- 1Department of Entomology, University of Georgia, Athens, GA, United States
- 2Department of Plant Pathology, Entomology, Microbiology Iowa State University, Ames, IA, United States
Neuropeptides are involved in almost all physiological activities of insects. Their classification is based on physiological function and the primary amino acid sequence. The pyrokinin (PK)/pheromone biosynthesis activating neuropeptides (PBAN) are one of the largest neuropeptide families in insects, with a conserved C-terminal domain of FXPRLamide. The peptide family is divided into two groups, PK1/diapause hormone (DH) with a WFGPRLa C-terminal ending and PK2/PBAN with FXPRLamide C-terminal ending. Since the development of cutting-edge technology, an increasing number of peptides have been sequenced primarily through genomic, transcriptomics, and proteomics, and their functions discovered using gene editing tools. In this review, we discussed newly discovered functions, and analyzed the distribution of genes encoding these peptides throughout different insect orders. In addition, the location of the peptides that were confirmed by PCR or immunocytochemistry is also described. A phylogenetic tree was constructed according to the sequences of the receptors of most insect orders. This review offers an understanding of the significance of this conserved peptide family in insects.
1 Introduction
Neuropeptides are very important in the physiology of insects. As the name indicates, they are peptides produced by the nervous system that act as endocrine and/or neuronal signals. They are synthesized in neurons and then travel down the axon to a synapse or can be released into the hemolymph for circulation through a neurohemal organ. Many neuropeptides have been identified from various insects and are involved in regulating physiology and behavior of insects (1, 2). Neuropeptides belong in families based on the primary amino acid sequence and physiological function. The major family of pyrokinin (PK)/pheromone biosynthesis activating neuropeptides (PBAN) is based on a conserved C-terminal motif of FXPRLamide which is the minimal requirement for biological activity (3, 4). The first PK was identified in the cockroach Leucophaea maderae, based on the stimulation of hindgut muscle contraction (5). The first PBAN peptide was isolated from the corn earworm moth, Helicoverpa zea, which regulates sex pheromone biosynthesis (4, 6). Since then, more peptides and their corresponding receptors have been identified through mass spectrometry, transcriptomics, proteomics, and genomics.
The gene encoding PBAN was first identified in H. zea (7). The H. zea pban gene can produce 5 neuropeptides including PBAN and diapause hormone (DH); the latter named because it induces embryonic diapause in the silkworm Bombyx mori (8). DH has also been referred to as trypto-PK due to the WFGPRLamide C-terminal ending (9). Here we will refer to DH- and PBAN-like peptides as PK1/DH and PK2/PBAN, respectively. The three other peptides have been named pyrokinins, subesophageal ganglion neuropeptides, or hugin peptides; the latter name is based on the hugin gene of Drosophila melanogaster (10). These peptides typically have a FXPRLamide C-terminal ending and will be referred to as PK2-1,2, and 3. Here we will align the neuropeptide gene sequences based on those of Lepidoptera where PK1/DH is the first peptide produced from the 5’ end followed by PK2-1 and 2, PBAN, and PK2-3 (Figure 1) (11). To date, the PK/PBAN family has been discovered in most insect orders and the function of these peptides has been characterized through different methods in many insects.
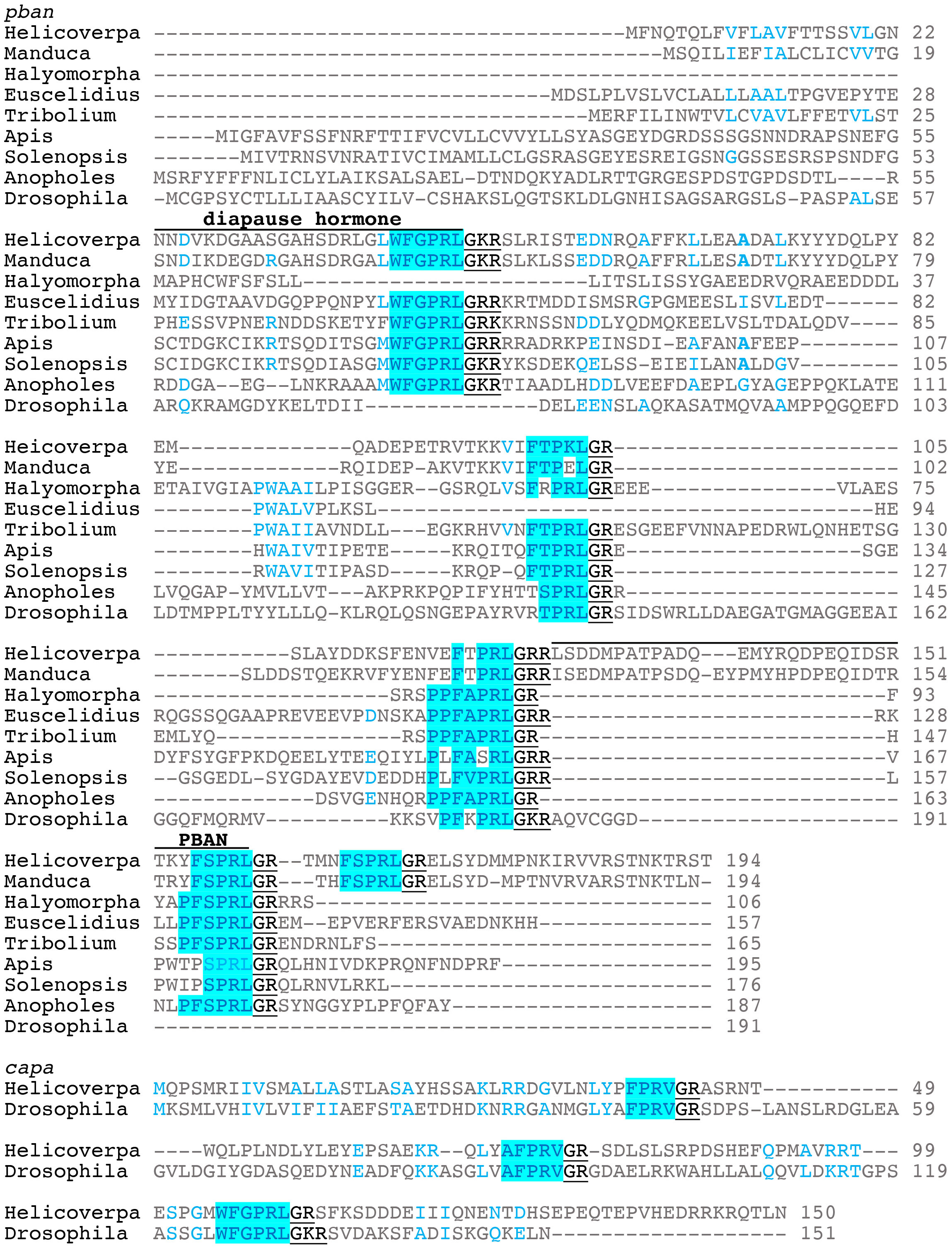
Figure 1 Representative gene sequence alignments for pban and capa. The diapause hormone and PBAN sequences as found in Lepidoptera are indicated on top of the sequences. The GK/R signal is underlined that produces the amide C-terminal ending. Helicoverapa zea and Manduca sexta are lepidopterans; Halymorpha halys and Euscelidius variegatus are hemipterans, Heteroptera and Auchenorrhyncha, respectively; Tribolium castaneum is a coleopteran; Apis mellifera and Solenopsis invicta are hymenopterans; Anopholes gambiae and Drosophila melanogaster are dipterans.
