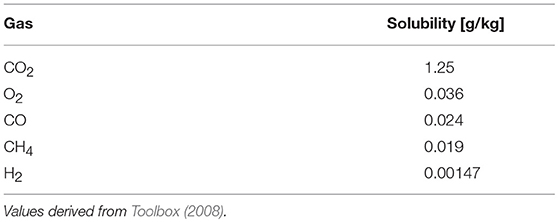- 1Universities Space Research Association at NASA Ames Research Center, Mountain View, CA, United States
- 2Department of Civil & Environmental Engineering, Stanford University, Stanford, CA, United States
The potent greenhouse gas methane presents a widely accessible resource, being the primary component in natural gas as well as in bio-gas from anaerobic digesters. Given its relatively low heating-value and several issues concerning its storage and transportation, methane upgrading to liquid fuels is of particular interest. Microbial methane conversion/utilization and upgrading is gaining increasing interest due to its high conversion efficiency. In this study we computationally compare aerobic and anaerobic microbial pathways for CH4-oxidation and discuss theoretically achievable biomass yields as well as the possibility for building synthetic biological production platforms for liquid fuels. Specifically, the presented in-silico work investigates the potential of microbial methane upgrading in a metabolic network analysis by means of elementary flux modes. Aerobic fixation of methane via conversion of methane to methanol by a methane monooxygenase (MMO) and different subsequent formaldehyde assimilation pathways (Serine-cycle, RuMP, XMP/DHA-pathway) is compared with anaerobic pathways for oxidation of methane (AOM) by means of reverse-methanogenesis or via a presumed glycyl-radical enzyme, which uses fumarate for activation of methane. The different pathways for aerobic and anaerobic methane oxidation are compared in different central carbon-metabolism envelopes in order to identify highest achievable carbon yields. The capability of efficient CO2 fixation, as well as energy preservation in form of reducing equivalents is identified as crucial to enable high yields, which ranged from 22 100%. The potential of the different microbes to grow on these gas streams is assessed by means of the maximum achievable biomass yield and the CO2/CH4 uptake ratio. CO2 co-utilization, by transferring reducing power between the two co-substrates, is highest, when combining reverse-methanogenesis with the Wood-Ljungdahl pathway, effectively replacing the need for H2 with CH4. Further, the possibility to upgrade methane into liquid (drop-in) bio-fuels is investigated. Established routes to methanol, ethanol, C4-alcoholes and farnesene are evaluated in the most promising substrate-pathway/organism combinations. Stoichiometric, thermodynamic and kinetic limitations are assessed and recommendations regarding potential industrial feasibility are given. The results presented here should guide future research efforts in search for feasible ways of (co)utilizing novel carbon substrates for sustainable production of fuels and chemicals.
Introduction
Two of the greatest challenges of today's society are represented by the development of sustainable replacement processes to produce chemicals and fuels from non-fossil resources while simultaneously reducing greenhouse gas emissions. Microbial gas fermentation offers a solution to both issues via organisms with the ability to utilize gaseous C1-compounds, such as CH4, CO2 and CO as feedstock for production of chemicals. Here, we propose, analyse and discuss different strategies for microbial methane upgrading, a challenging but auspicious approach.
Methane, the main component of natural gas, has several shortcomings as an energy carrier. It has low energy content (MJ/L) and economical storage requires liquefaction or at least compression (which is expensive, because energy intensive). The same applies to biogas, which is further often contaminated with large amounts of carbon dioxide (up to 50%), making it an even less efficient energy carrier (Miltner et al., 2017). Methane is also a very potent greenhouse gas; therefore, it is often flared when logistics are (economically) infeasible (Haynes and Gonzalez, 2014). An estimated amount of 5 quadrillion BTU, around 5% of the global natural gas production, is flared or vented annually (Fei et al., 2014). This “lost” methane does not only contribute to greenhouse gas emissions but also represents a considerable market value of around $13 billion per annum, which alone could satisfy 27% of the US electricity market if made accessible (Fei et al., 2014). Therefore, the industrial interest in methane upgrading is high and different approaches for its conversion into better energy carriers have been developed. The chemical transformation of methane into fuels is mainly realized in the Fischer-Tropsch process via activation with syngas. However, this process achieves maximum carbon efficiencies of <50% and is further limited by its intensive energy requirements for the generation and conversion of syngas as well as hydro-processing steps and has proven economically viable only at very large scale (Steynberg, 2004). Seeking more efficient and sustainable alternatives, biological conversion of methane into more readily transportable and valuable fuels via biocatalysts at ambient temperatures and pressures, termed “Bio-GTL,” receives increasing interest (biological gas-to-liquid).
The most extensively studied microorganisms for methane utilization are methanotrophic, aerobic α- and γ-proteobacteria, which are known to naturally metabolize methane as their only carbon and energy source. In these organisms the metabolism of methane starts with oxidation by O2 to methanol, which is assimilated after further oxidation to formaldehyde via different pathways, depending on the organism (serine-cycle in α-proteobacteria/type II methanotrophs; ribulose-monophosphate pathway in γ-proteobacteria/type I methanotrophs). Although extensively studied, most methanotrophs are genetically not very tractable, so that to date their industrial use remains limited to the production biomass (single-cell protein), which is used as a feedstock for livestock in agriculture (Kalyuzhnaya et al., 2015). The production of more valuable target compounds, such as methanol, formaldehyde, organic acids, ectoine, lipids and vitamin B12 has been demonstrated in natural and synthetically engineered methanotrophs (Strong et al., 2015). However, the industrial use of microbial methane oxidation via aerobic pathways has several major limitations. Genetic engineering approaches in natural hosts are challenging and the expression of the key enzyme, methane- monooxygenase (MMO) in heterologous hosts has had only limited success to date (Hwang et al., 2018). Further, the requirement of O2 as electron donor for methane oxidation has certain safety concerns at industrial scale due to explosive gas-mixtures. Additionally, the aerobic pathway via MMO has a limited maximum achievable carbon yield (67%) due to every third carbon being “lost” as CO2 in a decarboxylation reaction of the pathway (Conrado and Gonzalez, 2014).
Opposing to the aerobic pathways, a second, less-studied option for microbial oxidation of methane is based on anaerobic metabolism. Anaerobic oxidation of methane (AOM) requires a suitable electron acceptor, such as iron(III), nitrate or sulfate and has been observed as natural occurring phenomenon in several environments often including syntrophic consortia (Boetius et al., 2000; Valentine and Reeburgh, 2000). Majorly hindered by the unavailability of pure cultures, AOM has been significantly less studied compared to aerobic methane oxidation and as a result the exact pathways of AOM still involve several knowledge gaps (Scheller et al., 2017; Hwang et al., 2018). However, there is evidence that anaerobic methane oxidation coupled to the reduction of sulfate, iron(III), manganese, or nitrate is found in anaerobic methanotrophic archaea (ANME) and proceeds at least in part via reversed-methanogenesis involving the nickel enzyme methyl-coenzyme M reductase (Mcr) for methane activation (Thauer and Shima, 2008; Beal et al., 2009; Shima et al., 2011; Haroon et al., 2013; Ettwig et al., 2016; Scheller et al., 2017). At standard conditions this presents an endergonic reaction, and therefore will proceed inherently slow. Nevertheless, this pathway has higher conservation of energy and may thus achieve a carbon efficiency advantage over aerobic pathways.
A second, MCR-independent AOM pathway coupled to nitrite reduction was observed in bacteria (Ettwig et al., 2010; Scheller et al., 2017). The first step in this pathway is most likely the exergonic formation of 2-methyl-succinate from fumarate and methane catalyzed by a glycyl-radical activating enzyme (Thauer and Shima, 2008). The involvement of a radical enzyme in this first step would not allow a direct coupling to energy conservation, so that most, if not all, energy generated during methane activation would dissipate as heat in the first step of the pathway. Therefore, this would not leave enough energy to drive ADP phosphorylation in reactions further downstream if coupled to sulfate reduction. However, with nitrate or nitrite as electron acceptor the free energy change would be sufficient to allow formation of 2-methyl-succinate (Thauer and Shima, 2008). And indeed, the (kcat/Km) of AOM with nitrate was found more than 1,000 times higher than that of AOM with sulfate (Raghoebarsing et al., 2006). With no pure culture isolate available, the details of the proposed pathway for anaerobic methane oxidation via 2-methyl-succinate remain unknown to date.
Several synthetic biology and metabolic engineering approaches present new pieces to the puzzles of anaerobic methane oxidation pathways. Yan et al. successfully introduced the MCR of an unculturable ANME into Methanosarcina acetivorans, which enabled the genetically modified strain of anaerobic methanotrophic growth dependent on reduction of iron(III) resulting in a pathway remarkably similar to AOM pathways hypothesized for uncultured anaerobic methanotrophic archaea (Yan et al., 2018). Another recent study followed an enrichment approach, which identified an archaeon capable of iron-dependent AOM via reverse-methanogenesis (Cai et al., 2018). Interestingly, a high abundance of multi-heme c-type cytochromes was found in this culture, which are hypothesized to facilitate dissimilatory iron(III) reduction. The fast development of ~omics platforms and tools for genetic modification of non-model organisms gives reason to believe that significant progress regarding the fundamentals of aerobic and anaerobic oxidation of methane can be expected in the near future (Kalyuzhnaya et al., 2015). This system-level understanding of methanotrophic metabolism will lay the groundwork for metabolic engineering to generate value-added products from methane (Strong et al., 2015). However, it remains unknown which metabolic pathway for methane oxidation will prove most suitable for the development of this platform technology.
Here, we present a computational study to assess the potential of different pathways for the microbial oxidation of methane for formation of biomass and production of value added compounds. Using an in silico approach to calculate the metabolic capability of organisms to grow and produce chemicals from CH4 as only carbon and energy source, enables to theoretically evaluate the most promising routes while current knowledge gaps remain. First, the different discussed pathways for aerobic and anaerobic methane oxidation are implemented in a metabolic network of the model organisms for biotechnology, Escherichia coli. Based on stoichiometry, we compare the theoretical maximum achievable biomass yields of each pathway. In a second part, the possibility of simultaneous CO2 fixation enabled by the accumulation of reducing equivalents from the methane oxidizing pathway is discussed for different heterological host organisms. Finally, the different metabolic pathways for methane oxidation are paired with synthetic pathways for production of different (drop-in) fuels to evaluate most promising production routes. Benefits and limitations of the theoretical proposed scenarios are discussed critically.
