- 1Department of Orthopedics, Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi, China
- 2Department of Orthopedics, First Affiliated Hospital of Henan University of Science and Technology, Luoyang, Henan, China
- 3Department of Orthopedics, Xi’an Daxing Hospital, Xi’an, Shaanxi, China
Objectives: In-depth understanding of osteonecrosis of femoral head (ONFH) has revealed that degeneration of the hip cartilage plays a crucial role in ONFH progression. However, the underlying molecular mechanisms and susceptibility to environmental factors in hip cartilage that contribute to ONFH progression remain elusive.
Methods: We conducted a multiomics study and chemical−gene interaction analysis of hip cartilage in ONFH. The differentially expressed genes (DEGs) involved in ONFH progression were identified in paired hip cartilage samples from 36 patients by combining genome-wide DNA methylation profiling, gene expression profiling, and quantitative proteomics. Gene functional enrichment and pathway analyses were performed via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. Functional links between proteins were discovered through protein−protein interaction (PPI) networks. The ONFH-associated chemicals were identified by integrating the DEGs with the chemical−gene interaction sets in the Comparative Toxicogenomics Database (CTD). Finally, the DEGs, including MMP13 and CHI3L1, were validated via quantitative real-time PCR (qRT−PCR) and immunohistochemistry (IHC).
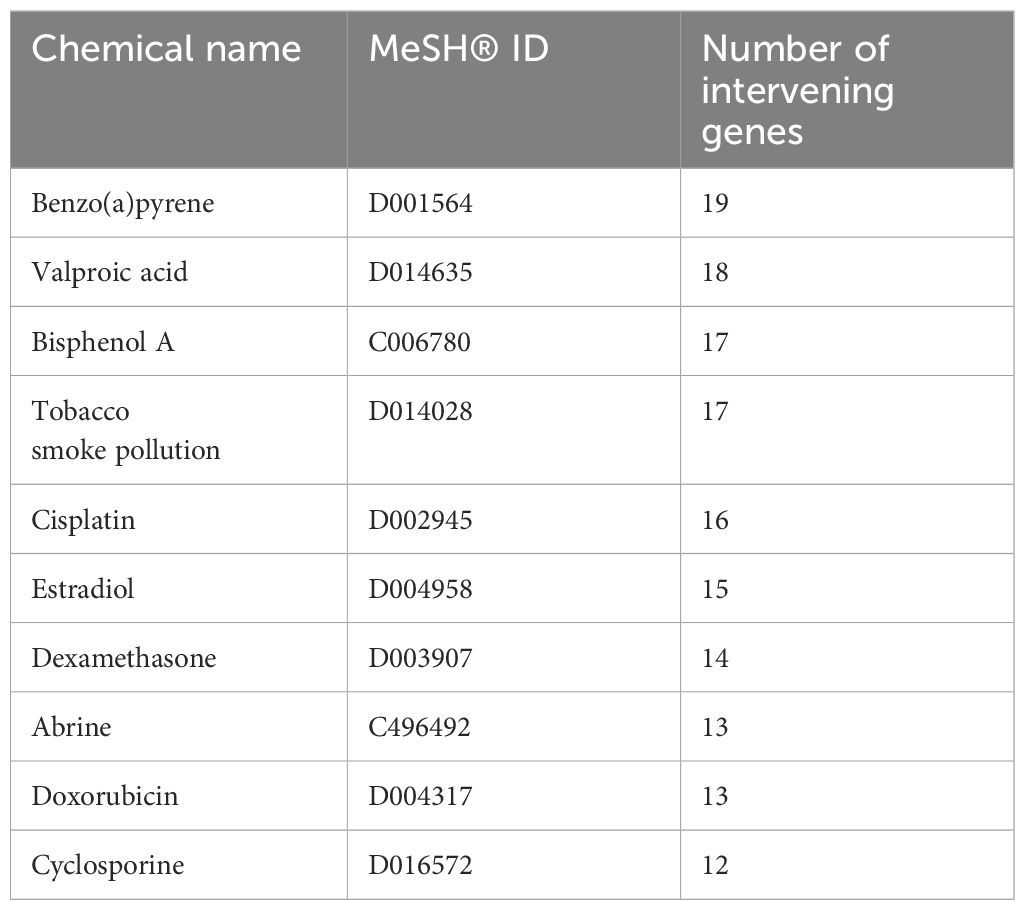
Results: Twenty-two DEGs were identified across all three omics levels in ONFH cartilage, 16 of which were upregulated and six of which were downregulated. The collagen-containing extracellular matrix (ECM), ECM structural constituents, response to amino acids, the relaxin signaling pathway, and protein digestion and absorption were found to be primarily involved in cartilage degeneration in ONFH. Moreover, ten major ONFH-associated chemicals were identified, including, benzo(a)pyrene, valproic acid, and bisphenol A.
Conclusion: Overall, our study identified several candidate genes, pathways, and chemicals associated with cartilage degeneration in ONFH, providing novel clues into the etiology and biological processes of ONFH progression.
1 Introduction
Osteonecrosis of femoral head (ONFH) is a severely disabling disease that usually affects young adults (1). As the pathogenesis of ONFH remains elusive, there is no authorized effective treatment for ONFH. Approximately 65% of ONFH patients ultimately need total hip replacement (2), resulting in heavy medical and economic burdens on these patients. However, the etiology and pathogenesis of hip cartilage degeneration in ONFH are currently unknown. Recent studies have demonstrated that hip cartilage degeneration plays a crucial role in ONFH development, with cartilage degeneration occurring early in ONFH (3). The destruction of hip cartilage increases hip instability, exacerbates femoral head collapse, and accelerates the development of ONFH (4). Thus, hip cartilage degeneration therapy may retard or even reverse the destruction of the hip joint caused by ONFH.
Recently, numerous studies have verified the impact of abnormal DNA methylation on the pathogenesis of bone and cartilage diseases (5, 6). Transcriptomics can reveal the mechanisms of ONFH progression at the genetic level (7). Proteomics is the study of the characterization of proteins at a large scale and can be applied to studies of cartilage pathophysiology (8). However, owing to the complexity and variability of molecules, individual omics analyses have difficulty providing a systematic and comprehensive understanding of complex biological processes (9). With their ability to study biological phenomena comprehensively, multiomics analyses can integrate the interactions of molecules at different layers and help bridge the gap from genotype to phenotype (10), improving the predictive accuracy of disease biological processes (11). Integrated proteomic and metabolomic analyses of synovial fluid have identified classical complement pathways and proinflammatory cytokines (IL-6, IL-8, and IL-18) as potential biomarkers of osteoarthritis (OA) and identified metabolically distinct OA subgroups (12). Multiomics exploration of therapeutic strategies for rheumatoid arthritis revealed that phellodendrine could effectively inhibit the activity of synovial cells and reduce the expression levels of inflammatory factors to alleviate joint inflammation and cartilage injury (13). Multiomics classification helps to distinguish the aggressiveness of cartilage tumors (14). Therefore, multiomics analyses of hip cartilage in ONFH comprehensively reveal the biological process of disease progression, which contributes to ONFH treatment and prevention.
The interactions between environmental factors and genes are involved in the development of chronic diseases by regulating physiological processes (15). However, to date, there are no epidemiologic reports of ONFH worldwide (16), and the environmental factors that predispose patients to ONFH are still unknown. The Comparative Toxicogenomics Database (CTD) is a publicly accessible database providing data about chemical−gene interactions (17). Therefore, integrating differentially expressed genes (DEGs) identified via multiomics analyses with chemical−gene interaction sets from the CTD will reveal the etiology and high-risk populations of ONFH patients.
In this study, we applied integrated multiomics (genome-wide DNA methylation profiling, gene expression profiling, and quantitative proteomics) in ONFH hip cartilage to obtain a comprehensive molecular portrait of cartilage degeneration. The ONFH-associated chemicals were subsequently identified by integrating the DEGs with the chemical−gene interaction sets in the CTD. Finally, the DEGs were validated via quantitative real-time PCR (qRT−PCR) and immunohistochemistry (IHC).
2 Methods
2.1 Patients and samples
A diagram of the research strategy, created with MedPeer (www.medpeer.cn), is shown in Figure 1. Briefly, human subjects were recruited from the Second Affiliated Hospital of Xi’an Jiaotong University. The clinical information of the study subjects is summarized in Supplementary Table 1. All the study subjects were Chinese Han individuals who were undergoing total hip replacement surgery. ONFH patients and control subjects were diagnosed according to clinical manifestations and radiologic imaging of the hip by two blinded ONFH experts. ONFH cartilage samples were harvested from patients with Ficat grade III ONFH (18). All cartilage samples were collected from anterosuperior portions of the femoral head (where the cartilage had collapsed in ONFH patients) within 2 h of total hip replacement. This study was approved by the Human Ethics Committees of Xi’an Jiaotong University. Informed consent documents were signed by all participants before they participated in this study. All methods were carried out in accordance with the relevant guidelines and regulations.