2 Function of the peptides
Previous research has demonstrated that the PK/PBAN family of peptides acts in a variety of functions, including sex pheromone biosynthesis in most moths (6, 12, 13) (but see 14, 15 for exceptions); melanization in moth larvae (16); embryonic diapause in B. mori (17); puparium formation in the higher dipteran, Neobellieria bullata (18); and breaking pupal diapause in heliothine moths (19, 20). More functions have been identified in recent years, indicating the pleiotropic nature of the peptide family. In Lepidoptera, several functions have been identified in addition to sex pheromone biosynthesis and embryonic diapause, including seasonal reproductive polyphenism in the tussock moth, Orgyia thyellina (21) and in addition to the breaking of pupal diapause in heliothine moths, the peptide could terminate larval diapause in Omphisa fuscidentalis (22). RNAi mediated knockdown of pban implicates other functions due to increased pupal mortality in H. zea (23) and delay of larval growth, interference of pupal development, and mortality in H. zea and H. armigera (24). In the fall armyworm, Spodoptera frugiperda, PBAN signaling regulates fecundity (25). Recently, the CRISPR/Cas9 gene editing tool was used to determine the role of pban in Mythimna separata (26). The study demonstrated that pban is necessary for density-dependent cuticular melanization during the late larval stages and is required for sex pheromone production in female adults.
Functions for the PK/PBAN family of peptides have been identified in other orders. In the fire ant, Solenopsis invicta, the knockdown of pban increases larval and adult mortality, and delays pupal development (23). It is also involved in production of the trail pheromone in the fire ant (23). DH-like peptides promote egg diapause in Locusta migratoria (27). The peptides produced by hugin are thought to be involved in regulating feeding behaviors in D. melanogaster (28). PKs were found to be involved in regulation of hindgut function in the adult mosquito, Aedes aegypti (29). Recently, PBAN from the western flower thrip, Frankliniella occidentalis, was shown to induce production of an aggregation pheromone (30).
3 PBAN-like gene encoding neuropeptides in insects
In most insects, two genes (pban and capa) encode several PK/PBAN-like peptides. The pban gene can encode up to five peptides, including a PBAN and a DH, was first identified in moths (7, 31). In Drosophila, the hugin gene is homologous to pban which encodes two PKs (10). The capa gene was first identified in D. melanogaster, encoding two periviscerokinin-like peptides with a PRV/Iamide and a PK with an WFGPRLamide C-terminal ending (32). Representative gene sequences for pban and capa are shown in Figure 1. Biological functions for the periviscerokinins include myotropic activation of muscles, including the heart, and control of diuretic action on the Malpighian tubules (33). So far, based on transcriptomics, the WFGPRLamide peptide produced by capa is found in almost all insects, with a few exceptions (11). It was not found in three species of Siphonaptera and only in the Symphyta of Hymenoptera. The capa of Apoidea and Formicidae could produce a peptide with a GYTPRLamide C-terminal ending in addition to the two periviscerokinins. This review will concentrate on the peptides produced by pban across Insecta (11).
The number of transcriptome analysis of insect species has greatly increased in the past few years. This has greatly increased the number of species in which pban can be identified in NCBI databases and with increased numbers better inferences can be made. The peptides can be identified based on the GR or GK signal at the C-terminal end where the glycine is used to make the amide of the mature peptide (Figure 1). The N-terminal start of the peptide is also determined by a signal sequence that is more variable and also produces a variable length peptide. Here we will concentrate on the C-terminal ending in comparing pban across the Insecta as shown in Figure 2. A gene sequence does not necessarily mean that the insect will produce the mature peptide, which requires processing of the prepropeptide, but a transcriptome sequence does indicate that at least the mRNA is produced and thus the potential to produce a mature peptide.

Figure 2 Consensus sequences of the C-terminal amino acids of peptides that could be produced by the pban gene of the indicated orders and groups of insects. All the peptides are C-terminal amidated using the glycine which precedes the R or K signal. The number of sequences used to generate the sequence logo is indicated as a subscript for each individual peptide. The sequence logo was created using Weblogo (34).
It is immediately apparent that the PK1/DH peptide has the a consistent C-terminal ending and was found in almost all insects. The exception is the Hemiptera in which only the suborder Heteroptera consistently had PK1/DH and was more variable in the suborders Sternorryncha and Auchenorryncha. However, the Heteroptera were found not to have a PK1/DH sequence as was discovered in the brown marmorated stink bug (35). Also, in some Miridae (Heteroptera) pban produces only PK1/DH and PK2-1 with PK1/DH having an atypical FQPRSa C-terminal ending (36). PK2-3 was found in only a relatively few Hemiptera in the suborder Sternorrhyncha.
In the Apterygota, the most primitive hexapods include Diplura, Collembola, Archaeognatha, and Thysanura. All five peptides could be produced but PK1/DH was not found in 4 species of Diplura (37, 38). As described in Derst et al. (37), the Archaeognatha and Collembola have a gene that can produce both CAPA- and PK-like peptide sequences. The Paleoptera consisted of 35 species of Odonata and 5 species of Ephemeroptera, with the latter having the PK1/DH sequence. All species of Odonata lacked the PK1/DH sequence. The Polyneoptera included many hemimetabolous orders. The Polyneoptera order that is missing from Figure 2 is the Orthoptera which apparently do not have a typical pban. All of the genes found in Orthoptera produce multiple copies of PK1/DH-like peptides that end in WFGPRXamide where X is variable but usually a valine. For example, in Locusta migratoria, five PK1/DH precursor genes were identified to produce five types of PK1/DH-like neuropeptides. A functional assay showed that four of the PK1/DH-like neuropeptides stimulated egg diapause under a short photoperiod, but no responses under a long photoperiod (27).
In the holometabolous insects, only the Lepidoptera have a pban that can produce all five peptides. The lower number of species in which PK2-3 was found in Lepidoptera is probably due to truncated sequences of transcriptome data rather than lack of a PK2-3 peptide. One notable difference in PK1/DH sequences is that the Tortricidae have a VFKPILa C-terminal ending (39). In the sister taxon to Lepidoptera, the pban of Trichoptera produced 4 peptides and all 24 species had a remarkable similar sequence structure.
The Neuroptera group included 42 species of Neuroptera, 17 species of Raphidoptera, and 11 species of Megaloptera. The gene structure of Neuroptera is different in that only the PK1/DH peptides were found. In addition, upstream of the PK1/DH sequence are two PRVamide sequences. These are similar to the perivicerokinins found in capa of other insects. Apparently, the peptides produced by pban and capa in other insects are consolidated together in one gene in Neuroptera. In Megaloptera and Raphidioptera, two pban-like genes were found; one similar to the Neuroptera and the other similar to other insects in that in addition to PK1/DH and PK2-1, PK2-2 could be produced. The PK2/PBAN sequence was found in only 4 species of Raphidoptera.
