
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Energy Res. , 22 April 2021
Sec. Bioenergy and Biofuels
Volume 9 - 2021 | https://doi.org/10.3389/fenrg.2021.616945
This article is part of the Research Topic Biomass, Bioenergy and Biofuels for Circular Bioeconomy View all 4 articles
 Andi Nuraliyah
Andi Nuraliyah Meka Saima Perdani
Meka Saima Perdani Dwini Normayulisa Putri
Dwini Normayulisa Putri Muhamad Sahlan
Muhamad Sahlan Anondho Wijanarko
Anondho Wijanarko Heri Hermansyah*
Heri Hermansyah*Adsorption-crosslinking is one of the immobilization methods to improve the reusability of lipase. It requires amino groups to reduce cross-link immobilization risk so that lipase–support interaction increases and the immobilization is attainable. Also, the amino group on the support is expected to increase lipase performance. This study aimed to analyze the effect of amino group addition on immobilized Aspergillus niger lipase by the adsorption-crosslinking using MP-64 macroporous anion resin and XAD-7HP macroporous nonionic resin that has been treated with chitosan. The chitosan-coated resin was characterized by Fourier transform infrared spectrometry (FTIR) and scanning electron microscope (SEM). Lipase immobilization was carried out by adding 10 ml lipase solution containing 0.75 g resins and shaken at 25°C for 150 rpm. Adsorption was achieved for 4 h, followed by cross-linking separately (adding 0.5% (v/v) glutaraldehyde and re-reacting for 20 min). Lipase activity was measured with the titrimetric of olive oil emulsion; mixed with Aspergillus niger lipase, emulsion, and a buffer solution (pH 6.5, ionic strength of 0.7); and incubated for 30 min at 37°C. The effect of amino-functional groups was investigated based on lipase loading and lipase activity. The best lipase loading and lipase activity of 83.79% and 29.41 U/g support were achieved in the adsorption-crosslinking using MP-64 resin coated with chitosan. After four cycles, biodiesel synthesis was maintained at 70.61% of the initial yield. These results indicated that chitosan as an affordable and readily available source of amino groups could be used to modify support for Aspergillus niger lipase immobilization.
Interest in substitute for conventional diesel fuel to biodiesel has occurred in several countries due to the massive use of nonrenewable diesel fuel. Biodiesel can be produced through transesterification or interesterification of triglycerides (long-chain fatty acids) derived from plant oils or animal fats into monoalkyl esters using a base catalyst (metal-based hydroxides, alkali metal–based hydroxides, and carbonates), acid catalyst (sulfonic and sulfuric acid), nanocatalyst, and biocatalyst (Hermansyah et al., 2011; Mishra and Goswami, 2017; Rizwanul Fattah et al., 2020). The use of catalysts for biodiesel production has its respective advantages and disadvantages. According to Sheldon and van Pelt (2013), Mohamad et al. (2015), and Reis et al. (2019), enzymatic processes are preferred because they are more environmentally friendly, cost-effective, and sustainable.
Lipase (EC. 3.1.1.3) is one of the enzymes applied in several industries, that is, biofuel, foods and flavors, pharmaceuticals, agrochemicals, and cosmetics (Mendes et al., 2012). Lipase acts as a biocatalyst for saponifications, esterifications, ester hydrolysis, and alcoholysis reaction (Manoel et al., 2015; Galvão et al., 2018). Biodiesel can be produced using lipase. The use of lipase is preferred because of its catalyst properties with superiority in selectivity, well specificity, and working under mild conditions (Garcia-Galan et al., 2011; Sheldon and van Pelt, 2013; Barbosa et al., 2015; Reis et al., 2019). The use of free lipases in some industries is certainly less economical, sometimes hampered by environmental instability, such as restriction on its reproducibility (Verma et al., 2013) so that homogenous lipases can be engineered into heterogeneous catalysts with several suitable immobilization techniques in order to recover, easily separate, and reuse (Cantone et al., 2013). Lipase is known to be immobilized by adsorption and cross-linking (Yang et al., 2010; Lage et al., 2016). Combining the adsorption and cross-linking methods develops a traditional process capable of increasing enzyme activity and stability during the reaction (Alamsyah et al., 2017).
Adsorption is an uncomplicated enzyme immobilization method due to low production cost and high enzyme activity. However, this process has weak bonds between the enzymes and their supports (Mohamad et al., 2015). Cross-linking is an enzyme immobilization method without support and used only for bifunctional cross-linkers such as glutaraldehyde. Glutaraldehyde is a strong cross-linker, giving intermolecular cross-linking in any supports, coping the enzyme leakage, providing the stabilization of enzyme, and also functions as an activator to allow an enzyme–support covalent bond, and can be used to bind the enzymes to the matrix that has a free amino group (Broun, 1976; Barbosa et al., 2012; Barbosa et al., 2014; Moreira et al., 2020). Also, glutaraldehyde acts as a spacer arm to decrease inadmissible enzyme–functional group carrier surface interaction and minimize the steric hindrance (Yang et al., 2010).
According to Elnashar (2010), support characteristics are also essential. Hydrophobic-macroporous organic support polymers, such as Lewatit MP-64 and Amberlite XAD-7HP, have a better protein affinity than inorganic support and suitable for lipase immobilization (Facin et al., 2019). However, lipase immobilization needs more improvements for activity, stability, and specificity (Manoel et al., 2015). One of the efforts is the attachment of amino groups to the hydrophobic support surface. The addition of amino groups enhances the bond strength between the enzyme and surface of the support, reduces leaching to produce a good reproducibility, and reuses high efficiency (Salis et al., 2005; Talbert and Goddard, 2012; Jesionowski et al., 2014). In this study, the addition of amino-functional groups was carried out through a resin-coating process using chitosan. Chitosan is a naturally occurring polysaccharide found in the crustacean exoskeleton, has a long-chain polymer with the NH2 functional group, and has excellent properties, such as nontoxicity, biocompatibility, chemical reactivity, and allowing easy enzyme fixation (Minovska et al., 2005; Pinheiro et al., 2019). Chitosan provides primary amino groups that play a role in compiling amino-glutaraldehyde–glutaraldehyde groups and reducing cross-link immobilization risk (Barbosa et al., 2013; Pinheiro et al., 2019). Also, it provides additional support of having one to two amino groups carrying space arms (Barbosa et al., 2013).
This study was conducted to evaluate the effect of amino-functional group addition derived from chitosan on Amberlite XAD-7HP and Lewatit MP-64 resins in the adsorption-crosslinking method. The evaluation was carried out by comparing the Aspergillus niger lipase loading and lipase activity on the chitosan-coated resin and chitosan-free resin, regardless of specific affinity and inhibitor activity factors. Immobilized lipase with the highest lipase loading was further tested to analyze its activity and reusability.
