
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Environ. Sci. , 23 November 2018
Sec. Soil Processes
Volume 6 - 2018 | https://doi.org/10.3389/fenvs.2018.00139
This article is part of the Research Topic Microbe-Mediated Processes in Soils View all 7 articles
The rhizosphere is the interface between plant roots and soil where intense, varied interactions between plants and microbes influence plants' health and growth through their influence on biochemical cycles, such as the carbon, nitrogen, and iron cycles. The rhizosphere is also a changing environment where oxygen can be rapidly limited and anaerobic zones can be established. Microorganisms successfully colonize the rhizosphere when they possess specific traits referred to as rhizosphere competence. Anaerobic respiration flexibility contributes to the rhizosphere competence of microbes. Indeed, a wide range of compounds that are available in the rhizosphere can serve as alternative terminal electron acceptors during anaerobic respiration such as nitrates, iron, carbon compounds, sulfur, metalloids, and radionuclides. In the presence of multiple terminal electron acceptors in a complex environment such as the rhizosphere and in the absence of O2, microorganisms will first use the most energetic option to sustain growth. Anaerobic respiration has been deeply studied, and the genes involved in anaerobic respiration have been identified. However, aqueous environment and paddy soils are the most studied environments for anaerobic respiration, even if we provide evidence in this review that anaerobic respiration also occurs in the plant rhizosphere. Indeed, we provide evidence by performing a BLAST analysis on metatranscriptomic data that genes involved in iron, sulfur, arsenate and selenate anaerobic respiration are expressed in the rhizosphere, underscoring that the rhizosphere environment is suitable for the establishment of anaerobic respiration. We thus focus this review on current research concerning the different types of anaerobic respiration that occur in the rhizosphere. We also discuss the flexibility of anaerobic respiration as a fundamental trait for the microbial colonization of roots, environmental and ecological adaptation, persistence and bioremediation in the rhizosphere. Anaerobic respiration appears to be a key process for the functioning of an ecosystem and interactions between plants and microbes.
Bacteria are ubiquitous and occupy a very wide variety of ecological niches in particular the plant rhizosphere, the volume of soil influenced by root exudates (Kai et al., 2016). The rhizosphere is a densely populated soil fraction rich in carbon, insofar as 10–40% of photosynthetic fixed carbon is released through the root into the surrounding soil named “root-adhering soil” (Marschner, 1995; Haichar et al., 2014). As a result of this carbon flux, bacteria are attracted to the surface of the roots that they successfully colonize if they possess specific genetic and physiologic traits named “rhizosphere competence.” These traits shape bacterial growth and activity and are influenced by environmental factors, such as the pH, soil structure, water content, oxygen flux and nutrient availability (Philippot et al., 2013). Oxygen flux is one of the most changing factors in the rhizosphere, as plants can be subjected to aerobic or microaerophilic conditions followed by alternating exclusively anoxic conditions or anaerobic conditions, as in soil aggregates (Philippot et al., 1996). This variation in oxygen concentration constrains bacteria to adapt their physiology and to develop flexible metabolic pathways, such as the respiration of compounds other than oxygen when it is lacking. Indeed, bacterial anaerobic respiration is one of the most flexible and diverse metabolic processes (Hong and Gu, 2009). Among microbes, a wide range of compounds can serve as terminal electron acceptors (TEAs), including oxygen (the most energetic one); nitrogen oxyanions and nitrogen oxides (Zumft, 1997; Kuypers et al., 2018); carbon compounds, such as fumarate (Kröger, 1978; Alves et al., 2015); transition metals, such as Fe(III) and Mn(IV) (Lovley and Phillips, 1987; Kasai et al., 2015); metalloid oxyanions, such as selenate and arsenate (Macy et al., 1996; Switzer Blum et al., 1998; Glasser et al., 2018); radionuclides, such as U(VI) (Lovley et al., 1991; Marshall et al., 2006); and elemental sulfur and sulfur oxyanions (Klimmek et al., 1991; Eller et al., 2018). These TEAs are more or less all available in the rhizosphere and can serve for microbial anaerobic respiration. The appearance, development and evolution of such respiration have been the subjects of a numerous studies (Lovley and Coates, 2000; Richardson, 2000; Philippot, 2002; Kuypers et al., 2018). Respiratory chains have already been deeply studied in several bacteria (Richardson, 2000). Some bacteria present high anaerobic respiration flexibility through the use of different TEAs for growth. For example, Deltaproteobacteria exhibit considerable flexibility in respiratory processes, including sulfate reduction, iron reduction, fermentation, and dehalogenation (Sanford et al., 2002). Indeed, strains of Geobacter can at minimum use Fe(III), Mn(IV), nitrate, fumarate, arsenic oxyanions, and U(VI) as TEAs (Lovley et al., 1991). Shewanella oneidensis is even more versatile than Geobacter and can use additional TEAs, such as sulfur oxyanions and selenium oxyanions (Beliaev et al., 2005; Klonowska et al., 2005). Due to the high flexibility of anaerobic respiration, microorganisms affect a number of biogeochemical cycles, such as those for N, C, Fe and Se (Richardson, 2000). Those processes may result to greenhouse gases release (CO2 and N2O) and nutrient loading and consumption, which coupled with changes of interacting spheres of the earth (Zheng et al., 2018). Globally, bacterial anaerobic respiration is well-known but remains an incomplete story in the rhizosphere. Indeed, aside from denitrification and Fe(III) respiration, little is known about other respiration processes, and only few model strains have been deeply studied, even if certain rhizospheric models possess genes involved in anaerobic respiratory pathways. Furthermore, the reasons for microbial respiratory diversity and flexibility in the rhizosphere has yet remain incompletely known, but this diversity appears key to rhizosphere competence, adaptation, and persistence belowground. For example, microbial respiratory flexibility is suggested to provide competitive advantage to Fe(III)-reducing bacteria to colonize iron oxide plaque on root surfaces (Somenahally et al., 2011) and confers rhizosphere competence to denitrifiers in colonizing different plant rhizospheres, such as those of maize and tomato plants (Philippot et al., 1995; Mirleau et al., 2001; Chèneby et al., 2004).
In this review, we detail for the first time, anaerobic respirations that occur in the rhizosphere. First, attention is paid to the environmental conditions that affect anaerobic respirations and the different respirations encountered in the rhizosphere. Second, research on the genes involved in anaerobic respiration from metatranscriptomic data on the rhizosphere are conducted to evidence how this trait is largely distributed among rhizobacteria. Finally, the ecological and environmental advantages of anaerobic respiration in the rhizosphere are discussed.
Plant roots modulate anaerobic respiration both indirectly and directly. Indeed, it is well-known that roots increase the anaerobic volume of the soil by consuming oxygen, exude different carbon sources that may act as TEAs, such as fumarate, and compete with the microbial community for certain elements, such as nitrate, thereby diminishing its availability to microbes. Therefore, elucidation of how the rhizospheric microbiota switches from aerobic to anaerobic respiration in the absence of oxygen and which alternative electron acceptors it uses holds great potential to understand population dynamics and evolutionary strategies for coping with oxygen depletion.
One of the most significant factors influencing anaerobic respiration is the lack of oxygen. Its flux has been extensively studied at the root level of submerged plants (Christensen et al., 1994). However, only a few studies are available regarding the rhizosphere. The spatial distribution of oxygen depends strongly on the investigated scale. At the pedon scale, a higher O2 partial pressure (pO2) is found in the topsoil and gradually decreases with depth (Stepniewski and Stepniewska, 2009) due to the greater diffusion distance from the free atmosphere. At the aggregate scale, pO2 decreases from the outside perimeter to the aggregate heart, which can reach anaerobic conditions (Sexstone et al., 1985; Zausig et al., 1993; Philippot et al., 1996). At the rhizosphere scale, O2 distribution from the root surface into the bulk soil is driven by its consumption due to root and microbiotic respiratory processes and diffusive O2 supply (Glinski and Stepniewski, 1985; Uteau et al., 2015). Oxygen flux in soil is very difficult to predict, as it depends on soil properties such as porosity, texture, organic matter content, moisture content, temperature and pH (Runkles, 1956). The soil structure is the most important soil property related to oxygen diffusion in the soil. Indeed, the modification of the soil structure due to compaction limits gas transport in the soil, since it decreases the pore spaces filled with air where gas diffusion occurs (Neira et al., 2015). In addition, soil organic matter impacts oxygen level by stabilizing soil aggregates (Uteau et al., 2013).
The basic anaerobic respiratory process involves the oxidation of a low-redox-potential electrons donor, such as carbon substrates, the transfer of electrons through a wide range of cytochromes and the reduction of a high-redox-potential electrons acceptor. Bacteria synthetize ATP through a membrane-bound ATP synthase complex driven by proton-motive force with the intermediary action of cytochromes (Richardson, 2000; Kartal et al., 2011). A wide range of metallic and non-metallic terminal electron acceptors (TEAs) is encountered in soils and can serve for bacterial anaerobic respiration (Figure 1, Table 1). Their availability is spatio-temporally variable (Jackson and Caldwell, 1993; Farley and Fitter, 1999; Lark et al., 2004) and depends on the constant interaction of processes such as weathering, atmospheric deposition, nutrient leaching, biological cycling, and plants nutrition, resulting in the formation of vertical and horizontal TEA gradients within the soil (Figure 1) (Giehl and Wirén, 2014). Additional processes, such as rainfall, the inflow of dust in agricultural soils, and the supply of manure and chemical fertilizers, introduce additional nutrients to the soil pool (Giehl and Wirén, 2014), which can act as TEAs.
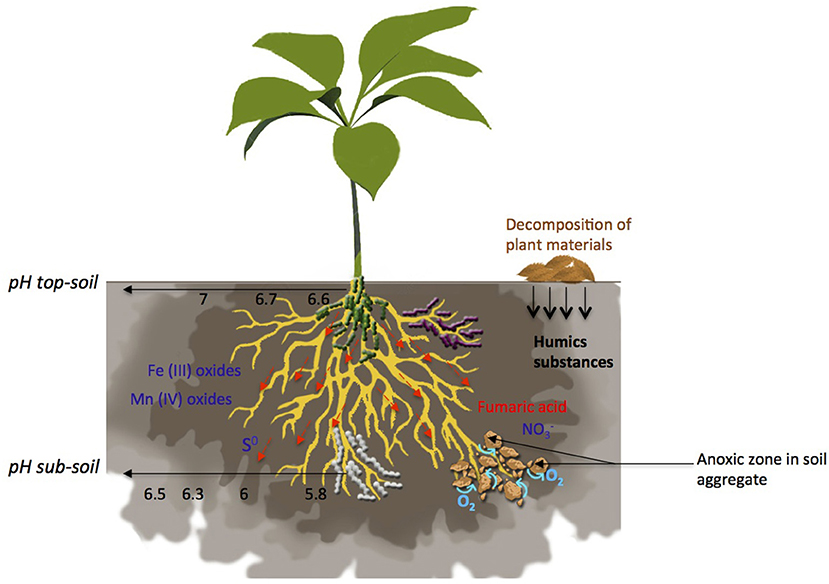
Figure 1. Terminal Electron Acceptors (TEAs) distribution, O2 flux and pH gradient in the rhizosphere. Many factors are involved in the formation of zones favorable for anaerobic respiration establishment such as TEAs availability, pH, water movement and soil aggregate formation. moves freely within the soil, with a vertical gradient of availability from sub-soil to top-soil. Decomposition of plant materials leads to the release of humic substances. Fumaric acid is exuded by plant roots. Availability of Fe(III), Mn(IV) and S0 is mostly driven by pH. Roots influence pH leading to a gradient from an acidic pH close to the roots to a neutral pH in the rhizosphere. Anoxic zones occur in soil aggregates and in soils submitted to water flow.
Nitrate, fumarate, and humic substances represent the most studied non-metallic TEAs. Their availability in the rhizosphere is variable and depends on different biotic and abiotic factors. The bioavailability of N forms in soils, such as nitrate (NO) and nitrite (NO) is a dynamic balance influenced by microbial nitrification and denitrification (Jones and Hallin, 2018). formation occurs near the surface and concentration decrease rapidly with deep (Arth and Frenzel, 2000). The spatial and temporal availability of soil is highly heterogeneous. At the centimeter scale or over the course of a day, may vary by an order of magnitude (Beliaev et al., 2005; Klonowska et al., 2005). This heterogeneity is derived from several factors that release or remove nitrates from the soil (Bloom et al., 2002). For example, a high rate of nitrification activity or fertilizer use in agricultural soils leads to an increase in nitrate concentration (Burger and Jackson, 2003; Galloway et al., 2008), whereas plant root uptake, which competes with soil microbiota, reduces nitrate in soils (Kuzyakov and Xu, 2013). Notably, some bacteria impact positively N availability for plants, such as Rhizobia that are able to provide N2 to plants by nitrogen fixation in nodules (Haichar et al., 2014). In the rhizosphere, nitrate move freely within the soil and reach the root surface through mass flow, which is driven by the nutrient concentration in the soil solution and the rate of plant transpiration (Giehl and Wirén, 2014). Their concentration varies from 0.04 to 5 g.kg−1 for soils deficient in nitrate to 80–100 g.kg−1 for soils rich in nitrate (unpublished data).