In Coleoptera, most species produced 3 peptides as illustrated in Tribolium castaneum (40). Notable exceptions include 4 species of Coccinellidae and 6 species of Lampyridae that lack the PK1/DH peptide sequence. The PK2-l and PK2-2 sequences were found in most Coleoptera. The corresponding PK2/PBAN sequence was found in fewer species but could be due to truncated sequences of transcriptomic data.
In Hymenoptera, all species had a very similar C-terminal sequence for the PK1/DH peptide. The remaining three peptides were not found in all Hymenoptera. Notably 8 species of the superfamily Chalcidoidea were found to lack the PK2-1 peptide. All 24 species of Vespidae only had the PK1/DH sequence and no other PK2 sequence.
In the lower Diptera (Nematocera), the PK1/DH sequence was found in Tipulidae, Chironomidae, and Culicidae. The PK1/DH sequence was lacking in all the other dipteran families including some lower Diptera like Psychodidae (41), Sciaridae and Keroplatidae. PK2-2 was found in the lower Diptera and in the lower Brachycera families. Only PK2-1 and PK2/PBAN-like sequences were found in the higher Brachycera families. Similar findings were found in a study by Farris (28).
4 PBAN location in CNS
The PK/PBAN family of peptides are detected in the brain, suboesophageal ganglion (SEG), certain cells of the corpora cardiaca, thoracic ganglia (TG), and abdominal ganglia (AG), based on immunohistochemistry using anti-PBAN in moths (42–44). The same sites were confirmed by RT-PCR in H. armigera (45). PVKs are typically found in the AG (46). In noctuid moths, the pban gene expression locations, the structure of PBAN precursors, and the processed neuropeptides are all very comparable. In other insects, such as Drosophila (47, 48), locusts (49), and cockroaches (50), similar expression profiles were found. Similar to other insects, PBAN immunoreactive neurons were detected in the fire ant central nervous system (CNS), but not in the last AG (51). The PK/PBAN-like peptides that could be released into the hemolymph were detected in the brain, SEG, TG, and AG of the mosquito nervous system by using immunohistochemistry (52). Products of the capa gene could be produced in the AG and the products of the pban gene could be produced in the SEG (52). This finding was supported by peptidomics studies in A. aegypti (53), Delia radicum (54), Phlebotomus papatasi (55) and Lucilia cuprina (56). In the stink bug, Halyomorpha halys (Hemiptera), the capa gene was highly expressed in the AG, whereas the pban gene was strongly expressed in the brain, specifically cerebral ganglia - gnathal ganglion (also known as the SEG) (35). The same results were found in silverfish, as in all Pterygota studied so far (38). It appears that in most insects pban is highly expressed in the SEG where PBAN could be released into circulation through the corpora cardiaca. Some of the PBAN containing neurons of the SEG also send axons down the ventral nerve cord to terminate in the last AG.
5 Receptors
Receptors for the PK/PBAN family of peptides are G-protein coupled receptors (GPCR) which are characterized by having seven transmembrane domains. The receptors (r) could be classified into two groups: PK2/PBANr and PK1/DHr based on whether they bind PK2/PBAN-like sequences or PK1/DH-like sequences. The PBAN/PK2r have been cloned and characterized in many insect species starting with D. melanogaster (57) and followed by H. zea (58) and B. mori (59) and later other moths (60). In addition, receptors have been detected in the transcriptome of pheromone glands from various female moths (61–65). In other insects the receptors have been identified in the mosquitoes Anopholes gambiae (66) and Aedes aegypti (67), and the beetle, Tribolium castaneum (40, 68).
The first PK1/DHrs were identified in Drosophila and confirmed using a PK1/DH peptide ligand (69), and then in B. mori (70). Their ligands are similar with a WFGPRLamide C-terminal ending. Previous research demonstrated that the WFGPRLamide peptide activated PK1/DHr in Drosophila, An. gambiae, and B. mori at low nM levels while the other PKs require significantly greater ligand concentrations (67, 69, 70). In R. prolixus, three PK1/DHr variants have been cloned, and they showed different dose responses to PK peptides (71). In H. zea, a PK1/DHr has been identified and has responses to both PK1/DH and PK2 peptides (72). A PKr was also identified in the southern cattle tick, Rhipicephalus (Boophilus) microplus (73) and was found to be basal to both the PK2/PBAN and PK1/DH receptors. The knockdown of the tick pkr caused an increase in mortality and decreased weight of both surviving females and subsequent egg masses indicating a function in reproduction (74).
The PBAN receptors have been detected in most insect orders. The phylogenetic tree shown in Figure 3 for PK2/PBANr was built with representative sequences from 19 orders and in Figure 4 for PK1/DHr was built with representative sequences from 18 orders. The sequences were selected using BLAST results to non-redundant protein sequences with known sequences from D. melanogaster, H. zea, A. mellifera, B. mori, and T. castaneum, and the remaining receptors were detected in the transcriptome shotgun assembly database but without annotation. As demonstrated in previous studies the PK2/PBANr and PK1/DHr are closely related but distinguished from each other in a phylogenetic tree (11, 57, 69). The PK2r and PBANr are grouped together since they have similar ligands with a FXPRLamide C-terminal ending, while PK1r and DHr are grouped together as they are activated by similar ligands with a WFGPRLamide C-terminal ending.