The materials used in this research were commercial Aspergillus niger lipase powder as a free enzyme in ∼200 U g−1 (SKU No. 62301), Lewatit MP-64, and Amberlite XAD-7HP (see Table 1 for specifications) obtained from Sigma-Aldrich (St. Louis, United States). Chitosan was obtained from PT. Monadon Group (Bogor, Indonesia), and commercial palm cooking oil was purchased from the local market under the production of PT. Salim Ivomas Pratama, Tbk. (Jakarta, Indonesia), while other chemicals used in analytical standards were obtained from Sigma-Aldrich (Singapore).
Resin modifications with the amino group were prepared based on the methodology described by Liu et al. (2011). Chitosan solution was formed by dissolving 1 g of chitosan in 1% acetic acid (glacial acetic acid) solution and stirring overnight. Two grams of resins were added to the chitosan solution and shaken at 150 rpm in a water bath at 30°C for 8 h. After removing excess chitosan solution through vacuum filtration, resins were immersed in 2 M NaOH in 20% ethanol and stirred for 3 h. The chitosan-coated resin solution was then filtered, washed with deionized water, dried, and stored at room temperature.
Fourier transform infrared spectroscopy (FTIR) was used to determine the functional groups in different resins, including chitosan-free resin (XAD-7HP and MP-64), chitosan-coated resin, and resin reacted with Aspergillus niger lipase and glutaraldehyde. The spectrums were recorded using FTIR Thermo Scientific Nicolet is5 with Aspergillus niger lipase immobilization to determine the morphology of resins with all the samples characterized by scanning electron microscopy (SEM), the Tescan Vega 3 LMU type. The samples characterized were resin without chitosan coating and those reacted with Aspergillus niger lipase and glutaraldehyde.
Aspergillus niger lipase solution with a concentration of 10 mg/ml was prepared using phosphate buffer 0.02 M with an ionic strength of 0.28 in pH 7, stirred at 300 rpm, and incubated at 4°C for 60 min. The enzyme was filtered first using Whatman No. 1 before use for immobilization.
XAD-7HP and MP-64 resins were used as support to immobilize Aspergillus niger lipase. According to Wang et al. (2011), lipase immobilization was carried out from the previous research method modification process by adding 10 mL enzyme solution to a 100-mL flask containing 0.75 g resins. The samples were incubated at 25°C and shaken for 150 rpm. Furthermore, the immobilization reaction was carried out for 4 h (adsorption) before adding 0.5% (v/v) glutaraldehyde and re-reacted for another 20 min to complete the cross-linking process. A supernatant sample was collected for lipolytic activity and lipase loading analysis. The support-contained immobilized lipase was rinsed using 25 mL phosphate buffer (0.02 M, pH 7, ionic strength of 0.28) and rinsed repeatedly using 25 mL distilled water.
The Bradford method was used to analyze the amount of enzyme (Bradford, 1976) using bovine serum albumin (BSA) as a protein standard. The amount of immobilized protein was determined from the difference between the initial amounts of pre-immobilization availability found in the filtrate solution and supernatant separated from the immobilized mixture (Osuna et al., 2015). The enzyme loading was calculated by Eq. 1.
Ci, Cs, and Cw are the initial enzyme, supernatant, and washing agent concentration (mg/ml), whereas Vi, Vs, and Vw are the volumes of the initial enzyme, supernatant, and washing agent (ml). The measurement was used for both the adsorption and adsorption-crosslinking processes, with and without the addition of chitosan.
The lipase activity was tested by a titrimetric method with olive oil emulsion used as a substrate by mixing 7 g Arabic gum, 50 mL olive oil, and 50 mL distilled water for approximately 5 min (Damaso et al., 2013). The reaction mixtures consisted of 4 ml phosphate buffer (0.05 M, pH 6.5, ionic strength of 0.7), 5 mL emulsion, and 1 mL lipase incubated for 30 min at 37°C. The reaction was stopped by adding a 10-mL solution mixture of acetone, water, and methanol in equal ratios. Furthermore, free fatty acids were titrated with 0.05 M sodium hydroxide solution in the presence of phenolphthalein indicator. A blank experiment was carried out in the same method by comparing it with Aspergillus niger lipase to produce 1 µmole of free fatty acid per minute under test conditions. The lipase activity was described by Li et al. (2010), followed by Eq. 2.
Where A0 and Ai are the lipase activity (U) in the supernatant measured before and after immobilization, respectively, and m is the mass of support (g).
Biodiesel was synthesized in a batch system using Erlenmeyer flasks following Hermansyah et al. (2011). Furthermore, palm cooking oil was used as a substrate with methyl acetate as an acyl acceptor in a ratio of 1:12. The biocatalyst was added to 4% (w/w) palm cooking oil, and the reaction was carried out in a water shaker bath and mixed at 150 rpm with a reaction time of 50 h. Biodiesel (methyl ester) formed was analyzed using HPLC-type Shimadzu HPLC 20-AD equipped with a diode array detector (DAD) and C-18 RP column, with a flow rate of 0.8 mL/min, methanol (solvent A), and 5:4 v/v isopropanol–hexane (solvent B) as the mobile phase that was described further in Meher et al. (2006) at a wavelength of 205 nm.
Analysis of statistics was conducted using Minitab 19 with one-way ANOVA and Tukey’s post hoc test. The measurements of enzyme loading and specific activity were in duplicate.
The characteristics of support have a strong influence on the immobilized enzyme (Tischer and Wedekind, 1999). FTIR is used to identify functional groups or chemical bonds on the surface of prepared materials with a specific energy absorption band (Silva et al., 2012; Osuna et al., 2015). It is also used to ascertain and estimate enzyme immobilization performance (Cipolatti et al., 2014; Nicoletti et al., 2015).
In Figures 1A,B, FTIR analysis was used to determine functional group change of XAD-7HP and MP-64. In general, there was a decrease in transmittance for chitosan-coated resin, particularly in the 3,300–3,500 cm−1 area, as shown in Figures 1A,B. A similar result was obtained from the chitosan-coated DSO3, ZGC351MB, and ZGC35 resins, which exhibited a resin bond with the NH2 group (Jin and Bai, 2002). The intensity was also changed due to the deformation of the C–H bond and the support’s alkyl chain (Urrutia et al., 2017). An immobilization process that occurred in the reaction was able to distract the chain of enzyme or support. Somehow, this change affected the functionalization of the enzyme. In our work, we coated lipase with chitosan to prevent deformation in the immobilization process. Glutaraldehyde was also used to maintain the pore of the support. As confirmed by the FTIR analysis, the NH2 group was shown in the wavelength of 3,300–3,500 cm−1 (Urrutia et al., 2017).
Change in the absorption band intensity was related to the formation of chemical bonds between nitrogen and oxygen, which indicated the amine and OH groups were involved in the reaction with glutaraldehyde to form the Schiff base reaction (Yang et al., 2010; Silva et al., 2012; Pal and Banat, 2014). FTIR results obtained by Silva et al. (2012) showed that the reaction between support and glutaraldehyde led to the loss of NH and OH functional groups. This trend was similar to Figure 1B for chitosan-coated MP-64 resin after cross-linking with lipase by glutaraldehyde. The change in peak wavelength intensity indicated a successful immobilization process (Osuna et al., 2012).
