
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 13 December 2023
Sec. Experimental Endocrinology
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.1292730
This article is part of the Research Topic Recent Insights into the Role of Hormones during Development and Their Functional Regulation View all 6 articles
Background: Interspecies hybridization is an important breeding method to generate fishes with heterosis in aquaculture. Using this method, hybrid Nile tilapia (Oreochromis niloticus, ♀) × blue tilapia (Oreochromis aureus, ♂) has been produced and widely farmed due to its growth and appetite superiorities. However, the genetic mechanism of these advanced traits is still not well understood. Ghrelin is a crucial gene that regulates growth and appetite in fishes. In the present study, we focused on the expression characteristics and its regulation of ghrelin in the hybrid.
Results: The tissue distribution analysis showed that ghrelin was predominantly expressed in the stomach in the hybrid. Ghrelin was more highly expressed in the stomach in the hybrid and Nile tilapia, compared to blue tilapia, showing a nonadditive pattern. Two single-nucleotide polymorphism (SNP) sites were identified including T/C and C/G from the second exon in the ghrelin gene from Nile tilapia and blue tilapia. By pyrosequencing based on the SNP sites, the allele-specific expression (ASE) of ghrelin in the hybrid was assayed. The result indicated that ghrelin in the hybrid showed higher maternal allelic transcript ratios. Fasting significantly increased ghrelin overall expression at 4, 8, 12, 24, and 48 h. In addition, higher maternal allelic transcript ratios were not changed in the fasting hybrids at 48 h. The cis and trans effects were determined by evaluating the overall expression and ASE values in the hybrid. The expression of ghrelin was mediated by compensating cis and trans effects in hybrid.
Conclusion: In summary, the present lines of evidence showed the nonadditive expression of ghrelin in the hybrid tilapia and its regulation by subgenomes, offering new insight into gene expression characteristics in hybrids.
Fish farming provides abundant proteins for people around the world. In recent years, genetic breeding has been accelerating the yields and quality of the farmed fish. Among the breeding technologies, selective breeding and hybridization have been widely used to improve the fish germplasm (1). Compared to selective breeding, hybridization combined the genetic resources from different lineages from the same species (intraspecific hybridization) or even different species (distant hybridization) (2). Hybridization may generate new fish germplasm or varieties in one generation; thus, the breeding time has been shortened (3, 4). Using this approach, intraspecific hybrids of Atlantic salmon (Salmo salar) (5), Chinook salmon (Oncorhynchus tshawytscha) (6), and interspecific hybrids of common carp (Cyprinus carpio) × red crucian carp (Carassius auratus, red var.) (7), Japanese crucian carp (Carassius cuvieri) × red crucian carp (8), and channel catfish (Ictalurus punctatus) × blue catfish (I. furcatus) (9) have been produced as new germplasm or varieties. These hybrids have the potential to be farmed as advanced parents for producing fry with productive advantages or used directly as farming fishes with heterosis (10). Thereinto, the hybrid combination is crucial for the heterosis (11). To date, the researchers could only obtain hybrids with advantages by attempts using different hybrid combinations. Therefore, understanding the genetic mechanism of heterosis is the way to improve the efficiency for hybrid breeding.
Heterosis is the phenomenon in which progeny has improved phenotypes such as higher growth rates, stronger ability to fight diseases, and higher fertility compared to its parents. Since hybrids contain subgenomes from different parents, there is high genetic variation of subgenome and transcriptomic changes (12, 13). In Arabidopsis allotetraploids, approximately 5.2%–5.6% genes had expression divergence from the midparent value (MPV) suggesting nonadditive expression (14). Further studies showed that several genes that participated in multiple biological processes including development, organism growth, and hormone regulation were nonadditively expressed (15, 16). In addition, these genes contributed to the heterosis in hybrid plants (17, 18). In hybrid fishes, similar results have been reported, such as Nile tilapia (Oreochromis niloticus) × blue tilapia (O. aureus) hybrid (15) and blunt snout bream (Megalobrama amblycephala) × topmouth culter (Culter alburnus) hybrid (19). Generally, it supposes that gene expression in hybrid is equal to MPV showing an additive effect while actually several genes related to growth and anti-disease are nonadditive expression genes. These nonadditive expression genes would affect phenotypes of hybrids leading to heterosis in transcriptome perspective (16). The transcription of gene is regulated by transcription initiation activity, which is controlled by cis and trans effects (20). In hybrids, the heterozygous genomes provide convenience for parsing the cis and trans effects. Cis-regulatory elements are located in the upstream region of the gene while trans-regulatory elements such as transcription factors control transcriptional activities in an intermolecular interaction way (21). Based on these features, the cis and trans effects could be evaluated by calculating the differential allelic expression value in hybrids and the differential expression between the parents (22). Interpreting the cis and trans effects of functional genes would provide clues for understanding the genetic mechanism of heterosis from the perspective of nonadditive expression (23).
Growth performance is the key trait for breeding researchers. The growth hormone (GH)/insulin like factor (IGF) axis is the well-known regulatory pathway for controlling growth in fish (24). Besides this axis, other regulators have also been reported recently. Among them, ghrelin secreted in enteroendocrine cells of the gastrointestinal tract has function in growth and appetite regulation (25). Ghrelin also regulates the GH/IGF axis via endocrine regulation (26). A previous study showed that GH had higher expression in Nile tilapia × blue tilapia hybrid compared to its parents (27). Additionally, the GH showed asymmetric allelic expression in the hybrid tilapia (27). The result showed new clues for the regulation of functional gene expression in hybrid tilapia from transcribing initiation activity level. To date, the connection between the heterozygous genome and expression patterns in hybrid tilapia is far away from systematically understand.
The Nile tilapia (♀) × blue tilapia (♂) hybrid is one of the most popular farmed tilapia varieties in China, due to its fast growth performance and strong resistance to low temperature (28). This tilapia variety also showed higher appetite (29). We suppose that in the hybrid tilapia, the fish may have a special expression pattern of ghrelin, which is attributed to the regulation by heterozygous genome. Herein, the ghrelin expression in the hybrid tilapia was assayed compared to its parents. The allele-specific expression (ASE) of ghrelin in the hybrid tilapia was analyzed. Furthermore, the cis and trans effects on transcription of ghrelin in the hybrid were determined based on the expression results. The present study demonstrated the ghrelin expression pattern and its transcriptional regulation, which would aid in understanding the genetic mechanism of advantages in the growth performance of hybrid tilapia.
The experimental fishes, including Nile tilapia (N), Blue tilapia (B), and their hybrid (NB) were obtained from National Tilapia Breeding Fields of Nanning from Guangxi Academy of Fishery Sciences (Nanning, China). The NB individuals were obtained by interspecific crossing between 20 female N and 10 male B. The experimental fish of N and B were generated by self-mating. All the tested fish were farmed in three adjacent ponds (N, B, and NB were reared in the separate ponds) in Guangxi Academy of Fishery Sciences (Nanning, China). The water and feeding conditions were similar among the ponds. For each kind of tilapia, 200 individuals were reared for 120 days post hatching to adulthood. Before tissue collection, the tested fish were anesthetized by MS-222. The muscle, pituitary, heart, spleen, intestine, brain, ovary, testis, kidney, liver, and stomach were dissected from the fish, frozen in liquid nitrogen, and restored at −80°C. In tilapia, males grow faster than females. Thus, most of the farmed individuals are males that are generated by breeding technology (30, 31). In the present study, considering the differences of growth rate in tilapia between males and females, we only used males for the comparison study for N, B, and NB. The present experiments have been approved by Ethics Committees of Hunan Normal University (Changsha, China).
