- 1Department of Biology, University of Ottawa, Ottawa, ON, Canada
- 2Department of Physiological Sciences, University of Florida, Gainesville, FL, United States
The detection and quantification of hormones are important to assess the reproductive and stress status of experimental models and for the diagnosis of diseases in human and veterinary clinics. Traditionally, steroid, peptide, and protein hormones are analyzed in individual experiments using different extraction methodologies. With the new advancement on HPLC sorbents, the simultaneous measurement of hormones from different categories becomes possible. In this study, we present a novel sample processing strategy for the simultaneous extraction and detection of peptides, steroids, and proteins using high-resolution liquid chromatography tandem mass spectrometry. We demonstrate the sensitivity of our method for small tissues by acquiring data from brain, pituitary gland, and gonads of single zebrafish samples. This approach promises to shed light on the hormonal pathways and their interrelationships, providing knowledge on the integration of hormone systems.
1 Introduction
Measuring peptides, steroids, and proteins from tissue extracts is essential for studying normal physiological processes and diagnosing various pathological states with underlying endocrine etiologies. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is the technique of choice for qualitative and quantitative hormone measurements because of critical advances in analytical chemistry. However, the diverse chemical structures of peptides, steroids, and proteins have traditionally required separate LC-MS/MS analyses of different biological samples, which limits project scope and the ability to simultaneously collect integrated data (1–3). For peptide and steroid methodologies, proteins are typically precipitated out of extracts and discarded, while for proteomics, the protein analytes are precipitated and preserved, and the supernatants are discarded (4–6). These incompatible approaches have limited comprehensive evaluation and pose significant challenges for data interpretation. Simultaneous measurement of these diverse analytes would represent a major advancement in endocrine research. Direct comparison of variations and interrelationships for numerous hormones in a single sample could reveal critical physiological mechanisms or underlying causes of disease.
The emergence of high-quality high-performance liquid chromatography (HPLC) sorbents has made it possible to address the long-standing issue of co-extraction and analysis. Our approach employs a unique extraction strategy, which uses a reverse phase sorbent initially intended for ultra-high-performance liquid chromatography (UHPLC). The homogenization solution consists of a high percentage of methanol and acetic acid to precipitate proteins from tissue homogenates, while keeping peptide and steroid analytes in solution. This enables the digestion of the protein pellet for bottom-up proteomics. The supernatant containing peptides and steroids can be safely stored for subsequent experiments (Figure 1).
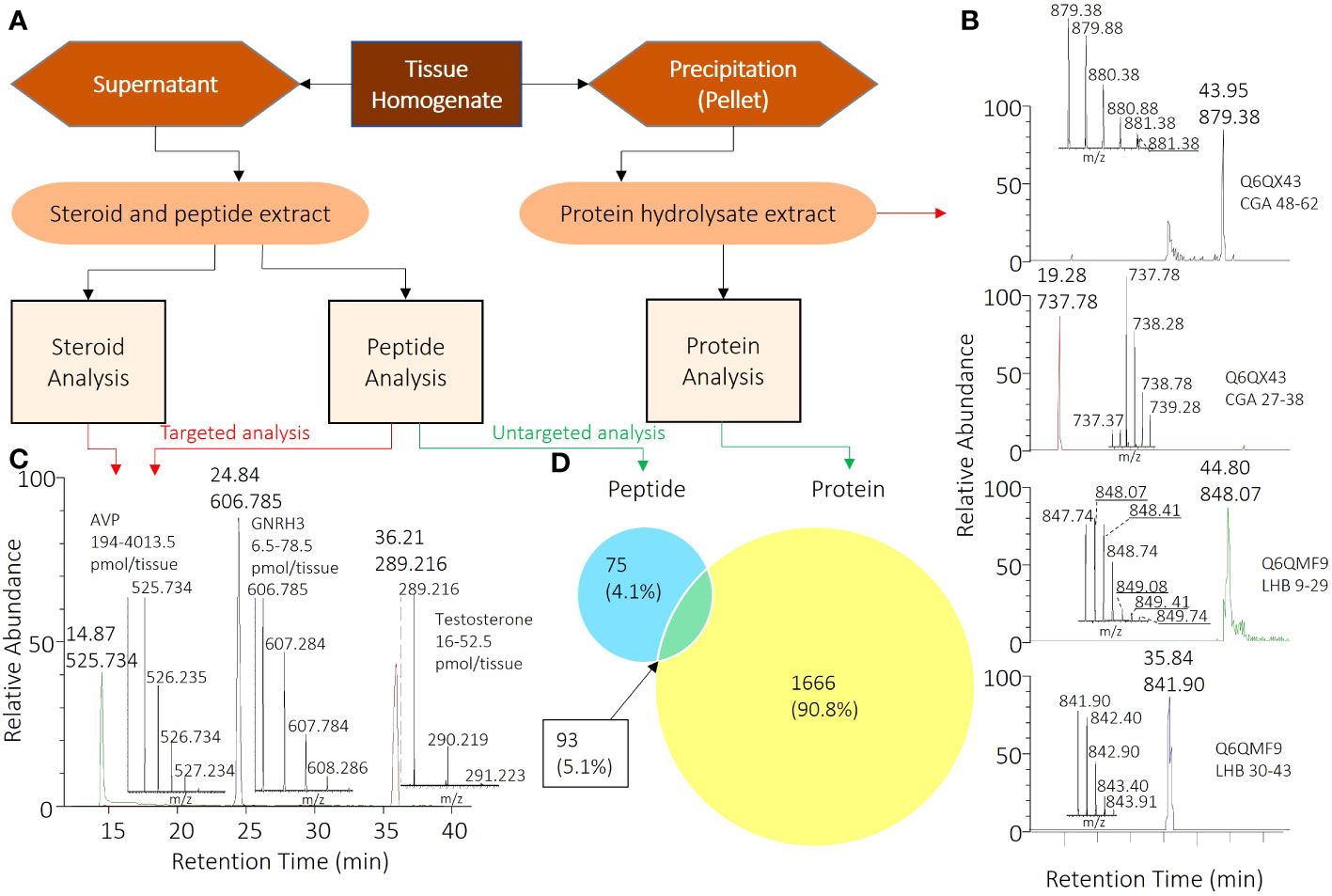
Figure 1. Comprehensive Workflow for Analyzing Peptide, Steroid, and Protein Components in Pituitary Samples. (A) Overall sample processing workflow from tissue homogenate to specific samples for corresponding analyses. (B) Chromatogram and mass spectrometry results for CGA and LHB) peptides selected for targeted proteome analysis. (C) Chromatogram and mass spectrometry results for selected peptide and steroid analyses, with AVP, GNRH3, and testosterone serving as model compounds among all analytes. (D) 93 pituitary proteins (5.1%) that were identified with both peptidomics and trypsin-digested proteomics. Also shown are the numbers of other peptides (75; 4.1%) and proteins (1666; 90.8%) identified.