The samples were characterized using SEM to determine morphological differences before and after the modification process. Furthermore, alteration in the resin’s surface was followed by adding enzymes and glutaraldehyde using SEM images, as shown in Figure 2. The surfaces of XAD-7HP and MP-64 resins without chitosan coating are shown in Figures 2A,C. After immobilization of XAD-7HP and MP-64 resin coated with chitosan, the resin’s surface became rough with different levels according to each resin’s absorption capacity, as shown in Figures 2B,D. It indicated that many protein molecules were attached in the support under chitosan addition.
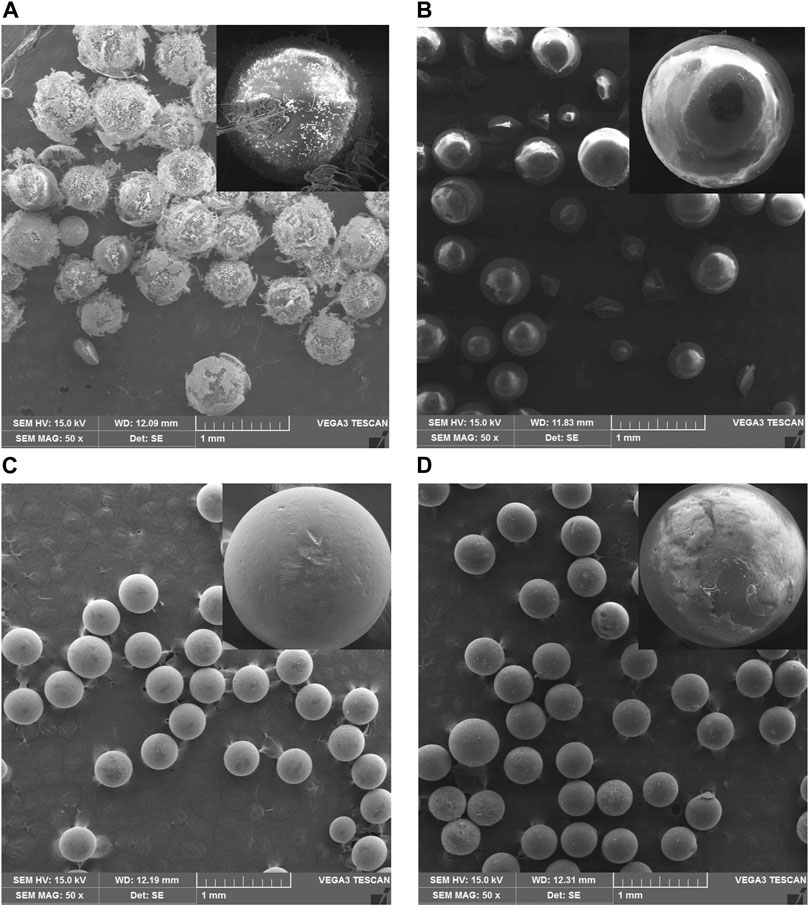
FIGURE 2. SEM results of (A) XAD-7HP resin without chitosan coating, (B) chitosan-coated XAD-7HP resin after being reacted with Aspergillus niger lipase and glutaraldehyde, (C) MP-64 resin without chitosan coating, and (D) chitosan-coated XAD-7HP resin after being reacted with Aspergillus niger lipase and glutaraldehyde.
As shown in Figure 2, the immobilized enzyme’s rough surface was caused by the presence of an aldehyde group from glutaraldehyde. The chitosan presence built a bulky particle form that agreed with values reported for other chitosan supports (Dos Santos et al., 2017; Urrutia et al., 2017). In the synthesis process, numerous researchers found a problematic way to prevent the particles from the racemic and bulky. Several methods include activator agents, changing solvent or even decreasing concentration (Dos Santos et al., 2017).
In the previous study, chitosan beads were no longer uniform and showed some irregularities in spherical and compact spheres. In our study, immobilized chitosan has also been spherical with some irregularities (Weber et al., 2019). Since the enzyme form is not explicitly affected by the catalysis properties, we have demonstrated the characteristics of activity analysis, enzyme loading, and biodiesel application.
The production of lipase enzyme had been developed and applied in many methods. Numerous researchers also studied catalytic loss. Immobilization is becoming a tool to improve catalyst stability, catalytic activity, and resistance to inhibitors. Previous research found that lipase’s immobilization in modified chitosan resulted in similar efficiency for the specific acid–base and could be performed for five cycles without catalytic activity loss (Akil et al., 2020). In our work, we used lipase and immobilized it with the specific resin. The immobilization process started with the lipase’s adsorption onto the supporting surface for a specific time, followed by a cross-linking process, by adding glutaraldehyde to link the reactive amino groups in the enzyme shown in Figure 3. The immobilization of lipase on hydrophobic support was based on the activity of the interface. Therefore, it can be used in the hydrolysis of fats and oils (Montero et al., 1993; Urrutia et al., 2017).
The effect of the amino-functional group on the enzyme loading of XAD-7HP and MP-64 resins has been observed in this study. The effect of amino-functional groups was carried out by comparing the chitosan-free resin (without amino-functional groups) and chitosan-coated resin (with amino-functional groups) through adsorption and adsorption–crosslinking method. Experiments with the adsorption method were only intended to prove whether the resin immobilized by cross-linking adsorption has a better enzyme loading ability or vice versa. The enzyme loading of XAD-7HP and MP-64 resins is shown in Figures 4A,B.
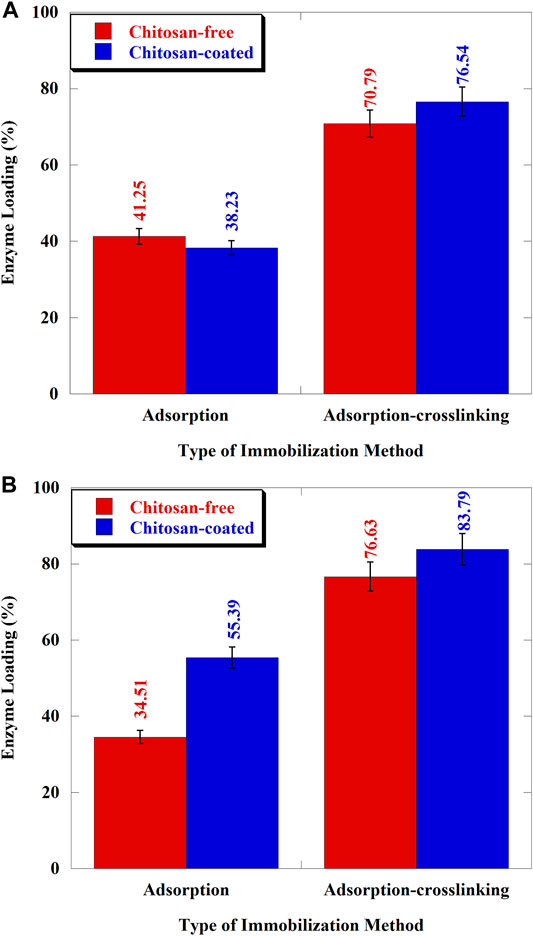
FIGURE 4. Enzyme loading of Aspergillus niger lipase immobilized on resins: (A) XAD-7HP and (B) MP-64.
