- 1The Orthopedic Center, Wenling First People’s Hospital (The Affiliated Wenling Hospital of Wenzhou Medical University), Wenling, China
- 2Hunan Provincial Engineering Research Center of Applied Microbial Resources Development for Livestock and Poultry, College of Bioscience and Biotechnology, Hunan Agricultural University, Changsha, China
- 3Department of Cardiology, Wenling First People’s Hospital (The Affiliated Wenling Hospital of Wenzhou Medical University), Wenling, China
Osteosarcoma is a common malignancy that often occurs in children, teenagers and young adults. Although the treatment strategy has improved, the results are still poor for most patients with metastatic or recurrent osteosarcomas. Therefore, it is necessary to identify new and effective prognostic biomarkers and therapeutic targets for diseases. Human genomes contain lncRNAs, transcripts with limited or insufficient capacity to encode proteins. They have been implicated in tumorigenesis, particularly regarding the onset, advancement, resistance to treatment, recurrence and remote dissemination of malignancies. Aberrant lncRNA expression in osteosarcomas has been reported by numerous researchers; lncRNAs have the potential to exhibit either oncogenic or tumor-suppressing behaviors and thus, to govern the advancement of this skeletal cancer. They are suspected to influence osteosarcoma cell growth, replication, invasion, migration, remote dissemination and programmed cell death. Additionally, they have been recognized as clinical markers, and may participate in the development of multidrug resistance. Therefore, the study of lncRNAs in the growth, metastasis, treatment and prognosis of osteosarcoma is very important for the active prevention and treatment of osteosarcoma. Consequently, this work reviews the functions of lncRNAs.
1 Introduction to the treatment and research of osteosarcoma
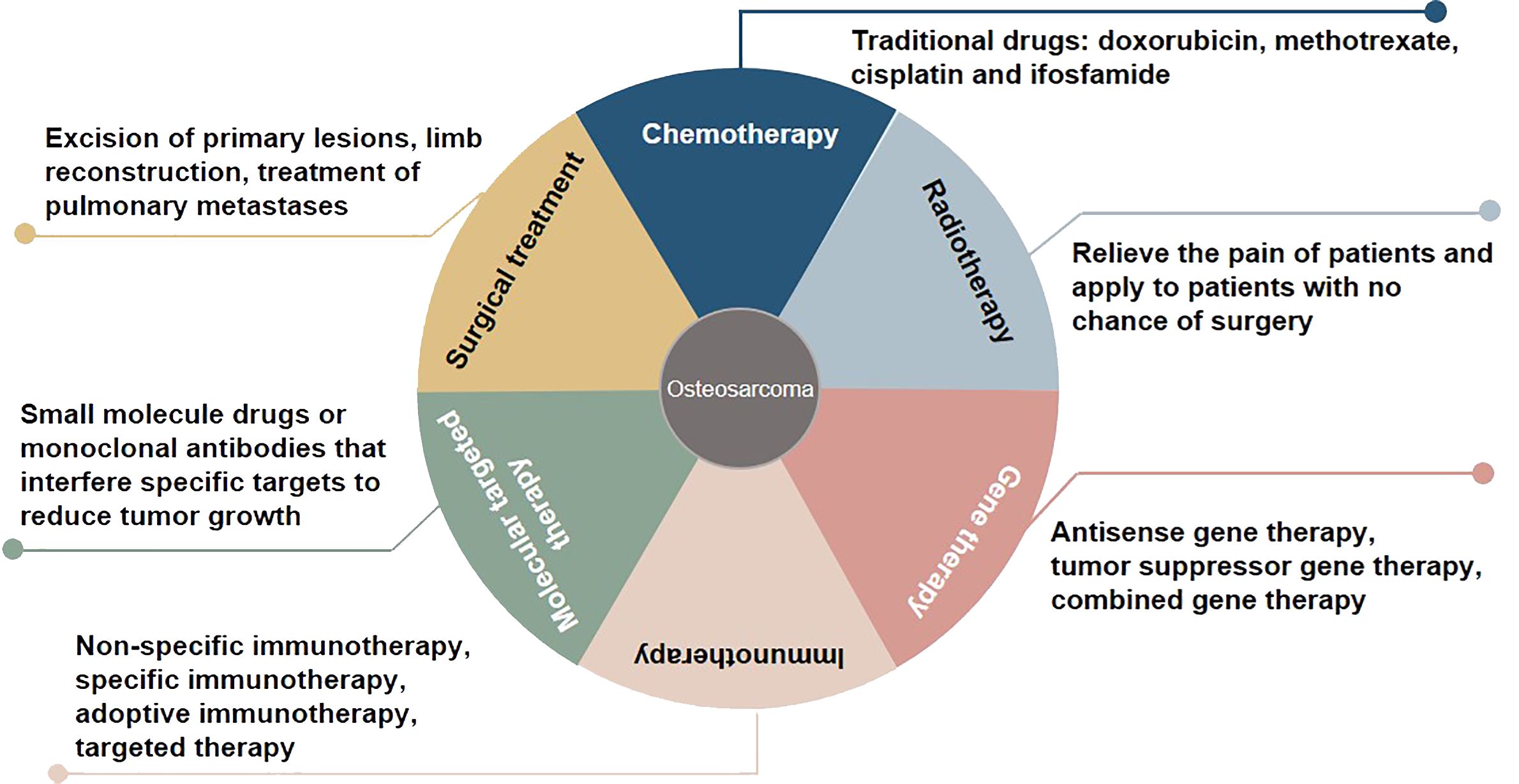
Osteosarcoma, which originates from mesenchymal cells, is one of the most frequently occurring bone tumor. It is responsible for a significant proportion of child and teenage mortalities occurring due to malignancy. Typical histological appearances of osteosarcoma include spindle cells and the abnormal generation of osteoid (1). The long-term survival rate of individuals who present without remote tumor dissemination is approximately 80%. Conversely, when metastatic deposits are detected at diagnosis, the likelihood of long-term survival is lower than 20%, even when aggressive treatment regimens are instituted (2). At present, some progress has been made in the treatment of osteosarcoma, including chemotherapy, radiotherapy, gene therapy, immunotherapy, molecular targeted therapy, and surgical treatment (3, 4) (Figure 1). Due to the limitations of early medical development, the early surgical treatment of osteosarcoma is mainly based on amputations to save lives. However, the appearance and function of residual limbs after amputations adversely affect patients’ quality of life, causing long-term psychological distress. With the rapid development of medical science and technology, limb salvage treatment for osteosarcoma has replaced most amputations in clinical practice (5). Doxorubicin, methotrexate, cisplatin and, ifosfamide are known as classic drugs for osteosarcoma chemotherapy that have significantly improved the curative effect, survival rate, and quality of life, and reduced the harm to the human body caused by the side effects of treatment (6). The sensitivity of osteosarcoma to radiotherapy is relatively poor, and it is recommended to relieve the pain of patients and apply it to patients with no chance of surgery (7). Gene therapy, immunotherapy, and molecular targeted therapy are new methods for treating osteosarcoma, and their application prospects and values are incalculable. In the studies of whole genome sequencing and whole exome sequencing of osteosarcoma, some genes usually changed, including tumor suppressor genes TP53, RB1, ATRX and DLG2 (8). Moreover, genomic instability of BRCA1/2-deficient tumors was observed in 80% of osteosarcomas (9). The combined genomic and transcriptome analysis of osteosarcoma samples also identified multiple fusion transcripts directly related to chromosome rearrangements. Although these fusion transcripts involve multiple loci in different cell lines and patient samples, the most common is affecting the TP53 locus (10). Tailor-made therapies using biomarkers that predict responses to specific molecular targeted therapies are central concepts in precision medicine. Osteosarcoma mostly exhibits somatic changes in cell cycle and/or DNA damage repair pathways. Targeted therapy based on this abnormal prediction is a potential precise medical method. For example, abnormal expression of TP53 impairs the G1 cell cycle checkpoint, thereby increasing the dependence of tumor cells on the G2 checkpoint to maintain DNA integrity and complete cell division (11). Therefore, drugs that disrupt the G2 checkpoint, such as WEE1 inhibitors, may enhance the activity of DNA damage agents and induce mitotic death in TP53-mutant osteosarcoma cells. However, most therapies are still in the stage of basic scientific research experiments, and only a few are used in clinical observation. In addition, the regulatory mechanism of osteosarcoma has yet to be clarified, so the therapeutic effect of targeted agents needs to be improved, which makes the use of contemporary treatments more challenging (12). Therefore, there is an urgent need to identify clinically relevant prognostic biomarkers for these patients.
Although several promising molecular markers have been detected in recent years, a diagnostic indicator particular to osteosarcoma has yet to be identified (13). Data are being accrued, which suggests that many malignancy-related genomic mutations occur in areas that, instead of encoding for proteins, frequently undergo transcription into IncRNAs. These are a cohort of non-protein transcripts of approximately 200 nucleotides in length (14).
LncRNAs contribute to the governance of gene expression via numerous mechanisms, for example, as epigenetic moderators, signals for the enhancement of transcription, transcription suppression decoys, or scaffolds for the generation of ribonucleoprotein complexes through engagement with proteins (15). The integrity and translation of mRNAs is influenced by lncRNAs; the latter also form antecedents to miRNAs and control their distribution. Overall, IncRNAs impact the expression of genes at both the transcriptional and post-transcriptional levels (15, 16). Additional pathways in which lncRNAs have been implicated encompass cell development, differentiation, and replication, together with the governance of the cell cycle and apoptosis (17, 18). They also play a significant part in the advancement and remote spread of a range of malignancies, for example, large intestine, hepatic, breast, bladder, and cervical neoplasias (19).
Even though research regarding the roles played by lncRNAs in osteosarcoma is still in its infancy, promising data imply that, in addition to their contribution to the control of numerous pathophysiological pathways, lncRNAs influence carcinogenesis as well as tumor advancement and remote dissemination (20, 21). They have also been utilized as biological markers, autonomous indicators of clinical prognosis and therapeutic targets, thus having notable diagnostic and therapeutic utility, as well as offering a method of tumor surveillance (22). Evidence suggests that the effect of chemotherapeutic drugs may be potentiated by lncRNAs, which influences patients’ prognosis (23). Therefore, this study reviewed the role of lncRNAs in osteosarcomas, which includes their influence on tumorigenesis, remote dissemination, clinical outcomes, and resistance to chemotherapeutic agents.
2 Classification and biological function of lncRNAs
LncRNAs were first identified in murine transcripts (24). Typically, their length extend in excess of 200 nucleotides, and they have no capacity to encode proteins (24). They are categorized into the following types: bidirectional, intergenic, intronic, antisense, sense, and enhancer lncRNAs, depending on the configurational relationship between the lncRNAs and the genes for protein coding within the genome (Figure 2A) (25). RNA polymerase II is responsible for the transcription of most of these molecules, with a 5’cap and polyadenylated tail are common (26). Owing to the myriad transcripts, they were first deemed to be “noise” associated with the transcription process (27). However, more detailed contemporary work has demonstrated that lncRNAs have an abundance of physiological roles, such as the moderation of the epigenetic, transcriptional, and post-transcriptional expression of genes (Figure 2B). Additionally, they have considerable intracellular functionality, contributing to cell replication, differentiation, cell cycle advancement, growth, and programmed cell death (28, 29). The alteration and remodeling of chromatin and histone, together with the localization of the nuclear body, are mechanisms via which gene expression can be modified by lncRNAs (30). Chromosomal configurations can also be adjusted by lncRNAs in concert with the SWI/SNF complex, a process that immediately impacts gene expression (31). Although lncRNAs are not attributed to coding protein, several are suspected of involvement in silencing protein encoding genes during transcription through modification of the structure of chromatin and the conscription of complexes that impact this process (32).
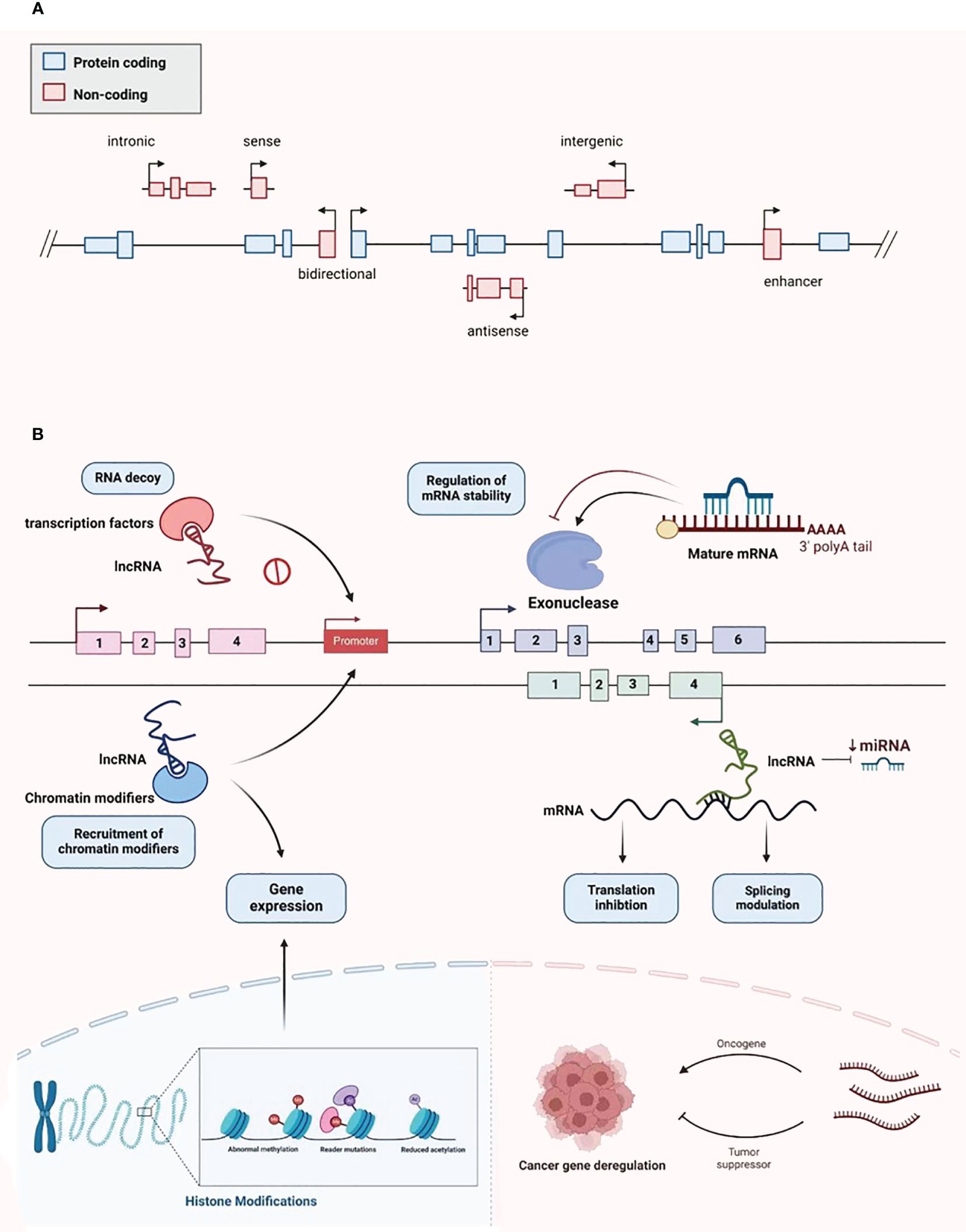
Figure 2 Classification and biological function of lncRNA. (A) LncRNA is divided into bidirectional, intergenic, intronic, antisense, sense, and enhancer lncRNA. (B) LncRNA plays various biological functions, including recruiting chromatin modifiers, regulating gene expression, participating in transcriptional silencing of protein coding genes, regulating mRNA stability, increasing mRNA expression, and being able to serve as key regulatory factors and oncogenes or tumor suppressors in tumor development and metastasis.