To test the method, we employed the zebrafish (Danio rerio), an emerging small model organism for the study of reproductive biology and endocrine diseases (7–9). We have been able to compare the levels of well-known hormones with those of the newest reproductive hormone secretoneurin, which we have reported on recently (10). We show that it is possible to perform both proteomics and peptidomics on the same small tissue sample, revealing the intricate relationships between precursor and product. In addition, we have succeeded in extracting peptides, steroids, and proteins from a single and very small tissue, the pituitary gland of the zebrafish. The processing procedures were optimized for small sample volume using a 96-well plate platform.
2 Materials and equipment
2.1 Reagents and materials
• 10 µm sorbent (Dr. Maisch HPLC GmbH, cat. no. r13.aq.0010)
• 3 µm sorbent (Dr. Maisch HPLC GmbH, cat. no. r13.aq.0003)
• Filtered 96-well plate (OROCHEM, cat. no. OF1100)
• Estrone (Cayman Chemical, cat. no. 10006485)
• Estradiol (Cayman Chemical, cat. no. 10006315)
• Estriol (Cayman Chemical, cat. no. 10006484)
• Testosterone (Cayman Chemical, cat. no. 15645)
• 11-Ketotestosterone (Cayman Chemical, cat. no. 9002564)
• Fmoc-Ala-OH (Cayman Chemical, cat. no. 41004)
• Fmoc-Arg(Pbf)-OH (P3 BioSystems, cat. no. 41002)
• Fmoc-Asn(Trt)-OH (P3 BioSystems, cat. no. 41007)
• Fmoc-Asp(OtBu)-OH (P3 BioSystems, cat. no. 41019)
• Fmoc-Cys(Trt)-OH (P3 BioSystems, cat. no. 41008)
• Fmoc-Cys(Acm)-OH (P3 BioSystems, cat. no. 41063)
• Fmoc-Glu(OtBu)-OH (P3 BioSystems, cat. no. 41005)
• Fmoc-Gln(Trt)-OH (P3 BioSystems, cat. no. 41011)
• Fmoc-Gly-OH (P3 BioSystems, cat. no. 41010)
• Fmoc-His(Trt)-OH (P3 BioSystems, cat. no. 41017)
• Fmoc-Ile-OH (P3 BioSystems, cat. no. 41018)
• Fmoc-Leu-OH (P3 BioSystems, cat. no. 41003)
• Fmoc-Lys(Boc)-OH (P3 BioSystems, cat. no. 41001)
• Fmoc-Met-OH (P3 BioSystems, cat. no. 41020)
• Fmoc-Phe-OH (P3 BioSystems, cat. no. 41013)
• Fmoc-Pro-OH (P3 BioSystems, cat. no. 41009)
• Fmoc-Ser(tBu)-OH (P3 BioSystems, cat. no. 41006)
• Fmoc-Thr(tBu)-OH (P3 BioSystems, cat. no. 41016)
• Fmoc-Trp(Boc)-OH (P3 BioSystems, cat. no. 41012)
• Fmoc-Tyr(tBu)-OH (P3 BioSystems, cat. no. 41014)
• Fmoc-Val-OH (P3 BioSystems, cat. no. 41015)
• Boc-Pyr-OH (P3 BioSystems, cat. no. 47222)
• (2- (1 H -benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) (P3 BioSystems, cat. no. 31001)
• Tentagel resin (Rapp Polymere GmBH, cat. no. S3023)
• N-methylmorpholine (NMM) (Millipore sigma, cat. no. M56557)
• Trifluoroacetic acid (TFA) (Millipore sigma, cat. no. T6508)
• Formic acid (FA) (Millipore sigma, cat. no. 695076)
• Triisopropylsilane (Millipore sigma, cat. no. 233781)
• Piperidine (Millipore sigma, cat. no. 104094)
• Iodine (Millipore sigma, cat. no. 207772)
• Acetonitrile (ACN) (Fisher Scientific, cat. no. A9984)
• Methanol (MeOH) (Fisher Scientific, cat. no. BP1105-4)
• Dimethylformamide (DMF) (Fisher Scientific, cat. no. BPD1194)
• N-methyl-2-pyrrolidone (NMP) (Millipore sigma, cat. no. 8060721000)
• Urea (Millipore sigma, cat. no. 9510-OP)
• Ammonium bicarbonate (Millipore sigma, cat. no. A6141)
• Dithiothreitol (DTT) (Millipore sigma, cat. no. D9163)
• Iodoacetamide (Millipore sigma, cat. no. A3221-10VL)
• Ammonium fluoride (NH4F) (Millipore sigma, cat. no. 338869-25G)
• Trypsin (Worthington-biochemical, cat. no. LS02120)
• Milli-Q water
2.2 Recipes for solutions used in the protocol
2.2.1 Homogenization solution
▪ 90% MeOH
▪ 9% water
▪ 1% acetic acid
2.2.2 Activation solution
▪ 0.2% FA
▪ 50% ACN
▪ 50% water
2.2.3 Equilibrium solution
▪ 0.2% FA
▪ 5% ACN
▪ 95% water
2.2.4 Elution solution
▪ 0.2% FA
▪ 75% ACN
▪ 25% water
2.2.5 Mobile phase A
▪ 0.1% FA in water
2.2.6 Mobile phase B
▪ 0.1% FA in ACN
2.2.7 Mobile phase A1
▪ 0.2% FA in water
2.2.8 Mobile phase B1
▪ 0.2% FA
▪ 80% ACN
▪ 20% water
2.2.9 Mobile phase A2
▪ 1 µM NH4F in water
2.2.10 Mobile phase B2
▪ 1 µM NH4F
▪ 65% MeOH
▪ 30% ACN
▪ 5% water
2.2.11 Digestion buffer
▪ 8 M urea
▪ 50 mM ammonium bicarbonate
▪ pH 8.0
2.2.12 Cleavage cocktail
▪ 95% TFA
▪ 2.5% triisopropylsilane
▪ 2.5% H2O
2.3 Equipment
• HPLC (for LC-MS/MS): Agilent 1100 system (stack with G1376A binary capillary pump, G1477A micro well-plate autosampler, and G1330A ALS thermostat).
• HPLC (for peptide purification): Agilent 1100 HPLC system (stack with G1322A vacuum degasser, G1311A quaternary pump, G1313A autosampler, G1316A column compartment, G1315B diode array detector, and G1364C analytical fraction collector).
• Mass spectrometer: Thermo Scientific, Thermo Scientific Orbitrap Velos
• Probe homogenizer: Fisherbrand, model no. FB120110
• Peptide synthesis machine: Intavis Multipeptide RSi
3 Methods
3.1 Sample preparation
Wildtype zebrafish (AB strain) were bred and kept in the University of Ottawa Aquatics core facility at 28°C on a 14:10h light/dark cycle. All the fish were fed twice a day. Sexually mature zebrafish were anaesthetized using buffered 4.5% tricaine, then sacrificed by spinal transection. Brain, pituitary and gonads from 42 females and 12 males were dissected and immediately transferred into 1.5 mL centrifuge tubes containing 200 µL of homogenization solution on ice. To homogenize the tissues, a probe homogenizer was utilized at a power output of 10 Watts for 20 seconds. The homogenization solution dissolves the peptide and steroid hormones, while precipitating the proteins to facilitate analysis. After centrifugation, the resulting homogenate was separated into pellet and supernatant fractions, which were then evaporated using a speed-vac.