The addition of amino-functional groups to the resin affected the enzyme loading values. Figure 4A showed that the enzyme loading of chitosan-free and chitosan-coated XAD-7HP resins was 41.25 and 38.23% for adsorption only, respectively. In this case, the chitosan-coated XAD-7HP had a lower enzyme loading than the chitosan-free XAD-7HP (p < 0.05). We hypothesized that some of the pore surfaces of the resins were covered with chitosan, thereby inhibiting the adsorption of Aspergillus niger lipase to the resin’s surfaces. Previous studies have reached comparable findings indicating that pore size affected the enzymatic loading capacity (Gao et al., 2010). Also, Binhayeeding et al. (2020) stated that the resin’s modification process also inhibits enzyme adsorption. With adsorption-crosslinking, the chitosan-coated support showed an admirable result than uncoated support (8.12% higher). The phenomenon occurred because of glutaraldehyde bonding to amino-functional groups of chitosan to reduce steric hindrance between enzyme–support surface interactions.
Figure 4B showed that the enzyme loading for MP-64 immobilized by adsorption only was improved to 60.5% higher using chitosan-coated resin (p < 0.05), surprisingly. However, for the adsorption-crosslinking, the increase in enzyme loading only reached 9.34%. The results indicated that the addition of amino-functional groups to the resin surface improved protein loading to MP-64. The adsorption without cross-linking give a significant result compare to the adsorption with cross-linking because MP-64 presented the tertiary/quaternary amino-functional group in its surface and bonded the chitosan to form anionic binding with the result that enhancing the enzyme loading. The interactions between lipases and resin surfaces containing amino-functional groups increased the amount of lipase adsorbed (Ye et al., 2013). Other studies on the addition of amino-functional groups have been carried out using aminopropyl graft on silica, increasing the lipase-loading proportion (Bai et al., 2012).
The optimum lipase-loading values for XAD-7HP and MP-64 resins were, respectively, obtained from chitosan coated with the adsorption-crosslinking process. The enzyme loading of chitosan-coated MP-64 increased up to 51.27% after the addition of glutaraldehyde (p < 0.05). Meanwhile, for chitosan-coated XAD-7HP resin, the lipase loading increased twofold with the same treatment. This phenomenon was predicted to occur due to lipases, which tended to form a monolayer rather than aggregates. Generally, adsorption-crosslinking provided a better result than adsorption only, which applied to both. The addition of amino groups from chitosan aided the bond between enzymes and support (Gao et al., 2010). The role of glutaraldehyde as a cross-linking agent is fulfilled when there are at least two reactive groups on the protein surface with sufficient spacing (Barbosa et al., 2014). Other than that, Vescovi et al. (2016) reported that glutaraldehyde and amino addition were able to enhance the protein immobilization up to 30.65%, and then only hydrophobic adsorption. In another study, enzyme loading with the addition of amino-functional groups was carried out by Bai et al. (2012) using the support of mesoporous silica nanotubes grafted with aminopropyl to immobilize the lipase of Candida sp. Research that has been conducted by Cheng et al. (2017) showed that the best enzyme activity and protein loading capacity were in the use of amino resins in the immobilization of AGE and NAL. Similar results were reported by Xu et al. (2011) that showed the attachment of 98% APTES as an amino-functional group to mesoporous SBA-15 in the covalent-binding immobilization increased the amount of lipase loading by 42.01% compared to lipase immobilization in SBA-15 without the addition of APTES. It occurred due to the SBA-15 surface undergoing a hydrophilic–hydrophobic phase transition after attachment of the amino groups from APTES. Based on works of literature and our results, it shows that the addition of an amino-functional group can increase the lipase–support attachment and lipase performance.
Furthermore, the enzyme loading in chitosan-coated MP-64 resin (84.94%) was higher than that in chitosan-coated XAD-7HP resin (76.54%). The SEM supports the difference in the enzyme loading values for these two support results in Figures 2B,D. The surface of MP-64 resin after adsorption-crosslinking (Figure 2D) appeared to be coarser, indicating that more enzymes were adsorbed on MP-64 resin than on XAD-7HP resin (Figure 2B). The diffusional limitations affected the enzymatic reaction after immobilization. Reducing the biocatalyst size is one way to prevent this (using low-particle diameter of support; Boudrant et al., 2020). MP resin was previously used as a support for lipase immobilization to produce medium-chain, oleic, and medium-chain fatty acid structure lipid. The result showed that the incorporation of medium-chain fatty acid in structured lipid over 20% needed a long reaction time of at least 40 h. This difference in lipase catalytic activity was caused by enzyme immobilization. The enzyme loading was also a factor in increasing the catalytic reaction time (Akil et al., 2020).
Lipase activity test was carried out on XAD-7HP and MP-64 resins immobilized by the adsorption-crosslinking process at 36oC and pH 6.5, as shown in Figure 5. Based on Figure 5, the lipase activity of chitosan-free XAD-7HP and MP-64 resins was 25.44 U/g support and 26.67 U/g support, respectively. After coating with chitosan, immobilized lipase activity values were increased up to 6.11 and 15.60%, respectively, for XAD-7HP and MP-64 resins.
In this research, the enzyme was immobilized by the addition of glutaraldehyde in the reaction. Glutaraldehyde can modify amino groups after first incubation. However, this modification will not be affected by the ionization of the amino group, but it will be affected by the enzyme activity. An addition of this cross-linker agent produced a hydrophobization on the enzyme (Weber et al., 2019).
The increase of the lipase activity value could be due to some of the possibilities during the lipolysis using chitosan and glutaraldehyde as a bridge unit and cross-linking agent, including intramolecular modification, interprotein cross-linking, and enzyme–support reactions (Gao et al., 2010; Barbosa et al., 2013). Another study from Barbosa et al. (2012) also showed that the immobilization of lipases using amino-glutaraldehyde allowed a high versatility and giving the best activity and enantioselectivity at pH 5. Therefore, the addition of amino-functional groups to the resin as the support through chitosan coating has a positive effect on the immobilized lipase activity. Moreover, the lipase activity of modified MP-64 was the greatest (29.41 U/g support) because the amino groups from chitosan activated the lipase catalytic center after lipase attachment to MP-64 resins coated with chitosan. The particle size of resins also facilitated the activation of the support interface to open the protective cover of the active site and made the soluble substrate (olive oil) easily access the lipase (Li et al., 2010). Similar results from the previous study reached 82.89% higher than no additional amino-functional group (APTES) to hydrophobic mesoporous SBA-15 in porcine pancreatic lipase (PPL) immobilization (Xu et al., 2011). PPL activity increases since the catalytic center is activated by the interaction between the hydrophobic lipase domain and the modified matrix surface hydrophobic group (Xu et al., 2011).