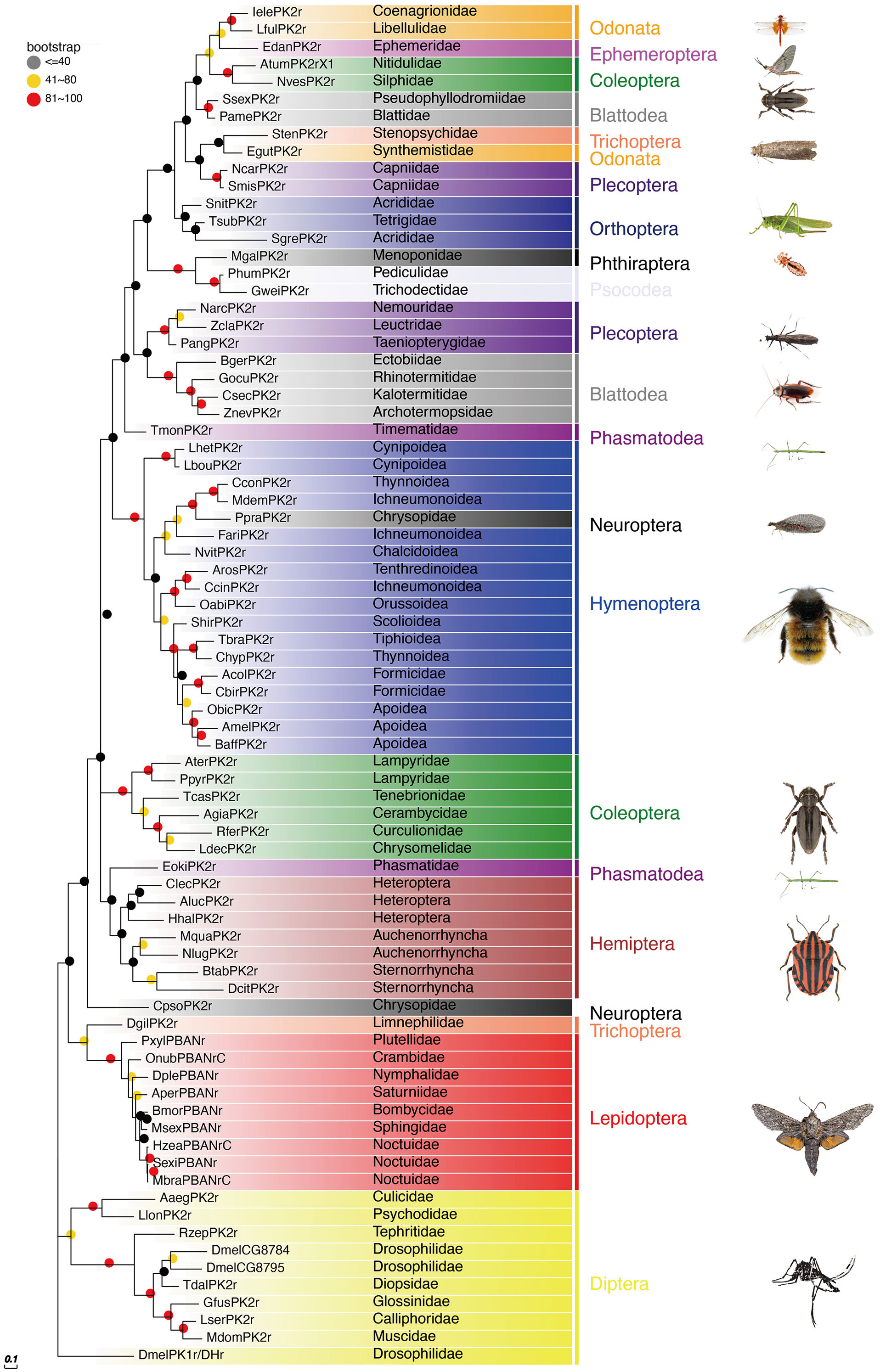
Figure 3 Phylogenetic relationship of PK2/PBANr representative sequences. C-terminal and N-terminal sequences of the receptors were removed then aligned using ClustalW, and the phylogeny calculated using the Maximum Likelihood algorithm with 100 bootstrap replicates. The family and order name are shown in the tree. The tree was modified using Evolview (75).
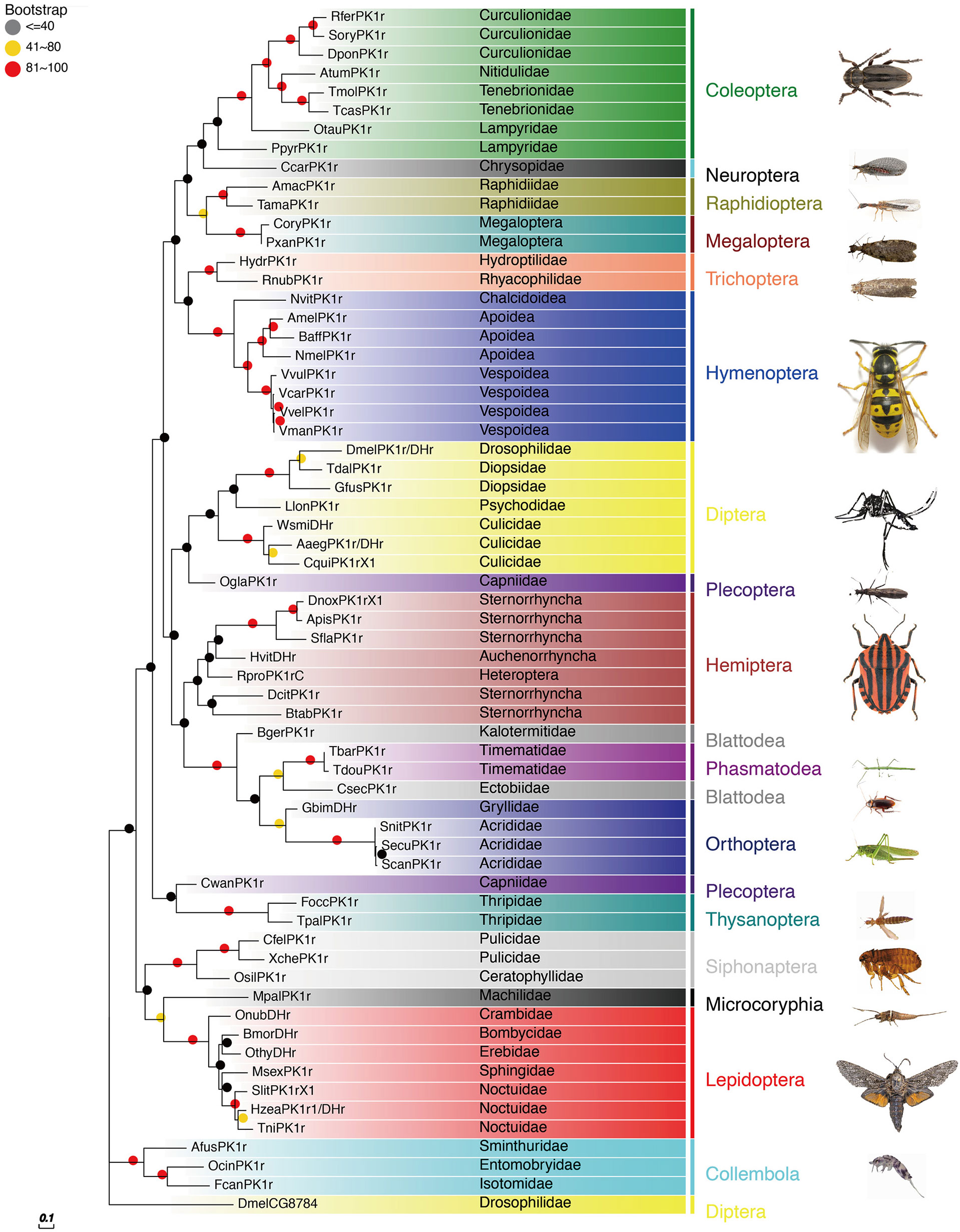
Figure 4 Phylogenetic relationship of PK1/DHr representative sequences. C-terminal and N-terminal sequences of the receptors were removed then aligned using ClustalW, and the phylogeny calculated using the Maximum Likelihood algorithm with 100 bootstrap replicates. The family and order name are shown in the tree. The tree was modified using Evolview (75).
Although representatives of most insect orders have a PK2/PBANr there are some families that do not have these receptors. One such family is the Vespidae in the Hymenoptera. This corresponds to the finding that all of the Vespidae have a pban that only has the PK1/DH sequence and does not have the PK2 sequences.
Due to the importance of PBAN in regulating sex pheromone biosynthesis in Lepidoptera, the PBANr has been extensively studied in moths. The first PBAN receptor was identified in H. zea (58) and followed by in B. mori (59). PBAN binds to its receptor, triggering a signal cascade that stimulates the sex pheromone biosynthetic pathway by activating calcium/calmodulin-dependent adenylate cyclase and cyclic AMP (cAMP) as shown in heliothine moths. The cAMP then activates acetyl-CoA carboxylase to initiate sex pheromone biosynthesis (76). Whereas, the PBAN signal promotes fatty acid removal from lipid droplets and activates fatty acid reductases in B. mori (76, 77).
