
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Environ. Chem. , 15 August 2023
Sec. Environmental Analytical Methods
Volume 4 - 2023 | https://doi.org/10.3389/fenvc.2023.1168328
 Agnieszka Gruszecka-Kosowska1*
Agnieszka Gruszecka-Kosowska1* Antonios Ampatzoglou2
Antonios Ampatzoglou2 Javier Pineda-Pampliega3,4
Javier Pineda-Pampliega3,4 Octavian Augustin Mihalache5
Octavian Augustin Mihalache5This perspective highlights the role of new-generation analytical techniques in future applications in environmental sciences since the shift to the One Health research paradigm. It reviews the interactions between the compartments of One Health and indicates the current challenges in traditional environmental research. The term One Health was first used at the beginning of the 20th century, yet much is still needed for the cross-disciplinary research collaboration required in this approach.
The natural environment and its compartments constitute the basis of life on Earth. It affects not only to human health but also that of animals and the environment. The concept of One Health is longstanding, first known as One Medicine, then One World, and finally One Health (Mackenzie and Jeggo, 2019). The term One Health was first used around 2003–2004 in relation to severe acute respiratory (SARS) and avian influenza H5N1 diseases (Mackenzie and Jeggo, 2019). But still, there is no unified definition of One Health. The University of California defines One Health as an approach to ensure the wellbeing of humans, animals, and the environment using collaborative problem-solving on local, national, and global scales (Mackenzie and Jeggo, 2019). This definition underlines that these three components must be treated equally and, more importantly, highlights their interconnections, which means that they can affect one another (Figure 1).
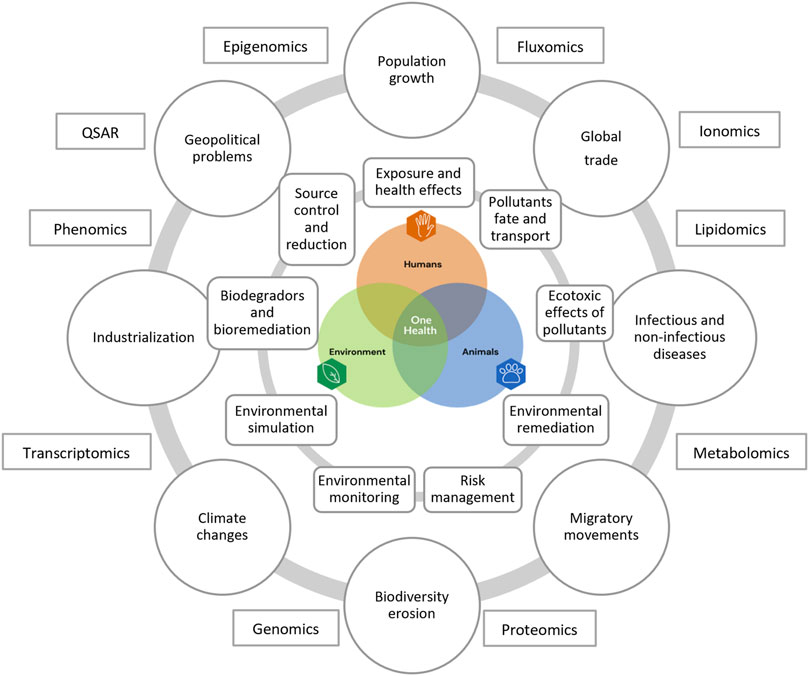
FIGURE 1. Environmental research challenges in the prism of the One Health approach and current life sciences analytical methods (redrawn from Liébana, 2022 and based on Ampatzoglou et al., 2022; Gruszecka-Kosowska et al., 2022; Destoumieux-Garzon et al., 2018; Zhu et al., 2021).
The intensive industrial and agricultural activities, along with urbanization, disrupt the natural balance of our planet (Zhang et al., 2023), leading to higher levels of known chemicals in the environment, as well as the introduction of new chemicals. The new chemicals introduced by anthropogenic activities are named contaminants of emerging concern (CECs) or emerging contaminants (ECs). CECs are likely to cause adverse health and environmental impacts and usually are not yet regulated under environmental laws (Bayabil et al., 2022). They are also recognized as xenobiotics, a term referring to all foreign chemicals (to the biological organism under consideration), and they may cause adverse effects (Ortiz et al., 2022). Among xenobiotics, currently, special attention is given to pharmaceuticals, personal care products, endocrine-disrupting chemicals (EDCs), and microbiota-disrupting chemicals (MDCs) (Ampatzoglou et al., 2022). The list is not exhaustive, and more will likely be added as adverse health effects are reported in case studies related to environmental exposure.
