
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 15 August 2023
Sec. Cellular Endocrinology
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.1235501
This article is part of the Research Topic Global Excellence in Natural Products for Endocrine Disorders View all 6 articles
Introduction: The action of environmental steroids on the human glucocorticoid receptor (hGR) has been pointed out with the risk to impair physiological immune and metabolic processes regulated by this nuclear receptor. However, there is still a lack of mechanistic information regarding their ability to interact with GR in aquatic species.
Methods: To investigate ligand activation differences between hGR and zebrafish GR (zfGR), we tested several natural and synthetic steroids using reporter cell lines expressing hGR or zfGR.
Results and discussion: Almost all the glucocorticoids tested (dexamethasone, cortisol, bimedrazol, medrol, cortivazol and fluticasone) are agonists of the two receptors with similar potencies. The dissociated glucocorticoids, RU24782 and RU24858 are agonists of both zfGR and hGR but with a better potency for the latter. On the other hand, the synthetic glucocorticoid forbimenol and the mineralocorticoid aldosterone are agonist on hGR but antagonist on zfGR. The other steroids tested, androgens and progestins, are all antagonists of both GRs with equal or lower potency on zfGR than on hGR. Surprisingly, the lower efficacy and potency on zfGR of aldosterone, forbimenol and the dissociated glucocorticoids is not related to their affinity for the receptors which would suggest that it could be related to less efficacious recruitment of coactivators by zfGR compared to hGR.
The glucocorticoid receptor (NR3C1, GR) is a ligand-activated transcription factor belonging to the family of the nuclear receptors (NRs) (1). GR is composed of three major domains: i) an N-terminal transactivation domain (NTD); ii) a small central DNA-binding domain (DBD) and iii) a C-terminal ligand binding domain (LBD) which hosts the ligand-dependent transcriptional activation function 2 (AF-2) (2). In the absence of ligand, GR is located in the cytosol and is affiliated with a large multiprotein complex that includes heat shock protein (HSP) 90, HSP70 and immunophilins (3, 4). Upon ligand binding with the ligand, the HSP complex disassociates and the receptor translocates into the nucleus to exert its transactivating effects, where it binds as a homodimer to glucocorticoid response elements (GREs) in the promoter regions of target genes. GR can also exert transrepressing effects by binding to negative GRE (nGREs) and probably by interfering with the binding of other transcription factors (5). Moreover, GR can also exert its transrepression activity independently of the DNA binding by interacting with transcription factors such as NF-kB and AP-1, which control the genes of many mediators of inflammation and immunity (6).
Glucocorticoids (GCs) are cholesterol-derived lipophilic steroid hormones produced by adrenal glands in response to external and internal signals. The main endogenous glucocorticoid hormone produced in human is cortisol, while synthetic GCs like dexamethasone and prednisolone are used extensively both in the treatment of chronic inflammatory diseases, such as rheumatoid arthritis and asthma, and for their immunosuppressant action in preventing organ rejection post transplantation (7, 8). As beneficial effects of GCs are limited by their undesirable side effects like diabetes, osteoporosis, hypertension and skin thinnings, synthetic GCs called “dissociated glucocorticoids” have been synthetized (9). These chemicals less able to maintain hGR in a conformation able to recruit coactivators than full agonists like dexamethasone displayed limited transactivation potency but strong transrepression activity (10).
Recent studies have reported that xenobiotic substances such as metals, bisphenols, vinclozolin metabolites, organotins, polybrominated diphenyl ethers and polychlorinated biphenyls can interfere with hGR (11–15). Moreover, due to the wide use of synthetic glucocorticoids and other steroids as medicaments and their incomplete removal in discharged water systems, they have been detected in the aquatic environment (16–18).
In the last decades, the presence in the environment of substances potentially interfering with NRs has been widely investigated using different in vitro assays expressing human NRs (19–21). However, it has been recently underlined that extrapolation of data from mammalian pharmacology and toxicology into fish species could not be appropriate due to interspecies differences between receptors (21–26) On the other hand, the use of zebrafish as model organism for aquatic toxicology is a reliable tool due to its specific features that made its use considerably grown in the last decades in scientific research. In this regard, we have established two reporter cell lines expressing respectively hGR and zfGR with the aim of improve Endocrine disrupting chemicals (EDCs) risk assessment in water quality monitoring. In this work, we tested several natural and pharmaceutical steroids on these established cell lines with the aim of identifying possible human and zebrafish differences in the capacity of these chemicals to transactivate GR, as long as we have recently pointed out species-specificity differences in the activation or inhibition of hNRs and zfNRs in water extracts (21). The findings of the current study provide new information on the activities of the chemicals on hGR and zfGR and allow the development of new biological tools to evaluate EDC in environmental samples.
Cell culture materials are from Life Technologies (Cergy-Pontoise, France). Luciferin (sodium salt) was purchased from Promega (Charbonnières, France). Chemical substances used in this study are presented in Table 1. Methyltrienolone (R1881), dexamethasone (DEX), mifepristone (RU486), aldosterone (ALDO), pregnenolone (P5), progesterone (P4), dydrogesterone (DYD), norethindrone (NET), tibolone (TIB), spironolactone (SPI), canrenone (CAN), fluticasone propionate (FT), deacetyl cortivazol/bimedrazole (DAC), cortivazol (CVZ), methylprednisolone (MPS), cortisol (CORT), dihydrotestosterone (DHT), drospirenone (DRO) and17α-hydroxyprogesterone (17-OHP) were obtained from Sigma-Aldrich (Saint-Quentin Fallavier, France). Promegestone (R5020), forbimenol, RU24782 and RU24858 are synthetic steroids non-commercially available and kindly gifts from Sanofi, Vertolaye, France.