PBAN has a relatively low threshold for activating the receptor. PBANr in H. zea could be activated at EC50 of 25 nM (58). Interestingly, PBAN activates PBANr from A. pernyi in CHO-K1 and HEK293 cells at two different concentrations (2.32 nM and 28.6 nM, respectively). However, the activation of PBANr does not activate the cAMP elevation pathway when expressed in the mammalian cell line HEK293 (78). Several variants from one to three were found in Lepidoptera (79). All three receptor sequences were identical except for C-terminal extensions. Only one PK/PBANr was detected in S. littoralis and it responded to both PBAN and DH with EC50 at 0.12 pM and 0.013 pM, respectively (80). In H. peltigera, the PBANr responses to PBAN at a similar concentration but was not activated by DH (80). Two PBANr variants were detected in the transcriptome of H. zea (61) but only one was confirmed by functional analysis (58). In M. brassicae, three PBANr variants were identified and all showed activity in inducing Ca2+, but only PBANr-B and PBANr-C caused ligand-induced internalization when expressed in Sf9 cells (81). In O. nubilalis, three PBANr variants and one DHr were detected in pheromone glands, but only variant C was activated by PBAN with an EC50 of 44 nM and variants A and B were inactive. The DHr was activated by DH with EC50 at 150 nM (82). Interestingly, PBANr was also detected in male H. armigera hair-pencil-aedaegus complexes. The knockdown of this receptor significantly reduced the amount of various male components including free fatty-acid components and alcohol components by 58%–74% (83). These results indicate that PBAN could be involved in regulating male hair-pencil components that are involved in courtship behaviors.
Studies using PKr from Drosophila and An. gambiae have revealed similar cross-reactivity of ligands across the PK1/DH and PK2/PBAN receptors, despite the fact that the receptors responded differently to PK1/DH and PK2/PBAN peptides (57, 66). Additionally, neuromedin U can activate the H. zea PBANr but not the D. melanogaster PK1r (84), indicating that these two receptors can be distinguished from one another. These findings show that the PK/PBANr can be activated by a variety of peptides with a PRLamide ending.
6 Conclusion
PK/PBAN family peptide sequences have been extensively investigated by genomics, transcriptome, and peptidomics, and the neuronal sites are examined by immunocytochemistry, peptidomics, and PCR. RNA interference (RNAi) and gene editing are also used to examine the function of these peptides. It is important to highlight that most insect orders include the genes encoding these peptides and their corresponding receptors. Some insect orders have conserved functions, however, the functions in many insects remain unknown. In this review, we analyzed the function of the peptides in different orders, several recent functions have been identified, such as cuticular melanization in the oriental armyworm larvae (26) and aggregation pheromone production in the western flower thrip (30), etc. Also, we analyzed the genes encoding these peptides. In most insects, the pban gene produced multiple peptides, including PBAN, DH, and several PK-like neuropeptides, while the capa gene produced one PK1/DH and multiple PVKs. Even though some receptors exhibit cross-reactivity to several ligands, these various C-terminal ending peptides are the ligands for activating their particular receptors. Since the importance of the PK/PBAN family peptides and the conserved nature across insects, it is important to integrate the molecular modeling and biochemical studies to make contribution to insect control strategies. The design of the antagonist to FXPRLa peptides and receptors has the potential application for pest management (85).
Author contributions
RJ: Writing – review & editing. XD: Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gade G, Hoffmann KH. Neuropeptides regulating development and reproduction in insects. Physiol Entomol (2005) 30:103–21. doi: 10.1111/j.1365-3032.2005.00442.x
2. Nassel DR, Zandawala M. Recent advances in neuropeptide signaling in Drosophila, from genes to physiology and behavior. Prog Neurobiol (2019) 179:101607. doi: 10.1016/j.pneurobio.2019.02.003
3. Nachman RJ, Holman GM, Cook BJ. Active fragments and analogs of the insect neuropeptide leucopyrokinin: structure-function studies. Biochem Bioph Res Commun (1986) 137:936–42. doi: 10.1016/0006-291x(86)90315-3
4. Raina AK, Kempe TG. Structure activity studies of PBAN of Helicoverpa zea (Lepidoptera, Noctuidae). Insect Biochem Molec Biol (1992) 22:221–5. doi: 10.1016/0965-1748(92)90058-M
5. Holman GM, Cook BJ, Nachman RJ. Primary structure and synthesis of a blocked myotropic neuropeptide isolated from the cockroach, Leucophaea maderae. Comp Biochem Phys C (1986) 85:219–24. doi: 10.1016/0742-8413(86)90077-0
6. Raina AK, Jaffe H, Kempe TG, Keim P, Blacher RW, Fales HM, et al. Identification of a neuropeptide hormone that regulates sex pheromone production in female moths. Science (1989) 244:796–8. doi: 10.1126/science.244.4906.796
7. Ma PWK, Knipple DC, Roelofs WL. Structural organization of the Helicoverpa zea gene encoding the precursor protein for pheromone biosynthesis-activating neuropeptide and other neuropeptides. Proc Natl Acad Sci USA (1994) 91:6506–10. doi: 10.1073/pnas.91.14.6506
8. Imai K, Konno T, Nakazawa Y, Komiya T, Isobe M, Koga K, et al. Isolation and structure of diapause hormone of the silkworm, Bombyx mori. Proc Jpn Acad B-Phys (1991) 67:98–101. doi: 10.2183/pjab.67.98
9. Veenstra JA. The contribution of the genomes of a termite and a locust to our understanding of insect neuropeptides and neurohormones. Front Physiol (2014) 5:454. doi: 10.3389/fphys.2014.00454
10. Meng XJ, Wahlstrom G, Immonen T, Kolmer M, Tirronen M, Predel R, et al. The Drosophila hugin gene codes for myostimulatory and ecdysis-modifying neuropeptides. Mech Dev (2002) 117:5–13. doi: 10.1016/S0925-4773(02)00175-2
11. Jurenka R. The prxamide neuropeptide signalling system: Conserved in animals. Adv Insect Physiol (2015) 49:123–70. doi: 10.1016/bs.aiip.2015.07.001
