- 1Department of Isotope Biogeochemistry, Helmholtz Centre for Environmental Research, Leipzig, Germany
- 2MicroPro GmbH, Gommern, Germany
- 3Department of Environmental Microbiology, Helmholtz Centre for Environmental Research, Leipzig, Germany
- 4Institute for Bioanalysis, Department of Applied Sciences, Coburg University of Applied Sciences and Arts, Coburg, Germany
- 5Bayreuth Center of Ecology and Environmental Research (BayCEER), University of Bayreuth, Bayreuth, Germany
While interest in underground gas storage (UGS) of hydrogen (H2) in salt caverns is increasing in a growing H2 economy, knowledge of the microbial communities inhabiting those hypersaline environments is still scarce. High salt concentrations and limited availability of carbon (C) sources in cavern environments reduce microbial growth rates and metabolic activities. Growth conditions potentially change once H2 is stored in salt caverns. H2 is a universal electron donor that can facilitate autotrophic growth and subsequently growth of heterotrophs. In this study, a mixed culture enriched from hypersaline UGS sites was investigated in microcosm experiments with H2 atmosphere, testing the effect of different salt concentrations and C sources on methanogenesis, sulfate reduction and homoacetogenesis. Sulfate, acetate and lactate concentrations were quantified throughout a time span of 125 days of incubation and correlations with the microbial community structure and function were explored through 16S rRNA gene-based amplicon sequencing at the end of the experiment. Many of the amplicon sequence variants (ASVs) were only assigned to family or order level, reflecting that a large number of ASVs belong to previously undescribed taxa. At 4.4 M NaCl, close to cavern brine salinity, members of the Desulfovibrionales were absent when no C source other than CO2 was offered as the sole C source. This is in line with the finding that no sulfate reduction occurred at these conditions. Acetogenic Halanaerobiia dominated in these high salinity levels. Based on metagenome sequencing of four selected samples, we found that acetogenesis at autotrophic conditions relies on the activity of a halophilic homoacetogen, Acetohalobium sp. and that sulfate reduction can most likely be associated with a so far undescribed member of the Desulfonatronovibrionales. We further discuss the discrepancy between sulfate reduction at heterotrophic conditions, while no sulfate reduction was observed under autotrophic conditions although acetate was produced through the homoacetogenic activity of Acetohalobium. For the application of UGS of H2, this means that the presence and activity of the aforementioned microorganisms must be investigated, as they can eventually lead to the formation of acetate and allow sulfate reduction at relevant concentrations.
1 Introduction
Hydrogen (H2) gas as a chemical energy carrier, is expected to play a significant role in the transition towards a renewable energy system. Using Power-to-Gas technology surplus power, generated from renewable energy sources, can be stored through electrolysis of water into gaseous oxygen and H2 (Götz et al., 2016). The produced H2 is used directly as an energy carrier or converted to methane (CH4) (Braga Nan et al., 2020). In addition, H2 is currently used in industry as a reactant for different chemical processes (Matos et al., 2019). As supply of renewable energy can fluctuate, large-scale storage facilities are essential for successful implementation of a H2 based energy system (Blanco and Faaij, 2018; Crotogino et al., 2018). To meet future H2 storage demands, underground gas storage (UGS) in geological formations has been proposed, as they allow for a safe, low-cost, large-scale and decentralized storage facility. These geological formations include depleted oil and gas reservoirs, artificial salt caverns, deep aquifers, hard rock caverns and abandoned mines (Matos et al., 2019). Out of these sites, artificial salt caverns are the most promising option, due to their low cushion gas requirement and high sealing properties (Caglayan et al., 2020). Experience for gas storage in artificial salt caverns already exists for natural gas, coal gas and carbon dioxide (CO2) (Panfilov, 2016). Different ancient salt deposits can be found in Europe, including Tertiary and Mesozoic deposits in Southern parts as well as a large Paleozoic deposit across northern Europe (Crotogino et al., 2010). Due to their thalassohaline origin, these salt deposits are primarily composed of halite, anhydrite and carbonate (Strohmenger et al., 1996). Within the Paleozic deposits, which originate from the precipitation series of the Zechstein sea, anhydrite makes up to 10% of the rock formation, and sulfate the third most abundant ion (Geluk, 2000; Schwab et al., 2022). Generally, dissolved sulfate in geological reservoir formations results from the dissolution of anhydrite; the reactive anhydrite is in solution equilibrium with the brine (Hemme and van Berk, 2018).
Although microbial diversity and activity are expected to be limited in these highly saline environments, salt caverns can still harbor a variety of halophilic microorganisms (Bordenave et al., 2013; Heinemann et al., 2021; Schwab et al., 2022) that could cause several undesired or harmful side-effects when H2 is being stored (Dopffel et al., 2021). Firstly, H2 is an ancient and important electron donor for microbial respiration, since activity of hydrogenases allow for efficient electron extraction (Vignais and Billoud, 2007). In the presence of sulfate, sulfate-reducing prokaryotes can produce the corrosive gas H2S, which is also toxic to humans (Rabus et al., 2015). This process has been already observed during UGS and in oil reservoirs and led to microbial induced corrosion and souring (Hamilton, 2003; Rabus et al., 2015; Hemme and van Berk, 2017). Secondly, the reduction of CO2 with H2 can result in the production of CH4 and acetate, thereby changing the composition of the gas mixture and producing an attractive C source for other members of the cavern microbiome.
Hypersaline conditions require strategies for microbial osmoregulation (Becker et al., 2014; Gunde-Cimerman et al., 2018). These either include the synthesis of organic molecules, such as ectoine, or the accumulation of monovalent ions, K+, Na+ and Cl− (Reistad, 1970; Deole et al., 2013; Oren, 2013). Both strategies function on the expense of ATP, provided by chemoorganotrophic or phototrophic metabolism (Gunde-Cimerman et al., 2018). High intracellular concentrations of ions were reported to result in proteomic adaptations in aerobic halophilic bacteria and archaea, shifting their proteome isoelectric point (pI) towards acidic values (Oren, 2013). This was not reported for anaerobic halophilic Firmicutes, which appear to accumulate ions without proteomic adaptations (Gunde-Cimerman et al., 2018). For halophilic sulfate-reducing bacteria, both accumulation of compatible solutes or ions has been reported (Roberts, 2005). Generally, increasing salinity levels are accompanied with a decrease in metabolic rates of halophilic microorganisms due to the high energetic expenses of the synthesis of compatible solutes, leading to slow growth rates especially at energy-limited strictly anoxic conditions (Oren, 2002; Sorokin et al., 2011a; Sorokin et al. 2015; Sorokin et al. 2017).
At UGS conditions, light-dependent reactions cannot be expected and C sources are limiting. Therefore, we hypothesize that extremely halophilic autotrophs, their metabolites and biomass form the basis for heterotrophs and that both groups can consume H2, leading to sulfidogenesis and potentially cause damage to the gas infrastructure. However, salt caverns which are currently being used as natural gas UGS are characterized by microbial cell numbers rarely exceeding 107 cells mL-1; furthermore, high-pressure air-locks are required to introduce sampling devices, restricting their size and ultimately the sample volume (Schwab et al., 2022). Thus, gaining active mixed cultures or even isolates from material of hypersaline aquifers or caverns is difficult, and not much information is known about the structure and function of microbial communities present in those habitats. In this study, we tested the effect of varying salt concentrations and C sources on H2 oxidation in mixed cultures enriched from material of various hypersaline aquifers and caverns. We focused on homoacetogenesis, methanogenesis and sulfate reduction as expected important metabolic processes driven by H2 oxidation. We used amplicon sequencing to identify the microbial key players in the community composition and metagenome sequencing to further explore their metabolic potential and osmoregulation strategies.
