
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 09 April 2015
Sec. Environmental Toxicology
Volume 3 - 2015 | https://doi.org/10.3389/fenvs.2015.00027
This article is part of the Research Topic Redox homeostasis managers in plants under environmental stresses View all 18 articles
 Ileana Vera-Reyes1
Ileana Vera-Reyes1 Ariana A. Huerta-Heredia1
Ariana A. Huerta-Heredia1 Teresa Ponce-Noyola1
Teresa Ponce-Noyola1 Carlos M. Cerda-García-Rojas2
Carlos M. Cerda-García-Rojas2 Gabriela Trejo-Tapia3
Gabriela Trejo-Tapia3 Ana C. Ramos-Valdivia1*
Ana C. Ramos-Valdivia1*Plants cells sense their environment through oxidative signaling responses and make appropriate adjustments to gene expression, physiology and metabolic defense. Root cultures of Uncaria tomentosa, a native plant of the Amazon rainforest, were exposed to stressful conditions by combined addition of the glutathione inhibitor, buthionine sulfoximine (0.8 mM) and 0.2 mM jasmonic acid. This procedure induced a synchronized two-fold increase of hydrogen peroxide and guaiacol peroxidases, while the glutathione content and glutathione reductase activity were reduced. Likewise, in elicited cultures, production of the antioxidant secondary metabolites, monoterpenoid oxindole, and glucoindole alkaloids, were 2.1 and 5.5-fold stimulated (704.0 ± 14.9 and 845.5 ± 13.0 μg/g DW, respectively) after 12 h, while phenols were three times increased. Upon elicitation, the activities and mRNA transcript levels of two enzymes involved in the alkaloid biosynthesis, strictosidine synthase and strictosidine β-glucosidase, were also enhanced. Differential proteome analysis performed by two-dimensional polyacrylamide gel electrophoresis of elicited and control root cultures showed that after elicitation several new protein spots appeared. Two of them were identified as thiol-related enzymes, namely cysteine synthase and methionine synthase. Proteins associated with antioxidant and stress responses, including two strictosidine synthase isoforms, were identified as well, together with others as caffeic acid O-methyltransferase. Our results propose that in U. tomentosa roots a signaling network involving hydrogen peroxide and jasmonate derivatives coordinately regulates the antioxidant response and secondary metabolic defense via transcriptional and protein activation.
Oxidative stress arises from disruption in redox balance due that the amount of reactive oxygen species (ROS) exceeds the ability of the cell to accomplish an effective antioxidant response. Unlike other ROS, hydrogen peroxide (H2O2) is a non-radical species, containing no net charge, with a relatively long half-life. Because of these properties, H2O2 acts as a long-distance signaling molecule and is a physiological indicator of the intensity of biotic and/or abiotic stress (Apel and Hirt, 2004). In turn, to prevent the harmful effects of ROS, plants have evolved coordinate antioxidant mechanisms that include superoxide dismutase, peroxidases, the ascorbate-glutathione cycle, and other antioxidant responses (Noctor and Foyer, 1998).
Glutathione is a low molecular weight tripeptide useful in protecting plant cells from oxidative injury due to its redox buffering capacity and relative abundance. In response to environmental stress through the ascorbate–glutathione pathway, the redox potential of the reduced glutathione (GSH) pool is altered and converted to the disulfide form (GSSG) without net consumption (Meyer and Fricker, 2002). It has been reported that H2O2, produced in response against various stimuli, would be acting as a signaling molecule, regulating the expression of selected genes, including those involved in the defense pathways and participating in the crosstalk between other metabolic signals (Quan et al., 2008). Several studies suggest that, as the result of adaptation responses of plants to oxidative stress, changes occur not only in the primary defense mechanisms but also in the profile of secondary metabolism (Apel and Hirt, 2004).
Alkaloids represent one of the most active natural product groups against a wide range of organisms. The main role of these substances is generally linked to plant defense mechanisms from predators, besides the important ecological factors associated to them. However, the close relationship between alkaloids and the oxido-reduction processes in plants containing them strongly suggests that these compounds play a fundamental role in protecting plants when they are subjected to oxidative stress (Ramos-Valdivia et al., 2012). Furthermore, polyphenols are the most abundant and widely distributed group of naturally occurring compounds. Their functions are critical to the maintenance of the plant, being relevant in the defense against herbivores, for protection to different types of biotic or abiotic stress, as well as signals in interactions either with other plants or with microbes (Buer et al., 2010).
GSH deficit may occur in plants as a consequence of increased cellular consumption and/or due to biosynthetic disorders. However, GSH depletion of GSH can occur by addition of L-buthionine-(S,R)-sulphoximine (BSO). This nontoxic substance is a specific inhibitor of γ-glutamylcysteine synthetase (Ruegsegger et al., 1990; May and Leaver, 1993). Treatment of plant tissue with BSO has been used as an elicitor of secondary metabolites since this substance weakens the antioxidant defense mechanisms, provoking endogenous accumulation of H2O2 and oxidative stress (Berglund and Ohlsson, 1993; Guo et al., 1993; Vera-Reyes et al., 2013).
Uncaria tomentosa, which belongs to the Rubiaceae family, is an Amazon rainforest species known as cat's claw. This plant produces the highly oxidized monoterpenoid oxindole alkaloids (MOA) isopteropodine, mitraphylline, isomitraphylline and rhynchophylline, which exhibits immunomodulatory, anti-AIDS, cytotoxic, and antileukemic properties (Laus, 2004). In previous work, it was found that root suspension cultures of this species produced MOA and accumulated 3α-dihydrocadambine (Huerta-Heredia et al., 2009), a glucoindole alkaloid with hypotensive and antioxidant activities (Endo et al., 1983) and dolichantoside (Luna-Palencia et al., 2013), a N-β-methylated strictosidine with potent anti-malarial effect (Frédérich et al., 2000). Moreover, the antioxidant response and alkaloid production stimulation have been correlated with oxidative stress (Trejo-Tapia et al., 2007) triggered by H2O2 treatment (Huerta-Heredia et al., 2009; Vera-Reyes et al., 2013) and by combined addition of the glutathione inhibitor, buthionine sulfoximine and jasmonic acid (Vera-Reyes et al., 2013). It has been suggested that monoterpenoid indole alkaloids (MIA) are precursors of MOA whose transformation may take place through oxidation of the indole ring system. The central precursor of the MIA pathway is the glycosylated indole alkaloid strictosidine, which is formed through the condensation of the indole precursor tryptamine with secologanin catalyzed by the enzyme strictosidine synthase (STR; EC 4.3.3.2). Then, strictosidine β-D-glucosidase (SGD; EC 3.2.1.105) hydrolyzes the glucose moiety present in strictosidine forming an aglycone, which is rapidly converted to a dialdehyde intermediate. In some plants such as Catharanthus roseus, this substance is reduced by NADPH to ajmalicine or their isomers through cathenamine (Kutchan, 1995). Strictosidine also participates in the biosynthesis of other glucoindole alkaloids characteristic of the Rubiaceae family such as isodihydrocadambine (Szabó, 2008).