12. Lu Q, Huang LY, Chen P, Yu JF, Xu J, Deng JY, et al. Identification and RNA interference of the pheromone biosynthesis activating neuropeptide (PBAN) in the common cutworm moth Spodoptera litura (Lepidoptera: Noctuidae). J Econ Entomol (2015) 108:1344–53. doi: 10.1093/jee/tov108
13. Fodor J, Koblos G, Kakai A, Karpati Z, Molnar BP, Danko T, et al. Molecular cloning, mRNA expression and biological activity of the pheromone biosynthesis activating neuropeptide (PBAN) from the european corn borer, Ostrinia nubilalis. Insect Molec Biol (2017) 26:616–32. doi: 10.1111/imb.12324
14. Foster SP. Reinvestigation of sex pheromone biosynthesis in the moth Trichoplusiani reveals novel quantitative control mechanisms. Insect Biochem Mol Biol (2022) 140:103700. doi: 10.1016/j.ibmb.2021.103700
15. Fujii T, Nakano R, Takubo Y, Qian S, Yamakawa R, Ando T, et al. Female sex pheromone of a lichen moth Eilema japonica (Arctiidae, Lithosiinae): components and control of production. J Insect Physiol (2010) 56:1986–91. doi: 10.1016/j.jinsphys.2010.08.024
16. Matsumoto S, Kitamura A, Nagasawa H, Kataoka H, Orikasa C, Mitsui T, et al. Functional diversity of a neurohormone produced by the subesophageal ganglion: molecular identity of melanization and reddish coloration hormone and pheromone biosynthesis activating neuropeptide. J Insect Physiol (1990) 36:427–32. doi: 10.1016/0022-1910(90)90060-S
17. Suwan S, Isobe M, Yamashita O, Minakata H, Imai K. Silkworm diapause hormone, structure-activity relationships indispensable role of C-terminal amide. Insect Biochem Molec Biol (1994) 24:1001–7. doi: 10.1016/0965-1748(94)90137-6
18. Verleyen P, Clynen E, Huybrechts J, Van Lommel A, Bosch LV, De Loof A, et al. Fraenkel's pupariation factor identified at last. Dev Biol (2004) 273:38–47. doi: 10.1016/j.ydbio.2004.05.021
19. Xu WH, Denlinger DL. Molecular characterization of prothoracicotropic hormone and diapause hormone in Heliothis virescens during diapause, and a new role for diapause hormone. Insect Molec Biol (2003) 12:509–16. doi: 10.1046/j.1365-2583.2003.00437.x
20. Zhang TY, Sun JS, Zhang QR, Xu J, Jiang RJ, Xu WH. The diapause hormone-pheromone biosynthesis activating neuropeptide gene of Helicoverpa armigera encodes multiple peptides that break, rather than induce, diapause. J Insect Physiol (2004) 50:547–54. doi: 10.1016/j.jinsphys.2004.03.011
21. Uehara H, Senoh Y, Yoneda K, Kato Y, Shiomi K. An FXPRLamide neuropeptide induces seasonal reproductive polyphenism underlying a life-history tradeoff in the tussock moth. PloS One (2011) 6:e24213. doi: 10.1371/journal.pone.0024213
22. Subta P, Suang S, Chantawannakul P, Manaboon M. Diapause hormone terminates larval diapause in the bamboo borer, Omphisa fuscidentalis (Hampson). J Asia-Pac Entomol (2017) 20:1014–8. doi: 10.1016/j.aspen.2017.07.015
23. Choi MY, Meer RKV, Coy M, Scharf ME. Phenotypic impacts of pban RNA interference in an ant, Solenopsis invicta, and a moth, Helicoverpa zea. J Insect Physiol (2012) 58:1159–65. doi: 10.1016/j.jinsphys.2012.06.005
24. Choi MY, Vander Meer RK. Phenotypic effects of PBAN RNAi using oral delivery of dsRNA to corn earworm (Lepidoptera: Noctuidae) and tobacco budworm larvae. J Econ Entomol (2019) 112:434–9. doi: 10.1093/jee/toy356
25. Park Y, Vatanparast M. Suppression of PBAN receptor expression reduces fecundity in the fall armyworm, Spodoptera frugiperda. Arch Insect Biochem (2022) 110:e21897. doi: 10.1002/arch.21897
26. Shirai Y, Ono H, Daimon T. Redundant actions of neuropeptides encoded by the dh-pban gene for larval color pattern formation in the oriental armyworm Mythimna separata. Insect Biochem Mol Biol (2023) 157:103955. doi: 10.1016/j.ibmb.2023.103955
27. Hao K, Tu XB, Ullah H, McNeill MR, Zhang ZH. Novel Lom-dh genes play potential role in promoting egg diapause of Locusta migratoria L. Front Physiol (2019) 10:767. doi: 10.3389/fphys.2019.00767
28. Farris SM. Insect PRXamides: Evolutionary divergence, novelty, and loss in a conserved neuropeptide system. J Insect Sci (2023) 23:3. doi: 10.1093/jisesa/ieac079
29. Lajevardi A, Paluzzi JPV. Receptor characterization and functional activity of pyrokinins on the hindgut in the adult mosquito, Aedes aEgypti. Front Physiol (2020) 11:490. doi: 10.3389/fphys.2020.00490
30. Khan F, Kim K, Sung J, Lim H, Kim SG, Choi MY, et al. A novel physiological function of pheromone biosynthesis-activating neuropeptide in production of aggregation pheromone. Sci Rep (2023) 13:5551. doi: 10.1038/s41598-023-32833-9
31. Kawano T, Kataoka H, Nagasawa H, Isogai A, Suzuki A. CDNA cloning and sequence determination of the pheromone biosynthesis activating neuropeptide of the silkworm, Bombyx mori. Biochem Biophys Res Commun (1992) 189:221–6. doi: 10.1016/0006-291x(92)91547-4
32. Kean L, Cazenave W, Costes L, Broderick KE, Graham S, Pollock VP, et al. Two nitridergic peptides are encoded by the gene capability in Drosophila melanogaster. Am J Physiol Regul Integr Comp Physiol (2002) 282:R1297–307. doi: 10.1152/ajpregu.00584.2001
33. Davies SA, Cabrero P, Povsic M, Johnston NR, Terhzaz S, Dow JA. Signaling by Drosophila capa neuropeptides. Gen Comp Endocrinol (2013) 188:60–6. doi: 10.1016/j.ygcen.2013.03.012
34. Crooks GE, Hon G, Chandonia JM, Brenner SE. Weblogo: A sequence logo generator. Genome Res (2004) 14:1188–90. doi: 10.1101/gr.849004
35. Ahn SJ, Choi MY. Identification and characterization of capa and pyrokinin genes in the brown marmorated stink bug, Halyomorpha halys (Hemiptera): Gene structure, immunocytochemistry, and differential expression. Arch Insect Biochem (2018) 99:21500. doi: 10.1002/arch.21500
36. Hull JJ, Brent CS, Choi MY, Miko Z, Fodor J, Fonagy A. Molecular and functional characterization of pyrokinin-like peptides in the western tarnished plant bug Lygus hesperus (Hemiptera: Miridae). Insects (2021) 12:914. doi: 10.3390/insects12100914
37. Derst C, Dircksen H, Meusemann K, Zhou X, Liu S, Predel R. Evolution of neuropeptides in non-pterygote hexapods. BMC Evol Biol (2016) 16:51. doi: 10.1186/s12862-016-0621-4