Fumaric acid or fumarate is an organic acid exuded by plants in the rhizosphere of various plant species such as maize, banana and Dactylis glomerata (Zhang et al., 2014; Guyonnet et al., 2017). Its concentration in the rhizosphere varies according to plant developmental stage (Petersen and Böttger, 1991). Recently, Guyonnet et al. (2017) evidenced that the relative abundance of fumaric acid was approximately 0.04% in the root-adhering soil of D. glomerata. Fumaric acid exudation was also observed with barley (Hordeum vulgare cv. Barke) and was likely involved in the tripartite interaction of beneficial microorganisms, plant and pathogens in the rhizosphere (Jousset et al., 2014). In addition, Liu et al. (2014) and Yuan et al. (2015) demonstrated that fumaric acid exudation is involved in improving plant growth-promoting rhizobacterial colonization by stimulating biofilm formation. Hence, fumaric acid has a role in plant-microbe interactions and in microbial anaerobic respiration in the rhizosphere.
In addition to fumaric acid, humic substances (HSs) have also been investigated as TEAs for microbial anaerobic respiration (Lovley et al., 1996; Hong and Gu, 2009). HS are highly polymerized natural polymers of high molecular weight and poor biodegradability that are formed during the decomposition of plant materials by microorganisms in soils and sediments (Hong and Gu, 2009). The remarkable recalcitrance of such materials is reflected by their long residence times in the environment, exceeding 500 years. Lovley et al. (1996) were the first to put forward the concept of humic respiration, which was supported by experiments in which both Geobacter metallireducens and Shewanella alga can grow with humus and/or anthraquinone-2,6-disulfonate (AQDS) as TEAs when a wide variety of organic substrates or H2 are oxidized.
The most-studied metallic TEA are iron [Fe (III)], manganese [Mn (IV)], arsenic [As (V)], selenium [Se (VI)], sulfur (S0), and uranium [U(VI)]. Their availability in the rhizosphere also depends on biotic and abiotic factors. Among those TEAs, Fe, Mn, and S also serve as plant nutrients (Giehl and Wirén, 2014); hence, mutualistic/competitive interactions may occur with rhizosphere microbiota for their uptake (Somenahally et al., 2011; Moreau et al., 2015).
Iron (Fe) is a relatively abundant element in many cultivated soils, with an average total concentration of approximately 20–40 g.kg−1 (Cornell and Schwertmann, 2003); however, the amount of its available fraction is generally very small (Colombo et al., 2014). Iron is considered an essential micronutrient for plants and microbes, and its availability is crucial for their growth. Thus, bacteria and plants both adopt strategies to acquire Fe for their metabolisms, such as the secretion of microbe- and phyto-siderophores (Colombo et al., 2014). Fe availability in the rhizosphere is mediated by two major factors: the redox potential (i.e., oxidizing or reducing conditions) and pH (Lemanceau et al., 2009; Colombo et al., 2014). In fact, soil pH can increase or decrease by up to two units in the rhizosphere owing to the release and uptake of ions by roots (Hinsinger et al., 2009). Among Fe species present in the soil environment, a wide variety of Fe(III) oxides and clay minerals can be used as a TEAs during microbial anaerobic respiration (Lovley et al., 2004; Weber et al., 2006; Colombo et al., 2014).
Manganese is, an essential trace element for life, usually present in relatively large amount in the plant rhizosphere (Uren, 2013), and its availability is governed by oxidation and reduction processes (Marschner, 1988; Posta et al., 1994). The balance between these two processes is a function of soil chemical, microbial and plant factors (Marschner, 1988). Theoretically, Mn may exist in any of the redox states ranging from 0 to +7, but it is primarily present under the three oxidation states +II, +III, and +IV in natural environments (Davison, 1993). In addition, various oxides and oxyhydroxides of Mn(IV) are present in soils and serve as sinks for bioavailable Mn (Post, 1999). These Mn oxides are typically generated during diagenetic processes or the oxidation of Mn(II) (Mouret et al., 2009) and serve as TEAs for microbial anaerobic respiration (Lovley et al., 2004).
In soils, sulfur occurs in inorganic and organic forms and is cycled between these forms via mobilization, mineralization, immobilization, oxidation, and reduction processes (Scherer, 2009). Whereas, organic sulfur compounds are largely immobile, inorganic sulfur is more mobile, with sulfate being the most mobile (Scherer, 2001). Sulfate is also the most important S source for plants, generally accounting for <5% of the total S in the soil (Scherer, 2009). In grassland soils, the total S content in the upper soil layer (0 to 20 cm) ranges from 0.21 to 1.7 g.kg−1 (Wang et al., 2006). Among sulfur compounds encountered in soils, sulfite, organic sulfoxides, elemental sulfur, polysulfide, tetrathionate, and organic disulfides are important TEAs in the biogeochemical cycling of sulfur anaerobically (Schauder and Kröger, 1993; Kertesz, 2000; Burns and DiChristina, 2009). Their availability, as for Fe(III) oxides, is also dependent on pH (Hedderich et al., 1998).
Selenium is an essential trace element available in different chemical and physical forms and is found in all natural environments, such as soils (Nancharaiah and Lens, 2015). In most soils, selenium concentrations are very low, in the range of 0.01–2 mg.kg−1. However, in seleniferous soils, selenium concentrations of up 1,200 mg.kg−1 have been reported (Fordyce, 2013). The selenium cycle has a wide range of oxidation states, from –II to +VI, and their distribution may vary in the environment depending on the prevailing redox conditions and pH (González et al., 2006). Generally, selenium oxyanions selenate [SeO or Se (VI)] and selenite [SeO or Se (Se IV)] are soluble and stable and exhibit high bioavailability and toxicity in oxic environments. In anoxic conditions, selenium oxyanions serve as TEAs for anaerobic microbial respiration and are replaced by elemental selenium (Se0).
Arsenic, despite its toxicity, is readily used by a great diversity of prokaryotes for cell growth and metabolism (Stolz et al., 2006). Arsenic, as is the case with selenium, is widely distributed in the environment, with an average As content in Earth's crust varying between 2 and 3 mg.kg−1 (Tanaka, 1988). Its concentration in soils depends on rock type, human activities, climate, As forms and soil redox conditions (Yan-Chu, 1994). Arsenic is found in four oxidation states, arsenate As(V), arsenite As(III), elemental As0 and arsenide As(-III) distributed according to oxygen availability (Stolz et al., 2006; Mohan et al., 2007). Among these species, As(V) is used as a TEA by microbes for anaerobic respiration called “arsenate respiration,” which is the dominant process in anaerobic conditions (Oremland et al., 2005).
Uranium is an important component of Earth's crust and is found in all environments from water to soils (Gavrilescu et al., 2009). Uranium is a radioactive element whose 3 dominant isotopes are 238U, 235U, and 234U. Thus, uranium is a toxic metal, and multiple studies are interested in the remediation of this element (Akob et al., 2007; Lee and Yang, 2010; Choudhary and Sar, 2011; Yan et al., 2016). Uranium is found in soil in concentrations ranging from 8.1 × 10−4 to 3.2 μg.g−1 soil (Tzortzis and Tsertos, 2004). It is available in the environment in the form of oxides such as uranyl ion (UO) and UO2OH+, which can be complexed with carbonate, nitrate or humic substances, for example, (Markich, 2002; Gavrilescu et al., 2009) and which can be used as TEAs for microbial anaerobic respiration.
As described above, the rhizosphere contains a large variety of compounds that can serve as terminal electron acceptors for microbial respiration in the anoxic zone of the rhizosphere. However, only a few studies have been interested in understanding anaerobic respiration in the rhizosphere (Philippot et al., 1995; Ghiglione et al., 2000; Mirleau et al., 2001). Denitrification represents the most studied one area, likely due to its large contribution to the increase of N2O emissions, a gas that has an approximately 300-fold greater impact on global warming than CO2 (Bakken et al., 2012; Domeignoz-Horta et al., 2015; Gaimster et al., 2018). In the next paragraph, we review the different mechanisms of anaerobic respiration encountered in the environment and we attempt to determine whether certain respiration processes studied in a given environment can take place in the rhizosphere by the microbiota associated with plants.
In anoxic conditions, bacteria have developed specific processes that allow them to use sufficiently abundant alternative TEAs to survive, proliferate and acclimate quickly in a rapidly changing environment such as the rhizosphere. Indeed, the rhizosphere can lead to prolonged anoxic conditions such as in rice field or aerobic conditions followed by alternating anoxic conditions such as soils aggregates (Figure 1) (Philippot et al., 1996; Kostka et al., 2002). In these conditions, the TEAs described above can be respired by rhizospheric microbiota. According to the acceptor, bacteria can grow more quickly, such as with nitrate, or less quickly, such as with sulfur (Nealson et al., 1994), due to the difference in free energy released during respiratory metabolism. Here, we summarize the different respiration processes, which potentially occur in the rhizosphere, and the energy they generated (Figure 1).
Denitrification is a part of nitrogen cycle and occurs in anaerobic conditions mainly by bacteria. Denitrification uses nitrate as a TEA and yields the highest energy to sustain growth among anaerobic respirations; it includes nitrate, nitrite, nitric oxide and nitrous oxide reductions (NO 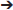
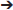 NO
NO 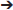 N2O
N2O 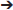 N2) (Zumft, 1997). Each reaction is catalyzed by a reductase that is coupled to energy-conserving electron-transport pathways and regulated by the transcriptional regulator “Fnr” (fumarate and nitrate reductase). Fnr is part of a superfamily of Crp-Fnr transcriptional regulators that includes Crp (catabolite repression), FnrN (nitrogen fixation/photosynthesis), FixK (nitrogen fixation), NnrR (response to NO in denitrification) and Dnr (dissimilatory nitrate respiration regulator) (Körner et al., 2003). Notably, in Bradirhizobium japonicum, the global response regulator that controls expression of denitrification gene is called RegR (Torres et al., 2014).
N2) (Zumft, 1997). Each reaction is catalyzed by a reductase that is coupled to energy-conserving electron-transport pathways and regulated by the transcriptional regulator “Fnr” (fumarate and nitrate reductase). Fnr is part of a superfamily of Crp-Fnr transcriptional regulators that includes Crp (catabolite repression), FnrN (nitrogen fixation/photosynthesis), FixK (nitrogen fixation), NnrR (response to NO in denitrification) and Dnr (dissimilatory nitrate respiration regulator) (Körner et al., 2003). Notably, in Bradirhizobium japonicum, the global response regulator that controls expression of denitrification gene is called RegR (Torres et al., 2014).
Unlike other anaerobic respirations, genes encoding the denitrifying reductases have been characterized in bacteria inhabiting the rhizosphere, such as Pseudomonas stutzeri, Pseudomonas fluorescens and Parococcus denitrificans (Zumft, 1997; Kuypers et al., 2018). The reductases involved in this respiratory process are described as dissimilatory nitrate reductase (membrane-bound “NAR, narGH” and periplasmic “NAP, napA”), nitrite reductase (Nir), nitric oxide reductase (cytochrome c-dependent “cNOR, cnorB”, quinol-dependent “qNOR, norZ” and copper-containing quinol-dependent nitric oxide reductases “CuANOR”) and nitrous oxide reductase (Nos) (Philippot, 2002; Kuypers et al., 2018). Two types of dissimilatory nitrate reductases are found in bacteria and even in the same bacteria as in Escherichia coli: a membrane-bound (Nar) and a periplasmic-bound (Nap) nitrate reductase (Moreno-Vivián et al., 1999; González et al., 2006; Kraft et al., 2011). Nar is composed of 3 subunits encoded by narGHI, as found in E. coli, P. fluorescens, P. stutzeri and B. subtilis (Philippot and Højberg, 1999; Lalucat et al., 2006), whereas Nap is composed of 2 subunits encoded by napAB, as found in Rhodabacter sphaeroides and B. japonicum (Reyes et al., 1998; Bedmar et al., 2005). The nar operon is induced under low oxygen partial pressure and in the presence of nitrogen oxides (Philippot et al., 2001; Giannopoulos et al., 2017), whereas the expression of the nap operon can be affected or not by anaerobiosis depending on the organism (Philippot, 2002; Bueno et al., 2017).