Both STR and SGD are encoded by single genes (McKnight et al., 1990), even though the STR from C. roseus has shown several isoforms due to post-translational modifications (De Waal et al., 1995; Jacobs et al., 2005). Vera-Reyes et al. (2013) reported that in U. tomentosa root cultures, the increase of oxindole and glucoindole alkaloids observed under oxidative stress, is provoked by the regulatory mechanisms at the level of enzyme activities and gene expression of STR and SGD. Thus, proteomics provides a promising approach for the study of the protein response to oxidative stress in general and its relation with the secondary metabolism production (Ramos-Valdivia et al., 2012). Particularly, comparative proteomic studies based on contrasting plant cultures on stressed and non-stressed conditions are essential for understanding the stress-related defense mechanisms.
In order the study the regulatory mechanisms functioning in the monoterpenoid indole alkaloid production in U. tomentosa root cultures, activities and mRNA transcript levels of two enzymes involved its alkaloid biosynthesis, antioxidant defense and comparative proteome analysis in response to oxidative stress were examined.
Root cultures of U. tomentosa (line Utr-3) arising from micropropagated plantlets (Luna-Palencia et al., 2013) were grown in 250-mL Erlenmeyer flasks (covered with aluminum foil) with 100-mL of MS medium (Murashige and Skoog, 1962), 2% sucrose without plant growth regulators and pH 6.4 adjusted prior to sterilization. The cultures were incubated at 25 ± 2°C, using orbital agitation at 110 rpm, and under continuous light intensity 13 μmol m−2 s−1. The cultures were sub-cultivated every 20 days and uniform inocula for the experiments were developed in 1000-mL Erlenmeyer flasks containing 400-mL of culture medium. A selection of 20-days-old roots were cut in pieces of ~5 cm length and kept in deionized water until they were inoculated (2 g FW) into 250-mL shaken flasks containing 100-mL culture medium. Roots were elicited at day 13 with simultaneous addition of 0.8 mM BSO and 0.2 mM jasmonic acid (BSO-JA) and were incubated as indicated above. Three control cultures and three elicited flask cultures were harvested after 12 h.
Ten grams of frozen roots were ground using a mortar and a pestle and were cooled with liquid N2. A solution (20 mL) of cold (−20°C) 10% TCA in acetone with 0.07% β-mercaptoethanol was poured over the sample (Jacobs et al., 2005). The mixture was kept at 20°C overnight to enable a complete precipitation. After centrifugation for 15 min at 3000 g, samples were washed twice with a cold solution (−20°C) of acetone and 0.07% β-mercaptoethanol for removing TCA. The precipitate was solubilized in ReadyPrep rehydration/sample buffer BioRad [8 M urea, 2% CHAPS, 50 mM dithiothreitol (DTT), 0.2% (w/v) Bio-Lyte® 3/10 ampholytes, and bromophenol blue (trace)] completed with 2 M thiourea. The mixture was vortexed and centrifuged (5 min, 16,000 g) several times during 1 h. The supernatant was recovered and cleaned up using a Micro Bio-Spin® column (BioRad, USA) and stored at −80°C. The concentration of protein was measured with a 2D Quant kit (Amersham Biosciences, USA).
About 250 μg of protein was loaded into 11-cm strips with a pH gradient between 4 and 7 (IPG, immobilized pH gradient, Bio-Rad) by in-gel rehydration during 12 h. Isoelectric focusing (IEF) was carried out on a Protean IEF apparatus (Bio-Rad, USA) at 20°C by application of a voltage gradient from 0 to 250 V for 1 h, 250 to 500 V for 1 h, 1000 to 8000 V for 1 h, from 8000 to 20,000 V for 2 h, and 500 V for 2 h. The protein IPG strips were equilibrated before applying a sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) procedure using an equilibration buffer I (50 mM Tris HCl [pH 8], 8 M urea, 30% glycerol, 2% SDS, and 0.3% DTT) for 10 min. The strips were then soaked for 10 min in the equilibration buffer II containing 50 mM Tris HCl (pH 8), 8 M urea, 30% glycerol, 2% SDS, and 4.5% iodoacetamide. SDS-PAGE was done using polyacrylamide 12% acrylamide gels. Electrophoresis was carried out at 25 mA for 45 min and 35 mA for 2.5 h (SE 600 Ruby™; GE Healthcare Life Science, USA). Protein samples were visualized by staining with Sypro Ruby (BioRad, USA).
At least three independent 2-D experiments were repeated at minimum four times to confirm reproducibility. Image analysis was achieved by visual inspection and the observed changes were qualitative using Melanie 7.0 gel analysis platform (GE Healthcare). The volume of each spot was normalized as a relative volume to compensate for the variability in gel staining. Manual editing was carried out after the automated detection and matching for each spot, achieving this procedure with a minimum of four gels for each sample. Only those spots that showed significant and reproducible changes (at least 1.3-fold) were taken in to account as differentially expressed proteins, ANOVA (p < 0.05). The Scaffold program (version 4.0.6.1 from Proteome Software Inc., Portland, OR) was employed for protein identification. The validation was done if the probability was greater than 99.0% and contained at least 2 identified peptides. Protein probabilities were allocated by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that could not be differentiated based on MS/MS analysis were grouped to satisfy the principles of parsimony. The estimated experimental Mr/pI was useful to rise the identification confidence.
Excised SYPRO®Ruby (BioRad)-stained protein gel spots following 2D SDS-PAGE were digested with trypsin (10 μg/mL) at 37°C for 12 h. Tandem mass spectrometry coupled to liquid chromatography (LC-MS/MS) analysis of in-gel trypsin digested-proteins (Shevchenko et al., 1996) was performed in a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, San Jose, CA) furnished with an Advion nanomate ESI source (Advion, Ithaca, NY). ZipTip (Millipore, Billerica, MA) C18 sample clean-up was achieved as indicated in the manufacturer's instructions. The peptide fraction was eluted from a C18 precolumn of 100-μm id × 2 cm (Thermo Fisher Scientific) and loaded onto an analytical C18 column of 75-μm ID × 10 cm C18 (Thermo Fisher Scientific) eluting with solvent A (water and 0.1% formic acid) and a 5–10% gradient of solvent B (acetonitrile, 0.1% formic acid) for 5 min, followed by a 10–35% gradient of solvent B for 35 min, 35–50% gradient of solvent B for 20 min, 50–95% gradient of solvent B for 5 min, and 95% solvent B for 5 min, all elutions were run at a flow rate of 400 nl/min. Data dependent scanning (m/z 400–1600) was carried out in the Orbitrap analyzer, followed by collision-induced dissociation (CID) tandem mass spectrometry (MS/MS) of the 14 most intense ions in the linear ion trap analyzer using the Xcalibur v 2.1.0 software (Andon et al., 2002) and a mass scan of 60,000 resolution. The precursor ions were chosen by the monoisotopic precursor selection (MIPS) setting the acceptance or rejection of ions thought a ±10 ppm window. Dynamic exclusion was established to place any selected m/z peak on an exclusion list for 45 s after a single MS/MS. All MS/MS spectra were explored against asterids proteins downloaded from Uniprot or from NCBI on October 09, 2012 or June 20, 2013, respectively, using Thermo Proteome Discoverer 1.3 (Thermo Fisher Scientific). The UniprotKB protein database of all species was also used in searching the data independently. Variable modifications considered during the search included methionine oxidation, adding 15.995 Da, and/or cysteine carbamidomethylation, adding 57.021 Da. At the time of the search, asterids database from Uniprot or NCBI contained 65,406 and 102,843 entries, respectively (UniprotKB database contained 452,768 entries as of October 10, 2012). Proteins were identified with a confidence level of 99% with XCorr score cut-offs (Qian et al., 2005) as determined by a reversed database search. The results were displayed with the Scaffold program v 3.6.1 (Proteome Software Inc., Portland OR) that depends on various search engines (Sequest, X!Tandem, MASCOT) using Bayesian statistics (Keller et al., 2002; Nesvizhskii et al., 2003).