Materials and Methods
Construction of Metabolic Networks
Metabolic networks of the different organisms (Komagataella phaffii formerly Pichia pastoris, Saccharomyces cerevisiae, Escherichia coli, Bacillus subtilis, Corynebacterium glutamicum, Cupriavidus necator formerly Ralstonia eutropha and Clostridium autoethanogenum) were modified from published stoichiometric network analyses (Melzer et al., 2009; Lopar et al., 2013, 2014; Ternon et al., 2014; Unrean, 2014; Kracke et al., 2016; Koch et al., 2017; Averesch and Krömer, 2018; Averesch et al., 2018) to fulfill the requirements of this elementary flux mode analysis. Specifically, the networks were integrated with methane assimilation pathways, compiled as follows:
The methanol/formaldehyde assimilation pathways Serine-cycle, Ribulose-Monophosphate Pathway (RuMP) and Xylulose-Monophosphate Pathway (XMP)/Dihydroxyacetone- (DHA-) pathway, as described in Fei et al. (2014), Hwang et al. (2018), and on MetaCyc (Caspi et al., 2014), were compiled into stoichiometric reactions. The DHA-pathway corresponds to the XMP where instead of the DHA synthase a formolase is used (Siegel et al., 2015). Further, an NAD dependent methanol dehydrogenase (MDH) (Bennett et al., 2018) was evaluated in comparison to O2 as electron acceptor for oxidation of methanol to formaldehyde, to determine the potential for increased carbon efficiency and energy conservation. NADH and NADPH dependent MMOs, which allow the initial oxidation of methane, completed the three fundamentally different pathways. Co-factor recycling allowed redox power to be derived from the oxidation of formaldehyde to CO2 and proceeded with tetrahydrofolate (THF) for the Serine-cycle, with tetrahydromethanopterin (H4MPT) in the RuMP and via glutathione (GSH) in the XMP/DHA-pathway (Marx et al., 2003). Figure 1 shows the three pathways for aerobic methane catabolism in detail.
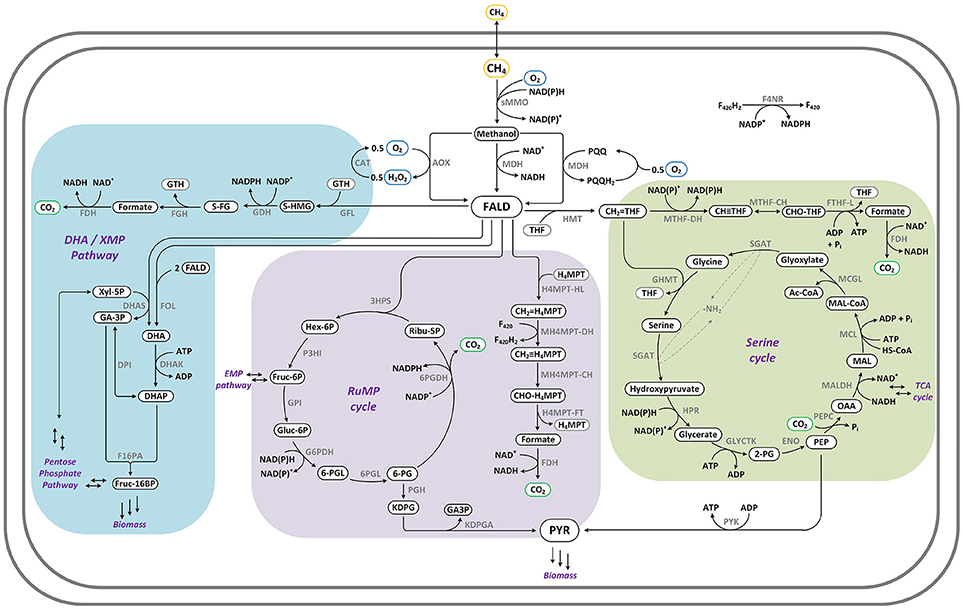
Figure 1. Pathways for aerobic methane oxidation via methane monooxygenase (MMO): Serine-cycle (green), RuMP (purple), XMP/DHA-pathway (blue). Enzymes: 3HPS: 3-hexulose-6-phosphate synthase; 6PGD: 6-phosphogluconate dehydrogenase; 6PGL: 6-phosphogluconolactonase; AOX: alcohol oxidase; CAT: catalase; DHAK: dihydroxyacetone kinase; DHAS: dihydroxyacetone synthase; DPI: triose-phosphate isomerase; F16PA: fructose 1,6-bisphosphate aldolase; F4NR: F420-dependent NADP reductase; FDH: formate dehydrogenase; FGH: S-formylglutathione hydralase; FOL: formolase; FTHF-L: formate:tetrahydrofolate ligase; G6PDH: glucose-6-phosphate dehydrogenase; GDH: S-(hydroxymethyl)glutathione dehydrogenase; GFL: S-(hydroxymethyl)glutathione formaldehyde-lyase; GHMT: glycine hydroxymethyltransferase; GLYCTK: glycerate 2-kinase; H4MPT-FT: methenyltetrahydromethanopterin formyltransferase/hydrolase complex; H4MPT-HL: 5,6,7,8-tetrahydromethanopterin hydro-lyase; HMT: hydroxymethyltransferase; HPR: hydroxypyruvate reductase; KDPGA: 2-keto-3-deoxygluconate-6-phosphate aldolase; MALDH: malate dehydrogenase; MCGL: malyl-CoA glyoxylate-lyase; MCL: malate:CoA ligase; MDH: methanol dehydrogenase; MH4MPT-CH: methenyltetrahydromethanopterin cyclohydrolase; MH4MPT-DH: methylenetetrahydromethanopterin dehydrogenase; MTHF-CH: methenyltetrahydrofolate cyclohydrolase; MTHF-DH: methylenetetrahydrofolate dehydrogenase; P3HI: 6-phospho-3-hexuloisomerase; PEPC: phosphoenolpyruvate carboxylase; PGH: phosphogluconate dehydratase; ENO: enolase/phosphopyruvate hydratase; PYK: pyruvate kinase; SGAT: serine:glyoxylate aminotransferase.
The proposed AOM by means of a glycyl-radical enzyme via methyl-succinate (Mss-AOM) was defined as proposed by previous studies (Thauer and Shima, 2008; Haynes and Gonzalez, 2014). The different options for regeneration of fumarate include a series of reactions via TCA-cycle analogous reactions or a combination of β-oxidation and ketogenesis/GABA-metabolism. Figure 2 gives a detailed overview of the different variations of this potential metabolic route for methane fixation. Here, nitrate respiration was assumed as a feasible way to complete the electron transport chain under anoxic conditions in the bacterial networks (Unden and Bongaerts, 1997; Nakano and Zuber, 1998; Nishimura et al., 2007; Tiemeyer et al., 2007).
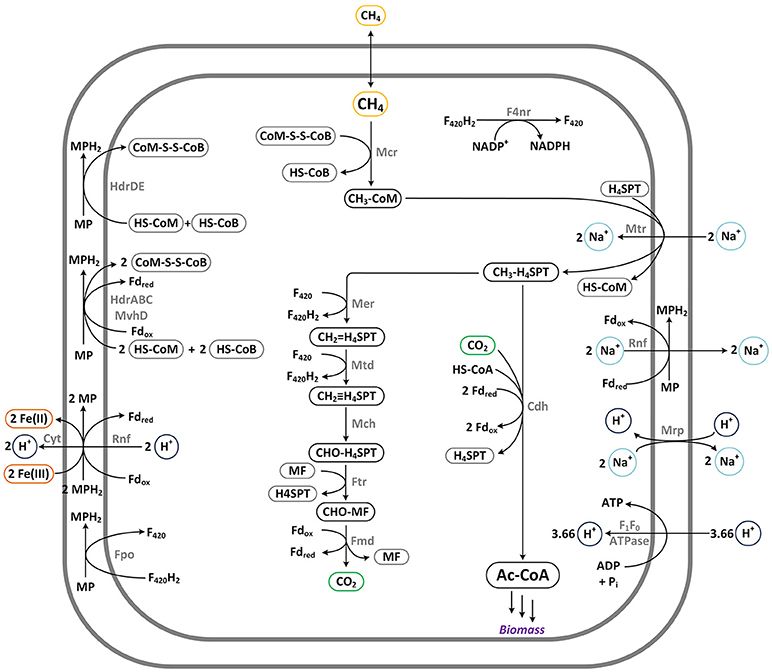
Figure 2. Pathways for anaerobic methane oxidation via activation to 2-methyl-succinate: Mss-AOM. Note that this pathway requires a final electron acceptor, such as nitrate via anaerobic respiratory chain (not shown). Enzymes: BCDH: butyryl-CoA dehydrogenase; BCS: butyryl-CoA synthetase; BCT: butyryl-CoA transferase; CCH: crotonyl-CoA hydratase; CCL: citramalyl-CoA lyase; CCS: citramalyl-CoA synthetase; EMCDC: ethylmalonyl-CoA decarboxylase; HBCDH: hydroxyacyl-CoA dehydrogenase; HBCH: hydroxybutyryl-CoA hydrolase; HBCI: hydroxybutyryl-CoA isomerase; HBDH: 4-hydroxybutyrate dehydrogenase; MF: methylfumarase; MSCI: methylsuccinyl-CoA isomerase; MSCS: methylsuccinyl-CoA synthetase; MSDC: methylsuccinate decarboxylase; MSDH: methylsuccinate dehydrogenase; MSS: methylsuccinate synthase; SSADH: succinate-semialdehyde dehydrogenase; β-KAT: β-ketoacyl-thiolase.
AOM via reverse-methanogenesis by means of methyl-coenzyme M reductase (Mcr-AOM) was implemented as proposed by Nazem-Bokaee et al. (2016) and Bennett et al. (2018). The two branches of the pathway, which rely on an electron transport chain with iron(III) as terminal acceptor, are depicted in Figure 3.
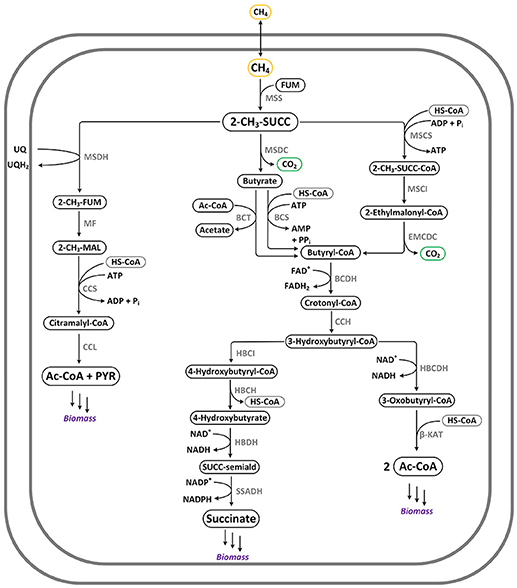
Figure 3. Pathway for anaerobic methane oxidation via reverse-methanogenesis: Mcr-AOM. This pathway requires a final electron acceptor, such as iron(III) shown here. Enzymes: ATPase: ATP synthase; Cdh: CO dehydrogenase/acetyl-CoA synthase; Cyt: c-type cytochrome; F4nr: F420-dependent NADP reductase; Fmd: formylmethanofuran dehydrogenase; Fpo: F420H2:phenazine oxidoreductase; Ftr: formylmethanofuran-H4MPT formyltransferase; HdrABC: hydrogenase; HdrDE: CoB-CoM heterodisulfide reductase; Mch: N5,N10-methenyl-H4MPT cyclohydrolase; Mcr: methyl-CoM reductase; Mer: N5,N10-methylene-H4MPT reductase; Mrp: sodium/proton antiporter; Mtd: F420H2-dependent methylene-H4MPT dehydrogenase; Mtr: N5-methyl-H4MPT:CoM methyltransferase; MvhD: heterodisulfide reductase (membrane-bound); Rnf: proton-translocating electron bifurcating oxidoreductase.