A recent study demonstrated that the two types of nitrite reductase can also be found in the same bacteria, contrary to what has been previously accepted (Sánchez and Minamisawa, 2018). The two nitrite reductases encoded by nirK and nirS are evolutionarily unrelated enzymes (Philippot, 2002). Both nitrite reductases encoded by nirS in P. fluorescens and P. stutzeri and nirK in R. sphaeroides are expressed under low O2 levels and in the presence of nitrate or nitrite (Tosques et al., 1997; Härtig and Zumft, 1999; Philippot et al., 2001). Moreover, it has been demonstrated that NO induces nirS expression in P. stutzeri (Vollack and Zumft, 2001). Nitric oxide reductase is composed of 2 subunits encoded by norCB and is expressed in the absence of O2 and in the presence of NO (Zumft, 1997; Vollack and Zumft, 2001; Hino et al., 2010). The enzyme involved in nitrous oxide respiration is encoded by nosZ (Zumft and Kroneck, 2007; Hartsock and Shapleigh, 2010; Thomson et al., 2012). This gene is lacking in many denitrifying bacteria and archaea, leading to greenhouse gas emissions (Hu et al., 2015). Recently, it has been demonstrated that non-denitrifying bacteria, such as Wolinella succinogenes, can grow with N2O as the sole electron acceptor, thereby allowing to the reduction of the N2O greenhouse gas (Simon et al., 2004). These N2O-reducing microorganisms are the subject of a recent review by Hallin et al. (2017) in which the two distinct groups of N2O reducers and their ecology are described. They underscore the fact that NirK denitrifying microorganisms are more lacking the nos cluster than are NirS denitrifying microorganisms. So far, no study has been conducted on the co-occurrence of the nar/nap operons and nirK and/or nirS denitrifying microorganisms.
Microorganisms contribute to N cycle not only through denitrification process also through anaerobic ammonium oxidation (anammox) process (van de Graaf et al., 1995). Anammox is responsible for a loss of total N from fertilized soils, such as paddy soils (Nie et al., 2015) but less significant than loss caused by denitrification. Anammox is the oxidation of ammonium to dinitrogen gas under anaerobic conditions with nitrite as TEA (Gori et al., 2011). Hydrazine (N2H4) and NO are intermediates of anammox process (Strous et al., 2006). Anammox activity is linked to the ammonium, nitrate and nitrite concentration in soils (Hu et al., 2011). So, this process depends on denitrification and ammonia oxidation rates to form the substrates of the reaction (Naeher et al., 2015). Anammox is also influenced by oxygen flux, soil pH, salinity and rhizosphere effect (Nie et al., 2018). Indeed, soil pH affects ammonium availability and hence plays an important role in shaping anammox bacterial community. Like denitrification, anammox respiration occurs in a larger variety of niches in rhizospheres and soils (Humbert et al., 2010; Li et al., 2016). The five predominant anammox bacterial genera are Kuenenia, Brocadia, Jettenia, Scalindua, and Anammoxoglobus and detected in many habitats, including soils and sediments (Humbert et al., 2010; Li et al., 2016). Genes encoding specific proteins involved in anammox were deciphered in Kuenenia stuttgartiensis (Strous et al., 2006; Kraft et al., 2011). The first step of anammox corresponding to the reduction of in NO is performed by the dentification protein NirS. The second step of the predicted anammox pathway would be the reduction of NO and its simultaneous condensation with ammonium to produce N2H4. In K. stuttgartiensis, two gene clusters potentially involved in this step may formed an enzymatic complex called hydrazine synthase, HZS (Kraft et al., 2011). The final step of annamox is the oxidation of N2H4 to N2. In K. stuttgartiensis, the dedicated enzyme involved in this step is the hydrazine dehydrogenase (HDH) encoded by kuste0694 (Kraft et al., 2011).
Anammox respiration is a sufficient energetic strategy to sustain growth during anaerobic conditions (Kraft et al., 2011). The overall reaction for anammox is NH + 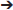 N2 + 2 H2O and the associated free energy is −358 kJ.mol−1 of NH (van de Graaf et al., 1995, 1996).
N2 + 2 H2O and the associated free energy is −358 kJ.mol−1 of NH (van de Graaf et al., 1995, 1996).
Electron acceptors such as Fe(III) and Mn(IV) are highly insoluble in most environments at pH 7. Thus, there is an important challenge for Fe(III) and Mn(IV) reducers to develop strategies for electron transfer to an insoluble and extracellular TEA (Lovley et al., 2004). Dissimilatory Fe(III)-reducing bacteria need (i) to transfer electrons from the central metabolism to a site of reduction somewhere outside the inner membrane and (ii) to express a reductase located on the outer membrane to reduce Fe(III) oxides to Fe (II). The same observations can be made for Mn (IV)-respiring bacteria (Lovley et al., 2004).
Fe (III) and Mn (IV) anaerobic respirations have been extensively studied in S. oneidensis MR-1 and Geobacter sulfurreducens models (Lovley et al., 2004). However, until recently, no study has been made available using rhizobacteria as model. In addition, the mechanisms responsible for these respirations are not fully understood, but a number of genes seem to be involved (Bretschger et al., 2007). In S. oneidensis MR-1, these include the mtrA, mtrB, mtrC, omcA and cymA genes. MtrABC serve as an electron conduit between the periplasm and the extracellular environments (Hartshorne et al., 2009). MtrC and OmcA have been hypothesized to serve as terminal Mn(IV) and Fe(III) reductases (Bretschger et al., 2007). CymA, cytoplasmic membrane-bound, tetraheme c-type cytochrome, is involved in electron flow during Fe(III) and Mn (IV) respiration (Myers and Myers, 1997). In G. sulfurreducens, different genes are also involved in each respiratory pathway (Aklujkar et al., 2013). Interestingly, OmcS, an outer-membrane protein, is required for both types of respiration (Mehta et al., 2005). Respiration of Fe(III) or Mn(IV) has the potential to yield sufficient energy to support growth, as does nitrate (Tables 1, 2). Interestingly, some anammox bacteria, such as K. stuttgartiensis are known to use Fe(III) and Mn(IV) oxides as TEA during anaerobic conditions (Strous et al., 2006; Zhao et al., 2014).
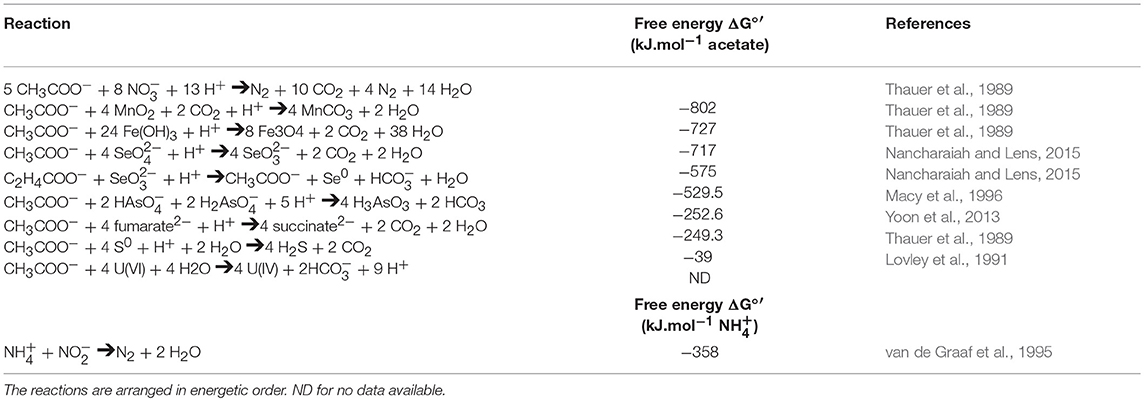
Table 2. Comparison of free energy ΔG°′ for various TEAs coupled to acetate oxidation and associated reactions.
Fumarate respiration has been well-known since the 1970s. However, it has been described in only a few bacteria, such as E. coli, W. succinogenes and Shewanella (Ackrell et al., 1992; Kröger et al., 1992; Arkhipova and Akimenko, 2005). Fumarate is reduced to succinate, the reverse reaction of succinate to fumarate that occurs aerobically in the tricarboxylic acid cycle
(TCA cycle). Under anaerobic conditions in the presence of fumarate in E. coli, a specific membrane quinol-bound fumarate reductase complex encoded by four genes frdA-D is induced (Jones and Gunsalus, 1987). Fumarate reductase is a distinct complex from the succinate dehydrogenase complex, encoded by sdhA-D genes. The regulation of these two enzymes is quite distinct since frd genes are expressed under anaerobic conditions in the presence of fumarate, whereas sdh genes are expressed under aerobic conditions and depend on glucose (Maklashina et al., 2013). In addition to the fumarate reductase complex, different global regulators are also involved in fumarate respiration, such as FNR and CymA. Indeed, in S. oneidensis MR-1, as in Fe(III) oxides and Mn(IV) respirations, CymA is also involved in anaerobic fumarate respiration (Myers and Myers, 1997).
Unlike the aforementioned respiration processes, fumarate respiration is less energetically favorable (see Tables 1, 2).
Anaerobic respiration with elemental sulfur/polysulfide or organic disulfides is performed by several bacteria and archaea but has only been investigated in detail in a few organisms, such as W. succinogenes and S. oneidensis MR-1. The electron transport chain that catalyzes the polysulfide reduction in W. succinogenes consists of polysulfide reductase (Psr) which is composed of 3 subunits (PsrA, B, C) encoded by psrABC genes, where PsrA seems to be the catalytic subunit (Hedderich et al., 1998), PsrB serves for electron transfer, and PsrC anchors the polysulfide reductase (PsrA) (Krafft et al., 1995).
The energy gain from sulfur respiration is the smallest compared to the previous respiration processes (Table 2; Thauer et al., 1989). The data suggest that this respiration process does not support bacterial growth and probably allows bacteria to persist until conditions are favorable.
Dissimilatory reduction of selenium oxyanions, viz., selenate (SeO) and selenite (SeO), is significantly important in the environment and involves the conservation of metabolic energy for microorganisms (Knight et al., 2002). Microbes that can reduce selenium oxyanions are not restricted to any particular group or subgroup of prokaryotes, and examples are polyphyletically found throughout the bacterial and archaeal domain (Watts et al., 2003). Selenium oxyanion respiration consists of 2 distinct respirations: selenate and selenite. Two successive reductions occur during anaerobic respiration: selenate to selenite and selenite to selenium. Anaerobic respiration of selenate is induced by its presence under anaerobic conditions, leading to the formation of selenite and then selenium (Debieux et al., 2011). It has been suggested that selenate reduction may be catalyzed in many cases by bacterial nitrate reductases, and selenate reductase activity of both membrane-bound nitrate reductase (NAR) and periplasmic nitrate reductase (NAP) has been reported (Avazéri et al., 1997; Sabaty et al., 2001). However, it is evident that nitrate reductases are poor reducers of selenate and may not contribute significantly to global selenate reduction, particularly in areas enriched with both selenate and nitrate (Watts et al., 2003). Consequently, novel enzyme systems that catalyze the reduction of selenate selectively have been sought, and to date, detailed biochemical studies have been limited mainly to species isolated from aqueous or contaminated environments, such as Thauera selenatis, Bacillus selenatarsenatis SF-1, Sulfurospirillum barnesii, and Enterobacter cloacae SLD1a-1 (Rabenstein and Tan, 1988; Losi and Frankenberger, 1997; Yamamura and Amachi, 2014). For example, the selenate reductase of T. selenatis is part of the SerABC operon (Rabenstein and Tan, 1988), whereas the selenate reductase respiratory activity in B. selenatarsenatis is conferred by the srdBCA operon (Kuroda et al., 2011). In T. selenatis, SerABC has a high affinity to selenate and does not use selenite or other electron acceptors as substrates (Stolz et al., 2006). In addition, SerABC shares several similarities with the respiratory nitrate reductases. Furthermore, genetic studies have shown that selenate reductase activity requires the global anaerobic regulatory gene FNR (Yee et al., 2007).
