggregates (Figure 5D), especially at lower pressures (0.1 and 0.45 MPa). The cooperative interaction between the ANME-2 and DSS is still under debate: Milucka et al. (2012) stated that a syntrophic partner might not be required for ANME-2 and that they can be decoupled by using external electron acceptors (Scheller et al., 2016), whereas recent studies have shown direct electron transfer between the two (ANME-2 and DSS) partners (McGlynn et al., 2015; Wegener et al., 2016). However, differently than ANME-3, ANME-2 prefers a higher CH4 partial pressure as shown by the higher abundance of these cells at 10 and 20 MPa (Figure 4). This result was similar to that found in previous studies (Timmers et al., 2015a; Bhattarai et al., 2018). This suggests that ANME-2 prefers high pressure over scarce CH4 availability and they are more piezo-tolerant than the ANME-3 in the studied marine Lake Grevelingen sediment. The number of counted ANME-2 cells at 10 and 20 MPa was quite high (0.10–0.28 of ANME-2 cells over the total counted DAPI cells, Figure 4), but the AOM rate was relatively low compared to the incubations at lower CH4 partial pressure (Table 1), suggesting that the ANME-2 were less active than the ANME-3. However, further studies need to be performed to confirm this hypothesis.
The CH4 partial pressure steers the sulfur cycling in the marine Lake Grevelingen sediment community (Figure 1). Only at 0.45 and 10 MPa, almost all sulfate was reduced to total dissolved sulfide. Differently, at 0.1 MPa CH4 pressure, the reduced sulfate was converted to both sulfide and zero-valent sulfur (Figure 1C). Elemental sulfur has been considered as intermediate in the AOM-SR process, which is consumed by ANME to generate energy (Milucka et al., 2012). Milucka et al. (2012) showed that ANME-2 cells could stand alone without the metabolic support of the bacterial partner, assuming that CH4 was oxidized to bicarbonate and sulfate was reduced to disulfide (S) through zero-valent sulfur as an intracellular intermediate. However, this mechanism cannot be supported by our results since S was hardly detected in any incubations (Supplementary Figure S1A).
In our study, elemental sulfur could have been formed either by sulfide oxidation due to the presence of ferric iron in the marine Grevelingen sediment (Bhattarai et al., 2017), but this does not fully explain why elemental sulfur is formed only at lower pressure (0.1 MPa). The highest abundance of Desulfobulbus spp. (DBB) was found at 0.1 MPa and in the control without CH4 (Figure 4). This genus is capable of partial sulfate reduction to elemental sulfur and its disproportionation to sulfide and sulfate (Finster, 2008), which could explain the sulfate, elemental sulfur and total dissolved sulfide concentrations (Figures 1A–C). Moreover, the relative sequence abundance of sulfur reducers (e.g., Desulforomonadales and Desulfovibrionales) was lower in the incubations at 0.1 MPa than at higher pressures, showing that the potentially produced elemental sulfur might have been reduced in the incubations at pressures higher than 0.1 MPa (Figure 3B).
Furthermore, the amount of disulfide or other polysulfides formed during the incubations (Figure 1D and Supplementary Figure S1) was very low, in most cases below the detection limit (0.1 μmol). Thus, at 20 and 40 MPa the sulfur mass balance was not completely closed (Table 1). However, the highest polysulfide concentrations were measured at pressures between 0.45 and 20 MPa, when the total dissolved sulfide concentration and the pH was higher (7.5–8) than at the start of the incubations (6.5–7.0). The long chain polysulfides, i.e., S, are more abundant at pH above 7.5 (Kamyshny et al., 2004), which is in line with what was found in this study (Figure 1A and Supplementary Figure S1E). The formation of these intermediate sulfur compounds in the ANME process needs nevertheless to be further elucidated using e.g., isotopic labeled sulfate (35S) and nanometre scale secondary ion mass spectrometry (NanoSIMS) analysis.
Methanogenic activity in marine Lake Grevelingen sediment was previously described by Egger et al. (2016) and confirmed in this study at low pressure (0.1 MPa) or when no CH4 was added in the incubation (Table 1 and Supplementary Figure S2B). Trace CH4 oxidation occurs during methanogenesis (Meulepas et al., 2010a; Timmers et al., 2015a), thus, the determined AOM at 0.1 MPa cannot account only for the AOM-dependent SR. In the incubations where methanogenesis occurred, AOM is partly due to the methanogenic activity and not due to SR.