It should be noted, that in Figures 1–3 pathways were drawn out until a central metabolism metabolite (e.g., fructose-1,6-bisphosphate, pyruvate, succinate, acetyl-coenzyme A) was reached and connections to other pathways in central metabolism are indicated with double arrows. The full metabolic networks, integrated with the pathways, can be found in Supplementary File 1.
For chapter Potential of Different Organisms to Assimilate Additional Carbon via CO2 Co-utilization the metabolic networks were amended with established product pathway(s) for the designated target products, as described previously (Jang et al., 2012; Peralta-Yahya et al., 2012) and/or according to records in databases like KEGG (Kanehisa and Goto, 2000; Kanehisa et al., 2016, 2017) and MetaCyc (Caspi et al., 2014). All pathways can be found in Supplementary File 1, including details regarding the enzymes catalyzing the respective reactions and origin of the pathway.
Elementary Flux Mode Analysis
Using MATLAB® (MathWorks®) (RRID:SCR_001622) the metabolic networks were parsed with EFMTool (Terzer and Stelling, 2008; RRID:SCR_016289) into stoichiometric matrices. Metabolic solutions for each network were calculated as elementary flux modes (EFMs) using the most recent implementation FluxModeCalculator (van Klinken and Willems Van Dijk, 2015; RRID:SCR_016290) and evaluated as described before (Averesch et al., 2016), calculating the yields by drawing carbon balances around the boundary reactions. The ratio of consumed or produced carbon dioxide to the (main) substrate was determined as the molar quotient of net CO2-flux to other available carbon sources (most frequently methane) according to Equation (1). All calculated data and calculations based on the data can be found in Supplementary File 1.
Results
Maximum Achievable Biomass Yields Via Different CH4 Fixation Pathways
To compare the maximum achievable biomass-yields (BMY in % C-mol/C-mol) from CH4 as sole electron and carbon source, the different pathways for aerobic and anaerobic methane oxidation were analyzed using the heterological host E. coli as model organism. The results are presented in Table 1.
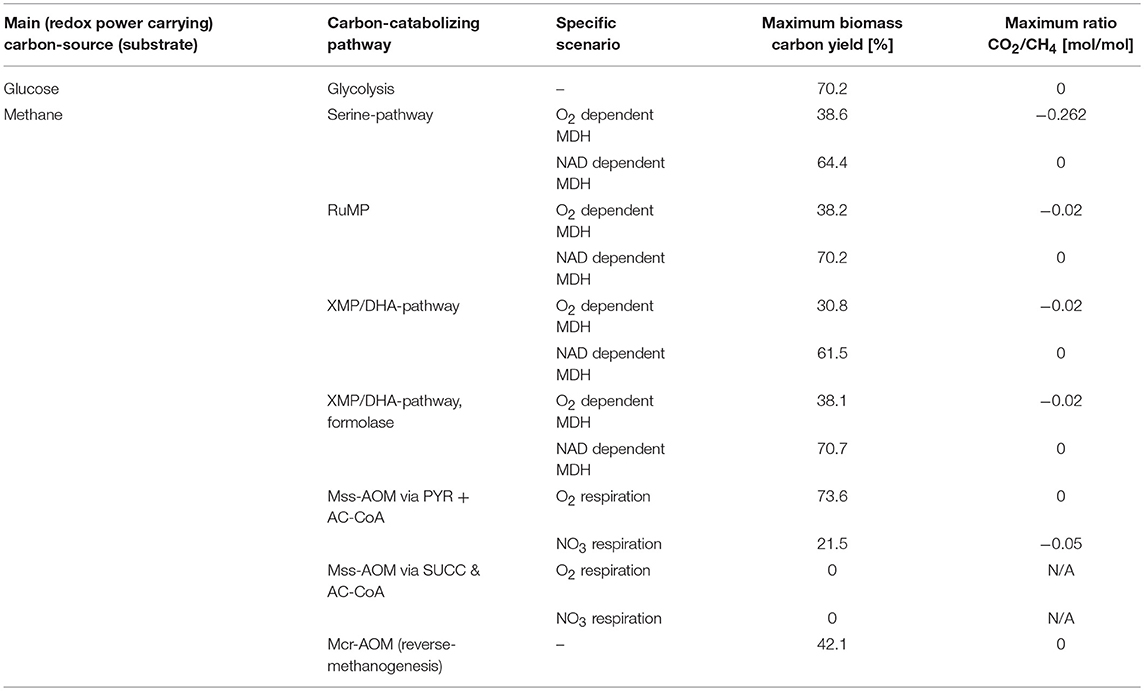
Table 1. Maximum theoretical biomass carbon yields and CO2/substrate uptake ratios of different methane-catabolizing pathways implemented in E. coli, compared to glucose as natural carbon-source.
When comparing the different pathways for methane catabolism pathways (see Figures 1–3), one universal feature can be identified: accompanying the reductive pathway for formation of carbon-carbon bonds from activated methane, there is always a second, oxidative pathway branch that ultimately forms CO2 and provides the redox equivalents required for the reductive pathway steps. Additionally, all aerobic pathways require providing reducing equivalents for the MMO in the first pathway step (see Figure 1). This requirement of the key enzyme for one reducing equivalent per CH4 limits the maximum achievable BMY via aerobic methane oxidation to 38% or lower. However, if the second pathway step, the formation of formaldehyde from methanol, could recover the electrons via a NAD-dependent enzyme (MDH) this limitation can be overcome, which is reflected in a theoretical maximum achievable BMY of 60–70% (see Table 1). In case of the XMP the initial step of dihydroxyacetone phosphate formation from formaldehyde is a bottleneck, as xylulose-5-phosphate needs to be regenerated. Replacing the DHA synthase with a formolase, as successfully demonstrated by Siegel et al. (Siegel et al., 2015), re-routes the pathway and eliminates this need, directly linking it to DHA, which benefits the theoretical maximum BMY.
For anaerobic methane oxidation the proposed pathway via 2-methyl-succinate (Mss-AOM; Figure 2) and reverse-methanogenesis (Mcr-AOM; Figure 3) were considered. For the Mss-AOM a theoretical yield linked to respiration is given for reference, even though a feasibility of the entire pathway in one organism is regarded infeasible since the proposed glycyl-radical enzyme will require strictly anaerobic environments. For the different pathway versions of Mss-AOM only the branch via TCA-cycle analogous reactions was found feasible. This is due to decarboxylation reactions in the β-oxidation analogous pathway options, which lead to production of one CO2 per fixed CH4. Biomass assimilation via these pathway branches would only be feasible in combination with an efficient mechanism for CO2 re-fixation (e.g., via RuBisCO, cf. C. necator BMY, Supplementary File 1). For the same reason, the Mss-AOM needs to recycle fumarate via the glyoxylic shunt, to compensate for the decarboxylation reactions during the TCA-cycle. The fact that this pathway needs to be operated with a different terminal electron acceptor than O2, in this case NO3, severely impacts the yield—otherwise it would present the most attractive option for CH4 utilization based on stoichiometry (see Table 1).
Reverse-methanogenesis (Mcr-AOM) resulted in a maximum achievable BMY of 42%, which is about twice as high as the BMY for Mss-AOM with nitrate as electron acceptor. More importantly, this theoretical maximum BMY is in the same range or higher than the results for aerobic methane oxidation via MMO and Serine-cycle, RuMP or XMP/DHA-pathways in case of the natural, O2-dependent MDH. This indicates that the AOM pathway via reverse-methanogenesis is not more restricted by stoichiometry than the aerobic options or the Mss-AOM.
Potential of Different Organisms to Assimilate Additional Carbon via CO2 Co-utilization
Since methane is fully reduced (degree of reduction(CH4) = 8), its assimilation in biomass as well as transformation into other hydrocarbons requires a parallel, electron accepting pathway. Here, we analyse several metabolic possibilities for microbial co-utilization of CO2 during methane oxidation. The previous chapter, including Figures 1–3, illustrates how aerobic, as well as anaerobic methane oxidation always requires a certain amount of substrate to be oxidized in order to provide sufficient reducing equivalents for methane activation. Often oxidation in this pathway branch is complete, resulting in emission of CO2 as by-product (degree of reduction(CO2) = 0). We illustrate this formation of CO2 as the ratio of net CO2 production to the uptake of methane—CO2:CH4 [mol/mol] (cf. Equation 1, section Elementary Flux Mode Analysis). The rightmost column in Table 1 shows the maximum ratios for the different pathways of microbial methane utilization applied to E. coli. A negative value refers to CO2 production, while a ratio of “0” indicates that no net-flux of CO2 is created; meaning that occurring decarboxylation reactions can be metabolically compensated for, e.g., via enzymes, such as pyruvate carboxylase. This is highly desirable, as the metabolic potential for carbon re-fixation is essential to maximize the carbon yield.
When comparing the different pathways, it appears that all aerobic pathway options inevitably will lead to CO2 formation as by-product (for O2-dependent MDH). The Serine cycle shows an especially high ratio of −0.262, translating to about 1 mol of CO2 formed per 4 mol of CH4 that are taken up. In the RuMP and XMP/DHA-pathway, potentially less carbon is “lost” as CO2, however, complete avoidance of CO2 formation (reflected in a ratio of “0”) is only observed in case of NADH-dependent MDH for any of the aerobic pathways. Of all pathways, Mcr-AOM, seems most beneficial in this aspect as the CO2 of its oxidative branch is directly re-fixated by adding it to the activated form of methane resulting in acetyl-CoA (see Figure 3). The complete reversal of methanogenesis is thermodynamically only feasible if coupled to an electron accepting pathway, in this case reduction of iron(III). If this electron accepting pathway could be provided via a carbon fixation pathway (fully or partially), the efficiency of microbial methane oxidation could potentially be further increased. Additionally, a co-fixation of CO2 would present great environmental potential and be of particular benefit for biogas upgrading applications (Weiland, 2010; Conrado and Gonzalez, 2014). Therefore, several metabolic options for additional CO2 uptake were investigated.
