- 1Julius L Chambers Biomedical/Biotechnology Institute (JLC-BBRI), North Carolina Central University, Durham, NC, United States
- 2Charles River Discovery Services, Morrisville, NC, United States
- 3Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, United States
Both cancer and cardio-metabolic disease disparities exist among specific populations in the US. For example, African Americans experience the highest rates of breast and prostate cancer mortality and the highest incidence of obesity. Native and Hispanic Americans experience the highest rates of liver cancer mortality. At the same time, Pacific Islanders have the highest death rate attributed to type 2 diabetes (T2D), and Asian Americans experience the highest incidence of non-alcoholic fatty liver disease (NAFLD) and cancers induced by infectious agents. Notably, the pathologic progression of both cancer and cardio-metabolic diseases involves innate immunity and mechanisms of inflammation. Innate immunity in individuals is established through genetic inheritance and external stimuli to respond to environmental threats and stresses such as pathogen exposure. Further, individual genomes contain characteristic genetic markers associated with one or more geographic ancestries (ethnic groups), including protective innate immune genetic programming optimized for survival in their corresponding ancestral environment(s). This perspective explores evidence related to our working hypothesis that genetic variations in innate immune genes, particularly those that are commonly found but unevenly distributed between populations, are associated with disparities between populations in both cancer and cardio-metabolic diseases. Identifying conventional and unconventional innate immune genes that fit this profile may provide critical insights into the underlying mechanisms that connect these two families of complex diseases and offer novel targets for precision-based treatment of cancer and/or cardio-metabolic disease.
1 Introduction
1.1 Double-edged swords: important factors connecting metabolic disorders and cancer development
The following perspective was written in response to an invited Frontiers research topic to explore methods, mechanisms, and hypotheses that may ultimately identify and exploit biological processes contributing to complex disease progression and molecular interactions enabling cross-talk between cancer and cardio-metabolic disease. Based on our hypothesis that innate immunity differences contribute to observed population disease disparities in cancer and metabolic disorders, we apply a functional genomics approach to identify specific innate immune genes as potential therapeutic targets at the intersection of these two complex disease families.
1.2 Framing precision drug target discovery in the context of health disparities
1.2.1 Defining health disparities
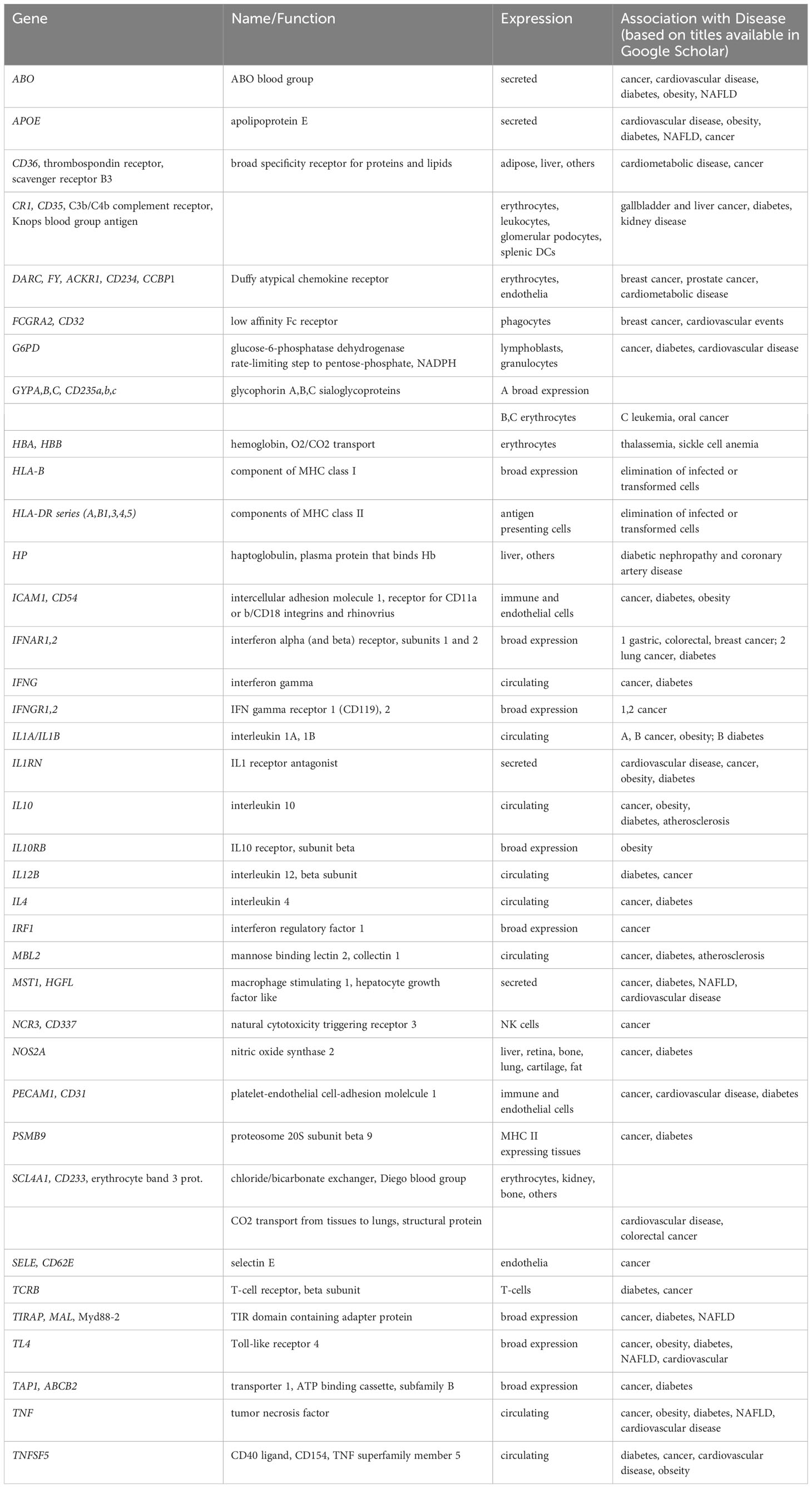
The US National Institute on Minority Health and Health Disparities (NIMHD) defines health disparities as “a health difference (compared with the general population), based on one or more health outcomes (such as the overall rate of disease incidence, prevalence, morbidity, mortality or survival) that adversely affect disadvantaged populations.” In the US, such populations include Blacks/African Americans, Hispanics/Latinos, Asians, American Indians/Alaska Natives, and Native Hawaiians/other Pacific Islanders) (1). Diverse sources, from sponsored websites (such as 2 and associated links) to peer-reviewed articles summarizing disparities in one or more diseases between two or more populations, provide ample evidence for differences in cancer (3), cardio-metabolic disease (4) and overall health risks and outcomes (5) based on ethnic background/geographic ancestry. By way of illustration, Tables 1, 2 summarize disparities in cancer incidence and mortality among US ethnic populations (adapted from 6) and population differences in overall mortality rates of cancer and cardio-metabolic diseases (adapted from 7), respectively.
Assessing health differences between populations is complicated because results may vary depending on the size and granular composition of the populations being compared. On the one hand, evaluating larger, more heterogeneous populations improves statistical reliability, but this approach may mask disparities among subpopulations. For example, among Asians in the US (8) and Asia (9), the incidence of liver cancer varies widely based on geography and/or geographic ancestry. Further, trends in incidence and/or mortality may change due to cohort variations in age, exposure to risk, and geographic location, as is the case for liver (10) and breast cancer incidence (11) in the US and for global cancer mortality rates (e.g., 12).
Defining/distinguishing populations is a critical aspect of evaluating health disparities. Many analyses have been based on self-identified ethnicity; it stands to reason that this approach is likely to align more closely with social determinants of health. In contrast, a relatively precise biological assessment of geographic ancestry can be obtained using genetic markers to identify ethnic origins. In this approach, selected ancestry informative markers (AIMs) were initially used to evaluate genetic admixture and geographic ancestry and provide valuable background information when comparing individuals representing different populations (13). Improved methods and more extensive and complete reference datasets have further refined admixture mapping (14).
For the purposes of this perspective, we will refer to populations as they are defined by individual authors; populations in Section 3 are defined according to Karczewski (15). The interested reader is referred to a recent book chapter entitled “Using Population Descriptors in Genetics and Genomics Research: A New Framework for an Evolving Field” written by the National Academies of Sciences’ Committee on the Use of Race, Ethnicity, and Ancestry as Population Descriptors in Genomics Research (16) for a thorough treatment of this subject.
1.2.2 Past and present challenges to advancing research in the biology of health disparities
Cancer and cardio-metabolic disease disparities have multifactorial etiologies, including biological, behavioral, environmental, and social components. There is ample evidence that these disparate etiological factors are not adequately understood in isolation from one another. The interested reader is referred to reviews on the impact of physical, social, and chemical environments on the biology of health disparities (17–19) and on the biological impacts of stress (20), including racism-induced stress and increased allostatic load (21–23), all of which are beyond the scope of this perspective.
The relative contribution of biology to cancer and cardio-metabolic disparities continues to be a matter of debate among scientists in various disciplines and even among biologists themselves (24). The hesitation to consider geographic ancestral differences in biology among some mainstream biomedical scientists is just one of several obstacles that have hindered a rigorous study of the biology of health disparities.
Social forces continue to hinder the participation of minority populations in medical research and to limit their access to medical care. For example, an entrenched and well-founded mistrust of the medical establishment in the US exists among minority populations due to a long history of abuses (25). Limited access to healthcare and subpar healthcare quality further exacerbate health disparities in minority populations, leading to lower life expectancy in American Hispanic and Black populations (26).
Traditional research approaches and the most widely available resources in the biomedical sciences have also unintentionally hindered a rigorous characterization of the biological differences that underlie health disparities. In vitro studies employ samples and cell lines obtained most often from individuals of European descent (27, 28) and the majority of clinical trials disproportionately enroll individuals from this same population (29, 30). Thus, at multiple stages in the drug research and development cycle, biases exist towards agents optimized for those of European ancestry. Fortunately, the need to increase the diversity of human samples and cell lines and to engage diverse study populations in biomedical research and clinical trials has recently gained the attention and enthusiastic support of pre-eminent scientists (29, 31–33) and the NIH (34).
1.2.3 Considering geographic ancestry in the development of effective treatments
The human genome possesses a high degree of variation. According to a 2016 meta-analysis of 60,706 individuals of diverse ancestries, an average of 1 in 8 bases of the coding sequence were variants, and 72% of these had not been previously identified and/or characterized (35). Wide genetic variations within populations are at least as diverse as genetic variations between populations (36). This finding implies that not all genetic variations contribute to putative biological differences between populations.
Genetic differences associated with geographic ancestry, such as AIMs, may result in the uneven among populations distribution of gene variants. In many cases, these variants are uncommon, and/or their impact on protein expression, function, or disease is either insignificant or unknown. However, an intriguing study by Ahsan et al. (37) identified 65 “minor” drug response alleles that were present in more than 50% of individuals in at least one population; in other words, in some populations, the variant was more common than the wild type/canonical protein. Consistent with this is a body of clinical evidence that specific drug responses vary according to geographic ancestry, with outcomes that range from lack of efficacy to drug-related pathology and death in one or more minority populations (38–40). Therefore, we sought to identify population-specific potential therapeutic targets at the intersection of cancer and cardio-metabolic disease, in part by hand-curating gene variants with “minor” alleles that were common in at least one major population (as defined by 15) but that were significantly less common in at least one other major population.
1.3 Innate immunity as a biological driver of health disparities
Gene variants that confer protective immunity are retained in each population to optimize survival. For example, in the case of those with African ancestry, gene variants retained in the pan-African genome have been identified that provide defense against indigenous pathogens such as malaria and trypanosomiasis (African sleeping sickness/Chagas disease). The selective pressure imposed by pathogens on gene variation is impressive; in the case of malaria, variants of at least 40 different genes are thought to protect against one or more species of Plasmodium (41, 42).
Unfortunately, immune protection frequently involves a trade-off where protective innate immune variants may introduce new pathologies. For example, among the gene variants that protect against malaria, HbS also promotes sickle cell anemia, HbE promotes thalassemia, G6PD variants promote hemolytic anemia, and Duffy antigen receptor (DARC) variants are associated with increased breast cancer metastasis and mortality (43, 44). Similarly, the same APOL1 variants shown to protect against severe trypanosomiasis are also associated with nephropathy (45, 46).
Several lines of evidence affirm that innate immune genes are highly adaptable and optimized to respond to local pathogens. First, within the human genome, genes associated with immunity are under the strongest selective pressure (47, 48). Second, selective pressure on immune genes is pathogen-driven (49, 50). Third, the geographic distribution of populations bearing the highest frequency of HbS (51) and DARC (52) gene variants closely resemble the geographic distribution of the malarial strains they protect against. Finally, according to their geographical ancestry, populations differ in their susceptibility to infectious disease (53), in their immune response to pathogens (54) and even in their macrophage function and circulating cytokine levels (55–57). All of these findings indicate that protective innate immune variants are distributed among individuals based on their geographic ancestry.
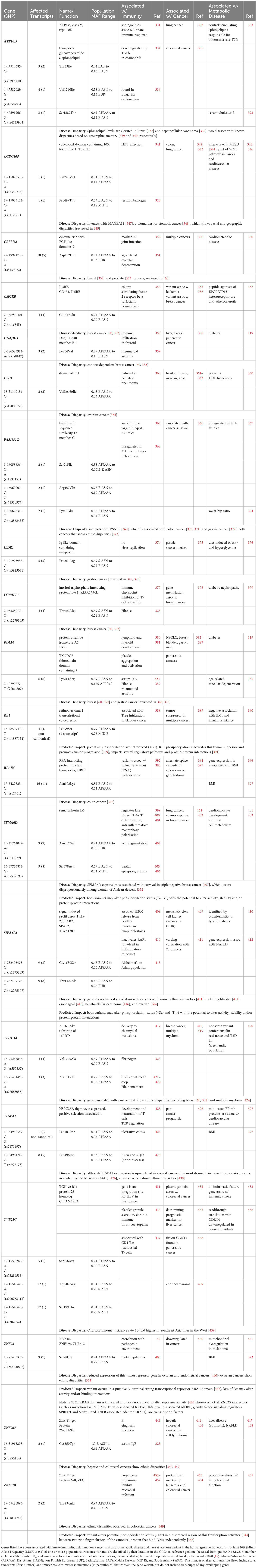
It is important to note that genes associated with innate immunity are structurally and functionally diverse. Some are well-characterized participants in inflammation, including but not limited to cytokines, chemokines, and pattern recognition receptors (lectins, Toll-like receptor (TLR) family members, and NLRs) and their related pathways. However, as illustrated by the variety of genes that protect against malaria (summarized in Table 3), others are pleiotropic, expressed in non-immune tissues and/or frequently better known for their “day jobs”. Most of the protective variants listed in Table 3 can be tied directly to immunity. Still, a few (such as APOE, G6PD, glycophorin (GYP), hemoglobin (HB), and haptoglobulin (HP)) would be considered unconventional innate immune genes.
1.4 Inflammation is a component of cancer and cardio-metabolic diseases
1.4.1 Cancer and inflammation
Hanahan and Weinberg, in their seminal review, describe six hallmarks of cancer, many of which are enabled by mechanisms of immunity, including inflammation (58). Their observations are particularly relevant to this perspective since further research in the field has established that reprogrammed energy metabolism and immune evasion are additional hallmarks (58, 59).
In a previously published perspective, we presented evidence for an association between breast and prostate cancer disparities in African Americans (AAs) and classic innate immune gene variants (interleukins, Toll-like receptors, monocyte activity) more commonly found in AAs (60). Since 2019, Google Scholar (accessed 4/18/23) has listed more than 18,000 publications with titles that include “cancer” and “inflammation,” “infection”, “immune,” “immunity,” or “innate”; these publications address a wide range of topics, including immune escape by cancer cells, the contribution of chronic inflammation to tumor progression, and immune-based cancer therapies, that are beyond the scope of this perspective. Notably, less than 40 of these publications (< 0.2%) include the terms “disparity” or “disparities” in their titles. Among this small set of publications are descriptions of population differences in tumor microenvironment and immune signatures in breast (61, 62), head and neck (63–65), lung (66, 67), and colorectal (68, 69) cancers, as well as cancer generally (70). Of particular interest is a recent exploration of the link between racial differences in mitochondrial metabolism and the tumor immune microenvironment (71).
1.4.2 Cardio-metabolic disease and inflammation
The constellation of inter-related cardio-metabolic diseases has been collectively referred to as metabolic syndrome (MetS), and their cumulative effect on global health is massive (reviewed in 72–74). Clinical definitions of MetS vary depending on which disease(s) are of primary interest (reviewed in 75–77). The National Heart Lung and Blood Institute (NHLBI) lists the following MetS risk factors as abdominal obesity and/or insulin resistance, elevated triglycerides and LDL-cholesterol, reduced HDL-cholesterol, hypertension, elevated glucose and pro-thrombotic or pro-inflammatory states (78). Several metabolic diseases have been associated with these risk factors, including hypertension, obesity, atherosclerotic cardiovascular disease, type 2 diabetes (T2D), non-alcoholic fatty liver disease (NAFLD), and stroke.
Genetic and environmental factors impact cardio-metabolic diseases, and their risk, morbidity, and mortality vary with age, gender, and race/ethnicity (4, 76). Unfortunately, the effects of MetS are not confined to cardio-metabolic co-morbidities, given that MetS is also associated with increases in the incidence and/or mortality of arthritis, chronic kidney disease, schizophrenia, depression and cancer, as noted in references (79, 80).
Inflammation is a key contributor to MetS and associated co-morbidities (81–83), just as MetS pathologies impact inflammation (c.f. 84). In general, low-grade chronic inflammation evoked during metabolic disease stimulates the production of pro-inflammatory cytokines, immuno-modulatory proteins, lipids, and other mediators of inflammation that impact systemic and/or localized tissue inflammation (82, 85). Unfortunately, the treatment of metabolic diseases is complicated by the cross-talk between pro- and anti-inflammatory mechanisms at work among MetS co-morbidities (c.f. 77, 86–88). Further, inflammation from one metabolic disease can also exacerbate other MetS co-morbidities.
As with almost all tissues, organs that regulate systemic metabolism possess innate immune response capabilities. Notably, some organs that regulate overall metabolic homeostasis also impact systemic inflammation. In the case of both adipose tissue (89–91) and liver (92–94), these organs harbor and partner with resident macrophages (ATMs and Kupffer cells, respectively) in inflammation. Further, adipose tissue and liver produce unique immunologically active biomolecules, such as adipokines (86, 95) and bile acids (96–99). Perhaps less appreciated are two additional organs associated with metabolic homeostasis that control systemic levels of immunologically active biomolecules: the gallbladder regulates bile acid levels and the pancreas controls insulin, which levels of insulin, with its known anti-inflammatory effects (100).
Just as mediators of metabolism can impact inflammation, mediators of immunity can impact metabolism. For example, innate immune receptors have demonstrated roles in metabolic disease progression (101), and pro-inflammatory cytokines produced in the adipose tissue of obese individuals contribute to the development of T2D (102). Significantly, biomolecules such as adipokines, insulin, and bile acids mediate metabolism and inflammation. Further, besides their widely recognized role in lipid transport and cellular metabolic homeostasis, serum lipids and lipoproteins also provide innate immune protection (103, 104).
2 A functional genomics approach to novel target discovery
Using functional genomics, we and others have observed associations between specific innate immune gene variants and cancer or metabolic disease risk or outcome that differ according to geographic ancestry (57, 60, 105). Given that immunity including inflammation contributes to the progression of both complex disease families, we have hypothesized that population differences in genetic (and epigenetic) innate immune programs contribute to complex disease disparities between populations. Based on this conceptual framework, this perspective seeks to identify innate immune gene candidates associated with both cancer and cardio-metabolic disease that differ between populations.
Genome wide association studies (GWAS) in general (106) and the Genome Aggregation Database (gnomAD) in particular (107) provide researchers with the capacity to compare thousands of complete genomes from individuals among all largely-grouped populations. These resources catalog gene variations called single nucleotide polymorphisms (SNPs) across the entire genome of each individual. SNPs are located not only in protein coding genes (including coding exons as well as non-coding introns and remote, up-, down-, and mid-stream regulatory sites), but also across regions associated with short and long non-coding RNAs, chromosomal architecture, and other essential functions that have been previously underappreciated and mislabeled as “junk DNA” (108). The number of genes and the percentage of the human genome they occupy varies depending on their definition (109). Notably, most SNPs associated with disease states or changes in phenotype (95%) are located outside coding exons (110).
Nevertheless, in this perspective, we will focus on widely occurring gene variants that code for changes in the canonical amino acid (aa) sequence, also referred to as missense variants or nonsynonymous SNPs, as a first step towards accelerating the development of optimally safe and active drugs that target understudied protein variants widely found in patients with diverse geographical ancestries. Importantly, nonsynonymous SNPs have the potential to impact protein conformation, activity and/or protein-protein interactions, potentially altering disease states and phenotypes. For simplicity, we have also excluded synonymous SNPs (exonic point mutations that do not alter aa sequence), in spite of mounting evidence that suggests they can function in isoform selection (protein size and sequence), transcript expression levels and stability, translational folding rate, overall conformation, and posttranslational modifications, all of which possess potential functional consequences on cell behavior and disease risk (111–113).
This perspective identifies conventional and unconventional innate immune genes (summarized in Section 3) that meet the following criteria. First, there is evidence that each gene participates in, is a target of, or is associated with innate immunity including inflammation. Second, there is evidence that each gene is associated with at least one form of cancer and at least one cardio-metabolic disease. Finally, each gene occurs among the global population as at least one population-enriched variant, which we define as a widely occurring missense variant distributed unevenly among populations.
