
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol., 27 January 2023
Sec. Gut Endocrinology
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.973624
This article is part of the Research TopicMicrobiota-Hormones-Gut Axis as a Therapeutic Target for Metabolic DisordersView all 6 articles
The gut microbiota, as a ‘new organ’ of humans, has been identified to affect many biological processes, including immunity, inflammatory response, gut-brain neural circuits, and energy metabolism. Profound dysbiosis of the gut microbiome could change the metabolic pattern, aggravate systemic inflammation and insulin resistance, and exacerbate metabolic disturbance and the progression of type 2 diabetes (T2D). The aim of this review is to focus on the potential roles and functional mechanisms of gut microbiota in the antidiabetic therapy. In general, antidiabetic drugs (α-glucosidase inhibitor, biguanides, incretin-based agents, and traditional Chinese medicine) induce the alteration of microbial diversity and composition, and the levels of bacterial component and derived metabolites, such as lipopolysaccharide (LPS), short chain fatty acids (SCFAs), bile acids and indoles. The altered microbial metabolites are involved in the regulation of gut barrier, inflammation response, insulin resistance and glucose homeostasis. Furthermore, we summarize the new strategies for antidiabetic treatment based on microbial regulation, such as pro/prebiotics administration and fecal microbiota transplantation, and discuss the need for more basic and clinical researches to evaluate the feasibility and efficacy of the new therapies for diabetes.
The gut microbiota is the largest human microecosystem, and is a complex community consisting of more than 500 microbial species (1, 2). The gut bacteria are mainly composed of five phyla, namely, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria and Verrucomicrobia, and more than 90% of the bacteria belong to Firmicutes and Bacteroidetes (3). Dysbiosis of the gut microbiota is associated with the occurrence and development of various diseases, such as immunological diseases (4, 5), inflamed intestinal diseases (6, 7), mental disorders (8, 9), and metabolic diseases (10–13). The homeostasis of gut microbiota could be affected by host genetics (14, 15), environmental factors (16), medicines (17, 18), and lifestyles (19). Kawano et al. found that high-fat, high-sugar diet aggravated glucose intolerance and insulin resistance in mice by depleting Th17 inducing microbes, particularly segmented filamentous bacteria (SFB) (20). Besides, one randomized-controlled trial on the effects of non-nutritive sweeteners (NNS) in human found that sucralose and saccharin supplementation significantly impaired glycemic response in healthy adults. Transplantation of microbiomes from top and bottom NNS responders into germ-free (GF) mice indicated that the alteration of gut microbiota induced by NNS could cause personalized glycemic alterations, as exemplified by sucralose (21). In the context of metabolic disorders, the gut microbiota is closely connected with the dysfunction of glucose and lipid metabolism, inflammation response and insulin resistance (22–24). Recent studies found that GF mice are protected against high-fat diet (HFD) induced glucose intolerance and obesity (25). In addition, colonization of GF mice with gut microbiota isolated from conventionally raised obese donors led to a significant increase in insulin resistance and body fat content (26, 27). Consequently, the gut microbiota was identified to be closely linked to the pathogenesis of diabetes and obesity.
Recently, numerous studies have shown that the gut microbiota is changed in both type 1 diabetes (T1D) and type 2 diabetes (T2D) patients, which indicates an etiological relationship between the gut microbiota and diabetes. The microbiome of T1D presented a decrease in diversity, an increase in the Firmicutes and Firmicutes/Bacteroidetes ratio, and a reduction in Proteobacteria and Bacteroidetes (28–30). Compared with controls, 16S rRNA data of individuals with T1D showed a lower proportion of butyrate-producing and mucin-degrading bacteria (28), and fecal microbiota transplantation (FMT) from healthy donors halted the decline in endogenous insulin production in new-onset T1D patients (31). Similarly, in metagenome-wide association studies, T2D patients were characterized by a moderate degree of gut microbial dysbiosis, an increase in various opportunistic pathogens and a decrease in the abundance of some universal butyrate-producing bacteria, such as Clostridium species (12, 32). Among the commonly and consistently reported findings, the genera of Bifidobacterium, Bacteroides, Faecalibacterium, Akkermansia and Roseburia were negatively associated with T2D, while the genera of Ruminococcus, Fusobacterium, and Blautia were positively associated with T2D (33). Moreover, it has been found that the altered gut microbiota in diabetes is associated with metabolic parameters. For instance, the ratio of Bacteroidetes to Firmicutes and the ratios of Bacteroides-Prevotella group to C. coccoides-E. rectale group were correlated significantly and positively with plasma glucose concentration (34). These studies suggest that dysbiosis of gut microbiota might greatly contribute to the occurrence and progression of diabetes.
In this review, we focus on the potential roles and mechanisms of gut microbiota in the antidiabetic therapy. Briefly, we summarize the contribution of microbiota to glucoregulatory therapy for T2D, and the demonstrated mechanism of microbial metabolites in regulating inflammation response, insulin resistance and glucose homeostasis, and discuss the new strategies for targeting intestinal bacteria in the treatment of diabetes.
Recently, it has been reported that the alteration of gut microbiota mediated by antidiabetic agents was involved in the regulation of glucose and lipid metabolism in diabetes. In this part, we concluded the role of commonly used glucoregulatory agents in the regulation of the composition and function of gut microbiota.
α-glucosidase inhibitor, as one fermentation product of bacterial strains derived from Actinoplanes sp. SE50 (35), inhibits α-glucosidase activity and slows carbohydrate uptake in the brush border of the small intestinal mucosa to reduce postprandial hyperglycemia (36, 37). This results in an increase in dietary carbohydrate in the distal intestine, where it becomes food for the gut bacterial community. Some studies have focused on the effect of α-glucosidase inhibitors on the gut microbial composition in diabetes. Acarbose, voglibose, and miglitol are pseudo-carbohydrates that competitively inhibit α-glucosidase activity. Acarbose is the most used glucoregulatory drug of this family. In T2D patients and rats, acarbose treatment significantly increased the content of Bifidobacterium (38–40), which has an anti-inflammatory effect, and has been reported to decrease in diabetes (41). The increased B. longum showed a positive correlation with the level of high-density lipoprotein cholesterol (HDL-C), a potent protective factor against atherosclerosis (38). One controlled crossover trial involving 52 Chinese prediabetic patients discovered that short chain fatty acids (SCFAs)-producing taxa, such as Lactobacillus, Faecalibacterium and Prevotella, were greatly increased after acarbose treatment (42). SCFAs, including acetate, propionate and butyrate, are fermented from indigestible dietary carbohydrates by intestinal bacteria. It has been demonstrated that SCFAs have important roles in stimulating glucagon-like peptide-1 (GLP-1) and insulin secretion, ameliorating inflammation response, and improving glucose intolerance and insulin resistance (43–45). Besides, some SCFAs-producing bacteria, such as Lactobacillus strains, have a broader spectrum of inhibitory activity than acarbose, effectively inhibiting α-glucosidase and β-glucosidase activities (46), and have beneficial effects on insulin resistance and weight loss (47, 48). Moreover, administration of acarbose to treatment-naïve T2D patients also changed the relative level of microbial genes involved in bile acid metabolism, and increased the ratio of primary bile acids to secondary bile acids. The correlation analysis demonstrated that patients with a higher baseline abundance of Bacteroides had lower levels of secondary bile acids, and exhibit more beneficial therapeutic responses to acarbose treatment (49). That is, the acarbose-changed microbiome was associated with its treatment efficiency for T2D. Additionally, voglibose, another α-glucosidase inhibitor, decreased the ratio of Firmicutes to Bacteroidetes, and increased the circulating levels of taurocholic and cholic acid, which ultimately has systemic effects on body weight and lipid metabolism in HFD mice (50).
