- 1Department of Earth and Environmental Sciences, Wright State University, Dayton, OH, United States
- 2Department of Ocean Sciences, University of California, Santa Cruz, Santa Cruz, CA, United States
- 3Department of Marine Science, University of Connecticut, Groton, CT, United States
- 4Oceanography Department, University of Alaska Fairbanks, Fairbanks, AK, United States
Methylmercury (MeHg) is a neurotoxin that bioaccumulates to potentially harmful concentrations in Arctic and Subarctic marine predators and those that consume them. Monitoring and modeling MeHg bioaccumulation and biogeochemical cycling in the ocean requires an understanding of the mechanisms behind net mercury (Hg) methylation. The key functional gene pair for Hg methylation, hgcAB, is widely distributed throughout ocean basins and spans multiple microbial phyla. While multiple microbially mediated anaerobic pathways for Hg methylation in the ocean are known, the majority of hgcA homologs have been found in oxic subsurface waters, in contrast to other ecosystems. In particular, microaerophilic Nitrospina, a genera of nitrite-oxidizing bacteria containing a hgcA-like sequence, have been proposed as a potentially important Hg methylator in the upper ocean. The objective of this work was therefore to examine the potential of nitrifiers as Hg methylators and quantify total Hg and MeHg across three Arctic and Subarctic seas (the Gulf of Alaska, the Bering Sea and the Chukchi Sea) in regions where Nitrospina are likely present. In Spring 2021, samples for Hg analysis were obtained with a trace metal clean rosette across these seas. Mercury methylation rates were quantified in concert with nitrification rates using onboard incubation experiments with additions of stable isotope-labeled Hg and NH4+. A significant correlation between Hg methylation and nitrification was observed across all sites (R2 = 0.34, p < 0.05), with the strongest correlation in the Chukchi Sea (R2 = 0.99, p < 0.001). Nitrospina-specific hgcA-like genes were detected at all sites. This study, linking Hg methylation and nitrification in oxic seawater, furthers understanding of MeHg cycling in these high latitude waters, and the ocean in general. Furthermore, these studies inform predictions of how climate and human interactions could influence MeHg concentrations across the Arctic in the future.
1 Introduction
The harmful bioaccumulating neurotoxin, methylmercury (MeHg), has increased over the past 150 years in Arctic and subarctic mammals, resulting in high concentrations in the Canadian Arctic and Northern Gulf of Alaska (NGA) marine predators (Beckmen et al., 2002; Dietz et al., 2009; Lehnherr et al., 2011; Wang et al., 2018). Consumption of high trophic level marine organisms serves as the main MeHg exposure route to humans (Sunderland and Mason, 2007). The health of indigenous communities that rely on marine predators, both fish and mammals, as a food source continues to be an area of concern as the MeHg blood concentrations of Arctic women remain high (AMAP, 2021). The largest source of inorganic mercury (HgII) to the global ocean is through atmospheric deposition (Mason et al., 2012). Rivers also serve as a large source of HgII to Arctic coastal zones compared to other ocean basins (Sørensen et al., 2016; Sonke et al., 2018; Dastoor et al., 2022; Zolkos et al., 2022). Mercury concentrations in ocean surface waters have been increasing since industrialization (Lamborg et al., 2014), with concurrent increases in marine MeHg concentrations (Médieu et al., 2022). Monitoring and modeling current and future MeHg production in the ocean requires an understanding of the mechanisms behind Hg methylation, but the pathways for MeHg formation are not clearly understood.
High MeHg concentrations have been observed just below the productive surface layer in the Arctic Ocean, Bering Sea, Northwest Atlantic and the North Pacific Ocean (Sunderland et al., 2009; Heimbürger et al., 2015; Schartup et al., 2015; Wang et al., 2018; Agather et al., 2019; Jonsson et al., 2022). These MeHg maxima residing within oxic layers are shallower than equatorial regions (Bowman et al., 2016; Kim et al., 2017) and result in a higher rate of MeHg bioaccumulation into the food chain (Heimbürger et al., 2015; Wang et al., 2018). Previous studies attribute higher MeHg concentrations in oxic layers to production in anoxic particle microenvironments or the resuspension and remineralization of MeHg bound to organic matter (Hammerschmidt and Fitzgerald, 2006; Ortiz et al., 2015; Motta et al., 2022). With low benthic-pelagic coupling in the central ocean, and given the rate of photo-degradation and demethylation of MeHg in the surface waters (DiMento and Mason, 2017; Munson et al., 2018), the question remains: by what mechanism is biotic Hg methylation occurring in the oxic surface waters at high latitudes? Lehnherr et al. (2011) developed a simple model based on their methylation and demethylation studies that estimated biotic MeHg formation accounts for a significant fraction of the MeHg present in polar marine waters but did not provide a mechanism for this process.
The presumed key functional gene pair for Hg methylation, hgcAB, is widely distributed throughout all ocean basins and spans multiple microbial phyla (Parks et al., 2013; Villar et al., 2020). All currently confirmed pathways for biotic marine MeHg production in marine water columns and sediment are anaerobic (Gilmour et al., 2013; Gionfriddo et al., 2020) and have been shown to coincide with oxygen minimum zones (Hammerschmidt and Bowman, 2012; Munson et al., 2018). However, across all ocean basins, 78% of discovered hgcA homologs have been found in oxic subsurface waters (Villar et al., 2020). In recent years, a new aerobic pathway for Hg methylation has been suggested in conjunction with nitrite-oxidation (Gionfriddo et al., 2016), the second, non-rate limiting reaction in nitrification (Delwiche, 1970). The putative Hg methylating microbes belong to two aerobic nitrite-oxidizing clades, Nitrospina and Nitrospira (Gionfriddo et al., 2016; Christiansen et al., 2019). While Nitrospira hgcAB genes have only been recovered in freshwater sediment and tidal marsh metagenomes (Gionfriddo et al., 2020), Nitrospina hgcA-like sequences have been found to be abundant across all ocean basins, including the Arctic (Bowman et al., 2020; Tada et al., 2020; Villar et al., 2020; Lin et al., 2021; Tada et al., 2021). As the most widely distributed microaerophilic putative Hg methylating microbe, it is hypothesized that Nitrospina could play a key role in global MeHg production in the ocean subsurface (Villar et al., 2020), although in situ measurements of Hg methylation rates by cultured Nitrospina have yet to be completed.