At pressures higher than 0.1 MPa, AOM-SR was preferred over methanogenesis (Table 1 and Supplementary Figure S2B). Methanogenesis becomes less thermodynamically favorable at high pressures, less free energy (12 kJ mol−1 less) is released upon changing the incubation pressure from 0.1 to 10 MPa (Meulepas et al., 2010a). Timmers et al. (2015a) found that at 10 MPa net AOM-SR occurred, while at 0.1 MPa methanogenesis and trace CH4 oxidation dominated. In this study, the optimal AOM-SR was 0.45 MPa: the SR activity decreased at pressures higher than 10 MPa, while AOM activity already decreased at pressures higher than 0.45 MPa (Figures 1B,C).
To conclude, this is the first study showing that the active ANME from the shallow marine Lake Grevelingen sediment preferred lower (0.1 and 0.45 MPa) over elevated (10, 20, 40 MPa) pressures, in contrast to previous studies that show strong positive correlations between the AOM-SR rates and the CH4 partial pressure. Pressure steered the abundance and structure of the different types of ANME and SRB. The ANME-3 type was predominantly enriched in incubations at low CH4 partial pressures, whereas high pressures enhanced ANME-2 proliferation. Similarly, DSS was more predominantly enriched at higher (10, 20, 40 MPa) than at lower (0.1 and 0.45 MPa) CH4 partial pressure. This research highlights that ANME-3 from marine Lake Grevelingen can be enriched at rather low CH4 partial pressures, which is important to further understand their metabolism and physiology. Marine Lake Grevelingen sediment is easily accessible and the ANME-3 cells present possess high specific AOM-SR rates, thus, the use of this sediment as inoculum can be of great potential for the desulfurization of wastewater. However, the SR rate with CH4 as electron donor should be around 100 mmol gvss−1 d−1 to be competitive with the SR rates achieved with other electron donors, such as hydrogen or ethanol (Meulepas et al., 2009a; Suarez-Zuluaga et al., 2016), which is still much higher than what was obtained in this study. Thus, further enrichment and research is required for potential biotechnological applications of ANME.
CC: experimental work, writing manuscript, and preparing tables and figures. YZ: writing manuscript, figure, and text on 16S rRNA sequence analysis. PL: writing manuscript, project design, and supervision.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge Filip Meysman from the Royal Netherlands Institute of Sea Research (NIOZ, Yerseke, The Netherlands) for providing the Lake Grevelingen sediment. The authors are grateful for using the analytical facilities of the Center for Chemical Microscopy (ProVIS) at the Helmholtz-Center for Environmental Research which is supported by European regional Development Funds (EFRE and Europe funds Saxony) and the Helmholtz Association. The authors acknowledge Dr. Niculina Musat (team leader of ProVIS) for her advice and practical support during FISH analysis. This research was funded by the Erasmus Mundus Joint Doctorate Programme ETeCoS3 (Environmental Technologies for Contaminated Solids, Soils and Sediments) under the grant agreement FPA no. 2010-0009 and the National Natural Science Foundation of China (grant number: 41476123). The manuscript has been released as preprint at BioRxiv (Cassarini et al., 2018) and HAL (Cassarini, 2017).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2018.00162/full#supplementary-material
Bhattarai, S., Cassarini, C., Gonzalez-Gil, G., Egger, M., Slomp, C. P., Zhang, Y., et al. (2017). Anaerobic methane-oxidizing microbial community in a coastal marine sediment: anaerobic methanotrophy dominated by ANME-3. Microb. Ecol. 74, 608–622. doi: 10.1007/s00248-017-0978-y
Bhattarai, S., Zhang, Y., and Lens, P. N. L. (2018). Effect of pressure and temperature on anaerobic methanotrophic activities of a highly enriched ANME-2a community. Environ. Sci. Pollut. Res. 25, 30031–30043. doi: 10.1007/s11356-018-2573-2
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Cassarini, C. (2017). Anaerobic Oxidation of Methane Coupled to the Reduction of Different Sulfur Compounds in Bioreactors. HAL [Preprint]. Available online at: https://tel.archives-ouvertes.fr/tel-01743834/file/TH2017PESC1051.pdf (Accessed July 05, 2018).
