- 1Department of Gastroenterology, First Affiliated Hospital of Henan University of Chinese Medicine, Zhengzhou, China
- 2The First Clinical Medical College of Henan University of Chinese Medicine, Zhengzhou, China
- 3Department of Gastroenterology, Changzhou Hospital of Traditional Chinese Medicine, Changzhou, China
Objective: Previous studies have suggested a potential association between gut microbiota and the development of alcohol-related liver disease (ALD). However, the causal relationship between gut microbiota and ALD, as well as the role of inflammatory cytokines as mediators, remains unclear. This study aims to explore the causal relationship between gut microbiota and ALD using Mendelian randomization (MR) methods, and to analyze the mediating role of inflammatory cytokines.
Methods: Gut microbiota, 91 inflammatory cytokines, and ALD were identified from summary data of large-scale genome-wide association studies (GWAS). MR was employed to investigate the causal relationship between gut microbiota, cytokines, and ALD, with the inverse variance-weighted method (IVW) as the primary statistical approach. Additionally, we examined whether inflammatory cytokines act as mediating factors in the pathway from gut microbiota to ALD.
Results: The IVW results confirmed two positive and one negative causal effect between genetic liability in the gut microbiota and ALD. Escherichia coli (P= 0.003) was identified as a protective factor for ALD, while Roseburia hominis (P=0.023) and Family Porphyromonadaceae (P=0.038) were identified as risk factors for ALD. Furthermore, there were five positive and two negative causal effects between inflammatory cytokines and ALD, with CUB domain-containing protein 1 (P= 0.035), Macrophage colony-stimulating factor 1 (P=0.047), Cystatin D (P = 0.035), Fractalkine (P=0.000000038), Monocyte chemoattractant protein-1 (P=0.004) positively associated with ALD onset. CD40L receptor (P= 0.044) and Leukemia inhibitory factor (P = 0.024) exhibited protective effects against ALD. Mediation MR analysis indicated that CUB domain-containing protein 1 (mediation proportion=1.6%, P=0.035), Cystatin D (mediation proportion=1.5%, P=0.012), and Monocyte chemoattractant protein-1 (mediation proportion=3.3%, P=0.005) mediated the causal effect of Roseburia hominis on ALD.
Conclusion: In conclusion, our study supports a causal relationship among gut microbiota, inflammatory cytokines and ALD, with inflammatory cytokines potentially acting as mediating factors in the pathway from gut microbiota to ALD.
1 Introduction
Alcohol-related liver disease (ALD) refers to liver diseases primarily caused by long-term heavy alcohol consumption, which includes asymptomatic hepatic steatosis, fibrosis, cirrhosis, alcoholic hepatitis, and their complications. Approximately 8-20% of chronic alcohol drinkers develop ALD with cirrhosis, and about 2% progress to hepatocellular carcinoma (1). Epidemiological research has demonstrated that ALD accounts for approximately 19% of global mortality attributable to alcohol-related liver cancer, and up to 25% of deaths from cirrhosis secondary to alcohol related liver disease (2, 3). Most patients with ALD do not exhibit overt symptoms during the early inflammatory phase prior to the development of cirrhosis. However, once the disease progresses to the cirrhotic stage, they may develop serious clinical manifestations including ascites, gastrointestinal bleeding, and edema, which are associated with a poor overall prognosis. The onset of ALD is typically insidious, characterized by nonspecific clinical symptoms in the early stages. Existing evidence suggests the pathogenesis of alcoholic liver disease involves complex interactions between various cell types and organ systems. Importantly, modulation of the gut microbiota has emerged as a potential therapeutic target for ALD (4).
The liver possesses remarkable self-repair capabilities that rely on the coordinated function of diverse cell types and extracellular factors (5). Additionally, there is a well-established connection between the liver and intestines via the portal vein, the biliary system and circulating mediators, allowing for bidirectional microbial interactions between the gut and liver (6). Consequently, the crosstalk between the gut microbiome and the liver has received increasing attention in context (7).
The human gut microbiome is characterized by vast diversity, with the majority of bacteria belonging to the phyla Firmicutes (60-80%), Bacteroidetes (20-40%), Proteobacteria, Actinobacteria, Verrucomicrobia, Fusobacteria, and Cyanobacteria (8). Alterations in the relative abundance of these bacterial phyla have been shown to impact various dimensions of human health (9). Studies indicated that Patients with alcohol-use disorder and liver disease exhibit reduced gut bacterial diversity, with decreased proportions of several beneficial bacteria, including Lactobacillus, Bifidobacterium, Prevotella, and Akkermansia muciniphila. In addition, these patients demonstrate shifts in gut microbiome composition, with decreased fungal diversity and increased proportions and quantities of Candida (10). Specifically, Grander et al. found a decreased abundance of the symbiont A. mucinip hila in patients with ALD, and supplementation of this bacterium significantly improved ethanol-induced intestinal and liver damage in mice (11). Furthermore, Duan et al. discovered a correlation between the severity of ALD and the cellulase activity of fecal Enterococcus, and targeting these bacteria with bacteriophages reduced cellulase levels in the liver and ameliorated liver disease in humanized mouse models (12). In ALD, the gut microbiota promotes inflammation along the gut-liver axis, while the presence of inflammatory cytokines may further accelerate disease progression (13). Establishing a causal relationship between ALD and gut microbiome alterations has proven challenging in previous studies, owing to the influence of confounding factors such as ethanol exposure, inflammation, and the complex interplay between these variables.
To overcome the limitations of confounding and reverse causality inherent in observational epidemiological studies, elucidate the causal relationships between multiple factors in the progression of the disease condition. The Mendelian randomization (MR) method, using single nucleotide polymorphisms (SNPs) strongly associated with exposure as instrumental variables (IV), has been increasingly accepted and utilized to assess causal effects between exposure and outcomes (14). The genetic variations used as instrumental variables in MR are fixed at conception, allowing researchers to make causal inferences about the impact of modifiable risk factors while overcoming the influence of certain confounding factors (15). Early genetic studies have suggested that host genetic variations can influence the composition of the gut microbiota, providing a foundation for employing the MR method to investigate the causal relationship between gut microbiota and ALD, as well as the potential mediating role of inflammatory processes. In this study, we aim to leverage summary data from genome-wide association studies (GWAS) and implement a bidirectional and mediation MR design across two independent samples. This approach will enable us to explore the causal relationship between gut microbiota and ALD, as well as elucidate the mediating role of inflammatory factors in this relationship.
2 Materials and methods
2.1 Study design
This study will consist of three main components. First, analyzing the causal effects of gut microbiota composition, comprising 207 distinct taxa, on the development of ALD using MR (Path A); second, investigating the causal impact of 91 inflammatory cytokines on the risk of ALD, also employing the MR approach (Path B); and third, examining the potential mediating role of the 91 cytokines in the causal pathway from gut microbiota composition to ALD (Path C). MR analysis will leverage SNPs as instrumental variables (IVs). This method relies on three core assumptions (16): (1) The selected SNPs must be strongly associated with the exposure factors of interest; (2) The SNPs must be independent of confounding factors; (3) The SNPs must influence the outcome only through their effect on the exposure, and not directly (Refer to Figure 1 for a detailed flowchart of the study design) (16).