Recently, in the Equatorial Atlantic, a strong positive correlation was found between nitrification and Hg methylation at the chlorophyll a maximum and in surface waters (Starr et al., 2022). While Nitrospina-specific 16S transcripts were recovered, Nitrospina-specific hgcA-like gene abundance was not assessed (Starr et al., 2022). Here, we further investigate the correlation between this putative microbially mediated aerobic Hg methylation pathway and nitrite oxidation across five nutrient-replete, polar regions (NGA, Unimak Pass, Bering Sea, Bering Strait and Chukchi Sea; Figure 1) where Nitrospina hgcA-like sequences have been previously reported (Bowman et al., 2020).
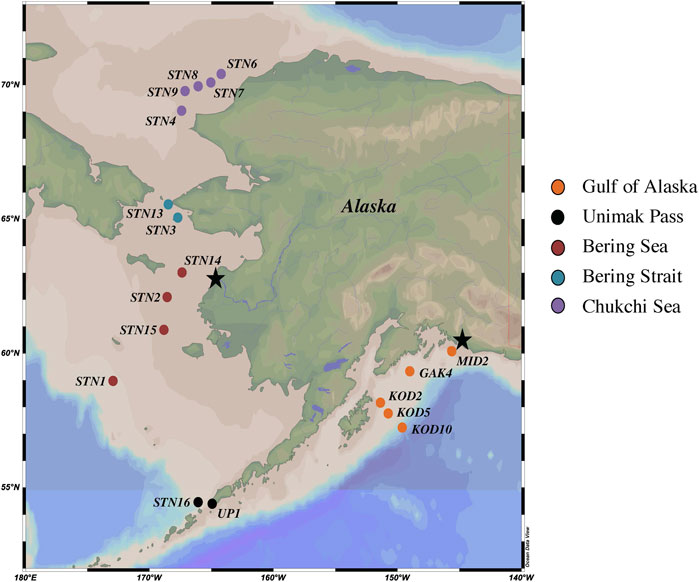
FIGURE 1. Map of NGA, Unimak Pass, Bering Sea, Bering Strait and Chukchi Sea 2021 sampling stations. Yukon and Copper River outflows denoted by black stars.
2 Methods
2.1 Sample collection
Seawater samples were collected between April and June 2021 in the NGA, Bering Sea, and Chukchi Sea aboard the R/V Sikuliaq during three oceanographic cruises: Northern Gulf of Alaska Long Term Ecological Research (NGA LTER), Bering and Aleutian Internal Tide Mixing, and Hg Cycling in the Arctic (Supplementary Table S1). All sampling stations were on the Alaskan continental shelf, except KOD10, which was on the slope in the NGA. The water column depth at the sampling locations in the Bering and Chukchi Seas were shallow (48 ± 6 m) compared to the NGA (610 ± 477 m) (Supplementary Table S2). Water samples were collected at a single depth, the chlorophyll a maximum (Figure 1; Supplementary Table S2), which was selected because previous studies have shown in situ Hg methylation at that depth (Munson et al., 2018; Starr et al., 2022). The ship’s SeaBird 911+ CTD rosette was used to assess the chlorophyll a maximum during the down cast, which determined the sampling depth. During the downcast, other parameters were measured such as salinity, dissolved oxygen, photosynthetically active radiation (PAR), temperature, and pressure. Due to limited water allowances, Hg methylation, nitrification, and genetic samples were collected from the ship’s regular CTD rosette (non-trace metal clean) in the NGA. Mercury methylation, nitrification, and genetic samples from the other 13 stations throughout the Bering and Chukchi Seas were collected using a trace metal clean SBE19plus V2 SeaCAT Profiler (SeaBird) CTD rosette equipped with Teflon-coated, external spring Niskin bottles (General Oceanics). Samples from the trace metal CTD rosette were transferred to acid washed sample bottles (Hammerschmidt et al., 2011) inside a positive-pressure, plastic enclosure (the bubble), constructed at the beginning of the NGA LTER cruise. Seawater samples for genetic analysis was collected in an acid washed 5-gallon collapsible polyethylene carboy. Using an Eppendorf peristaltic pump, 2 L of sample water was filtered through a 0.22 μM Sterivex filter pack (Millipore). The Sterivex was filled with RNAlater (ThermoFisher), stabilizing the nucleic acids, and stored at −80°C until extraction (Bowman et al., 2020). Seawater for Hg methylation rate experiments was collected in 4 2-L acid washed plastic bottles. Seawater samples for nitrification pool dilution experiments were collected into an acid washed 1-L and 3 125-mL polycarbonate incubation bottles. Incubation experiments occurred upon sample collection.
2.2 Mercury methylation experiments
Onboard dark incubation experiments were carried out with stable Hg isotope additions to quantify Hg methylation and demethylation rates. Four replicate 2-L unfiltered water samples were collected and promptly amended with 7.5 pM 200HgII (Oak Ridge National Laboratory) and 7.7 pM CH3201Hg by volume. MeHg synthesis was completed prior to the cruise with methyl-cobalamin and inorganic Hg isotopes (Oak Ridge National Laboratory). Initial sample volume was measured, and all samples were placed in a dark incubation chamber at in situ temperature (4°C) without further oxygenation. After 24 h, samples were removed, reduced to 1.5 L and immediately acidified to 1% H2SO4 by volume to stop biological transformations. Samples were stored unrefrigerated in a dark container until analysis.
Mercury methylation rate samples were analyzed at Wright State University (WSU) within 8 months of collection. Prior to analysis, these samples were neutralized to pH 4.9 ± 0.3 with 14 M potassium hydroxide, buffered with 14 mM acetate buffer by volume and amended with 0.1 mL of 2.5% ascorbic acid (Munson et al., 2014). Neutralized samples were derivatized with sodium tetraethylborate and purged with Hg-free N2 gas. During the bubbling process, methylethylmercury was concentrated onto Tenax traps and analyzed on an optimized gas chromatography inductively coupled plasma mass spectrometer (GC-ICPMS; Perkin Elmer Elan 9000; Hintelmann and Evans, 1997). Ambient Hg standards and enriched isotope standards were analyzed on both the Tekran and ICP-MS to quantify both concentration and isotope abundance. Mercury methylation and demethylation rates were calculated from replicates at one time point using matrix methodology (Hintelmann and Evans, 1997; Ouerdane et al., 2009), as CH3200Hg created and CH3201Hg lost since amendment respectively: Hg methylation rate (pM day−1) = km * [HgII] and MMHg demethylation rate (pM day−1) = kd * [MMHg]. For Hg methylation rate potentials, first order specific rates (km) were also calculated as specific rate (day−1) = Hg methylation rate * [HgII]−1.