Powdered roots (0.20 g) were frozen in liquid N2, pulverized and sonically extracted with 5 mL of methanol-water (8:2 v/v) and centrifuged. A supernatant aliquot of 0.2 mL was mixed with 0.2-mL Folin-Ciocalteu reagent diluted 1:1 (v/v) with water, 0.6 mL of sodium carbonate (Na2CO3) saturated solution and 4 mL of deionized water. The mixture was intensively shaken, left at room temperature for 25 min, and centrifuged at 5000 rpm for 10 min. The absorbance of supernatant was registered at 725 nm in a Genesys 10V spectrophotometer (Thermo Scientific). Total phenols were expressed in terms of D-catechin equivalents. Quantification of individual phenols was done by HPLC analysis according (Pavei et al., 2010) using a 3-caffeoylquinic acid (chlorogenic acid) calibration curve.
Alkaloid extraction and quantification were performed as described previously (Vera-Reyes et al., 2013). Briefly, frozen roots (liquid N2) were pulverized and sonically extracted with 5% hydrochloric acid. Alkaloids from the acid-solutions or culture media were extracted twice with chloroform adjusting the pH to 8-9 using a NH4OH solution. The organic layer was vacuum evaporated and the solid residue was dissolved in a 9:11 mixture of acetonitrile and 10 mM phosphate buffer at pH 7. The solutions were filtered and injected into a Varian ProStar 333 HPLC system equipped with a photodiode array detector (Varian, Walnut Creek, CA) using a reverse-phase C18 column (Waters Spherisorb 5 mm ODS2 of 250 mm length 4.6 mm i.d.). Elution was carried out with the same 9:11 mixture of acetonitrile and phosphate buffer at 0.7 mL/min flow rate and detecting at 244 nm. For quantification of MOA and glucoindole alkaloids, mitraphylline and 3α-dihydrocadambine respectively, were used as the standard compound to determine the calibration curve.
All measurements were done in triplicate and the statistical evaluation was achieved with Anova, taking p ≤ 0.05 as significant.
Roots (1 g) were homogenized in a pre-chilled mortar under liquid N2 with 1–2% (w/w) polyvinylpyrrolidone. Extraction buffer (0.1 M potassium phosphate pH 6.3, containing 3 mM EDTA and 6 mM DTT) was added in a 1:1 ratio (v/w) shaking to obtain a homogeneous mixture. For GR assay, the extraction buffer was 0.1 M potassium phosphate pH 7.5, with 1 mM EDTA. Centrifugation at 18,000 g was done for 10 min at 4°C and the supernatant was collected and desalted on Bio-Rad Micro Bio-Spin® P-30 columns. The eluted samples were employed for the enzymatic assays.
The protein fractions were kept frozen at −20°C until use. The total protein content was determined following the procedure described by Peterson (1977) with bovine serum albumin as the standard.
Guaiacol peroxidases were measured as oxidation of guaiacol (8.26 mM, ∈ = 26.6 mM−1 cm−1) according to Pütter (1974). Enzyme extract was incubated in 100 mM phosphate buffer pH 6.0 containing 3 mM H2O2. The reaction was started by addition of 15 mM guaiacol and the absorption was measured for 2 min at 470 nm using a Beckmann spectrophotometer (DU 7500, Munich). Rates were corrected by chemical control experiments. Peroxide activity was determined as the amount of protein that produces 1 μmol of oxidized guaiacol. The activity of glutathione reductase was measured using the Glutathione Reductase Assay Kit (Sigma-Aldrich, St. Louis, USA), which was determined by the absorbance decrease caused by NADPH oxidation at 340 nm. One enzyme unit (U) catalyzes the oxidation of 1 μmol of NADPH per min at 25°C.
The assay of strictosidine synthase (STR) activity depends on the enzymatic condensation of secologanin and tryptamine to produce strictosidine. Strictosidine formation was quantified by HPLC using a strictosidine standard (Phytoconsult, The Netherlands) for constructing the calibration curve. Strictosidine glucosidase (SGD) activity was determined by measuring the glucose release using Amplex Red® (Invitrogen) assay kit. Both enzyme assays were previously described (Vera-Reyes et al., 2013).
RNA isolation, DNA treatment, reverse transcription, and semiquantitative-PCR amplification were achieves as reported previously (Vera-Reyes et al., 2013), as well as the primers used for the genes: STR (strictosidine synthase), SGD (strictosidine glucosidase and the control 18S rRNA. The relative gene expression was analyzed using a Kodak Image Station 2200R, DU® 730 equipped with Molecular Imaging Software version 1.4 (Kodak) on a 1.2% agarose gel. The gene expression analysis is represented in arbitrary units employing average values of semi-quantitative RT-PCR assays in triplicate with respect to the corresponding non-treated cultures.
Roots (500 mg) were frozen and pulverized under liquid N2. The powder was extracted with 5 mL of 0.1% TCA (w/v), mixed with ice for 5 min, and pelleted by centrifugation at 10,000 g at 4°C for 10 min. The supernatant was neutralized with 0.2 M NH4OH to pH 8.0 and was centrifuged at 3000 g for 2 min to sediment the insoluble material. The quantification of H2O2 in the extracts was done with the Amplex Red Hydrogen Peroxide Assay kit (Molecular Probes, Invitrogen), according to the manufacturer instructions. A total of 50 μL of extract was combined with an equal volume of 50 mM sodium phosphate buffer pH 7.4 containing 0.1 U/mL of horseradish peroxidase and incubated for 1 h at room temperature, measuring the absorbance at 560 nm. The H2O2 concentration for each sample was determined with a standard curve obtained with known concentrations of H2O2.
The levels of total glutathione (GSH + GSSG) were determined with a glutathione assay kit (Sigma) following the manufacturer's protocol. Roots were frozen in liquid N2 and pulverized until obtaining fine particles. A solution of 5% 5-sulfosalicylic acid (500 μL) was added to 0.1 g of the powder to deproteinize the sample. Glutathione was measured in a kinetic assay based on the reduction of 5,5-dithiobis(2-nitrobenzoic acid) (DTNB) to yellow TNB, which was spectrophotometrically measured at 412 nm. The amount of total glutathione was determined with a standard curve of reduced glutathione.