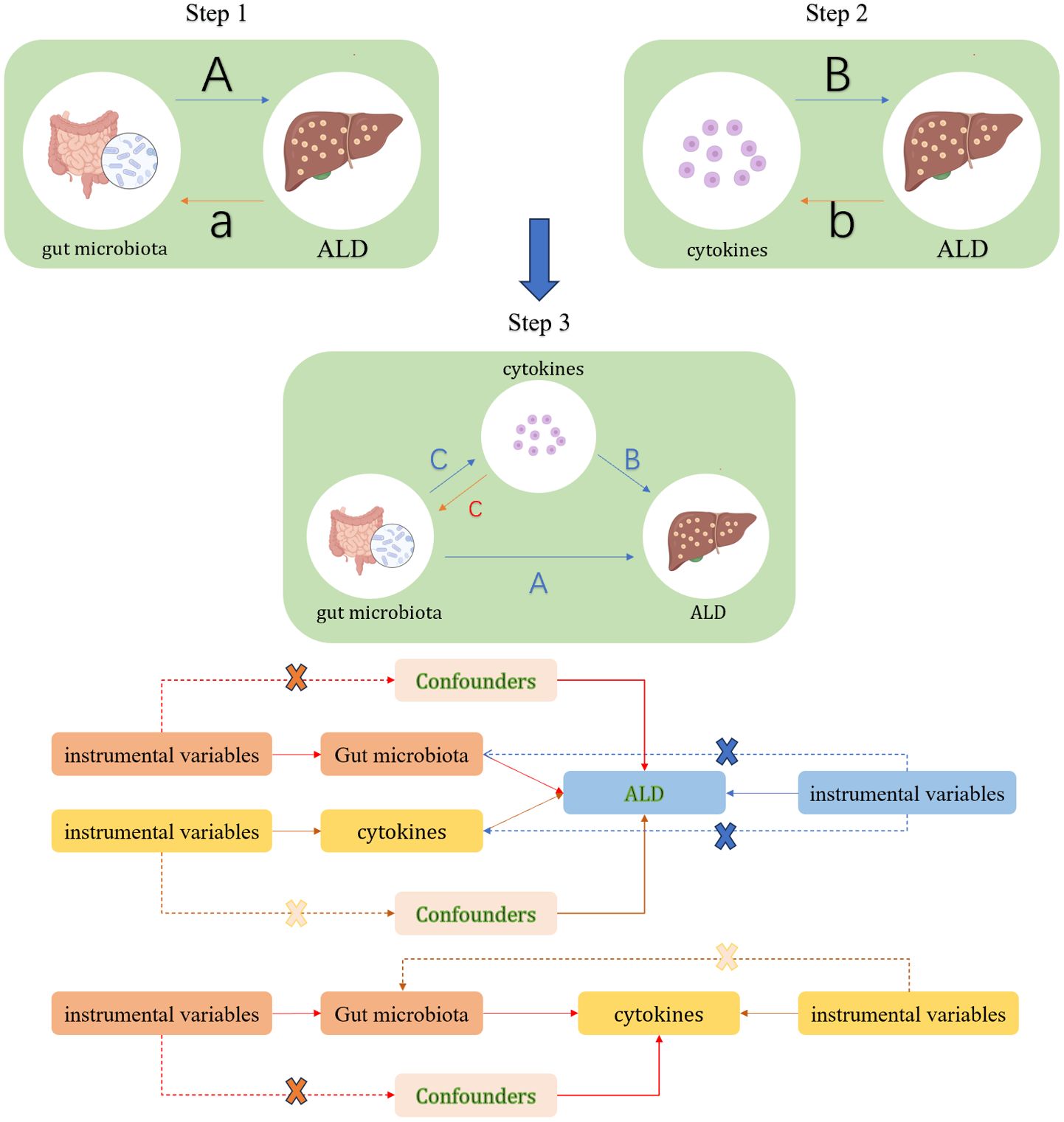
Figure 1. Study Overview. In Step 1, path A represents the causal impact of gut microbiota on ALD, while path a denotes the reverse causal effect of ALD on gut microbiota. In Step 2, path B signifies the causal impact of cytokines on ALD, with path b indicating the reverse causal effect between ALD and cytokines. Step 3 involves the mediation analysis of cytokines in the pathway from gut microbiota to ALD: Path A represents the overall impact of gut microbiota on ALD; Path B signifies the causal effect of cytokines on ALD; Path C indicates the causal effect of gut microbiota on cytokines, and path c denotes the reverse causal effect of cytokines on gut microbiota.
2.2 Data source
Genetic data on gut microbiome were obtained from the Dutch Microbiome Project (DMP) genome-wide association study (GWAS) of 7,738 individuals of Dutch ancestry (17, 18). The data included 207 taxonomic groups spanning 5 phyla, 10 classes, 13 orders, 26 families, 48 genera, and 105 species. Data on 91 inflammatory cytokines were sourced from a GWAS of 14,824 individuals of European descent across 11 cohorts, and these individuals are all from England (accession numbers GCST90274758 to GCST90274848) (19).
Large-scale GWAS meta-analysis data on ALD were obtained from the FinnGen consortium (https://www.finngen.fi/fi) (17). The FinnGen consortium is one of the largest GWAS databases globally, with a sample size of 2,408 available disease phenotypes, and it aims to study the genome and national health register data of 500,000 Finnish individuals. For this study, the ALD phenotype data were sourced from the FinnGen GWAS summary statistics, which are publicly available through the IEU OPEN GWAS PROJECT (https://gwas.mrcieu.ac.uk/), with the dataset coded as finn-b-K11_ALCOLIV.
This study involves secondary analysis of publicly available GWAS summary-level data, all of which have received the appropriate ethical approvals from the respective cohorts and consortia. As the study does not utilize individual-level participant data, no additional ethical review board approval is required.
2.3 Instrumental variables selection
Previous studies have demonstrated that independent SNPs form the basis of MR analyses (20). We selected SNPs with significant associations (P < 1×10-5) and an effect allele frequency (EAF) > 0.01 with gut microbiota and inflammatory protein factors to serve as instrumental variables. During the analysis, due to the limited number of inflammatory cytokines reaching the conventional genome-wide significance threshold of P < 5×10-8 for independent SNPs, the significance threshold was adjusted to P < 1×10-5. This was followed by linkage disequilibrium analysis (LDA) to ensure the independence of the selected SNPs, using a standard threshold of r2 < 0.1 within a 500kb window. An important step in MR analysis is ensuring that the effect allele for the association between the SNPs and the exposure corresponds to the effect allele for the association between the SNPs and the outcome. After this matching process, we removed any palindromic SNPs. (A palindromic SNP is an SNP with the A/T or G/C allele.) from the analysis.
2.4 MR analysis
2.4.1 Primary analysis
To assess the causal impact of gut microbiota and inflammatory cytokines on ALD, we conducted analyses using the Two Sample MR (21), MRPRESSO, and MR packages in R studio (version 4.3.2). Initially, we performed MR analyses on two separate samples (steps A and B in Figure 1). During the analysis, when the number of instrumental variable SNPs was greater than or equal to 2, we opted for the Inverse Variance Weighted (IVW) method to estimate the potential causal effects of gut microbiota or inflammatory cytokines on ALD. In cases where the number of instrumental variable SNPs was less than 2, we employed the Wald ratio method for sensitivity analysis, a common and precise approach in MR (22). The MR results were presented as Odds Ratios (OR) and their corresponding 95% Confidence Intervals (CI). Statistical significance was considered when the P-value of IVW was less than 0.05 and the direction of the IVW estimate was consistent with the MR-Egger direction.
2.4.2 Mediation analysis
In the mediation analysis, gut microbiota and inflammatory cytokines with significant causal effects on ALD from the two-sample MR analysis were included. We explored whether there was a causal relationship between gut microbiota and inflammatory cytokines (step 3, path C in Figure 1). If a causal effect was found between the two, multiple MR analyses would be conducted to investigate whether inflammatory cytokines act as a mediator in the pathway from gut microbiota to ALD.
2.4.3 Bidirectional causal analysis
To evaluate the bidirectional causal effects among gut microbiota, inflammatory cytokines, and ALD, we used ALD as the “exposure” and gut microbiota or inflammatory cytokines associated with ALD as the “outcome” (paths a and b in Figure 1). SNPs significantly associated with ALD (P < 1×10-5) were selected as instrumental variables (IVs).