2.3 Nitrification experiments
The collected 1 L seawater samples were amended with 20 mM 15NH4Cl and inverted three times. The 20 μM spiked sample was transferred into three 125-mL polycarbonate incubation bottles. An initial sample was taken immediately from each incubation replicate via a 60 mL plastic syringe and filtered through a 0.22 μm filter into one 15-mL polypropylene tube and two 20-mL scintillation vials. An initial control was sampled from the 125 mL unamended seawater sample following the same methodology. Initial samples were frozen upon collection and stored until analysis. The four 125-mL incubation bottles were incubated for 24 h in the onboard dark incubation chamber at in situ temperature. After the 24-h incubation, the four bottles were removed, final samples from the control and replicates were collected using the same methodology as the initial samples, and samples were frozen until analysis at WSU.
Cadmium (Cd) reductions (to reduce NO3− to NO2−) followed by sodium azide (NaN3) reductions were completed on each sample to quantify the production of nitrous oxide (N2O) from both nitrite and nitrate (NOx−; Heiss and Fulweiler, 2016; McIlvin and Altabet, 2005). To each sample, 100 mg and 6.6 g of dried magnesium and sodium chloride was added respectively and mixed. To complete the Cd reductions, 1 g of activated wet Cd powder was added to each sample and placed on a shaker table for 17 h. Samples were centrifuged for 15 min at 1,000 rpm to decant the reduced solution. 7.5 mL of supernatant was removed from the centrifuge tubes, transferred to a 12-mL Labco exetainer, and sealed with a single-wadded septa cap. Using a glass syringe with a needle, 0.25 mL of a solution containing 1:1 2 M sodium azide: 20% acetic acid purged with Argon gas was injected into the exetainer septa. Samples were shaken and incubated at 30°C for 1 h. Following incubation, the reaction was stopped by injecting 0.15 mL of 10 M NaOH through the exetainer septa with a clean needle and syringe (McIlvin and Altabet, 2005). Exetainers containing the reduced sample were stored upside down and shipped to the University of California—Davis Stable Isotope Facility for analysis of labeled 45,46N2O gas production using an isotope-ratio mass spectrometer (Hamilton and Ostrom, 2007). Incubation samples collected in falcon tubes were thawed and quantified for total NH4+ using colorimetric flow-injection analysis (Lachat Quikchem 8500) at WSU. Results from both analyses were used to calculate the 15NH4+ transformation and were corrected for NaN3 reductions using Eq. 1, modified from Heiss and Fulweiler (2016), Beman et al. (2008), and Hampel et al. (2020). Nitrification rate experiments were only completed in the NGA, Bering Sea, Bering Strait, and Chukchi Sea.
2.4 Molecular analysis
DNA was extracted using the DNeasy PowerWater Sterivex Kit (Qiagen) and assessed for quality using 260/280 and 260/230 ratios, with a NanoDrop Microvolume Spectrophotometer (ThermoFisher), verified by Qubit. Extractions were stored at −18°C until assay. Nitrospina-specific hgcA primers (Supplementary Table S3), Nitro_SP14_1F and Nitro_SP14_1R, designed from sequences found in Antarctic Sea ice were used for amplification (Gionfriddo et al., 2016). The SP8 sequence, 504 bp, from Gionfriddo et al. (2016) was used as a positive control for the real-time polymerase chain reaction (qPCR) assay. A set of seven serial dilutions (10–4–10–10 ng μL−1) were prepared from the SP8 sequence and used to form a standard curve (R2 > 0.90). With a reaction volume of 20 μL, the following chemical volumes were used for amplification: 10 μL Luna Universal qPCR Master Mix (New England BioLabs), 1 μL Nitro_SP14_1F and Nitro_SP14_1R, 1 μL of sample (5 ng DNA), diluted SP8, or Nuclease free water, and 7 μL of nuclease free water. Samples were amplified on a Mastercycler ep Realplex2 Real-Time PCR system (Eppendorf) with thermal cycling parameters (outlined in Gionfriddo et al., 2016) and a melting curve (Supplementary Table S3). Amplification efficiency was 95.6%, and sample amplicon concentration was calculated. Gene abundance was reported as Nitrospina-specific hgcA gene copies mL−1 and calculated using Eq. 2 and normalized to sample volume (mL).
3 Results and discussion
3.1 Physiochemical oceanography
The NGA is heavily influenced by the Alaskan Coastal Current (ACC), which is driven by alongshore winds and characterized by freshwater inputs derived from coastal runoff and the Copper River outflow (Stabeno et al., 2004). From fall to spring the NGA is dominated by downwelling currents and Ekman transport from off-shelf surface waters (Stabeno et al., 2004). The downwelling dissipates as winds decrease throughout the spring, allowing for the resuspension of key nutrients (such as nitrate, phosphate, and silicate) promoting primary production (Stabeno et al., 2004). During the 2021 Spring NGA LTER cruise there was a large early season diatom bloom from late April to early May 2021, which was thought to be induced by the increased sunlight and reduced winds. Chlorophyll a concentrations at the sampling depth (chlorophyll a maximum) across the NGA ranged from 0.3–10.1 mg m−3 (Supplementary Table S2). The highest value (10.1 mg m−3) was found at MID2, within the mixed waters under the Copper River plume. A small Copper River plume can be identified by a thin layer of nutrient deplete freshwater extending from the Copper River outflow. In the NGA, salinity increased with distance from the coastline, as the influence from runoff, freshwater inputs, and ACC wanes (Stabeno et al., 2016).
Water flows north across the Bering Sea shelf and Bering Strait, into the Chukchi Sea via the Bering Shelf Current and Chukchi Shelf current (Danielson et al., 2022). Higher concentrations of chlorophyll a and DO were seen in the surface waters in the southern Bering Sea near Unimak Pass (Supplementary Table S2). This can be attributed to the Aleutian North Slope current and deep mixing that resuspends key limited nutrients into the surface waters of this high nutrient low chlorophyll system (Aguilar-Islas et al., 2007; Danielson et al., 2022).
3.2 Mercury transformations
In situ Hg methylation occurred at all stations (0.003–0.008 d−1). Specific (Hg-normalized) Hg methylation rate potentials (km) across all regions were comparable with previous measurements from the chlorophyll a maxima across the Arctic (0.004–0.010 d−1; Lehnherr et al., 2011), Mediterranean Sea (0.004–0.038 d−1; Monperrus et al., 2007) and the central Pacific Ocean (0.004–0.017 d−1; Munson et al., 2018). Variability of km within each region is attributed to temporal and spatial heterogeneity between sampling stations impacting particle and nutrient distribution as a result of differing current dynamics (Stabeno et al., 2004; Strom et al., 2006). Differing assemblages of Hg methylating bacteria, Hg speciation, and demethylation rates also contribute to the variability of km within each sampled polar region. The highest km was measured at Unimak Pass (one-way ANOVA: p < 0.05; Table 1), which can be attributed to deep mixing, providing a substrate and microhabitat for Hg methylating anaerobic bacteria (Swift and Aagaard, 1976; Ortiz et al., 2015; Motta et al., 2022). The samples for incubation experiments in the NGA were collected in the early spring during a diatom bloom. This increase in productivity could decrease the amount of HgII available for Hg methylation. This could potentially explain the lower km across the NGA, when compared to other regions. Changes in water chemistry across the regions could result in differing contributions of instantaneous Hg methylation to the measured Hg methylation rate potentials. This could explain the variability seen in Hg methylation rate potentials within and across regions.