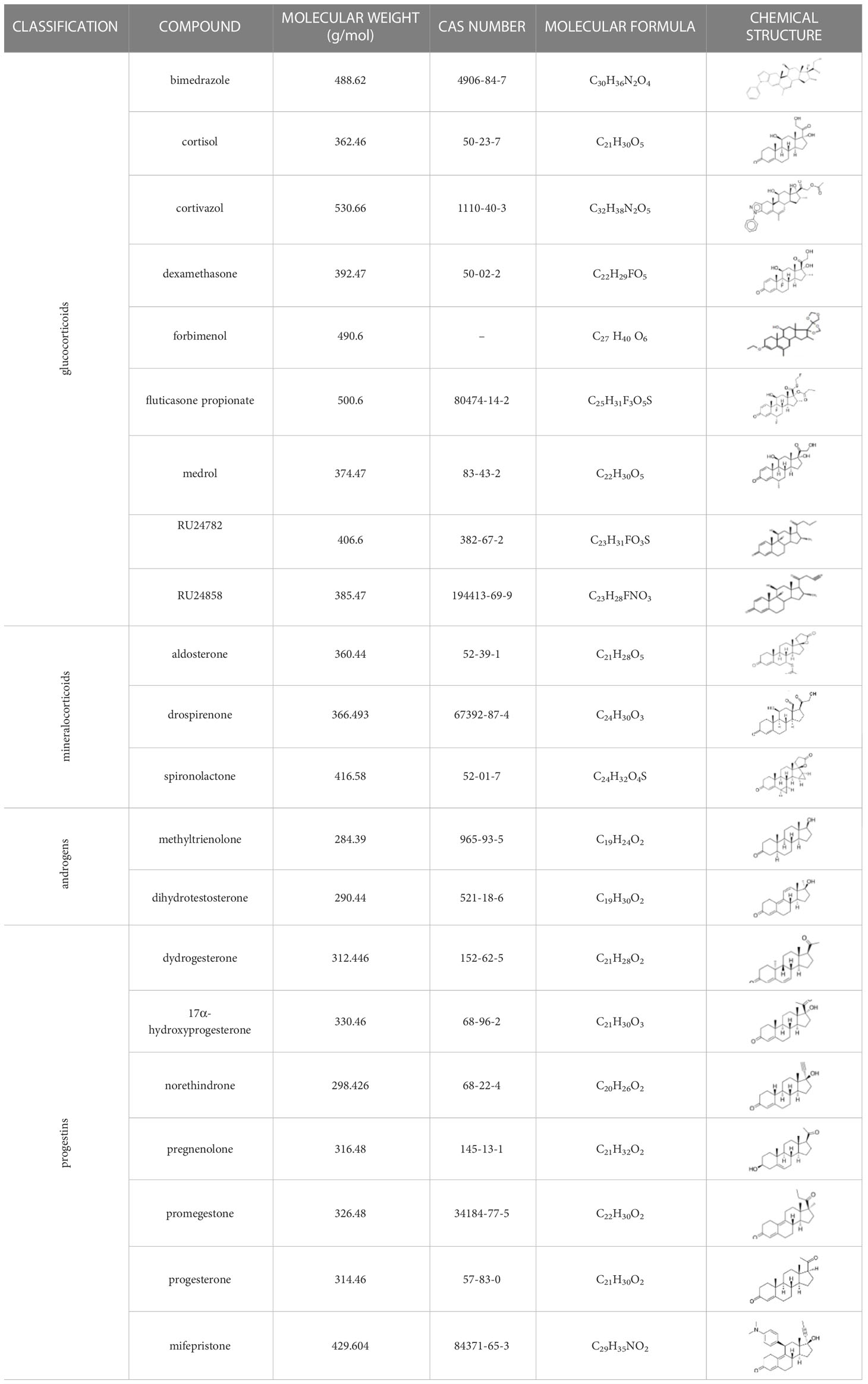
Table 1 Classification of natural and synthetic steroids tested on HMLN-hGR cells and UMLN-zfGR cells.
Stock solutions of chemicals were prepared in dimethyl sulfoxide (DMSO) and stored at -20°C. Fresh solution of test chemicals in test medium were prepared before each experiment. The final DMSO concentrations during treatment did not exceed 0.1% (v/v) of the test medium.
MMTV-luciferase-SV-neo plasmid was already described (27). pSG5-hGR puromycin and pSG5-puromycin are kind gifts of H Gronemeyer (IGBMC, Illkirch-Graffenstaden, France). pSG5-zfGR puromycin plasmid was obtained by cloning zfGR (M1-L746) in the BamHI site of pSG5-puromycin.
HMLN-hGR and UMLN-zfGR clonal cell lines were already described (14, 21). Briefly, HMLN-hGR cells were obtained by stable co-transfection of GR positive human HeLa cells with a glucocorticoid responsive gene (MMTV-Luciferase) and a hGR (pSG5-hGR-puromycin) expressing plasmid. To obtain UMLN-zfGR cells, human U2OS cells that mildly expressed hGR (28, 29) were stably co-transfected with the MMTV-Luciferase and a zfGR expressing plasmids (pSG5-zfGR-puromycin). 48 h after the transfection, cells were treated with G418 (1 mg/ml) and puromycin (0.5 μg/ml). Within 21 days, G418 and puromycin resistant clones appeared. For each cell line, 10 clones were chosen for their ligand-induced luciferase expression. The clones were amplified, and luciferase expression was checked at several passages. For each cell line, the clone with the best induction of luciferase activity was selected and used for the screening of the different steroids. The basal expression of luciferase is 1% and 11% for HMLN-hGR and UMLN-zfGR cell lines, respectively, of the maximal luciferase expression obtained in presence of dexamethasone 100 nM. The stability and the inducibility of luciferase expression were checked during at least 20 passages (20 weeks). The stability of the hGR and zfGR expression by ligand binding assay was also checked at different passages.
To obtain UMLN-hGR pool cells, U2OS cells were stably co-transfected with the MMTV-Luciferase and the hGR expressing plasmids. ZFL-zfGR pool cells were obtained by stable co-transfection of zebrafish ZFL cells with the MMTV-Luciferase and the zfGR expressing plasmids. The transfected cells were treated 7 days with G418 and puromycin but not cloned.
HMLN- and UMLN-GR cells were grown in a 5% CO2 humidified atmosphere at 37°C in Dulbecco’s Modified Eagle’s Medium: Nutrient Mixture F-12 (DMEM/F-12) containing phenol red, 1 g/L glucose, 10% fetal bovine serum (FBS), 100 units/mL of penicillin, 100 µg/mL of streptomycin, 1 mg/mL geneticin and 0.5 µg/mL puromycin (culture medium). Exposure was made in phenol red-free DMEM medium supplemented with 5% of dextran-coated charcoal FBS (DCC), 100 units/mL of penicillin and 100 µg/mL of streptomycin (HMLN hGR and UMLN GR test medium).
ZFL-zfGR cells were cultured at 28°C in humidified atmosphere with 5% CO2 in LDF medium (50% Leibovitz 15 culture medium L15, 35% DMEM high glucose and 15% Ham’s-F12 medium) with 0.15 g/L sodium bicarbonate, 15 mM 4-(2-hydroxy-ethyl)-1-piperazineethanesulfonic acid (HEPES), 0.01 mg/mL insulin, 50 ng/mL epidermal growth factor (EGF), 50 U/mL penicillin and streptomycin antibiotics, 10% (v/v) fetal bovine serum (FBS), 1 mg/mL geneticin and 0.5 µg/mL puromycin (culture medium). Exposure was made in the same culture medium excepted that 10% (v/v) fetal bovine serum (FBS) was replaced by 5% of dextran-coated charcoal FBS (DCC-FBS). The different reporter cell lines used in this study are summarized in the Supplementary Table 1.
In vitro transactivation assays were performed in 96-wells white opaque clear bottom culture plates (Greiner CellStar, Dutscher, Brumath, France). GR reporter cell lines were seeded at density of 5 x 104 cells per well in 150 µL culture medium and incubated at 37°C and 5% CO2 for 24 h. Then, medium was removed and cells were exposed to increasing dilutions of tested compounds in test medium (DMSO; final concentration 0.1% v/v). Cells were incubated for 16 h at 37°C and 5% CO2.
Results of transactivation activity were expressed as percentage of the maximum luciferase activity induced by dexamethasone at 10-7 M.
For antagonistic activity assessment, cells were exposed to different concentrations of the tested compounds and 3 nM dexamethasone. At this concentration, dexamethasone yields 60-80% of the maximal response. After the incubation period, medium was removed and replaced with 50 µL/well of test medium containing 0.3 mM luciferin. Luminescence signal was monitored in intact living cells for 2 s per well using a MicroBeta Trilux microplate scintillation and luminescence counter (PerkinElmer, Courtaboeuf, France).