The total RNA from stomach in N and B (3 samples from each species) was isolated using RNAiso Plus (TRIzol) reagent (Takara, Japan) according to the manufacturer’s instruction. The quality and concentration of the extracted RNA were detected by 1% agarose gel and SmartSpec™ Plus Spectrophotometer (Bio-Rad, USA). The cDNA was synthesized by the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan) using 1 μg of total RNA for each sample. The predicted cDNA sequences of ghrelin based on N and B genome data were obtained from NCBI. A pair of primers was designed for amplification of ghrelin open reading frame (ORF) (Supplementary Table 1). The PCR was performed as 94°C for 5 min for initial predenaturation, 30 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 45 s, and extension at 72°C for 10 min. The PCR products were analyzed by 1.2% agarose gels following extraction and sequenced by Shanghai Sangon Biotechnology Co. Ltd. (Shanghai, China).
The sequenced ORF was compared with ghrelin sequences of other species by NCBI blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The amino acids were predicted based on the ORF sequences and the alignment of ghrelin N and B was constructed using ClustalW (32). The phylogenetic tree was constructed by MRBAYES (Version 3.2.7) with the parameters as prset aamodelpr = VT, mcmcp ngen = 100000000, printfreq = 100, Samplefreq = 100 (33).
The cDNA from muscle, pituitary, heart, spleen, intestines, brain, ovary, testis, kidney, liver, and stomach in NB was synthesized as mentioned in Section 2.2. Reverse transcriptase PCR (RT-PCR) was conducted to detect ghrelin expression in these tissues. The primers of ghrelin and β-actin were designed for the PCR (Supplementary Table 1). The parameters of RT-PCR were as follows: 94°C for 5 min for initial predenaturation, 30 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 45 s, and extension at 72°C for 10 min. The PCR products were detected by 1.2% agarose gels and photographed using a Gel-Doc XR (Bio-Rad, USA).
To compare the ghrelin expression in NB and its parents in stomach, the total RNA of stomach from N, B, and NB (n = 5 for each kind of tilapia) was isolated and cDNA was synthesized as mentioned in Section 2.2. The quantitative PCR (qPCR) was conducted on a PikoReal 96 Real-Time PCR system (Thermo Fisher Scientific, USA). The reagents used for qPCR were TB Green™ Fast qPCR Mix (Takara, Japan) according to the manufacturer’s instruction. The qPCR cycles were as follows: 95°C for 7 min, 40 cycles of 95°C for 5 s, and 60°C for 30 s. After the reaction, the melting curve was used to determine the specificity of PCR amplification. To normalize the results, β-actin was introduced as internal reference gene. The ghrelin expression levels were calculated using the 2−ΔΔCT method (34). To identify the nonadditive expression, we compared the ghrelin expression level in NB and MPV (mean value of the expression level in N and B). If NB ≠ MPV with significant difference, the expression of ghrelin was determined as nonadditive expression.
By comparing the cDNA sequences of ghrelin, two single-nucleotide polymorphism (SNP) sites between the different species were identified. Subsequently, a pair of primers was designed to obtain the sequences with the SNP sites (Supplementary Table 1). The PCR was performed and the amplified fragments were sequenced by Shanghai Sangon Biotechnology Co. Ltd. (Shanghai, China). The SNP sites from three individuals of N, B, and NB were compared using the raw data of AB1 file from sequencer (ABI 3730xl, ABI, USA).
Pyrosequencing was used to determine the frequency of allele expression, which was performed by Shanghai Sangon Biotechnology Co. Ltd. (Shanghai, China). The stomach tissues from five different NB individuals were used for the experiment. The primers for targeted sequences containing the identified SNP sites (T/C, C/G) were designed (Supplementary Table 1). The PCR conditions were as follows: 95°C for 5 min, 35 cycles of 94°C for 30 s, 60°C for 25–30 s, 72°C for 50 s, and extension of 72°C for 7 min. The PCR production was pyrosequenced by a PyroMark Q96 ID instrument (Qiagen, USA) and the primers are shown in Supplementary Table 1.
In order to determine whether different feeding conditions have an effect on ghrelin expression in NB, 120 NB individuals (1 month old) were randomly divided into two groups: normal feeding group (control) and fasting group (60 individuals for each group). Before this experiment, the two groups had an adaptation period, during which the hybrids were fed commercial feed with 2% of their body weight. Then, the hybrids in the feeding group were fed with 2% of their weight, while the hybrids in the fasting group were not fed. The stomach of fish from the two groups were collected at 0, 4, 8, 12, 24, and 48 h (n = 5). All the individuals were analyzed by qPCR at 0, 4, 8, 12, 24 and 48h to evaluate the mRNA expression of ghrelin (n = 5). The expression of alleles was assayed by pyrosequencing using the individuals at 0 h and 48 h.
The determination of cis or trans effects is based on a previous study (22). The A value is defined as log2(NN/BB) (NN = the overall ghrelin expression of Nile tilapia, BB = the overall ghrelin expression of blue tilapia). Thus, A represents the expression difference between two parents, and also represents the sum of cis and trans effects. The B value is defined as log2 (allelic expression of Nile tilapia in hybrid/allelic expression of blue tilapia in hybrid); hence, B represents the cis effect. Thus, A-B represents the trans effect. If (A > 0 and A-B > 0) or (A < 0 and A-B < 0), there were enhancing cis and trans interactions, and if (A > 0 and A-B < 0) or (A < 0 and A-B > 0), there were compensating cis and trans interactions.
Mean value ± standard deviation was used to present all expression levels. All the statistical analyses were performed by SPSS 16.0 (SPSS Inc., Chicago, USA). The ghrelin expression change in stomach among N, B, and NB was compared by one-way ANOVA. The ASE between maternal and paternal sub-genomes was compared using t-test, and the ASE between different feeding conditions and times were compared by two-way ANOVA. According to the analysis result, p < 0.05 meant there were significant difference between the data.
The ORF sequences of preproghrelin from Nile tilapia and blue tilapia were the same in length, which contained 324 nucleotides encoding 107 amino acids. The signal peptides of N and B were both MLLKRNTCLLAFLLCSLTLWCKSTSA. The mature ghrelin peptides from N and B were identical: GSSFLSPSOKPONKVKSSRI. Only one amino acid was different comparing the N and B sequences, which was D/E at the 60th site of the preproghrelin (Figure 1A). The phylogenetic tree of preproghrelin suggested that all the Cichlidae fishes were clustered together. The Cyprinidae had the farthest distance to the tilapia. In addition, the species from Oreochromis showed higher similarity (Figure 1B).