As all environmental compartments are interlinked, there is a strong trend in research to take a global approach, adopting the perspective of ecosystems. An excellent example is the gut microbiome, with the state of health of the host organism (aka eubiosis) depending on the microorganisms living in the gut and the host-microorganism interactions. In this sense, it can be considered as ecosystem. The removal of xenobiotics from the organism via chemical transformation is divided into following steps: host absorption, distribution, metabolism, and excretion, called ADME approach (Tsaioun et al., 2016). Recent discoveries in the medical sciences revealed that the removal of xenobiotics from the organism is closely related with the human microbiota, that might play important roles in these processes (Silbergeld et al., 2017; Koppel et al., 2018; Abdelsalam et al., 2020; Dikeocha et al., 2022; Miglani et al., 2022).
Environmental analysis consist of three main stages: sampling and sample preparation, sample analysis, and result evaluation. To achieve reliable results, sampling needs to be representative, and measurements must be accurate (Hussain and Kecili, 2020). Depending on the environmental compartment, samples can be gaseous (ambient or soil air), liquid (water or wastewater samples, urine, or blood), or solid (soil, waste, or biological tissues). The state of matter as well as the target pollutants, investigated in the analyzed samples, determine the analytical methods of detection. Traditional environmental analysis use methods that determine the content of known pollutants. This implies that the pollutants or groups of pollutants of interest are known. To detect investigated contaminants, first extraction techniques must be applied to purify and concentrate the analytes in the medium intended to be analyzed using the analytical instrument. Currently, the most popular analytical techniques used for investigating environmental contaminants are gas chromatography (GC), high-performance liquid chromatography (HPLC), ion chromatography (IC), size-exclusion chromatography (SEC), and supercritical fluid chromatography (SFC), and spectroscopic techniques, such as UV-visible (UV-vis), infrared spectroscopy (IR), and Raman spectroscopy (Hussain and Kecili, 2020). However, as we shift from monitoring, i.e., measuring the concentration of the analyzed pollutants in a particular environmental compartment, to investigations on their fate and ecological and health effects due to their presence, exposure and cycling in the environment, brand-new methods and tools are required.
Since the exposome has become the new research paradigm in biomedical science (González-Domínguez et al., 2020) the link between environmental sciences and life sciences has tightened. The term exposome was originally introduced to represent everything that the genome could not explain in terms of chronic disease in developed countries, and thus revealed nongenetic causes of disease and gene-environment interactions throughout the lifetime of the population (Zhang et al., 2021). The original concept of the exposome, described in 2005 as all environmental exposures, including lifestyle factors and excluding genetic factors, throughout a lifetime from the prenatal period onwards, has evolved, with various definitions in existence (Wild, 2012). The latest expansion of the exposome definition has included the presence of chemicals in environmental media such as air, water, diet, and the built environment (Zhang et al., 2021).
The concept of exposome is challenging as it involves multiple measures of each environmental exposure to the chemical under investigation and its subsequent effects on the human body, integrating many external and internal exposures from different sources continuously over the life course (Escher et al., 2022). Thus, this presents an excellent opportunity to integrate new analytical methods, including omics techniques and new approach methodologies (NAMs) in toxicology, such as Adverse Outcome Pathways (AOPs) or Source-To-Outcome (STO), into classical environmental sciences.
The climate change is widely called a threat multiplier to the One Health approach (Black and Butler, 2014). As climate change we understand the changes of the climate caused by the anthropogenic activities and resulting in magnifying the frequency and intensity of the extreme weather events (Adamo et al., 2021; Ondiko et al., 2022).
In terms of environmental pollution, it is prognosed that climate change could cause such changes in the weather conditions that will amplify air pollution (Zhang et al., 2022), and the negative impact of air pollution on human health in undeniable (Manisalidis et al., 2020). Aligning One Health with climate change is suggested by Essack (2018) to result in entrenching the environment compartment in the One Health triad.
The climate change phenomenon affects not only the state of the natural environment but also have wider implications. Regarding the disease transmissions, climate change might significantly impact the spatial distribution of disease vectors, encroachment in the habitats of animals, disturbing the pathways of animal migration, and changes in the functioning and management of crops, animals, and their habitats (Zhang et al., 2022).
The impact of climate change on food safety is not well known yet, however the main issues that need to be addressed in relation to food production chain are as follows: soil quality and degradation, marine environment pollution and seafood production, changing weather conditions and its impact on livestock and husbandry production, increased antimicrobial resistance, water and food-borne diseases related with zoonosis pathogen transmissions, and food spoilage (Misiou and Koutsoumanis, 2023).
Climate change become the game-changer that indicated that even better connections than was thought were needed among various scientific disciplines, are required to tackle ecological systems threats (Stephen, 2023).
Antimicrobial resistance (AMR) is a global public health threat affecting human, animal, plant, and environmental health. Interestingly, microbiomes may play an important role in the course of AMR development, with the gut microbiome believed to be a reservoir for antibiotic resistance genes (Gibson et al., 2015; Anthony et al., 2021) and the role of the environment increasingly coming into focus (Larsson and Flach, 2022).