In stem cells, the differentiation of cells is influenced by lncRNAs; in somatic and pluripotent stem cells, sizeable intergenic non-coding RNAs can promote cellular reprogramming (33). Furthermore, in addition to immediately influencing target expression, lncRNAs also have the ability to govern expression through secondary pathways, for example, acting as ‘molecular sinks” for signaling molecules, such as RNA-binding proteins that moderate chromatin (32). A key mechanism for their ability to control gene expression is that they act as competing endogenous RNAs (ceRNAs) for miRNAs, reducing the bioavailability of miRNAs and enhancing target mRNA titers (34). Several studies have additionally demonstrated that lncRNAs work as key controlling molecules that behave as oncogenes or tumor-inhibiting agents. These are essential actors in numerous forms of malignancy, with the potential to promote the progression of carcinogenesis, remote tumor dissemination, and resistance to chemotherapeutic agents (35–38).
3 LncRNAs in osteosarcoma
3.1 Up-regulated lncRNAs in osteosarcoma
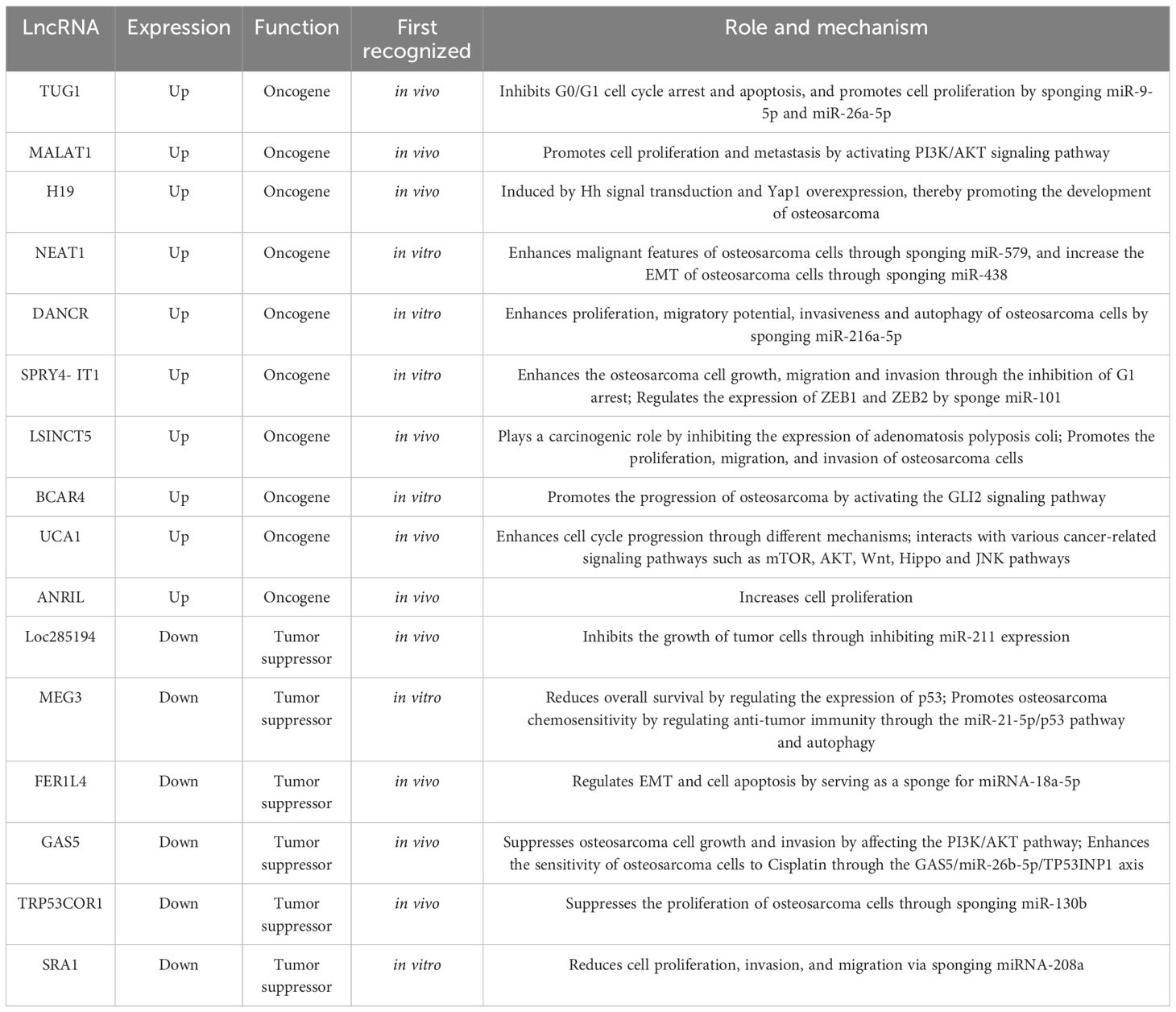
The potential oncogenic effects of lncRNAs in osteosarcoma described in previous studies are presented in Table 1. When samples from osteosarcomas were compared with neighboring normal tissues, the expression of the lncRNA TUG1, which is situated on chromosome 22q12.2 and is 7.1 kb in size, was noted to be amplified (2). This finding was related to adverse prognosis, forming an autonomous predictive indicator of overall survival (39). In vitro studies have demonstrated that cell replication is inhibited in the presence of TUG1 knockdown, which is associated with the arrest of the G0/G1 cell cycle and programmed cell death. Additionally, in vivo work has shown that TUG1 knockdown leads to a reduction in the growth of cancerous lesions (39). TUG1 can promote the proliferation of cancer cells. When TUG1 is inhibited, the proliferation of cancer cells decreases, and the replication efficiency of osteosarcoma cells decreases. Recent study has also found that TUG1 can act as a miR-26a-5p sponge and promote the progression of osteosarcoma by up-regulating ZBTB7C (40), so TUG1 can be used as a diagnostic marker for osteosarcoma, targeting TUG1 may be an effective strategy for the treatment of osteosarcoma.
Another lncRNA that exhibits excessive expression in osteosarcoma is metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which is also termed nuclear-enriched transcript 2 (NEAT2) and mascRNA. It comprises 8700 nucleotides and is present on a locus of chromosome 11q13 (41–43). MALAT1 is not polyadenylated but instead contains a highly unusual 3’-terminal structural motif designated as the stability element for nuclear expression (ENE), which is essential for the stability of mature transcripts and the subsequent accumulation and carcinogenic activity of MALAT1. Therefore, cells are free from the influence of cellular degradation pathways, leading to MALAT1’s sustained carcinogenic activity in a variety of cancer types (44, 45). It was originally identified as a biological indicator of metastatic disease in the initial phases of non-small cell lung cancer (46). In osteosarcoma samples, MALAT1 expression was increased, a finding that has a positive association with metastatic deposits in the lungs (47). Furthermore, the expression of phosphorylated PI3Kp85α, matrix metallopeptidase 9 (MMP-9), proliferating cell nuclear antigen (PCNA), and Akt were significantly inhibited in MALAT1-deficient cells. The replication and migration of osteosarcoma cells was inhibited following MALAT1 knockdown. Additionally, in MNNG/HOS and U2OS cells, the generation of tubular network configurations was suppressed, and stress fibers were fractured (48). The up-regulated expression of MALAT1 is closely related to the presence of distal osteosarcoma deposits, and the inhibition of MALAT1 can reduce the migration and invasion of osteosarcoma cells. In addition, it has been shown that, METTL3 in osteosarcoma cells promotes the m6A modification of MALAT1, and enhances the carcinogenic function of MALAT1 (49). One of the key oncogenic pathways in osteosarcomas affecting humans is the phosphatidylinositol 3-kinase (PI3K)/Akt pathway (50, 51). It has several major functions relating to cancer cell growth, apoptosis, and migratory and invasive properties (52). Diminishing MALAT1 expression led to lower levels of phosphorylated PI3K p85α and Akt (47). MALAT1 exhibits competitive bonding to miRNA-129-5p and therefore prevents this molecule from promoting the breakdown of RET. Subsequently, the rise in RET concentrations enhances the activity of the PI3K/Akt signaling pathway (53). Additionally, MALAT1 demonstrated oncogenic functions, increasing osteosarcoma cell growth potentially through the stimulation of the PI3K/AKT and RhoA/ROCK pathway (47, 48).
Initially described as a molecule that could inhibit epithelial cell differentiation, differentiation antagonizing non-protein coding RNA (DANCR or ANCR), which has its locus on human chromosome 4q12, has also been shown to enhance the stemness properties of cell lines from hepatocellular carcinoma (54, 55). DANCR can promote the pathogenesis of osteosarcoma, and the silencing of DANCR has been demonstrated to promote programmed cell death, using functional assays, to inhibit the replication, migratory ability, invasiveness, and autophagic capability of cells from osteosarcomas. The expression of SOX5 and levels of the receptor tyrosine kinase AXL are amplified by DANCR owing to its sponge-like function in relation to miR-216a-5p and miR-33a-5p (56), respectively. Moreover, METTL3 promotes osteosarcoma progression by increasing the stability of DANCR mRNA through m6A modification, which means that METTL3 may be a promising therapeutic target for osteosarcoma treatment (57).
Another lncRNA, H19, is 2.3 kb in size and is near the telomeric region of human chromosome 11p15.5. It is implicated in the governance of the expression of IGF2 (58). In osteosarcomas, an abnormal degree of H19 expression has been detected, which can be promoted by amplified Hedgehog (Hh) signaling and upregulated yes-associated protein 1 (Yap1) (59). This observation substantiates the theory that erroneous Hh signaling expression in osteoblasts plays a key role in the osteosarcoma associated with the overexpression of H19 and Yap1. Gallic acid inhibits tumor growth in osteosarcoma cells through the H19-mediated Wnt/β-catenin signaling regulatory axis (60).
The familial tumor syndrome multiple endocrine neoplasia type 1 locus, situated on chromosome 11, gives rise to the transcription of the lncRNA NEAT1 (61). Its oncogenic properties have been demonstrated to augment the neoplastic characteristics of cells from osteosarcomas (62). By sponging miR-579 and upregulating MMP13, the role of NEAT1 in the epithelial-mesenchymal transition (EMT) is promoted (63). NEAT1 can also sponge miR-438, increase the expression of STAT3, and inhibit that of STAT1, and then increase the EMT of osteosarcoma cells (64).
Another lncRNA, SPRY4-IT1, which has a size of 708 bp, originates from a SPRY4 intron on chromosome 5q31.3, which contains the coding for an intrinsic receptor-transduced mitogen-activated protein kinase pathway inhibitor (65, 66). The expression of this lncRNA was observed to be amplified in cell lines and specimens from osteosarcomas, while the knockdown of it inhibited the invasive, migratory, and replicative properties of the malignant cells through the generation of G1 cell cycle arrest and heightened apoptosis (67). SPRY4-IT1 can also regulate the expression of ZEB1 and ZEB2 by sponge miR-101 activity, thereby promoting the progression of osteosarcoma (68).
Compared with adjacent normal tissues, long stress-induced non-coding transcript 5 (LSINCT5) was significantly up-regulated in osteosarcoma tissues. The abnormal expression of LSINCT5 is usually associated with cancer progression and poor prognosis, and high LSINCT5 expression is associated with advanced Enneking stage, large tumor volume, high histological grade, and current distant metastasis (69). The overexpression of LSINCT5 can promote the proliferation, migration, and invasion of osteosarcoma cells in vitro, while the inhibition of its expression has the opposite effect (69). The exploration of the mechanism displayed that LSINCT5 can interact with EZH2 to inhibit the expression of adenomatosis polyposis coli, a negative regulator of the Wnt/β-catenin pathway, to play a carcinogenic role (70). In general, LSINCT5 plays a carcinogenic role in osteosarcoma cells and may be a predictor of the clinical outcomes of osteosarcoma patients, and a promising candidate for osteosarcoma prognosis and treatment.
In addition, there are some other common lncRNAs that are up-regulated in osteosarcoma. For example, the expression of BCAR4 (71), HULC (72), UCA1 (73), and ANRIL (74) in osteosarcoma tissues is significantly higher than that in osteoblasts and adjacent tissues. After knocking down BCAR4, the proliferation, invasion and migration of osteosarcoma cells were inhibited. In addition, BCAR4 promotes the progression of osteosarcoma by activating the GLI2 signaling pathway (75). UCA1 can act as a sponge for many tumor suppressor miRNAs (73), enhance cell cycle progression through different mechanisms (76), and interact with various cancer-related signaling pathways such as mTOR, AKT, Wnt, Hippo and JNK pathways (77). Overexpression of ANRIL increases cell proliferation, and higher ANRIL expression is significantly associated with death and metastasis (78).
3.2 Down-regulated lncRNAs in osteosarcoma
Table 1 illustrates several cancer-suppressing lncRNAs that exhibit diminished expression in osteosarcoma.
The lncRNA, Loc285194, which is also named the LSAMP antisense RNA 3, has its locus on chromosome 3q13.3. It comprises 4 exons and is 2105 nucleotides long (79). Its tumor-suppressing activity is moderated via p53 due to the inhibition of the expression of miR-211 (80). In specimens and cell lines from primary osteosarcomas, a lack of Loc285194 expression has been demonstrated. When Loc285194 was depleted, the replication of normal osteoblasts was enhanced because of its effects on the cell cycle, apoptotic transcripts, and the expression of VEGF receptor 1 (79).