2 Material and methods
2.1 Origin of samples
We chose halophilic enrichment cultures which were originally sampled from 2007 until 2013 during microbiological routine analysis at various European UGS and hydrocarbon storages in salt caverns and saline aquifers. Microbial communities from these samples were enriched and maintained with various electron donors and acceptors. The selection was based on microbial activity in general and their ability to grow at hypersaline and sulfate reducing conditions. The initial enrichment procedure is provided in Supplementary Table S1 of the Supplementary Material. These cultures were unified and cultivated for 90 days (Supplementary Table S2) for further enrichment. From the unified samples, three pre-cultures were subsequently prepared at three salinity levels (2.5, 3.4 and 4.4 M NaCl) and cultivated for another 90 days, before subsets with different C sources (acetate, lactate, methanol, no additional) were prepared. These subsets were used as inoculum for the microcosm studies which are described in 2.2. A modified mineral nutrient medium was used, and the composition per L was as follows: 0.5 g K2HPO4; 0.43 g MgCl2∙6H2O; 0.01 g CaCl2; 0.5 g (NH4)2Cl; 2.8 g Na2SO4, 0.5 ml resazurin solution (0.2%), 150 (2.5 M), 200 (3.4 M) or 260 g NaCl (4.4 M) depending on the culture. After autoclaving, the following solutions were added from sterile anoxic stock solutions: 1 ml trace element solution (Widdel et al., 1983), 2 ml vitamin solution (Wolin et al., 1963), 2 ml selenite-tungstate solution (0.02 mM), 30 ml bicarbonate solution (1 M) and 10 ml sodium dithionite solution (0.01 M). The final pH value was set to 7.2 using either anoxic HCl or NaOH. Hundred ml were subsequently dispensed anoxically to 375 ml serum bottles in a N2 atmosphere. Serum bottles were sealed with butyl rubber stoppers (Ochs Laborfachhandel e. K, Bovenden, Germany) and the headspace (275 ml) was changed to a gas mixture of H2:CO2 (8:2) set to 2000 mbar.
2.2 Experimental set-up
The microcosms (S4-S15) were cultivated at three salinity levels (low NaCl concentration with 2.5 M (S4, S7, S10, S13), medium NaCl concentration with 3.4 M (S5, S8, S11, S14), and high NaCl concentration with 4.4 M (S6, S9, S12, S15)), in minimal medium (see above) with 14 mM sulfate and a headspace gas mixture of 80% H2 and 20% CO2 (2000 mbar). Each enrichment culture was spiked with one organic C source: 10 mM acetate (S4, S5, S6), 8 mM lactate (S7, S8, S9), or 10 mM methanol (S10, S11, S12). One set was incubated without organic C source (S13, S14, S15). Acetate and lactate were prepared in 2 M stock solutions from their sodium salts at neutral pH value. Both stock solutions and methanol (≥99.8%) were degassed and subsequently sterile-filtered into 120 ml serum bottles using an anaerobic work bench. All cultures were kept in serum bottles, at a temperature of 30°C, and a pH value between 7.2–7.5 for an incubation period of 125 d. During this time, cell numbers, acetate, lactate and sulfate concentrations, as well as the composition of the gas phase were monitored in intervals of 20–25 d. After the incubation period, the microbial community composition was assessed by 16S rRNA gene-based amplicon sequencing. Once samples were subcultured, after a total incubation time of 1445 d, sulfate concentrations were measured again and four samples (S11, S12, S14 and S15) were selected for metagenome sequencing.
2.3 Ion chromatography
Anions (sulfate, acetate, and lactate) were analyzed throughout the experiment, at intervals of 20–25 d. Therefore, sub-samples of 1 mL were aseptically removed from the culture flasks using sterile, disposable syringes, filtered (13 mm syringe filter, PES, 0.22 µm) and stored at −18°C in 1.5 ml reaction tubes until measurement. They were analyzed using a Dionex ICS-1100 ion chromatograph equipped with a Dionex IonPac™ AS23 column (4 × 250 mm) and a conductivity detector (flow rate 1 ml min-1, sample volume 25 µl). Separation was achieved with gradient elution in 4.5 mM/0.8 mM Na2CO3. Organic acids (lactate, acetate) were separated on the same ion chromatograph but using a Dionex IonPac™ AS1 (9 × 250 mm) column and elution with 0.5 mM heptafluorobutyric acid. The volume of culture liquid that was removed for analysis was not refilled.
2.4 Gas chromatography
The composition of the gas phase was monitored throughout the experiment, at intervals of 20–25 d using sub-samples of 0.1–1 ml from the headspace that were aseptically removed from the culture flasks using gastight syringes. Gases were analyzed using a Gach 21.3 gas chromatograph (VEB Chromatron, Berlin) equipped with a thermal conductivity detector. For CO2 measurements, 100 µl were injected into a hand-packed Al2O3 (VEB Leuna-Werke) column (3 m) coated with 20% Oxidipropionitril, H2 served as carrier gas with a flow rate of 0.8 ml s-1. For analysis of CH4 and H2, a molecular sieve 5A column (3 m; Molsieb 5A 60/80 Mesh, Ziemer Chromatographie) was used with argon as carrier gas at a flow rate of 0.8 ml s-1. The sampled headspace volume was replaced with an equal volume of the initial gas composition, and gas consumption was calculated accordingly.
2.5 Cell enumeration
Samples were examined microscopically (Zeiss Axioscope A.1, Jena, Germany) to determine the total number of cells and to study cell morphology. Cell counts were determined by means of phase contrast microscopy (magnification 1:1.200) using a THOMA counting chamber.
2.6 Amplicon sequencing
After an incubation time of 125 days, 2 ml aliquots of each of the 12 microcosms were filtered onto sterile 0.1 μm polyethersulfone filters (PES, Sartorius, Göttingen). Genomic DNA (gDNA) extraction was performed using a previously described phenol-chloroform method (Schwab et al., 2022). Archaeal and bacterial 16S rRNA gene-based Illumina MiSeq amplicon analysis was performed in duplicates at LGC Genomics GmbH (Berlin, Germany) with 2 × 300 bp paired-end sequencing according to manufacturer’s instructions. Briefly, gDNA was amplified using bacterial primer-set Bac-341-fwd/Bac-785-rev (5′-CCTACGGGNGGCWGCAG-3′ and: 5′-GACTACHVGGGTATCTAATCC-3′) with a temperature profile of 95°C for 3 min, followed by 30 cycles of 95, 55°C and 72°C, each step for 30 s, and a final elongation at 72°C for 5 min (Klindworth et al., 2013). In addition, gDNA was amplified in a nested PCR approach, firstly pre-amplifying archaeal sequences with primer-set 340F-fwd/1000R-rev (5′-CCCTAYGGGGYGCASCAG-3′ and: 5′-GGCCATGCACYWCYTCTC-3′) with a temperature profile including an initial denaturation at 96°C for 1 min, followed by 20 cycles of 96°C for 15 s, 50°C for 30 s and 70°C for 90 s. Amplicons (1 μl) of this reaction mixture were taken as a template for the second PCR, using the same temperature profile and the universal primer-set 341F-fwd/806R-rev (5′-CCTAYGGGRBGCASCAG-3′ and 5′-GGACTACNNGGGTATCTAAT- 3′) (Gantner et al., 2011; Sundberg et al., 2013).
Raw adapter-trimmed reads were processed using QIIME2 v2021.2.0 (Bolyen et al., 2019). Briefly, the quality of the reads was assessed with FastQC v0.11.9 (Andrews, 2010) and summarized with MultiQC v1.10.1 (Ewels et al., 2016), before and after primer removal. Primers were removed with cutadapt v3.2 (Martin, 2011), discarding reads shorter than 50 bp. Sequences were denoised, truncated to 270 bases (forward reads) and 240 bases (reverse reads), de-replicated and filtered for chimeras (chimera-method consensus, min-fold-parent-over-abundance 2) using the QIIME2 plugin DADA2 (version q2-dada2 v2021.2.0) (Callahan et al., 2016). Taxonomy was assigned to the resulting amplicon sequence variants (ASVs) with the QIIME2 plugin feature-classifier using a pre-fitted sklearn-based taxonomy classifier (Pedregosa et al., 2011; Bokulich et al., 2018). This classifier was trained against the 16S rRNA gene reference database SILVA (release 138, 27.08.2020). Results were unified in a biom-formatted file (McDonald et al., 2012) analyzed using RStudio with the package phyloseq to combine all reads on the family level. Taxa with a relative sequence read counts below 3% were summarized as “other”. Finally, the results were visualized using OriginPro (OriginLab Corporation, Northampton, MA, United States).
