- Key Laboratory of Freshwater Fisheries Germplasm Resources Certificated by The Ministry of Agriculture and Rural Affairs/National Demonstration Center for Experimental Fisheries Science Education/Shanghai Engineering/Research Center of Aquaculture, Shanghai Ocean University, Shanghai, China
Purpose: Molting is a pivotal biological process regulated by the ecdysteroid signaling pathway that requires molecular coordination of two transcription factors, Ecdysone receptor (EcR) and ultraspiracle (USP) in arthropods. However, the molecular interplay of EcR and Retinoid X receptor (RXR), the crustacean homolog of USP in the ecdysteroid signaling pathway, is not well understood.
Methods: In this study, we conducted temporal and spatial expression, co-immunoprecipitation (CO-IP), and luciferase reporter assay experiments to investigate the molecular function and interplay of EcR and RXR during the molting process of the Chinese mitten crab, Eriocheir sinensis.
Results: The results showed that the expression level of RXR was more stable and significantly higher than EcR during the entire molting process. However, the expression level of EcR fluctuated dynamically and increased sharply at the premolt stage. The CO-IP and luciferase reporter assay results confirmed the molecular interplay of EcR and RXR. The heterodimer complex formed by the two transcription factors significantly induced the transcription of E75, an essential gene in the ecdysteroid signaling pathway.
Conclusions: Our study unveiled the diverse molecular function and molecular interplay of EcR and RXR; RXR is possibly a “constitutive-type” gene, and EcR is possibly a vital speed-limiting gene while both EcR and RXR are required to initiate the ecdysteroid signaling cascade, which may be indispensable for molting regulation in E. sinensis. The results provide a theoretical basis for the endocrine control of molting in E. sinensis and novel insights into the molecular mechanism of molting mediated by the ecdysteroid signaling pathway in crustaceans.
1 Introduction
Molting plays essential roles in growth and reproduction, which is regulated by the ecdysteroid signaling pathway in arthropods (1). The ecdysteroid signaling pathway is a signal cascade process that requires molecular regulation of two transcription factors, ecdysone receptor (EcR) and ultraspiracle (USP), ecdysteroid, and other factors (2). During the molting process, the response to ecdysteroid is mediated by a heterodimer complex receptor composed of USP and EcR (3), which can initiate the transcriptional activation of a cascade of downstream genes and induce the subsequent molting (4–7). In crustaceans, RXR is the homologous gene of USP in insects, and it is believed to have similar molecular functions in regulating molting (2). In insects, it has been reported that EcR binds to USP to form heterodimer complex, and then ecdysteroid binds to the heterodimer complex to activate the transcription of downstream genes in the ecdysteroid cassette signaling pathway (6–11). However, the extent to which RXR interacts with EcR in regulating molting in crustaceans remains incompletely understood.
While studies have revealed that the crystal structure of the heterodimer formed by EcR and RXR in crustaceans may resemble that of the EcR/USP complex in insects, some differences in structure and function have been identified between RXR and USP (12). The in vitro experiments of CV1 cells indicated the EcR/RXR heterodimer has different pharmacological and functional properties from the EcR/USP heterodimer (5). In CV1 and Hela cells, it was observed that the DNA binding activity of EcR/USP remained unaffected by hormones, whereas the DNA binding activity of EcR/RXR was found to be influenced by ecdysteroids or 9-cis retinoic acid (13). Juvenile hormones can directly regulate the activity of the EcR/USP heterodimer complex in insects, but exogenous 9-cis-retinoic acid and juvenile hormones are not able to activate EcR/RXR complex in cultured Sf 9 cells (14). Compared with RXR, USP is more effective and can lead to the nuclear localization EcR even without the ligands (15). Furthermore, it has been observed that USP is insensitive to the known RXR ligands, which suggests that the function of USP may differ from that of RXR (16, 17). As yet, it remains unclear whether EcR forms a heterodimer complex with RXR in crustaceans, and whether the interaction between EcR and RXR in crustaceans is analogous to that observed in insects.
Moreover, it has been indicated that EcR and USP can bind with the ecdysteroid response elements of the ecdysteroid inducible gene E75, Hsp27, Dronc, and other genes, thereby initiating their transcription (18–21). E75, E74, and BrC genes are the early responsive genes of the ecdysteroid signaling cascade pathway and play crucial roles in the molting process (22). However, in crustaceans, it has yet to be definitively established whether the classical ecdysteroid signaling cascade pathway, as inferred from insect research that EcR forms a heterodimer complex with RXR to initiate the transcription of early responsive genes such as E75 during molting, is applicable.
Eriocheir sinensis, belonging to the Arthropoda, Crustacca, Decapoda, Grapsidae, and Eriocheir genera, is widely distributed in lakes along the north and south coasts of China and one of the most important aquaculture and economic crabs in China (23). In its whole life, periodic molts will occurs 18-21 times and the ecdysone signaling pathway regulate the molting process (24). In this study, we deciphered the gene structure of EcR and RXR, conducted spatial and temporal expression experiments, co-immunoprecipitation, and luciferase reporter assay experiments of these two genes in E. sinensis. Our objective was to investigate the molecular function and molecular interplay of EcR and RXR in the molting regulation of E. sinensis.
2 Materials and methods
2.1 Sample collection and ethics statement
In this study, all the E. sinensis individuals were collected from the Aquatic Animal Germplasm Station of Shanghai Ocean University (Shanghai, China). The molting stage was observed and determined according to the setal developmental characters of the second maxilla (25). The hepatopancreas, muscle, and epidermis tissues were sampled at the premolt (PrM), intermolt (InM), and postmolt (PoM) stages with six biological replicates. Sampling procedures complied with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Shanghai Ocean University on the care and use of animals for scientific purposes. The experimental protocols were approved by the IACUC of Shanghai Ocean University.
2.2 RNA isolation and cDNA synthesis
Total RNA from each tissue was extracted using a MiniBEST Universal RNA Extraction Kit (Takara, Dalian, China) according to the manufacturer’s instructions. The quality of the extracted RNA was evaluated by 1.0% agarose gels and the quantity of RNA was measured by NanoDrop 2000 (Thermo Fisher, USA). A total of 1 μg RNA was used to synthesize complementary DNA (cDNA) using the PrimeSript™ RT reagent Kit (Takara, Dalian, China) in accordance with the manufacturer’s protocol.
2.3 Gene structure, cloning and sequence alignment
The gene sequence of RXR and exon-intron information was extracted from the assembled genome of E. sinensis (26). The gene structure of RXR was visualized using the gene structure display server 2 (http://gsds.gao-lab.org/) (27). To verify the open reading frame (ORF) sequence of RXR, primer was designed according to the extracted mRNA sequence of RXR (Table S1), and PCR was performed on the BioRad Thermal Cycler (California, USA). The expected PCR products were sequenced with an ABI3730 sequencer (Sangon, Shanghai, China). The amino acid sequence of RXR was translated according to the verified ORF sequence, and the protein domains were predicted using the CD-search software with the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (28). The amino acid sequences of RXR from Callinectes sapidus (HQ630860.1), Fenneropenaeus chinensis (FJ194479.1), Daphnia magna (DQ530508.1), Drosophila melanogaster (NM_057433.4), Homo sapiens (U38480.1), and Mus musculus (M84817.1) were downloaded from NCBI database. Multiple sequence alignment was conducted using Clustal X software.