Different microbial hosts were chosen as model organisms for CH4-CO2 co-utilization to evaluate the potential across several industrially relevant species: Komagatella phaffii (formerly Pichia pastoris) and Saccharomyces cerevisiae, Escherichia coli, Bacillus subtilis, Corynebacterium glutamicum, Cupriavidus necator (formerly Ralstonia eutropha), and Clostridium autoethanogenum. This includes eukaryotes, prokaryotes, Gram-positives and Gram-negatives, heterotrophic as well as autotrophic species (photoautotrophic organisms were not considered within this study). Exact details regarding the specific metabolism of each organism can be found in Supplementary File 1, while Table 2 presents the maximum possible CO2 uptake calculated for each case. This is again presented as maximum ratio of CO2:CH4 in mol/mol (positive ratio = CO2 is fixed, negative ratio = CO2 is produced, 0 = no net-CO2-flux). The scenarios (C-source and pathway) that are listed in the last column represent the specific pathways, which resulted in the highest achievable CO2 fixation in each case. A full list of results for each individual pathway and organism is included in Supplementary File 1.
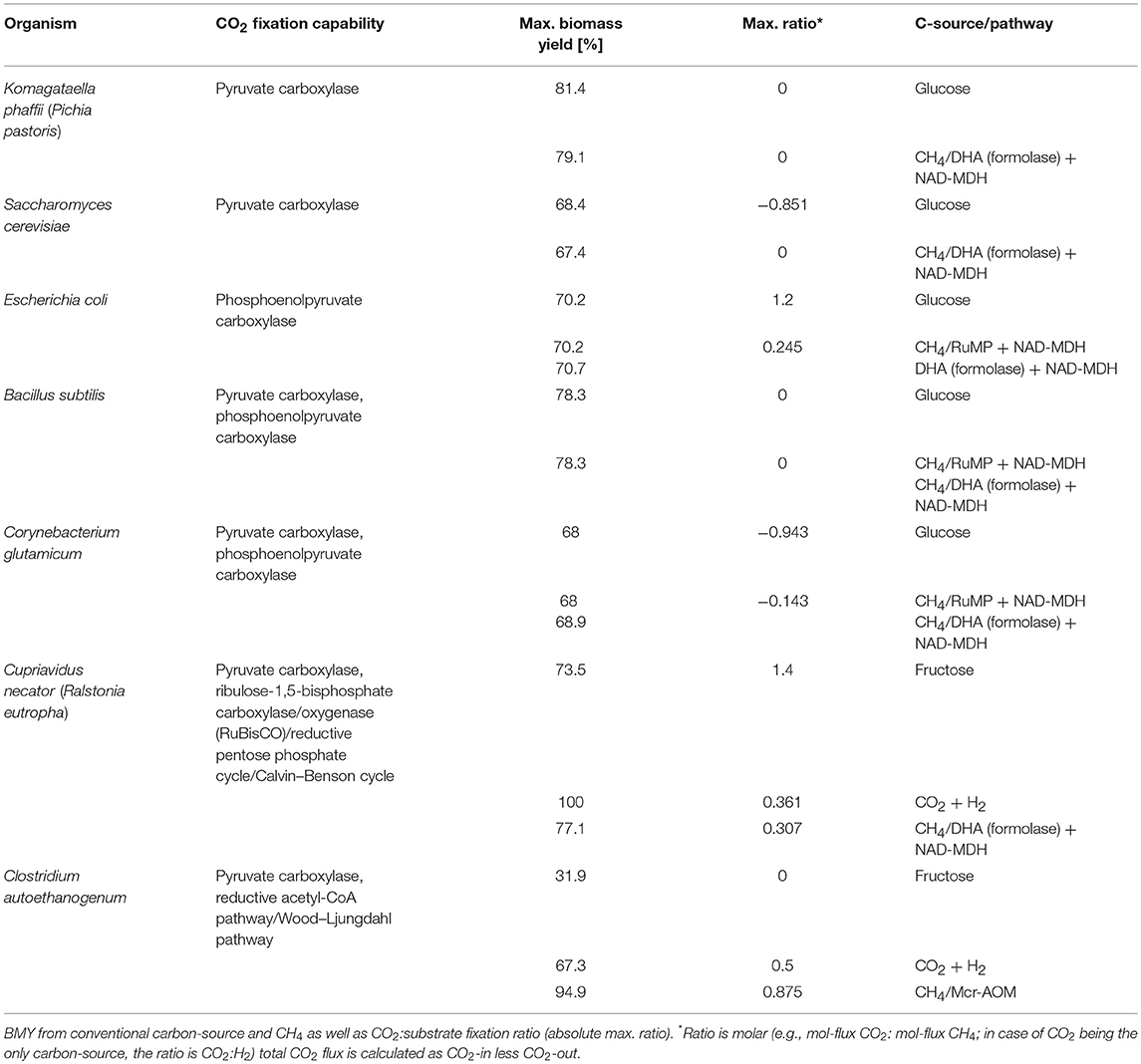
Table 2. Overview of organisms and pathways modeled, with information on CO2 fixation capability, including results: max.
For aerobic methane oxidation in E. coli, the most beneficial scenario identified above, MMO with NAD-dependent MDH, can be further improved in the case of CO2-co utilization. The maximum ratio of 0.245 indicates that reducing equivalents can potentially be re-distributed across the metabolism to allow for additional CO2 fixation at the theoretical maximum level of ~1 mol CO2 per 4 mol CH4. The same scenario (RuMP and DHA-pathway with NAD-dependent MDH) was also found most beneficial for B. subtilis where it theoretically enables for CO2-neutral CH4-utilization. Maybe not surprisingly, most interesting scenarios are represented by organisms, which naturally inherent CO2-fixation pathways as their major carbon metabolism: Cupriavidus necator and Clostridium autoethanogenum. The hydrogen-oxidizing bacterium C. necator has a very versatile metabolism, being able to switch between aerobic, anaerobic and nitrate respiration. Here, we found that if this metabolism could be paired with the ability for methane oxidation, high maximum biomass yields may be achieved. Further, CO2 co-fixation at a maximum ratio of 0.307 was determined for the DHA-pathway featuring an NAD-dependent MDH (cf. Table 2). The key enzyme, which enables this high CO2-fixation capacity is RuBisCO. The anaerobic acetogen C. autoethanogenum, on the other hand, uses the strict anaerobic Wood-Ljungdahl pathway for CO2 fixation. This particular pathway was the only option found in this analysis to efficiently enable CO2 co-utilization from reverse-methanogenesis. Since both pathways, Mcr-AOM and Wood-Ljungdahl pathway, share ferredoxin as redox carrier, electrons can be transferred most efficiently, resulting in a maximum substrate ratio of 0.875 CO2 per CH4 [mol/mol]. This theoretical transfer of electrons from methane to the carbon fixation pathway further allows a very high maximum BMY of 95% (cf. Table 2).
Bio-GTL: Production of (Drop-In) Fuels
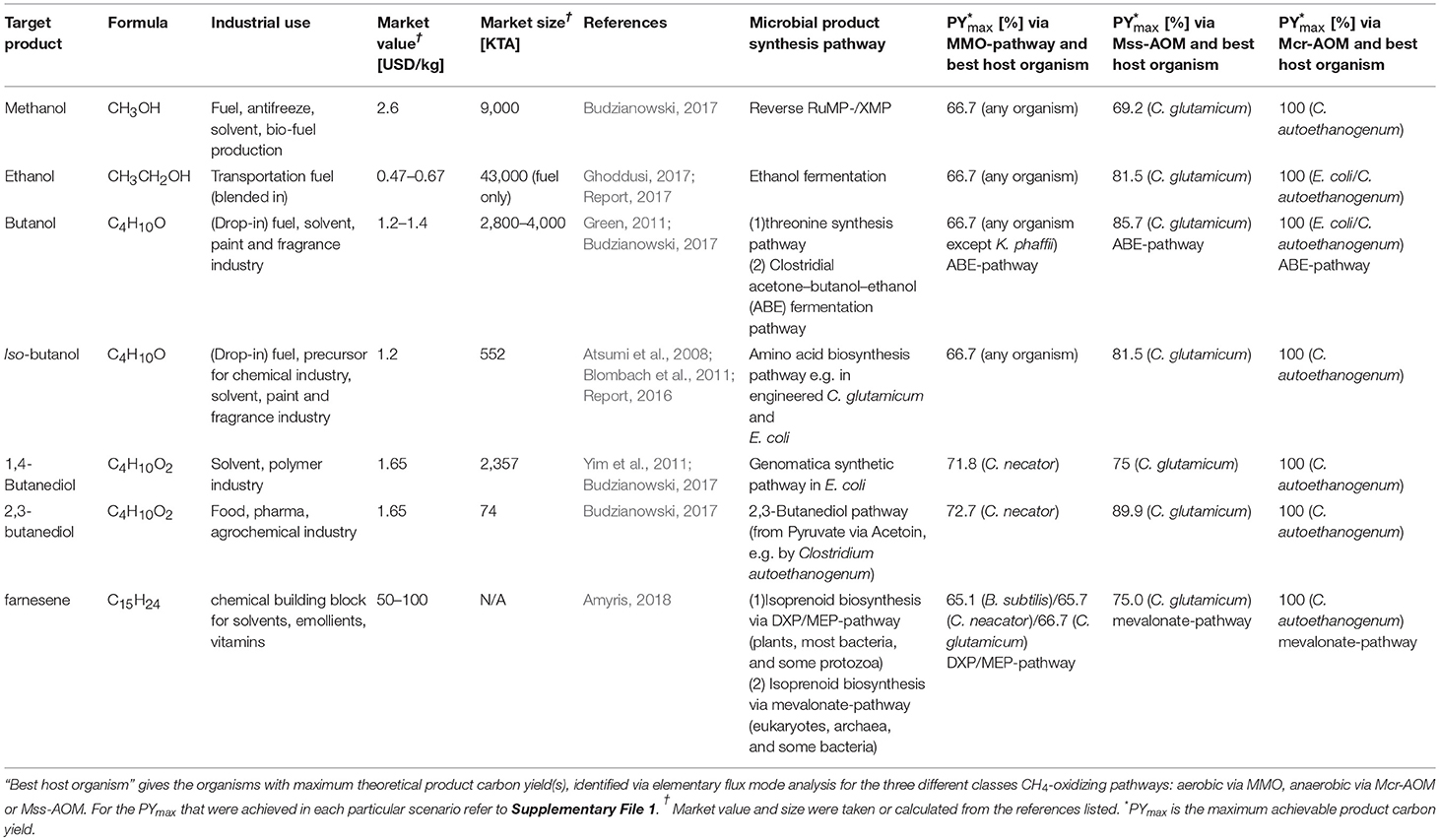
The ultimate benefit of microbial methane oxidation is the potential production of liquid fuels with high specificity and at ambient temperatures and pressures (Conrado and Gonzalez, 2014). Therefore, the next step of our analysis pairs the different microbial pathways for CH4-oxidation with production pathways for industrially relevant fuels to identify the most promising scenarios. The here investigated drop-in fuels are methanol, ethanol, butanol, iso-butanol, butanediol and farnesene. Table 3 summarizes information on each target product, including the corresponding synthetic pathway(s) for each compound, which were implemented in the different metabolic networks. The three columns on the right, “PYmax” and “best host organism,” list the most promising scenario that was determined by our analysis for each individual target product. Further, we distinguish between the different metabolic pathways for methane oxidation that were discussed in the previous chapters. The full report on all results from each individual combination is given in Supplementary File 1. Additionally, in Figure 4 the distribution of EFMs is displayed for selected scenarios in plots of the CO2:CH4-ratio vs. the PY of the individual EFMs.