Cassarini, C., Zhang, Y., and Lens, P. N. L. (2018). Pressure Sensitivity of ANME-3 Predominant Anaerobic Methane Oxidizing Community From Coastal Marine Lake Grevelingen Sediment. BioRxiv [Preprint]. Available online at: https://www.biorxiv.org/content/early/2018/04/25/307082 (Accessed July 05, 2018).
Daims, H., Brühl, A., Amann, R., Schleifer, K.-H., and Wagner, M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444. doi: 10.1016/S0723-2020(99)80053-8
Daly, K., Sharp, R. J., and McCarthy, A. J. (2000). Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiology 146, 1693–1705. doi: 10.1099/00221287-146-7-1693
Deusner, C., Meyer, V., and Ferdelman, T. G. (2009). High-pressure systems for gas-phase free continuous incubation of enriched marine microbial communities performing anaerobic oxidation of methane. Biotechnol. Bioeng. 105, 524–533. doi: 10.1002/bit.22553
Deutzmann, J. S., and Schink, B. (2011). Anaerobic oxidation of methane in sediments of Lake Constance, an oligotrophic freshwater lake. Appl. Environ. Microbiol. 77, 4429–4436. doi: 10.1128/AEM.00340-11
Duan, Z., and Mao, S. (2006). A thermodynamic model for calculating methane solubility, density and gas phase composition of methane-bearing aqueous fluids from 273 to 523K and from 1 to 2000bar. Geochim. Cosmochim. Ac. 70, 3369–3386. doi: 10.1016/j.gca.2006.03.018
Egger, M., Lenstra, W., Jong, D., Meysman, F. J. R., Sapart, C. J., van der Veen, C., et al. (2016). Rapid sediment accumulation results in high methane effluxes from coastal sediments. PloS ONE 11, e0161609. doi: 10.1371/journal.pone.0161609
Finster, K. (2008). Microbiological disproportionation of inorganic sulfur compounds. J. Sulfur Chem. 29, 281–292. doi: 10.1080/17415990802105770
Girguis, P. R., Cozen, A. E., and De Long, E. F. (2005). Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl. Environ. Microbiol. 71, 3725–3733. doi: 10.1128/AEM.71.7.3725-3733.2005
Gonzalez-Gil, G., Meulepas, R. J. W., and Lens, P. N. L. (2011). “Biotechnological aspects of the use of methane as electron donor for sulfate teduction,” in Comprehensive Biotechnology, ed. M. Y. Murray (Amsterdam: Elsevier B.V.), 419–434.
Hagens, M., Slomp, C. P., Meysman, F. J. R., Seitaj, D., Harlay, J., Borges, A. V., et al. (2015). Biogeochemical processes and buffering capacity concurrently affect acidification in a seasonally hypoxic coastal marine basin. Biogeosciences 12, 1561–1583. doi: 10.5194/bg-12-1561-2015
Hinrichs, K. U., Hayes, J. M., Sylva, S. P., Brewer, P. G., and DeLong, E. F. (1999). Methane-consuming archaebacteria in marine sediments. Nature 398, 802–805. doi: 10.1038/19751
Jebbar, M., Franzetti, B., Girard, E., and Oger, P. (2015). Microbial diversity and adaptation to high hydrostatic pressure in deep-sea hydrothermal vents prokaryotes. Extremophiles 19:721–740. doi: 10.1007/s00792-015-0760-3.