3.2 Pellet digestion
Dried pellets from the tissue homogenates were solubilized in 200 µL digestion buffer. Dithiothreitol was added to a final concentration of 10 mM and incubated at room temperature for 1hr for reduction. Iodoacetamide was added to a final concentration of 20 mM and incubated at room temperature for 40 min in dark for alkylation. In the end, the sample was diluted 5-fold by 50 mM ammonium bicarbonate and digested by 1 µg trypsin/20 µg sample protein overnight at room temperature.
3.3 Solid phase extraction
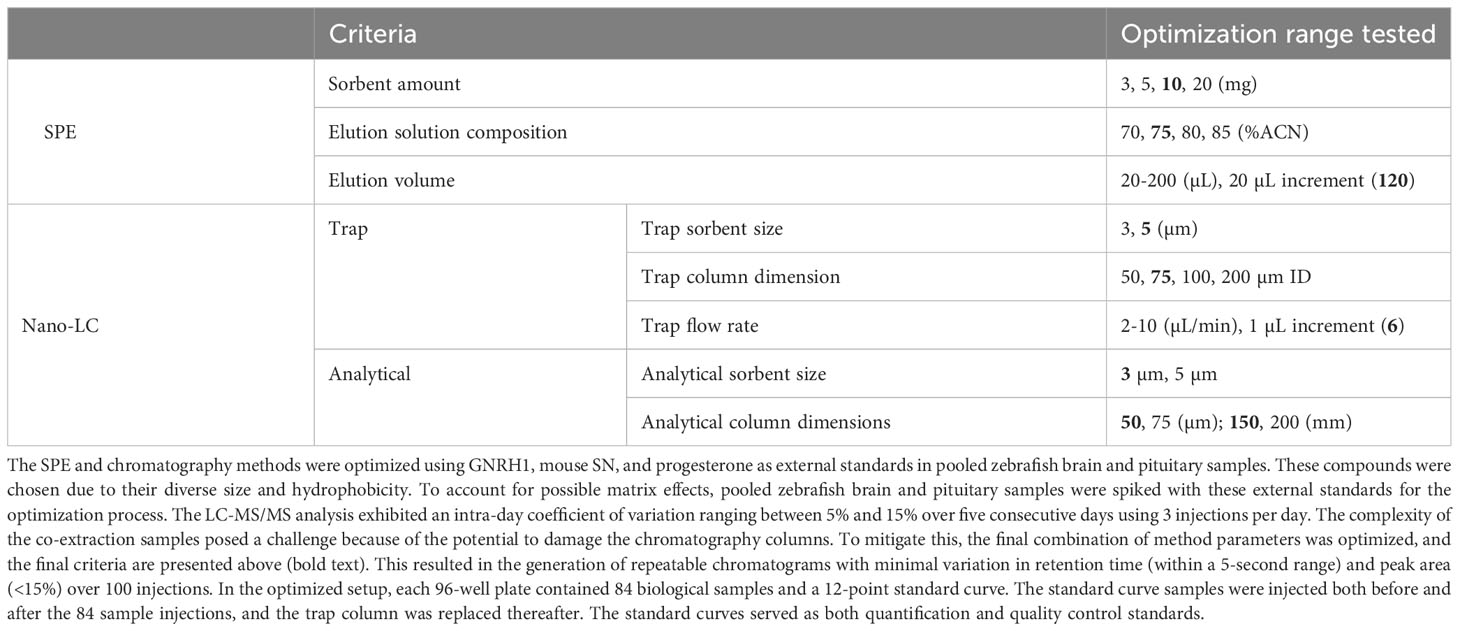
The solid phase extraction (SPE) plates were prepared for both peptide-steroid co-extraction and protein hydrolysate extraction by packing 10 mg of extraction sorbent into filtered 96-well plates. Prior to extraction, the pre-packed extraction plates were activated by adding 200 µL of activation solution for 5 times, followed by 5 times 200 µL of equilibrium solution. The samples were dissolved and loaded in the equilibrium solution, and the loaded SPE plates were washed another 5 times with 200 µL of the same solution. The elution was performed by adding 20 µL of elution solution for 6 times. The activation elution solutions were flushed through the packed SPE bed by centrifugation at 500 rcf, while the equilibrium solution was flushed at 1000 rcf. The stepwise procedure is presented in Supplementary File 2.
3.4 LC-MS/MS
For both untargeted proteomics and peptidomics, the system consisted of an Agilent 1100 micro-HPLC system coupled with a Thermofisher LTQ-Orbitrap mass spectrometer equipped with a nano-electrospray interface. Peptide separation was performed on an in-house packed 50 μm × 100 mm analytical column. For proteomics, the gradient was from 5% to 35% mobile phase B in 120 min. The mass spectrometry (MS) method consisted of one full MS scan, followed by data-dependent MS/MS scan of the 5 most intense ions using the LTQ-Orbitrap mass spectrometer. The full MS scan range was 150 to 1500 m/z for peptides and steroids, and 300 to 1700 m/z for digested samples. To improve mass accuracy, all measurements in the Orbitrap mass analyzer were performed with internal recalibration (“Lock Mass”). On the Orbitrap, the charge state rejection function was enabled, with single and “unassigned” charge states rejected for untargeted experiments. The raw files generated by the LTQ-Orbitrap were processed and analyzed using MaxQuant, Version 1.2.2.5 with the Uniprot protein FASTA database, including commonly observed contaminants. The following parameters were used: cysteine carbamidomethylation was selected as fixed modification; methionine oxidation, protein N-terminal acetylation, and enzyme specificity was set to trypsin. Up to two missing cleavages of trypsin were allowed to compensate insufficient enzyme digestion. Precursor ion mass tolerances were 7 ppm, and fragment ion mass tolerance was 0.8 Da for MS/MS spectra. Identified peptide sequences from one protein that were equal to or contained within another protein’s peptide set were grouped together and reported as one protein group. The false discovery rate (FDR) for peptide and protein was set at 1%, and a minimum length of six amino acids was used for peptide identification. Quantification was performed using normalized label-free quantitation (LFQ) intensity.
Untargeted peptidomics followed a similar analysis pipeline. However, we selected methionine oxidation, protein N-terminal acetylation, and no enzyme digestion as modifications, and cysteine carbamidomethylation was not fixed.
For targeted peptide and steroid quantification, the HPLC was run in micro mode with a pre-column splitter on load and elute modes, using an in-house-prepared 50 µm × 150 mm analytical column with 3 µm C18-AQ sorbent from Dr. Maisch. The sample was loaded on the column using 95% mobile phase A (A1 for positive mode, A2 for negative mode) at a flow rate of 6 μL/min for 3 min. Then, we used a gradient of 20-90% B1 for positive mode analysis and a gradient of 50-90% B2 for negative mode analysis. We set the mass spectrometer to run on data-dependent acquisition, with the orbitrap analyzer performing the master scan at resolution 60,000 (FWHM), and the linear ion-trap analyzer performing the fragmentation and secondary MS scan according to the mass list and master scan. The mass list collects all primary ions of analytes of interest. We used Thermo Xcalibur software, version 2.2, for data acquisition and analysis. We confirmed peptide structures by three MS2 ions and steroid structures by one MS2 ion. We plotted chromatograms by accurate MS1 with 10 ppm mass tolerance and 120 sec identification windows, using Genesis peak detection when there were over five data points to form a chromatographic peak. We determined the level by comparing the peak area to the standard curve generated by standard compounds. Steroid standards were commercially available. Peptide standards were prepared by solid phase peptide synthesis and purified by HPLC.