The effect of the tested chemicals on cell viability was assessed with the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, after luminescence detection, medium containing luciferin was removed and replaced with 100 µL/well of test medium containing 0,4 mg/ml MTT for 4 h. Colorimetric signal was monitored at 570 nM using a Pherastar microplate reader (BMG Labtech, Champigny s/Marne, france). Experiments were performed in quadruplicate and repeated three times.
For measurement of GR expression, HMLN-hGR, U2OS and UMLN-zfGR cells were seeded in 24-wells transparent plates at a density of 400,000 cells per well in culture medium. Cells were incubated for 24 h in a 5% CO2 humidified atmosphere at 37°C. Then, medium was then removed and replaced with test medium containing 10 nM [3H]-dexamethasone (84 Cu/mmol, Perkin Elmer), in the absence or presence of 10 μM of non-radioactive dexamethasone. After 3 h, unbound material was aspirated, and cells were washed three times with cold PBS in order to remove additional unbound material. Then, 0.4 ml of lysis buffer (250 mM Tris phosphate pH 7.8, 0.1% triton X-100) was added and plates were shaked for 5 min. 0.1 ml of the total cell lysate was mixed with of 0.1 ml of LSC-cocktail (Emulsifier-Safe, Perkin Elmer) and [3H] bound radioactivity was liquid scintillation counted (MicroBeta trilux, PerkinElmer). Protein concentrations of 0.1 ml of the total cell lysate were measured by Bio-Rad protein assay (Bio-Rad, Marnes-la-Coquette, France) and used to normalize bound radioactivity values expressed in dpm. Specific binding was determined by subtracting non-specific binding from total binding and enable to determine GR expression in fentomoles of protein per mg of protein. Experiments were performed in quadruplicate and repeated three times.
For ligand competition assays, HMLN-hGR and UMLN-zfGR cells were seeded in 96-wells white opaque clear bottom culture plates (Greiner CellStar, Dutscher) at density of 105 cells per well in 0.2 ml of culture medium and incubated at 37°C and 5% CO2 for 24 h. Then, medium was removed and cells were exposed to with 1 nM [3H]-dexamethasone in the absence or presence of increasing concentrations of non-radioactive competitive compounds. After 3 h, unbound material was aspirated, and cells were washed three times with cold PBS in order to remove additional unbound material. Then, 150 μl of lysis buffer (250 mM Tris phosphate pH 7.8, 0.1% triton X-100) was added and plates were shaked for 5 min. 50 μl of the total cell lysate was mixed with of 50 μl of LSC-cocktail (Emulsifier-Safe, Perkin Elmer) and [3H] bound radioactivity was liquid scintillation counted (MicroBeta trilux, Perkin Elmer). Protein concentration of 50 μl of the total cell lysate was measured by Bio-Rad protein assay and used to normalize bound radioactivity values expressed in dpm. Results were plotted as measured dpm versus concentration of tested compound. IC50 values were defined as compound concentration required to decrease maximum [3H]-dexamethasone binding by 50%. All the experiments were performed in quadruplicates and in at least three independent experiments.
Results of transactivation activity were expressed as percentage of the maximum luciferase activity induced by dexamethasone at 10-7 M. Dose-response curves were fitted using the sigmoidal dose-response function of a graphics and statistics software program (GraphPad Prism 8, GraphPad Software Inc.). We assumed a compound being agonist when an effect above 10% was noted as compared to the control cells, and antagonist when it decreased luciferase activity by more than 20% in the presence of the reference chemical at the concentration inducing 80% of the maximal response. Effective concentrations and inhibitory concentrations were derived from the Hill equation. For a given chemical, EC50 was defined as the concentration inducing 50% of its maximal effect and IC50 represented the concentration required for 50% inhibition. Lower and upper 95% confidence limits of EC50s of IC50s were calculated.
For ligand binding assays, dose-response curves were also fitted using GraphPad Prism. Inhibitory concentrations were derived from the Hill equation. Lower and upper 95% confidence limits of EC50s of IC50s were calculated.
The HMLN-hGR and UMLN-zfGR clonal cell lines were previously established in our laboratory (21). To obtain HMLN-hGR cells, we transfected HeLa cells which express endogenously hGR by a hGR expressing plasmid to overexpress hGR and the MMTV-Luc GR-responsive gene. The UMLN-zfGR cell line was established in U2OS cells which slightly express hGR (28, 29) by co-transfection of a zfGR expressing plasmid and the MMTV-Luc plasmid. As shown in Figure 1, receptor protein level (hGR or zfGR) expressed in each cell line was estimated by saturation ligand- binding assay (LBA) with 10 nM [3H]-DEX in a ‘‘whole-cell’’ experiment. These whole-cell LBAs confirmed the endogenous expression of GR in HeLa cells (217 fentomoles/mg protein) whereas in U2OS this receptor is very slightly expressed (17 fentomoles/mg protein). Stable transfection of an hGR expression plasmid enabled to increase the expression of hGR in HMLN-hGR cells (535 fentomoles/mg protein). Finally, transfection of an zfGR expression in UMLN-zfGR cells enabled to express 324 fentomoles of zfGR per mg of protein (Figure 1; Supplementary Table 2).