2.4 Construction of phylogenetic tree and prediction of protein structure
A phylogenetic tree was constructed by MEGA 11 software using maximum likelihood methods with 1,000 bootstraps based on the amino acid sequence of RXR open reading frame and the amino acid sequence of EcR was used as outgroup. The nucleotide sequences of RXR from M. musculus (NM_011305.3), H. sapiens (NM_001291920.2), Danio rerio (NM_131217.4), Salmo salar (XM_014132462.2), Oncorhynchus mykiss (XM_036977326.1), D. magna (AB274819.1), Portunus trituberculatus (KF061043.2), Scylla paramamosain (KT970086.1), Penaeus monodon (KX898397.1), Litopenaeus vannamei (KF234773.1), Fenneropenaeus chinensis (FJ194479.1), Manduca sexta (U44837.1), Bombyx mori (NM_001044005.1), Spodoptera litura (JQ730734.1), Bactrocera dorsalis (HM195185.1) involved in the construction of the phylogenetic tree were downloaded from NCBI database. The 3D protein structures of EcR and RXR were modeled by the Swiss-Model server (https://swissmodel.expasy.org/) based on 4nqa.1. B and 4nqa.1.A as template, respectively (29).
2.5 Relative expression profiles of EcR and RXR during the molting cycle
Quantitative real-time PCR (qRT-PCR) was used to quantify the relative expression levels of EcR and RXR in hepatopancreas, muscle, and epidermis during different molting stages. Beta-actin (β-actin) was used as the normalized gene for qRT-PCR (30). The primer for RXR was designed according to the verified gene sequence and the primer for EcR was designed according to our previously published paper (Table S1) (2). The amplification efficiency and specificity of primers were checked for validation, and the primers used in this study had an amplification efficiency between 95% and 105%. qRT-PCR for RXR and EcR was performed with a 25 μL reaction mixture that contained 12.5 μL SYBR Green Premix Ex Taq (Takara, Dalian, China), 1 μL primer (10 μM), 2 μL dilution cDNA, and 8.5 μL dd H2O. The PCR procedure was as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, and 60°C for 30 s. qRT-PCR was performed in triplicate for each sample and each sample had six biological replicates. The relative expression was estimated using the 2 -ΔΔCt method with the InM stage as calibration control (31), and statistical significance (P < 0.05) was determined using Student t-tests.
2.6 Absolute quantitation of EcR and RXR during the molting cycle
To quantify and compare the absolute expression level (number of gene copies) for EcR and RXR in various tissues among different molting stages, standard plasmid products of EcR and RXR were first prepared. The ORFs of EcR and RXR were cloned into the pMD-19-T vector (Takara, Dalian, China), and the vector was transformed into DH5α competent cell. Then, the plasmids were extracted as standards, and the copy number of EcR and RXR plasmids was calculated with the following formula: Plasmid copy number (copies/μL) = 6.02 × 1023 (copies/μL) × plasmid concentration (g/μL)/plasmid molecular weight (g/mol) (32). The standard curves for EcR and RXR plasmids were constructed using a 10-fold serially diluted standard plasmid with a known copy number calculated above. The correlation coefficient for the standard curve was greater than 0.99, and the amplification efficiency were between 0.95 and 1.05. The absolute quantitation PCR reactions of EcR and RXR at different molting stages were performed with a 25 μL reaction mixture, and the PCR procedure was conducted the same as the above relative quantitation experiment. cDNA of various tissues from different molting stages and standard plasmid products of EcR and RXR were quantified on the fluorescence quantitative PCR instrument at the same time and each sample had six biological replicates. The absolute quantitation expression level (copy numbers) of EcR and RXR from the collected tissues among different molting stages was measured based on the equation of linear regression of the standard curve.
2.7 Co-immunoprecipitation
To construct the required plasmids for the co-immunoprecipitation experiment, designed primers with Bam HI and Hind III restriction endonuclease sites were introduced according to the gene sequence of EcR and RXR (Table S1). FLAG-tag and HA-tag sequences were added to EcR and RXR, respectively. After PCR amplification, the EcR-FLAG and RXR-HA PCR products with Bam HI and Hind III endonuclease sites were obtained. Then, the PCR products of EcR-FLAG and RXR-HA were digested with double restriction enzymes (Bam HI and Hind III) and were cloned into pcDNA3.1(-) vector to obtain the pcDNA3.1-EcR-FLAG and pcDNA3.1-RXR-HA plasmids, respectively.
The constructed pcDNA3.1-EcR-FLAG and pcDNA3.1-RXR-HA plasmids were transfected into HEK293T cells. The transfected cells were harvested after 24 hours, washed with cold phos-phate-buffered saline (PBS) twice, and lysed with cell lysis buffer (Beyotime, Shanghai). The sampled cells were ultrasonic and centrifuged at the maximum speed for 10 mins. The supernatant was collected and incubated with FLAG beads at 4 °C overnight with gentle rotation. Then the FLAG beads were washed twice with lysis buffer, loading buffer was added and boiled for 10 mins. Finally, the western bolt experiment was conducted, and mouse anti-HA (Beyotime, AH158, 1:3000) and mouse anti-FLAG (Beyotime, AF519, 1:2000) monoclonal antibodies were used.
2.8 Dual-luciferase reporter assay
E75 has been identified as a downstream responsive gene in the ecdysteroid signaling cascade, which is initiated by the EcR/USP heterodimer complex. The coding region and DNA sequence of the E75 gene were obtained from the assembled genome of E. sinensis (26). Primer was designed to amplify the upstream 2,000 bp region of E75, which is usually considered as the promotor region. Then the E75 promotor was digested by the double enzyme (NheI and HindIII) and inserted into the pGL3 vector. The constructed pGL3-E75 plasmid was used as a reporter vector in the dual luciferase reporter assay. The whole ORF of EcR and RXR with HindIII and BamHI restriction endonuclease sites were amplified and then digested with HindIII and BamHI enzymes. Later, the digested fragments were inserted into the pcDNA3.1(-) vector to construct the pcDNA3.1-EcR and pcDNA3.1-RXR expression plasmids.
The constructed pcDNA3.1-EcR, pcDNA3.1-RXR plasmids, the normalized vector (phRL-TK), and the reporter vector (pGL3-E75) were transfected into HEK293T cells seeded in 48-well plates according to the Lipofectamine 2000 manual. We designed five experimental groups: Group 1, pcDNA3.1 vector, pGL3 vector, and phRL-TK plasmid were transfected into HEK293T cells with the molar ratio of 24: 24: 2; Group 2, pcDNA3.1 vector, pGL3-E75, and phRL-TK plasmid were transfected into HEK293T cells with the molar ratio of 24:24: 2; Group3, pGL3-E75, pcDNA3.1-EcR, and phRL-TK were transfected into HEK293T cells with the molar ratio of 24: 24: 2; Group4, pGL3-E75, pcDNA3.1-RXR, and phRL-TK were transfected into HEK293T cells with the mole ratio of 24: 24: 2; Group5, pGL3-E75, pcDNA3.1-RXR, pcDNA3.1-EcR, and phRL-TK were transfected into HEK293T cells with the molar ratio of 24: 12: 12: 2. 24 hours after transfection, muristerone A (10 μm, Cayman, USA) was added into cell culture medium (33). Forty-eight hours after muristerone A added, the cells were washed twice with PBS. The enzymatic activities were measured by using Dual-Luciferase® Reporter Assay System (Promega, USA) according to the instruction. The relative luciferase activity was determined by the normalized renilla luciferase activity (34). Each assay was performed in triplicate.
3 Results
3.1 The gene structure of EcR and RXR genes
EcR gene locates at Chromosome 26 of the E. sinensis genome, with 9 exons and 8 introns which were consistent with our previous study (accession number: KY303919) (35). RXR gene locates at the chromosome 28 of the E. sinensis genome, and contains 10 exons and 9 introns (Figure 1A, Table S2). The whole ORF sequence of RXR was verified by Sanger sequencing (accession number: MK604180).