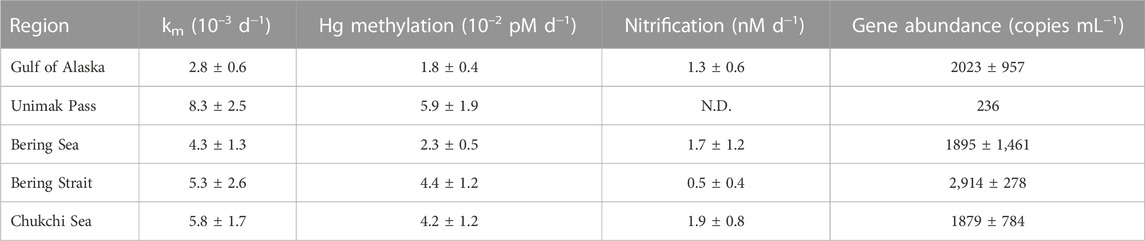
TABLE 1. Mean Nitrospina-specific hgcA-like gene abundance and rate potentials with standard error across five regions: NGA, Unimak Pass, Bering Sea, Bering Strait, and Chukchi Sea.
Across all regions, correlations between km and oxygen, ambient Hg, ambient NH4+, salinity, and temperature were weak and not significant. The difference between predicted and measured MeHg concentrations is attributed to MeHg demethylation in the surface waters and mixing throughout the Bering Strait and Chukchi Sea (DiMento and Mason, 2017; Munson et al., 2018; Wang et al., 2018).
Methylmercury demethylation rate potentials are not reported as the demethylation rates were unrealistically high. Instantaneous demethylation could explain the low CH3201Hg recovered in the samples (Munson et al., 2018). For future Hg rate experiments, adding additional time points, including a time zero, should be done as suggested by others (Munson et al., 2018).
3.3 Nitrification
Potential nitrification rates ranged 0.5–2.0 nM d−1 across all polar regions. These measurements are concurrent with in situ nitrification rates measured at deeper depths, 100–200 m, in oligotrophic regions such as the Sargasso Sea (2.0 ± 0.1 nM d−1; Newell et al., 2013). The mean specific nitrification rate across all stations, 7.2 × 10−5 d−1 is lower than the global ocean estimate 0.195 d−1 (Yool et al., 2007), which is thought to be driven by high 15NH4Cl amendment. Nitrite and nitrate produced during nitrification pool dilution experiments are reported as potential nitrification rates to account for any bias from the high amendment, but the rates are low, suggesting that the nitrifiers become ammonium-saturated at low concentrations (Horak et al., 2013; Newell et al., 2013). In the Chukchi Sea (1.9 ± 0.8 nM d−1), nitrification rate potentials were higher than previous measurements by Shiozaki et al. (2019) in the surface waters within the Chukchi and Beaufort Seas (<0.27 nM d−1), which is not surprising as those rates are low for the open ocean (Yool et al., 2007). Nitrification rate experiments were not conducted at Unimak Pass, but it is hypothesized that Unimak Pass would have a higher apparent nitrification rate as deep mixing in this region would provide remineralized NH4+ and metals (Cu, Mo, and Fe) for enzymes involved in both steps of nitrification, ammonia monooxygenase and nitrite oxidoreductase (Musiani et al., 2020; Chicano et al., 2021). It is likely that at the chlorophyll a maximum, competition between microbes and plankton taking up various forms of N are driving the lower nitrification rates in this study. The nitrite maximum is typically seen at the base of the euphotic zone as many known ammonia oxidizing bacteria have been shown to be photo-inhibited, resulting in higher nitrification rates with reduced light (Horrigan and Springer, 1990; Lomas and Lipschultz, 2006; Merbt et al., 2012). Previous studies in the North Pacific and Arctic Ocean show nitrification maximum occurring below the chlorophyll a maximum, around the nitrite maximum, under <1% PAR (Grundle et al., 2013; Shiozaki et al., 2016; Shiozaki et al., 2019). The potential nitrification rates measured at the chlorophyll a maximum across all subpolar marine regions reported in this study are therefore not expected to reflect the highest nitrification rates in the water column.
3.4 Mercury methylation and nitrification
A positive linear relationship (R2 = 0.34, p < 0.05; Figure 2) was seen between nitrification and Hg methylation rate potentials across all stations, suggesting a possible link between these two processes in surface waters surrounding Alaska (NGA, Bering Sea, Bering Strait and Chukchi Sea). The linear regression slope between nitrification and Hg methylation rate potentials across all regions (m = 0.0087 pM MeHg nM−1 NOx−; Figure 2) is the same order of magnitude as previous findings in the Amazon River Plume (m = 0.0028 pM MeHg nM−1 NOx−; Starr et al., 2022). This difference in slope can likely be attributed to differing ammonia oxidizing bacteria and archaea activities, substrate availability, and/or microbial assemblages including nitrifier community structure.
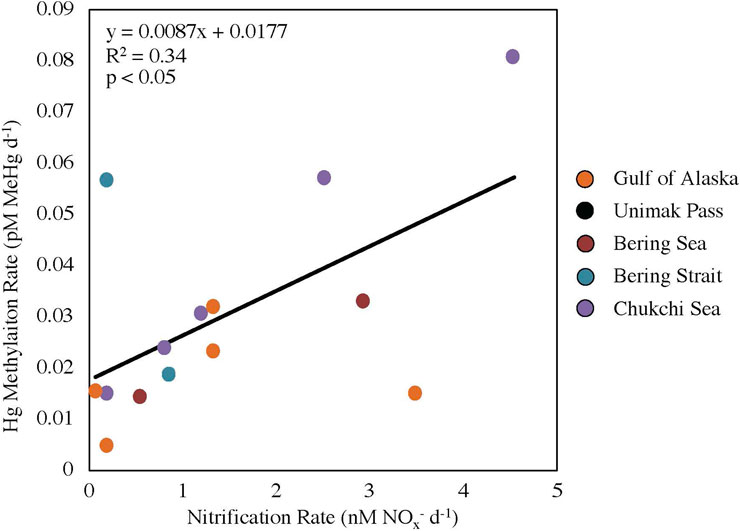
FIGURE 2. Linear regression between nitrification and mercury methylation rate potentials across five regions (p < 0.05, y = 0.0087x + 0.0177). Data points for Unimak Pass are not shown as nitrification rate experiments were not completed.