Kamyshny, A., Borkenstein, C. G., and Ferdelman, T. G. (2009). Protocol for quantitative detection of elemental sulfur and polysulfide zero-valent sulfur distribution in natural aquatic samples. Geostand. Geoanal. Res. 33, 415–435. doi: 10.1111/j.1751-908X.2009.00907.x
Kamyshny, A., Ekeltchik, I., Gun, J., and Lev, O. (2006). Method for the determination of inorganic polysulfide distribution in aquatic systems. Anal. Chem. 78, 2631–2639. doi: 10.1021/ac051854a
Kamyshny, A., Goifman, A., Gun, J., Rizkov, D., and Lev, O. (2004). Equilibrium distribution of polysulfide ions in aqueous solutions at 25 °C: a new approach for the study of polysulfides' equilibria. Environ. Sci. Technol. 38, 6633–6644. doi: 10.1021/es049514e
Kleindienst, S., Ramette, A., Amann, R., and Knittel, K. (2012). Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ. Microbiol. 14, 2689–2710. doi: 10.1111/j.1462-2920.2012.02832.x
Knittel, K., Lo, T., Boetius, A., Kort, R., Amann, R., and Lösekann, T. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479. doi: 10.1128/AEM.71.1.467-479.2005
Krüger, M., Treude, T., Wolters, H., Nauhaus, K., and Boetius, A. (2005). Microbial methane turnover in different marine habitats. Palaeogeogr. Palaeoecl. 227, 6–17. doi: 10.1016/j.palaeo.2005.04.031
Krüger, M., Wolters, H., Gehre, M., Joye, S. B., and Richnow, H.-H. (2008). Tracing the slow growth of anaerobic methane-oxidizing communities by 15N-labelling techniques. FEMS Microbiol. Ecol. 63, 401–411. doi: 10.1111/j.1574-6941.2007.00431.x.
Liamleam, W., and Annachhatre, A. P. (2007). Electron donors for biological sulfate reduction. Biotechnol. Adv. 25, 452–463. doi: 10.1016/j.biotechadv.2007.05.002
Lösekann, T., Knittel, K., Nadalig, T., Fuchs, B., Niemann, H., Boetius, A., et al. (2007). Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 73, 3348–3362. doi: 10.1128/AEM.00016-07
McGlynn, S. E., Chadwick, G. L., Kempes, C. P., and Orphan, V. J. (2015). Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535. doi: 10.1038/nature15512
Meulepas, R. J. W., Jagersma, C. G., Gieteling, J., Buisman, C. J. N., Stams, A. J. M., and Lens, P. N. L. (2009a). Enrichment of anaerobic methanotrophs in sulfate-reducing membrane bioreactors. Biotechnol. Bioeng. 104, 458–470. doi: 10.1002/bit.22412
Meulepas, R. J. W., Jagersma, C. G., Khadem, A. F., Buisman, C. J. N., Stams, A. J. M., and Lens, P. N. L. (2009b). Effect of environmental conditions on sulfate reduction with methane as electron donor by an Eckemförde Bay enrichment. Environ. Sci. Technol. 43, 6553–6559. doi: 10.1021/es900633c
Meulepas, R. J. W., Jagersma, C. G., Zhang, Y., Petrillo, M., Cai, H., Buisman, C. J. N., et al. (2010a). Trace methane oxidation and the methane dependency of sulfate reduction in anaerobic granular sludge. FEMS Microbiol. Ecol. 72, 261–271. doi: 10.1111/j.1574-6941.2010.00849.x
Meulepas, R. J. W., Stams, A. J. M., and Lens, P. N. L. (2010b). Biotechnological aspects of sulfate reduction with methane as electron donor. Rev. Environ. Sci. Biotechnol. 9, 59–78. doi: 10.1007/s11157-010-9193-8
Milucka, J., Ferdelman, T. G., Polerecky, L., Franzke, D., Wegener, G., Schmid, M., et al. (2012). Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 491, 541–546. doi: 10.1038/nature11656
Musat, N., Halm, H., Winterholler, B., Hoppe, P., Peduzzi, S., Hillion, F., et al. (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. PNAS 105, 17861–17866. doi: 10.1073/pnas.0809329105
Nadkarni, M. A., Martin, F. E., Jacques, N. A., and Hunter, N. (2002). Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148, 257–266. doi: 10.1099/00221287-148-1-257
Nauhaus, K., Boetius, A., Krüger, M., and Widdel, F. (2002). In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4, 296–305. doi: 10.1046/j.1462-2920.2002.00299.x