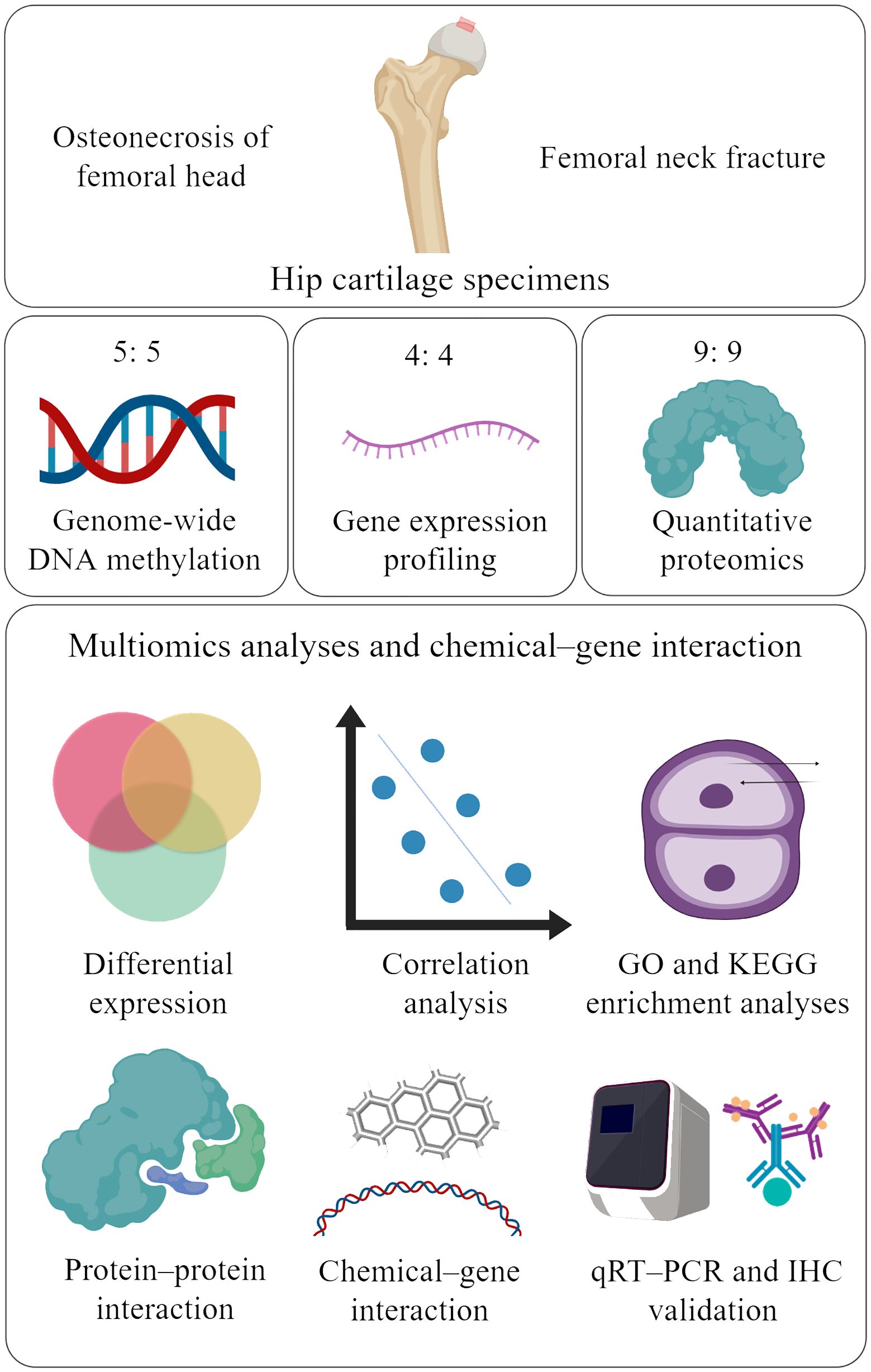
Figure 1. Study strategy (created with MedPeer). The red box shows the regions from which cartilage was collected from the femoral head. Cartilage samples from 18 ONFH patients and 18 femoral neck fracture controls were included in the study. Five, four, and nine ONFH control samples were selected for genome-wide DNA methylation profiling, gene expression profiling, and quantitative proteomics, respectively. The multiomics data were integrated to identify candidate genes, correlations, pathways, protein−protein interactions, and chemical−gene interactions. Finally, the differentially expressed proteins were validated via qRT−PCR and IHC. ONFH, osteonecrosis of femoral head; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; qRT−PCR, quantitative real-time PCR; IHC, immunohistochemistry.
Ten hip articular cartilage samples were used for genome-wide DNA methylation profiling. Eight hip articular cartilage samples were used for gene expression profiling. Eighteen hip articular cartilage samples were subjected to quantitative proteomics analysis. ONFH samples from the above groups were paired with control samples.
2.2 Genome-wide DNA methylation
Genome-wide DNA methylation profiling was conducted via the Illumina Infinium HumanMethylation 850 BeadChip according to the manufacturer’s standard procedure. The percentage of methylated cytosine at a given CpG site was calculated as a β value, ranging from 0.0 (completely unmethylated) to 1.0 (completely methylated). A generalized linear mixed model was used to adjust for age and sex as covariates. We excluded CpG probes that mapped to sex chromosomes, cross-reacted with sex chromosomes or contained genetic variants. A total of 844,832 CpG sites that passed the quality control procedure were analyzed in this study. The Pearson correlation coefficients of the β values were calculated to evaluate the correlations of the samples.
Differentially methylated CpG sites were identified via the empirical Bayes moderated t test of the limma package (version 3.1.2) of R via DNA methylation M values. The Benjamini–Hochberg method was used to obtain an adjusted P value for each CpG locus. CpG sites with adjusted P values < 0.05 and |β value difference|>1 were considered significantly differentially methylated loci. For quality control, the CpG sites with missing values or P values > 0.05 in > 95% of the articular cartilage samples were eliminated.
2.3 Gene expression profiling
Total RNA was isolated from the cartilage samples via an Agilent Total RNA Isolation Mini Kit (Agilent Technologies, Santa Clara, CA, USA) following the manufacturer’s instructions. The isolated total RNA and complementary RNA (cRNA) contents were determined with an Agilent ND-1000 (Agilent Technologies). One microgram of labeled cRNA was mixed with hybridization buffer and hybridized to the Agilent Human 4x44K Gene Expression Microarray (v2, Agilent Technologies). The microarray data have been deposited in the Gene Expression Omnibus database [GEO: GSE74089]. The genes presenting both fold changes > 1.0 and P values < 0.05 were considered significantly differentially expressed. The false discovery rate (FDR) values were calculated via the permutation-based analysis algorithm of significance analysis of microarrays (SAM) (19).
2.4 Quantitative proteomics analysis
Isobaric tandem mass tag (TMT)-based quantitative proteomics analysis was applied to investigate the altered proteins associated with ONFH. LC−MS/MS analysis was performed using an Easy-nLC nanoflow HPLC system connected to a Q Exactive instrument (Thermo Fisher Scientific, MA, USA). The mass spectrometer was operated in positive ion mode. MS spectra were acquired over a range of 350–2000 m/z. The results were filtered on the basis of peptide and protein FDRs ≤ 1%. The relative peak intensities of the TMT reporter ions released in each of the MS/MS spectra were used. Only unique peptides obtained with a confidence percentage of > 95% were included. A fold change value of > 1 or < 1 and a P value of < 0.05 were used to identify upregulated or downregulated proteins.
2.5 Pathway and functional enrichment analyses
Pathway and functional enrichment analyses of DEGs from the integration of at least two omics methods were performed via Gene Ontology (GO) annotation (20) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping (21). Genes annotated to the same term were treated as “pathways”. The enrichment analyses of DEGs according to up- or downregulation were also conducted. Empirical p values for the enrichments were obtained from randomizations.
2.6 Protein−protein interaction network construction
The internet tool STRING (https://string-db.org) was used to analyze the probable interactions of differential proteins across all three omics levels (22). Interactions with summed scores greater than 0.15 were extracted to construct the PPI network. The PPI network was then visualized and subjected to further evaluation via the Cytoscape program (www.cytoscape.org). The Cytoscape MCODE plugin was adopted to identify crucial modules in the PPI network. The pathways involved in the crucial modules are shown in combination with KEGG.
2.7 Chemical−gene expression interactions
The CTD (http://ctdbase.org/) is a publicly available online database that provides access to data on chemical−gene interactions, chemical−disease associations, chemical−pathway associations, and chemical−phenotype associations. In the present study, we uploaded the differentially expressed genes across all three omics levels into the “Batch Query” of the CTD, selected the output of chemical−gene interactions, and screened the H. sapiens tissue data to obtain the top ten chemicals associated with ONFH on the basis of the number of chemical-acting genes.
2.8 qRT−PCR validation
The expression levels of mRNAs (MMP13 and CHI3L1) identified via transcriptomic data were validated via qRT−PCR in a cohort comprising four ONFH patients and four control samples. Total RNA from cartilage and subchondral bone tissue was extracted with TRIzol Reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. Then, 1 μg of total RNA was reverse transcribed via the Evo M-MLV RT Kit (Agbio, China), and qRT−PCR was performed via a QuantGene 9600 Real-Time PCR System (Bioer Technology, China). SYBR Green-based three-step qRT−PCR was performed via a SYBR Green Pro Taq HS HS qPCR Kit (Agbio). The primer sequences were retrieved from the online PrimerBank database (https://pga.mgh.harvard.edu/primerbank/index.html). GAPDH was used as the internal reference gene. Detailed information about the primers used for qRT−PCR is summarized in Supplementary Table 2.
2.9 IHC verification
The expression of MMP13 and CHI3L1 proteins in cartilage samples from ONFH patients and controls was verified via IHC. Briefly, cartilage tissue was fixed with paraformaldehyde, decalcified, and embedded in paraffin. The paraffin-embedded tissue sections were immersed in xylene to remove the paraffin, hydrated with a series of ethanol solutions, exposed to a 3% hydrogen peroxide solution for 10 minutes to deparaffinize and hydrate the cartilage sections, and then rinsed with PBS. After being blocked with QuickBlock™ Buffer (Beyotime Biotechnology, China) for 1 h, the sections were incubated with an MMP13 antibody (dilution: 1:100, ProteinTech Group; Chicago, IL, USA, no. 18165-1-AP) and a CHI3L1 antibody (dilution: 1:200, ProteinTech Group; Chicago, IL, USA, no. 12036-1-AP) at 4°C overnight. Following rinsing with PBS, the sections were exposed to an alkaline phosphatase-labeled secondary antibody (ZHONGSHAN Golden Bridge Biotechnology, Beijing, China) for 15 minutes at 37°C. They were then treated with streptavidin-horseradish peroxidase for another 15 minutes at the same temperature. Finally, the sections were stained with 3,3’-diaminobenzidine (DAB). The percentage of positive areas (cells) in the field of interest was calculated in the superficial, middle, and deep zones. Finally, differences in the expression of MMP13 and CHI3L1 proteins in the cartilage of the three ONFH patients and the three normal controls were analyzed via two-tailed Student’s t tests. The participants received information about the study, and their written consent was obtained.
3 Results
The abundance of 629 proteins was found to vary: 112 were observed at higher abundance and 517 at lower abundance in ONFH cartilage (Figure 2).