We have employed a hand-curated discovery process to identify population-specific innate immune genes at the intersection of cancer and metabolic disease. From the primary and secondary literature, gene lists associated with innate immunity (49, 114, 115), cancer (116, 117), or cardio-metabolic disease (118, 119) were vetted for the following characteristics:
1) Evidence in the primary or secondary literature (accessed through Google Scholar) indicated that the candidate gene was involved in all three disease categories: innate immunity/inflammation, cancer, and cardio-metabolic disease.
2) Indication in gnomAD that the candidate gene occurs as at least one nonsynonymous SNP/missense variant with
a. a high minor allele frequency (MAF ≥ 0.2 in at least one of the six major populations defined by 15): African/African American (AFR/AA), East Asian (E ASN), non-Finnish European (EUR), Latino/Latina (LAT), Middle Eastern (MID E), and South Asian (S ASN),
b. a difference in MAF among significant populations of ≥ 0.2 from the highest to lowest frequency.
Note that among genes with missense variants, we chose only those with common variants that occur widely among individuals in one or more populations, i.e., missense variants that occurred in at least 20% of individuals in one or more populations (by definition, having a minor allele frequency (MAF) ≥ 0.20 and varying widely in the frequency of their occurrence among populations. This approach was based on our rationale that variants selected and retained in the human genome provide a survival benefit for the population(s) in which they occur, even as they may also paradoxically contribute to complex disease as discussed above for HbS and APOL1 variants (see Section 1.3).
3 Candidate innate immune genes at the intersection of cancer and cardio-metabolic disease disparities
Among the candidate innate immune genes that we identified at the intersection of cancer and cardio-metabolic disease, we found both “conventional” innate immune genes, such as cytokines and cytokine receptors, pattern recognition receptors, and other genes that have widely acknowledged roles in immune cell function, and “unconventional genes” with pleiotropic functions that include innate immunity, such apolipoproteins, biomolecule transporters, and transcription regulators. Using the approach described in Section 2, three lists of innate immune genes implicated in cancer and cardio-metabolic disease were generated. Each gene listed in the three tables below possesses at least one population-enriched variant with an amino acid replacement that differs in its distribution among populations, suggesting its potential role in both cancer and cardio-metabolic disparities. The 52 genes identified provide a representative but not exhaustive list of candidate genes, thus serving as preliminary data for further investigation.
Section 3.1 summarizes conventional innate immune genes and their corresponding population-enriched variants previously shown to impact disease or biological function. Similarly, Section 3.2 summarizes unconventional innate immune genes (better known for their non-immune functions) and their corresponding population-enriched variants that have been previously shown to impact disease or biological function. Finally, Section 3.3 summarizes genes associated with innate immunity, cancer, and cardio-metabolic diseases and their corresponding population-enriched variants whose impact on disease or biological function has not yet been established.
3.1 Conventional innate immune genes with previously characterized population-enriched variants
Table 4 includes 14 genes best known for their roles in immunity, including inflammation, that are present as at least one population-enriched variant shown to impact biological function. Among these are cytokines and cytokine receptors, including macrophage inhibitory cytokine 1 (MIC-1/GDF15), interleukin 3 and the alpha subunit of its receptor (IL3 and IL3RA), along with subunits for interleukin 4, 6 and 7 receptors (IL4R, IL6R, and IL7R), and the leptin adipokine receptor (LEPR). Additional immune receptors include the soluble receptor for MHC I antigens I (leukocyte Ig-like receptor A3, LILRA3/CD85E) and two pattern recognition receptors, the intracellular pattern recognition receptor nucleotide-binding oligomerization domain containing 2 (NOD2) and the five transmembrane stimulator of interferon response CGAMP interactor 1 (STING1/TMEM173). Also included were the catalytic enzyme in the rate-limiting step of the kynurenine pathway during inflammation indoleamine 2,3-dioxygenase 2 (IDO2), the temperature-sensitive cation channel TRPM8, and two adhesion molecules, one expressed in lymphocytes (integrin alpha L, ITGAL/LFA-1/CD11A) and the other expressed in leukocytes (junctional adhesion molecule-like, JAML/AMICA).
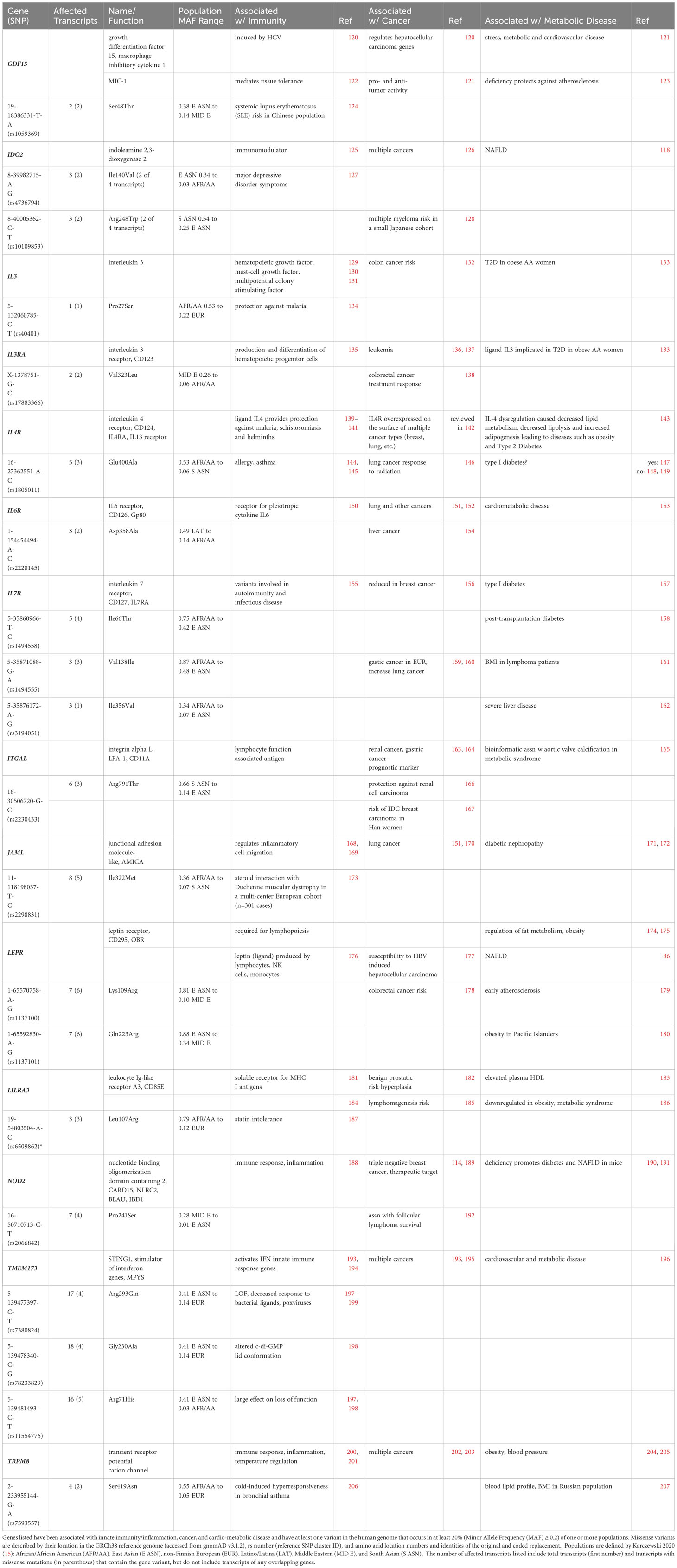
Table 4 Candidate Conventional Innate Immune Genes at the Intersection of Cancer and Cardio-Metabolic Disease.
3.1.1 Interleukin 3 and interleukin 3 receptor alpha chain
IL-3 is a growth factor produced by activated T-cells (129) that regulates the growth of hematopoietic progenitor cells and activates mature neutrophils and macrophages (208). IL-3 is also implicated in priming (131) and activating (130) basophils. Intriguingly, increased serum levels of IL-3 have recently been associated with the onset of type 2 diabetes in African American women as determined by serum levels of glucose and HbA1c (133). Genetic variations in IL3 have been noted in colon and rectal cancers (132). The Pro27Ser variant (5-132060785-C-T) has been associated with protection against malaria (134) but also with an increase in miscarriages following in vitro fertilization (IVF) in women of various populations (209).
The interleukin 3 receptor is a heterodimer comprised of an interleukin 3-specific alpha chain (IL-3RA, CD123) and the common cytokine beta chain CSF2RB, another candidate listed below in Section 3.3, that also forms dimers with the alpha chains of both GM-CSF and IL-5 receptors. High-affinity IL-3 binding induces hetero-dimerization of IL-3RA and CSF2RB, and subsequent disulfide linkage of these receptor chains is required for receptor activation and CSF2RB phosphorylation (210). IL-3RA expression varies among CD34+ hematopoietic cell types, with negative/low expression in primitive hematopoietic cells and little or no surface expression in early erythroid progenitors, but high expression in B-lymphoid and myeloid progenitors (135). The X-chromosome-linked IL3RA Val323Leu variant (X-1378751-G-C) was associated with non-complete response to neoadjuvant chemotherapy against locally advanced rectal cancer in Hong Kong patients (138).
3.1.2 Interleukin 4 receptor alpha chain
The IL-4R alpha chain (IL4R, CD124) forms heterodimers with at least two partners. Type 1 IL-4 receptors are composed of IL-4R complexed with the common cytokine receptor gamma chain (IL2RG, CD132), which may alternatively dimerize with IL-2, IL-7 and IL-21 cytokine receptors, so that IL-2, IL-7, and IL-21 receptors compete with IL-4R for binding to IL2RG. Type 2 IL-4 receptors are composed of IL-4R complexed with IL-13RA1 (IL13Rα1, CD213A1). Thus, IL-4 activates both Type 1 and Type 2 IL-4 receptors, while IL-13 activates Type 2 IL-4 receptors. Both IL-4 and IL-13 signaling through the IL-4R mediate type 2 (humoral, as opposed to type 1 cellular) immunity against helminths, toxins and tropical parasites such as plasmodium (malaria) and trypanosomes (African sleeping sickness/Chagas disease) (139–141, 211). Both IL-4Rα and IL13-Rα1 have also been implicated in cancer progression and were recently identified as prognostic indicators in soft-tissue sarcoma patients when present in the nucleus. IL-4 regulates lipid metabolism (143), and (142) recent findings highlight an intriguing relationship between non-hematopoietic IL-4Rα activation of a non-canonical signaling pathway that regulates a high-fat, high-carbohydrate diet-driven induction of obesity and impacts the severity of obesity-associated sequelae in mice (212). Numerous genetic epidemiological studies have also shown that IL4 and IL4R and their gene polymorphisms play important roles in asthma in various populations. Notably, individuals carrying one or two copies of the IL4R Glu400Ala (16-27362551-A-C) minor allele were at higher risk to suffer from allergy (145) and asthma (144, 213).
3.1.3 Interleukin 7 receptor alpha chain
The integral membrane interleukin 7 receptor (IL-7R) transmits pro-inflammatory signals initiated by IL-7 at the cell surface. The functional IL-7 receptor is a heterodimer comprised of the IL-7 receptor alpha chain (IL7R, IL7Rα, CD127) and the same common cytokine receptor gamma chain (IL2RG, CD132) that dimerizes with the IL-4R alpha chain. The assembled IL-7R recognizes not only IL-7 but also thymic stromal lymphopoietin (TSLP), both cytokines with 4 α-helical strands (214). Multiple transcriptional and post-transcriptional mechanisms exist to regulate expression of the IL-7R protein (215). Some of these mechanisms are homeostatic, molecular and cytokine-mediated, where IL7Rα transcription decreases in CD4+ and CD8+ cells once naïve T cells become activated. Notably, IL-7 binding to IL-7R activates the Janus kinase (JAK/STAT) pathway, which plays an essential role in lipid metabolism (216). However, peripheral blood mononuclear cells (PBMCs) in breast cancer patients show defects in STAT5 phosphorylation and altered expression of IL-7Rα that ultimately impacts memory T cell development (156).
Notably, compared to the canonical gene, the IL7R variants 5-35874473-C-T (rs6897932), 5-35860966-T-C (rs1494558) and 5-35871088-G-A (rs1494555) alter the pathology of autoimmune and infectious diseases due to their impact on IL7R expression and alternative splicing (155). Further, all three population-enriched missense variants of IL7R identified in Table 4 show an association with cardio-metabolic disease: Ile66Thr (5-35860966-T-C, rs1494558) with post-transplantation diabetes (158); Val138Ile (5-35871088-G-A, rs1494555) with body mass index (BMI) in lymphoma patients (161), and Ile356Val (5-35876172-A-G, rs3194051) with severe liver disease (162). However, to date only Val138Ile has been associated with increased cancer risk, both in lung (160) and stomach (159).
3.2 Unconventional innate immune genes with previously characterized population-enriched variants
Table 5 includes 18 genes representing several classes of proteins primarily associated with non-immune functions that occur as population-enriched variants shown to impact biological function. These genes include transport membrane proteins, consisting of the multidrug resistance pump (ABCB1), the Niemann-Pick cholesterol transporter 1 (NPC1, SLC65A1), and the Na+-dependent multivitamin transporter (SLC5A6). Among the class of regulatory metabolic enzymes are alcohol dehydrogenase (ADH1C), mitochondrial dihydroorotate dehydrogenase (DHODH), hydroxysteroid (17-beta) dehydrogenase 4 (HSD17B4) involved in peroxisomal fatty acid beta-oxidation, and glycogen phosphorylase B (PYGB) involved in regulating glycogen mobilization. Among the genes that participate in signal transduction are the membrane glycoprotein signaling co-receptor neuregulin (NRG1), phosphodiesterase 10A (PDE10A, which regulates cAMP concentrations), along with the small bioactive neuropeptide neuromedin B (NMB). Transcription factors and/or nucleic acid binding protein genes coded as population-enriched variants include hypoxia-inducible factor 2A (EPAS1, HIF2A), Iroquois homeobox 2 (IRX2), mismatch repair MutL homolog 3 (MLH3), the novel intracellular and extracellular ribonuclease T2 (RNASET2) and the SURP and G-Patch domain containing 1 (SUGP1) splicing factor. Also included are the lipid transport protein apolipoprotein B (APOB), the triacylglycerol lipase patatin-like phospholipase domain containing 3 (PNPLA3), and the adhesion cadherin family member desmoglein 2 (DSG2).
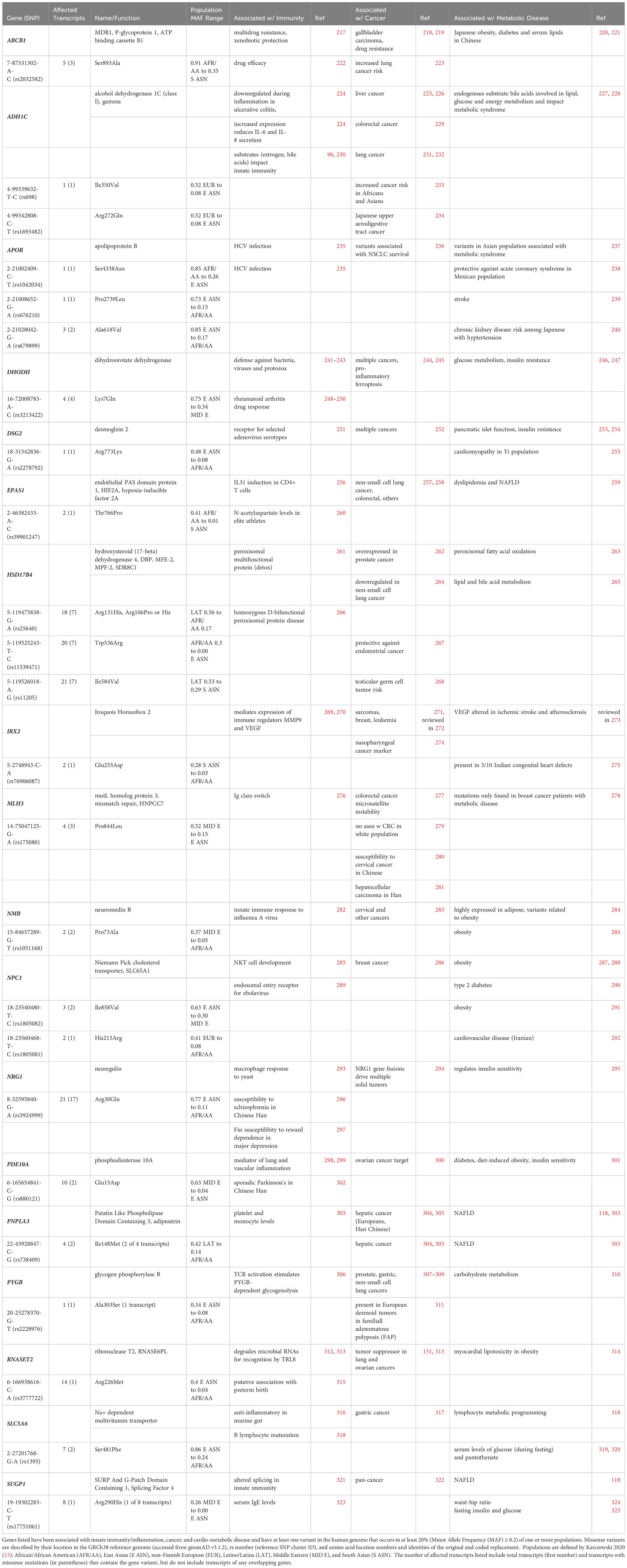
Table 5 Candidate Unconventional Innate Immune Genes at the Intersection of Cancer and Cardio-Metabolic Disease.
3.2.1 Multidrug resistance gene
The ATP binding cassette subfamily B member 1 (ABCB1) gene is commonly known as the first of two multidrug resistance (MDR1) genes in humans and is one of 48 ABC family members (217). ABCB1 functions at the plasma membrane as a 170 kDa monomer with 12 transmembrane domains (TMs), is glycosylated on the first extracellular loop (between TM1 and TM2), and has two intracellular ATP binding sites (one located between TMs 6 and 7, and the other in the carboxy terminus downstream of TM12). ABCB1 is expressed in a wide range of tissues (such as intestine, colon, placenta, liver, and blood-brain barrier) to protect against the intracellular build-up of xenobiotic molecules in vulnerable cells and organs by expelling toxins, including chemotherapeutics, from the cell interior. Thus, ABCB1 has become a widely-known source of and marker for chemoresistance (c.f. 219). ABCB1 also functions as a broad specificity lipid translocase (326). In a Chinese cohort, a variant in the ABCB1 promoter showed pleiotropic effects related to T2D and lipid metabolism (221). Notably, the ABCB1 Ser893Ala variant (7-87531302-A-C, rs2032582) has been correlated with obesity in a Japanese population (220) and with increased susceptibility to lung cancer in a Spanish cohort (223). This ABCB1 variant occurs in 91% of Africans/African Americans, but in only 35-62% of other populations (gnomAD) and was shown to impact drug (etanercept) efficacy in the treatment of Chinese Han patients with ankylosing spondylitis (222).
3.2.2 Mismatch repair protein MutL homolog 3
MLH3 is a homolog of the mismatch repair protein MutL. DNA mismatch repair (MMR) proteins play a vital role in maintaining genome integrity and in antibody maturation during class switch DNA recombination and somatic hypermutation (276). In cases of microsatellite instability, tumors often display somatic mutations in MLH3, while hereditary nonpolyposis colorectal cancer type 7 (HNPCC7) has been associated with germline mutations in the same gene (276, 327). Further, reduced MLH3 expression was observed in individuals diagnosed with grade II and III breast cancer, suggesting MLH3 may serve as a reliable susceptibility marker (278, 328). There was no correlation between the MLH3 Pro844Leu variant (14-75047125-G-A, rs175080, predominantly found in the Middle East) and susceptibility to colorectal cancer in a predominantly white cohort (279). However, in Chinese patients this variant was associated with both cervical cancer (280) and hepatocellular carcinoma (281).
3.2.3 Apolipoprotein B
Lipoproteins enclose otherwise insoluble lipid particles (made up of a central core of cholesterol esters and triglycerides and an outer layer of phospholipids, free cholesterol, and apolipoproteins) for transport through the blood to various tissues (329). Apolipoprotein B (APOB) serves as the primary carrier for several classes of serum lipid particles, including chylomicrons, low-density lipoprotein (LDL), very low-density lipoprotein (VLDL), intermediate-density lipoprotein, and lipoprotein. In LDL particles, APOB interacts with the apoB/E (LDL) receptor, facilitating the removal of LDL cholesterol from the circulation via cellular uptake followed by intracellular LDL breakdown. In a small Japanese study correlating variants of genes related to lipid regulation (including apolipoproteins), the population-enriched missense APOB variant 2-21002409-C-T (rs1042034) correlated with HCV infection (235) variant has an allele frequency of 0.85 in African American populations but only 0.26 in East Asian populations (gnomAD). Another population-enriched missense APOB variant, 2-21008652-G-A (rs676210) (present in 73% of East Asians vs. 15% of Africans/African Americans (gnomAD)) correlated with the occurrence of initial non-cardioembolic ischemic stroke in a small European cohort (239). A third population-enriched missense APOB variant, 2-21028042-G-A (rs679899) (present in 85% of East Asians vs. 17% of Africans/African Americans (gnomAD)) and was protective against acute coronary syndrome in a Mexican population (238). This was associated with both hypertension and chronic kidney disease in a cohort of 3696 Japanese individuals (240).
Functional effects of additional APOB missense variants have also been reported. The Arg3638Gln variant (2-21005955-C-T, rs1801701), which is present in no more than 10% of any population, was associated with survival outcomes in non-small cell lung cancer (NSCLC) patients (236). Additionally, two nonsynonymous variants unique to the Asian population, namely 2-21006289-G-A (rs144467873, MAF = 0.001253 and 0.0003594 in East and South Asians, respectively, but < 0.00008 for all other populations (gnomAD v2.1.1) and 2-21029662-G-A (rs13306194, MAF = 0.1343 in East Asians, MAF < 0.007 in all other populations) were evaluated for their association with lipid profiles, metabolic syndrome and risk of diabetes in a large Taiwan Biobank study (237). Both variants were independently associated with total, LDL, and non-HDL cholesterol levels, whereas rs144467873 (Arg3527Trp) was associated with elevated lipid levels and metabolic syndrome, while rs13306194 (Arg532Trp) was linked with serum triglyceride levels.