Niemann, H., Lösekann, T., de Beer, D., Elvert, M., Nadalig, T., Knittel, K., et al. (2006). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443, 854–858. doi: 10.1038/nature05227
Omoregie, E. O., Mastalerz, V., de Lange, G., Straub, K. L., Kappler, A., Røy, H., et al. (2008). Biogeochemistry and community composition of iron- and sulfur-precipitating microbial mats at the Chefren mud volcano (Nile Deep Sea Fan, Eastern Mediterranean). Appl. Environ. Microbiol. 74, 3198–3215. doi: 10.1128/AEM.01751-07
Orphan, V. J., House, C. H., Hinrichs, K.-U., McKeegan, K. D., and DeLong, E. F. (2002). Multiple archaeal groups mediate methane oxidation in an- oxic cold seep sediments. Proc. Natl. Acad. Sci.U. S. A. 99, 7663–7668. doi: 10.1073/pnas.072210299
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data com- patible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Reeburgh, W. S. (2007). Oceanic methane biogeochemistry. Chem. Rev. 107, 486–513. doi: 10.1021/cr050362v
Ruff, S. E., Felden, J., Gruber-Vodicka, H. R., Marcon, Y., Knittel, K., Ramette, A., et al. (2018). In situ development of a methanotrophic microbiome in deep-sea sediments. ISME J. doi: 10.1038/s41396-018-0263-1
Ruff, S. E., Kuhfuss, H., Wegener, G., Lott, C., Ramette, A., Wiedling, J., et al. (2016). Methane seep in shallow-water permeable sediment harbors high diversity of anaerobic methanotrophic communities, Elba, Italy. Front. Microbiol. 7:374. doi: 10.3389/fmicb.2016.00374
Scheller, S., Yu, H., Chadwick, G. L., McGlynn, S. E., and Orphan, V. J. (2016). Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351, 703–707. doi: 10.1126/science.aad7154
Schloss, P. D., and Westcott, S. L. (2011). Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77, 3219–3226. doi: 10.1128/AEM.02810-10
Shigematsu, T., Tang, Y., Kobayashi, T., Kawaguchi, H., Morimura, S., and Kida, K. (2004). Effect of dilution rate on metabolic pathway shift between aceticlastic and nonaceticlastic methanogenesis in chemostat cultivation. Appl. Environ. Microbiol. 70, 4048–4052. doi: 10.1128/AEM.70.7.4048-4052.2004
Siegert, M., Krüger, M., Teichert, B., Wiedicke, M., and Schippers, A. (2011). Anaerobic oxidation of methane at a marine methane seep in a forearc sediment basin off Sumatra, Indian Ocean. Front. Microbiol. 2:249. doi: 10.3389/fmicb.2011.00249
Sievert, S. M., Kiene, R. P., and Schulz-Vogt, H. N. (2007). The sulfur cycle. Oceanography 20, 117–123. doi: 10.5670/oceanog.2007.55
Snaidr, J., Amann, R., Huber, I., Ludwig, W., and Schleifer, K. H. (1997). Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63, 2884–2896.
Song, Z.-Q., Wang, F.-P., Zhi, X.-Y., Chen, J.-Q., Zhou, E.-M., Liang, F., et al. (2013). Bacterial and archaeal diversities in Yunnan and Tibetan hot springs, China. Environ. Microbiol. 15, 1160–1175. doi: 10.1111/1462-2920.12025
Stahl, D. A. (1991). “Development and application of nucleic acid probes,” in Nucleic Acid Techniques in Bacterial Systematics, eds. E. Stackebrandt and M. Goodfellow. (Chichester: John Wiley & Sons), 205–248.
Suarez-Zuluaga, D. A., Timmers, P. H. A., Plugge, C. M., Stams, A. J. M., Buisman, C. J. N., and Weijma, J. (2016). Thiosulphate conversion in a methane and acetate fed membrane bioreactor. Environ. Sci. Pollut. 23, 2467–2478. doi: 10.1007/s11356-015-5344-3
Sulu-Gambari, F., Seitaj, D., Meysman, F. J. R., Schauer, R., Polerecky, L., and Slomp, C. P. (2016). Cable bacteria control iron–phosphorus dynamics in sediments of a coastal hypoxic basin. Environ. Sci. Technol. 50, 1227–1233. doi: 10.1021/acs.est.5b04369
Takai, K., and Horikoshi, K. (2000). Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66, 5066–5072. doi: 10.1128/AEM.66.11.5066-5072.2000
Thauer, R. K., Jungermann, K., and Decker, K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacterial. Rev. 41, 100–180.