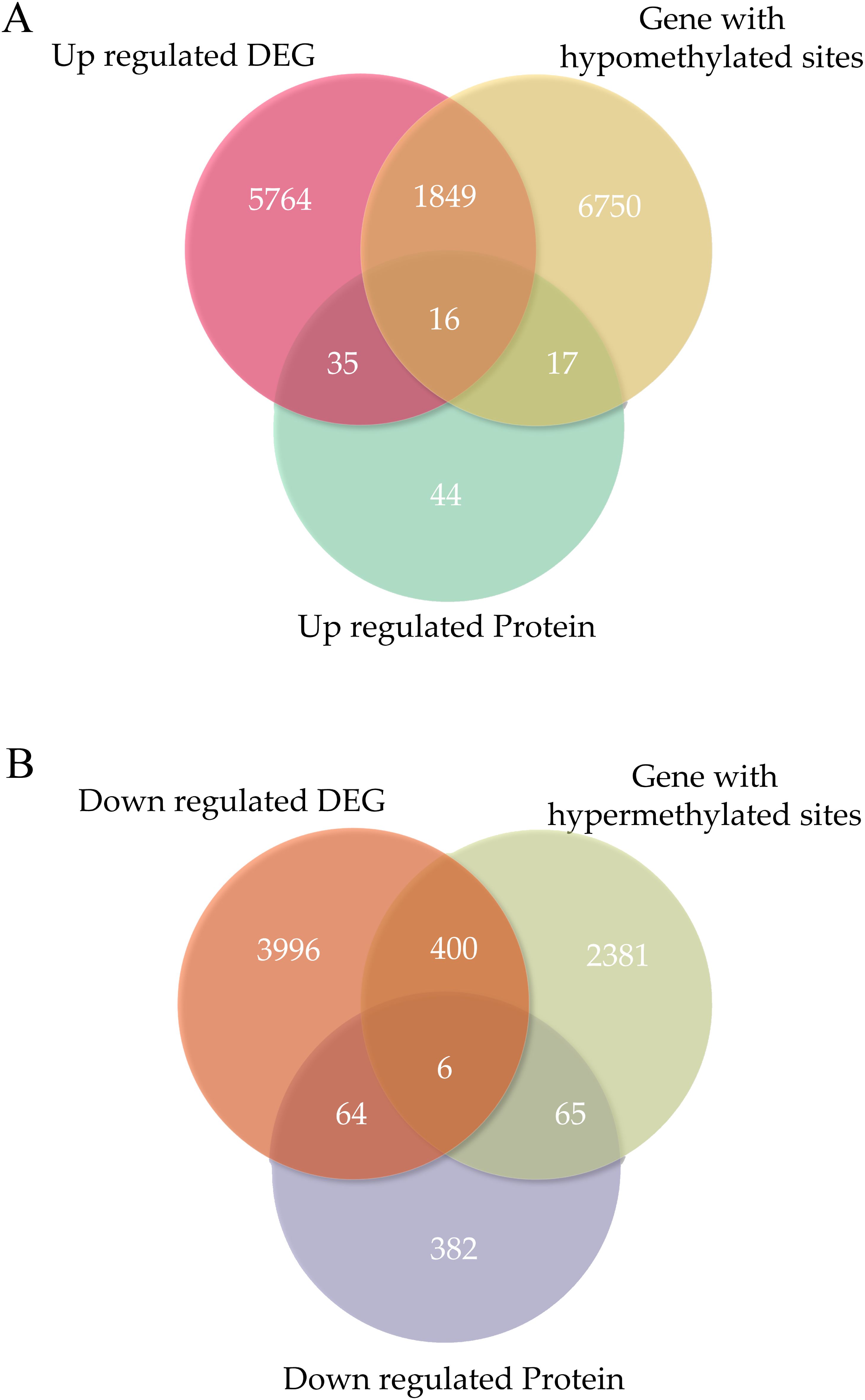
Figure 2. The overlapping genes were identified in each omics experiment. (A) Venn diagrams identifying the number of overlapping genes in each omics experiment for upregulation. (B) Venn diagrams identifying the number of overlapping genes in each -omics experiment for downregulation. DEGs, differentially expressed genes.
The expression of 12130 genes was significantly altered: 7664 genes were upregulated, and 4466 were downregulated in ONFH cartilage (Figure 2). The MMP13 was significantly upregulated in the ONFH cartilage than in the control cartilage.
The expression of 11484 methylated sites significantly changed: 8632 genes exhibited hypomethylated sites and 2852 exhibited hypermethylated sites in ONFH cartilage (Figure 2).
3.1 Integration across multiple omics levels: methylation, RNA sequencing, and proteomics
At the three-omics level, 22 DEGs (including 16 upregulated genes and 6 downregulated genes) were found to be involved in the degenerative process of ONFH cartilage (Supplementary Tables 3, 4). Among these, alterations in the expression of MMP13, MMP2, COL3A1, COL5A2, and CHI3L1 have been previously reported in both ONFH and cartilage. In addition, 17 DEGs have not been reported in studies of ONFH; furthermore, seven genes have not been reported in cartilage. MMP13, ECM1, and TPPP3, which are upregulated in ONFH cartilage, were the three genes associated with the greatest log-fold changes in protein expression across the three omics methods. CHI3L1, GDF10, and GNPDA1, which are downregulated in ONFH cartilage, were the top three genes in terms of log-fold changes in protein expression across the three omics methods. The differentially expressed genes identified in this study that have never been previously mentioned in ONFH are summarized in Supplementary Table 5.
3.2 Proteomics and RNA sequencing
Among the proteins with evidence of differential abundance, 121 genes were also differentially expressed between the ONFH and control samples at the RNA level, of which 51 were upregulated and 70 were downregulated at both the RNA and protein levels in ONFH cartilage. Considering the RNA and protein changes in ONFH cartilage samples, we identified a significant positive correlation (Figure 3A).
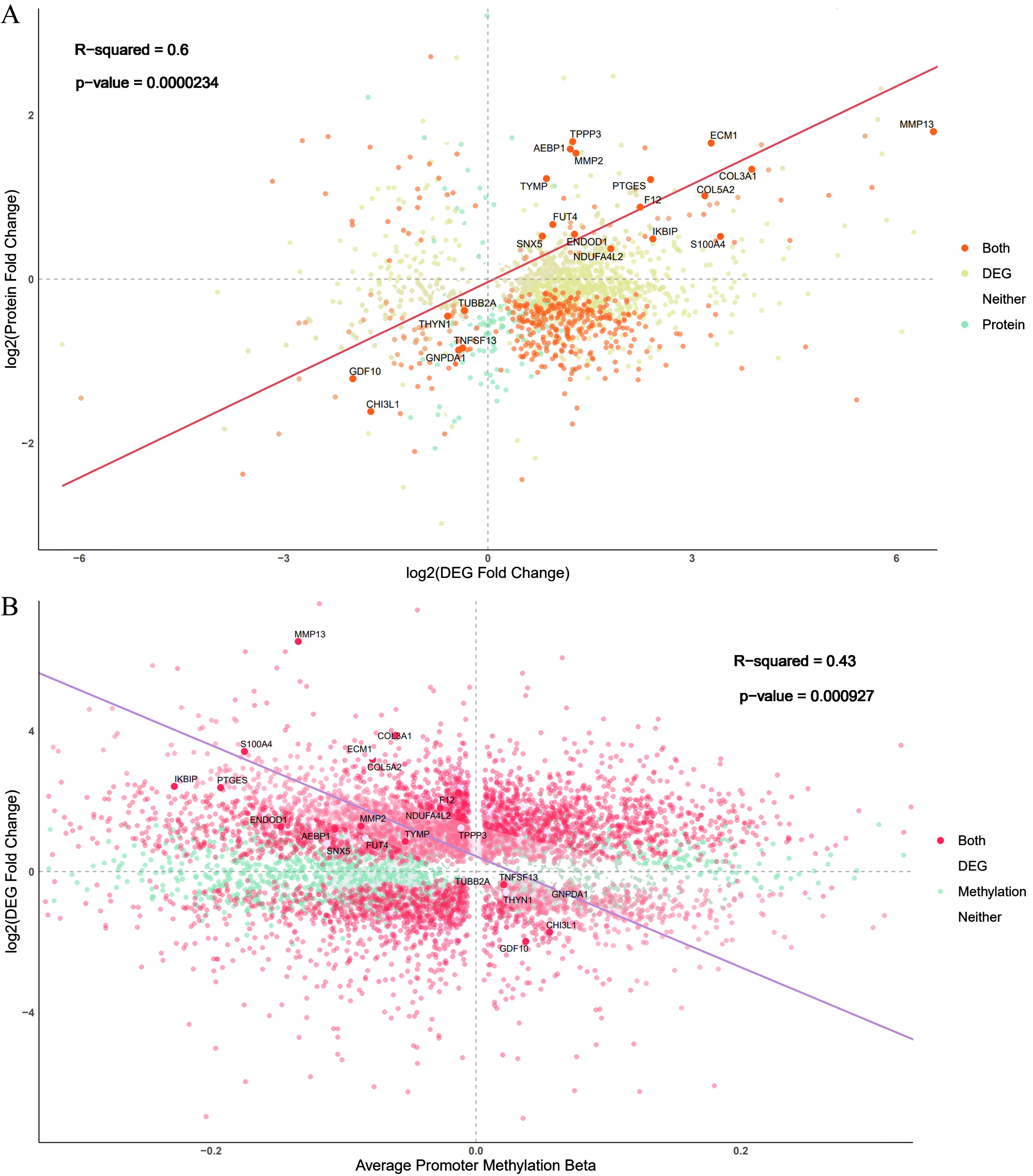
Figure 3. Comparison of changes identified in the omics experiments. (A) Comparison of the log-fold changes in both the proteomics and RNA-seq experiments. Each gene is represented as a single point, and the color corresponds to whether the gene is identified as differentially expressed via RNA-seq, proteomics analyses, or both. The screening threshold for the transcriptome and proteome was a P value less than 0.05. (B) Comparison of RNA-seq log-fold-change and average promoter region methylation changes in the differentially expressed genes. The screening threshold for methylation was a P value less than 0.05.
3.3 Methylation and RNA sequencing
Among the 8632 hypomethylated sites, 1865 genes were also upregulated at the RNA level in ONFH cartilage. Among the 2852 hypermethylated sites, 406 genes were also downregulated at the RNA level in ONFH cartilage. A direct comparison of promoter region methylation and gene expression revealed the expected negative correlation between promoter region methylation and gene expression (Figure 3B).
3.4 Proteomics and methylation
Among these upregulated proteins, 33 hypomethylated sites were identified at the methylation level. Moreover, 71 hypermethylated sites were identified in the downregulated proteins at the methylation level.
3.5 Enrichment analyses involved in ONFH progression
According to the results of the GO enrichment analysis (Figure 4), the DEGs at the three multiomics levels were primarily involved in the biological processes of the collagen-containing extracellular matrix, the extracellular matrix structural constituent, and the response to amino acids, which contributed to ONFH progression. Collagen-containing extracellular matrix, extracellular matrix structural constituent, external encapsulating structure organization, ossification, and cartilage development were enriched at the transcriptional and proteomic levels. Collagen-containing extracellular matrix and extracellular matrix structural constituents were also enriched at the methylation and proteomics levels. There was no common enriched pathway among the transcriptional and methylation levels. The detailed results of the GO enrichment analysis are shown in Supplementary Figure 1.