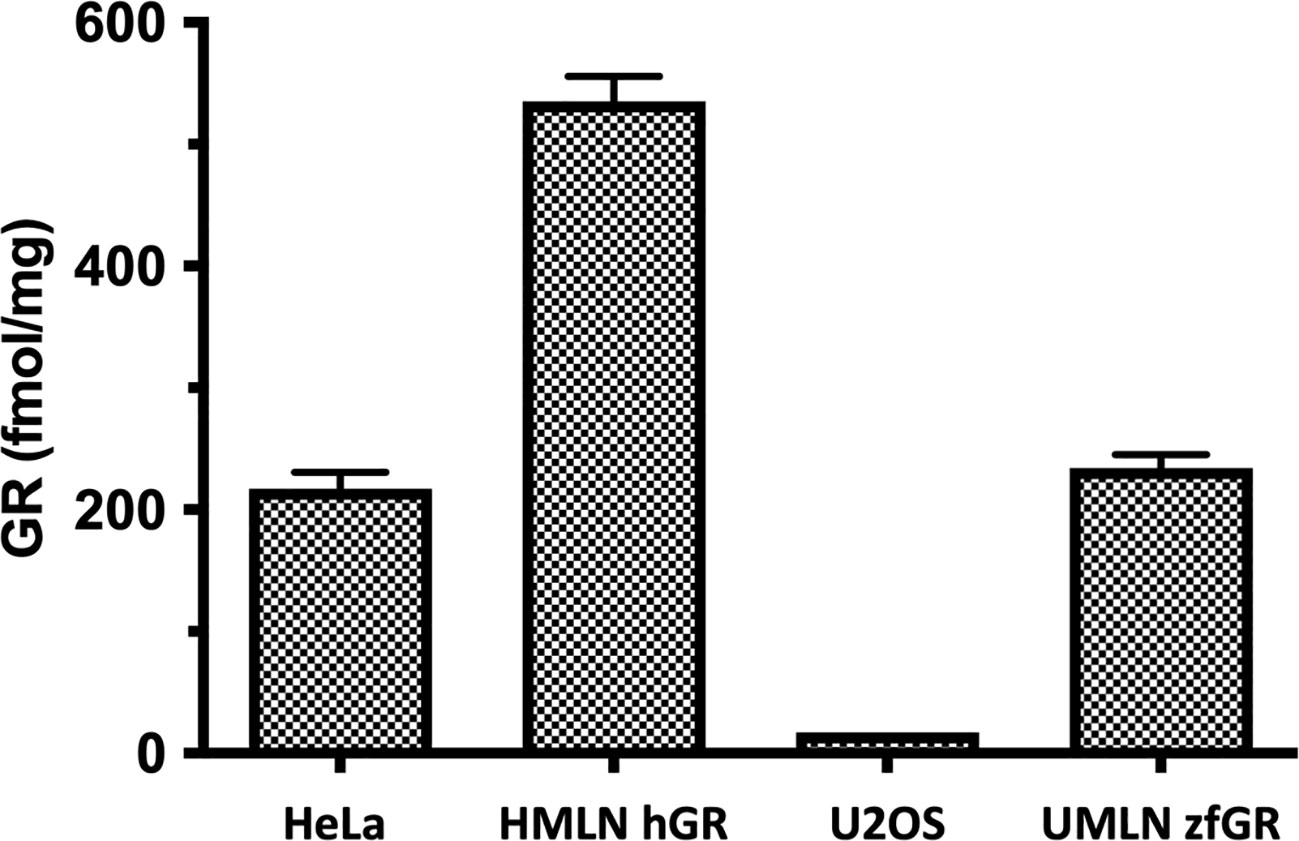
Figure 1 Expression of GR in HeLa, HMLN-hGR, U2OS and UMLN-zfGR cells. GR expression was measured by [3H]-DEX binding assay. Concentrations of GR are expressed in fentomoles/mg of protein. Values are means ± SD of 3 independent experiments.
The reference glucocorticoid agonist dexamethasone was tested in transactivation assays in both reporter cell models revealing close potency between the HMLN-hGR and UMLN-zfGR cell lines (EC50 of 1.38 and 2 nM for hGR and zfGR, respectively) (Table 2; Figure 2A). Almost all the chemicals belonging to the group of GCs, presented in Table 1, showed full agonistic activity both on hGR and on zfGR in transactivation assays. Bimedrazole, cortivazol, fluticasone propionate and medrol exhibited similar potency on the two nuclear receptors (Table 2) whereas cortisol was slighly more potent on zfGR than on hGR with EC50 of 40 nM and 15 nM for hGR and zfGR, respectively (Figure 2B; Table 2). On the contrary, the dissociated glucocorticoids RU24782 and RU24858 which were structurally designed to distinguish between the transrepression and transactivation (10) have showed better potency on hGR than zfGR (Table 2; Figures 2C, D). Interestingly, the synthetic glucocorticoid forbimenol only activated hGR (EC50 503 nM) (Table 2; Figure 2E). Similarly, the mineralocorticoid aldosterone, which is produced naturally in human but not in fish (30), was fully agonist on hGR but partial agonist on zfGR (maximal activity of 96 and 19% for hGR and zfGR respectively) (Table 2; Figure 2F). All the other chemicals tested lacked glucocorticoid agonist activity.
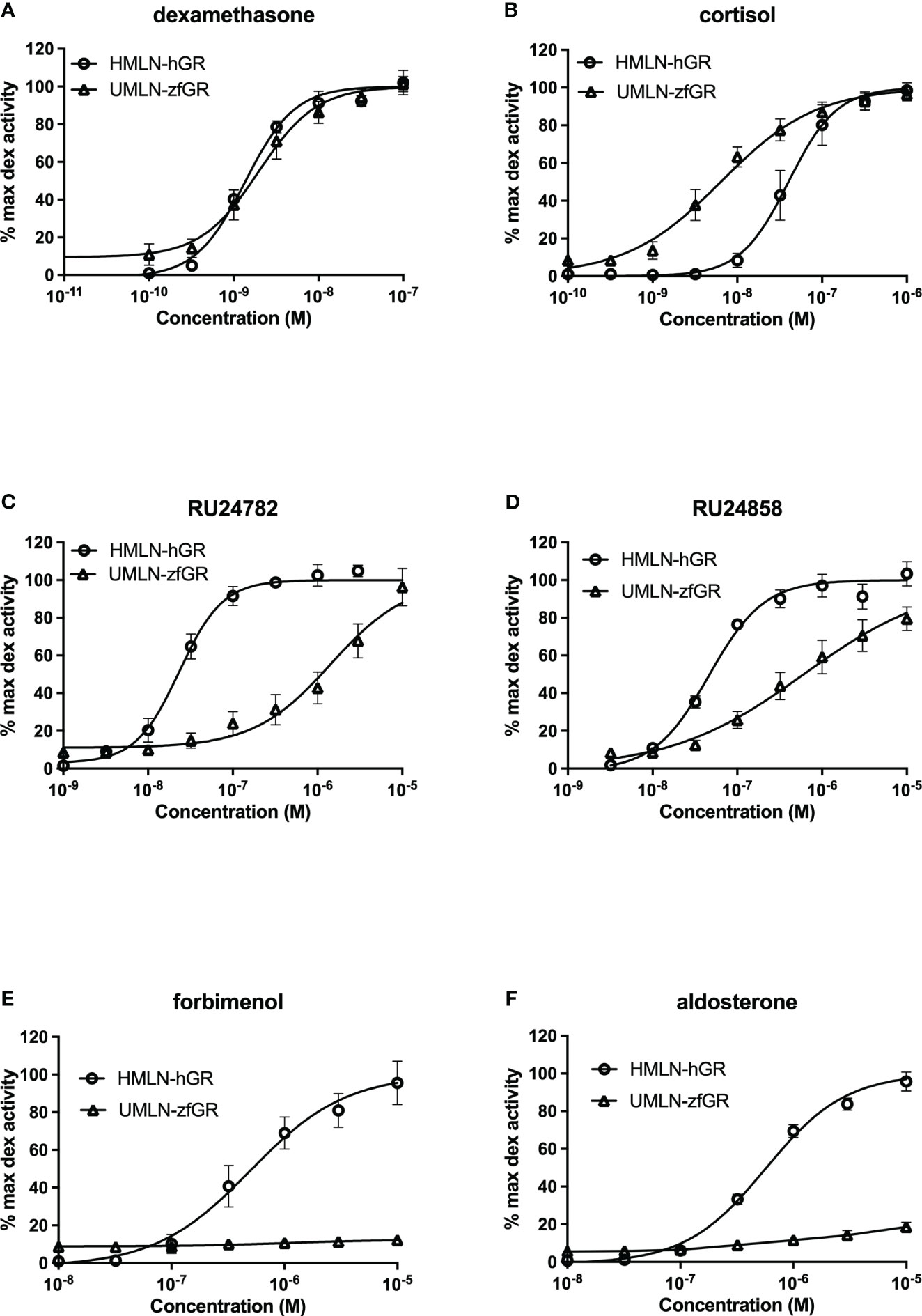
Figure 2 Dose-response curves of dexamethasone (A), cortisol (B), RU24782 (C), RU24858 (D), forbimenol (E) and aldosterone (F) activity in HMLN-hGR and UMLN-zfGR cells. Results are expressed as the percentage of the maximum luciferase activity induced by 100 nM dexamethasone. Error bars represent standard deviation.