Among biguanides, metformin is recommended by the American Association of Clinical Endocrinologists (AACE) as a first-line hypoglycemic treatment of T2D for its glucose lowering and insulin sensitizing effects (51), and is the most commonly used oral glucoregulatory agent. The hypoglycemic effect is ascribed to suppress hepatic gluconeogenesis, improve the uptake and utilization of glucose in peripheral tissues, promote the release of glucagon-like peptide-1 (GLP-1) and peptide YY (PYY) from L cells, and enhance insulin sensitivity (52–55). In T2D patients and murine models, metformin has been identified to modify gut microbial structure and diversity. Indeed, Shin et al. found that metformin-treated HFD mice showed significant differences in the abundance of Firmicutes and Bacteroidetes, when compared with non-treated mice (56). Similarly, Lee and Ryan also observed that metformin induced an increase in the abundance of Bacteroidetes and Verrucomicrobia, and a decrease in the abundance of Firmicutes in HFD mice (57) (58). At the genus level, the SCFAs-producing bacteria such as Blautia, Bacteroides, Butyricoccus and Phascolarctobacterium were enriched by metformin treatment (59). Particularly, the abundance of Akkermansia, which are mucin-degrading bacteria, was significantly increased in metformin treated mice (57, 58). Administration of cultured A. muciniphila to HFD mice significantly enhanced glucose tolerance, and attenuated serum lipopolysaccharide (LPS) and inflammation status by inducing Foxp3 regulatory T cells in visceral adipose tissue (56).
Similar with the results observed in rodents, administration of metformin to T2D patients increased the relative abundances of Lactobacillus and Akkermansia, as well as the SCFAs-producing bacteria such as Bifidobacterium, Prevotella, Megasphaera and Butyrivibrio (60). B. adolescentis has been shown to negatively correlate with HbA1c. Thus, this taxon might contribute to the glucose-lowering effect of metformin (61). Besides, metformin treatment decreased the relative abundance of Intestinibacter, and enriched Escherichia in human feces. Whereas Escherichia probably plays an important role in the side-effects of metformin (62). In addition to changing the microbial composition, metformin treatment also promoted a functional shift in the gut microbiome, increasing the levels of propionate, butyrate and bile acids in the gut of T2D patients (61–63). As reported by CT Jiang, metformin-increased glycoursodeoxycholic acid (GUDCA) was identified as an intestinal FXR antagonist that improved various metabolic endpoints in mice with established obesity (64). Even though the mechanism of hypoglycemia of metformin is not completely clear, its administration altered the gut microbial composition and function, which might be involved in the regulation of glucose metabolism.
Incretins are small peptide hormones secreted by intestinal endocrine cells responding to nutrient ingestion, mainly glucose and fat. There are two main incretin hormones in humans: GLP-1 and GIP (gastric inhibitory peptide). They stimulate insulin secretion in a glucose-dependent manner, regulate appetite and gastric emptying, and play a crucial role in the local gastrointestinal physiology (65). Additionally, PYY is one of the first hormones to be expressed in the developing fetal gastrointestinal tract. The primary role of PYY is acting as a potent anorectic hormone. PYY3-36, a major form of PYY in both the gut mucosal endocrine cells and the circulation, induces satiety by targeting the appetite-regulating system of the hypothalamus (66). Insulin-like peptide-5 (INSL5) is an orexigenic gut hormone, secreted by enteroendocrine L cells present in the ileum and colon together with GLP-1 and PYY. INSL5 modestly inhibited forskolin-stimulated GLP-1 secretion in NCI-H716 cells (67).
Naturally intact GLP-1 is degraded rapidly, mainly via enzymatic inactivity by dipeptidyl peptidase-4 (DPP-4). Clinically, incretin-based therapies for diabetes include GLP-1 receptor agonists (incretin mimetics) and DPP-4 inhibitors (DPP-4i, incretin enhancers). The GLP-1 receptor agonist, such as liraglutide, promotes the synthesis and secretion of insulin, and inhibits appetite, gastric emptying and food intake to control blood glucose level in diabetes (68, 69). In the liraglutide-treated obese and diabetic rodents, in addition to the obviously improved glucose and lipid metabolism, substantial rearrangements of bacterial structure were observed. It has been found that liraglutide decreased the obesity-related phylotypes (such as Erysipelotrichaceae, Roseburia, and Parabacteroides), and increased the lean-related phylotypes (such as Prevotella, Lactobacillus and Akkermansia) (70, 71). The bacteria positively correlated with hepatic steatosis associated parameters were decreased in HFD rats upon liraglutide intervention (72). Colonization of GF mice with gut microbiota from liraglutide-treated diabetic mice improved glucose-induced insulin secretion and regulated the intestinal immune system (73). Additionally, some SCFAs-producing bacteria, including Bacteroides, Lachnospiraceae and Bifidobacterium, were selectively enriched in liraglutide-treated diabetic rats (74). The liraglutide treatment was also associated with bile acid metabolism (75). SCFAs stimulates GLP-1 expression and secretion through binding with G-coupled protein receptor GPR43 and GPR41 (76), while bile acids via other receptors such as FXR and TGR5 (77). However, one randomized placebo-controlled trial in adults with T2D suggested that the beneficial effects of liraglutide on glucose metabolism are not linked to the intestinal microbiota composition, when used as add-on therapies to metformin or sulphonylureas (78). As mentioned in the study, the limitations including the lack of dietary monitoring/standardization and control of co-medication use might be the reason for the inconsistency with the results observed in animal experiments.
DPP-4i is one of the most extensively used oral glucoregulatory agent, and its target DPP-4 enzyme has a high expression level in the small intestine (79, 80). DPP-4i exerts a hypoglycemic effect by inhibiting the degradation activity of DPP-4 to increase the blood level of GLP-1 and GIP (81). Recently, some subjects have revealed that the gut microbial community is modified by DPP-4i administration. In obese and diabetic rats, sitagliptin treatment moderately restored the microbial composition to that in controls. At the phylum level, sitagliptin reduced the ratio of Firmicutes to Bacteroidetes (82). At the genus level, probiotics Bifidobacterium and Lactobacillus were obviously increased after sitagliptin treatment (39), while the abundance of SCFAs-producing bacteria (Roseburia and Clostridium) was not significantly altered by sitagliptin. Consistent with these findings, in our previous study, we found that DPP-4i (sitagliptin and saxagliptin) altered the gut microbial composition in T2D patients and HFD mice, especially reducing the ratio of Firmicutes to Bacteroidetes, and fecal microbiota transfer from sitagliptin-treated T2D patients to GF mice improved the glucose tolerance in recipients (83). Moreover, we have demonstrated that sitagliptin also changed the pattern of fecal and serum metabolites in HFD mice and T2D patients, including the levels of succinate, branched-chain amino acids and aromatic amino acids (83, 84). We are still exploring the molecular mechanism by which DPP-4i improves glucose homeostasis through gut microbiota-derived metabolites, especially indole derivatives.
Traditional Chinese medicine (TCM), generally also known as botanical medicine or phytomedicine, is an important scientific and technological resource with therapeutic or other human health benefits. Generally, the use of TCM herbal formula (FuFang in Chinese) is fundamental and includes several medicinal herbs. TCM has been reported to be an effective remedy for metabolic disorders, including T2D (85). In recent ten years, TCM with antidiabetic effects has also been identified to have effects on the structure of the gut microbiota (86, 87). Berberine is the major pharmacological component of the Chinese herb Coptis chinensis (Huanglian). The oral bioavailability of berberine is poor (below 5%), and over 95% stays in the gut (88, 89), which suggests a potential effect on the intestinal bacteria. Actually, it has been reported that berberine induced a marked shift in the structure of gut microbiota, some of which were selectively enriched in putative SCFAs-producing bacteria, including Blautia, Allobaculum, Bacteriodes and Butyricoccus, consequently leading to an increase in the fecal SCFAs concentration in obese and T2D rats (59, 90, 91). Berberine was also identified to promote the gene expression of ACK, MMD and BUT, which are the key enzymes in the synthetic pathway for SCFAs (92). In addition, berberine has been reported to have a hypoglycemic effect in T2D patients by inhibiting deoxycholic acid biotransformation by Ruminococcus bromii (89). Likewise, administration of Gegen Qinlian Decoction (GQD) to T2D rats decreased the levels of blood glucose and inflammatory cytokines, and altered the gut microbial composition, especially increasing the proportions of SCFAs-producing and anti-inflammatory bacteria, and decreasing the proportions of conditioned pathogenic bacteria associated with diabetic phenotype (93). Moreover, a double-blinded trial revealed a dose-dependent deviation of gut microbiota in response to GQD treatment in T2D patients, and the deviation occurred before significant improvement in T2D symptoms. The GQD enriched F. prausnitzii was positively correlated with β-cell function, and negatively correlated with FBG, HbA1c and postprandial blood glucose levels in T2D patients (94). In addition, a water extract of Ganoderma lucidum mycelium (WEGL), which is a medicinal mushroom used in TCM, decreased the ratio of Firmicutes to Bacteroidetes and endotoxin-bearing Proteobacteria levels, as well as improved inflammation response and insulin resistance in HFD mice. The effects were transmissible via FMT from WEGL-treated mice to HFD mice (95). Taken together, these studies indicate that the amelioration of hyperglycemia and insulin resistance by TCM in T2D is at least partially mediated by structural regulation of the gut microbiota.