U. tomentosa roots induce their antioxidant defense to scavenge excess of ROS in response to combined addition of BSO-JA. After 12 h of elicitation, a two-fold increase of H2O2 concentration (from 0.48 ± 0.05 to 0.96 ± 0.03 μmol/g FW) and POD activity (from 243.9 ± 15.4 to 370.8 ± 8.9 μM/mg.min protein) were found (Figures 1A,C). In these elicited cultures, glutathione concentration was significantly reduced in a 55%, while the GR activity was slightly lower (17%) than non-treated roots (Figures 1B,D). Noteworthy, biomass concentration (6.33 ± 0.20 g DW/L) and viability of roots after the elicitation remained essentially the same as in controls.
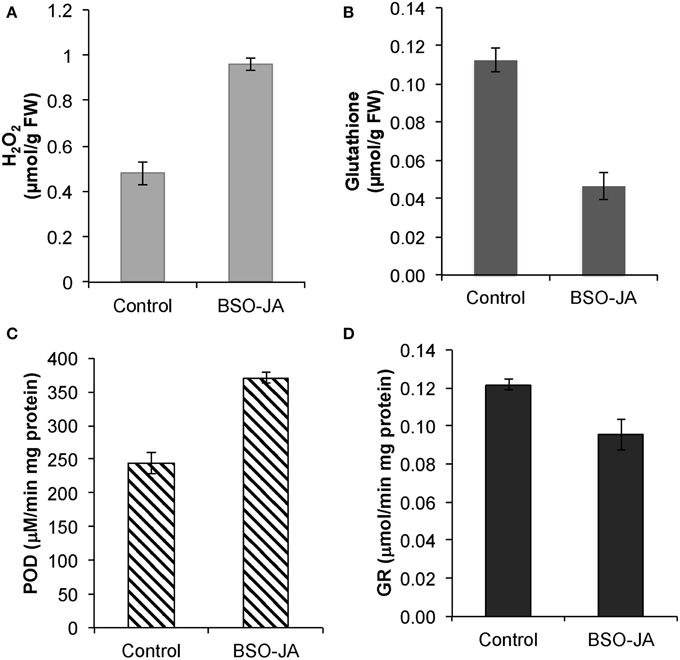
Figure 1. Responses of Uncaria tomentosa root cultures after 12 h of BSO-JA elicitor addition, (A) Changes in peroxide hydrogen (B) glutathione concentration. Antioxidant enzyme activities of (C) guaiacol peroxidase (POD) and (D) glutathione reductase (GR). The elicitor was added to 13 days-old root cultures (exponential growth phase). Error bars indicate standard deviation from the mean (n = 3).
After 12 h of elicitor treatment, MOA, 3α-dihydrocadambine and dolichantoside production (Figure 2A) were rapidly increased by 2.1-, 5.5-, and 2.6-fold, respectively, compared with control cultures (329.7 ± 39.8 μg/g DW; 152.4 ± 27.9 μg/g DW; 14.0 ± 1.8 μg/g DW). Concurrently, BSO-JA treatment increased STR activity by three times in relation to untreated roots (38.7 ± 4.0 pKat/mg protein), while SGD activity had 4.2 times more activity than the control (65.8 ± 2.9 pKat/mg protein) (Figure 2B). Upon elicitation, STR and SGD transcripts increased during the first 12 h after treatment reaching 5.8- and 9.7-fold higher, respectively, compared to the control levels (Figures 2C,D).
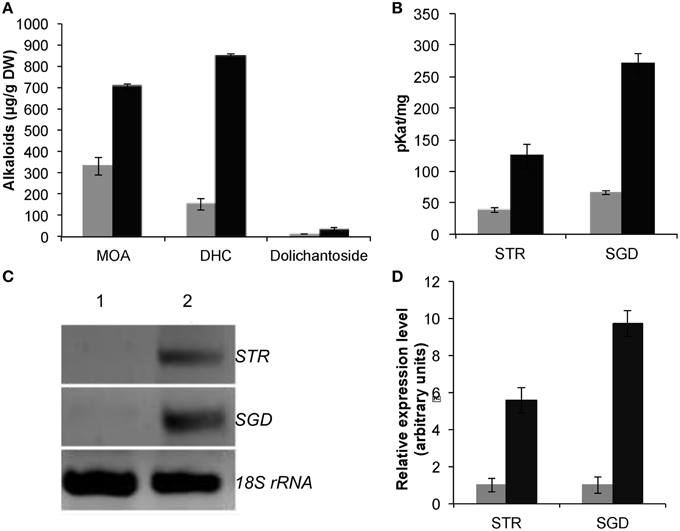
Figure 2. Effect of BSO-JA elicitation on U. tomentosa root cultures growing in Erlenmeyer flasks after elicitor addition (black bars). (A) Production of MOA, DHC, and dolichantoside. (B) Specific activities of strictosidine synthase (STR) and strictosidine β-glucosidase (SGD). Error bars indicate standard deviation from the mean (n = 3). Semiquantitative RT-PCR analysis of the elicitor effects in the STR and SGD mRNA transcript levels (C) Representative expression profile (The amplification products were analyzed by agarose gel electrophoresis (lane 1 control, lane 2 elicited) stained with ethidium bromide, and visualized by UV transillumination (306 nm). An inverse image of the stained gel is shown. (D) Schematic presentation of the RT-PCR results in arbitrary units. The analysis of gene expression is represented in the bar chart using average values of semiquantitative RT-PCR triplicate analysis. The results are relative to non-treated root cultures.
In correlation with the alkaloid induction after BSO-JA addition, total polyphenol content in U. tomentosa root cultures increased from 3.40 ± 0.12 mg/g to 11.45 ± 0.02 mg/g DW. In these elicited roots, the content of 3-caffeoylquinic acid, caffeic acid, catechin, and epicatechin were increased by 210.5, 50.8, 21.7, and 58.8%, respectively (Table 1).
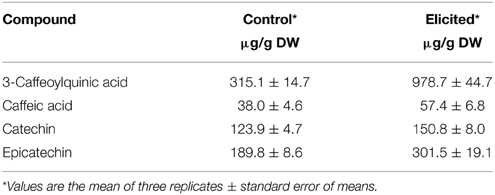
Table 1. Polyphenols accumulation in Uncaria tomentosa root cultures growing in Erlenmeyer flasks 12 h after BSO-JA elicitor addition.
One of the key approaches of proteomic analysis is to identify differential protein expression between control and experimental samples. Hence, four replicate gels of U. tomentosa protein extracts from 12 h after BSO-JA addition were compared with the same number of replicates from non-treated root cultures.