MEG3, is a member of the DLK1–MEG3 locus found on human chromosome 14q32.3 (81). Its functions include the accrual of protein p53, the transcription of p53-dependent promoters, and the exclusive moderation of the expression of p53 target genes (82, 83). When contrasted with neighboring benign tissue samples, there was an obviously reduced MEG3 expression in tissues from osteosarcomas. This observation was correlated with the disease stage and the presence of remote metastases (P < 0.05) (2). This gene is associated with the Notch, and TGF-β pathways (84); the amplified expression of MEG3 led to a suppression of TGF-β, Notch1, N-cadherin, and Hes1 expression, and the upregulation of the expression of E-cadherin (85). Mechanistically, MEG3 promotes osteosarcoma chemosensitivity by regulating anti-tumor immunity through the miR-21-5p/p53 pathway and autophagy, demonstrating that MEG3 may be a promising therapeutic target for osteosarcoma chemoresistance (86).
A range of malignant pathologies is influenced by the biological activities of FER-1 family member 4 (FER1L4), a lncRNA that has a size of 6.7 kb and is situated on chromosome 20q11.22 (87). Compared to normal tissues, titers of this molecule were notably reduced in samples and cell lines from osteosarcomas. Additionally, the osteosarcoma-enhancing miRNA, miRNA-18a-5p, which has been proposed to be the target molecule for FER1L4 in this malignancy, was recognized regarding the governance of programmed cell death and EMT (52).
Moreover, chromosome 1q25 is the location of the de novo tumor-suppressor, growth arrest-specific transcript 5 (GAS5). This lncRNA is a 5’-terminal oligopyrimidine RNA that comprises a dozen non-conserved exons and approximately 630 nucleotides (88). Its expression is diminished in breast, stomach, lung, and prostate tumors, as well as in additional neoplasias (89, 90). Its expression is also downregulated in osteosarcoma samples and cell lines in humans when judged against neighboring areas and normal osteoblast cells. Functionally, it acts as a ceRNA, sponging miR-23a-3p to amplify the expression of PTEN and inhibit osteosarcoma cell replication and invasion through its effect on the PI3K/AKT pathway (91). In addition, GAS5 also enhances the sensitivity of osteosarcoma cells to Cisplatin through the GAS5/miR-26b-5p/TP53INP1 axis, which may be a potential indicator for the treatment of osteosarcoma (92).
In addition, the replication of osteosarcoma cells is subject to modification by lncRNA-p21, which is also referred to as TRP53COR1. This is associated with amplified PTEN expression due to the replication of osteosarcoma cells being subject to modification by lncRNA-p21, which is also termed TRP53COR1. This is associated with amplified PTEN expression because of sponging miR-130b, an oncomiR (64). As a principal gene dictating cancer suppression in humans, PTEN is responsible for the dephosphorylation of AKT at Thr 308 and Ser 473, which leads to the inhibition of the signaling potency of AKT and, consequently, a reduction in cell growth and the enhancement of apoptosis (65).
A further lncRNA, which operates as a coactivator of RNA, SRA1 has been proposed to be a major actor in myogenesis, steroidogenesis, breast carcinogenesis, and cardiac muscle disorders. Anomalous levels of expression of SRA1 have been identified in several malignancies in humans (93). An anti-cancer function of SRA1 was observed in osteosarcomas, in which SRA1 diminished the migratory, replicative, and invasive properties of the tumor cells and also acted as a sponge for miRNA-208a, thus promoting apoptosis (94).
4 LncRNAs regulate the cell proliferation, invasion and metastasis of osteosarcomas
Ongoing cell growth and replication without regulation is a pathognomonic sign of tumorigenesis; promoting the removal of malignant cells is deemed the definitive goal of therapy (95). Carcinogenesis can be facilitated by activating oncogenes and inhibiting tumor suppressor genes (96). Various lncRNAs display these functions in osteosarcomas, modifying either the cell cycle or apoptosis, and regulating cellular replication or migration (Figure 3).
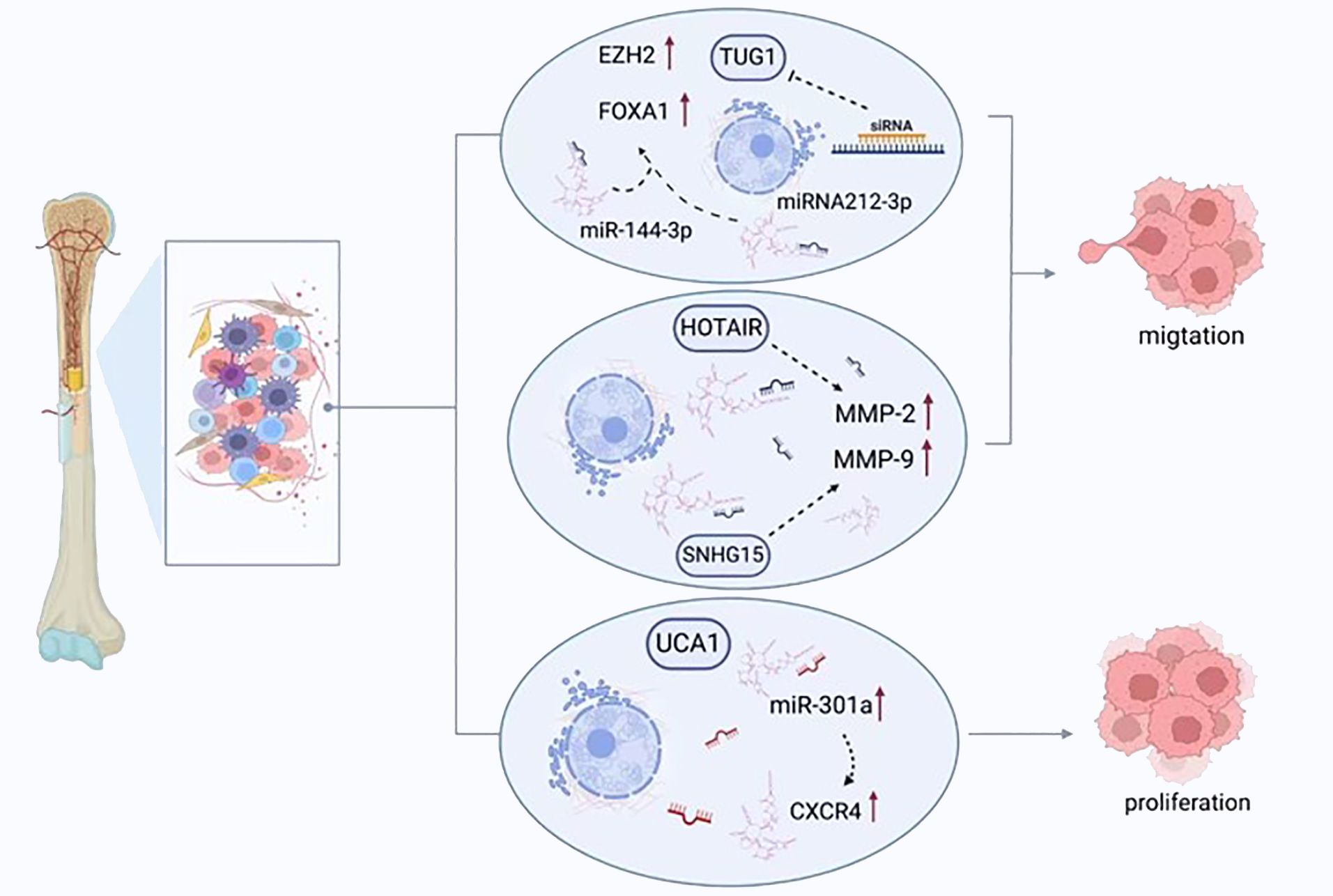
Figure 3 LncRNAs regulate the migration, proliferation, and apoptosis of osteosarcoma cells. TUG1 promotes the expression of FOXA1 and enhancer of zeste homolog 2 (EZH2) by competitively binding to miRNA-212-3p and miR-144-3p, thereby enhancing the proliferation of cancer cells. UCA1 enhances the migration of tumor cells by increasing the expression of miR-301a and chemokine receptor-C-X-C motif chemokine receptor 4 (CXCR4). HOTAIR and SNHG15 promote cell proliferation by regulating the expression of MMP-2/MMP-9.
4.1 LncRNAs regulate the cell proliferation and migration of osteosarcomas
In samples from osteosarcoma, a polymerase chain reaction was utilized to quantify the levels of hypoxia-inducible factor-2α (HIF2α) promoter upstream transcript (HIF2PUT). The results indicated that HIF2PUT inhibited tumor stem cells through its effects on the expression of HIF2α. Significant inhibitory influences were observed on cellular replication and migration in the presence of amplified HIF2PUT expression. The proportion of CD133-expressing cells was reduced and the MG63 cells evidenced a diminished capacity to form osteosarcoma stem spheres (19, 97). Conversely, when HIF2PUT was depleted via siRNA, there was a marked enhancement of cell growth and migration (97).
It has also been determined that replication and colony generation in cells from osteosarcomas can be suppressed by tumor suppressor candidate 7 (TUSC7) (43). Its knockdown with siRNA caused increased cellular replication and the presence of cell colonies, together with a decrease in programmed cell death. However, there was no impact on the cell cycle (98). The amplification of the expression of the angiomotin (AMOT) gene in cells from human osteosarcomas by SNHG12 also has been reported to accelerate cellular replication and migration, while the knockdown of SNHG12 had the opposite consequence but failed to influence programmed cell death (99). When SNGH12 and AMOT underwent knockdown, attenuated cell migration and growth were detected (99). Notch signaling is a pathway that is extremely conserved, underpins many biological activities, and contributes to the pathogenesis of numerous malignancies (100). Notch signaling is responsible for the governance of a number of lncRNAs and is regulated by them simultaneously. SNHG12 stimulates the Notch signaling pathway and precipitates enhanced oncogenesis and osteosarcoma dissemination by sponging miR-195-5p (101). Research has observed that the growth and replication of osteosarcoma cells was impeded by atypical lncRNA expression. A lack of TUG1 diminishes the proliferation of cancerous cells and pauses the cell cycle at the stage, G0/G1 (102), additionally, it also acts as a ceRNA to sponging miRNA-212-3p and miR-144-3p (103). Subsequently, the FOXA1 and enhancer of zeste homolog 2 (EZH2) expression was increased, which are target genes for the transcription of oncogenes (103). The potency of osteosarcoma cell replication was diminished, and the apoptosis was enhanced with the suppression of TUG1 with siRNA (104). The arrest of the cell cycle and apoptosis are both triggered following MALAT1 knockdown, which also leads to the suppression of the growth of osteosarcoma and its dissemination (48).
The continuation of an undifferentiated cell type is influenced by a de novo recognized oncogenic lncRNA, DANCR (19). In U2OS and SAOS cells, the replication of the cells was markedly reduced by DANCR knockdown, which also reduced colony generation within U20S cells and paused the cell cycle in the latter at stage, G0/G1. The endogenous levels of proteins associated with the cell cycle, such as p21, CDK2, and CDK4, are governed by DANCR (105). miRNA-335-5p and miRNA-1972 expression are also promoted by DANCR, which accelerates replication that is induced via ROCK1 and ceRNA network transfer and consequently promotes the pathogenesis of osteosarcomas (106).
The amplified expression of TMPO antisense RNA 1 (TMPO-AS1) has been observed in osteosarcomas. Conversely, samples and cell lines from osteosarcomas have evidenced the downregulation of miR-199a-5p (107). The inhibition of cellular replication and enhanced programmed cell death were identified following TMPO-AS1 knockdown, and the regulation of WNT7B occurred via the immediate sponging of miR-199a-5p, which suppressed Wnt/β-catenin activity (108). Furthermore, the WNT7B knockdown salvaged the miR-199-5p inhibitor’s suppressive effect on osteosarcomas, which could be eradicated by administering the Wnt pathway stimulator lithium chloride.
In individuals with osteosarcoma, a poor prognosis was suggested by the amplified expression of bladder cancer-associated transcript 1 (BLACAT1) (109). This was linked with promoted cellular replication and invasive properties, whereas a reduction in BLACAT1 expression was associated with the opposite effect. Interestingly, the enhancement of tumor cell replication and migratory function was achieved through an effect on STAT3 phosphorylation.
Another major influence on the onset and progression of osteosarcoma is the modified frailty index 2 (MFI2), which MMFI2 promotes the proliferation and migration of osteosarcoma cells by regulating the expression of forkhead box P4 (FOXP4) (110). Cellular replication within osteosarcomas has been demonstrated to be enhanced by P50-associated COX-2 extragenic RNA (PACER). The methylation of DNA has a regulatory influence on PACER, which arises through the NF-κB-dependent stimulation of the gene, COX-2 (111). Triggering ZEB1 transcription via ZEB1 Antisense 1 (ZEB1-AS1), which has oncogenic characteristics, also induces the replication of osteosarcoma cells (112). In cells from human osteosarcomas, oncogenic lncRNA SNHG12 additionally impacts cellular replication through angiomotin gene expression amplification (99). Osteosarcoma cell replication was suppressed by reduced HNF1A-antisense 1 (HNF1A-AS1) expression, an action mediated through Wnt/β-catenin cascade inactivation (113).
4.2 LncRNAs regulate the cell apoptosis of osteosarcomas
The interaction between EZHW and HOXD-AS1 reduces the expression of p57 and promotes the development of osteosarcoma (114). The most common arising Apo-ERα-regulated lncRNA (AER-lncRNA) in mammary tumors is down syndrome cell adhesion molecule anti-sense RNA 1 (DSCAM-AS1) (115), which has been found to have a high level of expression within cell lines from osteosarcomas. Apoptosis in osteosarcoma has been markedly accelerated through the DSCAM-AS1 knockdown and inhibition of the Wnt pathway (116).
4.3 LncRNAs regulate the invasion and metastasis of osteosarcoma
The main clinical issues relating to osteosarcomas are remote tumor dissemination and the likelihood of recurrence, which hinders the efficacy of therapies and leads to poor prognosis in individuals with this type of malignancy. Cancerous deposits distant to the primary lesion occur over several pathological stages via a complicated mechanism that comprises regional tumor invasion, intravasation, remote spread, extravasation, and colonization (22). At the original tumor site, the interplay between cells and the extracellular matrix (ECM) is changed by malignant cells. These cells break away from the primary location and extend into neighboring structures, and gain passage through the circulation to remote viscera, where they attach to the vasculature walls and leak into their target tissues before replicating from microscopic colonies to generate secondary neoplastic deposits (98). Genetic and epigenetic abnormalities usually underlie this clinical process (117, 118). Metastatic tumors in osteosarcomas frequently occur in the lungs; these are challenging to address and give rise to the most common mode of death, respiratory failure (119). It is thought that about 85% of individuals with skeletal malignancy present with remote tumor deposits (120).