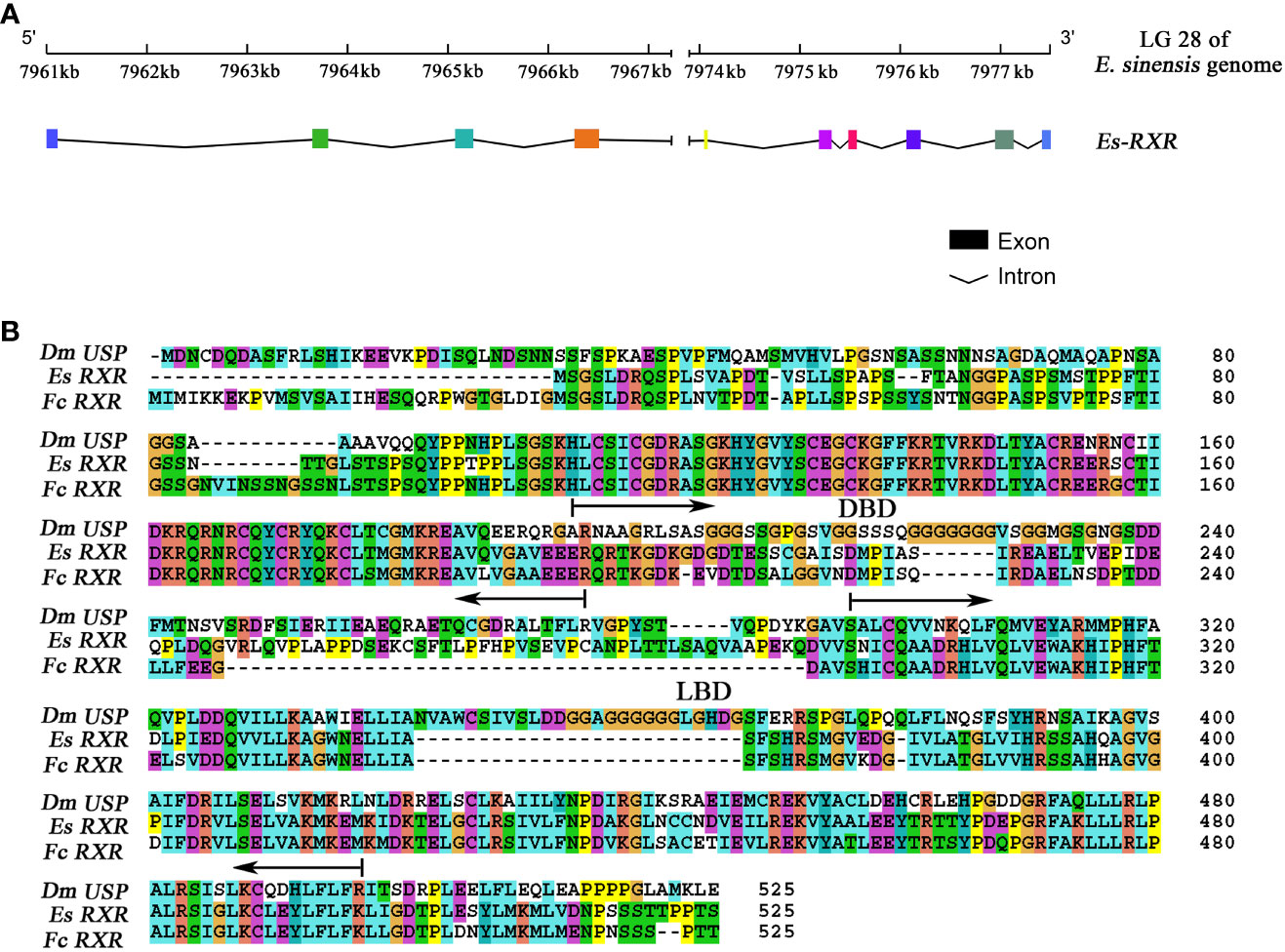
Figure 1 The gene structure characterization of RXR in E. sinensis. (A) The gene structure of RXR. Colored boxes indicated the different exons, and the black underline in the bottom figure denoted the intron boundary. (B) Amino acid sequence alignment of RXR with homologs from F. chinensis and D. melanogaster. Dm indicates D. melanogaster, Es indicates E. sinensis and Fc indicates F. chinensis. The following regions are underlined: DNA binding domain (DBD) (from 71 to 152) and ligand binding domain (LBD) (from 173 to 421).
The amino acid sequence alignment of RXR from E. sinensis with homologs from F. chinensis and D. melanogaster is presented in Figure 1B. Protein sequence alignment analysis showed that the sequence identity of RXR between E. sinensis and C. sapidus, F. chinensis, D. magna, D. melanogaster, H. sapiens, and M. musculus were 82.67%, 66.80%, 60.72%, 37.06%, 47.67%, and 47.39%, respectively. The DNA binding domain (DBD) and ligand binding domain (LBD) of RXR in E. sinensis were identified by searching the conserved domains in the NCBI database using CD-search algorithm (36). The lengths of DBD and LBD in E. sinensis were 82 (from 71 to 152) and 249 (from 173 to 421) amino acid residues, respectively. The sequence identity of RXR’s DBD between E. sinensis and C. sapidus, F. chinensis, D. magna, D. melanogaster, H. sapiens, and M. musculus was 92.68%, 95.12%, 89.02%, 89.02%, 79.27%, 81.71%, respectively. However, for the LBD, the sequence identity for the above comparison was 76.31%, 67.47%, 60.64%, 37.32%, 59.44%, and 58.23%, respectively.
3.2 Phylogenetic tree of RXR and protein structure prediction
The phylogenetic tree of RXR based on amino acid sequence could be separated into three clades, with RXR from crustacean and vertebrate species forming two clades and USP from insects forming another clade (Figure 2A). The phylogenetic tree may indicate the sequence divergence and functional difference of RXR and USP among crustaceans, insects, and vertebrates. Based on the predicted protein structure of EcR and RXR, typical features of nuclear receptors, such as the DNA binding domain and ligand binding domain, were identified (Figure 2B, C).
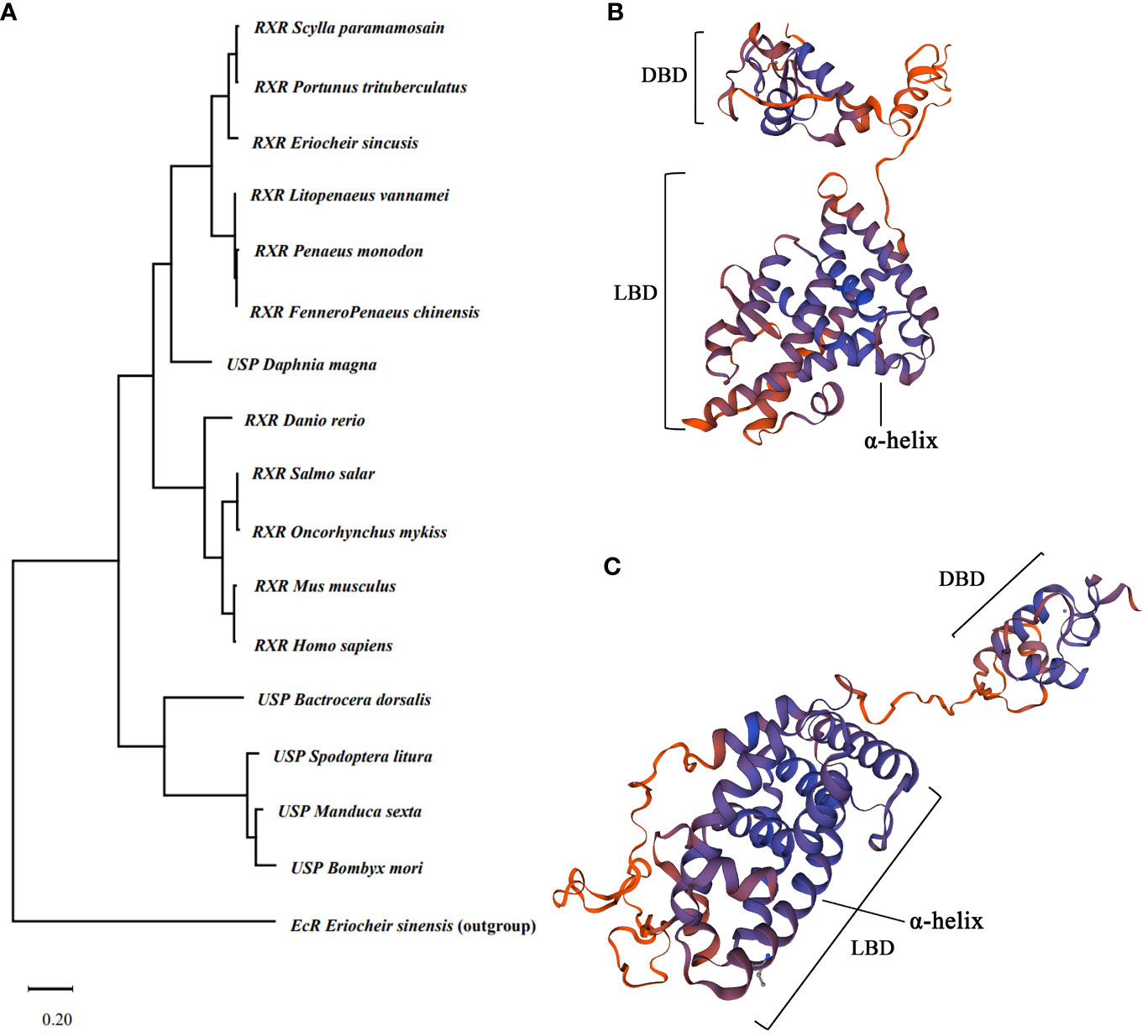
Figure 2 Evolutionary analysis of RXR gene and 3D protein structure prediction. (A) Maximum likelihood phylogenetic tree based on RXR amino acid sequences. The tree was generated by MEGA 11 with a bootstrap value of 1,000 bootstraps. The EcR of Ericheir sinensis was used as outgroup. (B) Predicted protein structure of EcR of E sinensis. (C) Predicted protein structure of RXR of E sinensis. DBD indicate DNA binding domain, and LBD indicate ligand binding domain.