Although the gels showed the same profile, the control gels exhibited more proteins than the elicited ones. The control gel with higher protein spots (480) was used for the analysis as standard reference gel. An 87% of the protein spots on the other three gels from untreated roots coincided with those found in the reference gel, whereas those from elicited extracts were 85% coincident. The new proteins that appeared after elicitation and those proteins from the region pI 5–6 and 30–35 kDa (Figure 3) that, as previously reported correspond to alkaloid biosynthesis enzymes (Jacobs, 2005), were selected for sequenciation. The 14 identified proteins (Table 2 and Supplementary Table 1) can be classified into several functional categories, including energy metabolism and photosynthesis: two triosephosphate isomerases (chloroplastic and cytoplasmic; spots 1, 2, and 8), as the same protein in multiple spots differing in pI and Mr, and ribulose 1,5-bisphosphate carboxylase (Rubisco) large chain (spot 4). Protein synthesis: some proteins involved in the sulpur amino acid biosynthesis such as cysteine synthase (spot 11) and methionine synthase (spot 10) were up-regulated in BSO-JA conditions. Secondary metabolism: oxidative stress increased the expression of protein spots 7 and 9, identified as strictosidine synthase (STR) isoforms, and spot 12 identified as caffeic acid O-methyltransferase. ROS scavenging, defense and stress: abundance of defense-related proteins as ascorbate peroxidase (spot 3), proteasome alpha subunit (spot 6), universal stress protein (spot 13), and pathogenesis-related protein (spot 14) were altered during oxidative stress condition.
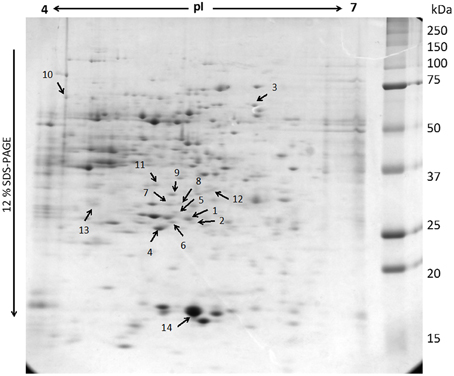
Figure 3. Representation of two-dimensional electrophoresis (2-D SDS-PAGE) profiles of the soluble proteins extracted from Uncaria tomentosa root cultures (250 μg). The proteins were separated on a pH 4-7 linear IPG strip, followed by 12% SDS-PAGE. The gel was visualized by Sypro Ruby staining. Number indicates the protein spots identified by MS analysis.
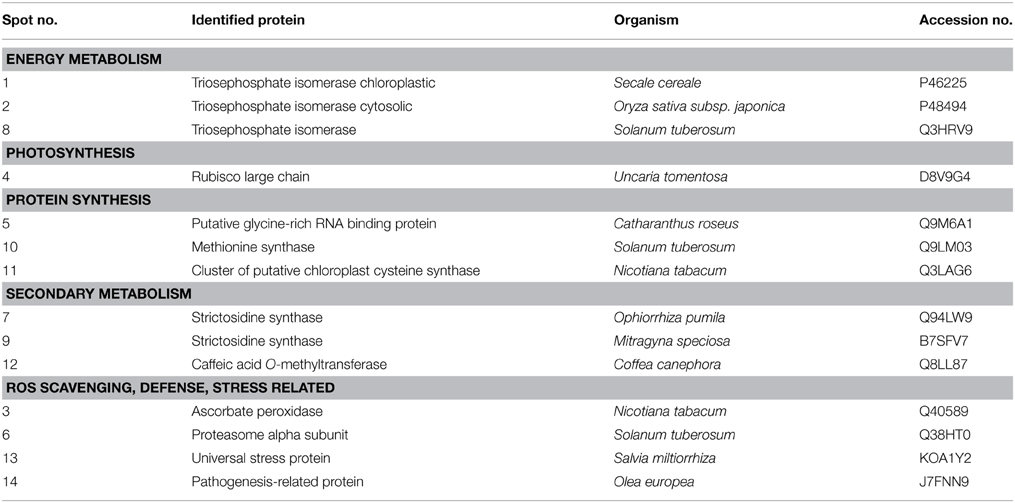
Table 2. Protein identification through MALDI-TOF from Uncaria tomentosa root cultures under BSO-JA treatment.
The high increase in H2O2 combined with reduction of glutathione concentration in U. tomentosa roots 12 h after addition of BSO-JA may reflect that oxidative stress conditions were present. The plant cell protection of reduced glutathione (GSH) against the oxidative injury is established by its redox buffering activity and abundance. Therefore, treatment of plant cell or tissue with the glutathione biosynthesis inhibitor BSO can lead to weakened antioxidative defenses; an increase in the concentration of endogenous H2O2; and secondary metabolites stimulation (Berglund and Ohlsson, 1993; Guo and Ohta, 1993; Guo et al., 1993). Furthermore, H2O2 is a secondary messenger that mediates hormonal responses, biotic/abiotic environmental stresses, and developmental signals (Neill et al., 2002). Thus, the jasmonate signaling is mediated by H2O2 (Orozco-Cárdenas et al., 2001), and is controlled via a suitable antioxidant response to neutralize its adverse effects. The increase of peroxidase activity in elicited cell and plant cultures has been found as a primary response to oxidative stress (Quan et al., 2008), whereas glutathione reductase plays a key role in the antioxidant defense processes by reducing oxidized glutathione (GSSG) to glutathione (GSH), thus allowing the maintenance of a high GSH/GSSG ratio (Foyer and Noctor, 2005). Roots grown in the presence of BSO would be unprotected by the glutathione diminution. Nevertheless, the non-induction of GR activity at 12 h after elicitation could be caused by the increase in polyphenol content as previously reported (Zhang et al., 1997).
Under the assayed conditions, BSO-JA addition did not affect the biomass concentration and root viability, probably because jasmonic acid would be inducing the biosynthesis and activity of other defense responses (Sasaki-Sekimoto et al., 2005) offsetting the antioxidant diminishing caused by BSO.
In cell or plant cultures, a synergistic effect of elicitors on secondary metabolites production may occur (Zhao et al., 2005). It has been reported that BSO induces oxidative stress by depletion of glutathione (Noctor and Foyer, 1998), JA can induce ROS production, and JA signaling is important for oxidative stress tolerance (Sasaki-Sekimoto et al., 2005; Pauwels et al., 2008). Separate application of JA or BSO in U. tomentosa roots also elicited the production of alkaloids but in smaller quantities (Vera-Reyes et al., 2013). An increase in secondary metabolite production was also obtained in carrot cells when BSO was used alone or in combination with a yeast glucan elicitor, stimulating an increase in the H2O2 at cellular level (Guo and Ohta, 1993). In U. tomentosa cell cultures growing in bioreactors, a positive correlation among the increment of endogenous H2O2 level, activities of NAD(P)H oxidase and peroxidases, and MOA production was reported (Trejo-Tapia et al., 2007). Moreover, H2O2 treatment induced oxidative stress and alkaloid production in U. tomentosa roots (Huerta-Heredia et al., 2009; Vera-Reyes et al., 2013).