2.4.4 Sensitivity analysis
To assess the robustness of the IVs, we conducted heterogeneity assessment for each SNP using Cochran’s Q test. Subsequently, we performed leave-one-out analysis by sequentially excluding each SNP and applying the IVW method to the remaining SNPs to evaluate the potential impact of specific variants on the estimates. Scatter plots of SNP-exposure associations and SNP-outcome associations, along with leave-one-out plots, were generated to visualize the MR results (23). Additionally, we employed MR-Egger intercept and MR-PRESSO methods to detect horizontal pleiotropy (24). In MR-Egger analysis, a p-value < 0.05 indicates the presence of pleiotropy (25). The MR-PRESSO method can identify potential outliers and correct for pleiotropic effects by removing these outliers.
In summary, our study utilized a two-step MR approach to assess the potential mediating role of inflammatory cytokines on the relationship between gut microbiota and ALD. We aimed to determine the microbial taxa showing a causal relationship with ALD through their impact on inflammatory cytokines. Univariable MR analyses were conducted to assess the causal effects of gut microbiota on inflammatory cytokines (Beta1), inflammatory cytokines on ALD (Beta2), and gut microbiota on ALD (Beta3). The proportion of the total effect mediated by inflammatory cytokines was estimated by dividing the indirect effect by the total effect (Beta1×Beta2/Beta3).
3 Results
3.1 Results of the weak instrumental variable test
To mitigate the bias risks stemming from weak instrumental variables, we computed the general F-statistics for each exposure factor. The F-statistics for all SNPs used as instruments for the gut microbiome exposure ranged from 19.51 to 60.95 (Supplementary Table S1), while the F-statistics for the SNPs used as instruments for the inflammatory cytokines exposure ranged between 19.51 and 2058.59 (Supplementary Table S2). Both sets of F-statistics were greater than 10, indicating a high correlation between the instrumental variables and the exposure factors (26).
3.2 Causal relationships between the gut microbiome, inflammatory cytokines and ALD
3.2.1 Impact of the gut microbiome on ALD
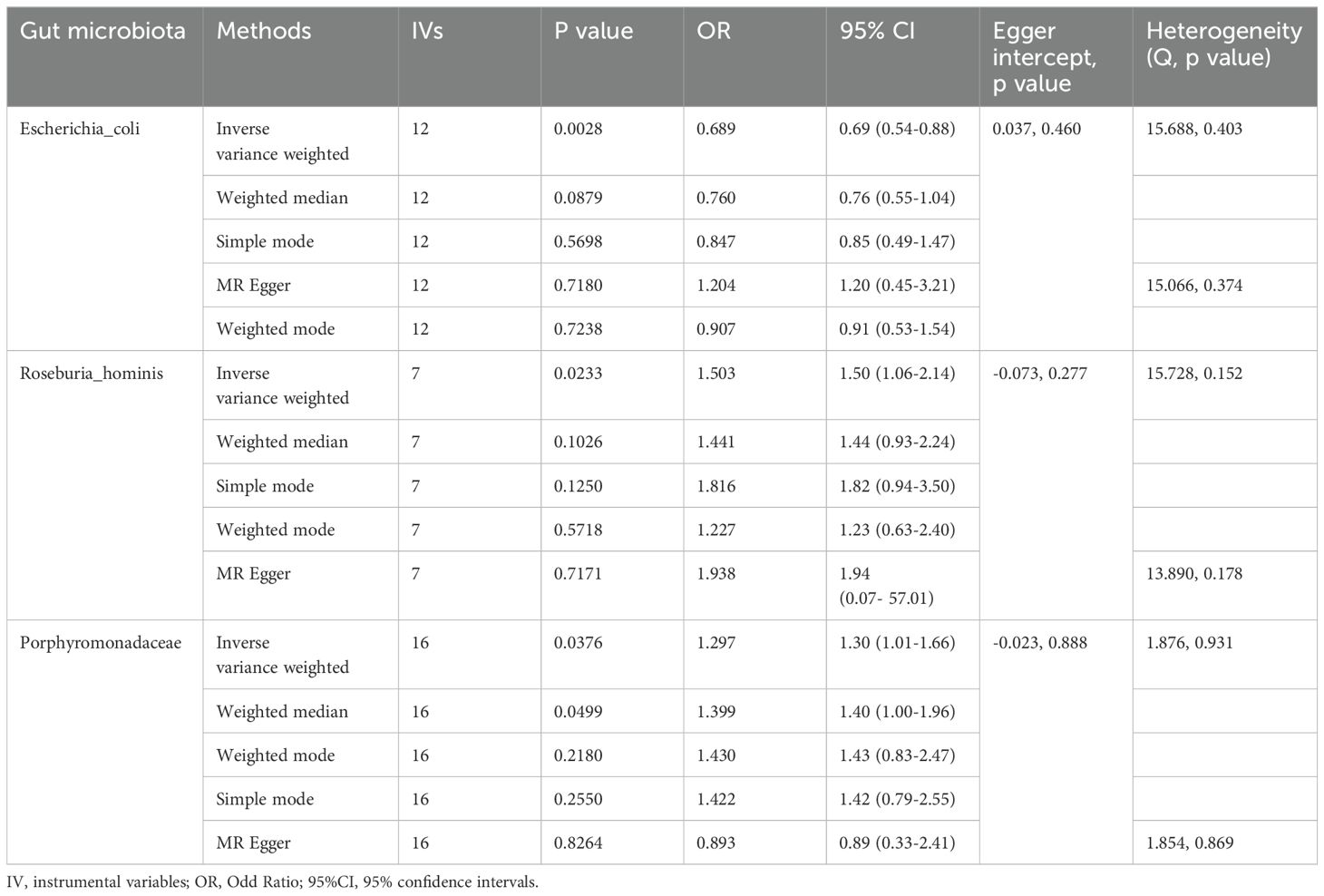
The IVW MR analysis revealed significant causal relationships between ALD and three specific gut microbial taxa: Escherichia coli (OR= 0.69, 95% CI: 0.54-0.88, P= 0.003), Roseburia hominis (OR=1.50, 95% CI: 1.06-2.14, P=0.023), and the bacterial Family Porphyromonadaceae (OR=1.30, 95% CI: 1.01-1.66, P=0.038). The results showed that Escherichia coli had a negative causal relationship with ALD (OR < 1), suggesting a protective effect of this gut microbiome component against the development of ALD (Table 1). On the other hand, Roseburia hominis and the bacterial Family Porphyromonadaceae showed positive causal relationships with ALD (OR > 1), indicating they may act as risk factors for ALD development (Table 1).
Through the MR-Egger regression intercept method, the analysis found no evidence of bias due to genetic pleiotropy in the results. Cochran’s Q test indicated there was no statistically significant heterogeneity (P > 0.05), and the MR-PRESSO analysis revealed no horizontal pleiotropy in this MR study (Figure 2).