3.3 Spatial and temporal expression profiles of EcR and RXR during the molting process
The qRT-PCR experiment indicated that both EcR and RXR were expressed in the hepatopancreas, epidermis, and muscle tissue during the whole molting process (Figure 3). Throughout the molting cycle, the expression of EcR exhibited dynamic fluctuations and increased significantly during the PrM stage in all three tissues. Specifically, the expression level of EcR was found to be 5.2 times, 8.3 times, and 20 times higher at the PrM stage compared to that at the InM stage in the hepatopancreas, epidermis, and muscle tissues, respectively (Figure 3). However, there was no significant expression difference for RXR in the three tissues during the molting process (p > 0.05) (Figure 3).
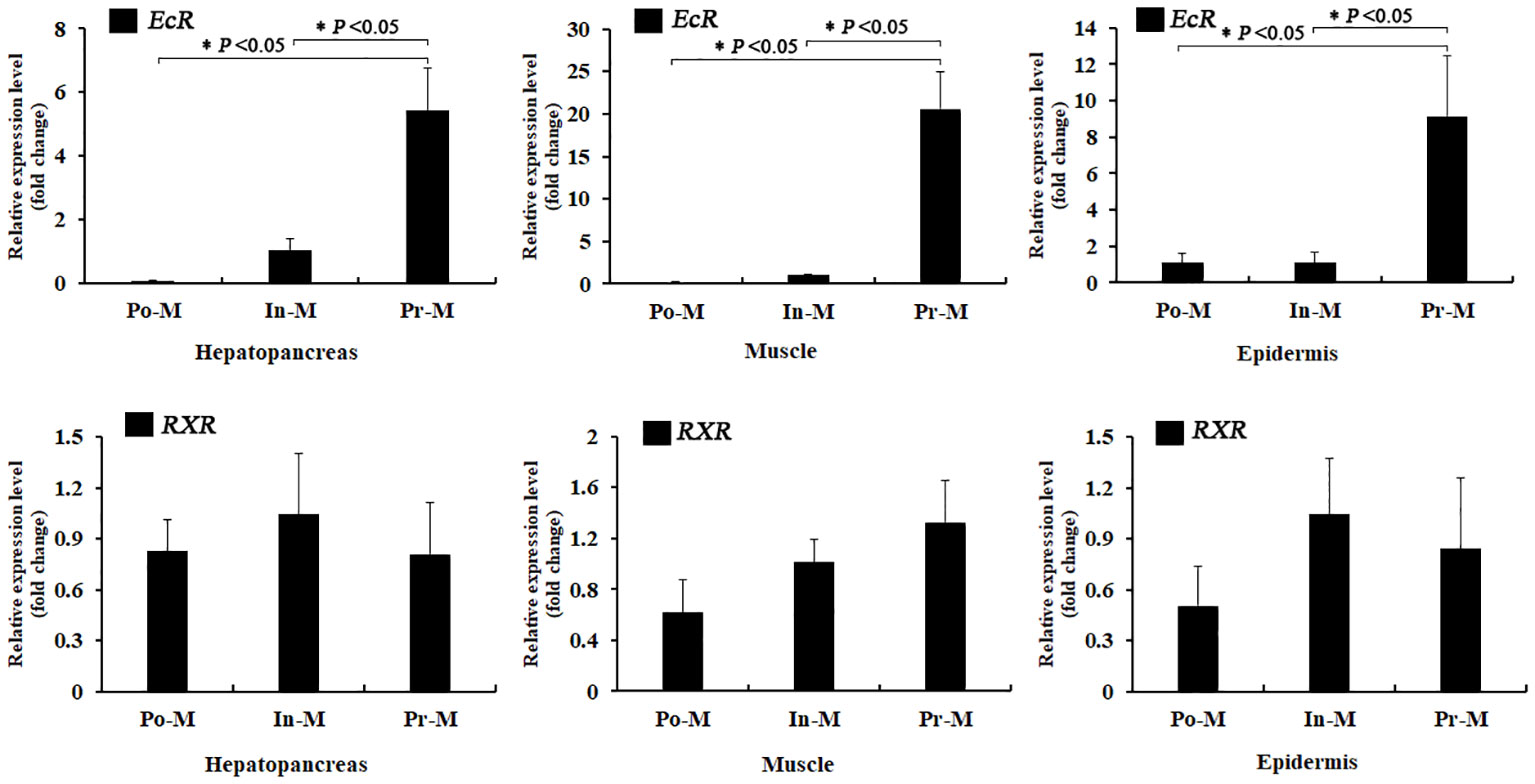
Figure 3 Relative expression analysis of EcR and RXR in different tissues and molt stages of E. sinensis. Po-M means postmolt, In-M means intermolt, and Pre-M mean premolt. Error bars represent standard error. “*” means significant difference (p < 0.05).
The absolute expression level of RXR was significantly higher than that of EcR in all three tissues and molting stages (Figure 4). Specifically, the fold change of the expression level of RXR/EcR was 289 at PoM and 5 at PrM in hepatopancreas tissue, was 179 at PoM and 34 at PrM in epidermis tissue, and was 289 at PoM and 10 at PrM in muscle tissue. During the molting process, the absolute expression level of EcR showed dynamic expression and was significantly upregulated at the PrM stage in the three tissues. However, the absolute expression level of RXR was relatively stable, consistent with qRT-PCR results (Figure 3).