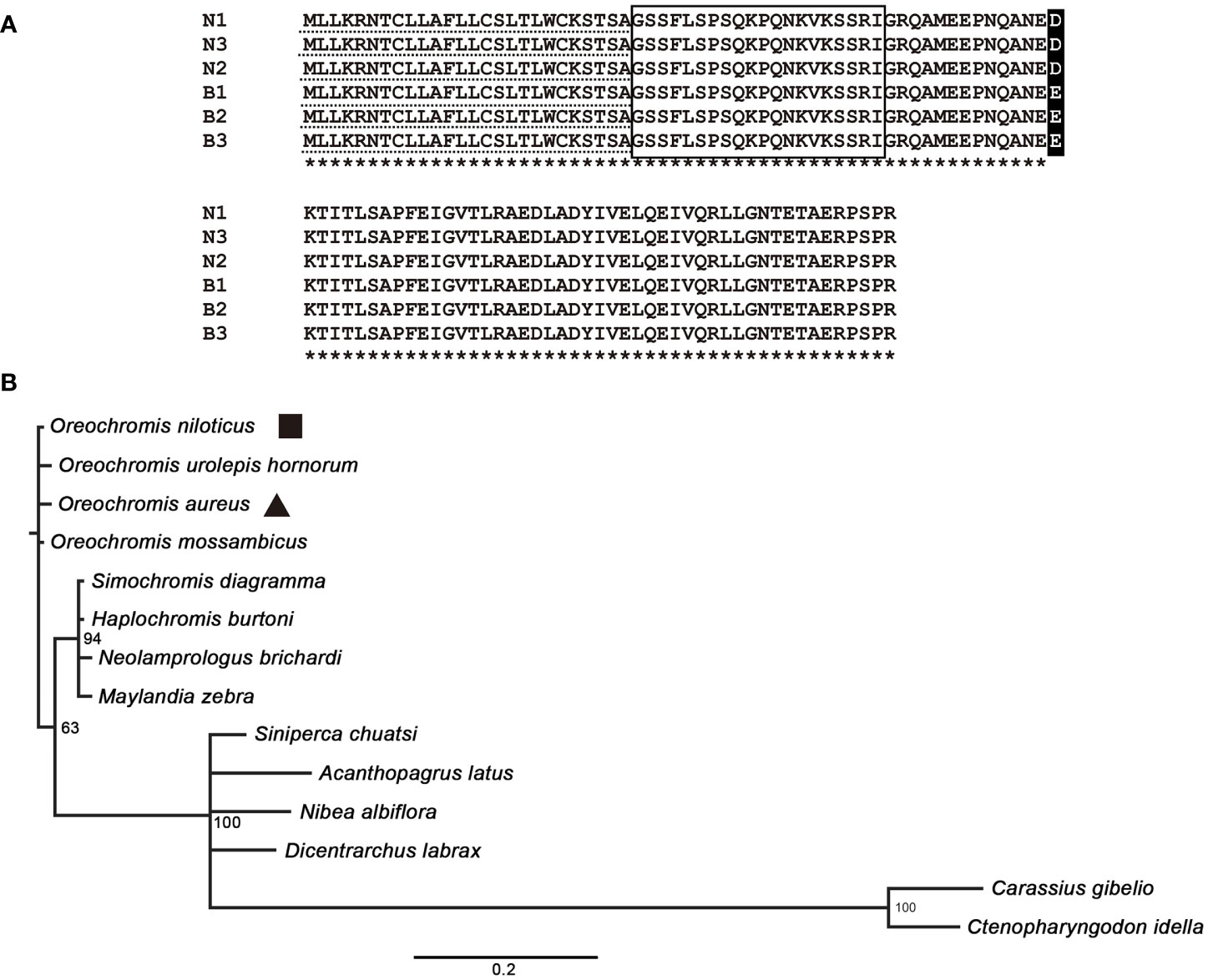
Figure 1 Analysis of preproghrelin protein in tilapia. (A) Amino acid sequence alignment of the Nile tilapia and blue tilapia. N1–N3 and B1–B3 represented the three biological replications of Nile tilapia and blue tilapia, respectively. The dotted line showed the signal peptides and the box showed the mature peptide of ghrelin. Asterisks represents the same amino acids. (B) Phylogenetic tree analysis using MrBayes 3.2. The sequences were obtained from the Genbank database with Accession Numbers of Oreochromis niloticus (XP_003441511.1), Oreochromis urolepis hornorum (ABN13418.1), Oreochromis aureus (XP_031613045.1), Oreochromis mossambicus (BAC55160.1), Simochromis diagramma (XP_039885313.1), Haplochromis burtoni (XP_005944008.1), Neolamprologus brichardi (XP_006807654.1), Maylandia zebra (XP_004544940.1), Siniperca chuatsi (XP_044033097.1), Acanthopagrus latus (XP_036957504.1), Nibea albiflora (KAG8007860.1), Dicentrarchus labrax (XP_051270674.1), Carassius gibelio (XP_052455871.1), and Ctenopharyngodon idella (XP_051752346.1).
The RT-PCR results showed that the ghrelin had the highest expression in stomach following testis and ovary while other tissues including muscle, pituitary, heart, spleen, intestine, brain, and liver had no significant signals in NB (Figure 2A). Furthermore, we compared the mRNA expression of ghrelin in NB and its parents N and B in stomach by qPCR. The results indicated that NB and N had higher expression levels compared to B (p < 0.05). Meanwhile, the expression levels of NB were significantly higher than MPV, indicating the nonadditive expression of ghrelin in NB (Figure 2B).
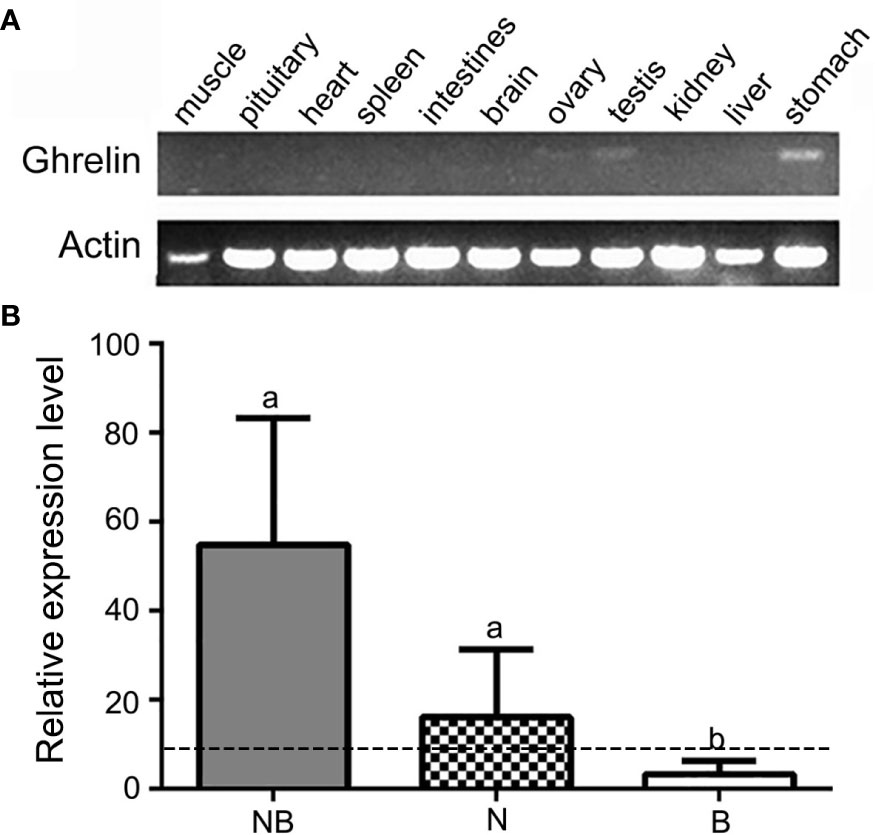
Figure 2 Overall expression of ghrelin in hybrid tilapia. (A) Tissue distribution of ghrelin in hybrid tilapia detected by RT-PCR. (B) qPCR analysis of ghrelin in the stomach from Nile tilapia (N), blue tilapia (B), and hybrid tilapia (NB). The different lowercase letters on each bar indicated the significant difference (p < 0.05) and the dashed line indicated the midparent value (MPV).