A group of proteolytic enzymes, MMPs, play a key role in the invasion and dissemination of cancerous cells through their capacity to breakdown the ECM and basement membrane, thus remodeling the microenvironment of the primary lesion and encouraging the formation of a malignant neovasculature. In many human malignancies, increased levels of MMP-2 and MMP-9 are released, an observation linked with an adverse clinical prognosis (121). Amplified HOTAIR expression accelerates osteosarcoma cell invasion through the elevated liberation of MMP-2 and MMP-9 (122). Conversely, reduced HOTAIR expression in cell lines from osteosarcomas, including U2OS, 143B, MNNG/HOS, and MG-63, suppresses malignant cell replication and invasion as well as inhibits the release of MMP-2 and MMP-9 (122). The growth of osteosarcoma can be inhibited in U20S cells with HOTAIR knockdown following implantation in xenograft models (122). SNHG15 can also promotes cell proliferation and invasion by regulating the expression of MMP-2/MMP-9. In addition, MMP-2 and MMP-9 expression can be enhanced by LINC00968 amplification, leading to ECM breakdown and encouraging migration and invasion by cancerous cells (123).
The evidence that lncRNAs are linked with signaling pathways, which are key actors in controlling osteosarcoma cell invasion, migration, and dissemination, is irrefutable. LncRNA plays a role in the development of osteosarcomas by affecting the wnt pathway. For example, when cell lines from osteosarcomas are contrasted with benign osteoblastic cells, the expression of the lncRNA gastric carcinoma proliferation enhancing transcript 1 (GHET1) is amplified. Knockdown of this molecule has been demonstrated to suppress the migration of osteosarcoma cells, together with their invasion and EMT. To some extent, these events are mediated through the control of the Wnt/β-catenin pathway. Compared with the control group, the expression levels of Wnt and β-catenin were decreased after GHET1 knockdown (124). Elevated levels of the lncRNA, CRNDE, have been identified in cell lines and tissue samples from osteosarcomas, with the knockdown of CRNDE leading to limited cellular invasion, the reduced expression of N-cadherin, vimentin, and snail, and amplified E-cadherin and ZO-1 expression (125). A potential mechanism for this process may be the activation of the Wnt/β-catenin signaling pathway via the increased phosphorylation of GSK-3β (125). The upregulation of MALAT1 expression strongly relates to the presence of remote osteosarcoma deposits. This lncRNA can bind to miR-144-3p and enhance ROCK1/2 levels which contribute significantly to the metastatic process (126). MALAT1 may inhibit tumor growth and metastasis through PI3K/AKT signaling pathway (47). The in vivo and in vitro lentivirus-mediated siRNA reduction of MALAT1 expression decreases in PCNA, MMP-9, p-PI3K, and RhoA/ROCKs expression, which then attenuates the growth, invasion, and dissemination of osteosarcoma cells (47, 48). MiR-184, as well as β-catenin, TCF4, and c-MYC, which are downstream Wnt signaling pathway factors, is negatively impacted by MEG3, which inhibits the replication and the migration of osteosarcoma cells in vitro and cancerous growth in vivo (127). The transition of epithelial cells into a mesenchymal phenotype is known as enforced EMT, which enhances tumor invasiveness and, clinically, leads to worse overall survival (128). This process can be recognized by a rise in mesenchymal indicators, including N-cadherin, Slug, Twist, Vimentin, and Fibronectin, and by a reduction in epithelial indicators such as E-cadherin (12). The Hh signaling exhibits anomalous activity in cell lines from osteosarcomas and samples from primary human osteosarcomas, which promotes migration and the onset of osteoblastic osteosarcoma (129). In mice with a heterozygous p53 background with amplified Hh pathway stimulation, skeletal cancers occurred freely, even at 7 months of age, in association with marked rises in the potent oncogenes Yap1 and H19 (59). Notably, cancer progression can be suppressed by silencing either of these genes (130).
Tumor cell migration and invasion can also be influenced by lncRNAs via miRNA. Cell survival, migration, and invasion were improved by urothelial carcinoma associated 1 (UCA1) (131), with the expression of this molecule having a positive correlation with chemokine receptor-C-X-C motif chemokine receptor 4 (CXCR4) and miR-301a. Additionally, the expression of miR-301a was amplified, which consequently heightened the expression of CXCR4 (107). Some studies have reported a robust link between the degree of CXCR4 expression and the invasive and metastatic properties of osteosarcoma (132, 133). Moreover, miR-301a has been demonstrated to display malignant functionality in osteosarcomas and additional human malignancies (134). Furthermore, amplified miR-301a expression can prevent the suppressive action of knockdown of UCA1 in cells from osteosarcomas, a process that can be reversed by inhibiting CXCR4. ATB has demonstrated an increase in cancer. In individuals with osteosarcomas, such raised levels lead to greater ZEB1 and SEB2 expression through the suppression of miRNA-200. These findings are associated with a late Enneking stage, remote tumor dissemination, and adverse survival statistics (135). When ATB is lacking, the use of shRNA has a marked inhibitory impact on the growth, migration, and invasion of osteosarcoma cells.
Augmented cancer dimensions, late Enneking stage, remote dissemination, and adverse survival rates were linked with FGFR3 antisense transcript 1 (FGFR3-AS1), which enhanced the stability of FGFR3 mRNA and amplified the expression of FGFR3 by coupling the antisense with FGFR3 3′-UTR (19). When FGFR3-AS1 underwent knockdown, the in vitro growth of osteosarcoma cells within a xenograft was inhibited, and similar findings were seen in vivo (136).
In contrast to the oncogenic function of many lncRNAs, some operate in a tumor-suppressive capacity. The depletion of NBAT1 accelerates tumor cell replication and invasion (137), while excessive NBAT1 expression suppresses these events and tumor cell migration by inhibiting the effects of miR-21 and impacting its targeted gene (138). Influencing the miR-34a-5p/Sirt1 network via the knockdown of CAMK2D-associated transcript 1 (C2dat1) has also been demonstrated to diminish the invasive and migratory abilities of osteosarcoma cells (139).
5 LncRNAs’ regulation of other signaling pathways in osteosarcoma
As a major contributor to cellular pathways, cell signaling has a significant influence on the modification of the expression of numerous genes in a spectrum of malignancies. A recent study has indicated that the control of many signaling cascades in osteosarcoma is impacted by lncRNAs, which results in changes to cell replication, differentiation, and programmed cell death (Figure 4). In advancing malignant disease in humans, Wnt signaling is a notable and well-recognized example (140). Changes in the constituents of this pathway, such as mutations, amplifications, deletions, hypermethylation of promoters, and changes in localization within the subcellular level, are considered some of the most significant mechanisms involved in the onset and development of osteosarcomas (141). In this malignancy, the regulation of several components, such as ligands, receptors, co-receptors, and antagonists, is disrupted, this finding suggests that this cascade plays a major role in promoting malignant characteristics and tumor metastasis (142). In individuals with osteosarcomas, there is marked enhancement of the activity of the Wnt signaling cascade, and the accrual of the downstream transcription factor, β-catenin, in tissue specimens (143). It has been suggested that this factor could be stabilized with the competitive blockade of miRNA in relation to mRNA binding, and through the conscription of RNA-binding protein to mRNA, processes facilitated by lncRNAs (144–146). There is further potential for lncRNAs to form a complex with DNA and to modify gene expression, such as amplifying the transcription of β-catenin and thus downregulating the expression of the Wnt pathway inhibitor (70). The amplified expression of the lncRNA, BE503655, has been identified in cell lines and samples from osteosarcomas (15), with the mRNA levels and protein expression of β-catenin markedly diminished in this tumor following BE503655 knockdown by regulating Wnt/β-catenin pathways (145). Equivalent findings are present for the lncRNA CAT 104, and the small nucleolar RNA host gene 1 (SNHG1), with a significant rise in their expression. They influenced the Wnt cascade by operating as ceRNAs sponging to miR-381 and miR-577, respectively (147, 148).
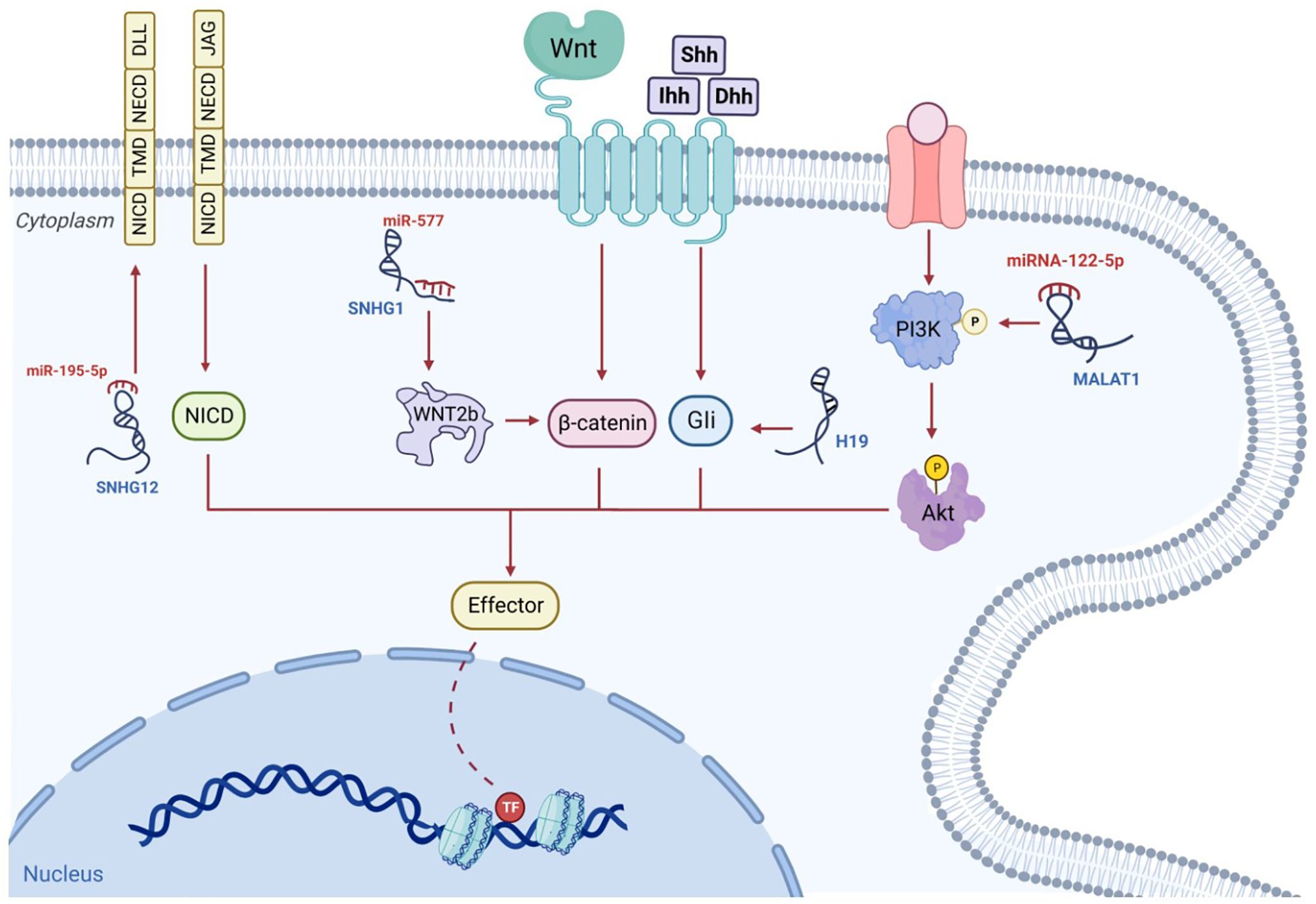
Figure 4 The signaling pathways associated with osteosarcoma are governed by lncRNAs. For example, the lncRNA, SNHG, behaves as a competitive miRNA blocker and increases the stability of β-catenin; subsequently, the triggered β-catenin is transported to the nucleus, where it instigates the transcriptive process. The competitive binding of MALAT1 to miRNA-122-5p can occur. This enhances phosphorylated PI3K and AKT levels which influence osteosarcoma progression. The hedgehog ligands, Shh, Ihh and Dhh, enhance Gli segregation from the microtube, after which Gli enters the nucleus to commence transcription. This mechanism may be enhanced by additional lncRNAs such as H19. NICD, TMD and NECD make up the lone transmembrane protein known as the Notch receptor. Ligands, including DLLs and JAGs, which are situated on adjacent cell walls, attach to their extracellular component, NECD, and switch on the Notch signaling cascade. The stimulated NICD is liberated into the cytoplasm to trigger transcription. Additional lncRNAs such as SNHG12, can increase Notch2 expression and facilitate this mechanism.
In addition to the signaling pathways mentioned above, there are some other signaling pathways associated with lncRNA in osteosarcoma. The NF-κB pathway plays a key role in cancer progression, which is regulated by a variety of mechanisms and controls cell growth, invasion and metastasis (149). The study found that the expression of NKILA, which reduced the proliferation, invasion and migration of osteosarcoma cells, was activated by the NF-κB pathway. NF-κB inhibitors can reverse the effect of knockdown of NKILA on cell migration and proliferation (150). In addition, targeting XIST can also inhibit cell proliferation and tumorigenesis by activating NF-kB and NF-kB-dependent PUMA signaling pathways (151). HIF-1α has been shown to regulate hypoxia gene expression through a signal transduction network. FOXD2-AS1 in the hypoxic tumor region can act as a miRNA sponge to regulate tumorigenesis. The expression of FOXD2-AS1 is up-regulated in osteosarcoma and is positively correlated with poor prognosis. HIF-1α can bind to the promoter region of FOXD2-AS, thereby increasing mRNA and protein levels (152). LINK-A may also act as an upstream activator of HIF1α and participate in the metastasis of osteosarcoma by up-regulating the HIF1α pathway (153).