RU24782, RU24858, forbimenol and aldosterone were also assessed for antagonism in UMLN-zfGR cells. Interestingly, RU24782, RU24858 partially repress dexamethasone-induced activity at sub-micromolar concentrations (Figure 3A). As these GCs were described as less able to recruit coactivators than dexamethasone to hGR (10), their zfGR antagonism at sub-micromolar concentrations and agonism at micromolar concentrations is probably relied to their lower efficacy for recruiting coactivators. Forbimenol and aldosterone also antagonized zfGR (IC50 3160 and 13980 nM, respectively) (Table 3; Figure 3B). Finally, the anti-progestin mifepristone was partial agonist of hGR and zfGR with higher efficacy on the last one (Table 2; Figures 4A, B). In presence of dexamethasone, it acts as partial antagonist with IC50 in the 10 to 40 nanomolar range (Table 3; Figures 4A, B). The other chemicals lacking glucocorticoid agonist activity were also tested for their antagonist activity. They all exhibited antagonist activity on hGR and zfGR. IC50 varied between 772 (promegestone) and 6101 (dihydrostestosterone) nM for hGR and 1066 (17α-hydroxyprogesterone) and 7711 (spironolactone) nM for zfGR (Table 3).
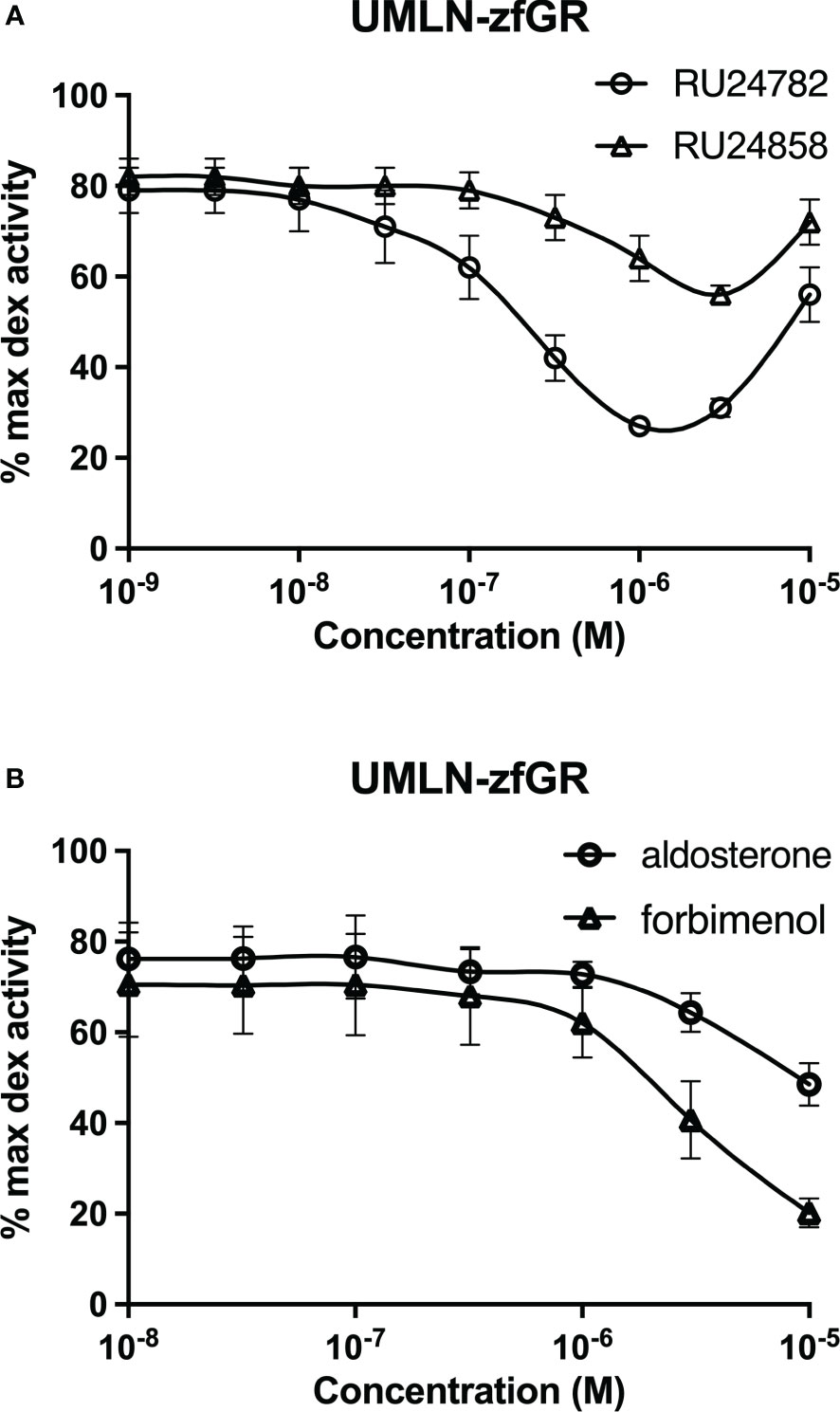
Figure 3 Dose-response curves of RU24782 and RU24858 (A), aldosterone and forbimenol (B) activity in UMLN-zfGR cells in presence of dexamethasone 3 nM. Results are expressed as percentage of the maximum luciferase activity induced by 100 nM M dexamethasone. Error bars represent standard deviation.