38. Diesner M, Blaser M, Eckardt S, Iliffe TM, Theile EB, Predel R. Expression pattern of capa/pyrokinin neuropeptide genes in remipedia and silverfish: Rapid differentiation after gene duplication in early hexapoda, followed by strong conservation of newly established features in insects. Peptides (2021) 144:170610. doi: 10.1016/j.peptides.2021.170610
39. Choi MY, Lee JM, Han KS, Boo KS. Identification of a new member of PBAN family and immunoreactivity in the central nervous system from Adoxophyes sp. (Lepidoptera: Tortricidae). Insect Biochem Molec Biol (2004) 34:927–35. doi: 10.1016/j.ibmb.2004.06.003
40. Jiang H, Wei Z, Nachman RJ, Adams ME, Park Y. Functional phylogenetics reveals contributions of pleitropic peptide action to ligand-receptor coevolution. Sci Rep (2014) 4:6800. doi: 10.1038/srep06800
41. Choi MY, Sanscrainte ND, Estep AS, Vander Meer RK, Becnel JJ. Identification and expression of a new member of the pyrokinin/pban gene family in the sand fly Phlebotomus papatasi. J Insect Physiol (2015) 79:55–62. doi: 10.1016/j.jinsphys.2015.06.001
42. Ajitha VS, Muraleedharan D. Tissue localization and partial characterization of pheromone biosynthesis activating neuropeptide in Achaea janata. J Biosci (2005) 30:191–200. doi: 10.1007/Bf02703699
43. Golubeva E, Kingan TG, Blackburn MB, Masler EP, Raina AK. The distribution of PBAN (pheromone biosynthesis activating neuropeptide)-like immunoreactivity in the nervous system of the gypsy moth, Lymantria dispar. Arch Insect Biochem (1997) 34:391–408. doi: 10.1002/(Sici)1520-6327(1997)34:4<391::Aid-Arch1>3.0.Co;2-W
44. Chen Y, Liu YY, Tian HJ, Chen YX, Lin S, Mao QZ, et al. Distribution of pheromone biosynthesis-activating neuropeptide in the central nervous system of Plutella xylostella (Lepidoptera: Plutellidae). J Econ Entomol (2019) 112:2638–48. doi: 10.1093/jee/toz192
45. Sun JS, Zhang TY, Zhang QR, Xu WH. Effect of the brain and suboesophageal ganglion on pupal development in Helicoverpa armigera through regulation of FXPRLamide neuropeptides. Regul Peptides (2003) 116:163–71. doi: 10.1016/j.regpep.2003.09.002
46. Ragionieri L, Ozbagci B, Neupert S, Salts Y, Davidovitch M, Altstein M, et al. Identification of mature peptides from pban and capa genes of the moths Heliothis peltigera and Spodoptera littoralis. Peptides (2017) 94:1–9. doi: 10.1016/j.peptides.2017.05.004
47. Choi MY, Rafaeli A, Jurenka RA. Pyrokinin/pban-like peptides in the central nervous system of Drosophila melanogaster. Cell Tissue Res (2001) 306:459–65. doi: 10.1007/s00441-001-0467-x
48. Melcher C, Pankratz MJ. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PloS Biol (2005) 3:e305. doi: 10.1371/journal.pbio.0030305
49. Braunig P, Bohme C, Staufer B. Morphology of locust neurosecretory cells projecting into the nervus corporis allati II of the suboesophageal ganglion. Microsc Res Techniq (1996) 35:230–41. doi: 10.1002/(Sici)1097-0029(19961015)35:3<230::Aid-Jemt4>3.0.Co;2-R
50. Predel R, Linde D, Rapus J, Vettermann S, Penzlin H. Periviscerokinin (pea-pvk): A novel myotropic neuropeptide from the perisympathetic organs of the American cockroach. Peptides (1995) 16:61–6. doi: 10.1016/0196-9781(94)00144-u
51. Choi MY, Raina A, Vander Meer RK. Pban/pyrokinin peptides in the central nervous system of the fire ant, Solenopsis invicta. Cell Tissue Res (2009) 335:431–9. doi: 10.1007/s00441-008-0721-6
52. Hellmich E, Nusawardani T, Bartholomay L, Jurenka R. Pyrokinin/PBAN-like peptides in the central nervous system of mosquitoes. Cell Tissue Res (2014) 356:39–47. doi: 10.1007/s00441-013-1782-8
53. Predel R, Neupert S, Garczynski SF, Crim JW, Brown MR, Russell WK, et al. Neuropeptidomics of the mosquito Aedes aEgypti. J Proteome Res (2010) 9:2006–15. doi: 10.1021/pr901187p
54. Zoephel J, Reiher W, Rexer KH, Kahnt J, Wegener C. Peptidomics of the agriculturally damaging larval stage of the cabbage root fly Delia radicum (Diptera: Anthomyiidae). PloS One (2012) 7:e41543. doi: 10.1371/journal.pone.0041543
55. Predel R, Neupert S, Russell WK, Hauser F, Russell DH, Li A, et al. Capa-gene products in the haematophagous sandfly Phlebotomus papatasi (scopoli): vector for leishmaniasis disease. Peptides (2013) 41:2–7. doi: 10.1016/j.peptides.2012.12.009
56. Rahman MM, Neupert S, Predel R. Neuropeptidomics of the Australian sheep blowfly Lucilia cuprina (wiedemann) and related diptera. Peptides (2013) 41:31–7. doi: 10.1016/j.peptides.2012.12.021
57. Park Y, Kim YJ, Adams ME. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc Natl Acad Sci U.S.A. (2002) 99:11423–8. doi: 10.1073/pnas.162276199
58. Choi MY, Fuerst EJ, Rafaeli A, Jurenka R. Identification of a G protein-coupled receptor for pheromone biosynthesis activating neuropeptide from pheromone glands of the moth Helicoverpa zea. Proc Natl Acad Sci USA (2003) 100:9721–6. doi: 10.1073/pnas.1632485100
59. Hull JJ, Ohnishi A, Moto K, Kawasaki Y, Kurata R, Suzuki MG, et al. Cloning and characterization of the pheromone biosynthesis activating neuropeptide receptor from the silkmoth, Bombyx mori: significance of the carboxyl terminus in receptor internalization. J Biol Chem (2004) 279:51500–7. doi: 10.1074/jbc.M408142200
60. Lee DW, Shrestha S, Kim AY, Park SJ, Yang CY, Kim Y, et al. RNA interference of pheromone biosynthesis-activating neuropeptide receptor suppresses mating behavior by inhibiting sex pheromone production in Plutella xylostella (L.). Insect Biochem Molec (2011) 41:236–43. doi: 10.1016/j.ibmb.2011.01.001
61. Dou XY, Liu SJ, Ahn SJ, Choi MY, Jurenka R. Transcriptional comparison between pheromone gland-ovipositor and tarsi in the corn earworm moth Helicoverpa zea. Comp Biochem Phys D (2019) 31:604. doi: 10.1016/j.cbd.2019.100604
62. Dou XY, Liu SJ, Soroker V, Harari A, Jurenka R. Pheromone gland transcriptome of the pink bollworm moth, Pectinophora gossypiella: Comparison between a laboratory and field population. PloS One (2019) 14:220187. doi: 10.1371/journal.pone.0220187
63. Cha WH, Jung JK, Lee DW. Identification of G protein-coupled receptors in the pheromone gland of Maraca vitrata by transcriptomic analysis. J Asia-Pac Entomol (2018) 21:1203–10. doi: 10.1016/j.aspen.2018.09.005