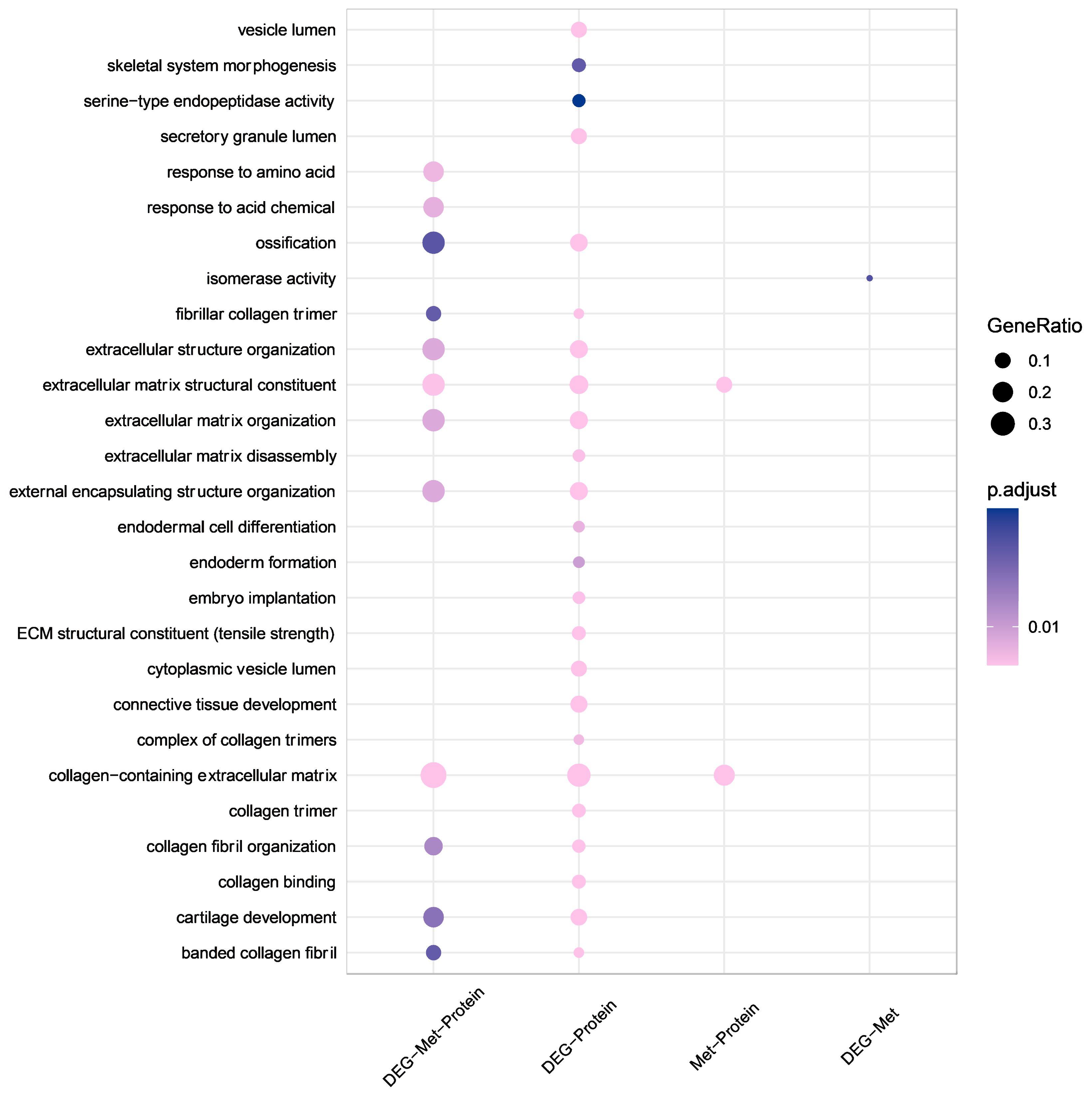
Figure 4. GO enrichment analysis at multiple omics levels. Larger bubble size represents a greater number of genes involved. The shapes are colored from pink to purple, representing low to high p.adjust. GO, Gene Ontology.
The results of the KEGG pathway analysis revealed that the relaxin and bladder cancer signaling pathways were enriched at three multiomics levels (Supplementary Figure 1). Amyotrophic lateral sclerosis, Huntington disease, Parkinson disease, and ribosomes were enriched at the transcriptional and methylation levels. Protein digestion and absorption, ECM-receptor interaction, and focal adhesion were enriched at the transcriptional and proteomic levels. Fatty acid degradation was enriched at the methylation and proteomics levels. The enrichment analysis results of the DEGs are shown in Supplementary Figures 2-4.
3.6 PPI network and KEGG pathway
The functional links between 22 differential proteins across all three omics levels were analyzed via a protein–protein interaction (PPI) network from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (Figure 5A). Eight genes, seven upregulated and one downregulated, were identified in ONFH cartilage on the basis of the degree of interaction in the PPI network. The KEGG pathways associated with the eight genes are shown in Figure 5B. The relaxin signaling pathway and protein digestion and absorption are involved mainly in the pathogenesis of ONFH in cartilage. MMP13 is associated mainly with the IL-17 signaling pathway, relaxin signaling pathway, and parathyroid hormone synthesis, secretion, and action. CHI3L1 is associated mainly with chitinase-like lectins, the innate immune system, and the mammary gland development pathway (Stage 4 of 4).
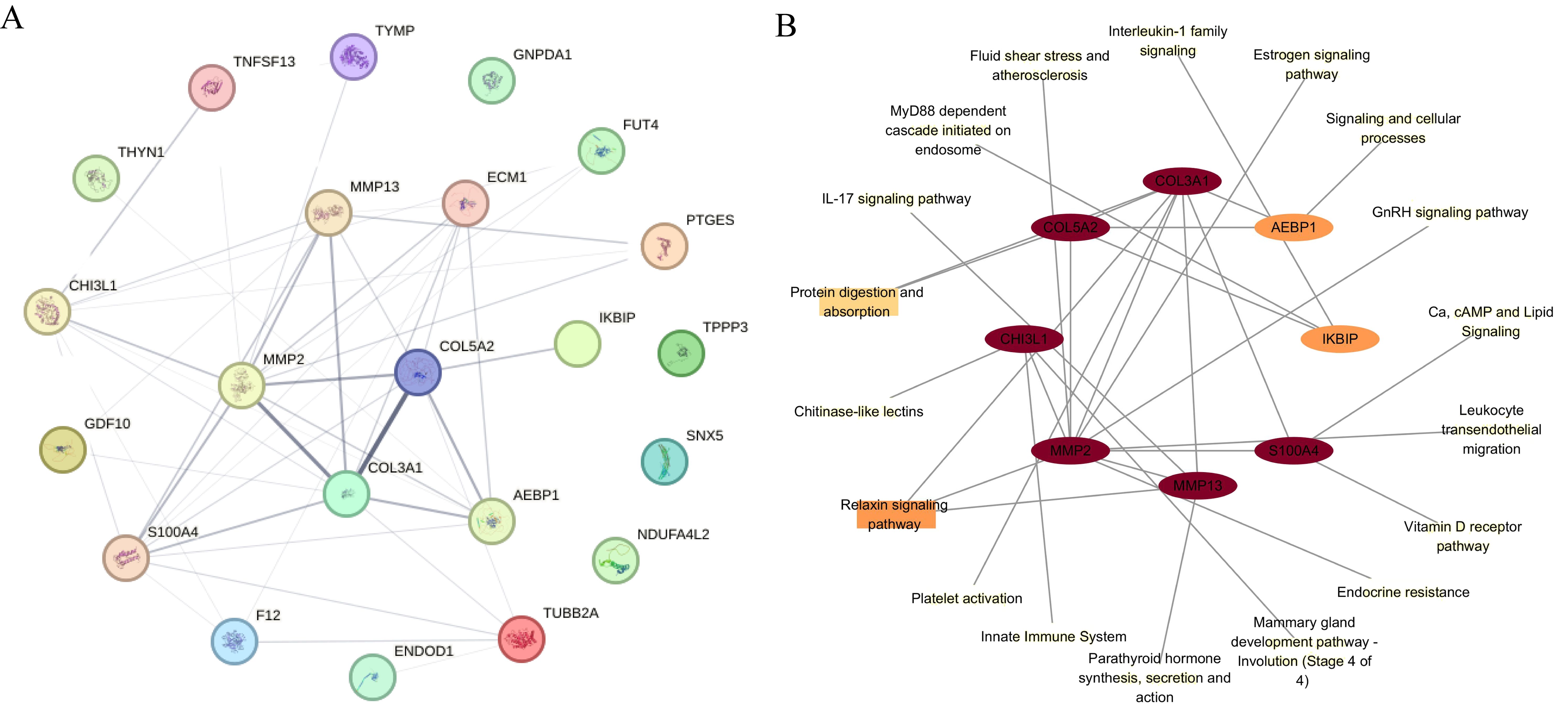
Figure 5. PPI analysis and KEGG pathway. (A) The PPI network of differentially regulated proteins across all three omics levels. (B) Eight proteins in ONFH cartilage were identified on the basis of their degree of interaction in the PPI network. The KEGG pathways associated with the eight proteins are indicated. The ellipse indicates proteins, and the rectangle indicates the KEGG pathway or biological process. The shapes are colored from yellow to red, representing low to high degrees. PPI, protein–protein interaction; KEGG, Kyoto Encyclopedia of Genes and Genomes.
3.7 Chemical−gene expression interactions in ONFH
Six hundred and seventy-six chemicals associated with DEGs across all three omics levels were identified. The top ten chemicals associated with ONFH were identified on the basis of the number of genes on which the chemicals act, in the order of benzo(a)pyrene (BaP), valproic acid, bisphenol A (BPA), tobacco smoke contamination, cisplatin, estradiol, dexamethasone (Dex), abrine, doxorubicin, and cyclosporine (Table 1).
3.8 qRT−PCR validation of mRNAs
To validate the reliability of the transcriptomic data, we quantified MMP13 and CHI3L1 expression in four ONFH patients and four control patients via qRT−PCR. The results of qRT−PCR showed that the expression levels of MMP13 and CHI3L1 were consistent with those of the transcriptomic analysis (Figure 6).