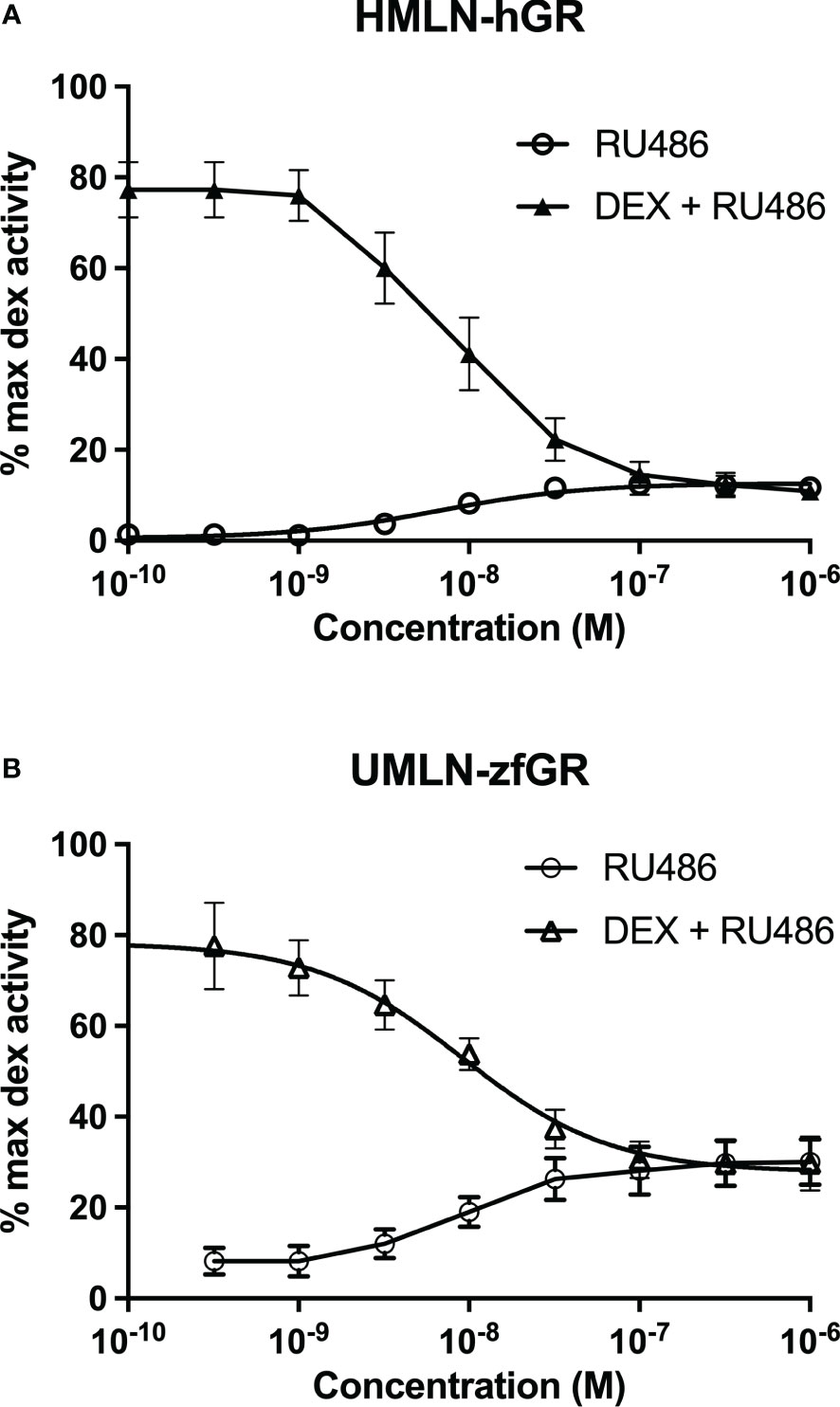
Figure 4 Dose-response curves of mifepristone activity in HMLN-hGR (A) and UMLN-zfGR (B) cells in absence or presence of dexamethasone 3 nM. Results are expressed as percentage of the maximum luciferase activity induced by 100 nM dexamethasone. Error bars represent standard deviation.
Whole-cell competitive binding assays were performed with HMLN-hGR and UMLN-zfGR cells to determine whether the different potencies observed in transactivation assays for some of the chemicals reflected their abilities to differently bind to hGR and zfGR. Table 4 summarized IC50 and RBA values for hGR and zfGR. IC50 of DEX of 5.8 and 3.1 nM for hGR and zfGR, respectively confirmed the similar potency of this chemical for the two receptors. By contrast, cortisol bound preferentially zfGR as shown by IC50s of 55.2 and 3.5 nM for hGR and zfGR, respectively (Table 4) and thus explained its higher potency on the fish receptor. RU24782 bound preferentially to hGR than to zfGR but these differences in affinity did not reflect the differences in potency reinforcing the hypothesis that that this chemical is less able to recruit coactivators for zfGR than for hGR. Finally, aldosterone and forbimenol which are agonists on hGR and antagonists on zfGR bound also slightly preferentially hGR than zfGR but these differences in affinity do not explain their differences in activities for the two receptors.
Since cellular context could influence the transcriptional activity of NRs and hGR and zfGR reporter cell lines are different (Hela for hGR and U2OS for zfGR), we established UMLN-hGR pool cells. We tested in these cells the activity of RU24782, RU24858, forbimenol and aldosterone. The response profiles obtained (Figure 5A) are very similar to those obtained with the HMLN-hGR cell line indicating that at least for hGR these chemicals had a similar potency. The four chemicals were agonists on these cells and the EC50 values for RU24782, RU24858, forbimenol and aldosterone were respectively of 3.1, 23.3, 54.5, 338 and 224 nM.
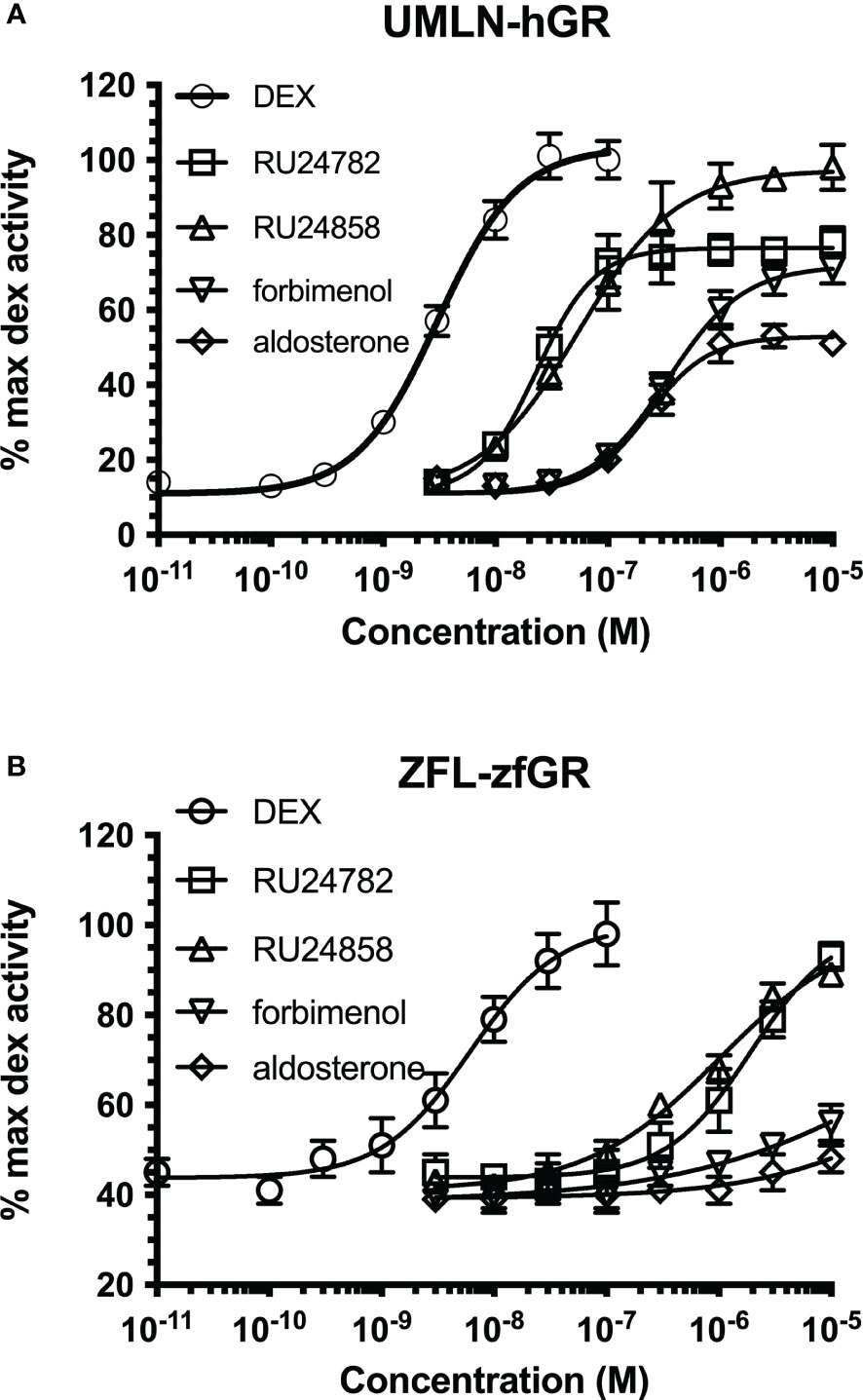
Figure 5 Dose-response curves of dexamethasone, RU24782, RU24858, aldosterone and forbimenol activity in UMLN-hGR (A) and ZFL-zfGR (B) cells. Results are expressed as percentage of the maximum luciferase activity induced by 100 nM dexamethasone. Error bars represent standard deviation.