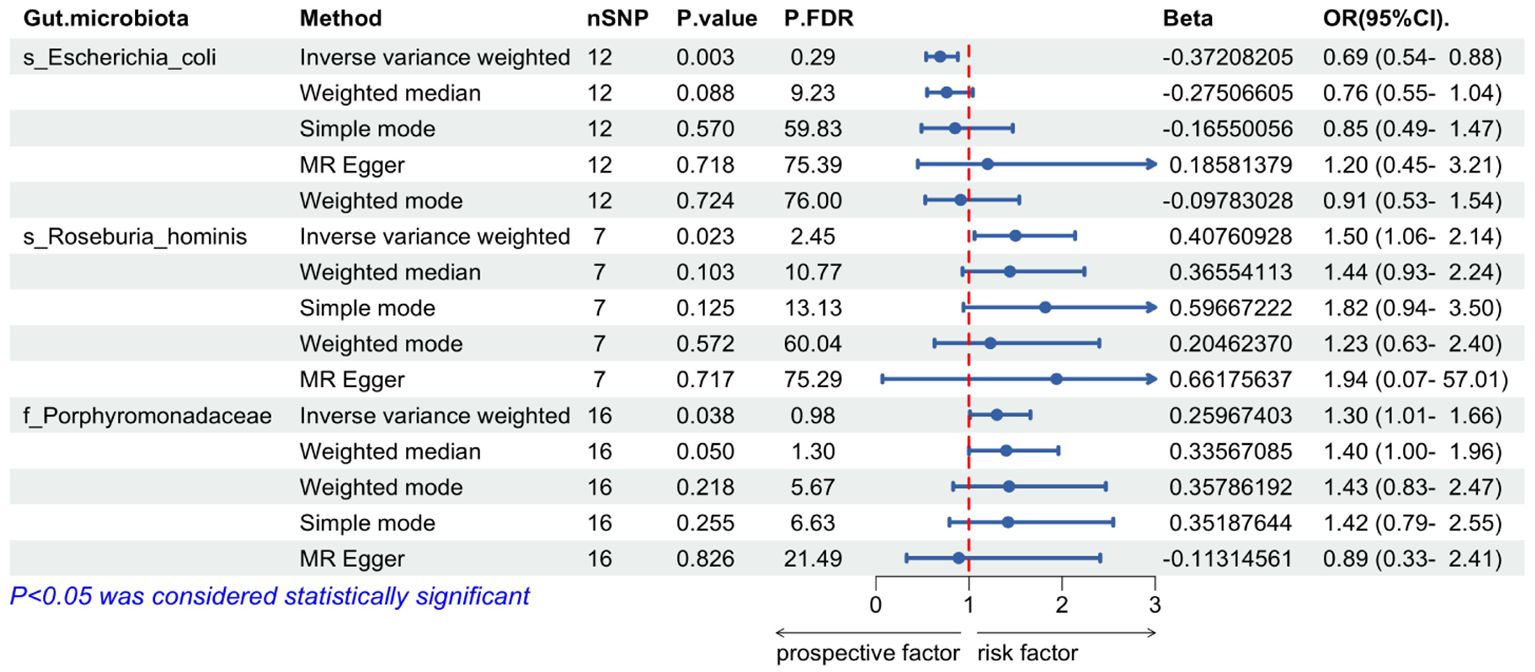
Figure 2. Mendelian randomization of causal effects between gut microbiota and ALD. P.FDR is the p-value adjusted for multiple comparisons using the False Discovery Rate (FDR) method; nSNP refers to the number of genetic variants used as instrumental variables in the MR analysis; Beta refers to the estimated causal effect of the exposure on the outcome.
3.2.2 Influence of inflammatory cytokines on ALD
In our IVW MR analysis of the impact of inflammatory cytokine levels on ALD, we identified seven inflammatory cytokines whose genetically-predicted expressions were significantly causally related to ALD. Among them, five cytokines showed positive causal relationships with ALD——CUB domain-containing protein 1(OR = 1.16, 95% CI: 1.01–1.32, P = 0.035), Macrophage colony-stimulating factor 1(OR = 1.23, 95% CI: 1.00–1.51, P = 0.047), Cystatin D (OR= 1.09, 95%CI: 1.01-1.18, P= 0.035), Fractalkine (OR=1.53, 95%CI: 1.32-1.78, P=0.000000038), Monocyte chemoattractant protein-1 (OR=1.24, 95%, CI: 1.07-1.44, P=0.004),suggesting their potential to increase the risk of ALD (Table 2). Conversely, the genetically-predicted expressions of two inflammatory cytokines, CD40L receptor(OR = 0.89, 95%CI: 0.794–0.997, P = 0.044), Leukemia inhibitory factor(OR = 0.73, 95%CI: 0.55–0.96, P = 0.024)exhibited negative causal relationships with ALD, indicating their potential to reduce the risk of ALD (Table 2).
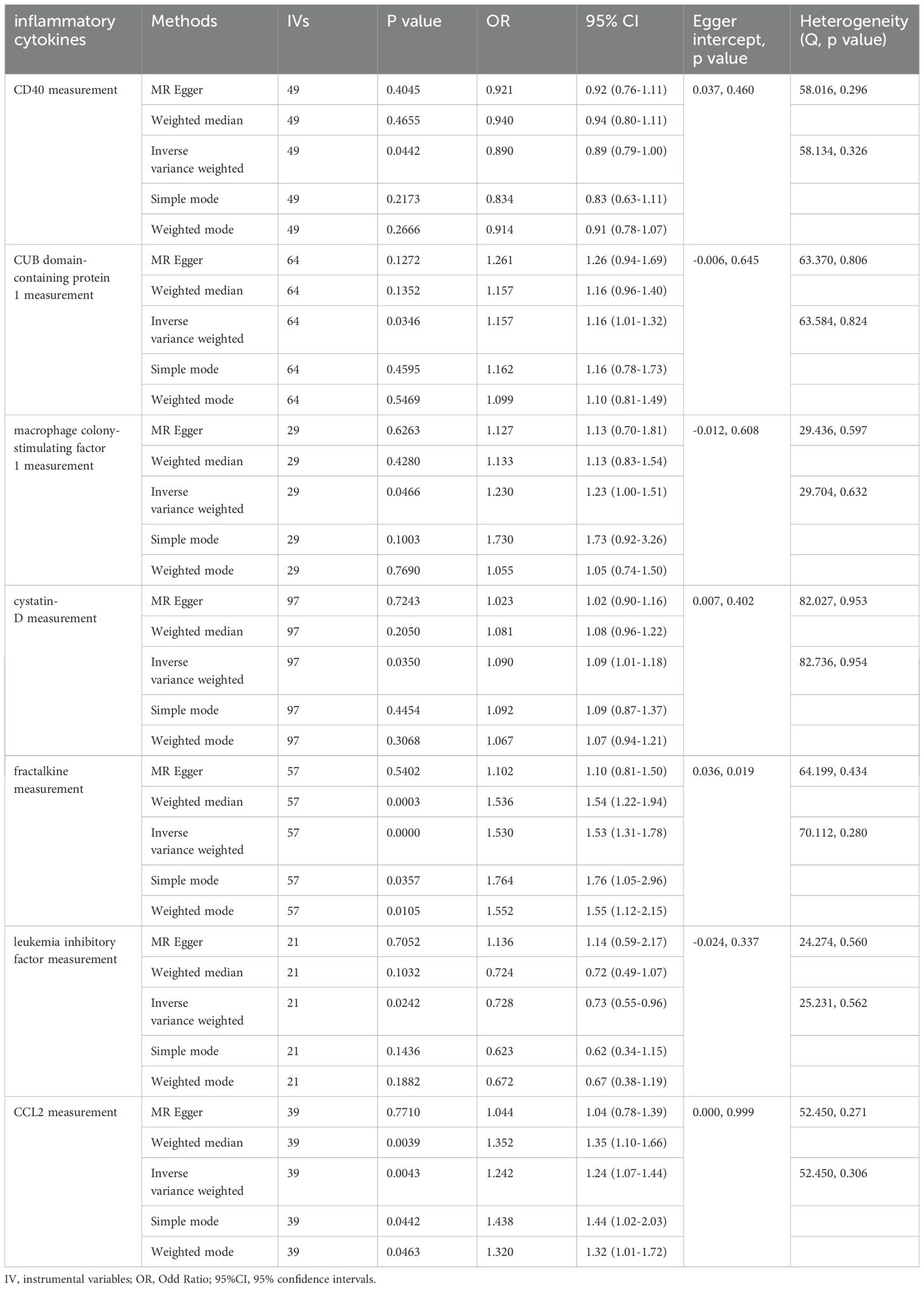
Table 2. Mendelian randomization analysis on the causal effect between inflammatory cytokines and ALD.
To validate these results, sensitivity and pleiotropy analyses were conducted using MR-Egger and Cochran’s Q test methods, which confirmed the absence of significant heterogeneity in the results. Additionally, MR-PRESSO analysis showed P>0.05, indicating no evidence of horizontal pleiotropy in the MR analysis (Figure 3).