In C. roseus, STR and strictosidine are confined inside the vacuole (McKnight et al., 1990) separated from the activity of the nuclear localized SGD (Guirimand et al., 2010). In U. tomentosa root cultures, a probable cell compartmentalization for alkaloids has been suggested (Vera-Reyes et al., 2013) as MOA were mainly found in the culture medium, while the glucoindole alkaloids 3α-dihydrocadambine and dolichantoside were always found inside the roots. Furthermore, alkaloid biosynthesis includes multiple oxidations catalyzed in a stereo- and regiospecific fashion, indicating that specific oxidases are involved in the in vivo biosynthesis. It has been found that peroxidases, microsomal cytochrome P-450-dependent enzymes, 2-oxoglutarate dependent dioxygenases and flavoproteins catalyze some of these oxidations with high substrate specificity enzymes (Kutchan, 1995). However, in vitro studies have revealed the ability of plant peroxidases to accept alkaloids as substrates as well as a number of vacuolar metabolites, such as phenols and flavonoids (Sottomayor et al., 2004; Takahama, 2004). In response to the BSO-JA elicitation, polyphenols production in U. tomentosa root cultures, mainly 3-O-caffeoylquinic acid and catechins, was highly stimulated due to the prevailing oxidative stress. Therefore, polyphenols, as flavonols and phenylpropanoids present in vacuoles and the apoplast, can metabolize H2O2 as an electron donor for phenol peroxidases. This change results in the formation of the respective phenoxyl radicals, which can be regenerated by a non-enzymatic reaction with ascorbate (Figure 4). Thus, in C. roseus it has been suggested that vacuolar alkaloids, peroxidases, and phenolic derivatives can function as a hydrogen peroxide scavenging system (Ferreres et al., 2011).
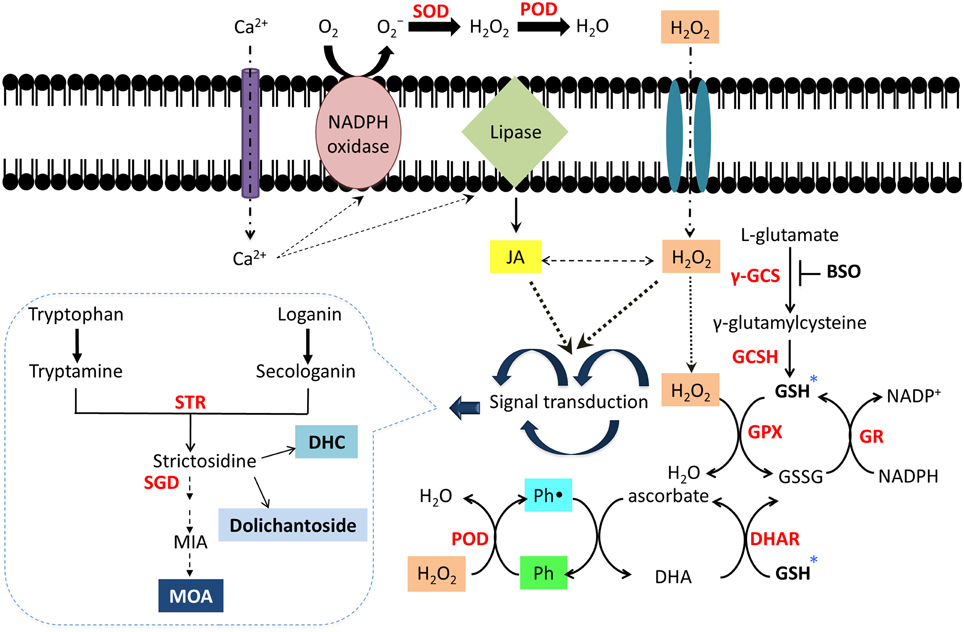
Figure 4. Schematic network of alkaloid and polyphenol activation through signal transduction caused by oxidative stress in Uncaria tomentosa root cultures. The elicitation with BSO-JA activates a signal cascade into the cell which include Ca2+ influx, cytosolic free Ca2+, stimulation of peroxidases, NADPH oxidases, and phospholipases, which further generates other signaling messengers, such as ROS (Quan et al., 2008) and JA together with inducible secondary metabolites production (Zhao et al., 2005; Peebles et al., 2009). *Inhibition of GSH biosynthesis by BSO could provoke an increase in ROS (H2O2) due to non-enzymatic scavengers such as ascorbate or GSH are unable to be regenerated into the cell (Guo and Ohta, 1993). In order to maintain the redox equilibrium in the cell, production of antioxidant compounds as polyphenols and alkaloids (secondary metabolites) could be induced (Ramos-Valdivia et al., 2012). Polyphenols (Ph) can detoxify H2O2 as electron donors by peroxidases (PDO), which results in formation of the respective phenolic radical (Ph•). This species can be regenerated by a non-enzymatic reaction with ascorbate. Dehydroascorbate (DHA) may be reduced back to ascorbate by a dehydroascorbic acid reductase (DHAR) (Ferreres et al., 2011). Superoxide dismutase (SOD); glutathione peroxidase (GPx).
Identification of proteins that differ in stressed and control plants has revealed groups of proteins that respond to oxidative stress conditions with different roles. Nevertheless, the crucial limitation for protein identification using mass spectrometry analysis is the lack of the sequence data of genes and proteins of U. tomentosa. The SWISS-PROT database (November 2014) only contains five protein entries for this species. Consequently, identification of proteins from 2D-gels requires the knowledge of the sequence data and not relying solely on peptide masses. Several studies reported that oxidative stress provoked different responses such as induction or more often repression of the enzymes involved in carbon metabolism. Therefore, plants must be required to make an economical use of their metabolites and energy to deal with adverse environments (Zhang et al., 2012).
It has also been reported that under conditions of oxidative stress, Rubisco was differentially regulated even though its activity decreased having transcriptional and translational repression thereof caused by jasmonates (Weidhase et al., 1987). Moreover, JA stimulates the glutathione, ascorbate and cysteine accumulation while increases dehydroascorbate reductase activity. This last is a relevant enzyme involved in the ascorbate recycling system (Sasaki-Sekimoto et al., 2005). Cysteine synthase is a key enzyme in cysteine biosynthesis, which constitutes one of the significant factors limiting GSH biosynthesis in plants (Vierling, 1991).
Proteolysis-related proteins like proteasome alpha subunit were also more abundant in stressed conditions because they are necessary for degradation of damaged proteins and for maintaining cellular protein homeostasis (Kurepa et al., 2009). Evidence obtained in U. tomentosa BSO-JA elicited cultures indicates post-translational modifications of STR proteins in correlation with the three times increase in the STR enzyme activity. Six isoforms of the glycosylated enzyme STR have been detected in C. roseus (De Waal et al., 1995), while in these cell cultures five STR isoforms were induced after elicitation with P. aphanidermun (Jacobs et al., 2005). It is known that jasmonic acid acts as a signal for the biosynthesis of MIA, and is involved in the activation of transcription factors such as ORCA, which have shown to activate transcription of the STR (Peebles et al., 2009). Another interesting protein identified as up-accumulated in the present study was caffeic acid-O-methyl transferase, one of the key enzymes that catalyzes O-methylation of the hydroxyl group at C5 in phenolic rings (Tu et al., 2010). In general, most methyltransferases possess a broad substrate permissiveness, which also includes several alkaloid N-methyltransferases (Zubieta et al., 2003; Nomura and Kutchan, 2010).
The ascorbate peroxidase, which constitutes one of the most important antioxidant systems for removal of H2O2 generated in the cell, was also up-expressed by BSO-JA addition. Deficiency of cytosolic ascorbate peroxidase occasioned accumulation of H2O2 and consequently damage in specific proteins of leaf cells (Davletova et al., 2005).