3.2.4 Dihydroorotate dehydrogenase
Dihydroorotate dehydrogenase (DHODH), which catalyzes the initial and rate-limiting step of the de novo pyrimidine pathway, is positioned on the inner mitochondrial membrane (330). DHODH has been a therapeutic target for the treatment of rheumatoid arthritis, psoriasis, autoimmune disorders, and Plasmodium, bacterial and fungal infections (241). For over five decades, elevated DHODH expression has been known to promote tumor progression. De novo pyrimidine synthesis becomes essential during increased demands for nucleic acid precursors in rapidly dividing cells making cancer cells highly dependent on DHODH and suggesting that this enzyme is a strategic target for cancer therapy (245). Recently, DHODH was also shown to protect against mitochondrial ferroptosis by preventing the lipid peroxidation that triggers this phenomenon (244). Notably, cancer cells exhibit low levels of glutathione peroxidase 4 (GPX4) and inhibition of DHODH hinders respiration, boosts glycolysis and enhances GLUT4 translocation to the plasma membrane (246). This is further supported by the activation of the tumor suppressor p53, which elevates the levels of GDF15/MIC1 (another candidate listed in Table 4), a cytokine known for its appetite-reducing effects and ability to extend lifespan. DHODH inhibition that depletes pyrimidine ribonucleotides is also thought to be responsible for reduced RNA virus replication and decelerated growth in rapidly dividing cells, such as activated T cells and, as just mentioned, cancer cells (243). Interestingly, uridine, a pyrimidine nucleoside present in RNA, has been shown to modulate insulin activity and glycogen synthesis through its interaction with uridine diphosphate (UDP)-glucose (247). The base sequence of the DHODH gene is remarkably conserved, with one exception being a prevalent Lys7Gln missense polymorphism (16-72008783-A-C, rs3213422) found in its first exon (248). This variant is found in 75% of individuals in East Asia vs. 34% of individuals in the Middle East (gnomAD) and has been linked with drug (leflunomide) response to rheumatoid arthritis (248–250).
3.3 Population-enriched variants with unknown/uncharacterized function
No known effect on gross phenotype or evidence of association with disease has yet been reported among the population-enriched variants identified with the 20 genes listed in Table 6. However, a newly released resource, GWAS Central (457), was accessed to provide phenotype associations with a subset of variants in Table 6. Further, disease disparities related to the parent gene and/or other variants of the gene were identified and/or the predicted impact of a population-enriched variant on the coded change in protein function were evaluated and listed in Table 6.
3.3.1 Understudied genes SIPA1L2 and TVP23C
Among the 20 genes in Table 6, six of these remain understudied, including the exosomal CCDC105/TEKTL1, the putative protein disulfide isomerase CRELD2, the FAM131C protein with unknown function, the putative immune checkpoint ITPRIPL1 membrane protein, the presumptive neural GTPase activator SIPA1L2, and the putative vesicular protein transporter TVP23C. Notably, evidence of an impact on function does exist for one of two population-enriched variants of SIPA1L2 and one of three population-enriched variants of TVP23C. In the case of SIPA1L2, both characterized and uncharacterized variants occur at the same high frequency (MAF = 0.48) in East Asians, but Gly1639Ser increases the number of potential phosphorylation sites, whereas Thr1322Ala reduces them, which may result in different functional outcomes (e.g. changes in activation status and/or protein-protein interactions). In both SIPA1L2 variants, eight of nine possible transcripts code for missense mutations, whereas with TVP23C, only in the canonical transcript does the variant result in a missense mutation among five (Ser256Arg) or twelve (Trp202Arg and Ser199Thr) possible isoforms, some of which are read-through fusions with CDRT4 (CMT1A Duplicated Region Transcript 4). It is likely that the TVP23C Trp202Arg and Ser199Thr variants commonly co-occur, given their proximity to one another on the gene and their matching frequency distribution, as both have MAFs that range from 0.54 in East Asians to 0.28 in South Asians. Thus, one might speculate that the unknown functional impact of Ser199Thr matches that of Trp202Arg, which was found in a choriocarcinoma patient (458). Notably, choriocarcinoma shows a geographical disparity as it occurs at a ten-fold greater frequency in Southeast Asia than in the West (reviewed in 439). The third TVP23C variant Ser256Arg is most common among Africans/African Americans (MAF = 0.24) and involves the loss of a potential phosphorylation site about 50 amino acid residues downstream of the other two TVP23C variants.
3.3.2 Additional representative genes of interest
The remaining 14 genes in Table 6 are better characterized; notably, many have pleiotropic functions beyond the functions initially attributed to them. ATPase Phospholipid Transporting 10D (ATP10D) codes for the catalytic subunit of a glycoslyceramide flippase complex at the endoplasmic reticulum (ER), nucleoplasm, and plasma membrane. DnaJ Heat Shock Protein Family (Hsp40) Member B11 (DNAJB11) codes for an ER-resident and secreted co-chaperone of BiP/GRP78/HSPA5. Desmocollin 1 (DSC1) codes for an adhesive glycoprotein cadherin family member. The Immunoglobulin Like Domain Containing Receptor 1 protein (ILDR1) maintains structural barriers in epithelia and auditory neurosensory hair cells (459), mediates fatty acid and lipoprotein-stimulated cholecystokinin secretion in the small intestine (460), regulates water homeostasis in kidney (461), and interferes with phospholipid scramblase (PLSCR1) anti-viral activity (374). Protein Disulfide Isomerase Family A Member 6 (PDIA6) inhibits intracellular aggregation of misfolded proteins and extracellular aggregation of platelets (381). Replication Protein A Interacting Protein (RPAIN) participates in DNA metabolism, nuclear import, and response to UV light. The Semaphorin 6D (SEMA6D) gene codes for an integral membrane protein member of the semaphorin family whose members collectively sculpt axonal paths, branches, conduction, and target selection; the distribution of nine SEMA6D transcript isoforms varies according to developmental stage and tissue type. Tre-2/BUB2/CDC16 (TBC) Domain Family Member 4 (TBC1D4, also referred to as Akt Substrate of 160 kD or AS160) is a Rab-GTPase activator with multiple transcript variants; isoform 2 promotes SLC2A4/GLUT4 presentation at the plasma membrane to increase cellular glucose uptake (344). Thymocyte Expressed, Positive Selection Associated 1 (TESPA1) interacts with COP9 and TCR signalsomes and participates in T cell differentiation and T cell receptor signaling. Three zinc finger (ZNF) proteins ZNF23, ZNF267, and ZNF628 localize to the nucleus and regulate transcription. Parent genes and the corresponding population-enriched variants of the common cytokine receptor beta chain CSF2RB and the transcription co-repressor RB1 are both discussed below.
3.3.2.1 CSF2RB
Colony stimulating factor 2 receptor beta (CSF2RB, CD131) forms dimers with the alpha receptor subunits for cytokines IL-3, IL-5, and GM-CSF (CSF2). As noted above, a population-enriched variant of the IL3RA subunit also exists, although the population distributions of these two variants are very different: the Val323Leu IL3RA variant is found least frequently among Africans/African Americans (MAF = 0.06, Table 4), whereas the Glu249Gln CSF2RB variant is more predominant in Africans/African Americans than any other population (MAF = 0.21).
CSF2RB is associated with pulmonary alveolar proteinosis (PAP), which involves the accumulation of surfactant and macrophage dysfunction in alveoli (reviewed in 462). Although studies so far have not suggested geographic or population differences in PAP occurrence, the most common PAP co-morbidities include cardiovascular disease, type 2 diabetes, and hypertension, all of which are unevenly distributed among populations. Further, a rare Arg461Cys CSF2RB variant (MAF< 0.001, not listed in Table 6) was found in individual patients with leukemia (355) and breast cancer (356). Notably, both of these cancers show racial and ethnic disparities [430 and 352 respectively].
3.3.2.2 RB1
Retinoblastoma (RB1) was one of the first tumor suppressors to be identified. Alterations in the expression and sequence of the RB1 gene have been implicated in several cancers besides retinoblastoma where they were originally characterized (reviewed in 391). More than 40 years of extensive research indicates that regulation of and by RB1 is highly complex, linked with multiple signaling pathways, and varies with context. Not surprisingly, the number of proteins shown to interact with RB1 is more than 30 as curated by UniProt (344) and more than 150 as curated in BioGRID (463) and IntAct (464). The functional diversity of the binding partners of RB1 is consistent with its pleiotropic effects, which extend beyond transcription and cell cycle control to include progenitor maturation, terminal differentiation, and immune evasion (391).
Five protein coding transcripts of RB1 have been identified. These include 1) the MANE select (canonical) protein composed of 27 exons encoding a total of 928 aa residues; 2) a closely related transcript that is 5 as shorter and differs from the canonical protein by 18 of its last 19 C-terminal residues; and 3) three much shorter transcripts (coding for 53, 103 or 110 aa peptides) which include all or portions of only 2 or 3 exons of the canonical protein. Of these shorter transcripts, the two shortest are derived from the N-terminal portion of RB1. In contrast, the 110 aa non-canonical transcript codes for an unidentified N-terminal residue equivalent to the Ser501 residue of the canonical protein and then aligns with all canonical residues up through Ser565; the remaining non-canonical aa residues 66-110 are located downstream of the canonical C-terminal residue 928. It is in this extra-exonic portion of the non-canonical 110 aa RB1 isoform that the Leu99Ser population-enriched variant, which introduces a potential phosphorylation site, is found. In spite of the high number of aa residues (n ≥ 105) in the canonical RB1 protein that are known to be post-translationally modified, within the aa 501-565 residue range that overlaps with the first 65 residues of the 110 aa isoforms, only two potential ubiquitination sites have been identified in the vicinity of aa 550) (391).
4 Conclusion
Population studies have traditionally focused on querying individual diseases or combinations of diseases, including cancer and cardio-metabolic disease, which frequently show disparate prevalence and/or severity in non-European populations. In this perspective, we have introduced a complementary approach that explores the intersection of innate immunity, cancer, and cardio-metabolic diseases. The effective elimination of disease disparities will involve not only addressing the profound social and behavioral determinants of health, but also identifying and treating the biological contributors of disease that include novel genes as well as previously characterized genes that participate in novel pathways.
We suggest that careful evaluation of population differences in conventional and unconventional innate immune genes and their related pathways will provide key insights into the underlying mechanisms that connect cancer and cardio-metabolic diseases. At the same time, the genes we have identified in this study that are associated with both cancer and cardio-metabolic diseases may play critical roles in under-appreciated facets of innate immunity and their contribution to disease disparities. Further, we predict that the geographic ancestral distribution of innate immune gene variants will match the geographical distribution of the environmental stressors (including but not limited to infectious agents) that they are designed to mitigate as described above for HbS and DARC variants with malaria (Section 1.3).
The genes we have identified serve as potential targets for diagnostics and/or therapeutic interventions. Notably, the development and clinical use of therapeutics targeting these candidate genes is likely to require a nuanced approach since variations in these genes across different global populations are likely to alter the activity and/or expression of their coded proteins, with the subsequent potential to impact therapeutic outcomes. Assessing the prevalence of specific target variants in one or more major populations and, more precisely, the presence of these specific target variants in individuals is a consequential step towards increasing the safety and effectiveness of emerging therapies. This perspective highlights the importance of 1) considering genetic diversity in identifying and developing treatments and 2) continuing to incorporate ongoing GWAS projects as they identify and characterize new or understudied genes and their population-enriched variants associated with complex and infectious diseases.
Author contributions
SY: Conceptualization, Data curation, Writing – original draft. DH: Data curation, Writing – original draft. KW: Data curation, Writing – original draft. NL: Writing – review & editing. KSK: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors are grateful for NIH support of the NCCU JLC-BBRI RCMI (U54MD007602), Morehouse School of Medicine (U54MD007602) and the Director’s Transformative R01 (R01MD017405) grants, both to KSK.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Alvidrez J, Castille D, Laude-Sharp M, Rosario A, Tabor D. The national institute on minority health and health disparities research framework. Am J Public Health (2019) 109(S1):S16–20. doi: 10.2105/AJPH.2018.304883
2. Lutz R. Health Disparities Among African-Americans (2022). Available at: https://www.pfizer.com/news/articles/health_disparities_among_african_americans.
3. Deshmukh SK, Azim S, Ahmad A, Zubair H, Tyagi N, Srivastava SK, et al. Biological basis of cancer health disparities: resources and challenges for research. Am J Cancer Res (2017) 7(1):1–12.
4. Zhang R, Sun J, Wang C, Wang X, Zhao P, Yuan Y, et al. The racial disparities in the epidemic of metabolic syndrome with increased age: A study from 28,049 Chinese and American adults. Front Public Health (2021) 9:797183. doi: 10.3389/fpubh.2021.797183
5. Cullen MR, Lemeshow AR, Russo LJ, Barnes DM, Ababio Y, Habtezion A. Disease-specific health disparities: A targeted review focusing on race and ethnicity. Healthc (Basel) (2022) 10(4):603. doi: 10.3390/healthcare10040603
6. American Cancer Society (ACS). Cancer Facts & Figures 2022. Atlanta: American Cancer Society (2022).
8. Pham C, Fong TL, Zhang J, Liu L. Striking racial/ethnic disparities in liver cancer incidence rates and temporal trends in California 1988-2012. J Natl Cancer Inst (2018) 110(11):1259–69. doi: 10.1093/jnci/djy051
9. Zhang CH, Cheng Y, Zhang S, Fan J, Gao Q. Changing epidemiology of hepatocellular carcinoma in Asia. Liver Int (2022) 42(9):2029–41. doi: 10.1111/liv.15251
10. Salvatore M, Jeon J, Meza R. Changing trends in liver cancer incidence by race/ethnicity and sex in the US: 1992-2016. Cancer Causes Control (2019) 30(12):1377–88. doi: 10.1007/s10552-019-01237-4
11. Ellington T, Miller J, Henley S, Wilson R, Wu M, Richardson L. Trends in breast cancer incidence, by race, ethnicity, and age among women aged ≥20 years — United States 1999–2018. Morbidity Mortality Weekly Rep (MMWR) (2022). doi: 10.15585/mmwr.mm7102a2
12. Hashim D, Boffetta P, La Vecchia C, Rota M, Bertuccio P, Malvezzi M, et al. The global decrease in cancer mortality: trends and disparities. Ann Oncol (2016) 27(5):926–33. doi: 10.1093/annonc/mdw027
13. Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat (2009) 30(1):69–78. doi: 10.1002/humu.20822
15. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature (2020) 581(7809):434–43. doi: 10.1038/s41586-020-2308-7
16. National Academies of Sciences, Engineering, and Medicine, Division of Behavioral and Social Sciences and Education, Health and Medicine Division, Committee on Population, Board on Health Sciences Policy, Committee on the Use of Race, Ethnicity, and Ancestry as Population Descriptors in Genomics Research. 1, Population Descriptors in Human Genetics Research: Genesis, Evolution, and Challenges. In: Using Population Descriptors in Genetics and Genomics Research: A New Framework for an Evolving Field. Washington (DC: National Academies Press (US (2023). Available at: https://www.ncbi.nlm.nih.gov/books/NBK592850/.
17. Olden K, White SL. Health-related disparities: influence of environmental factors. Med Clinics North America (2005) 89(4):721–38. doi: 10.1016/j.mcna.2005.02.001
18. Tung EL, Cagney KA, Peek ME, Chin MH. Spatial context and health inequity: reconfiguring race, place, and poverty. J Urban Health (2017) 94(6):757–63. doi: 10.1007/s11524-017-0210-x
19. Ruiz D, Becerra M, Jagai JS, Ard K, Sargis RM. Disparities in environmental exposures to endocrine-disrupting chemicals and diabetes risk in vulnerable populations. Diabetes Care (2018) 41:193–205. doi: 10.2337/dc16-2765
20. Vitlic A, Lord JM, Phillips AC. Stress, ageing and their influence on functional, cellular and molecular aspects of the immune system. Age (Dordr) (2014) 36(3):9631. doi: 10.1007/s11357-014-9631-6
21. Borrell LN, Rodriguez-Alvarez E, Dallo FJ. Racial/ethnic inequities in the associations of allostatic load with all-cause and cardiovascular-specific mortality risk in US adults. PloS One (2020) 15(2):e0228336. doi: 10.1371/journal.pone.0228336
22. Van Dyke ME, Baumhofer NK, Slopen N, Mujahid MS, Clark CR, Williams DR, et al. Pervasive discrimination and allostatic load in African American and white adults. Psychosom Med (2020) 82(3):316–23. doi: 10.1097/PSY.0000000000000788
23. Miller HN, LaFave S, Marineau L, Stephens J, Thorpe RJ. The impact of discrimination on allostatic load in adults: An integrative review of literature. J Psychosomatic Res (2021) 146:110434. doi: 10.1016/j.jpsychores.2021.110434
24. Fine MJ, Ibrahim SA, Thomas SB. The role of race and genetics in health disparities research. Am J Public Health (2005) 95(12):2125–8. doi: 10.2105/AJPH.2005.076588
25. Scharff DP, Mathews KJ, Jackson P, Hoffsuemmer J, Martin E, Edwards D. More than Tuskegee: understanding mistrust about research participation. J Health Care Poor Underserved (2010) 21(3):879–97. doi: 10.1353/hpu.0.0323
26. 2022 National Healthcare Quality and Disparities Report. Rockville, MD: Agency for Healthcare Research and Quality (2022). Available at: https://www.ahrq.gov/research/findings/nhqrdr/nhqdr22/index.html.
27. Dutil J, Chen Z, Monteiro AN, Teer JK, Eschrich SA. An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines. Cancer Res (2019) 79(7):1263–73. doi: 10.1158/0008-5472.CAN-18-2747
28. Hooker SE Jr., Woods-Burnham L, Bathina M, Lloyd S, Gorjala P, Mitra R, et al. Genetic ancestry analysis reveals misclassification of commonly used cancer cell lines. Cancer Epidemiol Biomarkers Prev (2019) 28(6):1003–9. doi: 10.1158/1055-9965.EPI-18-1132
29. Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell (2019) 177(1):26–31. doi: 10.1016/j.cell.2019.02.048
30. Cavazzoni P, Anagnostiadis E, Lolic M. 2020 Drug Trials Snapshots Summary Report. Center for Drug Evaluation and Research (CDER), editor. Silver Spring, MD: Administration USFaD (20b21). Available at: www.fda.gov.
31. Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature (2016) 538:161–4. doi: 10.1038/538161a
32. Rajman I, Knapp L, Morgan T, Masimirembwa C. African genetic diversity: implications for cytochrome P450-mediated drug metabolism and drug development. EBioMedicine (2017) 17:67–74. doi: 10.1016/j.ebiom.2017.02.017
33. Denny JC, Collins FS. Precision medicine in 2030-seven ways to transform healthcare. Cell (2021) 184(6):1415–9. doi: 10.1016/j.cell.2021.01.015
34. Collins FS, Adams AB, Aklin C, Archer TK, Bernard MA, Boone E, et al. Affirming NIH’s commitment to addressing structural racism in the biomedical research enterprise. Cell (2021) 184(12):3075–9. doi: 10.1016/j.cell.2021.05.014
35. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature (2016) 536(7616):285–91. doi: 10.1038/nature19057
36. Witherspoon DJ, Wooding S, Rogers AR, Marchani EE, Watkins WS, Batzer MA, et al. Genetic similarities within and between human populations. Genetics (2007) 176(1):351–9. doi: 10.1534/genetics.106.067355
37. Ahsan T, Urmi NJ, Sajib AA. Heterogeneity in the distribution of 159 drug-response related SNPs in world populations and their genetic relatedness. PloS One (2020) 15(1):e0228000. doi: 10.1371/journal.pone.0228000
38. Burroughs VJ, Maxey RW, Levy RA. Racial and ethnic differences in response to medicines: towards individualized pharmaceutical treatment. J Natl Med Assoc (2002) 94(10 Suppl):1–26.