Station STN13 in the Bering Strait had a higher relative Hg methylation rate potential compared to nitrification rate potential (Figure 2). Chlorophyll a concentrations at STN13 were four times higher than those of STN3, the other station within the Bering Strait sampled 12 days prior to STN13 (Table 1; Supplementary Table S1). A higher chlorophyll a concentration suggests a general increase in microbial activity or decreased grazing pressure. Data from the USGS Yukon River Pilot Station show little change in Yukon River discharge throughout the 12-day period between sampling, suggesting riverine inputs were not the main driver in differing rate potentials or the high chlorophyll a concentrations. First year ice melt could influence the rate potentials between the two stations in the Bering Strait. A reduction in ice cover could change current dynamics driven by winds promoting water column mixing and resuspension of particles inhabited by Hg methylators. Although these are potential reasons for why STN13 had a higher Hg methylation rate potential compared to nitrification rate potentials, further investigation is needed to conclusively determine the decoupling of these rate potentials in the Bering Sea.
Station MID2, in the NGA near the Copper River outflow, had a higher relative nitrification rate potential compared to the Hg methylation rate potential. This higher relative nitrification rate potential can be attributed to nutrient resuspension, such as NH4+ and Cu, and differing communities of nitrifiers that can withstand freshwater plume dynamics (Bernhard et al., 2010). Lower substrate availability for Hg methylation associated the NGA diatom bloom could result from increased binding of HgII to particulate material, potentially accounting for the lower Hg methylation relative to nitrification rate potentials in the NGA. Overall, it is likely that the larger temporal and spatial differences among stations accounts for the higher variability in this study compared with the previous measurements in the Amazon River Plume (R2 = 0.78; Starr et al., 2022).
The stations within the Chukchi Sea had less temporal and spatial variability than the other regions, as these stations were sampled within the shortest time period (6-day) and across the smallest distance (110 miles; Supplementary Table S1). The correlation between Hg methylation and nitrification rate potentials in the Chukchi Sea was highly significant (R2 = 0.99, p < 0.001; Figure 3). Although the correlation between nitrification and Hg methylation rate potentials were strong, it cannot be concluded that nitrification rates alone are driving Hg methylation rates throughout these polar regions, as the functionality of Nitrospina-specific hgcA-like genes have not yet been confirmed. It is possible that the measured correlation between Hg methylation and nitrification may also be a result of other biogeochemical processes, such as general bacterial productivity, acting as a proxy (although nitrifiers are autotrophs, while most known Hg methylators are heterotrophs; Yool et al., 2007; Monperrus et al., 2007).
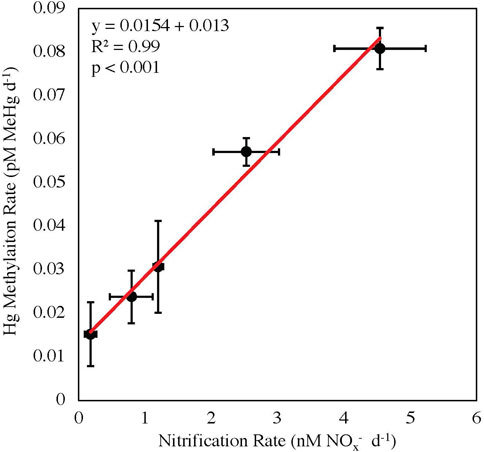
FIGURE 3. Linear regression between nitrification and mercury methylation rate potentials in the Chukchi Sea with standard errors (p < 0.001, y = 0.0154x + 0.013).
3.5 hgcA-like genes
Nitrospina-specific hgcA-like genes were present across all stations. The Bering Strait had the highest average gene copy numbers (2,914 copies mL−1) compared to the NGA (2023 copies mL−1), Bering Sea (1895 copies mL−1), and Chukchi Sea (1879 copies mL−1). Unimak Pass had the lowest gene abundance (236 copies mL−1), which was surprising as Unimak Pass had the highest km (Table 1). It is important to note that Hg methylation and nitrification rate potentials reported are a composite of the entire Hg methylator and nitrifier community, not just Nitrospina. Hg methylating microbes at Unimak Pass may be primarily anaerobic and particle-associated, as the station is characterized by deep water column mixing. We hypothesize that anaerobic and microaerophilic Hg methylating microbes inhabit settling or suspended particles, derived from sediment resuspension in the well mixed, shallow Bering Strait and Chukchi Sea, the well mixed waters of Unimak Pass, and the particle-rich Copper River plume. While this may include microaerophilic Nitrospina (Lücker et al., 2013), such particle microenvironments are likely dominated by anaerobic Hg methylating microbes such as Desulfobacteraceae and Methanomassiliicoccus, which have been found throughout the Bering Sea, Chukchi Sea, and Arctic Ocean water column (Bowman et al., 2020).
Across all sampled regions there was not a correlation between Nitrospina-specific hgcA-like genes and Hg methylation rate potentials. However, these Nitrospina hgcA-like amplicons only show gene presence, not enzyme function, so the lack of correlation may indicate that not all of these genes were active at the time of sampling. Despite a lack of correlation, the presence of Nitrospina-specific hgcA-like genes provides support for the potential for these organisms to be the primary methylators, given the relationship between Hg methylation and nitrification.
4 Conclusion
The link between Hg methylation and nitrification at the chlorophyll a maximum depth suggests a possible mechanism for Hg methylation in the oxic water column of these Alaskan Seas. The similarity between this link in subpolar waters and the Amazon River Plume further supports the hypothesis that nitrification and biotic Hg methylation are linked at the base of the oxic region of the euphotic (>1–9% PAR) ocean. As Nitrospina hgcA-like metagenomes have been recovered across all ocean basins (Villar et al., 2020), the presence of Nitrospina could be a major pathway for MeHg formation in the oxic ocean subsurface, provided these microbes are active (i.e., the conserved region of hgcA is functional). If these Nitrospina-specific hgcA-like genes are functional, then models utilizing current global specific nitrification rate estimates and Nitrospina hgcA-like amino acid sequence recoveries, could be used to predict global Hg methylation rate potentials in the ocean subsurface. However, until Nitrospina’s Hg methylating capabilities are confirmed in culture, more robust conclusions regarding Hg methylation by Nitrospina cannot be made. More research is needed to further examine the role of Nitrospina in the methylation of Hg in the global ocean.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
MD collected samples, carried out rate experiments, and completed analysis. RM and AA-I assisted in sample collection and contributed ambient Hg and nutrient data respectively. RM and CL helped with mercury rate experiments. SN aided in nitrification rate experiments. SN, CH, and MD conceived the project idea. All authors contributed to the article and approved the submitted version.