2.7 Metagenome sequencing and analysis
Metagenome sequencing was performed on gDNA extracts (3.5–7.5 ng μl-1) from the medium and high salinity level microcosms with methanol and no additional organic C source (S11, S12, S14, S15). Raw, demultiplexed reads were obtained from Azenta GmbH (Leipzig, Germany; using Illumina NovaSeq 2 × 150 bp sequencing) and analyzed with the metaWrap pipeline (Uritskiy et al., 2018). In brief, the reads were trimmed with trim-galore (Krüger, 2012) and quality was assessed using fastqc and quast (Andrews, 2010; Gurevich et al., 2013). Subsequently, contigs were assembled to a minimal length of 1000 bp using metaspades (Nurk et al., 2017) and binned with metabat2 (Kang et al., 2019), maxbin2 (Wu et al., 2016) and concoct (Alneberg et al., 2014). In case of maxbin2, parameters were set to 40 marker gene sets shared by bacteria and archaea. Finally, bins from all three binning tools were refined using binning_refiner (Song and Thomas, 2017), selecting for metagenome assembled genomes (MAGs) with at least 80% integrity (I) and a maximum contamination (X) of 5% which was assessed with CheckM (Parks et al., 2015). Taxonomy was assigned using GTDB-Tk-v1.7.0 with the GTDB release R06-RS202 (Chaumeil et al., 2019; Parks et al., 2021), and the genome was annotated using DRAM (Shaffer et al., 2020). The amino acid composition of the annotated proteins was used to calculate their pI in the post annotation, by employing the ExPASy´s Compute pI function within the R package seqinr (Gasteiger et al., 2005; Charif and Lobry, 2007). Phylogenetic relationships of the recovered MAGs were determined by multiple sequence alignment of 49 core genes using Fasttree plugin on KBase and visualized using ggtree in RStudio. The raw 16S rRNA gene sequences and metagenome sequences were deposited at the European Nucleotide Archive (ENA) under the primary accession number PRJEB57771.
3 Results
3.1 Nutrient cycling
At all salinity levels, an increase of cell number was observed during the incubation period of 125 d (Figures 1A, E, I, M). Without added organic C substrate, acetate (formed by acetogenesis) accumulated at all tested salinity levels with no clear trend with regard to salinity: after 125 days, most acetate was produced at medium salinity, and fewest at low salinity level (Figure 1O). In turn, lactate was converted to acetate (Figures 1G,H), and no enhanced correlation between sulfate consumption and lactate oxidation was observable at medium and high salinity level (Figures 1F,H).
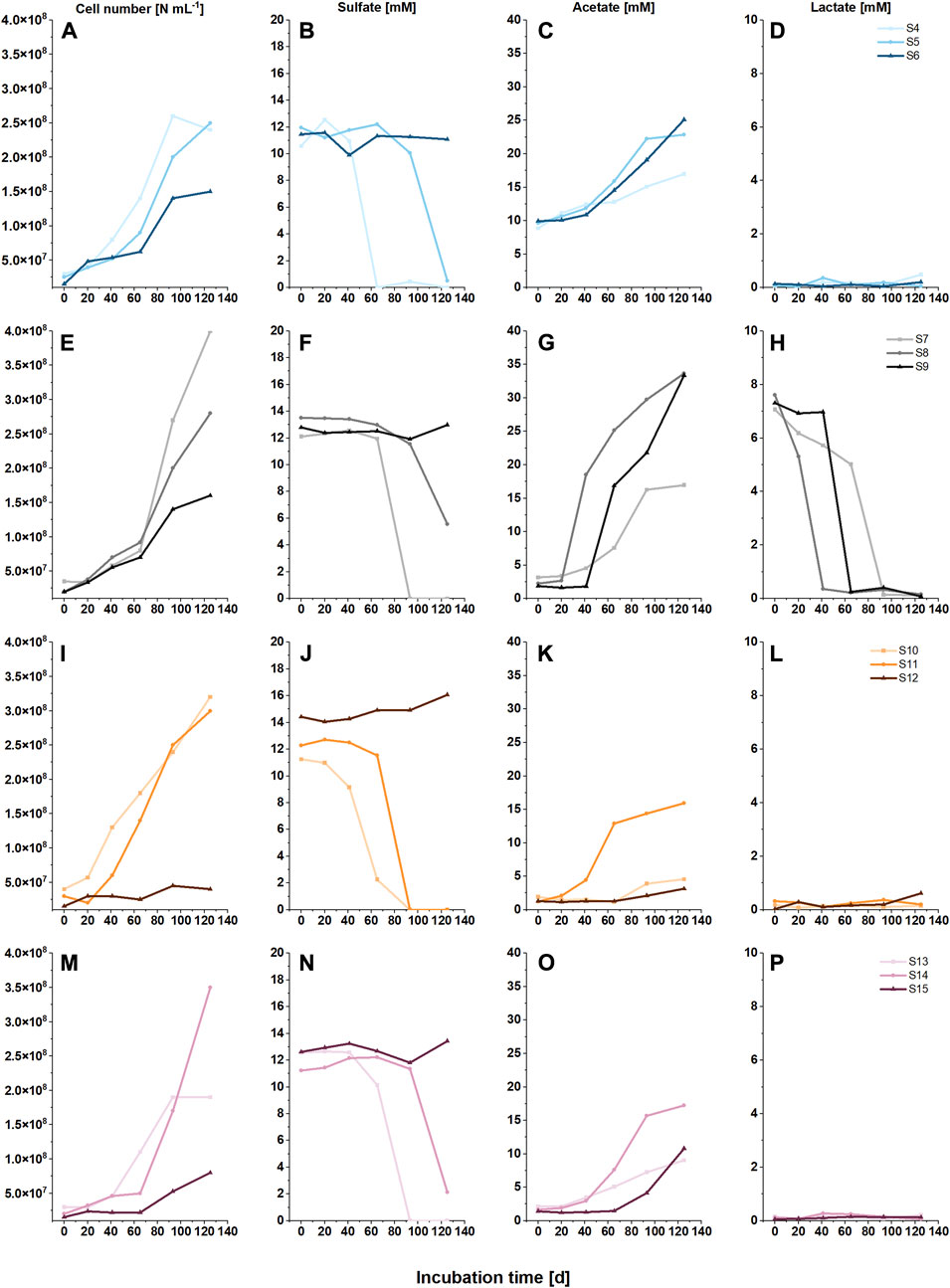
FIGURE 1. Cell numbers (A,E,I,M) and concentrations of sulfate (B, F, J, N), acetate (C, G, K, O) and lactate (D, H, L, P) of microcosms with three salinity levels, indicated by different color shades (2.6 M NaCl, light; 3.4 M NaCl, normal; and 4.4 M NaCl, dark), with either acetate (A, B, C, D, blue), lactate (E, F, G, H, black), methanol (I, J, K, L, orange) or no C source (M, N, O, P, pink) and a H2:CO2 gas (8:2) atmosphere.
Sulfate was added as potential electron acceptor in all microcosms, and sulfate reduction was observed at all low and medium salinity levels within 125 days (Figures 1B, F, J, N). The rate of sulfate reduction seemed to be affected by both, cell number and type of C substrate (Table 1). In brief, after a lag phase of 20–60 d, sulfate was fully consumed in all low salinity microcosms within 93 d; faster sulfate reduction was observed with acetate and methanol as C sources, than with CO2 and lactate (compare Figures 1B,J with Figures 1F,N). Complete and faster sulfate consumption was also achieved in medium salinity level with acetate and methanol as C sources. Incomplete sulfate reduction (final sulfate concentration 2 mM and 6 mM after 125 days of incubation) was achieved in medium saline level with CO2 or lactate as C substrate (Figures 1F,N), while no sulfate reduction was observable during 125 d at high salinity. However, after a total of 1445 days, sulfate was fully consumed in the high salinity setups with organic C source (Table 2, S6, S9, S12). In S15, with CO2 as sole C source and high salinity, the sulfate concentration was unchanged after 1445 days. Acetate production from CO2 was observed at low and medium salinity level (Figure 1O), indicating homoacetogenic activity. Furthermore, acetate accumulated at low and medium salinity level, illustrating that acetogenesis was faster than a putative consumption of acetate by other microorganisms in the community. Likewise, acetate was produced at high salinity level but without sulfate reduction (Figures 1N,O). Acetate was also produced from lactate; the data show that lactate was oxidized to acetate prior to sulfate reduction at medium and high salinity level, while at low salinity level lactate oxidation co-occurred with sulfate reduction (Figures 1F–H). Methane was solely produced from methanol at low and medium salinity level (Supplementary Figure S1, Table 1).