39. Johnson JA. Ethnic differences in cardiovascular drug response: potential contribution of pharmacogenetics. Circulation (2008) 118(13):1383–93. doi: 10.1161/CIRCULATIONAHA.107.704023
40. De T, Park CS, Perera MA. Cardiovascular pharmacogenomics: does it matter if you’re black or white? Annu Rev Pharmacol Toxicol (2019) 59:577–603. doi: 10.1146/annurev-pharmtox-010818-021154
41. Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet (2005) 77(2):171–92. doi: 10.1086/432519
42. Sirugo G, Hennig BJ, Adeyemo AA, Matimba A, Newport MJ, Ibrahim ME, et al. Genetic studies of African populations: an overview on disease susceptibility and response to vaccines and therapeutics. Hum Genet (2008) 123(6):557–98. doi: 10.1007/s00439-008-0511-y
43. Wang J, Ou ZL, Hou YF, Luo JM, Shen ZZ, Ding J, et al. Enhanced expression of Duffy antigen receptor for chemokines by breast cancer cells attenuates growth and metastasis potential. Oncogene (2006) 25(54):7201–11. doi: 10.1038/sj.onc.1209703
44. Mangano VD, Modiano D. An evolutionary perspective of how infection drives human genome diversity: the case of malaria. Curr Opin Immunol (2014) 30:39–47. doi: 10.1016/j.coi.2014.06.004
45. Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, et al. Association of trypanolytic apoL1 variants with kidney disease in African-Americans. Science (2010) 329(5993):841–5. doi: 10.1126/science.1193032
46. Freedman BI, Limou S, Ma L, Kopp JB. APOL1-associated nephropathy: A key contributor to racial disparities in CKD. Am J Kidney Dis (2018) 72(5 Suppl 1):S8–S16. doi: 10.1053/j.ajkd.2018.06.020
47. Quintana-Murci L. Human immunology through the lens of evolutionary genetics. Cell (2019) 177(1):184–99. doi: 10.1016/j.cell.2019.02.033
48. Deschamps M, Laval G, Fagny M, Itan Y, Abel L, Casanova JL, et al. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am J Hum Genet (2016) 98(1):5–21. doi: 10.1016/j.ajhg.2015.11.014
49. Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admetlla A, Pattini L, Nielsen R. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PloS Genet (2011) 7(11):e1002355. doi: 10.1371/journal.pgen.1002355
50. Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet (2010) 11(1):17–30. doi: 10.1038/nrg2698
51. Piel FB, Patil AP, Howes RE, Nyangiri OA, Gething PW, Williams TN, et al. Global distribution of the sickle cell gene and geographical confirmation of the malaria hypothesis. Nat Commun (2010) 1:104. doi: 10.1038/ncomms1104
52. Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, et al. The global distribution of the Duffy blood group. Nat Commun (2011) 2:266. doi: 10.1038/ncomms1265
53. Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nat Rev Genet (2014) 15(6):379–93. doi: 10.1038/nrg3734
54. Nedelec Y, Sanz J, Baharian G, Szpiech ZA, Pacis A, Dumaine A, et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell (2016) 167(3):657–669 e621. doi: 10.1016/j.cell.2016.09.025
55. Nahid P, Jarlsberg LG, Kato-Maeda M, Segal MR, Osmond DH, Gagneux S, et al. Interplay of strain and race/ethnicity in the innate immune response to M. tuberculosis. PloS One (2018) 13(5):e0195392. doi: 10.1371/journal.pone.0195392
56. Yao S, Hong CC, Ruiz-Narvaez EA, Evans SS, Zhu Q, Schaefer BA, et al. Genetic ancestry and population differences in levels of inflammatory cytokines in women: Role for evolutionary selection and environmental factors. PloS Genet (2018) 14(6):e1007368. doi: 10.1371/journal.pgen.1007368
57. Li Y, Oosting M, Smeekens SP, Jaeger M, Aguirre-Gamboa R, Le KTT, et al. A functional genomics approach to understand variation in cytokine production in humans. Cell (2016) 167(4):1099–1110 e1014. doi: 10.1016/j.cell.2016.10.017
58. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
59. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discovery (2022) 12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059
60. Yeyeodu ST, Kidd LR, Kimbro KS. Protective innate immune variants in racial/ethnic disparities of breast and prostate cancer. Cancer Immunol Res (2019) 7(9):1384–9. doi: 10.1158/2326-6066.CIR-18-0564
61. Chen CH, Lu YS, Cheng AL, Huang CS, Kuo WH, Wang MY, et al. Disparity in tumor immune microenvironment of breast cancer and prognostic impact: Asian versus Western populations. Oncologist (2020) 25(1):e16–23. doi: 10.1634/theoncologist.2019-0123
62. Zajac KK, Malla S, Babu RJ, Raman D, Tiwari AK. Ethnic disparities in the immune microenvironment of triple negative breast cancer and its role in therapeutic outcomes. Cancer Rep (2023) Suppl 1(Suppl 1):e1779. doi: 10.1002/cnr2.1779
63. Chaudhary S, Ganguly K, Muniyan S, Pothuraju R, Sayed Z, Jones DT, et al. Immunometabolic alterations by HPV infection: new dimensions to head and neck cancer disparity. JNCI: J Natl Cancer Institute (2019) 111(3):233–44. doi: 10.1093/jnci/djy207
64. Chaudhary S, Dam V, Ganguly K, Sharma S, Atri P, Chirravuri-Venkata R, et al. Differential mutation spectrum and immune landscape in African Americans versus Whites: A possible determinant to health disparity in head and neck cancer. Cancer Lett (2020) 492:44–53. doi: 10.1016/j.canlet.2020.07.029
65. Guerrero-Preston R, Lawson F, Rodriguez-Torres S, Noordhuis MG, Pirini F, Manuel L, et al. JAK3 variant, immune signatures, DNA methylation, and social determinants linked to survival racial disparities in head and neck cancer patients. Molecular disparities in black and non-latino white HNSCC. Cancer Prev Res (2019) 12(4):255–70. doi: 10.1158/1940-6207.CAPR-17-0356
66. Byrne CA, Gomez SL, Kim S, Oddo VM, Koh TJ, Fantuzzi G. Disparities in inflammation between non-Hispanic black and white individuals with lung cancer in the Greater Chicago Metropolitan area. Front Immunol (2022) 13:1008674. doi: 10.3389/fimmu.2022.1008674
67. Xu Y, Zhang L, Thaiparambil J, Mai S, Perera DN, Zhang J, et al. Patients with lung cancer of different racial backgrounds harbor distinct immune cell profiles. Cancer Res Commun (2022) 2(8):884–93. doi: 10.1158/2767-9764.CRC-22-0057
68. Curran T, Sun Z, Gerry B, Findlay VJ, Wallace K, Li Z, et al. Differential immune signatures in the tumor microenvironment are associated with colon cancer racial disparities. Cancer Med (2021) 10(5):1805–14. doi: 10.1002/cam4.3753
69. Ahmad S, Ashktorab H, Brim H, Housseau F. Inflammation, microbiome and colorectal cancer disparity in African-Americans: Are there bugs in the genetics? World J Gastroenterol (2022) 28(25):2782. doi: 10.3748/wjg.v28.i25.2782
70. Kiely M, Lord B, Ambs S. Immune response and inflammation in cancer health disparities. Trends Cancer (2022) 8(4):316–27. doi: 10.1016/j.trecan.2021.11.010
71. Kalyanaraman B. Exploiting the tumor immune microenvironment and immunometabolism using mitochondria-targeted drugs: Challenges and opportunities in racial disparity and cancer outcome research. FASEB J (2022) 36(4):e22226. doi: 10.1096/fj.202101862R
72. Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol (2018) 15(1):11–20. doi: 10.1038/nrgastro.2017.109
73. Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res Clin Pract (2019) 157:107843. doi: 10.1016/j.diabres.2019.107843
74. Malik VS, Willet WC, Hu FB. Nearly a decade on - trends, risk factors and policy implications in global obesity. Nat Rev Endocrinol (2020) 16(11):615–6. doi: 10.1038/s41574-020-00411-y
75. Huang PL. A comprehensive definition for metabolic syndrome. Dis Model Mech (2009) 2(5-6):231–7. doi: 10.1242/dmm.001180
76. Rochlani Y, Pothineni NV, Kovelamudi S, Mehta JL. Metabolic syndrome: pathophysiology, management, and modulation by natural compounds. Ther Adv Cardiovasc Dis (2017) 11(8):215–25. doi: 10.1177/1753944717711379
77. Godoy-Matos AF, Silva Junior WS, Valerio CM. NAFLD as a continuum: from obesity to metabolic syndrome and diabetes. Diabetol Metab Syndr (2020) 12:60. doi: 10.1186/s13098-020-00570-y
78. Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation (2005) 112(17):2735–52. doi: 10.1161/CIRCULATIONAHA.105.169404
79. Chan KL, Cathomas F, Russo SJ. Central and peripheral inflammation link metabolic syndrome and major depressive disorder. Physiol (Bethesda Md.) (2019) 34(2):123–33. doi: 10.1152/physiol.00047.2018
80. Akinyemiju T, Do AN, Patki A, Aslibekyan S, Zhi D, Hidalgo B, et al. Epigenome-wide association study of metabolic syndrome in African-American adults. Clin Epigenet (2018) 10:49. doi: 10.1186/s13148-018-0483-2
81. Al Rifai M, Silverman MG, Nasir K, Budoff MJ, Blankstein R, Szklo M, et al. The association of nonalcoholic fatty liver disease, obesity, and metabolic syndrome, with systemic inflammation and subclinical atherosclerosis: The Multi-Ethnic Study of Atherosclerosis (MESA). Atherosclerosis (2015) 239(2):629–33. doi: 10.1016/j.atherosclerosis.2015.02.011
82. Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature (2017) 542(7640):177–85. doi: 10.1038/nature21363
83. Denis GV, Sebastiani P, Bertrand KA, Strissel KJ, Tran AH, Slama J, et al. Inflammatory signatures distinguish metabolic health in African American women with obesity. PloS One (2018) 13(5):e0196755. doi: 10.1371/journal.pone.0196755
84. Berbudi A, Rahmadika N, Tjahjadi AI, Ruslami R. Type 2 diabetes and its impact on the immune system. Curr Diabetes Rev (2020) 16(5):442–9. doi: 10.2174/1573399815666191024085838
85. Brestoff JR, Artis D. Immune regulation of metabolic homeostasis in health and disease. Cell (2015) 161(1):146–60. doi: 10.1016/j.cell.2015.02.022
86. Adolph TE, Grander C, Grabherr F, Tilg H. Adipokines and non-alcoholic fatty liver disease: multiple interactions. Int J Mol Sci (2017) 18(8):1649. doi: 10.3390/ijms18081649
87. Zatterale F, Longo M, Naderi J, Raciti GA, Desiderio A, Miele C, et al. Chronic adipose tissue inflammation linking obesity to insulin resistance and type 2 diabetes. Front Physiol (2019) 10:1607. doi: 10.3389/fphys.2019.01607
88. Wu H, Ballantyne CM. Metabolic inflammation and insulin resistance in obesity. Circ Res (2020) 126(11):1549–64. doi: 10.1161/CIRCRESAHA.119.315896
89. Coats BR, Schoenfelt KQ, Barbosa-Lorenzi VC, Peris E, Cui C, Hoffman A, et al. Metabolically activated adipose tissue macrophages perform detrimental and beneficial functions during diet-induced obesity. Cell Rep (2017) 20(13):3149–61. doi: 10.1016/j.celrep.2017.08.096
90. Li Y, Yun K, Mu R. A review on the biology and properties of adipose tissue macrophages involved in adipose tissue physiological and pathophysiological processes. Lipids Health Dis (2020) 19(1):164. doi: 10.1186/s12944-020-01342-3
91. Tong X, Wei L, Wang T, Han R. Remodeling of Macrophages in White Adipose Tissue under the Conditions of Obesity as well as Lipolysis. Oxid Med Cell Longev (2021) 2021:9980877. doi: 10.1155/2021/9980877
93. Robinson MW, Harmon C, O’Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell Mol Immunol (2016) 13(3):267–76. doi: 10.1038/cmi.2016.3
94. Kubes P, Jenne C. Immune responses in the liver. Annu Rev Immunol (2018) 36:247–77. doi: 10.1146/annurev-immunol-051116-052415
95. Gonzalez FB, Villar SR, Toneatto J, Pacini MF, Marquez J, D’Attilio L, et al. Immune response triggered by Trypanosoma cruzi infection strikes adipose tissue homeostasis altering lipid storage, enzyme profile and adipokine expression. Med Microbiol Immunol (2019) 208(5):651–66. doi: 10.1007/s00430-018-0572-z
96. Fiorucci S, Biagioli M, Zampella A, Distrutti E. Bile acids activated receptors regulate innate immunity. Front Immunol (2018) 9:1853. doi: 10.3389/fimmu.2018.01853
97. Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity (2016) 45(4):802–16. doi: 10.1016/j.immuni.2016.09.008
98. Chen ML, Takeda K, Sundrud MS. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol (2019) 12(4):851–61. doi: 10.1038/s41385-019-0162-4
99. Camilleri M. Bile acid detergency: permeability, inflammation, and effects of sulfation. Am J Physiol Gastrointest Liver Physiol (2022) 322(5):G480–8. doi: 10.1152/ajpgi.00011.2022
100. Chang YW, Hung LC, Chen YC, Wang WH, Lin CY, Tzeng HH, et al. Insulin reduces inflammation by regulating the activation of the NLRP3 inflammasome. Front Immunol (2020) 11:587229. doi: 10.3389/fimmu.2020.587229
101. Jin C, Henao-Mejia J, Flavell RA. Innate immune receptors: key regulators of metabolic disease progression. Cell Metab (2013) 17(6):873–82. doi: 10.1016/j.cmet.2013.05.011
102. Burhans MS, Hagman DK, Kuzma JN, Schmidt KA, Kratz M. Contribution of adipose tissue inflammation to the development of type 2 diabetes mellitus. Compr Physiol (2018) 9(1):1–58. doi: 10.1002/cphy.c170040
103. Bernardi S, Marcuzzi A, Piscianz E, Tommasini A, Fabris B. The complex interplay between lipids, immune system and interleukins in cardio-metabolic diseases. Int J Mol Sci (2018) 19(12):4058. doi: 10.3390/ijms19124058
104. Pirillo A, Bonacina F, Norata GD, Catapano AL. The interplay of lipids, lipoproteins, and immunity in atherosclerosis. Curr Atheroscler Rep (2018) 20(3):12. doi: 10.1007/s11883-018-0715-0
105. Prohaska A, Racimo F, Schork AJ, Sikora M, Stern AJ, Ilardo M, et al. Human disease variation in the light of population genomics. Cell (2019) 177(1):115–31. doi: 10.1016/j.cell.2019.01.052
106. Uffelmann E, Huang QQ, Munung NS, de Vries J, Okada Y, Martin AR, et al. Genome-wide association studies. Nat Rev Methods Primers (2021) 1(1):59. doi: 10.1038/s43586-021-00056-9
107. Gudmundsson S, Singer-Berk M, Watts NA, Phu W, Goodrich JK, Solomonson M, et al. Variant interpretation using population databases: Lessons from gnomAD. Hum Mutat (2022) 43(8):1012–30. doi: 10.1002/humu.24309
108. Pennisi E. ENCODE project writes eulogy for junk DNA. Science (2012) 337(6099):1159–61. doi: 10.1126/science.337.6099.115
109. Salzberg SL. Open questions: How many genes do we have? BMC Biol (2018) 16(1):94. doi: 10.1186/s12915-018-0564-x
110. Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science (2012) 337(6099):1190–5. doi: 10.1126/science.1222794
111. Sharma Y, Miladi M, Dukare S, Boulay K, Caudron-Herger M, Gross M, et al. A pan-cancer analysis of synonymous mutations. Nat Commun (2019) 10(1):2569. doi: 10.1038/s41467-019-10489-2
112. Walsh IM, Bowman MA, Soto Santarriaga IF, Rodriguez A, Clark PL. Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc Natl Acad Sci U.S.A. (2020) 117(7):3528–34. doi: 10.1073/pnas.1907126117
113. Herreros E, Janssens X, Pepe D, Keersmaecker KD. SNPs ability to influence disease risk: breaking the silence on synonymous mutations in cancer. In: Single Nucleotide Polymorphisms. (Cham, Switzerland: Springer) (2022). p. 77–96.
114. Velloso FJ, Campos AR, Sogayar MC, Correa RG. Proteome profiling of triple negative breast cancer cells overexpressing NOD1 and NOD2 receptors unveils molecular signatures of Malignant cell proliferation. BMC Genomics (2019) 20(1):152. doi: 10.1186/s12864-019-5523-6
115. Mackinnon MJ, Ndila C, Uyoga S, Macharia A, Snow RW, Band G, et al. Environmental correlation analysis for genes associated with protection against malaria. Mol Biol Evol (2016) 33(5):1188–204. doi: 10.1093/molbev/msw004
116. Liang B, Ding H, Huang L, Luo H, Zhu X. GWAS in cancer: progress and challenges. Mol Genet Genomics (2020) 295(3):537–61. doi: 10.1007/s00438-020-01647-z
117. Nazarian A, Arbeev KG, Yashkin AP, Kulminski AM. Genome-wide analysis of genetic predisposition to common polygenic cancers. J Appl Genet (2022) 63(2):315–25. doi: 10.1007/s13353-021-00679-4
118. Anstee QM, Darlay R, Cockell S, Meroni M, Govaere O, Tiniakos D, et al. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort(☆). J Hepatol (2020) 73(3):505–15. doi: 10.1016/j.jhep.2020.04.003
119. Liu GM, Zeng HD, Zhang CY, Xu JW. Key genes associated with diabetes mellitus and hepatocellular carcinoma. Pathol Res Pract (2019) 215(11):152510. doi: 10.1016/j.prp.2019.152510
120. Si Y, Liu X, Cheng M, Wang M, Gong Q, Yang Y, et al. Growth differentiation factor 15 is induced by hepatitis C virus infection and regulates hepatocellular carcinoma-related genes. PloS One (2011) 6(5):e19967. doi: 10.1371/journal.pone.0019967
121. Emmerson PJ, Duffin KL, Chintharlapalli S, Wu X. GDF15 and growth control. Front Physiol (2018) 9:1712. doi: 10.3389/fphys.2018.01712
122. Luan HH, Wang A, Hilliard BK, Carvalho F, Rosen CE, Ahasic AM, et al. GDF15 is an inflammation-induced central mediator of tissue tolerance. Cell (2019) 178(5):1231–1244 e1211. doi: 10.1016/j.cell.2019.07.033
123. de Jager SC, Bermudez B, Bot I, Koenen RR, Bot M, Kavelaars A, et al. Growth differentiation factor 15 deficiency protects against atherosclerosis by attenuating CCR2-mediated macrophage chemotaxis. J Exp Med (2011) 208(2):217–25. doi: 10.1084/jem.20100370
124. Xu W-D, Huang Q, Yang C, Li R, Huang A-F. GDF-15: A potential biomarker and therapeutic target in systemic lupus erythematosus. Front Immunol (2022) 13:926373. doi: 10.3389/fimmu.2022.926373
125. Prendergast GC, Metz R, Muller AJ, Merlo LM, Mandik-Nayak L. IDO2 in immunomodulation and autoimmune disease. Front Immunol (2014) 5:585. doi: 10.3389/fimmu.2014.00585
126. Li P, Xu W, Liu F, Zhu H, Zhang L, Ding Z, et al. The emerging roles of IDO2 in cancer and its potential as a therapeutic target. Biomed Pharmacother (2021) 137:111295. doi: 10.1016/j.biopha.2021.111295
127. Cutler JA, Rush AJ, McMahon FJ, Laje G. Common genetic variation in the indoleamine-2,3-dioxygenase genes and antidepressant treatment outcome in major depressive disorder. J Psychopharmacol (2012) 26(3):360–7. doi: 10.1177/0269881111434622
128. Kasamatsu T, Hashimoto N, Sakaya N, Awata-Shiraiwa M, Ishihara R, Murakami Y, et al. IDO2 rs10109853 polymorphism affects the susceptibility to multiple myeloma. Clin Exp Med (2021) 21:323–9. doi: 10.1007/s10238-020-00681-w
129. Guba SC, Stella G, Turka LA, June CH, Thompson CB, Emerson SG. Regulation of interleukin 3 gene induction in normal human T cells. J Clin Invest (1989) 84(6):1701–6. doi: 10.1172/jci114352
130. Hong L, Tang Y, Pan S, Xu M, Shi Y, Gao S, et al. Interleukin 3-induced GITR promotes the activation of human basophils. Cytokine (2020) 136:155268. doi: 10.1016/j.cyto.2020.155268
131. Okayama Y, Begishvili TB, Church MK. Comparison of mechanisms of IL-3 induced histamine release and IL-3 priming effect on human basophils. Clin Exp Allergy (1993) 23(11):901–10. doi: 10.1111/j.1365-2222.1993.tb00274.x
132. Bondurant KL, Lundgreen A, Herrick JS, Kadlubar S, Wolff RK, Slattery ML. Interleukin genes and associations with colon and rectal cancer risk and overall survival. Int J Cancer (2013) 132(4):905–15. doi: 10.1002/ijc.27660
133. Williams A, Greene N, Kimbro K. Increased circulating cytokine levels in African American women with obesity and elevated HbA1c. Cytokine (2020) 128:154989. doi: 10.1016/j.cyto.2020.154989