By comparing the cDNA sequences of ghrelin in N and B, two SNP sites including T/C and C/G in the second exon of the gene were identified (Figure 3A). To determine these two SNP sites, the sequences of genome DNA containing the sites were confirmed from N, B, and NB. By using the raw data of the sequencing, at the T/C site, only T peaks were observed in N and C peaks were found in B. While the NB showed the heterozygosity that contain T and C peaks (Figure 3B). At the C/G site, N contained only C peaks and B contained only G peaks, whereas NB had both C and G peaks (Figure 3C). These results showed that the T/C and C/G as SNP sites could be used as allelic locus for analyzing allelic-specific expression.
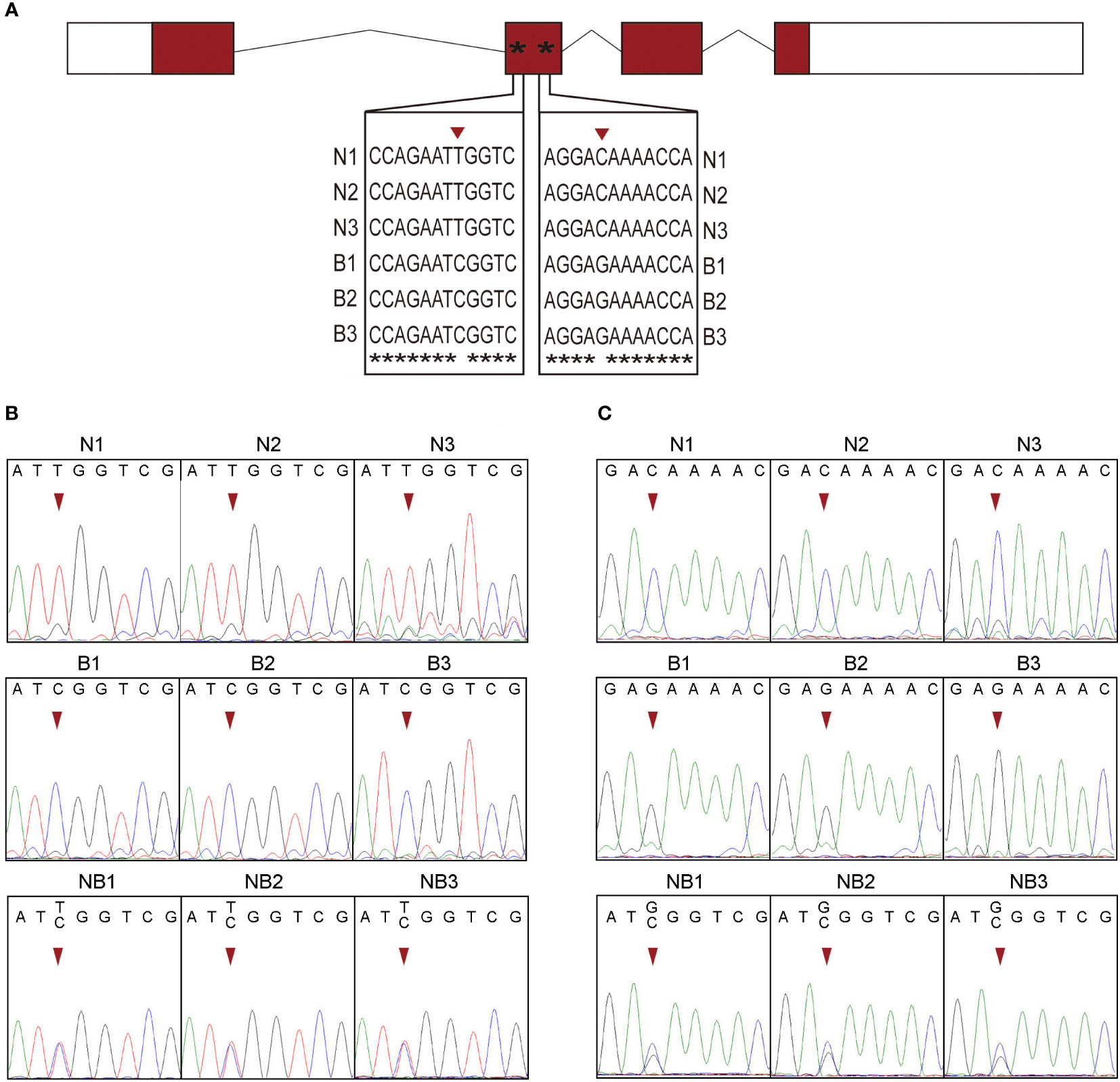
Figure 3 SNP identification form Nile tilapia (N) and blue tilapia (B). (A) The two SNP sites were identified from cDNA sequences and displayed using alignment result by ClustalW. Asterisks represents the same nucleotide. (B, C) Validation of the two SNP sites by DNA sequencing from N, B, and the hybrid. N1–N3, B1–B3, and NB1–NB3 represented the three biological replications of Nile tilapia, blue tilapia, and hybrid, respectively.
Pyrosequencing was performed to evaluate the ASE of ghrelin. Both the analysis of the two SNP sites suggested the significantly higher expression of maternal alleles than paternal alleles in NB (p < 0.05) (Figure 4). Furthermore, the feeding and fasting NB were used to determine whether the ASE depends on the feeding condition. For the mRNA expression, ghrelin had significantly lower expression in the feeding group compared to the fasting group at 4 h, 8 h, 12 h, 24 h, and 48 h (p < 0.05) (Figure 5A). The ASE expression of ghrelin in the fasting group at 0 h and 48 h was assayed. Both the samples at 0 h and 48 h in the fasting group showed the dominant maternal allele expression with a significant difference (p < 0.05) (Figures 5B, C).
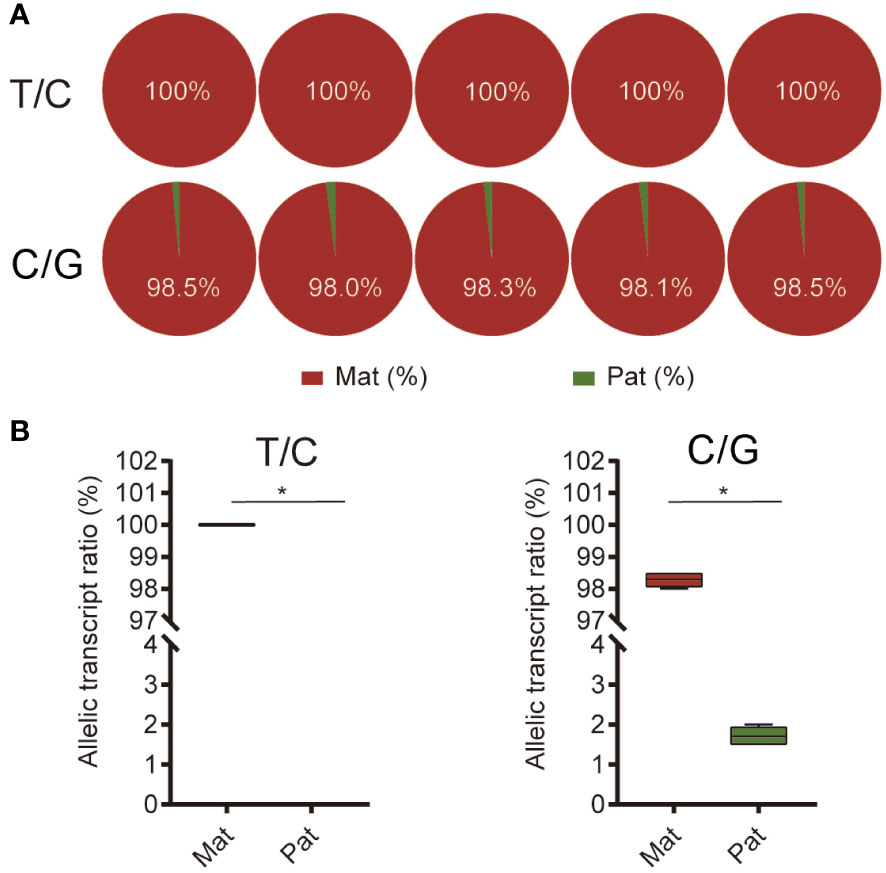
Figure 4 Allele-specific expression (ASE) of ghrelin in hybrid. (A) The pyrosequencing of ghrelin in stomach from hybrid based on the T/C and C/G SNP sites. The five pie plots for each site represented five individuals. (B) Allelic transcript ratios of T/C and C/G in hybrid. Mat, maternal; Pat, parental. Asterisks represent values significantly different.