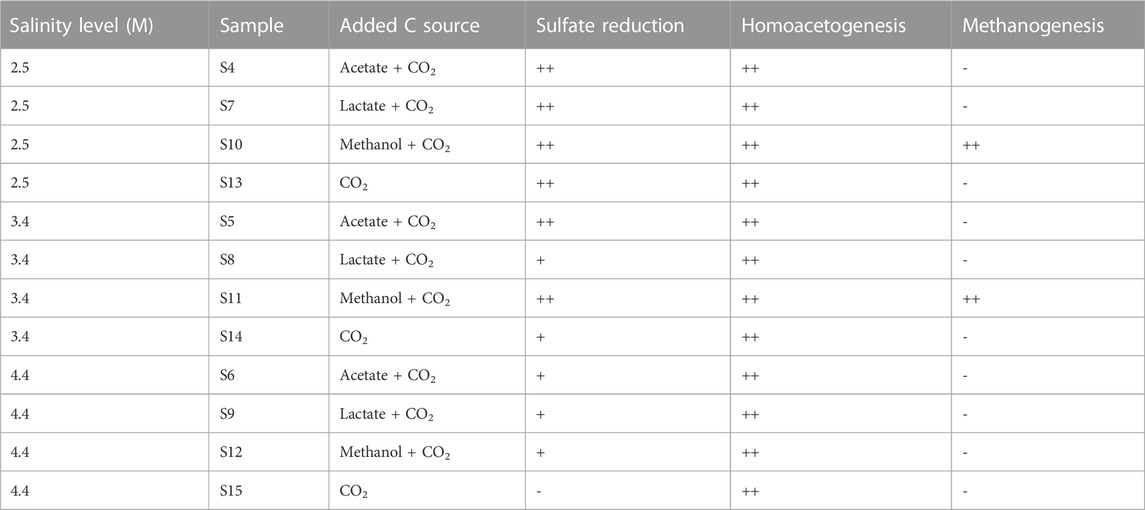
TABLE 1. Metabolic reactions at different salinity levels and with different C sources. The metabolic reactions shown in Figure 1 and Supplementary Figure S1 are listed. Complete sulfate consumption and occurrence of aceto- and methanogenesis within the incubation period of 125 d are indicated with (++), incomplete sulfate consumption and sulfate consumption post 125 d is indicated with (+). No occurrence within 1445 d is indicated with (−).
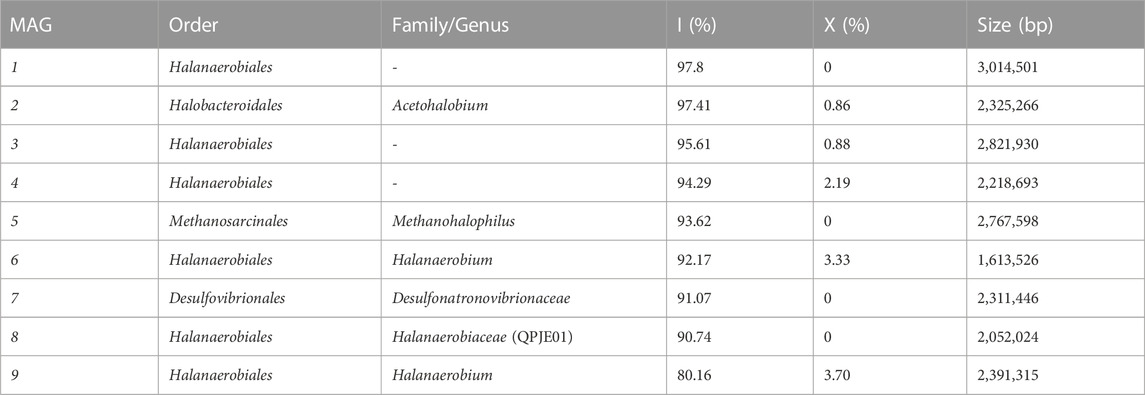
TABLE 2. Metagenome assembled genomes. Nine metagenome assembled genomes (MAGs) resulting from genomic DNA sequencing of samples S11, S12, S14 and S15 with integrity (I) higher than 80% and contamination rate (X) below 5%.
3.2 Proliferation and community structure
The cell numbers increased at all salinity levels over time, and amplicon sequencing data imply that members of the class Halanaerobiia had the highest sequence read abundances (Figure 2); Halanaerobiaceae and Halobacteroidaceae were found at all salinity levels. This finding was underlined with results from metagenome sequencing, which further revealed three MAGs that were assigned at the order level to Halanaerobiales (Table 1). Cell numbers ranged from 1.9 to 4.0∙108 cells ml-1 at low salinity level, with lactate and methanol being most beneficial for cell proliferation. As expected for slow growing microorganisms, stationary phase was not reached with these C sources during the incubation period of 125 days (Figures 1A,E,I, M). Cell numbers in the medium saline level varied less, ranging from 2.5 to 3.5∙108 cells ml-1. At low and medium salinity level, halophilic archaea of the family Methanosarcinaceae contributed largely to the sequence read abundance, indicating a high cell number. At the highest salinity level, acetate and lactate clearly supported proliferation, contrary to the same salinity level with the C sources methanol and CO2 (Figures 1A, E, I, M). Members of the family Desulfohalobiaceae were present at all salinity levels except at highest salinity and only CO2 as C source. Desulfohalobiaceae phylotypes were not found in the metagenome sequencing approach. Instead, sulfate reducing bacteria (SRB) belonging to the family Desulfonatronovibrionaceae sp. were identified (Figure 2; Table 2).
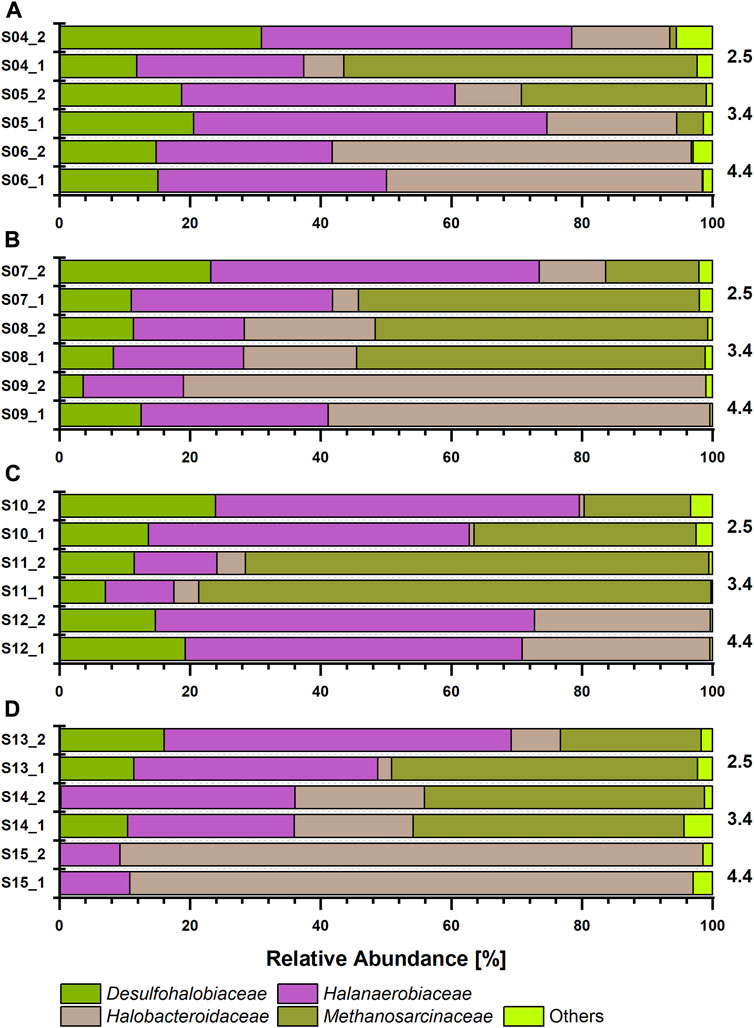
FIGURE 2. Prokaryotic relative sequence read abundances according to 16S rRNA gene-based amplicon sequencing after incubation for 125 days at 30°C grown at three salinity levels (2.5, 3.4 and 4.4 M NaCl, as indicated on the right), under H2:CO2 (8:2) atmosphere and with defined C substrates: (A) acetate (B) lactate, (C) methanol (D) no additional carbon source. Each sample was prepared in duplicates.