134. Meyer CG, Calixto Fernandes MH, Intemann CD, Kreuels B, Kobbe R, Kreuzberg C, et al. IL3 variant on chromosomal region 5q31-33 and protection from recurrent malaria attacks. Hum Mol Genet (2011) 20(6):1173–81. doi: 10.1093/hmg/ddq562
135. Huang S, Chen Z, Yu JF, Young D, Bashey A, Ho AD, et al. Correlation between IL-3 receptor expression and growth potential of human CD34+ Hematopoietic cells from different tissues. Stem Cells (Dayton Ohio) (1999) 17(5):265–72. doi: 10.1002/stem.170265
136. Agger K, Miyagi S, Pedersen MT, Kooistra SM, Johansen JV, Helin K. Jmjd2/Kdm4 demethylases are required for expression of Il3ra and survival of acute myeloid leukemia cells. Genes Dev (2016) 30(11):1278–88. doi: 10.1101/gad.280495.116
137. Yan J, Hou Z, Ren Y, Hu J. EP300-ZNF384 rearrangement drive B-cell acute lymphoblastic leukemia development by activating IL3-IL3RA signaling. Blood (2022) 140(Supplement 1):11508–8. doi: 10.1182/blood-2022-168535
138. Bagaria J, Kim K-O, Bagyinszky E, An SSA, Baek J-H. Discriminating potential genetic markers for complete response and non-complete response patients to neoadjuvant chemotherapy with locally advanced rectal cancer. Int J Environ Res Public Health (2022) 19(7):4008. doi: 10.3390/ijerph19074008
139. Carvalho LH, Sano GI, Hafalla JC, Morrot A, De Lafaille MAC, Zavala F. IL-4-secreting CD4+ T cells are crucial to the development of CD8+ T-cell responses against malaria liver stages. Nat Med (2002) 8(2):166–70. doi: 10.1038/nm0202-166
140. Burke ML, Jones MK, Gobert GN, Li YS, Ellis MK, McManus DP. Immunopathogenesis of human schistosomiasis. Parasite Immunol (2009) 31(4):163–76. doi: 10.1111/j.1365-3024.2009.01098.x
141. Harris NL. Recent advances in type-2-cell-mediated immunity: insights from helminth infection. Immunity (2017) 47(6):1024–36. doi: 10.1016/j.immuni.2017.11.015
142. Kim KM, Hussein UK, Park SH, Moon YJ, Zhang Z, Ahmed AG, et al. Expression of IL4Rα and IL13Rα1 are associated with poor prognosis of soft-tissue sarcoma of the extremities, superficial trunk, and retroperitoneum. Diagn Pathol (2021) 16(1):1–12. doi: 10.1186/s13000-020-01066-z
143. Tsao CH, Shiau MY, Chuang PH, Chang YH, Hwang J. Interleukin-4 regulates lipid metabolism by inhibiting adipogenesis and promoting lipolysis. J Lipid Res (2014) 55(3):385–97. doi: 10.1194/jlr.M041392
144. Battle NC, Choudhry S, Tsai HJ, Eng C, Kumar G, Beckman KB, et al. Ethnicity-specific gene–gene interaction between IL-13 and IL-4Rα among African Americans with asthma. Am J Respir Crit Care Med (2007) 175(9):881–7. doi: 10.1164/rccm.200607-992OC
145. Narożna B, Hoffmann A, Sobkowiak P, Schoneich N, Bręborowicz A, Szczepankiewicz A. Polymorphisms in the interleukin 4, interleukin 4 receptor and interleukin 13 genes and allergic phenotype: A case control study. Adv Med Sci (2016) 61(1):40–5. doi: 10.1016/j.advms.2015.07.003
146. Hildebrandt MA, Komaki R, Liao Z, Gu J, Chang JY, Ye Y, et al. Genetic variants in inflammation-related genes are associated with radiation-induced toxicity following treatment for non-small cell lung cancer. PloS One (2010) 5(8):e12402. doi: 10.1371/journal.pone.0012402
147. Erlich HA, Lohman K, Mack SJ, Valdes AM, Julier C, Mirel D, et al. Association analysis of SNPs in the IL4R locus with type I diabetes. Genes Immun (2009) 10 Suppl 1(Suppl 1):S33–41. doi: 10.1038/gene.2009.89
148. Maier LM, Chapman J, Howson JM, Clayton DG, Pask R, Strachan DP, et al. No evidence of association or interaction between the IL4RA, IL4, and IL13 genes in type 1 diabetes. Am J Hum Genet (2005) 76(3):517–21. doi: 10.1086/428387
149. Qu HQ, Tessier MC, Fréchette R, Bacot F, Polychronakos C. Lack of association of type 1 diabetes with the IL4R gene. Diabetologia (2006) 49(5):958–61. doi: 10.1007/s00125-006-0199-2
150. Kang S, Tanaka T, Narazaki M, Kishimoto T. Targeting interleukin-6 signaling in clinic. Immunity (2019) 50(4):1007–23. doi: 10.1016/j.immuni.2019.03.026
151. McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet (2017) 49(7):1126–32. doi: 10.1038/ng.3892
152. Lippitz BE, Harris RA. Cytokine patterns in cancer patients: A review of the correlation between interleukin 6 and prognosis. Oncoimmunology (2016) 5(5):e1093722. doi: 10.1080/2162402X.2015.1093722
153. Cupido AJ, Asselbergs FW, Natarajan P, CHARGE Inflammation Working Group, Ridker PM, Hovingh GK, et al. Dissecting the IL-6 pathway in cardiometabolic disease: A Mendelian randomization study on both IL6 and IL6R. Br J Clin Pharmacol (2022) 88(6):2875–84. doi: 10.1111/bcp.15191
154. Liao X, Yu L, Liu X, Han C, Yu T, Qin W, et al. Genome-wide association pathway analysis to identify candidate single nucleotide polymorphisms and molecular pathways associated with TP53 expression status in HBV-related hepatocellular carcinoma. Cancer Manage Res (2018) 10:953. doi: 10.2147/CMAR.S163209
155. Lundtoft C, Seyfarth J, Jacobsen M. IL7RA genetic variants differentially affect IL-7Rα expression and alternative splicing: a role in autoimmune and infectious diseases? Genes Immun (2020) 21(2):83–90. doi: 10.1038/s41435-019-0091-y
156. Vudattu NK, Magalhaes I, Schmidt M, Seyfert-Margolis V, Maeurer MJ. Reduced numbers of IL-7 receptor (CD127) expressing immune cells and IL-7-signaling defects in peripheral blood from patients with breast cancer. Int J Cancer (2007) 121(7):1512–9. doi: 10.1002/ijc.22854
157. Hoffmann M, Enczmann J, Balz V, Kummer S, Reinauer C, Döing C, et al. Interleukin-7 and soluble Interleukin-7 receptor levels in type 1 diabetes–Impact of IL7RA polymorphisms, HLA risk genotypes and clinical features. Clin Immunol (2022) 235:108928. doi: 10.1016/j.clim.2022.108928
158. Guad RM, Taylor-Robinson AW, Wu YS, Gan SH, Zaharan NL, Basu RC, et al. Clinical and genetic risk factors for new-onset diabetes mellitus after transplantation (NODAT) in major transplant centres in Malaysia. BMC Nephrol (2020) 21:1–8. doi: 10.1186/s12882-020-02052-9
159. Mahajan R, El-Omar EM, Lissowska J, Grillo P, Rabkin CS, Baccarelli A, et al. Genetic variants in T helper cell type 1, 2 and 3 pathways and gastric cancer risk in a Polish population. Japanese J Clin Oncol (2008) 38(9):626–33. doi: 10.1093/jjco/hyn075
160. Van Dyke AL, Cote ML, Wenzlaff AS, Chen W, Abrams J, Land S, et al. Cytokine and cytokine receptor single-nucleotide polymorphisms predict risk for non-small cell lung cancer among women. Cancer Epidemiol Biomarkers Prev (2009) 18(6):1829–40. doi: 10.1158/1055-9965.EPI-08-0962
161. Chen Y, Zheng T, Lan Q, Foss F, Kim C, Chen X, et al. Cytokine polymorphisms in Th1/Th2 pathway genes, body mass index, and risk of non-Hodgkin lymphoma. Blood J Am Soc Hematol (2011) 117(2):585–90. doi: 10.1182/blood-2010-07-295097
162. Guzmán-Fulgencio M, Berenguer J, Jiménez-Sousa MA, Pineda-Tenor D, Aldámiz-Echevarria T, García-Broncano P, et al. Association between IL7R polymorphisms and severe liver disease in HIV/HCV coinfected patients: a cross-sectional study. J Trans Med (2015) 13(1):1–9. doi: 10.1186/s12967-015-0577-y
163. Boguslawska J, Kedzierska H, Poplawski P, Rybicka B, Tanski Z, Piekielko-Witkowska A. Expression of genes involved in cellular adhesion and extracellular matrix remodeling correlates with poor survival of patients with renal cancer. J Urol (2016) 195(6):1892–902. doi: 10.1016/j.juro.2015.11.050
164. Zhang J, Wang H, Yuan C, Wu J, Xu J, Chen S, et al. ITGAL as a prognostic biomarker correlated with immune infiltrates in gastric cancer. Front Cell Dev Biol (2022) 10:808212. doi: 10.3389/fcell.2022.808212
165. Zhou Y, Shi W, Zhao D, Xiao S, Wang K, Wang J. Identification of immune-associated genes in diagnosing aortic valve calcification with metabolic syndrome by integrated bioinformatics analysis and machine learning. Front Immunol (2022) 13:937886. doi: 10.3389/fimmu.2022.937886
166. Pençe HH, Alsaadoni H, Çaykara B, Ötünçtemur A, Kuraş S, Tokat B. Investigation of LFA-1 rs2230433 variation in renal cell carcinoma tissues. Haydarpasa Numune Training Res Hosp Med J (2019) 59(3):216–9. doi: 10.14744/hnhj.2019.63325
167. Fu Z, Jiao M, Zhang M, Xu F, Yuan W, Pang D, et al. LFA-1 gene polymorphisms are associated with the sporadic infiltrative duct breast carcinoma in Chinese Han women of Heilongjiang Province. Breast Cancer Res Treat (2011) 127(1):265–71. doi: 10.1007/s10549-010-1203-6
168. Weber C, Fraemohs L, Dejana E. The role of junctional adhesion molecules in vascular inflammation. Nat Rev Immunol (2007) 7(6):467–77. doi: 10.1038/nri2096
169. Verdino P, Witherden DA, Havran WL, Wilson IA. The molecular interaction of CAR and JAML recruits the central cell signal transducer PI3K. Science (2010) 329(5996):1210–4. doi: 10.1126/science.1187996
170. Fang L, Yu W, Yu G, Zhong F, Ye B. Junctional adhesion molecule-like protein (JAML) is correlated with prognosis and immune infiltrates in lung adenocarcinoma. Med Sci Monitor (2022) 28:e933503. doi: 10.12659/MSM.933503
171. Fu Y, Sun Y, Wang M, Hou Y, Huang W, Zhou D, et al. Elevation of JAML promotes diabetic kidney disease by modulating podocyte lipid metabolism. Cell Metab (2020) 32(6):1052–62. doi: 10.1016/j.cmet.2020.10.019
172. Huang W, Wang B-O, Hou Y-F, Fu Y, Cui S-J, Zhu J-H, et al. JAML promotes acute kidney injury mainly through a macrophage-dependent mechanism. JCI Insight (2022) 7(14):e158571. doi: 10.1172/jci.insight.158571
173. Spitali P, Zaharieva I, Bohringer S, Hiller M, Chaouch A, Roos A, et al. TCTEX1D1 is a genetic modifier of disease progression in Duchenne muscular dystrophy. Eur J Hum Genet EJHG (2020) 28(6):815–25. doi: 10.1038/s41431-019-0563-6
174. Koerber-Rosso I, Brandt S, von Schnurbein J, Fischer-Posovszky P, Hoegel J, Rabenstein H, et al. A fresh look to the phenotype in mono-allelic likely pathogenic variants of the leptin and the leptin receptor gene. Mol Cell Pediatr (2021) 8(1):10. doi: 10.1186/s40348-021-00119-7
175. Loos RJF, Yeo GSH. The genetics of obesity: from discovery to biology. Nat Rev Genet (2022) 23(2):120–33. doi: 10.1038/s41576-021-00414-z
176. Yadav A, Deo N. Influence of leptin on immunity. Curr Immunol Rev (2013) 9(1):23–30. doi: 10.2174/1573395511309010004
177. Li Z, Yuan W, Ning S, Li J, Zhai W, Zhang S. Role of leptin receptor (LEPR) gene polymorphisms and haplotypes in susceptibility to hepatocellular carcinoma in subjects with chronic hepatitis B virus infection. Mol Diagnosis Ther (2012) 16(6):383–8. doi: 10.1007/s40291-012-0008-1
178. Lin J, Xie Z, Lan B, Guo Z, Tang WF, Liu C, et al. Investigation of Leptin and its receptor (LEPR) for single nucleotide polymorphisms in colorectal cancer: a case-control study involving 2,306 subjects. Am J Trans Res (2020) 12(7):3613–28. doi: 10.1200/JCO.2020.38.15_suppl.e16100
179. Saukko M, Kesäniemi YA, Ukkola O. Leptin receptor Lys109Arg and Gln223Arg polymorphisms are associated with early atherosclerosis. Metab Syndrome Related Disord (2010) 8(5):425–30. doi: 10.1089/met.2010.0004
180. Furusawa T, Naka I, Yamauchi T, Natsuhara K, Kimura R, Nakazawa M, et al. The Q223R polymorphism in LEPR is associated with obesity in Pacific Islanders. Hum Genet (2010) 127(3):287–94. doi: 10.1007/s00439-009-0768-9
181. Hirayasu K, Ohashi J, Tanaka H, Kashiwase K, Ogawa A, Takanashi M, et al. Evidence for natural selection on leukocyte immunoglobulin-like receptors for HLA class I in northeast asians. Am J Hum Genet (2008) 82(5):1075–83. doi: 10.1016/j.ajhg.2008.03.012
182. Jiao Y, Wang L, Gu X, Tao S, Tian L, Na R, et al. LILRA3 is associated with benign prostatic hyperplasia risk in a Chinese population. Int J Mol Sci (2013) 14(5):8832–40. doi: 10.3390/ijms14058832
183. Singaraja RR, Tietjen I, Hovingh GK, Franchini PL, Radomski C, Wong K, et al. Identification of four novel genes contributing to familial elevated plasma HDL cholesterol in humans. J Lipid Res (2014) 55(8):1693–701. doi: 10.1194/jlr.M048710
184. Ryu M, Chen Y, Qi J, Liu J, Fan Z, Nam G, et al. LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence. PloS One (2011) 6(4):e19245. doi: 10.1371/journal.pone.0019245
185. Argyriou E, Roussos P, Nezos A, Venetsanopoulou A, Boki KA, Tzioufas A, et al. Thu0204 association of lilra3 gene with lymphomagenesis risk in young ss patients. Ann Rheum Dis (2019) 78, 380. doi: 10.1136/annrheumdis-2019-eular.6054
186. Cox AJ, Zhang P, Evans TJ, Scott RJ, Cripps AW, West NP. Gene expression profiles in whole blood and associations with metabolic dysregulation in obesity. Obes Res Clin Pract (2018) 12(2):204–13. doi: 10.1016/j.orcp.2017.07.001
188. Mukherjee T, Hovingh ES, Foerster EG, Abdel-Nour M, Philpott DJ, Girardin SE. NOD1 and NOD2 in inflammation, immunity and disease. Arch Biochem Biophysics (2019) 670:69–81. doi: 10.1016/j.abb.2018.12.022
189. Wang D. NOD1 and NOD2 are potential therapeutic targets for cancer immunotherapy. Comput Intell Neurosci (2022) 2022. doi: 10.1155/2022/2271788
190. Carlos D, Pérez MM, Leite JA, Rocha FA, Martins LM, Pereira CA, et al. NOD2 deficiency promotes intestinal CD4+ T lymphocyte imbalance, metainflammation, and aggravates type 2 diabetes in murine model. Front Immunol (2020) 11:1265. doi: 10.3389/fimmu.2020.01265
191. Cavallari JF, Pokrajac NT, Zlitni S, Foley KP, Henriksbo BD, Schertzer JD. NOD2 in hepatocytes engages a liver-gut axis to protect against steatosis, fibrosis, and gut dysbiosis during fatty liver disease in mice. Am J Physiol-Endocrinol Metab (2020) 319(2):E305–14. doi: 10.1152/ajpendo.00181.2020
192. Cerhan JR, Wang S, Maurer MJ, Ansell SM, Geyer SM, Cozen W, et al. Prognostic significance of host immune gene polymorphisms in follicular lymphoma survival. Blood (2007) 109(12):5439–46. doi: 10.1182/blood-2006-11-058040
193. Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol (2015) 15(12):760–70. doi: 10.1038/nri3921
194. Zhang X, Bai XC, Chen ZJ. Structures and mechanisms in the cGAS-STING innate immunity pathway. Immunity (2020) 53(1):43–53. doi: 10.1016/j.immuni.2020.05.013
195. Corrales L, McWhirter SM, Dubensky TW Jr., Gajewski TF. The host STING pathway at the interface of cancer and immunity. J Clin Invest (2016) 126(7):2404–11. doi: 10.1172/JCI86892
196. Oduro PK, Zheng X, Wei J, Yang Y, Wang Y, Zhang H, et al. The cGAS-STING signaling in cardiovascular and metabolic diseases: Future novel target option for pharmacotherapy. Acta Pharm Sin B (2022) 12(1):50–75. doi: 10.1016/j.apsb.2021.05.011
197. Jin L, Xu L, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, et al. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun (2011) 12(4):263–9. doi: 10.1038/gene.2010.75
198. Yi G, Brendel VP, Shu C, Li P, Palanathan S, Cheng Kao C. Single nucleotide polymorphisms of human STING can affect innate immune response to cyclic dinucleotides. PloS One (2013) 8(10):e77846. doi: 10.1371/journal.pone.0077846
199. Kennedy RB, Haralambieva IH, Ovsyannikova IG, Voigt EA, Larrabee BR, Schaid DJ, et al. Polymorphisms in STING Affect Human Innate Immune Responses to Poxviruses. Front Immunol (2020) 11:567348.
200. Straub RH. TRPV1, TRPA1, and TRPM8 channels in inflammation, energy redirection, and water retention: role in chronic inflammatory diseases with an evolutionary perspective. J Mol Med (2014) 92:925–37. doi: 10.1007/s00109-014-1175-9
201. Kozyreva TV, Khramova GM. Effects of activation of skin ion channels TRPM8, TRPV1, and TRPA1 on the immune response. Comparison with effects of cold and heat exposure. J Thermal Biol (2020) 93:102729. doi: 10.1016/j.jtherbio.2020.102729
202. Yee NS. Roles of TRPM8 ion channels in cancer: proliferation, survival, and invasion. Cancers (2015) 7(4):2134–46. doi: 10.3390/cancers7040882
203. Liu Z, Wu H, Wei Z, Wang X, Shen P, Wang S, et al. TRPM8: a potential target for cancer treatment. J Cancer Res Clin Oncol (2016) 142:1871–81. doi: 10.1007/s00432-015-2112-1
204. Sanders OD, Rajagopal JA, Rajagopal L. Menthol to induce non-shivering thermogenesis via TRPM8/PKA signaling for treatment of obesity. J Obes Metab Syndrome (2021) 30(1):4–11. doi: 10.7570/jomes20038
205. Kozyreva TV, Kozaruk VP, Meyta ES. Effect of the peripheral TRPM8 ion channel activation on the cardiovascular parameters. Int Arch Clin Pharmacol (2019) 5:1–7. doi: 10.23937/2572-3987.1510019
206. Naumov DE, Perelman JM, Kolosov VP, Potapova TA, Maksimov VN, Zhou X. Transient receptor potential melastatin 8 gene polymorphism is associated with cold-induced airway hyperresponsiveness in bronchial asthma. Respirology (2015) 20(8):1192–7. doi: 10.1111/resp.12605
207. Potapova TA, Babenko VN, Kobzev VF, Romashchenko AG, Maksimov VN, Voevoda MI. Associations of cold receptor TRPM8 gene single nucleotide polymorphism with blood lipids and anthropometric parameters in Russian population. Bull Exp Biol Med (2014) 157(6):757–61. doi: 10.1007/s10517-014-2660-4
208. Mangi MH, Newland AC. Interleukin-3: promises and perspectives. Hematol (Amsterdam Netherlands) (1998) 3(1):55–66. doi: 10.1080/10245332.1998.11752123
209. Wu C-H, Lee T-H, Yang S-F, Tsao H-M, Chang Y-J, Chou C-H, et al. Interleukin-3 polymorphism is associated with miscarriage of fresh in vitro fertilization cycles. Int J Environ Res Public Health (2019) 16(6):995. doi: 10.3390/ijerph16060995
210. Stomski FC, Sun Q, Bagley CJ, Woodcock J, Goodall G, Andrews RK, et al. Human interleukin-3 (IL-3) induces disulfide-linked IL-3 receptor alpha- and beta-chain heterodimerization, which is required for receptor activation but not high-affinity binding. Mol Cell Biol (1996) 16(6):3035–46. doi: 10.1128/MCB.16.6.3035
211. Heeb LEM, Egholm C, Boyman O. Evolution and function of interleukin-4 receptor signaling in adaptive immunity and neutrophils. Genes Immun (2020) 21(3):143–9. doi: 10.1038/s41435-020-0095-7
212. Damen M, Stankiewicz TE, Park SH, Helsley RN, Chan CC, Moreno-Fernandez ME, et al. Non-hematopoietic IL-4Ralpha expression contributes to fructose-driven obesity and metabolic sequelae. Int J Obes (Lond) (2021) 45(11):2377–87. doi: 10.1038/s41366-021-00902-6
213. Zhu N, Gong Y, Chen XD, Zhang J, Long F, He J, et al. Association between the polymorphisms of interleukin-4, the interleukin-4 receptor gene and asthma. Chin Med J (Engl) (2013) 126(15):2943–51.
214. ElKassar N, Gress RE. An overview of IL-7 biology and its use in immunotherapy. J Immunotoxicol (2010) 7(1):1–7. doi: 10.3109/15476910903453296
215. Carrette F, Surh CD. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin Immunol (2012) 24(3):209–17. doi: 10.1016/j.smim.2012.04.010
216. Nguyen V, Mendelsohn A, Larrick JW. Interleukin-7 and immunosenescence. J Immunol Res (2017) 2017:4807853. doi: 10.1155/2017/4807853
217. Bossennec M, Di Roio A, Caux C, Ménétrier-Caux C. MDR1 in immunity: friend or foe? Oncoimmunology (2018) 7(12):e1499388. doi: 10.1080/2162402X.2018.1499388
218. Wang BL, Zhai HY, Chen BY, Zhai SP, Yang HY, Chen XP, et al. Clinical relationship between MDR1 gene and gallbladder cancer. Hepatobiliary Pancreat Dis Int (2004) 3(2):296–9.