6 Potential preventive and therapeutic effects of lncRNA in osteosarcoma
6.1 LncRNA in osteosarcoma diagnosis and prognosis
Although increasing numbers of current treatment alternatives are available for individuals with sarcomas, the efficacy of contemporary therapies has not changed over time. At present, the methods used to treat osteosarcoma include high-dose methotrexate, adriamycin and cisplatin before and after surgical resection. In addition, different combinations of etoposide, methotrexate, cisplatin, doxorubicin, and ifosfamide were also used to consolidate the treatment, but the results were not satisfactory (154, 155). Chemotherapy regimens consisting of ifosfamide and etoposide or gemcitabine and docetaxel have a certain effect on patients with unresectable recurrent diseases, but the overall event-free survival at 4 months is only 12% (156, 157). Adverse prognoses arise owing to the high risk of remote tumor dissemination and relapse. If malignancy were detected early and a precise prognosis was evident, there would be the potential for prompt targeted treatment and better overall survival statistics. Therefore, the presence of apposite clinical diagnostic or prognostic indicators would be extremely valuable. Numerous studies that have evaluated the role of lncRNAs in osteosarcoma have suggested that these molecules have promise for this application (Tables 2, 3).
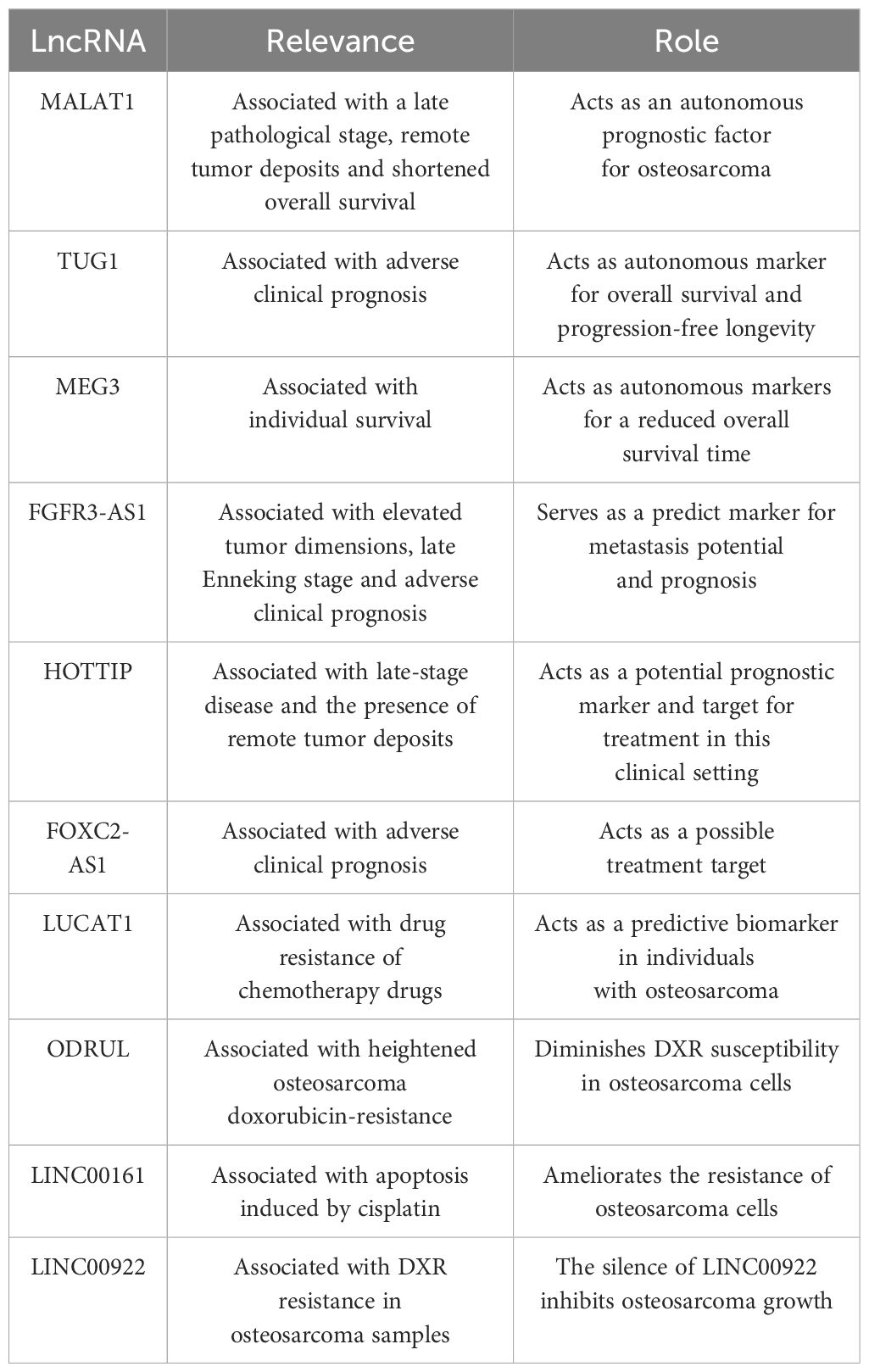
Elevated MALAT1 in osteosarcoma tissue have been found to be associated with a late pathological stage, remote tumor deposits, and shortened overall survival, suggesting that this lncRNA could act as an autonomous prognostic factor for this tumor type (158). Significantly, MALAT1 levels within the serum could also be utilized in this regard. The degree of MALAT1 expression had a negative association with 5-year survival figures. In the cohorts with low and high expression, the 5-year survival statistics were 56.5% and 39.1%, respectively, a difference additionally shown with Kaplan-Meier survival curves (159).
An adverse clinical prognosis was robustly linked with amplified TUG1 expression, which was determined to be an autonomous marker of overall survival and progression-free longevity. When judged against preoperative values, serum TUG1 levels were noted to be markedly diminished in individuals following surgery, with the increase of TUG1 in serum identified in those patients exhibiting advancing malignancy or recurrence (39). Moreover, TUG1 knockdown in the osteosarcoma cell lines, U20S and Saos-2, induced by selective siRNA, leads to a declining cell replication and colony generation, heightened programmed cell death, and the arrest of the cell cycle at the G1/S phase. It is also associated with reduced POU class 2 homeobox 1 (POU2F1) expression, indicating its influence on the TUG1/mir-p-5p/POU2F1-axis in these cell types (160). When these data are combined, TUG1 could be used as a marker for prognosis, treatment, diagnosis, and monitoring purposes.
Overall survival was less in individuals with reduced, as opposed to amplified MEG3 expression (161). In patients presenting with osteosarcomas, low levels of MEG3 expression, a late pathological stage, and the presence of remote tumor deposits were all determined to be autonomous markers of reduced overall survival time.
Elevated tumor dimensions, late Enneking stage, and adverse clinical prognosis have been linked to the amplified expression of FGFR3-AS1 (136). When identified within cells, FGFR3-AS1 promotes amplified FGFR3 expression through a mechanism involving the coupling of antisense with the 3’UTR section of FGFR3. Additionally, when the FGFR3-AS1 shRNA expression plasmid pGPU6/GFP/Neo is used to knockdown FDFR3-ASi in MG63 cell lines in vitro, advancement of the cell cycle and cell replication are inhibited (136). Correspondingly, prognosis may be predicted using FGFR3-AS1 in the clinical context of osteosarcoma.
The late-stage disease and the presence of remote tumor deposits have been linked with the amplified expression of HOTTIP in samples from osteosarcomas. Increased expression of HOTTIP has also been related to high mortality and is an autonomous prognostic indicator for overall survival in individuals with this tumor type. Hence, this lncRNA is a potential prognostic marker and target for treatment in this clinical setting (162).
Amplified MIR100HG, HOXD-AS1, EWSAT1, and LMCD1-AS1 expression, in addition to several further lncRNAs, have been determined by Kaplan-Meier curves to forecast adverse clinical outcomes in individuals with osteosarcoma (163, 164). Statistical analyses, such as univariate and multivariate Cox regression, have also indicated the prognostic value of lncRNAs, including LOXL1-AS1, EWSAT1, ILF3-AS1, CBR3-AS1, DLX6-AS1, UCA1, DICER1-AS1, and Ftx in this form of malignancy (164).
6.2 LncRNAs in osteosarcoma chemotherapy and chemotherapeutic drug resistance
The principal types of therapy applied to malignancy include operative resection, and treatment with either chemotherapy or radiation. However, many osteosarcomas fail to demonstrate significant regression following conventional chemotherapy or radiotherapy regimens and may develop resistance to therapies (165). Resistance to chemotherapeutic agents can be categorized as either acquired or primary (166). The latter is already present, whereas the former arises during treatment following the adaptation of malignant cells (167). This occurs via a complicated mechanism to which numerous factors contribute, together with impaired processes of programmed cell death and autophagy (168). In osteosarcoma, the lack of efficacy of chemotherapeutic endeavors predominantly arises owing to secondary resistance to drugs such as doxorubicin (DXR) and cisplatin (169). In some cases, using lncRNAs as targets for therapy has been shown to attenuate the issues of pharmaceutical resistance and enhance the susceptibility of the tumor to treatment (170) (Table 2).
An abundance of research has concentrated on utilizing genetic and molecular analytical techniques to investigate the mechanisms underpinning chemoresistance in osteosarcoma. The factors identified factors include a spectrum of intrinsic biological changes, including the heightened expression of members of the ATP-binding cassette (ABC) membrane transporter family, aberrant metabolic pathways, disruption to the governance of the cell cycle, and abnormal apoptotic processes (171, 172). These alterations can inactivate pharmaceutical agents, diminish the accrual of drugs within the cells, induce chemoresistance via the effects on malignant stem cells, and cause dysfunctional signal transduction pathways, the aberrant control of miRNAs and drug resistance due to aberrant cell death and autophagy (173, 174). The ABC transporter protein family members rely on ATP for the efflux of pharmaceutical agents (175). Their expression is influenced by lncRNAs, which consequently affect the resistance of malignant cells (176, 177).
In samples from human osteosarcomas and in the cell lines MG63 and KH-OS, which exhibit DXR resistance, there is excessive expression of long non-coding RNA Forkhead box protein C2 antisense 1 (FOXC2-AS1) (169). In human tissue samples from osteosarcomas, this finding is associated with an adverse clinical prognosis, and in vitro, it is linked with the enhancement of resistance to DXR in the same cell lines. Conversely, FOXC2-AS1 knockdown promotes the susceptibility of cells from osteosarcomas to DXR and reduces osteosarcoma cell resistance to DXR in cell lines, including MG63/DXR and KH-OS/DXR, through FOXC2 inhibition. ABCB1 expression is also promoted by FOXC2, which adds to the drug resistance seen in osteosarcomas (169). Thus, the amplification of ABCB1 expression is a common pathway to DXR resistance in cells from osteosarcomas caused by FOXC2-AS1 and FOXC2. FOXC2-AS1 appears to have two potential roles. Firstly, it may be a useful marker in patients with osteosarcoma to highlight potential DXR resistance, and secondly, it represents a possible treatment target for increasing tumor susceptibility to DXR.
Cell lines from osteosarcomas that display resistance to methotrexate, an extremely potent chemotherapeutic agent used in this type of neoplasia, have been demonstrated to contain amplified lung cancer-associated transcript 1 (LUCAT1) expression (178). In addition, LUCAT1 can interact with ABCB1 through miR-200c, which combines with the 3’UTR of ABCB1 and is regulated by LUCAT1. Accordingly, if elevated levels of LUCAT1 were detected in human osteosarcomas, it can be used as a predictive biomarker in individuals with osteosarcomas.
Patients with osteosarcomas and tumor dissemination to the lungs and a poor response to chemotherapy demonstrated heightened osteosarcoma doxorubicin-resistance related to up-regulated lncRNA (ODRUL) expression. Notably, the lncRNA ODRUL may diminish DXR susceptibility in osteosarcoma cells owing to its ability to amplify ABCB1 expression (179). Furthermore, several lncRNAs, such as ENST00000563280 and NR-036444, were recognized from a lncRNA-mRNA co-expression network. These engage with genes including ABCB1, HIF1A and FOXC2, which may contribute to elevated levels of osteosarcoma resistance to DXR (180).
Sustained cell survival necessitates prompt and apposite reparation of any injury to DNA. Many miRNAs might control the responsiveness of osteosarcoma cells to treatment with radiation by influencing these mechanisms. Apoptosis brought about by cisplatin is significantly influenced by lncRNA, long intergenic non-coding RNA 161 (LINC00161). Additionally, this lncRNA ameliorates the resistance of osteosarcoma cells by targeting the signaling axis comprising the miR-645-interferon-induced with tetratricopeptide repeats 2 (IFIT2) (181).
DXR resistance in osteosarcoma samples has been linked to amplified LINC00922 expression, the silence of which inhibits osteosarcoma growth. One of LINC00922’s functionalities is to sponge miR-424-5p, which decreases transcription factor TFAP2C expression. Consequently, it has been proposed that a feedback loop, including TFAP2C, LINC00922, and miR-424-5p, is important in DXR resistance (182).
CTA levels have been demonstrated to be diminished in cells that are resistant to DXR (130). Significantly, enhancing CTA levels led to a notable rise in two recognized miR-2010 targets, including caspase-8-associated protein 2 and apoptosis-inducing factor, mitochondrion-associated 3 (183). Correspondingly, the reparation of CTA may therefore potentially diminish cell survival through competitive bonding with miR-201, enhancing apoptosis and suppressing autophagy. Moreover, increased SNHG12 transcript levels have been noted in cells from osteosarcomas that demonstrated DXR resistance, as opposed to those that are susceptible to this drug at cytotoxic levels. The mechanism of action of SNHG12 in cells from osteosarcomas arises through its capacity to sponge miR-320a, which has an inhibitory influence on the expression of MCL1 (101).
Anomalies in drug breakdown form a second key pathway that brings about resistance to pharmaceutical reagent. Important enzymes within these metabolic pathways can be affected by several lncRNAs. Enzyme systems that are key to the activation and inactivation of many drugs include the cytochrome P450 (CYP) system, and the glutathione-S-transferase (GST) and the uridine diphosphoglucuronosyltransferase (UGT) superfamilies (184). Numerous pharmaceutical reagents used in the treatment of malignancy have to be activated by the body’s metabolism, which could lead to drug resistance if this process is impaired by neoplastic cells (167). Elevated CYP3A4/5 levels have been associated with adverse clinical outcomes in individuals with osteosarcomas (185). Furthermore, after the administration of DXR or cisplatin, glutathione-S-transferase P1 (GSTP1) levels were found to be elevated, and amplified GSTP1 expression in SAOS-2 cell lines from osteosarcomas was linked with heightened vulnerability to resistance to these two pharmaceutical reagents (186).