The exploration of the potential mechanisms of gut microbiota in the occurrence and progression of diabetes mainly focuses on microbial metabolites, especially LPS, SCFAs, bile acids, and indoles, which are involved in the regulation of gut barrier, inflammation response, insulin resistance and glucose homeostasis (96–98) (Figure 1).
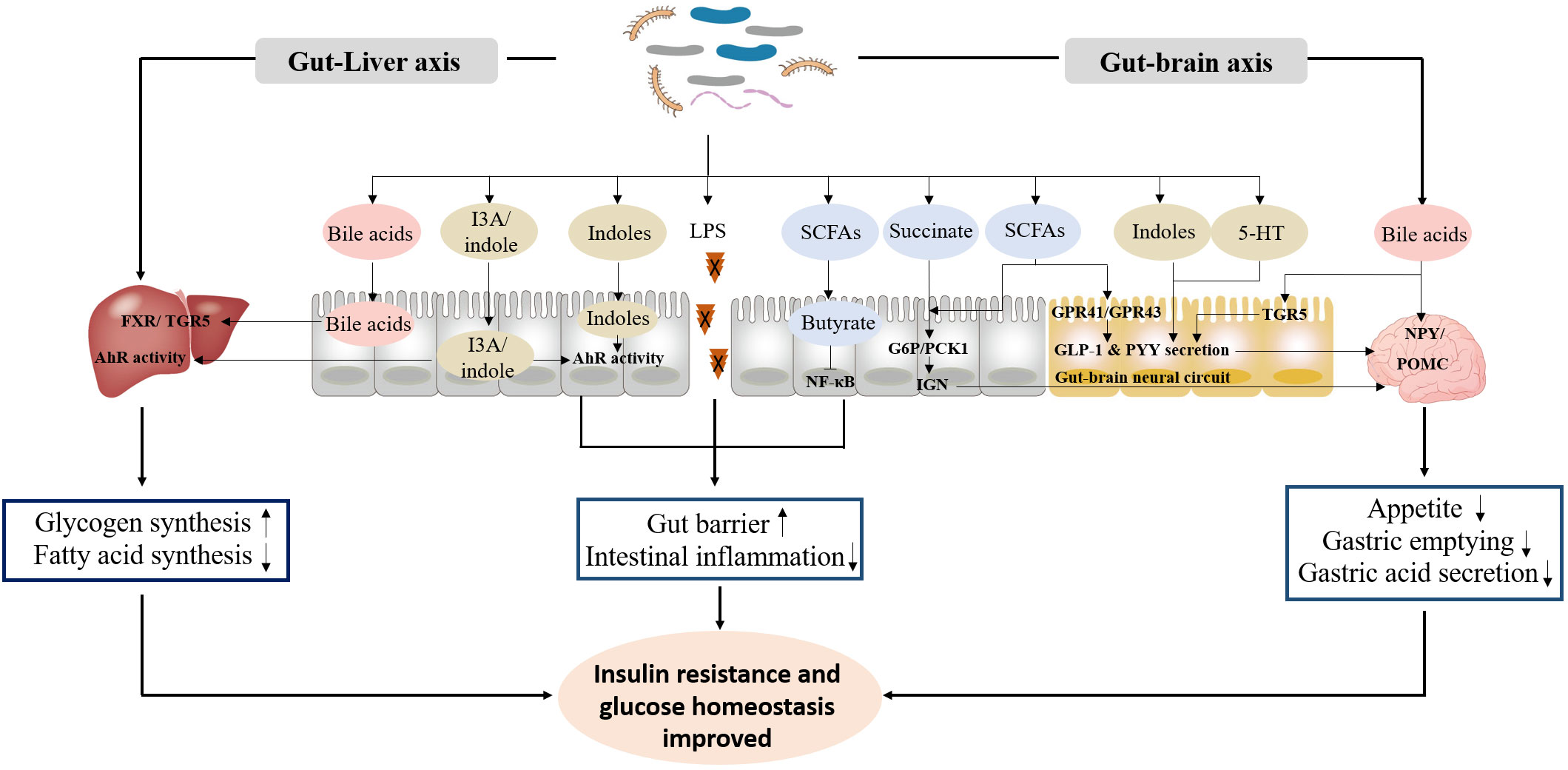
Figure 1 The primary functional mechanism of bacterial metabolites in the improvement of insulin resistance and glucose homeostasis. ① LPS, SCFAs and indoles improve gut barrier, and inhibit intestinal inflammation response. ② Bile acids, indoles, SCFAs, succinate and 5-HT promote GLP-1 and PYY secretion, and inhibit appetite, gastric emptying and gastric acid secretion through gut-brain axis. ③ Bile acids and indoles promote glycogen synthesis and insulin sensitivity, and inhibit fatty acid synthesis through gut-liver axis.
Dysbiosis of gut microbiota induces an increase in the levels of pathogens and endotoxins in the intestine. These toxic substances will induce intestinal macrophages to release proinflammatory factors such as TNF-α and histamine, and induce inflammatory response in intestinal cells, subsequently increasing the permeability of intestinal mucosa and damaging the intestinal barrier (99, 100). It has been recognized that low-grade inflammation is one of the most important pathophysiologic factors resulting in the progression of T2D (101). The intestinal bacteria produced LPS is a triggering factor for low-grade inflammation via the LPS-CD14 system, and further induces metabolic endotoxemia and insulin resistance (102). Two species (B. vulgatus and B. dorei) with potential benefits for T2D have been found to decrease gut microbial LPS production, and effectively suppress pro-inflammatory immune response in a mouse model (103). Similarly, administration of A. muciniphila to diabetic mice reduced the level of serum LPS, relieved intestinal inflammation, and improved glucose homeostasis (104). The butyrate-producing bacteria (such as Roseburia and Faecalibacterium) is known to eliminate constitutive NF-κB and suppress its activation, subsequently reducing colonic paracellular permeability and preventing inflammatory response (105, 106).
In addition, indole, as the most prevalent metabolite of tryptophan, is produced by various bacterial species, including those belonging to the genera Bacteroides, Escherichia, and Clostridium (107). Recent studies have revealed that indoles-induced aryl hydrocarbon receptor (AhR) activation may be an important way for bacteria to improve inflammatory status and insulin resistance (108). For example, indole derivates indole-3-carboxaldehyde upregulates the production of anti-inflammatory factor IL-22 by activating AhR in intestinal immune cells (109, 110).
Bacteria produced metabolites, such as SCFAs, bile acids and indoles, have effects on the intestinal endocrine cells to modulate the secretion of gut hormones, which play important roles in the gut-brain axis (111). SCFAs promote the secretion of GLP-1 and PYY by binding to membrane receptors GPR41 and GPR43 on endocrine L cells (112, 113). Likewise, depletion of Firmicutes and Bacteroidetes with antibiotics in diet-induced obesity mice increased production of gut-derived taurocholic acid, which promoted GLP-1 secretion from L-cells (114). Indole was also identified to promote GLP-1 secretion from L cells during short exposure, but reduce GLP-1 release over longer period. These different effects were due to whether indole enhance the acute secretion of GLP-1 by enhancing Ca2+ entry or reduce its release by blocking NADH dehydrogenase to slow ATP production (115). The secreted GLP-1 and PYY can inhibit appetite, gastric emptying and gastric acid secretion through acting on pro-opiomelanocortin (POMC) and neuropeptide Y (NPY) neurons in the hypothalamus (116). Similarly, bile acid reaches hypothalamus, specifically activates the expression of AgRP/NPY neuron membrane receptor TGR5, and then regulates the appetite of mice (117). Additionally, Gilles Mithieux and his colleagues reported that propionate, butyrate and succinate activated intestinal gluconeogenesis (IGN) to initiate a gut-brain neural circuit that has beneficial effects on glucose tolerance and insulin resistance (43, 118).
Additionally, gut-derived serotonin (5-hydroxytryptamine, 5-HT), as a metabolite of tryptophan, is released from enterochromaffin (EC) cells. 5-HT has been discovered to shaped the gut microbiota composition, and inhibited β-defensins production from colonic epithelial cells (119, 120). Besides, 5-HT signaling is reported to be involved in the increase of L-cell density in mouse and human intestine and elevation of GLP-1 secretory capacity (121). Moreover, the activation of GLP-1 leads to the release of 5-HT in brain, subsequently reducing appetite and body weight (122).