219. Lai J, Yang S, Lin Z, Huang W, Li X, Li R, et al. Update on chemoresistance mechanisms to first-line chemotherapy for gallbladder cancer and potential reversal strategies. Am J Clin Oncol (2023) 46(4):131. doi: 10.1097/COC.0000000000000989
220. Ichihara S, Yamada Y, Kato K, Hibino T, Yokoi K, Matsuo H, et al. Association of a polymorphism of ABCB1 with obesity in Japanese individuals. Genomics (2008) 91(6):512–6. doi: 10.1016/j.ygeno.2008.03.004
221. Wu J, Wang X, Chen H, Yang R, Yu H, Wu Y, et al. Type 2 diabetes risk and lipid metabolism related to the pleiotropic effects of an ABCB1 variant: A chinese family-based cohort study. Metabolites (2022) 12(9):875. doi: 10.3390/metabo12090875
222. Yan RJ, Lou TT, Wu YF, Chen WS. Single nucleotide polymorphisms of ABCB1 gene and response to etanercept treatment in patients with ankylosing spondylitis in a Chinese Han population. Medicine (2017) 96(5):e5929. doi: 10.1097/MD.0000000000005929
223. Gervasini G, Carrillo JA, Garcia M, San Jose C, Cabanillas A, Benitez J. Adenosine triphosphate-binding cassette B1 (ABCB1) (multidrug resistance 1) G2677T/A gene polymorphism is associated with high risk of lung cancer. Cancer (2006) 107(12):2850–7. doi: 10.1002/CNCR.22332
224. Song F, Zhang Y, Pan Z, Hu X, Zhang Q, Huang F, et al. The role of alcohol dehydrogenase 1C in regulating inflammatory responses in ulcerative colitis. Biochem Pharmacol (2021) 192:114691. doi: 10.1016/j.bcp.2021.114691
225. Chen Q, Li F, Gao Y, Xu G, Liang L, Xu J. Identification of energy metabolism genes for the prediction of survival in hepatocellular carcinoma. Front Oncol (2020) 10:1210. doi: 10.3389/fonc.2020.01210
226. Liu X, Li T, Kong D, You H, Kong F, Tang R. Prognostic implications of alcohol dehydrogenases in hepatocellular carcinoma. BMC Cancer (2020) 20(1):1204. doi: 10.1186/s12885-020-07689-1
227. Molinaro A, Wahlström A, Marschall HU. Role of bile acids in metabolic control. Trends Endocrinol Metab (2018) 29(1):31–41. doi: 10.1016/j.tem.2017.11.002
228. McGlone ER, Bloom SR. Bile acids and the metabolic syndrome. Ann Clin Biochem (2019) 56(3):326–37. doi: 10.1177/0004563218817798
229. Li M, Liu Z, Song J, Wang T, Wang H, Wang Y, et al. Identification of down-regulated ADH1C is associated with poor prognosis in colorectal cancer using bioinformatics analysis. Front Mol Biosci (2022) 9:791249. doi: 10.3389/fmolb.2022.791249
230. Jaillon S, Berthenet K, Garlanda C. Sexual dimorphism in innate immunity. Clin Rev Allergy Immunol (2019) 56(3):308–21. doi: 10.1007/s12016-017-8648-x
231. Wang P, Zhang L, Huang C, Huang P, Zhang J. Distinct prognostic values of alcohol dehydrogenase family members for non-small cell lung cancer. Med Sci Monit (2018) 24:3578–90. doi: 10.12659/MSM.910026
232. Shen XY, Liu XP, Song CK, Wang YJ, Li S, Hu WD. Genome-wide analysis reveals alcohol dehydrogenase 1C and secreted phosphoprotein 1 for prognostic biomarkers in lung adenocarcinoma. J Cell Physiol (2019) 234(12):22311–20. doi: 10.1002/jcp.28797
233. Xue Y, Wang M, Zhong D, Tong N, Chu H, Sheng X, et al. ADH1C Ile350Val polymorphism and cancer risk: evidence from 35 case–control studies. PloS One (2012) 7(5):e37227. doi: 10.1371/journal.pone.0037227
234. Oze I, Matsuo K, Suzuki T, Kawase T, Watanabe M, Hiraki A, et al. Impact of multiple alcohol dehydrogenase gene polymorphisms on risk of upper aerodigestive tract cancers in a Japanese population. Cancer Epidemiol Biomarkers Prev (2009) 18(11):3097–102. doi: 10.1158/1055-9965.Epi-09-0499
235. Harada R, Kimura M, Sato Y, Taniguchi T, Tomonari T, Tanaka T, et al. APOB codon 4311 polymorphism is associated with hepatitis C virus infection through altered lipid metabolism. BMC Gastroenterol (2018) 18(1):24. doi: 10.1186/S12876-018-0747-5
236. Deng W, Liu H, Luo S, Clarke J, Glass C, Su L, et al. APOB genotypes and CDH13 haplotypes in the cholesterol-related pathway genes predict non-small cell lung cancer survival. Cancer Epidemiol Biomarkers Prev (2020) 29(6):1204–13. doi: 10.1158/1055-9965.EPI-19-1262
237. Jang SJ, Tuan WL, Hsu LA, Er LK, Teng MS, Wu S, et al. Pleiotropic effects of APOB variants on lipid profiles, metabolic syndrome, and the risk of diabetes mellitus. Int J Mol Sci (2022) 23(23):14963. doi: 10.3390/IJMS232314963/S1
238. Aceves-Ramírez M, Valle Y, Casillas-Muñoz F, Martínez-Fernández DE, Parra-Reyna B, López-Moreno VA, et al. Analysis of the APOB Gene and Apolipoprotein B Serum Levels in a Mexican Population with Acute Coronary Syndrome: Association with the Single Nucleotide Variants rs1469513, rs673548, rs676210, and rs1042034. Genet Res (Camb) (2022) 2022:4901090. doi: 10.1155/2022/4901090
239. Shishkova VN, Adasheva TV, Stakhovskaya LV, Remennic AYU, Valyaeva VV. Assessment of twenty-five variants of polymorphic genes for lipid and carbohydrate metabolism, vascular inflammation and the neurotransmitter system in the first non-cardioembolic ischemic stroke. Eur Heart J (2020) 41(Supplement_2):ehaa946.2408. doi: 10.1093/EHJCI/EHAA946.2408
240. Yoshida T, Kato K, Yokoi K, Watanabe S, Metoki N, Satoh K, et al. Association of candidate gene polymorphisms with chronic kidney disease in Japanese individuals with hypertension. Hypertens Res (2009) 32(5):411–8. doi: 10.1038/hr.2009.22
241. Walse B, Dufe VT, Svensson B, Fritzson I, Dahlberg L, Khairoullina A, et al. The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites. Biochemistry (2008) 47(34):8929–36. doi: 10.1021/BI8003318
242. Evers DL, Wang X, Huong SM, Andreoni KA, Huang ES. Inhibition of human cytomegalovirus signaling and replication by the immunosuppressant FK778. Antiviral Res (2005) 65(1):1–12. doi: 10.1016/j.antiviral.2004.03.007
243. Luban J, Sattler RA, Mühlberger E, Graci JD, Cao L, Weetall M, et al. The DHODH inhibitor PTC299 arrests SARS-CoV-2 replication and suppresses induction of inflammatory cytokines. Virus Res (2021) 292:198246. doi: 10.1016/J.VIRUSRES.2020.198246
244. Mao C, Liu X, Zhang Y, Lei G, Yan Y, Lee H, et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature (2021) 593(7860):586–90. doi: 10.1038/S41586-021-03539-7
245. Zhou Y, Tao L, Zhou X, Zuo Z, Gong J, Liu X, et al. DHODH and cancer: promising prospects to be explored. Cancer Metab (2021) 9(1):22. doi: 10.1186/S40170-021-00250-Z
246. Zhang J, Teran G, Popa M, Madapura H, Ladds M, Lianoudaki D, et al. DHODH inhibition modulates glucose metabolism and circulating GDF15, and improves metabolic balance. iScience (2021) 24(5):102494. doi: 10.1016/j.isci.2021.102494
247. Liu K, Jin X, Zhang X, Lian H, Ye J. The mechanisms of nucleotide actions in insulin resistance. J Genet Genomics (2022) 49(4):299–307. doi: 10.1016/J.JGG.2022.01.006
248. Pawlik A, Herczynska M, Kurzawski M, Safranow K, Dziedziejko V, Drozdzik M. The effect of exon (19C>A) dihydroorotate dehydrogenase gene polymorphism on rheumatoid arthritis treatment with leflunomide. Pharmacogenomics (2009) 10(2):303–9. doi: 10.2217/14622416.10.2.303
249. Soukup T, Dosedel M, Nekvindova J. Genetic polymorphisms in metabolic pathways of leflunomide in the treatment of rheumatoid arthritis. Clin Exp Rheumatol (2015) 33:426–32.
250. Makarem YS, Hareedy MS, Hassanien M, Ahmed EA, Hetta HF, Mohamed AAA. Frequency and impact of DHODH, ABCG2 and CYP2C19 SNPs on the therapeutic efficacy, tolerability and toxicity of leflunomide. Pharmacogenomics (2021) 22(18):1201–9. doi: 10.2217/PGS-2020-0146
251. Wang H, Li ZY, Liu Y, Persson J, Beyer I, Möller T, et al. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat Med (2011) 17(1):96–104. doi: 10.1038/nm.2270
252. Wang J, Hao S, Gu J, Rudd SG, Wang Y. The prognostic and clinicopathological significance of desmoglein 2 in human cancers: a systematic review and meta-analysis. PeerJ (2022) 10:e13141. doi: 10.7717/peerj.13141
253. Myo Min KK, Rojas-Canales D, Penko D, DeNichilo M, Cockshell MP, Ffrench CB, et al. Desmoglein-2 is important for islet function and β-cell survival. Cell Death Dis (2022) 13(10):1–15. doi: 10.1038/s41419-022-05326-2
254. Choi H, Koh HW, Zhou L, Cheng H, Loh TP, Parvaresh Rizi E, et al. Plasma protein and microRNA biomarkers of insulin resistance: A network-based integrative-omics analysis. Front Physiol (2019) 10:379. doi: 10.3389/fphys.2019.00379
255. Liu K, Wang YB, Du JL, Qu PF, Ma L, Tang X, et al. Cardiac disease associated genetic variants in Yi nationality in regions with high incidence of Yunnan sudden unexplained death. Fa yi xue za zhi (2020) 36(4):497–501. doi: 10.12116/j.issn.1004-5619.2020.04.013
256. Yamamura K, Uruno T, Shiraishi A, Tanaka Y, Ushijima M, Nakahara T, et al. The transcription factor EPAS1 links DOCK8 deficiency to atopic skin inflammation via IL-31 induction. Nat Commun (2017) 8(1):13946. doi: 10.1038/ncomms13946
257. Putra AC, Eguchi H, Lee KL, Yamane Y, Gustine E, Isobe T, et al. The A Allele at rs13419896 of EPAS1 is associated with enhanced expression and poor prognosis for non-small cell lung cancer. PloS One (2015) 10(8):e0134496. doi: 10.1371/journal.pone.0134496
258. Islam F, Gopalan V, Lu CT, Pillai S, Lam AK. Identification of novel mutations and functional impacts of EPAS1 in colorectal cancer. Cancer Med (2021) 10(16):5557–73. doi: 10.1002/cam4.4116
259. Gao F, Yao Q, Zhu J, Chen W, Feng X, Feng B, et al. Dyslipidemia and non-alcoholic fatty liver disease due to a somatic mutation in HIF2A. Lancet (2023). (pre-print). doi: 10.2139/ssrn.4401718
260. Al-Khelaifi F, Diboun I, Donati F, Botrè F, Abraham D, Hingorani A, et al. Metabolic GWAS of elite athletes reveals novel genetically-influenced metabolites associated with athletic performance. Sci Rep (2019) 9(1):19889. doi: 10.1038/s41598-019-56496-7
261. Jiang LL, Kobayashi A, Matsuura H, Fukushima H, Hashimoto T. Purification and properties of human d-3-Hydroxyacyl-CoA dehydratase: Medium-chain enoyl-CoA hydratase is d-3-Hydroxyacyl-CoA Dehydratase1. J Biochem (1996) 120(3):624–32. doi: 10.1093/oxfordjournals.jbchem.a021458
262. Rasiah KK, Gardiner-Garden M, Padilla EJ, Möller G, Kench JG, Alles MC, et al. HSD17B4 overexpression, an independent biomarker of poor patient outcome in prostate cancer. Mol Cell Endocrinol (2009) 301(1-2):89–96. doi: 10.1016/j.mce.2008.11.021
263. Ferdinandusse S, Denis S, van Roermund CWT, Wanders RJA, Dacremont G. Identification of the peroxisomal β-oxidation enzymes involved in the degradation of long-chain dicarboxylic acids. J Lipid Res (2004) 45(6):1104–11. doi: 10.1194/jlr.M300512-JLR200
264. Zhang X, Yang H, Zhang J, Gao F, Dai L. HSD17B4, ACAA1, and PXMP4 in peroxisome pathway are down-regulated and have clinical significance in non-small cell lung cancer. Front Genet (2020) 11:273. doi: 10.3389/fgene.2020.00273
265. Ferdinandusse S, Houten SM. Peroxisomes and bile acid biosynthesis. Biochim Biophys Acta (BBA) - Mol Cell Res (2006) 1763(12):1427–40. doi: 10.1016/j.bbamcr.2006.09.001
266. Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res (2012) 40(Database issue):D1308–1312. doi: 10.1093/nar/gkr798
267. Karageorgi S, McGrath M, Lee IM, Buring J, Kraft P, De Vivo I. Polymorphisms in genes hydroxysteroid-dehydrogenase-17b type 2 and type 4 and endometrial cancer risk. Gynecologic Oncol (2011) 121(1):54–8. doi: 10.1016/j.ygyno.2010.11.014
268. Ferlin A, Ganz F, Pengo M, Selice R, Frigo AC, Foresta C. Association of testicular germ cell tumor with polymorphisms in estrogen receptor and steroid metabolism genes. Endocrine-Related Cancer (2010) 17(1):17. doi: 10.1677/ERC-09-0176
269. Cui N, Hu M, Khalil RA. Chapter one - biochemical and biological attributes of matrix metalloproteinases. In: Khalil RA, editor. Progress in Molecular Biology and Translational Science, vol. 147. Cambridge, MA: Academic Press (2017). p. 1–73.
270. Shibuya M. VEGF-VEGFR system as a target for suppressing inflammation and other diseases. Endocr Metab Immune Disord-Drug Targets (2015) 15(2):135–44. doi: 10.2174/1871530315666150316121956
271. Adamowicz M, Radlwimmer B, Rieker RJ, Mertens D, Schwarzbach M, Schraml P, et al. Frequent amplifications and abundant expression of TRIO, NKD2, and IRX2 in soft tissue sarcomas. Genes Chromosomes Cancer (2006) 45(9):829–38. doi: 10.1002/gcc.20343
272. Liu T, Zhou W, Zhang F, Shi G, Teng H, Xiao J, et al. Knockdown of IRX2 inhibits osteosarcoma cell proliferation and invasion by the AKT/MMP9 signaling pathway. Mol Med Rep (2014) 10(1):169–74. doi: 10.3892/mmr.2014.2215
273. Chen B, Zhang Y, Chen S, Xuran L, Dong J, Chen W, et al. The role of vascular endothelial growth factor in ischemic stroke. Pharmazie (2021) 76(4):127–31. doi: 10.1691/ph.2021.1315
274. Si J, Si Y, Zhang B, Lan G, Wei J, Huang B, et al. Up-regulation of the IRX2 gene predicts poor prognosis in nasopharyngeal carcinoma. Int J Clin Exp Pathol (2018) 11(8):4073.
275. Gholipoorfeshkecheh R, Agarwala S, Krishnappa S, Savitha MR, Doddaiah N, Ramachandra NB. Nuclear co-repressor 1: a potential candidate gene in the manifestation of congenital heart diseases. Int J Hum Genet (2020) 20(2):55–65. doi: 10.31901/24566330.2020/20.02.73
276. Wu X, Tsai CY, Patam MB, Zan H, Chen JP, Lipkin SM, et al. A role for the MutL mismatch repair Mlh3 protein in immunoglobulin class switch DNA recombination and somatic hypermutation. J Immunol (2006) 176(9):5426–37. doi: 10.4049/JIMMUNOL.176.9.5426
277. Wu Y, Berends MJ, Sijmons RH, Mensink RG, Verlind E, Kooi KA, et al. A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nat Genet (2001) 29(2):137–8. doi: 10.1038/ng1001-137
278. Jing W, Li L, Zhang X, Wu S, Zhao J, Hou Q, et al. Genetic profiling of breast cancer with and without preexisting metabolic disease. Trans Oncol (2020) 13(2):245–53. doi: 10.1016/J.TRANON.2019.09.008
279. de Jong MM, Hofstra RM, Kooi KA, Westra JL, Berends MJ, Wu Y, et al. No association between two MLH3 variants (S845G and P844L) and colorectal cancer risk. Cancer Genet Cytogenetics (2004) 152(1):70–1. doi: 10.1016/j.cancergencyto.2003.10.008
280. Ye F, Cheng Q, Shen J, Zhou C, Chen H. Mismatch repair gene MLH3 Pro844Leu and Thr942Ile polymorphisms and the susceptibility to cervical carcinoma and HPV infection: a case-control study in a Chinese population. PloS One (2014) 9(4):e96224. doi: 10.1371/journal.pone.0096224
281. Liu Y, Zhang X, Jia J, Tang L, Gao X, Yan L, et al. Correlation between polymorphisms in DNA mismatch repair genes and the risk of primary hepatocellular carcinoma for the Han population in northern China. Scand J Gastroenterol (2015) 50(11):1404–10. doi: 10.3109/00365521.2015.1045429
282. Yang G, Huang H, Tang M, Cai Z, Huang C, Qi B, et al. Role of neuromedin B and its receptor in the innate immune responses against influenza A virus infection in vitro and in vivo. Vet Res (2019) 50(1):80. doi: 10.1186/s13567-019-0695-2
283. Zeng R, Xiong X. Effect of NMB-regulated ERK1/2 and p65 signaling pathway on proliferation and apoptosis of cervical cancer. Pathol-Res Pract (2022) 238:154104. doi: 10.1016/j.prp.2022.154104
284. Kirac D, Kasimay Cakir O, Avcilar T, Deyneli O, Kurtel H, Yazici D, et al. Effects of MC4R, FTO, and NMB gene variants to obesity, physical activity, and eating behavior phenotypes. IUBMB Life (2016) 68(10):806–16. doi: 10.1002/iub.1558
285. Sagiv Y, Hudspeth K, Mattner J, Schrantz N, Stern RK, Zhou D, et al. Cutting edge: impaired glycosphingolipid trafficking and NKT cell development in mice lacking Niemann-Pick type C1 protein. J Immunol (2006) 177(1):26–30. doi: 10.4049/jimmunol.177.1.26
286. O’Neill KI, Kuo LW, Williams MM, Lind H, Crump LS, Hammond NG, et al. NPC1 confers metabolic flexibility in Triple Negative Breast Cancer. Cancers (2022) 14(14):3543. doi: 10.3390/cancers14143543
287. Lamri A, Pigeyre M, Garver WS, Meyre D. The extending spectrum of NPC1-related human disorders: From Niemann–Pick C1 disease to obesity. Endocr Rev (2018) 39(2):192–220. doi: 10.1210/er.2017-00176
288. Chiorean A, Garver WS, Meyre D. Signatures of natural selection and ethnic-specific prevalence of NPC1 pathogenic mutations contributing to obesity and Niemann–Pick disease type C1. Sci Rep (2020) 10(1):1–9. doi: 10.1038/s41598-020-75919-4
289. Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, et al. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature (2011) 477(7364):340–3. doi: 10.1038/nature10348
290. Al-Daghri NM, Cagliani R, Forni D, Alokail MS, Pozzoli U, Alkharfy KM, et al. Mammalian NPC1 genes may undergo positive selection and human polymorphisms associate with type 2 diabetes. BMC Med (2012) 10(1):1–11. doi: 10.1186/1741-7015-10-140
291. Meyre D, Delplanque J, Chèvre JC, Lecoeur C, Lobbens S, Gallina S, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet (2009) 41(2):157–9. doi: 10.1038/ng.301
292. Afzali M, Hashemi M, Tabatabaei SP, Fakheri KT, Nakhaee A. Association between the rs1805081 polymorphism of Niemann−Pick type C1 gene and cardiovascular disease in a sample of an Iranian population. Biomed Rep (2017) 6(1):83–8. doi: 10.3892/br.2016.802
293. Tavanti A, Campa D, Bertozzi A, Pardini G, Naglik JR, Barale R, et al. Candida albicans isolates with different genomic backgrounds display a differential response to macrophage infection. Microbes Infect (2006) 8(3):791–800. doi: 10.1016/j.micinf.2005.09.016
294. Zhang C, Mei W, Zeng C. Oncogenic Neuregulin 1 gene (NRG1) fusions in cancer: A potential new therapeutic opportunities. Biochim Biophys Acta (BBA)-Reviews Cancer (2022) 1877(3):188707. doi: 10.1016/j.bbcan.2022.188707
295. Gumà A, Díaz-Sáez F, Camps M, Zorzano A. Neuregulin, an effector on mitochondria metabolism that preserves insulin sensitivity. Front Physiol (2020) 11:696. doi: 10.3389/fphys.2020.00696
296. Yang JZ, Si TM, Ruan Y, Ling YS, Han YH, Wang XL, et al. Association study of neuregulin 1 gene with schizophrenia. Mol Psychiatry (2003) 8(7):706–9. doi: 10.1038/sj.mp.4001377
297. Andre K, Kampman O, Viikki M, Setälä-Soikkeli E, Illi A, Mononen N, et al. BDNF and NRG1 polymorphisms and temperament in selective serotonin reuptake inhibitor-treated patients with major depression. Acta Neuropsychiatr (2018) 30(3):168–74. doi: 10.1017/neu.2017.37
298. Hsu CG, Fazal F, Rahman A, Berk BC, Yan C. Phosphodiesterase 10A is a key mediator of lung inflammation. J Immunol (2021) 206(12):3010–20.
299. Cai Y, Cai Y, Yan C, Guzman RJ. Phosphodiesterase 10A regulates vascular inflammation and pathogenesis of abdominal aortic aneurysms. Circulation (2015) 132(Suppl 3):A18182–A.