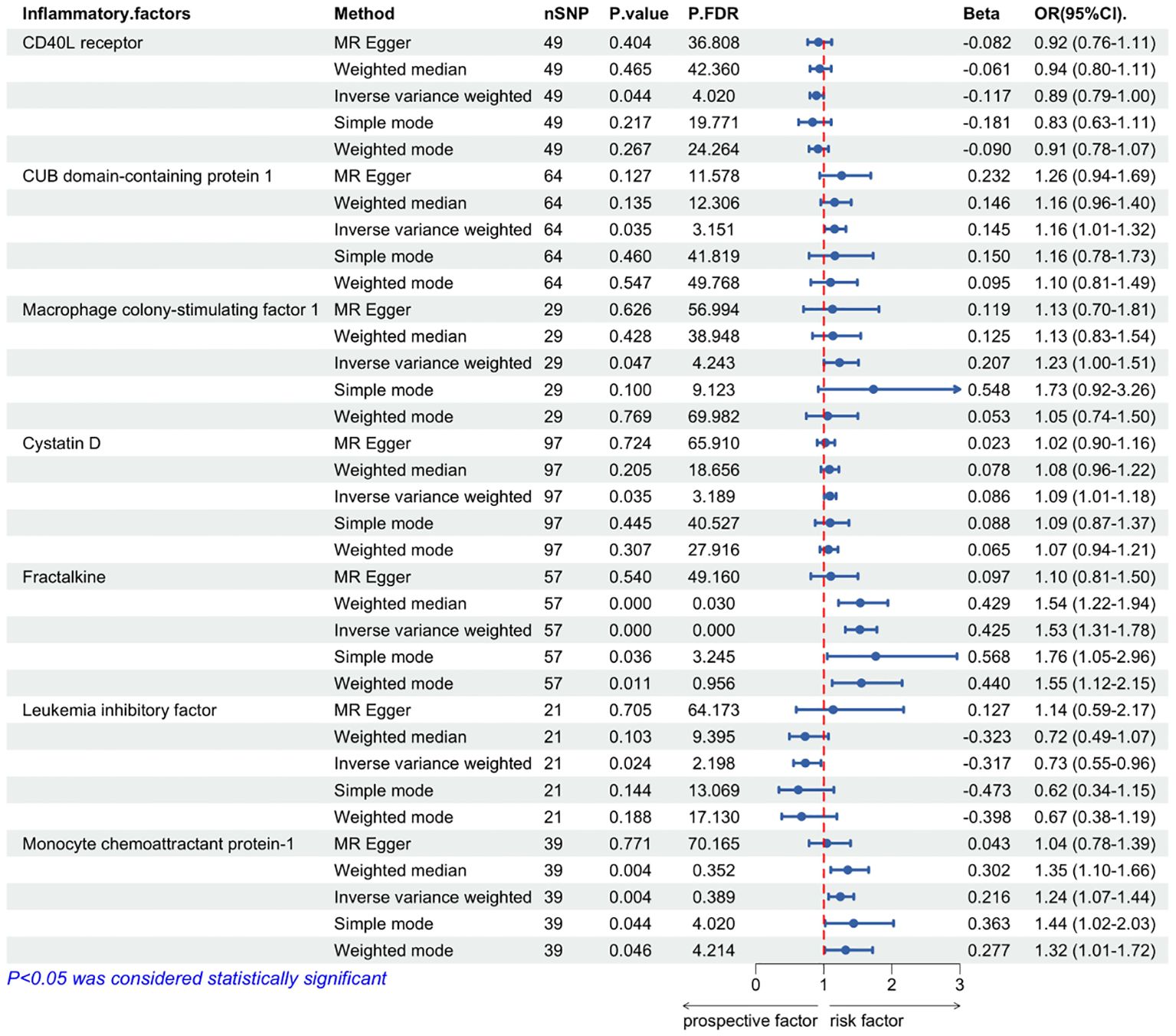
Figure 3. Mendelian randomization of causal effects between inflammatory cytokines and ALD. P.FDR is the p-value adjusted for multiple comparisons using the False Discovery Rate (FDR) method; nSNP refers to the number of genetic variants used as instrumental variables in the MR analysis; Beta refers to the estimated causal effect of the exposure on the outcome.
3.2.3 Influence of gut microbiota on inflammatory cytokines
In the preceding univariate MR analyses, a total of three gut microbiota species and seven inflammatory cytokines were found to have significant causal relationships with ALD, without evidence of heterogeneity or horizontal pleiotropy. Subsequently, we conducted two-sample MR analyses between these three gut microbiota species and seven inflammatory cytokines. The results revealed that two gut microbial species were causally related to four inflammatory cytokines. Specifically, only the genetically-predicted expression of CUB domain-containing protein 1 (OR = 1.07, 95% CI: 1.01–1.13, P = 0.032) showed a positive causal relationship with Escherichia coli, indicating that an increase in Escherichia coli may enhance the expression levels of CUB domain-containing protein 1. Additionally, four inflammatory cytokines were found to have causal relationships with the gut microbiome component Roseburia hominis: CD40L receptor (OR = 1.21, 95% CI: 1.07–1.36, P = 0.002), CUB domain-containing protein 1 (OR = 1.12, 95% CI: 1.01–1.24, P = 0.035), Cystatin D (OR = 1.19, 95% CI: 1.04–1.37, P = 0.012), and Monocyte chemoattractant protein-1 (OR = 1.17, 95% CI: 1.05–1.30, P= 0.005) (Table 3).
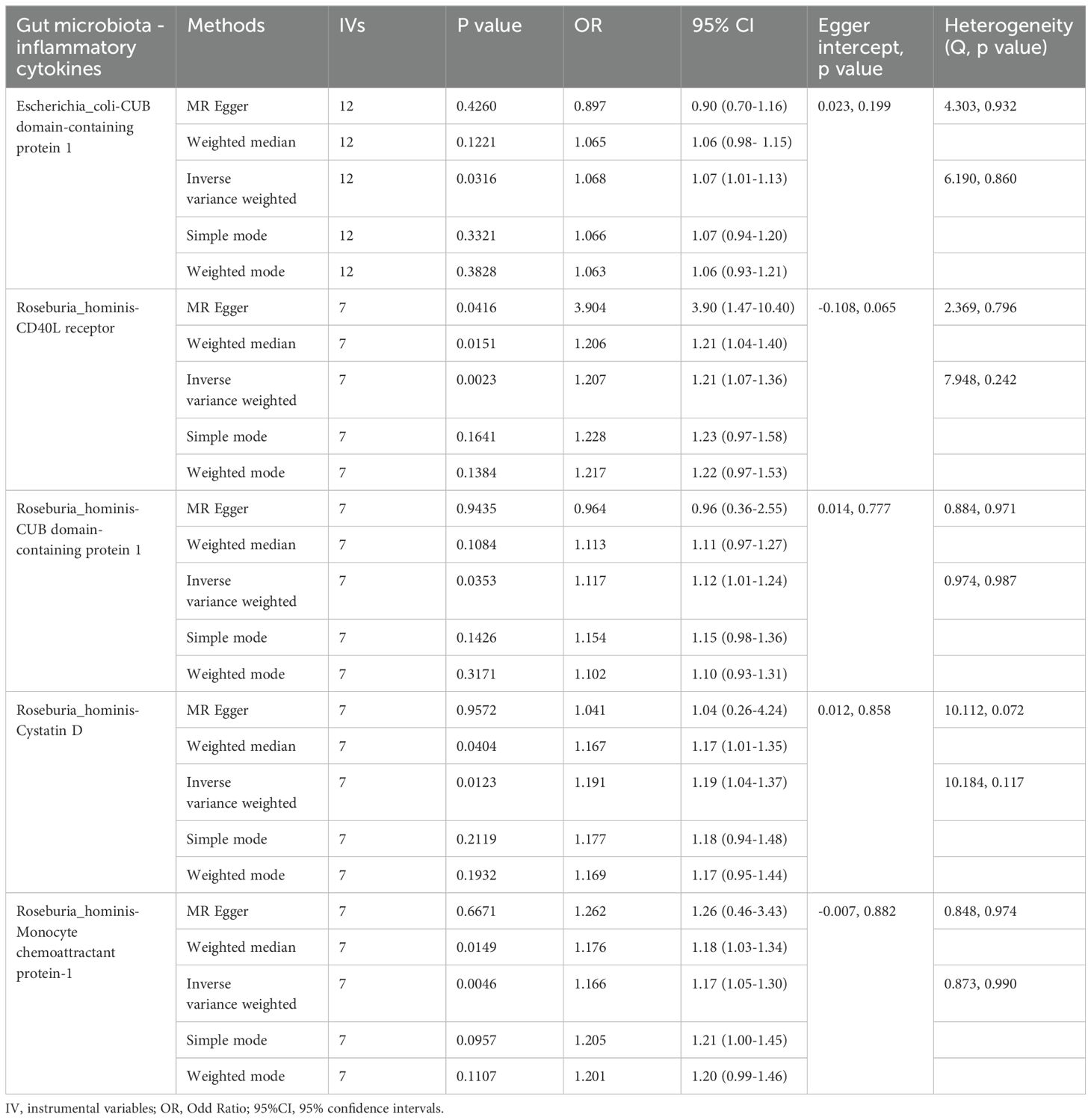
Table 3. Mendelian randomization analysis on the causal effect between gut microbiota and inflammatory cytokines.