Accumulating evidence demonstrated that bile acids as pleiotropic signaling molecules modulate specific host metabolic pathways and inflammatory response involving gut-liver crosstalk, via nuclear and G-Protein-coupled receptors, such as the farnesoid X-activated receptor (FXR) and TGR5 (64, 123). FXR signaling represents a core regulatory pathway involved in glucose and lipid metabolism, and maintains the barrier integrity and intestinal homeostasis (124, 125). TGR5 is a membrane receptor, mainly activated by lithocholic acid and deoxycholic, which are secondary bile acids produced by microbial metabolism. TGR5 has been discovered to have effects on inflammation, insulin pathway and glucose metabolism (126, 127).
Except for bile acids, tryptophan metabolite such as tryptamine and indole-3-acetate (I3A) attenuated inflammatory response and reduced fatty acid synthesis via activating receptor AhR in hepatocytes (128). SCFAs are readily absorbed by colonocytes upon produced. Butyrate is mainly utilized by colonic epithelium as an energy source, and a large amount of acetate enters systemic circulation, while propionate is primary utilized by the liver (129). Acetate and butyrate were found to be highly involved in liver palmitate and cholesterol synthesis, and propionate is a substrate for de novo gluconeogenesis in liver, not for lipogenesis (130).
In addition to gut-brain axis and gut-liver axis, microbial metabolites also play roles in adipose, pancreas and muscle, involved in the regulation of inflammatory response, insulin secretion, insulin sensitivity, etc. (44, 131, 132).
The molecular mechanism of gut microbiota in the pathophysiology of T2D is still not completely clear, while existing studies have indicated that some interventions, such as probiotics, prebiotics and FMT targeting gut bacteria improved glucose homeostasis and insulin resistance, which indicates that these interventions might be potential strategies for T2D therapy.
Probiotics, particularly Lactobacilli and Bifidobacterium, have recently emerged as prospective biotherapeutics in metabolic disease, which is supported by their established multifunctional roles in the prevention and treatment of metabolic disturbance (133, 134). In HFD mice, probiotics (L. rhamnosus, L. acidophilus and B. bifidum) treatment significantly reversed the impaired intestinal permeability, systemic inflammation and glucose intolerance (135). Administration of L. plantarum HAC01 to T2D mice obviously lowered blood glucose and HbA1c levels, and improved glucose tolerance and HOMA-IR. Notably, L. plantarum HAC01 increased the Akkermansiaceae family and increased SCFAs in serum (136). In randomized controlled trials, L. acidophilus and B. lactis or B. bifidum significantly ameliorated the FBG and antioxidant status in T2D patients (137, 138). Likewise, L. paracasei HII01 supplementation significantly decreased the levels of FBG, LPS, TNF-α, IL-6 and hsCRP in plasma of T2D patients compared with the placebo group (139).
Apart from Lactobacillus and Bifidobacterium, other bacteria associated with metabolic improvement may also be potential probiotics, such as Akkermansia muciniphila. Its abundance markedly increased after prebiotic feeding (140, 141), or metformin treatment in diabetic patients and mice (56, 61). A. muciniphila MucT administration to diabetic mice increased the intestinal levels of endocannabinoids, which modulate inflammation, gut barrier and gut peptide secretion (104). Plovier et al. revealed that Amuc_1100, a specific protein isolated from the outer membrane of A. muciniphila, interacted with Toll-like receptor 2 (TLR2) to improve the gut barrier, and partly recapitulated the beneficial effects of the bacterium. Furthermore, administration with live or pasteurized A. muciniphila grown on synthetic medium is safe for obese individuals (142), which paves the way for its potential clinical application in metabolic syndromes. Another potential probiotic, Parabacteroides goldsteinii, was also identified to enhance intestinal integrity, improve inflammation and insulin resistance, and promote adipose tissue thermogenesis after oral administered to HFD mice (143).
Prebiotics are nondigestible carbohydrates, selectively stimulating the growth and activity of some bacteria, such as Lactobacillus and Bifidobacterium (144). Oligofructose-enriched diet reduced the ratio of Firmicutes to Bacteroidetes, increased the abundance of probiotics, such as Bifidobacterium and Prevotella, and improved glucose tolerance and L-cell function in ob/ob mice (145). Oligofructose also improved fasting blood glucose and glucose-stimulated insulin secretion, and reduced body weight gain in HFD mice, and the effects were modulated in a GLP-1 receptor-dependent manner (145, 146). Moreover, supplementation with oligofructose-enriched inulin to T1D participants for 12 weeks increased C-peptide, and improved intestinal permeability (147). However, some studies on consumption of pro/prebiotics by individuals with T2D have provided conflicting results, which do not provide evidence for the role of pro/prebiotics in the treatment of diabetes (148, 149). Therefore, the effects of pre/probiotics need to be confirmed by conducting more reliable clinical trials.
FMT has been verified to be effective for the treatment of Clostridium difficile infection in immunocompromised patients (150) and subjects with depressive disorder (8). Intriguingly, FMT is also carried out in the treatment of metabolic syndrome, including diabetes and obesity. Transfer of fecal samples from metformin-treated T2D patients to GF mice showed that glucose tolerance was improved in recipient mice (61). Similarly, our study discovered that FMT from DPP-4i-treated T2D patients to GF mice also improved the glucose intolerance induced by HFD in recipient mice (83). Moreover, clinical studies showed that intestinal microbiota from lean donors to individuals with metabolic syndrome attenuated insulin resistance, increased gut microbial diversity, especially the levels of the butyrate-producing bacterium Roseburia intestinalis, and changed the plasma metabolites, such as γ-aminobutyric acid and tryptophan (151, 152). In addition, allogenic FMT using feces from metabolic syndrome donors decreased insulin sensitivity in metabolic syndrome recipients compared with using post-Roux-en-Y gastric bypass donors (153). However, we must emphasize that these studies did not report data on glucose levels during the intervention, and not all participants responded to FMT (152). The method is still very new and, thus, much more research must be dedicated to FMT, to explore the potential risks related to its impact on the physiological functions regulated by fecal microorganisms, most of which are unidentified and uncharacterized at present.
The gut microbiota is involved in the occurrence and development of diabetes, and responses to antidiabetic therapy both in intestinal tract and in extraintestinal tissues. In this review, we summarized the current studies about the effect and mechanism of gut microbiota in antidiabetic therapy, especially in improving inflammation, glucose homeostasis and insulin resistance modulated by antidiabetic agents. Although the molecular mechanism remains unclear, advances in sequencing technologies and bioinformatics have enabled the enormous complexity and number of metabolites derived from intestinal bacteria to be identified in biological samples. Current research suggests that LPS, SCFAs, bile acids and indoles are involved in regulating host metabolic homeostasis. However, which strains induce altered metabolites, and whether the strains can be cultured in vitro need to be investigated. Studies are also necessary to address where and how the altered metabolites regulate host metabolism. Given the many insights so far based on animal experiments, it will be key to determine which phenotypes observed in animal models are relevant to humans and could be repeated in humans. Therefore, more clinical trials should be summoned to clarify the potential therapeutic effect of gut bacteria and derived metabolites in diabetes.