3.3 Estimation of microbial metabolic capabilities
Metagenome sequencing allowed for closer characterization of the ASVs assigned to Halobacteroidaceae sp.; MAG 2 was identified as Acetohalobium sp. Automatic DRAM annotation and manual tblastN analysis revealed a complete Wood-Ljungdahl pathway for MAG 2 (Table 2). An incomplete Wood-Ljungdahl pathway was identified in MAG 4, which belongs to the Halanaerobiales (Table 2; Figure 3A). Genetic information for autotrophy were further found for the methanogen Methanohalophilus euhalobius (MAG 5), which was dominant at low and medium salinity levels. The lack of the acetate kinase gene and genes associated with H2 oxidation is in line with the result that methane was solely produced from methanol (Supplementary Figure SI2). In turn, MAG7, identified as a SRB due to the presence of the dissimilatory sulfate reduction pathway, did not fully encode genetic information for autotrophy or lactate-dehydrogenase (Table 2, Supplementary File S1, DRAM Energy), which is in line with the finding that the presence of lactate did not enhance sulfate reduction rates (Figures 1F, H). Lactate-dehydrogenase was found in MAGs 1, 2 and 6.
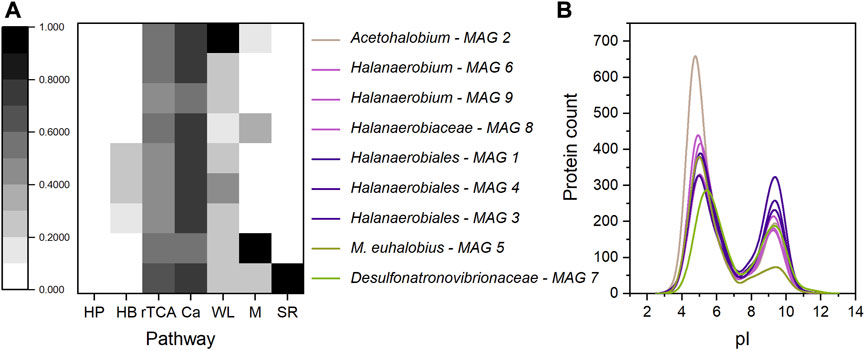
FIGURE 3. (A) Pathway completeness (represented through different shades of grey) of nine MAGs obtained through metagenome sequencing of samples S11, S14, S12 and S15. HP - 3-Hydroxypropionate bi-cycle; HB–Hydroxypropionate-hydroxybutyrate cycle; rTCA–reverse citric acid cycle; Ca–Calvin cycle; WL - Wood-Ljungdahl pathway; M–methanogenesis; SR–sulfate reduction; (B) Distribution of the protein isoelectric point (pI), obtained after MAG annotation using DRAM. The pI value was calculated using the expasy pI calculator plugin for RStudio. Violet: all members of Halanaerobiales, olive: Methanohalophilus euhalobius and green: Desulfonatronovibrionaceae sp. Mean pI-Value: MAG 1–6.7; MAG 2–6.1; MAG 3–6.9; MAG 4–7.0; MAG 5–5.9; MAG 6–6.4; MAG 7–7.1; MAG 8–6.6; MAG 9–6.6.
The proteomic pI value was analyzed to indirectly assess microbial fitness towards osmoadaptation (Figure 3B). Methanohalophilus euhalobius (MAG 5) had the lowest pI value (5.9), due to a small fraction of alkaline proteins (Figure 3B, olive). This low pI value indicates adaptation towards hypersaline conditions through K+ accumulation, however corresponding ASVs (Methanosarcinaceae) were absent at high salinity level. The SRB (MAG 7), in contrast, had the highest pI value of 7.1. The pI values of the other MAGs were within this range, but it has to be noted that a high count in acidic proteins was found in Acetohalobium (MAG 2), which exceeded the numbers of the other Halanaerobiia by more than 200 proteins and resulted in a pI value of 6.1.
Three MAGs assigned to the order Halanaerobiales (MAG 1, 3, 4) clustered on a separate phylogenetic branch, embedded between the Halanaerobiaceae and Acetohalobiaceae (Figure 4). Their pI values ranged from 6.7 to 7.0. A fourth MAG, MAG 8, was closely related to a recently described lineage from a saline offshore oil reservoir (Scheffer et al., 2021) and clustered with MAGs 6 and 9, all having similar pI values of 6.4–6.6.
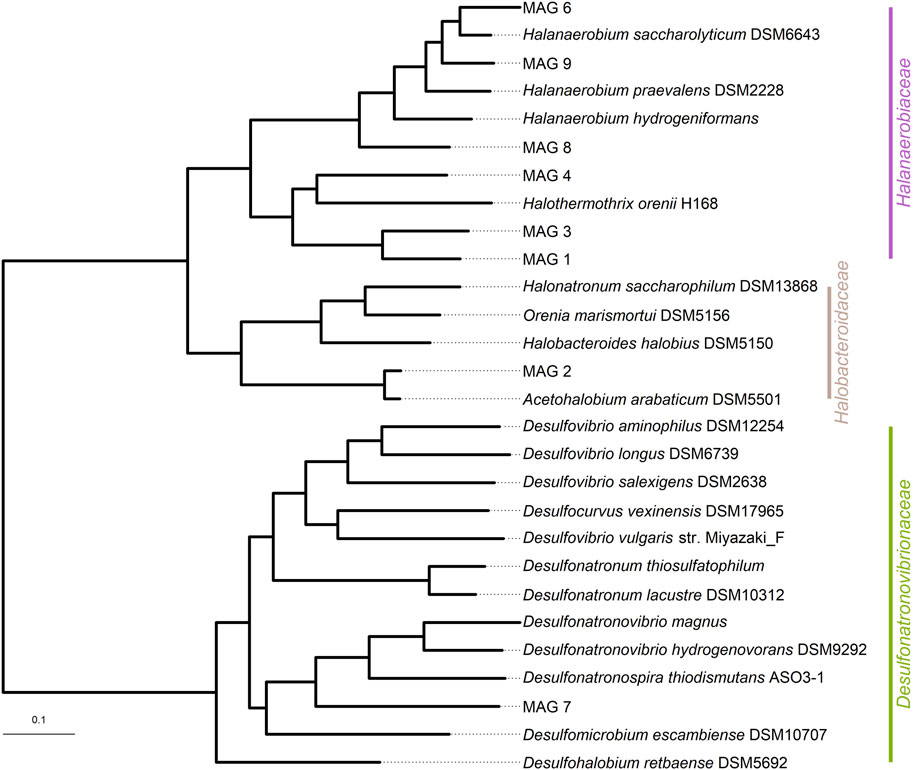
FIGURE 4. Phylogenetic tree based on 49 core, universal genes defined by COG (Clusters of Orthologous Groups) gene families using the species tree function of KBase. The bar represents the genetic distance.
4 Discussion
This study presents metabolic capabilities of mixed cultures enriched from material of saline underground gas storage sites towards H2 and additional C sources at three levels of hypersaline conditions. The cultures represent a mixed community of different sites and were chosen due to their metabolic activity and high cell numbers. With CO2 as sole C source, H2 was utilized regardless of the salt concentration and acetate was produced as metabolite. The accumulating acetate facilitates further heterotrophic processes to occur. The addition of organic C sources, such as methanol, acetate and lactate did not change the microbial community composition, but the rate at which sulfate was consumed and cells proliferated. In contrast, salinity affected both microbial community composition and dissimilatory sulfate reduction: at 4.4 M NaCl, methanogenic archaea disappeared and sulfate reduction rates decreased.