3.5 Solid phase peptide synthesis
Standard peptides were synthesized in-house using Fmoc peptide synthesis chemistry. Solid-phase synthesis was carried out on Tentagel resin. For deprotection of the Fmoc group on the N-terminus of each amino acid, 2 times 5 min 20% piperidine treatment was employed. Standard coupling reactions were performed with 4-fold HBTU (0.5 M), 4.4-fold amino acids (0.6 M), and 4-fold NMM (4 M) for 40 mins. Double coupling reactions were used to attach each amino acid for sequences with a length of 15 or below, while triple coupling reactions were used for sequences with a length greater than 15.
At the end of synthesis, the resin was treated with a cleavage cocktail for two hours to harvest crude peptides. To introduce cysteine into the peptide sequences of zebrafish oxytocin and vasopressin, Fmoc-Cys(Acm)-OH was used. The Acm-protected cysteine was oxidized on-resin by suspending the synthesis resin in a 4 M Iodine/DMF solution with mild mixing for 2.5 h, followed by washing with DMF 5 times and with DCM 3 times before cleavage. To introduce pyroglutamic acid (Pyr), Boc-Pyr-OH was used with a standard coupling protocol. The crude peptides were precipitated with ethyl ether on ice, dissolved in 50% ACN water, and lyophilized. Lyophilized crude peptide samples were stored at -20°C for purification.
Peptide purification was performed on an Agilent 1100 HPLC system with Luna Omega 5 µm 100 Å PS C18 RP-HPLC column (250×10 mm) at a flow rate of 4 ml/min. The column oven was set at 50°C. To purify each peptide, the HPLC gradients were adjusted to have the target peptide elute at least 7.5 min after the gradient started. The fraction collector was set to start collection 7.5 min after the gradient started on 400 mAU threshold, 500 mAU/s upslope, and 200 mAU/s downslope according to the 214 nm chromatogram. The collected fractions were chilled in liquid nitrogen for 5 min and lyophilized accordingly. Lyophilized purified peptide powders were stored at -20°C. The molecular weight of each peptide was confirmed on LC-MS.
4 Results
4.1 Peptide and steroid analysis
A total of 8 peptide and 5 steroid hormones were selected to evaluate the possibility of targeted analysis in unpooled, single tissues harvested from multiple females and males. The supernatants were resuspended and subjected to SPE using in-house prepared cartridges, as described in the methods and Table 1. Peptide standards were prepared in-house with Fmoc/tBu chemistry, purified, and confirmed by MS as described above. The level of each analyte was determined by comparing the chromatogram peak areas to standard curves. Chromatograms of 3 representative analytes (vasotocin, also known as fish arginine vasopressin (AVP), gonadotropin releasing hormone 3 (GNRH3) and testosterone) are shown in Figure 1C, and chosen for their diversity in size, structure, and hydrophilicity/lipophilicity. The limits of detection and maximum amounts of these and other analytes measured from tissues of both females and males are listed in Table 2. This new analysis strategy provides the possibility to measure diverse peptide and steroid hormones in tissue samples as small as a zebrafish pituitary. The performance of the in-house packed SPE cartridge was also tested with the LC-MS/MS system, and the overall recovery rates for the 3 representative analytes were over 85% on 0.01-10 nmol loading.
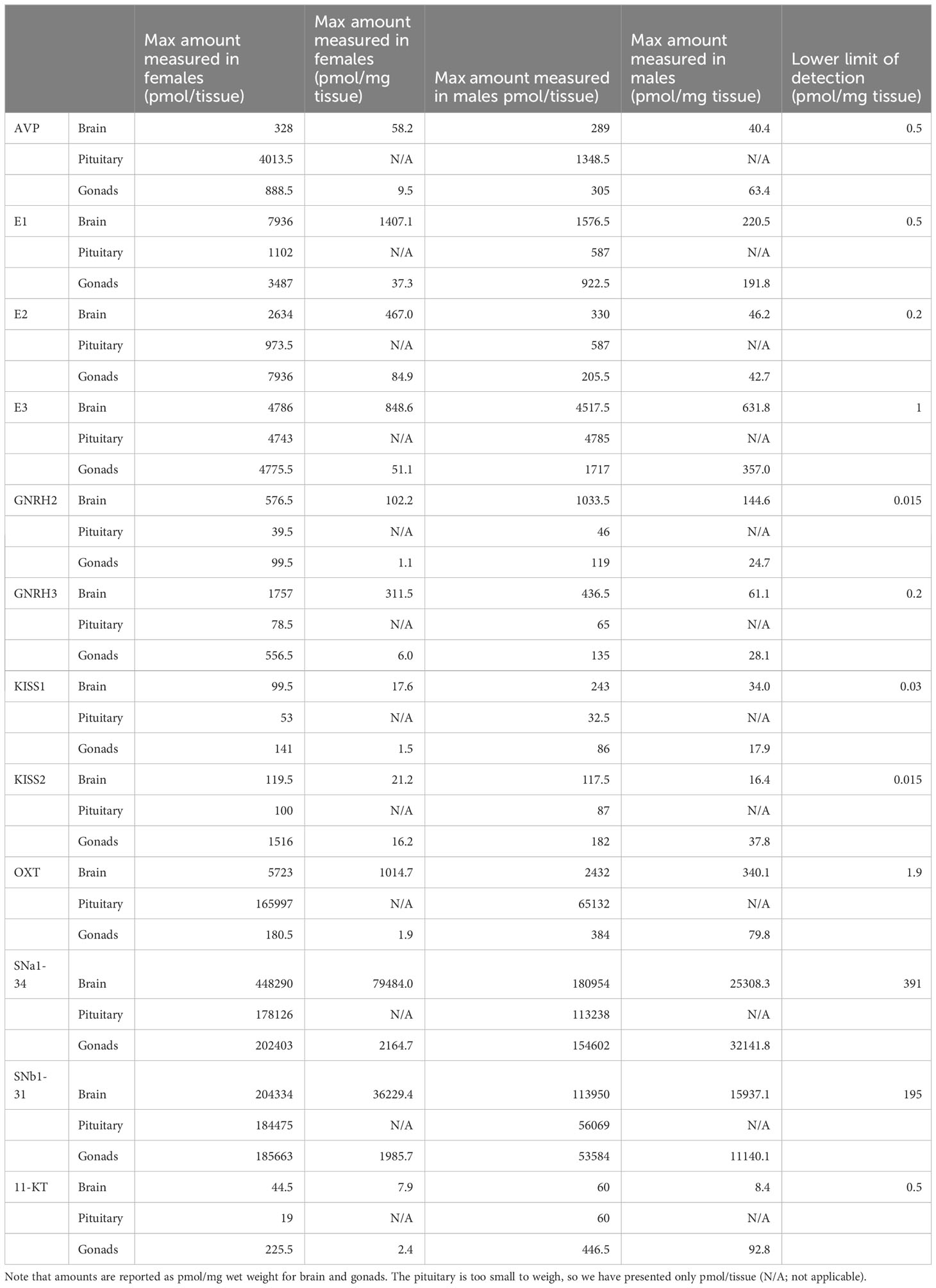
Table 2. The maximum levels measured for each compound in unpooled tissues from groups of female (N=42) and male (N=12) zebrafish. The lower limit of detection represents the lowest concentration point of the standard curve.