The irresponsible use of antibiotics is considered the major player in AMR (O’Neill, 2016; World Health Organization, 2021), and is well-established. Nevertheless, non-antibiotic antimicrobials, have previously been suggested as playing a role (Scientific Committee on Consumer Safety, 2010). Examples include MDCs triclosan and parabens (Aguilera et al., 2020). Their main contribution to AMR stems from the potential of microorganisms exposed to them over time to gradually develop resistance against them. However, another contribution may be more critical, albeit not yet conclusive. Specifically, microorganisms exposed to them may develop resistance against antibiotics (Scientific Committee on Consumer Safety, 2010; Ribado et al., 2017).
Exposure to non-antibiotic antimicrobials such as MDCs may apply selective pressure in favor of taxa with relevant metabolic pathways (López-Moreno et al., 2021; Torres-Sánchez et al., 2021). Although the selective pressure examples above would primarily be attributed to the direct antimicrobial effects of the MDCs in question, an alternative type of pressure is also applicable in the case of xenobiotics with no direct antimicrobial properties, such as those acting as nutrient sources (Frame et al., 2020) or altering the metabolism of (Anwar et al., 2018) selective taxa within the microbiome. In these cases, the non-antimicrobial xenobiotics may mediate directly or via the selected taxa, modulation of the composition and function of exposed microbiota, potentially reducing the microbiome’s diversity and inducing dysbiosis and adverse metabolic effects. Of course, these outcomes may be beneficial/desirable or detrimental/non-desirable from our perspective. Nevertheless, this highlights the potential for the concept of single-cell resistance to antimicrobials to be extended to xenobiotics, more generally, in the context of microbial communities. Under this perspective, the notion of xenobiotic resistance may warrant further consideration (Ampatzoglou et al., 2022).
Due to its global public health significance, increasing efforts have focused on combating AMR. Alternatives to antibiotics currently under investigation, include antibodies, probiotics, vaccines, phage lysins and antimicrobial peptides (Czaplewski et al., 2016; Alam et al., 2019), as well as versatile CRISPR-Cas9 antimicrobials, which are increasingly explored particularly in the context of multidrug-resistant microbial infections (Getahun et al., 2022). High-throughput methodologies are critical in the fight against AMR. For example, high-throughput qPCR screening can be used to simultaneously investigate the presence of antimicrobial resistance genes (ARGs) and mobile genetic elements, including plasmid replicons, integrons, and insertion sequences, in a wide range of bacteria (Delannoy et al., 2022), while single-cell spectroscopic techniques may detect signature changes in the spectra of pathogenic bacteria, including differentiating between antibiotic-sensitive and resistant strains (Lu et al., 2023; Xu, Cui, and Song, 2023). Moreover, high-throughput organoid-based novel model systems, e.g., air–liquid interface, can be used to screen antimicrobial peptides as potential drug candidates (Zhang et al., 2023). Additionally, bioinformatics is critical in the advancement of AMR research. A number of methods exist for detecting novel ARGs in silico, based on sequence or structural similarity, which, of course, can only be applied to known structural classes of ARGs. Functional metagenomics, e.g., shotgun metagenomic sequencing, overcome this limitation and can, thus, be used for the discovery of entirely new classes of ARGs (Bengtsson-Palme et al., 2023), although certain aspects, including the selection of appropriate bioinformatic methodology and databases, require careful consideration (Bengtsson-Palme, Larsson, and Kristiansson, 2017; Angers-Loustau et al., 2018). Finally, in recent years, machine learning has been increasingly applied to AMR research, including decision support systems, random forest, rapid detection, decision trees, high-throughput screening, and multivariate analysis, as well as more advanced deep learning models, such as artificial neural networks (Farhat et al., 2023).
As we all must eat, food safety and security have always been important issues, more so nowadays. All food originates from soils where edible plants are grown either for direct food production or for feed production, the latter leading to environmental exposure of food-producing animals. Thus, the quality and quantity of agricultural soil, the quality of water used for irrigation (including environment-friendly water treatment methods), the air quality (as it might be a source of contamination for crops), and the inorganic and organic contaminants that might be present in waste and wastewater used for fertilization and/or irrigation of crops (European Food Safety Authority, 2021) are all becoming burning topics, especially under current or developing geopolitical issues and global food shortages. As soil quality and quantity are under high pressure, remediation and restoration have become particular interests for researchers (Dessureault-Rompré, 2022). Looking at the significant achievements in research in the domain of living organisms under the remit of life sciences, similar approaches could be considered for adoption in environmental research. Although, in the case of soils, the concept of bioremediation is not new, such approaches have not been widely adopted due to limitations inherent to previous research and their effectiveness (Chandran et al., 2020). Nevertheless, as new techniques become potentially available, the opportunity arises to employ them in environmental sciences to enhance environmental research (Kim et al., 2022; Sharma et al., 2022). The “farm to fork” concept refers to the journey that food takes from the farm where it is grown or produced to the consumer’s plate. Consumer food safety behavior is a critical part of this journey as it plays a significant role in reducing the risk of foodborne illnesses (Fung et al., 2018; Mihalache et al., 2021).