We also tested in zebrafish liver ZFL-zfGR pool cells the activity of the three chemicals. Again, the response profiles obtained (Figure 5B) are very similar to those obtained with the UMLN-zfGR cell line. The EC50 value for DEX, RU24782 and RU24858 were respectively of 6, 1938 and 1027 nM were respectively whereas forbimenol and aldosterone only slightly activated luciferase expression (Figure 5B).
Previous studies investigating progestin receptor (PR) and mineralocorticoid (MR) receptor have showed strong differences in binding affinity and transactivation properties of steroids (24, 31). As few studies have been done on GR, in this work, we have evaluated the ability of 21 steroids to alter the transcriptional activity of hGR and zfGR.
Our data have showed that the majority of the steroids tested bind with a similar affinity to both GR. However, some of them presented marked differences in transactivation efficacies and potencies. The most different steroids are forbimenol and aldosterone. These chemicals have a full agonist profile on hGR while they do not induce luciferase activity in UMLN-zfGR cells. According to their ability to bind to zfGR, aldosterone and forbimenol were able to antagonize DEX-induced luciferase activity in a concentration-dependant manner in the zfGR assay. Strikingly, the dissociated glucocorticoids RU24782 and RU24858 which are synthetic GCs less able to recruit coactivators than full agonists (dexamethasone, bimedrazol) to hGR have similar affinities for hGR and zfGR but showed different potencies on the two receptors. Even more curiously, these compounds are zfGR antagonists at low concentrations (at which they bind to zfGR) and zfGR agonists at high concentrations.
As these four steroids have similar affinities for both GRs, their zfGR antagonism is probably relied to their lower efficacy for recruiting coactivators. Interestingly, we observed the same results in different cellular contexts (U2OS for hGR and ZFL for zfGR); indicating that these differences are probably not specific to the cellular context, but on the contrary reflected the intrinsic properties of steroids on zfGR.
Although we did not fully explain the differences of activity of the two GRs, we could point out that these differences could probably be due to different coactivators’ recruitment by hGR and zfGR. This should be further confirmed by using site-directed mutagenesis and analyses of the structures of the hGR and zfGR ligand binding domain which are currently lacking. These techniques have made it possible to explain that progesterone and spironolactone act as antagonists on hMR and agonists on zfMR due to a substitution of threonine in hMR by a leucine on zfMR (32). These techniques would be probably also valuable to explain why several progestins are antagonist on hPR while they are agonist on hPR (24).
Although few of the steroids tested exhibited differences in activation between human and zebrafish receptor for GR than for MR and PR, our results showed that they exist and confirm that investigating inter-species differences is important and can improve EDC risk assessment. According to our results, toxicological data extrapolated from mammalian models could not be always suitable for predicting possible hazard in other species such as zebrafish. Regarding this latter aspect, since in the last decades numerous studies have detected glucocorticoid activity in the environment raising concern about their distribution and their harmful effects on wildlife (16; 33), we consider that the use of our established cell lines could improve the endocrine disrupting assessment and environmental contamination monitoring.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
For reporter cell lines experiments (luciferase expression experiments, binding analysis), ethical approval was not required. The studies were conducted in accordance with local legislation and institutional requirements. The human samples used in this study were acquired from another research group. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
AT and AB performed experiments. AT and PB conceived the work. AT drafted the original manuscript and prepared the figures and tables. AE, CG and PB revised the manuscript. PB supervised the work. All authors contributed to the article and approved the submitted version.
This work was funded in part by the project ANSES TOXCHEM (2018/1/020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1235501/full#supplementary-material
1. Lu NZ, Wardell SE, Burnstein KL, Defranco D, Fuller PJ, Giguere V, et al. International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol Rev (2006) 58:782. doi: 10.1124/pr.58.4.9
2. Kumar R, Thompson EB. Gene regulation by the glucocorticoid receptor: structure: function relationship. J Steroid Biochem Mol Biol (2005) 94:383. doi: 10.1016/j.jsbmb.2004.12.046
3. Noddings CM, Wang RY, Johnson JL, Agard DA. Structure of Hsp90-p23-GR reveals the Hsp90 client-remodelling mechanism. Nature (2022) 601:465. doi: 10.1038/s41586-021-04236-1
4. Wang RY, Noddings CM, Kirschke E, Myasnikov AG, Johnson JL, Agard DA. Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism. Nature (2022) 601:460. doi: 10.1038/s41586-021-04252-1
5. Santos GM, Fairall L, Schwabe JW. Negative regulation by nuclear receptors: a plethora of mechanisms. Trends Endocrinol Metab (2011) 22:87. doi: 10.1016/j.tem.2010.11.004
6. Kadmiel M, Cidowski JA. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci (2013) 34:518. doi: 10.1016/j.tips.2013.07.003
7. Baschant U, Tuckermann J. The role of the glucocorticoid receptor in inflammation and immunity. J Steroid Biochem Mol Biol (2010) 120:69. doi: 10.1016/j.jsbmb.2010.03.058
8. Oakley RH, Cidlowski JA. The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J Allergy Clin Immunol (2013) 132:1033. doi: 10.1016/j.jaci.2013.09.007
9. Vayssière BM, Dupont S, Choquart A, Petit F, Garcia T, Marchandeau C, et al. Synthetic glucocorticoids that dissociate transactivation and AP-1 transrepression exhibit antiinflamatory activity in vivo. Mol Endocrinol (1997) 11:1245. doi: 10.1210/mend.11.9.9979