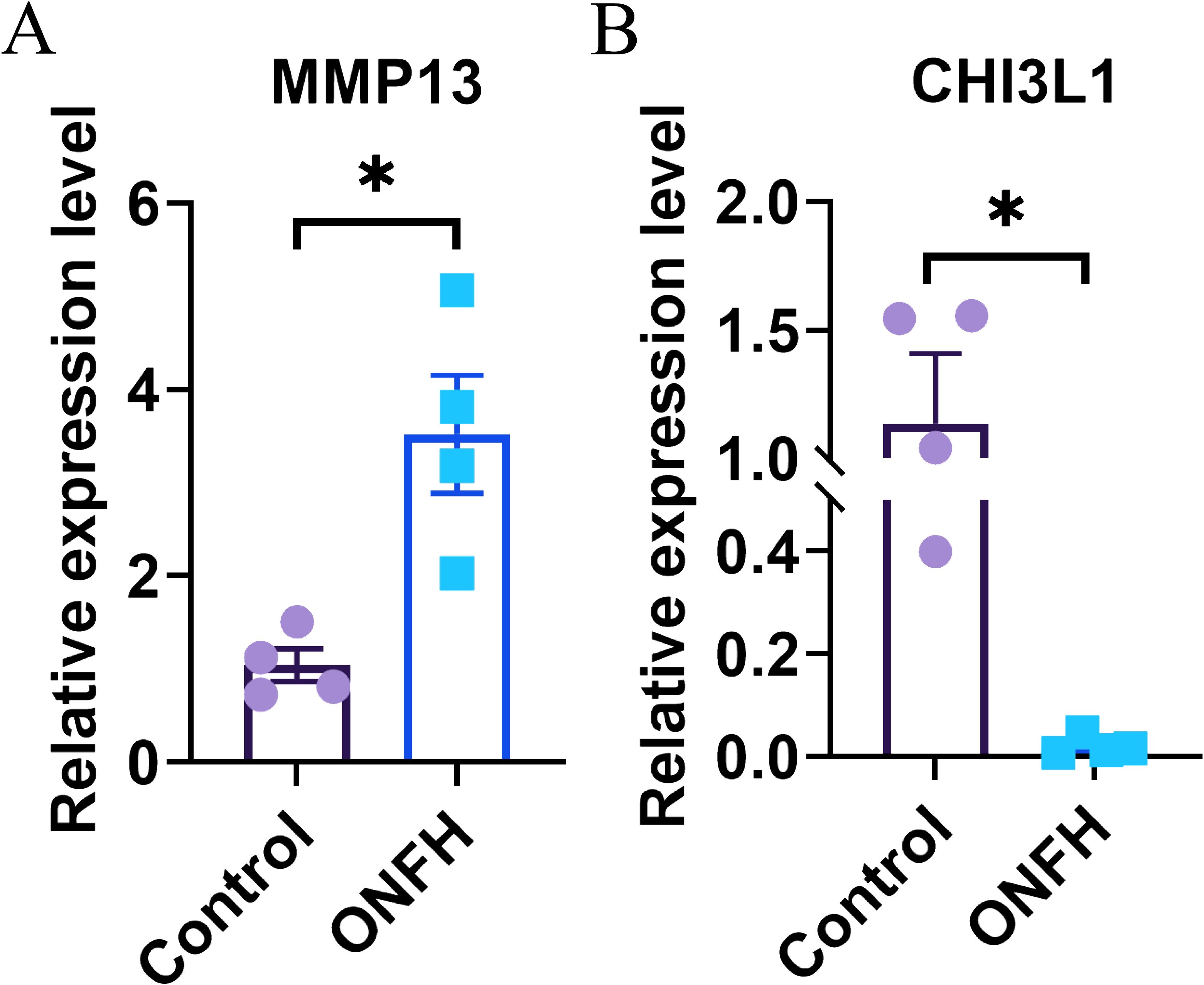
Figure 6. Results of qRT−PCR. (A, B) Confirmation of the transcriptomic results for mRNAs through qRT−PCR, with statistical significance at P < 0.05 (*). qRT−PCR, quantitative real-time PCR; ONFH, osteonecrosis of femoral head.
3.9 IHC verification
IHC was used to evaluate MMP13 and CHI3L1 protein expression in ONFH and control tissues (Figure 7). The protein expression levels of MMP13 in the superficial zone (SZ), middle zone (MZ), and deep zone (DZ) of ONFH cartilage were significantly greater than those in the control cartilage (all P values were less than 0.05). The protein expression levels of CHI3L1 in the SZ and MZ of ONFH cartilage were significantly lower than those in the control cartilage (all P values were less than 0.01). However, the protein expression levels of CHI3L1 in the DZ of ONFH cartilage were significantly greater than those in the control (P values were less than 0.01). The immunohistochemical results were consistent with the results of the multiomics analysis.
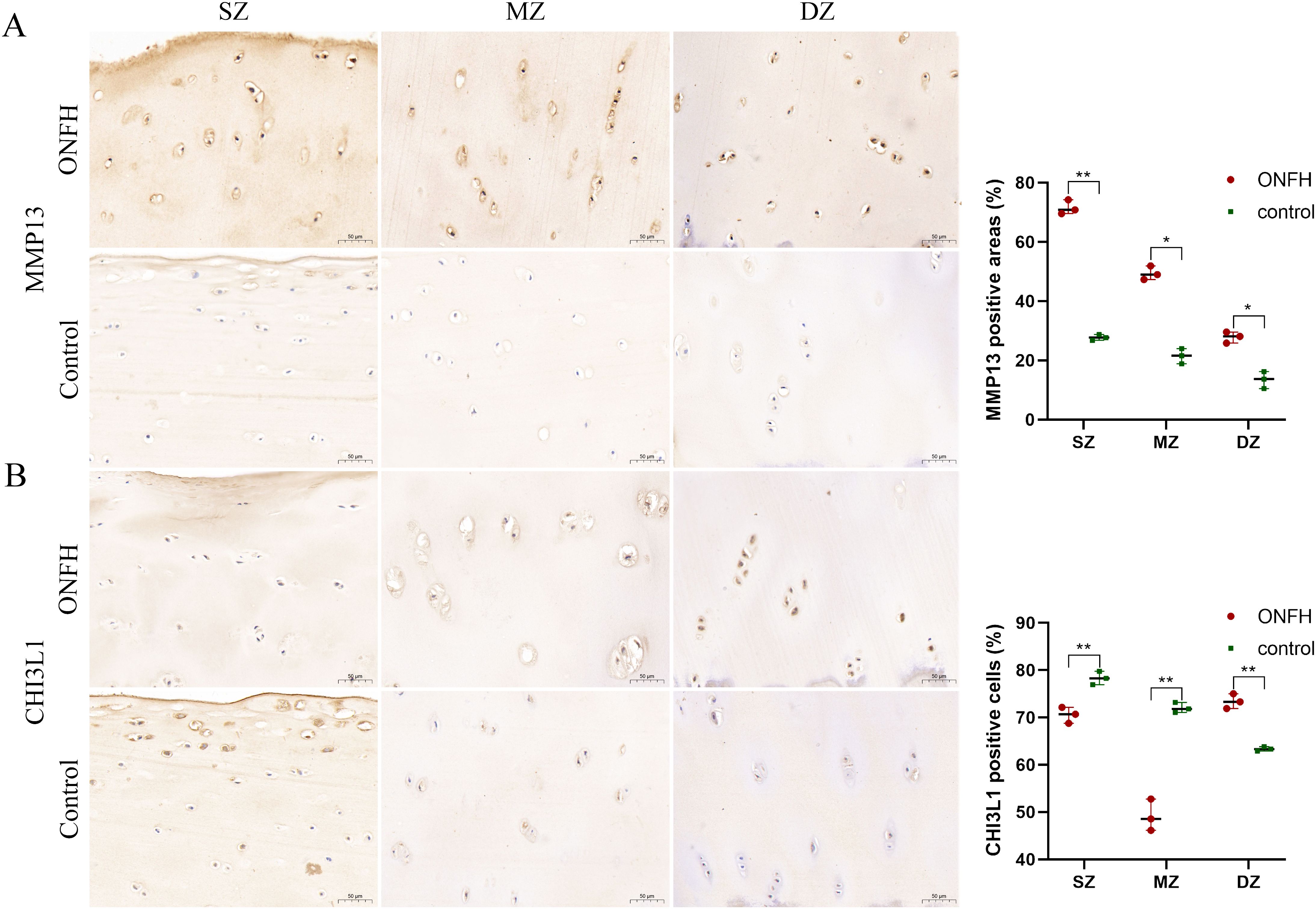
Figure 7. IHC results for the MMP13 (A) and CHI3L1 (B) proteins in hip cartilage from ONFH patients and fracture controls. Original magnification x300 of the SZ, MZ, and DZ. *P value < 0.05, **P value < 0.01. IHC, Immunohistochemistry; ONFH, osteonecrosis of femoral head; SZ, superficial zone; MZ, middle zone; DZ, deep zone. Scale bar: 50 μm.
4 Discussion
The pathophysiology of ONFH remains unclear. Recently, it has been increasingly recognized that cartilage degeneration plays a crucial role in ONFH progression (23). Previous studies of ONFH have focused mainly on single omics methods, such as methylation (24), transcription (25), or protein expression (26), or have combined up to two of these omics assays (27). However, single-omics analysis does not provide a systematic understanding of complex biological processes. This study provides comprehensive multiomics analyses of biological changes in ONFH cartilage across genome-wide DNA methylation profiling, gene expression profiling, and quantitative proteomics.
MMP13 breaks down the ECM, efficiently cleaves type II collagen and participates in cartilage pathophysiologic processes. MMP13 can degrade aggrecan, one of the most essential components of cartilage, and cause damage to cartilage (28). A previous study (28) confirmed increased levels of MMP13 in patients with ONFH, which is consistent with our omics and IHC results. During the development of ONFH, the expression of chondrogenesis-related genes (MMP13) is significantly and progressively elevated (29). Previous studies have shown that ECM1 binds to other extracellular matrix proteins, including collagen type IV, fibronectin, laminin 332, fibulin-1C/1D and MMP-9 (30). ECM1 plays a role in endochondral bone formation (31), promotes the proliferation of endothelial cells, and induces angiogenesis (32). TPPP3 enables the binding of tubulin, which is an early progenitor cell marker for both tendon and osteochondral cells. The heterotopic ossification-induced increase in TPPP3+ cells in a postoperative heterotopic ossification hip arthroplasty model led to ectopic cartilage formation (33). COL3A1 encodes the pro-alpha1 chain of type III collagen, a fibrillar collagen found in extensible connective tissues. Type III collagen is also a vital regulator of the collagen fibrillar structure and biomechanics of articular cartilage (34). A study by Wang et al. indicated that COL3A1 can be a biomarker for cartilage degradation in posttraumatic osteoarthritis (35). S100A4 encodes a small calcium-binding protein that is commonly overexpressed in a range of different tumor types, and a growing body of evidence suggests that S100A4 has an essential role in the process of cancer metastasis (36). The S100A4 protein binds to receptors for advanced glycation end products and mediates the induction of MMP13 in hip articular chondrocytes (37). Our findings indicate that COL3A1 and S100A4 are upregulated across multiple omics levels in ONFH cartilage and are significantly associated with ONFH cartilage degeneration. Moreover, additional studies should be conducted in the future.