Consumer food hygiene practices play a critical role in maintaining the One Health approach. By adopting safe food handling practices at home, such as washing their hands before handling food, properly storing food to prevent contamination, cooking food at the recommended temperature, and properly cleaning utensils and surfaces that come into contact with food, consumers can contribute to food safety (World Health Organization, 2006; Centers for Disease Control and Prevention, 2022). Consumers can also make informed purchasing decisions by looking for products that meet food safety standards, checking expiration dates, and avoiding products that are damaged or past their expiration date. Additionally, if consumers have any food safety concerns, they can report them to the relevant authorities to help prevent foodborne illnesses (Food and Agriculture Organization, 2023). It is important for consumers to be aware of the potential risks associated with different types of food and to take the necessary precautions to reduce those risks (Borda et al., 2021). By doing so, consumers can help ensure that the food they consume is microbiologically safe (Centers for Disease Control and Prevention, 2022). Following food hygiene practices reduces the risk of foodborne illnesses and contributes to the overall goal of the One Health approach, which is to protect and promote the health and wellbeing of humans, animals, and the environment.
The One Health approach applies to consumers’ dietary trends as well. EAT-Lancet recommendations include consuming a balanced diet, reducing the consumption of processed food, minimizing the consumption of animal-based products, and incorporating more plant-based foods in their diets (Willett et al., 2019). Shifting to alternative protein sources is part of the policy framework of the United Nations Sustainable Development Goals (2023). Therefore, consumers can contribute both to UN SDGs and the One Health system by adopting sustainable dietary trends.
The gut microbiome’s significance for human health has been well-established. A similar importance can be attributed to the environmental microbiome, where microorganisms play a crucial role in chemical processes (Naylor et al., 2022). The identification of various relevant organisms is an essential step to understanding the chemical processes occurring in the soil. Traditional cultivation methods in laboratory conditions can only detect less than 1% of soil microorganisms due to the limitation of laboratory media and conditions to support their growth (Wydro, 2022). However, omics techniques have overcome this limitation by analyzing nucleic acids (DNA and RNA) extracted from the soil microbiome to determine microbial biodiversity circumventing the need for cultivation, thus leading to a faster and more efficient process. This improved approach has also unlocked novel environmental applications of relevant taxa from the soil microbiome, ranging from improving crop development (e.g., disease resistance, reduced nutrient requirements) to remediation of contaminated soils (Wydro, 2022).
Pollution is a major environmental issue, and some pollutants are of a protein nature. Thus, the use of proteomics in soil research has been growing in recent years (Gouveia et al., 2019). Compared to classical techniques, proteomics offers several advantages in ecotoxicology. Proteomics allows for the comprehensive profiling of soil proteins, enabling a more precise determination of the proteins present and their quantification. This information provides a more comprehensive view of the molecular changes that occur in response to environmental stressors, enabling the construction of toxicity pathways and adverse outcome pathways (Gouveia et al., 2019). Additionally, this technique allows for the identification of novel biomarkers that can be used to assess environmental quality with advantages over classical biomarkers, such as easier detection and greater sensitivity to environmental changes. Overall, proteomics provides a more detailed and comprehensive approach to studying the impact of proteinic pollution on the environment, as well as a faster evaluation than classic methods. New Approach Methodologies (NAMs) encompass a wide range of technologies, methodologies, and approaches that provide information on risk assessment and chemical hazard without involving animal testing (Stucki et al., 2022). This includes in silico, in chemico, in vitro, and ex vivo approaches (Stucki et al., 2022). According to Andersen et al. (2019), NAMs for toxicity testing can be categorized into four levels: The first level relies on computational screening, the second involves high-throughput in vitro screening, the third focuses on fit-for-purpose assays selected based on presumptive modes of action, and the fourth includes more complex multi-dimensional or multi-cellular assays. However, in the context of environmental evaluation, NAMs are currently more commonly used at the first level, with Quantitative Structure Activity Relationship (QSAR) models being particularly emphasized.
These mathematical models rely on the principle that hazard is an inherent property of chemicals and is directly related to their molecular structure. They can predict the intrinsic hazard of chemicals by comparing them to large data sets (Gramatica et al., 2018). QSAR models offer several advantages in environmental studies: 1) They save time and resources in risk assessment by predicting the properties of new chemicals; 2) They help screen and prioritize chemicals’ impact, enabling regulators to make informed safety decisions; 3) They provide a comprehensive view of chemicals by combining multiple effects and properties into cumulative endpoints; 4) They eliminate the need for costly and time-consuming experimental data by utilizing existing chemical information.