3.2.4 Mediating role of inflammatory cytokines in the gut microbiota-ALD causal pathway
Within the aforementioned five pathways involving gut microbiota, inflammatory cytokines, and ALD causality, we also calculated the Mediated Proportion of these inflammatory cytokines. Through further analysis, we found that there were three mediatory pathways (Beta1×Beta2) that were consistent with the direction of the gut microbiota-ALD causal relationship (Beta3), and all of these mediatory pathways were related to the gut microbiome component Roseburia hominis. Roseburia hominis was found to increase the risk of ALD by mediating the levels of the risk factors CUB domain-containing protein 1, Cystatin D, or Monocyte chemoattractant protein-1. This suggests that in MR analysis, Roseburia hominis may promote the onset of ALD through the mediation of multiple inflammatory cytokines, indicating its potential key role in the development of ALD.
3.3 Bidirectional causal effects between ALD, gut microbiota, and inflammatory cytokines
There were no opposing causal effects detected between gut microbiota, inflammatory cytokines, and ALD. After matching the Functional Training Dataset (FTD) with gut microbiota or cytokines, no single nucleotide polymorphism (SNP) could be used as a valid instrumental variable.
4 Discussion
In the discussion, as far as we know, this study is the first to employ MR analysis to estimate the causal relationship between gut microbiota and ALD, as well as the mediating role of inflammatory cytokines. In this comprehensive mediating MR analysis, we identified multiple gut microbiota taxa that play crucial roles in the development of ALD, with three gut microbiota taxa (Escherichia coli, Roseburia hominis, Family Porphyromonadaceae) showing a causal relationship with ALD. The mediating MR results indicate that among these three microbiota taxa, only Roseburia hominis is involved in mediating causal pathways with inflammatory cytokines. Specifically, three inflammatory cytokines, including CUB domain-containing protein 1, Cystatin D, or Monocyte chemoattractant protein-1, may respectively account for 1.6%, 1.5%, and 3.3% of the impact of Roseburia hominis on ALD. Furthermore, all three types of inflammatory cytokines are risk factors for the occurrence and progression of ALD. This analysis highlights the connection between gut microbiota and ALD, emphasizing the mediating role of inflammatory cytokines. In addition, ALD itself may not affect changes in gut microbiota and inflammatory cytokines that we have described. This suggests that changes in the gut microbiota and inflammatory cytokines occur prior to the onset of ALD, rather than being a consequence of the disease process.
In most cases of chronic liver diseases, dysbiosis of gut microbiota serves as the cornerstone of gut-liver axis impairment. Changes in gut-liver axis characteristics in ALD patients can lead to compromised intestinal integrity, disrupted bile acid metabolism, translocation of pathogen-associated molecular patterns (PAMPs), live microbes, and microbial metabolites, further exacerbating liver inflammation (4, 10). Dysbiosis of gut microbiota results in intestinal inflammation, ethanol metabolites, intestinal bile acids, and potentially other metabolites, all contributing to the breakdown of the intestinal barrier, increasing susceptibility to ALD, and compromising the patient’s health (27). Roseburia hominis (R. hominis) is a representative species within the Roseburia genus, belonging to the Lachnospiraceae family within the Firmicutes phylum. Recent studies have shown that R. hominis can metabolize various short-chain fatty acids (SCFAs) including acetate, propionate, and butyrate, which are considered potential therapeutic factors in the treatment of neuroinflammation and colitis (28, 29).
Experiments by Patterson AM et al. suggested that the metabolites of R. hominis in the gut, particularly SCFAs, have multifaceted regulatory effects on gastrointestinal diseases (30). Through oral administration to colitis mice for 14 days, Patterson AM found that the group receiving R. hominis had lower levels of colonic mucosal inflammation compared to the untreated colitis mice, indicating a positive correlation between increased levels of R. hominis and reduced colonic mucosal inflammation. In a study by Song L et al. involving male Sprague-Dawley germ-free rats, oral administration of R. hominis resulted in a significant increase in melatonin levels in the gut. Interestingly, there was a significant increase in the concentrations of propionate and butyrate in the gut contents after gavage (31). In another study by the same team on the inhibitory effects of R. hominis on neuroinflammation, they also identified the presence of propionate and butyrate, derivatives of R. hominis (28). As R. hominis entered the bodies of germ-free rats through oral administration, the abundance of propionate and butyrate increased, leading to reduced activation of microglial cells in the hippocampus and decreased release of pro-inflammatory cytokines and chemokines. This suggests that R. hominis may improve gut inflammation and neuroinflammation through its derivatives, positioning R. hominis as a “multifunctional probiotic” in human life. This seems to contradict our analysis results that “R. hominis may exacerbate ALD through inflammatory factors”.
However, in a study on the correlation between gut microbiota composition at disease onset and major adverse cardiovascular events within 3.2 years, the molar ratio of acetate and butyrate in the feces of acute myocardial infarction (AMI) patients was higher, contradicting the role of SCFAs in intestinal inflammation (32). Concurrently, research by Ji et al. found that a high abundance of Romboutsia and Roseburia not only failed to improve symptoms in Parkinson’s disease patients but was also associated with an increased risk of Parkinson’s disease (33). These findings challenge the “probiotic” identity of R. hominis, as it and its derivatives appear to have varying effects on the human body. To our knowledge, no team has investigated the relationship between R. hominis and ALD or even liver diseases beyond its derivatives. Our research results suggest a causal relationship between the gut microbiome composition, particularly R. hominis, and the development of ALD. To explore potential mediating pathways, we further analyzed the mediating effects of inflammatory cytokines.
The CUB domain-containing protein 1 (CDCP1) is a type I transmembrane glycoprotein that is widely upregulated in the pathogenesis of various malignant tumors, including those of the liver and pancreas (34). CDCP1 serves as a marker for tumor progression and also accelerates tumor metastasis. Stimulation of CDCP1 expression promotes CDCP1-mediated cancer cell migration in vitro and exacerbates the pro-carcinogenic effects, possibly by promoting the formation of a tumor inflammatory microenvironment (35, 36). In studies related to non-alcoholic steatohepatitis (NASH), a significant decrease in circulating CDCP1 was observed after weight loss surgery, and CDCP1 levels showed a close correlation with liver injury markers ALT and AST, suggesting CDCP1 may be a risk factor for NASH (37). Our research indicates a positive correlation between CDCP1 and ALD, with CDCP1 being a risk factor for the onset of ALD. Combining with previous studies, we infer that CDCP1 may promote the transformation of ALD into alcohol-related liver cancer.