BL: consulted, analyzed and summarized the literatures, and drafted the manuscript. LZ consulted, analyzed and summarized the literatures. HY consulted the literatures. HZ: designed the study, and revised the manuscript. XL: designed the study, and drafted and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the grants from the National Science Fund for Distinguished Young Scholars of China (No. 81925007), the Natural Science Foundation of Chongqing (No. cstc2021jcyj-msxmX0465), the “Talent Project” of Army Medical University (2019R055), Special Program for Basic Research Frontier of Military Medicine of the Second Affiliated Hospital of Army Medical University (No. 2019YQYLY002 and No. 2018YQYLY006), and the Science Innovation Program Led by Academicians in Chongqing (No. cstc2017jcyj-yszxX0003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figure and legend:
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell (2006) 124(4):837–48. doi: 10.1016/j.cell.2006.02.017
2. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science (2005) 308(5728):1635–8. doi: 10.1126/science.1110591
3. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464(7285):59–65. doi: 10.1038/nature08821
4. Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol (2011) 7(10):569–78. doi: 10.1038/nrrheum.2011.121
5. Kishikawa T, Maeda Y, Nii T, Motooka D, Matsumoto Y, Matsushita M, et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann rheumatic Dis (2020) 79(1):103–11. doi: 10.1136/annrheumdis-2019-215743
6. Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science (2016) 352(6289):1116–20. doi: 10.1126/science.aad9948
7. Yilmaz B, Juillerat P, Oyas O, Ramon C, Bravo FD, Franc Y, et al. Microbial network disturbances in relapsing refractory crohn's disease. Nat Med (2019) 25(2):323–36. doi: 10.1038/s41591-018-0308-z
8. Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol Psychiatry (2016) 21(6):786–96. doi: 10.1038/mp.2016.44
9. Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol (2019) 4(4):623–32. doi: 10.1038/s41564-018-0337-x
10. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. New Engl J Med (2016) 375(24):2369–79. doi: 10.1056/NEJMra1600266
11. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature (2006) 444(7122):1022–3. doi: 10.1038/4441022a
12. Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature (2013) 498(7452):99–103. doi: 10.1038/nature12198
13. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol (2021) 19(1):55–71. doi: 10.1038/s41579-020-0433-9
14. Duparc T, Plovier H, Marrachelli VG, Van Hul M, Essaghir A, Stahlman M, et al. Hepatocyte Myd88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut (2017) 66(4):620–32. doi: 10.1136/gutjnl-2015-310904
15. Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, et al. Card9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med (2016) 22(6):598–605. doi: 10.1038/nm.4102
16. Bosman ES, Albert AY, Lui H, Dutz JP, Vallance BA. Skin exposure to narrow band ultraviolet (Uvb) light modulates the human intestinal microbiome. Front Microbiol (2019) 10:2410. doi: 10.3389/fmicb.2019.02410
17. Jackson MA, Goodrich JK, Maxan ME, Freedberg DE, Abrams JA, Poole AC, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut (2016) 65(5):749–56. doi: 10.1136/gutjnl-2015-310861
18. Vieira-Silva S, Falony G, Belda E, Nielsen T, Aron-Wisnewsky J, Chakaroun R, et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature (2020) 581(7808):310–5. doi: 10.1038/s41586-020-2269-x
19. Khan S, Waliullah S, Godfrey V, Khan MAW, Ramachandran RA, Cantarel BL, et al. Dietary simple sugars alter microbial ecology in the gut and promote colitis in mice. Sci Trans Med (2020) 12(567):eaay6218. doi: 10.1126/scitranslmed.aay6218
20. Kawano Y, Edwards M, Huang Y, Bilate AM, Araujo LP, Tanoue T, et al. Microbiota imbalance induced by dietary sugar disrupts immune-mediated protection from metabolic syndrome. Cell (2022) 185(19):3501–19 e20. doi: 10.1016/j.cell.2022.08.005
21. Suez J, Cohen Y, Valdes-Mas R, Mor U, Dori-Bachash M, Federici S, et al. Personalized microbiome-driven effects of non-nutritive sweeteners on human glucose tolerance. Cell (2022) 185(18):3307–28 e19. doi: 10.1016/j.cell.2022.07.016
22. Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci United States America (2004) 101(44):15718–23. doi: 10.1073/pnas.0407076101
23. Mazidi M, Rezaie P, Kengne AP, Mobarhan MG, Ferns GA. Gut microbiome and metabolic syndrome. Diabetes Metab syndr (2016) 10(2 Suppl 1):S150–7. doi: 10.1016/j.dsx.2016.01.024
24. Wu H, Tremaroli V, Schmidt C, Lundqvist A, Olsson LM, Kramer M, et al. The gut microbiota in prediabetes and diabetes: A population-based cross-sectional study. Cell Metab (2020) 32(3):379–90 e3. doi: 10.1016/j.cmet.2020.06.011
25. Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci United States America (2007) 104(3):979–84. doi: 10.1073/pnas.0605374104
26. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444(7122):1027–31. doi: 10.1038/nature05414
27. Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe (2008) 3(4):213–23. doi: 10.1016/j.chom.2008.02.015
28. Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One (2011) 6(10):e25792. doi: 10.1371/journal.pone.0025792
29. Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J (2011) 5(1):82–91. doi: 10.1038/ismej.2010.92
30. Pellegrini S, Sordi V, Bolla AM, Saita D, Ferrarese R, Canducci F, et al. Duodenal mucosa of patients with type 1 diabetes shows distinctive inflammatory profile and microbiota. J Clin Endocrinol Metab (2017) 102(5):1468–77. doi: 10.1210/jc.2016-3222
31. de Groot P, Nikolic T, Pellegrini S, Sordi V, Imangaliyev S, Rampanelli E, et al. Faecal microbiota transplantation halts progression of human new-onset type 1 diabetes in a randomised controlled trial. Gut (2021) 70(1):92–105. doi: 10.1136/gutjnl-2020-322630
32. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature (2012) 490(7418):55–60. doi: 10.1038/nature11450
33. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine (2020) 51:102590. doi: 10.1016/j.ebiom.2019.11.051
34. Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One (2010) 5(2):e9085. doi: 10.1371/journal.pone.0009085
35. Wehmeier UF, Piepersberg W. Biotechnology and molecular biology of the alpha-glucosidase inhibitor acarbose. Appl Microbiol Biotechnol (2004) 63(6):613–25. doi: 10.1007/s00253-003-1477-2
36. Ladas SD, Frydas A, Papadopoulos A, Raptis SA. Effects of alpha-glucosidase inhibitors on mouth to caecum transit time in humans. Gut (1992) 33(9):1246–8. doi: 10.1136/gut.33.9.1246
37. Hirsh AJ, Yao SY, Young JD, Cheeseman CI. Inhibition of glucose absorption in the rat jejunum: A novel action of alpha-D-Glucosidase inhibitors. Gastroenterology (1997) 113(1):205–11. doi: 10.1016/S0016-5085(97)70096-9
38. Su B, Liu H, Li J, Sunli Y, Liu B, Liu D, et al. Acarbose treatment affects the serum levels of inflammatory cytokines and the gut content of bifidobacteria in Chinese patients with type 2 diabetes mellitus. J Diabetes (2015) 7(5):729–39. doi: 10.1111/1753-0407.12232
39. Zhang M, Feng R, Yang M, Qian C, Wang Z, Liu W, et al. Effects of metformin, acarbose, and sitagliptin monotherapy on gut microbiota in zucker diabetic fatty rats. BMJ Open Diabetes Res Care (2019) 7(1):e000717. doi: 10.1136/bmjdrc-2019-000717
40. Takewaki F, Nakajima H, Takewaki D, Hashimoto Y, Majima S, Okada H, et al. Habitual dietary intake affects the altered pattern of gut microbiome by acarbose in patients with type 2 diabetes. Nutrients (2021) 13(6):2107. doi: 10.3390/nu13062107
41. Le KA, Li Y, Xu X, Yang W, Liu T, Zhao X, et al. Alterations in fecal lactobacillus and bifidobacterium species in type 2 diabetic patients in southern China population. Front Physiol (2012) 3:496. doi: 10.3389/fphys.2012.00496