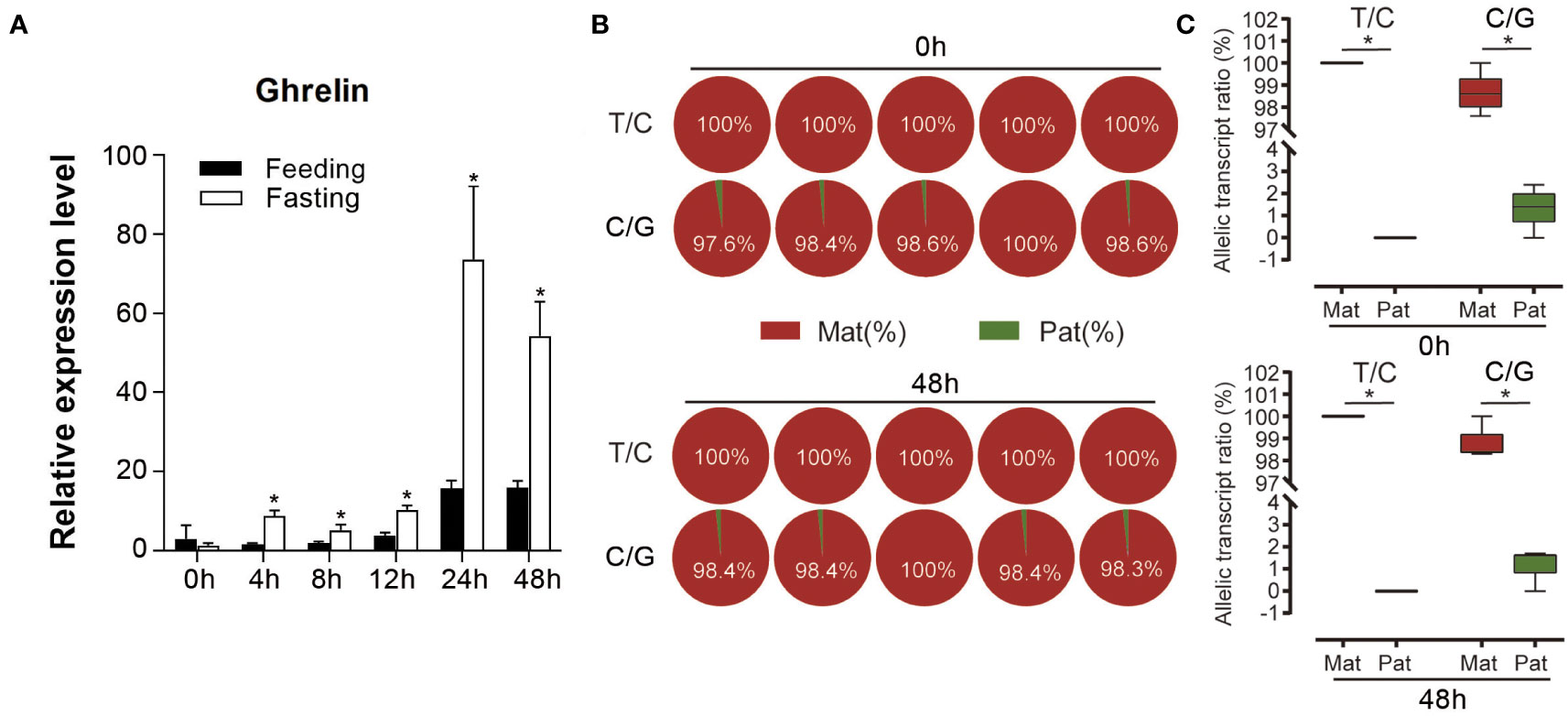
Figure 5 Expression of ghrelin in feeding and fasting hybrid. (A) Overall expression of ghrelin in feeding and fasting hybrid in different time points. Asterisks indicated the significant differences between feeding and fasting group (p < 0.05). (B) Allele-specific expression (ASE) of ghrelin in the fasting group at 0 h and 48 h by pyrosequencing in stomach from hybrid based on the T/C and C/G SNP sites. (C) Allelic transcript ratios of T/C and C/G in hybrid. Mat: maternal; Pat: parental.
Cis and trans effects could be determined by calculating the overall expression and ASE of hybrid and its parents. The gene expression divergence defined by the A value could be calculated as log2(NN/BB) and the ASE defined by the B value could be calculated as log2(N allele in NB/B allele in NB). The result showed that both the T/C and C/G alleles were compensating cis and trans effects (B > 0 and A-B < 0) in the hybrid, indicating the opposite direction of the regulation (Figure 6A).
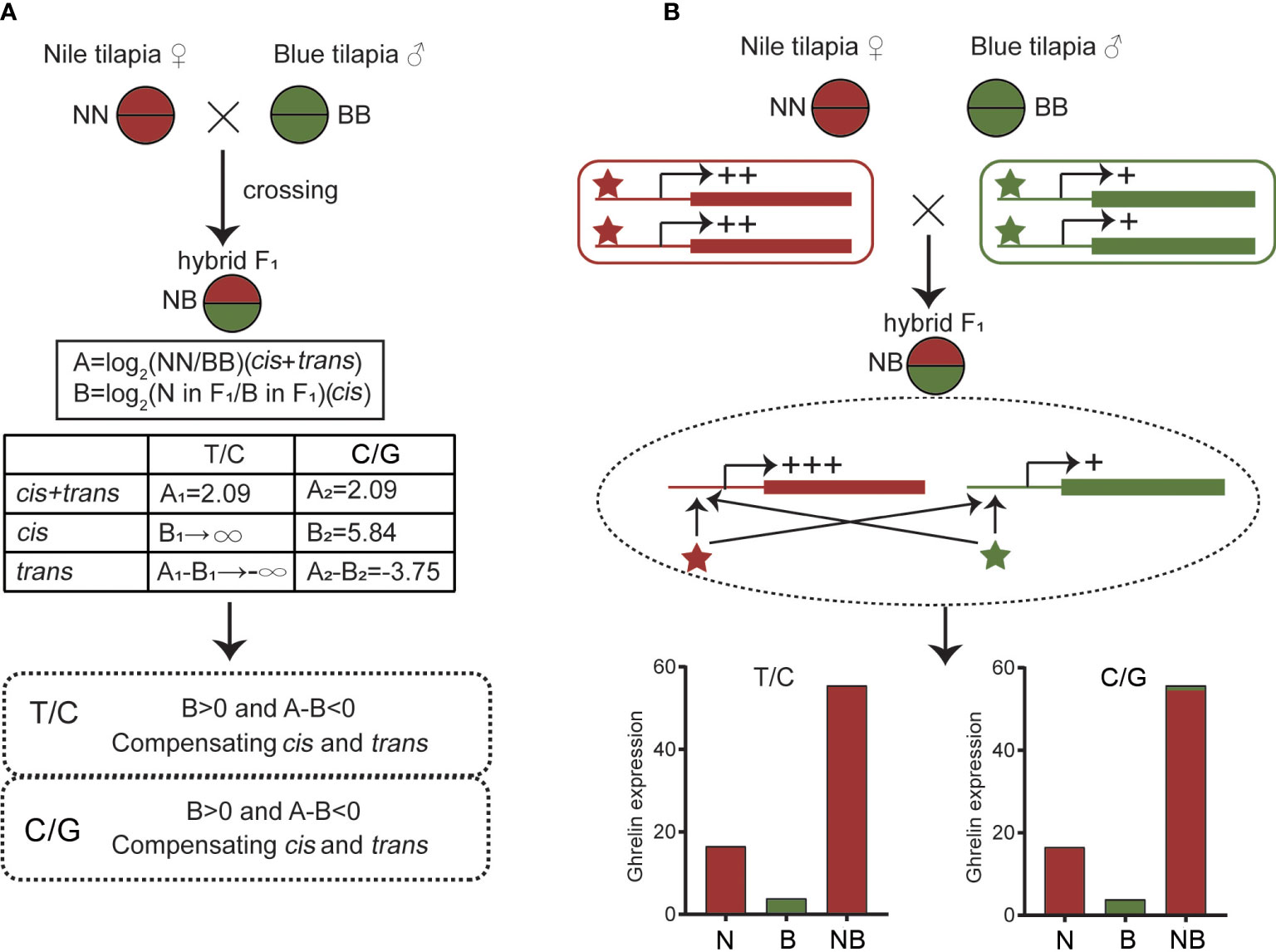
Figure 6 Cis and trans effects of ghrelin in hybrid. (A) Determination of cis and trans effects by calculating the overall expression and allele-specific expression (ASE) values. (B) Possible regulatory effects by cis- and trans-regulators of ghrelin in hybrid.