4.1 Initiation of acetogenic activities
In the microcosms, we observed a conversion of CO2/H2 to acetate (Figure 2; Table 2). This could be related to the presence of Acetohabium sp. as these species have been described to use the Wood Ljungdahl pathway to fix CO2 (Ljungdahl and Wood, 1969; Zhilina and Zavarzin, 1990; Sikorski et al., 2010). Our hydrochemical data (Figure 1) show that acetate was continuously produced at all salinity levels, indicating that acetogenesis was independent from the salinity level. A possible reason for the tolerance of acetogens to highest salt concentrations is that they require less energy for osmoregulation than sulfate reducers. Apparently, less energy can be expected from the thermodynamic calculations with the reduction of CO2 to acetate then from sulfate reduction (Thauer et al., 1977). In autotrophic homoacetogens, the net production of ATP from substrate level phosphorylation is zero and therefore has to be achieved through the activity of ATP synthase, either fueled by a H+ or Na+ gradient (Müller, 2003). So far, three types of energy-conserving complexes in acetogens coupled by ion pumping are known, either a ferredoxin:NAD+-oxidoreductase, an energy converting hydrogenase and the methyl viologen-reducing hydrogenase complex (Biegel and Müller, 2010; Schoelmerich and Müller, 2019; Kremp et al., 2022). Making use of an energy conserving complex, which at the same time regulates Na+ translocation, could be a reason for the efficient growth of Acetohalobium sp. The type-strain Acetohalobium arabaticum DSM 5501 was shown to be capable of producing 25 mM acetate when grown in pure culture at low salinity level, and the salinity range for growth was between 1.7 and 4.3 M NaCl (Zhilina and Zavarzin, 1990). The generation of a proton motive force is also necessary for the mode of osmo-adaptation. Some Halanaerobiales are known to belong to the salt-in strategists, which balance osmotic pressure with high intracellular concentrations of Na+, K+, Cl− ions, while not exhibiting an acidic proteome pI (Elevi Bardavid and Oren, 2012; Oren, 2013). Our data confirm a similar proteome pI and imply this mode of osmo-adaptation for the Halanaerobiia found in the dataset. This enables these microorganisms to be active even at extremely high salinity levels. However, the cell numbers at the highest salinity level tested in this study was four times lower than observed at the medium salinity level.
4.2 Initiation of sulfate reduction
All identified SRBs were affiliated as members of the order Desulfovibrionales (Figures 2 and 4), whose genetic information did not encode for a complete CO2 fixation pathway; thus, they probably rely on organic C sources (Table 2). Both, acetate and methanol had a beneficial effect on sulfate reduction, a finding which is supported by the fact that sulfate was fully consumed after 1445 days also at the highest salinity level tested with these organic C sources. Generally, a possible utilization of acetate as electron donor or C source by SRBs in the cultures during the 125 days incubation period is difficult to evaluate since acetate accumulated, although in different amounts, at all conditions (Figure 1) due to H2/CO2 driven acetogenesis or lactate oxidation. Several SRBs are known to oxidize acetate as electron donor or to assimilate acetate when using H2 as electron donor (Rabus et al., 2015). Lactate, being generally a favorable electron donor for SRBs (Santos et al., 2022), was consumed at all salinity levels, but this resulted in enhanced sulfate reduction rates only at low salinity level at which considerably less acetate accumulated compared to medium and high salinity level. High concentrations of acetate in the microcosms with lactate as C source at medium and high salinity level imply a conversion of lactate to acetate, which was not further used by SRBs. However, no DL-lactate dehydrogenases were encoded in the MAG of this SRB (Supplementary Figure S1), and no sulfate was reduced at high salinity level, hence lactate was seemingly not exclusively utilized by SRB, at least at high salinity level. A reduced sulfate reduction rate at high salt levels was previously shown, in which SRBs in anaerobic microbial co-culture belonged to Desulfonatronospira and underwent sulfate reduction at Na+ concentration up to 3.5 M and complete oxidation of 18 mM acetate during 300 d incubation (Sorokin et al., 2014). Other Desulfonatronovibrio spp. (D. thiodismutans, D. magnus and D. hydrogenovorans) grew at Na+ concentrations up to 3.0 M and with H2 as electron donor for sulfate reduction (Sorokin et al., 2011b). Growth at moderate salt concentrations was reported with H2 and formate or acetate, not CO2 (Zhilina, 1997; Sorokin et al., 2011b). The proteome pI value of the SRB was the highest of all MAGs found in the microcosms. For halophilic SRB both osmoadaptation strategies are reported and a differentiation was not possible here. However, sulfidogenesis from other S-species than sulfate, such as thiosulfate and elemental sulfur is common at salt concentrations close to saturation (Sorokin et al., 2010). A possibility remains that the SRB can grow as an autotroph, as the Calvin and reverse citric acid cycle were partially found in MAG 7 and the latter being a confirmed C fixation pathway in SRBs (Schauder et al., 1987).
4.3 Initiation of methanogenesis
Methanogenesis was only observed when methanol was offered as a C source at low and medium salinity level, a fact known from other saline systems (L’Haridon et al., 2020), as this compound needs less reducing power and is an energy efficient substrate for methanogens. In fact, other halophilic methanogens are reported to lack genes allowing for hydrogenotrophic and acetoclastic methanogenesis (Guan et al., 2019). The presence of Methanosarcinaceae in the low and medium salinity level microcosms without methanol suggests that these microorganisms entered a state of survival rather than growth, while inoculation at high salinity levels during the enrichment process decreased the number of methanogens, whose salt range is reported to be optimal below 3.4 M (Guan et al., 2019; L’Haridon et al., 2020). A potential relationship between the co-occurence of methanogens and SRBs can be drawn as sulfate reduction was accelerated when methanol as C substrate was present. Syntrophy between SRB and methanogens or an unknown sulfate reduction pathway in the methanogens themselves, has already been suggested for other archaea (Milucka et al., 2012).
5 Conclusion
H2 underground storage is already successfully performed in salt caverns in the UK and USA, and in subsurface salt formations, ionic strength even exceeds the salt concentrations tested in this study. If these concentrations eventually allow for sulfate reduction and associated production of corrosive and toxic H2S has to be carefully examined. Cultivation-based approaches can allow for the prediction of potential H2S generation if these specific microorganisms are isolated from these cavern environments. Our results show that microbial activity is strongly impaired by increasing salinity levels. High salinity level reduced sulfate reduction rates tremendously during 125 days of incubation and was only observed after 1445 days. To tolerate increasing salinity levels, microorganisms rely on energy-demanding ion transporters and in nutrient limited habitats, such as underground salt caverns, external C and energy sources are mandatory. The presence of added organic C substrates improved sulfate reduction rates at low and medium salinity level. However, even without added organic C substrate, homoacetogenic activity at high salinity level seemingly facilitated sulfate reduction. This makes the effect of various acetate concentrations on sulfate reducing activity in hypersaline systems a key question for further research.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, PRJEB57771.
Author contributions
GN performed the experiments. LS performed the molecular biological and bioinformatic work and analysed the data. Bioinformatic work was supported by LP and DP. GN, LS, MW, and CV designed the experiment. LS drafted the manuscript and CV, DP, GN, and MN revised it.
Funding
This study was funded by the Federal Ministry of Education and Research (BMBF) within the initiative ‘Zwanzig20 Partnerschaft für Innovation’, network “HYPOS”, project “H2-UGS”, Grant 03ZZ0721H, and by the BMBF-funded de. NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI) (031A532B, 031A533A, 031A533B, 031A534A, 031A535A, 031A537A, 031A537B, 031A537C, 031A537D, 031A538A).
Acknowledgments
We acknowledge Theresa Pretzien for selection and screening of the halophilic cultures of the culture collection. Furthermore, she performed the data acquisition and the gas chromatographic and ion chromatographic analyses.