300. Borneman RM, Gavin E, Musiyenko A, Richter W, Lee KJ, Crossman DK, et al. Phosphodiesterase 10A (PDE10A) as a novel target to suppress β-catenin and RAS signaling in epithelial ovarian cancer. J Ovarian Res (2022) 15(1):120.
301. Kilanowska A, Ziolkowska A. Role of phosphodiesterase in the biology and pathology of diabetes. Int J Mol Sci (2020) 21(21). doi: 10.3390/ijms21218244
302. Hu Y, Deng L, Zhang J, Fang X, Mei P, Cao X, et al. A pooling genome-wide association study combining a pathway analysis for typical sporadic Parkinson’s disease in the Han population of Chinese mainland. Mol Neurobiol (2016) 53:4302–18. doi: 10.1007/s12035-015-9331-y
303. Pirola CJ, Salatino A, Sookoian S. Pleiotropy within gene variants associated with nonalcoholic fatty liver disease and traits of the hematopoietic system. World J Gastroenterol (2021) 27(4):305–20. doi: 10.3748/wjg.v27.i4.305
304. Trépo E, Nahon P, Bontempi G, Valenti L, Falleti E, Nischalke HD, et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology (2014) 59(6):2170–7. doi: 10.1002/hep.26767
305. Bing H, Wang W, Li YL. [Correlation between PNPLA3 rs738409 and TM6SF2 rs58542926 gene polymorphism and primary liver cancer in the Han Population of China's Northeast region]. Zhonghua gan zang bing za zhi = Zhonghua ganzangbing zazhi = Chin J Hepatol (2021) 29(2):156–62. doi: 10.3760/cma.j.cn501113-20191021-00390
306. Zhang H, Liu J, Yang Z, Zeng L, Wei K, Zhu L, et al. TCR activation directly stimulates PYGB-dependent glycogenolysis to fuel the early recall response in CD8(+) memory T cells. Mol Cell (2022) 82(16):3077–3088 e3076. doi: 10.1016/j.molcel.2022.06.002
307. Wang Z, Han G, Liu Q, Zhang W, Wang J. Silencing of PYGB suppresses growth and promotes the apoptosis of prostate cancer cells via the NF−κB/Nrf2 signaling pathway. Mol Med Rep (2018) 18(4):3800–8. doi: 10.3892/mmr.2018.9388
308. Xia B, Zhang K, Liu C. PYGB promoted tumor progression by regulating Wnt/β-catenin pathway in gastric cancer. Technol Cancer Res Treat (2020) 19:1533033820926592. doi: 10.1177/1533033820926592
309. Xiao L, Wang W, Huangfu Q, Tao H, Zhang J. PYGB facilitates cell proliferation and invasiveness in non-small cell lung cancer by activating the Wnt–β-catenin signaling pathway. Biochem Cell Biol (2020) 98(5):565–74. doi: 10.1139/bcb-2019-0445
310. Newgard CB, Littman DR, van Genderen C, Smith M, Fletterick RJ. Human brain glycogen phosphorylase. Cloning, sequence analysis, chromosomal mapping, tissue expression, and comparison with the human liver and muscle isozymes. J Biol Chem (1988) 263(8):3850–7.
311. Walton SJ. Clinical and cellular studies in the pathogenesis of desmoid tumours in familial adenomatous polyposis (FAP). Department of Surgery & Cancer, Imperial College, London (2016).
312. Greulich W, Wagner M, Gaidt MM, Stafford C, Cheng Y, Linder A, et al. TLR8 is a sensor of RNase T2 degradation products. Cell (2019) 179(6):1264–1275 e1213. doi: 10.1016/j.cell.2019.11.001
313. Acquati F, Mortara L, De Vito A, Baci D, Albini A, Cippitelli M, et al. Innate immune response regulation by the human RNASET2 tumor suppressor gene. Front Immunol (2019) 10:2587. doi: 10.3389/fimmu.2019.02587
314. Sletten AC, Peterson LR, Schaffer JE. Manifestations and mechanisms of myocardial lipotoxicity in obesity. J Intern Med (2018) 284(5):478–91. doi: 10.1111/joim.12728
315. Zhang H, Baldwin DA, Bukowski RK, Parry S, Xu Y, Song C, et al. A genome-wide association study of early spontaneous preterm delivery. Genet Epidemiol (2015) 39(3):217–26. doi: 10.1002/gepi.21887
316. Sabui S, Skupsky J, Kapadia R, Cogburn K, Lambrecht NW, Agrawal A, et al. Tamoxifen-induced, intestinal-specific deletion of Slc5a6 in adult mice leads to spontaneous inflammation: involvement of NF-κB, NLRP3, and gut microbiota. Am J Physiol-Gastrointestinal Liver Physiol (2019) 317(4):G518–30. doi: 10.1152/ajpgi.00172.2019
317. Sun T, Wang D, Ping Y, Sang Y, Dai Y, Wang Y, et al. Integrated profiling identifies SLC5A6 and MFAP2 as novel diagnostic and prognostic biomarkers in gastric cancer patients. Int J Oncol (2020) 56(2):460–9. doi: 10.3892/ijo.2019.4944
318. Hsieh CH, Chen PC, Shieh CC. Novel SLC5A6 mutations lead to B lymphocyte maturation defects with metabolic abnormality rescuable by biotin replenishment. J Immunol (2022) 208(1_Supplement):168–03. doi: 10.4049/jimmunol.208.Supp.168.03
319. Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat Genet (2014) 46(6):543–50. doi: 10.1038/ng.2982
320. Shu X, Wu L, Khankari NK, Shu XO, Wang TJ, Michailidou K, et al. Associations of obesity and circulating insulin and glucose with breast cancer risk: a Mendelian randomization analysis. Int J Epidemiol (2019) 48(3):795–806. doi: 10.1093/ije/dyy201
321. Zhang J, Ali AM, Lieu YK, Liu Z, Gao J, Rabadan R, et al. Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol Cell (2019) 76(1):82–95. doi: 10.1016/j.molcel.2019.07.017
322. Liu Z, Zhang J, Sun Y, Perea-Chamblee TE, Manley JL, Rabadan R. Pan-cancer analysis identifies mutations in SUGP1 that recapitulate mutant SF3B1 splicing dysregulation. Proc Natl Acad Sci (2020) 117(19):10305–12. doi: 10.1073/pnas.1922622117
323. Strachan DP, Rudnicka AR, Power C, Shepherd P, Fuller E, Davis A, et al. Lifecourse influences on health among british adults: effects of region of residence in childhood and adulthood. Int J Epidemiol. (2007) 36(3):522–31. doi: 10.1093/ije/dyl309
324. Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet (2010) 42(11):949–60. doi: 10.1038/ng.685
325. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet (2010) 42(2):105–16. doi: 10.1038/ng.520
326. van Helvoort A, Smith AJ, Sprong H, Fritzsche I, Schinkel AH, Borst P, et al. MDR1 p-glycoprotein is a lipid translocase of broad specificity, while MDR3 p-glycoprotein specifically translocates phosphatidylcholine. Cell (1996) 87(3):507–17.
327. Edelmann L, Edelmann W. Loss of DNA mismatch repair function and cancer predisposition in the mouse: animal models for human hereditary nonpolyposis colorectal cancer. Am J Med Genet Part C Semin Med Genet (2004) 129C(1):91–9. doi: 10.1002/AJMG.C.30021
328. Khailany RA, Ozaslan M. Exonic sequencing and MLH3 gene expression analysis of breast cancer patients. Cell Mol Biol (Noisy-Le-Grand France) (2021) 67(3):35–43. doi: 10.14715/CMB/2021.67.3.5
329. Devaraj S, Semaan JR, Jialal I. Biochemistry, Apolipoprotein B. In: StatPearls. StatPearls Publishing (Treasure Island, FL) (2023). Available at: https://www.ncbi.nlm.nih.gov/books/NBK538139/.
330. Fang J, Uchiumi T, Yagi M, Matsumoto S, Amamoto R, Takazaki S, et al. Dihydro-orotate dehydrogenase is physically associated with the respiratory complex and its loss leads to mitochondrial dysfunction. Biosci Rep (2013) 33(2):217–27.
331. Huang FC. The role of sphingolipids on innate immunity to intestinal salmonella infection. Int J Mol Sci (2017) 18(8):1720. doi: 10.3390/ijms18081720
332. Fusco JP, Pita G, Pajares MJ, Andueza MP, Patino-Garcia A, de-Torres JP, et al. Genomic characterization of individuals presenting extreme phenotypes of high and low risk to develop tobacco-induced lung cancer. Cancer Med (2018) 7(7):3474–83. doi: 10.1002/cam4.1500
333. Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, et al. Genetic determinants of circulating sphingolipid concentrations in European populations. PloS Genet (2009) 5(10):e1000672. doi: 10.1371/journal.pgen.1000672
334. Shen ZJ, Hu J, Esnault S, Dozmorov I, Malter JS. RNA Seq profiling reveals a novel expression pattern of TGF-beta target genes in human blood eosinophils. Immunol Lett (2015) 167(1):1–10. doi: 10.1016/j.imlet.2015.06.012
335. Zhang N, Di J, Wang Z, Gao P, Jiang B, Su X. Genomic profiling of colorectal cancer with isolated lung metastasis. Cancer Cell Int (2020) 20:281. doi: 10.1186/s12935-020-01373-x
336. Ganev M, Balabanski L, Serbezov D, Karachanak-Yankova S, Vazharova R, Nesheva D, et al. Prioritization of genetic variants predisposing to coronary heart disease in the bulgarian population using centenarian exomes. Biotechnol Biotechnol Equipment. (2019) 33(1):1757–65. doi: 10.1080/13102818.2019.1700164
337. Al Gadban MM, German J, Truman JP, Soodavar F, Riemer EC, Twal WO, et al. Lack of nitric oxide synthases increases lipoprotein immune complex deposition in the aorta and elevates plasma sphingolipid levels in lupus. Cell Immunol (2012) 276(1-2):42–51. doi: 10.1016/j.cellimm.2012.03.007
338. Miura K, Nagahashi M, Prasoon P, Hirose Y, Kobayashi T, Sakata J, et al. Dysregulation of sphingolipid metabolic enzymes leads to high levels of sphingosine-1-phosphate and ceramide in human hepatocellular carcinoma. Hepatol Res (2021) 51(5):614–26. doi: 10.1111/hepr.13625
339. Fernández M, Alarcón GS, Calvo-alén J, Andrade R, McGwin G Jr., Vilá LM, et al. A multiethnic, multicenter cohort of patients with systemic lupus erythematosus (SLE) as a model for the study of ethnic disparities in SLE. Arthritis Care Res (2007) 57(4):576–84. doi: 10.1002/art.22672
340. Takeda H, Takai A, Inuzuka T, Marusawa H. Genetic basis of hepatitis virus-associated hepatocellular carcinoma: linkage between infection, inflammation, and tumorigenesis. J Gastroenterol (2017) 52(1):26–38. doi: 10.1007/s00535-016-1273-2
341. Elmessaoudi-Idrissi M, Windisch MP, Kettani A, Altawalah H, Pineau P, Benjelloun S, et al. An integrative gene expression microarray meta-analysis identifies host factors and key signatures involved in hepatitis B virus infection. Infect Disord Drug Targets (2020) 20(5):698–707. doi: 10.2174/1871526519666190807153901
342. Wolff RK, Hoffman MD, Wolff EC, Herrick JS, Sakoda LC, Samowitz WS, et al. Mutation analysis of adenomas and carcinomas of the colon: Early and late drivers. Genes Chromosomes Cancer (2018) 57(7):366–76. doi: 10.1002/gcc.22539
343. Liu Y, Xia J, McKay J, Tsavachidis S, Xiao X, Spitz MR, et al. Rare deleterious germline variants and risk of lung cancer. NPJ Precis Oncol (2021) 5(1):12. doi: 10.1038/s41698-021-00146-7
344. The UniProt Consortium. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res (2023) 51(D1):D523–31. doi: 10.1093/nar/gkac1052
345. Lu W, Liu CC, Thottassery JV, Bu G, Li Y. Mesd is a universal inhibitor of Wnt coreceptors LRP5 and LRP6 and blocks Wnt/beta-catenin signaling in cancer cells. Biochemistry (2010) 49(22):4635–43. doi: 10.1021/bi1001486
346. Gay A, Towler DA. Wnt signaling in cardiovascular disease: opportunities and challenges. Curr Opin Lipidol (2017) 28(5):387–96. doi: 10.1097/MOL.0000000000000445
347. Luck K, Kim D-K, Lambourne L, Spirohn K, Begg BE, Bian W, et al. A reference map of the human binary protein interactome. Nature (2020) 580(7803):402–8. doi: 10.1038/s41586-020-2188-x
348. Xiao C, Yang L, Jin L, Zhang F, Liu J, Yu C, et al. MAGEA11 as a STAD prognostic biomarker associated with immune infiltration. Diagnostics (2022) 12(10):2506. doi: 10.3390/diagnostics12102506
349. Gonzalez-Pons M, Torres-Cintrón CR, Soto-Salgado M, Vargas-Ramos Y, Perez-Portocarrero L, Morgan DR, et al. Racial/ethnic disparities in gastric cancer: A 15-year population-based analysis. Cancer Med (2023) 12(2):1860–8. doi: 10.1002/cam4.4997
350. Tang Q, Liu Q, Li Y, Mo L, He J. CRELD2, endoplasmic reticulum stress, and human diseases. Front Endocrinol (Lausanne) (2023) 14:1117414. doi: 10.3389/fendo.2023.1117414
351. Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, et al. Complement factor h polymorphism in age-related macular degeneration. Science (2005) 308(5720):385–9. doi: 10.1126/science.1109557
352. Jatoi I, Sung H, Jemal A. The emergence of the racial disparity in U.S. Breast-cancer mortality. New Engl J Med (2022) 386(25):2349–52. doi: 10.1056/NEJMp2200244
353. Karakas C, Wang C, Deng F, Huang H, Wang D, Lee P. Molecular mechanisms involving prostate cancer racial disparity. Am J Clin Exp Urol (2017) 5(3):34–48.
354. Dougan M, Dranoff G, Dougan SK. GM-CSF, IL-3, and IL-5 family of cytokines: regulators of inflammation. Immunity (2019) 50(4):796–811. doi: 10.1016/j.immuni.2019.03.022
355. Watanabe-Smith K, Tognon C, Tyner JW, Meijerink JP, Druker BJ, Agarwal A. Discovery and functional characterization of a germline, CSF2RB-activating mutation in leukemia. Leukemia (2016) 30(9):1950–3. doi: 10.1038/leu.2016.95
356. Rashid M, Ali R, Almuzzaini B, Song H, AlHallaj A, Abdulkarim AA, et al. Discovery of a novel potentially transforming somatic mutation in CSF2RB gene in breast cancer. Cancer Med (2021) 10(22):8138–50. doi: 10.1002/cam4.4106
357. Puchenkova OA, Nadezhdin SV, Soldatov VO, Zhuchenko MA, Korshunova DS, Kubekina MV, et al. Study of antiatherosclerotic and endothelioprotective activity of peptide agonists of EPOR/CD131 heteroreceptor. Pharm Pharmacol (2020) 8(2):100–11. doi: 10.19163/2307-9266-2020-8-2-100-111
358. Kim HY, Hong S. Multi-faceted roles of DNAJB protein in cancer metastasis and clinical implications. Int J Mol Sci (2022) 23(23):14970. doi: 10.3390/ijms232314970
359. Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis–a genomewide study. N Engl J Med (2007) 357(12):1199–209. doi: 10.1056/NEJMoa073491
360. Genest J, Schwertani A, Choi HY. Membrane microdomains and the regulation of HDL biogenesis. Curr Opin Lipidol (2018) 29(1):36–41. doi: 10.1097/MOL.0000000000000470
361. Zhang Z, Zhu H, Hu J. CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis (2021) 12(2):1–12. doi: 10.1038/s41419-021-03486-1
362. Wang Y, Chen C, Wang X, Jin F, Liu Y, Liu H, et al. Lower DSC1 expression is related to the poor differentiation and prognosis of head and neck squamous cell carcinoma (HNSCC). J Cancer Res Clin Oncol (2016) 142(12):2461–8. doi: 10.1007/s00432-016-2233-1
363. Myklebust MP, Fluge Ø, Immervoll H, Skarstein A, Balteskard L, Bruland O, et al. Expression of DSG1 and DSC1 are prognostic markers in anal carcinoma patients. Br J Cancer (2012) 106(4):756–62. doi: 10.1038/bjc.2011.548
364. Mei S, Chelmow D, Gecsi K, Barkley J, Barrows E, Brooks R, et al. Health disparities in ovarian cancer: report from the ovarian cancer evidence review conference. Obstet Gynecol (2023) 142(1):196–210. doi: 10.1097/AOG.0000000000005210
365. Hutchinson MA, Park HS, Zanotti KJ, Alvarez-Gonzalez J, Zhang J, Zhang L, et al. Auto-antibody production during experimental atherosclerosis in apoE(-/-) mice. Front Immunol (2021) 12:695220. doi: 10.3389/fimmu.2021.695220
366. Naderi A. Genomic and epigenetic aberrations of chromosome 1p36. 13 have prognostic implications in Malignancies. Chromosome Res (2020) 28(3):307–30. doi: 10.1007/s10577-020-09638-x
367. Ma Y, Kan C, Qiu H, Liu Y, Hou N, Han F, et al. Transcriptomic analysis reveals the protective effects of empagliflozin on lipid metabolism in nonalcoholic fatty liver disease. Front Pharmacol (2021) 12. doi: 10.3389/fphar.2021.793586
368. Ruggiero AD, Vemuri R, Block M, DeStephanis D, Davis M, Chou J, et al. Macrophage phenotypes and gene expression patterns are unique in naturally occurring metabolically healthy obesity. Int J Mol Sci (2022) 23(20):12680. doi: 10.3390/ijms232012680
369. Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, et al. A proteome-scale map of the human interactome network. Cell (2014) 159(5):1212–26. doi: 10.1016/j.cell.2014.10.050
370. He C, Liu W, Xiong Y, Wang Y, Pan L, Luo L, et al. VSNL1 promotes cell proliferation, migration, and invasion in colorectal cancer by binding with COL10A1. Ann Clin Lab Sci (2022) 52(1):60–72.