Funding
This research was partially supported by the U.S. National Science Foundation through OPP grant 1854454 to RM, and OCE grant 1656070 to AA-I.
Acknowledgments
We thank chief scientists, Russell Hopcroft and Tyler Hennon, the NGA LTER, and the Captain and crew of the R/V Sikuliaq. We also thank Yipeng He and Hannah Inman for help with sample collection and contributing ambient Hg data, Lauren Barrett for contributing nutrient data, Lindsay Starr and Justin Myers for help with analyses, Caitlin Gionfriddo for providing Nitrospina-specific hgcA amplicons, and the UC-Davis stable isotope laboratory.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvc.2023.1109537/full#supplementary-material
References
Agather, A. M., Bowman, K. L., Lamborg, C. H., and Hammerschmidt, C. R. (2019). Distribution of mercury species in the western Arctic Ocean (U.S. GEOTRACES GN01). Mar. Chem. 216. doi:10.1016/j.marchem.2019.103686
Aguilar-Islas, A. M., Hurst, M. P., Buck, K. N., Sohst, B., Smith, G. J., Lohan, M. C., et al. (2007). Micro- and macronutrients in the southeastern Bering Sea: Insight into iron-replete and iron-depleted regimes. Prog. Oceanogr. 73, 99–126. doi:10.1016/j.pocean.2006.12.002
Amap, (2021). AMAP assessment 2021: Human health in the arctic. Tromsø, Norway: Arctic Monitoring and Assessment Programme.
Beckmen, K. B., Duffy, L. K., Zhang, X., and Pitcher, K. W. (2002). Mercury concentrations is the Fur of Steller sea lions and northern Fur seals from Alaska. Mar. Pollut. Bull. 44, 1130–1135. doi:10.1016/S0025-326X(02)00167-4
Beman, J. M., Popp, B. N., and Francis, C. A. (2008). Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2, 429–441. doi:10.1038/ismej.2007.118
Bernhard, A. E., Landry, Z. C., Blevins, A., de la Torre, J. R., Giblin, A. E., and Stahl, D. A. (2010). Abundance of ammonia-oxidizing Archaea and Bacteria along an estuarine salinity gradient in relation to potential nitrification rates. Appl. Environ. Microbiol. 76. doi:10.1128/AEM.02018-09
Bowman, K. L., Hammerschmidt, C. R., Lamborg, C. H., Swarr, G. J., and Agather, A. M. (2016). Distribution of mercury species across a zonal section of the eastern tropical South Pacific Ocean (U.S. GEOTRACES GP16). Mar. Chem. 186, 156–166. doi:10.1016/j.marchem.2016.09.005
Bowman, K. L., Lamborg, C. H., and Agather, A. M. (2020). A global perspective on mercury cycling in the ocean. Sci. Total Environ. 710. doi:10.1016/j.scitotenv.2019.136166
Chicano, T. M., Dietrich, L., de Almeida, N. M., Akram, M., Hartmann, E., Leidreiter, F., et al. (2021). Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex. Nat. Microbiol. 6, 1129–1139. doi:10.1038/s41564-021-00934-8
Christensen, G. A., Gionfriddo, C. M., King, A. J., Moberly, J. G., Miller, C. L., Somenahally, A. C., et al. (2019). Determining the reliability of measuring mercury cycling gene abundance with correlations with mercury and methylmercury concentrations. Environ. Sci. Technol. 53, 8649–8663. doi:10.1021/acs.est.8b06389
Danielson, S. L., Grebmeier, J. M., Iken, K., Berchok, C., Britt, L., Dunton, K. H., et al. (2022). Monitoring Alaskan Arctic Shelf ecosystems through collaborative observation networks. Oceanography 35.
Dastoor, A., Angot, H., Bieser, J., Christensen, J. H., Douglas, T. A., Heimbürger-Boavida, L., et al. (2022). Arctic mercury cycling. Nat. Rev. Earth Environ. 3, 270–286. doi:10.1038/s43017-022-00269-w
Dietz, R., Outridge, P. M., and Hobson, K. A. (2009). Anthropogenic contributions to mercury levels in present-day Arctic animals – a review. Sci. Total Environ. 407, 6120–6131. doi:10.1016/j.scitotenv.2009.08.036
DiMento, B. P., and Mason, R. P. (2017). Factors controlling the photochemical degradation of methylmercury in coastal and oceanic waters. Mar. Chem. 196, 116–125. doi:10.1016/j.marchem.2017.08.006
Gilmour, C. C., Podar, M., Bullock, A. L., Graham, A. M., Brown, S. D., Somenahally, A. C., et al. (2013). Mercury methylation by novel microorganisms from new environments. Environ. Sci. Technol. 47, 11810–11820. doi:10.1021/es403075t
Gionfriddo, C. M., Stott, M. B., Power, J. F., Ogorek, J. M., Krabbenhoft, D. P., Wick, R., et al. (2020). Genome-resolved metagenomics and detailed geochemical speciation analyses yield new insights into microbial mercury cycling in geothermal springs. Appl. Environ. Microbiol. 86. doi:10.3389/fmicb.2020.541554
Gionfriddo, C. M., Tate, M. T., Wick, R. R., Schultz, M. B., Zemla, A., Thelen, M. P., et al. (2016). Microbial mercury methylation in Antarctic Sea ice. Nat. Microbiol. 1. doi:10.1038/nmicrobiol.2016.127
Grundle, D. S., Juniper, K., and Giesbrecht, K. E. (2013). Euphotic zone nitrification in the NE subarctic pacific: Implications for measurements of new production. Mar. Chem. 155, 113–123. doi:10.1016/j.marchem.2013.06.004
Hamilton, S. K., and Ostrom, N. E. (2007). Measurement of the stable isotope ratio of dissolved N2 in 15N tracer experiments. Limnol. Oceanogr. Methods 5, 233–240. doi:10.4319/lom.2007.5.233
Hammerschmidt, C. R., Bowman, K. L., Tabatchnick, M. D., and Lamborg, C. H. (2011). Storage bottles material and cleaning for determination of total mercury in seawater. Limnol. Oceanogr. 9, 426–431. doi:10.4319/lom.2011.9.426
Hammerschmidt, C. R., and Bowman, K. L. (2012). Vertical methylmercury distribution in the subtropical North Pacific ocean. Mar. Chem. 132-133, 77–82. doi:10.1016/j.marchem.2012.02.005