7 Summary and future prospects
Osteosarcoma, an extremely aggressive malignancy, is typically characterized by invasion at the site of origin, remote dissemination, and recurrence. There is an exigent requirement to comprehend the disease processes underlying this cancer type to facilitate the innovation of de novo clinical treatments. The above review has detailed the abundance of evidence that substantiates the possibility that lncRNAs are key actors in the onset, growth, invasion, systemic dissemination, and resistance to pharmaceutical agents exhibited by osteosarcomas. The expression amplification or downregulation of these lncRNAs means that they can display either oncogenic or tumor-suppressive properties, respectively. Examples include HOTAIR, MALA T1, H19, TUG1, MEG3, and TUSC7.
A range of pathways have been identified concerning lncRNAs, such as host gene targeting, signal pathway involvement, and ceRNA activity. Several lncRNAs have been determined to function as autonomous indicators of prognosis, including the likelihood of the presence of drug resistance to modern treatments such as DXR and cisplatin. Such data pertaining to lncRNAs are encouraging for the future innovation of clinically applicable biological indicators for the diagnosis and prediction of survival, as well as potential targets for treatment. LncRNA can be targeted by using small interfering RNA, antisense oligonucleotide, ribozyme, aptamer, and miRNA. These methods have long been evaluated for targeting key cancer-related genes, and they are at different stages of clinical trials. For example, flavonoids are negatively correlated with the risk of colorectal cancer. These bioactive compounds can target the lncRNA/Wnt pathway to reduce the side effects of anticancer drugs (187). Simvastatin, a potential therapeutic drug for colorectal cancer immunotherapy, promotes anti-tumor immunity by inhibiting lncRNA SNHG29-mediated YAP activation and inhibiting PD-L1 expression (188). In addition, due to the ability of oligonucleotides to enter cells and specifically target RNA that cannot be accessed by antibodies, oligonucleotide drugs show stronger target specificity than small molecule drugs and reduce potential side effects. Therefore, lncRNA-based oligonucleotide drugs are being studied and are expected to make further progress. Nevertheless, research investigating the role played by lncRNAs in osteosarcoma is still in its infancy and predominantly in the pre-clinical phase. Despite all this, recent experimental data are extremely promising regarding the utility of lncRNAs as future diagnostic and prognostic clinical indicators, and as treatment targets in patients with osteosarcoma.
Author contributions
SH: Writing – original draft, Writing – review & editing. XH: Writing – original draft, Writing – review & editing. GL: Supervision, Writing – review & editing. SW: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was supported by the grant from: National Natural Science Foundation of China (81900441), Natural Science Foundation of Zhejiang Province (LQ19H020002), Zhejiang Provincial Program for Medicine and Health (2022KY446, 2023KY411, 2023KY1347), Hunan Provincial Science and Technology Department (2020NK2004, 2019TP2004), Social Development Science and Technology Foundation of Taizhou (21ywb115, 21ywb118, 20ywb143), Social Development Science and Technology Foundation of Wenling (2020S0180083, 2020S0180127, 2021S00156, 2021S00197).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lu KH, Lu EW, Lin CW, Yang JS, Yang SF. New insights into molecular and cellular mechanisms of zoledronate in human osteosarcoma. Pharmacol Ther. (2020) 214:107611. doi: 10.1016/j.pharmthera.2020.107611
2. Li Z, Dou P, Liu T, He S. Application of long noncoding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cell Physiol Biochem. (2017) 42:1407–19. doi: 10.1159/000479205
3. Jafari F, Javdansirat S, Sanaie S, Naseri A, Shamekh A, Rostamzadeh D, et al. Osteosarcoma: A comprehensive review of management and treatment strategies. Ann Diagn Pathol. (2020) 49:151654. doi: 10.1016/j.anndiagpath.2020.151654
4. Chen C, Xie L, Ren T, Huang Y, Xu J, Guo W. Immunotherapy for osteosarcoma: Fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. (2021) 500:1–10. doi: 10.1016/j.canlet.2020.12.024
5. Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. (2016) 28:26–33. doi: 10.1097/MOP.0000000000000298
6. Aznab M, Hematti M. Evaluation of clinical process in osteosarcoma patients treated with chemotherapy including cisplatin, adriamycin, ifosfamide, and etoposide and determination of the treatment sequels in a long-term 11-year follow-up. J Cancer Res Ther. (2017) 13:291–6. doi: 10.4103/0973-1482.199447
7. Wong P, Houghton P, Kirsch DG, Finkelstein SE, Monjazeb AM, Xu-Welliver M, et al. Combining targeted agents with modern radiotherapy in soft tissue sarcomas. J Natl Cancer Instit. (2014) 106:dju329. doi: 10.1093/jnci/dju329
8. Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. (2014) 7:104–12. doi: 10.1016/j.celrep.2014.03.003
9. Kovac M, Blattmann C, Ribi S, Smida J, Mueller NS, Engert F, et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun. (2015) 6:8940. doi: 10.1038/ncomms9940
10. Lorenz S, Barøy T, Sun J, Nome T, Vodák D, Bryne JC, et al. Unscrambling the genomic chaos of osteosarcoma reveals extensive transcript fusion, recurrent rearrangements and frequent novel TP53 aberrations. Oncotarget. (2016) 7:5273–88. doi: 10.18632/oncotarget.v7i5
11. Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. (2021) 18:609–24. doi: 10.1038/s41571-021-00519-8
12. Danieau G, Morice S, Rédini F, Verrecchia F, Royer BB. New insights about the wnt/β-catenin signaling pathway in primary bone tumors and their microenvironment: A promising target to develop therapeutic strategies? Int J Mol Sci. (2019) 20:3751. doi: 10.3390/ijms20153751
13. Jiang M, Jike Y, Liu K, Gan F, Zhang K, Xie M, et al. Exosome-mediated miR-144-3p promotes ferroptosis to inhibit osteosarcoma proliferation, migration, and invasion through regulating ZEB1. Mol Cancer. (2023) 22:113. doi: 10.1186/s12943-023-01804-z
14. Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. (2014) 4:6088. doi: 10.1038/srep06088
15. Han J, Shen X. Long noncoding RNAs in osteosarcoma via various signaling pathways. J Clin Lab Anal. (2020) 34:e23317. doi: 10.1002/jcla.23317
16. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. (2017) 36:5661–7. doi: 10.1038/onc.2017.184
17. Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, et al. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation. (2016) 133:2050–65. doi: 10.1161/CIRCULATIONAHA.115.021019
18. Zhang Z, Zhou C, Chang Y, Zhang Z, Hu Y, Zhang F, et al. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/β-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. (2016) 376:62–73. doi: 10.1016/j.canlet.2016.03.022
19. Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. (2016) 7:e2389. doi: 10.1038/cddis.2016.272
20. Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. (2015) 12:1094–8. doi: 10.1080/15476286.2015.1063770
21. Han Li C, Chen Y. Small and long non-coding RNAs: novel targets in perspective cancer therapy. Curr Genomics. (2015) 16:319–26. doi: 10.2174/1389202916666150707155851
22. Xu S, Gong Y, Yin Y, Xing H, Zhang N. The multiple function of long noncoding RNAs in osteosarcoma progression, drug resistance and prognosis. Biomed pharmacother = Biomed pharmacother. (2020) 127:110141. doi: 10.1016/j.biopha.2020.110141
23. Hahne JC, Valeri N. Non-coding RNAs and resistance to anticancer drugs in gastrointestinal tumors. Front Oncol. (2018) 8:226. doi: 10.3389/fonc.2018.00226
24. Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. (2002) 420:563–73. doi: 10.1038/nature01266
25. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
26. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. (2009) 458:223–7. doi: 10.1038/nature07672
27. Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. (2007) 17:556–65. doi: 10.1101/gr.6036807
28. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. (2009) 10:155–9. doi: 10.1038/nrg2521
29. Li Z, Shen J, Chan MT, Wu WK. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell prolif. (2016) 49:471–5. doi: 10.1111/cpr.12269
30. Deans C, Maggert KA. What do you mean, "epigenetic"? Genetics. (2015) 199:887–96. doi: 10.1534/genetics.114.173492
31. Chen Z, Gao Y, Yao L, Liu Y, Huang L, Yan Z, et al. LncFZD6 initiates Wnt/β-catenin and liver TIC self-renewal through BRG1-mediated FZD6 transcriptional activation. Oncogene. (2018) 37:3098–112. doi: 10.1038/s41388-018-0203-6
32. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08.018
33. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. (2010) 42:1113–7. doi: 10.1038/ng.710
34. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B, et al. (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. (2019) 20:5758. doi: 10.3390/ijms20225758
35. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071–6. doi: 10.1038/nature08975
36. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumor biology. Nature. (2010) 465:1033–8. doi: 10.1038/nature09144
37. Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. (2013) 6:37. doi: 10.1186/1756-8722-6-37
38. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. (2016) 29:452–63. doi: 10.1016/j.ccell.2016.03.010
39. Ma B, Li M, Zhang L, Huang M, Lei JB, Fu GH, et al. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumor biol: J Int Soc Oncodevelop Biol Med. (2016) 37:4445–55. doi: 10.1007/s13277-015-4301-6
40. An X, Wu W, Wang P, Mahmut A, Guo J, Dong J, et al. Long noncoding RNA TUG1 promotes Malignant progression of osteosarcoma by enhancing ZBTB7C expression. Biomed J. (2023) 47:100651. doi: 10.1016/j.bj.2023.100651
41. Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. (2016) 1859:192–9. doi: 10.1016/j.bbagrm.2015.09.012
42. Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. (2015) 6:38005–15. doi: 10.18632/oncotarget.v6i35
43. Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ, et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. (2015) 6:41045–55. doi: 10.18632/oncotarget.v6i38
44. Yonkunas MJ, Baird NJ. A highly ordered, nonprotective MALAT1 ENE structure is adopted prior to triplex formation. RNA (New York NY). (2019) 25:975–84. doi: 10.1261/rna.069906.118
45. Torabi SF, DeGregorio SJ, Steitz JA. tRNA-like leader-trailer interaction promotes 3'-end maturation of MALAT1. RNA (New York NY). (2021) 27:1140–7. doi: 10.1261/rna.078810.121
46. Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac oncol: Off Publ Int Assoc Study Lung Cancer. (2011) 6:1984–92. doi: 10.1097/JTO.0b013e3182307eac
47. Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumor biol: J Int Soc Oncodevelop Biol Med. (2015) 36:1477–86. doi: 10.1007/s13277-014-2631-4
48. Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang S, et al. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J orthop res: Off Publ Orthop Res Soc. (2016) 34:932–41. doi: 10.1002/jor.23105
49. Zhang Y, Xu Y, Qiu G, Luo Y, Bao Y, Lu J, et al. METTL3 mediated MALAT1 m6A modification promotes proliferation and metastasis in osteosarcoma cells. Mol Biotechnol. (2023). doi: 10.1007/s12033-023-00953-2
50. Perry JA, Kiezun A, Tonzi P, Van Allen EM, Carter SL, Baca SC, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci United States America. (2014) 111:E5564–5573. doi: 10.1073/pnas.1419260111
51. Zhang J, Yu XH, Yan YG, Wang C, Wang WJ. PI3K/Akt signaling in osteosarcoma. Clinica chimica acta; Int J Clin Chem. (2015) 444:182–92. doi: 10.1016/j.cca.2014.12.041
52. Ye F, Tian L, Zhou Q, Feng D. LncRNA FER1L4 induces apoptosis and suppresses EMT and the activation of PI3K/AKT pathway in osteosarcoma cells via inhibiting miR-18a-5p to promote SOCS5. Gene. (2019) 721:144093. doi: 10.1016/j.gene.2019.144093
53. Chen Y, Huang W, Sun W, Zheng B, Wang C, Luo Z, et al. LncRNA MALAT1 promotes cancer metastasis in osteosarcoma via activation of the PI3K-akt signaling pathway. Cell Physiol Biochem. (2018) 51:1313–26. doi: 10.1159/000495550
54. Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. (2012) 26:338–43. doi: 10.1101/gad.182121.111
55. Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatol (Baltimore Md). (2016) 63:499–511. doi: 10.1002/hep.27893
56. Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. (2017) 405:46–55. doi: 10.1016/j.canlet.2017.06.009
57. Zhou X, Yang Y, Li Y, Liang G, Kang D, Zhou B, et al. METTL3 Contributes to Osteosarcoma Progression by Increasing DANCR mRNA Stability via m6A Modification. Front Cell Dev Biol. (2021) 9:784719. doi: 10.3389/fcell.2021.784719
58. Berteaux N, Aptel N, Cathala G, Genton C, Coll J, Daccache A, et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. (2008) 28:6731–45. doi: 10.1128/MCB.02103-07
59. Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene. (2014) 33:4857–66. doi: 10.1038/onc.2013.433
60. Pang F, Ding S, Li N, Li Z, Tian N, Shi C, et al. Gallic acid mediates tumor-suppressive effects on osteosarcoma through the H19-Wnt/β-catenin regulatory axis. J orthop translation. (2023) 39:34–42. doi: 10.1016/j.jot.2022.12.003
61. Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: A novel cancer-related long non-coding RNA. Cell prolif. (2017) 50:e12329. doi: 10.1111/cpr.12329
62. Li P, Huang R, Huang T, Cheng S, Chen Y, Wang Z. Long non-coding RNA NEAT1 promotes proliferation, migration and invasion of human osteosarcoma cells. Int J Med Sci. (2018) 15:1227–34. doi: 10.7150/ijms.25662
63. Wang L, Zhou J, Zhang Y, Hu T, Sun Y. Long Non-Coding RNA HCG11 Aggravates Osteosarcoma Carcinogenesis via Regulating the microRNA-579/MMP13 Axis. Int J Gen Med. (2020) 13:1685–95. doi: 10.2147/IJGM.S274641
64. Chen Y, Li J, Xiao JK, Xiao L, Xu BW, Li C. The lncRNA NEAT1 promotes the epithelial-mesenchymal transition and metastasis of osteosarcoma cells by sponging miR-483 to upregulate STAT3 expression. Cancer Cell Int. (2021) 21:90. doi: 10.1186/s12935-021-01780-8
65. Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. (2011) 71:3852–62. doi: 10.1158/0008-5472.CAN-10-4460
66. Li Z, Shen J, Chan MTV, Wu WKK. The long non-coding RNA SPRY4-IT1: An emerging player in tumorigenesis and osteosarcoma. Cell prolif. (2018) 51:e12446. doi: 10.1111/cpr.12446
67. Xu J, Ding R, Xu Y. Effects of long non-coding RNA SPRY4-IT1 on osteosarcoma cell biological behavior. Am J Trans Res. (2016) 8:5330–7.