371. Aiba T, Hijiya N, Akagi T, Tsukamoto Y, Hirashita Y, Kinoshita K, et al. Overexpression of VSNL1 enhances cell proliferation in colorectal carcinogenesis. Pathobiology (2023) 1–11. doi: 10.1159/000533877
372. Dai QQ, Wang YY, Jiang YP, Li L, Wang HJ. VSNL1 promotes gastric cancer cell proliferation and migration by regulating P2X3/P2Y2 receptors and is a clinical indicator of poor prognosis in gastric cancer patients. Gastroenterol Res Pract (2020) 2020:7241942. doi: 10.1155/2020/7241942
373. Bui A, Yang L, Soroudi C, May FP. Racial and ethnic disparities in incidence and mortality for the five most common gastrointestinal cancers in the United States. J Natl Med Assoc (2022) 114(4):426–9. doi: 10.1016/j.jnma.2022.04.001
374. Liu Y, Lin S, Xie Y, Zhao L, Du H, Yang S, et al. ILDR1 promotes influenza A virus replication through binding to PLSCR1. Sci Rep (2022) 12(1):1–13. doi: 10.1038/s41598-022-12598-3
375. Wang L, Zhai R, Song G, Wang Y. Analyses of the expression and prognosis of ILDR1 in human gastric cancer. Heliyon (2022) 8(9):e10253. doi: 10.1016/j.heliyon.2022.e10253
376. Chandra R, Aryal DK, Douros JD, Shahid R, Davis SJ, Campbell JE, et al. Ildr1 gene deletion protects against diet-induced obesity and hyperglycemia. PloS One (2022) 17(6):e0270329. doi: 10.1371/journal.pone.0270329
377. Deng S, Wang Y, Brosseau JP, Wang Y, Xu K, Chi H, et al. The ITPRIPL1-CD3ϵ axis: a novel immune checkpoint controlling T cells activation. bioRxiv (2022). doi: 10.1101/2022.02.25.481189
378. Wang SC, Liao LM, Ansar M, Lin SY, Hsu WW, Su CM, et al. Automatic detection of the circulating cell-free methylated DNA pattern of GCM2, ITPRIPL1 and CCDC181 for detection of early breast cancer and surgical treatment response. Cancers (2021) 13(6):1375. doi: 10.3390/cancers13061375
379. Joshi H, Vastrad B, Joshi N, Vastrad C. Integrated bioinformatics analysis reveals novel key biomarkers in diabetic nephropathy. SAGE Open Med (2022) 10:1–32. doi: 10.1177/20503121221137005
380. Choi JH, Zhong X, Zhang Z, Su L, McAlpine W, Misawa T, et al. Essential cell-extrinsic requirement for PDIA6 in lymphoid and myeloid development. J Exp Med (2020) 217(4):e20190006. doi: 10.1084/jem.20190006
381. Jordan PA, Stevens JM, Hubbard GP, Barrett NE, Sage T, Authi KS, et al. A role for the thiol isomerase protein ERP5 in platelet function. Blood (2005) 105(4):1500–7. doi: 10.1182/blood-2004-02-0608
382. Mao L, Wu X, Gong Z, Yu M, Huang Z. PDIA6 contributes to aerobic glycolysis and cancer progression in oral squamous cell carcinoma. World J Surg Oncol (2021) 19:1–9. doi: 10.1186/s12957-021-02190-w
383. Bai Y, Liu X, Qi X, Liu X, Peng F, Li H, et al. PDIA6 modulates apoptosis and autophagy of non-small cell lung cancer cells via the MAP4K1/JNK signaling pathway. EBioMedicine (2019) 42:311–25. doi: 10.1016/j.ebiom.2019.03.045
384. Ramos FS, Serino LT, Carvalho CM, Lima RS, Urban CA, Cavalli IJ, et al. PDIA3 and PDIA6 gene expression as an aggressiveness marker in primary ductal breast cancer. Genet Mol Res (2015) 14(2):6960–7. doi: 10.4238/2015.June.26.4
385. Cheng HP, Liu Q, Li Y, Li XD, Zhu CY. The inhibitory effect of PDIA6 downregulation on bladder cancer cell proliferation and invasion. Oncol Res Featuring Preclinical Clin Cancer Ther (2017) 25(4):587–93. doi: 10.3727/096504016X14761811155298
386. Yan C, Song X, Wang S, Wang J, Li L. Knockdown of PDIA6 inhibits cell proliferation and enhances the chemosensitivity in gastric cancer cells. Cancer Manage Res (2020) 12:11051. doi: 10.2147/CMAR.S267711
387. Ma Y, Xia P, Wang Z, Xu J, Zhang L, Jiang Y. PDIA6 promotes pancreatic cancer progression and immune escape through CSN5-mediated deubiquitination of β-catenin and PD-L1. Neoplasia (2021) 23(9):912–28. doi: 10.1016/j.neo.2021.07.004
388. Liu N, Wu T, Ma Y, Cheng H, Li W, Chen M. Identification and validation of RB1 as an immune-related prognostic signature based on tumor mutation burdens in bladder cancer. Anti-Cancer Drugs (2023) 34(2):269–80. doi: 10.1097/CAD.0000000000001399
389. Dyson NJ. RB1: a prototype tumor suppressor and an enigma. Genes Dev (2016) 30(13):1492–502. doi: 10.1101/gad.282145
390. Moreno-Navarrete JM, Petrov P, Serrano M, Ortega F, Garcia-Ruiz E, Oliver P, et al. Decreased RB1 mRNA, protein, and activity reflect obesity-induced altered adipogenic capacity in human adipose tissue. Diabetes (2013) 62(6):1923–31. doi: 10.2337/db12-0977
391. Janostiak R, Torres-Sanchez A, Posas F, de Nadal E. Understanding retinoblastoma post-translational regulation for the design of targeted cancer therapies. Cancers (Basel) (2022) 14(5):1265. doi: 10.3390/cancers14051265
392. Zuniga J, Buendia-Roldan I, Zhao Y, Jimenez L, Torres D, Romo J, et al. Genetic variants associated with severe pneumonia in A/H1N1 influenza infection. Eur Respir J (2012) 39(3):604–10. doi: 10.1183/09031936.00020611
393. Mehrbod P, Eybpoosh S, Farahmand B, Fotouhi F, Khanzadeh Alishahi M. Association of the host genetic factors, hypercholesterolemia and diabetes with mild influenza in an Iranian population. Virol J (2021) 18(1):1–11. doi: 10.1186/s12985-021-01486-3
394. Chen H, Luo J, Guo J. Identification of an alternative splicing signature as an independent factor in colon cancer. BMC Cancer (2020) 20(1):1–11. doi: 10.1186/s12885-020-07419-7
395. Sachamitr P, Ho JC, Ciamponi FE, Ba-Alawi W, Coutinho FJ, Guilhamon P, et al. PRMT5 inhibition disrupts splicing and stemness in glioblastoma. Nat Commun (2021) 12(1):1–17. doi: 10.1038/s41467-021-21204-5
396. Wu J, Wang M, Han L, Zhang H, Lei S, Zhang Y, et al. RNA modification-related variants in genomic loci associated with body mass index. Hum Genomics (2022) 16(1):25. doi: 10.1186/s40246-022-00403-1
397. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet (2010) 42(11):937–48. doi: 10.1038/ng.686
398. Augustus GJ, Ellis NA. Colorectal cancer disparity in African Americans: risk factors and carcinogenic mechanisms. Am J Pathol (2018) 188(2):291–303. doi: 10.1016/j.ajpath.2017.07.023
399. O'Connor BP, Eun SY, Ye Z, Zozulya AL, Lich JD, Moore CB, et al. Semaphorin 6D regulates the late phase of CD4+ T cell primary immune responses. Proc Natl Acad Sci (2008) 105(35):13015–20. doi: 10.1073/pnas.0803386105
400. Kanth SM, Gairhe S, Torabi-Parizi P. The role of semaphorins and their receptors in innate immune responses and clinical diseases of acute inflammation. Front Immunol (2021) 12:672441. doi: 10.3389/fimmu.2021.672441
401. Kang S, Nakanishi Y, Kioi Y, Okuzaki D, Kimura T, Takamatsu H, et al. Semaphorin 6D reverse signaling controls macrophage lipid metabolism and anti-inflammatory polarization. Nat Immunol (2018) 19(6):561–70. doi: 10.1038/s41590-018-0108-0
402. Baxter DE, Allinson LM, Al Amri WS, Poulter JA, Pramanik A, Thorne JL, et al. MiR-195 and Its Target SEMA6D regulate chemoresponse in breast cancer. Cancers (2021) 13(23):5979. doi: 10.3390/cancers13235979
403. Lu Q, Zhu L. The role of semaphorins in metabolic disorders. Int J Mol Sci (2020) 21(16):5641. doi: 10.3390/ijms21165641
404. Stokowski RP, Pant PV, Dadd T, Fereday A, Hinds DA, Jarman C, et al. A genomewide association study of skin pigmentation in a south asian population. Am J Hum Genet (2007) 81(6):1119–32. doi: 10.1086/522235
405. Kasperaviciūte D, Catarino CB, Heinzen EL, Depondt C, Cavalleri GL, Caboclo LO, et al. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain (2010) 133(Pt 7):2136–47. doi: 10.1093/brain/awq130
406. Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med (2010) 363(13):1211–21. doi: 10.1056/NEJMoa0906312
407. Chen D, Li Y, Wang L, Jiao K. SEMA6D expression and patient survival in breast invasive carcinoma. Int J Breast Cancer (2015) 2015:539721. doi: 10.1155/2015/539721
408. Attar H, Bedard K, Migliavacca E, Gagnebin M, Dupre Y, Descombes P, et al. Extensive natural variation for cellular hydrogen peroxide release is genetically controlled. PloS One (2012) 7(8):e43566. doi: 10.1371/journal.pone.0043566
409. Mendoza-Alvarez A, Guillen-Guio B, Baez-Ortega A, Hernandez-Perez C, Lakhwani-Lakhwani S, Maeso MDC, et al. Whole-exome sequencing identifies somatic mutations associated with mortality in metastatic clear cell kidney carcinoma. Front Genet (2019) 10:439. doi: 10.3389/fgene.2019.00439
410. Pena-Chilet M, Esteban-Medina M, Falco MM, Rian K, Hidalgo MR, Loucera C, et al. Using mechanistic models for the clinical interpretation of complex genomic variation. Sci Rep (2019) 9(1):18937. doi: 10.1038/s41598-019-55454-7
411. Wang Y, Li J, Li J, Li P, Wang L, Di L. An enhancer-based analysis revealed a new function of androgen receptor in tumor cell immune evasion. Front Genet (2020) 11:595550. doi: 10.3389/fgene.2020.595550
412. Pettinelli P, Arendt BM, Schwenger KJP, Sivaraj S, Bhat M, Comelli EM, et al. Relationship between hepatic gene expression, intestinal microbiota, and inferred functional metagenomic analysis in NAFLD. Clin Transl Gastroenterol (2022) 13(7):e00466. doi: 10.14309/ctg.0000000000000466
413. Canhong W, Jiarui LI, Jiahui C, Haizhu T. SIPA1L2 as a risk factor implicated in Alzheimer's disease. J Univ Sci Technol China (2021) 51(2):147. doi: 10.52396/JUST-2021-0008
414. Wang Y, Chang Q, Li Y. Racial differences in urinary bladder cancer in the United States. Sci Rep (2018) 8(12521):12521. doi: 10.1038/s41598-018-29987-2
415. Ghosh P, Campos VJ, Vo DT, Guccione C, Goheen-Holland V, Tindle C, et al. AI-assisted discovery of an ethnicity-influenced driver of cell transformation in esophageal and gastroesophageal junction adenocarcinomas. JCI Insight (2022) 7(18):e161334. doi: 10.1172/jci.insight.161334
416. Muhimpundu S, Conway RBN, Warren Andersen S, Lipworth L, Steinwandel MD, Blot WJ, et al. Racial differences in hepatocellular carcinoma incidence and risk factors among a low socioeconomic population. Cancers (2021) 13(15):3710. doi: 10.3390/cancers13153710
417. Capmany A, Gambarte Tudela J, Alonso Bivou M, Damiani MT. Akt/AS160 signaling pathway inhibition impairs infection by decreasing Rab14-controlled sphingolipids delivery to chlamydial inclusions. Front Microbiol (2019) 10:666. doi: 10.3389/fmicb.2019.00666
418. Jiang XH, Sun JW, Xu M, Jiang XF, Liu CF, Lu Y. Frequent hyperphosphorylation of AS160 in breast cancer. Cancer Biol Ther (2010) 10(4):362–7. doi: 10.4161/cbt.10.4.12426
419. Cheng JC, McBrayer SK, Coarfa C, Dalva-Aydemir S, Gunaratne PH, Carpten JD, et al. Expression and phosphorylation of the AS160_v2 splice variant supports GLUT4 activation and the Warburg effect in multiple myeloma. Cancer Metab (2013) 1(1):1–8. doi: 10.1186/2049-3002-1-14
420. Moltke I, Grarup N, Jørgensen ME, Bjerregaard P, Treebak JT, Fumagalli M, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature (2014) 512(7513):190–3. doi: 10.1038/nature13425
421. Chen MH, Raffield LM, Mousas A, Sakaue S, Huffman JE, Moscati A, et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell (2020) 182(5):1198–213. e14. doi: 10.1016/j.cell.2020.06.045
422. Sakaue S, Kanai M, Tanigawa Y, Karjalainen J, Kurki M, Koshiba S, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet (2021) 53(10):1415–24. doi: 10.1038/s41588-021-00931-x
423. Vuckovic D, Bao EL, Akbari P, Lareau CA, Mousas A, Jiang T, et al. The polygenic and monogenic basis of blood traits and diseases. Cell (2020) 182(5):1214–31.e11. doi: 10.1016/j.cell.2020.08.008
424. Kanapuru B, Fernandes LL, Fashoyin-Aje LA, Baines AC, Bhatnagar V, Ershler R, et al. Analysis of racial and ethnic disparities in multiple myeloma US FDA drug approval trials. Blood Adv (2022) 6(6):1684–91. doi: 10.1182/bloodadvances.2021005482
425. Lyu J, Wang P, Xu T, Shen Y, Cui Z, Zheng M, et al. Thymic-specific regulation of TCR signaling by Tespa1. Cell Mol Immunol (2019) 16(12):897–907. doi: 10.1038/s41423-019-0259-4
426. Liu L, Liu Y. Pan-cancer analysis of the prognostic and immunological role of thymocyte-expressed positive selection-associated protein 1 (TESPA1) in human tumors. Research Square pre-print (2022). doi: 10.21203/rs.3.rs-1249485/v1
427. Luan Y, Luan Y, Yuan RX, Feng Q, Chen X, Yang Y. Structure and function of mitochondria-associated endoplasmic reticulum membranes (MAMs) and their role in cardiovascular diseases. Oxid Med Cell Longevity (2021) 2021:4578809. doi: 10.1155/2021/4578809
428. Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet (2011) 43(3):246–52. doi: 10.1038/ng.764
429. Mead S, Uphill J, Beck J, Poulter M, Campbell T, Lowe J, et al. Genome-wide association study in multiple human prion diseases suggests genetic risk factors additional to PRNP. Hum Mol Genet (2012) 21(8):1897–906. doi: 10.1093/hmg/ddr607
430. Kirtane K, Lee SJ. Racial and ethnic disparities in hematologic Malignancies. Blood (2017) 130(15):1699–705. doi: 10.1182/blood-2017-04-778225
431. Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet (2012) 44(7):760–4. doi: 10.1038/ng.2291
432. Holm M, Joenväärä S, Saraswat M, Mustonen H, Tohmola T, Ristimäki A, et al. Identification of several plasma proteins whose levels in colorectal cancer patients differ depending on outcome. FASEB BioAdvances (2019) 1(12):723–30. doi: 10.1096/fba.2019-00062
433. Liu H, Bai N, Liu S, Liu W, Zhou Q, Zhang H. Exploring feature genes for ischemic stroke based on bioinformatics and machine learning. Atherosclerosis (2022) 355:238. doi: 10.1016/j.atherosclerosis.2022.06.920
434. Yucesan E, Hatirnaz Ng O, Yalniz FF, Yilmaz H, Salihoglu A, Sudutan T, et al. Copy-number variations in adult patients with chronic immune thrombocytopenia. Expert Rev Hematol (2020) 13(11):1277–87. doi: 10.1080/17474086.2020.1819786
435. Liu G, Tang H, Li C, Zhen H, Zhang Z, Sha Y. Prognostic gene biomarker identification in liver cancer by data mining. Am J Trans Res (2021) 13(5):4603.
436. Seyres D, Cabassi A, Lambourne JJ, Burden F, Farrow S, McKinney H, et al. Transcriptional, epigenetic and metabolic signatures in cardiometabolic syndrome defined by extreme phenotypes. Clin Epigenet (2022) 14(1):1–24. doi: 10.1186/s13148-022-01257-z
437. Masuda K, Kornberg A, Miller J, Lin S, Suek N, Botella T, et al. Multiplexed single-cell analysis reveals prognostic and nonprognostic T cell types in human colorectal cancer. JCI Insight (2022) 7(7):e154646. doi: 10.1172/jci.insight.154646
438. Liang WS, Craig DW, Carpten J, Borad MJ, Demeure MJ, Weiss GJ, et al. Genome-wide characterization of pancreatic adenocarcinoma patients using next generation sequencing. PloS One (2012) 7(10):e43192. doi: 10.1371/journal.pone.0043192
439. Lazare C, Zhi W, Dai J, Cao C, Sookha RR, Wang L, et al. A pilot study comparing the genetic molecular biology of gestational and non-gestational choriocarcinoma. Am J Transl Res (2019) 11(11):7049–62.
440. Huang C, Jia Y, Yang S, Chen B, Sun H, Shen F, et al. Characterization of ZNF23, a KRAB-containing protein that is downregulated in human cancers and inhibits cell cycle progression. Exp Cell Res (2007) 313(2):254–63. doi: 10.1016/j.yexcr.2006.10.009
441. Ruocco MR, Avagliano A, Granato G, Vigliar E, Masone S, Montagnani S, et al. Metabolic flexibility in melanoma: A potential therapeutic target. Semin Cancer Biol (2019) 59:187–207. doi: 10.1016/j.semcancer.2019.07.016
442. Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol (2003) 4(10):1–8. doi: 10.1186/gb-2003-4-10-231
443. Patel S, Howard D, Chowdhury N, Derieux C, Wellslager B, Yilmaz O, et al. Characterization of human genes modulated by porphyromonas gingivalis highlights the ribosome, hypothalamus, and cholinergic neurons. Front Immunol (2021) 12:646259. doi: 10.3389/fimmu.2021.646259
444. Schnabl B, Valletta D, Kirovski G, Hellerbrand C. Zinc finger protein 267 is up-regulated in hepatocellular carcinoma and promotes tumor cell proliferation and migration. Exp Mol Pathol (2011) 91(3):695–701. doi: 10.1016/j.yexmp.2011.07.006
445. Long X, Liu X, Deng T, Chen J, Lan J, Zhang S, et al. LARP6 suppresses colorectal cancer progression through ZNF267/SGMS2-mediated imbalance of sphingomyelin synthesis. J Exp Clin Cancer Res (2023) 42(1):33. doi: 10.1186/s13046-023-02605-4
446. Yang H, Wang L, Zheng Y, Hu G, Ma H, Shen L. Knockdown of zinc finger protein 267 suppresses diffuse large B-cell lymphoma progression, metastasis, and cancer stem cell properties. Bioengineered (2022) 13(1):1686–701. doi: 10.1080/21655979.2021.2014644
447. Schnabl B, Hu K, Mühlbauer M, Hellerbrand C, Stefanovic B, Brenner DA, et al. Zinc finger protein 267 is up-regulated during the activation process of human hepatic stellate cells and functions as a negative transcriptional regulator of MMP-10. Biochem Biophys Res Commun (2005) 335(1):87–96. doi: 10.1016/j.bbrc.2005.07.043
448. Schnabl B, Czech B, Valletta D, Weiss TS, Kirovski G, Hellerbrand C. Increased expression of Zinc finger protein 267 in non-alcoholic fatty liver disease. Int J Clin Exp Pathol (2011) 4(7):661.
449. Primm KM, Malabay AJ, Curry T, Chang S. Who, where, when: Colorectal cancer disparities by race and ethnicity, subsite, and stage. Cancer Med (2023) 12(13):14767–80. doi: 10.1002/cam4.6105
450. Gustafson EA, Seymour KA, Sigrist K, Rooij DGDE, Freiman RN. ZFP628 is a TAF4b-interacting transcription factor required for mouse spermiogenesis. Mol Cell Biol (2020) 40(7):e00228-19. doi: 10.1128/MCB
451. Aspedon A, Groisman EA. The antibacterial action of protamine: evidence for disruption of cytoplasmic membrane energization in Salmonella typhimurium. Microbiology (1996) 142(12):3389–97. doi: 10.1099/13500872-142-12-3389
452. Pink DA, Hasan FM, Quinn BE, Winterhalter M, Mohan M, Gill TA. Interaction of protamine with gram-negative bacteria membranes: possible alternative mechanisms of internalization in Escherichia coli, Salmonella typhimurium and Pseudomonas aeruginosa. J Pept Sci (2014) 20(4):240–50. doi: 10.1002/psc.2610
453. Meklat F, Zhang Y, Shahriar M, Ahmed SU, Li W, Voukkalis N, et al. Identification of protamine 1 as a novel cancer-testis antigen in early chronic lymphocytic leukaemia. Br J Haematol (2009) 144(5):660–6. doi: 10.1111/j.1365-2141.2008.07502.x
454. Ren S, Chen X, Tian X, Yang D, Dong Y, Chen F, et al. The expression, function, and utilization of Protamine1: a literature review. Transl Cancer Res (2021) 10(11):4947–57. doi: 10.21037/tcr-21-1582
455. Ramzan R, Michels S, Weber P, Rhiel A, Irqsusi M, Rastan AJ, et al. Protamine sulfate induces mitochondrial hyperpolarization and a subsequent increase in reactive oxygen species production. J Pharmacol Exp Ther (2019) 370(2):308–17. doi: 10.1124/jpet.119.257725
456. Chen GY, Muramatsu H, Ichihara-Tanaka K, Muramatsu T. ZEC, a zinc finger protein with novel binding specificity and transcription regulatory activity. Gene (2004) 340(1):71–81. doi: 10.1016/j.gene.2004.06.016
457. Beck T, Rowlands T, Shorter T, Brookes AJ. GWAS central: an expanding resource for finding and visualising genotype and phenotype data from genome-wide association studies. Nucleic Acids Res (2023) 51(D1):D986–D93. doi: 10.1093/nar/gkac1017
458. Hirata Y, Yanaihara N, Yanagida S, Fukui K, Iwadate K, Kiyokawa T, et al. Molecular genetic analysis of nongestational choriocarcinoma in a postmenopausal woman: a case report and literature review. Int J Gynecological Pathol (2012) 31(4):364–8. doi: 10.1097/PGP.0b013e318241d556
459. Higashi T, Tokuda S, Kitajiri S, Masuda S, Nakamura H, Oda Y, et al. Analysis of the A’ngulin’ proteins LSR, ILDR1 and ILDR2–tricellulin recruitment, epithelial barrier function and implication in deafness pathogenesis. J Cell Sci (2013) 126(Pt 4):966–77. doi: 10.1242/jcs.116442
460. Chandra R, Wang Y, Shahid RA, Vigna SR, Freedman NJ, Liddle RA. Immunoglobulin-like domain containing receptor 1 mediates fat-stimulated cholecystokinin secretion. J Clin Invest (2013) 123(8):3343–52. doi: 10.1172/jci68587
461. Gong Y, Himmerkus N, Sunq A, Milatz S, Merkel C, Bleich M, et al. ILDR1 is important for paracellular water transport and urine concentration mechanism. Proc Natl Acad Sci U.S.A. (2017) 114(20):5271–6. doi: 10.1073/pnas.1701006114
462. Trapnell BC, Nakata K, Bonella F, Campo I, Griese M, Hamilton J, et al. Pulmonary alveolar proteinosis. Nat Rev Dis Primers (2019) 5(1):16. doi: 10.1038/s41572-019-0066-3
463. Oughtred R, Rust J, Chang C, Breitkreutz BJ, Stark C, Willems A, et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci (2021) 30(1):187–200. doi: 10.1002/pro.3978
Keywords: innate immune variants, pleiotropic actions, cancer disparities, cardio-metabolic disparities, population-enriched variants, candidate protein targets
Citation: Yeyeodu S, Hanafi D, Webb K, Laurie NA and Kimbro KS (2024) Population-enriched innate immune variants may identify candidate gene targets at the intersection of cancer and cardio-metabolic disease. Front. Endocrinol. 14:1286979. doi: 10.3389/fendo.2023.1286979
Received: 01 September 2023; Accepted: 07 December 2023;
Published: 21 March 2024.
Edited by:
Irene Bertolini, Wistar Institute, United StatesReviewed by:
Ken Batai, University at Buffalo, United StatesAyo Priscille Doumatey, National Institutes of Health (NIH), United States
Copyright © 2024 Yeyeodu, Hanafi, Webb, Laurie and Kimbro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: K. Sean Kimbro, c2tpbWJyb0Btc20uZWR1
 Susan Yeyeodu
Susan Yeyeodu Donia Hanafi
Donia Hanafi Kenisha Webb
Kenisha Webb Nikia A. Laurie1
Nikia A. Laurie1 K. Sean Kimbro
K. Sean Kimbro