Hammerschmidt, C. R., and Fitzgerald, W. F. (2006). Methylmercury in freshwater fish linked to atmospheric mercury deposition. Environ. Sci. Technol. 40, 7764–7770. doi:10.1021/es061480i
Hampel, J. J., McCarthy, M. J., Aalto, S. L., and Newell, S. E. (2020). Hurricane disturbance stimulated nitrification and altered ammonia oxidizer community structure in Lake Okeechobee and St. Lucie Estuary (Florida). Front. Microbiol. 11. doi:10.3389/fmicb.2020.01541
Heimbürger, L., Sonke, J. E., Cossa, D., Point, D., Lagane, C., Laffont, L., et al. (2015). Shallow methylmercury production in the marginal sea ice zone of the central Arctic Ocean. Sci. Rep. 5. doi:10.1038/srep10318
Heiss, E. M., and Fulweiler, R. W. (2016). Coastal water column ammonium and nitrite oxidation are decoupled in summer. Estuar. Coast. Shelf Sci. 178, 110–119. doi:10.1016/j.ecss.2016.06.002
Hintelmann, H., and Evans, R. D. (1997). Application of stable isotopes in environmental tracer studies – measurement of monomethylmercury by isotope dilution ICP-MS and detection of species transformation. Fresenius J. Anal. Chem. 358, 378–385. doi:10.1007/s002160050433
Horak, R. E. A., Qin, W., Schauer, A. J., Armbrust, E. V., Ingalls, A. E., Moffett, J. W., et al. (2013). Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J. 7, 2023–2033. doi:10.1038/ismej.2013.75
Horrigan, S. G., and Springer, A. L. (1990). Oceanic and estuarine ammonium oxidation: Effects of light. Limnol. Oceanogr. 35, 479–482. doi:10.4319/lo.1990.35.2.0479
Jonsson, S., Mastomonaco, M. G. N., Gardfeldt, K., and Mason, R. P. (2022). Distribution of total mercury and methylated mercury species in Central Arctic Ocean water and ice. Mar. Chem. 242. doi:10.1016/j.marchem.2022.104105
Kim, H., Sørensen, A. L., Hur, J., Heimbürger, L., Hahm, D., Siek Rhee, T., et al. (2017). Methylmercury mass budgets and distribution characteristics in the Western Pacific Ocean. Environ. Sci. Technol. 51, 1186–1194. doi:10.1021/acs.est.6b04238
Lamborg, C. H., Hammerschmidt, C. R., Bowman, K. L., Swarr, G. J., Munson, K. M., Ohnemus, D. C., et al. (2014). A global ocean inventory of anthropogenic mercury based on water column measurements. Nature 512, 65–68. doi:10.1038/nature13563
Lehnherr, I., Louis, V. L., Hintelmann, H., and Kirk, J. L. (2011). Methylation of inorganic mercury in polar marine waters. Nat. Geosci. 4, 298–302. doi:10.1038/ngeo1134
Lin, H., Ascher, D. B., Myung, Y., Lamborg, C. H., Hallam, S. J., Gionfriddo, C. M., et al. (2021). Mercury methylation by metabolically versatile cosmopolitan marine bacteria. ISME J. 15, 1810–1825. doi:10.1038/s41396-020-00889-4
Lomas, M. W., and Lipschultz, F. (2006). Forming the primary nitrite maximum: Nitrifiers or phytoplankton? Limnol. Oceanogr. 51, 2453–2467. doi:10.4319/lo.2006.51.5.2453
Lücker, S., Nowka, B., Rattei, T., Spieck, E., and Daims, H. (2013). The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front. Microbiol. 4. doi:10.3389/fmicb.2013.00027
Mason, R. P., Choi, A. L., Fitzgerald, W. F., Hammerschmidt, C. R., Lamborg, C. H., Sørensen, A. L., et al. (2012). Mercury biogeochemical cycling in the ocean and policy implications. Environ. Res. 119, 101–117. doi:10.1016/j.envres.2012.03.013
McIlvin, M. R., and Altabet, M. A. (2005). Chemical conversion of nitrate and nitrite to nitrous oxide for nitrogen and oxygen isotopic analysis in freshwater and seawater. Anal. Chem. 77, 5589–5595. doi:10.1021/AC050528S
Médieu, A., Point, D., Itai, T., Angot, H., Buchanan, P. J., Allain, V., et al. (2022). Evidence that Pacific tuna mercury levels are driven by marine methylmercury production and anthropogenic inputs. PNAS 199. doi:10.1073/pnas.2113032119
Merbt, S. N., Stahl, D. A., Casamayor, E. O., Martí, E., Nicol, G. W., and Prosser, J. I. (2012). Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol. Lett. 327, 41–46. doi:10.1111/j.1574-6968.2011.02457.x
Monperrus, M., Tessier, E., Amouroux, D., Leynaert, A., Huonnic, P., and Donard, O. F. X. (2007). Mercury methylation, demethylation and reduction rates in coastal and marine surface waters of the Mediterranean Sea. Mar. Chem. 107, 49–63. doi:10.1016/j.marchem.2007.01.018
Motta, L. C., Blum, J. D., Popp, B. N., and Drazen, J. C. (2022). Mercury isotopic evidence for the importance of particles as a source of mercury to marine organisms. PNAS 119. doi:10.1073/pnas.2208183119
Munson, K. M., Babi, D., and Lamborg, C. H. (2014). Determination of monomethylmercury from seawater with ascorbic acid-assisted direct ethylation. Limnol. Oceanogr. 12, 1–9. doi:10.4319/lom.2014.12.1
Munson, K. M., Lamborg, C. H., Boiteau, R. M., and Saito, M. A. (2018). Dynamic mercury methylation and demethylation in oligotrophic marine water. Biogeosciences 15, 6451–6460. doi:10.5194/bg-15-6451-2018
Musiani, F., Broll, V., Evangelisti, E., and Ciurli, S. (2020). The model structure of the copper-dependent ammonia monooxygenase. J. Biol. Inorg. Chem. 25, 995–1007. doi:10.1007/s00775-020-01820-0
Newell, S. E., Fawcett, S. E., and Ward, B. B. (2013). Depth distribution of ammonia oxidation rates and ammonia-oxidizer community composition in the Sargasso Sea. Limnol. Oceanogr. 58, 1491–1500. doi:10.4319/lo.2013.58.4.1491