Environmental protection, environmental epidemiology, and biodiversity preservation have become major challenges in the current world. The One Health approach underlines the necessity of connection and cooperation. Thus, also in the upcoming tasks of the environmental sciences research field, cooperation between traditional approaches and new achievements from various research disciplines is required.
Omics and NAM techniques have established a permanent presence in environmental research. The speed and level of detail at which the information is generated using these techniques far outperform traditional methods. Although there is room for improvement, incorporating these techniques, and even using several of them simultaneously, is crucial for well-designed environmental research.
Among environmental sciences, all aspects related to food safety are under special concern. Consumers’ food safety practices are crucial as they can prevent the risk of foodborne illnesses. A combination of proper food hygiene practices and the integration of sustainable dietary trends will help improve consumers, animals, and the environment health. Hence, contributing to the main scope of the One Health approach is to achieve optimal health for the above-mentioned triad. Finally, AMR, one of the main threats to One Health, and particularly its interaction with environmental microbiomes, is one of the fields that warrants further research.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
AG-K, AA, JP-P, and OAM worked together to develop and write the manuscript. All authors contributed to the article and approved the submitted version.
This material is based upon work supported by the EU-FORA Program Cycle, 2021/2022. JP-P was supported by a postdoctoral grant Margarita Salas from the European Union—NextGenerationEU through the Complutense University of Madrid.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelsalam, N. A., Ramadan, A. T., ElRakaiby, M. T., and Aziz, R. K. (2020). Toxicomicrobiomics: the human microbiome vs. Pharmaceutical, dietary, and environmental xenobiotics. Front. Pharmacol. 11, 390. doi:10.3389/fphar.2020.00390
Adamo, N., Al-Ansari, N., and Sissakian, V. (2021). Review of climate change impacts on human environment: past, present and future projections. Engineering 13, 605–630. doi:10.4236/eng.2021.1311044
Aguilera, M., Gálvez-Ontiveros, Y., and Rivas, A. (2020). Endobolome, a new concept for determining the influence of microbiota disrupting chemicals (MDC) in relation to specific endocrine pathogenesis. Front. Microbiol. 11, 578007. doi:10.3389/fmicb.2020.578007
Alam, M. M., Islam, M., Wahab, A., and Billah, M. (2019). Antimicrobial resistance crisis and combating approaches. J. Med. 20 (1), 38–45. doi:10.3329/jom.v20i1.38842
Ampatzoglou, A., Gruszecka-Kosowska, A., and Aguilera-Gómez, M. (2022). Microbiota analysis for risk assessment of xenobiotics: toxicomicrobiomics, incorporating the gut microbiome in the risk assessment of xenobiotics and identifying beneficial components for one health. EFSA J. 20, e200915. doi:10.2903/j.efsa.2022.e200915
Andersen, M. E., McMullen, P. D., Phillips, M. B., Yoon, M., Pendse, S. N., Clewell, H. J., et al. (2021). Developing context appropriate toxicity testing approaches using new alternative methods (NAMs). ALTEX 36, 523–534. doi:10.14573/altex.1906261
Angers-Loustau, A., Petrillo, M., Bengtsson-Palme, J., Berendonk, T., Blais, B., Chan, K. G., et al. (2018). The challenges of designing a benchmark strategy for bioinformatics pipelines in the identification of antimicrobial resistance determinants using next generation sequencing technologies. F1000Res. 13 (7), 459. doi:10.12688/f1000research.14509.2
Anthony, W. E., Burnham, C.-A. D., Dantas, G., and Kwon, J. H. (2021). The gut microbiome as a reservoir for antimicrobial resistance. J. Infect. Dis. 223, 209–213. doi:10.1093/infdis/jiaa497
Anwar, S., Bhandari, U., Panda, B. P., Dubey, K., Khan, W., and Ahmad, S. (2018). Trigonelline inhibits intestinal microbial metabolism of choline and its associated cardiovascular risk. J. Pharm. Biomed. Anal. 159, 100–112. doi:10.1016/j.jpba.2018.06.027
Bayabil, H. K., Teshome, F. T., and Li, Y. C. (2022). Emerging contaminants in soil and water. Front. Environ. Sci. 10, 873499. doi:10.3389/fenvs.2022.873499
Bengtsson-Palme, J., Abramova, A., Berendonk, T. U., Coelho, L. P., Forslund, S. K., Gschwind, R., et al. (2023). Towards monitoring of antimicrobial resistance in the environment: for what reasons, how to implement it, and what are the data needs? Env. Int. 178, 108089. doi:10.1016/j.envint.2023.108089
Bengtsson-Palme, J., Larsson, D. G. J., and Kristiansson, E. (2017). Using metagenomics to investigate human and environmental resistomes. J. Antimicrob. Chemother. 72 (10), 2690–2703. doi:10.1093/jac/dkx199
Black, P. F., and Butler, C. D. (2014). One Health in a world with climate change. Rev. Sci. Tech. 33 (2), 465–473. doi:10.20506/rst.33.2.2293
Borda, D., Mihalache, O. A., Dumitraşcu, L., Gafiţianu, D., and Nicolau, A. I. (2021). Romanian consumers’ food safety knowledge, awareness on certified labelled food and trust in information sources. Food control. 120, 107544. doi:10.1016/j.foodcont.2020.107544
Centers for Disease Control and Prevention (2022). When and how to wash your hands. Available at: https://www.cdc.gov/handwashing/when-how-handwashing.html#:∼:text=Lather%20your%20hands%20by%20rubbing,Need%20a%20timer%3F (Accessed February 7, 2023).