4.2 Untargeted peptidomics
To perform untargeted peptidomics, we applied the peptide-steroid extracts from pituitary to LC-MS/MS analysis using a shallow gradient and data dependent MS acquisition. We used the acquired MS/MS spectra to search against the zebrafish International Protein Index protein sequence database (Version 3.85) with Maxquant and the LFQ option. Our analysis successfully identified 260 different peptides derived from 168 different proteins (Supplementary File 1, Figure 1D).
4.3 Untargeted proteomics
The protein pellets were subjected to reduction, alkylation, and digestion, and the extracts were further processed using the same SPE procedure as described in the Supplementary. The LC-MS/MS analysis and data processing followed a similar protocol, with trypsinization and alkylation being used as the default methods. Analysis of the resulting data revealed the identification of 1758 proteins, of which 93 were found to be shared with the peptidomics results (Supplementary File 1, Figure 1D).
4.4 Gene set enrichment analysis
We conducted gene set enrichment analysis (GSEA) using precursor proteins and associated peptides from both proteomics and the peptidomics results (Supplementary File 1). This analysis led to the identification of 393 pathways from the pituitary protein lysate (P<0.05) and 202 pathways from the peptide (P<0.05) datasets (Supplementary File 1). This approach allowed us to investigate biological processes and molecular pathways more comprehensively, providing valuable information for further exploration and interpretation of our results. Interestingly, 123 pathways were discovered in both peptidomics and proteomics experiments, suggesting their importance in the same pituitary tissue. The details of all pathways are in Supplementary File 1, and specific examples of hormonal pathways enriched in both datasets are presented in Table 3. Given the importance of the pituitary hormones in homeostasis and physiological regulation, detection of some of these were expected.
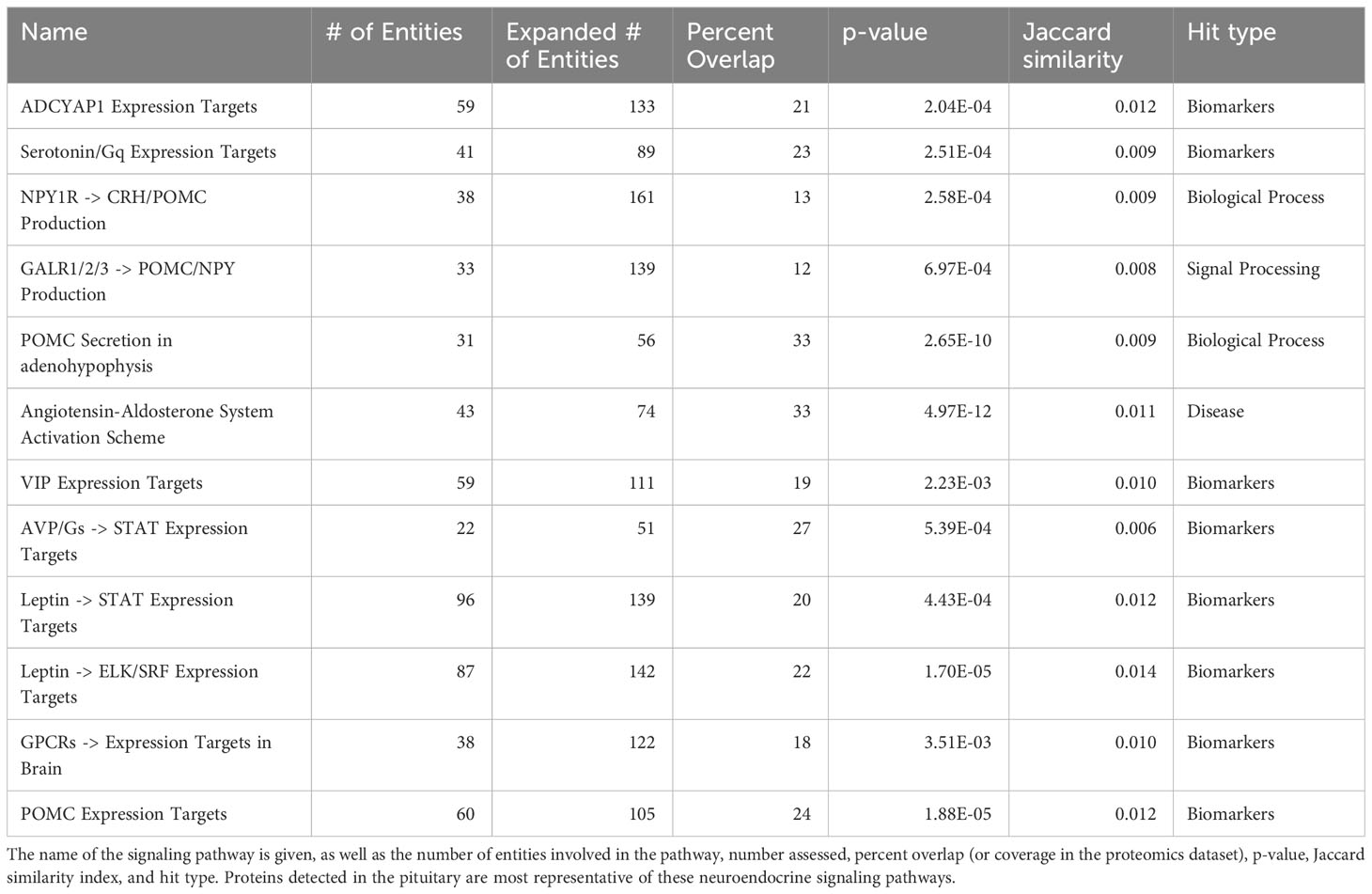
Table 3. Examples of gene sets enriched in the proteomics dataset that are also enriched in the peptidome dataset.
For example, pathways related to proopiomelanocortin (POMC) production, secretion and action are well represented in the data (Figure 2, Table 3). A lesser studied aspect of pituitary function is that of leptin action via STAT, ELK/SRF expression targets (Table 3). In fish, leptin can regulate prolactin (PRL) (11) and the gonadotropins (12). We also noted that enzymatic processing of classical hormones such as PRL can give rise to angiogenic peptides (13, 14) that were also found in the pituitary gland.