68. Yao H, Hou G, Wang QY, Xu WB, Zhao HQ, Xu YC. LncRNA SPRY4−IT1 promotes progression of osteosarcoma by regulating ZEB1 and ZEB2 expression through sponging of miR−101 activity. Int J Oncol. (2020) 56:85–100. doi: 10.3892/ijo.2019.4910
69. He W, Lu M, Xiao D. LSINCT5 predicts unfavorable prognosis and exerts oncogenic function in osteosarcoma. Biosci Rep. (2019) 39:BSR20190612. doi: 10.1042/BSR20190612
70. Kong D, Li C, Yang Q, Wei B, Wang L, Peng C. Long noncoding RNA LSINCT5 acts as an oncogene via increasing EZH2-induced inhibition of APC expression in osteosarcoma. Biochem Biophys Res Commun. (2018) 507:193–7. doi: 10.1016/j.bbrc.2018.11.005
71. Ju L, Zhou YM, Yang GS. Up-regulation of long non-coding RNA BCAR4 predicts a poor prognosis in patients with osteosarcoma, and promotes cell invasion and metastasis. Eur Rev Med Pharmacol Sci. (2016) 20:4445–51.
72. Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. (2015) 8:2994–3000.
73. Wen JJ, Ma YD, Yang GS, Wang GM. Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. (2017) 21:498–503.
74. Sanchez A, Lhuillier J, Grosjean G, Ayadi L, Maenner S. The long non-coding RNA ANRIL in cancers. Cancers. (2023) 15:4160. doi: 10.3390/cancers15164160
75. Chen F, Mo J, Zhang L. Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription. Tumor Biol. (2016) 37:13403–12. doi: 10.1007/s13277-016-5256-y
76. Guo N, Sun Q, Fu D, Zhang Y. Long non-coding RNA UCA1 promoted the growth of adrenocortical cancer cells via modulating the miR-298-CDK6 axis. Gene. (2019) 703:26–34. doi: 10.1016/j.gene.2019.03.066
77. Ghafouri-Fard S, Taheri M. UCA1 long non-coding RNA: An update on its roles in Malignant behavior of cancers. Biomed Pharmacother. (2019) 120:109459. doi: 10.1016/j.biopha.2019.109459
78. Lee AM, Ferdjallah A, Moore E, Kim DC, Nath A, Greengard E, et al. Long non-coding RNA ANRIL as a potential biomarker of chemosensitivity and clinical outcomes in osteosarcoma. Int J Mol Sci. (2021) 22:11168. doi: 10.3390/ijms222011168
79. Pasic I, Shlien A, Durbin AD, Stavropoulos DJ, Baskin B, Ray PN, et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. (2010) 70:160–71. doi: 10.1158/0008-5472.CAN-09-1902
80. Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. (2013) 41:4976–87. doi: 10.1093/nar/gkt182
81. da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends gene: TIG. (2008) 24:306–16. doi: 10.1016/j.tig.2008.03.011
82. Sun M, Xia R, Jin F, Xu T, Liu Z, De W, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumor Biol. (2014) 35:1065–73. doi: 10.1007/s13277-013-1142-z
83. Lv D, Sun R, Yu Q, Zhang X. The long non-coding RNA maternally expressed gene 3 activates p53 and is downregulated in esophageal squamous cell cancer. Tumor Biol. (2016) 37:16259–67. doi: 10.1007/s13277-016-5426-y
84. Guo Q, Qian Z, Yan D, Li L, Huang L. LncRNA-MEG3 inhibits cell proliferation of endometrial carcinoma by repressing Notch signaling. Biomed pharmacother = Biomed pharmacother. (2016) 82:589–94. doi: 10.1016/j.biopha.2016.02.049
85. Zhang SZ, Cai L, Li B. MEG3 long non-coding RNA prevents cell growth and metastasis of osteosarcoma. Bratislavske lekarske listy. (2017) 118:632–6. doi: 10.4149/BLL_2017_121
86. Huang X, Zhang W, Pu F, Zhang Z. LncRNA MEG3 promotes chemosensitivity of osteosarcoma by regulating antitumor immunity via miR-21-5p/p53 pathway and autophagy. Genes Dis. (2023) 10:531–41. doi: 10.1016/j.gendis.2021.11.004
87. Mou J, Wang B, Liu Y, Zhao F, Wu Y, Xu W, et al. FER1L4: A long non-coding RNA with multiple roles in the occurrence and development of tumors. Curr Pharm design. (2022) 28:1334–41. doi: 10.2174/1381612828666220324141016
88. Yang X, Xie Z, Lei X, Gan R. Long non-coding RNA GAS5 in human cancer. Oncol Lett. (2020) 20:2587–94. doi: 10.3892/ol
89. Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. (2014) 14:319. doi: 10.1186/1471-2407-14-319
90. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. (2013) 1832:1613–23. doi: 10.1016/j.bbadis.2013.05.005
91. Liu J, Chen M, Ma L, Dang X, Du G. LncRNA GAS5 Suppresses the Proliferation and Invasion of Osteosarcoma Cells via the miR-23a-3p/PTEN/PI3K/AKT Pathway. Cell Transplant. (2020) 29:963689720953093. doi: 10.1177/0963689720953093
92. Li G, Yan X. Long non-coding RNA GAS5 promotes cisplatin-chemosensitivity of osteosarcoma cells via microRNA-26b-5p/TP53INP1 axis. J orthop Surg Res. (2023) 18:890. doi: 10.1186/s13018-023-04387-z
93. Luo W, He H, Xiao W, Liu Q, Deng Z, Lu Y, et al. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget. (2016) 7:54733–43. doi: 10.18632/oncotarget.v7i34
94. Guo W, Jiang H, Li H, Li F, Yu Q, Liu Y, et al. LncRNA-SRA1 suppresses osteosarcoma cell proliferation while promoting cell apoptosis. Technol Cancer Res Treat. (2019) 18:1533033819841438. doi: 10.1177/1533033819841438
95. Sneeggen M, Guadagno NA, Progida C. Intracellular transport in cancer metabolic reprogramming. Front Cell Dev Biol. (2020) 8:597608. doi: 10.3389/fcell.2020.597608
96. Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discovery. (2015) 14:130–46. doi: 10.1038/nrd4504
97. Wang Y, Yao J, Meng H, Yu Z, Wang Z, Yuan X, et al. A novel long non-coding RNA, hypoxia-inducible factor-2α promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep. (2015) 11:2534–40. doi: 10.3892/mmr.2014.3024
98. Cong M, Li J, Jing R, Li Z. Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumor Biol. (2016) 37:9441–50. doi: 10.1007/s13277-015-4414-y
99. Ruan W, Wang P, Feng S, Xue Y, Li Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumor Biol. (2016) 37:4065–73. doi: 10.1007/s13277-015-4256-7
100. Takebe N, Nguyen D, Yang SX. Targeting notch signaling pathway in cancer: clinical development advances and challenges. Pharmacol Ther. (2014) 141:140–9. doi: 10.1016/j.pharmthera.2013.09.005
101. Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. (2018) 495:1822–32. doi: 10.1016/j.bbrc.2017.12.047
102. Xie C, Chen B, Wu B, Guo J, Cao Y. LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in osteosarcoma by regulating miR-212-3p/FOXA1 axis. Biomed pharmacother = Biomed pharmacother. (2018) 97:1645–53. doi: 10.1016/j.biopha.2017.12.004
103. Cao J, Han X, Qi X, Jin X, Li X. TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via miR-144-3p. Int J Oncol. (2017) 51:1115–23. doi: 10.3892/ijo.2017.4110
104. Zhang Q, Geng PL, Yin P, Wang XL, Jia JP, Yao J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pacific J Cancer prevent: APJCP. (2013) 14:2311–5. doi: 10.7314/APJCP.2013.14.4.2311
105. Min L, Hong S, Duan H, Zhou Y, Zhang W, Luo Y, et al. Antidifferentiation noncoding RNA regulates the proliferation of osteosarcoma cells. Cancer biother radiopharma. (2016) 31:52–7. doi: 10.1089/cbr.2015.1888
106. Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. (2018) 17:89. doi: 10.1186/s12943-018-0837-6
107. He J, Ling L, Liu Z, Ren X, Wan L, Tu C, et al. Functional interplay between long non-coding RNAs and the Wnt signaling cascade in osteosarcoma. Cancer Cell Int. (2021) 21:313. doi: 10.1186/s12935-021-02013-8
108. Cui H, Zhao J. LncRNA TMPO-AS1 serves as a ceRNA to promote osteosarcoma tumorigenesis by regulating miR-199a-5p/WNT7B axis. J Cell Biochem. (2020) 121:2284–93. doi: 10.1002/jcb.29451
109. Dong Z, Wang Y. LncRNA BLACAT1 accelerates the proliferation and migration of osteosarcoma cells through regulating STAT3. Pathol Res Pract. (2019) 215:571–9. doi: 10.1016/j.prp.2019.01.017
110. Yin Z, Ding H, He E, Chen J, Li M. Overexpression of long non-coding RNA MFI2 promotes cell proliferation and suppresses apoptosis in human osteosarcoma. Oncol Rep. (2016) 36:2033–40. doi: 10.3892/or.2016.5013
111. Qian M, Yang X, Li Z, Jiang C, Song D, Yan W, et al. P50-associated COX-2 extragenic RNA (PACER) overexpression promotes proliferation and metastasis of osteosarcoma cells by activating COX-2 gene. Tumor Biol. (2016) 37:3879–86. doi: 10.1007/s13277-015-3838-8
112. Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Trans Res. (2016) 8:4095–105.
113. Zhao H, Hou W, Tao J, Zhao Y, Wan G, Ma C, et al. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/β-catenin signaling pathway. Am J Trans Res. (2016) 8:3503–12.
114. Gu W, Zhang E, Song L, Tu L, Wang Z, Tian F, et al. Long noncoding RNA HOXD-AS1 aggravates osteosarcoma carcinogenesis through epigenetically inhibiting p57 via EZH2. Biomed pharmacother = Biomed pharmacother. (2018) 106:890–5. doi: 10.1016/j.biopha.2018.06.173
115. Miano V, Ferrero G, Reineri S, Caizzi L, Annaratone L, Ricci L, et al. Luminal long non-coding RNAs regulated by estrogen receptor alpha in a ligand-independent manner show functional roles in breast cancer. Oncotarget. (2016) 7:3201–16. doi: 10.18632/oncotarget.v7i3
116. Zhang S, Ding L, Gao F, Fan H. Long non-coding RNA DSCAM-AS1 upregulates USP47 expression through sponging miR-101-3p to accelerate osteosarcoma progression. Biochem Cell Biol = Biochimie biol cellulaire. (2020) 98:600–11. doi: 10.1139/bcb-2020-0031
117. Mirabello L, Koster R, Moriarity BS, Spector LG, Meltzer PS, Gary J, et al. A genome-wide scan identifies variants in NFIB associated with metastasis in patients with osteosarcoma. Cancer Discovery. (2015) 5:920–31. doi: 10.1158/2159-8290.CD-15-0125
118. Lu J, Song G, Tang Q, Zou C, Han F, Zhao Z, et al. IRX1 hypomethylation promotes osteosarcoma metastasis via induction of CXCL14/NF-κB signaling. J Clin Invest. (2015) 125:1839–56. doi: 10.1172/JCI78437
119. Hu F, Wang W, Zhou HC, Shang XF. High expression of periostin is dramatically associated with metastatic potential and poor prognosis of patients with osteosarcoma. World J Surg Oncol. (2014) 12:287. doi: 10.1186/1477-7819-12-287
120. Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. (2002) 20:776–90. doi: 10.1200/JCO.2002.20.3.776
121. Lamora A, Mullard M, Amiaud J, Brion R, Heymann D, Redini F, et al. Anticancer activity of halofuginone in a preclinical model of osteosarcoma: inhibition of tumor growth and lung metastases. Oncotarget. (2015) 6:14413–27. doi: 10.18632/oncotarget.v6i16
122. Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang K, et al. Overexpression of long non-coding RNA HOTAIR promotes tumor growth and metastasis in human osteosarcoma. Molecules Cells. (2015) 38:432–40. doi: 10.14348/molcells.2015.2327
123. Liu G, Yuan D, Sun P, Liu W, Wu PF, Liu H, et al. LINC00968 functions as an oncogene in osteosarcoma by activating the PI3K/AKT/mTOR signaling. J Cell Physiol. (2018) 233:8639–47. doi: 10.1002/jcp.26624
124. Chen X, Zhao W, Fan W. Long non−coding RNA GHET1 promotes osteosarcoma development and progression via Wnt/β−catenin signaling pathway. Oncol Rep. (2020) 44:349–59. doi: 10.3892/or
125. Ding Q, Mo F, Cai X, Zhang W, Wang J, Yang S, et al. LncRNA CRNDE is activated by SP1 and promotes osteosarcoma proliferation, invasion, and epithelial-mesenchymal transition via Wnt/β-catenin signaling pathway. J Cell Biochem. (2020) 121:3358–71. doi: 10.1002/jcb.29607
126. Wang Y, Zhang Y, Yang T, Zhao W, Wang N, Li P, et al. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. (2017) 8:59417–34. doi: 10.18632/oncotarget.v8i35
127. Li L, Pei S, Sun N. MEG3 targets miR-184 and Wnt/β-catenin and modulates properties of osteosarcoma. Front biosci (Landmark edition). (2020) 25:1901–12. doi: 10.2741/4884
128. Sun X, Wang M, Wang M, Yao L, Li X, Dong H, et al. Exploring the metabolic vulnerabilities of epithelial-mesenchymal transition in breast cancer. Front Cell Dev Biol. (2020) 8:655. doi: 10.3389/fcell.2020.00655
129. Kumar RM, Fuchs B. Hedgehog signaling inhibitors as anti-cancer agents in osteosarcoma. Cancers. (2015) 7:784–94. doi: 10.3390/cancers7020784
130. Zhao J, Ma ST. Downregulation of lncRNA H19 inhibits migration and invasion of human osteosarcoma through the NF-κB pathway. Mol Med Rep. (2018) 17:7388–94. doi: 10.3892/mmr
131. Zhu G, Liu X, Su Y, Kong F, Hong X, Lin Z. Knockdown of urothelial carcinoma-associated 1 suppressed cell growth and migration through regulating miR-301a and CXCR4 in osteosarcoma MHCC97 cells. Oncol Res. (2018) 27:55–64. doi: 10.3727/096504018X15201143705855
132. Gong C, Sun K, Xiong HH, Sneh T, Zhang J, Zhou X, et al. Expression of CXCR4 and MMP-2 is associated with poor prognosis in patients with osteosarcoma. Histol histopathol. (2020) 35:863–70. doi: 10.14670/HH-18-219
133. Xi Y, Qi Z, Ma J, Chen Y. PTEN loss activates a functional AKT/CXCR4 signaling axis to potentiate tumor growth and lung metastasis in human osteosarcoma cells. Clin Exp metastasis. (2020) 37:173–85. doi: 10.1007/s10585-019-09998-7
134. Yin J, Chen D, Luo K, Lu M, Gu Y, Zeng S, et al. Cip2a/miR-301a feedback loop promotes cell proliferation and invasion of triple-negative breast cancer. J Cancer. (2019) 10:5964–74. doi: 10.7150/jca.35704
135. Han F, Wang C, Wang Y, Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am J Cancer Res. (2017) 7:770–83.
136. Sun J, Wang X, Fu C, Wang X, Zou J, Hua H, et al. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol Biol Rep. (2016) 43:427–36. doi: 10.1007/s11033-016-3975-1
137. Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. (2014) 26:722–37. doi: 10.1016/j.ccell.2014.09.014
138. Yang C, Wang G, Yang J, Wang L. Long noncoding RNA NBAT1 negatively modulates growth and metastasis of osteosarcoma cells through suppression of miR-21. Am J Cancer Res. (2017) 7:2009–19.