42. Zhang X, Fang Z, Zhang C, Xia H, Jie Z, Han X, et al. Effects of acarbose on the gut microbiota of prediabetic patients: A randomized, double-blind, controlled crossover trial. Diabetes Ther res Treat Educ Diabetes related Disord (2017) 8(2):293–307. doi: 10.1007/s13300-017-0226-y
43. De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell (2014) 156(1-2):84–96. doi: 10.1016/j.cell.2013.12.016
44. Pingitore A, Chambers ES, Hill T, Maldonado IR, Liu B, Bewick G, et al. The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro. Diabetes Obes Metab (2017) 19(2):257–65. doi: 10.1111/dom.12811
45. Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-Gut-Brain communication. Nat Rev Gastroenterol Hepatol (2019) 16(8):461–78. doi: 10.1038/s41575-019-0157-3
46. Panwar H, Calderwood D, Grant IR, Grover S, Green BD. Lactobacillus strains isolated from infant faeces possess potent inhibitory activity against intestinal alpha- and beta-glucosidases suggesting anti-diabetic potential. Eur J Nutr (2014) 53(7):1465–74. doi: 10.1007/s00394-013-0649-9
47. Naito E, Yoshida Y, Makino K, Kounoshi Y, Kunihiro S, Takahashi R, et al. Beneficial effect of oral administration of lactobacillus casei strain shirota on insulin resistance in diet-induced obesity mice. J Appl Microbiol (2011) 110(3):650–7. doi: 10.1111/j.1365-2672.2010.04922.x
48. Kang JH, Yun SI, Park HO. Effects of lactobacillus gasseri Bnr17 on body weight and adipose tissue mass in diet-induced overweight rats. J Microbiol (2010) 48(5):712–4. doi: 10.1007/s12275-010-0363-8
49. Gu Y, Wang X, Li J, Zhang Y, Zhong H, Liu R, et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun (2017) 8(1):1785. doi: 10.1038/s41467-017-01682-2
50. Do HJ, Lee YS, Ha MJ, Cho Y, Yi H, Hwang YJ, et al. Beneficial effects of voglibose administration on body weight and lipid metabolism Via gastrointestinal bile acid modification. Endocr J (2016) 63(8):691–702. doi: 10.1507/endocrj.EJ15-0747
51. Rodbard HW, Jellinger PS. Comment on: Inzucchi et al. management of hyperglycemia in type 2 diabetes: A patient-centered approach. position statement of the American diabetes association (Ada) and the European association for the study of diabetes (Easd). Diabetes Care (2012) 35:1364–79. doi: 10.2337/dc12-0768
52. Sun EW, Martin AM, Wattchow DA, de Fontgalland D, Rabbitt P, Hollington P, et al. Metformin triggers pyy secretion in human gut mucosa. J Clin Endocrinol Metab (2019) 104(7):2668–74. doi: 10.1210/jc.2018-02460
53. Pernicova I, Korbonits M. Metformin–mode of action and clinical implications for diabetes and cancer. Nat Rev Endocrinol (2014) 10(3):143–56. doi: 10.1038/nrendo.2013.256
54. Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic amp. Nature (2013) 494(7436):256–60. doi: 10.1038/nature11808
55. Gunton JE, Delhanty PJ, Takahashi S, Baxter RC. Metformin rapidly increases insulin receptor activation in human liver and signals preferentially through insulin-receptor substrate-2. J Clin Endocrinol Metab (2003) 88(3):1323–32. doi: 10.1210/jc.2002-021394
56. Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut (2014) 63(5):727–35. doi: 10.1136/gutjnl-2012-303839
57. Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol (2014) 80(19):5935–43. doi: 10.1128/AEM.01357-14
58. Ryan PM, Patterson E, Carafa I, Mandal R, Wishart DS, Dinan TG, et al. Metformin and dipeptidyl peptidase-4 inhibitor differentially modulate the intestinal microbiota and plasma metabolome of metabolically dysfunctional mice. Can J Diabetes (2020) 44(2):146–55 e2. doi: 10.1016/j.jcjd.2019.05.008
59. Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X, et al. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep (2015) 5:14405. doi: 10.1038/srep14405
60. de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, et al. Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care (2017) 40(1):54–62. doi: 10.2337/dc16-1324
61. Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med (2017) 23(7):850–8. doi: 10.1038/nm.4345
62. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature (2015) 528(7581):262–6. doi: 10.1038/nature15766
63. Sansome DJ, Xie C, Veedfald S, Horowitz M, Rayner CK, Wu T. Mechanism of glucose-lowering by metformin in type 2 diabetes: Role of bile acids. Diabetes Obes Metab (2020) 22(2):141–8. doi: 10.1111/dom.13869
64. Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X, et al. Gut microbiota and intestinal fxr mediate the clinical benefits of metformin. Nat Med (2018) 24(12):1919–29. doi: 10.1038/s41591-018-0222-4
65. Campbell JE, Drucker DJ. Pharmacology, physiology, and mechanisms of incretin hormone action. Cell Metab (2013) 17(6):819–37. doi: 10.1016/j.cmet.2013.04.008
66. le Roux CW, Bloom SR. Peptide yy, appetite and food intake. Proc Nutr Soc (2005) 64(2):213–6. doi: 10.1079/pns2005427
67. Ang SY, Evans BA, Poole DP, Bron R, DiCello JJ, Bathgate RAD, et al. Insl5 activates multiple signalling pathways and regulates glp-1 secretion in nci-H716 cells. J Mol Endocrinol (2018) 60(3):213–24. doi: 10.1530/JME-17-0152
68. Astrup A, Rossner S, Van Gaal L, Rissanen A, Niskanen L, Al Hakim M, et al. Effects of liraglutide in the treatment of obesity: A randomised, double-blind, placebo-controlled study. Lancet (2009) 374(9701):1606–16. doi: 10.1016/S0140-6736(09)61375-1
69. Ahren B, Hirsch IB, Pieber TR, Mathieu C, Gomez-Peralta F, Hansen TK, et al. Efficacy and safety of liraglutide added to capped insulin treatment in subjects with type 1 diabetes: The adjunct two randomized trial. Diabetes Care (2016) 39(10):1693–701. doi: 10.2337/dc16-0690
70. Wang L, Li P, Tang Z, Yan X, Feng B. Structural modulation of the gut microbiota and the relationship with body weight: Compared evaluation of liraglutide and saxagliptin treatment. Sci Rep (2016) 6:33251. doi: 10.1038/srep33251
71. Moreira GV, Azevedo FF, Ribeiro LM, Santos A, Guadagnini D, Gama P, et al. Liraglutide modulates gut microbiota and reduces nafld in obese mice. J Nutr Biochem (2018) 62:143–54. doi: 10.1016/j.jnutbio.2018.07.009
72. Zhang N, Tao J, Gao L, Bi Y, Li P, Wang H, et al. Liraglutide attenuates nonalcoholic fatty liver disease by modulating gut microbiota in rats administered a high-fat diet. BioMed Res Int (2020) 2020:2947549. doi: 10.1155/2020/2947549
73. Charpentier J, Briand F, Lelouvier B, Servant F, Azalbert V, Puel A, et al. Liraglutide targets the gut microbiota and the intestinal immune system to regulate insulin secretion. Acta diabetol (2021) 58(7):881–97. doi: 10.1007/s00592-020-01657-8
74. Zhang Q, Xiao X, Zheng J, Li M, Yu M, Ping F, et al. Featured article: Structure moderation of gut microbiota in liraglutide-treated diabetic Male rats. Exp Biol Med (Maywood) (2018) 243(1):34–44. doi: 10.1177/1535370217743765
75. Jendle J, Hyotylainen T, Oresic M, Nystrom T. Pharmacometabolomic profiles in type 2 diabetic subjects treated with liraglutide or glimepiride. Cardiovasc Diabetol (2021) 20(1):237. doi: 10.1186/s12933-021-01431-2
76. Drucker DJ, Nauck MA. The incretin system: Glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet (2006) 368(9548):1696–705. doi: 10.1016/S0140-6736(06)69705-5
77. Zheng X, Chen T, Jiang R, Zhao A, Wu Q, Kuang J, et al. Hyocholic acid species improve glucose homeostasis through a distinct Tgr5 and fxr signaling mechanism. Cell Metab (2021) 33(4):791–803 e7. doi: 10.1016/j.cmet.2020.11.017
78. Smits MM, Fluitman KS, Herrema H, Davids M, Kramer MHH, Groen AK, et al. Liraglutide and sitagliptin have no effect on intestinal microbiota composition: A 12-week randomized placebo-controlled trial in adults with type 2 diabetes. Diabetes Metab (2021) 47(5):101223. doi: 10.1016/j.diabet.2021.101223
79. Bae EJ. Dpp-4 inhibitors in diabetic complications: Role of dpp-4 beyond glucose control. Arch pharmacal Res (2016) 39(8):1114–28. doi: 10.1007/s12272-016-0813-x