Chandran, H., Meena, M., and Sharma, K. (2020). Microbial biodiversity and bioremediation assessment through omics approaches. Front. Environ. Chem. 1, 570326. doi:10.3389/fenvc.2020.570326
Czaplewski, L., Bax, R., Clokie, M., Dawson, M., Fairhead, H., Fischetti, V. A., et al. (2016). Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 16 (2), 239–251. doi:10.1016/S1473-3099(15)00466-1
Delannoy, S., Hoffer, C., Youf, R., Dauvergne, E., Webb, H. E., Brauge, T., et al. (2022). High throughput screening of antimicrobial resistance genes in gram-negative seafood bacteria. Microorganisms, 10(6):6. doi:10.3390/microorganisms10061225
Dessureault-Rompré, J. (2022). Restoring soil functions and agroecosystem services through phytotechnologies. Front. Soil Sci. 2:927148. doi:10.3389/fsoil.2022.927148
Destoumieux-Garzón, D., Mavingui, P., Boetsch, G., Boissier, J., Darriet, F., Duboz, P., et al. (2018). The one health concept: 10 Years old and a long road ahead. Front. Vet. Sci. 5, 14. doi:10.3389/fvets.2018.00014
Dikeocha, I. J., Al-Kabsi, A. M., Miftahussurur, M., and Alshawsh, M. A. (2022). Pharmacomicrobiomics: influence of gut microbiota on drug and xenobiotic metabolism. FASEB J. 36 (6), e22350. doi:10.1096/fj.202101986R
Escher, S. E., Partosch, F., Konzok, S., Jennings, P., Luijten, M., Kienhuis, A., et al. (2022). Development of a roadmap for action on new approach methodologies in risk assessment. doi:10.2903/sp.efsa.2022.EN-7341
Essack, S. Y. (2018). Environment: the neglected component of the one health triad. Lancet Planet Health 2 (6), e238–e239. doi:10.1016/S2542-5196(18)30124-4
Farhat, F., Athar, M. T., Ahmad, S., Madsen, D. Ø., and Sohail, S. S. (2023). Antimicrobial resistance and machine learning: past, present, and future. Front. Microbiol. 14, 1179312. doi:10.3389/fmicb.2023.1179312
Food and Agriculture Organization (2023). One health. Available at: https://www.fao.org/one-health/en (Accessed February 7, 2023).
Frame, L. A., Costa, E., and Jackson, S. A. (2020). Current explorations of nutrition and the gut microbiome: a comprehensive evaluation of the review literature. Nutr. Rev. 78, 798–812. doi:10.1093/nutrit/nuz106
Fung, F., Wang, H.-S., and Menon, S. (2018). Food safety in the 21st century. Biomed. J. 41 (2), 88–95. doi:10.1016/j.bj.2018.03.003
Getahun, Y. A., Ali, D. A., Taye, B. W., and Alemayehu, Y. A. (2022). Multidrug-resistant microbial therapy using antimicrobial peptides and the CRISPR/Cas9 system. Vet. Med. (Auckl). 11 (13), 173–190. doi:10.2147/VMRR.S366533
Gibson, M. K., Crofts, T. S., and Dantas, G. (2015). Antibiotics and the developing infant gut microbiota and resistome. Curr. Opin. Microbiol. Antimicrob. - Microb. Syst. Biol. 27, 51–56. doi:10.1016/j.mib.2015.07.007
González-Domínguez, R., Jáuregui, O., Queipo-Ortuño, M. I., and Andrés-Lacueva, C. (2020). Characterization of the human exposome by a comprehensive and quantitative large-scale multianalyte metabolomics platform. Anal. Chem. 92, 13767–13775. doi:10.1021/acs.analchem.0c02008
Gouveia, D., Almunia, C., Cogne, Y., Pible, O., Degli-Esposti, D., Salvador, A., et al. (2019). Ecotoxicoproteomics: a decade of progress in our understanding of anthropogenic impact on the environment. J. Proteomics 198, 66–77. doi:10.1016/j.jprot.2018.12.001
Gramatica, P., Papa, E., and Sangion, A. (2018). QSAR modeling of cumulative environmental end-points for the prioritization of hazardous chemicals. Environ. Sci. Process. Impacts. 20, 38–47. doi:10.1039/c7em00519a
Gruszecka-Kosowska, A., Ampatzoglou, A., and Aguilera, M. (2022). Integration of omics approaches enhances the impact of scientific research in environmental applications. Int. J. Env. Res. Pub He 19 (14), 8758. doi:10.3390/ijerph19148758
Hussain, C. M., and Kecili, R. (2020). Modern environmental analysis techniques for pollutants. Berlin, Germany: Elsevier. doi:10.1016/C2018-0-01639-4