Ghrelin is the crucial hormone that controls growth and appetite performance (35). Understanding the expression regulation of ghrelin may provide valuable information for human health and animals. In fishes, hybridization has been proven as an efficient method for breeding. The hybrids with heterosis could be used for aquaculture, which contributes to the improvement of productivity (36). Hybrid tilapia (NB) is the progeny from intercrossing of N (♀) × B (♂), which has greater growth performance, stronger ability to fight diseases, and anti-low temperature (37, 38). The higher growth rate was reported in previous studies and the non-additive expression of GH had been reported before (27). In the present study, we analyzed the ghrelin expression and its regulation in NB hybrid. The non-additive expression and ASE suggested that both cis- and trans- regulation contribute to the appetite and growth advances in NB.
The sequences of preproghrelin shared high similarity in tilapia species including N and B. Only one deduced amino acid was different, suggesting the conserved structure and function. Preproghrelin would be processed to mature ghrelin to perform the function. Intriguingly, the mature ghrelin from N and B were identical. It is known that the ghrelin was secreted by stomach in animals (39). The present results also supported the idea that ghrelin is a gastrointestinal hormone. In addition, our results showed that ghrelin could be produced by ovary and testis. This result suggested the potential function of ghrelin in gonadal development. Ghrelin is a hormone that participates in multiple biological processes including growth controlling and reproductive regulation (40). Previous studies showed that ghrelin interacts with the hypothalamic–pituitary–gonadal axis (HPG axis) to regulate gamete maturation (41, 42). The present study focused on the growth and appetite advances; thus, we further assayed the expression of ghrelin in the stomach. In the present study, we found that fasting upregulated ghrelin expression in the stomach of NB. Fasting upregulated ghrelin expression in fishes has been reported widely, such as in goldfish (Carassius auratus), common carp, and grass carp (Ctenopharyngodon idellus) (43, 44). The result of the present study also supported the idea that fasting stimulates ghrelin secretion in fish gastrointestinal tract, which is similar to the results in other species. As a hormone that rises during fasting, it is reasonable for ghrelin to be upregulated by fasting in NB. Furthermore, nonadditive expression of ghrelin was found in NB, which is in accordance with the stronger ability to ingest and faster growth in NB. Theoretically, the expression of gene in hybrid is equal to the average of its parents. However, several genes have been found to not abide by this theory, namely, nonadditive expression (17). Nonadditive expression may lead to heterosis (45). This phenomenon could be attributed to the interaction of the two subgenome in hybrid. The subgenome expresses each allele and contributes to the total expression together. In this case, the nonadditive expression of ghrelin in NB may be related to allelic gene expression of ghrelin.
Two subgenomes were contained in NB as a hybrid. Theoretically, both of the two subgenomes could be expressed genes while the present result showed that ASE of ghrelin was found in the stomach from NB. In NB, ghrelin from the N subgenome was predominantly expressed while ghrelin from the B subgenome could hardly be detected. ASE is common in hybrid, which has been considered as a mechanism for heterosis (46). In addition, ASE provides valuable information for unveiling the mechanism of heterosis for hybrid breeding. The ASE of functional genes has been reported in several plants such as rice (46) and maize hybrids (47). In these studies, the genes that participated in multiple agronomic traits including organism growth, development, and reproduction were ASE, suggesting the underlying effects of ASE on heterosis (46, 47). In contrast, the studies of ASE in hybrid animals were less because it is difficult for interspecific crossing to generate hybrid in higher vertebrates. One of the most well-studied hybrid animals is Drosophila (48). Fish interspecific hybrids were also common and ASE in fish has been reported in hybrid Xiphophorus (49), cichlid fishes (50), and tilapia (20). In NB hybrid tilapia, GH and IGF-1 predominantly expressed by one subgenome from the parents (27, 37). This is similar to the IGF-2 in human during embryogenesis (51). In human, IGF-2 is the most famous imprint gene, which is related to embryo development, and loss of the expression pattern leads to overgrowth or even death (52). In addition, the feeding conditions did not change the expression trends for NB whether comparing to its parents or the two subgenomes in hybrid. All these clues suggested that the expression pattern of ghrelin in NB may lead to the growth advances, which is determined by the genetic rather than the feeding condition.
Since transcriptional activities are regulated by cis- and trans- regulators (53), the specific expression model of ghrelin in NB may due to the changes of the regulators. The present study showed that both cis and trans effect s were observed in NB. The different cis- and trans- regulators from parents together affected the expression of ghrelin in hybrid. In addition, the compensating cis and trans effects were found, suggesting the comprehensive regulations. In the hybrids, it has been reported that compensating cis and trans effects was more common than enhancing cis and trans effects, only cis effects, or only trans effects. This may due to the adaptability for heterozygous genome in hybrid. At the beginning of hybridization, two subgenomes from different species were instable. A high probability of recombination, indel, and structural variation exists in the genome from hybrid (54). The variations of genomes lead to changes in cis and trans factors that contribute to the regulatory changes (55). The ghrelin expression pattern in NB is similar, which is affected by the heterozygous genome. Then, trans- regulators promoted the transcription of ghrelin in the N subgenome while fewer effects were observed in the B subgenome resulting in ASE (Figure 6B). Meanwhile, the compressive effects promoted the overall expression of ghrelin in NB, indicating the non-additive expression.
In summary, the present findings suggested the higher expression of ghrelin in the hybrid compared to its parents. The maternal bias of ghrelin ASE was found in the stomach from NB tilapia, which was not dependent on feeding conditions. The elevated expression and ASE in NB were mediated by compensating cis and trans effects. The present lines of evidence suggested that the subgenomes in the hybrid contribute to changes of cis- and trans- regulators, leading to the specific expression pattern in NB.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
All the experiments have been approved by Ethics Committees of Hunan Normal University (Changsha, China). The study was conducted in accordance with the local legislation and institutional requirements.