Timmers, P. H., Gieteling, J., Widjaja-Greefkes, H. A., Plugge, C. M., Stams, A. J. M., Lens, P. N. L., et al. (2015a). Growth of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a high-pressure membrane capsule bioreactor. Appl. Environ. Microbiol. 81, 1286–1296. doi: 10.1128/AEM.03255-14
Timmers, P. H. A., Suarez-Zuluaga, D. A., van Rossem, M., Diender, M., Stams, A. J. M., and Plugge, C. M. (2015b). Anaerobic oxidation of methane associated with sulfate reduction in a natural freshwater gas source. ISME J. 10, 1400–1412. doi: 10.1038/ismej.2015.213
Treude, T., Knittel, K., Blumenberg, M., Seifert, R., and Boetius, A. (2005). Subsurface microbial methanotrophic mats in the Black Sea Appl. Environ. Microbiol. 71, 6375–6378. doi: 10.1128/AEM.71.10.6375-6378.2005
Valentine, D. L., Reeburgh, W. S., and Hall, R. (2000). New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2, 477–484. doi: 10.1046/j.1462-2920.2000.00135.x.
Vigneron, A., Cruaud, P., Pignet, P., Caprais, J.-C., Cambon-Bonavita, M.-A., Godfroy, A., et al. (2013). Archaeal and anaerobic methane oxidizer communities in the Sonora Margin cold seeps, Guaymas Basin (Gulf of California). ISME J. 7, 1595–1608. doi: 10.1038/ismej.2013.18
Wagner, M., Amann, R., Lemmer, H., and Schleifer, K. H. (1993). Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure Appl. Environ. Microbiol. 59, 1520–1525.
Wallner, G., Amann, R., and Beisker, W. (1993). Optimizing fluorescent in situ hybridization with rRNA-targeted oligo- nucleotide probes for flow cytometric identification of microorganisms. Cytometry 14, 136–143. doi: 10.1002/cyto.990140205
Wegener, G., Krukenberg, V., Ruff, S. E., Kellermann, M. Y., and Knittel, K. (2016). Metabolic capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane. Front. Microbiol. 7:46. doi: 10.3389/fmicb.2016.00046
Weijma, J., Veeken, A., Dijkman, H., Huisman, J., and Lens, P. N. L. (2006). “Heavy metal removal with biogenic sulphide: advancing to full-scale,” in Advanced Biological Treatment Processes for Industrial Wastewaters, Principles and Applications, eds. F. Cervantes, S. Pavlostathis and A. van Haandel. (London: IWA publishing), 321–333.
Yamamoto, S., Alcauskas, J. B., and Crozier, T. E. (1976). Solubility of methane in distilled water and seawater. J. Chem. Eng. Data 21, 78–80. doi: 10.1021/je60068a029
Zhang, Y., Henriet, J. P., Bursens, J., and Boon, N. (2010). Stimulation of in vitro anaerobic oxidation of methane rate in a continuous high-pressure bioreactor. Biores. Technol. 101, 3132–3138. doi: 10.1016/j.biortech.2009.11.103
Zhang, Y., Li, X., Bartlett, D. H., and Xiao, X. (2015). Current developments in marine microbiology: high-pressure biotechnology and the genetic engineering of piezophiles. Curr. Opin. Microbiol. 33, 157–164. doi: 10.1016/j.copbio.2015.02.013
Keywords: anaerobic oxidation of methane, sulfate reduction, anaerobic methanotrophic archaea (ANME), coastal sediments, pressure
Citation: Cassarini C, Zhang Y and Lens PNL (2019) Pressure Selects Dominant Anaerobic Methanotrophic Phylotype and Sulfate Reducing Bacteria in Coastal Marine Lake Grevelingen Sediment. Front. Environ. Sci. 6:162. doi: 10.3389/fenvs.2018.00162
Received: 04 July 2018; Accepted: 24 December 2018;
Published: 14 January 2019.
Edited by:
Marc Mussmann, Universität Wien, AustriaReviewed by:
Diana Vasquez Cardenas, Delft University of Technology, NetherlandsCopyright © 2019 Cassarini, Zhang and Lens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiara Cassarini, Y2hpYXJhLmNhc3NhcmluaUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.