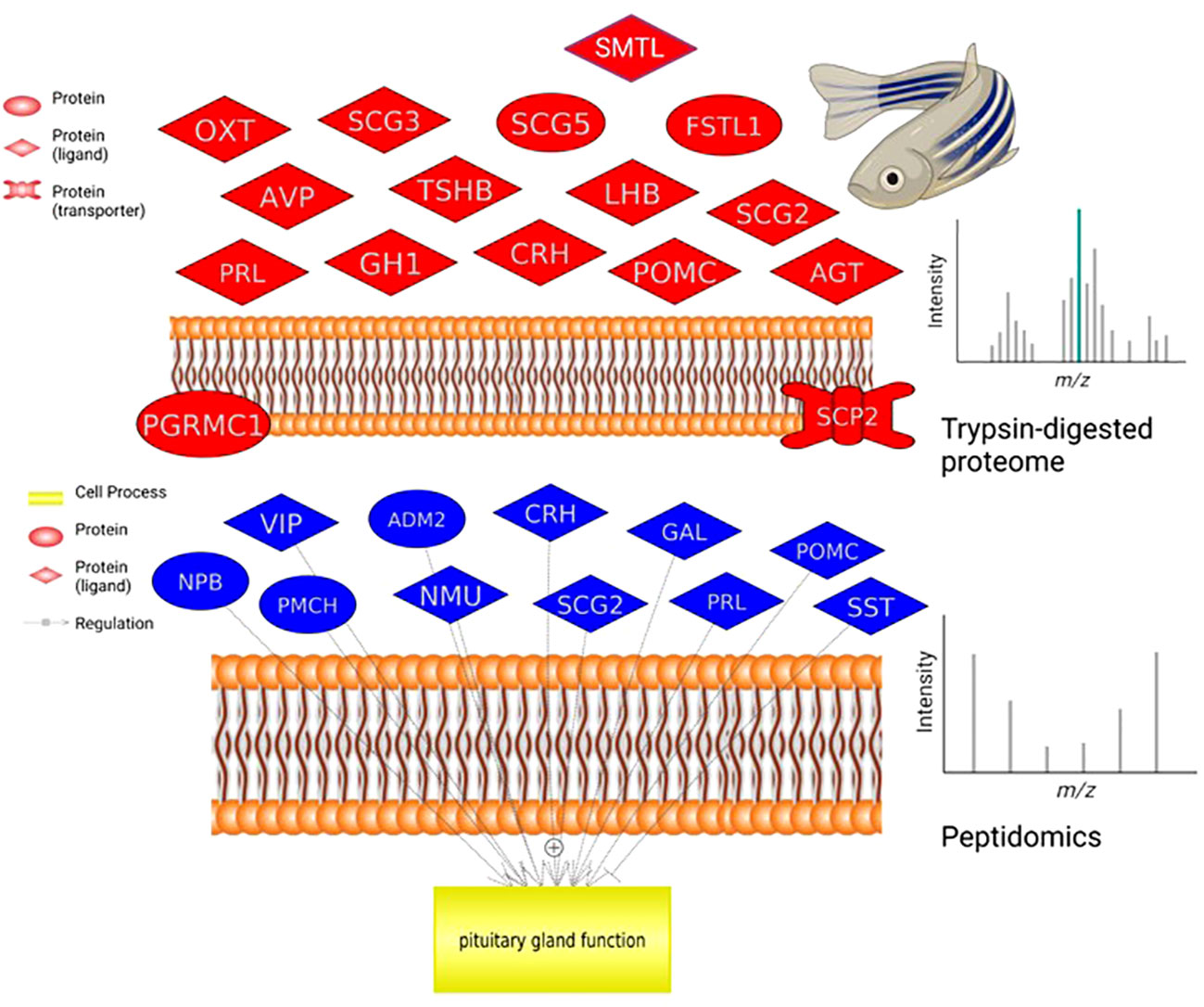
Figure 2. Pituitary proteome and peptidome comparison. Neurohormones identified by proteomics in a single zebrafish that regulate pituitary hormone release (top graph). Neuroendocrine proteins identified by peptidome analysis in a single zebrafish pituitary (bottom graph). For analysis and ease of comparison, we followed nomenclature and abbreviations conventions of HUGO (https://www.genenames.org/): ADM2, adrenomedullin 2; AGT, angiotensinogen; AVP, arginine vasopressin; CRH, corticotropin releasing hormone; FSTL1, follistatin like 1; GAL, galanin and GMAP prepropeptide; GH1, growth hormone 1; LHB, luteinizing hormone subunit beta; NMU, neuromedin U; NPB, neuropeptide B; OXT, oxytocin/neurophysin I prepropeptide; PGRMC1, progesterone receptor membrane component 1; PMCH, pro-melanin concentrating hormone; POMC, proopiomelanocortin; PRL, prolactin; SCG2, secretogranin II; SCG3, secretogranin III; SCG5, secretogranin V; SCP2, sterol carrier protein 2; SST, somatostatin; SMTL, somatolactin; TSHB, thyroid stimulating hormone subunit beta; VIP, vasoactive intestinal peptide.
A key example of the utility of the methodology is the detection and quantification of the emerging reproductive hormone secretoneurin (SN). It is derived from prohormone convertase-mediated processing of the larger secretogranin-2 (SCG2) precursor which was detected in the proteomic samples (Figure 2). In zebrafish and other teleosts there are 2 precursor forms, SCG2a and SCG2b, respectively giving rise to SNa1-34 and SNb1-31 which we synthesized and then quantified in brain, pituitary and gonads of both females and males (Table 2). TALEN-mediated frameshift mutation of the zebrafish SCG2a and SCG2b genes led to reduced levels of sexual activity, suboptimal ovulation and egg laying, fertility, and embryonic survival 24-hr postfertilization (10). The SN peptides stimulate LH production and release (15). Gene set enrichment analysis (GSEA) was conducted in Pathway Studio 12.0 (Elsevier). A total number of 2,095 proteins (proteomics) and 202 proteins (based on peptidomics) were mapped into the program for zebrafish using the official gene name. GeneCards (www.genecards.org/) was used to manually retrieve mammalian homologs for mapping. Gene set enrichment analysis proceeded with 1000 permutations to generate the distributions for statistical testing (permutation test) using a Kolmogorov-Smirnov Distribution test. Parameters were as follows: p-value ≤ 0.05, maximum number of networks shown in result = 1000 with a minimum network overlap of 5 entities. The enrichment identifies cell processes and signaling pathways most represented in the data sets.
4.5 Targeted protein analysis
We also analyzed pituitary protein lysates using targeted proteomics to investigate the presence of specific proteins, namely the subunits of luteinizing hormone (LH). Critically, LH is a dimeric pituitary hormone responsible for driving ovulation in all vertebrate animals (16) and inappropriate LH secretion can lead to various fertility problems (7). It is traditionally measured by radiometric or enzyme-liked immunoassays, with associated challenges (3, 17, 18). There is currently no specific immunoassay for zebrafish LH. As an alternative, we optimized proteomic digestion steps and then identified two fragments from the glycoprotein hormones alpha polypeptide (CGA) (CGA 48-62 and CGA 27-38) and two fragments from the luteinizing hormone subunit beta (LHB) (LHB 9-29 and LHB 30-43) for the detection of LH (Figure 1B).
5 Discussion
Our method enables the analysis of peptides, steroids and proteins at the same time. This lays the foundation for comprehensive assessment of hormonal pathways in a single sample, allowing for direct comparisons and improved data interpretation. The performance of the optimized method for simultaneously measuring both peptides and steroids was similar to previous reports on peptide or steroid analysis individually (19–21). The proposed strategy effectively addresses the problematic issue of separately isolating and detecting small lipophilic versus hydrophilic endogenous hormones.