The findings obtained were similar to those in the study by Liu et al. (2012) that conducted lipase immobilization from Thermomyces lanuginosus on macroporous resin, with and without chitosan coating. The lipase activity immobilized on the raw macroporous resin (without chitosan coating) was 58.4 U/g support. Meanwhile, lipase activity immobilized on the chitosan-coated resin ranged from 73.8 to 87.3 U/g support. The study revealed that macroporous resin coated with chitosan produce higher lipase activity than raw macroporous resin. The lipase activity increased around 26–49%, depending on chitosan’s molecular weight (Liu et al., 2012). However, the lipase activity of this study was relatively low compared with that in the study by Liu et al. (2012). It is due to the effect of chitosan’s molecular weight, glutaraldehyde concentration, and type of resins used.
Based on Figure 5, the lipase activity for XAD-7HP and MP-64 resin was almost comparable. However, the loading values for the two resins had large differences, as shown in Figure 4. According to Salis et al. (2009), high enzyme loading indicates a high affinity between support and enzymes. Strong interaction on the enzyme surface can distort the enzyme’s tertiary structure with partial deactivation, thereby decreasing the activity. The possibility of enzyme conformation caused by the immobilization or substrate diffuses into an active site capable of decreasing the enzyme activity despite large loading values (Cheng et al., 2017). The use of supports with smaller pore sizes may increase the diffusion limitations, notably, if the substrates have giant molecules (Dos Santos et al., 2015). For industrial biocatalyst, it is necessary to make the highly loaded enzymes in supports to regulate repeated use over a long operational period. Another study also proved that immobilized lipase activity decreases with an increase in enzyme loading (Gao et al., 2010). It is presumably because more lipase diffuses into the pore support, thereby partially aggregating and not distributing as a layer on the channel’s inner surface. It tends to prevent the contact between substrate and lipase in the lower layer of support, thus decreasing the enzyme activity. This result is in line with Wu et al. (2009), who immobilized the enzyme using magnetic support and labeled it as Fe3O4-chitosan nanoparticles. The type of chemical bond between support and enzymes also affected flexibility and limited the dynamics needed by catalytic activity (Salis et al., 2009). This result can be proven since MP-64 resin is an anion resin with an enormous particle size compared with nonionic XAD-7HP resin, as shown in Table 1.
The biodiesel synthesis test was only carried out on the best resins with the highest lipase activity capability. In this study, immobilized Aspergillus niger lipase in MP-64 resin was used to analyze the results of interesterification biodiesel synthesis. The advantages of interesterification for biodiesel synthesis are the formation of a by-product in the form of triacylglycerol (triacetin) and improving biodiesel properties as an additive to fuel (Hermansyah et al., 2011). This study’s interesterification reaction was carried out at a temperature of 40°C with a molar ratio of methyl acetate and oil (12:1) (Hermansyah et al., 2011). The results of the reusability test through the biodiesel synthesis reaction are shown in Figure 6.
For industrial areas, the reusability of the immobilized enzyme is a crucial thing. Strong interaction between enzyme and its support environment and washing steps gives the best result of reusability (resulting in low-yield differences per cycle) (Lage et al., 2016). Based on Figure 6, that the biodiesel yield in cycles I, II, III, and IV was 69, 54, 51, and 48%, respectively. The decrease in biodiesel yield from cycle I to II (21%) was significant compared to that in biodiesel yield from cycles II to III (5%) and III to IV (6%). It is probably due to the inhibition of substrate diffusion (cooking oil) caused by lipase aggregates in the resin pores. Therefore, the lipase that worked only outside the chitosan-coated MP-64 resin decreased the catalytic activity because it was used in the first cycle. This result is in line with the study carried out by Gao et al. (2010), which stated that the lipase aggregate outside the support experience leaching at the end of cycle II and opened the porosity of the macropore support; therefore, the decrease in biodiesel yield in the next cycle was not as significant as that in biodiesel yield in the previous cycle. In general, Figure 6 shows that repeated use of the Aspergillus niger enzyme for biodiesel synthesis results produced lower biodiesel yield due to an increase in the number of lipase molecules released from the support and a change in the enzyme conformation (Li et al., 2010).
A previous work was found with ten cycles of Aspergillus niger lipase reusability in the reaction condition of soybean oil and methanol for 12 h reaction time. The first-run biodiesel yield reached 93%. A fantastic biodiesel yield value is caused by a short reaction time (56 h). In our work, we monitored the reaction from 6 to 96 h. The short-term reaction was unable to break the alkyl chain in the biodiesel synthesis (Hu et al., 2018).
The reaction time is one of the factors that affect the biodiesel yield, as evidenced by the research carried out by Orçaire et al. (2006) and Usai et al. (2010) with 56 and 86% percentage values and reaction time of 6 and 96 h. However, the biodiesel yield in this study was 69%, with a reaction time of 50 h. This research’s interesterification reaction uses 4% (w/w) lipase, which is reacted with palm cooking oil for biodiesel synthesis. The same number of enzymes obtained a biodiesel yield of 63% using castor oil as a substrate and immobilized Burkhodilderia cepacia enzymes in attapulgite (You et al., 2013). However, this study’s biodiesel synthesis activity produces a higher result. This research’s reusability test was also carried out through the biodiesel synthesis reaction up to the fourth cycle to produce a 48% yield rate. The results showed a decrease in biodiesel yield by 29.39% against immobilized enzymes when first used. This value was more excellent in the same cycle than in other studies using the immobilized Aspergillus niger on Fe3O4.SiO2 support with a yield of 40% (Thangaraj et al., 2019). These data indicated that Aspergillus niger lipase immobilized in Lewatit M-64–layered chitosan could be used as a biocatalyst for biodiesel synthesis. However, several strategies (stability improvement, prevention of diffusion limitation, and leaching process) are needed to increase the immobilized enzyme’s usefulness, not to experience a significant decrease in yield in each cycle.
Modification of the support using chitosan-coated resins has been shown to increase the amino-functional groups on the surface. The results also showed that the lipase loading and lipase activity after the addition of functional groups were higher than those without chitosan coating. Through chitosan-coated MP-64, it has better lipase loading and activity values than Amberlite XAD-7HP. A potential natural material that is easily obtained at an affordable price, such as chitosan, is proven to be used to modify the support that will be used as Aspergillus niger lipase support. Immobilized Aspergillus niger in chitosan-coated MP-64 was applied through the interesterification reaction of biodiesel synthesis.
Furthermore, the biodiesel synthesis activity until the fourth cycle can still be maintained up to 70% of the initial cycle. The use of lipase, glutaraldehyde, chitosan, and its combination at a specific concentration range is needed to evaluate the current immobilization results so that the best immobilization modification results are obtained. Also, the stability of lipase on modified resin needs to be studied further to produce immobilized enzymes that are more stable at their optimum conditions.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
AN wrote and revised the manuscript under the guidance of MS, AW, and HH. MP and DP helped to draw the figures and write the manuscript. All the authors contributed to the article and accepted the version submitted.