Kim, S., Hollinger, H., and Radke, E. G. (2022). Omics in environmental epidemiological studies of chemical exposures: a systematic evidence map. Environ. Int. 164, 107243. doi:10.1016/j.envint.2022.107243
Koppel, N., Maini Rekdal, V., and Balskus, E. P. (2018). Chemical transformation of xenobiotics by the human gut microbiota. Science 356 (6344), eaag2770. doi:10.1126/science.aag2770
European Food Safety Authority Panel on Biological Hazards Koutsoumanis, K., Allende, A., Álvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., Chemaly, M., et al. (2021). Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 19 (6), e06651. doi:10.2903/j.efsa.2021.6651
Larsson, D. G. J., and Flach, C.-F. (2022). Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269. doi:10.1038/s41579-021-00649-x
Liébana, E. (2022). One health in EFSA: Animal health and food safety. Brussels: Consumers and Environment Category. EESC 20 June 2022.
López-Moreno, A., Ruiz-Moreno, Á., Pardo, J., Cerk, K., Torres-Sánchez, A., Ortíz, P., et al. (2021). Human gut microbial taxa metabolizing dietary obesogens: a bpa 1 directed-culturing and bioinformatics combined approach. doi:10.21203/rs.3.rs-754318/v1
Lu, W., Li, H., Qiu, H., Wang, L., Feng, J., and Fu, Y. V. (2023). Identification of pathogens and detection of antibiotic susceptibility at single-cell resolution by Raman spectroscopy combined with machine learning. Front. Microbiol. 4 (13), 1076965. doi:10.3389/fmicb.2022.1076965
Mackenzie, J,S., and Jeggo, M. (2019). The one health approach-why is it so important? Trop. Med. Infect. Dis. 4 (2), 88. doi:10.3390/tropicalmed4020088
Manisalidis, I., Stavropoulou, E., Stavropoulos, A., and Bezirtzoglou, E. (2020). Environmental and health impacts of air pollution: a review. Front. Public Health 8, 14. doi:10.3389/fpubh.2020.00014
Miglani, R., Parveen, N., Kumar, A., Ansari, M. A., Khanna, S., Rawat, G., et al. (2022). Degradation of xenobiotic pollutants: an environmentally sustainable approach. Metabolites 12 (9), 818. doi:10.3390/metabo12090818
Mihalache, O. A., Dumitraşcu, L., Nicolau, A. I., and Borda, D. (2021). Food safety knowledge, food shopping attitude and safety kitchen practices among Romanian consumers: a structural modelling approach. Food control. 120, 107545. doi:10.1016/j.foodcont.2020.107545
Misiou, O., and Koutsoumanis, K. (2023). Climate change and its implications for food safety and spoilage. Trends Food Sci. Technol. 126, 142–152. doi:10.1016/j.tifs.2021.03.031
Naylor, D., McClure, R., and Jansson, J. (2022). Trends in microbial community composition and function by soil depth. Microorganisms 10 (3), 540. doi:10.3390/microorganisms10030540
Ondiko, J. H., Karanja, A. M., and Ombogo, O. (2022). A review of the anthropogenic effects of climate change on the physical and social environment. Open Access Libr. J. 9, 1–14. doi:10.4236/oalib.1107751
O’Neill, J. (2016). Tackling drug-resistant infections globally: final report and recommendations (report). United Kingdom: Government of the United Kingdom.
Ortiz, P., Torres-Sánchez, A., López-Moreno, A., Cerk, K., Ruiz-Moreno, Á., Monteoliva-Sánchez, M., et al. (2022). Impact of cumulative environmental and dietary xenobiotics on human microbiota: risk assessment for One Health. J. Xenobiot. 12, 56–63. doi:10.3390/jox12010006
Ribado, J. V., Ley, C., Haggerty, T. D., Tkachenko, E., Bhatt, A. S., and Parsonnet, J. (2017). Household triclosan and triclocarban effects on the infant and maternal microbiome. EMBO Mol. Med. 9, 1732–1741. doi:10.15252/emmm.201707882
Scientific Committee on Consumer Safety (2010). Opinion on triclosan (antimicrobial resistance) (No. SCCP/1251/09. European: directorate-General for Health and Consumers. European Commission.