In U. tomentosa root cultures, BSO-JA elicitation induced intracellular JA and H2O2 accumulation by glutathione depletion (Figure 4). They act as a signal transducers and secondary messengers, triggering signaling cascades and activating certain late genes that regulate the activity of detoxifying enzymes associated with antioxidant compounds. Therefore, production of alkaloids and specific phenylpropanoids is also activated, protecting roots from oxidative stress damage. Identification of proteins with diverse roles that are present in oxidative stress conditions evidences the complexity of the responses. This approach contributes to the understanding of the metabolic mechanisms operating in U. tomentosa subjected to oxidative stress and the manner how this plant produces the appropriate adjustments for tolerating them.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was financed by CINVESTAV-IPN and CONACYT-Mexico (222097). IV thank CONACYT-Mexico for a doctoral (173034) fellowship. Authors wish to thank C. Fontaine for technical support. We appreciate the proteomics analytical support of Dr. George Tsaprailis (Arizona Proteomics Consortium). Mass spectrometry and proteomics data were acquired by the Arizona Proteomics Consortium supported by NIEHS grant ES06694 to the SWEHSC, NIH/NCI grant CA023074 to the UA Cancer Center and by the BIO5 Institute of the University of Arizona.
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fenvs.2015.00027/abstract
Andon, N. L., Hollingworth, S., Koller, A., Greenland, A. J., Yates, J. R., and Hanes, P. A. (2002). Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics 2, 1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Berglund, T., and Ohlsson, A. (1993). The glutathione biosynthesis inhibitor buthionine sulfoximine (BSO) induces cardenolide accumulation in Digitalis lanata tissue culture. J. Plant Physiol. 142, 248–250. doi: 10.1016/S0176-1617(11)80973-9
Buer, C. S., Nijat, I., and Djordjevic, M. A. (2010). Flavonoids: new roles for old molecules. J. Integr. Plant. Biol. 52, 98–111. doi: 10.1111/j.1744-7909.2010.00905.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Davletova, S., Rizhsky, L., Liang, H., Shengqiang, Z., Oliver, D. J., Coutu, J., et al. (2005). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17, 268–281. doi: 10.1105/tpc.104.026971
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
De Waal, A., Meijer, A. H., and Verpoorte, R. (1995). Strictosidine synthase from Catharanthus roseus: purification and characterization of multiple forms. Biochem. J. 306, 571–580.
Endo, K., Oshima, Y., Kikuchi, H., Koshihara, Y., and Hikino, H. (1983). Hypotensive principles of Uncaria hooks. Planta Med. 49, 188–190. doi: 10.1055/s-2007-969846
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ferreres, F., Figueiredo, R., Bettencourt, S., Carqueijeiro, I., Oliveira, J., Gil-Izquierdo, A., et al. (2011). Identification of phenolic compounds in isolated vacuoles of the medicinal plant Catharanthus roseus and their interaction with vacuolar class III peroxidase: an H2O2 affair? J. Exp. Bot. 62, 2841–2854. doi: 10.1093/jxb/erq458
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Foyer, C. H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875. doi: 10.1105/tpc.105.033589
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Frédérich, M., De Pauw, M. C., Llabrès, G., Tits, M., Hayette, M. P., Brandt, V., et al. (2000). New antimalarial and cytotoxic sungucine derivatives from Strychnos icaja roots. Planta Med. 66, 262–269. doi: 10.1055/s-2000-8559
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Guirimand, G., Courdavault, V., Lanoue, A., Mahroug, S., Guihur, A., Blanc, N., et al. (2010). Strictosidine activation in Apocynaceae: towards a “nuclear time bomb”? BMC Plant Biol. 10:182. doi: 10.1186/1471-2229-10-182
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Guo, Z. J., Nakagawara, S., Sumitani, K., and Ohta, Y. (1993). Effect of intracellular glutathione level on the production of 6-methoxymellein in cultured carrot (Daucus carota) cells. Plant Physiol. 102, 45–51. doi: 10.1104/pp.102.1.45
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Guo, Z. J., and Ohta, Y. (1993). A synergistic effect of glutathione depletion and elicitation on the production of 6-methoxymellein in carrot cells. Plant Cell Rep. 12, 617–620. doi: 10.1007/BF00232810
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Huerta-Heredia, A. A., Marín-López, R., Ponce-Noyola, T., Cerda-García-Rojas, C. M., Trejo-Tapia, G., and Ramos-Valdivia, A. C. (2009). Oxidative stress induces alkaloid production in Uncaria tomentosa root and cell cultures in bioreactors. Eng. Life Sci. 3, 211–221. doi: 10.1002/elsc.200800118
Jacobs, D. (2005). Proteome Analysis of the Medicinal Plant Catharanthus Roseus. Ph.D. Thesis. Leiden University, Leiden.
Jacobs, D. I., Gaspari, M., van der Greef, J., van der Heijden, R., and Verpoorte, R. (2005). Proteome analysis of the medicinal plant Catharanthus roseus. Planta 221, 690–704. doi: 10.1007/s00425-004-1474-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392. doi: 10.1021/ac025747h
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kurepa, J., Wang, S., Li, Y., and Smalle, J. (2009). Proteasome regulation, plant growth and stress tolerance. Plant Signal. Behav. 4, 924–927. doi: 10.4161/psb.4.10.9469
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kutchan, T. M. (1995). Alkaloid biosynthesis the basis for metabolic engineering of medicinal plants. Plant Cell 7, 1059–1070. doi: 10.2307/3870057
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Laus, G. (2004). Advances in chemistry and bioactivity of the genus Uncaria. Phytother. Res. 18, 259–274. doi: 10.1002/ptr.1469
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Luna-Palencia, G. R., Huerta-Heredia, A. A., Cerda-García-Rojas, C. M., and Ramos-Valdivia, A. C. (2013). Differential alkaloid profile in Uncaria tomentosa micropropagated plantlets and root cultures. Biotechnol. Lett. 35, 791–797. doi: 10.1007/s10529-012-1128-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
May, M. J., and Leaver, C. J. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103, 621–627.