80. Mentzel S, Dijkman HB, Van Son JP, Koene RA, Assmann KJ. Organ distribution of aminopeptidase a and dipeptidyl peptidase iv in normal mice. J Histochem Cytochem Off J Histochem Soc (1996) 44(5):445–61. doi: 10.1177/44.5.8627002
81. Doupis J, Veves A. Dpp4 inhibitors: A new approach in diabetes treatment. Adv Ther (2008) 25(7):627–43. doi: 10.1007/s12325-008-0076-1
82. Yan X, Feng B, Li P, Tang Z, Wang L. Microflora disturbance during progression of glucose intolerance and effect of sitagliptin: An animal study. J Diabetes Res (2016) 2016:2093171. doi: 10.1155/2016/2093171
83. Liao X, Song L, Zeng B, Liu B, Qiu Y, Qu H, et al. Alteration of gut microbiota induced by dpp-4i treatment improves glucose homeostasis. EBioMedicine (2019) 44:665–74. doi: 10.1016/j.ebiom.2019.03.057
84. Liao X, Liu B, Qu H, Zhang L, Lu Y, Xu Y, et al. A high level of circulating valine is a biomarker for type 2 diabetes and associated with the hypoglycemic effect of sitagliptin. Mediators Inflammation (2019) 2019:8247019. doi: 10.1155/2019/8247019
85. Hu C, Jia W. Therapeutic medications against diabetes: What we have and what we expect. Adv Drug del Rev (2019) 139:3–15. doi: 10.1016/j.addr.2018.11.008
86. Nie Q, Chen H, Hu J, Fan S, Nie S. Dietary compounds and traditional Chinese medicine ameliorate type 2 diabetes by modulating gut microbiota. Crit Rev Food Sci Nutr (2019) 59(6):848–63. doi: 10.1080/10408398.2018.1536646
87. Lin TL, Lu CC, Lai WF, Wu TS, Lu JJ, Chen YM, et al. Role of gut microbiota in identification of novel tcm-derived active metabolites. Protein Cell (2021) 12(5):394–410. doi: 10.1007/s13238-020-00784-w
88. Chen W, Miao YQ, Fan DJ, Yang SS, Lin X, Meng LK, et al. Bioavailability study of berberine and the enhancing effects of tpgs on intestinal absorption in rats. AAPS PharmSciTech (2011) 12(2):705–11. doi: 10.1208/s12249-011-9632-z
89. Zhang Y, Gu Y, Ren H, Wang S, Zhong H, Zhao X, et al. Gut microbiome-related effects of berberine and probiotics on type 2 diabetes (the premote study). Nat Commun (2020) 11(1):5015. doi: 10.1038/s41467-020-18414-8
90. Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One (2012) 7(8):e42529. doi: 10.1371/journal.pone.0042529
91. Zhao JD, Li Y, Sun M, Yu CJ, Li JY, Wang SH, et al. Effect of berberine on hyperglycaemia and gut microbiota composition in type 2 diabetic goto-kakizaki rats. World J Gastroenterol (2021) 27(8):708–24. doi: 10.3748/wjg.v27.i8.708
92. Wang LL, Guo HH, Huang S, Feng CL, Han YX, Jiang JD. Comprehensive evaluation of scfa production in the intestinal bacteria regulated by berberine using gas-chromatography combined with polymerase chain reaction. J Chromatogr B Anal Technol Biomed Life Sci (2017) 1057:70–80. doi: 10.1016/j.jchromb.2017.05.004
93. Tian J, Bai B, Gao Z, Yang Y, Wu H, Wang X, et al. Alleviation effects of gqd, a traditional Chinese medicine formula, on diabetes rats linked to modulation of the gut microbiome. Front Cell infect Microbiol (2021) 11:740236. doi: 10.3389/fcimb.2021.740236
94. Xu J, Lian F, Zhao L, Zhao Y, Chen X, Zhang X, et al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J (2015) 9(3):552–62. doi: 10.1038/ismej.2014.177
95. Chang CJ, Lin CS, Lu CC, Martel J, Ko YF, Ojcius DM, et al. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat Commun (2015) 6:7489. doi: 10.1038/ncomms8489
96. Tilg H, Moschen AR. Microbiota and diabetes: An evolving relationship. Gut (2014) 63(9):1513–21. doi: 10.1136/gutjnl-2014-306928
97. Han JL, Lin HL. Intestinal microbiota and type 2 diabetes: From mechanism insights to therapeutic perspective. World J Gastroenterol (2014) 20(47):17737–45. doi: 10.3748/wjg.v20.i47.17737
98. Udayappan SD, Hartstra AV, Dallinga-Thie GM, Nieuwdorp M. Intestinal microbiota and faecal transplantation as treatment modality for insulin resistance and type 2 diabetes mellitus. Clin Exp Immunol (2014) 177(1):24–9. doi: 10.1111/cei.12293
99. Ran Y, Fukui H, Xu X, Wang X, Ebisutani N, Tanaka Y, et al. Alteration of colonic mucosal permeability during antibiotic-induced dysbiosis. Int J Mol Sci (2020) 21(17):6108. doi: 10.3390/ijms21176108
100. Fajstova A, Galanova N, Coufal S, Malkova J, Kostovcik M, Cermakova M, et al. Diet rich in simple sugars promotes pro-inflammatory response Via gut microbiota alteration and Tlr4 signaling. Cells (2020) 9(12):2701. doi: 10.3390/cells9122701
101. Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes (2012) 3(4):279–88. doi: 10.4161/gmic.19625
102. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes (2007) 56(7):1761–72. doi: 10.2337/db06-1491
103. Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T, et al. Bacteroides vulgatus and bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation (2018) 138(22):2486–98. doi: 10.1161/CIRCULATIONAHA.118.033714
104. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci United States America (2013) 110(22):9066–71. doi: 10.1073/pnas.1219451110
105. Inan MS, Rasoulpour RJ, Yin L, Hubbard AK, Rosenberg DW, Giardina C. The luminal short-chain fatty acid butyrate modulates nf-kappab activity in a human colonic epithelial cell line. Gastroenterology (2000) 118(4):724–34. doi: 10.1016/s0016-5085(00)70142-9
106. Kinoshita M, Suzuki Y, Saito Y. Butyrate reduces colonic paracellular permeability by enhancing ppargamma activation. Biochem Biophys Res Commun (2002) 293(2):827–31. doi: 10.1016/S0006-291X(02)00294-2
107. Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev (2010) 34(4):426–44. doi: 10.1111/j.1574-6976.2009.00204.x
108. Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R, et al. Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab (2018) 28(5):737–49 e4. doi: 10.1016/j.cmet.2018.07.001
109. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity Via interleukin-22. Immunity (2013) 39(2):372–85. doi: 10.1016/j.immuni.2013.08.003
110. Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, et al. Plant-derived exosomal micrornas shape the gut microbiota. Cell Host Microbe (2018) 24(5):637–52 e8. doi: 10.1016/j.chom.2018.10.001
111. van de Wouw M, Schellekens H, Dinan TG, Cryan JF. Microbiota-Gut-Brain axis: Modulator of host metabolism and appetite. J Nutr (2017) 147(5):727–45. doi: 10.3945/jn.116.240481
112. Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion Via the G-Protein-Coupled receptor Ffar2. Diabetes (2012) 61(2):364–71. doi: 10.2337/db11-1019
113. Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci United States America (2008) 105(43):16767–72. doi: 10.1073/pnas.0808567105
114. Hwang I, Park YJ, Kim YR, Kim YN, Ka S, Lee HY, et al. Alteration of gut microbiota by vancomycin and bacitracin improves insulin resistance Via glucagon-like peptide 1 in diet-induced obesity. FASEB J (2015) 29(6):2397–411. doi: 10.1096/fj.14-265983
115. Chimerel C, Emery E, Summers DK, Keyser U, Gribble FM, Reimann F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine l cells. Cell Rep (2014) 9(4):1202–8. doi: 10.1016/j.celrep.2014.10.032
116. Zanchi D, Depoorter A, Egloff L, Haller S, Mahlmann L, Lang UE, et al. The impact of gut hormones on the neural circuit of appetite and satiety: A systematic review. Neurosci Biobehav Rev (2017) 80:457–75. doi: 10.1016/j.neubiorev.2017.06.013
117. Perino A, Velazquez-Villegas LA, Bresciani N, Sun Y, Huang Q, Fenelon VS, et al. Central anorexigenic actions of bile acids are mediated by Tgr5. Nat Metab (2021) 3(5):595–603. doi: 10.1038/s42255-021-00398-4
118. De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Backhed F, Mithieux G. Microbiota-produced succinate improves glucose homeostasis Via intestinal gluconeogenesis. Cell Metab (2016) 24(1):151–7. doi: 10.1016/j.cmet.2016.06.013
119. Kwon YH, Wang H, Denou E, Ghia JE, Rossi L, Fontes ME, et al. Modulation of gut microbiota composition by serotonin signaling influences intestinal immune response and susceptibility to colitis. Cell Mol Gastroenterol Hepatol (2019) 7(4):709–28. doi: 10.1016/j.jcmgh.2019.01.004