At present, no gene has been identified for the second reduction of selenate respiration from selenite into elemental selenium in T. selenatis. Anaerobic respiration of selenite is induced by its presence under anaerobic conditions, leading to the formation of S0. It is well-established that selenite reduction can be categorized into anaerobic respiration or a microbial detoxification strategy (Turner et al., 1998). The genetic mechanism of selenite respiration has been investigated only in S. oneidensis MR-1, and some mutagenesis analyses have been performed on respiratory or trans-membrane reductase (Taratus et al., 2000). It is still unclear whether S. oneidensis MR-1 is able to grow on selenite as the sole electron acceptor or if it is just a fortuitous detoxification process during anaerobic growth on fumarate, where fccA gene may be involved (Li et al., 2014).
Arsenic respiration has been well-studied in Mono Lake, resulting in the isolation of a number of arsenate-respiring bacteria (Oremland et al., 2000; Hollibaugh et al., 2005). The respiratory arsenate reductase (Arr) from Chrysiogenes arsenatis, Bacillus selenitireducens, Shewanella, and Alkalilimnicola ehlichii was identified and characterized (Krafft and Macy, 1998; Afkar et al., 2003; Richey et al., 2009). The arsenate reductase is a heterodimer, with a large catalytic subunit (ArrA) and a smaller electron transfer protein (ArrB). Stolz et al. (2006) performed a phylogenetic analysis of available sequences from arsenate reductases (Arr) and showed a high similarity between them. Remarkably, a complete genome analysis of A. ehlichii indicated that it lacks a conventional arsenite oxidase, but it instead possesses two operons that each encodes a putative respiratory arsenate reductase (Arr) expressed under different conditions acting as a bidirectional enzyme (Richey et al., 2009).
Only few known strains are able to anaerobically reduce uranium to sustain their growth such as Shewanella putrefaciens strain 200, S. oneidensis MR-1, G. metallireducens, and Desulfotomaculum reducens (Lovley et al., 1991; Tebo and Obraztsova, 1998). The reduction of U(VI) anaerobically was particularly deeply investigated in both Shewanella strains (Lovley et al., 1991; Wade and DiChristina, 2000; Marshall et al., 2006). To date, no uranium-specific reductase has been reported, but certain c-type cytochromes implicated in Fe(III) and Mn(IV) anaerobic respirations have been found to be involved in uranium reduction, such as MtrA, MtrB, MtrC, and OmcA (Beliaev and Saffarini, 1998; Beliaev et al., 2001; Marshall et al., 2006). Notably, in S. putrefaciens strain 2000, anaerobic respiration of U(VI) seems to be related to denitrification, as the U(VI) reduction-deficient mutant (Urr) lacks the ability to growth using U(VI) and nitrite.
Humic substances (HSs) are the major component of soil's organic matter and are derived from the partial degradation of plant material (Trevisan et al., 2010). The biogeochemical cycle of HSs is driven through oxidation and reduction performed by various microorganisms that can use HSs as electron donors and/or acceptors. Even if HS respiration has been known since the 1990s (Lovley et al., 1996), only few models have been studied for their ability to reduce HSs, such as Shewanella and G. sulfurreduscens (Lloyd et al., 2003; Hong et al., 2007). Certain genes involved in this respiration process were evidenced in G. sulfurreducens (Voordeckers et al., 2010) through the mutagenesis of five c-type cytochromes, OmcB, OmcS, OmcT, OmcE, and OmcZ. The authors concluded that AQDS and HS reduction might occur at the outer surface through these five c-type cytochromes. It is well-accepted that priming the soil with HS causes an increase in microbial activity due increased availability of energy required by microbes (Shah et al., 2018). However, to our knowledge, no information is available on the energy yielded by the anaerobic reduction of HSs.
As seen above, some bacteria are able to respire several TEAs, such as S. oneidensis or G. sulfurreducens. Remarkably, electron flows are shared by several anaerobic respiration processes. Indeed, the Mtr pathway of S. oneidensis is involved at least in Fe(III), Mn(IV) and U(VI) respiration (Beliaev et al., 2005; Marshall et al., 2006). Thus, in the presence of multiple TEAs in a complex environment, such as the rhizosphere in absence of O2, microorganisms will use TEAs in specific order (Achtnich et al., 1995). Even if this order is specific to each microorganism, they generally prefer to use the more energetic option first (Achtnich et al., 1995; Table 2).
As described below, the majority of anaerobic respiration models leverage aqueous environments or paddy-rice fields (Somenahally et al., 2011), with only a few having been derived from the plant rhizosphere, except for denitrification respiration (Zumft, 1997).
Here, to evidence whether genes involved in iron, sulfur, selenate and arsenate respirations are expressed in the plant rhizosphere, we used available rhizosphere metatranscriptomic data for BLAST analysis. Ten protein sequences from 6 model strains were selected as templates: MtrC and OmcA from S. oneidensis MR-1 for Fe(III) respiration; PsrA from W. succinogenes and S. oneidensis MR-1 for sulfur respiration; ArrA from Shewanella ANA-3, B. selenitireducens, and C. arsenatis for arsenate respiration; and SerA, SerB, and SerC from T. selenatis for selenate respiration. Eleven rhizosphere metatranscriptomic bioprojects published on NCBI (https://www.ncbi.nlm.nih.gov/sra/docs/), retrieved from the rhizospheric soil and/or the roots of Citrus (Zhang et al., 2017), Arabidopsis thaliana, Vellozia epidendroides, Populus, Miscanthus, corn, switchgrass, maize, canola, and sorghum were analyzed. The raw reads from these data were filtered, trimmed, and quality-controlled to generate the unique and clean reads. Then, we compared the sequence similarity by BLAST analysis (at least 40% identity and 80% coverage) of reference protein sequences against these unique reads. Interestingly, we found that genes involved in the selected anaerobic respirations are expressed in the rhizospheres of Citrus, Arabidopsis thaliana, Vellozia epidendroides, Populus, Miscanthus, corn, switchgrass and sorghum, confirming that the anaerobic respiration of iron, sulfur, selenate and arsenate occur in the plant rhizosphere and that such an environment is favorable for the establishment of these respiration processes by the rhizospheric microbial community (Figure 2). Notably, genes encoding Fe(III) respiration enzymes were less expressed in the various plant rhizospheres tested, suggesting that this is due either to a lower conservation of these genes between S. oneidensis MR-1 and rhizobacteria, or to Fe oxides being less available for anaerobic respiration.
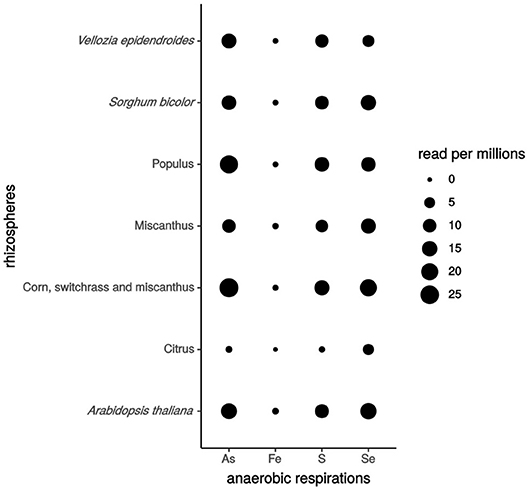
Figure 2. Anaerobic respiration gene expression in different plant rhizospheres. The number of hits per 106 reads expressed the level of genes expression. As for Arsenate, Fe for iron, S for sulfur and Se for selenate. Rhizospheric metatranscriptomics bioprojects accession numbers: Vellozia epidendroides (PRJNA441428), Sorghum bicolor (PRJNA406786), Populus (PRJNA375667), Miscanthus (PRJNA337035), [corn, switchgrass and Miscanthus] (PRJNA365487), Citrus (PRJNA324090), Arabidopsis thaliana (PRJNA366978, PRJNA366977, PRJNA336798).
Further studies are needed to identify and isolate model strains of the rhizosphere expressing anaerobic respiration genes to better elucidate the importance and extent of anaerobic respiration processes in this environment. We suggest that microbial anaerobic respiration appears to be a key process for ecosystem functioning and plant-microbe interactions that need to be put in the spotlight.
As reported above, rhizobacteria present a flexible respiratory process that may allow them to colonize and acclimate rapidly to a changing environment represented by the rhizosphere, where oxygen can be rapidly limited. In addition, certain respirations, such Fe(III) oxides by Fe(III)-reducing bacteria (FeRB), participate in Fe(II) availability for plants. Consequently, under anaerobic conditions and in the presence of Fe(III) oxides, FeRB are more abundant on the root surface of plants, as shown by Somenahally et al. (2011), suggesting a specific selection of this functional group by plants on the root surface, the most selective and rich habitat (Haichar et al., 2008). Under anaerobic conditions, Fe(III) oxide respiration may be considered a trait that allows bacteria to colonize plant roots, and a mutualistic interaction may take place whereby the plant provides root exudates for bacterial growth while the bacteria in participating iron availability for the plant.
With regard to iron, performing a denitrification process also offers competitive advantage for bacteria to colonize plant roots (Chèneby et al., 2004). Indeed, several authors have reported on the selection of the denitrifying community by plant roots (Henry et al., 2008; Guyonnet et al., 2017). However, only a few studies have evaluated the role of denitrification's function in the selection of microorganisms in the rhizosphere (Philippot et al., 1995; Ghiglione et al., 2000). Genes involved in denitrification have been evidenced to be involved in (i) the fitness of bacteria during anaerobic growth and (ii) rhizospheric competence. Indeed, nir or nor mutants of Nitrosomonas european presented limited growth under denitrification conditions (Schmidt et al., 2004), whereas nir and nor mutants of Parococcus denitrificans and Rhodobacter sphaeroides are unable to grow (De Boer et al., 1996; Bartnikas et al., 1997). Moreover, competition assays performed by Philippot et al. (1995) and Ghiglione et al. (2000, 2002) between wild type and nir or nar mutants of P. fluorescens YT101 on maize roots demonstrated that nirS and narG genes confer to the P. fluorescens YT101 strain a competitive advantage in rhizosphere colonization. In addition, Mirleau et al. (2001) demonstrated the role of nitrate reductase in the fitness of P. fluorescens C7R12 strain during in vitro and in planta assays (Mirleau et al., 2001). All these studies tend to evidence the role of denitrification genes (reductases) in rhizosphere competence.
Overall, to understand more deeply the advantage proffered by the reduction of TEAs, such as Fe(III) oxides in the rhizosphere for bacterial root colonization, a mutant of a model rhizobacterium affected in Fe(III) oxide reduction should be constructed,. As done previously for denitrification, an experiment based on comparing the mutant's and the wild type's (WT) fitness in the rhizosphere could be an easy and accurate way to evidence the importance of TEAs reduction in the rhizosphere competence. Further investigations are needed to prove the competitive advantage of TEA reducers within a complex rhizospheric microbial community.
Anaerobic processes by microorganisms are involved in the degradation of a wide range of pollutants named “detoxification” (Harwood et al., 1998). Bacterial anaerobic respiration is capable of using selective-priority pollutants as TEAs and reducing them to non-toxic compounds through the respiratory electron transfer chain (Williams et al., 2013; Nancharaiah and Lens 2015). This process plays an important role in the removal of contaminants and the remediation of soils. Several forms of bacterial anaerobic respiration and electron transfer related to the biotransformation of pollutants, including respiration with humic substances, sulfonates, halogenated chemicals, azo compounds, metallic elements, and non- metallic elements, were deeply reviewed by Hong and Gu (2009).
Non-metallic TEAs, such as As(V) and Se(IV), can be rendered unavailable from contaminated soils by anaerobic bacterial respiration. Indeed, As(V) is the major species of adsorbed arsenic found in contaminated-soils (Bissen and Frimmel, 2000). The reduction of this absorbed As(V) in more mobile and less adsorptive As(III) by dissimilar arsenate-reducing bacteria can facilitate As removal from soil's solid phase to an aqueous one and thus contribute to soil decontamination (Yamamura et al., 2007). In selenium-contaminated soils, highly toxic Se(V) can be reduced to less toxic Se(III) by certain selenium-reducing bacteria (Gerhardt et al., 1991). They so participate in the bioremediation of soil from Se(V). For certain bacteria, anaerobic reduction of toxic compounds does not always support growth in anaerobic conditions. In this case, anaerobic reduction of toxic oxyanions is coupled with another anaerobic respiration, such as Fe(III) and denitrification (Hunter and Kuykendall, 2007; Lee et al., 2012; Subedi et al., 2017).