Cystatin D (CST5) belongs to cystatin family II, which is from the cystatin superfamily. Studies have shown that CST5 profoundly affects cell phenotypes, increasing cell adhesion and inhibiting cell proliferation and migration. Researchers have observed that the expression of CST5 in human colon cancer cells is the opposite of CDCP1, significantly reducing the tumorigenic potential in immunodeficient mice (38). However, in hepatitis B virus-related liver cancer, the levels of CST5 in tumors are lower than in normal tissues (39). Our analysis results show that CST5 is also a risk factor for disease progression in ALD, suggesting that the role and mechanisms of CST5 may vary in different organs, and it may play a detrimental role in liver diseases.
Monocyte chemoattractant protein-1(MCP-1) is a member of the CC chemotaxis family, also known as Chemokine (CC-motif) ligand 2 (CCL2). MCP-1 is widely implicated in various diseases, including NASH, neuroinflammatory disorders, rheumatoid arthritis, cardiovascular diseases, and cancer, playing a role in attracting or enhancing the expression of other inflammatory factors and cells during these processes (40, 41). MCP-1 can recruit additional liver cells by regulating inflammatory cells, leading to liver fibrosis. Moreover, MCP-1 can also induce liver cell necrosis, fibronectin deposition, and DNA changes by modulating the expression of immune cells, contributing to the occurrence and development of liver tumors and liver cancer (42). In studies related to ALD, Umhau et al. conducted a multiple regression analysis of MCP-1 levels in the cerebrospinal fluid of alcoholics and the liver enzymes gamma-glutamyl transferase (GGT) and aspartate aminotransferase/glutamic-oxaloacetic transaminase (AST/GOT) in the blood, finding a positive correlation between MCP-1 and GGT as well as AST/GOT (43). This suggests a close association between MCP-1 and ALD, with MCP-1 potentially being a risk factor for the development of ALD. The researchers’ genetic validation confirmed a causal relationship between the two. However, further research is needed to elucidate the mechanisms by which MCP-1 contributes to ALD.
In the analysis process, we utilized a variety of common sensitivity analyses and excluded the influence of confounding factors and reverse causation. The preliminary research findings suggest a causal relationship between gut microbiota and ALD, as well as the involvement of inflammatory cytokines, providing further theoretical support and new directions for the treatment and prevention of ALD. Interestingly, the data analysis revealed that one gut bacterium (R. hominis) can regulate ALD by modulating multiple inflammatory cytokines (CDCP1, CST5, MCP-1). Although the individual effects of these inflammatory cytokines are relatively low, they offer a research path for the multi-target regulation of ALD by a single gut bacterium.
While the study has many strengths, there are some limitations. Firstly, the participants were exclusively of European descent, which limit the generalizability of the findings to other regions or ethnicities. Secondly, the results are based on theoretical data analysis and have not been validated through clinical or animal experiments, necessitating further elucidation through additional cellular, animal, and clinical studies. Nonetheless, the present study leveraged the largest available data collection, utilizing the most recently curated data source, and involving a substantial number of participants. Furthermore, rigorous sensitivity analyses have demonstrated the stability of our results, lending a high degree of reliability to the findings. Thirdly, apart from the 91 inflammatory cytokines studied, there may be other cytokines that were not included in this research, indicating the possibility of additional cytokines participating in the causal effects of gut microbiota on ALD (44). Lastly, although three inflammatory cytokines that mediate the causal relationship between gut microbiota and ALD have been identified and analyzed, their specific mechanisms in influencing the progression of ALD still warrant further investigation.
5 Conclusion
In this study, we comprehensively explored the causal effects between gut microbiota, inflammatory cytokines, and ALD. There were two positive and one negative causal effect between genetic liability in the gut microbiota and ALD. There were five positive correlations and two negative causal effects between inflammatory cytokines and ALD. In addition, we found one type of gut microbiota can separately regulate ALD through three different inflammatory cytokines.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Ethics committee of the UK Biobank. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
SZL: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. CZ: Writing – original draft. TL: Writing – original draft. LZ: Writing – original draft. STL: Writing – original draft. QZ: Writing – original draft. JL: Writing – original draft. WZ: Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (No. 82205086, 81473651), Henan Province Chinese medicine scientific research special topic (No. 2022JDZX006), Henan Province Traditional Chinese medicine “double first-class” to create a scientific research project of Henan University of Chinese Medicine (No. HSRP-DFCTCM-2023-3-13, HSRP-DFCTCM-2023-8-36, HSRP-DFCTCM-2023-3-23) and “National famous Traditional Chinese Medicine inheritance studio construction project (No: Teaching Letter of the State Office of Traditional Chinese Medicine (2022) No. 245)”.
Acknowledgments
The authors thank the investigators of the original studies for sharing the GWAS summary statistics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2024.1442603/full#supplementary-material
References
1. Seitz HK, Bataller R, Cortez-Pinto H, Gao B, Gual A, Lackner C, et al. Alcoholic liver disease. Nat Rev Dis Primers. (2018) 4:16. doi: 10.1038/s41572-018-0014-7
2. Huang DQ, Singal AG, Kono Y, Tan D, El-Serag HB, Loomba R. Changing global epidemiology of liver cancer from 2010 to 2019: nash is the fastest growing cause of liver cancer. Cell Metab. (2022) 34:969–977.E2. doi: 10.1016/j.cmet.2022.05.003
3. Collaborators, G. D. A. I. Global burden of 369 diseases and injuries in 204 countries and territories1990-2019: A systematic analysis for the global burden of disease study 2019. Lancet. (2020) 396:1204–22. doi: 10.1016/S0140-6736(20)30925-9
4. Wu X, Fan X, Miyata T, Kim A, Cajigas-Du RC, Ray S, et al. Recent advances in understanding of pathogenesis of alcohol-associated liver disease. Annu Rev Pathol. (2023) 18:411–38. doi: 10.1146/annurev-pathmechdis-031521-030435
5. Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Transl Res. (2014) 163:352–62. doi: 10.1016/j.trsl.2014.01.005
6. Lang S, Schnabl B. Microbiota and fatty liver disease-the known, the unknown, and the future. Cell Host Microbe. (2020) 28:233–44. doi: 10.1016/j.chom.2020.07.007
7. Natividad JM, Verdu EF. Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacol Res. (2013) 69:42–51. doi: 10.1016/j.phrs.2012.10.007
8. Allam-Ndoul B, Castonguay-Paradis S, Veilleux A. Gut microbiota and intestinal trans-epithelial permeability. Int J Mol Sci. (2020) 21:17. doi: 10.3390/ijms21176402
9. Xu Z, Jiang N, Xiao Y, Yuan K, Wang Z. The role of gut microbiota in liver regeneration. Front Immunol. (2022) 13:1003376. doi: 10.3389/fimmu.2022.1003376