CHI3L1 is expressed and secreted by various cell types, including macrophages, chondrocytes, fibroblast-like synovial cells, and vascular smooth muscle cells (38). CHI3L1 was localized in chondrocytes in the superficial and middle layers of the cartilage. CHI3L1 reflects not only the degree of inflammation but also the metabolism of cartilage. Kawasaki et al. (39) reported that a high abundance of CHI3L1 was correlated with Ficat stage III disease with collapsed femoral head. However, no other studies have reported a role for this gene in ONFH. We found that the protein expression of CHI3L1 was lower in chondrocytes and the superficial and middle layers of ONFH patients compared with controls. However, elevated expression in the deeper layers may occur because bone tissue necrosis first leads to inflammatory changes in the deeper layers of the cartilage. GDF10 (also called BMP-3b), a member of the bone morphogenetic protein (BMP) family and the transforming growth factor-beta superfamily, has been reported to be an essential regulator of critical events in the processes of articular chondrocyte differentiation and bone formation (40, 41). However, the role of GDF10 in ONFH is not fully understood. Our results revealed that GDF10, which is associated with cartilage degeneration, was downregulated at the omics level in ONFH patients. GNPDA1 is involved in the pathways of glycolysis and glycosaminoglycan metabolism. The downregulation of GNPDA1 leads to a decreased energy supply, which is one of the mechanisms of cartilage degeneration in ONFH. TNFSF13 (also called APRIL) is produced by myeloid cells and their precursors in the bone marrow (42) and is involved in the induction of the immunoglobulin switch and the survival of plasmocytes (43). It is involved in the development of systemic autoimmune diseases such as lupus erythematosus and rheumatoid arthritis. Moreover, it strongly promotes the proliferation of cancer cells and can be secreted by these cells (44). The downregulation of TNFSF13 in ONFH cartilage may be the reason for corticosteroid-induced ONFH, and most patients with this condition experienced autoimmune diseases in the past and required long-term glucocorticoid treatment.
Homeostasis of the cartilage matrix is maintained by cartilage cells. The ECM, which is mainly composed of type II collagen and proteoglycans, is a significant component of cartilage and is essential for chondrocyte function. In addition to supporting chondrocyte growth, the ECM responds to external environmental stimuli through dynamic regulation to maintain articular cartilage homeostasis (45). The key enzymes involved in cartilage destruction are matrix-degrading enzymes, which include matrix metalloproteinases (MMPs) and disintegrin and metalloproteinase with thrombospondin-like motifs (ADAMTSs) (46). In current studies, aggrecan is mainly degraded by MMPs (1, 3 and 13) and ADAMTSs (4 and 5), whereas collagen II is degraded by MMPs, mainly MMP13 (47, 48). The results of this study revealed that in ONFH cartilage, MMP13, MMP2, and ADAMTS5 are upregulated, which disrupts the homeostasis of the ECM and thus causes cartilage degeneration. Stabilizing the ECM in the future could delay the progression of ONFH. Amino sugars are important compounds required to form chondrocytes and represent one of the elementary units of the cartilage matrix and joint fluid (49). CCN2 may regulate the levels of free amino acids in the extracellular matrix of cartilage under physiological conditions (50). A study revealed a lower amino acid content in OA articular and meniscal cartilage than in normal articular cartilage (51). The levels of amino sugars were reduced in the present study, which may also be a potential factor in ONFH cartilage degeneration.
Additionally, among the identified chemicals, Dex is a recognized exposure factor that contributes to ONFH and is commonly used to construct experimental models of ONFH (52). Treating traumatic joint injuries with Dex prevents the initial damage caused by the release of inflammatory cytokines and maintains the cartilage structure (53). However, repeated use of Dex at high doses over a long period of time may result in more harm than beneficial effects to the joint (54). Dex results in increased expression of MMP13 (55) and F12 mRNA (56). Long-term administration of Dex resulted in decreased subchondral bone mass and bone density in mice. Research has shown that Dex hardens the extracellular matrix of chondrocytes, partly by activating AKT, and accelerates the death of chondrocytes in the layer of hardened cartilage. In addition, Dex attenuates stress-responsive autophagy over time (57). Steroids produced through the OPG/RANK/RANKL signaling pathway can lead to an imbalance in the bone remodeling process of the femoral head, inhibiting bone formation and causing osteonecrosis (58). An association was found between the methylation of the OPG, RANK and RANKL genes and steroid-related ONFH (59). Tobacco smoke contamination results in increased expression of CHI3L1 mRNA (60) and GNPDA1 protein (61), leading to decreased expression of MMP13 (62), S100A4 (63), and FUT4 mRNAs (64). CHI3L1 can be a potential marker for the severity and progression of osteoarthritis cartilage degeneration (65). CHI3L1 enhances mitosis in chondrocytes through MAP kinase and PI3 kinase-mediated signaling and induces SOX9 and type II collagen synthesis (66). Smoking increases the risk of ONFH (67), which aligns with our findings. Estradiol not only has positive effects on bone growth and bone remodeling (68) but can also lead to adverse effects of venous thromboembolism (69). Altered DNA methylation has more recently been implicated in BaP-mediated toxicity. DNA methylation, gene mutations and DNA damage can influence alternative splicing during gene transcription (70). Selective splicing is a significant process that generates diverse proteins in more complex organisms (71). BaP causes abnormal selective splicing of specific genes (72). Analysis of the changes in gene expression revealed that various important pathways, such as drug metabolism signaling, NRF2-mediated oxidative stress responses, and glutathione-mediated detoxification, are affected by BaP. Furthermore, the activation of several disease pathways, including organism death, growth failure, and abnormal development of embryonic tissues, is predicted (73). NRF2 is a major regulator of intracellular antioxidants, and upregulation of its expression provides femoral protection in glucocorticoid-induced ONFH (74). BaP significantly suppressed early and late osteogenic differentiation and downregulated runt-related transcription factor 2, osteocalcin, and osteopontin during the induction of BMP2 in mesenchymal stem cells (75). Moreover, BaP can impair the expression and function of the sonic hedgehog signaling pathway, perturbing the proliferation of chondrocytes and disturbing craniofacial skeletal development (76). Valproic acid decreases the expression of MMP2 and MMP9 mRNAs in osteosarcoma cells (77). Valproic acid results in increased methylation of the FUT4 and F12 genes (78). Previous studies have shown that valproic acid has a teratogenic effect on human chondrogenesis (79) and causes thrombocytopenia (80). Valproic acid counteracts the suppressive impact of glucocorticoids on the proliferation and osteogenesis of bone marrow−derived mesenchymal stem cells by suppressing apoptosis and enhancing the expression of proteins linked to osteogenesis. This mechanism may contribute to the prevention of glucocorticoid-induced ONFH in rats (81). BPA, a high-volume chemical with endocrine-disrupting properties, has been extensively studied in the context of bone metabolism. BPA results in decreased expression of FUT4 mRNA (82). BPA inhibits osteoblast differentiation and bone formation by activating retinoic acid-related orphan receptor α (83), resulting in osteoporosis (84).
5 Conclusion
Our study identified 22 DEGs across all three omics levels and several crucial pathways in ONFH cartilage through multiomics analyses, which may help to elucidate the pathogenesis of cartilage degeneration in ONFH. Moreover, we identified ten major environmental factors associated with ONFH progression, which can aid the early prevention and treatment of ONFH.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the Human Ethics Committees of Xi’an Jiaotong University (No. 2017-196). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
XL: Conceptualization, Formal Analysis, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. XW: Formal Analysis, Software, Validation, Visualization, Writing – original draft. PW: Investigation, Methodology, Validation, Writing – original draft. YZ: Methodology, Validation, Writing – review & editing. JL: Resources, Supervision, Writing – review & editing. GL: Resources, Writing – original draft. RL: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The study was supported by National Natural Scientific Fund of China (No. 81772411) and Natural Science Foundation of Shaanxi, China (No. 2023-YBSF-094).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2024.1419742/full#supplementary-material
Supplementary Figure 1 | GO and KEGG enrichment analyses at the two- or three-omics level. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Supplementary Figure 2 | Integrated enrichment analyses on the basis of adjustment direction at the two- or three-omics levels. (A) GO pathways of the upregulated genes. (B) KEGG pathway analysis of the upregulated genes. (C) GO pathways of the downregulated genes. (D) KEGG pathway analysis of the downregulated genes. The results of two- or three-omics enrichment analyses on the basis of reorientation. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Supplementary Figure 3 | GO and KEGG enrichment analyses of the upregulated gene sets at the two- or three-omics levels. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Supplementary Figure 4 | GO and KEGG enrichment analyses of the downregulated gene sets at the two- or three-omics levels. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Abbreviations
ONFH, osteonecrosis of femoral head; OA, osteoarthritis; DEGs, differentially expressed genes; TMT, tandem mass tag; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein−protein interaction; CTD, Comparative Toxicogenomics Database; IHC, immunohistochemistry; ECM, extracellular matrix; SZ, superficial zone; MZ, middle zone; DZ, deep zone.
References
1. Malizos KN, Karantanas AH, Varitimidis SE, Dailiana ZH, Bargiotas K, Maris T. Osteonecrosis of the femoral head: etiology, imaging and treatment. Eur J Radiol. (2007) 63:16–28. doi: 10.1016/j.ejrad.2007.03.019
2. Fukushima W, Fujioka M, Kubo T, Tamakoshi A, Nagai M, Hirota Y. Nationwide epidemiologic survey of idiopathic osteonecrosis of the femoral head. Clin Orthop Relat Res. (2010) 468:2715–24. doi: 10.1007/s11999-010-1292-x
3. Xu R, Wei B, Li J, Huang C, Lin R, Tang C, et al. Investigations of cartilage matrix degeneration in patients with early-stage femoral head necrosis. Med Sci Monit. (2017) 23:5783–92. doi: 10.12659/msm.907522
4. Magnussen RA, Guilak F, Vail TP. Cartilage degeneration in post-collapse cases of osteonecrosis of the human femoral head: altered mechanical properties in tension, compression, and shear. J Orthop Res. (2005) 23:576–83. doi: 10.1016/j.orthres.2004.12.006
5. Rushton MD, Reynard LN, Barter MJ, Refaie R, Rankin KS, Young DA, et al. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol (Hoboken NJ). (2014) 66:2450–60. doi: 10.1002/art.38713
6. Wang W, Yu Y, Hao J, Wen Y, Han J, Hou W, et al. Genome-wide DNA methylation profiling of articular cartilage reveals significant epigenetic alterations in Kashin-Beck disease and osteoarthritis. Osteoarthritis Cartilage. (2017) 25:2127–33. doi: 10.1016/j.joca.2017.08.002
7. Li L, Ding Y, Liu B, Wang Z, Carlone DL, Yu X, et al. Transcriptome landscape of the late-stage alcohol-induced osteonecrosis of the human femoral head. Bone. (2021) 150:116012. doi: 10.1016/j.bone.2021.116012
8. Hermansson M, Sawaji Y, Bolton M, Alexander S, Wallace A, Begum S, et al. Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin betaA (activin A), a regulatory molecule for chondrocytes. J Biol Chem. (2004) 279:43514–21. doi: 10.1074/jbc.M407041200
9. Subramanian I, Verma S, Kumar S, Jere A, Anamika K. Multi-omics data integration, interpretation, and its application. Bioinf Biol Insights. (2020) 14:1177932219899051. doi: 10.1177/1177932219899051
10. Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Briefings In Bioinf. (2018) 19:1370–81. doi: 10.1093/bib/bbx066
11. Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. (2017) 18:83. doi: 10.1186/s13059-017-1215-1
12. Vicenti G, Bizzoca D, Carrozzo M, Solarino G, Moretti B. Multi-omics analysis of synovial fluid: a promising approach in the study of osteoarthritis. J Biol Regulators Homeostatic Agents. (2018) 32:9–13.