The persistence of microorganisms relies on the ability to survive in the environment at low population density until conditions are more favorable (Kluepfel, 1993). Indeed, Fierer et al. (2012) suggested that adaptive mechanisms that facilitate better persistence for bacteria in adverse abiotic conditions are more important than competitive ones in shaping the microbial communities. In this respect, the ability of bacteria to grow in anaerobic conditions can be considered a persistence trait. Indeed, denitrification has been shown to contribute to bacterial persistence in extreme environments, such as the desert. Also, denitrifying enzymes are capable of tolerating extended periods of desiccation, allowing denitrifiers to better persist and to quickly respond to favorable conditions (Peterjohn, 1991). Regarding these results and the important differences of pO2 occurring in the rhizospheres, it is of great interest to demonstrate in what extant the anaerobic respiration of available TEAs contributes to bacterial persistence in this particular biotope.
The diversification of respiration in prokaryotes has made a major contribution to the ability of these microbes to colonize a wide range of environments from the oceans to the earth's crust and to adapt to changing environments, such as the rhizosphere. Improving our knowledge of bacterial respiratory flexibility in the rhizosphere is a major challenge, requiring a combination of a wide range of scientific expertise. Currently, the emergence of molecular approaches offers great potential for understanding respiratory processes at a rhizosphere scale. Approaches such as TnSeq (Perry and Yost, 2014) and CRISPR cas9 (Hsu et al., 2014) can be used for characterizing respiratory pathways in rhizospheric model strains, whereas metagenomics (Delgado-Baquerizo et al., 2018) and RNAseq (Newman et al., 2016) approaches can be applied to rhizospheric bacterial communities. Gaining a greater understanding of how bacterial anaerobic respiration participates in root colonization and environmental adaptation and persistence is an exciting research challenge. Finally, to better understand anaerobic respiration in the rhizosphere, it is crucial to consider all the participants, such as fungi and archaea.
SL and FH wrote the review, DA performed metatranscriptomic analyses and WA, TH, and XN reviewed and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We knowledge funding from the French National Research Agency (ANR-18-CE32-0005, DIORE). The authors gratefully thank Hamid BAYA for the design of the figures.
Achtnich, C., Bak, F., and Conrad, R. (1995). Competition for electron donors among nitrate reducers, ferric iron reducers, sulfate reducers, and methanogens in anoxic paddy soil. Biol. Fertil. Soils 19, 65–72. doi: 10.1007/BF00336349
Ackrell, B. A. C., Johnson, M. K., Gunsalus, R. P., and Cecchini, G. (1992). “Structure and function of succinate dehydrogenase and fumarate reductase,” in Chemistry and Biochemistry of Flavoenzymes, Vol. 3, ed F. Muller (Boca Raton, FL: CRC Press), 229–297.
Afkar, E., Lisak, J., Saltikov, C., Basu, P., Oremland, R. S., and Stolz, J. F. (2003). The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol. Lett. 226, 107–112. doi: 10.1016/S0378-1097(03)00609-8
Aklujkar, M., Coppi, M. V., Leang, C., Kim, B. C., Chavan, M. A., Perpetua, L. A., et al. (2013). Proteins involved in electron transfer to Fe(III) and Mn(IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 159, 515–535. doi: 10.1099/mic.0.064089-0
Akob, D. M., Mills, H. J., and Kostka, J. E. (2007). Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59, 95–107. doi: 10.1111/j.1574-6941.2006.00203.x
Alves, M. N., Neto, S. E., Alves, A. S., Fonseca, B. M., Carrêlo, A., Pacheco, I., et al. (2015). Characterization of the periplasmic redox network that sustains the versatile anaerobic metabolism of Shewanella oneidensis MR-1. Front. Microbiol. 6:665. doi: 10.3389/fmicb.2015.00665
Arkhipova, O. V., and Akimenko, V. K. (2005). Unsaturated organic acids as terminal electron acceptors for reductase chains of anaerobic bacteria. Microbiology 74, 629–639. doi: 10.1007/s11021-005-0116-6
Arth, I., and Frenzel, P. (2000). Nitrification and denitrification in the rhizosphere of rice: the detection of processes by a new multi-channel electrode. Biol. Fertil. Soils 31, 427–435. doi: 10.1007/s003749900190
Avazéri, C., Turner, R. J., Pommier, J., Weiner, J. H., Giordano, G., and Verméglio, A. (1997). Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology 143, 1181–1189. doi: 10.1099/00221287-143-4-1181
Bakken, L. R., Bergaust, L., Liu, B., and Frostegård, Å. (2012). Regulation of denitrification at the cellular level: a clue to the understanding of N2O emissions from soils. Philos. Transact. R. Soc. B 367, 1226–1234. doi: 10.1098/rstb.2011.0321
Bartnikas, T. B., Tosques, I. E., Laratta, W. P., Shi, J., and Shapleigh, J. P. (1997). Characterization of the nitric oxide reductase-encoding region in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 179, 3534–3540. doi: 10.1128/jb.179.11.3534-3540.1997
Bedmar, E. J., Robles, E. F., and Delgado, M. J. (2005). The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem. Soc. Transact. 33(Pt 1), 141–144. doi: 10.1042/BST0330141
Beliaev, A. S., Klingeman, D. M., Klappenbach, J. A., Wu, L., Romine, M. F., Tiedje, J. M., et al. (2005). Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J. Bacteriol. 187, 7138–7145. doi: 10.1128/JB.187.20.7138-7145.2005
Beliaev, A. S., and Saffarini, D. A. (1998). Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180, 6292–6297.
Beliaev, A. S., Saffarini, D. A., McLaughlin, J. L., and Hunnicutt, D. (2001). MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39, 722–730. doi: 10.1046/j.1365-2958.2001.02257.x
Bissen, M., and Frimmel, F. H. (2000). Speciation of As(III), As(V), MMA and DMA in contaminated soil extracts by HPLC-ICP/MS. Fresenius' J. Anal. Chem. 367, 51–55. doi: 10.1007/s002160051597
Bloom, A. J., Meyerhoff, P. A., Taylor, A. R., and Rost, T. L. (2002). Root development and absorption of ammonium and nitrate from the rhizosphere. J. Plant Growth Regul. 21, 416–431. doi: 10.1007/s00344-003-0009-8
Bretschger, O., Obraztsova, A., Sturm, C. A., Chang, I. S., Gorby, Y. A., Reed, S. B., et al. (2007). Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl. Environ. Microbiol. 73, 7003–7012. doi: 10.1128/AEM.01087-07
Bueno, E., Robles, E. F., Torres, M. J., Krell, T., Bedmar, E. J., Delgado, M. J., et al. (2017). Disparate response to microoxia and nitrogen oxides of the Bradyrhizobium japonicum napEDABC, nirK and norCBQD denitrification genes. Nitric Oxide Biol. Chem. 68, 137–149. doi: 10.1016/j.niox.2017.02.002
Burger, M., and Jackson, L. E. (2003). Microbial immobilization of ammonium and nitrate in relation to ammonification and nitrification rates in organic and conventional cropping systems. Soil Biol. Biochem. 35, 29–36. doi: 10.1016/S0038-0717(02)00233-X
Burns, J. L., and DiChristina, T. J. (2009). Anaerobic respiration of elemental sulfur and thiosulfate by Shewanella oneidensis MR-1 requires psrA, a homolog of the phsA Gene of Salmonella enterica serovar typhimurium LT2. Appl. Environ. Microbiol. 75, 5209–5217. doi: 10.1128/AEM.00888-09
Chèneby, D., Perrez, S., Devroe, C., Hallet, S., Couton, Y., Bizouard, F., et al. (2004). Denitrifying bacteria in bulk and maize-rhizospheric soil: diversity and N2O-reducing abilities. Can. J. Microbiol. 50, 469–474. doi: 10.1139/w04-037
Choudhary, S., and Sar, P. (2011). Uranium biomineralization by a metal resistant Pseudomonas aeruginosa strain isolated from contaminated mine waste. J. Hazard. Mater. 186, 336–343. doi: 10.1016/j.jhazmat.2010.11.004
Christensen, P. B., Revsbech, N. P., and Sand-Jensen, K. (1994). Microsensor analysis of oxygen in the rhizosphere of the aquatic macrophyte Littorella uniflora (L.) Ascherson. Plant Physiol. 105, 847–852. doi: 10.1104/pp.105.3.847
Colombo, C., Palumbo, G., He, J.-Z., Pinton, R., and Cesco, S. (2014). Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J. Soils Sediments 14, 538–548. doi: 10.1007/s11368-013-0814-z
Cornell, R. M., and Schwertmann, U. (2003). The Iron Oxides: Structure, Properties, Reactions, Occurrences and Uses. Weinheim: John Wiley & Sons.
Davison, W. (1993). Iron and manganese in lakes. Earth Sci. Rev. 34, 119–163. doi: 10.1016/0012-8252(93)90029-7
De Boer, A. P. N. D., Oost, J. V. D., Reijnders, W. N. M., Westerhoff, H. V., Stouthamer, A. H., and Spanning, R. J. M. V. (1996). Mutational analysis of the Nor gene cluster which encodes nitric-oxide reductase from Paracoccus denitrificans. Eur. J. Biochem. 242, 592–600. doi: 10.1111/j.1432-1033.1996.0592r.x
Debieux, C. M., Dridge, E. J., Mueller, C. M., Splatt, P., Paszkiewicz, K., Knight, I., et al. (2011). A bacterial process for selenium nanosphere assembly. Proc. Natl. Acad. Scie.U.S.A. 108, 13480–13485. doi: 10.1073/pnas.1105959108
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-González, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Science 359, 320–325. doi: 10.1126/science.aap9516
Domeignoz-Horta, L. A., Spor, A., Bru, D., Breuil, M.-C., Bizouard, F., Léonard, J., et al. (2015). The diversity of the N2O reducers matters for the N2O:N2 denitrification end-product ratio across an annual and a perennial cropping system. Front. Microbiol. 6:971. doi: 10.3389/fmicb.2015.00971
Eller, J., Hein, S., and Simon, J. (2018). Significance of MccR, MccC, MccD, MccL and 8-methylmenaquinone in sulfite respiration of Wolinella succinogenes. Biochim. Biophys. Acta. doi: 10.1016/j.bbabio.2018.10.002. [Epub ahead of print].
Farley, R. A., and Fitter, A. H. (1999). Temporal and spatial variation in soil resources in a deciduous woodland. J. Ecol. 87, 688–696. doi: 10.1046/j.1365-2745.1999.00390.x
Fierer, N., Leff, J. W., Adams, B. J., Nielsen, U. N., Bates, S. T., Lauber, C. L., et al. (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Scie.U.S.A. 109, 21390–21395. doi: 10.1073/pnas.1215210110
Fordyce, F. M. (2013). Selenium Deficiency and Toxicity in the Environment. In O. Selinus (Éd.), Essentials of Medical Geology: Revised Edition (p. 375–416). Dordrecht: Springer Netherlands. doi: 10.1007/978-94-007-4375-5_16
Gaimster, H., Alston, M., Richardson, D., Gates, A., and Rowley, G. (2018). Transcriptional and environmental control of bacterial denitrification and N2O emissions. FEMS Microbiol. Lett. 365:fnx277 doi: 10.1093/femsle/fnx277
Galloway, J. N., Townsend, A. R., Erisman, J. W., Bekunda, M., Cai, Z., Freney, J. R., et al. (2008). Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892. doi: 10.1126/science.1136674
Gavrilescu, M., Pavel, L. V., and Cretescu, I. (2009). Characterization and remediation of soils contaminated with uranium. J. Hazard. Mater. 163, 475–510. doi: 10.1016/j.jhazmat.2008.07.103
Gerhardt, M. B., Green, F. B., Newman, R. D., Lundquist, T. J., Tresan, R. B., and Oswald, W. J. (1991). Removal of selenium using a novel algal-bacterial process. Res. J. Water Poll. Contr. Feder. 63, 799–805.