In this proof-of-principle experiment, we targeted 6 well-known peptide and 5 steroid hormones plus the reproductive peptides SNa1-13 and SNb1-31 in brain, pituitary and gonads. We were able to quantify these 13 hormones in most samples, given the dynamic range of our standard curves. There were sex differences both in the total amounts produced in a tissue, or when adjusted for wet weight. While sex differences in steroids may be somewhat expected based on published literature (e.g., 22, 23), the peptides presented have never been measured in zebrafish tissues to our knowledge. The SNa and OXT peptides were found in high quantities, especially in the pituitary. This supports previous anatomical observations on the colocalization of OXT and SNa in goldfish magnocellular neurons and their projections to the anterior and posterior pituitary (24). In rat, the highest levels of SN were quantified in the median eminence (25), the functional equivalent to direct innervation of the anterior pituitary of teleost (26).
An impressive number of other pituitary peptides were evident from the untargeted peptidomics analysis. Our method was also designed for the comparison of the proteome and peptidome of small tissues, such as the pituitary. We were able to match precursor proteins with some of the peptides generated. As precursor processing by prohormone convertases is physiologically regulated, our method will be useful to study the generation on known hormonal peptides, but also the discovery of novel fragments. Gene set enrichment analyses identified signaling pathways common to both the proteomic and peptidome datasets. The main limiting factor for quantification of novel hormones is the availability of commercial standards, and but in many cases, as we have shown with SN and other teleost peptides, this is easily overcome with high quality synthesis and purification steps. Given that there are so far, ~20 stimulatory and ~10 inhibitory neuropeptides, 3 amines, 3 amino acid neurotransmitters and many steroids implicated in the control of teleost reproduction (16), the method holds promise for comprehensive assessment of natural variations, or in response to gene editing and environmental perturbations.
Peptide and steroid measurements are important not only in biomedical research but also in industries such as food safety evaluation and environmental assessment. For example, in dietary supplements, collagen hydrolysate peptide profile consistency is important for nutrition, while determining steroid levels is important for safety (27, 28). If the peptide profile and steroid levels are tested at the same time, food safety authorities could make inspections more efficient and less costly. Monitoring the levels of steroid hormones, especially synthetic and potent analogs, is crucial to monitoring pollution and endocrine-disrupting chemicals in the aquatic environment (29). Additionally, formylmethionyl-peptides are key indicators of the level of bacteria (30). Measuring these two categories of compounds together would permit ecotoxicologists to better understand chemical threats to ecosystems and human health. All of these will require further development.
The SPE method can be efficiently used to extract peptides and steroid samples for other assay systems. We have shown it to be compatible with a commercial estradiol ELISA kit, being validated using aromatase mutant zebrafish lines (31). Having reusable SPE plates in a small-scale research laboratory that serves multiple different ELISA tests can not only save on research funds but also significantly decrease the plastic waste it generates. In addition, it can decrease the variability between different batches of SPE kits. The extraction and analysis methods employ the 96-well plate platform, thus making it feasible to motorize sample processing procedure and downstream analysis. With minimum optimization, this package of methods can be used for high throughput routine analysis in research institutes or clinical test centers.
Data availability statement
All data are presented in the main article or in the supplemental files. Any other reasonable queries should be directed at the corresponding authors.
Ethics statement
The animal study was approved by Animal Care and Veterinary Services, University of Ottawa. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CL: Conceptualization, Methodology, Validation, Writing – original draft, Writing – review & editing. DP: Conceptualization, Data curation, Methodology, Writing – review & editing. WE: Data curation, Writing – review & editing. KM: Data curation, Writing – review & editing. CM: Data curation, Writing – review & editing. VT: Conceptualization, Funding acquisition, Resources, Writing – original draft, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the China Scholarship Council (DP), University of Ottawa Research Chair in Neuroendocrinology (VLT), and the Natural Sciences and Engineering Research Council of Canada Discovery Grant (RGPIN-2021-03174 to V.L.T).
Acknowledgments
The advice of Drs. Z. Ning and D. Figeys from the University of Ottawa Proteomics Resource Center is also acknowledged. We also recognize that we work and live on the unceded Algonquin Territory of the Three Fire Confederacy, Anishinaabewaki.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1266985/full#supplementary-material
References
1. Collado-Romero M, Aguilar C, Arce C, Lucena C, Codrea M, Morera L, et al. Quantitative proteomics and bioinformatic analysis provide new insight into the dynamic response of porcine intestine to Salmonella Typhimurium. Front Cell Infection Microbiol (2015) 5:64. doi: 10.3389/fcimb.2015.00064
2. Kay RG, Challis BG, Casey RT, Roberts GP, Meek CL, Reimann F, et al. Peptidomic analysis of endogenous plasma peptides from patients with pancreatic neuroendocrine tumours. Rapid Commun Mass Spectrom (2018) 32:1414–24. doi: 10.1002/rcm.8183
3. Burow S, Fontaine R, Von Krogh K, Mayer I, Nourizadeh-Lillabadi R, Hollander-Cohen L, et al. Establishment of specific enzyme-linked immunosorbent assay (ELISA) for measuring Fsh and Lh levels in medaka (Oryzias latipes), using recombinant gonadotropins. MethodsX (2019) 6:1473–9. doi: 10.1016/j.mex.2019.06.011
4. Olesti E, Boccard J, Visconti G, González-Ruiz V, Rudaz S. From a single steroid to the steroidome: Trends and analytical challenges. J Steroid Biochem Mol Biol (2021) 206:105797. doi: 10.1016/j.jsbmb.2020.105797
5. Rozanova S, Barkovits K, Nikolov M, Schmidt C, Urlaub H, Marcus K. Quantitative mass spectrometry-based proteomics: an overview. Methods Mol Biol (2021) 2228:85–116. doi: 10.1007/978-1-0716-1024-4_8