HZ: Funding acquisition, Supervision, Writing – original draft. BR: Methodology, Software, Writing – review & editing. CL: Data curation, Software, Writing – review & editing. YZ: Formal Analysis, Funding acquisition, Writing – original draft, Writing – review & editing. YL: Resources, Writing – review & editing. JX: Investigation, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key R&D Program of China (2022YFD2400102), the National Natural Science Foundation of China (31760756), the Hunan Provincial Natural Science Foundation of China (2021JJ20033 and 2022JJ30386), the State Key Laboratory of Developmental Biology of Freshwater Fish (2021KF006), and the Hunan Normal University undergraduates innovative experiment project and entrepreneurship program (2022269).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1292730/full#supplementary-material
1. Hu FZ, Zhong HT, Wu C, Wang S, Guo ZJ, Tao M, et al. Development of fisheries in China. Reprod Breed (2021) 1:64–79. doi: 10.1016/j.repbre.2021.03.003
2. Wang S, Tang C, Tao M, Qin Q, Zhang C, Luo K, et al. Establishment and application of distant hybridization technology in fish. Sci China Life Sci (2019) 62:22–45. doi: 10.1007/s11427-018-9408-x
3. Luo KK, Wang S, Fu YQ, Zhou P, Huang XX, Gu QH, et al. Rapid genomic DNA variation in newly hybridized carp lineages derived from Cyprinus carpio (♀) × Megalobrama amblycephala (♂). BMC Genet (2019) 20:87. doi: 10.1186/s12863-019-0784-2
4. Zhang ZH, Chen J, Li L, Tao M, Zhang C, Qin QB, et al. Research advances in animal distant hybridization. Sci China Life Sci (2014) 57:889–902. doi: 10.1007/s11427-014-4707-1
5. Houde ALS, Fraser DJ, Hutchings JA. Reduced anti-predator responses in multi-generational hybrids of farmed and wild Atlantic salmon (Salmo salar L.). Conserv Genet (2010) 11:785–94. doi: 10.1007/s10592-009-9892-2
6. Semeniuk CAD, Capelle PM, Dender MGE, Devlin R, Dixon B, Drown J, et al. Domestic-wild hybridization to improve aquaculture performance in Chinook salmon. Aquaculture (2019) 511:734255. doi: 10.1016/j.aquaculture.2019.734255
7. Liu SJ, Liu Y, Zhou GJ, Zhang XJ, Luo C, Feng H, et al. The formation of tetraploid stocks of red crucian carp × common carp hybrids as an effect of interspecific hybridization. Aquaculture (2001) 192:171–86. doi: 10.1016/S0044-8486(00)00451-8
8. Liu SJ, Liu QF, Zhou Y, Wang J, Zhang XY. The formation and biological characteristics of the hybrids derived from the hybridization hybridization of Japanese white crucian carp × red crucian carp. In: Liu SJ, editor. Fish Distant Hybridization. Singapore: Springer Nature Singapore (2022). p. 271–88.
9. Argue BJ, Kuhlers DL, Liu Z, Dunham RA. Growth of channel catfish (Ictalurus punctatus), blue catfish (I. furcatus), and their F1, F2, F3, and F1 reciprocal backcross hybrids in earthen ponds. J Anim Sci (2014) 92:4297–305. doi: 10.2527/jas.2013-7549
10. Xiao W, Chen BL, Wang J, Zou ZY, Wang CH, Li DY, et al. Integration of mRNA and miRNA profiling reveals heterosis in Oreochromis niloticus × O. aureus hybrid tilapia. Animals (2022) 12:640. doi: 10.3390/ani12050640
11. Lippman ZB, Zamir D. Heterosis: revisiting the magic. Trends Genet (2007) 23:60–6. doi: 10.1016/j.tig.2006.12.006
12. Liu SJ, Wang S, Liu QF, Wu C, Zhou Y, Tao M, et al. The research advances in animal distant hybridization and polyploid organisms. In: Liu SJ, editor. Fish Distant Hybridization. Singapore: Springer Nature Singapore (2022). p. 1–37.
13. Liu QF, Luo KK, Zhang XY, Liu FL, Qin QB, Tao M, et al. A new type of triploid fish derived from the diploid hybrid crucian carp (♀) × autotetraploid fish (♂). Reprod Breed (2021) 1:122–7. doi: 10.1016/j.repbre.2021.07.003
14. Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, et al. Genomewide nonadditive gene regulation in Arabidopsis Allotetraploids. Genetics (2006) 172:507–17. doi: 10.1534/genetics.105.047894
15. Zhou Y, Zhang XJ, Xu Q, Yan JP, Yu F, Xiao J, et al. Nonadditive expression of lipid metabolism pathway-related genes in intestine of hybrids of Nile tilapia females (Oreochromis niloticus) and blue tilapia males (Oreochromis aureus). Mol Biol Rep (2019) 46:425–32. doi: 10.1007/s11033-018-4490-3
16. Berbel-Filho WM, Pacheco G, Lira MG, Garcia de Leaniz C, Lima SMQ, Rodríguez-López CM, et al. Additive and non-additive epigenetic signatures of natural hybridization between fish species with different mating systems. Epigenetics (2022) 17:2356–65. doi: 10.1080/15592294.2022.2123014
17. Yoo M-J, Liu X, Pires JC, Soltis PS, Soltis DE. Nonadditive gene expression in polyploids. Annu Rev Genet (2014) 48:485–517. doi: 10.1146/annurev-genet-120213-092159
18. Auger DL, Gray AD, Ream TS, Kato A, Coe EH Jr., Birchler JA. Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics (2005) 169:389–97. doi: 10.1534/genetics.104.032987
19. Zhou Y, Ren L, Xiao J, Zhong H, Wang J, Hu J, et al. Global transcriptional and miRNA insights into bases of heterosis in hybridization of Cyprinidae. Sci Rep (2015) 5:13847. doi: 10.1038/srep13847
20. Zhong H, Zhou Y, Zhang H, Xiao W. DNA methylation pattern is associated with elevated expression of DGAT2 in hybrid tilapia. Aquaculture Nutr (2021) 27:1750–60. doi: 10.1111/anu.13312
21. Shi X, Ng DWK, Zhang C, Comai L, Ye W, Jeffrey Chen Z. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat Commun (2012) 3:950. doi: 10.1038/ncomms1954
22. Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci (2010) 15:57–71. doi: 10.1016/j.tplants.2009.12.003
23. Bassene JB, Froelicher Y, Dhuique-Mayer C, Mouhaya W, Ferrer RM, Ancillo G, et al. Non-additive phenotypic and transcriptomic inheritance in a citrus allotetraploid somatic hybrid between C. reticulata and C. limon: the case of pulp carotenoid biosynthesis pathway. Plant Cell Rep (2009) 28:1689–97. doi: 10.1007/s00299-009-0768-1
24. Sheridan MA, Hagemeister AL. Somatostatin and somatostatin receptors in fish growth. Gen Comp Endocrinol (2010) 167:360–5. doi: 10.1016/j.ygcen.2009.09.002
25. Zhong H, Hu Y, Yu F. A review on ghrelin and fish reproduction. Reprod Breed (2021) 1:128–35. doi: 10.1016/j.repbre.2021.07.004
26. Jönsson E. The role of ghrelin in energy balance regulation in fish. Gen Comp Endocrinol (2013) 187:79–85. doi: 10.1016/j.ygcen.2013.03.013
27. Zhong H, Zhang XJ, Xu Q, Yan JP, Han ZJ, Zheng HF, et al. Nonadditive and asymmetric allelic expression of growth hormone in hybrid tilapia. Front Genet (2019) 10:961. doi: 10.3389/fgene.2019.00961