Conflict of interest
Authors GN and MW were employed by the company MicroPro GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2023.1125619/full#supplementary-material
References
Alneberg, J., Bjarnason, B. S., de Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11 (11), 1144–1146. doi:10.1038/nmeth.3103
Andrews, S. (2010). FastQC: A quality control analysis tool for high throughput sequencing data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Becker, E. A., Seitzer, P. M., Tritt, A., Larsen, D., Krusor, M., Yao, A. I., et al. (2014). Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLOS Genet. 10 (11), e1004784. doi:10.1371/journal.pgen.1004784
Biegel, E., and Müller, V. (2010). Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc. Natl. Acad. Sci. 107 (42), 18138–18142. doi:10.1073/pnas.1010318107
Blanco, H., and Faaij, A. (2018). A review at the role of storage in energy systems with a focus on Power to Gas and long-term storage. Renew. Sustain. Energy Rev. 81, 1049–1086. doi:10.1016/j.rser.2017.07.062
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6 (1), 90. doi:10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi:10.1038/s41587-019-0209-9
Bordenave, S., Chatterjee, I., and Voordouw, G. (2013). Microbial community structure and microbial activities related to CO2 storage capacities of a salt cavern. Int. Biodeterior. Biodegrad. 81, 82–87. doi:10.1016/j.ibiod.2012.08.001
Braga Nan, L., Trably, E., Santa-Catalina, G., Bernet, N., Delgenès, J-P., and Escudié, R. (2020). Biomethanation processes: New insights on the effect of a high H2 partial pressure on microbial communities. Biotechnol. Biofuels 13 (1), 141. doi:10.1186/s13068-020-01776-y
Caglayan, D. G., Weber, N., Heinrichs, H. U., Linßen, J., Robinius, M., Kukla, P. A., et al. (2020). Technical potential of salt caverns for hydrogen storage in Europe. Int. J. Hydrogen Energy 45 (11), 6793–6805. doi:10.1016/j.ijhydene.2019.12.161
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13 (7), 581–583. doi:10.1038/nmeth.3869
Charif, D., and Lobry, J. R. (2007). Structural approaches to sequence evolution: Molecules, networks, populations. New York: Springer-Verlag.
Chaumeil, P-A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2019). GTDB-tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36 (6), 1925–1927. doi:10.1093/bioinformatics/btz848
Crotogino, F., Donadei, S., and Bünger, U. (2010). “Landinger H large-scale hydrogen underground storage for securing future energy supplies,” in Proceedings of the 18th World Hydrogen Energy Conference 2010-WHEC 2010, May 16.-21. 2010.
Crotogino, F., Schneider, G-S., and Evans, D. J. (2018). Renewable energy storage in geological formations. Proc. Institution Mech. Eng. Part A J. Power Energy 232 (1), 100–114. doi:10.1177/0957650917731181
Deole, R., Challacombe, J., Raiford, D. W., and Hoff, W. D. (2013). An extremely halophilic proteobacterium combines a highly acidic proteome with a low cytoplasmic potassium content. J. Biol. Chem. 288 (1), 581–588. doi:10.1074/jbc.m112.420505
Dopffel, N., Jansen, S., and Gerritse, J. (2021). Microbial side effects of underground hydrogen storage - knowledge gaps, risks and opportunities for successful implementation. Int. J. Hydrogen Energy 46 (12), 8594–8606. doi:10.1016/j.ijhydene.2020.12.058 .
Elevi Bardavid, R., and Oren, A. (2012). The amino acid composition of proteins from anaerobic halophilic bacteria of the order Halanaerobiales. Extremophiles 16 (3), 567–572. doi:10.1007/s00792-012-0455-y
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32 (19), 3047–3048. doi:10.1093/bioinformatics/btw354
Gantner, S., Andersson, A. F., Alonso-Saez, L., and Bertilsson, S. (2011). Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 84 (1), 12–18. doi:10.1016/j.mimet.2010.10.001
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M., Appel, R., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The proteomics protocols handbook. Editor J. M. Walker (Totowa, NJ: Humana Press), 571–607.
Geluk, M. C. (2000). Late permian (Zechstein) carbonate-facies maps, The Netherlands. Neth. J. Geosciences - Geol. en Mijnbouw 79 (1), 17–27. doi:10.1017/s0016774600021545
Götz, M., Lefebvre, J., Mörs, F., McDaniel Koch, A., Graf, F., Bajohr, S., et al. (2016). Renewable power-to-gas: A technological and economic review. Renew. Energy 85, 1371–1390. doi:10.1016/j.renene.2015.07.066
Guan, Y., Ngugi, D. K., Vinu, M., Blom, J., Alam, I., Guillot, S., et al. (2019). Comparative Genomics of the genus Methanohalophilus, including a newly isolated strain from kebrit deep in the red sea. Front. Microbiol. 10, 839. doi:10.3389/fmicb.2019.00839
Gunde-Cimerman, N., Plemenitaš, A., and Oren, A. (2018). Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 42 (3), 353–375. doi:10.1093/femsre/fuy009
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). Quast: Quality assessment tool for genome assemblies. Bioinformatics 29 (8), 1072–1075. doi:10.1093/bioinformatics/btt086
Hamilton, W. A. (2003). Microbially influenced corrosion as a model system for the study of metal microbe interactions: A unifying electron transfer hypothesis. Biofouling 19 (1), 65–76. doi:10.1080/0892701021000041078
Heinemann, N., Alcalde, J., Miocic, J. M., Hangx, S. J. T., Kallmeyer, J., Ostertag-Henning, C., et al. (2021). Enabling large-scale hydrogen storage in porous media – The scientific challenges. Energy and Environ. Sci. 14, 853–864. doi:10.1039/d0ee03536j
Hemme, C., and van Berk, W. (2018). Hydrogeochemical modeling to identify potential risks of underground hydrogen storage in depleted gas fields. Appl. Sci. 8, 2282. doi:10.3390/app8112282
Hemme, C., and van Berk, W. (2017). Potential risk of H2S generation and release in salt cavern gas storage. J. Nat. Gas Sci. Eng. 47, 114–123. doi:10.1016/j.jngse.2017.09.007
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359. doi:10.7717/peerj.7359
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41 (1), e1. doi:10.1093/nar/gks808
Kremp, F., Roth, J., and Müller, V. (2022). A third way of energy conservation in acetogenic bacteria. Microbiol. Spectr. 10 (4), e0138522–22. doi:10.1128/spectrum.01385-22
Krüger, F. (2012). Trim galore. Available at: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
L’Haridon, S., Haroun, H., Corre, E., Roussel, E., Chalopin, M., Pignet, P., et al. (2020). Methanohalophilus profundi sp. nov., a methylotrophic halophilic piezophilic methanogen isolated from a deep hypersaline anoxic basin. Syst. Appl. Microbiol. 43 (5), 126107. doi:10.1016/j.syapm.2020.126107
Ljungdahl, L. G., and Wood, H. G. (1969). Total synthesis of acetate from CO2 BY heterotrophic bacteria. Annu. Rev. Microbiol. 23 (1), 515–538. doi:10.1146/annurev.mi.23.100169.002503
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17 (1), 10. doi:10.14806/ej.17.1.200
Matos, C. R., Carneiro, J. F., and Silva, P. P. (2019). Overview of large-scale underground energy storage technologies for integration of renewable energies and criteria for reservoir identification. J. Energy Storage 21, 241–258. doi:10.1016/j.est.2018.11.023
McDonald, D., Clemente, J. C., Kuczynski, J., Rideout, J. R., Stombaugh, J., Wendel, D., et al. (2012). The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. GigaScience 1 (1), 2047–2217. doi:10.1186/2047-217x-1-7
Milucka, J., Ferdelman, T. G., Polerecky, L., Franzke, D., Wegener, G., Schmid, M., et al. (2012). Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 491 (7425), 541–546. doi:10.1038/nature11656
Müller, V. (2003). Energy conservation in acetogenic bacteria. Appl. Environ. Microbiol. 69 (11), 6345–6353. doi:10.1128/aem.69.11.6345-6353.2003 .
Nurk, S., Meleshko, D., Korobeynikov, A., and Pevzner, P. A. (2017). metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27 (5), 824–834. doi:10.1101/gr.213959.116
Oren, A. (2002). Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications. J. Industrial Microbiol. Biotechnol. 28, 56–63. doi:10.1038/sj/jim/7000176
Oren, A. (2013). Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front. Microbiol. 4 10.3389/fmicb.2013.00315 4, 315. doi:10.3389/fmicb.2013.00315
Panfilov, M. (2016). Underground and pipeline hydrogen storage. In: Compendium of Hydrogen Energy, Hydrogen Storage, Distribution and Infrastructure. Sawston, UK: Woodhead Publishing 2, 91–115. doi:10.1016/B978-1-78242-362-1.00004-3
Parks, D. H., Chuvochina, M., Rinke, C., Mussig, A. J., Chaumeil, P-A., and Hugenholtz, P. (2021). Gtdb: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794. doi:10.1093/nar/gkab776
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25 (7), 1043–1055. doi:10.1101/gr.186072.114
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., and Thirion, B. (2011). Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12, 2825–2830.