Sharma, P., Singh, S. P., Iqbal, H. M. N., and Tong, Y. W. (2022). Omics approaches in bioremediation of environmental contaminants: an integrated approach for environmental safety and sustainability. Environ. Res. 211, 113102. doi:10.1016/j.envres.2022.113102
Silbergeld, E. K. (2017). The microbiome: modulator of pharmacological and toxicological exposures and responses. Toxicol. Pathol. 45, 190–194. doi:10.1177/0192623316672073
Stephen, C. (2023). Strengthening ties with public health for joint action on global health threats. Part 2. Can. Vet. J. 64 (4), 385–387.
Stucki, A. O., Clippinger, A. J., Henry, T. R., Hirn, C., Stedeford, T. J., and Terry, C. (2022). Editorial: chemical testing using new approach methodologies (NAMs). Front. Toxicol. 4, 1048900. doi:10.3389/ftox.2022.1048900
Torres-Sánchez, A., Pardo-Cacho, J., López-Moreno, A., Ruiz-Moreno, Á., Cerk, K., and Aguilera, M. (2021). Antimicrobial effects of potential probiotics of Bacillus spp. isolated from human microbiota: in vitro and in silico methods. Microorganisms 9, 1615. doi:10.3390/microorganisms9081615
Tsaioun, K., Blaauboer, B. J., and Hartung, T. (2016). Evidence-based absorption, distribution, metabolism, excretion (ADME) and its interplay with alternative toxicity methods. ALTEX - Altern. animal Exp. 33 (4), 343–358. doi:10.14573/altex.1610101
United Nations Sustainable Development Goals (2023). Available at: https://www.un.org/sustainabledevelopment/ (Accessed February 8, 2023).
Wild, C. P. (2012). The exposome: from concept to utility. Int. J. Epidemiol. 41, 24–32. doi:10.1093/ije/dyr236
Willett, W., Rockström, J., Loken, B., Springmann, M., Lang, T., Vermeulen, S., et al. (2019). Food in the anthropocene: the EAT–lancet commission on healthy diets from sustainable food systems. Lancet 393 (10170), 447–492. doi:10.1016/S0140-6736(18)31788-4
World Health Organization (2021). Antimicrobial resistance. Antimicrob. Resist. Available at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed January 1, 2023).
Wydro, U. (2022). Soil microbiome study based on DNA extraction: a review. Water 14, 3999. doi:10.3390/w14243999
Xu, J., Cui, L., and Song, Y. (2023). Editorial: single-cell tools to combat antimicrobial resistance. Front. Microbiol. 14:1256107. doi:10.3389/fmicb.2023.1256107
Zhang, P., Carlsten, C., Chaleckis, R., Hanhineva, K., Huang, M., Isobe, T., et al. (2021). Defining the scope of exposome studies and research needs from a multidisciplinary perspective. Environ. Sci. Tech. Let. 8 (10), 839–852. doi:10.1021/acs.estlett.1c00648
Zhang, R., Tang, X., Liu, J., Visbeck, M., Guo, H., Murray, V., et al. (2022). From concept to action: a united, holistic and one health approach to respond to the climate change crisis. Infect. Dis. Poverty 11, 17. doi:10.1186/s40249-022-00941-9
Zhang, S., Han, Y., Peng, J., Chen, Y., Zhan, L., and Li, J. (2023a). Human health risk assessment for contaminated sites: a retrospective review. Environ. Int. 171, 107700. doi:10.1016/j.envint.2022.107700
Zhang, S., Shen, J., Wang, X., Sun, X., Wu, Y., Zhang, M. R., et al. (2023b). Integration of organoids in peptide drug discovery: rise of the high-throughput screening. View, 20230010. doi:10.1002/VIW.20230010
Keywords: One Health, environment, bioremediation, microbiome engineering, antimicrobial resistance, food safety
Citation: Gruszecka-Kosowska A, Ampatzoglou A, Pineda-Pampliega J and Mihalache OA (2023) How new generation techniques are shaping the future of environmental sciences. Front. Environ. Chem. 4:1168328. doi: 10.3389/fenvc.2023.1168328
Received: 17 February 2023; Accepted: 03 August 2023;
Published: 15 August 2023.
Edited by:
Subhankar Singha, JIS University, IndiaReviewed by:
Monisha Singha, Postdoctoral Research Associate, United KingdomCopyright © 2023 Gruszecka-Kosowska, Ampatzoglou, Pineda-Pampliega and Mihalache. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnieszka Gruszecka-Kosowska, YWduaWVzemthLmdydXN6ZWNrYUBhZ2guZWR1LnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.