Ortiz, V. L., Mason, R. P., and Ward, J. E. (2015). An examination of the factors influencing mercury and methylmercury particulate distributions, methylation and demethylation rates in laboratory generated marine snow. Mar. Chem. 177, 753–762. doi:10.1016/j.marchem.2015.07.006
Ouerdane, L., Mester, Z., and Meija, J. (2009). General equation for multiple spiking isotope dilution mass spectrometry. Anal. Chem. 81. doi:10.1021/ac900205b
Parks, J. M., Johs, A., Podar, M., Bridou, R., Hurt, R. A., Smith, S. D., et al. (2013). The genetic basis for bacterial mercury methylation. Science 339, 1332–1335. doi:10.1126/science.1230667
Schartup, A. T., Ndu, U., Balcom, P. H., Mason, R. P., and Sunderland, E. M. (2015). Contrasting effects of marine and terrestrially derived dissolved organic matter on mercury speciation and bioavailability in seawater. Environ. Sci. Technol. 49, 5965–5972. doi:10.1021/es506274x
Shiozaki, T., Ijichi, M., Fujiwara, A., Makabe, A., Nishino, S., Yoshikawa, C., et al. (2019). Factors regulating nitrification in the Arctic Ocean: Potential impact of sea ice reduction and ocean acidification. Glob. Biogeochem. Cy. 33, 1085–1099. doi:10.1029/2018GB006068
Shiozaki, T., Ijichi, M., Isobe, K., Hashihama, F., Nakamura, K., Ehama, M., et al. (2016). Nitrification and its influence on biogeochemical cycles from the equatorial Pacific to the Arctic Ocean. ISME J. 10, 2184–2197. doi:10.1038/ismej.2016.18
Sonke, J. E., Teisserenc, R., Heimbürger-Boavida, L., Petrova, M. V., Marusczak, N., Le Dantec, T., et al. (2018). Eurasian river spring flood observations support net Arctic Ocean mercury export to the atmosphere and Atlantic Ocean. PNAS 115. doi:10.1073/pnas.1811957115
Sørensen, A. L., Jacob, D. J., Schartup, A. T., Fisher, J. A., Lehnherr, I., Louis, St.V. L., et al. (2016). A mass budget for mercury and methylmercury in the Arctic Ocean. Glob. Biogeochem. Cy. 30, 560–575. doi:10.1002/2015GB005280
Stabeno, P. J., Bell, S., Cheng, W., Danielson, S., Kachel, N., and Mordy, C. W. (2016). Long-term observations of Alaska coastal current in the northern Gulf of Alaska. Deep Sea Res. II Top. Stud. Oceanogr. 132, 24–40. doi:10.1016/j.dsr2.2015.12.016
Stabeno, P. J., Bond, N. A., Hermann, A. J., Kachel, N. B., Mordy, C. W., and Overland, J. E. (2004). Meteorology and oceanography of the northern Gulf of Alaska. Cont. Shelf Res. 24, 859–897. doi:10.1016/j.csr.2004.02.007
Starr, L. D., McCarthy, M. J., Hammerschmidt, C. R., Subramaniam, A., Despins, M. C., Montoya, J. P., et al. (2022). Mercury methylation linked to nitrification in the tropical North Atlantic Ocean. Mar. Chem. 247. doi:10.1016/j.marchem.2022.104174
Strom, S. L., Brady Olson, M., Macri, E. L., and Mordy, C. W. (2006). Cross-shelf gradients in phytoplankton community structure, nutrient utilization, and growth rate in the coastal Gulf of Alaska. Mar. Ecol. Prog. 328, 75–92.
Sunderland, E. M., Krabbenhoft, D. P., Moreau, J. W., Strode, S. A., and Landing, W. M. (2009). Mercury sources, distribution, and bioavailability in the North Pacific Ocean: Insights from data and models. Glob. Biogeochem. Cy 23. doi:10.1029/2008GB003425
Sunderland, E. M., and Mason, R. P. (2007). Human impacts on open ocean mercury concentrations. Glob. Biogeochem. Cy. 21. doi:10.1029/2006GB002876
Swift, J. H., and Aagaard, K. (1976). Upwelling near samalga pass. Limnol. Oceanogr. 21, 339–408. doi:10.4319/lo.1976.21.3.0399
Tada, Y., Marumoto, K., and Takeuchi, A. (2021). Nitrospina-like bacteria are dominant potential mercury methylators in both the Oyashio and Kuroshio regions of the Western North Pacific. Environ. Microbiol. 9. doi:10.1128/Spectrum.00833-21
Tada, Y., Marumoto, K., and Takeuchi, A. (2020). Nitrospina-like bacteria are potential mercury methylators in the mesopelagic zone in the East China Sea. Front. Microbiol. 11. doi:10.3389/fmicb.2020.01369
Villar, E., Cabrol, L., and Heimbürger-Boavida, L. (2020). Widespread microbial mercury methylation genes in the global ocean. Environ. Microbiol. Rep. 12, 277–287. doi:10.1111/1758-2229.12829
Wang, K., Munson, K. M., Beaupre-Laperriere, A., Mucci, A., Macdonald, R. W., and Wang, F. (2018). Subsurface seawater methylmercury maximum explains biotic mercury concentrations in the Canadian Arctic. Sci. Rep. 8. doi:10.1038/s41598-018-32760-0
Yool, A., Martin, A. P., Fernández, C., and Clark, D. R. (2007). The significance of nitrification for oceanic new production. Nature 447, 999–1002. doi:10.1038/nature05885
Keywords: mercury, mercury methylation, nitrification, Arctic, Subarctic, hgcA, Nitrospina
Citation: Despins MC, Mason RP, Aguilar-Islas AM, Lamborg CH, Hammerschmidt CR and Newell SE (2023) Linked mercury methylation and nitrification across oxic subpolar regions. Front. Environ. Chem. 4:1109537. doi: 10.3389/fenvc.2023.1109537
Received: 27 November 2022; Accepted: 17 May 2023;
Published: 25 May 2023.
Edited by:
Marc Amyot, Montreal University, CanadaReviewed by:
Dario Acha, Universidad Mayor de San Andrés, BoliviaIgor Lehnherr, University of Toronto Mississauga, Canada
Copyright © 2023 Despins, Mason, Aguilar-Islas, Lamborg, Hammerschmidt and Newell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marissa C. Despins, bWRlc3BpbnNAdWNzYy5lZHU=
 Marissa C. Despins
Marissa C. Despins Robert P. Mason
Robert P. Mason Ana M. Aguilar-Islas
Ana M. Aguilar-Islas Carl H. Lamborg
Carl H. Lamborg Chad R. Hammerschmidt1
Chad R. Hammerschmidt1 Silvia E. Newell
Silvia E. Newell