10. Wang R, Tang R, Li B, Ma X, Schnabl B, Tilg H. Gut microbiome, liver immunology, and liver diseases. Cell Mol Immunol. (2021) 18:4–17. doi: 10.1038/s41423-020-00592-6
11. Grander C, Adolph TE, Wieser V, Lowe P, Wrzosek L, Gyongyosi B, et al. Recovery of ethanol-induced akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. (2018) 67:891–901. doi: 10.1136/gutjnl-2016-313432
12. Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. (2019) 575:505–11. doi: 10.1038/s41586-019-1742-x
13. Tilg H, Adolph TE, Trauner M. Gut-liver axis: pathophysiological concepts and clinical implications. Cell Metab. (2022) 34:1700–18. doi: 10.1016/j.cmet.2022.09.017
14. Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. Jama. (2017) 318:1925–6. doi: 10.1001/jama.2017.17219
15. Davies NM, Holmes MV, Davey SG. Reading mendelian randomisation studies: A guide, glossary, and checklist for clinicians. Bmj. (2018) 362:601–6. doi: 10.1136/bmj.k601
16. Bowden J, Holmes MV. Meta-analysis and mendelian randomization: A review. Res Synth Methods. (2019) 10:486–96. doi: 10.1002/jrsm.v10.4
17. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. Finngen provides genetic insights from A well-phenotyped isolated population. Nature. (2023) 613:508–18. doi: 10.1038/s41586-022-05473-8
18. Lopera-Maya EA, Kurilshikov A, van der Graaf A, Hu S, Andreu-Sánchez S, Chen L, et al. Effect of host genetics on the gut microbiome in 7,738 participants of the dutch microbiome project. Nat Genet. (2022) 54:143–51. doi: 10.1038/s41588-021-00992-y
19. Zhao JH, Stacey D, Eriksson N, Macdonald-Dunlop E, Hedman Å.K, Kalnapenkis A, et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Immunol. (2023) 24:1540–51. doi: 10.1038/s41590-023-01588-w
20. Sanna S, Van Zuydam NR, Mahajan A, Kurilshikov A, Vich VA, Võsa U, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. (2019) 51:600–5. doi: 10.1038/s41588-019-0350-x
21. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The mr-base platform supports systematic causal inference across the human phenome. Elife. (2018) 7. doi: 10.7554/eLife.34408
22. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. (2013) 37:658–65. doi: 10.1002/gepi.2013.37.issue-7
23. Burgess S, Thompson SG. Interpreting findings from mendelian randomization using the mr-egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
24. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
25. Bowden J, Davey SG, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
26. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for mendelian randomization studies using multiple genetic variants. Int J Epidemiol. (2011) 40:740–52. doi: 10.1093/ije/dyq151
27. Bluemel S, Williams B, Knight R, Schnabl B. Precision medicine in alcoholic and nonalcoholic fatty liver disease via modulating the gut microbiota. Am J Physiol Gastrointest Liver Physiol. (2016) 311:G1018–36. doi: 10.1152/ajpgi.00245.2016
28. Song L, Sun Q, Zheng H, Zhang Y, Wang Y, Liu S, et al. Roseburia hominis alleviates neuroinflammation via short-chain fatty acids through histone deacetylase inhibition. Mol Nutr Food Res. (2022) 66:E2200164. doi: 10.1002/mnfr.202200164
29. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species roseburia hominis and faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. (2014) 63:1275–83. doi: 10.1136/gutjnl-2013-304833
30. Patterson AM, Mulder IE, Travis AJ, Lan A, Cerf-Bensussan N, Gaboriau-Routhiau V, et al. Human gut symbiont roseburia hominis promotes and regulates innate immunity. Front Immunol. (2017) 81166. doi: 10.3389/fimmu.2017.01166
31. Song L, He M, Sun Q, Wang Y, Zhang J, Fang Y, et al. Roseburia hominis increases intestinal melatonin level by activating P-creb-aanat pathway. Nutrients. (2021) 14:117–22. doi: 10.3390/nu14010117
32. Liu C, Sun Z, Shali S, Mei Z, Chang S, Mo H, et al. The gut microbiome and microbial metabolites in acute myocardial infarction. J Genet Genomics. (2022) 49:569–78. doi: 10.1016/j.jgg.2021.12.007
33. Ji D, Chen WZ, Zhang L, Zhang ZH, Chen LJ. Gut microbiota, circulating cytokines and dementia: A mendelian randomization study. J Neuroinflamm. (2024) 21:2. doi: 10.1186/s12974-023-02999-0
34. Khan T, Kryza T, Lyons NJ, He Y, Hooper JD. The cdcp1 signaling hub: A target for cancer detection and therapeutic intervention. Cancer Res. (2021) 81:2259–69. doi: 10.1158/0008-5472.CAN-20-2978
35. Dong Y, He Y, De Boer L, Stack MS, Lumley JW, Clements JA, et al. The cell surface glycoprotein cub domain-containing protein 1 (Cdcp1) contributes to epidermal growth factor receptor-mediated cell migration. J Biol Chem. (2012) 287:9792–803. doi: 10.1074/jbc.M111.335448
36. He Y, Harrington BS, Hooper JD. New crossroads for potential therapeutic intervention in cancer - intersections between cdcp1, egfr family members and downstream signaling pathways. Oncoscience. (2016) 3:5–8. doi: 10.18632/oncoscience.v3i1
37. Jia X, Song E, Liu Y, Chen J, Wan P, Hu Y, et al. Identification and multicentric validation of soluble cdcp1 as A robust serological biomarker for risk stratification of nash in obese chinese. Cell Rep Med. (2023) 4:101257. doi: 10.1016/j.xcrm.2023.101257
38. Alvarez-Díaz S, Valle N, García JM, Peña C, Freije JM, Quesada V, et al. Cystatin D is A candidate tumor suppressor gene induced by vitamin D in human colon cancer cells. J Clin Invest. (2009) 119:2343–58. doi: 10.1172/JCI37205
39. Zhou X, Wang X, Huang K, Liao X, Yang C, Yu T, et al. Investigation of the clinical significance and prospective molecular mechanisms of cystatin genes in patients with hepatitis B virus−Related hepatocellular carcinoma. Oncol Rep. (2019) 42:189–201. doi: 10.3892/or.2019.7154
40. Singh S, Anshita D, Ravichandiran V. Mcp-1: function, regulation, and involvement in disease. Int Immunopharmacol. (2021) 101:107598. doi: 10.1016/j.intimp.2021.107598
41. Kang J, Postigo-Fernandez J, Kim K, Zhu C, Yu J, Meroni M, et al. Notch-mediated hepatocyte mcp-1 secretion causes liver fibrosis. JCI Insight. (2023) 8. doi: 10.1172/jci.insight.165369
42. Fantuzzi L, Tagliamonte M, Gauzzi MC, Lopalco L. Dual ccr5/ccr2 targeting: opportunities for the cure of complex disorders. Cell Mol Life Sci. (2019) 76:4869–86. doi: 10.1007/s00018-019-03255-6
43. Umhau JC, Schwandt M, Solomon MG, Yuan P, Nugent A, Zarate CA, et al. Cerebrospinal fluid monocyte chemoattractant protein-1 in alcoholics: support for A neuroinflammatory model of chronic alcoholism. Alcohol Clin Exp Res. (2014) 38:1301–6. doi: 10.1111/acer.2014.38.issue-5
Keywords: Mendelian randomization, gut microbiota, inflammatory cytokines, alcoholic liver disease, causal relationship
Citation: Li S, Zhou C, Liu T, Zhang L, Liu S, Zhao Q, Liu J and Zhao W (2024) Causal relationships between the gut microbiota, inflammatory cytokines, and alcoholic liver disease: a Mendelian randomization analysis. Front. Endocrinol. 15:1442603. doi: 10.3389/fendo.2024.1442603
Received: 02 June 2024; Accepted: 01 October 2024;
Published: 21 October 2024.
Edited by:
Jeff M. P. Holly, University of Bristol, United KingdomReviewed by:
Matthew Vincent, Washington University in St. Louis, United StatesMercedes G. López, National Polytechnic Institute of Mexico (CINVESTAV), Mexico
Copyright © 2024 Li, Zhou, Liu, Zhang, Liu, Zhao, Liu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxia Zhao, Wmhhby13ZW54aWFAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Shanzheng Li
Shanzheng Li Cheng Zhou
Cheng Zhou Tong Liu
Tong Liu Lihui Zhang
Lihui Zhang Sutong Liu
Sutong Liu Qing Zhao1
Qing Zhao1 Wenxia Zhao
Wenxia Zhao