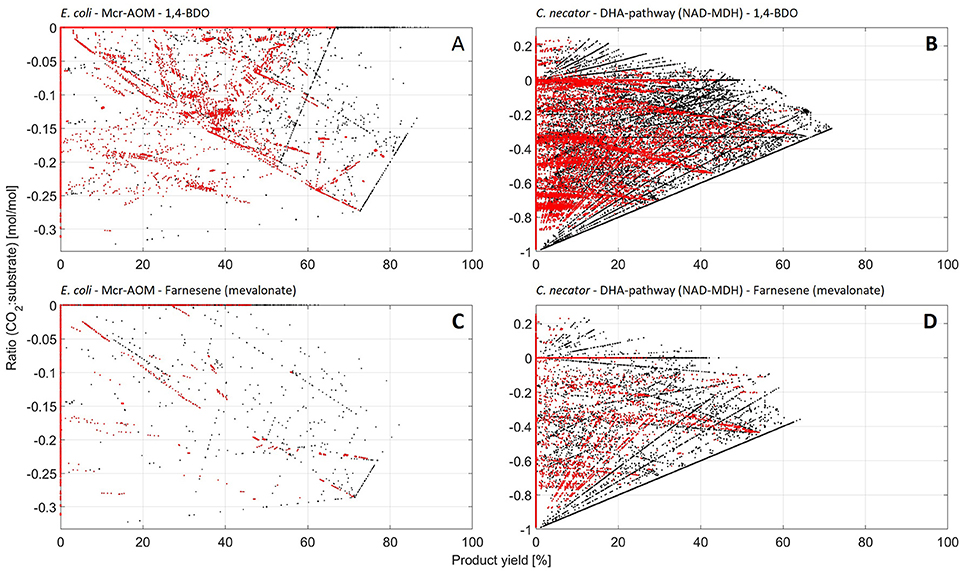
Figure 4. (A–D) Distribution of elementary flux modes (EFMs) by CO2:CH4 uptake-ratio vs. PY for selected scenarios. (A): Mcr-AOM in E. coli, target product 1,4-BDO; (B): DHA-pathway (NAD-dependent MDH) in C. necator, target product 1,4-BDO; (C): Mcr-AOM in E. coli, target product farnesene (mevalonate-pathway); (D): DHA-pathway (NAD-dependent MDH) in C. necator, target product farnesene (mevalonate-pathway). Each dot in a plot represents one feasible steady-state flux distribution. EFMs that also allow for formation of biomass (growth) are highlighted in red.
Under the aspect of stoichiometric constrains as applied here, one overall best scenario and host can be identified: All here investigated compounds can be produced with a theoretical PYmax of 100% from methane and CO2 via Mcr-AOM in C. autoethanogenum. The unique advantage of a common intracellular ferredoxin pool, accessible between the natural CO2 fixation pathway (Wood-Ljungdahl) as well as the AOM, facilitates optimum usage of redox equivalents. As discussed under 3.2, this enables for high CO2:CH4 fixation ratios, which are largely in the positive range (and never lower than 0!). However, high ratios of CO2 co-utilization and high PY are mutually exclusive. Nevertheless, this unique combination of AOM and Wood-Ljungdahl pathway presents an attractive option as production platform. Further, production via reverse-methanogenesis, Mcr-AOM, in E. coli shows an interesting pattern (cf. Figure 4), with maximum product yields of up to 100% (possible for ethanol and butanol via acetoacetyl-CoA pathway). Given the simultaneously rather low biomass yields of Mcr-AOM in E. coli (cf. Table 1; Supplementary File 2), this could imply that a favorable distribution of carbon between production pathways and biomass formation may be achieved.
The next best scenario for bio-GTL, evaluated by stoichiometry, is again presented by anaerobic oxidation of methane: Mss-AOM in C. glutamicum. Here, the PYmax for methanol is 69%, while for all other products PYmax of 75% or higher were calculated. The CO2:CH4 ratio for this scenario is always negative (CO2 is being produced), however, the highest PY is always obtained at the lowest CO2 output flux (ratio ≤ −0.1). Another promising host organism for production of fuels via the Mss-AOM is presented by the hydrogen-oxidizing bacterium C. necator. Theoretical achievable PYmax are slightly lower than in C. glutamicum (52% for farnesene, all others >60%), but the CO2:CH4 ratio can be positive, providing a promising platform for CO2-CH4-co-utilization. However, PYmax can only be reached at a negative ratio between −0.5 and −0.3 (cf. Figure 4).
Production via aerobic methane utilization pathways are usually limited to a maximum achievable carbon yield of 67% due to the decarboxylation steps as discussed earlier. This is also reflected in the presented analysis: the PYmax of aerobic methane oxidation to methanol, ethanol, butanol, iso-butanol and (in most cases) butanediol is limited to 67%, independent of organisms or formaldehyde assimilation pathway (cf. Supplementary File 1). Only C. necator may achieve PYmax above 70%, in the scenarios of butanediol production (cf. Figure 4), due to its efficient CO2-re-fixation mechanism. For the production of the high value hydrocarbon building block farnesene, our analysis shows higher PYmax of the DXP/MEP-pathway opposing to the mevalonate-pathway (cf. Table 3; Supplementary File 1), which was expected due to the noted higher carbon efficiency of the non-mevalonate pathway (Kirby et al., 2016). Nevertheless, C. necator achieves an almost equivalent PYmax via the mevalonate-pathway (64%), again due to its effective CO2 (re-)fixation capability. Similar, as seen for the maximum achievable biomass yields under chapter 3.1, all PYmax in aerobic methane oxidation scenarios feature the NAD-dependent MDH. Given that the O2-dependent MDH is thermodynamically greatly favored; pathways proceeding via O2-dependent MDH will likely have higher rates. Most promising target compound here is methanol, with a PYmax of 50%, while all other target compounds, show PYmax below 36% in this scenario.
Discussion
The presented analysis highlights the potential of different metabolic pathways for microbial methane utilization, which will determine future Bio-GTL processes. While the discoveries are intended to guide research efforts, it should be stressed that the presented data is theoretical and based on stoichiometry only. Therefore, the following sections discuss our results in the context of kinetic and thermodynamic limitations as well as challenges related to metabolic engineering approaches, which adds to a holistic interpretation of the study.
Challenges for Construction of Synthetic Methanotrophs, Pathway Engineering and Stoichiometric Limitations
The combination of aerobic or anaerobic pathways for methane oxidation with different anabolic pathways as discussed here, requires substantial metabolic engineering of either native methanotrophs or synthetic hosts, which present a significant challenge. Past approaches of various companies and research institutes have so far focused on aerobic methanotrophs for production applications. However, metabolic engineering of production pathways in native methylotrophs remains restricted by the limited toolset for genetic modification (Strong et al., 2015; Bennett et al., 2018). The transformation of a CH4-oxidation pathway into an industrial organism would thus provide great advantages but has proven equally challenging. Microbial hosts, which are used in industrial scale production processes like Escherichia coli, Corynebacterium glutamicum, and Saccharomyces cerevisiae have been successfully engineered to utilize methanol, paving the way toward a C1-based industrial biotechnology (Schrader et al., 2009; Haynes and Gonzalez, 2014; Strong et al., 2015; Meyer et al., 2018). Introduction of a methane monooxygenase (MMO) could make them methanotrophic and open the door to many established production routes for biofuels. However, the crucial missing link, expression of fully active MMO in heterologous hosts, has not been accomplished to date (Kalyuzhnaya et al., 2015; Hwang et al., 2018). MMOs are complex proteins, soluble MMOs consist of a reductase, a hydroxylase, and a regulatory protein and despite many attempts, heterologous expression yielded only a partially active sMMO with a functional hydroxylase (West et al., 1992; Strong et al., 2015). An alternative approach is presented by P450 monooxygenases, which have been heterologically expressed to mimic the function of the MMO, however, with similarly limited success (Hwang et al., 2018). Nevertheless, an engineered BM-3 cytochrome P450 monooxygenase from Bacillus megaterium has been patented (Arnold et al., 2005). With the implementation of emerging new technologies that enable rapid advances in synthetic biology (CRISPR on the molecular side, and lab-automation on the operational side), breakthroughs can be expected that 1 day may allow the metabolic engineering, which is necessary for the development and construction of synthetic methylotrophs in biotechnology.
Given that the major hurdle of initial activation of methane would be overcome, our results indicate that aerobically C. necator theoretically allows the highest maximum CO2:CH4 fixation ratios. This aligns well with the observation that a Methylacidiphilum fumariolicum strain fixates CO2 by means of the Calvin-cycle, in parallel to methane assimilation (Fei et al., 2014). The potential to aerobically use reducing power obtained from methane-oxidation to simultaneously fixate CO2, has recently been demonstrated in vivo when Methylobacterium extorquens AM1 was engineered toward autotrophy to fixate CO2 through a heterologous Calvin-cycle while growing on methanol (Schada von Borzyskowski et al., 2018). Further, our finding that the highest aerobic CO2:CH4-fixation ratio is obtained with the Serine-cycle, aligns with reports that α-proteobacteria can assimilate up to 50% of their biomass from CO2, while the γ-proteobacteria can assimilate up to 15% (Trotsenko and Murrell, 2008). These co-fixation levels of CO2 are only possible with NAD-dependent MDH (cf. Supplementary File 2), which is feasible in case of coupling of the MDH to pMMO via direct transfer of electrons (cf. section Thermodynamic Limitations of Aerobic and Anaerobic Oxidation of Methane; de la Torre et al., 2015).
Opposing to the efforts for engineering aerobic systems for CH4 utilization, little research has focused on the potential of AOM as pathway for bio-GTL, due to several knowledge gaps. Even though trace methane oxidation by reverse-methanogenesis has been successfully demonstrated, optimization of the pathway remains limited due to the unavailability of pure cultures (Moran et al., 2005; Scheller et al., 2010). However, a recent synthetic biology approach successfully demonstrated anaerobic production of chemicals from methane in an engineered Methanosarcina acetivorans (Soo et al., 2016). Introduction of the Mcr of an unculturable ANME resulted in the first (synthetic) pure culture capable of reverse-methanogenesis. This represents a significant breakthrough toward Bio-GTL technologies since AOM pathways offer a significant carbon efficiency advantage over aerobic pathways, as shown in our analysis. A follow-up study from the same group further demonstrated co-utilization of methane and bicarbonate through the reversal of the acetoclastic-pathway in the engineered M. acetivorans (Nazem-Bokaee et al., 2016). This finding underlines our results regarding the benefits of possible CH4-CO2 co-utilization via AOM. In particular, our analysis identified the Wood-Ljungdahl pathway as potential parallel pathway to AOM. The fact that electrons from CH4 could theoretically be efficiently conserved to act as electron carrier for CO2 reduction presents a very promising aspect and should attract further research efforts especially since C. autoethanogenum has emerged as a model organism for gas fermentation and is used in industrial scale production applications (Liew et al., 2016).