6. Zhou CX, Gao M, Han B, Cong H, Zhu XQ, Zhou HY. Quantitative peptidomics of mouse brain after infection with cyst-forming toxoplasma gondii. Front Immunol (2021) 12:681242. doi: 10.3389/fimmu.2021.681242
7. Anderson RC, Newton CL, Anderson RA, Millar RP. Gonadotropins and their analogs: current and potential clinical applications. Endocr Rev (2018) 39:911–37. doi: 10.1210/er.2018-00052
8. Li J, Ge W. Zebrafish as a model for studying ovarian development: Recent advances from targeted gene knockout studies. Mol Cell Endocrinol (2020) 507:110778. doi: 10.1016/j.mce.2020.110778
9. Ghaddar B, Diotel N. Zebrafish: A new promise to study the impact of metabolic disorders on the brain. Int J Mol Sci (2022) 23(10):5372. doi: 10.3390/ijms23105372
10. Mitchell K, Zhang WS, Lu C, Tao B, Chen L, Hu W, et al. Targeted mutation of secretogranin-2 disrupts sexual behavior and reproduction in zebrafish. Proc Natl Acad Sci (2020) 117:12772. doi: 10.1073/pnas.2002004117
11. Yan A, Chen Y, Chen S, Li S, Zhang Y, Jia J, et al. Leptin Stimulates Prolactin mRNA Expression in the Goldfish Pituitary through a Combination of the PI3K/Akt/mTOR, MKK(3/6)/p(38)MAPK and MEK(1/2)/ERK(1/2) Signalling Pathways. Int J Mol Sci (2017) 18(12):2781. doi: 10.3390/ijms18122781
12. Bagivalu Lakshminarasimha ,A, Puvvada M, Hammerschmidt M, Michel M. Leptin modulates oocyte maturation by a central and a direct pathway in zebrafish. J Endocrinol (2022) 254:1–12. doi: 10.1530/JOE-22-0026
13. Nguyen NQ, Tabruyn SP, Lins L, Lion M, Cornet AM, Lair F, et al. Prolactin/growth hormone-derived antiangiogenic peptides highlight a potential role of tilted peptides in angiogenesis. Proc Natl Acad Sci U.S.A. (2006) 103:14319–24. doi: 10.1073/pnas.0606638103
14. Friedrich C, Neugebauer L, Zamora M, Robles JP, Martínez De La Escalera G, Clapp C, et al. Plasmin generates vasoinhibin-like peptides by cleaving prolactin and placental lactogen. Mol Cell Endocrinol (2021) 538:111471. doi: 10.1016/j.mce.2021.111471
15. Mitchell K, Mikwar M, Da Fonte D, Lu C, Tao B, Peng D, et al. Secretoneurin is a secretogranin-2 derived hormonal peptide in vertebrate neuroendocrine systems. Gen Comp Endocrinol (2020) 299:113588. doi: 10.1016/j.ygcen.2020.113588
16. Trudeau VL. Neuroendocrine control of reproduction in teleost fish: concepts and controversies. Annu Rev Anim Biosci (2022) 10:107–30. doi: 10.1146/annurev-animal-020420-042015
17. Stanislaus D, Andersson H, Chapin R, Creasy D, Ferguson D, Gilbert M, et al. Society of toxicologic pathology position paper: review series: assessment of circulating hormones in nonclinical toxicity studies: general concepts and considerations. Toxicol Pathol (2012) 40:943–50. doi: 10.1177/0192623312444622
18. Kreisman MJ, Mccosh RB, Breen KM. A modified ultra-sensitive ELISA for measurement of LH in mice. Endocrinology (2022) 163(9):bqac109. doi: 10.1210/endocr/bqac109
19. Nouri M-Z, Kroll KJ, Webb M, Denslow ND. Quantification of steroid hormones in low volume plasma and tissue homogenates of fish using LC-MS/MS. Gen Comp Endocrinol (2020) 296:113543. doi: 10.1016/j.ygcen.2020.113543
20. Erikson DW, Blue SW, Kaucher AV, Shnitko TA. LC-MS/MS measurement of endogenous oxytocin in the posterior pituitary and CSF of macaques: A pilot study. Peptides (2021) 140:170544. doi: 10.1016/j.peptides.2021.170544
21. Li Y, Yan Z, Li X, Yin X, Li K. UPLC-TOF-MS method for simultaneous quantification of steroid hormones in tissue homogenates of zebrafish with solid-phase extraction. Molecules (2021) 26(20):6213. doi: 10.3390/molecules26206213
22. Pasmanik M, Callard GV. Changes in brain aromatase and 5α-reductase activities correlate significantly with seasonal reproductive cycles in goldfish (Carassius auratus)*. Endocrinology (1988) 122:1349–56. doi: 10.1210/endo-122-4-1349
23. Dai X, Pradhan A, Liu J, Liu R, Zhai G, Zhou L, et al. Zebrafish gonad mutant models reveal neuroendocrine mechanisms of brain sexual dimorphism and male mating behaviors of different brain regions. Biol Sex Dif (2023) 14:53. doi: 10.1186/s13293-023-00534-7
24. Canosa LF, Lopez GC, Scharrig E, Lesaux-Farmer K, Somoza GM, Kah O, et al. Forebrain mapping of secretoneurin-like immunoreactivity and its colocalization with isotocin in the preoptic nucleus and pituitary gland of goldfish. J Comp Neurol (2011) 519:3748–65. doi: 10.1002/cne.22688
25. Marksteiner J, Kirchmair R, Mahata SK, Mahata M, Fischer-Colbrie R, Hogue-Angeletti R, et al. Distribution of secretoneurin, a peptide derived from secretogranin II, in rat brain: an immunocytochemical and radioimmunological study. Neuroscience (1993) 54:923–44. doi: 10.1016/0306-4522(93)90585-4
26. Trudeau VL, Somoza GM. Multimodal hypothalamo-hypophysial communication in the vertebrates. Gen Comp Endocrinol (2020) 293:113475. doi: 10.1016/j.ygcen.2020.113475
27. Cho SH, Park HJ, Lee JH, Kim HJ, Cho S, Yoon CY, et al. Monitoring of 35 illegally added steroid compounds in foods and dietary supplements. Food Addit Contam Part A Chem Anal Control Expo Risk Assess (2014) 31:1470–5. doi: 10.1080/19440049.2014.946100
28. López-Morales CA, Vázquez-Leyva S, Vallejo-Castillo L, Carballo-Uicab G, Muñoz-García L, Herbert-Pucheta JE, et al. Determination of peptide profile consistency and safety of collagen hydrolysates as quality attributes. J Food Sci (2019) 84:430–9. doi: 10.1111/1750-3841.14466
29. Ojoghoro JO, Scrimshaw MD, Sumpter JP. Steroid hormones in the aquatic environment. Sci Total Environ (2021) 792:148306. doi: 10.1016/j.scitotenv.2021.148306
30. Siegel PD, Ronk EA, Clark PR, Shahan TA, Castranova V. Measurement of environmental formylmethionyl-peptides. J Toxicol Environ Health (1994) 42:275–88. doi: 10.1080/15287399409531879
Keywords: protein measurement, peptide quantification, steroid detection, LCMS (liquid chromatography-mass spectrometry), solid phase extraction
Citation: Lu C, Peng D, Erandani WCKU, Mitchell K, Martyniuk CJ and Trudeau VL (2023) Simultaneous extraction and detection of peptides, steroids, and proteins in small tissue samples. Front. Endocrinol. 14:1266985. doi: 10.3389/fendo.2023.1266985
Received: 25 July 2023; Accepted: 14 September 2023;
Published: 09 October 2023.
Edited by:
Paula Gabriela Vissio, University of Buenos Aires, ArgentinaReviewed by:
Radha Chaube, Banaras Hindu University, IndiaJohn Postlethwait, University of Oregon, United States
Copyright © 2023 Lu, Peng, Erandani, Mitchell, Martyniuk and Trudeau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vance L. Trudeau, dHJ1ZGVhdXZAdU90dGF3YS5jYQ==
†These authors share first authorship
 Chunyu Lu
Chunyu Lu Di Peng1†
Di Peng1† Christopher J. Martyniuk
Christopher J. Martyniuk Vance L. Trudeau
Vance L. Trudeau