64. Ding BJ, Lofstedt C. Analysis of the Agrotis segetum pheromone gland transcriptome in the light of sex pheromone biosynthesis. BMC Genomics (2015) 16:711. doi: 10.1186/s12864-015-1909-2
65. Antony B, Soffan A, Jakse J, Alfaifi S, Sutanto KD, Aldosari SA, et al. Genes involved in sex pheromone biosynthesis of Ephestia cautella, an important food storage pest, are determined by transcriptome sequencing. BMC Genomics (2015) 16:532. doi: 10.1186/s12864-015-1710-2
66. Olsen SS, Cazzamali G, Williamson M, Grimmelikhuijzen CJP, Hauser F. Identification of one capa and two pyrokinin receptors from the malaria mosquito Anopheles Gambiae. Biochem Bioph Res (2007) 362:245–51. doi: 10.1016/j.bbrc.2007.06.190
67. Choi MY, Estep A, Sanscrainte N, Becnel J, Meer RKV. Identification and expression of pban/diapause hormone and GPCRs from Aedes aEgypti. Mol Cell Endocrinol (2013) 375:113–20. doi: 10.1016/j.mce.2013.05.019
68. Jiang H, Wei Z, Nachman RJ, Kaczmarek K, Zabrocki J, Park Y. Functional characterization of five different PRXamide receptors of the red flour beetle Tribolium castaneum with peptidomimetics and identification of agonists and antagonists. Peptides (2015) 68:246–52. doi: 10.1016/j.peptides.2014.11.004
69. Cazzamali G, Torp M, Hauser F, Williamson M, Grimmelikhuijzen CJP. The Drosophila gene CG9918 codes for a pyrokinin-1 receptor. Biochem Bioph Res Co (2005) 335:14–9. doi: 10.1016/j.bbrc.2005.07.038
70. Homma T, Watanabe K, Tsurumaru S, Kataoka H, Imai K, Kamba M, et al. G protein-coupled receptor for diapause hormone, an inducer of Bombyx embryonic diapause. Biochem Bioph Res (2006) 344:386–93. doi: 10.1016/j.bbrc.2006.03.085
71. Paluzzi JP, O'Donnell MJ. Identification, spatial expression analysis and functional characterization of a pyrokinin-1 receptor in the Chagas' disease vector, Rhodnius prolixus. Mol Cell Endocrinol (2012) 363:36–45. doi: 10.1016/j.mce.2012.07.007
72. Jiang HB, Wei ZJ, Nachman RJ, Park Y. Molecular cloning and functional characterization of the diapause hormone receptor in the corn earworm Helicoverpa zea. Peptides (2014) 53:243–9. doi: 10.1016/j.peptides.2013.11.005
73. Yang YL, Nachman RJ, Pietrantonio PV. Molecular and pharmacological characterization of the Chelicerata pyrokinin receptor from the southern cattle tick, Rhipicephalus (Boophilus) microplus. Insect Biochem Molec Biol (2015) 60:13–23. doi: 10.1016/j.ibmb.2015.02.010
74. Wulff JP, Temeyer KB, Tidwell JP, Schlechte KG, Xiong CX, Lohmeyer KH, et al. Pyrokinin receptor silencing in females of the southern cattle tick Rhipicephalus (Boophilus) microplus is associated with a reproductive fitness cost. Parasite Vector (2022) 15:252. doi: 10.1186/s13071-022-05349-w
75. Zhang HK, Gao SH, Lercher MJ, Hu SN, Chen WH. Evolview, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res (2012) 40:W569–W72. doi: 10.1093/nar/gks576
76. Jurenka R. Regulation of pheromone biosynthesis in moths. Curr Opin Insect Sci (2017) 24:29–35. doi: 10.1016/j.cois.2017.09.002
77. Matsumoto S. Molecular mechanisms underlying sex pheromone production in moths. Biosci Biotechnol Biochem (2010) 74:223–31. doi: 10.1271/bbb.90756
78. Jiang L, Zhang F, Hou Y, Thakur K, Hu F, Zhang JG, et al. Isolation and functional characterization of the pheromone biosynthesis activating neuropeptide receptor of chinese oak silkworm, Antheraea pernyi. Int J Biol Macromolecules (2018) 117:42–50. doi: 10.1016/j.ijbiomac.2018.05.145
79. Lee JM, Hull JJ, Kawai T, Goto C, Kurihara M, Tanokura M, et al. Re-evaluation of the PBAN receptor (PBANR) molecule: characterization of PBANR variants expressed in the pheromone glands of moths. Front Endocrinol (2012) . 3:6. doi: 10.1073/pnas.1424386112
80. Hariton-Shalev A, Shalev M, Adir N, Belausov E, Altstein M. Structural and functional differences between pheromonotropic and melanotropic PK/PBAN receptors. BBA-Gen (2013) 1830:5036–48. doi: 10.1016/j.bbagen.2013.06.041
81. Fodor J, Hull JJ, Koblos G, Jacquin-Joly E, Szlanka T, Fonagy A. Identification and functional characterization of the pheromone biosynthesis activating neuropeptide receptor isoforms from Mamestra brassicae. Gen Comp Endocr (2018) 258:60–9. doi: 10.1016/j.ygcen.2017.05.024
82. Nusawardani T, Kroemer JA, Choi MY, Jurenka RA. Identification and characterization of the pyrokinin/pheromone biosynthesis activating neuropeptide family of G protein-coupled receptors from Ostrinia nubilalis. Insect Mol Biol (2013) 22:331–40. doi: 10.1111/imb.12025
83. Bober R, Rafaeli A. Gene-silencing reveals the functional significance of pheromone biosynthesis activating neuropeptide receptor (PBAN-r) in a male moth. Proc Natl Acad Sci USA (2010) 107:16858–62. doi: 10.1073/pnas.1008812107
84. Choi MY, Fuerst EJ, Rafaeli A, Jurenka R. Role of extracellular domains in pban/pyrokinin GPCRs from insects using chimera receptors. Insect Biochem Mol Biol (2007) 37:296–306. doi: 10.1016/j.ibmb.2006.12.004
85. Nachman RJ. Insect GPCRs and development of mimetic analogs of the insect kinin, pyrokinin-like, and sulfakinin neuropeptide classes as pest management tools. In: Saleuddin S, Lange AB, Orchard I, editors. Advances in Invertebrate (Neuro)Endocrinology: A collection of reviews in the post-genomic era, vol. 2 . Boca Raton: Apple Academic Press (2020). arthropoda49 p.
Keywords: neuropeptides, PK/PBAN peptides, PK1/DH peptides, PK2/PBAN peptides, receptors
Citation: Dou X and Jurenka R (2023) Pheromone biosynthesis activating neuropeptide family in insects: a review. Front. Endocrinol. 14:1274750. doi: 10.3389/fendo.2023.1274750
Received: 08 August 2023; Accepted: 30 November 2023;
Published: 14 December 2023.
Edited by:
Qisheng Song, University of Missouri, United StatesReviewed by:
Stephen Foster, North Dakota State University, United StatesMan-Yeon Choi, United States Department of Agriculture, United States
Takeshi Fujii, Setsunan University, Japan
Qisheng Song, University of Missouri, United States
Copyright © 2023 Dou and Jurenka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Russell Jurenka, cmp1cmVua2FAaWFzdGF0ZS5lZHU=
 Xiaoyi Dou
Xiaoyi Dou Russell Jurenka
Russell Jurenka