Regarding the proposed pathway of Mss-AOM many knowledge gaps remain. However, the here presented analysis can allow conclusions to be drawn toward the potential stoichiometry of the pathway: in 2008 Thauer and Shima proposed two different options for carbon-cycling and regeneration of fumarate after activation of methane (Thauer and Shima, 2008). In the present study, only one of the proposed Mss-pathways has proven feasible under the given assumptions, unless a simultaneous pathway for re-fixation of CO2 exists (e.g., RuBisCO). This finding also questions the conclusions made in a study on environmental samples of a bio-corrosive consortium, where the detection of butyric acid was interpreted as indication for activity of AOM via Mss (Duncan et al., 2009): in the proposed options for Mss-AOM, butyric acid presents an intermediate of the β-oxidation-analogous pathway branch, which has been identified as stoichiometrically infeasible (cf. Figure 3) in most cases. Given the predicted high carbon efficiency achievable via Mss-AOM, elucidation of the exact in vivo pathway is of particular interest for Bio-GTL applications.
Rate Requirements, Kinetic Considerations and Rate/Yield Trade-Off
It has previously been stated that the activity of Mcr is one to two orders of magnitude lower than that of pMMO and sMMO, respectively (Mueller et al., 2015). To achieve industrial feasibility [methane activation rate of 1 gCH4/(L × h)] in a bio-GTL process, and under the assumption that, in case of reverse-methanogenesis, Mcr comprises at least 20% of cellular protein (Mueller et al., 2015), this would translate to a requirement of an average cell density of 32 gCDW/L. These assumptions are likely to be a fair bit too optimistic, as the below considerations illustrate:
The maximum reaction rate of limiting steps of reverse-methanogenesis have been elucidated, the lowest and therefore the bottleneck being the transfer of the methyl-group from CH3-SCoM to THF/H4MPT, at 9.8 ± 1.2 nmol/(min × mg) ∧ 0.6 mmol/(h × g) (Yan et al., 2018). In a rough calculation, a specific maximum rate of substrate consumption for a microbial system can be estimated: assuming a total protein concentration of 0.5 g/gCDW and a maximum concentration of the respective enzyme of 1%, the maximum fraction of any given enzyme can be estimated to be 5 mg/gCDW (Averesch et al., 2018). With that, a maximum specific rate of 3 μmol/(gCDW × h) can be determined. This can be compared to established values for minimum rates to suffice standards in industry for production of biotech products. Specifically, these are a productivity in the single-digit g/(L × h) range and a minimum specific rate of 0.01 mol/(gCDW × h) (Averesch and Krömer, 2018). Measured on these, the Mcr-AOM can be evaluated as “the 10,000-fold amount of biomass is needed,” which means that reverse-methanogenesis is about four orders of magnitude away from operating in the range of industrial applications. However, it should be kept in mind that the thresholds presented above are accepted in the context of product formation in white biotechnology. Here, we apply these to the rate of substrate (CH4) uptake, hence the actual product formation rate may be even lower (equal only at 100% carbon yield, while lower in any other case to account for carbon partitioning depending on the yield of the pathway/efficiency of energy conservation). On the other hand, the production rate could potentially also be higher, if the CO2/CH4 ratio is positive (i.e., CO2 is a significant additional carbon-source). Nevertheless, common biotech processes rely on sugar-based carbon-sources, which, compared to methane, rank in a different price-segment, so that a gas-based processes might not have to suffice these strict standards. Additionally, utilization of a waste-stream as carbon-source, which in some cases even might be associated with a negative cost value, has the potential to change the picture, making reverse-methanogenesis still an attractive pathway for methane utilization and upgrading.
In the aerobic pathways the MMO is believed to be the rate-limiting step (Hwang et al., 2018). According to experimental data collected in BRENDA (Placzek et al., 2017), measured specific activities of sMMO span several orders of magnitude, from as low as 0.11 nmol/(min × mg) to 26.1 μmol/(min × mg) (Brenda, 2018). However, in most cases the substrate in these studies was not methane, but a longer unsaturated hydrocarbon (e.g., C3Hx). Further, most more-recent studies report activities higher than 0.1 μmol/(min × mg), with only few publications reporting activities higher than 10 μmol/(min × mg) ∧ 0.6 mol/(h × g), which was therefore used as a “best-case scenario” to compare the aerobic pathways to reverse-methanogenesis. Based on that, the aerobic pathways are potentially three orders of magnitude faster than the Mcr-AOM and only one order of magnitude away from the established requirements for industrial viability and therefore within an achievable range. However, a further improvement of MMO activity, without changing e.g., environmental parameters, is constraint by thermodynamic limitations (section Rate Requirements, Kinetic Considerations and Rate/Yield Trade-Off).
Finally, a more global kinetic constraint of application of methanotrophs at industrial scale relates to rate issues of gas-fermentations due to poor solubility of gases and thus limited mass-transfer, outlined as one of its greatest challenges (Strong et al., 2015). Comparing solubilities of gases participating in gas-fermentations (Table 4), it appears that CH4 and O2 have similar solubilities, with the one of H2 being orders of magnitude lower. Thus, aerobic processes would still be limited by the solubility of CH4, however, if in an anaerobic gas fermentation H2 could be replaced with CH4, severe mass-transfer limitations might be overcome. Further, these constraints are not as critical for AOM (due to its metabolic rate limitations) as they are for aerobic methane oxidation, which would favor large-scale applications using AOM (Bennett et al., 2018).
Thermodynamic Limitations of Aerobic and Anaerobic Oxidation of Methane
Thermodynamically, the energy change that is associated with the activation of methane with oxygen to methanol via MMO could theoretically phosphorylate up to 14 ATP without the ΔrG′° becoming ≥0 [ΔrG′m, which is the more relevant value for biological systems, becomes positive already at 9 ATP, as determined with equilibrator (Flamholz et al., 2012)]—however, this energy remains unused for some reason. Potentially, it is dissipated into heat, which could be an explanation for the low rates of MMOs: avoidance of overheating. This is backed by the largely negative ΔH′° of methanotrophic bioprocesses (−2,464 kJ/mol for butanol production via aerobic oxidation of methane), where most of the energy is lost in the form of heat, resulting in increased cooling demand (Haynes and Gonzalez, 2014).
Functionality of the NAD-dependent MDH has been evaluated in comparison to an MDH which relies on pyrroloquinoline quinone (and ultimately O2) as electron acceptor for oxidation of methanol to formaldehyde, to determine the potential for increased carbon efficiency and energy conservation, but is thermodynamically hampered (Bennett et al., 2018). With an estimated ΔrG′m of 34.2 kJ/mol [determined with equilibrator (Flamholz et al., 2012)] the NAD-dependent MDH has limitations in the same order of magnitude as the Mcr-AOM. Here, however, a similar argument could be made as compared to the Mcr, where it has been argued that substrate concentrations, are likely orders of magnitude higher since it is a gas, thus shifting the ΔrG into the feasible range (Thauer and Shima, 2008). For the MDH a similar gradient may be achieved, since the product concentration will always have to be very low, since formaldehyde is very toxic. Effectively this means, that when operated at highest yield, aerobic pathways may be subject to similar thermodynamic constrains as Mcr-AOM, bringing the respective maximum rates closer together. While it might be possible to further improve kinetics of the NAD-dependent MDH to a certain degree, thermodynamic limitations of an unfavored cannot be altered, unless coupled with a thermodynamically highly favored reaction, like the MMO: reportedly direct coupling of electron transfer between MDH and pMMO is possible (de la Torre et al., 2015), which could shift the ΔrG of a NAD-dependent enzyme in the feasible range. Further, thermodynamics of the NAD-dependent MDH improve with increased temperatures, which is likely a reason for thermophily of many methanotrophs (Hwang et al., 2018).
Regarding the anaerobic pathways, it has been stated that protein engineering efforts could potentially improve the catalytic activity of the key enzyme Mcr into the range of pMMO (Mueller et al., 2015). However, this statement has to be considered with care, respecting thermodynamic constraints. Opposing to aerobic oxidation of methane via MMO, reverse-methanogenesis has the opposite issue: the ΔrG′s of the first two steps (the initial activation of CH4 with CoM and transfer to H4MPT) have rather largely positive values of 30 kJ/mol [with an uncertainty of ±10 (Thauer and Shima, 2008)], while the recycling of the Coenzyme M—Coenzyme B dimer even has a ΔrG of 40 kJ/mol (Mueller et al., 2015). Further, many of the subsequent steps have a positive ΔrG or values close enough to 0 to impose additional bottlenecks [at a ΔrG of −1 kJ/mol, the flux-force efficacy is only 20% (Noor et al., 2014)].
Thermodynamic considerations can also be used to assess the likeliness of Mss-AOM. While there is strong indication that an alternative anaerobic pathway exists, which is based on fumarate to activate methane (Duncan et al., 2009), to date the responsible enzyme has not been identified nor has the reaction it catalyses been documented. While the reaction catalyzed by Mss has a ΔrG′° of −15 kJ/mol, and is thus in general thermodynamically feasible (Haynes and Gonzalez, 2014), the difference in dissociation energies of the methyl-radical and the glycyl-radical of almost 90 kJ/mol is technically too high to be overcome in a biological system (Thauer and Shima, 2008). Nevertheless, similar to Mcr glycyl-radical enzymes are functional dimers, which show half-of-the-site reactivity. Therefore, the relatively large difference might be overcome by coupling of the endergonic steps in one active site with exergonic steps of the second active site (Thauer and Shima, 2008).
Prospects of Bio-GTL Compared to Chemical GTL Technologies
In light of the slow kinetics of microbial methane oxidation, chemical and electrochemical processes are often regarded as a more promising route for methane utilization. Large-scale industrial processes for converting methane to liquid hydrocarbons are limited to two inorganic technologies: methanol-to-gasoline and Fischer–Tropsch synthesis, both of which rely on the expensive (because energy intense) intermediate production of syngas. Other chemical routes for methane activation include the direct oxidation of methane to methanol and formaldehyde, oxidative coupling of methane to ethylene, and direct conversion to aromatics and hydrogen in the absence of oxygen (Lunsford, 2000). Even though a direct activation of methane should have a distinct economic advantage over indirect syngas routes, these processes currently remain limited by low selectivity for target reactions, low conversion rates and dilute product streams (Holmen, 2009; Alvarez-Galvan et al., 2011). Biological processes for methane activation, on the other hand, may offer significant advantage by accessing high selectivity and specificity. Despite the general higher volumetric productivity of a chemical process, a biological solution could provide advantages especially for decentralized solutions due to the smaller required footprint, measured by area, which is required for product synthesis (Haynes and Gonzalez, 2014). Due to the integrated nature of bioprocesses, fewer unit operations are required, which enables profitability at smaller scales and therefore offers opportunities for new solutions especially at remote locations. This could allow for the 5% of currently flared global natural gas production to be utilized.