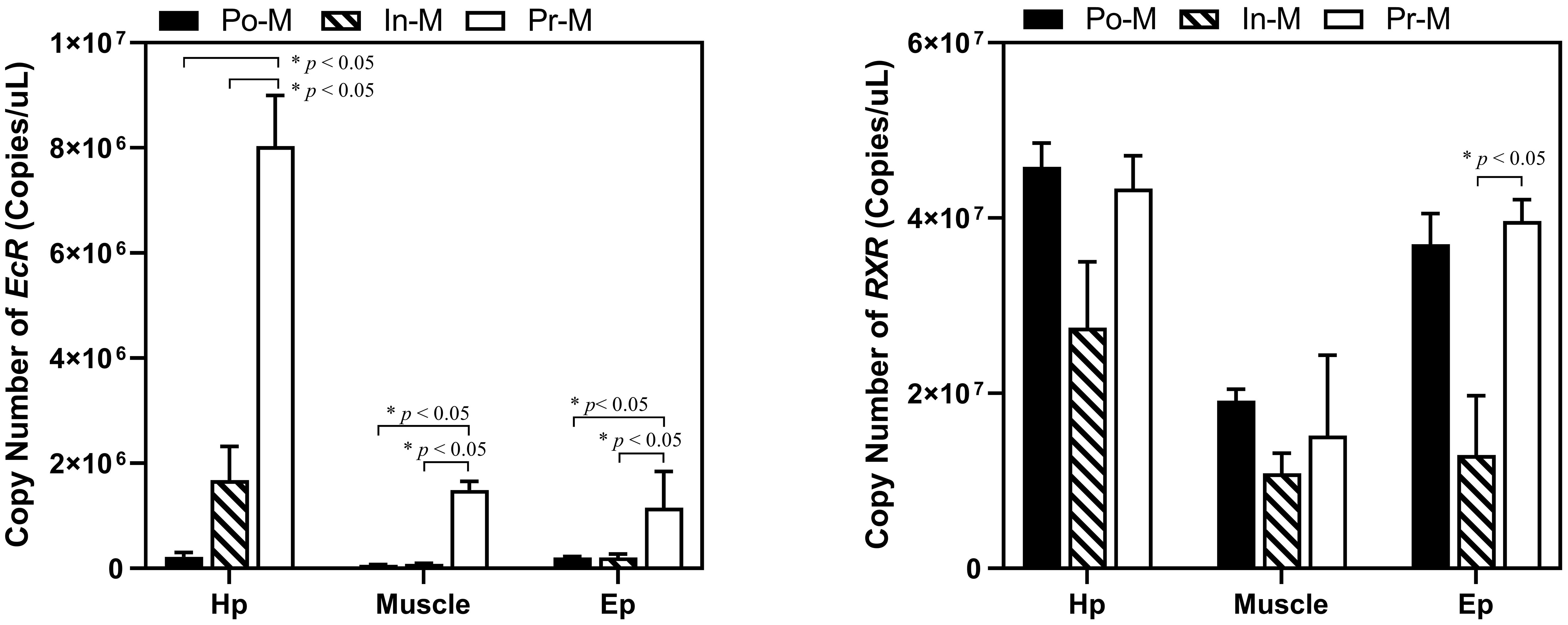
Figure 4 Absolute expression analysis of EcR and RXR in different tissues and molt stages of E. sinensis. Po-M means postmolt, In-M means intermolt, and Pre-M mean premolt. Error bars represent standard error. “*” means significant difference (p < 0.05).
3.4 Molecular interplay of EcR and RXR proteins
The overexpression experiment indicated that both EcR-FLAG and RXR-HA proteins were successfully expressed in 293T cells (Figure 5A). Regarding the Co-IP experiments, FLAG and HA proteins were successfully detected in the input group after overexpression, indicating successful overexpression of EcR-FLAG (~62 kDa) and RXR-HA (~52 kDa) in the 293T cells. In the IP group, flag protein was detected, indicating successful binding of flag beads with EcR-FLAG protein. Additionally, HA was also successfully detected, indicating that RXR-HA protein was pulled down by EcR-FLAG protein. (Figure 5B).
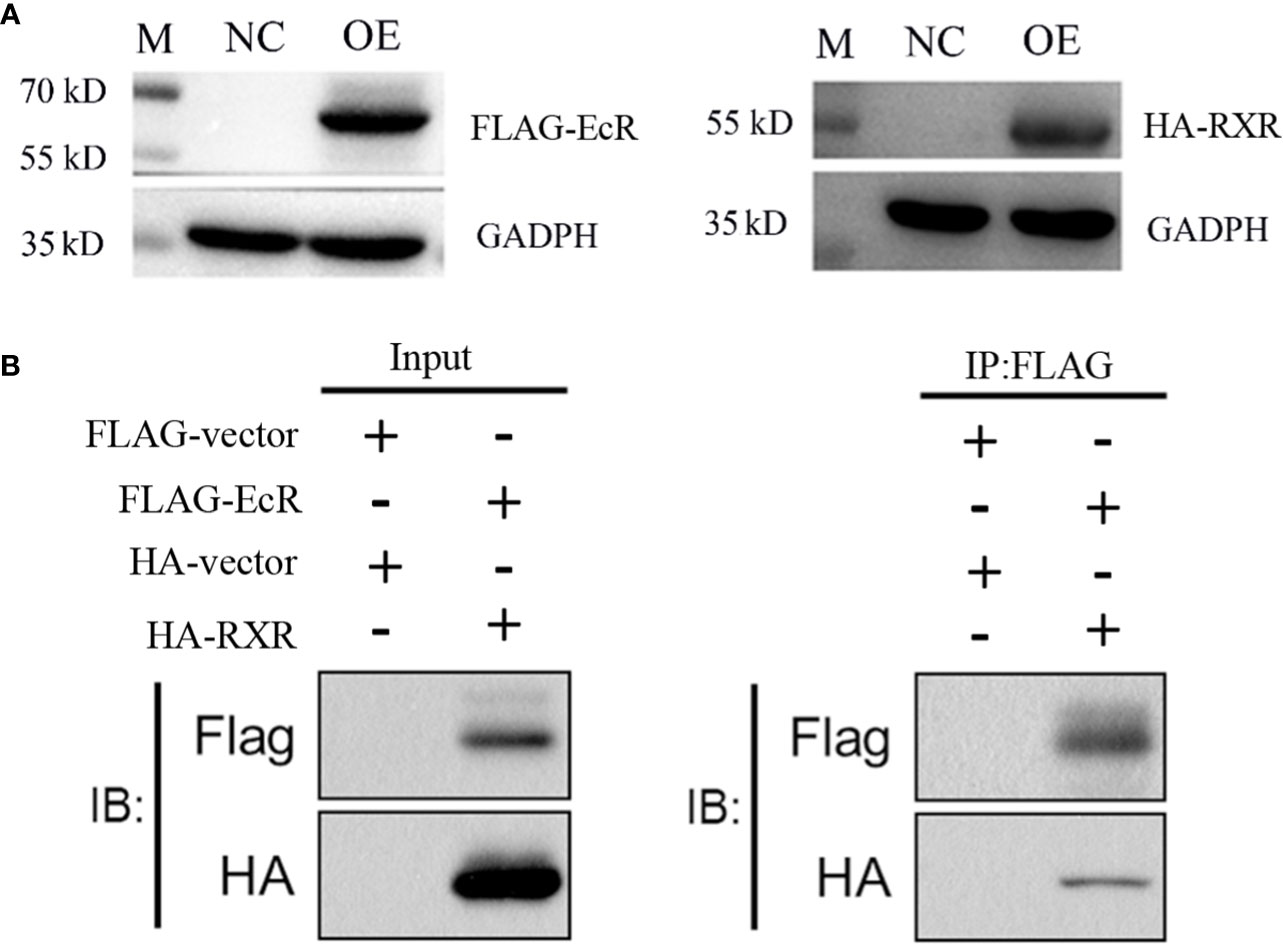
Figure 5 EcR interacts with RXR confirmed by a Co-IP assay. (A) Overexpression detection of EcR and RXR in 293T cells by Western blot. EcR-FLAG, RXR-HA, and control plasmids were overexpressed in 293T cells. M, marker. NC: negative control group. OE: overexpression group. The cells were harvested to verify the expression of EcR and RXR by Western blot. (B) The Co-IP experiment of EcR and RXR. The input and IP: Flag are labeled.