120. Fung TC, Vuong HE, Luna CDG, Pronovost GN, Aleksandrova AA, Riley NG, et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol (2019) 4(12):2064–73. doi: 10.1038/s41564-019-0540-4
121. Lund ML, Sorrentino G, Egerod KL, Kroone C, Mortensen B, Knop FK, et al. L-cell differentiation is induced by bile acids through Gpbar1 and paracrine glp-1 and serotonin signaling. Diabetes (2020) 69(4):614–23. doi: 10.2337/db19-0764
122. Anderberg RH, Richard JE, Eerola K, Lopez-Ferreras L, Banke E, Hansson C, et al. Glucagon-like peptide 1 and its analogs act in the dorsal raphe and modulate central serotonin to reduce appetite and body weight. Diabetes (2017) 66(4):1062–73. doi: 10.2337/db16-0755
123. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab (2016) 24(1):41–50. doi: 10.1016/j.cmet.2016.05.005
124. Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest (2006) 116(4):1102–9. doi: 10.1172/JCI25604
125. Wang K, Liao M, Zhou N, Bao L, Ma K, Zheng Z, et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions Via production of succinate and secondary bile acids. Cell Rep (2019) 26(1):222–35 e5. doi: 10.1016/j.celrep.2018.12.028
126. Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res (2009) 50(8):1509–20. doi: 10.1194/jlr.R900007-JLR200
127. Chavez-Talavera O, Tailleux A, Lefebvre P, Staels B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology (2017) 152(7):1679–94 e3. doi: 10.1053/j.gastro.2017.01.055
128. Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, et al. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep (2018) 23(4):1099–111. doi: 10.1016/j.celrep.2018.03.109
129. Lin HV, Frassetto A, Kowalik EJ Jr., Nawrocki AR, Lu MM, Kosinski JR, et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones Via free fatty acid receptor 3-independent mechanisms. PLoS One (2012) 7(4):e35240. doi: 10.1371/journal.pone.0035240
130. den Besten G, Lange K, Havinga R, van Dijk TH, Gerding A, van Eunen K, et al. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am J Physiol Gastrointest liver Physiol (2013) 305(12):G900–10. doi: 10.1152/ajpgi.00265.2013
131. Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. The gut microbiota suppresses insulin-mediated fat accumulation Via the short-chain fatty acid receptor Gpr43. Nat Commun (2013) 4:1829. doi: 10.1038/ncomms2852
132. Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes (2009) 58(7):1509–17. doi: 10.2337/db08-1637
133. Panwar H, Rashmi HM, Batish VK, Grover S. Probiotics as potential biotherapeutics in the management of type 2 diabetes - prospects and perspectives. Diabetes/metab Res Rev (2013) 29(2):103–12. doi: 10.1002/dmrr.2376
134. Bock PM, Telo GH, Ramalho R, Sbaraini M, Leivas G, Martins AF, et al. The effect of probiotics, prebiotics or synbiotics on metabolic outcomes in individuals with diabetes: A systematic review and meta-analysis. Diabetologia (2021) 64(1):26–41. doi: 10.1007/s00125-020-05295-1
135. Bagarolli RA, Tobar N, Oliveira AG, Araujo TG, Carvalho BM, Rocha GZ, et al. Probiotics modulate gut microbiota and improve insulin sensitivity in dio mice. J Nutr Biochem (2017) 50:16–25. doi: 10.1016/j.jnutbio.2017.08.006
136. Lee YS, Lee D, Park GS, Ko SH, Park J, Lee YK, et al. Lactobacillus plantarum Hac01 ameliorates type 2 diabetes in high-fat diet and streptozotocin-induced diabetic mice in association with modulating the gut microbiota. Food Funct (2021) 12(14):6363–73. doi: 10.1039/d1fo00698c
137. Moroti C, Souza Magri LF, de Rezende Costa M, Cavallini DC, Sivieri K. Effect of the consumption of a new symbiotic shake on glycemia and cholesterol levels in elderly people with type 2 diabetes mellitus. Lipids Health Dis (2012) 11:29. doi: 10.1186/1476-511X-11-29
138. Ejtahed HS, Mohtadi-Nia J, Homayouni-Rad A, Niafar M, Asghari-Jafarabadi M, Mofid V. Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition (2012) 28(5):539–43. doi: 10.1016/j.nut.2011.08.013
139. Toejing P, Khampithum N, Sirilun S, Chaiyasut C, Lailerd N. Influence of lactobacillus paracasei Hii01 supplementation on glycemia and inflammatory biomarkers in type 2 diabetes: A randomized clinical trial. Foods (2021) 10(7):1455. doi: 10.3390/foods10071455
140. Anhe FF, Roy D, Pilon G, Dudonne S, Matamoros S, Varin TV, et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased akkermansia spp. Popul Gut Microb Mice Gut (2015) 64(6):872–83. doi: 10.1136/gutjnl-2014-307142
141. Catry E, Bindels LB, Tailleux A, Lestavel S, Neyrinck AM, Goossens JF, et al. Targeting the gut microbiota with inulin-type fructans: Preclinical demonstration of a novel approach in the management of endothelial dysfunction. Gut (2018) 67(2):271–83. doi: 10.1136/gutjnl-2016-313316
142. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med (2017) 23(1):107–13. doi: 10.1038/nm.4236
143. Wu TR, Lin CS, Chang CJ, Lin TL, Martel J, Ko YF, et al. Gut commensal parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from hirsutella sinensis. Gut (2019) 68(2):248–62. doi: 10.1136/gutjnl-2017-315458
144. Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, Masoumi SJ, et al. Prebiotics: Definition, types, sources, mechanisms, and clinical applications. Foods (2019) 8(3):92. doi: 10.3390/foods8030092
145. Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes (2011) 60(11):2775–86. doi: 10.2337/db11-0227
146. Cani PD, Knauf C, Iglesias MA, Drucker DJ, Delzenne NM, Burcelin R. Improvement of glucose tolerance and hepatic insulin sensitivity by oligofructose requires a functional glucagon-like peptide 1 receptor. Diabetes (2006) 55(5):1484–90. doi: 10.2337/db05-1360
147. Ho J, Nicolucci AC, Virtanen H, Schick A, Meddings J, Reimer RA, et al. Effect of prebiotic on microbiota, intestinal permeability, and glycemic control in children with type 1 diabetes. J Clin Endocrinol Metab (2019) 104(10):4427–40. doi: 10.1210/jc.2019-00481
148. Hove KD, Brons C, Faerch K, Lund SS, Rossing P, Vaag A. Effects of 12 weeks of treatment with fermented milk on blood pressure, glucose metabolism and markers of cardiovascular risk in patients with type 2 diabetes: A randomised double-blind placebo-controlled study. Eur J Endocrinol (2015) 172(1):11–20. doi: 10.1530/EJE-14-0554
149. Pedersen C, Gallagher E, Horton F, Ellis RJ, Ijaz UZ, Wu H, et al. Host-microbiome interactions in human type 2 diabetes following prebiotic fibre (Galacto-oligosaccharide) intake. Br J Nutr (2016) 116(11):1869–77. doi: 10.1017/S0007114516004086
150. Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, et al. Fecal microbiota transplant for treatment of clostridium difficile infection in immunocompromised patients. Am J Gastroenterol (2014) 109(7):1065–71. doi: 10.1038/ajg.2014.133
151. Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology (2012) 143(4):913–6 e7. doi: 10.1053/j.gastro.2012.06.031
152. Kootte RS, Levin E, Salojarvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab (2017) 26(4):611–9 e6. doi: 10.1016/j.cmet.2017.09.008
Keywords: gut microbiota, metabolites, type 2 diabetes, antidiabetic therapy, glucoregulatory agents
Citation: Liu B, Zhang L, Yang H, Zheng H and Liao X (2023) Microbiota: A potential orchestrator of antidiabetic therapy. Front. Endocrinol. 14:973624. doi: 10.3389/fendo.2023.973624
Received: 20 June 2022; Accepted: 17 January 2023;
Published: 27 January 2023.
Edited by:
Anouar Abidi, Facuty of Medicine of Tunis, TunisiaReviewed by:
Saber Jedidi, Chief of laboratory of silvo-pastoral resources, TunisiaCopyright © 2023 Liu, Zhang, Yang, Zheng and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyu Liao, bGlhb3hpYW95dTIwMTBAMTYzLmNvbQ==; Hongting Zheng, Zm5mNzcwM0Bob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.