13. Wang Y, Zhang F, Li X, Li X, Wang J, He J, et al. Integrated multi-omics techniques and network pharmacology analysis to explore the material basis and mechanism of simiao pill in the treatment of rheumatoid arthritis. ACS Omega. (2023) 8:11138–50. doi: 10.1021/acsomega.2c07959
14. Nicolle R, Ayadi M, Gomez-Brouchet A, Armenoult L, Banneau G, Elarouci N, et al. Integrated molecular characterization of chondrosarcoma reveals critical determinants of disease progression. Nat Commun. (2019) 10:4622. doi: 10.1038/s41467-019-12525-7
15. Olden K, Wilson S. Environmental health and genomics: visions and implications. Nat Rev Genet. (2000) 1:149–53. doi: 10.1038/35038586
16. Zhao D, Zhang F, Wang B, Liu B, Li L, Kim S-Y, et al. Guidelines for clinical diagnosis and treatment of osteonecrosis of the femoral head in adults (2019 version). J Orthopaedic Translation. (2020) 21:100–10. doi: 10.1016/j.jot.2019.12.004
17. Mattingly CJ, Colby GT, Rosenstein MC, Forrest JN, Boyer JL. Promoting comparative molecular studies in environmental health research: an overview of the comparative toxicogenomics database (CTD). Pharmacogenomics J. (2004) 4:5–8. doi: 10.1038/sj.tpj.6500225
18. Ficat RP. Idiopathic bone necrosis of the femoral head. Early diagnosis and treatment. J Bone Joint Surg Br. (1985) 67:3–9. doi: 10.1302/0301-620X.67B1.3155745
19. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. (2001) 98:5116–21. doi: 10.1073/pnas.091062498
20. Binns D, Dimmer E, Huntley R, Barrell D, O’Donovan C, Apweiler R. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. (2009) 25:3045–6. doi: 10.1093/bioinformatics/btp536
21. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. (2000) 28:27–30. doi: 10.1093/nar/28.1.27
22. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. (2021) 49:D605–D12. doi: 10.1093/nar/gkaa1074
23. Cao H, Guan H, Lai Y, Qin L, Wang X. Review of various treatment options and potential therapies for osteonecrosis of the femoral head. J Orthop Translat. (2016) 4:57–70. doi: 10.1016/j.jot.2015.09.005
24. Zhou M, Xi J, Cheng Y, Sun D, Shu P, Chi S, et al. Reprogrammed mesenchymal stem cells derived from iPSCs promote bone repair in steroid-associated osteonecrosis of the femoral head. Stem Cell Res Ther. (2021) 12:175. doi: 10.1186/s13287-021-02249-1
25. Liao Z, Jin Y, Chu Y, Wu H, Li X, Deng Z, et al. Single-cell transcriptome analysis reveals aberrant stromal cells and heterogeneous endothelial cells in alcohol-induced osteonecrosis of the femoral head. Commun Biol. (2022) 5:324. doi: 10.1038/s42003-022-03271-6
26. Song J, Wu J, Poulet B, Liang J, Bai C, Dang X, et al. Proteomics analysis of hip articular cartilage identifies differentially expressed proteins associated with osteonecrosis of the femoral head. Osteoarthritis Cartilage. (2021) 29:1081–92. doi: 10.1016/j.joca.2021.04.005
27. Ma M, Li P, Liu L, Cheng S, Cheng B, Liang CJ, et al. Integrating transcriptome-wide association study and mRNA expression profiling identifies novel genes associated with osteonecrosis of the femoral head. Front Genet. (2021) 12:663080. doi: 10.3389/fgene.2021.663080
28. Chen B, Liu Y, Cheng L. IL-21 enhances the degradation of cartilage through the JAK-STAT signaling pathway during osteonecrosis of femoral head cartilage. Inflammation. (2018) 41:595–605. doi: 10.1007/s10753-017-0715-1
29. Chen G, Zhong L, Wang Q, Li Z, Shang J, Yang Q, et al. The expression of chondrogenesis-related and arthritis-related genes in human ONFH cartilage with different Ficat stages. PeerJ. (2019) 7:e6306. doi: 10.7717/peerj.6306
30. Sercu S, Zhang L, Merregaert J. The extracellular matrix protein 1: its molecular interaction and implication in tumor progression. Cancer Invest. (2008) 26:375–84. doi: 10.1080/07357900701788148
31. Deckers MM, Smits P, Karperien M, Ni J, Tylzanowski P, Feng P, et al. Recombinant human extracellular matrix protein 1 inhibits alkaline phosphatase activity and mineralization of mouse embryonic metatarsals in vitro. Bone. (2001) 28:14–20. doi: 10.1016/s8756-3282(00)00428-2
32. Mongiat M, Fu J, Oldershaw R, Greenhalgh R, Gown AM, Iozzo RV. Perlecan protein core interacts with extracellular matrix protein 1 (ECM1), a glycoprotein involved in bone formation and angiogenesis. J Biol Chem. (2003) 278:17491–9. doi: 10.1074/jbc.M210529200
33. Yea JH, Gomez-Salazar M, Onggo S, Li Z, Thottappillil N, Cherief M, et al. Tppp3(+) synovial/tendon sheath progenitor cells contribute to heterotopic bone after trauma. Bone Res. (2023) 11:39. doi: 10.1038/s41413-023-00272-x
34. Wang C, Brisson BK, Terajima M, Li Q, Hoxha KH, Han B, et al. Type III collagen is a key regulator of the collagen fibrillar structure and biomechanics of articular cartilage and meniscus. Matrix Biol: J Int Soc For Matrix Biol. (2020) 85-86:47–67. doi: 10.1016/j.matbio.2019.10.001
35. Wang Y, Li Y, Khabut A, Chubinskaya S, Grodzinsky AJ, Önnerfjord P. Quantitative proteomics analysis of cartilage response to mechanical injury and cytokine treatment. Matrix Biol: J Int Soc For Matrix Biol. (2017) 63:11–22. doi: 10.1016/j.matbio.2016.12.004
36. Aisenbrey EA, Bryant SJ. The role of chondroitin sulfate in regulating hypertrophy during MSC chondrogenesis in a cartilage mimetic hydrogel under dynamic loading. Biomaterials. (2019) 190-191:51–62. doi: 10.1016/j.biomaterials.2018.10.028
37. Sparvero LJ, Asafu-Adjei D, Kang R, Tang D, Amin N, Im J, et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J Trans Med. (2009) 7:17. doi: 10.1186/1479-5876-7-17
38. Tizaoui K, Yang JW, Lee KH, Kim JH, Kim M, Yoon S, et al. The role of YKL-40 in the pathogenesis of autoimmune diseases: a comprehensive review. Int J Biol Sci. (2022) 18:3731–46. doi: 10.7150/ijbs.67587
39. Kawasaki M, Hasegawa Y, Kondo S, Iwata H. Concentration and localization of YKL-40 in hip joint diseases. J Rheumatol. (2001) 28:341–5.