3.5 EcR interacts with RXR and regulates the transcription of E75
The enzymatic activity of luciferase driven by the E75 promoter was significantly higher than the control group (the empty pGL3 basic vector). Furthermore, in comparison to the transfection of pcDNA3.1-EcR (group 3) or pcDNA3.1-RXR (group 4) plasmid alone, co-transfection of pcDNA3.1-EcR and pcDNA3.1-RXR plasmids into HEK293T cells resulted in a significant increase in luciferase activity, indicating that EcR and RXR together induce the transcription of E75 (Figure 6).
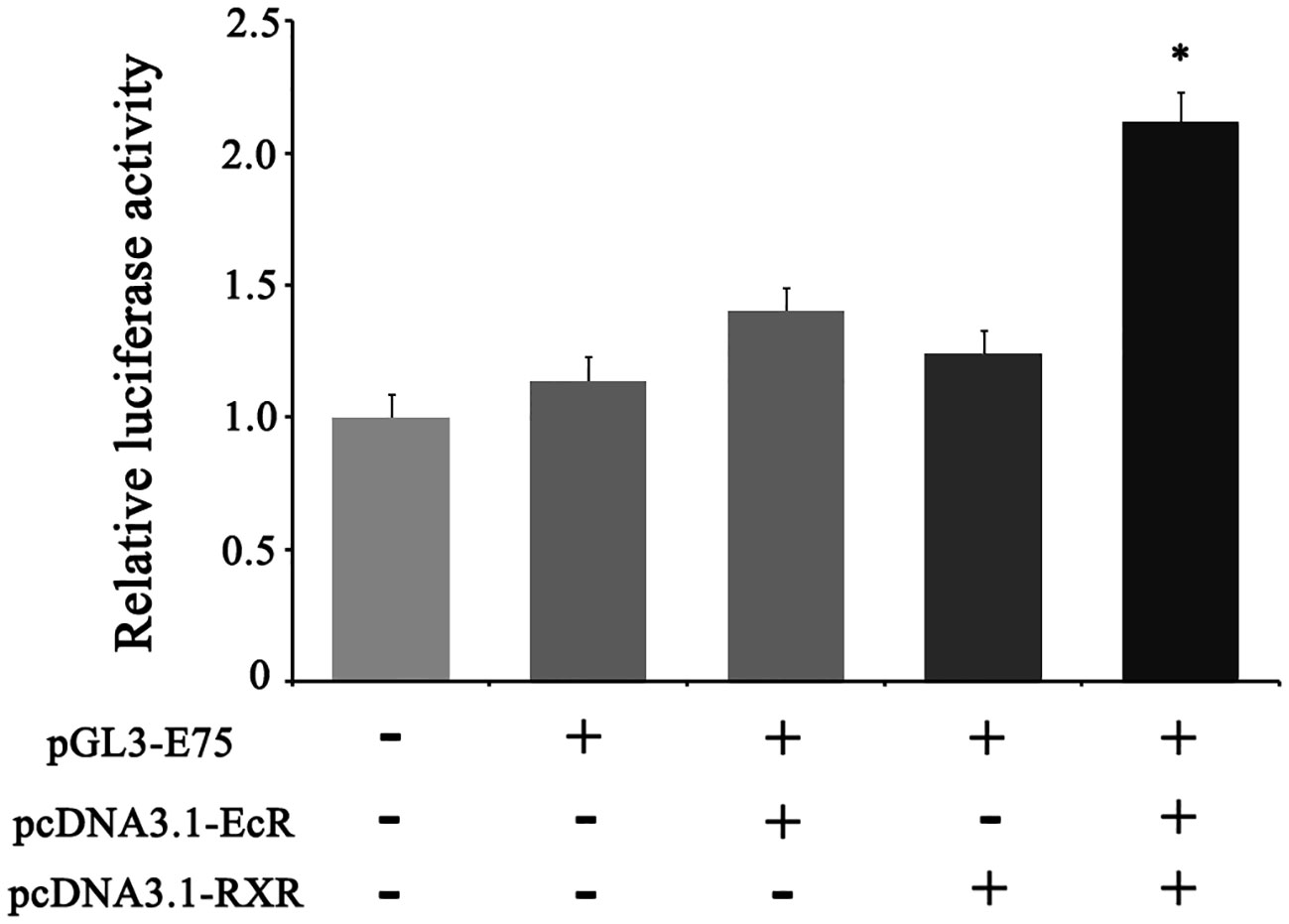
Figure 6 Transcriptional activity identification of E75 in the HEK-293T cell line by dual-luciferase reporter assay. phRL-TK (internal control), pGL3-E75 (reporter plasmid), pcDNA3.1-EcR (overexpression plasmid of EcR), and pcDNA3.1-RXR (overexpression plasmid of RXR) were used in the transfection assay. The mole ratio of pGL3-E75, pcDNA3.1-EcR/pcDNA3.1-RXR, phRL-TK was 24: 24: 2 in the third and fourth column, and the mole ratio of pGL3-E75, pcDNA3.1-EcR, pcDNA3.1-RXR, phRL-TK was 24: 12: 12: 2 in the fifth column. Equal mole of empty pGL3 was used in the first column and its activity was defined as 1. Data represent mean ± SEM with three replicates. “*” means significant difference with other groups (p < 0.05).
4 Discussion
Comparing the amino acid sequences of RXR in E. sinensis to those of other species, a higher sequence identity for the DBD of RXR than for the LBD was observed. This may illuminate why the target genes of RXR are almost identical, but the ligands of RXR are different among different organisms (37). For example, in insects, the USP, a homolog of RXR in crustaceans, does not respond to 9-cis-retinoic acid in cultured cells and tissues, while RXR can (38, 39). Moreover, phytanic acid and docosahexaenoic acid were identified as ligands of RXR, but not of USP (40, 41). These findings suggest that RXR and USP may have different LBD structures and ligands, indicating that the functions of EcR/RXR in crustaceans and EcR/USP in insects may differ to some extent.
In this study, the expression level of EcR increased sharply at the premolt stage, whereas the RXR presented a relatively stable expression level throughout the whole molting process (Figure 3). A consistent result was also identified in D. melanogaster, P. japonicus, and F. chinensis where the expression of EcR in hepatopancreas increased significantly from intermolt to premolt stage; however, the expression of RXR did not change much during the molting cycle (42–44). The research on the structure and function of the human RXR gene also indicated that RXR exhibits characteristics of a housekeeping gene (45). Moreover, the expression level of RXR was always much higher than that of EcR during the molting cycle (Figure 3). These results indicated that RXR may be a “constitutive-type” gene, similar to housekeeping genes, while EcR is a critical speed-limiting gene that plays an essential role in the molting process. Additionally, research on the structure of the promoter of the human RXRA gene has also indicated that RXR exhibits features of a housekeeping gene (45). It has been reported that RXR can dimerize with diverse nuclear receptors, such as FXR, PPARs and RARs, not only EcR, thus perform different functions other than molting regulation (46, 47). Regarding E. sinensis, RXR may also dimerize with other nuclear receptors to play different roles, which may explain the abundant and stable expression of RXR during the whole molting cycle. Taken together, our results suggest that RXR may be classified as a “constitutive-type” gene, similar to housekeeping genes, while EcR is a critical speed-limiting gene that plays an essential role in the molting process.