28. Zhu J, Zou Z, Li D, Xiao W, Yu J, Chen B, et al. Transcriptome profiling revealed basis for growth heterosis in hybrid tilapia (Oreochromis niloticus × O. aureus). Fishes (2022) 7(1):43. doi: 10.3390/fishes7010043
29. Tung PH, Shiau SY. Effects of meal frequency on growth performance of hybrid tilapia, Oreochromis niloticus × O. aureus, fed different carbohydrate diets. Aquaculture (1991) 92:343–50. doi: 10.1016/0044-8486(91)90039-A
30. Wu X, Zhao L, Fan Z, Lu B, Chen J, Tan D, et al. Screening and characterization of sex-linked DNA markers and marker-assisted selection in blue tilapia (Oreochromis aureus). Aquaculture (2021) 530:735934. doi: 10.1016/j.aquaculture.2020.735934
31. Curzon AY, Shirak A, Benet-Perlberg A, Naor A, Low-Tanne SI, Sharkawi H, et al. Absence of figla-like gene is concordant with femaleness in cichlids harboring the LG1 sex-determination system. Int J Mol Sci (2022) 23:7636. doi: 10.3390/ijms23147636
32. Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinf (2002) Chapter 2:2.3.1-2.3.22. doi: 10.1002/0471250953.bi0203s00
33. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. (2003) 19:1572–4. doi: 10.1093/bioinformatics/btg180
34. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
35. Cummings DE, Shannon MH. Roles for ghrelin in the regulation of appetite and body weight. Arch Surg (2003) 138:389–96. doi: 10.1001/archsurg.138.4.389
36. Rahman MA, Bhadra A, Begum N, Islam MS, Hussain MG. Production of hybrid vigor through cross breeding between Clarias batrachus Lin. and Clarias gariepinus Bur. Aquaculture (1995) 138:125–30. doi: 10.1016/0044-8486(95)01076-9
37. Zhou Y, Zhang XJ, Xu Q, Yan JP, Yu F, Wang FH, et al. Nonadditive and allele-specific expression of insulin-like growth factor 1 in Nile tilapia (Oreochromis niloticus, ♀) × blue tilapia (O. aureus, ♂) hybrids. Comp Biochem Physiol Part B: Biochem Mol Biol (2019) 232:93–100. doi: 10.1016/j.cbpb.2019.03.002
38. Cai WQ, Li SF, Ma JY. Diseases resistance of Nile tilapia (Oreochromis niloticus), blue tilapia (Oreochromis aureus) and their hybrid (female Nile tilapia × male blue tilapia) to Aeromonas sobria. Aquaculture (2004) 229:79–87. doi: 10.1016/S0044-8486(03)00357-0
39. Unniappan S, Peter RE. Structure, distribution and physiological functions of ghrelin in fish. Comp Biochem Physiol Part A: Mol Integr Physiol (2005) 140:396–408. doi: 10.1016/j.cbpb.2005.02.011
40. Unniappan S. Ghrelin: An emerging player in the regulation of reproduction in non-mammalian vertebrates. Gen Comp Endocrinol (2010) 167:340–3. doi: 10.1016/j.ygcen.2009.12.003
41. Tena-Sempere M. Ghrelin and reproduction: ghrelin as novel regulator of the gonadotropic axis. Vitam Horm. (2007) 77:285–300. doi: 10.1016/S0083-6729(06)77012-1
42. Sominsky L, Hodgson DM, McLaughlin EA, Smith R, Wall HM, Spencer SJ. Linking stress and infertility: a novel role for ghrelin. Endocrine Rev (2017) 38:432–67. doi: 10.1210/er.2016-1133
43. Feng KE, Zhang GR, Wei KJ, Xiong BX. Molecular cloning, tissue distribution, and ontogenetic expression of ghrelin and regulation of expression by fasting and refeeding in the grass carp (Ctenopharyngodon idellus). J Exp Zoology Part A: Ecol Genet Physiol (2013) 319:202–12. doi: 10.1002/jez.1784
44. Blanco AM, Gómez-Boronat M, Redondo I, Valenciano AI, Delgado MJ. Periprandial changes and effects of short-and long-term fasting on ghrelin, GOAT, and ghrelin receptors in goldfish (Carassius auratus). J Comp Physiol B (2016) 186:727–38. doi: 10.1007/s00360-016-0986-0
45. Hochholdinger F, Hoecker N. Towards the molecular basis of heterosis. Trends Plant Sci (2007) 12:427–32. doi: 10.1016/j.tplants.2007.08.005
46. Shao L, Xing F, Xu C, Zhang Q, Che J, Wang X, et al. Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc Natl Acad Sci (2019) 116:5653–8. doi: 10.1073/pnas.1820513116
47. Strogantsev R, Krueger F, Yamazawa K, Shi H, Gould P, Goldman-Roberts M, et al. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol (2015) 16:112. doi: 10.1186/s13059-015-0672-7
48. Graze RM, McIntyre LM, Main BJ, Wayne ML, Nuzhdin SV. Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression. Genetics (2009) 183:547–61. doi: 10.1534/genetics.109.105957
49. Shen Y, Garcia T, Pabuwal V, Boswell M, Pasquali A, Beldorth I, et al. Alternative strategies for development of a reference transcriptome for quantification of allele specific expression in organisms having sparse genomic resources. Comp Biochem Physiol Part D: Genomics Proteomics (2013) 8:11–6. doi: 10.1016/j.cbd.2012.10.006
50. York RA, Patil C, Abdilleh K, Johnson ZV, Conte MA, Genner MJ, et al. Behavior-dependent cis regulation reveals genes and pathways associated with bower building in cichlid fishes. Proc Natl Acad Sci (2018) 115:E11081–E90. doi: 10.1073/pnas.1810140115
51. Chao W, D’Amore PA. IGF2: Epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev (2008) 19:111–20. doi: 10.1016/j.cytogfr.2008.01.005
52. Tabano S, Colapietro P, Cetin I, Grati FR, Zanutto S, Mandò C, et al. Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra-embryonic compartments and its possible role in fetal growth restriction. Epigenetics (2010) 5:313–24. doi: 10.4161/epi.5.4.11637
53. Cheung VG, Nayak RR, Wang IX, Elwyn S, Cousins SM, Morley M, et al. Polymorphic cis- and trans-regulation of human gene expression. PloS Biol (2010) 8:e1000480. doi: 10.1371/journal.pbio.1000480
54. Göbel U, Arce AL, He F, Rico A, Schmitz G, de Meaux J. Robustness of transposable element regulation but no genomic shock observed in interspecific Arabidopsis hybrids. Genome Biol Evol (2018) 10:1403–15. doi: 10.1093/gbe/evy095
Keywords: ghrelin, hybrid tilapia, nonadditive expression, gene expression, heterosis
Citation: Zhong H, Ren B, Lou C, Zhou Y, Luo Y and Xiao J (2023) Nonadditive and allele-specific expression of ghrelin in hybrid tilapia. Front. Endocrinol. 14:1292730. doi: 10.3389/fendo.2023.1292730
Received: 12 September 2023; Accepted: 07 November 2023;
Published: 13 December 2023.
Edited by:
Zhan Yin, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Hongjuan Shi, Guangdong Ocean University, ChinaCopyright © 2023 Zhong, Ren, Lou, Zhou, Luo and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huan Zhong, emhvbmdodWFuemhAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.