40. Kaihara S, Bessho K, Okubo Y, Sonobe J, Komatsu Y, Miura M, et al. Over expression of bone morphogenetic protein-3b (BMP-3b) using an adenoviral vector promote the osteoblastic differentiation in C2C12 cells and augment the bone formation induced by bone morphogenetic protein-2 (BMP-2) in rats. Life Sci. (2003) 72:1683–93. doi: 10.1016/S0024-3205(02)02477-3
41. Lafont JE, Talma S, Hopfgarten C, Murphy CL. Hypoxia promotes the differentiated human articular chondrocyte phenotype through SOX9-dependent and -independent pathways. J Biol Chem. (2008) 283:4778–86. doi: 10.1074/jbc.M707729200
42. Matthes T, Dunand-Sauthier I, Santiago-Raber M-L, Krause K-H, Donze O, Passweg J, et al. Production of the plasma-cell survival factor a proliferation-inducing ligand (APRIL) peaks in myeloid precursor cells from human bone marrow. Blood. (2011) 118:1838–44. doi: 10.1182/blood-2011-01-332940
43. Vincent FB, Morand EF, Schneider P, Mackay F. The BAFF/APRIL system in SLE pathogenesis. Nat Rev Rheumatol. (2014) 10:365–73. doi: 10.1038/nrrheum.2014.33
44. Nowacka KH, Jabłońska E. Role of the APRIL molecule in solid tumors. Cytokine Growth Factor Rev. (2021) 61:38–44. doi: 10.1016/j.cytogfr.2021.08.001
45. Mouw JK, Ou G, Weaver VM. Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Biol. (2014) 15:771–85. doi: 10.1038/nrm3902
46. Zou Z-L, Sun M-H, Yin W-F, Yang L, Kong L-Y. Avicularin suppresses cartilage extracellular matrix degradation and inflammation via TRAF6/MAPK activation. Phytomed: Int J Phytother Phytopharmacol. (2021) 91:153657. doi: 10.1016/j.phymed.2021.153657
47. Wang M, Sampson ER, Jin H, Li J, Ke QH, Im H-J, et al. MMP13 is a critical target gene during the progression of osteoarthritis. Arthritis Res Ther. (2013) 15:R5. doi: 10.1186/ar4133
48. Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. (2005) 434:648–52. doi: 10.1038/nature03417
49. Ma Q, Gao X. Categories and biomanufacturing methods of glucosamine. Appl Microbiol Biotechnol. (2019) 103:7883–9. doi: 10.1007/s00253-019-10084-x
50. Murase Y, Hattori T, Aoyama E, Nishida T, Maeda-Uematsu A, Kawaki H, et al. Role of CCN2 in amino acid metabolism of chondrocytes. J Cell Biochem. (2016) 117:927–37. doi: 10.1002/jcb.25377
51. Stabler TV, Byers SS, Zura RD, Kraus VB. Amino acid racemization reveals differential protein turnover in osteoarthritic articular and meniscal cartilages. Arthritis Res Ther. (2009) 11:R34. doi: 10.1186/ar2639
52. Li Z, Shao W, Lv X, Wang B, Han L, Gong S, et al. Advances in experimental models of osteonecrosis of the femoral head. J Orthopaedic Translation. (2023) 39:88–99. doi: 10.1016/j.jot.2023.01.003
53. Liu Y, Cui Y, Chen Y, Gao X, Su Y, Cui L. Effects of dexamethasone, celecoxib, and methotrexate on the histology and metabolism of bone tissue in healthy Sprague Dawley rats. Clin Interv Aging. (2015) 10:1245–53. doi: 10.2147/cia.S85225
54. Black R, Grodzinsky AJ. Dexamethasone: chondroprotective corticosteroid or catabolic killer? Eur Cell Mater. (2019) 38:246–63. doi: 10.22203/eCM.v038a17
55. Wu G, Wang L, Li H, Ke Y, Yao Y. Function of sustained released resveratrol on IL-1beta-induced hBMSC MMP13 secretion inhibition and chondrogenic differentiation promotion. J Biomater Appl. (2016) 30:930–9. doi: 10.1177/0885328215614425
56. Xu S, He X, Shi J, Li Z, Song J, Wang J, et al. Interaction between retinoic acid and FGF/ERK signals are involved in Dexamethasone-induced abnormal myogenesis during embryonic development. Toxicology. (2021) 461:152917. doi: 10.1016/j.tox.2021.152917
57. Chen L, Ni Z, Huang J, Zhang R, Zhang J, Zhang B, et al. Long term usage of dexamethasone accelerating accelerates the initiation of osteoarthritis via enhancing chondrocyte apoptosis and the extracellular matrix calcification and apoptosis of chondrocytes. Int J Biol Sci. (2021) 17:4140–53. doi: 10.7150/ijbs.64152
58. He MC, Zhang J, Chen XJ, Shen YS, Fang B, Qin YX, et al. Osteoclastic activity was associated with the development of steroid-induced osteonecrosis of femoral head. Artif Cells Nanomed Biotechnol. (2020) 48:1036–46. doi: 10.1080/21691401.2020.1774596
59. Sun M, Cao Y, Yang X, An F, Wu H, Wang J. DNA methylation in the OPG/RANK/RANKL pathway is associated with steroid-induced osteonecrosis of the femoral head. BMC Musculoskelet Disord. (2021) 22:599. doi: 10.1186/s12891-021-04472-6
60. Titz B, Kogel U, Martin F, Schlage WK, Xiang Y, Nury C, et al. A 90-day OECD TG 413 rat inhalation study with systems toxicology endpoints demonstrates reduced exposure effects of the aerosol from the carbon heated tobacco product version 1.2 (CHTP1.2) compared with cigarette smoke. II. Systems toxicology assessment. Food Chem Toxicol. (2018) 115:284–301. doi: 10.1016/j.fct.2018.02.058
61. Jagadapillai R, Chen J, Canales L, Birtles T, Pisano MM, Neal RE. Developmental cigarette smoke exposure: kidney proteome profile alterations in low birth weight pups. Toxicology. (2012) 299:80–9. doi: 10.1016/j.tox.2012.04.015
62. Szostak J, Boue S, Talikka M, Guedj E, Martin F, Phillips B, et al. Aerosol from Tobacco Heating System 2.2 has reduced impact on mouse heart gene expression compared with cigarette smoke. Food Chem Toxicol. (2017) 101:157–67. doi: 10.1016/j.fct.2017.01.013
63. Wright WR, Parzych K, Crawford D, Mein C, Mitchell JA, Paul-Clark MJ. Inflammatory transcriptome profiling of human monocytes exposed acutely to cigarette smoke. PloS One. (2012) 7:e30120. doi: 10.1371/journal.pone.0030120
64. Xiong R, Wu Y, Wu Q, Muskhelishvili L, Davis K, Tripathi P, et al. Integration of transcriptome analysis with pathophysiological endpoints to evaluate cigarette smoke toxicity in an in vitro human airway tissue model. Arch Toxicol. (2021) 95:1739–61. doi: 10.1007/s00204-021-03008-0
65. Szychlinska MA, Trovato FM, Di Rosa M, Malaguarnera L, Puzzo L, Leonardi R, et al. Co-expression and co-localization of cartilage glycoproteins CHI3L1 and lubricin in osteoarthritic cartilage: morphological, immunohistochemical and gene expression profiles. Int J Mol Sci. (2016) 17:359. doi: 10.3390/ijms17030359
66. Jacques C, Recklies AD, Levy A, Berenbaum F. HC-gp39 contributes to chondrocyte differentiation by inducing SOX9 and type II collagen expressions. Osteoarthritis Cartilage. (2007) 15:138–46. doi: 10.1016/j.joca.2006.07.003
67. Takahashi S, Fukushima W, Kubo T, Iwamoto Y, Hirota Y, Nakamura H. Pronounced risk of nontraumatic osteonecrosis of the femoral head among cigarette smokers who have never used oral corticosteroids: a multicenter case-control study in Japan. J Orthopaedic Sci: Off J Japanese Orthopaedic Assoc. (2012) 17:730–6. doi: 10.1007/s00776-012-0293-x
68. Börjesson AE, Lagerquist MK, Windahl SH, Ohlsson C. The role of estrogen receptor α in the regulation of bone and growth plate cartilage. Cell Mol Life Sci: CMLS. (2013) 70:4023–37. doi: 10.1007/s00018-013-1317-1
69. Daly E, Vessey MP, Hawkins MM, Carson JL, Gough P, Marsh S. Risk of venous thromboembolism in users of hormone replacement therapy. Lancet (London England). (1996) 348:977–80. doi: 10.1016/S0140-6736(96)07113-9
70. Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. (2013) 23:1256–69. doi: 10.1038/cr.2013.110
71. Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. (2007) 8:749–61. doi: 10.1038/nrg2164
72. Yan C, Wu W, Li H, Zhang G, Duerksen-Hughes PJ, Zhu X, et al. Benzo a pyrene treatment leads to changes in nuclear protein expression and alternative splicing. Mutat Res. (2010) 686:47–56. doi: 10.1016/j.mrfmmm.2010.01.015
73. Fang X, Corrales J, Thornton C, Clerk T, Scheffler BE, Willett KL. Transcriptomic changes in zebrafish embryos and larvae following benzo a pyrene exposure. Toxicol Sci. (2015) 146:395–411. doi: 10.1093/toxsci/kfv105
74. Yang N, Sun H, Xue Y, Zhang W, Wang H, Tao H, et al. Inhibition of MAGL activates the Keap1/Nrf2 pathway to attenuate glucocorticoid-induced osteonecrosis of the femoral head. Clin Transl Med. (2021) 11:e447. doi: 10.1002/ctm2.447
75. An L, Shi Q, Fan M, Huang G, Zhu M, Zhang M, et al. Benzo a pyrene injures BMP2-induced osteogenic differentiation of mesenchymal stem cells through AhR reducing BMPRII. Ecotoxicol Environ Safety. (2020) 203:110930. doi: 10.1016/j.ecoenv.2020.110930
76. He C, Zuo Z, Shi X, Li R, Chen D, Huang X, et al. Effects of benzo(a)pyrene on the skeletal development of Sebastiscus marmoratus embryos and the molecular mechanism involved. Aquat Toxicol (Amsterdam Netherlands). (2011) 101:335–41. doi: 10.1016/j.aquatox.2010.11.008
77. Yamanegi K, Yamane J, Kobayashi K, Ohyama H, Nakasho K, Yamada N, et al. Downregulation of matrix metalloproteinase-9 mRNA by valproic acid plays a role in inhibiting the shedding of MHC class I-related molecules A and B on the surface of human osteosarcoma cells. Oncol Rep. (2012) 28:1585–90. doi: 10.3892/or.2012.1981
78. van Breda SGJ, Claessen SMH, van Herwijnen M, Theunissen DHJ, Jennen DGJ, de Kok T, et al. Integrative omics data analyses of repeated dose toxicity of valproic acid in vitro reveal new mechanisms of steatosis induction. Toxicology. (2018) 393:160–70. doi: 10.1016/j.tox.2017.11.013
79. Aulthouse AL, Hitt DC. The teratogenic effects of valproic acid in human chondrogenesis in vitro. Teratology. (1994) 49:208–17. doi: 10.1002/tera.1420490317
80. Kumar R, Vidaurre J, Gedela S. Valproic acid-induced coagulopathy. Pediatr Neurol. (2019) 98:25–30. doi: 10.1016/j.pediatrneurol.2019.04.019
81. Zhou D, Chen YX, Yin JH, Tao SC, Guo SC, Wei ZY, et al. Valproic acid prevents glucocorticoid−induced osteonecrosis of the femoral head of rats. Int J Mol Med. (2018) 41:3433–47. doi: 10.3892/ijmm.2018.3534
82. Tait S, Tassinari R, Maranghi F, Mantovani A. Bisphenol A affects placental layers morphology and angiogenesis during early pregnancy phase in mice. J Appl Toxicol. (2015) 35:1278–91. doi: 10.1002/jat.3176
83. Turan S. Endocrine disrupting chemicals and bone. Best Pract Res Clin Endocrinol Metab. (2021) 35:101495. doi: 10.1016/j.beem.2021.101495
Keywords: osteonecrosis of femoral head, cartilage, methylation, transcriptomics, proteomics, chemical-gene interaction
Citation: Lu X, Wang X, Wang P, Zhu Y, Liu J, Liu G and Liu R (2024) Identification of candidate genes and chemicals associated with osteonecrosis of femoral head by multiomics studies and chemical-gene interaction analysis. Front. Endocrinol. 15:1419742. doi: 10.3389/fendo.2024.1419742
Received: 18 April 2024; Accepted: 08 August 2024;
Published: 26 August 2024.
Edited by:
Giacomina Brunetti, University of Bari Aldo Moro, ItalyReviewed by:
Bouchra Edderkaoui, Loma Linda Veterans Association for Research and Education, United StatesChaofan Zhang, First Affiliated Hospital of Fujian Medical University, China
Copyright © 2024 Lu, Wang, Wang, Zhu, Liu, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruiyu Liu, bGl1cnl1QDEyNi5jb20=
†These authors have contributed equally to this work
 Xueliang Lu1,2†
Xueliang Lu1,2† Xu Wang
Xu Wang Yingkang Zhu
Yingkang Zhu Ruiyu Liu
Ruiyu Liu