Ghiglione, J.-F., Richaume, A., Philippot, L., and Lensi, R. (2002). Relative involvement of nitrate and nitrite reduction in the competitiveness of Pseudomonas fluorescens in the rhizosphere of maize under non-limiting nitrate conditions. FEMS Microbiol. Ecol. 39, 121–127. doi: 10.1111/j.1574-6941.2002.tb00913.x
Ghiglione, J. F., Gourbiere, F., Potier, P., Philippot, L., and Lensi, R. (2000). Role of respiratory nitrate reductase in ability of Pseudomonas fluorescens YT101 to colonize the rhizosphere of maize. Appl. Environ. Microbiol. 66, 4012–4016. doi: 10.1128/AEM.66.9.4012-4016.2000
Giannopoulos, G., Sullivan, M. J., Hartop, K. R., Rowley, G., Gates, A. J., Watmough, N. J., et al. (2017). Tuning the modular Paracoccus denitrificans respirome to adapt from aerobic respiration to anaerobic denitrification. Environ. Microbiol. 19, 4953–4964. doi: 10.1111/1462-2920.13974
Giehl, R. F. H., Wirén, N., and von (2014). Root nutrient foraging. Plant Physiol. 114:245225. doi: 10.1104/pp.114.245225
Glasser, N. R., Oyala, P. H., Osborne, T. H., Santini, J. M., and Newman, D. K. (2018). Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations. Proc. Natl. Acad. Scie.U.S.A. 115, E8614–E8623. doi: 10.1073/pnas.1807984115
Glinski, J., and Stepniewski, W. (1985). Soil Aeration and Its Role for Plants. Boca Raton, FL: CRC Press. Available online at: http://agris.fao.org/agris-search/search.do?recordID=US880872588
González, A. Z. I., Krachler, M., Cheburkin, A. K., and Shotyk, W. (2006). Spatial distribution of natural enrichments of arsenic, selenium, and uranium in a minerotrophic peatland, Gola di Lago, Canton Ticino, Switzerland. Environ. Sci. Technol. 40, 6568–6574. doi: 10.1021/es061080v
González, P. J., Correia, C., Moura, I., Brondino, C. D., and Moura, J. J. G. (2006). Bacterial nitrate reductases: Molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 100, 1015–1023. doi: 10.1016/j.jinorgbio.2005.11.024
Gori, F., Tringe, S. G., Kartal, B., Machiori, E., and Jetten, M. S. M. (2011). The metagenomic basis of anammox metabolism in Candidatus ‘Brocadia fulgida’. Biochem. Soc. Trans. 39, 1799–1804. doi: 10.1042/BST20110707
Guyonnet, J. P., Vautrin, F., Meiffren, G., Labois, C., Cantarel, A. A. M., Michalet, S., et al. (2017). The effects of plant nutritional strategy on soil microbial denitrification activity through rhizosphere primary metabolites. FEMS Microbiol. Ecol. 93:4. doi: 10.1093/femsec/fix022
Haichar, F., el, Z., Marol, C., Berge, O., Rangel-Castro, J. I., Prosser, J. I., et al. (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2, 1221–1230. doi: 10.1038/ismej.2008.80
Haichar, F., el, Z., Santaella, C., Heulin, T., and Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80. doi: 10.1016/j.soilbio.2014.06.017
Hallin, S., Philippot, L., Löffler, F. E., Sanford, R. A., and Jones, C. M. (2017). Genomics and ecology of novel N2O-reducing microorganisms. Trends Microbiol. 26, 43–55. doi: 10.1016/j.tim.2017.07.003
Härtig, E., and Zumft, W. G. (1999). Kinetics of nirS expression (cytochrome cd1 nitrite reductase) in Pseudomonas stutzeri during the transition from aerobic respiration to denitrification: evidence for a denitrification-specific nitrate- and nitrite-responsive regulatory system. J. Bacteriol. 181, 161–166.
Hartshorne, R. S., Reardon, C. L., Ross, D., Nuester, J., Clarke, T. A., Gates, A. J., et al. (2009). Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci.U.S.A. 106, 22169–22174. doi: 10.1073/pnas.0900086106
Hartsock, A., and Shapleigh, J. P. (2010). Identification, functional studies, and genomic comparisons of new members of the NnrR regulon in Rhodobacter sphaeroides. J. Bacteriol. 192, 903–911. doi: 10.1128/JB.01026-09
Harwood, C. S., Burchhardt, G., Herrmann, H., and Fuchs, G. (1998). Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22, 439–458. doi: 10.1111/j.1574-6976.1998.tb00380.x
Hedderich, R., Klimmek, O., Kröger, A., Dirmeier, R., Keller, M., and Stetter, K. O. (1998). Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol. Rev. 22, 353–381. doi: 10.1111/j.1574-6976.1998.tb00376.x
Henry, S., Texier, S., Hallet, S., Bru, D., Dambreville, C., Chèneby, D., et al. (2008). Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ. Microbiol. 10, 3082–3092. doi: 10.1111/j.1462-2920.2008.01599.x
Hino, T., Matsumoto, Y., Nagano, S., Sugimoto, H., Fukumori, Y., Murata, T., et al. (2010). Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 330, 1666–1670. doi: 10.1126/science.1195591
Hinsinger, P., Bengough, A. G., Vetterlein, D., and Young, I. M. (2009). Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321, 117–152. doi: 10.1007/s11104-008-9885-9
Hollibaugh, J. T., Carini, S., Gürleyük, H., Jellison, R., Joye, S. B., LeCleir, G., et al. (2005). Arsenic speciation in Mono Lake, California: response to seasonal stratification and anoxia. Geochim. Cosmochim. Acta 69, 1925–1937. doi: 10.1016/j.gca.2004.10.011
Hong, Y., and Gu, J.-D. (2009). Bacterial anaerobic respiration and electron transfer relevant to the biotransformation of pollutants. Int. Biodeterior. Biodegr. 63, 973–980. doi: 10.1016/j.ibiod.2009.08.001
Hong, Y. G., Guo, J., Xu, Z. C., Xu, M. Y., and Sun, G. P. (2007). Humic substances act as electron acceptor and redox mediator for microbial dissimilatory azoreduction by Shewanella decolorationis S12. J. Microbiol. Biotechnol. 17, 428–437.
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Hu, B., Shen, L., Xu, X., and Zheng, P. (2011). Anaerobic ammonium oxidation (anammox) in different natural ecosystems. Biochem. Soc. Trans. 39, 1811–1816. doi: 10.1042/BST20110711
Hu, H.-W., Chen, D., and He, J.-Z. (2015). Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol. Rev. 39, 729–749. doi: 10.1093/femsre/fuv021
Humbert, S., Tarnawski, S., Fromin, N., Mallet, M.-P., Aragno, M., and Zopfi, J. (2010). Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. ISME J. 4, 450–454. doi: 10.1038/ismej.2009.125
Hunter, W. J., and Kuykendall, L. D. (2007). Reduction of selenite to elemental red selenium by Rhizobium sp. strain B1. Curr. Microbiol. 55, 344–349. doi: 10.1007/s00284-007-0202-2
Jackson, R. B., and Caldwell, M. M. (1993). The scale of nutrient heterogeneity around individual plants and its quantification with geostatistics. Ecology 74, 612–614. doi: 10.2307/1939320
Jones, C. M., and Hallin, S. (2018). Geospatial variation in co-occurrence networks of nitrifying microbial guilds. Mol. Ecol. doi: 10.1111/mec.14893. [Epub ahead of print].
Jones, H. M., and Gunsalus, R. P. (1987). Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J. Bacteriol. 169, 3340–3349.
Jousset, A., Becker, J., Chatterjee, S., Karlovsky, P., Scheu, S., and Eisenhauer, N. (2014). Biodiversity and species identity shape the antifungal activity of bacterial communities. Ecology 95, 1184–1190. doi: 10.1890/13-1215.1
Kai, M., Effmert, U., and Piechulla, B. (2016). Bacterial-plant-interactions: approaches to unravel the biological function of bacterial volatiles in the rhizosphere. Front. Microbiol. 7:108. doi: 10.3389/fmicb.2016.00108
Kartal, B., Maalcke, W. J., de Almeida, N. M., Cirpus, I., Gloerich, J., Geerts, W., et al. (2011). Molecular mechanism of anaerobic ammonium oxidation. Nature 479, 127–130. doi: 10.1038/nature10453
Kasai, T., Kouzuma, A., Nojiri, H., and Watanabe, K. (2015). Transcriptional mechanisms for differential expression of outer membrane cytochrome genes omcA and mtrC in Shewanella oneidensis MR-1. BMC Microbiol. 15:68. doi: 10.1186/s12866-015-0406-8
Kertesz, M. A. (2000). Riding the sulfur cycle – metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 24, 135–175. doi: 10.1016/S0168-6445(99)00033-9
Klimmek, O., Kröger, A., Steudel, R., and Holdt, G. (1991). Growth of Wolinella succinogenes with polysulphide as terminal acceptor of phosphorylative electron transport. Arch. Microbiol. 155, 177–182. doi: 10.1007/BF00248614
Klonowska, A., Heulin, T., and Vermeglio, A. (2005). Selenite and Tellurite Reduction by Shewanella oneidensis. Appl. Environ. Microbiol. 71, 5607–5609. doi: 10.1128/AEM.71.9.5607-5609.2005
Kluepfel, D. A. (1993). The behavior and tracking of bacteria in the rhizosphere. Annu. Rev. Phytopathol. 31, 441–472. doi: 10.1146/annurev.py.31.090193.002301
Knight, V. K., Nijenhuis, I., Kerkhof, L. J., and Häggblom, M. M. (2002). Degradation of aromatic compounds coupled to selenate reduction. Geomicrobiol. J. 19, 77–86. doi: 10.1080/014904502317246183
Körner, H., Sofia, H. J., and Zumft, W. G. (2003). Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27, 559–592. doi: 10.1016/S0168-6445(03)00066-4
Kostka, J. E., Dalton, D. D., Skelton, H., Dollhopf, S., and Stucki, J. W. (2002). Growth of iron(III)-reducing bacteria on clay minerals as the sole electron acceptor and comparison of growth yields on a variety of oxidized iron forms. Appl. Environ. Microbiol. 68, 6256–6262. doi: 10.1128/AEM.68.12.6256-6262.2002
Krafft, T., Gross, R., and Kröger, A. (1995). The Function of Wolinella succinogenes psr genes in electron transport with polysulphide as the terminal electron acceptor. Eur. J. Biochem. 230, 601–606. doi: 10.1111/j.1432-1033.1995.0601h.x
Krafft, T., and Macy, J. M. (1998). Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur. J. Biochem. 255, 647–653. doi: 10.1046/j.1432-1327.1998.2550647.x
Kraft, B., Strous, M., and Tegetmeyer, H. E. (2011). Microbial nitrate respiration–genes, enzymes and environmental distribution. J. Biotechnol. 155, 104–117. doi: 10.1016/j.jbiotec.2010.12.025
Kröger, A., Geisler, V., Lemma, E., Theis, F., and Lenger, R. (1992). Bacterial fumarate respiration. Arch. Microbiol. 158, 311–314. doi: 10.1007/BF00245358
Kröger, A. (1978). Fumarate as terminal acceptor of phosphorylative electron transport. Biochim. Biophys. Acta 505, 129–145. doi: 10.1016/0304-4173(78)90010-1
Kuroda, M., Yamashita, M., Miwa, E., Imao, K., Fujimoto, N., Ono, H., et al. (2011). Molecular cloning and characterization of the srdbca operon encoding the respiratory selenate reductase complex from the selenate-reducing Bacterium, Bacillus selenatarsenatis SF-1. J. Bacteriol. 193, 2141–2148. doi: 10.1128/JB.01197-10
Kuypers, M. M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Kuzyakov, Y., and Xu, X. (2013). Competition between roots and microorganisms for nitrogen: mechanisms and ecological relevance. New Phytol. 198, 656–669. doi: 10.1111/nph.12235
Lalucat, J., Bennasar, A., Bosch, R., García-Valdés, E., and Palleroni, N. J. (2006). Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 70, 510–547. doi: 10.1128/MMBR.00047-05
Lark, R. M., Milne, A. E., Addiscott, T. M., Goulding, K. W. T., Webster, C. P., and O'Flaherty, S. (2004). Scale- and location-dependent correlation of nitrous oxide emissions with soil properties: an analysis using wavelets. Eur. J. Soil Sci. 55, 611–627. doi: 10.1111/j.1365-2389.2004.00620.x
Lee, K.-Y., Bosch, J., and Meckenstock, R. U. (2012). Use of metal-reducing bacteria for bioremediation of soil contaminated with mixed organic and inorganic pollutants. Environ. Geochem. Health 34, 135–142. doi: 10.1007/s10653-011-9406-2
Lee, M., and Yang, M. (2010). Rhizofiltration using sunflower (Helianthus annuus L.) and bean (Phaseolus vulgaris L. var. vulgaris) to remediate uranium contaminated groundwater. J. Hazard. Mater. 173, 589–596. doi: 10.1016/j.jhazmat.2009.08.127
Lemanceau, P., Bauer, P., Kraemer, S., and Briat, J.-F. (2009). Iron dynamics in the rhizosphere as a case study for analyzing interactions between soils, plants and microbes. Plant Soil 321, 513–535. doi: 10.1007/s11104-009-0039-5
Li, D.-B., Cheng, Y.-Y., Wu, C., Li, W.-W., Li, N., Yang, Z.-C., et al. (2014). Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm. Sci. Rep. 4:3735. doi: 10.1038/srep03735
Li, H., Yang, X., Weng, B., Su, J., Nie, S., Gilbert, J. A., et al. (2016). The phenological stage of rice growth determines anaerobic ammonium oxidation activity in rhizosphere soil. Soil Biol. Biochem. 100, 59–65. doi: 10.1016/j.soilbio.2016.05.015
Liu, Y., Zhang, N., Qiu, M., Feng, H., Vivanco, J. M., Shen, Q., et al. (2014). Enhanced rhizosphere colonization of beneficial Bacillus amyloliquefaciens SQR9 by pathogen infection. FEMS Microbiol. Lett. 353, 49–56. doi: 10.1111/1574-6968.12406
Lloyd, J. R., Leang, C., Myerson, A. L. H., Coppi, M. V., Cuifo, S., Methe, B., et al. (2003). Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369, 153–161. doi: 10.1042/bj20020597
Losi, M. E., and Frankenberger, W. T. (1997). Reduction of selenium oxyanions by Enterobacter cloacae SLD1a-1: isolation and growth of the bacterium and its expulsion of selenium particles. Appl. Environ. Microbiol. 63, 3079–3084.