Research has indicated that EcR/USP complexes activate downstream gene expression in insects (48, 49). In D. melanogaster, several researchers have demonstrated that USP forms heterodimers with EcR to activate downstream gene expression, and USP is critical for EcR function (48, 50). As a dynamic complex, the activity of EcR can only be activated by binding with its ligands (5, 6, 8, 13, 51, 52). Studies in epithelial cells of Chironomus tentans have shown that EcR and USP form a unique complex that acts on palindromic structure and induce the transcription of Hsp27 (10). In this study, EcR of E. sinensis can interact with RXR of E. sinensis, which was verified by the CO-IP experiment. Studies on Neomysis integer presented that EcR can also form heterodimer with RXR in GAL4 cell reporting system (53) and EcR and RXR of Uca pugilator formed heterodimer in S9 cell (54). These results strongly indicate that EcR may bind with RXR to form a functional heterodimer complex in crustaceans. Moreover, the transcriptional activity of downstream gene E75 was significantly higher when RXR and EcR were both overexpressed than when only EcR or RXR was overexpressed in cells (Figure 6). Our result strongly indicates that the transcription of downstream genes, such as E75, in E. sinensis during the molting process can be activated by the functional heterodimer complex formed by EcR and RXR.
In conclusion, we clearly deciphered the gene structure of EcR and RXR and investigated the molecular interplay of them in the molting process in E. sinensis. Our results indicate that EcR is a key speed-limiting gene and RXR is a “constitutive-type” gene involved in the ecdysteroid signaling pathway. Moreover, EcR and RXR bind to form heterodimers to activate the transcription of downstream response gene E75 to regulate the molting process of E. sinensis (Figure 7). Our research provides novel insights into the molecular mechanism of molting mediated by the ecdysteroid signaling pathway in crustaceans.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee (IACUC) of Shanghai Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XC, JW and CW contributed to design the study. XC and XH contributed most analysis for the study. HY and HL helped to organized data. XC wrote the draft. XH, JW and CW contributed to draft review and editing. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Project of China (2022YFD2400701).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1251723/full#supplementary-material
Supplementary Table 1 | Primers used in this study for cloning and expression analysis of Eriocheir sinensis EcR and RXR.
Supplementary Table 2 | The detailed position information of EcR and RXR in genome.
References
1. Yamanaka N. Chapter One - Ecdysteroid signalling in insects-From biosynthesis to gene expression regulation. In: Adams ME, editor. Advances in Insect Physiology. Cambridge, MA, USA: Academic Press (2021). 60 1–36.
2. Hyde CJ, Elizur A, Ventura T. The crustacean ecdysone cassette: A gatekeeper for molt and metamorphosis. J Steroid Biochem Mol Biol (2019) 185:172–83. doi: 10.1016/j.jsbmb.2018.08.012
3. Hult EF, Huang J, Marchal E, Lam J, Tobe SS. RXR/USP and EcR are critical for the regulation of reproduction and the control of JH biosynthesis in Diploptera punctata. J Insect Physiol (2015) 80:48–60. doi: 10.1016/j.jinsphys.2015.04.006
4. Iwema T, Chaumot A, Studer RA, Robinson-Rechavi M, Billas IML, Moras D, et al. Structural and evolutionary innovation of the heterodimerization interface between USP and the ecdysone receptor ECR in Insects. Mol Biol Evolution (2009) 26(4):753–68. doi: 10.1093/molbev/msn302
5. Yao T-P, Forman BM, Jiang Z, Cherbas L, Chen JD, McKeown M, et al. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature (1993) 366:476. doi: 10.1038/366476a0
6. Hu X, Cherbas L, Cherbas P. Transcription activation by the ecdysone receptor (EcR/USP): identification of activation functions. Mol Endocrinol (2003) 17(4):716–31. doi: 10.1210/me.2002-0287
7. Antoniewski C, Laval M, Dahan A, Lepesant JA. The ecdysone response enhancer of the Fbp1 gene of Drosophila melanogaster is a direct target for the EcR/USP nuclear receptor. Mol Cell Biol (1994) 14(7):4465. doi: 10.1128/MCB.14.7.4465
8. Yao T-P, Segraves WA, Oro AE, McKeown M, Evans RM. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell (1992) 71(1):63–72. doi: 10.1016/0092-8674(92)90266-F
9. Antoniewski C, Mugat B, Delbac F, Lepesant JA. Direct repeats bind the EcR/USP receptor and mediate ecdysteroid responses in Drosophila melanogaster. Mol Cell Biol (1996) 16(6):2977. doi: 10.1128/MCB.16.6.2977
10. Elke C, Rauch P, Spindler-Barth M, Spindler KD. DNA-binding properties of the ecdysteroid receptor-complex (EcR/USP) of the epithelial cell line from Chironomus tentans. Arch Insect Biochem Physiol (1999) 41:124–33. doi: 10.1002/(SICI)1520-6327(1999)41:3<124::AID-ARCH3>3.0.CO;2-C
11. Swevers L, Cherbas L, Cherbas P, Iatrou K. Bombyx EcR (BmEcR) and Bombyx USP (BmCF1) combine to form a functional ecdysone receptor. Insect Biochem Mol Biol (1996) 26(3):217–21. doi: 10.1016/0965-1748(95)00097-6
12. Devarakonda S, Harp JM, Kim Y, Ożyhar A, Rastinejad F. Structure of the heterodimeric ecdysone receptor DNA-binding complex. EMBO J (2003) 22(21):5827. doi: 10.1093/emboj/cdg569
13. Thomas HE, Stunnenberg HG, Stewart AF. Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature (1993) 362:471. doi: 10.1038/362471a0
14. Fang F, Xu Y, Jones D, Jones G. Interactions of ultraspiracle with ecdysone receptor in the transduction of ecdysone- and juvenile hormone-signaling. FEBS J (2005) 272(7):1577–89. doi: 10.1111/j.1742-4658.2005.04578.x
15. Nieva C, Spindler-Barth M, Spindler KD. Impact of heterodimerization on intracellular localization of the ecdysteroid receptor (EcR). Arch Insect Biochem Physiol (2008) 68(1):40–8. doi: 10.1002/arch.20234
16. Jones G, Sharp PA. Ultraspiracle: An invertebrate nuclear receptor for juvenile hormones. Proc Natl Acad Sci (1997) 94(25):13499. doi: 10.1073/pnas.94.25.13499
17. Nakagawa Y, Henrich VC. Arthropod nuclear receptors and their role in molting. FEBS J (2009) 276(21):6128–57. doi: 10.1111/j.1742-4658.2009.07347.x
18. Antoniewski C, Laval M, Lepesant J-A. Structural features critical to the activity of an ecdysone receptor binding site. Insect Biochem Mol Biol (1993) 23(1):105–14. doi: 10.1016/0965-1748(93)90088-A
19. Cakouros D, Daish TJ, Kumar S. Ecdysone receptor directly binds the promoter of the Drosophila caspase dronc, regulating its expression in specific tissues. J Cell Biol (2004) 165(5):631–40. doi: 10.1083/jcb.200311057
20. Bernardo TJ, Dubrovskaya VA, Xie X, Dubrovsky EB. A view through a chromatin loop: insights into the ecdysone activation of early genes in Drosophila. Nucleic Acids Res (2014) 42(16):10409–24. doi: 10.1093/nar/gku754
21. Ożyhar A, Pongs O. Mutational analysis of the interaction between ecdysteroid receptor and its response element. J Steroid Biochem Mol Biol (1993) 46(2):135–45. doi: 10.1016/0960-0760(93)90288-8
22. Zhou K, Zhou F, Jiang S, Huang J, Yang Q, Yang L, et al. Ecdysone inducible gene E75 from black tiger shrimp Penaeus monodon: Characterization and elucidation of its role in molting. Mol Reprod Dev (2019) 86(3):265–77. doi: 10.1002/mrd.23101
23. Wang C, Li C, Li S. Mitochondrial DNA-inferred population structure and demographic history of the mitten crab (Eriocheir sensu stricto) found along the coast of mainland China. Mol Ecol (2008) 17(15):3515–27. doi: 10.1111/j.1365-294X.2008.03850.x
24. Wang W, Wang C, Zhouxu M. Ecological Aquaculture of Chinese mitten crab. 2 edn. BeiJing, China: China Agriculture Press (2013).