Conclusion
The specifics of a successful future Bio-GTL process remain to be elucidated, however they will inevitably depend on the microbial host organisms and its metabolic pathway features. This study presents a comprehensive, stoichiometry-based analysis of microbial pathways for aerobic and anaerobic conversion of methane to liquid fuels. The proposed combination of pathways for methane activation, CO2 fixation and product formation require in any scenario extensive metabolic engineering, which remains a major limitation. However, recent technological advances in the field of synthetic biology give reason to believe that eventually synthetic pathways for methane utilization will be constructed, regardless of the final host organism being a native methanotroph or a model biotechnology organism.
Our analysis shows, that the low carbon efficiency of methane activation via MMO could be mitigated via a NAD-dependent MDH. However, highest product carbon yields are achievable via anaerobic pathways for methane oxidation, which proceed without de-carboxylation reactions. Especially promising seems the pairing of AOM and Wood-Ljungdahl pathway, which could allow for the efficient co-utilization of CH4 and CO2 for production of bio-fuels. Given the substantial knowledge gaps around the fundamentals of anaerobic methane oxidation, we deem future research efforts in this direction most necessary and auspicious.
Author Contributions
NA and FK jointly designed the study, researched the data and wrote the manuscript. NA constructed the metabolic networks and performed the calculations and computational analysis. Both authors approved the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was not supported by any particular funding. Thus, the given affiliations for both authors reflect their current positions only. We would like to thank Dr. Wenyu Gu for valued feedback on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2018.00106/full#supplementary-material
Abbreviations
ANME, anaerobic methanotrophic archaea; AOM, anaerobic oxidation of methane; BDO, butanediol; Bio-GTL, biological gas-to-liquid (microbial conversion of methane into liquid fuels); BMY, biomass carbon yield; BTU, British thermal unit; DHA, dihydroxyacetone; DXP, 1-deoxy-D-xylulose 5-phosphate; EFM, elementary flux mode; GSH, glutathione; H4MPT, tetrahydromethanopterin; MCR, methyl-coenzyme M reductase; MDH, methanol dehydrogenase; MEP, 2-C-methylerythritol 4-phosphate; MMO, methane monooxygenase; MSS, methyl-succinate synthase; PY, product carbon yield; RuBisCO, ribulose-1,5-bisphosphate carboxylase/oxygenase; RuMP, ribulose mmonophosphate pathway; THF, tetrahydrofolate; XMP, xylulose-monophosphate; ΔrG, Gibbs free energy of a chemical reaction; ΔrG°, Gibbs free energy of a chemical reaction at standard conditions (not accounting for pH or ionic strength); ΔrG′°, Gibbs free energy of a chemical reaction at a particular pH and ionic strength at 1 M standard concentrations; ΔrG′m, Gibbs free energy of a chemical reaction at a particular pH and ionic strength at 1 mM (physiologically relevant) concentrations
References
Alvarez-Galvan, M. C., Mota, N., Ojeda, M., Rojas, S., Navarro, R. M., and Fierro, J. L. G. (2011). Direct methane conversion routes to chemicals and fuels. Catal. Today 171, 15–23. doi: 10.1016/j.cattod.2011.02.028
Amyris (2018). Farnesene. The renewable Hydrocarbon Building Block. Available online at: http://farnesene.net/shop/ (Accessed May 21, 2018).
Arnold, F., Meinhold, P., Peters, M. W., Fasan, R., and Chen, M. M. Y. (2005). Alkane Oxidation by Modified Hydroxylases. USA patent application US20160024482A1.
Atsumi, S., Hanai, T., and Liao, J. C. (2008). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. doi: 10.1038/nature06450
Averesch, N. J., Winter, G., and Krömer, J. O. (2016). Production of para-aminobenzoic acid from different carbon-sources in engineered Saccharomyces cerevisiae. Microb. Cell Fact. 15:89. doi: 10.1186/s12934-016-0485-8
Averesch, N. J. H., and Krömer, J. O. (2018). Metabolic engineering of the shikimate pathway for production of aromatics and derived compounds – present and future strain construction strategies. Front. Bioeng. Biotechnol. 6:32. doi: 10.3389/fbioe.2018.00032
Averesch, N. J. H., Martínez, V. S., Nielsen, L. K., and Krömer, J. O. (2018). Toward synthetic biology strategies for adipic acid production: an in silico tool for combined thermodynamics and stoichiometric analysis of metabolic networks. ACS Synth. Biol. 7, 490–509. doi: 10.1021/acssynbio.7b00304
Beal, E. J., House, C. H., and Orphan, V. J. (2009). Manganese- and iron-dependent marine methane oxidation. Science 325, 184–187. doi: 10.1126/science.1169984
Bennett, R. K., Steinberg, L. M., Chen, W., and Papoutsakis, E. T. (2018). Engineering the bioconversion of methane and methanol to fuels and chemicals in native and synthetic methylotrophs. Curr. Opin. Biotechnol. 50, 81–93. doi: 10.1016/j.copbio.2017.11.010
Blombach, B., Riester, T., Wieschalka, S., Ziert, C., Youn, J.-W., Wendisch, V. F., et al. (2011). Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 77, 3300–3310. doi: 10.1128/AEM.02972-10
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Brenda (2018). Information on EC 1.14.13.25–Methane Monooxygenase (Soluble). Available online at: https://www.brenda-enzymes.org/enzyme.php?ecno=1.14.13.25 (Accessed May 22, 2018).
Budzianowski, W. M. (2017). High-value low-volume bioproducts coupled to bioenergies with potential to enhance business development of sustainable biorefineries. Renew. Sustain. Energy Rev. 70, 793–804. doi: 10.1016/j.rser.2016.11.260
Cai, C., Leu, A. O., Xie, G. -J., Guo, J., Feng, Y., Zhao, J. -X., et al. (2018). A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J. 12, 1929–1939. doi: 10.1038/s41396-018-0109-x
Caspi, R., Altman, T., Billington, R., Dreher, K., Foerster, H., Fulcher, C. A., et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 42, D459–D471. doi: 10.1093/nar/gkt1103
Conrado, R. J., and Gonzalez, R. (2014). Envisioning the bioconversion of methane to liquid fuels. Science 343, 621–623. doi: 10.1126/science.1246929
de la Torre, A., Metivier, A., Chu, F., Laurens, L. M., Beck, D. A., Pienkos, P. T., et al. (2015). Genome-scale metabolic reconstructions and theoretical investigation of methane conversion in Methylomicrobium buryatense strain 5G(B1). Microb. Cell Fact. 14:188. doi: 10.1186/s12934-015-0377-3
Duncan, K. E., Gieg, L. M., Parisi, V. A., Tanner, R. S., Tringe, S. G., Bristow, J., et al. (2009). Biocorrosive thermophilic microbial communities in alaskan north slope oil facilities. Environ. Sci. Technol. 43, 7977–7984. doi: 10.1021/es9013932
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M., et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548. doi: 10.1038/nature08883
Ettwig, K. F., Zhu, B., Speth, D., Keltjens, J. T., Jetten, M. S. M., and Kartal, B. (2016). Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. U.S.A. 113, 12792–12796. doi: 10.1073/pnas.1609534113
Fei, Q., Guarnieri, M. T., Tao, L., Laurens, L. M., Dowe, N., and Pienkos, P. T. (2014). Bioconversion of natural gas to liquid fuel: opportunities and challenges. Biotechnol. Adv. 32, 596–614. doi: 10.1016/j.biotechadv.2014.03.011
Flamholz, A., Noor, E., Bar-Even, A., and Milo, R. (2012). eQuilibrator—the biochemical thermodynamics calculator. Nucleic Acids Res. 40, D770–D775. doi: 10.1093/nar/gkr874
Ghoddusi, H. (2017). Blending under uncertainty: real options analysis of ethanol plants and biofuels mandates. Energy Econ. 61, 110–120. doi: 10.1016/j.eneco.2016.11.007
Green, E. M. (2011). Fermentative production of butanol—the industrial perspective. Curr. Opin. Biotechnol. 22, 337–343. doi: 10.1016/j.copbio.2011.02.004
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
Haynes, C. A., and Gonzalez, R. (2014). Rethinking biological activation of methane and conversion to liquid fuels. Nat. Chem. Biol. 10, 331–339. doi: 10.1038/nchembio.1509
Holmen, A. (2009). Direct conversion of methane to fuels and chemicals. Catal. Today 142, 2–8. doi: 10.1016/j.cattod.2009.01.004
Hwang, I. Y., Nguyen, A. D., Nguyen, T. T., Nguyen, L. T., Lee, O. K., and Lee, E. Y. (2018). Biological conversion of methane to chemicals and fuels: technical challenges and issues. Appl. Microbiol. Biotechnol. 102, 3071–3080. doi: 10.1007/s00253-018-8842-7
Jang, Y. S., Kim, B., Shin, J. H., Choi, Y. J., Choi, S., Song, C. W., et al. (2012). Bio-based production of C2–C6 platform chemicals. Biotechnol. Bioeng. 109, 2437–2459. doi: 10.1002/bit.24599
Kalyuzhnaya, M. G., Puri, A. W., and Lidstrom, M. E. (2015). Metabolic engineering in methanotrophic bacteria. Metab. Eng. 29, 142–152. doi: 10.1016/j.ymben.2015.03.010
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. doi: 10.1093/nar/gkw1092
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Kirby, J., Dietzel, K. L., Wichmann, G., Chan, R., Antipov, E., Moss, N., et al. (2016). Engineering a functional 1-deoxy-D-xylulose 5-phosphate (DXP) pathway in Saccharomyces cerevisiae. Metab. Eng. 38, 494–503. doi: 10.1016/j.ymben.2016.10.017
Koch, C., Kuchenbuch, A., Kracke, F., Bernhardt, P. V., Krömer, J., and Harnisch, F. (2017). Predicting and experimental evaluating bio-electrochemical synthesis — a case study with Clostridium kluyveri. Bioelectrochemistry 118, 114–122. doi: 10.1016/j.bioelechem.2017.07.009
Kracke, F., Virdis, B., Bernhardt, P. V., Rabaey, K., and Krömer, J. O. (2016). Redox dependent metabolic shift in Clostridium autoethanogenum by extracellular electron supply. Biotechnol. Biofuels 9:249. doi: 10.1186/s13068-016-0663-2