Lovley, D. R., and Coates, J. D. (2000). Novel forms of anaerobic respiration of environmental relevance. Curr. Opin. Microbiol. 3, 252–256. doi: 10.1016/S1369-5274(00)00085-0
Lovley, D. R., Coates, J. D., Blunt-Harris, E. L., Phillips, E. J. P., and Woodward, J. C. (1996). Humic substances as electron acceptors for microbial respiration. Nature 382, 445–448. doi: 10.1038/382445a0
Lovley, D. R., Holmes, D. E., and Nevin, K. P. (2004). Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49, 219–286. doi: 10.1016/S0065-2911(04)49005-5
Lovley, D. R., and Phillips, E. J. P. (1987). Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53, 1536–1540.
Lovley, D. R., Phillips, E. J. P., Gorby, Y. A., and Landa, E. R. (1991). Microbial reduction of uranium. Nature 350, 413–416. doi: 10.1038/350413a0
Macy, J. M., Nunan, K., Hagen, K. D., Dixon, D. R., Harbour, P. J., Cahill, M., et al. (1996). Chrysiogenes arsenatis gen. nov., sp. nov., a new arsenate-respiring bacterium isolated from gold mine wastewater. Int. J. Syst. Bacteriol. 46, 1153–1157. doi: 10.1099/00207713-46-4-1153
Maklashina, E., Cecchini, G., and Dikanov, S. A. (2013). Defining a direction: electron transfer and catalysis in Escherichia coli complex II enzymes. Biochim. Biophys. Acta 1827, 668–678. doi: 10.1016/j.bbabio.2013.01.010
Markich, S. J. (2002). Uranium speciation and bioavailability in aquatic systems: an overview. Sci. World J. 2, 707–729. doi: 10.1100/tsw.2002.130
Marschner, H. (1988). “Mechanisms of manganese acquisition by roots from soils,” in Manganese in Soils and Plants: Proceedings of the International Symposium on ‘Manganese in Soils and Plants’ held at the Waite Agricultural Research Institute, The University of Adelaide, Glen Osmond, South Australia, August 22–26, 1988 as an Australian Bicentennial Event, eds R. D. Graham, R. J. Hannam, and N. C. Uren (Dordrecht: Springer Netherlands), 191–204. doi: 10.1007/978-94-009-2817-6_14
Marshall, M. J., Beliaev, A. S., Dohnalkova, A. C., Kennedy, D. W., Shi, L., Wang, Z., et al. (2006). c-type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 4:e268. doi: 10.1371/journal.pbio.0040268
Mehta, T., Coppi, M. V., Childers, S. E., and Lovley, D. R. (2005). Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in geobacter sulfurreducens. Appl. Environ. Microbiol.71, 8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005
Mirleau, P., Philippot, L., Corberand, T., and Lemanceau, P. (2001). Involvement of nitrate reductase and pyoverdine in competitiveness of Pseudomonas fluorescens Strain C7R12 in Soil. Appl. Environ. Microbiol. 67, 2627–2635. doi: 10.1128/AEM.67.6.2627-2635.2001
Mohan, D., Pittman, C. U., Bricka, M., Smith, F., Yancey, B., Mohammad, J., et al. (2007). Sorption of arsenic, cadmium, and lead by chars produced from fast pyrolysis of wood and bark during bio-oil production. J. Colloid Interface Sci. 310, 57–73. doi: 10.1016/j.jcis.2007.01.020
Moreau, D., Pivato, B., Bru, D., Busset, H., Deau, F., Faivre, C., et al. (2015). Plant traits related to nitrogen uptake influence plant-microbe competition. Ecology 96, 2300–2310. doi: 10.1890/14-1761.1
Moreno-Vivián, C., Cabello, P., Martínez-Luque, M., Blasco, R., and Castillo, F. (1999). Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 181, 6573–6584.
Mouret, A., Anschutz, P., Lecroart, P., Chaillou, G., Hyacinthe, C., Deborde, J., et al. (2009). Benthic geochemistry of manganese in the Bay of Biscay, and sediment mass accumulation rate. Geo Mar. Lett. 29, 133–149. doi: 10.1007/s00367-008-0130-6
Myers, C. R., and Myers, J. M. (1997). Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol. 179, 1143–1152.
Naeher, S., Huguet, A., Roose-Amsaleg, C. L., Laverman, A. M., Fosse, C., Lehmann, M. F., et al. (2015). Molecular and geochemical constraints on anaerobic ammonium oxidation (anammox) in a riparian zone of the Seine Estuary (France). Biogeochemistry 123, 237–250. doi: 10.1007/s10533-014-0066-z
Nancharaiah, Y. V., and Lens, P. N. L. (2015). Ecology and biotechnology of selenium-respiring bacteria. Microbiol. Mol. Biol. Rev. 79, 61–80. doi: 10.1128/MMBR.00037-14
Nealson, K., and H Saffarini, D. (1994). Iron and manganese in anaerobic respiration: environmental significance, physiology, and regulation. Annu. Rev. Microbiol. 48, 311–343. doi: 10.1146/annurev.mi.48.100194.001523
Neira, J., Ortiz, M., Morales, L., and Acevedo, E. (2015). Oxygen diffusion in soils: understanding the factors and processes needed for modeling. Chil. J. Agr. Res. 75, 35–44. doi: 10.4067/S0718-58392015000300005.
Newman, M. M., Lorenz, N., Hoilett, N., Lee, N. R., Dick, R. P., Liles, M. R., et al. (2016). Changes in rhizosphere bacterial gene expression following glyphosate treatment. Sci. Total Environ. 553, 32–41. doi: 10.1016/j.scitotenv.2016.02.078
Nie, S., Li, H., Yang, X., Zhang, Z., Weng, B., Huang, F., et al. (2015). Nitrogen loss by anaerobic oxidation of ammonium in rice rhizosphere. ISME J. 9, 2059–2067. doi: 10.1038/ismej.2015.25
Nie, S., Zhu, G.-B., Singh, B., and Zhu, Y.-G. (2018). Anaerobic ammonium oxidation in agricultural soils-synthesis and prospective. Environ. Pollut. 244, 127–134. doi: 10.1016/j.envpol.2018.10.050
Oremland, R. S., Dowdle, P. R., Hoeft, S., Sharp, J. O., Schaefer, J. K., Miller, L. G., et al. (2000). Bacterial dissimilatory reduction of arsenate and sulfate in meromictic Mono Lake, California. Geochim. Cosmochim. Acta 64, 3073–3084. doi: 10.1016/S0016-7037(00)00422-1
Oremland, R. S., Kulp, T. R., Blum, J. S., Hoeft, S. E., Baesman, S., Miller, L. G., et al. (2005). A microbial arsenic cycle in a salt-saturated, extreme environment. Science 308, 1305–1308. doi: 10.1126/science.1110832
Perry, B. J., and Yost, C. K. (2014). Construction of a mariner-based transposon vector for use in insertion sequence mutagenesis in selected members of the Rhizobiaceae. BMC Microbiol. 14:298. doi: 10.1186/s12866-014-0298-z
Peterjohn, W. T. (1991). Denitrification: enzyme content and activity in desert soils. Soil Biol. Biochem. 23, 845–855. doi: 10.1016/0038-0717(91)90096-3
Petersen, W., and Böttger, M. (1991). Contribution of organic acids to the acidification of the rhizosphere of maize seedlings. Plant Soil 132, 159–163. doi: 10.1007/BF00010396
Philippot, L. (2002). Denitrifying genes in bacterial and Archaeal genomes. Biochim. Biophys. Acta 1577, 355–376. doi: 10.1016/S0167-4781(02)00420-7
Philippot, L., Clays-Josserand, A., and Lensi, R. (1995). Use of Tn5 mutants to assess the role of the dissimilatory nitrite reductase in the competitive abilities of two pseudomonas strains in soil. Appl. Environ. Microbiol. 61, 1426–1430.
Philippot, L., and Højberg, O. (1999). Dissimilatory nitrate reductases in bacteria. Biochim. Biophys. Acta 1446, 1–23.
Philippot, L., Mirleau, P., Mazurier, S., Siblot, S., Hartmann, A., Lemanceau, P., et al. (2001). Characterization and transcriptional analysis of Pseudomonas fluorescens denitrifying clusters containing the nar, nir, nor and nos genes. Biochim. Biophys. Acta 1517, 436–440. doi: 10.1016/S0167-4781(00)00286-4
Philippot, L., Raaijmakers, J. M., Lemanceau, P., Putten, W. H., and van der (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Philippot, L., Renault, P., Sierra, J., Hénault, C., Clays-Josserand, A., Chenu, C., et al. (1996). Dissimilatory nitrite-reductase provides a competitive advantage to Pseudomonas sp. RTC01 to colonise the centre of soil aggregates. FEMS Microbiol. Ecol. 21, 175–185. doi: 10.1111/j.1574-6941.1996.tb00345.x
Post, J. E. (1999). Manganese oxide minerals: crystal structures and economic and environmental significance. Proc. Natl Acad. Sci.U.S.A. 96, 3447–3454. doi: 10.1073/pnas.96.7.3447
Posta, K., Marschner, H., and Römheld, V. (1994). Manganese reduction in the rhizosphere of mycorrhizal and nonmycorrhizal maize. Mycorrhiza 5, 119–124. doi: 10.1007/BF00202343
Rabenstein, D. L., and Tan, K.-S. (1988). 77Se NMR studies of bis(alkylthio)selenides of biological thiols. Mag. Res. Chem. 26, 1079–1085. doi: 10.1002/mrc.1260261209
Reyes, F., Gavira, M., Castillo, F., and Moreno-Vivián, C. (1998). Periplasmic nitrate-reducing system of the phototrophic bacterium Rhodobacter sphaeroides DSM 158: transcriptional and mutational analysis of the napKEFDABC gene cluster. Biochem. J. 331, 897–904. doi: 10.1042/bj3310897
Richardson, D. J. (2000). Bacterial respiration: a flexible process for a changing environment. Microbiology 146, 551–571. doi: 10.1099/00221287-146-3-551
Richey, C., Chovanec, P., Hoeft, S. E., Oremland, R. S., Basu, P., and Stolz, J. F. (2009). Respiratory arsenate reductase as a bidirectional enzyme. Biochem. Biophys. Res. Commun. 382, 298–302. doi: 10.1016/j.bbrc.2009.03.045
Runkles, J. R. (1956). Diffusion, Sorption and Depth Distribution of Oxygen in Soils. Retrospective theses and dissertations, Iowa State University.