10. Dezitter X, Fagart J, Taront S, Fay M, Masselot B, Hétuin D, et al. A structural explanation of the effects of dissociated glucocorticoids on glucocorticoid receptor transactivation. Mol Pharmacol (2014) 85:226. doi: 10.1124/mol.113.085860
11. Molina-Molina JM, Hillenweck A, Jouanin I, Zalko D, Cravedi JP, Fernández M, et al. Steroid receptor profiling of vinclozolin and its primary metabolites. Toxicol Appl Pharmacol (2006) 216:44. doi: 10.1016/j.taap.2006.04.005
12. Gumy C, Chandsawangbhuwana C, Dzyakanchuk AA, Kratschmar DV, Baker ME, Odermatt A. Dibutyltin disrupts glucocorticoid receptor function and impairs glucocorticoid-induced suppression of cytokine production. PloS One (2008) 3:e3545. doi: 10.1371/journal.pone.0003545
13. Gulliver LSM. Xenobiotics and the glucocorticoid receptor. Toxicol Appl Pharmacol (2017) 319:69. doi: 10.1007/978-1-4939-9195-2_4
14. Grimaldi M, Boulahtouf A, Toporova L, Balaguer P. Functional profiling of bisphenols for nuclear receptors. Toxicology (2019) 420:39. doi: 10.1016/j.tox.2019.04.003
15. Zhang J, Yang Y, Liu W, Schlenk D, Liu J. Glucocorticoid and mineralocorticoid receptors and corticosteroid homeostasis are potential targets for endocrine-disrupting chemicals. Environ Int (2019) 133(Pt A):105133. doi: 10.1016/j.envint.2019.105133
16. Weizel A, Schlüsener MP, Dierkes G, Ternes TA. Occurrence of glucocorticoids, mineralocorticoids, and progestogens in various treated wastewater, rivers, and streams. Environ Sci Technol (2018) 52:5296. doi: 10.1021/acs.est.7b06147
17. Hashmi MAK, Krauss M, Escher BI, Teodorovic I, Brack W. Effect-directed analysis of progestogens and glucocorticoids at trace concentrations in river water. Environ Toxicol Chem (2020) 39:189. doi: 10.1016/j.scitotenv.2017.12.187
18. Shen X, Chang H, Sun Y, Wan Y. Determination and occurrence of natural and synthetic glucocorticoids in surface waters. Environ Int (2020) 134:105278. doi: 10.1016/j.envint.2019.105278
19. Leusch FDL, Neale PA, Arnal C, Aneck-Hahn NH, Balaguer P, Bruchet A, et al. Analysis of endocrine activity in drinking water, surface water and treated wastewater from six countries. Water Res (2018) 139:10. doi: 10.1016/j.watres.2018.03.056
20. Alygizakis NA, Besselink H, Paulus GK, Oswald P, Hornstra LM, Oswaldova M, et al. Characterization of wastewater effluents in the Danube River Basin with chemical screening, in vitro bioassays and antibiotic resistant genes analysis. Environ Int (2019) 127:420. doi: 10.1016/j.envint.2019.03.060
21. Neale PA, Grimaldi M, Boulahtouf A, Leusch FDL, Balaguer P. Assessing species-specific differences for nuclear receptor activation for environmental water extracts. Water Res (2020) 185:116247. doi: 10.1016/j.watres.2020.116247
22. Pinto C, Grimaldi M, Boulahtouf A, Pakdel F, Brion F, Cavaillès V, et al. Selectivity of natural and environmental estrogens for zebrafish estrogen receptors. Toxicol Appl Pharmacol (2014) 280:29. doi: 10.1016/j.taap.2014.07.020
23. Pinto C, Hao R, Grimaldi M, Thrikawala S, Boulahtouf A, Aït-Aïssa S, et al. Differential activity of BPA, BPAF and BPC on zebrafish estrogens receptors in vitro and in vivo. Toxicol Appl Pharmacol (2019) 380:114709. doi: 10.1016/j.taap.2019.114709
24. Garoche C, Aït-Aïssa S, Boulahtouf A, Creusot N, Hinfray N, Bourguet W, et al. Human and zebrafish nuclear progesterone receptors are differently activated by manifold progestins. Environ Sci Technol (2020) 54:9510. doi: 10.1021/acs.est.0c02056
25. Creusot N, Garoche C, Grimaldi M, Boulahtouf A, Chiavarina B, Bourguet W, et al. A Comparative study of uman and zebrafish pregnane X receptor activities of pesticides and steroids using in vitro reporter gene assays. Front Endocrinol (Lausanne) (2021) 12:665521. doi: 10.3389/fendo.2021.665521
26. Garoche C, Boulahtouf A, Grimaldi M, Chiavarina B, Toporova L, den Broeder MJ, et al. Interspecies differences in activation of peroxisome proliferator-activated receptor γ by pharmaceutical and environmental chemicals. Environ Sci Technol (2021) 55:16489. doi: 10.1021/acs.est.1c04318
27. Térouanne B, Tahiri B, Georget V, Belon C, Poujol N, Avances C, et al. A stable prostatic bioluminescent cell line to investigate androgen and antiandrogen effects. Mol Cell Endocrinol (2000) 160:39. doi: 10.1016/s0303-7207(99)00251-8
28. Sonneveld E, Jansen HJ, Riteco JA, Brouwer A, van der Burg B. Development of androgen- and estrogen-responsive bioassays, members of a panel of human cell line-based highly selective steroid-responsive bioassays. Toxicol Sci (2005) 83:136. doi: 10.1093/toxsci/kfi005
29. Milcamps A, Liska R, Langezaal I, Casey W, Dent M, Odum J. Reliability of the AR-CALUX®. In vitro method used to detect chemicals with (anti)androgen activity: results of an international ring trial. Toxicol Sci (2021) 184:170. doi: 10.1093/humupd/7.3.248
30. Cruz SA, Lin CH, Chao PL, Hwang P. Glucocorticoid receptor, but not mineralocorticoid receptor, mediates cortisol regulation of epidermal ionocyte development and ion transport in zebrafish (Danio rerio). PloS One (2013) 81:219. doi: 10.5935/0004-2749.20180044
31. Baker ME. Divergent evolution of progesterone and mineralocorticoid receptors in terrestrial vertebrates and fish influences endocrine disruption. Biochem Pharmacol (2022) 198:114951. doi: 10.1016/j.bcp.2022.114951
32. Fuller PJ, Yao YZ, Jin R, He S, Martín-Fernández B, Young MJ, et al. Molecular evolution of the switch for progesterone and spironolactone from mineralocorticoid receptor agonist to antagonist. Proc Natl Acad Sci U S A. (2019) 116:18578. doi: 10.1073/pnas.1903172116
Keywords: human GR, zebrafish GR, steroids, pharmaceuticals, reporter cell lines
Citation: Toso A, Boulahtouf A, Escande A, Garoche C and Balaguer P (2023) A comparative study of human and zebrafish glucocorticoid receptor activities of natural and pharmaceutical steroids. Front. Endocrinol. 14:1235501. doi: 10.3389/fendo.2023.1235501
Received: 06 June 2023; Accepted: 31 July 2023;
Published: 15 August 2023.
Edited by:
Honoo Satake, Suntory Foundation for Life Sciences, JapanReviewed by:
Patricia I. S. Pinto, University of Algarve, PortugalCopyright © 2023 Toso, Boulahtouf, Escande, Garoche and Balaguer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick Balaguer, cGF0cmljay5iYWxhZ3VlckBpbnNlcm0uZnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.