McKnight, T. D., Roessner, C. A., Devagupta, R., Scott, A. I., and Nessler, C. L. (1990). Nucleotide sequence of a cDNA encoding the vacuolar protein strictosidine synthase from Catharanthus roseus. Nucleic Acids Res. 18, 4939–4939. doi: 10.1093/nar/18.16.4939
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Meyer, A. J., and Fricker, M. (2002). Control of demand-driven biosynthesis of glutathione in green Arabidopsis suspension culture cells. Plant Physiol. 130, 1927–1937. doi: 10.1104/pp.008243
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Neill, S. J., Desikan, R., Clarke, A., Hurst, R. D., and Hancock, J. T. (2002). Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 53, 1237–1247. doi: 10.1093/jexbot/53.372.1237
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658. doi: 10.1021/ac0341261
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Noctor, G., and Foyer, C. H. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. doi: 10.1146/annurev.arplant.49.1.249
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nomura, T., and Kutchan, T. M. (2010). Three new O-methyltransferases are sufficient for all O-methylation reactions of ipecac alkaloid biosynthesis in root culture of Psychotria ipecacuanha. J. Biol. Chem. 285, 7722–7738. doi: 10.1074/jbc.M109.086157
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Orozco-Cárdenas, M. L., Narváez-Vásquez, J., and Ryan, C. A. (2001). Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13, 179–191. doi: 10.1105/tpc.13.1.179
Pauwels, L., Morreel, K., De Witte, E., Lammertyn, F., Van Montagu, M., Boerjan, W., et al. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. U.S.A. 29, 1380–1385. doi: 10.1073/pnas.0711203105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pavei, C., Kaiser, S., Borré, G. L., and Ortega, G. G. (2010). Validation of a LC method for polyphenols assay in cat's claw (Uncaria tomentosa). J. Liq. Chromatogr. Relat. Technol. 33, 1551–1561. doi: 10.1080/10826076.2010.503753
Peebles, C. A., Hughes, E. H., Shanks, J. V., and San, K. Y. (2009). Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metab. Eng. 11, 76–86. doi: 10.1016/j.ymben.2008.09.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Peterson, G. L. (1977). A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83, 346–356. doi: 10.1016/0003-2697(77)90043-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pütter, J. (1974). “Peroxidases,” in Methods of Enzymatic Analysis, ed H. U. Bergmeyer (New York, NY: Verlag Chemie-Academic Press), 685–690. doi: 10.1016/B978-0-12-091302-2.50033-5
Qian, W. J., Liu, T., Monroe, M. E., Strittmatter, E. F., Jacobs, J. M., Kangas, L. J., et al. (2005). Probability-Based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis:? the human proteome. J. Proteome Res. 4, 53–62. doi: 10.1021/pr0498638
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Quan, L. J., Zhang, B., Shi, W. W., and Li, H. Y. (2008). Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 50, 2–18. doi: 10.1111/j.1744-7909.2007.00599.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ramos-Valdivia, A. C., Huerta-Heredia, A. A., Trejo-Tapia, G., and Cerda-García-Rojas, C. M. (2012). “Secondary metabolites as non-enzymatic plant protectors from oxidative stress” in Oxidative Stress in Plants: Causes, Consequences and Tolerance, N. A. Anjum, S. Umar, and A. Ahmad (New Delhi: IK International Publishers), 413–441.
Ruegsegger, A., Schmutz, D., and Brunold, C. (1990). Regulation of glutathione synthesis by cadmium in Pisum sativum. Plant Physiol. 93, 1579–1584. doi: 10.1104/pp.93.4.1579
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sasaki-Sekimoto, Y., Taki, N., Obayashi, T., Aono, M., Matsumoto, F., Sakurai, N., et al. (2005). Coordinated activation of metabolic pathways for antioxidants and defense compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J. 44, 653–668. doi: 10.1111/j.1365-313X.2005.02560.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. doi: 10.1021/ac950914h
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sottomayor, M., Lopes Cardoso, I., Pereira, L. G., and Ros Barceló, A. (2004). Peroxidase and the biosynthesis of terpenoid indole alkaloids in the medicinal plant Catharanthus roseus (L.) G. Don. Phytochem. Rev. 3, 159–171. doi: 10.1023/B:PHYT.0000047807.66887.09
Szabó, L. F. (2008). Rigorous biogenetic network for group of indole alkaloids derived from strictosidine. Molecules 13, 1875–1896. doi: 10.3390/molecules13081875
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Takahama, U. (2004). Oxidation of vacuolar and apoplastic phenolic substrate by peroxidase: physiological significance of the oxidation reactions. Phytochem. Rev. 3, 207–219. doi: 10.1023/B:PHYT.0000047805.08470.e3
Trejo-Tapia, G., Sepúlveda-Jiménez, G., Trejo-Espino, J. L., Cerda-García-Rojas, C. M., de la Torre, M., Rodríguez-Monroy, M., et al. (2007). Hydrodynamic stress induces monoterpenoid oxindole alkaloid accumulation by Uncaria tomentosa (Willd) D. C. cell suspension cultures via oxidative burst. Biotechnol. Bioeng. 98, 230–238. doi: 10.1002/bit.21384
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tu, Y., Rochfort, S., Liu, Z., Ran, Y., Griffith, M., Badenhorst, P., et al. (2010). Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 22, 3357–3373. doi: 10.1105/tpc.109.072827
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vera-Reyes, I., Huerta-Heredia, A. A., Ponce-Noyola, T., Flores-Sanchez, I. J., Esparza-García, F., Cerda-García-Rojas, C. M., et al. (2013). Strictosidine-related enzymes involved in the alkaloid biosynthesis of Uncaria tomentosa root cultures grown under oxidative stress. Biotechnol. Prog. 29, 621–630. doi: 10.1002/btpr.1723
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vierling, E. (1991). The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 579–620. doi: 10.1146/annurev.pp.42.060191.003051
Weidhase, R. A., Kramell, H. M., Lehmann, J., Liebisch, H. W., Lerbs, W., and Parthier, B. (1987). Methyl jasmonate-induced changes in the polypeptide pattern of senescing barley leaf segments. Plant Sci. 51, 177–186. doi: 10.1016/0168-9452(87)90191-9
Zhang, H., Han, B., Wang, T., Chen, S., Li, H., Zhang, Y., et al. (2012). Mechanisms of plant salt response: insights from proteomics. J. Proteome Res. 11, 49–67. doi: 10.1021/pr200861w
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, K., Yang, E. B., Tang, W. Y., Wong, K. P., and Mack, P. (1997). Inhibition of glutathione reductase by plant polyphenols. Biochem. Pharmacol. 54, 1047–1053. doi: 10.1016/S0006-2952(97)00315-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhao, J., Davis, L. C., and Verpoorte, R. (2005). Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 23, 283–333. doi: 10.1016/j.biotechadv.2005.01.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zubieta, C., Ross, J. R., Koscheski, P., Yang, Y., Pichersky, E., and Noel, J. P. (2003). Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15, 1704–1716. doi: 10.1105/tpc.014548
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: oxidative stress, Uncaria tomentosa, proteome, antioxidant responses, glutathione
Citation: Vera-Reyes I, Huerta-Heredia AA, Ponce-Noyola T, Cerda-García-Rojas CM, Trejo-Tapia G and Ramos-Valdivia AC (2015) Monoterpenoid indole alkaloids and phenols are required antioxidants in glutathione depleted Uncaria tomentosa root cultures. Front. Environ. Sci. 3:27. doi: 10.3389/fenvs.2015.00027
Received: 06 January 2015; Accepted: 20 March 2015;
Published: 09 April 2015.
Edited by:
Rene Kizek, Central European Institute of Technology in Brno, Czech RepublicReviewed by:
Naser A. Anjum, University of Aveiro, PortugalCopyright © 2015 Vera-Reyes, Huerta-Heredia, Ponce-Noyola, Cerda-García-Rojas, Trejo-Tapia and Ramos-Valdivia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana C. Ramos-Valdivia, Departamento de Biotecnología y Biongeniería, Centro de Investigación y de Estudios Avanzados, Instituto Politécnico Nacional, A. P. 14-740, México, D. F. 07000, MexicoYXJhbW9zQGNpbnZlc3Rhdi5teA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.