25. Hou X, Chen X, Yang H, Yue WA-O, Wang JA-O, Han H, et al. V-ATPase subunit B plays essential roles in the molting process of the Chinese mitten crab, Eriocheir sinensis. Biol Open (2020) 9(5):bio048926. doi: 10.1242/bio.048926
26. Wang J, Chen X, Hou X, Wang J, Yue W, Huang S, et al. “Omics” data unveil early molecular response underlying limb regeneration in the Chinese mitten crab, Eriocheir sinensis. Sci Adv (2022) 8(37):eabl4642. doi: 10.1126/sciadv.abl4642
27. Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics (2015) 31(8):1296–7. doi: 10.1093/bioinformatics/btu817
28. Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res (2020) 48:265–8. doi: 10.1093/nar/gkz991
29. Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res (2018) 46:296–303. doi: 10.1093/nar/gky427
30. Huang S, Chen X, Wang J, Chen J, Yue W, Lu W, et al. Selection of appropriate reference genes for qPCR in the Chinese mitten crab, Eriocheir sinensis (Decapoda, Varunidae). Crustaceana (2017) 90(3):275–96. doi: 10.1163/15685403-00003651
31. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc (2008) 3(6):1101–8. doi: 10.1038/nprot.2008.73
32. Huo Y-Y, Li G-F, Qiu Y-H, Li W-M, Zhang Y-J. Rapid detection of prunus necrotic ringspot virus by reverse transcription-cross-priming amplification coupled with nucleic acid test strip cassette. Sci Rep (2017) 7(1):16175. doi: 10.1038/s41598-017-16536-6
33. Saez E, Nelson MC, Eshelman B, Banayo E, Koder A, Cho GJ, et al. Identification of ligands and coligands for the ecdysone-regulated gene switch. Proc Natl Acad Sci (2000) 97(26):14512–7. doi: 10.1073/pnas.260499497
34. He A, Liu C, Chen L, Ning L, Qin J, Li J, et al. Molecular characterization, transcriptional activity and nutritional regulation of peroxisome proliferator activated receptor gamma in Nile tilapia (Oreochromis niloticus). Gen Comp Endocrinol (2015) 223:139–47. doi: 10.1016/j.ygcen.2015.05.008
35. Chen X, Wang J, Yue W, Huang S, Chen J, Chen Y, et al. Structure and function of the alternatively spliced isoforms of the ecdysone receptor gene in the Chinese mitten crab, Eriocheir sinensis. Sci Rep (2017) 7(1):12993. doi: 10.1038/s41598-017-13474-1
36. Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res (2017) 45(D1):D200–D3. doi: 10.1093/nar/gkw1129
37. Evans RM. Mangelsdorf DJJC. Nuclear receptors, RXR, and the big bang. Cell (2014) 157(1):255–66. doi: 10.1016/j.cell.2014.03.012
38. Iwema T, Billas IML, Beck Y, Bonneton F, Nierengarten H, Chaumot A, et al. Structural and functional characterization of a novel type of ligand-independent RXR-USP receptor. EMBO J (2007) 26(16):3770. doi: 10.1038/sj.emboj.7601810
39. Ruehl R, Krzyzosiak A, Niewiadomska-Cimicka A, Rochel N, Szeles L, Vaz B, et al. 9-cis-13,14-Dihydroretinoic acid is an endogenous retinoid acting as RXR ligand in mice. PloS Genet (2015) 11(6):e1005213. doi: 10.1371/journal.pgen.1005213
40. Kitareewan S, Burka LT, Tomer KB, Parker CE, Deterding LJ, Stevens RD, et al. Phytol metabolites are circulating dietary factors that activate the nuclear receptor RXR. Mol Biol Cell (1996) 7(8):1153–66. doi: 10.1091/mbc.7.8.1153
41. Urquiza A, Liu S, Sjöberg M, Zetterström RH, Griffiths W, Sjövall J, et al. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science (2000) 290(5499):2140. doi: 10.1126/science.290.5499.2140
42. Asazuma H, Nagata S, Kono M, Nagasawa H. Molecular cloning and expression analysis of ecdysone receptor and retinoid X receptor from the kuruma prawn, Marsupenaeus japonicus. Comp Biochem Physiol Part B: Biochem Mol Biol (2007) 148(2):139–50. doi: 10.1016/j.cbpb.2007.05.002
43. Priya TAJ, Li F, Zhang J, Wang B, Zhao C, Xiang J. Molecular characterization and effect of RNA interference of retinoid X receptor (RXR) on E75 and chitinase gene expression in Chinese shrimp Fenneropenaeus chinensis. Comp Biochem Physiol Part B: Biochem Mol Biol (2009) 153(1):121–9. doi: 10.1016/j.cbpb.2009.02.009
44. Sullivan AA, Thummel CS. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol Endocrinol (2003) 17(11):2125–37. doi: 10.1210/me.2002-0430
45. Li G, Walch E, Yang X, Lippman SM, Clifford JL. Cloning and characterization of the human retinoid X receptor alpha gene: conservation of structure with the mouse homolog. Biochem Biophys Res Commun (2020) 269:54–7. doi: 10.1006/bbrc.2000.2244
46. Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell (2006) 126(4):801–10. doi: 10.1016/j.cell.2006.06.050
47. Lefebvre P, Benomar Y, Staels B. Retinoid X receptors: common heterodimerization partners with distinct functions. Trends Endocrinol Metab (2010) 21(11):676–83. doi: 10.1016/j.tem.2010.06.009
48. Billas IML, Iwema T, Garnier J-M, Mitschler A, Rochel N, Moras D. Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor. Nature (2003) 426:91. doi: 10.1038/nature02112
49. Sun X, Song Q, Barrett B. Effects of ecdysone agonists on the expression of EcR, USP and other specific proteins in the ovaries of the codling moth (Cydiapomonella L.). Insect Biochem Mol Biol (2003) 33(8):829–40. doi: 10.1016/S0965-1748(03)00082-1
50. Billas IML, Moras DJV. Ligand-binding pocket of the ecdysone receptor. Vitamins Hormones (2005) 73:101–29. doi: 10.1016/S0083-6729(05)73004-1
51. Buszczak M, Segraves WA. Drosophila metamorphosis: The only way is USP? Curr Biol (1998) 8(24):879–82. doi: 10.1016/S0960-9822(07)00550-7
52. Kozlova T, Thummel CS. Steroid regulation of postembryonic development and reproduction in Drosophila. Trends Endocrinol Metab (2000) 11(7):276–80. doi: 10.1016/S1043-2760(00)00282-4
53. De Wilde R, Swevers L, Soin T, Christiaens O, Rougé P, Cooreman K, et al. Cloning and functional analysis of the ecdysteroid receptor complex in the opossum shrimp Neomysis integer (Leach, 1814). Aquat Toxicol (2013) 130-131:31–40. doi: 10.1016/j.aquatox.2012.12.011
Keywords: molting, ecdysteroid signaling pathway, molecular interplay, ecdysone receptor, retinoid X receptor
Citation: Chen X, Hou X, Yang H, Liu H, Wang J and Wang C (2023) Molecular interplay between ecdysone receptor and retinoid X receptor in regulating the molting of the Chinese mitten crab, Eriocheir sinensis. Front. Endocrinol. 14:1251723. doi: 10.3389/fendo.2023.1251723
Received: 02 July 2023; Accepted: 05 October 2023;
Published: 19 October 2023.
Edited by:
Shaojun Liu, Hunan Normal University, ChinaReviewed by:
Naoki Okamoto, University of Tsukuba, JapanYuichiro Suzuki, Wellesley College, United States
Copyright © 2023 Chen, Hou, Yang, Liu, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenghui Wang, d2FuZ2NoQHNob3UuZWR1LmNu
†These authors have contributed equally to this work
 Xiaowen Chen
Xiaowen Chen Xin Hou†
Xin Hou† Jun Wang
Jun Wang Chenghui Wang
Chenghui Wang