This research was funded by Universitas Indonesia through PUTI Doktor Grant (number NKB-668/UN2.RST/HKP.05.00/2020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge the research and publication support provided by the Ministry of Education and Culture Republic of Indonesia and Universitas Indonesia through PUTI Doktor Grant (number NKB-668/UN2.RST/HKP.05.00/2020).
Ahn, H. H., and Lee, M. S. (2019). Recoveries of Ru(III) and Co(II) by solvent extraction and ion exchange from tungsten carbide-cobalt scrap through a HCl leaching solution. Metals 9, 858. doi:10.3390/met9080858
Akil, E., Pereira, A. S., El-Bacha, T., Amaral, P. F. F., and Torres, A. G. (2020). Efficient production of bioactive structured lipids by fast acidolysis catalyzed by Yarrowia lipolytica lipase,free and immobilized in chitosan-alginate beads, in solvent-free medium. Int. J. Biol. Macromol. 163, 910–918. doi:10.1016/j.ijbiomac.2020.06.282
Alamsyah, G., Albels, V. A., Sahlan, M., and Hermansyah, H. (2017). Effect of chitosan’s amino group in adsorption-crosslinking immobilization of lipase enzyme on resin to catalyze biodiesel synthesis. Energy Procedia 136, 47–52. doi:10.1016/j.egypro.2017.10.278
Bai, W., Yang, Y.-J., Tao, X., Chen, J.-F., and Tan, T. (2012). Immobilization of lipase on aminopropyl-grafted mesoporous silica nanotubes for the resolution of (R, S)-1-phenylethanol. J. Mol. Catal. B: Enzym. 76, 82–88. doi:10.1016/j.molcatb.2011.11.005
Barbosa, O., Ortiz, C., Berenguer-Murcia, Á., Torres, R., Rodrigues, R. C., and Fernandez-Lafuente, R. (2014). Glutaraldehyde in bio-catalysts design: a useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 4, 1583–1600. doi:10.1039/C3RA45991H
Barbosa, O., Ortiz, C., Berenguer-Murcia, Á., Torres, R., Rodrigues, R. C., and Fernandez-Lafuente, R. (2015). Strategies for the one-step immobilization-purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 33, 435–456. doi:10.1016/j.biotechadv.2015.03.006
Barbosa, O., Torres, R., Ortiz, C., Berenguer-Murcia, Á., Rodrigues, R. C., and Fernandez-Lafuente, R. (2013). Heterofunctional supports in enzyme immobilization: from traditional immobilization protocols to opportunities in tuning enzyme properties. Biomacromolecules 14, 2433–2462. doi:10.1021/bm400762h
Barbosa, O., Torres, R., Ortiz, C., and Fernandez-Lafuente, R. (2012). Versatility of glutaraldehyde to immobilize lipases: effect of the immobilization protocol on the properties of lipase B from Candida antarctica. Process Biochem. 47, 1220–1227. doi:10.1016/j.procbio.2012.04.019
Binhayeeding, N., Yunu, T., Pichid, N., Klomklao, S., and Sangkharak, K. (2020). Immobilisation of Candida rugosa lipase on polyhydroxybutyrate via a combination of adsorption and crosslinking agents to enhance acylglycerol production. Process Biochem. 95, 174–185. doi:10.1016/j.procbio.2020.02.007
Boudrant, J., Woodley, J. M., and Fernandez-Lafuente, R. (2020). Parameters necessary to define an immobilized enzyme preparation. Process Biochem. 90, 66–80. doi:10.1016/j.procbio.2019.11.026
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72 (1-2), 248–254. doi:10.1016/0003-2697(76)90527-3
Broun, G. B. (1976). Chemically aggregated enzymes. Methods Enzymol. 44, 263–280. doi:10.1016/s0076-6879(76)44022-3
Cantone, S., Ferrario, V., Corici, L., Ebert, C., Fattor, D., Spizzo, P., et al. (2013). Efficient immobilisation of industrial biocatalysts: criteria and constraints for the selection of organic polymeric carriers and immobilisation methods. Chem. Soc. Rev. 42, 6262–6276. doi:10.1039/c3cs35464d
Cheng, J., Zhuang, W., Tang, C., Chen, Y., Wu, J., Guo, T., et al. (2017). Efficient immobilization of AGE and NAL enzymes onto functional amino resin as recyclable and high-performance biocatalyst. Bioprocess Biosyst. Eng. 40, 331–340. doi:10.1007/s00449-016-1700-z
Cipolatti, E. P., Silva, M. J. A., Klein, M., Feddern, V., Feltes, M. M. C., Oliveira, J. V., et al. (2014). Current status and trends in enzymatic nanoimmobilization. J. Mol. Catal. B: Enzym. 99, 56–67. doi:10.1016/j.molcatb.2013.10.019
Damaso, M. C. T., Salum, T. F. C., Terzi, S., and Couri, S. (2013). Assay methods for lipase activity. Brasilia, Brazil: Bentham Books, 161–194.
Dos Santos, J. C. S., Bonazza, H. L., de Matos, L. J. B. L., Carneiro, E. A., Barbosa, O., Fernandez-Lafuente, R., et al. (2017). Immobilization of CALB on activated chitosan: application to enzymatic synthesis in supercritical and near-critical carbon dioxide. Biotechnol. Rep. 14, 16–26. doi:10.1016/j.btre.2017.02.003
Dos Santos, J. C. S., Rueda, N., Barbosa, O., Millán-Linares, M. C., Pedroche, J., Yuste, M. M., et al. (2015). Bovine trypsin immobilization on agarose activated with divinylsulfone: improved activity and stability via multipoint covalent attachment. J. Mol. Catal. B: Enzym. 117, 38–44. doi:10.1016/j.molcatb.2015.04.008
Elnashar, M. M. J. (2010). Immobilized molecules using biomaterials and nanobiotechnology. J. Biomater. Nanobiotechnol. 1, 61–77. doi:10.4236/jbnb.2010.11008
Facin, B. R., Melchiors, M. S., Valério, A., Oliveira, J. V., and Oliveira, D. (2019). Driving immobilized lipases as biocatalysts: 10 years state of the art and future prospects. Ind. Eng. Chem. Res. 58 (14), 5358–5378. doi:10.1021/acs.iecr.9b00448
Galvão, W. S., Pinheiro, B. B., Golçalves, L. R. B., de Mattos, M. C., Fonseca, T. S., Regis, T., et al. (2018). Novel nanohybrid biocatalyst: application in the kinetic resolution of secondary alcohols. J. Mater. Sci. 53, 14121–14137. doi:10.1007/s10853-018-2641-5
Gao, S., Wang, Y., Diao, X., Luo, G., and Dai, Y. (2010). Effect of pore diameter and crosslinking method on the immobilization efficiency of Candida rugosa lipase in SBA-15. Bioresour. Technol. 101 (11), 3830–3837. doi:10.1016/j.biortech.2010.01.023
Garcia-Gallan, C., Berenguer-Mliuurcia, A., Fernandez-Lafuente, R., and Rodrigues, R. C. (2011). Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 353, 2885–2904. doi:10.1002/adsc.201100534
Hermansyah, H., Arbianti, R., Prameshwari, D. A., and Environment, (2011). Non-alcohol route of biodiesel synthesis from used cooking oil using immobilized biocatalyst in packed bed reactor. J. Sustain. Dev. Energy Environ. 2, 1–5.