Sabaty, M., Avazeri, C., Pignol, D., and Vermeglio, A. (2001). Characterization of the reduction of selenate and tellurite by nitrate reductases. Appl. Environ. Microbiol. 67, 5122–5126. doi: 10.1128/AEM.67.11.5122-5126.2001
Sánchez, C., and Minamisawa, K. (2018). Redundant roles of Bradyrhizobium oligotrophicum Cu-type (NirK) and cd1-type (NirS) nitrite reductase genes under denitrifying conditions. FEMS Microbiol. Lett. 365:5. doi: 10.1093/femsle/fny015
Sanford, R. A., Cole, J. R., and Tiedje, J. M. (2002). Characterization and description of anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 68, 893–900. doi: 10.1128/AEM.68.2.893-900.2002
Schauder, R., and Kröger, A. (1993). Bacterial sulphur respiration. Arch. Microbiol. 159, 491–497. doi: 10.1007/BF00249025
Scherer, H W. (2009). Sulfur in soils. J. Plant Nutr. Soil Sci. 172, 326–335. doi: 10.1002/jpln.200900037
Scherer, H. W. (2001). Sulphur in crop production — invited paper. Eur. J. Agronom. 14, 81–111. doi: 10.1016/S1161-0301(00)00082-4
Schmidt, I., van Spanning, R. J. M., and Jetten, M. S. M. (2004). Denitrification and ammonia oxidation by Nitrosomonas europaea wild-type, and NirK- and NorB-deficient mutants. Microbiology 150, 4107–4114. doi: 10.1099/mic.0.27382-0
Sexstone, A. J., Revsbech, N. P., Parkin, T. B., and Tiedje, J. M. (1985). Direct measurement of oxygen profiles and denitrification rates in soil aggregates 1. Soil Sci. Soc. Am. J. 49, 645–651. doi: 10.2136/sssaj1985.03615995004900030024x
Shah, Z. H., Rehman, H. M., Akhtar, T., Alsamadany, H., Hamooh, B. T., Mujtaba, T., et al. (2018). Humic substances: determining potential molecular regulatory processes in plants. Front. Plant Sci. 9:263. doi: 10.3389/fpls.2018.00263
Simon, J., Einsle, O., Kroneck, P. M. H., and Zumft, W. G. (2004). The unprecedented nos gene cluster of Wolinella succinogenes encodes a novel respiratory electron transfer pathway to cytochrome c nitrous oxide reductase. FEBS Lett. 569:7–12. doi: 10.1016/j.febslet.2004.05.060
Somenahally, A. C., Hollister, E. B., Yan, W., Gentry, T. J., and Loeppert, R. H. (2011). Water management impacts on arsenic speciation and iron-reducing bacteria in contrasting rice-rhizosphere compartments. Environ. Sci. Technol. 45, 8328–8335. doi: 10.1021/es2012403
Stepniewski, W., and Stepniewska, Z. (2009). Selected oxygen-dependent process—Response to soil management and tillage. Soil Tillage Res. 102, 193–200. doi: 10.1016/j.still.2008.07.006
Stolz, J. F., Basu, P., Santini, J. M., and Oremland, R. S. (2006). Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60, 107–130. doi: 10.1146/annurev.micro.60.080805.142053
Strous, M., Pelletier, E., Mangenot, S., Rattei, T., Lehner, A., Taylor, M. W., et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794. doi: 10.1038/nature04647
Subedi, G., Taylor, J., Hatam, I., and Baldwin, S. A. (2017). Simultaneous selenate reduction and denitrification by a consortium of enriched mine site bacteria. Chemosphere,183, 536–545. doi: 10.1016/j.chemosphere.2017.05.144
Switzer Blum, S. J., Burns Bindi, A., Buzzelli, J., Stolz, J. F., and Oremland, R. S. (1998). Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171, 19–30.
Tanaka, T. (1988). Distribution of arsenic in the natural environment with emphasis on rocks and soils. Appl. Organomet. Chem. 2, 283–295. doi: 10.1002/aoc.590020403
Taratus, E. M., Eubanks, S. G., and Dichristina, T. J. (2000). Design and application of a rapid screening technique for isolation of selenite reduction-deficient mutants of Shewanella putrefaciens. Microbiol. Res. 155, 79–85. doi: 10.1016/S0944-5013(00)80041-5
Tebo, B. M., and Obraztsova, A. Y. (1998). Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162, 193–198. doi: 10.1111/j.1574-6968.1998.tb12998.x
Thauer, R. K., Zinkhan, D. M., and Spormann, A. M. (1989). Biochemistry of acetate catabolism in anaerobic chemotrophic bacteria. Annu. Rev. Microbiol. 43, 43–67. doi: 10.1146/annurev.mi.43.100189.000355
Thomson, A. J., Giannopoulos, G., Pretty, J., Baggs, E. M., and Richardson, D. J. (2012). Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos. Transact. R. Soc. B Biol. Sci. 367, 1157–1168. doi: 10.1098/rstb.2011.0415
Torres, M. J., Argandoña, M., Vargas, C., Bedmar, E. J., Fischer, H. M., Mesa, S., et al. (2014). The global response regulator RegR controls expression of denitrification genes in Bradyrhizobium japonicum. PLoS ONE, 9:e99011. doi: 10.1371/journal.pone.0099011
Tosques, I. E., Kwiatkowski, A. V., Shi, J., and Shapleigh, J. P. (1997). Characterization and regulation of the gene encoding nitrite reductase in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 179, 1090–1095. doi: 10.1128/jb.179.4.1090-1095.1997
Trevisan, S., Francioso, O., Quaggiotti, S., and Nardi, S. (2010). Humic substances biological activity at the plant-soil interface. Plant Signal. Behav. 5, 635–643. doi: 10.4161/psb.5.6.11211
Turner, R. J., Weiner, J. H., and Taylor, D. E. (1998). Selenium metabolism in Escherichia coli. Biometals 11, 223–227.
Tzortzis, M., and Tsertos, H. (2004). Determination of thorium, uranium and potassium elemental concentrations in surface soils in Cyprus. J. Environ. Radioact. 77, 325–338. doi: 10.1016/j.jenvrad.2004.03.014
Uren, N. C. (2013). “Cobalt and manganese,” inHeavy Metals in Soils: Trace Metals and Metalloids in Soils and their Bioavailability ed B. J. Alloway (Dordrecht: Springer Netherlands), 335–366. doi: 10.1007/978-94-007-4470-7_12
Uteau, D., Hafner, S., Pagenkemper, S. K., Peth, S., Wiesenberg, G. L. B., Kuzyakov, Y., et al. (2015). Oxygen and redox potential gradients in the rhizosphere of alfalfa grown on a loamy soil. J. Plant Nutr. Soil Sci. 178, 278–287. doi: 10.1002/jpln.201300624
Uteau, D., Pagenkemper, S. K., Peth, S., and Horn, R. (2013). Root and time dependent soil structure formation and its influence on gas transport in the subsoil. Soil Tillage Res. 132, 69–76. doi: 10.1016/j.still.2013.05.001
van de Graaf, A. A., de Bruijn, P., Robertson, L. A., Jetten, M. S. M., and Kuenen, J. G. (1996). Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology 142, 2187–2196. doi: 10.1099/13500872-142-8-2187
van de Graaf, A. A., Mulder, A., de Bruijn, P., Jetten, M. S., Robertson, L. A., and Kuenen, J. G. (1995). Anaerobic oxidation of ammonium is a biologically mediated process. Appl. Environ. Microbiol. 61, 1246–1251.
Vollack, K.-U., and Zumft, W. G. (2001). Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J. Bacteriol. 183, 2516–2526. doi: 10.1128/JB.183.8.2516-2526.2001
Voordeckers, J. W., Kim, B.-C., Izallalen, M., and Lovley, D. R. (2010). Role of geobacter sulfurreducens outer surface c-type cytochromes in reduction of soil humic acid and anthraquinone-2,6-disulfonate. Appl. Environ. Microbiol. 76, 2371–2375. doi: 10.1128/AEM.02250-09
Wade, R., and DiChristina, T. J. (2000). Isolation of U(VI) reduction-deficient mutants of Shewanella putrefaciens. FEMS Microbiol. Lett. 184, 143–148. doi: 10.1111/j.1574-6968.2000.tb09005.x
Wang, J., Solomon, D., Lehmann, J., Zhang, X., and Amelung, W. (2006). Soil organic sulfur forms and dynamics in the Great Plains of North America as influenced by long-term cultivation and climate. Geoderma 133, 160–172. doi: 10.1016/j.geoderma.2005.07.003
Watts, C. A., Ridley, H., Condie, K. L., Leaver, J. T., Richardson, D. J., and Butler, C. S. (2003). Selenate reduction by Enterobacter cloacae SLD1a-1 is catalysed by a molybdenum-dependent membrane-bound enzyme that is distinct from the membrane-bound nitrate reductase. FEMS Microbiol. Lett. 228, 273–279. doi: 10.1016/S0378-1097(03)00782-1
Weber, K. A., Achenbach, L. A., and Coates, J. D. (2006). Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 4, 752–764. doi: 10.1038/nrmicro1490
Williams, K. H., Wilkins, M. J., N'Guessan, A. L., Arey, B., Dodova, E., Dohnalkova, A., et al. (2013). Field evidence of selenium bioreduction in a uranium-contaminated aquifer. Environ. Microbiol. Rep. 5, 444–452. doi: 10.1111/1758-2229.12032
Yamamura, S., and Amachi, S. (2014). Microbiology of inorganic arsenic: from metabolism to bioremediation. J. Biosci. Bioeng. 118, 1–9. doi: 10.1016/j.jbiosc.2013.12.011
Yamamura, S., Yamashita, M., Fujimoto, N., Kuroda, M., Kashiwa, M., Sei, K., et al. (2007). Bacillus selenatarsenatis sp. nov., a selenate- and arsenate-reducing bacterium isolated from the effluent drain of a glass-manufacturing plant. Int. J. Syst. Evol. Microbiol. 57, 1060–1064. doi: 10.1099/ijs.0.64667-0
Yan, X., Luo, X., and Zhao, M. (2016). Metagenomic analysis of microbial community in uranium-contaminated soil. Appl. Microbiol. Biotechnol. 100, 299–310. doi: 10.1007/s00253-015-7003-5
Yan-Chu, H. (1994). “Arsenic distribution in soils,” in Arsenic in the Environment, Part 1, ed J. O. Nriagu, 17–49.
Yee, N., Ma, J., Dalia, A., Boonfueng, T., and Kobayashi, D. Y. (2007). Se(VI) Reduction and the precipitation of Se(0) by the facultative Bacterium Enterobacter cloacae SLD1a-1 Are Regulated by FNR. Appl. Environ. Microbiol., 73, 1914–1920. doi: 10.1128/AEM.02542-06
Yoon, S., Sanford, R. A., and Löffler, F. E. (2013). Shewanella spp. use acetate as an electron donor for denitrification but not ferric iron or fumarate reduction. Appl. Environ. Microbiol. 79, 2818–2822. doi: 10.1128/AEM.03872-12
Yuan, J., Zhang, N., Huang, Q., Raza, W., Li, R., Vivanco, J. M., et al. (2015). Organic acids from root exudates of banana help root colonization of PGPR strain Bacillus amyloliquefaciens NJN-6. Sci. Rep. 5:13438. doi: 10.1038/srep13438
Zausig, J., Stepniewski, W., and Horn, R. (1993). Oxygen concentration and redox potential gradients in unsaturated model soil aggregates. Soil Sci. Soc. Am. J. 57, 908–916. doi: 10.2136/sssaj1993.03615995005700040005x
Zhang, N., Wang, D., Liu, Y., Li, S., Shen, Q., and Zhang, R. (2014). Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil 374, 689–700. doi: 10.1007/s11104-013-1915-6
Zhang, Y., Xu, J., Riera, N., Jin, T., Li, J., and Wang, N. (2017). Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 5 :97. doi: 10.1186/s40168-017-0304-4
Zhao, R., Zhang, H., Li, Y., Jiang, T., and Yang, F. (2014). Research of iron reduction and the iron reductase localization of Anammox Bacteria. Curr. Microbiol. 69, 880–887. doi: 10.1007/s00284-014-0668-7
Zheng, B., Zhu, Y., Sardans, J., Peñuelas, J., and Su, J. (2018). QMEC: a tool for high-throughput quantitative assessment of microbial functional potential in C, N, P, and S biogeochemical cycling. Sci. China Life Sci. doi: 10.1007/s11427-018-9364-7. [Epub ahead of print].
Zumft, W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616.
Keywords: rhizosphere competence, anaerobic respiration, terminal electron acceptors, respiratory pathways, rhizobacteria, root colonization, adaptation
Citation: Lecomte SM, Achouak W, Abrouk D, Heulin T, Nesme X and Haichar FZ (2018) Diversifying Anaerobic Respiration Strategies to Compete in the Rhizosphere. Front. Environ. Sci. 6:139. doi: 10.3389/fenvs.2018.00139
Received: 27 September 2018; Accepted: 01 November 2018;
Published: 23 November 2018.
Edited by:
Luiz Fernando Wurdig Roesch, Federal University of Pampa, BrazilReviewed by:
Alice Checcucci, Università degli Studi di Firenze, ItalyCopyright © 2018 Lecomte, Achouak, Abrouk, Heulin, Nesme and Haichar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feth el Zahar Haichar, emFoYXIuaGFpY2hhckB1bml2LWx5b24xLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.