Liew, F., Martin, M. E., Tappel, R. C., Heijstra, B. D., Mihalcea, C., and Köpke, M. (2016). Gas fermentation—a flexible platform for commercial scale production of low-carbon-fuels and chemicals from waste and renewable feedstocks. Front. Microbiol. 7:694. doi: 10.3389/fmicb.2016.00694
Lopar, M., Špoljarić, I. V., Atlić, A., Koller, M., Braunegg, G., and Horvat, P. (2013). Five-step continuous production of PHB analyzed by elementary flux, modes, yield space analysis and high structured metabolic model. Biochem. Eng. J. 79, 57–70. doi: 10.1016/j.bej.2013.07.003
Lopar, M., Špoljarić, I. V., Cepanec, N., Koller, M., Braunegg, G., and Horvat, P. (2014). Study of metabolic network of Cupriavidus necator DSM 545 growing on glycerol by applying elementary flux modes and yield space analysis. J. Ind. Microbiol. Biotechnol. 41, 913–930. doi: 10.1007/s10295-014-1439-y
Lunsford, J. H. (2000). Catalytic conversion of methane to more useful chemicals and fuels: a challenge for the 21st century. Catal. Today 63, 165–174. doi: 10.1016/S0920-5861(00)00456-9
Marx, C. J., Chistoserdova, L., and Lidstrom, M. E. (2003). Formaldehyde-detoxifying role of the tetrahydromethanopterin-linked pathway in methylobacterium extorquens AM1. J. Bacteriol. 185, 7160–7168. doi: 10.1128/JB.185.23.7160-7168.2003
Melzer, G., Esfandabadi, M. E., Franco-Lara, E., and Wittmann, C. (2009). Flux design: In silico design of cell factories based on correlation of pathway fluxes to desired properties. BMC Syst. Biol. 3:120. doi: 10.1186/1752-0509-3-120
Meyer, F., Keller, P., Hartl, J., Gröninger, O. G., Kiefer, P., and Vorholt, J. A. (2018). Methanol-essential growth of Escherichia coli. Nat. Commun. 9:1508. doi: 10.1038/s41467-018-03937-y
Miltner, M., Makaruk, A., and Harasek, M. (2017). Review on available biogas upgrading technologies and innovations towards advanced solutions. J. Clean. Prod. 161, 1329–1337. doi: 10.1016/j.jclepro.2017.06.045
Moran, J. J., House, C. H., Freeman, K. H., and Ferry, J. G. (2005). Trace methane oxidation studied in several Euryarchaeota under diverse conditions. Archaea 1, 303–309. doi: 10.1155/2005/650670
Mueller, T. J., Grisewood, M. J., Nazem-Bokaee, H., Gopalakrishnan, S., Ferry, J. G., Wood, T. K., et al. (2015). Methane oxidation by anaerobic archaea for conversion to liquid fuels. J. Ind. Microbiol. Biotechnol. 42, 391–401. doi: 10.1007/s10295-014-1548-7
Nakano, M. M., and Zuber, P. (1998). Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52, 165–190. doi: 10.1146/annurev.micro.52.1.165
Nazem-Bokaee, H., Gopalakrishnan, S., Ferry, J. G., Wood, T. K., and Maranas, C. D. (2016). Assessing methanotrophy and carbon fixation for biofuel production by Methanosarcina acetivorans. Microb. Cell Fact. 15:10. doi: 10.1186/s12934-015-0404-4
Nishimura, T., Vertès, A. A., Shinoda, Y., Inui, M., and Yukawa, H. (2007). Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Appl. Microbiol. Biotechnol. 75, 889–897. doi: 10.1007/s00253-007-0879-y
Noor, E., Bar-Even, A., Flamholz, A., Reznik, E., Liebermeister, W., and Milo, R. (2014). Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput. Biol. 10:e1003483. doi: 10.1371/journal.pcbi.1003483
Peralta-Yahya, P. P., Zhang, F., Del Cardayre, S. B., and Keasling, J. D. (2012). Microbial engineering for the production of advanced biofuels. Nature 488, 320–328. doi: 10.1038/nature11478
Placzek, S., Schomburg, I., Chang, A., Jeske, L., Ulbrich, M., Tillack, J., et al. (2017). BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 45, D380–D388. doi: 10.1093/nar/gkw952
Raghoebarsing, A. A., Pol, A., Van De Pas-Schoonen, K. T., Smolders, A. J. P., Ettwig, K. F., Rijpstra, W. I., et al. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921. doi: 10.1038/nature04617
Report, M. R. (2016). Isobutanol Market Analysis by Product (Synthetic, Bio-Based), Application (Oil & Gas, Solvents & Coatings, Chemical Intermediates) and Segment Forecasts to 2022. Available online at: https://www.grandviewresearch.com/industry-analysis/isobutanol-market (Accessed January 10, 2018).
Report, M. R. (2017). Fuel Ethanol Market Analysis by Product (Starch-Based, Sugar-Based, Cellulosic), by Application (Conventional Vehicles, Flexible Fuel Vehicles), by Region, and Segment Forecasts, 2018–2025. Available online at: https://www.grandviewresearch.com/industry-analysis/fuel-ethanol-market (Accessed January 10, 2018).
Schada von Borzyskowski, L., Carrillo, M., Leupold, S., Glatter, T., Kiefer, P., Weishaupt, R., et al. (2018). An engineered Calvin-Benson-Bassham cycle for carbon dioxide fixation in Methylobacterium extorquens AM1. Metab. Eng. 47, 423–433. doi: 10.1016/j.ymben.2018.04.003
Scheller, S., Ermler, U., and Shima, S. (2017). “Catabolic pathways and enzymes involved in anaerobic methane oxidation,” in Anaerobic Utilization of Hydrocarbons, Oils, and Lipids. Handbook of Hydrocarbon and Lipid Microbiology, ed M. Boll (Cham: Springer), 1–29. Available online at: https://link.springer.com/referenceworkentry/10.1007%2F978-3-319-33598-8_3-1
Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K., and Jaun, B. (2010). The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608. doi: 10.1038/nature09015
Schrader, J., Schilling, M., Holtmann, D., Sell, D., Villela Filho, M., Marx, A., et al. (2009). Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol. 27, 107–115. doi: 10.1016/j.tibtech.2008.10.009
Shima, S., Krueger, M., Weinert, T., Demmer, U., Kahnt, J., Thauer, R. K., et al. (2011). Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature 481, 98–101. doi: 10.1038/nature10663
Siegel, J. B., Smith, A. L., Poust, S., Wargacki, A. J., Bar-Even, A., Louw, C., et al. (2015). Computational protein design enables a novel one-carbon assimilation pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 3704–3709. doi: 10.1073/pnas.1500545112
Soo, V. W., Mcanulty, M. J., Tripathi, A., Zhu, F., Zhang, L., Hatzakis, E., et al. (2016). Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb. Cell Fact. 15:11. doi: 10.1186/s12934-015-0397-z
Steynberg, A. (2004). “Chapter 1 – Introduction to fischer-tropsch technology,” in Studies in Surface Science and Catalysis, Vol. 52. eds A. Steynberg and M. Dry (Elsevier), 1–63. Available online at: https://www.sciencedirect.com/science/article/pii/S0167299104804580
Strong, P. J., Xie, S., and Clarke, W. P. (2015). Methane as a resource: can the methanotrophs add value? Environ. Sci. Technol. 49, 4001–4018. doi: 10.1021/es504242n
Ternon, C., Grousseau, E., Gunther, J., Gorret, N., Guillouet, S., Sinskey, A., et al. (2014). Dynamic model for isopropanol production by Cupriavidus necator. IFAC Proc. Vol. 47, 4388–4393. doi: 10.3182/20140824-6-ZA-1003.02267
Terzer, M., and Stelling, J. (2008). Large-scale computation of elementary flux modes with bit pattern trees. Bioinformatics 24, 2229–2235. doi: 10.1093/bioinformatics/btn401
Thauer, R. K., and Shima, S. (2008). Methane as fuel for anaerobic microorganisms. Ann. N. Y. Acad. Sci. 1125, 158–170. doi: 10.1196/annals.1419.000
Tiemeyer, A., Link, H., and Weuster-Botz, D. (2007). Kinetic studies on autohydrogenotrophic growth of Ralstonia eutropha with nitrate as terminal electron acceptor. Appl. Microbiol. Biotechnol. 76, 75–81. doi: 10.1007/s00253-007-0983-z
Toolbox (2008). Solubility of Gases in Water. Available online at: https://www.engineeringtoolbox.com/gases-solubility-water-d_1148.html (Accessed May 04, 2018).
Trotsenko, Y. A., and Murrell, J. C. (2008). “Metabolic aspects of aerobic obligate methanotrophy,” in Advances in Applied Microbiology, Vol. 63. (Academic Press), 183–229. Available online at: https://www.sciencedirect.com/science/article/pii/S0065216407000056?via%3Dihub
Unden, G., and Bongaerts, J. (1997). Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320, 217–234. doi: 10.1016/S0005-2728(97)00034-0
Unrean, P. (2014). Pathway analysis of Pichia pastoris to elucidate methanol metabolism and its regulation for production of recombinant proteins. Biotechnol. Prog. 30, 28–37. doi: 10.1002/btpr.1855
Valentine, D. L., and Reeburgh, W. S. (2000). New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2, 477–484. doi: 10.1046/j.1462-2920.2000.00135.x
van Klinken, J. B., and Willems Van Dijk, K. (2015). FluxModeCalculator: an efficient tool for large-flux mode computation. Bioinformatics 32, 1265–1266. doi: 10.1093/bioinformatics/btv742
Weiland, P. (2010). Biogas production: current state and perspectives. Appl. Microbiol. Biotechnol. 85, 849–860. doi: 10.1007/s00253-009-2246-7
West, C. A., Salmond, G. P. C., Dalton, H., and Murrell, J. C. (1992). Functional expression in Escherichia coli of proteins B and C from soluble methane monooxygenase of Methylococcus capsulatus (Bath). Microbiology 138, 1301–1307.
Yan, Z., Joshi, P., Gorski, C. A., and Ferry, J. G. (2018). A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration. Nat. Commun. 9:1642. doi: 10.1038/s41467-018-04097-9
Keywords: gas fermentation, methane upgrading, bio-GTL, elementary flux mode analysis, metabolic modeling, anaerobic methane oxidation, microbial CO2 fixation
Citation: Averesch NJH and Kracke F (2018) Metabolic Network Analysis of Microbial Methane Utilization for Biomass Formation and Upgrading to Bio-Fuels. Front. Energy Res. 6:106. doi: 10.3389/fenrg.2018.00106
Received: 02 June 2018; Accepted: 24 September 2018;
Published: 15 October 2018.
Edited by:
Andrea Schievano, Università degli Studi di Milano, ItalyReviewed by:
Andrea Franzetti, Università degli studi di Milano Bicocca, ItalyMohanakrishna Gunda, Qatar University, Qatar
Copyright © 2018 Averesch and Kracke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nils J. H. Averesch, bmlscy5hdmVyZXNjaEB1cS5uZXQuYXU=
†These authors have contributed equally to this work
 Nils J. H. Averesch
Nils J. H. Averesch Frauke Kracke
Frauke Kracke