Hu, Y., Dai, L., Liu, D., and Du, W. (2018). Rationally designing hydrophobic UiO-66 support forthe enhanced enzymatic per formance of immobilized lipase. Green Chem. 20, 4500–4506. doi:10.1039/C8GC01284A
Jesionowski, T., Zdarta, J., and Krajewska, B. J. A. (2014). Enzyme immobilization by adsorption: a review. Adsorption 20 (5-6), 801–821. doi:10.1007/s10450-014-9623-y
Jin, L., and Bai, R. J. L. (2002). Mechanisms of lead adsorption on chitosan/PVA hydrogel beads. Langmuir 18, 9765–9770. doi:10.1021/la025917l
Lage, F. A. P., Bassi, J. J., Corradini, M. C. C., Todero, L. M., Luiz, J. H. H., and Mendes, A. A. (2016). Preparation of a biocatalyst via physical adsorption of lipase from Thermomyces lanuginosus on hydrophobic support to catalyze biolubricant synthesis by esterification reaction in a solvent-free system. Enzyme Microb. Technol. 84, 56–67. doi:10.1016/j.enzmictec.2015.12.007
Li, Y., Gao, F., Wei, W., Qu, J. B., Ma, G. H., and Zhou, W. Q. J. (2010). Pore size of macroporous polystyrene microspheres affects lipase immobilization. J. Mol. Catal. B: Enzym. 66 (1-2), 182–189. doi:10.1016/j.molcatb.2010.05.007
Liu, K., Zhao, G., He, B., Chen, L., and Huang, L. (2012). Immobilization of pectinase and lipase on macroporous resin coated with chitosan for treatment of whitewater from papermaking. Bioresour. Technol. 123, 616–619. doi:10.1016/j.biortech.2012.07.074
Liu, T., Liu, Y., Wang, X., Li, Q., Wang, J., and Yan, Y. (2011). Improving catalytic performance of Burkholderia cepacia lipase immobilized on macroporous resin NKA. J. Mol. Catal. B: Enzym. 71, 45–50. doi:10.1016/j.molcatb.2011.03.007
Manoel, E. A., Ribeiro, M. F. P., dos Santos, J. C. S., Coelho, M. A. Z., Simas, A. B. C., Fernandez-Lafuente, R., et al. (2015). Accurel MP 1000 as a support for the immobilization of lipase from Burkholderia cepacia: application to the kinetic resolution of myo-inositol derivatives. Process Biochem. 50, 1557–1564. doi:10.1016/j.procbio.2015.06.023
Meher, L. C., Vidyasagar, D., and Naik, S. N. (2006). Technical aspects of biodiesel production by transesterification–a review. Renew. Sustain. Energy Rev. 10, 248–268. doi:10.1016/j.rser.2004.09.002
Mendes, A. A., Oliveira, P. C., Vélez, A. M., Giordano, R. C., and de Castro, H. F. (2012). Evaluation of immobilized lipase on poly-hydroxybutyrate beads to catalyze biodiesel synthesis. Int. J. Biol. Macromol. 50, 503–511. doi:10.1016/j.ijbiomac.2012.01.020
Minovska, V., Winkelhausen, E., and Kuzmanova, S. (2005). Lipase immobilized by different techniques on various support materials applied in oil hydrolysis. J. Serbian Chem. Soc. 70, 609–624. doi:10.2298/JSC0504609M
Mishra, V. K., and Goswami, R. (2017). A review of production, properties and advantages of biodiesel. Biofuels 9, 273–289. doi:10.1080/17597269.2017.1336350
Mohamad, N. R., Marzuki, N. H. C., Buang, N. A., Huyop, F., and Wahab, R. A. (2015). An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 29, 205–220. doi:10.1080/13102818.2015.1008192
Montero, S., Blanco, A., Virto, M. D., Landeta, L. C., Agud, I., Solozabal, R., et al. (1993). Immobilization of Candida rugosa lipase and some properties of the immobilized enzyme. 15, 239–247. doi:10.1016/0141-0229(93)90144-Q
Moreira, K. S., de Oliveira, A. L. B., Junior, L. S. M., Monteiro, R. R. C., da Rocha, T. N., Menezes, T. L., et al. (2020). Lipase from Rhizomuchor miehei immobilized on magnetic nanoparticles: performance in fatty acid ethyl ester (FAEE) optimized production by the Taguchi method. Front. Bioeng. Biotechnol. 8, 693. doi:10.3389/fbioe.2020.00693
Nicoletti, G., Cipolatti, E. P., Valério, A., Carbonera, N. G., Soares, N. S., Theilacker, E., et al. (2015). Evaluation of different methods for immobilization of Candida antarctica lipase B (CalB lipase) in polyurethane foam and its application in the production of geranyl propionate. Bioprocess Biosyst. Eng. 38, 1739–1748. doi:10.1007/s00449-015-1415-6
Orçaire, O., Buisson, P., and Pierre, A. C. (2006). Application of silica aerogel encapsulated lipases in the synthesis of biodiesel by transesterification reactions. J. Mol. Catal. B: Enzym. 42 (3–4), 106–113. doi:10.1016/j.molcatb.2006.08.002
Osuna, Y., Gregorio-Jauregui, K. M., Gaona-Lozano, J. G., de la Garza-Rodríguez, I. M., Ilyna, A., Barriga-Castro, E. D., et al. (2012). Chitosan-coated magnetic nanoparticles with low chitosan content prepared in one-step. J. Nanomater. 2012, 327562. doi:10.1155/2012/327562
Osuna, Y., Sandoval, J., Saade, H., López, R. G., Martinez, J. L., Colunga, E. M., et al. (2015). Immobilization of Aspergillus niger lipase on chitosan-coated magnetic nanoparticles using two covalent-binding methods. Bioprocess Biosyst. Eng. 38, 1437–1445. doi:10.1007/s00449-015-1385-8
Pal, P., and Banat, F. (2014). Comparison of heavy metal ions removal from industrial lean amine solvent using ion exchange resins and sand coated with chitosan. J. Nat. Gas Sci. Eng. 18, 227–236. doi:10.1016/j.jngse.2014.02.015
Pinheiro, B. B., Rios, N. S., Rodríguez Aguado, E., Fernandez-Lafuente, R., Freire, T. M., Fechine, P. B. A., et al. (2019). Chitosan activated with divinyl sulfone: a new heterofunctional support for enzyme immobilization. Application in the immobilization of lipase B from Candida antarctica. Int. J. Biol. Macromol. 130, 798–809. doi:10.1016/j.ijbiomac.2019.02.145
Reis, C. L. B., de Sousa, E. Y. A., Serpa, J. F., Oliveira, R. C., and dos Santos, J. C. S. (2019). Design of immobilized enzyme biocatalyst: drawbacks and opportunities. Quim. Nova 42, 768–783. doi:10.21577/0100-4042.20170381