Rabus, R., Venceslau, S. S., Wohlbrand, L., Voordouw, G., Wall, J. D., and Pereira, I. A. (2015). A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv. Microb. Physiol. 66, 55–321. doi:10.1016/bs.ampbs.2015.05.002
Reistad, R. (1970). On the composition and nature of the bulk protein of extremely halophilic bacteria. Arch. Mikrobiol. 71 (4), 353–360. doi:10.1007/bf00417131
Roberts, M. F. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 1 (1), 5. doi:10.1186/1746-1448-1-5
Santos, A. M. D., Costa, J. M., Braga, J. K., Flynn, T. M., Brucha, G., Sancinetti, G. P., et al. (2022). Lactate as an effective electron donor in the sulfate reduction: Impacts on the microbial diversity. Environ. Technol. 43 (20), 3149–3160. doi:10.1080/09593330.2021.1916092
Schauder, R., Widdel, F., and Fuchs, G. (1987). Carbon assimilation pathways in sulfate-reducing bacteria II. Enzymes of a reductive citric acid cycle in the autotrophic Desulfobacter hydrogenophilus. Archives Microbiol. 148 (3), 218–225. doi:10.1007/bf00414815
Scheffer, G., Hubert, C. R. J., Enning, D. R., Lahme, S., Mand, J., and de Rezende, J. R. (2021). Metagenomic investigation of a low diversity, high salinity offshore oil reservoir. Microorganisms 9 (11), 2266. doi:10.3390/microorganisms9112266
Schoelmerich, M. C., and Müller, V. (2019). Energy conservation by a hydrogenase-dependent chemiosmotic mechanism in an ancient metabolic pathway. Proc. Natl. Acad. Sci. 116 (13), 6329–6334. doi:10.1073/pnas.1818580116
Schwab, L., Popp, D., Nowack, G., Bombach, P., Vogt, C., and Richnow, H. H. (2022). Structural analysis of microbiomes from salt caverns used for underground gas storage. Int. J. Hydrog. Energy. doi:10.1016/j.ijhydene.2022.04.170
Shaffer, M., Borton, M. A., McGivern, B. B., Zayed, A. A., La Rosa Sabina, L., Solden, L. M., et al. (2020). DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 48 (16), 8883–8900. doi:10.1093/nar/gkaa621
Sikorski, J., Lapidus, A., Chertkov, O., Lucas, S., Copeland, A., Glavina Del Rio, T., et al. (2010). Complete genome sequence of Acetohalobium arabaticum type strain (Z-7288T). Stand Genomic Sci. 3 (1), 57–65. doi:10.4056/sigs.1062906
Song, W-Z., and Thomas, T. (2017). Binning_refiner: Improving genome bins through the combination of different binning programs. Bioinformatics 33 (12), 1873–1875. doi:10.1093/bioinformatics/btx086
Sorokin, D. Y., Abbas, B., Tourova, T. P., Bumazhkin, B. K., Kolganova, T. V., and Muyzer, G. (2014). Sulfate-dependent acetate oxidation under extremely natron-alkaline conditions by syntrophic associations from hypersaline soda lakes. Microbiology-Sgm 160, 723–732. doi:10.1099/mic.0.075093-0
Sorokin, D. Y., Chernyh, N. A., and Poroshina, M. N. (2015). Desulfonatronobacter acetoxydans sp nov., a first acetate-oxidizing, extremely salt-tolerant alkaliphilic SRB from a hypersaline soda lake. Extremophiles 19 (5), 899–907. doi:10.1007/s00792-015-0765-y
Sorokin, D. Y., Kuenen, J. G., and Muyzer, G. (2011a). The microbial sulfur cycle at extremely haloalkaline conditions of soda lakes. Front. Microbiol. 2 10.3389/fmicb.2011.00044 2, 44. doi:10.3389/fmicb.2011.00044
Sorokin, D. Y., Makarova, K. S., Abbas, B., Ferrer, M., Golyshin, P. N., Galinski, E. A., et al. (2017). Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat. Microbiol. 2, 81. doi:10.1038/nmicrobiol.2017.81
Sorokin, D. Y., Rusanov, , Pimenov, N. V., Tourova, T. P., Abbas, B., and Muyzer, G. (2010). Sulfidogenesis under extremely haloalkaline conditions in soda lakes of Kulunda Steppe (Altai, Russia). Fems Microbiol. Ecol. 73 (2), 278–290. doi:10.1111/j.1574-6941.2010.00901.x
Sorokin, D. Y., Tourova, T. P., Kolganova, T. V., Detkova, E. N., Galinski, E. A., and Muyzer, G. (2011b). Culturable diversity of lithotrophic haloalkaliphilic sulfate-reducing bacteria in soda lakes and the description of Desulfonatronum thioautotrophicum sp. nov., Desulfonatronum thiosulfatophilum sp. nov., Desulfonatronovibrio thiodismutans sp. nov., and Desulfonatronovibrio magnus sp. nov. Extremophiles 15 (3), 391–401. doi:10.1007/s00792-011-0370-7
Strohmenger, C., Voigt, E., and Zimdars, J. (1996). Sequence stratigraphy and cyclic development of Basal Zechstein carbonate-evaporite deposits with emphasis on Zechstein 2 off-platform carbonates (Upper Permian, Northeast Germany). Sediment. Geol. 102 (1), 33–54. doi:10.1016/0037-0738(95)00058-5
Sundberg, C., Al-Soud, W. A., Larsson, M., Alm, E., Yekta, S. S., Svensson, B. H., et al. (2013). 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 85 (3), 612–626. doi:10.1111/1574-6941.12148
Thauer, R. K., Jungermann, K., and Decker, K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41 (1), 100–180. doi:10.1128/br.41.1.100-180.1977
Uritskiy, G. V., DiRuggiero, J., and Taylor, J. (2018). MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6 (1), 158. doi:10.1186/s40168-018-0541-1
Vignais, P. M., and Billoud, B. (2007). Occurrence, classification, and biological function of hydrogenases: An overview. Chem. Rev. 107 (10), 4206–4272. doi:10.1021/cr050196r
Widdel, F., Kohring, G-W., and Mayer, F. (1983). Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. Archives Microbiol. 134 (4), 286–294. doi:10.1007/bf00407804
Wolin, E. A., Wolin, M. J., and Wolfe, R. S. (1963). Formation of methane by bacterial extracts. J. Biol. Chem. 238, 2882–2886. doi:10.1016/s0021-9258(18)67912-8
Wu, Y-W., Simmons, B. A., and Singer, S. W. (2016). MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32 (4), 605–607. doi:10.1093/bioinformatics/btv638
Zhilina, T. N., and Zavarzin, G. A. (1990). Extremely halophilic, methylotrophic, anaerobic bacteria. FEMS Microbiol. Rev. 7 (3-4), 315–322. doi:10.1111/j.1574-6968.1990.tb04930.x
Keywords: halophiles, hydrogen, salt cavern, sulfate reduction, homoacetogenesis
Citation: Schwab L, Prinsen L, Nowack G, Popp D, Noll M, Vogt C and Wagner M (2023) Sulfate reduction and homoacetogenesis at various hypersaline conditions: Implications for H2 underground gas storage. Front. Energy Res. 11:1125619. doi: 10.3389/fenrg.2023.1125619
Received: 16 December 2022; Accepted: 13 March 2023;
Published: 05 April 2023.
Edited by:
Diana Z Sousa, Wageningen University and Research, NetherlandsReviewed by:
Suman Bajracharya, Luleå University of Technology, SwedenEvgenii N. Frolov, Winogradsky Institute of Microbiology (RAS), Russia
Copyright © 2023 Schwab, Prinsen, Nowack, Popp, Noll, Vogt and Wagner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carsten Vogt, Y2Fyc3Rlbi52b2d0QHVmei5kZQ==
†Present address: Denny Popp, Institute of Human Genetics, University of Leipzig Medical Center, Leipzig, Germany
 Laura Schwab
Laura Schwab Lilian Prinsen
Lilian Prinsen Guido Nowack
Guido Nowack Denny Popp
Denny Popp Matthias Noll
Matthias Noll Carsten Vogt
Carsten Vogt Martin Wagner
Martin Wagner