139. Jia D, Niu Y, Li D, Liu Z. lncRNA C2dat1 promotes cell proliferation, migration, and invasion by targeting miR-34a-5p in osteosarcoma cells. Oncol Res. (2018) 26:753–64. doi: 10.3727/096504017X15024946480113
140. Singla A, Wang J, Yang R, Geller DS, Loeb DM, Hoang BH. Wnt signaling in osteosarcoma. Adv Exp Med Biol. (2020) 1258:125–39. doi: 10.1007/978-3-030-43085-6_8
141. Adamopoulos C, Gargalionis AN, Basdra EK, Papavassiliou AG. Deciphering signaling networks in osteosarcoma pathobiology. Exp Biol Med (Maywood NJ). (2016) 241:1296–305. doi: 10.1177/1535370216648806
142. McQueen P, Ghaffar S, Guo Y, Rubin EM, Zi X, Hoang BH. The Wnt signaling pathway: implications for therapy in osteosarcoma. Expert Rev Anticancer Ther. (2011) 11:1223–32. doi: 10.1586/era.11.94
143. Haydon RC, Deyrup A, Ishikawa A, Heck R, Jiang W, Zhou L, et al. Cytoplasmic and/or nuclear accumulation of the beta-catenin protein is a frequent event in human osteosarcoma. Int J Cancer. (2002) 102:338–42. doi: 10.1002/ijc.10719
144. Dai J, Xu LJ, Han GD, Jiang HT, Sun HL, Zhu GT, et al. Down-regulation of long non-coding RNA ITGB2-AS1 inhibits osteosarcoma proliferation and metastasis by repressing Wnt/β-catenin signaling and predicts favorable prognosis. Artif cells nanomed Biotechnol. (2018) 46:S783–s790. doi: 10.1080/21691401.2018.1511576
145. Huang Q, Shi SY, Ji HB, Xing SX. LncRNA BE503655 inhibits osteosarcoma cell proliferation, invasion/migration via Wnt/β-catenin pathway. Biosci Rep. (2019) 39:BSR20182200. doi: 10.1042/BSR20182200
146. Zhang RM, Tang T, Yu HM, Yao XD. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem Biophys Res Commun. (2018) 507:260–6. doi: 10.1016/j.bbrc.2018.11.019
147. Xia B, Wang L, Feng L, Tian B, Tan Y, Du B. Knockdown of long noncoding RNA CAT104 inhibits the proliferation, migration, and invasion of human osteosarcoma cells by regulating microRNA-381. Oncol Res. (2018) 27:89–98. doi: 10.3727/096504018X15199511344806
148. Jiang Z, Jiang C, Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin pathway. Biochem Biophys Res Commun. (2018) 495:238–45. doi: 10.1016/j.bbrc.2017.11.012
149. Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. (2018) 18:309–24. doi: 10.1038/nri.2017.142
150. Zhang GD, Li Y, Liao GJ, Qiu HW. LncRNA NKILA inhibits invasion and migration of osteosarcoma cells via NF-κB/Snail signaling pathway. Eur Rev Med Pharmacol Sci. (2019) 23:4118–25. doi: 10.26355/eurrev_201905_17913
151. Gao W, Gao J, Chen L, Ren Y, Ma J. Targeting XIST induced apoptosis of human osteosarcoma cells by activation of NF-kB/PUMA signal. Bioengineered. (2019) 10:261–70. doi: 10.1080/21655979.2019.1631104
152. Ren Z, Hu Y, Li G, Kang Y, Liu Y, Zhao H. HIF-1α induced long noncoding RNA FOXD2-AS1 promotes the osteosarcoma through repressing p21. Biomed pharmacother = Biomed pharmacother. (2019) 117:109104. doi: 10.1016/j.biopha.2019.109104
153. Zhao B, Liu K, Cai L. LINK-A lncRNA functions in the metastasis of osteosarcoma by upregulating HIF1α. Oncol Lett. (2019) 17:5005–11. doi: 10.3892/ol
154. Marina NM, Smeland S, Bielack SS, Bernstein M, Jovic G, Krailo MD, et al. Comparison of MAPIE versus MAP in patients with a poor response to preoperative chemotherapy for newly diagnosed high-grade osteosarcoma (EURAMOS-1): an open-label, international, randomized controlled trial. Lancet Oncol. (2016) 17:1396–408. doi: 10.1016/S1470-2045(16)30214-5
155. Gaspar N, Occean BV, Pacquement H, Bompas E, Bouvier C, Brisse HJ, et al. Results of methotrexate-etoposide-ifosfamide based regimen (M-EI) in osteosarcoma patients included in the French OS2006/sarcome-09 study. Eur J Cancer (Oxford England: 1990). (2018) 88:57–66. doi: 10.1016/j.ejca.2017.09.036
156. Palmerini E, Jones RL, Marchesi E, Paioli A, Cesari M, Longhi A, et al. Gemcitabine and docetaxel in relapsed and unresectable high-grade osteosarcoma and spindle cell sarcoma of bone. BMC Cancer. (2016) 16:280. doi: 10.1186/s12885-016-2312-3
157. Lagmay JP, Krailo MD, Dang H, Kim A, Hawkins DS, Beaty O 3rd, et al. Outcome of patients with recurrent osteosarcoma enrolled in seven phase II trials through children's cancer group, pediatric oncology group, and children's oncology group: learning from the past to move forward. J Clin Oncol. (2016) 34:3031–8. doi: 10.1200/JCO.2015.65.5381
158. Gao KT, Lian D. Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur Rev Med Pharmacol Sci. (2016) 20:3561–5.
159. Huo Y, Li Q, Wang X, Jiao X, Zheng J, Li Z, et al. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. (2017) 8:46993–7006. doi: 10.18632/oncotarget.v8i29
160. Xie CH, Cao YM, Huang Y, Shi QW, Guo JH, Fan ZW, et al. Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR-9-5p and regulating POU2F1 expression. Tumor Biol. (2016) 37:15031–41. doi: 10.1007/s13277-016-5391-5
161. Liu T, Ma Q, Zhang Y, Ke S, Yan K, Chen X, et al. Interleukin-11 receptor α is overexpressed in human osteosarcoma, and near-infrared-labeled IL-11Rα imaging agent could detect osteosarcoma in mouse tumor xenografts. Tumor Biol. (2015) 36:2369–75. doi: 10.1007/s13277-014-2844-6
162. Li F, Cao L, Hang D, Wang F, Wang Q. Long non-coding RNA HOTTIP is up-regulated and associated with poor prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. (2015) 8:11414–20.
163. Su X, Teng J, Jin G, Li J, Zhao Z, Cao X, et al. ELK1-induced upregulation of long non-coding RNA MIR100HG predicts poor prognosis and promotes the progression of osteosarcoma by epigenetically silencing LATS1 and LATS2. Biomed pharmacother = Biomed pharmacother. (2019) 109:788–97. doi: 10.1016/j.biopha.2018.10.029
164. Ghafouri-Fard S, Shirvani-Farsani Z, Hussen BM, Taheri M. The critical roles of lncRNAs in the development of osteosarcoma. Biomed pharmacother = Biomed pharmacother. (2021) 135:111217. doi: 10.1016/j.biopha.2021.111217
165. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. (2013) 13:714–26. doi: 10.1038/nrc3599
166. Brown HK, Tellez-Gabriel M, Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. (2017) 386:189–95. doi: 10.1016/j.canlet.2016.11.019
167. Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, et al. Drug resistance in cancer: an overview. Cancers. (2014) 6:1769–92. doi: 10.3390/cancers6031769
168. Kim M, Jung JY, Choi S, Lee H, Morales LD, Koh JT, et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. (2017) 13:149–68. doi: 10.1080/15548627.2016.1239676
169. Zhang CL, Zhu KP, Ma XL. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. (2017) 396:66–75. doi: 10.1016/j.canlet.2017.03.018
170. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. (2019) 179:1033–55. doi: 10.1016/j.cell.2019.10.017
171. He H, Ni J, Huang J. Molecular mechanisms of chemoresistance in osteosarcoma (Review). Oncol Lett. (2014) 7:1352–62. doi: 10.3892/ol.2014.1935
172. Chen R, Wang G, Zheng Y, Hua Y, Cai Z. Long non-coding RNAs in osteosarcoma. Oncotarget. (2017) 8:20462–75. doi: 10.18632/oncotarget.v8i12
173. Martins-Neves SR, Lopes ÁO, do Carmo A, Paiva AA, Simões PC, Abrunhosa AJ, et al. Therapeutic implications of an enriched cancer stem-like cell population in a human osteosarcoma cell line. BMC Cancer. (2012) 12:139. doi: 10.1186/1471-2407-12-139
174. Gougelet A, Pissaloux D, Besse A, Perez J, Duc A, Dutour A, et al. Micro-RNA profiles in osteosarcoma as a predictive tool for ifosfamide response. Int J Cancer. (2011) 129:680–90. doi: 10.1002/ijc.25715
175. Kathawala RJ, Gupta P, Ashby CR Jr., Chen ZS. The modulation of ABC transporter-mediated multidrug resistance in cancer: a review of the past decade. Drug resist updates: Rev comment antimicrobial Anticancer chemother. (2015) 18:1–17. doi: 10.1016/j.drup.2014.11.002
176. Hang Q, Sun R, Jiang C, Li Y. Notch 1 promotes cisplatin-resistant gastric cancer formation by upregulating lncRNA AK022798 expression. Anti-cancer Drugs. (2015) 26:632–40. doi: 10.1097/CAD.0000000000000227
177. Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signaling pathway via suppression of activator protein 2α. Gut. (2016) 65:1494–504. doi: 10.1136/gutjnl-2014-308392
178. Han Z, Shi L. Long non-coding RNA LUCAT1 modulates methotrexate resistance in osteosarcoma via miR-200c/ABCB1 axis. Biochem Biophys Res Commun. (2018) 495:947–53. doi: 10.1016/j.bbrc.2017.11.121
179. Zhang CL, Zhu KP, Shen GQ, Zhu ZS. A long non-coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumor Biol. (2016) 37:2737–48. doi: 10.1007/s13277-015-4130-7
180. Zhu KP, Zhang CL, Shen GQ, Zhu ZS. Long noncoding RNA expression profiles of the doxorubicin-resistant human osteosarcoma cell line MG63/DXR and its parental cell line MG63 as ascertained by microarray analysis. Int J Clin Exp Pathol. (2015) 8:8754–73.
181. Wang Y, Zhang L, Zheng X, Zhong W, Tian X, Yin B, et al. Long non-coding RNA LINC00161 sensitizes osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett. (2016) 382:137–46. doi: 10.1016/j.canlet.2016.08.024
182. Gu Z, Zhou Y, Cao C, Wang X, Wu L, Ye Z. TFAP2C-mediated LINC00922 signaling underpins doxorubicin-resistant osteosarcoma. Biomed pharmacother = Biomed pharmacother. (2020) 129:110363. doi: 10.1016/j.biopha.2020.110363
183. Wang Z, Liu Z, Wu S. Long non-coding RNA CTA sensitizes osteosarcoma cells to doxorubicin through inhibition of autophagy. Oncotarget. (2017) 8:31465–77. doi: 10.18632/oncotarget.v8i19
184. Michael M, Doherty MM. Tumoral drug metabolism: overview and its implications for cancer therapy. J Clin Oncol. (2005) 23:205–29. doi: 10.1200/JCO.2005.02.120
185. Dhaini HR, Thomas DG, Giordano TJ, Johnson TD, Biermann JS, Leu K, et al. Cytochrome P450 CYP3A4/5 expression as a biomarker of outcome in osteosarcoma. J Clin Oncol. (2003) 21:2481–5. doi: 10.1200/JCO.2003.06.015
186. Huang G, Mills L, Worth LL. Expression of human glutathione S-transferase P1 mediates the chemosensitivity of osteosarcoma cells. Mol Cancer Ther. (2007) 6:1610–9. doi: 10.1158/1535-7163.MCT-06-0580
187. Han S, Cao Y, Guo T, Lin Q, Luo F. Targeting lncRNA/Wnt axis by flavonoids: A promising therapeutic approach for colorectal cancer. Phytother res: PTR. (2022) 36:4024–40. doi: 10.1002/ptr.7550
Keywords: osteosarcoma, lncRNA, cancer cell, biomarkers, indicators
Citation: Hu S, Han X, Liu G and Wang S (2024) LncRNAs as potential prognosis/diagnosis markers and factors driving drug resistance of osteosarcoma, a review. Front. Endocrinol. 15:1415722. doi: 10.3389/fendo.2024.1415722
Received: 11 April 2024; Accepted: 17 June 2024;
Published: 02 July 2024.
Edited by:
Changjun Li, Central South University, ChinaReviewed by:
Yanqing Jiang, University of Pennsylvania, United StatesBo Jiang, Rice University, United States
Shuang Wu, The University of Chicago, United States
Copyright © 2024 Hu, Han, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Liu, Z2FuZ2xlLmxpdUBnbWFpbC5jb20=; Shuangshuang Wang, d2FuZ3NzMTAyM0AxMjYuY29t
†These authors have contributed equally to this work
 Siwang Hu
Siwang Hu Xuebing Han
Xuebing Han Gang Liu
Gang Liu Shuangshuang Wang
Shuangshuang Wang