Rizwanul Fattah, I., Ong, H., Mahlia, T., Mofijur, M., Silitonga, A., Rahman, S., et al. (2020). State of the art of catalysts for biodiesel production. Front. Energy Res. 8, 101. doi:10.3389/fenrg.2020.00101
Salis, A., Bhattacharyya, M. S., Monduzzi, M., and Solinas, V. J. (2009). Role of the support surface on the loading and the activity of Pseudomonas fluorescens lipase used for biodiesel synthesis. J. Mol. Catal. B: Enzym. 57 (1–4), 262–269. doi:10.1016/j.molcatb.2008.09.015
Salis, A., Meloni, D., Ligas, S., Casula, M. F., Monduzzi, M., Solinas, V., et al. (2005). Physical and chemical adsorption of Mucor javanicus lipase on SBA-15 mesoporous silica. synthesis, structural characterization, and activity performance. Langmuir 21, 5511–5516. doi:10.1021/la047225y
Sheldon, R. J., and van Pelt, S. (2013). Enzyme immobilisation in biocatalysis: why, what and how. Chem. Soc. Rev. 42, 6223–6235. doi:10.1039/C3CS60075K
Silva, J. A., Macedo, G. P., Rodrigues, D. S., Giordano, R. L. C., and Gonçalves, L. R. B. (2012). Immobilization of Candida antarctica lipase B by covalent attachment on chitosan-based hydrogels using different support activation strategies. Biochem. Eng. J. 60, 16–24. doi:10.1016/j.bej.2011.09.011
Talbert, J. N., and Goddard, J. M. (2012). Enzymes on material surfaces. Colloids Surf. B Biointerfaces 93, 8–19. doi:10.1016/j.colsurfb.2012.01.003
Thangaraj, B., Jia, Z., Dai, L., Liu, D., and Du, W. (2019). Effect of silica coating on Fe3O4 magnetic nanoparticles for lipase immobilization and their application for biodiesel production. Arab. J. Chem. 12, 4694–4706. doi:10.1016/j.arabjc.2016.09.004
Tischer, W., and Wedekind, F. (1999). “Immobilized enzymes: methods and applications,” in Biocatalysis-from discovery to application. Editor W. D. Fessner (Berlin, Germany: Springer), 95–126. doi:10.1007/3-540-68116-7_4
Urrutia, P., Arrieta, R., Alvarez, L., Cardenas, C., Mesa, M., and Wilson, L. (2017). Immobilization of lipases in hydrophobic chitosan for selective hydrolysis of fish oil: the impact of support functionalization on lipase activity, selectivity and stability. Int. J. Biol. Macromol. 108, 674–686. doi:10.1016/j.ijbiomac.2017.12.062
Usai, E., Gualdi, E., Solinas, V., and Battistel, E. (2010). Simultaneous enzymatic synthesis of FAME and triacetyl glycerol from triglycerides and methyl acetate. Bioresour. Technol. 101 (20), 7707–7712. doi:10.1016/j.biortech.2010.05.044
Verma, M. L., Naebe, M., Barrow, C. J., and Puri, M. (2013). Enzyme immobilisation on amino-functionalised multi-walled carbon nanotubes: structural and biocatalytic characterisation. PLoS One 8, e73642. doi:10.1371/journal.pone.0073642
Vescovi, V., Kopp, W., Guisán, J. M., Giordano, R. L. C., Mendes, A. A., and Tardioli, P. W. (2016). Improved catalytic properties of Candida antarctica lipase B multi-attached on tailor-made hydrophobic silica containing octyl and multifunctional amino- glutaraldehyde spacer arms. Process Biochem. 51, 2055–2066. doi:10.1016/j.procbio.2016.09.016
Wang, W., Jiang, Y., Zhou, L., and Gao, J. (2011). Comparison of the properties of lipase immobilized onto mesoporous resins by different methods. Appl. Biochem. Biotechnol. 164, 561–572. doi:10.1007/s12010-010-9157-z
Weber, D., da Graça Nascimento, M., and Parize, A. L. (2019). Immobilization of Burkholderia cepacia lipase on crosslinked chitosan-based support for the synthesis of geranyl acetate. Biocatal. Agric. Biotechnol. 19, 101133. doi:10.1016/j.bcab.2019.101133
Wu, Y., Wang, Y., Luo, G., and Dai, Y. (2009). In situ preparation of magnetic Fe3O4-chitosan nanoparticles for lipase immobilization by crosslinking and oxidation in aqueous solution. Bioresour. Technol. 100, 3459–3464. doi:10.1016/j.biortech.2009.02.018
Xu, Y.-q., Zhou, G.-W., Wu, C. -c., Li, T.-d., and Song, H.-b. (2011). Improving adsorption and activation of the lipase immobilized in amino-functionalized ordered mesoporous SBA-15. Solid State Sci. 13, 867–874. doi:10.1016/j.solidstatesciences.2011.03.003
Yang, J., Ma, X., Zhang, Z., Chen, B., Li, S., and Wang, G. (2010). Lipase immobilized by modification-coupled and adsorption-crosslinking methods: a comparative study. Biotechnol. Adv. 28, 644–650. doi:10.1016/j.biotechadv.2010.05.014
Ye, P., Han, Z.-P., Xu, Y.-J., Hu, P.-C., and Tong, J.-J. (2013). Effect of support surface chemistry on lipase adsorption and activity. J. Mol. Catal. B: Enzym. 94, 69–76. doi:10.1016/j.molcatb.2013.04.015
Keywords: adsorption-crosslinking, amino group, Aspergillus niger, biodiesel, chitosan, lipase, macroporous resin
Citation: Nuraliyah A, Perdani MS, Putri DN, Sahlan M, Wijanarko A and Hermansyah H (2021) Effect of Additional Amino Group to Improve the Performance of Immobilized Lipase From Aspergillus niger by Adsorption-Crosslinking Method. Front. Energy Res. 9:616945. doi: 10.3389/fenrg.2021.616945
Received: 13 October 2020; Accepted: 10 February 2021;
Published: 22 April 2021.
Edited by:
Nidia S. Caetano, Instituto Superior de Engenharia do Porto, PortugalReviewed by:
José Cleiton Sousa Dos Santos, University of International Integration of Afro-Brazilian Lusophony, BrazilCopyright © 2021 Nuraliyah, Perdani, Putri, Sahlan, Wijanarko and Hermansyah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heri Hermansyah, aGVyaUBjaGUudWkuYWMuaWQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.